
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Microbiol., 08 January 2024
Sec. Systems Microbiology
Volume 14 - 2023 | https://doi.org/10.3389/fmicb.2023.1323346
 Nitin Agrawal1,2
Nitin Agrawal1,2 Federico Fontana3,4
Federico Fontana3,4 Chiara Tarracchini3
Chiara Tarracchini3 Sohvi Lommi1,2
Sohvi Lommi1,2 Marco Ventura3,5
Marco Ventura3,5 Christian Milani3,5
Christian Milani3,5 Heli Viljakainen1,2*
Heli Viljakainen1,2*Background: Variation in diversity and composition of saliva microbiota has been linked to weight status, but findings have been inconsistent. Focusing on clinically relevant conditions such as central obesity and using advanced sequencing techniques might fill in the gaps of knowledge.
Aims: We investigated saliva microbiota with shallow metagenome sequencing in children with (n = 14) and without (n = 36) central obesity. Additionally, we examined the role of habitual food consumption on microbial enzymatic repertoire.
Methods: Data comprised 50 children (50% male) with a mean age of 14.2 (SD 0.3) years, selected from the Finnish Health in Teens (Fin-HIT) cohort. Dietary scores for consumption frequency of sweet treats (STI), dairy products (DCI) and plants (PCI) were derived based on a self-administered food frequency questionnaire. Central obesity was defined based on waist–height ratio using the cut-off 0.5. Saliva samples were subjected to whole-metagenome shotgun sequencing, and taxonomic and functional profiling was achieved with METAnnotatorX2 bioinformatics platform.
Results: Groups had an average 20 (95% CI 14–27) cm difference in waist circumference. We identified the lack of Pseudomonas guguagenesis and Prevotella scopos, oulorum and oris as putative biomarkers associated with central obesity and observed a total of 16 enzymatic reactions differing between the groups. DCI was associated with the highest number of enzyme profiles (122), followed by STI (60) and DCI (25) (Pearson correlation p < 0.05). Intriguingly, STI showed a high positive/negative correlation ratio (5.09), while DCI and PCI showed low ratios (0.54 and 0.33, respectively). Thus, the main driver of enzymatic reactions was STI, and the related pathways involved nitrate metabolism induced by Haemophilus parainfluenzae and Veilonella dispar among others.
Conclusion: Clinically relevant differences in central obesity were only modestly reflected in the composition of saliva microbiota. Habitual consumption of sweet treats was a strong determinant of enzymatic reactions of saliva microbiota in children with and without central obesity. The clinical relevance of these findings warrants further studies.
The prevalence of childhood overweight/obesity and obesity-related co-morbidities have been increasing at an alarming rate worldwide, posing a challenge to our well-being and healthcare system, even causing premature death (World Health Organization, 2022). The link between human microbiome and health conditions, obesity included, has been established but most of the microbiome research has focused on gut microbiota that likely contributes to human health by modifying nutrient intakes, preventing invading pathogens, and interacting with the immune system (Nishida et al., 2018; Lu et al., 2022). However, microbes reside in different niches of the human body: the oral cavity harboring the second most diverse microbiota after the gut (Deo and Deshmukh, 2019).
Saliva microbiota, in recent times, is gaining more attention as a first line of defense with similar contributions to human health as gut microbiota. Furthermore, as 1.5 liters of saliva is ingested daily, its role as an enhancer of gut colonization has also been proposed (Wade, 2021). Some studies have investigated saliva microbiota in overweight/obese children (Raju et al., 2019; Coker et al., 2022), but their results have been discordant with each other possibly due to differences in the study design, age range, and methodology. Moreover, none of these studies have considered central obesity that is mirrored by waist circumference or waist–height ratio (WHtR). WHtR is a better indicator of whole-body obesity than the traditionally used body mass index (McCarthy et al., 2005; Chrzanowska and Suder, 2010; Garnett et al., 2011), and intra-abdominal adipose tissue is known to function as an active organ producing hormones and cytokines which could potentially disrupt metabolic and inflammatory processes in the body (Mokha et al., 2010). In addition, the role of diet in oral microbiota has been scarcely studied in children. For example, frequent sugar consumption appears to play a role in the composition and functionality of the saliva microbiota (Lommi et al., 2022) but the associations between diet and enzymatic pathways of saliva microbial communities have so far not been examined.
While 16S rRNA gene profiling has been widely used to examine the human microbiome, it does not provide a reliable species-level identification. To overcome this shortcoming, metagenome sequencing methods are used that provide up to strain-level identification and thus, a deeper insight into the microbial population and its functional potential. In this study, we investigated central obesity and habitual food consumption as determinants of saliva microbiota composition. We compared composition and enzymatic properties of the saliva microbiota in participants with and without central obesity. Furthermore, we examined the enzymatic classes encoded by the salivary microbiome and the key taxa behind them. Saliva microbiota was subjected to shallow metagenome sequencing.
For this study, we utilized questionnaire data, saliva samples, and anthropometric measurements available from the prospective Finnish Health in Teens cohort (Fin-HIT) which started in 2011 as a school-based cohort study, initially comprising of 11,407 Finnish children aged between 9 and 12 years living in large cities or their surrounding areas. The details of the Fin-HIT cohort are described elsewhere (Figueiredo et al., 2019). In 2015–2016, 54% of the children participated in the first follow-up by filling in an online health survey and providing a saliva sample. Here, we used data exclusively from the follow-up. The Coordinating Ethics Committee of the Hospital District of Helsinki and Uusimaa approved the study protocol (169/13/03/00/10), and written informed consent was obtained from all participants and their parents.
Children self-reported their height (cm), waist circumference (cm), and weight (kg) with instructions that the measurements should be done with the help from an adult. We have previously reported the validity of home-measured anthropometry among 113 children (Sarkkola et al., 2016). Waist–height ratio (WHtR) was calculated by dividing waist circumference (cm) with height (cm) after which children were categorized into those without central obesity (WHtR < 0.5) and those with central obesity (WHtR ≥ 0.5).
Three dietary scores were derived based on a self-administered food frequency questionnaire (FFQ) measuring consumption frequencies of 16 foods and drinks during the preceding month of data collection. The sweet treat index (STI) measures the weekly consumption of chocolate/sweets, ice cream, sweet pastries, biscuits/cookies, sugary juice drinks and sugary soft drinks (Lommi et al., 2020), the plant consumption index (PCI) that of vegetables, fruits and berries (Räisänen et al., 2022), and finally, the dairy consumption index (DCI) measured the weekly consumption of milk/buttermilk and ice cream. Our FFQ was adapted from the FFQ used in the WHO’s International Health Behaviour in School-Aged Children study, which was validated and retested among school-age children in Europe (Vereecken and Maes, 2003; Vereecken et al., 2008).
Unstimulated saliva samples were collected using the Oragene® DNA (OG-500) Self-Collection Kit (DNA Genotek Inc., Ottawa, Ontario, Canada). Saliva samples were mixed with a stabilizing reagent within the collection tube and stored at room temperature per the manufacturer’s instructions. After an intensive lysis and mechanical disruption protocol of microbial cells, genomic DNA was extracted using a CMG-1035 saliva kit and Chemagic MSM1 nucleic acid extraction robot (PerkinElmer) (Raju et al., 2018). Sampling was based on an equal number of normal-and overweight children with equal sex distribution. In total, 50 saliva samples were subjected to whole-metagenome shotgun (WMGS) sequencing on Illumina HiSeq 2500 system (Illumina Inc., San Diego, CA, USA) at the Institute of Molecular Medicine Finland (FIMM). The raw data in fastq format were submitted to quality filtering (quality score < 25) and for the removal of reads derived from the host. Subsequently, taxonomic profiling of the reads which passed filtering was achieved with the METAnnotatorX2 bioinformatics platform (Milani et al., 2021) using MEGABLAST employing the curated non-redundant sequence database of genomes retrieved from the National Center for Biotechnology Information (NCBI).
IBM SPSS v29 (IBM Corp., Armonk, NY, USA) and R studio (version 4.2.2)1 were used for comparing the Shannon index between the groups. Similarities between samples (beta-diversity) were assessed by the Bray–Curtis dissimilarity index based on species abundance. PCoA representation of beta-diversity was performed using OriginPro v2021 (OriginLab Corporation, Northampton, MA, USA), and analysed with permutational multivariate analysis of variance [PERMANOVA].
Microbial community functional analyses were performed using METAnnotatorX2, giving us enzymatic classification numbers by using the Metacyc database as a reference (Caspi et al., 2014). OriginPro v2021 was used to perform the statistical tests in each comparison of differentially abundant taxa and differentially encoded functional enzymes.
Background characteristics, dietary scores, and mean abundance of genera and species were compared between groups with and without central obesity with Mann–Whitney U test or Chi-Square using IBM SPSS v29. Results are presented as means with standard deviations (SD) or as counts (n) with percentages (%).
STI, PCI, and DCI correlation with enzyme repertoire was examined with Pearson correlation on R studio (version 4.2.2). Only significant correlations with correlation scores >0.2 or < −0.2 were retained to remove weak correlations from the results. Force-driven Network was used to represent correlation scores through Gephi (Version 0.9.6) software (Bastian et al., 2009) and ForceAtlas2 algorithm (Jacomy et al., 2014). Statistical significance was set at p < 0.05.
The participants’ background factors except for waist circumference did not differ between the groups (Table 1). Of the three dietary summary scores, only the dairy consumption index seemed to be non-significantly lower in children with central obesity than those without central obesity (p = 0.101).
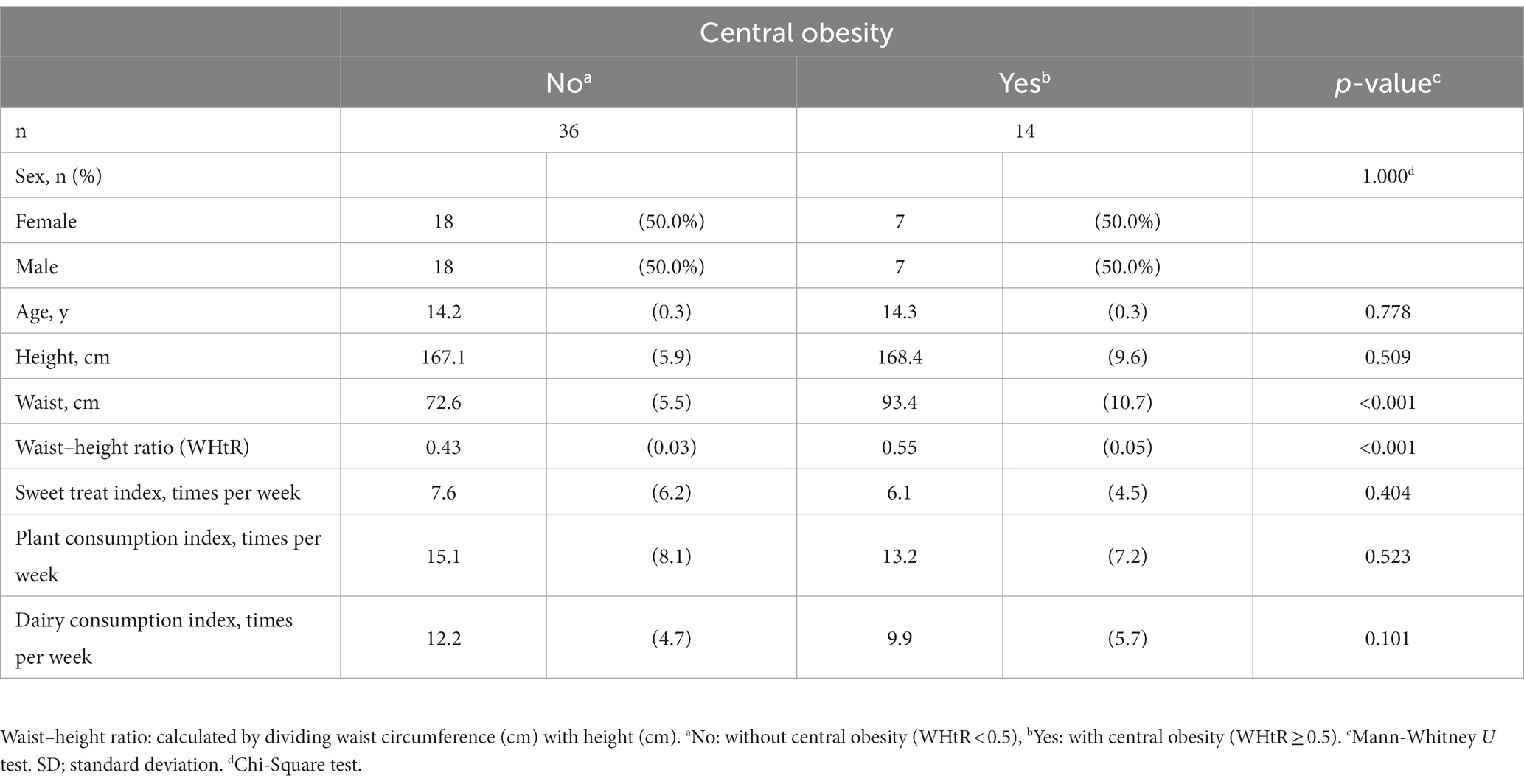
Table 1. Background characteristics of groups without (no) and with (yes) central obesity with mean (SD), if not indicated otherwise.
Whole-metagenome shotgun (WMGS) sequencing of the saliva microbiomes produced an average of 1.97 million ± 329,000 paired-end (2 × 150 bp) reads per sample. Following quality filtering and removal of reads that map on the Homo sapiens genome, an average of 285,567 microbial reads per sample, with a max of 1,427,437 reads were retained and subjected to downstream analyses (Supplementary Table 1).
In total, 43 genera were identified. On average, the participants had 15 genera present. The core microbiota consisted of seven genera based on the highest relative abundance across the samples as shown in Figure 1A, accounting for 81% of the total saliva microbiota.
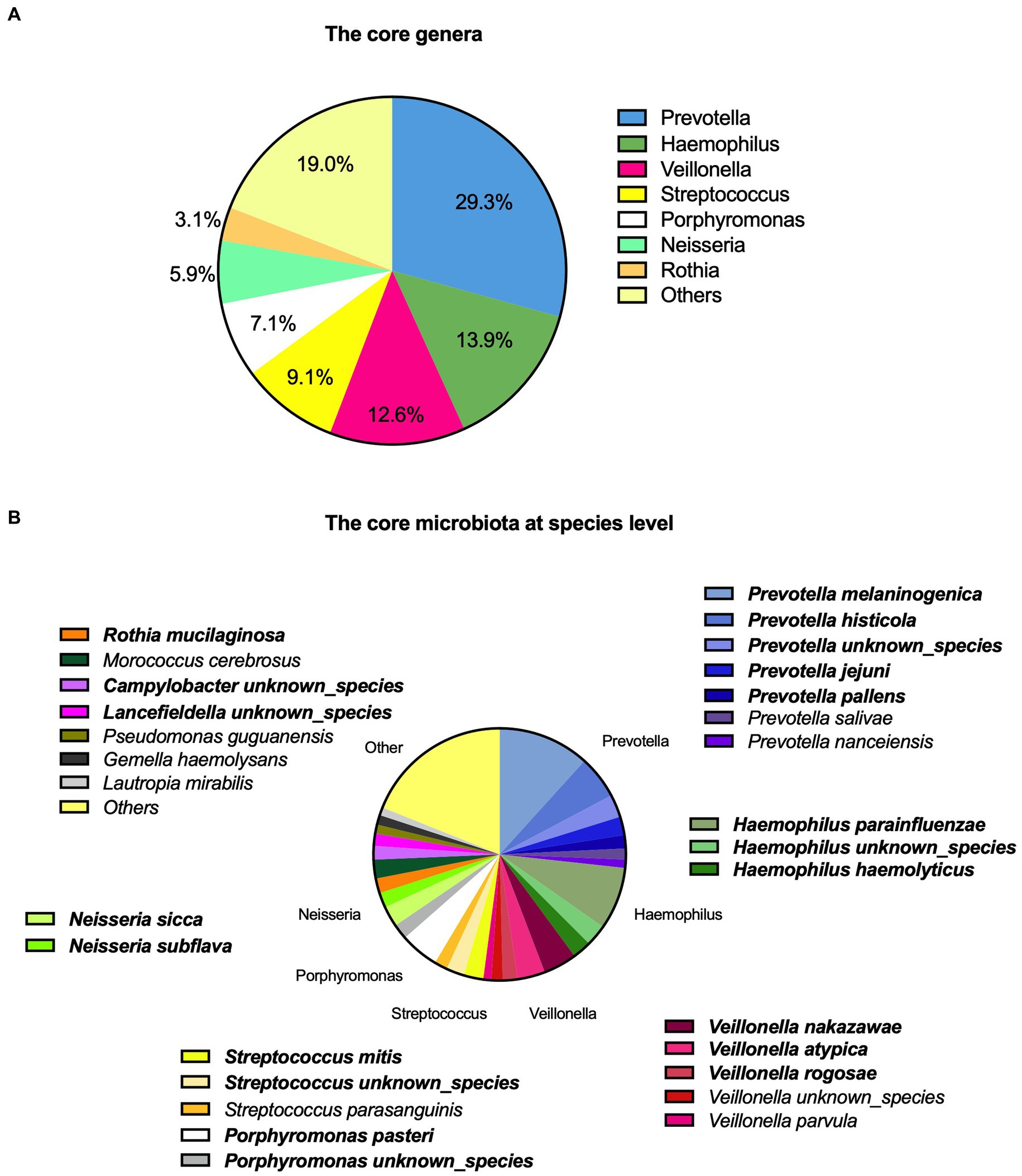
Figure 1. Description of saliva microbiota with (A) the core genera and (B) at a species level. The top 20 prevalent species are highlighted with bold.
We identified a total of 124 microbial species among the participants (Figure 1B). The top 20 species accounted for 69.5% of the total saliva microbiota, the top five being Prevotella melaninogenica (11.7%), Haemophilus parainfluenzae (8.0%), Prevotella histicola (5.5%), Porphyromonas pasteri (5.0%), and Veillonella nakazawae (4.3%). These accounted for 34.5% of the total. A complete description of the top 20 species is also shown in Supplementary Table 2.
The Shannon index (SD) (alpha diversity) was similar between the groups: 2.99 (0.4) vs. 3.06 (0.3) (p = 0.53) with and without central obesity, respectively. The analysis of inter-sample variability (beta-diversity) did not reveal any compositional differences at genus level between the groups (p = 0.744). Notably, some taxa-specific differences were detected. Pseudomonas showed lower mean abundance in the group with central obesity than those without central obesity (0.7% vs. 1.4%, p = 0.041; Figure 2A and Supplementary Figure 1).
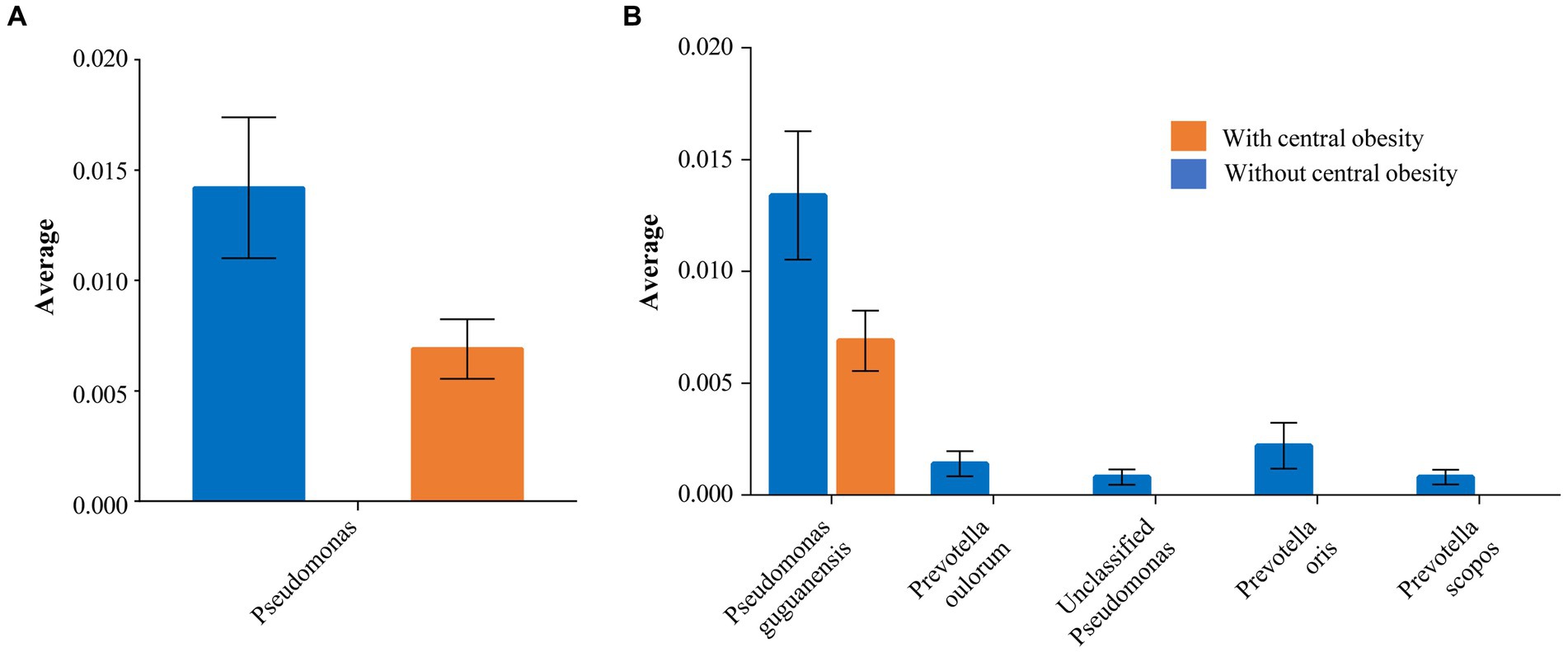
Figure 2. The taxa significantly differing between the groups with (orange) and without (blue) central obesity are shown with mean abundance for (A) genus and (B) species levels. The error bars represent SEM.
Similarly, a species-level investigation revealed that Pseudomonas guguanensis had a lower abundance among participants with central obesity compared with those without central obesity (0.7% vs. 1.3%, p = 0.046). Moreover, unclassified Pseudomonas and several Prevotella species, e.g., scopos, oulorum and oris, although present among those without central obesity, were completely absent in the group with central obesity (p < 0.05 for all; Figure 2B).
To assess whether and how saliva bacterial community-derived metabolites differed between the groups, we analyzed the potential microbiota-encoded enzymatic profiles based on the MetaCyc (Caspi et al., 2014) database and the bioinformatic suite METAnnotatorX2 (Milani et al., 2021).
A total of 16 enzymatic classes differed between the groups. Compared to the group without central obesity, group with central obesity showed enrichment in 15 enzymatic classes (Figure 3), while one enzymatic class was diminished. Through functional back-tracing of taxonomic information, we linked these classes to the microbial species that encode these enzymes. Overall, the analysis revealed that Staphylococcus aureus, Prevotella histicola, and Prevotella melaninogenica were the major taxa with the highest genetic potential for these enzyme-coding genes (Figure 3).
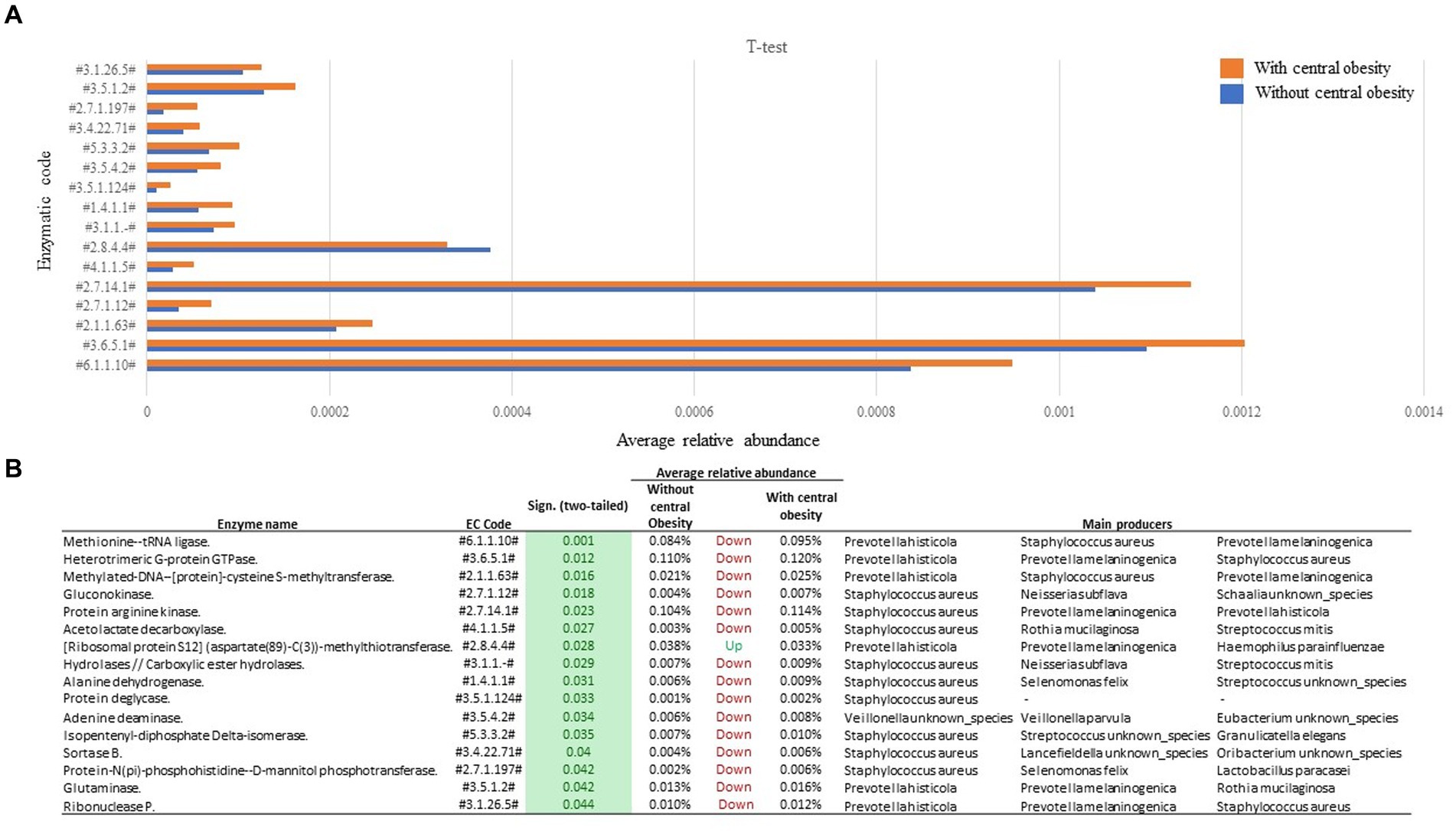
Figure 3. Functional analysis of saliva samples from participants with (orange, n = 14) and without central obesity (blue, n = 36) (A) revealed several differences in microbiota-encoded enzymatic potential profiles. (B) Table represents main producers of each enzymatic class.
To look for additional drivers for the enzymatic profiles, we examined correlations of DCI, PCI, and STI with relative EC numbers. DCI correlated with 122 enzymes, STI with 60, and PCI with 25 (p < 0.05). STI had a correlation ratio (positive/negative) of 5.09, DCI that of 0.54, and PCI that of 0.33 (Figure 4A). Figure 4B also shows the correlations of DCI, PCI, and STI with enzymes in the network representation. Thus, STI, with the highest correlation ratio and the most positive correlations with several enzymes, held a central position in the network.
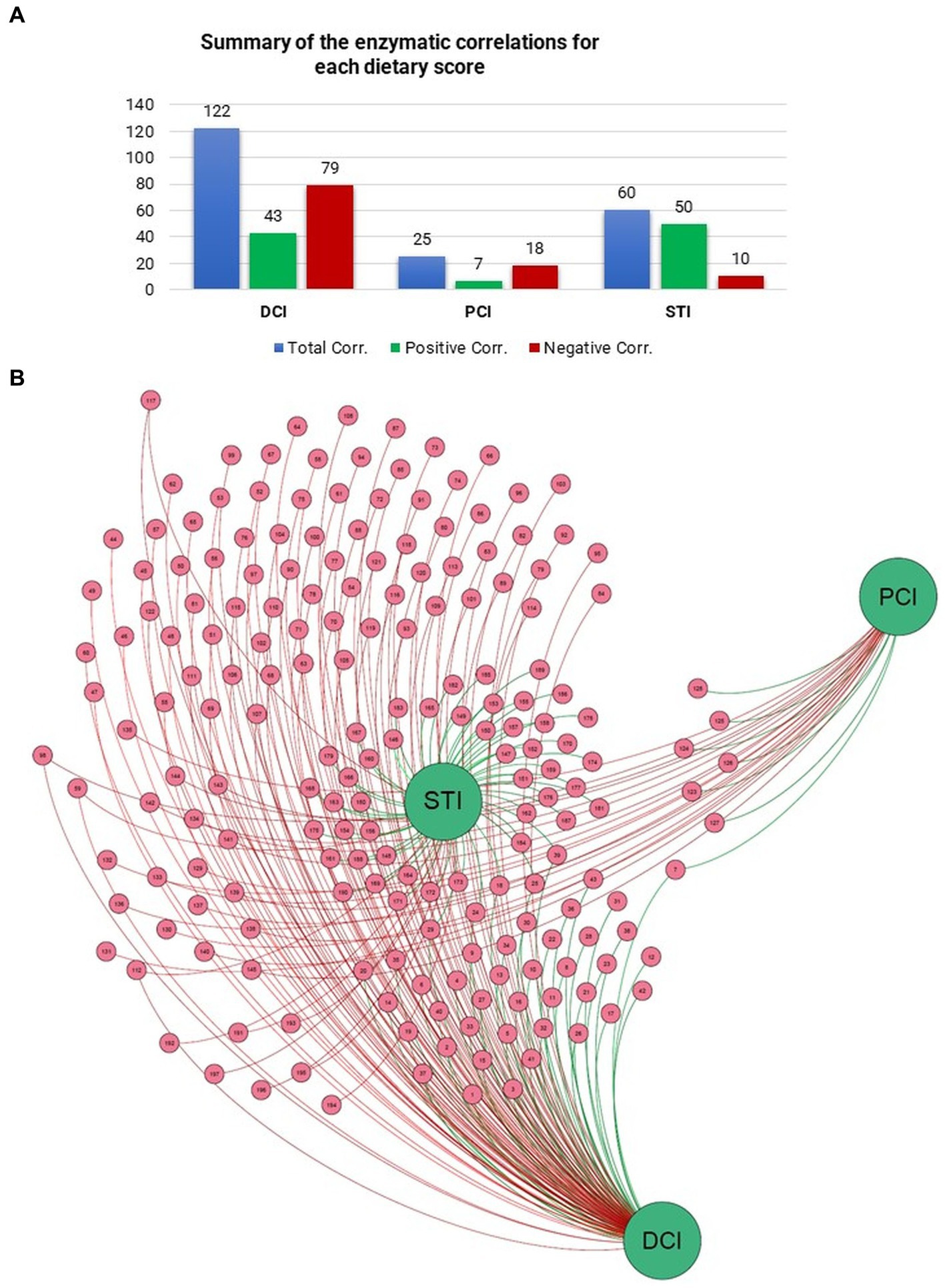
Figure 4. Number of Pearson correlations between (A) dietary scores and enzymes are presented in the table. (B) Force-driven network represents the entity of the correlation scores between dietary scores and enzymes through Gephi (Version 0.9.6) software and ForceAtlas2 algorithm.
A subsequent back-tracing analysis was performed among the reactions encoded by these enzymes, showing the highest correlations with PCI, DCI and STI (five unique reactions for each; Figure 5). The enzyme L-fucose isomerase (EC 5.3.1.25) correlated positively with both DCI and PCI, and is hence listed twice in the figure. The back-tracing analysis showed that Haemophilus genus has a key role in defining the enzymes associated with PCI, DCI and STI, acting as the main taxa encoding for eight out of 16 of the enzymes analyzed (Figure 5B). Other main taxa encoding for some of these enzymes belong to Pseudomonas, Veillonella, Rothia and Granulicatella genera, followed by Neisseria and Streptococcus genera.
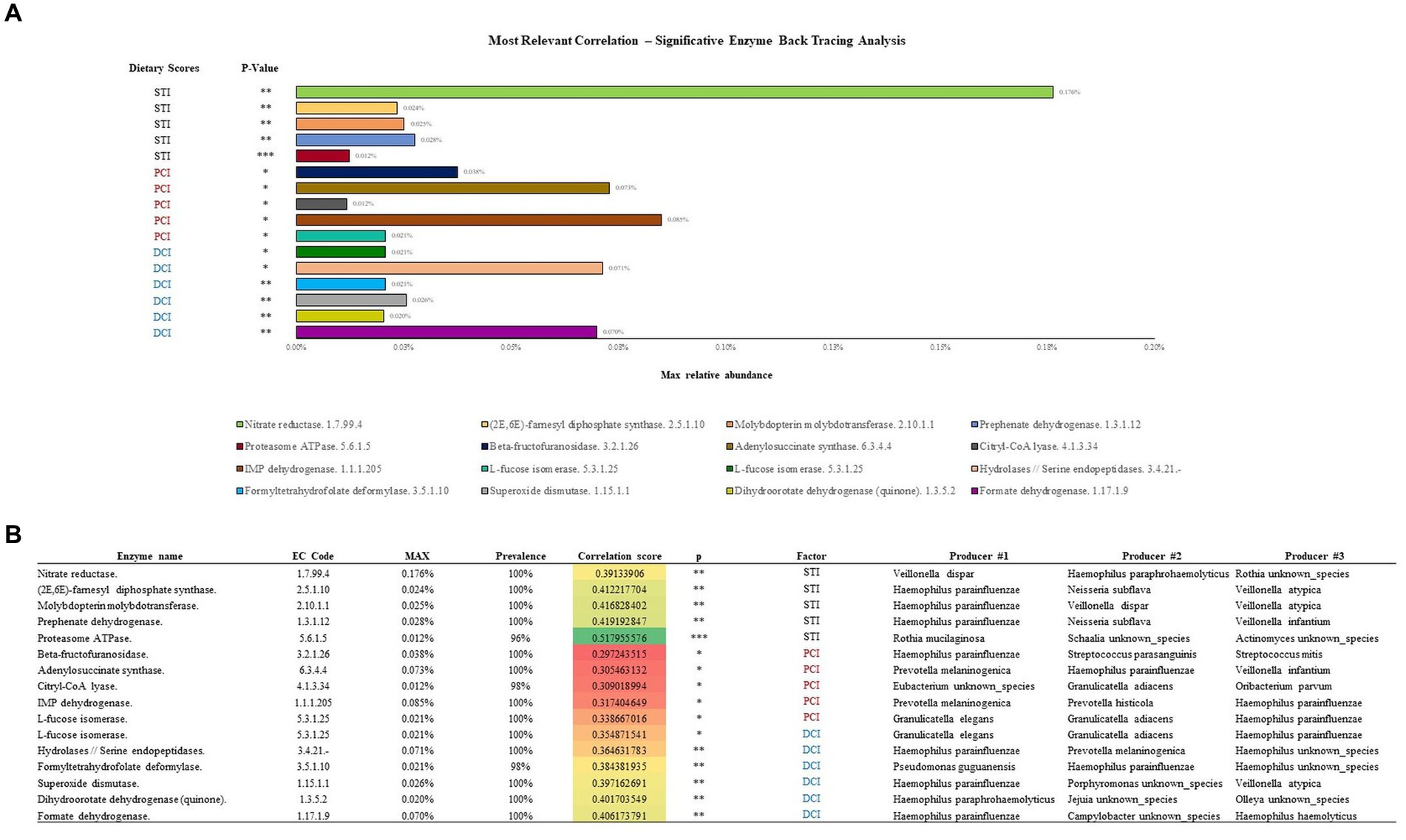
Figure 5. Enzymatic classes showing the highest correlations with STI, PCI and DCI are described in (A). (B) Shows the bacterial species responsible for the enzymatic classes.
The enzymatic class correlating with STI with maximum relative abundance (0.176%) was Nitrate reductase (EC 1.7.99.4) encoded by Veilonella dispar. Similarly, IMP dehydrogenase (EC 1.1.1.205) correlated (0.085%) with PCI, encoded by Prevotella melaninogenica, the most common species in our samples, and Hydrolases/Serine endopeptidases (EC 3.4.21.-) was most abundantly correlated (0.071%) with DCI, encoded by Haemophilus parainfluenzae. Overall, Haemophilus parainfluenzae encoded the most enzymatic classes in relation to the dietary scores.
Our findings based on saliva metagenome sequencing show a high inter-individual variability at the species level, which could be attributed to individual dietary habits, but less to central obesity in this age group. Central obesity is a clinically relevant marker of adiposity (Ross et al., 2020), yet, in our sample, bacterial composition differed only very modestly between groups with and without central obesity. In total, we observed 16 enzymatic classes that differed significantly between the groups. Contrary to this, habitual food consumption was highlighted as a potential modifier of bacterial enzymatic reactions in saliva. To our knowledge, no previous studies have addressed this topic.
Pseudomonas was the only genus that differed between the groups and was less abundant among children with central obesity compared to those without central obesity. The species-level analyses further verified that Pseudomonas guguanensis was less common and unclassified Pseudomonas fully absent in individuals with central obesity. In addition, the back-tracing analysis suggested that P. guguanensis contributes to formyltetrahydrofolate deformylase enzymatic class, which is a key enzyme in the pathway providing formate to the cell under aerobic growth conditions. This enzyme was also associated with dairy consumption in our sample, perhaps activating the same pathway. Indeed, slightly lower consumption of dairy products was marked among the group with central obesity compared with others, although this difference was not statistically significant.
Prevotella was the most common genus with prevalence of 29%, with similar abundances in the groups. However, certain species such as P. oris, P. oulorum, and P. scopos were fully absent in participants with central obesity. The most dominant species Prevotella melaninogenica and P. histicola were linked to the same enzymatic class IMP dehydrogenase which catalyzes the first committed, rate-limiting step in de novo guanine nucleotide biosynthesis in most organisms. Besides, P. melaninogenica also contributes to adenylosuccinate synthase, which catalyzes AMP’s de novo synthesis; both these enzymes were linked with the consumption of vegetables, fruits, and berries in our study. Previously, a high abundance of Prevotella was observed in the saliva of participants following plant-rich diets (Hansen et al., 2018; Daniele et al., 2021). In particular, the presence of Prevotella species correlated with high intake of dietary fiber (Daniele et al., 2021), which could be generally lower in individuals with central obesity (Parikh et al., 2012).
The novelty of our study is that we demonstrated associations of habitual food consumption with the bacterial enzymatic profiles in saliva. We measured habitual diet using three dietary indices indicating weekly consumption frequencies of sweet treats, plants, and dairy.
Although dairy consumption associated with a very large enzymatic repertoire both positively (n = 43) and negatively (n = 88), the strongest modulator was the consumption of sweet treats, which had the highest correlation ratio among the three dietary scores as well as the most positive correlations. It also contributed to the most abundant nitrate reduction pathway through nitrate reductase. These findings resemble our recent results in another Fin-HIT sample employing 16S profiling on saliva microbiota; that study also highlighted the nitrate reduction pathway in frequent sugar consumers (Lommi et al., 2022). The pathway converts nitrate to nitrite, allowing further metabolism to nitric oxide (Rosier et al., 2020). Nitric oxide has physiological effects like vasodilation, and it contributes to innate immunity responses (Gantner et al., 2020), while in the mouth it increases the nitrate reduction capacity of the oral microbiota and is related with lower prevalence of caries (Doel et al., 2005). Specifically, nitrate reduction prevents acidification and cariogenic bacteria’s overgrowth by increasing lactate and ammonia production (Rosier et al., 2020). Based on the back-tracing analysis in our study, species responsible for nitrate reduction were Veillonella dispar, Haemophilus paraprohaemolyticus and unclassified Rothia.
Plant consumption was associated with the lowest number of enzymatic classes and showed a positive correlation with only seven enzymes, among them beta-fructofuranosidase and L-fucose isomerase, which are utilized to break down certain complex sugars from plant sources (Vanhooren and Vandamme, 1999; Kotwal and Shankar, 2009). This suggests that the enzymatic profiles of saliva microbiome adapt to the prevailing diet.
Haemophilus parainfluenzae was among the key producers of the enzymatic reactions related to these three dietary summary scores. Typically, Haemophilus parainfluenzae is considered pathogenic to humans and is related to infections in urban environment (Roslund et al., 2021). In our sample of children from densely populated areas of Finland, it was the second most common species in saliva microbiome.
No difference in the beta diversity was observed between groups with and without central obesity, and only marginal differences in the abundance of some Pseudomonas and Prevotella species were noted. This could be due to small sample size and nonequal group sizes, but other explanations may exist as well. Initially, we selected samples with equal gender distribution with and without obesity based on BMI. However, since central obesity is considered a clinically more relevant measure of obesity than BMI, we divided the groups into with and without central obesity, leading to nonequal group sizes. Although we witnessed on average 20 cm larger waist circumference in the group with central obesity, the association with microbiome was superficial and likely explained by other factors instead. Here, we offer a plausible explanation with the habitual diet, but also other factors may contribute such as oral health. However, we have recently shown that the history of caries in this age group was not a determinant of saliva microbiome diversity nor composition (Manzoor et al., 2021) but associated positively with sugar-metabolizers such as Leptorichia and Paludibacter in sex-specific analyses. Nevertheless, our analysis was limited by the lack of information on active caries process.
Utilizing metagenomic sequencing is a strength of this study. It provides more detailed information than 16S profiling; classifying the taxa more accurately and allowing to reflect microbiome functions in terms of enzymatic classes and related pathways based on a detailed database on microbial species and their functions.
Our dietary summary scores rely on self-reported consumption of indicatory food items that do not provide a complete picture of the children’s diet. Furthermore, we had no exclusion criteria for participants; thus, some residual confounding may exist. Overall, the study should be considered a pilot study that provides observational data without full explanatory power.
Despite clinically relevant differences in waist circumference in participants, the saliva microbiota exhibited very modest differences in bacterial abundance and some in enzymatic classes according to central obesity. By contrast, habitual food consumption was associated with several enzymatic classes. Especially the role of sweet treat consumption was highlighted, as it contributed to nitrate reduction pathway through nitrate reductase produced by species Veillonella dispar and Haemophilus parainfluenzae among others. In adolescence, the contribution of lifestyle factors such as diet may have a bigger role on the saliva microbiome than central obesity per se.
The data presented in the study are deposited in the NCBI BioProject repository, accession number PRJNA1052614 (https://www.ncbi.nlm.nih.gov/bioproject/PRJNA1052614).
The studies involving humans were approved by the Coordinating Ethics Committee of the Hospital District of Helsinki and Uusimaa approved the study protocol (169/13/03/00/10). The studies were conducted in accordance with the local legislation and institutional requirements. Written informed consent for participation in this study was provided by the participant and his/her parent or legal guardian.
NA: Conceptualization, Investigation, Writing – original draft, Writing – review & editing. FF: Formal analysis, Investigation, Visualization, Writing – original draft, Writing – review & editing. CT: Formal analysis, Writing – review & editing. SL: Investigation, Visualization, Writing – review & editing. MV: Formal analysis, Writing – review & editing. CM: Conceptualization, Writing – review & editing, Formal analysis, Investigation, Supervision. HV: Conceptualization, Formal analysis, Funding acquisition, Supervision, Writing – original draft, Writing – review & editing, Investigation, Visualization.
The author(s) declare financial support was received for the research, authorship, and/or publication of this article. This project has received financial support from the Päivikki and Sakari Sohlberg Foundation, Swedish Cultural Foundation in Finland, Minerva Foundation, and Folkhälsan Research Foundation governed by HV. FF and CM have received funding under the National Recovery and Resilience Plan (NRRP), Mission 4 Component 2 Investment 1.3 – Call for tender no. 341 of 15 March 2022 of Italian Ministry of University and Research funded by the European Union – NextGenerationEU; Award Number: Project code PE00000003, Concession Decree no. 1550 of 11 October 2022 adopted by the Italian Ministry of University and Research, CUP D93C22000890001, Project title “ON Foods – Research and innovation network on food and nutrition Sustainability, Safety and Security – Working ON Foods.” There were no other specific grants from any public, commercial, or non-profit sectors relevant to this article to disclose. The funding sources had no role in the design and conduct of the study; collection, management, analysis, and interpretation of the data; preparation, review, or approval of the manuscript; and decision to submit the manuscript for publication.
We thank all the families for participating in the study.
FF was employed by GenProbio srl.
The remaining authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
The author(s) declared that they were an editorial board member of Frontiers, at the time of submission. This had no impact on the peer review process and the final decision.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2023.1323346/full#supplementary-material
Bastian, M., Heymann, S., and Jacomy, M. (2009). Gephi: an open source software for exploring and manipulating networks. Proceedings of the International AAAI Conference on Web and Social Media, San Jose, CA, United States.
Caspi, R., Altman, T., Billington, R., Dreher, K., Foerster, H., Fulcher, C. A., et al. (2014). The Meta Cyc database of metabolic pathways and enzymes and the BioCyc collection of pathway/genome databases. Nucleic Acids Res. 42, D459–D471. doi: 10.1093/NAR/GKT1103
Chrzanowska, M., and Suder, A. (2010). Changes in central fatness and abdominal obesity in children and adolescents from Cracow, Poland 1983-2000. Ann. Hum. Biol. 37, 243–253. doi: 10.3109/03014460903193237
Coker, M. O., Lebeaux, R. M., Hoen, A. G., Moroishi, Y., Gilbert-Diamond, D., Dade, E. F., et al. (2022). Metagenomic analysis reveals associations between salivary microbiota and body composition in early childhood. Sci. Rep. 12:13075. doi: 10.1038/S41598-022-14668-Y
Daniele, S., Scarfò, G., Ceccarelli, L., Fusi, J., Zappelli, E., Biagini, D., et al. (2021). The Mediterranean diet positively affects resting metabolic rate and salivary microbiota in human subjects: a comparison with the vegan regimen. Biology 10:1292. doi: 10.3390/biology10121292
Deo, P. N., and Deshmukh, R. (2019). Oral microbiome: unveiling the fundamentals. J. Oral Maxillofacial Pathol. 23, 122–128. doi: 10.4103/JOMFP.JOMFP_304_18
Doel, J. J., Benjamin, N., Hector, M. P., Rogers, M., and Allaker, R. P. (2005). Evaluation of bacterial nitrate reduction in the human oral cavity. Eur. J. Oral Sci. 113, 14–19. doi: 10.1111/J.1600-0722.2004.00184.X
Figueiredo, R. A. D. O., Simola-Ström, S., Rounge, T. B., Viljakainen, H., Eriksson, J. G., Roos, E., et al. (2019). Cohort profile: the Finnish health in teens (fin-HIT) study: a population-based study. Int. J. Epidemiol. 48, 23–24H. doi: 10.1093/ije/dyy189
Gantner, B. N., LaFond, K. M., and Bonini, M. G. (2020). Nitric oxide in cellular adaptation and disease. Redox Biol. 34:101550. doi: 10.1016/j.redox.2020.101550
Garnett, S. P., Baur, L. A., and Cowell, C. T. (2011). The prevalence of increased central adiposity in Australian school children 1985 to 2007. Obesity Rev. 12, 887–896. doi: 10.1111/J.1467-789X.2011.00899.X
Hansen, T. H., Kern, T., Bak, E. G., Kashani, A., Allin, K. H., Nielsen, T., et al. (2018). Impact of a vegan diet on the human salivary microbiota. Sci. Rep. 8:5847. doi: 10.1038/s41598-018-24207-3
Jacomy, M., Venturini, T., Heymann, S., and Bastian, M. (2014). ForceAtlas2, a continuous graph layout algorithm for Handy network visualization designed for the Gephi software. PLoS One 9:e98679. doi: 10.1371/JOURNAL.PONE.0098679
Kotwal, S. M., and Shankar, V. (2009). Immobilized invertase. Biotechnol. Adv. 27, 311–322. doi: 10.1016/j.biotechadv.2009.01.009
Lommi, S., Figueiredo, R. A. D. O., Tuorila, H., and Viljakainen, H. (2020). Frequent use of selected sugary products associates with thinness, but not overweight during preadolescence: a cross-sectional study. Br. J. Nutr. 124, 631–640. doi: 10.1017/S0007114520001361
Lommi, S., Manzoor, M., Engberg, E., Agrawal, N., Lakka, T. A., Leinonen, J., et al. (2022). The composition and functional capacities of saliva microbiota differ between children with low and high sweet treat consumption. Front. Nutr. 9:543. doi: 10.3389/FNUT.2022.864687/BIBTEX
Lu, Y., Yuan, X., Wang, M., He, Z., Li, H., Wang, J., et al. (2022). Gut microbiota influence immunotherapy responses: mechanisms and therapeutic strategies. J. Hematol. Oncol. 15:47. doi: 10.1186/S13045-022-01273-9
Manzoor, M., Lommi, S., Furuholm, J., Sarkkola, C., Engberg, E., Raju, S., et al. (2021). High abundance of sugar metabolisers in saliva of children with caries. Sci. Rep. 11, 4424–4410. doi: 10.1038/s41598-021-83846-1
McCarthy, H. D., Jarrett, K. V., Emmett, P. M., and Rogers, I. (2005). Trends in waist circumferences in young British children: a comparative study. Int. J. Obesity 29, 157–162. doi: 10.1038/SJ.IJO.0802849
Milani, C., Lugli, G. A., Fontana, F., Mancabelli, L., Alessandri, G., Longhi, G., et al. (2021). METAnnotatorX2: a comprehensive tool for deep and shallow metagenomic data set analyses. mSystems 6:e0058321. doi: 10.1128/MSYSTEMS.00583-21
Mokha, J. S., Srinivasan, S. R., DasMahapatra, P., Fernandez, C., Chen, W., Xu, J., et al. (2010). Utility of waist-to-height ratio in assessing the status of central obesity and related cardiometabolic risk profile among normal weight and overweight/obese children: the Bogalusa heart study. BMC Pediatr. 10:73. doi: 10.1186/1471-2431-10-73
Nishida, A., Inoue, R., Inatomi, O., Bamba, S., Naito, Y., and Andoh, A. (2018). Gut microbiota in the pathogenesis of inflammatory bowel disease. Clin. J. Gastroenterol. 11, 1–10. doi: 10.1007/S12328-017-0813-5
Parikh, S., Pollock, N. K., Bhagatwala, J., Guo, D.-H., Gutin, B., Zhu, H., et al. (2012). Adolescent fiber consumption is associated with visceral fat and inflammatory markers. J. Clin. Endocrinol. Metab. 97, E1451–E1457. doi: 10.1210/jc.2012-1784
Räisänen, L., Lommi, S., Engberg, E., Kolho, K. L., and Viljakainen, H. (2022). Central obesity in school-aged children increases the likelihood of developing paediatric autoimmune diseases. Pediatric Obesity 17:e12857. doi: 10.1111/IJPO.12857
Raju, S. C., Lagström, S., Ellonen, P., de Vos, W. M., Eriksson, J. G., Weiderpass, E., et al. (2018). Reproducibility and repeatability of six high-throughput 16S rDNA sequencing protocols for microbiota profiling. J. Microbiol. Methods 147, 76–86. doi: 10.1016/j.mimet.2018.03.003
Raju, S. C., Lagström, S., Ellonen, P., de Vos, W. M., Eriksson, J. G., Weiderpass, E., et al. (2019). Gender-specific associations between saliva microbiota and body size. Front. Microbiol. 10:767. doi: 10.3389/fmicb.2019.00767
Rosier, B. T., Moya-Gonzalvez, E. M., Corell-Escuin, P., and Mira, A. (2020). Isolation and characterization of nitrate-reducing bacteria as potential probiotics for oral and systemic health. Front. Microbiol. 11:555465. doi: 10.3389/FMICB.2020.555465
Roslund, M. I., Puhakka, R., Nurminen, N., Oikarinen, S., Siter, N., Grönroos, M., et al. (2021). Long-term biodiversity intervention shapes health-associated commensal microbiota among urban day-care children. Environ. Int. 157:106811. doi: 10.1016/J.ENVINT.2021.106811
Ross, R., Neeland, I. J., Yamashita, S., Shai, I., Seidell, J., Magni, P., et al. (2020). Waist circumference as a vital sign in clinical practice: a consensus statement from the IAS and ICCR working group on visceral obesity. Nat. Rev. Endocrinol. 16, 177–189. doi: 10.1038/S41574-019-0310-7
Sarkkola, C., Rounge, T. B., Simola-Ström, S., Von Kraemer, S., Roos, E., and Weiderpass, E. (2016). Validity of home-measured height, weight and waist circumference among adolescents. Eur. J. Pub. Health 26, 975–977. doi: 10.1093/EURPUB/CKW133
Vanhooren, P., and Vandamme, E. (1999). L-Fucose: occurrence, physiological role, chemical, enzymatic and microbial synthesis. J. Chem. Technol. Biotechnol. 74, 479–497. doi: 10.1002/(SICI)1097-4660(199906)74:6<479::AID-JCTB76>3.0.CO;2-E
Vereecken, C. A., and Maes, L. (2003). A Belgian study on the reliability and relative validity of the health behaviour in school-aged children food-frequency questionnaire. Public Health Nutr. 6, 581–588. doi: 10.1079/PHN2003466
Vereecken, C. A., Rossi, S., Giacchi, M. V., and Maes, L. (2008). Comparison of a short food-frequency questionnaire and derived indices with a seven-day diet record in Belgian and Italian children. Int. J. Public Health 53, 297–305. doi: 10.1007/S00038-008-7101-6
Wade, W. G. (2021). Resilience of the oral microbiome. Periodontol. 86, 113–122. doi: 10.1111/PRD.12365
Keywords: metagenomics, sugar, dairy, fruit and vegetables, bacterial species, oral microbiome
Citation: Agrawal N, Fontana F, Tarracchini C, Lommi S, Ventura M, Milani C and Viljakainen H (2024) Associations of central obesity and habitual food consumption with saliva microbiota and its enzymatic profiles – a pilot study in Finnish children. Front. Microbiol. 14:1323346. doi: 10.3389/fmicb.2023.1323346
Received: 17 October 2023; Accepted: 13 December 2023;
Published: 08 January 2024.
Edited by:
Clara G. De Los Reyes-Gavilan, Spanish National Research Council (CSIC), SpainReviewed by:
Nadia Andrea Andreani, Max Planck Institute for Evolutionary Biology, GermanyCopyright © 2024 Agrawal, Fontana, Tarracchini, Lommi, Ventura, Milani and Viljakainen. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Heli Viljakainen, aGVsaS52aWxqYWthaW5lbkBoZWxzaW5raS5maQ==
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.