- 1ICMR-National JALMA Institute for Leprosy and Other Mycobacterial Diseases, Agra, Uttar Pradesh, India
- 2Department of Biotechnology, Institute of Applied Sciences & Humanities, GLA University, Mathura, Uttar Pradesh, India
- 3Department of Chest and Tuberculosis, S. N. Medical College & Hospital, Agra, Uttar Pradesh, India
- 4Department of Agricultural Statistics and Computer Centre, N. M. College of Agriculture, Navsari Agricultural University, Navsari, Gujarat, India
Objectives: The objective of this study is to analyze the association between TLR2 deletion (−196 to −174) and TLR1 743 A > G gene polymorphism with drug resistant tuberculosis (PTB, MDR-TB, and XDR-TB) in a population from Agra, Uttar Pradesh.
Methods: The present case–control study included 101 pulmonary TB patients, 104 multidrug-resistant TB patients, 48 extremely drug-resistant TB patients, and 130 healthy and unrelated controls residing in the same locality. The genotyping method for TLR2 deletion (−196 to −174) was carried out by allele-specific polymerase chain reaction (PCR), and TLR1 743 A > G gene polymorphism was performed by hybridization probe chemistry in Roche Real-Time PCR. Genotype and allele frequencies were analyzed by the chi-square test. Cytokine levels were measured by ELISA and compared using Mann–Whitney and Kruskal–Wallis tests.
Results: The frequency of heterozygous (Ins/del) genotypes for TLR2 (−196 to −174) polymorphism was predominant in XDR-TB patients (0.57), whereas heterozygous A/G genotype for TLR1 743 A > G single nucleotide polymorphism (SNP) was predominant in healthy controls (0.57) for TLR1 743 A > G gene polymorphism. The heterozygous genotype of TLR2 deletion polymorphism was found to be significantly higher in XDR-TB (p = 0.0001). TLR1 743 A > G SNP, AG genotypes were found to be significantly associated with healthy controls than PTB (p = 0.047). The level of serum cytokines (IL-6, TNF-α, and IFN-γ) was also found to be significantly different among TB patients and healthy controls.
Conclusion: The findings suggested that in the present population, the heterozygous (Ins/Del) genotype and deletion allele of TLR2 deletion (−196 to −174) polymorphism are associated with the risk for the development of drug-resistant TB. Furthermore, for TLR1 743 A > G gene polymorphism, A/G genotype, and G allele are found associated with healthy controls, suggesting the protective role against TB.
1 Introduction
According to epidemiological data provided by the World Health Organization, the global count of newly reported Tuberculosis (TB) cases in 2021 reached 10.6 million cases, reflecting a 4.5% rise from the 10.1 million cases recorded in 2020. Between 2020 and 2021, there is an estimated 3.6% rise in the Tuberculosis (TB) incidence rate. In 2021, an estimated rate of DR-TB was observed to be 450,000 cases, which is reported to have increased 3.1% from 437,000 in 2020 (World Health Organization, 2022). The diverse host immune response to Mycobacterium tuberculosis infection is influenced by both host genetic factors and the reaction of the host to the infection (Berrington and Hawn, 2007). The host cell recognizes Mycobacteria through various receptors, and one of them is Toll-like receptor (TLR). TLR recognizes molecules such as lipoprotein and lipopolysaccharides known as PAMPs that are present in bacteria and bind to host pattern recognition receptors (PRRs) (Jahantigh et al., 2013). TLR1 recognizes tri-acylated lipopeptide, and TLR6 recognizes di-acyl lipopeptide (Buwitt-Beckmann et al., 2005; Omueti et al., 2005). TLR1 recognizes 19 kD lipoprotein of M.tb (Takeuchi et al., 2002), and TLR2 recognizes 19 kD lipoproteins and lipoarabinomann and other PAMPs molecule of M.tb (Brightbill et al., 1999). Dendritic cells, macrophages, natural killer cells, and T and B lymphocytes express TLRs (Vidya et al., 2018). The human TLR2 gene is situated on chromosome 4q32, and TLR2 molecules consist of two non-coding exons and one coding exon (Haehnel et al., 2002). Toll-like receptors involved in recognizing M.tb include TLR1, TLR2, TLR4, TLR6, TLR8, and TLR9 (Thada et al., 2013). The initial defense of our immune system is the innate immune response (Chen et al., 2010). In the previous study by Mittal et al., the frequency of −196 to −174 del polymorphism of the TLR2 gene was compared between PTB cases and healthy controls in population of Agra (Uttar Pradesh), and the similar allele frequencies were reported in both cases and controls; no significant difference was observed in genotype frequencies of TB patients and healthy controls (Mittal et al., 2018). In addition, the previous study by Sinha et al., where they examined the frequency of the 743 A > G gene polymorphism in the TLR1 gene, compared the cases of pulmonary tuberculosis (PTB) with healthy controls from the Agra region. The study revealed that AG genotypes were significantly more prevalent in healthy controls than in PTB cases. Additionally, the research indicated that the heterozygous condition at the 743 A > G locus conferred a degree of resistance to the disease by restricting bacterial growth (Sinha et al., 2014).
TLR2 (−196 to −174) del polymorphism is studied in various populations, suggesting the role in susceptibility toward TB. Khan et al. in their study has reported the higher allele frequency of the deletion allele of TLR2 (−196 to −174) (Ins/del) polymorphism in TB-infected patients in the Pakistani population (Khan et al., 2014). The study conducted by Ma et al. revealed an association between alleles encoding the 743 A > G variant and cases of pulmonary tuberculosis (PTB) in the African-American population (Ma et al., 2007). Uciechowski et al. discovered that the 743A > G variant was linked to susceptibility to tuberculosis (TB) in a low-endemic country, and at the same time, they did not observe any association in the Ghanaian population (Uciechowski et al., 2011). The role of these polymorphisms is unexplored in drug-resistant tuberculosis. Drug resistance develops due to some possible causes such as (1) drug compliance, (2) bacterial genotype, and (3) host genetic factors (Ben-Kahla and Sahal, 2016). The prevalence of DR-TB is high in India and is listed as one of the 30 high burden countries for DR-TB (World Health Organization, 2022). Observing the prevalence rate and the severity of the disease, it becomes imperative to conduct such studies which look into the cause of the disease.
2 Materials and methods
2.1 Ethics statement
The study protocol was approved by the Institute Human Ethics Committee and established following the guidelines outlined by the Indian Council of Medical Research (2017).
2.2 Study population
The tuberculosis patients were recruited from the Outpatient Department (OPD) of the Department of Chest and Tuberculosis, Sarojini Naidu Medical College, Agra, and ICMR-National JALMA Institute for Leprosy and Other Mycobacterial Diseases (NJIL & OMD), who were registered in the OPD from Monday to Friday from 2019 to 2021. Informed consent was obtained from all participants. Only those patients were included who consented to participate in the study. Most of these patients were inhabitants of Agra or neighboring areas within the state of Uttar Pradesh. The patients were clinically examined by the chest physician and evaluated following the guidelines of TB India (2017). Based on the findings of AFB smear positivity in sputum, CBNAAT, radiography, LPA and DST, the participants were categorized as follows.
2.2.1 Pulmonary tuberculosis (PTB) patients
PTB patients were those who were diagnosed based on smear positivity for AFB, CBNAAT (rifampicin resistance not detected), and clinical manifestations, following the guidelines of NTEP. A new TB patient is defined as someone with pulmonary tuberculosis (PTB) has never undergone treatment for TB or has taken anti-TB drugs for less than 1 month. Patients with seropositive HIV infection or suffering from diabetes or any other immunosuppressive disorders were excluded from the study.
2.2.2 Multi drug-resistant tuberculosis (MDR-TB) patients
Patients classified as MDR are those whose biological specimen demonstrates resistance to both isoniazid and rifampicin, with or without resistance to other first-line anti-TB drugs. Diagnosis of MDR-TB patients was confirmed based on the CBNAAT test (resistant to rifampicin) and FL-LPA, SL-LPA, and LC DST, following the guidelines (Guidelines on Programmatic Management of Drug-Resistant Tuberculosis in India, 2017). Patients who were showing co-infection with HIV or any other immunosuppressive disease were excluded from the study.
2.2.3 Extensively drug-resistant tuberculosis (XDR-TB) patients
XDR patients meet the criteria for MDR/RR-TB and additionally showed resistance to any fluoroquinolone (levofloxacin or moxifloxacin) and at least one additional Group A drug (either bedaquiline or linezolid or both). XDR patients were diagnosed based on FL-LPA, SL-LPA, and LC DST following the guidelines (Guidelines on Programmatic Management of Drug-Resistant Tuberculosis in India, 2017).
2.2.4 Healthy controls
Healthy unrelated individuals from the same socioeconomic background, including those accompanying patients to hospitals and students visiting our institute for training and project work, were enlisted as healthy controls. Schedules containing subject details were filled, and individuals with a recent history of fever, viral infection, other illnesses, or any immunological diseases, as well as those who had received treatment for tuberculosis or leprosy in the past or had a family history of tuberculosis, were excluded from the study. The current study included individuals aged 18–60 years who were in overall good health.
2.3 Selection of gene polymorphism
TLR2 (−196 to −174) del polymorphism is reported to alter the function of TLR2 promoter and decrease transcription-responsive promoters (Tahara et al., 2008). The TLR1 gene polymorphism 743 A > G located in the extracellular domain is associated with ligand-binding activity (Ma et al., 2007). Meta-analysis revealed that TLR2 (−196 to −174) del polymorphism and TLR1 743 A > G gene polymorphism were significantly associated with TB disease (Varshney et al., 2022). Although there are reports related to frequencies and association between TLR2 (−196 to −174) del polymorphism (Mittal et al., 2018) and TLR1 743 A > G gene polymorphism (Sinha et al., 2014), from Agra population, no information is available about the host gene polymorphism among the drug-resistant tuberculosis. The present study will be the first report on the frequencies of TLR2 (−196 to −174) del and TLR1 743 A > G gene polymorphisms in drug-resistant tuberculosis patients. Hence, TLR2 (−196 to −174) del polymorphism and TLR1 743 A > G gene polymorphism were selected.
2.4 Genotyping for the TLR2 gene
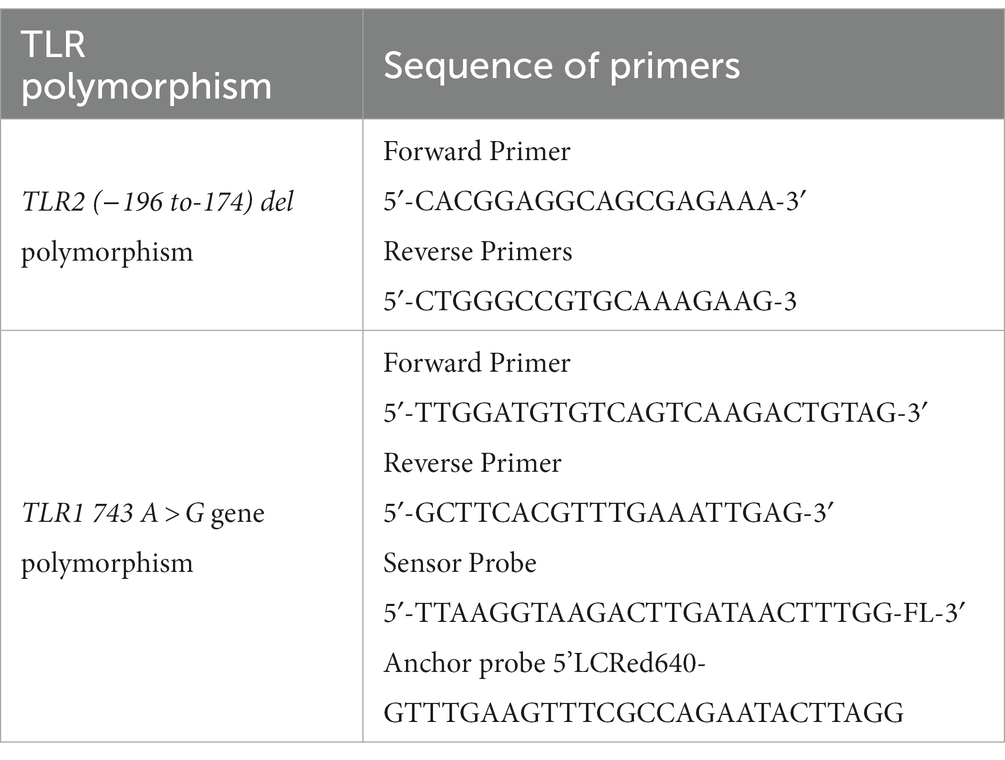
DNA extraction was performed using the salting out procedure from 1 mL of blood collected in tubes containing acid citrate dextrose from each subject (Miller et al., 1988) or promega genomic DNA isolation kit. Genotyping of this polymorphism at TLR2 (−196 to −174) del was performed by allele-specific polymerase chain reaction (PCR). The sequence of forward and reverse primers was adopted from the study by Tahara et al. (2008). The details of sequence of primers are presented in Table 1. The PCR products were observed through electrophoresis on a 3% agarose gel and stained with ethidium bromide for visualization. The observation was finally recorded after assessment of three independent observers. The amplified product of a single band of 286 bp was judged as wild type (insertion/insertion), and a single band of 264 bp was judged as homozygous mutant (deletion/deletion), whereas two bands of 284 and 264 bp were judged as heterozygous (insertion/deletion) for TLR2 (−196 to −174) del polymorphism.
2.5 Genotyping for the TLR1 gene
DNA extraction was performed as described above in section 2.4. Genotyping of TLR1 743 A > G gene polymorphism was performed by hybridization probe chemistry in Roche real-time PCR machine. The sequence of TLR1 gene (743 A > G, rs 4833095), forward primer, reverse primer, sensor probe, and anchor probe was adopted from the study by Sinha et al. (2014). Primers and probes were synthesized from TIB Mol biol, Berlin, Germany. The LightCycler 480 genotyping master mix was procured from Roche Company. The primer and probe sequences used for amplification and the FRET mechanism in detecting TLR1 743 A > G gene polymorphism are presented in Table 1. Real-time PCR was performed using 1.5 μL DNA, 2.5 Mm Mgcl2, TLR1 primers at 1.25 pmol, and 250 nm of the TLR1 743 A > G sensor probe and anchor probe. The sensor probe was labeled with fluorescein at the 3’end. The anchor probe was labeled with LightCycler red 640 at the 5’end. The amplification was conducted in the LightCycler 480 system (Roche Diagnostics) with the following parameters: initial denaturation at 95°C for 10 min, followed by 40 cycles of amplification with denaturation at 95°C for 10 s, annealing at 60°C for 20 s, and extension at 72°C for 15 s. Subsequently, a melting curve analysis was performed with 1 cycle at 95°C for 1 min, 40°C for 30 s, followed by a temperature increase to 80°C, and final cooling at 40°C.
2.6 Stimulation of the peripheral blood mononuclear cells (PBMCs)
Heparin-containing tubes were used to collect eight milliliters of blood samples from eleven PTB, ten MDR-TB, and eleven healthy controls. Density gradient centrifugation was used to isolate PBMCs. One million cells were stimulated with 1 μg/mL of Pam3CSK4 molecule, which is a ligand of TLR2/TLR1 agonist (Hook et al., 2020) of M.tb for 24, 48, and 72 h and incubated at 37°C in 5% CO2 in a water-jacketed CO2 incubator (Thermo Electron Corporation). At each time point, the supernatant was collected and stored at −20°C for cytokine assay.
2.7 Measurement of TNF-α, IL-6, and IFN-γ in sera samples and culture supernatant of patients and healthy controls
Cytokines were estimated by sandwich ELISA system using the commercially available reagents from R&D System, United States, following the instructions of the manufacturer. Initially, standardization of PBMC culture was carried out to find out the optimum duration for the production of TNF-α and IL-6 in culture supernatant, which was observed to be highest in 24 h. Cytokines were estimated in culture supernatant of PBMCs and sera samples. The analysis of cytokines in sera samples was performed in 97 PTB, 98 MDR, 45 XDR, and 98 healthy controls.
2.8 Statistical analysis
Frequency of genotypes and alleles was counted. Hardy–Weinberg Equilibrium of genotype and allelic frequencies were checked by the chi-square test. Genotype and allele frequencies were compared among the participants by the chi-square test. All statistical analyses were performed by STATA/MP 16.1 software. Comparison of level of cytokine in sera samples and production in culture supernatant was performed by non-parametric analyses (Mann–Whitney test) using GraphPad Prism version 8.01 for Windows (GraphPad software, San Diego, CA) and STATA. The results are depicted in the form of a scattered plots. Comparisons of groups were performed using Kruskal–Wallis equality of proportion rank test. Data were expressed as Mean + _SD or median. p < 0.05 was considered as significant.
3 Results
3.1 Analysis of demographic parameters
A total of 101 PTB, 104 MDR, 48 XDR patients, and 130 healthy controls were included in the study. The mean age of TB patients and healthy controls was found to be significantly different (p = 0.007). The male-to-female ratio was similar (p = 0.49) among drug-resistant TB patients. On the analysis of various risk factors (smoking and alcoholic habits), among PTB, MDR and XDR cases, no significant difference was observed for smoking (p = 0.49) and alcoholic habits (p = 0.52), although a significant difference was observed related to consumption (p = 0.02) of tobacco among TB patients and healthy controls. A detailed analysis of demographic factors is shown in Supplementary Table S1.
3.2 TLR2 del polymorphism genotypic analysis
In the present study, genetic polymorphism of the TLR2 gene was analyzed by allele-specific polymerase chain reaction. The frequencies of genotypes for TLR2 del polymorphism did not deviate from Hardy–Weinberg equilibrium in PTB and XDR patients (p > 0.05) but were found to have deviated in healthy controls (p < 0.05) and MDR cases (p < 0.05). The wild-type genotype (Ins/Ins) was observed to be predominant in healthy controls (0.71), PTB (0.73), and MDR-TB participants (0.70), and heterozygous genotypes (Ins/del) were predominant in XDR TB patients (0.57). Table 2 displays the genotype and allele frequency distributions among the different TB patient categories. A pairwise comparison was made between PTB versus MDR, MDR versus XDR, and XDR versus PTB and is presented in Table 3, and TB patients with healthy controls are presented in Table 4. A significant difference was observed in allele and genotypic frequencies of PTB, MDR, and XDR patients for TLR2 deletion polymorphism (p = 0.0001), and no significant difference was observed in TLR1 743 A > G gene polymorphism (Table 2). On analyzing genotype and allele frequencies between MDR-TB and XDR and PTB and XDR-TB, a significant difference was observed between the groups of dominant and over-dominant models (p = 0.0001), as shown in Table 3, whereas no significant difference was observed between PTB and MDR-TB with healthy controls (Table 4).
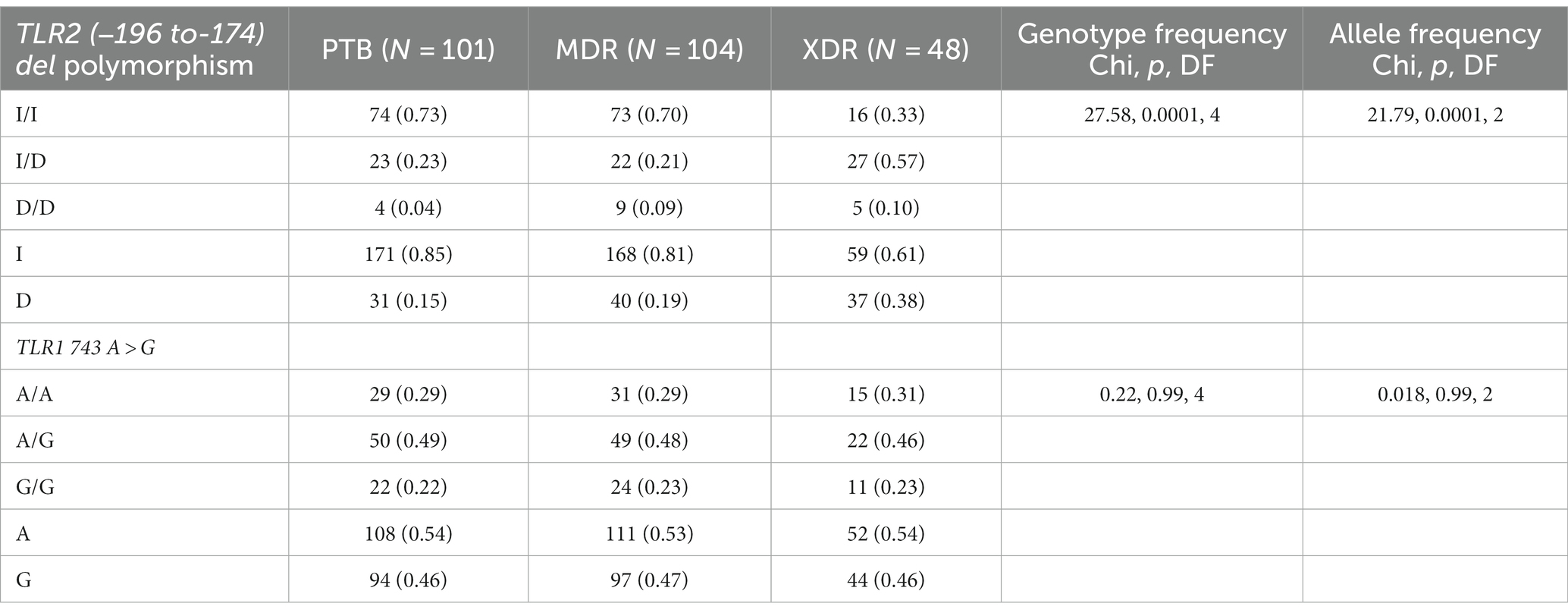
Table 2. TLR2 gene (−196 to −174) del polymorphism and TLR1 743 A > G genotype and allele frequencies among drug resistant TB patients (PTB, MDR and XDR).
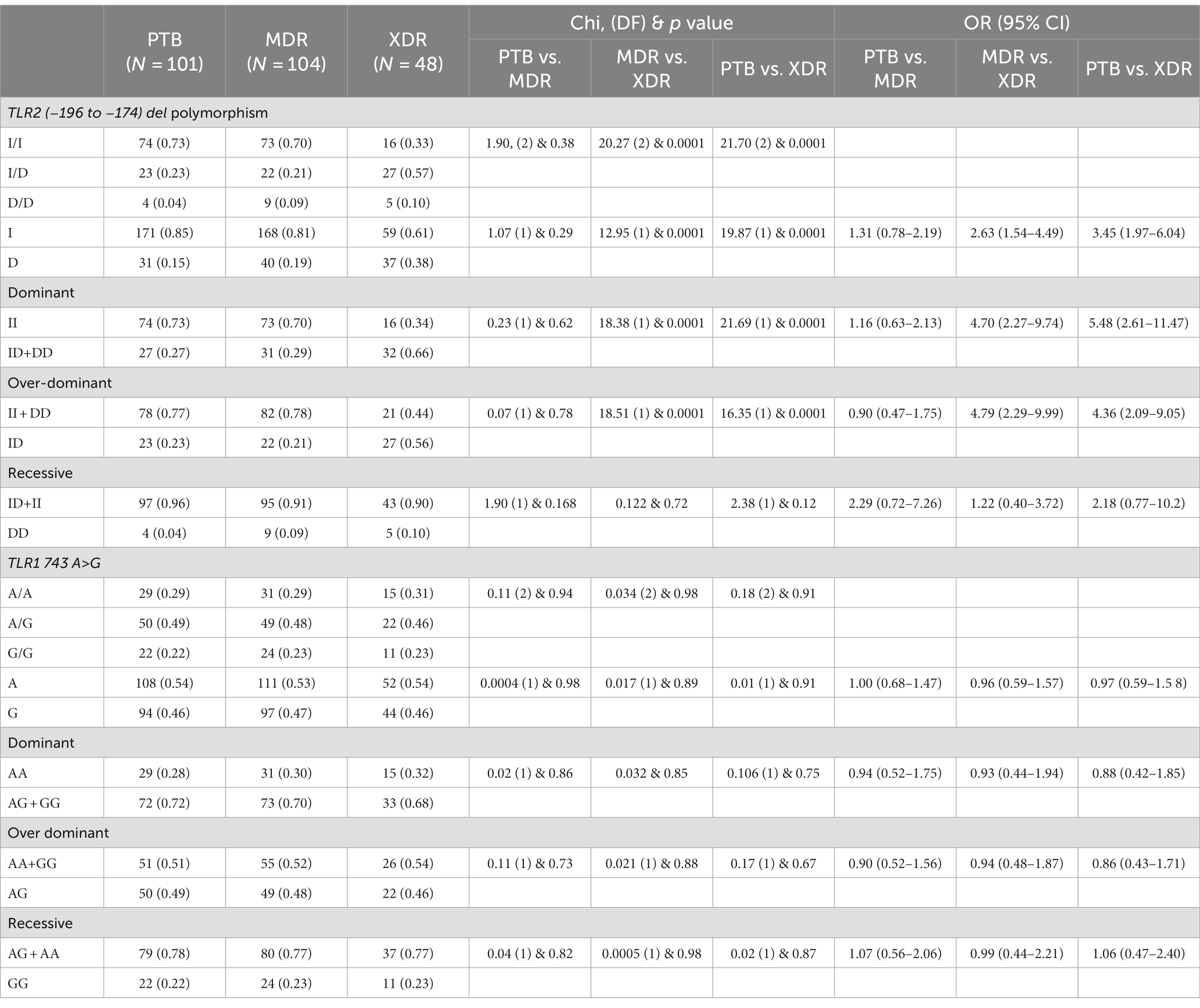
Table 3. TLR2 gene (−196 to −174) del polymorphism and TLR1 743 A > G genotype and allele frequencies among PTB and MDR, MDR and XDR, PTB and XDR.
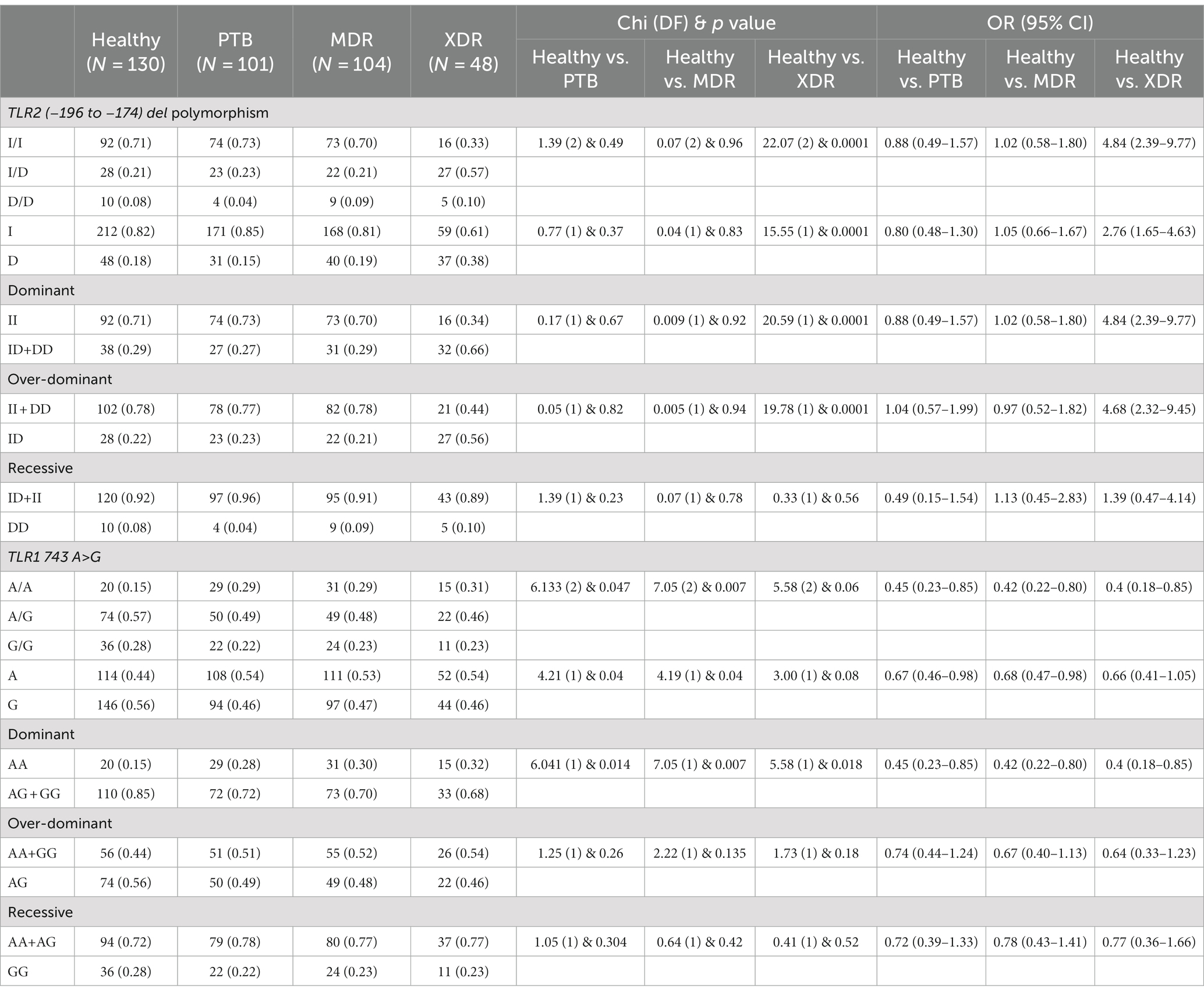
Table 4. TLR2 gene (−196 to −174) del polymorphism and TLR1 743 A> G genotype and allele frequencies between healthy controls and PTB, healthy controls and MDR, and healthy controls and XDR patients.
3.3 TLR1 743 (A > G) gene polymorphism genotypic analysis
In the present study, TLR1 743 A > G allele and genotype frequencies were analyzed by hybridization probe chemistry. TLR1 743 A > G gene polymorphism was analyzed in 101 PTB, 104 MDR, and 48 XDR patients and 130 healthy controls. The genotypic and allele frequencies did not deviate from Hardy–Weinberg equilibrium in any of the studied groups. The heterozygous genotype was observed to be predominant in healthy controls (0.57). Furthermore, analyses of genotype frequencies were carried out among the drug-resistant TB patients (PTB, MDR-TB, and XDR-TB). There was no significant difference in frequencies of alleles and genotypes (p = 0.99), as shown in Table 2. A significant difference in the frequencies of alleles and genotypes was observed between healthy controls and PTB patients, as well as between healthy controls and MDR-TB patients (p = 0.047), (p = 0.007) and also in the dominant model (p = 0.01), (p = 0.007), respectively, as shown in Table 4.
3.4 Estimation of TNF-α in relation to severity of tuberculosis disease (PTB, MDR-TB, XDR-TB, and HC) in sera samples and culture supernatant
The TNF-α level in sera samples among all categories is graphically presented in Figure 1A. The median level of TNF-α in sera samples was observed to be higher in PTB cases (97) (p < 0.0001) as compared with healthy controls (98), MDR-TB (98), and XDR cases (45). The level was higher in healthy controls compared with MDR (p < 0.0001) and XDR (p = 0.0006). There was no significant difference in TNF-α level between MDR and XDR TB patients, as shown in Figure 1A. The participants were stratified as per their TLR2 and TLR1 genotypes, and the level of TNF-α was compared among them. No significant difference was observed in the median level of cytokine among persons having various TLR2 genotypes within healthy controls and all disease types (PTB, MDR, and XDR cases). However, the level of serum TNF-α was higher in healthy controls having Ins/Ins and Ins/Del genotypes compared to XDR TB patients having similar genotypes. The observations are presented in Supplementary Figure S1. When the TNF-α level was analyzed in healthy controls and PTB cases having various TLR1 genotypes, a significant difference was observed in A/A (p = 0.003), A/G genotypes (p = 0.011), and G/G genotype (p = 0.003). There was no difference in the level of TNF-α among three TLR1 genotypes within the group (healthy controls, PTB cases, MDR cases, and XDR cases). The observations are presented in Supplementary Figure S2. The median level of TNF-α in culture supernatant was not significantly different in PTB cases and healthy controls in unstimulated samples as well as in samples stimulated with Pam3CSK4. When the level was compared in MDR and PTB cases, the median level was detected to be higher in MDR cases in unstimulated as well as in stimulated samples. The observations are presented in Supplementary Figure S3.
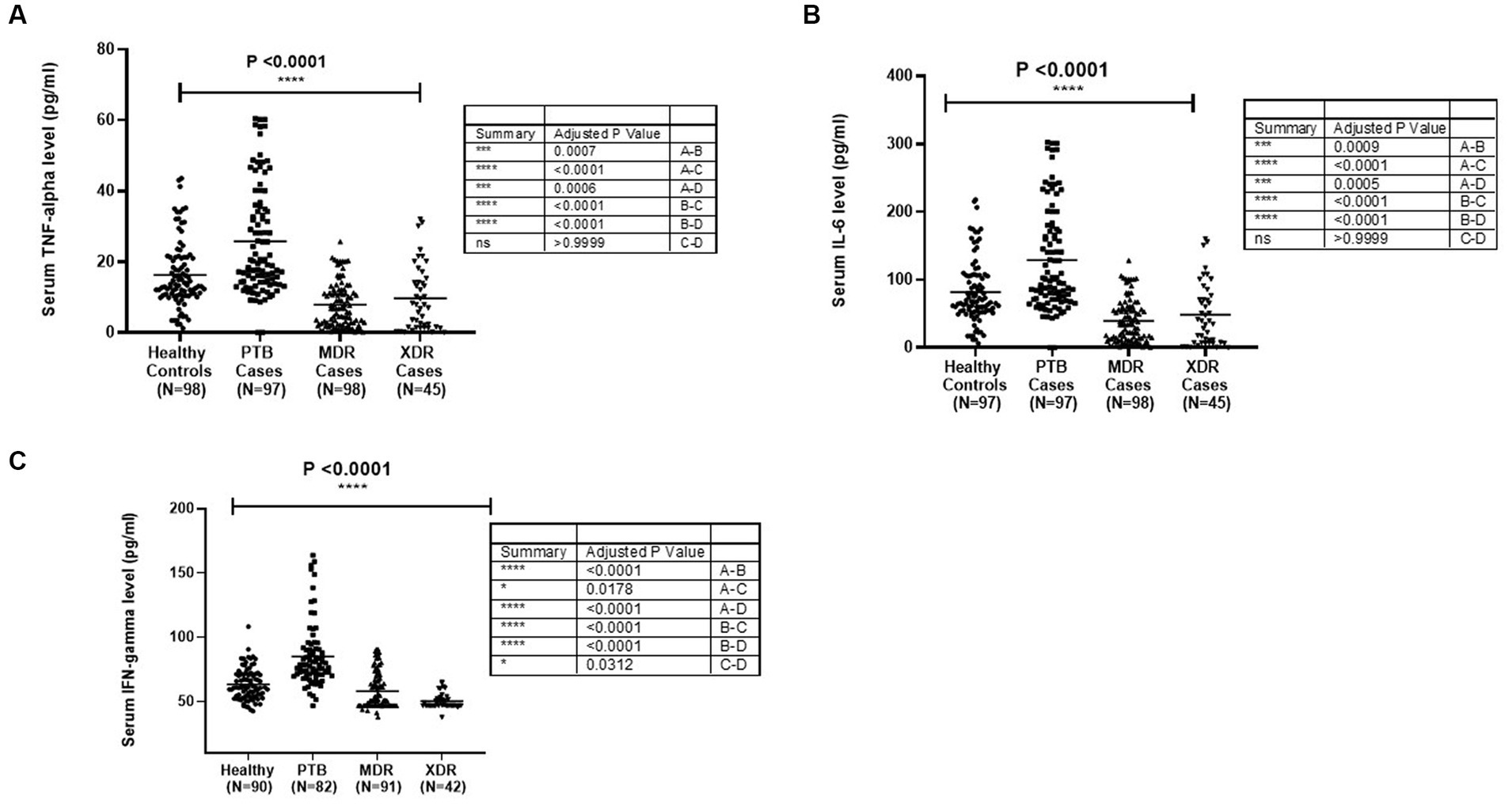
Figure 1. Cytokines [(A) TNF-α, (B) IL6 and (C) IFNγ] level in sera samples of healthy controls PTB, MDR-TB and XDR-TB patients. Each dot represents the level in pg/mL in the serum of one individual. The horizontal bar represents the median level. The level was compared among groups using the Kruskal-Wallis and Mann–Whitney test for pairwise comparison.
3.5 Estimation of IL-6 with the severity of tuberculosis disease (PTB, MDR-TB, XDR-TB, and HC) in sera samples and culture supernatant
The median level of IL-6 was detected to be higher in PTB cases as compared with healthy controls (p < 0.0001) and MDR-TB and XDR-TB cases. The level was higher in healthy controls compared with MDR (p < 0.0001) and XDR cases (p = 0.0005). The graphical representation of IL-6 level in sera samples is shown in Figure 1B. The participants were stratified as per their TLR1 and TLR2 genotypes, and the level of IL-6 was compared among them. No significant difference was observed in the mean level of cytokine among various genotypes within healthy controls and all disease types (PTB, MDR, and XDR cases). However, the level of serum IL-6 was higher in healthy controls having Ins/Ins and Ins/Del genotypes compared to XDR TB patients having similar genotypes. The observations are presented in Supplementary Figure S4. In the TLR1 gene, a significant difference was observed between healthy controls and PTB cases having A/A (0.006) and A/G (0.01) genotypes. The observations are presented in Supplementary Figure S5. The median level of IL-6 in culture supernatant was detected to be higher in PTB cases as compared with healthy controls in unstimulated and stimulated (Pam3CSK4) samples. The observations are presented in Supplementary Figure S6.
3.6 Estimation of IFN-γ in relation to severity of tuberculosis disease (PTB, MDR-TB, XDR-TB, and HC) in sera samples and culture supernatant
The graphical representation of IFN-γ level in sera samples is shown in Figure 1C. The median level of IFN-γ was detected to be higher in PTB cases as compared with healthy controls (p < 0.0001). The level was higher in healthy controls as compared with MDR (p = 0.0178) and XDR cases (p < 0.0001), as shown in Figure 1C. The participants were stratified as per their TLR1 and TLR2 genotypes, and the level of IFN-γ was compared among them. No significant difference was observed in the mean level of IFN-γ level among various genotypes within healthy controls and all disease types (PTB, MDR, and XDR cases). The observations are presented in Supplementary Figure S7. When the IFN-γ level was analyzed in healthy controls and PTB cases having various TLR1 genotypes, significantly higher level was observed in PTB patients A/A (p = 0.0021), A/G genotypes (p < 0.0001), and G/G genotype (p < 0.0001) as compared with healthy controls. No significant difference was observed in the median level of cytokine among various genotypes within healthy controls and all disease types (PTB, MDR, and XDR cases). The observations are presented in Supplementary Figure S8.
4 Discussion
TLRs play a crucial role in both innate and adaptive immune responses (Wu et al., 2015). The interaction between TLRs and their corresponding ligands initiates the recruitment of TIR domain-containing adaptor proteins, initiating signaling pathways that protect the host from microbial infections (Ogus et al., 2004). Human TLRs are a family of 10 proteins that selectively recognize microbes and initiate signaling pathways, resulting in the activation of the innate immune response, cytokine production, and the formation of an adaptive immune response (Guan et al., 2010).
The present study addressed the TLR2 (−196 to −174) del polymorphism and TLR1 743 A > G gene polymorphism among PTB, MDR, XDR, and healthy controls. The frequency of TLR2 (−196 to −174) ins/del genotype is observed to be higher among XDR-TB cases (0.57), whereas the frequency of TLR2 (−196 to −174) ins/del genotype in MDR-TB and PTB cases is almost equal (0.21 and 0.23, respectively). As a result, a significant difference is observed between XDR-TB and MDR-TB. The allele frequency of del allele of TLR2 (−196 to −174) polymorphism was observed to be higher among XDR-TB cases (0.38). When the genotypes carrying all del allele (ins/del + del/del) were compared against the ins allele (ins/ins), a significant difference was observed in both XDR-TB and MDR-TB (p = 0.0001) and XDR-TB and PTB (p = 0.0001), pointing toward the role of del polymorphism in developing XDR-TB. Velez et al. have reported the most robust association in TLR2 del polymorphism with TB in both Caucasian and African populations. This study indicated that TLR2 del is the susceptible gene for TB (Velez et al., 2010). According to the report by Khan et al., del polymorphism TLR2 (−196 to −174) increased the chance of TB in the Pakistani population (Khan et al., 2014). Tahara et al. identified the effect of this TLR2 (−196 to −174) del polymorphism on the severity of H. pylori-induced gastritis (Tahara et al., 2008). Noguchi et al. reported that the ins/ins and ins/del genotypes of this TLR2 (−196 to −174) del polymorphism were associated with highly transcriptional activity (Noguchi et al., 2004).
All research studies conducted up to this point has revealed information regarding the susceptibility of a gene polymorphism to either extrapulmonary or pulmonary tuberculosis. TLR2 (−196 to −174) del polymorphism may contribute to drug resistance in tuberculosis given that it is associated with disease severity and is found in a higher proportion of drug-resistant cases of the disease. This study represents the first attempt to comprehend the impact of this polymorphism.
The heterozygous genotype (AG) of the TLR1 743 A > G polymorphism is predominant in all tuberculosis cases, including PTB (49%), MDR-TB (48%), and XDR-TB cases (46%) and healthy controls (57%). When tuberculosis cases were compared with healthy controls, there was a significant difference found in PTB (p = 0.047) and MDR-TB cases (p = 0.007). The AG genotype may have a protective role against tuberculosis (TB) since it was more common in healthy controls than in PTB, MDR-TB, or XDR-TB cases.
According to Dittrich et al., the TLR1 743 A > G gene is linked to TB protection (Dittrich et al., 2015). According to Salie et al., TLR1 rs 4833095 was found to be significantly associated with tuberculosis diseases (Salie et al., 2015). Moreover, after being stimulated with M. tuberculosis, transfected HEK cells carrying the rs 4833095 genotype showed an increased NF-kβ induction (Dittrich et al., 2015). In the meta-analysis conducted by Schurz et al. and Zhou et al., the AG heterozygous genotype of TLR1 rs 4833095 was found to be associated with resistance to tuberculosis in various ethnic groups (Schurz et al., 2015; Zhou and Zhang, 2020). TLR2 plays a significant role in recognizing gram-positive bacteria and other microbial components, such as peptidoglycan and lipopolysaccharides (Akira et al., 2006). TLR1 is a 786 residue type I transmembrane protein having an extracellular domain that is 581 amino acids long and leucine-rich, a transmembrane domain that is 23 amino acids long, and a cytoplasmic toll homology signaling protein that is 181 amino acids long (Zarember and Godowski, 2002). Certain studies have suggested that mutations in the TLR1 gene, which change the amount of IL-10 and tumor necrosis factor in macrophages, raise the risk of leprosy in Brazil and India (Schuring et al., 2009). The rs 4833095 mutation changes the DNA sequence to replace an adenine with a guanine. It is a missense mutation that causes the asparagine to turn into serine at amino acid position 248 of the protein, lowering TLR1 receptor expression in immune system cells (Uciechowski et al., 2011).
All these studies provide information about the susceptibility of gene polymorphisms with pulmonary TB or extrapulmonary TB. Our observations suggested that a significant difference in frequencies of genotypes among TB patients and heterozygous genotype (Ins/del) of TLR2 (−196 to −174) del polymorphism was more prevalent in drug-resistant patients. When the allele was considered, the del allele was predominant in drug-resistant patients (XDR-TB). TLR2 (−196 to −174) del polymorphism may have a role in contributing to drug resistance in tuberculosis, though the exact mechanism is still unknown, given its correlation with disease severity and the higher frequency in drug-resistant tuberculosis cases. The present study is the maiden effort to understand the effect of this polymorphism.
We have analyzed various demographic and environmental factors that may contribute to the severity of the disease. A significant difference was observed among various tuberculosis cases related to age, so it was adjusted in all the analyses. No significant difference was observed related to smoking and alcoholism, but a significant difference was observed in relation to tobacco chewing (p = 0.02). In the study by Yu et al., they found a positive association of smoking with pulmonary TB (Yu et al., 1988). According to Ghasemian et al., smokers were 2.9 times more susceptible to suffering from pulmonary TB (Ghasemian et al., 2009). The inhalation of smoke activates the alveolar macrophage and produces an inflammatory response, and the longer use of these risk factors diminishes the expression of surface proteins related to antigen presentation by these macrophages (Pankow et al., 1991).
Cytokines play a pivotal role in the host defense mechanism against M.tb infection (Anoosheh et al., 2009). Multifunctional cytokines are responsible for the activation, shaping, and response of host body functions. However, a proper balance of cytokines is essential for the right protective responses (Cicchese et al., 2018), since hypo-reactions or hyper-reactions can be harmful and worsen the illness. TNF-α is a proinflammatory cytokine that showed multiple biological effects (Barnes et al., 1993). TNF-α stimulates apoptosis of macrophages, resulting in cell death and subsequent presentation of mycobacterial infection by dendritic cells (Keane et al., 2000). Nakya et al. studied that serum level of TNF-α in pulmonary tuberculosis patients is greater than the control group (Nakaya et al., 1995). The cytokine level is likely to be lower in MDR TB patients (Donnelly et al., 1995). Many investigators have documented that TNF-α level is higher in newly diagnosed TB patients (Juffermans et al., 1998). The result of our study showed that TNF-α level in sera, IL-6, and IFN-γ in PTB patients was greater than the healthy controls, MDR patients, and XDR patients.
Cytokines work in a cascade fashion during TB, where IL-12 controls T1 cytokine (IL-2/IFN-γ) production (Cooper and Khader, 2008; Cooper, 2009; Kumar et al., 2019). The IFN-γ, in turn, activates macrophages (Tötemeyer et al., 2006; Basingnaa et al., 2018) that induce TNF-α secretion for infection containment and mycobacterial growth restriction (Mattos et al., 2010; Nemeth et al., 2011; Basingnaa et al., 2018). TNF-α and IL-1α are the predominant contributors of granuloma formation (Hernandez-Pando et al., 1997; Yao et al., 2017). However, their protective effect was downregulated by the high IL-6/IL-10 levels in association with pSTAT3/SOCS3 expression, which ultimately led to immune suppression and impaired T-cell function (Harling et al., 2019). Moreover, type 1 cytokines (TNF-α and IFN-γ) change their protective nature to promote the spread of disease and use their distinct association with bacterial density to indicate the severity of an infection (Kumar et al., 2019). A significant difference was observed between PTB and MDR cases for TNF-α and healthy and PTB cases for IL-6 cytokine in stimulated samples in culture supernatant samples. It can be deduced that the increased cytokine levels in cases (TNF-α, IL-6 and IFN-γ) by M. tuberculosis components are the effect of the defense mechanism mediated through TLR2 del and TLR1 743 A > G gene polymorphism.
There was a significant difference observed between healthy controls and PTB cases having A/G genotypes for TLR1 743 A > G gene polymorphism and healthy controls and XDR cases having Ins/del genotypes for TLR2 del polymorphism. So it could be suggested that TLR genotypes may be just one of several factors controlling the cytokine level as TLR2 signaling is known to regulate the activation of various transcription factors (Grassin-Delyle et al., 2020) which in turn regulates the cytokine levels in sera. A variety of factors, in addition to specific genotypes or mutations, may modulate host responses in a way that affects the host’s ability to fight off the continuously multiplying drug-resistant tuberculosis bacteria by up-regulating or down-regulating immunity-related genes and pathways (Ben-Kahla and Sahal, 2016). TLRs, which are a component of the immune system’s physiological barrier, are responsible for identifying mycobacteria and determining which ligands, such as PAM, are linked to them. Any mutation in the TLR gene would prevent it from recognizing the ligand and, as a result, prevent the signaling mechanism from being triggered (Basu et al., 2012). Altered TLR function might lead to reduced recognition of bacteria, contributing to a host environment that is more permissive to the growth of a resistant M. tuberculosis stain. It is the complex interplay between host genetics and the pathogen’s ability to evade the immune system (Vijay, 2018; Sia and Rengarajan, 2019). However, further study is required to establish the essential role of TLR2 del and TLR1 743 A > G gene polymorphism in the recognition of M.tb and induction of TNF-α and other cytokines. This may be the first study to analyze the immune response to drug-resistant tuberculosis patients by analyzing TLR2 (−196 to −174) del and TLR1 743 A > G gene polymorphism.
Data availability statement
The original contributions presented in the study are included in the article/Supplementary material, further inquiries can be directed to the corresponding author.
Ethics statement
The studies involving humans were approved by Institute Human Ethics Committee, ICMR-National JALMA Institute for Leprosy and Other Mycobacterial Diseases, Agra. The studies were conducted in accordance with the local legislation and institutional requirements. The participants provided their written informed consent to participate in this study.
Author contributions
DV: Formal analysis, Methodology, Writing – original draft, Writing – review & editing, Data curation, Software. SS: Writing – review & editing. KM: Conceptualization, Supervision, Funding acquisition, Formal analysis, Writing – review & editing, Software, Project administration. SK: Writing – review & editing, Investigation, Methodology, Validation. NV: Formal analysis, Writing – review & editing. ES: Writing – review & editing. SKB: Writing – review & editing.
Funding
The author(s) declare financial support was received for the research, authorship, and/or publication of this article. This study is funded by intramural research funding of ICMR-National JALMA Institute for Leprosy and Other Mycobacterial Diseases, Agra. Raj Pal Suman is acknowledged for technical help. DV was financially supported as PhD scholar by the Department of Science and Technology (DST), New Delhi (Award letter code. IF180309).
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2023.1305974/full#supplementary-material
Abbreviations
M.tb, Mycobacterium tuberculosis; TLR, Toll-like receptor; PTB, Pulmonary Tuberculosis; MDR-TB, Multi-drug resistant-TB; XDR-TB, Extensively drug resistant tuberculosis; PAMP, Pathogen associated molecular pattern; AFB, Acid fast bacteria; CBNAAT, Cartridge-based nucleic acid amplification test; LPA, Line probe assay; LC DST, Liquid culture Drug susceptibility testing; FL-LPA, First line-line probe assay; SL-LPA, Second line-line probe assay; TNF-α, Tumour necrosis factor-alpha; IL-6, Interleukin-6; IFN-γ, Interferon-gamma.
References
Akira, S., Uematsu, S., and Takeuchi, O. (2006). Pathogen recognition and innate immunity. Cells 124, 783–801. doi: 10.1016/j.cell.2006.02.015
Anoosheh, S., Farnia, P., Kargar, M., Kazem Poor, M., Seif, S., Noroozi, J., et al. (2009). Relationship between TNF-αGene polymorphisms and susceptibility to pulmonary tuberculosis. J. Adv. Med. Biomed. Res. 17, 45–54.
Barnes, P. F., Lu, S., Abrams, J. S., Wang, E., Yamamura, M., and Modlin, R. (1993). Cytokine production at the site of disease in human tuberculosis. Infect. Immun. 61, 3482–3489. doi: 10.1128/iai.61.8.3482-3489.1993
Basingnaa, A., Antwi-Bafour, S., Nkansah, D. O., Afutu, E., and Owusu, E. (2018). Plasma levels of cytokines (IL-10, IFN-γ and TNF-α) in multidrug resistant tuberculosis and drug responsive tuberculosis patients in Ghana. Diseases 7:2. doi: 10.3390/diseases7010002
Basu, J., Shin, D. M., and Jo, E. K. (2012). Mycobacterial signaling through toll-like receptors. Front. Cell. Infect. Microbiol. 2, 301–747. doi: 10.3389/fcimb.2012.00145
Ben-Kahla, I., and Sahal, A. H. (2016). Drug-resistant tuberculosis viewed from bacterial and host genomes. Int. J. Antimicrob. Agents 48, 353–360. doi: 10.1016/j.ijantimicag.2016.07.010
Berrington, W. R., and Hawn, T. R. (2007). Mycobacterium tuberculosis, macrophages, and the innate immune response: does common variation matter. Immunol. Rev. 219, 167–186. doi: 10.1111/j.1600-065X.2007.00545.x
Brightbill, H. D., Libraty, D. H., Krutzik, S. R., Yang, R. B., Belisle, J. T., Bleharski, J. R., et al. (1999). Host defense mechanisms triggered by microbial lipoproteins through toll-like receptors. Science 285, 732–736. doi: 10.1126/science.285.5428.732
Buwitt-Beckmann, U., Heine, H., Wiesmüller, K. H., Jung, G., Brock, R., Akira, S., et al. (2005). Toll-like receptor 6-independent signaling by diacylated lipopeptides. Eur. J. Immunol. 35, 282–289. doi: 10.1002/eji.200424955
Chen, Y. C., Hsiao, C. C., Chen, C. J., Chin, C. H., Liu, S. F., Wu, C. C., et al. (2010). Toll-like receptor 2 gene polymorphisms, pulmonary tuberculosis, and natural killer cell counts. BMC Med. Genet. 11, 1–10. doi: 10.1186/1471-2350-11-17
Cicchese, J. M., Evans, S., Hult, C., Joslyn, L. R., Wessler, T., Millar, J. A., et al. (2018). Dynamic balance of pro- and anti-infammatory signals controls disease and limits pathology. Immunol. Rev. 285, 147–167. doi: 10.1111/imr.12671
Cooper, A. M. (2009). Cell-mediated immune responses in tuberculosis. Annu. Rev. Immunol. 27, 393–422. doi: 10.1146/annurev.immunol.021908.132703
Cooper, A. M., and Khader, S. A. (2008). The role of cytokines in the initiation, expansion, and control of cellular immunity to tuberculosis. Immunol. Rev. 226, 191–204. doi: 10.1111/j.1600-065X.2008.00702.x
Dittrich, N., Berrocal-Almanza, L. C., Thada, S., Goyal, S., Slevogt, H., Sumanlatha, G., et al. (2015). Toll-like receptor 1 variations influence susceptibility and immune response to Mycobacterium-tuberculosis. Tuberculosis 95, 328–335. doi: 10.1016/j.tube.2015.02.045
Donnelly, R. P., Freeman, S. L., and Hayes, M. P. (1995). Inhibition of IL-10 expression by IFN-gamma up-regulates transcription of TNF-alpha in human monocytes. J. Immunol. 155, 1420–1427. doi: 10.4049/jimmunol.155.3.1420
Ghasemian, R., Najafi, N., Yadgarinia, D., and Alian, S. (2009). Association between cigarette smoking and pulmonary tuberculosis in men: a case-control study in Mazandaran, Iran. Iran. J. Clin. Infect. Dis. 4, 135–141.
Grassin-Delyle, S., Abrial, C., Salvator, H., Brollo, M., Naline, E., and Devillier, P. (2020). The role of toll-like receptors in the production of cytokines by human lung macrophages. J. Innate Immun. 12, 63–73. doi: 10.1159/000494463
Guan, Y., Ranoa, D. R. E., Jiang, S., Mutha, S. K., Li, X., Baudry, J., et al. (2010). Human TLRs 10 and 1 share common mechanisms of innate immune sensing but not signaling. J. Immunol. 184, 5094–5103. doi: 10.4049/jimmunol.0901888
Haehnel, V., Schwarz Fischer, L., Fenton, M. J., and Rehli, M. (2002). Transcriptional regulation of the human toll-like receptor 2 gene in monocytes and macrophages. J. Immunol. 168, 5629–5637. doi: 10.4049/jimmunol.168.11.5629
Harling, K., Adankwah, E., Güler, A., Afum-Adjei Awuah, A., Adu-Amoah, L., Mayatepek, E., et al. (2019). Constitutive STAT3 phosphorylation and IL-6/IL-10 co-expression are associated with impaired T-cell function in tuberculosis patients. Cell. Mol. Immunol. 16, 275–287. doi: 10.1038/cmi.2018.5
Hernandez-Pando, R., Orozco, H., Arriaga, K., Sampieri, A., Larriva-Sahd, J., and Madrid-Marina, V. (1997). Analysis of the local kinetics and localization of interleukin-1 alpha, tumour necrosis factor-alpha and transforming growth factor-beta, during the course of experimental pulmonary tuberculosis. Immunology 90, 607–617. doi: 10.1046/j.1365-2567.1997.00193.x
Hook, J. S., Cao, M., Weng, K., Kinnare, N., and Moreland, J. G. (2020). Mycobacterium tuberculosis lipoarabinomannan activates human neutrophils via a TLR2/1 mechanism distinct from Pam3CSK4. J. Immunol. 204, 671–681. doi: 10.4049/jimmunol.1900919
Indian Council of Medical Research. Ethical guidelines for biomedical research on human subjects. New Delhi: Indian Council of Medical Research; (2017).
Jahantigh, D., Salimi, S., Alavi-Naini, R., Emamdadi, A., Owaysee Osquee, H., and Farajian Mashhadi, F. (2013). Association between TLR4 and TLR9 gene polymorphisms with development of pulmonary tuberculosis in Zahedan, southeastern Iran. Sci. World J. 2013:534053. doi: 10.1155/2013/534053
Juffermans, N. P., Verbon, A., Van Deventer, S. J., Van Deutekom, H. E. N. K., Speelman, P., and Van Der Poll, T. O. M. (1998). Tumor necrosis factor and interleukin-1 inhibitors as markers of disease activity of tuberculosis. Am. J. Respir. Crit. Care Med. 157, 1328–1331. doi: 10.1164/ajrccm.157.4.9709126
Keane, J., Remold, H. G., and Kornfeld, H. (2000). Virulent Mycobacterium tuberculosis strains evade apoptosis of infected alveolar macrophages. J. Immunol. 164, 2016–2020. doi: 10.4049/jimmunol.164.4.2016
Khan, A. U. H., Aslam, M. A., Hussain, I., Naz, A. G., Rana, I. A., Ahmad, M. M., et al. (2014). Role of toll-like receptor 2 (−196 to –174) polymorphism in susceptibility to pulmonary tuberculosis in Pakistani population. Int. J. Immunogenet. 41, 105–111. doi: 10.1111/iji.12086
Kumar, N. P., Moideen, K., Banurekha, V. V., Nair, D., and Babu, S. (2019). Plasma proinfammatory cytokines are markers of disease severity and bacterial burden in pulmonary tuberculosis. Open Forum Infect. Dis. 6:257. doi: 10.1093/ofid/ofz257
Ma, X., Liu, Y., Gowen, B. B., Graviss, E. A., Clark, A. G., and Musser, J. M. (2007). Full-exon resequencing reveals toll-like receptor variants contribute to human susceptibility to tuberculosis disease. PLoS One 2, 1–9. doi: 10.1371/journal.pone.0001318
Mattos, A. M. M., Almeida, C. S., Franken, K. L. M. C., Alves, C. C. S., Abramo, C., de Souza, M. A., et al. (2010). Increased IgG1, IFN-gamma, TNF-alpha and IL-6 responses to Mycobacterium tuberculosis antigens in patients with tuberculosis are lower after chemotherapy. Int. Immunol. 22, 775–782. doi: 10.1093/intimm/dxq429
Miller, S. A., Dykes, D., and Polesky, H. (1988). A simple salting out procedure for extracting DNA from human nucleated cells. Nucleic Acids Res. 16:1215. doi: 10.1093/nar/16.3.1215
Mittal, M., Biswas, S. K., Singh, V., Arela, N., Katoch, V. M., Das, R., et al. (2018). Association of Toll like receptor 2 and 9 gene variants with pulmonary tuberculosis: exploration in a northern Indian population. Mol. Biol. Rep. 45, 469–476. doi: 10.1007/s11033-018-4182-z
Nakaya, M., Yoneda, T., Yoshikawa, M., Tsukaguchi, K., Tokuyama, T., Fu, A., et al. (1995). The evaluation of interleukin-8 (IL-8) and tumor necrosis factor-α(TNF-α) level in peripheral blood of patients with active pulmonary tuberculosis. Kekkaku (Tuberculosis) 70, 461–466. doi: 10.11400/kekkaku1923.70.461
Guidelines on Programmatic Management of Drug-Resistant Tuberculosis in India. (2017). Revised National Tuberculosis Control Programme. Central TB Division, Directorate General of Health Services.
Nemeth, J., Winkler, H. M., Boeck, L., Adegnika, A. A., Clement, E., Mve, T. M., et al. (2011). Specific cytokine patterns of pulmonary tuberculosis in Central Africa. Clin. Immunol. 138, 50–59. doi: 10.1016/j.clim.2010.09.005
Noguchi, E., Nishimura, F., Fukai, H., Kim, J., Ichikawa, K., Shibasaki, M., et al. (2004). An association study of asthma and total serum immunoglobin E levels for toll-like receptor polymorphisms in a Japanese population. Clin Exp Allergy 34, 177–183. doi: 10.1111/j.1365-2222.2004.01839.x
Ogus, A. C., Yoldas, B., Ozdemir, T., Uguz, A., Olcen, S., Keser, I., et al. (2004). The Arg753GLn polymorphism of the human toll-like receptor 2 gene in tuberculosis disease. Eur. Respir. J. 23, 219–223. doi: 10.1183/09031936.03.00061703
Omueti, K. O., Beyer, J. M., Johnson, C. M., Lyle, E. A., and Tapping, R. I. (2005). Domain exchange between human toll-like receptors 1 and 6 reveals a region required for lipopeptide discrimination. J. Biol. Chem. 280, 36616–36625. doi: 10.1074/jbc.M504320200
Pankow, W., Neumann, K., Rüschoff, J., Schröder, R., and von Wichert, P. (1991). Reduction in HLA-DR antigen density on alveolar macrophages of smokers. Lung 169, 255–262. doi: 10.1007/BF02714161
Salie, M., Daya, M., Lucas, L. A., Warren, R. M., van der Spuy, G. D., van Helden, P. D., et al. (2015). Association of toll-like receptors with susceptibility to tuberculosis suggests sex-specific effects of TLR8 polymorphisms. Infect. Genet. Evol. 34, 221–229. doi: 10.1016/j.meegid.2015.07.004
Schuring, R. P., Hamann, L., Faber, W. R., Pahan, D., Richardus, J. H., Schumann, R. R., et al. (2009). Polymorphism N248S in the human toll-like receptor 1 gene is related to leprosy and leprosy reactions. J. Infect. Dis. 199, 1816–1819. doi: 10.1086/599121
Schurz, H., Daya, M., Möller, M., Hoal, E. G., and Salie, M. (2015). TLR1, 2, 4, 6 and 9 variants associated with tuberculosis susceptibility: a systematic review and meta-analysis. PLoS One 10:e0139711. doi: 10.1371/journal.pone.0139711
Sia, J. K., and Rengarajan, J. (2019). Immunology of Mycobacterium tuberculosis infections. Microbiol. Spectr. 7, 10–1128. doi: 10.1128/microbiolspec.gpp3-0022-2018
Sinha, E., Biswas, S. K., Mittal, M., Bajaj, B., Singh, V., Arela, N., et al. (2014). Toll-like receptor 1 743 A>G, 1805 T>G & Toll-like Receptor 6 745 C>T gene polymorphism and tuberculosis: a case control study of north Indian population from Agra (India). Hum. Immunol. 75, 880–886. doi: 10.1016/j.humimm.2014.06.014
Tahara, T., Arisawa, T., Wang, F., Shibata, T., Nakamura, M., Sakata, M., et al. (2008). Toll-like receptor 2 (TLR) -196 to -174 del polymorphism in gastro-duodenal diseases in Japanese population. Dig. Dis. Sci. 53, 919–924. doi: 10.1007/s10620-007-9950-x
Takeuchi, O., Sato, S., Horiuchi, T., Hoshino, K., Takeda, K., Dong, Z., et al. (2002). Cutting edge: role of toll-like receptor 1 in mediating immune response to microbial lipoproteins. J. Immunol. 169, 10–14. doi: 10.4049/jimmunol.169.1.10
TB India. (2017). Revised National Tuberculosis Control Programme, Annual Status Report. Central TB Division, Director General of Health Services, Ministry of Health and Family Welfare, New Delhi. www.tbcindia.gov.in
Thada, S., Valluri, V. L., and Gaddam, S. L. (2013). Influence of toll-like receptor gene polymorphisms to tuberculosis susceptibility in humans. Scand. J. Immunol. 78, 221–229. doi: 10.1111/sji.12066
Tötemeyer, S., Sheppard, M., Lloyd, A., Roper, D., Dowson, C., Underhill, D., et al. (2006). IFN-gamma enhances production of nitric oxide from macrophages via a mechanism that depends on nucleo-tide oligomerization domain-2. J. Immunol. 176, 4804–4810. doi: 10.4049/jimmunol.176.8.4804
Uciechowski, P., Imhoff, H., Lange, C., Meyer, C. G., Browne, E. N., Kirsten, D. K., et al. (2011). Susceptibility to tuberculosis is associated with TLR1 polymorphisms resulting in a lack of TLR1 cell surface expression. J. Leukoc. Biol. 90, 377–388. doi: 10.1189/jlb.0409233
Varshney, D., Singh, S., Sinha, E., Mohanty, K. K., Kumar, S., Barik, S. K., et al. (2022). Systematic review and meta-analysis of human toll-like receptors genetic polymorphisms for susceptibility to tuberculosis infection. Cytokine 152:155791. doi: 10.1016/j.cyto.2021.155791
Velez, D. R., Wejse, C., Stryjewski, M. E., Abbate, E., Hulme, W. F., Myers, J. L., et al. (2010). Variants in toll-like receptors 2 and 9 influence susceptibility to pulmonary tuberculosis in Caucasians, African-Americans, and west Africans. Hum. Genet. 127, 65–73. doi: 10.1007/s00439-009-0741-7
Vidya, M. K., Kumar, V. G., Sejian, V., Bagath, M., Krishnan, G., and Bhatta, R. (2018). Toll-like receptors: significance, ligands, signaling pathways, and functions in mammals. Int. Rev. Immunol. 37, 20–36. doi: 10.1080/08830185.2017.1380200
Vijay, K. (2018). Toll-like receptors in immunity and inflammatory diseases: past, present, and future. Int. Immunopharmacol. 59, 391–412. doi: 10.1016/j.intimp.2018.03.00
Wu, L., Hu, Y., Li, D., Jiang, W., and Xu, B. (2015). Screening toll-like receptor markers to predict latent tuberculosis infection and subsequent tuberculosis disease in a Chinese population. BMC Med. Genet. 16, 19–11. doi: 10.1186/s12881-015-0166-1
Yao, X., Liu, Y., Liu, Y., Liu, W., Ye, Z., Zheng, C., et al. (2017). Multiplex analysis of plasma cytokines/chemokines showing different immune responses in active TB patients, latent TB infection and healthy participants. Tuberculosis (Edinb) 107, 88–94. doi: 10.1016/j.tube.2017.07.013
Yu, G. P., Hsieht, C. C., and Peng, J. (1988). Risk factors associated with the prevalence of pulmonary tuberculosis among sanitary workers in Shanghai. Tubercle 69, 105–112. doi: 10.1016/0041-3879(88)90072-4
Zarember, K. A., and Godowski, P. J. (2002). Tissue expression of human toll-like receptors and differential regulation of toll-like receptor mRNAs in leukocytes in response to microbes, their products, and cytokines. J. Immunol. 168, 554–561. doi: 10.4049/jimmunol.168.2.554
Keywords: Toll-like receptors, drug-resistant tuberculosis, genotyping, TNF-α, IL-6, IFN-γ
Citation: Varshney D, Singh SV, Mohanty KK, Kumar S, Varshney N, Sinha E and Barik SK (2024) Toll-like receptor 2 (−196 to −174) del and TLR1 743 A > G gene polymorphism—a possible association with drug-resistant tuberculosis in the north Indian population. Front. Microbiol. 14:1305974. doi: 10.3389/fmicb.2023.1305974
Edited by:
Vishwanath Venketaraman, Western University of Health Sciences, United StatesReviewed by:
Shibali Das, Washington University in St. Louis, United StatesLuis Horacio Gutiérrez-González, National Institute of Respiratory Diseases-Mexico (INER), Mexico
Copyright © 2024 Varshney, Singh, Mohanty, Kumar, Varshney, Sinha and Barik. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Keshar Kunja Mohanty, a2VzaGFyNjNAeWFob28uY29t; bW9oYW50eS5ra0BpY21yLmdvdi5pbg==
†Present address: Sushanta Kumar Barik,Institute of Biosciences and Technology, MGM University, Chhatrapati Sambhajinagar, Aurangabad, Maharashtra, India
 Deepika Varshney1
Deepika Varshney1 Shoor Vir Singh
Shoor Vir Singh Keshar Kunja Mohanty
Keshar Kunja Mohanty Santosh Kumar
Santosh Kumar Sushanta Kumar Barik
Sushanta Kumar Barik