- 1Department of Biotechnology, University of Chemistry and Technology Prague, Prague, Czechia
- 2Department of Biomedical Engineering, Faculty of Electrical Engineering and Communication, Brno University of Technology, Brno, Czechia
- 3Department of Informatics, Ludwig-Maximilians-Universität München, Munich, Germany
- 4Department of Internal Medicine—Hematology and Oncology, University Hospital Brno, Brno, Czechia
- 5Department of Internal Medicine–Hematology and Oncology, Faculty of Medicine, Masaryk University, Brno, Czechia
- 6Department of Biochemistry and Microbiology, University of Chemistry and Technology Prague, Prague, Czechia
- 7Ecofuel Laboratories s.r.o., Prague, Czechia
- 8Hop Research Institute, Co. Ltd., Zatec, Czechia
Background: This paper brings new information about the genome and phenotypic characteristics of Pantoea agglomerans strain DBM 3797, isolated from fresh Czech hop (Humulus lupulus) in the Saaz hop-growing region. Although P. agglomerans strains are frequently isolated from different materials, there are not usually thoroughly characterized even if they have versatile metabolism and those isolated from plants may have a considerable potential for application in agriculture as a support culture for plant growth.
Methods: P. agglomerans DBM 3797 was cultured under aerobic and anaerobic conditions, its metabolites were analyzed by HPLC and it was tested for plant growth promotion abilities, such as phosphate solubilization, siderophore and indol-3-acetic acid productions. In addition, genomic DNA was extracted, sequenced and de novo assembly was performed. Further, genome annotation, pan-genome analysis and selected genome analyses, such as CRISPR arrays detection, antibiotic resistance and secondary metabolite genes identification were carried out.
Results and discussion: The typical appearance characteristics of the strain include the formation of symplasmata in submerged liquid culture and the formation of pale yellow colonies on agar. The genetic information of the strain (in total 4.8 Mb) is divided between a chromosome and two plasmids. The strain lacks any CRISPR-Cas system but is equipped with four restriction-modification systems. The phenotypic analysis focused on growth under both aerobic and anaerobic conditions, as well as traits associated with plant growth promotion. At both levels (genomic and phenotypic), the production of siderophores, indoleacetic acid-derived growth promoters, gluconic acid, and enzyme activities related to the degradation of complex organic compounds were found. Extracellular gluconic acid production under aerobic conditions (up to 8 g/l) is probably the result of glucose oxidation by the membrane-bound pyrroloquinoline quinone-dependent enzyme glucose dehydrogenase. The strain has a number of properties potentially beneficial to the hop plant and its closest relatives include the strains also isolated from the aerial parts of plants, yet its safety profile needs to be addressed in follow-up research.
1 Introduction
The Czech Republic is famous for its Pilsner beer, in which hops (Humulus lupulus L.) is irreplaceable feedstock. Hops (Humulus lupulus L.) is a perennial, dioecious, climbing plant belonging to the Cannabaceae family and order of Rosales (Zhang et al., 2011). Hop cones of the female plant contain in lupulin glands a lot of secondary metabolites which are mostly used in beer production. Hop resins, essential oils and their transformation products impart beer its typical bitter taste and hoppy aroma (Jaskula et al., 2008). A number of substances contained in hops have at the same time many biologically active effects. 8-Prenylnaringenin is known to be the most potent phytoestrogen to date (Milligan et al., 2000). Beta acids are characterized by strong antimicrobial effects against some groups of bacteria (Sleha et al., 2021; Fahle et al., 2022). Xanthohumol from the group of prenylated flavonoids has anticarcinogenic effects against certain types of cancer (Miranda et al., 1999).
Till now, hop plant research has focused on topics other than the natural colonization of the hop plant by endo- and epiphytic bacteria, with only a few exceptions (Goryluk-Salmonovicz et al., 2016; Allen et al., 2019; Micci et al., 2022). On the contrary, it was rather assumed that hop would not be colonized by bacteria because many of its metabolites have antimicrobial properties (for a review see, Bocquet et al., 2018). The considerable resistance of hops to bacteria is also evidenced by the fact that bacterial diseases of hops are rare compared to viruses or diseases caused by fungi. Nevertheless, some bacteria have already been isolated from hops—bacteria of the genus Streptomyces from the rhizosphere of hops (Koçak, 2019), Pseudomonas stutzeri and Pseudomonas fluorescens from hop cones (Sevigny et al., 2019) and Pantoea agglomerans from hop cones (Sevigny et al., 2019) and from dried hop pellets (Kolek et al., 2021).
Various Pantoea species and strains have been isolated as free-living bacteria from different habitats or from different hosts, having loose or tighter relationships to the host, i.e., many of them being plant epiphytes/endophytes, sometimes plant pathogens, others being insect symbionts or facultative human pathogens (Walterson and Stavrinides, 2015). Specifically, different strains of P. agglomerans were isolated from different plants (Walterson and Stavrinides, 2015) or were found as clinical isolates causing various health problems (Soutar and Stavrinides, 2019). In the same time, other P. agglomerans strains had beneficial properties in medicine (such as macrophage activation or to combat Plasmodium parasites) while yet other strains can become biocontrol agents or mediate improved plant nutrition, which might be very useful for future sustainable agricultural practice (Dutkiewicz et al., 2016). To identify differences between plant and clinical isolates through their genomes is difficult (Rezzonico et al., 2009) and is complicated by changing taxonomy and frequent misidentification of P. agglomerans clinical isolates (Rezzonico et al., 2009; Soutar and Stavrinides, 2019). Regarding taxonomy, into the species name P. agglomerans were transferred all older species originally called Enterobacter agglomerans or Erwinia herbicola and these species names are used as synonyms now. However, older isolates of E. agglomerans or E. herbicola frequently differ from P. agglomerans (Soutar and Stavrinides, 2019).
Despite a number of published materials, little attention has been devoted to the fact that P. agglomerans, as a facultative anaerobic bacterium, behaves very differently under different conditions and can switch between different metabolic pathways. In particular its anaerobic metabolism has been neglected, but can harbor surprises. P. agglomerans DBM 3696 was identified as the probable causative agent of inflation (production of CO2) in bags filled with dried hop pellets stored in a modified atmosphere (Kolek et al., 2021). This study aims to demonstrate the versatile metabolism of P. agglomerans DBM 3697 isolated from fresh green hop cones in the Steknik hopyards (Czech Republic) along with identification of significant metabolites, as well as the complete genome and its comprehensive analysis, stressing potentially beneficial properties that may be used in agriculture (e.g., phosphate solubilisation, siderophore and auxin (indol-3-acetic acid and its derivatives) productions and others).
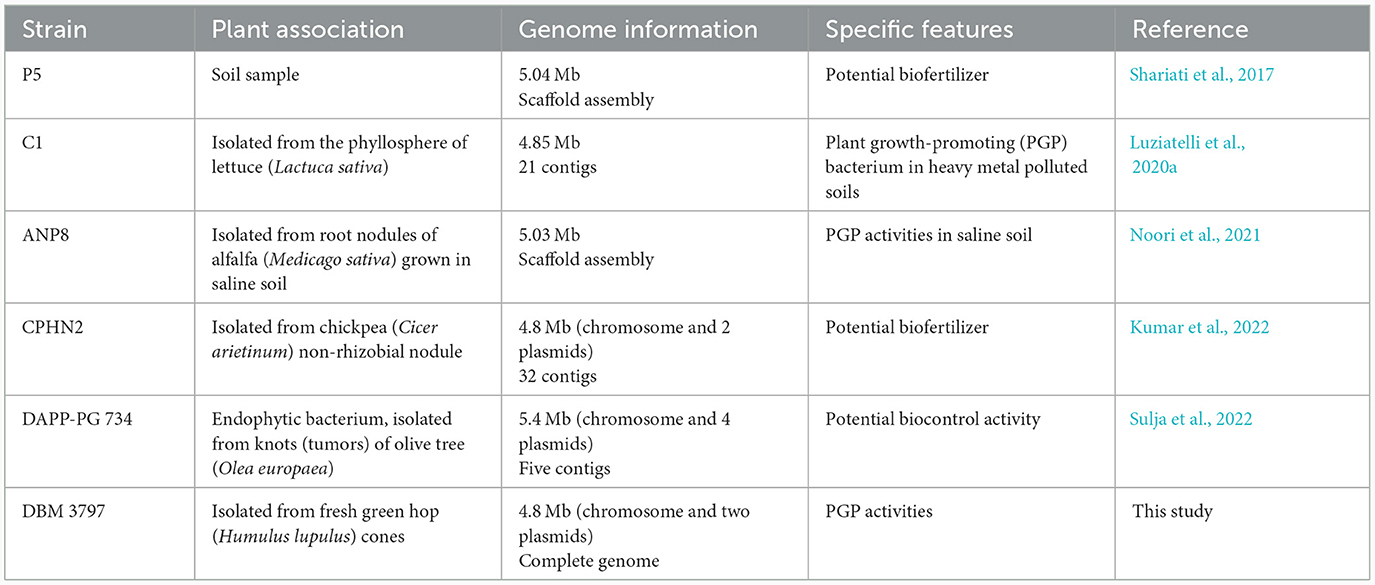
Currently, about 139 P. agglomerans genome reports can be found in the NCBI GenBank/RefSeq database, but in only 31 cases, complete genome sequences have been published.1 Regarding plant associated P. agglomerans, not showing pathogenesis, comprehensive genome analyses were performed for only five strains shown in Table 1. The complete available genomic dataset of different P. agglomerans strains, although it may seem extensive at first glance, is in fact insufficient for differential analysis of the genomes to find significant differences between beneficial and pathogenic strains. To be able to do so, it is necessary to expand this dataset to include strains that do not show pathogenesis to plants or humans, as well as clinical isolates or strains associated with plant pathogenesis.
2 Material and methods
2.1 The strain isolation and its storage
The strain Pantoea agglomerans DBM 3797, deposited in the culture collection of the Department of Biochemistry and Microbiology (DBM) of the UCT Prague, was stored at −80°C. The strain was isolated from fresh hop cones grown in Steknik hopyards in the Czech Republic (altitude 192 m,_latitude and longitude: 50.3166292 N 13.6102039 E). The plant material was collected under aseptic conditions in a sterile plastic bag, surface sterilization with ethanol was performed in the laboratory, the material was ground and suspended in sterile physiological solution (0.9% NaCl). The solid particles were then filtered under aseptic conditions through sterile folded filter paper and the filtrate was diluted 10 ×, 100 × and 1,000 ×. From each dilution, 0.1 ml was inoculated onto the surface of the solidified LB medium. The plates were incubated for 24 or 48 h at 30°C. From the initial growth, the culture was plated several times on the surface of the agar medium and individual colonies were isolated.
2.2 Culture conditions
All chemicals for preparation of culture media, as well as for microbiological assays of plant growth promoting activities, were purchased from Merck if not stated otherwise. The strain was cultured in Lysogeny Broth (LB) culture medium containing (g/l): tryptone 10, yeast extract 5 and NaCl 5 or in Pantoea glucose medium (PGM) containing (g/l): glucose 10 or 20, MgSO4.7H2O 0.4, NaCl 1, CaCl2.2H2O 0.2, NH4NO3 1.5; yeast extract 0.2, KCl 0.2, peptone 0.5. The ability to utilize different carbon sources was tested in PGM, where glucose was changed for xylose, cellulose (Avicel) or lignin, always at a concentration of 10 g/l. For bioreactor culture, the glucose concentration was 20 g/l. In some experiments, LB culture medium was supplemented with glucose at a concentration of 10 g/l. For growth in Petri dishes, culture medium was supplemented with 20 g/l of agar. For indole-3-acetic acid production, tryptophan was added to the culture medium at a concentration of 5 g/l. Culture experiments were performed at 30°C for 24–48 h. Each inoculum for culture experiments was prepared by overnight growth in LB liquid medium.
Cultivation experiments were run in Erlenmeyer shake flasks on a rotary shaker (150 rpm), in a 1 l bioreactor (Infors HT), both aerobically and anaerobically, in a thermostat (the case of growth on solidified medium in Petri dishes) or in an anaerobic chamber (Concept 400, UK). For bioreactor experiments, PGM culture medium with 20 g/l glucose was used. The working volume of the 1 l bioreactor was 700 ml (630 ml of fresh culture medium and 70 ml of inoculum) and pH monitoring were used. In aerobic culture, the filtered air rate was 1 VVM and oxygen saturation was measured using an oxygen electrode. Details of anaerobic bioreactor culture were described previously by Sedlar et al. (2021).
2.3 Analyses
Growth was monitored as an optical density (OD) at 600 nm using a spectrophotometer (Agilent Cary 60 UV-VIS) against the respective medium without inoculation as a blank. Microscopic control of the culture was performed using phase contrast microscopy (Olympus BX51; Olympus).
The concentration of substrate (glucose) and metabolites (ethanol, lactic, acetic and gluconic acids) were determined by HPLC (Agilent Series 1200 HPLC; Agilent) with refractive index detection. The parameters of the HPLC analysis were as follows: injection sample volume of 20 μl, 5 mM H2SO4 as a mobile phase, a flow rate of 1 ml/min, IEX H+ polymer column (Watrex) and a column temperature of 60°C.
Statistical analysis of different growth conditions was performed in R (v4.3.1). Data normality was checked with Shapiro-Wilk test (p-value < 0.05) and homogeneity of variance was verified using Bartlett's test (p-value < 0.05). One-way ANOVA with post-hoc Tukey test was performed at p-adjusted value < 0.05 to identify statistically significant changes under different cultivation conditions.
2.4 DNA extraction and sequencing
For short-read sequencing, genomic DNA was extracted and purified using the GenElute Bacterial Genomic DNA Kit (Sigma-Aldrich, St. Louis, MI, USA) following the manufacturer's protocols. The purity of the DNA was assessed using a NanoDrop spectrophotometer (Thermo Scientific, Wilmington, DE, USA), while the concentration was determined using the Qubit 3.0 (Thermo Scientific, Wilmington, DE, USA). DNA library construction was carried out using the KAPA HyperPlus kit, following the standard protocol. Subsequently, sequencing was performed on the Illumina MiSeq platform (Illumina, San Diego, CA, USA) using the MiSeq Reagent Kit v2 (500 cycles).
For long-read sequencing, high molecular weight genomic DNA was extracted using the MagAttract HMW DNAKit (Qiagene, Venlo. NL). The purity of the extracted DNA was assessed with the NanoDrop (Thermo Fisher Scientific, Waltham, MA, USA), while the concentration was determined using the Qubit 3.0 (Thermo Scientific, Wilmington, DE, USA). The DNA length was confirmed using the Agilent 4200 TapeStation (Agilent Technologies, Santa Clara, CA, USA). Ligation sequencing 1D Kit (Oxford Nanopore Technologies, Oxford, UK) was used for library preparation, and sequenced on the MinION platform (Oxford Nanopore Technologies) with the R9.4.1 flowcell.
2.5 Genome assembly
Long Oxford Nanopore Technologies (ONT) reads were basecalled with Guppy v3.4.4 and used for the initial de novo assembly performed with Flye v2.8.1. The assembly was polished with Racon v1.4.13 (Vaser et al., 2017) and Medaka v1.2.5 using ONT reads quality checked with MinIONQC (Lanfear et al., 2019). Auxiliary PAF files were generated using minimap2 (Li, 2018). Short Illumina paired reads were quality trimmed with Trimomatic v1.36 (Bolger et al., 2014), checked with FastQC v0.11.5 and MultiQC v1.7 (Ewels et al., 2016), and used for additional rounds of polishing with Pilon v1.24 (Walker et al., 2014). For that purpose, short reads were mapped onto ONT assembly with BWA v07.17 (Li and Durbin, 2009) and auxiliary BAM files were processed with SAMtools (Li et al., 2009). Finally, the resulting chromosomal sequence was rearranged according to the origin of replication (oriC) to dnaA, being the first gene on the sense strand, using the Ori-finder (Luo et al., 2019) and both plasmid sequences were rearranged in a similar manner to the repB gene coding for plasmid replication initiator, being the first gene on the sense strand, using manual BLAST searches (Altschul et al., 1990).
2.6 Genome annotation and analysis
Genome annotation was performed by NCBI Prokaryotic Genome Annotation Pipeline (PGAP) (Tatusova et al., 2016). The functional annotation of protein coding genes was extended by classification into categories of clusters of orthologous groups (COG). Overall three sources of COG categories were used, namely eggNOG-mapper (Cantalapiedra et al., 2021) (v2.1.9), Operon-mapper (Taboada et al., 2018) and Batch CD-Search (Marchler-Bauer and Bryant, 2004) tools. Results were further processed by COGtools (v1.0.0) (https://github.com/xpolak37/COGtools) to merge them and create a final improved COG annotation. Assigned COG categories were visualized as circular plots by DNAplotter (Carver et al., 2009), which is a part of the Artemis (Carver et al., 2012) (v2.18.0) software. Selected pathogenic and non-pathogenic chromosomal sequences were compared and visualized as a circular graph in BRIG (v0.95) software (Alikhan et al., 2011). Pan-genome analysis was performed using BPGA v1.3 (Chaudhari et al., 2016), with amino acid sequences clustered using USEARCH (Edgar, 2010), with an identity cut-off of 90%. In total, 139 genomes of P. agglomerans were obtained from the NCBI RefSeq database (30th October 2023) (O'Leary et al., 2016) to define the core genome and to perform a phylogenomic analysis, i.e., concatenated sequences of core genes were aligned with MUSCLE and resulting multiple sequence alignment was used to reconstruct phylogeny with Neighbor-Joining algorithm using Kimura distance implemented in BPGA.
The genome was searched for clustered regularly interspaced short palindromic repeat (CRISPR) arrays using the CRISPRDetect (Biswas et al., 2016) (v2.4) tool and cas genes were searched in the genome manually. Components of restriction-modification (RM) systems were identified using REBASE (v307) database (Roberts et al., 2023). Prophage DNA was searched with the online version of PHASTER (Arndt et al., 2016). Antibiotic-resistant genes search was performed using Resistance Gene Identifier (RGI) 6.0.0 included in the Comprehensive Antibiotic Resistance Database (CARD) 3.2.5 (Alcock et al., 2020) by submitting protein sequences of CDSs. Virulence factors were searched using online version of VFAnalyzer against the virulence factor database (VFDB) (Liu et al., 2019) with default parameters and using Klebsiella pneumoniae as the closest annotated reference for P. aglomerans. Homologs of genes involved in biosynthetic pathways, putatively contributing to plant growth promotion and other activities were identified with tBLASTn, with the use of target protein sequences from closely related species. The length of initial seeds was set to 5 and BLOSUM62 matrix was used for scoring the alignments while gap introduction and extension was set to 11 and 1, respectively. Finally, identification of secondary metabolite biosynthesis gene clusters was performed with antiSMASH v7.1.0 (Blin et al., 2023) through its web service using relaxed detection strictness parameter.
2.7 Plant growth promoting activities
Screening of PGP activities was performed by established microbiological assays combined with spectrophotometric or visual detection and frequently (if not stated otherwise) at a semi-quantitative level (low, medium, or high).
Siderophore production was tested on blue agar chrome azurol S medium containing chrome azurol S and hexadecyltrimethylammonium bromide as indicators. Development of a yellowish orange halo around the colonies was taken as indicative of siderophore production; for details see Schmidt et al. (2018).
Phosphate solubilization was detected as a clear zone, i.e., the ability to solubilize calcium phosphate using Pikovskaya medium see Schmidt et al. (2018) or was tested in liquid NBRIP medium where the concentration of phosphate was determined spectrophotometrically by the ammonium molybdate-ascorbic acid method (Stranska et al., 2021).
Nitrogen fixation ability was tested in NFGM medium and evaluated spectrophotometrically; details are presented in Stranska et al. (2021).
Amylase, lipase, pectinase, protease/peptidase, and cellulase production were tested for in the appropriate solidified culture medium (Hawar, 2022) and evaluated as a halo or colored zone around a colony.
Ammonium release was detected after 24 h growth in LB medium by the Quantofix rapid test following instructions of the producer (Quantofix).
Indole-3-acetic acid (IAA) or IAA-like compound production was tested by Salkowski reagent (0.01 M FeCl3 in 35% HClO4) in LB culture medium supplemented with tryptophan after 48 h growth on a rotary shaker; for details of the procedure see Gilbert et al. (2018).
Indole production/release was tested by reaction with Kovacs reagent (Merck) in LB culture medium after 24 h growth on a rotary shaker.
3 Results
3.1 Genome and pan-genome
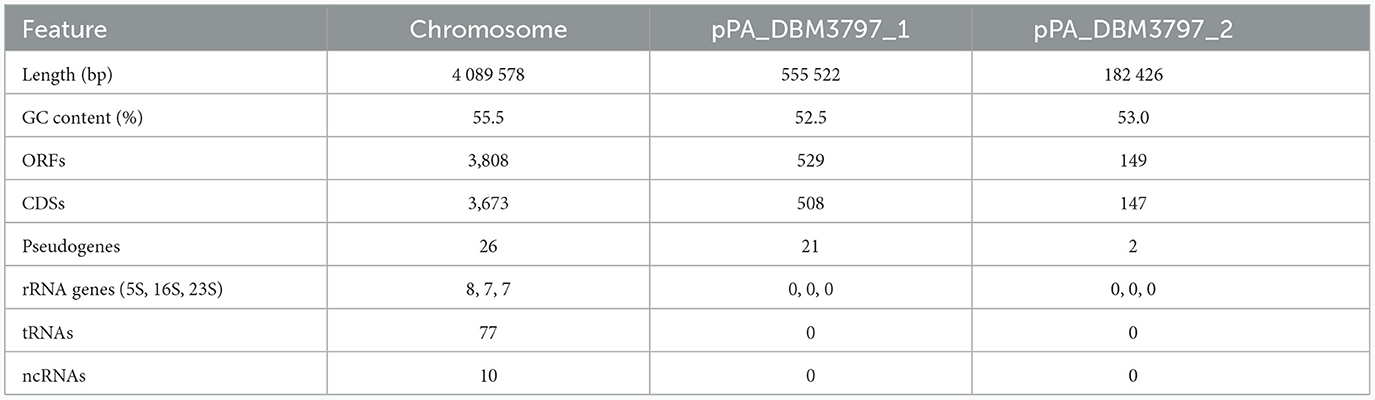
The genome of P. agglomerans DBM 3797 comprises a circular chromosome (size 4,089 kb) and two circular plasmids (pPA_DBM3797_1 size 555 kb and pPA_DBM3797_2 size 182 kb) assembled using both long reads and short reads in a hybrid approach with an overall coverage of 584 × and deposited at the DDBJ/EMBL/GenBank under accession numbers CP086133.1, CP086134.1, and CP086135.1, respectively. The overall genome length is 4,827,556 bp and contains 4,486 open reading frames (ORFs). While 4,328 ORFs present protein-coding sequences (CDSs), 49 genes had corrupted ORFs and formed pseudogenes. The remaining loci corresponded to RNA coding genes. Statistics for chromosome and both plasmids are summarized in Table 2. While most genes putatively corresponding to phenotypic traits were found on the chromosome, some of them were located on the large pPA_DBM3797_1 plasmid.
Functional annotation of the genome was done by classifying protein coding genes and pseudogenes into 26 categories of clusters of orthologous genes (COG), see Figure 1. For the chromosomal sequence, 3,280 genes were assigned a COG while 419 genes (11.33%) remained unannotated. Three most abundant categories were categories: E (Amino acid transport and metabolism) with 323 genes (8.73%), G (Carbohydrate transport and metabolism) with 321 genes (8.68%), and M (Cell wall/membrane/envelope biogenesis) with 268 genes (7.25%). For plasmid pPA_DBM3797_1 sequence, 66 genes (12.48%) remained unannotated and from 463 remaining genes, 78 genes (14.74%) were assigned category K (Transcription) and 69 genes (13.04%) category G (Carbohydrate transport and metabolism), forming the two most abundant groups. Moreover, 130 genes found on plasmid pPA_DBM3797_2 were assigned a COG while the most frequent was category T (Signal transduction mechanism) with 20 genes (13.42 %). Other abundant groups were formed by genes assigned to K (Transcription) and V (Defense mechanisms), both containing 15 genes (10.07%). Nineteen genes (12.75%) remained unannotated. For a complete summary of COG statistics see Supplementary Table 1. Additionally, seven secondary metabolite biosynthetic gene clusters were found. While five regions corresponding to redox-cofactor (2,192,097–2,214,263), arylpolyene and homoserine lactone (hserlactone) (2,614,136–2,673,970), thiopeptide (2,739,933–2,766,189), hserlactone (3,555,694–3,576,332), and NRP (non-ribosomal peptide)-metallophore and NRPS (non-ribosomal peptide synthase) (3,660,174–3,713,865) were located on the chromosome, remaining two regions corresponding to NI-siderophore (165,106–195,460) and terpene (376,789–400,350) were located on the pPA_DBM3797_1 plasmid (Supplementary Figure 1).
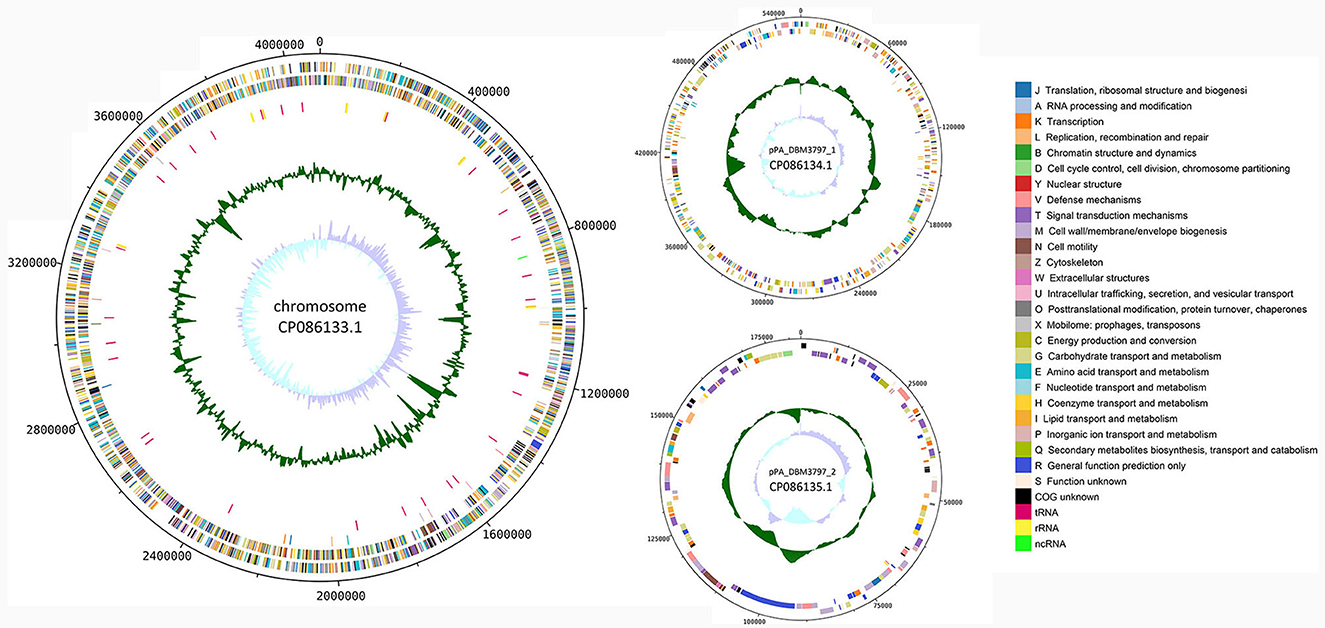
Figure 1. A circular map of the chromosome (on the left) and both plasmid (on the right) sequences of P. agglomerans DBM 3797. From outside to center: CDS on the forward strand (color-coded by COG categories), CDS on the reverse strand (color-coded by COG categories), pseudogenes (color-coded by COG categories), RNA genes (tRNA, rRNA, ncRNA), GC content and GC skew.
The P. agglomerans genome was missing any clustered regularly interspaced short palindromic repeat (CRISPR) arrays and similarly, no cas genes are present. Furthermore, we found four restriction-modification (R-M) systems, one was of type I and the remaining three were of type II. A type I R-M system consisted of one restriction enzyme Pag3797ORF16840P, two methyltransferases M1.Pag3797ORF16840P and M2.Pag3797ORF16840P, and one specificity subunit S.Pag3797ORF16840P. In all three type II systems, we found methyltransferases: M.Pag3797DamP, M.Pag3797ORF3130P and M.Pag3797DcmP, while the last enzyme was also coupled with nicking enzyme V.Pag3797DcmP. Complete results for R-M systems can be found in Supplementary Table 2. Only a single intact prophage of length 41.8 kbp corresponding to phage PHAGE_Erwini_ENT90_NC_019932 was found on the chromosome, within region 648492-690301. The whole region contained 57 proteins in total while 53 of these genes corresponded to phage DNA.
Last but not least, the genome was searched for antibiotic resistance and virulence genes. In total, 12 strict hits were found in the Comprehensive Antibiotic Resistance Database. While 11 genes were localized on chromosome, the remaining gene was found on plasmid pPA_DBM3797_2, see Supplementary Table 3. Six of these genes corresponded to antibiotic efflux resistance mechanisms, five were predicted to be responsible for antibiotic target alternation and one for antibiotic inactivation. The presence of these genes was confirmed by searching for virulence factors in general using the Virulence Factor Database. The presence of other virulence factors remained inconclusive as only partial hits to other secretion system or endotoxin genes were detected. The only complete system corresponded to gene machinery responsible for flagella construction, however, the cell motility is not necessarily connected to virulence.
The chromosomal sequence of P. agglomerans DBM 3797 was compared with chromosomal sequences of selected pathogenic and non-pathogenic strains downloaded from GenBank database under further mentioned accession numbers. These included the only pathogenic available strain isolated from clinical, FDAARGOS 1447 (CP077366.1); a plant pathogenic strain, BH6c (CP134744.1); and three non-pathogenic strains isolated from the same plant part (above-ground part) as DBM 3797, namely DAPP-PG734 (OW970315.1), CPHN 2(CP098414.1), and CFSAN047154 (CP034474.1). The results of a comparative analysis revealed no significant differences among the chromosomal sequences. All analyzed sequences were aligned to the reference strain DBM 3797 with 100% identity along almost the entire length of the sequence (see Supplementary Figure 2).
The pan-genome analysis showed that all currently available genomes of P. agglomerans strains with successful taxonomy check shared 2,399 genes that formed the core genome of the species. Phylogenomic tree reconstructed using concatenated sequences of all core genes showed that P. agglomerans DBM 3797 presented a well-distinguished strain with strains AB378 and CFBP8784 being the closest relatives, see Figure 2. The complete list of the strains included into the Figure 2 is shown in Supplementary Table 4.
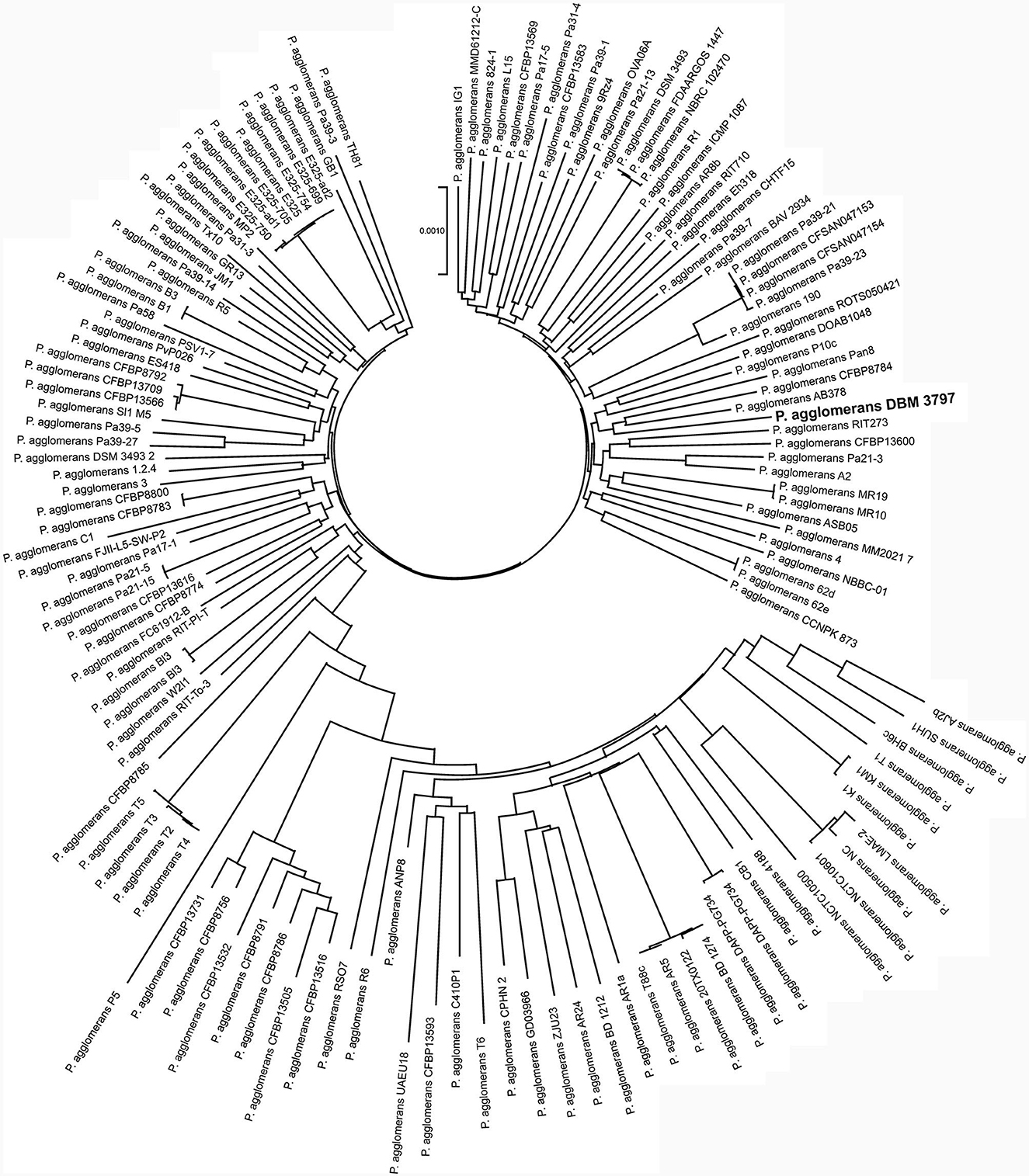
Figure 2. Phylogenomic analysis based on concatenated sequences of 2,399 core genes of 139 genomes of P. agglomerans strains. The tree was reconstructed using the Neighbor-Joining method using the Bacterial Pan Genome Analysis tool (BPGA).
3.2 Growth, metabolite formation, and putative corresponding genes
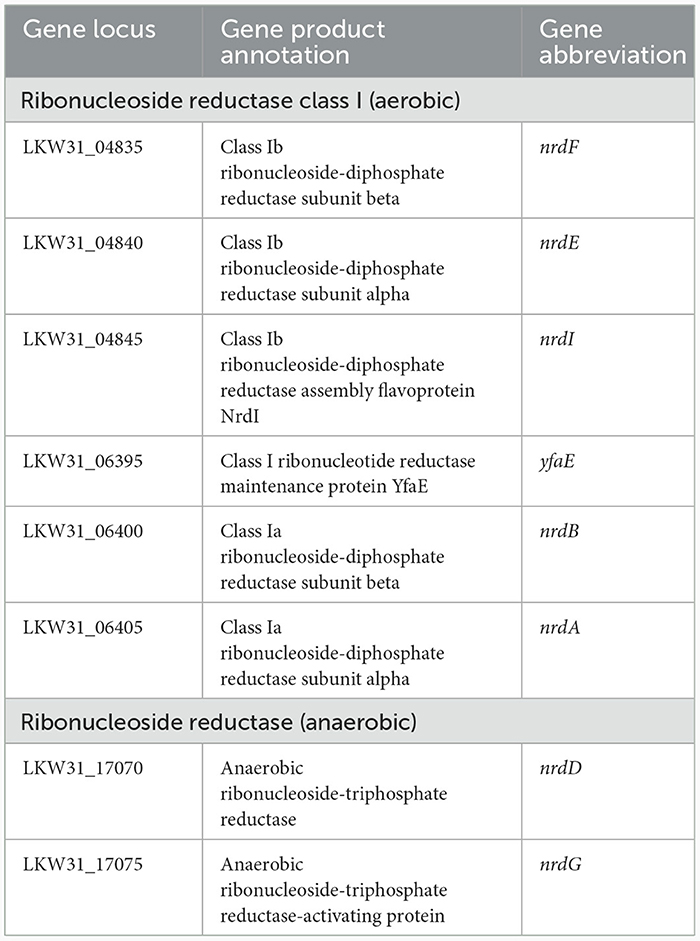
P. agglomerans DBM 3797 was grown under aerobic and anaerobic conditions in liquid medium. Ability to grow in the presence/absence and limitation of oxygen requires security of basic life functions under both conditions, such as ability to synthesize deoxyribonucleotides by ribonucleotide reductases (RNR). RNRs mediate the reduction of nucleotides differently under aerobic/anaerobic conditions and for this, different enzymes are required. The corresponding RNR genes located on the chromosome are shown in Table 3. Under aerobic conditions, the strain preferred LB medium, which did not contain saccharides. Young cells (5 h after inoculation) were highly motile while in older LB-medium, in the grown population (after 24 h), symplasmata formation was observed (Supplementary Figure 3). Putative genes responsible for motility and symplasmata formation (biofilm like structure) were found on the chromosome and are shown in Supplementary Table 5. It was also tested whether symplasmata formation might be initiated by indole release during tryptophan degradation (tryptophan presence was assumed in LB medium) by the reaction of culture medium supernatant with Kovacs reagent, but the reaction was negative. In addition, the gene for tryptophanase was not found on the chromosome, nor on the plasmids. An interesting feature of growth in LB medium was alkalization of the culture medium (up to pH 8.7, see Table 4), which was caused by the release of ammonium ions from amino acids, serving as a carbon source in the medium. Ammonium ion concentration was about 100–200 mg/l. Aerobically, the culture was able to utilize glucose, xylose, cellulose (Avicel) and lignin, however compared to LB medium growth, the cells were shorter, the amount of biomass formed in 24 h was about 10 times lower and no symplasmata were observed. Anaerobic growth required saccharides for a fermentative way of obtaining energy and therefore was not possible in simple LB medium not containing glucose. On solidified LB medium under aerobic conditions, the strain formed round, convex, slimy looking, cream to yellowish colored colonies (see Supplementary Figure 4). Genes for carotenoid pigment production, giving the colony a yellowish color, were found on plasmid pPA_DBM3797_1, see Supplementary Table 5 and Supplementary Figure 1 (terpene).
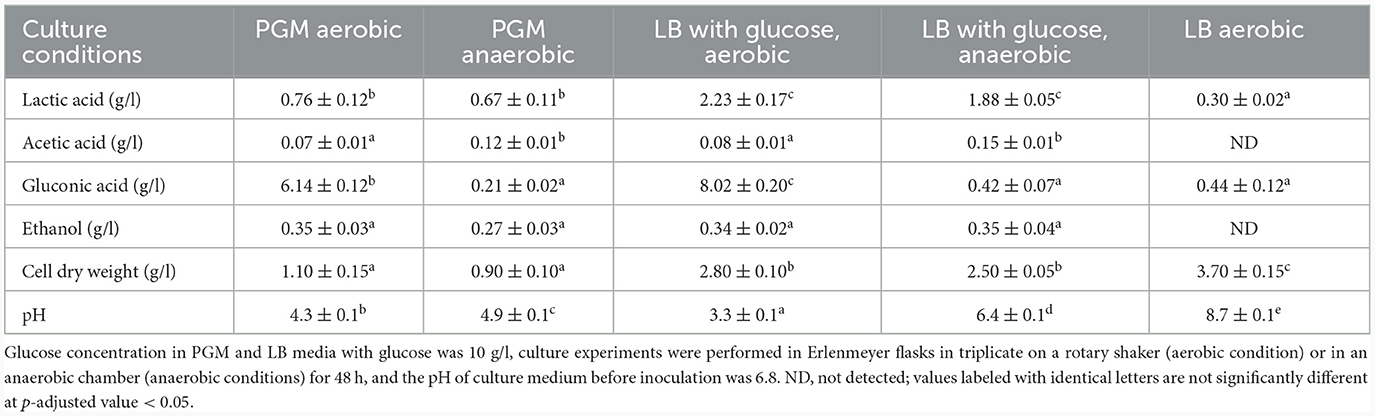
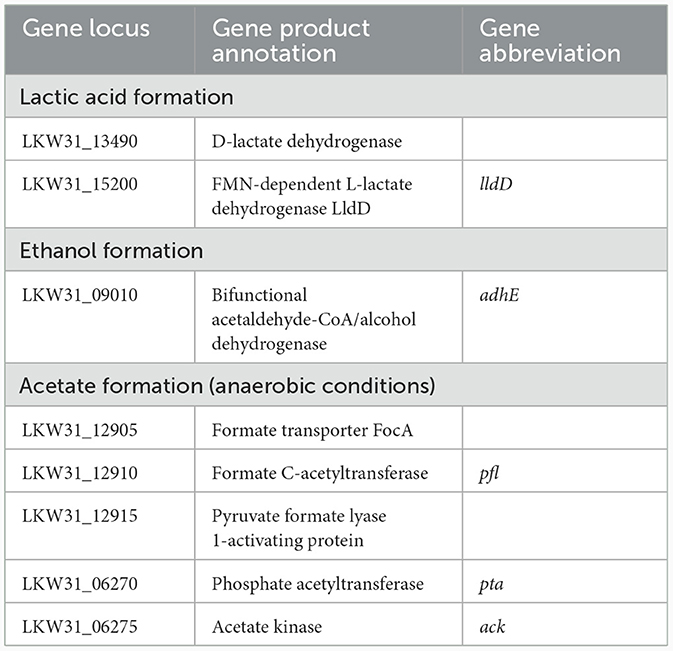
Under all conditions, acids were formed as the main primary metabolites, together with a small amount of ethanol. Acid formation resulted in a pH drop that caused growth to slow down and finally stop. While under anaerobic conditions, the main metabolites were acetic and lactic acids and ethanol, whereas under aerobic conditions, most of the glucose was oxidized to gluconic acid, plus the formation of lactic and acetic acids and ethanol. The concentration of lactic acid and the cell dry weight have rather significantly changed based on cultivation medium than based on aerobic/anaerobic conditions. However, statistically significant change based on aerobic/anaerobic conditions was observed for acetic acid and gluconic acid (with exception for LB aerobic cultivation condition). Ethanol level was similar under all conditions with no statistically significant change. Comparison of acid and ethanol production under different culture conditions is shown in Table 4 while the candidate genes coding for pyruvate processing into lactic and acetic acids and ethanol are shown in Table 5.
Cultivations under different oxygen availability were also compared during bioreactor cultivations using PGM with 20 g/l of glucose (Supplementary Figure 5). Under aerobic conditions in a bioreactor, glucose consumption was double that under anaerobic conditions, but a substantial fraction of glucose was oxidized to gluconic acid. Oxygen limitation was observed during aerobic bioreactor culture, demonstrated as zero oxygen saturation from the 4th to the 14th hour of cultivation. The extracellular concentration of gluconic acid achieved was about 6 g/l in the bioreactor experiment using PGM and up to 8 g/l in shake flask experiments where LB medium supplemented with glucose (10 g/l) was used (see Table 5). A scheme demonstrating the putative gluconic acid metabolic pathways was created, see Figure 3, and respective candidate genes are shown in Table 6. While it seems that gluconic acid production is mostly mediated by membrane bound enzymes and is extracellular, gluconate can be transported into a bacterial cell by a specific gluconate transporter and this transport might be coupled with phosphorylation. The resulting 6-phospho-gluconate may be processed to metabolites entering either the Entner-Doudoroff or Pentose Phosphate pathways, see Figure 3.
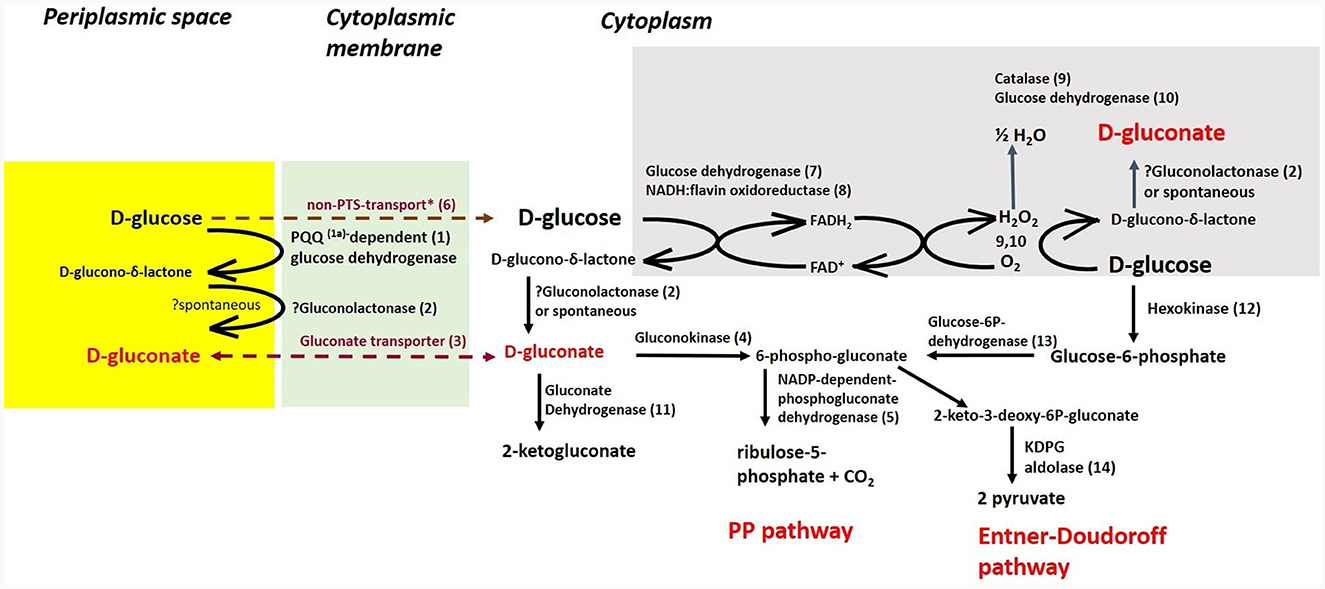
Figure 3. Putative pathways associated with gluconic acid production and its processing in P. agglomerans DBM 3797. The candidate genes for enzyme activities (1–14), as well as biosynthesis of pyrroloquinoline quinone (PQQ) (1a) are shown in Table 6. While the yellow highlighted part, which results in formation of extracellular gluconic acid, was confirmed by HPLC analysis, other proposed pathways require further confirmation. The cytoplasmic membrane is highlighted in green, intracellular gluconic acid production from glucose, atypical for bacteria, is highlighted in gray. PP stands for pentose phosphate. *Stands for supposed glucose transport by passive diffusion.
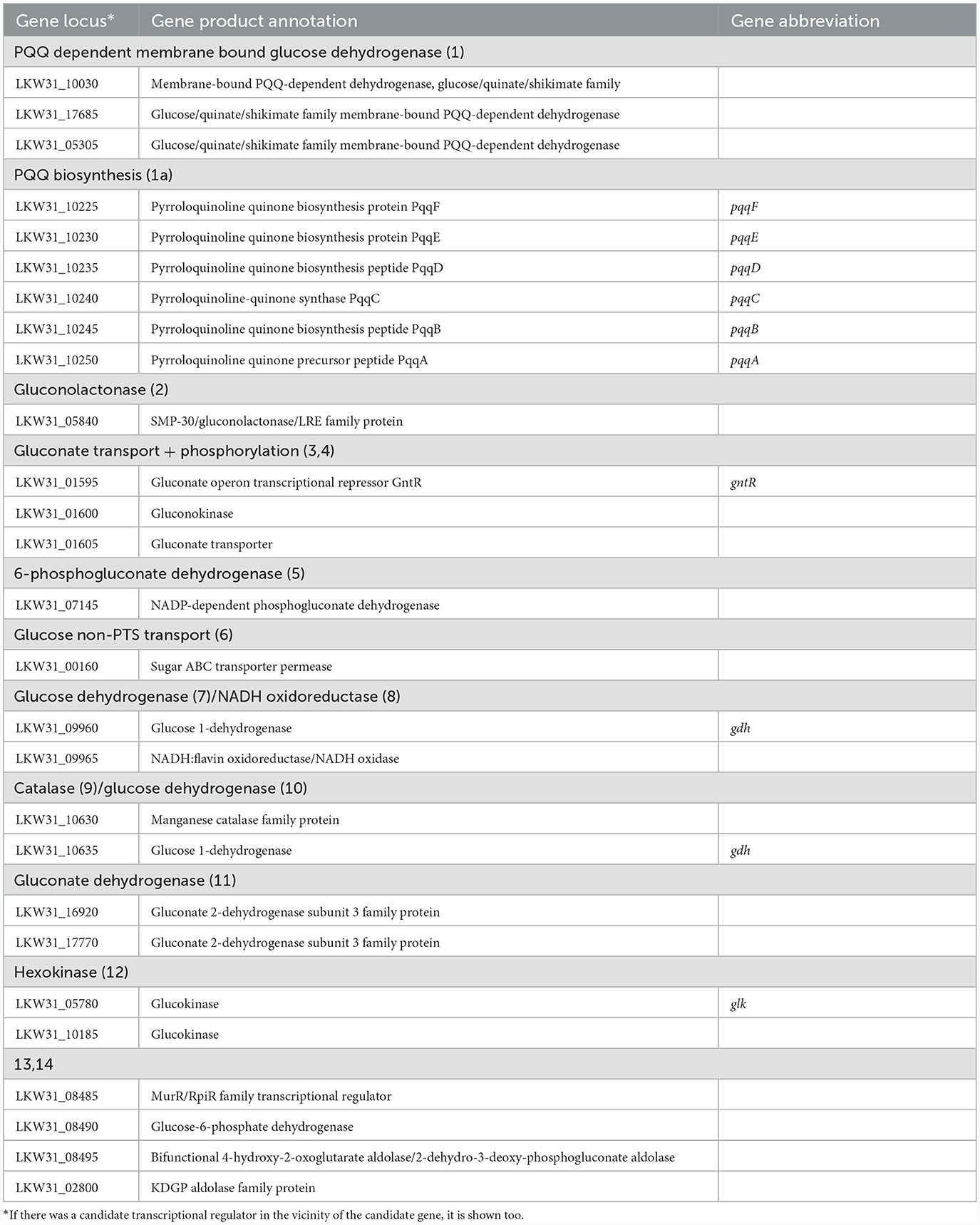
Table 6. Candidate genes for gluconate metabolism found on the chromosome of P. agglomerans DBM 3797.
3.3 Plant growth promotion
Plant growth promoting activities were tested in a series of traditional microbiological assays and the candidate genes for all PGP activities are shown in Supplementary Table 5. There were confirmed high proteolytic/peptidase and cellulase activities, medium siderophore and IAA related compounds productions, weak amylolytic, lipolytic and pectinase activities. The ability to form indole acetic acid (IAA) or IAA-like compounds was tested in culture medium supplemented with the precursor compound, tryptophan and a positive reaction with Salkowski reagent was obtained. As the color was distinct compared to standard (IAA) as well as its retention time in UHPLC analysis (not shown), it is probable that not directly IAA, but a similar compound is formed. The most well-known gene of the IAA pathway, indolepyruvate decarboxylase, ipdC, was found in the genome (Supplementary Table 5). Further, symplasmata (biofilm like structure), carotenoid pigment formation and the ability to release ammonium mentioned above, can be considered PGP activities too. The ability to degrade ethylene was not tested, however the putative gene for 1-aminocyclopropane-1-carboxylate (ACC) deaminase was found on the chromosome. Phosphate solubilisation (PS) was tested in different types of tests but was not confirmed even if the genes for phosphonate metabolism and phosphate transporters were found in the genome (Supplementary Table 5) and gluconic acid formation was demonstrated.
4 Discussion
The P. agglomerans DBM 3797 strain isolated from fresh hop has a somewhat different phenotype from the similar strain P. agglomerans DBM 3796 isolated from dried hop (Kolek et al., 2021) and differed mainly in the low production of CO2 associated with low production of ethanol and acetic acid. The strain has two plasmids, whose circularity was proven during de novo assembly that produced circular contigs. Moreover, both plasmids contained the repB gene coding for plasmid replication initiator, suggesting that both plasmids formed integral parts of the P. agglomerans genome rather than foreign DNA. Additionally, no intact prophage sequences were found on plasmids. The first plasmid pPA_DBM3797_1, of size 555 kb and harboring genes for carotenoid biosynthesis and siderophores (Supplementary Table 5), as well as thiamine biosynthesis (not shown) genes; these seem to meet the criteria for a large universal Pantoea plasmid (De Maayer et al., 2012). Growth under both aerobic and anaerobic conditions correlates with the possibility of synthesizing deoxyribonucleotides for DNA replication during growth by class I (aerobic) and class III (anaerobic) ribonucleotide reductases (Torrents, 2014). The second plasmid, pPA_DBM3797_2 is, according to a functional annotation, responsible for signal transduction and defense mechanisms rather than metabolism and carries one gene responsible for antibiotic resistance from pmr phosphoethanolamine transferase gene family. This gene might be involved in polymyxin resistance (Huang et al., 2018) and there is a potential risk for its spreading by horizontal gene transfer. Nevertheless, the risk assessment requires further study. Other antibiotic resistance genes are of lower risk as they are located on chromosome and in addition, a lot of them are efflux pumps genes which might be attributed to the need to resist the action of antimicrobial substances produced by the host hop plant. The antimicrobial active substances of hops include, for example, beta-acids, effective against methicillin resistant Staphylococcus aureus strains (Sleha et al., 2021). The absence of a native CRISPR-Cas system that can serve as a form of bacterial immunity (Sorek et al., 2013) is compensated for by the presence of numerous R-M systems. At least some of these systems are probably active, as P. agglomerans DBM 3797 contains only a minimum of foreign DNA, particularly only one intact prophage PHAGE_Erwini_ENT90_NC_019932. The presence of such foreign DNA is not unique for P. agglomerans as the very same prophage was previously identified in the genome of the strain P. agglomerans C1 (Luziatelli et al., 2019).
The most closely related strains, AB378 and CFBP8784, were, like strain DBM 3797, isolated from the phyllosphere of plants, specifically from Red Topaz apple blossom (AB378) and from radish Raphanus sativus flower (CFBP8784). Together with other neighboring strains (Figure 2), i.e., Pan8 (isolated from Pisum sativum phyllosphere), P10c (isolated from apple tree), and DOAB1048 (isolated from wheat leaves) they form a group of P. agglomerans environmental strains isolated from above-ground plant parts and differ from strains P5, ANP8, CPHN2, and DAPP-PG744 (Table 1) isolated from plant roots or from soil.
The ability to form a biofilm is considered to be an advantage for the bacteria colonizing the plants, as the biofilm protects both the bacterial population from adverse environmental influences and the colonized plant surface. In addition, the ability to communicate between the microbial community and the plant cells is enhanced by signal amplification during biofilm formation (Seneviratne et al., 2010). Symplasmata i.e., multicellular round aggregates mimicking colonies in a liquid medium, probably gave the original name to the species “agglomerans” (Tecon and Leveau, 2016) and were described in detail e.g., in the rice epiphyte P. agglomerans YS19 (Yu et al., 2016; Zheng et al., 2019). Although these formations were firstly described by M. W. Beijerinck in 1888 (Tecon and Leveau, 2016), it is still not completely clear why they are formed. They provide protection to the bacteria, comparable to biofilm formation, and are considered to be an advantage for colonization of rice roots (Achouak et al., 1994). Symplasmata formation seems to be initiated by culture conditions but different reports differ in descriptions of which factor is decisive. While Pantoea eucalypti best formed symplasmata in glucose medium not in LB medium, and at a pH higher than 7.6 (Tecon and Leveau, 2016), our strain P. agglomerans DBM 3797 formed them only in LB medium at pH 8.5. Our findings are in accordance with those for strain YS19 (Jiang et al., 2015; Yu et al., 2016; Zheng et al., 2019) and nitrogen-fixing E. agglomerans NO30 (Achouak et al., 1994). Symplasmata of strain YS19 were probably regulated by indole (Yu et al., 2016) and it was hypothesized that indole originated from tryptophan degradation and was considered by the P. agglomerans indolenon-producing strain as a marker of starvation (Jia et al., 2017). In addition, in the same strain YS19, an acyl-homoserine lactone quorum sensing system was also involved in symplasmata formation (Jiang et al., 2015). While the genes for acyl-homoserine lactone were confirmed in our strain by antiSMASH analysis, indole release was not confirmed. Symplasmata formation was also found in an anaerogenic group of Enterobacter clinical isolates (Gilardi and Bottone, 1971), which corresponds to the finding that genes for anaerobic metabolism were active during symplasmata formation in P. eucalypti aerobic culture (Tecon and Leveau, 2016) and it was deduced that there is an oxygen limitation inside these formations. The ability to cope with anaerobic conditions was proven in our strain.
The ability to utilize peptides in LB medium corresponds with the number of peptidase genes present in the genome (not shown). This type of fermentation is typical for certain fermented foods such as natto (fermented soybeans) or pidan (fermented eggs) rich in proteins and peptides (Wang and Fung, 1996) and was also described in detail for E. coli (Sezonov et al., 2007). Aerobic growth in LB medium is a special type of alkaline fermentation where amino acids are used as carbon sources instead of saccharides. Generally, it is supposed that amino acids are processed by oxidative deamination, generating α-keto acids and ammonia. It is believed that the key role in the process is played by glutamate dehydrogenase, but its respective candidate gene was not found in the genome. Thus, it seems more likely that metabolism of each amino acid is unique and while the ammonium cation is released or transferred to other compounds, the rest of the molecule can be transformed to pyruvate, acetyl CoA, acetoacetyl CoA and intermediates of the citric acid cycle under aerobic conditions, as described by Li et al. (2018). Part of the ammonium ion is used by the bacterium but its release is excessive which results in an increase in pH. Actually, the release of ammonium ions from available amino acids and peptides can be one of the advantageous features of the DBM 3797 strain, which might be used for controlled ammonia release if the strain was applied together with organic fertilizers.
In many aerobic Gram-negative bacteria, gluconic acid is formed by glucose dehydrogenase through D-glucono-δ-lactone in the periplasmic space (Ma et al., 2022). Further, it is expected that transport of gluconate from the periplasmic space through the outer membrane is mediated by porins. Based on knowledge gathered for Gluconobacter oxydans (Pronk et al., 1989), extracellular gluconic acid production is probably a result of the membrane bound PQQ-dependent glucose dehydrogenase, which was described in detail for Pantoea ananatis (Andreeva et al., 2011). Candidate genes for this enzyme activity, as well as the complete pqqABCDEF biosynthetic operon, were found in the genome. In Gram-negative aerobic bacteria, there are frequently found other membrane bound enzymes, such as PQQ-dependent 5-keto-gluconate dehydrogenase, and flavin/heme dependent gluconate and 2-keto-gluconate dehydrogenases (Ma et al., 2022), but the candidate genes were not found in the genome. Microbial gluconic acid production was reviewed by Ramachandran et al. (2006) and Ma et al. (2022) and it is obvious that fungi and bacteria differ in metabolic pathways for gluconic acid production. In fungi, such as in Aspergillus niger, its main industrial producer, gluconic acid is produced by FAD+-dependent glucose oxidase, which is coupled with catalase (Ramachandran et al., 2006) and surprisingly, similar candidate genes coding for this option, which are atypical for bacteria, were also found in our genome (see Figure 3; Table 5). Further, it appears that the catabolic pathway of glucose in the studied strain may use parts of known metabolic pathways such as the Entner-Doudoroff, Embden-Meyer-Parnas or Pentose Phosphate pathways, which are interconnected, similar to what has been found and described for Pseudomonas putida KT2440 (Nikel et al., 2015).
Gluconic acid production may be associated with defense against soil protozoa such as Vahlkampfia sp. or Neobodo designis (Gómez et al., 2010), however mostly it is exploited together with other organic acids in the solubilization of inorganic phosphate. Phosphate solubilization is a significant PGP feature that facilitates plant growth by increasing its accessibility from both inorganic and organic phosphate-containing compounds and complexes (Rawat et al., 2021). Typical phosphate solubilizing microorganisms, in addition to the formation of organic acids, may form siderophores, exopolysaccharides, phosphatases, phosphonatases and others (Liang et al., 2020; Rawat et al., 2021), and the genes for these functions were found in our strain too but actual PS activity was not confirmed in any type of applied test. Siderophore as well as metalophore formation gene clusters were also identified by antiSMASH. This finding is consistent with recently published information (Elhaissoufi et al., 2023) that even bacteria not showing PS capability in a given test can in fact contribute significantly to plant phosphorus supply.
The thiopeptide formation ability revealed by the antiSMASH analysis (Supplementary Figure 1) may indicate microcin(s), bioactive peptide(s) production [namely class IIa microcin having disulphide bond(s) in their structure (Parker and Davies, 2022)]. Microcin production was confirmed in P. agglomerans Eh252 (Vanneste et al., 2002) and the P. agglomerans E325 producing microcins was even applied for biocontrol of fire blight disease of apple, caused by Erwinia amylovora (Kim et al., 2012).
Aerobic metabolism of the strain is also associated with the production of potential auxin compounds, IAA-like substances, in the culture medium supplemented with tryptophan. In P. agglomerans, IAA biosynthesis begins with the formation of indole-3-pyruvic acid, mediated by aminotransferase, continues with indole-3-acetaldehyde formation by indolepyruvate decarboxylase, coded by ipdC, and ends with IAA formation catalyzed by indole-3-acetaldehyde dehydrogenase (Luziatelli et al., 2020b). In our strain, the ipdC gene was found, several candidate aminotransferase genes (not shown), but not the gene for indole-3-acetaldehyde dehydrogenase. Since the traditional method for detection of IAA using the Salkowski reagent resulted in the formation of an orange color with an absorption maximum of 450 nm rather than a pink color with a maximum of 530 nm, indole-3-butyric acid (IBA) was tested as a possible previously described (Gilbert et al., 2018) product of this reaction. Unfortunately, IBA was not confirmed as the reaction product. Gilbert et al. (2018) demonstrated that different bacterial isolates produced different compounds with potential auxin activity from tryptophan and we concluded that our strain probably belongs to this IAA-like compound producers' group.
P. agglomerans, strain DBM 3797, isolated from hops has a number of properties potentially beneficial to the hop plant, but its safety profile needs to be addressed in follow-up research. In particular, the possibility of horizontal transfer of antibiotic resistance genes, which has been little studied in the genus Pantoea, and virulence genes that may lead to pathogenicity in plants or animals, and humans in some strains of the species (Guevarra et al., 2021), need to be focused on. Unfortunately, there are not enough complete genome assemblies yet for a detailed comparison of particular strains. Although there are some specific inserts in the genome of P. agglomerans DBM 3797 in comparison to additional five strains (Supplementary Figure 2), no specific feature distinguishing pathogens from harmless strains isolated from above-ground parts of plants.
Data availability statement
The whole-genome sequence and plasmid sequences were deposited in the DDBJ/ENA/GenBank under accession numbers CP086133.1, CP086134.1, and CP086135.1, respectively. The NCBI BioProject and BioSample IDs are PRJNA774971 and SAMN22600026. The raw reads were deposited in the NCBI SRA database under accession numbers SRR25382413 (paired-end Illumina) and SRR25382412 (Oxford Nanopore Technologies).
Author contributions
PP: Conceptualization, Funding acquisition, Writing – original draft, Writing – review & editing. MV: Investigation, Writing – review & editing. KS: Conceptualization, Data curation, Funding acquisition, Writing – original draft, Writing – review & editing. KJ: Investigation, Visualization, Writing – review & editing. MB: Formal analysis, Investigation, Writing – review & editing. PL: Investigation, Writing – review & editing. BB: Investigation, Writing – review & editing. PK: Funding acquisition, Writing – review & editing. KK: Conceptualization, Investigation, Writing – review & editing.
Funding
The author(s) declare financial support was received for the research, authorship, and/or publication of this article. The research was supported by the TACR NPO project no. TN02000044/01. This work is a part of the project that has received funding from the European Union's Horizon 2020 research and innovation programme under the Marie Sklodowska-Curie grant agreement no. 101023766.
Conflict of interest
PK was employed by EcoFuel Laboratories s.r.o. KK was employed by Hop Research Institute, Co. Ltd.
The remaining authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher's note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2024.1305338/full#supplementary-material
Abbreviations
DBM, Department of Biochemistry and Microbiology of the University of Chemistry and Technology in Prague; EMP, Embden-Meyer-Parnas; IAA, indole-3-acetic acid; IBA, indole-3-butyric acid; LB, Lysogeny Broth; OD, optical density; PGM, Pantoea glucose medium; PGP, plant growth promoting; PP, pentose phosphate; PQQ, pyrroloquinoline quinone; PS, phosphate solubilization; PTS, phosphotransferase system; rNDP, ribonucleoside diphosphate reductase; RNR, ribonucleotide reductase.
Footnotes
1. ^NIH, National Center for Biotechnology Information (2024). Available online at: https://www.ncbi.nlm.nih.gov/datasets/genome/?taxon=549&annotated_only=true&refseq_annotation=true&genbank_annotation=true&assembly_level=3%3A3.
References
Achouak, W., Heulin, T., Villemin, G. J., and Balandreau, J. (1994). Root colonization by symplasmata-forming Enterobacter agglomerans. FEMS Microbiol. Ecol. 13, 287–294. doi: 10.1111/j.1574-6941.1994.tb00075.x
Alcock, B. P., Raphenya, A. R., Lau, T. T. Y., Tsang, K. K., Bouchard, M., Edalatmand, A., et al. (2020). CARD 2020: antibiotic resistome surveillance with the comprehensive antibiotic resistance database. Nucleic Acids Res. 48, D517–D525. doi: 10.1093/nar/gkz935
Alikhan, N. F., Petty, N. K., Ben Zakour, N. L., and Beatson, S. A. (2011). BLAST Ring Image Generator (BRIG): simple prokaryote genome comparisons. BMC Genom. 12:402. doi: 10.1186/1471-2164-12-402
Allen, M. E., Piefer, A. J., Cole, S. N., Werner, J. J., Benziger, P. T., Grieneisen, L., et al. (2019). Characterization of microbial communities populating the inflorescences of Humulus lupulus L. J. Am. Soc. Brew. Chem. 77, 243–250. doi: 10.1080/03610470.2019.1667739
Altschul, S. F., Gish, W., Miller, W., Myers, E. W., and Lipman, D. J. (1990). Basic local alignment search tool. J. Mol. Biol. 215, 403–410. doi: 10.1016/S0022-2836(05)80360-2
Andreeva, I. G., Golubeva, L. I., Kuvaeva, T. M., Gak, E. R., Katashkina, J. I., and Mashko, S. V. (2011). Identification of Pantoea ananatis gene encoding membrane pyrroloquinoline quinone (PQQ)-dependent glucose dehydrogenase and pqqABCDEF operon essential for PQQ biosynthesis. FEMS Microbiol. Lett. 318, 55–60. doi: 10.1111/j.1574-6968.2011.02240.x
Arndt, D., Grant, J., Marcu, A., Sajed, T., Pon, A., Liang, Y., et al. (2016). PHASTER: a better, faster version of the PHAST phage search tool. Nucleic Acids Res. 44, W16–W21. doi: 10.1093/nar/gkw387
Biswas, A., Staals, R. H. J., Morales, S. E., Fineran, P. C., and Brown, C. M. (2016). CRISPRDetect: a flexible algorithm to define CRISPR arrays' BMC Genom. 17:356. doi: 10.1186/s12864-016-2627-0
Blin, K., Shaw, S., Augustijn, H. E., Reitz, Z. L., Biermann, F., Alanjary, M., et al. (2023). antiSMASH 7.0: new and improved predictions for detection, regulation, chemical structures and visualisation. Nucl. Acids Res. 51. W46–W50. doi: 10.1093/nar/gkad344
Bocquet, L., Sahpaz, S., and Rivière, C. (2018). “An overview of the antimicrobial properties of hop,” in Natural Antimicrobial Agents. Sustainable Development and Biodiversity, Vol 19, eds J. M. Mérillon, and C. Riviere (Cham: Springer).
Bolger, A. M., Lohse, M., and Usadel, B. (2014). Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics 30, 2114–2120. doi: 10.1093/bioinformatics/btu170
Cantalapiedra, C. P., Hernández-Plaza, A., Letunic, I., Bork, P., and Huerta-Cepas, J. (2021). EggNOG-Mapper v2: functional annotation, orthology assignments, and domain prediction at the metagenomic scale. Mol. Biol. Evol. 38, 5825–5829. doi: 10.1093/molbev/msab293
Carver, T., Harris, S. H., Berriman, M., Parkhill, J., and McQuillan, J. A. (2012). Artemis: an integrated platform for visualization and analysis of high-throughput sequence-based experimental data. Bioinformatics 28, 464–469. doi: 10.1093/bioinformatics/btr703
Carver, T., Thomson, N., Bleasby, A., Berriman, M., and Parkhill, J. (2009). DNAPlotter: circular and linear interactive genome visualization. Bioinformatics 25, 119–120. doi: 10.1093/bioinformatics/btn578
Chaudhari, N. M., Gupta, V. K., and Dutta, C. (2016). BPGA-an ultra-fast pan-genome analysis pipeline. Sci. Rep. 6:24373. doi: 10.1038/srep24373
De Maayer, P., Chan, W. Y., Blom, J., Venter, S. N., Duffy, B., Smits, T. H. M., et al. (2012). The large universal Pantoea plasmid LPP-1 plays a major role in biological and ecological diversification. BMC Genom. 13:625. doi: 10.1186/1471-2164-13-625
Dutkiewicz, J., Mackiewicz, B., Lemieszek, M. K., Golec, M., and Milanowski, J. (2016). Pantoea agglomerans: a mysterious bacterium of evil and good. Part IV. Beneficial effects. Ann. Agric. Environ. Med. 23, 206–222. doi: 10.5604/12321966.1203879
Edgar, R. C. (2010). Search and clustering orders of magnitude faster than BLAST. Bioinformatics 26, 2460–2461. doi: 10.1093/bioinformatics/btq461
Elhaissoufi, W., Ibnyasser, A., Haddine, M., Zeroual, Y., Ghani, R., Barakat, A., et al. (2023). Screening of potential phosphate solubilizing bacteria inoculants should consider the contrast in phosphorus bio-solubilization rate along with plant growth promotion and phosphorus use efficiency. J. Appl. Microbiol. 134:lxac077. doi: 10.1093/jambio/lxac077
Ewels, P., Magnusson., M, Lundin., S, and Käller, M. (2016). MultiQC: summarize analysis results for multiple tools and samples in a single report. Bioinformatics 32, 3047–3048. doi: 10.1093/bioinformatics/btw354
Fahle, A., Bereswill, S., and Heimesaat, M. M. (2022). Antibacterial effects of biologically active ingredients in hop provide promising options to fight infections by pathogens including multi-drug resistant bacteria. Eur. J. Microbiol. Immunol. 12, 22–30. doi: 10.1556/1886.2022.00006
Gilardi, G. L., and Bottone, E. (1971). Erwinia and yellow-pigmented Enterobacter isolates from human sources. Antonie Van Leeuwenh. 37, 529–535. doi: 10.1007/BF02218523
Gilbert, S., Xu, J., Acosta, K., Poulev, A., Lebeis, S., and Lam, E. (2018). Bacterial production of indole related compounds reveals their role in association between duckweeds and endophytes. Front. Chem. 6:265. doi: 10.3389/fchem.2018.00265
Gómez, W., Buela, L., Castro, L. T., Chaparro, V., Ball, M. M., and Yarzábal, L. A. (2010). Evidence for gluconic acid production by Enterobacter intermedium as an efficient strategy to avoid protozoan grazing. Soil Biol. Biochem. 42, 822–830. doi: 10.1016/j.soilbio.2010.01.019
Goryluk-Salmonovicz, A., Piórek, M., Rekosz-Burlaga, H., Studnicki, M., and Blasczyk, M. (2016). Endophytic detection in selected European herbal plants. Polish J. Microbiol. 65, 369–375. doi: 10.5604/17331331.1215617
Guevarra, R. B., Magez, S., Peeters, E., Chung, M. S., Kim, K. H., and Radwanska, M. (2021). Comprehensive genomic analysis reveals virulence factors and antibiotic resistance genes in Pantoea agglomerans KM1, a potential opportunistic pathogen. PLoS ONE 16:e0239792. doi: 10.1371/journal.pone.0239792
Hawar, S. N. (2022). Extracellular enzyme of endophytic fungi isolated from Ziziphus spina leaves as medicinal plant. Int. J. Biomater. 2022:2135927. doi: 10.1155/2022/2135927
Huang, J., Zhu, Y., Han, M. L., Li, M., Song, J., Velkov, T., et al. (2018). Comparative analysis of phosphoethanolamine transferases involved in polymyxin resistance across 10 clinically relevant Gram-negative bacteria. Int. J. Antimicrob. Agents 51, 586–593. doi: 10.1016/j.ijantimicag.2017.12.016
Jaskula, B., Kafarski, P., Aerts, G., and De Cooman, L. (2008). A kinetic study on the isomerization of hop alpha acids. J. Agric. Food Chem. 56, 6408–6415. doi: 10.1021/jf8004965
Jia, M., Yu, X., Jiang, J., Li, Z., and Feng, Y. (2017). The cytidine repressor participates in the regulatory pathway of indole in Pantoea agglomerans. Res. Microbiol. 7, 636–643. doi: 10.1016/j.resmic.2017.04.006
Jiang, J., Wu, S., Wang, J., and Feng, Y. (2015). AHL-type quorum sensing and its regulation on symplasmata formation in Pantoea agglomerans YS19. J. Basic Microbiol. 55, 607–616. doi: 10.1002/jobm.201400472
Kim, I. J., Pusey, P. L., Zhao, Y., Korban, S. S., Choi, H., and Kim, K. K. (2012). Controlled release of Pantoea agglomerans E325 for biocontrol of fire blight disease of apple. J. Controll. Release 161, 109–115. doi: 10.1016/j.jconrel.2012.03.028
Koçak, O. F. (2019). Identification of Streptomyces strains isolated from Humulus lupulus rhizosphere and determination of plant growth promotion potential of selected strains. Turk. J. Biol. 43, 391–403. doi: 10.3906/biy-1906-37
Kolek, J., Patakova, P., Junkova, P., Krofta, K., Hynek, R., and Dostalek, P. (2021). Isolation and identification of Pantoea agglomerans from the inflated bag with dried hop pellets stored under a modified atmosphere. J. Appl. Microbiol. 131, 281–287. doi: 10.1111/jam.14970
Kumar, P., Rani, S., Dahiya, P., Kumar, A., Dang, A. S., and Pooja, S. (2022). Whole genome analysis for plant growth promotion profiling of Pantoea agglomerans CPHN2, a non-rhizobial nodule endophyte. Front. Microbiol. 13:998821. doi: 10.3389/fmicb.2022.998821
Lanfear, R., Schalamun, M., Kainer, D., Wang, W., and Schwessinger, B. (2019). MinIONQC: fast and simple quality control for MinION sequencing data. Bioinformatics 35, 523–525. doi: 10.1093/bioinformatics/bty654
Li, H. (2018). Minimap2: pairwise alignment for nucleotide sequences. Bioinformatics 34, 3094–3100. doi: 10.1093/bioinformatics/bty191
Li, H., and Durbin, R. (2009). Fast and accurate short read alignment with Burrows–Wheeler transform. Bioinformatics 25, 1754–1760. doi: 10.1093/bioinformatics/btp324
Li, H., Handsaker, B., Wysoker, A., Fennel,l, T., Ruan, J., Homer, N., et al. (2009). The sequence alignment/map format and SAMtools. Bioinformatics 25, 2078–2079. doi: 10.1093/bioinformatics/btp352
Li, S. Y., Ng, I. S., Chen, P. T., Chiang, C. J., and Chao, Y. P. (2018). Biorefining of protein waste for production of sustainable fuels and chemicals. Biotechnol. Biofuels 11:256. doi: 10.1186/s13068-018-1234-5
Liang, J. L., Liu, J., Jia, P., Yang, T. T., Zeng, Q. W., et al. (2020). Novel phosphate-solubilizing bacteria enhance soil phosphorus cycling following ecological restoration of land degraded by mining. ISME J. 14, 1600–1613. doi: 10.1038/s41396-020-0632-4
Liu, B., Zheng, D., Jin, Q., Chen, L., and Yang, J. (2019). VFDB 2019: a comparative pathogenomic platform with an interactive web interface. Nucl. Acids Res. 47, D687–D692. doi: 10.1093/nar/gky1080
Luo, H., Quan, C.-L., Peng, C., and Gao, F. (2019). Recent development of Ori-Finder system and DoriC database for microbial replication origins. Brief. Bioinform. 20, 1114–1124. doi: 10.1093/bib/bbx174
Luziatelli, F., Ficca, A. G., Bonini, P., Muleo, R., Gatti, L., Meneghin.i, M., Tronati, M., Melini, F., and Ruzzi, M (2020b). Genetic and metabolomic perspective on the production of indole-3-acetic acid by Pantoea agglomerans and use of their metabolites as biostimulants in plant nurseries. Front. Microbiol. 11:1475. doi: 10.3389/fmicb.2020.01475
Luziatelli, F., Ficca, A. G., Cardarelli, M., Melini, F., Cavalieri, A., and Ruzzi, M. (2020a). Genome sequencing of Pantoea agglomerans C1 provides insights into molecular and genetic mechanisms of plant growth-promotion and tolerance to heavy metals. Microorganisms 8:153. doi: 10.3390/microorganisms8020153
Luziatelli, F., Ficca, A. G., Melini, F., and Ruzzi, M. (2019). Genome sequence of the plant growth-promoting rhizobacterium Pantoea agglomerans C1. Microbiol. Resour. Announ. 8, e00828–e00819. doi: 10.1128/MRA.00828-19
Ma, Y., Li, B., Zhang, X., Wang, C., and Chen, W. (2022). Production of gluconic acid and its derivatives by microbial fermentation: Process improvement based on integrated routes. Front. Bioeng. Biotechnol. 10:864787. doi: 10.3389/fbioe.2022.864787
Marchler-Bauer, A., and Bryant, S. H. (2004). CD-search: protein domain annotations on the fly. Nucl. Acids Res. 32, W327–W331. doi: 10.1093/nar/gkh454
Micci, A., Zhang, Q., Chang, X., Kingsley, K., Park, L., Chiaranunt, P., et al. (2022). Histochemical evidence for nitrogen-transfer endosymbiosis in non-photosynthetic cells of leaves and inflorescence bracts of angiosperms. Biology 11:876. doi: 10.3390/biology11060876
Milligan, S. R., Kalita, J. C., Pocock, V., Van de Kauter, V., Stevens, J. F., Deinzer, M. L., et al. (2000). The endocrine activities of 8-prenylnaringenin and related hop (Humulus lupulus L.) flavonoids. J. Clin. Endocrinol. Metab. 85, 4912–4915. doi: 10.1210/jcem.85.12.7168
Miranda, C. L., Stevens, J. F., Helmrich, A., Henderson, M. C., Rodriguez, R. J., Yang, Y. H., et al. (1999). Antiproliferative and cytotoxic effects of prenylated flavonoids from hops (Humulus lupulus) in human cancer cell lines. Food Chem. Toxicol. 37, 271–285. doi: 10.1016/S.0278-6915(99)00019-8
Nikel, P. I., Chavarría, M., Fuhrer, T., Sauer, U., and de Lorenzo, V. (2015). Pseudomonas putida KT2440 strain metabolizes glucose through a cycle formed by enzymes of the Entner-Doudoroff, Embden-Meyerhof-Parnas, and pentose phosphate pathways. J. Biol. Chem. 290, 25920–25932. doi: 10.1074/jbc.M115.687749
Noori, F., Etesami, H., Noori, S., Forouzan, E., Jouzani, G. S., and Malboobi, M. A. (2021). Whole genome sequence of Pantoea agglomerans ANP8, a salinity and drought stress–resistant bacterium isolated from alfalfa (Medicago sativa L.) root nodules. Biotechnol. Rep. 29:e00600. doi: 10.1016/j.btre.2021.e00600
O'Leary, N. A., Wright, M. W., Brister, J. R., Ciufo, S., Haddad, D., McVeigh, R., et al. (2016). Reference sequence (RefSeq) database at NCBI: current status, taxonomic expansion, and functional annotation. Nucl. Acids Res. 44, D733–745. doi: 10.1093/nar/gkv1189
Parker, J. K., and Davies, B. W. (2022). Microcins reveal natural mechanisms of bacterial manipulation to inform therapeutic development. Microbiology 168:001175. doi: 10.1099/mic.0.001175
Pronk, J. T., Levering, P. R., Olijve, W., and van Dijken, J. P. (1989). Role of NADP-dependent and quinoprotein glucose dehydrogenases in gluconic acid production by Gluconobacter oxydans. Enzyme Microb. Technol. 11, 160–164. doi: 10.1016/0141-0229(89)90075-6
Ramachandran, S., Fontanille, P., Pandey, A., and Larroche, C. (2006). Gluconic acid: properties, applications and microbial production. Food Technol. Biotechnol. 44, 185–195. Available online at: https://hrcak.srce.hr/file/161891
Rawat, P., Das, S., Shankhdhar, D., and Shankhdar, S. C. (2021). Phosphate-solubilizing microorganisms: mechanism and their role in phosphate solubilization and uptake. J. Soil Sci. Plant Nutr. 21, 49–68. doi: 10.1007/s42729-020-00342-7
Rezzonico, F., Smits, T. H., Montesinos, E., Frey, J. E., and Duffy, B. (2009). Genotypic comparison of Pantoea agglomerans plant and clinical strains. BMC Microbiol. 9:204. doi: 10.1186/1471-2180-9-204
Roberts, R. J., Vincze, T., Posfai, J., and Macelis, D. (2023). REBASE: a database for DNA restriction and modification: enzymes, genes and genomes. Nucl. Acids Res. 51, D629–D630. doi: 10.1093/nar/gkac975
Schmidt, C. S., Lovecka, P., Mrnka, L., Vychodilova, A., Strejcek, M., Fenclova, M., et al. (2018). Distinct communities of poplar endophytes on an unpolluted and a risk element-polluted site and their plant growth-promoting potential in vitro. Microb. Ecol. 75, 955–969. doi: 10.1007/s00248-017-1103-y
Sedlar, K., Vasylkivska, M., Musilova, J., Branska, B., Provaznik, I., and Patakova, P. (2021). Phenotypic and genomic analysis of isopropanol and 1,3-propanediol producer Clostridium diolis DSM 15410. Genomics 113, 1109–1119. doi: 10.1016/j.ygeno.2020.11.007
Seneviratne, G., Weerasekara, M. L. M. A.W., Seneviratne, C., Zavahir, J. S., Kcskés, M. L., and Kennedy, I. R. (2010). “Importance of biofilm formation in plant growth promoting rhizobacterial action,” in Plant Growth and Health Promoting Bacteria, ed D. K. Maheshawari (Berlin: Springer-Verlag), 81–95.
Sevigny, J. L., Lloyd, B., McComish, C., Ramsey, A., and Koziol, L. (2019). Whole-genome sequences of Pantoea agglomerans BL3, Pseudomonas fluorescens BL, and Pseudomonas stutzeri CM14, isolated from Hops (Humulus lupulus). Microbiol. Resour. Announ. 8, e00545–e00519. doi: 10.1128/MRA.00545-19
Sezonov, G., Joseleau-Petit, D., and D'Ari, R. (2007). Escherichia coli physiology in Luria-Bertani broth. J. Bacteriol. 189, 8746–8749. doi: 10.1128/JB.01368-07
Shariati, J. V, Malboobi, M. A., Tabrizi, Z., Tevakol, E., Owlia, P., et al. (2017). Comprehensive genomic analysis of a plant growth-promoting rhizobacterium Pantoea agglomerans strain P5. Sci. Rep. 7:15610. doi: 10.1038/s41598-017-15820-9
Sleha, R., Radochova, V., Malis, J., Mikyska, A., Houska, M., Krofta, K., et al. (2021). Strong antimicrobial and healing effects of Beta-Acids from Hops in methicillin-resistant Staphylococcus aureus-infected external wounds in vivo. Antibiotics 10:708. doi: 10.3390/antibiotics10060708
Sorek, R., Lawrence, C. M., and Wiedenheft, B. (2013). CRISPR-mediated adaptive immune systems in bacteria and archaea. Annu. Rev. Biochem. 82, 237–266. doi: 10.1146/annurev-biochem-072911-172315
Soutar, C. D., and Stavrinides, J. (2019). Molecular validation of clinical Pantoea isolates identified by MALDI-TOF. PLoS ONE 14:e0224731. doi: 10.1371/journal.pone.0224731
Stranska, M., Lovecka, P., Vrchotova, B., Uttl, L., Bechynska, K., Behner, A., et al. (2021). Bacterial endophytes from Vitis vinifera L – Metabolomics characterization of plant-endophyte crosstalk. Chem. Biodivers. 18:e2100516. doi: 10.1002/cbdv.202100516
Sulja, A., Pothier, J. F., Blom, J., Moretti, C., Buonario, R., Rezzonico, F., et al. (2022). Comparative genomics to examine the endophytic potential of Pantoea agglomerans DAPP-PG 734. BMC Genom. 23:742. doi: 10.1186/s12864-022-08966-y
Taboada, B., Estrada, K., Ciria, R., and Merino, E. (2018). Operon-mapper: a web server for precise operon identification in bacterial and archaeal genomes. Bioinformatics 34, 118–120. doi: 10.1093/bioinformatics/bty496
Tatusova, T., DiCuccio, M., Badretdin, A., Chetvernin, V., Nawrocki, E. P., Zaslavsky, L., et al. (2016). NCBI prokaryotic genome annotation pipeline. Nucl. Acids Res. 44, 6614–6624. doi: 10.1093/nar/gkw569
Tecon, R., and Leveau, J. (2016). Symplasmata are a clonal, conditional, and reversible type of bacterial multicellularity. Sci. Rep. 6:31914. doi: 10.1038/srep31914
Torrents, E. (2014). Ribonucleotide reductases: essential enzymes for bacterial life. Front. Cell. Infect. Microbiol. 4:52. doi: 10.3389/fcimb.2014.00052
Vanneste, J. L., Cornish, D. A., Yu, J., and Voyle, M. D. (2002). The peptide antibiotic produced by Pantoea agglomerans EH252 is a microcin. Acta Hortic. 590, 285–290. doi: 10.17660/ActaHortic.2002.590.42
Vaser, R., Sović, I., Nagarajan, N., and Šikić, M (2017). Fast and accurate de novo genome assembly from long uncorrected reads. Genome Res. 27, 737–746. doi: 10.1101/gr.214270.116
Walker, B. J., Abeel, T., Shea, T., Priest, M., Abouellie,l, A., Sakthikumar, S., et al. (2014). Pilon: an integrated tool for comprehensive microbial variant detection and genome assembly improvement. PLoS ONE 9:e112963. doi: 10.1371/journal.pone.0112963
Walterson, A. M., and Stavrinides, J. (2015). Pantoea: insights into a highly versatile and diverse genus within the Enterobacteriaceae. FEMS Microbiol. Rev. 39, 968–984. doi: 10.1093/femsre/fuv027
Wang, J., and Fung, D. Y. C. (1996). Alkaline-fermented foods: a review with emphasis on pidan fermentation. Crit. Rev. Microbiol. 2, 101–138. doi: 10.3109/10408419609106457
Yu, X., Jiang, J., Liang, C., Zhang, X., Wang, J., Shen, D., et al. (2016). Indole affects the formation of multicellular aggregate structures in Pantoea agglomerans YS19. J. Gen. Appl. Microbiol. 62, 31–37. doi: 10.2323/jgam.62.31
Zhang, S., Soltis, D. E., Yang, Y., Li, D., and Yi, T. (2011). Multi-gene analysis provides a well-supported phylogeny of Rosales. Mol. Phylogenet. Evol. 60, 21–28. doi: 10.1016/j.ympev.2011.04.008
Keywords: Pantoea agglomerans, hops endophyte, plant growth promotion, genome characterization, gluconic acid
Citation: Patakova P, Vasylkivska M, Sedlar K, Jureckova K, Bezdicek M, Lovecka P, Branska B, Kastanek P and Krofta K (2024) Whole genome sequencing and characterization of Pantoea agglomerans DBM 3797, endophyte, isolated from fresh hop (Humulus lupulus L.). Front. Microbiol. 15:1305338. doi: 10.3389/fmicb.2024.1305338
Received: 01 October 2023; Accepted: 29 January 2024;
Published: 08 February 2024.
Edited by:
Pragya Tiwari, Yeungnam University, Republic of KoreaReviewed by:
Dibya Jyoti Hazarika, AAU-Zonal Research Station, IndiaSudipta Sankar Bora, Assam Agricultural University, India
Copyright © 2024 Patakova, Vasylkivska, Sedlar, Jureckova, Bezdicek, Lovecka, Branska, Kastanek and Krofta. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Petra Patakova, cGV0cmEucGF0YWtvdmFAdnNjaHQuY3o=
 Petra Patakova
Petra Patakova Maryna Vasylkivska
Maryna Vasylkivska Karel Sedlar
Karel Sedlar Katerina Jureckova
Katerina Jureckova Matej Bezdicek
Matej Bezdicek Petra Lovecka
Petra Lovecka Barbora Branska
Barbora Branska Petr Kastanek7
Petr Kastanek7