
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Microbiol. , 29 January 2024
Sec. Food Microbiology
Volume 15 - 2024 | https://doi.org/10.3389/fmicb.2024.1307610
This article is part of the Research Topic Microbial Food Safety in Retail Stores and Restaurants View all 8 articles
 Nirosha Ruwani Amarasekara1
Nirosha Ruwani Amarasekara1 Amrita Subramanya Swamy1
Amrita Subramanya Swamy1 Sumit Kumar Paudel1
Sumit Kumar Paudel1 Wentao Jiang2
Wentao Jiang2 KaWang Li2
KaWang Li2 Cangliang Shen2
Cangliang Shen2 Yifan Zhang1*
Yifan Zhang1*Introduction: This study aimed to determine the prevalence and virulome of Listeria in fresh produce distributed in urban communities.
Methods: A total of 432 fresh produce samples were collected from farmer’s markets in Michigan and West Virginia, USA, resulting in 109 pooled samples. Listeria spp. were isolated and L. monocytogenes was subjected to genoserogrouping by PCR and genotyping by pulsed-field gel electrophoresis (PFGE). Multi-locus sequence typing (MLST) and core-genome multi-locus sequence typing (cgMLST) were conducted for clonal identification.
Results: Forty-eight of 109 samples (44.0%) were contaminated with Listeria spp. L. monocytogenes serotype 1/2a and 4b were recovered from radishes, potatoes, and romaine lettuce. Four clonal complexes (CC) were identified and included hypervirulent CC1 (ST1) and CC4 (ST219) of lineage I as well as CC7 (ST7) and CC11 (ST451) of lineage II. Clones CC4 and CC7 were present in the same romaine lettuce sample. CC1 carried Listeria pathogenicity island LIPI-1 and LIPI-3 whereas CC4 contained LIPI-1, LIPI-3, and LIPI-4. CC7 and CC11 had LIPI-1 only.
Discussion: Due to previous implication in outbreaks, L. monocytogenes hypervirulent clones in fresh produce pose a public health concern in urban communities.
Listeria is widely distributed in the environment, such as soil, manure, and water (Zhu et al., 2017). Due to the high mortality and hospitalization rate of listeriosis (20–30%) (Radoshevich and Cossart, 2018) and the increasing popularity of farmer’s markets in urban communities, it is important to understand the prevalence and public health significance of Listeria spp. in fresh produce associated with this sector of agriculture. Recent studies have reported 3.1% to 3.9% of fresh produce contaminated with Listeria at farmer’s markets and supermarkets (Li K. et al., 2017; Scheinberg et al., 2017; Roth et al., 2018). While the prevalence of L. monocytogenes was generally below 1.9% (Johnston et al., 2006; Korir et al., 2016; Li K. et al., 2017; Scheinberg et al., 2017), none of the research provided molecular information or virulence characteristics of the pathogen from these types of products, which is critical for microbial risk assessment of food contamination.
Of 14 serotypes of L. monocytogenes, most are in lineage I (serotypes 1/2b, 3b, 3c, 4b) and lineage II (serotypes 1/2a, 1/2c, and 3a). Serotypes 4b and 1/2a predominate in human and food/environment isolates, respectively (Poimenidou et al., 2018). Lineage III and lineage IV are common in animal sources (Shen et al., 2013) and comprise serotypes 4a, some 4b strains, and 4c. L. monocytogenes can be further categorized into clonal complexes (CCs) based on multi-locus sequence typing (MLST). CC1, CC2, CC4, and CC6 of serotype 4b are remarkably more common in human listeriosis than food isolates and considered hypervirulent clones (Maury et al., 2016). Targeting hundreds of core genes of the entire L. monocytogenes genome, core genomic MLST (cgMLST) can determine the relatedness of isolates to epidemic clones (EC) and demonstrate the clinical relevance of bacteria (Chen et al., 2016; Li Z. et al., 2017). So far four major ECs (ECI, ECII, ECIII, and ECIV) have been identified in L. monocytogenes. ECI, ECII, and ECIV belong to serotype 4b while ECIII belongs to serotype 1/2a (Chen and Knabel, 2007).
Listeria pathogenicity island (LIPI) is a key virulence determinant in Listeria (Disson et al., 2021). LIPI-1 is listeriolysin O (LLO) island and necessary for intracellular survival and spread of L. monocytogenes; LIPI-2 is specific to L. ivanovii; LIPI-3 is listeriolysin S island, important for gastrointestinal colonization, and commonly associated with lineage I; LIPI-4 is the most recently identified neurovirulence/placental infection island (Maury et al., 2016) and strongly associated with the hypervirulence of CC4. LIPI-4 has been detected in CC4 as well as emerging clones of human and environmental origin (Lee et al., 2018). Another virulence feature that is crucial for the infectious potential of Listeria is the gene clusters responsible for teichoic acid biosynthesis. Teichoic acids are the most abundant glycopolymers in Listeria cell wall and play an important role in biofilm formation, phage susceptibility, antimicrobial resistance, and virulence (Sumrall et al., 2020). Serotype-specific glycosylation of teichoic acids mediated by different genes can lead to variation in virulence potential across clonal groups. Of the most common genes reported, gltA and gltB are specific for serotype 4b (Lei et al., 2001). gtcA has been found in serotype 4b and 1/2a (Cheng et al., 2008). Listeria internalin A encoded by inlA is a key factor mediating bacterial adhesion and invasion of host cells. Full-length inlA confers stronger virulence than Listeria harboring a premature stop codon (PMSC), the latter of which is 10,000-fold less invasive (Chen et al., 2011). While inlA and inlB are both important in intestinal and placental barrier invasion, inlB also plays a critical role in neuroinvasion and bacterial persistence (Maudet et al., 2022).
Considering the increasing popularity of farmer’s markets in urban communities, it is critical to understand Listeria contamination in locally grown fresh produce and their public health significance. Even though several studies reported the prevalence of Listeria in fresh produce obtained from farmer’s markets as discussed above, none described the virulence characteristics of bacteria. Therefore, this study aimed to determine the clonal identity and virulome of L. monocytogenes in fresh produce obtained from multiple farmer’s markets as related to human infections based on whole-genome sequencing (WGS) and molecular analysis.
A total of 432 fresh produce samples were collected from six farmer’s markets (A through F) in Michigan and West Virginia in the summer of 2019. The sites in Michigan were in the metro Detroit area and those in West Virginia in the Morgantown area. The two areas were approximately 400 miles apart. Markets A, C, D, and E mainly sold plant-based products, such as produce, herbs, beans etc. Markets B and F had a diverse collection of vendors that sold a large variety of items, including produce, meats, eggs, spices, etc. Convenient samples were taken from different produce vendors each week for the entire summer. For Listeria isolation, 50 grams of each vegetable sample was mixed with 450 mL of buffered listeria enrichment broth (BD, Sparks, MD, USA) and manually homogenized for 2 min. Three to six individual rinses of the same category were pooled and resulted in 109 pooled samples (Salazar et al., 2021; Hitchins et al., 2022). These included 41 leafy greens (romaine lettuce, collards, celery, basil, and kale), 39 root vegetables (radish, turnip, carrot, potato, beet, and onion), and 29 seeded vegetables (cantaloupe, tomato, cucumber, and zucchini).
Bacteria isolation followed the FDA Bacteriological Analytical Manual (BAM) method. One hundred milliliters of vegetable rinse were incubated at 30°C for 4 h for pre-enrichment, followed by the addition of selective agents, acriflavine (10 mg/L) and nalidixic acid (40 mg/L), and further incubated for 48 h at 30°C. The enriched broth was spread on PALCAM Listeria agar (Remel Inc., Lenexa, KS, USA) at 24 h and 48 h of incubation. Plates were incubated at 30°C for 24 h before presumptive Listeria colonies were streaked onto Rapid L’ mono differentiation agar (Bio-Rad, Hercules, CA, USA). PCR was conducted to identify Listeria spp., L. monocytogenes, and L. monocytogenes serotypes 1/2a, 1/2b, and 4b. L. monocytogenes ATCC 51773 was used as positive control. A no template control was included as well. PCR parameters and primer sequences were followed as described by Chen et al. (2017). The Chi-Square test of independence and the Post-Hoc tests were performed using SPSS v29.0.1 to compare Listeria prevalence in different sites and the significant association at 0.05 significance level.
Pulsed-field gel electrophoresis was carried out following the standardized protocol for L. monocytogenes (Graves and Swaminathan, 2001). Genomic DNA was prepared by mixing 240 μl of standardized cell suspension and 60 μl of 20 mg/ml lysozyme solution (ThermoFisher Scientific, Wilmington, DE, USA) followed by incubation at 37°C for 10 min. Sample plugs were digested with 25 U of AscI (New England Biolabs, Ipswich, MA, USA) at 37°C for 4 h. Plugs were then loaded on 1.2% Megabase agarose gel (Bio-rad) and electrophoresed on a CHEF- DR III apparatus (Bio-rad) using the following parameters: initial switch time, 4 s; final switch time, 40 s; run time, 22 h; angle, 120 ; gradient, 6 V/cm; temperature, 14°C; ramping factor, linear. PFGE patterns were analyzed using Bionumerics (version 6.6; Applied Maths, Austin, TX, USA). Indistinguishable profiles were defined as those demonstrating the same number and sizes of DNA fragments.
Whole genome sequencing was conducted at Omega Bioservices (Norcross, GA, USA). Briefly, DNA was extracted using the E.Z.N.A.® Bacterial DNA Kit (Omega Bio-tek, Norcross, GA, USA). The concentration was measured using the QuantiFluor dsDNA System on a Quantus Fluorometer (Promega, Madison, WI, USA). A Kapa Biosystems HyperPlus kit (Kapa Biosystems, Wilmington, MA, USA) was used for the whole-genome library construction. DNA was fragmented, and ends were repaired, 3’ adenylated, and ligated to adapters. The resulting adapter-ligated libraries were PCR-amplified. After Illumina indexes were added, they were pooled for multiplexed sequencing on an Illumina X10 platform (Illumina, San Diego, CA, USA) using the pair-end 150 bp run format. de novo assembly was performed on Ridom SeqSphere + v9.0 (Ridom GmbH, Germany) using SKESA 2.4.0 (Souvorov et al., 2018).
In silico MLST was performed to identify clonal complex (CC) and sequence type (ST) on CLC genomic workbench (Qiagen, Redwood City, CA, USA) using seven housekeeping genes of L. monocytogenes, including ABC transporter (abcZ), beta-glucosidase (bglA), catalase (cat), succinyl diaminopimelate dessucinylase (dapE), d-amino acid aminotransferase (dat), l-lactate dehydrogenase (ldh), and histidine kinase (lhkA). An allele number was assigned to the sequence of each allele and CC determined based on the definition of Ragon et al. (2008) and the Pasteur MLST database.1 WGS data were further subjected to cgMLST cluster identification on Ridom SeqSphere + v9.0 (Ridom GmbH, Germany) (L. monocytogenes cgMLST scheme, 1701 loci) (Ruppitsch et al., 2015). The cluster distance threshold for the core genome was 10 allele differences. Epidemic clones of ECI, ECII, ECIII and ECIV (Chen and Knabel, 2007) were downloaded from the NCBI website.
Virulence genes and antibiotic resistance genes were identified against the BIGSdb-Lm database (Moura et al., 2016). The heatmap was generated based on the presence or absence of a virulence gene using Morpheus matrix visualization and analysis software from the Broad Institute.2 The WGS data were also screened for stress islands as well as biocide tolerance genes for benzalkonium chloride (bcrABC, emrCE) and quaternary ammonium compounds (qacA), which are the most common biocides used for L. monocytogenes. The coding sequence of inlA was examined to ascertain whether the isolates carried a complete sequence or a mutation leading to a PMSC. Nucleotide sequences were aligned with inlA of L. monocytogenes EGD-e (NCBI: NC_003210.1) as reference.
All six markets had at least one sample positive for Listeria (Table 1). Site A had the highest Listeria prevalence (100%) (p < 0.05) and site B the lowest (28.6%, p < 0.05). Listeria spp. were recovered from 48 samples (44.0%), of which 32 were contaminated by L. welshimeri, 13 by L. innocua, and three by L. monocytogenes. All isolates from sites A, D, and E were L. welshimeri. Sites B and F had three Listeria species identified. L. monocytogenes was detected in sites B and F, the two markets that sold both vegetables and animal products such as meat and eggs. The two sites also had the highest number of samples collected, 49 from B and 34 from F, total accounting for more than 76% of 109 pooled samples. L. monocytogenes isolates included five each from potato and radish on site B and two from romaine lettuce on site F. All five isolates from radish (43a-e) and one from romaine lettuce (V11c) belonged to serotype 4b and the remaining six from potato (52a-e) and romaine lettuce (V11a) were identified as serotype 1/2a.
Four unique PFGE patterns were identified in L. monocytogenes (Figure 1). Isolates of the same serotype fell into the same cluster. Radish isolates 43a-e of serotype 4b shared the same DNA pattern and were clustered together with V11c from romaine lettuce. Similarly, 52a-e of serotype 1/2a from potato were indistinguishable from each other and clustered together with V11a.
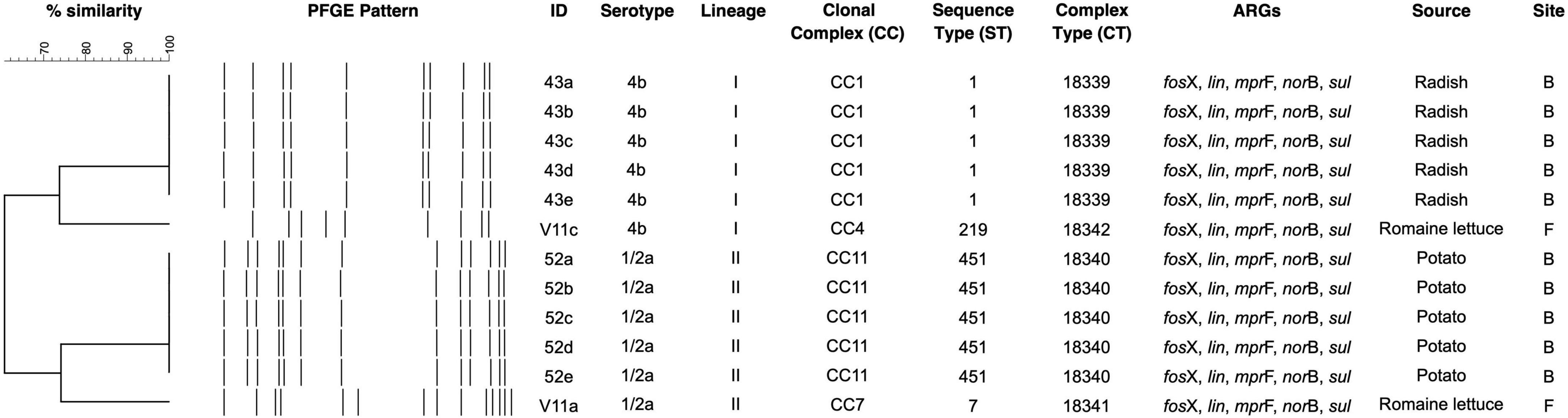
Figure 1. PFGE dendrogram of L. monocytogenes from fresh produce. ARG denotes antibiotic resistance gene.
Twelve isolates that were subjected to WGS had a sequencing coverage of 35 × to 50 × and a quality score above 30 (or base call accuracy 99.9%). Gene sequence data were used for downstream MLST and cgMLST analysis. In silico MLST revealed four CCs and STs (Figure 2). A 43a-e and V11c of serotype 4b (lineage I) belonged to CC1 (ST1) and CC4 (ST219), respectively, whereas V11a and 52a-e of serotype 1/2a (lineage II) fell under CC7 (ST7) and CC11 (ST451), respectively. The two isolates from romaine lettuce were designated as different CCs (CC4 and CC7). The hypervirulent CC1 and CC4 differed by five alleles out of seven target genes. The four CCs identified by MLST corresponded to four complex types (CT) by cgMLST (Figures 1, 3). Isolates 43a-e and 52a-e belonged to CT18339 and CT18340, respectively. Two isolates from romaine lettuce were split into CT18341 (isolate V11a) and CT18342 (isolate V11c) and belonged to two distinct clusters (Figure 3). V11a and 52a-e were in cluster 1. Isolates 52a-e were further divided into two groups by one allelic difference. V11c and 43a-e were in cluster 2, along with epidemic clones ECI, ECII and ECIV. Belonging to different CTs, 43a-e and ECI were closely related bearing only 68 allelic differences out of 1701 target loci. V11c and ECI differed by 969 alleles.
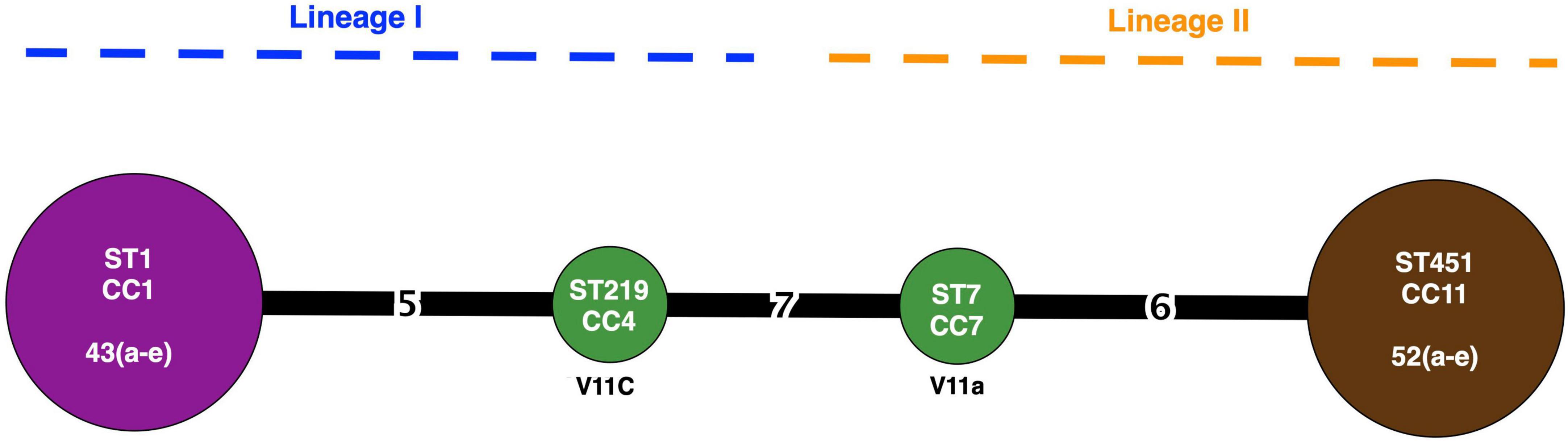
Figure 2. CC, ST, and lineage distribution of L. monocytogenes based on MLST. Node size represents number of isolates. Number on the connecting line represents allelic differences. Node color represents source of isolates: purple (radish), green (romaine lettuce), and brown (potato).
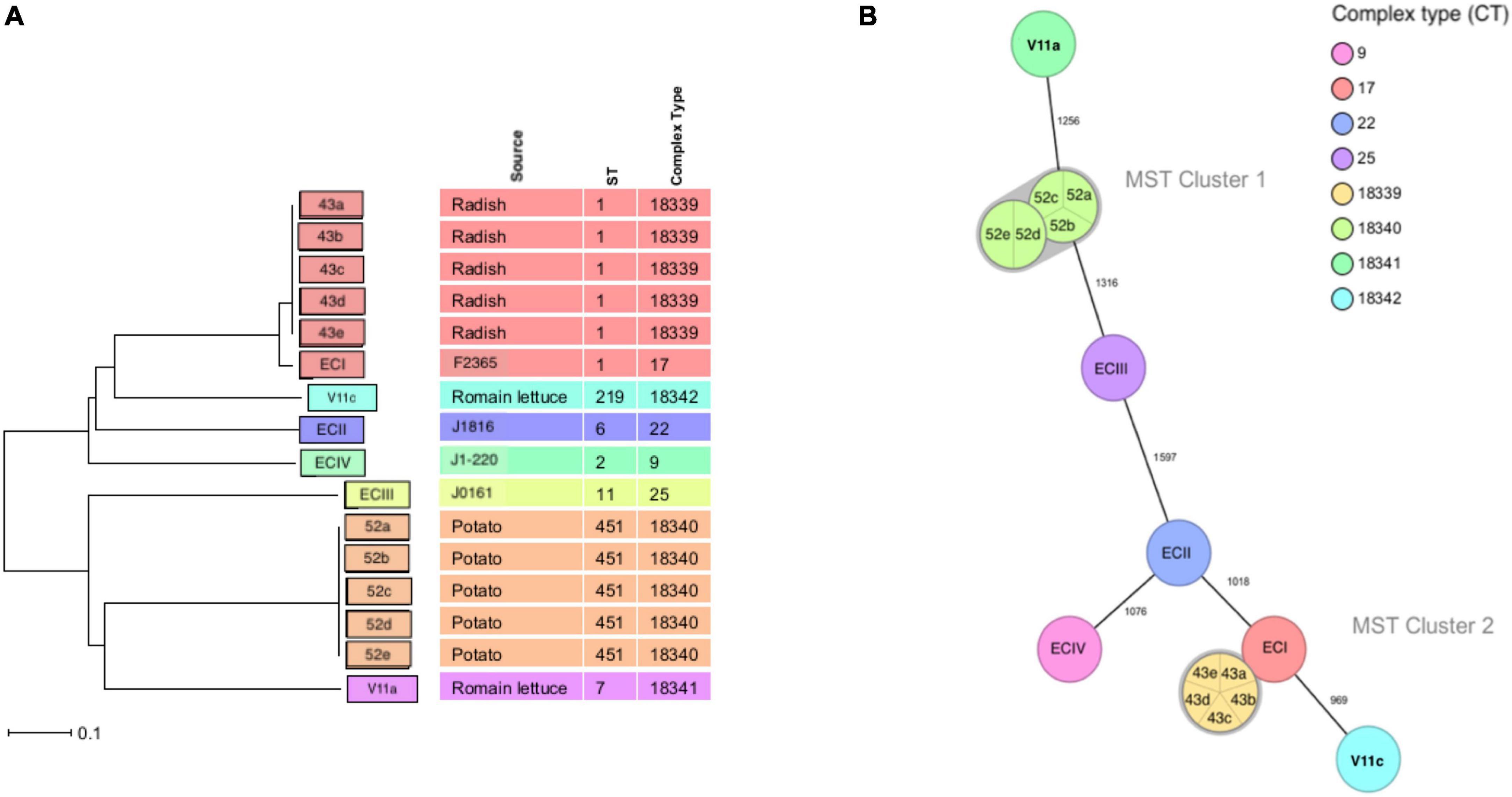
Figure 3. Minimum spanning tree of cgMLST analysis. (A) Minimum Spanning Tree (MST) and (B) Neighbor Joining Tree (NJT) illustrating the phylogenetic relationship based on allelic profiles of L. monocytogenes. MST cluster threshold is 10. Numbers on connecting lines represent the number of allelic differences between two strains.
The number of virulence genes ranged from 53 to 70 per isolate (Figure 4). Identical gene profile existed in isolates 43a-e, and so did in 52a-e. LIPI-1 was detected in all isolates. The major difference between lineage I and II was in LIPI-3, LIPI-4, and genes for teichoic acid biosynthesis. LIPI-3 was detected in serotype 4b CC1 and CC4. LIPI-4 was carried by CC4 only. Of the four genes for teichoic acid biosynthesis, gtcA was identified in both serotypes. gltA and gltB were found in serotype 4b, whereas tagB was contained by serotype 1/2a. Other virulence factors included internalin genes and those involved in various steps of infection such as adherence, invasion, intracellular survival, regulation of transcription and translation, surface protein anchoring, peptidoglycan modification, immune modification, bile resistance, and biofilm formation. All isolates had internalin genes inlABCEFIJKP. No PMSC was detected in inlA. inlG was found in serotype 1/2a but not 4b. CC7 (isolate V11a) was the only one carrying inlL but lacking inlC2DH. This isolate also had ami for adherence that was missing from all other isolates. Another notable difference was the invasion gene aut that was found in lineage II whereas its variant (aut_IVb) was carried by lineage I. Serotype 4b isolates also contained comK, a gene presumably for biofilm formation and persistence in food processing facilities (Hurley et al., 2019). V11a was found to harbor stress survival islet-1 (SSI-1), while none of the other isolates exhibited any stress islands. Biocide tolerance genes were not detected in any isolates. All isolates shared the same antibiotic resistance gene profile that included fosX (fosfomycin), lin (lincomycin), mprF (defensin), norB (fluoroquinolone), and sul (sulfonamide).
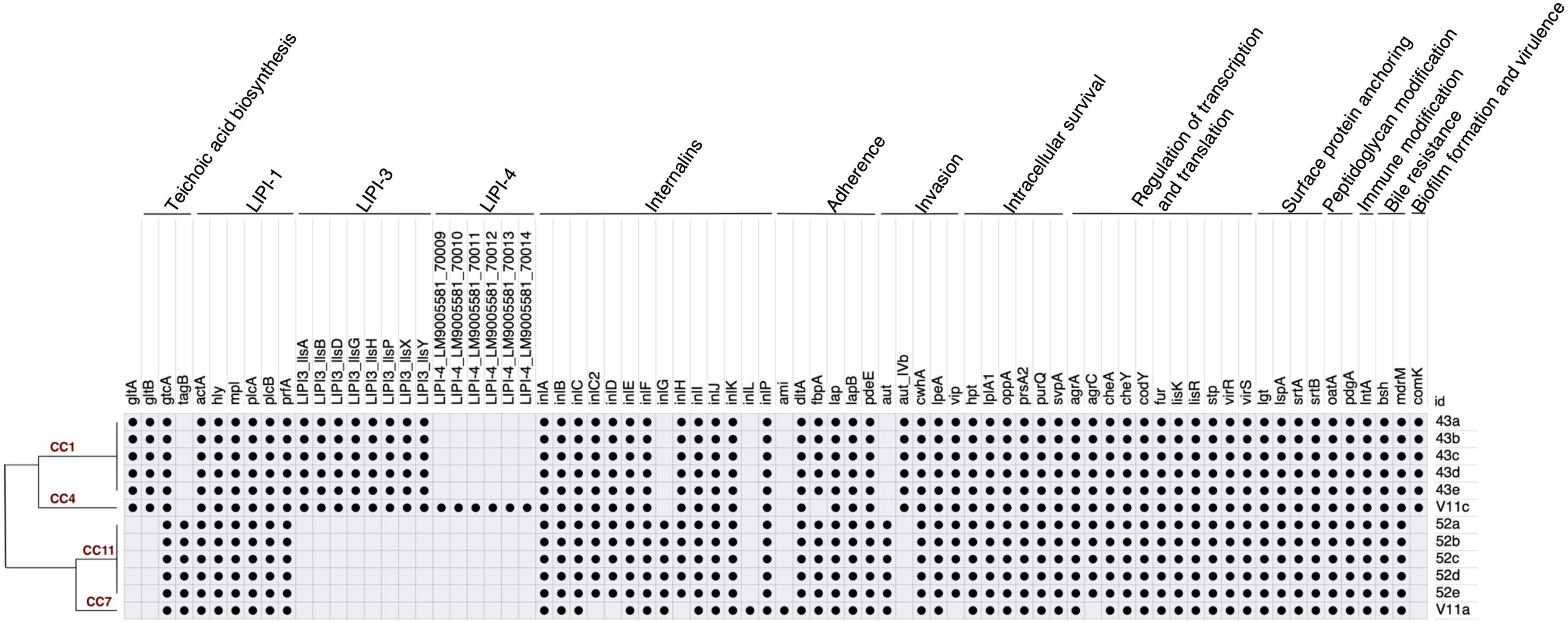
Figure 4. Virulence gene profile of L. monocytogenes from fresh produce.  denotes the presence of virulence gene.
denotes the presence of virulence gene.
Listeria are commonly present in the soil of various agricultural landscapes, thus freshly harvested produce is at a high risk of contamination (Chapin et al., 2014). Because vegetables sold at farmer’s markets have gone through limited processing and packaging procedures, any microbial contamination at farm level would end up in the final product. The recovery of CC4 (ST219) in fresh produce in this study is of public health concern. Considering the hypervirulence potential of this clonal group and the ready-to-eat nature of fresh produce, it is important to monitor the prevalence of hypervirulent clones of L. monocytogenes in urban communities and understand their clinical relevance.
Listeria prevalence in fresh produce varied across previous studies. Research on farmer’s markets in West Virginia and Kentucky (Li K. et al., 2017) as well as Pennsylvania (Scheinberg et al., 2017) both revealed the prevalence of slightly below 4%. An earlier study conducted in the Washington D.C. area demonstrated 19.7% of fresh produce contaminated with Listeria (Thunberg et al., 2002). In comparison, our study found Listeria at 44.0% (48 of 109 samples). This high prevalence can be due to the isolation procedure as most studies (Li K. et al., 2017; Scheinberg et al., 2017) used individual samples for bacteria isolation whereas others (Thunberg et al., 2002) and our study used pooled samples. The overall 2.8% of L. monocytogenes prevalence in this study is in line with previous research where the contamination level in fresh produce ranged from 0.66% to 4.7% (Thunberg et al., 2002; Li K. et al., 2017; Scheinberg et al., 2017; Roth et al., 2018). The market variation on L. monocytogenes prevalence (p > 0.05) can be explained by the predominance of samples in markets B and F, although cross contamination from meat cannot be excluded at large-scale markets with a diversity of vendors. In fact, multiple commodities sold at farmer’s markets, including meat, have been found contaminated with L. monocytogenes (Scheinberg et al., 2017; Kim et al., 2021). L. welshimeri and L. innocua appeared to be the most common Listeria species in fresh produce and have been reported previously (Thunberg et al., 2002).
The detection of L. monocytogenes 1/2a and 4b in fresh produce indicates a food safety concern given that 1/2a, 1/2b, and 4b are the top three serotypes responsible for human listeriosis (Liu, 2006). Isolates from radish and potato can be considered identical clones due to demonstrating indistinguishable profiles. The one-allelic difference between 52a-c and 52d-e from potato based on cgMLST should be negligible as the isolates shared otherwise identical profiles, including virulence genes and antibiotic resistance genes. Two genetically distinct clones in romaine lettuce may suggest different sources of contamination and the variation in virulence potential, although it is also possible that the two isolates were from two individual samples before being pooled for bacteria isolation. All four clonal groups identified in this study have been implicated in foodborne outbreaks worldwide (Chen et al., 2016). CC1 was formerly designated ECI because of its previous outbreak association. Their close connection is reflected by isolates 43a-e of this study differing from ECI by only 68 alleles. CC4 seemed to have mainly caused outbreaks in Europe (Dorozynski, 2000; Tasara et al., 2015; Chen et al., 2016), yet its recovery from local fresh produce should be highlighted because of its strong association with the central nervous system and maternal-neonatal listeriosis (Maury et al., 2016). Future surveillance at a larger scale would benefit our understanding of the extent of hypervirulent clones in urban communities. Moreover, while the hypervirulence potential itself could be a factor for the overrepresentation of CC4 in human isolates, non-food exposure should be considered as well because CC4 has been documented in various environmental sources such as animals and surface water (Lee et al., 2018; Raschle et al., 2021). CC7 and CC11 have caused multiple US outbreaks (Olsen et al., 2005; Centers for Disease Control and Prevention, 2011; Jackson et al., 2011). CC11 in association with maternal-neonatal listeriosis has been suggested from outbreaks involving pregnant, newborn, and elderly patients from consuming contaminated deli meat (Olsen et al., 2005) and Mexican-style cheese (Jackson et al., 2011). Taken together, fresh produce distributed in urban communities can be potential sources of Listeria clones with serious public health implication and requires close monitoring.
At the gene level, LIPI-1 is present in all L. monocytogenes and considered core virulence genes (Disson et al., 2021). LIPI-3 and LIPI-4 are critical for lineage I strains. LIPI-3 was carried by CC1 and CC4 of this study. It comprised genes encoding hemolysin listeriolysin S (llsABDGHPXY) and important for gastrointestinal colonization. Strongly associated with the hypervirulence of CC4, LIPI-4 is the most recently discovered pathogenicity island that encodes cellobiose-type phosphotransferase systems (PTS) and enhances neural and placental infection. The distribution of genes involved in teichoic acid biosynthesis is consistent with Listeria in Swiss surface water (Raschle et al., 2021) where CC1 and CC4 contained serotype 4b-specific gltA and gltB but lacked tagB, a gene most characterized in Bacillus subtilis (Brown et al., 2013). Because tagB was one of the genes unique to strong biofilm producers of L. monocytogenes (Park et al., 2022), the presence in serotype 1/2a in this study indicates the high potential of biofilm formation by these isolates. Despite that PMSC is commonly found in Listeria from food and food production environment (Nightingale et al., 2008; Li Z. et al., 2017), none of the isolates in this study carried PMSC, suggesting a full virulence potential.
The five antibiotic resistance genes detected in this study (fosX, lin, mprF, norB, sul) have been reported from various sources worldwide, including ready-to-eat meat (Mafuna et al., 2021; Centorotola et al., 2023), food processing environment (Hurley et al., 2019; Wieczorek et al., 2020), surface water (Raschle et al., 2021), and wildlife (Brown et al., 2023). This is not surprising as Listeria are intrinsically resistant to fosfomycin, many modern cephalosporins, oxacillin, sulfonamide, and nalidixic acid (Troxler et al., 2000; Mota et al., 2020; Brown et al., 2023). The genes detected in this study were either known (fosX) or strongly suggested (lin, norB, sul) to be intrinsic resistance genes. Because the isolates in this study were recovered from geographically and temporally separate occasions, it remains to be tested as to their antibiotic susceptibility phenotypes as genotypes and phenotypes do not always coincide. Although antibiotic resistance in general is not a serious issue in Listeria, multidrug-resistant Listeria do occur in food (Kayode and Okoh, 2022a,b). Thus, it is important to monitor the prevalence of antibiotic resistance genes, especially in hypervirulence clonal groups.
In conclusion, Listeria species are prevalent in vegetables distributed in urban communities. The presence of L. monocytogenes clones carrying a range of virulence genes indicates a potential public health concern. It is critical for urban food producers and distributors to control microbial transmission from the environment and other sources at urban gardens and farmer’s markets. Given the remarkable clinical significance of the hypervirulent clones and their recovery in the environment and animals, future research should focus on understanding the source of contamination, including wildlife and animal fertilizers, as well as the contribution of food, animal, and environmental exposure to listeriosis.
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/supplementary material. The L. monocytogenes WGS data presented in the study are deposited in the NCBI repository, BioProject PRJNA1006699.
NA: Data curation, Formal analysis, Investigation, Methodology, Validation, Writing – original draft. AS: Investigation, Writing – review and editing. SP: Investigation, Writing – review and editing. WJ: Investigation, Writing – review and editing. KL: Investigation, Writing – review and editing. CS: Conceptualization, Resources, Supervision, Validation, Writing – review and editing. YZ: Conceptualization, Funding acquisition, Project administration, Resources, Supervision, Validation, Writing – review and editing.
The author(s) declare financial support was received for the research, authorship, and/or publication of the article. The project was supported by the US Department of Agriculture, National Institute of Food and Agriculture [Grant No. 2018-70001-28938].
We would like to thank Fay Hansen for coordinating sampling sites.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Brown, P., Hernandez, K., Parsons, C., Chen, Y., Gould, N., DePerno, C. S., et al. (2023). Tetracycline resistance in Listeria monocytogenes and L. innocua from wild black bears (Ursus americanus) in the United States is mediated by novel transposable elements. Appl. Environ. Microbiol. 89:e0120523. doi: 10.1128/aem.01205-23
Brown, S., Santa Maria, J. P. Jr., and Walker, S. (2013). Wall teichoic acids of gram-positive bacteria. Annu. Rev. Microbiol. 67, 313–336. doi: 10.1146/annurev-micro-092412-155620
Centers for Disease Control and Prevention (2011). Multistate outbreak of listeriosis associated with jensen farms cantaloupe–united states, august-september 2011. MMWR Morb. Mortal. Wkly. Rep. 60, 1357–1358.
Centorotola, G., Ziba, M. W., Cornacchia, A., Chiaverini, A., Torresi, M., Guidi, F., et al. (2023). Listeria monocytogenes in ready to eat meat products from Zambia: Phenotypical and genomic characterization of isolates. Front. Microbiol. 14:1228726. doi: 10.3389/fmicb.2023.1228726
Chapin, T. K., Nightingale, K. K., Worobo, R. W., Wiedmann, M., and Strawn, L. K. (2014). Geographical and meteorological factors associated with isolation of Listeria species in New York State produce production and natural environments. J. Food Protect. 77, 1919–1928. doi: 10.4315/0362-028X.JFP-14-132
Chen, J.-Q., Healey, S., Regan, P., Laksanalamai, P., and Hu, Z. (2017). PCR-based methodologies for detection and characterization of Listeria monocytogenes and Listeria ivanovii in foods and environmental sources. Food Sci. Hum. Wellness 6, 39–59. doi: 10.1016/j.fshw.2017.03.001
Chen, Y., Gonzalez-Escalona, N., Hammack, T. S., Allard, M. W., Strain, E. A., and Brown, E. W. (2016). Core genome multilocus sequence typing for identification of globally distributed clonal groups and differentiation of outbreak strains of Listeria monocytogenes. Appl. Environ. Microbiol. 82, 6258–6272. doi: 10.1128/AEM.01532-16
Chen, Y., and Knabel, S. J. (2007). Multiplex PCR for simultaneous detection of bacteria of the genus Listeria, Listeria monocytogenes, and major serotypes and epidemic clones of L. monocytogenes. Appl. Environ. Microbiol. 73, 6299–6304. doi: 10.1128/AEM.00961-07
Chen, Y., Ross, W. H., Whiting, R. C., Van Stelten, A., Nightingale, K. K., Wiedmann, M., et al. (2011). Variation in Listeria monocytogenes dose responses in relation to subtypes encoding a full-length or truncated internalin A. Appl. Environ. Microbiol. 77, 1171–1180. doi: 10.1128/AEM.01564-10
Cheng, Y., Promadej, N., Kim, J.-W., and Kathariou, S. (2008). Teichoic acid glycosylation mediated by gtcA is required for phage adsorption and susceptibility of Listeria monocytogenes serotype 4b. Appl. Environ. Microbiol. 74, 1653–1655. doi: 10.1128/AEM.01773-07
Disson, O., Moura, A., and Lecuit, M. (2021). Making sense of the biodiversity and virulence of Listeria monocytogenes. Trends Microbiol. 29, 811–822. doi: 10.1016/j.tim.2021.01.008
Dorozynski, A. (2000). Seven die in French Listeria outbreak. BMJ 320:601. doi: 10.1136/sbmj.000355b
Graves, L. M., and Swaminathan, B. (2001). PulseNet standardized protocol for subtyping Listeria monocytogenes by macrorestriction and pulsed-field gel electrophoresis. Int. J. Food Microbiol. 65, 55–62. doi: 10.1016/S0168-1605(00)00501-8
Hitchins, A. D., Jinneman, K., and Chen, Y. (2022). Bacteriological Analytical Manual (BAM) Chapter 10: Detection of Listeria monocytogenes in Foods and Environmental Samples, and Enumeration of Listeria monocytogenes in Foods. Silver Spring, MA: US Food and Drug Administration.
Hurley, D., Luque-Sastre, L., Parker, C. T., Huynh, S., Eshwar, A. K., Nguyen, S. V., et al. (2019). Whole-genome sequencing-based characterization of 100 Listeria monocytogenes isolates collected from food processing environments over a four-year period. mSphere 4:e00252-19. doi: 10.1128/mSphere.00252-19
Jackson, K., Biggerstaff, M., Sweat, D., Klos, R., Nosari, J., Garrison, O., et al. (2011). Multistate outbreak of Listeria monocytogenes associated with Mexican-style cheese made from pasteurized milk among pregnant, Hispanic women. J. Food Protect. 74, 949–953. doi: 10.4315/0362-028X.JFP-10-536
Johnston, L. M., Jaykus, L.-A., Moll, D., Anciso, J., Mora, B., and Moe, C. L. (2006). A field study of the microbiological quality of fresh produce of domestic and Mexican origin. Int. J. Food Microbiol. 112, 83–95. doi: 10.1016/j.ijfoodmicro.2006.05.002
Kayode, A. J., and Okoh, A. I. (2022a). Assessment of multidrug-resistant Listeria monocytogenes in milk and milk product and One Health perspective. PLoS One 17:e0270993. doi: 10.1371/journal.pone.0270993
Kayode, A. J., and Okoh, A. I. (2022b). Incidence and genetic diversity of multi-drug resistant Listeria monocytogenes isolates recovered from fruits and vegetables in the Eastern Cape Province, South Africa. Int. J. Food Microbiol. 363:109513. doi: 10.1016/j.ijfoodmicro.2021.109513
Kim, C., Almuqati, R., Fatani, A., Rahemi, A., Kaseloo, P., Wynn, C., et al. (2021). Prevalence and antimicrobial resistance of foodborne pathogens in select fresh produce procured from farmers’ markets in Central Virginia. J. Food Saf. 41:e12895. doi: 10.1111/jfs.12895
Korir, R. C., Parveen, S., Hashem, F., and Bowers, J. (2016). Microbiological quality of fresh produce obtained from retail stores on the Eastern Shore of Maryland, United States of America. Food Microbiol. 56, 29–34. doi: 10.1016/j.fm.2015.12.003
Lee, S., Chen, Y., Gorski, L., Ward, T. J., Osborne, J., and Kathariou, S. (2018). Listeria monocytogenes source distribution analysis indicates regional heterogeneity and ecological niche preference among serotype 4b clones. mBio 9:e00396-18. doi: 10.1128/mBio.00396-18
Lei, X.-H., Fiedler, F., Lan, Z., and Kathariou, S. (2001). A novel serotype-specific gene cassette (gltA-gltB) is required for expression of teichoic acid-associated surface antigens in Listeria monocytogenes of serotype 4b. J. Bacteriol. 183, 1133–1139. doi: 10.1128/JB.183.4.1133-1139.2001
Li, K., Weidhaas, J., Lemonakis, L., Khouryieh, H., Stone, M., Jones, L., et al. (2017). Microbiological quality and safety of fresh produce in West Virginia and Kentucky farmers’ markets and validation of a post-harvest washing practice with antimicrobials to inactivate Salmonella and Listeria monocytogenes. Food Control 79, 101–108. doi: 10.1016/j.foodcont.2017.03.031
Li, Z., Pérez-Osorio, A., Wang, Y., Eckmann, K., Glover, W. A., Allard, M. W., et al. (2017). Whole genome sequencing analyses of Listeria monocytogenes that persisted in a milkshake machine for a year and caused illnesses in Washington State. BMC Microbiol. 17:134. doi: 10.1186/s12866-017-1043-1
Liu, D. (2006). Identification, subtyping and virulence determination of Listeria monocytogenes, an important foodborne pathogen. J. Med. Microbiol. 55, 645–659. doi: 10.1099/jmm.0.46495-0
Mafuna, T., Matle, I., Magwedere, K., Pierneef, R. E., and Reva, O. N. (2021). Whole genome-based characterization of Listeria monocytogenes isolates recovered from the food chain in South Africa. Front. Microbiol. 12:669287. doi: 10.3389/fmicb.2021.669287
Maudet, C., Kheloufi, M., Levallois, S., Gaillard, J., Huang, L., Gaultier, C., et al. (2022). Bacterial inhibition of Fas-mediated killing promotes neuroinvasion and persistence. Nature 603, 900–906. doi: 10.1038/s41586-022-04505-7
Maury, M. M., Tsai, Y.-H., Charlier, C., Touchon, M., Chenal-Francisque, V., Leclercq, A., et al. (2016). Uncovering Listeria monocytogenes hypervirulence by harnessing its biodiversity. Nat. Genet. 48, 308–313. doi: 10.1038/ng.3501
Mota, M. I., Vázquez, S., Cornejo, C., D’Alessandro, B., Braga, V., Caetano, A., et al. (2020). Does Shiga toxin-producing Escherichia coli and Listeria monocytogenes contribute significantly to the burden of antimicrobial resistance in Uruguay? Front. Vet. Sci. 7:583930. doi: 10.3389/fvets.2020.583930
Moura, A., Criscuolo, A., Pouseele, H., Maury, M. M., Leclercq, A., Tarr, C., et al. (2016). Whole genome-based population biology and epidemiological surveillance of Listeria monocytogenes. Nat. Microbiol. 2, 1–10. doi: 10.1038/nmicrobiol.2016.185
Nightingale, K., Ivy, R., Ho, A., Fortes, E., Njaa, B., Peters, R., et al. (2008). inlA premature stop codons are common among Listeria monocytogenes isolates from foods and yield virulence-attenuated strains that confer protection against fully virulent strains. Appl. Environ. Microbiol. 74, 6570–6583. doi: 10.1128/AEM.00997-08
Olsen, S. J., Patrick, M., Hunter, S. B., Reddy, V., Kornstein, L., MacKenzie, W. R., et al. (2005). Multistate outbreak of Listeria monocytogenes infection linked to delicatessen turkey meat. Clin. Infect. Dis. 40, 962–967. doi: 10.1086/428575
Park, M., Kim, J., Horn, L., Haan, J., Strickland, A., Lappi, V., et al. (2022). Sugar modification of wall teichoic acids determines serotype-dependent strong biofilm production in Listeria monocytogenes. Microbiol. Spectr. 10:e0276922. doi: 10.1128/spectrum.02769-22
Poimenidou, S. V., Dalmasso, M., Papadimitriou, K., Fox, E. M., Skandamis, P. N., and Jordan, K. (2018). Virulence gene sequencing highlights similarities and differences in sequences in Listeria monocytogenes serotype 1/2a and 4b strains of clinical and food origin from 3 different geographic locations. Front. Microbiol. 9:1103. doi: 10.3389/fmicb.2018.01103
Radoshevich, L., and Cossart, P. (2018). Listeria monocytogenes: Towards a complete picture of its physiology and pathogenesis. Nat. Rev. Microbiol. 16, 32–46. doi: 10.1038/nrmicro.2017.126
Ragon, M., Wirth, T., Hollandt, F., Lavenir, R., Lecuit, M., Le Monnier, A., et al. (2008). A new perspective on Listeria monocytogenes evolution. PLoS Pathog. 4:e1000146. doi: 10.1371/journal.ppat.1000146
Raschle, S., Stephan, R., Stevens, M. J., Cernela, N., Zurfluh, K., Muchaamba, F., et al. (2021). Environmental dissemination of pathogenic Listeria monocytogenes in flowing surface waters in Switzerland. Sci. Rep. 11:9066. doi: 10.1038/s41598-021-88514-y
Roth, L., Simonne, A., House, L., and Ahn, S. (2018). Microbiological analysis of fresh produce sold at Florida farmers’ markets. Food Control 92, 444–449. doi: 10.1016/j.foodcont.2018.05.030
Ruppitsch, W., Pietzka, A., Prior, K., Bletz, S., Fernandez, H. L., Allerberger, F., et al. (2015). Defining and evaluating a core genome multilocus sequence typing scheme for whole-genome sequence-based typing of Listeria monocytogenes. J. Clin. Microbiol. 53, 2869–2876. doi: 10.1128/JCM.01193-15
Salazar, J. K., Fay, M., Eckert, C., Stewart, D., Cranford, V., and Tortorello, M. L. (2021). Evaluation of methods of enrichment and compositing of environmental samples for detection of Listeria monocytogenes. J. Food Protect. 84, 639–646. doi: 10.4315/JFP-20-276
Scheinberg, J. A., Dudley, E. G., Campbell, J., Roberts, B., DiMarzio, M., DebRoy, C., et al. (2017). Prevalence and phylogenetic characterization of Escherichia coli and hygiene indicator bacteria isolated from leafy green produce, beef, and pork obtained from farmers’ markets in Pennsylvania. J. Food Protect. 80, 237–244. doi: 10.4315/0362-028X.JFP-16-282
Shen, J., Rump, L., Zhang, Y., Chen, Y., Wang, X., and Meng, J. (2013). Molecular subtyping and virulence gene analysis of Listeria monocytogenes isolates from food. Food Microbiol. 35, 58–64. doi: 10.1016/j.fm.2013.02.014
Souvorov, A., Agarwala, R., and Lipman, D. J. (2018). SKESA: Strategic k-mer extension for scrupulous assemblies. Genome Biol. 19:153. doi: 10.1186/s13059-018-1540-z
Sumrall, E. T., Keller, A. P., Shen, Y., and Loessner, M. J. (2020). Structure and function of Listeria teichoic acids and their implications. Mol. Microbiol. 113, 627–637. doi: 10.1111/mmi.14472
Tasara, T., Ebner, R., Klumpp, J., and Stephan, R. (2015). Complete genome sequence of Listeria monocytogenes N2306, a strain associated with the 2013-2014 listeriosis outbreak in Switzerland. Genome Announc. 3:e00553-15. doi: 10.1128/genomeA.00553-15
Thunberg, R. L., Tran, T. T., Bennett, R. W., Matthews, R. N., and Belay, N. (2002). Microbial evaluation of selected fresh produce obtained at retail markets. J. Food Protect. 65, 677–682. doi: 10.4315/0362-028X-65.4.677
Troxler, R., Von Graevenitz, A., Funke, G., Wiedemann, B., and Stock, I. (2000). Natural antibiotic susceptibility of Listeria species: L. grayi, L. innocua, L. ivanovii, L. monocytogenes, L. seeligeri and L. welshimeri strains. Clin. Microbiol. Infect. 6, 525–535. doi: 10.1046/j.1469-0691.2000.00168.x
Wieczorek, K., Bomba, A., and Osek, J. (2020). Whole-genome sequencing-based characterization of Listeria monocytogenes from fish and fish production environments in Poland. Int. J. Mol. Sci. 21:9419. doi: 10.3390/ijms21249419
Keywords: Listeria, hypervirulence, clonal complex, fresh produce, farmer’s market
Citation: Amarasekara NR, Swamy AS, Paudel SK, Jiang W, Li K, Shen C and Zhang Y (2024) Hypervirulent clonal complex (CC) of Listeria monocytogenes in fresh produce from urban communities. Front. Microbiol. 15:1307610. doi: 10.3389/fmicb.2024.1307610
Received: 10 October 2023; Accepted: 08 January 2024;
Published: 29 January 2024.
Edited by:
Lucilla Iacumin, University of Udine, ItalyReviewed by:
Alexandre Leclercq, Institut Pasteur, FranceCopyright © 2024 Amarasekara, Swamy, Paudel, Jiang, Li, Shen and Zhang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Yifan Zhang, eWlmYW4uemhhbmdAd2F5bmUuZWR1
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.