- 1School of Ecology and Environmental Sciences, East China Normal University, Shanghai, China
- 2School of Geographic and Oceanographic Sciences, Nanjing University, Nanjing, China
The discovery of the lanthanide requiring enzymes in microbes was a significant scientific discovery that opened a whole new avenue of biotechnological research of this important group of metals. However, the ecological impact of lanthanides on microbial communities utilizing methane (CH4) remains largely unexplored. In this study, a laboratory microcosm model experiment was performed using rice field soils with different pH origins (5.76, 7.2, and 8.36) and different concentrations of La3+ in the form of lanthanum chloride (LaCl3). Results clearly showed that CH4 consumption was inhibited by the addition of La3+ but that the response depended on the soil origin and pH. 16S rRNA gene sequencing revealed the genus Methylobacter, Methylosarcina, and Methylocystis as key players in CH4 consumption under La3+ addition. We suggest that the soil microbiome involved in CH4 consumption can generally tolerate addition of high concentrations of La3+, and adjustments in community composition ensured ecosystem functionality over time. As La3+ concentrations increase, the way that the soil microbiome reacts may not only differ within the same environment but also vary when comparing different environments, underscoring the need for further research into this subject.
1 Introduction
Methane (CH4) is an important greenhouse gas after carbon dioxide (CO2) with a global warming potential of 85 or 32 for 20- or 100-y time horizons, respectively (IPCC, 2021). Rice fields stand out as one of the major anthropogenic contributors to atmospheric CH4, accounting for 14.1 to 48% of total agricultural CH4 emissions (Carlson et al., 2017; Saunois et al., 2020; Gupta et al., 2021; Li et al., 2022; Duan et al., 2023; Qian et al., 2023).
Methylotrophs constitute a microbial group that can utilize single-carbon or multi-carbon compounds without carbon–carbon bonds, including methanol and methylamine, for growth and as energy source (Vu et al., 2021). CH4-oxidizing bacteria (methanotrophs) are a special case in this group because they are additionally capable of utilizing CH4 as their primary carbon and energy source (Hanson and Hanson, 1996). In rice fields, methanotrophs can oxidize up to 90% of the CH₄ before it reaches the atmosphere (Conrad and Rothfuss, 1991). Consequently, methanotrophs play a vital role as a biofilter, significantly contributing to the regulation of the global climate in agro-ecosystems like rice fields.
Lanthanides, a group of 15 elements from lanthanum (La3+, Z = 57) to lutetium (Z = 71) on the periodic table, along with scandium and yttrium, collectively known as Rare Earth Elements (REEs), have garnered increased interest over the past four decades. Their pivotal role in modern technology, serving as essential components in portable electronic devices and clean energy technologies, has fueled this growing interest (Leal Filho et al., 2023). Notably, REEs are also of interest in agriculture, being used as additives in animal feed to enhance livestock yield and as components in fertilizers to boost crop productivity (Balaram, 2019; Tommasi et al., 2021). REEs can be introduced to the soil through phosphate fertilizers, contributing to their accumulation in the soil (Silva et al., 2019; Dushyantha et al., 2020). For instance, the concentration of La3+ in the soil can vary widely from 1.1 to 413 mg/kg, but also can reach exceptionally high levels in areas close to mining areas, ranging from 1800.44 to 6905.24 mg/kg (Tyler, 2004; Li et al., 2010; Liang et al., 2014; Ramos et al., 2016).
Lanthanides were long considered biologically inactive due to their low bioavailability (Bulman, 2003) but a biological significance has recently been discovered. In the first step of CH4-oxidation, CH4 is converted to methanol which is then further oxidized to formaldehyde by an enzyme called methanol dehydrogenase (MDH). Calcium (II) ions have been thought of as the cofactor of MDHs (Anthony, 2000), but a novel form of MDHs is requiring lanthanides (Hibi et al., 2011; Pol et al., 2014; Skovran and Martinez-Gomez, 2015; Picone and op den Camp, 2019; Kato et al., 2020). It was demonstrated that the newly discovered functional MDH is primarily expressed even in the presence of 95 μM Ca2+ in the cultivation medium and with as little as 1 μM supplemental La3+ (Chu and Lidstrom, 2016). Despite these discoveries in physiology, there is still a knowledge gap for the role and function of La3+ in natural and agroecosystems. It is largely unknown whether methanotrophic communities in soils are inhibited or stimulated by the addition of La3+.
Because the diversity of methanotrophs is limited, elevated levels of La3+ in the soil may lead to a loss of function that could not be compensated by other microbes with dramatic consequences for atmospheric CH4 concentrations and global warming. However, La3+ is an important element in the metabolism of methanotrophs, and increasing concentrations in the environment could be beneficial with a positive effect on ecosystem processes, bioremediation, and biotechnological applications (Skovran and Martinez-Gomez, 2015; Dance, 2023). To the best of our knowledge, only Kravchenko and colleagues (Kravchenko et al., 2022) have examined the implications of La3+ addition on the composition and activity of methanotrophs in sod-podzolic soil (pH = 5.4). The addition of 5 μg La3+ stimulated the growth of the methanotrophic genus Methylobacter and the non-methanotrophic genus Methylotenera. In addition, a stimulating effect on CH4 oxidation was observed at 200 parts per million (ppm) in the headspace of their experimental setup (Kravchenko et al., 2023).
In this study, we focused on the impact of increasing amounts of La3+ in rice field soil. Therefore, we performed a comprehensive lab incubation study combining gas chromatography, mass spectrometry, sequencing, and quantitative real-time polymerase chain reaction (qPCR) to get a better understanding of the ecological implications of the increasing amounts of REEs on the soil microbiome, which plays a crucial role in mitigating atmospheric CH4 emissions. We selected three rice field soils with different pH conditions to explore the relationship between the soil microbiome and the bioavailability and mobility of La3+ within this soil ecosystem. Previous studies clearly demonstrated the direct impact of pH on the solubility, mobility, and bioavailability of La in the soil (Iftekhar et al., 2018; Kotelnikova et al., 2021; Zhao et al., 2021). We hypothesized that La3+ in the form of lanthanum chloride (LaCl3) will stimulate methanotrophic activity and growth due to their availability to utilize lanthanides in their enzyme machinery. We further hypothesized that the soil origin and their specific soil properties (e.g., pH) will influence the La3+ retention capacity and consequently the effect on the soil microbiome.
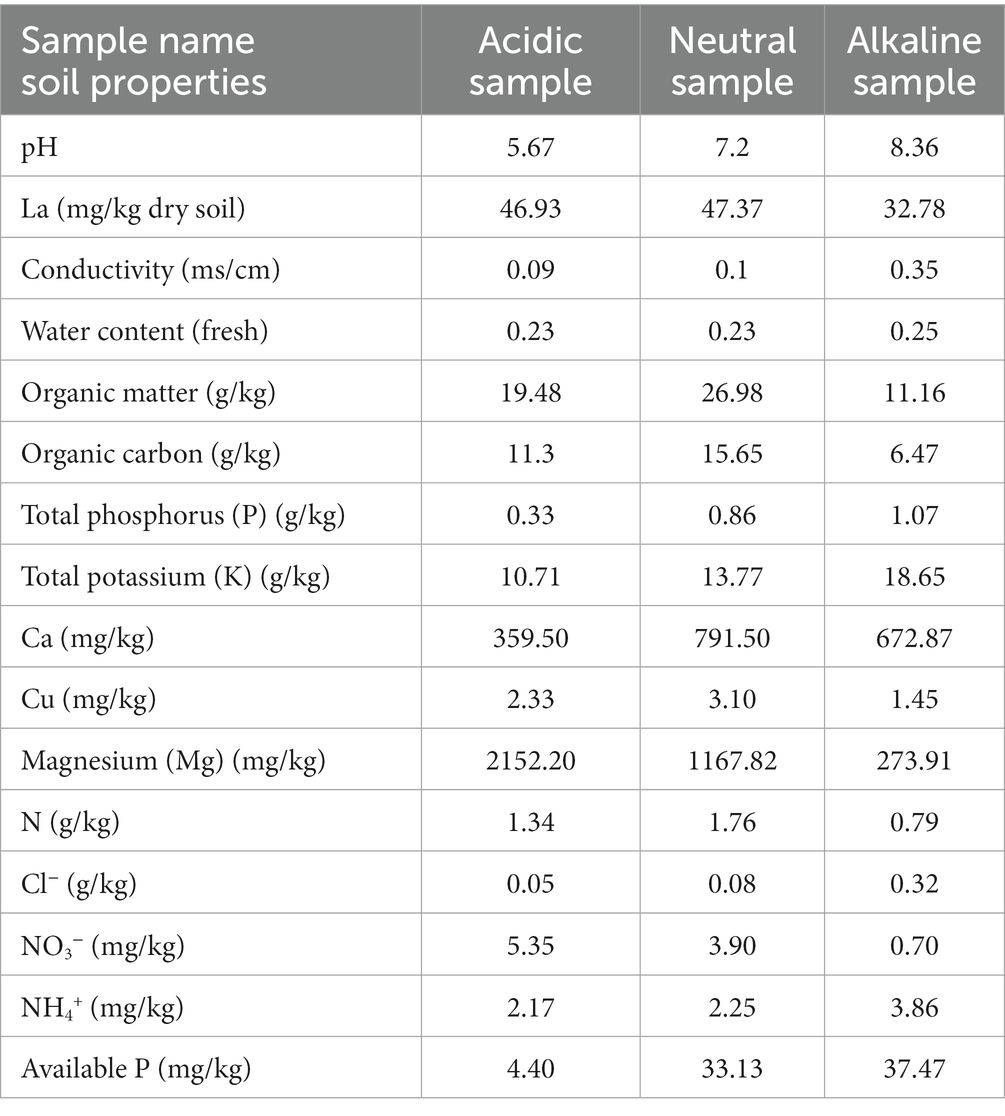
2 Materials and methods
2.1 Study sites
In October 2020, three rice field soil samples with different pH (0–20 cm) were collected from Jiangsu Province, China, each originating from varying pH levels of 5.76, 7.2, and 8.36, representing acidic, neutral, and alkaline soils, respectively. Acidic soil samples were collected from Yangzhou City (32°24′ N, 119°2′ E). Neutral soil samples were collected from Xuzhou City (34°15′ N, 118°8′ E). Alkaline soil samples were collected from Yancheng City (33°17′ N, 120°34′ E). For incubation experiments, soil samples were stored at 4°C. However, soil physicochemical properties were determined from fresh or frozen soil (−20°C) except for the determination of La content in the original soil samples. All measured properties are summarized in Table 1. Detailed description of methods was published in a previous study (Guo et al., 2022) and carried out by Nanjing innovation biotechnology Co., LTD. The determination of La was carried out by TÜV Rheinland (Shanghai) Co., Ltd., China using Inductively Coupled Plasma Mass Spectrometry (ICP-MS).
2.2 Experimental setup and sampling
In August 2022, 2 grams (g) of rice field soils and 25 milliliters (mL) of diluted nitrate mineral salt (NMS) Ca2+ medium (0.1 × strength, Dedysh and Dunfield, 2017) were mixed in 100 mL transparent serum bottles, these bottles were then placed in the shaker (MQT-60R, Shanghai Minquan Instrument Co., Ltd., China) and incubated in the dark at 20°C and 120 revolutions per minute (rpm) for 4 weeks. The diluted NMS medium consisted of 22.5 mL different concentrations of La3+ in the form of LaCl3 (Sangon Biotech (Shanghai) Co., Ltd., China) and 2.5 mL NMS (Ca2+) medium. To find suitable concentrations of La3+ for our experiment, we first searched the literature for ecotoxicological studies that applied La3+ (e.g., Chu et al., 2003; Li et al., 2018; Kotelnikova et al., 2019). We then chose LaCL3 concentrations ranging from 2 mg/L to 1,200 mg/L to determine the levels that depicted the strongest response in CH4 oxidation. Finally, the chosen concentrations included no added La3+ (control), 300 mg/L (low), 600 mg/L (intermediate), and 1,200 mg/L (high), denoted as control CK, L, M, and H levels, respectively.
All chemical reagents were purchased through Sangon Biotech (Shanghai) Co., Ltd., China. Each soil sample has four La3+ treatments in triplicate, 36 bottles in total. All bottles for the experiment were thoroughly washed with 1 mol/L hydrochloric acid (HCl) overnight and autoclaved before use, sealed with butyl rubber stoppers, and capped with plastic aluminum shells.
All 36 bottles were pre-incubated on a shaker at 20°C and 120 rpm in the dark for 24 h. During the experiment, on the first day of each week, the headspace of the bottles was flushed three times with air using a 60 mL syringe. Subsequently, 1.5 mL of 99.999% high-purity CH4 (Shanghai Jiaya Chemical Co., Ltd., China) was added resulting in a starting CH4 concentration of approximately 10,000 parts per million volume (ppmV) (~1%) in the headspace reflecting on typical in-situ methane concentrations in rice fields.
On the seventh day of each week, 2.5 mL of a well-mixed cultivation solution was extracted from the bottles. The extracted solution was then transferred to centrifuge at 7,800 rpm for 12 min using an Eppendorf Microcentrifuge 5,430 R (Eppendorf, Germany). Subsequently, the resulting supernatant underwent analysis for pH (S220-K, METTLER TOLEDO, Switzerland), and the remaining samples were stored in a − 80°C refrigerator for subsequent molecular analyses and La3+, NH4+, Ca2+, NO3−, and PO43− determination. The CH4 concentration in the headspace was determined using GC (Nexis GC-2030, Shimadzu, Japan) equipped with a Flame Ionization Detector (FID).
2.3 Determination of La3+ in the soil
The analysis of La3+ levels in the soil during the fourth week was carried out by Shanghai WEIPU Testing Technology Group Co., Ltd. (Shanghai, China). In brief, approximately 0.1 g of each soil sample was placed into a microwave digestion vessel and 4 mL of nitric acid was added for microwave digestion. Subsequently, each sample was tested using the Inductively Coupled Plasma-Optical Emission Spectrometer (ICP-OES) (PerkinElmer Avio 500, United States).
2.4 Determination of La3+, NH4+, Ca2+, NO3−, and PO43− ions in the supernatant
All analyses were performed by the Shanghai WEIPU Testing Technology Group Co., Ltd. (Shanghai, China). In brief, for the La3+ determination 0.1 mL of the supernatant was transferred into digestion tubes, followed by the addition of 4 mL of nitric acid. The tubes were then heated at 120°C for 4 h until the solution became clear and transparent. Subsequently, the digestive solution was adjusted to a constant volume of 10 mL. La3+ was determined using either ICP-MS (PerkinElmer NexION 2000, United States) or ICP-OES (PerkinElmer AvioVIO 500, United States), depending on the concentration range. NH4+, Ca2+, NO3−, and PO43− concentrations were determined by diluting each sample 10–20 times with water, and then the aqueous solution was filtered through a 0.22 μM membrane into a 2 mL Agilent vial and an injection tube for ion chromatography analysis using the Dionex ICS-5000+ ion chromatography system (Thermo Fisher Scientific, USA).
2.5 Soil DNA extraction and 16S rRNA gene analysis
The DNeasy PowerSoil® Pro Kit (Qiagen, Germany) was used to extract total soil community genomic DNA from 0.2–0.25 g of each sample, following the manufacturer’s steps. The concentration of the DNA solution was checked through NanoDrop™ One Microvolume UV–Vis Spectrophotometer (Thermo Fisher Scientific, United States). The variable regions V1-V3 of bacterial 16S rRNA gene were amplified using the primers of 27F (AGRGTTTGATCMTGGCTCAG) (Lane, 1991) and 519R (GWATTACCGCGGCKGCTG) (Lane et al., 1985). The process of PCR amplification of the 16S rRNA gene, followed by library preparation, quantification, and sequencing was executed using the Illumina MiSeq platform (Illumina MiSeq, United States) at Sangon Biotech (Shanghai) Co., Ltd., China. The reaction system of PCR was used as follows: microbial DNA (10–20 ng/μL) 2 μL; amplicon PCR forward primer (10 μM) 1 μL; amplicon PCR reverse primer (10 μM) 1 μL; 2 × Hieff® Robust PCR Master Mix (Yeasen, 10105ES03, China) 15 μL; 30 μL in total. The following PCR program was used: 1 cycle of denaturing at 94°C for 3 min, first 5 cycles of denaturing at 94°C for 30 s, annealing at 45°C for 20 s, elongation at 65°C for 30°C for 30 s, then 20 cycles of denaturing at 94°C for 20 s, annealing at 55°C for 20 s, elongation at 72°C for 30 s and a final extension at 72°C for 5 min.
16S rRNA gene high-throughput raw sequencing data was analyzed with the package of DADA2 version 1.26.0 (Callahan et al., 2016) in R 4.2.2 (Team, R. C, 2022). In brief, 16S rRNA gene forward and reverse reads were trimmed at positions 290 and 250 from the end or any position. Error models were constructed with random sample picking. The resulting amplicon sequence variances (ASVs) were taxonomically classified by using the SILVA ribosomal RNA database SSU version 138 (Pruesse et al., 2012). ASVs that were identified as mitochondrial or chloroplast sequences, or not classified into prokaryotic phyla were removed from the dataset. Removing ASVs less than 0.001% left 5,794 ASVs in the final dataset, 95.12% of total reads were retained.
2.6 qPCR of the pmoA gene
This experiment was conducted by Sangon Biotech (Shanghai) Co., Ltd., China. In brief, the pmoA gene was quantified using a specific qPCR assay to enumerate the total abundance of methanotrophs (MTOT assay; Kolb et al., 2003). The pmoA gene encodes the β-subunit of the particulate methane monooxygenase (pMMO) which catalyzes the first step in CH4-oxidation. It has been widely used as a functional marker to identify methanotrophs from the environment (Dedysh and Knief, 2018). The primer sequences were A189 F (GGN GAC TGG GAC TTC TGG) and mb661R (CCG GMG CAA CGT CYT TAC C) (Holmes et al., 1995; Costello and Lidstrom, 1999). In addition, this primer set was shown to have a broad coverage of methanotroph diversity (Bourne et al., 2001). The qPCR protocols for the pmoA gene were executed as follows: initial pre-denaturation at 95°C for 4 min; followed by denaturation at 95°C for 15 s; annealing at 67°C for 20 s; and extension at 72°C for 30 s. The previous steps comprised 10 cycles, with the annealing temperature decreasing by 1°C after each cycle. Subsequently, denaturation was carried out at 95°C for 15 s, followed by annealing at 57°C for 20 s, and extension at 72°C for 30 s, repeated for a total of 35 cycles. Copy numbers of target genes were derived from plasmid DNA. The PCR efficiency was on average 92.8% with an R2 value of 0.9972. Amplicon specificity was determined from melt curve analysis and confirmed by 1.5% agarose gel electrophoresis.
2.7 Statistical analysis
The CH4 concentration in the headspace was calculated by using a CH4 standard gas (1,002 ppmV) measured on the GC before sample measurements. pmoA gene copy numbers were expressed as per gram dry weight.
The Shannon index was calculated using the R package vegan 2.6.4 (Oksanen et al., 2022), with prior sequence flattening. The variation in microbial community compositions with different La3+ treatments and time points were assessed through non-metric multidimensional scaling (NMDS). First, ASVs with relative abundance below 0.05% were removed, reducing sparsity from 94 to 62.73%. Next, Bayesian-Multiplicative replacement of count zeros (cmultRepl) in the R package zCompositions package version 1.4.0.1 (Palarea-Albaladejo and Martín-Fernández, 2015) was employed to impute zeros in compositional count data. Then, a centered log-ratio (CLR) transformation was applied using R package compositions version 2.0.4 (van den Boogaart et al., 2022). For assessing the dissimilarity among various treatments and time points, Permutational multivariate analysis of variance (PERMANOVA) was performed using the function of adonis2 in the R package vegan version 2.6.4. NMDS plots at a 95% confidence level were generated using the “stat_ellipse” function in the R package ggplot2 version 3.4.1 (Wickham, 2016). R package reshape2 version 1.4.4 (Wickham, 2007), dplyr version 1.1.0 (Wickham et al., 2023), and tidyverse version 2.0.0 (Wickham et al., 2019) were utilized to process the data to calculate relative abundance plots. The ddply function in the R package plyr version 1.8.8 (Wickham, 2011) was employed to calculate the mean value ± standard deviation of pH in the supernatant. The leveneTest function in the R package car version 3.1.2 (Fox and Weisberg, 2019) was used to conduct the homogeneity of variance test of data. Analysis of variance (ANOVA) was performed using the stat_compare_means function in the R package ggpubr version 0.6.0 (Kassambara, 2023). Following that, post hoc tests were performed using the Least Square Difference (LSD) test in R package agricolae version 1.3.5 (de Mendiburu, 2021).
3 Results
3.1 CH4 Consumption is affected by the addition of La3+
We observed a consistent trend of inhibited CH4 consumption across all treatments, with the degree of inhibition corresponding to the concentrations of La3+ added to the soil slurries. It is important to note that the extent of inhibition correlated with the pH of the soil slurries, each derived from rice fields with distinct acidity levels: acidic, neutral, and alkaline (Figure 1).
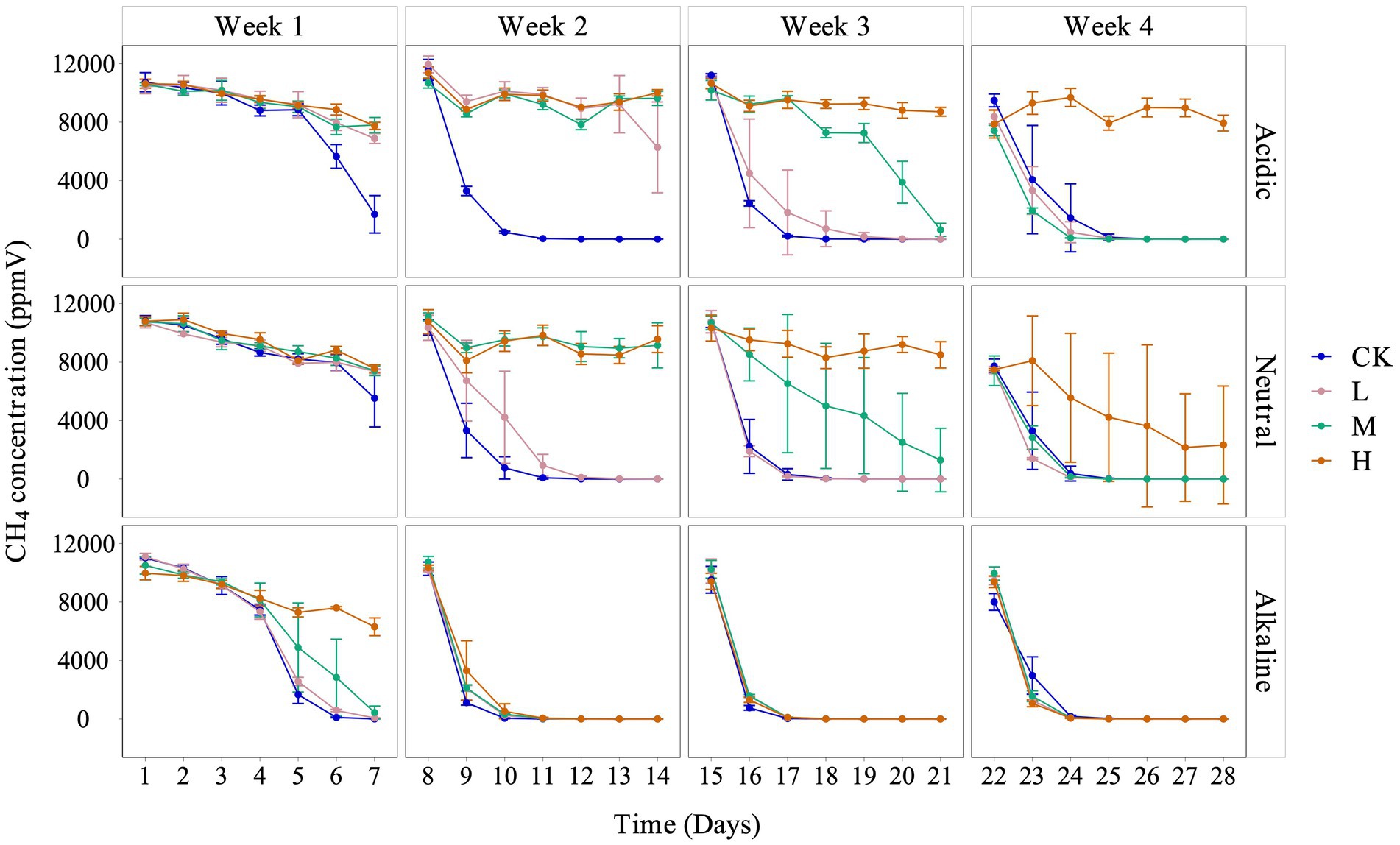
Figure 1. Variation of CH4 concentration in the headspace with different pH conditions for control (CK), 300 (L), 600 (M), and 1200 (H) mg/L La3+ treatments, from top to bottom are acidic, neutral, and alkaline samples, respectively. Error bars indicate the standard deviation (n = 3).
Although there was a clear inhibition of CH4 consumption, the data revealed an interesting trend: during the fourth week, in soil slurries from acidic and neutral rice fields, the low and intermediate treatments exhibited a faster rate of CH4 consumption between 22 and 24 days. This trend was also observed across all La treatments throughout the entire fourth week cultivation period, in soil slurries from alkaline rice fields (Figure 1).
3.2 The effect of La3+ on the methanotrophic community composition, microbial diversity, and methanotroph abundance
We identified a total of 13 genera of methylotrophs using Illumina sequencing of the 16S rRNA gene (Figure 2). These types were distributed among five families: Beijerinckiaceae (comprising Methylocella, Methylorosula); Crenotrichaceae (Crenothrix); Hyphomicrobiaceae (Methyloceanibacter); Methylococcaceae (including Methyloparacoccus. Methylobacter, Methylomicrobium, Methylomonas, and Methylosarcina); Methylocystaceae (comprising Methylocystis and Methylosinus); and Methylophilaceae (Methylotenera and Methylovorus sp. strain MM2). Among these, Methyloceanibacter, Methylorosula, Methylotenera, and Methylovorus sp. strain MM2 were identified as facultative methylotrophs, while the remaining 9 genera were classified as obligate methanotrophs. Notably, Methylocella, Methyloparacoccus, and Methylorosula were absent from the neutral and alkaline soil samples.
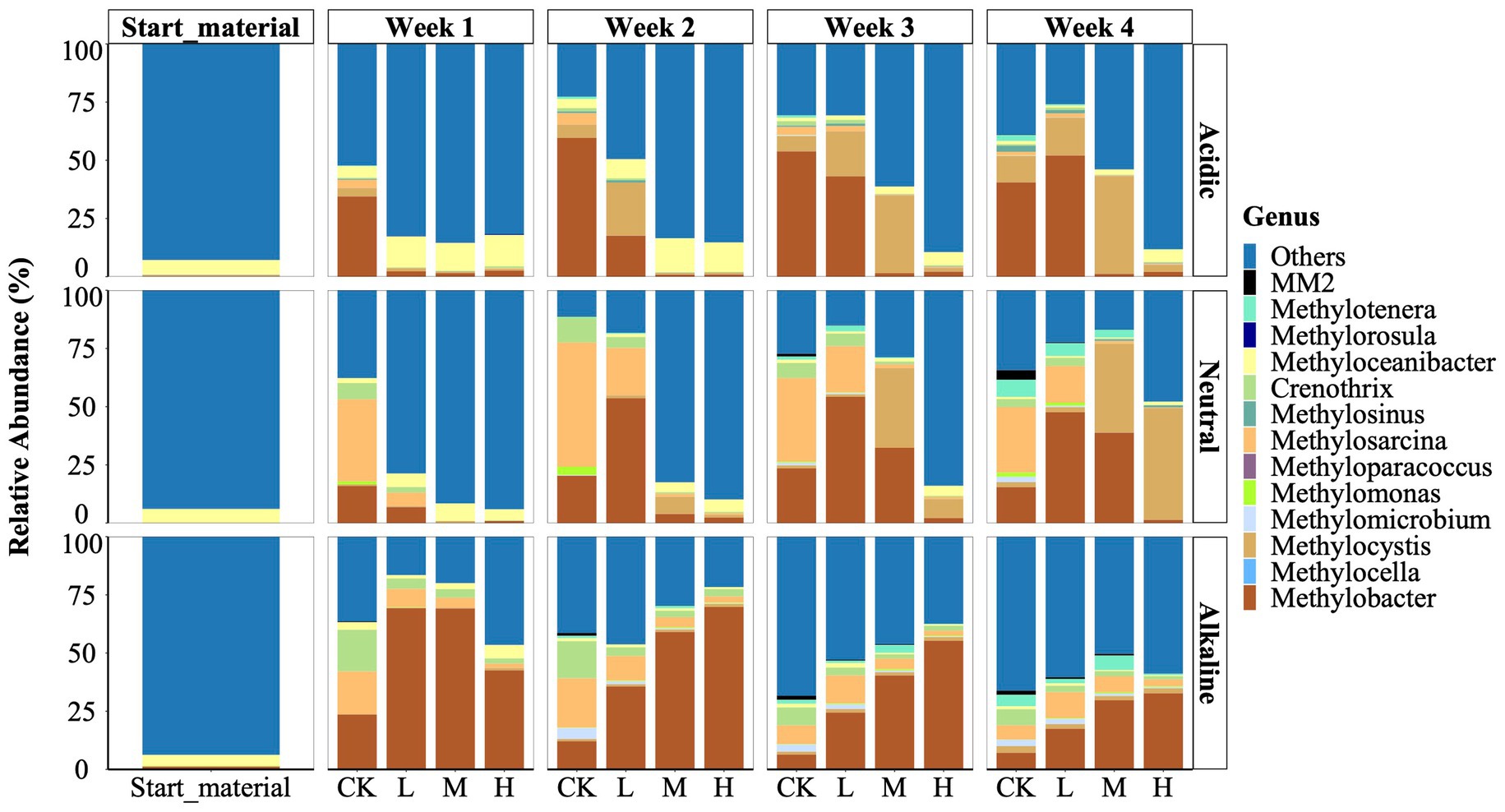
Figure 2. Relative abundance of the methylotrophs of starting material, control (CK), 300 (L), 600 (M), and 1200 (H) mg/L La3+ treatments in varying pH rice field soil samples, from top to bottom, respectively, are acidic, neutral, alkaline samples. Data is based on 16 s rRNA gene sequencing data (see Material and Methods for details).
We observed that the genus Methyloceanibacter was a prevalent microbe across the three initial rice field soil samples with a relative abundance of 6.3, 6.0, and 4.6% in the acidic, neutral, and alkaline samples, respectively (Figure 2). Representatives of Methyloceanibacter have been described to also oxidize CH4, but possess solely a SMMO, and were mainly found in marine environments (Vekeman et al., 2016).
We further observed that during the incubation period, the genera Methylobacter, Methylocystis, and Methylosarcina emerged as the dominant methanotrophs in the control treatment. However, their response to La3+ addition displayed variations among soil slurries with different initial pH levels: acidic, neutral, and alkaline (Figure 2). In both soil slurries from acidic and neutral rice fields, the application of higher doses of La3+ consistently resulted in lower relative abundances of the genus Methylobacter. For instance, in the treatments with high amount of added La3+ (referred to as the “H treatments”), Methylobacter’s relative abundance ranged from 0.9 to 2.6%. In contrast, in the corresponding control treatments (referred to as the “CK treatments”) for acidic samples, Methylobacter’s relative abundance remained notably higher, ranging from 34.4 to 59.6% (Figure 2). Once CH4 consumption was resumed (Figure 1) we also observed higher relative abundance of the genus Methylobacter (Figure 2). Intriguingly, when examining soil slurries from alkaline soil, we observed a distinct relationship between the relative abundance of the genus Methylobacter and the amount of added La3+. Specifically, in the CK and treatments with low, intermediate, and high levels of added La3+, the relative abundance of Methylobacter increased progressively between treatments, such as its abundance was 12.1% (CK), 35.7% (L), 59.1% (M), and 69.8% (H) in week 2 (Figure 2).
Next, we noticed a significant shift in the predominant methanotroph population from the genus Methylobacter (Type I) to the genus Methylocystis (Type II) in soil slurries from acidic and neutral rice field soils at higher La3+ concentrations (Figure 2). In soil slurries from the acidic rice field, Methylobacter dominated the CK treatments consistently throughout the cultivation process, while Methylocystis gradually took over in the M treatment after the second week. For instance, in the M treatments, Methylocystis showed a notably higher relative abundance during week 3 (33.4%) and week 4 (41.9%) compared to Methylobacter, which only accounted for 1.5% during week 3 and 1.2% during week 4. In soil slurries from the neutral rice field, Methylocystis progressively established as the dominant species within the M and H treatments. For instance, in week 4, Methylocystis showed a significantly higher relative abundance (48%) in the H treatment compared to Methylobacter, which had only 1.3% relative abundance.
In general, the relative abundance of non-methanotrophic taxa such as Afipia, Pseudolabrys, and Pseudarthrobacter increased with La3+ addition in acidic soil slurry (Supplementary Figure S1A). Noteworthy, in alkaline soil slurries, the ASV6-Bdellovibrionaceae-Bdellovibrio emerged as the most predominant non-methanotrophic organism. Notably, the Bdellovibrio genus is recognized as a bacterial predator (Feng et al., 2017). The relative abundance of the ASV6-Bdellovibrionaceae-Bdellovibrio in L treatment was higher than that in the CK treatment, Specifically, in the L treatments, its relative abundance was 12.9, 8.3, and 11.7% from week 2 to week 4, while in the CK treatments it was 7.2, 5.2, and 4.3%, respectively (Supplementary Figure S1C). Finally, in soil slurries that displayed inhibition of CH4 consumption (Figure 1) we observed that the community composition resembled its original inoculum suggesting that the community did not exhibit any growth (Supplementary Figure S1).
We used NMDS and PERMANOVA to provide further evidence for the observed shifts in community composition. NMDS plots clearly illustrated a significant effect of added La3+ on the composition of microbial communities in soil slurries from the acidic (p < 0.05), neutral (p < 0.001), and alkaline (p < 0.001) rice fields (Supplementary Figure S2).
The Shannon diversity index (SDI) was used to describe the response of the total microbial diversity to the addition of different amounts of La3+. The results showed that, in all experimental treatments including the control, the complexity of microbial communities simplified when compared to the starting material over the course of the experiment (Supplementary Figure S3). However, we observed that the diversity values for the H treatments of soil slurries from the acidic rice field in week 3 (SDI = 5.70) and week 4 (SDI = 5.52) nearly matched that of the initial material (SDI = 5.59). A similar trend was observed for the diversity values for the M (SDI = 5.37) and H (SDI = 5.48) in week 2 and the H (SDI = 5.38) treatment in week 3 of soil slurries from the neutral rice field. This similarity to the starting material indicated and artifact and suggested that the addition of La3+ likely inhibited the total microbial community (see Supplementary Figure S3).
Overall, pmoA gene copy numbers (per g) did not vary significantly by the end of the experiment (Figure 3). However, in soil slurries from the acidic rice field, a significant inhibitory effect of added La3+ was observed but this diminished in the last week of the experiment. For instance, during the second week, the pmoA gene copy numbers (per g) in the CK, L, M, and H groups were 7.12 × 107, 2.35 × 107, 2.38 × 106, and 2.20 × 106, respectively. In soil slurries from the neutral rice field, we observed no significant differences but observed a similar trend in the second week of the experiment. In the second week, pmoA gene copy numbers (per g) for the CK, L, M, and H groups were 1.27 × 108, 7.33 × 107, 8.49 × 106, and 5.15 × 105, respectively. In soil slurries from the alkaline rice field, we only observed a significant inhibition of added La3+ in the second week, the pmoA gene copy numbers (per g) in the CK, L, M, and H groups were 1.01 × 108, 2.63 × 107, 1.69 × 107, and 3.32 × 107, respectively (Figure 3).
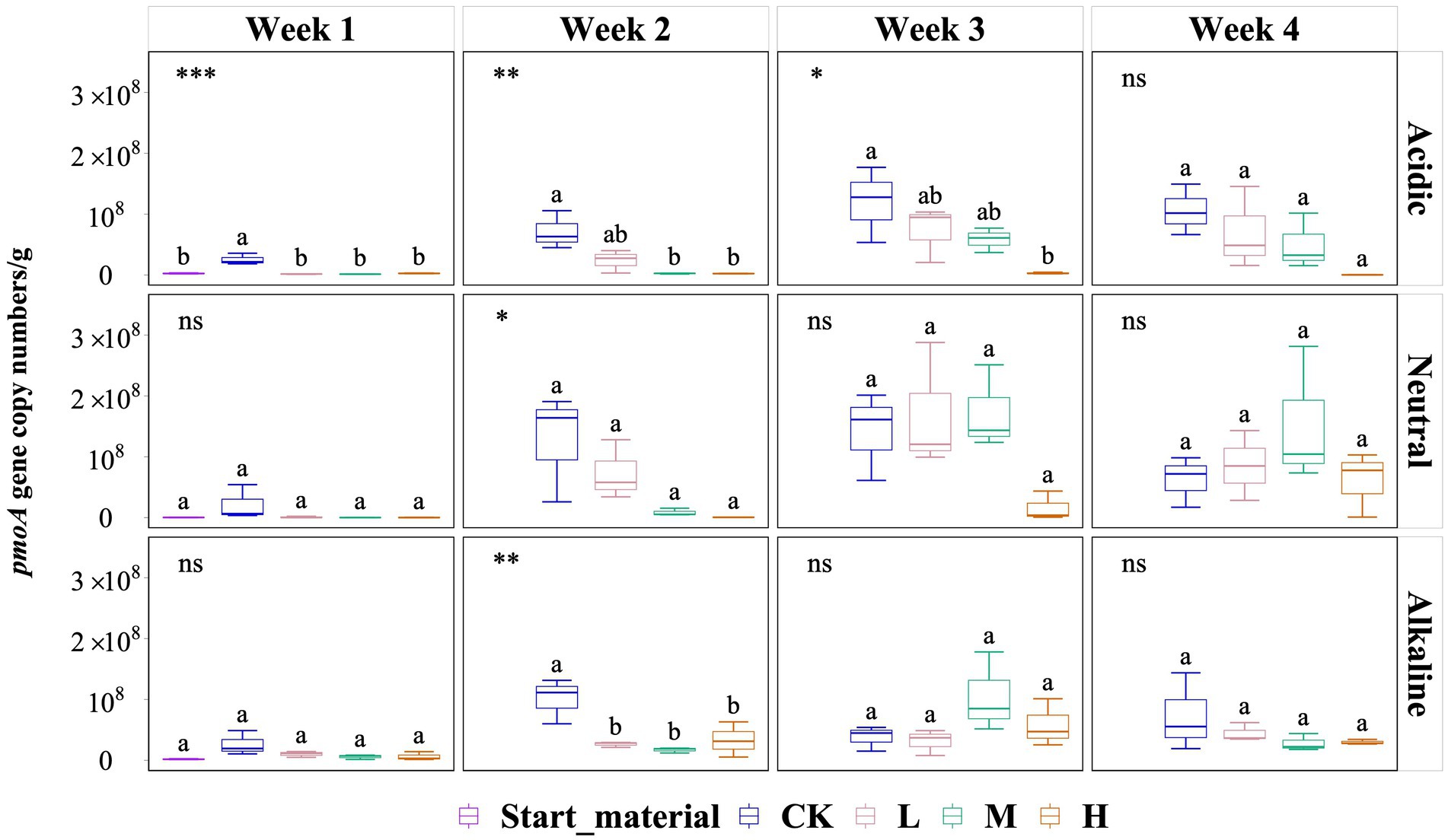
Figure 3. Variation of pmoA gene copy numbers (per g) in three samples with varied pH rice field soil conditions for start material, control (CK), 300 (L), 600 (M), and 1200 (H) mg/L La3+ treatments. Error bars indicate the standard deviation (n = 3) (*, p < 0.05; **, p < 0.01; ***, p < 0.001; ****, p < 0.0001; ns, no significance). Distinct letters in the plot indicate statistically significant differences among different La3+ treatments (p < 0.05).
3.3 Added La3+ affected pH in soil slurries and the retention of La3+ in soil particles
The pH in the supernatant indicated a clear relationship between added La3+ and acidity of the soil slurries from acidic and neutral rice fields (Table 2). In contrast, soil slurries from the alkaline rice field did not show any effect of added La3+ on the pH indicating a higher buffer capacity of that soil (Table 2).
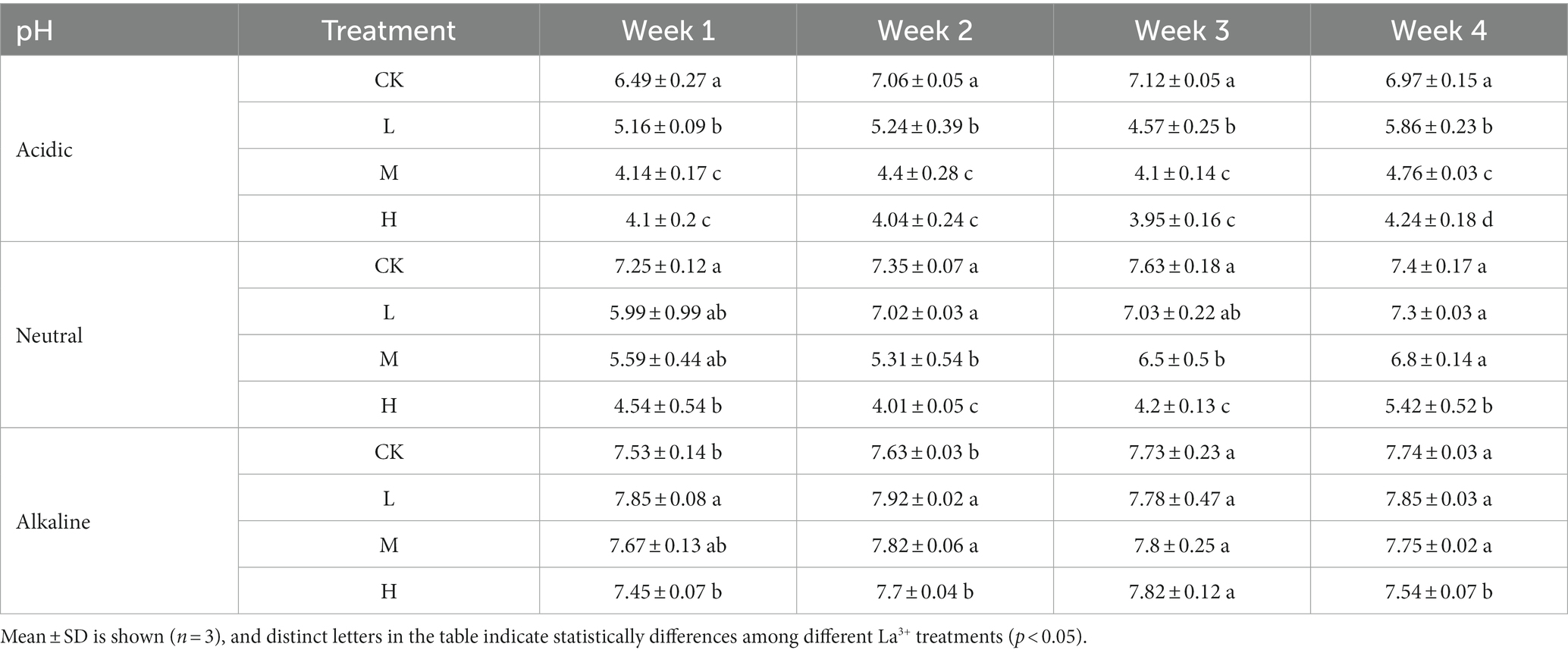
Table 2. pH of the supernatant for control (CK), 300 (L), 600 (M), and 1200 (H) mg/L La3+ treatments in samples with different pH levels.
The concentration of La3+ in soil particles at the end of the experiment was similar across soil slurries from rice fields with different pH values (Figure 4). There were no significant differences in La3+ absorption capacity between CK, L, and M treatments from all soil types, except for soil slurries from the acidic rice field in the H treatment (p < 0.05). In this case the La3+ content reached values of 4,560 mg/kg (acidic soil condition), 7,830 mg/kg (neutral soil condition), and 6,982 mg/kg (alkaline soil condition).
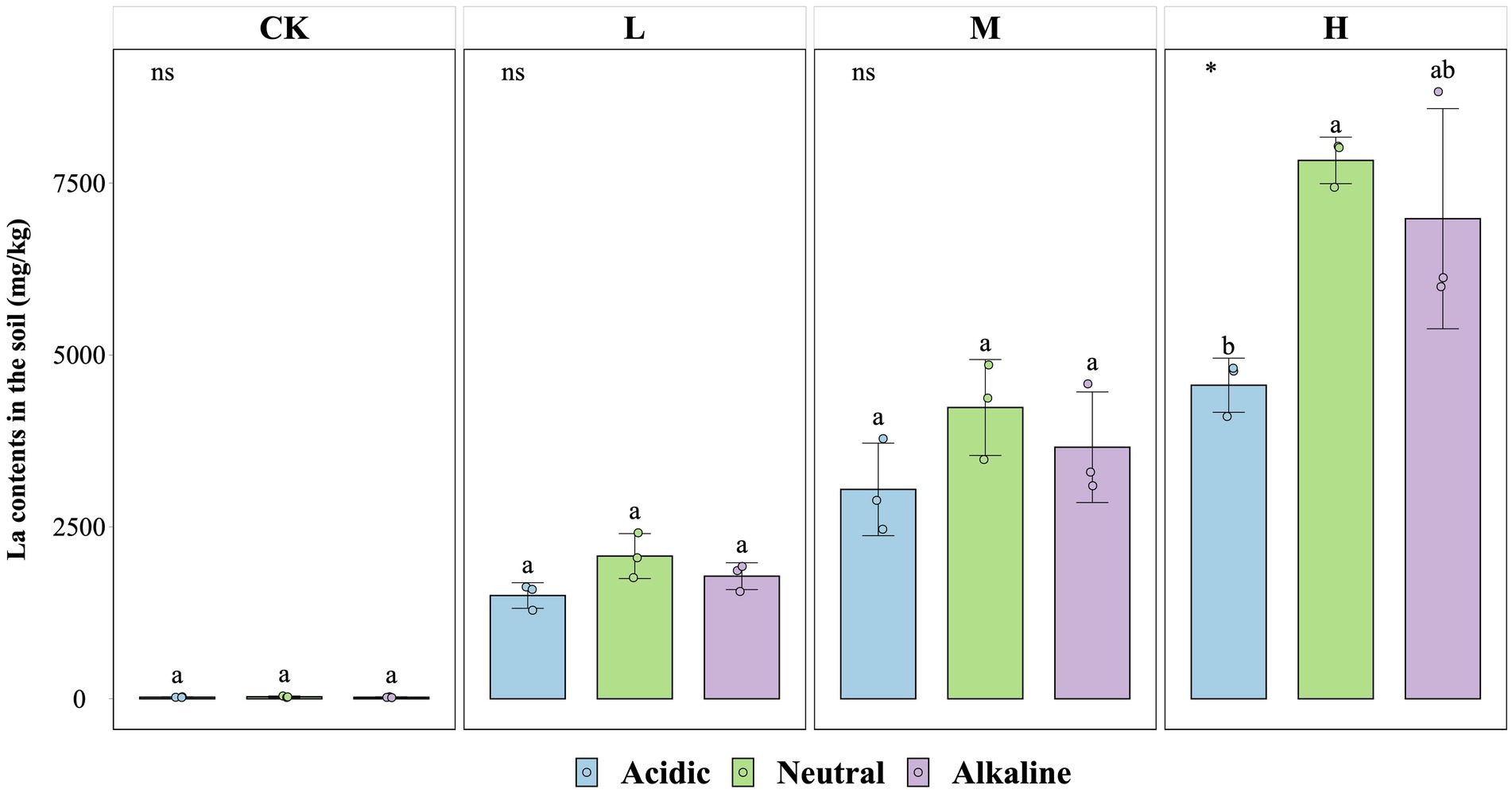
Figure 4. La contents in soil for the fourth week for control (CK), 300 (L), 600 (M), and 1200 (H) mg/L La3+ treatments (*, p < 0.05; **, p < 0.01; ***, p < 0.001; ****, p < 0.0001; ns, no significance). Error bars indicate SE (n = 3). Distinct letters in the plot indicate statistically differences among different pH samples (p < 0.05).
We also measured the La3+ content in the supernatant at the end of each week of the experiment to identify patterns in the bioavailable La3+ (Supplementary Figure S4). No consistent patterns were observed. However, a notable discrepancy in La3+ concentration was observed between the H and the L treatments in soil slurries from acidic rice fields during weeks 2, 3, and 4. Specifically, the La3+ concentration in the H treatments measured 140.73 mg/L, 133.53 mg/L, 143.27 mg/L, respectively, surpassing that of the L treatments in week 2 (64.93 μg/L), week 3 (171.83 μg/L), and week 4 (191.33 μg/L).
In soil slurries from the neutral rice field, the La3+ concentrations in the CK and L treatments were below the detection limit of the machine (5 μg/L) but we observed a similar trend as seen before that the contents of La3+ in the H treatments far greater than the M treatments (Supplementary Figure S4). The La3+ concentrations in the H treatments were 16.77 mg/L (week 1), 14.98 mg/L (week 2), 13.67 mg/L (week 3), and 16.04 mg/L (week 4), whereas in the M treatments, the corresponding concentrations were 346 μg/L (week 1), 200.23 μg/L (week 2), 100.97 μg/L (week 3), and 326.87 μg/L (week 4). Moreover, when analyzing soil slurries from the alkaline rice field, it was evident that the La3+ content was lower compared to acidic and neutral samples. The concentrations of La3+ in week 1, in the L treatments in week 2 and 3, and in the M treatments in week 3, and CK treatments in week 4 were all below the detectable limit of the machine (5 μg/L). Interestingly, the La3+ concentration in solution displayed a positive correlation with the amount of added La3+ in week 4 (76.60, 170.83, and 228.93 μg/L for L, M, and H treatments, respectively).
3.4 NH4+, Ca2+, NO3−, and PO43− concentration changes in supernatant of soil slurries of different soil types
We also measured important soil chemical data in the first and last week of the experiment (Supplementary Figure S5). Firstly, concentration levels of NH4+ and PO43− in the supernatant were found to be below the detection limit of the machine (0.1 mg/kg and 0.5 mg/kg, respectively, Supplementary Figure S5). Secondly, NO3− concentration levels did not show any notable differences in the first week. By week 4 an overall decrease was observed in all setups. In the supernatant of soil slurries from acidic rice fields the NO3− concentration in week 4 decreased by 49.89% (CK), 44.58% (L), 31.85% (M), and 3.28% (H) compared to their respective concentrations in week 1. In the supernatant of soil slurries from neutral rice fields the NO3− concentration decreased by 31.30, 50.96, 36.73, and 14.46% for CK, L, M, and H treatments, respectively. In the supernatant of soil slurries from alkaline rice fields the NO3− concentration decreased by roughly 22.55, 45.63, 44.07, and 43.79% for CK, L, M, and H treatments, respectively. Finally, we observed a positive correlation between the Ca2+ concentration in the supernatant of soil slurries and La3+ dosage in week 1 and 4 of the experiment (Supplementary Figure S5). In week 4, Ca2+ concentration in the supernatant of soil slurries from the acidic rice field reached the following levels: 1.80 mg/kg (CK), 33.83 mg/kg (L), 72.29 mg/kg (M), and 103.77 mg/kg (H). In neutral rice field soil slurries, its contents were 28.60 mg/kg (CK), 49.74 mg/kg (L), 93.77 mg/kg (M), and 175.18 mg/kg (H). Ca2+ concentration in the supernatant of soil slurries from the alkaline samples reached the following levels in week 4: 40.48 mg/kg (CK), 56.40 mg/kg (L), 98.56 mg/kg (M) and 193.65 mg/kg (H) (Supplementary Figure S5).
4 Discussion
In this laboratory microcosm study, we aimed to gain a better understanding of the ecological implications of increasing amounts of REEs, in terms of the soil microbiome which plays a crucial role in mitigating CH4 emissions in agricultural soils. We specifically chose three rice field soils with different pH conditions because the pH is an important regulating factor for the bioavailability of La3+ in aquatic and soil ecosystems (Ran and Liu, 1993; Cao et al., 2001).
The addition of external La3+ changed the activity of CH4-consuming microbial communities in the soil slurries. We observed an inhibition of CH4 consumption based on the amount of added La3+, time, and soil origin. In line with the CH4 consumption data (Figure 1), the response of the soil microbiome followed a similar pattern with no significant changes in community composition from the starting material at higher concentrations of added La3+, especially in slurries with soil from acidic rice fields (Supplementary Figure S1). As such, our experimental data did not support our hypothesis that the soil microbiome, involved in CH4 consumption, will be stimulated by the addition of La3+ (Figure 2). Our data rather suggests that there are complex interactions between the soil origin, environmental conditions (e.g., pH), and the soil microbiome, involved in CH4 consumption. Hence, the soil microbiome’s response to increasing concentrations of La3+ may not only vary within a single ecosystem but also between ecosystems, calling for additional research on this topic.
One possible explanation for the inhibitory effect of La3+ on the soil microbiome in our experiment may be attributed to chemical interactions between this element and other significant elements important in microbial cellular functions. For instance, Ca2+ are versatile signaling molecules that impact almost every aspect of cellular life and play crucial roles in physiological functions such as cell migration and growth (Clapham, 2007; Giorgi et al., 2018). Numerous studies have reported analogous chemical properties between La3+ and Ca2+ owing to their closely matched ionic radius, charge, ligand specificities, and biological roles (e.g., Reeves and Condrescu, 2003). These similarities suggest that La3+ can effectively substitute Ca2+, with its trivalent ion displaying a higher affinity for given binding sites compared to Ca2+ (Peng et al., 2004; Dudev et al., 2005; Kotelnikova et al., 2021).
First, the La3+ initially present in the solution could have facilitated an exchange with the Ca2+ present in the soil, resulting in the release of Ca2+ into the solution while La3+ integrated into the soil. Most of the available Ca2+ from the soil might have been replaced this way. Our data supports this idea, because we observed an increasing concentration of available Ca2+ in the supernatant of soil slurries when higher amounts of La3+ were added, particular in alkaline soil slurry in week 1 (Supplementary Figure S5); and most of the La3+ was found in the soil particles of the soil slurries (Figure 4) and less La3+ in the supernatant (Supplementary Figure S4).
Second, Ca2+ ions in biological cells could have been replaced by La3+ ions which has been shown to inhibit cellular functions of microorganisms (Aussel et al., 1996; Mu et al., 2006; Jiang et al., 2008). In addition, the excessive amount of La3+, may have blocked Ca2+ channels on the cell membrane and subsequently interacting with membrane related enzymes and impairing the functionality of Ca2+-dependent processes (Aussel et al., 1996; Wang et al., 2007). These findings suggest that the described processes involving La3+ and Ca2+ exchange may similarly apply to methanotrophs, potentially influencing their cellular functions and disrupting Ca2+-dependent processes.
The low pH in soil slurries from the acidic and neutral rice fields is another possible driving factor of the observed activity and community composition of CH4 consuming communities in our laboratory microcosm system. The soil microbiome mainly consuming CH4 faced challenges in adapting to acidic conditions (pH = 5.76), limiting the activity of their main MMO through changes in available metal ions such as copper and iron (Yao et al., 2023). However, low pH conditions did not correlate with the observed CH4 consumption patterns and are likely not the main explanation for the observed inhibition patterns. For instance, we observed a pH of 4.57 in week 3, but in week 4, the pH in the L treatments in the supernatant of soil slurries from acidic rice fields was 5.86 (see Table 2). However, the CH4 consumption activity and community composition remained similar. Therefore, we suggest that the effect of La3+ perhaps had a greater impact on CH4 consumption at lower pH levels, possibly due to alterations in the solubility and bioavailability of La3+.
Next, to pH, other soil properties are important contributing factors that can influence the effect of La3+ on the soil microbiome. The mineral composition of the parent material can affect soil cation exchange capacity, which determines the soil’s ability to retain and release nutrients, including La3+. In addition, organic matter content plays a crucial role in buffering pH and retaining nutrients. More importantly, organic matter can also bind to La3+ ions, affecting their availability to soil microbes (Luo et al., 2021). In our three rice field soils the organic matter content differed substantially, with neutral samples (26.98 g/kg) having the highest concentration, followed by acidic samples (19.48 g/kg), and the lowest in alkaline samples (11.16 g/kg) (Table 1). Soil origin can also influence the availability of essential nutrients, such as nitrogen, potassium, and phosphorus. It is well known that the addition of small quantities of LaCl3 forms small precipitates of LaPO4 in the presence of phosphates (Ding et al., 2007; Phuruangrat et al., 2012; Silva et al., 2019). As such, higher available phosphorus content in alkaline (37.47 mg/kg) and neutral (33.13 mg/kg) rice field samples may have contributed to the precipitation of La3+ thereby changing the bioavailability of La3+.
It should be noted that CH4 was the sole carbon source added to the soil slurries over the course of the experiment. Our data demonstrated that the soil microbiome simplified in their community composition, with the Methylobacter genus as the dominant methanotroph but also non-methanotrophic methylotrophs such as the genus Methylotenera and Methylovorus. As such, our findings align with previous metagenomic studies that have examined the distribution of carbon derived from CH4 across various bacterial populations (Kalyuzhnaya et al., 2008; Hernandez et al., 2015; Oshkin et al., 2015). As shown by Krause et al. (2017) the Methylobacter genus can provide carbon in the form methanol to non-methanotrophic methylotrophs such as the Methylotenera genus in the presence of La3+. Hence, our results further provide evidence for the concept that these communities collectively contribute to CH4 cycling, rather than a single type of microorganism. In addition, both methylotrophs are known to possess lanthanide (Ln)-dependent alcohol dehydrogenases (ADH) which may confer an advantage in the presence of high La3+ concentrations (Krause et al., 2017; Huang et al., 2018). In this context, we observed that the Methylocystis genus, a type II methanotroph, depicted higher relative abundance than the Methylobacter genus (Type I) in soil slurries with acidic and neutral rice field soil at higher La3+ concentrations (Figure 2). Previous studies already indicated that Methylocystis is more flexible than Methylobacter, mainly exhibited by their substrate utilization, oxygen requirements, and environmental adaptability (Krause et al., 2015; Hakobyan and Liesack, 2020; Nguyen et al., 2021). Interestingly, the Methylocystis genus has been demonstrated to harbor Ln-dependent ADHs (Jung et al., 2020). Our results indicate that Methylocystis genus also exhibits increased flexibility under high La3+ conditions as well. Hence, we suggest that the soil microbiome involved in CH4 consumption can generally cope with the addition of excessive amount of La3+ and changes in community composition ensured ecosystem functionality.
While our primary hypothesis lacked support from the data, we did make several noteworthy observations. Firstly, in the fourth week of the experiment, the CH4 consumption data revealed a faster decrease in CH4 concentration in the L and M treatments when compared to the control treatment for all soil slurries with different initial pH levels (Figure 1). We believe that the experimental design, utilizing only a 1% CH4 concentration in the headspace, may have constrained the CH4 oxidation potential of these communities and hindered the identification of clear stimulatory effects.
Secondly, we observed an increase in the relative abundance of the genus Methylobacter in response to varying levels of added La3+ in soil slurries from alkaline rice fields (Figure 2). We conducted qPCR analysis of the pmoA gene using the A189F and mb661R primer set (Holmes et al., 1995; Costello and Lidstrom, 1999); however, we were unable to confirm this trend with this primer set. A possible explanation lies in the coverage of the used primer set that fails to detect phylogenetic clusters that fall between pmoA and amoA (subunit of ammonium monooxygenase) sequences like pxmA cluster and high-affinity CH4 oxidizers cluster. Previously, the genera Methylomonas, Methylobacter and Methylomicrobium have been shown to a encode a sequence-divergent particulate monooxygenase (pXMO) (Tavormina et al., 2011). In addition, members of the family Beijerinckiaceae are not well amplified (Bourne et al., 2001; Knief, 2015). Hence, considering our pmoA gene copy number data using A189F/mb661R, it appears that a single primer might not be enough to fully investigate the abundance of methanotrophs in these rice field samples during our experiments, which might not cover all the active methanotrophs.
More importantly, we also did not pick methanotrophs that only have soluble form of the methane monooxygenase (SMMO) next to the pMMO such as Methyloceanibacter, which was an essential member of the total microbial community in all our initial soil slurries. Although it is not categorized as a methanotroph, this microbe has been identified to possess a SMMO and gene copies of Ln-dependent PQQ ADHs in marine environments, making it a putative methanotroph that can benefit from the addition of La3+ (Vekeman et al., 2016).
It is worth noting another contributing factor to our findings. As mentioned before, the presence of La3+ can enhance the activity of methanotrophs by increasing the production of MDH, particularly XoxF, and higher conversion of CH3OH into HCHO (Chu and Lidstrom, 2016). However, the metabolites produced in this process may have an impact on the activity of formaldehyde dehydrogenase (FDH) and other enzymes, inhibiting the conversion of CH4 into biomass. Consequently, certain intermediates in the CH4 oxidation pathway could potentially increase the cell toxicity of the methanotrophs and inhibit overall cell growth.
While our study contributes valuable insights to the ecological effect of added La3+ on methanotrophic communities in a microcosm model system, it is essential to acknowledge certain limitations that may impact the interpretation of our findings. One notable constraint is the detection of only approximately 37% of La3+ retained in the soil (Supplementary Figure S6). Together with the presence of negligible amounts in the supernatant (approximately 1%), it raises the question about potential precipitation and sequestration mechanisms for the remaining unaccounted fraction (Supplementary Figure S6). As mentioned before La3+ has a strong tendency to bind phosphates. Interestingly, we did not detect any PO43− in the supernatant of samples during the experiment (Supplementary Figure S5) supporting this as a mechanism for La3+ precipitation. In addition, the portion that remains unaccounted for could also be ascribed to potential interactions within microbial cells, possibly through bioaccumulation. For instance, at an exposure concentration of 20 μM, the La3+ concentration within bacteria was measured at a level of 10832 μg g−1 dry weight (Zhang et al., 2010). These limitations highlight a need for further studies into the fate and behavior of La3+ in soil ecosystems. Rather than relying solely on La3+ in the supernatant as a proxy for its bioavailability, employing sequential extractions followed by ICP-MS could enhance our understanding of the unaccounted portion of La3+.
5 Conclusion
This study offers valuable insights into the potential impacts of increasing amounts of REEs in the form La3+ on the soil microbiome originating from rice fields. Our findings imply that the consumption of CH4 in soil is influenced by intricate relationships between the soil origin, environmental factors like pH, and the community of microorganisms inhabiting the soil. As such, these findings contribute to a better understanding of the crucial role that microbial communities play in mitigating greenhouse gas emissions and combating climate change. In future research, it is crucial to better account for potential precipitation and bioaccumulation effects and concentrate on the cellular responses of the CH4-consuming community. This involves obtaining metagenome-assembled genomes coupled with transcriptomics to elucidate the biological and chemical components during the observed interactions with La3+.
Data availability statement
The data presented in the study are deposited in the National Center for Biotechnology Information (NCBI) repository, BioProject accession number PRJNA1014707.
Author contributions
RL: Conceptualization, Data curation, Formal analysis, Investigation, Methodology, Validation, Visualization, Writing – original draft, Writing – review & editing. ZW: Writing – original draft, Writing – review & editing. WD: Methodology, Formal analysis, Writing – review & editing. RW: Writing – original draft, Writing – review & editing. JA: Resources, Writing – review & editing. LY: Resources, Writing – review & editing. SK: Conceptualization, Formal analysis, Funding acquisition, Investigation, Methodology, Project administration, Resources, Supervision, Validation, Visualization, Writing – original draft, Writing – review & editing.
Funding
The author(s) declare financial support was received for the research, authorship, and/or publication of this article. SK was funded by National Natural Science Foundation of China (NSFC) International (Regional) Cooperation and Exchange Program (grant number 32050410288). JA and LY were supported by the Fundamental Research Funds for the Central Universities (grant number 020914380079). ChatGPT version 3.5 was used to improve English grammar and language.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
The author(s) declared that they were an editorial board member of Frontiers, at the time of submission. This had no impact on the peer review process and the final decision.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2024.1298154/full#supplementary-material
References
Anthony, C. (2000). Methanol dehydrogenase, a PQQ-containing quinoprotein dehydrogenase. Subcell. Biochem. 35, 73–117. doi: 10.1007/0-306-46828-x_3
Aussel, C., Marhaba, R., Pelassy, C., and Breittmayer, J. P. (1996). Submicromolar La3+ concentrations block the calcium release-activated channel, and impair CD69 and CD25 expression in CD3- or thapsigargin-activated Jurkat cells. Biochem. J. 313, 909–913. doi: 10.1042/bj3130909
Balaram, V. (2019). Rare earth elements: a review of applications, occurrence, exploration, analysis, recycling, and environmental impact. Geosci. Front. 10, 1285–1303. doi: 10.1016/j.gsf.2018.12.005
Bourne, D., McDonald, I., and Murrell, J. (2001). Comparison of pmoA PCR primer sets as tools for investigating Methanotroph diversity in three Danish soils. Appl. Environ. Microbiol. 67, 3802–3809. doi: 10.1128/AEM.67.9.3802-3809.2001
Bulman, R. (2003). Mobilization of lanthanides through the terrestrial biosphere. Met. Ions Biol. Syst. The lanthanides and their interrelations with biosystems eds A. A. Sigel and H. Sigel (Marcel Dekker, Inc, New York, NY) 4, 39–67.
Callahan, B. J., McMurdie, P. J., Rosen, M. J., Han, A. W., Johnson, A. J. A., and Holmes, S. P. (2016). DADA2: high-resolution sample inference from Illumina amplicon data. Nat. Methods 13, 581–583. doi: 10.1038/nmeth.3869
Cao, X., Chen, Y., Wang, X., and Deng, X. (2001). Effects of redox potential and pH value on the release of rare earth elements from soil. Chemosphere 44, 655–661. doi: 10.1016/S0045-6535(00)00492-6
Carlson, K. M., Gerber, J. S., Mueller, N. D., Herrero, M., MacDonald, G. K., Brauman, K. A., et al. (2017). Greenhouse gas emissions intensity of global croplands. Nat. Clim. Chang. 7, 63–68. doi: 10.1038/nclimate3158
Chu, F., and Lidstrom, M. E. (2016). XoxF acts as the predominant methanol dehydrogenase in the type I Methanotroph Methylomicrobium buryatense. J. Bacteriol. 198, 1317–1325. doi: 10.1128/JB.00959-15
Chu, H. Y., Zhu, J. G., Xie, Z. B., Zeng, Q., Li, Z. G., and Cao, Z. H. (2003). Availability and toxicity of exogenous lanthanum in a haplic acrisols. Geoderma 115, 121–128. doi: 10.1016/S0016-7061(03)00081-8
Conrad, R., and Rothfuss, F. (1991). Methane oxidation in the soil surface layer of a flooded rice field and the effect of ammonium. Biol. Fertil. Soils 12, 28–32. doi: 10.1007/BF00369384
Costello, A. M., and Lidstrom, M. E. (1999). Molecular characterization of functional and phylogenetic genes from natural populations of methanotrophs in lake sediments. Appl. Environ. Microbiol. 65, 5066–5074. doi: 10.1128/aem.65.11.5066-5074.1999
Dance, A. (2023). Microbial miners take on rare-earth metals. Nature 623, 876–878. doi: 10.1038/d41586-023-03611-4
de Mendiburu, F. (2021). Agricolae: Statistical procedures for agricultural research. Available at: https://CRAN.R-project.org/package=agricolae (accessed June 05, 2021).
Dedysh, S. N., and Dunfield, P. F. (2017). “Cultivation of Methanotrophs” in Hydrocarbon and Lipid Microbiology Protocols: Isolation and Cultivation. eds. T. J. McGenity, K.-N. Timmis, and B. Nogales (Cham: Springer Berlin Heidelberg)
Dedysh, S. N., and Knief, C. (2018). “Diversity and phylogeny of described aerobic Methanotrophs” in Methane biocatalysis: Paving the way to sustainability. eds. M. G. Kalyuzhnaya and X.-H. Xing (Cham: Springer International Publishing)
Ding, S., Liang, T., Yan, J., Zhang, Z., Huang, Z., and Xie, Y. (2007). Fractionations of rare earth elements in plants and their conceptive model. Sci. China Life Sci. 50, 47–55. doi: 10.1007/s11427-007-2040-7
Duan, Y., Gao, Y., Zhao, J., Xue, Y., Zhang, W., Wu, W., et al. (2023). Agricultural methane emissions in China: inventories, driving forces and mitigation strategies. Environ. Sci. Technol. 57, 13292–13303. doi: 10.1021/acs.est.3c04209
Dudev, T., Chang, L.-Y., and Lim, C. (2005). Factors governing the substitution of La3+ for Ca2+ and Mg2+ in Metalloproteins: a DFT/CDM study. J. Am. Chem. Soc. 127, 4091–4103. doi: 10.1021/ja044404t
Dushyantha, N., Batapola, N., Ilankoon, I. M. S. K., Rohitha, S., Premasiri, R., Abeysinghe, B., et al. (2020). The story of rare earth elements (REEs): occurrences, global distribution, genesis, geology, mineralogy and global production. Ore Geol. Rev. 122:103521. doi: 10.1016/j.oregeorev.2020.103521
Feng, S., Tan, C. H., Constancias, F., Kohli, G. S., Cohen, Y., and Rice, S. A. (2017). Predation by Bdellovibrio bacteriovorus significantly reduces viability and alters the microbial community composition of activated sludge flocs and granules. FEMS Microbiol. Ecol. 93. doi: 10.1093/femsec/fix020
Fox, J., and Weisberg, S. (2019). An R companion to applied regression, Thousand Oaks, CA: Sage Publications.
Giorgi, C., Marchi, S., and Pinton, P. (2018). The machineries, regulation and cellular functions of mitochondrial calcium. Nat. Rev. Mol. Cell Biol. 19, 713–730. doi: 10.1038/s41580-018-0052-8
Guo, Y., Song, B., Li, A., Wu, Q., Huang, H., Li, N., et al. (2022). Higher pH is associated with enhanced co-occurrence network complexity, stability and nutrient cycling functions in the rice rhizosphere microbiome. Environ. Microbiol. 24, 6200–6219. doi: 10.1111/1462-2920.16185
Gupta, K., Kumar, R., Baruah, K. K., Hazarika, S., Karmakar, S., and Bordoloi, N. (2021). Greenhouse gas emission from rice fields: a review from Indian context. Environ. Sci. Pollut. Res. 28, 30551–30572. doi: 10.1007/s11356-021-13935-1
Hakobyan, A., and Liesack, W. (2020). Unexpected metabolic versatility among type II methanotrophs in the Alphaproteobacteria. J. Biol. Chem. 401, 1469–1477. doi: 10.1515/hsz-2020-0200
Hanson, R. S., and Hanson, T. E. (1996). Methanotrophic bacteria. Microbiol. Rev. 60, 439–471. doi: 10.1128/mr.60.2.439-471.1996
Hernandez, M. E., Beck, D., Lidstrom, M., and Chistoserdova, L. (2015). Oxygen availability is a major factor in determining the composition of microbial communities involved in methane oxidation. PeerJ. 3:e801. doi: 10.7717/peerj.801
Hibi, Y., Asai, K., Arafuka, H., Hamajima, M., Iwama, T., and Kawai, K. (2011). Molecular structure of La3+−induced methanol dehydrogenase-like protein in Methylobacterium radiotolerans. J. Biosci. Bioeng. 111, 547–549. doi: 10.1016/j.jbiosc.2010.12.017
Holmes, A. J., Costello, A., Lidstrom, M. E., and Murrell, J. C. (1995). Evidence that particulate methane monooxygenase and ammonia monooxygenase may be evolutionarily related. FEMS Microbiol. Lett. 132, 203–208. doi: 10.1111/j.1574-6968.1995.tb07834.x
Huang, J., Yu, Z., and Chistoserdova, L. (2018). Lanthanide-dependent methanol dehydrogenases of XoxF4 and XoxF5 clades are differentially distributed among methylotrophic Bacteria and they reveal different biochemical properties. Front. Microbiol. 9:1366. doi: 10.3389/fmicb.2018.01366
Iftekhar, S., Ramasamy, D. L., Srivastava, V., Asif, M. B., and Sillanpää, M. (2018). Understanding the factors affecting the adsorption of lanthanum using different adsorbents: a critical review. Chemosphere 204, 413–430. doi: 10.1016/j.chemosphere.2018.04.053
IPCC (2021). The physical science basis. Contribution of working group I to the sixth assessment report of the intergovernmental panel on climate change, Cambridge: Cambridge University Press.
Jiang, Z., Weng, B., Huang, Y., Jiang, Z., and Wang, Y. (2008). Effects of lanthanum on soil microorganism. J. Chin. Rare Earth Soc. 26, 498–502. doi: 10.3321/j.issn:1000-4343.2008.04.021
Jung, G. Y., Rhee, S. K., Han, Y. S., and Kim, S. J. (2020). Genomic and physiological properties of a facultative methane-oxidizing bacterial strain of Methylocystis sp. from a Wetland. Microorganisms 8:1719. doi: 10.3390/microorganisms8111719
Kalyuzhnaya, M. G., Lapidus, A., Ivanova, N., Copeland, A. C., McHardy, A. C., Szeto, E., et al. (2008). High-resolution metagenomics targets specific functional types in complex microbial communities. Nat. Biotechnol. 26, 1029–1034. doi: 10.1038/nbt.1488
Kassambara, A. (2023). Ggpubr: ‘ggplot2’ based publication ready plots. Available at: https://CRAN.R-project.org/package=ggpubr (accessed June 05, 2021).
Kato, S., Takashino, M., Igarashi, K., and Kitagawa, W. (2020). Isolation and genomic characterization of a Proteobacterial Methanotroph requiring lanthanides. Microbes Environ. 35:ME19128. doi: 10.1264/jsme2.ME19128
Knief, C. (2015). Diversity and habitat preferences of cultivated and uncultivated aerobic Methanotrophic Bacteria evaluated based on pmoA as molecular marker. Front. Microbiol. 6:1346. doi: 10.3389/fmicb.2015.01346
Kolb, S., Knief, C., Stubner, S., and Conrad, R. (2003). Quantitative detection of methanotrophs in soil by novel pmoA-targeted real-time PCR assays. Appl. Environ. Microbiol. 69, 2423–2429. doi: 10.1128/aem.69.5.2423-2429.2003
Kotelnikova, A., Fastovets, I., Rogova, O., Volkov, D. S., and Stolbova, V. (2019). Toxicity assay of lanthanum and cerium in solutions and soil. Ecotoxicol. Environ. Saf. 167, 20–28. doi: 10.1016/j.ecoenv.2018.09.117
Kotelnikova, A. D., Rogova, O. B., and Stolbova, V. V. (2021). Lanthanides in the soil: routes of entry, content, effect on plants, and genotoxicity (a review). Eurasian Soil Sci. 54, 117–134. doi: 10.1134/S1064229321010051
Krause, S. M. B., Johnson, T., Samadhi Karunaratne, Y., Fu, Y., Beck, D. A. C., Chistoserdova, L., et al. (2017). Lanthanide-dependent cross-feeding of methane-derived carbon is linked by microbial community interactions. Proc. Nat. Acad. Sci. U. S. A. 114, 358–363. doi: 10.1073/pnas.1619871114
Krause, S., Niklaus, P. A., Badwan Morcillo, S., Meima Franke, M., Lüke, C., Reim, A., et al. (2015). Compositional and functional stability of aerobic methane consuming communities in drained and rewetted peat meadows. FEMS Microbiol. Ecol. 91:fiv119. doi: 10.1093/femsec/fiv119
Kravchenko, I. K., Sizov, L. R., and Lysak, L. V. (2023). Laboratory study of the effect of ammonium and lanthanum salts on methane oxidation and composition of microbial communities in Soddy-Podzolic soil. Eurasian Soil Sci. 56, 573–583. doi: 10.1134/S1064229322602773
Kravchenko, I. K., Sizov, L. R., Tikhonova, E. N., and Lysak, L. V. (2022). The effect of lanthanum on the composition of Methanotrophic Community of sod-Podzolic Soil. Microbiology 91, 599–603. doi: 10.1134/S002626172260135X
Lane, D. J. (1991). “16S/23S rRNA sequencing” in Nucleic acid techniques in bacterial systematic. ed. E. G. S. Michael (New York, USA: John Wiley & Sons), 115–175.
Lane, D. J., Pace, B., Olsen, G. J., Stahl, D. A., Sogin, M. L., and Pace, N. R. (1985). Rapid determination of 16S ribosomal RNA sequences for phylogenetic analyses. PNAS 82, 6955–6959. doi: 10.1073/pnas.82.20.6955
Leal Filho, W., Kotter, R., Özuyar, P. G., Abubakar, I. R., Eustachio, J. H. P. P., and Matandirotya, N. R. (2023). Understanding rare earth elements as critical raw materials. Sustainability 15:1919. doi: 10.3390/su15031919
Li, S., Chen, L., Han, X., Yang, K., Liu, K., Wang, J., et al. (2022). Rice cultivar renewal reduces methane emissions by improving root traits and optimizing photosynthetic carbon allocation. Agriculture 12:2134. doi: 10.3390/agriculture12122134
Li, J., Hong, M., Yin, X., and Liu, J. (2010). Effects of the accumulation of the rare earth elements on soil macrofauna community. J. Rare Earths 28, 957–964. doi: 10.1016/S1002-0721(09)60233-7
Li, J., Verweij, R. A., and van Gestel, C. A. M. (2018). Lanthanum toxicity to five different species of soil invertebrates in relation to availability in soil. Chemosphere 193, 412–420. doi: 10.1016/j.chemosphere.2017.11.040
Liang, T., Li, K., and Wang, L. (2014). State of rare earth elements in different environmental components in mining areas of China. Environ. Monit. Assess. 186, 1499–1513. doi: 10.1007/s10661-013-3469-8
Luo, Y., Yuan, H., Zhao, J., Qi, Y., Cao, W.-W., Liu, J.-M., et al. (2021). Multiple factors influence bacterial community diversity and composition in soils with rare earth element and heavy metal co-contamination. Ecotoxicol. Environ. Saf. 225:112749. doi: 10.1016/j.ecoenv.2021.112749
Mu, K., Zhao, X., Hu, L., Zhang, F., Zhang, W., and Cui, J. (2006). Toxicity of lanthanum to pathogenic Fungi and its morphological characteristics. J. Rare Earths 24, 607–612. doi: 10.1016/S1002-0721(06)60173-7
Nguyen, D. T. N., Lee, O. K., Nguyen, T. T., and Lee, E. Y. (2021). Type II methanotrophs: a promising microbial cell-factory platform for bioconversion of methane to chemicals. Biotechnol. Adv. 47:107700. doi: 10.1016/j.biotechadv.2021.107700
Oksanen, J., Simpson, G. L., Blanchet, F. G., Kindt, R., Legendre, P., Minchin, P. R., et al.. (2022). Vegan: Community ecology package. Available at: https://CRAN.R-project.org/package=vegan
Oshkin, I. Y., Beck, D. A. C., Lamb, A. E., Tchesnokova, V., Benuska, G., McTaggart, T. L., et al. (2015). Methane-fed microbial microcosms show differential community dynamics and pinpoint taxa involved in communal response. ISME J. 9, 1119–1129. doi: 10.1038/ismej.2014.203 (accessed June 05, 2021).
Palarea-Albaladejo, J., and Martín-Fernández, J. (2015). ZCompositions - R package for multivariate imputation of left-censored data under a compositional approach. Chemometr. Intell. Lab. Syst. 143, 85–96. doi: 10.1016/j.chemolab.2015.02.019
Peng, L., Yi, L., Zhexue, L., Juncheng, Z., Jiaxin, D., Daiwen, P., et al. (2004). Study on biological effect of La3+ on Escherichia coli by atomic force microscopy. J. Inorg. Biochem. 98, 68–72. doi: 10.1016/j.jinorgbio.2003.08.012
Phuruangrat, A., Ekthammathat, N., Thongtem, S., and Thongtem, T. (2012). Preparation of LaPO4 nanowires with high aspect ratio by a facile hydrothermal method and their photoluminescence. Res. Chem. Intermed. 39, 1363–1371. doi: 10.1007/s11164-012-0692-9
Picone, N., and op den Camp, H. J. M. (2019). Role of rare earth elements in methanol oxidation. Curr. Opin. Chem. Biol. 49, 39–44. doi: 10.1016/j.cbpa.2018.09.019
Pol, A., Barends, T. R. M., Dietl, A., Khadem, A. F., Eygensteyn, J., Jetten, M. S. M., et al. (2014). Rare earth metals are essential for methanotrophic life in volcanic mudpots. Environ. Microbiol. 16, 255–264. doi: 10.1111/1462-2920.12249
Pruesse, E., Peplies, J., and Glöckner, F. O. (2012). SINA: accurate high-throughput multiple sequence alignment of ribosomal RNA genes. Bioinformatics 28, 1823–1829. doi: 10.1093/bioinformatics/bts252
Qian, H., Zhu, X., Huang, S., Linquist, B., Kuzyakov, Y., Wassmann, R., et al. (2023). Greenhouse gas emissions and mitigation in rice agriculture. Nat. Rev. Earth Environ. 4, 716–732. doi: 10.1038/s43017-023-00482-1
Ramos, S. J., Dinali, G. S., Oliveira, C., Martins, G. C., Moreira, C. G., Siqueira, J. O., et al. (2016). Rare earth elements in the soil environment. Curr. Pollut. Rep. 2, 28–50. doi: 10.1007/s40726-016-0026-4
Ran, Y., and Liu, Z. (1993). The characteristics of adsorption and desorption of rare earth elements by the Main types of soils of China. J. Rare Earths 1, 46–51.
Reeves, J. P., and Condrescu, M. (2003). Lanthanum is transported by the sodium/calcium exchanger and regulates its activity. Am. J. Physiol. Cell Physiol. 285, C763–C770. doi: 10.1152/ajpcell.00168.2003
Saunois, M., Stavert, A., Poulter, B., Bousquet, P., Canadell, J., Jackson, R., et al. (2020). The global methane budget 2000–2017. Earth Syst. Sci. Data. 12, 1561–1623. doi: 10.5194/essd-12-1561-2020
Silva, F. B. V., Nascimento, C. W. A., Alvarez, A. M., and Araújo, P. R. M. (2019). Inputs of rare earth elements in Brazilian agricultural soils via P-containing fertilizers and soil correctives. J. Environ. Manag. 232, 90–96. doi: 10.1016/j.jenvman.2018.11.031
Skovran, E., and Martinez-Gomez, N. C. (2015). Just add lanthanides. Science 348, 862–863. doi: 10.1126/science.aaa9091
Tavormina, P. L., Orphan, V. J., Kalyuzhnaya, M. G., Jetten, M. S., and Klotz, M. G. (2011). A novel family of functional operons encoding methane/ammonia monooxygenase-related proteins in gammaproteobacterial methanotrophs. Environ. Microbiol. Rep. 3, 91–100. doi: 10.1111/j.1758-2229.2010.00192.x
Team, R. C (2022). R: a language and environment for statistical computing. R Foundation for Statistical Computing patent application. Available at: https://www.R-project.org/ (accessed June 05, 2022).
Tommasi, F., Thomas, P. J., Pagano, G., Perono, G. A., Oral, R., Lyons, D. M., et al. (2021). Review of rare earth elements as fertilizers and feed additives: a knowledge gap analysis. Arch. Environ. Contam. Toxicol. 81, 531–540. doi: 10.1007/s00244-020-00773-4
Tyler, G. (2004). Rare earth elements in soil and plant systems - a review. Plant Soil 267, 191–206. doi: 10.1007/s11104-005-4888-2
van den Boogaart, K. G., Tolosana-Delgado, R., and Bren, M. (2022). Compositions: compositional data analysis. Available at: https://CRAN.R-project.org/package=compositions (accessed June 05, 2021).
Vekeman, B., Kerckhof, F.-M., Cremers, G., de Vos, P., Vandamme, P., Boon, N., et al. (2016). New Methyloceanibacter diversity from North Sea sediments includes methanotroph containing solely the soluble methane monooxygenase. Environ. Microbiol. 18, 4523–4536. doi: 10.1111/1462-2920.13485
Vu, H. N., Subuyuj, G. A., Crisostomo, R. V., and Skovran, E. (2021). Chapter seven - transposon mutagenesis for methylotrophic bacteria using Methylorubrum extorquens AM1 as a model system. Methods Enzymol. J. 650, 159–184. doi: 10.1016/bs.mie.2021.01.015
Wang, X., Shi, G. X., Xu, Q. S., Xu, B. J., and Zhao, J. (2007). Lanthanum- and cerium-induced oxidative stress in submerged Hydrilla verticillata plants. Russ. J. Plant Physiol. 54, 693–697. doi: 10.1134/S1021443707050184
Wickham, H. (2007). Reshaping data with the reshape package. J. Stat. Softw. 21, 1–20. doi: 10.18637/jss.v021.i12
Wickham, H. (2011). The Split-apply-combine strategy for data analysis. J. Stat. Softw. 40, 1–29. doi: 10.18637/jss.v040.i01
Wickham, H., Averick, M., Bryan, J., Chang, W., McGowan, L., François, R., et al. (2019). Welcome to the Tidyverse. J. Open Source Softw. 4:1686. doi: 10.21105/joss.01686 (accessed January 29, 2023).
Wickham, H., François, R., Henry, L., Müller, K., and Vaughan, D. (2023). Dplyr: a grammar of data manipulation. Available at: https://CRAN.R-project.org/package=dplyr
Yao, X., Wang, J., and Hu, B. (2023). How methanotrophs respond to pH: a review of ecophysiology. Front. Microbiol. 13:1034164. doi: 10.3389/fmicb.2022.1034164
Zhang, H., He, X., Bai, W., Guo, X., Chai, Z., and Zhao, Y. (2010). Ecotoxicological assessment of lanthanum with Caenorhabditis elegans in liquid medium. Metallomics 2, 806–810. doi: 10.1039/C0MT00059K
Keywords: methane, soil health, rare earth elements, ecotoxicology, microbial communities, lanthanum, global change
Citation: Liu R, Wei Z, Dong W, Wang R, Adams JM, Yang L and Krause SMB (2024) Unraveling the impact of lanthanum on methane consuming microbial communities in rice field soils. Front. Microbiol. 15:1298154. doi: 10.3389/fmicb.2024.1298154
Edited by:
Marika Pellegrini, University of L’Aquila, ItalyReviewed by:
Carl-Eric Wegner, Friedrich Schiller University Jena, GermanyRihab Djebaili, Consorzio Interuniversitario Nazionale per le Scienze Ambientali, Italy
Beatrice Farda, University of L’Aquila, Italy, in collaboration with reviewer RD
Copyright © 2024 Liu, Wei, Dong, Wang, Adams, Yang and Krause. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Sascha M. B. Krause, c3NhQGRlcy5lY251LmVkdS5jbg==
 Ruyan Liu
Ruyan Liu Ziting Wei1
Ziting Wei1 Jonathan M. Adams
Jonathan M. Adams Lin Yang
Lin Yang Sascha M. B. Krause
Sascha M. B. Krause