
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Microbiol. , 27 February 2024
Sec. Food Microbiology
Volume 15 - 2024 | https://doi.org/10.3389/fmicb.2024.1348159
This article is part of the Research Topic How the Application of Antimicrobial Hurdles in Meat Processing Facilities Shapes Microbial Ecology View all 6 articles
Introduction: Targeted amplicon sequencing of the 16S rRNA delineates the complex microbial interactions that occur during food spoilage, providing a tool to intensively screen microbiota response to antimicrobial processing aids and interventions. The current research determines the microbiota and spoilage indicator (total aerobes and lactic acid bacteria; LAB) response to inorganic and organic antimicrobial intervention use on the shelf-life of fresh, never-frozen, skin-on, bone-in chicken wings.
Methods: Wings (n=200) were sourced from local processor and either not treated (NT) or treated with 15-s dips of tap water (TW), organic (peracetic acid; PAA), inorganic acids (sodium bisulfate; SBS), and their combination (SBS + PAA). Wings were stored (4°C) and rinsed in neutralizing Buffered Peptone Water (BPW) for 1 min on d 0, 7, 14, and 21 post-treatment. Spoilage indicators, aerobic mesophiles and LAB, were quantified from rinsates. Genomic DNA of d 14 and 21 rinsates were extracted, and V4 of 16S rRNA gene was sequenced. Sequences were analyzed using QIIME2.2019.7. APC and LAB counts were reported as Log10 CFU/g of chicken and analyzed in R Studio as a General Linear Model using ANOVA. Pairwise differences were determined using Tukey’s HSD (P£0.05).
Results: Spoilage was indicated for all products by day 21 according to APC counts (>7 Log10 CFU/g); however, wings treated with SBS and SBS + PAA demonstrated a 7-day extended shelf-life compared to those treated with NT, TW, or PAA. The interaction of treatment and time impacted the microbial diversity and composition (p < 0.05), with those treated with SBS having a lower richness and evenness compared to those treated with the controls (NT and TW; p < 0.05, Q < 0.05). On d 14, those treated with SBS and SBS + PAA had lower relative abundance of typical spoilage population while having a greater relative abundance of Bacillus spp. (~70 and 50% of population; ANCOM p < 0.05). By d 21, the Bacillus spp. populations decreased below 10% of the population among those treated with SBS and SBS + PAA.
Discussion: Therefore, there are differential effects on the microbial community depending on the chemical intervention used with organic and inorganic acids, impacting the microbial ecology differently.
Spoilage is a significant cost and concern for the poultry industry (Manning et al., 2007). In congruence with pathogen mitigation strategies, the poultry industry employs multiple technologies to extend the shelf life of poultry products (Souza et al., 2018; Baltic et al., 2019; Karaca et al., 2023). Antimicrobial agents, such as organic and inorganic acids, chlorine, and bromide-based chemicals, are used throughout poultry processing as sprays or immersion dips on whole birds or parts (Dittoe et al., 2019a,b; Feye et al., 2019; Micciche et al., 2019). However, due to the multiple selective pressures created by differing modes of action associated with each varying antimicrobial treatment, the frequent application of these chemical interventions used within a multi-hurdle approach alters or modifies the microbial ecology of poultry products (Rahman, 2015). In addition to selective pressures endured at the time of application, residues resulting from the extensive half-life of the processing antimicrobials on meat exist (Walsh et al., 2018).
With current methodologies employed by the poultry industry and the United States Department of Agriculture – Food Safety Inspection Service (USDA – FSIS), the impact these antimicrobial extension efforts have on the microbial ecology of the raw meat product is not a routine measurement of efficacy (Ricke et al., 2018). Traditionally, these routine measurements of shelf-life extension efforts include microbial plating of indicator organisms such as total mesophilic aerobic bacteria (>7 Log10 CFU/ml of poultry carcass rinsates) and lactic acid bacteria (LAB; > 6.8 Log10 CFU/ml of carcass rinsates; Balamatsia et al., 2007). However, these measurements do not provide insight into the early and late onset changes that impact the microbiota due to the chemical interventions utilized during or after processing. Due to the chemical composition and mode of action with these interventions, certain microorganisms associated with spoilage may be increased but not necessarily detected in mesophilic aerobic bacteria and LAB counts. For example, specific acid-tolerant spoilage populations, acetic acid bacteria, and acid-tolerant spore-forming spoilage bacteria may remain after treatment with acidic compounds, such as peracetic acid (Tran et al., 2020; Sun et al., 2021); however, these population differences will not be demonstrated in the indicator microorganisms, mesophilic aerobic bacteria, and LAB. Therefore, elucidating the microbiota composition of treated products over time and determining the microbiota’s stability may provide a more accurate understanding of how these chemical interventions impact shelf-life extension.
Recently, the use of sodium bisulfate (SBS) has been evaluated in previous studies for its ability to reduce experimentally inoculated foodborne pathogens on processed poultry parts (Dittoe et al., 2019a). The use of SBS has also been demonstrated to reduce premature browning and extend the shelf-life of apples (Fan et al., 2009; Kim et al., 2018). While it is an efficacious antimicrobial, the effect it has on the shelf-life of poultry as compared with the industry standard is not defined. Therefore, the objective of the current study was to determine the microbial quality of poultry wings over an extended period (21 days) after a short duration (15 s) treatment of either peroxyacetic acid (PAA), an organic acid and peroxide, or sodium bisulfate (SBS), an inorganic acid, as defined by traditional microbial plating (LAB and APC) and microbiome sequencing. Traditional microbial plating of spoilage bacteria (LAB) coupled with total microbial load (APC) may indicate microbial shifts that proceed to spoilage that correspond with the underlying microbiota dynamics. Ultimately, this study hopes to preliminarily evaluate the antimicrobial effects of SBS and PAA and their combination on the microbial ecology of poultry products and how these antimicrobials may impact the corresponding shelf-life using traditional microbiological testing methods.
A total of 200 wings (5 treatments, 4 time points, and 10 replicates) were obtained from a local chicken processor in northwest Arkansas. Wings were procured no longer than 24 h before the onset of the study. The bone-in, skin-on-whole chicken wings had a mean weight of 114.74 g with a standard error of 5.16 g.
The tap water used in the current experiment was not treated (un-sterilized) and was obtained from the Center for Food Safety at the University of Arkansas, which is supplied by the Beaver Water District (Lowell, AR, United States) that distributes water across northwest Arkansas. The water was reported to have less than 1 ppm of chlorine, fluoride, and nitrates and < 1 ppm of lead and copper (City of Fayetteville Arkansas, 2020). Therefore, the quality of the water should not have interfered with the chemistry of antimicrobial treatments. However, the water was not sterilized, which could have allowed the introduction of microorganisms. The tap water was not altered or sterilized in order to mimic industry settings.
The five experimental antimicrobial treatments were created by combining 15 L of tap water with either 3% sodium bisulfate (w/v; SBS, Jones-Hamilton Co., Walbridge, OH, United States), 500 ppm of peroxyacetic acid (Spectrum® 22, PeroxyChem, Philadelphia, PA, United States), or the combination of them to create the following treatments: a no treatment control (NT), Tap Water alone (TW), TW with the inclusion of 3% (w/v) SBS (SBS), TW with the addition of 500 ppm of PAA (PAA), and the combination of TW, SBS, and PAA (SBS + PAA). The treatment TW was included to demonstrate the rinsing effect of TW without the addition of antimicrobial treatments. A SympHony pH meter and probe (VWR International, Radnor, PA, United States) were used to determine the pH of the solutions (TW: 8.52 pH; SBS: 1.20 pH; PAA: 3.62 pH; SBS + PAA: 1.20 pH).
Subsequently, 350 mL of the treatments was aliquoted to sterile whirlpak bags for treatment application. The weight of the whole chicken wings was recorded and then independently dipped into sterile collection bags for 15 s (VWR International, Radnor, PA, United States) containing the five previously described treatments. Treatments were applied at ambient room temperature (20–22°C).
After the wings were treated in antimicrobial dips for 15 s, they were aseptically shaken (10–15 s) within the sterile collection bag to allow excess treatments to drip off prior to storage in new sterile collection bags. On d 0, the treated wings were allowed to rest for 2 min in the new collection bags prior to microbial analysis (Dittoe et al., 2019a). Immediately following the rest period, the wings were either evaluated immediately on d 0 or maintained at 4°C until d 7, 14, or 21 and analyzed for a total load of mesophilic aerobic bacteria and LAB per gram of wing. At each time point after treatment, d 0, 7, 14, and 21, the wings were rinsed with 150 mL of sterile neutralizing Buffered Peptone Water (nBPW; 20.0 g of buffered peptone, 7 g of refined soy lecithin or equivalent, 1.0 g of sodium thiosulfate, 12.5 g of sodium bicarbonate, per 1 L of DI water; 20–22°C; USDA FSIS, 2019). Wings were manually agitated for 1 min, with the resulting rinsates being collected for downstream analysis while the wings were discarded. Rinsates were aliquoted (1.0 mL) in two 1.5 mL microcentrifuge tubes and either maintained at 4°C for mesophilic aerobes and LAB enumeration or at −80°C for 16S rRNA gene sequencing.
Whole chicken wing rinsates were subsequently serially diluted to 10−7 (1:10 dilution factor) in 96-well plates (25 μL of rinsate in 225 μL of 1 × Phosphate Buffered Saline, PBS). After dilution, 10 μL of each dilution was dot plated onto Tryptic Soy Agar (TSA, BD Difco™, Franklin Lakes, NJ, United States) and De Man, Rogosa, and Sharpe agar (MRS, BD Difco™, Franklin Lakes, NJ, United States) in duplicate (Jett et al., 1997; Herigstad et al., 2001; Naghili et al., 2013). The plates were dried, inverted, and incubated aerobically for 24 h at 37°C or 48 h anaerobically at 37°C, respectively. Plated dilutions with CFU counts between 6 and 60 were enumerated.
The CFU of bacteria per gram of treated whole chicken wings on days 0, 7, 14, and 21 was calculated using the equation described by Dittoe et al. (2019a):
On 14 and 21 d post-treatment, rinsates were aliquoted to 1.5 mL microcentrifuge tubes and stored at −80°C until DNA extraction could occur. Following the Gram-Negative Bacteria protocol of the QIAGEN DNeasy Blood and Tissue Kit (QIAGEN, Hilden, Germany), a subset of aliquoted rinsates (n = 5) was thawed, centrifuged for 10 min at 5,000 × g, and decanted to remove fat and buffer from the pelleted microorganisms. The pellet was resuspended in 180 μL of ATL buffer. Following, the DNA was extracted using the standard protocol with DNA eluted in 30 L of Buffer AE. The DNA purity and quality were assessed using a Nanodrop™ 1,000 spectrophotometer (Thermo Scientific, Waltham, MA, United States). Subsequently, DNA was diluted to 10 ng/μl in Buffer AE.
Libraries were constructed according to Kozich et al. (2013), where dual-index paired-end primers targeting V34, V4, and V45 regions of the bacterial 16S rRNA gene and a high-fidelity polymerase (AccuPrime Pfx SuperMix, Invitrogen, Carlsbad, CA, United States) were used. Amplicons in uniform concentrations (18 μL) were normalized using SequalPrep Normalization Kit (Invitrogen, Carlsbad, CA, United States) and quantified using a Qubit fluorometer and kit (Invitrogen, Carlsbad, CA, United States). Following, normalized amplicons were pooled together in equimolar concentrations into a 1.5 mL microcentrifuge tube and quantified again using the Qubit system. The final concentration of the library was verified through the use of quantitative PCR (qPCR) with the KAPA library quantification kit for Illumina platforms (KAPA Biosystems, Wilmington, MA, United States) using SYBR green technology. Amplicon size was determined using a bioanalyzer (Agilent, Santa Clara, CA, United States).
The library was diluted to 20 μM in HT1 buffer (Illumina, San Diego, California, United States) and denatured with NaOH (0.2 N). The diluted library was mixed with 10% PhiX (Illumina, San Diego, California, United States) and loaded into a MiSeq v2 500 cartridge (Illumina, San Diego, California, United States). The cartridge was then loaded into an Illumina MiSeq (Illumina, San Diego, California, United States) sequencer with subsequent sequences uploaded to BaseSpace (Illumina, San Diego, California, United States), NCBI Sequence Read Archive (PRJNA847567), and GitHub.1
Before the onset of the experiment, a treatment and a time point were randomly assigned to each wing. Only the CFU of total mesophilic aerobes and LAB between 6 and 60 were recorded, with those below the limit of detection (6 × 102 CFU/mL) being recorded as 6 × 102 CFU/mL. Total mesophilic aerobes and LAB (CFU/ml) were calculated as CFU per gram of wing and, subsequently, Log10 transformed. The data were assessed for normality, satisfying the assumptions of a linear model in R Studio (R version 4.3.2; R Core Team, 2023). As the same wing was not continuously sampled throughout time, the microbial count data were analyzed as a randomized complete block design with day designated as the block. The main effects of day and treatment and the interaction were analyzed using a general linear model. Pairwise differences were determined using Tukey’s protected HSD at a p ≤ 0.05 level of significance. Figures of the microbiological count data were generated in R Studio with a linear line at 7 Log CFU/g added to demonstrate the onset of spoilage (Balamatsia et al., 2007).
Demultiplexed amplicon sequences were downloaded from BaseSpace and imported into QIIME2 2019.7 (Bolyen et al., 2019) using Casava 1.8 paired-end demultiplexed format (via qiime tools import), where microbiome informatics were performed on rinsates collected on d 14 and 21. Quality filtering and denoising of demultiplexed sequences were performed using DADA2 (Callahan et al., 2016). The amplicon sequence variants (ASVs) were aligned with SEPP (via q2-fragment insertion) (Matsen et al., 2010; Eddy, 2011; Janssen et al., 2018). Using fasttreee2 (via q2-phylogeny), a rooted phylogenetic tree was generated (Price et al., 2010). The ASVs were putatively identified using SILVA (Quast et al., 2013; Yilmaz et al., 2014; Glöckner et al., 2017) with the sk-learn Bayesian algorithm, which accounts for the error rate associated with sequencing, alignment, and upstream plug-ins (via q2-feature-classifier) (Bokulich et al., 2018a). Populations were rarified to the average sequencing depth (100,000) and visualized for saturation purely to select the point at which alpha and beta diversity analysis could be performed while maintaining all samples (via q2-diversity). As such, the core alpha and beta metrics were performed at a sequencing depth of 200, which retained 10,000 (0.19%) features in all 50 (100.00%) samples at the specified sampling depth (Supplementary Figure S1). Alpha diversity was analyzed using the traditional pipeline, with the main effects and interactions identified using ANOVA (via q2-longitudinal) (Bokulich et al., 2018b) and pairwise comparisons using Kruskal–Wallis for Shannon’s Diversity Index and (Shannon, 1948; Pielou, 1966). Beta diversity metrics, Weighted and Unweighted Unifrac, were also analyzed using the traditional pipeline with ANOSIM (Lozupone and Knight, 2005; Lozupone et al., 2007; Oksanen et al., 2018), with the main effects and interactions identified using ADONIS (Anderson, 2001). Volatility plots were calculated via q2-longitudinal. Compositional differences were evaluated using ANCOM (via q2-composition) (Mandal et al., 2015). Final ANCOM tables were visualized in Microsoft Excel (Microsoft, Redmond, WA, United States).
There was an interaction between treatment and time on the aerobic mesophiles recovered from non-treated and treated chicken wings (p < 0.05; Figure 1; Supplementary Table S1). At day 0, there were no differences between treatments (p > 0.05). On d 7, chicken wings treated with PAA, SBS, and SBS + PAA had significantly lower concentrations of aerobic mesophiles than those treated as NT or TW (p < 0.05). In addition, those treated with PAA had higher concentrations of aerobic mesophiles on d 7 than those treated with SBS and SBS + PAA (p < 0.05). On d 14, there were fewer aerobic mesophiles recovered from those treated with PAA, SBS, and SBS + PAA than those treated with the controls, NT, and TW (p < 0.05). On d 21, there were no differences in the concentration of recovered aerobic mesophiles (p > 0.05).
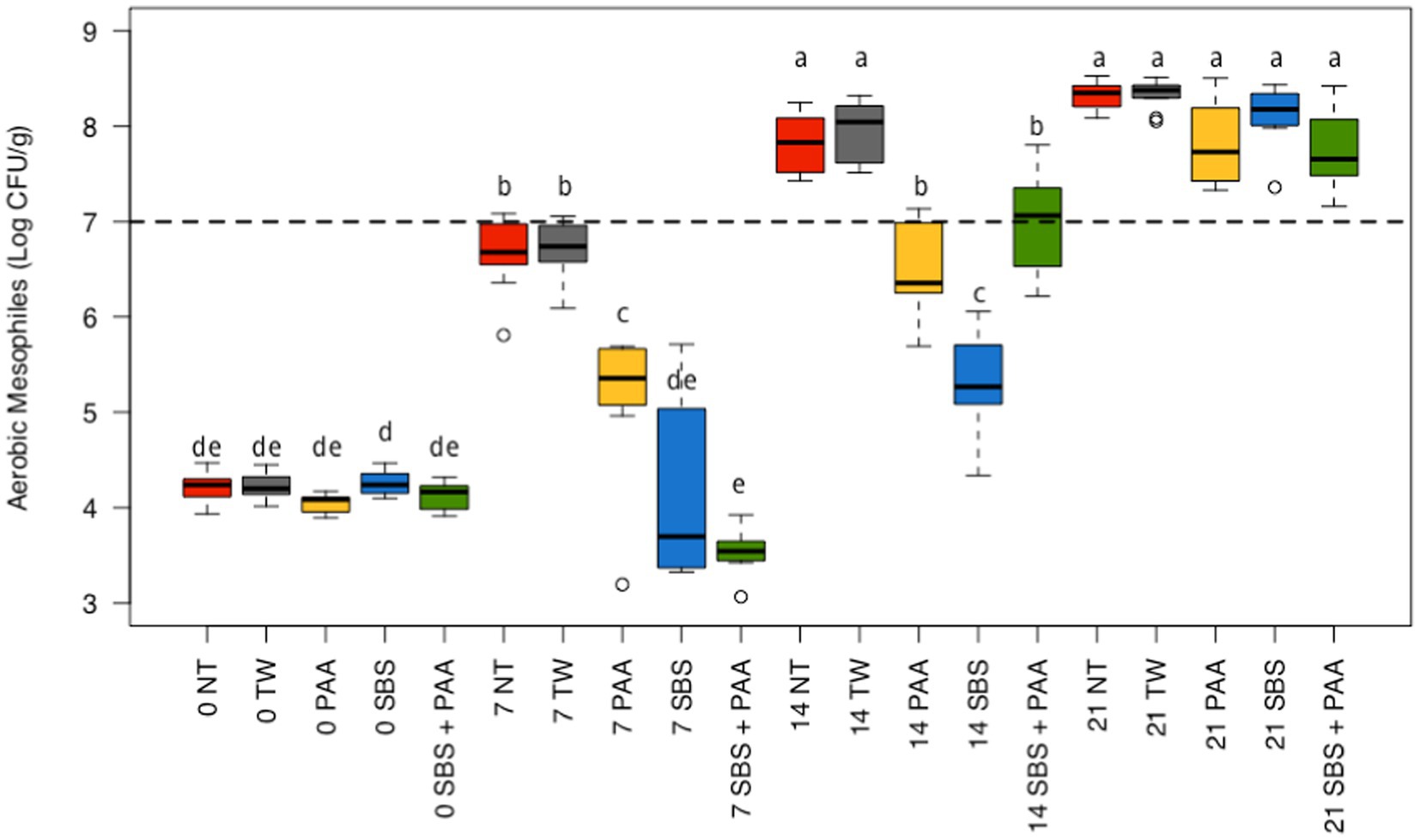
Figure 1. The interaction between treatment and day on the total load of mesophilic aerobes on chicken wings treated with organic and inorganic acids over a 21-d period (p < 0.05). The distribution of the data was captured using boxplots with outliers. Treatments were applied as 15-s dips into either with no treatment (NT), tap water (TW), TW + 500 ppm peracetic acid (PAA), TW + 3% Sodium Bisulfate (SBS), and the combination of TW + SBS + PAA (SBS + PAA) on d 0. The dashed line represents the theoretical onset of spoilage, 7 Log10 CFU/g. Those treated as NT, TW, PAA, SBS, and SBS + PAA are represented as red, grey, yellow, blue, and green boxplots, respectively. Those with different connecting letters (a-e) are considered significantly different (p < 0.05). Exact mean and standard error of the mean Log10 CFU/g values are presented in Supplementary Table S1.
Those treated with NT and TW demonstrated an increase in aerobic mesophiles from d 0 to d 7 (~ + 2.50 Log10 CFU/g) and d 7 to d 14 (~ + 1.10 Log10 CFU/g) and did not differ from d 14 to d 21 (p < 0.05; Figure 1). Those treated with PAA had an increase in recovered aerobic mesophiles at each time-point until d 21 (~ + 1 Log10 CFU/g per 7 d). Chicken wings treated with SBS and SBS + PAA did not have an increase in aerobic mesophiles until d 7 and then increased until d 21 (p < 0.05).
There was also an interaction between treatment and time on the LAB recovered from chicken wings (p < 0.05; Figure 2; Supplementary Table S1). There was no difference between those not treated, NT and TW, and those treated, SBS, PAA, and SBS + PAA, on d 0 (p > 0.05). On d 7, those treated, SBS, PAA, and SBS + PAA, had less recoverable total LAB than those not treated, NT and TW (p < 0.05). On d 14, those treated also had less recoverable LAB than those treated as controls. On d 14, those treated with SBS and SBS + PAA had lower recoverable LAB than those treated with PAA (p < 0.05). On d 21, only those treated with PAA or SBS + PAA had lower LAB recovered than both of the controls; however, those treated with SBS did have lower LAB than those treated with TW (p < 0.05). Although wings treated with PAA and SBS + PAA did not have different LAB levels (Log10 CFU/g) on d 21 (p > 0.05), those treated with SBS + PAA had the lowest LAB load on d 21.
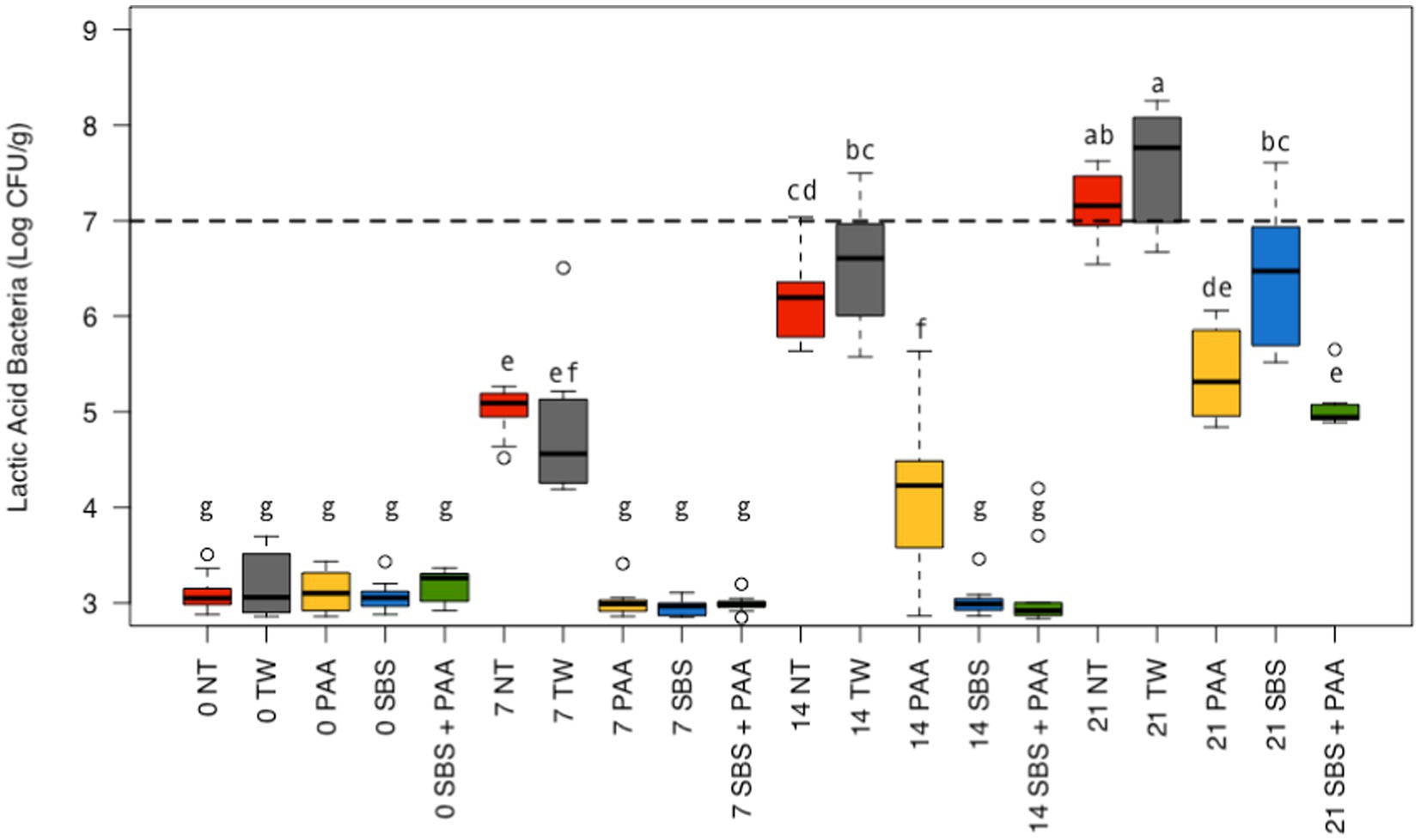
Figure 2. The interaction between treatment and day on the total load of lactic acid bacteria of chicken wings treated with organic and inorganic acids over a 21-d period (p < 0.05). Treatments were applied as 15-s dips into either with no treatment (NT), tap water (TW), TW + 500 ppm peracetic acid (PAA), TW + 3% Sodium Bisulfate (SBS), and the combination of TW + SBS + PAA (SBS + PAA) on d 0. The distribution of the data was captured using boxplots with outliers with the dashed line representing the theoretical onset of spoilage, 7 Log10 CFU/g. Those treated as NT, TW, PAA, SBS, and SBS + PAA are represented as red, grey, yellow, blue, and green boxplots, respectively. Those with different connecting letters (a-e) are considered significantly different (p < 0.05). Exact mean and standard error of the mean Log10 CFU/g values are presented in Supplementary Table S1.
Over time, all the levels of LAB increased among all treated wings; however, those treated with SBS and SBS + PAA did not increase until d 21, with recovered LAB being consistent from d 0 to d 14 (Figure 2). Those treated with PAA did not have an increase in LAB from d 0 to d 7, but from d 7 to d 14 and d 14 to d 21, there was an increase in LAB (~ + 1 Log10 CFU/g). The controls increased at each time-point (p < 0.05).
There was neither any interaction between treatment and day nor the main effect of day on the richness (Shannon’s Entropy) and evenness (Pielou’s Evenness) of the rinsates of bone-in, skin-on chicken wings (Supplementary Table S2; p > 0.05). However, there was a treatment effect on the richness and evenness of the wings (Supplementary Table S2; p < 0.001 and p = 0.02). Those treated with PAA and SBS (2.02 ± 0.11 and 1.60 ± 0.18 Shannon’s Entropy) had a lower richness than those not treated (2.85 ± 0.05 and 2.74 ± 0.08 Shannon’s Entropy) (p < 0.05, Q < 0.05), with those treated with SBS + PAA (2.24 ± 0.23 Shannon’s Entropy) not being different than those untreated or treated (Figure 3A; Supplementary Table S3). Those treated with SBS and SBS + PAA (0.47 ± 0.04 and 0.58 ± 0.03 Pielou’s Evenness) had a lower evenness than those untreated (0.70 ± 0.02 and 0.69 ± 0.02 Pielou’s Evenness). Those treated with SBS + PAA did not differ in evenness from those treated with PAA (0.64 ± 0.02 Pielou’s Evenness; (p > 0.05; Q > 0.05) Figure 3B).
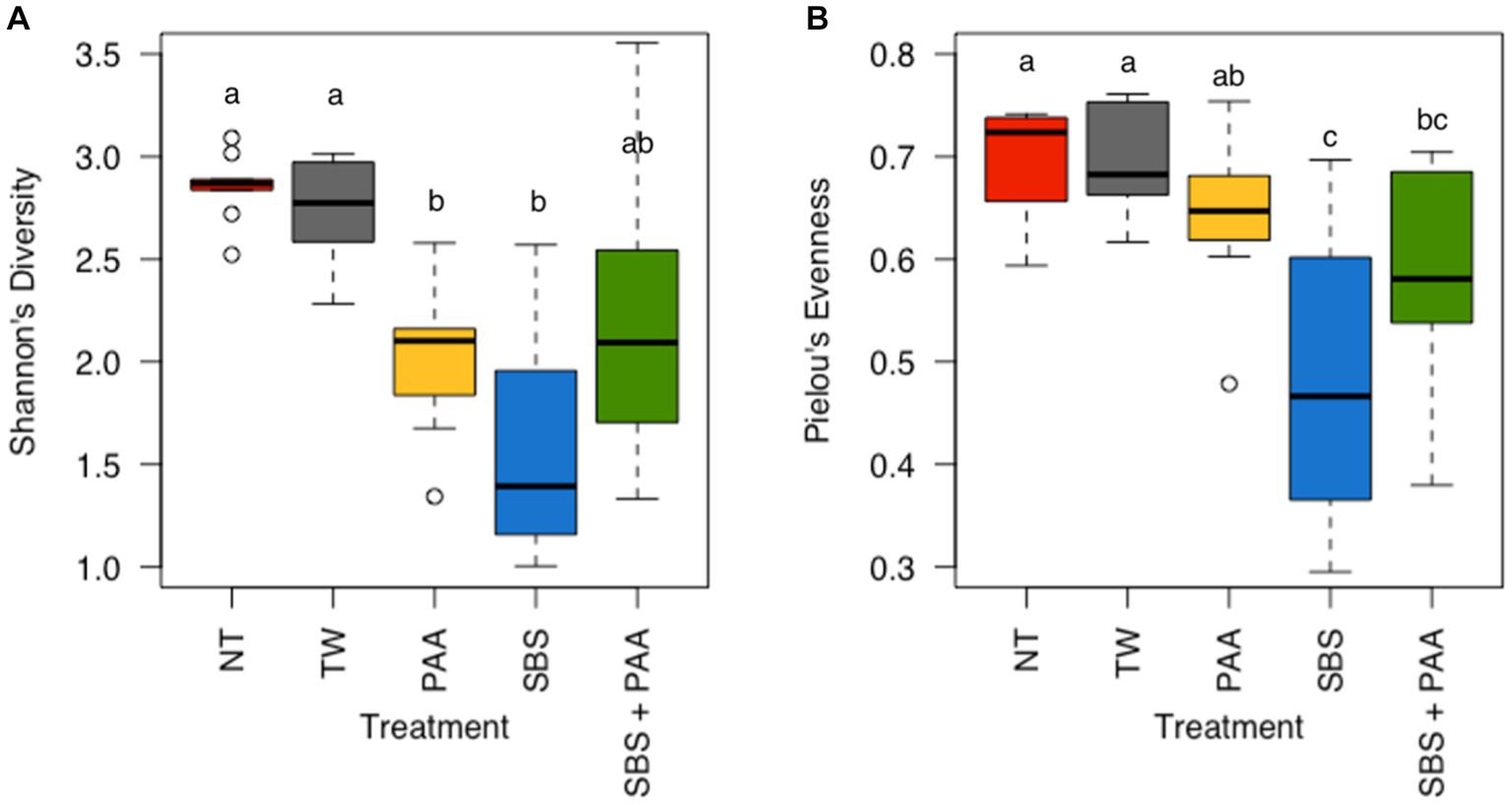
Figure 3. The richness and evenness of the microbiota of the rinsates of bone-in, skin-on chicken wings treated with no treatment (NT), tap water (TW), TW + 500 ppm peracetic acid (PAA), TW + 3% Sodium Bisulfate (SBS), and the combination of TW + SBS + PAA (SBS + PAA). There was a main effect of treatment on both Shannon’s Entropy (A) and Pielou’s Evenness (B) of the microbiota recovered from the chicken wing poultry rinsates (N = 50, n = 10, k = 5, p < 0.05). Those treated as NT, TW, PAA, SBS, and SBS + PAA are represented as red, grey, yellow, blue, and green boxplots, respectively. Those with different connecting letters (a-c) are considered significantly different (Q < 0.05; Supplementary Tables S2, S3).
There was an interaction between treatment and day on the Bray–Curtis, Jaccard Dissimilarity, and Unweighted Unifrac (p < 0.05; Supplementary Table S4). Treatment and time had a main effect on Weighted Unifrac (p > 0.05). To explore this interaction, the volatility of the beta diversity metrics was determined on d 14 and 21 (Figure 4). Those treated with SBS had a stable volatility over time with a higher Bray–Curtis, Jaccard, and Unweighted Unifrac volatility than wings treated with NT, TW, PAA, and SBS + PAA. Those treated with PAA increased in volatility in all beta diversity metrics, whereas those treated with the combination of SBS + PAA increased in Bray–Curtis, Jaccard Dissimilarity, and Unweighted Unifrac volatility over time (Figures 4A–C). Wings designated as NT had a decrease in Bray–Curtis and Weighted Unifrac volatility from d 14 to d 21 (Figures 4A,C). Those designated as TW did not change in beta diversity volatility over time.
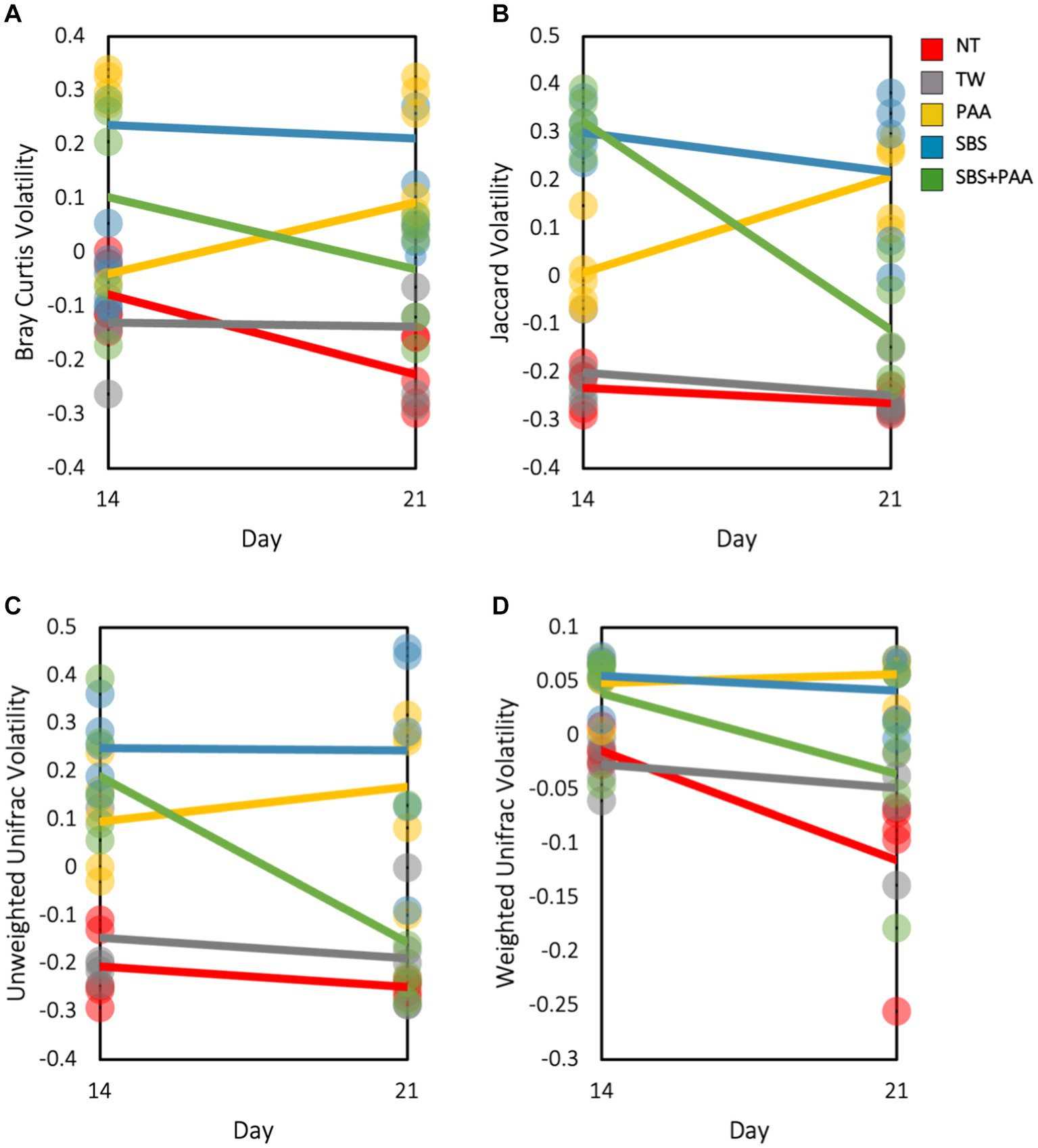
Figure 4. The beta volatility of the microbiota of the rinsates of bone-in, skin-on chicken wings treated with no treatment (NT), tap water (TW), TW + 500 ppm peracetic acid (PAA), TW + 3% Sodium Bisulfate (SBS), and the combination of TW + SBS + PAA (SBS + PAA) on d 14 and 21 post-treatment. An interaction between treatment × day on Bray–Curtis (A), Jaccard Dissimilarity Index (B), Unweighted Unifrac (C), and Weighted Unifrac (D) of the microbiota recovered from the chicken wing poultry rinsates was observed (N = 50, n = 10, k = 5; p = 0.045, 0.004, 0.025, and 0.154).
With treatment being a driving factor on the beta diversity of the untreated and treated bone-in, skin-in chicken wings, 3-dimensional plots (PCoA) were utilized to depict the effect of treatments on the beta diversity of the wings with pairwise differences being determined by ANOSIM (Figure 5; Supplementary Table S5). In all beta diversity metrics, those designated as the controls, NT, and TW did differ and were different than all wings treated with PAA, SBS, and SBS + PAA (p > 0.05; Q > 0.05; Figure 5; Supplementary Table S5). The Bray–Curtis and Jaccard Dissimilarity of those treated with PAA and SBS were different (p < 0.05, Q < 0.05; Figures 5A,B). There was no difference between the Unweighted and Weighted Unifrac of those treated with PAA and SBS (p > 0.05, Q > 0.05; Figures 5C,D). The Bray–Curtis and Weighted Unifrac of those treated with SBS + PAA were different than PAA, but only the Bray–Curtis of those treated with SBS + PAA was different than SBS (p < 0.05, Q < 0.05; Figures 5A,D; Supplementary Table S5).

Figure 5. The beta diversity of the microbiota of the rinsates of bone-in, skin-on chicken wings treated with no treatment (NT), tap water (TW), TW + 500 ppm peracetic acid (PAA), TW + 3% Sodium Bisulfate (SBS), and the combination of TW + SBS + PAA (SBS + PAA) as represented in a three-dimensional plot, PcoA. A main effect of treatment on Bray–Curtis (A), Jaccard Dissimilarity Index (B), Unweighted Unifrac (C), and Weighted Unifrac (D) of the microbiota recovered from the chicken wing poultry rinsates was observed (N = 50, n = 10, k = 5, p < 0.05). Pairwise differences are represented in Supplementary Table S5.
There were 54 unique taxa identified at the class level, and 479 unique taxa were identified at the species level. There were 4 differentially abundant taxa (10 < W > 8) at the class level and 14 differentially abundant taxa (70 < W > 59) at the species level on d 14 and d21 due to treatment application when using ANCOM, the analysis of compositions of microbiomes (p < 0.05; Figure 6). At the class level, Bacilli, Clostridia, Bacteroidia, and Gracilibacteria were differentially abundant than 10, 10, 9, and 8 other taxa at the class level (p < 0.05; Figure 6A). At the species level, Aneurinibacillus thermoaerophilus, Monocercomonoides sp. PA203, Bacillus, Bacillales, Acinetobacter, Shewanella, Escherichia-Shigella, Janthinobacterium, Klebsiella, Psychrobacter, Aeromonas, Hafnia-Obesumbacterium, Bacteroides, and Enterobacteriaceae were differentially abundant than, 70, 69, 68, 68, 68, 67, 66, 65, 62, 62, 61, 60, 60, and 59 other taxa at the species level (p < 0.05; Figure 6B). Those treated with SBS and SBS + PAA had a higher relative abundance of Bacilli at the class level on d 14 than those not treated or those treated with PAA, which translated to higher levels of Bacillales and Bacillus spp. at the species level (Figure 6). By d 21, though there were still populations of bacilli, the populations of Bacteroidia at the class level and populations of Shewanella spp. at the species level were highly prevalent among the significantly differentially abundant taxa (Figure 6).
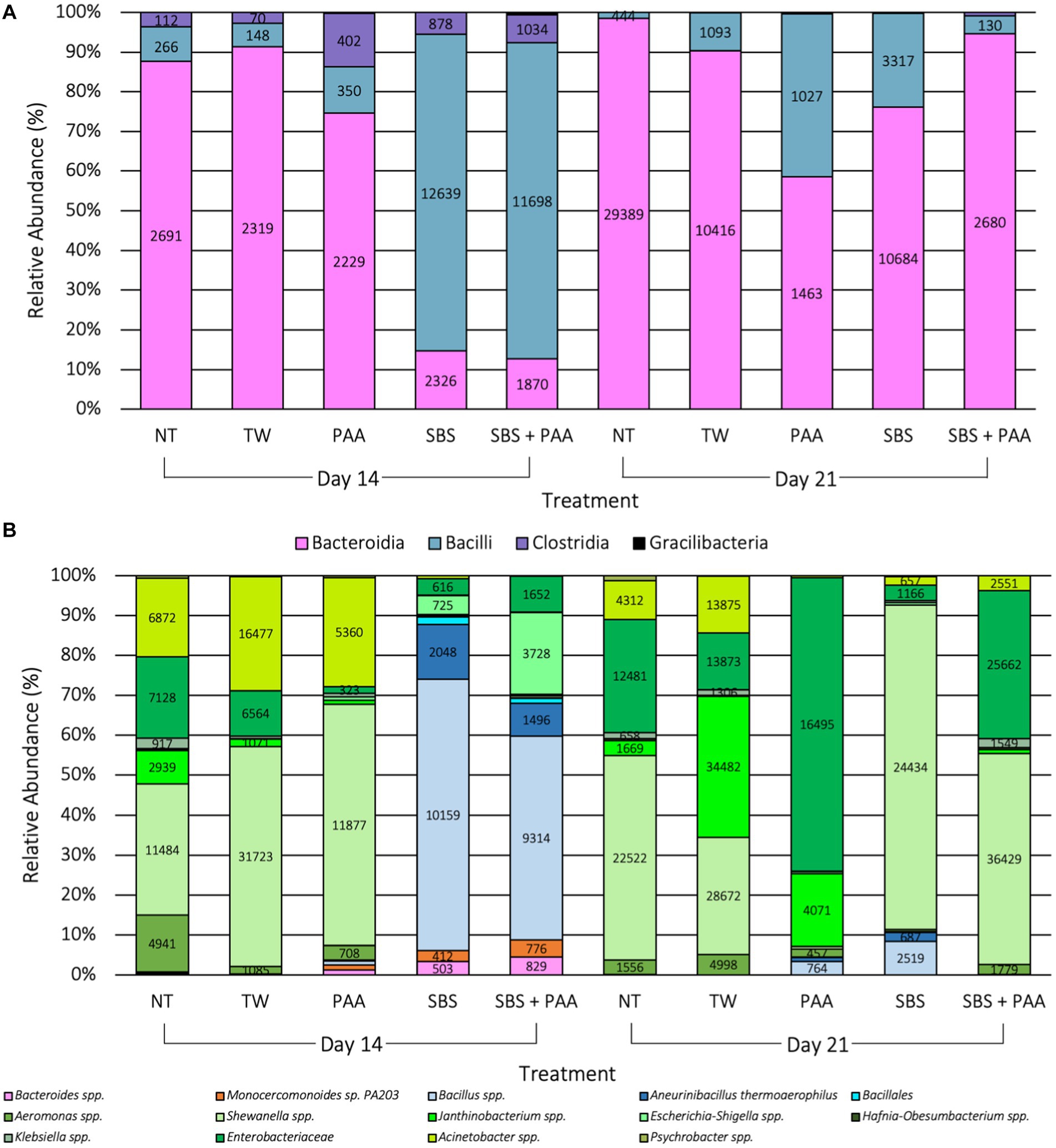
Figure 6. Significant taxa at the class (A) and species (B) levels among the microbiota of the rinsates of bone-in, skin-on chicken wings treated with no treatment (NT), tap water (TW), TW + 500 ppm peracetic acid (PAA), TW + 3% Sodium Bisulfate (SBS), and the combination of TW + SBS + PAA (SBS + PAA) on d 14 and 21 post-treatment (p < 0.05, W > 59). Taxa belonging to Bacteroidetes are pink, Bacilli are blue, Clostridia are purple, Alphaproteobacteria are yellow, Verrucomicrobia are beige, and Gammaproteobacteria are green.
The noticeable spoilage (off odors, discoloration, and slime production) of poultry typically occurs when the total mesophilic aerobic bacteria and LAB populations reach approximately 7 Log10 CFU/g of product (Mead, 2007; Nychas et al., 2008). Commonly, the measurement of total aerobic bacteria and LAB (< 7 Log10 CFU/g) to determine the shelf-life of poultry is utilized with the application of this metric as the preliminary step to identify spoilage. In the current study, culture-based quantification of these indicator organisms was the first step in determining the shelf-life of bone-in, skin-on chicken wings with the end of shelf-life microbiota being explored on d 14 and 21 of refrigeration. When interpreted together, the indicator organisms and the microbiome have the potential to provide a more thorough description of shelf-life extension through the use of inorganic acids. Although the use of pH, colorimetric, and texture measurements would have provided additional physiochemical variables, these metrics were reserved for future and more expansive studies, where regression analyses could be implemented as a final validation step.
Using the standard indicators of spoilage, total aerobic mesophiles, and LAB, spoilage, as defined by 7 Log10 CFU/g of product, occurred between days 7 and 14 (Figures 1, 2). The typical shelf-life of raw bone-in, skin-on broiler wings is between d 7 and 14. Therefore, the occurrence of spoilage (> 7 Log10 CFU/g) in the current project is within the projected range of shelf-life. When evaluating total mesophiles alone (Figure 1), the treatment of wings with short-duration (15 s) antimicrobial dips in PAA, SBS, and SBS + PAA resulted in the extended shelf-life of the wings to day 14, while those treated with NT and TW were spoiled by d 7 as indicated by the level of total aerobic mesophiles recovered. When only considering the mitigation of LAB bacteria, the treatment of wings with SBS and SBS + PAA had the most profound effect on the reduction of LAB over the 21-day period (p < 0.05; Figure 2).
It is apparent that there are minimal differences in the efficacy of treatments reducing total aerobic mesophiles, but there may be an advantage of including SBS as a short duration dip (15-s) alone or in combination with PAA as it may decrease LAB levels more effectively past d 14 of shelf-life. PAA has previously been demonstrated to extend the shelf-life of broiler carcasses when used during immersion chilling (1 h) at 200 ppm (d 15: 5.89 ± 0.31 Log10 CFU/sample) compared with those treated with tap water (d 15: 6.88 ± 0.78 Log10 CFU/ml), despite not being significantly different (Oksanen et al., 2018). As no comparison between PAA and SBS on these shelf-life microorganisms of raw poultry products exists beyond the current study, the current research demonstrates a potentially selective efficacy against LAB compared with aerobic mesophiles.
A combinatorial effect of SBS + PAA was evident based on the current data; however, more analyses evaluating the combinatorial effects are required. These results are in congruence with Dittoe et al. (2019a), who determined that the use of 3% SBS, 2% SBS with the addition of 200 ppm PAA, and 3% SBS with the addition of 200 ppm PAA had a significant effect on foodborne pathogens, such as Salmonella Enteritidis among raw bone-in poultry drumsticks. Therefore, Dittoe et al. (2019a) summarized that there could be a synergistic effect of adding SBS with PAA. If proven true with more research studies, this combinatorial effect could be due to the independent modes of action having an added benefit effect. Likely, SBS leads to a loss in the ability to maintain cellular osmolarity due to its extremely low pKa of 1.9 (Knueven, 1999), resulting in cytolysis, whereas PAA is primarily thought to work as an oxidizer, resulting in cell death by disrupting cell wall permeability (Middleton et al., 1997; Block, 2001). The use of PAA, an oxidizer, may also result in the denaturation of proteins and oxidation of sulfhydryl and sulfur bonds in proteins, enzymes, and other metabolites (Middleton et al., 1997; Block, 2001).
Additionally, the use of an inorganic acid such as SBS may also be able to counteract the buffering capacity of chicken skin which has the potential to reduce the efficacy of different antimicrobial hurdles (Tan et al., 2014a,b). Thus, the current research employed skin-on poultry parts rather than skinless to demonstrate the protective effect of the skin, allowing bacterial attachment and subsequently protecting those attached cells (Tan et al., 2014a,b). Tan et al. (2014b) demonstrated the use of organic acids, such as acetic acid was capable of reducing Salmonella Typhimurium levels better on meat and fat (~1.5 to 7 Log10 CFU/g) than that of skin remnants (~1 Log10 CFU/g) as the skin remnants had a stronger buffering capacity (13 mmol H+/(pH*kg)) than chicken meat and fat (7 mmol H+/(pH*kg) and 6.9 mmol H+/(pH*kg), respectively). Therefore, in the current study, the use of SBS was demonstrated to be an effective antimicrobial hurdle on skin-on poultry parts in comparison to the industry standard, PAA, with both extending shelf-life 7 days past the NT and TW controls.
One of the first avenues for understanding microbial ecology is through the evaluation of alpha diversity, which is the evenness and richness of a microbial community. The treatment groups induced significant changes in community richness and evenness but not in time (p < 0.05). The richness of the wings treated with NT and TW was higher than that of those treated with PAA and SBS, with the richness not differing between those treated with PAA and SBS (Figure 3A). These results are in congruence with Kim et al. (2017), who determined no difference in richness between broiler carcasses treated post-chill with PAA and Amplon (Zoetis, Parsippany, New Jersey, United States). Amplon is a product similar to SBS in that it is comprised of sulfuric acid and sodium sulfate (4.5–5.5% sodium sulfate, 38.5–39.5% sulfuric acid). Weinroth et al. (2019) determined the use of Amplon, at a pH of 1.2, reduced Faith’s PD, another measure of richness, of ground beef under MAP packaging compared to those treated as the control or with 350 ppm of PAA. Weinroth et al. (2019) attributed these differences to the ability of these different antimicrobials to target different microorganisms by altering the pH of the ground beef. Although pH was not measured in the current study, the buffering capacity of the poultry skin (Weinroth et al., 2019) and the increase in PAA to 500 ppm may have been attributed to the lack of significance between the use of PAA and SBS on the richness of the microbial community. Pielou’s Evenness paralleled the richness findings, with rinsates of wings treated with SBS and SBS + PAA having the least amount of evenness as compared with the other groups (p < 0.05; Q < 0.05; Figure 3B).
Beta diversity indices provide information on the non-descriptive compositional variation of that community structure and variance across a population. Using ADONIS, a multivariate analysis can determine that if main or interactive effects are driving beta diversity metrics, there was an interaction between treatment and time on the beta diversity of the microbial communities, with time being a significant driver for those effects (p < 0.05; Supplementary Table S3). As such, volatility plots were created to visualize shifts over time (Figure 4). As expected, the most readily spoiled poultry parts, the control groups, did not change drastically over time (d 14 to 21). However, those treated demonstrated differences over time, with PAA slightly increasing in diversification over time and SBS and SBS + PAA sustaining or reducing the microbial compositional diversity. Although the current study was not intended to compare culture-dependent and culture-independent analyses, the aerobic mesophile and LAB counts correspond well with the stability demonstrated through the volatility plots. As the controls, NT and TW, were on the verge of spoilage on d 7, the d 14 and 21 microbial composition should not have shifted significantly. However, those treated did not reach the end of their shelf-life until d 14 or 21. Therefore, the stability of a population could be indicative of spoilage. Unlike the current study, Weinroth et al. (2019) did not demonstrate differences in the beta diversity, Weighted Unifrac, or ground beef before (15 d post-grind) and after retail display (26 d post-grind). However, Weinroth et al. (2019) demonstrated distinct Weighted Unifrac Distances between ground beef treated as the control, with 350 ppm PAA or with sulfuric acid, and sodium sulfate blend at a pH of 1.2 irrespective of time.
At the end of shelf-life (d 12 and 16), raw poultry products have been reported to be comprised of Brochothrix, Carnobacterium, Vagococcus, and Janthinobacterium when using culture-independent methods, such as microbiome sequencing (Lauritsen et al., 2019). Additionally, at 10 d of shelf-life under aerobic conditions, broiler legs treated with water or PAA (13.67 mM) have been demonstrated to be comprised of Pseudomonas, Psychrobacter, Carnobacterium, Enterobacteriaceae, and Serratia (Zhang et al., 2022). The reported findings are congruent with the current research, where 74 taxa were identified at the species level, with 4 and 14 differentially abundant taxa at the class and species levels (Figure 6).
In the current study, it was evident that the use of antimicrobials such as PAA or SBS as short-duration dips does alter the microbial composition. As observed with Kim et al. (2017), the current study saw the emergence of taxa belonging to Bacteroides (Monocercomonoides sp. PA203) and Bacillus (Aneurinibacillus thermophilus, Bacillales, and Janthinobacterium spp.), emerging in the PAA and SBS treatment groups. Those treated with SBS and SBS + PAA had a greater abundance of Bacilli, such as Bacillus spp., Aneurinibacillus thermoaerophilus, and Bacillales, than other groups. The addition of PAA had similar relative abundance of species belonging to Gammaproteobacteria as those treated with NT and TW on d 14. Additionally, those treated with PAA had a greater abundance of Shewanella spp. and Acinetobacter spp. on d 14; however, on d 21, those treated with PAA were predominated by Enterobacteriaceae and Janthinobacterium spp. Meanwhile, those treated with SBS were more abundant with Shewanella spp., and those treated with SBS + PAA were more abundant in Shewanella spp. and Enterobacteriaceae on d 21. Although the NT and TW groups demonstrated changes in composition, these changes were not as dramatic as the changes were observed in composition among those treated, reinforcing the beta and alpha diversity results observed in the current study.
Weinroth et al. (2019) demonstrated that the spray treatment of trim prior to grinding significantly impacted the families such as Enterobacteriaceae, Lactobacillaceae, and Leuconostocaceae with the ground beef treated with Amplon and the blend of sulfuric acid and sodium sulfate, having a higher normalized sequence count of Lactobacillaceae and Leuconostocaceae than those treated as the control or with 350 ppm PAA (ANCOM, W = 8). Additionally, those treated with Amplon or PAA had lower Enterobacteriaceae normalized counts than those treated as the control (Weinroth et al., 2019). Weinroth et al. (2019) hypothesized that the use of weak organic acids, such as Amplon, is more effective against Gram-negative microorganisms compared with Gram-positives and, thus, was the reason for the inability of Amplon to reduce these populations. Although Amplon and SBS are similar in chemical composition, the application in the current study did not result in significant differences in these populations. Additionally, Weinroth postulated that the pH of the Amplon treatment (1.2 pH) would have altered the pH of the ground beef, and the LAB populations would be altered. Although the pH of the meat was not measured in the current study, the buffering capacity of chicken skin would likely inhibit the LAB effects, which was demonstrated in ground beef (Tan et al., 2014a,b). The use of SBS and SBS + PAA on chicken wings decreased culture-dependent LAB levels compared with those treated as the controls or treated with PAA. Therefore, the observations made by Weinroth et al. (2019) may not apply to poultry products with beef.
As previously mentioned, the current study did not observe an increase in LAB, according to Weinroth et al. (2019); however, the current study observed an increase in a gram-positive population within the class of Bacilli among wings treated with SBS and SBS + PAA on d 14 and 21. Within the class of Bacilli are several acid-resistant spore-forming spoilage bacteria, such as those within the species of Bacillus subtilis (Sun et al., 2021). During poultry processing, spore formers such as Bacillus spp. are prevalent among the carcass microbiome (Kim et al., 2017). However, the use of an inorganic acid with an extremely low pKa, such as SBS, may inhibit typical spoilage-associated microorganisms such as Aeromonas, Shewanella, Enterobacteriaceae, Janthinobacterium, Acinetobacter, Escherichia-Shigella, and Psychrobacter spp. may not be able to mitigate acid-tolerant spore formers. With no competitors, these acid-tolerant spore-forming spoilage bacteria are capable of sporulating and dominating the microbiome of chicken wings. This phenomenon is becoming more evident with multiple food matrices (André et al., 2017; Sagdic et al., 2017) and has been gathering increased awareness across the food industry (Zhang and Mathys, 2019).
Spoilage of poultry products has generally been defined by physical characteristics (slime, off-odor, and texture) and microbial abundances (LAB and aerobic mesophiles). All these metrics have proven to be essential components for determining the onset of spoilage and measuring the impact of shelf-life extension efforts. However, as new technologies emerge, the ability to improve the understanding of how shelf-life extension efforts impact the microbial ecology of poultry products and subsequent spoilage has surfaced. By discerning the impact different antimicrobials have on shelf-life in terms of microbial ecology and stability, poultry processing personnel can make more informed decisions at the facility level.
Although the current study is a preliminary evaluation of the antimicrobial effects of SBS and the combination of SBS and PAA on the microbial ecology of poultry products using culture-dependent and independent means, the treatment of bone-in, skin-on chicken wings with 3% SBS increased the shelf-life of these poultry products for 7 days more than those treated with NT and TW by microbiological standards, with spoilage occurring between d 14 and day 21 post-treatment. In fact, the wings treated with SBS and SBS + PAA exhibited LAB levels approximately 3 Log10 CFU/g on d 7 and 14. Regardless, by d 21, there was a significant increase in microbial populations among all groups. These changes in traditional microbiological plating were in parallel to the changes identified among the microbiota.
While microbiome sequencing may not be a rapid screen for spoilage, understanding antimicrobial effects on that population and how spoilage may be delayed or impacted is essential for designing a targeted multi-hurdle approach. In the current study, microbiome sequencing revealed the selective enrichment of Bacilli, a class comprised of acid-resistant spore-forming spoilage bacteria, with the use of SBS as a short-duration antimicrobial dip (15 s) on skin-on, bone-in poultry wings while reducing other spoilage-associated microorganisms. Between d 14 and 21 of shelf-life, the microbial ecology of the treated wings was collapsing with both volatility and ANCOM results, demonstrating the increase in spoilage microorganisms among treated wings, whereas the controls maintained static from d 14 to 21 as the shelf-life on untreated wings had already been reached by d 14. Additionally, those treated with SBS alone or in combination with PAA demonstrated a loss of Clostridia and Bacilli selective enrichment and the emergence and predominance of spoilage populations on d 21 of shelf-life.
In conclusion, using SBS as a short-duration (15 s) antimicrobial dip is an effective shelf-life extension tool on bone-in, skin-on chicken wings compared with the industry standard, PAA. More specifically, the use of SBS as an antimicrobial: (1) resulted in the increase in shelf-life by 7 days compared to untreated chicken wings; (2) is more effective in reducing culture-dependent (LAB plate data) and culture-independent (microbiome data) populations compared to the industry standard PAA on d 14 of shelf-life but these effects were gone by the end of shelf-life, d-21; and (3) potentially selects for acid-tolerant spore-forming spoilage bacteria within the class of Bacilli on d 14. Therefore, the use of SBS as a short-duration (15 s) antimicrobial dip on poultry products is an attractive shelf-life extension hurdle that warrants further research and eventual integration into poultry processing facilities.
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found at: https://www.ncbi.nlm.nih.gov/, PRJNA847567; https://github.com, https://github.com/RickeLab-UW/Microbiome-of-Treated-Chicken-Wings.
Ethical approval was not required for the studies on animals in accordance with the local legislation and institutional requirements because carcass parts were procured from a commercial processor.
DD: Writing – original draft, Writing – review & editing, Conceptualization, Data curation, Formal analysis, Investigation, Methodology, Project administration, Visualization. KF: Conceptualization, Formal analysis, Project administration, Supervision, Writing – original draft, Writing – review & editing. CO: Conceptualization, Funding acquisition, Resources, Writing – review & editing. HT: Data curation, Project administration, Writing – review & editing. SR: Conceptualization, Funding acquisition, Project administration, Resources, Supervision, Writing – review & editing.
The author(s) declare financial support was received for the research, authorship, and/or publication of this article. Jones-Hamilton Co. funded the current research. Their financial support did not influence the design, execution, and results of the current work.
All authors would like to acknowledge Jones-Hamilton Co. for their technical assistance in applying the antimicrobial treatments in the current research.
CO was employed by Jones-Hamilton Co. (Walbridge, OH, United States). Jones-Hamilton Co. did provide financial support for this work. However, their financial support did not influence the design, execution, and results of the current work.
The remaining authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
The author(s) declared that they were editorial board members of Frontiers, at the time of submission. This had no impact on the peer review process and the final decision.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2024.1348159/full#supplementary-material
Anderson, M. J. (2001). A new method for non-parametric multivariate analysis of variance. Austral Ecol. 26, 32–46. doi: 10.1111/j.1442-9993.2001.01070.pp.x
André, S., Vallaeys, T., and Planchon, S. (2017). Spore-forming bacteria responsible for food spoilage. Res. Microbiol. 168, 379–387. doi: 10.1016/j.resmic.2016.10.003
Balamatsia, C. C., Patsias, A., Kontominas, M. G., and Savvaidis, I. N. (2007). Possible role of volatile amines as quality-indicating metabolites in modified atmosphere-packaged chicken fillets: correlation with microbiological and sensory attributes. Food Chem. 104, 1622–1628. doi: 10.1016/j.foodchem.2007.03.013
Baltic, T., Ciric, J., Brankovic Lazic, I., Ljubojevic Pelic, D., Mitrovic, R., Djordjevic, V., et al. (2019). Packaging as a tool to improve the shelf life of poultry meat. IOP Conf. Ser. Earth Environ. Sci. 333:012044. doi: 10.1088/1755-1315/333/1/012044
Block, S. S. (2001). Disinfection, sterilization, and preservation. Philadelphia, PA: Lippincott Williams & Wilkins.
Bokulich, N. A., Dillon, M. R., Zhang, Y., Rideout, J. R., Bolyen, E., Li, H., et al. (2018b). q2-longitudinal: longitudinal and paired-sample analyses of microbiome data. mSystems. 3, e00219–e00318. doi: 10.1128/mSystems.00219-18
Bokulich, N. A., Kaehler, B. D., Rideout, J. R., Dillon, M., Bolyen, E., Knight, R., et al. (2018a). Optimizing taxonomic classification of marker-gene amplicon sequences with QIIME 2’s q2-feature-classifier plug-in. Microbiome 6:90. doi: 10.1186/s40168-018-0470-z
Bolyen, E., Rideout, J. R., Dillon, M. R., Bokulich, N. A., Abnet, C. C., Al-Ghalith, G. A., et al. (2019). Reproducible, interactive, scalable and extensible microbiome data science using QIIME 2. Nat. Biotechnol. 37, 852–857. doi: 10.1038/s41587-019-0209-9
Callahan, B. J., McMurdie, P. J., Rosen, M. J., Han, A. W., Johnson, A. J., Holmes, S. P., et al. (2016). DADA2: high-resolution sample inference from Illumina amplicon data. Nat. Methods 13, 581–583. doi: 10.1038/nmeth.3869
City of Fayetteville Arkansas. (2020). Annual drinking water quality report. Available at: https://www.fayetteville-ar.gov/ (Accessed January 10, 2024).
Dittoe, D. K., Atchley, J. A., Feye, K. M., Lee, J., Knueven, C. J., and Ricke, S. C. (2019a). The efficacy of sodium bisulfate salt (SBS) as an antimicrobial on whole chicken drumsticks artificially inoculated with Salmonella enteritidis. Front. Vet. Sci. 6:6. doi: 10.3389/fvets.2019.00006
Dittoe, D. K., Feye, K. M., Peyton, B., Worlie, D., Draper, M. J., and Ricke, S. C. (2019b). The addition of Viriditec™ aqueous ozone to peracetic acid as an antimicrobial spray increases air quality while maintaining Salmonella Typhimurium, non-pathogenic Escherichia coli, and Campylobacter jejuni reduction on whole carcasses. Front. Microbiol. 9:3180. doi: 10.3389/fmicb.2018.03180
Eddy, S. R. (2011). Accelerated profile HMM searches. PLoS Comput. Biol. 7:e1002195. doi: 10.1371/journal.pcbi.1002195
Fan, X., Sokorai, K. J., Liao, C. H., Cooke, P., and Zhang, H. Q. (2009). Antibrowning and antimicrobial properties of sodium acid sulfate in apple slices. J. Food Sci. 74, M485–M492. doi: 10.1111/j.1750-3841.2009.01362.x
Feye, K. M., Dittoe, D. K., Shi, Z., Woitte, J., Owens, C. M., Kogut, M. H., et al. (2019). The reduction of pathogen load on Ross 708 broilers when using different sources of commercial peracetic acid sanitizers in a pilot processing plant. Microorganisms 7:503. doi: 10.3390/microorganisms7110503
Glöckner, F. O., Yilmaz, P., Quast, C., Gerken, J., Beccati, A., Ciuprina, A., et al. (2017). 25 years of serving the community with ribosomal RNA gene reference databases and tools. J. Biotechnol. 261, 169–176. doi: 10.1016/j.jbiotec.2017.06.1198
Herigstad, B., Hamilton, M., and Heersink, J. (2001). How to optimize the drop plate method for enumerating bacteria. J. Microbiol. Methods 44, 121–129. doi: 10.1016/S0167-7012(00)00241-4
Janssen, S., McDonald, D., Gonzalez, A., Navas-Molina, J. A., Jiang, L., Xu, Z. Z., et al. (2018). Phylogenetic placement of exact amplicon sequences improves associations with clinical information. mSystems 3:e00021. doi: 10.1128/mSystems.00021-18
Jett, B. D., Hatter, K. L., Huycke, M. M., and Gilmore, M. S. (1997). Simplified agar plate method for quantifying viable bacteria. BioTechniques 23, 648–650. doi: 10.2144/97234bm22
Karaca, I. M., Haskaraca, G., Ayhan, Z., and Gültekin, E. (2023). Development of real time-pH sensitive intelligent indicators for monitoring chicken breast freshness/spoilage using real packaging practices. Food Res. Int. 173:113261. doi: 10.1016/j.foodres.2023.113261
Kim, S. A., Park, S. H., Knueven, C., Basel, R., and Ricke, S. C. (2018). A decontamination approach using a combination of bisulfate soda and peracetic acid against Listeria innocua inoculated on whole apples. Food Contr. 84, 106–110. doi: 10.1016/j.foodcont.2017.07.036
Kim, S. A., Park, S. H., Lee, S. I., Owens, C. M., and Ricke, S. C. (2017). Assessment of chicken carcass microbiome responses during processing in the presence of commercial antimicrobials using a next generation sequencing approach. Sci. Rep. 7:43354. doi: 10.1038/srep43354
Knueven, C. J. (1999). Sodium bisulfate as Acidulant in foods. U.S. Patent No 5,958,491. Washington, DC: Patent US, and Trademark Office.
Kozich, J. J., Westcott, S. L., Baxter, N. T., Highlander, S. K., and Schloss, P. D. (2013). Development of a dual-index sequencing strategy and curation pipeline for analyzing amplicon sequence data on the MiSeq Illumina sequencing platform. Appl. Environ. Microbiol. 79, 5112–5120. doi: 10.1128/AEM.01043-13
Lauritsen, C. V., Kjeldgaard, J., Ingmer, H., Bisgaard, M., and Christensen, H. (2019). Microbiota encompassing putative spoilage bacteria in retail packaged broiler meat and commercial broiler abattoir. Int. J. Food Microbiol. 300, 14–21. doi: 10.1016/j.ijfoodmicro.2019.04.003
Lozupone, C. A., Hamady, M., Kelley, S. T., and Knight, R. (2007). Quantitative and qualitative beta diversity measures lead to different insights into factors that structure microbial communities. Appl. Environ. Microbiol. 73, 1576–1585. doi: 10.1128/AEM.01996-06
Lozupone, C., and Knight, R. (2005). UniFrac: a new phylogenetic method for comparing microbial communities. Appl. Environ. Microbiolol. 71, 8228–8235. doi: 10.1128/AEM.71.12.8228-8235.2005
Mandal, S., Treuren, W. V., White, R. A., Eggesbø, M., Knight, R., and Peddada, S. D. (2015). Analysis of composition of microbiomes: a novel method for studying microbial composition. Microb. Ecol. Health Dis. 26:27663. doi: 10.3402/mehd.v26.27663
Manning, L., Baines, R. N., and Chad, S. A. (2007). Trends in the global poultry meat supply chain. Br. Food J. 109, 332–342. doi: 10.1108/00070700710746759
Matsen, F. R., Kodner, R. B., and Ambrust, E. V. (2010). Pplacer: linear time maximum-likelihood and Bayesian phologenetic placement of sequences onto a fixed reference tree. BMC Bioinformatics 11:538. doi: 10.1186/1471-2105-11-538
Mead, G. C. (2007). “Sampling methods for poultry-meat products” in Microbiological analysis of red meat, poultry and eggs. ed. G. Mead (Sawston, UK: Woodhead Publishing), 148–164.
Micciche, A. C., Feye, K. M., Rubinelli, P. M., Lee, J., Knueven, C. J., and Ricke, S. C. (2019). Comparison of acid sanitizers on Salmonella Typhimurium inoculated commercial poultry processing reuse water. Front. Sustain. Food Syst. 2:90. doi: 10.3389/fsufs.2018.00090
Middleton, A. M., Chadwick, M. V., and Gaya, H. (1997). Disinfection of bronchoscopes, contaminated in vitro with Mycobacterium tuberculosis, Mycobacterium avium-intracellulare and Mycobacterium chelonae in sputum, using stabilized, buffered peracetic acid solution (‘nu-Cidex’). J. Hosp. Infect. 37, 137–143. doi: 10.1016/S0195-6701(97)90183-3
Naghili, H., Tajik, H., Mardani, K., Razavi Rouhani, S. M., Ehsani, A., and Zare, P. (2013). Validation of drop plate technique for bacterial enumeration by parametric and nonparametric tests. Vet. Res. Forum 4, 179–183.
Nychas, G. J. E., Skandamis, P. N., Tassou, C. C., and Koutsoumanis, K. P. (2008). Meat spoilage during distribution. Meat Sci. 78, 77–89. doi: 10.1016/j.meatsci.2007.06.020
Oksanen, J., Blanchet, F., Friendly, M., Kindt, R., Legendre, P., Minchin, P., et al. (2018). Vegan: community ecology package. R package version 2.5-3. Available at: https://CRAN.R-project.org/package=vegan.
Pielou, E. C. (1966). The measurement of diversity in different types of biological collections. J. Theor. Biol. 13, 131–144. doi: 10.1016/0022-5193(66)90013-0
Price, M. N., Dehal, P. S., and Arkin, A. P. (2010). FastTree 2–approximately maximum-likelihood trees for large alignments. PLoS ONE 5:e9490. doi: 10.1371/journal.pone.0009490
Quast, C., Pruesse, E., Yilmaz, P., Gerken, J., Schweer, T., Yarza, P., et al. (2013). The SILVA ribosomal RNA gene database project: improved data processing and web-based tools. Nucleic Acids Res. 41, D590–D596. doi: 10.1093/gks1219
R Core Team. (2023). R: A language and environment for statistical computing. Vienna, Austria: R Foundation for Statistical Computing.
Rahman, M. S. (2015). “Hurdle Technology in Food Preservation” in Minimally processed foods. Food engineering series. eds. M. Siddiqui and M. Rahman (Cham: Springer)
Ricke, S. C., Hacker, J. C., Yearkey, K. L., Shi, Z., Park, S. H., and Rainwater, C. E. (2018). “Unraveling food production microbiomes: concepts and future directions” in Food and feed safety systems and analysis. eds. S. C. Ricke, G. G. Atungulu, S. H. Park, and C. E. Rainwater (Cambridge, MA: Academic Press), 347–374.
Sagdic, O., Tornuk, F., Karasu, S., Durak, M. Z., and Arici, M. (2017). Microbial ecology of mayonnaise, margarine, and sauces. In: A. De Souza Sant’Ana Quantitative Microbiology in Food Processing: Modeling the Microbial Ecology. New York: John Wiley & Sons, pp. 519–532.
Shannon, C. A. (1948). A mathematical theory of communication. Bell Syst. Tech. J. 27, 623–656. doi: 10.1002/j.1538-7305.1948.tb01338.x
Souza, V. G., Pires, J. R. A., Vieiera, É. T., Coelhoso, I. M., Duarte, M. P., and Fernando, A. L. (2018). Shelf life assessment of fresh poultry meat packaged in novel bionanocomposite of chitosan/montmorillonite incorporated with ginger essential oil. Coatings 8:177. doi: 10.3390/coatings8050177
Sun, R., Vermeulen, A., Wieme, A. D., Vandamme, P., and Devlieghere, F. (2021). Identification and characterization of acid-tolerant spore-forming spoilage bacteria from acidified and low-acid pasteurized sauces. LWT 152:112378. doi: 10.1016/j.wt.2021.112378
Tan, S. M., Lee, S. M., and Dykes, G. A. (2014a). Buffering effect of chicken skin and meat protects Salmonella enterica strains against hydrochloric acid but not organic acid treatment. Food Control 42, 329–334. doi: 10.1016/j.foodcont.2014.02.031
Tan, S. M., Lee, S. M., and Dykes, G. A. (2014b). Fat contributes to the buffering capacity of chicken skin and meat but enhances the vulnerability of attached Salmonella cells to acetic acid treatment. Food Res. Int. 66, 417–423. doi: 10.1016/j.foodres.2014.10.007
Tran, T., Grandvalet, C., Verdier, F., Martin, A., and Alexandre, H. (2020). Microbial dynamics between yeasts and acetic acid bacteria in Kombucha: impacts on the chemical composition of the beverage. Food Secur. 9:963. doi: 10.3390/foods9070963
Walsh, R. J., White, B., Hunker, L., Leishman, O., Hilgren, J., and Klein, D. (2018). Peracetic acid and hydrogen peroxide post-dip decay kinetics on red meat and poultry. Food Prot. Trends. 38, 96–103.
Weinroth, M. D., Britton, B. C., McCullough, K. R., Martin, J. N., Geornaras, I., Knight, R., et al. (2019). Ground beef microbiome changes with antimicrobial decontamination interventions and product storage. PLoS ONE. 14:e0217947. doi: 10.1371/journal.pone.0217947
Yilmaz, P., Parfrey, L. W., Yarza, P., Gerken, J., Pruesse, E., Quast, C., et al. (2014). The SILVA and “all-species living tree project (LTP)” taxonomic frameworks. Nucleic Acids Res. 42, D643–D648. doi: 10.1093/nar/gkt1209
Zhang, Y., and Mathys, A. (2019). Superdormant spores as a hurdle for gentle germination- inactivation based spore control strategies. Front. Microbiol. 9:3163. doi: 10.3389/fmicb.2018.03163
Keywords: shelf life, microbiome, poultry, peracetic acid, sodium bisulfate
Citation: Dittoe DK, Feye KM, Ovall C, Thompson HA and Ricke SC (2024) Exploiting the microbiota of organic and inorganic acid-treated raw poultry products to improve shelf-life. Front. Microbiol. 15:1348159. doi: 10.3389/fmicb.2024.1348159
Received: 01 December 2023; Accepted: 06 February 2024;
Published: 27 February 2024.
Edited by:
Rong Wang, Agricultural Research Service (USDA), United StatesReviewed by:
Monica Ponder, Virginia Tech, United StatesCopyright © 2024 Dittoe, Feye, Ovall, Thompson and Ricke. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Dana K. Dittoe, ZGRpdHRvZUB1d3lvLmVkdQ==; Steven C. Ricke, c3JpY2tlQHdpc2MuZWR1
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.