
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
REVIEW article
Front. Microbiol., 06 February 2024
Sec. Infectious Agents and Disease
Volume 15 - 2024 | https://doi.org/10.3389/fmicb.2024.1331508
This article is part of the Research TopicRecent Advancements in mycobacterial diseases researchView all 10 articles
Mycobacterium abscessus, a leading cause of severe lung infections in immunocompromised individuals, poses significant challenges for current therapeutic strategies due to resistance mechanisms. Therefore, understanding the intrinsic and acquired antibiotic resistance of M. abscessus is crucial for effective treatment. This review highlights the mechanisms employed by M. abscessus to sustain antibiotic resistance, encompassing not only conventional drugs but also newly discovered drug candidates. This comprehensive analysis aims to identify novel entities capable of overcoming the notorious resistance exhibited by M. abscessus, providing insights for the development of more effective therapeutic interventions.
Non-tuberculous mycobacteria (referred to as NTMs hereafter) have emerged as a significant public health concern, with steadily increasing morbidity and mortality rates worldwide, eventually surpassing those of tuberculosis (Howard et al., 2006; Wassilew et al., 2016). NTM infections are opportunistic diseases primarily affecting individuals with compromised immune systems, such as patients with cystic fibrosis (CF), chronic obstructive pulmonary disease, renal failure, transplant recipients with chronic corticosteroid use, TNF-α, and leukemia (Faria et al., 2015). While NTM infections most commonly occur in the lungs, they can also develop in other organs. Importantly, NTM infections are rarely contagious, signifying that they do not spread from person to person, distinguishing them from other types of respiratory infections (Swenson et al., 2018; Lipman et al., 2021).
Mycobacterium abscessus (referred to as Mab hereafter) is the second most significant pathogen in NTM-induced pulmonary disease, and it is increasingly emerging as the most prominent and concerning pathogen in hospitals and CF centers worldwide (Degiacomi et al., 2019). Mab was firstly isolated in 1952 by Moore and Frerichs from a 63-year-old woman’s knee abscess and it was classified as Mycobacterium chelonae subsp. abscessus (MOORE and FRERICHS Moore and Frerichs, 1953; Victoria et al., 2021). However, Mab was recognized as an independent species from M. chelonae based on DNA hybridization and two new species Mycobacterium massiliense and Mycobacterium bolletii were described as novel and closely related to Mab based on the rpoB gene sequence (Lee et al., 2015; Lopeman et al., 2019; Victoria et al., 2021). However, since all these thee species share more than 70% relatedness based on DNA–DNA hybridization, M. massiliense, M. bolletii, and M. abscessus were presented as subspecies such as Mab subsp. abscessus, Mab subsp. bolletti, and Mab subsp. massiliense (hereafter referred to as M. abscessus, M. bolletii, and M. massiliense) and the combinations of the three subspecies were known as Mab complex (Lopeman et al., 2019; Victoria et al., 2021). The genome of Mab (CIP 104536 T) comprise 5,067,172-bp circular chromosome including 4,920 predicted coding sequences (CDS), an 81-kb full-length prophage and 5 IS elements, and a 23-kb mercury resistance plasmid almost identical to pMM23 from Mycobacterium marinum (Ripoll et al., 2009). Mab complex is responsible for 2.6–13.0% of all NTM pulmonary infections (Dedrick et al., 2023). The natural habitat of Mab is in soil and water sources, leading to a high rate of human-pathogen contact. Furthermore, nosocomial outbreaks and the transmission of Mab have been continuously occurring in clinics that conduct cosmetic surgery, liposuction, mesotherapy, or intravenous infusion of cell therapy (Lee et al., 2015; Desai and Hurtado, 2018). Nosocomial outbreaks of Mab through Mab contaminated surgical materials and hospital tap water, have also been reported as well in patients without CF (Baker et al., 2017; Fernandes Garcia de Carvalho et al., 2018). While it was previously believed that a significant portion of Mab infections in CF patients originated from exposure to sources such as soil, household dust, or water, potentially through contact with contaminated objects (fomites) or airborne particles (aerosols) (Falkinham, 2011), recent studies indicate that individuals with CF can also be infected through person-to-person transmission through hospital-based (Bryant et al., 2016). Additionally, a study by Ruis et al. suggests that dominant Mab circulating clones initially emerged within non-CF populations and were later amplified and spread within the CF community. Consequently, individuals with CF might be more permissible hosts, while non-CF individuals play a crucial role in transmission networks, potentially facilitating long-distance spread. This conclusion was drawn from an evolutionary phylogenetic analysis employing whole-genome sequences of clinical isolates from 1,178 CF and non-CF individuals across five continents (Ruis et al., 2021).
For the aspect of Mab diagnosis, there is frequent misdiagnosis of Mab as M. tuberculosis (referred to as Mtb), primarily due to the visual similarities observed in sputum samples under microscopic analysis (Wu et al., 2018). These circumstances not only lead to incorrect treatments but also have significant consequences, including the underestimation of NTM incidence and the inefficient allocation of budgetary resources dedicated to combating the disease. Moreover, it’s crucial to recognize that monotherapies alone are insufficient to fully eradicate the microbiological infection. According to the latest 2020 ATS/ERS/ESCMID/IDSA clinical practice guidelines, the treatment for Mab pulmonary disease is categorized based on macrolide susceptibility. For macrolide-susceptible cases, the guidelines recommend an initial phase with 1–2 parenteral drugs (amikacin; AMK, imipenem; IMP, cefoxitin; CFX, and tigecycline; TGC) and two oral drugs (azithromycin; AZM, clofazimine; CFZ, and linezolid; LZ), along with inhaled AMK. In the case of macrolide-resistant organisms, the recommendations include an initial phase with 2–3 parenteral drugs (AMK, IMP, CFX, and TGC) and 2–3 oral drugs (AZM, CFZ, and LZ), supplemented with inhaled AMK. The addition of AZM is for its immunomodulatory effect, although adherence to these guidelines may have adverse effects on NTM patients (Moguillansky et al., 2023). However, Mab has demonstrated resistance to a broad spectrum of antibiotics, including the aforementioned treatment regimen, and patients experience multiple relapses with low cure rates, making it challenging to achieve a complete cure (Victoria et al., 2021). This discouraging success rate primarily stems from the rapid development of drug resistance, which can be attributed to both intrinsic and acquired multidrug resistance to antibiotics. Notably, even first-line anti-TB medications, such as isoniazid (INH) and rifampicin (RFP), lack efficacy against Mab (Zheng et al., 2023). As a result, the majority of Mab treatment protocols involve extended multi-antibiotic regimens that can last up to 24 months (Wu et al., 2018; Ratnatunga et al., 2020). However, the effectiveness of these treatments remains limited, with disease remission rates reaching only 30% (Wu et al., 2018; Ratnatunga et al., 2020). Additionally, in cases of pulmonary infections, no class of antibiotics has demonstrated the ability to achieve long-term sputum smear conversion (Wu et al., 2018; Ratnatunga et al., 2020).
The three subspecies of Mab shows distinct clinical outcomes (Blauwendraat et al., 2012; Harada et al., 2012; Shin et al., 2013; Jeong et al., 2017; Abate et al., 2019). Firstly, M. abscessus exhibits resistance to macrolides such as AZM and clarithromycin (CLR) due to an adaptive resistance mechanism involving the inducible erythromycin ribosomal methyltransferase, erm(41) (Stout and Floto, 2012; Rubio et al., 2015; Christianson et al., 2016; Abate et al., 2019). Consequently, the use of macrolides in treating M. abscessus infections should be approached with great caution (Maurer et al., 2014). Secondly, M. massiliense is the most recent subspecies within this group and has a broader geographical distribution compared to the other subspecies. Notably, this subspecies tends to yield more favorable clinical outcomes than the other two, primarily because it lacks the functional erm gene. Lastly, M. bolletii represents the rarest among the three subspecies and is also resistant to CLR.
However, our understanding of the intrinsic or acquired antibiotic resistance of Mab remains limited. Therefore, alongside the ongoing efforts to discover novel alternative compounds for Mab treatment, it is crucial to elucidate the resistance mechanisms employed by Mab against existing antibiotics. This endeavor not only aids in enhancing the effectiveness of current antibiotics to overcome these resistance barriers but also provides valuable insights for the development of new compounds. This review aims to offer a comprehensive overview of the current knowledge of antibiotic resistance mechanisms in Mab, with the goal of clarifying the molecular components contributing to its significant resistance to chemotherapy and facilitating the development of a drug pipeline for Mab.
The quantity of initiatives in antimicrobial drug development has significantly diminished since the remarkable era of antibiotic discovery, and several factors contribute to this decline. Firstly, the increasing prevalence of drug-resistant bacteria limits the effectiveness of new antibiotics, making it challenging to recoup investments in antibiotic development. Secondly, antibiotics are typically prescribed for short durations, in contrast to drugs for chronic conditions like hypertension or diabetes, which may render antibiotics less financially appealing to pharmaceutical companies. Thirdly, novel effective drugs are often preserved as last-resort treatments for highly-resistant bacterial infections. The goal is to mitigate the development of further resistance by limiting their widespread use. Overuse or misuse of these potent drugs can accelerate the emergence of resistant strains, making them ineffective sooner. Overexposure can diminish their efficacy over time, making it crucial to reserve them for cases where no other options are viable (Baker et al., 2017). Lastly, the discovery of new antibiotics presents scientific challenges (Ventola, 2015; Quang and Jang, 2021). Finding compounds that are both effective against bacteria and safe for humans is a complex process, and success is not guaranteed (Chopra et al., 2011). The identification and development of innovative and potent medications for combating NTMs are of paramount importance in the medical field. In comparison to the tuberculosis drug pipeline, which features a significant number of compounds undergoing clinical trials, the NTMs drug pipeline lags considerably (Wu et al., 2018). Notably, there is a critical need for the development of new treatments targeting Mab as there are currently no antibiotics approved by the Food and Drug Administration (FDA) for Mab infection.
Two primary strategies exist to bolster the development of effective Mab-targeting medicines. The first strategy follows the conventional drug development process, which encompasses the identification of novel chemical compounds. This process commences with drug screening using chemical and natural product libraries, progressing through hit identification, lead optimization, target identification, comprehensive preclinical investigations, and ultimately clinical trials (Egorova et al., 2021). Various screening methods have been employed in this pursuit for Mab drug discovery, including reporter-based assays, resazurin-based microplate assays, and image-based phenotypic screens (Gupta et al., 2017; Jeong et al., 2018; Richter et al., 2018; Kim et al., 2019; Malin et al., 2019; Hanh et al., 2020a,b). Nevertheless, despite these efforts, promising new chemical leads ready for clinical trials and market release remain scarce (Hanh et al., 2020a). This challenge may be attributed to the intrinsic drug-resistant mechanisms of Mab, resulting in a low hit rate for compounds targeting this bacterium (Malin et al., 2019). It’s noteworthy that the hit rate achieved in Mab screens is significantly lower than what is typically observed in screens targeting Mtb (Malin et al., 2019; Hanh et al., 2020a). Moreover, recent Mab drug screens have relied on conventional libraries composed of known antimycobacterial or antibacterial agents (Malin et al., 2019), diminishing the likelihood of identifying novel compounds targeting new mechanisms of action. Hence, there is an urgent need to develop new libraries with expanded chemical diversity to discover unique compounds. Additionally, reliable cell-based or in vivo assessment/screening methods that accurately mimic the host environment infected with Mab are imperative to advance our understanding and discovery of effective treatments for Mab-induced human infections (Carvalho et al., 2011; Bernut et al., 2014, 2015). The second strategy involves repurposing or repositioning existing medications for novel therapeutic indications. Most contemporary antibiotics and potential prospects against Mab have origins in the repurposing of pre-existing drugs or the cross-testing of compounds with activity and various mechanism of action against Mtb (Egorova et al., 2021). This method is particularly intriguing in the field of antibacterials, as the rapid evolution of resistance often outpaces the pace of medication development (Egorova et al., 2021). Repurposing previously approved pharmaceuticals can expedite the development process and reduce expenses (Egorova et al., 2021). Regrettably, the Mab drug pipeline remains underpopulated (Ganapathy and Dick, 2022). Currently, there are four recruiting, four completed, one terminated, and two non-recruiting clinical trials evaluating drug efficacy in Mab infection (NIH ClinicalTrials, 2023). However, these clinical trials have primarily utilized existing antibiotics through various drug delivery methods, including inhalation, novel drug encapsulation using biocompatible liposomes, and the exploration of new drug combinations (Quang and Jang, 2021). The primary reason for the limited success in anti-Mab drug discovery is the remarkable intrinsic resistance capabilities of Mab and its rapid acquired resistance against currently available active drugs (Wu et al., 2018).
Inherent drug resistance in NTMs is responsible for their limited susceptibility to a wide range of medicines and chemicals (Wu et al., 2018). This inherent resistance in Mab and other mycobacterial species can be attributed to several factors, including the presence of a waxy impermeable cell wall that acts as both a physical (size exclusion) and a chemical (hydrophobic) barrier, drug export systems, enzymes capable of modifying drugs or target enzymes, and genetic polymorphisms in target genes (Nessar et al., 2012) (Figure 1). In addition to harboring numerous intrinsic resistance mechanisms, Mab possesses the ability to acquire novel resistance through genetic changes that can be passed down to subsequent generations. Acquired resistance is not linked to genes introduced by transmissible genetic elements like plasmids and transposons (Martin et al., 1990). Instead, resistance arises due to spontaneous mutations occurring at specific genes in response to the presence of antibiotics following extended courses of treatment (Johansen et al., 2020b). This allows bacteria to undergo genetic changes in the target gene or other associated genes, resulting in the acquisition of significant levels of resistance, rendering the medicine ineffective. However, species or subspecies may exhibit variations in their antibiotic resistance phenotype and genotype, emphasizing the need for research on accurately identified strains (Nessar et al., 2012). In this section, we focus on intrinsic and acquired resistance to essential drugs and new drug candidates that have demonstrated efficacy against Mab.
Mycobacteria’s cell wall is primarily composed of lipids, specifically mycolic acids, constituting a significant portion (up to 60%) of the bacteria’s overall dry mass (Brennan and Nikaido, 1995). This cell wall features a waxy composition that serves as a physical barrier (Nessar et al., 2012), rendering mycobacteria less permeable than the outer membranes of gram-negative bacteria. In more detail, the mycobacterial envelope consists of three distinct layers: a typical plasma membrane, a complex cell wall, and an outer layer. The cell wall, notably, comprises a thick peptidoglycan layer covalently linked to arabinogalactan, which is esterified by mycolic acids, forming the inner leaflet of the mycomembrane (Figure 2). This unique structure inherently makes mycobacteria resistant to many antimicrobials. Hydrophilic drugs penetrate the mycobacterial cell wall slowly due to the inefficiency of mycobacterial porin in allowing antibiotic permeation, resulting in low antibiotic concentrations within the bacteria. The dense mycobacterial cell wall not only shields the bacterium from stressors but also poses challenges in nutrient uptake from the environment. To address this, mycobacteria often produce porins, proteins that create limited pathways for nutrient absorption. The expression of these porins is closely tied to the growth rate of NTMs, and they provide a conduit for certain antimicrobial agents to enter the mycobacterial cell (Saxena et al., 2021). Lipophilic agents may be hindered by the lipid bilayer, which has unusually low fluidity and thickness (Jarlier and Nikaido, 1994; Gutiérrez et al., 2018). Intriguingly, Mycobacterium chelonae, a species closely related to Mab due to its nearly identical biochemical features, has a cell envelope that is about 10–20 times less permeable than that of Mtb (Jarlier and Nikaido, 1994). Similar to Mtb, Mab possesses a mycobacterial cell wall with low permeability, which contributes to its drug resistance. Notably, like Mtb, Mab is thought to regulate cell wall structure and homeostasis through lipoprotein glycosylation. For example, the absence of protein-O-mannosyltransferase Pmt (MAB_1122c) in Mab leads to increased cell wall permeability and greater susceptibility to antibiotics such as RFP (Ganapathy et al., 2019). Furthermore, glycosylation of lipoproteins limits cell wall permeability to antibiotics like β-lactam agents that inhibit peptidoglycan synthesis. In β-lactam drug resistance, mycobacterial porins also play a role by facilitating the transport of small hydrophilic drugs across the membrane. Once antibiotics are internalized, they can reach their target in the cytoplasm and activate potential internal drug resistance mechanisms, collectively known as the “intrinsic resistome.” This resistome includes efflux pumps, antibiotic-modifying/inactivating enzymes, target-modifying enzymes, and genes conferring metal resistance (Nguyen and Thompson, 2006; Nessar et al., 2012).
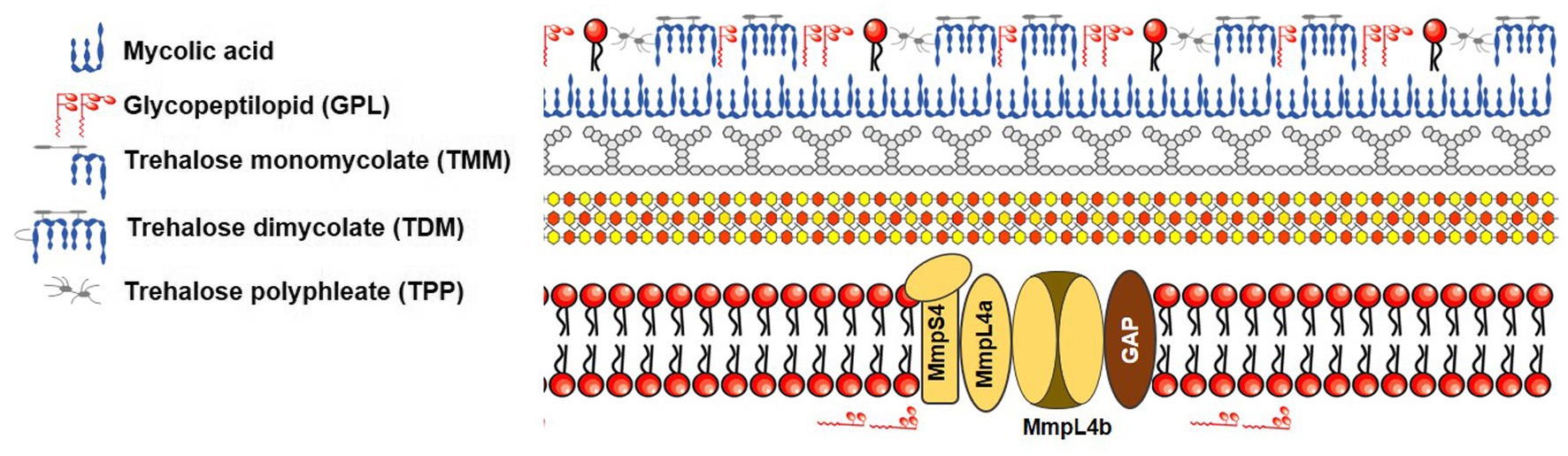
Figure 2. Schematic representation of mycobacterial cell envelope based on the figure of Gutiérrez et al.
Remarkably, unlike Mtb, Mab exhibits two distinct colony morphotypes: smooth non-cording (S) and rough cording morphotype (R). These differences in morphotypes depend on the presence or absence of cell surface-associated glycopeptidolipids (GPL), respectively (Howard et al., 2006). This distinctive property, associated with GPL status, affects sliding motility, biofilm formation, and drug susceptibility. For example, S morphotype strains that contain GPL, such as M. abscessus and M. bolletii, facilitate sliding across the surface and biofilm formation. Indeed, the Mab growing inside biofilms become tolerant to antibiotics due to physical barrier that can prevent the intracellular penetration of compounds. In fact, in vitro biofilm models of Mab have been exhibited to have decreased susceptibility to several first-line antibiotics, such as cefoxitin, amikacin, and clarithromycin (Greendyke and Byrd, 2008; Marrakchi et al., 2014). Conversely, R morphotype strains exhibit aggregation and cording. Recent studies suggest that Mab R strains are capable of growing in biofilm-like structures, which, similar to S biofilms, show greater tolerance than planktonic cultures to acidic pH, hydrogen peroxide, and treatment with antibiotics like AMK, AZM, and β-lactams (Gutiérrez et al., 2018; Story-Roller et al., 2018; Daher et al., 2022; López-Roa et al., 2022). Furthermore, biofilms formed by R colony types display higher mechanical resistance compared to those formed by S colony types (López-Roa et al., 2022). Biofilms also inhibit oxygen and nutrients from entering the cell, which causes a reduction Mab metabolism and, consequently, an increased tolerance to a harsh environment such as antibiotics treatment. Therefore, compounds that specifically target biofilm formation during antibiotic therapy are a new therapeutic strategy for clearance of Mab.
In invasive infections causing pulmonary colonization, Mab R strains are accountable for producing higher levels of trehalose dimycolate, consequently leading to the formation of massive bacterial cords. With the rough variant, the entire phagosome quickly merges with the lysosome, inducing phagosomal acidification and activating apoptosis and autophagy (Roux et al., 2016; López-Roa et al., 2022). This robust apoptosis-driven cell-death activity facilitates the extracellular replication of the R variant through rapid cord formation, preventing the engulfment of bacilli by neutrophils and macrophages. This process leads to abscess formation, tissue destruction, and acute infection (Jönsson et al., 2013; Bernut et al., 2016; López-Roa et al., 2022).
Mab produces enzymes capable of modifying antibiotics by cleaving, altering their structure, and adding or removing chemical groups (Luthra et al., 2018). These modifications can render antibiotics ineffective by either preventing their binding to their target or increasing their susceptibility to hydrolysis by the bacteria (Luthra et al., 2018). The efficacy of antibiotics was restored when these modifying genes were knocked down (Nessar et al., 2012). AG antibiotics are composed of amino carbohydrates linked by glycoside bonds (Mingeot-Leclercq et al., 1999). Among antibacterial drugs, AMK has shown the most efficacy against Mab (Tsai et al., 2015). AGs can diffuse through porins and interact with 30S ribosomes (e.g., streptomycin), 50S ribosomes (others), or both 30S and 50S ribosomes (Chulluncuy et al., 2016). This interaction prevents the initiation of protein synthesis, the continuation of translation, or the incorporation of incorrect proteins (Bhattacharjee, 2016). Unfortunately, Mab has developed resistance to aminoglycosides. Mab’s genome annotation suggests the presence of various AG-modifying enzymes, including AG phosphotransferases, AG nucleotidyltransferases, and AG acetyltransferases (AACs). Among these, AG AAC (2′-N-acetyltransferase) and AG phosphotransferases render AG antibiotics inactive by transferring acetyl or phosphate residues to crucial positions within the antibiotic (Nessar et al., 2012) (Supplementary Figures S1, S2). Mab’s genome analysis and AG drug susceptibility testing indicate the presence of several putative AACs, which acetylate aminoglycosides with a 2′ amino group, such as gentamicin, tobramycin, and KM. ORF MAB_4395 is annotated as a putative AG 2′-N-acetyltransferase [aac(2′)], and aac(2′) deletion mutants increased Mab’s susceptibility to KM-B, tobramycin, dibekacin, and gentamicin C (Rominski et al., 2017b). Furthermore, MAB_2385, which serves as the main determinant of resistance to the first-discovered aminoglycoside, streptomycin, functions as a 3″-O-phosphotransferase. Deletion of MAB_2385 in Mab increases susceptibility to streptomycin, while introducing MAB_2385 in M. smegmatis (Msm) confers resistance to streptomycin (Dal Molin et al., 2017). In addition to MAB_4395 and MAB_2385, Mab possesses additional AG-modifying enzymes named Eis (enhanced intracellular survival), including MAB_4124 (also known as eis1; sharing 33% identity with Mtb Rv2416c) and MAB_4532c (eis2), which are involved in AG resistance. MAB_4532c significantly enhances Mab’s intracellular survival and has been shown to modify KM, hygromycin, and AMK in vitro (Ung et al., 2019). Deletion of MAB_4532c strains increased Mab’s susceptibility to AGs and capreomycin (Rominski et al., 2017b; Johansen et al., 2020b). Furthermore, MAB_4532c is responsible for the lack of bactericidal activity of AMK in vitro and affected AMK activity in vivo (Lorè et al., 2022; Selchow et al., 2022).
Clinically acquired pan-AG resistance is linked to mutations in ribosomal RNA genes, specifically rrs, which encode the 16S rRNA molecule as acquired resistance (Rominski et al., 2017b). The prolonged use of AGs can lead to genetic modifications in rrs (Wallace et al., 1996; Prammananan et al., 1998; Maurer et al., 2012). It’s worth noting that the substitution of adenine with guanine at position 1408 (A1408G) in rrs significantly increases resistance to KM, AMK, and tobramycin (Nessar et al., 2011). Recently, two novel rrs mutations, C1496T and T1498A, were also identified from Mab-pulmonary disease patients (Young et al., 2021). Additionally, mutations at locations T1406A, C1409T, and G1491T in rrs could potentially confer a high level of resistance to KM, AMK, and gentamicin (Nessar et al., 2011).
Apramycin (AP; also known as Nebramycin II) is presently authorized by the Veterinary Medicines Directorate in the UK for use in pigs, cattle, rabbits, and chickens. It is available either as (i) a premix for medicated feedstuff (200 g/kg, 100,000 IU/g, 100 g/kg) or (ii) a soluble powder for oral solution, with a concentration of 10% or less (Moore et al., 2018). AP has a distinctive AG structure and demonstrates potent activity against Mab (Selchow et al., 2022). Furthermore, it displays minimal cross-resistance to other aminoglycosides and exhibits favorable therapeutic lung exposure and a low toxicity profile (Matt et al., 2012; Ishikawa et al., 2019; Juhas et al., 2019; Becker et al., 2021; Selchow et al., 2022). Recently, Selchow et al., reported that AP is not modified by Eis2 or Aac(2′) and is not affected by the multidrug resistance regulator WhiB7. This favorable feature of apramycin is reflected in a mouse model of pulmonary Mab infection, which demonstrates superior activity, compared with amikacin (Selchow et al., 2022). At present, there are no established antibiotic breakpoints for AP against Mab provided by organizations like EUCAST (The European Committee on Antimicrobial Susceptibility Testing) or CLSI (Clinical & Laboratory Standards Institute). Pharmacokinetic/pharmacodynamics studies are essential to address this gap for AP in treating chronic Mab infections in humans. This evaluation process holds significant importance for approval by the FDA and the European Medicines Agency (EMA) (Moore et al., 2018). Albeit, AP is currently promising agent as Mab treatment option together with other candidates such as RFB and omadacycline (OMC).
Beta-lactams are a class of antibiotics characterized by a four-atom beta-lactam ring (De Rosa et al., 2021). They are among the most widely prescribed antibiotics due to their broad spectrum of activity against bacteria (De Rosa et al., 2021). Beta-lactam antibiotics are bactericidal because they inhibit the cross-linking or transpeptidation of the peptidoglycan layer in bacterial cell walls by covalently binding to penicillin-binding proteins (PBPs). Bacterial enzymes that hydrolyze peptidoglycan cross-links continue to function even when PBPs are inactivated by beta-lactam antibiotics, leading to further degradation of the cell wall. The buildup of peptidoglycan precursors activates cell wall hydrolases, ultimately causing the cells to burst (Bush and Bradford, 2016). To counteract the effects of beta-lactam antibiotics, Mab possesses a beta-lactamase-encoding gene, namely blaMab, which serves to break down beta-lactam antibiotics, rendering them ineffective (Dubée et al., 2015a) (Supplementary Figure S3). BlaMab efficiently degrades multiple β-lactams, surpassing the activity of BlaC, the principal β-lactamase of Mtb. Deletion of blaMab in a recombinant Mab strain increased its susceptibility to β-lactams, making it responsive to antibiotics like amoxicillin and ceftaroline (Lefebvre et al., 2016). Moreover, BlaMab exhibits reduced susceptibility to common β-lactamase inhibitors, such as clavulanate, tazobactam, and sulbactam, unlike inhibitors of BlaC in Mtb (Soroka et al., 2017). Additionally, M. massiliense harbors an additional β-lactamase, BlaMmas (Ramírez et al., 2017).
However, recent combination studies have shown that non-β-lactam-based β-lactamase inhibitors known as diazabicyclooctane (DBO) inhibitors, including avibactam, effectively inhibit BlaMab. This inhibition leads to a reduction in the minimum inhibitory concentration (MIC) of carbapenems and cephalosporins against Mab to clinically achievable levels. Currently, avibactam is exclusively marketed in combination with the cephalosporin ceftazidime under the name Avycaz in the United States (Dubée et al., 2015a; Lefebvre et al., 2017; Meir et al., 2018; Le Run et al., 2019; Kaushik et al., 2019b; Dousa et al., 2022). Furthermore, Dousa et al. have demonstrated the effectiveness of two new non-β-lactam-based β-lactamase DBO inhibitors, relebactam and vaborbactam, when evaluated in combination with various commercially available β-lactams against clinical isolates of Mab. In their study, both relebactam and vaborbactam significantly enhanced the anti-Mab activity of several carbapenems (IMP and meropenem) and cephalosporins (cefepime, ceftaroline, and cefuroxime) (Dubée et al., 2015b). Currently, the IMP-relebactam combination is undergoing phase III trials, and the meropenem-vaborbactam combination is already available in the market. This established and effective combination opens the door to using potent β-lactams for the treatment of Mab infections (Kaushik et al., 2019b). Furthermore, more recent DBO class β-lactamase inhibitors, including nacubactam, zidebactam, and durlobactam, have been suggested as potent β-lactamase inhibitors that can restore susceptibility to β-lactams against Mab in vitro (Dubée et al., 2015a; Lefebvre et al., 2017; Meir et al., 2018; Le Run et al., 2019; Kaushik et al., 2019b; Dousa et al., 2022).
Rifampicin (RFP) stands as the first-line treatment for Mtb, primarily due to its ability to halt transcription by binding to the beta-subunit of RNA polymerase encoded by rpoB. This enzyme is pivotal for bacterial transcription (Piccaro et al., 2014). This interaction prevents the bacterium from transcribing essential genetic material, ultimately leading to its demise. However, RFP is notably ineffective against Mab. In Mab, RFP’s efficacy is nullified due to the presence of the Arr gene (MAB_0591) (Rominski et al., 2017a). This gene produces an RFP ADP-ribosyltransferase homolog, which inactivates rifamycins by catalyzing ADP-ribosylation at position C23 (Rominski et al., 2017a) (Supplementary Figure S4). ADP-ribosylation confers innate rifamycin resistance in Mab (Ganapathy et al., 2019). Deletion of MAB_0591 in Mab has proven to not only decrease the MIC for RFP but also enhance susceptibility to RFP analogs like rifaximin rifabutin (RFB) and rifapentine (Rominski et al., 2017a; Schäfle et al., 2021). For instance, the removal of this gene results in a significant increase in Mab’s susceptibility to RFP, rifapentine, and rifaximin, with a 100-fold reduction in RFP’s MIC (Johansen et al., 2020b; Zheng and Lupoli, 2021). Moreover, recent research has highlighted the potential of a rifamycin analogue known as RFB following its identification through drug screening. However, although being a substrate of Arr, RFB has demonstrated promising anti-Mab effects both in vitro and in vivo. In these studies, RFB not only inhibited Mab growth but also exhibited bactericidal properties against all three Mab subspecies. Particularly, RFB displayed comparable activity to clarithromycin (CLR) against Mab K21 in NOD.CB17-Prkdcscid/NCrCrl mice. These in vitro and in vivo findings suggest that RFB may enhance cure rates and shorten treatment duration for the predominantly challenging Mab lung disease. Hence, it should be considered a viable clinical candidate for patients with Mab infections (Dick et al., 2020; Johansen et al., 2020a).
Recently, 25-O-desacetyl-25-O-nicotinoylrifabutin (RFB-5 m), a new rifabutin analogue, overcomes inherent rifamycin resistance caused by Arr (Ganapathy et al., 2023b). RFB-5 m prevents enzymatic oxidation by maintaining rifabutin’s naphthoquinone core (Lan et al., 2022; Ganapathy et al., 2023b). Importantly, RFB-5 m’s unique C25 group prevents Arr Mab ADP-ribosylation (Lan et al., 2022; Ganapathy et al., 2023b). Compared to rifabutin, RFB-5 m is 50 times more effective against Mab (Lan et al., 2022; Ganapathy et al., 2023b). Moreover, RFB-5 m was observed to display bactericidal properties against the persisters of Mab in caseum (Lan et al., 2022). RFB-5 m also had strong enhanced potency against all members of the Mab complex, other clinically relevant rapidly and slowly growing NTM, all of which encode Arr and block ADP-ribosylation (Ganapathy et al., 2023b). Recently, Paulowski et al. reported that benzyl piperidine rifamycin derivative known as 5j, which possesses a morpholino substituted C3 position and a naphthoquinone core, does not undergo any modifications when exposed to pure Arr (Paulowski et al., 2022). The thermal characterization of Arr in the presence of 5j, RMP, or RFB reveals that 5j exhibits no binding affinity towards Arr (Paulowski et al., 2022) and 5j also has substantial antibiotic efficacy against Mab within human macrophages, and exhibits synergistic effects when combined with AMK and AZM (Paulowski et al., 2022).
Additionally, Hanh et al. recently shed light on the activity of Rifamycin O, a derivative of rifamycin resulting from the oxidation of natural rifamycin B, against Mab. In their study, Rifamycin O exhibited promising in vitro activity (MIC90 = 4.0–6.2 μM) and demonstrated comparable in vivo efficacy to RFB using a zebrafish (Danio rerio) infection model (Hanh et al., 2020b). This suggests that certain rifamycin analogs like RFB and Rifamycin O can evade MAB_0591-mediated rifamycin resistance mechanisms. Of note, RFB and Rifamycin O exhibit distinct chemical structures at positions C1 and C4, setting them apart from other rifamycin analogs such as RFP, rifapentine, rifamycin SV, and rifaximin. These other analogs contain hydroquinone, which can readily oxidize into RFP quinone in the presence of oxygen and divalent cations. In contrast, RFB and Rifamycin O lack hydroquinone, granting them resistance to autoxidation and thereby ensuring their efficacy against Mab even under oxidative conditions. Consequently, it is conjectured that the unique structural attributes at C1 and C4 of rifamycin analogs are pivotal for their effectiveness against Mab (Hanh et al., 2020b; Ganapathy et al., 2021b). Recently, RFB was redesigned strategically to enhance its potency against Mab. Modifications at the C-25 position yielded analogs over a hundred times more powerful than RFP and resistant to Mab ADP-ribosylation. Molecular studies highlighted additional interactions, contributing to their superior on-target effectiveness. Validated in vitro, these compounds effectively countered Arr-mediated resistance, displaying potent in vivo efficacy comparable to clarithromycin against Mab. The compound 5 m, exemplary candidate in aspect of antibacterial activity, excellent drug disposition, and significantly improved in vivo pharmacokinetic traits is ongoing investigations to unveil its in vivo efficacy (Lan et al., 2022).
Macrolides represent an antibiotic class that can effectively combat a wide range of bacterial types, including staphylococci, streptococci, mycoplasma, and more. Macrolides are characterized by their structure, comprising amino sugar and/or neutral sugar components linked to a lactone ring, forming macrolides with 12-, 14-, 15-, or 16-membered rings through glycosidic connections. Their mode of action involves binding to the 50S ribosomal subunit in bacteria, which results in the inhibition of protein synthesis (Dinos, 2017). Macrolides remain the core drugs for treating Mab infections (Guo et al., 2021). In certain NTMs, exposure to macrolides triggers the production of specific enzymes that modify the drug’s target binding site. For example, resistance to clarithromycin (CLR), whether intrinsic or acquired, is associated with erm(41) and mutations in the gene rrl encoding a 23S peptidyl transferase in the large 23S ribosomal subunit, respectively (Nessar et al., 2012). In M. abscessus and M. bolletii, inducible macrolide resistance occurs due to a T-C polymorphism in erm(41) at position 28 (only isolates with T28 develop resistance) (Nash et al., 2009). Interestingly, M. massiliense isolate exhibits susceptibility to macrolides due to a non-functional erm(41) caused by a 274-bp deletion. As a result, determining subspecies and macrolide susceptibility is crucial for guiding appropriate treatment. More recently, Guo et al. identified another macrolide-resistant gene in Mab. The gene MAB_2355c exhibits ATP hydrolysis activity and contributes to macrolide resistance by protecting ribosomes (Guo et al., 2021). Expression of MAB_2355c mRNA is significantly upregulated after exposure to macrolides compared to other ribosome-targeting antibiotics. Deletion of MAB_2355c in Mab strains resulted in increased sensitivity to macrolides, while complemented strains exhibited reduced sensitivity to macrolides (Guo et al., 2021).
Acquired resistance to macrolides in a clinical setting often arises from spontaneous mutations at positions 2058 and 2059 on the rrl gene. Additionally, Vester et al. demonstrated that mutations at positions 2057 and 2,611 on rrl can lead to low-level resistance although these mutations are located outside the primary site of macrolide interaction (Vester and Douthwaite, 2001).
Mab possesses a family of transcriptional regulators that may play a role in conferring drug resistance, particularly the WhiB gene family (Morris et al., 2005). WhiB7, a transcriptional activator belonging to the WhiB family of transcriptional regulators, is conserved in actinomycetes and regulates critical cellular processes, including cell division, pathogenesis, and oxidative stress responses. The presence of a helix-turn-helix motif indicates its DNA-binding function (Soliveri et al., 2000; Burian et al., 2012; Nessar et al., 2012). In Mab, 128 genes, including erm(41) and eis2, have been identified in the WhiB7 regulon, indicating their induction through a WhiB7-dependent mechanism. Deletion of Mab whiB7 (MAB_3508c) renders the bacteria more susceptible to antibiotics, such as erythromycin, CLR, streptomycin, spectinomycin (SPC), AMK, and tetracycline, although it does not affect resistance to RFP or INH (Hurst-Hess et al., 2017). Significantly, exposing Mab to sub-inhibitory concentrations of CLR leads to the activation of whiB7 gene expression (Pryjma et al., 2017). This activation subsequently results in the development of resistance to AMK and CLR due to increased expression of erm(41) and eis2 genes (Pryjma et al., 2017; Johansen et al., 2020b).
Tetracycline molecules feature a linear fused tetracyclic core to which several functional groups are attached. These compounds are known to inhibit bacterial growth by preventing the binding of charged aminoacyl-tRNA to the ribosomal A site (Chopra and Roberts, 2001). Tigecycline (TGC) became the first glycylcycline antibiotic approved by the US FDA (Stein and Craig, 2006). While tetracyclines have been one of the most successful classes of antibiotics, their widespread use has led to extensive drug resistance, necessitating their discontinuation in treating various bacterial infections (Rudra et al., 2018). Occasionally, tetracyclines become ineffective due to TetX enzymes, also known as tetracycline destructases. In the past, the limited tolerance of Msm and Mtb to tetracycline was attributed to the WhiB7-dependent TetV/Tap efflux pump. However, Mab exhibits a resistance level approximately 500-fold higher than that of Msm and Mtb. Recently, Rudra and colleagues revealed that this heightened resistance in Mab to the tetracycline class is conferred by a WhiB7-independent tetracycline-inactivating monooxygenase, MAB_1496c (MabTetX). Exposure to sublethal doses of tetracycline and doxycycline leads to a more than 200-fold induction of MabTetX. Conversely, an isogenic deletion strain shows high sensitivity to both antibiotics. The authors also demonstrated that MabTetX’s expression is suppressed by MabTetRx. This finding highlights the potential use of an inhibitor to potentially reinstate the effectiveness of tetracycline and doxycycline (Rudra et al., 2018). As for acquired drug resistance, mutations in the sigH-rshA genes, which regulate heat shock and oxidative-stress responses, have also been found to be involved in TGC resistance or decreased sensitivity in Mab (Ng and Ngeow, 2022). Overexpression of the sigH gene, resulting from the C51R mutation in rshA, causes resistance to or decreased susceptibility to TGC (Ng and Ngeow, 2022).
Recently, OMC, which is classified as a tetracycline antibiotic, has exhibited favorable MIC against various Mab species (Singh et al., 2023). The activity and efficacy of OMC against Mab infection have been proven in both in vitro and in vivo (Shoen et al., 2019; Watkins and Deresinski, 2019; Kaushik et al., 2019a; Brown-Elliott and Wallace, 2021; Nicklas et al., 2022). OMC, like other tetracycline derivatives via binding to the tetracycline-binding site of the bacterial 16S ribosomal RNA, inhibiting bacterial protein synthesis (Brown-Elliott and Wallace, 2021). In contrast to first-generation tetracyclines, OMC has been intentionally engineered to bypass ribosomal protection and tetracycline efflux mechanisms. In Mab, the production of a monooxygenase enzyme may degrade tetracyclines, such as minocycline and doxycycline, but does not affect OMC (Luthra et al., 2018; El Ghali et al., 2023). This feature, in part, revitalizes its activity against recalcitrant MAB and may serve as a deterrent against the development of resistance during treatment. For example, studies by Bax et al. revealed that, except for a 1.5% occurrence at an OMC concentration of 4 mg/L, no instances of drug resistance selection exceeded the spontaneous mutation frequency (Bax et al., 2019).
Active efflux mechanisms have been identified as potential contributors to antibiotic resistance in mycobacteria. The primary role of efflux pump (EP) systems is to protect bacteria from harmful substances, maintain cellular homeostasis, and uphold physiological equilibrium by expelling toxins or metabolites into the extracellular environment (Louw et al., 2009; Remm et al., 2022). Mab possesses genetic sequences that encode protein constituents belonging to the major facilitator family ABC transporters and mycobacterial membrane protein large (MmpL) families (Ripoll et al., 2009), although the precise contribution of EPs to antibiotic resistance in Mab is not fully elucidated. ABC-type multidrug transporters utilize ATP energy to actively remove compounds from the cellular environment. MmpL transporters are multidrug EPs that play a crucial role in transporting various substrates from the periplasmic space to the extracellular environment (Kerr, 2002; Kerr et al., 2005). In further detail, the MmpL transporter family encodes proteins belonging to the resistance, nodulation, and cell division (RND) category. These proteins function as multidrug resistance pumps with the ability to transport a wide range of compounds, including cationic, anionic, and neutral substances, such as drugs, metals, and fatty acids (Domenech et al., 2005).
SPC is an aminocyclitol antibiotic that robustly inhibits bacterial protein synthesis by binding to the 30S subunit of the ribosome. However, its effectiveness against mycobacteria is restricted due to inherent resistance mechanisms (Hurst-Hess K. R. et al., 2023). Mab exhibits a notable inherent resistance to SPC, with MIC exceeding 1,000 μg/mL, rendering it unsuitable for therapeutic applications (Hurst-Hess K. R. et al., 2023). The whiB7 is responsible for Mab resistance for SPC because sublethal exposure to SPC strongly induces whiB7 and its regulon, and a ΔMab_whiB7 strain shows SPC sensitive (Hurst-Hess K. R. et al., 2023). Furthermore, MAB_2780c, a TetV-like efflux pump, provides high-level SPC resistance in Mab (Hurst-Hess K. et al., 2023). For instance, the elimination of MAB_2780c resulted in a significant enhancement of susceptibility to SPC, approximately 150 times greater than that observed in the wildtype bacteria (Hurst-Hess K. R. et al., 2023). The inclusion of the efflux pump inhibitor (EPI), verapamil, leads to a reduction in the MIC of SPC by over 100-fold in bacteria that express MAB_2780c, bringing it down to levels comparable to those observed for the deletion mutant of MAB_2780c (Hurst-Hess K. R. et al., 2023).
Recent studies have attributed the drug resistance function of the Mab MmpL family. The MmpS-MmpL protein complex provides significant resistance to thiacetazone (TAC) analogs, bedaquiline (BDQ), and CFZ (Gutiérrez et al., 2019). Point mutations occurring in the MAB_4384 gene of Mab, a transcriptional repressor belonging to the TetR family, have been linked to resistance to various drugs. These mutations lead to elevated expression levels of the MAB_4383c (mmpS5) and MAB_4382c (mmpL5) genes, ultimately resulting in drug resistance, including resistance to TAC analogs (Halloum et al., 2017; Richard et al., 2018a). In a separate study, Li et al. identified mutations in MAB_4384 in clinically isolated strains resistant to BDQ (Li et al., 2018). Additionally, Negatu et al. reported an EP’s involvement in LZ resistance. They induced high-level LZ resistance in the Mab reference strain in vitro and identified resistance mutations in MAB_4384, resulting in a lower level of antibiotics resistance for drugs such as Sutezolid (STZ), Tedizolid (TDZ), and TBI-223 (Negatu et al., 2023). This MAB_4384-associated lower level of antibiotic resistance was also observed with SPR719 and TAC analogs as well (Halloum et al., 2017; Richard et al., 2018a; Aragaw et al., 2022). The repression of transcriptional expression of two MmpS-MmpL EPs is also facilitated by MAB_2299c, which mediates the production of MmpT5 (Alexander et al., 2017). MmpT5 is a member of the TetR family that regulates the expression of the adjacent mmpS-mmpL (MAB_2300–2,301) genes (Alexander et al., 2017). Mutations in the DNA-binding domain of MAB_2299c lead to upregulated EPs, increased drug efflux, and resistance to CFZ and BDQ (Alexander et al., 2017; Richard et al., 2018b). Furthermore, Gutiérrez et al. identified a new target of MAB_2299c named mmpS-mmpL (MAB_1135c-MAB_1134c), which encodes a new MmpS-MmpL EP system involved in intrinsic resistance to CFZ and BDQ (Gutiérrez et al., 2019).
Apart from the above-mentioned EPs, various EP-related genes exist in Mab. Two EP-encoding genes, MAB_1409 and MAB_3142, were consistently overexpressed upon exposure to CLR (Vianna et al., 2019). Guo et al. also identified six clinical isolates with CLR resistance among 194 whole-genome sequenced isolates. These resistant isolates, which lacked the common rrl 2270/2271 mutation and showed no mutations in the rrl, rplC, rplD, rplV, or erm(41) genes, exhibited elevated expression of EP genes, specifically MAB_2355c, MAB_1409c, and MAB_1846 (Guo et al., 2020). Additionally, Gorzynski et al. reported the upregulation of MAB_0937c, MAB_1137c, MAB_4117c, and MAB_4237c, all of which encode EPs and transporter systems when exposed to AMK (Gorzynski et al., 2021). A promising strategy to enhance drug susceptibility involves inhibiting EP activity using EPIs. EPIs are compounds designed to act on EPs and block their efflux function (Song and Wu, 2016; Remm et al., 2022). Several experimental examples demonstrate the use of EPIs to increase the susceptibility of anti-Mab agents. In a study by Vianna et al., the crucial role of efflux activity in Mab resistance to CLR was highlighted. This was evident in the increased mRNA expression levels of MAB_1409 and MAB_3142 in Mab after exposure to CLR. Moreover, the researchers discovered that verapamil (VP), an FDA-approved EPI with potential as adjunctive chemotherapy for tuberculosis, significantly enhanced susceptibility to CLR. This effect was observed across Mab clinical isolates belonging to the T28 erm(41) sequevar., known for inducible resistance to CLR (Vianna et al., 2019). In addition, Guo et al. reported that the presence of EPIs, such as phenylalanine-arginine β-naphthylamide (PAβN), a peptidomimetic compound, carbonyl cyanide m-chlorophenylhydrazone (CCCP), and VP, significantly decreased the MIC of CLR for Mab resistant isolates that exhibited no rrl 2270/2271 mutation (Guo et al., 2020).
Genetic polymorphisms in highly conserved genes targeted by pharmaceutical medications have been associated with variations in sensitivity to pharmacological effects in Mab infections (Nessar et al., 2012).
Ethambutol and fluoroquinolone resistance serve as examples of genetic polymorphism influencing drug resistance. Ethambutol is chemically derived from ethylenediamine, possessing the stereochemical structure S,S, achieved by substituting a hydrogen atom on each nitrogen atom of ethane-1,2-diamine with a 1-hydroxybutan-2-yl group (Jahangir et al., 2016). Ethambutol functions as a bacteriostatic medication against mycobacterial infections by inhibiting cell wall formation in these microorganisms (Sreevatsan et al., 1997). However, Mab exhibits substantial inherent resistance to ethambutol, primarily due to variant nucleotides within the conserved ethambutol resistance-determining region (ERDR) of the embB gene (Alcaide et al., 1997). Mab shows amino acid substitutions, particularly the replacement of isoleucine by glutamine at position 303 and leucine by methionine at position 304 (referred to as I303Q and L304M, respectively) (Sreevatsan et al., 1997).
Another example of genetic polymorphism influencing drug resistance involves fluoroquinolones. These antibiotics encompass over 20 medications originating from the identification of nalidixic acid. Derived from the quinolone family, fluoroquinolones are synthetic compounds formed by modifying 1-alkyl-1,8-naphthyridin-4-one-3-carboxylic acid. Fluoroquinolones strongly inhibit bacterial enzymes, DNA gyrase, and topoisomerase, critical for processes like DNA replication (Redgrave et al., 2014). They are secondary therapeutic agents for multi-drug resistant tuberculosis (MDR-TB), working by inhibiting DNA gyrase’s supercoiling activity, a specific target of fluoroquinolones (Pantel et al., 2011). NTM resistance to fluoroquinolones is predominantly due to genetic factors, particularly the analysis of conserved sections called quinolone resistance-determining regions (QRDRs) within DNA gyrase subunits GyrA and GyrB, which are the primary targets of quinolone drugs. In Mab, resistance results from the presence of alanine at position 83 (Ala-83) in the GyrA QRDR. Furthermore, resistance is conferred by arginine at position 447 (Arg-447) and asparagine at position 464 (Asn-464) within the GyrB QRDR (Matrat et al., 2008).
Telacebec (Q203) is a groundbreaking anti-tuberculosis drug designed to inhibit the cytochrome bc1 complex, affecting cellular energy production in Mtb. This inhibition reduces ATP synthesis, halting bacterial growth (Pethe et al., 2013; de Jager et al., 2020). Notably, Q203 lacks inhibitory activity against Mab. A recent study by Sorayah et al. revealed that naturally occurring polymorphisms within Mab QcrB are responsible for its increased resistance to Q203. To confirm this resistance mechanism, they engineered a Mycobacterium bovis BCG strain, integrating the chimeric Mab qcrCAB operon, where four amino acids (D311E, L314A, G179S, and C393A on QcrB) were modified to match their counterparts in Mtb. This genetic adjustment rendered the chimeric M. bovis BCG strain susceptible to Q203, indicating that Mab’s resistance to Q203 is attributable to naturally occurring polymorphisms in the drug target, QcrB, rather than other inherent resistance mechanisms such as efflux pumps, cell wall permeability, or target-modifying enzymes (Sorayah et al., 2019).
In recent years, aminoacyl-tRNA synthetases (AARSs) have become significant targets for new antibacterial interventions. AARSs facilitate the acylation process, linking amino acids and tRNA. Inhibiting AARS activity halts protein synthesis, impeding bacterial growth. Benzoxaboroles, known as boron-heterocyclic antibiotics, inhibit leucyl-tRNA synthetase (LeuRS) (Rock et al., 2007). These drugs obstruct protein synthesis via the oxaborole tRNA-trapping mechanism, forming adducts with uncharged tRNALeu molecules that bind to the LeuRS editing domain (Rock et al., 2007). Multiple reports discuss the anti-Mab effects of LeuRS inhibitors, particularly a new class of LeuRS inhibitors such as epetraborole, DS86760016, EC/11770, MRX-6038, and GSK656 (Kim et al., 2021; Wu et al., 2022; Ganapathy et al., 2023a). High-level epetraborole resistance may be attributed to mutations in Mab at locations S303L, T322I, T323P, F321V, G393V, and Y421D within the LeuS gene (Ganapathy et al., 2021a). Among the LeuRS inhibitors, DS86760016 exhibits an improved pharmacokinetic profile, lower plasma clearance, longer plasma half-life, and higher renal excretion than epetraborole in animal models. A recent study by Nguyen et al. demonstrated that DS86760016 displayed similar activity to epetraborole treatment against Mab in vitro, intracellularly, and in zebrafish infection models, with a significantly lower mutation frequency. Laboratory-induced DS86760016-resistant strains included D284G, Q345R, Y420C, I426T, V468L, N469Y, and E524K, which were not found on the LeuS gene in epetraborole-resistant mutants (Nguyen et al., 2023).
Mycolic acids (MA) exist in various forms, including trehalose monomycolates (TMMs), trehalose dimycolates (TDMs), and mycolates covalently linked to arabinogalactan (AG) polysaccharides (McNeil et al., 2020). In the process of MA synthesis, MmpL3 plays a crucial role by transporting MA across the inner membrane, making its contribution to cell wall production indispensable (McNeil et al., 2020). Inhibition of the MmpL3 transporter leads to the accumulation of TMM intracellularly, causing a decrease in mycolyl arabinogalactan peptidoglycan (mAGP) and TDM levels (McNeil et al., 2020). Consequently, MmpL3 represents a versatile drug target, and inhibiting MmpL3 disrupts cell wall biosynthesis (Li et al., 2014). Multiple MmpL3 inhibitors have been recently identified through phenotypic screenings of chemical libraries against Mab. Promisingly, PIPD1, a piperidinol-based molecule, has shown potent in vitro and in vivo activity against clinical Mab strains. Treatment of infected zebrafish with PIPD1 increased embryo survival and reduced bacterial burden (Dupont et al., 2016). Major resistance to PIPD1 and other MmpL3 inhibitors, such as EJMCh-6 (2-(2-cyclohexylethyl)-5,6-dimethyl-1H-benzo[d]imidazole) and BMC-2i, is attributed to mutations in the MmpL3 gene (MAB_4508) of Mab at location A309P. The overexpression of MmpL3, containing the Ala309Pro mutation in Mab wild-type bacteria, results in significant drug resistance to MmpL3 inhibitors, confirming MmpL3 as their target (Dupont et al., 2016; Kozikowski et al., 2017).
LZ, a representative oxazolidinone, disrupts protein synthesis by inhibiting the peptidyl transferase activity of the 23S rRNA in the 50S ribosomal subunit, leading to ribosome stalling. Unfortunately, most clinical Mab isolates exhibit poor susceptibility to LZ (Negatu et al., 2023). However, the effectiveness of LZ against NTMs varies among different derivatives, and its clinical use in patients with NTMs can sometimes result in adverse events, such as peripheral neuropathy and cytopenias. Recently, Kim et al. introduced a novel oxazolidinone with a cyclic amidrazone named DPZ (LCB01-0371). DPZ demonstrated effective inhibition of Mab growth, both in vitro and in mouse lungs in vivo compared to LZ. Furthermore, DPZ exhibited bactericidal activity against all bacterial strains, irrespective of their resistance to AMK, CFX, or CLR. Kim et al. generated laboratory-induced resistant mutants to DPZ and identified mutations in rplC (encoding 50S ribosomal protein L3) at T424C and G419A, along with a nucleotide insertion at position 503 through sequencing analysis (Kim et al., 2017).
The aminobenzimidazole SPR719 targets the ATPase located on Gyrase B in Mtb. SPR719 also demonstrates activity against NTM and has recently entered clinical trials for lung diseases caused by NTM (Aragaw et al., 2022). Brown-Elliott et al. (2018) demonstrated in vitro activity of SPR719 against M. abscessus, M. massiliense, and related subspecies, with an observed MIC50 (MIC required to inhibit the growth of 50% of Mab) value of ~2.0 μg/mL. Additionally, Rubio et al. reported that the phosphate prodrug SPR720 (of SPR719) exhibited favorable in vivo efficacy at a dose of 100 mg/kg/day in an SCID mouse model infected with Mab (Egorova et al., 2021). To identify the molecular target for Mab, Aragaw et al. recently induced two different morphotypes of SPR719-resistant mutants on agar containing 16 x MIC of SPR719, named large and small colonies. Interestingly, the small colony phenotype reflected a lower level of resistance, while large colonies showed high-level SPR719 resistance (>16-fold MIC increase). All strains contained a single amino acid polymorphism, Thr169Asn, in the ATPase domain of Gyrase B, and non-gyrB DNA sequence polymorphisms were revealed by whole-genome sequencing. Thr169 in Mab DNA gyrase corresponds to Ser169 in Mtb GyrB, causing resistance to SPR719 in Mtb (Aragaw et al., 2022).
Mab has emerged as a significant threat to human health, posing challenges in treatment due to its resistance to currently available commercial medications (Quang and Jang, 2021). The rising number of publications on NTM, especially Mab, signifies exponential growth. However, the level of attention dedicated to this issue remains insufficient to effectively address the problem. The primary challenge in developing drugs for Mab is attributed to its exceptional innate and acquired resistance capabilities (Wu et al., 2018). High drug resistance discourages investments by both pharmaceutical companies and governments in this area, leading to passive involvement. As a result, small- and medium-sized organizations, along with academic institutions, are currently the primary sources of knowledge regarding Mab infections and antibiotics. Antibiotic resistance in mycobacterial species can occur through natural or acquired mechanisms (Nessar et al., 2012). Development of natural drug resistance in Mab can be attributed to several factors, including a waxy impermeable cell wall acting as both a physical and chemical barrier (Nessar et al., 2012). Additionally, drug export systems, enzymes capable of modifying drugs or target enzymes, and genetic polymorphism in target genes contribute to this phenomenon (Nessar et al., 2012). Acquired resistance arises from spontaneous mutations at specific genes in response to antibiotics following extended treatment (Johansen et al., 2020b). Such mutations alter the target gene or other related genes, rendering the medication ineffective (Johansen et al., 2020b). Our understanding of natural or acquired antibiotic resistance in Mab remains limited, highlighting the importance of further research into resistance mechanisms against current antibiotics and the discovery of new compounds to overcome resistance hurdles and develop novel drugs.
TN: Investigation, Methodology, Validation, Writing – original draft. BH: Investigation, Methodology, Validation, Writing – review & editing. SJ: Investigation, Methodology, Validation, Writing – review & editing. AA: Investigation, Methodology, Validation, Writing – review & editing. HL: Investigation, Methodology, Validation, Writing – review & editing. CM: Investigation, Methodology, Validation, Writing – review & editing. JJ: Conceptualization, Funding acquisition, Methodology, Validation, Writing – original draft, Writing – review & editing.
The author(s) declare financial support was received for the research, authorship, and/or publication of this article. This work is supported by a grant of the Ministry of Health & Welfare, Republic of Korea (HI22C136100 and 2020ER520601), funded by the grant 2020R1A2C100407714 and RS-2023-00266419 from the National Research Foundation (NRF) of Republic of Korea. This research was also supported by “Regional Innovation Strategy (RIS)” through the NRF funded by the Ministry of Education (MOE) (2021RIS001, 1345370811). TN, BH, SJ, and AA were supported by the BK21 Four Program. AA was sponsored by the Global Korea Scholarship program under the MOE in Korea.
The authors would like to thank all scientists who are developing novel anti-Mab agents.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2024.1331508/full#supplementary-material
Abate, G., Hamzabegovic, F., Eickhoff, C. S., and Hoft, D. F. (2019). BCG vaccination Induces M. Avium and M. abscessus cross-protective immunity. Front. Immunol. 10:234. doi: 10.3389/FIMMU.2019.00234
Alcaide, F., Pfyffer, G. E., and Telenti, A. (1997). Role of embB in natural and acquired resistance to ethambutol in mycobacteria. Antimicrob. Agents Chemother. 41, 2270–2273. doi: 10.1128/AAC.41.10.2270
Alexander, D. C., Vasireddy, R., Vasireddy, S., Philley, J. V., Brown-Elliott, B. A., Perry, B. J., et al. (2017). Emergence of mmpT5 variants during bedaquiline treatment of Mycobacterium intracellulare lung disease. J. Clin. Microbiol. 55, 574–584. doi: 10.1128/JCM.02087-16
Aragaw, W. W., Cotroneo, N., Stokes, S., Pucci, M., Critchley, I., Gengenbacher, M., et al. (2022). In vitro resistance against DNA gyrase inhibitor SPR719 in Mycobacterium avium and Mycobacterium abscessus. Microbiol. Spectr. 10:e0132121. doi: 10.1128/SPECTRUM.01321-21
Baker, A. W., Lewis, S. S., Alexander, B. D., Chen, L. F., Wallace, R. J., Brown-Elliott, B. A., et al. (2017). Two-phase hospital-associated outbreak of Mycobacterium abscessus: investigation and mitigation. Clin. Infect. Dis. 64, 902–911. doi: 10.1093/CID/CIW877
Bax, H. I., De Vogel, C. P., Mouton, J. W., and De Steenwinkel, J. E. M. (2019). Omadacycline as a promising new agent for the treatment of infections with Mycobacterium abscessus. J. Antimicrob. Chemother. 74, 2930–2933. doi: 10.1093/JAC/DKZ267
Becker, K., Aranzana-Climent, V., Cao, S., Nilsson, A., Shariatgorji, R., Haldimann, K., et al. (2021). Efficacy of EBL-1003 (apramycin) against Acinetobacter baumannii lung infections in mice. Clin. Microbiol. Infect. 27, 1315–1321. doi: 10.1016/J.CMI.2020.12.004
Bernut, A., Dupont, C., Sahuquet, A., Herrmann, J. L., Lutfalla, G., and Kremer, L. (2015). Deciphering and imaging pathogenesis and cording of Mycobacterium abscessus in zebrafish embryos. J. Vis. Exp. 103:53130. doi: 10.3791/53130
Bernut, A., Le Moigne, V., Lesne, T., Lutfalla, G., Herrmann, J. L., and Kremer, L. (2014). In vivo assessment of drug efficacy against Mycobacterium abscessus using the embryonic zebrafish test system. Antimicrob. Agents Chemother. 58, 4054–4063. doi: 10.1128/AAC.00142-14
Bernut, A., Nguyen-Chi, M., Halloum, I., Herrmann, J. L., Lutfalla, G., and Kremer, L. (2016). Mycobacterium abscessus-induced granuloma formation is strictly dependent on TNF signaling and neutrophil trafficking. PLoS Pathog. 12:e1005986. doi: 10.1371/JOURNAL.PPAT.1005986
Bhattacharjee, M. K. (2016). “Antibiotics that inhibit protein synthesis” in Chemistry of antibiotics and related drugs (Cham: Springer), 129–151. Available at: https://link.springer.com/book/10.1007/978-3-319-40746-3
Blauwendraat, C., Dixon, G. L. J., Hartley, J. C., Foweraker, J., and Harris, K. A. (2012). The use of a two-gene sequencing approach to accurately distinguish between the species within the Mycobacterium abscessus complex and Mycobacterium chelonae. Eur. J. Clin. Microbiol. Infect. Dis. 31, 1847–1853. doi: 10.1007/S10096-011-1510-9
Brennan, P. J., and Nikaido, H. (1995). The envelope of mycobacteria. Annu. Rev. Biochem. 64, 29–63. doi: 10.1146/ANNUREV.BI.64.070195.000333
Brown-Elliott, B. A., Rubio, A., and Wallace, R. J. (2018). In vitro susceptibility testing of a novel Benzimidazole, SPR719, against nontuberculous mycobacteria. Antimicrob. Agents Chemother. 62, e01503-18. doi: 10.1128/AAC.01503-18
Brown-Elliott, B. A., and Wallace, R. J. (2021). In vitro susceptibility testing of omadacycline against nontuberculous mycobacteria. Antimicrob. Agents Chemother. 65, e01947-20. doi: 10.1128/AAC.01947-20
Bryant, J. M., Grogono, D. M., Rodriguez-Rincon, D., Everall, I., Brown, K. P., Moreno, P., et al. (2016). Emergence and spread of a humantransmissible multidrug-resistant nontuberculous mycobacterium. Science 354, 751–757. doi: 10.1126/science.aaf8156
Burian, J., Ramón-García, S., Howes, C. G., and Thompson, C. J. (2012). WhiB7, a transcriptional activator that coordinates physiology with intrinsic drug resistance in Mycobacterium tuberculosis. Expert Rev. Anti-Infect. Ther. 10, 1037–1047. doi: 10.1586/ERI.12.90
Bush, K., and Bradford, P. A. (2016). β-Lactams and β-lactamase inhibitors: an overview. Cold Spring Harb. Perspect. Med. 6:a025247. doi: 10.1101/CSHPERSPECT.A025247
Carvalho, R., de Sonneville, J., Stockhammer, O. W., Savage, N. D. L., Veneman, W. J., Ottenhoff, T. H. M., et al. (2011). A high-throughput screen for tuberculosis progression. PLoS One 6:e16779. doi: 10.1371/JOURNAL.PONE.0016779
Chopra, S., Matsuyama, K., Hutson, C., and Madrid, P. (2011). Identification of antimicrobial activity among FDA-approved drugs for combating Mycobacterium abscessus and Mycobacterium chelonae. J. Antimicrob. Chemother. 66, 1533–1536. doi: 10.1093/jac/dkr154
Chopra, I., and Roberts, M. (2001). Tetracycline antibiotics: mode of action, applications, molecular biology, and epidemiology of bacterial resistance. Microbiol. Mol. Biol. Rev. 65, 232–260. doi: 10.1128/MMBR.65.2.232-260.2001
Christianson, S., Grierson, W., Kein, D., Tyler, A. D., Wolfe, J., and Sharma, M. K. (2016). Time-to-detection of inducible macrolide resistance in Mycobacterium abscessus subspecies and its association with the Erm(41) Sequevar. PLoS One 11:e0158723. doi: 10.1371/JOURNAL.PONE.0158723
Chulluncuy, R., Espiche, C., Nakamoto, J. A., Fabbretti, A., and Milón, P. (2016). Conformational response of 30S-bound IF3 to A-site binders streptomycin and kanamycin. Antibiotics 5:38. doi: 10.3390/ANTIBIOTICS5040038
Daher, W., Leclercq, L. D., Johansen, M. D., Hamela, C., Karam, J., Trivelli, X., et al. (2022). Glycopeptidolipid glycosylation controls surface properties and pathogenicity in Mycobacterium abscessus. Cell Chem. Biol. 29, 910–924.e7. doi: 10.1016/J.CHEMBIOL.2022.03.008
Dal Molin, M., Gut, M., Rominski, A., Haldimann, K., Becker, K., and Sander, P. (2017). Molecular mechanisms of intrinsic streptomycin resistance in Mycobacterium abscessus. Antimicrob. Agents Chemother. 62, e01427-17. doi: 10.1128/AAC.01427-17
de Jager, V. R., Dawson, R., van Niekerk, C., Hutchings, J., Kim, J., Vanker, N., et al. (2020). Telacebec (Q203), a new antituberculosis agent. N. Engl. J. Med. 382, 1280–1281. doi: 10.1056/NEJMC1913327
De Rosa, M., Verdino, A., Soriente, A., and Marabotti, A. (2021). The odd couple(s): an overview of Beta-lactam antibiotics bearing more than one pharmacophoric group. Int. J. Mol. Sci. 22:617. doi: 10.3390/IJMS22020617
Dedrick, R. M., Abad, L., Storey, N., Kaganovsky, A. M., Smith, B. E., Aull, H. A., et al. (2023). The problem of Mycobacterium abscessus complex: multi-drug resistance, bacteriophage susceptibility and potential healthcare transmission. Clin. Microbiol. Infect. 29, 1335.e9–1335.e16. doi: 10.1016/J.CMI.2023.06.026
Degiacomi, G., Sammartino, J. C., Chiarelli, L. R., Riabova, O., Makarov, V., and Pasca, M. R. (2019). Mycobacterium abscessus, an emerging and worrisome pathogen among cystic fibrosis patients. Int. J. Mol. Sci. 20:5868. doi: 10.3390/IJMS20235868
Desai, A. N., and Hurtado, R. M. (2018). Infections and outbreaks of nontuberculous mycobacteria in hospital settings. Curr. Treat. Options Infect. Dis. 10, 169–181. doi: 10.1007/S40506-018-0165-9
Dick, T., Shin, S. J., Koh, W. J., Dartois, V., and Gengenbacher, M. (2020). Rifabutin is active against Mycobacterium abscessus in mice. Antimicrob. Agents Chemother. 64, e01943-19. doi: 10.1128/AAC.01943-19
Dinos, G. P. (2017). The macrolide antibiotic renaissance. Br. J. Pharmacol. 174, 2967–2983. doi: 10.1111/BPH.13936
Domenech, P., Reed, M. B., and Barry, C. E. (2005). Contribution of the Mycobacterium tuberculosis MmpL protein family to virulence and drug resistance. Infect. Immun. 73, 3492–3501. doi: 10.1128/IAI.73.6.3492-3501.2005
Dousa, K. M., Nguyen, D. C., Kurz, S. G., Taracila, M. A., Bethel, C. R., Schinabeck, W., et al. (2022). Inhibiting Mycobacterium abscessus cell wall synthesis: using a novel diazabicyclooctane β-lactamase inhibitor to augment β-lactam action. MBio 13:e0352921. doi: 10.1128/MBIO.03529-21
Dubée, V., Bernut, A., Cortes, M., Lesne, T., Dorchene, D., Lefebvre, A. L., et al. (2015a). β-Lactamase inhibition by avibactam in Mycobacterium abscessus. J. Antimicrob. Chemother. 70, 1051–1058. doi: 10.1093/JAC/DKU510
Dubée, V., Soroka, D., Cortes, M., Lefebvre, A. L., Gutmann, L., Hugonnet, J. E., et al. (2015b). Impact of β-lactamase inhibition on the activity of ceftaroline against Mycobacterium tuberculosis and Mycobacterium abscessus. Antimicrob. Agents Chemother. 59, 2938–2941. doi: 10.1128/AAC.05080-14
Dupont, C., Viljoen, A., Dubar, F., Blaise, M., Bernut, A., Pawlik, A., et al. (2016). A new piperidinol derivative targeting mycolic acid transport in Mycobacterium abscessus. Mol. Microbiol. 101, 515–529. doi: 10.1111/MMI.13406
Egorova, A., Jackson, M., Gavrilyuk, V., and Makarov, V. (2021). Pipeline of anti-Mycobacterium abscessus small molecules: repurposable drugs and promising novel chemical entities. Med. Res. Rev. 41, 2350–2387. doi: 10.1002/MED.21798
El Ghali, A., Morrisette, T., Alosaimy, S., Lucas, K., Tupayachi-Ortiz, M. G., Vemula, R., et al. (2023). Long-term evaluation of clinical success and safety of omadacycline in nontuberculous mycobacteria infections: a retrospective, multicenter cohort of real-world health outcomes. Antimicrob. Agents Chemother. 67:e0082423. doi: 10.1128/AAC.00824-23
Falkinham, J. O. (2011). Nontuberculous mycobacteria from household plumbing of patients with nontuberculous mycobacteria disease. Emerg. Infect. Dis. 17, 419–424. doi: 10.3201/eid1703.101510
Faria, S., Joao, I., and Jordao, L. (2015). General overview on nontuberculous mycobacteria, biofilms, and human infection. J. Pathog. 2015, 1–10. doi: 10.1155/2015/809014
Fernandes Garcia de Carvalho, N., Rodrigues Mestrinari, A. C., Brandão, A., Souza Jorge, L., Franco, C., da Silveira Paro Pedro, H., et al. (2018). Hospital bronchoscopy-related pseudo-outbreak caused by a circulating Mycobacterium abscessus subsp. massiliense. J. Hosp. Infect. 100, e138–e141. doi: 10.1016/J.JHIN.2018.07.043
Ganapathy, U. S., Dartois, V., and Dick, T. (2019). Repositioning rifamycins for Mycobacterium abscessus lung disease. Expert Opin. Drug Discov. 14, 867–878. doi: 10.1080/17460441.2019.1629414
Ganapathy, U. S., Del Rio, R. G., Cacho-Izquierdo, M., Ortega, F., Lelièvre, J., Barros-Aguirre, D., et al. (2023a). A Leucyl-tRNA synthetase inhibitor with broad-spectrum anti-mycobacterial activity. Antimicrob. Agents Chemother. 95, e02420-20. doi: 10.1128/AAC.02420-20
Ganapathy, U. S., and Dick, T. (2022). Why matter matters: fast-tracking Mycobacterium abscessus drug discovery. Molecules 27:6948. doi: 10.3390/MOLECULES27206948
Ganapathy, U. S., Gengenbacher, M., and Dick, T. (2021a). Epetraborole is active against Mycobacterium abscessus. Antimicrob. Agents Chemother. 65:e0115621. doi: 10.1128/AAC.01156-21
Ganapathy, U. S., Lan, T., Dartois, V., Aldrich, C. C., and Dick, T. (2023b). Blocking ADP-ribosylation expands the anti-mycobacterial spectrum of rifamycins. Microbiol. Spectr. 11:e0190023. doi: 10.1128/SPECTRUM.01900-23
Ganapathy, U. S., Lan, T., Krastel, P., Lindman, M., Zimmerman, M. D., Ho, H. P., et al. (2021b). Blocking bacterial naphthohydroquinone oxidation and ADP-ribosylation improves activity of rifamycins against Mycobacterium abscessus. Antimicrob. Agents Chemother. 65:e0097821. doi: 10.1128/AAC.00978-21
Gorzynski, M., Week, T., Jaramillo, T., Dzalamidze, E., and Danelishvili, L. (2021). Mycobacterium abscessus genetic determinants associated with the intrinsic resistance to antibiotics. Microorganisms 9:2527. doi: 10.3390/MICROORGANISMS9122527
Greendyke, R., and Byrd, T. F. (2008). Differential antibiotic susceptibility of Mycobacterium abscessus variants in biofilms and macrophages compared to that of planktonic bacteria. Antimicrob. Agents Chemother. 52, 2019–2026. doi: 10.1128/AAC.00986-07
Guo, Q., Chen, J., Zhang, S., Zou, Y., Zhang, Y., Huang, D., et al. (2020). Efflux pumps contribute to intrinsic clarithromycin resistance in clinical, Mycobacterium abscessus Isolates. Infect. Drug Resist. 13, 447–454. doi: 10.2147/IDR.S239850
Guo, Q., Zhang, Y., Fan, J., Zhang, H., Zhang, Z., Li, B., et al. (2021). MAB_2355c confers macrolide resistance in Mycobacterium abscessus by ribosome protection. Antimicrob. Agents Chemother. 65:e0033021. doi: 10.1128/AAC.00330-21
Gupta, R., Netherton, M., Byrd, T. F., and Rohde, K. H. (2017). Reporter-based assays for high-throughput drug screening against Mycobacterium abscessus. Front. Microbiol. 8:2204. doi: 10.3389/FMICB.2017.02204
Gutiérrez, A. V., Richard, M., Roquet-Banères, F., Viljoen, A., and Kremer, L. (2019). The TetR family transcription factor MAB_2299c regulates the expression of two distinct MmpS-MmpL efflux pumps involved in cross-resistance to clofazimine and bedaquiline in Mycobacterium abscessus. Antimicrob. Agents Chemother. 63, e01000-19. doi: 10.1128/AAC.01000-19
Gutiérrez, A. V., Viljoen, A., Ghigo, E., Herrmann, J. L., and Kremer, L. (2018). Glycopeptidolipids, a double-edged sword of the Mycobacterium abscessus complex. Front. Microbiol. 9:1145. doi: 10.3389/FMICB.2018.01145
Halloum, I., Viljoen, A., Khanna, V., Craig, D., Bouchier, C., Brosch, R., et al. (2017). Resistance to thiacetazone derivatives active against Mycobacterium abscessus involves mutations in the MmpL5 transcriptional repressor MAB_4384. Antimicrob. Agents Chemother. 61, e02509-16. doi: 10.1128/AAC.02509-16
Hanh, B. T. B., Kim, T. H., Park, J. W., Lee, D. G., Kim, J. S., Du, Y. E., et al. (2020a). Etamycin as a novel Mycobacterium abscessus inhibitor. Int. J. Mol. Sci. 21:6908. doi: 10.3390/IJMS21186908
Hanh, B. T. B., Park, J. W., Kim, T. H., Kim, J. S., Yang, C. S., Jang, K., et al. (2020b). Rifamycin O, an alternative anti-Mycobacterium abscessus agent. Molecules 25:1597. doi: 10.3390/MOLECULES25071597
Harada, T., Akiyama, Y., Kurashima, A., Nagai, H., Tsuyuguchi, K., Fujii, T., et al. (2012). Clinical and microbiological differences between Mycobacterium abscessus and Mycobacterium massiliense lung diseases. J. Clin. Microbiol. 50, 3556–3561. doi: 10.1128/JCM.01175-12
Howard, S. T., Rhoades, E., Recht, J., Pang, X., Alsup, A., Kolter, R., et al. (2006). Spontaneous reversion of Mycobacterium abscessus from a smooth to a rough morphotype is associated with reduced expression of glycopeptidolipid and reacquisition of an invasive phenotype. Microbiology 152, 1581–1590. doi: 10.1099/mic.0.28625-0
Hurst-Hess, K., McManaman, C., Yang, Y., Gupta, S., and Ghosh, P. (2023). Hierarchy and interconnected networks in the WhiB7 mediated transcriptional response to antibiotic stress in Mycobacterium abscessus. PLoS Genet. 19:e1011060. doi: 10.1371/JOURNAL.PGEN.1011060
Hurst-Hess, K. R., Phelps, G. A., Wilt, L. A., Lee, R. E., and Ghosh, P. (2023). Mab2780c, a TetV-like efflux pump, confers high-level spectinomycin resistance in Mycobacterium abscessus. Tuberculosis (Edinb.) 138:102295. doi: 10.1016/J.TUBE.2022.102295
Hurst-Hess, K., Rudra, P., and Ghosh, P. (2017). Mycobacterium abscessus WhiB7 regulates a species-specific repertoire of genes to confer extreme antibiotic resistance. Antimicrob. Agents Chemother. 61, e01347-17. doi: 10.1128/AAC.01353-17
Ishikawa, M., García-Mateo, N., Čusak, A., López-Hernández, I., Fernández-Martínez, M., Müller, M., et al. (2019). Lower ototoxicity and absence of hidden hearing loss point to gentamicin C1a and apramycin as promising antibiotics for clinical use. Sci. Rep. 9:2410. doi: 10.1038/S41598-019-38634-3
Jahangir, M., Farwa, U., Mazhar, F., Malik, A., and Ahmad, E. (2016). Pak. J. Pharm. Sci. 29:Metal II complexes of ethambutol as good enzyme inhibitor and promising antioxidant, 1601–1608.
Jarlier, V., and Nikaido, H. (1994). Mycobacterial cell wall: structure and role in natural resistance to antibiotics. FEMS Microbiol. Lett. 123, 11–18. doi: 10.1111/J.1574-6968.1994.TB07194.X
Jeong, S. H., Kim, S. Y., Huh, H. J., Ki, C. S., Lee, N. Y., Kang, C. I., et al. (2017). Mycobacteriological characteristics and treatment outcomes in extrapulmonary Mycobacterium abscessus complex infections. Int. J. Infect. Dis. 60, 49–56. doi: 10.1016/J.IJID.2017.05.007
Jeong, J., Kim, G., Moon, C., Kim, H. J., Kim, T. H., and Jang, J. (2018). Pathogen box screening for hit identification against Mycobacterium abscessus. PLoS One 13:e0195595. doi: 10.1371/JOURNAL.PONE.0195595
Johansen, M. D., Daher, W., Roquet-Banères, F., Raynaud, C., Alcaraz, M., Maurer, F. P., et al. (2020a). Rifabutin is bactericidal against intracellular and extracellular forms of Mycobacterium abscessus. Antimicrob. Agents Chemother. 64, e00363-20. doi: 10.1128/AAC.00363-20
Johansen, M. D., Herrmann, J. L., and Kremer, L. (2020b). Non-tuberculous mycobacteria and the rise of Mycobacterium abscessus. Nat. Rev. Microbiol. 18, 392–407. doi: 10.1038/S41579-020-0331-1
Jönsson, B. E., Bylund, J., Johansson, B. R., Telemo, E., and Wold, A. E. (2013). Cord-forming mycobacteria induce DNA meshwork formation by human peripheral blood mononuclear cells. Pathog. Dis. 67, 54–66. doi: 10.1111/2049-632X.12007
Juhas, M., Widlake, E., Teo, J., Huseby, D. L., Tyrrell, J. M., Polikanov, Y. S., et al. (2019). In vitro activity of apramycin against multidrug-, carbapenem- and aminoglycoside-resistant enterobacteriaceae and Acinetobacter baumannii. J. Antimicrob. Chemother. 74, 944–952. doi: 10.1093/JAC/DKY546
Kaushik, A., Ammerman, N. C., Martins, O., Parrish, N. M., and Nuermberger, E. L. (2019a). In vitro activity of new tetracycline analogs omadacycline and eravacycline against drug-resistant clinical isolates of Mycobacterium abscessus. Antimicrob. Agents Chemother. 63, e00470-19. doi: 10.1128/AAC.00470-19
Kaushik, A., Ammerman, N. C., Parrish, N. M., and Nuermberger, E. L. (2019b). New β-lactamase inhibitors nacubactam and zidebactam improve the in vitro activity of β-lactam antibiotics against Mycobacterium abscessus complex clinical isolates. Antimicrob. Agents Chemother. 63, e00733-19. doi: 10.1128/AAC.00733-19
Kerr, I. D. (2002). Structure and association of ATP-binding cassette transporter nucleotide-binding domains. Biochim. Biophys. Acta-Biomembr. 1561, 47–64. doi: 10.1016/S0304-4157(01)00008-9
Kerr, I. D., Reynolds, E. D., and Cove, J. H. (2005). ABC proteins and antibiotic drug resistance: is it all about transport? Biochem. Soc. Trans. 33, 1000–1002. doi: 10.1042/BST0331000
Kim, T. H., Bich Hanh, B. T., Kim, G., Lee, D. G., Park, J. W., Lee, S. E., et al. (2019). Thiostrepton: a novel therapeutic drug candidate for Mycobacterium abscessus infection. Molecules 24:4511. doi: 10.3390/MOLECULES24244511
Kim, T. S., Choe, J. H., Kim, Y. J., Yang, C. S., Kwon, H. J., Jeong, J., et al. (2017). Activity of LCB01-0371, a novel oxazolidinone, against Mycobacterium abscessus. Antimicrob. Agents Chemother. 61, e02752-16. doi: 10.1128/AAC.02752-16
Kim, T., Hanh, B. T. B., Heo, B., Quang, N., Park, Y., Shin, J., et al. (2021). A screening of the MMV pandemic response box reveals epetraborole as a new potent inhibitor against Mycobacterium abscessus. Int. J. Mol. Sci. 22:5936. doi: 10.3390/IJMS22115936
Kozikowski, A. P., Onajole, O. K., Stec, J., Dupont, C., Viljoen, A., Richard, M., et al. (2017). Targeting mycolic acid transport by indole-2-carboxamides for the treatment of Mycobacterium abscessus infections. J. Med. Chem. 60, 5876–5888. doi: 10.1021/ACS.JMEDCHEM.7B00582
Lan, T., Ganapathy, U. S., Sharma, S., Ahn, Y. M., Zimmerman, M., Molodtsov, V., et al. (2022). Redesign of rifamycin antibiotics to overcome ADP-ribosylation-mediated resistance. Angew. Chem. Int. Ed. Engl. 61:e202211498. doi: 10.1002/ANIE.202211498
Le Run, E., Arthur, M., and Mainardia, J. L. (2019). In vitro and intracellular activity of imipenem combined with tedizolid, rifabutin, and avibactam against Mycobacterium abscessus. Antimicrob. Agents Chemother. 63, e01915-18. doi: 10.1128/AAC.01915-18
Lee, M. R., Sheng, W. H., Hung, C. C., Yu, C. J., Lee, L. N., and Hsueh, P. R. (2015). Mycobacterium abscessus complex infections in humans. Emerg. Infect. Dis. 21, 1638–1646. doi: 10.3201/EID2109.141634
Lefebvre, A. L., Dubée, V., Cortes, M., Dorchêne, D., Arthur, M., and Mainardi, J. L. (2016). Bactericidal and intracellular activity of β-lactams against Mycobacterium abscessus. J. Antimicrob. Chemother. 71, 1556–1563. doi: 10.1093/JAC/DKW022
Lefebvre, A. L., Le Moigne, V., Bernut, A., Veckerlé, C., Compain, F., Herrmann, J. L., et al. (2017). Inhibition of the β-lactamase BlaMab by avibactam improves the in vitro and in vivo efficacy of imipenem against Mycobacterium abscessus. Antimicrob. Agents Chemother. 61, e02440-16. doi: 10.1128/AAC.02440-16
Li, W., Upadhyay, A., Fontes, F. L., North, E. J., Wang, Y., Crans, D. C., et al. (2014). Novel insights into the mechanism of inhibition of MmpL3, a target of multiple pharmacophores in Mycobacterium tuberculosis. Antimicrob. Agents Chemother. 58, 6413–6423. doi: 10.1128/AAC.03229-14
Li, B., Ye, M., Guo, Q., Zhang, Z., Yang, S., Ma, W., et al. (2018). Determination of MIC distribution and mechanisms of decreased susceptibility to bedaquiline among clinical isolates of Mycobacterium abscessus. Antimicrob. Agents Chemother. 62, e00175-18. doi: 10.1128/AAC.00175-18
Lipman, M., Kunst, H., Loebinger, M. R., Milburn, H. J., and King, M. (2021). Non tuberculous mycobacteria pulmonary disease: patients and clinicians working together to improve the evidence base for care. Int. J. Infect. Dis. 113, S73–S77. doi: 10.1016/J.IJID.2021.03.064
Lopeman, R. C., Harrison, J., Desai, M., and Cox, J. A. G. (2019). Mycobacterium abscessus: environmental bacterium turned clinical nightmare. Microorganisms 7:90. doi: 10.3390/MICROORGANISMS7030090
López-Roa, P., Esteban, J., and Muñoz-Egea, M. C. (2022). Updated review on the mechanisms of pathogenicity in Mycobacterium abscessus, a rapidly growing emerging pathogen. Microorganisms 11:90. doi: 10.3390/MICROORGANISMS11010090
Lorè, N. I., Saliu, F., Spitaleri, A., Schäfle, D., Nicola, F., Cirillo, D. M., et al. (2022). The aminoglycoside-modifying enzyme Eis2 represents a new potential in vivo target for reducing antimicrobial drug resistance in Mycobacterium abscessus complex. Eur. Respir. J. 60:2201541. doi: 10.1183/13993003.01541-2022
Louw, G. E., Warren, R. M., Gey Van Pittius, N. C., McEvoy, C. R. E., Van Helden, P. D., and Victor, T. C. (2009). A balancing act: efflux/influx in mycobacterial drug resistance. Antimicrob. Agents Chemother. 53, 3181–3189. doi: 10.1128/AAC.01577-08
Luthra, S., Rominski, A., and Sander, P. (2018). The role of antibiotic-target-modifying and antibiotic-modifying enzymes in Mycobacterium abscessus drug resistance. Front. Microbiol. 9:2179. doi: 10.3389/FMICB.2018.02179
Malin, J. J., Winter, S., Van Gumpel, E., Plum, G., and Rybniker, J. (2019). Extremely low hit rate in a diverse chemical drug screen targeting Mycobacterium abscessus. Antimicrob. Agents Chemother. 63, e01008-19. doi: 10.1128/AAC.01008-19
Marrakchi, H., Lanéelle, M. A., and Daffé, M. (2014). Mycolic acids: structures, biosynthesis, and beyond. Chem. Biol. 21, 67–85. doi: 10.1016/J.CHEMBIOL.2013.11.011
Martin, C., Timm, J., Rauzier, J., Gomez-Lus, R., Davies, J., and Gicquel, B. (1990). Transposition of an antibiotic resistance element in mycobacteria. Nat 345, 739–743. doi: 10.1038/345739a0
Matrat, S., Aubry, A., Mayer, C., Jarlier, V., and Cambau, E. (2008). Mutagenesis in the α3α4 GyrA helix and in the toprim domain of GyrB refines the contribution of Mycobacterium tuberculosis DNA gyrase to intrinsic resistance to quinolones. Antimicrob. Agents Chemother. 52, 2909–2914. doi: 10.1128/AAC.01380-07
Matt, T., Ng, C. L., Lang, K., Sha, S. H., Akbergenov, R., Shcherbakov, D., et al. (2012). Dissociation of antibacterial activity and aminoglycoside ototoxicity in the 4-monosubstituted 2-deoxystreptamine apramycin. Proc. Natl. Acad. Sci. U. S. A. 109, 10984–10989. doi: 10.1073/PNAS.1204073109
Maurer, F. P., Castelberg, C., Quiblier, C., Böttger, E. C., and Somoskövi, A. (2014). Erm(41)-dependent inducible resistance to azithromycin and clarithromycin in clinical isolates of Mycobacterium abscessus. J. Antimicrob. Chemother. 69, 1559–1563. doi: 10.1093/JAC/DKU007
Maurer, F. P., Rüegger, V., Ritter, C., Bloemberg, G. V., and Böttger, E. C. (2012). Acquisition of clarithromycin resistance mutations in the 23S rRNA gene of Mycobacterium abscessus in the presence of inducible erm(41). J. Antimicrob. Chemother. 67, 2606–2611. doi: 10.1093/JAC/DKS279
McNeil, M. B., O’Malley, T., Dennison, D., Shelton, C. D., Sunde, B., and Parish, T. (2020). Multiple mutations in Mycobacterium tuberculosis MmpL3 increase resistance to MmpL3 inhibitors. mSphere 5, e00985-20. doi: 10.1128/MSPHERE.00985-20
Meir, M., Bifani, P., and Barkan, D. (2018). The addition of avibactam renders piperacillin an effective treatment for Mycobacterium abscessus infection in an in vivo model. Antimicrob. Resist. Infect. Control 7:151. doi: 10.1186/S13756-018-0448-4
Mingeot-Leclercq, M. P., Glupczynski, Y., and Tulkens, P. M. (1999). Aminoglycosides: activity and resistance. Antimicrob. Agents Chemother. 43, 727–737. doi: 10.1128/AAC.43.4.727
Moguillansky, N., DeSear, K., and Dousa, K. M. (2023). A 40-year-old female with Mycobacterium abscessus successfully treated with a dual beta-lactam combination. Cureus 15:e40993. doi: 10.7759/CUREUS.40993
Moore, M., and Frerichs, J. B. (1953). An unusual acid-fast infection of the knee with subcutaneous, abscess-like lesions of the gluteal region; report of a case with a study of the organism, Mycobacterium abscessus, n. sp. J. Invest. Dermatol. 20, 133–169. doi: 10.1038/jid.1953.18
Moore, J. E., Koulianos, G., Hardy, M., Misawa, N., and Millar, B. C. (2018). Antimycobacterial activity of veterinary antibiotics (apramycin and framycetin) against Mycobacterium abscessus: implication for patients with cystic fibrosis. Int. J. Mycobacteriol. 7, 265–267. doi: 10.4103/IJMY.IJMY_73_18
Morris, R. P., Nguyen, L., Gatfield, J., Visconti, K., Nguyen, K., Schnappinger, D., et al. (2005). Ancestral antibiotic resistance in Mycobacterium tuberculosis. Proc. Natl. Acad. Sci. U. S. A. 102, 12200–12205. doi: 10.1073/PNAS.0505446102
Nash, K. A., Brown-Elliott, A. B., and Wallace, R. J. (2009). A novel gene, erm(41), confers inducible macrolide resistance to clinical isolates of Mycobacterium abscessus but is absent from Mycobacterium chelonae. Antimicrob. Agents Chemother. 53, 1367–1376. doi: 10.1128/AAC.01275-08
Negatu, D. A., Aragaw, W. W., Dartois, V., and Dick, T. (2023). Characterization of in vitro resistance to linezolid in Mycobacterium abscessus. Microbiol. Spectr. 11:e0219923. doi: 10.1128/SPECTRUM.02199-23
Nessar, R., Cambau, E., Reyrat, J. M., Murray, A., and Gicquel, B. (2012). Mycobacterium abscessus: a new antibiotic nightmare. J. Antimicrob. Chemother. 67, 810–818. doi: 10.1093/JAC/DKR578
Nessar, R., Reyrat, J. M., Murray, A., and Gicquel, B. (2011). Genetic analysis of new 16S rRNA mutations conferring aminoglycoside resistance in Mycobacterium abscessus. J. Antimicrob. Chemother. 66, 1719–1724. doi: 10.1093/JAC/DKR209
Ng, H. F., and Ngeow, Y. F. (2022). Genetic determinants of tigecycline resistance in Mycobacteroides abscessus. Antibiotics 11:572. doi: 10.3390/ANTIBIOTICS11050572
Nguyen, T. Q., Heo, B. E., Hanh, B. T. B., Jeon, S., Park, Y., Choudhary, A., et al. (2023). DS86760016, a Leucyl-tRNA synthetase inhibitor, is active against Mycobacterium abscessus. Antimicrob. Agents Chemother. 67:e0156722. doi: 10.1128/AAC.01567-22
Nguyen, L., and Thompson, C. J. (2006). Foundations of antibiotic resistance in bacterial physiology: the mycobacterial paradigm. Trends Microbiol. 14, 304–312. doi: 10.1016/j.tim.2006.05.005
Nicklas, D. A., Maggioncalda, E. C., Story-Roller, E., Eichelman, B., Tabor, C., Serio, A. W., et al. (2022). Potency of omadacycline against Mycobacteroides abscessus clinical isolates in vitro and in a mouse model of pulmonary infection. Antimicrob. Agents Chemother. 66:e0170421. doi: 10.1128/AAC.01704-21
NIH ClinicalTrials (2023). Available at: https://classic.clinicaltrials.gov/ct2/results?cond=abscessus&term=&cntry=&state=&city=&dist=.
Pantel, A., Petrella, S., Matrat, S., Brossier, F., Bastian, S., Reitter, D., et al. (2011). DNA gyrase inhibition assays are necessary to demonstrate fluoroquinolone resistance secondary to gyrB mutations in Mycobacterium tuberculosis. Antimicrob. Agents Chemother. 55, 4524–4529. doi: 10.1128/AAC.00707-11
Paulowski, L., Beckham, K. S. H., Johansen, M. D., Berneking, L., Van, N., Degefu, Y., et al. (2022). C25-modified rifamycin derivatives with improved activity against Mycobacterium abscessus. PNAS Nexus 1:pgac130. doi: 10.1093/PNASNEXUS/PGAC130
Pethe, K., Bifani, P., Jang, J., Kang, S., Park, S., Ahn, S., et al. (2013). Discovery of Q203, a potent clinical candidate for the treatment of tuberculosis. Nat. Med. 19, 1157–1160. doi: 10.1038/NM.3262
Piccaro, G., Pietraforte, D., Giannoni, F., Mustazzolu, A., and Fattorini, L. (2014). Rifampin induces hydroxyl radical formation in Mycobacterium tuberculosis. Antimicrob. Agents Chemother. 58, 7527–7533. doi: 10.1128/AAC.03169-14
Prammananan, T., Sander, P., Brown, B. A., Frischkorn, K., Onyi, G. O., Zhang, Y., et al. (1998). A single 16S ribosomal RNA substitution is responsible for resistance to amikacin and other 2-deoxystreptamine aminoglycosides in Mycobacterium abscessus and Mycobacterium chelonae. J. Infect. Dis. 177, 1573–1581. doi: 10.1086/515328
Pryjma, M., Burian, J., Kuchinski, K., and Thompson, C. J. (2017). Antagonism between front-line antibiotics clarithromycin and amikacin in the treatment of Mycobacterium abscessus infections is mediated by the whiB7 gene. Antimicrob. Agents Chemother. 61, e01353-17. doi: 10.1128/AAC.01347-17
Quang, N. T., and Jang, J. (2021). Current molecular therapeutic agents and drug candidates for Mycobacterium abscessus. Front. Pharmacol. 12:2117. doi: 10.3389/FPHAR.2021.724725
Ramírez, A., Ruggiero, M., Aranaga, C., Cataldi, A., Gutkind, G., De Waard, J. H., et al. (2017). Biochemical characterization of β-lactamases from Mycobacterium abscessus complex and genetic environment of the β-lactamase-encoding gene. Microb. Drug Resist. 23, 294–300. doi: 10.1089/MDR.2016.0047
Ratnatunga, C. N., Lutzky, V. P., Kupz, A., Doolan, D. L., Reid, D. W., Field, M., et al. (2020). The rise of non-tuberculosis mycobacterial lung disease. Front. Immunol. 11:303. doi: 10.3389/FIMMU.2020.00303
Redgrave, L. S., Sutton, S. B., Webber, M. A., and Piddock, L. J. V. (2014). Fluoroquinolone resistance: mechanisms, impact on bacteria, and role in evolutionary success. Trends Microbiol. 22, 438–445. doi: 10.1016/J.TIM.2014.04.007
Remm, S., Earp, J. C., Dick, T., Dartois, V., and Seeger, M. A. (2022). Critical discussion on drug efflux in Mycobacterium tuberculosis. FEMS Microbiol. Rev. 46:fuab050. doi: 10.1093/FEMSRE/FUAB050
Richard, M., Gutiérrez, A. V., Viljoen, A. J., Ghigo, E., Blaise, M., and Kremer, L. (2018a). Mechanistic and structural insights into the unique TetR-dependent regulation of a drug efflux pump in Mycobacterium abscessus. Front. Microbiol. 9:649. doi: 10.3389/FMICB.2018.00649
Richard, M., Gutiérrez, A. V., Viljoen, A., Rodriguez-Rincon, D., Roquet-Baneres, F., Blaise, M., et al. (2018b). Mutations in the MAB_2299c TetR regulator confer cross-resistance to clofazimine and bedaquiline in Mycobacterium abscessus. Antimicrob. Agents Chemother. 63, e01316-18. doi: 10.1128/AAC.01316-18
Richter, A., Strauch, A., Chao, J., Ko, M., and Av-Gay, Y. (2018). Screening of preselected libraries targeting Mycobacterium abscessus for drug discovery. Antimicrob. Agents Chemother. 62, e00828-18. doi: 10.1128/AAC.00828-18
Ripoll, F., Pasek, S., Schenowitz, C., Dossat, C., Barbe, V., Rottman, M., et al. (2009). Non mycobacterial virulence genes in the genome of the emerging pathogen Mycobacterium abscessus. PLoS One 4:e5660. doi: 10.1371/JOURNAL.PONE.0005660
Rock, F. L., Mao, W., Yaremchuk, A., Tukalo, M., Crépin, T., Zhou, H., et al. (2007). An antifungal agent inhibits an aminoacyl-tRNA synthetase by trapping tRNA in the editing site. Science 316, 1759–1761. doi: 10.1126/science.1142189
Rominski, A., Roditscheff, A., Selchow, P., Böttger, E. C., and Sander, P. (2017a). Intrinsic rifamycin resistance of Mycobacterium abscessus is mediated by ADP-ribosyltransferase MAB_0591. J. Antimicrob. Chemother. 72, 376–384. doi: 10.1093/JAC/DKW466
Rominski, A., Selchow, P., Becker, K., Brülle, J. K., Dal Molin, M., and Sander, P. (2017b). Elucidation of Mycobacterium abscessus aminoglycoside and capreomycin resistance by targeted deletion of three putative resistance genes. J. Antimicrob. Chemother. 72, 2191–2200. doi: 10.1093/JAC/DKX125
Roux, A. L., Viljoen, A., Bah, A., Simeone, R., Bernut, A., Laencina, L., et al. (2016). The distinct fate of smooth and rough Mycobacterium abscessus variants inside macrophages. Open Biol. 6:160185. doi: 10.1098/RSOB.160185
Rubio, M., March, F., Garrigó, M., Moreno, C., Español, M., and Coll, P. (2015). Inducible and acquired clarithromycin resistance in the Mycobacterium abscessus complex. PLoS One 10:e0140166. doi: 10.1371/JOURNAL.PONE.0140166
Rudra, P., Hurst-Hess, K., Lappierre, P., and Ghosha, P. (2018). High levels of intrinsic tetracycline resistance in Mycobacterium abscessus are conferred by a tetracycline-modifying monooxygenase. Antimicrob. Agents Chemother. 62, e00119-18. doi: 10.1128/AAC.00119-18
Ruis, C., Bryant, J. M., Bell, S. C., Thomson, R., Davidson, R. M., Hasan, N. A., et al. (2021). Dissemination of Mycobacterium abscessus via global transmission networks. Nat. Microbiol. 6, 1279–1288. doi: 10.1038/s41564-021-00963-3
Saxena, S., Spaink, H. P., and Forn-Cuní, G. (2021). Drug resistance in nontuberculous mycobacteria: mechanisms and models. Biology (Basel). 10, 1–22. doi: 10.3390/BIOLOGY10020096
Schäfle, D., Selchow, P., Borer, B., Meuli, M., Rominski, A., Schulthess, B., et al. (2021). Rifabutin is inactivated by Mycobacterium abscessus Arr. Antimicrob. Agents Chemother. 65, e02215-20. doi: 10.1128/AAC.02215-20
Selchow, P., Ordway, D. J., Verma, D., Whittel, N., Petrig, A., Hobbie, S. N., et al. (2022). Apramycin overcomes the inherent lack of antimicrobial bactericidal activity in Mycobacterium abscessus. Antimicrob. Agents Chemother. 66:e0151021. doi: 10.1128/AAC.01510-21
Shin, S. J., Choi, G. E., Cho, S. N., Woo, S. Y., Jeong, B. H., Jeon, K., et al. (2013). Mycobacterial genotypes are associated with clinical manifestation and progression of lung disease caused by Mycobacterium abscessus and Mycobacterium massiliense. Clin. Infect. Dis. 57, 32–39. doi: 10.1093/CID/CIT172
Shoen, C., Benaroch, D., Sklaney, M., and Cynamon, M. (2019). In vitro activities of Omadacycline against rapidly growing mycobacteria. Antimicrob. Agents Chemother. 63, e02522-18. doi: 10.1128/AAC.02522-18
Singh, S., Wang, J. Y., Heysell, S. K., McShane, P. J., Wadle, C., Shankar, P., et al. (2023). Omadacycline pharmacokinetics/pharmacodynamics in the hollow fiber model and clinical validation of efficacy to treat pulmonary Mycobacterium abscessus disease. Int. J. Antimicrob. Agents 62:106847. doi: 10.1016/J.IJANTIMICAG.2023.106847
Soliveri, J. A., Gomez, J., Bishai, W. R., and Chater, K. F. (2000). Multiple paralogous genes related to the Streptomyces coelicolor developmental regulatory gene whiB are present in streptomyces and other actinomycetes. Microbiology 146, 333–343. doi: 10.1099/00221287-146-2-333
Song, L., and Wu, X. (2016). Development of efflux pump inhibitors in antituberculosis therapy. Int. J. Antimicrob. Agents 47, 421–429. doi: 10.1016/J.IJANTIMICAG.2016.04.007
Sorayah, R., Manimekalai, M. S. S., Shin, S. J., Koh, W. J., Grüber, G., and Pethe, K. (2019). Naturally-occurring polymorphisms in QcrB are responsible for resistance to telacebec in Mycobacterium abscessus. ACS Infect. Dis. 5, 2055–2060. doi: 10.1021/ACSINFECDIS.9B00322
Soroka, D., Ourghanlian, C., Compain, F., Fichini, M., Dubée, V., Mainardi, J. L., et al. (2017). Inhibition of β-lactamases of mycobacteria by avibactam and clavulanate. J. Antimicrob. Chemother. 72, dkw546–dkw1088. doi: 10.1093/JAC/DKW546
Sreevatsan, S., Stockbauer, K. E., Pan, X., Kreiswirth, B. N., Moghazeh, S. L., Jacobs, W. R., et al. (1997). Ethambutol resistance in Mycobacterium tuberculosis: critical role of embB mutations. Antimicrob. Agents Chemother. 41, 1677–1681. doi: 10.1128/AAC.41.8.1677
Stein, G. E., and Craig, W. A. (2006). Tigecycline: a critical analysis. Clin. Infect. Dis. 43, 518–524. doi: 10.1086/505494
Story-Roller, E., Maggioncalda, E. C., Cohen, K. A., and Lamichhane, G. (2018). Mycobacterium abscessus and β-lactams: emerging insights and potential opportunities. Front. Microbiol. 9:2273. doi: 10.3389/FMICB.2018.02273
Stout, J. E., and Floto, R. A. (2012). Treatment of Mycobacterium abscessus: all macrolides are equal, but perhaps some are more equal than others. Am. J. Respir. Crit. Care Med. 186, 822–823. doi: 10.1164/RCCM.201208-1500ED
Swenson, C., Zerbe, C. S., and Fennelly, K. (2018). Host variability in NTM disease: implications for research needs. Front. Microbiol. 9:2901. doi: 10.3389/FMICB.2018.02901
Tsai, S. H., Lai, H. C., and Hu, S. T. (2015). Subinhibitory doses of aminoglycoside antibiotics induce changes in the phenotype of Mycobacterium abscessus. Antimicrob. Agents Chemother. 59, 6161–6169. doi: 10.1128/AAC.01132-15
Ung, K. L., Alsarraf, H. M. A. B., Olieric, V., Kremer, L., and Blaise, M. (2019). Crystal structure of the aminoglycosides N-acetyltransferase Eis2 from Mycobacterium abscessus. FEBS J. 286, 4342–4355. doi: 10.1111/FEBS.14975
Ventola, C. L. (2015). The antibiotic resistance crisis: part 1: causes and threats. Pharm. Ther. 40:277.
Vester, B., and Douthwaite, S. (2001). Macrolide resistance conferred by base substitutions in 23S rRNA. Antimicrob. Agents Chemother. 45, 1–12. doi: 10.1128/AAC.45.1.1-12.2001
Vianna, J. S., Machado, D., Ramis, I. B., Silva, F. P., Bierhals, D. V., Abril, M. A., et al. (2019). The contribution of efflux pumps in Mycobacterium abscessus complex resistance to clarithromycin. Antibiotics (Basel, Switzerland) 8:153. doi: 10.3390/ANTIBIOTICS8030153
Victoria, L., Gupta, A., Gómez, J. L., and Robledo, J. (2021). Mycobacterium abscessus complex: a review of recent developments in an emerging pathogen. Front. Cell. Infect. Microbiol. 11:659997. doi: 10.3389/FCIMB.2021.659997
Wallace, R. J., Meier, A., Brown, B. A., Zhang, Y., Sander, P., Onyi, G. O., et al. (1996). Genetic basis for clarithromycin resistance among isolates of Mycobacterium chelonae and Mycobacterium abscessus. Antimicrob. Agents Chemother. 40, 1676–1681. doi: 10.1128/AAC.40.7.1676
Wassilew, N., Hoffmann, H., Andrejak, C., and Lange, C. (2016). Pulmonary disease caused by non-tuberculous mycobacteria. Respiration 91, 386–402. doi: 10.1159/000445906
Watkins, R. R., and Deresinski, S. (2019). Omadacycline: a novel tetracycline derivative with oral and intravenous formulations. Clin. Infect. Dis. 69, 890–896. doi: 10.1093/CID/CIZ242
Wu, M. L., Aziz, D. B., Dartois, V., and Dick, T. (2018). NTM drug discovery: status, gaps and the way forward. Drug Discov. Today 23, 1502–1519. doi: 10.1016/J.DRUDIS.2018.04.001
Wu, W., He, S., Li, A., Guo, Q., Tan, Z., Liu, S., et al. (2022). A novel leucyl-tRNA synthetase inhibitor, MRX-6038, expresses anti-Mycobacterium abscessus activity in vitro and in vivo. Antimicrob. Agents Chemother. 66:e0060122. doi: 10.1128/AAC.00601-22
Young, S., Kim, D. H., Moon, S. M., Song, J. Y., Huh, H. J., Lee, N. Y., et al. (2021). Association between 16S rRNA gene mutations and susceptibility to amikacin in Mycobacterium avium complex and Mycobacterium abscessus clinical isolates. Sci. Rep. 11:6108. doi: 10.1038/s41598-021-85721-5
Zheng, M., and Lupoli, T. J. (2021). Modulation of a mycobacterial ADP-ribosyltransferase to augment rifamycin antibiotic resistance. ACS Infect. Dis. 7, 2604–2611. doi: 10.1021/ACSINFECDIS.1C00297
Zheng, L., Qi, X., Zhang, W., Wang, H., Fu, L., Wang, B., et al. (2023). Efficacy of PBTZ169 and pretomanid against Mycobacterium avium, Mycobacterium abscessus, Mycobacterium chelonae, and Mycobacterium fortuitum in BALB/c mice models. Front. Cell. Infect. Microbiol. 13:1115530. doi: 10.3389/FCIMB.2023.1115530
Keywords: Mycobacterium abscessus, mycobacterium drug efficacy, drug resistance, efflux pump, intrinsic-extrinsic drug resistance
Citation: Nguyen TQ, Heo BE, Jeon S, Ash A, Lee H, Moon C and Jang J (2024) Exploring antibiotic resistance mechanisms in Mycobacterium abscessus for enhanced therapeutic approaches. Front. Microbiol. 15:1331508. doi: 10.3389/fmicb.2024.1331508
Received: 01 November 2023; Accepted: 17 January 2024;
Published: 06 February 2024.
Edited by:
Matt Johansen, University of Technology Sydney, AustraliaReviewed by:
Peter Sander, University of Zurich, SwitzerlandCopyright © 2024 Nguyen, Heo, Jeon, Ash, Lee, Moon and Jang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Jichan Jang, amljaGFuamFuZ0BnbnUuYWMua3I=
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.