
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Microbiol., 11 June 2024
Sec. Virology
Volume 15 - 2024 | https://doi.org/10.3389/fmicb.2024.1346251
This article is part of the Research TopicThe Role of Viruses in Marine EnvironmentsView all 9 articles
A correction has been applied to this article in:
Corrigendum: The trade-off of Vibrio parahaemolyticus between bacteriophage resistance and growth competitiveness
Vibrio parahaemolyticus is a food-borne pathogen, which is often isolated from various seafood products. In this study, two kinds of bacteriophages was isolated from the offshore sediments samples. The anti-phage mutant strain were obtained after seventeen rounds of co-culture of Vibrio parahaemolyticus and mixed bacteriophage, multigroup sequencing was carried out on spontaneous the anti-phage mutant strain and the wild-type strain. We used the Sanger sequencing to verify the accuracy of the mutation sites. Biolog GEN III MicroPlates were used to evaluate the metabolic capacity of wild-type strains and the anti-phage mutant strain. In this study, we found that with flaG gene (slight homology to N terminus of multiple flagellins) mutated, making the bacteriophage unable to absorb to the cell surface of the host. And, the growth competitiveness of the anti-phage mutant strain is lower than the wild-type strain. These results indicated that the fitness cost, including loss of the growth competitiveness, constitutes a barrier to the prevalence of these defense mechanisms. And the selection pressure on different anti-phage strategies depends on the trade-off between mortality imposed by bacteriophages and fitness cost of the defense strategy under the given environmental conditions. In conclusion, this study provides valuable insights into the phage-host interaction and phage resistance in Vibrio parahaemolyticus. Our study provided knowledge for the evolutionary adaption of bacteria against the bacteriophage, which could add more information to understand the phage resistance mechanism before applying in the industry.
Vibrio parahaemolyticus is a Gram-negative bacterium, which is widely distributed in seawater, seabed sediments and seafood, eating fresh food containing Vibrio parahaemolyticus will cause human infection (Raszl et al., 2016; Kim et al., 2017). Antimicrobial agents have been an important attempt at animal therapy since penicillin was discovered in the 1920s (Aarestrup and Wegener, 1999). In order to control the harm of Vibrio parahaemolyticus, antibiotics have to be used in aquaculture. However, antibiotic-resistant Vibrio parahaemolyticus appeared because of the overuse of antibiotics and antibiotic-resistant Vibrio parahaemolyticus will cause economic losses and threaten human health (Lesmana et al., 2001; Ottaviani et al., 2013; Elmahdi et al., 2016; Kang et al., 2017). Therefore, it is necessary to find a new way to control the spread of antibiotic-resistant Vibrio parahaemolyticus.
With the emergence of drug-resistant bacteria, the antibacterial effect of antibiotics has failed and new antibiotics have not been found. Bacteriophage therapy has returned to people’s field of vision because bacteriophage destroys the potential of bacteria (Carlton, 1999; Lu and Collins, 2007, 2009; Sharma et al., 2017; Jamal et al., 2019). The mechanism of bacteriophage therapy is different from that of antibiotics, bacteriolytic bacteriophage can control and kill bacteria in the process of host proliferation (Yang et al., 2019). Importantly, these viruses were nontoxic to humans, bacteriophages cannot survive in the environment alone, and they are aimed at specific host strains (Chan et al., 2018). As early as 1919, bacteriophage therapy treated chickens infected with Salmonella gallinarum (Sulakvelidze et al., 2001). Bacteriophage therapy could control bacterial infections in fish, shrimp, and other aquatic products in aquaculture (Matamp and Bhat, 2019; Nikapitiya et al., 2020). Karunasagar et al. (2007) have confirmed that bacteriophage therapy is an effective disease prevention method for aquaculture. Therefore, bacteriophage therapy is an eco-friendly treatment and can effectively kill Vibrio parahaemolyticus in aquaculture.
At present, there have been a lot of reports about the isolation and identification of Vibrio bacteriophage and its application in animals (Rong et al., 2014; Alagappan et al., 2016; Raszl et al., 2016; Kalatzis et al., 2018; Ahmmed et al., 2019; Ren et al., 2019; Liang et al., 2022). However, this method has not yet reached the stage of clinical application, and it is still in the exploratory stage (Plaza et al., 2018). Before clinical application, several limiting problems (such as effect, mode of administration, bacteriophage resistance, etc.) must be solved (Kalatzis et al., 2018). In this study, two lytic bacteriophage (PGA and PGB) infecting Vibrio parahaemolyticus, was isolated from a large-scale aquaculture areas, and their morphological size, biological characteristics and genetic characteristics were identified. Then the inhibition of PGA and PGB on the growth of Vibrio parahaemolyticus were studied and the evolutionary trade-offs between bacteriophage and bacteria were studied. This identification and analysis will deepen our understanding of Vibrio bacteriophage and provide a theoretical basis for controlling Vibrio parahaemolyticus.
A total of 10 bacterial strains were used in this study (Table 1). A strain of Vibrio parahaemolyticus (MCCC 1A16298) came from China Marine Culture Collection Center (MCCC). Other strains were provided by Third Institute of Oceanography of China State Oceanic Administration. All the bacteria were grown in 2216E liquid medium at 28°C.
The offshore sedimentse (100 g) was collected from an aquaculture areas. Bacteriophages were isolated using the double-layer agar plate method (Clokie, 2009). Briefly, approximately 100 g of offshore sedimentse was mixed with 300 mL of 2216E liquid culture and 50 mL of Vibrio parahaemolyticus and cultivated at 28°C, 180 rpm for 5 days. Samples (10 mL) were collected at 24, 72, and 120 h (Ding et al., 2020). After centrifugation, the supernatant was filtered with 0.22-μm membrane and diluted in sterile PBS. Then 100 μL of diluted supernatant was mixed with 100 μL of Vibrio parahaemolyticus by incubation at 28°C for 10 min. Finally, add 6 mL 2216E semisolid medium, pour it on the surface of hard agar plate and incubate overnight at 37°C. The plaque was purified 6 times until the plaque with the same size and shape was obtained to ensure the purity of the phage stock (Wang et al., 2019; Ding et al., 2020).
To identify the morphological characteristics, the morphology of bacteriophage PGA and PGB were characterized using the TEM. Briefly, the purified bacteriophage supernatants were added onto the surface of a copper grid and adsorbed at room temperature for 15 min. The bacteriophages were negatively stained with 2% phosphotungstic acid in darkness for 30 s and the morphological characteristics were observed under TEM.
Genomic DNA of bacteriophage was extracted using a Genomic DNA Mini Kit. DNA concentration and purity were measured using NanoDrop One at the same time. ALFA-SEQ DNA Library Prep kit was used for Library-building operations. The library quality was assessed by the Qubit 4.0 Fluorometer and Qsep400 High-Throughput Nucleic Acid Protein Analysis system. Then prepared DNA samples were initially fragmented randomly to generate DNA fragments of desired lengths. The sticky ends resulting from the fragmentation were then repaired to create blunt ends. Subsequently, a specific adapter with a 3′ end containing a “T” base was ligated to the repaired DNA fragments by adding a base “A” at the 3′ end of the fragments. Finally, PCR amplification was performed to amplify the DNA fragments with the ligated adapters at both ends, completing the construction of the entire library. The constructed and qualified library was subjected to cluster preparation and sequencing on an Illumina Novaseq 6000 platform. To control the quality, Soapnuke was employed to eliminate low-quality sequencing data and duplicate data generated during PCR (Chen et al., 2018). Then BWA removed the host sequences from the reads (Li and Durbin, 2009). The trimmed reads were assembled by metaviralspade. In addition, ORFs of the bacteriophage genome were predicted by the GeneMarks online server and ORF Finder (Stalin and Srinivasan, 2017). In addition, the GeneMarks online server and ORF Finder were used to predict ORFs in the bacteriophage genome (Stalin and Srinivasan, 2017). Translated ORFs are annotated against NCBI’s non-redundant protein database by BLASTP algorithm (E value < 0.001). To predict the prophage and antibiotics resistance, we used the web tools VirulenceFinder (Joensen et al., 2014) and ResFinder server (Zankari et al., 2012). The two bacteriophage genomes were uploaded to the ViPTree for phylogenetic analysis (Nishimura et al., 2017). We compared the bacteriophages with other reported bacteriophages by progressive Mauve algorithm (Darling et al., 2004; Li et al., 2022).
In the prediction of bacteriophage hosts, BLASTn was used for preliminary analysis (Altschul et al., 1990), followed by the refinement of the analysis results using DeepHost (Ruohan et al., 2022). The dependent database is NCBI taxonomy database1 (Federhen, 2012). Graphage was used to predict the lytic activity of bacteriophages (Wang et al., 2022) with its database derived from TemPhD2 (Zhang X. et al., 2022; Zhang M. et al., 2022) and the NCBI RefSeq database3 (O’Leary et al., 2016). The genes conferring bacteriophage resistance against bacterial CRISPR systems were analyzed by MMseqs2 (Steinegger and Söding, 2017) and AcRanker (Eitzinger et al., 2020), and reference data was from Anti-CRISPRdb4 (Dong et al., 2018). MMseqs2 was also used to analyze the safety of bacteriophages (Steinegger and Söding, 2017). It involved searching the bacteriophage genome for antibiotic resistance genes and virulence genes in the CARD5 (McArthur et al., 2013) and VFDB6 (Chen et al., 2005) databases.
The host range of bacteriophage PGA and PGB on the Vibrio parahaemolyticus strains was determined by the double-layer agar plate method for 10 bacteria strains (Table 1; Pujato et al., 2017; Moodley et al., 2019; Feng et al., 2021). For this, 100 μL of Vibrio parahaemolyticus was added into 6 mL of 2216E semisolid medium, pour it on the surface of hard agar plate. After curing, 5 μL bacteriophage liquid was dripped onto the plate and incubated overnight at 37°C to allow cell lysis by the isolated bacteriophages, then their plaque formation is monitored. Check whether lysis has occurred, judging from the clarity of lysis: (−) no lysis; (+) weak lysis; (++) lysis; (+++) strong lysis (Liang et al., 2022).
The one-step growth experiment was carried out as mentioned above, and some modifications were made (Hagens and Loessner, 2010). In brief, bacteriophage was mixed with Vibrio parahaemolyticus at MOI of 0.1 and incubated at 28°C for 20 min. After centrifugation at 8,000 rpm for 10 min to remove unabsorbed free bacteriophage, the mixture of bacteriophage and bacteria was washed with 2216E for three times. When they were harvested by centrifugation, the sediments of bacterial cells and bacteriophages were suspended with 200 mL of fresh 2216E, and incubated with shaking at 28°C at a speed of 150 rpm. The moment was defined as t = 0 min, and samples are collected at intervals of 10 or 30 min (0, 10, 20, 30, 40, 50, 60, 90, and 120 min) before the bacteriophage titer of each sample is determined by the double-layer method. The outbreak size was calculated as the ratio of the final count of bacteriophage particles released during the incubation period to the initial count of infected bacterial cells. One-step growth curves was drawn, and the incubation period, rising period and bacteriophage burst size of bacteriophage PGA and PGB were calculated as mentioned above (Pujato et al., 2015).
The antibacterial effect of bacteriophage PGA and PGB against Vibrio parahaemolyticus was detected in a 96-well plate (Liang et al., 2022). Briefly, bacteriophage was added into the Vibrio parahaemolyticus as MOI 0.01, 0.1, 1, and 10. Then 200 μL of the mixture was added into a 96-well plate. The blank group was 2216E liquid medium, and the control group was Vibrio parahaemolyticus. The 96-well plate was placed in the Microplate Reader (28°C, low speed) for culture. Starting from 0 min, the absorbance (OD600) was detected for each sample every 10 min.
To obtain Vibrio parahaemolyticus mutants resistant to bacteriophage, following the method described (Yang et al., 2020) as described previously with some modifications. Briefly, 100 μL of bacteriophage was added into 10 mL of exponential phase Vibrio parahaemolyticus (OD600 = 0.1), the culture was cultivated on a shaker at 28°C with shaking for 3 days and the OD600 was monitored.
Bacteriophage adsorption test was carried out by Vibrio parahaemolyticus (Le et al., 2014). Briefly, bacteriophage was added with an MOI of 0.01 and adsorbed at 28°C for 20 min. Then the bacteriophage and bacteria mixture were washed three times with PBS. Finally, the morphology of mixture was characterized using the transmission electron microscopy (Yang et al., 2020).
Genome Sequencing was performed by Magigene Biotechnology Co., Ltd. (Guangzhou, China). The expression level of transcripts were quantified by Salmon (Patro et al., 2017). DEGs were performed using the edgeR (Robinson et al., 2010). GO and KEGG enrichment analysis of DEGs were implemented by the clusterProfiler.
Wild strains and the anti-phage mutant strain of Vibrio parahaemolyticus were analyzed by Biolog GEN III MicroPlates. The growth assays of bacteriophages PGA and PGB was tested according to the method described by Williams et al. (2017).
In order to calculate the average value and standard deviation of the numerical data of three independent experiments, the variance analysis method is used. The differences between treatments were analyzed by t-test.
The complete genome sequence of bacteriophage PGA and PGB have been deposited in the GenBank database under the accession number PP001175 and PP001176. Transcriptional sequencing of Vibrio parahaemolyticus have been deposited in the BioProject database under the accession number PRJNA1055304.
We induced and isolated two bacteriophage that could target the Vibrio parahaemolyticus, obtained from the offshore sedimentse of a large-scale aquaculture areas. Bacteriophage PGA and PGB formed clear plaques (about 1.6 mm in diameter of PGA and about 1 mm in diameter of PGB) on the double-layered agar plate (Figures 1A,B), indicating that the bacteriophage was lytic. After staining with 2% PTA negatively, the TEM image showed that PGA (Figure 1C) and PGB (Figure 1D) consist of an icosahedral head and a long non-contractile tail. The measurements of length of head, diameter of head, and length of contractile tails of PGA were 66.5 ± 2, 61 ± 1, and 100 ± 5 nm while those of PGB were 72.5 ± 2, 70.5 ± 1, and 136 ± 5 nm. By the International Virus Classification and Nomenclature, bacteriophages PGA and PGB belong to the family Myoviridae, order Caudovirales (Lefkowitz et al., 2018). Bacteriophages PGA and PGB belonged to the order Myoviridae and then were named as vB_VpaS_PGA (bacteriophage PGA) and vB_VpaS_PGB (bacteriophage PGB).
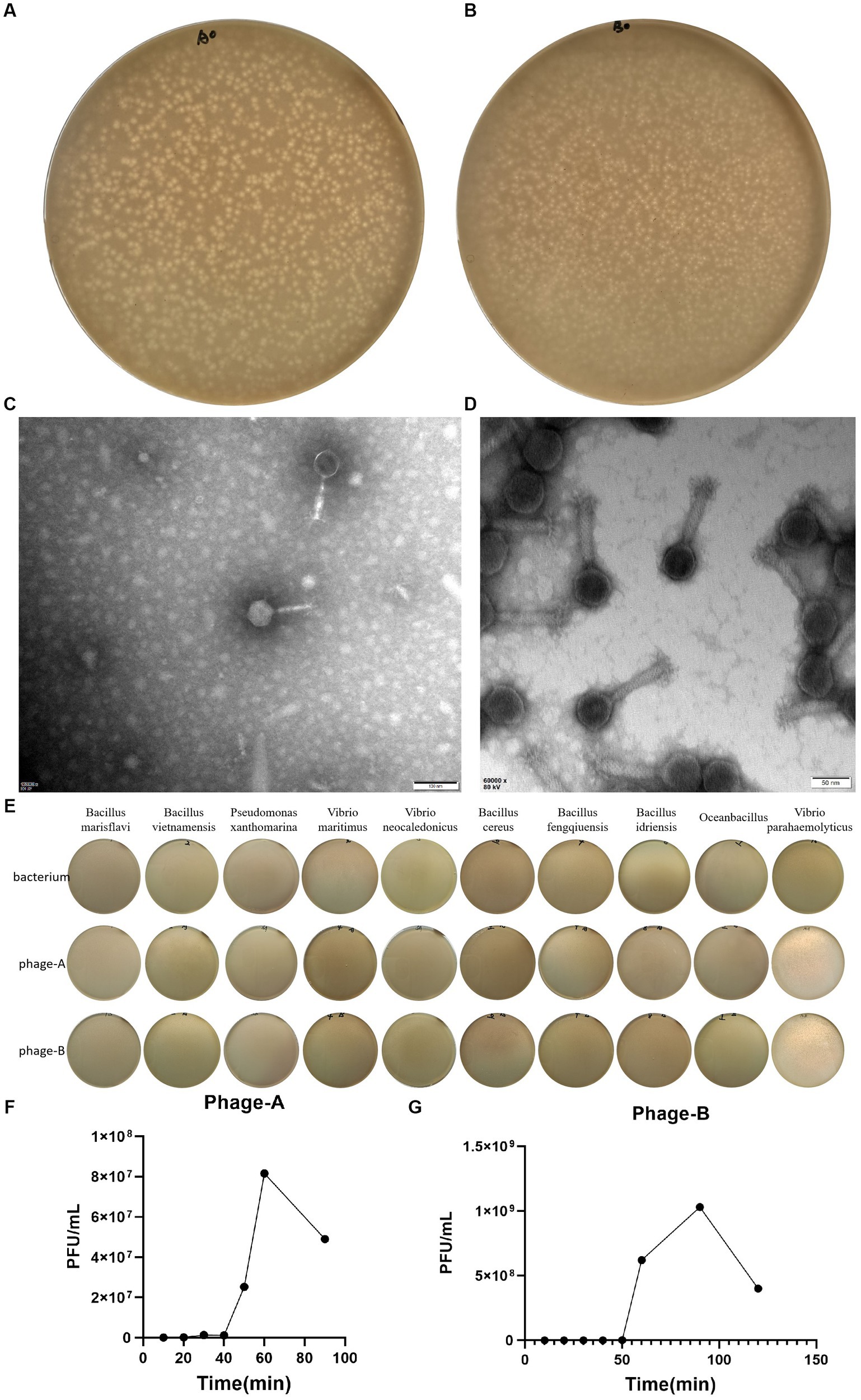
Figure 1. Characterization of Bacteriophage. (A) Plaques of bacteriophage vB_VpaS_PGA. (B) Plaques of bacteriophage vB_VpaS_PGB. (C) Transmission electron microscopy of bacteriophage vB_VpaS_PGA. (D) Transmission electron microscopy of bacteriophage vB_VpaS_PGB. (E) Host range of bacteriophage vB_VpaS_PGA and vB_VpaS_PGB. (F) One-step growth curve of PGA. (G) One-step growth curve of PGB.
To determine the host range of bacteriophages PGA and PGB, the double-layer agar plate method was used for 10 bacteria strains. As the results (Figure 1E; Table 1), bacteriophage PGA and PGB can only lyse Vibrio parahaemolyticus. These results indicated that these phages had a narrow host range which can specifically target Vibrio parahaemolyticus strains, suggesting the potential of the bacteriophage PGA and PGB to be a candidate for bacteriophage therapy (Liang et al., 2022).
The life cycle of bacteriophage, including the latent period, explosive phase, and plateau phase, was quantified using a one-step growth curve. As shown in Figure 1F, PGA was characterized by a short incubation period of 20 min, an outbreak period of 60 min and a burst size of 88.4 PFUs/infected cell, indicating that phages grew efficiently and rapidly after adsorption on the host surface. The latent period and burst period of phage PGB were 50 and 90 min, and the burst size was about 222.0 PFUs/infected cell (Figure 1G). Taking these together, bacteriophages PGA and PGB have a good lysis effect to Vibrio parahaemolyticus (Liang et al., 2022).
In order to understand the Genetic characteristics of bacteriophages, the genome of bacteriophage PGA and PGB were sequenced and analyzed (Yang et al., 2020). Genomic characterization of the two bacteriophages indicated (Figures 2A,B) that the genomes of PGA and PGB are circular double-stranded DNA (of 40.27 and 40.27 bp, respectively). The GC content of bacteriophage PGA (42.26%) was similar to PGB (42.28%; Li et al., 2022). In addition, the results (Table 2) show that although 63 and 62 ORFs were identified in PGA and PGB respectively, only 28.57% and 27.42% of the ORFs were assigned to specific functions (DNA replication, DNA metabolism, DNA packaging, and structure formation).
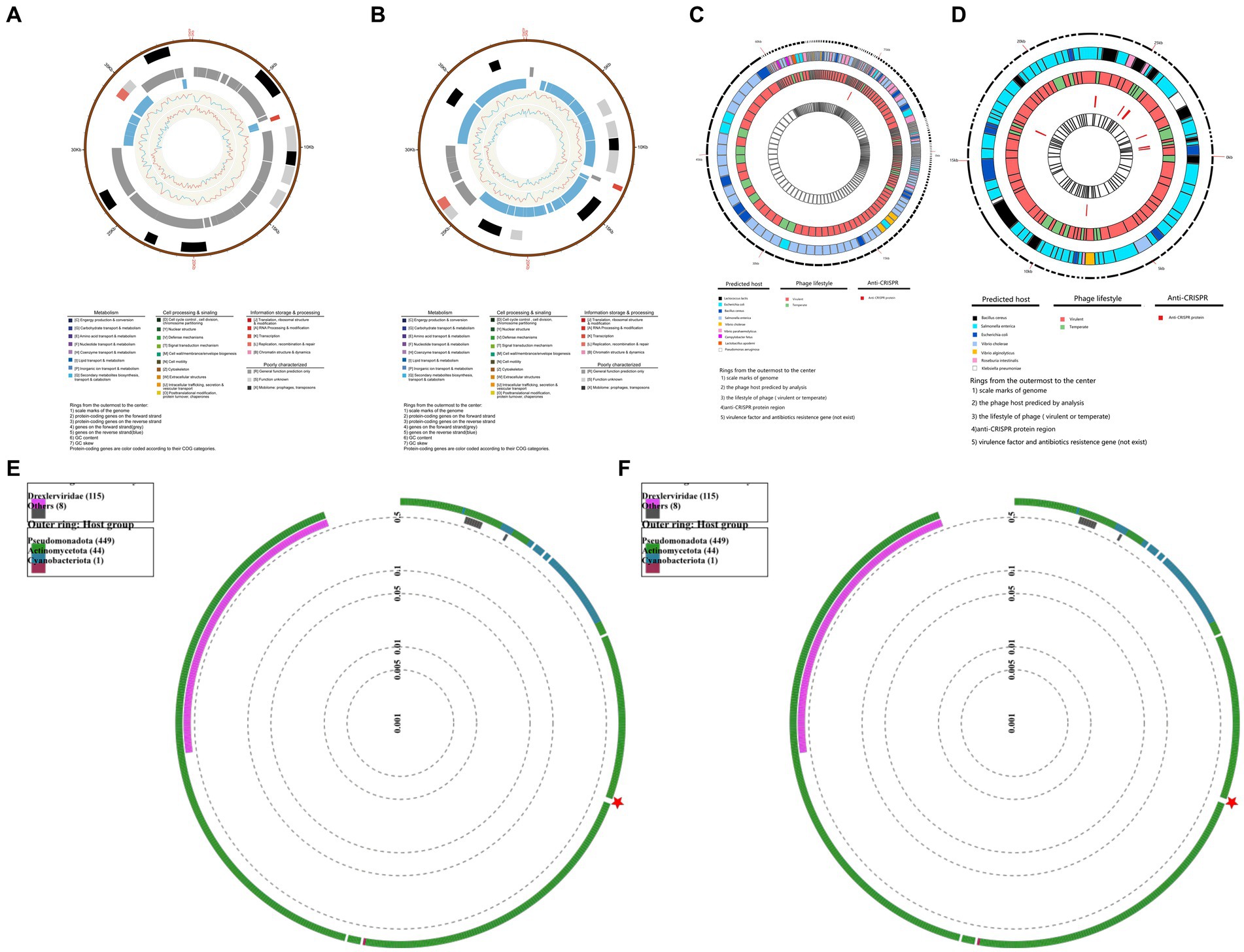
Figure 2. Genomic characterization of bacteriophages. (A) Genomic characterization of vB_VpaS_PGA. Rings from the outermost to the center: (1) scale marks of the genome; (2) protein-coding genes on the forward strand; (3) protein-coding genes on the reverse strand; (4) genes on the forward strand (gray); (5) genes on the reverse strand (blue); (6) GC content; (7) GC skewProtein-coding genes are color coded according to their COG categories. (B) Genomic characterization of vB_VpaS_PGB. Rings from the outermost to the center: (1) scale marks of the genome; (2) protein-coding genes on the forward strand; (3) protein-coding genes on the reverse strand; (4) genes on the forward strand (gray); (5) genes on the reverse strand (blue); (6) GC content; (7) GC skewProtein-coding genes are color coded according to their COG categories. (C) Security and efficacy of vB_VpaS_PGA. Rings from the outermost to the center: (1) scale marks of genome; (2) the phage host predicted by analysis; (3) the lifestyle of phage (virulent or temperate); (4)anti-CRISPR protein region; (5) virulence factor and antibiotics resistance gene (not exist). (D) Security and efficacy of vB_VpaS_PGA. Rings from the outermost to the center: (1) scale marks of genome; (2) the phage host predicted by analysis; (3) the lifestyle of phage (virulent or temperate); (4)anti-CRISPR protein region; (5) virulence factor and antibiotics resistance gene (not exist). (E) Phylogenetic analysis of vB_VpaS_PGA (circular). The phylogenetic tree was constructed using the Maximum Likelihood method with 1,000 bootstrap replicates. Amino acid sequences of the terminase large subunit. (F) Phylogenetic analysis of vB_VpaS_PGB (circular). The phylogenetic tree was constructed using the Maximum Likelihood method with 1,000 bootstrap replicates. Amino acid sequences of the terminase large subunit.
The potential applications of bacteriophages were analyzed via bioinformatics tools, and the results were visualized using TBtools-II in Figures 2C,D (Chen et al., 2023). The genome structure and ORFs of PGA and PGB were determined from previous whole genome sequencing analysis. BLASTn and DeepHost were utilized to analyze potential hosts for the bacteriophage and map them to their corresponding positions on the genome. From the analysis of bacteriophage lifestyle, it is evident that both PGA and PGB demonstrate pronounced bacteriolytic activity against their respective hosts. AcRanker and MMseq2 revealed a higher abundance of anti-CRISPR system genes in PGB compared to PGA, which exhibited a lower presence of such genes. Based on the alignment and comparison results using MMseqs2, the presence of virulence genes and antibiotic resistance genes was not detected in both PGA and PGB (as shown in the innermost circles in Figures 2C,D). PGA and PGB meet the prerequisites (non-virulence genes, antimicrobial resistance genes or lysogenic genes) of bacteriophage therapy candidates, so they can be used as specific lytic bacteriophages (Bardina et al., 2016; Li et al., 2021). In summary, the analyses have demonstrated the strong lytic activity of PGA and PGB toward their host, as well as the absence of virulence genes and drug resistance genes, thus ensuring the genetic safety of these phages for applications. The bioinformatics-based predictions and validations contribute to assessing the security and efficacy of utilizing these two phage strains for therapeutic interventions.
We found that the phylogenetic analysis (Figures 2E,F) revealed that PGA and PGB had the closest relationship withVibrio bacteriophage vB_VpP_BT-1011 (NC_070774). The homology of PGA and PGB with vB_VpP_BT-1011 was 75.86% and 76.35% respectively, only two Vibrio bacteriophages appear in the corresponding phylogenetic tree and it shows that they are new species of this genus (Li et al., 2022).
To assess the growth-inhibitory effect of bacteriophages PGA and PGB on Vibrio parahaemolyticus, the growth curve was measure by bacteriophage PGA/PGB. As the results (Figures 3A,B), when the MOI was 1 and 10 respectively, the growth of Vibrio parahaemolyticus was obviously inhibited within 8 h, and then increased exponentially. Meanwhile Vibrio parahaemolyticus regrew after 4 h at MOI of 0.01 and 0.1. This means that Vibrio parahaemolyticus regrows after adding single bacteriophage. Then we determine the growth-inhibitory effect of the mixed bacteriophage on Vibrio parahaemolyticus. The results (Figure 3C) again revealed spontaneous mutation of Vibrio parahaemolyticus occurred during co-culture. Based on the bacteriophage co-culture assay, we hypothesized that spontaneous mutation of Vibrio parahaemolyticus may lead to the inhibition of bacteriophage adsorption. In order to prove this hypothesis, the anti-phage mutant strain were obtained after 17 rounds of co-culture of Vibrio parahaemolyticus and mixed bacteriophage. The anti-phage mutant strain was designated as the anti-phage mutant strain VP-17. Then whether the bacteriophage was adsorbed on Vibrio parahaemolyticus was observed by transmission electron microscope. The results (Figures 4A–D) confirmed that spontaneous mutation of Vibrio parahaemolyticus result in the inhibition of bacteriophage adsorption.
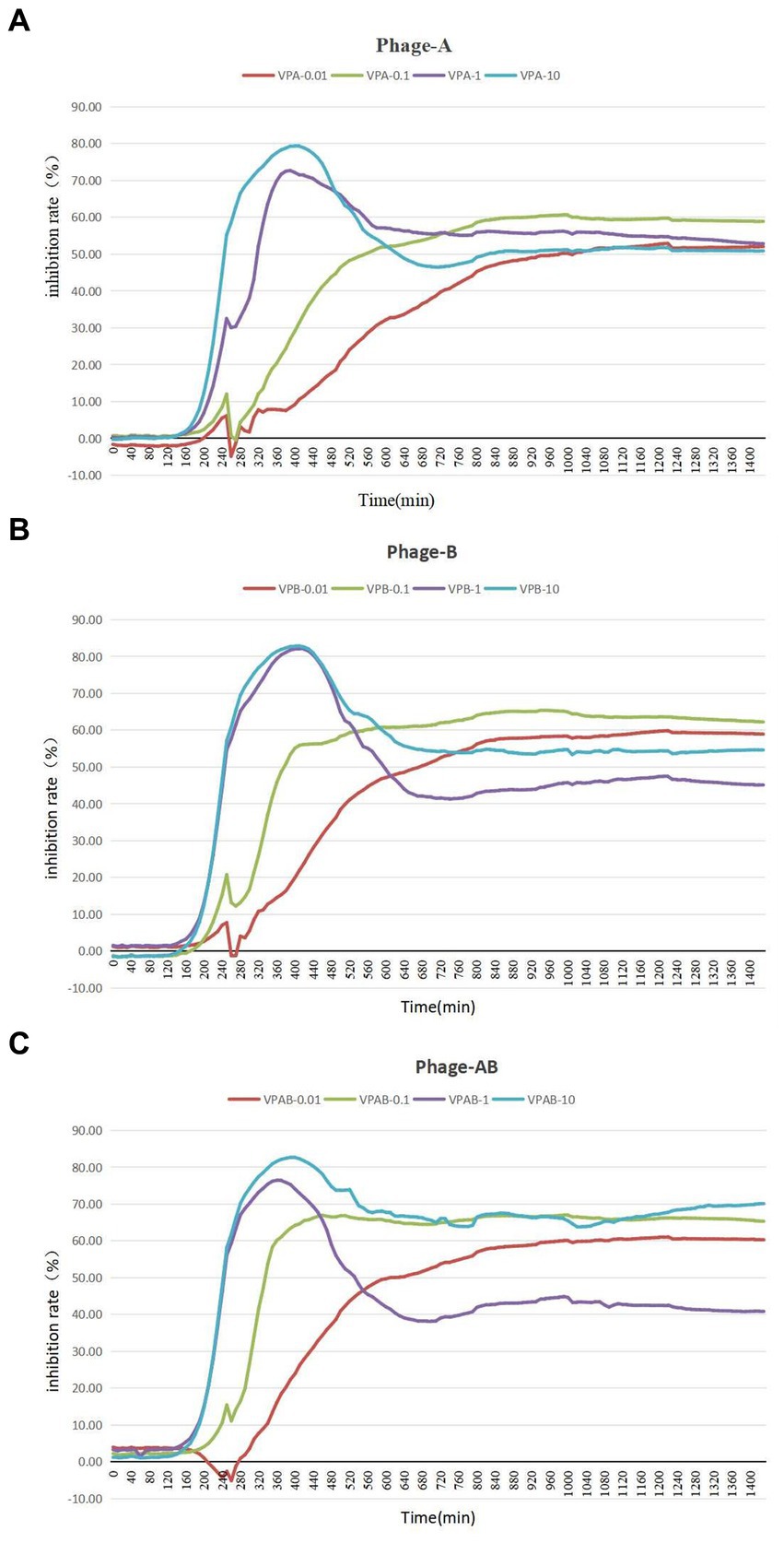
Figure 3. Lysis of Vibrio parahaemolyticus strains by vibriophage vB_VpaS_PGA and vB_VpaS_PGB. vB_VpaS_PGA and vB_VpaS_PGB infected different Vibrio parahaemolyticus strains of infection (MOI), and the densities of the cultures were determined by measuring absorbance at 600 nm. (A) Bacteriophage vB_VpaS_PGB. (B) Bacteriophage vB_VpaS_PGB. (C) Bacteriophage vB_VpaS_PGA and bacteriophage vB_VpaS_PGB.
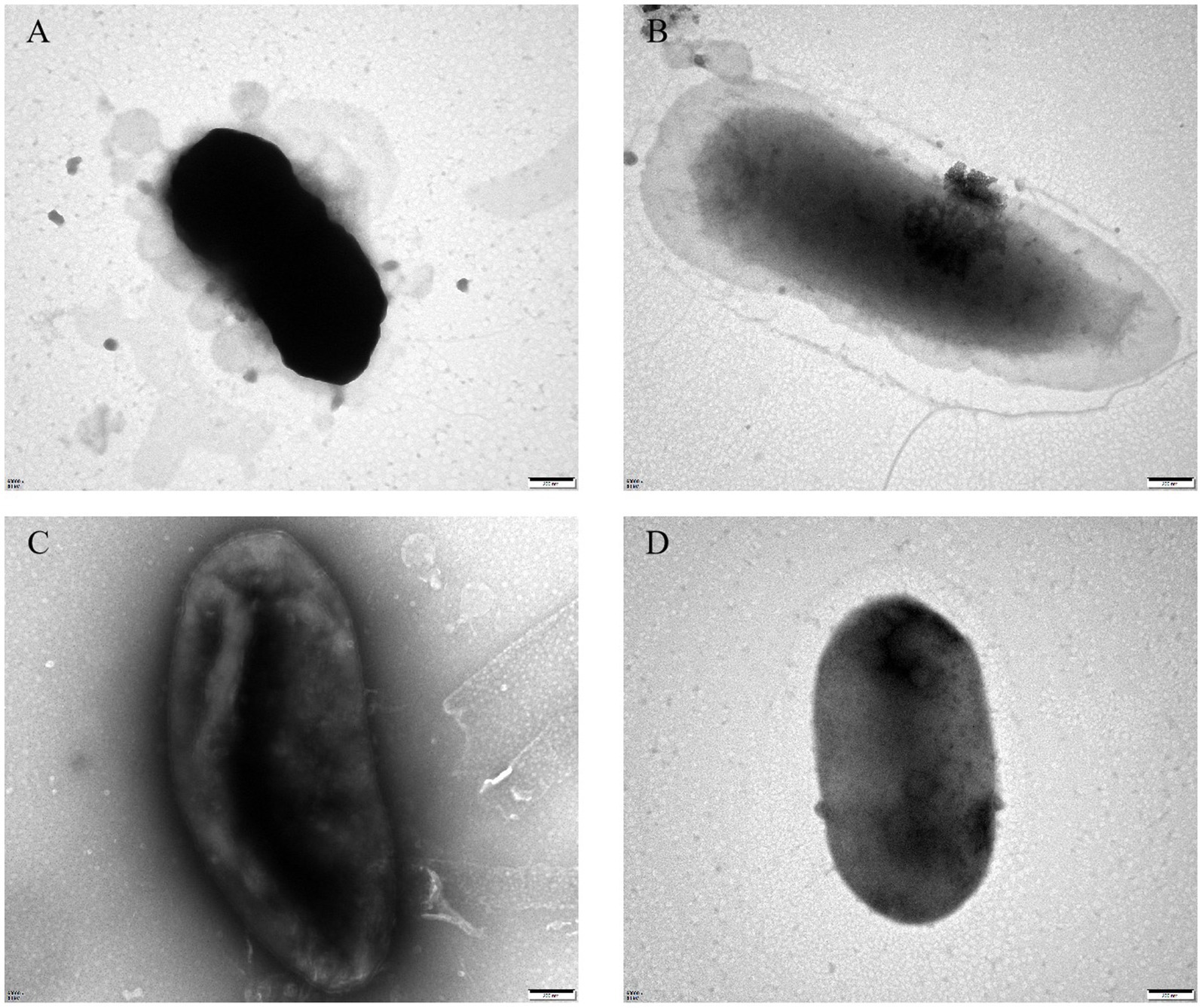
Figure 4. Transmission electron microscopy of bacteriophage adsorbing Vibrio parahaemolyticus. (A) Bacteriophage vB_VpaS_PGA adsorption of the wild-type strain. (B) Bacteriophage vB_VpaS_PGA adsorption of the anti-phage mutant strain. (C) Bacteriophage vB_VpaS_PGB adsorption of the wild-type strain. (D) Bacteriophage vB_VpaS_PGB adsorption of the anti-phage mutant strain.
To determine the genomic changes associated with bacteriophage adsorption, multigroup sequencing was carried out on spontaneous the anti-phage mutant strain (VP-17) and the wild-type strain (MCCC 1A16298; Martin et al., 2018). We found 25 SNP differences by comparing the core genomes of the wild-type strains and the anti-phage mutant strain (Supplementary Table S2). Since an spontaneous mutation was observed in the co-culture by Vibrio parahaemolyticus (Figure 3C), it is expected that the mutant gene of bacteriophage resistance can be identified by differential RNA-seq. Specifically, the results of differential expression analysis showed that compared with wild-type strains, 316 genes were up-regulated and 466 genes were down-regulated in phage-resistant mutant (Figure 5A). On the whole, genes related to pyruvate metabolism, propanoate metabolism, glycolysis/gluconeogenesis and flagellar assembly (Figure 5B) were found to be more differentially expressed when spontaneous mutation of Vibrio parahaemolyticus occurred during co-culture. The function of these genes are closely related to the survival of bacteria and their adaptation to stimuli (Kim and McCarter, 2000), and the results of GO enrichment analysis also confirmed this result (Figure 5C). The analysis of the interaction between the gene where the mutation site is located and the differentially expressed gene shows that the gene 1_orf 01980 where the mutation site is located has an interaction relationship with 90 differential genes (Figure 5D). The gene 1_orf 01980 prediction is related to flaG gene, flaG gene is a flagella gene, which has chemotaxis and motility (Ahmmed et al., 2019). In order to verify the accuracy of the mutation site, we used the first generation sequencing to sequence SNPs. The result (Figure 5E) showed that the anti-phage mutant strain did exist SNP mutation sites.
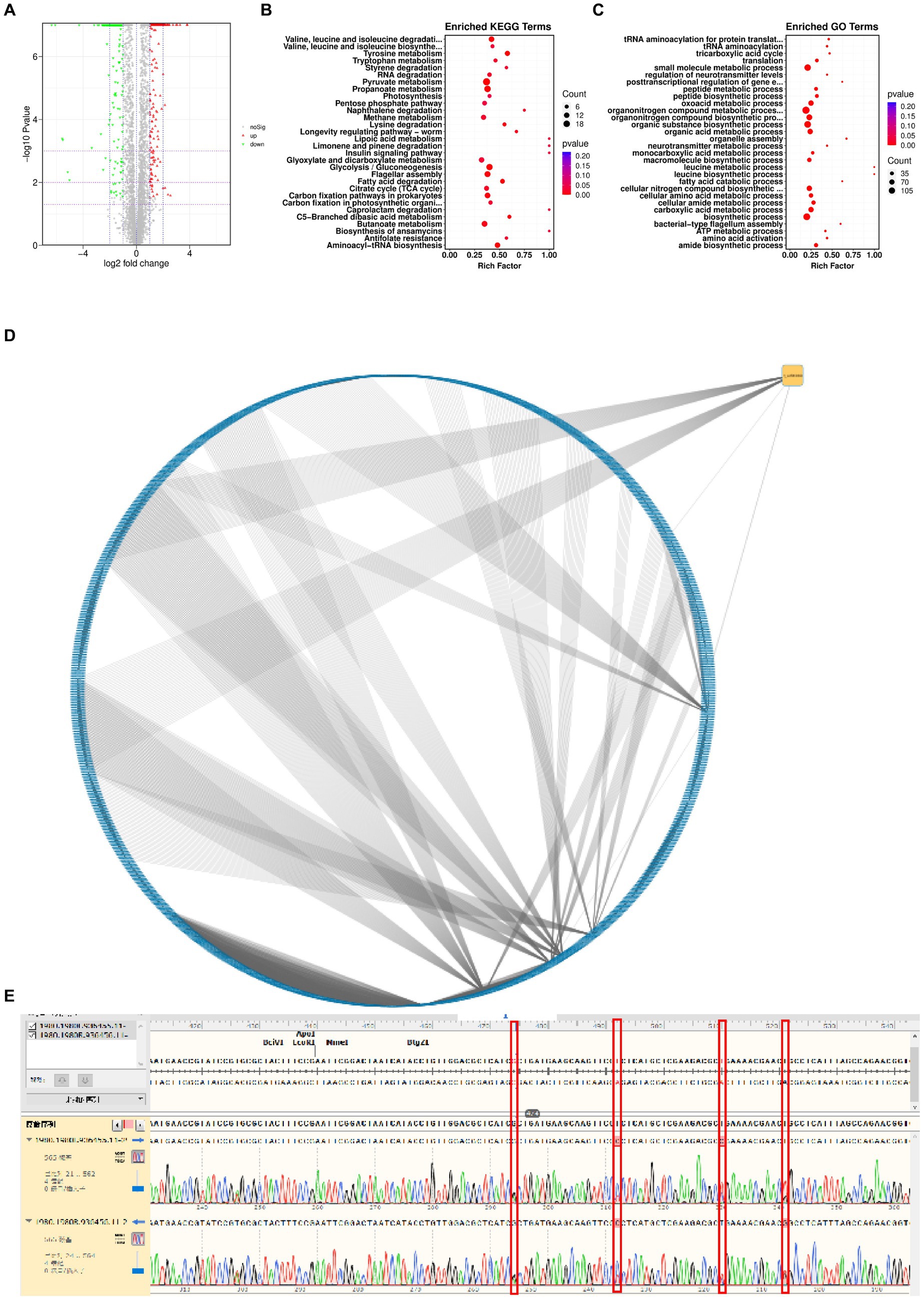
Figure 5. Genetic changes from the wild-type strain to the anti-phage mutant strain. (A) Differential gene volcanic map. (B) KEGG function enrichment analysis statistical chart. (C) GO function enrichment analysis statistical chart. (D) Network diagram of interaction relationship between genes with mutation sites and differentially expressed genes. Blue is the differentially expressed gene, and yellow is the gene with mutation site. From an interactive point of view, there is variation information. Gene 1_orf 01980 interacts with 90 different genes. (E) SNP comparison.
In order to evaluate the metabolic capacity of the wild-type strain and the anti-phage mutant strain, Biolog GEN III MicroPlates were used to analyze this capacity. After 24 h of incubation, MCCC 1A16298 and VP-17 strains had differences in carbon source utilization on the Biolog GEN III MicroPlate (Figures 6A–F) by naked eye observation, it shows that there are differences between the two kinds of bacteria in their ability to use carbon sources. Statistical analysis via a t-test (Figures 7A,B) showing that every negative control of MCCC 1A16298 and VP-17 did not increase significantly, and no statistically significant differences in the absorbance of the positive control, it shows that the growth rate in complete culture medium was similar. At these four time points (6, 12, 18, and 24 h), the data trend of all carbon source were consistent. Therefore, we use data of 24 h time point for statistical analysis. Of particular surprise, the results (Figures 7C–J) showed that there were no statistically significant differences between VP-17 and MCCC 1A16298. Then the growth competitiveness of the wild strains and the anti-phage mutant strain was also tested, the two kinds of bacteria were mixed in a certain proportion, and samples were taken and sequenced at different time points (60, 260, 610 and 1,010 min). The results (Figure 7K) show that with the passage of time, the proportion of the anti-phage mutant strain is decreasing, while the proportion of the wild-type strains is increasing. In other words, the growth competitiveness of the wild-type strain is stronger than that of the anti-phage mutant strain. The above data demonstrated that the anti-phage mutant strain obtained bacteriophage resistance at the cost of growth competitiveness.
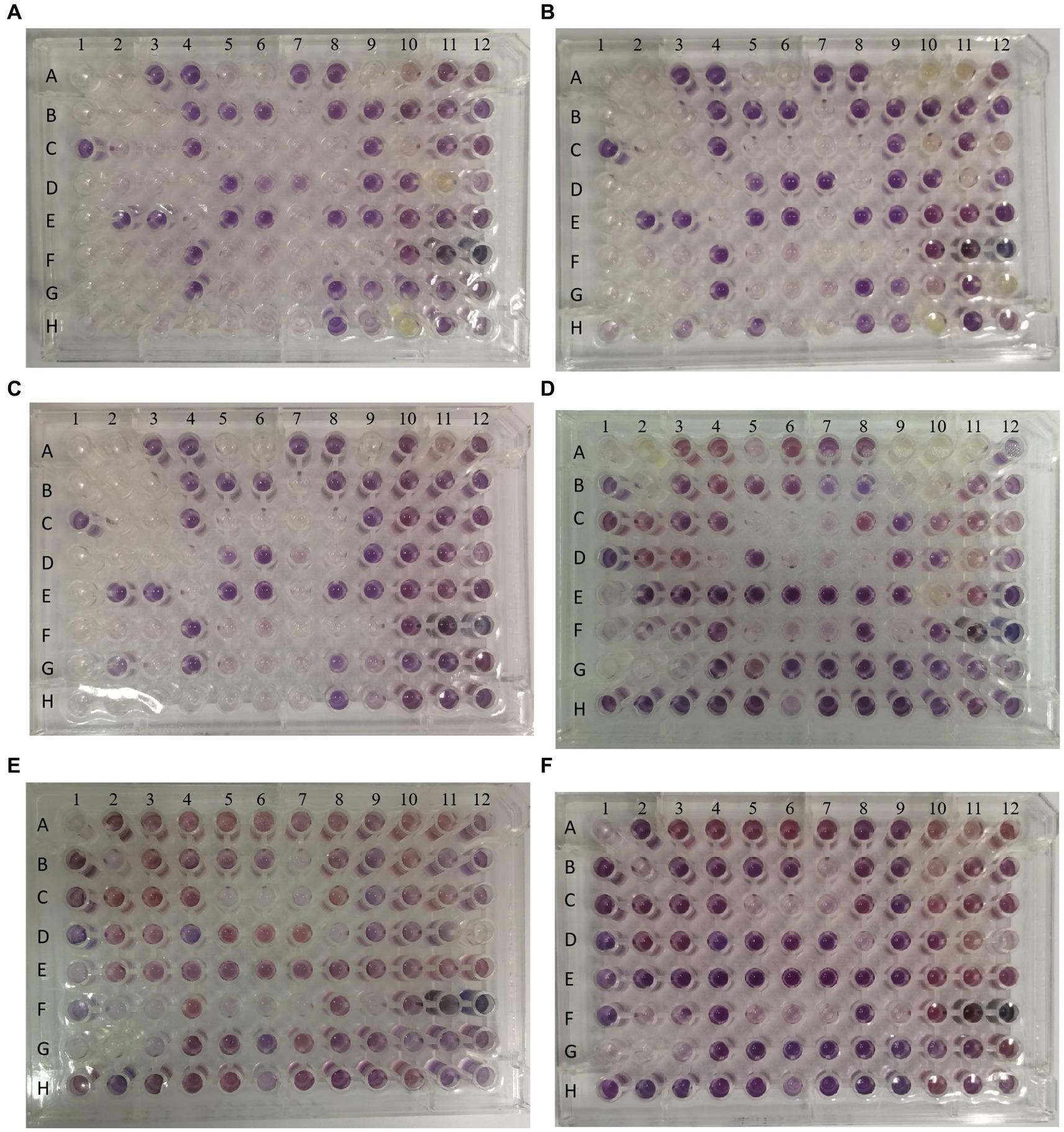
Figure 6. Picture of one representative set of Biolog plates after 24 h of incubation. From left to right: Wells A1 and A9 of each plate are the negative and positive controls. Chemical indicator assays are in columns 10–12. The names of carbon sources in each well are provided in Table 3. (A–C) The three independent biological replicates of the wild-type strain. (D–F) The three independent biological replicates of the anti-phage mutant strain.
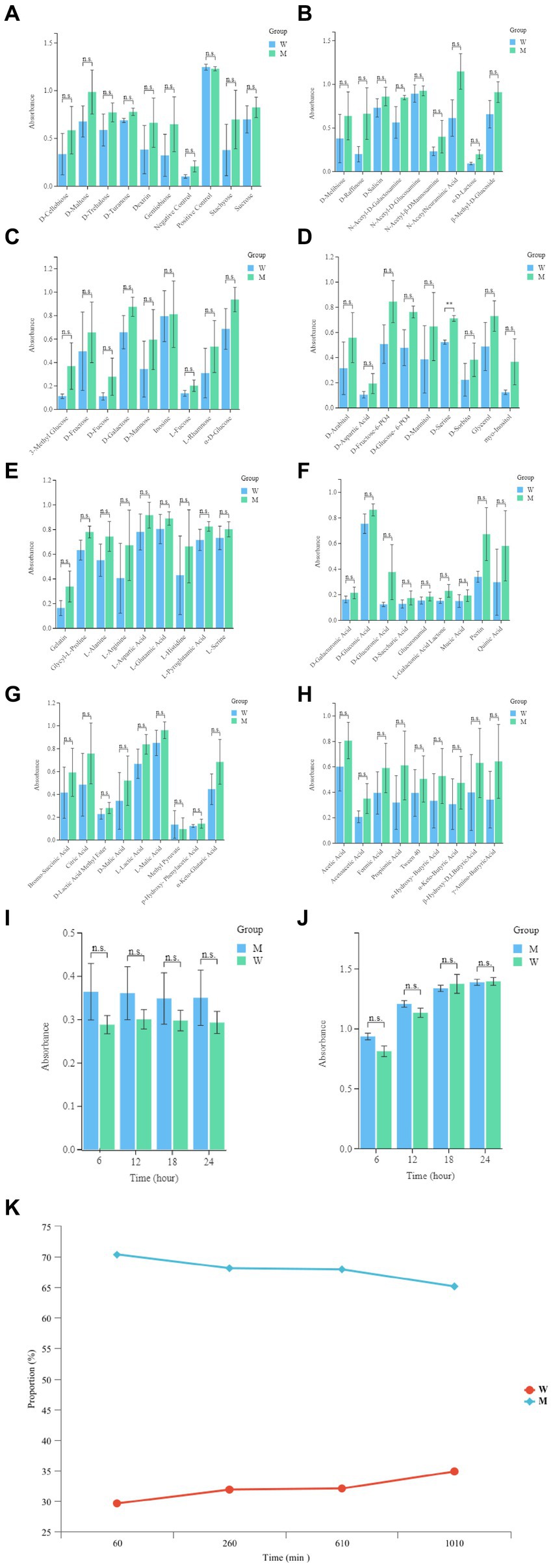
Figure 7. Comparative analysis of growth of the wild-type strain and the anti-phage mutant strain. (A) The absorbance readings of the negative control wells, the raw spectrophotometric triplicate data collected from each experimental well at 6, 12, 18, and 24 h was directly analyzed. (B) The absorbance readings of the positive control well, the raw spectrophotometric triplicate data collected from each experimental well at 6, 12, 18 and 24 h was directly analyzed. (C–J) Each panel corresponds to a row in the Biolog plate, respectively. The raw data absorbance values for triplicate samples from the 24 h time point were averaged. The names of the carbon sources in wells 1–9 within each row are provided on the x-axis. Each set of data represent the wild-type strain and the anti-phage mutant strain from left to right, respectively. Error bars were calculated using the standard error across the three independent biological replicates. (K) The growth competitiveness of the wild strains and the anti-phage mutant strain, Compare the proportions of the wild strains and the anti-phage mutant strain at different time points (60, 260, 610, and 1,010 min).
With the continuous deterioration of antibiotic resistance and the continuous growth of drug-resistant pathogens, bacteriophage therapy is favored (Carlton, 1999; Lu and Collins, 2007, 2009). As early as 1919, bacteriophage therapy treated chickens infected with Salmonella gallinarum (Sulakvelidze et al., 2001). In the past 20 years, many researchers have studied the identification and clinical application of bacteriophage (Dedrick et al., 2019; Jault et al., 2019). At present, most of the research on bacteriophage is based on sequencing to predict its function. For bacteriophage therapy, it is necessary to solve the development of bacteriophage resistance to the host. However, the mechanism of bacteriophage resistance is rarely reported, we lack detailed knowledge about their ecological and evolutionary interactions (Kortright et al., 2019; Markwitz et al., 2021; North and Brown, 2021; Tang et al., 2022).
The interactions between bacteriophages and the anti-phage mutants are complex. To gain insight into such relationships, we screened a collection of 2 vibriophages (PGA and PGB) from the offshore sedimentse samples. It is evident that both PGA and PGB demonstrate pronounced bacteriolytic activity against their respective hosts. Most importantly, no virulence gene was identified showing the safety of vibriophages PGA and PGB in biocontrol applications. To resist bacteriophage infection, host genes sometimes evolve mutations and have a significant impact on their own metabolic activities. In this study, it was found that the anti-phage mutants can not be adsorbed by bacteriophage because of the mutation of flaG gene (slight homology to N terminus of multiple flagellins) mutated (Figure 5E; Sar et al., 1990; Ahmmed et al., 2019). It is reported that through modifying cell surface to prevent bacteriophage attachment and entry, bacteriophage adsorption is blocked, resulting in bacteriophage resistance (Destoumieux-Garzón et al., 2005; Samson et al., 2013; Hill et al., 2018). Specifically, the mutation of cell surface receptors (fimbriae, flagella, outer membrane proteins, and lipopolysaccharide) is very important to inhibit the binding of bacteriophage to the host bacteria (Qimron et al., 2006; Westra et al., 2015; Houte et al., 2016; Shen and Loessner, 2021).
In addition, the growth competitiveness of wild strains and the anti-phage mutants was also tested, it was found that the growth competitiveness of the anti-phage mutant strain is lower than the wild-type strain. It is reported that under the pressure of bacteriophage selection, bacteria evolve bacteriophage resistance through bacterial defense systems (Bikard and Marraffini, 2012; Samson et al., 2013). Anti-phage mutant strain may make trade-offs with bacterial growth rate, toxicity, drug resistance, utilization of carbon source and formation of biofilm (Mangalea and Duerkop, 2020; Castledine et al., 2022), which is consistent with our results. In the natural environment, the bacterial community are diverse and the bacterial population are dynamic. Bacteria compete for resources and follow the principle of competition and exclusion, and those bacteria with slow growth rates will be eliminated. Although the anti-phage mutants acquired resistance to phage, their growth competitiveness also declined. Under the condition of competitive exclusion principle, the mutant strain is not competitive and may be eliminated, which opens up a new idea for phage therapy to produce drug resistance. In the natural environment, few living things exist in a constant environment, and trade-offs will be different in different environments. This study also provides an insight into the co-evolution of trade-offs from the perspective of the evolution of microbial laboratory.
Overall, this characterization and analysis provides a theoretical basis for the application of bacteriophage therapy. Bacteriophage treatment can not only enhance the resistance to bacteriophage, but also enhance the sensitivity to antibiotics (German and Misra, 2001; Chan et al., 2016). The positive effect of bacteriophage resistance can improve the sensitivity of antibiotics, which can make up for the deficiency of bacteriophage therapy (Wang et al., 2021). This provides an idea for treatment and can be used together with other reagents (such as antibiotics and lyases). This study provides a basis for further study on the relationship between related bacteriophages of Vibrio parahaemolyticus. In the future, it is necessary to further study the diversity of bacteriophage and its biological control mechanism against bacteria, especially pathogens, and more work is needed to understand the complexity of pleiotropic interaction between various bacteriophages and bacteria. However, this research has its limitations. The environment of this experiment is too single simple, the medium used to isolate and cultivate microorganisms in the laboratory has a strong selection function. This method can not fully reflect the ecological function of marine microorganisms (Bacteria and bacteriophages), for the dynamic changes during phage therapy, it is very important to better understand the community evolution and ecological variability.
The authors acknowledge that the data presented in this study must be deposited and made publicly available in an acceptable repository, prior to publication. Frontiers cannot accept a manuscript that does not adhere to our open data policies.
XZ: Data curation, Methodology, Writing – original draft. SL: Writing – original draft, Resources. GG: Resources, Writing – review & editing, Software. YH: Writing – review & editing, Conceptualization. YS: Writing – review & editing, Supervision, Funding acquisition. JD: Writing - review & editing, Data curation.
The author(s) declare financial support was received for the research, authorship, and/or publication of this article. This work was supported by the Project of Ningbo No.2 Hospital (Project Number: 2023-Y07), the Natural Science Foundation of National (42006093), the Natural Science Foundation of Ningbo (2021J323, 2023J057, 2021J324, and 2021J326), research initiation project of Ningbo Institute of Life and Health Industry, Chinese Academy of Sciences (2021YJY1010 and 2022YJY0204), the Natural Science Foundation of Zhejiang Province (ZCLTGY24H1901), the “Bei’an essence” Talent Project of Ningbo City (2023RC003), Zhejiang Province and Ningbo City Co-constructed Project of Leading Medical & Health Discipline (2016-S04), and the second batch of excellent health professionals in Ningbo-outstanding young and middle-aged health technicians (2012008).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2024.1346251/full#supplementary-material
1. ^https://www.ncbi.nlm.nih.gov/taxonomy
2. ^https://phage.deepomics.org/
3. ^https://www.ncbi.nlm.nih.gov/refseq/
Aarestrup, F. M., and Wegener, H. C. (1999). The effects of antibiotic usage in food animals on the development of antimicrobial resistance of importance for humans in campylobacter and Escherichia coli. Microbes Infect. 1, 639–644. doi: 10.1016/s1286-4579(99)80064-1
Ahmmed, S., Khan, M. A. A. K., Eshik, M. M. E., Punom, N. J., Islam, A. B. M. M. K., and Rahman, M. S. (2019). Genomic and evolutionary features of two AHPND positive Vibrio parahaemolyticus strains isolated from shrimp (Penaeus monodon) of south-West Bangladesh. BMC Microbiol. 19, 1–14. doi: 10.21203/rs.2.14490/v2
Alagappan, K., Karuppiah, V., and Deivasigamani, B. (2016). Protective effect of phages on experimental Vibrio parahaemolyticus infection and immune response in shrimp (Fabricius, 1798). Aquaculture 453, 86–92. doi: 10.1016/j.aquaculture.2015.11.037
Altschul, S. F., Gish, W., Miller, W., Myers, E. W., and Lipman, D. J. (1990). Basic local alignment search tool[J]. J. Mol. Biol. 215, 403–410. doi: 10.1016/S0022-2836(05)80360-2
Bardina, C., Colom, J., Spricigo, D. A., Otero, J., Sánchez-Osuna, M., Cortés, P., et al. (2016). Genomics of three new bacteriophages useful in the biocontrol of salmonella. Front. Microbiol. 7:545. doi: 10.3389/fmicb.2016.00545
Bikard, D., and Marraffini, L. A. (2012). Innate and adaptive immunity in bacteria: mechanisms of programmed genetic variation to fight bacteriophages. Curr. Opin. Immunol. 24, 15–20. doi: 10.1016/j.coi.2011.10.005
Carlton, R. M. (1999). Phage therapy: past history and future prospects. Arch. Immunol. Ther. Exp. (Warsz.) 47, 267–274.
Castledine, M., Padfield, D., Sierocinski, P., Pascual, J. S., Hughes, A., Mäkinen, L., et al. (2022). Parallel evolution of Pseudomonas aeruginosa phage resistance and virulence loss in response to phage treatment in vivo and in vitro. eLife 11:e73679. doi: 10.7554/eLife.73679
Chan, B. K., Sistrom, M., Wertz, J. E., Kortright, K. E., Narayan, D., and Turner, P. E. (2016). Phage selection restores antibiotic sensitivity in MDR Pseudomonas aeruginosa. Sci. Rep. 6:26717. doi: 10.1038/srep26717
Chan, B. K., Turner, P. E., Kim, S., Mojibian, H. R., Elefteriades, J. A., and Narayan, D. (2018). Phage treatment of an aortic graft infected with Pseudomonas aeruginosa. Evolut Med Public Health 2018, 60–66. doi: 10.1093/emph/eoy005
Chen, Y., Chen, Y., Shi, C., Huang, Z., Zhang, Y., Li, S., et al. (2018). SOAPnuke: a MapReduce acceleration-supported software for integrated quality control and preprocessing of high-throughput sequencing data. Gigascience. 7, 1–6. doi: 10.1093/gigascience/gix120
Chen, C., Wu, Y., Li, J., Wang, X., Zeng, Z., Xu, J., et al. (2023). TBtools-II: a "one for all, all for one" bioinformatics platform for biological big-data mining. Mol. Plant 16, 1733–1742. doi: 10.1016/j.molp.2023.09.010
Chen, L., Yang, J., Yu, J., Yao, Z., Sun, L., Shen, Y., et al. (2005). VFDB: a reference database for bacterial virulence factors. Nucleic Acids Res. 33, D325–D328. doi: 10.1093/nar/gki008
Clokie, R. J. (2009). Kropinski Bacteriophages: methods and protocols. Methods Mol. Biol. 501:307. doi: 10.1007/978-1-60327-164-6
Darling, A. C., Mau, B., Blattner, F. R., and Perna, N. T. (2004). Mauve: multiple alignment of conserved genomic sequence with rearrangements. Genome Res. 14, 1394–1403. doi: 10.1101/gr.2289704
Dedrick, R. M., Guerrero-Bustamante, C. A., Garlena, R. A., Russell, D. A., Ford, K., Harris, K., et al. (2019). Engineered bacteriophages for treatment of a patient with a disseminated drug-resistant Mycobacterium abscessus. Nat. Med. 25, 730–733. doi: 10.1038/s41591-019-0437-z
Destoumieux-Garzón, D., Duquesne, S., Peduzzi, J., Goulard, C., Desmadril, M., Letellier, L., et al. (2005). The iron–siderophore transporter FhuA is the receptor for the antimicrobial peptide microcin J25: role of the microcin Val11–Pro16 β-hairpin region in the recognition mechanism. Biochem. J. 389, 869–876. doi: 10.1042/BJ20042107
Ding, T., Sun, H., Pan, Q., Zhao, F., Zhang, Z., Ren, H., et al. (2020). Isolation and characterization of Vibrio parahaemolyticus bacteriophage vB_VpaS_PG07. Virus Res. 286:198080. doi: 10.1016/j.virusres.2020.198080
Dong, C., Hao, G. F., Hua, H. L., Liu, S., Labena, A. A., Chai, G., et al. (2018). Anti-CRISPRdb: a comprehensive online resource for anti-CRISPR proteins. Nucleic Acids Res. 46, D393–D398. doi: 10.1093/nar/gkx835
Eitzinger, S., Asif, A., Watters, K. E., Iavarone, A. T., Knott, G. J., Doudna, J. A., et al. (2020). Machine learning predicts new anti-CRISPR proteins. Nucleic Acids Res. 48, 4698–4708. doi: 10.1093/nar/gkaa219
Elmahdi, S., DaSilva, L. V., and Parveen, S. (2016). Antibiotic resistance of Vibrio parahaemolyticus and Vibrio vulnificus in various countries: a review. Food Microbiol. 57, 128–134. doi: 10.1016/j.fm.2016.02.008
Federhen, S. (2012). The NCBI taxonomy database. Nucleic Acids Res. 40, D136–D143. doi: 10.1093/nar/gkr1178
Feng, T., Leptihn, S., Dong, K., Loh, B., Zhang, Y., Stefan, M. I., et al. (2021). JD419, a Staphylococcus aureus phage with a unique morphology and broad host range. Front. Microbiol. 12:602902. doi: 10.3389/fmicb.2021.602902
German, G. J., and Misra, R. (2001). The TolC protein of Escherichia coli serves as a cell-surface receptor for the newly characterized TLS bacteriophage. J. Mol. Biol. 308, 579–585. doi: 10.1006/jmbi.2001.4578
Hagens, S., and Loessner, J. M. (2010). Bacteriophage for biocontrol of foodborne pathogens: calculations and considerations. Curr. Pharm. Biotechnol. 11, 58–68. doi: 10.2174/138920110790725429
Hill, C., Mills, S., and Ross, R. P. (2018). Phages & antibiotic resistance: are the most abundant entities on earth ready for a comeback? Future Microbiol. 13, 711–726. doi: 10.2217/fmb-2017-0261
Houte, S. V., Ekroth, A. K., Broniewski, J. M., Chabas, H., Ashby, B., Bondy-Denomy, J., et al. (2016). The diversity-generating benefits of a prokaryotic adaptive immune system. Nature 532, 385–388. doi: 10.1038/NATURE17436
Jamal, M., Bukhari, S. M., Andleeb, S., Ali, M., Raza, S., Nawaz, M. A., et al. (2019). Bacteriophages: an overview of the control strategies against multiple bacterial infections in different fields. J. Basic Microbiol. 59, 123–133. doi: 10.1002/jobm.201800412
Jault, P., Leclerc, T., Jennes, S., Pirnay, J. P., Que, Y. A., Resch, G., et al. (2019). Efficacy and tolerability of a cocktail of bacteriophages to treat burn wounds infected by Pseudomonas aeruginosa (PhagoBurn): a randomised, controlled, double-blind phase 1/2 trial. Lancet Infect. Dis. 19, 35–45. doi: 10.1016/S1473-3099(18)30482-1
Joensen, K. G., Scheutz, F., Lund, O., Hasman, H., Kaas, R. S., Nielsen, E. M., et al. (2014). Real-time whole-genome sequencing for routine typing, surveillance, and outbreak detection of verotoxigenic Escherichia coli. J. Clin. Microbiol. 52, 1501–1510. doi: 10.1128/JCM.03617-13
Kalatzis, P. G., Castillo, D., Katharios, P., and Middelboe, M. (2018). Bacteriophage interactions with marine pathogenic vibrios: implications for phage therapy. Antibiotics 7, 1–23. doi: 10.3390/antibiotics7010015
Kang, C. H., Shin, Y., Jang, S., Yu, H., Kim, S., An, S., et al. (2017). Characterization of Vibrio parahaemolyticus isolated from oysters in Korea: resistance to various antibiotics and prevalence of virulence genes. Mar Pollut Bull, 118 261–266. doi: 10.1016/j.marpolbul.2017.02.070
Karunasagar, I., Shivu, M. M., Girisha, S. K., Krohne, G., and Karunasagar, I. (2007). Biocontrol of pathogens in shrimp hatcheries using bacteriophages. Aquaculture 268, 288–292. doi: 10.1016/j.aquaculture.2007.04.049
Kim, H. W., Hong, Y. J., Jo, J. I., Ha, S. D., Kim, S. H., Lee, H. J., et al. (2017). Raw ready-to-eat seafood safety: microbiological quality of the various seafood species available in fishery, hyper and online markets. Lett. Appl. Microbiol. 64, 27–34. doi: 10.1111/lam.12688
Kim, Y. K., and McCarter, L. L. (2000). Analysis of the polar flagellar gene system of Vibrio parahaemolyticus. J. Bacteriol. 182, 3693–3704. doi: 10.1128/jb.182.13.3693-3704.2000
Kortright, K. E., Chan, B. K., Koff, J. L., and Turner, P. E. (2019). Phage therapy: a renewed approach to combat antibiotic-resistant bacteria. Cell Host Microbe 25, 219–232. doi: 10.1016/j.chom.2019.01.014
Le, S., Yao, X., Lu, S., Tan, Y., Rao, X., Li, M., et al. (2014). Chromosomal DNA deletion confers phage resistance to Pseudomonas aeruginosa. Sci. Rep. 4:4738. doi: 10.1038/srep04738
Lefkowitz, E. J., Dempsey, D. M., Hendrickson, R. C., Orton, R. J., Siddell, S. G., and Smith, D. B. (2018). Virus taxonomy: the database of the international committee on taxonomy of viruses (ICTV). Nucleic Acids Res. 46, D708–D717. doi: 10.1093/nar/gkx932
Lesmana, M., Subekti, D., Simanjuntak, C. H., Tjaniadi, P., Campbell, J. R., Oyofo, B. A., et al. (2001). Vibrio parahaemolyticus associated with cholera-like diarrhea among patients in North Jakarta, Indonesia. Diagn Microbiol Infect Dis, 39 71–75. doi: 10.1016/s0732-8893(00)00232-7
Li, H., and Durbin, R. (2009). Fast and accurate short read alignment with burrows-wheeler transform. Bioinformatics 25, 1754–1760. doi: 10.1093/bioinformatics/btp324
Li, C., Shi, T., Sun, Y., and Zhang, Y. (2022). A novel method to create efficient phage cocktails via use of phage-resistant bacteria. Appl. Environ. Microbiol. 88:e0232321. doi: 10.1128/aem.02323-21
Li, C., Wang, Z., Zhao, J., Wang, L., Xie, G., Huang, J., et al. (2021). A novel vibriophage vB_VcaS_HC containing lysogeny-related gene has strong lytic ability against pathogenic bacteria. Virol. Sin. 36, 281–290. doi: 10.1007/s12250-020-00271-w
Liang, X., Wang, Y., Hong, B., Li, Y., Ma, Y., and Wang, J. (2022). Isolation and characterization of a lytic Vibrio parahaemolyticus phage vB_VpaP_GHSM17 from sewage samples. Viruses 14:1601. doi: 10.3390/v14081601
Lu, T. K., and Collins, J. J. (2007). Dispersing biofilms with engineered enzymatic bacteriophage. Proc. Natl. Acad. Sci. 104, 11197–11202. doi: 10.1073/pnas.0704624104
Lu, T. K., and Collins, J. J. (2009). Engineered bacteriophage targeting gene networks as adjuvants for antibiotic therapy. Proc. Natl. Acad. Sci. 106, 4629–4634. doi: 10.1073/pnas.0800442106
Mangalea, M. R., and Duerkop, B. A. (2020). Fitness trade-offs resulting from bacteriophage resistance potentiate synergistic antibacterial strategies. Infect. Immun. 88, 10–1128. doi: 10.1128/iai.00926-19
Markwitz, P., Olszak, T., Gula, G., Kowalska, M., Arabski, M., and Drulis-Kawa, Z. (2021). Emerging phage resistance in Pseudomonas aeruginosa PAO1 is accompanied by an enhanced heterogeneity and reduced virulence. Viruses 13:1332. doi: 10.3390/v13071332
Martin, J., Schackwitz, W., and Lipzen, A. (2018). Genomic sequence variation analysis by resequencing. Fungal Genom 1775:18. doi: 10.1007/978-1-4939-7804-5_18
Matamp, N., and Bhat, S. G. (2019). Phage endolysins as potential antimicrobials against multidrug resistant vibrio alginolyticus and Vibrio parahaemolyticus: current status of research and challenges ahead. Microorganisms 7:84. doi: 10.3390/microorganisms7030084
McArthur, A. G., Waglechner, N., Nizam, F., Yan, A., Azad, M. A., Baylay, A. J., et al. (2013). The comprehensive antibiotic resistance database. Antimicrob. Agents Chemother. 57, 3348–3357. doi: 10.1128/AAC.00419-13
Moodley, A., Kot, W., Nälgård, S., Jakociune, D., Neve, H., Hansen, L. H., et al. (2019). Isolation and characterization of bacteriophages active against methicillin-resistant Staphylococcus pseudintermedius. Res. Vet. Sci. 122, 81–85. doi: 10.1016/j.rvsc.2018.11.008
Nikapitiya, C., Chandrarathna, H. P. S. U., Dananjaya, S. H. S., De Zoysa, M., and Lee, J. (2020). Isolation and characterization of phage (ETP-1) specific to multidrug resistant pathogenic Edwardsiella tarda and its in vivo biocontrol efficacy in zebrafish (Danio rerio). Biologicals 63, 14–23. doi: 10.1016/j.biologicals.2019.12.006
Nishimura, Y., Yoshida, T., Kuronishi, M., Uehara, H., Ogata, H., and Goto, S. (2017). ViPTree: the viral proteomic tree server. Bioinformatics 33, 2379–2380. doi: 10.1093/bioinformatics/btx157
North, O. I., and Brown, E. D. (2021). Phage–antibiotic combinations: a promising approach to constrain resistance evolution in bacteria. Ann. N. Y. Acad. Sci. 1496, 23–34. doi: 10.1111/nyas.14533
O’Leary, N. A., Wright, M. W., Brister, J. R., Ciufo, S., Haddad, D., McVeigh, R., et al. (2016). Reference sequence (RefSeq) database at NCBI: current status, taxonomic expansion, and functional annotation. Nucleic Acids Res. 44, D733–D745. doi: 10.1093/nar/gkv1189
Ottaviani, D., Leoni, F., Talevi, G., Masini, L., Santarelli, S., Rocchegiani, E., et al. (2013). Extensive investigation of antimicrobial resistance in Vibrio parahaemolyticus from shellfish and clinical sources, Italy. Int J Antimicrob Agents, 42 191–193. doi: 10.1016/j.ijantimicag.2013.05.003
Patro, R., Duggal, G., Love, M. I., Irizarry, R. A., and Kingsford, C. (2017). Salmon provides fast and bias-aware quantification of transcript expression. Nat. Methods 14, 417–419. doi: 10.1038/nmeth.4197
Plaza, N., Castillo, D., Pérez-Reytor, D., Higuera, G., García, K., and Bastías, R. (2018). Bacteriophages in the control of pathogenic vibrios. Electron. J. Biotechnol. 31, 24–33. doi: 10.1016/j.ejbt.2017.10.012
Pujato, S. A., Guglielmotti, D. M., Martinez-Garcia, M., Quiberoni, A., and Mojica, F. J. (2017). Leuconostoc mesenteroides and Leuconostoc pseudomesenteroides bacteriophages: genomics and cross-species host ranges. Int. J. Food Microbiol. 257, 128–137. doi: 10.1016/j.ijfoodmicro.2017.06.009
Pujato, S. A., Mercanti, D. J., Guglielmotti, D. M., Rousseau, G. M., Moineau, S., Reinheimer, J. A., et al. (2015). Phages of dairy Leuconostoc mesenteroides: genomics and factors influencing their adsorption. Int. J. Food Microbiol. 201, 58–65. doi: 10.1016/j.ijfoodmicro.2015.02.016
Qimron, U., Marintcheva, B., Tabor, S., and Richardson, C. C. (2006). Genomewide screens for Escherichia coli genes affecting growth of T7 bacteriophage. Proc. Natl. Acad. Sci. 103, 19039–19044. doi: 10.1073/pnas.0609428103
Raszl, S. M., Froelich, B. A., Vieira, C. R. W., Blackwood, A. D., and Noble, R. T. (2016). Vibrio parahaemolyticus and Vibrio vulnificus in South America: water, seafood and human infections. J. Appl. Microbiol. 121, 1201–1222. doi: 10.1111/jam.13246
Ren, H., Li, Z., Xu, Y., Wang, L., and Li, X. (2019). Protective effectiveness of feeding phage cocktails in controlling Vibrio parahaemolyticus infection of sea cucumber Apostichopus japonicus. Aquaculture 503, 322–329. doi: 10.1016/j.aquaculture.2019.01.006
Robinson, M. D., McCarthy, D. J., and Smyth, G. K. (2010). edgeR: a bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 26, 139–140. doi: 10.1093/bioinformatics/btp616
Rong, R., Lin, H., Wang, J., Khan, M. N., and Li, M. (2014). Reductions of Vibrio parahaemolyticus in oysters after bacteriophage application during depuration. Aquaculture 418-419, 171–176. doi: 10.1016/j.aquaculture.2013.09.028
Ruohan, W., Xianglilan, Z., Jianping, W., and Shuai Cheng, L. I. (2022). DeepHost: phage host prediction with convolutional neural network. Brief. Bioinform. 23:385. doi: 10.1093/bib/bbab385
Samson, J. E., Magadán, A. H., Sabri, M., and Moineau, S. (2013). Revenge of the phages: defeating bacterial defences. Nat. Rev. Microbiol. 11, 675–687. doi: 10.1038/nrmicro3096
Sar, N. E. C. H. E. M. I. A., McCARTER, L. I. N. D. A., Simon, M. E. L. V. I. N., and Silverman, M. I. C. H. A. E. L. (1990). Chemotactic control of the two flagellar systems of Vibrio parahaemolyticus. J. Bacteriol. 172, 334–341. doi: 10.1128/jb.172.1.334-341.1990
Sharma, S., Chatterjee, S., Datta, S., Prasad, R., Dubey, D., Prasad, R. K., et al. (2017). Bacteriophages and its applications: an overview. Folia Microbiol. 62, 17–55. doi: 10.1007/s12223-016-0471-x
Shen, Y., and Loessner, M. J. (2021). Beyond antibacterials–exploring bacteriophages as antivirulence agents. Curr. Opin. Biotechnol. 68, 166–173. doi: 10.1016/j.copbio.2020.11.004
Stalin, N., and Srinivasan, P. (2017). Efficacy of potential phage cocktails against Vibrio harveyi and closely related vibrio species isolated from shrimp aquaculture environment in the south east coast of India. Vet. Microbiol. 207, 83–96. doi: 10.1016/j.vetmic.2017.06.006
Steinegger, M., and Söding, J. (2017). MMseqs2 enables sensitive protein sequence searching for the analysis of massive data sets. Nat. Biotechnol. 35, 1026–1028. doi: 10.1038/nbt.3988
Sulakvelidze, A., Alavidze, Z., and Morris, J. G. Jr. (2001). Bacteriophage therapy. Antimicrob. Agents Chemother. 45, 649–659. doi: 10.1128/aac.45.3.649-659.2001
Tang, Y., Li, J., Wang, Y., Song, Z., Ying, H., Kong, L., et al. (2022). Campylobacter jejuni developed the resistance to bacteriophage CP39 by phase variable expression of 06875 encoding the CGPTase. Viruses 14:485. doi: 10.3390/v14030485
Wang, J., Zhao, F., Sun, H., Wang, Q., Zhang, C., Liu, W., et al. (2019). Isolation and characterization of the Staphylococcus aureus bacteriophage vB_SauS_SA2. AIMS Microbiol. doi: 10.3934/microbiol.2019.3.285
Wang, X., Loh, B., Gordillo Altamirano, F., Yu, Y., Hua, X., and Leptihn, S. (2021). Colistin-phage combinations decrease antibiotic resistance in Acinetobacter baumannii via changes in envelope architecture. Emerg Microbes Infect 10, 2205–2219. doi: 10.1080/22221751.2021.2002671
Wang, R., Ng, Y. K., Zhang, X., Wang, J., and Li, S. C. (2022). A graph representation of gapped patterns in phage sequences for graph convolutional network. bioRxiv [Preprint], 2022.
Westra, E. R., van Houte, S., Oyesiku-Blakemore, S., Makin, B., Broniewski, J. M., Best, A., et al. (2015). Parasite exposure drives selective evolution of constitutive versus inducible defense. Curr. Biol. 25, 1043–1049. doi: 10.1016/j.cub.2015.01.065
Williams, S. L., Jensen, R. V., Kuhn, D. D., and Stevens, A. M. (2017). Analyzing the metabolic capabilities of a Vibrio parahaemolyticus strain that causes early mortality syndrome in shrimp. Aquaculture 476, 44–48. doi: 10.1016/j.aquaculture.2017.03.030
Yang, Y., Shen, W., Zhong, Q., Chen, Q., He, X., Baker, J. L., et al. (2020). Development of a bacteriophage cocktail to constrain the emergence of phageresistant Pseudomonas aeruginosa. Front. Microbiol. 11:327. doi: 10.3389/fmicb.2020.00327
Yang, Z., Yin, S., Li, G., Wang, J., Huang, G., Jiang, B., et al. (2019). Global transcriptomic analysis of the interactions between phage φAbp1 and extensively drug-resistant acinetobacter baumannii. mSystems 4, 10–1128. doi: 10.1128/mSystems.00068-19
Zankari, E., Hasman, H., Cosentino, S., Vestergaard, M., Rasmussen, S., Lund, O., et al. (2012). Identification of acquired antimicrobial resistance genes. J. Antimicrob. Chemother. 67, 2640–2644. doi: 10.1093/jac/dks261
Zhang, M., Qian, J., Xu, X., Ahmed, T., Yang, Y., Yan, C., et al. (2022). Resistance of Xanthomonas oryzae pv. Oryzae to lytic phage X2 by spontaneous mutation of lipopolysaccharide synthesis-related glycosyltransferase. Viruses 14:1088. doi: 10.3390/v14051088
Keywords: vibriophage, bacteriophage therapy, anti-phage mutant, bacteriophage resistance, trade-off
Citation: Zeng X, Liang S, Dong J, Gao G, Hu Y and Sun Y (2024) The trade-off of Vibrio parahaemolyticus between bacteriophage resistance and growth competitiveness. Front. Microbiol. 15:1346251. doi: 10.3389/fmicb.2024.1346251
Received: 29 November 2023; Accepted: 10 April 2024;
Published: 11 June 2024.
Edited by:
Yi Gong, Nanchang University, ChinaReviewed by:
Zengmeng Wang, Chinese Academy of Sciences (CAS), ChinaCopyright © 2024 Zeng, Liang, Dong, Gao, Hu and Sun. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Yuechao Sun, MTE2MDcwMTBAemp1LmVkdS5jbg==
†These authors share first authorship
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.