
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Microbiol. , 14 February 2024
Sec. Infectious Agents and Disease
Volume 15 - 2024 | https://doi.org/10.3389/fmicb.2024.1349163
This article is part of the Research Topic Insights in Infectious Agents and Disease: 2023/2024 View all 31 articles
 Deborah M. Cooke1
Deborah M. Cooke1 Charlene Clarke2
Charlene Clarke2 Tanya J. Kerr1
Tanya J. Kerr1 Robin M. Warren1
Robin M. Warren1 Carmel Witte1,3
Carmel Witte1,3 Michele A. Miller1†
Michele A. Miller1† Wynand J. Goosen1*†
Wynand J. Goosen1*†Animal tuberculosis, caused by Mycobacterium bovis, presents a significant threat to both livestock industries and public health. Mycobacterium bovis tests rely on detecting antigen specific immune responses, which can be influenced by exposure to non-tuberculous mycobacteria, test technique, and duration and severity of infection. Despite advancements in direct M. bovis detection, mycobacterial culture remains the primary diagnostic standard. Recent efforts have explored culture-independent PCR-based methods for identifying mycobacterial DNA in respiratory samples. This study aimed to detect M. bovis in nasal swabs from goats (Capra hircus) cohabiting with M. bovis-infected cattle in KwaZulu-Natal, South Africa. Nasal swabs were collected from 137 communal goats exposed to M. bovis-positive cattle and 20 goats from a commercial dairy herd without M. bovis history. Swabs were divided into three aliquots for analysis. The first underwent GeneXpert® MTB/RIF Ultra assay (Ultra) screening. DNA from the second underwent mycobacterial genus-specific PCR and Sanger sequencing, while the third underwent mycobacterial culture followed by PCR and sequencing. Deep sequencing identified M. bovis DNA in selected Ultra-positive swabs, confirmed by region-of-difference (RD) PCR. Despite no other evidence of M. bovis infection, viable M. bovis was cultured from three communal goat swabs, confirmed by PCR and sequencing. Deep sequencing of DNA directly from swabs identified M. bovis in the same culture-positive swabs and eight additional communal goats. No M. bovis was found in commercial dairy goats, but various NTM species were detected. This highlights the risk of M. bovis exposure or infection in goats sharing pastures with infected cattle. Rapid Ultra screening shows promise for selecting goats for further M. bovis testing. These techniques may enhance M. bovis detection in paucibacillary samples and serve as valuable research tools.
Mycobacterium bovis (M. bovis), a member of the Mycobacterium tuberculosis complex (MTBC), is the causative agent of animal tuberculosis (Borham et al., 2022). Although cattle are considered the primary host, M. bovis is known to have the widest host range of all members of the MTBC, with the ability to cause disease in domestic animals, wildlife, and humans (Mostowy et al., 2005; Palmer et al., 2012). The World Organization for Animal Health (WOAH) lists animal tuberculosis (bTB) as a notifiable disease (World Organization for Animal Health, 2023) and many developed countries have programs to manage and control this disease in livestock, primarily to prevent spread to humans (Reviriego Gordejo and Vermeersch, 2006; Palmer and Waters, 2011; World Health Organization, 2017; More, 2019).
Tuberculosis in domestic goats (Capra hircus) is mostly attributed to Mycobacterium bovis (M. bovis) and M. caprae, which are capable of infecting other animals, including humans (Rodríguez et al., 2009; Pesciaroli et al., 2014; Bezos et al., 2015). Zoonotic TB remains a considerable global challenge; in 2019, an estimated 140,000 new cases and 11,400 deaths were reported (World Health Organization, 2020). In South Africa, M. bovis is endemic in communal cattle and some wildlife populations with TB control programs predominantly focusing on cattle (Renwick et al., 2007; Arnot and Michel, 2020). The epidemiology and diagnosis of goat TB are similar to cattle, which are both natural hosts (Quintas et al., 2010; Pesciaroli et al., 2014). Although there are published reports of M. bovis testing of goats in other countries (Quintas et al., 2010), M. bovis in domestic goats has not been thoroughly investigated in South Africa (Nyoni, 2019). The traditional approach to keeping livestock in South African communities involves goats sharing communal pastures and water sources with cattle, and occasionally with wildlife; therefore, the lack of surveillance may lead to under-recognition of goats as a potential source of M. bovis spread to cattle or other livestock as well as humans and wildlife.
Tests that accurately identify M. bovis infected individuals and herds are the foundation of bTB control programs. Most M. bovis diagnostic tests for livestock rely on detecting host antigen-specific cell-mediated immune (CMI) responses to mycobacterial antigens, typically the in vivo tuberculin skin test (Welsh et al., 2005; Bernitz et al., 2021). Currently, the official South African (SA) guidelines for TB testing in livestock and African buffaloes (Syncerus caffer) advocate the use of the single intradermal comparative tuberculin test (SICTT; Department of Agriculture, Forestry and Fisheries of SA, 2016; Department of Agriculture, Forestry and Fisheries of SA, 2017; Department of Agriculture, Land Reform and Rural Development, 2018; Arnot and Michel, 2020). However, the interpretation of the SICTT can be confounded by several factors including exposure to environmental non-tuberculous mycobacteria (NTM), which may cause cross-reactivity (Michel et al., 2011). Since NTMs are ubiquitous in the environment (Falkinham, 2021), with a high diversity occurring in SA (Gcebe et al., 2013), their presence may lead to M. bovis false-positive reactions in tested animals. This, in turn, may cause unnecessary expenses due to additional testing, loss of income, and loss of animals (Vordermeier et al., 2007; Bolaños et al., 2017). This is especially problematic for rural farmers in SA, who depend on small-scale livestock farming as a source of income (Sichewo et al., 2020).
While the SICTT is routinely used for antemortem screening, definitive diagnosis of bTB is based on the direct detection of M. bovis from animal tissue samples using mycobacterial culture, followed by speciation using region-of-difference (RD) PCRs (Warren et al., 2006; Bernitz et al., 2021). However, conventional mycobacterial culture has suboptimal performance, is slow, laborious (Ghodbane et al., 2014), introduces in vitro bacterial selection pressure, and may lead to false negative results (especially with paucibacillary samples) due to harsh sample decontamination steps (de Boer et al., 2002). Although technical advances have improved direct detection of pathogenic Mycobacteria spp., most applications still heavily rely on culture to obtain sufficient organisms to confirm infection (Bernitz et al., 2021). Therefore, it is important to continuously explore new methods, in addition to culture, for the direct detection of M. bovis infected livestock.
Recently, culture-independent PCR-based sequencing methods have been investigated for direct detection and identification of important mycobacterial organisms in antemortem respiratory samples and postmortem tissues (Clarke et al., 2022a; Goosen et al., 2022a). Studies have shown that the GeneXpert® MTB/RIF Ultra assay (Ultra) can detect MTBC DNA in animal samples and provides a rapid sensitive screening test (Goosen et al., 2020; Clarke et al., 2021). Furthermore, conventional, and real-time PCR assays, followed by amplicon sequencing, have also shown promise for detecting and characterizing both MTBC and NTM species present in cultures, as well as directly from raw specimens (Warren et al., 2006; Deggim-Messmer et al., 2016; Jung et al., 2016; Goosen et al., 2020, 2022a; Clarke et al., 2022a,b). Amplicons from these PCRs can be used for targeted next generation sequencing, which facilitates the accurate detection and characterization of multiple mycobacterial species present, especially in paucibacillary oronasal swabs, other respiratory samples, as well as fecal samples (Adékambi et al., 2003; Lee et al., 2020; Goosen et al., 2022a). Combining culture with these techniques will enable confirmation of infection in animals with positive host CMI test results.
Culture-independent detection of pathogenic mycobacterial species using extracted DNA can enhance individual diagnosis, identify infected herds, and improve disease management, especially when samples for culture cannot be transported due to sample movement restrictions, or where there is no laboratory capacity for mycobacterial culture. In SA, this is especially relevant when testing rural livestock in areas where the presence of controlled diseases, such as Foot and Mouth Disease (FMD), restrict movement of animals and samples, unless they are heat-inactivated (Brückner et al., 2002). Therefore, the aim of this study was to perform MTBC- and Mycobacterium genus-specific PCRs and sequencing, using DNA extracted directly from swabs and from swab cultures, to determine the presence and species of MTBC in goat respiratory samples.
The Stellenbosch University Animal Care and Use Committee granted ethical approval for this project (ACU-2020-14560) and Section 20 approval was issued by the Department of Agriculture, Land Reform, and Rural Development (DALRRD) (12/11/1/7/2 (16045S)). Consent was obtained from all goat owners prior to testing.
In 2019, nasal swabs were collected from 157 goats (Capra hircus) from the KwaZulu-Natal midlands, SA. This included opportunistic sampling from 137 communal domestic goats in an area with confirmed M. bovis infected cattle (Umkhanyakude district of Northern Zululand, KwaZulu-Natal) and sampling from 20 goats in a closed commercial dairy herd consisting of stud Saanen goats with no known exposure to M. bovis, a 20-year long history of negative annual SICCT tests, and a high level of management and biosecurity practices, as previously described (Cooke et al., 2023; Figure 1). Nasal swabs were taken using sterile OmniSwabs (Whatman®, Qiagen, Germantown, MD, United States). Swab heads were placed directly into cryovials containing ~1.5 mL sterile saline solution, transported with ice bricks in a cooler box, and subsequently frozen at −80°C within 8 h of collection, before being transported to Stellenbosch University for further downstream processing.
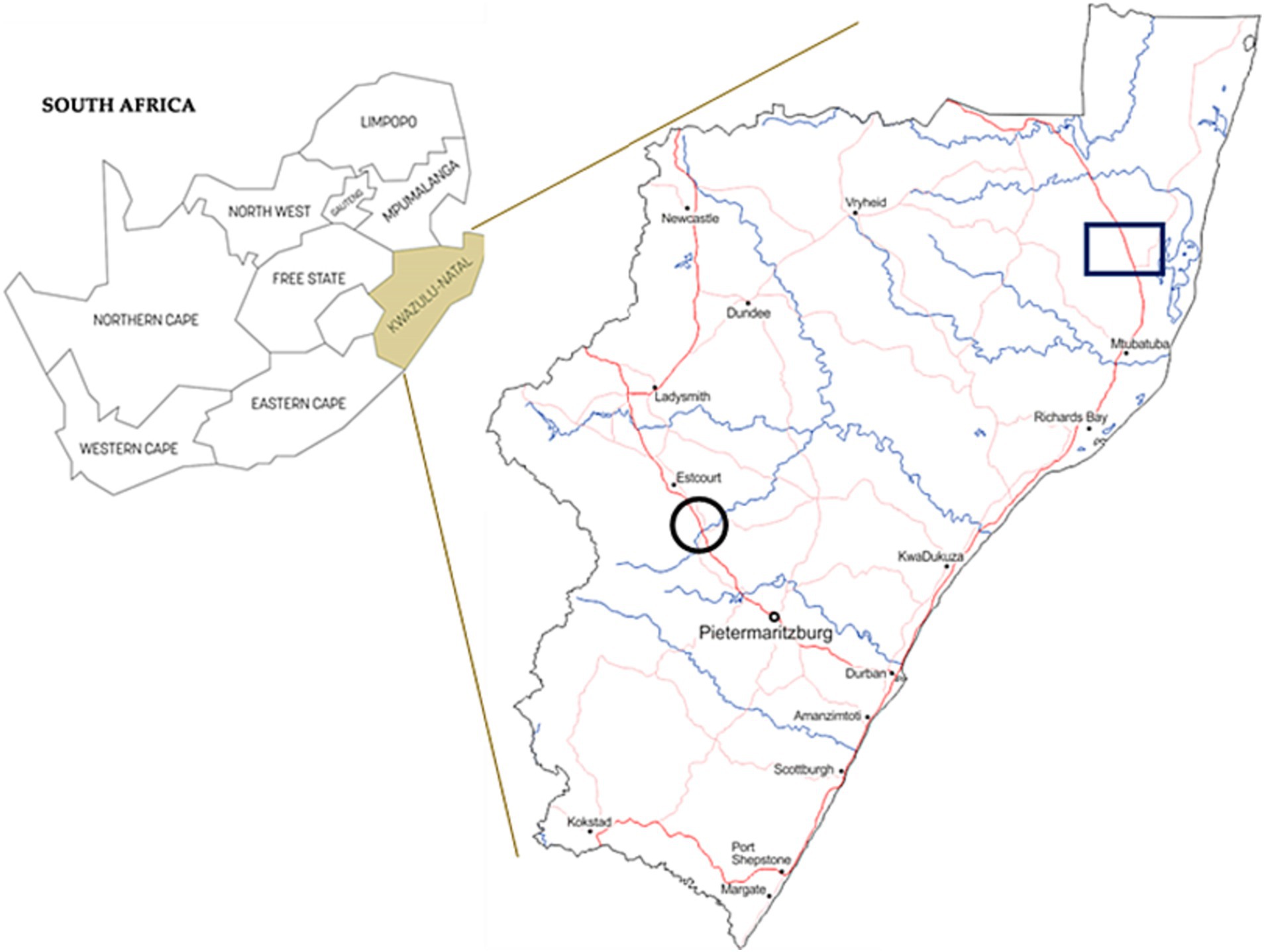
Figure 1. Map of South Africa with province of KwaZulu-Natal shown in insert. Sampling area of communal domestic goat herds are indicated by a black square. Black circles show the location of the commercial dairy goat herd.
After transport of frozen nasal swabs, all samples (n = 157) were split into three aliquots of ~500 μL each in a biosafety level 3 (BSL-3) facility (Figure 2). One aliquot was processed directly for MTBC DNA detection using the GeneXpert® MTB/RIF Ultra assay (Cepheid, Sunnyvale, CA, United States), as previously described (Goosen et al., 2020). A second aliquot underwent DNA extraction, using the DNeasy Blood and Tissue kit (Qiagen, Hilden, Germany) as instructed by the manufacturer. The extracted DNA was used for Mycobacterium genus-specific PCRs (hsp65 and rpoB), subsequent Mycobacteria spp. identification by amplicon sequencing using Sanger and a NGS platform as well as RD-PCR speciation of all MTBC DNA positive samples, as described below (Goosen et al., 2022a). The third aliquot was decontaminated by using a 1:1 volume of MycoPrep to sample volume, incubated for 15 min and then neutralized with sterile PO4 buffer (at the same volume of sample plus MycoPrep). The sample was then shaken to produce a homogeneous solution and then centrifuged for 15 min at 2,000× g for MGIT inoculation, after which, the supernatant was aspirated off, down to a remaining 1 mL buffer volume just above the pellet. The pellet was then thoroughly reconstituted and transferred to Mycobacterial Growth Indicator Tubes (MGIT) and incubated in the BACTECTM MGITTM 960 TB system (Becton Dickinson, Franklin Lakes, NJ, United States), as previously described, with a minor modification (Goosen et al., 2022b).
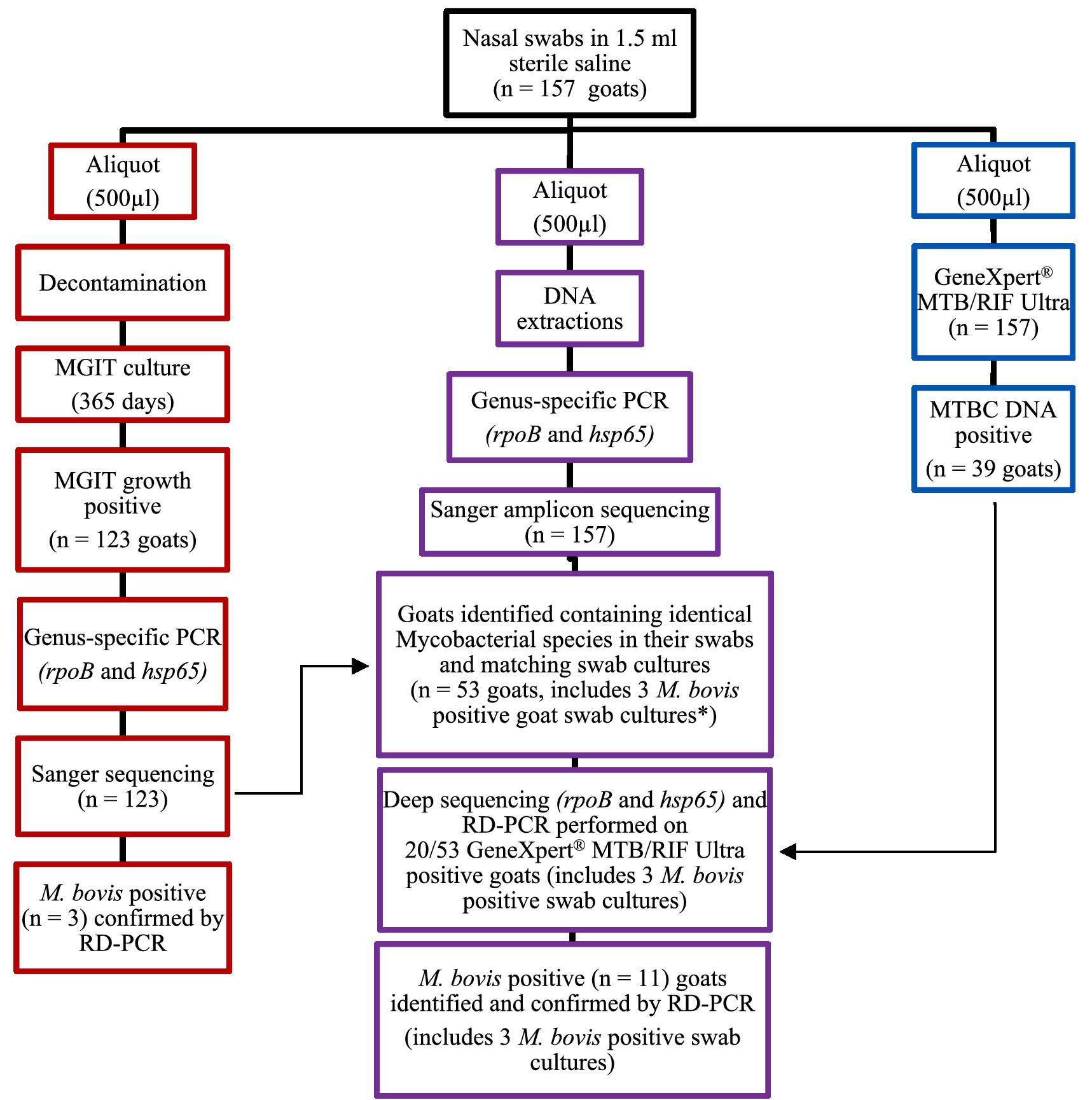
Figure 2. Flow chart for goat nasal swab (n = 157) sample processing for PCR testing and mycobacterial identification. The red section (left) shows the process of aliquot 3 for mycobacterial culture and PCR; the purple section (middle) shows the process followed for the raw swab samples (aliquot 2); and the blue section (right) shows how aliquot 1 was processed directly for MTBC DNA detection using the GeneXpert® MTB/RIF Ultra assay. *Three Mycobacterium bovis culture-positive goats also had M. bovis DNA detected using culture-independent processes for the raw swabs; these were goats (C139, F91, and N112).
Briefly, all MGIT tubes, including two uninoculated MGIT tubes designated as negative controls, underwent aseptic replenishment with 1.5 mL fresh media sourced from new MGIT tubes (Becton Dickinson, United States). This procedure was conducted in batches of 5 MGITs at a time within a Class II Biological Safety Cabinet located in a BSL-3 facility every 56 days, spanning up to 365 days of incubation. At each 56-day interval, RD-PCRs were performed on 1.5 mL boiled MGIT homogenates (20 min at 98°C followed by centrifugation at 2,000× g for 20 min to ensure maximum release of DNA and to reduce the possibility of discarding some paucibacillary bacilli) from all MGIT tubes to check for the presence of MTBC DNA. Only at the final 365-day time point, DNA was extracted from 1.5 mL aliquots from all MGIT cultures, followed by a genus-specific PCR, subsequent Sanger amplicon sequencing and MTBC speciation by RD-PCR of all samples identified as containing MTBC DNA following amplicon sequencing, as described below. Furthermore, spoligotyping were performed on all M. bovis-positive MGIT samples (confirmed by RD-PCR) following an established protocol as described by Kamerbeek et al. (1997). It is noteworthy that the MGIT negative control tubes consistently maintained a culture-negative status throughout the entire duration of this process.
The Ultra assay (Cepheid) was performed on all raw nasal swab aliquots (n = 157; Goosen et al., 2020). Briefly, Ultra sample lysis reagent was added to the swab sample aliquot at a ratio of 2:1, thoroughly vortexed for 10 s, incubated at room temperature for 10 min, vortexed for 10 s, and incubated for a final 5 min at room temperature in a BSL-3 laboratory. Thereafter, the solution was transferred into the sample chamber of the Ultra cartridge. Samples were analyzed for the presence of MTBC DNA using the automated GeneXpert module PCR system (Cepheid). The read-out of the Ultra assay was recorded as MTB detected high, medium, low, very low, MTB trace detected, or MTB not detected (Goosen et al., 2020). The “MTB not detected” read-out was regarded as an Ultra negative result (no MTBC DNA present) and all other read-outs were considered a positive Ultra result.
Mycobacterium genus-specific rpoB (764 bp) and hsp65 (436 bp) Q5 HiFi Taq (New England Biolabs, Ipswich, MA, United States) PCRs were performed to detect the presence of any Mycobacteria spp. These PCRs used DNA extracted from (a) 1.5 mL boiled aliquots from MGIT cultures (n = 157) after 365 days and (b) a 500 μL aliquot directly from raw nasal swabs (n = 157), as previously described (Clarke et al., 2022b). The PCR amplicons for rpoB and hsp65 (referred to as genus-specific PCR) from cultures were pooled for each goat and sent to the Central Analytical Facility (CAF, Stellenbosch University, Stellenbosch, SA) for Sanger sequencing. Similarly, genus-specific PCR amplicons (hsp65 and rpoB) using DNA extracted from raw swabs (b) were also pooled for each goat and further deep sequenced as a separate sample for a select few animals using the Ion S5™ next generation sequencing platform (Thermo Fisher, Waltham, MA, United States) at CAF.
Briefly, genus-specific amplicon pools (hsp65 and rpoB) of each animal’s raw swabs were selected to undergo deep sequencing if they had Ultra positive results and if they had concordant Mycobacteria spp. results identified through Sanger sequencing between raw swabs and swab cultures. Furthermore, all MTBC positive results, as indicated by Sanger and/or deep sequencing, were further speciated using RD-PCR to confirm the presence or absence of M. bovis, as previously described (Warren et al., 2006). Controls included: (1) DNA extraction controls, and (2) PCR amplification controls (positive and negative). All controls were included during each PCR and subsequent sequencing events.
For all Sanger sequences generated, mycobacterial species identification was performed by NCBI BLAST analysis using a species identity match threshold ≥99 and 100% coverage as previously described (Clarke et al., 2022b). For Ion Torrent sequencing, Flow space calibration and BaseCaller analyses were performed using default analysis parameters in the Torrent Suite Version 5.16.1 software (Thermo Fisher). Deep sequences generated by the Ion Torrent platform reference-free species level identification, down to taxonomic level 7, were identified using QIIME2’s database and QIIME2view,1 as previously described (Shi et al., 2019; Figure 2).
Frequency distributions of identified mycobacterial species in swab samples, determined by Sanger sequencing, were grouped, and reported by the number of goats tested. The Ion Torrent amplicon deep sequencing results were reported as the percentage of high-quality sequenced reads assigned to a specific Mycobacteria spp.; this provided a description of the relative abundance of identified mycobacteria in the polymicrobial samples (goat nasal swabs; Deurenberg et al., 2017).
Of the 157 swab samples processed for mycobacterial culture, MGIT growth was detected in 123 (78%). Based on genus-specific PCR amplicon Sanger results, matching Mycobacteria spp. were identified in paired raw swab DNA and swab cultures from 53 goats (Figure 3; Supplementary Table 1; i.e., complete concordance). Moreover, raw swab aliquots from all goats were screened for MTBC DNA with the GeneXpert® MTB/RIF Ultra. Of the 53 goats of interest, 20 (38%) had positive results for MTBC DNA using the Ultra. This included one swab from a commercial goat herd (Supplementary Table 2).
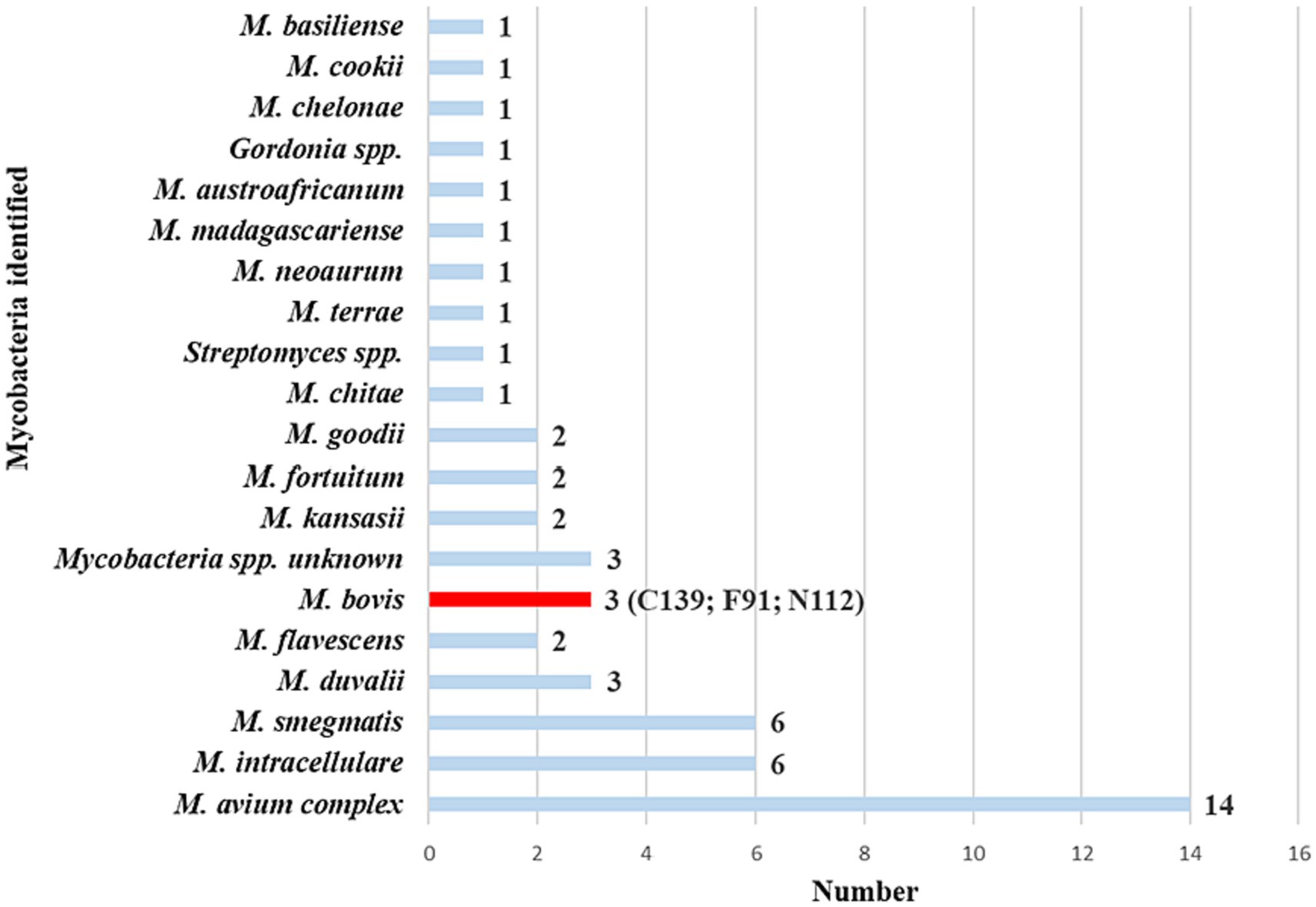
Figure 3. Mycobacteria spp. identified from 53 goat nasal swabs using genus-specific PCR targeting hsp65 and rpoB. The PCR amplicons from the two targets were pooled from raw swab DNA and matching swab culture samples, then subjected to Sanger sequencing. There was complete concordance of identified mycobacterial species between both gene targets and sample types for each animal. This provided confidence in the results, leading to the selection of these animals as the focus group for further investigation. Subsequently, samples were further speciated using region-of-difference (RD) PCR to confirm the presence or absence of Mycobacterium bovis. The number of goats with each species of Mycobacterium is reported next to each species name. Goat IDs for the three M. bovis positive goats were C139, F91, and N112.
Swab samples from the 20 goats with positive Ultra results were further evaluated by performing a genus-specific PCR with Sanger sequencing and RD-PCR to confirm and speciate any MTBC present. Results showed that 3 goats had M. bovis DNA in paired cultured and raw swab samples. Unfortunately, further speciation by spoligotyping was unsuccessful for all M. bovis-positive MGIT samples. All three goats were from communal herds. Figure 4 shows the diversity of all mycobacterial species identified in the 20 Ultra positive goats.
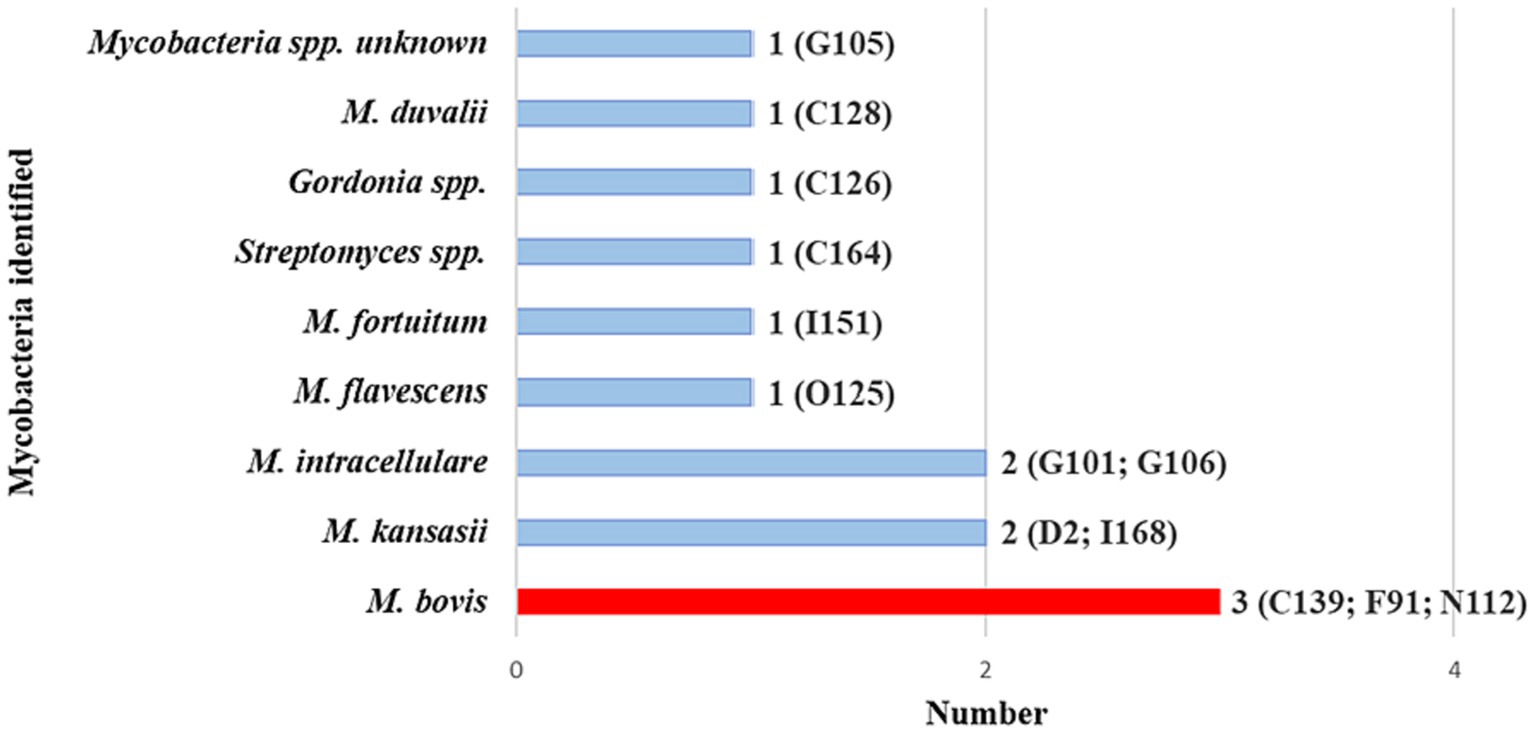
Figure 4. Mycobacterial species identified by Sanger sequencing of pooled hsp65 and rpoB amplicons generated from DNA extracted from culture and raw swabs in the 20 GeneXpert® MTB/RIF Ultra positive goats. These goats are a subset of the 53 with the same Mycobacteria spp. in both the raw samples and cultured samples, as per Sanger sequencing results (shown in Figure 3). Individual goat ID and number of goats with a specific Mycobacterium spp. are shown. Goat IDs for the three Mycobacterium bovis-positive goats were C139, F91, and N112.
In addition, the amplicon pools derived from the 20 raw swab samples, were subjected to PCR amplicon deep sequencing. Mycobacterium bovis DNA was identified in 11 out of 20 goat swab samples (Supplementary Table 1). The positive group included the 3 goats (goat IDs: C139, F91 and N112) whose cultured samples were also M. bovis positive, indicating the presence of viable M. bovis. Deep sequencing of amplicon targets produced high numbers of total reads assigned to Mycobacterium genus, as shown at the top of Figure 5. The relative abundance (i.e., percentage of reads) of M. bovis DNA among all Mycobacteria spp. identified in the 20 goats is also shown in Figure 5. Results ranged from 0.8 to 97.5%. The three goat samples that were also culture-positive for M. bovis had a high percentage of M. bovis-specific reads, with goat C139 having 64.1%, F91 having 93.4%, and N112 with 68%. Interestingly, there were other goat samples with high percentages of M. bovis reads that did not have M. bovis detected in cultures, for example, goat O125 with 97.5%, and C126 with 65.8%. Raw samples from three goats with very low M. bovis abundance (E72 with 1.2%, G106 with 4.7%, I166 with 0.8%) were associated with having Ultra positive, but negative culture results. Although 20 goats had Ultra positive results, only 11 were confirmed to have M. bovis DNA using amplicon deep sequencing, and only 3 were considered M. bovis positive using culture.
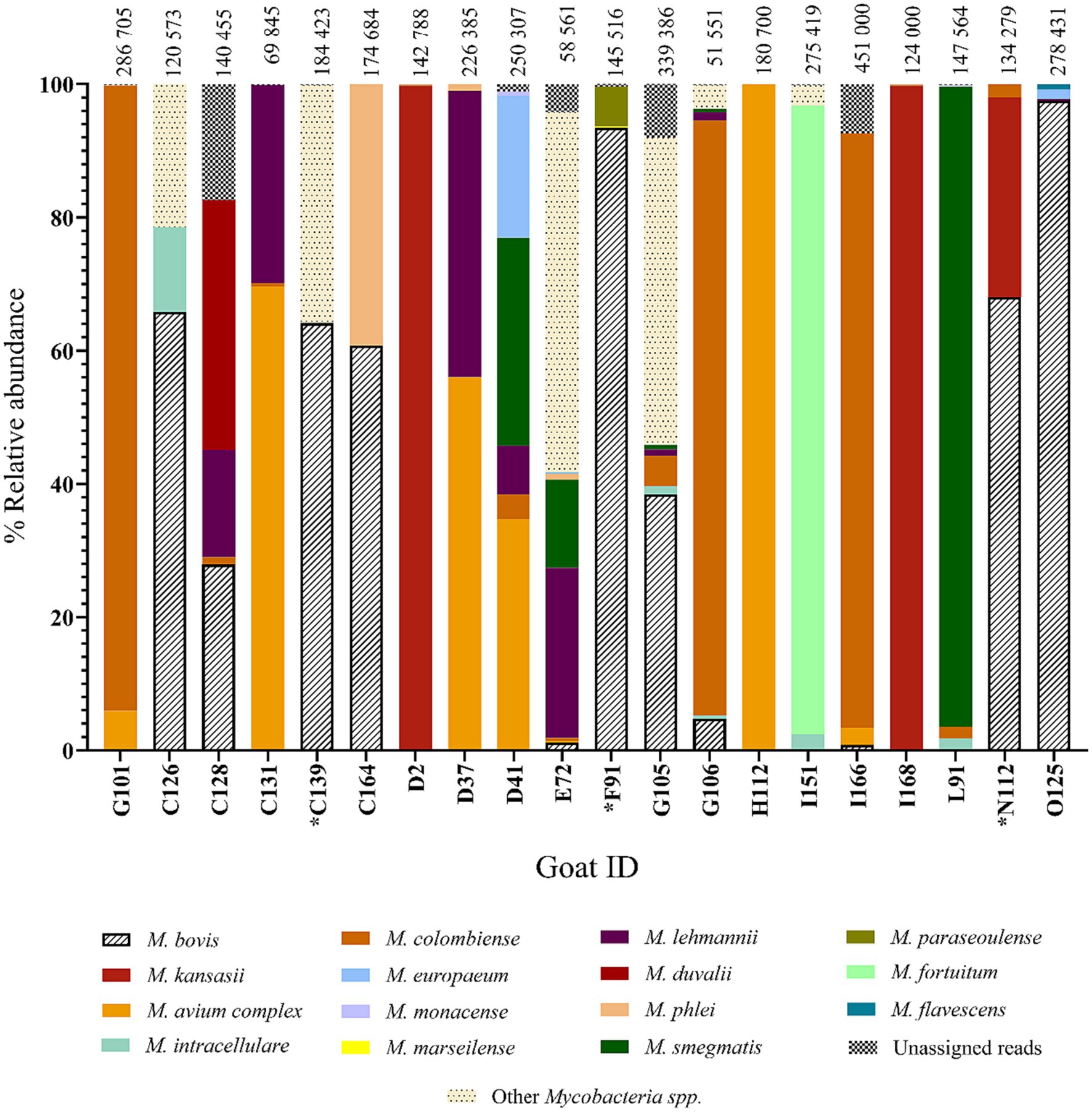
Figure 5. Relative abundance of Mycobacteria spp. by individual goat ID (n = 20). Data represent the amplicon deep sequencing read for rpoB and hsp65 combined that aligned to various Mycobacteria spp. identified from the raw swab DNA. The 20 goats shown here are also represented in Figure 4 and include all animals that were GeneXpert® MTB/RIF Ultra positive that had the same Mycobacteria spp. Sanger sequencing results for their raw swab DNA and the matching swab culture. *Mycobacterium bovis was detected by all three approaches (M. bovis DNA discovered by genus-specific PCR and Sanger sequencing of both culture and raw samples, as well as by deep sequencing of amplicon targets, Goat IDs: C139, F91, and N112).
The swab samples also contained a range of NTM species, which were identified based on the deep sequencing results. The relative abundances of different NTM species and M. bovis in the 20 Ultra positive goats are shown in Figure 5. The NTM species identified included M. avium complex, as well as M. kansasii, M. fortuitum, M. goodii, M. flavescens, M. terrae, and M. virginiense (Supplementary Table 2).
This study aimed to determine whether M. bovis could be detected in nasal swabs from goats sharing pastures and water points with M. bovis infected cattle in KwaZulu-Natal, SA. Using mycobacterial culture and culture-independent techniques, 53 out of 157 were identified with matched Mycobacterium spp. These goat samples were further characterized using the Ultra as a rapid sensitive screening method, which resulted in 20 goat swabs suspected to contain MTBC DNA. Based on PCR results from DNA extracted from cultures, 3 of the 20 Ultra positive goat samples were identified to have viable M. bovis present in respiratory secretions. However, deep sequencing of PCR amplicons using DNA extracted directly from swabs detected an additional eight goats with M. bovis DNA, as well as the three culture positive goats. Based on the presence of M. bovis DNA in 11 goat nasal samples, it appeared that communal goats were exposed and possibly infected. This is not surprising since it was hypothesized that these goats were at risk since they shared grazing and water sources with M. bovis infected cattle, could have undetected goat to goat transmission, and a low likelihood of interaction with infected wildlife in nearby game parks, as has been previously observed (Ciaravino et al., 2021). Results also highlight the potential value of culture-independent detection methods for epidemiological investigations as well as an ancillary method for antemortem diagnosis of M. bovis in goats.
A large portion of the nasal swabs were positive for growth in MGIT cultures (78%; 123/157). This was expected since nasal swabs were likely to contain environmental contamination with NTMs and other organisms. Non-tuberculous mycobacteria are ubiquitous in the environment and studies have confirmed a high diversity of environmental NTMs in SA (Gcebe et al., 2013). This was likely why there was a high percentage (43%; 53/123) of positive cultures that contained matching mycobacterial species in both the culture isolate and DNA extracted directly from swabs. The observed diversity of NTMs identified suggests that these goats were exposed to environmental mycobacteria, many of which have been recently proven to confound M. bovis diagnosis in guinea pigs, especially when using tests based on host immune responses (Fernández-Veiga et al., 2023). Further studies are needed to investigate the association between presence of NTMs and potential cross-reactivity in M. bovis specific cell-mediated immunological tests in goats.
Although matched Mycobacterium spp. was identified in culture and directly from swabs in 53 goats, Sanger sequencing results could not distinguish NTM from MTBC. Therefore, the Ultra was used as a rapid, sensitive method to screen raw swabs for the presence of MTBC in these samples. One goat from the commercial stud herd was among the 20 Ultra positive goats. Since the herd history supported the assumption that commercial stud goats had no previous exposure to M. bovis, and further deep sequencing did not confirm the presence of M. bovis, this finding was most likely a false positive result. Although high bacterial loads of NTMs have been suspected as a cause of false positive Ultra results, one study determined that M. abscessus, M. aurum, M. marinum, M. phlei, and M. smegmatis do not lead to cross-reactivity (Huh et al., 2019). Furthermore, a positive Ultra result does not differentiate between dead or non-viable mycobacteria, or residual DNA within the sample, as has been observed in human patients (Miotto et al., 2012; Theron et al., 2018). Despite this, the Ultra’s diagnostic appeal, for use on animal specimens suspected of having MTBC infections, is increasing (Chakravorty et al., 2017), and has provided rapid detection of MTBC DNA in tissue and respiratory samples collected from infected African buffaloes (Clarke et al., 2021, 2022a), African elephants (Goosen et al., 2020), and white rhinoceros (Goosen et al., 2020). The positive Ultra results in the 19 communal goats were likely true positives, particularly since they share grazing and water sources with M. bovis infected cattle (Sichewo et al., 2020). Therefore, further analyses of Ultra positive samples were performed to speciate and confirm the presence of M. bovis.
Mycobacterial culture is the cornerstone for detecting viable MTBC in animal samples (Bernitz et al., 2021). Therefore, one of the swab aliquots from each goat was processed for mycobacterial culture to detect the presence of viable MTBC. Of the 20 Ultra positive goats, 3 were confirmed to have M. bovis growth in culture from their nasal swabs, based on genus-specific PCR results and confirmed by RD-PCR. All three samples were also Ultra positive, which shows promise as a screening tool to identify potential infected animals. Since it is known that nasal secretions may be paucibacillary and to overcome the suboptimal sensitivity of culture (Corner et al., 2012), the duration of culture for the goat nasal swabs was extended from the conventional 56 days to 365 days. The presence of viable M. bovis, in at least three samples suggests that these goats either were infected and shedding or had respiratory colonization. Although further testing would be required to differentiate between these conditions, this finding supports the hypothesis that communal goats in this area were likely exposed and may become infected with M. bovis.
There were eight additional goats identified with M. bovis DNA, based on amplicon deep sequencing. Since these were only detected using DNA extracted directly from swabs, it suggests that these samples were paucibacillary, in a non-culturable form, or represented residual M. bovis DNA. These results may be attributed to several factors. The number of viable bacteria in nasal samples can differ depending on the stage of disease and intermittent shedding (de la Rua-Domenech et al., 2006; de Souza Figueiredo et al., 2010), affecting the culture outcome. A sample’s viable bacterial load may also be reduced during the collection, handling and storage of samples (Corner et al., 2012; Goosen et al., 2022b). The culture of MTBC also involves a decontamination process, which can reduce the amount of viable MTBC (Burdz et al., 2003; Steingart et al., 2006; Madigan, 2012). Nasal swabs are expected to contain a high number of contaminating microbes, including environmental non-tuberculous mycobacteria, which may outcompete or inhibit growth of slow growing MTBC (Robbe-Austerman et al., 2013). This was supported by the high proportion of MGIT cultures with positive growth. Since the swab sample diluent was split for different analyses, it is also possible that the presence and number of MTBC bacilli varied between the aliquots. The most likely explanations for the discordant results between direct and culture detection were that MTBC were present in low numbers, had variable viability, and the high level of contamination inhibited growth of MTBC in culture. Therefore, MTBC DNA detection should employ more than one technique to increase confidence in results.
The relative abundance of M. bovis DNA, determined by amplicon deep sequencing, varied in the 11 positive goat samples, which is not surprising given the complex nature of the sample. The 3 M. bovis culture positive goats had higher numbers of M. bovis-specific reads (range 91,310–135,956). Six of the 11 goats had M. bovis-specific sequence reads >79,000, increasing confidence in these results, despite not finding viable bacilli by culture in some cases. However, we were unable to discern whether the M. bovis was inhaled from the environment vs. being secreted by an infected individual. Despite this limitation, this finding is important since it suggests M. bovis was present in this system. A study in humans suggested that presence of M. tuberculosis DNA in nasal swabs might precede development of pulmonary infection (Balcells et al., 2016). We speculated that goats with higher numbers of M. bovis reads might indicate that these goats were infected and shedding, vs. simple contamination since lower numbers of bacilli would likely be present in the environment (Santos et al., 2015). However, without ancillary testing, such as M. bovis specific immunological responses in these goats, it is difficult to discern the source of the mycobacteria. Further studies should explore the association between numbers of sequence reads and likelihood of infection.
The use of whole genome and targeted next-generation sequencing (tNGS) for diagnosis and detection of drug resistance in Mycobacterium tuberculosis in clinical samples has grown exponentially in recent years (Nimmo et al., 2019; Cabibbe et al., 2020). The advantages are rapid culture-independent results to inform patient care. In addition, this approach has shown that there is greater genomic diversity in sputum than sequences derived from culture (Nimmo et al., 2019). Similar to this goat study, Kambli et al. (2021) screened sputum with Ultra and then used tNGS to diagnose patients and determine drug resistance profiles (Kambli et al., 2021). Bacterial load, as assessed by Ultra, appeared to correlate with hsp65 gene coverage depth. In livestock and wildlife, PCR-based tools have been used with oronasal swabs to detect Mycobacterium bovis infection and shedding (McCorry et al., 2005; Clarke et al., 2022b). In naturally infected wild boar, nasal shedding of MTBC DNA was detected in 40.8% of TB-affected animals and 73.6% of these had generalized TB lesions in head lymph nodes and lungs (Risco et al., 2019). Therefore, it is likely that at least some of the goats with positive nasal swabs were infected, although this could not be confirmed due to limitations of the tests performed.
This study had several limitations, including the inability to confirm infection/disease using antemortem tests to detect M. bovis-specific host immune responses, or postmortem techniques including mycobacterial culture and histopathology. Although three goats did have M. bovis isolated by culture, it was only after an extended (365 days) period of incubation and was likely a very paucibacillary sample, which may not represent true infection. The presence of NTMs in the complex samples could have also obscured the presence of M. bovis by rapid overgrowth. Most positive goats had only M. bovis DNA detected and this could represent contamination from environmental sources or residual DNA rather than infection. Variability between PCR amplification and culturability among different mycobacteria species may also contribute to some of the discrepancies observed. Furthermore, attempts to obtain further genomic data for the M. bovis positive samples through both culture and raw aliquots, were unsuccessful, with spoligotyping returning indistinguishable patterns. Additional techniques will be pursued to characterize these isolates. In addition, the process for identifying M. bovis DNA in these 11 goats required several labor-intensive and costly steps, including multiple PCRs and deep sequencing. Therefore, although this approach may be valuable for research investigations and routine surveillance in developed countries, at the moment, it is a bit more challenging for developing countries.
The detection of viable M. bovis and DNA in goat nasal passages provides evidence that goats were exposed and potentially infected. Although one dairy goat had an Ultra positive result, the majority of goats with M. bovis DNA detected were from communal herds, which suggests that there may be a greater risk of infection in goats that share an environment with infected cattle. The Ultra appeared to be a useful screening tool to detect MTBC and select nasal swabs for additional analyses. The genus-specific PCR with amplicon Sanger sequencing and RD-PCR could identify animals with higher abundance of M. bovis, but miss animals with lower abundance, which could be found with targeted deep sequencing. This culture-independent approach has promise for improved detection of M. bovis in paucibacillary samples from goats.
The datasets presented in this study can be found in an online repository available at: https://www.ebi.ac.uk/ena/browser/search under project reference number PRJEB70955. Moreover, samples-, experiment- and run accession numbers can be found under Supplementary material.
The animal studies were approved by Stellenbosch University Animal Care and Use Committee (ACU‐2020‐14560) and the Department of Agriculture, Land Reform, and Rural Development [12/11/1/7/2 (16045S)]. The studies were conducted in accordance with the local legislation and institutional requirements. Written informed consent was obtained from the owners for the participation of their animals in this study.
DC: Formal Analysis, Investigation, Methodology, Visualization, Writing – original draft. CC: Investigation, Methodology, Resources, Writing – review & editing. TK: Data curation, Methodology, Writing – review & editing. RW: Formal Analysis, Methodology, Supervision, Writing – review & editing. CW: Data curation, Formal Analysis, Investigation, Methodology, Writing – review & editing. MM: Conceptualization, Data curation, Formal Analysis, Funding acquisition, Investigation, Methodology, Project administration, Resources, Supervision, Visualization, Writing – original draft, Writing – review & editing. WG: Conceptualization, Data curation, Formal Analysis, Funding acquisition, Investigation, Methodology, Project administration, Resources, Supervision, Validation, Visualization, Writing – original draft, Writing – review & editing.
The author(s) declare financial support was received for the research, authorship, and/or publication of this article. This research was funded by the Wellcome Foundation (grant #222941/Z/21/Z), the South African Medical Research Council Centre for TB research, American Association of Zoo Veterinarians Wild Animal Health Fund (S005651 and S007355), the National Research Foundation South African Research Chair Initiative (grant #86949), and the European Union Global Health EDCTP3 Joint undertaking and its members (project 101103171).
The authors wish to thank David Mayida and Warren McCall from the Hluhluwe State Veterinary office for their assistance with this study. In addition, we acknowledge the assistance provided by Animal Health Technicians Alicia McCall and Simon Mncube, who helped with the sourcing as well as the sampling of the goats used in this study.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Views and opinions expressed are however those of the author(s) only, and do not necessarily reflect those of the funders.
The Supplementary material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2024.1349163/full#supplementary-material
Adékambi, T., Colson, P., and Drancourt, M. (2003). rpoB-based identification of nonpigmented and late-pigmenting rapidly growing mycobacteria. J. Clin. Microbiol. 41, 5699–5708. doi: 10.1128/JCM.41.12.5699-5708.2003
Arnot, L. F., and Michel, A. (2020). Challenges for controlling bovine tuberculosis in South Africa. Onderstepoort J. Vet. Res. 87, e1–e8. doi: 10.4102/ojvr.v87i1.1690
Balcells, M. E., Huilcamán, M., Peña, C., Castillo, C., Carvajal, C., Scioscia, N., et al. (2016). M. tuberculosis DNA detection in nasopharyngeal mucosa can precede tuberculosis development in contacts. Int. J. Tuberc. Lung Dis. 20, 848–852. doi: 10.5588/ijtld.15.0872
Bernitz, N., Kerr, T. J., Goosen, W. J., Chileshe, J., Higgitt, R. L., Roos, E. O., et al. (2021). Review of diagnostic tests for detection of Mycobacterium bovis infection in south African wildlife. Front. Vet. Sci. 8:697. doi: 10.3389/fvets.2021.588697
Bezos, J., Casal, C., Díez-Delgado, I., Romero, B., Liandris, E., Álvarez, J., et al. (2015). Goats challenged with different members of the Mycobacterium tuberculosis complex display different clinical pictures. Vet. Immunol. Immunopathol. 167, 185–189. doi: 10.1016/j.vetimm.2015.07.009
Bolaños, C. A. D., Paula, C. L., Guerra, S. T., Franco, M. M. J., and Ribeiro, M. G. (2017). Diagnosis of mycobacteria in bovine milk: an overview. Rev. Inst. Med. Trop. Sao Paulo 59:e40. doi: 10.1590/S1678-9946201759040
Borham, M., Oreiby, A., El-Gedawy, A., Hegazy, Y., Khalifa, H. O., Al-Gaabary, M., et al. (2022). Review on bovine tuberculosis: an emerging disease associated with multidrug-resistant Mycobacterium species. Pathogens 11:715. doi: 10.3390/pathogens11070715
Brückner, G. K., Vosloo, W., Du Plessis, B. J. A., Kloeck, P. E. L. G., Connoway, L., Ekron, M. D., et al. (2002). Foot and mouth disease: the experience of South Africa. Rev. Sci. Tech. 21, 751–764. doi: 10.20506/rst.21.3.1368
Burdz, T. V. N., Wolfe, J., and Kabani, A. (2003). Evaluation of sputum decontamination methods for Mycobacterium tuberculosis using viable colony counts and flow cytometry. Diagn. Microbiol. Infect. Dis. 47, 503–509. doi: 10.1016/s0732-8893(03)00138-x
Cabibbe, A. M., Spitaleri, A., Battaglia, S., Colman, R. E., Suresh, A., Uplekar, S., et al. (2020). Application of targeted next-generation sequencing assay on a portable sequencing platform for culture-free detection of drug-resistant tuberculosis from clinical samples. J. Clin. Microbiol. 58, e00632–e00620. doi: 10.1128/JCM.00632-20
Chakravorty, S., Simmons, A. M., Rowneki, M., Parmar, H., Cao, Y., Ryan, J., et al. (2017). The new Xpert MTB/RIF ultra: improving detection of Mycobacterium tuberculosis and resistance to rifampin in an assay suitable for point-of-care testing. MBio 8, e00812–e00817. doi: 10.1128/mBio.00812-17
Ciaravino, G., Vidal, E., Cortey, M., Martín, M., Sanz, A., Mercader, I., et al. (2021). Phylogenetic relationships investigation of Mycobacterium caprae strains from sympatric wild boar and goats based on whole genome sequencing. Transbound. Emerg. Dis. 68, 1476–1486. doi: 10.1111/tbed.13816
Clarke, C., Cooper, D. V., Miller, M. A., and Goosen, W. J. (2022a). Detection of Mycobacterium tuberculosis complex DNA in oronasal swabs from infected African buffaloes (Syncerus caffer). Sci. Rep. 12, 1834–1836. doi: 10.1038/s41598-022-05982-6
Clarke, C., Kerr, T. J., Warren, R. M., Kleynhans, L., Miller, M. A., and Goosen, W. J. (2022b). Identification and characterisation of nontuberculous mycobacteria in African buffaloes (Syncerus caffer), South Africa. Microorganisms 10:1861. doi: 10.3390/microorganisms10091861
Clarke, C., Smith, K., Goldswain, S. J., Helm, C., Cooper, D. V., Kerr, T. J., et al. (2021). Novel molecular transport medium used in combination with Xpert MTB/RIF ultra provides rapid detection of Mycobacterium bovis in African buffaloes. Sci. Rep. 11, 7061–7066. doi: 10.1038/s41598-021-86682-5
Cooke, D. M., Goosen, W. J., Burgess, T., Witte, C., and Miller, M. A. (2023). Mycobacterium tuberculosis complex detection in rural goat herds in South Africa using Bayesian latent class analysis. Vet. Immunol. Immunopathol. 257:110559. doi: 10.1016/j.vetimm.2023.110559
Corner, L. A. L., Gormley, E., and Pfeiffer, D. U. (2012). Primary isolation of Mycobacterium bovis from bovine tissues: conditions for maximising the number of positive cultures. Vet. Microbiol. 156, 162–171. doi: 10.1016/j.vetmic.2011.10.016
de Boer, A. S., Blommerde, B., de Haas, P. E. W., Sebek, M. M. G. G., Lambregts-van Weezenbeek, K. S. B., Dessens, M., et al. (2002). False-positive Mycobacterium tuberculosis cultures in 44 laboratories in the Netherlands (1993 to 2000): incidence, risk factors, and consequences. J. Clin. Microbiol. 40, 4004–4009. doi: 10.1128/JCM.40.11.4004-4009.2002
de la Rua-Domenech, R., Goodchild, A. T., Vordermeier, H. M., Hewinson, R. G., Christiansen, K. H., and Clifton-Hadley, R. S. (2006). Ante mortem diagnosis of tuberculosis in cattle: a review of the tuberculin tests, gamma-interferon assay, and other ancillary diagnostic techniques. Res. Vet. Sci. 81, 190–210. doi: 10.1016/j.rvsc.2005.11.005
de Souza Figueiredo, E. E., Carvalho, R. C. T., Silvestre, F. G., Lilenbaum, W., Fonseca, L. S., Silva, J. T., et al. (2010). Detection of Mycobacterium bovis DNA in nasal swabs from tuberculous cattle by a multiplex PCR. Braz. J. Microbiol. 41, 386–390. doi: 10.1590/S1517-838220100002000020
Deggim-Messmer, V., Bloemberg, G. V., Ritter, C., Voit, A., Hömke, R., Keller, P. M., et al. (2016). Diagnostic molecular mycobacteriology in regions with low tuberculosis Endemicity: combining real-time PCR assays for detection of multiple mycobacterial pathogens with line probe assays for identification of resistance mutations. EBioMedicine 9, 228–237. doi: 10.1016/j.ebiom.2016.06.016
Department of Agriculture, Forestry and Fisheries of SA . (2016). Bovine tuberculosis manual. Available at: https://nahf.co.za/updated-tuberculosis-manual/ (Accessed 7 November 2023).
Department of Agriculture, Forestry and Fisheries of SA . (2017). Abstract of agricultural statistics 2015. Available at: http://webapps.daff.gov.za/AmisAdmin/upload/2017%20Abstract%20%20of%20Agricultural%20Statistics.pdf (Accessed 3 November 2023).
Department of Agriculture, Land Reform and Rural Development . (2018). Tuberculosis testing in sheep and goat’s manual. Available at: https://nahf.co.za/tuberculosis-testing-in-sheep-and-goats-2018-11-29/ (Accessed 7 November 2023).
Deurenberg, R. H., Bathoorn, E., Chlebowicz, M. A., Couto, N., Ferdous, M., García-Cobos, S., et al. (2017). Application of next generation sequencing in clinical microbiology and infection prevention. J. Biotechnol. 243, 16–24. doi: 10.1016/j.jbiotec.2016.12.022
Falkinham, J. O. (2021). Ecology of nontuberculous mycobacteria. Microorganisms 9:2262. doi: 10.3390/microorganisms9112262
Fernández-Veiga, L., Fuertes, M., Geijo, M. V., Pérez de Val, B., Vidal, E., Michelet, L., et al. (2023). Differences in skin test reactions to official and defined antigens in guinea pigs exposed to non-tuberculous and tuberculous bacteria. Sci. Rep. 13:2936. doi: 10.1038/s41598-023-30147-4
Gcebe, N., Rutten, V., Gey van Pittius, N. C., and Michel, A. (2013). Prevalence and distribution of non-tuberculous mycobacteria (NTM) in cattle, African buffaloes (Syncerus caffer) and their environments in South Africa. Transbound. Emerg. Dis. 60, 74–84. doi: 10.1111/tbed.12133
Ghodbane, R., Raoult, D., and Drancourt, M. (2014). Dramatic reduction of culture time of Mycobacterium tuberculosis. Sci. Rep. 4:4236. doi: 10.1038/srep04236
Goosen, W. J., Clarke, C., Kleynhans, L., Kerr, T. J., Buss, P., and Miller, M. A. (2022a). Culture-independent PCR detection and differentiation of mycobacteria spp. in antemortem respiratory samples from African elephants (Loxodonta africana) and rhinoceros (Ceratotherium simum, Diceros bicornis) in South Africa. Pathogens 11:709. doi: 10.3390/pathogens11060709
Goosen, W. J., Kerr, T. J., Kleynhans, L., Warren, R. M., van Helden, P. D., Persing, D. H., et al. (2020). The Xpert MTB/RIF ultra assay detects Mycobacterium tuberculosis complex DNA in white rhinoceros (Ceratotherium simum) and African elephants (Loxodonta africana). Sci. Rep. 10:14482. doi: 10.1038/s41598-020-71568-9
Goosen, W. J., Kleynhans, L., Kerr, T. J., van Helden, P. D., Buss, P., Warren, R. M., et al. (2022b). Improved detection of Mycobacterium tuberculosis and M. bovis in African wildlife samples using cationic peptide decontamination and mycobacterial culture supplementation. J. Vet. Diagn. Invest. 34, 61–67. doi: 10.1177/10406387211044192
Huh, H. J., Song, D. J., Ki, C. S., and Lee, N. Y. (2019). Is cross-reactivity with nontuberculous mycobacteria a systematic problem in the Xpert MTB/RIF assay? Tuberc. Respir. Dis. 82, 88–89. doi: 10.4046/trd.2018.0075
Jung, Y. J., Kim, J.-Y., Song, D. J., Koh, W.-J., Huh, H. J., Ki, C.-S., et al. (2016). Evaluation of three real-time PCR assays for differential identification of Mycobacterium tuberculosis complex and nontuberculous mycobacteria species in liquid culture media. Diagn. Microbiol. Infect. Dis. 85, 186–191. doi: 10.1016/j.diagmicrobio.2016.03.014
Kambli, P., Ajbani, K., Kazi, M., Sadani, M., Naik, S., Shetty, A., et al. (2021). Targeted next generation sequencing directly from sputum for comprehensive genetic information on drug resistant Mycobacterium tuberculosis. Tuberculosis 127:102051. doi: 10.1016/j.tube.2021.102051
Kamerbeek, J., Schouls, L., Kolk, A., van Agterveld, M., van Soolingen, D., Kuijper, S., et al. (1997). Simultaneous detection and strain differentiation of Mycobacterium tuberculosis for diagnosis and epidemiology. J. Clin. Microbiol. 35, 907–914. doi: 10.1128/jcm.35.4.907-914.1997
Lee, R. S., Proulx, J.-F., McIntosh, F., Behr, M. A., and Hanage, W. P. (2020). Previously undetected super-spreading of Mycobacterium tuberculosis revealed by deep sequencing. Elife 9:e53245. doi: 10.7554/eLife.53245
Madigan, G. (2012). Evaluation of different methods for the detection of Mycobacterium bovis in lymph node tissue. Thesis of Master of Science, National University of Ireland Maynooth. 37.
McCorry, T., Whelan, A. O., Welsh, M. D., McNair, J., Walton, E., Bryson, D. G., et al. (2005). Shedding of Mycobacterium bovis in the nasal mucus of cattle infected experimentally with tuberculosis by the intranasal and intratracheal routes. Vet. Rec. 157, 613–618. doi: 10.1136/vr.157.20.613
Michel, A. L., Cooper, D., Jooste, J., de Klerk, L.-M., and Jolles, A. (2011). Approaches towards optimizing the gamma interferon assay for diagnosing Mycobacterium bovis infection in African buffalo (Syncerus caffer). Prev. Vet. Med. 98, 142–151. doi: 10.1016/j.prevetmed.2010.10.016
Miotto, P., Bigoni, S., Migliori, G. B., Matteelli, A., and Cirillo, D. M. (2012). Early tuberculosis treatment monitoring by Xpert® MTB/RIF. Eur. Respir. J. 39, 1269–1271. doi: 10.1183/09031936.00124711
More, S. J. (2019). Can bovine TB be eradicated from the Republic of Ireland? Could this be achieved by 2030? Ir. Vet. J. 72:3. doi: 10.1186/s13620-019-0140-x
Mostowy, S., Inwald, J., Gordon, S., Martin, C., Warren, R., Kremer, K., et al. (2005). Revisiting the evolution of Mycobacterium bovis. J. Bacteriol. 187, 6386–6395. doi: 10.1128/JB.187.18.6386-6395.2005
Nimmo, C., Shaw, L. P., Doyle, R., Williams, R., Brien, K., Burgess, C., et al. (2019). Whole genome sequencing Mycobacterium tuberculosis directly from sputum identifies more genetic diversity than sequencing from culture. BMC Genomics 20:389. doi: 10.1186/s12864-019-5782-2
Nyoni, G. , (2019). Questionnaire-based study to determine the state of tuberculosis testing in goats in South Africa. (MSc (Tropical Animal Health) mini dissertation), University of Pretoria. Available at: http://hdl.handle.net/2263/76754 (Accessed 20 January 2024).
Palmer, M. V., Thacker, T. C., Waters, W. R., Gortázar, C., and Corner, L. A. L. (2012). Mycobacterium bovis: a model pathogen at the interface of livestock, wildlife, and humans. Vet Med Int 2012:236205. doi: 10.1155/2012/236205
Palmer, M. V., and Waters, W. R. (2011). Bovine tuberculosis and the establishment of an eradication program in the United States: role of veterinarians. Vet Med Int 2011, 1–12. doi: 10.4061/2011/816345
Pesciaroli, M., Alvarez, J., Boniotti, M. B., Cagiola, M., Di Marco, V., Marianelli, C., et al. (2014). Tuberculosis in domestic animal species. Res. Vet. Sci. 97, S78–S85. doi: 10.1016/j.rvsc.2014.05.015
Quintas, H., Reis, J., Pires, I., and Alegria, N. (2010). Tuberculosis in goats. Vet. Rec. 166, 437–438. doi: 10.1136/vr.c1678
Renwick, A. R., White, P. C., and Bengis, R. G. (2007). Bovine tuberculosis in southern African wildlife: a multi-species host-pathogen system. Epidemiol. Infect. 135, 529–540. doi: 10.1017/S0950268806007205
Reviriego Gordejo, F. J., and Vermeersch, J. P., (2006). Towards eradication of bovine tuberculosis in the European Union. Veterinary microbiology, In: 4th International Conference on Mycobacterium bovis. 112, 101–109.
Risco, D., Martínez, R., Bravo, M., Fernández Llario, P., Cerrato, R., Garcia-Jiménez, W. L., et al. (2019). Nasal shedding of Mycobacterium tuberculosis in wild boar is related to generalized tuberculosis and concomitant infections. Vet. Rec. 185:629. doi: 10.1136/vr.105511
Robbe-Austerman, S., Bravo, D. M., and Harris, B. (2013). Comparison of the MGIT 960, BACTEC 460 TB and solid media for isolation of Mycobacterium bovis in United States veterinary specimens. BMC Vet. Res. 9:74. doi: 10.1186/1746-6148-9-74
Rodríguez, E., Sánchez, L. P., Pérez, S., Herrera, L., Jiménez, M. S., Samper, S., et al. (2009). Human tuberculosis due to Mycobacterium bovis and M. caprae in Spain, 2004–2007. Int. J. Tuberc. Lung Dis. 13, 1536–1541. Available at: https://pubmed.ncbi.nlm.nih.gov/19919773/.
Santos, N., Santos, C., Valente, T., Gortázar, C., Almeida, V., and Correia-Neves, M. (2015). Widespread environmental contamination with Mycobacterium tuberculosis complex revealed by a molecular detection protocol. PLoS One 10:e0142079. doi: 10.1371/journal.pone.0142079
Shi, W., Hu, Y., Zheng, X., Ning, Z., Wu, M., Xia, F., et al. (2019). Longitudinal profiling of gut microbiome among tuberculosis patients under anti-tuberculosis treatment in China: protocol of a prospective cohort study. BMC Pulm. Med. 19:211. doi: 10.1186/s12890-019-0981-9
Sichewo, P. R., Etter, E. M. C., and Michel, A. L. (2020). Wildlife-cattle interactions emerge as drivers of bovine tuberculosis in traditionally farmed cattle. Prev. Vet. Med. 174:104847. doi: 10.1016/j.prevetmed.2019.104847
Steingart, K. R., Ng, V., Henry, M., Hopewell, P. C., Ramsay, A., Cunningham, J., et al. (2006). Sputum processing methods to improve the sensitivity of smear microscopy for tuberculosis: a systematic review. Lancet Infect. Dis. 6, 664–674. doi: 10.1016/S1473-3099(06)70602-8
Theron, G., Venter, R., Smith, L., Esmail, A., Randall, P., Sood, V., et al. (2018). False-positive Xpert MTB/RIF results in retested patients with previous tuberculosis: frequency, profile, and prospective clinical outcomes. J. Clin. Microbiol. 56:17. doi: 10.1128/JCM.01696-17
Vordermeier, H. M., Brown, J., Cockle, P. J., Franken, W. P. J., Drijfhout, J. W., Arend, S. M., et al. (2007). Assessment of cross-reactivity between Mycobacterium bovis and M. kansasii ESAT-6 and CFP-10 at the T-cell epitope level. Clin. Vaccine Immunol. 14, 1203–1209. doi: 10.1128/CVI.00116-07
Warren, R. M., Gey van Pittius, N. C., Barnard, M., Hesseling, A., Engelke, E., de Kock, M., et al. (2006). Differentiation of Mycobacterium tuberculosis complex by PCR amplification of genomic regions of difference. Int. J. Tuberc. Lung Dis. 10, 818–822.
Welsh, M. D., Cunningham, R. T., Corbett, D. M., Girvin, R. M., McNair, J., Skuce, R. A., et al. (2005). Influence of pathological progression on the balance between cellular and humoral immune responses in bovine tuberculosis. Immunology 114. doi: 10.1111/j.1365-2567.2004.02003
World Health Organization . (2017). Roadmap for zoonotic tuberculosis. Available at: https://www.who.int/publications/i/item/9789241513043. (Accessed 6 June 2023).
World Health Organization . (2020). Global Tuberculosis report 2020, Geneva, Switzerland. Available at: https://www.who.int/publications/i/item/9789240013131 (Accessed 20 January 2024).
World Organization for Animal Health . (2023). Available at: https://www.woah.org/en/what-we-do/animal-health-and-welfare/animal-diseases/old-classification-of-diseases-notifiable-to-the-oie-list-b/ (Accessed 5 June 2023).
Keywords: animal tuberculosis, culture-independent detection, GeneXpert® MTB/RIF Ultra, Capra hircus, hsp65, rpoB, Ion Torrent Genexus sequencing, Mycobacterium bovis
Citation: Cooke DM, Clarke C, Kerr TJ, Warren RM, Witte C, Miller MA and Goosen WJ (2024) Detection of Mycobacterium bovis in nasal swabs from communal goats (Capra hircus) in rural KwaZulu-Natal, South Africa. Front. Microbiol. 15:1349163. doi: 10.3389/fmicb.2024.1349163
Received: 04 December 2023; Accepted: 01 February 2024;
Published: 14 February 2024.
Edited by:
Axel Cloeckaert, Institut National de recherche pour l’agriculture, l’alimentation et l’environnement (INRAE), FranceReviewed by:
Bernat Pérez de Val, IRTA-CReSA, Centre for Research on Animal Health, SpainCopyright © 2024 Cooke, Clarke, Kerr, Warren, Witte, Miller and Goosen. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Wynand J. Goosen, d2pnb29zZW5Ac3VuLmFjLnph
†These authors share senior authorship
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.