- 1Department of Biological Science and Technology, Jinzhong University, Jinzhong, China
- 2College of Food Science and Engineering, Shanxi Agriculture University, Jinzhong, China
Introduction: Proso millet, a high-quality fermentation material used for Chinese yellow wine production, can produce special flavored substances; however, its role in improving the flavor and altering microbial communities of light-flavored Baijiu during fermentation remain unknown. Thus, we aimed to investigate the effect of proso millet on improving the flavor of light-flavored Baijiu and altering microbial communities during different fermentation stages.
Methods: The dynamic changes in the microbial communities and flavor of proso millet (50%) + sorghum (50%) mixed fermentation samples were analyzed through intermittent sampling on days 7, 14, 21, and 28 of the fermentation process. Microbial high-throughput sequencing and the analysis of flavor characteristics were conducted through 16S DNA/ ITS amplicon sequencing and gas chromatography (multi-capillary column)-ion mobility spectrometry, respectively.
Results: Proso millet significantly changed the core flavor compound composition of traditional light-flavored Baijiu from ethyl acetate, ethyl hexanoate, ethyl hexanoate dimer, ethyl butanoate, ethyl lactate, and butyl acetate to oct-2-ene, 2-butanol, propyl propanoate, 2-pentenal, and 4-methylpentanal. The amplicon sequencing analysis revealed that the alpha diversity parameters of bacterial and fungal communities, including the Chao1, Pielou_e, Shannon, and Simpson indices, for proso millet–sorghum mixed fermentation samples were significantly higher than those for sorghum fermentation samples (p < 0.05). Of the 40 most significant microbial genera in two treatments, proso millet significantly increased the abundance of 12 bacterial and 18 fungal genera. Among the 40 most significant bacterial and fungal species, 23 bacterial species belonged to the Lactobacillus genus, whereas the 30 primary fungal species belonged to 28 different genera. The analysis of the relationship between microbial changes and the main flavor compounds of light-flavored Baijiu showed that bacteria from the Weissella, Acinetobacter, Bacteroides, Psychrobacter, Pseudarthrobacter, Lactococcus, Chloroplast, Saccharopolyspora, Psychrobacter, Saccharopolyspora, Pseudonocardiaceae, Bacteroides genera and fungi from the Thermoascus, Aspergillus, Pichia, Rhizomucor, Papiliotrema, Hyphopichia, and Mucor genera significantly inhibited the synthesis of ethyl hexanoate, ethyl butanoate, ethyl lactate ethyl lactate, and butyl acetate but increased the synthesis of ethyl acetate (p < 0.05). Moreover, these microbes exhibited a significantly greater abundance in proso millet–sorghum mixed fermentation samples than in sorghum samples. The synthesis of special flavored compounds in proso millet Baijiu was significantly positively correlated with the presence of fungi from the Rhizopus, Papiliotrema, Wickerhamomyces, Aspergillus, and Thermoascus genera but negative correlated with the presence of bacteria from the Weissella, Acinetobacter, Psychrobacter, Pseudarthrobacter, Bacteroides, and Saccharopolyspora genera. Regarding ethanol content, the low alcohol content of Fenjiu may be due to the significantly high abundance of fungi from the Psathyrella genus and bacteria from the Staphylococcus, Kroppenstedtia, Brevibacterium, and Acetobacter genera during fermentation. In summary, proso millet significantly altered the flavor of light-flavored Baijiu by inducing the formation of a special microbial community; however, it did not increase alcohol concentration.
Discussion: This study lays the foundation for future research on Baijiu fermentation. Additionally, the study findings may help improve the production efficiency and elevate the quality and flavor of the final product.
1 Introduction
In China, Baijiu has evolved from being a mere beverage to a symbol of culture. The Baijiu culture in China has a rich historical background, and companies that produce distilled Baijiu operate on a large scale with huge profits. Fenjiu, traditional Chinese Baijiu primarily made from non-glutinous sorghum, is highly popular light aroma-style beverage that has been consumed for over 1,500 years (Ma et al., 2013). However, companies that produce light-flavored Baijiu face several challenges, including low yield, limited high-end product options, declining quality, and a stagnant market due to a narrow flavor profile, which have hindered their development (Yifan et al., 2021). Currently, researchers are actively exploring methods to enhance their understanding of light-flavored Baijiu and improve its quality, with research focusing on aspects such as the microbial community structure in light-flavored Daqu, flavor chemistry of light-flavored Baijiu, influence of sorghum varieties on the flavor of light-flavored Baijiu, and the relationship between light-flavored Baijiu and microorganisms (Liu and Sun, 2018; Huang et al., 2020; Zhang et al., 2022; Xiang et al., 2023; Zhang et al., 2023). In addition, studies have examined the physicochemical properties of Chinese light-flavored Baijiu during storage and explored ways to improve sorghum varieties for Baijiu production (Ma et al., 2013; Kang et al., 2020). Furthermore, to improve the quality of light-flavored Baijiu, some other traditional brewing materials such as rice, barley, and glutinous sorghum, have been currently added to the raw materials used for its production. However, it remains unclear whether proso millet, which is the primary raw material for Chinese yellow wine production, can improve the flavor of light-flavored Baijiu. In addition, the microbial communities and flavor profiles of sorghum-based light-flavored Baijiu during fermentation have not yet been determined.
Proso millet (Panicum miliaceum L.) is an annual herbaceous cereal crop, commonly known as “huangmi” in North and Northwest China; it has a high nutritional value and is rich in starch, dietary fibers, proteins, phenolic compounds, vitamins, amino acids, and trace elements (Kalam Azad et al., 2019; Wiedemair et al., 2020; Yuan et al., 2022). It is extensively cultivated in arid or semi-arid regions across North China, Asia, Australia, North America, Europe, and Africa (Sanderson et al., 2017; Yang et al., 2018; Kučera et al., 2019; Gong et al., 2021; Narciso and Nyström, 2023). Moreover, proso millet has demonstrated antioxidant, anti-proliferative, antimicrobial, and anti-carcinogenic properties. It has also been associated with alleviating celiac disease, regulating blood glucose and cholesterol levels, and preventing diabetes and cardiovascular disorders (Kim et al., 2011; Sanderson et al., 2017; Shen et al., 2018; Yang et al., 2018; Kalam Azad et al., 2019; Agarwal and Chauhan, 2019). Consequently, proso millet is a valuable resource for the development of healthy foods, and has recently been used to produce popular products, such as flour, bread, couscous, biscuits, porridge, extruded snacks, and fermented beverages (Sanderson et al., 2017; McSweeney et al., 2017a; Wang J. et al., 2020). To promote the development of new products, increase awareness regarding the functional properties of proso millet, and determine the potential applications of proso millet in pasta processing or other cooking processes, significant research attention has been devoted to topics such as drought resistance, cultivation methods, yield improvement, breeding, functional properties, physicochemical characteristics, nutritional composition, protein properties, starch physicochemical properties, and digestibility of the crop, as well as its biological structure and microstructure analyses (Gulati et al., 2017, 2018; Hasseldine et al., 2017; McSweeney et al., 2017b; Balkrishna and Visvanathan, 2019; Johnson et al., 2019; Yang et al., 2019; Liu et al., 2020; Wiedemair et al., 2020; Gong et al., 2021; Chang et al., 2023; Kumar et al., 2023). However, limited research has been carried out on its fermentation characteristics and its use in the development of fermented products. Despite the development of healthy proso millet wines (yellow wine or fermented alcoholic drinks), beer, and vinegar, with recent studies shedding light on their flavor composition and fermentation processes (Liu et al., 2018; Santra et al., 2019; Zheng et al., 2020; Bian et al., 2022), there is limited research on new fermentation products, particularly hard liquor made from proso millet. Moreover, studies on the relevant fermentation processes, flavor profiles, and mechanisms underlying quality formation for proso millet liquor are limited.
Therefore, in recent years, our research team has optimized the fermentation formula and technology based on Fenjiu Daqu, leading to the successful development of a new product (ZL201410293583.1). In this study, we aimed to investigate the effects of proso millet on improving the flavor of light-flavored Baijiu by studying the microbial composition and flavor differences between the new product and sorghum-based light-flavored Baijiu during fermentation. In addition, we aimed to reveal the mechanistic correlations between microbial community composition and flavor formation. Our findings may help expand the application range of proso millet and provide a reference basis for improving the flavor of Fenjiu or other light-flavored Baijiu beverages.
2 Materials and methods
2.1 Experimental design and sampling
Sorghum (Jinza No. 22) and proso millet (Jinshu No. 9) were procured from Shanxi Jinliang Agricultural Development Company, Xiyang County, Jinzhong City, Shanxi Province, China. Two fermentation tests, sorghum and mixed material (composition of 50% sorghum and 50% proso millet) fermentation, were conducted under commercial conditions at the Xinghua Village Fen Distillery in Shanxi Province. Samples of fermented grains (FG) were collected at four different time points during alcoholic fermentation in the pit: days 7, 14, 21, and 28. To ensure comprehensive data and representativeness, each sample at every time point comprised nine subsamples collected from various spatial positions within the underground vats, with each subsample weighing 100 g (Supplementary Figure S1). Three parallel samples (from three random cellars) were collected for each sample type. Subsequently, all 24 samples were transported to the laboratory on ice and stored at −80°C until further analysis. The mixed proso millet fermentation samples collected on days 7, 14, 21, and 28 of fermentation were designated as MA, MB, MC, and MD, respectively. The sorghum fermentation samples collected on days 7, 14, 21, and 28 of fermentation were designated as GA, GB, GC, and GD, respectively.
2.2 Microbiological analysis
2.2.1 DNA extraction and high-throughput sequencing analysis
Total genomic DNA was extracted from 200 mg of FG samples using Fast DNA SPIN extraction kits (MP Biomedicals, Santa Ana, CA, USA), following the manufacturer’s protocol. The quantity and quality of the extracted DNA were assessed using an ND-1000 spectrophotometer (Thermo Fisher Scientific, Waltham, MA, USA) and agarose gel electrophoresis, respectively.
For bacterial analysis, the V3-V4 hypervariable region of 16S rRNA genes was amplified using the universal primer pair, 338F (5′-ACTCCTACGGGAGGCAGCA-3′) and 806R (5′-GGACTACHVGGGTWTCTAAT-3′), each with unique barcodes (Lin et al., 2018). Sample-specific 7-bp barcodes were incorporated into the primers for multiplex sequencing. Polymerase chain reactions (PCRs) were conducted in a 25-μL reaction volume, consisting of 5 μL of Q5 reaction buffer (5×), 5 μL of Q5 high-fidelity GC buffer (5×), 0.25 μL of Q5 high-fidelity DNA polymerase (5 U/μL), 2 μL (2.5 mM) of deoxynucleotide triphosphates, 1 μL (10 μM) of each forward and reverse primer, 2 μL of DNA template, and 8.75 μL of double distilled water. The thermal cycle profile included an initial 30 s of denaturation at 98°C, followed by denaturation at 98°C for 15 s, annealing at 50°C for 30 s, and extension at 72°C for 30 s, with final extension at 72°C for 5 min.
For fungal analysis, the ITS1 region was targeted using the primer ITS5 (5′-GGAAGTAAAAGTCGTAACAAGG-3′), and the reverse primer, ITS2 (5′-GCTGCGTTCTTCATCGATGC-3′), following a previously published method (Huang et al., 2016). The PCR volume and conditions were the same as those for the bacterial analysis.
PCR amplicons were purified using Agencourt AMPure Beads (Beckman Coulter, Indianapolis, IN, USA) and quantified using the PicoGreen dsDNA Assay Kit (Invitrogen, Carlsbad, CA, USA). Following individual quantification, amplicons were pooled in equal amounts, and pair-end 2 × 300 bp sequencing was conducted on the Illumina MiSeq platform using MiSeq Reagent Kit v3, carried out by Shanghai Personal Biotechnology Co., Ltd. (Shanghai, China).
2.2.2 Bioinformatics and statistical analysis
The Quantitative Insights Into Microbial Ecology (QIIME, v1.8.0) pipeline was employed to process sequencing data, following established procedures (Caporaso et al., 2010). In summary, raw sequencing reads that precisely matched the barcodes were assigned to their respective samples and identified as valid sequences. Paired-end DNA fragments were initially assembled using FLASH (Zhang et al., 2014). After chimera detection, the remaining high-quality sequences were clustered into operational taxonomic units (OTUs) with 97% sequence identity using UCLUST (Jiang et al., 2012). Taxonomic assignments for representative sequences from each OTU were determined using the Silva database (16S rDNA) and Unite database (ITS) within the QIIME platform (Caporaso et al., 2010). Data analysis was primarily conducted using the QIIME and R packages (v3.2.0). Alpha diversity indices at the OTU level, including the Chao1 richness estimator, the abundance-based coverage estimator metric, the Shannon diversity index, and the Simpson’s index, were calculated using the OTU table in QIIME. Differences in microbiota structure among groups were assessed using permutational multivariate analysis of variance and analysis of similarities using the “vegan” function in R (Takeshita et al., 2012; Kelly et al., 2015). Taxonomic compositions and abundance were visualized using MEGAN and GraPhlAn (Li et al., 2019). Taxa abundances at the phylum, class, order, family, genus, and species levels were statistically compared among samples or groups using Metastats (Elokil et al., 2020) and visualized using violin plots. Raw sequence data have been deposited in the NCBI Sequence Read Archive under the accession numbers, PRJNA1015004, PRJNA1014983, PRJNA1013705, and PRJNA1014132.
2.3 Flavor analysis
The FG samples used for flavor analysis were the same as those mentioned in Section 2.1. Volatile compounds were detected using FlavourSpec® (G.A.S., Germany) with an FS-SE-54-CB-1 column (15 m length × 0.53 mm inner diameter) via GC-IMS. Briefly, 2 g of each fermented sample was placed in a 20-mL headspace bottle, incubated at 60°C, and centrifuged at 500 rpm for 15 min. Subsequently, 200 μL of liquid was extracted from the headspace bottle using an 85°C syringe and automatically injected into the injector of the GC-IMS equipment. Nitrogen gas with 99.999% purity was used as the carrier gas, and volatile organic compounds (VOCs) in the FG samples were separated using a 60°C quartz capillary column. The GC flow conditions were as follows: 2 mL/min for 2 min, 100 mL/min for 20 min, 150 mL/min for 30 min, and then the flow was halted. VOC separation was carried out at a column temperature of 60°C, with detection at 45°C in the IMS ionization chamber at a drift gas flow rate of 150 mL/min. All experiments were conducted in triplicate.
2.4 Statistical analyses
Statistical analyses were performed using one-way analysis of variance in the SPSS version 17.0 software (SPSS, Inc., Chicago, IL, USA). Differences between groups were evaluated using Duncan’s multiple comparison tests, and p < 0.05 was considered statistically significant.
3 Results
3.1 Microbiological analysis
3.1.1 Overall analysis of Illumina MiSeq data
In total, 324,421 and 292,751 high-quality sequences were obtained from the bacterial 16S rRNA V3-V4 sequences, whereas 514,202 and 470,916 high-quality sequences were obtained from the fungal ITS sequences for sorghum and proso millet FG samples, respectively. These sequences were clustered into 3,133 and 371 (sorghum FG samples) and 7,255 and 574 (proso millet FG samples) amplicon sequence variants (ASVs), respectively, at a 97% similarity level. The coverage estimator for bacterial and fungal ASVs exceeded 99% for all samples, indicating that the obtained sequence reads adequately represented the microbial diversity.
Microbial community richness, diversity, and evenness in sorghum and proso millet FG samples were assessed using the Chao1, Shannon, Simpson, and Pielou’s evenness indices (Table 1). Microbial diversity in both sorghum and proso millet FG samples decreased as fermentation time increased. Despite showing a lower number of high-quality sequences, proso millet FG samples exhibited a higher microbial diversity than sorghum FG samples as the bacterial Chao1, Shannon, Pielou’s evenness, and Simpson indices for proso millet FG samples at different stages of fermentation were approximately twice as those for sorghum FG samples; in addition, the fungal Chao1 index was considerably higher for proso millet FG samples than for sorghum FG samples (Table 1).
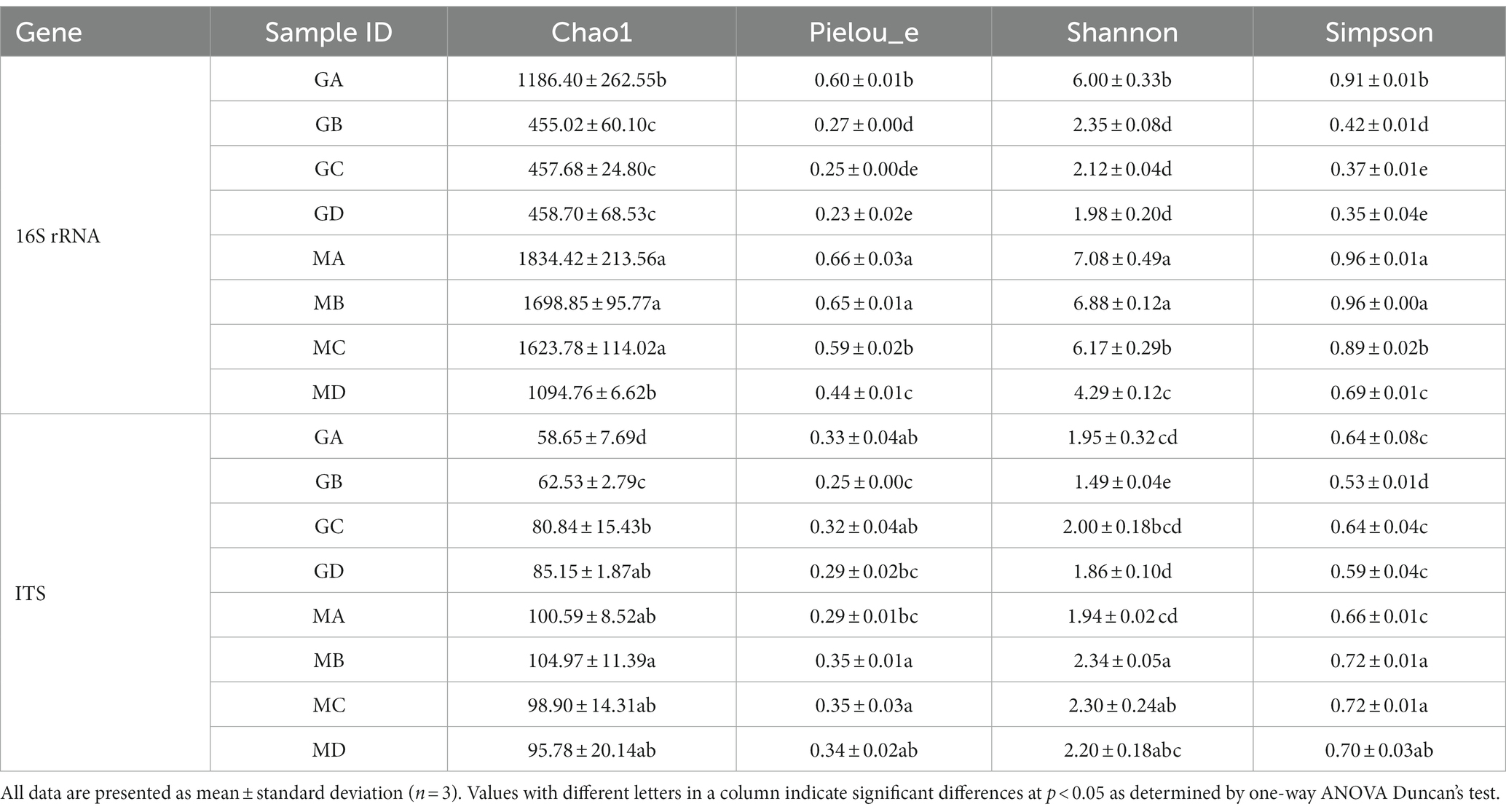
Table 1. Summary of diversity indices for microbial communities based on 16S rRNA and ITS1 gene sequencing data.
3.1.2 Bacterial community composition during proso millet Baijiu fermentation
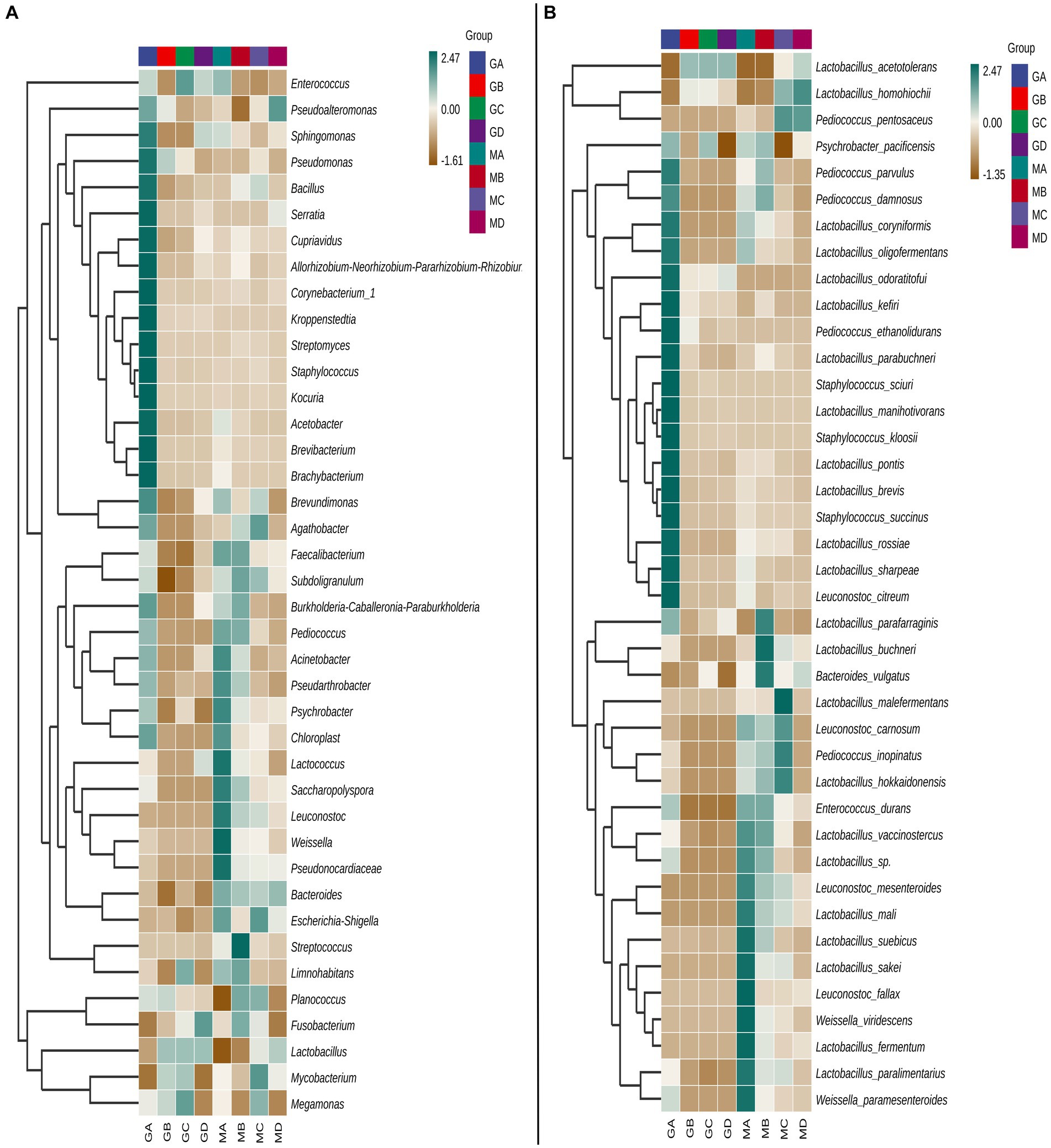
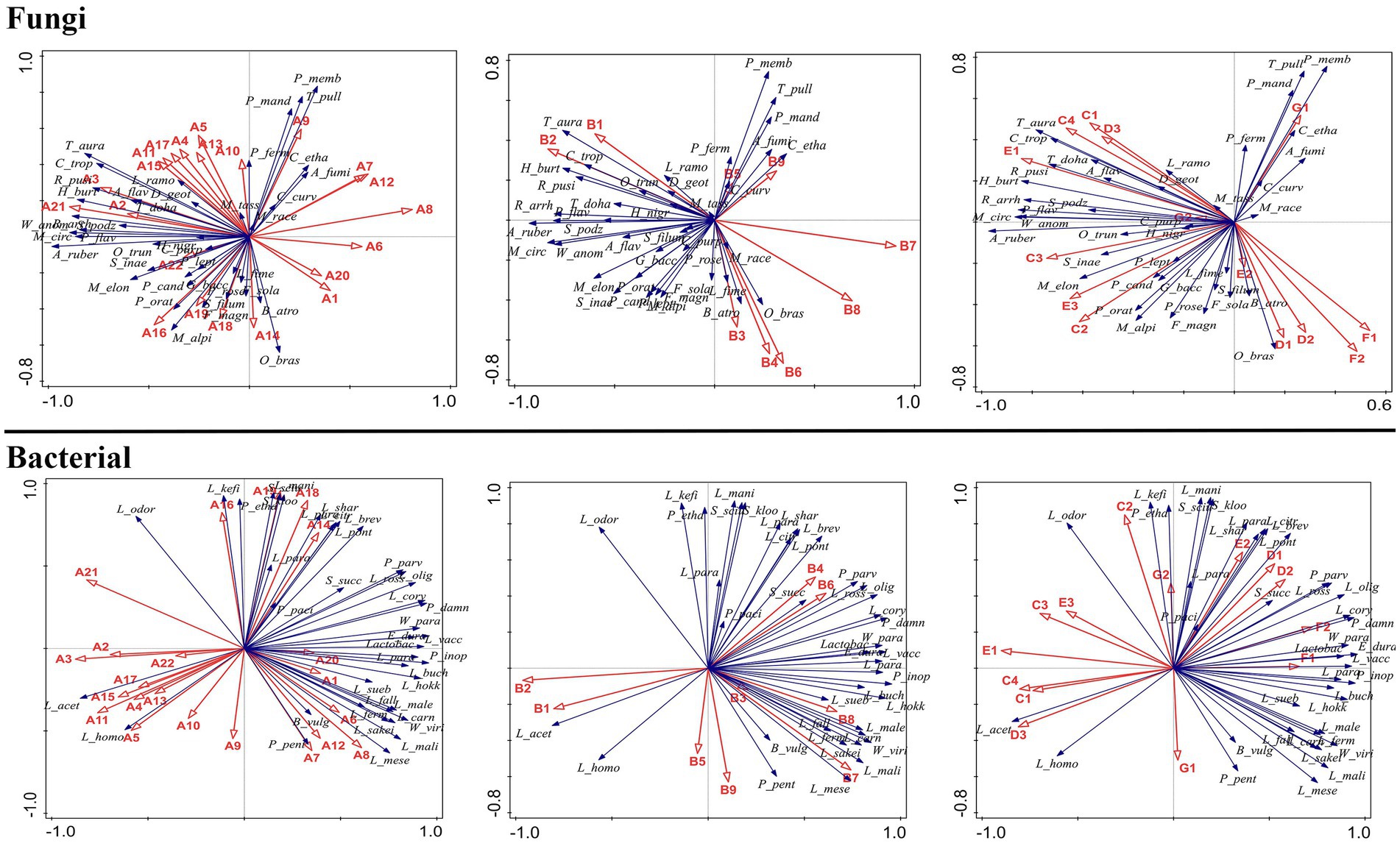
At the genus level, the bacterial community composition in samples MA–MD differed significantly from that in samples GA–GD (Figure 1A). Compared to those in sorghum fermentation samples (GA–GD), the core bacteria genera in proso millet fermentation samples changed from Lactobacillus, Megamonas, and Enterococcus to Bacteroides, Escherichia-Shigella, Leuconostoc, Subdoligranulum, and Faecalibacterium. However, Lactobacillus species played a major role in both proso millet and sorghum Baijiu fermentation. For MA samples, the abundance of bacteria from the genera Acinetobacter, Pseudarthrobacter, Psychrobacter, Chloroplast, Lactococcus, Saccharopolyspora, Leuconostoc, Weissella, and Pseudonocardiaceae was considerably higher than that of bacteria from other genera. However, the abundance of bacteria belonging to these nine genera gradually decreased in MB–MD samples. The Acinetobacter, Pseudarthrobacter, Psychrobacter, and Chloroplast genera were predominant in GA samples; however, their abundance was lower than that of other genera, such as the Sphingomonas, Pseudomonas, Bacillus, Serratia, Streptomyces, Staphylococcus, Cupriavidus, Allorhizobium-Neorhizobium-Pararhizobium-Rhizobium, Corynebacterium, Kocuria, Kroppenstedtia, Acetobacter, Brevibacterium, and Brachybacterium. In MB samples, bacteria from the Streptococcus, Limnohabitans, Planococcus, Fusobacterium, Faecalibacterium, Subdoligranulum, Burkholderia-Caballeronia-Paraburkholderia, and Pediococcus genera dominated, with their abundance surpassing that in GB samples. Genera such as Agathobacter, Subdoligranulum, Escherichia-Shigella, Planococcus, Mycobacterium, and Megamonas were prevalent in MC samples. MC samples were mainly characterized by Megamonas, Limnohabitans, Lactobacillus, Mycobacterium, and Enterococcus species. In contrast, MD samples exhibited a dominance of the genera, Pseudoalteromonas, Bacteroides, and Lactobacillus. GD samples were characterized by the genera, Fusobacterium, and Lactobacillus.
At the species level, the core bacterial composition in mixed fermentation samples was significantly different from that in sorghum fermentation samples. Species such as Leuconostoc carnosum, Pediococcus inopinatus, Lactobacillus hokkaidonensis, and Leuconostoc mesenteroides were the core bacteria identified in mixed fermentation samples, whereas species such as Lactobacillus acetotolerans and Lactobacillus homohiochii were the core bacteria identified in sorghum fermentation samples at different fermentation stages (Figure 1B).
Of the 40 most significant species, 23 belonged to the Lactobacillus genus. However, species composition differed between proso millet and sorghum fermentation samples (Figure 1B). In MA samples, species such as L. carnosum, Enterococcus durans, Lactobacillus vaccinostercus, L. mesenteroides, Lactobacillus mali, Lactobacillus suebicus, Lactobacillus sakei, Leuconostoc fallax, Weissella viridescens, Lactobacillus fermentum, Lactobacillus paralimentarius, and Weissella paramesenteroides were predominant. In contrast, GA samples were characterized by species such as Pediococcus parvulus, Pediococcus damnosus, Lactobacillus coryniformis, Lactobacillus oligofermentans, Lactobacillus odoratitofui, Lactobacillus kefiri, Pediococcus ethanolidurans, Lactobacillus parabuchneri, Staphylococcus sciuri, Lactobacillus manihotivorans, Staphylococcus kloosii, Lactobacillus pontis, Lactobacillus brevis, Staphylococcus succinus, Lactobacillus rossiae, Lactobacillus sharpeae, and Leuconostoc citreum, which was significantly different from the composition of MA samples. In MA samples, in total, six, three, two, and one species belonged to Lactobacillus, Leuconostoc, Weissella, and Enterococcus genera, respectively, were identified. In MB samples, the abundance of species such as Lactobacillus parafarraginis, Lactobacillus buchneri, and Bacteroides vulgatus was significantly higher than that of other species and that in GB samples. In contrast, species such as Lactobacillus malefermentans, L. carnosum, P. inopinatus, L. hokkaidonensis, L. homohiochii, and Pediococcus pentosaceus were the primary species in MC samples. In MD samples, L. homohiochii and P. pentosaceus dominated. L. acetotolerans was the core species in samples GB, GC, and GD (Figure 1B).
3.1.3 Fungal community composition during proso millet Baijiu fermentation
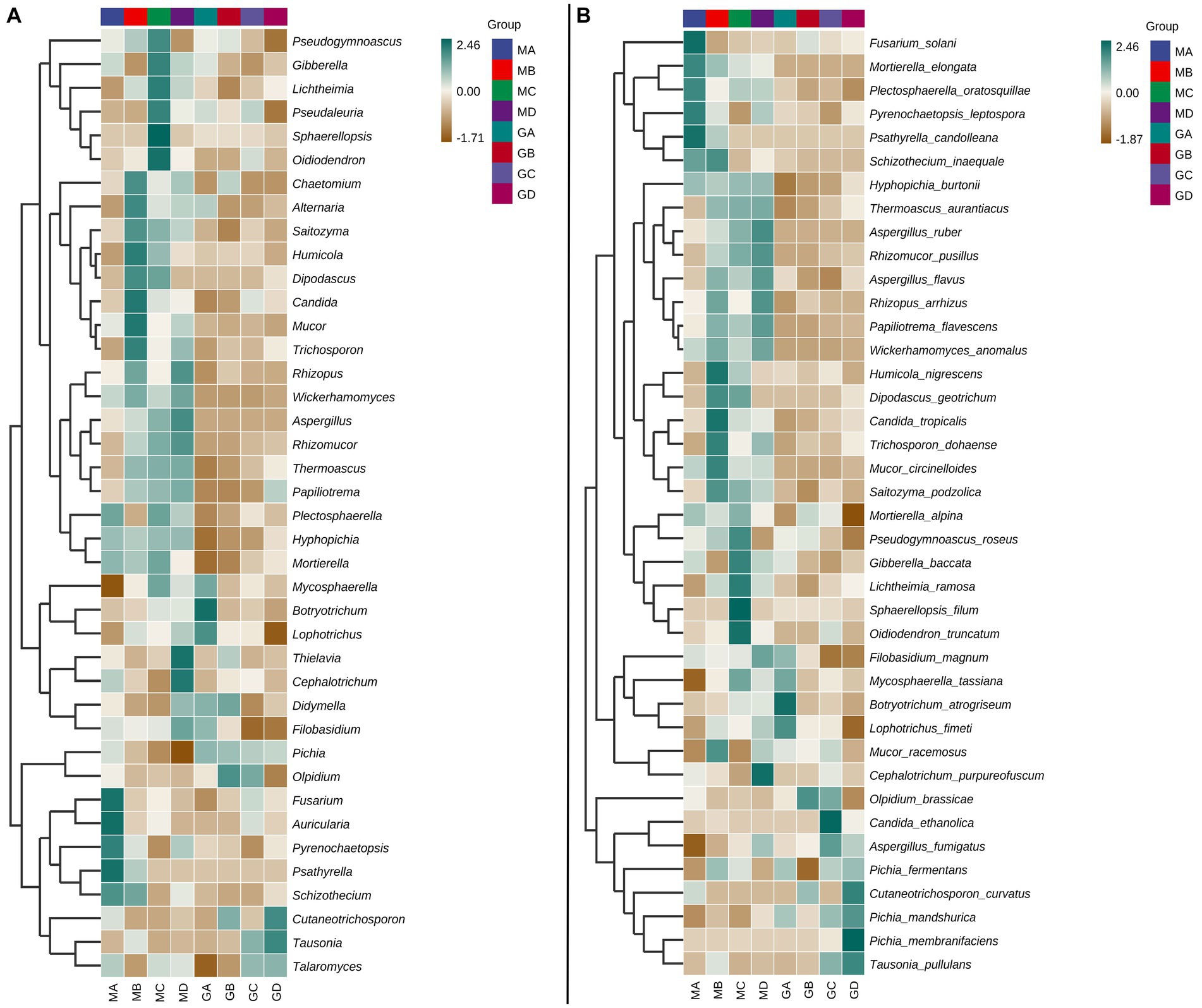
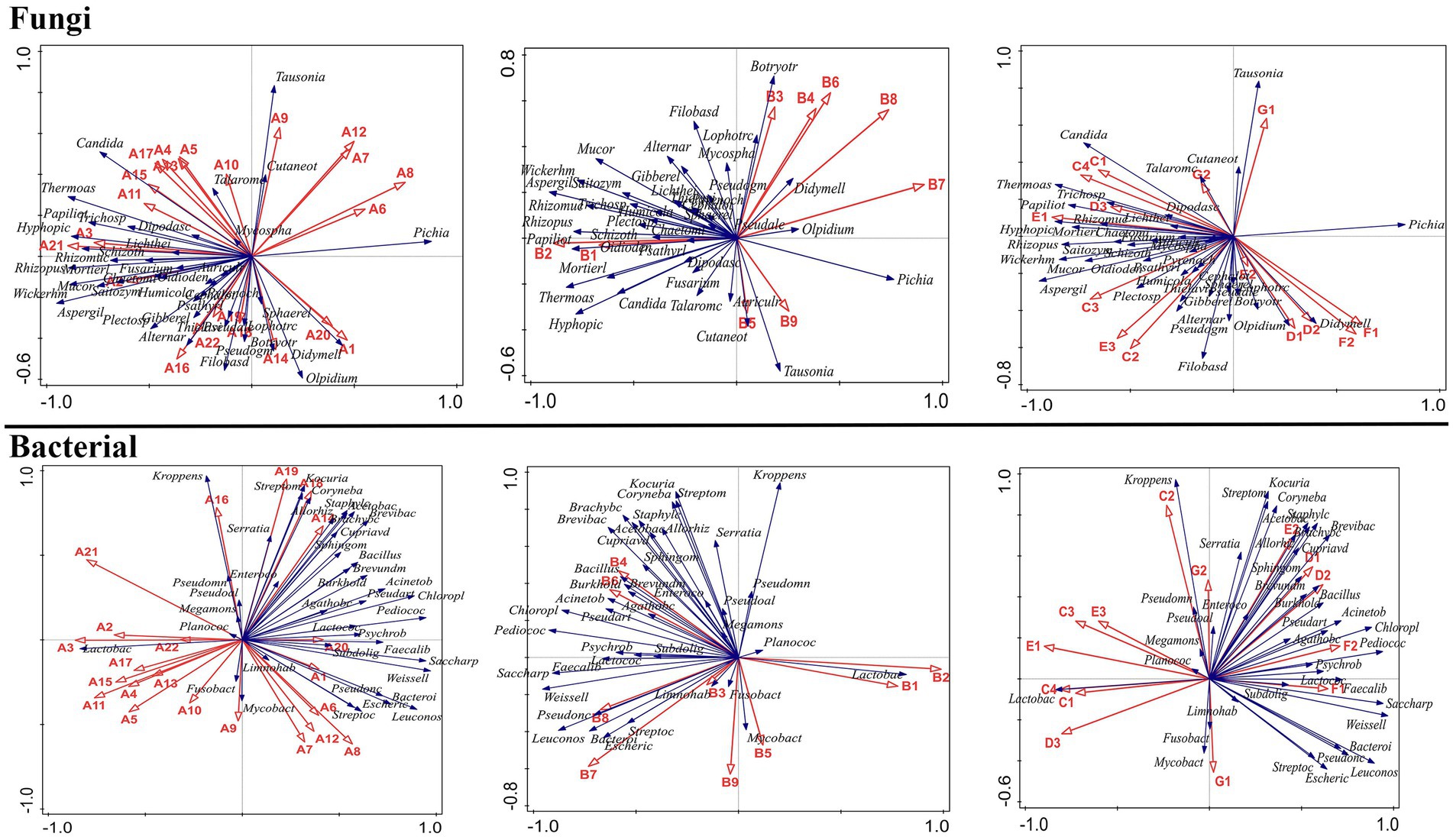
At the genus level, the fungal community composition of MA–MD samples exhibited significant differences from that of GA–GD samples (Figure 2A). Compared to those in sorghum fermentation samples (GA–GD), core fungal genera in proso millet fermentation samples changed from Pichia, Olpidium, Tausonia, Talaromyces, Cutaneotrichosporon, and Didymella to Wickerhamomyces, Rhizopus, Aspergillus, Rhizomucor, Hyphopichia, Mortierella, Papiliotrema, Plectosphaerella, Mucor, Trichosporon, Candida, Chaetomium, Gibberella, and Pseudogymnoascus. In addition, the main genera identified in MA samples were Fusarium, Auricularia, Pyrenochaetopsis, Psathyrella, and Schizothecium; however GA samples were characterized by dominant genera such as Mycosphaerella, Botryotrichum, and Lophotrichus. MB samples included Chaetomium, Alternaria, Saitozyma, Humicola, Dipodascus, Candida, Mucor, and Trichosporon as the main genera. However, GB samples contained fungi from the Olpidium, Didymella, and Cutaneotrichosporon genera. In contrast, MC samples exhibited a different profile, with genera such as Pseudogymnoascus, Gibberella, Lichtheimia, Pseudaleuria, Sphaerellopsis, and Oidiodendron being predominant. At the time of GC sample collection, the dominant genera had shifted to Olpidium, Tausonia, and Talaromyces. Finally, MD samples exhibited Rhizopus, Wickerhamomyces, Aspergillus, Rhizomucor, Thielavia, Cephalotrichum, and Filobasidium as main genera. The Cutaneotrichosporon and Tausonia genera were the dominant genera in GD samples (Figure 2A).
At the species level, specific species were associated with different stages of fermentation. For proso millet + sorghum mixed fermentation samples, there were 23 typical core species: Hyphopichia burtonii, Thermoascus aurantiacus, Plectosphaerella oratosquillae, Pyrenochaetopsis leptospora, Mortierella elongate, Aspergillus ruber, Rhizomucor pusillus, Aspergillus flavus, Rhizopus arrhizus, Papiliotrema flavescens, and Wickerhamomyces anomalus. However, only Pichia fermentans, Pichia mandshurica, Pichia membranifaciens, Tausonia pullulans, and Olpidium brassicae were identified as core genera in sorghum fermentation samples based on the analysis of relative abundance (Figure 2B). Fusarium solani, Mortierella elongata, P. oratosquillae, P. leptospora, Psathyrella candolleana, and Schizothecium inaequale were prominent species in MA samples. T. aurantiacus, A. flavus, R. arrhizus, P. flavescens, W. anomalus, Humicola nigrescens, Dipodascus geotrichum, Candida tropicalis, Trichosporon dohaense, Mucor circinelloides, Saitozyma podzolica, and Mucor racemosus were the main species in MB samples, with S. inaequale and M. elongata present in both MA and MB samples. MC samples exhibited a different set of dominant species, including S. podzolica, Mortierella alpine, Pseudogymnoascus roseus, Gibberella baccata, Lichtheimia ramosa, Sphaerellopsis filum, and Oidiodendron truncatum. Furthermore, MD samples featured T. aurantiacus, A. ruber, R. pusillus, A. flavus, R. arrhizus, P. flavescens, W. anomalus, Filobasidium magnum, and Cephalotrichum purpureofuscum. Across samples MA–MD, 30 dominant species belonging to 28 genera were identified. In contrast, GA samples featured different dominant species, including F. magnum, Mycosphaerella tassiana, Botryotrichum atrogriseum, Lophotrichus fimeti, P. fermentans, and P. mandshurica. GB samples were characterized by O. brassicae and Cutaneotrichosporon curvatus as main species, whereas GC samples exhibited a different profile, with species such as O. brassicae, Candida ethanolica, Aspergillus fumigatus, P. mandshurica, and T. pullulans being identified. GD samples exhibited dominant species such as P. fermentans, C. curvatus, P. mandshurica, P. membranifaciens, and T. pullulans (Figure 2B). Significant differences in high-abundance species were identified between sorghum and millet fermentation samples, with the number of high-abundance species in millet fermentation samples (MA–MD) exceeding that in sorghum fermentation samples (GA–GD) (Figure 2B).
3.2 Flavor compound analysis
3.2.1 Flavor compound composition of proso millet Baijiu fermentation samples
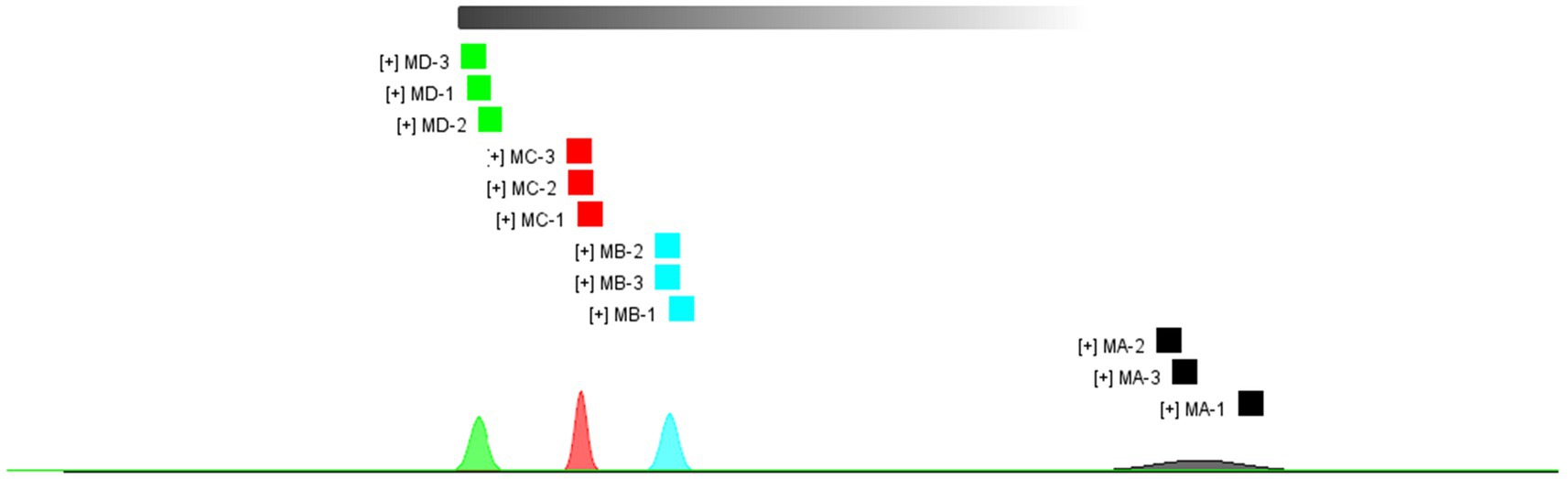
As shown in Figure 3, there were significant differences among the four samples (MA–MD). The dominant compositions of flavor compounds and their abundances in the four samples were also different. Moreover, compared with MA samples, MD samples exhibited evident differences in the abundances of compounds (Figure 4).

Figure 4. Difference diagram showing the gas phase ion mobility spectrum of the MA, MB, MC, and MD samples. The MA sample was selected as the reference. The spectra of the other samples were obtained from a previous study. If two volatile organic compounds are the same, the deducted background is white, red indicates that the concentration of the substance is higher than that of the reference, and blue indicates that the concentration of the substance is lower than that of the reference.
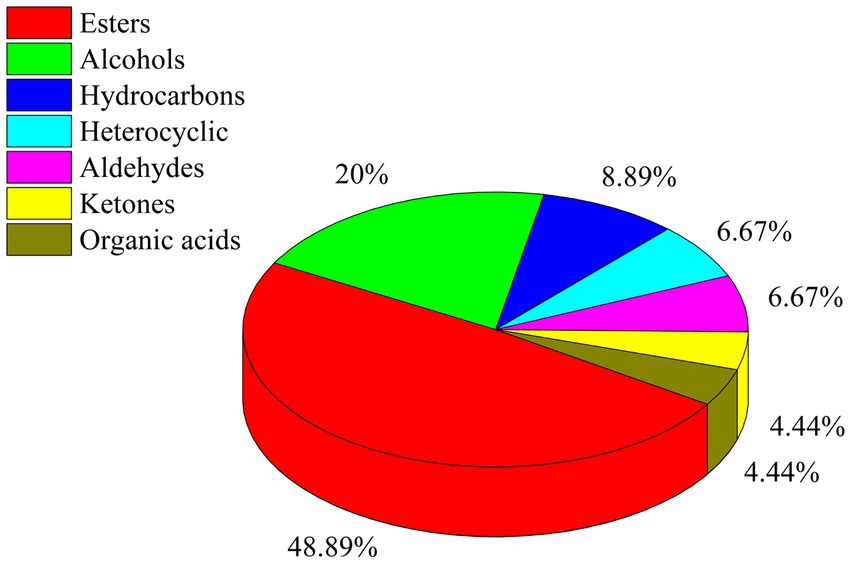
In total, 75 flavor compounds were detected through GC-IMS; however, only 45 of them could be identified based on parameters such as molecular weight, retention index (Ri), retention time (Rt), and mobility time (Dt) (Table 2). Twenty compounds remained unidentified due to similarities in Ri, Rt, and Dt (Supplementary Table S1). Among these 45 compounds, esters were the dominant flavor compounds, constituting 48.89% (22 compounds). Alcohols (nine compounds), hydrocarbons (four compounds), heterocyclic compounds (three compounds), aldehydes (three compounds), ketones (two compounds), and organic acids (two compounds) accounted for 20.00, 8.89, 6.67, 6.67, 4.44, and 4.44% of the compounds, respectively (Figure 5; Table 2).
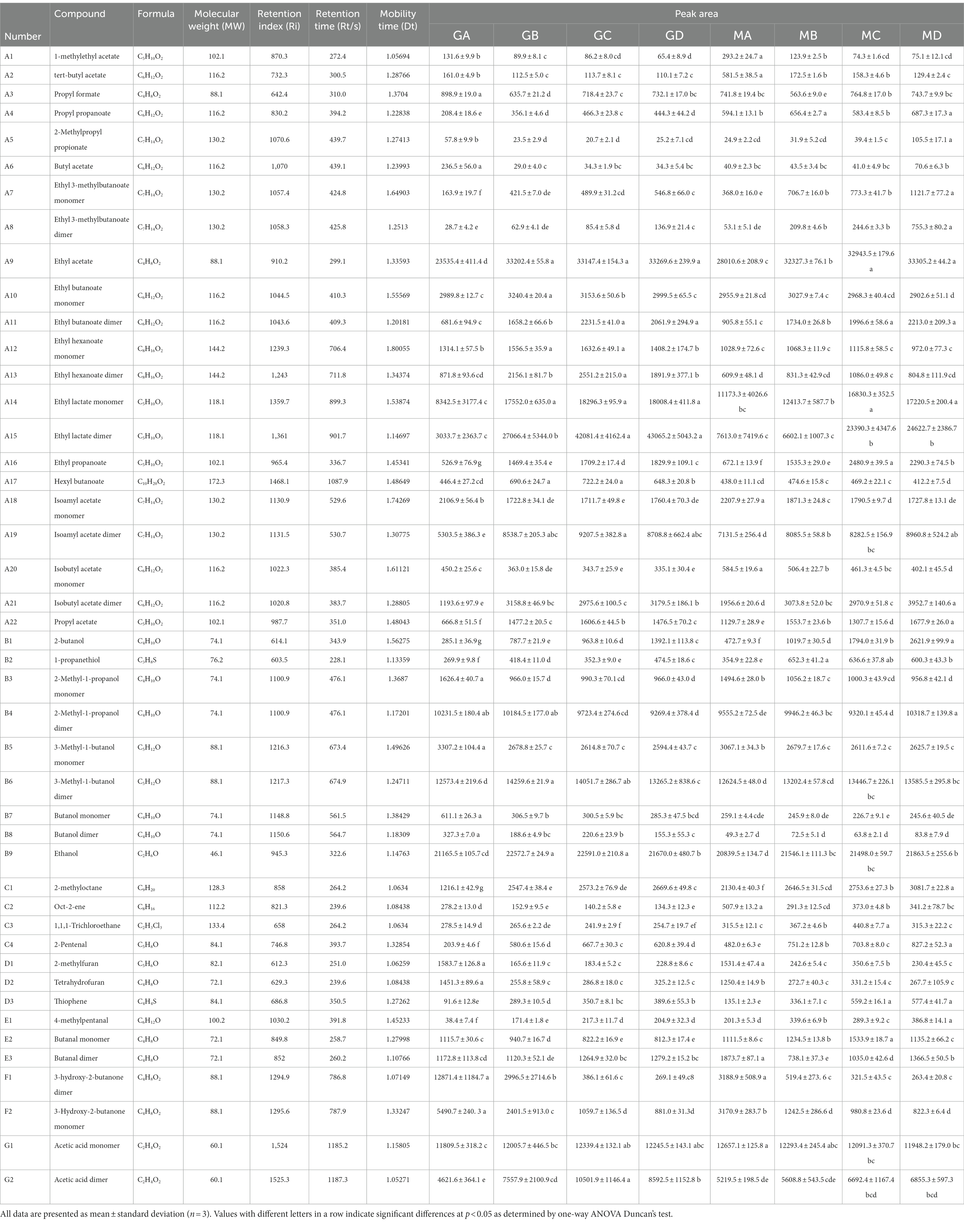
Table 2. Identification of flavor compounds in fermented grain samples at different stages of fermentation based on gas chromatography–ion mobility spectrometry.
As shown in Figure 6, main flavor compound composition and abundance varied not only between MA–MD samples but also significantly varied between MA–MD samples and samples GA–GD. Although 46 compounds were identified across all samples, their abundances differed. In MA samples, 18 compounds showed significantly higher abundances as compared to their levels in GA samples, with compounds such as 2-butanol, oct-2-ene, 1-methylethyl acetate, tert-butyl acetate, propyl propanoate, 2-pentenal, and 4-methylpentanal being particularly prominent (Figure 6). However, the abundance of the 2-methyl-1-propanol monomer, the 3-hydroxy-2-butanone monomer, the 3-methyl-1-butanol monomer, the butanol dimer, the butanol monomer, and butyl acetate was higher in GA samples than in MA samples. Eleven compounds, i.e., the ethyl 3-methylbutanoate dimer, the ethyl 3-methylbutanoate monomer, 1-propanethiol, oct-2-ene, 1,1,1-trichloroethane, 1-methylethyl acetate, tert-butyl acetate, 2-butanol, propyl propanoate, 2-pentenal, and 4-methylpentanal, exhibited significantly higher abundances in MB samples than in GB samples; however, the abundance of the ethyl hexanoate dimer, ethyl hexanoate monomer, ethyl lactate dimer, butanol dimer, butanol monomer, 3-hydroxy-2-butanone dimer, and 3-hydroxy-2-butanone monomer was higher in GB samples than in MB samples. The abundance of the ethyl 3-methylbutanoate dimer, the ethyl 3-methylbutanoate monomer, ethyl propanoate, 2-methylpropyl propionate, 2-methylfuran, 1-propanethiol, oct-2-ene, 1,1,1-trichloroethane, 1-methylethyl acetate, thiophene, and 2-butanol was significantly higher in GC samples than in GA samples; however the abundance of hexyl butanoate, the ethyl hexanoate dimer, the ethyl hexanoate monomer, the butanol dimer, and the butanol monomer was significantly lower in GC samples than in GA samples. The abundance of the ethyl 3-methylbutanoate dimer, the ethyl 3-methylbutanoate monomer, ethyl propanoate, 2-butanol, 2-methyloctane, 1-propanethiol, oct-2-ene, thiophene, propyl propanoate, 2-pentenal, 4-methylpentanal, and 2-methylpropyl propionate was significantly higher in MD samples than in GD samples; however, the abundance of hexyl butanoate, the ethyl hexanoate dimer, the ethyl hexanoate monomer, the butanol dimer, and the butanol monomer was significantly lower in MD samples than in GD (Figure 6; Table 2).
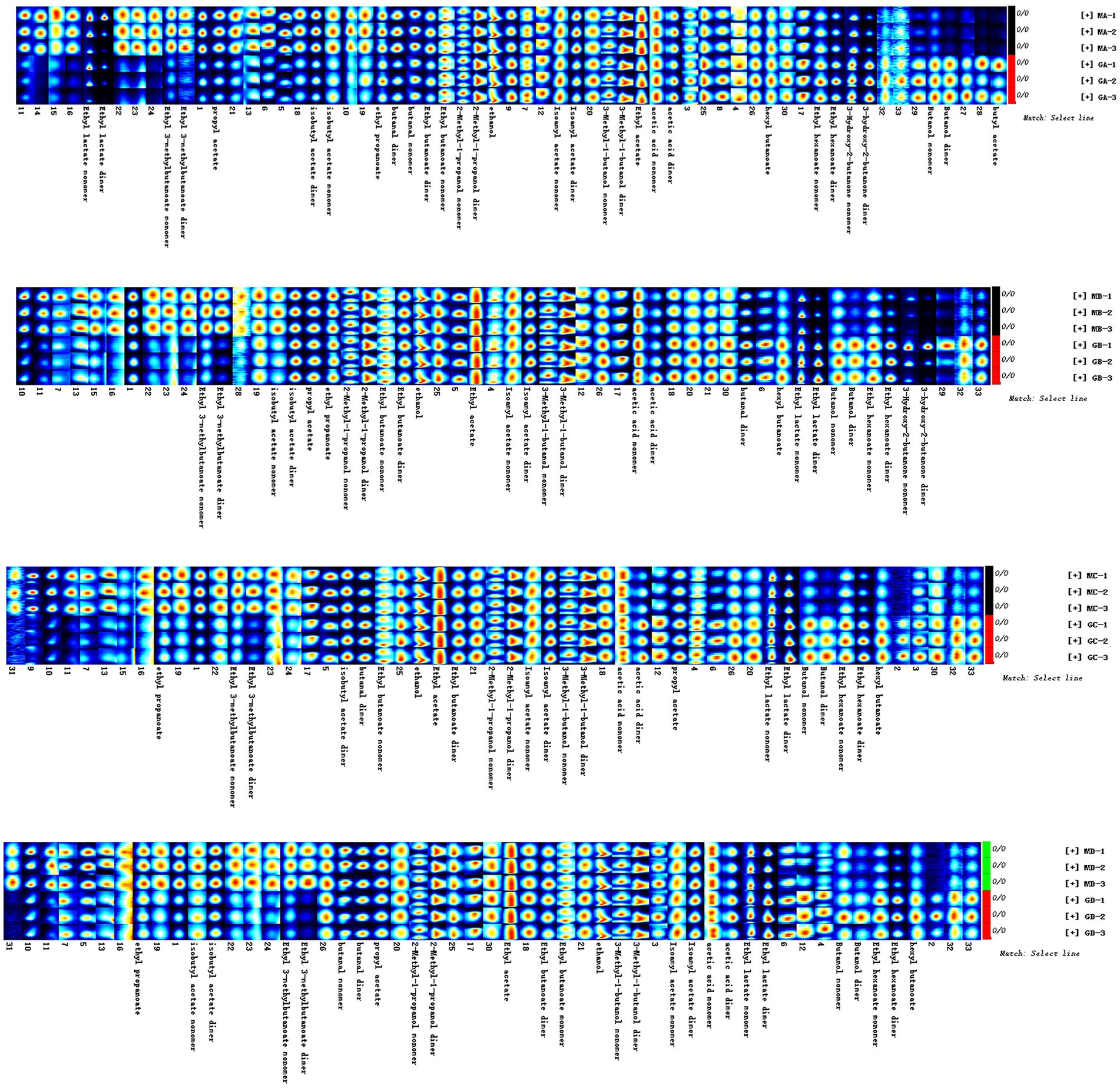
Figure 6. Gallery plot of the volatile flavor compounds based on the gas phase ion mobility spectrum. The numbers represent unidentified compounds in the mobility library.
In addition, as shown in Figure 7, the abundance of compounds in areas 1 and 3 was higher than that in other samples, and these compounds could be used as characteristic flavor compounds in MA and MD samples, respectively. In area 2, the levels of compounds such as ethyl propionate, ethyl lactate, acetic acid, and ethyl butyrate gradually increased and attained a maximum abundance during fermentation. In area 4, compound abundance remained relatively constant during fermentation. Flavor compounds in MA and MD samples were significantly different, whereas those in MB and MC samples were relatively similar. In area 5, the abundance of compounds such as butyl acetate, 3-hydroxy-2-butanone, and butyraldehyde was significantly higher in GA samples than in GB–GD samples. In area 6, compound abundance initially increased and then decreased. However, in area 7, the abundance of compounds such as ethyl 3-methylbutyrate, ethyl caproate, ethyl butyrate, ethyl propionate, ethyl lactate, propyl acetate, isoamyl acetate, and isobutyl acetate, gradually increased during fermentation. In area 8, compound abundance remained relatively constant throughout the fermentation process.
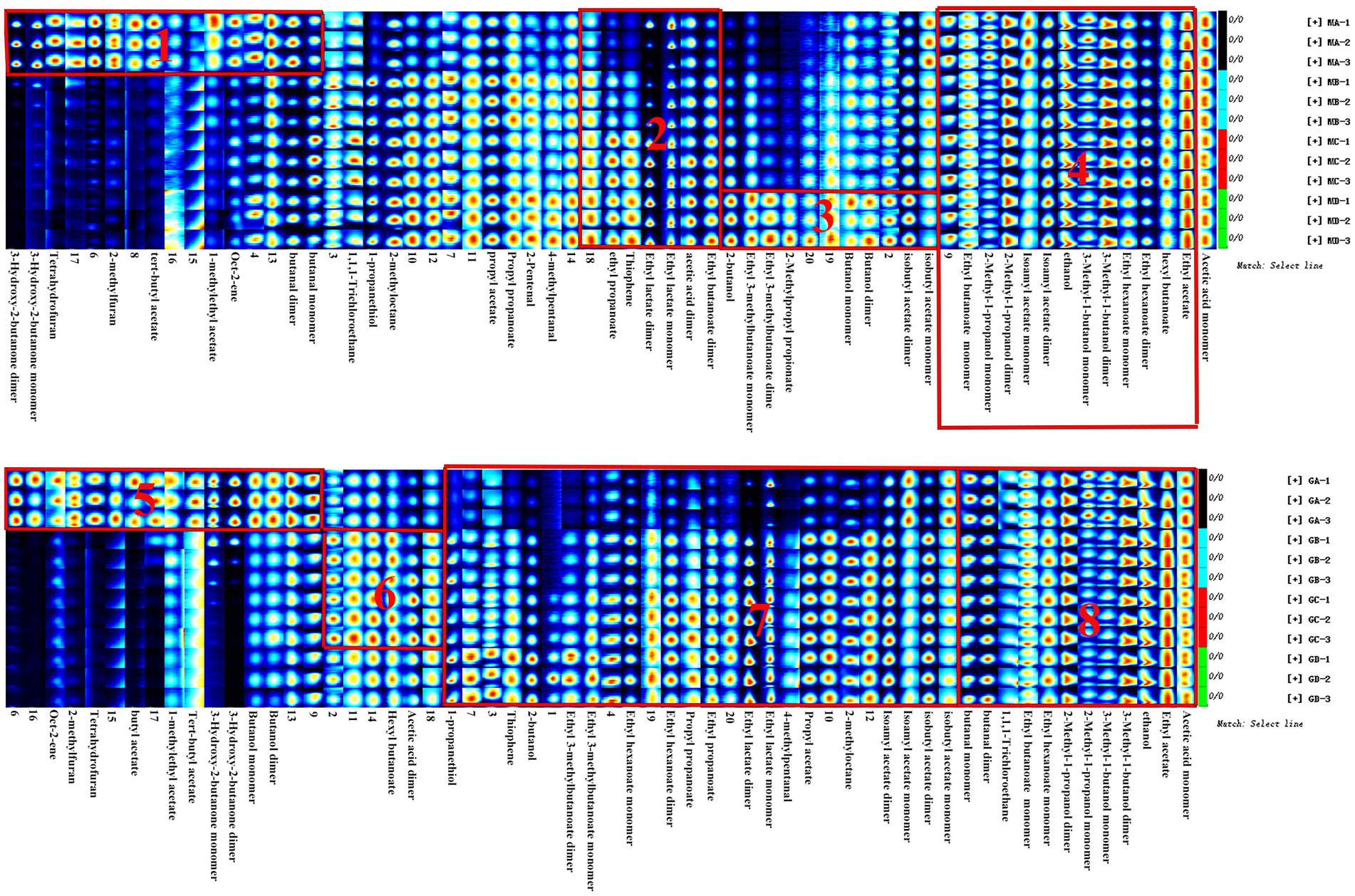
Figure 7. Gallery plot of volatile flavor compounds based on the gas phase ion mobility spectrum at different stages of proso millet and sorghum fermentation. The numbers represent unidentified compounds in the mobility library.
3.3 Relationship between microbial composition changes and flavor compound formation
The RDA analysis result showed that bacteria from the Weissella, Acinetobacter, Bacteroides, Psychrobacter, Pseudarthrobacter, Lactococcus, Chloroplast, Saccharopolyspora, Psychrobacter, Saccharopolyspora, Pseudonocardiaceae, and Bacteroides genera and fungi from the Thermoascus, Aspergillus, Pichia, Rhizomucor, Papiliotrema, Hyphopichia, and Mucor genera significantly inhibited the synthesis of the flavor compounds, such as ethyl hexanoate, ethyl butanoate, ethyl lactate ethyl lactate, and butyl acetate, but improved the abundance of ethyl acetate (p < 0.05) (Figures 8, 9). Moreover, these microbes exhibited a greater abundance in proso millet–sorghum mixed fermentation samples than in sorghum samples. The synthesis of special flavor compounds in proso millet Baijiu was significantly promoted in the presence of fungi from the Rhizopus, Papiliotrema, Wickerhamomyces, Aspergillus, and Thermoascus genera but inhibited by bacteria from the Weissella, Acinetobacter, Psychrobacter, Pseudarthrobacter, Bacteroides, and Saccharopolyspora genera (Figures 8, 9).
Among the 40 fungal and bacterial species with the highest abundance, all bacterial and 30 fungal species were found to be significantly associated with changes in the abundance of flavor compounds (Supplementary Table S2). More specifically, among these 30 fungal species, P. fermentans, C. tropicalis, D. geotrichum, S. inaequale, P. membranifaciens, C. curvatus, C. ethanolica, and R. arrhizus played significant roles in altering the abundance of flavor compounds, and were significantly correlated with changes in the abundance of 18, 18, 18, 15, 13, 11, 10, and 9 types of flavor compounds, respectively. In addition, eight main fungal species exerted a significant effect on changes in the abundance of 41 compounds, with the exception of the ethyl 3-methyl butanoate monomer, the ethyl 3-methyl butanoate dimer, 2-methylfuran, and thiophene. The ethyl 3-methyl butanoate monomer and the ethyl 3-methyl butanoate dimer were found to be significantly positively correlated with the presence of F. solani and P. candolleana (p < 0.05). In addition, the abundance of 2-methylfuran was significantly positively correlated with F. solani abundance (correlation coefficient R = 0.475), but negatively correlated with R. arrhizus abundance (correlation coefficient R = −0.443) (p < 0.05). Thiophene abundance was significantly positively correlated with P. fermentans, T. pullulans, and C. ethanolica abundance (p < 0.01), with correlation coefficients of 0.701, 0.600, and 0.741, respectively; however, thiopene abundance was negatively correlated with G. baccata abundance (p < 0.05) (correlation coefficient R = −0.431).
Among the 40 most significant bacterial species, L. oligofermentans, L. acetotolerans, L. coryniformis, W. paramesenteroides, L. mali, E. durans, L. paralimentarius, L. vaccinostercus, L. mesenteroides, L. homohiochii, L. fallax, W. viridescens, L. fermentum, L. suebicus, L. sakei, and L. citreum were the main species the abundance of which was significantly correlated with changes in the abundance of flavor compounds, with these species inducing significant changes in the abundance of 31, 30, 30, 29, 29, 29, 28, 28, 28, 27, 27, 27, 26, 26, 25, and 25 types of flavor compounds, respectively. Fifteen of these main bacterial species exerted significant positive or negative effects on 44 flavor compounds, except for 2-methylpropyl propionate. Furthermore, the abundance of 2-methylpropyl propionate was significantly negatively correlated with the presence of L. buchneri (R = −0.406; p < 0.05). Furthermore, the formation of this flavor compound was significantly associated with the presence of the fungal species, F. solani, P. candolleana, M. elongat, S. inaequale, and P. leptospora, with correlation coefficients of 0.631 (p < 0.01), 0.741 (p < 0.01), 0.512 (p < 0.05), 0.690 (p < 0.01), and 0.453 (p < 0.05), respectively.
Furthermore, based on a redundancy analysis and Pearson’s correlation coefficients (Figures 8, 9), we found that the relative abundance of esters was significantly correlated with the presence of fungal species such as P. fermentans, A. ruber, C. tropicalis, R. arrhizus, A. flavus, P. membranifaciens, O. brassicae, M. circinelloides, F. solani, B. atrogriseum, S. podzolica, T. dohaense, H. nigrescens, T. pullulans, C. ethanolica, P. candolleana, D. geotrichum, M. elongata, G. baccata, F. magnum, A. fumigatus, C. curvatus, O. truncatum, L. fimeti, and S. inaequale. In addition, the abundance of esters was also correlated with the presence of 38 bacterial species, excluding L. parafarraginis and P. pacificensis. Changes in the relative abundance of alcohols were significantly correlated with the presence of several fungal species, including P. fermentans, A. ruber, C. tropicalis, R. arrhizus, A. flavus, P. membranifaciens, O. brassicae, F. solani, B. atrogriseum, S. podzolica, C. ethanolica, P. candolleana, D. geotrichum, P. flavescens, F. magnum, C. curvatus, O. truncatum, and S. inaequale. Changes in alcohol abundance were also correlated with the presence of 37 bacterial species, excluding L. parafarraginis, P. pacificensis, and S. succinus. Hydrocarbon abundance was significantly correlated with the presence of specific fungal species, including P. fermentans, C. tropicalis, R. arrhizus, A. flavus, P. membranifaciens, O. brassicae, B. atrogriseum, T. pullulans, C. ethanolica, G. baccata, and C. curvatus. In addition, hydrocarbon abundance was correlated with the presence of 37 bacterial species, excluding L. parafarraginis, P. pacificensis, and S. succinus. Heterocyclic compound abundance was significantly correlated with the presence of the fungal species, D. geotrichum and S. inaequale, as well as that of 34 bacterial species, excluding L. hokkaidonensis, L. parafarraginis, P. pacificensis, S. succinus, P. pentosaceus, and B. vulgatus. Aldehyde abundance was significantly correlated with the presence of fungal species such as P. fermentans, T. aurantiacus, R. arrhizus, P. membranifaciens, F. solani, M. tassiana, T. dohaense, H. nigrescens, T. pullulans, C. ethanolica, G. baccata, and C. curvatus. In addition, aldehyde abundance was correlated with the presence of 36 bacterial species, excluding L. parafarraginis, P. pacificensis, L. homohiochii, and B. vulgatus. Ketone abundance was correlated with the presence of the fungal species, C. tropicalis, R. arrhizus, S. podzolica, D. geotrichum, and S. inaequale, as well as that of bacterial species such as L. acetotolerans, P. parvulus, L. mesenteroides, L. coryniformis, P. damnosus, L. homohiochii, W. viridescens, L. oligofermentans, L. rossiae, L. paralimentarius, W. paramesenteroides, L. vaccinostercus, L. sakei, L. mali, E. durans, L. fermentum, Lactobacillus sp., L. sharpeae, L. citreum, L. suebicus, and L. fallax. Organic acid abundance was correlated with the presence of fungal species such as P. fermentans, C. tropicalis, P. membranifaciens, L. ramosa, M. circinelloides, S. podzolica, H. nigrescens, G. baccata, and A. fumigatus, as well as that of bacterial species such as L. brevis, L. parabuchneri, P. inopinatus, L. kefiri, L. homohiochii, W. viridescens, L. malefermentans, L. manihotivorans, L. odoratitofui, L. oligofermentans, L. rossiae, L. hokkaidonensis, L. pontis, S. kloosii, P. ethanolidurans, L. mali, P. pentosaceus, L. fermentum, S. sciuri, L. sharpeae, L. citreum, L. suebicus, and L. fallax.
4 Discussion
Proso millet, a small miscellaneous grain rich in β-carotene and multiple vitamins, is often utilized in the production of rice wine and other viscous foods due to its high viscosity. However, its application in Chinese Baijiu fermentation faces significant challenges. In a previous study, to effectively utilize millet in Chinese Baijiu fermentation, we optimized the fermentation process and successfully developed a new Chinese Baijiu product. Moreover, sensory evaluation indicated significant differences in taste and flavor between the new Chinese Baijiu product and traditional variants. Therefore, this study aimed to describe differences between traditional and new Chinese Baijiu products through the analysis of microbial communities and flavor at various fermentation stages.
4.1 Changes in core microbial communities in proso millet Chinese Baijiu at different fermentation stages
Chinese Baijiu is produced through microbial fermentation and its quality is related to the microbial communities involved in its production. Analysis of changes in microbial communities revealed that the core microbial communities in samples MA–MD differed from those in samples GA–GD. Thus, proso millet may be an effective material for improving the quality of Baijiu. Compared to sorghum samples, proso millet samples exhibited a significant increase in microbial diversity during fermentation, as well as a change in the core microbial community composition. Among the core microbes involved in proso millet Baijiu production, only fungi of the genera, Mucor, Thermoascus, Aspergillus, Rhizomucor, Rhizopus, Pichia, and Candida and bacteria of the genera, Bacillus, Lactobacillus, Lactococcus, Weissella, Staphylococcus, Saccharopolyspora, Streptococcus, and Pediococcus, have been previously reported to be involved in traditional Chinese Baijiu production (Yang et al., 2017; Zou et al., 2018; Gan et al., 2019; Xie et al., 2020). At the species level, aside from the fungal species, L. ramosa, C. tropicalis, A. flavus, M. racemosus, and W. anomalus, and the bacterial species, L. brevis, L. buchneri, P. parvulus, L. odoratitofui, L. citreum, Lactobacillus sp., L. pontis, P. pentosaceus, and L. homohiochii, the rest of the core microbes identified in our study have not been reported in other traditional Chinese Baijiu beverages (Huang et al., 2020; Sakandar et al., 2020; Wu et al., 2021; Xie et al., 2021).
In addition, the core microbes identified at different stages of proso millet Chinese Baijiu fermentation in this study are different from those identified in other traditional Chinese Baijiu beverages. For instance, for Xiaoqu Baijiu (Jing Brand Co., Ltd.), core microbes identified at the beginning of the fermentation process included Leuconostoc lactis, W. paramesenteroides, Lactococcus lactis, Lactobacillus delbrueckii, Gluconacetobacter sacchari, Rhizopus oryzae, and Hypocreales sp., and those identified at the end of the fermentation process included Acetobacter pasteurianus, L. pontis, L. acetotolerans, W. anomalus, Monascus purpureus, C. tropicalis, and A. flavus (Xie et al., 2021). Similarly, in light-flavored Daqu Baijiu (Xinghuacun Fenjiu Distillery Co., Ltd.), the core microbes shifted from species such as P. kudravzerii, L. ramosa, L. citreum, and B. licheniformis on day 1 of fermentation to L. acetotolerans, L. buchneri, and L. hilgardii at the end of fermentation (Huang et al., 2020). In Chinese strong-flavored Baijiu, the main microbes identified during the early stages of fermentation (days 1–23) included Lactobacillus, Thermoascus, Aspergillus, Emericella, Monascus, Candida, unidentified Saccharomycetales, Rhizopus, and Pichia species; however, during the mid-stage of fermentation (days 23–48), Thermoascus, Aspergillus, Emericella, Candida, and unidentified Saccharomycetales species were the primary microbes identified (Zhang et al., 2017). In MA samples, at the beginning of fermentation, the main core fungal species identified included F. solani, M. elongata, P. oratosquillae, P. leptospora, P. candolleana, and S. inaequale (abundance >1%), and the primary bacterial species identified included L. vaccinostercus, L. mesenteroides, L. mali, L. suebicus, L. sakei, L. fallax, W. viridescens, L. fermentum, L. paralimentarius, and W. paramesenteroides. The core microbes identified in MB and MC samples during mid-stage fermentation included S. inaequale, H. nigrescens, D. geotrichum, C. tropicalis, T. dohaense, M. circinelloides, S. podzolica, P. roseus, G., L. ramose, S. filum, O. truncatum, L. parafarraginis, L. buchneri, B. vulgatus, L. malefermentans, L. carnosum, P. inopinatus, and L. hokkaidonensis, and core microbes identified in MD samples during the end of fermentation included C. purpureofuscum, F. magnum, A. ruber, R. pusillus, A. flavus, R. arrhizus, P. flavescens, W. anomalus, L. homohiochii, and P. pentosaceus (Figures 2, 3). These differences in core microbial communities across different Baijiu products highlight the impact of fermentation raw materials, technology, and Daqu on microbial communities.
4.2 Flavor compound formation in proso millet Chinese Baijiu and differences between proso millet Chinese Baijiu and other products
Compared to sorghum fermentation, proso millet fermentation induced a change in the core flavor compounds of traditional light-flavored Baijiu from ethyl acetate, ethyl hexanoate, ethyl hexanoate dimer, ethyl butanoate, ethyl lactate, and butyl acetate to oct-2-ene, 2-butanol, propyl propanoate, 2-pentenal, and 4-methylpentanal. However, further evaluation is required to determine the contribution of each volatile flavor compound to the overall flavor of Baijiu. In this study, we detected a total of 22 esters, 9 alcohols, 4 hydrocarbons, 3 heterocyclic compounds, 3 aldehydes, 2 ketones, and 2 organic acids in proso millet Chinese Baijiu through GC-IMS. Based on their relative peak areas (above 1800), the key flavor compounds in proso millet Chinese Baijiu were identified to be ethyl acetate (A9), ethyl butanoate dimer (A11), ethyl lactate monomer (A14), ethyl lactate dimer (A15), isoamyl acetate monomer (A18), isoamyl acetate dimer (A19), isobutyl acetate monomer (A20), 2-methyl-1-propanol monomer (B3), 3-methyl-1-butanol monomer (B5), 3-methyl-1-butanol dimer (B6), 2-methyloctane (C1), butanal monomer (E2), 3-hydroxy-2-butanone dimer (F1), 3-hydroxy-2-butanone monomer (F2), acetic acid monomer (G1), and acetic acid dimer (G2) (Figure 7; Table 2).
However, other traditional Chinese Baijiu formulas have different flavor compound compositions. For instance, strong-aroma Baijiu contains 11 main flavor compounds: ethyl hexanoate, ethyl acetate, ethyl lactate, ethyl butyrate, ethyl valerate, ethyl heptanoate, hexanoic acid, butanoic acid, heptanoic acid, furfural, and phenylethyl alcohol. Light-aroma Baijiu mainly contains 6 compounds: ethyl acetate, ethyl lactate, acetic acid, 2-methylpropanoic acid, β-damascenone, and terpenoids. Ten main flavor compounds (ethyl hexanoate, hexanoic acid, 3-methylbutanoic acid, 3-methylbutanol, tetramethylpyrazine, ethyl 2-phenylacetate, 2-phenylethyl acetate, ethyl 3-phenylpropanoate, 4-methylguaiacol, and γ-decalactone) were detected in sauce-aroma Baijiu. Ethyl acetate, ethyl caproate, and isoamyl alcohol were identified as the main flavor compounds in feng-aroma Baijiu (Liu et al., 2018; Fang et al., 2019). In addition, 1874 types of volatile flavor compounds have been identified in Chinese spirits, and Fenjiu alone is marketed in nine series and 113 products, graded by flavor and quality level. The new product, proso millet Baijiu, stands out as it contains significantly different flavor compounds as compared to other light-flavored Baijiu products; these include the ethyl butanoate dimer (A11), ethyl lactate monomer (A14), ethyl lactate dimer (A15), isoamyl acetate monomer (A18), isoamyl acetate dimer (A19), 2-methyloctane (C1), butanal monomer (E2), 3-hydroxy-2-butanone dimer (F1), and 3-hydroxy-2-butanone monomer (F2), which are not found in other Fenjiu products (Li et al., 2020). Furthermore, the relative abundance of the same flavor compounds varies between different Chinese spirits, indicating the distinct flavor characteristics of Fenjiu series products, including proso millet light-flavored Baijiu, as compared to other Baijiu formulas (Li et al., 2020; Wang J.-Y. et al., 2020). Studies have shown that raw materials, Daqu, and the fermentation process may be the primary parameters underlying flavor differences in different products. Thus, proso millet Baijiu is a new light-flavored Baijiu product and an effective material for enhancing the flavor of Baijiu. Moreover, modifying the fermentation and Daqu-making technology is an important approach for improving the flavor of Baijiu. Finally, although 45 flavor compounds were identified through GC-IMS, 20 other flavor compounds need to be further identified through alternative detection methods.
4.3 Relationship between core microbes and flavor compound formation
The RDA analysis showed that proso millet induced a change in the core flavor compounds of sorghum Baijiu by increasing the abundance of flavor-related microbes, such as bacteria from the Weissella, Acinetobacter, Bacteroides, Psychrobacter, Pseudarthrobacter, Lactococcus, Chloroplast, Saccharopolyspora, Psychrobacter, Saccharopolyspora, Pseudonocardiaceae, and Bacteroides genera and fungi from the Thermoascus, Aspergillus, Pichia, Rhizomucor, Papiliotrema, Hyphopichia, and Mucor genera, the abundance of which was significantly greater in proso millet–sorghum mixed fermentation samples than in sorghum samples. In addition, among the core fungal species, F. solani was significantly positively correlated with the formation of the main flavor compound, A20. Furthermore, P. candolleana was significantly positively correlated with the formation of the main flavor compound, B3. S. inaequale was significantly negatively correlated with the formation of the main flavor compound, A9, but was significantly positively correlated with the formation of the main flavor compounds, B3, B4, B6, F1, and F2. Furthermore, T. aurantiacus and M. tassiana were significantly negatively correlated with the formation of the main flavor compound, E2. In addition, A. ruber was significantly positively correlated with the formation of the main flavor compounds, A11 and B9. R. arrhizus was significantly negatively correlated with the formation of the main flavor compound, C1, but was significantly positively correlated with the formation of the main flavor compound, F1. H. nigrescens was significantly negatively correlated with the formation of the main flavor compounds, E2 and G2. Furthermore, D. geotrichum was significantly negatively correlated with the formation of the main flavor compounds, A9, A15, A18, and A20, but was significantly positively correlated with the formation of the main flavor compounds, A19, B4, B6, and F2. C. tropicalis was significantly negatively correlated with the formation of the main flavor compounds, A9, A14, A18, A20, C1, G1, and G2, but was significantly positively correlated with the formation of the main flavor compounds, B4, B6, F1, and F2. T. dohaense was significantly negatively correlated with the formation of the main flavor compounds, A15 and E2. M. circinelloides was significantly negatively correlated with the formation of the main flavor compounds, A14, A15, and G2. S. podzolica was significantly negatively correlated with the formation of the main flavor compound, G2, but was significantly positively correlated with the formation of the main flavor compound, F1. G. baccata and A. fumigatus were significantly positively correlated with the formation of the main flavor compound, G1. L. ramosa was significantly negatively correlated with the formation of the main flavor compound, G2. O. truncatum was significantly positively correlated with the formation of the main flavor compounds, B5 and B9. F. magnum was significantly negatively correlated with the formation of the main flavor compounds, A19 and B3. Furthermore, B. atrogriseum was significantly negatively correlated with the formation of the main flavor compound, B3, but was significantly positively correlated with the formation of the main flavor compound, A14. P. fermentans was significantly negatively correlated with the formation of the main flavor compounds, A14, B9, and G1, but was significantly positively correlated with the formation of the main flavor compounds, A19 and G2. C. curvatus was significantly negatively correlated with the formation of the main flavor compound, A11, but was significantly positively correlated with the formation of the main flavor compound, E2. T. pullulans was significantly negatively correlated with the formation of the main flavor compound, A14.
Among the core bacterial species, L. acetotolerans was significantly negatively correlated with the formation of the main flavor compounds, A19, B4, B6, E2, F1, and F2, but was significantly positively correlated with the formation of the main flavor compounds, A9, A15, A18, A20, and C1. L. homohiochii was significantly negatively correlated with the formation of the main flavor compounds, A19, B3, B4, B6, E2, F1, and F2, but was significantly positively correlated with the formation of the main flavor compounds, A9, A14, A15, A18, A20, C1, and G1. P. pentosaceus was significantly positively correlated with the formation of the main flavor compounds, A14, A15, B9, and G1. P. parvulus was significantly positively correlated with the formation of the main flavor compounds, A9, A19, A20, B4, B6, E2, and F1. P. damnosus was significantly negatively correlated with the formation of the main flavor compounds, A9, A19, and C1, but was significantly positively correlated with the formation of the main flavor compounds, A19, B4, B6, E2, and F2. L. coryniformis was significantly negatively correlated with the formation of the main flavor compounds, A9, A14, A15, A18, A20, B5, B9, and C1, but was significantly positively correlated with the formation of the main flavor compounds, A19, B4, B6, E2, F1, and F2. L. oligofermentans was significantly negatively correlated with the formation of the main flavor compounds, A14, A15, A18, A20, B5, B9, C1, and G1, but was significantly positively correlated with the formation of the main flavor compounds, A9, A19, B4, B6, E2, F1, and F2. L. buchneri and B. vulgatus were significantly positively correlated with the formation of the main flavor compounds, A11, B5, and B9. Furthermore, L. malefermentans and L. hokkaidonensis were significantly positively correlated with the formation of the main flavor compounds, A11, A14, B5, B9, and G1. L. carnosum was significantly positively correlated with the formation of the main flavor compound, A11, but was negatively correlated with the formation of the main flavor compound, C1. P. inopinatus was significantly positively correlated with the formation of the main flavor compounds, A11, B9, and G1. E. durans was significantly positively correlated with the formation of the main flavor compounds, A19, B4, B6, F1, and F2, but was negatively correlated with the formation of the main flavor compounds, A9, A18, A20, and C1. L. vaccinostercus and L. mesenteroides were significantly positively correlated with the formation of the main flavor compounds, A11, B3, B4, B6, F1, and F2, but were negatively correlated with the formation of the main flavor compounds, A9, A18, A20, and C1. L. mali was significantly positively correlated with the formation of the main flavor compounds, B3, B4, B6, F1, and F2, but was negatively correlated with the formation of the main flavor compounds, A9, A18, A20, C1, and G2. L. suebicus was significantly positively correlated with the formation of the main flavor compounds, B3, B6, F1, and F2, but was negatively correlated with the formation of the main flavor compounds, A9, A18, A20, C1, and G2. L. sakei was significantly positively correlated with the formation of the main flavor compounds, B4, B6, F1, and F2, but was negatively correlated with the formation of the main flavor compounds, A9, A15, A18, A20, and C1. W. viridescens, W. paramesenteroides, L. fermentum, L. paralimentarius, and L. fallax were significantly positively correlated with the formation of the main flavor compounds, B4, B6, F1 and F2, but were negatively correlated with the formation of the main flavor compounds, A9, A15, A18, A20, C1, and G2.
In 2017, FG samples were collected from Shanxi Xinghuacun Fenjiu Distillery Co., Ltd. (Fenyang, China) during light-flavored Baijiu production (Huang et al., 2020), and L. brevis was found to be positively correlated with A9 formation; however, L. paralimentarius, Hanseniaspora uvarum, and Bacillus licheniformis were negatively correlated with A9 formation. In addition, L. buchneri, L. odoratitofui, and L. parabuchneri were positively correlated with A14 formation. L. brevis was positively correlated with A20 and B3 formation. Wu et al., 2021 found that esters present in Chinese liquor were produced by A. oryzae, S. cerevisiae, S. pombe, S. fibuligera, and W. anomalus, and that the main alcohol- and acid-producing microbes were A. oryzae, A. hennebergii, S. cerevisiae, Z. bailii, P. kudriavzevii, S. pombe, L. homohiochii, and L. buchneri. Furthermore, for the production of strong-flavored Baijiu, the most common hexanoate-producing Clostridium species identified included C. kluyveri, C. lushun, Clostridium spp. W1, and C. celerecrescens. Ethyl lactate-, ethyl acetate-, and ethyl butanoate-producing microbes in the strong-flavored Baijiu ecosystem were identified to be Corynebacterium xerosis, Staphylococcus auricularis, Bacillus subtilis, Bacillus megaterium, Bacillus cereus group, and Paenibacillus macerans (Zou et al., 2018).
4.4 Effect of proso millet on ethanol production and the abundance of key microbial communities for ethanol synthesis
Ethanol concentration is a direct reflection of the Baijiu yield and is significantly affected by sorghum variety and the microbial communities present during fermentation. A comparison of sorghum fermentation and mixed fermentation showed no significant difference in ethanol concentration at the final fermentation stage (28 days); however, ethanol concentration at the fermentation stages on days 14 and 21 were significantly lower for sorghum fermentation than for mixed fermentation (Table 2). The RDA analysis results showed that the presence of fungal species of the genera Olipidium was significantly positively correlated with ethanol production; however, the presence of fungi from the Psathyrella and Schizothecium genera was significantly negatively correlated with ethanol production (Supplementary Table S2). In total, 12 bacterial genera were significantly negatively correlated with ethanol production, with the negative correlation coefficient for the genus, Brachybacterium, being higher than that for other genera (−0.582; p < 0.01). In addition, the relative abundance of the bacteria of the genus, Olipidium, in mixed fermentation samples was lower than that in sorghum samples; however, the relative abundance of microbes that inhibit ethanol synthesis was higher in mixed fermentation samples than in sorghum samples (Figures 2, 3). Therefore, we speculated that proso millet could improve the flavor of light-flavored Baijiu but could not improve its yield. Furthermore, L. buchneri, P. inopinatus, L. malefermentans, L. hokkaidonensis, P. pentosaceus, B. vulgatus, A. ruber, and O. truncatum played important roles in ethanol production in proso millet Baijiu. Conversely, during the production of light-flavored Xiaoqu Baijiu, ethanol production was significantly positively correlated with the presence of the microbe, Saccharomyces cerevisiae, and negatively correlated with the presence of A. pasteurianus (Xie et al., 2021). L. ramosa, Saccharomycopsis fibuligera, B. licheniformis, S. cerevisiae, and Pichia kudriavzevii play significant roles in starch degradation and ethanol production (Huang et al., 2020). Furthermore, S. cerevisiae, Z. bailii, and S. pombe were found to play important roles in ethanol production in Daqu Chinese Baijiu (Wu et al., 2021); in addition, S. cerevisiae was the main ethanol-producing microbe identified during zaopei fermentation (Zou et al., 2018).
While the effects of proso millet on dynamic changes in microbial communities and flavor during different stages of light-flavored Baijiu fermentation using the light-flavored Baijiu technology of Xinghua Village, Shanxi, have been studied and some key microorganisms closely related to flavor changes identified, several important aspects still require further research. These include determining the optimal quantity of proso millet to be added, selecting superior proso millet varieties, investigating the interaction between proso millet quality and Baijiu flavor, and determining whether the addition of key flavor microorganisms can further enhance Baijiu quality. In addition, differences in flavor analysis results determined using GC × GC-TOF-MS and GC-IMS need to be further investigated.
5 Conclusion
In this study, we evaluated the changes in microbial communities and volatile flavor compounds in proso millet Baijiu, a new Baijiu product of Shanxi light-flavored Baijiu, as well as the relationship between them. Proso millet significantly changed the main flavor compound composition of light-flavored Baijiu from ethyl acetate, ethyl hexanoate, ethyl hexanoate dimer, ethyl butanoate, ethyl lactate, and butyl acetate to oct-2-ene, 2-butanol, propyl propanoate, 2-pentenal, and 4-methylpentanal. In addition, proso millet significantly changed the microbial communities in light-flavored Baijiu during fermentation, particularly during the early stages of fermentation (0–14 days). Furthermore, the formation of special flavor compounds in proso millet Baijiu was significantly correlated with the presence of fungi from the Rhizopus, Papiliotrema, Wickerhamomyces, Aspergillus, and Thermoascus genera but negatively correlated with the presence of bacteria from the Weissella, Acinetobacter, Psychrobacter, Pseudarthrobacter, Bacteroides, and Saccharopolyspora genera. The low alcohol production rate observed in Fenjiu may be due to the presence of fungi from Psathyrella genus and bacteria from the Staphylococcus, Kroppenstedtia, Brevibacterium, and Acetobacter genera, which exhibit a significantly high abundance during fermentation. Proso millet significantly changed the flavor of light-flavored Baijiu by inducing the formation of special microbial communities but did not increase alcohol concentration. This study lays a foundation for future research on improving the flavor of light-flavored Baijiu by modifying fermentation materials.
Data availability statement
The original contributions presented in the study are publicly available. This data can be found at: https://www.ncbi.nlm.nih.gov/, PRJNA1014983, PRJNA1015004, PRJNA1013705, PRJNA1014132.
Author contributions
JZ: Conceptualization, Data curation, Formal analysis, Funding acquisition, Methodology, Project administration, Writing – original draft, Writing – review & editing. ZG: Conceptualization, Data curation, Investigation, Methodology, Project administration, Supervision, Writing – review & editing.
Funding
The author(s) declare financial support was received for the research, authorship, and/or publication of this article. This study was supported financially by the Shanxi Provincial Higher Education Science and Technology Innovation Project (2022L498), the Shanxi Province Science Foundation for Youths (20210302124512), the Doctoral Foundation of Shanxi Academy of Agricultural Sciences (YBSJJ2001, YBSJJ2002, and YBSJJ2013), the Agricultural Science and Technology Innovation Research Project of Shanxi Academy of Agricultural Sciences (YCX2020BH3), and an earmarked fund for the China Agriculture Research System (CARS-06-14.5-A30).
Acknowledgments
The authors thank Editage for language editing.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2024.1333466/full#supplementary-material
References
Agarwal, S., and Chauhan, E. S. (2019). The Nutritional Composition, various processing and health benefits of Proso Millet: A Review. RJPT 12, 4013–4017. doi: 10.5958/0974-360X.2019.00691.7
Balkrishna, S. P., and Visvanathan, R. (2019). Hydration kinetics of little millet and proso millet grains: effect of soaking temperature. J. Food Sci. Technol. 56, 3534–3539. doi: 10.1007/s13197-019-03789-4
Bian, X., Chen, J. R., Yang, Y., Yu, D. H., Ma, Z. Q., Ren, L. K., et al. (2022). Effects of fermentation on the structure and physical properties of glutinous proso millet starch. Food Hydrocoll. 123:107144. doi: 10.1016/j.foodhyd.2021.107144
Caporaso, J. G., Kuczynski, J., Stombaugh, J., Bittinger, K., Bushman, F. D., Costello, E. K., et al. (2010). QIIME allows analysis of high-throughput community sequencing data. Nat. Methods 7, 335–336. doi: 10.1038/nmeth.f.303
Chang, L., Zhao, N., Jiang, F., Ji, X., Feng, B., Liang, J., et al. (2023). Structure, physicochemical, functional and in vitro digestibility properties of non-waxy and waxy proso millet starches. Int. J. Biol. Macromol. 224, 594–603. doi: 10.1016/j.ijbiomac.2022.10.149
Elokil, A. A., Abouelezz, K. F. M., Ahmad, H. I., Pan, Y., and Li, S. (2020). Investigation of the impacts of antibiotic exposure on the diversity of the gut microbiota in chicks. Animals 10:896. doi: 10.3390/ani10050896
Fang, C., Du, H., Zheng, X., Zhao, A., Jia, W., and Xu, Y. (2019). Solid-state fermented Chinese alcoholic beverage (Baijiu) and ethanol resulted in distinct metabolic and microbiome responses. FASEB J. 33, 7274–7288. doi: 10.1096/fj.201802306R
Gan, S. H., Yang, F., Sahu, S. K., Luo, R. Y., Liao, S. L., Wang, H. Y., et al. (2019). Deciphering the composition and functional profile of the microbial communities in Chinese Moutai liquor starters. Front. Microbiol. 10:1540. doi: 10.3389/fmicb.2019.01540
Gong, X., Dang, K., Liu, L., Zhao, G., Lv, S., Tian, L., et al. (2021). Intercropping combined with nitrogen input promotes proso millet (Panicum miliaceum L.) growth and resource use efficiency to increase grain yield on the loess plateau of China. Agric. Water Manag. 243:106434. doi: 10.1016/j.agwat.2020.106434
Gulati, P., Li, A., Holding, D., Santra, D., Zhang, Y., and Rose, D. J. (2017). Heating reduces proso millet protein digestibility via formation of hydrophobic aggregates. J. Agric. Food Chem. 65, 1952–1959. doi: 10.1021/acs.jafc.6b05574
Gulati, P., Zhou, Y., Elowsky, C., and Rose, D. J. (2018). Microstructural changes to proso millet protein bodies upon cooking and digestion. J. Cereal Sci. 80, 80–86. doi: 10.1016/j.jcs.2018.02.001
Hasseldine, B. P. J., Gao, C., and Li, Y., (2017). “Damage initiation and evolution of Panicum Miliaceum seeds under compression”, in ASME 2017 International Mechanical Engineering Congress and Exposition.
Huang, X., Fan, Y., Lu, T., Kang, J., Pang, X., Han, B., et al. (2020). Composition and metabolic functions of the microbiome in fermented grain during light-flavor Baijiu fermentation. Microorganisms 8:1281. doi: 10.3390/microorganisms8091281
Huang, Y., Kuang, Z., Wang, W., and Cao, L. (2016). Exploring potential bacterial and fungal biocontrol agents transmitted from seeds to sprouts of wheat. Biol. Control 98, 27–33. doi: 10.1016/j.biocontrol.2016.02.013
Jiang, X.-T., Zhang, H., Sheng, H.-F., Wang, Y., He, Y., Zou, F., et al. (2012). Two-stage clustering (TSC): a pipeline for selecting operational taxonomic units for the high-throughput sequencing of PCR amplicons. PLoS One 7:e30230. doi: 10.1371/journal.pone.0030230
Johnson, M., Deshpande, S., Vetriventhan, M., Upadhyaya, H. D., and Wallace, J. G. (2019). Genome-wide population structure analyses of three minor millets: Kodo millet, little millet, and proso millet. Plant Genome 12, 1–9. doi: 10.3835/plantgenome2019.03.0021
Kalam Azad, M. O., Jeong, D. I., Adnan, M., Salitxay, T., Heo, J. W., Naznin, M. T., et al. (2019). Effect of different processing methods on the accumulation of the phenolic compounds and antioxidant profile of broomcorn millet (Panicum miliaceum L.) flour. Foods 8, 8:230. doi: 10.3390/foods8070230
Kang, C., Luo, F., and Sun, S. (2020). Research status and prospect of QTL mapping for tillers and other traits in forage sorghum. Mol Plant Breed. 11:18. doi: 10.5376/mpb.cn.2020.18.0018
Kelly, B. J., Gross, R., Bittinger, K., Sherrill-Mix, S., Lewis, J. D., Collman, R. G., et al. (2015). Power and sample-size estimation for microbiome studies using pairwise distances and PERMANOVA. Bioinformatics 31, 2461–2468. doi: 10.1093/bioinformatics/btv183
Kim, J.-S., Hyun, T. K., and Kim, M.-J. (2011). The inhibitory effects of ethanol extracts from sorghum, foxtail millet and proso millet on α-glucosidase and α-amylase activities. Food Chem. 124, 1647–1651. doi: 10.1016/j.foodchem.2010.08.020
Kučera, L., Peška, J., Fojtík, P., Barták, P., Kučerová, P., Pavelka, J., et al. (2019). First direct evidence of broomcorn millet (Panicum miliaceum) in Central Europe. Archaeol. Anthropol. Sci. 11, 4221–4227. doi: 10.1007/s12520-019-00798-4
Kumar, S. R., Tangsrianugul, N., Sriprablom, J., Wongsagonsup, R., Wansuksri, R., and Suphantharika, M. (2023). Effect of heat-moisture treatment on the physicochemical properties and digestibility of proso millet flour and starch. Carbohydr. Polym. 307:120630. doi: 10.1016/j.carbpol.2023.120630
Li, M., Fan, W., and Xu, Y. (2020). Volatile compounds sorption during the aging of Chinese liquor (Baijiu) using pottery powder. Food Chem. 345:128705. doi: 10.1016/j.foodchem.2020.128705
Li, J., Li, Z., Deng, X., Song, C., Raghavan, V., and Xie, Y. (2020). A flavor uniformity evaluation and improvement of Chinese spirit by electronic nose. Food Sci. Nutr. 8, 1509–1521. doi: 10.1002/fsn3.1436
Li, S., Qi, Y., Chen, L., Qu, D., Li, Z., Gao, K., et al. (2019). Effects of Panax ginseng polysaccharides on the gut microbiota in mice with antibiotic-associated diarrhea. Int. J. Biol. Macromol. 124, 931–937. doi: 10.1016/j.ijbiomac.2018.11.271
Lin, Z., Ye, W., Zu, X., Xie, H., Li, H., Li, Y., et al. (2018). Integrative metabolic and microbial profiling on patients with spleen-yang-deficiency syndrome. Sci. Rep. 8:6619. doi: 10.1038/s41598-018-24130-7
Liu, C., Gong, X., Wang, H., Dang, K., Deng, X., and Feng, B. (2020). Low-nitrogen tolerance comprehensive evaluation and physiological response to nitrogen stress in broomcorn millet (Panicum miliaceum L.) seedling. Plant Physiol. Biochem. 151, 233–242. doi: 10.1016/j.plaphy.2020.03.027
Liu, H., and Sun, B. (2018). Effect of fermentation processing on the flavor of Baijiu. J. Agric. Food Chem. 66, 5425–5432. doi: 10.1021/acs.jafc.8b00692
Liu, J., Zhao, W., Li, S., Zhang, A., Zhang, Y., and Liu, S. (2018). Characterization of the key aroma compounds in proso millet wine using headspace solid-phase microextraction and gas chromatography-mass spectrometry. Molecules 23:462. doi: 10.3390/molecules23020462
Ma, Y., Qiao, H., Wang, W., Chen, T., Du, X., Zhai, X., et al. (2013). Variations in physicochemical properties of Chinese Fenjiu during storage and high-gravity Technology of Liquor Aging. Int. J. Food Prop. 17, 923–936. doi: 10.1080/10942912.2012.678536
McSweeney, M. B., Ferenc, A., Smolkova, K., Lazier, A., Tucker, A., Seetharaman, K., et al. (2017a). Glycaemic response of proso millet-based (Panicum miliaceum) products. Int. J. Food Sci. Nutr. 68, 873–880. doi: 10.1080/09637486.2017.1301890
McSweeney, M. B., Seetharaman, K., Ramdath, D. D., and Duizer, L. M. (2017b). Chemical and physical characteristics of proso millet (Panicum miliaceum)-based products. Cereal Chem. J. 94, 357–362. doi: 10.1094/cchem-07-16-0185-r
Narciso, J. O., and Nyström, L. (2023). The genetic diversity and nutritional quality of proso millet (Panicum miliaceum) and its Philippine ecotype, the ancient grain “kabog millet”: a review. J. Agricult. Food Res. 11:100499. doi: 10.1016/j.jafr.2023.100499
Sakandar, H. A., Hussain, R., Farid Khan, Q., and Zhang, H. (2020). Functional microbiota in Chinese traditional Baijiu and Mijiu Qu (starters): a review. Food Res. Int. 138:109830. doi: 10.1016/j.foodres.2020.109830
Sanderson, E., Duizer, L. M., and McSweeney, M. B. (2017). Descriptive analysis of a new proso millet product. Int. J. Gastron. Food Sci. 8, 14–18. doi: 10.1016/j.ijgfs.2017.02.001
Santra, D. K., Khound, R., and Das, S. (2019). “Proso millet (Panicum miliaceum L.) breeding: Progress, challenges and opportunities” in Advances in plant breeding strategies: Cereals. eds. J. M. Al-Khayri, S. M. Jain, and D. V. Johnson, vol. 5 (Cham: Springer International Publishing), 223–257.
Shen, R., Ma, Y., Jiang, L., Dong, J., Zhu, Y., and Ren, G. (2018). Chemical composition, antioxidant, and antiproliferative activities of nine Chinese proso millet varieties. Food Agric. Immunol. 29, 625–637. doi: 10.1080/09540105.2018.1428283
Takeshita, T., Suzuki, N., Nakano, Y., Yasui, M., Yoneda, M., Shimazaki, Y., et al. (2012). Discrimination of the oral microbiota associated with high hydrogen sulfide and methyl mercaptan production. Sci. Rep. 2:215. doi: 10.1038/srep00215
Wang, J.-Y., Meng, Q.-H., Jin, X.-W., and Sun, Z.-H. (2020). Design of Handheld Electronic Nose Bionic Chambers for Chinese liquors recognition. Measurement 172:108856. doi: 10.1016/j.measurement.2020.108856
Wang, J., Yuan, C., Gao, X., Kang, Y., Huang, M., Wu, J., et al. (2020). Characterization of key aroma compounds in Huangjiu from northern China by sensory-directed flavor analysis. Food Res. Int. 134:109238. doi: 10.1016/j.foodres.2020.109238
Wiedemair, V., Scholl-Bürgi, S., Karall, D., and Huck, C. W. (2020). Amino acid profiles and compositions of different cultivars of Panicum miliaceum L. Chromatographia 83, 829–837. doi: 10.1007/s10337-020-03899-8
Wu, Q., Zhu, Y., Fang, C., Wijffels, R. H., and Xu, Y. (2021). Can we control microbiota in spontaneous food fermentation? – Chinese liquor as a case example. Trends Food Sci. Technol. 110, 321–331. doi: 10.1016/j.tifs.2021.02.011
Xiang, F., Cai, W., Hou, Q., Gai, J., Dong, X., Li, L., et al. (2023). Comparative analysis of the microbial community structure in light-flavor Daqu in Taiyuan and Suizhou regions, China. LWT 177:114599. doi: 10.1016/j.lwt.2023.114599
Xie, M., Lv, F., Ma, G., Farooq, A., Li, H., Du, Y., et al. (2020). High throughput sequencing of the bacterial composition and dynamic succession in Daqu for Chinese sesame flavour liquor. J. Inst. Brew. 126, 98–104. doi: 10.1002/jib.592
Xie, Z.-B., Zhang, K.-Z., Kang, Z.-H., and Yang, J.-G. (2021). Saccharomycopsis fibuligera in liquor production: a review. Eur. Food Res. Technol. 247, 1569–1577. doi: 10.1007/s00217-021-03743-9
Yang, J.-G., Dou, X., Han, P.-J., Bai, F.-Y., Zhou, J., Zhang, S.-Y., et al. (2017). Microbial diversity in Daqu during production of Luzhou flavored liquor. J. Am. Soc. Brew. Chem. 75, 136–144. doi: 10.1094/ASBCJ-2017-2879-01
Yang, Q., Liu, L., Zhang, W., Li, J., Gao, X., and Feng, B. (2019). Changes in morphological and physicochemical properties of waxy and non-waxy proso millets during cooking process. Foods 8:583. doi: 10.3390/foods8110583
Yang, Q., Zhang, P., Qu, Y., Gao, X., Liang, J., Yang, P., et al. (2018). Comparison of physicochemical properties and cooking edibility of waxy and non-waxy proso millet (Panicum miliaceum L.). Food Chem. 257, 271–278. doi: 10.1016/j.foodchem.2018.03.009
Yifan, N., Zhaoqi, Z., and Zhuoran, W., (2021). Study on the profitability of Fenjiu group based on the financial index system. Proceedings of the 2021 International Conference on Financial Management and Economic Transition (FMET 2021), Amsterdam, Netherlands. pp. 568–573.
Yuan, Y., Xiang, J., Zheng, B., Sun, J., Luo, D., Li, P., et al. (2022). Diversity of phenolics including hydroxycinnamic acid amide derivatives, phenolic acids contribute to antioxidant properties of proso millet. LWT 154:112611. doi: 10.1016/j.lwt.2021.112611
Zhang, L., Guo, B., Yang, H., and Wang, C. (2022). A novel strain of moderately thermophilic Streptomyces from the Fenjiu-flavor Daqu. OALib 09, 1–9. doi: 10.4236/oalib.1108388
Zhang, J., Kobert, K., Flouri, T., and Stamatakis, A. (2014). PEAR: a fast and accurate Illumina paired-end reAd mergeR. Bioinformatics 30, 614–620. doi: 10.1093/bioinformatics/btt593
Zhang, L., Xu, J., Ding, Y., Cao, N., Gao, X., Feng, Z., et al. (2023). GWAS of grain color and tannin content in Chinese sorghum based on whole-genome sequencing. Theor. Appl. Genet. 136:77. doi: 10.1007/s00122-023-04307-z
Zhang, Y., Zhu, X., Li, X., Tao, Y., Jia, J., and He, X. (2017). The process-related dynamics of microbial community during a simulated fermentation of Chinese strong-flavored liquor. BMC Microbiol. 17:196. doi: 10.1186/s12866-017-1106-3
Zheng, N., Jiang, S., He, Y., Chen, Y., Zhang, C., Guo, X., et al. (2020). Production of low-alcohol Huangjiu with improved acidity and reduced levels of higher alcohols by fermentation with scarless ALD6 overexpression yeast. Food Chem. 321:126691. doi: 10.1016/j.foodchem.2020.126691
Keywords: proso millet, Baijiu, fermentation, microbial community, flavor
Citation: Zhao J and Gao Z (2024) Dynamic changes in microbial communities and flavor during different fermentation stages of proso millet Baijiu, a new product from Shanxi light-flavored Baijiu. Front. Microbiol. 15:1333466. doi: 10.3389/fmicb.2024.1333466
Edited by:
Antonio Alfonzo, University of Palermo, ItalyReviewed by:
Bin-Bin Hu, Yunnan Academy of Tobacco Agricultural Sciences, ChinaZhilei Fu, Hebei Normal University for Nationalities, China
Copyright © 2024 Zhao and Gao. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Zhenfeng Gao, ODYyMTU3MTY4QDE2My5jb20=
 Jia Zhao1
Jia Zhao1 Zhenfeng Gao
Zhenfeng Gao