- 1The Third Affiliated Hospital of Zhejiang Chinese Medical University (Zhongshan Hospital of Zhejiang Province), Hangzhou, Zhejiang, China
- 2The Third School of Clinical Medicine (School of Rehabilitation Medicine), Zhejiang Chinese Medical University, Hangzhou, Zhejiang, China
- 3Hangzhou Hospital of Traditional Chinese Medicine, Zhejiang Chinese Medical University, Hangzhou, Zhejiang, China
- 4Research Institute of Tuina (Spinal disease), Zhejiang Chinese Medical University, Hangzhou, Zhejiang, China
Background: Observational studies have hinted at a correlation between the gut microbiota and spinal pain (SP). However, the impact of the gut microbiota on SP remains inconclusive.
Methods: In this study, we employed a two-sample Mendelian randomization (MR) analysis to explore the causal relationship between the gut microbiota and SP, encompassing neck pain (NP), thoracic spine pain (TSP), low back pain (LBP), and back pain (BP). The compiled gut microbiota data originated from a genome-wide association study (GWAS) conducted by the MiBioGen consortium (n = 18,340). Summary data for NP were sourced from the UK Biobank, TSP from the FinnGen Biobank, and LBP from both the UK Biobank and FinnGen Biobank. Summary data for BP were obtained from the UK Biobank. The primary analytical approach for assessing causal relationships was the Inverse Variance Weighted (IVW) method, supplemented by various sensitivity analyses to ensure result robustness.
Results: The IVW analysis unveiled 37 bacterial genera with a potential causal relationship to SP. After Benjamini-Hochberg corrected test, four bacterial genera emerged with a strong causal relationship to SP. Specifically, Oxalobacter (OR: 1.143, 95% CI 1.061–1.232, P = 0.0004) and Tyzzerella 3 (OR: 1.145, 95% CI 1.059–1.238, P = 0.0007) were identified as risk factors for LBP, while Ruminococcaceae UCG011 (OR: 0.859, 95% CI 0.791–0.932, P = 0.0003) was marked as a protective factor for LBP, and Olsenella (OR: 0.893, 95% CI 0.839–0.951, P = 0.0004) was recognized as a protective factor for low back pain or/and sciatica. No significant heterogeneity or horizontal pleiotropy was observed through alternative testing methods.
Conclusion: This study establishes a causal relationship between the gut microbiota and SP, shedding light on the “gut-spine” axis. These findings offer novel perspectives for understanding the etiology of SP and provide a theoretical foundation for potential interventions targeting the gut microbiota to prevent and treat SP.
1 Introduction
Spinal pain (SP) manifests as a prevalent symptom in various spine-related disorders, encompassing neck pain (NP) and back pain (BP), further categorized into thoracic spine pain (TSP) and low back pain (LBP). The prevalence in the general population is noteworthy, with NP affecting approximately 44%, TSP 15%, and LBP 56% (Manchikanti et al., 2004). The impact of SP on daily life and occupational functions surpasses that of other musculoskeletal disorders due to the spine’s role as the central axis and core support of the human skeleton (Haldeman et al., 2012). The 2015 Global Burden of Disease study underscored SP as a paramount contributor to disability globally, especially affecting individuals aged 25–64 (GBD 2015 Disease and Injury Incidence and Prevalence Collaborators, 2016). Consequently, healthcare costs for SP treatment soared, reaching an estimated $134.5 billion in the United States in 2020 (de Luca et al., 2023). The pressing need for effective prevention and treatment strategies for SP is a critical concern in the realm of global public health.
The gut microbiota, comprising over 10 trillion microorganisms, exists harmoniously within the human intestinal tract, outnumbering human cells tenfold (Clark and Coopersmith, 2007; Zhang et al., 2015). The colon, serving as the primary habitat, hosts an astounding 10∧11–10∧12 microorganisms per milliliter (Ley et al., 2006). This rich, diverse, and stable microbiota plays a crucial role in sustaining various physiological functions, including nutrient metabolism, xenobiotic and drug metabolism, immunomodulation, and antimicrobial defense (Jandhyala et al., 2015). Mounting evidence links ecological destabilization of the gut microbiota to the development of various diseases, including intestinal (Quaglio et al., 2022), hepatic (Chassaing et al., 2014), metabolic (Lin et al., 2020), allergic (Akagawa and Kaneko, 2022), and autoimmune diseases (Yang et al., 2023).
Recent studies propose a potential relationship between the gut microbiota and the development of SP. Patients with NP exhibited higher levels of Bacteroidetes and Proteobacteria and significantly lower levels of Firmicutes compared to healthy controls (Qin, 2022). Similarly, individuals with BP showed a distinct gut microbiota composition, characterized by elevated concentrations of Adlercreutzia, Roseburia, and Uncl. Christensenellaceae (Dekker Nitert et al., 2020). These findings underscore a compelling relationship between gut microbiota and SP. While degenerative spinal diseases traditionally bear the blame for SP, a notable shift toward a younger demographic experiencing SP suggests alternative causative factors beyond structural spinal degeneration (Lei et al., 2015). Consequently, gut microbiota dysbiosis emerges as a significant potential contributor to SP.
In light of these observations, MR offers a promising avenue for exploring causal relationships. MR, first proposed by Smith and Ebrahim (2003), utilizes genetic variation as an instrumental variable, enabling the elimination of confounding bias and reverse causation (Emdin et al., 2017; Soremekun et al., 2022). In this study, we collected data on gut microbiota and SP from the genome-wide association study (GWAS) database, employing MR analysis to discern the causal relationship between them. The study aims to provide innovative insights for the prevention and treatment of SP by unraveling the intricate interplay between gut microbiota dynamics and SP.
2 Materials and methods
2.1 Study design
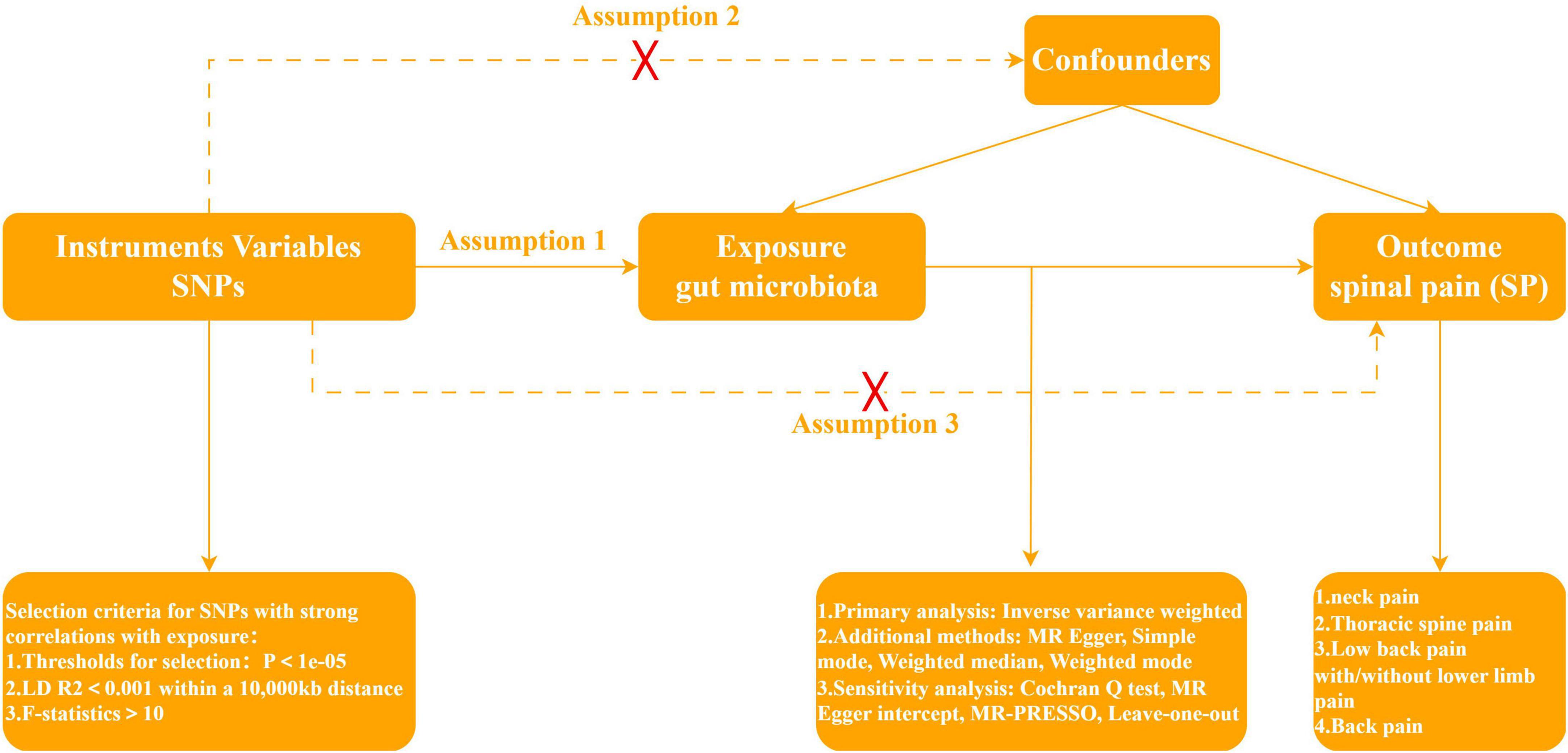
Mendelian Randomization, a pivotal method for evaluating causal relationships between exposure and outcome, stands as the cornerstone of our investigative framework. In quest of discerning whether gut microbiota assumes a protective or facilitative role in SP development, our study strategically designates gut microbiota as the exposure variable, with four SP types—NP, TSP, LBP, and BP—comprising the outcome variables. According to Davies et al. (2018), the implementation of a two-sample MR protocol necessitated strict adherence to three fundamental assumptions. First, the instrumental variables, genetic variation, should exhibit a robust relationship with gut microbiota. Second, genetic variation must remain unlinked to confounding factors, excluding those pertaining to gut microbiota and the four SP types. Third, the effects of genetic variation on the four SP types must be channeled through a singular pathway—namely, gut microbiota—with the exclusion of any alternative pathways. In Figure 1, the specific flowchart detailing the MR analysis is presented.
2.2 Exposure data
The summary statistics for gut microbiota utilized in this investigation originated from a GWAS conducted by the MiBioGen consortium in 2021 (Kurilshikov et al., 2021). The study encompassed a substantial cohort of 18,340 individuals, with over 70% representing European ancestry (n = 13,266). Within this cohort, participants were stratified into 24 distinct groups based on factors such as background, sex, and age. The composition of their fecal microbiota was meticulously examined through the application of 16S rRNA gene sequencing. This comprehensive analysis led to the identification of 211 taxa, spanning across 131 genera, 35 families, 20 orders, 16 classes, and 9 phyla. Recognizing the significance of genus-level precision in characterizing the microbiome, we opted to exclusively retain data at the genus level for subsequent phases of our study and detailed analysis. This strategic decision ensures a more granular and accurate exploration of each potentially pathogenic microbiome.
2.3 Outcome data
All GWAS summary statistics pertaining to SP were meticulously sourced from the EU Open GWAS platform.1 This comprehensive repository comprises data packages for the four primary types of SP: NP, TSP, LBP, and BP. After a thorough review of pertinent datasets, eight were judiciously selected for our analysis. Specifically, two GWAS summary statistics for NP, considering disease durations of 1 month and 3+ months, were incorporated into our study, both sourced from the UK Biobank (ukb-e). The GWAS summary statistics for TSP were obtained from the FinnGen Biobank. Similarly, the GWAS summary statistics for LBP were procured from the FinnGen Biobank. Recognizing that patients with LBP often exhibit radiating pain in the lower limbs (Koes et al., 2006), we expanded our dataset to include two GWAS summary statistics: one for low back pain with radicular pain from the UK Biobank (ukb-e) and another for low back pain with sciatica from the FinnGen Biobank. For the BP category, two GWAS summary statistics for BP, comparing disease durations of 1 month versus 3+ months, were selected from the UK Biobank (ukb-e). For a detailed breakdown of outcome summary statistics, please refer to Supplementary Table 1.
2.4 Instrumental variable (IV)
In order to ensure the precision and robustness of the study outcomes, we incorporated single nucleotide polymorphisms (SNPs) demonstrating robust relationships with the exposure factors. While the conventional threshold for controlling the false positive rate is typically set at P < 5e−08, this study opted for a more inclusive threshold of P < 1e−05 (Xie et al., 2023). This adjustment was made to augment the pool of screened SNPs, thereby enhancing result credibility, given the limited number of SNPs obtained under the more stringent condition. Subsequently, to acquire mutually independent instrumental variables, SNPs within a 10,000 kb clumping window were excluded to mitigate Linkage Disequilibrium (LD), specifically retaining SNPs with an R2 < 0.001. Concurrently, F-statistics of the SNPs were computed, leading to the exclusion of weak instrumental variables characterized by F < 10 (Burgess and Thompson, 2011). Finally, a meticulous harmonization process was applied to align the effector alleles of the exposure and outcome SNPs. This involved the exclusion of alleles that were incompatible (e.g., A/C paired with A/G) or exhibited intermediate allele frequencies.
2.5 Statistical analysis
The causal relationship between the gut microbiota and SP was primarily analyzed using the Inverse Variance Weighted (IVW) method. A potential causal relationship between the gut microbiota and SP was considered when the IVW result yielded a P < 0.05. Following multiple testing correction of the IVW results using the Benjamini-Hochberg False Discovery Rate (FDR) method, gut microbiota with P < 0.05 were deemed to exhibit a significant causal relationship with SP. However, the validity of this conclusion relies on the absence of horizontal pleiotropy and heterogeneity (Burgess et al., 2016). Therefore, various additional methods were incorporated to scrutinize the potential presence or absence of horizontal pleiotropy and heterogeneity.
Initially, Cochrane’s Q test was employed to assess heterogeneity in the IVs. A significance level of P < 0.05 denoted heterogeneity, necessitating the use of the random-effects IVW model; conversely, the fixed-effects IVW model was applied (Greco et al., 2015). In the IVW regression, where the intercept term is not considered, results become inaccurate in the presence of horizontal pleiotropy (Burgess et al., 2017). To address this, the MR-Egger regression method and the Weighted Median (WM) method were employed to correct for horizontal pleiotropy. MR-Egger assumes that the IVs of horizontal pleiotropy is greater than 50%, while WM assumes it to be less than 50%, both re-estimating results accordingly (Bowden et al., 2016). Both methods complemented the IVW method, correcting for bias introduced by horizontal pleiotropy and enhancing result credibility (Bowden et al., 2015; Slob and Burgess, 2020). MR-Egger and MR Pleiotropy RESidual Sum and Outlier (MR-PRESSO) were employed to test horizontal pleiotropy. An MR-Egger P-value of less than 0.05 indicated a non-zero intercept, signifying the presence of horizontal pleiotropy. In addition to testing horizontal pleiotropy, MR-PRESSO identified and corrected for outliers, thus improving the accuracy of the analysis results. Finally, a leave-one-out sensitivity analysis was conducted to assess the stability of the MR results and detect the presence of outliers in the SNPs.
All the aforementioned statistical analyses were performed using the R program (version 4.3.1), with the “VariantAnnotation,” “mrcieu/gwasglue,” “mrcieu/ieugwasr,” and “MRCIEU/TwoSampleMR” packages loaded and executed. The specific MR analysis codes are detailed in the Supplementary material.
3 Results
3.1 SNP selection
From a pool of 119 bacterial genera, we meticulously screened and identified 1531 SNPs to serve as IVs in our analysis. Crucially, all of these SNPs exhibited F-statistics surpassing 10, with values ranging from 14.58 to 88.42. This range signifies a robust F-statistic profile, indicating the absence of bias arising from weak IVs. This stringent selection process ensures the reliability and strength of the IVs utilized in our study.
3.2 Causal influence of gut microbiota on the risk of spinal pain
3.2.1 Causal influence of gut microbiota on the risk of neck pain
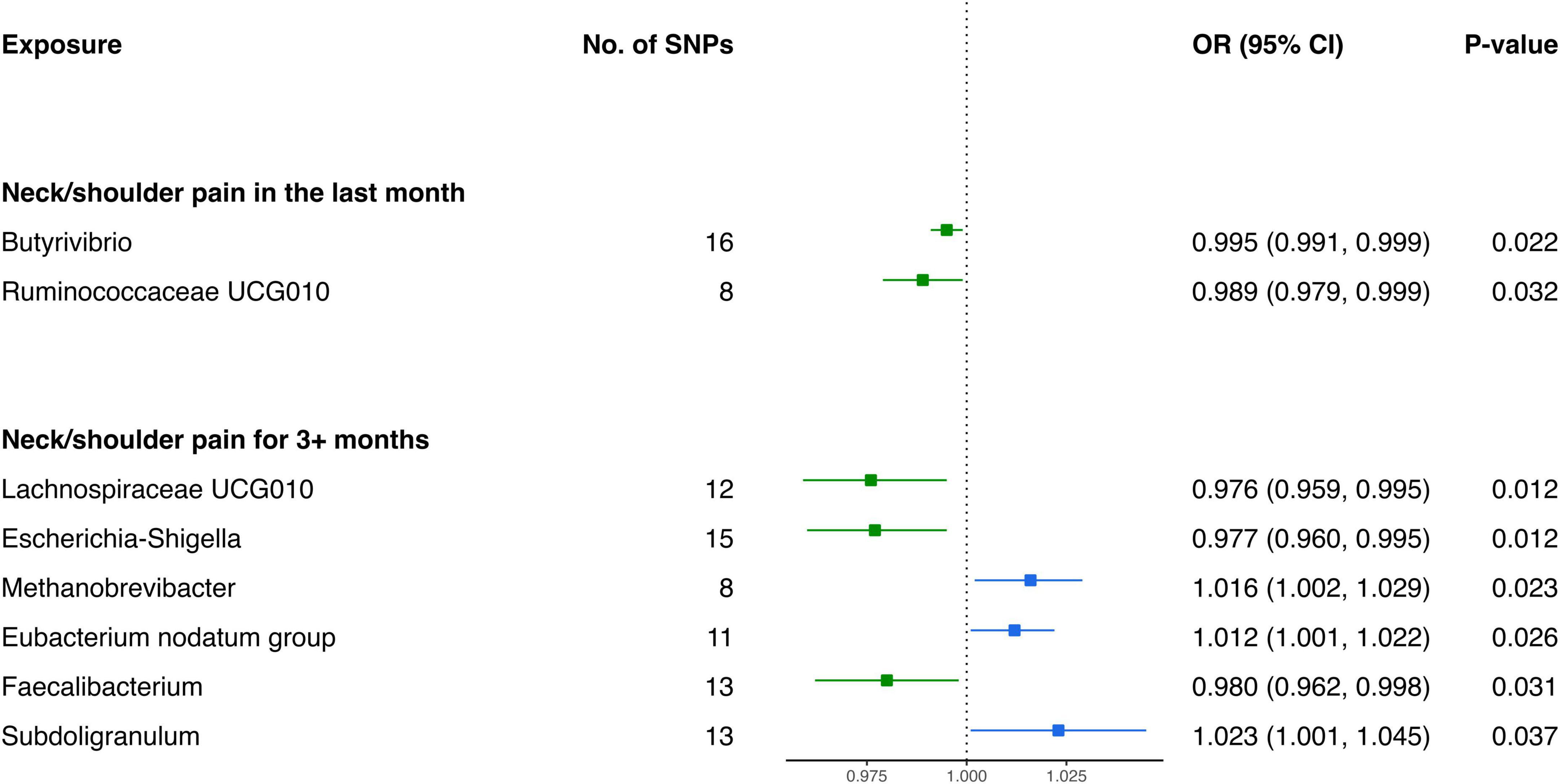
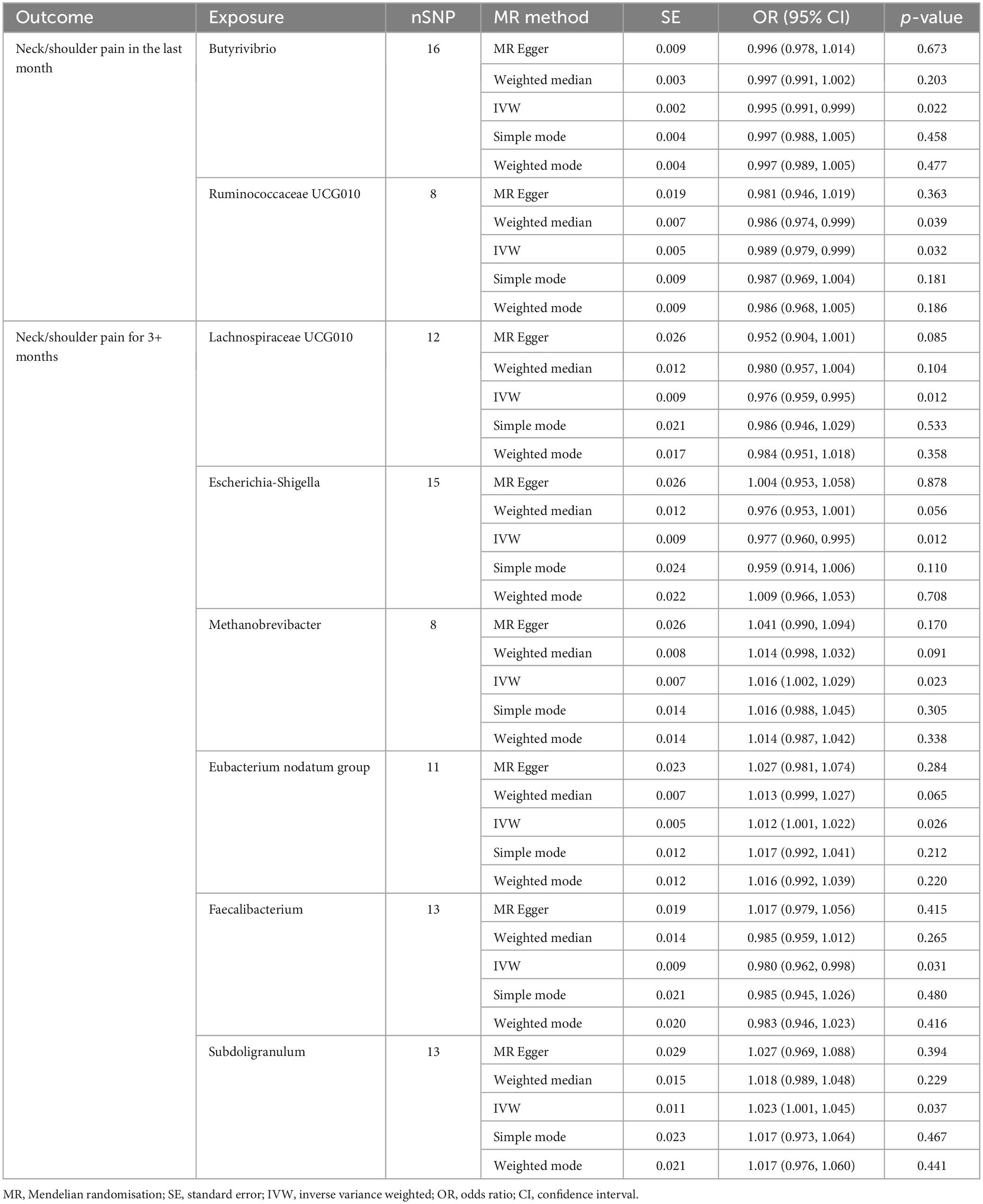
Based on the outcomes of the IVW analysis, we identified two bacterial genera demonstrating protective effects against NP in the last month (Figure 2 and Table 1): Ruminococcaceae UCG010 (OR: 0.989, 95% CI 0.979–0.999, P = 0.032) and Butyrivibrio (OR: 0.995, 95% CI 0.991–0.999, P = 0.022).
Furthermore, the IVW estimates indicated that Methanobrevibacter (OR: 1.016, 95% CI 1.002–1.029, P = 0.023), Eubacterium nodatum group (OR: 1.012, 95% CI 1.001–1.022, P = 0.026), and Subdoligranulum (OR: 1.023, 95% CI 1.001–1.045, P = 0.037) were positively associated with the risk of NP for 3+ months (Figure 2). In contrast, the genetically predicted abundance of Escherichia-Shigella (OR: 0.977, 95% CI 0.960–0.995, P = 0.012), Faecalibacterium (OR: 0.980, 95% CI 0.962–0.998, P = 0.031), and Lachnospiraceae UCG010 (OR: 0.976, 95% CI 0.959–0.995, P = 0.012) exhibited a negative association with the risk of NP for 3+ months (Table 1).
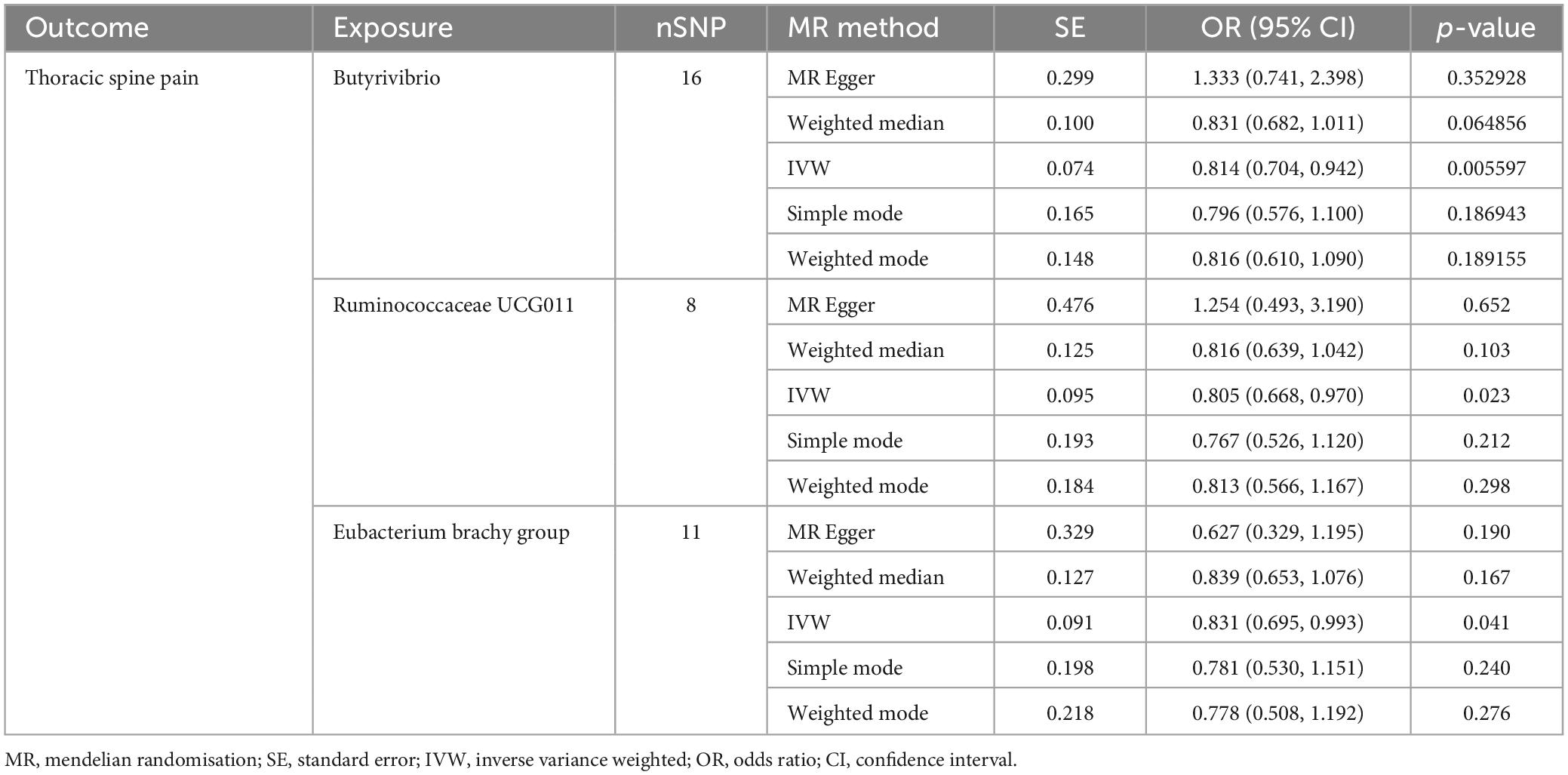
3.2.2 Causal influence of gut microbiota on the risk of thoracic spine pain
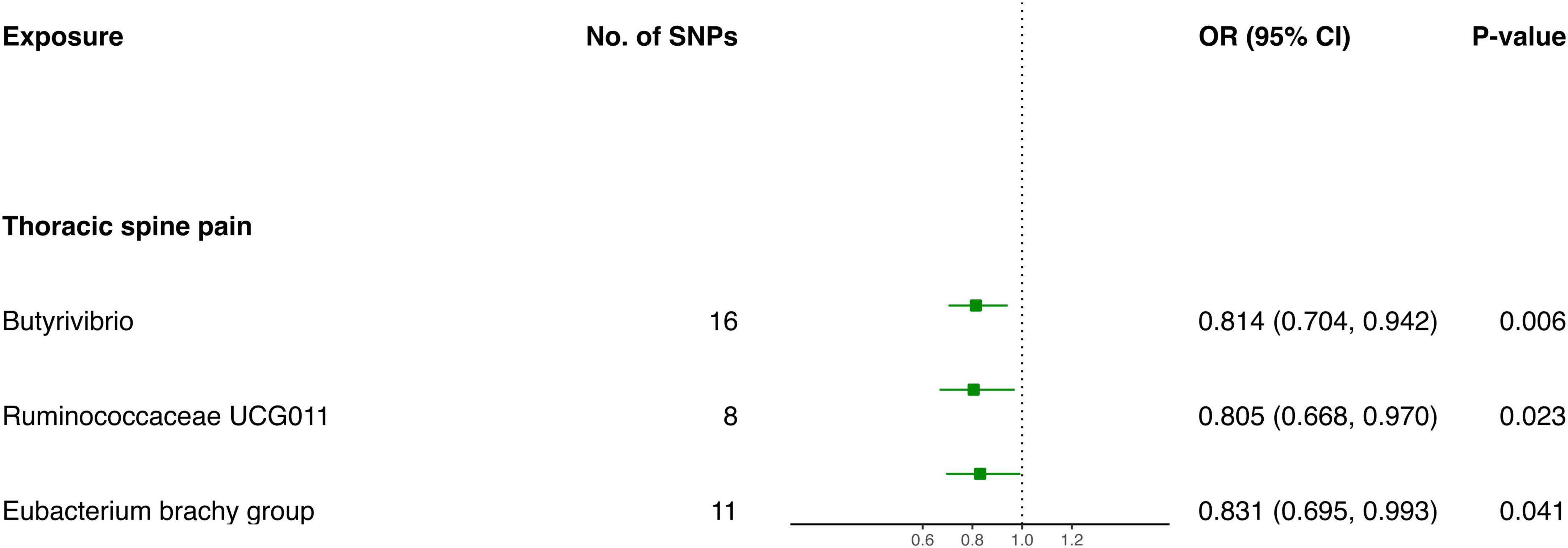
The IVW screening revealed three bacterial genera exhibiting a causal relationship with TSP (Figure 3 and Table 2). The outcomes indicated a genetically predicted low relative abundance for Butyrivibrio (OR: 0.814, 95% CI 0.704–0.942, P = 0.006), Ruminococcaceae UCG011 (OR: 0.805, 95% CI 0.668–0.970, P = 0.023), and Eubacterium brachy group (OR: 0.831, 95% CI 0.695–0.993, P = 0.041), which were associated with a reduced risk of TSP.
3.2.3 Causal influence of gut microbiota on the risk of low back pain
The results of the IVW analysis delineate distinct associations between bacterial genera and the risk of LBP. Genetically predicted Oxalobacter (OR: 1.143, 95% CI 1.061–1.232, P < 0.001) and Tyzzerella 3 (OR: 1.145, 95% CI 1.059–1.238, P < 0.001) were found to be correlated with an elevated risk of LBP (Figure 4). In contrast, Ruminococcaceae UCG011 (OR: 0.859, 95% CI 0.791–0.932, P < 0.001), Olsenella (OR: 0.894, 95% CI 0.829–0.963, P = 0.003), Eisenbergiella (OR: 0.882, 95% CI 0.810–0.960, P = 0.004), and Roseburia (OR: 0.845, 95% CI 0.741–0.963, P = 0.011) were associated with a reduced risk of LBP (Table 3).
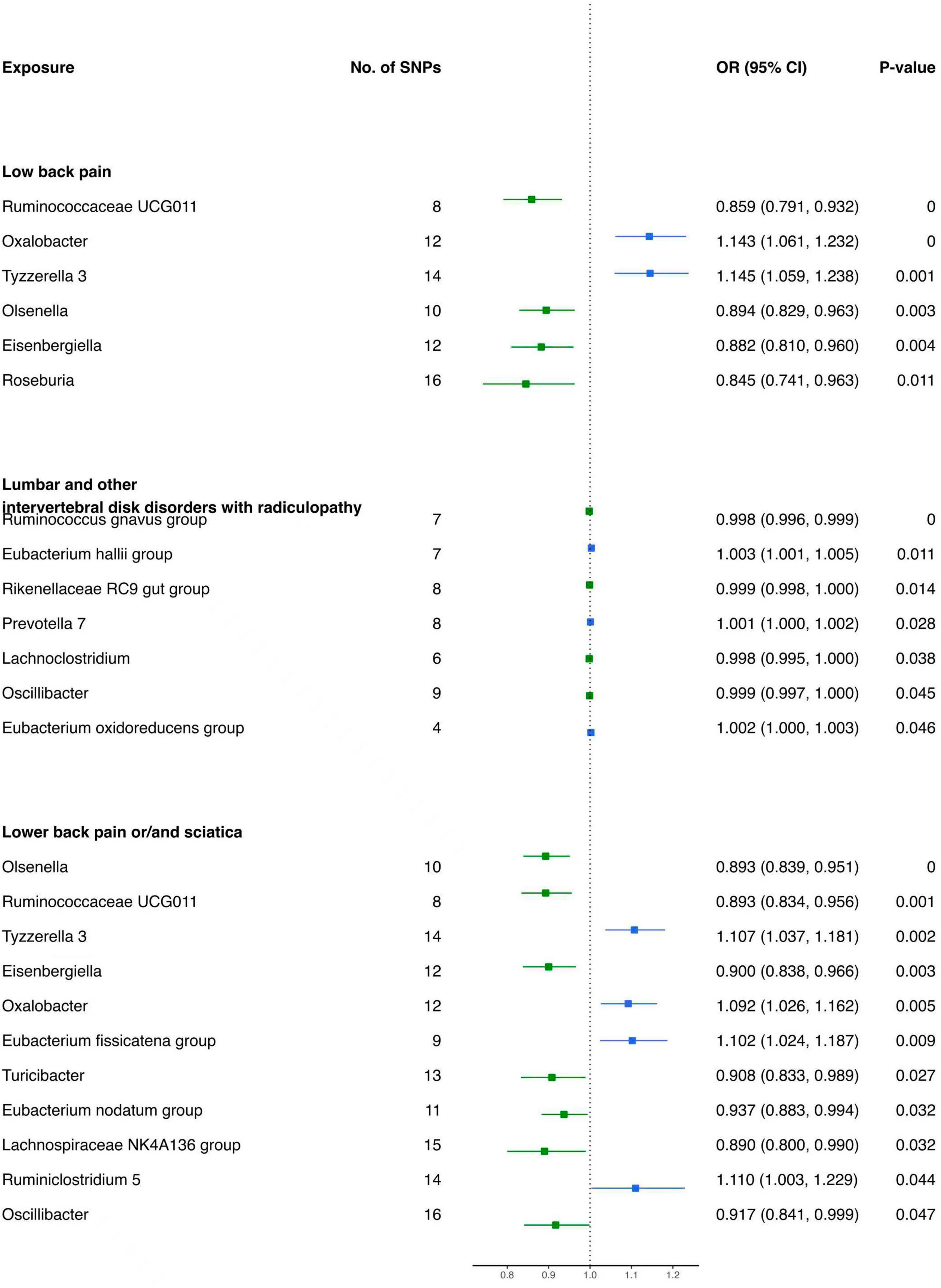
Figure 4. Forest plot of causality between gut microbiota and risk of low back pain with/without lower limb pain.
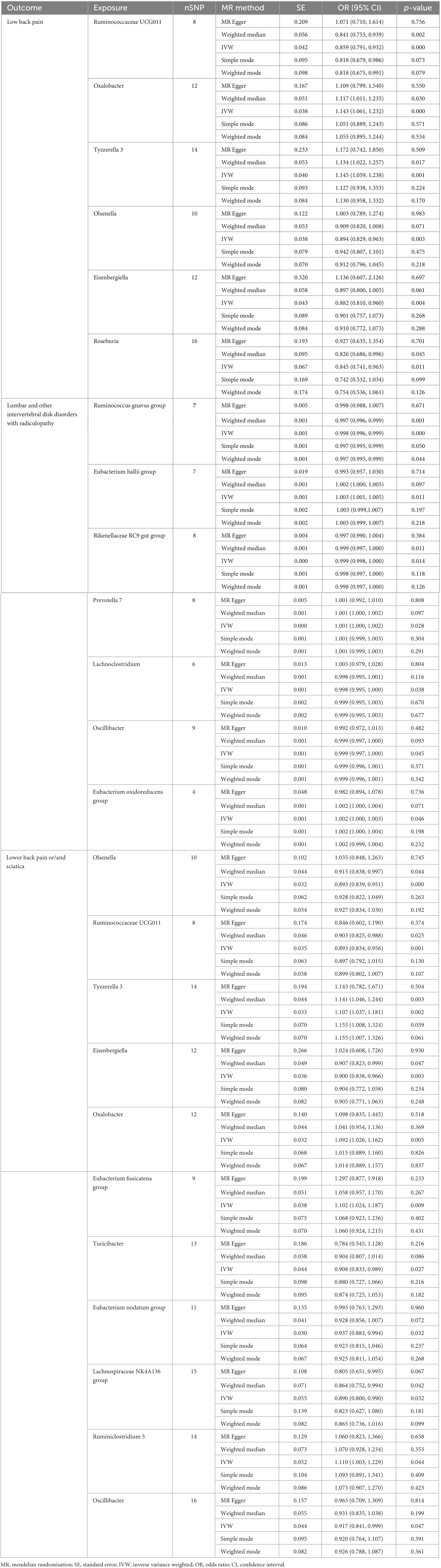
Table 3. MR results of causal effects between gut microbiome and low back pain with/without lower limb pain.
For lumbar and other intervertebral disk disorders with radiculopathy, specific bacterial genera demonstrated distinct associations. Prevotella 7 (OR: 1.001, 95% CI 1.000–1.002, P = 0.028), Eubacterium hallii group (OR: 1.003, 95% CI 1.001–1.005, P = 0.011), and Eubacterium oxidoreducens group (OR: 1.002, 95% CI 1.000–1.003, P = 0.046) were positively associated with the risk (Figure 4). Conversely, Lachnoclostridium (OR: 0.998, 95% CI 0.995–1.000, P = 0.038), Ruminococcus gnavus group (OR: 0.998, 95% CI 0.996–0.999, P < 0.001), Rikenellaceae RC9 gut group (OR: 0.999, 95% CI 0.998–1.000, P = 0.014), and Oscillibacter (OR: 0.999, 95% CI 0.997–1.000, P = 0.045) were negatively associated with the risk of these disorders (Table 3).
Moreover, for low back pain or/and sciatica, Tyzzerella 3 (OR: 1.107, 95% CI 1.037–1.181, P = 0.002), Oxalobacter (OR: 1.092, 95% CI 1.026–1.162, P = 0.005), Eubacterium fissicatena group (OR. 1.102, 95% CI 1.024–1.187, P = 0.009), and Ruminiclostridium 5 (OR: 1.110, 95% CI 1.003–1.229, P = 0.044) were identified as having contributory effects (Figure 4). Conversely, Olsenella (OR: 0.893, 95% CI 0.839–0.951, P < 0.001), Ruminococcaceae UCG011 (OR: 0.893, 95% CI 0.834–0.956, P = 0.001), Eisenbergiella (OR: 0.900, 95% CI 0.838–0.966, P = 0.003), Turicibacter (OR: 0.908, 95% CI 0.833–0.989, P = 0.027), Eubacterium nodatum group (OR: 0.937, 95% CI 0.883–0.994, P = 0.032), Lachnospiraceae NK4A136 group (OR: 0.890, 95% CI 0.800–0.990, P = 0.032), and Oscillibacter (OR: 0.917, 95% CI 0.841–0.999, P = 0.047) were found to be protective (Table 3).
3.2.4 Causal influence of gut microbiota on the risk of back pain
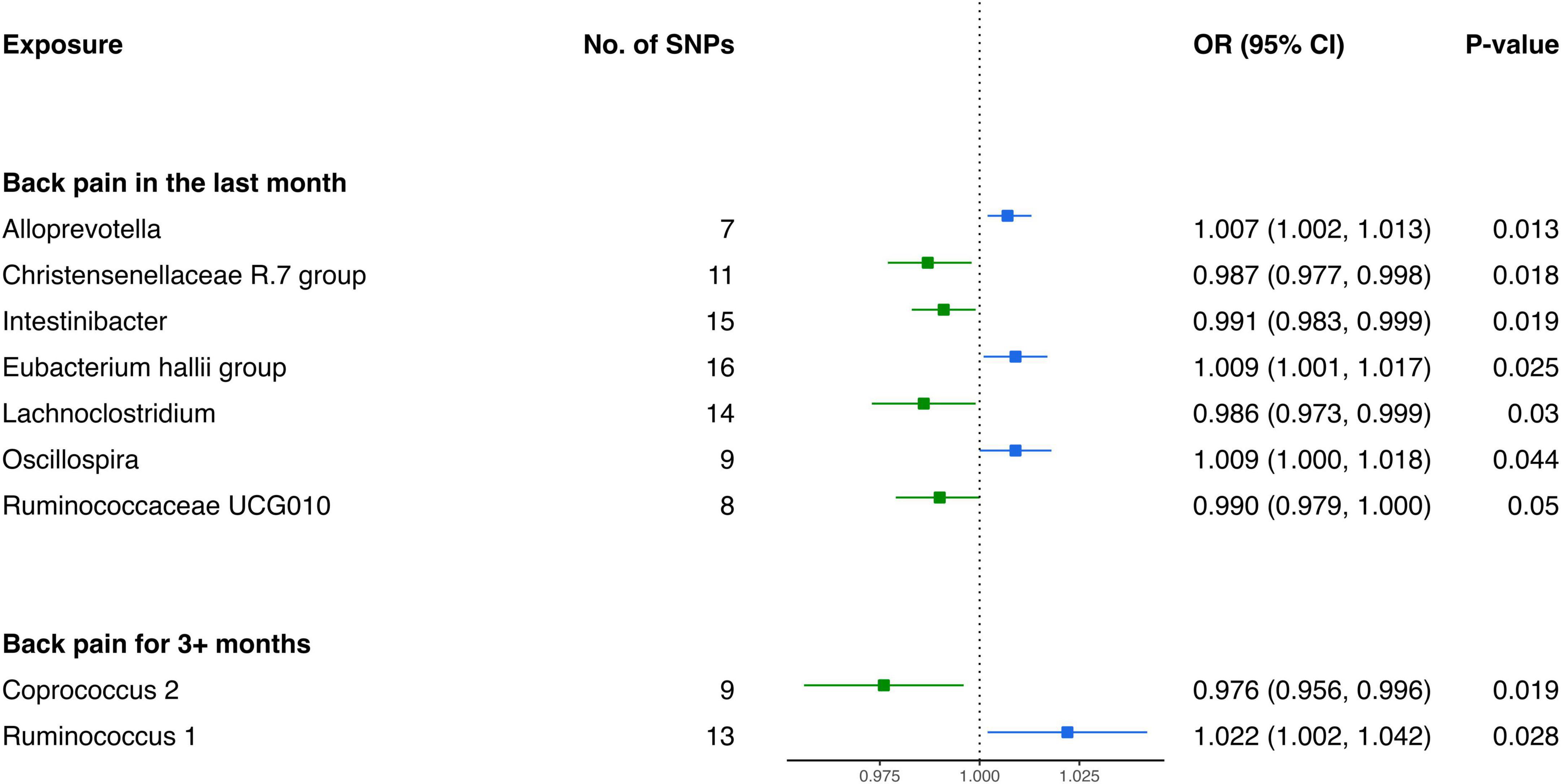
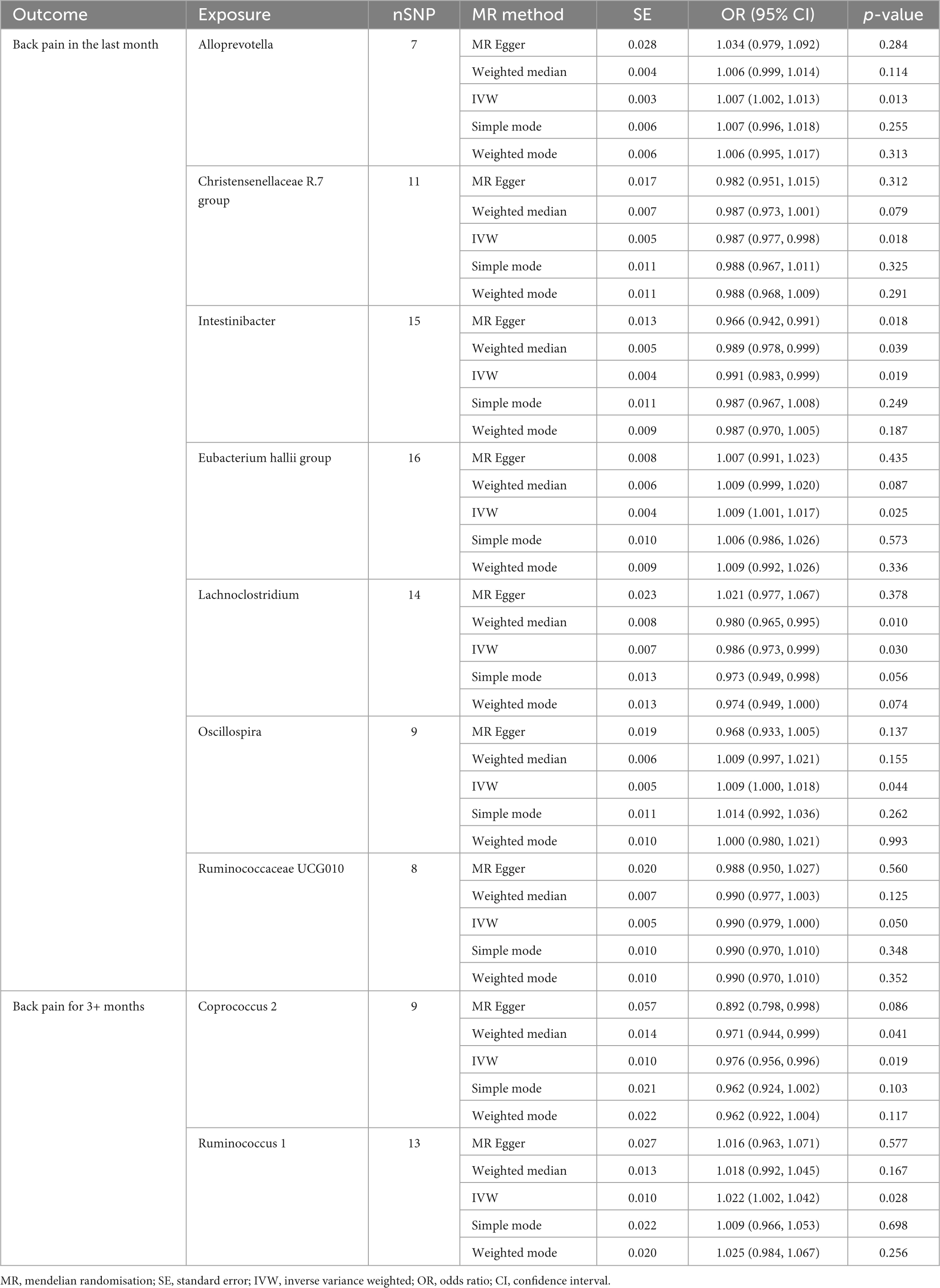
Analysis from the IVW test has revealed distinctive associations between specific bacterial genera and the risk of BP across varying durations. For BP in the last month, genetically predicted relative abundance of Alloprevotella (OR: 1.007, 95% CI 1.002–1.013, P = 0.013), Oscillospira (OR: 1.009, 95% CI 1.000–1.018, P = 0.044), and Eubacterium hallii group (OR: 1.009, 95% CI 1.001–1.017, P = 0.025) exhibited positive relationships with increased risk (Figure 5). Contrarily, Christensenellaceae R.7 group (OR: 0.987, 95% CI 0.977–0.998, P = 0.018), Intestinibacter (OR: 0.991, 95% CI 0.983–0.999, P = 0.019), Ruminococcaceae UCG010 (OR: 0.990, 95% CI 0.979–1.000, P = 0.050), and Lachnoclostridium (OR: 0.986, 95% CI 0.973–0.999, P = 0.030) were inversely associated with the risk of BP in the last month (Table 4).
Furthermore, in the context of BP for 3+ months, two bacterial genera exhibited causal relationships. The genetically predicted relative abundance of Ruminococcus 1 (OR: 1.022, 95% CI 1.002–1.042, P = 0.028) was linked to an increased risk of BP for 3+ months. Conversely, Coprococcus 2 (OR: 0.976, 95% CI 0.956–0.996, P = 0.019) demonstrated a negative association, suggesting a potential protective effect against BP for 3+ months.
3.2.5 Sensitivity analysis and Benjamini–Hochberg corrected test
The results of our MR analysis were subjected to a comprehensive set of sensitivity tests to ensure the robustness and reliability of the findings. We employed the IVW test, MR-Egger’s intercept test, and the MR-PRESSO global test (Supplementary Table 10). Importantly, the vast majority of IVW test, relying on Cochran’s Q test, demonstrated a lack of heterogeneity (P > 0.05), indicating consistency across our analyses. Furthermore, neither MR-Egger’s intercept test nor the MR-PRESSO global test detected evidence of horizontal pleiotropy (P > 0.05), reinforcing the reliability of our findings. Scatterplots illustrating the outcomes of the five MR analysis methods have been presented in Supplementary Figures 1–8. In addition, a leave-one-out analysis, as illustrated in Supplementary Figures 9–16, demonstrated that the exclusion of any single SNP did not significantly alter the overall results, confirming the stability and reliability of our findings. To control for false positives, we applied the Benjamini–Hochberg corrected test. Corrected for multiple testing, Oxalobacter (OR: 1.143, 95% CI 1.061–1.232, P = 0.0004) and Tyzzerella 3 (OR: 1.145, 95% CI 1.059–1.238, P = 0.0007) were identified as risk factors for LBP, while Ruminococcaceae UCG011 (OR: 0.859, 95% CI 0.791–0.932, P = 0.0003) was identified as a protective factor against LBP. Additionally, Ruminococcus gnavus group (OR: 0.998, 95% CI 0.996–0.999, P = 0.0004) was revealed as a protective factor specifically against lumbar and other intervertebral disk disorders with radiculopathy, and Olsenella (OR: 0.893, 95% CI 0.839–0.951, P = 0.0004) emerged as a protective factor against low back pain or/and sciatica.
4 Discussion
To the best of our knowledge, this study stands as the inaugural and thorough investigation employing publicly available GWAS databases to scrutinize the causal relationship between the gut microbiota and four prevalent types of SP—specifically, NP, TSP, LBP, and BP. Through a meticulous analysis of the data, we observed that all IVW results within the dataset for ‘ lumbar and other intervertebral disk disorders with radiculopathy ’ were close to 1, with narrow 95% confidence intervals. This suggests the absence of a significant causal effect between the gut microbiota and this particular type of LBP. Consequently, we opted to exclude the data related to this specific type of LBP. Following a stringent selection process, we successfully identified a total of 37 bacterial genera exhibiting potential causal relationships with SP. Subsequently, through the application of the Benjamini–Hochberg corrected test, we corroborated the strong causal relationship of four bacterial genera with SP. The discoveries emerging from this study serve as an invaluable reference for subsequent research endeavors aimed at preventing and managing SP through precise interventions in the gut microbiota.
The study demonstrates that over 90% of cases involving SP lack a definitive diagnosis related to identifiable causes such as fractures, infections, tumors, etc (Haldeman et al., 2012). Consequently, clinical interventions face challenges in delivering precise and effective targeted treatments, often relying on symptomatic relief measures such as analgesics and soft tissue relaxation (Cheng, 2021). In recent years, growing attention has been directed toward understanding the intricate relationship between the gut microbiota and pain. The gut microbiota, with its array of metabolites, neurotransmitters, by-products, and other mediators, possesses the ability to modulate pain (Guo et al., 2019). In this context, we conducted a comprehensive exploration of the causal relationship between the gut microbiota and SP using Mendelian randomization analysis. Upon subjecting the MR results to the Benjamini–Hochberg corrected test, a strong causal relationship emerged between the gut microbiota and low back pain or/and lower limb pain. However, no significant causal relationship was discerned between the gut microbiota and NP, TSP, or BP. The outcomes of our study align with the observations of Arai et al. (2018), who reported a noteworthy positive relationship between the severity of constipation and the intensity of pain in individuals experiencing low back pain or/and lower limb pain. Notably, the severity of constipation is known to exert a significant influence on the composition of the gut microbiota (Zhu et al., 2014).
We observed that Oxalobacter and Tyzzerella 3 act as risk factors for low back pain or/and lower limb pain, while Ruminococcaceae UCG011 and Olsenella exhibit protective effects. Rajasekaran et al. (2020) identified a higher abundance of Oxalobacter in degenerated intervertebral disks. Intervertebral disk degeneration is widely recognized as a crucial factor in the onset and progression of LBP (Meng et al., 2023). Hence, we posit that Oxalobacter might contribute to LBP by potentially inducing intervertebral disk degeneration. Currently, there is limited clinical or experimental research on Tyzzerella 3 concerning LBP, and direct evidence establishing Tyzzerella 3 as a risk factor is lacking. Some studies found that Tyzzerella can produce aromatic amines and is positively correlated with the occurrence of diabetes (Ruuskanen et al., 2022; Sugiyama et al., 2022). Therefore, Tyzzerella 3 might promote the development of LBP by influencing endocrine or endothelial functions. Regarding the protective bacterial genera identified in the development of LBP, relevant studies on changes in the abundance of Ruminococcaceae UCG011 and Olsenella in the feces of LBP patients or animal models are currently unavailable. Ruminococcaceae UCG011 belongs to the Ruminococcaceae family, and evidence indicates a significant reduction in Ruminococcaceae family bacteria in the feces of mice with lumbar disk herniation compared to the control group (Wang et al., 2021). Butyrate produced by Ruminococcaceae family bacteria can promote the synthesis of vitamin D in the body (Murdaca et al., 2021). Studies have found that vitamin D has a regulatory effect on the immune function of the body, as it can inhibit the differentiation of pro-inflammatory cells such as T helper (Th) 17 cells, thereby reducing the secretion of pro-inflammatory factors such as IL-17 (Murdaca et al., 2011; Yamamoto and Jørgensen, 2020). Therefore, we hypothesize that improving local chronic inflammation in the body may be a potential mechanism for the protective effect of Ruminococcaceae UCG011. Additionally, research indicates that Short-chain fatty acids (SCFAs) produced by Olsenella can mediate the immune balance between Th17 cells and regulatory T cells (Tregs) by activating Tregs’ activity, thus balancing the levels of pro-inflammatory factors (derived from Th17 cells) and anti-inflammatory factors (derived from Treg cells) in the body (Kim, 2021). This could be a potential mechanism by which Olsenella resists the development of LBP. The involvement of the gut microbiota in the development of LBP is an exceedingly complex process. Therefore, the specific mechanisms by which the four identified positive gut microbiota affect low back pain or/and lower limb pain, require further experimental validation in subsequent studies.
In addition to the four bacterial genera strongly associated with SP previously mentioned, our study identified 33 other bacterial genera exhibiting potential causal relationship with SP. To ensure a comprehensive exploration of gut microbiota potentially linked to SP, we incorporated data from all gut microbiota groups with Benjamini–Hochberg corrected test p-values below 0.05. Among the eight bacterial genera potentially causally related to NP, the majority belonged to the Firmicutes phylum. Qin et al. (2019) reported symptom improvement in cervical spondylosis patients following manual therapy, accompanied by a significant increase in the proportions of Lactobacillus and Bifidobacterium in feces, both belonging to the Firmicutes phylum. Unfortunately, limited clinical and foundational research exists on the gut microbiota’s association with NP, and there is no evidence supporting the relationship between Lachnospiraceae UCG010, Escherichia-Shigella, Methanobrevibacter, Eubacterium nodatum group, Faecalibacterium, Subdoligranulum, Butyrivibrio, Ruminococcaceae UCG010, and NP. Potential protective factors for TSP include Butyrivibrio, Ruminococcaceae UCG011, and Eubacterium brachy group. However, similar to NP, research on the gut microbiota and TSP is scarce, lacking supporting evidence. Interestingly, through a comparative analysis of potential causally related bacterial genera in NP and TSP, Butyrivibrio was identified as a shared potential protective genus. Its production of butyrate, propionate, and other SCFAs inhibiting histone deacetylase activity may be one of the potential mechanisms underlying Butyrivibrio’s analgesic effects (Descalzi et al., 2015; Tang et al., 2022). Research on the relationship between gut microbiota and low back pain or/and lower limb pain is more abundant. Yao et al. (2023) found a significant decrease in Lachnospiraceae NK4A136 group, Turicibacter, and Roseburia in the feces of rats in a LBP model compared to the control group. In various LBP animal models, Wang et al. (2021) also found lower abundance of Lachnospiraceae NK4A136 group in rats with lumbar disk herniation compared to the control group. A cross-sectional study revealed a significant reduction in skeletal muscle mass index in patients with LBP compared to normal individuals (Park et al., 2023). Sugimura et al. (2022) found a positive relationship between the abundance of Eisenbergiella and skeletal muscle mass, suggesting a reasonable hypothesis that Eisenbergiella may prevent LBP by increasing lumbar skeletal muscle mass. Consistent with the above findings, our study identified Lachnospiraceae NK4A136 group, Turicibacter, Roseburia, and Eisenbergiella as potential protective factors for LBP. In studies investigating the relationship between BP and gut microbiota, Dekker Nitert et al. (2020) found a higher abundance of the genus Uncl. Christensenellaceae in patients with BP. However, contrary to this conclusion, our results indicate that Christensenellaceae R.7 group is a potential protective factor for BP, a finding supported by Hollister et al. (2020). In summary, this study experimentally validated some of the bacterial genera with potential causal relationships to SP that were screened. This underscores the significance of exploring bacterial genera with potential causal relationships to SP, and the unvalidated genera warrant further investigation.
Whether it is NP, TSP, LBP, or BP, they represent distinct segments of SP. Fundamentally, these pains originate in the spinal column, suggesting a certain degree of interconnectedness among pains occurring in different spinal segments. To dissect this phenomenon, we conducted a comparative analysis of bacterial genera potentially influencing SP across different segments. Our investigation revealed that Ruminococcaceae UCG010 exhibits potential protective properties, common to both NP in the last month and BP in the last month. Similarly, Ruminococcaceae UCG011 emerges as a potential safeguard against TSP, LBP, and the amalgamation of low back pain or/and sciatica. Notably, Butyrivibrio surfaces as a potential protective agent against NP in the last month and TSP. Intriguingly, the Eubacterium nodatum group assumes a potential risk factor role in NP for 3+ months while concurrently acting as a potential protective factor for low back pain or/and sciatica. This nuanced outcome suggests that a singular bacterial genus might exert counteractive regulatory influences on pains manifesting in different spinal segments. In summation, the intricate modulation of pains across different spinal segments by the gut microbiota underscores a complex process. Shared or divergent mechanisms may underscore these phenomena, necessitating further exploration to unveil the underlying intricacies.
Furthermore, to scrutinize potential disparities in causally related bacterial genera associated with SP across varying durations, we incorporated datasets reflecting distinct timeframes— 1 month and 3 months. Regrettably, datasets for different durations of TSP and low back pain or/and lower limb pain, were unavailable. Consequently, our analysis encompassed datasets for 1 month and 3 months of NP and BP. MR outcomes reveal an absence of shared potential causally related bacterial genera between 1 month and 3 months of NP, a trend also mirrored in the context of BP. This suggests a dynamic nature, signifying that bacterial genera with potential causal relationship with SP may evolve during the course of its progression. Contrary to the findings of Dekker Nitert et al. (2020), who observed heightened abundance of Adlercreutzia, Roseburia, and Uncl. Christensenellaceae in patients with 1 month of BP, our MR results challenge this observation. Specifically, Adlercreutzia (P = 0.53) and Roseburia (P = 0.92) do not emerge as potential causally related bacterial genera for 1 month of BP. Instead, the Christensenellaceae R.7 group surfaces as a potential protective bacterial genus for this duration of BP. We posit that discrepancies in results may be attributed to the relatively modest sample size employed in the clinical study. In conclusion, it is imperative to delineate causally related bacterial genera associated with SP at distinct developmental stages. This endeavor holds paramount significance for tailoring precise interventions targeting SP across varying durations.
The robustness of this investigation stems from the application of MR to evaluate the causal relationship between gut microbiota and SP. By leveraging genetic variations as IVs, MR effectively mitigates external confounding factors, thereby bolstering internal validity owing to the inherent randomness and independence of these genetic determinants. Notably, subgroup analyses conducted for distinct durations of spinal pain (i.e., 1 month and 3 months) contribute valuable insights, offering implications for future interventions targeting specific gut microbiota to address SP across varying timeframes. However, our study contends with several limitations. Firstly, the exclusivity of data from European countries prompts the necessity for further experimental validation to ascertain the generalizability of MR results to populations in other regions, such as Asia and Africa. Secondly, the adherence to the conventional significance threshold for SNPs in MR studies (P < 5e−08) restricts the number of selected SNPs, potentially influencing subsequent sensitivity analyses and control for horizontal pleiotropy. Thirdly, the constraint of exposure data to the genus level hampers a more nuanced analysis of the causal relationship between higher taxonomic levels of gut microbiota and SP. Lastly, relying on public databases rather than original sources limits the scope of available datasets, resulting in the omission of subgroup analyses for TSP and low back pain or/and lower limb pain during SP subgroup analyses— a notable constraint in our study.
5 Conclusion
Through our two-sample Mendelian randomization (MR) analysis, we have successfully identified a comprehensive set of 37 bacterial genera potentially causally related to SP, encompassing NP, TSP, LBP, and BP. Employing the rigorous Benjamini-Hochberg corrected test, we have discerned four bacterial genera that exhibit robust causal relationships with SP. Notably, our analysis has unveiled a potential common regulatory mechanism, suggesting that certain bacterial genera may influence spinal pain across different segments. Furthermore, the dynamic nature of these potential causal relationships has been illuminated, indicating that, as the duration of SP progresses, the involved bacterial genera undergo changes. In conclusion, this study contributes valuable insights into the intricate interplay between the gut microbiota and SP. Such revelations not only serve as a significant reference for understanding these complex connections but also pave the way for future research endeavors aimed at preventing and treating SP through targeted interventions in the gut microbiota.
Data availability statement
The original contributions presented in the study are included in the article/Supplementary material, further inquiries can be directed to the corresponding author/s.
Author contributions
SH: Conceptualization, Writing – original draft. LC: Conceptualization, Writing – original draft. XZ: Formal analysis, Writing – review and editing. YH: Data curation, Writing – review and editing. YT: Writing – review and editing. HH: Writing – review and editing. HW: Validation, Writing – review and editing. CY: Validation, Writing – review and editing. ZL: Funding acquisition, Project administration, Writing – review and editing. LL: Funding acquisition, Validation, Writing – review and editing. BY: Writing – review and editing.
Funding
The author(s) declare financial support was received for the research, authorship, and/or publication of this article. This work was supported by the “Pioneer” and “Leading Goose” R&D Program of Zhejiang (No. 2022C03123), the General Program of the National Natural Science Foundation of China (81774442 and 82274672), the National Natural Science Foundation of China Youth Fund Project (82305426), Key Discipline Project of High-level TCM of National Administration of Traditional Chinese Medicine (GJXK2023-85), the Zhejiang Provincial Natural Science Foundation Project (Q23H270025), the Scientific research project of Zhejiang Provincial Department of Education (Y202351266), and the School-Level Scientific Research Project of Zhejiang Chinese Medicine University (2021RCZXZK03, 2022FSYYZQ13, and 2022GJYY045).
Acknowledgments
We thank the UK Biobank, FinnGen Biobank, and MiBioGen consortium for publicly sharing GWAS summary data for related diseases.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2024.1357303/full#supplementary-material
Supplementary Data Sheet 1 | Genetically predicted causal effects of gut microbiota on spinal pain: a two-sample Mendelian Randomization analysis.
Supplementary Data Sheet 2 | Scatterplot results for five analysis methods and leave-one-out analysis results.
Supplementary Data Sheet 3 | Analysis codes for MR analysis.
Footnotes
References
Akagawa, S., and Kaneko, K. (2022). Gut microbiota and allergic diseases in children. Allergol. Int. 3, 301–309. doi: 10.1016/j.alit.2022.02.004
Arai, Y., Shiro, Y., Funak, Y., Kasugaii, K., Omichi, Y., and Sakurai, H. (2018). The association between constipation or stool consistency and pain severity in patients with chronic pain. Anesth. Pain Med. 4:e69275. doi: 10.5812/aapm.69275
Bowden, J., Davey Smith, G., and Burgess, S. (2015). Mendelian randomization with invalid instruments: effect estimation and bias detection through Egger regression. Int. J. Epidemiol. 2, 512–525. doi: 10.1093/ije/dyv080
Bowden, J., Davey Smith, G., Haycock, P., and Burgess, S. (2016). Consistent estimation in mendelian randomization with some invalid instruments using a weighted median estimator. Genet. Epidemiol. 4, 304–314. doi: 10.1002/gepi.21965
Burgess, S., Bowden, J., Fall, T., Ingelsson, E., and Thompson, S. (2017). Sensitivity analyses for robust causal inference from mendelian randomization analyses with multiple genetic variants. Epidemiology 1, 30–42. doi: 10.1097/EDE.0000000000000559
Burgess, S., Dudbridge, F., and Thompson, S. (2016). Combining information on multiple instrumental variables in mendelian randomization: comparison of allele score and summarized data methods. Stat. Med. 11, 1880–1906. doi: 10.1002/sim.6835
Burgess, S., and Thompson, S. (2011). Bias in causal estimates from Mendelian randomization studies with weak instruments. Stat. Med. 11, 1312–1323. doi: 10.1002/sim.4197
Chassaing, B., Etienne-Mesmin, L., and Gewirtz, A. (2014). Microbiota-liver axis in hepatic disease. Hepatology 1, 328–339. doi: 10.1002/hep.26494
Cheng, Z. (2021). The treatment strategies of spinal pain. Chin. J. Painol. 4, 339–341. doi: 10.3760/cma.j.cn101658-20210719-00101
Clark, J., and Coopersmith, C. (2007). Intestinal crosstalk: a new paradigm for understanding the gut as the “motor” of critical illness. Shock 4, 384–393. doi: 10.1097/shk.0b013e31805569df
Davies, N., Holmes, M., and Davey Smith, G. (2018). Reading Mendelian randomisation studies: a guide, glossary, and checklist for clinicians. BMJ 362:k601. doi: 10.1136/bmj.k601
de Luca, K., Tavares, P., Yang, H., Hurwitz, E., Green, B., Dale, H., et al. (2023). Spinal pain, chronic health conditions and health behaviors: data from the 2016-2018 national health interview survey. Int. J. Environ. Res. Public Health 7:5369. doi: 10.3390/ijerph20075369
Dekker Nitert, M., Mousa, A., Barrett, H., Naderpoor, N., and de Courten, B. (2020). Altered gut microbiota composition is associated with back pain in overweight and obese individuals. Front. Endocrinol. 11:605. doi: 10.3389/fendo.2020.00605
Descalzi, G., Ikegami, D., Ushijima, T., Nestler, E., Zachariou, V., and Narita, M. (2015). Epigenetic mechanisms of chronic pain. Trends Neurosci. 4, 237–246. doi: 10.1016/j.tins.2015.02.001
Emdin, C., Khera, A., and Kathiresan, S. (2017). Mendelian Randomization. JAMA 19, 1925–1926. doi: 10.1001/jama.2017.17219
GBD 2015 Disease and Injury Incidence and Prevalence Collaborators (2016). Global, regional, and national incidence, prevalence, and years lived with disability for 310 diseases and injuries, 1990-2015: a systematic analysis for the Global Burden of Disease Study 2015. Lancet 10053, 1545–1602. doi: 10.1016/S0140-6736(16)31678-6
Greco, M., Minelli, C., Sheehan, N., and Thompson, J. (2015). Detecting pleiotropy in Mendelian randomisation studies with summary data and a continuous outcome. Stat. Med. 21, 2926–2940. doi: 10.1002/sim.6522
Guo, R., Chen, L., Xing, C., and Liu, T. (2019). Pain regulation by gut microbiota: molecular mechanisms and therapeutic potential. Br. J. Anaesth. 5, 637–654. doi: 10.1016/j.bja.2019.07.026
Haldeman, S., Kopansky-Giles, D., Hurwitz, E., Hoy, D., Mark Erwin, W., Dagenais, S., et al. (2012). Advancements in the management of spine disorders. Best Pract. Res. Clin. Rheumatol. 2, 263–280. doi: 10.1016/j.berh.2012.03.006
Hollister, E., Cain, K., Shulman, R., Jarrett, M., Burr, R., Ko, C., et al. (2020). Relationships of microbiome markers with extraintestinal, psychological distress and gastrointestinal symptoms, and quality of life in women with irritable bowel syndrome. J. ClinGastroenterol. 2, 175–183. doi: 10.1097/MCG.0000000000001107
Jandhyala, S., Talukdar, R., Subramanyam, C., Vuyyuru, H., Sasikala, M., and Nageshwar Reddy, D. (2015). Role of the normal gut microbiota. World J Gastroenterol. 29, 8787–8803. doi: 10.3748/wjg.v21.i29.8787
Kim, C. (2021). Control of lymphocyte functions by gut microbiota-derived short-chain fatty acids. Cell Mol. Immunol. 5, 1161–1171. doi: 10.1038/s41423-020-00625-0
Koes, B., van Tulder, M., and Thomas, S. (2006). Diagnosis and treatment of low back pain. BMJ 7555, 1430–1434. doi: 10.1136/bmj.332.7555.1430
Kurilshikov, A., Medina-Gomez, C., and Bacigalupe, R. (2021). Large-scale association analyses identify host factors influencing human gut microbiome composition. Nat Genet. 2, 156–165. doi: 10.1038/s41588-020-00763-1
Lei, T., He, Y., and Xiao, Y. (2015). Application research of Miao medicine liquid dispensing the drug stick therapy in spinal pain. Chin. J. Biochem. Pharmaceutics 2, 113–115.
Ley, R., Peterson, D., and Gordon, J. (2006). Ecological and evolutionary forces shaping microbial diversity in the human intestine. Cell 4, 837–848. doi: 10.1016/j.cell.2006.02.017
Lin, K., Zhu, L., and Yang, L. (2020). Gut and obesity/metabolic disease: focus on microbiota metabolites. MedComm 3:e171. doi: 10.1002/mco2.171
Manchikanti, L., Boswell, M., Singh, V., Pampati, V., Damron, K., and Beyer, C. (2004). Prevalence of facet joint pain in chronic spinal pain of cervical, thoracic, and lumbar regions. BMC Musculoskelet Disord. 5:15. doi: 10.1186/1471-2474-5-15
Meng, Q., Liu, K., Liu, Z., Liu, J., Tian, Z., Qin, S., et al. (2023). Digoxin protects against intervertebral disc degeneration via TNF/NF-κB and LRP4 signaling. Front. Immunol. 14:1251517. doi: 10.3389/fimmu.2023.1251517
Murdaca, G., Colombo, B., and Puppo, F. (2011). The role of Th17 lymphocytes in the autoimmune and chronic inflammatory diseases. Intern. Emerg. Med. 6, 487–495. doi: 10.1007/s11739-011-0517-7
Murdaca, G., Gerosa, A., Paladin, F., Petrocchi, L., Banchero, S., and Gangemi, S. (2021). Vitamin D and microbiota: is there a link with allergies? Int. J. Mol. Sci. 8:4288. doi: 10.3390/ijms22084288
Park, M., Park, S., and Chung, S. (2023). Relationships between skeletal muscle mass, lumbar lordosis, and chronic low back pain in the elderly. Neurospine 3, 959–968. doi: 10.14245/ns.2346494.247
Qin, Y. (2022). Pueraria lobata targeted preparation improves the clinical symptoms of cervical spondylosis by regulating the balance of gut microbiota. Comput. Math Methods Med. 2022:2136807. doi: 10.1155/2022/2136807
Qin, Y., Wu, Y., Lin, F., Yang, X., and Hao, F. (2019). Clinical study on the therapy of rubbing abdomen and pinching along the spine for cervical spondylosis of vertebral artery type. J. New Chin. Med. 1, 217–220. doi: 10.13457/j.cnki.jncm.2019.01.057
Quaglio, A., Grillo, T., De Oliveira, E., Di Stasi, L., and Sassaki, L. (2022). Gut microbiota, inflammatory bowel disease and colorectal cancer. World J. Gastroenterol. 30, 4053–4060. doi: 10.3748/wjg.v28.i30.4053
Rajasekaran, S., Soundararajan, D., Tangavel, C., Muthurajan, R., Sri Vijay, Anand, K., et al. (2020). Human intervertebral discs harbour a unique microbiome and dysbiosis determines health and disease. Eur. Spine J. 7, 1621–1640. doi: 10.1007/s00586-020-06446-z
Ruuskanen, M., Erawijantari, P., Havulinna, A., Liu, Y., Méric, G., and Tuomilehto, J. (2022). Gut microbiome composition is predictive of incident type 2 diabetes in a population cohort of 5,572 finnish adults. Diabetes Care 4, 811–818. doi: 10.2337/dc21-2358
Slob, E., and Burgess, S. (2020). A comparison of robust Mendelian randomization methods using summary data. Genet. Epidemiol. 4, 313–329. doi: 10.1002/gepi.22295
Smith, G., and Ebrahim, S. (2003). ‘Mendelian randomization’: can genetic epidemiology contribute to understanding environmental determinants of disease? Int. J. Epidemiol. 1, 1–22. doi: 10.1093/ije/dyg070
Soremekun, O., Slob, E., Burgess, S., Romuald Boua, P., Fatumo, S., and Gill, D. (2022). Genetically predicted lipid traits, diabetes liability, and carotid intima-media thickness in african ancestry individuals: a mendelian randomization study. Circ. Genom. Precis. Med. 6:e003910. doi: 10.1161/CIRCGEN.122.003910
Sugimura, Y., Kanda, A., Sawada, K., Wai, K., Tanabu, A., Ozato, N., et al. (2022). Association between gut microbiota and body composition in japanese general population: a focus on gut microbiota and skeletal muscle. Int. J. Environ. Res. Public Health 12:7464. doi: 10.3390/ijerph19127464
Sugiyama, Y., Mori, Y., Nara, M., Kotani, Y., Nagai, E., and Kawada, H. (2022). Gut bacterial aromatic amine production: aromatic amino acid decarboxylase and its effects on peripheral serotonin production. Gut Microbes 1:2128605. doi: 10.1080/19490976.2022.2128605
Tang, Y., Du, J., Wu, H., Wang, M., Liu, S., and Tao, F. (2022). Potential therapeutic effects of short-chain fatty acids on chronic pain. Curr. Neuropharmacol. 22, 191–203. doi: 10.2174/1570159X20666220927092016
Wang, Z., Wu, H., Chen, Y., Chen, H., Wang, X., and Yuan, W. (2021). Lactobacillus paracasei S16 alleviates lumbar disc herniation by modulating inflammation response and gut microbiota. Front. Nutr. 8:701644. doi: 10.3389/fnut.2021.701644
Xie, L., Zhao, H., and Chen, W. (2023). Relationship between gut microbiota and thyroid function: a two-sample Mendelian randomization study. Front. Endocrinol. 14:1240752. doi: 10.3389/fendo.2023.1240752
Yamamoto, E., and Jørgensen, T. (2020). Relationships between vitamin D, gut microbiome, and systemic autoimmunity. Front. Immunol. 10:3141. doi: 10.3389/fimmu.2019.03141
Yang, R., Chen, Z., and Cai, J. (2023). Fecal microbiota transplantation: emerging applications in autoimmune diseases. J. Autoimmun. 141:103038. doi: 10.1016/j.jaut.2023.103038
Yao, B., Cai, Y., Wang, W., Deng, J., Zhao, L., Han, Z., et al. (2023). The effect of gut microbiota on the progression of intervertebral disc degeneration. Orthop. Surg. 3, 858–867. doi: 10.1111/os.13626
Zhang, Y., Li, S., Gan, R., Zhou, T., Xu, D., and Li, H. (2015). Impacts of gut bacteria on human health and diseases. Int. J. Mol. Sci. 4, 7493–7519. doi: 10.3390/ijms16047493
Keywords: spinal pain, neck pain, thoracic spine pain, low back pain, back pain, gut microbiota, Mendelian randomization, causality
Citation: Hong S, Chen L, Zhou X, Huang Y, Tian Y, Hu H, Yu B, Wu H, Yang C, Lv Z and Lv L (2024) Genetically predicted causal effects of gut microbiota on spinal pain: a two-sample Mendelian randomization analysis. Front. Microbiol. 15:1357303. doi: 10.3389/fmicb.2024.1357303
Received: 05 January 2024; Accepted: 07 March 2024;
Published: 25 March 2024.
Edited by:
Luca Posa, Cornell University, United StatesReviewed by:
Joseph Atia Ayariga, Alabama State University, United StatesGiuseppe Murdaca, University of Genoa, Italy
Copyright © 2024 Hong, Chen, Zhou, Huang, Tian, Hu, Yu, Wu, Yang, Lv and Lv. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Zhizhen Lv, bHZ6aGl6aGVuMTk5MkAxNjMuY29t; Lijiang Lv, bHZsaWppYW5nMDI4OEAxNjMuY29t
†These authors have contributed equally to this work
 Shuangwei Hong
Shuangwei Hong Longhao Chen
Longhao Chen Xingchen Zhou
Xingchen Zhou Yuanshen Huang1,2†
Yuanshen Huang1,2† Zhizhen Lv
Zhizhen Lv