- Department of Molecular Biology, Graduate School of Pharmaceutical Sciences, Kyushu University, Fukuoka, Japan
Timely initiation of chromosomal DNA replication in Escherichia coli is achieved by cell cycle-coordinated regulation of the replication origin, oriC, and the replication initiator, ATP-DnaA. Cellular levels of ATP-DnaA increase and peak at the time for initiation at oriC, after which hydrolysis of DnaA-bound ATP causes those to fall, yielding initiation-inactive ADP-DnaA. This hydrolysis is facilitated by the chromosomal locus datA located downstream of the tRNA-Gly (glyV-X-Y) operon, which possesses a cluster of DnaA-binding sequences and a single binding site (IBS) for the DNA bending protein IHF (integration host factor). While IHF binding activates the datA function and is regulated to occur specifically at post-initiation time, the underlying regulatory mechanisms remain obscure. Here, we demonstrate that datA-IHF binding at pre-initiation time is down-regulated depending on the read-through transcription of datA IBS initiated at the glyV-X-Y promoter. During the cell cycle, the level of read-through transcription, but not promoter activity, fluctuated in a manner inversely related to datA-IHF binding. Transcription from the glyV-X-Y promoter was predominantly interrupted at datA IBS by IHF binding. The terminator/attenuator sequence of the glyV-X-Y operon, as well as DnaA binding within datA overall, contributed to attenuation of transcription upstream of datA IBS, preserving the timely fluctuation of read-through transcription. These findings provide a mechanistic insight of tRNA transcription-dependent datA-IHF regulation, in which an unidentified factor is additionally required for the timely datA-IHF dissociation, and support the significance of datA for controlling the cell cycle progression as a connecting hub of tRNA production and replication initiation.
Introduction
The E. coli chromosome is a circular double-strand 4.6 Mb DNA molecule that forms a compact structure, the nucleoid, condensed by binding of various nucleoid-associated proteins (NAPs) and a DNA superstructure, such as supercoiled DNA (Dillon and Dorman, 2010; Hołówka and Zakrzewska-Czerwińska, 2020; Molan and Žgur Bertok, 2022). The E. coli chromosome has a single replication origin, oriC (located at 84.6 min of the genome map), and initiation of replication at oriC is regulated precisely during the cell cycle so that, even in rapidly growing cells, initiation occurs simultaneously at multiple sister oriCs (Frimodt-Møller et al., 2017; Hansen and Atlung, 2018; Grimwade and Leonard, 2021; Kasho et al., 2023). In E. coli, the initiation reactions of DNA replication occur in a higher-order complex that includes oriC, the initiator protein DnaA, and a NAP IHF (integration host factor). The oriC region comprises a duplex-unwinding element (DUE) and a flanking DnaA oligomerization region (DOR), which contains a cluster of DnaA-binding sites (DnaA boxes). ATP-bound DnaA, but not ADP-bound DnaA, forms specific oligomers with the DOR that promote DUE unwinding in concert with IHF or its homologue HU (Cassler et al., 1995; Shimizu et al., 2016; Sakiyama et al., 2017; Kasho et al., 2021; Yoshida et al., 2023). IHF specifically binds to a consensus sequence (TAAnnnnTTGATW, where W is A or T, and n is any nucleotides) at a DUE-proximal site within the DOR and induces sharp DNA bending (120°–180°) (Figure 1; Aeling et al., 2006). This promotes specific binding of a DOR-bound DnaA oligomer to the single-stranded (ss) DUE, thus stabilizing the unwound state of DUE. The stably unwound DUE causes subsequent loading of the replicative DNA helicase, DnaB, onto the ssDUE (Hayashi et al., 2020), thereby initiating DNA synthesis by the DnaG primase and the DNA polymerase III holoenzyme.
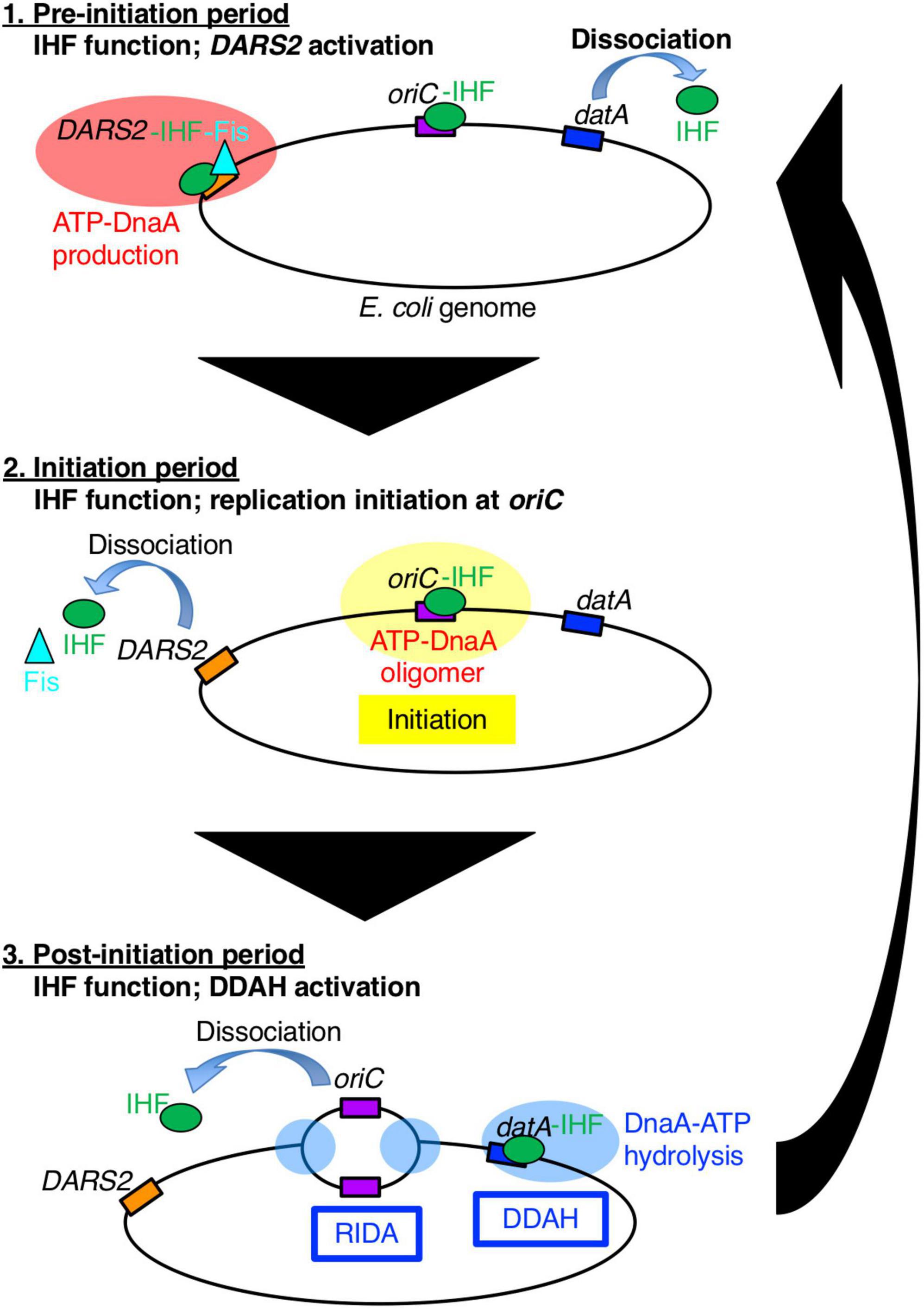
Figure 1. A schematic view of the timely binding/dissociation of IHF and Fis at oriC, datA, and DARS2 during the cell cycle. At the pre-initiation stage, during which the ADP-DnaA level is high, the DARS2–IHF–Fis complex increases the level of ATP-DnaA. Concurrently, IHF dissociates from datA. Next, at the initiation stage, the elevated ATP-DnaA level results in initiation of replication at the IHF-bound oriC and Fis dissociation from DARS2. Concurrently, IHF dissociates from DARS2 via an unknown mechanism. After initiation, IHF temporarily binds to datA and activates DDAH to inactivate DnaA. For simplicity, IHF and Fis proteins are drawn as a green circle and a light blue triangle, respectively, and DnaA protein is omitted.
E. coli DnaA is composed of four distinct domains, whose basic features are conserved among other bacteria (Katayama et al., 2017). The N-terminal domain I is involved in interactions with DnaB and DiaA, a stimulatory factor for ATP-DnaA assembly on oriC (Sutton et al., 1998; Keyamura et al., 2009; Zawilak-Pawlik et al., 2017). Domain II is a flexible linker (Nozaki and Ogawa, 2008). Domain III contains AAA+ motifs involved in ATP/ADP binding (Kawakami et al., 2006), ATP hydrolysis (Nishida et al., 2002), and domain III–domain III interactions (Kawakami et al., 2005; Erzberger et al., 2006; Noguchi et al., 2015), in addition to a specific motif for ssDUE binding (Ozaki et al., 2008; Duderstadt et al., 2011; Yoshida et al., 2023). Domain IV binds specifically to the 9-mer DnaA box consensus sequence, TTAWnCACA (where W is A and T) (Fujikawa et al., 2003).
Integration host factor exists as a heterodimer of α- and β-subunits and is abundant in E. coli cells (approximately 5,000 molecules per cell at log phase and 10,000 molecules per cell at stationary phase) (Ali Azam et al., 1999). In addition to replication initiation, IHF regulates chromosome conformation and transcription of hundreds of genes by inducing DNA bending, or modulating DNA supercoiling (Prieto et al., 2012). To achieve precise regulation of replication initiation timing, IHF plays essential roles not only in the timely activation of oriC, but also in the timely regulation of DnaA activity via the chromosomal loci DARS2 and datA (Figure 1; Kasho and Katayama, 2013; Kasho et al., 2014). DARS2 catalytically stimulate the release of ADP from DnaA to produce ATP-DnaA (Fujimitsu et al., 2009; Sugiyama et al., 2019). DARS2 contains specific binding sites for IHF and another NAP, Fis (factor for inversion stimulation), and is activated during the cell cycle in a manner dependent on the temporal binding of IHF and Fis (Kasho et al., 2014). A recent study revealed that DARS2 is regulated by a negative feedback mechanism (Figure 1; Miyoshi et al., 2021): at the pre-initiation stage, the increased ATP-DnaA molecules form oligomers at the Fis binding site, competitively dissociate Fis. This simple negative feedback regulation of DARS2 is fundamental for the timely repression of DARS2 activity; however, it is currently unclear how IHF binding/dissociation at the DARS2 locus is regulated during the cell cycle.
The main system for inactivation of DnaA is called RIDA (regulatory inactivation of DnaA), in which DnaA-bound ATP is catalytically hydrolyzed by the complex of the AAA+ Hda protein and the DNA-loaded form of the clamp (β subunit) of the DNA polymerase III holoenzyme in a manner coupled with DNA replication (Katayama et al., 2010, 2017). However, for strict regulation that represses untimely initiation, the specific chromosomal locus datA (located at 94.7 min) is required: datA efficiently inactivates ATP-DnaA by DnaA-ATP hydrolysis in a manner dependent on IHF and independent of RIDA. This regulatory system is termed DDAH (datA-dependent DnaA-ATP hydrolysis) (Figure 1; Kasho and Katayama, 2013). Deletion of datA causes untimely initiations in growing cells (Nozaki et al., 2009; Kasho et al., 2017). The minimal datA region contains a single IBS and four DnaA boxes, of which three (DnaA boxes 2, 3, and 7) are essential and one (DnaA box 4) is stimulatory for datA function (Figure 2A; Kasho et al., 2017). The datA-IHF complex interacts with ATP-DnaA, catalytically promoting the hydrolysis of DnaA-bound ATP. Timely regulation of DDAH depends on the temporal binding of IHF to datA (Kasho and Katayama, 2013). IHF is dissociated from datA ahead of initiation and binds to datA only after initiation, ensuring the timely activation of DDAH. However, the mechanism for ensuring the timing of datA-IHF binding/dissociation remains unknown.
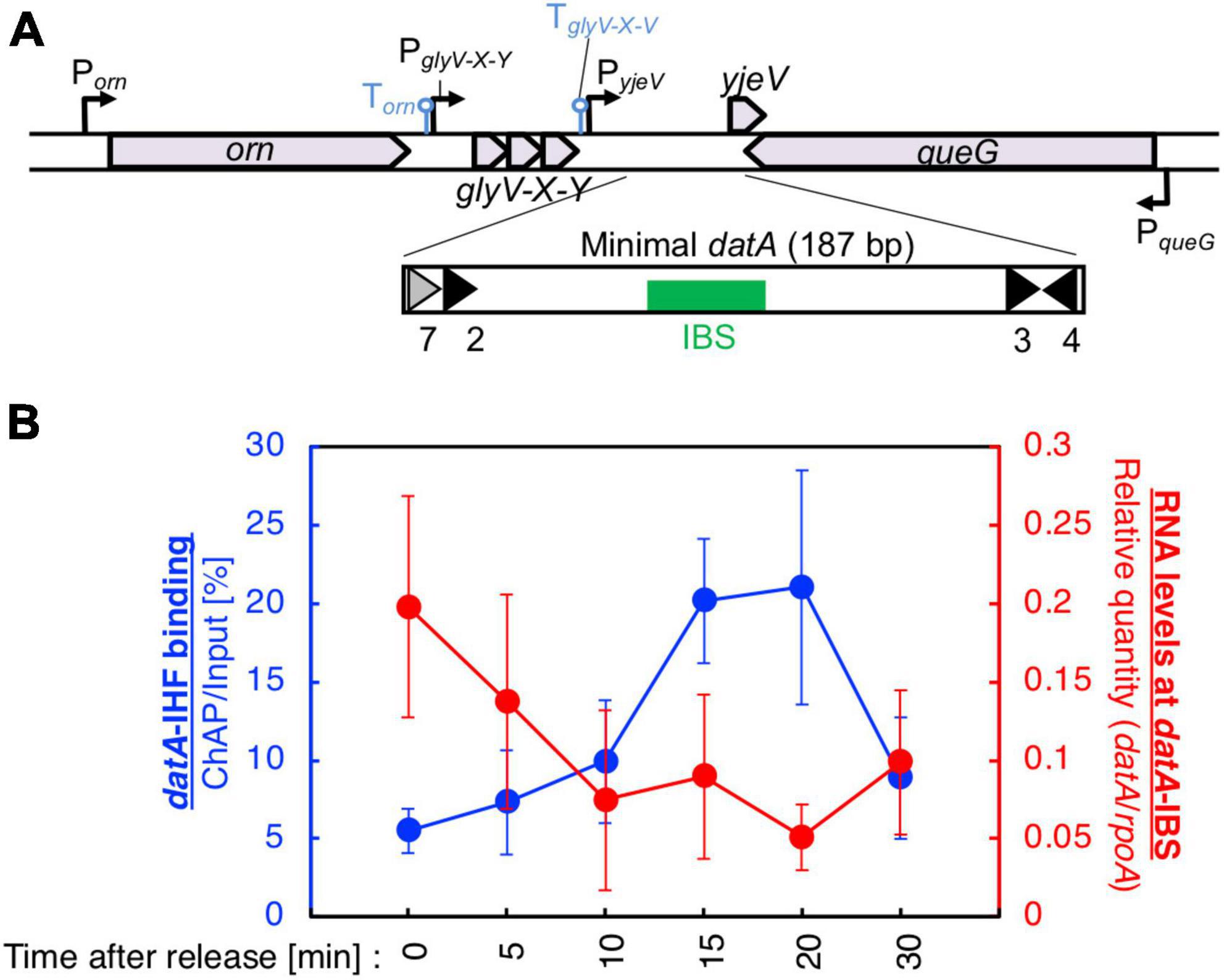
Figure 2. Cell cycle-dependent oscillation of datA-IHF binding and datA transcription level. (A) Functional structures of datA and surrounding regions are shown. The location and direction of transcriptional promoters and genes around the chromosomal datA and DARS2 loci are indicated by thin arrows and pentagons. DnaA boxes 3–4 and 7 within the minimal datA are indicated by gray triangles (low-affinity boxes with 2–3 mismatches) and black triangles (high affinity boxes with 0–1 mismatch). IBS is indicated by a green box. (B) Cell cycle-coordinated correlation between datA-IHF binding (blue line) and datA transcription (red line). datA-IHF binding was analyzed by ChAP-qPCR. SH022 (dnaC2 ihfA-cHis12) cells growing in LB medium at 30°C were transferred to 37°C and incubated for 80 min. The cells were then transferred to 30°C (Time 0) and further incubated for 5–30 min at 30°C. The relative levels of datA before and after Ni-affinity purification were determined using real-time qPCR, and yield was calculated (expressed as %). Cell cycle-coordinated oscillation of the RNA level of datA IBS was analyzed by RT-qPCR. SH022 cells were cultured as described above before the total RNA was extracted. RNA levels of datA IBS relative to those of the rpoA gene were determined using RT-qPCR. Data represent means (± S.D.) of at least four independent experiments. Based on the Student’s t-test, the 0 min vs. 20 min difference (p < 0.02) of RT-qPCR is statistically significant, and 0 min vs. 15 min (p < 0.01), 0 min vs. 20 min (p < 0.05), and 15 min vs. 30 min (p < 0.05) differences of IHF ChAP are statistically significant.
Previous studies have presented three clues relating to regulatory mechanisms for datA function. (1) Chromosomal positional effects of datA, i.e., when the locus of datA is moved to a position distant from oriC, datA function is moderately inhibited, resulting in precocious initiations (Kitagawa et al., 1998; Frimodt-Møller et al., 2015). The mechanisms responsible for this effect remain to be investigated. Local structure of the chromosome, transcriptional activities surrounding datA etc. in addition to distance itself from oriC are suggested to possibly influence datA function as described below. (2) DNA supercoiling-dependent stimulation of DDAH, i.e., negative DNA supercoiling stabilizes datA-IHF binding in vitro and stimulates DDAH activity (Kasho et al., 2017). In support of this hypothesis, inhibition of negative DNA supercoiling by the DNA gyrase inhibitor novobiocin decreases the ability of datA to repress replication initiation in vivo. IHF preferentially binds to curved DNA rather than to straight-shaped DNA (Watson et al., 2022). (3) Transcription of datA, i.e., the minimal region of datA required for full DDAH activity is located at the intergenic region downstream of the tRNA-Gly (glyV-X-Y) operon and queG gene (Figure 2; Kitagawa et al., 1996; Kasho and Katayama, 2013), which enables a hypothesis that datA-IHF binding/dissociation is regulated by transcription of the datA locus. The transcriptional promoter of the putative yjeV gene (PyjeV) partly overlaps with datA DnaA box 7, which is essential for DDAH activity. Consistent with this hypothesis, the transcription inhibitor rifampicin inhibits timely binding and dissociation of datA-IHF complexes, and datA activity is inhibited by the translocation of datA to a highly transcribed position (Kasho and Katayama, 2013; Frimodt-Møller et al., 2015). However, it is currently unclear which mechanism is predominant for regulating the datA-IHF binding/dissociation.
In this study, we addressed the requirement of transcription of the datA region for timely regulation of IHF binding/dissociation. We found that datA-IHF binding is down-regulated depending on the read-through transcription of datA IBS initiated at the promoter of tRNA-Gly operon, which ensures timely datA-IHF binding. Cell cycle analyses suggest that the levels of read-through transcripts of datA IBS and datA-IHF binding were inversely correlated. We next addressed the mechanism of cell cycle-coordinated switching between read-through transcription and premature transcription termination. Multifaceted analysis using various mutants suggested that transcription initiation from the glyV-X-Y promoter is constant and premature transcription termination at the datA IBS region occurs in a temporal manner. Timely transcription termination predominantly requires IHF binding to datA IBS. Also, the terminator/attenuator sequence of glyV-X-Y operon and DnaA binding at the datA DnaA box 2 contribute to controlling the basal level of transcription termination. Based on these findings, we propose a model and significance of tRNA transcription-dependent regulation for timely datA-IHF binding/dissociation for ensuring precise timing of replication initiation.
Materials and methods
Bacterial strains and cultures
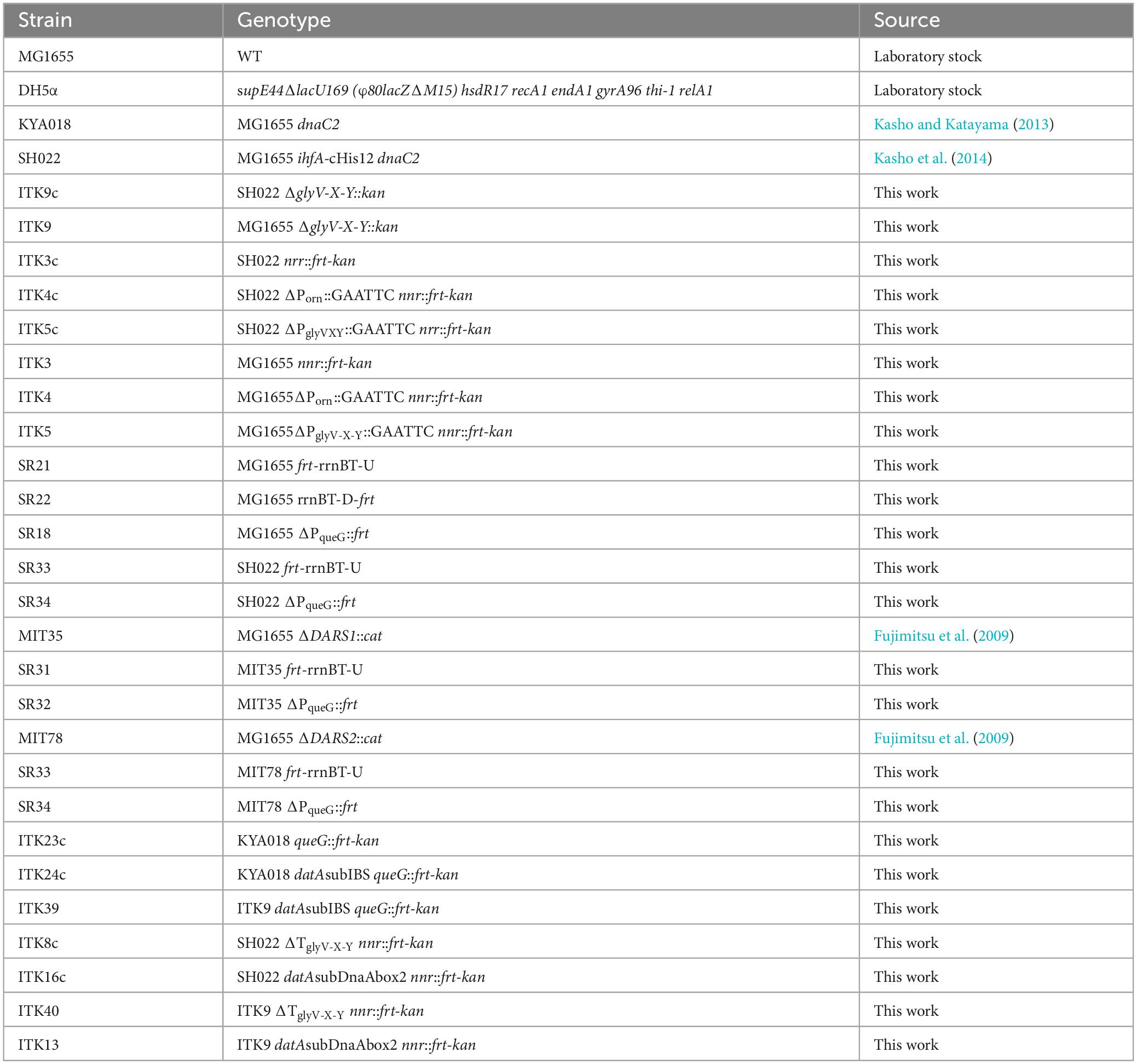
The E. coli strains used in this study are listed in Table 1. The λRED system was used to construct chromosomal mutations (Datsenko and Wanner, 2000). Briefly, PCR fragments including mutated gly-V-X-Y operon or datA and frt-kan gene was transformed into the strain bearing pKD46 (λ RED expression plasmid). To verify the presence of specific mutations in each strain constructed in this study, the chromosomal DNA regions of interest were PCR amplified and analyzed by Sanger sequencing. The detail of λRED system was described below. All bacterial strains were grown in LB medium or M9 medium supplemented with 0.2% casamino acids, 5 μg/mL thiamine, and 0.2% glucose.
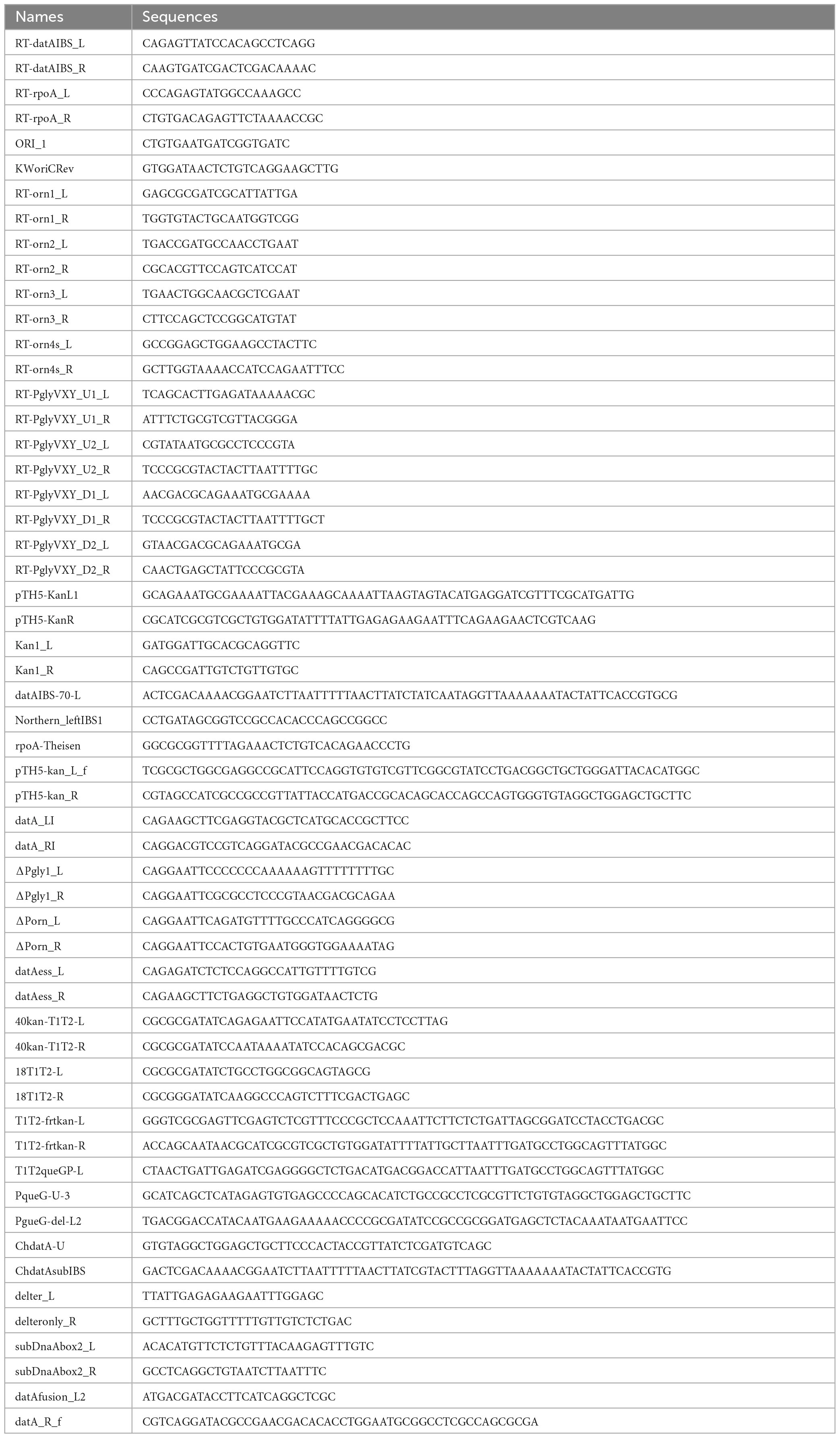
A datA fragment spanning Porn to PqueG was amplified by PCR from MG1655 genomic DNA and primers datA_LI and datA_RI (see Table 2 for primer sequences), digested with AatII and HindIII-HF, and inserted into the pACYC177 to yield pITK1. pITK1-derivatives for construction of genomic mutants were generated using inverse PCR with the following primers (Table 2): ΔPorn_L and ΔPorn_R for pITK2 (ΔPorn), ΔPgly1_L and ΔPgly1_R for pITK3 (ΔPglyV-X-Y), delter_L and delteronly_R for pITK12 (ΔTglyV-X-Y), and subDnaAbox2_L and subDnaAbox2_R for pITK15 (datAsubDnaAbox2). A glyV-X-Y fragment was amplified by PCR from MG1655 genomic DNA using primers datAess_L and datAess_R, digested with BglII and HindIII-HF, and inserted into the pKP1673 to yield pITK6. A frt-kan fragment was amplified by PCR using pTH5 and primers 40kan-T1T2-L and 40kan-T1T2-R, digested with EcoRI-HF, and ligated with a EcoRI-HF-digested vector fragment that was amplified by inverse PCR using pBAD18 and primers 18-T1T2-L and 18-T1T2-R to yield pSR14 carrying frt-kan instead of the bla gene.
For the construction of mutant strains in Figures 3, 5, 6, datA fragments carrying the desired mutation were amplified from mutant plasmids (pITK2 for ΔPorn, pITK3 for ΔPglyV-X-Y, pITK12 for ΔTglyV-X-Y and pITK15 for datA subDnaAbox2) with primers datAfusion_L2 and datA_R_f (Table 2). Using the overlap extension (OE)-PCR method (Hilgarth and Lanigan, 2020), these fragments were joined with a frt-kan fragment amplified from pTH5 using primers pTH5-kan_L_f and pTH5-kan_R. Fragments were transformed into MG1655 or SH022 cells bearing pKD46 by electroporation, and the frt-kan was removed by introduction of the pCP20 plasmid, yielding strains ITK4 (ΔPorn), ITK5 (ΔPglyV-X-Y), ITK8 (ΔTglyV-X-Y), ITK16 (datAsubDnaAbox2), ITK4c (dnaC2ΔPorn), ITK5c (dnaC2ΔPglyV-X-Y), ITK8c (dnaC2ΔTglyV-X-Y), and ITK16c (dnaC2 datAsubDnaAbox2). For generation of the ITK3 control strain, a frt-kan fragment amplified from the pTH5 plasmid with primers pTH5-kan_L_f and pTH5-kan_R was transformed into MG1655 cells bearing pKD46. For the construction of mutant strains in Figure 4, fragments carrying a transcription terminator sequence of the rrnB operon (rrnBT) were amplified from pSR14 using primers T1T2-frtkan-L and T1T2-frtkan-R for rrnBT-U, and T1T2-queGP-L and T1T2-frtkan-R for rrnBT-D, and were transformed into MG1655 or SH022 cells bearing pKD46 by electroporation. The frt-kan was removed by introduction of the pCP20 plasmid, yielding strains SR21 (rrnBT-U), SR22 (rrnBT-D), and SR33 (dnaC2 rrnBT-U). A fragment carrying frt-kan was amplified from pTH5 with primers PqueG-U-3 and PgueG-del-L2, and was transformed into MG1655 or SH022 cells bearing pKD46 by electroporation. The frt-kan was removed by introduction of the pCP20 plasmid, yielding strains SR18 (ΔPqueG) and SR34 (dnaC2 ΔPqueG). A fragment carrying the datAsubIBS mutation and frt-kan was amplified from pKX40kan (Kasho et al., 2017) with primers ChdatA-U and ChdatAsubIBS, and transformed into MG1655 or SH022 cells bearing pKD46 using electroporation. The frt-kan was removed by introduction of the pCP20 plasmid, yielding strains ITK24 (datAsubIBS) and ITK24c (dnaC2 datAsubIBS), respectively. For checking the introduction of specific mutations in each strains constructed in this study, the sequences of chromosomal datA locus were confirmed by DNA sequencing.
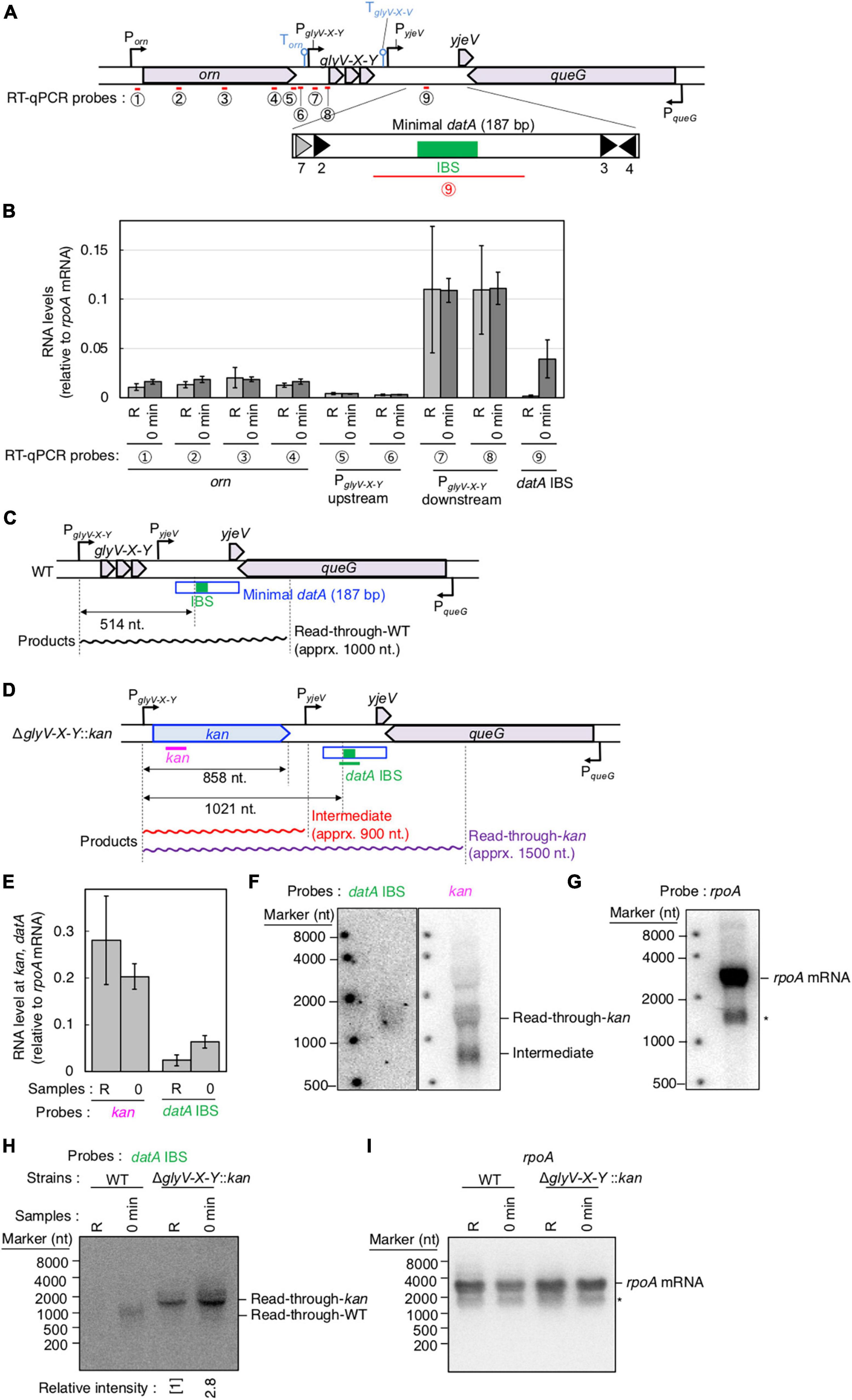
Figure 3. Determination of the transcriptional promoter for datA transcription. (A) Functional structures of datA and surrounding regions are shown as in Figure 2A. The amplified regions at orn gene, glyV-X-Y operon, and datA IBS locus are indicated by red bars: Each region is indicated by the encircled number. (B) SH022 cells growing in LB medium at 30°C (R; random) were transferred to 37°C and incubated for 80 min (0 min; synchronized), and then the total RNA was extracted. The RNA levels of each region relative to those of the rpoA gene were determined using RT-qPCR. Data represent means (± S.D.) of at least four independent experiments. The encircled numbers correspond to the regions shown in panel (A). Based on the Student’s t-test, the R vs. 0 min difference (p < 0.007) using datA IBS probe is statistically significant. The effect of the dnaC2-dependent cell cycle synchronization was confirmed by checking the decrease of mioC transcript (Supplementary Figure 2A). (C,D) Construction of the ΔglyV-X-Y::kan mutant. The wild-type (C) and mutant (D) structures are illustrated. The probes at kan gene and datA IBS locus for northern blotting are shown as pink lines. The read-through datA RNA or its intermediate observed in northern blotting are shown as wave lines. (E) ITK9c (dnaC2ΔglyV-X-Y::kan) cells growing in LB medium at 30°C (R; random) were transferred to 37°C and incubated for 80 min (0 min; synchronized). The total RNAs were extracted at each time. The RNA levels of kan gene and datA IBS locus as well as rpoA gene were determined using RT-qPCR, and calculated as the relative value compared with those of the rpoA gene. Data represent means (± S.D.) of three independent experiments. Based on the Student’s t-test, the R vs. 0 min difference (p < 0.01) using datA IBS probe is statistically significant. The effect of the dnaC2-dependent cell cycle synchronization was confirmed by checking the decrease of mioC transcript (Supplementary Figure 2B). (F,G) Northern blotting using total RNA prepared from ITK9 (ΔglyV-X-Y::kan) cells and the probes for datA IBS [(F); left], kan gene [(F); right], and rpoA gene (G). (H,I) Northern blotting using total RNA prepared from randomly-growing (R) or synchronized (0 min) dnaC2 strains SH022 (WT) or ITK9c (ΔglyV-X-Y::kan). The probes for datA IBS (H) and rpoA gene (I) were used. The asterisk means unidentified bands.
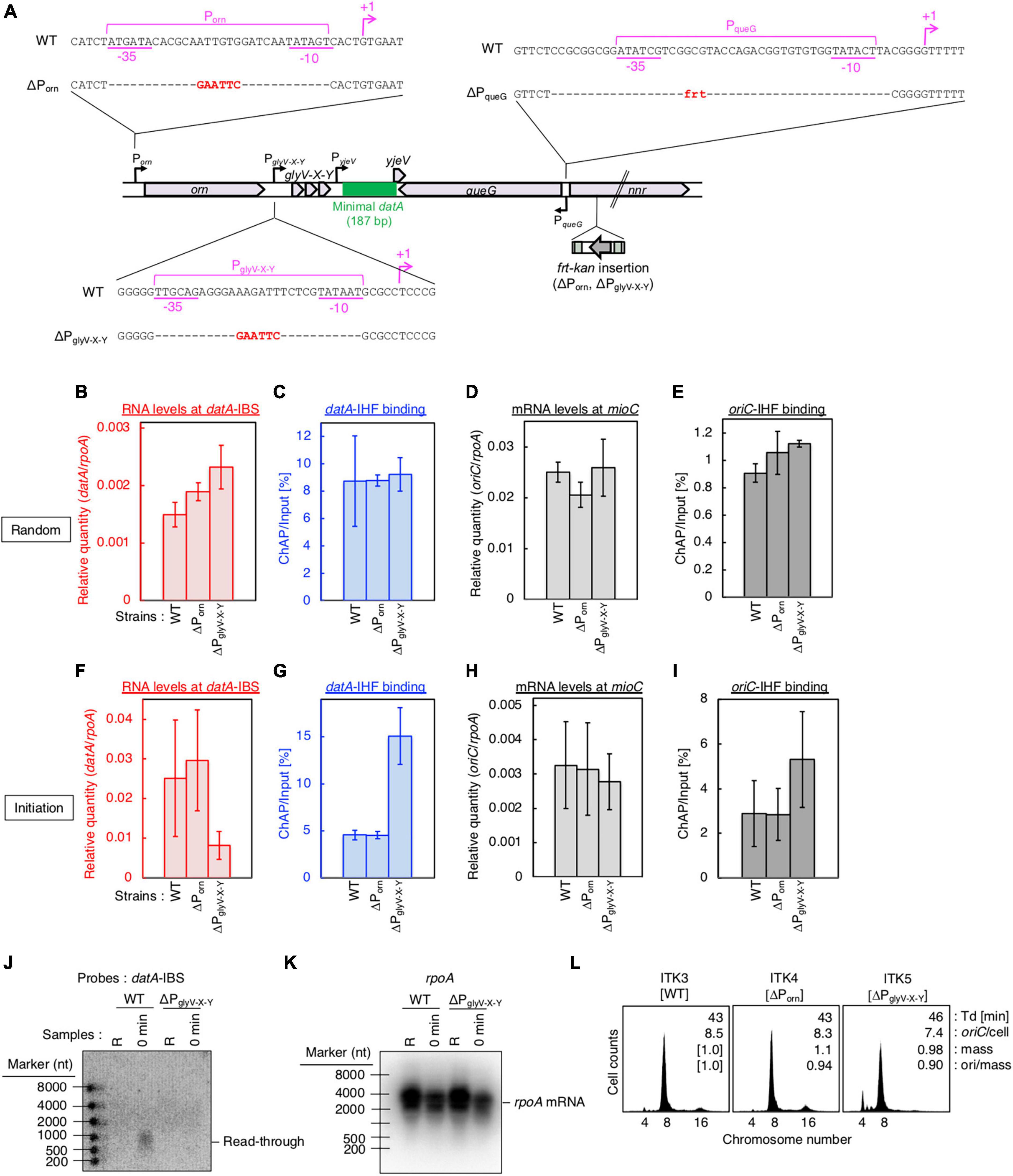
Figure 4. PglyV-X-Y is required for transcription-dependent datA regulation. (A) Construction of transcriptional promoter mutant cells (ΔPorn and ΔPglyV-X-Y harboring an frt-kan insertion at the nrr gene, and ΔPqueG::frt). (B–I) ITK3c (dnaC2) (WT), ITK4c (dnaC2ΔPorn) (ΔPorn), and ITK5c (dnaC2ΔPglyV-X-Y) (ΔPglyV-X-Y) cells growing in LB medium at 30°C [random culture; panels (B–E)] were transferred to 37°C and incubated for 80 min [synchronized at the initiation period; panels (F–I)]. The RNA levels of datA IBS (B,F) or the mioC gene (D,H) relative to those of the rpoA gene were determined using RT-qPCR. IHF binding at datA (C,G) and oriC (D,I) were determined by ChAP-qPCR, and yield was calculated (expressed as %). Data represent means (± S.D.) of at least two independent experiments. Based on the Student’s t-test, the WT vs. ΔPglyV-X-Y differences in panel (F) (p < 0.12) and 4G (p < 0.01) are statistically significant. (J,K) Northern blotting using total RNA prepared from randomly-growing (R) or synchronized (0 min) ITK3c (WT) or ITK5c (ΔPglyV-X-Y) cells, and the probes for datA IBS (J) and rpoA gene (K). (L) Flow cytometry analysis of ITK3 (WT), ITK4 (ΔPorn), and ITK5c (ΔPglyV-X-Y) cells. Cells were grown at 30°C in LB medium, followed by further incubation with rifampicin and cephalexin for run-out replication. DNA content was quantified using flow cytometry. Cell size (mass) at the time of drug addiction was measured using a Coulter counter. The oriC/cell, mean mass, and ori/mass ratio of each cell are indicated in each panel.
Chromatin affinity precipitation combined with quantitative PCR (ChAP-qPCR)
Chromatin affinity precipitation (ChAP, a modified version of ChIP assay) and qPCR experiments were performed according to a previously described method (Kasho et al., 2014, 2021; Inoue et al., 2016). In brief, SH022 (dnaC2 ihfA-cHis12) or its derivatives were grown in LB medium at 30°C, a permissive temperature, until A660 of 0.04 was reached, after which cells were incubated at 37°C, a restrictive temperature, for 80 min (“0 min” sample). Cells were then incubated at 30°C for 5 min to initiate replication (“5 min” sample), followed by further incubation at 38°C for 5–25 min to allow a single round replication to proceed (“10–30 min” samples). For preparing random culture (“R”) samples, SH022 cells or their derivatives were grown in LB medium at 30°C until the A660 reached 0.15. Samples were cross-linked with 3% (final) formaldehyde at room temperature for 5 min. The reactions were quenched by incubation in 125 mM glycine for an additional 5 min. Then, the cells were collected by centrifugation, washed twice with 1 mL of ice-cold TBS [50 mM Tris–HCl (pH7.5) and 500 mM NaCl], resuspended in 500 μL of binding buffer [50 mM Tris–HCl (pH 7.5), 500 mM NaCl, 1% (vol/vol) Triton X-100, 5 mM imidazole, and 0.1 mM PMSF], and sonicated six times for 20 s each. The resulting size of the chromosomal DNA was about 1 kb. Cell debris was then removed by centrifugation at 16,000 × g for 15 min at 4 °C, and a portion (400 μL) of the resulting supernatant (450 μL) was mixed with 10 μL of Dynabeads His-tag Isolation and Pulldown (lifetechnologies), followed by incubation at 4°C for 1 h with a gentle rotation. Beads and bound materials were washed four times with wash buffer [50 mM Tris–HCl (pH 7.5), 500 mM NaCl, 1% (vol/vol) Triton X-100, and 5 mM imidazole], resuspended in elution buffer [50 mM Tris–HCl (pH 7.5), 500 mM imidazole, 1% SDS, 10 mM EDTA, and 10 mM dithiothreitol], and incubated at 65°C for 12 h to allow de-crosslinking. DNA in the samples before (Input) and after (ChAP) pull down was purified using a Wizard SV Gel and PCR Clean-Up System (Promega). The levels of oriC, datA, and ylcC were quantified by real-time qPCR using TB Green Premix Ex TaqII (Tli RNaseH Plus) (TaKaRa) and primers ORI_1 and KWoriCRev for oriC; RT-datAIBS-L and RT-datAIBS-R for datA; and RTYLCC-L and RTYLCC-R for ylcC.
Total RNA purification
Total cellular RNA samples used in Figures 2B, 3B, 5, and Supplementary Figure 1 were purified using NucleoSpin™ RNA (Macherey-Nagel) according to the manufacturer’s protocol. Total cellular RNA samples used in Figures 3E–I, 4, 6, 7 were purified using the hot phenol method (Su’etsugu et al., 2003). In brief, KYA018 (dnaC2), SH022, or their derivative cells were grown in LB medium at 30°C until they reached A660 of 0.04, followed by further incubation at 37°C for 80 min. Cells were then incubated at 30°C for 5 min, followed by further incubation at 38°C for 5–25 min. Samples were withdrawn at each of the indicated time-points and immediately mixed with equal volume of Stop buffer (20 mM sodium acetate, pH 5.2, 2% 30 mM sodium acetate-saturated phenol, pH 5.0, 2 mM EDTA, and 75% ethanol in diethylpyrocarbonate-treated MilliQ) to inactivate cellular RNase activity. The cells were then collected by centrifugation and resuspended with 400 μL of Resuspension buffer (20 mM sodium acetate, pH 5.2, 2% sodium dodecyl sulfate in diethylpyrocarbonate-treated MilliQ). Total RNA was then extracted twice by treatment with 400 μL 30 mM sodium acetate-saturated phenol (pH 5.0) and three times with 400 μL phenol/chloroform/isoamyl alcohol (25:24:1), collected by ethanol precipitation, and suspended with 50 μL RNase free dH2O (TaKaRa). The concentration of purified total RNA samples was determined using a NanoDrop™ Lite (Thermo Scientific).
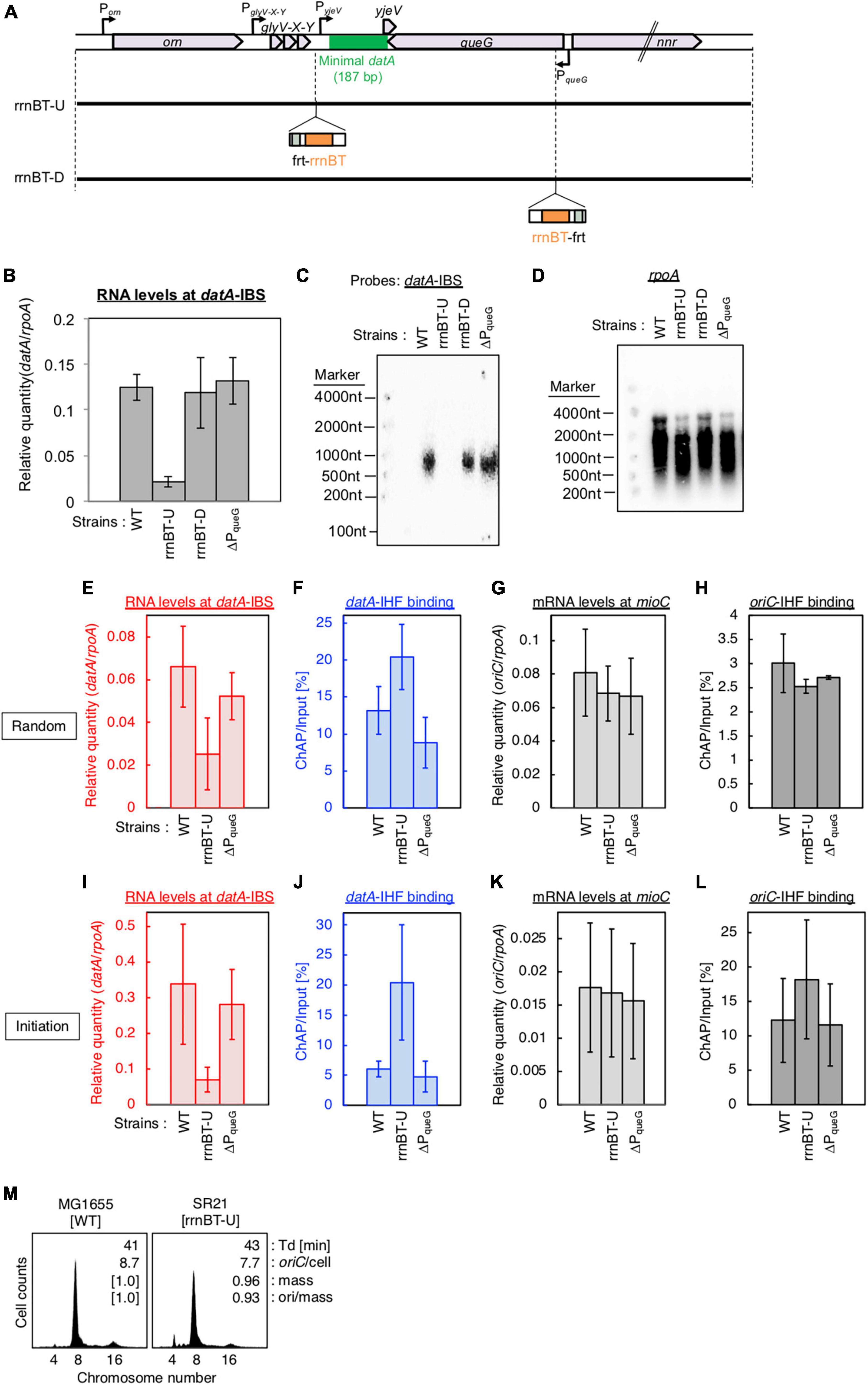
Figure 5. Insertion of a transcriptional terminator inhibits transcription-dependent datA regulation. (A) Construction of transcriptional terminator (rrnBT) insertion mutant strains. (B) MG1655 (WT), SR21 (rrnBT-U), SR22 (rrnBT-D), and SR18 (ΔPqueG) were cultivated in LB medium at 37°C and then the total RNA was extracted. The RNA levels of datA IBS relative to those of the rpoA gene were determined using RT-qPCR. Data represent means with error bars of two independent experiments. Based on the Student’s t-test, the WT vs. rrnBT-U difference (p < 0.01) is statistically significant. (C,D) Northern blotting using total RNA prepared from MG1655, SR21, SR22, and SR18 cells and the probes for datA IBS (C) and rpoA gene (D). (E–L) SH022 (dnaC2 ihfA-cHis12), SR35 (dnaC2 ihfA-cHis12 rrnBT-U), and SR36 (dnaC2 ihfA-cHis12 rrnBT-D) cells growing in LB medium at 30°C [random cultures; (E–H)] were transferred to 37°C and incubated for 80 min [synchronized; (I–L)]. The RNA levels of datA IBS (E,I) and mioC gene (G,K) relative to those of the rpoA gene were determined using RT-qPCR. IHF binding at datA (F,J) and oriC (H,L) were determined by ChAP-qPCR, and yield was calculated (expressed as %). Data represent means with error bars of two independent experiments. Based on the Student’s t-test, the WT vs. rrnBT-U differences in panel (F) (p < 0.04), 5I (p < 0.1) and 5J (p < 0.03) are statistically significant. (M) Flow cytometry analysis of rrnBT-U mutant cell. Cells were grown at 30°C in LB medium, followed by flow cytometry analysis.
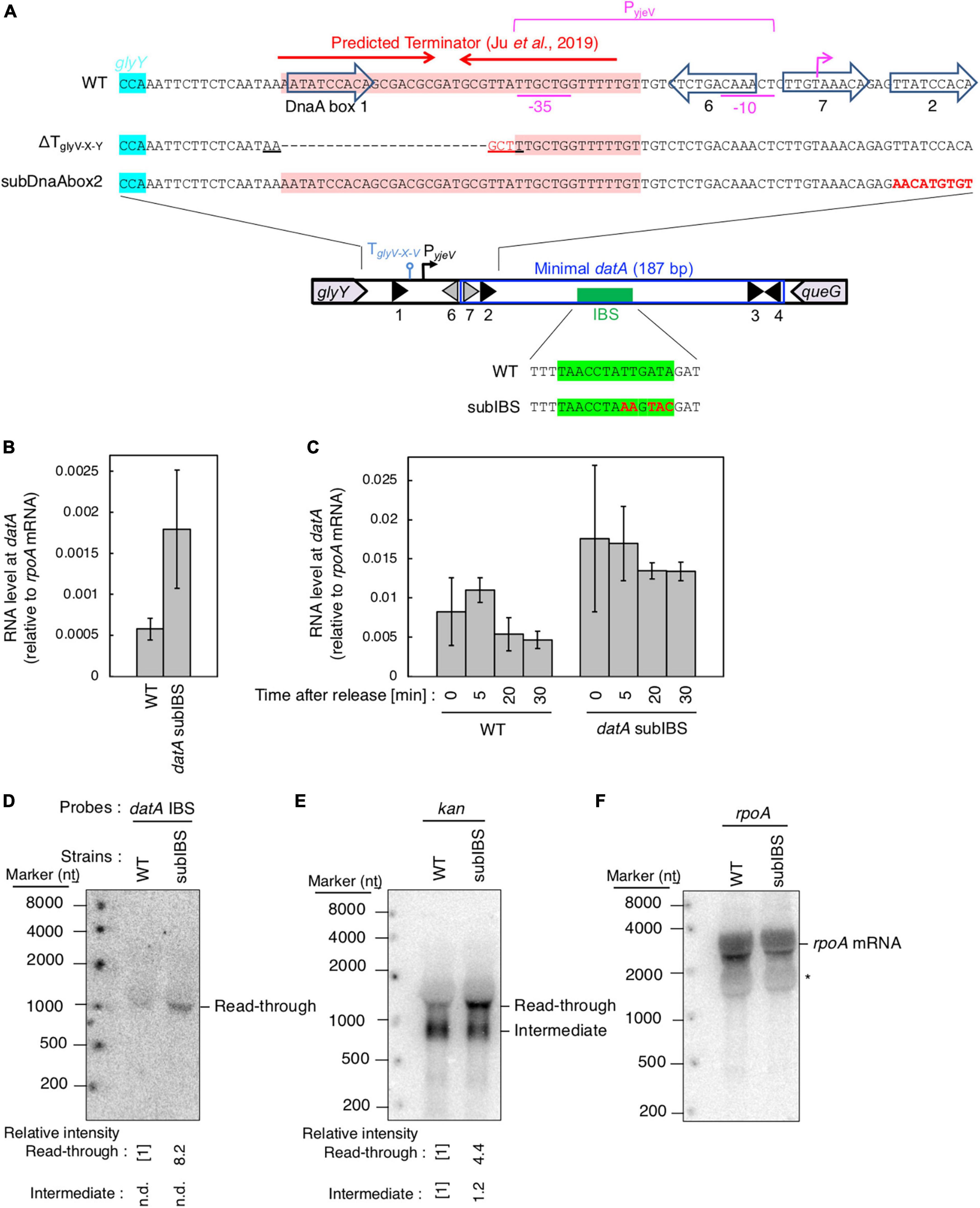
Figure 6. datA-IHF binding inhibits progression of datA transcription. (A) Construction of the mutant cells lacking transcriptional terminator/attenuator sequence (ΔTglyV-X-Y) and substitution mutant cells of datA DnaA box 2 or IBS (datAsubDnaAbox2 or subIBS). (B,C) ITK23c (dnaC2 queG::frt) (WT) and ITK24c (ITK23c datAsubIBS) (datAsubIBS) cells growing in LB medium at 30°C [random cultures; (B)] were transferred to 37°C and incubated for 80 min [synchronized; (C)]. The cells were transferred to 30°C (Time 0) and further incubated for 5–15 min at 30°C, before total RNA was extracted. The RNA levels of datA IBS locus relative to those of the rpoA gene were determined using RT-qPCR. Data represent means (± S.D.) of at least three independent experiments. Based on the Student’s t-test, the WT vs. datA subIBS difference (p < 0.02) in panel (B) and 5 min vs. 20 min (p < 0.02) and 5 min vs. 30 min (p < 0.06) of the WT samples in panel (C) are statistically significant. The effect of the dnaC2-dependent cell cycle synchronization was confirmed by checking the decrease of mioC transcript (Supplementary Figure 2C). (D–F) Northern blotting using total RNA prepared from ITK9 (ΔglyV-X-Y::kan datA WT) and ITK39 (ΔglyV-X-Y::kan datAsubIBS) cells, and the probes for datA IBS (D), kan gene (E), and rpoA gene (F). The asterisk means unidentified bands.
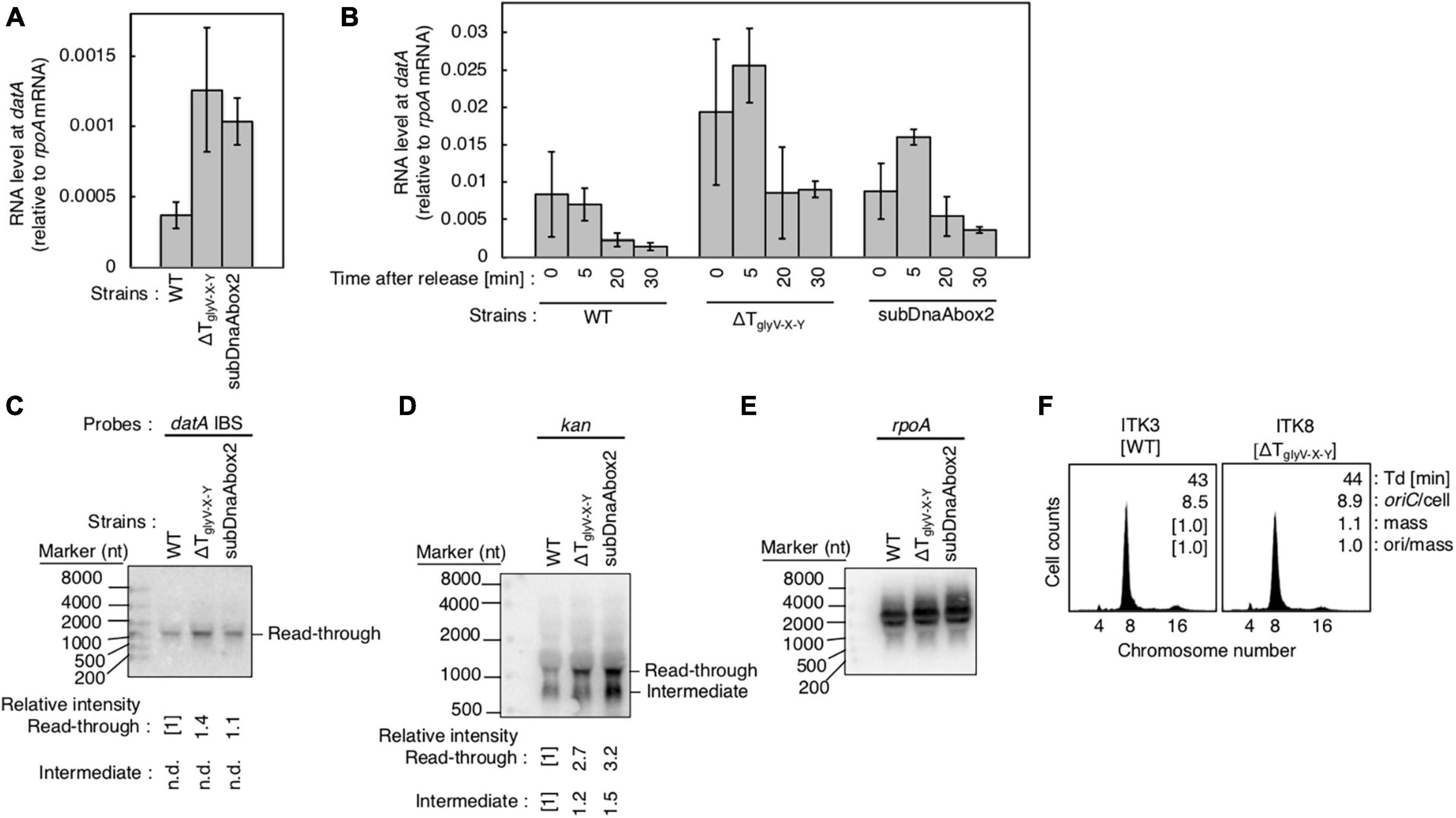
Figure 7. Analysis of transcriptional terminator/attenuator sequence of the glyV-X-Y operon and datA DnaA box 2 on transcription-dependent datA regulation. (A,B) ITK3c (dnaC2) (WT), ITK8c (dnaC2ΔTglyV-X-Y) (ΔTglyV-X-Y), and ITK16c (dnaC2 datAsubDnaAbox2) (subDnaAbox2) cells growing in LB medium at 30°C (random cultures; A) were transferred to 37°C and incubated for 80 min (B). The cells were transferred to 30°C (Time 0) and further incubated for 5–30 min at 30°C, and then the total RNA were extracted. The RNA levels of datA IBS locus relative to those of the rpoA gene were determined using RT-qPCR. Data represent means with error bars of at least two independent experiments. Based on the Student’s t-test, the WT vs. ΔTglyV-X-Y (p < 0.04) and WT vs. subDnaAbox2 difference (p < 0.006) in panel (A), 5 min vs. 20 min (p < 0.02) and 5 min vs. 30 min (p < 0.007) of the WT samples, 5 min vs. 30 min (p < 0.09) of the ΔTglyV-X-Y samples, and 5 min vs. 20 min (p < 0.07) and 5 min vs. 30 min (p < 0.008) of the subDnaAbox2 samples in panel (B) are statistically significant. The effect of the dnaC2-dependent cell cycle synchronization was confirmed by checking the decrease of mioC transcript (Supplementary Figure 2D). (C–E) Northern blotting using total RNA prepared from randomly-growing ITK9 (glyV-X-Y::kan datA WT), ITK40 (glyV-X-Y::kan ΔTglyV-X-Y), or ITK13 (glyV-X-Y::kan datAsubDnaAbox2) cells and the probes for datA IBS (C), kan gene (D), and rpoA gene (E). (F) Flow cytometry analysis of ΔTglyV-X-Y mutant cells. Cells were grown at 30°C in LB medium, followed by flow cytometry analysis.
RT (reverse transcription)-qPCR
For RT-qPCR, cDNA samples were prepared using 1 μg of each total RNA sample and PrimeScript™ RT reagent Kit with gDNA Eraser (Perfect Real Time) (TaKaRa). The RNA levels were quantified by real-time qPCR using TB Green Premix Ex Taq™II (Tli RNaseH Plus) (TaKaRa) and primers ORI_1 and KWoriCRev for oriC; RT-datAIBS_L and RT-datAIBS_R for datA; RTrpoA-L and RTrpoA-R for rpoA; and RTmioC-L and RTmioC-R for mioC.
Northern blotting
Northern blotting experiments were performed according to a previously described method (Su’etsugu et al., 2003), with the following minor modifications. Each transcript in purified total RNA samples was separated by electrophoresis using 2% agarose/2.2 M formaldehyde gel in 1x MOPS running buffer, transferred to Hybond™ N+ membrane (Cytiva) using the capillary blotting method, dried at 80°C for 2 h, and then fixed by exposure to UV (120 mJ/cm2) for 5 min using FUNA-UV-Crosslinker FS-800 (Funakoshi). Oligonucleotide probes with complementary sequences to either the datA IBS locus or the rpoA gene were end-labeled with 32P, and this labeled probe was hybridized with RNA on the membrane in the presence of ULTRAhyb™ Ultrasensitive Hybridization Buffer (Invitrogen). The membrane was then washed with 2x SSC (300 mM sodium chloride, 30 mM sodium citrate), 2x SSC+0.1% sodium dodecyl sulfate, 0.2x SSC (30 mM sodium chloride, 3 mM sodium citrate) +0.1% sodium dodecyl sulfate, and 0.2x SSC, dried, and analyzed using the Typhoon™ 7500 imaging analyzer (Cytiva).
Results
The levels of datA IBS transcription and datA-IHF binding fluctuate during the cell cycle
The minimal region of datA required for full DDAH activity is located at the intergenic region downstream of the tRNA-Gly (glyV-X-Y) operon between the yjeV gene and queG gene (Figure 2A). Indeed, the datA IBS was transcribed in exponentially-growing MG1655 (WT) cells when cultivated at 37°C in LB or supplemented M9 medium (Supplementary Figure 1A). Thus, we hypothesized that datA-IHF binding/dissociation is regulated by transcription of the datA locus. This theory is in accordance with previous observations that rifampicin inhibits timely dissociation of datA-IHF complexes (Kasho and Katayama, 2013), and that datA activity is inhibited by the translocation of datA to a highly transcribed position (Frimodt-Møller et al., 2015).
To analyze the cell cycle-dependent correlation between datA-IHF binding and transcription of the datA locus, we combined RT-qPCR and ChAP assays using temperature sensitive dnaC2 cells, which allow synchronization of the cell cycle in LB medium (Withers and Bernander, 1998). DnaC is the helicase loader, and the dnaC2 mutation inhibits replication initiation at oriC specifically at restrictive temperatures (37–42°C). Thus, the cell cycle of the dnaC2 cells can be synchronized by incubation at 37°C for 80 min just before replication initiation (0 min), followed by a temperature down shift to 30°C for 5 min to release the cell cycle by activating DnaC, and induce a single round of replication initiation. An upshift back to 37°C for 10–30 min allows DNA synthesis to proceed while inhibiting additional replication initiation. This dnaC2-based synchronization system has previously been used to identify the cell cycle-coordinated IHF binding/dissociation pattern at oriC, datA, and DARS2 (Kasho and Katayama, 2013; Kasho et al., 2014), as well as the cell cycle-specific oscillation of expression levels of genes including dnaA, mioC, and gidA (Theisen et al., 1993; Su’etsugu et al., 2003); thus, this well-established method is suitable for the purpose of this study.
To validate our assay, initial control experiments were performed in which the cell cycle-coordinated binding/dissociation of IHF at the datA and oriC loci was confirmed using ChAP-qPCR. In accordance with previous studies (Kasho and Katayama, 2013), at the oriC locus IHF was stably bound prior to initiation (0 min) and immediately dissociated after initiation (5–10 min) (Supplementary Figure 1B), while at the datA locus, IHF was not bound before initiation (0 min) and became bound after initiation (15–20 min) (Figure 2B).
Next, under the same experimental conditions as in Figure 2B, we prepared total RNA samples and performed RT-qPCR using primers recognizing datA IBS, the mioC gene as a control for cell cycle synchronization, or the housekeeping gene rpoA gene as a loading control (Su’etsugu et al., 2003). Consistent with a previous report, the mRNA level of the mioC gene relative to that of rpoA was lower at 0 min (Supplementary Figure 1D), indicating that the dnaC2 cells were properly synchronized before initiation. By contrast, the relative RNA level at datA IBS was very low in the non-synchronized cells, dramatically increased at the initiation period (0–5 min), and decreased after initiation (10–20 min) (Figure 2B; Supplementary Figure 1C). No dramatic increase in the datA RNA level or decrease in the mioC mRNA level was observed when MG1655 (dnaC WT) cells were incubated at 37°C (Supplementary Figures 1C, D), indicating that transcription of datA is regulated in a cell cycle-dependent manner. A similar oscillation pattern of datA RNA level was observed using supplemented M9 medium (Supplementary Figure 1E). Thus, these results indicated that transcription of datA is regulated in a cell cycle-dependent manner.
In dnaC2 cells, the level of datA RNA was approximately 8–40-fold higher at initiation (0 min) than in the random culture (R) sample (Figure 2B; Supplementary Figure 1C). The datA RNA levels were still higher at the post-initiation period in dnaC2 cells than in random culture samples, which can also be explained by the accumulation of datA RNA as a result of cell cycle synchronization. Furthermore, the accumulation of datA RNA might affect the overall high baseline datA RNA level after releasing the cell cycle, i.e., during 5–10 min incubation, it is possible that datA RNA is not significantly degraded but is also newly produced. Thus, we concluded that the levels of IHF binding and transcription at the datA locus are inversely correlated with each other, supporting the idea that the datA-IHF binding/dissociation is regulated in a manner dependent on transcription.
glyV-X-Y promoter is predominant for read-through transcription covering datA-IHF
To determine the transcriptional start site of datA RNA, we performed RT-qPCR using four primer sets for the orn gene, two sets each for the region upstream or downstream of the transcriptional promoter for glyV-X-Y operon (PglyV-X-Y), and one for datA IBS (Figure 3A). While the RNA levels of the orn gene and the glyV-X-Y transcription units remained constant, the relative RNA level of datA IBS fluctuated during the cell cycle and significantly increased at initiation, consistent with Supplementary Figure 1C (Figure 3B; probe 9). The relative mRNA levels of orn gene (probes 1–4) and the region upstream of the transcriptional promoter PglyV-X-Y (probes 5 and 6) were much lower than those of the glyV-X-Y operon (probes 7 and 8) or datA IBS (probe 9), suggesting that transcription of the orn gene is substantially terminated at the PglyV-X-Y upstream locus.
To confirm whether datA transcription is initiated at PglyV-X-Y, northern blotting was used to analyze the length of the datA-containing RNA products. The relative amounts of datA-IBS RNA were low (Figure 3B), which may result from degradation of the 3′-region of RNA products including tRNA-Gly during the tRNA maturation processes (Wellner et al., 2018). Thus, to efficiently analyze full-length RNA products that read through the datA IBS locus, we constructed the ΔglyV-X-Y::kan strain, in which the entire glyV-X-Y operon was replaced by the kan gene (Figures 3C, D). To avoid cell lethality resulting from lack of tRNA-Gly, tRNA-Gly was supplemented from the pITK6 plasmid-encoded glyV-X-Y in all the ΔglyV-X-Y::kan mutant cells constructed in this study.
In the ITK9c (dnaC2 ΔglyV-X-Y::kan) cells, the level of datA RNA was significantly lower than that of the kan gene (Figures 3B, E; “R” sample), suggesting that transcription inhibition occurs at the datA-upstream region. Furthermore, the datA RNA level was 3-fold higher at initiation compared with randomly-growing cells (Figure 3E; “0 min” sample). Consistent with the RT-qPCR experiment, the northern blotting experiments revealed that the read-through RNA products, including datA IBS (approximately 1,500 nt), were observed using either the datA IBS probe or the kan probe (Figures 3F, G) and were increased at the initiation period (Figures 3H, I). Notably, the shorter RNA intermediate (approximately 800 nt) was only observed using the kan probe (Figure 3F). Given the observed fluctuation of datA-IBS RNA, these data suggest that the transcripts initiated from PglyV-X-Y pass through datA IBS specifically at the initiation period and are terminated in front of datA IBS after initiation. We thus hypothesized that the specific DNA element located between the region downstream of glyV-X-Y and datA IBS is required for the cell cycle-coordinated datA-IBS transcription. Consistently, the relative RNA levels initiated from PglyV-X-Y were not increased at the initiation period in either WT or ΔglyV-X-Y::kan cells (Figures 3B, E), supporting our hypothesis. The relative datA-IBS transcription levels in Figure 3B were moderately lower than those in Figure 2B, which might be due to subtle differences in cell-cultivation conditions affecting rates of transcription and processing of the full-length datA RNA products, including tRNA-Gly.
Transcription-dependent regulation for datA-IHF binding
Next, to address the requirement of PglyV-X-Y for transcription-dependent regulation of datA-IHF binding, we constructed ITK4c (dnaC2 ΔPorn) and ITK5c (dnaC2 ΔPglyV-X-Y) strains by inserting frt-kan into the nnr gene located close to, but outside of, the datA region (Figure 4A). In random cultures (Figures 4B–E), the nnr::frt-kan mutation had little effect on cell cycle-coordinated datA-IHF binding and datA transcription (Figures 4B, C), or oriC-IHF binding and mioC mRNA level (Figures 4D, E). We thus used ITK3c (dnaC2 nnr::frt-kan) cells as a WT control for analyzing cell cycle-coordinated oscillation of datA RNA and datA-IHF binding/dissociation.
In cells synchronized at the initiation period (Figures 4F, G), introduction of the ΔPglyV-X-Y mutation greatly decreased the datA-IBS RNA level compared with that of ΔPorn and the WT control and, notably, diminished the initiation period-specific decrease in datA-IHF binding (Figures 4F, G). Low residual RNA levels could be due to read-through RNA products from Porn. In contrast to datA IBS RNA, the mioC mRNA level was not substantially changed by introduction of the ΔPglyV-X-Y mutation (Figure 4H). There may be a minor increase in the oriC-IHF binding level in ΔPglyV-X-Y mutant cells, which could be an indirect consequence of a moderate delay in replication initiation induced by increased datA-IHF binding (see below). Consistent with qPCR analysis, northern blotting supports the requirement of PglyV-X-Y for the initiation period-specific production of read-through datA RNA (Figures 4J, K).
To address the impact of the ΔPglyV-X-Y mutation on regulation of replication initiation, we performed flow cytometry analysis. Cells were exponentially grown in LB medium and further incubated in the presence of rifampicin and cephalexin, which inhibit replication initiation and cell division, respectively, to complete replication. Cell mass (cell size) and chromosomal DNA content were then analyzed by flow cytometry to allow determination of the oriC/cell and the ori/mass, and thus the frequency of replication initiation. While WT cells predominantly showed peaks corresponding to four and eight origins, ΔPglyV-X-Y, but not ΔPorn, cells showed a significant increase in the peak corresponding to four origins and a decrease in the eight origins peak (Figure 4L). This means that the ΔPglyV-X-Y mutation specifically decreased initiation frequency (ori/cell or ori/mass) (Figure 4L), consistent with our observation that PglyV-X-Y is required for production of datA RNA and transcription-dependent regulation of datA-IHF binding. Given that IHF dissociates from oriC immediately after initiation, delay of initiation could result in an increased level of oriC-IHF complexes (Figure 4I).
To confirm the requirement of PglyV-X-Y for regulation of datA function, we inserted the frt site-flanking strong transcriptional terminator element (rrnBT; terminator of E. coli rrnB operon) (Orosz et al., 1991) either upstream or downstream of datA IBS (rrnBT-U or -D, respectively) using the inserting frt site (Figure 5A). The rrnBT-U cells, but not the rrnBT-D or ΔPqueG cells, had significantly decreased levels of datA-IBS RNA compared with WT cells (Figure 5B). Consistent results were obtained by Northern blotting where the read-through RNA products including datA IBS (500–1,000 nt) were observed in WT, rrnBT-D, and ΔPqueG cells, but not in rrnBT-U cells (Figures 5C, D). Importantly, insertion of rrnBT-U specifically impaired timely datA-IHF dissociation and datA transcription at the initiation period, while stable oriC-IHF binding was maintained in rrnBT-U cells (Figures 5E–L). An observed slight increase in levels of oriC-IHF binding in rrnBT-U cells could be due to delayed initiation as for ΔPglyV-X-Y (Figure 5L). Consistently, rrnBT-U cells showed moderately delayed initiation frequency in WT, ΔDARS1, or ΔDARS2 backgrounds, whereas ΔPqueG cells did not (Figure 5G; Supplementary Figure 3). Also, in the rrnBT-U cells, rrnBT was inserted at the region upstream of TglyV-X-Y to avoid affecting PyjeV transcription, thus suggesting that PglyV-X-Y has a predominant role in transcription-dependent datA-IHF regulation and the requirement of PyjeV is limited. Taken together, these observations support the idea that PglyV-X-Y is critical for the transcriptional regulation of timely datA-IHF binding.
datA-proximal DNA elements are required for the timely oscillation of datA transcription
To address the regulatory mechanism and the requirement of the specific DNA element located between the region downstream of the glyV-X-Y operon and the datA IBS for transcription-dependent datA regulation, we used substitution mutants at datA DnaA box2 and IBS (subDnaAbox2 and subIBS, respectively) and a deletion mutant in which a predicted terminator/attenuator sequence TglyV-X-Y of the glyV-X-Y operon was removed (ΔTglyV-X-Y) (Figure 6A; Ju et al., 2019). Because a previous study suggested that IHF binding at the promoter-attenuator region of the ilvG-M-E-D-A operon induces the pausing of transcription machinery (Pagel and Hatfield, 1991), we first tested whether datA-IHF binding interrupts transcription. For this purpose, we analyzed transcription levels of the datA IBS region in dnaC2 or dnaC2 datAsubIBS double mutant cells using RT-qPCR and primer set No. 9 from Figure 3A because these primers hybridize to sites outside of the datA IBS.
In dnaC2 datA WT cells, datA RNA was abundant during the initiation period (0–5 min) and decreased after initiation (20–30 min) (Figures 6B, C). By contrast, introduction of the datAsubIBS mutation diminished this oscillation and the level of datA RNA remained constant. The constant increase of datA RNA throughout cell cycle resulted in accumulation of transcript and, therefore, an overall increase in datA RNA levels relative to WT cells in random culture samples (Figure 6B). Consistent with Figures 6B, C, introduction of the datAsubIBS mutation into dnaC2 ΔglyV-X-Y::kan cells resulted in increased total datA RNA levels (Figure 6D), and notably also increased the level of read-through datA RNA (datA IBS or kan probes; approximately 1,500 nt) relative to that of the RNA intermediate (kan probe; approximately 900 nt) (Figures 6D–F). This suggests that datA-IHF binding at the post-initiation period is stable enough to significantly interrupt transcription. However, a slight increase in the datA RNA level at the initiation period was observed, even in the dnaC2 datAsubIBS cells (Figures 6B, C), implying that other potential regulatory elements may exist to interrupt datA transcription.
To address the possible role of TglyV-X-Y for regulating transcription of the datA locus, we constructed a deletion mutant of TglyV-X-Y (ΔTglyV-X-Y; Figure 6A) (Ju et al., 2019). TglyV-X-Y overlaps with datA DnaA box 1 and the −35 sequence of a putative promoter, PyjeV (Kitagawa et al., 1996). Therefore, we chose to delete datA DnaA box 1, which is dispensable for DDAH activity (Kasho and Katayama, 2013; Kasho et al., 2017), but not PyjeV. In addition, because previous studies suggested that DnaA binding around the terminator sequence of the asnC gene inhibits the passage of transcription machinery (Schaefer and Messer, 1988, 1989), we also addressed the possibility that the DnaA binding at the upstream region of datA IBS can act as a roadblock for transcription from PglyV-X-Y. Since datA DnaA box 2 is essential for ATP-DnaA-specific oligomer formation at datA, we constructed a datA mutant with DnaA box 2 substitution (datAsubDnaAbox2) (Figure 6A), which lacks the DDAH activity of datA (Kasho and Katayama, 2013; Kasho et al., 2017).
Introduction of ΔTglyV-X-Y or the datAsubDnaAbox2 mutation resulted in increased levels of datA-IBS RNA at the initiation period (0–5 min) (Figures 7A, B). In contrast to the datAsubIBS mutant cells (Figures 6B, C), both the ΔTglyV-X-Y and datAsubDnaAbox2 mutant cells sustained cell cycle-coordinated oscillation of datA RNA levels, although datA RNA levels at the post-initiation period (20–30 min) were also increased compared with those of WT cells. This suggests that either TglyV-X-Y or datA DnaA box 2 may assist in regulation of the overall transcription level of datA but are not essential for the timely dissociation of the datA-IHF complex. Consistently, northern blotting analysis of datA-containing transcripts showed that the level of read-through datA RNA (IBS or kan probes; approximately 1,500 nt) relative to the intermediate (kan probe; approximately 900 nt) was only moderately increased in ΔTglyV-X-Y and datAsubDnaAbox2 cells compared with WT cells (Figures 7C–E). Despite the increase in basal datA transcription levels, the ΔTglyV-X-Y mutation had little effect on initiation timing (Figure 7F). These results suggest that transcription block by TglyV-X-Y or the DnaA binding at datA DnaA boxes 2 play specific roles in regulating the basal level of datA transcription but is dispensable for its cell cycle-coordinated oscillation and for controlling the timing of initiation. Taken together, these results suggest that another regulatory factor may play a role in timely datA-IHF binding in a manner independent of datA transcription.
Discussion
Chromosomal DNA replication in bacteria is initiated at the origin of replication oriC and its timing is tightly regulated to ensure that firing at the replication origin occurs only once per cell cycle. The timely initiation is achieved through the activities of the replication initiator ATP-DnaA, whose cellular levels peak prior to initiation. And immediately after initiation, DnaA-bound ATP is hydrolyzed by RIDA and DDAH depending on datA-IHF complex to produce ADP-DnaA. To further understand how the timely IHF binding/dissociation at the datA locus is regulated, we investigated the requirement for transcription of the neighboring tRNA-Gly (glyV-X-Y) operon. Cell cycle analysis revealed that datA-IHF binding and read-through transcription of datA IBS are highly correlated during the cell cycle, i.e., IHF is not associated with datA at the initiation period and temporarily binds after initiation, while transcription of datA IBS is significantly decreased after initiation (Figure 2B). The timing of datA-IHF binding largely corresponds to that of datA duplication (Kasho and Katayama, 2013), implying the possibility that the regulation of datA-IHF binding is related to the progression of replication forks. Deletion of PglyV-X-Y or insertion of a transcriptional terminator at the region downstream of glyY gene resulted in inhibition of datA transcription and enhanced datA-IHF binding (Figures 4, 5), indicating that transcription derived from PglyV-X-Y and read-through transcription of datA IBS is required for timely dissociation of the datA-IHF complex (Figure 8). These mutations moderately inhibit replication initiation in vivo (Figures 4L, 5M), supporting the idea that PglyV-X-Y-derived transcription is required for down-regulation of datA function by reducing the chance of datA-IHF binding and that the chromosomal locus spanning tRNA-Gly operon to datA could function as a connecting hub for balancing the tRNA transcription level and the timing of replication initiation by sensing the cellular growth environment or stresses. Transcription of E. coli tRNA is globally regulated by Fis in a growth phase-dependent manner, and peaks at the exponential phase (Bosch et al., 1990; Nilsson et al., 1992). Other studies have suggested that some specific stringent conditions, such as starvation of isoleucine or folate, decrease the transcription level of most tRNAs (Ikemura and Dahlberg, 1973; Irving et al., 2021). A decrease in datA read-through transcription will stimulate constitutive datA-IHF binding, enhancing inactivation of DnaA. Taken together, we point to the possibility that E. coli cells fine-tune the DnaA activity through a regulatory mechanism that coordinates the tRNA-Gly transcription levels and concomitantly the basal amounts of read-through datA transcripts upon adaptation to various growing conditions such as exponential phase of cell growth or stringent condition.
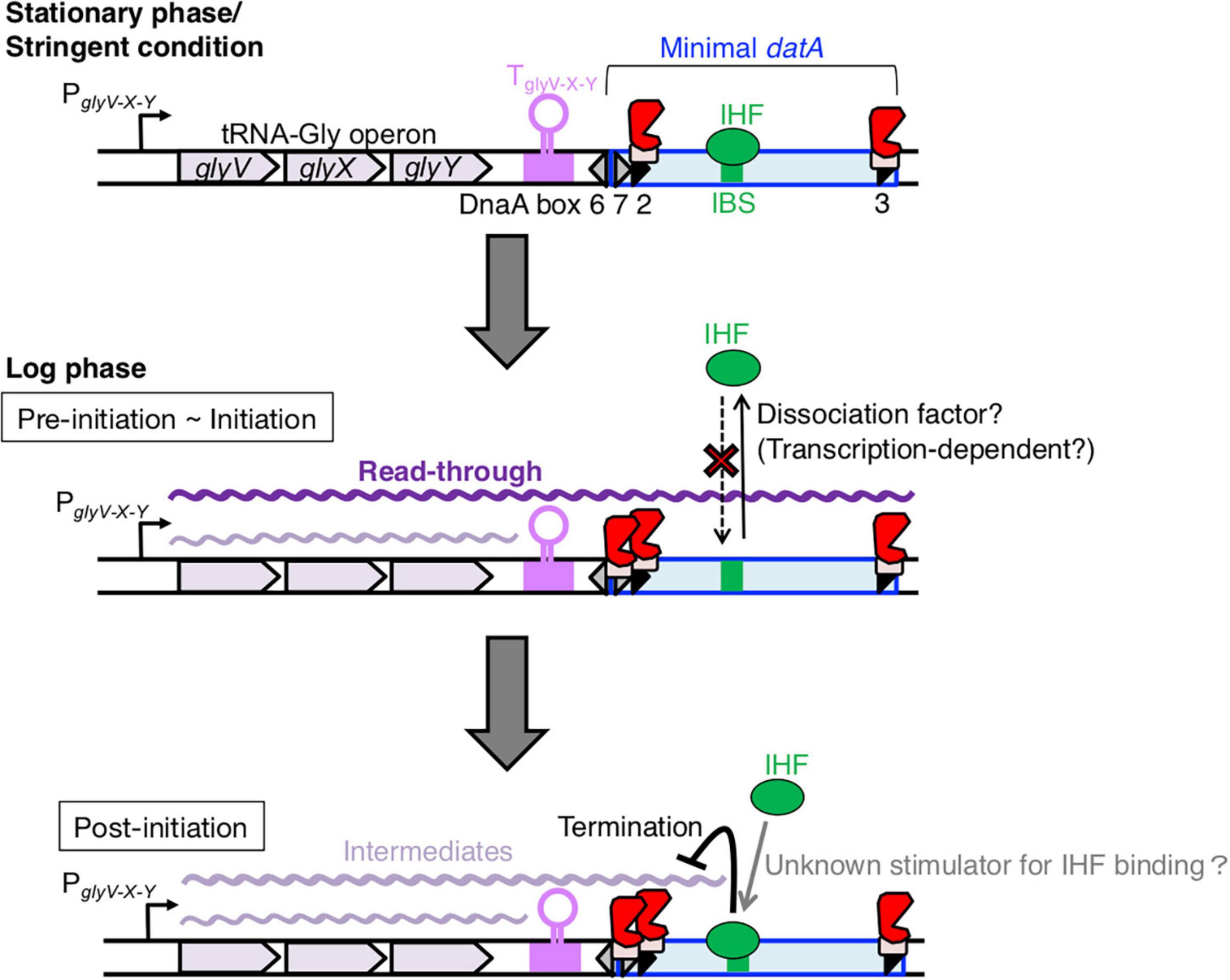
Figure 8. Model of tRNA transcription-dependent regulation for datA-IHF binding and dissociation during the cell cycle. In stationary phase cells, transcription of the tRNA-Gly (glyV-X-Y) operon is repressed (the top panel). In the pre-initiation and initiation periods of growing cells (the middle panel), the read-through transcription of the whole datA region is initiated from PglyV-X-Y of the tRNA-Gly operon and is prerequisite for inhibiting datA-IHF binding or stimulating datA-IHF dissociation. An unknown factor might participate in this inhibition. The level of read-through datA transcription is constantly down-regulated by DnaA binding at datA DnaA box 2 and by the transcription terminator/attenuator sequence TglyV-X-Y of the tRNA-Gly operon. In the post-initiation period (the bottom panel), IHF binding to datA IBS is stimulated/stabilized by an unknown mechanism and strongly inhibits read-through datA transcription at the site, resulting in timely activation of DDAH for repressing excess initiations.
Because the cell cycle analysis suggested that transcriptional initiation of the glyV-X-Y operon does not greatly increase in a replication initiation-specific manner (Figures 3A, B), we next addressed requirements of the specific DNA elements located between the region downstream of glyY and datA (IBS, DnaA box 2, and its proximal terminator/attenuator sequence TglyV-X-Y of the glyV-X-Y operon) for the transcription termination. Importantly, in the datAsubIBS mutant cells, the datA RNA level increased at the initiation period and did not substantially decrease during the ensuing 30 min after initiation, which is markedly different to WT datA cells (Figure 6C). Taken together, we suggest that the datA-IHF complex regulates cell cycle-coordinated attenuation of datA transcription and interrupts read-through datA transcription in a timely manner after initiation (Figure 8). This is consistent with previous studies showing the timely binding of IHF to datA and the transcription-termination activity of DNA-bound IHF (Pagel and Hatfield, 1991).
In datAsubDnaAbox2 mutant cells, the level of datA RNA increased at the initiation period and decreased shortly after initiation, like that of the WT datA cells, and the overall level of datA transcription was increased compared with that of WT cells (Figures 7A, B). The role of datA DnaA box 2 for transcription termination could be consistent with previous observations that the DnaA oligomer formed at the specific region of asnC terminator sequence inhibits the passage of transcription machinery and causes transcription pausing (Schaefer and Messer, 1988, 1989). RNA polymerase might directly interact with DnaA (Flåtten et al., 2009). Another possible role of the datA DnaA box 2 could be as a riboswitch for transcription attenuation. A recent study suggested that DnaA binds to the specific RNA sequence rDnaA box and plays a regulatory role for transcription termination (Yao et al., 2023).
In addition, we addressed the role of TglyV-X-Y which should be considered as an attenuator because considerable read-through is allowed (Figure 3F; Ju et al., 2019). In E. coli, the transcription units of each tRNA include a typical Rho-independent terminator sequence just 10–25 bp away from their tRNA-coding regions (Komine et al., 1990; Ju et al., 2019), and the unique feature of glyV-X-Y is that the DnaA box and the −35 sequence of PyjeV partly overlap with the TglyV-X-Y sequence (Kitagawa et al., 1996). Our results suggest that partial deletion of TglyV-X-Y which also loses datA DnaA box 1, as well as datAsubDnaAbox2 mutation, increased the overall level of read-through transcription of datA, but allowed a decrease after replicational initiation (Figure 7B). Despite the increase in basal datA transcription levels, the ΔTglyV-X-Y mutation little affected the initiation timing (Figure 7F), suggesting that TglyV-X-Y is also important for the overall transcriptional attenuation but not required for the timely attenuation of datA transcription.
Based on these observations, we propose a model whereby read-through datA transcription is required to down-regulate datA-IHF binding and is likely terminated by the timely assembly of the datA-IHF complex after replication initiation (Figure 8). In this model, datA transcription is only a prerequisite for IHF dissociation. datA transcription by itself is not sufficient for controlling the timing of datA-IHF binding, suggesting that an unknown regulatory factor is simultaneously required for the timely datA-IHF dissociation. As tRNA-Gly transcription initiation from PglyV-X-Y and transcriptional attenuation by datA DnaA box 2 and TglyV-X-Y are likely to operate constitutively during cell cycle, regulation of the timing of IHF dissociation would be due to the timely function of this unknown factor.
Previous studies have shown that the expression of most tRNA operons is regulated according to growth conditions, and that the level of tRNA-Gly (anticodon: GCC) derived from the glyV-X-Y operon is higher in richer medium (Emilsson et al., 1993). In this study, the transcription levels of datA IBS in exponentially-growing cells were broadly similar under different growth conditions (LB or supplemented M9 medium; Supplementary Figure 1A), yet stringent conditions would decline tRNA transcription initiation from PglyV-X-Y. Therefore, the transcription attenuation might modulate the transcription level of datA IBS under different growth conditions. Since some bacterial species such as Bacillus subtilis and Streptomyces coelicolor also have DnaA box clusters analogous to datA for repressing untimely initiations (Smulczyk-Krawczyszyn et al., 2006; Okumura et al., 2012), our findings and further future studies may provide a universal concept for transcription-coupled regulation of initiator proteins.
Data availability statement
The original contributions presented in this study are included in this article/Supplementary material, further inquiries can be directed to the corresponding author.
Author contributions
KK: Conceptualization, Data curation, Funding acquisition, Investigation, Methodology, Project administration, Supervision, Visualization, Writing—original draft, Writing—review and editing. RyS: Conceptualization, Data curation, Investigation, Methodology, Visualization, Writing—review and editing. KI: Data curation, Investigation, Methodology, Visualization, Writing—review and editing. WN: Investigation, Writing—review and editing. RiS: Investigation, Writing—review and editing. TJ: Investigation, Writing—review and editing. SO: Supervision, Writing—review and editing. TK: Conceptualization, Funding acquisition, Project administration, Supervision, Visualization, Writing—review and editing.
Funding
The author(s) declare financial support was received for the research, authorship, and/or publication of this article. This work was supported by the Japan Society for the Promotion of Science, JSPS KAKENHI (Grant Numbers JP15K18479, JP21K19233, and JP23H02438) and Gushinkai, 2023 Samuro Kakiuchi Memorial Research Award for Young Scientists.
Acknowledgments
We thank the Research Support Center, Graduate School of Medical Sciences, Kyushu University for DNA sequencing.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2024.1360108/full#supplementary-material
References
Aeling, K. A., Opel, M. L., Steffen, N. R., Tretyachenko-Ladokhina, V., Hatfield, G. W., Lathrop, R. H., et al. (2006). Indirect recognition in sequence-specific DNA binding by Escherichia coli integration host factor: The role of DNA deformation energy. J. Biol. Chem. 281, 39236–39248. doi: 10.1074/jbc.M606363200
Ali Azam, T., Iwata, A., Nishimura, A., Ueda, S., and Ishihama, A. (1999). Growth phase-dependent variation in protein composition of the Escherichia coli nucleoid. J. Bacteriol. 181, 6361–6370. doi: 10.1128/JB.181.20.6361-6370.1999
Bosch, L., Nilsson, L., Vijgenboom, E., and Verbeek, H. (1990). FIS-dependent trans-activation of tRNA and rRNA operons of Escherichia coli. Biochim. Biophys. Acta 1050, 293–301. doi: 10.1016/0167-4781(90)90184-4
Cassler, M. R., Grimwade, J. E., and Leonard, A. C. (1995). Cell cycle-specific changes in nucleoprotein complexes at a chromosomal replication origin. EMBO J. 14, 5833–5841. doi: 10.1002/j.1460-2075.1995.tb00271.x
Datsenko, K. A., and Wanner, B. L. (2000). One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc. Natl. Acad. Sci. U.S.A. 97, 6640–6645. doi: 10.1073/pnas.120163297
Dillon, S. C., and Dorman, C. J. (2010). Bacterial nucleoid-associated proteins, nucleoid structure and gene expression. Nat. Rev. Microbiol. 8, 185–195. doi: 10.1038/nrmicro2261
Duderstadt, K. E., Chuang, K., and Berger, J. M. (2011). DNA stretching by bacterial initiators promotes replication origin opening. Nature 478, 209–213. doi: 10.1038/nature10455
Emilsson, V., Näslund, A. K., and Kurland, C. G. (1993). Growth-rate-dependent accumulation of twelve tRNA species in Escherichia coli. J. Mol. Biol. 230, 483–491. doi: 10.1006/jmbi.1993.1165
Erzberger, J. P., Mott, M. L., and Berger, J. M. (2006). Structural basis for ATP-dependent DnaA assembly and replication-origin remodeling. Nat. Struct. Mol. Biol. 13, 676–683. doi: 10.1038/nsmb1115
Flåtten, I., and Morigen, Skarstad, K. (2009). DnaA protein interacts with RNA polymerase and partially protects it from the effect of rifampicin. Mol. Microbiol. 71, 1018–1030. doi: 10.1111/j.1365-2958.2008.06585.x
Frimodt-Møller, J., Charbon, G., and Løbner-Olesen, A. (2017). Control of bacterial chromosome replication by non-coding regions outside the origin. Curr. Genet. 63, 607–611. doi: 10.1007/s00294-016-0671-6
Frimodt-Møller, J., Charbon, G., Krogfelt, K. A., and Løbner-Olesen, A. (2015). Control regions for chromosome replication are conserved with respect to sequence and location among Escherichia coli strains. Front. Microbiol. 6:1011. doi: 10.3389/fmicb.2015.01011
Fujikawa, N., Kurumizaka, H., Nureki, O., Terada, T., Shirouzu, M., Katayama, T., et al. (2003). Structural basis of replication origin recognition by the DnaA protein. Nucleic Acids Res. 31, 2077–2086. doi: 10.1093/nar/gkg309
Fujimitsu, K., Senriuchi, T., and Katayama, T. (2009). Specific genomic sequences of E. coli promote replicational initiation by directly reactivating ADP-DnaA. Genes Dev. 23, 1221–1233. doi: 10.1101/gad.1775809
Grimwade, J. E., and Leonard, A. C. (2021). Blocking, bending, and binding: Regulation of initiation of chromosome replication during the Escherichia coli cell cycle by transcriptional modulators that interact with origin DNA. Front. Microbiol. 12:732270. doi: 10.3389/fmicb.2021.732270
Hansen, F. G., and Atlung, T. (2018). The DnaA Tale. Front. Microbiol. 9:319. doi: 10.3389/fmicb.2018.00319
Hayashi, C., Miyazaki, E., Ozaki, S., Abe, Y., and Katayama, T. (2020). DnaB helicase is recruited to the replication initiation complex via binding of DnaA domain I to the lateral surface of the DnaB N-terminal domain. J. Biol. Chem. 295, 1131–1143. doi: 10.1074/jbc.ra120.014235
Hilgarth, R. S., and Lanigan, T. M. (2020). Optimization of overlap extension PCR for efficient transgene construction. MethodsX 7:100759. doi: 10.1016/j.mex.2019.12.001
Hołówka, J., and Zakrzewska-Czerwińska, J. (2020). Nucleoid associated proteins: The small organizers that help to cope with stress. Front. Microbiol. 11:590. doi: 10.3389/fmicb.2020.00590
Ikemura, T., and Dahlberg, J. E. (1973). Small ribonucleic acids of Escherichia coli. II. Noncoordinate accumulation during stringent control. J. Biol. Chem. 248, 5033–5041.
Inoue, Y., Tanaka, H., Kasho, K., Fujimitsu, K., Oshima, T., and Katayama, T. (2016). Chromosomal location of the DnaA-reactivating sequence DARS2 is important to regulate timely initiation of DNA replication in Escherichia coli. Genes Cells 21, 1015–1023. doi: 10.1111/gtc.12395
Irving, S. E., Choudhury, N. R., and Corrigan, R. M. (2021). The stringent response and physiological roles of (pp)pGpp in bacteria. Nat. Rev. Microbiol. 19, 256–271. doi: 10.1038/s41579-020-00470-y
Ju, X., Li, D., and Liu, S. (2019). Full-length RNA profiling reveals pervasive bidirectional transcription terminators in bacteria. Nat. Microbiol. 4, 1907–1918. doi: 10.1038/s41564-019-0500-z
Kasho, K., and Katayama, T. (2013). DnaA binding locus datA promotes DnaA-ATP hydrolysis to enable cell cycle-coordinated replication initiation. Proc. Natl. Acad. Sci. U.S.A. 110, 936–941. doi: 10.1073/pnas.1212070110
Kasho, K., Fujimitsu, K., Matoba, T., Oshima, T., and Katayama, T. (2014). Timely binding of IHF and Fis to DARS2 regulates ATP-DnaA production and replication initiation. Nucleic Acids Res. 42, 13134–13149. doi: 10.1093/nar/gku1051
Kasho, K., Oshima, T., Chumsakul, O., Nakamura, K., Fukamachi, K., and Katayama, T. (2021). Whole-genome analysis reveals that the nucleoid protein IHF predominantly binds to the replication origin oriC specifically at the time of initiation. Front. Microbiol. 12:697712. doi: 10.3389/fmicb.2021.697712
Kasho, K., Ozaki, S., and Katayama, T. (2023). IHF and Fis as Escherichia coli cell cycle regulators: Activation of the replication origin oriC and the regulatory cycle of the DnaA initiator. Int. J. Mol. Sci. 24:11572. doi: 10.3390/ijms241411572
Kasho, K., Tanaka, H., Sakai, R., and Katayama, T. (2017). Cooperative DnaA binding to the negatively supercoiled datA locus stimulates DnaA-ATP hydrolysis. J. Biol. Chem. 292, 1251–1266. doi: 10.1074/jbc.M116.762815
Katayama, T., Kasho, K., and Kawakami, H. (2017). The DnaA cycle in Escherichia coli: Activation, function and inactivation of the initiator protein. Front. Microbiol. 8:2496. doi: 10.3389/fmicb.2017.02496
Katayama, T., Ozaki, S., Keyamura, K., and Fujimitsu, K. (2010). Regulation of the replication cycle: Conserved and diverse regulatory systems for DnaA and oriC. Nat. Rev. Microbiol. 8, 163–170. doi: 10.1038/nrmicro2314
Kawakami, H., Keyamura, K., and Katayama, T. (2005). Formation of an ATP-DnaA-specific initiation complex requires DnaA Arginine 285, a conserved motif in the AAA+ protein family. J. Biol. Chem. 280, 27420–27430. doi: 10.1074/jbc.M502764200
Kawakami, H., Ozaki, S., Suzuki, S., Nakamura, K., Senriuchi, T., Su’etsugu, M., et al. (2006). The exceptionally tight affinity of DnaA for ATP/ADP requires a unique aspartic acid residue in the AAA+ sensor 1 motif. Mol. Microbiol. 62, 1310–1324. doi: 10.1111/j.1365-2958.2006.05450.x
Keyamura, K., Abe, Y., Higashi, M., Ueda, T., and Katayama, T. (2009). DiaA dynamics are coupled with changes in initial origin complexes leading to helicase loading. J. Biol. Chem. 284, 25038–25050. doi: 10.1074/jbc.M109.002717
Kitagawa, R., Mitsuki, H., Okazaki, T., and Ogawa, T. (1996). A novel DnaA protein-binding site at 94.7 min on the Escherichia coli chromosome. Mol. Microbiol. 19, 1137–1147. doi: 10.1046/j.1365-2958.1996.453983.x
Kitagawa, R., Ozaki, T., Moriya, S., and Ogawa, T. (1998). Negative control of replication initiation by a novel chromosomal locus exhibiting exceptional affinity for Escherichia coli DnaA protein. Genes Dev. 12, 3032–3043. doi: 10.1101/gad.12.19.3032
Komine, Y., Adachi, T., Inokuchi, H., and Ozeki, H. (1990). Genomic organization and physical mapping of the transfer RNA genes in Escherichia coli K12. J. Mol. Biol. 212, 579–598. doi: 10.1016/0022-2836(90)90224-A
Miyoshi, K., Tatsumoto, Y., Ozaki, S., and Katayama, T. (2021). Negative feedback for DARS2–Fis complex by ATP–DnaA supports the cell cycle-coordinated regulation for chromosome replication. Nucleic Acids Res. 49, 12820–12835. doi: 10.1093/nar/gkab1171
Molan, K., and Žgur Bertok, D. (2022). Small prokaryotic DNA-binding proteins protect genome integrity throughout the life cycle. Int. J. Mol. Sci. 23:4008. doi: 10.3390/ijms23074008
Nilsson, L., Verbeek, H., Vijgenboom, E., van Drunen, C., Vanet, A., and Bosch, L. (1992). FIS-dependent trans activation of stable RNA operons of Escherichia coli under various growth conditions. J. Bacteriol. 174, 921–929. doi: 10.1128/jb.174.3.921-929.1992
Nishida, S., Fujimitsu, K., Sekimizu, K., Ohmura, T., Ueda, T., and Katayama, T. (2002). A nucleotide switch in the Escherichia coli DnaA protein initiates chromosomal replication: Evidnece from a mutant DnaA protein defective in regulatory ATP hydrolysis in vitro and in vivo. J. Biol. Chem. 277, 14986–14995. doi: 10.1074/jbc.M108303200
Noguchi, Y., Sakiyama, Y., Kawakami, H., and Katayama, T. (2015). The Arg fingers of key DnaA protomers are oriented inward within the replication origin oriC and stimulate DnaA subcomplexes in the initiation complex. J. Biol. Chem. 290, 20295–20312. doi: 10.1074/jbc.M115.662601
Nozaki, S., and Ogawa, T. (2008). Determination of the minimum domain II size of Escherichia coli DnaA protein essential for cell viability. Microbiology 154, 3379–3384. doi: 10.1099/mic.0.2008/019745-0
Nozaki, S., Yamada, Y., and Ogawa, T. (2009). Initiator titration complex formed at datA with the aid of IHF regulates replication timing in Escherichia coli. Genes Cells 14, 329–341. doi: 10.1111/j.1365-2443.2008.01269.x
Okumura, H., Yoshimura, M., Ueki, M., Oshima, T., Ogasawara, N., and Ishikawa, S. (2012). Regulation of chromosomal replication initiation by oriC-proximal DnaA-box clusters in Bacillus subtilis. Nucleic Acids Res. 40, 220–234. doi: 10.1093/nar/gkr716
Orosz, A., Boros, I., and Venetianer, P. (1991). Analysis of the complex transcription termination region of the Escherichia coli rrnB gene. Eur. J. Biochem. 201, 653–659. doi: 10.1111/j.1432-1033.1991.tb16326.x
Ozaki, S., Kawakami, H., Nakamura, K., Fujikawa, N., Kagawa, W., Park, S.-Y., et al. (2008). A common mechanism for the ATP-DnaA-dependent formation of open complexes at the replication origin. J. Biol. Chem. 283, 8351–8362. doi: 10.1074/jbc.M708684200
Pagel, J. M., and Hatfield, G. W. (1991). Integration host factor-mediated expression of the ilvGMEDA operon of Escherichia coli. J. Biol. Chem. 266, 1985–1996.
Prieto, A. I., Kahramanoglou, C., Ali, R. M., Fraser, G. M., Seshasayee, A. S. N., and Luscombe, N. M. (2012). Genomic analysis of DNA binding and gene regulation by homologous nucleoid-associated proteins IHF and HU in Escherichia coli K12. Nucleic Acids Res. 40, 3524–3537. doi: 10.1093/nar/gkr1236
Sakiyama, Y., Kasho, K., Noguchi, Y., Kawakami, H., and Katayama, T. (2017). Regulatory dynamics in the ternary DnaA complex for initiation of chromosomal replication in Escherichia coli. Nucleic Acids Res. 45, 12354–12373. doi: 10.1093/nar/gkx914
Schaefer, C., and Messer, W. (1988). Termination of the Escherichia coli asnC transcript. The DnaA protein/dnaA box complex blocks transcribing RNA polymerase. Gene 73, 347–354. doi: 10.1016/0378-1119(88)90499-4
Schaefer, C., and Messer, W. (1989). Directionality of DnaA protein/DNA interaction. Active orientation of the DnaA protein/dnaA box complex in transcription termination. EMBO J. 8, 1609–1613. doi: 10.1002/j.1460-2075.1989.tb03545.x
Shimizu, M., Noguchi, Y., Sakiyama, Y., Kawakami, H., Katayama, T., and Takada, S. (2016). Near-atomic structural model for bacterial DNA replication initiation complex and its functional insights. Proc. Natl. Acad. Sci. U.S.A. 113, E8021–E8030. doi: 10.1073/pnas.1609649113
Smulczyk-Krawczyszyn, A., Jakimowicz, D., Ruban-Osmialowska, B., Zawilak-Pawlik, A., Majka, J., Chater, K., et al. (2006). Cluster of DnaA boxes involved in regulation of Streptomyces chromosome replication: From in silico to in vivo studies. J. Bacteriol. 188, 6184–6194. doi: 10.1128/JB.00528-06
Su’etsugu, M., Emoto, A., Fujimitsu, K., Keyamura, K., and Katayama, T. (2003). Transcriptional control for initiation of chromosomal replication in Escherichia coli: Fluctuation of the level of origin transcription ensures timely initiation. Genes Cells 8, 731–745. doi: 10.1046/j.1365-2443.2003.00671.x
Sugiyama, R., Kasho, K., Miyoshi, K., Ozaki, S., Kagawa, W., Kurumizaka, H., et al. (2019). A novel mode of DnaA-DnaA interaction promotes ADP dissociation for reactivation of replication initiation activity. Nucleic Acids Res. 47, 11209–11224. doi: 10.1093/nar/gkz795
Sutton, M. D., Carr, K. M., Vicente, M., and Kaguni, J. M. (1998). Escherichia coli DnaA protein. The N-terminal domain and loading of DnaB helicase at the E. coli chromosomal origin. J. Biol. Chem. 273, 34255–34262. doi: 10.1074/jbc.273.51.34255
Theisen, P. W., Grimwade, J. E., Leonard, A. C., Bogan, J. A., and Helmstetter, C. E. (1993). Correlation of gene transcription with the time of initiation of chromosome replication in Escherichia coli. Mol. Microbiol. 10, 575–584. doi: 10.1111/j.1365-2958.1993.tb00929.x
Watson, G. D., Chan, E. W., Leake, M. C., and Noy, A. (2022). Structural interplay between DNA-shape protein recognition and supercoiling: The case of IHF. Comput. Struct. Biotechnol. J. 20, 5264–5274. doi: 10.1016/j.csbj.2022.09.020
Wellner, K., Czech, A., Ignatova, Z., Betat, H., and Mörl, M. (2018). Examining tRNA 3’-ends in Escherichia coli: Teamwork between CCA-adding enzyme, RNase T, and RNase R. RNA 24, 361–370. doi: 10.1261/rna.064436.117
Withers, H. L., and Bernander, R. (1998). Characterization of dnaC2 and dnaC28 mutants by flow cytometry. J. Bacteriol. 180, 1624–1631.
Yao, Y., Sun, H., Wurihan, Gegeheng, Gezi, Skarstad, K., et al. (2023). A DnaA-dependent riboswitch for transcription attenuation of the his operon. mLife 2, 126–140. doi: 10.1002/mlf2.12075
Yoshida, R., Ozaki, S., Kawakami, H., and Katayama, T. (2023). Single-stranded DNA recruitment mechanism in replication origin unwinding by DnaA initiator protein and HU, an evolutionary ubiquitous nucleoid protein. Nucleic Acids Res. 51, 6286–6306. doi: 10.1093/nar/gkad389
Keywords: E. coli, DnaA, datA, IHF, tRNA transcription, cell cycle
Citation: Kasho K, Sakai R, Ito K, Nakagaki W, Satomura R, Jinnouchi T, Ozaki S and Katayama T (2024) Read-through transcription of tRNA underlies the cell cycle-dependent dissociation of IHF from the DnaA-inactivating sequence datA. Front. Microbiol. 15:1360108. doi: 10.3389/fmicb.2024.1360108
Received: 22 December 2023; Accepted: 05 February 2024;
Published: 28 February 2024.
Edited by:
Feng Gao, Tianjin University, ChinaReviewed by:
Morigen Morigen, Inner Mongolia University, ChinaAnders Løbner-Olesen, University of Copenhagen, Denmark
Ian Grainge, The University of Newcastle, Australia
Copyright © 2024 Kasho, Sakai, Ito, Nakagaki, Satomura, Jinnouchi, Ozaki and Katayama. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Kazutoshi Kasho, a2F6dXRvc2hpLmthc2hvQHBoYXIua3l1c2h1LXUuYWMuanA=
†These authors have contributed equally to this work
 Kazutoshi Kasho
Kazutoshi Kasho Ryuji Sakai†
Ryuji Sakai† Shogo Ozaki
Shogo Ozaki Tsutomu Katayama
Tsutomu Katayama