- 1College of Food and Bioengineering, Henan University of Science and Technology, Luoyang, China
- 2College of Horticulture and Plant Protection, Henan University of Science and Technology, Luoyang, China
Introduction: Arbuscular mycorrhizal fungi (AMF) have been demonstrated their ability to enhance the arsenic (As) tolerance of host plants, and making the utilization of mycorrhizal plants a promising and practical approach for remediating As-contaminated soils. However, comprehensive transcriptome analysis to reveal the molecular mechanism of As tolerance in the symbiotic process between AMF and host plants is still limited.
Methods: In this study, transcriptomic analysis of Gossypium seedlings was conducted with four treatments: non-inoculated Gossypium under non-As stress (CK0), non-inoculated Gossypium under As stress (CK100), F. mosseae-inoculated Gossypium under non-As stress (FM0), and F. mosseae-inoculated Gossypium under As stress (FM100).
Results: Our results showed that inoculation with F. mosseae led to a reduction in net fluxes of Ca2+, while increasing Ca2+ contents in the roots and leaves of Gossypium under the same As level in soil. Notably, 199 and 3129 differentially expressed genes (DEGs) were specially regulated by F. mosseae inoculation under As stress and non-As stress, respectively. Through Gene Ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) annotation and enrichment analyses, we found that under As stress, F. mosseae inoculation up-regulated a significant number of genes related to the Ca2+ signaling pathway genes, involved in cellular process, membrane part, and signal transduction. This suggests a potential role in mitigating As tolerance in Gossypium seedlings. Furthermore, our analysis identified specific DEGs in transcription factor families, including ERF, MYB, NAC, and WRKY, that were upregulated by F. mosseae inoculation. Conversely, MYB and HB-other were down-regulated. The ERF and MYB families exhibited the highest number of up- and down-regulated DEGs, respectively, which were speculated to play an important role in alleviating the As toxicity of Gossypium.
Discussion: Our findings provided valuable insights into the molecular theoretical basis of the Ca2+ signaling pathway in improving As tolerance of mycorrhizal plants in the future.
1 Introduction
Arsenic (As), a hazardous metalloid element in the earth’s crust, poses a significant environmental threat worldwide due to its widespread contamination in groundwater and soil. This contamination has garnered increasing attention regarding the global security of agricultural products (Li et al., 2018a, 2020). As also enriched in orpiment, realgar, and other ores in nature (Sharma et al., 2020). Over the past century, industrialization and anthropogenic activities, such as mining As ores, using As-based agrochemicals and fertilizer, smelting metal and glass, irrigating As-contaminated groundwater, and burning fossil fuels, have exacerbated As contamination (Chen et al., 2007; Salim et al., 2009; Srivastava et al., 2009). The entry of As and its inorganic compounds into the human food chain can lead to serious health issues, including cancers, cardiovascular diseases, diabetes, compromised immunity, and infertility (Sharma et al., 2020; Liu et al., 2021). In plants, high concentrations of As interfere with metabolic processes, leading to physiological and morphological disorders (Spagnoletti et al., 2016; Sharma et al., 2017; Martínez-Castillo et al., 2022). The symptoms of As phytotoxicity include the damage of cell structure and function, inhibition of photosynthesis and respiration, retardation of germination and growth, reduction of production, and even senescence and death, which ultimately cause vegetation degradation in the As-polluted areas (Spagnoletti et al., 2017; Surbhi et al., 2017). Therefore, some measures of biotechnological remediation need to be applied to prevent environmental deterioration due to As contamination (Martínez-Castillo et al., 2022).
To adapt to heavy metal (HM)-polluted environment, plants have evolved the intrinsic strategies of resistance to HM stress, which involves calcium (Ca) signaling responses (Sharma et al., 2020). As ubiquitous intracellular second messengers, Ca2+ ions regulate diverse cellular processes, which convey a wide range of environmental and developmental stimuli (Lecourieux et al., 2006; Kurusu et al., 2010). Changes in cytosolic-free Ca2+ concentration, which is tightly regulated by active transport (Ca2+−antiporters and Ca2+-ATPases) and passive fluxes (Ca2+ channels) across membranes, couple a series of signal responses in plants (Bush, 1995; Sanders et al., 1999). Under As stress, plants experience an increase in cytoplasmic Ca2+ concentrations, relying on transcriptional changes and the kinase activity of Ca2+ sensors (Sharma et al., 2020). Differential gene expression of Ca2+ sensors suggests their involvement of Ca2+ −dependent signaling responses to As toxicity (Chakrabarty et al., 2009). However, there is no direct experimental evidence supporting a relationship between As stress and Ca2+ signal transduction pathways.
Arbuscular mycorrhizal fungi (AMF), belonging to the phylum Glomeromycota, are recognized as phytobeneficial obligate symbiotic fungi (Schüßler and Walker, 2010). These rhizosphere endophytes form mutualistic relationships with over 80% of terrestrial higher plants, representing the most widespread beneficial plant–microbe interactions through a complex and coordinated network (Smith and Read, 2008). AMF acquire photoassimilates from plant roots, and, in return, transport essential mineral nutrients (potassium (K), phosphorus (P), nitrogen (N), etc.) from rhizosphere soil to plant roots through their arbuscules and hyphal coils (Riley and Corradi, 2013; Song et al., 2014). Mycorrhizal hyphae attach to the root surface, infiltrate into the epidermis, and further spread into inner cortical cells to develop a highly branched arbuscular structure, where more efficient uptake of water and nutrients takes place (Handa et al., 2015). Additionally, AMF colonization improves plant resistance against various biotic and abiotic stresses, including As stress (Smith and Read, 2008). In recent years, the As tolerance mechanism of host plants enhanced by AMF symbiosis was gradually investigated (Gonzalez-Chavez et al., 2002; Zhang et al., 2020, 2022). The widely accepted mechanism is “growth dilution effect”; phosphorus (P) element, sharing chemical properties with As, is acquired more P nutrition for host plants via their hyphae, leading to increased plant biomass and reduced As concentration in plant tissues (Spagnoletti and Lavado, 2015; Li et al., 2018a). Furthermore, AMF deposit As ions in their cell walls, detoxify As ions outside their hyphae by sequestering siderophores, and export them more efficiently than plant cells (Ouziad et al., 2005).
Hymenoscyphus ericae, a species of ericoid mycorrhizal fungi, improved the As resistance of Calluna vulgaris by reducing cellular arsenate to arsenite, effluxing arsenite from cells, and assimilating phosphate (Sharples et al., 2000). Similarly, two strains of arbuscular mycorrhizal fungi (AMF), Funneliformis mosseae and Glomus caledonium, inhibited high-affinity arsenate/phosphate transfer into Holcus lanatus roots and decreased the As phytotoxicity of host plant (Gonzalez-Chavez et al., 2002). Rhizophagus intraradices alleviated the As toxicity of Robinia pseudoacacia by improving plant growth, root morphology, phytohormones, and the concentration of soil glomal (Zhang et al., 2020). Furthermore, inoculation with R. intraradices enhanced the As tolerance of Sophora davidii, which was closely related to AMF, improving their growth, gas exchange, and chlorophyll fluorescence parameter, reactive oxygen species, and antioxidant enzymes while decreasing As accumulation in S. davidii tissues (Zhang et al., 2022). The above studies investigated the positive effects of AMF inoculation on As tolerance of host plants by using phenomic approaches. The establishment of AMF symbiosis involves complex signaling events that often involve the recruitment and modification of gene expression patterns in host plants to fulfill the symbiotic requirements, and this process ultimately leads to changes in the expression of mycorrhiza-regulated genes in the host plants (Song et al., 2014). RNA sequencing (RNA-Seq) is an effective method for investigating the transcriptome-wide response of plants and elucidating the underlying molecular mechanisms (Li et al., 2020). This method generates vast annotation information on differentially expressed genes, providing opportunities for function analysis of genes regulated by specific factors (Ma et al., 2018). However, a comprehensive transcriptome analysis, to elucidate the molecular mechanism involved in the symbiotic process of AMF and host plants under As stress, is still not completely understood.
The phytoremediation potential of Pteris vittata L. (P. vittata), an As hyperaccumulator with strong As accumulation capabilities, demonstrated in various remediation projects in As-contaminated areas of China (Wan et al., 2016; Sun et al., 2023). The inoculation of AMF (Claroideoglomus etunicatum) increased the phytoremediation efficiency of P. vittata in As-contaminated soils through hyphal uptake, promotion of plant growth, and alteration of rhizosphere environment (Pan et al., 2024). As absorption by mycorrhizal hyphae can be transported and relayed to mycorrhizae via symbiotic structures, such as arbuscular, accounting for approximately one-third of As in symbionts (Pan et al., 2024). Despite these advancements, challenges such as slow plant growth and low biomass of P. vittata hindered the overall phytoremediation efficiency. In recent years, China actively promoted the advancement of soil As pollution technologies, including the intercropping remediation approach that integrated hyperaccumulated plants, microorganisms, and economic crops on As-contaminated farmland (Ministry of Environmental Protection, 2015). This approach aims to enhance phytoremediation outcomes while also promoting sustainable agricultural practices and economic viability. Cotton (Gossypium spp.) is a fiber crop with large biomass, a developed root system, and high economic value, and it also shows more excellent tolerance (Bharwana et al., 2014; Li et al., 2020). Xinjiang Autonomous Region is the main cotton producing region in China; in Kuitun Prefecture of Xinjiang, there are 134,700 hectares of cotton planting area, and groundwater is the main source of irrigation water. However, the groundwater in Kuitun is contained with a natural As level of 800 ug/L, which exceeds the set limit of 50 ug/L for drinking water and 100 ug/L for irrigation (Chen et al., 2018). As a non-edible crop, Gossypium is in no risk of transferring HM into the food chain of humans; laboratory experiments show that the HM contents in the fiber of Gossypium grown in HM-polluted soils are negligible, making it a promising candidate for phytoremediation in HM-contaminated areas (Li et al., 2020). Planting cotton has been considered for potentially remediating HM-polluted soils in southern China (Ma et al., 2017). To gain a deeper understanding of the molecular mechanisms underlying the alleviation of As phytotoxicity by AMF symbiosis in cotton seedlings, a pot experiment was conducted for investigating the effect of F. mosseae inoculation on Ca2+ flux and concentration in Gossypium seedlings under As stress. Subsequently, transcriptomic analysis was used to elucidate the specific molecular mechanism of the Ca2+ signaling pathway, which was induced by F. mosseae inoculation under As stress. Our findings provide novel insights into their As tolerance mechanism of plants regulated by AMF and also offer a molecular theoretical basis for the ecological restoration function of mycorrhizal plants grown in As-contaminated soil in the future.
2 Materials and methods
2.1 Plant materials, AMF inoculum, and pot experiment
Seeds of cotton (Gossypium spp. cultivar: Dalingmian 69) were sourced from the Institute of Cotton Research, Chinese Academy of Agricultural Sciences. The cotton seeds were sterilized in 75% ethanol for 20 min, followed by deionized water three times. Subsequently, they were germinated in autoclaved wet sand at 25°C for 10 days. The healthy seedlings were selected and transplanted into plastic containers. Funneliformis mosseae (BGC XZ02A) was purchased from the Institute of Plant Nutrition and Resources, Beijing Academy of Agricultural and Forestry Sciences, China. F. mosseae inoculum was placed 3 cm below in each pot of mycorrhizal treatment, half of the pots received 30 g F. mosseae inoculum and the other half received 50 mL suspensions of 30 g of unsterilized F. mosseae inoculum filtrated by 10 μm ultrafiltration membrane.
2.2 Experimental procedure and design
The seedlings were cultivated in a greenhouse with a temperature of 20–35°C and relative humidity of 60–85% from April to July 2020. Each pot was supplemented with 100 mL of Hoagland’s solution (2.0 mmoL/L) weekly (Hoagland and Arnon, 1950; Liu et al., 2020), and the soil moisture was kept at 75% field capacity with deionized water by regular weighing. According to Risk Control Standard of Soil Pollution in Agricultural Land in China (GB 15618–2018), when the As concentration in farmland soil was higher than 100 mg/kg, it was considered high risk and was forbidden to plant agricultural products. The As5+ addition levels (100 mg·kg −1 dry soil) were applied as Na3AsO4 •12H2O solution which was thoroughly mixed with soils, in order to ensure uniform distribution of As in the soil. All Gossypium seedlings were harvested 3 months after AMF inoculation.
The design of the present study was set up in a completely randomized block arrangement with two factors: (1) AMF treatments, i.e., F. mosseae and a non-AMF inoculated control; (2) two As levels in soils, i.e., 0 and 100 mg kg −1 dry soil, respectively. Thus, this experiment included four treatments: (1) non-inoculated Gossypium under non-As stress (CK0), (2) non-inoculated Gossypium under As stress (CK100), (3) F. mosseae-inoculated Gossypium under non-As stress (FM0), and (4) F. mosseae-inoculated Gossypium under As stress (FM100). Hence, each of the four treatments had three replicates, making 12 pots (one seedling per pot).
2.3 Assessment of AMF colonization
Approximately 1 cm long segments of fine capillary root were cleared in 10% KOH (w/v) at 90°C for 60 min and acidified in 1% HCl (v/v) for 5 min. Then, the root samples were stained with 0.05% trypan blue (w/v) at 90°C for 30 min and destained overnight in a lactoglycerol solution (lactic acid, glycerol, and water, 1:1:1) (Phillips and Hayman, 1970). Forty root segments were randomly selected from each root sample, mounted in glycerin on microscope slides with coverslips, and examined at ×100–400 magnification under a microscope (Nikon YS100, Japan). The stained sub-samples were microscopically examined to determine the percentage of root pieces containing arbuscules, vesicles, and hyphae. Total colonization (T%), arbuscular colonization (A%), vesicular colonization (V%), and hyphal colonization (H%) were defined as the percent of mycorrhizal colonization in fine root segments at 15, 30, 45, 60, 75, and 90 days post-inoculation (dpi), respectively. The rates of mycorrhizal colonization were calculated using the method by Giovannetti and Mosse (1980). The percentage of AM fungal colonization was assessed and calculated as follows: Fungal colonization = 100% × (number of intersections with fungal structure/total number of counted intersections) (Ban et al., 2023).
2.4 Detection of Ca2+ fluxes in Gossypium leaves and roots
Ca2+ fluxes were measured by using the non-invasive micro-test technique (NMT-YG-100, Younger USA LLC, Amherst, MA, United States). After 3 months of growth, the Gossypium root tips (1–2 cm) and the middle of the third leaves were used for measuring steady ion fluxes. Each ion was measured by a specific selective micro-electrode for Ca2+. A continuous recording of ion flux was taken for 300 s for each sample. After drying at 75°C for 24 h, Gossypium leaves and roots were ground into powder. Ca2+ ions were extracted by H2O2-H2SO4 using a microwave digestion instrument (Bayue BYWB-40, Changsha, China) and, subsequently, assayed using an atomic absorption spectrophotometer (PerkinElmer PE800, Waltham, MA, USA).
2.5 Total RNA extraction, library preparation, and illumina HiSeq sequencing
The total RNA of Gossypium entire plant was extracted from 12 Gossypium samples using the tTRIzol® Reagent, according to the manufacturer’s protocol (Invitrogen, CA). The concentration and purity of extracted RNA were analyzed using a NanoDrop spectrophotometer ND-2000 (Thermo Fisher Scientific Inc., Waltham, MA, USA). Only high-quality RNA sample was selected for constructing the sequencing library. After purification, end repair, and ligation to sequencing adapters, RNA-seq transcriptome library was prepared by TruSeq RNA Sample Preparation Kit (Illumina Inc., San Diego, USA), according to the manufacturer’s instructions. Finally, libraries generated from the plants were sequenced by using the Illumina NovaSeq 6,000 platform at BIOZERON Co., Ltd (Shanghai, China). The raw reads are available in the NCBI SRA database under the accession number PRJNA1062123 (Release date: 2028-01-06).
2.6 De novo assembly, functional annotation, and identification of differential expression genes
Before transcript assembly, Cutadapt v2.10 software was employed to cut off adapter sequences (Martin, 2011). To obtain more reliable clean reads, greater than 10% undetermined bases, low-quality reads (Q-value ≤ 20), and poly-N were discarded. Trinity v2.10.0 software was operated for assembling with the filtered data and constructing the unigene set of sequences (Grabherr et al., 2011). Clean data from each sample were mapped back onto the assembly transcripts (mismatch = 2), and according to the mapping results, the read count for each gene was received using RSEM software. The corresponding fragments per kilobase of transcript per million mapped reads (FPKM) were obtained by calculating the number of mapped and filtered reads of each unigene (Trapnell et al., 2010). All unigenes were annotated through BLASTX against the databases NCBI protein nonredundant (NR), Swiss-Prot, Clusters of Orthologous Groups of proteins (COG), Gene Ontology (GO), and Kyoto Encyclopedia of Genes and Genomes (KEGG) at an E-value cutoff of 1e−5.
To detect differential expression genes (DEGs) between two different samples, the expression levels of each transcript were measured by the method of reads per kilobase of exon per million mapped reads (RPKM). RSEM online software (http://deweylab.biostatwisc.edu/rsem/) was used to quantify transcript abundances. Gene differential expression analysis was carried out by using R statistical package software EdgeR (http://www.bioconductor.org/packages/2.12/bioc/html/edgeR.html). To determine the transcriptional changes in the two species under As stress, DEGs were identified by comparing the expression levels at CK0 versus Fm0 and CK100 versus Fm100, respectively. To fully determine which genes were specifically modulated during AMF inoculation and/or As stress, transcripts with p-value < 0.05 and |log2FoldChange| ≥ 2 were identified as DEGs between any of the groups.
2.7 Functional enrichment of DEGs and identification of transcription factor
To gain insight into the functional categories of DEGs regulated by As stress and F. mosseae inoculation in Gossypium seedlings, GO and KEGG enrichment analyses were performed for DEGs in CK0 versus FM0 and CK100 versus FM100 comparisons. These analyses aimed to identify significantly enriched DEGs in GO terms and KEGG metabolic pathways at P -value ≤ 0.05 under the whole-transcriptome background by Goatools (https://github.com/tanghaibao/Goatools) and KOBAS (http://kobas.cbi.pku.edu.cn/home.do). TFs were identified with the Plant Transcription Factor Database v5.0 (http://planttfdb.cbi.pku.edu.cn/family).
2.8 Co-expression network analysis
A gene co-expression network analysis was conducted for CBLs and CIPKs in Gossypium using the Pearson correlation coefficient (PCC), which was calculated using SPSS 25 software (SPSS Statistics, IBM, United States). All CBL and CIPK gene pairs with PCC of p < 0.05 were collected using Cytoscape 3.10.1 software, to create the co-expression networks (Shannon et al., 2003). Similarly, a co-expression network of transcription factors and Ca2+ signaling pathway-related genes of Gossypium was also performed and displayed by the above method. Only TFs with a sub-network of at least two genes were considered for further analysis.
3 Results
3.1 Mycorrhizal colonization
In non-inoculated Gossypium roots, no structures of arbuscular mycorrhizal fungi (AMF), including hyphae, arbuscules, and vesicles, were found. However, in F. mosseae-inoculated roots, total colonization rate (T%), arbuscular colonization rate (A%), vesicular colonization rate (V%), and hyphal colonization rate (H%) increased significantly with time of post-inoculation (Figure 1). As stress in soils had a negative influence on F. mosseae colonization in Gossypium roots. Rapid T%, V%, and H% increase began on 30 days post-inoculation (dpi), and A% increase began on on 45 dpi. T%, A%, V%, and H% remained stable from 75 dpi. At 90 dpi, under non-As stress, T%, A%, V%, and H% were 57.12, 15.3, 27.12, and 36.50%, respectively. Under As stress, these values were lower at 46.32, 13.2, 20.45, and 28.42%, respectively.
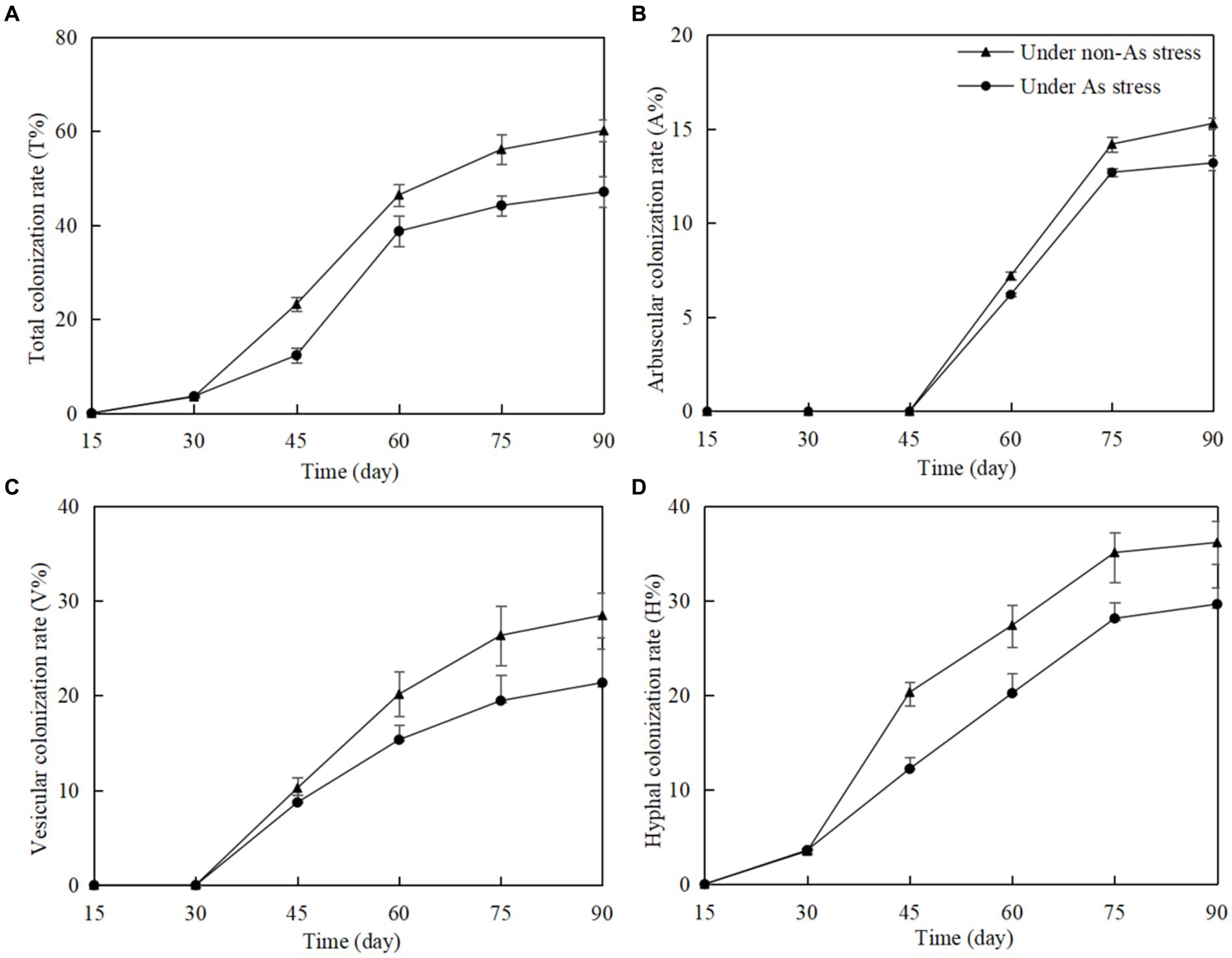
Figure 1. Funneliformis mosseae colonization rate in Gossypium roots at different growth time under non-As stress and As stress (A) total colonization rate; (B) arbuscular colonization rate; (C) vesicular colonization rate; (D) hyphal colonization rate.
3.2 Ca2+ Fluxes and contents in Gossypium roots and leaves
Ion kinetics investigations revealed the net fluxes of Ca2+ in Gossypium roots and leaves (Figure 2). As stress increased the net fluxes of Ca2+ in Gossypium roots (Figure 2A) and leaves (Figure 2B) under consistent inoculation conditions. However, F. mosseae inoculation decreased the net fluxes of Ca2+ in Gossypium roots and leaves under the same As level in soils. Under non-As stress condition, the mean net rates of Ca2+ fluxes in roots and leaves of non-inoculated seedlings were 38.58 and 18.15 pmol/(cm−2·s). In contrast, roots and leaves of F. mosseae-inoculated Gossypium were − 3.18 and 2.26 pmol/(cm−2·s), respectively (Figure 2C). Under As stress condition, the mean net rates of Ca2+ fluxes were 38.58 and 18.15 pmol/(cm−2·s) in the roots and leaves of non-inoculated seedlings and were − 3.18 and 2.26 pmol/(cm−2·s) in the roots and leaves of F. mosseae-inoculated seedlings, respectively (Figure 2C). The Ca2+ contents in Gossypium leaves were always higher than that in the roots under the same growth conditions. As stress decreased the the Ca2+ contents in Gossypium roots and leaves under the same inoculation conditions, but F. mosseae inoculation increased the Ca2+ contents in roots and leaves under the same As level in soils (Figure 2D).
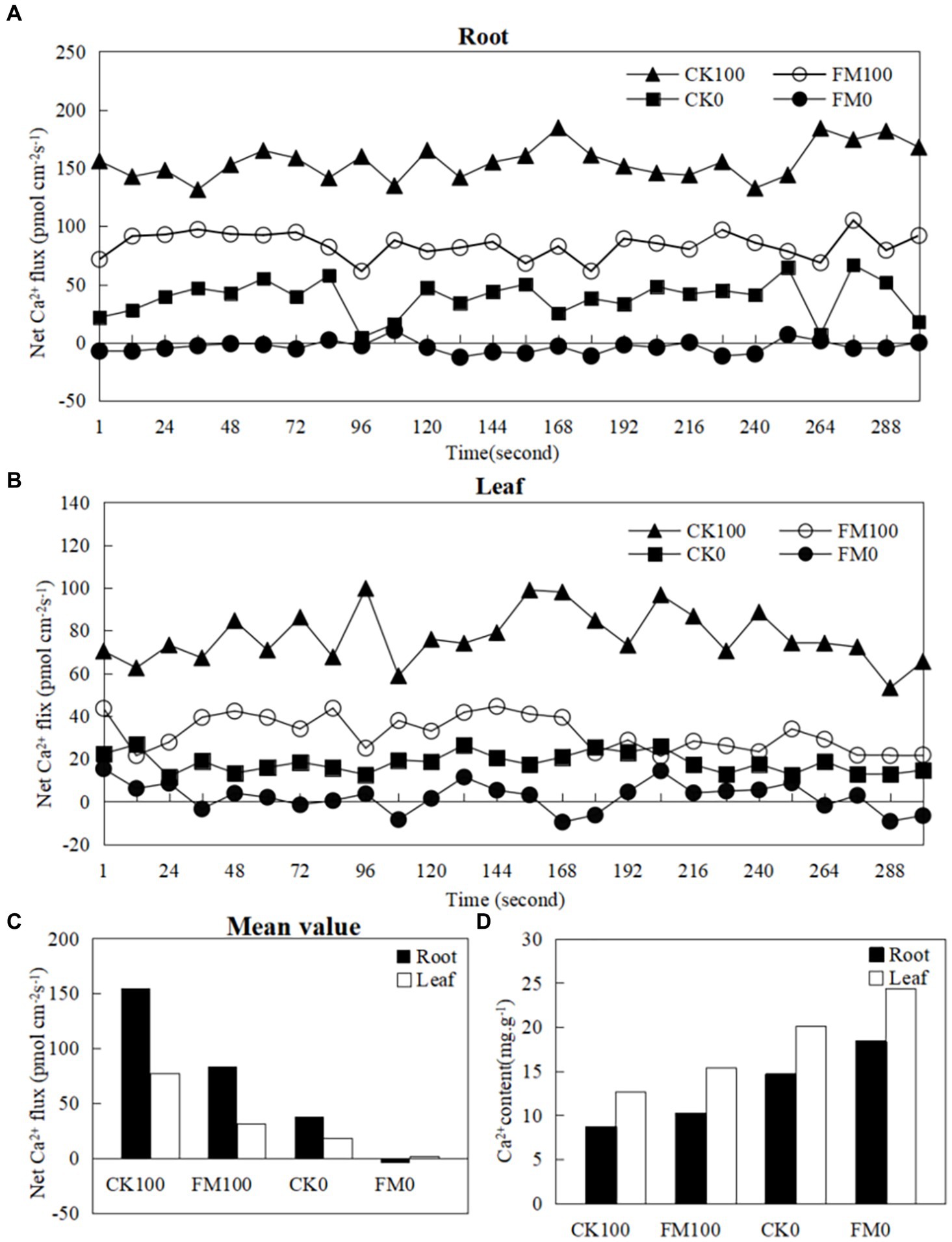
Figure 2. Effects of As stress and F. mosseae on the net Ca2+ fluxes and Ca2+ content in Gossypium roots and leaves. The flux rates of Ca2+ were measured for 300 s. (A) The root of net Ca2+ fluxes; (B) the leaf of net Ca2+ fluxes; (C) the mean value of net Ca2+ fluxes in Gossypium roots and leaves; (D) the Ca2+ content in Gossypium roots and leaves.
3.3 Transcriptome sequencing and gene annotation
The method of RNA-seq was employed for investigating the transcriptional responses of F. mosseae-inoculated Gossypium under As stress. A total of 12 cDNA libraries, i.e., CK0, CK100, FM0, and FM100, were constructed and sequenced in triplicates for each treatment. After deleting the low-quality raw reads (Q-value < 20) and trimming adaptor sequences, the percentage of Q30 base was > 92% and GC content was > 43% (Supplementary Table S1A). Clean reads were assembled into 73,746 expressed unigenes. The total number of expressed transcripts, all unigenes, and all transcripts were 124,000, 86,620, and 140,451, respectively (Supplementary Table S1B).
To obtain the annotation information of expressed unigenes and transcripts, BLAST search was used for comparing the assembled sequences with the GO, KEGG, COG, NR, Swiss-Prot, and Pfam databases. The obtained unigenes were functionally annotated with gene function, and there were 65,488, 31,198, 61,825, 82,630, 58,267, and 62,158 sequences annotated to GO, KEGG, COG, NR, Swiss-Prot, and Pfam databases, respectively. The total annotated number of expressed unigenes, expressed transcripts, all unigenes, and all transcripts in all six databases were 70,556, 120,016, 82,677, and 135,694, respectively (Supplementary Table S1B).
3.4 Identification of differential expression genes (DEGs) related to Funneliformis mosseae inoculation and as stress
The identification of DEGs linked to F. mosseae inoculation and As stress was achieved through a pairwise comparison of their expression levels. This analysis provided novel insights into the role of AMF symbiosis with Gossypium under As stress (Figure 3). Specifically, F. mosseae inoculation induced 3,129 DEGs under non-As stress condition; the upregulated and downregulated DEGs were 2,321 and 808, respectively (Figure 3A). When exposed to As stress, F. mosseae inoculation induced 1791 DEGs, and the upregulated and downregulated DEGs were 1,394 and 397, respectively (Figure 3B).
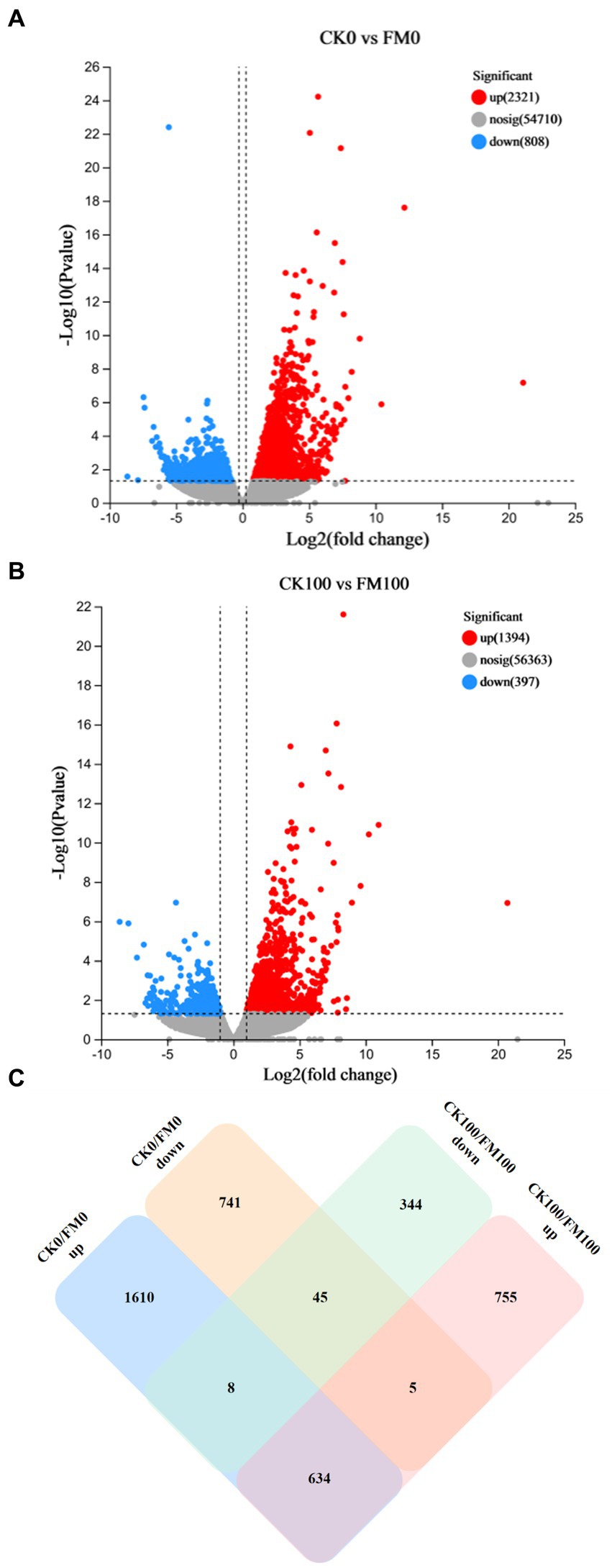
Figure 3. Analysis of upregulated and downregulated DEGs of Gossypium in the different comparisons. Volcano plot showed differentially expressed genes (A,B). Green and red dots indicated the significantly downregulated and upregulated genes, respectively. Grey dot indicated the non-significantly different genes. (C) Venn diagram.
A Venn diagram was generated to further explore the relationships among 4,142 DEGs from 4 different comparisons (Figure 3C). The comparison between CK100 and FM100 had 1,394 upregulated DEGs (categorized as 755, 5, and 634) and 397 downregulated DEGs (344, 45, and 8), and these above DEGs induced by F. mosseae inoculation may help Gossypium seedlings to alleviate As toxicity under As stress. In total, 1,099 DEGs (344 upregulated and 755 downregulated) in CK100 versus FM100 were different from those in CK0 versus FM0 (Figure 3C), and the above DEGs may be specific to F. mosseae-inoculated seedlings subjected to As stress. In total, 2,351 DEGs (741 upregulated and 1,610 downregulated) in CK0 versus FM0 comparison were different from those in CK100 versus FM100 comparison, and these DEGs may be specific DEGs in F. mosseae-inoculated seedlings under non-As stress conditions. In total, 679 DEGs (45 downregulated and 634 upregulated) were not regulated by As stress to maintain the same original trend, and the above genes are speculated to be key genes, to maintain the symbiotic relationship. Additionally, 13 DEGs (8 and 5) exhibited opposite trends under both As stress and non-As stress, implicating their involvement in regulating both stress responses and symbiotic relationships. Collectively, 1,112 DEGs (comprising 1,099 specific DEGs under As stress and 13 oppositely trending DEGs) were speculated to be specifically involved in the response of F. mosseae-inoculated Gossypium to As stress.
3.5 Functional annotation of DEGs induced by Funneliformis mosseae inoculation under as stress
To elucidate the mechanisms underlying the effect of F. mosseae inoculation on Gossypium seedlings under As stress, 1,791 upregulated and downregulated DEGs in the CK100 versus FM100 comparison were picked for GO and KEGG annotation evaluation (Figure 4). Out of these 1,791 DEGs, 1,441 were successfully categorized into at least one GO term, to characterize the biological processes (BP), cellular components (CC), and molecular function (MF) groups (Figure 4A). Within the BP group, genes were mainly annotated with ‘cellular process’ (311), ‘metabolic process’ (262), ‘biological regulation’ (176), and ‘response to stimulus’ (124). Within the CC group, ‘membrane part’ (508), ‘cell part’ (442), and ‘organelle’ (323) were the most represented. Within the MF group, the ‘binding’ (734), ‘catalytic activity’ (699), and ‘transcription regulator activity’ (132) were predominantly matched terms. Furthermore, the annotation analysis of the KEGG pathways revealed that 383 of 1,791 DEGs were subdivided into at least one metabolic pathway categories: metabolism (M), environmental information processing (EIP), organismal systems (OS), cellular processes (CP), and genetic information processing (GIP) groups (Figure 4B). DEGs mostly annotated with ‘folding, sorting, and degradation’ (15) in the GIP group, ‘transport and catabolism’ (16) in the CP group, ‘environmental adaptation’ (38) in the OS group, and ‘signal transduction’ (67) in the EIP group. In the M group, DEGs mainly annotated with ‘carbohydrate metabolism’ (70), ‘amino acid metabolism’ (35), ‘lipid metabolism’ (27), ‘biosynthesis of other secondary metabolites’ (25), ‘metabolism of terpenoids and polyketides’ (22), ‘metabolism of other amino acids’ (19), and ‘energy metabolism’ (18).
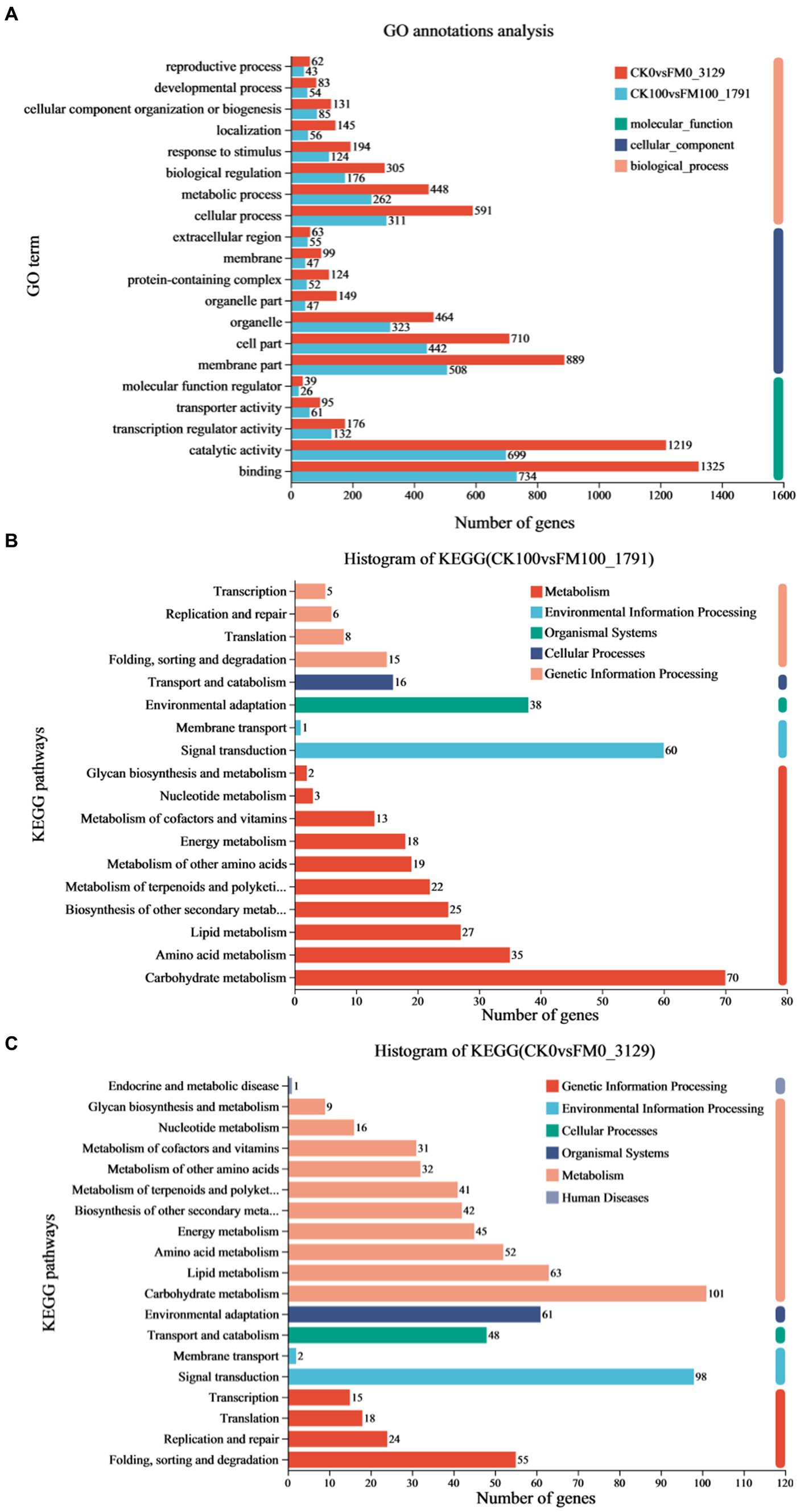
Figure 4. Analysis of functional annotation of DEGs of Gossypium induced by Funneliformis mosseae inoculation under As stress. (A) Significantly enriched biological process, molecular function, and cellular component of GO terms of DEGs in CK100 versus FM100 and CK0 versus FM0 comparison. (B) The histogram of KEGG in CK100 versus FM100 comparison. (C) The histogram of KEGG in CK0 versus FM0 comparison.
A comprehensive functional analysis of 3,129 differentially expressed genes (DEGs) was conducted in the CK0 versus FM0 comparison, utilizing GO and KEGG annotations. Among these DEGs, 2,521 were successfully categorized into at least one GO term across three functional categories to describe their BP, CC, and MF (Figure 4A). Within the BP group, DEGs mainly annotated with ‘cellular process’ (591), ‘metabolic process’ (448), ‘biological regulation’ (305), ‘response to stimulus’ (194), ‘localization’ (145), and ‘cellular component organization or biogenesis’ (131). Within the CC group, the dominant subcategories were ‘membrane part’ (889), ‘cell part’ (710), ‘organelle’ (464), ‘organelle part’ (149), and ‘protein-containing complex’ (124). Within the MF group, the ‘binding’ (1325), ‘catalytic activity’ (1219), and ‘transcription regulator activity’ (176) were predominantly matched terms. The results of KEGG analysis indicated that 754 of 3,129 DEGs in the CK0 versus FM0 comparison were subdivided into at least one metabolic pathway category: M, EIP, OS, CP, GIP, and Human disease (H) groups (Figure 4C). The M group, ‘carbohydrate metabolism’ (14) and ‘lipid metabolism’ (10), the OS group with ‘environmental adaptation’ (61), the CP group with ‘transport and catabolism’ (48), and the EIP group with ‘signal transduction’ were the most represented (98). Within the GIP group, the dominant subcategories were ‘folding, sorting, and degradation’ (55), ‘replication and repair’ (24), ‘translation’ (18), and ‘transcription’ (15).
3.6 Functional enrichment of DEGs induced by Funneliformis mosseae inoculation under as stress
GO enrichment analysis was used for elucidating the functions of DEGs, which can be classified under biological process, cellular component, and molecular function (Figure 5). The top 20 significantly enriched GO terms for DEGs (p-value ≤ 0.05) are shown in Figure 5. In the CK0 versus FM0 comparison, the most significantly enriched GO terms were ‘xyloglucan: xyloglucosyl transferase activity’, ‘xyloglucan metabolic process’, and ‘cell wall biogenesis’. A larger number of enriched GO terms were ‘membrane-bounded organelle’ (438) and ‘intracellular membrane-bounded organelle’ (431) (Figure 5A). In contrast, for the CK100 versus FM100 comparison, the most significantly enriched GO terms were ‘hemicellulose metabolic process’, ‘xyloglucan metabolic process’, and ‘glucosyltransferase activity’, and a larger number of enriched GO terms were ‘molecular function’ (1186), ‘organelle’ (323), and ‘intracellular organelle’ (322) (Figure 5B).
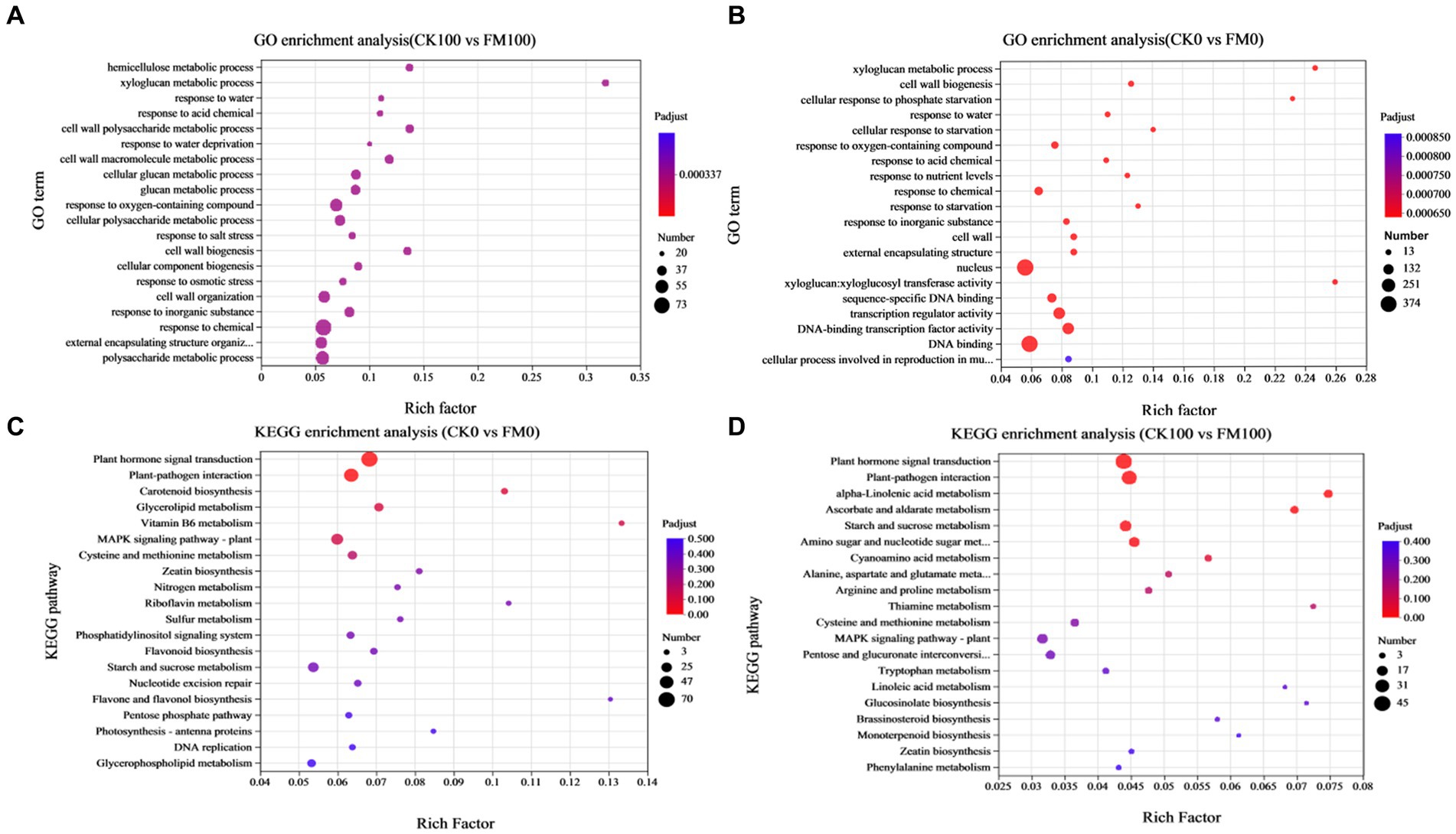
Figure 5. Enrichment analysis of DEGs of Gossypium induced by Funneliformis mosseae inoculation under As stress. (A) The GO enrichment bubble plot of DEGs in CK0 versus FM0. (B) The GO enrichment bubble plot of DEGs in CK100 versus FM100. (C) The KEGG pathway enrichment bubble plot of DEGs in CK0 versus FM0. (D) The KEGG enrichment bubble plot of DEGs in CK100 versus FM100.
According to the enrichment results of KEGG pathway analysis, 1,143 (CK0 vs. FM0) and 612 (CK100 vs. FM100) exclusive DEGs were enriched, and the top 20 significantly enriched KEGG pathways for DEGs (p-value ≤ 0.05) are shown in Figure 5, which are divided into six KEGG pathways of the first category. For the CK0 versus FM0 comparison, the top pathway of the first category involving the largest number of DEGs was ‘plant hormone signal transduction’ (70), ‘plant–pathogen interaction’ (54), and ‘MAPK signaling pathway-plant’ (36) (Figure 5C), and the most significant enriched pathways of first category were ‘vitamin B6 metabolism’, ‘flavone and flavonol biosynthesis’, and ‘riboflavin metabolism’. For the CK100 versus FM100 comparison, the pathways of the first category involving the largest number of DEGs were ‘plant hormone signal transduction’ (45), ‘plant–pathogen interaction’ (38), and ‘starch and sucrose metabolism’ (23), and the most significant enriched pathways of the first category were ‘alpha-linolenic acid metabolism’, ‘thiamine metabolism’, and ‘glucosinolate biosynthesis’ (Figure 5D).
3.7 Expression patterns of DEGs involved in ABC transport protein, calcineurin B-like (CBL)-interacting protein kinase (CIPK) protein, and Ca2+-transporting ATPase (CA) regulated by Funneliformis mosseae inoculation under as stress
To further elucidate the molecular mechanisms by which AMF mitigate the adverse effects of As stress, we examined the DEG expression patterns associated with Ca2+ signaling pathway genes, such as ABC, CBL, CIPK, and CA. Cluster analysis of gene sets found that 15 ABC genes were expressed in Gossypium seedlings, and these genes showed differential expression patterns (Figure 6A). F. mosseae inoculation downregulated the expression of ABCG3, ABCG9, ABCG14, ABCG17, and ABCG31, and upregulated the expression of 10 ABC genes (ABCG2, ABCG5, ABCG7, ABCG8, ABCG11, ABCG22, ABCG24, ABCG32, ABCG38, and ABCG39) under non-As stress. Under As stress, F. mosseae inoculation also triggered the expression of 12 ABC genes (ABCG2, ABCG3, ABCG5, ABCG7, ABCG8, ABCG11, ABCG14, ABCG22, ABCG24, ABCG31, ABCG32, and ABCG39) and repressed the expression of ABCG9 and ABCG17. As stress downregulated the expression of 11 ABC genes (ABCG2, ABCG3, ABCG5, ABCG8, ABCG9, ABCG14, ABCG22, ABCG24, ABCG31, ABCG32, and ABCG38) and promoted ABCG7, ABCG11, ABCG17, and ABCG39 under non-inoculaion condition. It also induced the expression of ABCG7, ABCG11, ABCG17, ABCG22, and ABCG39 and repressed the expression of 10 ABC genes (ABCG2, ABCG3, ABCG5, ABCG8, ABCG9, ABCG14, ABCG24, ABCG31, ABCG32, and ABCG38) under F. mosseae-inoculaion condition.
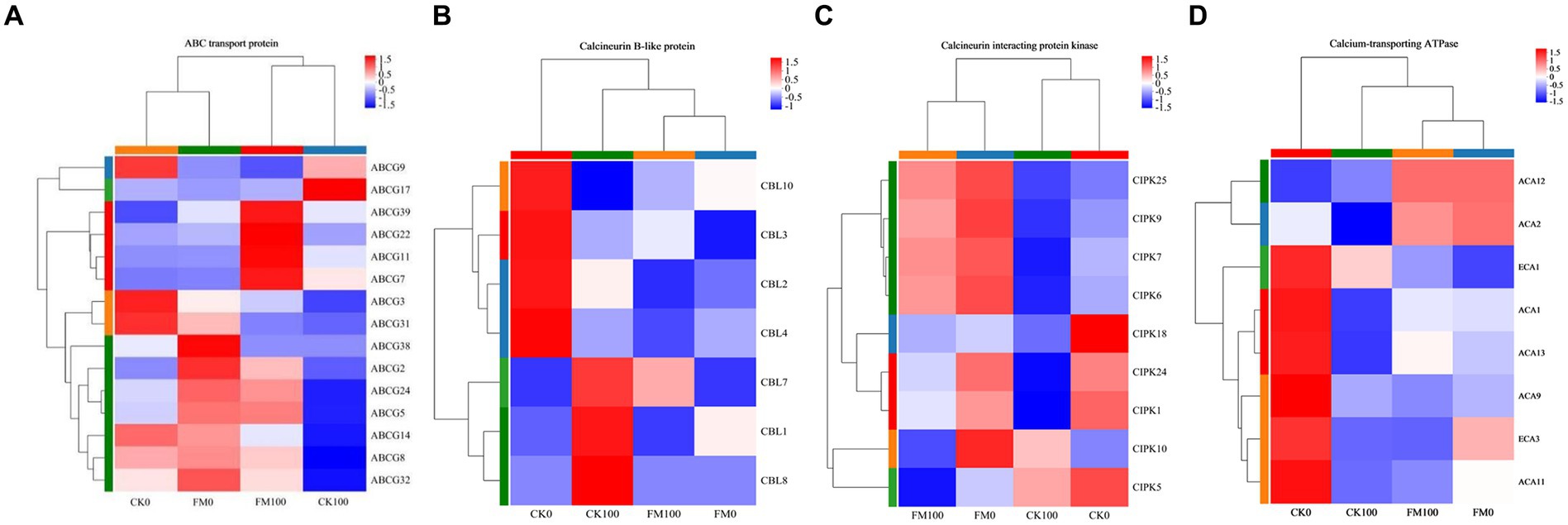
Figure 6. Heatmap of the expression patterns of DEGs of Gossypium involved in ABC transport protein, calcineurin B-like protein, calcineurin interacting protein kinase, and calcium-transporting ATPase protein regulated by AMF inoculation under As stress. Each column represents a sample, each row represents a gene, and the colors in the graph represent the expression level of the gene after standardization in each sample.
Cluster analysis of gene sets found that seven CBL genes were expressed in Gossypium seedlings, and these genes showed differential expression patterns (Figure 6B). F. mosseae inoculation downregulated the expression of CBL2, CBL3, CBL4, and CBL10 and upregulated the CBL1 expression under non-As stress. F. mosseae inoculation also induced the expression of CBL3 and CBL10 and repressed the expression of CBL1, CBL2, CBL4, CBL7, and CBL8 under As stress. As stress downregulated the expression of CBL2, CBL3, CBL4, and CBL10 and promoted CBL1, CBL7, and CBL8 under non-inoculaion condition. Under F. mosseae-inoculaion condition, As stress also induced the expression of CBL3 and CBL7 and repressed the expression of CBL1, CBL2, CBL4, and CBL10.
The nine CIPK genes of Gossypium were found to show differential expression patterns by cluster analysis of gene sets (Figure 6C). Under non-As stress, F. mosseae inoculation downregulated the expression of CIPK1, CIPK5, and CIPK18 and upregulated the expression of CIPK6, CIPK7, CIPK9, CIPK10, CIPK24, and CIPK25. Under As stress, F. mosseae inoculation also triggered the expression of CIPK1, CIPK6, CIPK7, CIPK9, CIPK18, CIPK24, and CIPK25 and repressed the expression of CIPK5 and CIPK10. Under non-inoculaion condition, As stress downregulated the expression of CIPK1, CIPK5, CIPK6, CIPK7, CIPK9, CIPK18, CIPK24, and CIPK25 and promoted CIPK10. It also induced the expression of CIPK1, CIPK5, CIPK6, CIPK7, CIPK9, CIPK10, CIPK18, CIPK24, and CIPK25 under F. mosseae-inoculaion condition.
Cluster analysis of gene sets found that eight calcium-transporting ATPase genes were expressed in Gossypium, and these genes showed differential expression patterns (Figure 6D). Under non-As stress, F. mosseae inoculation downregulated the expression of ECA1, ECA3, ACA1, ACA9, ACA11, and ACA13 and upregulated the expression of ACA2 and ACA12. Under As stress, F. mosseae inoculation also triggered the expression of ACA1, ACA2, ACA11, ACA12, and ACA13 and repressed the expression of ECA1, ECA3, and ACA9. As stress downregulated the expression of ECA1, ECA3, ACA1, ACA2, ACA9, ACA11, and ACA13 and upregulated ACA12 under non-inoculaion condition. It also increased the expression of ECA1, ACA1, and ACA13 and repressed the expression of ECA3, ACA2, ACA9, and ACA11 under F. mosseae-inoculaion condition.
Co-expression analysis identified gene pairs of CBL-CIPK signal transduction, responding to As stress and F. mosseae-inoculaion. Within this network, 63 regulatory edges represented 63 co-expression gene pairs of CBLs-CIPKs which are linked by their PCCs. In terms of relatedness, 6 out of 63 had positive and significant correlations. CBL7 and CBL10 showed the largest association with CIPKs. Among these, CBL2-CIPK5, CBL4-CIPK5, CBL7-CIPK7, CBL7-CIPK9, CBL10-CIPK1, and CBL10-CIPK7 had significant positive correlations in response to both As stress and F. mosseae-inoculaion (Figure 7).
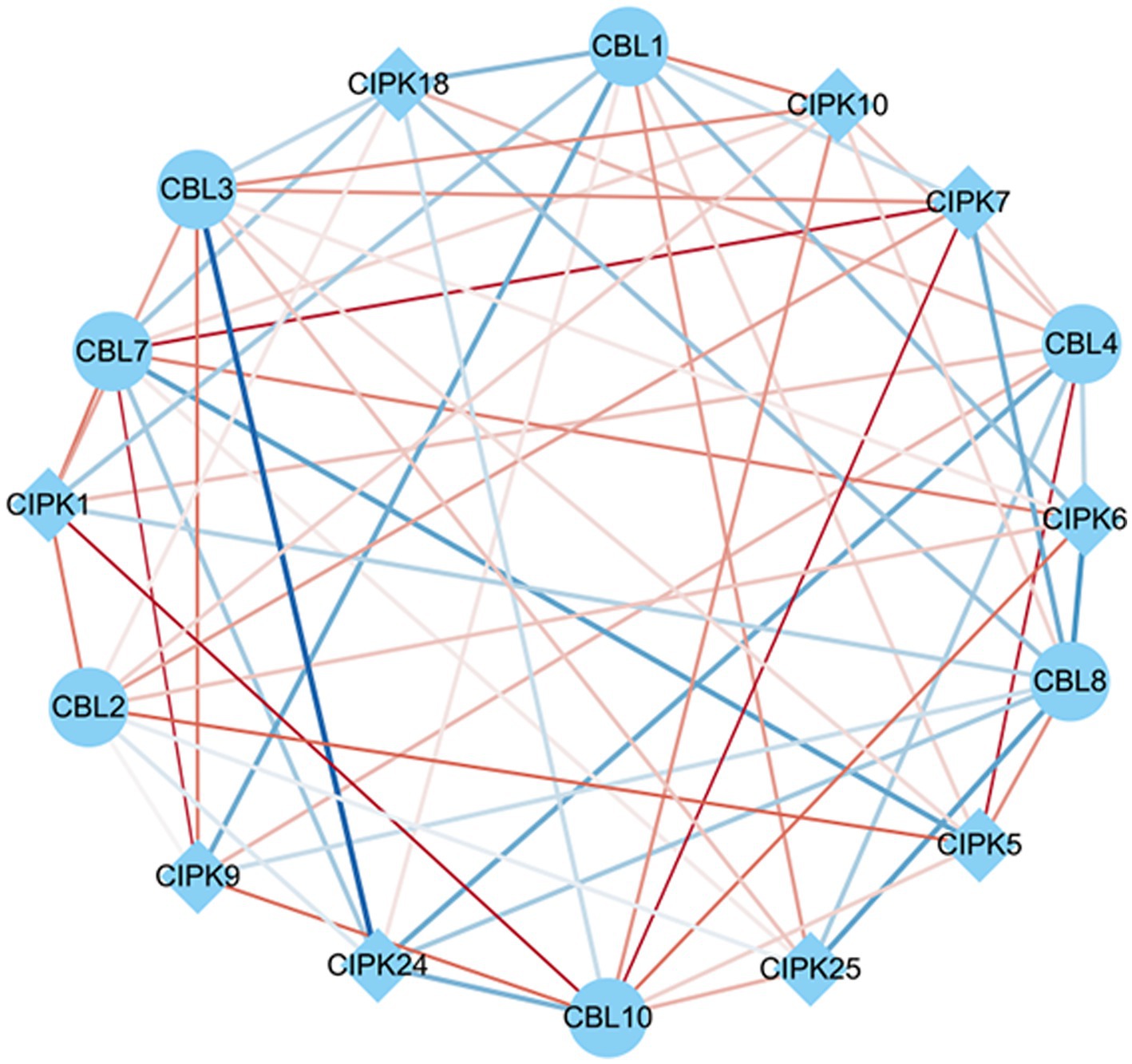
Figure 7. Co-expression networks of calcineurin B-like proteins (CBLs) and calcineurin B-like protein interacting protein kinases (CIPKs) in response to Funneliformis mosseae inoculation under As stress. The co-expression networks were established based on the Pearson correlation coefficients (PCC) of the CBL and CIPK pairs. The different line sizes indicate different relevance levels of co-expression gene pairs. Positive correlations are indicated with red lines (PCC ≥ 0.6), whereas blue lines indicate a negative correlations (PCC ≤ −0.6).
3.8 Identification and co-expression network analysis of transcription factors (TFs)
To gain insight into the mechanisms by which AMF inoculation alleviates As stress in Gossypium seedlings, 336 and 276 DEGs encoding TF family proteins were found to differentially express in CK0 versus FM0 and CK100 versus FM100 comparisons, respectively. The expression of TF genes in Gossypium belonged to 34 families, primarily belonging to the TF families of bHLH, HB-other, ERF, MYB, MYB-related, NAC, and WRKY (Figure 8A). The number of upregulated DEGs was much higher than that of downregulated DEGs in both comparisons, respectively. In the CK100 versus FM100 comparison, F. mosseae inoculation led to the upregulation of most DEGs in the ERF, MYB, NAC, and WRKY families, while it downregulated DEGs in the MYB and HB-other families. Among these, ERF and MYB had the highest number of upregulated and downregulated DEGs, respectively. Similarly, in CK0 versus FM0 comparison, F. mosseae inoculation upregulated DEGs in the ERF, MYB, NAC, WRKY, MYB-related, and bHLH families, while F. mosseae inoculation downregulated MYB. ERF and MYB had the highest number of upregulated and downregulated DEGs, respectively.
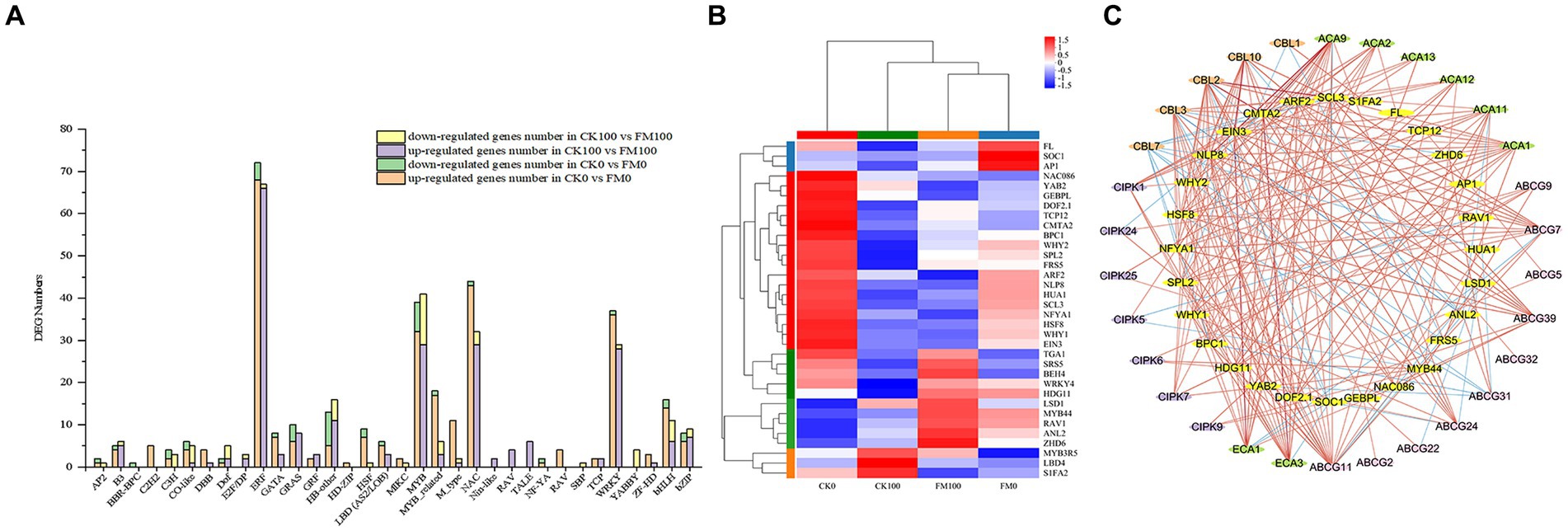
Figure 8. Identification and co-expression network analysis of transcription factors in Gossypium. (A) Analysis of DEG numbers of transcription factors (TFs) in CK100 versus FM100 and CK0 versuss FM0. X-axis represents TF family, and Y-axis represents DEG numbers. (B) Heatmap of the expression patterns of the 34 TFs regulated by AMF inoculation under As stress. (C) Co-expression network analyses of 28 TFs regulated by AMF inoculation under As stress. Red and blue lines denote positive correlation and negative correlation, respectively.
Characterizing the expression profiles of TFs was a crucial step in transcriptome analyses, 34 TFs were screened and found to significantly express among 4 treatments in this study (Figure 8B). Under As stress, F. mosseae inoculation resulted in the upregulation of 23 TFs and downregulation of 11 TFs, respectively. Many of the upregulated TFs, such as WRKY4, MYB44, and HSF8, are known to be involved in abiotic stress response. Downregulated TFs, including MYB3R5, NAC086, and ARF2, were also identified.
The co-expression network analysis was conducted to highlight specific interactions between key TFs and genes related to the Ca2+ signaling pathway. Only the TFs with a sub-network of at least two genes were considered for further analysis (6 out of 34 TFs and 9 out of 39 genes). The co-expressed network contained 30 genes associated with the Ca2+ signaling pathway and 28 TFs, forming the main network (Figure 8C). The network showed positive correlations between several co-expressed genes and TFs, suggesting TF-inductive regulations. SCL3 TFs demonstrated the highest number of connections within the main co-expressed network. Additionally, several previously mentioned DEGs with important roles were found to co-express together with specific TFs. For instance, CBL10 and ECA1 were co-expressed with EIN3, while both TFs CMAT2 were connected to ECA3 and ACA9. Furthermore, CBL2, CBL2, ACA9, and ECA3 were co-expressed with SCL3.
4 Discussion
In our previous studies, Gossypium (Cultivar: Dalingmian 69) seedlings were inoculated with R. intraradices and F. mosseae separately under As stress; the results demonstrated that both AMF strains formed symbiotic relationships with Gossypium roots. Furthermore, single R. intraradices or F. mosseae inoculation individually enhanced the Gossypium tolerance to As toxicity by increasing the net photosynthetic rate and light energy utilization efficiency, improving the osmotic regulation and antioxidant capacity, alleviating the As inhibition on root development, and ultimately reducing the arsenic concentration in Gossypium seedlings (Liu et al., 2021; Gong et al., 2022). This present study aimed to investigate the transcriptional changes induced by F. mosseae inoculation in Gossypium seedlings under As stress. Our results revealed that both As stress and F. mosseae inoculation induced a large number of changes in gene expression, with 4,998 DEGs identified through transcriptome sequencing.
4.1 AMF-induced Ca2+ fluxes and contents under as stress
In plants, Ca2+ ions serve as a crucial secondary messenger responding to various environmental stimuli, such as heat, salt, cold, drought, and heavy metal stresses (Sanders et al., 1999). Under abiotic stress conditions, plants release Ca2+ from both extracellular and intracellular sources into cytoplasm through Ca2+ channels, leading to a rapid increase in cytosolic-free Ca2+ concentration (Zhu et al., 2020). Ca2+ fluxes play a pivotal role in the early defense responses of plants, exhibiting complex spatiotemporal patterns (Kurusu et al., 2010). Positive values of Ca2+ fluxes indicate an outflow, while negative values suggested an inflow (Zhu et al., 2020). In this study, the Ca2+ fluxes in Gossypium leaves and roots were investigated by using non-invasive micro-test technique; As stress increased the net fluxes of Ca2+ in both leaves and roots under the same inoculation condition. However, F. mosseae inoculation significantly decreased the net fluxes of Ca2+ in Gossypium leaves and roots exposed to the same As level. This indicated that F. mosseae inoculation reduced the Ca2+ outflow and increased the Ca2+ inflow in Gossypium seedling compared with non-inoculated seedling. The increased Ca2+ inflow activated Ca2+ −binding proteins, upregulated the genes of Na+/H+ antiporter, and eventually improved the stress resistance of plants (Jiang et al., 2019). Similarly, ectomycorrhizal Paxillus involutus-inoculated roots retained higher Ca2+ influx than non-inoculated Populus × canescens roots under cadmium stress. The enrichment of Ca2+ in ectomycorrhizal roots was associated with the high capacity of P. involutus fungal hyphae for the uptake of Ca2+ (Zhang et al., 2017). In our study, As stress decreased Ca2+ contents in Gossypium roots and leaves under the same inoculation conditions. However, F. mosseae inoculation increased Ca2+ contents in Gossypium roots and leaves exposed to the same As level in soil. Wang et al. (2022) also reported that R. irregularis inoculation increased Ca2+ contents, reduced the Na+/Ca2+ ratio, and maintained ion balance in Casuarina glauca roots under NaCl stress.
4.2 Functional annotation and enrichment of DEGs induced by Funneliformis mosseae inoculation under as stress
The establishment of symbiosis between AMF and host plants often led to transcriptional changes in host plants, conferring multiple benefits to plant growth, development, and stress tolerance (Bucher et al., 2014). Annotating and enriching DEGs identified in CK0 versus FM0 and CK100 versus FM100 comparisons can elucidate the similarities and differences of transcriptome expression between non-inoculated and F. mosseae-inoculated Gossypium seedlings under As stress. Our analysis revealed that both comparisons were significantly enriched in 353 and 90 GO terms, respectively, which indicated that F. mosseae inoculation has a broad effect on the gene expression of Gossypium seedlings under both As stress and non-As stress conditions. According to Wang et al. (2022), similar transcriptome analysis in Casuarina glauca identified 110 and 119 significantly enriched GO terms in RN0 versus RN600 and RA0 versus RA600 comparisons, respectively (where N represents non-inoculated with AMF, A represents inoculated with R. irregularis, 0 represents no NaCl stress, and 600 represents 600 mM NaCl stress). Most GO terms appeared in CK100 versus FM100 and were primarily related to biological processes (such as ‘metabolic process’ and ‘cellular process’), cellular components (such as ‘cell part’, ‘organelle’, and ‘membrane part’), and molecular functions (such as ‘binding’, ‘catalytic activity’, and ‘transcription regulator activity’), which indicated that F. mosseae inoculation regulated these key pathways in Gossypium seedlings to alleviate As toxicity. In these major pathways, F. mosseae inoculation could regulate related DEGs, enabling Gossypium seedlings to better cope with As stress.
4.3 AMF-induced CBL-CIPK gene expression under as stress
The calcineurin B-like protein (CBL)-interacting protein kinase (CIPK) complex was a primary Ca2+ sensor in perceiving a range of signals associated with biotic and abiotic stresses (Shu et al., 2020). Stress-induced Ca2+ signals were decoded by many Ca2+ sensors, including the CBL-CIPK signaling pathway, and in turn, downstream cascades were activated, finally leading to plant physiological and developmental response (Chen et al., 2011; Aslam et al., 2019). MeCIPK23 interacted with MeCBL1 and MeCBL9 in Manihot esculenta; over-expression of three genes improved the plant’s defense against Xanthomonas axonopodis pv manihotis infection (Yan et al., 2018). Similarly, overexpressing ScCBL genes in sugarcane (Saccharum spp.) resulted in altered expression of immunity genes in Nicotiana benthamiana leaves, bolstering the plant’s resistance to Ralstonia solanacearum infection (Su et al., 2020). AMF colonization also downregulated three CIPKs in wheat under higher salt stress (Puccio et al., 2023). In Citrus sinensis, eight CsCBLs exhibited differential expression patterns in the roots; AMF colonization induced the expression of CsCBL4, 5, 6, and 7 and repressed the expression of CsCBL1, 2, 3, and 8 under both well-watered and drought conditions; furthermore, drought significantly downregulated the expression of CsCBL8 and upregulated CsCBL7 under non-AMF condition (Shu et al., 2020). The expression profile of Ananas comosus AcCBL and AcCIPK genes expressed differentially in different tissues and development stages (Aslam et al., 2019). In our study, 7 CBLs and 9 CIPK genes were identified and characterized the expression in F. mosseae-inoculated Gossypium under As stress, which indicated that these genes play vital roles in the CBL-CIPK signaling pathway under As stress. Not all 7 CBLs and 9 CIPKs genes were consistently upregulated or downregulated by F. mosseae inoculation or As stress, which may be attributed to the integration of gene regulatory elements, such as cis-elements and trans-acting factors.
4.4 AMF-induced Ca2+-transporting ATPase gene expression under as stress
Ca2+−transporting ATPase play a crucial role in plants by transporting Ca2+ across cell membranes against an electrochemical gradient using the energy released by the hydrolysis of ATP molecules, which is one of the important physiological processes of the Ca2+ signaling pathways in plants (Geisler et al., 2000; Axelsen and Palmgren, 2001). The Ca2+-ATPases participated in all the stages of the plant life cycle including growth, development, immunity, and responses to environmental stress (Kamrul et al., 2013). Bago et al. (1997) found that ATPase activities of plasmalemma, mitochondria, and vacuolae were increased in mycorrhizal roots at the later colonization stage. The relative expression levels of both Ca2+−transporting ATPase genes (CA1 and CA9) were analyzed in F. mosseae-inoculated cucumber roots under chilling stress by using quantitative real-time PCR; the expression levels of both genes in F. mosseae-inoculated treatment were increased by 1.60 and 0.74 folds compared with non-inoculated treatment, respectively, which indicated that Ca2+ -ATPase genes played an important role in AMF-mediated chilling tolerance of cucumber (Liu et al., 2014). Similarly, in wheat, moss, soybean, and rice seedlings, two genes of P2B Ca2+ATPase were upregulated by AMF inoculation under salt stress (Puccio et al., 2023). The expression of some Ca2+−transporting ATPase in blueberry seedlings was upregulated by F. mosseae inoculation under high pH conditions (Yang et al., 2020). In this study, As stress downregulated 7 of 8 Ca2+ −transporting ATPase genes under non-inoculation condition, but F. mosseae inoculation upregulated the 5 of 8 Ca2+−transporting ATPase genes under As stress, and these findings suggested that F. mosseae inoculation positively impacted the tolerance of Gossypium seedlings to arsenic by enhancing the gene expression of Ca2+−transporting ATPase and improving Ca2+ signaling pathway.
4.5 AMF-induced ABC transporter gene expression under as stress
The ATP binding cassette (ABC) transporters constitute the most extensive conserved families of integral membrane proteins in plants and play essential roles in the detoxification of HMs in plants by facilitating the transport of phytochelatin-heavy metal (PC-HM) complexes to the vacuoles (Ban et al., 2023). ABC transporter proteins are a group of transmembrane transport proteins commonly found in prokaryotes and eukaryotes, which have a large and diverse range of functions. In plants, ABC transporters can enhance the host plant’s heavy metal tolerance (Guo et al., 2022). As ions with thiol-rich compounds, such as glutathione or PCs, reduced their mobility and sequestered into the vacuole by the ABC transporters (Zhao et al., 2010). Two ABC transporters (AtABCC1 and AtABCC2) in Arabidopsis were proven to improve As (V) tolerance in plants (Song et al., 2010). The reports suggest that ABC transporter genes were predominantly expressed in plant cells containing arbuscules, and expression of the upregulated ABC genes was stimulated during the signal communication between AMF and roots of host plants under HM stress (Shabani et al., 2016; Li et al., 2018b). When Populus × canescens seedlings were inoculated with ectomycorrhizas Paxillus Involutus, cadmium stress led to the downregulation of the ABCC1 gene in mycorrhizal roots (Ma et al., 2013). However, under CuO-NPs and/or flood stress, the gene expression of ABC transporters in Phragmites Australis was upregulated by F. mosseae inoculation, which indicated that upregulating gene expression of AMF-regulated ABC transporters played a key role in plant response to abiotic stress (Ban et al., 2023). Similarly, ABC transporter transcripts were accumulated to significantly higher levels in F. mosseae-inoculated tall fescue compared with non-mycorrhizal plants under nickel (Ni) stress, and these upregulated ABC genes alleviated Ni-induced stress in tall fescue (Shabani et al., 2016). Zhang et al. (2021) further verified that AMF symbiosis enhanced water and nutrient uptake and homeostasis under salinity stress, which induced the expression of DEG-encoding ABC transporter proteins. In our study, under As stress, 15 genes encoding ABC transport protein were screened from the transcriptome gene set and were found to be significantly induced by F. mosseae inoculation, and 12 ABC genes were upregulated by F. mosseae inoculation, which indicated that AMF symbiosis positively enhanced As tolerance in Gossypium seedlings due to the upregulated ABC gene expression.
4.6 Regulation of transcription factors by Funneliformis mosseae inoculation under as stress
The transcription factors constituted a category of protein sequences that bound to a specific template chain of DNA, thereby governing the transcription from DNA to mRNA (Du et al., 2016; Wang et al., 2022). Many transcription factors, including NAC, MYB, bHLH, ERF, BBX, and WRKY, played pivotal roles in plant growth and development, and they were implicated in the regulation of certain plant genes related to physiological and biochemical processes under abiotic stresses (Cheng et al., 2018). In this study, Gossypium seedlings exhibited expression of TF genes belonging to 34 families, with prominent representation from bHLH, ERF, MYB, MYB-related, NAC, and WRKY families. A large number of TF genes (MYB and NAC) related to cell wall pathways were enriched during inoculation with F. mosseae under As stress, which suggests that F. mosseae inoculation possibly increased the As tolerance of Gossypium seedlings by improving the gene expression of TFs related to the development of cell walls and the degree of lignification. Similar results were also found in R. irregularis-inoculated Asparagus officinalis and Casuarina glauca under NaCl stress (Zhang et al., 2021; Wang et al., 2022). The upregulation of MYB, WRKY, ERF, and bHLH TF genes in plant roots was also considered to be an important trigger of the gene expression, underlying the biotic or abiotic stress response (Corso et al., 2018). In this study, the bHLH transcription factors were mainly downregulated by As stress but upregulated by F. mosseae inoculation, which was similar to the study by Zhang et al. (2021) on R. irregularis-inoculated Asparagus officinalis L. under salt stress. Puccio et al. (2023) identified 17 wheat TFs regulated by AMF inoculation, which included two WRKY70, one NAC68, and two bHLH. Overexpression of WRKY, NAC, and bHLH members was proven to improve abiotic and biotic stress responses in Oryza sativa L., Arabidopsis, and Nicotiana benthamiana (Ouziad et al., 2006; Aroca et al., 2007; Corso et al., 2018). Wang et al. (2009) found that NAC TFs regulated defense responses against necrotrophic fungal and bacterial pathogens in tomato seedlings. The WRKY6 TF repressed the expression of PHT1:1 gene and simultaneously restricted As-induced transposon activation, which decreased As (V) uptake and increased As tolerance of Arabidopsis thaliana (Castrillo et al., 2013).
5 Conclusion
In our previous studies, F. mosseae-inoculated Gossypium seedlings exhibited superior physiological performance under As stress, which was attributed to improved photosynthetic, osmotic regulation, and antioxidant capacity, alleviated the As inhibition on root development, and reduced the As concentration in Gossypium seedlings (Liu et al., 2021; Gong et al., 2022). In the present study, the inoculation of F. mosseae was found to decrease the net fluxes of Ca2+ while increased the Ca2+ contents in roots and leaves of Gossypium under the same As level in soil. To elucidate molecular mechanism related to the Ca2+ signaling pathway on how AM fungi could help to alleviate As toxicity, AMF-inoculated and non-inoculated Gossypium seedlings under distinct concentrations of Na3AsO4 •12H2O were subjected to high throughput RNA sequencing, respectively. Based on transcriptome data, we provided a global gene profile of F. mosseae-inoculated Gossypium seedlings under As stress. Specifically, 199 and 3,129 differentially expressed genes (DEGs) were uniquely regulated by F. mosseae inoculation under As stress and non-As stress, respectively. Notably, a significant enrichment of genes related to the Ca2+ signaling pathway was observed. Furthermore, transcription factors were identified as potentially crucial players in facilitating the relief of As stress. The enrichment of GO (Gene Ontology) and KEGG (Kyoto Encyclopedia of Genes and Genomes) revealed that F. mosseae inoculation under As stress upregulated the expression of several Ca2+ signaling pathway genes associated with cellular process, membrane part, and signal transduction, which may contribute to alleviating the As toxicity of Gossypium seedlings. Additionally, F. mosseae inoculation was found to regulate specific DEGs belonging to the transcription factor families ERF, MYB, NAC, WRKY, MYB, and HB-other, which played important roles in mitigating the detrimental effect of As stress on Gossypium. Overall, this study provides valuable insights into the molecular theoretical basis underlying the improvement in As tolerance in host plants induced by AMF inoculation, particularly focusing on the Ca2+ signaling pathway.
Data availability statement
The original contributions presented in the study are included in the article/Supplementary material, further inquiries can be directed to the corresponding author.
Author contributions
MG: Writing – review & editing, Writing – original draft, Funding acquisition. NB: Writing – original draft, Methodology, Formal analysis. JS: Writing – review & editing, Methodology. YuW: Writing – original draft, Validation. YaW: Writing – review & editing, Data curation. QZ: Writing – review & editing, Funding acquisition.
Funding
The author(s) declare that financial support was received for the research, authorship, and/or publication of this article. This study was supported by the National Natural Science Foundation of China (Nos. 31870093, and 31800096), and the Natural Science Foundation of Henan Province, China (No. 182300410050).
Acknowledgments
The authors would like to thank all contributing authors for their submissions to this Research Topic and all the reviewers for their valuable comments and inputs.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2024.1362296/full#supplementary-material
References
Aroca, R., Porcel, R., and Ruiz-Lozano, J. M. (2007). How does arbuscular mycorrhizal symbiosis regulate root hydraulic properties and plasma membrane aquaporins in Phaseolus vulgaris under drought, cold or salinity stresses? New Phytol. 173, 808–816. doi: 10.1111/j.1469-8137.2006.01961.x
Aslam, M., Fakher, B., Jakada, B. H., Zhao, L. H., Cao, S. J., Cheng, Y., et al. (2019). Genome-wide identification and expression profiling of CBL-CIPK gene family in pineapple (Ananas comosus) and the role of AcCBL1 in abiotic and biotic stress response. Biomol. Ther. 9:293. doi: 10.3390/biom9070293
Axelsen, K. B., and Palmgren, M. G. (2001). Inventory of the superfamily of P-type ion pumps in Arabidopsis. Plant Physiol. 126, 696–706. doi: 10.1104/pp.126.2.696
Bago, B., Donaire, J. P., and AZCÓN-AGUILAR, C. (1997). Atpase activities of root microsomes from mycorrhizal sunflower (Helianthus annuus) and onion (allium cepd) plants. New Phytol. 136, 305–311. doi: 10.1046/j.1469-8137.1997.00741.x
Ban, Y. H., Tan, J. Y., Xiong, Y., Mo, X. T., Jiang, Y. H., and Xu, Z. Y. (2023). Transcriptome analysis reveals the molecular mechanisms of Phragmites australis tolerance to CuO-nanoparticles and/or flood stress induced by arbuscular mycorrhizal fungi. J. Hazard. Mater. 442:130118. doi: 10.1016/j.jhazmat.2022.130118
Bharwana, S. A., Ali, S., Farooq, M. A., Ali, B., Iqbal, N., Abbsa, F., et al. (2014). Hydrogen sulfide ameliorates lead-induced morphological, photosynthetic, oxidative damages and biochemical changes in cotton. Environ. Sci. Pollut. Res. 21, 717–731. doi: 10.1007/s11356-013-1920-6
Bucher, M., Hause, B., Krajinski, F., and Küster, H. (2014). Through the doors of perception to function in arbuscular mycorrhizal symbioses. New Phytol. 204, 833–840. doi: 10.1111/nph.12862
Bush, D. S. (1995). Calcium regulation in plant cells and its role in signalling. New Phytol. 171, 249–269. doi: 10.1111/j.1469-8137.2006.01777.x
Castrillo, G., Sanchez-Bermejo, E., De Lorenzo, L., Crevillen, P., Fraile-Escanciano, A., Tc, M., et al. (2013). WRKY6 transcription factor restricts arsenate uptake and transposon activation in Arabidopsis. Plant Cell 25, 2944–2957. doi: 10.1105/tpc.113.114009
Chakrabarty, D., Trivedi, P. K., Misra, P., Tiwari, M., Shri, M., Shukla, D., et al. (2009). Comparative transcriptome analysis of arsenate and arsenite stresses in rice seedlings. Chemosphere 74, 688–702. doi: 10.1016/j.chemosphere.2008.09.082
Chen, B., Xiao, X. Y., Zhu, Y. G., Smith, F. A., Xie, Z. M., and Smith, S. E. (2007). The arbuscular mycorrhizal fungus glomus mosseae gives contradictory effects on phosphorus and arsenic acquisition by Medicago sativa Linn. Sci. Total Environ. 379, 226–234. doi: 10.1016/j.scitotenv.2006.07.038
Chen, T., Su, Y., and Yuan, X. (2018). Influx and efflux of arsenic in cotton fields irrigated with arsenic-contaminated groundwater. Biorem. J. 22, 103–111. doi: 10.1080/10889868.2018.151661
Chen, X., Gu, Z., Liu, F., Ma, B., and Zhang, H. (2011). Molecular analysis of Rice CIPKs involved in both biotic and abiotic stress responses. Rice Sci. 18, 1–9. doi: 10.1016/S1672-6308(11)60001-2
Cheng, P. L., Gao, J. J., Feng, Y. T., Zhang, Z. X., Liu, Y. N., Fang, W. M., et al. (2018). The chrysanthemum leaf and root transcript profiling in response to salinity stress. Gene 674, 161–169. doi: 10.1016/j.gene.2018.06.075
Corso, M., Doccula, F. G., de Melo, J. R. F., Costa, A., and Verbruggen, N. (2018). Endoplasmic reticulum-localized CCX2 is required for osmotolerance by regulating ER and cytosolic Ca2+ dynamics in Arabidopsis. Proc. Natl. Acad. Sci. 115, 3966–3971. doi: 10.1073/pnas.1720422115
Du, X. L., Wang, G., Ji, J., Shi, L. P., Guan, C. F., and Jin, C. (2016). Comparative transcriptome analysis of transcription factors in different maize varieties under salt stress conditions. Plant Growth Regul. 81, 183–195. doi: 10.1007/s10725-016-0192-9
Geisler, M., Axelsen, K. B., Harper, J. F., and Palmgren, M. G. (2000). Molecular aspects of higher plant P-type Ca2+-ATPases. Biochim. Biophys. Acta 1465, 52–78. doi: 10.1016/S0005-2736(00)00131-0
Giovannetti, M., and Mosse, B. (1980). An evaluation of techniques for measuring vesicular arbuscular mycorrhizal infection in roots. New Phytol. 84, 489–500. doi: 10.1111/j.1469-8137.1980.tb04556.x
Gong, M. G., Liu, K. Y., Wei, Y. N., Bai, N., Qiu, Z. J., and Zhang, Q. M. (2022). Effects of arbuscular mycorrhizal fungi on photosynthetic characteristics and mesophyll cell ultrastructure of cotton under arsenic stress. Cotton. Sci. 34, 256–266. doi: 10.11963/cs20220022
Gonzalez-Chavez, C., Harris, P. J., Dodd, J., and Meharg, A. A. (2002). Arbuscular mycorrhizal fungi confer enhanced arsenate resistance on Holcus lanatus. New Phytol. 155, 163–171. doi: 10.1046/j.1469-8137.2002.00430.x
Grabherr, M. G., Haas, B. J., Yassour, M., Levin, J. Z., Thompson, D. A., Amit, I., et al. (2011). Full-length transcriptome assembly from RNA-Seq data without a reference genome. Nat. Biotechnol. 29, 644–652. doi: 10.1038/nbt.1883
Guo, Z. L., Yuan, X. Q., Li, L. Y., Zeng, M., Yang, J., Tang, H., et al. (2022). Genome-wide analysis of the ATP-binding cassette (ABC) transporter family in Zea mays L. and its response to heavy metal stresses. Int. J. Mol. Sci. 23:2109. doi: 10.3390/ijms23042109
Handa, Y., Nishide, H., Takeda, N., Suzuki, Y., Kawaguchi, M., and Saito, K. (2015). RNA-seq transcriptional profiling of an arbuscular mycorrhiza provides insights into regulated and coordinated gene expression in Lotus japonicus and Rhizophagus irregularis. Plan. Cell. Physiol. 56, 1490–1511. doi: 10.1093/pcp/pcv071
Hoagland, M., and Arnon, D. (1950). The water culture method for growing plants without soil. Calif. Agricul. Experim. Station. 347:1950.
Jiang, Z. Z., Zhou, X. P., Tao, M., Yuan, F., Liu, L. L., Wu, F. H., et al. (2019). Plant cell-surface GIPC sphingolipids sense salt to trigger Ca2+ influx. Nature 572, 341–346. doi: 10.1038/s41586-019-1449-z
Kamrul, H. K. M., Akhter, B. M. S., Renu, T., and Narendra, T. (2013). Global calcium transducer P-type Ca2+-ATPases open new avenues for agriculture by regulating stress signalling. J. Exp. Bot. 64, 3099–3109. doi: 10.1093/jxb/ert182
Kurusu, T., Hamada, J., Nokajima, H., Kitagawa, Y., Kiyoduka, M., Takahashi, A., et al. (2010). Regulation of microbe-associated molecular pattern-induced hypersensitive cell death, phytoalexin production, and Defense gene expression by calcineurin B-like protein-interacting protein kinases, OsCIPK14/15, in Rice Cultured Cells. Plant. Physiol. 5:678692, 1045–1047. doi: 10.4161/psb.5.8.12407
Lecourieux, D., Ranjeva, R., and Pugin, A. (2006). Calcium in plant defence-signaling pathways. New. Phytot. 171, 249–269.
Li, C. F., Zheng, C. S., Zhou, K. H., Han, W. B., Tian, C. J., Ye, S. H., et al. (2020). Toleration and accumulation of cotton to heavy metal - potential use for phytoremediation. Soil Sediment Contam. 29, 516–531. doi: 10.1080/15320383.2020.1747979
Li, J. L., Sun, Y. Q., Zhang, X., Hu, Y. J., Li, T., Zhang, X. M., et al. (2018a). A methyltransferase gene from arbuscular mycorrhizal fungi involved in arsenic methylation and volatilization. Chemosphere 209, 392–400. doi: 10.1016/j.chemosphere.2018.06.092
Li, M., Wang, R., Tian, H., and Gao, Y. (2018b). Transcriptome responses in wheat roots to colonization by the arbuscular mycorrhizal fungus Rhizophagus irregularis. Mycorrhiza 28, 747–759. doi: 10.1007/s00572-018-0868-2
Liu, A., Chen, S. C., Chang, R., Liu, D. L., Chen, H. R., Ahammed, G. J., et al. (2014). Arbuscular mycorrhizae improve low temperature tolerance in cucumber via alterations in H2O2 accumulation and ATPase activity. J. Plant Res. 127, 775–785. doi: 10.1007/s10265-014-0657-8
Liu, C. Y., Zhang, F., Zhang, D. J., Zou, Y. N., Shu, B., and Wu, Q. S. (2020). Transcriptome analysis reveals improved root hair growth in trifoliate orange seedlings by arbuscular mycorrhizal fungi. Plant Growth Regul. 92, 195–203. doi: 10.1007/s10725-020-00630-3
Liu, K. Y., Qiu, Z. J., Zhang, Q. M., Zhang, A. R., and Gong, M. G. (2021). Effect of arbuscular mycorrhizal fungi on root morphological and physiological characteristics of cotton under arsenic stress. Acta Botan. Boreali-Occiden. Sin. 41, 1188–1198. doi: 10.7606/j.issn.1000-4025.2021.07.1188
Ma, J., Sun, C., Bai, L., Dong, R., Yan, Y., Yu, X., et al. (2018). Transcriptome analysis of cucumber roots reveals key cold-resistance genes induced by AM fungi. Plant Mol. Biol. Report. 36, 135–148. doi: 10.1007/s11105-018-1066-2
Ma, X. F., Zheng, C. S., Li, W., Ai, S. Y., Zhang, Z. G., Zhou, X. J., et al. (2017). Potential use of cotton for remediating heavy metal-polluted soils in southern China. J. Soils Sediments 17, 2866–2872. doi: 10.1007/s11368-017-1697-1
Ma, Y., He, J., Ma, C., Luo, J., Li, H., Liu, T., et al. (2013). Ectomycorrhizas with Paxillus involutus enhance cadmium uptake and tolerance in populus × canescens. Plant Cell Environ. 37, 627–642. doi: 10.1111/pce.12183
Martin, M. (2011). Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnet. J. 17:10. doi: 10.14806/ej.17.1.200
Martínez-Castillo, J. I., Saldaa-Robles, A., and Ozuna, C. (2022). Arsenic stress in plants: a metabolomic perspective. Plant Stress. 3:100055. doi: 10.1016/j.stress.2022.100055
Ministry of Environmental Protection (2015). Arsenic pollution control technology policy. People's Republic of China. Available from: https://www.mee.gov.cn/gkml/hbb/bgg/201512/W020151228354146522922.pdf
Ouziad, F., Hildebrandt, U., Schmelzer, E., and Bothe, H. (2005). Differential gene expressions in arbuscular mycorrhizal-colonized tomato grown under heavy metal stress. J. Plant Physiol. 162, 634–649. doi: 10.1016/j.jplph.2004.09.014
Ouziad, F., Wilde, P., Schmelzer, E., Hildebrandt, U., and Bothe, H. (2006). Analysis of expression of aquaporins and Na+/H+ transporters in tomato colonized by arbuscular mycorrhizal fungi and afected by salt stress. Environ. Exp. Bot. 57, 177–186. doi: 10.1016/j.envexpbot.2005.05.011
Pan, G. F., Li, W. Z., Huang, L. K., Mo, G. Z., and Wang, X. L. (2024). Arbuscular mycorrhizal fungi promote arsenic accumulation in Pteris vittata L. through arsenic solubilization in rhizosphere soil and arsenic uptake by hyphae. J. Hazard. Mater. 466:133579. doi: 10.1016/j.jhazmat.2024.133579
Phillips, J. M., and Hayman, D. S. (1970). Improved procedures for clearing roots and staining parasitic and vesicular arbuscular mycorrhizal fungi for rapid assessment of infection. Trans. Br. Mycol. Soc. 55, 158–IN18. doi: 10.1016/S0007-1536(70)80110-3
Puccio, G., Ingrafa, R., Mercati, F., Amato, G., Giambalvo, D., Martinelli, F., et al. (2023). Transcriptome changes induced byrbuscular mycorrhizal symbiosis in leaves of durum wheat (Triticum durum Desf.) promote higher salt tolerance. Sci. Rep. 13:116. doi: 10.1038/s41598-022-26903-7
Riley, R., and Corradi, N. (2013). Searching for clues of sexual reproduction in the genomes of arbuscular mycorrhizal fungi. Fungal Ecol. 6, 44–49. doi: 10.1016/j.funeco.2012.01.010
Salim, N. A. A., Wood, A. K., Yusof, A. M., Hamzah, M. S., Elias, M. S., and Rahman, S. A. (2009). A study of arsenic and chromium contamination in sediments of freshwater bodies. Fresenius Environ. Bull. 18, 1618–1623.
Sanders, D., Brownlee, C., and Harper, J. F. (1999). Communicating with calcium. Plant. Cell 11, 691–706.
Schüßler, A., and Walker, C. (2010). The Glomeromycota: A Species List with New Families and New Genera. Available at: www.amf-phylogeny.com
Shabani, L., Sabzalian, M. R., and Mostafavi pour, S. (2016). Arbuscular mycorrhiza affects nickel translocation and expression of ABC transporter and metallothionein genes in Festuca arundinacea. Mycorrhiza 26, 67–76. doi: 10.1007/s00572-015-0647-2
Shannon, P., Markiel, A., Ozier, O., Baliga, N. S., Wang, J. T., Ramage, D., et al. (2003). Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res. 13, 2498–2504. doi: 10.1101/gr.1239303
Sharma, S. S., Kumar, V., and Dietz, K. J. (2020). Emerging Trends in Metalloid-Dependent Signaling in Plants. Trends Plant Sci. 26, 452–471. doi: 10.1016/j.tplants.2020.11.003
Sharma, S., Anand, G., Singh, N., and Kapoor, R. (2017). Arbuscular mycorrhiza augments arsenic tolerance in wheat (Triticum aestivum L.) by strengthening antioxidant Defense system and thiol metabolism. Front. Plant Sci. 8, 1–21.
Sharples, J. M., Meharg, A. A., Chambers, S. M., and Cairney, J. W. (2000). Evolution: symbiotic solution to arsenic contamination. Nature 404, 951–952. doi: 10.1038/35010193
Shu, B., Cai, D., Zhang, F., Zhang, D. J., Liu, C. Y., Wu, Q. S., et al. (2020). Identifying citrus CBL and CIPK gene families and their expressions in response to drought and arbuscular mycorrhizal fungi colonization. Bio. plantarum. 64, 773–783. doi: 10.32615/BP.2020.123
Smith, S. E., and Read, D. J. (2008). Mycorrhizal symbiosis, thirded. Academic Press, New York, London. 787.
Song, F., Li, J., and Zhang, X. (2014). Characterization of expressed genes in the establishment of arbuscular mycorrhiza between Amorpha fruticosa and glomus mosseae. J. Forestry. Res. 25, 541–548. doi: 10.1007/s11676-014-0493-7
Song, W. Y., Park, J., Mendoza-Cózatl, D. G., Suter-Grotemeyer, M., Shim, D., Hörtensteiner, S., et al. (2010). Arsenic tolerance in Arabidopsis is mediated by two ABCC-type phytochelatin transporters. PNAS Nexus 107, 21187–21192. doi: 10.1073/pnas.1013964107
Spagnoletti, F. N., Balestrasse, K., Lavado, R. S., and Giacometti, R. (2016). Arbuscular mycorrhiza detoxifying response against arsenic and pathogenic fungus in soybean. Ecotoxicol. Environ. Saf. 133, 47–56. doi: 10.1016/j.ecoenv.2016.06.012
Spagnoletti, F., and Lavado, R. S. (2015). The arbuscular mycorrhiza Rhizophagus intraradices reduces the negative effects of arsenic on soybean plants. Agronomy 5, 188–199. doi: 10.3390/agronomy5020188
Spagnoletti, F., Carmona, M., Gómez, N. E. T., Chiocchio, V., and Lavado, R. S. (2017). Arbuscular mycorrhiza reduces the negative effects of M-phaseolina on soybean plants in arsenic-contaminated soils. Appl. Soil Ecol. 121, 41–47. doi: 10.1016/j.apsoil.2017.09.019
Srivastava, S., Srivastava, A. K., Suprasanna, P., and D’Souza, S. F. (2009). Comparative biochemical and transcriptional profiling of two contrasting varieties of Brassica juncea L. in response to arsenic exposure reveals mechanisms of stress perception and tolerance. J. Exp. Bot. 60, 3419–3431. doi: 10.1093/jxb/erp181
Su, W., Huang, L., Ling, H., Mao, H., Huang, N., Su, Y., et al. (2020). Sugarcane calcineurin B-like (CBL) genes play important but versatile roles in regulation of responses to biotic and abiotic stresses. Sci. Rep. 10, 167–113. doi: 10.1038/s41598-019-57058-7
Sun, D., Zhang, X., Yin, Z., Feng, H., Hu, C., Guo, N., et al. (2023). As-hyperaccumulator Pteris vittata and non-hyperaccumulator Pteris ensiformis under low as-exposure: transcriptome analysis and implication for as hyperaccumulation. J. Hazard. Mater. 458:132034. doi: 10.1016/j.jhazmat.2023.132034
Surbhi, S., Garima, A., Neeraja, S., and Rupam, K. (2017). Arbuscular mycorrhiza augments arsenic tolerance in wheat (Triticum aestivum L.) by strengthening antioxidant defense system and thiol metabolism. Front. Plant Sci. 8, 1–21. doi: 10.3389/fpls.2017.00906
Trapnell, C., Williams, B. A., Pertea, G., Mortazavi, A., Kwan, G., van Baren, M. J., et al. (2010). Transcript assembly and quantifcation by RNA-Seq reveals unannotated transcripts and isoform switching during cell diferentiation. Nat. Biotechnol. 28, 511–515. doi: 10.1038/nbt.1621
Wan, X., Lei, M., and Chen, T. (2016). Cost-benefit calculation of phytoremediation technology for heavy-metal-contaminated soil. Sci. Total Environ. 563-564, 796–802. doi: 10.1016/j.scitotenv.2015.12.080
Wang, X. E., Basnayake, B. S., Zhang, H. J., Li, G., Li, W., Virk, N., et al. (2009). The Arabidopsis ATAF1, a NAC transcription factor, is a negative regulator of defense responses against necrotrophic fungal and bacterial pathogens. MolPlant-Microbe Interact 22, 1227–1238. doi: 10.1094/MPMI-22-10-1227
Wang, Y., Dong, F., and Tang, M. (2022). Transcriptome analysis of arbuscular mycorrhizal Casuarina glauca in damage mitigation of roots on NaCl stress. Microorganisms 10:15. doi: 10.3390/microorganisms10010015
Yan, Y., He, X. Y., Hu, W., Liu, G. Y., Wang, P., He, C. Z., et al. (2018). Functional analysis of MeCIPK23 and MeCBL1/9 in cassava defense response against Xanthomonas axonopodis pv. Manihotis. Plant Cell Rep. 37, 887–900. doi: 10.1007/s00299-018-2276-7
Yang, L., Li, Q. Q., Yang, Y., Chen, Q., and Xiao, J. X. (2020). Comparative transcriptome analysis reveals positive effects of arbuscular mycorrhizal fungi inoculation on photosynthesis and high-ph tolerance in blueberry seedlings. Trees 34, 433–444. doi: 10.1007/s00468-019-01926-2
Zhang, Q. M., Gong, M. G., Xu, S. S., Wei, Y., Yuan, J. F., Chang, Q. S., et al. (2022). Rhizophagus intraradices improves arsenic tolerance in Sophora viciifolia hance. Ann. Microbiol. 72, 1–14. doi: 10.1186/s13213-022-01668-6
Zhang, Q. M., Gong, M. G., Liu, K. Y., Chen, Y. N., and Chang, Q. S. (2020). Rhizoglomus intraradices improves plant growth, root morphology and phytohormone balance of Robinia pseudoacacia in arsenic-contaminated soils. Front. Microbiol. 11:1428. doi: 10.3389/fmicb.2020.01428
Zhang, Q. M., Gong, M. G., Yuan, J. F., Hou, Y., Zhang, H. M., Wang, Y., et al. (2017). Dark septate endophyte improves drought tolerance in sorghum. Int. J. Agric. Biol. 19, 53–60. doi: 10.17957/IJAB/15.0241
Zhang, X. H., Gao, H. M., Liang, Y. Q., and Cao, Y. P. (2021). Full-length transcriptome analysis of asparagus roots reveals the molecular mechanism of salt tolerance induced by arbuscular mycorrhizal fungi. Envmiron. Exp. Bot. 185:104402. doi: 10.1016/j.envexpbot.2021.104402
Zhao, F. J., McGrath, S. P., and Meharg, A. A. (2010). Arsenic as a food chain contaminant: mechanisms of plant uptake and metabolism and mitigation strategies. Annu. Rev. Plant Biol. 61, 535–559. doi: 10.1146/annurev-arplant-042809-112152
Keywords: cotton seedling, differentially expressed genes, Funneliformis mosseae, arsenic stress, RNA-Seq
Citation: Gong M, Bai N, Su J, Wang Y, Wei Y and Zhang Q (2024) Transcriptome analysis of Gossypium reveals the molecular mechanisms of Ca2+ signaling pathway on arsenic tolerance induced by arbuscular mycorrhizal fungi. Front. Microbiol. 15:1362296. doi: 10.3389/fmicb.2024.1362296
Edited by:
Shengqiang Wang, Guangxi University, ChinaReviewed by:
Xueli Wang, Guangxi University, ChinaZhen Li, Shanxi Datong University, China
Xueguang Sun, Guizhou University, China
Copyright © 2024 Gong, Bai, Su, Wang, Wei and Zhang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Qiaoming Zhang, emhhbmdxbTEwMTNAMTYzLmNvbQ==
 Minggui Gong
Minggui Gong Na Bai1
Na Bai1