
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Microbiol. , 12 March 2024
Sec. Food Microbiology
Volume 15 - 2024 | https://doi.org/10.3389/fmicb.2024.1304325
This article is part of the Research Topic Zoonotic Antimicrobial Resistance and Virulence: One Health Integrated Approaches to Monitor and Reduce Food Chain Hazards View all 8 articles
 Xuchuan Ma1
Xuchuan Ma1 Marcel H. Tempelaars1
Marcel H. Tempelaars1 Marcel H. Zwietering1
Marcel H. Zwietering1 Sjef Boeren2
Sjef Boeren2 Conor P. O’Byrne3
Conor P. O’Byrne3 Heidy M. W. den Besten1
Heidy M. W. den Besten1 Tjakko Abee1*
Tjakko Abee1*Microbial population heterogeneity leads to different stress responses and growth behavior of individual cells in a population. Previously, a point mutation in the rpsU gene (rpsUG50C) encoding ribosomal protein S21 was identified in a Listeria monocytogenes LO28 variant, which leads to increased multi-stress resistance and a reduced maximum specific growth rate. However, the underlying mechanisms of these phenotypic changes remain unknown. In L. monocytogenes, the alternative sigma factor SigB regulates the general stress response, with its activation controlled by a series of Rsb proteins, including RsbR1 and anti-sigma factor RsbW and its antagonist RsbV. We combined a phenotype and proteomics approach to investigate the acid and heat stress resistance, growth rate, and SigB activation of L. monocytogenes EGDe wild type and the ΔsigB, ΔrsbV, and ΔrsbR1 mutant strains. While the introduction of rpsUG50C in the ΔsigB mutant did not induce a SigB-mediated increase in robustness, the presence of rpsUG50C in the ΔrsbV and the ΔrsbR1 mutants led to SigB activation and concomitant increased robustness, indicating an alternative signaling pathway for the SigB activation in rpsUG50C mutants. Interestingly, all these rpsUG50C mutants exhibited reduced maximum specific growth rates, independent of SigB activation, possibly attributed to compromised ribosomal functioning. In summary, the increased stress resistance in the L. monocytogenes EGDe rpsUG50C mutant results from SigB activation through an unknown mechanism distinct from the classical stressosome and RsbV/RsbW partner switching model. Moreover, the reduced maximum specific growth rate of the EGDe rpsUG50C mutant is likely unrelated to SigB activation and potentially linked to impaired ribosomal function.
Listeria monocytogenes is a ubiquitous foodborne pathogen, which can cause the disease listeriosis typically caused by ingestion of contaminated food (Radoshevich and Cossart, 2018). Listeria monocytogenes is well adapted to survive exposure to severe environmental challenges including high salt concentrations, a wide range of temperatures and extreme pH supporting its transmission from soil to food and ultimately to the human body (NicAogáin and O’Byrne, 2016; Quereda et al., 2021). Understanding these specific protective strategies including the impact of L. monocytogenes population heterogeneity, is crucial in designing effective food processing and preservation methods aimed at minimizing the risk this pathogen poses to consumers (Abee et al., 2016). Population heterogeneity includes genetic and non-genetic population variability, and both can generate phenotypic variation in a population and contribute to the overall fitness, adaptation, and survival capacity of the population (Smits et al., 2006; Davidson and Surette, 2008; Ryall et al., 2012). Pathogens may be inactivated during food processing, and differences in stress resistance between individual cells can result in a higher-than-expected number of surviving cells and selection of stress-resistant variants (Metselaar et al., 2016).
Previously, 23 stable stress resistance L. monocytogenes variants have been isolated upon acid treatment of L. monocytogenes strain LO28 (Metselaar et al., 2013). These variants showed a trade-off between reduced maximum specific growth rate and increased resistance against acid, heat, high hydrostatic pressure and benzalkonium chloride (Metselaar et al., 2013, 2015). Whole genome sequencing analysis showed that 11 of the 23 variants had mutations in the rpsU gene locus, which encodes the ribosome 30S small sub-unit protein S21 (RpsU) (Metselaar et al., 2015). Two variants have been selected for further research, namely, variant V14 and variant V15 (Koomen et al., 2018). Variant V14 has a deletion of the whole rpsU and yqeY genes and half of phoH gene, while variant V15 has a nucleotide substitution from G to C in rpsU at position 50 [NC_003210.1:g.1501930G > C p.(Arg17Pro), designated as rpsUG50C in this study], which may lead to an amino acid substitution from arginine to proline in the RpsU protein (marked as RpsU17Arg-Pro in this study) (Metselaar et al., 2015; Koomen et al., 2021). The introduction of an extra proline-associated turn conceivably results in loss of functionality and/or exclusion of RpsU17Arg-Pro from the 30S ribosome in V15 (Koomen et al., 2021). Comparative transcriptomic and phenotypic studies showed that variants V14 and V15 have a large overlap in the gene expression profiles and similar phenotypic results including increased freeze–thaw resistance, higher glycerol utilization rates, flagella absence and higher Caco-2 cells attachment and invasion levels compared to the wild type (Koomen et al., 2018). These results suggest that the deletion of the whole rpsU and point mutation rpsUG50C may affect the phenotype by the same mechanism (Koomen et al., 2018). Additional studies following introduction of the rpsUG50C mutation into L. monocytogenes LO28 and EGDe wild type strains, confirmed that this mutation results in heat and acid resistance and reduced maximum specific growth rate in both mutant strains (Koomen et al., 2021).
SigB is considered as the regulator of general stress response and controls the transcription of approximately 300 genes that contribute to the stress response and virulence of L. monocytogenes (O’Byrne and Karatzas, 2008; Toledo-Arana et al., 2009; Liu et al., 2019; Guerreiro et al., 2020a). Indeed, previous transcriptomic and proteomic analyses showed that many SigB regulon genes and proteins were strongly upregulated in the rpsU variants, which suggests that the activation of SigB-mediated stress may explain the multiple stress resistance phenotype of rpsU variants (Koomen et al., 2018, 2021). Generally, the activation of SigB is controlled at the post-translation level through the stressosome and a series of other Rsb proteins (Supplementary Figure S1) (Becker et al., 1998; Guerreiro et al., 2020a, 2022a). Briefly, RsbT is captured by the stressosome, which is composed of RsbS, RsbR1, and RsbR1 paralogues in unstressed cells. Upon environmental stress, RsbR1 and RsbS are phosphorylated, and RsbT is released from the stressosome. The free RsbT can bind to RsbU and stimulate its phosphatase function. Then anti-sigma factor antagonist RsbV is dephosphorylated by RsbU and binds to anti-sigma factor RsbW, which releases the previously bound SigB, which is then free to bind to RNA polymerase and initiate the transcription of the SigB regulon. Once stress is removed, RsbX, which is co-expressed with SigB, can dephosphorylate RsbR1 and RsbS, and RsbT binds back to the stressosome and inactivates the signal transduction (Guerreiro et al., 2020a; Oliveira et al., 2022).
To date, although the rpsU mutation is confirmed to confer stress resistance and decrease growth rate in L. monocytogenes, the role of SigB in these phenotypic changes, and the involvement of the stressosome-mediated signaling pathway in SigB activation in the L. monocytogenes rpsUG50C mutant under unstressed conditions, remain uncertain. Therefore, in the current study we aim to investigate first the involvement of SigB in the stress resistance of rpsU variants and whether the stressosome and/or the anti-sigma factor antagonist RsbV are involved in the activation of SigB in the rpsUG50C mutant, or if other factors may contribute to (indirect) activation of SigB in this mutant. Second, we sought to evaluate whether the activation of SigB and its regulon lead to reduced fitness of the rpsUG50C mutant. To address these questions, the rpsUG50C mutation was introduced in L. monocytogenes EGDe wild type (WT), and in the RsbR1, RsbV, and SigB deletion mutants, previously used to study stressosome structure and functionality (Utratna et al., 2012; Dessaux et al., 2020; Guerreiro et al., 2020b). Comparative phenotypic and proteomic study of the L. monocytogenes EGDe WT, rpsUG50C, ΔrsbR1, ΔrsbV, and ΔrsbV single mutant strains, and ΔrsbR1-rpsUG50C, ΔrsbV-rpsUG50C, and ΔsigB-rpsUG50C double mutant strains will shed light on the interaction between the ribosome and stressosome-dependent SigB activation and the fitness effect in cells with and without functional RpsU, and whether additional factors are involved.
It has been reported that the rpsUG50C mutation in L. monocytogenes can lead to a multi-stress resistance phenotype (Koomen et al., 2021). To confirm that the rpsUG50C mutation can lead to increased acid stress resistance of the L. monocytogenes EGDe strain used in the current study, the wild-type strain EGDe WT and the EGDe-rpsUG50C mutant were exposed to pH 3.0 for 15 min. As expected, the EGDe WT had a significantly (p value < 0.05) higher log-reduction (~ 4 log10CFU/mL) than the EGDe-rpsUG50C mutant (~ 0.5 log10CFU/mL) after exposure to acid, which indicates that the EGDe WT had lower acid resistance than the EGDe-rpsUG50C mutant (Figure 1A). Then, to explore the effect of SigB on the acid stress resistance of rpsUG50C mutants, the EGDe ΔsigB mutant and the ΔsigB-rpsUG50C double mutant were exposed to acid stress. No significant differences were observed between the ΔsigB and the ΔsigB-rpsUG50C mutants, which indicates that SigB is essential, to a large extent, for the increased acid resistance of the rpsUG50C mutant (Figure 1A). To test whether the stressosome was also involved in the SigB-mediated stress resistance of the rpsUG50C mutant, we also introduced the rpsUG50C mutation into the EGDe WT and the ΔrsbR1 mutant. The ΔrsbR1 mutant does not have a functional stressosome, and therefore the signaling pathway is interrupted. Acid stress resistance data show that the ΔrsbR1-rpsUG50C double mutant is significantly more acid-stress resistant than the ΔrsbR1 mutant, with comparable stress resistance as the single rpsUG50C mutant (Figure 1A). This indicates that the stressosome is not involved in the increased acid stress resistance of the rpsUG50C mutant. Apart from the stressosome, there are several other regulators in the SigB activation pathway, in which anti-sigma factor antagonist RsbV is the most downstream positive regulator. To investigate whether RsbV and upstream SigB activation pathway regulators were involved in the increased acid stress resistance of the rpsUG50C mutant, the rpsUG50C mutation was also introduced in the EGDe ΔrsbV mutant. Interestingly, the EGDe ΔrsbV-rpsUG50C double mutant still had higher acid stress resistance than the ΔrsbV mutant, indicating that the SigB-related acid stress resistance of the rpsUG50C mutant was independent of RsbV. The EGDe WT strain and the single and double mutant strains were also tested for heat stress resistance by exposure to 60°C for 5 min. Again, the rpsUG50C mutant strains except the ΔsigB-rpsUG50C mutant were more resistant than their counterpart, underlining that the mutation confers SigB-dependent resistance to multiple stresses (Figure 1B). Combining the results, we can conclude that the rpsUG50C mutation can lead to increased multi-stress resistance of L. monocytogenes, which requires SigB but not RsbR1 nor RsbV. This suggests that an additional signaling pathway is involved in preventing binding of anti-sigma factor RsbW to SigB in the rpsUG50C mutant.
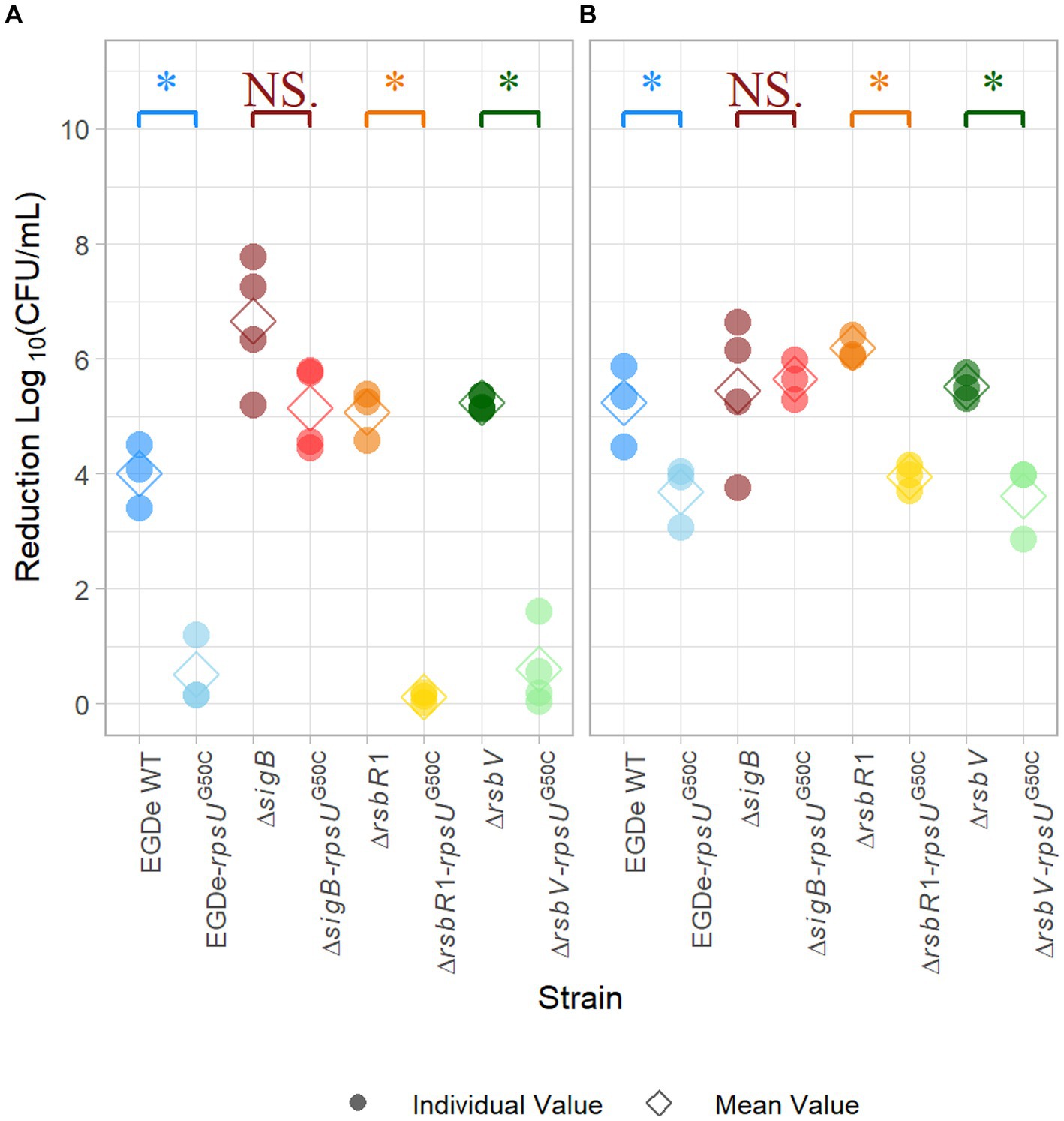
Figure 1. Stress resistance of late-exponential phase cells of Listeria monocytogenes EGDe WT, ΔsigB, ΔrsbV, and ΔrsbR1 mutants and their rpsUG50C mutants in BHI broth. Late-exponential phase cells were exposed to pH 3.0 for 15 min at 37°C (A), and 5 min at 60°C (B). Results are expressed as reduction in log10 (CFU/mL) after exposure compared to log10 (CFU/mL) before exposure. The mean values are represented by diamonds, while individual replicates are represented by circles. Significant differences (p < 0.05) between each pair of rpsUG50C mutants and parent strains are indicated by an asterisk, and no significant differences are indicated by NS.
In previous research, rpsUG50C mutants showed increased stress resistance and lower maximum specific growth rates (Metselaar et al., 2013, 2015, 2016; Koomen et al., 2021). Previous research suggested that the reduced growth ability might be the trade-off for the increased resistance (Metselaar et al., 2015). To further investigate this trade-off, the maximum specific growth rate (μmax) of EGDe WT, the ΔsigB mutant, the ΔrsbR1 mutant and the ΔrsbV mutant and their rpsUG50C mutants were estimated. Since the previous stress resistance experiments were performed using 30°C-grown cultures and 37°C is the optimal growth temperature of L. monocytogenes, the μmax was estimated in BHI at both 30 and 37°C. As expected, the EGDe WT had higher μmax than the EGDe-rpsUG50C mutant at both temperatures, although the difference was not statistically significant at 37°C (p value > 0.05) (Figure 2). This lack of significance could be attributed to increased variability associated with adaptation of the cells following the transition from 30 to 37°C. In addition, the ΔrsbR1-rpsUG50C and the ΔrsbV-rpsUG50C mutants, which both had increased stress resistance, had significantly lower μmax than the ΔrsbR1 and the ΔrsbV mutants at 30 and 37°C (Figure 2). However, the ΔsigB-rpsUG50C mutant, which had similar low stress resistance levels as the ΔsigB mutant, still had significantly lower μmax than the ΔsigB mutant. This observation provides evidence that the rpsUG50C mutation leads to reduced growth rate independently from RsbR1, RsbV, and SigB.
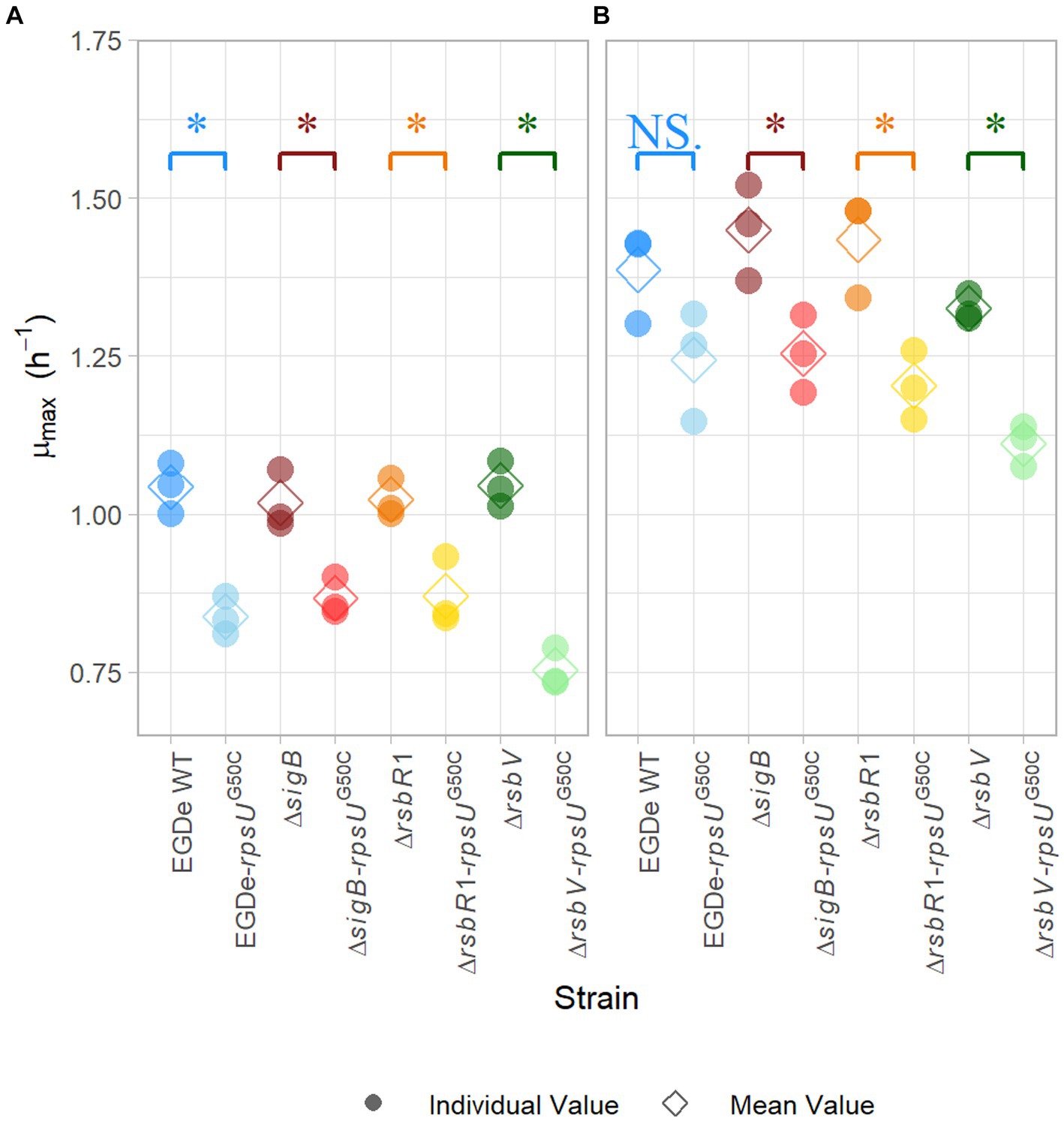
Figure 2. Maximum specific growth rate of Listeria monocytogenes EGDe WT, ΔsigB, ΔrsbV, and ΔrsbR1 mutants and their rpsUG50C mutants in BHI broth at 30°C (A) and 37°C (B), determined by the 2-fold dilution method. The mean values are represented by diamonds, while individual replicates are represented by circles. Significant differences (p < 0.05) between each pair of rpsUG50C mutants and parent strains are indicated by an asterisk, and no significant differences are indicated by NS.
Our proteomic data showed that 106 proteins were significantly higher expressed in the EGDe-rpsUG50C mutant compared to EGDe WT, and 54 of these higher expressed proteins belonged to SigB regulon (Figure 3A; Supplementary Table S1). For these 106 proteins, the Kyoto Encyclopedia of Genes and Genomes (KEGG) ABC transporter system was significantly enriched but no Gene Ontology (GO) term was overrepresented. The GO terms of these 106 proteins were shown in Supplementary Figure S2. In addition, the proteomic data showed that only two SigB regulon proteins were significantly upregulated in the ΔsigB-rpsUG50C mutant compared to the ΔsigB mutant (Figure 3B; Supplementary Table S1). Since the EGDe-rpsUG50C mutant, but not the ΔsigB-rpsUG50C mutant, has increased stress resistance (Figure 1), these proteomic data confirmed that the rpsUG50C mutation resulted in SigB activation and the upregulation of SigB regulon proteins, which caused the increased multi-stress resistance of the rpsUG50C mutant. For the ΔrsbV-rpsUG50C mutant, which lacks the anti-sigma factor antagonist RsbV, SigB should not be activated in this mutant and the SigB regulon should not be upregulated. However, our phenotypic data showed that the ΔrsbV-rpsUG50C mutant still had increased stress resistance, which implies an RsbV-independent SigB activation in the ΔrsbV-rpsUG50C mutant (Figure 1). Indeed, of the 113 proteins that were significantly higher expressed in the ΔrsbV-rpsUG50C mutant compared to the ΔrsbV mutant, 65 proteins belonged to the SigB regulon (Figure 3C; Supplementary Table S1). No GO term or specific KEGG pathways were significantly overrepresented among these 113 proteins. The GO terms of these proteins were shown in Supplementary Figure S2. These results provide further evidence that in contrast to the traditional model, RsbV is not involved in the SigB activation and upregulation of regulon members in the rpsUG50C mutant.
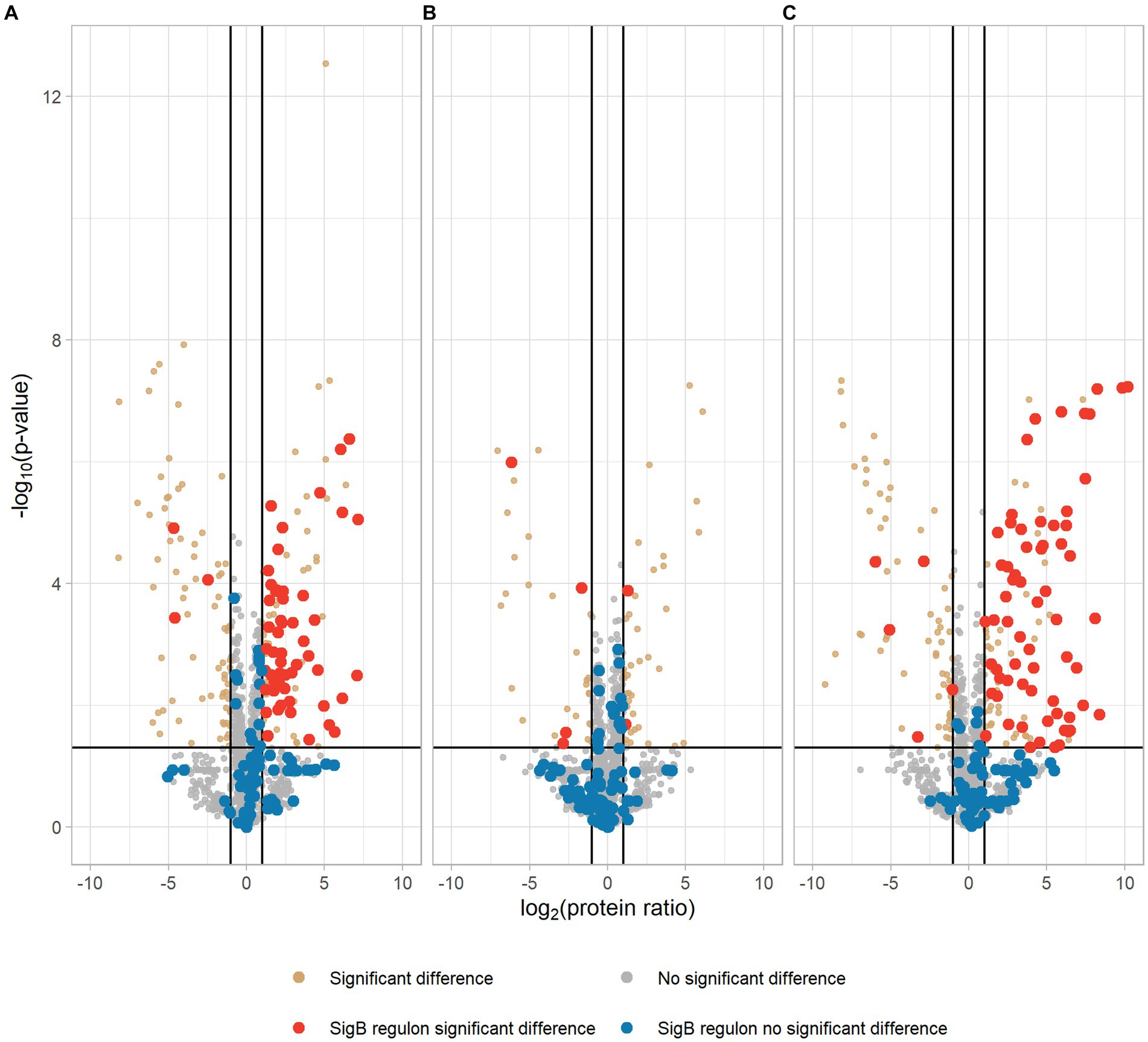
Figure 3. Volcano plot of proteomic data comparing Listeria monocytogenes EGDe WT (A), ΔsigB (B), and ΔrsbV (C) mutants with their rpsUG50C mutants EGDe-rpsUG50C, ΔsigB-rpsUG50C, and ΔrsbV-rpsUG50C, respectively. The −log10 (p value) is plotted against the log2 (protein ratio: rpsUG50C mutants over parent strains). The horizontal line represents the cutoff of the p value (0.05), and the vertical lines represent the cutoff of log2 (protein ratio) (±1). Red dots and blue dots indicate significantly regulated and not significantly regulated SigB regulon proteins, respectively. Brown dots and gray dots represent other significantly regulated and not significantly regulated proteins, respectively.
To further investigate these significantly upregulated or downregulated proteins, the numbers of differentially expressed proteins in each rpsUG50C mutant are shown in Figure 4. There were 65 proteins that were upregulated in both EGDe-rpsUG50C and ΔrsbV-rpsUG50C mutants compared to their parent strains, of which 46 proteins belonged to the SigB regulon (Figure 4A). Also, there were 36 proteins that were downregulated in both EGDe-rpsUG50C and ΔrsbV-rpsUG50C mutants compared to their parent strains (Figure 4B). KEGG pathway over-representation analysis (p value < 0.05) of these 36 proteins showed that three enriched terms were found including flagellar assembly, bacterial chemotaxis, and two-component systems. The ΔsigB-rpsUG50C mutant had less proteins that were significantly upregulated or downregulated compared to the EGDe-rpsUG50C and the ΔrsbV-rpsUG50C mutants (Figure 4), indicating that the ΔsigB-rpsUG50C mutant had a rather similar proteomic profile as its parent strain the ΔsigB mutant, and this is in line with the observed similar reduced stress resistant phenotype.
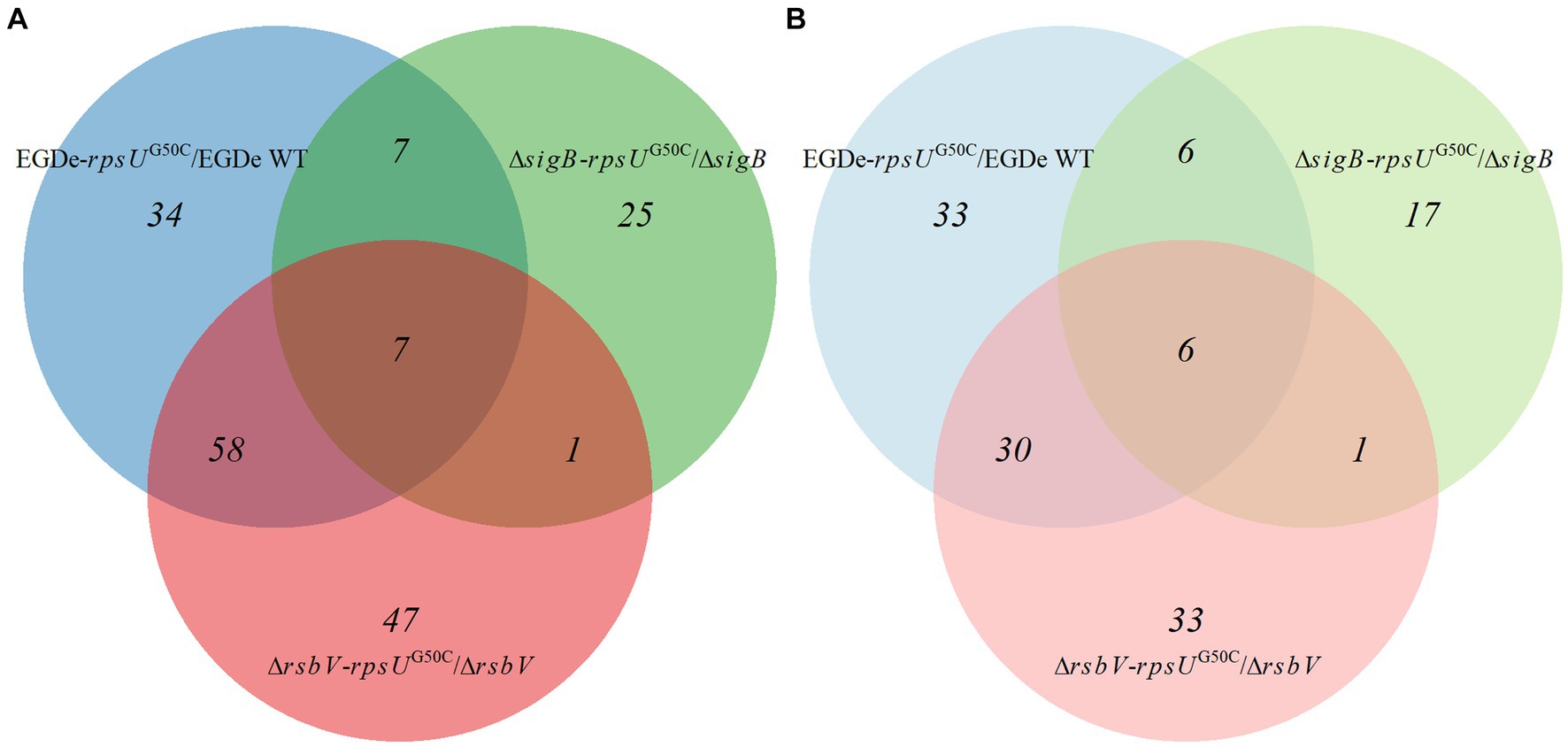
Figure 4. Venn graph of differentially expressed proteins by comparing Listeria monocytogenes EGDe WT, ΔsigB, and ΔrsbV mutants with their rpsUG50C mutants EGDe-rpsUG50C, ΔsigB-rpsUG50C, and ΔrsbV-rpsUG50C, respectively. The panels (A,B) represent the upregulated and downregulated proteins, respectively. (Light) blue, (light) green, and (light) red circles represent the upregulated or downregulated proteins when comparing EGDe-rpsUG50C, ΔsigB-rpsUG50C, and ΔrsbV-rpsUG50C mutants to their parent strains, respectively.
The activation of SigB requires the release of SigB from the anti-SigB factor RsbW. Interestingly, our proteomic data showed that both RsbW and SigB were upregulated in the EGDe-rpsUG50C and the ΔrsbV-rpsUG50C mutants, but to slightly different levels, which might lead to changes in the protein abundance ratio between RsbW and SigB (Supplementary Table S1; Supplementary Figure S3). A possible lower ratio of RsbW:SigB in the rpsUG50C mutant strains may make SigB more available for binding with the RNA polymerase. To evaluate this, the LFQ data from MaxQuant ProteinGroups file were used to calculate the protein ratio of RsbW:SigB for the EGDe WT, the EGDe-rpsUG50C, the ΔrsbV and the ΔrsbV-rpsUG50C mutants (Figure 5). The RsbW:SigB ratio was not significantly lower in the EGDe-rpsUG50C mutant than the EGDe WT, and additionally, the ratio was still 2:1, which is the ratio previously determined for the RsbW:SigB complex in B. subtilis (Pathak et al., 2020). With the deletion of RsbV, there should be more RsbW available for SigB in ΔrsbV-rpsUG50C. However, the ΔrsbV-rpsUG50C mutant had an even higher RsbW:SigB ratio than the EGDe-rpsUG50C mutant. Therefore, the RsbV-independent SigB activation in the rpsUG50C mutant could not be explained by a reduced RsbW:SigB ratio in the rpsUG50C mutant.
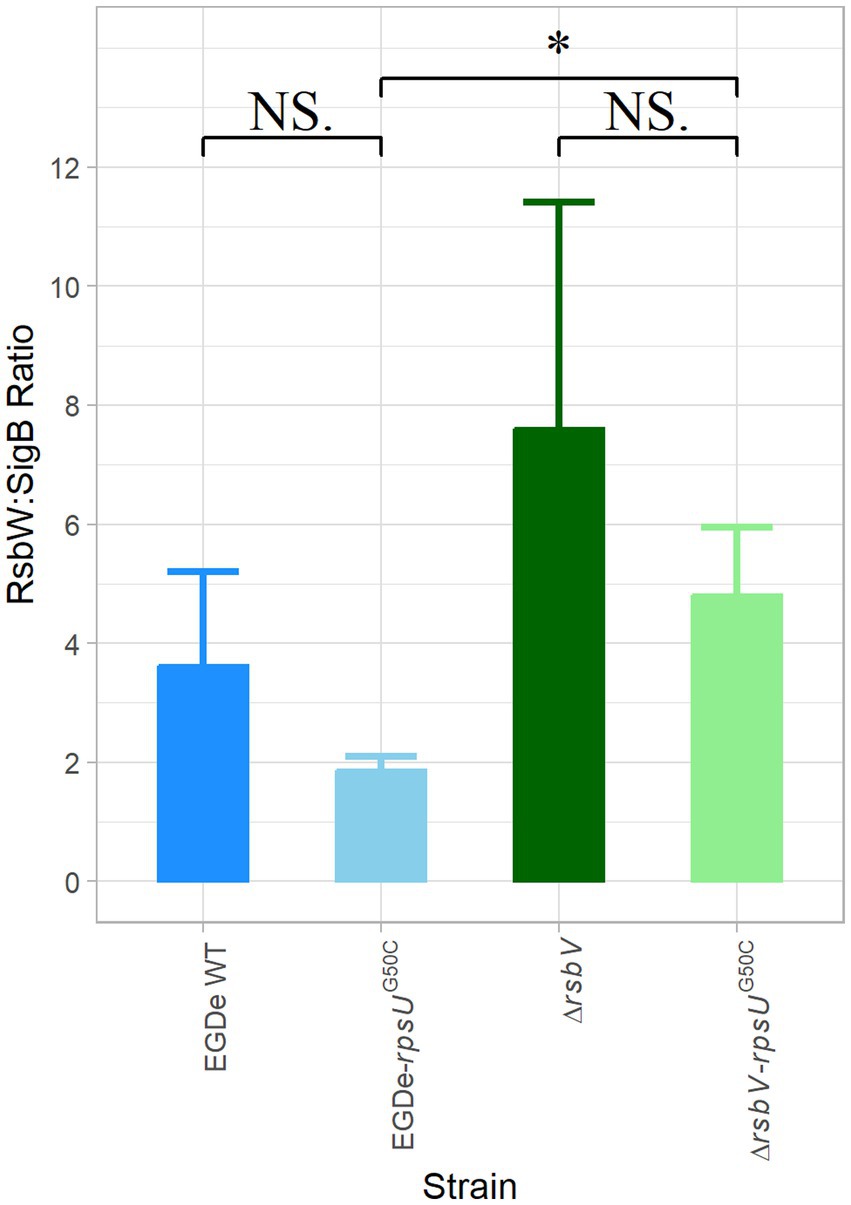
Figure 5. RsbW:SigB protein abundance ratio. The protein abundance ratio between RsbW:SigB in Listeria monocytogenes EGDe WT, ΔrsbV mutant, and their rpsUG50C mutants, which is calculated based on proteomic data. Significant differences are indicated by an asterisk, and no significant differences are indicated by NS.
Since the combined data on RsbW:SigB ratios could not explain the SigB activation in the rpsUG50C mutant, proteins whose expression levels were affected by the rpsUG50C mutation but independent from the presence of SigB or RsbV should be considered. In all three rpsUG50C mutants, seven proteins were significantly upregulated and six proteins were significantly downregulated (Figure 4; Supplementary Tables S2, S3). Among these proteins, Lmo2499, encoding a putative phosphate ABC transporter substrate-binding protein (PstS), was the highest differentially expressed. The corresponding additional transporter components (lmo2495-Lmo2498) constitute an uptake system that is generally activated in bacteria as part of the phosphate starvation response (Allenby et al., 2005). To further analyze the possible role of PstS in SigB activation in the rpsUG50C mutant in the tested conditions, the pstS gene was deleted in the EGDe WT and the EGDe-rpsUG50C mutant (Table 1). The acid and heat stress resistance and the maximum specific growth rate of the ΔpstS mutant and the ΔpstS-rpsUG50C double mutant were then tested. The acid resistance and heat resistance of the ΔpstS mutant was lower compared to the ΔpstS-rpsUG50C mutant, although the difference was not significant for heat resistance (Supplementary Figure S4). Also, the ΔpstS mutant had a significantly higher μmax than the ΔpstS-rpsUG50C mutant at 30°C and not significantly higher μmax at 37°C (Supplementary Figure S5). Comparative WGS showed an additional mutation in the double mutant (Supplementary Table S4), but based on the observed stress resistance and fitness phenotypes, it can be concluded that PstS did not directly contribute to increased robustness and reduced fitness of the rpsU mutants. rpsU mutants.
The aim of this study was to examine how the rpsUG50C mutation influences the stress resistance and the maximum specific growth rate of L. monocytogenes. The phenotypic and proteomic data showed that SigB was activated in the rpsUG50C mutant, which led to SigB regulon upregulation and concomitant increased stress resistance. Based on the current knowledge of the SigB controlling pathway, the activation of SigB requires the presence of RsbR1 and RsbV (Supplementary Figure S1) (Guerreiro et al., 2020a). However, both the ΔrsbR1-rpsUG50C and the ΔrsbV-rpsUG50C mutants surprisingly had higher stress resistance than their parent strains, indicating that the SigB-mediated increased stress resistance in the rpsUG50C mutant was independent of RsbR1, i.e., a functional stressosome, and the anti-sigma factor antagonist RsbV. The proteomic analysis also shows that the SigB regulon was still induced in the ΔrsbV-rpsUG50C mutant, in which SigB was expected to be inactive due to binding to RsbW. As shown in Supplementary Figure S1, RsbW is the only SigB regulator downstream of RsbV in the SigB regulation pathway. Hence, the activation signal in the rpsUG50C mutant must enter the SigB activation pathway downstream from RsbV, so the mutation in the ribosome may induce an alternative signaling pathway that reduces or prevents the binding between RsbW and SigB, which leads to the RsbV-independent SigB activation.
Previously, activation of SigB at low or high temperature has been observed in growing cells of B. subtilis (16 or 51°C) and L. monocytogenes (4°C) wild type and respective rsbV mutants (Brigulla et al., 2003; Holtmann et al., 2004; Utratna et al., 2014). It was hypothesized that key physical interactions between RsbW and SigB or between SigB and core RNA polymerase might change at low or high temperatures, but this hypothesis cannot explain the RsbV-independent SigB activation in the current study, since the L. monocytogenes strains were cultured at 30°C in rich media (BHI). Another explanation may involve changes in the RsbW:SigB ratio of 2:1, which was previously determined in B. subtilis based on protein quantification and 3D structural modeling (Pathak et al., 2020). Based on our proteomic results, the respective RsbW:SigB ratios were 2:1 or even higher in the tested rpsUG50C mutants (Figure 5). Therefore, RsbV-independent SigB activation could not be explained by a decrease in the RsbW:SigB ratio.
Another hypothesis suggested in previous studies was that signaling proteins acting independently from RsbV to RsbW could disrupt the inhibitory RsbW-SigB complex and allow activation of SigB (Brigulla et al., 2003). In the current study, Lmo2499, a protein homologous to the periplasmic phosphate sensory binding protein PstS, has been investigated, since the proteomic data showed that PstS was upregulated more than 4-fold with a p value less than 0.01 in all three rpsUG50C mutants, namely, the EGDe-rpsUG50C, the ΔsigB-rpsUG50C, and the ΔrsbV-rpsUG50C mutants (Supplementary Table S1). PstS is involved in Pi transport and Pho regulon regulation (Hsieh and Wanner, 2010; Vaestermark and Saier, 2014; Santos-Beneit, 2015). In B. subtilis, both the Pho regulon and the SigB regulon can be activated by Pi starvation, and the signal of Pi starvation is transmitted to SigB via SigB regulator RsbP (Allenby et al., 2005). For B. subtilis SigB activation, RsbP is also required in response to energy stress, and another SigB regulator, RsbU, is required for response to environmental stress (Vijay et al., 2000). Listeria monocytogenes only has RsbU but not RsbP, and the energy stress-triggered activation pathway remains to be elucidated (Shin et al., 2010). To our knowledge, there is no research about the L. monocytogenes Pi starvation reaction or the activation mechanism of SigB by Pi starvation yet. Since SigB can be activated by Pi starvation in B. subtilis, it is possible that SigB can also be activated by Pi starvation in L. monocytogenes. However, the phenotypic characterization of the ΔpstS and the ΔpstS-rpsUG50C mutants showed that the ΔpstS-rpsUG50C mutant still had higher acid and heat stress resistance than the ΔpstS mutant (Supplementary Figure S4); excluding a direct link of PstS with SigB activation in the mutant strains for the tested conditions. Whether the upregulation of PstS signifies changes in intracellular Pi concentrations in rpsUG50C mutant strains, resulting in possible effects on (cross-reacting) kinase activity in other regulatory networks (Shi et al., 2014), that subsequently affect RsbW and SigB interaction in rpsUG50C mutants, remains to be studied.
Apart from the stress resistance, we have also tested the fitness of each strain to investigate the stress resistance-fitness trade-off of the rpsUG50C mutant. Generally, there is a trade-off between stress resistance and growth rate for bacteria, and this phenomenon has also been reported in rpsUG50C mutants in previous studies (Nystrom, 2004; Metselaar et al., 2013, 2016; Koomen et al., 2018, 2021). This may be due to the competition between SigB and housekeeping SigA for the RNA polymerase, with the latter responsible for the transcription of growth-related genes (Nystrom, 2004; Österberg et al., 2011). In addition, activation of SigB and its regulon conceivably consumes energy, resulting in a negative impact on growth (Xia et al., 2016; Guerreiro et al., 2020a). Indeed, studies have shown that mutations in SigB can increase fitness under sub-optimal conditions, including 0.5 M NaCl, 42°C, and blue light (Abram et al., 2008; O’Donoghue et al., 2016; Guerreiro et al., 2020b, 2022b). However, our previous evolution experiments with rpsUG50C mutants resulted in the selection of evolved variants with enhanced fitness (Koomen, 2022). The fact that no variants were obtained with mutations in sigB or genes of the SigB operon suggested that the major negative effect on fitness did not derive from SigB activation. Indeed, in the current study, all these rpsUG50C mutants, including the ΔsigB-rpsUG50C mutant, had lower maximum specific growth rates than their respective parent strains in BHI at 30°C (Figure 2). Therefore, the growth rate decrease of the rpsUG50C mutant is independent of SigB activation and SigB-mediated stress response. In addition, the ΔpstS-rpsUG50C mutant also had a lower specific growth rate than the ΔpstS mutant (Supplementary Figure S5). Thus, the upregulation of pstS in the rpsUG50C mutant did not contribute to the reduced fitness either.
In Escherichia coli and B. subtilis, RpsU (ribosomal protein S21) is involved in translation initiation (Van Duin and Wijnands, 1981; Berk et al., 2006; Sohmen et al., 2015). Combined with the results above, it is conceivable that reduced fitness of L. monocytogenes rpsUG50C mutants is linked to decreased translation efficacy and/or the availability of functional 70S ribosomes (Koomen, 2022). The L. monocytogenes Lmo0762 protein, HflXr, a homolog of a ribosome-splitting factor, HflX, was also upregulated in all three rpsUG50C mutants (Supplementary Table S2). HflX belongs to the GTPase OBG-HflX-like superfamily. Another member of this superfamily, Obg (Lmo1537/ObgE) that was detected in the EGDe WT and mutant proteomes, has been reported to play a role in the activation of SigB in B. subtilis (Scott and Haldenwang, 1999; Verstraeten et al., 2011; Kint et al., 2014). Whether HflXr and/or ObgE play a role in L. monocytogenes RsbV-independent SigB activation and/or fitness modulation in rpsUG50C mutants remains to be elucidated. Furthermore, additional biochemical studies are needed to elucidate the phosphorylation and acetylation status of the RsbW protein, as well as to understand its impact on protein–protein interactions with SigB and RsbV.
While our study provides valuable insights into the characteristics of the rpsUG50C mutants under controlled laboratory conditions, it is important to acknowledge that the prevalence and significance of the rpsU mutations, including rpsUG50C, in the naturally circulating bacterial populations remain largely unexplored. Investigating the prevalence of these mutations in L. monocytogenes publicly available genome database will be essential for assessing their role in the pathogen’s epidemiology.
We recognize the limitation posed by the absence of complemented strains in our study, which if available could have provided further confirmation of the SigB activation by these genetic modifications. However, the complexity of generating complemented strains, particularly for the double mutants, presented significant technical challenges that were beyond the scope of this initial investigation. The use of whole-genome sequencing and subsequent SNP analysis of our mutants and parental strains, helps to mitigate this limitation since it provides the full genetic context in which these mutations are operating.
Additionally, our use of the L. monocytogenes EGDe strain, chosen for its well-documented genetic background, may limit the broader applicability of our results. Given that the EGDe strain is from lineage II, CC9, it might not fully represent the genetic diversity and pathogenic potential prevalent in strains commonly associated with clinical infections. This limitation highlights the need for future studies to examine the effects of the rpsUG50C across a broader range of L. monocytogenes strains to confirm the universality of our findings across different genetic backgrounds of L. monocytogenes.
Taken together, the current study shows that the activation of SigB in the L. monocytogenes rpsUG50C mutant resulting in multi-stress robustness and lower maximum specific growth rate is independent of the stressosome protein RsbR1 and anti-sigma factor antagonist RsbV. Although there is generally a trade-off between stress resistance and growth rate for bacteria, we observed that the reduced growth rate is independent of the activation of SigB and its regulon members and conceivably due to reduced ribosomal functioning. Further studies are needed to elucidate the mechanism of RsbV-independent SigB activation and the fitness modulation in rpsUG50C mutants. A deeper understanding of these specific protective strategies including the impact of L. monocytogenes population heterogeneity, can contribute to further improve efficacy of food processing and preservation methods aimed at minimizing the risk this pathogen poses to consumers.
The bacterial strains, plasmids, and primers used in this study are described in Tables 1, 2. The model strain EGDe has been used in this study, since it is the most used model strain in lab with in vitro and in animal research. It also has been proved to have heat and acid stress resistance with rpsUG50C mutation (Koomen et al., 2021). The shuttle vector pAULA-rpsUG50C and pKSV7-ΔpstS were used for introducing the pstS gene deletion and the rpsUG50C point mutation in the target L. monocytogenes strains, respectively. The pKSV7-ΔpstS was constructed as described previously with modification (Rychli et al., 2021). The upstream and down region from pstS gene was amplified from gDNA of EGDe WT using KAPA HiFi Hotstart ReadyMix (KAPA biosystems, United States) with the up region primers (pstS-Up-EcoRI-F and pstS-Up-NotI-R) and the down region primers (pstS-Down-NotI-F and pstS-Down-SalI-R), respectively. The resulting fragments were fused and ligated into the pKSV7 multiple cloning site. The resulting construct was confirmed by PCR and sequencing using primers M13-F and M13-R. To construct rpsUG50C mutants, pAULA-rpsUG50C was transformed into L. monocytogenes competent cells by electroporation (2.5 kV, 25 μF, 200 D) and plated on Brain Heart Infusion (BHI, Oxoid, Ltd., Basingstoke, England) agar (1.5% (w/w), bacteriological agar no. 1 Oxoid) plates at 30°C with 5 μg/mL erythromycin to select for transformants. The erythromycin-resistant colonies were inoculated in BHI broth with 5 μg/mL erythromycin and grown at 42°C overnight. The 42°C-grown overnight cultures were inoculated into fresh BHI for overnight culture at 30°C and subsequently plated on BHI agar plates at 30°C. The resulting colonies were replica plated on BHI with and without 5 μg/mL erythromycin and incubated at 30°C. The erythromycin-sensitive colonies were selected and the rpsUG50C point mutation was verified by PCR and Sanger sequencing (BaseClear B.V. Leiden, The Netherlands) with primers rpsU-EcoRI-F and rpsU-SalI-R. To construct ΔpstS mutants, the same process has been performed with pKSV7-ΔpstS, and the colonies were selected by chloramphenicol (10 μg/mL) and verified by PCR and sequencing with primers pstS-Flank-F, pstS-Flank-R pstS-Flank-F, pstS-Flank-R, pstS-Up-Check-F, and pstS-Down-Check-R. Single nucleotide polymorphism (SNP) analysis was performed on all constructed mutants as described in the following section and confirmed the absence of any other significant undesired mutations (Supplementary Table S4).
The genomic DNA was isolated for sequencing using DNeasy Blood and tissue kit (Qiagen, Hilden, Germany). Two times 2 mL of overnight culture was centrifuged (17,000 × ), washed by 1 mL PPS and resuspended in 1 mL lysis buffer [20 mM Tris–HCl, 2 mM EDTA, 1.2% (w/v) Triton X-100, 20 mg/mL lysozyme, pH 8.0]. The suspension was incubated at 37°C for 1 h under gentle shaking in an Eppendorf Thermomixer 5,436 (Eppendorf AG, Hamburg, Germany). Then, 10 μL RNAse (10 mg/mL, Qiagen, Hilden, Germany) was added and incubated for 5 min at room temperature. Subsequently, 62.5 μL proteinase K and 500 μL AL buffer (provided by the manufacturer) were added and incubated at 56°C for 1 h under gentle shaking. Then, 500 μL absolute ethanol was added and the suspension was transferred to a spin column provided by the kit and incubated for 10 min. After incubation, the columns were centrifuged for 1 min at 6,000 × . The filters were washed two times with 500 μL buffer AW1 and two times with 500 μL buffer AW2 at 6,000 × . The AW1 and AW2 are provided by the kit. After washing steps, the columns were centrifuged at 17,000 × for 3 min. Subsequently, 50 μL of AE buffer was added to the center of the column. The column was incubated for 10 min and centrifuged at 6,000 × to collect the target sample. Samples were stored at −20°C until sequencing. Library preparation and paired-end 2 × 150 bp short-reads were generated using the INVIEW resequencing of bacteria service from Eurofins GmbH (Constance, Germany) using Illumina technology. On Galaxy platform, read quality control was performed using FastQC (0.73 + galaxy0), and SNPs were identified using snippy (4.6.0 + galaxy0) with reference genome of EGDe (ASM19603v1) (Andrews, 2010; Torsten, 2015; Galaxy Community, 2022).
For stress resistance experiments and proteomics experiments, the L. monocytogenes strains were cultured as described previously (Metselaar et al., 2013). Briefly, stock cultures were grown for 1–2 days at 30°C on BHI agar plates. One single colony was then inoculated in 20 mL BHI broth and cultured at 30°C overnight under shaking at 160 rpm. A 0.5% (v/v) inoculum was added to fresh BHI broth and cells were grown at 30°C at 160 rpm until the late-exponential growth phase (Optical Density at 600 nm OD600 = 0.4–0.5).
Acid and heat inactivation experiments were performed as described before (Metselaar et al., 2013). Briefly, 100 mL late-exponential phase culture was harvested by centrifuging for 5 min at 2,880 × g, followed by resuspension in 10 mL BHI broth, centrifugation again for 5 min at 2,880 × g, and resuspension in 1.1 mL 0.1% peptone physiological salt solution (PPS, Tritium Microbiologie B.V., the Netherlands). For acid inactivation, 1 mL suspension was added to a 100 mL Erlenmeyer flask with 9 mL BHI broth, which was pre-warmed to 37°C and adjusted to a pH of 3.00 ± 0.01 using 10 M HCl to simulate the acidic conditions encountered by pathogens in the human stomach. The flask was placed in a shaking water bath at 37°C. At the beginning and after 15 min, 100 μL samples were taken. For heat inactivation, 0.1 mL suspension was diluted in PPS and plated to determine the concentration before inactivation, and the remaining 1 mL suspension was added to 19 mL BHI broth, which was preheated to 60°C and sampled 1 mL after 5 min. All the samples were decimally diluted and plated on BHI agar plates in duplicate, using a spiral plater (Eddy Jet, IUL S.A.) or by spread plating when no dilution steps were needed. Plates were incubated at 30°C and counted after 4–6 days to allow recovery of all cells. The experiment was done with at least three independent biological replicates.
The maximum specific growth rate μmax was determined by using the 2-fold dilution method as described previously (Biesta-Peters et al., 2010). Briefly, the overnight culture was diluted, plated on BHI agar plate, and incubated at 30°C for 2 days. In parallel, the culture was 10,000 times diluted, and 400 μL of the diluted culture was added to the first well of a 100-well honeycomb plate in duplicate. Subsequently, four times 2-fold dilution series was made by mixing 200 μL diluted bacterial culture and 200 μL fresh BHI in honeycomb plates. The plates were incubated in the Bioscreen (Oy Growth Curves AB Ltd.) at 30 or 37°C with constant medium shaking. The OD600 was measured every 10 min to determine the time-to-detection (TTD) of each well, which was defined as the time OD600 reaching 0.2. The μmax (h−1) of each culture was calculated by taking the negative reciprocal of the slope between the TTD and the natural logarithm of the initial concentration N0 (ln(N0)) of the five wells. The experiment was done with independent biological triplicates.
The strains for proteomic analysis were cultured as described in Section 4.3. For proteomic analysis, 4-mL aliquots of late-exponential phase culture were centrifuged for 1 min at 12,800 × g in two 2-mL LoBind Eppendorf tubes, resuspended in 200 μL ice-cold 100 mM Tris (pH 8), pooled together in one tube, and centrifuged again for 1 min at 12,800 × g. The pellets were washed using 100 mM Tris, centrifuged for 1 min at 12,800 × g, resuspended in 50 μL 100 mM Tris (pH 8), and lysed by sonication for 45 s on ice at maximum power twice (MSE Soniprep 150). Samples were prepared according to the universal solid-phase protein preparation protocol (Dagley et al., 2019) with doubled washing steps (washing with 70% ethanol and 100% acetonitrile). For each prepared peptide sample, 5 μL sample was injected into a nanoLC-MS/MS (Thermo nLC1000 connected to an Exploris 480 with FAIMS at CV = −45 V) for further analyzing as described previously (Wendrich et al., 2017; Feng et al., 2022). nLC-MSMS system quality was checked with PTXQC using the MaxQuant result files (Bielow et al., 2016). LCMS data with all MS/MS spectra were analyzed with the MaxQuant quantitative proteomics software package as described before (Cox et al., 2014; Bielow et al., 2016). The reference proteome database used for L. monocytogenes EGD-e (Proteome ID: UP000000817) was downloaded from UniProt. Perseus was used for filtering and further bioinformatics and statistical analysis of the MaxQuant ProteinGroups file (Tyanova et al., 2016). Reverse hits and contaminants were filtered out. Significant upregulation or downregulation was defined as a change in protein abundance relative to the parent strains of at least 2-fold with a p value less than 0.05. The proteins that belonged to SigB regulon were identified according to previous research (Kazmierczak et al., 2003; Hain et al., 2008; Ollinger et al., 2009; Oliver et al., 2010; Liu et al., 2017; Guariglia-Oropeza et al., 2018; Mattila et al., 2020). Data visualization was performed using the statistical programming language R (4.0.3). By using R package “clusterProfiler” (4.1.2) with setting “pvalueCutoff = 0.05,” KEGG and GO analysis were performed by using “enrichKEGG” and “compareCluster” functions, respectively (Wu et al., 2021).
Statistical significance analysis of phenotypic data analysis was performed in JASP (0.11.1) by using an independent samples t-test.
The datasets presented in this study can be found in the online repository PRIDE (Perez-Riverol et al., 2022). The accession number is PXD045800.
XM: Data curation, Methodology, Visualization, Writing – original draft, Writing – review & editing. MT: Methodology, Writing – review & editing. MZ: Project administration, Supervision, Writing – review & editing. SB: Data curation, Methodology, Writing – review & editing. CO’B: Writing – review & editing. HB: Project administration, Supervision, Writing – review & editing. TA: Project administration, Supervision, Writing – review & editing.
The author(s) declare that financial support was received for the research, authorship, and/or publication of this article. XM was supported by a grant from the China Scholarship Council (File No. 201907720086).
We thank Jeroen Koomen (Food Microbiology, Wageningen University) for mutant construction support. We thank Chendi Zhang and Kirsten van Kooten (Food Microbiology, Wageningen University) for data collection assistance. We thank Jasper Bannenberg (Food Microbiology, Wageningen University) for proteomic data analysis assistance. We also thank Oscar van Mastrigt (Food Microbiology, Wageningen University) for SNP analysis support.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2024.1304325/full#supplementary-material
Abee, T., Koomen, J., Metselaar, K. I., Zwietering, M. H., and Den Besten, H. M. W. (2016). Impact of pathogen population heterogeneity and stress-resistant variants on food safety. Annu. Rev. Food Sci. Technol. 7, 439–456. doi: 10.1146/annurev-food-041715-033128
Abram, F., Starr, E., Karatzas, K. A. G., Matlawska-Wasowska, K., Boyd, A., Wiedmann, M., et al. (2008). Identification of components of the sigma B regulon in Listeria monocytogenes that contribute to acid and salt tolerance. Appl. Environ. Microbiol. 74, 6848–6858. doi: 10.1128/AEM.00442-08
Allenby, N. E. E., O’Connor, N., Prágai, Z., Ward, A. C., Wipat, A., and Harwood, C. R. (2005). Genome-wide transcriptional analysis of the phosphate starvation stimulon of Bacillus subtilis. J. Bacteriol. 187, 8063–8080. doi: 10.1128/JB.187.23.8063-8080.2005
Andrews, S . (2010). FastQC A Quality Control tool for High Throughput Sequence Data. Available at: http://www.bioinformatics.babraham.ac.uk/projects/fastqc/
Becker, L. A., Çetin, M. S., Hutkins, R. W., and Benson, A. K. (1998). Identification of the gene encoding the alternative sigma factor sigma(B) from Listeria monocytogenes and its role in osmotolerance. J. Bacteriol. 180, 4547–4554. doi: 10.1128/JB.180.17.4547-4554.1998
Berk, V., Zhang, W., Pai, R. D., and Cate, J. H. D. (2006). Structural basis for mRNA and tRNA positioning on the ribosome. Proc. Natl. Acad. Sci. USA 103, 15830–15834. doi: 10.1073/pnas.0607541103
Bielow, C., Mastrobuoni, G., and Kempa, S. (2016). Proteomics quality control: quality control software for MaxQuant results. J. Proteome Res. 15, 777–787. doi: 10.1021/acs.jproteome.5b00780
Biesta-Peters, E. G., Reij, M. W., Joosten, H., Gorris, L. G. M., and Zwietering, M. H. (2010). Comparison of two optical-density-based methods and a plate count method for estimation of growth parameters of Bacillus cereus. Appl. Environ. Microbiol. 76, 1399–1405. doi: 10.1128/AEM.02336-09
Brigulla, M., Hoffmann, T., Krisp, A., Völker, A., Bremer, E., and Völker, U. (2003). Chill induction of the SigB-dependent general stress response in Bacillus subtilis and its contribution to low-temperature adaptation. J. Bacteriol. 185, 4305–4314. doi: 10.1128/JB.185.15.4305-4314.2003
Cox, J., Hein, M. Y., Luber, C. A., Paron, I., Nagaraj, N., and Mann, M. (2014). Accurate proteome-wide label-free quantification by delayed normalization and maximal peptide ratio extraction, termed MaxLFQ *. Mol. Cell. Proteomics 13, 2513–2526. doi: 10.1074/mcp.M113.031591
Dagley, L. F., Infusini, G., Larsen, R. H., Sandow, J. J., and Webb, A. I. (2019). Universal solid-phase protein preparation (USP3) for bottom-up and top-down proteomics. J. Proteome Res. 18, 2915–2924. doi: 10.1021/acs.jproteome.9b00217
Davidson, C. J., and Surette, M. G. (2008). Individuality in Bacteria. Annu. Rev. Genet. 42, 253–268. doi: 10.1146/annurev.genet.42.110807.091601
Dessaux, C., Guerreiro, D. N., Pucciarelli, M. G., O’Byrne, C. P., and García-del Portillo, F. (2020). Impact of osmotic stress on the phosphorylation and subcellular location of Listeria monocytogenes stressosome proteins. Sci. Rep. 10:20837. doi: 10.1038/s41598-020-77738-z
Feng, Y., Bui, T. P. N., Stams, A. J. M., Boeren, S., Sánchez-Andrea, I., and de Vos, W. M. (2022). Comparative genomics and proteomics of Eubacterium maltosivorans: functional identification of trimethylamine methyltransferases and bacterial microcompartments in a human intestinal bacterium with a versatile lifestyle. Environ. Microbiol. 24, 517–534. doi: 10.1111/1462-2920.15886
Galaxy Community (2022). The galaxy platform for accessible, reproducible, and collaborative biomedical analyses: 2022 update. Nucleic Acids Res. 50, W345–W351. doi: 10.1093/nar/gkac247
Guariglia-Oropeza, V., Orsi, R. H., Guldimann, C., Wiedmann, M., and Boor, K. J. (2018). The Listeria monocytogenes bile stimulon under acidic conditions is characterized by strain-specific patterns and the upregulation of motility, cell wall modification functions, and the PrfA regulon. Front. Microbiol. 9:120. doi: 10.3389/fmicb.2018.00120
Guerreiro, D. N., Arcari, T., and O’Byrne, C. P. (2020a). The σ(B)-mediated general stress response of Listeria monocytogenes: life and death decision making in a pathogen. Front. Microbiol. 11:1505. doi: 10.3389/fmicb.2020.01505
Guerreiro, D. N., Pucciarelli, M. G., Tiensuu, T., Gudynaite, D., Boyd, A., Johansson, J., et al. (2022a). Acid stress signals are integrated into the σB-dependent general stress response pathway via the stressosome in the food-borne pathogen Listeria monocytogenes. PLoS Pathog. 18:e1010213. doi: 10.1371/journal.ppat.1010213
Guerreiro, D. N., Wu, J., Dessaux, C., Oliveira, A. H., Tiensuu, T., Gudynaite, D., et al. (2020b). Mild stress conditions during laboratory culture promote the proliferation of mutations that negatively affect sigma B activity in Listeria monocytogenes. J. Bacteriol. 202, e00751–e00819. doi: 10.1128/JB.00751-19
Guerreiro, D. N., Wu, J., McDermott, E., Garmyn, D., Dockery, P., Boyd, A., et al. (2022b). In vitro evolution of Listeria monocytogenes reveals selective pressure for loss of SigB and AgrA function at different incubation temperatures. Appl. Environ. Microbiol. 88:e0033022. doi: 10.1128/aem.00330-22
Hain, T., Hossain, H., Chatterjee, S. S., Machata, S., Volk, U., Wagner, S., et al. (2008). Temporal transcriptomic analysis of the Listeria monocytogenes EGD-e sigmaB regulon. BMC Microbiol. 8:20. doi: 10.1186/1471-2180-8-20
Holtmann, G., Brigulla, M., Steil, L., Schütz, A., Barnekow, K., Völker, U., et al. (2004). RsbV-independent induction of the SigB-dependent general stress regulon of Bacillus subtilis during growth at high temperature. J. Bacteriol. 186, 6150–6158. doi: 10.1128/JB.186.18.6150-6158.2004
Hsieh, Y.-J., and Wanner, B. L. (2010). Global regulation by the seven-component pi signaling system. Curr. Opin. Microbiol. 13, 198–203. doi: 10.1016/j.mib.2010.01.014
Kazmierczak, M. J., Mithoe, S. C., Boor, K. J., and Wiedmann, M. (2003). Listeria monocytogenes sigma B regulates stress response and virulence functions. J. Bacteriol. 185, 5722–5734. doi: 10.1128/JB.185.19.5722-5734.2003
Kint, C., Verstraeten, N., Hofkens, J., Fauvart, M., and Michiels, J. (2014). Bacterial Obg proteins: GTPases at the nexus of protein and DNA synthesis. Crit. Rev. Microbiol. 40, 207–224. doi: 10.3109/1040841X.2013.776510
Koomen, J. (2022). On the role of ribosomal proteins in stress resistance and fitness of Listeria monocytogenes: a laboratory evolution approach. Wageningen, the Netherlands: Wageningen University. Available at: https://edepot.wur.nl/557306
Koomen, J., Den Besten, H. M. W., Metselaar, K. I., Tempelaars, M. H., Wijnands, L. M., Zwietering, M. H., et al. (2018). Gene profiling-based phenotyping for identification of cellular parameters that contribute to fitness, stress-tolerance, and virulence of Listeria monocytogenes variants. Int. J. Food Microbiol. 283, 14–21. doi: 10.1016/j.ijfoodmicro.2018.06.003
Koomen, J., Huijboom, L., Ma, X., Tempelaars, M. H., Boeren, S., Zwietering, M. H., et al. (2021). Amino acid substitutions in ribosomal protein RpsU enable switching between high fitness and multiple-stress resistance in Listeria monocytogenes. Int. J. Food Microbiol. 351:109269. doi: 10.1016/j.ijfoodmicro.2021.109269
Liu, Y., Orsi, R. H., Boor, K. J., Wiedmann, M., and Guariglia-Oropeza, V. (2017). Home alone: elimination of all but one alternative sigma factor in Listeria monocytogenes allows prediction of new roles for σ(B). Front. Microbiol. 8:1910. doi: 10.3389/fmicb.2017.01910
Liu, Y., Orsi, R. H., Gaballa, A., Wiedmann, M., Boor, K. J., and Guariglia-Oropeza, V. (2019). Systematic review of the Listeria monocytogenes B regulon supports a role in stress response, virulence, and metabolism. Future Microbiol. 14, 801–828. doi: 10.2217/fmb-2019-0072
Mattila, M., Somervuo, P., Korkeala, H., Stephan, R., and Tasara, T. (2020). Transcriptomic and phenotypic analyses of the sigma B-dependent characteristics and the synergism between sigma B and sigma L in Listeria monocytogenes EGD-e. Microorganisms 8:1644. doi: 10.3390/microorganisms8111644
Metselaar, K. I., Abee, T., Zwietering, M. H., and Den Besten, H. M. W. (2016). Modeling and validation of the ecological behavior of wild-type Listeria monocytogenes and stress-resistant variants. Appl. Environ. Microbiol. 82, 5389–5401. doi: 10.1128/AEM.00442-16
Metselaar, K. I., Den Besten, H. M. W., Abee, T., Moezelaar, R., and Zwietering, M. H. (2013). Isolation and quantification of highly acid resistant variants of Listeria monocytogenes. Int. J. Food Microbiol. 166, 508–514. doi: 10.1016/j.ijfoodmicro.2013.08.011
Metselaar, K. I., Den Besten, H. M. W., Boekhorst, J., Van Hijum, S. A. F. T., Zwietering, M. H., and Abee, T. (2015). Diversity of acid stress resistant variants of Listeria monocytogenes and the potential role of ribosomal protein S21 encoded by rpsU. Front. Microbiol. 6:422. doi: 10.3389/fmicb.2015.00422
NicAogáin, K., and O’Byrne, C. P. (2016). The role of stress and stress adaptations in determining the fate of the bacterial pathogen Listeria monocytogenes in the food chain. Front. Microbiol. 7:1865. doi: 10.3389/fmicb.2016.01865
Nystrom, T. (2004). Growth versus maintenance: a trade-off dictated by RNA polymerase availability and sigma factor competition? Mol. Microbiol. 54, 855–862. doi: 10.1111/j.1365-2958.2004.04342.x
O’Byrne, C. P., and Karatzas, K. A. G. (2008). “Chapter 5—the role of sigma B ( B) in the stress adaptations of Listeria monocytogenes: overlaps between stress adaptation and virulence” in Advances in Applied Microbiology. eds. A. I. Laskin, S. Sariaslani, and G. M. Gadd (United States: Academic Press), 115–140.
O’Donoghue, B., NicAogáin, K., Bennett, C., Conneely, A., Tiensuu, T., Johansson, J., et al. (2016). Blue-light inhibition of Listeria monocytogenes growth is mediated by reactive oxygen species and is influenced by σB and the blue-light sensor Lmo0799. Appl. Environ. Microbiol. 82, 4017–4027. doi: 10.1128/AEM.00685-16
Oliveira, A. H., Tiensuu, T., Guerreiro, D. N., Tükenmez, H., Dessaux, C., García-Del Portillo, F., et al. (2022). Listeria monocytogenes requires the RsbX protein to prevent SigB activation under nonstressed conditions. J. Bacteriol. 204:e0048621. doi: 10.1128/JB.00486-21
Oliver, H. F., Orsi, R. H., Wiedmann, M., and Boor, K. J. (2010). Listeria monocytogenes {sigma}B has a small core regulon and a conserved role in virulence but makes differential contributions to stress tolerance across a diverse collection of strains. Appl. Environ. Microbiol. 76, 4216–4232. doi: 10.1128/AEM.00031-10
Ollinger, J., Bowen, B., Wiedmann, M., Boor, K. J., and Bergholz, T. M. (2009). Listeria monocytogenes sigmaB modulates PrfA-mediated virulence factor expression. Infect. Immun. 77, 2113–2124. doi: 10.1128/IAI.01205-08
Österberg, S., Peso-Santos, T.Del, and Shingler, V. (2011). Regulation of alternative sigma factor use. Ann. Rev. Microbiol. 65, 37–55. doi: 10.1146/annurev.micro.112408.134219
Pathak, D., Jin, K. S., Tandukar, S., Kim, J. H., Kwon, E., and Kim, D. Y. (2020). Structural insights into the regulation of SigB activity by RsbV and RsbW. IUCrJ 7, 737–747. doi: 10.1107/S2052252520007617
Perez-Riverol, Y., Bai, J., Bandla, C., García-Seisdedos, D., Hewapathirana, S., Kamatchinathan, S., et al. (2022). The PRIDE database resources in 2022: a hub for mass spectrometry-based proteomics evidences. Nucleic Acids Res. 50, D543–D552. doi: 10.1093/nar/gkab1038
Quereda, J. J., Morón-García, A., Palacios-Gorba, C., Dessaux, C., García-del Portillo, F., Pucciarelli, M. G., et al. (2021). Pathogenicity and virulence of Listeria monocytogenes: a trip from environmental to medical microbiology. Virulence 12, 2509–2545. doi: 10.1080/21505594.2021.1975526
Radoshevich, L., and Cossart, P. (2018). Listeria monocytogenes: towards a complete picture of its physiology and pathogenesis. Nat. Rev. Microbiol. 16, 32–46. doi: 10.1038/nrmicro.2017.126
Ryall, B., Eydallin, G., and Ferenci, T. (2012). Culture history and population heterogeneity as determinants of bacterial adaptation: the adaptomics of a single environmental transition. Microbiol. Mol. Biol. Rev. 76, 597–625. doi: 10.1128/MMBR.05028-11
Rychli, K., Wagner, E., Guinane, C. M., Daly, K., Hill, C., and Cotter, P. D. (2021). “Generation of nonpolar deletion mutants in Listeria monocytogenes using the ‘SOEing’ method” in Listeria monocytogenes: Methods and Protocols Methods in Molecular Biology. eds. E. M. Fox, H. Bierne, and B. Stessl, vol. 2220 (New York, NY: Springer US), 165–175.
Santos-Beneit, F. (2015). The pho regulon: a huge regulatory network in bacteria. Front. Microbiol. 6:402. doi: 10.3389/fmicb.2015.00402
Scott, J. M., and Haldenwang, W. G. (1999). Obg, an essential GTP binding protein of Bacillus subtilis, is necessary for stress activation of transcription factor B. J. Bacteriol. 181, 4653–4660. doi: 10.1128/JB.181.15.4653-4660.1999
Shi, L., Pigeonneau, N., Ravikumar, V., Dobrinic, P., Macek, B., Franjevic, D., et al. (2014). Cross-phosphorylation of bacterial serine/threonine and tyrosine protein kinases on key regulatory residues. Front. Microbiol. 5:495. doi: 10.3389/fmicb.2014.00495
Shin, J.-H., Brody, M. S., and Price, C. W. (2010). Physical and antibiotic stresses require activation of the RsbU phosphatase to induce the general stress response in Listeria monocytogenes. Microbiology 156, 2660–2669. doi: 10.1099/mic.0.041202-0
Smith, K., and Youngman, P. (1992). Use of a new integrational vector to investigate compartment-specific expression of the Bacillus subtilis spollM gene. Biochimie 74, 705–711. doi: 10.1016/0300-9084(92)90143-3
Smits, W. K., Kuipers, O. P., and Veening, J.-W. (2006). Phenotypic variation in bacteria: the role of feedback regulation. Nat. Rev. Microbiol. 4, 259–271. doi: 10.1038/nrmicro1381
Sohmen, D., Chiba, S., Shimokawa-Chiba, N., Innis, C. A., Berninghausen, O., Beckmann, R., et al. (2015). Structure of the Bacillus subtilis 70S ribosome reveals the basis for species-specific stalling. Nat. Commun. 6:6941. doi: 10.1038/ncomms7941
Toledo-Arana, A., Dussurget, O., Nikitas, G., Sesto, N., Guet-Revillet, H., Balestrino, D., et al. (2009). The Listeria transcriptional landscape from saprophytism to virulence. Nature 459, 950–956. doi: 10.1038/nature08080
Torsten, S . (2015). Snippy: fast bacterial variant calling from NGS reads. Available at: https://github.com/tseemann/snippy
Tyanova, S., Temu, T., Sinitcyn, P., Carlson, A., Hein, M. Y., Geiger, T., et al. (2016). The Perseus computational platform for comprehensive analysis of (prote)omics data. Nat. Methods 13, 731–740. doi: 10.1038/nmeth.3901
Utratna, M., Cosgrave, E., Baustian, C., Ceredig, R., and O’Byrne, C. (2012). Development and optimization of an EGFP-based reporter for measuring the general stress response in Listeria monocytogenes. Bioengineered 3, 93–103. doi: 10.4161/bbug.19476
Utratna, M., Cosgrave, E., Baustian, C., Ceredig, R. H., and O’Byrne, C. P. (2014). Effects of growth phase and temperature on σB activity within a Listeria monocytogenes population: evidence for RsbV-independent activation of σB at refrigeration temperatures. Biomed. Res. Int. 2014:641647. doi: 10.1155/2014/641647
Vaestermark, A., and Saier, M. H. (2014). The involvement of transport proteins in transcriptional and metabolic regulation. Curr. Opin. Microbiol. 18, 8–15. doi: 10.1016/j.mib.2014.01.002
Van Duin, J., and Wijnands, R. (1981). The function of ribosomal protein S21 in protein synthesis. Eur. J. Biochem. 118, 615–619. doi: 10.1111/j.1432-1033.1981.tb05563.x
Verstraeten, N., Fauvart, M., Versées, W., and Michiels, J. (2011). The universally conserved prokaryotic GTPases. Microbiol. Mol. Biol. Rev. 75, 507–542. doi: 10.1128/MMBR.00009-11
Vijay, K., Brody, M. S., Fredlund, E., and Price, C. W. (2000). A PP2C phosphatase containing a PAS domain is required to convey signals of energy stress to the sigma(B) transcription factor of Bacillus subtilis. Mol. Microbiol. 35, 180–188. doi: 10.1046/j.1365-2958.2000.01697.x
Wendrich, J. R., Boeren, S., Möller, B. K., Weijers, D., and De Rybel, B. (2017). “In vivo identification of plant protein complexes using IP-MS/MS” in Plant Hormones: Methods and Protocols Methods in Molecular Biology. eds. J. Kleine-Vehn and M. Sauer (New York, NY: Springer), 147–158.
Wu, T., Hu, E., Xu, S., Chen, M., Guo, P., Dai, Z., et al. (2021). clusterProfiler 4.0: a universal enrichment tool for interpreting omics data. Innovation 2:100141. doi: 10.1016/j.xinn.2021.100141
Xia, Y., Xin, Y., Li, X., and Fang, W. (2016). To modulate survival under secondary stress conditions, Listeria monocytogenes 10403S employs RsbX to downregulate sigma(B) activity in the poststress recovery stage or stationary phase. Appl. Environ. Microbiol. 82, 1126–1135. doi: 10.1128/AEM.03218-15
Keywords: population heterogeneity, pathogen, variant, stress resistance, fitness
Citation: Ma X, Tempelaars MH, Zwietering MH, Boeren S, O’Byrne CP, den Besten HMW and Abee T (2024) A single point mutation in the Listeria monocytogenes ribosomal gene rpsU enables SigB activation independently of the stressosome and the anti-sigma factor antagonist RsbV. Front. Microbiol. 15:1304325. doi: 10.3389/fmicb.2024.1304325
Received: 02 October 2023; Accepted: 21 February 2024;
Published: 12 March 2024.
Edited by:
Ioana Cristina Marinas, University of Bucharest, RomaniaReviewed by:
Qingli Dong, University of Shanghai for Science and Technology, ChinaCopyright © 2024 Ma, Tempelaars, Zwietering, Boeren, O’Byrne, den Besten and Abee. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Tjakko Abee, dGpha2tvLmFiZWVAd3VyLm5s
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.