
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Microbiol., 04 January 2024
Sec. Microbiotechnology
Volume 14 - 2023 | https://doi.org/10.3389/fmicb.2023.1333777
Introduction: Furfural, a main inhibitor produced during pretreatment of lignocellulose, has shown inhibitory effects on S. cerevisiae.
Method: In the present study, new strains named 12–1 with enhanced resistance to furfural were obtained through adaptive laboratory evolution, which exhibited a shortened lag phase by 36 h, and an increased ethanol conversion rate by 6.67% under 4 g/L furfural.
Results and Discussion: To further explore the mechanism of enhanced furfural tolerance, ADR1_1802 mutant was constructed by CRISPR/Cas9 technology, based on whole genome re-sequencing data. The results indicated that the time when ADR1_1802 begin to grow was shortened by 20 h compared with reference strain (S. cerevisiae CEN.PK113-5D) when furfural was 4 g/L. Additionally, the transcription levels of GRE2 and ADH6 in ADR1_ 1802 mutant were increased by 53.69 and 44.95%, respectively, according to real-time fluorescence quantitative PCR analysis. These findings suggest that the enhanced furfural tolerance of mutant is due to accelerated furfural degradation. Importance: Renewable carbon worldwide is vital to achieve “zero carbon” target. Bioethanol obtained from biomass is one of them. To make bioethanol price competitive to fossil fuel, higher ethanol yield is necessary, therefore, monosaccharide produced during biomass pretreatment should be effectively converted to ethanol by Saccharomyces cerevisiae. However, inhibitors formed by glucose or xylose oxidation could make ethanol yield lower. Thus, inhibitor tolerant Saccharomyces cerevisiae is important to this process. As one of the main component of pretreatment hydrolysate, furfural shows obvious impact on growth and ethanol production of Saccharomyces cerevisiae. To get furfural tolerant Saccharomyces cerevisiae and find the underlying mechanism, adaptive laboratory evolution and CRISPR/Cas9 technology were applied in the present study
The wide development and application of lignocellulosic materials in biofuel industry represent a pivotal approach to provide sufficient alternative fuels for fossile fuels (Lin and Lu, 2021). Nevertheless, pretreatment before fermentation is a necessity to enhance hexose and pentose yield from lignocellulose during enzymatic hydrolysis process, as it generates a substantial amount of by-products simultaneously that severely hinder the growth and metabolism of microorganisms (Rabemanolontsoa and Saka, 2016). The content and type of generated inhibitors are closely related with applied pretreatment method. During dilute acid pretreatment, most of hemicellulose is dissolved to generate hexose and pentose, which are degraded further to form inhibitors for microorganism, such as 5-HMF, furfural, formic acid and levulinic acid. The concentrations of these inhibitors are enhanced with increased concentration of applied acid. After alkali pretreatment, phenolic inhibitors in the hydrolysate are significantly increased, of which ferulic acid shows strong cytotoxic effect on cells (Alves et al., 2020). The content of inhibitors varies for different lignocellulose hydrolysates. The acetic acid concentration in rice straw and softwood hydrolysate ranges from 0.49 to 6.5 g/L. The content of furfural, 5-HMF and vanillin after hydrothermal pretreatment is 0.2–2.0, 0.1–1.0, and 0.5–2.0 g/L, respectively (He et al., 2016).
Liu et al. investigated the impact of various inhibitors derived from biomass on the growth of Rhodosporon, including furfural, 5-hydroxymethylfurfural, acetic acid, vanillic acid, and vanillin. The results demonstrated that furfural showed the strongest inhibitory effect, followed by vanillin and 5-HMF (Liu et al., 2021). Moreover, it has been proven that aldehydes can be detoxified by Saccharomyces cerevisiae in situ to corresponding alcohols. This reduction reaction is catalyzed by NAD (P)H-dependent aldehyde reductase (Wang et al., 2018). Saccharomyces cerevisiae can convert 100% HMF to furfural at a concentration of 60 mM, and 100% furfural to furfuryl alcohol at 30 mM (Liu et al., 2005).
Therefore, enhancing the tolerance of microorganisms to inhibitors is a crucial approach. Recently, a strain improvement strategy called “adaptive laboratory evolution” (ALE) has gained widespread application (Wu et al., 2022). Adaptive laboratory evolution is the process of obtaining new microbial phenotypes through natural selection. Compared with other technologies, ALE is not restricted by physiological mechanisms behind microbial phenotypes to achieve improvement. The combination of ALE and DNA sequencing technology has been widely applied to identify key gene mutations (Sandberg et al., 2019). But it requires a longer time for forced selection of specific phenotypes, and negative mutations may also occur (Hemansi et al., 2022). Through adaptive laboratory evolution, a robust mutant with enhanced inhibitor tolerance for Kluyveromyces marxianus 1727 was obtained in mixed inhibitors with increased concentrations by Du and his coworkers. The mutant showed significantly increased uptake rates of acetate, formate, furfural, and vanillin (Du et al., 2022). Earlier studies revealed that the ethanol yield of Zymomonas mobilis was severely inhibited by the phenolic aldehyde produced by lignocellulosic pretreatment. But following a process of laboratory adaptive evolution, the obtained Zymomonas mobilis Z198 showed a significant increase in the conversion rate of vanillin by 6.3 times, and an increase in ethanol production by 21.6% (Yan et al., 2021).
Researchers tried to obtain enhanced stress tolerant strains for ethanol fermentation, but the underlying mechanism are still not fully understood. Several genes related to furfural tolerance of yeast were identified already, including SFP1, sigI, araR, YAP1, GSH1, GLR1, YNL134C, YML131W, ADH6 (Petersson et al., 2006; Kim and Hahn, 2013; Li et al., 2014; Zhao et al., 2015; Chen et al., 2016; Alves et al., 2020). ALE can improve the robustness of microorganisms to specific selection pressures, and can be useful to find genes responsible for further metabolic engineering (Jin et al., 2019).
Characteristics of Saccharomyces cerevisiae includes low fermentation cost, high ethanol production, high safety, and is widely applied in industrial fermentation (Yu and Zhang, 2004). Microorganism with higher tolerance, higher ethanol production, and clear genetic background is a better choice for exploration of genes related to furfural tolerance enhancement and underlying mechanisms. A deeper understanding of the furfural tolerance mechanism of yeast can reduce the production cost of ethanol (Sharma et al., 2022). In this study, ALE was first applied to obtain genetically stable and resistant strains to furfural stress, followed by complete genome sequencing and comparative genomic analysis to identify changes in genes related to enhanced furfural stress. Furthermore, a mutant strain was constructed using CRISPR/Cas9 to explore the potential mechanism of increased furfural tolerance. This study has shed light on the molecular mechanisms related to furfural tolerance in Saccharomyces cerevisiae through genome sequencing and reverse engineering.
Saccharomyces cerevisiae CEN.PK113-7D, S. cerevisiae CEN.PK113-5D, S. cerevisiae 12–1 (obtained after ALE) were maintained in slants. Escherichia coli JM110 was applied for plasmid construction.
Precultures were grown overnight at 30°C and 200 rpm in YEPD until the exponential phase (about 12 h).
The wild type S. cerevisiae CEN.PK113-7D strain was subjected to serial batch culture in YEPD with increased furfural stress (from 1 to 3.8 g/L), as shown in Figure 1. 1 g/L furfural was the initial stress concentration. When the yeast grows to 80% of the maximum OD600 value, inoculation is carried out. The culture is transferred to fresh 50 mL medium with OD600 of 0.5. The furfural concentration is increased by 0.2 g/L each time, the passage is repeated three times in each cycle. After more than 20 rounds of passage in 98 days, concentration of furfural reached 3.8 g/L finally. After 98 days of adaptive laboratory evolution, the culture medium was spread on YEPD medium containing 1 g/L furfural for preliminary screening.
The concentration of biomass, glucose, ethanol and furfural was same as previously published studies (Dai et al., 2020; Jia et al., 2022).
0, 0.5, 0.8, 1, and 1.5 g/L of furfural were added to the YEPD culture medium that had just been sterilized. The OD600 of wild-type strain and adapted strain was diluted to 0.6, which then was diluted to 10−1, 10−2, 10−3. Take 1 μL of each concentration and place them in the corresponding marked area of the plate and kept in a 30°C incubator for 60 h.
The Saccharomyces cerevisiae CEN.PK113-7D and Saccharomyces cerevisiae 12–1 were cultivated overnight in YEPD medium. The yeast cells were obtained after centrifuge at 12,000 rpm for 1 min. Omega Fungal DNA Kit was applied for DNA extraction (D3390-02, Tiangen Biochemical Technology Co., Ltd.). Quantitative purification was performed using a TBS-380 fluorometer (Turner BioSystems Inc., Sunnyvale, CA). Then, the DNA was sent to Shanghai Majorbio Biopharm Biotechnology Co., Ltd. for sequencing.
The analysis process was referred to published literatures (Xia et al., 2022).
CRISPR/Cas9 technology was applied to construct a mutated ADR1 based on pML104 plasmid, the process was shown in Supplementary Figure S1. Primers used in the construction of strains were listed in Table 1.
Ten microliter preculture of E.coli JM110 containing pML104 plasmid (stored in our laboratory) was incubated at 37°C and 200 rpm for 12–16 h in LB medium containing 100 μg/mL ampicillin sodium. pML104 plasmid was extracted following the FastPure@ Plasma Mini Kit instructions (Nanjing Novozan Biotechnology Co., Ltd.). Then linearized pML104 plasmid was constructed for the cloning of sgRNA.
gRNA was designed near the mutation site, which was then hybridized as described previously (Xia et al., 2022). Then it was directly ligated into linearized pML104 plasmids. The recombinant plasmid pML104-adr1gRNA was obtained and transformed to select positive transformants. Insertion validation of gRNA into the pML104 plasmid was confirmed using T3 primers.
Upstream (825 bp) and downstream (612 bp) homologous arms of ADR1 were obtained by PCR amplification of ADR1-UF/ADR1-UR and ADRI-DF/ADR1-DR from Saccharomyces cerevisiae CEN.PK113-5D genomic DNA. After that, a donor fragment contains a mutation in the ADRI gene at position 1802 (A changes to G) was obtained. After transformation, PCR was used to identify positive colonies were tested by PCR, which was confirmed by sequencing (Laughery et al., 2015).
Take 1 mL of fermentation broth, centrifuge at 12,000 r/min for 5 min to get the cells, which then was washed 3 times with deionized water. The ratio of 104 cells to the volume of the extraction solution (mL) was 500–1,000:1. Ultrasound (power 20%, ultrasound for 3 s, interval of 10 s, repeat 30 times) was applied to get the cell lysate, and the supernatant for detection of catalase activity and superoxide dismutase was obtained after centrifuge at 4°C for 10 min at 8,000 r/min.
Take 1 mL of fermentation broth, centrifuge at 12,000 r/min for 10 min to get the cells, which then was washed 3 times with deionized water. After adding 1 mL of ultrapure water and an appropriate amount of glass beads, the disrupted cells were obtained after shaking and crushing with a vortex meter for 20 min. The supernatant obtained after centrifuge at 12,000 r/min for 10 min was applied for intracellular glycerol content determination.
The concentration of glycerol is detected by a glycerol detection kit (E1002, Beijing Pulilai Gene Technology Co., Ltd.). Catalase activity was measured by a catalase detection kit (A007-1-1, Nanjing Jiancheng Biotechnology Research Institute). Superoxide dismutase was measured by a superoxide dismutase detection kit (A001-3-1, Nanjing Jiancheng Biotechnology Research Institute).
S. cerevisiae CEN.PK113-5D and ADR1_ 1802 were incubated at 30°C at 200 rpm. When the OD600 reached approximately 1.0, 4 g/L of furfural was added to the medium, and the time was defined as 0 h. After incubation for 2 h, the cells were collected by centrifuging at 4,000 r/min at room temperature for 2 min. Cells obtained from cultures free of furfural at 0 and 2 h were taken as controls. The primers used for qRT-PCR analysis were listed in Table 2. Total RNA was isolated according to previously described protocol (Liu and Slininger, 2007), and then purified using the RNA clean kit (Tiangen Biotechnology Co., Ltd., Beijing, China). The reverse transcription reaction was carried out using the procedure described by Lewis et al. (2009). HiScript II QRT SuperMix (Norwich Biotechnology Co., Ltd.) and ChamQ universal SYBR qPCR Master Mix (Norwich Biotechnology Co., Ltd.) were applied for qRT-PCR. The qRT-PCR data were analyzed using the method described by Liu et al. (2009). The reference gene used for quantitative PCR is ENO1.
OriginPro 2019b software (Northampton, MA, United States) was used for diagrams drawing. GraphPad Prism 8 and Statistica 23.0 software (SPSS Inc., Chicago, IL, United States) were applied for statistical analysis. All experiments were conducted with triplicate.
The Adaptive Laboratory Evolution (ALE) strategy is used to obtain robust Saccharomyces cerevisiae cells that can tolerate high concentrations of furfural inhibitors. S. cerevisiae CEN PK 113-7D is widely used in metabolic engineering and systems biology research, and it shows excellent growth characteristics in industrial production. It is widely used in metabolic and evolutionary engineering research, such as fermentation of pentose, the production of ethanol, lactic acid and pyruvate (Nijkamp et al., 2012). Thus, it was applied in the present study as the starting strain. Prior to ALE, it is important to determine the appropriate initial furfural concentration that will inhibit yeast cells to a certain extent, but not severely damage yeast cells. Our preliminary results showed that 1 g/L of furfural was determined as the initial concentration for evolution.
The endpoint concentration of furfural reached 3.8 g/L. A total of 18 colonies were isolated, which were applied for further analysis of furfural tolerance. As shown in Supplementary Figure S3, under the stress of 4 g/L furfural, the cells of wild-type strains were elongated larger, flattened, irregular. The surface was unsmooth, and vacuoles could be observed. While cells of mutant strains were regular, round, plump. The surface was smooth, and a small amount of sprouting could be observed.
By comparing the differences in tolerance to furfural stress between the adapted strain and the wild-type strain, it was found that the starting strain did not grow on a YEPD solid plate containing 1.5 g/L furfural, while the evolutionary strain 12–1 could (Supplementary Figure S4). The results showed 12–1 strain showed the best performance under 4.0 g/L of furfural stress (Supplementary Figure S5). Compared with the original strain (Supplementary Figure S6; Supplementary Table S1), under 4.0 g/L of furfural treatment, the time to get to exponential phase was reduced by 36 h, and 7.02% more ethanol was obtained (Figure 2 and Table 3). The furfural was completely degraded after 48 h, indicating that the time required to degrade furfural was significantly shortened. Furthermore, the specific growth rate of 12–1 was 32.43% higher than that of S. cerevisiae CEN.PK113-7D, and the ethanol conversion rate was 6.67% higher. 1 g/L of furfural could reduce ethanol productivity of 12–1 strain obviously. 4 g/L of furfural could decrease ethanol productivity by 82%, indicating ethanol productivity was significantly decreased with the increase of furfural concentration. The reason was that during the lag phage, yeast cells needs to convert furfural into less toxic furfuryl alcohol, hardly produces ethanol, resulting in a decrease in ethanol productivity.
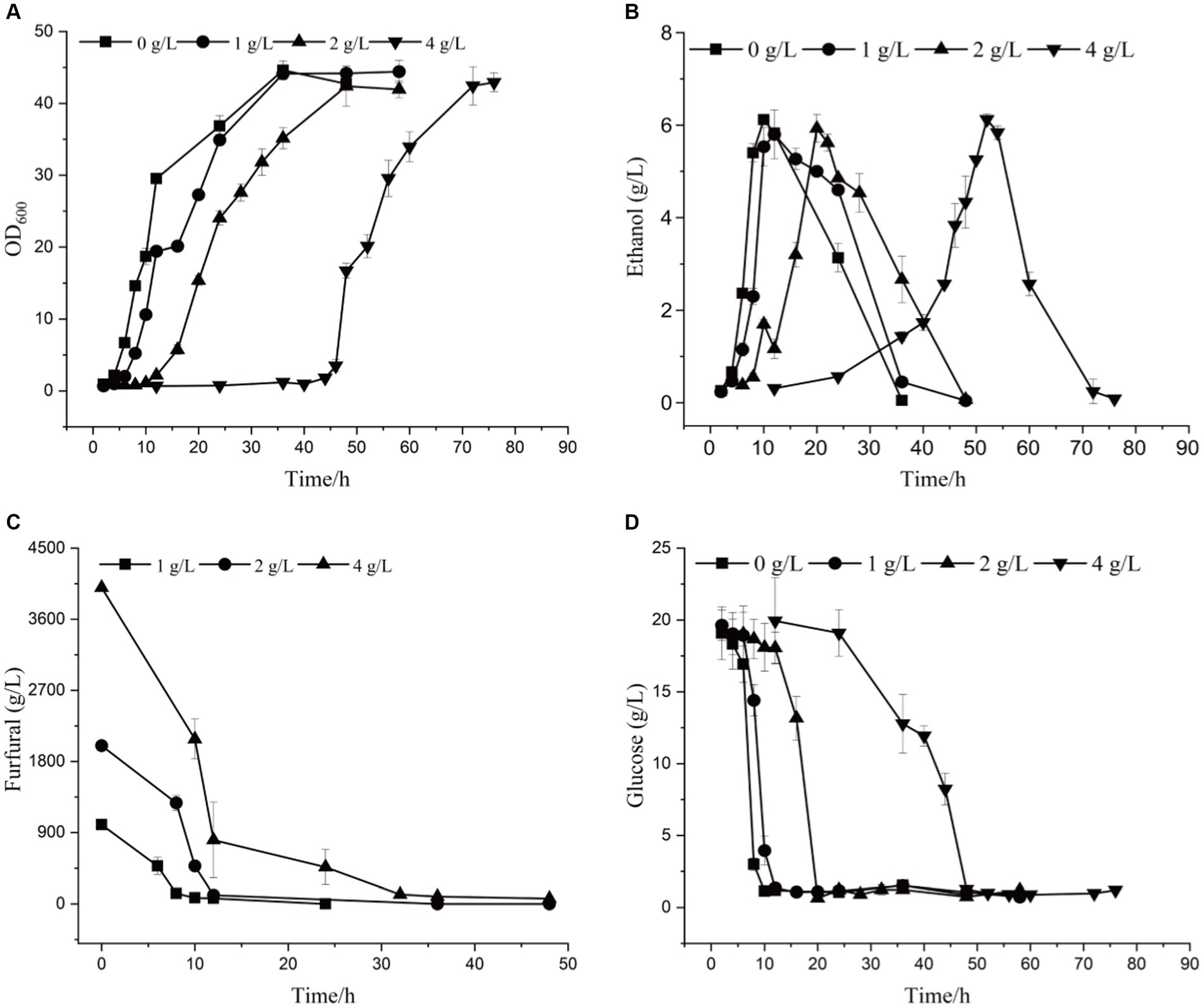
Figure 2. The growth curve (A), ethanol yield (B), furfural concentration (C), and glucose content (D) of 12–1 strain under different furfural stress.
In order to explore the changes in genes that enhance tolerance of 12–1 strain to furfural stress, the entire genome sequencing of 12–1 and CEN.PK113-7D strain were sequenced. A total of 1,219 mutations were obtained, including 403 single nucleotide insertion or deletion mutations (Indel) and 816 single nucleotide polymorphisms (SNP) (Figures 3A,B). It was suggested that the adaptation of Saccharomyces cerevisiae CEN.PK113-7D to furfural stress may be closely related to gene expression regulation. In addition, Saccharomyces cerevisiae can also adapt to furfural stress by changing the function of some proteins to promote their degradation (Zhao et al., 2015). The specific mutation sites of each mutanted gene were shown in Figure 3C. The genes with missense mutations were UTH1, RET1, HDA1, HYP2, ADR1, TFC1, and COS8, and the genes with frameshift mutations were PAU20 and CDC14, respectively. In summary, strain 12–1 may enhance the adaptation to furfural stress by prolonging cell life, affecting autophagy, changing the structure and function of cell walls, and improving the viability of cells.
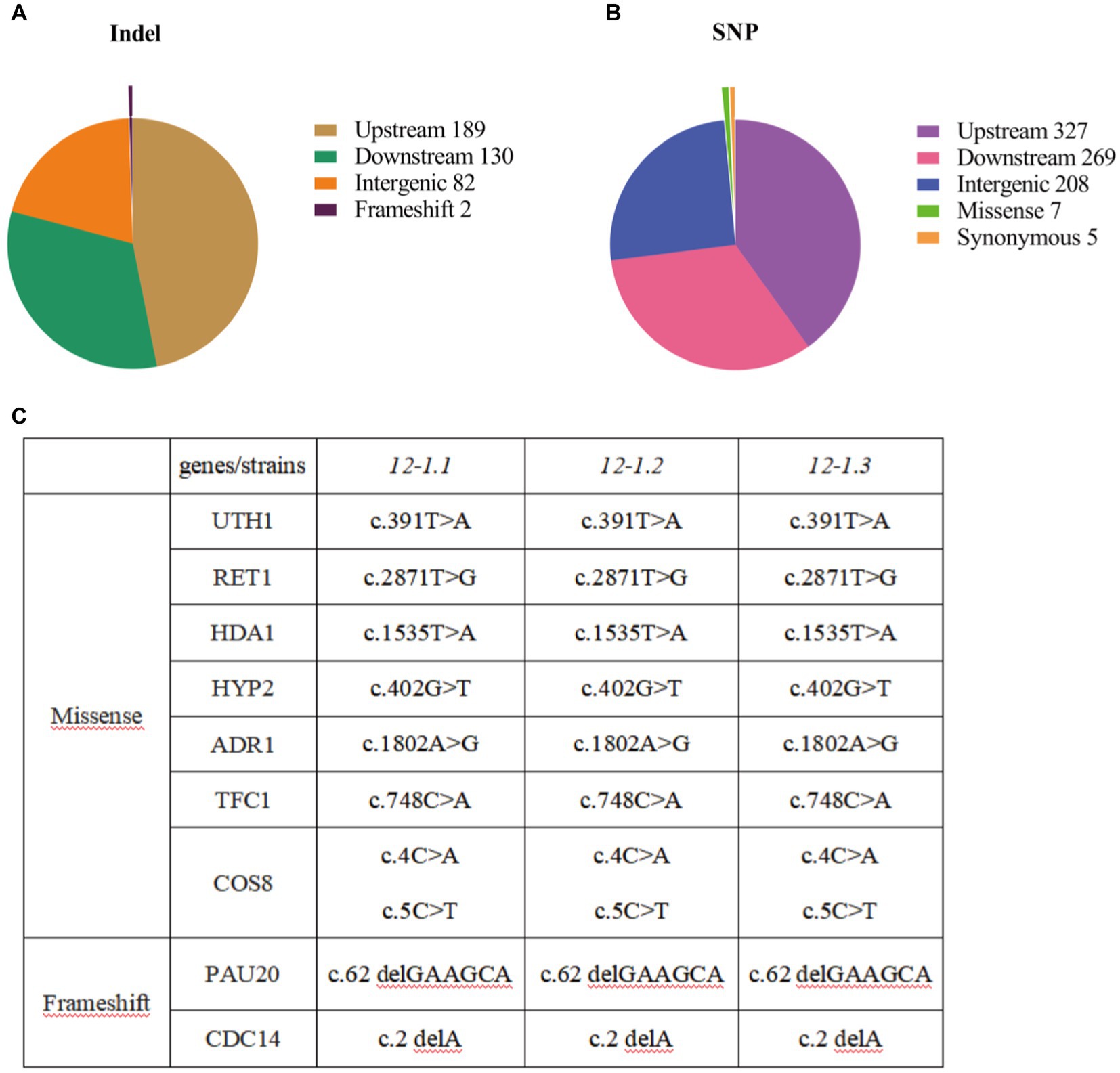
Figure 3. Results of whole genome sequencing. (A) Statistical results of single nucleotide insertion or deletion mutations in whole genome sequencing. (B) Statistical results of single nucleotide mutations in whole genome sequencing. (C) Statistical results of mutation types and mutation genes in whole genome sequencing.
Based on the whole genome sequencing results, the ADR1 gene mutation was constructed to obtain the engineered strain ADR1_ 1802, to explore the effect of ADR1 mutation on the growth and fermentation of Saccharomyces cerevisiae under furfural stress. Effects of different concentrations of furfural on the growth and ethanol production of ADR1_1802 mutant were determined, as shown in Figure 4, Supplementary Figure S7, and Table 4. Compared with the reference strain (S. cerevisiae CEN. PK113-5D), under 4 g/L furfural treatment, the lag phase was shortened by 20 h.
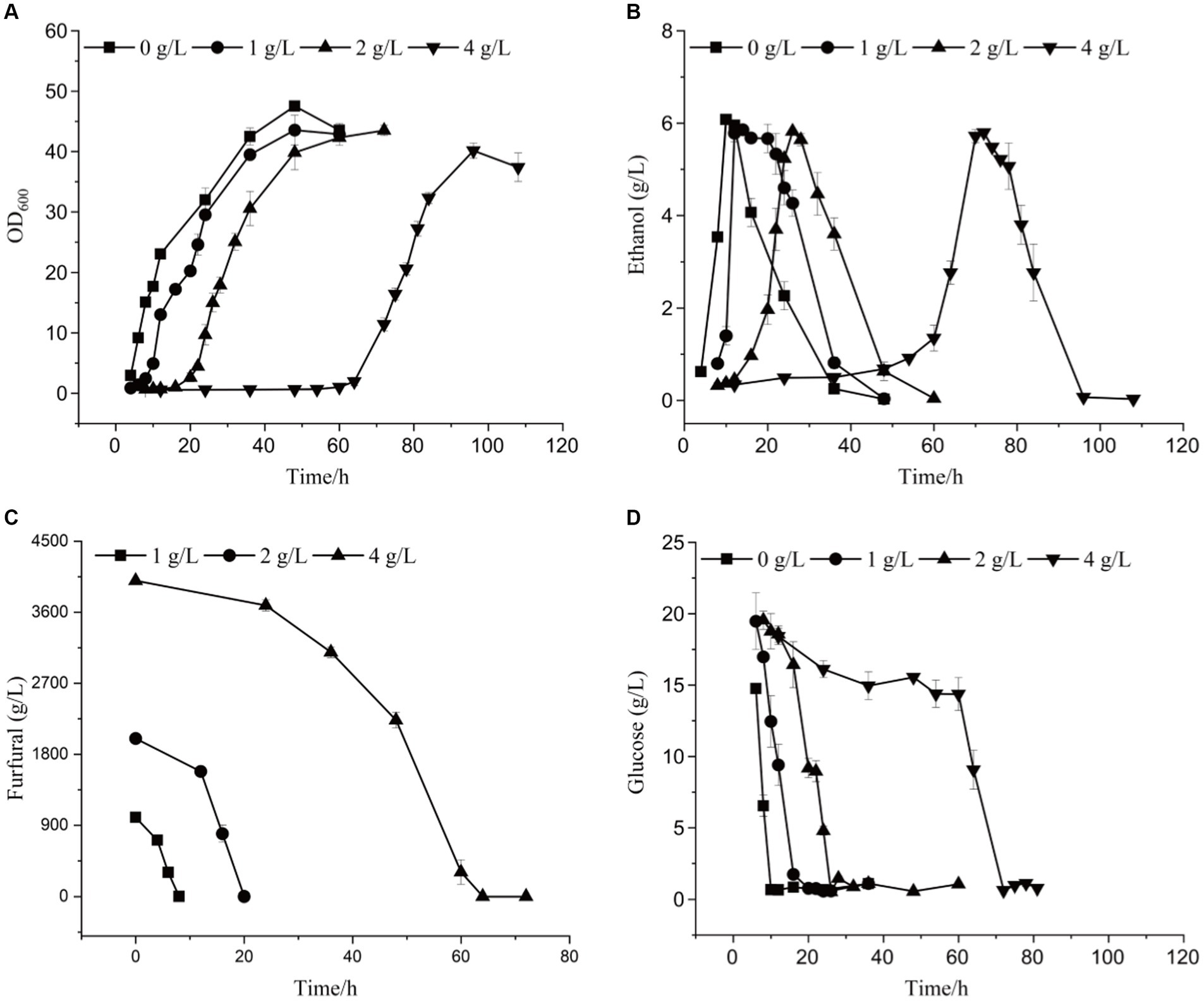
Figure 4. The growth curve (A), ethanol yield (B), furfural concentration (C), and glucose content (D) of ADR1_1802 mutant under different furfural stress.
Adr1p is essential for the transcription of genes required for glycerol utilization, and glycerol is the main osmotic protector for Saccharomyces cerevisiae (Hohmann, 2002). Glycerol plays an important role in yeast adaptation to changing conditions (Påhlman et al., 2000). As shown in Figure 5A, during the lag phase, the intracellular glycerol content of the ADR1_1802 mutant was 87.74% lower than that of the 5D strain, and there was no significant difference in the logarithmic phase. Based on this result, it was indicated that intracellular glycerol was applied to resist the toxicity of furfural in ADR1_ 1802 mutant.
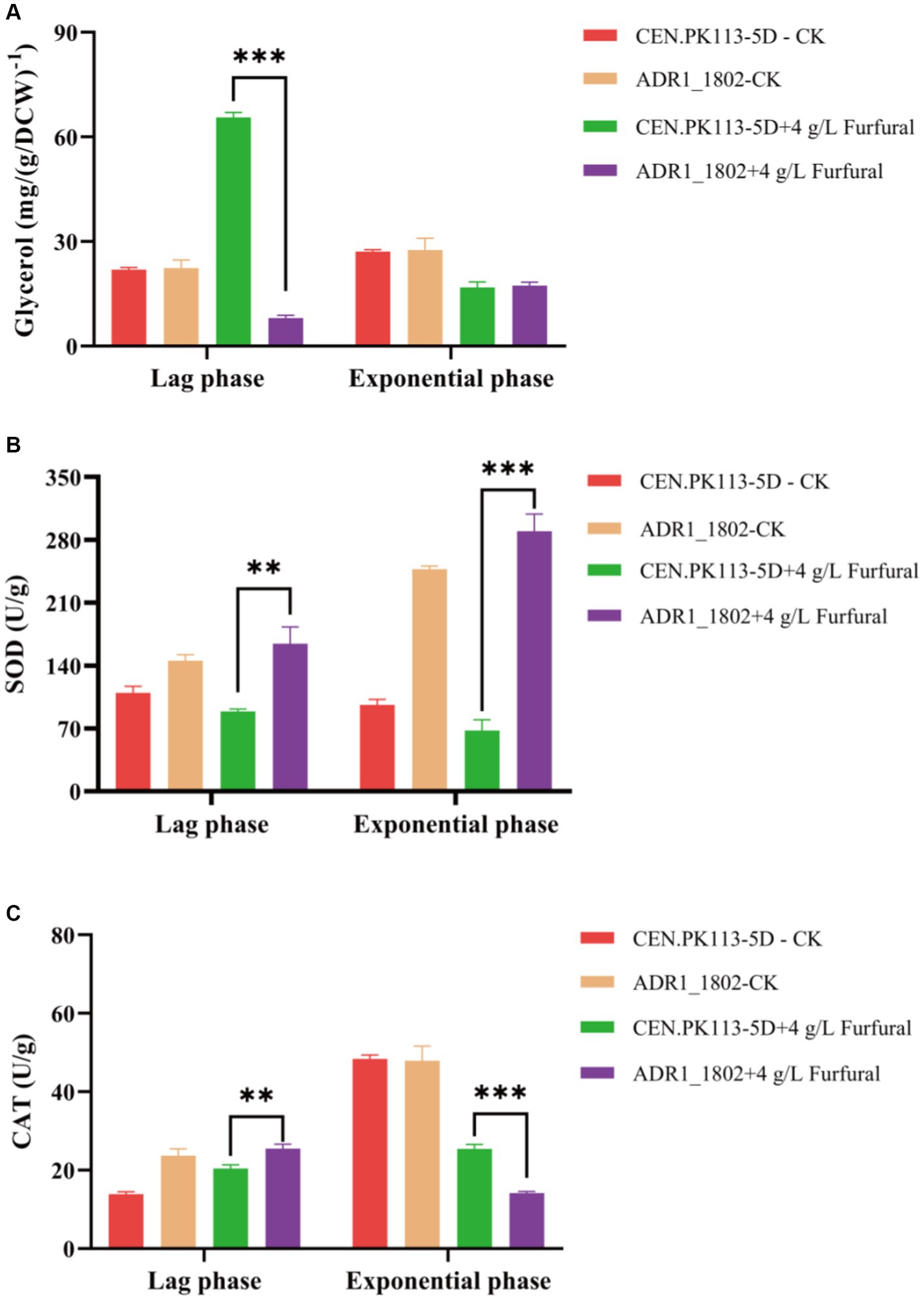
Figure 5. Comparison of mutant strain ADR1_1802 with reference strain of intracellular glycerol (A), SOD (B) and CAT (C) enzyme activity under furfural stress. “**” (0.001 ≤ p < 0.01), “***” (p < 0.001).
Due to involvement of ADR1 in the expression of peroxisome genes, under high concentration of furfural (4 g/L) stress, the enzyme activities of CAT and SOD in ADR_1802 mutant and reference strain were determined and compared. As shown in Figures 5B,C, compared with reference strain, SOD enzyme activity was increased by 84.52% and CAT enzyme activity was increased by 24.72% during the lag phase in ADR_1802 mutant. Nakazawa et al. found that due to the high activity of CAT and SOD of strain IB1306, this strain effectively resisted the increase in ROS levels induced by H2O2 (Nakazawa et al., 2018). Overexpression of catalase increases tolerance to furfural and HMF (Kim and Hahn, 2013).
ADR1_1802 mutant strain utilizes the intracellular glycerol, high activity of CAT, and SOD to resist the toxicity of high concentrations of furfural.
Furfural could be reduced to furfuryl alcohol in Saccharomyces cerevisiae with NAD (P) H (Liu et al., 2008). From above results, it can be seen that the time required for the degradation of furfural by ADR1_1802 strain was significantly shorter than that of the reference strain, and the lag phase was shortened obviously. Therefore, the transcription level of genes in ADR1_1802 strain and reference strain related to the furfural degradation was analyzed and compared (Supplementary Figure S8).
As shown in Figure 6, the results of RT-PCR showed that, the transcription levels of ADH6, ALD4, GRE2, GRE3, YPR1, and ARI1 genes in S. cerevisiae CEN. PK113-5D strain increased by 2.87, 1.69, 20.04, 4.37, 6.06, and 2.01 times, respectively, compared with control. While in ADR1_1802 mutant strain, they were upregulated by 4.16, 1.71, 35.80, 3.83, 3.15, and 2.84 times, respectively. It could conclude that GRE2 was a key gene for the reduction of furfural. Under 4 g/L furfural treatment, compared with reference strain, transcription level of GRE2 in ADR1_1802 mutant was increased by 53.69%, and that of ADH6 was increased by 44.95%.
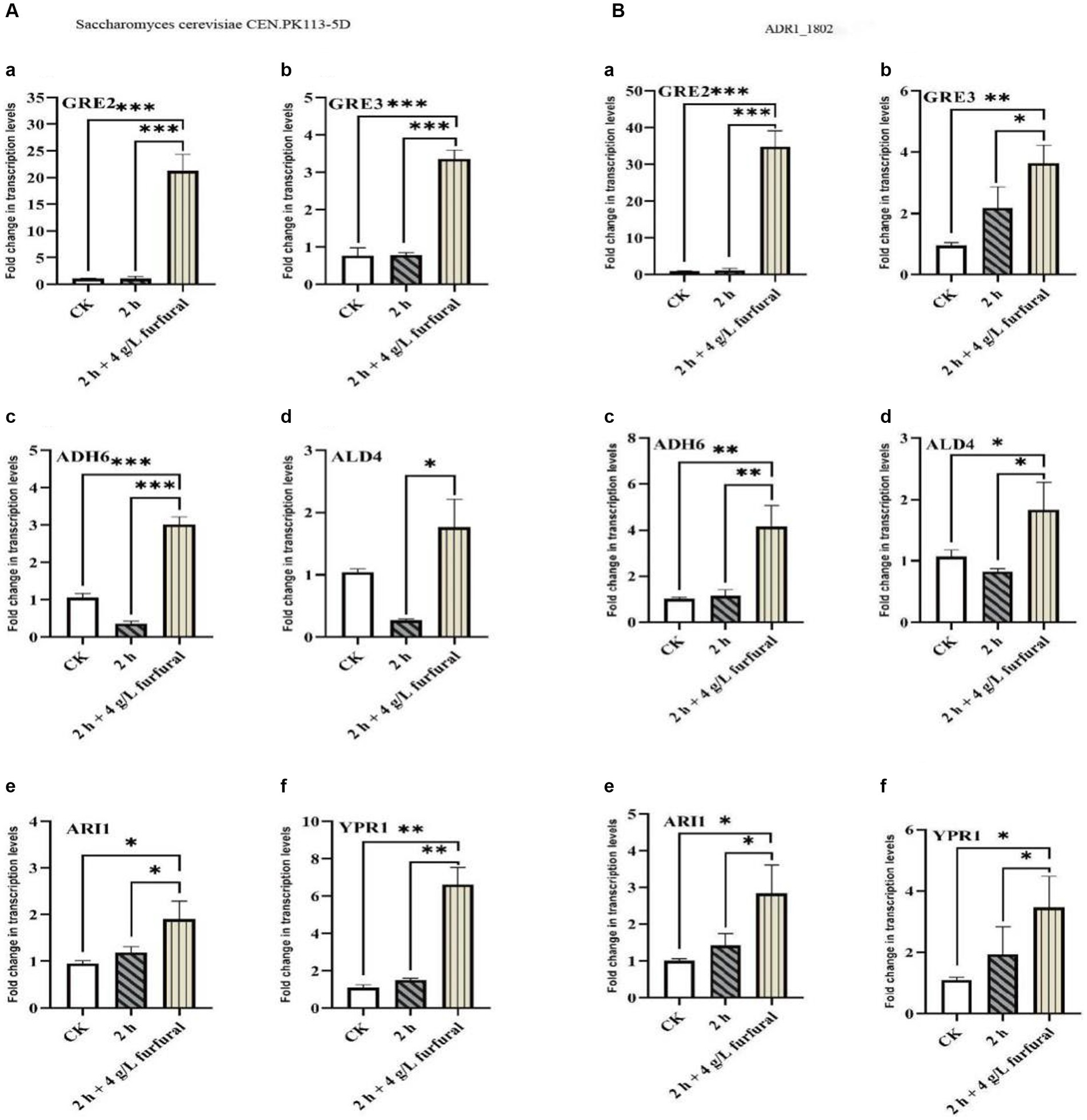
Figure 6. Transcription levels of ADH6, ALD4, GRE2, GRE3, YPR1, and ARI1 in response to furfural. “*” (0.01 ≤ p < 0.05), “**” (0.001 ≤ p < 0.01), and “***” (p < 0.001).
After ALE by accumulating spontaneous mutations generation after generation under specific pressure of furfural, a modified microbial strain is obtained through forced selection of specific phenotypes. After a long period of adaptive laboratory evolution, this strain continuously adapts and evolves in a culture medium with gradually increasing furfural concentration, resulting in stains with faster growth and enhanced tolerance to furfural stress.
It was suggested that the adaptation of Saccharomyces cerevisiae CEN.PK113-7D to furfural stress may be closely related to gene expression regulation. For Saccharomyces cerevisiae, Pau protein may play a role in adapting to stress, which could be induced to express under low temperature and anaerobic conditions (Luo and Van Vuuren, 2009). The regulation of sphingolipids synthesis by COS8 could help cells obtain the ability to integrate signals from the pheromone, osmolarity, and TOR pathways to modify membrane structure (Bailly-Bechet et al., 2011). Genes TFC1, HYP2, and RET1 are mainly involved in cell proliferation and growth, and are crucial for cell survival (Swanson et al., 1991; Bobkova et al., 1999). The proteins Uth1p, Hda1p, Cdc14p, and Adr1p are closely related with oxidative stress responses, cell lifespan, and autophagy in yeast (Valenzano et al., 2006). Mutations in these genes may alter intracellular oxidative stress levels, extend cell lifespan, and change the structure and function of cell walls, thereby improving the adaptability of Saccharomyces cerevisiae to furfural stress.
Adr1p is transcription factor that is required for the transcription of glucose inhibitory gene ADH2, peroxisome protein gene, and genes required for ethanol, glycerol, and fatty acid utilization. When glucose is absent, Saccharomyces cerevisiae can also effectively grow in carbon sources such as glycerol, acetate, ethanol, or oleate. This requires metabolic rearrangement, and involves several transcription factors, of which Adr1 is the most critical one (Moreno-Cermeño et al., 2013). Young et al. found that peroxisome genes were significantly upregulated under derepressing conditions due to the loss of Med14 activity, which were dependent on the Adr1 and Oaf1/Pip2 transcription complexes (Young et al., 2009). The sequencing analysis results in this study showed that there was a missense mutation in the ADR1 gene of 12–1 strain (lysine at 601 changed to arginine).
After ADR1p mutation on Saccharomyces cerevisiae, it was found that during the lag phase, the intracellular glycerol content of the ADR1_1802 mutant was 87.74% lower than that of reference strain. Previous studies by Ask et al. found that the addition of 3.9 g/L HMF and 1.2 g/L furfural to Saccharomyces cerevisiae resulted in significant physiological reactions. The specific growth rate and glycerol production was decreased significantly (Ask et al., 2013). The physiological characteristics of a new heat-resistant strain (LBGA-01) was found by Cleiton et al., which can grow at 40°C and is more resistant to sucrose, furfural, and ethanol than industrial strains. At 40°C, gene expression related to the formation of glycerol (GPD2) is reduced (Prado et al., 2020). Furthermore, Reactive oxygen species (ROS) generated during metabolic process, includes hydrogen peroxide (H2O2), hydroxyl radicals, and superoxide anions, can damage proteins, lipids, carbohydrates, and DNA (Bora et al., 2018). Under high concentration of furfural (4 g/L) stress, ADR1_1802 mutant strain showed higher CAT and SOD enzyme activity than the wild-type strain in lag phage, which is consistent with previous studies (Nakazawa et al., 2018). Although CAT activity in the mutant strain in exponential phase was lower than that of the wild-type, lag phage is more important, during which furfural was degraded by cells metabolism to less toxic compounds.
Earlier studies indicated that overexpression of ADH6 in Saccharomyces cerevisiae enhances the in situ detoxification furfural produced during biomass pretreatment (Quarterman et al., 2018). Furthermore, the HMF conversion rate in vivo of S. cerevisiae CEN.PK 113-5D overexpressing ADH6 was significantly increased, indicating that overexpression indeed increased reduction capacity (Petersson et al., 2006). The GRE2 gene of Scheffersomyces (Pichia) stipitis is highly induced under furfural and HMF stress, which can enhance tolerant ability of S. cerevisiae to furfural after overexpression (Wang et al., 2016).
In summary, the expression of genes related to furfural tolerance such as GRE2 and ADH6 were increased for the reduction of furfural to furfural alcohol in ADR1 mutation strain, providing sufficient NAD (P)(+) for NAD (P) H regeneration, thereby enhancing resistance of ADR1_1802 mutant to furfural stress.
The present study involved screening 12–1 strains of Saccharomyces cerevisiae by laboratory adaptive evolution to improve ethanol production and shorten the lag phase. Whole genome sequencing results demonstrated thatADR1 genecan be mutated to get ADR1_1802 mutant by CRISPR/Cas9 technology, which exhibited increased resistance to furfural stress comparable fermentation performance with reference strain. These findings suggested the potential for genetic engineering to increase the furfural tolerance of Saccharomyces cerevisiae.
The original contributions presented in the study are included in the article/Supplementary material, further inquiries can be directed to the corresponding author. Sequence data associated with this project have been deposited in the NCBI (PRJNA1049636).
LY: Writing – original draft. YJ: Formal analysis, Methodology, Writing – original draft. QZ: Investigation, Writing – review & editing. XZ: Writing – review & editing. HY: Writing – review & editing. JD: Writing – review & editing. XC: Funding acquisition, Supervision, Writing – review & editing.
The author(s) declare financial support was received for the research, authorship, and/or publication of this article. This work was supported by the National Natural Science Foundation of China (Nos. 21978074 and 31871789) and Key Project of Hubei Provincial Department of Education (Nos. D20211404 and T2022011).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2023.1333777/full#supplementary-material
Alves, R., Zetty-Arenas, A., Demirci, H., Dias, O., Rocha, I., Basso, T., et al. (2020). Enhancing acetic acid and 5-hydroxymethyl furfural tolerance of C. saccharoperbutylacetonicum through adaptive laboratory evolution. Process Biochem. 101, 179–189. doi: 10.1016/j.procbio.2020.11.013
Ask, M., Bettiga, M., Duraiswamy, V., and Olsson, L. (2013). Pulsed addition of HMF and furfural to batch-grown xylose-utilizing Saccharomyces cerevisiae results in different physiological responses in glucose and xylose consumption phase. Biotechnol. Biofuels 6:181. doi: 10.1186/1754-6834-6-181
Bailly-Bechet, M., Borgs, C., and Braunstein, A. (2011). Finding undetected protein associations in cell signaling by belief propagation. Proc. Natl. Acad. Sci. U. S. A. 108, 882–887. doi: 10.1073/pnas.1004751108
Bobkova, E., Habib, N., Alexander, G., and Hall, B. (1999). Mutational analysis of the hydrolytic activity of yeast RNA polymerase III. J. Biol. Chem. 274, 21342–21348. doi: 10.1074/JBC.274.30.21342
Bora, P., Chauhan, P., Manna, S., and Chakrapani, H. (2018). A vinyl coronate Ester based Persulfide donor controllable by hydrogen peroxide, a reactive oxygen specifications (ROS). Org. Lett. 20, 7916–7920. doi: 10.1021/acs.orglett.8b03471
Chen, Y., Sheng, J., Jiang, T., Stevens, J., Feng, X., and Wei, N. (2016). Transcriptional profiling reveals molecular basis and novel genetic targets for improved resistance to multiple fermentation inhibitors in Saccharomyces cerevisiae. Biotechnol. Biofuels 9, 9–18. doi: 10.1186/s13068-015-0418-5
Dai, J., Li, K., Song, N., Yao, W., Xia, H., Yang, Q., et al. (2020). Zygosaccharomyces rouxii, an aromatic yeast isolated from chili sauce, is able to biosynthesize 2-phenylethanol via the shikimate or Ehrlich pathways. Front. Microbiol. 11:597454. doi: 10.3389/fmicb.2020.597454
Du, C., Li, Y., and Xiang, R. (2022). Co-utilization of multiple lignocellulose-derived sugars and inhibitors by the robust mutant Kluyveromyces marxianus through adaptive laboratory evolution. Fuel 315:122816. doi: 10.1016/j.fuel.2021.122816
He, J., Zhang, W., Liu, X., Xu, N., and Xiong, P. (2016). Optimization of prehydrolysis time and substrate feeding to improve ethanol production by simultaneous saccharification and fermentation of furfural process residue. J. Biosci. Bioeng. 122, 563–569. doi: 10.1016/j.jbiosc.2016.04.012
Hemansi,, Himanshu,, Patel, A., Saini, J., and Singhania, R. (2022). Development of multiple inhibitor tolerance year via adaptive laboratory evolution for sustainable bioethanol production. Bioresour. Technol. 344, 126247–126255. doi: 10.1016/j.biortech.2021.126247
Hohmann, S. (2002). Osmotic stress signaling and Osmoadaptation in yeasts. Microbiol. Mol. Biol. R. 66, 300–372. doi: 10.1128/MMBR.66.2.300-372.2002
Jia, Y., Zhang, Q., Dai, J., Zheng, X., Meng, X., Zhou, R., et al. (2022). Ergosterol supplementation improves furfural tolerance of Saccharomyces cerevisiae to produce ethanol and its underlying mechanism. Bio Resoures. 18, 228–246. doi: 10.15376/biores.18.1.228-246
Jin, C., Hou, W., Yao, R., Zhou, P., Zhang, H., and Bao, J. (2019). Adaptive evolution of Gluconobacter oxydans accelerates the conversion rate of non-glucose sugars derived from lignocellulose biomass. Bioresour. Technol. 289:121623. doi: 10.1016/j.biortech.2019.121623
Kim, D., and Hahn, J. (2013). Roles of the yap 1 transcription factor and antioxidants in Saccharomyces cerevisiae's tolerance to furfural and 5-Hydroxymethylfurfural, which function as thiol-reactive electrophiles generating oxidative stress. Appl. Environ. Microb. 79, 5069–5077. doi: 10.1128/AEM.00643-13
Laughery, M., Hunter, T., Brown, A., Hoopes, J., Ostbye, T., Shumaker, T., et al. (2015). New vectors for simple and streamlined CRISPR-Cas 9 genome editing in Saccharomyces cerevisiae. Yeast 32, 711–720. doi: 10.1002/yea.3098
Lewis, L., Ma, M., and Song, M. (2009). Evolutionarily engineered ethanologenic yeast detoxifies lignocellulosic biomass conversion inhibitors by reprogrammed pathways. Mol. Gen. Genomics. 282, 233–244. doi: 10.1007/s00438-009-0461-7
Li, X., Yang, R., Ma, M., Wang, X., Tang, J., Zhao, X., et al. (2014). A novel aldehyde reductase encoded by YML131W from Saccharomyces cerevisiae confers tolerance to furfural derived from lignocellulosic biomass conversion. Bioenerg. Res. 8, 119–129. doi: 10.1007/s12155-014-9506-9
Lin, C., and Lu, C. (2021). Development perspectives of promising lignocellulose feedstocks for production of advanced generation biofuels: a review. Renew. Sust. Energ. Rev. 136:110445. doi: 10.1016/j.rser.2020.110445
Liu, Z., Fels, M., Dragone, G., and Mussatto, S. I. (2021). Effects of inhibitory compounds derived from lignocellulosic biomass on the growth of the wild-type and evolved oleaginous yeast Rhodosporidium toruloides. Ind. Crop. Prod. 170:113799. doi: 10.1016/J.INDCROP.2021.113799
Liu, Z., Moon, J., Andersh, B., Slininger, P., and Weber, S. (2008). Multiple gene mediated NAD (P)H-dependent aldehyde reduction is a mechanism of in situ detoxification of furfural and 5-hydroxymethylfurfural by Saccharomyces cerevisiae. Appl. Microbiol. Biotechnol. 81, 743–753. doi: 10.1007/s00253-008-1702-0
Liu, Z., Palmquist, D., Ma, M., Liu, J., and Alexander, N. (2009). Application of a master equation for quantitative mRNA analysis using qRT-PCR. J. Biotechnol. 143, 10–16. doi: 10.1016/j.jbiotec.2009.06.006
Liu, Z., and Slininger, P. (2007). Universal external RNA controls for microbial gene expression analysis using microarray and qRT-PCR. J. Microbiol. Methods 68, 486–496. doi: 10.1016/j.mimet.2006.10.014
Liu, Z., Slininger, P., and Gorsich, S. (2005). Enhanced biotransformation of furfural and Hydroxymethylfurfural by newly developed Ethanologenic yeast strains. Appl. Biochem. Biotechnol. 121, 451–460. doi: 10.1385/ABAB:121:1-3:0451
Luo, Z., and Van Vuuren, H. (2009). Functional analyses of PAU genes in Saccharomyces cerevisiae. Microbiology 155, 4036–4049. doi: 10.1099/mic.0.030726-0
Moreno-Cermeño, A., Alsina, D., Cabiscol, E., Tamarit, J., and Ros, J. (2013). Metabolic remodeling in frataxin-deficient yeast is mediated by Cth 2 and Adr 1. Biochim. Biophys. Acta 1833, 3326–3337. doi: 10.1016/j.bbamcr.2013.09.019
Nakazawa, N., Yanata, H., Ito, N., Kaneta, E., and Takahashi, K. (2018). Oxidative stress tolerance of a spore clone isolated from Shirakami Kodama yeast depends on altered regulation of msn 2 leading to enhanced expression of ROS-degrading enzymes. J Gen Appl Microb. 64, 149–157. doi: 10.2323/jgam.2017.11.002
Nijkamp, J., Van, D., Datema, E., De, K., Bosman, L., Luttik, M., et al. (2012). De novo sequencing, assembly and analysis of the genome of the laboratory strain Saccharomyces cerevisiae CEN.PK113-7D, a model for modern industrial biotechnology. Microb. Cell Factories 11, 36–52. doi: 10.1186/1475-2859-11-36
Påhlman, A., Granath, K., Ansell, R., Hohmann, S., and Adler, L. (2000). The yeast glycerol 3-phosphatases Gpp 1p and Gpp 2p are required for glycerol biosynthesis and differentially involved in the cellular responses to osmotic, anaerobic, and oxidative stress. J. Biol. Chem. 276, 3555–3563. doi: 10.1074/jbc.M007164200
Petersson, A., Almeida, J. R., and Modig, T. (2006). A 5-hydroxymethyl furfural reducing enzyme encoded by the Saccharomyces cerevisiae ADH6 gene conveys HMF tolerance. Yeast 23, 455–464. doi: 10.1002/yea.1370
Prado, C., Mandrujano, G., Souza, J., Sgobbi, F., Novaes, H., da Silva, J., et al. (2020). Physiological characterization of a new thermotolerant yeast strain isolated during Brazilian ethanol production, and its application in high-temperature fermentation. Biotechnol. Biofuels 13:178. doi: 10.21203/rs.3.rs-40885/v1
Quarterman, J. C., Slininger, P. J., Hector, R. E., and Dien, B. S. (2018). Engineering Candida phangngensis - an oleaginous yeast from the Yarrowia clade – for enhanced detoxification of lignocellulose-derived inhibitors and lipid overproduction. FEMS Yeast Res. 18:foy102. doi: 10.1093/femsyr/foy102
Rabemanolontsoa, H., and Saka, S. (2016). Various pretreatments of lignocellulosics. Bioresour. Technol. 199, 83–91. doi: 10.1016/j.biortech.2015.08.029
Sandberg, T., Salazar, M., Wen, L., Palsson, B., and Feist, A. (2019). The emergency of adaptive laboratory evolution as an effective tool for biological discovery and industrial biotechnology. Metab. Eng. 56, 1–16. doi: 10.1016/j.ymben.2019.08.004
Sharma, J., Kumar, V., Prasad, R., and Gaur, N. (2022). Engineering of Saccharomyces cerevisiae as a consolidated bioprocessing host to produce cellulosic ethanol: recent advancements and current challenges. Biotechnol. Adv. 56:107925. doi: 10.1016/j.biotechadv.2022.107925
Swanson, R., Conesa, C., Lefebvre, O., Carles, C., Ruet, A., Quemeneur, E., et al. (1991). Isolation of TFC1, a gene encoding one of two DNA-binding subunits of yeast transcription factor tau (TFIIIC). Proc. Natl. Acad. Sci. U. S. A. 88, 4887–4891. doi: 10.1073/pnas.88.11.4887
Valenzano, D., Terzibasi, E., Genade, T., Cattaneo, A., Domenici, L., and Cellerino, A. (2006). Resveratrol prolongs lifespan and retards the onset of age-related markers in a short-lived vertebrate. Curr. Biol. 16, 296–300. doi: 10.1016/j.cub.2005.12.038
Wang, H., Li, Q., Kuang, X., Xiao, D., Han, X., Hu, X., et al. (2018). Functions of aldehyde reductases from Saccharomyces cerevisiae in detoxification of aldehyde inhibitors and their biotechnological applications. Appl. Microbiol. Biot. 102, 10439–10456. doi: 10.1007/s00253-018-9425-3
Wang, X., Ma, M., Liu, Z., Xiang, Q., Li, X., Liu, N., et al. (2016). GRE2 from Scheffersomyces stipitis as an aldehyde reductase contributes tolerance to aldehyde inhibitors derived from lignocellulosic biomass. Appl. Microbiol. Biot. 100, 6671–6682. doi: 10.1007/s00253-016-7445-4
Wu, Y., Jameel, A., Xing, X., and Zhang, C. (2022). Advanced strategies and tools to facilitate and streamline microbial adaptive laboratory evolution. Trends Biotechnol. 40, 38–59. doi: 10.1016/j.tibtech.2021.04.002
Xia, H., Kang, Y., Ma, Z., Hu, C., Yang, Q., Zhang, X., et al. (2022). Evolutionary and reverse engineering in Saccharomyces cerevisiae reveals a Pdr 1p mutation-dependent mechanism for 2-phenylethanol tolerance. Microb. Cell Factories 21:269. doi: 10.1186/s12934-022-01996-x
Yan, Z., Zhang, J., and Bao, J. (2021). Increasing cellulosic ethanol production by enhancing phenolic tolerance of Zymomonas mobilis in adaptive evolution. Bioresour. Technol. 329:124926. doi: 10.1016/j.biortech.2021.124926
Young, E., Yen, K., Dombek, K., Law, G., Chang, E., and Arms, E. (2009). Snf 1-independent, glucose-resistant transcription of Adr 1-dependent genes in a mediator mutant of Saccharomyces cerevisiae. Mol. Microbiol. 74, 364–383. doi: 10.1111/j.1365-2958.2009.06866.x
Yu, Z., and Zhang, H. (2004). Ethanol fermentation of acid-hydrolyzed cellulosic pyrolysate with Saccharomyces cerevisiae. Bioresour. Technol. 90, 95–100. doi: 10.1016/S0960-8524(03)00093-2
Keywords: adaptive laboratory evolution, Saccharomyces cerevisiae, furfural tolerance, bioethanol, CRISPR/Cas9
Citation: Yao L, Jia Y, Zhang Q, Zheng X, Yang H, Dai J and Chen X (2024) Adaptive laboratory evolution to obtain furfural tolerant Saccharomyces cerevisiae for bioethanol production and the underlying mechanism. Front. Microbiol. 14:1333777. doi: 10.3389/fmicb.2023.1333777
Received: 06 November 2023; Accepted: 05 December 2023;
Published: 04 January 2024.
Edited by:
Ming Jun Zhu, South China University of Technology, ChinaReviewed by:
Pornthap Thanonkeo, Khon Kaen University, ThailandCopyright © 2024 Yao, Jia, Zhang, Zheng, Yang, Dai and Chen. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Xiong Chen, Y3gxNjNfcXhAMTYzLmNvbQ==
†These authors have contributed equally to this work
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.