- 1Laboratory of Molecular Biology and Genetic Engineering, School of Biotechnology, Jawaharlal Nehru University, New Delhi, India
- 2Divacc Research Laboratories Pvt. Ltd., incubated under Atal Incubation Centre, Jawaharlal Nehru University, New Delhi, India
The prevalence of bacterial persisters is related to their phenotypic diversity and is responsible for the relapse of chronic infections. Tolerance to antibiotic therapy is the hallmark of bacterial persistence. In this study, we have screened a transposon library of Mycobacterium smegmatis mc2155 strain using antibiotic tolerance, survival in mouse macrophages, and biofilm-forming ability of the mutants. Out of 10 thousand clones screened, we selected ten mutants defective in all the three phenotypes. Six mutants showed significantly lower persister abundance under different stress conditions. Insertions in three genes belonging to the pathways of oxidative phosphorylation msmeg_3233 (cydA), biotin metabolism msmeg_3194 (bioB), and oxidative metabolism msmeg_0719, a flavoprotein monooxygenase, significantly reduced the number of live cells, suggesting their role in pathways promoting long-term survival. Another group that displayed a moderate reduction in CFU included a glycosyltransferase, msmeg_0392, a hydrogenase subunit, msmeg_2263 (hybC), and a DNA binding protein, msmeg_2211. The study has revealed potential candidates likely to facilitate the long-term survival of M. smegmatis. The findings offer new targets to develop antibiotics against persisters. Further, investigating the corresponding genes in M. tuberculosis may provide valuable leads in improving the treatment of chronic and persistent tuberculosis infections.
Introduction
Mycobacterium tuberculosis, the etiologic agent of tuberculosis, is reported to infect 2.6 million people in India alone (World Health Organization, 2021). Majority of the cases (90%) constitute a latent or persistent form of infection with no apparent clinical signs or symptoms (Fauvart et al., 2011; World Health Organization, 2021) and comprise the transmissible form of infectious bacteria with the ability to reactivate the disease. Tolerance and resistance are other phenotypes that are sometimes confused with the persistence (Brauner et al., 2016; Schrader et al., 2020). The latter population presents a formidable challenge in eradicating the disease (Zhang, 2014). The molecular mechanisms leading to the development of persistence are not fully understood due to several reasons. They constitute a small, heterogeneous population that is transient in nature and changes with the environment. Secondly, lack of a biological model that can mimic persistence (Zhang, 2014), and finally, due to the redundancy of mechanisms that select persisters. They constitute a subpopulation of bacteria with non-heritable characteristics that enable them to adapt to stressful conditions for long-term survival. They have the same genetic makeup as the drug-sensitive parent population, indicating that bacterial persistence is an epigenetic trait (Bigger, 1944; Keren et al., 2004; Lewis, 2010; Zhang, 2014; Torrey et al., 2016).
Persister cells are classified into three main categories: spontaneous persisters, triggered persisters, and specialized persisters (Urbaniec et al., 2022). Spontaneous persisters are slow-growing cells formed during the logarithmic phase, independent of any environmental trigger. These persisters are pre-existing in bacterial culture. The triggered persisters constitute non-growing cells produced in the stationary phase of growth due to environmental stress and exhibit an extended lag phase upon inoculation into fresh medium (Balaban et al., 2019). The specialized persisters evolve in response to specific antibiotics (Urbaniec et al., 2022). In Escherichia coli, tolerance to fluoroquinolones occurs by extension of the lag phase through growth-inhibiting toxin components (MazF or HipA) of the toxin-antitoxin (TA) systems (Mok and Brynildsen, 2018). Similarly, exposure of E. coli to ciprofloxacin reduces the proton motive force (PMF) and ATP levels in the cell, which decreases antibiotic transport, inducing drug-specific persistence (Dörr et al., 2009). In mycobacteria, variation in the levels of the catalase-peroxidase enzymes, necessary for activation of isoniazid, causes a reduction in the effective concentration of the drug, leading to drug-specific persistence (Wakamoto et al., 2013).
Biofilm formation by pathogens has emerged as one of the essential attributes contributing to the establishment of persistent infections in the host (Helaine et al., 2014). The shreds of evidence from in vitro experiments revealed an abundance of persisters in the biofilms; a link between the two is established, suggesting a significant role in relapses and recalcitrance of pathogenic bacteria (Keren et al., 2004; Lafleur et al., 2010; Conlon et al., 2013). The presence of persisters is believed to be responsible for antibiotic treatment failure (Van den Bergh et al., 2016; Levin-Reisman et al., 2017). It is widely accepted now that the underlying reasons behind the emergence of antibiotic-tolerant or persister cells are most likely multifactorial, especially within the host, where different interconnected metabolic processes influence each other.
Persisters have been identified in the host-dependent bacterial species, e.g., Pseudomonas aeruginosa, Salmonella enterica serovar Typhimurium, Streptococcus suis, E. coli, and M. tuberculosis, etc., (Helaine et al., 2014; Michiels et al., 2016). To counter host defense mechanisms, M. tuberculosis is reported to express various phenotypes such as biofilm growth (Chakraborty et al., 2021), tolerance to antibiotics (Ciofu et al., 2017), granuloma formation (Barry et al., 2009), survival in the host macrophage (Helaine et al., 2014), and immune evasion (Domingue et al., 2020). The hostile and restrictive environments encountered within the host macrophages and granulomatous lesions force manifestation of phenotypic heterogeneity, which enables a small population to survive/persist longer (Helaine et al., 2014; Mouton et al., 2016; Fisher et al., 2017). In-vitro models simulating the stress conditions experienced by mycobacteria inside the host were shown to change the metabolic state of the bacteria and generate a quiescent reservoir of bacterial cells promoting latent infection (Zhang, 2014; Lipworth et al., 2016; Joshi et al., 2021).
To identify genes regulating persistence in different bacterial species, mutagenesis is employed, which results in either decreased (e.g., ybaL, relA, phoU, sucB, ubiF, and lamA) or increased (e.g., hipA, metG, tktA, glpD, and hupB) survival propensity (Li and Zhang, 2007; Ma et al., 2010; Rego et al., 2017; Hingley-Wilson et al., 2020). These genes regulate various bacterial pathways such as TA systems, SOS response, signalling pathways, stringent response, antioxidant defence, alternative energy production, enhanced efflux or transporter activity, and phosphate metabolism, etc., (Zhang, 2014). Apparently, persisters are formed through independent parallel mechanisms, providing a significant adaptive advantage to the organism. This redundant design works in favor of the organism, as no single compound can abolish persister formation.
Latency or persistence is frequently observed in pathogenic and non-pathogenic species of mycobacteria. The fast-growing, non-pathogenic Mycobacterium smegmatis, which shares 70% homology with the pathogenic M. tuberculosis, is often used as a model to study persistence genes (Malhotra et al., 2017; Ranjitha et al., 2020). The present study is an attempt at enhancing our understanding of mycobacterial persistence by screening a transposon mutant library of M. smegmatis mc2155 strain using three key phenotypes commonly associated with persistence - antibiotic tolerance, biofilm formation, and survival in macrophages to identify genes that influence the survival of the bacteria. Our findings have revealed novel targets, which might play a role in the adaptation of M. smegmatis leading to persistence.
Materials and methods
Bacterial strains, media, growth conditions, and plasmids
The M. smegmatis mc2155 strain was used to construct and screen a transposon library. Cultures were grown in Middlebrook 7H9 liquid Broth or 7H10 Agar (Difco) solid medium enriched with 10% oleic acid-albumin-dextrose-catalase (OADC) supplement, glycerol (0.5%), and Tween-80 (0.05%). E. coli DH5α λ pir strain was used for all the cloning experiments and as the host for sequencing the transposon insertions. The chemicals used in the study were purchased from Sigma-Aldrich. A stock of antibiotics was prepared (Andrews, 2001), and dilutions were made using a 7H9 broth medium. All the strains were cultured in triplicate, for each experiment.
Determination of minimum inhibitory concentration of antibiotics
The MIC was determined using resazurin end point microtiter assay (REMA) (Taneja and Tyagi, 2007) with some modifications. The culture was grown till the mid-exponential phase (OD ∼ 0.6−0.8) and diluted 1:100 for the assay. The initial dilutions of the antibiotics were prepared in either DMSO or sterile deionized water, and successive 2-fold dilutions were done in 100 μl of 7H9 medium supplemented with 0.05% glycerol (without Tween-80) in the microtiter plates. Antibiotic concentrations ranged from 0.005 to 4 μg/ml. In a total volume of 200 μl, each well contained 100 μl of culture containing 1 x 104 CFU and 100 μl of medium containing the antibiotic. A positive control containing bacteria alone and a negative antibiotic control without culture was used to determine the bacterial viability. The plates were sealed with Parafilm and incubated at 37°C for 1−2 days. After 2 days, 30 μl of 0.02% resazurin and 12.5 μl of 20% Tween-80 were added to each well. The colour of the medium changed from blue to pink after 2 days. The fluorescence was determined using a spectrofluorimeter by excitation at 530 nm and emission at 590 nm. The lowest concentration of the antibiotic causing 90% inhibition was considered as the MIC. The percentage inhibition of viability was calculated as shown below:
Viability inhibition (%) = 1-(test well fluorescence/mean fluorescence of positive control) x 100
The MIC was tested on solid media also. A standard culture (1 x 104 CFU) was plated onto antibiotic-containing (ranging from 0.005 to 4 μg/ml) and antibiotic-free control 7H10 agar plates in duplicate. The culture was spread with a sterile spreader, sealed with Parafilm, and incubated at 37°C for 3 days. The number of CFUs was recorded after 3 days. The number of live bacteria (CFU) was counted to calculate the MIC.
Antibiotics selection for tolerant cell generation in the wild-type M. smegmatis
Tolerant cells generated by wild-type (WT) M. smegmatis were evaluated by replica plating the colonies obtained after exposure of the culture to specific antibiotics. M. smegmatis was treated with the antibiotics at 10X and 50X MIC (minimum inhibitory concentration), isoniazid (25 and 125 μg/ml), rifampicin (6.25 and 31.25 μg/ml), levofloxacin (0.78 and 3.90 μg/ml), and moxifloxacin (0.039 and 0.195 μg/ml) for 3 days. Samples were taken at different stages of growth, and plated on 7H10 agar medium. The colonies obtained in the previous step were replica plated with and without the drug to obtain the number of tolerant cells. The difference between the total number of colonies and the colonies able to grow in the presence of the antibiotic on sub-culturing were considered as the tolerant population for plotting a time-kill curve.
Construction of transposon mutant library
A mutant library of M. smegmatis mc2155 was constructed by random transposon insertion mutagenesis using a φMycomarT7 phage carrying Himar1 transposon. The transposon was transferred to M. smegmatis by transduction according to Lamichhane et al. (2003) with minor changes – M. smegmatis was grown in 7H9 containing ADC to OD600 of 1.2 (Lamichhane et al., 2003). The bacterial cells were centrifuged, washed two times in mycobacteriophage (MP) buffer, and the pellet was resuspended in 8 ml of the MP buffer. An aliquot was saved and used as a control for CFU counts. The cell suspension was pre-warmed to 37°C in a water bath and was mixed with approximately 2 ml of 1 x 1011 PFU (Plaque forming unit) or MP buffer control. The mixture was plated onto 15 cm 7H10-OADC agar plates (five to ten in number) containing 0.05% Tween-80 and 25 μg/ml kanamycin and incubated at 37°C for 2−3 days. The titer of transduction in M. smegmatis was ∼1.36 x 105 kanamycin-resistant colonies. After harvesting the mutant colonies from the plates, they were resuspended in 7H9-OADC media containing Tween-80 (0.05%) and glycerol (15%). For storage, the cells were resuspended by sonication in water baths for two cycles of 5 s each and stored in aliquots of 1 ml at −80°C. Transposon insertion was checked by colony PCR, using forward and reverse primers (Table 1).
Selection of transposon mutants
A glycerol stock of the mutant library was inoculated in a 7H9 medium supplemented with kanamycin (25 μg/ml), grown at 37°C for 24 h, and spread on 7H10 agar plates. Based on their size, the colonies were classified as small, medium, and large. In the initial elimination stage, the segregated colonies were screened based on biofilm formation and antibiotic tolerance as follows. The colonies were grown in OADC supplemented 7H9 medium containing kanamycin in 96-well microtiter plates at 37°C for 2 days. The cultures were diluted 100 times into fresh 7H9 medium in 96-well plates. The plates were incubated at 37°C for 3 days till the OD600 reached the stationary phase. Levofloxacin was added to each well at a concentration of 3.9 μg/ml (50X), and incubation continued. Growth was monitored by measuring OD600 on days 3 and 6 and simultaneously streaked on 7H10 supplemented agar plates with no antibiotic to observe the extent of growth inhibition. The antibiotic tolerance assay was repeated with a second fluoroquinolone antibiotic, moxifloxacin. The mutants showing higher susceptibility to both levofloxacin and moxifloxacin after 3 or 6 days of exposure were selected. The antibiotic exposure was repeated to confirm the sensitivity of the mutant phenotype. Next, the selected mutants from the previous step were subjected to two cycles of biofilm formation as the second selection criteria. Those showing a defect in both criteria were subjected to the next round of tests.
Identification of transposon insertion sites
Genomic DNA was digested using SacII restriction enzyme, and the fragments were self-ligated as plasmid for cloning. The competent cells of E. coli DH5α λ pir strain were transformed with the ligated mixture and spread on LB agar plates containing 50 μg/ml of kanamycin. The plates were incubated overnight at 37°C, and individual colonies were grown in 5 ml LB media with shaking at 50 μg/ml kanamycin overnight. Plasmid DNA was isolated from the cultures and sequenced with a primer designed to anneal with the 3’ end of the aph gene cassette in the transposon to determine the insertion site (Table 1). BLAST was used to identify the genes disrupted by insertion, with M. smegmatis mc2155 as the reference genome.
Preparation of plasmids for genetic complementation
For complementation, plasmids were constructed by amplifying the selected genes with promoter sequence using M. smegmatis genomic DNA as a template with specific primers containing a 5’-BamHI restriction site and a HindIII restriction site at the 3’ end. The amplified products were digested with BamHI and HindIII restriction enzymes and ligated in pMV261 vector. The plasmids were electroporated in the respective mutant strains of M. smegmatis. The WT strain containing the pMV261 plasmid alone was used as a control.
Growth determination
M. smegmatis and its transposon mutants were grown in triplicate in standard 7H9 liquid medium, and samples were removed at regular intervals for measuring the absorbance at 600 nm. The data presented are the mean values from three individual experiments with standard deviations.
Influence of antibiotic time and dose on tolerance development
The strains were grown in the presence of inhibitory concentrations of levofloxacin and isoniazid and variation in the number of live/tolerant cells with time was measured. The primary cultures were grown in 10 ml of 7H9 medium containing kanamycin (25 μg/ml) at 37°C for 3 days with shaking at 150 rpm. The cultures were diluted 1:100 in 10 ml 7H9 medium and allowed to grow with shaking until their OD600 reached ∼ 1.8. These growth conditions were adopted for all the following experiments. An aliquot was removed to determine the CFU as a zero time point of untreated bacterial cultures. 10X MIC of levofloxacin (0.78 μg/ml) and isoniazid (25 μg/ml) were added to all the M. smegmatis variant strains and incubated for 6 days at 37°C with shaking. For CFU counts, aliquots were removed every 24 h and plated on 7H10 agar plates supplemented with OADC. The live cells were replica plated with and without the respective antibiotics on 7H10 agar plates and incubated at 37°C.
In another experiment, the WT M. smegmatis, the mutants, and their complemented strains were grown in triplicate to stationary phase, as described above. An aliquot was plated to record the CFU count of the untreated cultures. To measure the dose dependence of the strains, levofloxacin and isoniazid were added at different concentrations (10X, 20X, 30X, 40X, and 50X MIC) to all the M. smegmatis cultures. After incubation for 3 days at 37°C, the cultures were harvested and plated after serial dilution in 7H10 agar plates supplemented with OADC. The live cells were replica plated with and without antibiotics as described above, and the CFU counts were recorded.
Biofilm formation assay
To examine biofilm formation, M. smegmatis strains were cultured in Sauton’s medium (without Tween-80) (Ojha et al., 2008). 10 μl of 3 days saturated cultures were inoculated in 1 ml of the medium and grown in 24-well, flat bottom culture plates at 37°C under stationary conditions. After 4 days of incubation, the medium and the non-adherent bacteria were removed carefully by suction, and the wells were washed three times with sterile distilled water. The attached bacteria forming the biofilm were stained for 30 min with 500 μl of 1% crystal violet (CV) at room temperature, rinsed three times with distilled water, and allowed to air dry. The stained biofilm was dissolved in 500 μl of 95% ethanol at 37°C for 30 min and used to measure optical density at 595 nm in an ELISA plate reader (TECAN, Sunrise AZreader). The assay was repeated at least three times with all the strains.
Intracellular survival of the M. smegmatis variants in murine macrophages
To measure the rate of clearance of the mutants in cultured macrophages, RAW 264.7 macrophages were seeded at a density of 1 x 105 cells per well in 24-well culture plates and incubated for 24 h. Mid-logarithmic phase bacterial strains were centrifuged, rinsed in PBS twice, and resuspended in DMEM containing 10% FBS medium to achieve OD600 ∼0.1. The bacterial suspension was kept in an ultrasonic water bath for 15 min to disaggregate the clumps and used to infect the macrophages at an MOI (multiplicity of infection) of 25:1. After 2 h, the supernatant containing floating bacterial cells was removed, and the wells were rinsed with fresh medium. Gentamicin (100 μg/ml) was added to each well and incubated for 1 h to kill bacteria attached to the surface. The infected murine macrophages were removed at different time points (0, 24, and 72 h), rinsed with PBS, and lysed in autoclaved water containing 0.1% Triton X-100. The bacterial cells in the lysate were serially diluted in PBS, followed by plating on a 7H10 agar medium enriched with OADC. The plates were incubated for 3 days at 37°C. The assay was repeated at least three times.
Stress tolerance of M. smegmatis variants under in vitro conditions
To measure the ability of the mutant strains to tolerate stress conditions encountered in the host, the following tests were performed. Tolerance to oxidative stress was evaluated by treatment with hydrogen peroxide. The M. smegmatis strains were grown in supplemented 7H9 media till the stationary phase as described above. Fresh 50 ml media were inoculated with the corresponding stationary phase cultures and incubated at 37°C for 3 days. Aliquots of 5 ml were removed at 24 h intervals and treated with 10 mM hydrogen peroxide at 37°C for 2 h. An untreated culture was run in parallel as a control for comparison. At each time point, the cultures were plated and CFUs were determined.
For hypoxic stress, the M. smegmatis strains were grown in 10 ml 7H9 medium in glass tubes and incubated at 37°C with shaking at 150 rpm till the OD600 reached ∼ 0.6−0.8. The cultures were diluted 100 times in 10 ml 7H9 medium in screw-capped, flat-bottom culture tubes with 5 ml ambient air (using a headspace ratio of 0.5). Methylene blue at a concentration of 1.5 μg/ml was added to the medium before sealing to measure oxygen consumption. The culture tubes were sealed well to make them airtight and incubated with shaking at low speed at 37°C for 10 days. The depletion of oxygen was observed with the decoloration of methylene blue. Cells were harvested, plated, and the survival of the cells was determined by CFU counting.
Growth and survival of M. smegmatis transposon mutants in mice
Female C57BL/6J mice were sourced from Rodent Research India Pvt. Ltd (Haryana, India). The mice were housed in sterile autoclaved micro-isolator cages maintained at temperature (26°C), light & dark cycles (12 h each), and constant humidity (30%). The animals were provided with free access to food and water during the study. The Institutional Animal Ethics Committee and Biosafety Committees of Jawaharlal Nehru University, New Delhi, India, authorized the protocols. All the experiments were performed following the animal welfare regulations of the World Organization for Animal Health. Qualified personnel performed all experiments in a biosafety level II (BSL-II) facility at the School of Biotechnology, Jawaharlal Nehru University.
To evaluate the in vivo survival of the WT, the transposon mutants, and their complemented strains, 6-week-old female C57BL/6J mice (three per group) were used. All the bacterial strains were grown at 37°C with shaking for 2 days, harvested, and resuspended in 0.05% PBST. The mice were injected with 1 x 106 CFUs of the M. smegmatis strains intravenously via the tail vein. At each time point (1, 4, 7, 14, and 21 days), the mice were euthanized, and the lungs and spleen were removed aseptically and homogenized in sterile 0.05% PBST. The tissue homogenates were serially diluted in PBST and grown on 7H10 agar plates supplemented with 10% OADC and 25 μg/ml kanamycin. The plates were incubated at 37°C, and bacterial colonies were counted after 3−4 days.
Statistical analysis
All the experiments in this study were repeated at least three times. Graphs were prepared using Graph Pad Prism 8 (GraphPad Software, La Jolla, CA). In the graphs, the data points represent the average of three independent experiments, while the error bars represent the standard deviations. Appropriate statistical tests (i.e., One-way ANOVA and Two-way ANOVA) were performed for multiple comparisons, and a P-value of *P < 0.05, **P < 0.01, ***P < 0.005, and ****P < 0.001 was considered statistically significant.
Results
Generation and screening of transposon mutants
A library of 1.36 x 105 transposon mutants of the M. smegmatis mc2155 strain was generated by random insertion of Himar1 transposon present in φMycomarT7 phage. The Himar1 transposon requires TA dinucleotide sites in the genome for insertion. A total of 1.36 x 105 random transposon mutants generated in this study suggested an approximate 19-fold targeted transposon insertions at 4.89-log10 TA sites (Foreman et al., 2020), in 6938 ORFs present in the genome of M. smegmatis. This indicated that the constructed library is likely to be saturated. From the library, 10,000 aph-positive clones were screened, and ∼20 were randomly checked by colony PCR for aph gene (data not shown). Further screening was based on the chosen phenotypes, such as antibiotic tolerance and biofilm formation. Of the total number of selected mutants, 134 showed reduced tolerance to levofloxacin and moxifloxacin, while 107 mutants were defective in biofilm formation, and ten mutants common in the above two groups, were defective in both the phenotypes. Since the probability of persistence with a defects in two of the typical features is likely to be higher, the latter were selected for detailed studies.
Evaluation of tolerance to antibiotics in wild-type M. smegmatis
To generate the antibiotic sensitivity profile of WT M. smegmatis to select drugs for later experiments, multiple antibiotics (two first-line anti-TB drugs, isoniazid and rifampicin, and two second-line drugs, levofloxacin and moxifloxacin) were tested. After exposure of the culture to the antibiotics, response of the strain was evaluated by plating on 7H10 agar medium. Next, the colonies obtained in the previous step were replica plated to determine the number of tolerant cells. M. smegmatis cells that survived the initial exposure to isoniazid and rifampicin were less sensitive, as the number of live cells remained relatively high after 3 days (Supplementary Table 1). However, in the case of isoniazid, the percentage of resistant cells was relatively much smaller than the tolerant population, while treatment with rifampicin produced 80−90% of resistant cells in all the growth phases (Supplementary Table 1). Levofloxacin and moxifloxacin antibiotics were more effective in bacterial killing, but like isoniazid, they resulted in a higher proportion of tolerant than resistant cells (Supplementary Table 1). The number of surviving bacteria at 10X or 50X MICs was almost similar in all the antibiotics, suggesting that the peak antibiotic sensitivity was attained at 10X level (data not shown). Based on the above observations, isoniazid and levofloxacin at 10X MIC were selected for screening of the transposon mutants.
Identification of genes involved in defective phenotypes
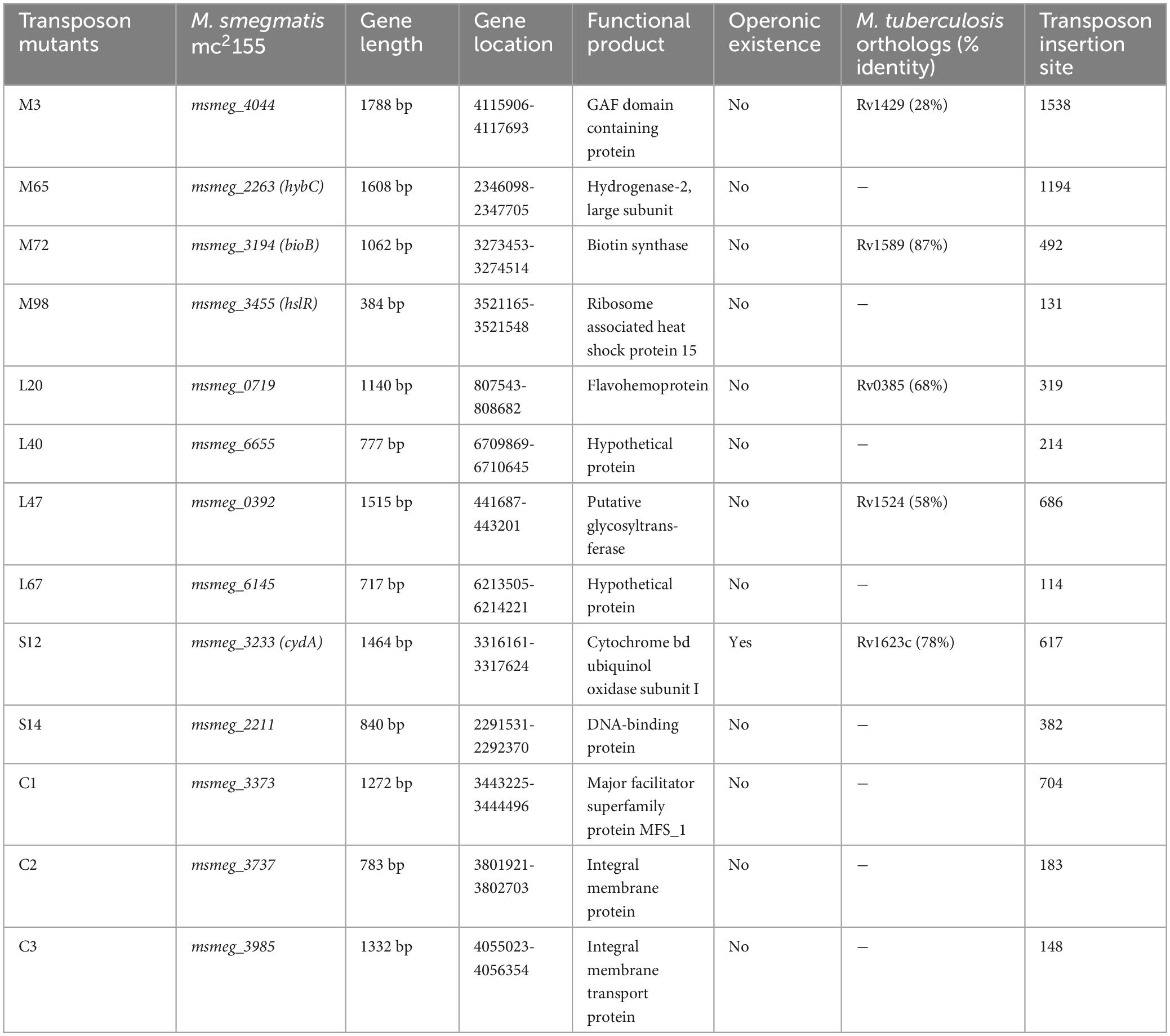
DNA sequencing of the mutants revealed the genes disrupted by transposon insertion (Table 2). The information on gene length, its location within the genome, and the putative functional product of the identified genes are shown in Table 2. Mutant strains C1, with phenotypes (antibiotic resistant, biofilm negative), C2 (antibiotic resistant, biofilm positive), and C3 (antibiotic sensitive, biofilm positive) were used as controls in the stress endurance assays. Some of the genes belonged to different pathways, such as biotin biosynthesis, cellular metabolism, glycan modifications, oxidative phosphorylation, etc. The functional aspect of the genes was derived from the KEGG (Kyoto Encyclopaedia of Genes and Genomes) database.
General properties of the selected transposon mutants
To characterize the transposon mutants, the growth profile of the strains was examined, which showed an identical pattern with WT M. smegmatis (Supplementary Figure 1), and the MIC values of the antibiotics - isoniazid, rifampicin, levofloxacin, and moxifloxacin, were also not significantly different from the parental strain (Supplementary Table 2). These results suggest that the reduction in tolerance of the transposon mutants is not due to reduced growth rate or greater antibiotic sensitivity, but independent of the growth phase, antibiotic type, and antibiotic concentration.
Kinetics of variation of antibiotic tolerance with time
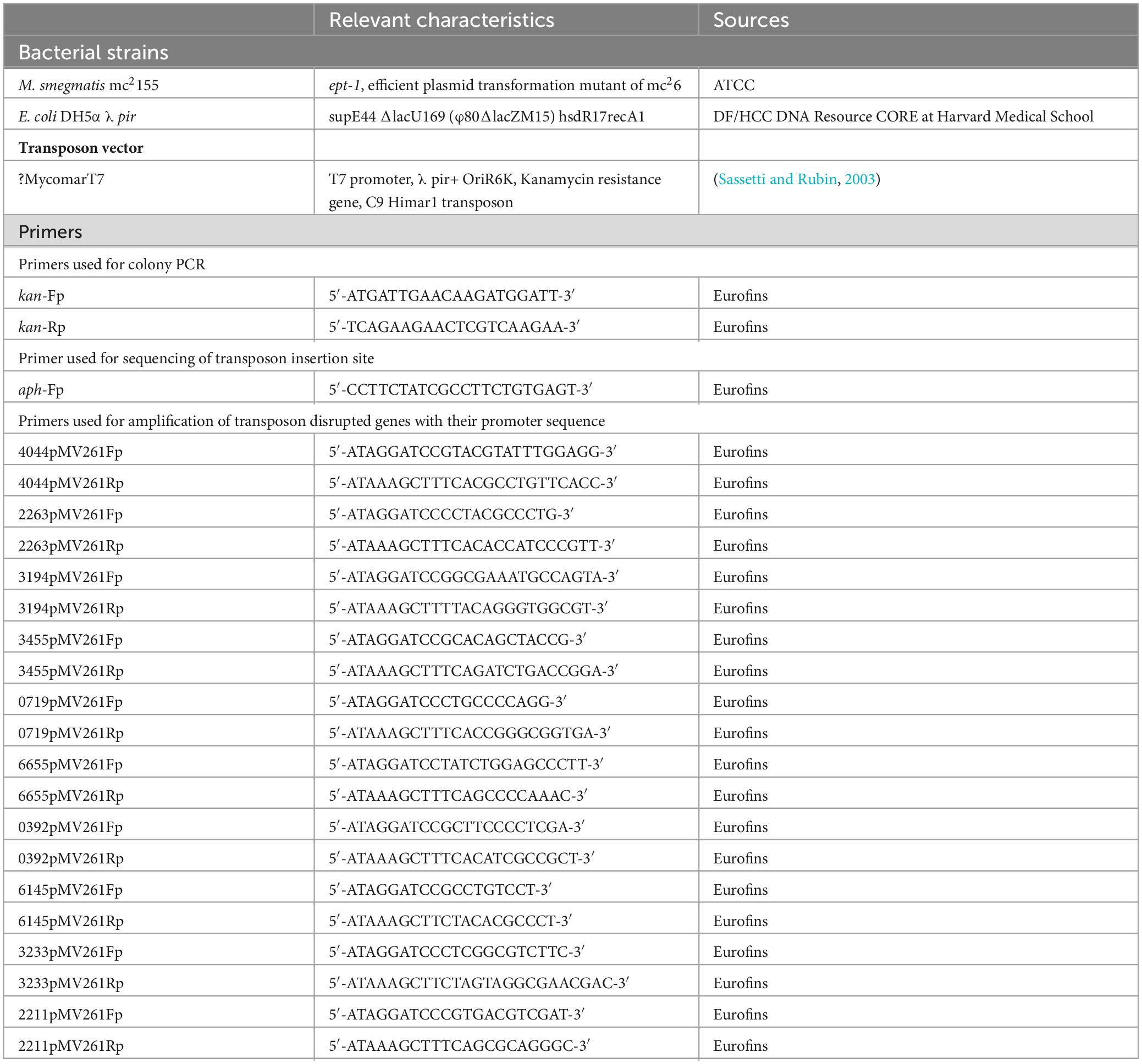
To evaluate the antibiotic tolerance of the strains with time, the live cell count of the selected mutants was measured for 6 days in the presence of 10X MIC of the antibiotics. Isoniazid and levofloxacin were used followed by replica plating to exclude the resistant from the tolerant population. A time-kill curve of levofloxacin is shown in Figure 1. Out of the ten mutants, CFU counts in the msmeg_3233 (cydA), msmeg_0392, msmeg_6655, msmeg_2211, msmeg_0719, msmeg_3194 (bioB), msmeg_6145, msmeg_3455 (hslR), and msmeg_2263 (hybC) strains were 2.19-log, 1.91-log, 1.02-log, 0.84-log, 0.83-log, 0.76-log, 0.76-log, 0.7-log, and 0.66-log, respectively, fold lower than the parental strain. The complemented strains showed the same growth kinetics as the WT strain. Likewise, time-kill curves at 10X MIC of isoniazid after 3 days showed a reduction in the number of live/tolerant cells of 1.97-log, 0.95-log, 0.92-log, 0.89-log, 0.83-log, 0.71-log, 0.71-log, 0.65-log, and 0.43-log, in the cydA, msmeg_0392, msmeg_6655, bioB, hslR, msmeg_2211, msmeg_6145, msmeg_0719, and hybC mutant strains, respectively, compared to the parental strain (Figure 2). No significant difference in the number of live cells was observed in the msmeg_4044 mutant compared to the parental strain in response to either antibiotic. In most cases, the maximum reduction was recorded on day 3 of incubation of the strains. Taken together, the above results demonstrate that the number of tolerant cells was reduced in all the mutants. The decrease in the number of tolerant cells was significant in cydA, msmeg_0392, and msmeg_6655 mutants relative to the WT strain and may play a role in the development of persistence in M. smegmatis.
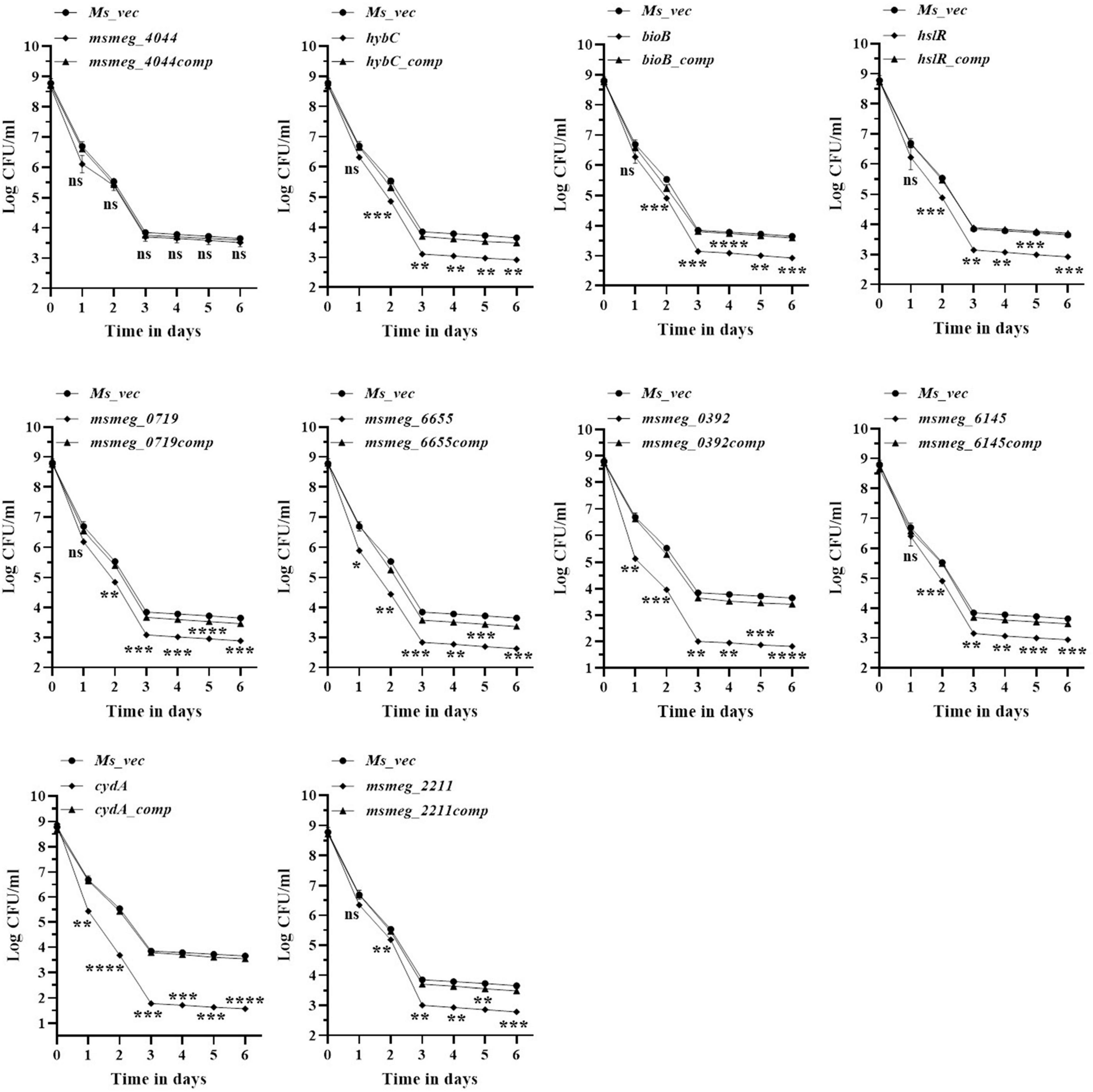
Figure 1. Time-dependent tolerance of M. smegmatis mutants against levofloxacin. Cultures were grown till OD600 ∼1.8, and levofloxacin was added. Samples were removed on day 1, 2, 3, 4, 5, and 6 and tolerant cell numbers were obtained by replica plating as described in Methods. The error bars represent the mean ± SD from three independent experiments. The P-values were calculated by two-way ANOVA with Dunnett’s multiple comparison tests. *P < 0.05, **P < 0.01, ***P < 0.005, and ****P < 0.001 in all the experiments.
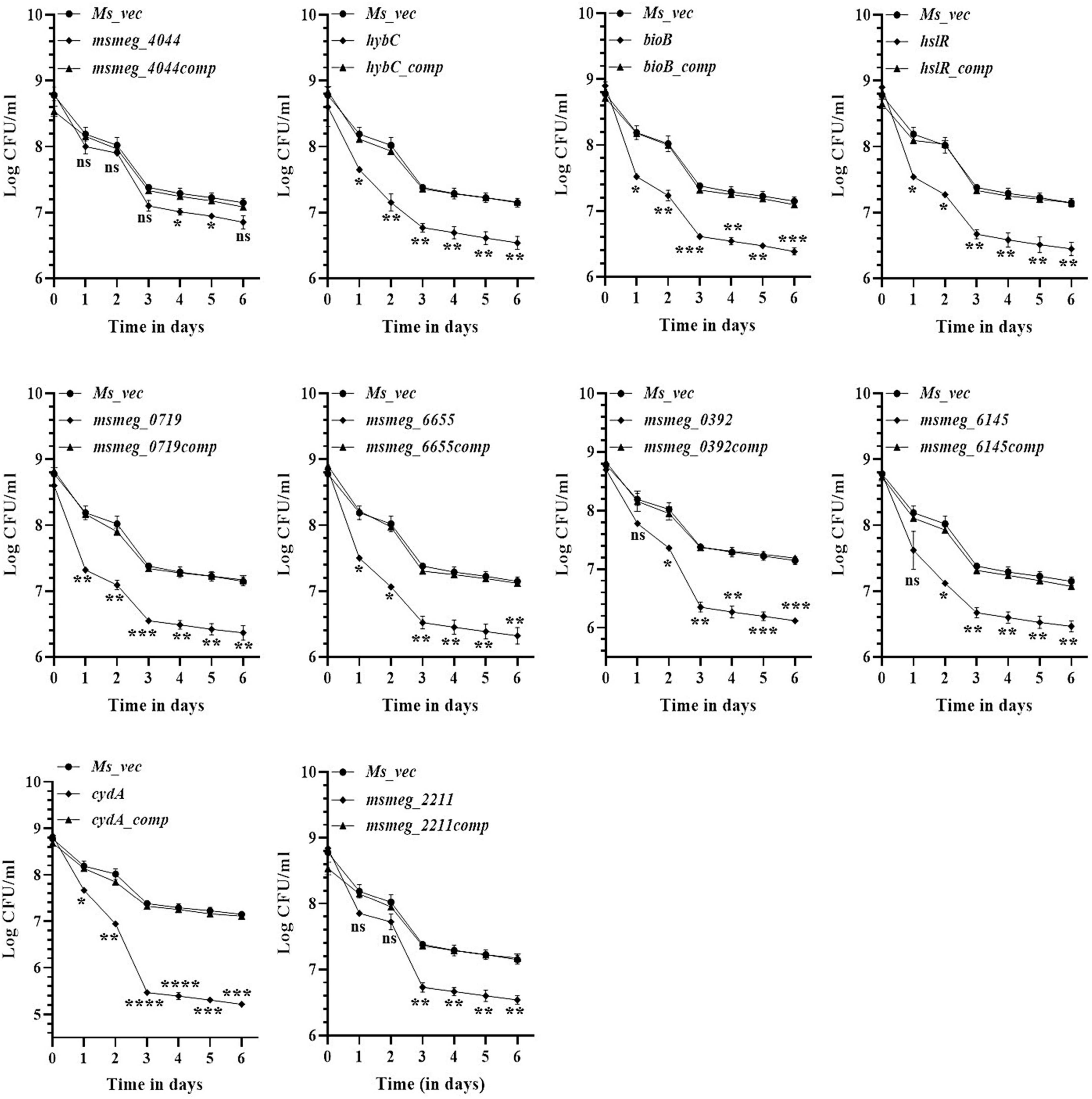
Figure 2. Time-dependent tolerance of M. smegmatis mutants against isoniazid. Cultures were grown to stationary phase (OD600 ∼1.8) and isoniazid was added. Samples were removed on day 1, 2, 3, 4, 5, and 6 and tolerant cell numbers were obtained by replica plating as described in Methods. The error bars represent the mean ± SD from three independent experiments. The P-values were calculated by two-way ANOVA with Dunnett’s multiple comparison tests. *P < 0.05, **P < 0.01, ***P < 0.005, and ****P < 0.001 in all the experiments.
Kinetics of variation of antibiotic dose with tolerance
The number of tolerant cell counts at varying antibiotic concentrations (10X−50X) was measured after 3 days of exposure. The change in the number of live cells with increasing concentrations of levofloxacin is shown in Figure 3. Maximum antibiotic susceptibility with respect to the WT was observed between 10X and 20X MIC in most of the mutants. The fold decrease in the live cell counts of cydA, msmeg_0392, and msmeg_6655 mutants was 2.17-log, 2.21-log, and 1.21-log, respectively, relative to the parent strain, over the entire range of antibiotic concentrations (Figure 3). The mutant strains complemented with the respective WT genes showed full recovery in the number of surviving cells. The reduction in the levels of live cells in the remaining transposon mutant ranged between 0.5-log to 1-log compared to the WT and complemented strains (Figure 3). At higher levofloxacin concentrations, the trend in the sensitivity of the strains followed the same pattern as before. In the case of isoniazid, the maximum reduction in the counts of drug-tolerant cells occurred at 10X MIC in all the mutants, as shown in Figure 4. The cydA mutant recorded a maximum reduction of ∼ 2-log, while in the remaining mutants, the decrease in the number of tolerant cell counts varied between 0.5-log and 1-log. Further, similar results were obtained at higher concentrations of isoniazid (Figure 4). The sensitivity of msmeg_4044 mutant strain to both levofloxacin and isoniazid was not significantly different from the WT strain.
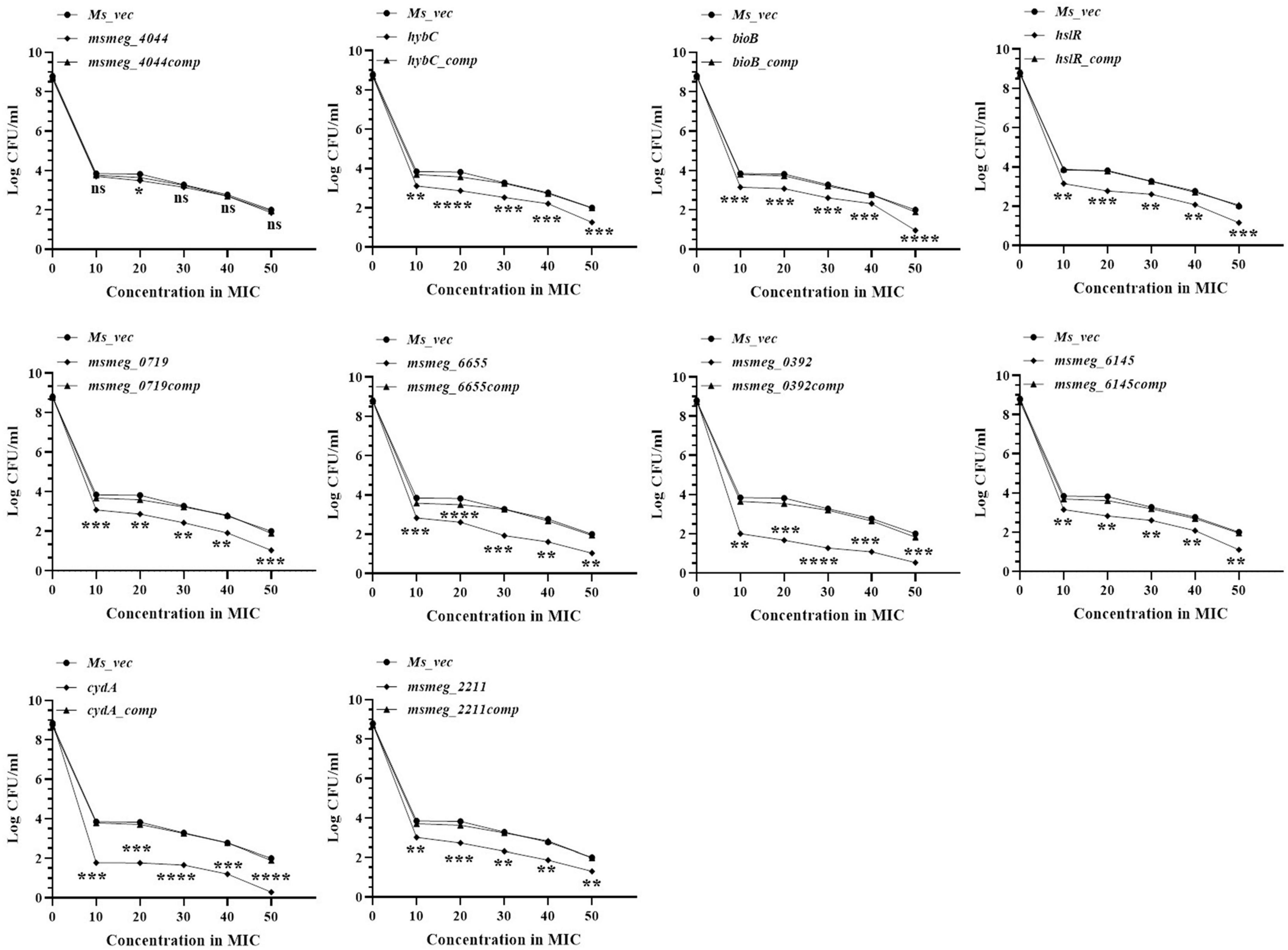
Figure 3. Dose-dependent tolerance of M. smegmatis mutants against levofloxacin. Stationary phase cultures were exposed to different concentrations of levofloxacin and incubated at 37°C for 3 days. Samples were removed on day 3 and tolerant cell numbers were obtained by replica plating as described in Methods. The error bars represent the mean ± SD from three independent experiments. The P-values were calculated by two-way ANOVA with Dunnett’s multiple comparison tests. *P < 0.05, **P < 0.01, ***P < 0.005, and ****P < 0.001 in all the experiments.
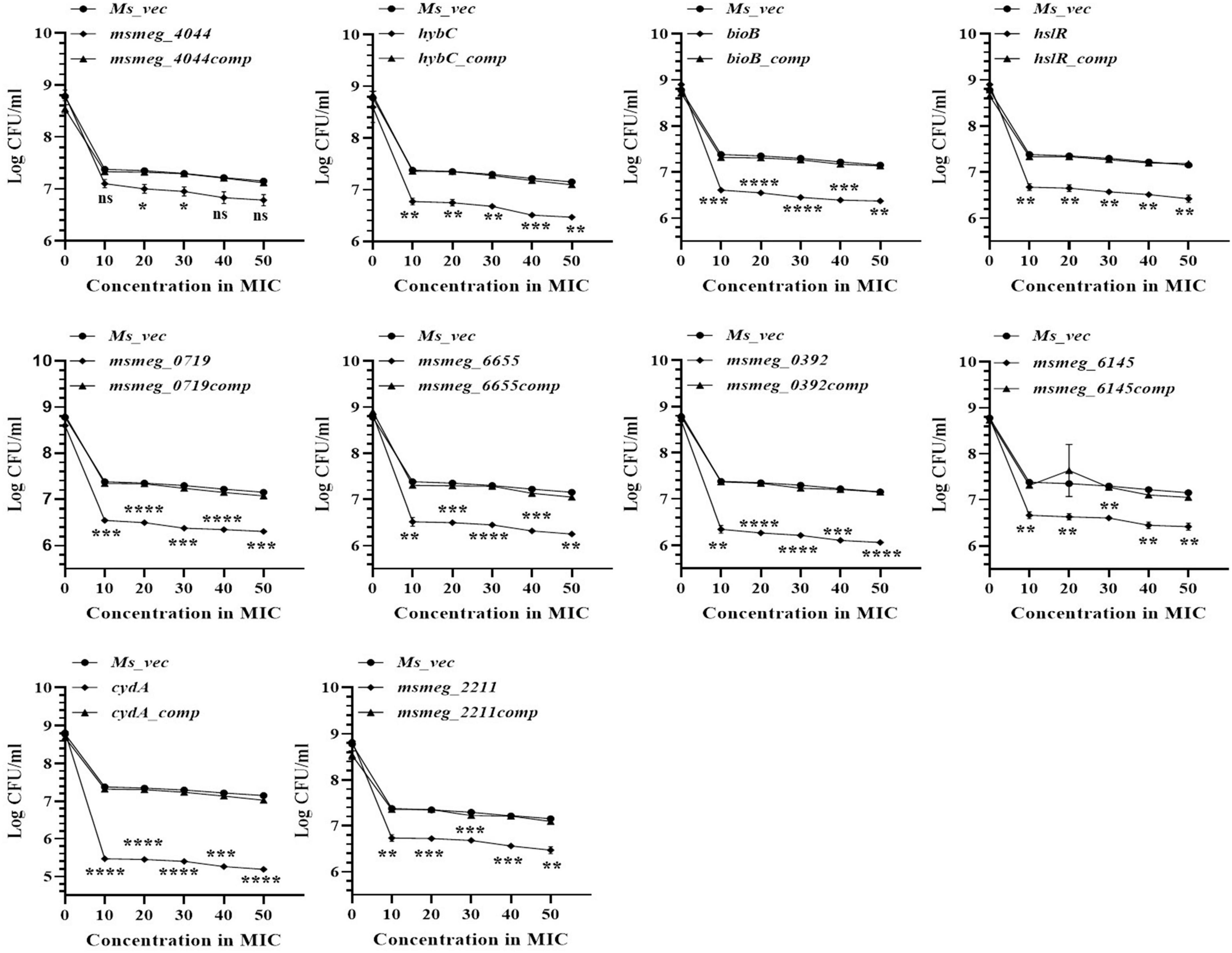
Figure 4. Dose-dependent tolerance of M. smegmatis mutants against isoniazid. Stationary phase cultures were exposed to different concentrations of isoniazid and incubated at 37°C for 3 days. Samples were removed on day 3 and tolerant cell numbers were obtained by replica plating as described in Methods. The error bars represent the mean ± SD from three independent experiments. The P-values were calculated by two-way ANOVA with Dunnett’s multiple comparison tests. *P < 0.05, **P < 0.01, ***P < 0.005, and ****P < 0.001 in all the experiments.
Effect on Biofilm formation by M. smegmatis strains
The ability to form biofilm is a hallmark of persistent pathogens causing chronic infections. Hence, it was used as a marker to measure tolerance of M. smegmatis. Biofilm development on hydrophobic, plastic surface by the mutants is shown in Figure 5. The CV staining confirmed variable degrees of defect in biofilm formation by all the mutants relative to M. smegmatis or the complemented strains. The msmeg_4044, bioB, msmeg_0719, msmeg_6655, and msmeg_0392 variants showed a significant reduction in CV binding, while hybC, hslR, and msmeg_2211 mutant strains were affected to a lesser degree. The control mutant stains C1 showed ∼50% reduction, while C2 and C3 showed no defect in biofilm development, as expected (Figure 5). Our results suggest that the disrupted genes in the selected transposon mutants are directly or indirectly involved in the process of biofilm formation in M. smegmatis, eventually helping the cell to survive.
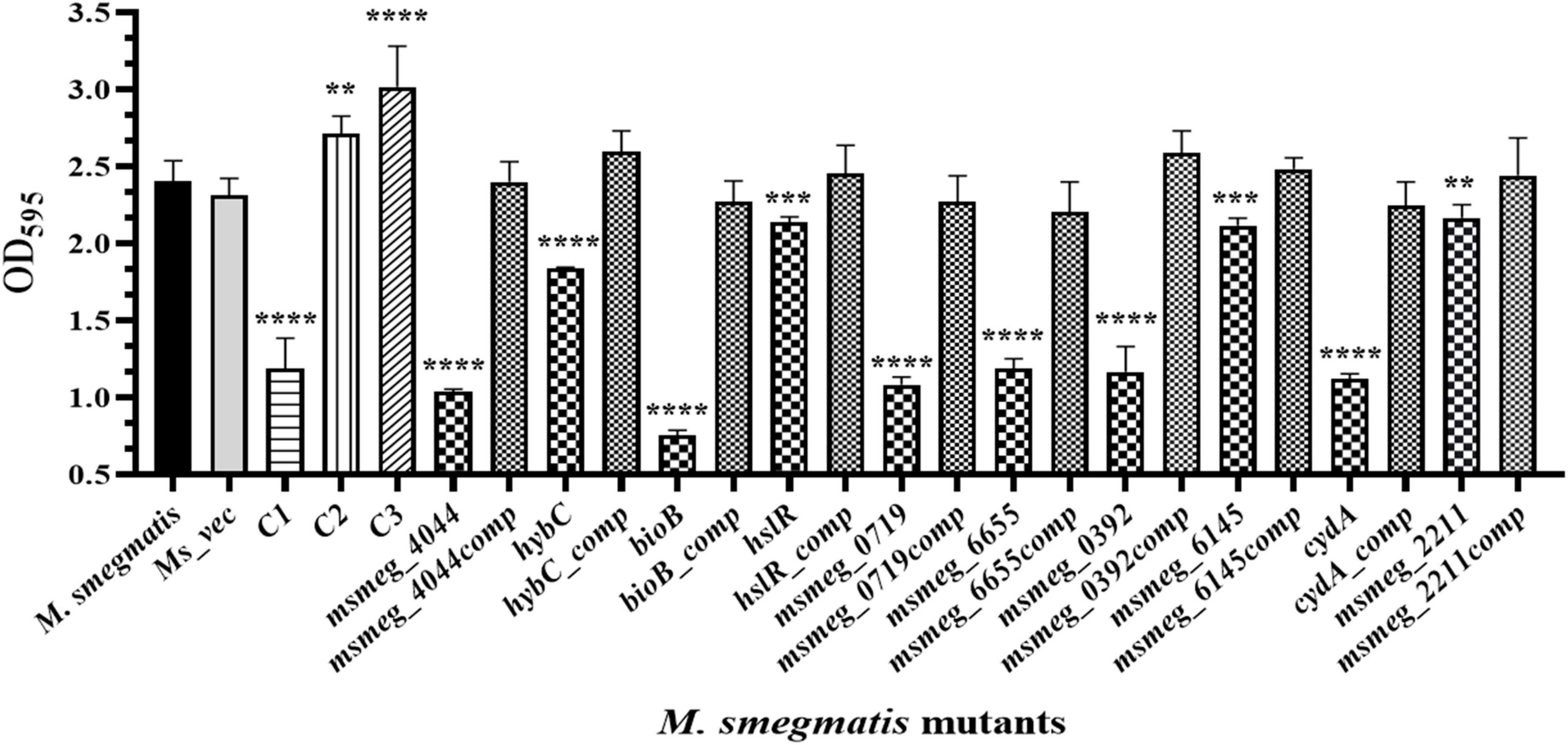
Figure 5. Biofilm formation by transposon mutants. Quantitative representation of the biofilm formed by the mutants. The stationary phase saturated culture was inoculated in Sauton’s medium as above and incubated at 37°C for 4 days. The medium was removed carefully, and adherent biofilms were stained with crystal violet for optical density measurement at 595 nm. The error bars indicate mean ± SD from three independent experiments. *P < 0.05, **P < 0.01, ***P < 0.005, and ****P < 0.001 in all the experiments.
Survival of the mutants in mouse macrophages
To evaluate the ability of the mutants to tolerate the hostile conditions in vivo, they were infected in the murine macrophage cell line. The clearance rate of all the mutant strains from the RAW 264.7 macrophages was faster than the WT strain from 24 h onwards. After 72 h, msmeg_0719 recorded more than 2-log; msmeg_6655, cydA, msmeg_4044, bioB, and msmeg_0392 between 1-log to 2-log; and msmeg_6145, hybC, hslR, and msmeg_2211 mutants recorded about 0.5-log, lower counts than the WT and the complemented strains (Figure 6). Overall, all ten transposon mutants displayed a variable degree of defect in intracellular survival within the murine macrophages. The clearance time of the control strains C1, C2, and C3 were similar to the WT strain, suggesting that a defect in single phenotype is not enough to cause a significant reduction in the survival of M. smegmatis.
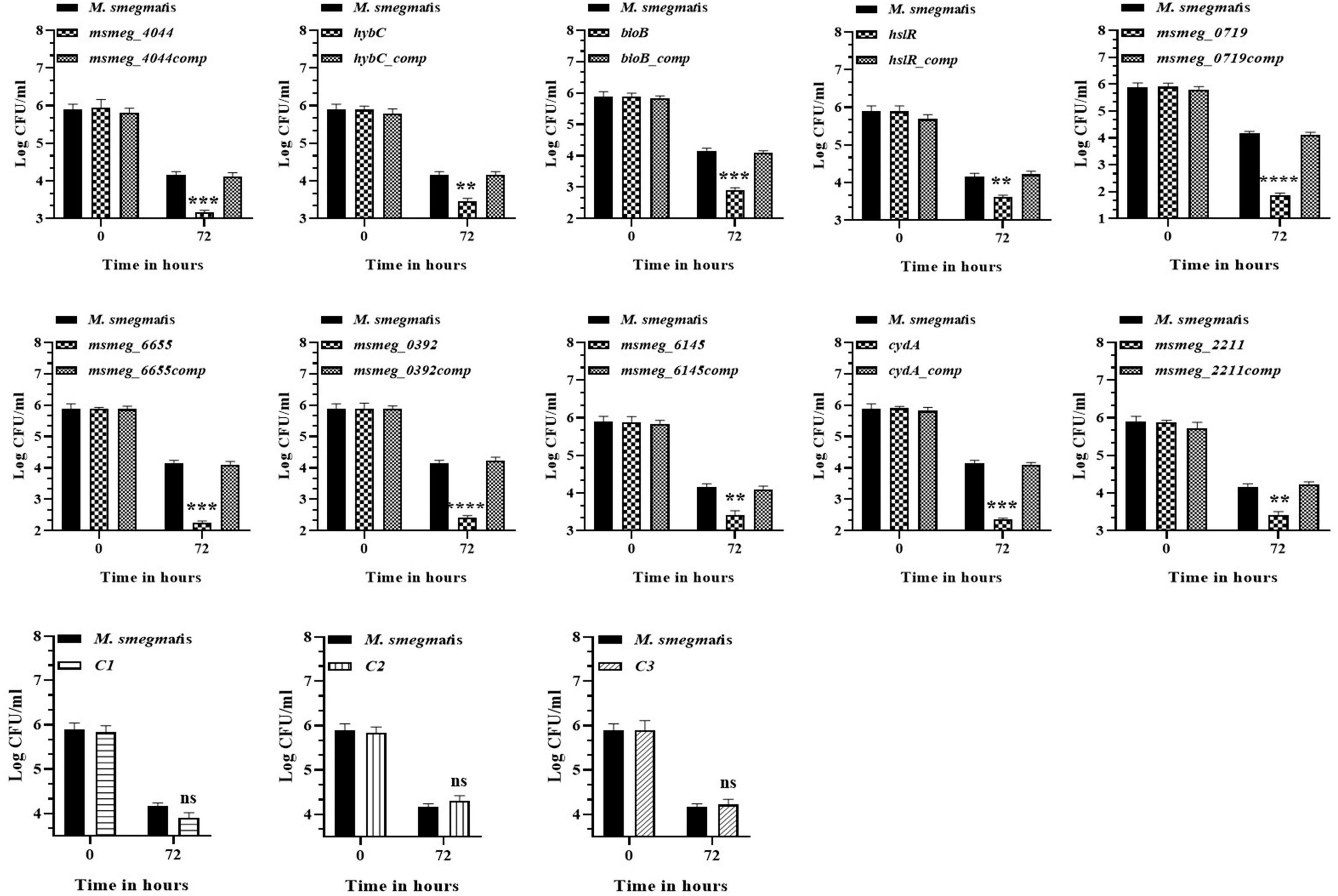
Figure 6. Survival of M. smegmatis mutants in murine macrophages. Cultured RAW 264.7 cells were infected with M. smegmatis strains at m.o.i of 25:1. The cells were harvested at 0, 24, and 72 h post-infection and CFUs counts were obtained by plating appropriate dilutions on 7H10 agar plates supplemented with kanamycin (25 μg/ml). The error bars represent mean ± SD from three individual experiments. The P-values were calculated by two-way ANOVA with Dunnett’s multiple comparison tests. *P < 0.05, **P < 0.01, ***P < 0.005, and ****P < 0.001 in all the experiments.
Stress response of M. smegmatis variants
To examine the stress response of the mutants, the latter were exposed to conditions encountered by mycobacteria during infection, e.g., oxidative stress and hypoxic environment, which reduced the number of survivors in all the mutants compared to the WT strain. All the strains displayed greater sensitivity to oxidative stress at the peak of the growth phase after 48 h. The msmeg_0719 and cydA mutants appear more sensitive than others in the group, showing maximum reduction of ∼ 2-log in CFU compared to the WT strain (Figure 7A). While ∼ 1 log-fold reduction was observed in the hybC, bioB, msmeg_0392, and msmeg_6145 mutants over the WT strain (Figure 7A). Of the nine transposon mutants tested, msmeg_4044, hslR, and msmeg_6655 mutants, were not affected significantly during exposure to oxidative stress. The number of live cells in the control mutant strains was similar to WT strain, reiterating the synergistic action of multiple genetic loci in adaptation and long-term survival of M. smegmatis (Figure 7A).
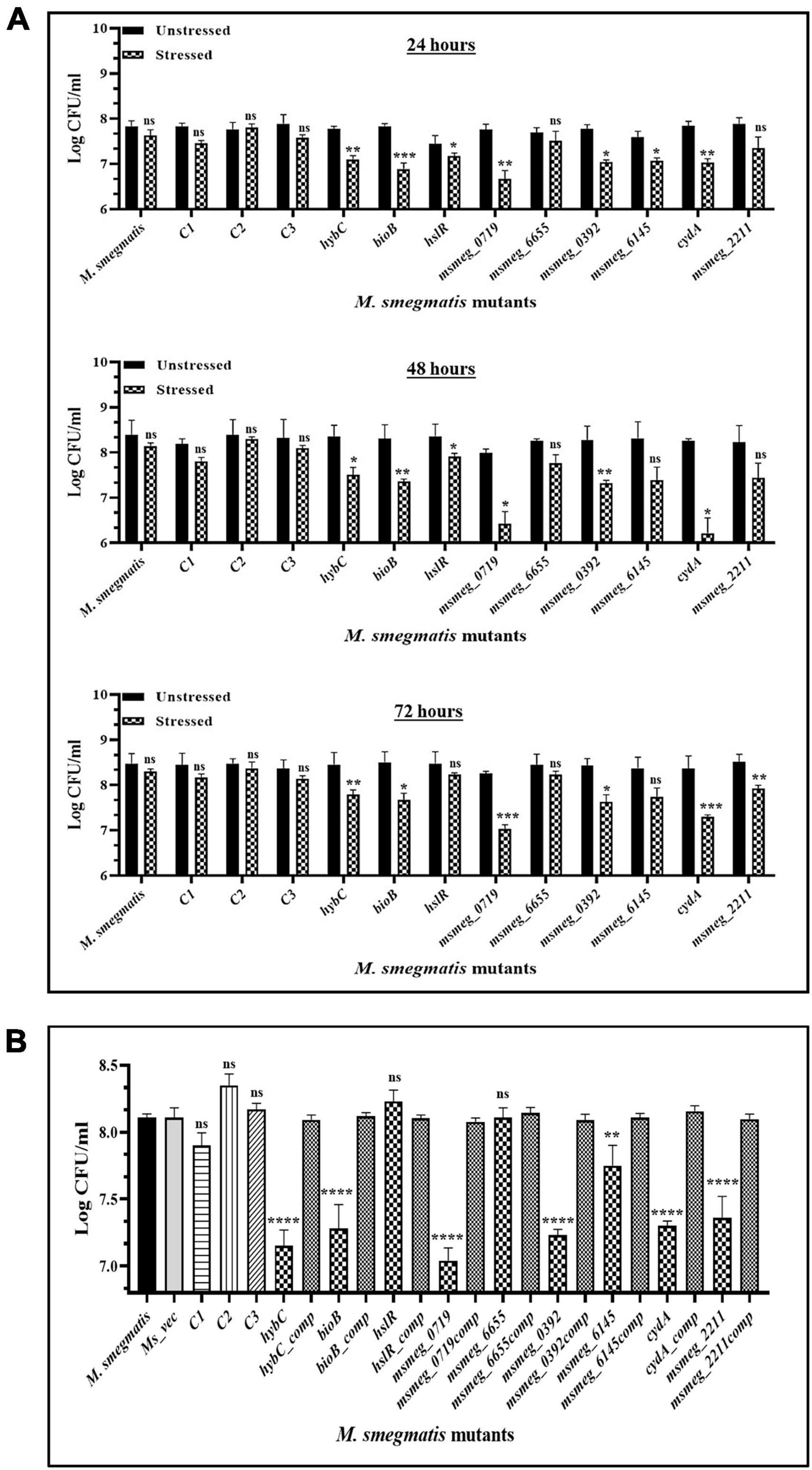
Figure 7. Survival of M. smegmatis mutants under in vitro stress conditions. (A) Stationary phase cells of bacterial strains were washed and exposed to 10 mM hydrogen peroxide at 37°C for 2 h at different time intervals. (B) All the bacterial strains were incubated in hypoxic conditions for 10 days at 37°C. The CFUs were recorded by plating on 7H10 agar plates supplemented with kanamycin. The error bars represent the mean ± SD from three independent experiments. The P-values were calculated by two-way ANOVA with Dunnett’s multiple comparison tests. *P < 0.05, **P < 0.01, ***P < 0.005, and ****P < 0.001 in all the experiments.
Some of the mutants showed higher sensitivity to hypoxic stress (Figure 7B). The strain msmeg_0719 showed maximum reduction of ∼1-log in CFU counts, while hybC, bioB, msmeg_0392, cydA, and msmeg_2211 were all affected adversely with lower than 1-log-fold change under hypoxic conditions. As in oxidative conditions, the hslR and msmeg_6655 mutants, were not affected significantly (Figure 7B).
Survival of the transposon mutants in mice
The degree of tolerance of the strains in vivo was tested in mice. C57BL/6J mice were injected with M. smegmatis strains, and CFU in the lungs and spleen were enumerated. In general, the bacterial burden was substantially reduced in the organs by 7 days and was fully cleared at 21 days of infection. After 7 days of infection, the bacterial counts in the lungs and the spleen of several mutants, namely msmeg_0719, hybC, bioB, and cydA, were ∼1-log lower than the WT and the complemented strains. The CFU counts decreased further at 14 days in the lungs of msmeg_0719, msmeg_0392, hybC, bioB, cydA, and msmeg_2211 mutants (ranging between 1.7-log to 0.66-log) with respect to the parent strain, while msmeg_6145 was not significantly different from the M. smegmatis control (Figure 8A). Likewise, the number of surviving cells in the spleens of msmeg_0719, msmeg_0392, hybC, bioB, cydA, and msmeg_2211, mutants was 1.52-log, 1.48-log, 1.04-log, 0.97-log, 0.6-log, and 0.53-log lower than the WT and complemented strains after 14 days (Figure 8B). Overall, initially (7 days) the rate of clearance of WT bacteria in the spleen was higher (2.35-log) than in the lungs (1.82-log), but at later time point (14 days), the clearance rate was nearly same in both the organs. These results indicate that due to the disruption of some genes, the six mutants mentioned above displayed a significantly lower propensity to survive under in vivo conditions than the parental strain, and the genes affected in the latter may play a role in bacterial persistence in the host tissues.
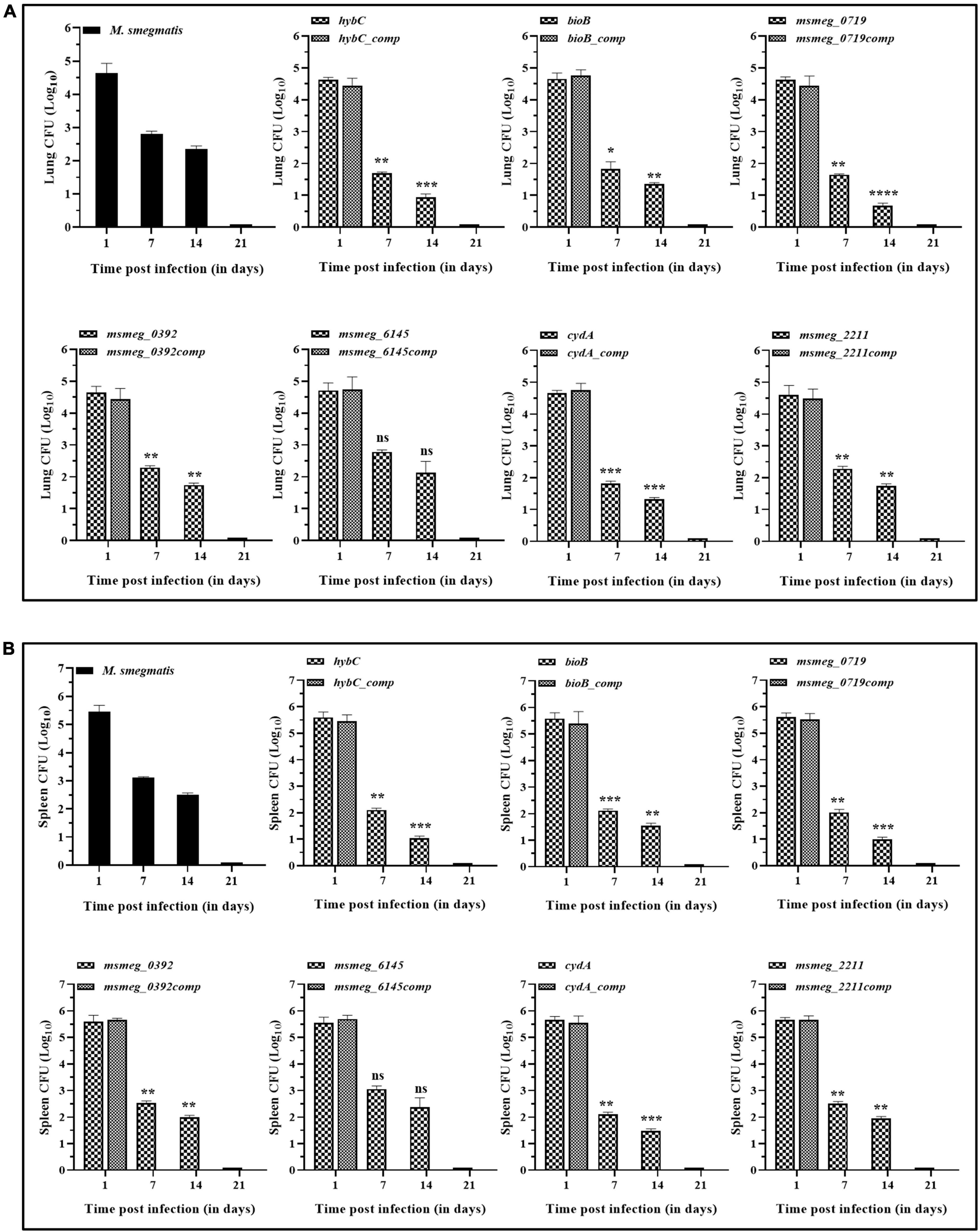
Figure 8. Persistence of M. smegmatis mutants in lungs and spleen of mice. C57BL/6J mice were injected intravenously with 1 x 106 CFUs of actively growing cells. Survival of the bacteria in the (A) lungs and (B) spleen was determined on day 1, 7, 14, and 21 by plating appropriate dilutions on 7H10 agar plates containing kanamycin (25 μg/ml). At day 21 complete bacterial clearance was observed. The data for complemented strain was plotted for day 1, as it indicates the gene function was restored. The statistical significance was determined by comparing mutant strains with the wild type at different time points of infection. The error bars represent the mean ± SD from three independent experiments. The P-values were calculated by two-way ANOVA with Dunnett’s multiple comparison tests. *P < 0.05, **P < 0.01, ***P < 0.005, and ****P < 0.001 in all the experiments.
Discussion
It has become abundantly clear that there is no single mechanism to explain the phenomenon of persistence in bacteria (Hu and Coates, 2005; Zhang, 2014; Urbaniec et al., 2022). The ability of M. tuberculosis and M. smegmatis to form persister cells in vitro and in vivo has been demonstrated in several studies (Grant et al., 2012; Manina et al., 2015; Torrey et al., 2016). Here, we have adopted three different screening criteria intimately linked to bacterial persistence, e.g., antibiotic tolerance, biofilm formation, and survival in vivo in the macrophages, to screen a transposon mutant library of M. smegmatis mc2155. The rationale for using multiple phenotypes for selection was based on the assumption that a gene may control multiple phenotypes indirectly by participating in different metabolic pathways. Hence, disruption of a genetic locus, manifesting defects in multiple phenotypes is likely to be involved in persistence development in a strain. Also, supported by our observations that multiple disabilities in a strain increased the susceptibility of the organism to host clearance mechanisms.
The msmeg_3233 gene encodes the CydA subunit of cytochrome bd oxidase. In mycobacteria, the electron transport chain (ETC) terminates through the actions of two-terminal oxidases: an aa3-type cytochrome c oxidase and cytochrome bd oxidase. The latter is involved in the generation of the PMF to fuel energetic processes in the cell membrane (Cook et al., 2014). The cytochrome bd oxidase has been implicated in the adaptation of M. tuberculosis to host immunity-induced stress microenvironments during infection (Giuffrè et al., 2012; Forte et al., 2016). In M. smegmatis, although not essential under normal oxygen conditions, maintaining mycobacterial respiration is indispensable during hypoxic conditions (Aung et al., 2014; Jeong et al., 2018). In different mycobacterial species, cydA expression was induced by cell wall inhibitors, e.g., Q203, bedaquiline, nitroimidazoles, augmentin, potassium cyanide, and clofazimine, emphasizing its importance in stress alleviation and survival (Koul et al., 2014; Boot et al., 2017; Kalia et al., 2017). Based on these reports, cytochrome bd oxidase appears to relieve the pressure on the ETC and allow the bacteria to persist longer by maintaining membrane potential in the absence of cytochrome bcc1-aa3 complex and ATP synthase activities. In the light of previous studies, the defects displayed by cydA mutant in all the parameters tested, make a strong case for a key role in global stress management and survival of M. smegmatis.
Disruption of the msmeg_0719 gene is attributed to faster clearance of the mutant, making it sensitive to intracellular stress conditions; the most important among them were oxidative stress, hypoxia, and survival in the mouse macrophages. Oxidative stress is one of the major causes of death of intracellular pathogens. The reactive oxygen species released within macrophages cause lipid peroxidation of the lipids-rich cell walls of mycobacteria and produce toxic methylglyoxal and D-lactate (Rachman et al., 2006). In M. tuberculosis, the corresponding ortholog Rv0385 encodes a peripheral membrane-associated flavohemoglobin, which catalyzes D-lactate to pyruvate and protects the respiratory membranes from toxic ROS-mediated killing (Gupta et al., 2012). Secondly, under hypoxic conditions, the heme domain of the protein transfers the electrons acquired from D-lactate to the electron transport chain in the cell membrane to produce energy at a low level, keeping the bacteria viable in a non-dividing state within the host (Gupta et al., 2012). The precise mechanism of Rv0385 in the persistence of M. tuberculosis would require further investigation. Since it has been conserved in the genome of a number of the pathogenic and non-pathogenic mycobacterial strains (Gupta et al., 2011), it could be a potential target for combating long-term, non-replicative survival of mycobacteria.
In biological systems, biotin acts as a cofactor for enzymes that catalyze metabolic reactions e.g., membrane lipid synthesis, tricarboxylic acid cycle, and amino acid metabolism etc., (Salaemae et al., 2016). Biotin is synthesized from pimeloyl-CoA using four enzymes encoded by bioF, bioA, bioD, and bioB genes organized in an operon in M. tuberculosis and in two separate ones in M. smegmatis (Tang et al., 2014; Salaemae et al., 2016). BioB gene plays a key role in biotin synthesis in both the mycobacterial species (Fan et al., 2015; Lazar et al., 2017). Development of auxotrophy in the bioB gene mutant (Supplementary Figure 2) and significant reduction in the live cell counts under different stress conditions, e.g., bactericidal antibiotics, oxidative stress, intracellular survival, and hypoxia, reiterate the importance of biotin synthesis in growth and global stress management in M. smegmatis.
The reduction in biofilm growth of the bioB mutant is most likely a direct effect of disruption in biotin synthesis by the strain. Further, in a bioB mutant of M. bovis, increased susceptibility to Para-amino salicylic acid and rifampicin in biotin-limiting conditions was attributed to lack of biotin, implying its significance in antibiotics tolerance in mycobacteria (Howe et al., 2018). Thus, mutation in the bioB gene appears to be responsible for the defects in both the phenotypes selected as our screening criteria in M. smegmatis. Biotin is known to play a critical role in the biofilm growth of bacteria, which protects them from the hostile conditions in the host (Chakraborty et al., 2021). Ojha and Hatfull (2007) have reported activation of bioB gene transcription in the late stages of biofilm formation in M. smegmatis. Considering the degree of attenuation of the mutant, a potential role of the bioB gene in the slow-growing, non-replicating, persistent state of M. tuberculosis cannot be ignored.
The msmeg_0392 gene encodes a putative glycosyltransferase enzyme involved in carbohydrate transport and metabolism (Vera-Cabrera et al., 2007). It belongs to the Gtf3 family of proteins, necessary for synthesizing glycosylated glycopeptidolipids (GPLs) in M. smegmatis (Deshayes et al., 2005). It has been reported that M. smegmatis produces triglycosylated GPLs during the late stationary phase under low carbon conditions (Ojha et al., 2002), implicating their role in shielding the cells in unfavorable conditions. Thus, the enhanced susceptibility of the msmeg_0392 gene mutant to multiple stress conditions merits further investigations to learn about its mechanism of action in the context of persistence. Additionally, the study of M. tuberculosis ortholog (rv1524) may also reveal necessary attributes for the long-term survival of the pathogen.
The msmeg_2263 gene is involved in molecular hydrogen (H2) metabolism in mycobacteria (Berney et al., 2014). The msmeg_2263 (hybC) and hybA genes encode the large and small subunits, respectively, of the nickel and iron-dependent hydrogenase-2 enzyme in M. smegmatis. The hydrogenases catalyze the conversion of molecular hydrogen into electrons and protons. The protons are responsible for generating the PMF, whereas the electrons enter aerobic or anaerobic respiratory chains (Benoit et al., 2020). The hydrogenases maintain the flow of reducing equivalents to the ETC during conditions of energy limitation. Genetic studies have shown that the msmeg_2263 gene is induced during starvation and hypoxia, which promotes survival by oxidizing H2 in M. smegmatis (Berney and Cook, 2010; Cook et al., 2014). Further, under hypoxia, mycobacteria switch on hydrogenases to adapt and, like anaerobic bacteria, transfer electrons in the absence of exogenous electron acceptors through the unique electron carriers—ferredoxins (Berney and Cook, 2010). Based on the available information, it is clear that the role of hydrogenases in facilitating long-term survival under low carbon/energy and oxygen conditions is crucial for M. smegmatis, making it a good target for preventing persister formation. Furthermore, it would be interesting to identify counterparts of the gene in M. tuberculosis and study their role in persistence.
The emerging scenario from this study highlights the cooperative role of (i) alternate routes of energy generation in low carbon and hypoxic situations and (ii) protection from free radicals generated by oxidative burst in the phagocytic cells, as the key factors that enable mycobacteria to outlive the hostile environment in the host. Presently we are unable to propose a mechanism of action of other genes that were picked up by our screen e.g., hslR, a ribosome-associated heat shock protein15; msmeg_6655, and msmeg_6145, hypothetical proteins; msmeg_2211, a DNA-binding protein; and msmeg_4044, a GAF domain-containing protein in mycobacteria, due to lack of comprehensive information about their functional nature and will be the subject of our future studies.
In comparison to the pathogenic mycobacterial strains, the faster growth rate of M. smegmatis may raise questions about its suitability as a model to study tolerance/ persistence. However, the long-term survival of an organism requires multiple attributes other than growth rate to tolerate the hostile host environment. The literature is replete with studies performed in M. smegmatis, which provide valuable insight into the unique characteristics of the genus Mycobacterium (Sharbati-Tehrani et al., 2005; Malhotra et al., 2017; Ranjitha et al., 2020; Sparks et al., 2023). Using M. smegmatis as a model has the advantage of acquiring a large volume of data in a short time for studying the pathogenic traits of a species. This study was aimed to do the groundwork for identifying potential genes (with orthologs in M. tuberculosis) involved in the long-term survival of mycobacteria. We assumed that investigating the conserved stress-inducible housekeeping genes of M. smegmatis may reveal information applicable to M. tuberculosis persistence.
It is challenging to identify genes involved in persistence development due to the transient nature of the phenotypes and the redundancy of mechanisms promoting long-term survival. Each effort reveals new targets, providing insight to understand the process. Likewise, this study has identified some target genes in M. smegmatis. To our knowledge, the five genes have not been described in the context of the persistence of mycobacteria. These genes participate in vital metabolic pathways, including alternate methods of energy production, hydrogen metabolism, biotin synthesis, biofilm formation, and carbohydrate metabolism. Some of these genes have orthologs in M. tuberculosis and offer possibilities for developing novel antibiotics to treat chronic and persistent tuberculosis infections.
Data availability statement
The original contributions presented in this study are included in the article/Supplementary material, further inquiries can be directed to the corresponding authors.
Ethics statement
The animal study was approved by the Institutional Animal Ethics Committee of Jawaharlal Nehru University. The study was conducted in accordance with the local legislation and institutional requirements.
Author contributions
HJ: Conceptualization, Data curation, Formal Analysis, Investigation, Methodology, Validation, Writing – original draft, Writing – review & editing. DK: Conceptualization, Formal Analysis, Methodology, Writing – review & editing. SM: Supervision, Visualization, Writing – review & editing. RB: Funding acquisition, Resources, Supervision, Visualization, Writing – review & editing. NB: Conceptualization, Data curation, Formal Analysis, Investigation, Methodology, Supervision, Writing – review & editing.
Funding
The author(s) declare financial support was received for the research, authorship, and/or publication of this article. This financial assistance by the Department of Science & Technology (DST), Government of India (Grant Sanction no. NSC/2021/000026) is greatly acknowledged. Department of Biotechnology (DBT), Government of India is also acknowledged for funding (Grant Sanction nos. BT/PR22952/NER/95/572/2017 and DBT/2017/JNU/849).
Conflict of interest
NB and DK were employed by the Divacc Research Laboratories Pvt Ltd.
The remaining authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2024.1302883/full#supplementary-material
References
Andrews, J. (2001). Determination of minimum inhibitory concentrations. J. Antimicrob. Chemother. 48, 5–16.
Aung, H., Berney, M., and Cook, G. (2014). Hypoxia-activated cytochrome bd expression in Mycobacterium smegmatis is cyclic AMP receptor protein dependent. J. Bacteriol. 196, 3091–3097. doi: 10.1128/JB.01771-14
Balaban, N., Helaine, S., Lewis, K., Ackermann, M., Aldridge, B., Andersson, D., et al. (2019). Definitions and guidelines for research on antibiotic persistence. Nat. Rev. Microbiol. 17, 441–448.
Barry, C., Boshoff, H., Dartois, V., Dick, T., Ehrt, S., Flynn, J., et al. (2009). The spectrum of latent tuberculosis: rethinking the biology and intervention strategies. Nat. Rev. Microbiol. 7, 845–855. doi: 10.1038/nrmicro2236
Benoit, S., Maier, R., Sawers, R., and Greening, C. (2020). Molecular hydrogen metabolism: a widespread trait of pathogenic bacteria and protists. Microbiol. Mol. Biol. Rev. 84:e00092-19. doi: 10.1128/MMBR.00092-19
Berney, M., and Cook, G. (2010). Unique flexibility in energy metabolism allows mycobacteria to combat starvation and hypoxia. PLoS One 5:e8614. doi: 10.1371/journal.pone.0008614
Berney, M., Greening, C., Hards, K., Collins, D., and Cook, G. (2014). Three different [NiFe] hydrogenases confer metabolic flexibility in the obligate aerobe Mycobacterium smegmatis. Environ. Microbiol. 16, 318–330.
Bigger, J. (1944). Treatment of staphyloeoeeal infections with penicillin by intermittent sterilisation. Lancet 244, 497–500.
Boot, M., Jim, K., Liu, T., Commandeur, S., Lu, P., Verboom, T., et al. (2017). A fluorescence-based reporter for monitoring expression of mycobacterial cytochrome bd in response to antibacterials and during infection. Sci Rep. 7:10665. doi: 10.1038/s41598-017-10944-4
Brauner, A., Fridman, O., Gefen, O., and Balaban, N. (2016). Distinguishing between resistance, tolerance and persistence to antibiotic treatment. Nat. Rev. Microbiol. 14, 320–330.
Chakraborty, P., Bajeli, S., Kaushal, D., Radotra, B., and Kumar, A. (2021). Biofilm formation in the lung contributes to virulence and drug tolerance of Mycobacterium tuberculosis. Nat. Commun. 12:1606. doi: 10.1038/s41467-021-21748-6
Ciofu, O., Rojo-Molinero, E., Macià, M., and Oliver, A. (2017). Antibiotic treatment of biofilm infections. APMIS 125, 304–319.
Conlon, B., Nakayasu, E., Fleck, L., LaFleur, M., Isabella, V., Coleman, K., et al. (2013). Activated ClpP kills persisters and eradicates a chronic biofilm infection. Nature 503, 365–370.
Cook, G., Hards, K., Vilchèze, C., Hartman, T., and Berney, M. (2014). Energetics of respiration and oxidative phosphorylation in Mycobacteria. Microbiol. Spectr. 2, 1–30. doi: 10.1128/microbiolspec.MGM2-0015-2013
Deshayes, C., Laval, F., Montrozier, H., Daffé, M., Etienne, G., and Reyrat, J. (2005). A glycosyltransferase involved in biosynthesis of triglycosylated glycopeptidolipids in Mycobacterium smegmatis: impact on surface properties. J. Bacteriol. 187, 7283–7291.
Domingue, J., Drewes, J., Merlo, C., Housseau, F., and Sears, C. (2020). Host responses to mucosal biofilms in the lung and gut. Mucosal Immunol. 13, 413–422. doi: 10.1038/s41385-020-0270-1
Dörr, T., Lewis, K., and Vulić, M. (2009). SOS response induces persistence to fluoroquinolones in Escherichia coli. PLoS Genet. 5:e1000760. doi: 10.1371/journal.pgen.1000760
Fan, S., Li, D., Wang, D., Fleming, J., Zhang, H., Zhou, Y., et al. (2015). Structure and function of Mycobacterium smegmatis 7-keto-8-aminopelargonic acid (KAPA) synthase. Int. J. Biochem. Cell Biol. 58, 71–80. doi: 10.1016/j.biocel.2014.11.006
Fauvart, M., De Groote, V., and Michiels, J. (2011). Role of persister cells in chronic infections: clinical relevance and perspectives on anti-persister therapies. J. Med. Microbiol. 60(Pt 6), 699–709. doi: 10.1099/jmm.0.030932-0
Fisher, R., Gollan, B., and Helaine, S. (2017). Persistent bacterial infections and persister cells. Nat. Rev. Microbiol. 15, 453–464.
Foreman, M., Gershoni, M., and Barkan, D. (2020). A simplified and efficient method for Himar-1 transposon sequencing in bacteria, demonstrated by creation and analysis of a saturated transposon-mutant library in Mycobacterium abscessus. mSystems 5:e00976-920. doi: 10.1128/mSystems.00976-20
Forte, E., Borisov, V., Falabella, M., Colaço, H., Tinajero-Trejo, M., Poole, R., et al. (2016). The terminal oxidase cytochrome bd promotes sulfide-resistant bacterial respiration and growth. Sci. Rep. 6:23788. doi: 10.1038/srep23788
Giuffrè, A., Borisov, V., Mastronicola, D., Sarti, P., and Forte, E. (2012). Cytochrome bd oxidase and nitric oxide: from reaction mechanisms to bacterial physiology. FEBS Lett. 586, 622–629. doi: 10.1016/j.febslet.2011.07.035
Grant, S., Kaufmann, B., Chand, N., Haseley, N., and Hung, D. (2012). Eradication of bacterial persisters with antibiotic-generated hydroxyl radicals. Proc. Natl. Acad. Sci. U S A. 109, 12147–12152. doi: 10.1073/pnas.1203735109
Gupta, S., Pawaria, S., Lu, C., Hade, M., Singh, C., Yeh, S., et al. (2012). An unconventional hexacoordinated flavohemoglobin from Mycobacterium tuberculosis. J. Biol. Chem. 287, 16435–16446. doi: 10.1074/jbc.M111.329920
Gupta, S., Pawaria, S., Lu, C., Yeh, S., and Dikshit, K. (2011). Novel flavohemoglobins of mycobacteria. IUBMB Life 63, 337–345.
Helaine, S., Cheverton, A., Watson, K., Faure, L., Matthews, S., and Holden, D. (2014). Internalization of Salmonella by macrophages induces formation of nonreplicating persisters. Science 343, 204–208. doi: 10.1126/science.1244705
Hingley-Wilson, S., Ma, N., Hu, Y., Casey, R., Bramming, A., Curry, R., et al. (2020). Loss of phenotypic inheritance associated with ydcI mutation leads to increased frequency of small, slow persisters in Escherichia coli. Proc. Natl. Acad. Sci. U S A. 117, 4152–4157. doi: 10.1073/pnas.1914741117
Howe, M., Kordus, S., Cole, M., Bauman, A., Aldrich, C., Baughn, A., et al. (2018). Methionine antagonizes para-aminosalicylic acid activity via affecting folate precursor biosynthesis in Mycobacterium tuberculosis. Front. Cell Infect. Microbiol. 8:399. doi: 10.3389/fcimb.2018.00399
Hu, Y., and Coates, A. (2005). Transposon mutagenesis identifies genes which control antimicrobial drug tolerance in stationary-phase Escherichia coli. FEMS Microbiol. Lett. 243, 117–124. doi: 10.1016/j.femsle.2004.11.049
Jeong, J., Park, S., Yoon, D., Kim, S., Kang, H., and Oh, J. (2018). Roles of alanine dehydrogenase and induction of its gene in Mycobacterium smegmatis under respiration-inhibitory conditions. J. Bacteriol. 200:e00152-18. doi: 10.1128/JB.00152-18
Joshi, H., Kandari, D., and Bhatnagar, R. (2021). Insights into the molecular determinants involved in Mycobacterium tuberculosis persistence and their therapeutic implications. Virulence 12, 2721–2749.
Kalia, N., Hasenoehrl, E., Ab Rahman, N., Koh, V., Ang, M., Sajorda, D., et al. (2017). Exploiting the synthetic lethality between terminal respiratory oxidases to kill Mycobacterium tuberculosis and clear host infection. Proc. Natl. Acad. Sci. U S A. 114, 7426–7431. doi: 10.1073/pnas.1706139114
Keren, I., Kaldalu, N., Spoering, A., Wang, Y., and Lewis, K. (2004). Persister cells and tolerance to antimicrobials. FEMS Microbiol. Lett. 230, 13–18.
Koul, A., Vranckx, L., Dhar, N., Göhlmann, H., Özdemir, E., Neefs, J., et al. (2014). Delayed bactericidal response of Mycobacterium tuberculosis to bedaquiline involves remodelling of bacterial metabolism. Nat. Commun. 5:3369. doi: 10.1038/ncomms4369
Lafleur, M., Qi, Q., and Lewis, K. (2010). Patients with long-term oral carriage harbor high-persister mutants of Candida albicans. Antimicrob. Agents Chemother. 54, 39–44. doi: 10.1128/AAC.00860-09
Lamichhane, G., Zignol, M., Blades, N., Geiman, D., Dougherty, A., Grosset, J., et al. (2003). A postgenomic method for predicting essential genes at subsaturation levels of mutagenesis: application to Mycobacterium tuberculosis. Proc. Natl. Acad. Sci. U S A. 100, 7213–7218. doi: 10.1073/pnas.1231432100
Lazar, N., Fay, A., Nandakumar, M., Boyle, K., Xavier, J., Rhee, K., et al. (2017). Control of biotin biosynthesis in mycobacteria by a pyruvate carboxylase dependent metabolic signal. Mol. Microbiol. 106, 1018–1031. doi: 10.1111/mmi.13865
Levin-Reisman, I., Ronin, I., Gefen, O., Braniss, I., Shoresh, N., and Balaban, N. (2017). Antibiotic tolerance facilitates the evolution of resistance. Science 355, 826–830.
Li, Y., and Zhang, Y. (2007). PhoU is a persistence switch involved in persister formation and tolerance to multiple antibiotics and stresses in Escherichia coli. Antimicrob. Agents Chemother. 51, 2092–2099. doi: 10.1128/AAC.00052-07
Lipworth, S., Hammond, R., Baron, V., Hu, Y., Coates, A., and Gillespie, S. (2016). Defining dormancy in mycobacterial disease. Tuberculosis 99, 131–142.
Ma, C., Sim, S., Shi, W., Du, L., Xing, D., and Zhang, Y. (2010). Energy production genes sucB and ubiF are involved in persister survival and tolerance to multiple antibiotics and stresses in Escherichia coli. FEMS Microbiol. Lett. 303, 33–40. doi: 10.1111/j.1574-6968.2009.01857.x
Malhotra, S., Vedithi, S., and Blundell, T. (2017). Decoding the similarities and differences among mycobacterial species. PLoS Negl. Trop. Dis. 11:e0005883. doi: 10.1371/journal.pntd.0005883
Manina, G., Dhar, N., and McKinney, J. (2015). Stress and host immunity amplify Mycobacterium tuberculosis phenotypic heterogeneity and induce nongrowing metabolically active forms. Cell Host Microbe 17, 32–46. doi: 10.1016/j.chom.2014.11.016
Michiels, J., Van den Bergh, B., Verstraeten, N., and Michiels, J. (2016). Molecular mechanisms and clinical implications of bacterial persistence. Drug Resist. Updat. 29, 76–89.
Mok, W., and Brynildsen, M. (2018). Timing of DNA damage responses impacts persistence to fluoroquinolones. Proc. Natl. Acad. Sci. U S A. 115, E6301–E6309. doi: 10.1073/pnas.1804218115
Mouton, J., Helaine, S., Holden, D., and Sampson, S. (2016). Elucidating population-wide mycobacterial replication dynamics at the single-cell level. Microbiology 162, 966–978. doi: 10.1099/mic.0.000288
Ojha, A., Baughn, A., Sambandan, D., Hsu, T., Trivelli, X., Guerardel, Y., et al. (2008). Growth of Mycobacterium tuberculosis biofilms containing free mycolic acids and harbouring drug-tolerant bacteria. Mol. Microbiol. 69, 164–174. doi: 10.1111/j.1365-2958.2008.06274.x
Ojha, A., and Hatfull, G. (2007). The role of iron in Mycobacterium smegmatis biofilm formation: the exochelin siderophore is essential in limiting iron conditions for biofilm formation but not for planktonic growth. Mol. Microbiol. 66, 468–483. doi: 10.1111/j.1365-2958.2007.05935.x
Ojha, A., Varma, S., and Chatterji, D. (2002). Synthesis of an unusual polar glycopeptidolipid in glucose-limited culture of Mycobacterium smegmatis. Microbiology 148(Pt 10), 3039–3048. doi: 10.1099/00221287-148-10-3039
Rachman, H., Kim, N., Ulrichs, T., Baumann, S., Pradl, L., Nasser Eddine, A., et al. (2006). Critical role of methylglyoxal and AGE in mycobacteria-induced macrophage apoptosis and activation. PLoS One 1:e29. doi: 10.1371/journal.pone.0000029
Ranjitha, J., Rajan, A., and Shankar, V. (2020). Features of the biochemistry of Mycobacterium smegmatis, as a possible model for Mycobacterium tuberculosis. J. Infect. Public Health 13, 1255–1264.
Rego, E., Audette, R., and Rubin, E. (2017). Deletion of a mycobacterial divisome factor collapses single-cell phenotypic heterogeneity. Nature 546, 153–157. doi: 10.1038/nature22361
Salaemae, W., Booker, G., and Polyak, S. (2016). The role of biotin in bacterial physiology and virulence: a novel antibiotic target for Mycobacterium tuberculosis. Microbiol. Spectr. 4, 797–822. doi: 10.1128/microbiolspec.VMBF-0008-2015
Sassetti, C., and Rubin, E. (2003). Genetic requirements for mycobacterial survival during infection. Proc. Natl. Acad. Sci. U S A. 100, 12989–12994.
Schrader, S., Vaubourgeix, J., and Nathan, C. (2020). Biology of antimicrobial resistance and approaches to combat it. Sci. Transl. Med. 12:eaaz6992.
Sharbati-Tehrani, S., Stephan, J., Holland, G., Appel, B., Niederweis, M., and Lewin, A. (2005). Porins limit the intracellular persistence of Mycobacterium smegmatis. Microbiology 151(Pt 7), 2403–2410. doi: 10.1099/mic.0.27969-0
Sparks, I., Derbyshire, K., Jacobs, W., and Morita, Y. (2023). Mycobacterium smegmatis: the vanguard of mycobacterial research. J. Bacteriol. 205:e0033722. doi: 10.1128/jb.00337-22
Taneja, N., and Tyagi, J. (2007). Resazurin reduction assays for screening of anti-tubercular compounds against dormant and actively growing Mycobacterium tuberculosis, Mycobacterium bovis BCG and Mycobacterium smegmatis. J. Antimicrob. Chemother. 60, 288–293. doi: 10.1093/jac/dkm207
Tang, Q., Li, X., Zou, T., Zhang, H., Wang, Y., Gao, R., et al. (2014). Mycobacterium smegmatis BioQ defines a new regulatory network for biotin metabolism. Mol. Microbiol. Online ahead of print. doi: 10.1111/mmi.12817
Torrey, H., Keren, I., Via, L., Lee, J., and Lewis, K. (2016). High persister mutants in Mycobacterium tuberculosis. PLoS One 11:e0155127. doi: 10.1371/journal.pone.0155127
Urbaniec, J., Xu, Y., Hu, Y., Hingley-Wilson, S., and McFadden, J. (2022). Phenotypic heterogeneity in persisters: a novel ‘hunker’ theory of persistence. FEMS Microbiol. Rev. 46:fuab042. doi: 10.1093/femsre/fuab042
Van den Bergh, B., Michiels, J., Wenseleers, T., Windels, E., Boer, P., Kestemont, D., et al. (2016). Frequency of antibiotic application drives rapid evolutionary adaptation of Escherichia coli persistence. Nat. Microbiol. 1:16020. doi: 10.1038/nmicrobiol.2016.20
Vera-Cabrera, L., Molina-Torres, C., Hernández-Vera, M., Barrios-García, H., Blackwood, K., Villareal-Treviño, L., et al. (2007). Genetic characterization of Mycobacterium tuberculosis clinical isolates with deletions in the plcA-plcB-plcC locus. Tuberculosis 87, 21–29. doi: 10.1016/j.tube.2006.01.023
Wakamoto, Y., Dhar, N., Chait, R., Schneider, K., Signorino-Gelo, F., Leibler, S., et al. (2013). Dynamic persistence of antibiotic-stressed mycobacteria. Science 339, 91–95.
World Health Organization (2021). Global Tuberculosis Report 2021. Geneva: World Health Organization.
Keywords: persisters, Mycobacterium smegmatis, antibiotic tolerance, biofilm formation, in vivo survival
Citation: Joshi H, Kandari D, Maitra SS, Bhatnagar R and Banerjee N (2024) Identification of genes associated with persistence in Mycobacterium smegmatis. Front. Microbiol. 15:1302883. doi: 10.3389/fmicb.2024.1302883
Received: 27 September 2023; Accepted: 22 January 2024;
Published: 12 February 2024.
Edited by:
Rajni Garg, Amity University Punjab, IndiaReviewed by:
Tiago Beites, Universidade do Porto, PortugalBen Gold, Weill Cornell Medicine, United States
Copyright © 2024 Joshi, Kandari, Maitra, Bhatnagar and Banerjee. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Nirupama Banerjee, bmlydXBhbWFiYW5AeWFob28uY29t; Rakesh Bhatnagar, cmFrZXNoYmhhdG5hZ2FyQGpudS5hYy5pbg==
 Hemant Joshi
Hemant Joshi Divya Kandari
Divya Kandari Subhrangsu Sundar Maitra1
Subhrangsu Sundar Maitra1 Rakesh Bhatnagar
Rakesh Bhatnagar Nirupama Banerjee
Nirupama Banerjee