- 1Yunnan Provincial Key Laboratory of Entomological Biopharmaceutical R&D, Dali University, Dali, China
- 2College of Pharmacy, Dali University, Dali, China
Wasps play important roles as predators and pollinators in the ecosystem. The Jingpo minority residing in Yunnan Province, China, has a traditional practice of using wine infused with mature wasps as a customary remedy for managing rheumatoid arthritis. The larva of the wasp is also a tasteful folk dish that has created a tremendous market. There is a paucity of survival knowledge, which has greatly restricted their potential applications in food and healthcare. Recent research has highlighted the importance of gut microbiota in insect growth. Nevertheless, there is still a lack of understanding regarding the composition, changes, and functions of the gut microbiota in Vespa mandarinia during development. In this research, the gut microbiota were investigated across three growth stages of Vespa mandarinia using a metagenomic technology. The result revealed that there are significant variations in the proportion of main gut microbes during the metamorphosis of Vespa mandarinia. Tenericutes were found to dominate during the larval stage, while Proteobacteria emerged as the dominant group post-pupation. Through a comprehensive analysis of the gut microbiota metagenome, this study revealed functional differences in the wasp gut microbiota at various growth stages. During the larval stage, the gut microbiota plays a central role in promoting metabolism. Following pupation, the gut microbiota exhibited diversified functions, likely due to the complex environments and diverse food sources encountered after metamorphosis. These functions included amino acid metabolism, compound degradation, and defense mechanisms. This research provides an extensive dataset on the gut microbiota during the metamorphosis of Vespa mandarinia, contributing to a deeper understanding of the influence of gut microbiota on wasp growth. Furthermore, this study uncovers a unique microbial treasure within insect guts, which is important for advancing the application of wasps in the fields of food and medicine.
1 Introduction
Insects are the most abundant and diverse animals, distributed in almost all niches on the earth (Stork, 2007). Many of these insects have exhibited remarkably medicinal properties or economic importance (Dimarcq and Hunneyball, 2003). Hence, insects have become an indispensable part of human life. Vespa mandarinia (V. mandarinia), a member of the Hymenoptera Vespidae, is a special entomological resource of Yunnan province, China (Zhou et al., 2019). As a folk medicine of the Jingpo minority, the wine made by V. mandarinia has been used to treat rheumatoid arthritis for centuries. The living V. mandarinia is soaked in an alcohol product named ‘Baijiu' that contains the toxin of wasp, along with a variety of bioactive components from its body. This empirical formula has been listed in the Chinese Pharmacopeia as having an obvious therapeutic effect (Zhou et al., 2019). Moreover, the larva of the wasp is a delicious ethnic food that won the favor of local people (Feng et al., 2018). Wasps have been consumed by people worldwide for a long time and used as the chosen delicacy for celebration in Japan (Dobermann et al., 2017). Recent scientific data indicated that the wasp larva can offer sufficient and balanced essential nutrients due to its low fat, high protein, and carbohydrates (Jeong et al., 2020). The increasing market demand has made the cultivation of V. mandarinia become a main source of income for villagers. However, reliable breeding systems are still deficient, and more knowledge about the growth and development of wasps is needed.
As the main place for digestion and absorption, the distinctive environments of the insect digestive tract have attracted the colonization of abundant and various microbes. Some symbiotic bacteria could affect the host's physiology (Wang et al., 2020). Previous research has indicated that the gut microbiota of insects shows diversified symbiotic functions, such as increasing oviposition, shortening larval development, assisting in food digestion, providing essential nutrition, and protecting against pathogens (Engel and Moran, 2013; Jang and Kikuchi, 2020). Some specific antimicrobial symbionts of wasps can even transfer to the brood cells to assist their larvae in fighting against pathogens (Kroiss et al., 2010). The Wolbachia of the parasitic wasp can reportedly interfere with programmed cell death for the production of mature oocytes to maintain reproduction (Dedeine et al., 2001). The type and extent of benefits provided by gut microbiota are highly variable across insects and mainly depend on the host species, as the great distinction in ecology, dietary habits, morphology, and lifecycle of the host. A typical example is the differences in gut bacterial communities between adult and larval dung beetles, which possess disparate genes involved in cellulose degradation and nitrogen fixation that are correlated with their distinct diet composition (Shukla et al., 2016). The gut microbiome of adult V. mandarinia was first reported in 2019, providing novel insights into how food habits affect the gut microbiota of social insects (Suenami et al., 2019). Despite this, some articles have reported the diversity of gut microbiota in a few species of wasps (Suenami et al., 2019; Cini et al., 2020). Their variation during growth as well as their role in metamorphosis are still unknown.
With the advances in microbial technologies, it has now become clear that the fungi, protists, bacteria, and archaea are the major components of the microorganisms in insect guts (Schmidt and Engel, 2021). For such extremely heterogeneous microbes, it is an impossible task to discover important ones and identify their function with conventional culture-dependent technologies, as the distinctive environments of the digestive tract make many of these bacteria uncultivable with the existing technologies. The advanced culture-independent technologies, including 16S rRNA gene analysis, DNA fingerprinting, and denaturing gradient gel electrophoresis, especially those genome-based “omics” technologies, have broadened our understanding of the communities of gut bacteria (Nayfach and Pollard, 2016; Bobay et al., 2020). Among the technologies, shotgun metagenomics can simultaneously provide an integrated overview of microbial composition as well as their potential functions at the genetic level (Nayfach and Pollard, 2016).
Herein, this research used shotgun metagenomic sequencing to provide hitherto undocumented evidence of the gut microbiota landscape at different growth stages of V. mandarinia. Relevant gene sequencing results revealed that the relative abundance of gut microbiota has changed significantly during the metamorphosis of V. mandarinia, and these symbiotic bacteria play growth-promoting roles in the development of the host.
This research provides a comprehensive landscape of the gut microbiota between adults and larvae of V. mandarinia through metagenomic analysis. This knowledge could be helpful for the technical improvement in wasp farming. Moreover, this understanding uncovers a unique treasure that can explore a special microorganism resource in insect guts.
2 Materials and methods
2.1 Insect collection
The samples of V. mandarinia were collected from Tengchong district, Yunnan Province, China, in September 2020. These samples of V. mandarinia were randomly caught in the hive (Figures 1A, B). According to the growth stages, these samples were divided into three groups: larva, nymph, and adult (Figures 1C–E), frozen immediately, and stored in liquid nitrogen.
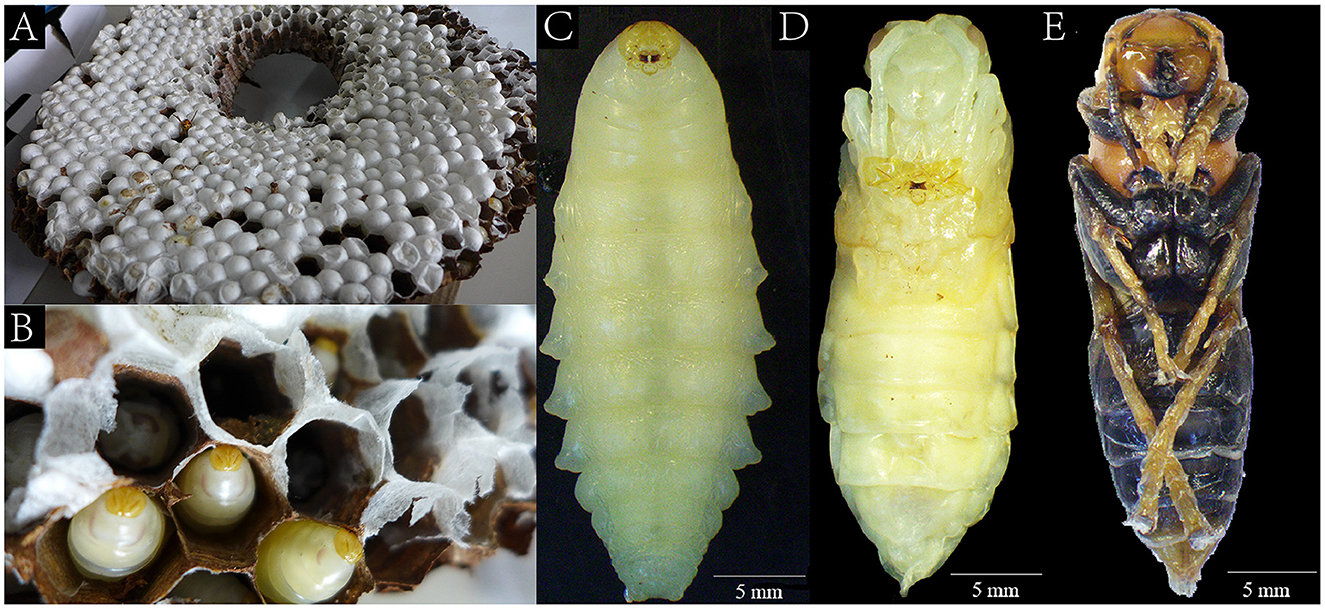
Figure 1. The living environment of the wasps and the wasp morphology in three growth stages. The landscape of transection in wasp hives (A, B). The larva (C) of the wasp lived in the hive that was fed by the adult (E), and the nymph (D) was developed in the cocoon.
2.2 Dissection and DNA extraction of the Vespa mandarinia guts
The samples of V. mandarinia in their three distinct forms were randomly selected. Subsequently, 75% alcohol and 0.5% sodium hypochlorite were sequentially used to sterilize the body surface of V. mandarinia for 1 min and then washed twice for 1 min in sterile distilled water. Moreover, the anatomical tools were sterilized by autoclaving at 121°C for 30 min. The samples were disintegrated under an aseptic condition, and the gut portions of larval and adult wasps were transferred, respectively, to sterile 1.5 ml microcentrifuge tubes. The TruSeq DNA sample prep kit was used to extract the total meta-genomic DNA from the entomic gut, and then the extracts were analyzed using 1% agarose gel electrophoresis (Qian et al., 2021). An Illumina PE library was constructed: “Y” sticky joints were ligated, eliminating self-ligated fragments through magnetic bead purification, enriching template libraries via PCR, and generating single-stranded DNA fragments using sodium hydroxide (Guo et al., 2021). Single-stranded DNA sequences are amplified into clusters by bridge PCR.
2.3 Original metagenomic sequencing of the Vespa mandarinia gut
Three paired-end libraries were constructed according to different growth stages, and the extracted metagenomic DNA was sequenced using the Illumina HiSeq technology (Holm et al., 2019). The raw sequencing data were first cleaned and filtered to improve the quality and reliability of the subsequent analysis. The processing method was conducted as follows: (1) filtering these N basic reads with the tail mass below 20, (2) removing below 50 bp reads, (3) removing the read containing adapter and exceeding 2 N bases, and (4) removing reads that are highly similar to the host. The reference wasp genome's assembly accession on NCBI was GCF 014083535.2 (Miller et al., 2022). BWA software aligned the reads to the host DNA sequence and removed highly similar contaminant reads (Oliva et al., 2021). These high-quality reads were used for splicing assembly and gene prediction.
2.4 The gene prediction and construction of non-redundant genes
Based on the principles of interactive De Bruijn graphs, the assembly of reads was executed using MEGAHIT software across varying sequencing depths. By progressively increasing the k-mer size, long contigs with few interruptions and branches are achieved through iterative processes (Huang et al., 2020). Contigs ≥100 bp were selected, and MetaGeneMark software (Version 2.10) was used to predict the open reading frame (Wu et al., 2019). The related results were translated into the amino acid sequence. Due to many microbial genes being repeated in the sample, gene sets were obtained by clustering to remove redundancy and enhance data quality. The variation in the abundance of different genes among samples can reflect their general characteristics and discrepancies. The non-redundant gene catalog was constructed to describe the overall information of the genes in these environments. The predicted genes from all samples were clustered by CD-HIT software (95% identity and 90% coverage), and the longest gene of each class was used to build a non-redundant gene database.
2.5 Metagenomic analysis
To clarify the composition of the gut microbiome and their variation during the metamorphosis of V. mandarinia, the non-redundant genes were compared using BLAST to the non-redundant protein database of the NCBI as a reference database with a set e-value threshold of 10−5 (Gao et al., 2017). The function of the gut microbiome was annotated with the eggNOG and KEGG databases. The corresponding Clusters of Orthologous Groups (COG) of proteins were compared to the eggNOG database using BLAST (Huerta-Cepas et al., 2016). The abundance of each COG was calculated by summing the gene abundances associated with it. The gene sets were compared to the KEGG genes database using BLAST (Kanehisa et al., 2017). The resulting alignments of KEGG were then used for functional annotation using KOBAS 2.0 (Xie et al., 2011). The abundance of each functional category, such as pathway, was calculated by summing the gene abundances associated with it.
3 Results
3.1 Initial sequencing reads and genome filtering results
The whole guts of samples (larvae, nymph, and adults, three groups of each) were extracted to assess their associated microbial communities. A total of 64.47 Gb of metagenomics raw data was obtained by using the Illumina HiSeq. These gut data included genetic information from both host and microbial communities. After quality assessment and host data filtering, the gut microbial metagenomic datasets contained 1.17 Gb. The assembly and binning of profiles are shown in Supplementary Sheet S1. These filtering data were used to analyze the gut microbial diversity and functions in the host and had been uploaded to the NCBI's SRA database with accession number PRJNA751718.
3.2 Identification and community diversity of gut microbiota from Vespa mandarinia
Before taxonomic diversity analysis, rarefaction curves of species richness estimates reflected a saturated sampling (Figure 2A). The average Good's coverage was >95%, demonstrating that the sample data were enough for follow-up analyses (Supplementary Sheet S1). The open reading frames (ORFs) were analyzed and predicted from filtered contigs by MetaGeneMark software. On average, the larva had 155 unique ORFs per sample, the nymph had 1,397, and the adult had 1,260 (Supplementary Sheet S1). The taxonomic annotations of those reads were identified using the Non-Redundant protein sequence database (NR database). The species richness of gut bacteria from V. mandarinia was compared among different growth stages (Figure 2B). The results exhibited that the gut bacteria were almost the same (one-way ANOVA, LSD post-hoc test, p = 0.753 > 0.05) in the nymph and adult. However, the gut bacteria in larvae were quite distinctive compared with other stages (one-way ANOVA, LSD post-hoc test, p = 1.31e-08 < 0.01). The same trend was also observed in the Chao1, Ace richness estimators, Shannon, and Simpson diversity indexes (Supplementary Sheet S1).
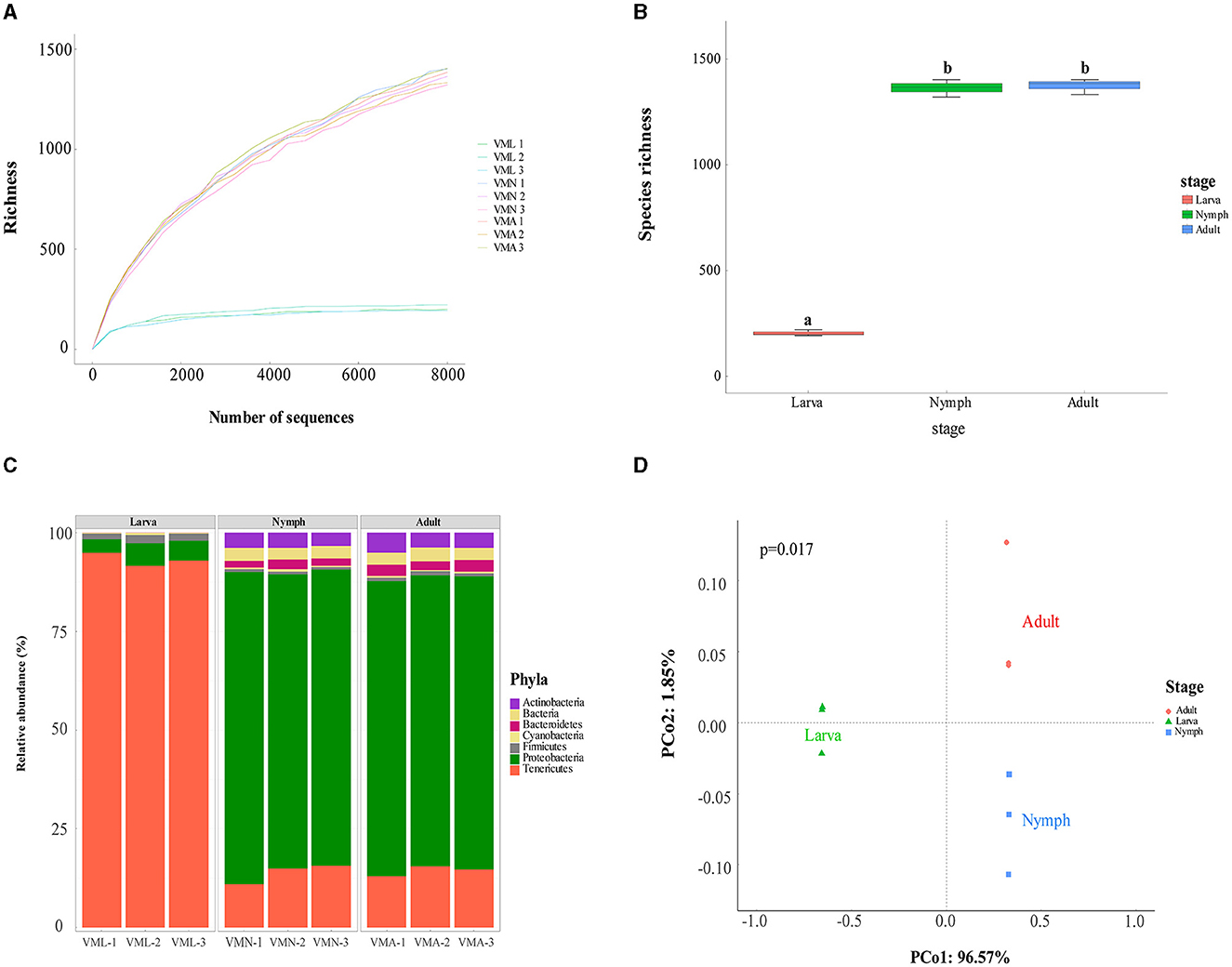
Figure 2. Gut bacterial community dynamics among Vespa mandarinia's three growth stages. VMA represents adult, VMN represents nymph, and VML represents larva. (A) The rarefaction curve of species richness, drawn based on the abundance of microbial communities in the samples represented (the sampling gradient was 100 sequences until the maximum depth). These legends were listed in a descending order of richness. (B) Boxplot of species richness determined by observed species. Letters above each host species indicate significant differences in the mean values (one-way ANOVA, LSD post-hoc test, p = 1.31e-08 < 0.01). (C) Relative abundance of microbial phyla in different samples. (D) PCoA plot based on Bray–Curtis distance for the effect of host development stages on taxonomic diversity (PERMANOVA test with 999 permutations, p = 0.017 < 0.05).
The taxonomic analysis revealed that there were seven phyla of microbes distributed in the gut of V. mandarinia, which were categorized into 9 classes, 14 orders, 16 families, 19 genera, and 30 species. At the phylum level (Figure 2C and Supplementary Sheet S2), the gut of larvae was occupied mainly by three types of microorganisms, namely, Tenericutes (93.14 ± 1.65%), Proteobacteria (4.76 ± 1.20%), and Firmicutes (1.78 ± 0.25%). However, in nymph and adult groups, the four high-ranking phyla were Proteobacteria (76.23 ± 2.61% in nymph and 74.19 ± 0.61% in adult), Tenericutes (13.84 ± 2.58% and 14.43 ± 1.29%), Actinobacteria (3.69 ± 0.28% and 4.21 ± 0.74%), and Bacteroidetes (1.95 ± 0.37% and 2.60 ± 0.35%). These data indicated that the dominant microbe in the gut of the wasp was rapidly replaced by Proteobacteria after pupation. These changes in the abundance of main gut microbes between different stages of V. mandarinia were statistically significant (one-way ANOVA, LSD post-hoc test, p(Tenericutes) = 3.558e-07 < 0.01, p(Proteobacteria) = 0.001052 < 0.01, p(Firmicutes) = 7.024e-07 < 0.01). PCoA ordination also suggested that larvae possess a distinct microbiota from other stages. In the scatter plot, the first two principal coordinates, PCO1 and PCO2, explained 96.57% and 1.85% of the data variation, clearly separating each group (Figure 2D).
A heatmap was prepared using the spearman index to compare the characteristics of the bacterial communities between groups at the genus level (Figure 3A and Supplementary Sheet S3). In the larval groups, the dominant taxa are o_Entomoplasmatales, Nitratireductor, and Christensenella. The nymph and adult share a similar microbial composition with larvae. The representative bacteria in the nymph and adult wasp mainly belonged to four genera, namely p_Proteobacteria, o_Entomoplasmatales, Escherichia, and Mycobacterium. The representative sequences of operational taxonomic units showed that larvae have different core strains of intestinal bacteria and contain fewer microbial species. A further analysis was conducted on the bacterial community composition among experimental samples at the species level (Figure 3B and Supplementary Sheet S4). The result revealed that the proportion of gut microbial species showed significant heterogeneity across different life stages. Although gene sequences can provide genetic information about microorganisms, they may not be sufficient to determine the species level. The aligned result would provide guidance for the subsequent functional analysis. In the larval group, the main microbe is associated with a bacterium called EntAcro10 and accounts for the highest proportion (Supplementary Sheet S4), which is also symbiotic with ants (Sapountzis et al., 2018), followed by Nitratireductor basaltis, Christensenella timonensis. In the adult and nymph groups, the main microbe is associated with Solemya velum gill symbiont, which accounts for half of gut microbes, followed by EntAcro10, Escherichia coli (Supplementary Sheet S4). The genetic sequences of the two dominant microbial species in the samples and their corresponding results in the NR database are shown in Supplementary Sheet S9.
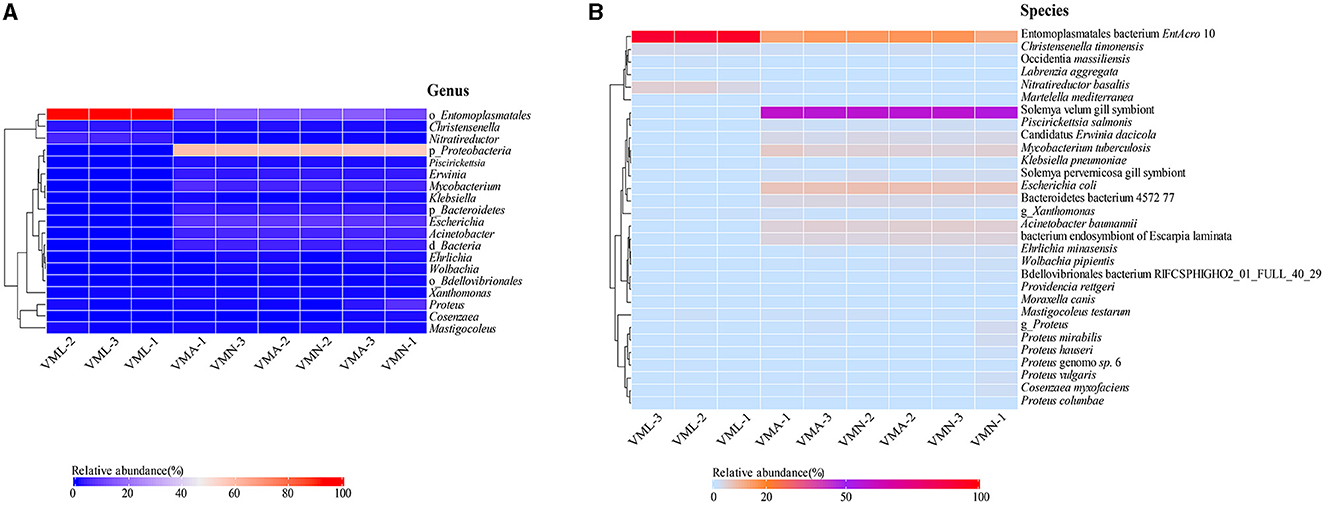
Figure 3. Heatmap of taxa over the Vespa mandarinia life cycle at the genera and species level. (A) Heatmap showing the relative abundance of genus taxa. (B) Heatmap showing the relative abundance of species taxa. Microbial cluster analysis was based on the Spearman distance with the complete-linkage method. The greater the species richness, the darker the color.
3.3 Functional analysis of gut microbes in Vespa mandarinia
Shotgun metagenomic profiles were frequently used to evaluate the functional diversity and potential of microbial communities (Sim et al., 2022). Accordingly, this work analyzed the capabilities of gut microbials from V. mandarinia and found their functional divergence with the host growth stage. By searching the KEGG database (Kanehisa et al., 2017), 3,369 genes were aligned from all gut samples, and 2,354 genes could be annotated into 6 categories according to the different functions (Supplementary Sheet S5). Significantly, metabolism was the primary function in larval gut microbes, accounting for 99.85 ± 0.04% of overall annotated sequences. The other functional homologs were present in a very small quantity compared to metabolism. Among adults and nymph groups, metabolism was also enriched, followed by human diseases, environmental information processing, cellular processes, genetic information processing, and organismal systems.
A further analysis of the 10 most abundant functions at metabolic level 3 was conducted (Supplementary Sheet S6). Oxidative phosphorylation had the highest functional richness in the larva, which was significantly differentiated from other stages (one-way ANOVA, LSD post-hoc test, p = 1.68e-08 < 0.01). Purine, pyrimidine, nicotinate, and nicotinamide metabolism were enriched in nymphs and adults. Beyond the aforementioned major roles, gut microbes in adult wasps exhibited a greater diversity of functions than larvae. The results of the functional alignments of KEGG pathways and the gene-pathway matching in the samples were presented (Supplementary Sheets S10, S11). For instance, drug metabolism (ko00982), the degradation of chloroalkane and chloroalkene (ko00625), and naphthalene (ko00626). The pathways involved in those main functions were matched on the KEGG database, and their expression in the host was compared (Figures 4A–D). Pathway ko00190 was associated with oxidative phosphorylation, ko00230 was associated with purine metabolism, ko00240 was associated with pyrimidine metabolism, and ko00760 was associated with nicotinate and nicotinamide metabolism.
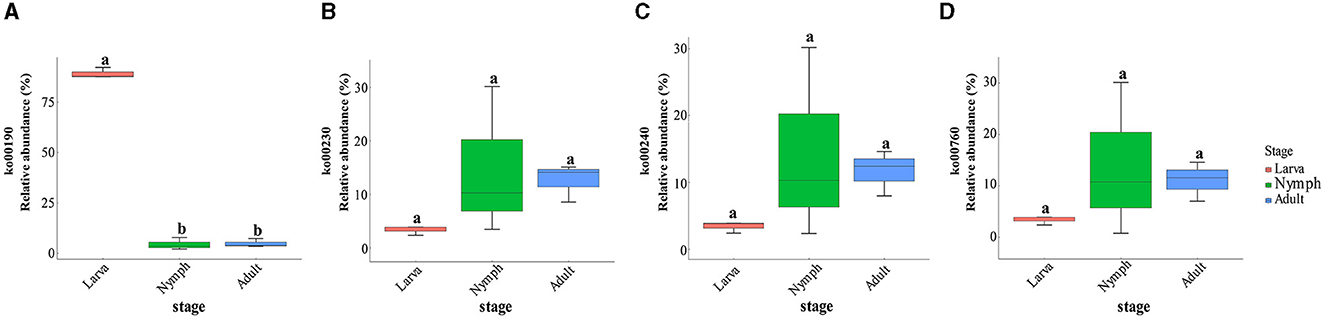
Figure 4. (A) Relative abundance and variation of ko00190 (one-way ANOVA, LSD post-hoc test, p = 2.58e-08 < 0.01). (B) Relative abundance and variation of ko00230 (one-way ANOVA, LSD post-hoc test, p = 0.281 > 0.05). (C) Relative abundance and variation of ko00240 (one-way ANOVA, LSD post-hoc test, p = 0.331 > 0.05). (D) Relative abundance and variation of ko00760 (one-way ANOVA, LSD post-hoc test, p = 0.387 > 0.05).
The EggNOG database is a hierarchical and phylogenetically annotated orthology resource that has collected numerous protein sequences from various organisms (Huerta-Cepas et al., 2016). Therefore, the metagenomic data from the gut microbiota of V. mandarinia were analyzed for functional classification through sequence alignment with the cluster of orthologous groups (COGs). In total, 631 COGs were annotated by function, which were assigned into 23 kinds of COG categories (Figure 5A and Supplementary Sheet S7). The top 10 abundant COGs are listed in Supplementary Sheet S8. Among them, COG1008 was the most abundant protein appearing in the larva and significantly differentiated from other stages (Figure 5B, p = 2.3e-07 < 0.01). COG1008 was ubiquinone oxidoreductase subunit 4 of chain M (NADH) and extensively involved in energy production and conversion. The other assigned COGs also contributed to the growth and development of larvae, such as COG0733 and COG5176. ENOG410Y65V was the most expressed in nymphs and adults and different from larval stages (Figure 5C). This COG plays significant functions in the retrotransposon protein. The content of other proteins with similar or related functions was also increased after pupation, including ENOG410XQYK and ENOG410XS82.
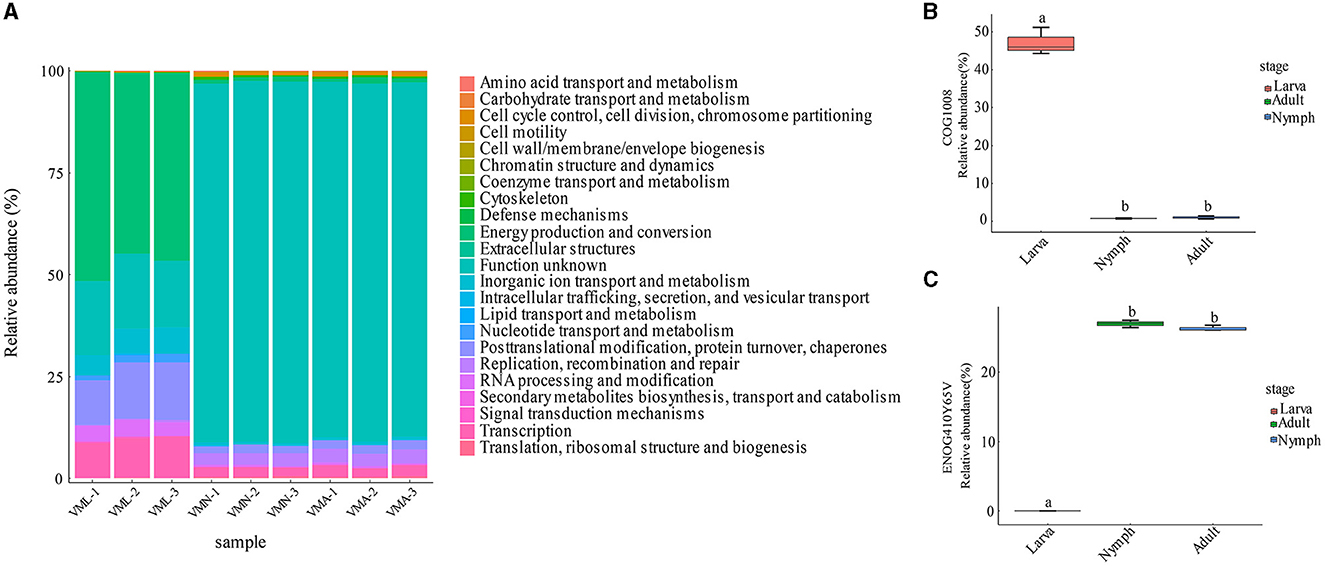
Figure 5. The characteristics of wasp gut microbial homologous genes based on the eggNOG database. (A) The functional proteins contained in the wasp gut microbes. (B) Relative abundance of the gene COG1008 in the wasp three stages. (C) Relative abundance of the gene ENOG410Y65V in the wasp three stages.
To sum up, the abundance and function distribution of gut microbial COGs were altered with host growth. Energy production and conversion were central categories in larval gut microbes. However, the gut microbes in adults and nymphs contained many predicted proteins with unknown functions. In addition to these unknown COGs, the preponderant functions are related to DNA replication, recombination, and repair. Following that is genic transcription and posttranslational modification of proteins.
4 Discussion
As an important economic insect in Yunnan Province (China), current cultivation technology of wasp cannot meet the increasing demands of food and medicinal usage. Some gut microorganisms contribute to the host's development by regulating the microenvironment of the gut (Zhang J. et al., 2022). To gain more knowledge about the role that gut microbiota play in the growth of wasps, a metagenomic analysis of the three developmental stages of V. mandarinia is conducted in this research. The results substantiated that the composition of gut microflora in V. mandarinia exhibits diversity. They are changing with the growth of the wasp to meet the needs of development and help their host adapt to a new survival environment.
In current experiments, the results of metagenomic sequencing indicated that Tenericutes, Proteobacteria, Firmicutes, Cyanobacteria, Actinobacteria, Bacteria, and Bacteroidetes are the main species of gut microbiota in V. mandarinia. Plenty of studies have shown that those microorganisms have a significant effect on the survival, behavior, oviposition, longevity, and larval development of host insects (Suenami et al., 2019). Moreover, the unique enzyme from insect gut microbiota can facilitate the breakdown of diversified food resources, and those versatile antimicrobial molecules produced by gut microbiota can help their host survive in harsh environments (Poulsen et al., 2011; Jaffar et al., 2022). The discovery of those promising leads with amazing structural architecture and various bioactivities from intestinal microbes have attracted great interest from chemical and pharmaceutical scientists (Um et al., 2016). As a consequence, in the context of the search for new antibiotics and therapeutic strategies from unique microbes in peculiar environments (Engel and Moran, 2013), this work reveals a previously unexplored isolation source from the insect gut.
Research has demonstrated that the composition of the gut microbiota is closely related to the food source of the host insect and their lifestyle (Xia et al., 2020). The gut microbiota communities of insects widely vary according to ecology. V. mandarinia is a social insect, effected by the natural ecological environment, and they can consume a wide variety of food types, such as plants and insects. Therefore, adult wasps are generally considered to be “feeding generalists.” There is a certain difference between the diets of larvae and adults. Adult wasps feed on nectar and other sugary substances, while larvae primarily consume protein-rich prey, which provides them with essential nutrients.
In this work, metagenomic data indicated that the gut of V. mandarinia was occupied mainly by two core microbial communities, and their abundance was changed with its cyclogeny. Tenericutes occupy a dominant percentage of larvae and was replaced by Proteobacteria after pupation. Previous studies have demonstrated that Tenericutes are directly associated with metabolism, and hosts with a high abundance of Tenericutes exhibit increased insulin sensitivity (Yuan et al., 2021). Consequently, their high concentration is helpful for the nutrient intake of larvae, and this phenomenon has been documented in other insects (Montalban et al., 2022). By comparison, the ability to use diversified food is an effective way to increase the survival rate for adult wasps. It is relatively easier for wasps to acquire food from plants than from animals. Reports have indicated that the degradation ability of Proteobacteria usually has a high degree of diversity; they can ferment carbohydrates (Delalibera et al., 2005), leading to the accumulation of extracellular enzymes for digesting food. The enzymes produced by Proteobacteria play important roles in the digestion of starches and hemicellulose. The aforementioned function can enhance the ability of adult wasps to utilize multiple foods. In addition, Proteobacteria can provide nutrients such as amino acids and nitrogen (Pinto-Tomás et al., 2009). Hence, the dominant position of Proteobacteria after pupation can increase the environmental adaptation of wasps (Zhou et al., 2020). The dominant species of larvae and adults of the wasp were associated with EntAcro10 and Solemya velum gill symbiont, respectively. EntAcro10 can utilize glycerol from ant host cells and monosaccharides (Sapountzis et al., 2018). Solemya velum gill symbiont is a sulfur-oxidizing chemosynthetic bacterium. The symbionts are contained within specialized gill cells, giving them access to electron acceptor oxygen in addition to sulfide and carbon dioxide for carbon fixation, fueling both the host and the symbionts (Russell et al., 2018). To sum up, as a special organ and huge gene resource for the host, the variable species richness of the gut microbiota is an adaptive regulatory mechanism of V. mandarinia. This mechanism can balance the food supply and nutritional requirements for development by adjusting the ratio of gut microbes with different metabolic capabilities.
Further KEGG analysis also confirms that metabolism is the predominant function of the gut microbes in V. mandarinia. In larvae, oxidative phosphorylation is the most abundant function, followed by nicotinate and nicotinamide metabolism, purine metabolism, and pyrimidine metabolism. The oxidative phosphorylation genes were expressed at a high level during the larval stage of V. mandarinia, which has been observed in other insects (Ren et al., 2022). Sufficient ATP was produced through oxidative phosphorylation for energy supply to promote the rapid growth of insect larvae (Bretscher and O'Connor, 2020). Entomological research has revealed that it only takes about 40 days for a wasp egg to grow into an adult wasp. Considering the limited time for growth and development of V. mandarinia and the scarcity of food resources, optimal conditions are necessary for the completion of their life cycle seasonally. The rapid and plentiful energy supply is essential for the survival of wasp larvae (Tan et al., 2015). Despite energy metabolism is still a core mission of the gut microbes after metamorphosis. Other diverse functions of intestinal microbials from adult wasp are also worth attention, especially the metabolic capacity of different substances, including drug metabolism, chloroalkane and chloroalkene degradation, and naphthalene degradation. The degradability of chloroalkene and naphthalene, which are also found in some insects, would enhance insect defense capacity (Zhang Q. et al., 2022). In Supplementary Sheet S6, we observed an increasing trend in the degradation abundance of chloroalkane, chloroalkene, and naphthalene with the lifecycle. Based on this observation, these functionalities would be acquired by wasp adults when foraging in a certain ecological environment. These capacities may generate unique secondary metabolites and help the adult wasp deal with complicated challenges from various environments.
The eggNOG analysis indicated that larval gut bacteria had abundant COG1008 gene, which was associated with the NADH respiratory complex and oxidative phosphorylation to produce ATP (Getz et al., 2018). Whereafter, with the downregulation of oxidative phosphorylation, retrotransposon proteins were significantly increased in the nymph and adult groups. The high expression of retrotransposons proteins has also been found in other wasps (Ye et al., 2022). The transposable elements may inevitably lead to genomic instability, including breaks and gene disruptions, causing cell death (Ayarpadikannan and Kim, 2014). The high expression and expansion of retranscription proteins in wasp would promote transposable element silencing and reduce cellular damage (Ye et al., 2022).
Retrotransposons and viruses are two major types of genetic elements; even some viruses show a common evolutionary origin with retrotransposons (Koonin and Krupovic, 2017). The symbiotic relationships between wasps and viruses have ancient origins, and some species have mutualistic viral relationships with viruses (Roossinck, 2011). Some wasps have formed phylogenetic symbioses with specific viruses, as revealed by viral metagenomes (Leigh et al., 2018). The typical representative of it is the wasp Diadromus pulchellus, which carries a virus that delays the replication of other viruses to protect the host (Sylvaine et al., 2005). Although the burst of retrotransposons has been found in many wasps, their reasons and roles are still not entirely revealed. This research found the high expression of retrotransposon proteins in gut microbes from V. mandarinia after pupation, providing new direction for further research.
5 Conclusion
This work enriched our knowledge about the gut microbiota during the multiple stages of V. mandarinia. Based on these data, three aspects of knowledge can be observed: First, the gut microbiota of V. mandarinia has a diverse composition; second, the abundance of gut microbes has changed significantly during metamorphosis; and third, the variation of gut microbes in different developmental stages represents an adaptive selection of V. mandarinia to utilize their specific functions for survival. Based on these findings, this work reveals the impact of gut microbiota on the survival capacity of V. mandarinia. V. mandarinia is a member of the Hymenoptera Vespidae. This work has the potential to contribute to the optimization of wasp cultivation techniques in order to meet food and health market demand. Additionally, this study sheds light on the symbiotic relationship between gut microbes and host and uncovers fewer microorganism resources from insect gut.
Data availability statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found at: https://www.ncbi.nlm.nih.gov/, PRJNA751718.
Author contributions
P-KY: Data curation, Formal analysis, Writing – original draft, Writing – review & editing. HX: Formal analysis, Writing – review & editing. Z-BY: Resources, Writing – review & editing. D-SY: Conceptualization, Funding acquisition, Project administration, Writing – review & editing. Y-HY: Conceptualization, Supervision, Writing – review & editing.
Funding
The author(s) declare that financial support was received for the research, authorship, and/or publication of this article. This research was funded by the Reserve Talents Project for Young and Middle-Aged Academic and Technical Leaders of Yunnan Province (Grant nos. 202105AC160062 and 202305AC160035), the Yunnan Fundamental Research Projects (Grant nos. 202101AT070031 and 202201AT070007), Team Project of Yunnan Revitalization Talent Support Program (202305AS350001), the National Natural Science Foundation of China (Grant no. 82060765), the Yunnan Provincial Universities Joint Special Project (Grant no. 202001BA070001-186), and the Special Program of Science and Technology of Yunnan Province (Grant no. 202002AA100007).
Acknowledgments
The authors acknowledge the institution for supplying specimens for analyses: samples from the insect sample room of the Key Laboratory of Entomological Biopharmaceutical R&D, Dali University.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher's note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2024.1288051/full#supplementary-material
Supplementary Sheet S1. Assembly and binning of metagenomic data of Vespa mandarinia gut.
Supplementary Sheet S2. The community and abundance of Vespa mandarinia gut microbes at the phylum level.
Supplementary Sheet S3. The community and abundance of Vespa mandarinia gut microbes at the genus level.
Supplementary Sheet S4. The community and abundance of Vespa mandarinia gut microbes at the species level.
Supplementary Sheet S5. The functional characters in the wasp's different growth stages based on KEGG level 1.
Supplementary Sheet S6. The functional characters in the wasp's different growth stages based on KEGG level 3.
Supplementary Sheet S7. The gut microbial functions of Vespa mandarinia based on the eggNOG database.
Supplementary Sheet S8. Those COG genes that ranked in the top ten in terms of abundance at different growth stages.
Supplementary Sheet S9. The alignment results of gene data from the two major microorganisms against the NR database.
Supplementary Sheet S10. The functional definition and functional links of the KEGG pathway in the samples.
Supplementary Sheet S11. The table of functional alignment between KEGG pathways and the genes in the samples.
References
Ayarpadikannan, S., and Kim, H. S. (2014). The impact of transposable elements in genome evolution and genetic instability and their implications in various diseases. Genomics Inform. 12, 98–104. doi: 10.5808/GI.2014.12.3.98
Bobay, L. M., Wissel, E. F., and Raymann, K. (2020). Strain structure and dynamics revealed by targeted deep sequencing of the honey bee gut microbiome. mSphere 5, e00694–e00620. doi: 10.1128/mSphere.00694-20
Bretscher, H., and O'Connor, M. B. (2020). The role of muscle in insect energy homeostasis. Front. Physiol. 11, 580687. doi: 10.3389/fphys.2020.580687
Cini, A., Meriggi, N., Bacci, G., Cappa, F., Vitali, F., Cavalieri, D., et al. (2020). Gut microbial composition in different castes and developmental stages of the invasive hornet Vespa velutina nigrithorax. Sci. Total Environ. 745, 140873. doi: 10.1016/j.scitotenv.2020.140873
Dedeine, F., Vavre, F., Fleury, F., Loppin, B., Hochberg, M. E., Bouletreau, M., et al. (2001). Removing symbiotic Wolbachia bacteria specifically inhibits oogenesis in a parasitic wasp. Proc. Natl. Acad. Sci. U. S. A. 98, 6247–6252. doi: 10.1073/pnas.101304298
Delalibera, I. Jr, Handelsman, J., and Raffa, K. F. (2005). Contrasts in cellulolytic activities of gut microorganisms between the wood borer, Saperda vestita (Coleoptera: Cerambycidae), and the bark beetles, Ips pini and Dendroctonus frontalis (Coleoptera: Curculionidae). Environ. Entomol. 34, 541–547. doi: 10.1603/0046-225X-34.3.541
Dimarcq, J. L., and Hunneyball, L. (2003). Pharma-entomology: when bugs become drugs. Drug Discov. Today 8, 107–110. doi: 10.1016/S1359-6446(02)02582-5
Dobermann, D., Swift, J. A., and Field, L. M. (2017). Opportunities and hurdles of edible insects for food and feed. Nutr. Bullet. 42, 293–308. doi: 10.1111/nbu.12291
Engel, P., and Moran, N. A. (2013). The gut microbiota of insects - diversity in structure and function. FEMS Microbiol. Rev. 37, 699–735. doi: 10.1111/1574-6976.12025
Feng, Y., Chen, X. M., Zhao, M., He, Z., Sun, L., Wang, C. Y., et al. (2018). Edible insects in china: utilization and prospects. Insect Sci. 25, 184–198. doi: 10.1111/1744-7917.12449
Gao, G., Xiong, H., Chen, K., Chen, J., Chen, P., Yu, C., et al. (2017). Gene expression profiling of ramie roots during hydroponic induction and adaption to aquatic environment. Genom. Data. 14, 32–35. doi: 10.1016/j.gdata.2017.08.002
Getz, E. W., Tithi, S. S., Zhang, L., and Aylward, F. O. (2018). Parallel evolution of genome streamlining and cellular bioenergetics across the marine radiation of a bacterial phylum. mBio 9, e01089–e01018. doi: 10.1128/mBio.01089-18
Guo, Y., Wang, L., Lu, J., Jiao, J., Yang, Y., Zhao, H., et al. (2021). Ginsenoside Rg1 improves cognitive capability and affects the microbiota of large intestine of tree shrew model for Alzheimer's disease. Mol. Med. Rep. 23, 291. doi: 10.3892/mmr.2021.11931
Holm, J. B., Humphrys, M. S., Robinson, C. K., Settles, M. L., Ott, S., Fu, L., et al. (2019). Ultrahigh-throughput multiplexing and sequencing of > 500-base-pair amplicon regions on the illumina hiseq 2500 platform. mSystems 4, e00029–e00019. doi: 10.1128/mSystems.00029-19
Huang, Z., Zeng, S., Xiong, J., Hou, D., Zhou, R., Xing, C., et al. (2020). Microecological Koch's postulates reveal that intestinal microbiota dysbiosis contributes to shrimp white feces syndrome. Microbiome 8, 32. doi: 10.1186/s40168-020-00802-3
Huerta-Cepas, J., Szklarczyk, D., Forslund, K., Cook, H., Heller, D., Walter, M. C., et al. (2016). EggNOG 4.5: a hierarchical orthology framework with improved functional annotations for eukaryotic, prokaryotic and viral sequences. Nucleic Acids Res. 44, D286–D293. doi: 10.1093/nar/gkv1248
Jaffar, S., Ahmad, S., and Lu, Y. (2022). Contribution of insect gut microbiota and their associated enzymes in insect physiology and biodegradation of pesticides. Front. Microbiol. 13, 979383. doi: 10.3389/fmicb.2022.979383
Jang, S., and Kikuchi, Y. (2020). Impact of the insect gut microbiota on ecology, evolution, and industry. Curr. Opin. Insect Sci. 41, 33–39. doi: 10.1016/j.cois.2020.06.004
Jeong, H., Kim, J. M., Kim, B., Nam, J. O., Hahn, D., Choi, M. B., et al. (2020). Nutritional value of the larvae of the alien invasive wasp vespa velutina nigrithorax and amino acid composition of the larval saliva. Foods 9, 885. doi: 10.3390/foods9070885
Kanehisa, M., Furumichi, M., Tanabe, M., Sato, Y., and Morishima, K. (2017). KEGG: new perspectives on genomes, pathways, diseases and drugs. Nucleic Acids Res. 45, D353–D361. doi: 10.1093/nar/gkw1092
Koonin, E. V., and Krupovic, M. (2017). Polintons, virophages and transpovirons: a tangled web linking viruses, transposons and immunity. Curr. Opin. Virol. 25, 7–15. doi: 10.1016/j.coviro.2017.06.008
Kroiss, J., Kaltenpoth, M., Schneider, B., Schwinger, M. G., Hertweck, C., Maddula, R. K., et al. (2010). Symbiotic streptomycetes provide antibiotic combination prophylaxis for wasp offspring. Nat. Chem. Biol. 6, 261–263. doi: 10.1038/nchembio.331
Leigh, B. A., Bordenstein, S. R., Brooks, A. W., Mikaelyan, A., and Bordenstein, S. R. (2018). Finer-scale phylosymbiosis: insights from insect viromes. mSystems 3, e00131–e00118. doi: 10.1128/mSystems.00131-18
Miller, S. E., Legan, A. W., Uy, F. M. K., and Sheehan, M. J. (2022). Highly contiguous genome assemblies of the Guinea paper wasp (Polistes exclamans) and Mischocyttarus mexicanus. Genome Biol. Evol. 14, evac110. doi: 10.1093/gbe/evac110
Montalban, A., Sanchez, C. J., Hernandez, F., Schiavone, A., Madrid, J., Martinez-Miro, S., et al. (2022). Effects of agro-industrial byproduct-based diets on the growth performance, digestibility, nutritional and microbiota composition of mealworm (Tenebrio molitor L.). Insects 13, 323. doi: 10.3390/insects13040323
Nayfach, S., and Pollard, K. S. (2016). Toward accurate and quantitative comparative metagenomics. Cell. 166, 1103–1116. doi: 10.1016/j.cell.2016.08.007
Oliva, A., Tobler, R., Cooper, A., Llamas, B., and Souilmi, Y. (2021). Systematic benchmark of ancient DNA read mapping. Brief. Bioinform. 22, 1–12. doi: 10.1093/bib/bbab076
Pinto-Tomás, A. A., Anderson, M. A., Suen, G., Stevenson, D. M., Chu, F. S. T., Cleland, W. W., et al. (2009). Symbiotic nitrogen fixation in the fungus gardens of leaf-cutter ants. Science 326, 1120–1123. doi: 10.1126/science.1173036
Poulsen, M., Oh, D. C., Clardy, J., and Currie, C. R. (2011). Chemical analyses of wasp-associated streptomyces bacteria reveal a prolific potential for natural products discovery. PLoS ONE 6, e16763. doi: 10.1371/journal.pone.0016763
Qian, T., Chen, C., Li, C., Gong, Q., Liu, K., Wang, G., et al. (2021). A novel 4.25 kb heterozygous deletion in PAX6 in a Chinese Han family with congenital aniridia combined with cataract and nystagmus. BMC Ophthalmol. 21, 353. doi: 10.1186/s12886-021-02120-0
Ren, L., Shang, Y., Zhang, X., Chen, S., Heng, Y., Zou, Y., et al. (2022). Temporal expression profiles reveal potential targets during postembryonic development of forensically important sarcophaga peregrina (Diptera: Sarcophagidae). Insects 13, 453. doi: 10.3390/insects13050453
Roossinck, M. J. (2011). The good viruses: viral mutualistic symbioses. Nat. Rev. Microbiol. 9, 99–108. doi: 10.1038/nrmicro2491
Russell, S. L., McCartney, E., and Cavanaugh, C. M. (2018). Transmission strategies in a chemosynthetic symbiosis: detection and quantification of symbionts in host tissues and their environment. Proc. Biol. Sci. 285, 20182157. doi: 10.1098/rspb.2018.2157
Sapountzis, P., Zhukova, M., Shik, J. Z., Schiott, M., and Boomsma, J. J. (2018). Reconstructing the functions of endosymbiotic Mollicutes in fungus-growing ants. Elife 7, e39209. doi: 10.7554/eLife.39209
Schmidt, K., and Engel, P. (2021). Mechanisms underlying gut microbiota-host interactions in insects. J. Exp. Biol. 224, jeb207696. doi: 10.1242/jeb.207696
Shukla, S. P., Sanders, J. G., Byrne, M. J., and Pierce, N. E. (2016). Gut microbiota of dung beetles correspond to dietary specializations of adults and larvae. Mol. Ecol. 25, 6092–6106. doi: 10.1111/mec.13901
Sim, M., Lee, J., Wy, S., Park, N., Lee, D., Kwon, D., et al. (2022). Generation and application of pseudo-long reads for metagenome assembly. Gigascience 11, giac044. doi: 10.1093/gigascience/giac044
Suenami, S., Nobu, M. K., and Miyazaki, R. (2019). Community analysis of gut microbiota in hornets, the largest eusocial wasps, Vespa mandarinia and V. simillima. Sci. Rep. 9, 9830. doi: 10.1038/s41598-019-46388-1
Sylvaine, R., Stasiak, K., Federici, B., and Bigot, Y. (2005). Commensal and mutualistic relationships of reoviruses with their parasitoid wasp hosts. J. Insect Physiol. 51, 137–148. doi: 10.1016/j.jinsphys.2004.08.002
Tan, J. L., Achterberg, C. V., and Chen, X. X. (2015). Potentially lethal social wasps, fauna of the Chinese Vespidae (Hymenoptera: Vespidae). Beijing: Science Press.
Um, S., Bach, D. H., Shin, B., Ahn, C. H., Kim, S. H., Bang, H. S., et al. (2016). Naphthoquinone-oxindole alkaloids, coprisidins a and b, from a gut-associated bacterium in the dung beetle, copris tripartitus. Org. Lett. 18, 5792–5795. doi: 10.1021/acs.orglett.6b02555
Wang, S., Wang, L., Fan, X., Yu, C., Feng, L., Yi, L., et al. (2020). An insight into diversity and functionalities of gut microbiota in insects. Curr. Microbiol. 77, 1976–1986. doi: 10.1007/s00284-020-02084-2
Wu, W., Lei, J., Hussain, M., Cao, S., Du, B., Wang, R., et al. (2019). Structure and function of the fruit microbiome in healthy and diseased kiwifruit. Pakistan J. Agric. Sci. 56, 577–585. doi: 10.21162/PAKJAS/19.8820
Xia, X., Lan, B., Tao, X., Lin, J., and You, M. (2020). Characterization of Spodoptera litura gut bacteria and their role in feeding and growth of the host. Front. Microbiol. 11, 1492. doi: 10.3389/fmicb.2020.01492
Xie, C., Mao, X., Huang, J., Ding, Y., Wu, J., Dong, S., et al. (2011). KOBAS 2.0: a web server for annotation and identification of enriched pathways and diseases. Nucleic Acids. Res.39, W316–322. doi: 10.1093/nar/gkr483
Ye, X., Yang, Y., Zhao, C., Xiao, S., Sun, Y. H., He, C., et al. (2022). Genomic signatures associated with maintenance of genome stability and venom turnover in two parasitoid wasps. Nat. Commun. 13, 6417. doi: 10.1038/s41467-022-34202-y
Yuan, X., Chen, R., McCormick, K. L., Zhang, Y., Lin, X., Yang, X., et al. (2021). The role of the gut microbiota on the metabolic status of obese children. Microb. Cell Fact 20, 53. doi: 10.1186/s12934-021-01548-9
Zhang, J., Gao, S., Zheng, F., and Wang, N. (2022). Intestinal bacterial diversity and functional analysis of three lepidopteran corn ear worm larvae. Insects 13, 740. doi: 10.3390/insects13080740
Zhang, Q., Wang, S., Zhang, X., Zhang, K., Li, Y., Yin, Y., et al. (2022). Beneficial bacteria in the intestines of housefly larvae promote larval development and humoral phenoloxidase activity, while harmful bacteria do the opposite. Front. Immunol. 13, 938972. doi: 10.3389/fimmu.2022.938972
Zhou, S. T., Luan, K., Ni, L. L., Wang, Y., Yuan, S. M., Che, Y. H., et al. (2019). A strategy for quality control of Vespa Magnifica (smith) venom based on hplc fingerprint analysis and multi-component separation combined with quantitative analysis. Molecules 24, 2920. doi: 10.3390/molecules24162920
Keywords: Vespa mandarinia, gut microbiota, metagenomics, WASP, metamorphosis, metabolism
Citation: Yin P-K, Xiao H, Yang Z-B, Yang D-S and Yang Y-H (2024) Shotgun metagenomics reveals the gut microbial diversity and functions in Vespa mandarinia (Hymenoptera: Vespidae) at multiple life stages. Front. Microbiol. 15:1288051. doi: 10.3389/fmicb.2024.1288051
Received: 03 September 2023; Accepted: 12 February 2024;
Published: 11 March 2024.
Edited by:
Panagiota Stathopoulou, University of Patras, GreeceReviewed by:
Tomas Erban, Crop Research Institute (CRI), CzechiaBo Chen, Zhejiang University, China
Copyright © 2024 Yin, Xiao, Yang, Yang and Yang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Yin-He Yang, eXloODYxMkAxMjYuY29t; Da-Song Yang, eWFuZ2RzQGNwdS5lZHUuY24=
 Peng-Kai Yin
Peng-Kai Yin Huai Xiao1,2
Huai Xiao1,2 Zhi-Bin Yang
Zhi-Bin Yang Da-Song Yang
Da-Song Yang Yin-He Yang
Yin-He Yang