
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Microbiol., 24 January 2024
Sec. Antimicrobials, Resistance and Chemotherapy
Volume 15 - 2024 | https://doi.org/10.3389/fmicb.2024.1336387
 Mulatu Gashaw1,2*
Mulatu Gashaw1,2* Esayas Kebede Gudina3
Esayas Kebede Gudina3 Solomon Ali4
Solomon Ali4 Liegl Gabriele5
Liegl Gabriele5 Thomas Seeholzer6
Thomas Seeholzer6 Bikila Alemu1
Bikila Alemu1 Guenter Froeschl2,7
Guenter Froeschl2,7 Arne Kroidl2,7,8
Arne Kroidl2,7,8 Andreas Wieser5,6,7,8
Andreas Wieser5,6,7,8Background: In resource-constrained settings, limited antibiotic options make treating carbapenem-resistant bacterial infections difficult for healthcare providers. This study aimed to assess carbapenemase expression in Gram-negative bacteria isolated from clinical samples in Jimma, Ethiopia.
Methods: A cross-sectional study was conducted to assess carbapenemase expression in Gram-negative bacteria isolated from patients attending Jimma Medical Center. Totally, 846 Gram-negative bacteria were isolated and identified using matrix-assisted laser desorption ionization-time of flight mass spectrometry (MALDI-TOF MS). Phenotypic antibiotic resistance patterns were determined using the Kirby-Bauer disk diffusion method and Etest strips. Extended-spectrum β-lactamase phenotype was determined using MAST disks, and carbapenemases were characterized using multiplex polymerase chain reactions (PCR).
Results: Among the isolates, 19% (157/846) showed phenotypic resistance to carbapenem antibiotics. PCR analysis revealed that at least one carbapenemase gene was detected in 69% (107/155) of these strains. The most frequently detected acquired genes were blaNDM in 35% (37/107), blaVIM in 24% (26/107), and blaKPC42 in 13% (14/107) of the isolates. Coexistence of two or more acquired genes was observed in 31% (33/107) of the isolates. The most common coexisting acquired genes were blaNDM + blaOXA-23, detected in 24% (8/33) of these isolates. No carbapenemase-encoding genes could be detected in 31% (48/155) of carbapenem-resistant isolates, with P. aeruginosa accounting for 85% (41/48) thereof.
Conclusion: This study revealed high and incremental rates of carbapenem-resistant bacteria in clinical samples with various carbapenemase-encoding genes. This imposes a severe challenge to effective patient care in the context of already limited treatment options against Gram-negative bacterial infections in resource-constrained settings.
Gram-negative bacteria (GNB), such as Escherichia coli, Klebsiella pneumoniae, Acinetobacter baumannii, and Pseudomonas aeruginosa, are common culprits in healthcare-associated infections (Sikora and Zahra, 2020). Carbapenem resistance is increasing at alarming rates in these organisms (Beshah et al., 2023). The resistance can arise from various mechanisms, including the production of carbapenemase enzymes, decreased permeability of the bacterial cell wall, increased efflux pump activity, alterations in outer membrane porins, and target site mutations that reduce affinity to carbapenems (Aurilio et al., 2022). These mechanisms can act individually or in combination, leading to the development of multidrug-resistant strains that pose significant challenges in treating infections caused by these bacteria (Das, 2023). GNB have the ability to acquire and express a variety of carbapenemase genes (Dwomoh et al., 2022; Tenover et al., 2022; Tilahun et al., 2022). These genes can spread within or between different bacterial species through horizontal transfer of plasmids, conjugative transposons, or integrons (Hammoudi Halat and Ayoub Moubareck, 2020). As a result, carbapenem resistance in GNB is a major public-health concern worldwide. The most common carbapenemases identified in GNB include oxacillinases (OXA), Klebsiella pneumoniae carbapenemase (KPCs), and metallo-beta-lactamases (MBLs), including New Delhi metallo-β-lactamase (NDM) and Verona integron-encoded metallo-beta-lactamase imipenemase (VIM) (Rabaan et al., 2022). These enzymes can break down carbapenem antibiotics, and develop resistance not only to carbapenems, but also to many other beta-lactam antibiotics, such as penicillins, cephalosporins, and monobactams (Jean et al., 2022).
Infections with these pathogens are associated with high rates of mortality and morbidity since treatment options are limited to a few last-resort antibiotics that often come with many side effects (Caston et al., 2022). Furthermore, infections with carbapenem-resistant GNBs increase healthcare cost and the length of hospital stays (Van Duin, 2017). Such infections are major concerns for critically ill patients, immunocompromised individuals, and those with comorbidities (Aleidan et al., 2021; Di Carlo et al., 2021). In resource-constrained countries, including Ethiopia, the public health impact is even worse due to the lack of reserve treatment options (Alemayehu et al., 2023; Beshah et al., 2023).
Rapid and reliable detection of carbapenem-resistant GNB is critical for appropriate laboratory-guided patient management, for surveillance, and for applying effective evidence-based infection prevention and control practices (Nordmann and Poirel, 2019; Shanmugakani et al., 2020). A combination of phenotypic detection and genotypic confirmation of carbapenemase-expressing genes by polymerase chain reaction (PCR) is recommended (Rabaan et al., 2022).
However, due to lack of technical expertise, specialized equipment, and reagents, detecting and tracking the molecular epidemiology of carbapenem-resistant bacterial isolates is difficult in low-income countries (Nordmann and Poirel, 2019; Shanmugakani et al., 2020). As a result, data on the burden of infections with carbapenem-resistant bacterial species and associated outcomes is scarce in Sub-Saharan African countries, including Ethiopia (Stewardson et al., 2019). Therefore, this study aimed to determine the extent of carbapenemases among GNBs obtained from clinical samples using both phenotypic and genotypic techniques.
A cross-sectional study was conducted to detect the carbapenemase genes in carbapenem-resistant GNB obtained from patients treated at Jimma Medical Center (JMC). JMC is an 800-bed teaching hospital in southwest Ethiopia with a catchment population of over 20 million. All patients from whom samples were sent for culture and antibiotic susceptibility test as part of routine clinical care were recruited prospectively for the study.
Clinical samples (blood, cerebrospinal fluid [CSF], wound swabs, ascitic fluid, pleural fluid, abscess, peritoneal fluid, and synovial fluid) were collected aseptically by the clinicians, nurses or laboratory professionals. Other clinical samples such as urine, stool, and sputum were collected by the patients themselves after proper instruction was provided. Samples were then transported within 1 h after collection to the JMC microbiology laboratory for analysis.
All clinical specimens, except for blood, were inoculated on 5% Colombia Sheep Blood, Chocolate, and MacConkey agars and incubated aerobically at 35–37°C for 18–22 h. Blood samples were collected and added to BD BACTEC bottles (Becton Dickinson, Sparks, MD, USA) and then incubated for 5 days at 35–37°C in the BD BACTEC™ FX40 (Becton Dickinson, Sparks, MD, USA) automated culture machine. If growth was observed, it was sub-cultured on 5% Colombia Sheep Blood, Chocolate, and MacConkey agars in similar environmental conditions for further analysis. Subsequently, all positive pure cultures were tested for antimicrobial susceptibility. Isolates were picked off the plates and kept at −80°C in storage media containing skimmed milk, tryptone soya, glucose, glycerol, and distilled water until they were transported to Max von Pettenkofer Institute, Hospital Hygiene, and Medical Microbiology Laboratory in Munich, Germany. There, the isolates were re-identified using matrix-assisted laser desorption ionization-time of flight mass spectrometry (MALDI-TOF MS, Bruker, Germany).
Antimicrobial susceptibility testing was carried out according to the Kirby-Bauer disk diffusion technique using 16 antibiotics (Bio-Rad, France) (Supplementary Table S1). Reading of the results was done using the ADAGIO 93400 automated system (Bio-Rad, France) and interpreted as resistant (R), intermediate (I), and susceptible (S) based on the respective breakpoints for specific organisms in the European Committee on Antimicrobial Susceptibility Testing (EUCAST, 2021).
ESBL phenotype identification was carried out using MAST disks (Mast Group, UK) on all isolates (n = 648) that were non-susceptible to beta-lactam antibiotics such as cefotaxime, cefoxitin, cefepime, piperacillin/tazobactam, or meropenem in the Kirby-Bauer disk diffusion technique. The results were interpreted using the Mast Disks Combi D68C ESBL/AmpC calculator spreadsheet (Mast Group, UK) and reported as negative, positive, or inconclusive for ESBL or/and AmpC. Isolates with reports of “Further work required” or “Equivocal” or that grew toward all disks with below 9 mm of inhibition zone were grouped together as “inconclusive.”
All bacterial isolates that were intermediate or resistant to meropenem in the Kirby-Bauer disk diffusion method were tested with ertapenem Etest strips for Enterobacterales and meropenem Etest strips (both BioMérieux Deutschland GmbH) for non-lactose fermenting Gram-negative rods. According to EUCAST’s breakpoints for meropenem, an isolate was considered intermediate if the MIC value was between 2 and 8 mg/L and resistant when the MIC was greater than 8 mg/L. Bacterial isolates with MIC values greater than 0.5 mg/L were interpreted as resistant to ertapenem. Otherwise, all the remaining strains were considered susceptible to meropenem or ertapenem, respectively (EUCAST, 2021).
The DNA was extracted from 3 to 5 fresh pure colonies of the respective bacterial isolate and extracted using High Pure PCR template preparation kit (Roche, Germany) following the manufacturer’s instruction. The quantity, purity, and concentration of the extracted DNA were measured by Nano-Drop ND-100 (Thermo Fisher Scientific, Wilmington, USA). Excluding the intrinsic carbapenem-resistant S. maltophilia, all the remaining isolates (n = 155) that were resistant to carbapenem antibiotics and/or showed inconclusive results in ESBL phenotypes by Mast disks (Mast Group, UK) were characterized by multiplex PCR to detect the carbapenemase encoding genes using specific primers and probes (Supplementary Table S2) used in previous studies (Kruttgen et al., 2011; Huang et al., 2012) and kindly provided by the molecular diagnostics of the Max von Pettenkofer Institute by Schubert S. and Gross B. Reference strains carrying blaOXA-48 (K. pneumoniae ATCC-BAA-2524), blaKPC (E. coli ATCC-1101362), and blaNDM (K. pneumoniae ATCC-BAA-2146) were used as positive controls.
The data was entered and analyzed using Microsoft Office 2016 excel sheets and GraphPad Prism version 8.4.3. Tables and graphs were used to display the frequency of phenotypic antibiotic resistance patterns and the distribution of carbapenemase encoding genes among phenotypically carbapenem-resistant bacterial pathogens.
The study was carried out with the approval of both Jimma University Institute of Health Institutional Review Board, Ethiopia (protocol numbers: IHRPGO/495/2018 & IHRPGO/1087/21) and the Ethics Committee of the Medical Faculty of Ludwig-Maximilians-Universität of Munich, Germany (Opinion No: 21–0157). Written informed consent was obtained from study participants and parents or guardians in case of neonates, infants, and children before enrollment in the study. All the information was kept confidential and recorded anonymously. The culture results were sent back timely to the treating physicians to provide the recommended medical attention to the respective patients.
A total of 1,794 clinical specimens were processed during the study period. Of these, 953 specimens collected from 894 patients were positive resulting in the isolation of 1,010 bacterial strains. The majority of isolates (846/1,010) were GNB, which were the only one included in the current study. A single bacterial pathogen was identified in 896 specimens, while two and three isolates were detected in the remaining 55 and 2 clinical samples, respectively. Overall, more than 30 different species of GNB were identified. The most commonly identified bacterial pathogen was E. coli accounting for 27% (231/846) of the GNB isolates, followed by K. pneumoniae 19% (163/846), A. baumannii complex 15% (126/846), and E. cloacae complex 13% (108/846) (Supplementary Table S3). More than 75% (643/846) of the GNB were isolated from admitted patients. Of these, 32% (206/643) were from the neonatal intensive care unit (NICU), 27% (184/643) from surgical, 27% (173/643) from pediatric, and 12% (80/643) from medical wards.
In Kirby-Bauer disk diffusion technique, a remarkable prevalence of non-susceptibility was observed against cefuroxime, ampicillin, and piperacillin, with rates reaching 100% (846/846), 92% (763/827), and 91% (655/720) respectively. Among the tested antibiotics, meropenem and amikacin showed the least resistance, 18% (149/846) and 12% (97/846), respectively. The isolates also exhibited a high rate of resistance to trimethoprim-sulfamethoxazole (60%), aminoglycosides (11–57.4%), and fluoroquinolones (55.3–61.1%) (Figure 1).
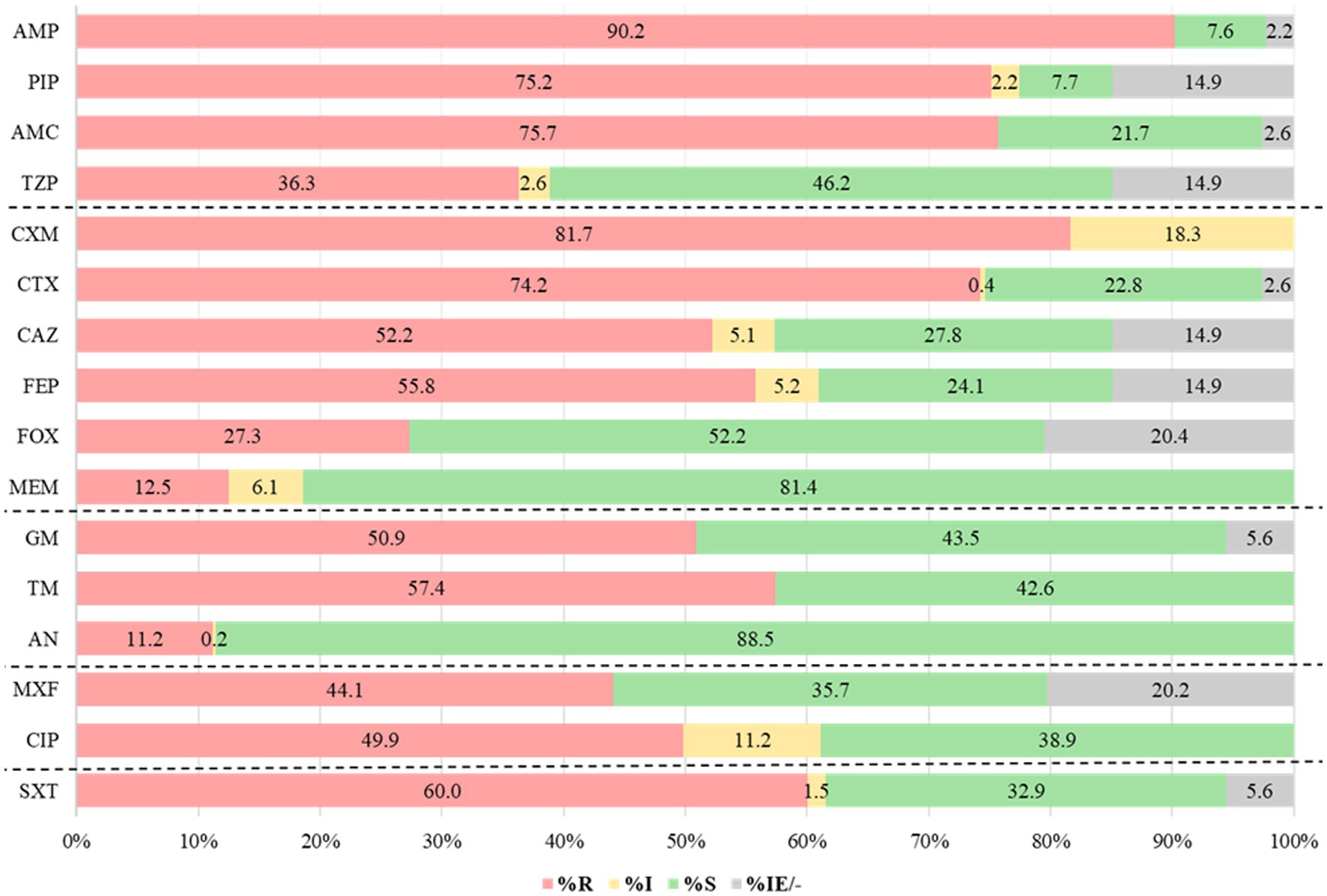
Figure 1. Antibiotic resistance patterns for Gram-negative bacteria (n = 846). AMP, ampicillin; PIP, piperacillin; AMC, amoxicillin-clavulanic acid; TZP, piperacillin-tazobactam; CXM, cefuroxime; CTX, cefotaxime; CAZ, ceftazidime; FEP, cefepime; FOX, cefoxitin; MEM, meropenem; GM, gentamicin; TM, tobramycin; AN, amikacin; MXF, moxifloxacin; CIP, ciprofloxacin; SXT, Trimethoprim- sulfamethoxazole; R, resistant; I, intermediate; S, susceptible, IE: insufficient evidence; and “–” No breakpoints.
All 648 bacterial isolates that were non-susceptible (tested intermediate or resistant) to one of the β-lactam antibiotics were further analyzed for ESBL phenotypes using Mast disks (MAST group UK). The analysis revealed that 66% (425/648) of the isolates produced extended-spectrum beta-lactamases (ESBL), 7% (47/648) had both ESBL and AmpC phenotypes, and 3% (19/648) showed only an AmpC phenotype (Figure 2). The remaining 24% (157/648) of the isolates showed inconclusive results when read with Mast disks combi D68C ESBL/AmpC calculator spreadsheets (Mast group, UK).
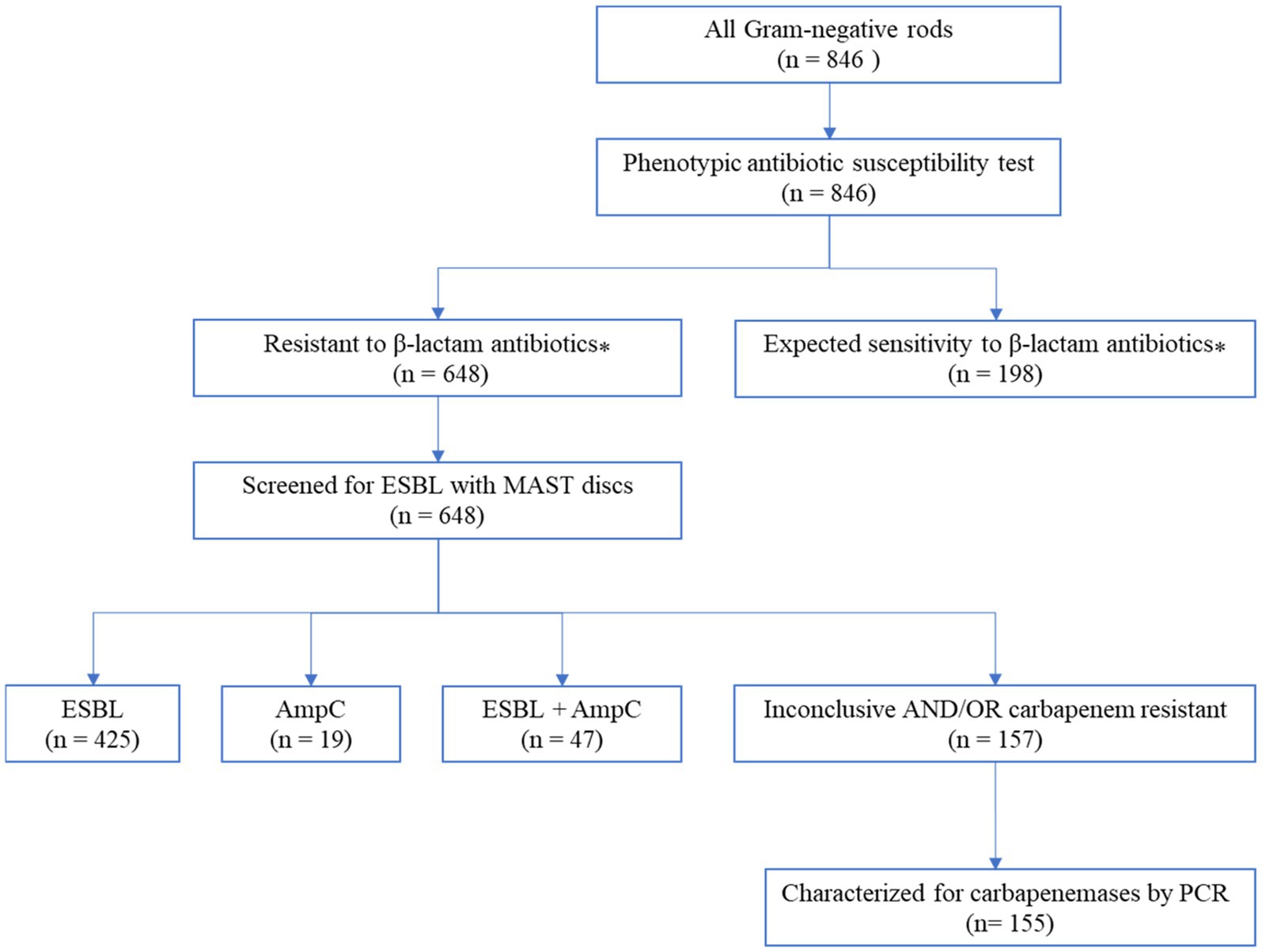
Figure 2. Flow diagram of the laboratory analysis to detect carbapenem-resistant Gram-negative bacteria. ⁎The antibiotic susceptibility test result to selected beta-lactam antibiotics such as cefotaxime, cefoxitin, cefepime, piperacillin/tazobactam, or meropenem; intrinsic resistances according to EUCAST are considered as expected; values with insufficient evidence according to EUCAST were not taken into account (EUCAST, 2021).
More than 75% (491/648) of the isolates that showed resistance to β-lactam antibiotics in the disk diffusion technique were confirmed as ESBL and/or AmpC phenotypes by Mast disks (Mast group, UK). As shown in Table 1, all Citrobacter species, K. oxytoca, Proteus species, S. marcescens, M. morganii, C. sakazakii, L. adecarboxylata, M. odoratimimus, and P. stuartii were confirmed as ESBL producers. Furthermore, the prevalence of ESBL production was observed in 93% (127/137) of K. pneumoniae, 94% (134/142) of E. coli, and 97% (98/101) of Enterobacter isolates. The remaining 24% (157/648) of the isolates showed inconclusive results, primarily A. baumannii complex, and P. aeruginosa which accounted for 71% (87/122) and 98% (42/43) of the respective isolates as shown in Table 1.
The minimum inhibitory concentrations (MIC) of carbapenem antibiotics, specifically ertapenem for Enterobacterales and meropenem for non-lactose fermenting GNB, was determined using Etest strips. This was done for all isolates (n = 155) that were tested carbapenem-resistant in the Kirby-Bauer disk diffusion method and/or showed inconclusive results in the Mast disk analysis. Accordingly, 79% (105/133) of non-lactose fermenting isolates and 100% (24/24) of the lactose fermenting isolates showed intermediate or resistant phenotypes against meropenem or ertapenem Etest strip, respectively (Figure 3).
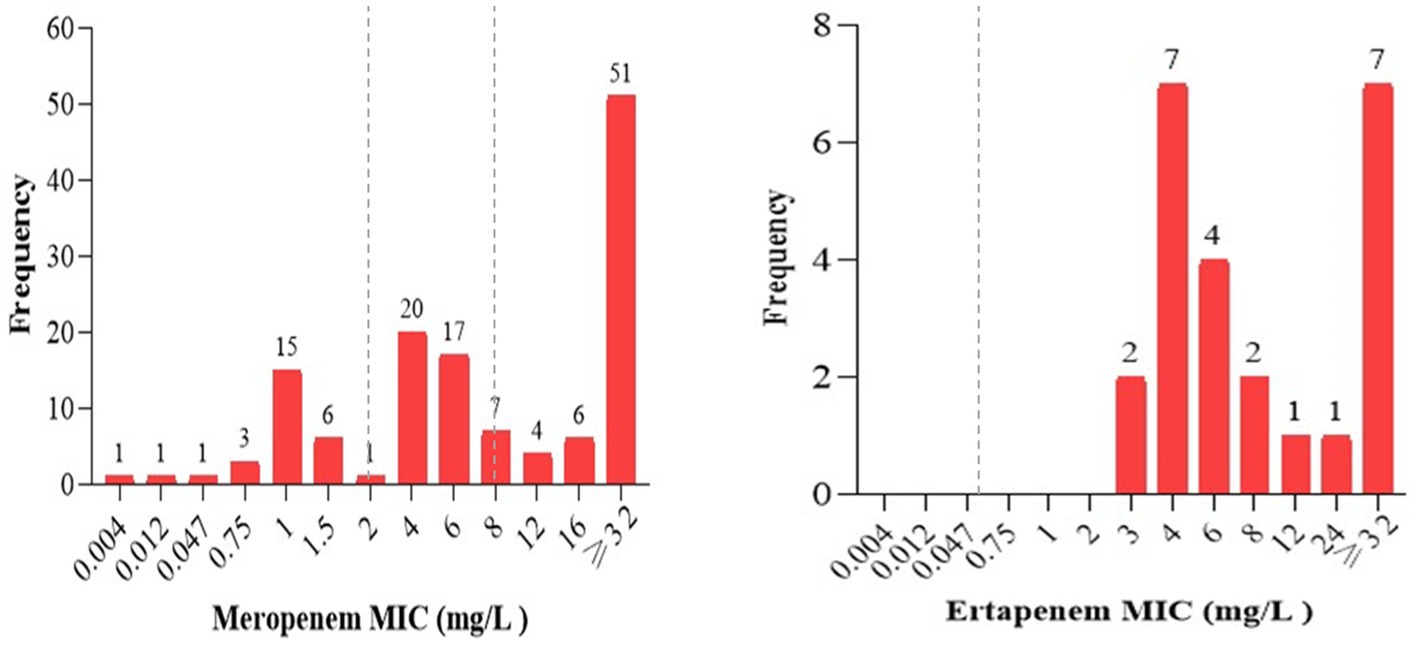
Figure 3. The frequency of carbapenem minimum inhibitory concentrations of all strains tested resistant in Kirby-Bauer disk diffusion or having inconclusive results in the Mast Disk assay. The MICs of meropenem ranging from (0–2 mg/L), (2–8 mg/L), and > 8 mg/L were interpreted as sensitive, intermediate, and resistant; ertapenem MIC values ≤0.5 mg/L and > 0.5 mg/L were interpreted as sensitive and resistant, respectively, as indicated in the broken lines according to EUCAST breakpoints (EUCAST, 2021).
The PCR analysis revealed that 69% (107/155) of the carbapenem non-susceptible isolates carried at least one carbapenemase-encoding gene, including both inherent and acquired genes. Among the acquired carbapenemase genes, the most frequently identified gene was blaNDM, constituting 21% (37/179) of the total detected genes. This was followed by blaVIM and blaKPC42, accounting for 15% (26/179), and 8% (14/179) respectively (Figure 4A). Regarding the distribution of carbapenemase-encoding genes, blaNDM was detected in various strains including A. baumannii (24), E. coli (6), K. pneumoniae (3), K. variicola (1), P. aeruginosa (1), P. mendocina (1) and A. haemolyticus (1). On the other hand, due to its intrinsic presence in A. baumannii, the blaOXA-51-like gene was exclusively found in A. baumannii strains (79) (Figure 4B). Conversely, no carbapenemase-encoding genes could be detected in 31% (48/155) of carbapenem-resistant isolates. P. aeruginosa was the most common, accounting for 85% (41/48) of them (Figure 4B).
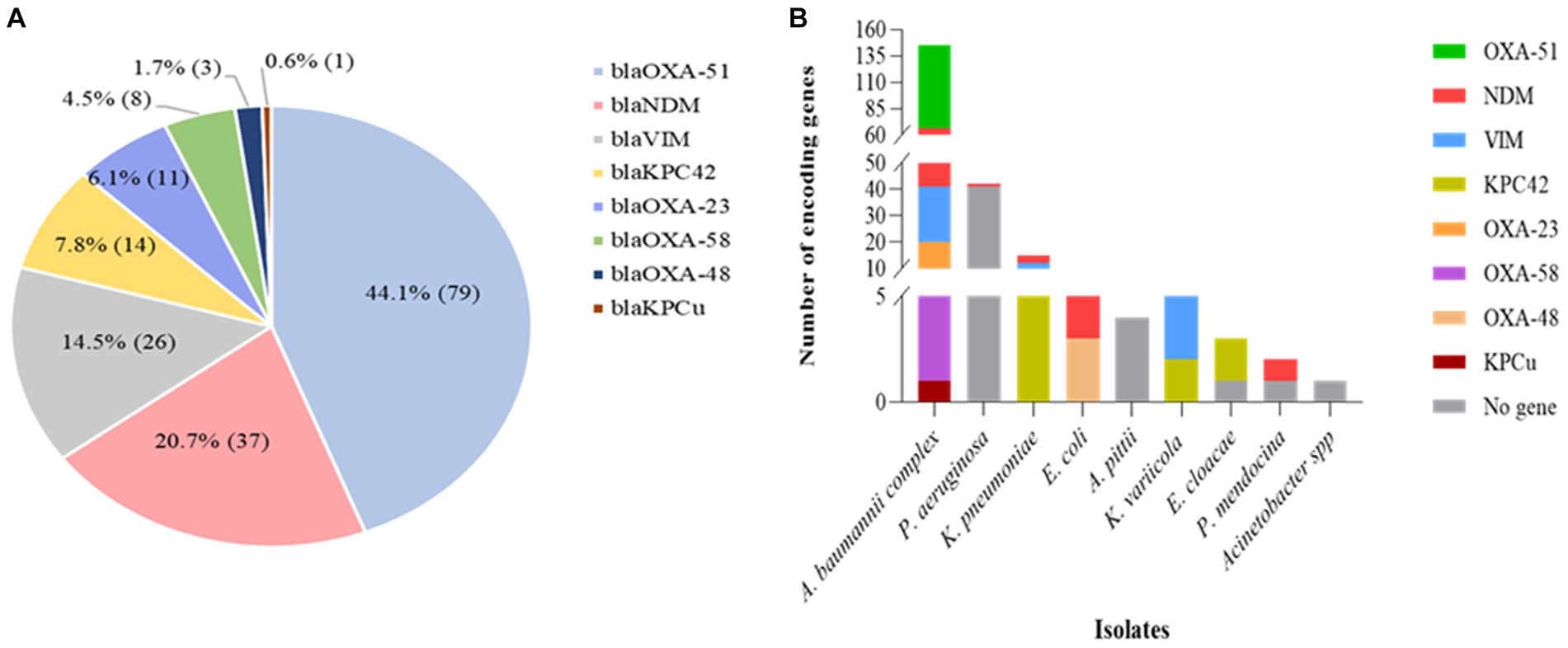
Figure 4. Distribution of carbapenemase encoding genes in various Gram-negative bacterial isolates with phenotypic resistance against carbapenems, as determined by PCR analysis. (A) The relative proportion of carbapenemase encoding genes (n = 179) as indicated in the pie chart. (B) The distribution of carbapenemase determinants in carbapenem-resistant isolates (n = 155). The PCR analysis revealed the presence of several types of carbapenemase determinants in many of the bacterial species. As a result, more than one carbapenemase determinant or mechanism of resistance was identified in 49 of the isolates.
Co-harboring of two or more acquired genes was observed in 31% (33/107) of the isolates, with A. baumannii being the predominant strain, accounting for 70% (23/33) of those isolates. Multiple gene coexistence was also detected in A. haemolyticus (1), E. coli (1), K. pneumoniae (5), and K. variicola (3) strains. The most common acquired coexisting genes were blaNDM + blaOXA-23, observed in 24% (8/33) of the isolates (Table 2).
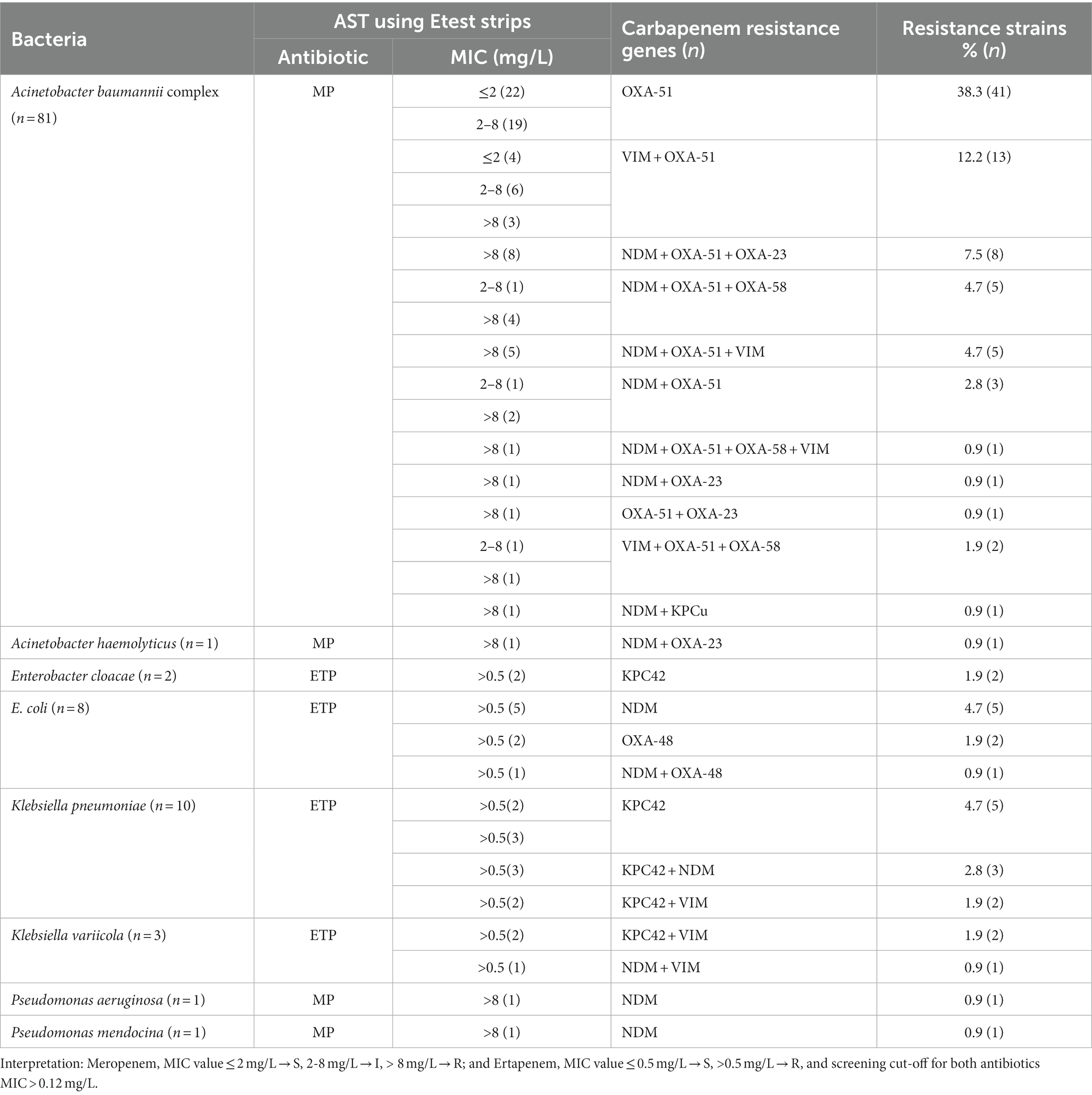
Table 2. Frequency and distribution of carbapenemase-coding genes among Gram-negative bacteria (n = 107).
Our study revealed high proportions of ESBL and carbapenemase producing Gram-negative pathogens, primarily E. coli, K. pneumoniae, E. cloacae complex, A. baumannii complex, and P. aeruginosa in comparison to previous studies conducted worldwide (Chen et al., 2021; Jean et al., 2022). In most low-income countries, carbapenems are considered the last-resort antibiotics, as other antibiotics like colistin and polymyxin B are not available. Carbapenem-resistant infections are increasing at alarming rates worldwide (Hammoudi Halat and Ayoub Moubareck, 2020), and this trend is even worse in low-income countries (Stewardson et al., 2019) including Ethiopia (Sewunet et al., 2022; Tilahun et al., 2022). Inadequate infection prevention and control measures, lack of proper hand hygiene, insufficient isolation precautions, and limited regular AMR surveillance (Ali et al., 2018; Eshetu et al., 2019) contribute to this problem.
More than three-fourths (76.6%, 648) of the isolates were tested resistant to one or more beta-lactam antibiotics such as cefotaxime, cefoxitin, cefepime, piperacillin/tazobactam, or meropenem. Among all isolates, 59% (499/846) showed ESBL phenotypes, and 19% (157/846) were carbapenem-resistant phenotypically. Our findings indicate an increase in ESBL phenotypes in Jimma compared to previous reports of 50–51% in 2016 (Gashaw et al., 2018; Zeynudin et al., 2018). The observed high prevalence of ESBL-producing isolates could be explained by the high rate of nosocomial infections among hospitalized patients (Ali et al., 2018). The lack of proper infection prevention and control practices (Sastry et al., 2017; Maki and Zervos, 2021), along with horizontal gene transfer (Da Silva and Domingues, 2016) and the spread of resistant genes within local microbial populations may contribute to the high rate of beta-lactam resistance. Additionally, the high rates of Acinetobacter and Pseudomonas species which are intrinsically resistant to many beta-lactam antibiotics could explain this increase.
In previous studies conducted in Ethiopia, the rate of carbapenem resistance among Gram-negative rods was low ranging 1.7–15.1% (Misha et al., 2021; Tekele et al., 2021; Seman et al., 2022; Tilahun et al., 2022; Alemayehu et al., 2023). However, our findings showed an increase in resistance to carbapenems (18.6%). Our current study revealed high rates of phenotypic carbapenem resistance among Acinetobacter (71.3%) and Pseudomonas species (97.7%, 42/43), compared to a previous study conducted in the same area in 2016, where resistance rates were 56.4 and 7.3% for Acinetobacter and Pseudomonas isolates, respectively (Sewunet et al., 2022). This increase in resistance may be attributed to the increasing use of carbapenems at the hospital and poor infection control measures. Infections caused by such resistant isolates greatly limit the treatment options. Therefore, addressing the rising threat of carbapenemase-producing Acinetobacter and Pseudomonas species requires a multifaceted approach including the implementation of effective infection prevention and control measures, promotion of antimicrobial stewardship programs to ensure appropriate antibiotics use, and development of new antibiotics effective against these resistant strains (Mulani et al., 2019; Jean et al., 2022).
Additionally, it is important to identify the determinants of carbapenem resistance in bacterial pathogens. While many isolates express a carbapenemase, others may develop resistance due to other mechanisms such as porin loss (Atrissi et al., 2021). In our study, we investigated both the phenotypic resistance and the presence of carbapenemase genes. In A. baumannii, we found the presence of intrinsically encoded blaOXA-51-like genes, as well as the acquired blaNDM and blaKPC encoding genes. We did not investigate any regulatory phenotypes involved in increased expression of blaOXA-51-like enzymes, so we can only speculate on their role in the phenotypically resistant isolates, possibly in combination with permeability issues or efflux pumps. Nevertheless, in the case of P. aeruginosa, the observed carbapenem resistance could not be linked to the carbapenemases tested in the study. Instead, it is more likely that the resistance is due to porin loss as suggested by a previous study (Atrissi et al., 2021).
Similar to previous studies conducted in Egypt (Abouelfetouh et al., 2019) and South Africa (Anane et al., 2020), PCR analysis revealed that all A. baumannii isolates carried the blaOXA-51-like genes. In 13.6% (11/82) and 9.9% (8/82) of Acinetobacter strains, blaOXA-23-like and blaOXA-58-like genes were detected, respectively. The prevalence of blaOXA-51-like gene in our study was higher than reported in a previous study in Jimma (63.1%) (Sewunet et al., 2022). This can be explained by the higher proportion of A. baumannii strains that currently dominate nosocomial infections as compared to previous studies. All 79 A. baumannii isolates carried the intrinsic blaOXA-51-like gene, but 22 of them were phenotypically susceptible to meropenem according to the MIC values. This can be explained by the intrinsic low efficiency of blaOXA-51, which is not easily detected by phenotypic methods, as reported in previous studies (Hu et al., 2007; Nigro and Hall, 2018).
The New Delhi metallo-beta-lactamase (NDM), classified as group B in the Ambler classification, is an enzyme that can break down a wide range of beta-lactam antibiotics, including carbapenems. It was first reported in Ethiopia in 2017 in A. baumannii strains (Pritsch et al., 2017). Back then, it could only be detected in some isolates of Acinetobacter baumannii, with no evidence of its presence in other isolates. However, NDM is no longer limited to Acinetobacter species and has been found in various GNB, such as K. pneumoniae, K. variicola, E. coli, P. aeruginosa, and P. mendocina (Legese et al., 2022; Seman et al., 2022; Sewunet et al., 2022; Tufa et al., 2022). This enzyme is particularly concerning because it can rapidly spread between different bacterial species through horizontal gene transfer, leading to the emergence of extensively drug-resistant infections (Da Silva and Domingues, 2016). It is also frequently associated with other antibiotic resistance determinants and may be transferred alongside them. Our study detected the blaNDM gene in 34.6% of carbapenemase positive isolates, which is comparable to a study conducted in Kenya where 30% of the isolates carried the NDM gene (Villinger et al., 2022). The other commonly acquired carbapenemase gene identified in our study was blaKPC42, which was found in all carbapenem-resistant K. pneumoniae (10) and two of the three carbapenem resistant K. variicola strains. It has not been previously reported in Ethiopia but has been frequently reported in other parts of the world (Miranda et al., 2018).
Most of the A. baumannii isolates in our study harbored two (19) or three (21) carbapenemase genes. Moreover, five K. pneumoniae and three K. variicola isolates carried two carbapenemase genes. In total, 50 of the isolates carried multiple carbapenemase genes (blaOXA-51, blaNDM, blaVIM, blaOXA-23, blaOXA-58, blaKPC42, blaOXA-48, and blaKPCu), which is consistent with other studies conducted in Ethiopia where multiple carbapenemase determinants have been reported (Legese et al., 2022; Sewunet et al., 2022). In general, the prevalence of NDM in Acinetobacter and other GNB has been increasing globally in recent years (Sands et al., 2021; Awoke et al., 2022; Seman et al., 2022).
There are certain limitations to our study that should be considered when interpreting the results. First, the study was conducted in a single tertiary level facility, which may not fully represent the diversity of antimicrobial resistance patterns in the broader community or other healthcare settings in the region. Second, the PCR analysis was performed on isolates that were phenotypically resistant to carbapenems in the disk diffusion method and/or showed inconclusive results in the Mast disk analysis. This approach may have excluded some isolates with reduced carbapenem susceptibility that were not detected by the phenotypic resistance, potentially underestimating the true burden of carbapenem resistance in the study area. Third, we did not investigate if the resistance against carbapenems observed in some A. baumannii strains was due to overexpression of OXA-51 or other metabolic or regulatory changes such as loss of permeability or increased efflux.
Our study demonstrated a high rate of carbapenem resistance among GNB, primarily in Acinetobacter species. The majority of this resistance was attributed to carbapenemases, probably along with other factors. Consequently, treating infections caused by these pathogens in this region may prove challenging due to limited treatment options. To address this issue, it is essential to revise treatment strategies in order to effectively manage infections caused by resistant strains. Moreover, it is imperative to uphold diligent surveillance, apply optimal infection prevention and control strategies, and promote antimicrobial stewardship practices to effectively manage and combat the dissemination of carbapenem-resistant bacteria.
The original contributions presented in the study are included in the article/Supplementary material, further inquiries can be directed to the corresponding author/s.
This study was approved by the Institutional Review Board (IRB) of Jimma University Institute of Health, Ethiopia and The Ethics Committee at the Medical Faculty of LMU Munich, Germany. Written informed consent was also obtained from patients, parents, or guardians prior to recruitment in the study.
MG: Conceptualization, Data curation, Formal analysis, Investigation, Methodology, Project administration, Supervision, Writing – original draft, Writing – review & editing. EG: Conceptualization, Data curation, Funding acquisition, Supervision, Writing – review & editing. SA: Conceptualization, Supervision, Writing – review & editing. LG: Data curation, Writing – review & editing. TS: Writing – review & editing. BA: Data curation, Writing – review & editing. GF: Conceptualization, Funding acquisition, Supervision, Writing – review & editing. AK: Conceptualization, Funding acquisition, Supervision, Writing – review & editing. AW: Conceptualization, Data curation, Funding acquisition, Investigation, Methodology, Supervision, Writing – original draft, Writing – review & editing.
The author(s) declare financial support was received for the research, authorship, and/or publication of this article. The study was funded by the German Academic Exchange Service (DAAD) through its Exceed program and by the University & Hospital Partnerships in Africa initiative of the German Gesellschaft für International Zusammenarbeit (GIZ) (grant number 81248133) in cooperation with the German Ministry of Economic Cooperation and Development (BMZ). The funder did not have any role in the design of the study and collection, analysis, and interpretation of data or in the development of the manuscript.
We are grateful to acknowledge the German Academic Exchange Service (DAAD) particularly its Exceed program and the GIZ University & Hospital Partnerships in Africa initiative for funding this work. We would also like to thank all the data collectors who are working at Jimma Medical Center, Jimma, Ethiopia and the Max von Pettenkofer Institute, Munich, Germany.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2024.1336387/full#supplementary-material
Abouelfetouh, A., Torky, A. S., and Aboulmagd, E. (2019). Phenotypic and genotypic characterization of Carbapenem-resistant Acinetobacter baumannii isolates from Egypt. Antimicro. Resist. Infect. Control 8, 1–9. doi: 10.1186/s13756-019-0611-6
Aleidan, F. A., Alkhelaifi, H., Alsenaid, A., Alromaizan, H., Alsalham, F., Almutairi, A., et al. (2021). Incidence and risk factors of carbapenem-resistant Enterobacteriaceae infection in intensive care units: a matched case–control study. Expert Rev. Anti-Infect. Ther. 19, 393–398. doi: 10.1080/14787210.2020.1822736
Alemayehu, E., Fiseha, T., Gedefie, A., Alemayehu Tesfaye, N., Ebrahim, H., Ebrahim, E., et al. (2023). Prevalence of Carbapenemase-producing Enterobacteriaceae from human clinical samples in Ethiopia: a systematic review and Meta-analysis. BMC Infect. Dis. 23:277. doi: 10.1186/s12879-023-08237-5
Ali, S., Birhane, M., Bekele, S., Kibru, G., Teshager, L., Yilma, Y., et al. (2018). Healthcare associated infection and its risk factors among patients admitted to a tertiary hospital in Ethiopia: longitudinal study. Antimicrob. Resist. Infect. Control 7, 1–9. doi: 10.1186/s13756-017-0298-5
Anane, Y. A., Apalata, T., Vasaikar, S., Okuthe, G. E., and Songca, S. (2020). Molecular detection of carbapenemase-encoding genes in multidrug-resistant Acinetobacter baumannii clinical isolates in South Africa. Int. J. Microb. 2020, 1–10. doi: 10.1155/2020/7380740
Atrissi, J., Milan, A., Bressan, R., Lucafò, M., Petix, V., Busetti, M., et al. (2021). Interplay of Opdp Porin and chromosomal carbapenemases in the determination of carbapenem resistance/susceptibility in Pseudomonas aeruginosa. Microbiol. Spectrum 9, E01186–E01121. doi: 10.1128/Spectrum.01186-21
Aurilio, C., Sansone, P., Barbarisi, M., Pota, V., Giaccari, L. G., Coppolino, F., et al. (2022). Mechanisms of action of Carbapenem resistance. Antibiotics 11:421. doi: 10.3390/antibiotics11030421
Awoke, T., Teka, B., Aseffa, A., Sebre, S., Seman, A., Yeshitela, B., et al. (2022). Detection of Bla Kpc and Bla Ndm carbapenemase genes among Klebsiella pneumoniae isolates in Addis Ababa, Ethiopia: dominance of Bla Ndm. PLoS One 17:E0267657. doi: 10.1371/journal.pone.0267657
Beshah, D., Desta, A. F., Woldemichael, G. B., Belachew, E. B., Derese, S. G., Zelelie, T. Z., et al. (2023). High burden of Esbl and carbapenemase-producing gram-negative Bacteria in bloodstream infection patients at a tertiary care hospital in Addis Ababa, Ethiopia. PLoS One 18:E0287453. doi: 10.1371/journal.pone.0287453
Caston, J. J., Cano, A., Perez-Camacho, I., Aguado, J. M., Carratala, J., Ramasco, F., et al. (2022). Impact of ceftazidime/avibactam versus best available therapy on mortality from infections caused by Carbapenemase-producing Enterobacterales (Cavicor study). J. Antimicrob. Chemother. 77, 1452–1460. doi: 10.1093/jac/dkac049
Chen, H., Jean, S., Lee, Y., Lu, M., Ko, W., and Liu, P. (2021). Carbapenem-resistant Enterobacterales in long-term care facilities: a global and narrative review. Front. Cell. Infect. Microbiol. 11:601968. doi: 10.3389/fcimb.2021.601968
Da Silva, G. J., and Domingues, S. (2016). Insights on the horizontal gene transfer of Carbapenemase determinants in the opportunistic pathogen Acinetobacter baumannii. Microorganisms 4:29. doi: 10.3390/microorganisms4030029
Das, S. (2023). The crisis of carbapenemase-mediated carbapenem resistance across the human–animal–environmental Interface in India. Infect Diseases Now 53:104628. doi: 10.1016/j.idnow.2022.09.023
Di Carlo, P., Serra, N., Lo Sauro, S., Carelli, V. M., Giarratana, M., Signorello, J. C., et al. (2021). Epidemiology and pattern of resistance of gram-negative Bacteria isolated from blood samples in hospitalized patients: a single center retrospective analysis from southern Italy. Antibiotics 10:1402. doi: 10.3390/antibiotics10111402
Dwomoh, F. P., Kotey, F. C., Dayie, N. T., Osei, M.-M., Amoa-Owusu, F., Bannah, V., et al. (2022). Phenotypic and genotypic detection of carbapenemase-producing Escherichia Coli and Klebsiella pneumoniae in Accra, Ghana. PLoS One 17:E0279715. doi: 10.1371/journal.pone.0279715
Eshetu, B., Gashaw, M., Berhane, M., Abdissa, A., Mcclure, E. M., Goldenberg, R. L., et al. (2019). Intravenous fluid contaminated with Klebsiella oxytoca as a source of Sepsis in a preterm newborn: case report. Am. J. Infect. Control 47, 840–842. doi: 10.1016/j.ajic.2018.12.025
EUCAST (2021). European committee on antimicrobial susceptibility testing, breakpoint tables for interpretation of Mics and zone diameters. European Society Of Clinical Microbiology and Infectious Diseases: Basel
Gashaw, M., Berhane, M., Bekele, S., Kibru, G., Teshager, L., Yilma, Y., et al. (2018). Emergence of high drug resistant bacterial isolates from patients with health care associated infections at Jimma University medical center: a cross sectional study. Antimicrob. Resist. Infect. Control 7, 1–8. doi: 10.1186/s13756-018-0431-0
Hammoudi Halat, D., and Ayoub Moubareck, C. (2020). The current burden of carbapenemases: review of significant properties and dissemination among gram-negative Bacteria. Antibiotics 9:186. doi: 10.3390/antibiotics9040186
Hu, W. S., Yao, S.-M., Fung, C.-P., Hsieh, Y.-P., Liu, C.-P., and Lin, J.-F. (2007). An Oxa-66/Oxa-51-like carbapenemase and possibly an efflux pump are associated with resistance to imipenem in Acinetobacter baumannii. Antimicrob. Agents Chemother. 51, 3844–3852. doi: 10.1128/AAC.01512-06
Huang, X.-Z., Cash, D. M., Chahine, M. A., Nikolich, M. P., and Craft, D. W. (2012). Development and validation of a multiplex Taqman real-time Pcr for rapid detection of genes encoding four types of class D carbapenemase in Acinetobacter baumannii. J. Med. Microbiol. 61, 1532–1537. doi: 10.1099/jmm.0.045823-0
Jean, S.-S., Harnod, D., and Hsueh, P.-R. (2022). Global threat of Carbapenem-resistant gram-negative Bacteria. Front. Cell. Infect. Microbiol. 12:236. doi: 10.3389/fcimb.2022.823684
Kruttgen, A., Razavi, S., Imohl, M., and Ritter, K. (2011). Real-time Pcr assay and a synthetic positive control for the rapid and sensitive detection of the emerging resistance Gene new Delhi Metallo-Β-Lactamase-1 (Bla Ndm-1). Med. Microbiol. Immunol. 200, 137–141. doi: 10.1007/s00430-011-0189-y
Legese, M. H., Asrat, D., Mihret, A., Hasan, B., Mekasha, A., Aseffa, A., et al. (2022). Genomic epidemiology of Carbapenemase-producing and Colistin-resistant Enterobacteriaceae among Sepsis patients in Ethiopia: a whole-genome analysis. Antimicrob. Agents Chemother. 66, E00534–E00522. doi: 10.1128/aac.00534-22
Maki, G., and Zervos, M. (2021). Health care-acquired infections in low- and middle-income countries and the role of infection prevention and control. Infect. Dis. Clin. 35, 827–839. doi: 10.1016/j.idc.2021.04.014
Miranda, I. F., Dos Santos, M. L., Oliveira, W. C. S., and Oliveira, M. C. (2018). Klebsiella pneumoniae Produtora De Carbapenemase Do Tipo Kpc: Disseminação Mundial E Situação Atual No Brasil. Brazil. J. Surg. Clin. Res. 25, 113–119.
Misha, G., Chelkeba, L., and Melaku, T. (2021). Bacterial profile and antimicrobial susceptibility patterns of isolates among patients diagnosed with surgical site infection at a tertiary teaching hospital in Ethiopia: a prospective cohort study. Ann. Clin. Microbiol. Antimicrob. 20, 1–10. doi: 10.1186/s12941-021-00440-z
Mulani, M. S., Kamble, E. E., Kumkar, S. N., Tawre, M. S., and Pardesi, K. R. (2019). Emerging strategies to combat Eskape pathogens in the era of antimicrobial resistance: a review. Front. Microbiol. 10:539. doi: 10.3389/fmicb.2019.00539
Nigro, S. J., and Hall, R. M. (2018). Does the intrinsic Oxaab (Bla Oxa-51-like) gene of Acinetobacter baumannii confer resistance to Carbapenems when activated by Isaba1? J. Antimicrob. Chemother. 73, 3518–3520. doi: 10.1093/jac/dky334
Nordmann, P., and Poirel, L. (2019). Epidemiology and diagnostics of Carbapenem resistance in gram-negative Bacteria. Clin. Infect. Dis. 69, S521–S528. doi: 10.1093/cid/ciz824
Pritsch, M., Zeynudin, A., Messerer, M., Baumer, S., Liegl, G., Schubert, S., et al. (2017). First report on Bla Ndm-1-producing Acinetobacter baumannii in three clinical isolates from Ethiopia. BMC Infect. Dis. 17, 1–7. doi: 10.1186/s12879-017-2289-9
Rabaan, A. A., Eljaaly, K., Alhumaid, S., Albayat, H., Al-Adsani, W., Sabour, A. A., et al. (2022). An overview on phenotypic and genotypic characterisation of carbapenem-resistant enterobacterales. Medicina 58:1675. doi: 10.3390/medicina58111675
Sands, K., Carvalho, M. J., Portal, E., Thomson, K., Dyer, C., Akpulu, C., et al. (2021). Characterization of antimicrobial-resistant gram-negative Bacteria that cause neonatal Sepsis in seven low-and middle-income countries. Nat. Microbiol. 6, 512–523. doi: 10.1038/s41564-021-00870-7
Sastry, S., Masroor, N., Bearman, G., Hajjeh, R., Holmes, A., Memish, Z., et al. (2017). The 17th international congress on infectious diseases workshop on developing infection prevention and control resources for low-and middle-income countries. Int. J. Infect. Dis. 57, 138–143. doi: 10.1016/j.ijid.2017.01.040
Seman, A., Mihret, A., Sebre, S., Awoke, T., Yeshitela, B., Yitayew, B., et al. (2022). Prevalence and molecular characterization of extended Spectrum Β-lactamase and carbapenemase-producing Enterobacteriaceae isolates from bloodstream infection suspected patients in Addis Ababa, Ethiopia. Infect. Drug Resist. 15, 1367–1382. doi: 10.2147/IDR.S349566
Sewunet, T., Asrat, D., Woldeamanuel, Y., Aseffa, A., and Giske, C. G. (2022). Molecular epidemiology and antimicrobial susceptibility of Pseudomonas Spp. and Acinetobacter Spp. from clinical samples at Jimma Medical Center, Ethiopia. Front. Microbiol. 13:951857. doi: 10.3389/fmicb.2022.951857
Shanmugakani, R. K., Srinivasan, B., Glesby, M. J., Westblade, L. F., Cárdenas, W. B., Raj, T., et al. (2020). Current state of the art in rapid diagnostics for antimicrobial resistance. Lab Chip 20, 2607–2625. doi: 10.1039/D0LC00034E
Sikora, A., and Zahra, F. (2020). Nosocomial infections. StatPearl. Available at: https://www.ncbi.nlm.nih.gov/books/NBK559312
Stewardson, A. J., Marimuthu, K., Sengupta, S., Allignol, A., El-Bouseary, M., Carvalho, M. J., et al. (2019). Effect of Carbapenem resistance on outcomes of bloodstream infection caused by Enterobacteriaceae in low-income and middle-income countries (panorama): a multinational prospective cohort study. Lancet Infect. Dis. 19, 601–610. doi: 10.1016/S1473-3099(18)30792-8
Tekele, S. G., Teklu, D. S., Legese, M. H., Weldehana, D. G., Belete, M. A., Tullu, K. D., et al. (2021). Multidrug-resistant and Carbapenemase-producing Enterobacteriaceae in Addis Ababa, Ethiopia. Biomed. Res. Int. 2021, 1–10. doi: 10.1155/2021/9999638
Tenover, F. C., Nicolau, D. P., and Gill, C. M. (2022). Carbapenemase-producing Pseudomonas aeruginosa–an emerging challenge. Emerg. Microbes Infect. 11, 811–814. doi: 10.1080/22221751.2022.2048972
Tilahun, M., Gedefie, A., Bisetegn, H., and Debash, H. (2022). Emergence of high prevalence of extended-spectrum beta-lactamase and carbapenemase producing acinetobacter species and Pseudomonas aeruginosa among hospitalized patients at Dessie comprehensive specialized hospital, north-East Ethiopia. Infect. Drug Resist. 15, 895–911. doi: 10.2147/IDR.S358116
Tufa, T. B., Mackenzie, C. R., Orth, H. M., Wienemann, T., Nordmann, T., Abdissa, S., et al. (2022). Prevalence and characterization of antimicrobial resistance among gram-negative Bacteria isolated from febrile hospitalized patients in Central Ethiopia. Antimicrob. Resist. Infect. Control 11:8. doi: 10.1186/s13756-022-01053-7
Van Duin, D. (2017). Carbapenem-resistant Enterobacteriaceae: what we know and what we need to know, vol. 8. Oxfordshire, UK: Taylor & Francis. 379–382.
Villinger, D., Schultze, T. G., Musyoki, V. M., Inwani, I., Aluvaala, J., Okutoyi, L., et al. (2022). Genomic transmission analysis of multidrug-resistant gram-negative Bacteria within a newborn unit of a Kenyan tertiary hospital: a four-month prospective colonization study. Front. Cell. Infect. Microbiol. 12:892126. doi: 10.3389/fcimb.2022.892126
Zeynudin, A., Pritsch, M., Schubert, S., Messerer, M., Liegl, G., Hoelscher, M., et al. (2018). Prevalence and antibiotic susceptibility pattern of Ctx-M type extended-Spectrum Β-lactamases among clinical isolates of gram-negative Bacilli in Jimma, Ethiopia. BMC Infect. Dis. 18, 1–10. doi: 10.1186/s12879-018-3436-7
Keywords: carbapenem-resistant, carbapenemases, blaOXA, blaNDM, ESBL, Jimma
Citation: Gashaw M, Gudina EK, Ali S, Gabriele L, Seeholzer T, Alemu B, Froeschl G, Kroidl A and Wieser A (2024) Molecular characterization of carbapenem-resistance in Gram-negative isolates obtained from clinical samples at Jimma Medical Center, Ethiopia. Front. Microbiol. 15:1336387. doi: 10.3389/fmicb.2024.1336387
Received: 10 November 2023; Accepted: 08 January 2024;
Published: 24 January 2024.
Edited by:
Nabil Karah, Umeå University, SwedenReviewed by:
Anna Giammanco, University of Palermo, ItalyCopyright © 2024 Gashaw, Gudina, Ali, Gabriele, Seeholzer, Alemu, Froeschl, Kroidl and Wieser. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Mulatu Gashaw, bXVsYXR1Lmdhc2hhd0BqdS5lZHUuZXQ=
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.