
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Microbiol., 30 April 2024
Sec. Antimicrobials, Resistance and Chemotherapy
Volume 15 - 2024 | https://doi.org/10.3389/fmicb.2024.1291876
This article is part of the Research TopicAntimicrobial Peptides and their Druggability, Bio-safety, Stability, and ResistanceView all 23 articles
 Yijia Han1,2
Yijia Han1,2 Yi Zhang2
Yi Zhang2 Xiaodong Zhang1
Xiaodong Zhang1 Zeyu Huang1
Zeyu Huang1 Jingchun Kong2
Jingchun Kong2 Xiuxiu Wang1
Xiuxiu Wang1 Lijiang Chen1
Lijiang Chen1 Yue Wang1
Yue Wang1 Jianming Cao2
Jianming Cao2 Tieli Zhou1*
Tieli Zhou1* Mo Shen1*
Mo Shen1*Introduction: Antibiotic misuse and overuse have led to the emergence of carbapenem-resistant bacteria. The global spread of resistance to the novel antibiotic combination ceftazidime-avibactam (CZA) is becoming a severe problem. Antimicrobial peptide PAM-1 offers a novel approach for treating infections caused by antibiotic-resistant bacteria. This study explores its antibacterial and anti-biofilm activities and mechanisms against CZA-resistant Escherichia. Coli (E. coli), evaluating its stability and biosafety as well.
Methods: The broth microdilution method, growth curve analysis, crystal violet staining, scanning electron microscopy, and propidium iodide staining/N-phenyl-1-naphthylamine uptake experiments were performed to explore the antibacterial action and potential mechanism of PAM-1 against CZA-resistant E. coli. The biosafety in diverse environments of PAM-1 was evaluated by red blood cell hemolysis, and cytotoxicity tests. Its stability was further assessed under different temperatures, serum concentrations, and ionic conditions using the broth microdilution method to determine its minimum inhibitory concentration (MIC). Galleria mellonella infection model and RT-qPCR were used to investigate the in vivo antibacterial and anti-inflammatory effects.
Results and discussion: In vitro antibacterial experiments demonstrated that the MICs of PAM-1 ranged from 2 to 8 μg/mL, with its effectiveness sustained for a duration of 24 h. PAM-1 exhibited significant antibiofilm activities against CZA-resistant E. coli (p < 0.05). Furthermore, Membrane permeability test revealed that PAM-1 may exert its antibacterial effect by disrupting membrane integrity by forming transmembrane pores (p < 0.05). Red blood cell hemolysis and cytotoxicity tests revealed that PAM-1 exerts no adverse effects at experimental concentrations (p < 0.05). Moreover, stability tests revealed its effectiveness in serum and at room temperature. The Galleria mellonella infection model revealed that PAM-1 can significantly improve the survival rate of Galleria mellonella (>50%)for in vivo treatment. Lastly, RT-qPCR revealed that PAM-1 downregulates the expression of inflammatory cytokines (p < 0.05). Overall, our study findings highlight the potential of PAM-1 as a therapeutic agent for CZA-resistant E. coli infections, offering new avenues for research and alternative antimicrobial therapy strategies.
Global public health is increasingly threatened by antibiotic resistance, with carbapenem-resistant Enterobacteriaceae (CRE) posing a significant challenge by elevating infection rates and mortality (Satlin et al., 2017; van Duin et al., 2020). The limited effectiveness of traditional antibiotics led to the 2015 FDA (Food and Drug Administration) approval of ceftazidime-avibactam (CZA), a novel combination of a cephalosporin and a β-lactamase inhibitor. This innovative combination exhibits in vitro activity against Enterobacterales harboring β-lactamases of Ambler class A [including extended-spectrum β-lactamases (ESBLs) and Klebsiella pneumoniae carbapenemases (KPC)], class C (AmpC cephalosporinases), and some class D enzymes (e.g., OXA-48-type, many of which also possess ESBLs) (Morrill et al., 2015; Falcone et al., 2016; Livermore et al., 2018; Zhang et al., 2018). However, the subsequent global increase in CZA usage has gradually increased CZA resistance, with new cases of CZA-resistant E. coli strains being frequently reported (Shields et al., 2016, 2017; Hernández-García et al., 2021). Therefore, developing novel antimicrobial drugs to combat CZA-resistant E. coli infections is urgently warranted.
Antimicrobial peptides (AMPs), an intrinsic component of the innate immune system in several animals, confer protection against foreign microorganisms and infectious threats (Pasupuleti et al., 2012). In general, these peptides are short and positively charged and exhibit broad-spectrum antibacterial activity as well as effects on biofilms (Spencer et al., 2018; Luo and Song, 2021). Furthermore, AMPs can regulate inflammatory responses and promote wound healing (Nguyen et al., 2020; Thapa et al., 2020). To enhance the application of antimicrobial peptides (AMPs) in clinical antimicrobial treatments, researchers are actively engaged in identifying and screening AMPs that not only demonstrate potent antimicrobial effects but also exhibit a high level of safety.
PAM-1, an antimicrobial peptide sourced from the cathelicidin gene family in the platypus genome, exhibits significant antibacterial effects against standard strains of Bacillus subtilis, Staphylococcus aureus, Streptococcus uberis, Streptococcus pyogenes, E. coli, Salmonella cholerae, and Pseudomonas aeruginosa (Wang et al., 2011). Through bioinformatics analysis, it is predicted that PAM-1 possesses a wide-ranging antibacterial activity against various bacteria. However, regrettably, the antibacterial and antibiofilm activities and the mechanisms of action of PAM-1 against clinical strains remain unelucidated in previous studies.
In the present study, a series of experiments were conducted to investigate the antibacterial, antibiofilm, and anti-inflammatory activities of PAM-1 against CZA-resistant E. coli strains. The potential mechanisms by which PAM-1 exerts its antibacterial effects were elucidated, and its stability and safety were thoroughly assessed. Additionally, the in vivo efficacy of PAM-1 was evaluated using the Galleria mellonella (G. mellonella) infection model. The findings of this study suggest that PAM-1 holds promise as a novel antibacterial agent for combating CZA-resistant E. coli infections.
PAM-1 (95.66%, 100 mg) was synthesized by Nanjing Yuanpeptide Biotechnology Co., Ltd. (Nanjing, China). The amino acid sequence of PAM-1 is RTKRRIKLIKNGVKKVKDILKNNNIIILPGSNEK (Wang et al., 2011). To prepare PAM-1 for the experiments, it was dissolved in double distilled water and stored at −20°C. Ceftazidime (≥ 99%), avibactam (≥ 99%), meropenem (MEM, ≥ 98.0%), and imipenem (IPM, ≥ 98.0%) used in this study were purchased from Wenzhou Kangtai Biotechnology Co., Ltd. (Zhejiang, China).
PAM-1 was analyzed using a predictive analysis tool in ProtParam of Expasy (https://web.expasy.org/protparam/). The physical and chemical properties of PAM-1, including net charge and relative molecular mass, were determined. The antibacterial activity of PAM-1 was predict using the online software CAMPR3 (http://www.camp3.bicnirrh.res.in/).
Six CZA-susceptible E. coli and six CZA-resistant E. coli isolates were selected from the First Affiliated Hospital of Wenzhou Medical University. All isolates were identified using MALDI-TOF MS (Quintilla et al., 2018; Ashfaq et al., 2022) (BioMerieux, France). These clinically relevant E. coli strains were preserved by freezing them at −80°C in Luria–Bertani (LBT, hermo Fisher Scientific, America) broth containing 30% glycerol. E. coli ATCC 25922 was used as the quality control strain.
DNA was extracted from the 12 E. coli strains using a bacterial genome extraction kit (Solarbio, Beijing, China) according to the manufacturer’s instructions (Zeng et al., 2022). The resistant determinants, including carbapenem genes (blaKPC, blaNDM, blaIMP, blaVIM, blaOXA − 23, and blaOXA − 48) and ESBL genes (blaSHV, blaTEM, blaCTX − M − 1, blaCTX − M − 9, and blaCTX − M − 14), were examined via PCR using specific primers (Supplementary Table S1). Subsequently, the positive PCR products were sequenced and aligned using the BLAST tool in NCBI (https://blast.ncbi.nlm.nih.gov/Blast.cgi).
The broth microdilution method was used to determine the minimum inhibitory concentrations (MICs) of CZA, MEM, IPM, and PAM-1 against the 12 E. coli strains (García-Cobos et al., 2008). The 0.5 McFarland bacterial suspension was 1:100 diluted by Mueller Hinton broth (MHB, Thermo Fisher Scientific, America). Then, 100 μL of the bacterial solution was added to a 96-well microplate containing various concentrations of the antibiotics: CZA, MEM, IPM, and PAM-1. The microplates were incubated at 37°C for 16–18 h. The results were interpreted based on the 2020 Clinical and Laboratory Standards Institute guidelines (CLSI). MIC was defined as the lowest concentration of an antibiotic that completely inhibited bacterial growth.
To determine the CRE (MBC), 100 μL of the bacterial suspension from the well corresponding to the MIC and three subsequent wells was collected and spread onto LB agar plates. Then, the plates were incubated at 37°C for 16–20 h. MBC was determined as the drug concentration that resulted in no bacterial colony formation on the plate.
Each experiment was independently repeated three times to ensure the accuracy and reproducibility of the results.
The growth curve was determined according to a previously described method, with some modifications (Jayathilaka et al., 2021). The diluted E. coli culture was added to a 96-well plate along with PAM-1 at concentrations corresponding to its 1/2 MIC, 1 MIC, 2 MIC, and 4 MIC (ranging from 1–32 μg/mL). The plate was incubated at 37°C for 24 h. Bacterial growth was monitored by measuring the absorbance at 600 nm at different time intervals (0, 2, 4, 6, 8, 10, 12, and 24 h). Each experiment was repeated three times to ensure accuracy and reproducibility.
The effect of PAM-1 on biofilm formation and eradication was determined by performing crystal violet staining (O’Loughlin et al., 2013). The bacterial culture was added to a 96-well plate with the corresponding concentration of PAM-1. After incubation for 24 h at 37°C, the 96-well plate was gently washed with phosphate-buffered saline (PBS, Beijing Solarbio Science & Technology Co., Ltd., Beijing, China) to remove any planktonic bacteria. Then, the plate was allowed to naturally dry at room temperature. Subsequently, a 0.1% crystal violet solution was added to each well, followed by incubation at 37°C for 15 min. After washing the plate with PBS and allowing it to naturally dry, 200 μL of a solution containing 95% ethanol and 5% acetic acid was added to dissolve the crystal violet. Biofilm biomass was calculated by measuring the absorbance at 595 nm. Two separate tests were performed: one to evaluate the effect of PAM-1 on biofilm formation (where the drug was added before biofilm formation) and another to evaluate the eradication of mature biofilms (where the drug was added after biofilm formation). Each test was conducted three times, and each experiment within a test was repeated three times for reliability and consistency.
The SEM was determined according to a previously described method, with some modifications (Zhang et al., 2019). Silicon wafers were placed in the wells and 10 μL of the bacterial suspension was added to 990 μL of MHB containing PAM-1. Then, the plates were incubated for 24 h. Thereafter, the wafers were cleaned three times with PBS, fixed with 2.5% glutaraldehyde, and dehydrated for 5 min using an ethanol concentration gradient (30, 50, 70, 80, 90, 95, and 100%). The final samples were air-dried, gold-sprayed, and observed under a scanning electron microscope (SU8010, Hitachi, Japan).
To investigate the mechanism of antimicrobial activity of PAM-1, membrane permeability tests were conducted as described previously, with some modifications (Liu et al., 2017). The bacteria suspension was shaken in the medium to the logarithmic stage and divided into equal parts. Then, different concentrations of PAM-1 were added to the bacterial suspensionand was continued to be shaken for 2 h. The sample after centrifugation were resuspended with propidium iodide (PI, Beijing Solarbio Science & Technology Co., Ltd., Beijing, China) and N-phenyl-1-naphthylamine (NPN, Beijing Solarbio Science & Technology Co., Ltd., Beijing, China) solution and incubated in a water bath at 37°C for 30 min. Then, the samples were scanned at excitation wavelengths of 535 nm and 350 nm and emission wavelengths of 615 nm (PI) and 420 nm (NPN), respectively, using a microplate reader (BioTek Synergy NEO2, United States).
To detect reactive oxygen species (ROS), 500 μL of the bacterial suspension containing dichlorodihydrofluorescein diacetate (DCFH-DA, Shanghai Generay Biotech Co., Ltd) was incubated at 37°C for 60 min. After removing excess DCFH-DA probes, the cells were then treated with PAM-1 at 30°C for 2 h. Sterile distilled water was used as the blank control. All samples were scanned at an excitation wavelength of 488 nm and an emission wavelength of 535 nm using a microplate reader (BioTek Synergy NEO2). This assay aimed to investigate the mechanism of action of PAM-1. Each experiment was independently repeated three times.
To further evaluate the effect of PAM-1 on the expression of proinflammatory factors, reverse transcription quantitative PCR (RT-qPCR) was performed using a previously described method, with slight modifications (de Campos et al., 2020). RAW 264.7(Procell Life Science and Technology Co., Ltd., China) cells were cultured overnight and Dulbecco’s modified Eagle medium (DMEM, Thermo Fisher Scientific, USA) supplemented with 10% fetal bovine serum (FBS, Gibco, USA) was used to culture the cells. The cells were washed three times with PBS, and the culture medium was discarded. Bacteria at a multiplicity of infection (MOI) of 10 were added to infect the cells, followed by treatment with different concentrations of PAM-1for 2 h. After removing the culture medium and washing with PBS, total RNA was extracted from RAW 264.7 cells using the Trizol total RNA extraction method.
Total RNA was extracted from RAW 264.7 cells using the Trizol total RNA extraction method (Tang et al., 2017; Trendel et al., 2019). After measuring the RNA concentration (DeNovix ultramicro-UV–visible spectrophotometer DS-11, USA), the extracted mRNA was converted into cDNA using the PrimeScript™ RT Reagent Kit according to the manufacturer’s instructions. The expression of the inflammatory factors interleukin (IL)-1β and tumor necrosis factor (TNF)-α was assessed using an RT-qPCR system (S100™ Thermal Cycler PCR machine, BIO-RAD, USA). Table S1 lists the specific primers used in this analysis. The 2−ΔΔCT method was used to evaluate the data (Xu et al., 2020), with all values normalized to the expression of the housekeeping gene β-actin. The experiment was conducted three times to ensure reliability and reproducibility.
Next, based on a previous study (Dong et al., 2021), we evaluated whether PAM-1 activity is affected by various factors. E. coli DC 8873 and DC 8466 were selected as the test strains to explore the effect of temperature. The prepared drug was incubated at different temperatures (−80°C, −20°C, 4°C, 25°C, and 37°C) for 3, 12, and 24 h. The MIC of PAM-1 was determined using the broth microdilution method after the incubation period.
Next, we evaluated the effect of serum on PAM-1 activity (Zhang et al., 2022). PAM-1 was incubated at 37°C for 24 h with various serum concentrations (0, 5, and 10%). After co-incubating the bacterial culture with serum, the MIC value of PAM-1 was evaluated using the same method mentioned above.
The effects of the presence of salts on the antimicrobial activity were evaluated as previously described (Kim et al., 2018). PAM-1 was incubated at 37°C with different concentrations of NaCl (0, 50, 100, and 150 mM), CaCl2 (0, 1.25, 2.5, and 5 mM) or MgCl2 (0, 0.5, 1, and 2 mM) for 24 h. Subsequently, the MIC of PAM-1 was determined using the broth microdilution method.
Using RAW 264.7 cells, the safety of PAM-1 was assessed using a previously described method with some modifications (da Luz et al., 2020). The cell suspension (100 μL) containing 1 × 105 cells was inoculated into the wells of a 96-well plate. Then, 10 μL of PAM-1 at various concentrations (1–256 μg/mL) was added to the medium. The plate was incubated at 37°C with 5% CO2 for 12 h. Thereafter, 10 μL of Cell Counting Kit-8 solution (CCK-8, Beyotime, China) was added to each well, followed by incubation at room temperature in the dark for 1 h. The absorbance of the wells was measured at 450 nm using the SpectraMax iD5 enzyme-labeled meter (BECKMAN, USA). This experiment was repeated three times.
The hemolytic activity of PAM-1 was determined as previously described (Dong et al., 2012). Female BALB/c mice were used according to the Chinese National Standards for Laboratory Animals (GB 14925–2010). The Zhejiang Association for Science and Technology SYXK approved these analyses (ID: SYXK [Zhejiang] 2018–0017) and were consistent with the Wenzhou Laboratory Animal Welfare and Ethics standards. Fresh whole blood samples were collected and centrifuged. The RBCs obtained were washed with normal saline and then resuspended with normal saline to form a 5% RBC suspension. PAM-1 at various concentrations (1–80 μg/mL) was added to the 5% RBC suspension in a total volume of 2 mL. After centrifugation, the supernatant was collected and transferred to a 96-well plate. The absorbance of the supernatant was measured at 540 nm to reflect the hemolysis degree. The hemolysis rate was calculated using the following formula: Hemolysis rate (%) = (ODexperimental group − ODnegative control group)/(ODpositive control group − ODnegative control group), where OD represents the absorbance at 540 nm. The negative control group comprised only normal saline, whereas the positive control group comprised 0.1% Triton X-100. This experiment was performed three times.
The effectiveness of PAM-1 was evaluated in vivo by measuring the survival rate of the G. mellonella model, which is commonly used as a model organism for studying microbial infections (Tsai et al., 2016; Borjan et al., 2020). The larvae of G. mellonella used in this experiment were opalescent and weighed 250–300 mg. Overnight cultures of the bacteria were diluted to a density of 1.5 × 107 CFU/mL. Then, the samples were divided into two groups: control and treatment groups. Furthermore, the treatment group was divided into subgroups, with each subgroup receiving different concentrations of PAM-1 (1/2MIC × 7, 1 MIC × 7, and 2MIC × 7). Using a microinjector, 10 μL of the bacterial suspension (CZA-susceptible E. coli DC 8873 and CZA-resistant E. coli DC 8466) was injected into the rear left proleg of each G. mellonella larva. After 2 h, the control group was administered 10 μL of sterile saline, whereas the treatment group received an appropriate concentration of the PAM-1 injection. To compare the efficacy of PAM-1 with standard agents, CZA was used. The larvae were then incubated at 37°C to measure their survival after 7 days. Larvae were considered dead if they repeatedly failed to respond to physical stimuli. The main experimental outcome was the analysis of the survival rate of G. mellonella using Kaplan–Meier analysis and the log-rank test.
All experiments were conducted in triplicate. Data analysis was performed using Prism 8 (GraphPad Software Inc., CA, USA), and the data are presented as mean and standard deviation. Significance was determined by using the two-sample t-test and log-rank test and indicated as *p < 0.05, ∗*p < 0.01, and ***p < 0.001.
The properties and antibacterial activity of PAM-1 were determined using the Bioanalysis website. Table 1 presents the physical and chemical properties obtained via ProtParam analysis. PAM-1 comprises 34 amino acid residues, carries 9 net charges, and has an isoelectric point of 11.24. The GRAVY index of PAM-1 was −0.774 (negative value represents hydrophilicity), indicating that it exhibits good solubility.
Next, the antibacterial activity of PAM-1 was predicted via biogenic analysis using CAMPR3. Both algorithms (Random Forest Classifier and Discriminant Analysis classifier) revealed that PAM-1 had better antibacterial activity (Table 2).
Twelve nonrepetitive E. coli clinical isolates were selected. PCR was performed to determine the carbapenemase types of the experimental strains. Table 3 summarizes the MICs of PAM-1, CZA, and MEM and the MBC of PAM-1. These results suggest the effective antibacterial activity of PAM-1 against both CZA-resistant and susceptible E. coli strains. The MICs ranged from 2 to 8 μg/mL, whereas the MBCs ranged from 2 to 16 μg/mL. This indicates that PAM-1 exerts a bactericidal effect at the MBC.
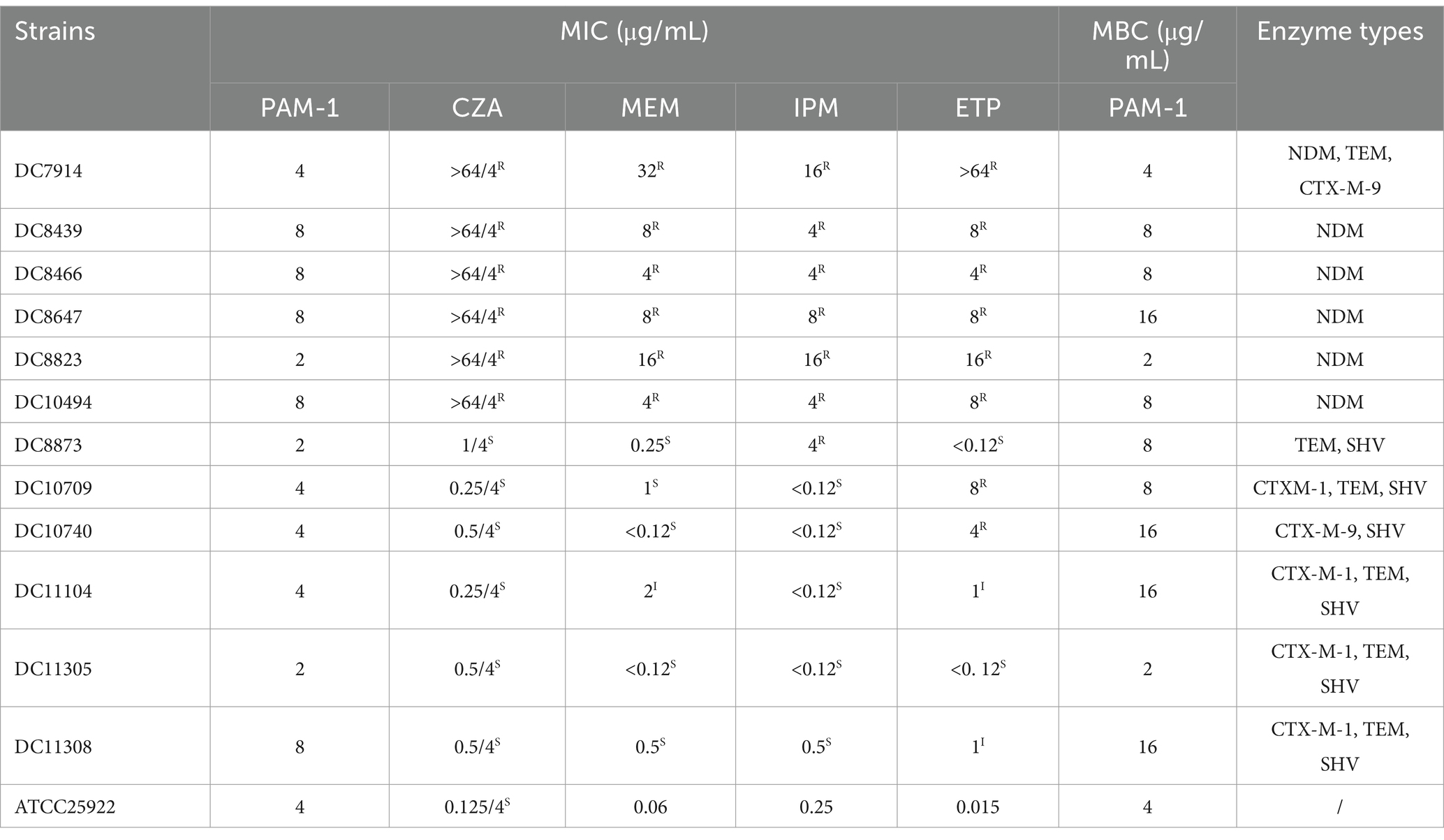
Table 3. MICs of ceftazidime/avibactam, carbapenems, and PAM-1 and MBCs of PAM-1 against Escherichia coli and the carbapenemase types of strains.
The effect of PAM-1 on bacterial growth was assessed by measuring OD600 changes over 24 h. Figure 1 demonstrates that the growth of the experimental strains was inhibited within 24 h when PAM-1 concentration exceeded or was equal to the 1 MIC. This finding suggests that PAM-1 exhibits a prolonged antibacterial effect on E. coli at the 1 MIC, and it even demonstrates a 12 h antibacterial effect on DC 11308 at 1/2 MIC concentration.
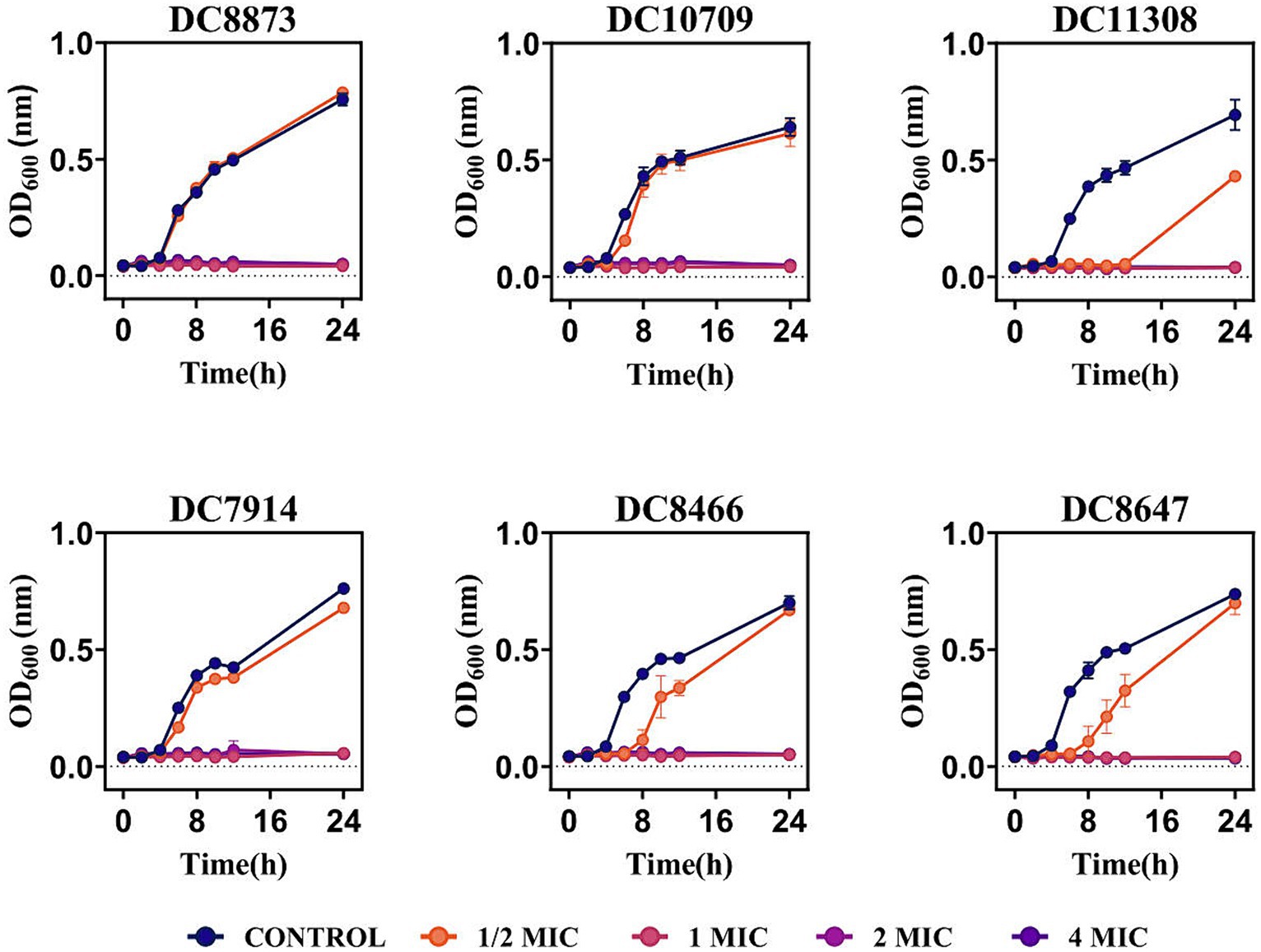
Figure 1. Growth curves of PAM-1 treatment against 3 CZA-susceptible E. coli and 3 CZA-resistant E. coli.
Crystal violet staining was used to determine the effect of PAM-1 treatment on inhibiting and eradicating biofilm formation. Figure 2A illustrates a noteworthy reduction in biofilm formation in the PAM-1-treated group compared to the control group. This observation suggests that1/2 MIC concentration PAM-1 possesses the ability to inhibit the formation of E. coli biofilm. Furthermore, it has an analogous efficacy in eradicating the pre-formed biofilms. Additionally, the eradication of established biofilms is achieved in a concentration-dependent manner, as illustrated in Figure 2B.
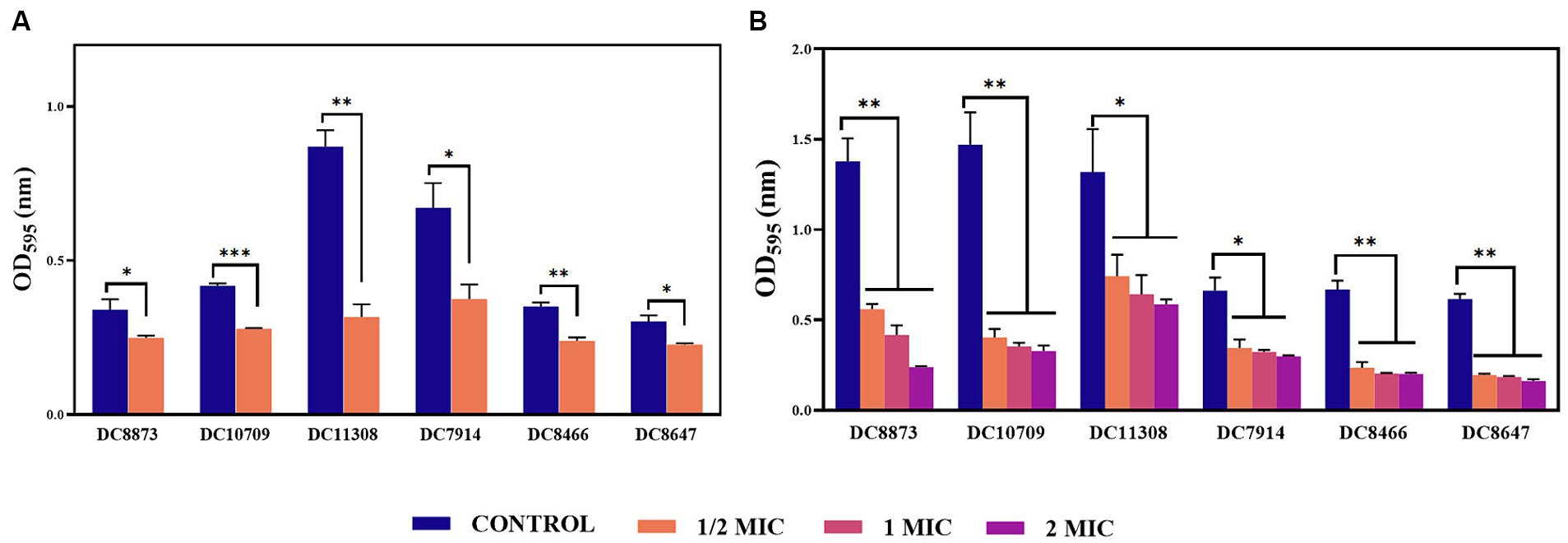
Figure 2. The effects of PAM-1 on E. coli biofilm. (A) The effect of PAM-1 on biofilm formation. (B) The effect of PAM-1 on the eradication of established biofilms. *p < 0.05, **p < 0.01, ***p < 0.001 were analyzed by Student’s t-test. The experiments were conducted thrice.
SEM analysis revealed significant morphological changes of E. coli biofilm treated with various concentrations of PAM-1 as compared with the untreated control group (Figure 3). In detail, SEM images exhibited a fully formed and dense biofilm in the control group, with no noticeable bacterial destruction. However, a significant decrease in its number and density and changes in its structure were observed after PAM-1 treatment. And some bacteria also showed damage to some extent.
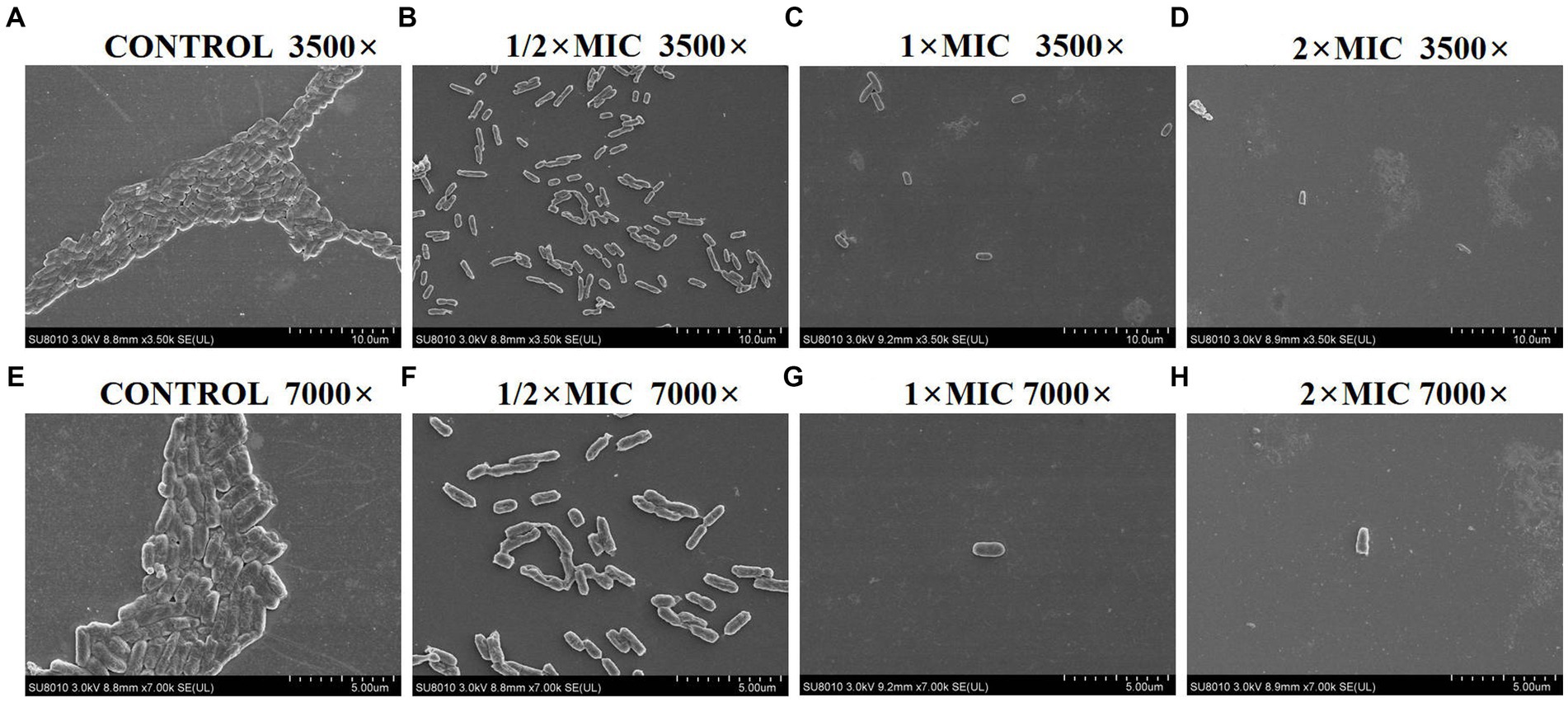
Figure 3. SEM images of E. coli DC 8466 biofilm and bacterial morphology results among various groups. (A) The broth control group, 3,500×, (E) 7,000×; (B) PAM-1 (1/2 MIC), 3,500×, (F) 7,000×; (C) PAM-1 (1 MIC), 3,500×, (G) 7,000×; (D) PAM-1 (2 MIC), 3,500×, (H) 7,000 ×.
Figure 4 illustrates that PAM-1 increased bacterial membrane permeability. To investigate the changes in cell membrane permeability induced by PAM-1, two different assays were performed: PI staining and NPN uptake assay. PI is a probe that cannot penetrate the cell membrane and can only bind to the nucleic acid of bacteria with damaged membranes. On the other hand, NPN can help assess the permeability of the outer membrane. Normally, NPN is hindered by the intact outer membrane. However, if the outer membrane structure is compromised and its permeability is altered, NPN can enter the interior of the cell and emit fluorescence. Both PI staining and the NPN uptake assay revealed that the fluorescence intensity of PAM-1 treated group was significantly higher than that of untreated group, indicating that PAM-1 induced a dose-dependent increase in membrane permeability.
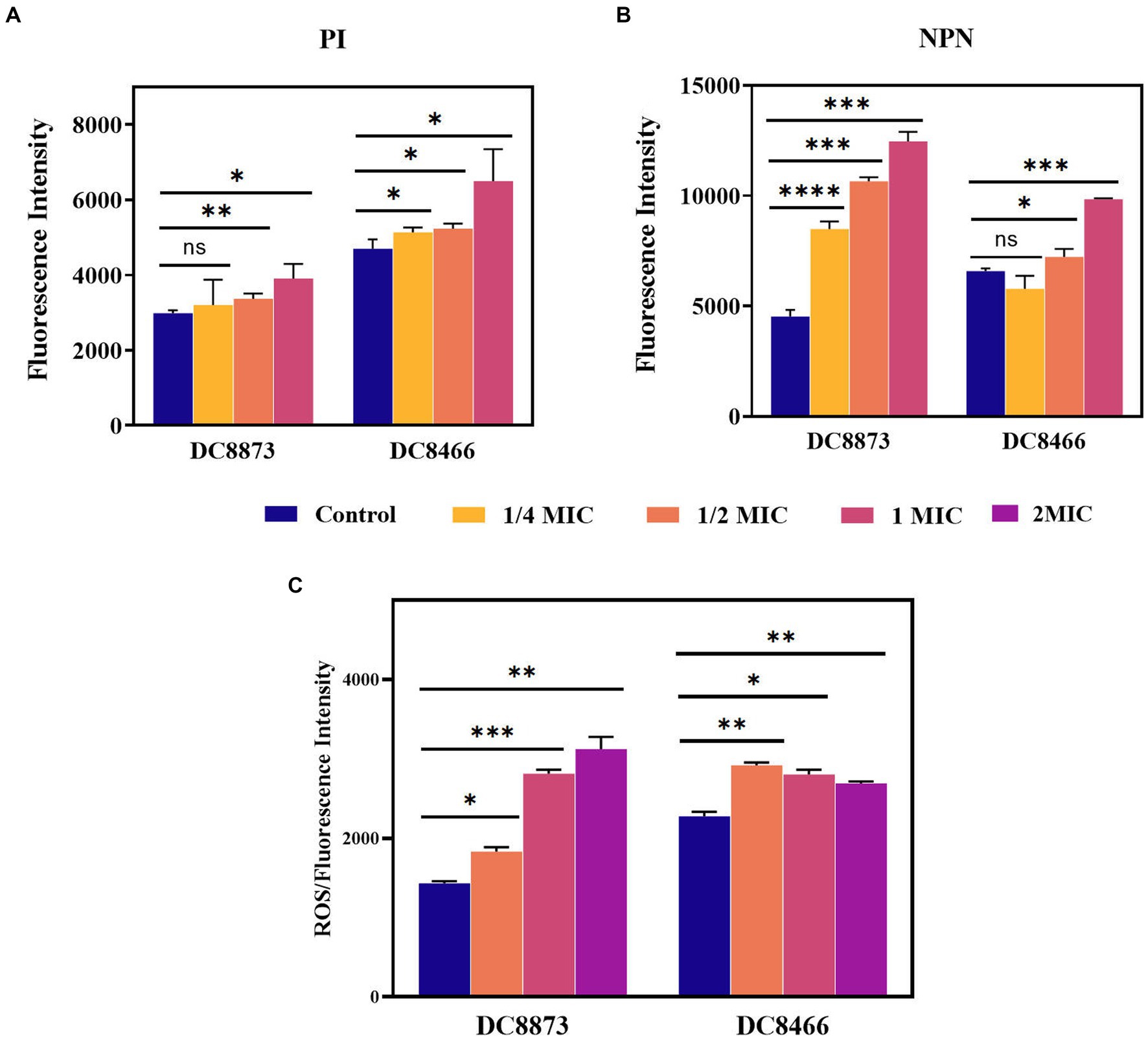
Figure 4. The antimicrobial mechanism of PAM-1 was investigated by PI (A), NPN (B), and (C) ROS. The fluorescence intensity of DC 8873 and DC 8466 treated with different concentrations of PAM-1. ns, not statistically significant; *p < 0.05, **p < 0.01, ***p < 0.001, ****p < 0.0001.
Furthermore, PAM-1 significantly increased ROS levels (Figure 4C); this may have played a role in its antibacterial activity. The level of ROS is closely correlated with the physiological activities of cells. When intracellular ROS production surpasses the cells’ natural antioxidant defenses, it induces oxidative stress, resulting in cellular damage and apoptosis (Azad et al., 2009). Collectively, these findings suggest that even at low concentrations, PAM-1 can effectively damage bacterial membranes, increase cell membrane permeability, and ultimately lead to bacterial death.
RT-qPCR was performed to elucidate the anti-inflammatory effects of PAM-1. Initially, using a cytotoxicity assay, we confirmed that an MOI of 10 did not affect cell viability. Therefore, RAW 264.7 cells were infected with bacterial cultures with an MOI of 10; then, the expression of IL-1β and TNF-α was analyzed. Figure 5 demonstrates that DC 8873 and DC 8466 significantly promoted IL-1β and TNF-α expression compared with the negative control group. However, PAM-1 dose-dependently inhibited IL-1β and TNF-α expression. The effect was more pronounced in DC 8466, which exhibited more significant inhibition of inflammatory cytokine. This indicates that PAM-1 possesses anti-inflammatory properties.
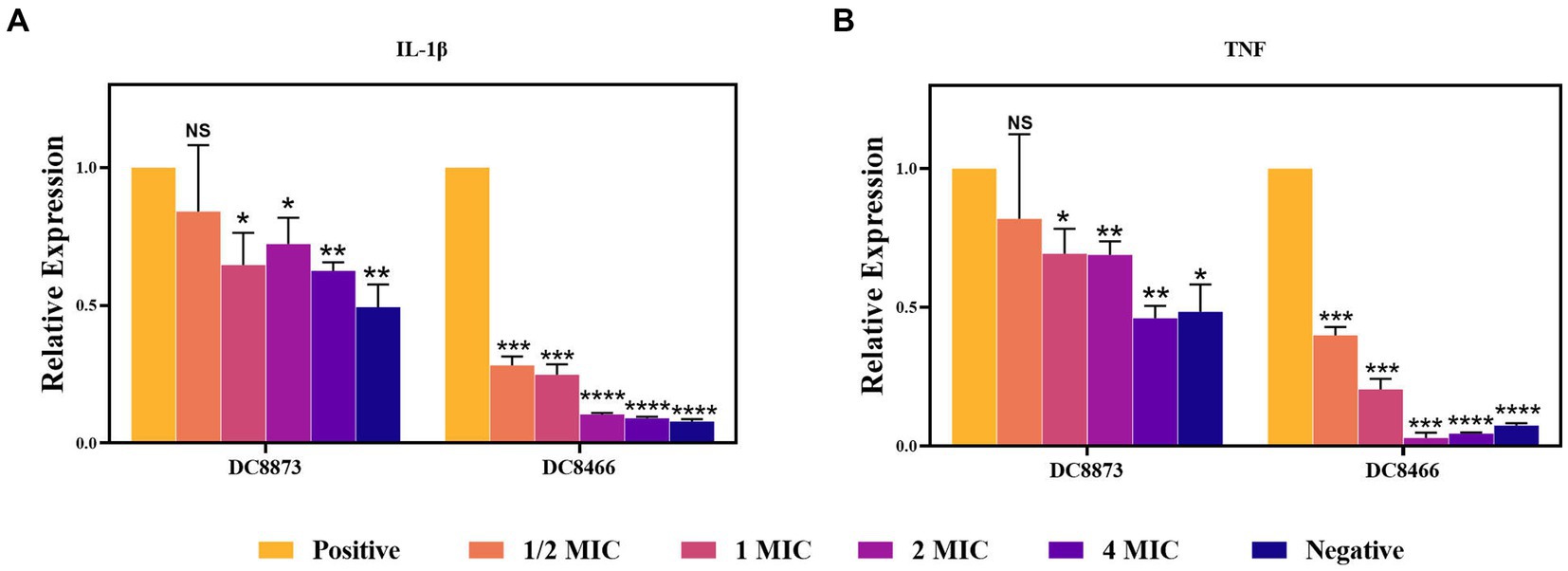
Figure 5. The effect of PAM-1 on RAW 264.7 inflammatory factor after E. coli infection. Negative, only cells are added; Positive, strain and cell co-infection (without PAM-1); NS, not statistically significant; *p < 0.05, **p < 0.01, ***p < 0.001, ****p < 0.0001.
We investigated the stability of PAM-1 under different conditions, including changes in temperature, serum exposure, and presence of salt ions. Table 4 demonstrates that neither ambient nor low temperatures affected the antibacterial activity of PAM-1. Furthermore, based on the provided instructions, it can be stored at −20°C for extended periods. Moreover, we observed that PAM-1 maintained significant antibacterial activity even in the presence of 10% serum (Table 5). Unfortunately, PAM-1 exhibited poor performance in a saline environment, with the presence of NaCl, CaCl2, and MgCl2 considerably decreasing its antibacterial efficacy (Table 6). This phenomenon could be attributed to the initial interaction of the positively charged cationic peptide as it interacts with and neutralizes the negatively charged bacterial outer membrane through electrostatic interaction (Bin Hafeez et al., 2021). Moreover, the presence of salt ions not only establishes a salt bridge with lipopolysaccharides to stabilize the outer membrane but also leads to the need for higher concentrations of AMPs to compete with and replace the salt ions on the outer membrane.
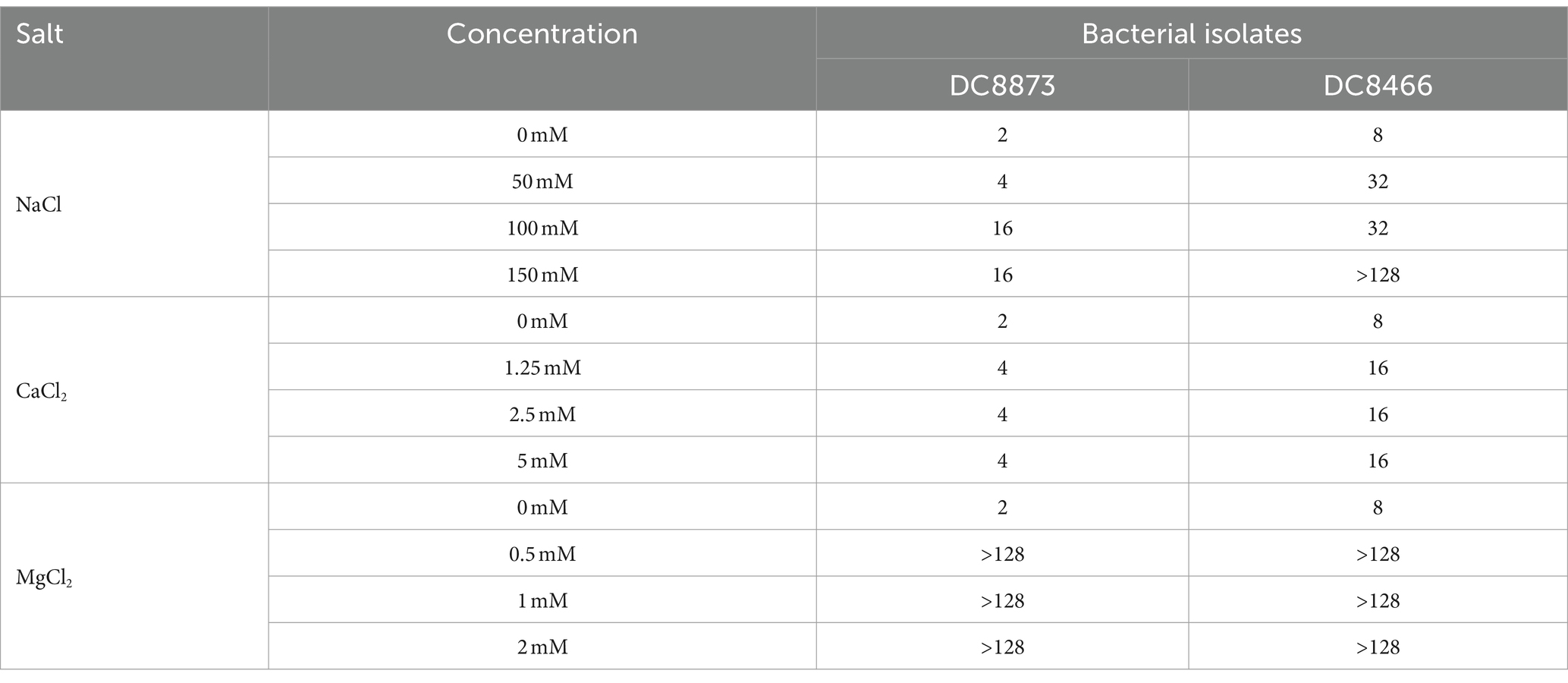
Table 6. The effect of conditions mimicking physiological concentrations of salt on the antibacterial activity of PAM-1 [MIC (μg/mL)].
To further investigate the security of PAM-1, RBC hemolysis and cytotoxicity were assessed (Figure 6). The hemolysis rate was <3% even when PAM-1 concentration reached 80 μg/mL; furthermore, no visible hemolysis was observed (Figure 6A). In addition, PAM-1 exhibited no cytotoxicity in RAW 264.7 cells at a concentration of 256 μg/mL (Figure 6B). Collectively, these findings suggest that PAM-1 holds promising prospects for in vivo applications.
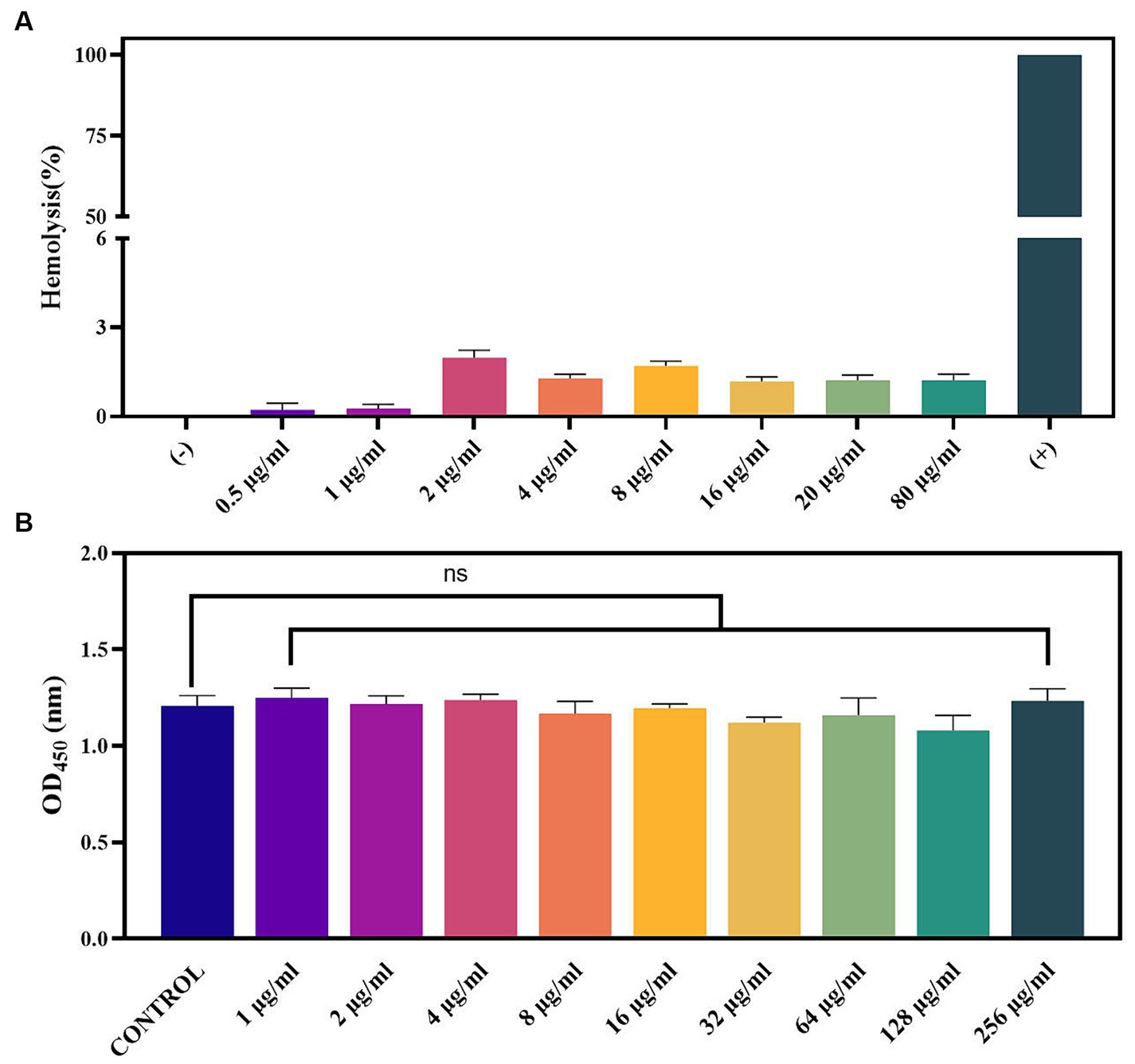
Figure 6. Toxicity of PAM-1. (A) The effect of PAM-1 on erythrocytes; (B) cytotoxicity of PAM-1 with different concentrations against RAW 264.7 murine macrophage cell line. ns, not statistically significant.
Considering the findings from in vitro antibacterial studies, conducting in vivo experiments is vital to confirm the antibacterial activity of PAM-1 in living organisms. To this end, a G. mellonella infection model was developed to assess the in vivo bactericidal activity of PAM-1 and compare it with that of CZA (Figure 7). Compared with the control group, PAM-1 significantly improved the survival rate of G. mellonella. After 7 days of PAM-1 therapy, the survival rates of DC 8873 (CZA-susceptible E. coli) and DC 8466(CZA-resistant E. coli) were > 60 and > 50%, respectively; however, all control subjects died at 7 and 4 days, respectively. Importantly, the survival rate of the PAM-1 treatment groups was either comparable with or higher than that of the CZA treatment groups; this suggests the superior efficacy of PAM-1 over CZA. Collectively, these results suggest the potential efficacy of PAM-1 in treating E. coli infections.
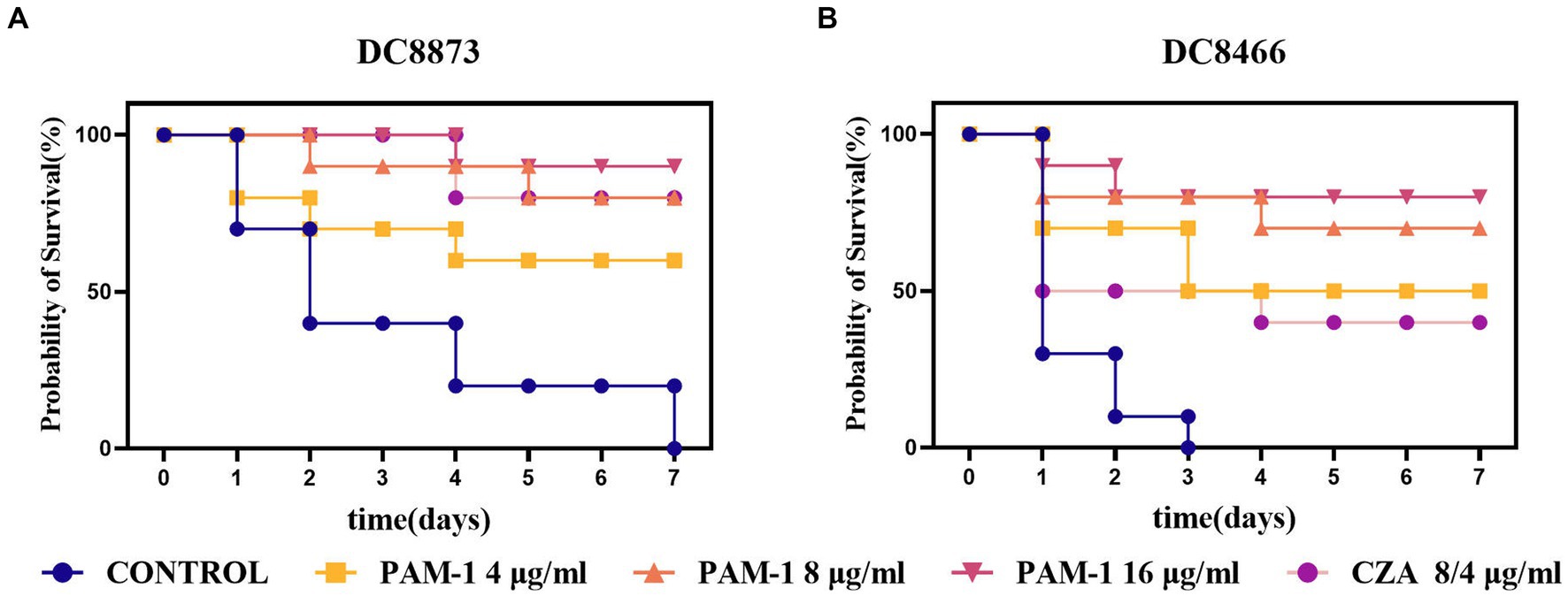
Figure 7. The survival rate of G. mellonella for different therapies. DC 8873 and DC 8466 as the experimental strains, and the survival rate record of G. mellonella in 7 days.
Owing to the worldwide emergence of antibiotic resistance and the declining rate of new antibiotic discovery, exploring novel antimicrobial agents has become an urgent issue. The increase in multidrug-resistant microorganisms poses a challenge for doctors and leads to higher rates of infections and deaths among seriously ill patients (Peleg and Hooper, 2010; Cornejo-Juárez et al., 2015). AMPs have been identified as promising candidates for antimicrobial therapy. They exert broad-spectrum antimicrobial activity and anti-inflammatory effects, have a rapid mode of action, and have minimal drug resistance development in pathogens (Yeaman and Yount, 2003).
AMPs derived from various natural sources, including plants, animals, and humans, provide valuable compositional and structural information. For example, WAM-1 from marsupials exhibits broad-spectrum antimicrobial activity (Wang et al., 2011); furthermore, AMPs from scorpion venom can kill multidrug-resistant pathogens (Harrison et al., 2014; Hong et al., 2021). However, some AMPs have inherent limitations, including low abundance, cytotoxicity, and instability under some conditions (Mahlapuu et al., 2020).
Although the in vitro antibacterial activity of PAM-1, an AMP isolated from the platypus, against standard strains of B. subtilis, S. aureus, and E. coli has been studied (Wang et al., 2011), studies on clinical strains and further assessment of its antibacterial effects and mechanisms in both in vitro and in vivo settings are lacking. Therefore, in the present study, we elucidated the antibacterial, antibiofilm, and anti-inflammatory activities of PAM-1 and its underlying mechanisms as well as evaluated its safety and efficacy in vivo.
First, PAM-1 exhibited significant antibacterial activity against both CZA-sensitive and resistant E. coli isolates (Table 3). PI staining and the NPN uptake assay revealed that PAM-1 may kill bacteria by destroying the bacterial membrane (Figure 4). Previous studies have reported that positively charged PAM-1 may disrupt the integrity of the bacterial membrane and exert antibacterial effects by interacting with and neutralizing the negatively charged bacterial outer membrane via electrostatic interactions, thereby forming transmembrane pores via the “barrel-stave” model (Oren and Shai, 1998; Bin Hafeez et al., 2021; Han et al., 2022). Based on this mechanism of action, PAM-1 can sustain antibacterial effects over an extended period, making it a promising therapeutic agent. During aerobic metabolism, cells continuously produce ROS, and protect against ROS overproduction. However, oxidative stress is induced when ROS production overwhelms the natural antioxidant defenses of the cell. PAM-1 may induce bacterial cell damage and apoptosis by significantly increasing ROS levels (Figure 4).
Biofilms are microbial communities attached to solid surfaces; they enhance bacterial adaptability to the environment, impede the penetration of conventional antibiotics, and contribute to recurrent infections (Longo et al., 2014; Flemming et al., 2016). PAM-1 exhibited a notable inhibitory effect on biofilm growth and mature biofilm eradication (Figure 2). SEM analysis revealed that PAM-1 effectively destroys biofilms and inflicts significant damage to the bacteria (Figure 3).
Inflammatory responses play a vital role in defending against pathogens (Kaczyńska et al., 2016). For example, TNF-α actively participates in the pathogenesis of skin infections (Mittal et al., 2010). Bacterial infections often trigger inflammation and associated complications, making it necessary to regulate proinflammatory factor production. RT-qPCR revealed that PAM-1 can effectively alleviate inflammatory responses by decreasing IL-1β and TNF-α production (Figure 5).
To evaluate the clinical potential of PAM-1, cytotoxicity and RBC hemolysis assays were conducted (Figure 6). Unlike most AMPs, PAM-1 maintained its antibacterial activity at 37°C as well as in the presence of serum (Tables 4, 5). However, Table 6 indicate PAM-1 exhibited instability in the presence of salt ions. This may be because cationic AMPs initially interact with and neutralize the negatively charged bacterial outer membrane via electrostatic interactions (Bin Hafeez et al., 2021). The presence of salt ions not only stabilizes the outer membrane by establishing a salt bridge with lipopolysaccharides but also increases the demand for AMP to replace the salt ions on the outer membrane (Han et al., 2022). Therefore, additional studies are warranted to explain the competition and stabilization mechanisms between AMP and salt ions and to solve the application of cationic AMPs in high-salt environments.
Based on the findings of the abovementioned experiments, we used the G. mellonella larval infection model to further verify the in vivo therapeutic effect of PAM-1. Compared with the treatment effect of CZA, the survival rate of G. mellonella larvae treated with PAM-1 was similar or better (Figure 7). This suggests that PAM-1 had a good application prospect in vivo and can exert a better therapeutic effect than CZA in vivo.
This is the first study to demonstrate the antibacterial effect of PAM-1 against CZA-resistant E. coli both in vitro and in vivo. We observed that PAM-1 is a promising agent for treating CZA-resistant E. coli infections. In addition to antibacterial and antibiofilm activities, PAM-1 also exhibits potent anti-inflammatory effects (Figure 8). Although the mechanism underlying the effect of salt ions on its activity deserves further investigation, PAM-1 exhibits good stability and effects in serum, at room temperature, and in vivo. Collectively, these findings indicate that PAM-1 is a promising antibacterial agent against drug-resistant E. coli infections.
The original contributions presented in the study are included in the article/Supplementary material, further inquiries can be directed to the corresponding authors.
The animal study was approved by the Zhejiang Association for Science and Technology SYXK. The study was conducted in accordance with the local legislation and institutional requirements.
YH: Data curation, Formal analysis, Investigation, Methodology, Software, Writing – original draft. YZ: Investigation, Methodology, Validation, Writing – review & editing. XZ: Conceptualization, Supervision, Writing – review & editing. ZH: Investigation, Software, Writing – review & editing. JK: Investigation, Writing – review & editing. XW: Investigation, Project administration, Resources, Writing – review & editing. LC: Investigation, Resources, Writing – review & editing. YW: Methodology, Software, Supervision, Validation, Writing – review & editing. JC: Conceptualization, Data curation, Supervision, Writing – review & editing. TZ: Conceptualization, Funding acquisition, Resources, Supervision, Writing – review & editing. MS: Conceptualization, Resources, Supervision, Writing – review & editing.
The author(s) declare financial support was received for the research, authorship, and/or publication of this article. This work was funded by the Key Laboratory of Clinical Laboratory Diagnosis and Translational Research of Zhejiang Province (2022E10022).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2024.1291876/full#supplementary-material
Ashfaq, M. Y., da'na, D. A., and al-Ghouti, M. A. (2022). Application of MALDI-TOF MS for identification of environmental bacteria: a review. J. Environ. Manag. 305:114359. doi: 10.1016/j.jenvman.2021.114359
Azad, M. B., Chen, Y., and Gibson, S. B. (2009). Regulation of autophagy by reactive oxygen species (ROS): implications for Cancer progression and treatment. Antioxid. Redox Signal. 11, 777–790. doi: 10.1089/ars.2008.2270
Bin Hafeez, A., Jiang, X., Bergen, P. J., and Zhu, Y. (2021). Antimicrobial peptides: An update on classifications and databases. Int. J. Mol. Sci. 22:11691. doi: 10.3390/ijms222111691
Borjan, J., Meyer, K. A., Shields, R. K., and Wenzler, E. (2020). Activity of ceftazidime-avibactam alone and in combination with polymyxin B against carbapenem-resistant Klebsiella pneumoniae in a tandem in vitro time-kill/in vivo galleria mellonella survival model analysis. Int. J. Antimicrob. Agents 55:105852. doi: 10.1016/j.ijantimicag.2019.11.009
Cornejo-Juárez, P., Vilar-Compte, D., Pérez-Jiménez, C., Ñamendys-Silva, S. A., Sandoval-Hernández, S., and Volkow-Fernández, P. (2015). The impact of hospital-acquired infections with multidrug-resistant bacteria in an oncology intensive care unit. Int. J. Infect. Dis. 31, 31–34. doi: 10.1016/j.ijid.2014.12.022
da Luz, J. Z., Machado, T. N., Bezerra, A. G., de Oliveira Ribeiro, C. A., and Neto, F. F. (2020). Cytotoxicity of bismuth nanoparticles in the murine macrophage cell line RAW 264.7. J. Mater. Sci. Mater. Med. 31:95. doi: 10.1007/s10856-020-06427-0
de Campos, G. Y., Oliveira, R. A., Oliveira-Brito, P. K. M., Roque-Barreira, M. C., and da Silva, T. A. (2020). Pro-inflammatory response ensured by LPS and pam 3CSK4 in RAW 264.7 cells did not improve a fungistatic effect on Cryptococcus gattii infection. PeerJ 8:e10295. doi: 10.7717/peerj.10295
Dong, M., Kwok, S. H., Humble, J. L., Liang, Y., Tang, S. W., Tang, K. H., et al. (2021). BING, a novel antimicrobial peptide isolated from Japanese medaka plasma, targets bacterial envelope stress response by suppressing cpx R expression. Sci. Rep. 11:12219. doi: 10.1038/s41598-021-91765-4
Dong, N., Ma, Q., Shan, A., Lv, Y., Hu, W., Gu, Y., et al. (2012). Strand length-dependent antimicrobial activity and membrane-active mechanism of arginine- and valine-rich β-hairpin-like antimicrobial peptides. Antimicrob. Agents Chemother. 56, 2994–3003. doi: 10.1128/AAC.06327-11
Falcone, M., Russo, A., Iacovelli, A., Restuccia, G., Ceccarelli, G., Giordano, A., et al. (2016). Predictors of outcome in ICU patients with septic shock caused by Klebsiella pneumoniae carbapenemase–producing K. pneumoniae. Clin. Microbiol. Infect. 22, 444–450. doi: 10.1016/j.cmi.2016.01.016
Flemming, H.-C., Wingender, J., Szewzyk, U., Steinberg, P., Rice, S. A., and Kjelleberg, S. (2016). Biofilms: an emergent form of bacterial life. Nat. Rev. Microbiol. 14, 563–575. doi: 10.1038/nrmicro.2016.94
García-Cobos, S., Campos, J., Román, F., Carrera, C., Pérez-Vázquez, M., Aracil, B., et al. (2008). Low β-lactamase-negative ampicillin-resistant Haemophilus influenzae strains are best detected by testing amoxicillin susceptibility by the broth microdilution method. Antimicrob. Agents Chemother. 52, 2407–2414. doi: 10.1128/AAC.00214-08
Han, W., Wei, Z., and Camesano, T. A. (2022). New antimicrobial peptide-antibiotic combination strategy for Pseudomonas aeruginosa inactivation. Biointerphases 17:041002. doi: 10.1116/6.0001981
Harrison, P. L., Abdel-Rahman, M. A., Miller, K., and Strong, P. N. (2014). Antimicrobial peptides from scorpion venoms. Toxicon 88, 115–137. doi: 10.1016/j.toxicon.2014.06.006
Hernández-García, M., Sánchez-López, J., Martínez-García, L., Becerra-Aparicio, F., Morosini, M. I., Ruiz-Garbajosa, P., et al. (2021). Emergence of the new KPC-49 variant conferring an ESBL phenotype with resistance to ceftazidime-avibactam in the ST131-H30R1 Escherichia coli high-risk clone. Pathogens 10:67. doi: 10.3390/pathogens10010067
Hong, M. J., Kim, M. K., and Park, Y. (2021). Comparative antimicrobial activity of Hp404 peptide and its analogs against Acinetobacter baumannii. Int. J. Mol. Sci. 22:5540. doi: 10.3390/ijms22115540
Jayathilaka, E. H. T. T., Rajapaksha, D. C., Nikapitiya, C., De Zoysa, M., and Whang, I. (2021). Antimicrobial and anti-biofilm peptide Octominin for controlling multidrug-resistant Acinetobacter baumannii. Int. J. Mol. Sci. 22:5353. doi: 10.3390/ijms22105353
Kaczyńska, K., Kogut, E., Zając, D., Jampolska, M., Andrzejewski, K., Sulejczak, D., et al. (2016). Neurotensin-based hybrid peptide’s anti-inflammatory activity in murine model of a contact sensitivity response. Eur. J. Pharm. Sci. 93, 84–89. doi: 10.1016/j.ejps.2016.08.012
Kim, M. K., Kang, H. K., Ko, S. J., Hong, M. J., Bang, J. K., Seo, C. H., et al. (2018). Mechanisms driving the antibacterial and antibiofilm properties of Hp1404 and its analogue peptides against multidrug-resistant Pseudomonas aeruginosa. Sci. Rep. 8:1763. doi: 10.1038/s41598-018-19434-7
Liu, Y., Ding, S., Dietrich, R., Märtlbauer, E., and Zhu, K. (2017). A biosurfactant-inspired Heptapeptide with improved specificity to kill MRSA. Angew. Chem. Int. Ed. Engl. 56, 1486–1490. doi: 10.1002/anie.201609277
Livermore, D. M., Meunier, D., Hopkins, K. L., Doumith, M., Hill, R., Pike, R., et al. (2018). Activity of ceftazidime/avibactam against problem Enterobacteriaceae and Pseudomonas aeruginosa in the UK, 2015–16. J. Antimicrob. Chemother. 73, 648–657. doi: 10.1093/jac/dkx438
Longo, F., Vuotto, C., and Donelli, G. (2014). Biofilm formation in Acinetobacter baumannii. New Microbiol. 37, 119–127.
Luo, Y., and Song, Y. (2021). Mechanism of antimicrobial peptides: antimicrobial, anti-inflammatory and Antibiofilm activities. Int. J. Mol. Sci. 22:11401. doi: 10.3390/ijms222111401
Mahlapuu, M., Björn, C., and Ekblom, J. (2020). Antimicrobial peptides as therapeutic agents: opportunities and challenges. Crit. Rev. Biotechnol. 40, 978–992. doi: 10.1080/07388551.2020.1796576
Mittal, D., Saccheri, F., Vénéreau, E., Pusterla, T., Bianchi, M. E., and Rescigno, M. (2010). TLR4-mediated skin carcinogenesis is dependent on immune and radioresistant cells. EMBO J. 29, 2242–2252. doi: 10.1038/emboj.2010.94
Morrill, H. J., Pogue, J. M., Kaye, K. S., and LaPlante, K. L. (2015). Treatment options for Carbapenem-resistant Enterobacteriaceae infections. Open forum. Infect. Dis. 2:ofv 050. doi: 10.1093/ofid/ofv050
Nguyen, H. L. T., Trujillo-Paez, J. V., Umehara, Y., Yue, H., Peng, G., Kiatsurayanon, C., et al. (2020). Role of antimicrobial peptides in skin barrier repair in individuals with atopic dermatitis. Int. J. Mol. Sci. 21:7607. doi: 10.3390/ijms21207607
O’Loughlin, C. T., Miller, L. C., Siryaporn, A., Drescher, K., Semmelhack, M. F., and Bassler, B. L. (2013). A quorum-sensing inhibitor blocks Pseudomonas aeruginosa virulence and biofilm formation. Proc. Natl. Acad. Sci. USA 110, 17981–17986. doi: 10.1073/pnas.1316981110
Oren, Z., and Shai, Y. (1998). Mode of action of linear amphipathic α-helical antimicrobial peptides. Biopolymers 47, 451–463. doi: 10.1002/(SICI)1097-0282(1998)47:6<451::AID-BIP4>3.0.CO;2-F
Pasupuleti, M., Schmidtchen, A., and Malmsten, M. (2012). Antimicrobial peptides: key components of the innate immune system. Crit. Rev. Biotechnol. 32, 143–171. doi: 10.3109/07388551.2011.594423
Peleg, A. Y., and Hooper, D. C. (2010). Hospital-acquired infections due to gram-negative Bacteria. N. Engl. J. Med. 362, 1804–1813. doi: 10.1056/NEJMra0904124
Quintilla, R., Kolecka, A., Casaregola, S., Daniel, H. M., Houbraken, J., Kostrzewa, M., et al. (2018). MALDI-TOF MS as a tool to identify foodborne yeasts and yeast-like fungi. Int. J. Food Microbiol. 266, 109–118. doi: 10.1016/j.ijfoodmicro.2017.11.016
Satlin, M. J., Chen, L., Patel, G., Gomez-Simmonds, A., Weston, G., Kim, A. C., et al. (2017). Multicenter clinical and molecular epidemiological analysis of bacteremia due to Carbapenem-resistant Enterobacteriaceae (CRE) in the CRE epicenter of the United States. Antimicrob. Agents Chemother. 61, e02349–e02316. doi: 10.1128/AAC.02349-16
Shields, R. K., Chen, L., Cheng, S., Chavda, K. D., Press, E. G., Snyder, A., et al. (2017). Emergence of ceftazidime-avibactam resistance due to plasmid-borne Bla KPC-3 mutations during treatment of Carbapenem-resistant Klebsiella pneumoniae infections. Antimicrob. Agents Chemother. 61, e02097–e02016. doi: 10.1128/AAC.02097-16
Shields, R. K., Potoski, B. A., Haidar, G., Hao, B., Doi, Y., Chen, L., et al. (2016). Clinical outcomes, drug toxicity, and emergence of ceftazidime-avibactam resistance among patients treated for Carbapenem-resistant Enterobacteriaceae infections. Clin. Infect. Dis. 63, 1615–1618. doi: 10.1093/cid/ciw636
Spencer, J. J., Pitts, R. E., Pearson, R. A., and King, L. B. (2018). The effects of antimicrobial peptides WAM-1 and LL-37 on multidrug-resistant Acinetobacter baumannii. Pathog Dis 76. doi: 10.1093/femspd/fty007
Tang, Y.-T., Huang, Y.-Y., Zheng, L., Qin, S.-H., Xu, X.-P., An, T.-X., et al. (2017). Comparison of isolation methods of exosomes and exosomal RNA from cell culture medium and serum. Int. J. Mol. Med. 40, 834–844. doi: 10.3892/ijmm.2017.3080
Thapa, R. K., Diep, D. B., and Tønnesen, H. H. (2020). Topical antimicrobial peptide formulations for wound healing: current developments and future prospects. Acta Biomater. 103, 52–67. doi: 10.1016/j.actbio.2019.12.025
Trendel, J., Schwarzl, T., Horos, R., Prakash, A., Bateman, A., Hentze, M. W., et al. (2019). The human RNA-binding proteome and its dynamics during translational arrest. Cell 176, 391–403.e19. doi: 10.1016/j.cell.2018.11.004
Tsai, C. J.-Y., Loh, J. M. S., and Proft, T. (2016). Galleria mellonella infection models for the study of bacterial diseases and for antimicrobial drug testing. Virulence 7, 214–229. doi: 10.1080/21505594.2015.1135289
van Duin, D., Arias, C. A., Komarow, L., Chen, L., Hanson, B. M., Weston, G., et al. (2020). Molecular and clinical epidemiology of Carbapenem-resistant Enterobacteriaceae in the United States: a prospective cohort study. Lancet Infect. Dis. 20, 731–741. doi: 10.1016/S1473-3099(19)30755-8
Wang, J., Wong, E. S. W., Whitley, J. C., Li, J., Stringer, J. M., Short, K. R., et al. (2011). Ancient antimicrobial peptides kill antibiotic-resistant pathogens: Australian mammals provide new options. PLoS One 6:e24030. doi: 10.1371/journal.pone.0024030
Xu, W., Chen, T., Wang, H., Zeng, W., Wu, Q., Yu, K., et al. (2020). Molecular mechanisms and epidemiology of Fosfomycin resistance in Staphylococcus aureus isolated from patients at a teaching Hospital in China. Front. Microbiol. 11:1290. doi: 10.3389/fmicb.2020.01290
Yeaman, M. R., and Yount, N. Y. (2003). Mechanisms of antimicrobial peptide action and resistance. Pharmacol. Rev. 55, 27–55. doi: 10.1124/pr.55.1.2
Zeng, W., Liao, W., Zhao, Y., Wang, L., Shu, H., Jia, H., et al. (2022). A selective medium for screening ceftazidime/avibactam resistance in Carbapenem-resistant Enterobacterales. Front. Microbiol. 13:956044. doi: 10.3389/fmicb.2022.956044
Zhang, W., Guo, Y., Li, J., Zhang, Y., Yang, Y., Dong, D., et al. (2018). In vitro and in vivo bactericidal activity of ceftazidime-avibactam against Carbapenemase–producing Klebsiella pneumoniae. Antimicrob. Resist. Infect. Control 7:142. doi: 10.1186/s13756-018-0435-9
Zhang, X., Shi, S., Yao, Z., Zheng, X., Li, W., Zhang, Y., et al. (2022). Antimicrobial peptide WAM-1: a promising antibacterial and anti-inflammatory drug against carbapenem-resistant Klebsiella pneumoniae. J. Antimicrob. Chemother. 77, 1903–1911. doi: 10.1093/jac/dkac128
Keywords: Escherichia coli, biofilm, CZA -resistant, PAM-1, antimicrobial peptide
Citation: Han Y, Zhang Y, Zhang X, Huang Z, Kong J, Wang X, Chen L, Wang Y, Cao J, Zhou T and Shen M (2024) PAM-1: an antimicrobial peptide with promise against ceftazidime-avibactam resistant Escherichia coli infection. Front. Microbiol. 15:1291876. doi: 10.3389/fmicb.2024.1291876
Received: 10 September 2023; Accepted: 10 April 2024;
Published: 30 April 2024.
Edited by:
Mona I. Shaaban, Mansoura Universiy, EgyptReviewed by:
Bijit Bhowmik, Croda Inc, United StatesCopyright © 2024 Han, Zhang, Zhang, Huang, Kong, Wang, Chen, Wang, Cao, Zhou and Shen. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Tieli Zhou, d3l6dGxpQDE2My5jb20=; Mo Shen, c2hlbm1vNjAxQDE2My5jb20=
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.