
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Microbiol. , 01 May 2024
Sec. Microorganisms in Vertebrate Digestive Systems
Volume 15 - 2024 | https://doi.org/10.3389/fmicb.2024.1323842
This article is part of the Research Topic The Intricate Web of Gastrointestinal Virome, Mycome and Archaeome: Implications for Gastrointestinal Diseases View all 5 articles
Objective: Gastric and intestinal diseases possess distinct characteristics although they are interconnected. The primary objective of this study was to investigate the pathogenesis of gastrointestinal diseases through different analyses of clinical characteristics, serum immunology, and gut microbiota in patients with gastrointestinal diseases.
Methods: We collected serum samples from 89 patients with gastrointestinal diseases and 9 healthy controls for immunological assessment, stool samples for DNA extraction, library construction, sequencing, as well as clinical data for subsequent analysis.
Results: Regarding clinical characteristics, there were significant differences between the disease group and the healthy control (HC) group, particularly in terms of age, cancer antigen 125 (CA125), cancer antigen 199 (CA199), alpha-fetoprotein (AFP), total bilirubin (TBIL) and indirect bilirubin (IBIL). The intestinal disease (ID) group exhibited the highest IL-6 level, which significantly differed from the stomach disease (SD) group (p < 0.05). In comparing the HC with the ID groups, significant differences in abundance were detected across 46 species. The HC group displayed a greater abundance of Clostridiales, Clostridia, Firmicutes, Bifidobacterium, Bifidobacteriaceae, Bifidobacteriales, Actinobacteria, Veillonellaceae, Longum, Copri, Megamonas and Callidus than other species. Similarly, when comparing the HC with the SD groups, significant differences in abundance were identified among 49 species, with only one species that the Lachnospiraceae in the HC group exhibited a higher abundance than others. Furthermore, certain clinical characteristics, such as CA125, CA199, glucose (Glu), creatine kinase-MB (CKMB) and interleukin-22 (IL-22), displayed positive correlations with enriched gut species in the ID and SD groups, while exhibiting a negative correlation with the HC group.
Conclusion: The disturbance in human gut microbiota is intimately associated with the development and progression of gastrointestinal diseases. Moreover, the gut microbiota in the HC group was found more diverse than that in the ID and SD groups, and there were significant differences in microbial species among the three groups at different classification levels. Notably, a correlation was identified between specific clinical characteristics (e.g., CA125, CA199, Glu, CKMB and IL-22) and gut microbiota among patients with gastrointestinal diseases.
The human gastrointestinal (GI) tract contains a rich microbial community, aggregating about 100 trillion microbes. In addition to bacteria and fungi, the human gut microbiota also includes viruses, bacteriophages and archaea. The gut microbiota is considered as one of the most important types of microorganisms for maintaining human health (Hou et al., 2022a), such as food fermentation, protecting the body from pathogenic harm, stimulating immune responses and producing vitamins (Heintz-Buschart and Wilmes, 2018; Adak and Khan, 2019; de Vos et al., 2022). Regarding the immune system, the human microbiota can not only protect the host from external pathogens by producing antibacterial substances, but also play a significant role in the development of intestinal mucosa and immune system (Sittipo et al., 2018). Thus, the maturation of the immune system requires the development of symbiotic microbes while the GI tract has numerous immune cells. For example, one mechanism by which gut microbiota affects the immune system is mediating neutrophil migration, thereby influencing T cell differentiation into diverse types, such as helper T cells (Th1, Th2, and Th17 cells) and regulatory T (Treg) cells (Wu et al., 2021; Shim et al., 2023). Interestingly, the composition and diversity of gut microbiota can predict responses to immunotherapy with immune checkpoint inhibitors (ICIs) (Morkunas et al., 2020). For instance, gut microbiota can affect adoptive T-cell transfer (ACT) immunotherapy strategies, CpG oligodeoxynucleotide (CpG ODN) immunotherapy strategies and cell-based immunotherapies by regulating innate immunity, adaptive immunity and tumor antigens to improve ICI response (Lu et al., 2022). Additionally, adaptive immune response is another important part of maintaining a healthy microbiota and immune balance, achieved through the differentiation and maturation of B and T cells, as well as the establishment of immune tolerance to the microbiota. According to the type of bacteria, CD4T cell responses vary greatly, leading to differentiation into different subgroups and subsequent release of pro-inflammatory cytokines, such as interferon-γ and interleukin-17A (IL-17A) (Mazmanian et al., 2005; Weaver et al., 2006; Alnahas et al., 2017).
To date, a growing body of evidence has confirmed that the microbiota is associated with the development of cardiovascular diseases (Lau et al., 2017; Sanchez-Rodriguez et al., 2020), cancer (Kostic et al., 2013; Jin et al., 2019; Sami et al., 2020; Vyhnalova et al., 2021), respiratory diseases (Segal et al., 2013; Bassis et al., 2015; Cait et al., 2018; Sokolowska et al., 2018), diabetes (Larsen et al., 2010; Vatanen et al., 2018; Hou et al., 2022b), inflammatory bowel disease (IBD) (Manichanh et al., 2012; Ahmed et al., 2016; Lucas Lopez et al., 2017), brain diseases (de Theije et al., 2014; Gilbert, 2015; Allen et al., 2016; Zhang et al., 2022), chronic kidney disease (Chung et al., 2019; Lun et al., 2019; Pluznick, 2020), liver disease (Mouzaki et al., 2013; Tripathi et al., 2018) and osteoporosis (Qin et al., 2021). Thus, the manipulation of human microbiota may be the key to disease treatment. It is noteworthy that the relative distribution of microbes within the gut is individual-specific and can even vary within the same individual (Hou et al., 2022a). Numerous factors have been identified as regulators of the host’s gut-brain axis, both internally and externally, encompassing genetics, socioeconomic status, dietary habits, medication use and environmental influences.
Regarding cancers, Helicobacter pylori is regarded as a pathogenic factor in gastric cancer (Hu et al., 2022), while Fusobacterium nucleatum is recognized as a contributing factor in colorectal cancer (CRC) (Bullman et al., 2017). Escherichia coli strains that possess Polyketide synthase (PKS) genotoxins have the potential to promote colorectal tumorigenesis. Bacteroides fragilis, on the other hand, can generate reactive oxygen species (ROS), which can lead to DNA damage. Although these bacteria have a direct impact on the occurrence of tumors, some microorganisms can promote inflammation or weaken immune surveillance, indirectly promoting the development of cancer. These microbial immune regulatory activities are known as the immune-tumor-microbe axis (Ting et al., 2022). Gut microbiome can induce epithelial barrier deterioration, triggering tumor-induced inflammation, as well as driving progression of CRC through influencing certain signaling pathways (e.g., E-cadherin/β-catenin, TLR4/MYD88 and SMO/RAS/p38MAPK) (Li et al., 2021). The elevated expression levels of circular RNAs, specifically circRNA ciRS-7, circNSUN2 and circ-ERBIN, have been identified to be contributed to the progression of CRC. Meanwhile, CircIL4R has been found to enhance the proliferation, migration and invasion of CRC cells by modulating the PI3K/AKT signaling pathway (Jiang et al., 2021). In addition to bacteria and fungi, viruses exhibit distinctive disease characteristics within CRC. The presence of viruses indirectly influences CRC by altering the associated bacterial ecosystem (Li et al., 2022).
Gut microbiota is highly associated with the development of IBD. Mechanistically, dysbiosis of the microbiota is correlated with IBD through influencing inflammation and intestinal barriers (Konig et al., 2016). As reported by Kleessen et al. (2002), a higher prevalence of mucosal bacterial infiltration was found in the IBD group compared to the control. Notably, in patients with Crohn’s disease (CD), there was a significant elevation in the presence of adherent-invasive E. coli, indicating that pathogens could influence intestinal permeability, the composition of the gut microbiota and ultimately contribute to intestinal inflammation (Ahmed et al., 2016). The pathophysiology of IBD is multifactorial, involving complex interactions among genetics (Marcuzzi et al., 2013), environment (Leone et al., 2013), microbiome (Manichanh et al., 2012; Wang et al., 2014), immunity (Kaistha and Levine, 2014) and potential other risk factors.
There are many existing analyses on the differences in gut microbiota among patients with gastrointestinal diseases, but there are few analyses on the differences in gut microbiota among patients with gastric diseases, and there is a lack of analysis on the differences in serum immunology and gut microbiota among patients with gastrointestinal diseases, which hinders further research on the role of microbiota. Although stomach and intestinal diseases share some common risk factors, they have different phenotypes and disease manifestations. This cross-sectional cohort study analyzed the clinical characteristics, serum immunology and differences in gut microbiota of patients with gastrointestinal diseases, aiming to explore the pathogenesis of gastrointestinal diseases, further prevent and reduce the risk of gastrointestinal diseases in China through customized intervention measures, and develop personalized multimodal treatment strategies.
Serum and fecal samples: serum and fecal samples were collected from 89 patients with gastrointestinal diseases who were admitted to the Affiliated Hospital of Jiaying Medical College (Meizhou, China, which was established in November 2009 and is the first university affiliated hospital in Meizhou City. It is under the management of Jiaying College Medical College and is a public non-profit comprehensive hospital) from March 2021 to March 2022, and 9 healthy individuals were also involved as healthy controls. A total of 500 μL of serum samples were collected in 2 mL sterile centrifuge tubes, while 50 g of fecal samples were gathered in specialized fecal collection tubes. These samples were subsequently stored at −80°C for future utilization.
Inclusion criteria for patients with gastrointestinal diseases: patients diagnosed with stomach or/and intestinal diseases (such as inflammation, tumors, functional disorders, etc.). Exclude patients with other tumors, liver disease, kidney disease or other traumatic bleeding at the same time.
Inclusion criteria for the healthy control group: volunteers who have passed physical examinations and do not have any stomach or/and intestinal diseases, tumors, liver diseases, kidney disease or other traumatic bleeding are recognized as healthy.
Clinical data: patients’ and healthy controls’ data, including date of admission, age, gender, routine blood test results and biochemical indicators were recorded.
The levels of relevant cytokines, including IL-6, IL-9 and IL-22 in the serum, were determined using human interleukin 6, 9 and 22 ELISA kits (Jiangsu Meimian, China). Additionally, the levels of cytokines, such as IL-8, IFN-γ and TNF-α were measured using human interleukin 8, IFN-γ and TNF-α ELISA kits (Guangzhou Daan, China). The specific procedures for these assessments were carried out in accordance with manufacturers’ instructions.
Among 89 patients with gastrointestinal diseases, 16S rDNA amplifier sequencing was performed on 4 patients with gastric diseases, 17 patients with intestinal diseases and 9 individuals undergoing routine medical examinations. Universal primers designed for conserved regions were utilized for PCR amplification, followed by double-end (Paired-End) sequencing on the MISEQ/Hiseq platform, generating one or more high-variability 16S rDNA sequences. These sequences were then subjected to operational taxonomic unit (OTU) clustering, species annotations and abundance analysis, along with assessments of α- and β-diversity, as well as other analyses.
Total DNA extraction from the microbial community in stool samples was carried out using the E.Z.N.A® Stool DNA kit (Omega Bio-tek, Norcross, GA, United States), and the DNA was quantified using a Thermo NanoDrop 2000 (Thermo Fisher Scientific, Waltham, MA, United States).
For PCR amplification, the target sequence was amplified with specific primers (341F: 5′-CCTACGGGNGGCWGCAG-3′, 805R: 5′-GACTACHVGGGTATCTAATCC-3′). The resulting PCR product was confirmed through electrophoresis on a 2% agarose gel. Subsequently, PCR products were purified using AMPure XT beads (Beckman Coulter Genomics, Danvers, MA, United States) and quantified with Qubit (Invitrogen, Carlsbad, CA, United States). The amplicon pool was utilized for sequencing. The size and quantity of the amplified library were assessed via the Agilent 2100 Bioanalyzer (Agilent Technologies Inc., Santa Clara, CA, United States) and the Illumina Library Quantitative Kit (Kapa Biosciences, Woburn, MA, United States). Sequencing of the library was conducted on the Illumina NovaSeq PE250 platform (Illumina Inc., San Diego, United States).
Statistical analysis was performed using SPSS 22.0.0.0.202 software (IBM, Armonk, NY, United States). Descriptive statistics for continuous variables following a normal distribution were expressed as mean ± standard deviation (SD). Multivariate analysis of variance (ANOVA) was utilized to compare multiple samples. In cases of abnormally distributed continuous variables, descriptive statistics were presented as median and interquartile range (IQR), and the Mann–Whitney U test was employed for making comparisons between two groups. Variations of species between groups were analyzed using t-tests, nonparametric rank sum tests (U tests), and linear discriminant analysis effect size (LEfSe). The Analysis of similarities (ANOSIM) (Chapman and Underwood, 1999) was applied to assess differences in community structure between groups, and Spearman’s rank correlation was utilized to evaluate associations between microbial diversity and biochemical indicators. The significance level was set at 5%.
According to the specific type of disease, 89 patients were categorized into distinct groups, namely the stomach disease (SD) group consisting of 13 cases, the intestinal disease (ID) group encompassing 60 cases, the gastrointestinal disease (GD) group comprising 16 cases, and healthy control (HC) group involving 9 cases. In terms of age, the mean age of the HC group was the youngest at 33.11 years old, which was notably different from that in the SD and ID groups (p < 0.01). When comparing the results of routine blood tests and biochemical indicators, it was noted that the levels of cancer antigen 125 (CA125) and cancer antigen 199 (CA199) in the HC group were significantly different from those in the three disease groups (p < 0.01). The median levels of CA125 and CA199 were the lowest in the HC group (1.21, 3.28), with the highest median level of CA125 observed in the SD group (19.50) and the highest median level of CA199 found in the GD group (29.04). There was also a significant difference in CA125 level between the SD group and the ID & GD groups (p < 0.01). Alpha-fetoprotein (AFP) yielded its highest median level in the GD group (4.44), demonstrating a highly significant difference compared with the SD & HC groups (p < 0.01). Similarly, the highest mean neutrophil (N) count was identified in the GD group (7.27), which exhibited a significant difference compared with the SD group (p < 0.05). Total bilirubin (TBIL) and indirect bilirubin (IBIL) reached their highest mean levels in the SD group (15.64, 10.35), demonstrating significant differences compared with the HC group (p < 0.05). Regarding other characteristics, no significant differences were found among all the groups (Table 1).
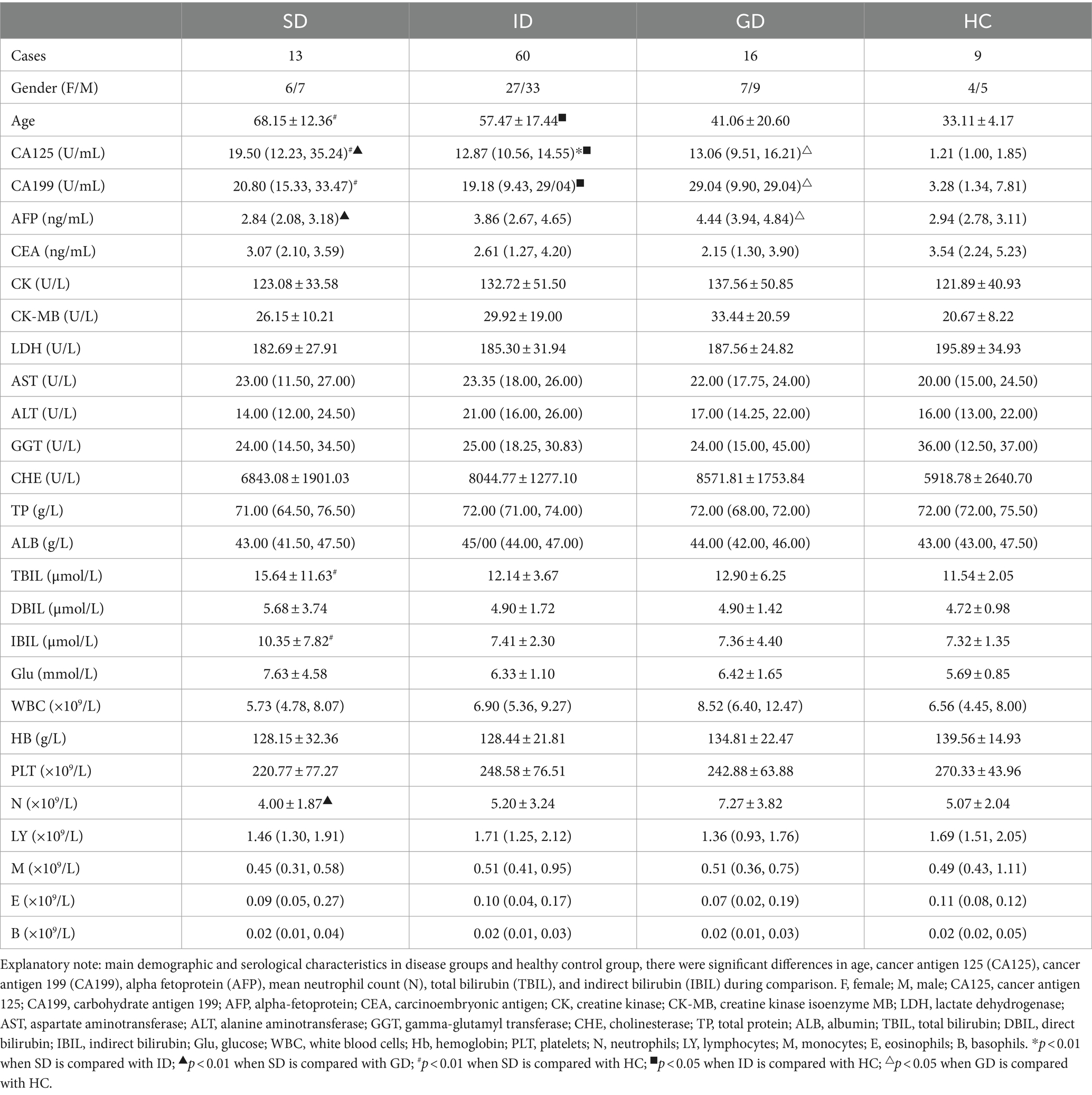
Table 1. Main demographic and serological characteristics in disease groups and healthy control group.
Among the 6 cytokines examined in this study, the serum level of IL-6 was highest in the ID group, with an average concentration of 5.20 pg./mL, which was significantly different from that in the SD group (p < 0.05), and there were no significant differences among other groups, as shown in Figure 1.
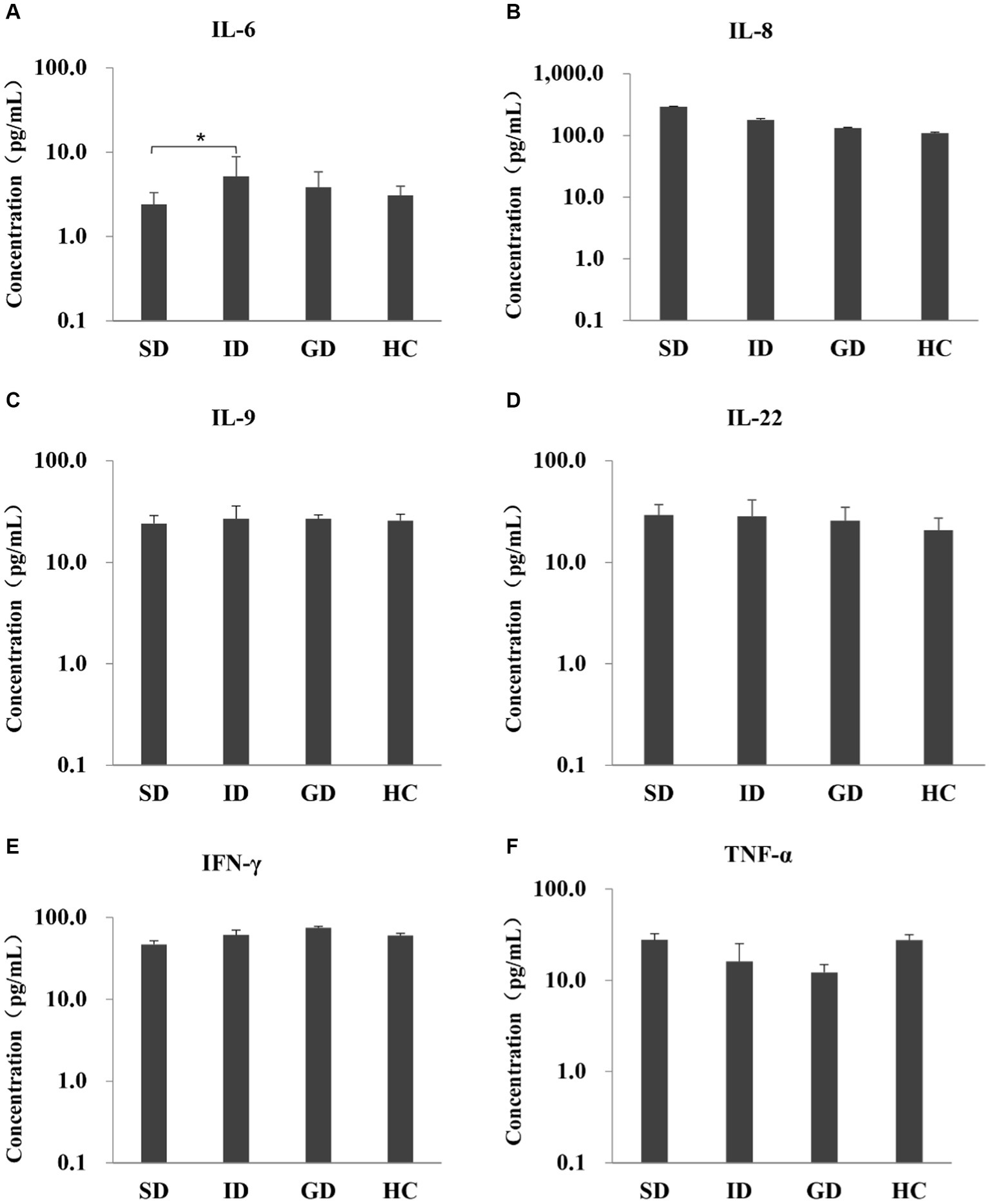
Figure 1. Evaluate the immune function of the disease and the HC groups by measuring the levels of six serum cytokines.
The species annotation results obtained from fecal samples of 4 patients in the SD group, 17 patients in the ID group, and 9 cases in the HC group revealed that the gut microbiota types in the SD and ID groups are more similar, with differences compared to the HC group. The gut microbiota in the SD, ID and HC groups primarily consisted of four predominant phyla: Firmicutes, Bacteroidetes, Proteobacteria, and Actinobacteria. The difference in Firmicutes between the ID and HC groups was significant. At the class level, Clostridia, Gammaproteobacteria, Bacteroidia and Bacilli were prevalent in both SD and ID groups, while Clostridia, Gammaproteobacteria, Bacteroidia and Actinobacteria were dominated in the HC group. Concerning the order level, Clostridiales, Enterobacteriales, Bacteroidales and Lactobacillales were dominant in both the SD and ID groups, whereas Clostridiales, Enterobacteriales, Bacteroidales and Bifidobacteriales were dominated in the HC group. The difference in Clostridia between the ID and HC groups was significant in class and order. On the family level, Enterobacteriaceae, Bacteroidaceae, Lachnospiraceae and Ruminococcaceae were more abundant in both ID and HC groups, while Enterobacteriaceae and Streptococcaceae were more abundant in the SD group. At the genus level, Escherichia, Bacteroids and Ruminococcus were more prevalent in the ID group, whereas Escherichia, Streptococcus and Klebsiella were more abundant in the SD group, and Bacteroids, Faecalibacterium and Bifidobacteria were more prominent in the HC group (Figures 2A,B). Furthermore, we conducted through α diversity analysis (Figure 2C) on the community diversity within the sample, using Shannon and Simpson indices to characterize the richness and evenness of microbial communities, indicated no significant differences between any two groups among the SD, ID and HC groups (t-test and nonparametric rank-sum test, U test, p > 0.05 for all; Figure 2C). Simultaneously through β diversity (Figure 2D) analyzed the differences in community structure between groups, and ANOSIM analysis was used to test whether the differences between groups were significantly greater than those within groups. We conducted a significance test for the differences between groups based on the rank of Bray–Curtis distance values. As determined by Bray–Curtis distance values (ANOSIM analysis, p > 0.05 for all), there were no significant distinctions among microbial communities between any two groups among the SD, ID and HC groups (Figure 2D).
Comparison of microbial communities. (A,B) The gut microbiota in SD, ID and HC groups are abundant at the levels of phylum, class, order, family, genus and species. And t-tests were conducted on different classification levels to obtain p-value, which was corrected using the FDR method. We selected only microbial communities with a p < 0.05 for each classification level to draw a forest map. (C) Comparison of α-diversity (Shannon and Simpson indices) between any two groups among SD, ID and HC groups. (D) Comparison of β-diversity (determined by Bray–Curtis distance values) between any two groups among SD, ID and HC groups.
As depicted in Figure 3, a comparative analysis between the HC group and both the ID and SD groups, respectively, involved the identification of species significantly contributing to intergroup distinctions and species exhibiting notable differences in abundance across different groups (as indicated by the LDA score), according to the results of LEfSe analysis. The results illustrated in Figure 3A revealed an enrichment of 6 species classified under the phylum Synergistetes, including Pyramidobacter, Piscolens, Synergistetes, Synergistia, Synergistales and Dethiosulfovibrionaceae, within the ID group. Conversely, 19 species from Firmicutes and 12 species from Actinobacteria exhibited enrichment in the HC group. It was indicated that the SD group was accompanied by an enrichment of 15 species associated with Proteobacteria and 6 species from Firmicutes, while the HC group demonstrated enrichment in 14 species from Firmicutes and 6 species from Verrucomicrobia (Figure 3B). It can be seen that the microbial community of the HC group exhibits higher taxonomic diversity.
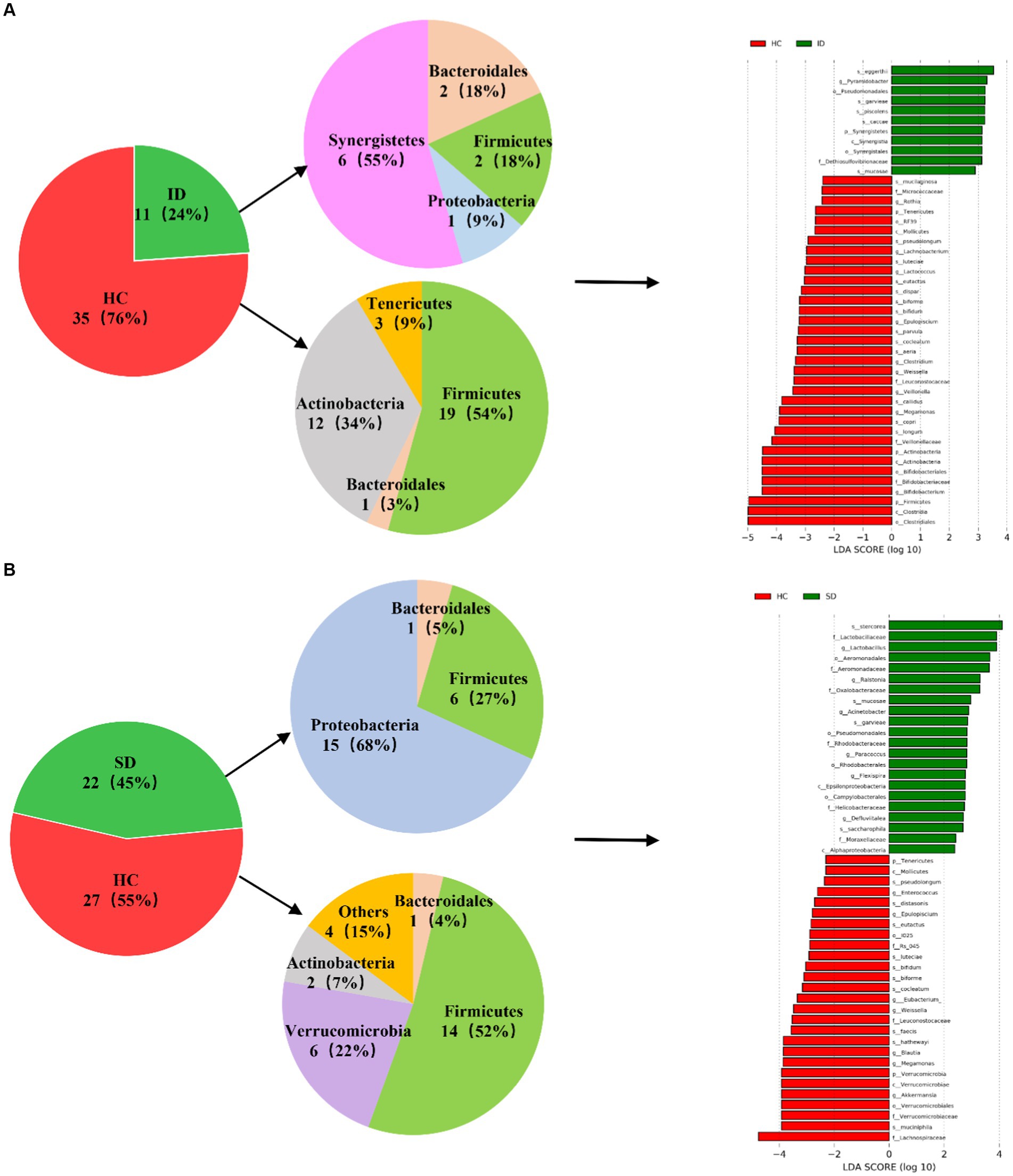
Figure 3. Analysis of microbial communities contributing significantly to intergroup differences. (A) Differences in species abundance between ID and HC groups. (B) Differences in species abundance between SD and HC groups.
In the present study, Spearman correlation analysis was employed to examine the relationship between clinical attributes and species abundance in the ID group versus the HC group, as well as the SD group versus the HC group. The analysis comparing the ID group with the HC group (illustrated in Figure 4A) revealed that most of the gut microbes enriched in the ID group significantly exhibited positive correlations with 15 clinical characteristics, including creatine kinase-MB (CKMB), interleukin-22 (IL-22), CHE, CA125, CA199, E, L, IL-6, N, white blood cells (WBC), IFN-γ, alanine aminotransferase (ALT), aspartate aminotransferase (AST), AFP and glucose (Glu), among which CA125 and CA199 show a strong positive correlation. Conversely, they displayed negative correlations with 16 other clinical characteristics. In contrast, the HC group displayed the opposite pattern, in which the enriched microbes were significantly negatively correlated with the aforementioned 15 clinical characteristics and positively correlated with the other 16 clinical characteristics. Among them, the three indicators CHE, CA125 and CA199 showed a strong negative correlation, while DBIL showed a strong positive correlation. Similarly, in the SD and HC groups (Figure 4B), most gut microbes enriched in the SD group exhibited significantly positive correlations with 12 clinical characteristics, with CA125, CA199 and IL-22 showing a strong positive correlation, while demonstrating negative correlations with 19 additional clinical characteristics. Conversely, in the HC group, the microbes that were enriched displayed a contrasting pattern, in which they were significantly negatively correlated with the aforementioned 12 clinical characteristics, with CA125 and IL-22 showing a strong negative correlation, and positively correlated with other 19 clinical characteristics, with DBIL showing a strong positive correlation.

Figure 4. Association of microbial communities with clinical characteristics. (A) Correlation matrix of species and clinical parameters in the ID group vs. HC group. Red indicates a positive correlation, while blue indicates a negative correlation. *, ** and *** represent p < 0.05, p < 0.01, and p < 0.001, respectively; the number of significant correlations for the 5 species with the most frequent correlations and 5 clinical characteristics with the most frequent correlations (only the top five are listed). (B) Correlation matrix of species and clinical parameters in the SD group vs. HC group. Red indicates a positive correlation, while blue indicates a negative correlation. *, ** and *** represent p < 0.05, p < 0.01, and p < 0.001, respectively; the number of significant correlations for the 5 species with the most frequent correlations and 5 clinical characteristics with the most frequent correlations (only the top five are listed).
Among 46 species analyzed in the ID and HC groups, the species “longum” exhibited the strongest correlation with clinical characteristics (n = 7, positive correlation: M, LDH, DBIL and N; negative correlation: CA199, E and L, p < 0.05), followed by “bifidum” (n = 6, positive correlation: DBIL; negative correlation: CKMB, CHE, CA125, CA199 and IL-6, p < 0.05), “copri,” “caccae,” and “eutactus” (n = 5, p < 0.05, Figure 4A). Among them, LDH and DBIL are positively correlated with copri, while CHE, CA125 and CA199 are negatively correlated; CKMB, IL-22, CHE, CA125 and CA199 are positively correlated with caccae; TNF-α is positively correlated with eutactus, while CA125, CA199, ALT and AST are negatively correlated; Notably, only “caccae” was enriched in the ID group. Among 49 species analyzed in the SD and HC groups, “bifidum” and “stercorea” displayed the strongest correlation with clinical characteristics (n = 4, p < 0.05). Among them, AST and DBIL are positively correlated with bifidum, while CA125 and IL-22 are negatively correlated; Glu, CA125 and CA199 are positively correlated with stercorea, while AST is negatively correlated, followed by “biforme,” “cocleatum” and “garvieae” (n = 3, p < 0.05, Figure 4B); “stercorea” and “garvieae” were enriched in the SD group. Importantly, in both the ID and SD groups, the most pronounced correlation was identified between CA125/CA199 and bacterial species (ID group: n = 10/11, p < 0.05; SD group: n = 5/5, p < 0.05), followed by IL-22 in the SD group (n = 6, p < 0.05).
The human microbiota and its host maintain a sophisticated symbiotic relationship, exerting significant impacts on host health. The balance and stability of the gut microbiota play a pivotal role in preserving human health. Dietary and healthy lifestyle habits are considered principal regulators of the gut microbiome, both in the short-term and long-term. Common clinical interventions to regulate the microbiome include microbiome modulation, fecal microbiota transplantation (FMT) and bacterial engineering (Lu et al., 2022). Furthermore, in addition to a well-balanced diet, supplementation with plant extracts and increased physical activity can also effectively regulate the gut microbiota (Zhou et al., 2023). Gastrointestinal diseases can be triggered by diverse factors, mainly manifesting with multiple symptoms, such as nausea, vomiting and diarrhea. Severe symptoms, including fever and dehydration, can also develop, potentially leading to carcinogenic changes. Thus, early prevention and treatment are imperative. However, conventional diagnostic methods as gastroscopy or tissue biopsy are invasive, expensive, painful and risky. These factors often hinder patients from undergoing these procedures, potentially delaying diagnosis and treatment (Dai et al., 2019; Wong and Yu, 2023).
The human gastrointestinal tract, in contrast to other body parts, hosts a substantial microbial community. Gut microbiota composition may vary with factors, such as age, environmental influences (e.g., medication use) and anatomical locations. These microbial communities play a vital role in human nutrient processing, metabolism and immunity (Bouskra et al., 2008). Notably, the overall diversity of human gut microbiota is constantly changing throughout life, as individuals age, the diversity of intestinal microbes decreases (Lynch and Pedersen, 2016), accompanying by a reduction in proportions of Firmicutes and Actinobacteria and an increase in the abundance of Proteobacteria. The diversity of gut microbiota in the elderly can significantly vary due to individual differences (Maynard and Weinkove, 2018; Adak and Khan, 2019; Shi et al., 2022). In the present study, significant differences were found in clinical characteristics among the disease groups versus the HC group. These differences pertained to age, CA125, CA199, AFP, TBIL and IBIL. We detected six cytokines closely related to the gut microbiota or its metabolites, hoping to further explore the serum immune function of patients with gastrointestinal diseases. The expression of inflammatory cytokines IL-6, IL-8 and TNF-α is related to the metabolism of dietary nutrients involved in intestinal microbiota. The metabolic end products trimethylamine N-oxide (TMAO) and short-chain fatty acids affect inflammation and immune response by affecting the expression of various receptors, signaling and apoptosis genes, and lead to the expression of these three inflammatory cytokines. IL-22 is also affected by short-chain fatty acids (Liu et al., 2017; Li et al., 2022). Similarly, the upregulation of IFN-γ is influenced by inosine, a purine metabolite of Akkermansiamucophila and Bifidobacterium pseudolongum, which activates immune cells through the inosine-A2AR-cAMP-PKA signaling pathway and stimulates phosphorylation of cAMP response element binding protein (pCREB). Thereby upregulating IFN-γ transcription (Mager et al., 2020). Interestingly, valeric acid and butyric acid also promote the expression of effector molecules such as IFN-γ and TNF-α (Luu et al., 2021). Then, IL-9 is a pleiotropic cytokine that affects a variety of cells, and IL-9 and its receptors contribute to the pathogenesis of ulcerative colitis, which can further develop into cancer if left untreated (Matusiewicz et al., 2017). Among the six cytokines examined, IL-6 exhibited the highest serum level in the ID group and demonstrated a significant difference compared to the SD group (p < 0.05). Research has indicated that IL-6 level significantly increased in patients with CRC and IBD. However, treatment with Bifidobacterium and mixed lactic acid bacteria reduced IL-6 level (Li et al., 2022; Zhong et al., 2022). Imbalance of gut microbiota can lead to activation of tumor associated macrophages (TAMs), thereby promoting the secretion of IL-6. IL-6 accelerates the development of CRC by promoting epithelial mesenchymal transition (EMT) formation (Wan et al., 2018). Notably, the release of IL-6 is also reported to be associated with the presence of diacyl phosphatidylethanolamine on the cell membrane of Akkermansiamuciniphila (Akk bacterium) (Bae et al., 2022). A growing body of evidence suggested that an imbalance in the human gut microbiota is closely correlated with the development and progression of gastrointestinal diseases. Consequently, it is necessary to conduct cross-sectional cohort studies on the relationship between gut microbiota, serum immunology and clinical characteristics to analyze the differences in gut microbiota in patients with gastrointestinal diseases, and explore the correlation between gut microbiota and clinical characteristics. The in-depth analysis of this study will provide insights into the pathogenesis of gastrointestinal disease patients.
The intestinal flora typically includes six phyla: Firmicutes, Bacteroidetes, Actinobacteria, Proteobacteria, Fusobacteria and Verrucomicrobia, with Firmicutes and Bacteroidetes as the predominant species (Manichanh et al., 2012). Consistent with the findings of this study, the gut microbiota of the SD, ID and HC groups were mainly composed of Firmicutes, Proteobacteria, Bacteroidetes and Actinobacteria, but gradually showed differences at the levels of class, order, family, genus and species (Figures 2A,B). As in adults, 60–70% of the gut microbiome is stable, but the degree of stability varies among different phyla (Faith et al., 2013). When the gut microbiota becomes imbalanced, its characteristics are changes in composition or abnormal fluctuations in quantity (He et al., 2023). This study was conducted through α diversity analysis (Figure 2C) and β diversity analysis (Figure 2D), we found that there were no significant differences in the diversity of α and β between the three groups, but there were significant differences in the types and quantities of microorganisms at different taxonomic levels (Figures 2A,B). For example, there are significant differences in Firmicutes and Clostridia between the ID group and the HC group. The reason is that the substances metabolized at different anatomical sites are different, resulting in different types of gut microbiota in different parts of the gastrointestinal tract (Flint et al., 2012; de Vos et al., 2022). And in various diseases, including irritable bowel syndrome (IBS), abnormalities in the gut microbiota can also lead to dysbiosis of the gut microbiota (Konig and Brummer, 2014). Compared with the HC group, the mucosal and fecal microbial diversity of IBS patients decreased after infection (Sundin et al., 2015). Existing research has consistently shown differences in gut microflora composition between IBD patients and HCs. Specifically, IBD patients tend to exhibit a decrease in the proportion of Bacteroidetes and Firmicutes, accompanied by an increase in γ-proteobacteria (Morgan et al., 2012). Compared with HC, the microbial community of CRC patients is mainly composed of pathogenic bacteria related to metabolic disorders, while the abundance of probiotics (such as butyrate producing bacteria) is reduced, such as Rothia, Clostridium, Fecal and Bifidobacterium, which are usually lower in abundance (Yu et al., 2017; Thomas et al., 2019). Main bacteria present in CRC patients include E. faecalis, E. coli, B. fragilis, S. bovis, F. nucleatum and H. pylori (Dai et al., 2019). Among them, F. nucleatum, which is enriched in CRC, exhibited a close relationship with CRC-associated Aspergillus rambellii. This co-enrichment suggests that combined biomarkers involving both fungi and bacteria have more potential as diagnostic markers for CRC than pure bacterial groups (Lin et al., 2022). In addition, the gut microbiota can interact with CRC cancer cells, affecting the tumor microenvironment, thereby enhancing tumor invasiveness (Chen et al., 2022). Through synergistic inflammatory tumor promotion mechanisms, it promotes invasive mesenchymal transition and facilitates distant metastasis of invasive CRC in susceptible tissue environments (Slowicka et al., 2020), such as colorectal cancer liver metastasis (CRLM) (Wei et al., 2024). In the present study, when comparing the HC group with the ID group, significant differences were found in 46 species, with 35 belonging to the HC group and 11 to the ID group. The primary contributors to the HC group were Firmicutes and Actinobacteria, while Synergistetes played a predominant role in the ID group. Among these 46 species, 13 exhibited higher abundances than the others, including Clostridiales, Clostridia, Firmicutes, Bifidobacterium, Bifidobacteriaceae, Bifidobacterales, Actinobacteria, Actinomyces, Veillonellaceae, Longum, Copri, Megamonas and Callidus. Notably, Bifidobacterium is known for its role in stimulating the immune system and is associated with worsening inflammatory status (Guigoz et al., 2008). Furthermore, the richness of intestinal flora in the HC group was higher versus that in the ID group. When comparing the HC and SD groups, significant differences were identified in 49 species, with 27 belonging to the HC and 22 to the SD groups. Among these 49 species, only Lachnospiraceae in the HC group displayed a higher abundance than others. Therefore, from the perspective of microbial quantity differences between the HC and SD groups, the variations were not substantial. However, after gastrectomy, gastric cancer (GC) patients experience a significant increase in specific aerobic bacteria (Streptococcus I and Enterococcus) and facultative anaerobic bacteria (Escherichia coli, Enterobacterium and Streptococcus) due to increased intestinal oxygen content and translocation of oral microbiota (Palleja et al., 2016; Ilhan et al., 2017; He et al., 2023). So, it is essential to consider the distribution of species richness within the gut microflora, where the HC group maintained a higher diversity compared with the SD group. By comparing the species abundance of these three groups, we found that the microbial community of the HC group exhibited high taxonomic diversity, high microbial gene richness, and stable core microbial community, which is consistent with previous research (Fan and Pedersen, 2021). The significance of exploring the differences in the composition of gut microbiota between patients with gastrointestinal diseases and healthy individuals is that these results may contribute to the development of strategies for restoring microbial profiling in patients with gastrointestinal diseases. Microbial communities are powerful targets for treating gastrointestinal diseases. A detailed analysis of the diversity and richness of gut microbiota in patients with gastrointestinal diseases will provide new strategies for future prevention of gastrointestinal diseases and potential clinical applications of gut microbiota in treatment, including as anti-tumor drugs and affecting the efficacy of immunotherapy.
Moreover, it was found that some clinical characteristics (e.g., CA125, CA199, Glu, CKMB, IL-22, etc.) in the ID and SD groups were significantly positively correlated with enriched gut species and negatively correlated with the HC group. CA125 and CA199 are major tumor markers related to gastrointestinal cancer. Gastrointestinal cancer cells produce CA199 and CA125 proteins, which enter the blood stream and can be identified through marker tests. The positivity rates of CA125 and CA199 are relatively low in early-stage gastric cancer (Feng et al., 2017), the positivity rates of CA199 and CA125 in stages III/IV gastric cancer are higher than those in stages I/II, and preoperative serum positivity for both is associated with poor prognosis; they can be utilized to predict recurrence or metastasis of gastric cancer (Zhou et al., 2018). In the present study, there were significant differences in CA125 and CA199 levels when comparing the ID group and SD group with the HC group, and they were significantly positively correlated with Firmicutes’ Garvieae, Mucosae, and Saccharophila, in which Bacteroidetes’ Eggerthii, Caccae and Stercorea were among the enriched gut species. This indicates that combined detection of clinical characteristics, such as CA125 and CA199 along with microbial analysis can improve diagnostic efficiency for gastrointestinal diseases. Similarly, Glu and CKMB are frequently utilized clinical indicators. It was revealed that Glu and CKMB in the ID and SD groups showed no significant difference compared with HC group, while they were significantly positively correlated with Bacteroidetes’ Eggerthii, Caccae, Stercorea and Firmicutes’ Saccharophila. The gastrointestinal tract plays a crucial role in digesting sugars from food. For instance, Bacteroides species located within the colon are responsible for sugar uptake (Martens et al., 2008), while pathogenic Enterobacteriaceae can also utilize sugars and amino acids present within the intestine (Ducarmon et al., 2019). The cytokine IL-22 is of paramount importance in the intestinal defense mechanisms against microbiota. In the present study, the results showed that IL-22 level within the ID and SD groups did not significantly differ from that in the HC group. However, IL22 exhibited a significantly positive correlation with specific species within the Firmicutes phylum, namely Garvieae, Mucosae and Saccharophila, as well as with Bacteroidetes’ Caccae among the enriched species. Consistent with this study, Lactobacillus plantarum belonging to the Firmicutes can induce NK cells to secrete IL-22 (Suzuki, 2013). Existing research suggests that certain bacterial products, such as LPS, Pam2 and Pam3, can stimulate the production of IL-22 within T cells, and this response is further enhanced by macrophage-derived TNFα-induced expression (Klooster et al., 2021). Furthermore, short chain fatty acids (SCFAs) can promote IL-22 production in CD4 T cells and ILCs through GPR41 and inhibition of histone deacetylase (Yang et al., 2020). It is noteworthy that the present study revealed correlations between gut microbiota and specific clinical characteristics among patients suffering from gastrointestinal diseases. However, it is crucial to notice that there is currently limited research in this area. As a result, this research findings will carry valuable clinical reference significance and provide additional opportunities and possibilities for the diagnosis and treatment of patients with gastrointestinal diseases.
Therefore, comprehending the association and underlying mechanisms connecting clinical characteristics, serum immunology, and gut microbiota in individuals afflicted by gastrointestinal conditions can significantly enhance diagnostic and therapeutic approaches. This insight can be pivotal in exploring the potential role of microbial communities within the stomach and intestines. Notably, gastric and intestinal diseases, while closely intertwined, exhibit distinctive characteristics. Tailoring interventions for regulating gut microbiota to influence disease progression holds promise in averting and mitigating the incidence of gastrointestinal disorders, leading to develop personalized multimodal therapeutic strategies. Given the limited sample size in this study, future investigations should encompass larger cohorts to more deeply assess into the intricate interplay among clinical attributes, serum immunology, and gut microbiota in patients with gastrointestinal diseases, but the results of the present work may constitute a good starting point for this purpose. And we should focus on exploring new ideas for prevention, early intervention and treatment of gastrointestinal diseases by intervening in the microbial community of antibiotics or blocking the production of bacterial components. The future research will shed light on the underlying mechanisms driving these conditions.
In conclusion, this study, employing different analyses of clinical characteristics, serum immunology and gut microbiota in patients with gastrointestinal diseases, revealed a close association between imbalances in human gut microbiota and the onset and advancement of gastrointestinal diseases. Notably, the gut microbiota in the HC group exhibited a greater abundance versus the ID and SD groups, and there were significant differences in microbial species among the three groups at different classification levels. Additionally, a discernible correlation was established between certain clinical characteristics, including CA125, CA199, IL-22, among others and gut microbiota among patients with gastrointestinal diseases. Therefore, in the diagnosis and management of gastrointestinal diseases, combining clinical features with serum immunology and gut microbiota will greatly promote the diagnosis and treatment of gastrointestinal disease patients by clinical doctors, especially in terms of personalized multi-mode precise treatment strategies. In addition, the results of this study will provide new insights into the pathogenesis of gastrointestinal diseases in patients, and provide new ideas for the prevention and early intervention of gastrointestinal diseases.
The data presented in the study are deposited in the NCBI SRA repository, accession number PRJNA1073875.
The studies involving humans were approved by Medical Research Ethics Committee of Jiaying University. The studies were conducted in accordance with the local legislation and institutional requirements. The participants provided their written informed consent to participate in this study.
HC: Conceptualization, Methodology, Writing – original draft, Writing – review & editing. YZ: Investigation, Methodology, Writing – review & editing. GC: Data curation, Formal analysis, Writing – review & editing. WG: Resources, Writing – review & editing. YY: Funding acquisition, Project administration, Writing – review & editing.
The author(s) declare that financial support was received for the research, authorship, and/or publication of this article. This study was supported by Guangdong Provincial Health Commission, Medical Science and Technology Research Fund Project (B2021083) and Key Projects of Jiaying College School of Medicine (323E0168).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2024.1323842/full#supplementary-material
Adak, A., and Khan, M. R. (2019). An insight into gut microbiota and its functionalities. Cell. Mol. Life Sci. 76, 473–493. doi: 10.1007/s00018-018-2943-4
Ahmed, I., Roy, B. C., Khan, S. A., Septer, S., and Umar, S. (2016). Microbiome, metabolome and inflammatory bowel disease. Microorganisms 4:20. doi: 10.3390/microorganisms4020020
Allen, A. P., Hutch, W., Borre, Y. E., Kennedy, P. J., Temko, A., Boylan, G., et al. (2016). Bifidobacterium longum 1714 as a translational psychobiotic: modulation of stress, electrophysiology and neurocognition in healthy volunteers. Transl. Psychiatry 6:e939. doi: 10.1038/tp.2016.191
Alnahas, S., Hagner, S., Raifer, H., Kilic, A., Gasteiger, G., Mutters, R., et al. (2017). IL-17 and TNF-α are key mediators of Moraxella catarrhalis triggered exacerbation of allergic airway inflammation. Front. Immunol. 8:1562. doi: 10.3389/fimmu.2017.01562
Bae, M., Cassilly, C. D., Liu, X., Park, S. M., Tusi, B. K., Chen, X., et al. (2022). Akkermansia muciniphila phospholipid induces homeostatic immune responses. Nature 608, 168–173. doi: 10.1038/s41586-022-04985-7
Bassis, C. M., Erb-Downward, J. R., Dickson, R. P., Freeman, C. M., Schmidt, T. M., Young, V. B., et al. (2015). Analysis of the upper respiratory tract microbiotas as the source of the lung and gastric microbiotas in healthy individuals. mBio 6:e00097. doi: 10.1128/mBio.00037-15
Bouskra, D., Brezillon, C., Berard, M., Werts, C., Varona, R., Boneca, I. G., et al. (2008). Lymphoid tissue genesis induced by commensals through NOD1 regulates intestinal homeostasis. Nature 456, 507–510. doi: 10.1038/nature07450
Bullman, S., Pedamallu, C. S., Sicinska, E., Clancy, T. E., Zhang, X., Cai, D., et al. (2017). Analysis of Fusobacterium persistence and antibiotic response in colorectal cancer. Science 358, 1443–1448. doi: 10.1126/science.aal5240
Cait, A., Hughes, M. R., Antignano, F., Cait, J., Dimitriu, P. A., Maas, K. R., et al. (2018). Microbiome-driven allergic lung inflammation is ameliorated by short-chain fatty acids. Mucosal Immunol. 11, 785–795. doi: 10.1038/mi.2017.75
Chapman, M. G., and Underwood, A. J. (1999). Ecological patterns in multivariate assemblages: information and interpretation of negative values in ANOSIM tests. Mar. Ecol. Prog. Ser. 180, 257–265. doi: 10.3354/meps180257
Chen, S., Zhang, L., Li, M., Zhang, Y., Sun, M., Wang, L., et al. (2022). Fusobacterium nucleatum reduces METTL3-mediated m6A modification and contributes to colorectal cancer metastasis. Nat. Commun. 13:1248. doi: 10.1038/s41467-022-28913-5
Chung, S., Barnes, J. L., and Astroth, K. S. (2019). Gastrointestinal microbiota in patients with chronic kidney disease: a systematic review. Adv. Nutr. 10, 888–901. doi: 10.1093/advances/nmz028
Dai, Z., Zhang, J., Wu, Q., Chen, J., Liu, J., Wang, L., et al. (2019). The role of microbiota in the development of colorectal cancer. Int. J. Cancer 145, 2032–2041. doi: 10.1002/ijc.32017
de Theije, C. G., Wopereis, H., Ramadan, M., Van Eijndthoven, T., Lambert, J., Knol, J., et al. (2014). Altered gut microbiota and activity in a murine model of autism spectrum disorders. Brain Behav. Immun. 37, 197–206. doi: 10.1016/j.bbi.2013.12.005
de Vos, W. M., Tilg, H., Van Hul, M., and Cani, P. D. (2022). Gut microbiome and health: mechanistic insights. Gut 71, 1020–1032. doi: 10.1136/gutjnl-2021-326789
Ducarmon, Q. R., Zwittink, R. D., Hornung, B. V. H., Van Schaik, W., Young, V. B., and Kuijper, E. J. (2019). Gut microbiota and colonization resistance against bacterial enteric infection. Microbiol. Mol. Biol. Rev. 83:e00007. doi: 10.1128/MMBR.00007-19
Faith, J. J., Guruge, J. L., Charbonneau, M., Subramanian, S., Seedorf, H., Goodman, A. L., et al. (2013). The long-term stability of the human gut microbiota. Science 341:1237439. doi: 10.1126/science.1237439
Fan, Y., and Pedersen, O. (2021). Gut microbiota in human metabolic health and disease. Nat. Rev. Microbiol. 19, 55–71. doi: 10.1038/s41579-020-0433-9
Feng, F., Tian, Y., Xu, G., Liu, Z., Liu, S., Zheng, G., et al. (2017). Diagnostic and prognostic value of CEA, CA19-9, AFP and CA125 for early gastric cancer. BMC Cancer 17:737. doi: 10.1186/s12885-017-3738-y
Flint, H. J., Scott, K. P., Louis, P., and Duncan, S. H. (2012). The role of the gut microbiota in nutrition and health. Nat. Rev. Gastroenterol. Hepatol. 9, 577–589. doi: 10.1038/nrgastro.2012.156
Guigoz, Y., Dore, J., and Schiffrin, E. J. (2008). The inflammatory status of old age can be nurtured from the intestinal environment. Curr. Opin. Clin. Nutr. Metab. Care 11, 13–20. doi: 10.1097/MCO.0b013e3282f2bfdf
He, J., Li, H., Jia, J., Liu, Y., Zhang, N., Wang, R., et al. (2023). Mechanisms by which the intestinal microbiota affects gastrointestinal tumours and therapeutic effects. Mol. Biomed. 4:45. doi: 10.1186/s43556-023-00157-9
Heintz-Buschart, A., and Wilmes, P. (2018). Human gut microbiome: function matters. Trends Microbiol. 26, 563–574. doi: 10.1016/j.tim.2017.11.002
Hou, K., Wu, Z. X., Chen, X. Y., Wang, J. Q., Zhang, D., Xiao, C., et al. (2022a). Microbiota in health and diseases. Signal Transduct. Target. Ther. 7:135. doi: 10.1038/s41392-022-00974-4
Hou, K., Zhang, S., Wu, Z., Zhu, D., Chen, F., Lei, Z. N., et al. (2022b). Reconstruction of intestinal microecology of type 2 diabetes by fecal microbiota transplantation: why and how. Bosn. J. Basic Med. Sci. 22, 315–325. doi: 10.17305/bjbms.2021.6323
Hu, Y., Zhuang, Y., Gou, H. Y., Xie, C., and Ge, Z. M. (2022). Editorial: the interactions between gastrointestinal microbiota and Helicobacter pylori in diseases. Front. Cell. Infect. Microbiol. 12:1043906. doi: 10.3389/fcimb.2022.1043906
Ilhan, Z. E., Dibaise, J. K., Isern, N. G., Hoyt, D. W., Marcus, A. K., Kang, D. W., et al. (2017). Distinctive microbiomes and metabolites linked with weight loss after gastric bypass, but not gastric banding. ISME J. 11, 2047–2058. doi: 10.1038/ismej.2017.71
Jiang, T., Wang, H., Liu, L., Song, H., Zhang, Y., Wang, J., et al. (2021). CircIL4R activates the PI3K/AKT signaling pathway via the miR-761/TRIM29/PHLPP1 axis and promotes proliferation and metastasis in colorectal cancer. Mol. Cancer 20:167. doi: 10.1186/s12943-021-01474-9
Jin, C., Lagoudas, G. K., Zhao, C., Bullman, S., Bhutkar, A., Hu, B., et al. (2019). Commensal microbiota promote lung Cancer development via Gammadelta T cells. Cell 176:e16. doi: 10.1016/j.cell.2018.12.040
Kaistha, A., and Levine, J. (2014). Inflammatory bowel disease: the classic gastrointestinal autoimmune disease. Curr. Probl. Pediatr. Adolesc. Health Care 44, 328–334. doi: 10.1016/j.cppeds.2014.10.003
Kleessen, B., Kroesen, A. J., Buhr, H. J., and Blaut, M. (2002). Mucosal and invading bacteria in patients with inflammatory bowel disease compared with controls. Scand. J. Gastroenterol. 37, 1034–1041. doi: 10.1080/003655202320378220
Klooster, J. P. T., Bol-Schoenmakers, M., van Summeren, K., van Vliet, A. L. W., de Haan, C. A. M., van Kuppeveld, F. J. M., et al. (2021). Enterocytes, fibroblasts and myeloid cells synergize in anti-bacterial and anti-viral pathways with Il22 as the central cytokine. Commun. Biol. 4:631. doi: 10.1038/s42003-021-02176-0
Konig, J., and Brummer, R. J. (2014). Alteration of the intestinal microbiota as a cause of and a potential therapeutic option in irritable bowel syndrome. Benef. Microbes 5, 247–261. doi: 10.3920/BM2013.0033
Konig, J., Wells, J., Cani, P. D., Garcia-Rodenas, C. L., Macdonald, T., Mercenier, A., et al. (2016). Human intestinal barrier function in health and disease. Clin. Transl. Gastroenterol. 7:e196. doi: 10.1038/ctg.2016.54
Kostic, A. D., Chun, E., Robertson, L., Glickman, J. N., Gallini, C. A., Michaud, M., et al. (2013). Fusobacterium nucleatum potentiates intestinal tumorigenesis and modulates the tumor-immune microenvironment. Cell Host Microbe 14, 207–215. doi: 10.1016/j.chom.2013.07.007
Larsen, N., Vogensen, F. K., Van Den Berg, F. W., Nielsen, D. S., Andreasen, A. S., Pedersen, B. K., et al. (2010). Gut microbiota in human adults with type 2 diabetes differs from non-diabetic adults. PLoS One 5:e9085. doi: 10.1371/journal.pone.0009085
Lau, K., Srivatsav, V., Rizwan, A., Nashed, A., Liu, R., Shen, R., et al. (2017). Bridging the gap between gut microbial dysbiosis and cardiovascular diseases. Nutrients 9:859. doi: 10.3390/nu9080859
Leone, V., Chang, E. B., and Devkota, S. (2013). Diet, microbes, and host genetics: the perfect storm in inflammatory bowel diseases. J. Gastroenterol. 48, 315–321. doi: 10.1007/s00535-013-0777-2
Li, S., Liu, J., Zheng, X., Ren, L., Yang, Y., Li, W., et al. (2021). Tumorigenic bacteria in colorectal cancer: mechanisms and treatments. Cancer Biol. Med. 19, 147–162. doi: 10.20892/j.issn.2095-3941.2020.0651
Li, J., Zhang, A. H., Wu, F. F., and Wang, X. J. (2022). Alterations in the gut microbiota and their metabolites in colorectal cancer: recent progress and future prospects. Front. Oncol. 12:841552. doi: 10.3389/fonc.2022.1085432
Lin, Y., Lau, H. C., Liu, Y., Kang, X., Wang, Y., Ting, N. L., et al. (2022). Altered mycobiota signatures and enriched pathogenic Aspergillus rambellii are associated with colorectal cancer based on multicohort fecal metagenomic analyses. Gastroenterology 163, 908–921. doi: 10.1053/j.gastro.2022.06.038
Liu, T., Zhang, L., Joo, D., and Sun, S. C. (2017). NF-κB signaling in inflammation. Signal Transduct. Target. Ther. 2:17023. doi: 10.1038/sigtrans.2017.23
Lu, Y., Yuan, X., Wang, M., He, Z., Li, H., Wang, J., et al. (2022). Gut microbiota influence immunotherapy responses: mechanisms and therapeutic strategies. J. Hematol. Oncol. 15:47. doi: 10.1186/s13045-022-01273-9
Lucas Lopez, R., Grande Burgos, M. J., Galvez, A., and Perez Pulido, R. (2017). The human gastrointestinal tract and oral microbiota in inflammatory bowel disease: a state of the science review. APMIS 125, 3–10. doi: 10.1111/apm.12609
Lun, H., Yang, W., Zhao, S., Jiang, M., Xu, M., Liu, F., et al. (2019). Altered gut microbiota and microbial biomarkers associated with chronic kidney disease. Microbiologyopen 8:e00678. doi: 10.1002/mbo3.678
Luu, M., Riester, Z., Baldrich, A., Reichardt, N., Yuille, S., Busetti, A., et al. (2021). Microbial short-chain fatty acids modulate CD8+ T cell responses and improve adoptive immunotherapy for cancer. Nat. Commun. 12:4077. doi: 10.1038/s41467-021-24331-1
Lynch, S. V., and Pedersen, O. (2016). The human intestinal microbiome in health and disease. N. Engl. J. Med. 375, 2369–2379. doi: 10.1056/NEJMra1600266
Mager, L. F., Burkhard, R., Pett, N., Cooke, N. C. A., Brown, K., Ramay, H., et al. (2020). Microbiome-derived inosine modulates response to checkpoint inhibitor immunotherapy. Science 369, 1481–1489. doi: 10.1126/science.abc3421
Manichanh, C., Borruel, N., Casellas, F., and Guarner, F. (2012). The gut microbiota in IBD. Nat. Rev. Gastroenterol. Hepatol. 9, 599–608. doi: 10.1038/nrgastro.2012.152
Marcuzzi, A., Bianco, A. M., Girardelli, M., Tommasini, A., Martelossi, S., Monasta, L., et al. (2013). Genetic and functional profiling of Crohn’s disease: autophagy mechanism and susceptibility to infectious diseases. Biomed. Res. Int. 2013:297501. doi: 10.1155/2013/297501
Martens, E. C., Chiang, H. C., and Gordon, J. I. (2008). Mucosal glycan foraging enhances fitness and transmission of a saccharolytic human gut bacterial symbiont. Cell Host Microbe 4, 447–457. doi: 10.1016/j.chom.2008.09.007
Matusiewicz, M., Neubauer, K., Bednarz-Misa, I., Gorska, S., and Krzystek-Korpacka, M. (2017). Systemic interleukin-9 in inflammatory bowel disease: association with mucosal healing in ulcerative colitis. World J. Gastroenterol. 23, 4039–4046. doi: 10.3748/wjg.v23.i22.4039
Maynard, C., and Weinkove, D. (2018). The gut microbiota and ageing. Subcell. Biochem. 90, 351–371. doi: 10.1007/978-981-13-2835-0_12
Mazmanian, S. K., Liu, C. H., Tzianabos, A. O., and Kasper, D. L. (2005). An immunomodulatory molecule of symbiotic bacteria directs maturation of the host immune system. Cell 122, 107–118. doi: 10.1016/j.cell.2005.05.007
Morgan, X. C., Tickle, T. L., Sokol, H., Gevers, D., Devaney, K. L., Ward, D. V., et al. (2012). Dysfunction of the intestinal microbiome in inflammatory bowel disease and treatment. Genome Biol. 13:R79. doi: 10.1186/gb-2012-13-9-r79
Morkunas, E., Skieceviciene, J., and Kupcinskas, J. (2020). The impact of modulating the gastrointestinal microbiota in cancer patients. Best Pract. Res. Clin. Gastroenterol. 48–49:101700. doi: 10.1016/j.bpg.2020.101700
Mouzaki, M., Comelli, E. M., Arendt, B. M., Bonengel, J., Fung, S. K., Fischer, S. E., et al. (2013). Intestinal microbiota in patients with nonalcoholic fatty liver disease. Hepatology 58, 120–127. doi: 10.1002/hep.26319
Palleja, A., Kashani, A., Allin, K. H., Nielsen, T., Zhang, C., Li, Y., et al. (2016). Roux-en-Y gastric bypass surgery of morbidly obese patients induces swift and persistent changes of the individual gut microbiota. Genome Med. 8:67. doi: 10.1186/s13073-016-0312-1
Pluznick, J. L. (2020). The gut microbiota in kidney disease. Science 369, 1426–1427. doi: 10.1126/science.abd8344
Qin, Q., Yan, S., Yang, Y., Chen, J., Yan, H., Li, T., et al. (2021). The relationship between osteoporosis and intestinal microbes in the Henan province of China. Front. Cell Dev. Biol. 9:752990. doi: 10.3389/fcell.2021.752990
Sami, A., Elimairi, I., Stanton, C., Ross, R. P., and Ryan, C. A. (2020). The role of the microbiome in oral squamous cell carcinoma with insight into the microbiome-treatment axis. Int. J. Mol. Sci. 21:8061. doi: 10.3390/ijms21218061
Sanchez-Rodriguez, E., Egea-Zorrilla, A., Plaza-Diaz, J., Aragon-Vela, J., Munoz-Quezada, S., Tercedor-Sanchez, L., et al. (2020). The gut microbiota and its implication in the development of atherosclerosis and related cardiovascular diseases. Nutrients 12:605. doi: 10.3390/nu12030605
Segal, L. N., Alekseyenko, A. V., Clemente, J. C., Kulkarni, R., Wu, B., Gao, Z., et al. (2013). Enrichment of lung microbiome with Supraglottic taxa is associated with increased pulmonary inflammation. Microbiome 1:19. doi: 10.1186/2049-2618-1-19
Shi, X., Ma, T., Sakandar, H. A., Menghe, B., and Sun, Z. (2022). Gut microbiome and aging nexus and underlying mechanism. Appl. Microbiol. Biotechnol. 106, 5349–5358. doi: 10.1007/s00253-022-12089-5
Shim, J. A., Ryu, J. H., Jo, Y., and Hong, C. (2023). The role of gut microbiota in T cell immunity and immune mediated disorders. Int. J. Biol. Sci. 19, 1178–1191. doi: 10.7150/ijbs.79430
Sittipo, P., Lobionda, S., Lee, Y. K., and Maynard, C. L. (2018). Intestinal microbiota and the immune system in metabolic diseases. J. Microbiol. 56, 154–162. doi: 10.1007/s12275-018-7548-y
Slowicka, K., Petta, I., Blancke, G., Hoste, E., Dumas, E., Sze, M., et al. (2020). Zeb2 drives invasive and microbiota-dependent colon carcinoma. Nat. Cancer 1, 620–634. doi: 10.1038/s43018-020-0070-2
Sokolowska, M., Frei, R., Lunjani, N., Akdis, C. A., and O’Mahony, L. (2018). Microbiome and asthma. Asthma Res. Pract. 4:1. doi: 10.1186/s40733-017-0037-y
Sundin, J., Rangel, I., Fuentes, S., Heikamp-De Jong, I., Hultgren-Hornquist, E., De Vos, W. M., et al. (2015). Altered faecal and mucosal microbial composition in post-infectious irritable bowel syndrome patients correlates with mucosal lymphocyte phenotypes and psychological distress. Aliment. Pharmacol. Ther. 41, 342–351. doi: 10.1111/apt.13055
Suzuki, T. (2013). Regulation of intestinal epithelial permeability by tight junctions. Cell. Mol. Life Sci. 70, 631–659. doi: 10.1007/s00018-012-1070-x
Thomas, A. M., Manghi, P., Asnicar, F., Pasolli, E., Armanini, F., Zolfo, M., et al. (2019). Metagenomic analysis of colorectal cancer datasets identifies cross-cohort microbial diagnostic signatures and a link with choline degradation. Nat. Med. 25, 667–678. doi: 10.1038/s41591-019-0405-7
Ting, N. L., Lau, H. C., and Yu, J. (2022). Cancer pharmacomicrobiomics: targeting microbiota to optimise cancer therapy outcomes. Gut 71, 1412–1425. doi: 10.1136/gutjnl-2021-326264
Tripathi, A., Debelius, J., Brenner, D. A., Karin, M., Loomba, R., Schnabl, B., et al. (2018). The gut-liver axis and the intersection with the microbiome. Nat. Rev. Gastroenterol. Hepatol. 15, 397–411. doi: 10.1038/s41575-018-0011-z
Vatanen, T., Franzosa, E. A., Schwager, R., Tripathi, S., Arthur, T. D., Vehik, K., et al. (2018). The human gut microbiome in early-onset type 1 diabetes from the teddy study. Nature 562, 589–594. doi: 10.1038/s41586-018-0620-2
Vyhnalova, T., Danek, Z., Gachova, D., and Linhartova, P. B. (2021). The role of the oral microbiota in the etiopathogenesis of oral squamous cell carcinoma. Microorganisms 9:1549. doi: 10.3390/microorganisms9081549
Wan, G., Xie, M., Yu, H., and Chen, H. (2018). Intestinal dysbacteriosis activates tumor-associated macrophages to promote epithelial-mesenchymal transition of colorectal cancer. Innate Immun. 24, 480–489. doi: 10.1177/1753425918801496
Wang, Z. K., Yang, Y. S., Chen, Y., Yuan, J., Sun, G., and Peng, L. H. (2014). Intestinal microbiota pathogenesis and fecal microbiota transplantation for inflammatory bowel disease. World J. Gastroenterol. 20, 14805–14820. doi: 10.3748/wjg.v20.i40.14805
Weaver, C. T., Harrington, L. E., Mangan, P. R., Gavrieli, M., and Murphy, K. M. (2006). Th17: an effector Cd4 T cell lineage with regulatory T cell ties. Immunity 24, 677–688. doi: 10.1016/j.immuni.2006.06.002
Wei, P., Han, W., Zhang, Z., Tian, X., Yang, C., Wang, Q., et al. (2024). Microbiota in colorectal cancer related to liver metastasis. Chin. J. Cancer Res. 36, 17–24. doi: 10.21147/j.issn.1000-9604.2024.01.02
Wong, C. C., and Yu, J. (2023). Gut microbiota in colorectal cancer development and therapy. Nat. Rev. Clin. Oncol. 20, 429–452. doi: 10.1038/s41571-023-00766-x
Wu, D., Zhang, Y., Dong, S., and Zhong, C. (2021). Mutual interaction of microbiota and host immunity during health and diseases. Biophys. Rep. 7, 326–340. doi: 10.52601/bpr.2021.200045
Yang, W., Yu, T., Huang, X., Bilotta, A. J., Xu, L., Lu, Y., et al. (2020). Intestinal microbiota-derived short-chain fatty acids regulation of immune cell Il-22 production and gut immunity. Nat. Commun. 11:4457. doi: 10.1038/s41467-020-18262-6
Yu, J., Feng, Q., Wong, S. H., Zhang, D., Liang, Q. Y., Qin, Y., et al. (2017). Metagenomic analysis of faecal microbiome as a tool towards targeted non-invasive biomarkers for colorectal cancer. Gut 66, 70–78. doi: 10.1136/gutjnl-2015-309800
Zhang, H., Chen, Y., Wang, Z., Xie, G., Liu, M., Yuan, B., et al. (2022). Implications of gut microbiota in neurodegenerative diseases. Front. Immunol. 13:785644. doi: 10.3389/fimmu.2022.1091188
Zhong, Y., Fu, D., Deng, Z., Tang, W., Mao, J., Zhu, T., et al. (2022). Lactic acid bacteria mixture isolated from wild pig alleviated the gut inflammation of mice challenged by Escherichia coli. Front. Immunol. 13:822754. doi: 10.3389/fimmu.2022.1081889
Zhou, H., Dong, A., Xia, H., He, G., and Cui, J. (2018). Associations between CA19-9 and CA125 levels and human epidermal growth factor receptor 2 overexpression in patients with gastric cancer. Oncol. Lett. 16, 1079–1086. doi: 10.3892/ol.2018.8731
Keywords: gastrointestinal diseases, serum, immunology, gut, microbiota
Citation: Chen HZ, Zeng YY, Cai GX, Gu WD and Yang Y (2024) Differential analysis of serum immunology and gut microbiota in patients with gastrointestinal diseases. Front. Microbiol. 15:1323842. doi: 10.3389/fmicb.2024.1323842
Received: 18 October 2023; Accepted: 05 April 2024;
Published: 01 May 2024.
Edited by:
Mohammad Ashfaq, Chandigarh University, IndiaReviewed by:
Veeranoot Nissapatorn, Walailak University, ThailandCopyright © 2024 Chen, Zeng, Cai, Gu and Yang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: YaLi Yang, eXlhbGkxOTg2MjJAMTYzLmNvbQ==
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.