- 1College of Life and Environment Science, Minzu University of China, Beijing, China
- 2Ministry of Education Key Laboratory for Ecology of Tropical Islands, Key Laboratory of Tropical Animal and Plant Ecology of Hainan Province, College of Life Sciences, Hainan Normal University, Haikou, China
A “grazing ban” policy has been implemented in some pastoral areas in China to fence degraded grasslands for restoration. However, fencing increased grazing pressures in unmanaged grasslands. Based on the mechanism of negative edge effect, we investigated whether overgrazing on unmanaged grassland interfered with the restoration of adjacent grazing-banned grassland by affecting soil properties and microbial community using a sample in Hulun Buir of Inner Mongolia, in order to optimize the “grazing ban” policy. Plant and soil were sampled in areas 30 m away from the fence in unmanaged grassland (UM) and in areas 30 m (adjacent to UM) and 30–60 m (not adjacent to UM) away from the fence in the grazing-banned grassland (F-30 m and F-60 m). The species richness and diversity of plant communities and the ASV number of fungal communities significantly decreased in F-30 m and UM, and the Simpson index of the bacterial community significantly decreased in F-30 m compared with F-60 m. The abundance of fungi involved in soil organic matter decomposition significantly decreased and the abundance of stress-resistant bacteria significantly increased, while the abundance of bacteria involved in litter decomposition significantly decreased in UM and F-30 m compared with F-60 m. The simplification of plant communities decreased in soil water and total organic carbon contents can explain the variations of soil microbial communities in both UM and F-30 m compared with F-60 m. The results of PLS-PM show that changes in plant community and soil microbial function guilds in UM may affect those in F-30 m by changing soil water and total organic carbon contents. These results indicate that overgrazing on unmanaged grassland interfered with the restoration of adjacent grazing-banned grassland by affecting soil properties and microbial community. The grazing-banned grasslands should be adjusted periodically in order to avoid negative edge effects.
1 Introduction
Grassland is an ecosystem where humans and nature coexist and interact (Wang et al., 2018; Sun et al., 2022). Grasslands provide grazing resources for humans, and moderate grazing promotes decomposition and regulates the structure of plant communities, maintaining the dynamic balance of grasslands (Xun et al., 2018). However, the ongoing impact of overgrazing and global climate change has resulted in grassland degradation, which has become a major factor threatening the stability of the global terrestrial ecosystem (Boval and Dixon, 2012; Gibbs and Salmon, 2015; Liu et al., 2019; Lyu et al., 2020; Bardgett et al., 2021; Török et al., 2021). To curb grassland degradation, a series of restoration policies have been implemented in some pastoral areas of China (Kolås, 2014; Sun et al., 2020). “Grazing ban” is one such policy to fence some grasslands from grazing. The implementation of the policy significantly improved the plant diversity, total organic carbon, soil water content, and total nitrogen in the grazing-banned areas (Pei et al., 2008; Cheng et al., 2016; Wang et al., 2016; Xiong et al., 2016; Li et al., 2017). However, fencing increased grazing pressure in the unmanaged areas which are usually adjacent to the grazing-banned areas (Sun et al., 2020). In some long-term fenced grasslands, the areas adjacent to unmanaged grasslands have degraded. Therefore, it is necessary to understand whether the overgrazing on unmanaged grasslands interfered with the restoration of grazing-banned grasslands.
Fencing formed a clear boundary between unmanaged and grazing-banned grasslands. Edge effect refers to the changes in community structure and ecological processes caused by ecological factors or system-attributed changes at the edge zone of two adjacent ecosystems (Fonseca and Joner, 2007; Eldegard et al., 2015). In anthropogenic interference-induced edge zones, the biological community structures within the edge zones changed rapidly and had weak anti-interference abilities (Kark, 2013; Smith and Goetz, 2021). Light, energy utilization rate, soil temperature, and soil physicochemical properties in such areas changed with the changes in the biological community (Schmidt et al., 2017; Koelemeijer et al., 2023). Thus, edge effects induced by anthropogenic interference have been confirmed to negatively influence species diversity, community dynamics, and ecosystem functions (Guerra et al., 2017; Krishnadas et al., 2018; Fischer et al., 2021; Blanchard et al., 2023; Lapola et al., 2023).
It is well known that the anthropogenic interference-induced edge effect is caused by disrupting the interaction between soil microbial and plant communities in the edge zone (Pennanen, 2001; Ettema and Wardle, 2002; Malmivaara-Lämsä et al., 2008). The interaction between plants and soil microorganisms is important for maintaining the stability and productivity of ecosystems (van der Heijden et al., 2008; Bardgett and van der Putten, 2014). Plants serve as specific hosts for soil microorganisms (Zhang et al., 2018), influence the microbial community structure through root exudates (Meier et al., 2017; Canarini et al., 2019), and provide energy and nutrients for microbial metabolism (van der Heijden et al., 2008; Luo et al., 2022). Soil microorganisms regulate nutrient cycling between plants and soil, aiding the formation of soil structures that are conducive to plant growth (Bissett and Burke, 2007; Coban et al., 2022; Hartmann and Six, 2023; Li et al., 2023). The soil microbial community structure and function in the edge zone change with the changes in soil properties and plant community, and these changes influence litter decomposition rates and nutrient cycling in the edge zone (Pennanen, 2001). Changes in nutrient and water supply may in turn affect the structure and diversity of plant communities in adjacent areas, ultimately leading to vegetation degradation in these areas (Ettema and Wardle, 2002). For example, forest fragmentation-induced edge effect changed soil microbial communities by affecting the soil properties in the forest edge zone (Malmivaara-Lämsä et al., 2008). Based on the mechanism of edge effect induced by anthropogenic interference, overgrazing on unmanaged grassland may deteriorate the interaction between soil microbial and plant communities in adjacent grazing-banned grassland, reducing the restoration effectiveness in grazing-banned grassland. Therefore, it is necessary to understand whether such a negative edge effect exists in order to optimize the “grazing ban” policy.
The Inner Mongolia grassland is one of the pastoral areas in China where the “grazing ban” policy is implemented (Hao et al., 2014; Kolås, 2014; Gao et al., 2022). Grasslands designated by the local government as under “restoration” cover approximately 70% of the total grassland. Of the total area under “restoration”, pastures fenced under “grazing ban” cover approximately 20%, and pastures subjected to unspecified restoration measures cover approximately 20% (Kolås, 2014). Due to the reduction in grazing grasslands, herders overgrazed on the unmanaged grasslands, resulting in severe degradation of these areas. Many such unmanaged areas are adjacent to grazing-banned areas. Based on the known mechanisms of edge effect induced by anthropogenic interference described above, this study aims to investigate whether overgrazing on unmanaged grasslands interfered with the restoration of adjacent grazing-banned grassland by affecting soil properties and microbial communities. We predicted that (1) in grazing-banned grassland, the composition and the interaction of soil microbial and plant communities of the area adjacent to unmanaged grassland were similar to those of the unmanaged grassland but different from those of the area not adjacent to the unmanaged grassland; and (2) the changes of plant and soil microbial communities in the unmanaged grassland may change those in adjacent grazing-banned grassland by changing soil properties.
2 Materials and methods
2.1 Study site
The study site was located in the Xin Barag Right Banner of Hulun Buir, in the Inner Mongolia Autonomous Region (47°36′00″N ~ 49°50′0″N and 115°31′00″E ~ 117°43′00″E), and is an example of a typical temperate grassland. The average annual precipitation is 243.9 mm, with the majority of rainfall occurring during the summer months. The average annual temperature is 1.6°C. The primary soil type is Calcic Luvisols (FAO, 1988), and the dominant plant species include Leymus chinensis, Stipa capillata, Poa annua, and Caragana stenophylla, among others.
2.2 Experimental design
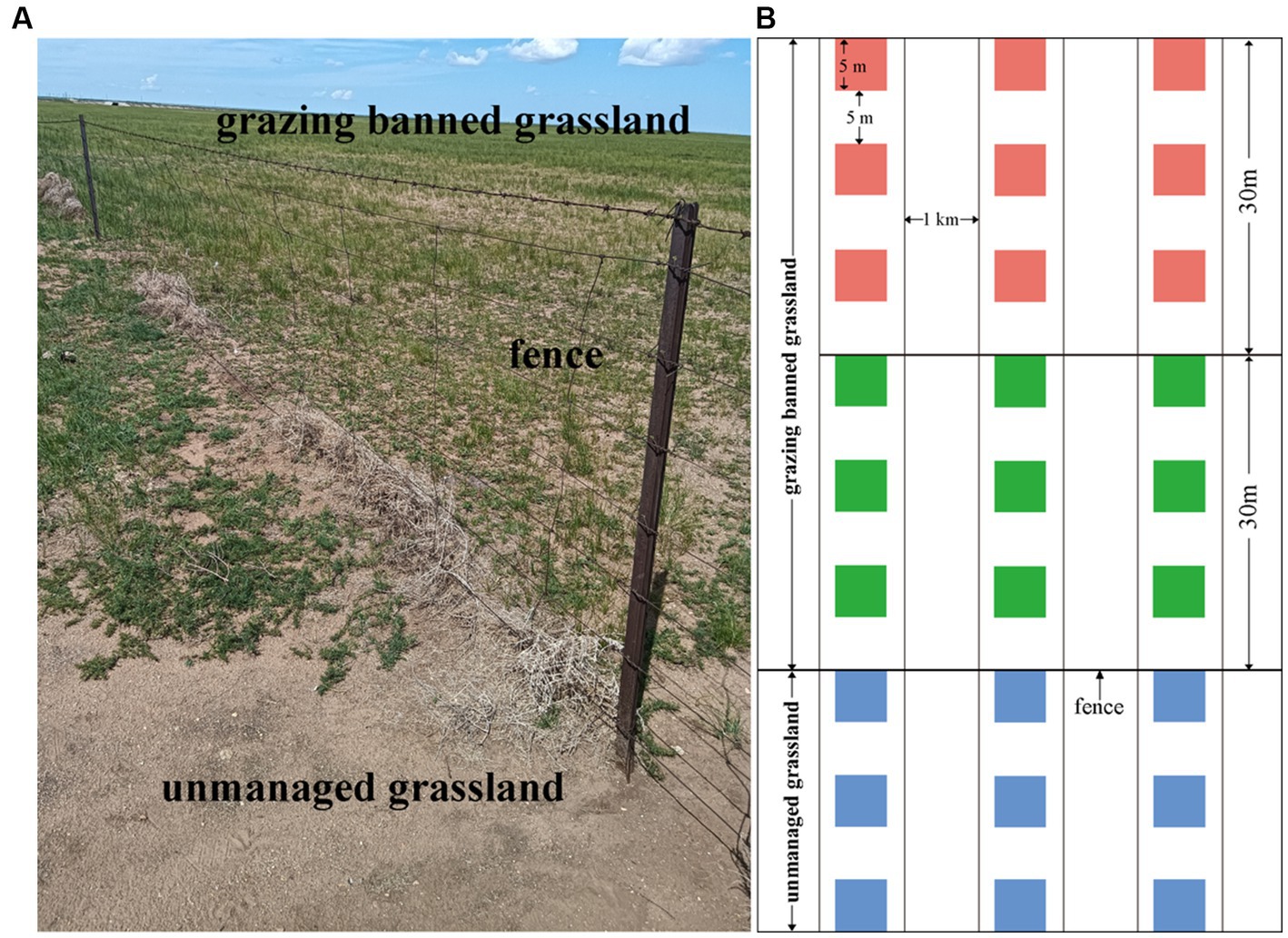
The grazing-banned grassland selected in this study was fenced in 2008 and is adjacent to an unmanaged grassland (Figure 1A). Three belt transects spaced 1 km apart were set up perpendicular to the fence. According to the observation that degradation area in the grazing-banned grassland is approximately 30 m away from the fence, we defined 30 m as a distance unit from the fence (Figure 1A). In each belt transect, three 5 m × 5 m plots, spaced 5 m apart, were set up 30 m away from the fence in unmanaged grassland (hereafter UM) and the areas 30 m (adjacent to the unmanaged grassland) and 30–60 m (not adjacent to the unmanaged grassland) away from the fence in grazing-banned grassland (hereafter F-30 m and F-60 m), respectively. The sampling diagram is shown in Figure 1B. Within each plot, the plant community was surveyed, and the soil was sampled. To test our two predictions, we compared the composition of the plant and soil microbial communities, soil microbial functions, and soil physicochemical properties in the three types of sampling grasslands, and then we analyzed the relationships between plant community, soil physicochemical properties, and soil microbial community in UM and F-30 m, respectively; finally, we estimated the direct and indirect effects of plant community, soil property, and soil microbial community in UM on those in F-30 m.
2.3 Plant community survey and soil sampling
The plant community survey and soil sampling were conducted in July 2022. Plant species were identified, and the number of each species in each plot was counted. Five soil samples, 5 cm in diameter and 20 cm deep, were taken from the four corners and centers of each plot. Samples collected from the same plot were combined and passed through a 2 mm sieve before being divided into three parts for analysis. One portion was stored at 4°C for analysis of pH and soil water content (SWC). Another portion was air dried for analysis of soil element content, and the final portion was stored at −80°C for Illumina sequencing of soil microorganisms.
2.4 Analysis of soil physicochemical properties
The soil samples were immediately weighed after collection, before being placed in an oven for 24 h at 60°C and weighed again. The difference between the wet and dry soil weights was used to determine the soil water content (SWC). The soil pH values were measured in the soil suspension (soil:water = 1:2.5) using a pH meter (Sartorius PB-10, Gottingen, Germany). We measured the total organic carbon (TOC) and total nitrogen (TN) contents using a carbon-nitrogen analyzer (Vario Max CN, Elementar, Germany), total phosphorus (TP) content using ammonium molybdate spectrophotometry, and available potassium (AK) content using flame atomic absorption spectroscopy.
2.5 Soil microbial analysis
We extracted DNA from different samples using Omega Bio-Tek (Soil DNA Kit D5625, United States), amplified the V4 region of the bacterial 16S rRNA gene using 515F (5’-GTGYCAGCMGCGGTAA-3′) and 806R (5’-GACTACHVGGGTWTCTAAT-3′) primers, and then amplified the ITS2 region of fungi using ITS1FI2 (5’-GTGARTCATC GAATCTTTG-3′) and ITS2 (5’-TCCTCTTATTGC-3′) primers. PCR amplification was performed in a 25 μl reaction mixture containing 50 ng template DNA, 12.5 μL Phusion Hot start flex 2X Master Mix, and 2.5 μL forward and reverse primers, and ddH2O was used to adjust the volume to 25 μL. The PCR amplification conditions were as follows: initial denaturation at 98°C for 30 s, followed by 32 cycles consisting of denaturation at 98°C for 10 s, annealing at 54°C for 30 s, and extension at 72°C for 45 s. The bacteria and fungi were amplified 35 times and 32 times, respectively, followed by a final extension step at 72°C for 10 min. PCR products were purified and quantified using AMPure XT beads (Beckman Coulter Genomics, Danvers, MA, United States) and Qubit (Invitrogen, United States), respectively. Amplicon library was prepared, and its size and quantity were assessed using an Agilent 2,100 Bioanalyzer (Agilent, United States) and the Library Quantification Kit for Illumina (Kappa Biosciences, Woburn, MA, United States). Finally, we used the NovaSeq PE250 platform to sequence the library.
2.6 Bioinformatics analysis on microbial community
We separated the sample data based on barcode information, removed joints and barcode sequences, and then concatenated and filtered the data. The Differential Amplicon Denoising Algorithm 2 (DADA2) was used for length filtering and denoising, and single base precision biological sequences were obtained and renamed as amplified subsequence variants (ASVs), and singleton ASVs were removed. The feature sequences of each ASV were obtained through taxonomic annotation. Based on the obtained ASV feature sequence and ASV abundance table, the Shannon and Simpson indices of bacteria and fungi in each sample were calculated, and the dominant phyla were determined (relative abundance >1%). The prediction of the functional guilds of bacteria and fungi was performed in FAPROTAX and FUNGuild, respectively.
2.7 Calculation
We defined the number of plant species in each sample plot as plant species richness and measured plant species diversity using the Shannon−Weiner index (Spellerberg and Fedor, 2003). Based on the Z-score of the average abundance of each plant species, a cluster analysis was carried out on the composition of plant species. The microbial community alpha diversity was indicated by the Shannon−Weiner and Simpson indices (Wang et al., 2020). Based on the Bray−Curtis distance, we performed clustering analyses on the composition of microorganisms at the phylum and genus levels. We calculated the abundance of dominant phyla and functional guilds of bacteria and fungi.
2.8 Statistical analysis
One-way ANOVA and Tukey’s HSD multiple comparison tests were used to determine the significance of differences in the soil physicochemical properties and the alpha diversity index of plant communities among different types of grassland. The Kruskal–Wallis test and Steel–Dwass multiple comparison tests were used to determine the significance of differences in the alpha diversity index of microbial community and abundance of dominant phyla and functional guilds of soil microbial community among different types of grassland. Redundant analysis (RDA) was used to identify the effects of plant community composition and soil physicochemical properties on the soil microbial community composition in UM and F-30 m, respectively, taking F-60 m as a reference group. Pearson correlation was used to analyze the relationship between plant community composition, soil physicochemical properties, and the relative abundance of functional guilds of microorganisms in UM and F-30 m, respectively. Finally, we used partial least squares path modeling (PLS-PM) to estimate the influence paths among plant communities, soil properties, and soil microbial communities in UM and F-30 m. The PLS-PM (“plspm” package in R) used PLS regression to estimate the direction and strength of linear correlations between multiple variables. GoF > 0.5 is used as the indicator to determine the fitness of a path model. One-way ANOVA and Kruskal–Wallis tests were completed in SPSS 20.0, while RDA analysis, Pearson correlation, and PLS-PM were conducted in R4.2.2.
3 Results
3.1 Plant and soil microbial community composition and soil physicochemical properties
Plant species richness and Shannon–Weiner index were significantly lower in F-30 m and UM than in F-60 m (one-way ANOVA and Tukey’s HSD test, p < 0.05; Figures 2A,B). Leymus chinensis, Caragana stenophylla, Poa annua, and Cleistogenes squarrosa are the dominant species in three types of grassland (Figure 2C). According to the results of cluster analysis, there were more Gramineae plants in F-60 m than in F-30 m and UM (Figure 2C). The Simpson index of bacterial community in F-30 m significantly decreased (Kruskal–Wallis and Steel–Dwass tests, p < 0.05, Figure 3C). The numbers of ASVs of fungal communities in UM and F-30 m significantly decreased (Kruskal–Wallis and Steel–Dwass tests, p < 0.05, Figure 3D). The Shannon–Weiner index of fungal community significantly decreased in UM (Kruskal–Wallis and Steel–Dwass tests, p < 0.05, Figure 3E). The bacteria genus, fungal genus, and phylum in F-30 m are similar to UM and different from F-60 m (Figures 4A,C,D), while the bacteria phylum in F-30 m is similar to F-60 m and different from UM (Figure 4B). There was no significant difference in the numbers of ASVs and Shannon–Weiner index of bacterial community and Simpson index of fungal community among different types of grassland (Figures 3A,B,F, Kruskal–Wallis, p > 0.05). The abundances of Verrucomirobiota and Methlomirabiota significantly increased, but the abundance of Proteobacteria significantly decreased in UM and F-30 m (Kruskal–Wallis and Steel–Dwass tests, p < 0.05, Figure 5A). The abundances of Chloroflexi and Bacteroidota significantly increased and decreased in UM, respectively (Kruskal–Wallis and Steel–Dwass tests, p < 0.05, Figure 5A). The abundances of Ascomycetes and Fungi_unclassified in UM and F-30 m significantly decreased and increased, respectively (Kruskal–Wallis and Steel–Dwass test, p < 0.05, Figure 5B). The abundance of Basidiomycota significantly increased in UM (Kruskal–Wallis and Steel–Dwass tests, p < 0.05, Figure 5B). The TOC content and SWC in UM and F-30 m significantly decreased. The TN content significantly decreased in F-30 m. The TP content significantly increased in UM (one-way ANOVA and Tukey’s HSD test, p < 0.05, Table 1).
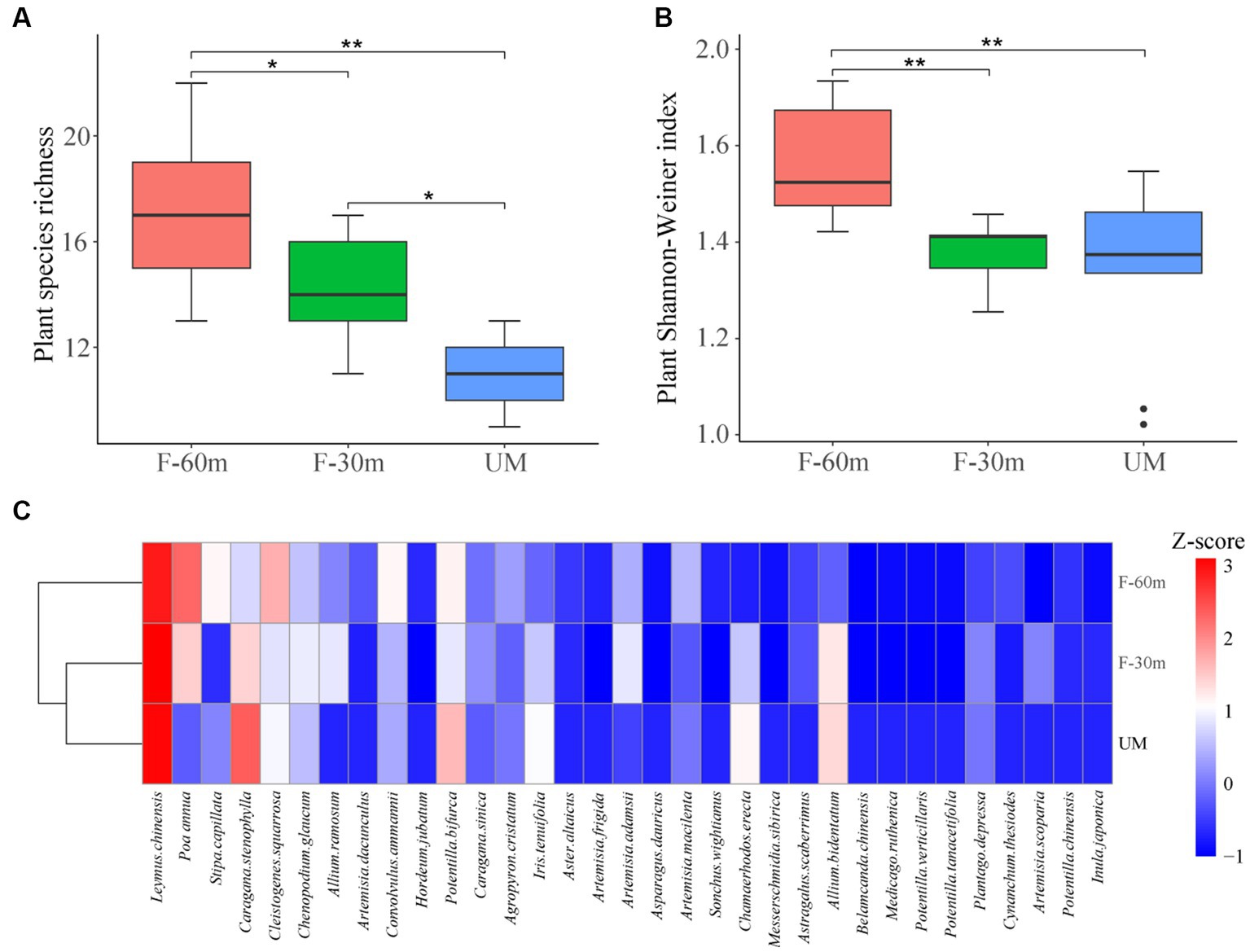
Figure 2. Plant species richness (A), Shannon–Weiner Index (B), and cluster analysis (C) of unmanaged grassland (UM) and the areas 30 m and 30–60 m away from the fence in grazing-banned grassland (F-30 m and F-60 m). ** and * mean one-way ANOVA and Tukey’s HSD test, p < 0.01 and p < 0.05. Z-score is the average abundance of each plant species.
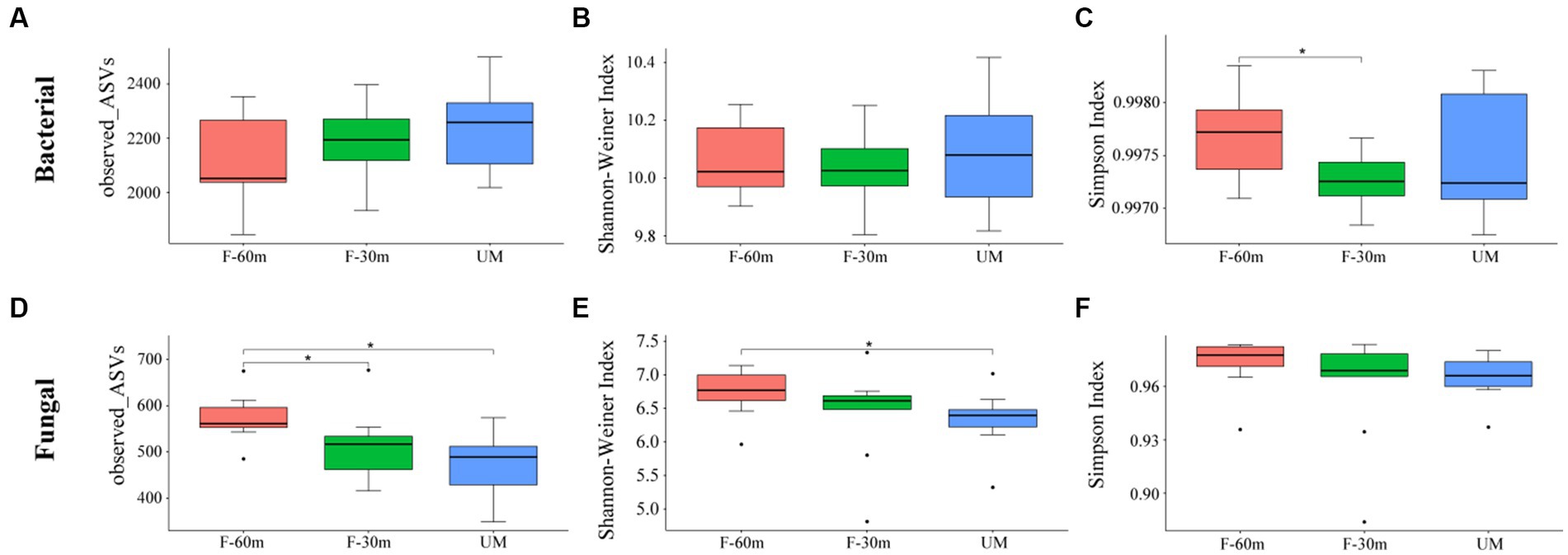
Figure 3. The alpha diversity of bacteria and fungi in unmanaged grassland (UM) and the areas 30 m and 30–60 m away from the fence in grazing-banned grassland (F-30 m and F-60 m). * means Kruskal–Wallis and Steel–Dwass tests p < 0.05. (A) bacterial ASV number; (B) bacterial Shannon–Weiner index; (C) bacterial Simpson index; (D) fungal ASV number; (E) fungal Shannon–Weiner index; (F) fungal Simpson index.
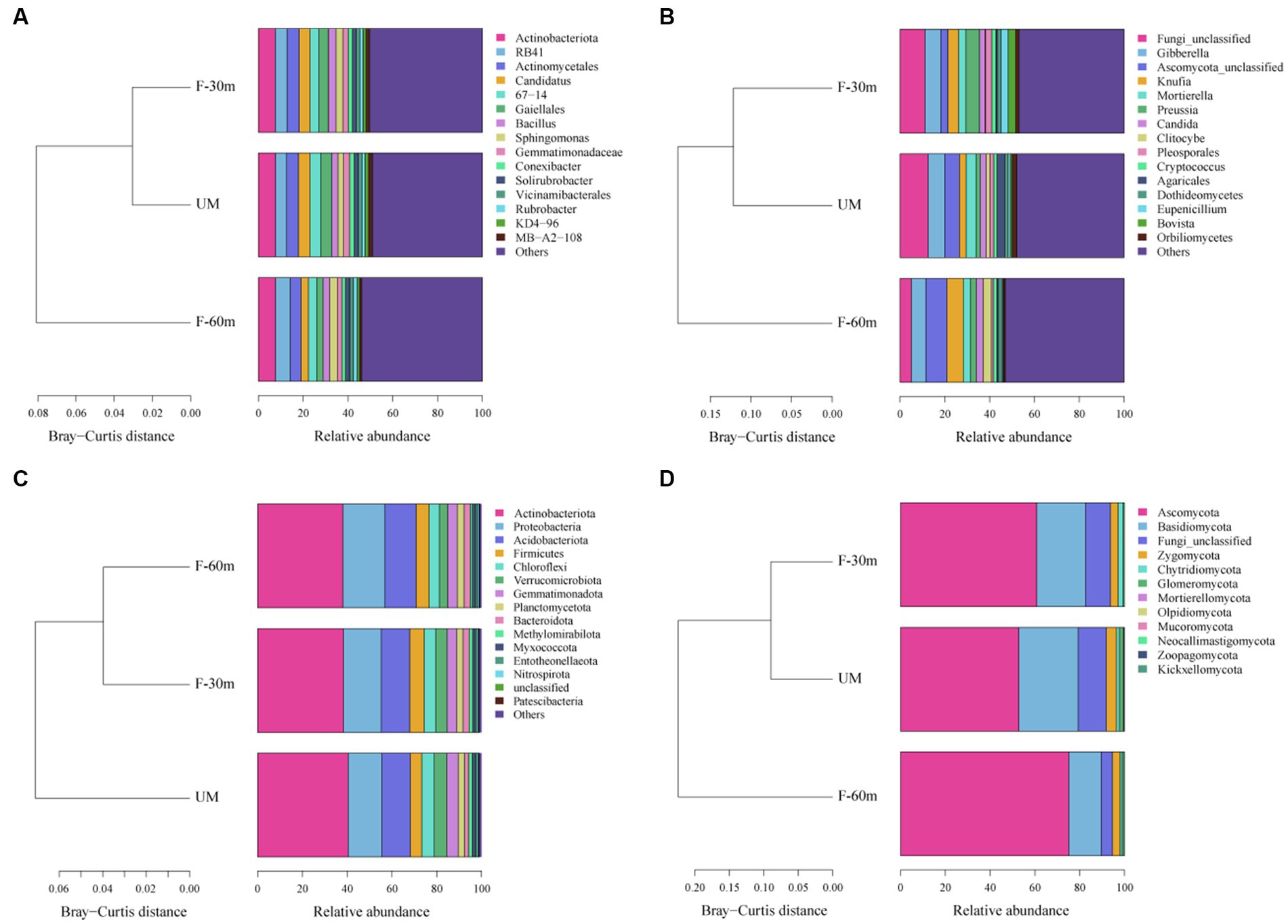
Figure 4. Cluster analyses based on Bray–Curtis distance for dominant bacterial genera (A), fungal genera (B), bacterial phyla (C), and fungal phyla (D) in unmanaged grassland (UM) and the areas 30 m and 30–60 m away from the fence in grazing-banned grassland (F-30 m and F-60 m).
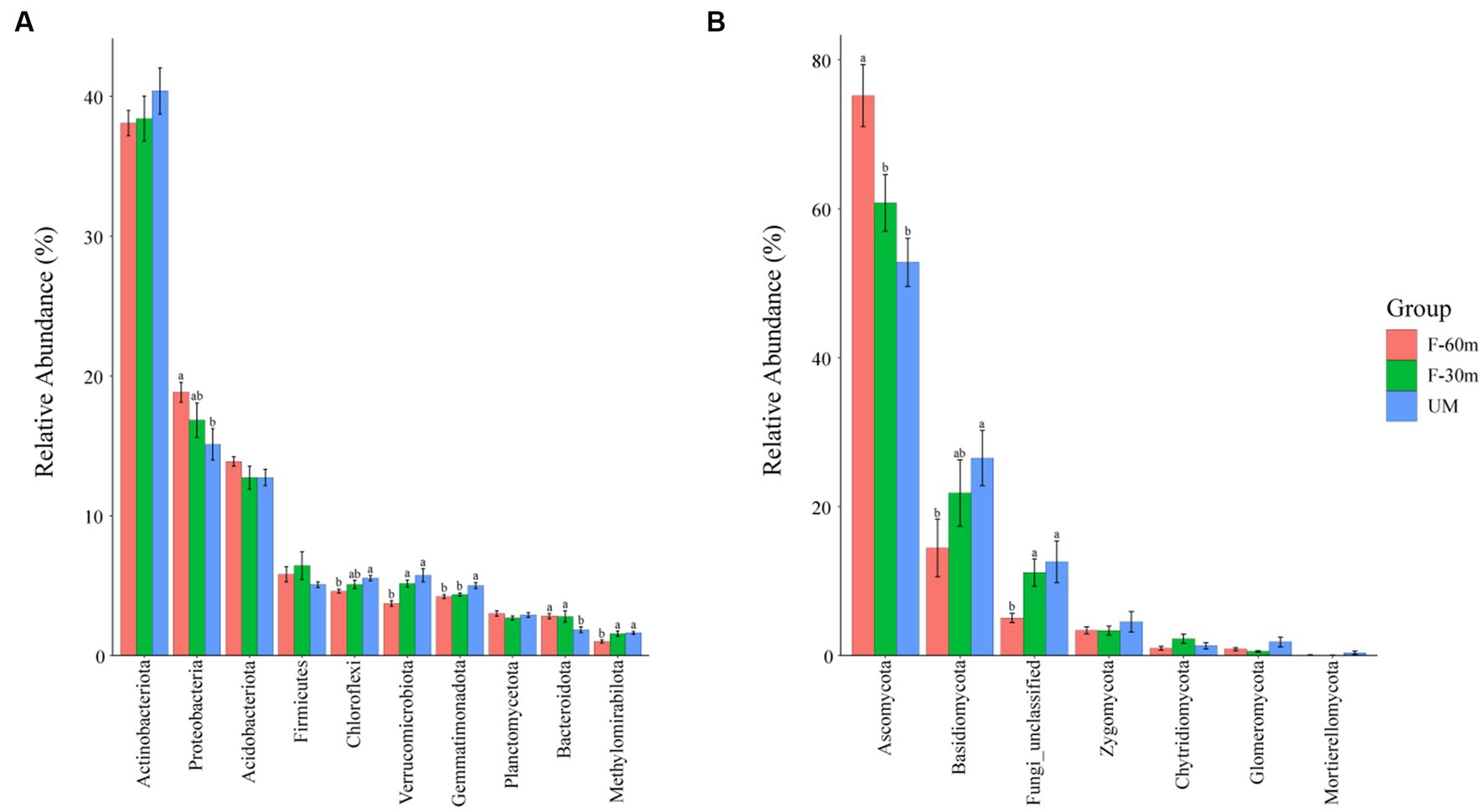
Figure 5. The relative abundance of bacterial dominant phyla (A) and fungal dominant phyla (B) in unmanaged grassland (UM) and the areas 30 m and 30–60 m away from the fence in grazing-banned grassland (F-30 m and F-60 m). The relative abundance of dominant phyla is mean ± standard deviation. The lowercase letters (a, b) on the bar chart indicate significant differences between different grasslands (Kruskal–Wallis and Steel–Dwass tests, p < 0.05).
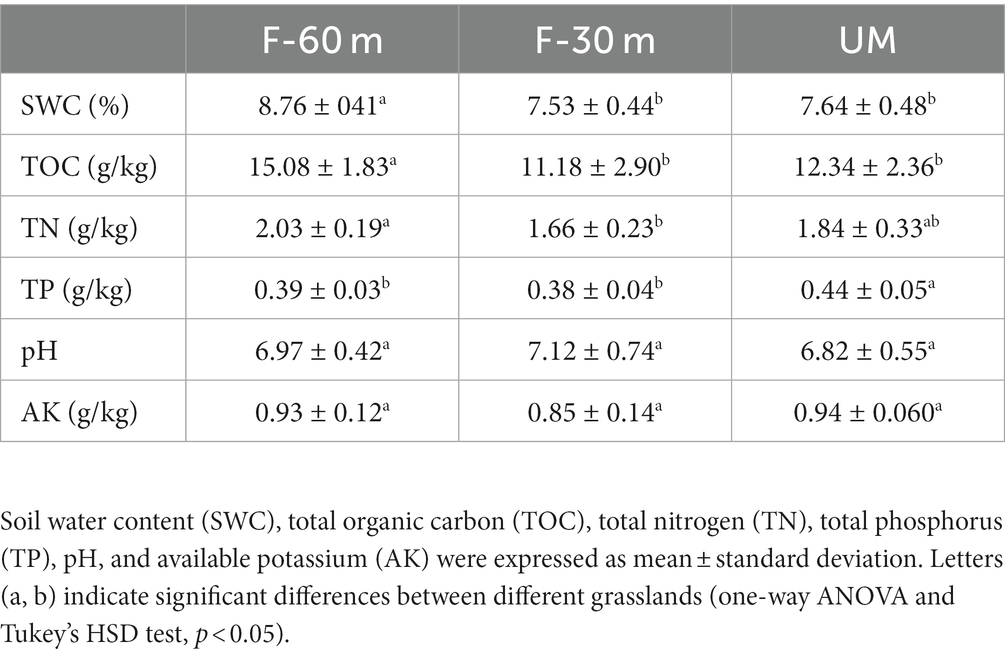
Table 1. Soil physicochemical properties in the unmanaged grassland (UM) and the areas 30 m and 30–60 m away from the fence in grazing-banned grassland (F-30 m and F-60 m).
3.2 Soil microbial functional guilds
Among the bacterial functional guilds, the abundance of chemoheterotrophy and aerobic chemoheterotrophy in UM was significantly lower than in F-60 m (Kruskal–Wallis and Steel–Dwass tests, p < 0.05; Figure 6A). Among the fungal functional guilds, the abundance of undefined saprotroph in F-30 m and UM was significantly lower than in F-60 m (Kruskal–Wallis and Steel–Dwass tests, p < 0.05), while the abundance of dung saprotroph was significantly higher in F-30 m and UM than in F-60 m (Kruskal–Wallis and Steel–Dwass tests, p < 0.05). The abundance of fungal parasites in F-60 m was significantly higher than in UM (Kruskal–Wallis and Steel–Dwass tests, p < 0.05, Figure 6B).
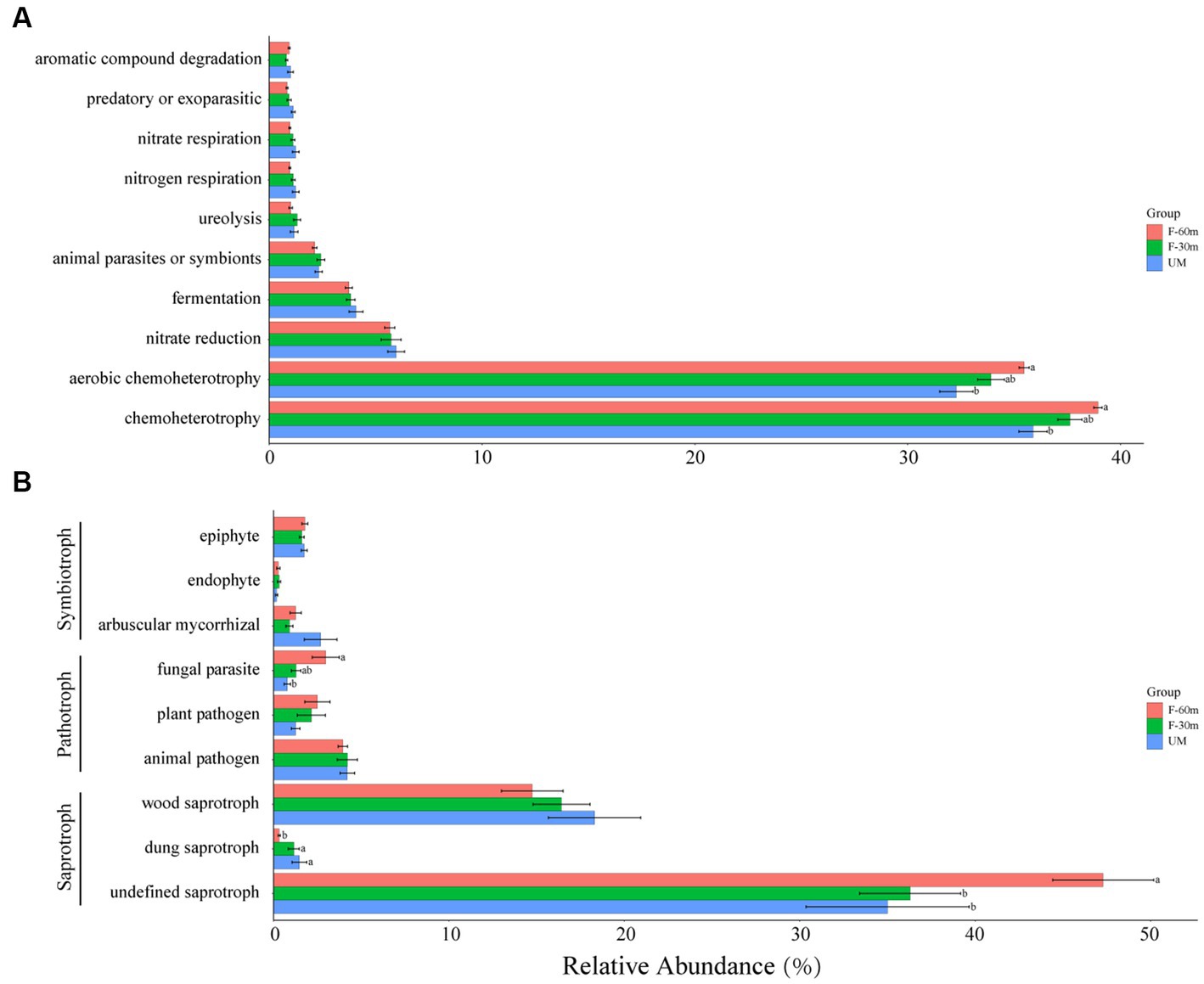
Figure 6. The relative abundance of bacterial functional guilds (A) and fungal functional guilds (B) in unmanaged grassland (UM) and the areas 30 m and 30–60 m away from the fence in grazing-banned grassland (F-30 m and F-60 m). The relative abundance of functional guilds is mean ± standard deviation. The lowercase letters (a, b) on the bar chart indicate significant differences between different grasslands (Kruskal–Wallis and Steel–Dwass tests, p < 0.05).
3.3 Effects of plant community and soil properties on soil microbial community
Plant species richness, plant Shannon–Weiner index, SWC, and the abundance of L. chinensis, P. annua, and C. squarrosa were the main factors affecting the bacterial community composition in UM compared with F-60 m, which explained 53.87% of the variation in the composition of the bacterial communities (Figure 7A). Plant Shannon–Weiner index, SWC, TN, and the abundance of P. annua were the main factors affecting the bacterial community composition in F-30 m compared with F-60 m, which explained 40.98% of the variation in the composition of the bacterial communities (Figure 7B). Plant Shannon–Weiner index, SWC, TOC, TN, and the abundance of P. annua and C. squarrosa were the main factors affecting the fungal community composition in UM compared with F-60 m, which explained 42.54% of the variation in the composition of the fungal communities (Figure 7C). Plant Shannon–Weiner index, TOC, TN, and the abundance of P. annua and C. squarrosa were the main factors affecting the fungal community composition in F-30 m compared with F-60 m, which explained 34.77% of the variation in the composition of the fungal communities (Figure 7D).
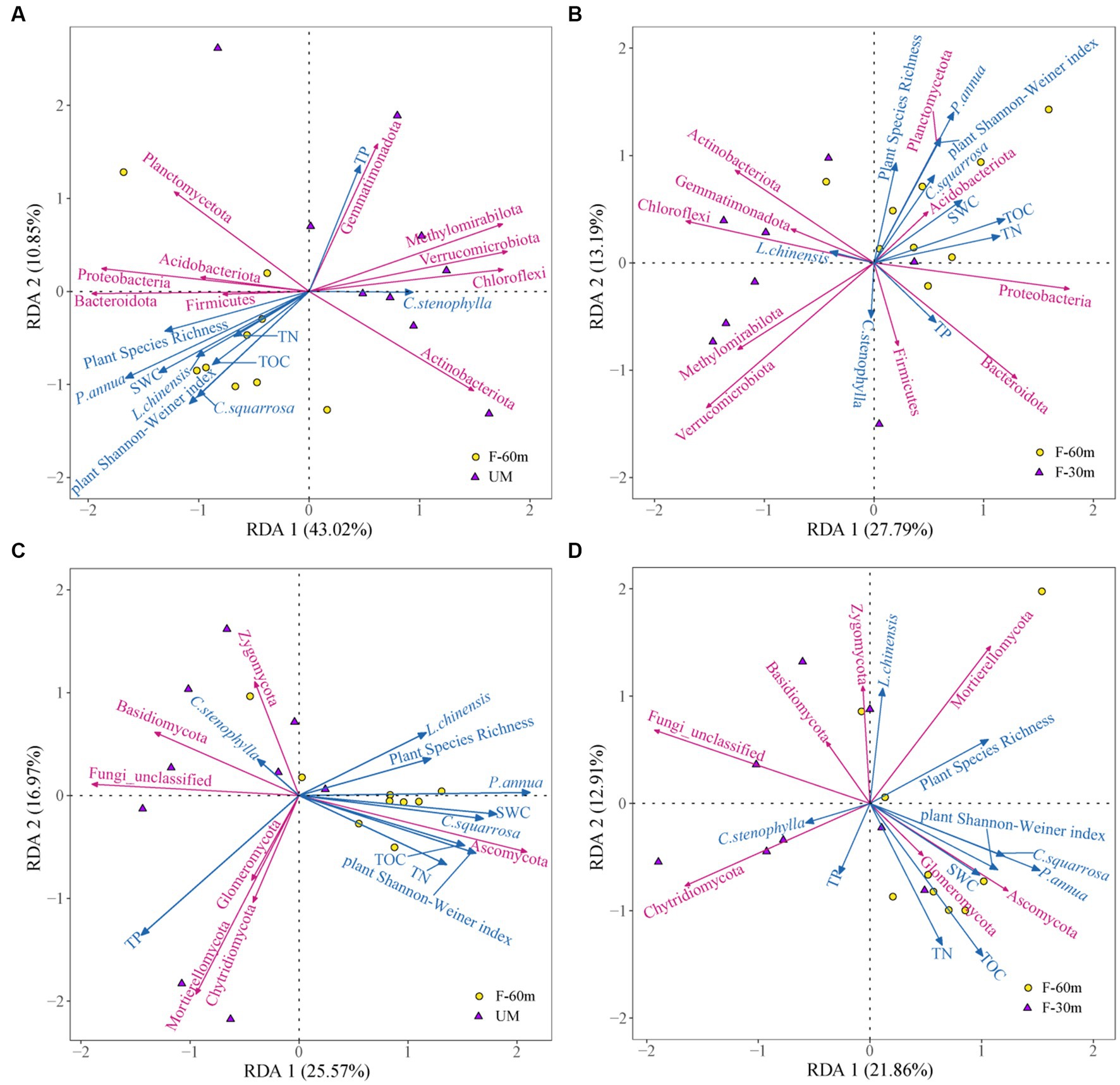
Figure 7. Redundancy analysis on the effect of plant diversity, plant dominant species abundance, and soil properties on soil bacterial communities in UM (A) and F-30 m (B) and soil fungal communities in UM (C) and F-30 m (D) compared with F-60 m.
3.4 Effects of plant community and soil properties on soil microbial functional guilds
In UM and F-60 m, plant species richness, plant Shannon–Weiner index, SWC, and TOC positively correlated with the abundance of chemoheterotroph and aerobic chemoheterotroph bacteria (p < 0.05, Figure 8A). Three plant species positively or negatively correlated with the abundance of different bacteria functional guilds (p < 0.05, Figure 8A). SWC positively correlated with the abundance of parasite fungi (p < 0.05, Figure 8C). Two plant species positively or negatively correlated with the abundance of different fungal functional guilds (p < 0.05, Figure 8C). In F-30 m and F-60 m, plant Shannon–Weiner index and SWC positively correlated with the abundance of chemoheterotroph and aerobic chemoheterotroph bacteria (p < 0.05, Figure 8B). TOC positively and negatively correlated with the abundance of undefined saprotroph fungi and dung saprotroph fungi, respectively (p < 0.05, Figure 8D); SWC positively correlated with the abundance of parasite fungi (p < 0.05, Figure 8D). Three plant species positively or negatively correlated with the abundance of different fungal functional guilds (p < 0.05, Figure 8D).
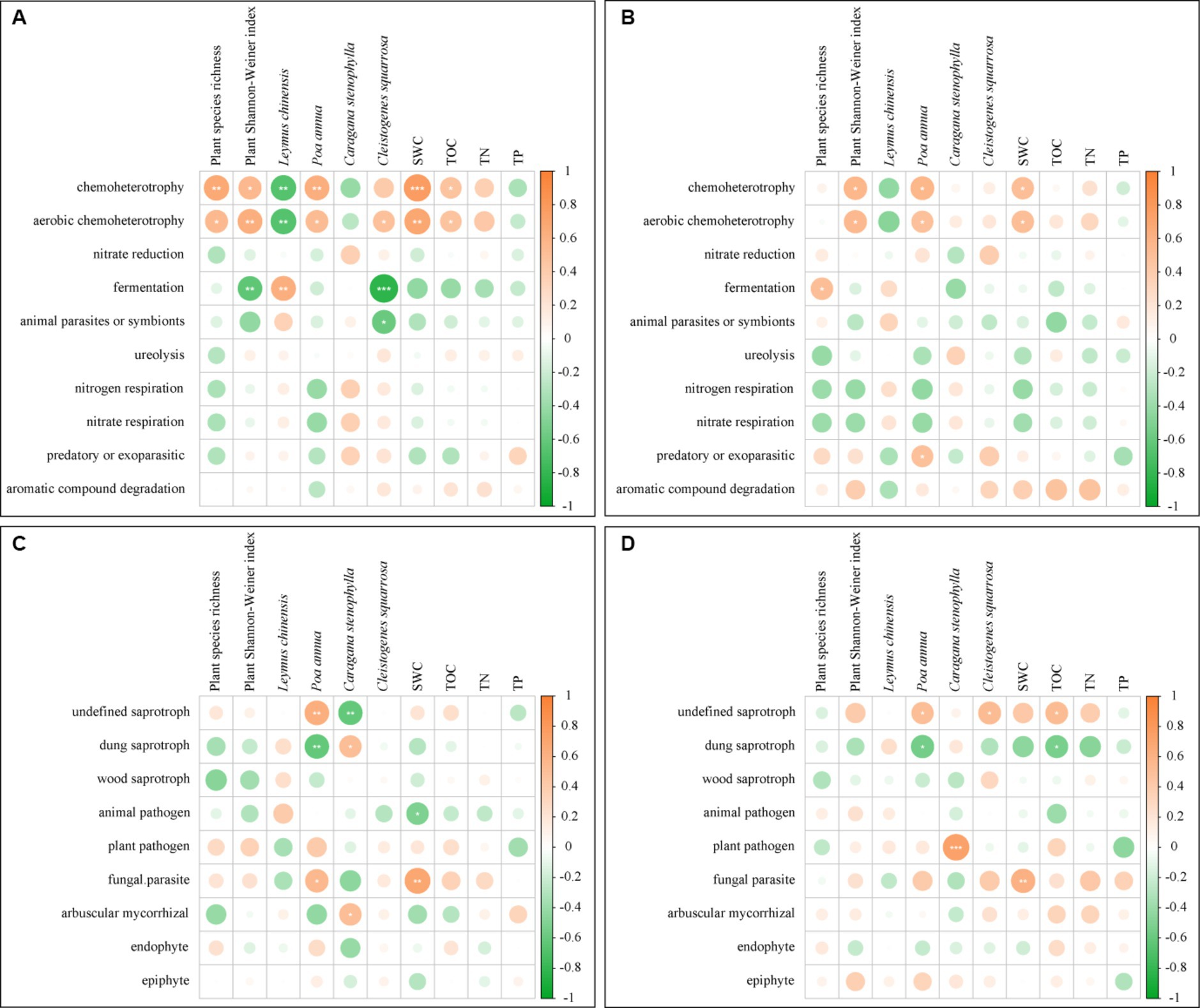
Figure 8. The Pearson correlation between the bacterial and fungal functional guilds and plant diversity, relative abundance of dominant plants, and soil physicochemical properties [(A) bacterial guilds in UM and F-60 m samples; (B) bacterial guilds in F-30 m and F-60 m samples; (C) fungal guilds in UM and F-60 m samples; (D) fungal guilds in F-30 m and F-60 m samples]. SWC, soil water content; TOC, total organic carbon; TN, total nitrogen; and TP, total phosphorus. * p < 0.05, ** p < 0.01, and *** p < 0.001.
3.5 Path model of the plant community, soil microbial community, and soil properties in UM and F-30 m
According to the results in 3.4, we selected factors that significantly affect soil microbial function guilds to construct three variables, including plant diversity (plant species richness and Shannon–Weiner index), dominant species abundance (Leymus chinensis, Caragana stenophyll, Poa annua, and Cleistogenes squarrosa), and soil properties (SWC and TOC). The PLS-PM estimation was performed between these three variables and soil microbial functions. The path model with GoF = 0.56 is shown in Figure 9. Plant diversity in UM had a positive effect on the bacterial function in UM, while the abundance of dominant species in UM had a negative effect on the fungal function in UM. The bacterial and fungal functions in UM had positive effects on soil properties in UM. The soil properties in UM had a direct positive effect on the soil properties of F-30 m. The soil properties in F-30 m had positive effects on bacterial and fungal functions in F-30 m. Bacterial function in F-30 m had positive effects on plant diversity and dominant species abundance in F-30 m. Fungal function in F-30 m had a positive effect on the abundance of dominant species in F-30 m.
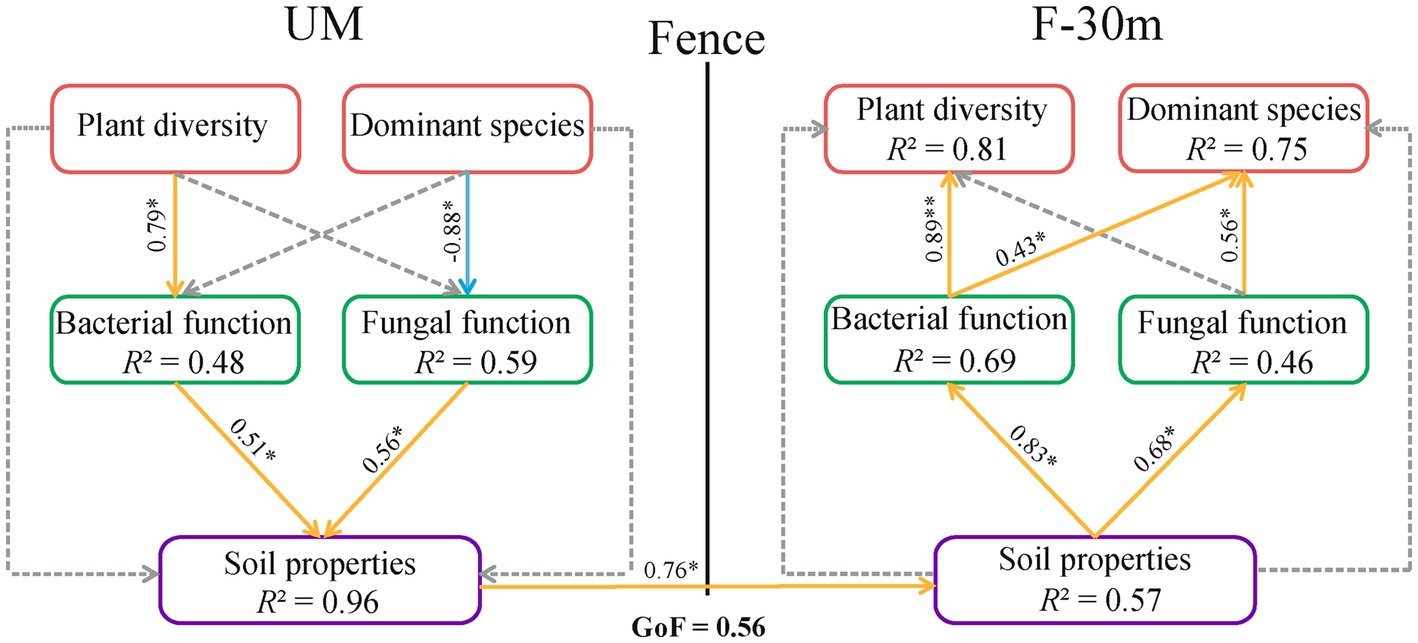
Figure 9. The partial least squares path model (PLS-PM) of the plant diversity, the abundance of dominant species, soil microbial function, and soil properties in UM and F-30 m. The orange and blue arrows indicate positive and negative effects (p < 0.05), respectively. The dashed line represents a non-significant (p > 0.05) relationship. Plant diversity includes plant species richness and Shannon–Weiner index; dominant species include Leymus chinensis, Caragana stenophyll, Poa annua, and Cleistogenes squarrosa; Soil properties include SWC (soil water content) and TOC (total organic carbon).* means p < 0.05.
4 Discussion
Numerous studies have demonstrated that long-term repeated grazing can reduce plant diversity and vegetation productivity (Eldridge and Delgado-Baquerizo, 2017; Xun et al., 2018). The results showed that UM had significantly lower plant richness and diversity, indicating the degradation of unmanaged areas. The results of the plant Shannon index and species cluster analysis showed that the plant community in F-30 m was similar to that in UM and different from that in F-60 m (Figure 2), indicating that the area adjacent to UM in the grazing-banned grassland exhibited similar degradation to unmanaged grassland, although there was no grazing in this area. Plant community degradation can cause significant changes in microbial communities (Zhou et al., 2019; Wang et al., 2022). The results of fungi ASVs, Shannon index, and cluster analysis showed that the fungi community in F-30 m was also similar to UM and different from F-60 m. The bacterial community at the genus level in the F-30 m was similar to UM. The abundance of stress-resistant microorganisms significantly increased in UM and F-30 m areas, such as Verrucomirobiota and Methylomirabilota, appearing in drought and hypoxic environments (Chowdhury et al., 2019; Zhu et al., 2022). The abundance of bacteria phylum Proteobacteria involved in litter decomposition and fungal phylum Ascomycota involved in degrading lignin and cellulose (Fierer et al., 2007; Wang et al., 2020; Zheng et al., 2021) significantly decreased in UM and F-30 m areas. These results support our first prediction that the structures of plant and soil microbial communities in the areas adjacent to unmanaged grassland in grazing-banned grassland are similar to those in unmanaged grassland but different from those in the areas not adjacent to unmanaged grassland.
The bacterial functional guild analysis reveals that the levels of chemoheterotroph and aerobic chemoheterotroph, which are involved in the transformation of organic matter into soil (Lian et al., 2021; Zheng et al., 2023), are significantly lower in UM and F-30 m than in F-60 m. Thus, the decomposition of soil organic matter in the unmanaged grassland and the edge area in the grazing-banned grassland is weak, which is detrimental to the soil carbon cycling in these areas. The fungal parasite and saprophytic fungal guilds contributed to the majority of differences observed among the three types of grassland. The significant decrease in fungal parasites in UM and F-30 m is related to a decrease in plant diversity, which leads to a decrease in the host population (Maurice et al., 2021). There was a significant decrease in the abundance of undefined saprotrophs in UM and F-30 m. Saprophytic fungi play a crucial role in organic matter decomposition, facilitating the cycling of key soil nutrient elements, affecting species coexistence by altering soil nutrients, and maintaining stable vegetation productivity (Crowther et al., 2012; Chen et al., 2019; Liu et al., 2022). The bacterial and fungal functional guild analysis jointly suggest that soil nutrient cycling in F-30 m and UM may be inhibited.
Long-term overgrazing can reduce plant diversity, dominant species abundance, soil nutrients, and water content in grassland (Wu et al., 2014; Ji et al., 2022; Luo et al., 2022), which is reflected by the plant Shannon–Weiner index, SWC, TOC, and TN reduction in the UM grassland in our study. Human interference can negatively alter soil properties in edge areas, such as TOC, TN, and C/N (Malmivaara-Lämsä et al., 2008). Our results showed that the SWC, TOC, and TN in the F-30 m are similar to those in UM, which indicate that the soil properties in the grazing-banned area adjacent to unmanaged grassland degrade. Degradation in both plant community and soil properties can alter the soil bacterial and fungal composition (Parton et al., 1995; Manzoni et al., 2012; Steinauer et al., 2015; Zhou et al., 2019; Huang et al., 2021; Wang et al., 2021; Yu et al., 2021), especially reducing the microorganisms related to nutrient cycling (Pennanen, 2001; Malmivaara-Lämsä et al., 2008). Changes in plant diversity can alter the composition and function of soil microorganisms by affecting litter input (Steinauer et al., 2015). SWC can directly affect the transformation of soil nutrients and soil microbial metabolism, thereby affecting the composition and function of soil microbial communities (Parton et al., 1995; Manzoni et al., 2012). TOC can provide energy and carbon sources for soil microorganisms (Zhou et al., 2019; Wang et al., 2021). In our RDA and Pearson correlation results, compared with F-60 m, the plant Shannon–Weiner index and the abundance of Poa annua influence soil microbial composition and functional guilds in both UM and F-30 m, while the soil TOC and SWC influence the soil microbial functional guilds in both UM and F-30 m. The decrease in plant Shannon–Weiner index, Poa annua, TOC, and SWC in UM and F-30 m results in the decrease in chemoheterotroph and aerobic chemoheterotroph bacteria, saprotroph, and parasite fungi in UM and F-30 m, which may reduce nutrient cycling efficiency. These results indicate that the grazing-banned area adjacent to unmanaged grassland exhibits a degraded interaction between plant and microbial communities just like unmanaged grassland.
It has been known that plant community degradation can reduce the abundance of microorganisms related to nutrient cycling and degrade the soil properties of the adjacent area (Pennanen, 2001; Malmivaara-Lämsä et al., 2008). Changes in soil water and nutrient supply may in turn affect the structure and diversity of plant community and ultimately lead to the vegetation degradation of the adjacent area (Ettema and Wardle, 2002). Our PLS path model indicates that plant diversity or dominant species in UM can affect the soil water and organic carbon contents in UM by affecting the bacterial and fungal functional guilds, and the changes of soil properties in UM influence the soil properties in F-30 m. Subsequently, the changes in soil properties in F-30 m affect the bacterial and fungal functional guilds in F-30 m and then affect the plant diversity and dominant species in F-30 m. These results support our second prediction that the degradation of plant community, soil properties, and soil microbial community in unmanaged grassland interfere with soil microbial communities and, consequently, the plant restoration in the adjacent grazing-banned grassland. Such edge effects may gradually interfere with more areas in grazing-banned grassland.
In summary, our results showed that soil microbial composition, soil properties, and plant community composition in the grazing-banned grassland adjacent to unmanaged grassland are similar to those in degraded unmanaged grassland, indicating that the degradation of unmanaged grasslands can affect the restoration of the grazing-banned grasslands via negative edge effect. In our study area, the grazing-banned grassland has been fenced for 15 years. Long-term fencing increased grazing pressure in unmanaged grasslands, and degradation in these areas interfered with the restoration of fenced areas. Traditional nomadism in the Inner Mongolia grassland is distinguished by mobile grazing which helps prevent grassland degradation caused by long-term sedentary grazing on the same grassland (Yamamura et al., 2013). Thus, we suggest combining “grazing ban” with traditional nomadism to avoid excessive use of unmanaged grassland. The fence should be removed according to the vegetation restoration status. The study on the Tibetan plateau showed that degraded grassland can be restored after 4 to 8 years of fencing (Sun et al., 2020); thus, grazing-banned areas should be adjusted after such a period in order to avoid the negative edge effect.
5 Conclusion
The soil microbial and plant communities in the edge zone of grazing-banned grassland have been similar to those in adjacent unmanaged overgrazed grassland. The abundance of microorganisms was related to nutrient cycling reduced in both unmanaged and adjacent grazing-banned grasslands. The plant diversity, dominant plant species, soil organic carbon, and water content can explain the soil microbial community variation in both unmanaged and adjacent grazing-banned grasslands. Changes in soil water and organic carbon contents in unmanaged grassland affect these soil properties in adjacent grazing-banned grassland and subsequently affect soil microbial functions and plant community. Degradation in unmanaged grassland interferes with the restoration of grazing-banned grassland via negative edge effects.
Data availability statement
Microbial raw sequence data that support the findings of this study are openly available in NCBI at Sequence Read Archive (SRA), reference number PRJNA1047307 (bacteria) and PRJNA1047293 (fungi).
Author contributions
MF: Investigation, Software, Writing – original draft. GL: Writing – review & editing, Investigation. SZ: Writing – review & editing, Conceptualization, Supervision. WL: Writing – review & editing, Conceptualization.
Funding
The author(s) declare financial support was received for the research, authorship, and/or publication of this article. This study was supported by the Key laboratory of Ecology and Environment in Minority Area, National Ethnic Affairs Commission (KLEEMA202207) for the research and received no specific grant from any public and commercial funding agencies.
Acknowledgments
The authors are grateful to Shuling Yan, Batu Wang, Muren Wu, and Songtao Liu in Hulun Lake National Nature Reserve for their help with the fieldwork.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
Bardgett, R. D., Bullock, J. M., Lavorel, S., Manning, P., Schaffner, U., Ostle, N., et al. (2021). Combatting global grassland degradation. Nat. Rev. Earth Environ. 2, 720–735. doi: 10.1038/s43017-021-00207-2
Bardgett, R. D., and van der Putten, W. H. (2014). Belowground biodiversity and ecosystem functioning. Nature 515, 505–511. doi: 10.1038/nature13855
Bissett, A., and Burke, C. (2007). Bacterial community shifts in organically perturbed sediments. Environ. Microbiol. 9, 46–60. doi: 10.1111/j.1462-2920.2006.01110.x
Blanchard, G., Barbier, N., Vieilledent, G., Ibanez, T., Hequet, V., McCoy, S., et al. (2023). UAV-Lidar reveals that canopy structure mediates the influence of edge effects on forest diversity, function and microclimate. J. Ecol. 111, 1411–1427. doi: 10.1111/1365-2745.14105
Boval, M., and Dixon, R. M. (2012). The importance of grasslands for animal production and other functions: a review on management and methodological progress in the tropics. Animal 6, 748–762. doi: 10.1017/S1751731112000304
Canarini, A., Kaiser, C., Merchant, A., Richter, A., and Wanek, W. (2019). Root exudation of primary metabolites: mechanisms and their roles in plant responses to environmental stimuli. Front. Plant Sci. 10:157. doi: 10.3389/fpls.2019.00157
Chen, L., Swenson, N., Ji, N. N., Mi, X. C., Ren, H. B., Guo, L. D., et al. (2019). Differential soil fungus accumulation and density dependence of trees in a subtropical forest. Science 366, 124–128. doi: 10.1126/science.aau1361
Cheng, J. M., Jing, G. H., Wei, L., and Jing, Z. B. (2016). Long-term grazing exclusion effects on vegetation characteristics, soil properties and bacterial communities in the semi-arid grasslands of China. Ecol. Eng. 97, 170–178. doi: 10.1016/j.ecoleng.2016.09.003
Chowdhury, T. R., Lee, J., Bottos, E. M., Brislawn, C. J., White, R. A. III, Bramer, L. M., et al. (2019). Metaphenomic responses of a native prairie soil microbiome to moisture perturbations. mSystems 4, e00061–e00019. doi: 10.1128/mSystems.00061-19
Coban, O., De Deyn, G. B., and van der Ploeg, M. (2022). Soil microbiota as game-changers in restoration of degraded lands. Science 375:abe0725. doi: 10.1126/science.abe0725
Crowther, T. W., Boddy, L., and Hefin, J. T. (2012). Functional and ecological consequences of saprotrophic fungus-grazer interactions. ISME J. 6, 1992–2001. doi: 10.1038/ismej.2012.53
Eldegard, K., Totland, Ø., and Moe, S. R. (2015). Edge effects on plant communities along power line clearings. J. Appl. Ecol. 52, 871–880. doi: 10.2307/43869259
Eldridge, D. J., and Delgado-Baquerizo, M. (2017). Continental-scale impacts of livestock grazing on ecosystem supporting and regulating services. Land Degrad. Dev. 28, 1473–1481. doi: 10.1002/ldr.2668
Ettema, C. H., and Wardle, D. A. (2002). Spatial soil ecology. Trends Ecol. Evol. 17, 177–183. doi: 10.1016/S0169-5347(02)02496-5
FAO . (1988). Soil map of the world. Revised Legend. World soil resources Report No. 60. Rome: Food and Agriculture Organization of the United Nations.
Fierer, N., Bradford, M. A., and Jackson, R. B. (2007). Toward an ecological classification of soil bacteria. Ecology 88, 1354–1364. doi: 10.1890/05-1839
Fischer, R., Taubert, F., Müller, M. S., Groeneveld, J., Lehmann, S., Wiegand, T., et al. (2021). Accelerated forest fragmentation leads to critical increase in tropical forest edge area. Sci. Adv. 7:eabg7012. doi: 10.1126/sciadv.abg7012
Fonseca, C. R., and Joner, F. (2007). Two-sided edge effect studies and the restoration of endangered ecosystems. Restor. Ecol. 15, 613–619. doi: 10.1111/j.1526-100X.2007.00273.x
Gao, W. B., Jiang, H. T., Zhang, S., Hai, C. X., and Liu, B. Y. (2022). Vegetation characteristics and soil properties in grazing exclusion areas of the inner Mongolia desert steppe. Int. Soil Water Conserv. Res. 11, 549–560. doi: 10.1016/j.iswcr.2022.11.005
Gibbs, H. K., and Salmon, J. M. (2015). Mapping the world’s degraded lands. Appl. Geogr. 57, 12–21. doi: 10.1016/j.apgeog.2014.11.024
Guerra, T. N. F., Araújo, E. L., Sampaio, E. V. S. B., and Ferraz, E. M. N. (2017). Urban or rural areas: which types of surrounding land use induce stronger edge effects on the functional traits of tropical forests plants? Appl. Veg. Sci. 20, 538–548. doi: 10.1111/avsc.12315
Hao, L., Sun, G., Liu, Y. Q., Gao, Z. Q., He, J. J., Shi, T. T., et al. (2014). Effects of precipitation on grassland ecosystem restoration under grazing exclusion in Inner Mongolia, China. Landsc. Ecol. 29, 1657–1673. doi: 10.1007/s10980-014-0092-1
Hartmann, M., and Six, J. (2023). Soil structure and microbiome functions in agroecosystems. Nat. Rev. Earth Environ. 4, 4–18. doi: 10.1038/s43017-022-00366-w
Huang, R. L., Crowther, T. W., Sui, Y. Y., Sun, B., and Liang, Y. T. (2021). High stability and metabolic capacity of bacterial community promote the rapid reduction of easily decomposing carbon in soil. Commun. Biol. 4:1376. doi: 10.1038/s42003-021-02907-3
Ji, G. X., Hu, G. F., Liu, G. F., Bai, Z. J., Li, B., Li, D. D., et al. (2022). Response of soil microbes to Carex meyeriana meadow degeneration caused by overgrazing in inner Mongolia. Acta Oecol. 117:103860. doi: 10.1016/j.actao.2022.103860
Kark, S. (2013). “Ecotones and ecological gradients” in Ecological systems. ed. R. Leemans (New York, NY: Springer)
Koelemeijer, I. A., Ehrlé, J., De Frenne, P., Jönsson, M. T., Berg, P., and Hylander, K. (2023). Forest edge effects on moss growth are amplified by drought. Ecol. Appl. 33:e2851. doi: 10.1002/eap.2851
Kolås, Å. (2014). Degradation discourse and green governmentality in the Xilinguole grasslands of inner Mongolia. Dev. Change 45, 308–328. doi: 10.1111/dech.12077
Krishnadas, M., Bagchi, R., Sridhara, S., and Comita, L. S. (2018). Weaker plant-enemy interactions decrease tree seedling diversity with edge-effects in a fragmented tropical forest. Nat. Commun. 9:4523. doi: 10.1038/s41467-018-06997-2
Lapola, D., Pinho, P., Barlow, J., Aragão, L. E. O. C., Berenguer, E., Carmenta, R., et al. (2023). The drivers and impacts of Amazon forest degradation. Science 379:eabp8622. doi: 10.1126/science.abp8622
Li, W., Cao, W. X., Wang, J. L., Li, X. L., Xu, C. L., and Shi, S. L. (2017). Effects of grazing regime on vegetation structure, productivity, soil quality, carbon and nitrogen storage of alpine meadow on the Qinghai-Tibetan plateau. Ecol. Eng. 98, 123–133. doi: 10.1016/j.ecoleng.2016.10.026
Li, C., Xiao, C. W., Li, M. X., Xu, L., and He, N. P. (2023). The quality and quantity of SOM determines the mineralization of recently added labile C and priming of native SOM in grazed grasslands. Geoderma 432:116385. doi: 10.1016/j.geoderma.2023.116385
Lian, J. S., Wang, H. Y., Xu, M. G., Wei, W. L., Duan, Y. H., and Liu, S. T. (2021). Diversity and function prediction of bacteria community in fluvo-aquic soils as affected by long-term organic fertilization. J. Plant Nutr. Fertil. 27, 2073–2082. doi: 10.11674/zwyf.2021225
Liu, S. G., García-Palacios, P., Tedersoo, L., Guirado, E., Van der Heijden, M. G. A., Wagg, C., et al. (2022). Phylotype diversity within soil fungal functional groups drives ecosystem stability. Nat. Ecol. Evol. 6, 900–909. doi: 10.1038/s41559-022-01756-5
Liu, Y. Y., Wang, Q., Zhang, Z. Y., Tong, L. J., Wang, Z. Q., and Li, J. L. (2019). Grassland dynamics in responses to climate variation and human activities in China from 2000 to 2013. Sci. Total Environ. 690, 27–39. doi: 10.1016/j.scitotenv.2019.06.503
Luo, Z. M., Liu, J. X., He, L., Du, J. Q., Wang, L. X., Jia, T., et al. (2022). Degradation-induced microbiome alterations may aggravate soil nutrient loss in subalpine meadows. Land Degrad. Dev. 33, 2699–2712. doi: 10.1002/ldr.4289
Lyu, X., Li, X. B., Dang, D. L., Dou, H. S., Xuan, X. J., Liu, S. Y., et al. (2020). A new method for grassland degradation monitoring by vegetation species composition using hyperspectral remote sensing. Ecol. Indic. 114:106310. doi: 10.1016/j.ecolind.2020.106310
Malmivaara-Lämsä, M., Hamberg, L., Haapamäki, E., Liski, J., Kotze, J., Lehvävirta, S., et al. (2008). Edge effects and trampling in boreal urban forest fragments – impacts on the soil microbial community. Soil Biol. Biochem. 40, 1612–1621. doi: 10.1016/j.soilbio.2008.01.013
Manzoni, S., Schimel, J. P., and Porporato, A. (2012). Responses of soil microbial communities to water stress: results from a meta-analysis. Ecology 93, 930–938. doi: 10.1890/11-0026.1
Maurice, S., Arnault, G., Nordén, J., Botnen, S. S., Miettinen, O., and Kauserud, H. (2021). Fungal sporocarps house diverse and host-specific communities of fungicolous fungi. ISME J. 15, 1445–1457. doi: 10.1038/s41396-020-00862-1
Meier, I. C., Finzi, A. C., and Phillips, R. P. (2017). Root exudates increase N availability by stimulating microbial turnover of fast-cycling N pools. Soil Biol. Biochem. 106, 119–128. doi: 10.1016/j.soilbio.2016.12.004
Parton, W. J., Scurlock, J. M. O., Ojima, D. S., Schimel, D. S., Hall, D. O., Scopegram Group Members, et al. (1995). Impact of climate change on grassland production and soil carbon worldwide. Glob. Chang. Biol. 1, 13–22. doi: 10.1111/j.1365-2486.1995.tb00002.x
Pei, S. F., Fu, H., and Wan, C. G. (2008). Changes in soil properties and vegetation following exclosure and grazing in degraded Alxa desert steppe of Inner Mongolia, China. Agric. Ecosyst. Environ. 124, 33–39. doi: 10.1016/j.agee.2007.08.008
Pennanen, T. (2001). Microbial communities in boreal coniferous forest humus exposed to heavy metals and changes in soil pH – a summary of the use of phospholipid fatty acids, biolog® and 3H-thymidine incorporation methods in field studies. Geoderma 100, 91–126. doi: 10.1016/S0016-7061(00)00082-3
Schmidt, M., Jochheim, H., Kersebaum, K. C., Lischeid, G., and Nendel, C. (2017). Gradients of microclimate, carbon and nitrogen in transition zones of fragmented landscapes–a review. Agric. For. Meteorol. 232, 659–671. doi: 10.1016/j.agrformet.2016.10.022
Smith, A. J., and Goetz, E. M. (2021). Climate change drives increased directional movement of landscape ecotones. Landsc. Ecol. 36, 3105–3116. doi: 10.1007/s10980-021-01314-7
Spellerberg, I. F., and Fedor, P. J. (2003). A tribute to Claude Shannon (1916–2001) and a plea for more rigorous use of species richness, species diversity and the “Shannon–wiener” index. Glob. Ecol. Biogeogr. 12, 177–179. doi: 10.1046/j.1466-822X.2003.00015.x
Steinauer, K., Tilman, D., Wragg, P. D., Cesarz, S., Cowles, J. M., Pritsch, K., et al. (2015). Plant diversity effects on soil microbial functions and enzymes are stronger than warming in a grassland experiment. Ecology 96, 99–112. doi: 10.1890/14-0088.1
Sun, J., Liu, M., Fu, B. J., Kemp, D., Zhao, W. W., Liu, G. H., et al. (2020). Reconsidering the efficiency of grazing exclusion using fences on the Tibetan plateau. Sci. Bull. 65, 1405–1414. doi: 10.1016/j.scib.2020.04.035
Sun, J., Wang, Y., Piao, S. L., Liu, M., Han, G. D., Li, J. R., et al. (2022). Toward a sustainable grassland ecosystem worldwide. Innovations 3:100265. doi: 10.1016/j.xinn.2022.100265
Török, P., Brudvig, L. A., Kollmann, J., Price, J., and Tóthmérész, B. (2021). The present and future of grassland restoration. Restor. Ecol. 29:e13378. doi: 10.1111/rec.13378
van der Heijden, M. G. A., Bardgett, R. D., and van Straalen, N. M. (2008). The unseen majority: soil microbes as drivers of plant diversity and productivity in terrestrial ecosystems. Ecol. Lett. 11, 296–310. doi: 10.1111/j.1461-0248.2007.01139.x
Wang, K. B., Deng, L., Ren, Z. P., Li, J. P., and Shangguan, Z. P. (2016). Grazing exclusion significantly improves grassland ecosystem C and N pools in a desert steppe of Northwest China. Catena 137, 441–448. doi: 10.1016/j.catena.2015.10.018
Wang, F., Gao, L. J., and Zhang, S. P. (2020). Effects of bird aggregation on the soil properties and microbial community diversity of urban forest fragments. Sci. Total Environ. 737:140250. doi: 10.1016/j.scitotenv.2020.140250
Wang, C., Qu, L. R., Yang, L. M., Liu, D. W., Morrissey, E., Miao, R. H., et al. (2021). Large-scale importance of microbial carbon use efficiency and necromass to soil organic carbon. Glob. Chang. Biol. 27, 2039–2048. doi: 10.1111/gcb.15550
Wang, D. L., Wang, L., Liu, J. S., Zhu, H., and Zhong, Z. W. (2018). Grassland ecology in China: perspectives and challenges. Front. Agric. Sci. Eng. 5, 24–43. doi: 10.15302/J-FASE-2018205
Wang, D. J., Zhou, H. K., Zuo, J., Chen, P., She, Y. D., Yao, B. Q., et al. (2022). Responses of soil microbial metabolic activity and community structure to different degraded and restored grassland gradients of the Tibetan plateau. Front. Plant Sci. 13:770315. doi: 10.1016/j.agee.2017.07.012
Wu, G. L., Ren, G. H., Dong, Q. M., Shi, J. J., and Wang, Y. L. (2014). Above – and belowground response along degradation gradient in an alpine grassland of the Qinghai-Tibetan plateau. Clean-Soil Air Water 42, 319–323. doi: 10.1002/clen.201200084
Xiong, J. B., Peng, F., Sun, H. B., Xue, X., and Chu, H. Y. (2016). Effects of grazing exclusion on carbon sequestration and plant diversity in grasslands of China a meta-analysis. Ecol. Eng. 94, 647–655. doi: 10.1016/j.ecoleng.2016.06.124
Xun, W. B., Yan, R. R., Ren, Y., Jin, D. Y., Xiong, W., Zhang, G. S., et al. (2018). Grazing-induced microbiome alterations drive soil organic carbon turnover and productivity in meadow steppe. Microbiome 6:170. doi: 10.1186/s40168-018-0544-y
Yamamura, N., Fujita, N., and Maekawa, A. (2013). The Mongolian ecosystem network. Environmental issues under climate and social changes Springer Available at: https://link.springer.com/book/10.1007/978-4-431-54052-6.
Yu, Y., Zheng, L., Zhou, Y. J., Sang, W. G., Zhao, J. N., Liu, L., et al. (2021). Changes in soil microbial community structure and function following degradation in a temperate grassland. J. Plant Ecol. 14, 384–397. doi: 10.1093/jpe/rtaa102
Zhang, C., Liu, G. B., Song, Z. L., Wang, J., and Guo, L. (2018). Interactions of soil bacteria and fungi with plants during long-term grazing exclusion in semiarid grasslands. Soil Biol. Biochem. 124, 47–58. doi: 10.1016/j.soilbio.2018.05.026
Zheng, Y. F., Cao, X. W., Zhou, Y. N., Li, Z., Yang, Y. Z., Zhao, D. L., et al. (2023). Effect of planting salt-tolerant legumes on coastal saline soil nutrient availability and microbial communities. J. Environ. Manag. 345:118574. doi: 10.1016/j.jenvman.2023.118574
Zheng, H. P., Yang, T. J., Bao, Y. Z., He, P. P., Yang, K. M., Mei, X. L., et al. (2021). Network analysis and subsequent culturing reveal keystone taxa involved in microbial litter decomposition dynamics. Soil Biol. Biochem. 157:108230. doi: 10.1016/j.soilbio.2021.108230
Zhou, H., Zhang, D. G., Jiang, Z. H., Sun, P., Xiao, H. L., Wu, Y. X., et al. (2019). Changes in the soil microbial communities of alpine steppe at Qinghai-Tibetan plateau under different degradation levels. Sci. Total Environ. 651, 2281–2291. doi: 10.1016/j.scitotenv.2018.09.336
Keywords: grazing ban, overgrazing, restoration, soil properties, soil microbial community, edge effect
Citation: Fang M, Lu G, Zhang S and Liang W (2024) Overgrazing on unmanaged grassland interfered with the restoration of adjacent grazing-banned grassland by affecting soil properties and microbial community. Front. Microbiol. 14:1327056. doi: 10.3389/fmicb.2023.1327056
Edited by:
Jianming Wang, Beijing Forestry University, ChinaCopyright © 2024 Fang, Lu, Zhang and Liang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Shuping Zhang, emhhbmdzaHVwaW5nQG11Yy5lZHUuY24=
 Mengchao Fang1
Mengchao Fang1 Shuping Zhang
Shuping Zhang Wei Liang
Wei Liang