- College of Life Sciences, Sichuan Agricultural University, Ya’an, China
Tartary buckwheat (Fagopyrum tataricum) is a significant medicinal crop, with flavonoids serving as a crucial measure of its quality. Presently, the artificial cultivation of Tartary buckwheat yields low results, and the quality varies across different origins. Therefore, it is imperative to identify an effective method to enhance the yield and quality of buckwheat. Endophytic fungi reside within plants and form a mutually beneficial symbiotic relationship, aiding plants in nutrient absorption, promoting host growth, and improving secondary metabolites akin to the host. In this study, high-throughput sequencing technology was employed to assess the diversity of endophytic fungi in Tartary buckwheat. Subsequently, a correlation analysis was performed between fungi and metabolites, revealing potential increases in flavonoid content due to endophytic fungi such as Bipolaris, Hymenula, and Colletotrichum. Additionally, a correlation analysis between fungi and phenotypic traits unveiled the potential influence of endophytic fungi such as Bipolaris, Buckleyzyma, and Trichosporon on the phenotypic traits of Tartary buckwheat. Notably, the endophytic fungi of the Bipolaris genus exhibited the potential to elevate the content of Tartary buckwheat metabolites and enhance crop growth. Consequently, this study successfully identified the resources of endophytic fungi in Tartary buckwheat, explored potential functional endophytic fungi, and laid a scientific foundation for future implementation of biological fertilizers in improving the quality and growth of Tartary buckwheat.
1 Introduction
Tartary buckwheat [Fagopyrum tataricum (L.) Gaertn.] is a traditional cereal crop in China, originating from the southwestern part of the country (Zhou et al., 2023). Its cultivation has expanded across various Asian countries, including China, India, Nepal, and others (Kreft et al., 2023). China ranks first globally in terms of both cultivation area and yields for Tartary buckwheat, with primary cultivation regions located in Sichuan, Guizhou, Yunnan, Xizang, Qinghai, and other areas (Li N. et al., 2022). Notably, historical records in the “Compendium of Materia Medica” highlight the digestive, heat-alleviating, and swelling-reducing properties of Tartary buckwheat (Yan et al., 2021). Scientific research has further revealed the high nutritional value of Tartary buckwheat, including its abundance of carbohydrates, proteins, polyphenols, and minerals (Zou et al., 2021). Moreover, Tartary buckwheat stands out among other food crops due to its rich content of bioactive compounds such as rutin, quercetin, and other flavonoids, which contribute to its noteworthy therapeutic effects in antioxidant, anticancer, blood sugar-lowering, blood pressure-lowering, and lipid-lowering aspects (Zhu, 2016).
Tartary buckwheat is primarily cultivated in arid and semi-arid regions at altitudes ranging from 1,000 to 3,000 meters (Fan et al., 2019). However, challenging cultivation environments often negatively impact seed germination, leading to lower yields and reduced quality of Tartary buckwheat (Weng et al., 2022). Furthermore, the current cultivation area of Tartary buckwheat in China, approximately 705,000 hm2, lags significantly behind other traditional cereal crops such as rice and wheat (Chen, 2018). However, with improving living standards, there is a growing emphasis on health and well-being, resulting in an increased demand for whole grains, including Tartary buckwheat. This surge in demand necessitates an emphasis on higher-quality Tartary buckwheat (Qu, 2021). Therefore, in addition to enhancing cultivation techniques and expanding cultivation areas for Tartary buckwheat, there should be a focus on increasing the content of secondary metabolites to enhance its overall quality and align it with the modern demand for health-conscious food choices. Research suggests that the production of plant secondary metabolites is influenced by environmental factors such as altitude, light exposure, and temperature (Večeřová et al., 2021). Furthermore, it is also influenced by inherent plant characteristics, including endophytic fungi and enzymes (Aktar et al., 2022).
Endophytic fungi are non-pathogenic fungal organisms that reside within plant tissues for extended periods, establishing mutualistic relationships with their hosts (Sun et al., 2022). Previous studies indicate that endophytic fungi play a crucial role in nutrient absorption, host immunity enhancement, and overall growth and development promotion (Poudel et al., 2021; Ding et al., 2022). Moreover, specific endophytic fungi can aid plants in resisting pathogens and adversity, enhancing their environmental adaptability and resilience (Waller et al., 2005). Additionally, certain endophytic fungi contribute to the degradation and recycling of organic substances in the environment, thereby promoting soil health and ecosystem stability (Rouydel et al., 2021). Furthermore, extensive research demonstrates that endophytic fungi significantly influence the production of host secondary metabolites. For instance, the diversity and abundance of endophytic fungi in Gentiana officinalis exhibit a notable correlation with the content of four medicinal secondary metabolites (Hou et al., 2022). Similarly, four endophytic fungal strains were screened from rice (Oryza sativa L.) that effectively promoted rice growth as well as significantly increased the accumulation of phenolic compounds and anthocyanins in rice (Gateta et al., 2023). Notably, the co-fermentation of the endophytic fungus Ilyonectria cyclaminicola with the residue of Epimedii folium during the fermentation process enhances the content of total flavonoids and flavonols in the fermentation broth (Guo et al., 2023). Under alkaline stress, endophytic fungi promote the production of secondary metabolites such as phosphorus, polyphenols, and alkaloids in Hordeum bogdanii, thus enhancing its alkaline tolerance (Han et al., 2022). In other words, endophytic fungi play a crucial role in helping plants endure adversities by stimulating the production of host metabolites. Notably, studies by Harrison et al. demonstrated the influence of endophytic fungi Alternaria on plant phenotypes, such as the promotion of leaf area and plant height in Astragalus lentiginosus (Harrison et al., 2021). Despite the plethora of research reports on endophytic fungi, the functions of many of these organisms remain unclear.
Traditional methods in microbial research have historically relied on direct cultivation using culture media, which involves the formation of visible microbial colonies followed by purification and molecular characterization (Wei et al., 2021). However, these methods have significant drawbacks, including time-consuming procedures, operational complexity, sample contamination risks, and notable limitations. It is worth noting that cultivable microorganisms only represent a small fraction (approximately one-tenth) of the total microbial community within a sample (Lahlali et al., 2021; Long et al., 2021). Fortunately, with the rapid advancement of bioinformatics technology, DNA sequencing has achieved remarkable progress, particularly through the utilization of high-throughput sequencing. This innovative approach has become the mainstream technology for studying microbial communities due to its advantages, including cost-effectiveness, shorter processing times, and higher data accuracy, overcoming the limitations associated with traditional methods (Jo et al., 2020). In this study, high-throughput sequencing technology was implemented for the first time to examine the diversity of endophytic fungi in various regions and tissues of Tartary buckwheat. Subsequently, correlation analyses were conducted between endophytic fungi and flavonoid secondary metabolites, as well as phenotypic traits. Through this comprehensive analysis of the approach to find a target endophytic fungal strains with the potential to promote the secretion of flavonoids in Tartary buckwheat and with the potential to promote the growth of Tartary buckwheat. This approach establishes a solid theoretical foundation for the potential utilization of microbial fertilizers and provides new insights for further enhancing the yield and quality of Tartary buckwheat.
2 Materials and methods
2.1 Acquisition of experimental materials and phenotypic assessment
In June 2022, seven wild Fagopyrum tataricum species with distinct phenotypes were collected from the Wild Buckwheat Germplasm Resource Nursery located in Liangshan Yi Autonomous Prefecture, Sichuan Province (longitude: 102°49′E, latitude: 27°59′N, altitude: 2,118 m). These seven materials originate from diverse habitats in five different provinces across China (Figure 1; Supplementary Table S1). Each species consisted of 10 plants, resulting in 70 Tartary buckwheat samples.
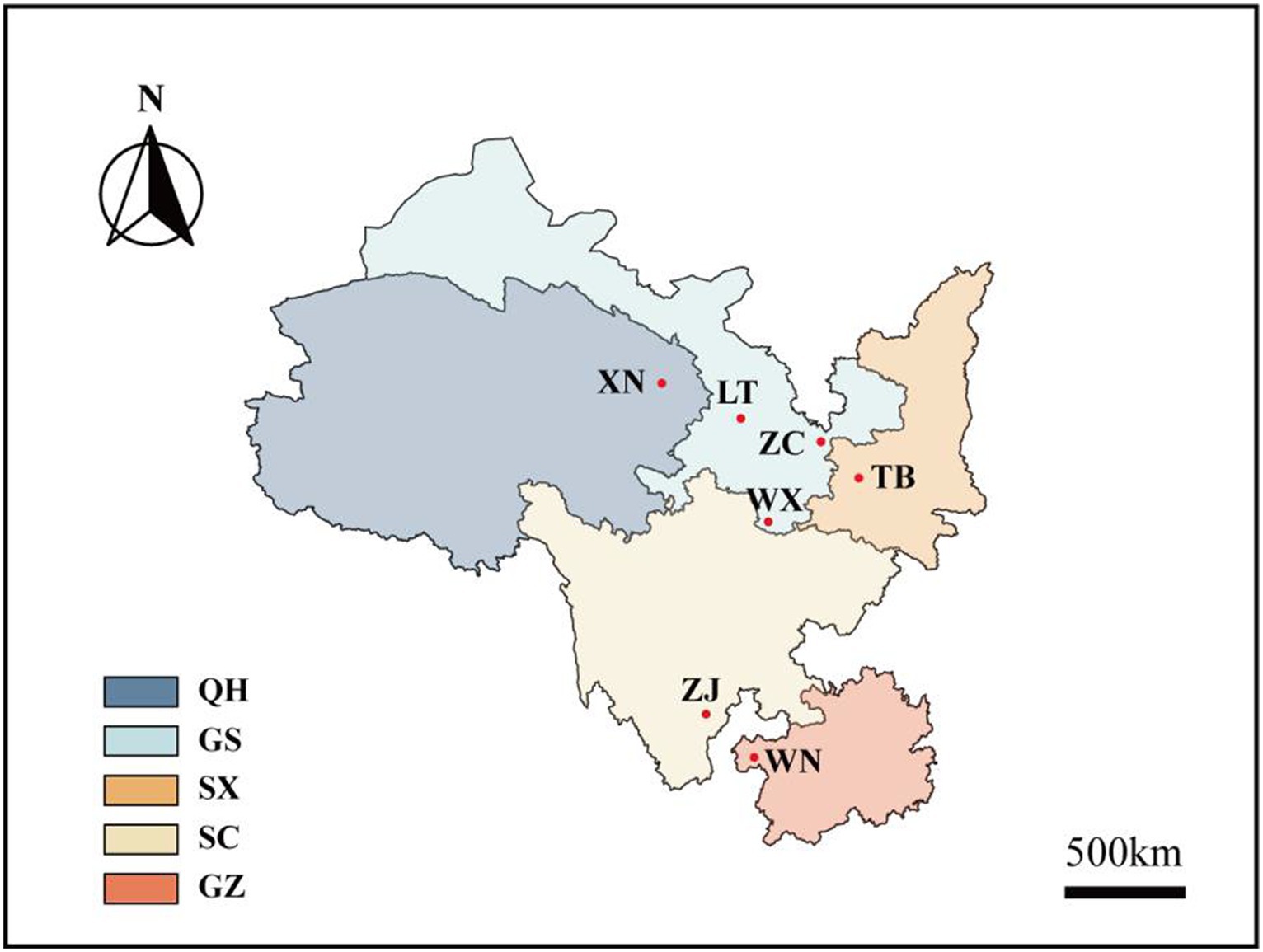
Figure 1. Distribution map of Tartary buckwheat. Xining City (XN), Lintao County (LT), Zhangchuan County (ZC), Wen County (WX), Taibai County (TB), Zhaojue County (ZJ), and Weining County (WN) are the seven wild buckwheat collection sites. Among them, XN belongs to China’s Qinghai Province (QH), LT, ZC, and WX belongs to China’s Gansu Province (GS), TB belongs to China’s Shaanxi Province (SX), ZJ belongs to China’s Sichuan Province (SC), WN belongs to China’s Guizhou Province (GZ).
A total of eight phenotypic traits were identified in the sample. Plant height and length of middle branches were measured using a tape measure, whereas stem thickness, leaf length, and leaf width were measured using calipers. The branching angle was determined using a protractor. The number of main stem nodes and branches on the main stem were directly observed and counted. Initially, the eight phenotypic traits of Tartary buckwheat samples were determined. Plant height and length of middle branches were measured using a tape measure, while stem thickness, leaf length, and leaf width were measured using calipers. The branching angle was determined using a protractor, and the number of main stem nodes and branches on the main stem were directly observed and counted. Following this, the roots, stems, and leaves of the sample were separated and subjected to surface sterilization treatment within 48 h. This involved soaking them in 75% alcohol for 1 min, followed by immersion in a 2.5% sodium hypochlorite solution for 5–10 min, and rinsing five to six times with sterile water. The Tartary buckwheat roots, stems, and leaves were then divided equally into two parts. One part was stored in sterile self-sealing bags for high-throughput sequencing, while the other part was placed in an oven at a constant temperature of 60°C for drying. Once dried, the material was ground and passed through a 40 mesh sieve to obtain a uniform powder. This powder was used for extracting and quantifying the amount of secondary metabolites in Tartary buckwheat.
2.2 Total DNA extraction and high throughput sequencing of Tartary buckwheat
Total genomic DNA samples were extracted utilizing the OMEGA Soil DNA Kit (Omega Bio-Tek, Norcross, GA, United States). Subsequently, the DNA samples were stored at −20°C for further analysis. The quality of the DNA was assessed utilizing a NanoDrop NC2000 spectrophotometer (Thermo Fisher Scientific, Waltham, MA, United States) as well as agarose gel electrophoresis. The fungal internal transcribed spacer (ITS rDNA) was amplified using the primers ITS1F (5′-CTTGGTCATTTAGAGGAAGTAA-3′) and ITS2R (5′-GCTGCGTTCTTCATCGATGC-3′). The resulting PCR amplicons were purified using Vazyme VAHTSTM DNA Clean Beads (Vazyme, Nanjing, China) and quantified utilizing the Quant-iT PicoGreen dsDNA Assay Kit (Invitrogen, Carlsbad, CA, United States). Following individual quantification, the amplicons were pooled in equal amounts and subjected to pair-end 2 × 250 bp sequencing utilizing the Illumina NovaSeq platform with the NovaSeq 6000 SP Reagent Kit (500 cycles) (Shanghai Personal Biotechnology Co., Ltd., Shanghai, China). The raw DNA sequence data were deposited in the National Center for Biotechnology Information Sequence Read Archive database (BioProject accession number: PRJNA1043747).
2.3 Determination of total flavonoids and polyphenols in the roots, stems and leaves of Tartary buckwheat
The content of total flavonoids was determined using the sodium nitrite-aluminum nitrate method (Zhao et al., 2016). Tartary buckwheat root and stem leaves weighing 0.3–0.4 g were extracted with 30% ethanol under ultrasonic conditions for 40 min. The resulting supernatant was utilized as the test solution for measurement. To this solution, 0.7 mL of a 5% NaNO2 solution was added, followed by the immediate addition of 0.7 mL of a 10% Al(NO3)3 solution, with subsequent shaking. After 5 min, 5 mL of a 4% NaOH solution was added, mixed, and left to stand for another 5 min. The flavonoid content was determined by measuring the D510 nm value using a UV spectrophotometer and calculating it against a standard curve. The total phenol content was determined using the Folin phenol method (You and Cao, 2013). Specifically, 1 mL of the test solution was mixed with 1 mL of Folin Phenol chromogenic agent and 3 mL of 20% Na2CO3. The mixture was thoroughly combined and allowed to react in a water bath at 50°C for 30 min. The absorbance was measured at 765 nm, and the total phenol content was calculated against a gallic acid standard curve. All concentration determinations were performed in triplicate, and the average values were taken.
2.4 Extraction and content determination of 40 flavonoids from roots, stems, and leaves of ZC and LT
Following the study conducted by Gao et al. (2016), the extraction method was optimized. First, 0.5 g of the crushed dry sample was weighed and mixed with 4 mL of a 1% hydrochloric acid methanol solution. The resulting mixture was vortexed for 1 min and then sonicated in an ice water bath for 30 min. Subsequently, it was centrifuged at 11,000 rpm for 30 min, and the supernatant was carefully removed. This extraction process was repeated once, and the supernatant was combined twice and diluted to a volume of 10 mL. The resulting solution was subjected to quantitative and qualitative analysis after passing through a 0.22-μm filter membrane.
The Agilent 1260 high-performance liquid chromatograph, in series with a 6420A mass spectrometer system, was utilized for the analysis. Various bioactive compounds were separated using a reverse chromatography column, the Agilent ZORBAX Eclipse Plus C18 (3.5 μm, 2.1*150 mm). The chromatographic conditions were based on the method outlined by Aguirre-Hernández et al. (2009). The mobile phase consisted of 0.3% phosphoric acid (A) and acetonitrile (B), with the elution gradient as follows: 0 ∼ 1 min, 80% ~ 10% A; 1 ∼ 5 min, 10% A; 5 ∼ 5.1 min, 10% ~ 80% A; 5.1 ∼ 10 min, 80% A. The flow rate was set at 0.3 mL/min, the injection volume at 3 μL, and the column temperature at 35°C. To establish standard curves of 40 flavonoids, the mobile phase was used to achieve final concentrations of 2 ng/mL, 5 ng/mL, 20 ng/mL, 50 ng/mL, 200 ng/mL, and 500 ng/mL. Following analysis under the aforementioned chromatographic conditions, a standard curve was constructed (Supplementary Figure S1). Qualitative analysis was then conducted by comparing the retention time of the standards with the subsequent detection results. Furthermore, quantitative determination was performed by comparing the concentration and peak area of the reference standard in the chromatography with the constructed standard curve.
The method described by Ma et al. (2022) formed the basis for the detection using mass spectrometry, albeit with slight optimizations. LC–MS analysis was conducted employing a 6420A mass spectrometer (Agilent Technologies, Santa Clara, CA, United States). Details of the selection reaction detection conditions for the protonation and deprotonation of the samples are displayed in Supplementary Table S2. To confirm the presence of flavonoid metabolites, the mass spectra of the total ion chromatography peaks obtained from the extract of Tartary buckwheat roots, stems, and leaves were compared to the mass spectra of standard substances such as rutin, hesperidin, and quercetin.
2.5 Bioinformatics and statistical analysis
The raw sequence data were demultiplexed using the demux plugin, followed by primer cutting using the cutadapt plugin (Martin, 2011). Subsequently, the sequences underwent quality filtering, denoising, merging, and chimera removal using the DADA2 plugin (Callahan et al., 2016). The sequence data was analyzed using QIIME2 and R packages (v3.2.0).
To assess ASV-level alpha diversity indices, including the Chao1 richness estimator, Observed species, Shannon diversity index, Simpson index, Pielou’s evenness, and Good’s coverage, the ASV table in QIIME2 was utilized. Beta diversity analysis was conducted using Bray-Curtis metrics to investigate the structural variation of microbial communities among the samples, with the results visualized through principal coordinate analysis (PCoA). The R package “VennDiagram” was employed to generate Venn diagrams, allowing visualization of shared and unique ASVs across samples or groups based on their occurrence, regardless of their relative abundance (Zaura et al., 2009). To detect differentially abundant taxa across groups, Linear Discriminant Analysis Effect Size (LEfSe) was employed, utilizing default parameters (Segata et al., 2011). Additionally, a co-occurrence network was constructed based on the correlation coefficient and visualized using the R packages igraph and ggraph.
3 Results
3.1 Diversity analysis of endophytic fungi in Tartary buckwheat
Through high-throughput sequencing, 4,605,862 effective reads, 4,575,604 high-quality sequences, and 823 operational taxonomic units (OTUs) were obtained from 21 Tartary buckwheat tissue samples. The length of the sequences ranged from 146 bp to 438 bp. The smoothness of the sparse curves indicated that sequencing depth influenced the diversity observed in the samples. When the sequencing depth increased, the curve depicting the number of OTUs in the 21 samples flattened out (Figure 2A). This indicated that the sequencing results captured the diversity present in the current sample, and further increasing the sequencing depth did not reveal new OTUs. The Venn diagram of OTUs (Figure 2B) showed 73 common OTUs among the 7 different varieties of Tartary buckwheat. However, each variety had unique OTUs, with ZC, XN, LT, TB, WN, ZJ, and WX having 113, 81, 92, 141, 218, 32, and 73 unique OTUs, respectively. Additionally, there were 179 OTUs shared between the root, stem, and leaf samples, with 253 unique OTUs in the root, 218 unique OTUs in the stem, and 317 unique OTUs in the leaf (Figure 2C).
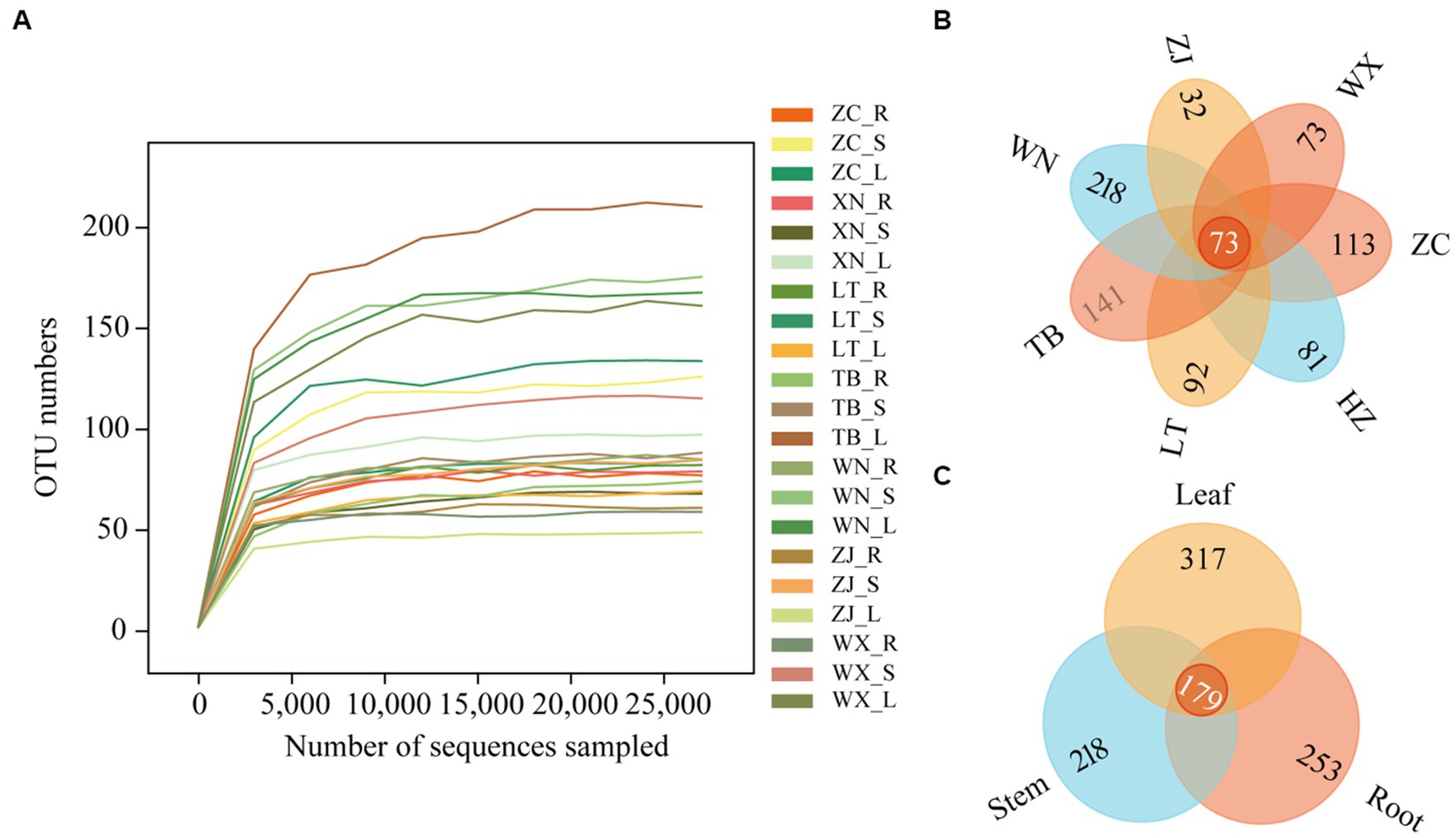
Figure 2. Sparse curves and Venn diagram of 21 samples. The abscissa represents the ranking of operational taxonomic units (OTUs), while the ordinate represents the relative percentage of species at the classification level (A). The position of the abscissa of the extension end point of the sample curve represents the number of species in each sample. (B) The Venn diagram describes different and common OTUs in 7 different Tartary buckwheat materials (Zhaojue, ZJ; Wenxian, WX; Zhangchuan, ZC; Xining, XN; Lintao, LT; Taibai, TB; Weining, WN). (C) Different and common OTUs in roots, stems, and leaves.
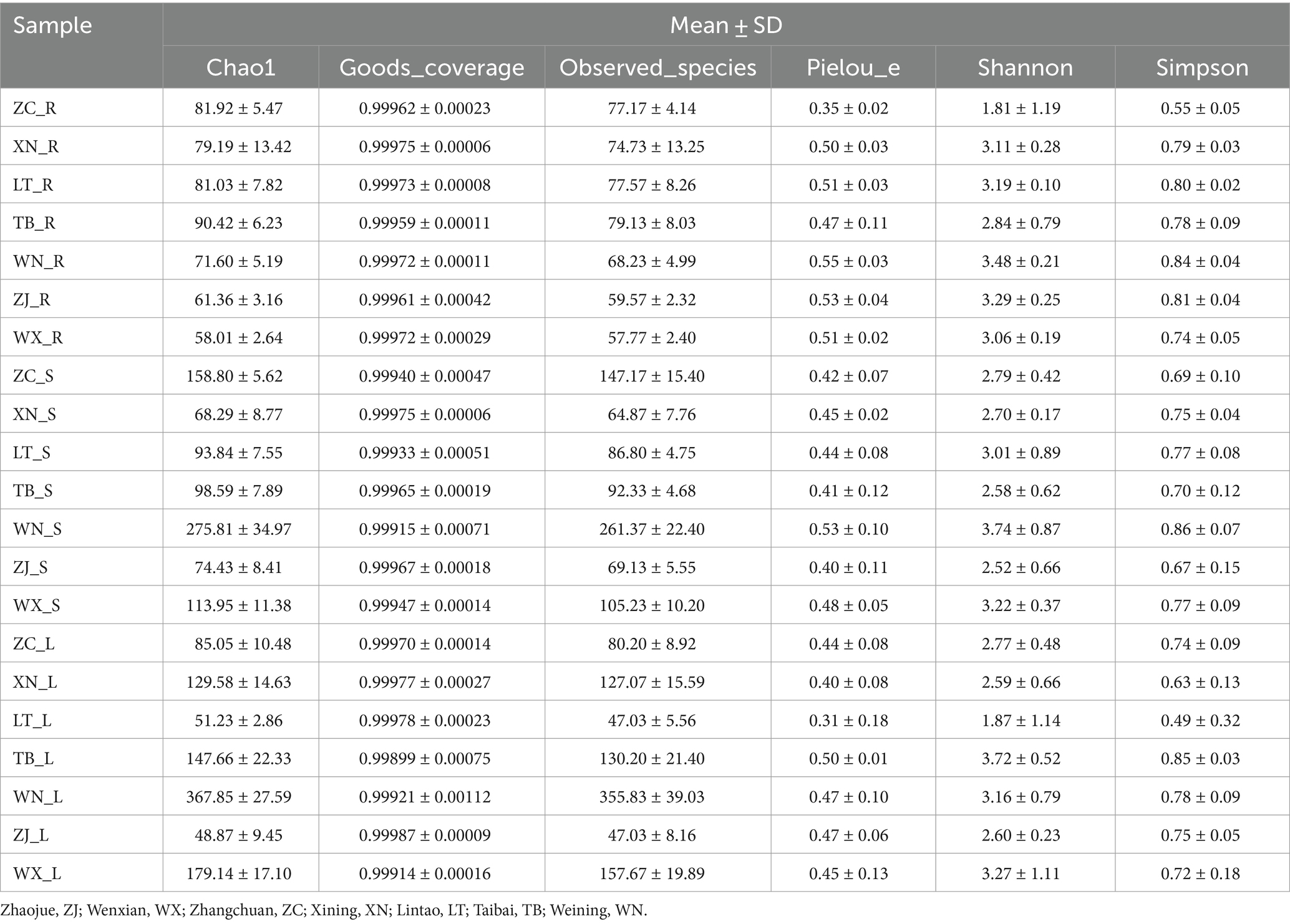
The α diversity of endophytic fungi in Tartary buckwheat was assessed in seven sampling areas using measures of community richness (Chao 1 index, Observed species), coverage (Good coverage index), diversity (Shannon index, Simpson index), and evenness (Pielou’s index). As displayed in Table 1, it was evident that the roots of all seven materials had the lowest Chao 1 index and Observed_species index, while the stems and leaves exhibited higher values. Among the different samples, WN had the highest Chao 1 index and Observed_species index, whereas ZJ had the lowest Chao 1 index. The Good’s Coverage values for all 21 samples exceeded 99.8%, indicating that the sequencing achieved a high fungal coverage rate and accurately reflected the fungal communities in the samples. There were no significant differences in evenness among the 21 samples. In terms of community diversity, the roots, stems, and leaves of WN displayed high diversity, whereas ZC showed relatively low diversity.
Beta diversity is a measure used to assess the differences in species composition across various habitats. Higher beta diversity indicates greater dissimilarity in species composition among habitats. In this study, ANOSIM analysis was conducted using the Bray-Curtis distance algorithm to cluster the endophytic fungal communities of Tartary buckwheat in seven sample plots. The results of the ANOSIM analysis revealed that the R-value of Tartary buckwheat from different collection sites was 0.210 (p = 0.001), indicating a significant difference in species composition between the sample groups compared to within the group (Figure 3A). Similarly, the ANOSIM analysis of different tissue parts (Figure 3B) yielded an R-value of 0.216 (p = 0.001), indicating significant variation in endophytic fungal diversity among different tissue parts. PCoA analysis further demonstrated differences in the endophytic fungal communities of Tartary buckwheat among regions, with the PCo1 axis explaining 14.1% of the data and the PCo2 axis explaining 9.8% of the data (Figure 3C). Specifically, the samples from WX and ZJ displayed a relatively compact clustering pattern, suggesting similar endophytic fungal communities. ZC and XN samples also exhibited a compact grouping. However, the samples from other plots displayed a looser clustering pattern, indicating differences in endophytic fungal diversity. Similarly, there were differences observed in the distribution of endophytic fungi among different tissue parts of Tartary buckwheat (Figure 3D). The distribution of endophytic fungi in the roots appeared compact, with a relatively large distance observed between the roots and the stems or leaves, indicating significant differences in endophytic fungi between the root and the stem/leaf. Conversely, the distribution of endophytic fungi in the stems and leaves appeared looser and closer, suggesting a similar diversity of endophytic fungi within the stems and leaves.
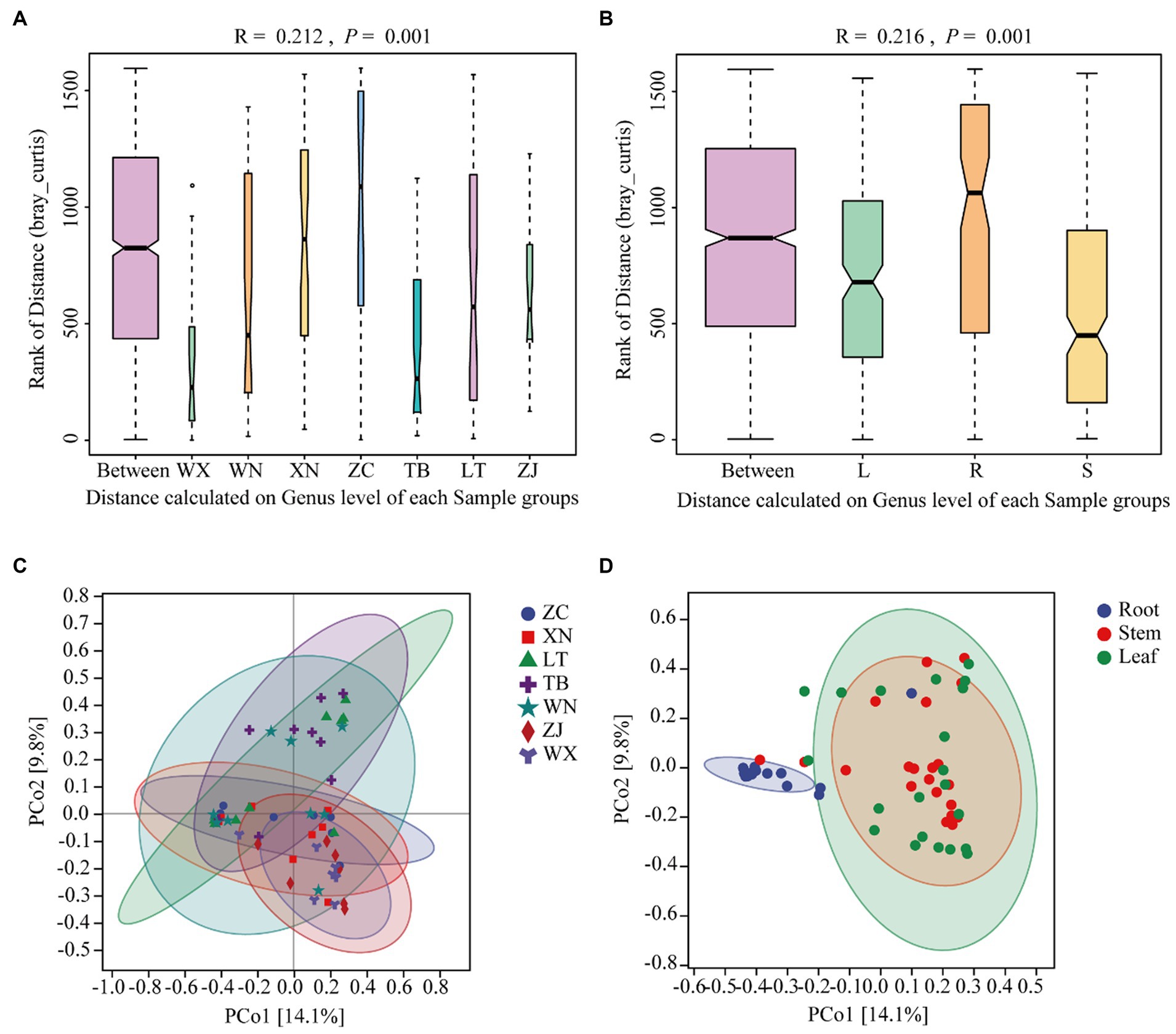
Figure 3. Similarity ANOSIM analysis and PCoA analysis of the endophytic fungal community in Tartary buckwheat based on Bray-Curtis distance. (A,C) ANOSIM analysis and PCoA analysis of 7 different Tartary buckwheat materials. (B,D) ANOSIM analysis and PCOA analysis of different tissue parts (roots, stems, and leaves).
3.2 Composition of endophytic fungal communities in Tartary buckwheat
The classification chart of the top 10 phyla levels of abundance revealed the dominance of Ascomycota as the most prevalent phylum among endophytic fungi in Tartary buckwheat, with relative abundances ranging from 57.47 to 86.21% (Figure 4A), followed by Basidiomycota. These two phyla encompassed the majority of the endophytic fungi, while other phyla had minimal representation. At the class level (Figure 4B), Sortariomycetes was the dominant class in ZC, XN, and WN, accounting for 29.26, 29.35, and 38.76% of the relative abundances, respectively. Conversely, Dothideomycetes was the dominant class in LT, TB, and WX, comprising 35.81, 40.97, and 34.40% of the relative abundances, respectively. In ZJ, Leotiomycetes was the dominant class, with a relative abundance of 50.75%. At the order level (Figure 4C), Hypocreales was the dominant order in ZC, XN, and WN, with relative abundances of 25.41, 25.87, and 31.46%, respectively. Meanwhile, Capnodiales prevailed in LT, TB, and WX, representing 26.52, 27.38, and 24.92% of the relative abundances, respectively. In ZJ, the dominant order was Helotiales, with a relative abundance of 50.75%. At the genus level (Figure 4D), Ilyonectria was the dominant genus in ZC, XN, and WN, with relative abundances of 21.38, 19.485, and 17.87%, respectively. Caryophylloseptoria prevailed in LT and TB, accounting for 25.86 and 26.46% of the relative abundances, respectively. In ZJ and WX, Monilinia dominates, representing 37.06 and 20.99% of the relative abundances, respectively.
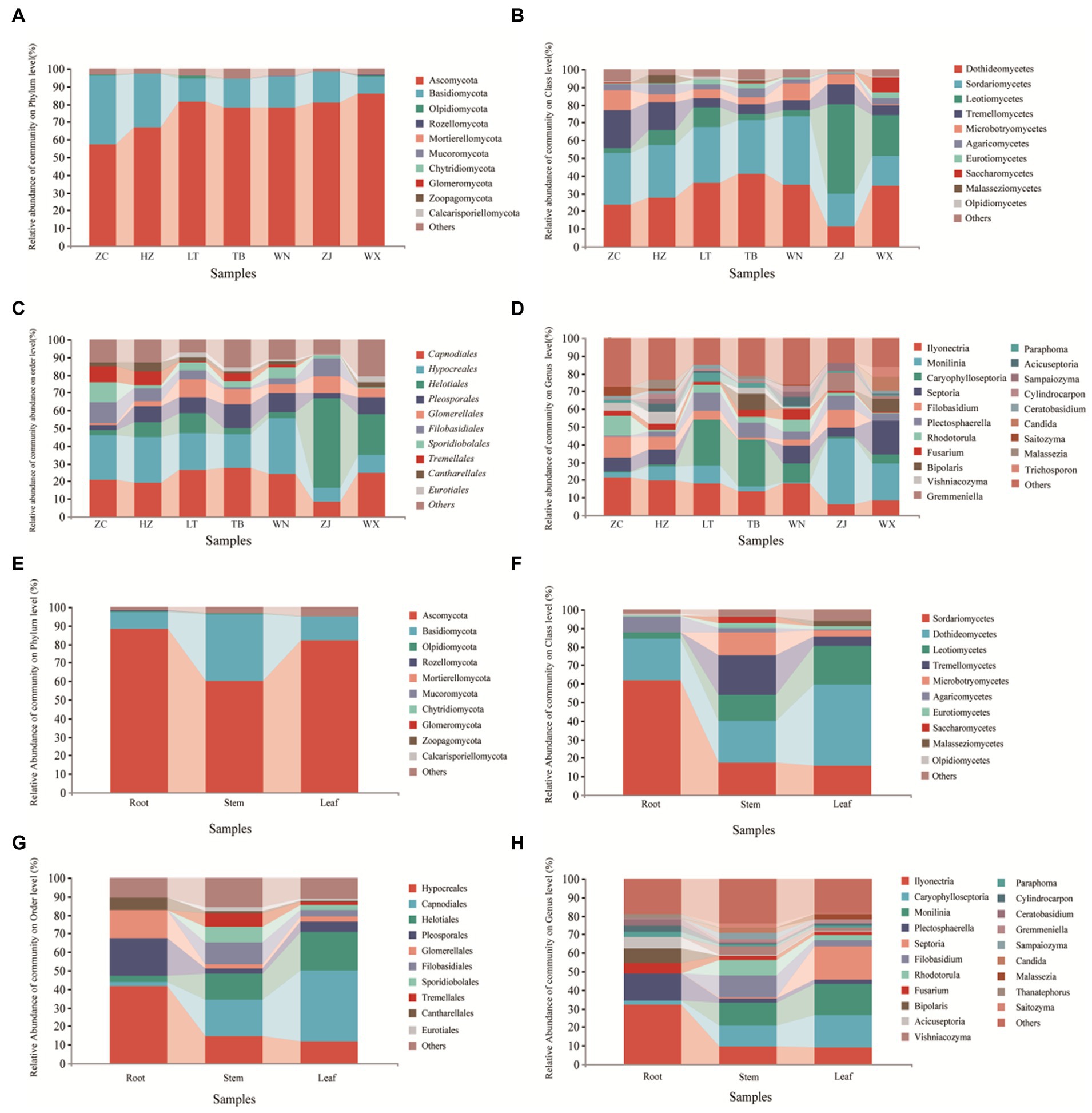
Figure 4. Composition of endophytic fungal community of Tartary buckwheat. The first four figures represent the composition of endophytic fungi at the phylum level (A), class level (B), order level (C), and genus level (D) for different samples. The following four figures represent the composition of endophytic fungi at the phylum level (E), class level (F), order level (G), and genus level (H) in different tissue parts.
Upon examination of different tissue parts of Tartary buckwheat, it was observed that the dominant phylum in the roots, stems, and leaves was Ascomycota, with relative abundances of 88.47, 60.24, and 81.92%, respectively (Figure 4E). At the class level (Figure 4F), Sortariomycetes prevailed in the roots, accounting for 61.60% of the relative abundance. In the stems and leaves, Dothideomycetes was the dominant class, representing relative abundances of 22.34 and 44.10%, respectively. When considering the order level (Figure 4G), Hypocreales emerged as the dominant order in the roots, with a relative abundance of 41.49%. In contrast, Capnodiales dominated in the stems and leaves, exhibiting relative abundances of 19.66 and 38.18%, respectively. At the genus level (Figure 4H), Ilyonectria prevailed in the roots, accounting for 31.89% of the relative abundance. Monilinia emerged as the dominant genus in the stems, constituting 12.43% of the relative abundance. For the leaves, Caryophylloseptoria demonstrated dominance with a relative richness of 17.44%.
3.3 The effect of endophytic fungi on the content of flavonoid metabolites in Tartary buckwheat
The total flavonoid content in the roots, stems, and leaves of the seven samples was determined using the NaNO2-Al(NO3)3 method, as illustrated in Figure 5A. The distribution of total flavonoid content was summarized as follows: leaves>roots>stems. Among the various leaf parts, ZC, LT, and TB displayed the highest total flavonoid content, with values of 1.830, 1.787, and 1.714%, respectively, while XN and WX showed slightly lower levels of 1.030 and 1.011%, respectively. WN and ZJ exhibited the lowest content, measuring less than 1%. In the roots, ZC, XN, LT, WN, and WX displayed relatively high total flavonoid content, with values of 0.519, 0.492, 0.549, 0.591, and 0.616%, respectively, showing no significant difference. Conversely, TB and ZJ showed lower levels of total flavonoid content, measuring less than 0.4%. The stems generally exhibited low total flavonoid content, ranging from 0.156 to 0.232%, with no significant differences observed among the seven types of Tartary buckwheat stems. The total phenolic content in the roots, stems, and leaves of the seven samples was determined using the Folin phenol method. Figure 5B presents the results, illustrating that the approximate distribution of total phenolic content followed a similar pattern: leaves>roots>stems. Among the leaves, ZC and TB presented the highest total phenolic content, with values of 6.234 and 6.313%, respectively, while LT exhibited a slightly lower level of 4.279%. The total phenolic content of XN, WN, ZJ, and WX ranged from 2.997 to 3.594%, indicating the lowest values. In the roots, ZC, XN, LT, WN, and WX displayed relatively high total phenolic content, measuring 1.986, 2.009, 2.142, 2.412, and 2.620%, respectively. Conversely, TB and ZJ showed lower levels of total phenolic content at 1.683 and 1.084%, respectively. The stems demonstrated generally low levels of polyphenols, ranging from 0.909 to 1.076%. Overall, the trends in total flavonoid and total phenolic content among the different parts of the seven samples were consistent. Specifically, ZC exhibited the highest total flavonoid and phenolic content in its roots, stems, and leaves. The LT leaves displayed higher total flavonoid content, but lower total phenolic content. WN and ZJ generally showed low levels of total flavonoid and phenolic content.
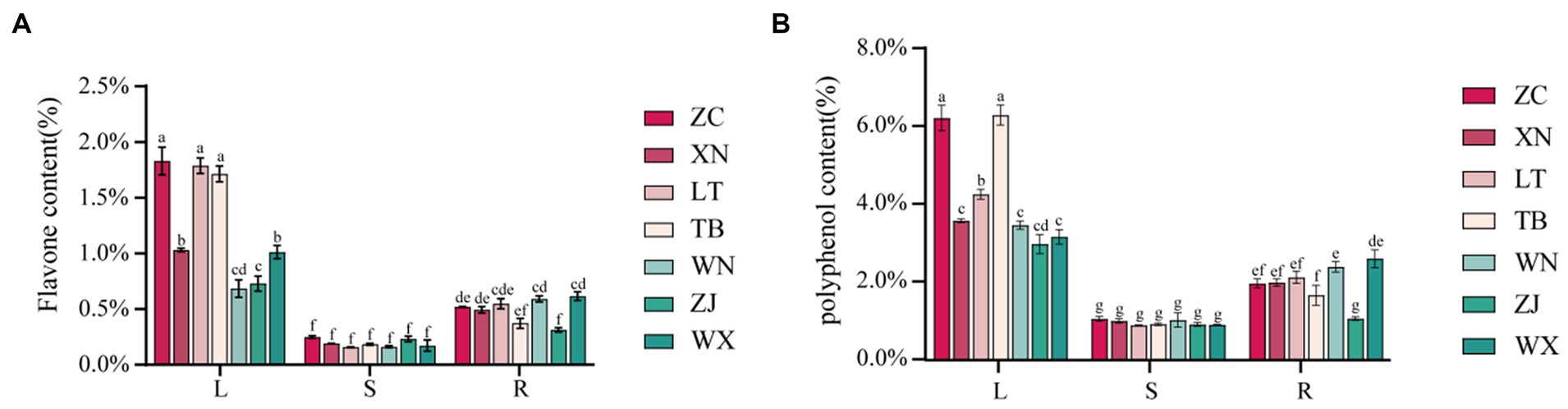
Figure 5. Content map of flavonoids and polyphenols in 21 Tartary buckwheat samples (Zhaojue, ZJ; Wenxian, WX; Zhangchuan, ZC; Xining, XN; Lintao, LT; Taibai, TB; Weining, WN). (A) Flavone content in 21 Tartary buckwheat samples. (B) Polyphenol content in 21 Tartary buckwheat samples.
Numerous studies have demonstrated the influence of endophytic fungi on the secondary metabolite content in plants (Shen et al., 2022; Lu et al., 2023). The results of this study indicated significant differences in the composition and abundance of endophytic fungi among the seven distinct collection sites of Tartary buckwheat materials. These differences also translated into variations in the total flavonoid and phenol content within the plants. Therefore, we hypothesized that specific endophytic fungi in Tartary buckwheat contributed to the discrepancies in secondary metabolite content. To validate this hypothesis and identify the precise metabolites affected by endophytic fungi, we selected ZC and LT, which displayed the greatest discrepancy in fungi composition as well as significant differences in flavonoid and polyphenol content. Subsequently, the content of 40 flavonoids in the roots, stems, and leaves of these two materials was examined. As depicted in Figure 6A, 22 flavonoids were detected from the roots, stems, and leaves of ZC and LT (Tartary buckwheat does not contain the other 18 flavonoid metabolites, Supplementary Table S3). The levels of these flavonoids ranged from highest to lowest as follows: Rutin, Hesperidin, Quercetin, Epicatechin, Kaempferol 3-rutinoside, Catechin, Protocatechuic acid, Neohesperidin, Gallic acid, Hyperoside, Protocatechualdehyde, Kaempferol, Emodin, Chlorogenic acid, Caffeic acid, Naringenin, Homoorientin, Vitexin, Umbelliferone, Isovitexin, Genistin, and Apigenin. The heat map demonstrated significant variations in metabolite content across the roots, stems, and leaves of Tartary buckwheat (Figure 6B). For instance, the content of Genistin, Neohesperidin, Hyperoside, Hesperidin, Kaempferol 3-rutinoside, Kaempferol, Quercetin, Rutin, Emodin, and Protocatechuic acid in the leaves was notably higher than in the stems and roots. Furthermore, Chlorogenic acid, Umbelliferone, and Caffeic acid exhibited the highest content in the stems, while Naringenin and Protocatechualdehyde was significantly higher in the roots compared to the leaves and stems. Notably, there were distinctive differences in the content of flavonoid metabolites among different Tartary buckwheat materials. For example, ZC leaves displayed higher levels of Caffeic acid, Vitexin, Catechin, Epicatechin, Apigenin, Isovitexin, Homoorientin, and Gallic acid compared to LT. Additionally, ZC roots also contained greater amounts of Apigenin and Isovitexin. The results of the ANOSIM analysis revealed that the R-value of different tissue samples of ZC and LT was 1 (p = 0.001), indicating that the differences in flavonoid metabolite content between tissue samples were highly significantly greater than within samples (Figure 6C). PCA analysis visualized this intergroup difference, with the PC1 axis explaining 99.5% of the data and the PC2 explaining 0.4% of the data (Figure 6D). It is easy to see in the figure that flavonoid metabolite content is more similar between different samples of the same tissue, such as within roots, stems, and leaves. On the contrary, the content of flavonoid metabolites varied very much between different tissue samples of the same sample, especially in leaves compared to roots and stems.
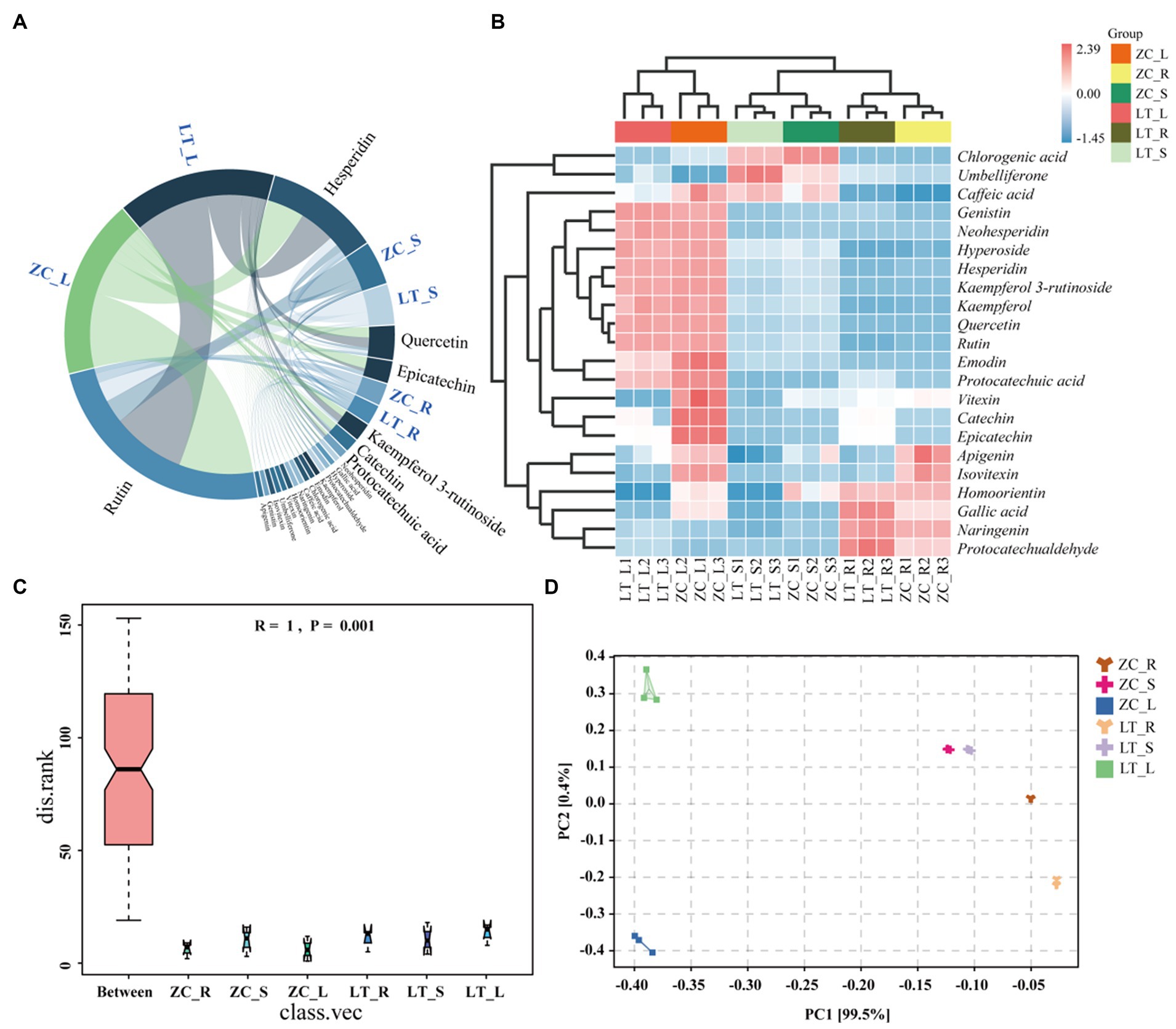
Figure 6. Chord diagram, heat maps, ANOSIM analyses, and PCA analyses of 22 flavonoid metabolites in ZC and LT roots, stems, and leaves. (A) The chord diagram shows the proportion of 22 flavonoids in the roots, stems, and leaves of ZC and TL. (B) The heat map intuitively reflects the differences in the content of 22 flavonoids in different samples. In the heat map, red indicates a content higher than the average, while blue indicates a content lower than the average. The darker the color, the more significant the difference. (C,D) ANOSIM and PCA analyses of flavonoid metabolite content in roots (R), stems (S), and leaves (L) of tissue samples from ZC and LT.
To examine the variations in endophytic fungi between ZC and LT, we performed LEfSe analysis on their respective populations. LEfSe analysis is a differential analysis method that enables simultaneous analysis across all classification levels. Additionally, it emphasizes the identification of robust differential species, referred to as biomarkers. The results of the LEfSe analysis (Figure 7A) revealed significant differences in the endophytic fungi of ZC and LT at the class, order, family, and genus levels. Notably, there were nine significantly different genus-level classifications, including unclassified_Liptosphaeriaceae, Bipolaris, Hymenula, Colletotrichum, Hydroptosphaera, Dactylonectria, Chaetomium, unclassified_Sortariales, and one unidentified taxonomic unit (g_identified). Among these nine distinct genus-level taxonomic units, unclassified_Sordariales accounted for the largest proportion at 29.9%, followed by Chaetomium (25.1%), Colletotrichum (14.2%), Bipolaris (10.4%), Hymenula (7.6%), unclassified_Lettosphaeriaceae (6.4%), Hydroptosphaera (5.0%), and Dactylonectria (1.5%) (Figure 7B). Furthermore, Pearson correlation analysis was conducted to examine the relationship between endophytic fungi and flavonoid metabolite content at the genus level. Supplementary Figure S2 displays the complete correlation heatmap, while Figure 7C presents a network diagram highlighting the significant correlations between fungi and flavonoid metabolites. The correlation network diagram distinctly demonstrated the impact of different genus-level endophytic fungi on flavonoid content. Bipolaris, Hymenula, Colletotrichum, Dactylonectria, and unclassified_Lettosphaeriaceae exhibited significant positive correlations with the content of 11 or more flavonoids, including Rutin, Hesperidin, Quercetin, Epicatechin, Kaempferol 3-rutinoside, Catechin, Protocatechuic acid, and others. Hydroptosphaera and Chaetomium displayed significant positive correlations with the content of four and three flavonoid metabolites, respectively. Additionally, Chaetomium exhibited a significant negative correlation with the content of Caffeic acid. Unclassified_Sordariales demonstrated a significant positive correlation with the content of two flavonoid metabolites.
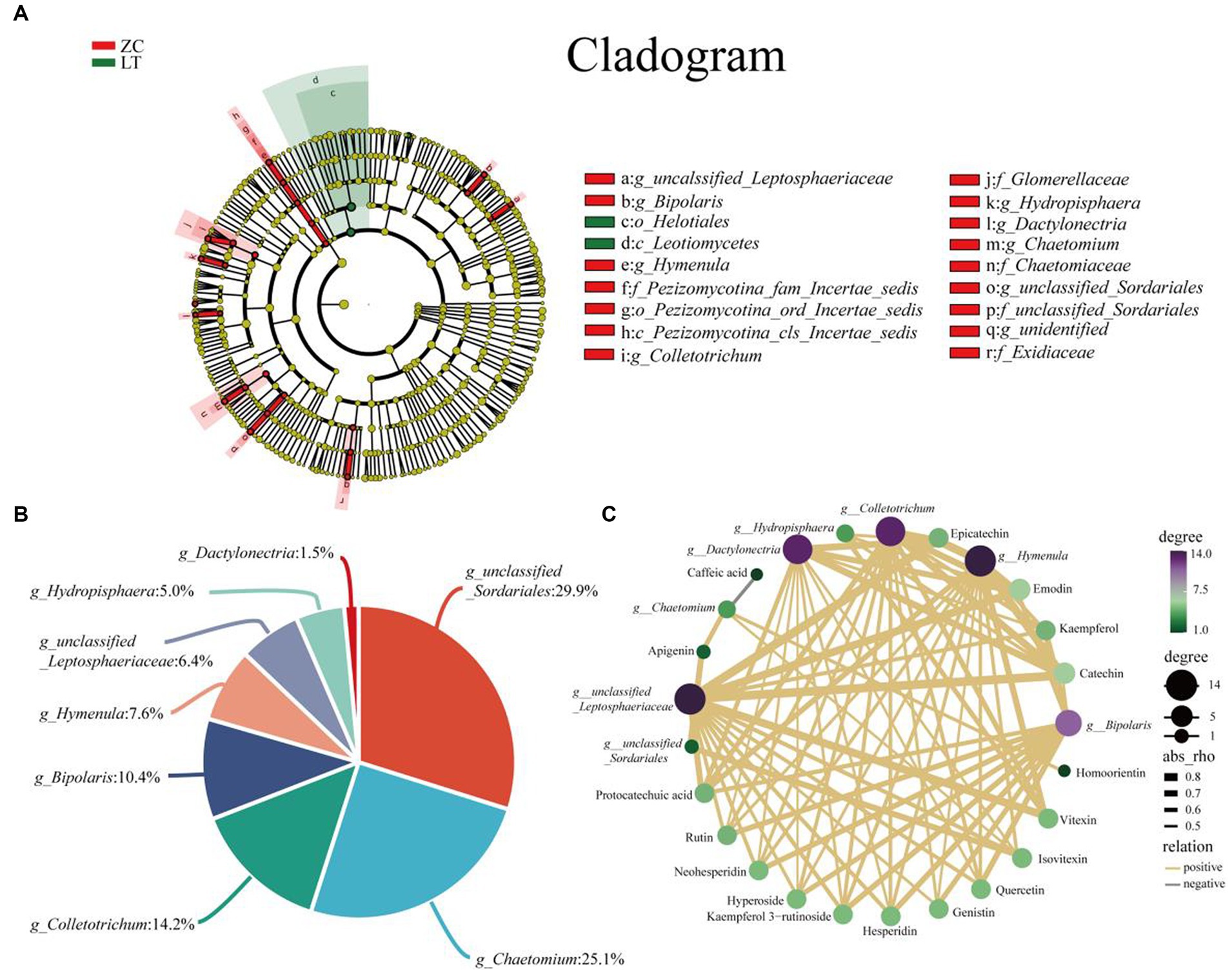
Figure 7. Analysis of differences in endophytic fungi between ZC and LT, as well as correlation between flavonoids and different endophytic fungi at the genus level. (A) LEfSe analysis shows the hierarchical relationships of the main taxonomic units in the sample community from phylum to genus (from inner circle to outer circle). The node size corresponds to the average relative abundance of the classification unit; A node with a red background indicates a higher abundance of the classification unit in ZC, while a node with a green background indicates a higher abundance of the classification unit in LT. The letters indicate the names of taxonomic units with significant differences between groups. (B) Percentage of endophytic fungi with genus level differences. (C) Association network diagram of endophytic fungi and flavonoid metabolites with significant correlation.
3.4 The effect of endophytic fungi on the phenotypic characters of Tartary buckwheat
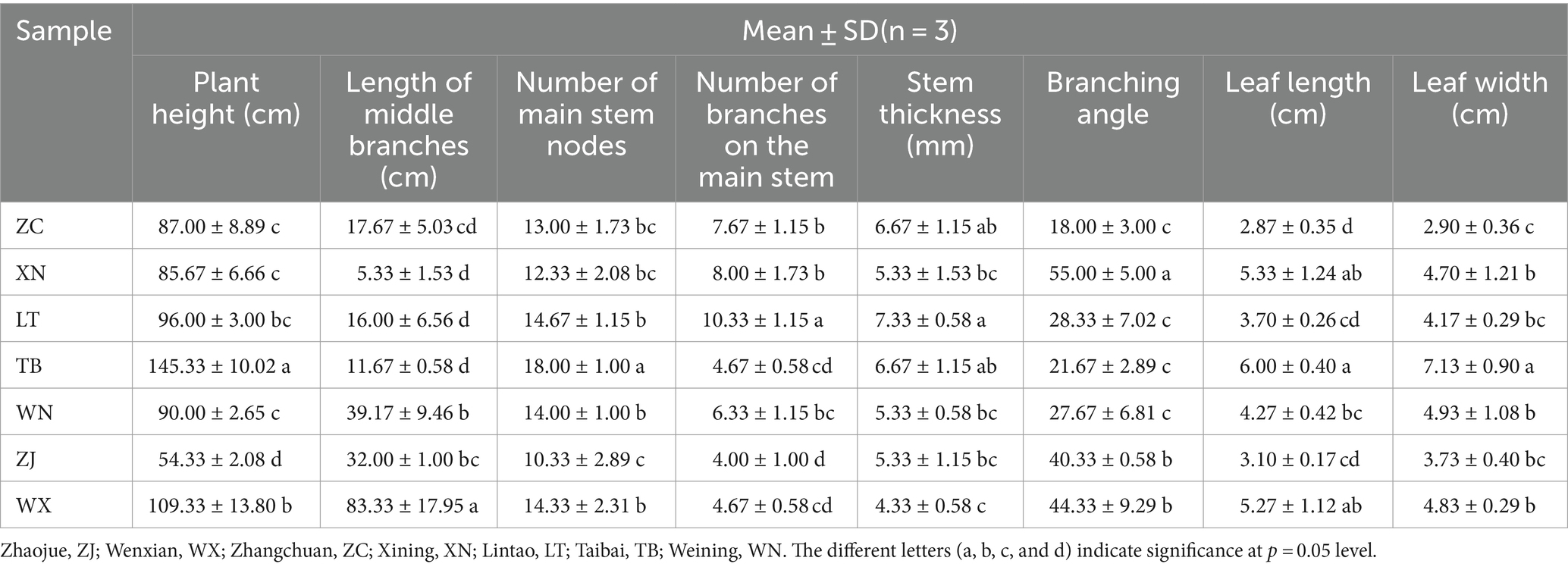
Various biological characteristics of 7 plant samples were measured, namely plant height, middle branch length, number of main stem nodes, number of main stem branches, stem thickness, branch angle, leaf length, and leaf width (Table 2). Plant height serves as an intuitive indicator of plant growth and speed (Zhao et al., 2022). Notably, significant differences in plant height were observed among the seven collection locations of Tartary buckwheat. Specifically, TB and WX exhibited higher plant heights at 145.33 cm and 109.33 cm, respectively. Furthermore, TB demonstrated significantly taller plants compared to the other six samples. Conversely, ZJ plants exhibited the shortest height at only 54.33 cm. The length of the middle branch, number of main stem nodes, number of main stem branches, and stem thickness are indicative of plant characteristics along the stem. Notably, WX had the longest middle branch, measuring 83.33 cm, which was significantly longer than the other six species. The remaining Tartary buckwheat samples displayed middle branch lengths ranging from 5.33 cm to 39.17 cm. Additionally, TB boasted the highest number of main stem nodes, averaging 18 nodes per plant. The distribution of node numbers among the other plants was relatively uniform, ranging from 10.33 to 14.66. As for the number of branches on the main stem, LT exhibited the highest count, with an average of 10.33 branches per plant. In contrast, ZJ and WX had the lowest branch counts, averaging only 4 branches per plant. Similarly, LT showcased the greatest stem thickness at 7.33 mm, while WX displayed a stem thickness of only 4.33 mm. In terms of branching angles, XN and WX boasted the highest angles, measuring 55.00° and 44.33°, respectively. ZJ, LT, WN, and TB followed with branching angles of 30.33°, 28.33°, 27.67°, and 21.67°, respectively. Remarkably, ZC exhibited the smallest branch angle, measuring only 18.00°. Regarding leaf characteristics, TB possessed the largest leaves, with lengths and widths measuring 6.00 cm and 7.13 cm, respectively. WX and XN closely followed, followed by ZJ, LT, and WN, with ZC exhibiting the smallest leaf size.
Endophytic fungi play a crucial role throughout the entire life cycle of plants, influencing their growth, development, and metabolic processes (Kandar et al., 2018; Yan et al., 2019; Wang et al., 2022). Therefore, it is highly plausible that the endophytic fungi found in Tartary buckwheat have an impact on various phenotypic traits, including plant height, stem diameter, and number of branches. Similarly, LEfse analysis was employed to identify endophytic fungal species exhibiting significant differences among the 7 samples. The distinct distribution of endophytic fungi across the 7 samples was observed at various taxonomic levels, including 1 phylum, 3 classes, 7 orders, 13 families, and 24 genera, with the inclusion of several unidentified endophytic fungal taxonomic units (Supplementary Figure S3). Moreover, a correlation analysis was conducted between the 21 different genus-level endophytic fungi and 8 biological traits of Tartary buckwheat (Supplementary Figure S4). Initially, we discovered a significant positive correlation between plant height and Buckleyzyma and Bipolaris (Figure 8). Furthermore, the length of the middle branch exhibited a significant positive correlation with multiple genera of endophytic fungi, such as Penicillium, Minimelanolocus, Neurospora, Trichosporon, and one unidentified endophytic fungal taxon. The number of main stem branches showed a significant negative correlation with Neosetophoma while displaying a significant positive correlation with an unidentified endophytic fungal taxon. Leaf width displayed a significant positive correlation with Buckleyzyma and Cystofilobasidium. Notably, no significant correlation was observed between endophytic fungi and stem diameter, number of main stem nodes, leaf length, and branching angle at the genus level.
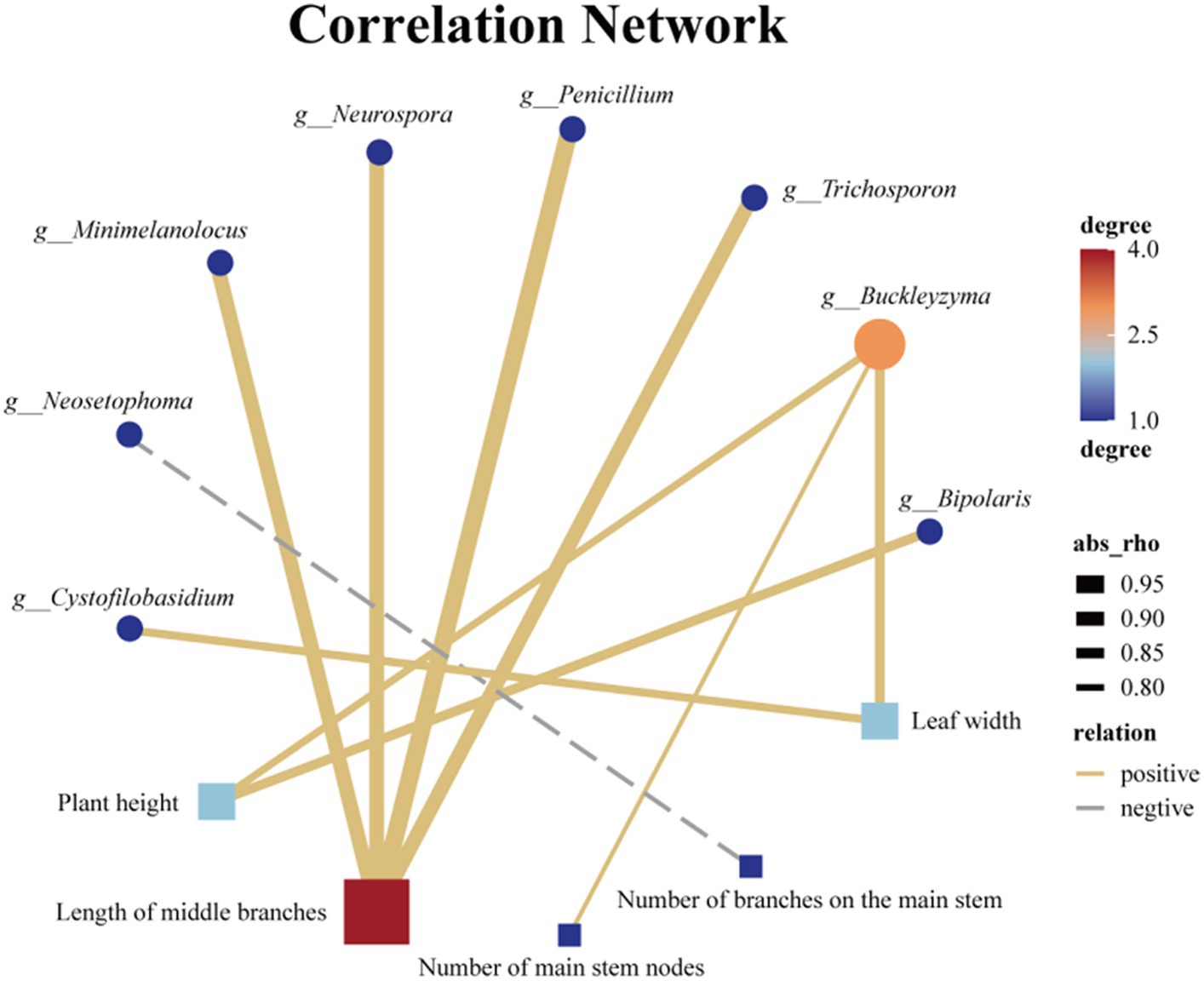
Figure 8. Association network diagram of Endophytic fungi with differences at the genus level and phenotypic traits with significant correlation.
4 Discussion
Extensive research on fungal plant interactions has focused on endophytic fungi due to their ability to increase the content of secondary metabolites, enhance biocontrol capabilities, and promote host growth and development (Kumar and Verma, 2018; Mishra et al., 2021; Zhang et al., 2022). For instance, Alternaria in S. alopecuroides seeds has been identified as a potential promoter for the accumulation of bioactive compounds in the host (Ju et al., 2022). Wild halophytic rice Oryza coarctata showed significant enhancement in root length, stem length, and total tiller number after inoculation with the endophytic fungus Aspergillus welwitschiae Ocstreb1 (AwOcstreb1) (Airin et al., 2023). In other crop plants such as maize, bean, and soybean, plant height and dry weight were also significantly increased after inoculation with the fungi Purpureocillium spp. and M. marquandii strains (Baron et al., 2020). Furthermore, inoculation with specific endophytic fungi, such as Alternaria chlamydospora, Piriformospora indica, and Trichoderma longibrachiatum, has been shown to impact the plant height, stem diameter, number of branches, and root length of the host (Kandar et al., 2018; Abdolmaleki et al., 2022; Wang et al., 2022; Zuo et al., 2022). In the current study, a comprehensive correlation analysis was conducted between endophytic fungi in Tartary buckwheat and both flavonoid metabolite content and phenotypic traits. Notably, we observed a strong positive correlation between the Bipolaris genus and the content of 11 flavonoid metabolites, including key metabolites like rutin and quercetin. Additionally, a significant positive correlation with plant height was uncovered. These findings suggest that endophytic fungi belonging to the Bipolaris genus play a crucial role in the modulation of Tartary buckwheat. Consequently, this provides new avenues for exploring functional strains that produce flavonoids from Fagopyrum tataricum, utilizing wild resources, and implementing biological fertilizer strategies to enhance its quality and yield.
High-throughput sequencing technology has found extensive utilization in microbial sequencing, enabling the comprehension of microorganism species composition and abundance across different plants, thereby shedding light on microbial diversity and its dynamic patterns. Additionally, it provides insights into the functional potential of microorganisms within the communities, aiding the understanding of their roles and interrelationships within ecosystems (Baldrian et al., 2022). In this study, MiSeq high-throughput technology was employed to sequence the ITS rDNA genes of endophytic fungi in 21 samples of Tartary buckwheat from 7 distribution plots in central and southern China. This approach comprehensively captured the community structure and diversity of endophytic fungi present in Tartary buckwheat. The findings revealed that WN exhibited the highest richness of endophytic fungal communities in both the stems and leaves, significantly surpassing other groups. Additionally, WN demonstrated high uniformity and diversity. Conversely, ZJ generally displayed lower community richness, as well as lower evenness and diversity within its stems and leaves. No significant differences in diversity indices were observed among the other samples, potentially attributable to variations in climate, soil composition, and surrounding plant diversity across different geographical locations. Research conducted by Wang et al. (2023) highlighted the importance of temperature thresholds in shaping changes in root-associated endophytic fungal diversity. Beyond these thresholds, the abundance of OTUs colonizing root tissue rapidly decreases. Remarkably, habitats characterized by high soil pH and low nutrient availability tend to exhibit greater fungal diversity. Moreover, the species composition and quantity of accompanying plants also impact the diversity and richness of endophytic fungi within the host plant (Yokoya et al., 2017; Fu et al., 2022).
The distribution of endophytic fungi is tissue-specific, as observed in Azadirachta Indica where the richness and diversity of endophytic fungi vary across the roots, leaves, stems, bark, and branches (Chutulo and Chalannavar, 2018). Hence, this study aimed to comprehensively assess the diversity and richness of endophytic fungi in Tartary buckwheat by selecting seven different sampling locations and three distinct tissues. Regardless of the sampling location or tissue, the predominant classification at the phylum level was Ascomycota, which is the largest classification level among fungi and encompasses approximately 64,000 known species (Bennett and Turgeon, 2016). The dominant genera of endophytic fungi found in various sampling sites and tissues included Ilyonectria, Monilinia, and Caryophylloseptoria. Ilyonectria is associated with root rot symptoms in diverse herbaceous and woody host plants such as ginseng, Panax notoginseng, and apple (Zhu et al., 2019; Popp et al., 2020; Bischoff Nunes and Goodwin, 2022). Interestingly, it can also exist as an endophytic fungus in seemingly healthy plant roots, where it can inhibit other fungal root pathogens and maintain the health of the host (Yang et al., 2019). Based on this, we speculated that Ilyonectria functioned similarly in Tartary buckwheat, as no significant lesions were observed in the roots. Monilinia primarily inflicts damage on orchards, causing fruit brown rot (Pereira et al., 2019; Akhoon et al., 2023), while Caryophylloseptoria mainly leads to leaf spot disease (Yang et al., 2022). However, despite being dominant in Tartary buckwheat, these genera are hypothesized to share a similar mechanism with Ilyonectria. Takahiro Noda comprehensively reviewed the differences in flavonoid metabolites, such as rutin, vitexin, isovitexin, and quercetin, in various parts of Tartary buckwheat, including the roots, stems, leaves, flowers, and seeds (Takahiro et al., 2023). Notably, rutin constitutes 90% of the total phenolic content, with the highest concentration in the leaves. This finding aligns with the results of our research. The correlation analysis between flavonoids and differential fungi is presented in Figure 7.
Colletotrichum exhibits a significant positive correlation with the content of 13 flavonoid metabolites, suggesting its close association with flavonoid metabolism in Tartary buckwheat and its potential involvement in crucial steps of various flavonoid metabolic pathways. Fungal metabolites are typically similar to those of their hosts, indicating a close relationship between host and fungal metabolites (Johny et al., 2021). Likewise, Kim and Shim (2019) identified 25 phenolic compounds, including vitexin and skullcapflavone II, along with various terpenoids, pyranones, and other metabolites in the metabolome of Colletotrichum. Furthermore, two Colletotrichum strains isolated from Andrographis paniculata exhibited substantial antioxidant activity and produced high yields of total phenols and flavonoids (Li Q. et al., 2022). Another study found that after exogenous inoculation with a Colletotrichum strain, the content of total flavonoids, soluble total phenols, coumarins, and kaempferol increased in the body of Annona muricata (Rubio-Melgarejo et al., 2020). Based on the aforementioned findings, we speculated that Colletotrichum, as an endophytic fungus in Tartary buckwheat, was capable of producing abundant levels of flavonoids and other polyphenols. On the other hand, Colletotrichum can stimulate Tartary buckwheat to increase its secretion of flavonoids. Although the first aspect is easily understood, the underlying reason why Colletotrichum induces the release of flavonoids from Tartary buckwheat remains unclear. However, a significant amount of research has focused on Colletotrichum’s ability to cause anthracnose in plants, thereby posing a threat to their survival (Camiletti et al., 2022; de Aguiar Carraro et al., 2022; Willie Anderson et al., 2022). When plants encounter abiotic or biotic stress, they tend to accumulate substantial amounts of phenolic compounds in their tissues (Pedras et al., 2011). Previous studies have shown that flavonoids and other polyphenols provide some protection against certain pathogenic bacteria in the genus Colletotrichum (Roy et al., 2018; Tugizimana et al., 2019). For instance, during the initial stages of Colletotrichum infection, as mentioned earlier in Phaseolus vulgaris and Annona muricata, the levels of phenolic compounds in their tissues are upregulated. Upon infection with Colletotrichum in olive plants, an increase in PAL enzyme activity and PAL gene expression occurs, indicating that these genes play a role in olive’s defense response against anthracnose (Gouvinhas et al., 2019). PAL enzyme, the key enzyme in flavonoid synthesis and the initial enzyme in the phenylalanine pathway, catalyzes the conversion of phenylalanine to aminophenylpyruvate and ammonia (Liu et al., 2021). A second plausible speculation is that although Colletotrichum acts as an endophytic fungus of Tartary buckwheat rather than as an exogenous pathogen, as a parasitic fungus within the genus Colletotrichum, it is equally likely to trigger a certain level of chemical defense response in the plant, causing the host to secrete more phenolic compounds, a response that already exists as a norm.
Additionally, we also identified a notable positive correlation between Bipolaris and the content of 11 flavonoids, suggesting a potential relationship between the presence of Bipolaris and flavonoid levels. Bipolaris, specifically Bipolaris oryzae, is renowned for causing rice blast (Dorneles et al., 2020) as well as spot disease in diverse crops, including wheat (Zaccaron and Bluhm, 2017; Meng et al., 2020; Manzar et al., 2022). It is noteworthy that Bipolaris secondary metabolites are rich in flavonoids and other polyphenols, although their applications in this area still necessitate further development (Das et al., 2017; Muthukrishnan et al., 2022). Additionally, Bipolaris infection in wheat triggers its own SA signal transduction and WRKY33 transcription factor, enhancing the expression of phenylpropanoid pathway genes, which potentially leads to the accumulation of phenolic defense metabolites (Sahu et al., 2016). Wanke et al. (2023) have discovered that plants deposit phenolic substances at sites invaded by fungi to reinforce their cell walls and impede further fungal development. Consequently, it is plausible that Bipolaris may induce the host plant to secrete higher levels of polyphenols following invasion. The aforementioned studies partially elucidate the significant positive correlation between Bipolaris and the various flavonoid content observed in Tartary buckwheat.
Plant phenotypic traits play a vital role in the collection of wild resources (Melouk et al., 2023). Environmental factors such as altitude (Bonanomi et al., 2016), precipitation (Sun et al., 2016), and temperature (Zahedi and Sarikhani, 2016) significantly influence most morphological traits. This study focused on seven wild Tartary buckwheat samples from various habitats within the wild buckwheat germplasm resource nursery. To assess the differences, eight relevant phenotypic traits, including plant height, stem diameter, and number of main stem nodes, were measured. The aim was to examine the impact of endophytic fungi, which are consistently associated with the plant in its physiological context, by eliminating the influence of environmental factors from their native habitats. The correlation between the differential endophytic fungi and phenotypic traits in these seven Tartary buckwheat samples is shown in Figure 8. Notably, there was a significant positive correlation between Bipolaris and the height of Tartary buckwheat plants. Previous analyses have shown that Bipolaris is also significantly and positively correlated with 11 flavonoids. Under salt stress, inoculation with Bipolaris sp. significantly increases soybean stem length, root length, root fresh weight, and chlorophyll content (Lubna et al., 2022). The culture filtrate of Bipolaris is rich in indoleacetic acid, demonstrating strong free radical scavenging ability and anti-lipid peroxidation activity (Khan et al., 2017). Thus, we speculate that the endophytic fungi of the Bipolaris genus in Tartary buckwheat may promote its growth by secreting indoleacetic acid. Additionally, other endophytic fungi affect the plant phenotype of Tartary buckwheat, such as Buckleyzyma, which affects plant height and leaf width, and Minimelanolocus, Neurospora, and Trichosporon, which impact the length of middle branches. Limited research has been conducted on these aforementioned endophytic fungi, and currently, there are no studies discussing their impact on plant phenotypes. However, this study provides new insights into the multifaceted exploration and utilization of these endophytic fungi.
5 Conclusion
This study demonstrated significant differences in the plant phenotype, endophytic fungal community diversity, and flavonoid metabolite content of Tartary buckwheat from different regions. Pearson correlation analysis revealed that several endophytic fungi, including Bipolaris, Hymenula, Colletotrichum, Dactylonectria, and unclassified_ Liptosphaeriaceae, had the potential to enhance the content of more than 10 flavonoids, such as Rutin, Hesperidin, Quercetin, Epicatechin, Kaempferol 3-rutinoside, Catechin, and others. Moreover, Bipolaris, Buckleyzyma, Trichosporon, Penicillium, Neurospora, Minimelanolocus, and Cystofilobasidium exhibited the potential to promote Tartary buckwheat growth, including plant height, length of middle branches, number of main stem nodes, number of branches on the main stem, and leaf width. Particularly, Bipolaris not only enhanced the content of flavonoids but also stimulated growth in Tartary buckwheat. Further validation of Bipolaris’ functionality and utilization as a key candidate fungus for the development of biological fertilizers for Tartary buckwheat should be pursued in future research.
Data availability statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found at: https://www.ncbi.nlm.nih.gov/, PRJNA1043747.
Author contributions
MC: Conceptualization, Writing – original draft. ZD: Writing – original draft, Formal analysis. MZ: Investigation, Writing – review & editing. YS: Methodology, Writing – review & editing, Conceptualization. CL: Visualization, Writing – review & editing, Software. QL: Visualization, Writing – review & editing, Data curation. TB: Validation, Writing – review & editing, Formal analysis. ZT: Validation, Writing – review & editing. HC: Resources, Supervision, Writing – original draft.
Funding
The author(s) declare financial support was received for the research, authorship, and/or publication of this article. The authors are grateful for the support from the National Key R&D Program of China (2021YFD1200105).
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2024.1360988/full#supplementary-material
References
Abdolmaleki, A. K., Pirdashti, H., Yaghoubian, Y., Abbasian, A., and Shiade, S. R. G. (2022). Endophytic fungi improve growth and yield of wheat (Triticum aestivum L.) under limited light conditions. Gesunde Pflanz. 75, 1517–1529. doi: 10.1007/s10343-022-00816-x
Aguirre-Hernández, E., González-Trujano, M. E., Martínez, A. L., Moreno, J., Kite, G., Terrazas, T., et al. (2009). HPLC/MS analysis and anxiolytic-like effect of quercetin and kaempferol flavonoids from Tilia americana var. mexicana. J. Ethnopharmacol. 127, 91–97. doi: 10.1016/j.jep.2009.09.044
Airin, A. A., Arafat, M. I., Begum, R. A., Islam, M. R., and Seraj, Z. I. (2023). Plant growth-promoting endophytic fungi of the wild halophytic rice Oryza coarctata. Ann. Microbiol. 73:36. doi: 10.1186/s13213-023-01738-3
Akhoon, B. A., Gupta, S. K., and Dhar, M. K. (2023). Dissecting the genome, secretome, and effectome repertoires of Monilinia spp.: The causal agent of brown rot disease: A comparative analysis. Postharvest Biol. Technol. 195:112120. doi: 10.1016/j.postharvbio.2022.112120
Aktar, S., Bai, P., and Wang, L. (2022). Identification of a BAHD acyltransferase gene involved in plant growth and secondary metabolism in tea plants. Plants (Basel) 11:2483. doi: 10.3390/plants11192483
Baldrian, P., Větrovský, T., Lepinay, C., and Kohout, P. (2022). High-throughput sequencing view on the magnitude of global fungal diversity. Fungal Divers. 114, 539–547. doi: 10.1007/s13225-021-00472-y
Baron, N. C., Pollo, A. S., and Rigobelo, E. C. (2020). Purpureocillium lilacinum and Metarhizium marquandii as plant growth-promoting fungi. PeerJ 8:e9005. doi: 10.7717/peerj.9005
Bennett, R. J., and Turgeon, B. G. (2016). Fungal sex: The Ascomycota. Microbiol. Spectr. 4:0005. doi: 10.1128/microbiolspec.funk-0005-2016
Bischoff Nunes, I., and Goodwin, P. H. (2022). Interaction of ginseng with Ilyonectria root rot pathogens. Plan. Theory 11:2152. doi: 10.3390/plants11162152
Bonanomi, G., Stinca, A., Chirico, G. B., Ciaschetti, G., Saracino, A., and Incerti, G. (2016). Cushion plant morphology controls biogenic capability and facilitation effects of Silene acaulis along an elevation gradient. Funct. Ecol. 30, 1216–1226. doi: 10.1111/1365-2435.12596
Callahan, B. J., McMurdie, P. J., Rosen, M. J., Han, A. W., Johnson, A. J. A., and Holmes, S. P. (2016). DADA2:High-resolution sample inference from Illumina amplicon data. Nat. Methods 13, 581–583. doi: 10.1038/nmeth.3869
Camiletti, B. X., Lichtemberg, P. S. F., Paredes, J. A., Carraro, T. A., Velascos, J., and Michailides, T. J. (2022). Characterization of colletotrichum isolates causing colletotrichum dieback of citrus in california. Phytopathology 112, 1454–1466. doi: 10.1094/PHYTO-10-21-0434-R
Chen, Q. F. (2018). Buckwheat production status and new types of cultivated buckwheat breeding research recent progress. J. Guizhou Normal Univ. (Nat. Sci. Ed.) 36:1-7+131. doi: 10.16614/j.gznuj.zrb.2018.03.001
Chutulo, E. C., and Chalannavar, R. K. (2018). Endophytic mycoflora and their bioactive compounds from Azadirachta Indica: A Comprehensive Review. J. Fungi 4:42. doi: 10.3390/jof4020042
Das, M., Prakash, H. S., and Nalini, M. S. (2017). Antioxidative properties of phenolic compounds isolated from the fungal endophytes of Zingiber nimmonii (J. Graham) Dalzell. Front. Microbiol. 12, 151–162. doi: 10.1007/s11515-016-1441-z
de Aguiar Carraro, T., Lichtemberg, P. D. S. F., Michailides, T. J., Miranda Borges, M. I., Pereira, W. V., and May de Mio, L. L. (2022). Identification and characterization of colletotrichum species associated with anthracnose on persimmon in Brazil. Fungal Biol. 126, 235–249. doi: 10.1016/j.funbio.2021.12.003
Ding, C., Wang, S., and Li, J. (2022). Transcriptomic analysis reveals the mechanism of host growth promotion by endophytic fungus of Rumex gmelinii Turcz. Arch. Microbiol. 204:443. doi: 10.1007/s00203-022-03072-9
Dorneles, K. R., Refatti, J. P., Pazdiora, P. C., Avila, L. A., Deuner, S., and Dallagnol, L. J. (2020). Biochemical defenses of rice against Bipolaris oryzae increase with high atmospheric concentration of CO2. Physiol. Mol. Plant Pathol. 110:101484. doi: 10.1016/j.pmpp.2020.101484
Fan, Y., Ding, M. Q., Zhang, K. X., Yang, K. L., Tang, Y., Zhang, Z. W., et al. (2019). Overview of buckwheat germplasm resources. J. Plant Genet. Resour. 20, 813–828. doi: 10.13430/j.cnki.jpgr.20190428002
Fu, H., Song, Z., Li, S., Lan, S., Zeng, X., and Huang, W. (2022). Effects of bamboo forest type and density on the growth of Bletilla striata and root endophytic fungi. Diversity 14:391. doi: 10.3390/d14050391
Gao, Z., Wang, Y.-C, and Chang, Y.-X. (2016). Determination of flavonoids and anthocyanins in nitraria tangutorum by high performance liquid chromatography coupled with tandem mass spectrometry. Protein Pept. Lett. 23, 424–432.
Gateta, T., Nacoon, S., Seemakram, W., Ekprasert, J., Theerakulpisut, P., Sanitchon, J., et al. (2023). The potential of endophytic fungi for enhancing the growth and accumulation of phenolic compounds and anthocyanin in maled phai rice (Oryza sativa L.). J. Fungi 9:937. doi: 10.3390/jof9090937
Gouvinhas, I., Martins-Lopes, P., Carvalho, T., Barros, A., and Gomes, S. (2019). Impact of Colletotrichum acutatum pathogen on olive phenylpropanoid metabolism. Agriculture 9:173. doi: 10.3390/agriculture9080173
Guo, J. J., Ning, X., and Zhuang,. (2023). Effect of solid-state fermentation by the endophytic bacterium Ilyonectria cyclaminicola on the efficacy components of Epimedium medicinal dregs. Shi Zhen Guo Yi Guo Yao 34, 428–431. doi: 10.3969/j.issn.1008-0805.2023.02.50
Han, D., Wang, K., Long, F., Zhang, W. B., Yao, X., and Chen, S. H. (2022). Effects of endophytic fungi on the secondary metabolites of Hordeum bogdanii under alkaline stress. AMB Express 12:73. doi: 10.1186/s13568-022-01414-w
Harrison, J. G., Beltran, L. P., Buerkle, C. A., Cook, D., Gardner, D. R., Parchman, T. L., et al. (2021). A suite of rare microbes interacts with a dominant, heritable, fungal endophyte to influence plant trait expression. ISME J. 15, 2763–2778. doi: 10.1038/s41396-021-00964-4
Hou, Q., Chen, D., Wang, Y.-P., Ehmet, N., Ma, J., and Sun, K. (2022). Analysis of endophyte diversity of Gentiana officinalis among different tissue types and ages and their association with four medicinal secondary metabolites. Peer J 10:e13949. doi: 10.7717/peerj.13949
Jo, J., Oh, J., and Park, C. (2020). Microbial community analysis using high-throughput sequencing technology: a beginner’s guide for microbiologists. J. Microbiol. 58, 176–192. doi: 10.1007/s12275-020-9525-5
Johny, L., Cahill, D. M., and Adholeya, A. (2021). AMF enhance secondary metabolite production in ashwagandha, licorice, and marigold in a fungi-host specific manner. Rhizosphere 17:100314. doi: 10.1016/j.rhisph.2021.100314
Ju, M. X., Zhang, Q. C., Wang, R. T., Yan, S. Y., Li, Z. N., Li, P., et al. (2022). Correlation in endophytic fungi community diversity and bioactive compounds of Sophora alopecuroides. Front. Microbiol. 13:955647. doi: 10.3389/fmicb.2022.955647
Kandar, M., Suhandono, S., and Aryantha, I. N. P. (2018). Growth Promotion of Rice Plant by Endophytic Fungi. J. Pure. Appl. Microbiol. 12, 1569–1577. doi: 10.22207/JPAM.12.3.62
Khan, A. L., Gilani, S. A., Waqas, M., Al-Hosni, K., Al-Khiziri, S., Kim, Y.-H., et al. (2017). Endophytes from medicinal plants and their potential for producing indole acetic acid, improving seed germination and mitigating oxidative stress. J Zhejiang Univ Sci B 18, 125–137. doi: 10.1631/jzus.B1500271
Kim, J. W., and Shim, S. H. (2019). The fungus Colletotrichum as a source for bioactive secondary metabolites. Arch. Pharm. Res. 42, 735–753. doi: 10.1007/s12272-019-01142-z
Kreft, I., Golob, A., Vombergar, B., and Germ, M. (2023). Tartary buckwheat grain as a source of bioactive compounds in husked groats. Plants (Basel) 12:1122. doi: 10.3390/plants12051122
Kumar, A., and Verma, J. P. (2018). Does plant—Microbe interaction confer stress tolerance in plants:A review? Microbiol. Res. 207, 41–52. doi: 10.1016/j.micres.2017.11.004
Lahlali, R., Ibrahim, D. S. S., Belabess, Z., Roni, M. Z. K., Radouane, N., Vicente, C. S. L., et al. (2021). High-throughput molecular technologies for unraveling the mystery of soil microbial community: challenges and future prospects. Heliyon 7:e08142. doi: 10.1016/j.heliyon.2021.e08142
Li, N., Xu, D., Huang, R. H., Zheng, J. Y., Liu, Y. Y., Hu, B. S., et al. (2022). A New Source of Diterpene Lactones From Andrographis paniculata (Burm. f.) Nees—Two Endophytic Fungi of Colletotrichum sp. With Antibacterial and Antioxidant Activities. Front. Microbiol. 13:819770. doi: 10.3389/fmicb.2022.819770
Li, Q., Zhao, J. L., and Xiang, D. B. (2022). Buckwheat industry integration model research and study. Sichuan Agricult. Sci. Technol. 6, 10–14. doi: 10.3969/j.issn.1004-1028.2022.06.005
Liu, W., Feng, Y., Yu, S., Fan, Z., Li, X., Li, J., et al. (2021). The flavonoid biosynthesis network in plants. Int. J. Mol. Sci. 22:12824. doi: 10.3390/ijms222312824
Long, N., Liu, J., Liao, X., Jia, B., Liu, J., Zhou, L., et al. (2021). Fungal communities in Nelumbinis semen characterized by high-throughput sequencing. Int. J. Food Microbiol. 359:109428. doi: 10.1016/j.ijfoodmicro.2021.109428
Lu, J., Wang, J., Zhang, J., Zhu, Y., Qin, L., and Zhu, B. (2023). Diversity of culturable endophytic fungi in crocus sativus and their correlation with crocin content. Curr. Microbiol. 80:73. doi: 10.1007/s00284-023-03177-4
Lubna, K. M. A., Asaf, S., Jan, R., Waqas, M., Kim, K. M., and Lee, I. J. (2022). Endophytic fungus Bipolaris sp. CSL-1 induces salt tolerance in Glycine max. L via modulating its endogenous hormones, antioxidative system and gene expression. J. Plant Interact. 17, 319–332. doi: 10.1080/17429145.2022.2036836
Ma, X., Wang, F., Hang, T., and Dramou, P. (2022). Degradation study of rutin as template from magnetic composite molecularly imprinted polymer supernatant samples by liquid chromatography-mass spectrometry. J. Chromatogr. A 1673:463199. doi: 10.1016/j.chroma.2022.463199
Manzar, N., Kashyap, A. S., Maurya, A., Rajawat, M. V. S., Sharma, P. K., Srivastava, A. K., et al. (2022). Multi-gene phylogenetic approach for identification and diversity analysis of bipolaris maydis and curvularia lunata isolates causing foliar blight of Zea mays. J. Fungi (Basel) 8:802. doi: 10.3390/jof8080802
Martin, M. (2011). Cutadapt removes adapter sequences from high-through-put sequencing reads. Embnet J. 17, 10–12. doi: 10.14806/ej.17.1.200
Melouk, S. A. M., Hassan, M. A., Elwan, M. W. M., El-Seifi, S. K., Habib, E. S., and Yousef, E. A. A. (2023). Horticultural, chemical and genetic diversity using SSR markers in Leek germplasm collection. Sci. Hortic. 311:111782. doi: 10.1016/j.scienta.2022.111782
Meng, Y., Wang, J., Bai, B., Wang, L., Yao, L., Ma, Z., et al. (2020). Genome sequence resource for pathogen Bipolaris sorokiniana shoemaker GN1 causing spot blotch of barley (Hordeum vulgare L.). Plant Dis. 104, 1574–1577. doi: 10.1094/PDIS-12-19-2582-A
Mishra, S., Sahu, P. K., Agarwal, V., and Singh, N. (2021). Exploiting endophytic microbes as micro-factories for plant secondary metabolite production. Appl. Microbiol. Biotechnol. 105, 6579–6596. doi: 10.1007/s00253-021-11527-0
Muthukrishnan, S., Prakathi, P., Sivakumar, T., Thiruvengadam, M., Jayaprakash, B., Baskar, V., et al. (2022). Bioactive components and health potential of endophytic micro-fungal diversity in medicinal plants. Antibiotics (Basel) 11:1533. doi: 10.3390/antibiotics11111533
Pedras, M. S. C., Yaya, E. E., and Glawischnig, E. (2011). The phytoalexins from cultivated and wild crucifers: Chemistry and biology. Nat. Prod. Rep. 28, 1381–1405. doi: 10.1039/c1np00020a
Pereira, W. V., Padilha, A. C. N., Kaiser, J. A. O., Nesi, C. N., Fischer, J. M. M., and May-De-Mio, L. L. (2019). Monilinia spp. from imported stone fruits may represent a risk to Brazilian fruit production. Trop. Plant Pathol. 44, 120–131. doi: 10.1007/s40858-018-0243-z
Popp, C., Wamhoff, D., Winkelmann, T., Maiss, E., and Grunewaldt-Stöcker, G. (2020). Molecular identification of Nectriaceae in infections of apple replant disease affected roots collected by Harris Uni-Core punching or laser microdissection. J. Plant Dis. Prot. 127, 571–582. doi: 10.1007/s41348-020-00333-x
Poudel, M., Mendes, R., Costa, L. A. S., Bueno, C. G., Meng, Y., Folimonova, S. Y., et al. (2021). The role of plant-associated bacteria, fungi, and viruses in drought stress mitigation. Front. Microbiol. 12:743512. doi: 10.3389/fmicb.2021.743512
Qu, J. J. (2021). Study on the supply and demand of miscellaneous grains in China. Master Chinese Academy of Agricultural Sciences (CASA). doi: 10.27630/d.cnki.gznky.2021.000493
Rouydel, Z., Barin, M., Rasouli-Sadaghiani, M. H., Khezri, M., Vetukuri, R. R., and Kushwaha, S. (2021). Harnessing the potential of symbiotic endophytic fungi and plant growth-promoting rhizobacteria to enhance soil quality in saline soils. PRO 9:1810. doi: 10.3390/pr9101810
Roy, S., Nuckles, E., and Archbold, D. D. (2018). Effects of Phenolic Compounds on growth of Colletotrichum spp. in vitro. Curr. Microbiol. 75, 550–556. doi: 10.1007/s00284-017-1415-7
Rubio-Melgarejo, A., Balois-Morales, R., Palomino-Hermosillo, Y. A., López-Guzmán, G. G., Ramírez-Ramírez, J. C., Cervantes-García, E., et al. (2020). Phytochemical and antioxidant dynamics of the soursop fruit (Annona muricata L.) in response to Colletotrichum spp. J. Food Qual. :3180634. doi: 10.1155/2020/3180634
Sahu, R., Sharaff, M., Pradhan, M., Sethi, A., Bandyopadhyay, T., Mishra, V. K., et al. (2016). Elucidation of defense-related signaling responses to spot blotch infection in bread wheat (Triticum aestivum L.). Plant J. 86, 35–49. doi: 10.1111/tpj.13149
Segata, N., Izard, J., Waldron, L., Gevers, D., Miropolsky, L., Garrett, W. S., et al. (2011). Metagenomic biomarker discovery and explanation. Genome Biol. 12:R60. doi: 10.1186/gb-2011-12-6-r60
Shen, Z., Liu, X., Yang, J., Wang, Y., Yao, K., Huo, Q., et al. (2022). The temporal and spatial endophytic fungal community of Huperzia serrata: diversity and relevance to huperzine A production by the host. BMC Microbiol. 22:281. doi: 10.1186/s12866-022-02702-y
Sun, R.-T., Feng, X.-C., Zhang, Z.-Z., Zhou, N., Feng, H.-D., Liu, Y.-M., et al. (2022). Root endophytic fungi regulate changes in sugar and medicinal Compositions of Polygonum cuspidatum. Front. Plant Sci. 13:818909. doi: 10.3389/fpls.2022.818909
Sun, M., Su, T., Zhang, S.-B., Li, S.-F., Anberree-Lebreton, J., and Zhou, Z.-K. (2016). Variations in leaf morphological traits of Quercus guyavifolia (Fagaceae) were mainly influenced by water and ultraviolet irradiation at high elevations on the qinghai-tibet plateau. China. Int. J. Agric. Biol. 18, 266–273. doi: 10.17957/IJAB/15.0074
Takahiro, N., Koji, I., Tatsuro, S., and Toshikazu, M. (2023). Tartary buckwheat bran: a review of its chemical composition, processing methods and food uses. Plants (Basel) 12:1965. doi: 10.3390/plants12101965
Tugizimana, F., Djami-Tchatchou, A. T., Steenkamp, P. A., Piater, L. A., and Dubery, L. A. L. (2019). Metabolomic analysis of defense-related reprogramming in Sorghum bicolor in response to colletotrichum sublineolum infection reveals a functional metabolic web of phenylpropanoid and flavonoid pathways. Front. Plant Sci. 9:01840. doi: 10.3389/fpls.2018.01840
Večeřová, K., Klem, K., Veselá, B., Holub, P., Grace, J., and Urban, O. (2021). Combined effect of altitude, season and light on the accumulation of extractable terpenes in norway spruce needles. Forests 12:1737. doi: 10.3390/f12121737
Waller, F., Achatz, B., Baltruschat, H., Fodor, J., Becker, K., Fischer, M., et al. (2005). The endophytic fungus Piriformospora indica reprograms barley to salt-stress tolerance, disease resistance, and higher yield. Proc. Natl. Acad. Sci. USA 102, 13386–13391. doi: 10.1073/pnas.0504423102
Wang, B., Chen, C., Xiao, Y., Chen, K., Wang, J., and Zhou, G. (2023). Temperature thresholds drive the biogeographic pattern of root endophytic fungal diversity in the Qinghai-Tibet Plateau. Sci. Total Environ. 889:164270. doi: 10.1016/j.scitotenv.2023.164270
Wang, X., Tian, Z., Xi, Y., and Guo, Y. (2022). Identification of endophytic fungi with ACC deaminase-producing isolated from halophyte Kosteletzkya. Plant Signal. Behav. 17:2152224. doi: 10.1080/15592324.2022.2152224
Wanke, A., Van Boerdonk, S., Mahdi, L. K., Wawra, S., Neidert, M., Chandrasekar, B., et al. (2023). A GH81-type β-glucan-binding protein facilitates colonization by mutualistic fungi in barley. BioRxiv 2023:536646. doi: 10.1101/2023.04.12.536646
Wei, L. Q., Cheong, I. H., Yang, G. H., Li, X. G., Kozlakidis, Z., Ding, L., et al. (2021). The application of high-throughput technologies for the study of microbiome and cancer. Front. Genet. 12:699793. doi: 10.3389/fgene.2021.699793
Weng, W., Lu, X., Zhou, M., Gao, A., Yao, X., Tang, Y., et al. (2022). FtbZIP12 positively regulates responses to osmotic stress in tartary buckwheat. Int. J. Mol. Sci. 23:13072. doi: 10.3390/ijms232113072
Willie Anderson, V., Veloso, J. S., Silva, A. C., Nunes, A. D. S., Doyle, V. P., Castlebury, L. A., et al. (2022). Elucidating the Colletotrichum spp. diversity responsible for papaya anthracnose in Brazil. Fungal Biol. 126, 623–630. doi: 10.1016/j.funbio.2022.08.001
Yan, W. J., Duan, H., Lv, Y. N., and Wang, J. (2021). Buckwheat in China’s health food application progress. Food Sci. Technol. 46, 55–61. doi: 10.13684/j.cnki.spkj.2021.06.010
Yan, L., Zhu, J., Zhao, X., Shi, J., Jiang, C., and Shao, D. (2019). Beneficial effects of endophytic fungi colonization on plants. Appl. Microbiol. Biotechnol. 103, 3327–3340. doi: 10.1007/s00253-019-09713-2
Yang, K., He, S. H., Ye, C., Zhang, S., Chen, Z. H., Li, Y., et al. (2022). First report of leaf spot on Panax notoginseng caused by Caryophylloseptoria pseudolychnidis in Yunnan. China. Plant Dis. 106:2525. doi: 10.1094/PDIS-12-21-2817-PDN
Yang, M., Li, Q., Chen, C., Huang, W., and Liu, C. (2019). The complete mitochondrial genome of Ilyonectria sp. (Hypocreales: Hypocreomycetidae). Mitochondrial DNA B Resour. 4, 2385–2386. doi: 10.1080/23802359.2018.1536490
Yokoya, K., Postel, S., Fang, R., and Sarasan, V. (2017). Endophytic fungal diversity of Fragaria vesca, a crop wild relative of strawberry, along environmental gradients within a small geographical area. PeerJ 5:e2860. doi: 10.7717/peerj.2860
You, J. M., and Cao, X. Z. (2013). Determination of the distribution level of tea polyphenols in tea plants by forintol method. Hubei Agricult. Sci. 52, 2417–2419. doi: 10.14088/j.cnki.issn0439-8114.2013.10.050
Zaccaron, A. Z., and Bluhm, B. H. (2017). The genome sequence of Bipolaris cookei reveals mechanisms of pathogenesis underlying target leaf spot of sorghum. Sci. Rep. 7:17217. doi: 10.1038/s41598-017-17476-x
Zahedi, S. M., and Sarikhani, H. (2016). Effect of far-red light, temperature, and plant age on morphological changes and induction of flowering of a ‘June-bearing’ strawberry. Hortic. Environ. Biotechnol. 57, 340–347. doi: 10.1007/s13580-016-0018-8
Zaura, E., Keijser, B. J. F., Huse, S. M., and Crielaard, W. (2009). Defining the healthy “core microbiome” of oral microbial communities. BMC Microbiol. 9:259. doi: 10.1186/1471-2180-9-259
Zhang, X., Peng, X., Yang, G., Chen, Q., and Jin, D. (2022). The colonization and effect of Isaria Cateinannulata on buckwheat sprouts. Plan. Theory 12:145. doi: 10.3390/plants12010145
Zhao, X., Sun, X.-F., Zhao, L.-L., Huang, L.-J., and Wang, P.-C. (2022). Morphological, transcriptomic and metabolomic analyses of Sophora davidii mutants for plant height. BMC Plant Biol. 22:144. doi: 10.1186/s12870-022-03503-1
Zhao, C., Zhao, X., Zhang, J., Zou, W., Zhang, Y., Li, L., et al. (2016). Screening of Bacillus strains from sun vinegar for efficient production of flavonoid and phenol. Indian J. Microbiol. 56, 498–503. doi: 10.1007/s12088-016-0602-8
Zhou, Q., Tang, J., Liu, C., Huang, K., and Huang, X. (2023). Effects of phosphate fertilizer application on the growth and yield of Tartary Buckwheat under low-nitrogen condition. Agronomy 13:1886. doi: 10.3390/agronomy13071886
Zhu, F. (2016). Chemical composition and health effects of Tartary buckwheat. Food Chem. 203, 231–245. doi: 10.1016/j.foodchem.2016.02.050
Zhu, B., Wang, S., Mi, C.-Y., Yang, R.-H., Zen, G.-H., and Hu, X.-F. (2019). Genome sequence resource for Ilyonectria mors-panacis, causing rusty root rot of Panax notoginseng. Mol. Plant-Microbe Interact. 32, 1468–1471. doi: 10.1094/MPMI-05-19-0118-A
Zou, L., Wu, D., Ren, G., Hu, Y., Peng, L., Zhao, J., et al. (2021). Bioactive compounds, health benefits, and industrial applications of Tartary buckwheat (Fagopyrum tataricum). Crit. Rev. Food Sci. Nutr. 63, 657–673. doi: 10.1080/10408398.2021.1952161
Zuo, Y.-L., Hu, Q.-N., Qin, L., Liu, J.-Q., and He, X.-L. (2022). Species identity and combinations differ in their overall benefits to Astragalus adsurgens plants inoculated with single or multiple endophytic fungi under drought conditions. Front. Plant Sci. 13:933738. doi: 10.3389/fpls.2022.933738
Keywords: Tartary buckwheat, endophytic fungi, high-throughput sequencing, flavonoids, phenotypic traits
Citation: Chen M, Ding Z, Zhou M, Shang Y, Li C, Li Q, Bu T, Tang Z and Chen H (2024) The diversity of endophytic fungi in Tartary buckwheat (Fagopyrum tataricum) and its correlation with flavonoids and phenotypic traits. Front. Microbiol. 15:1360988. doi: 10.3389/fmicb.2024.1360988
Edited by:
Francisca Suárez-Estrella, University of Almeria, SpainReviewed by:
Juan Hua, Shenyang Agricultural University, ChinaSudeep Tiwari, University of Nebraska-Lincoln, United States
Sophon Boonlue, Khon Kaen University, Thailand
Copyright © 2024 Chen, Ding, Zhou, Shang, Li, Li, Bu, Tang and Chen. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Hui Chen, Y2hlbmh1aUBzaWNhdS5lZHUuY24=
†These authors have contributed equally to this work
 Meiqi Chen†
Meiqi Chen† Hui Chen
Hui Chen