- State Key Laboratory for Animal Disease Control and Prevention, Harbin Veterinary Research Institute, Chinese Academy of Agricultural Sciences, Harbin, China
Antibiotic resistance in Enterococcus faecium, Enterococcus faecalis, and Staphylococcus aureus remains a major public health concern worldwide. Furthermore, these microbes frequently co-exist in biofilm-associated infections, largely nullifying antibiotic-based therapy. Therefore, it is imperative to develop an efficient therapeutic strategy for combating infections caused by polymicrobial biofilms. In this study, we investigated the antibacterial and antibiofilm activity of the bacteriophage endolysin Ply113 in vitro. Ply113 exhibited high and rapid lytic activity against E. faecium, E. faecalis, and S. aureus, including vancomycin-resistant Enterococcus and methicillin-resistant S. aureus isolates. Transmission electron microscopy revealed that Ply113 treatment led to the detachment of bacterial cell walls and considerable cell lysis. Ply113 maintained stable lytic activity over a temperature range of 4–45°C, over a pH range of 5.0–8.0, and in the presence of 0–400 mM NaCl. Ply113 treatment effectively eliminated the mono-species biofilms formed by E. faecium, E. faecalis, and S. aureus in a dose-dependent manner. Ply113 was also able to eliminate the dual-species biofilms of E. faecium–S. aureus and E. faecalis–S. aureus. Additionally, Ply113 exerted potent antibacterial efficacy in vivo, distinctly decreasing the bacterial loads in a murine peritoneal septicemia model. Our findings suggest that the bacteriophage endolysin Ply113 is a promising antimicrobial agent for the treatment of polymicrobial infections.
1 Introduction
The Gram-positive bacteria Enterococcus faecium, Enterococcus faecalis, and Staphylococcus aureus are important opportunistic pathogens affecting both humans and animals, seriously threatening public health worldwide (Kadariya et al., 2014; García-Solache and Rice, 2019). These pathogens can cause severe and diverse clinical manifestations, such as bacteremia, endocarditis, urinary tract infections, skin wounds, and soft tissue infections (García-Solache and Rice, 2019; Cheung et al., 2021). Currently, an increasing number of antibiotic-resistant strains of E. faecium, E. faecalis, and S. aureus are being found in hospital and community-related infections, causing a huge economic burden globally (Jaradat et al., 2020; Van Hal et al., 2022). The emergence of vancomycin-resistant Enterococcus (VRE) and methicillin-resistant S. aureus (MRSA) poses significant challenges to clinical treatment. In recent years, MRSA infections have shifted from healthcare-related infections to community-related infections. These community-acquired strains carry the staphylococcal chromosome cassette mec IV, which is easily transferred horizontally, exacerbating the prevalence of MRSA (Ma et al., 2002; Long, 2020). Similarly, the prevalence of invasive VRE infections has recently increased worldwide, and they are difficult to treat, resulting in a high risk of mortality (Papanicolaou et al., 2019). Both VRE and MRSA have been highlighted by the World Health Organization, which has listed them on its most recent list of high-priority pathogens for which new therapies are urgently needed (Tacconelli et al., 2018).
Enterococcus species and S. aureus are known for their propensity to produce biofilms, which aids in their survival in hostile environments and persistence on host tissue (Moormeier and Bayles, 2017; Ch'ng et al., 2019). Biofilm-associated infections are not only difficult to eradicate, but they also serve as a breeding ground for bacteria. In comparison to infection with mono-species biofilms, infection with polymicrobial biofilms usually results in more severe clinical signs with a high mortality rate (Peters et al., 2012). Enterococcus species and S. aureus, especially VRE and MRSA, are frequently co-isolated from intestinal samples, chronic wounds, and device insertion sites of patients (Furuno et al., 2005; Heinze et al., 2019; Yoon et al., 2019). In the microenvironment of polymicrobial biofilms, VRE has been observed to promote the transfer of antibiotic resistance genes and the formation of vancomycin-resistant S. aureus (Weigel et al., 2007). Additionally, S. aureus could offer heme to boost E. faecalis growth and the formation of dual-species biofilms (Ch'ng et al., 2022). Therefore, new antibacterial agents need to be active not just against antibiotic-resistant bacteria, but also against infections caused by polymicrobial biofilms.
Bacteriophages and their derived lytic proteins are an intriguing class of therapeutic agents with significant potential in treating infections caused by antibiotic-resistant pathogens. Bacteriophages, also known as phages, are viruses that can infect and replicate in bacteria. When the lytic bacteriophages infect their host bacteria, they inject their genome into the host bacteria and use their host’s biosynthetic machinery to replicate and produce progeny virions (Rodriguez-Rubio et al., 2016). At the last phase of the lytic cycle, bacteriophage endolysins are generated to break the bacterial cell walls and allow the progeny to be released (Roach and Donovan, 2015). Whereas the lytic cycle results in bacterial cell lysis, the lysogenic cycle involves the phage genome integrating into the host bacterial genome and replicating alongside the host (Erez et al., 2017). Bacteriophage endolysins are widely regarded as a promising antibacterial alternative for treating antibiotic-resistant bacteria in clinical settings. Furthermore, several endolysins have been demonstrated to be effective in the control of biofilm formation, highlighting their potential as antibiofilm agents that could be used in the treatment of biofilm-associated infection (Sass and Bierbaum, 2007; Poonacha et al., 2017; Lusiak-Szelachowska et al., 2020).
In this study, we investigated the antibacterial activity of Ply113, an endolysin derived from an E. faecium bacteriophage, against antibiotic-resistant strains of E. faecium, E. faecalis, and S. aureus. The efficacy of Ply113 was also evaluated in eradicating mono-species and polymicrobial biofilms produced by Enterococcus species and S. aureus.
2 Materials and methods
2.1 Bacterial strains and growth conditions
The E. faecium, E. faecalis, and S. aureus strains used in this study are listed in Supplementary Table S1. All E. faecium and E. faecalis strains were routinely cultured in Brain Heart Infusion (BHI) broth (Difco, Detroit, MI, United States) at 37°C, and all S. aureus strains were grown in Tryptic Soy Broth (TSB) medium (Difco) at 37°C. Escherichia coli strains DH5α and BL21 (DE3) were cultured in Luria–Bertani (LB) broth (Difco) to clone and express the Ply113 protein, respectively.
2.2 Bioinformatics analysis
The conserved domains of Ply113 were identified using SMART (Letunic et al., 2021). Three-dimensional (3D) structural homology modeling of Ply113 was conducted with the automated SWISS-MODEL (Bienert et al., 2017). Molecular models were analyzed and presented by PyMOL (DeLano Scientific, Palo Alto, CA, United States).
2.3 Protein expression and purification
The gene fragment encoding Ply113 (GenBank Accession No. MZ147816.1) was synthesized by BGI Biotech (Beijing, China) and cloned into the pET22b expression vector (Novagen, San Diego, CA, USA). The recombinant plasmid containing the correct insert was transformed into E. coli BL21 (DE3) competent cells. To induce Ply113 protein expression, the cells were grown in LB broth to an optical density at 600 nm (OD600) of 0.6. Then, Isopropyl β-D-1-thiogalactopyranoside (IPTG) (Sigma-Aldrich, St. Louis, MO, United States) was added at a final concentration of 0.8 mM, and the cells were incubated at 16°C for 12 h. Bacterial cells were harvested by centrifugation at 6,000 × g for 10 min, washed twice, and then resuspended in 20 mL of lysis buffer (20 mM Tris–HCl, 500 mM NaCl, pH 8.0). After sonication on ice, the lysates were centrifuged at 13,000 × g for 10 min. The Ply113 protein was purified by loading the collected supernatant onto a Ni-Sepharose™ 6 Fast Flow column (GE Healthcare, Amersham, Uppsala, Sweden) according to the manufacturer’s instructions. Phosphate buffer saline (PBS) was used in place of elution buffer using a PD-10 desalination column (GE Healthcare). The purified Ply113 protein was verified using sodium dodecyl sulfate-polyacrylamide gel electrophoresis.
2.4 Determination of minimal inhibitory concentrations
The minimum inhibitory concentrations (MICs) of Ply113 and antibiotics were determined in triplicate by the standard broth microdilution method (Clinical and Laboratory Standards Institute [CLSI], 2012). Briefly, 5.0 × 105 colony-forming units (CFU)/mL of freshly cultured E. faecium, E. faecalis, or S. aureus strains were distributed into the wells of 96-well polypropylene microtiter plates. Ply113, vancomycin, daptomycin, linezolid, and oxacillin were added to the wells in two-fold serial dilutions with final concentrations ranging from 0.5 to 256 μg/mL. The plate was incubated overnight at 37°C, and the MIC value was defined as the lowest concentration that inhibited visible growth.
2.5 Bactericidal assay
The bactericidal activity of Ply113 was determined by CFU reduction assays as previously described (Schuch et al., 2014). Briefly, the E. faecium, E. faecalis, and S. aureus strains were grown to an OD600 of 0.6 and collected by centrifugation at 3,000 × g for 10 min. The bacterial cells were washed three times with PBS, and then their concentration was adjusted to approximately 108 CFU/mL. Ply113 was added to the bacterial suspension at a final concentration of 8 μg/mL. PBS buffer was used as a control. After treatment at 37°C for 1 h, the mixture was serially diluted and plated on BHI or TSB agar in triplicate. The CFU reduction was calculated following incubation at 37°C for 20 h. For the time-killing assay, E. faecium strain ATCC700221, E. faecalis strain ATCC51299, and S. aureus strain ATCC33591 were separately incubated with Ply113 and antibiotics (1 × MIC) at 37°C for 5 h. PBS buffer was used as a control. After incubation, each sample was diluted and spread onto BHI or TSB agar plates to count CFU. All assays were performed in triplicate.
2.6 Transmission electron microscopy
E. faecium strain ATCC700221, E. faecalis strain ATCC51299, and S. aureus strain ATCC33591 were grown in 10 mL of BHI or TSB medium at 37°C to the mid-logarithmic phase. The bacterial cells were washed with PBS and then treated with 4 μg/mL Ply113 for 1 h at 37°C. Bacteria cells treated with PBS were the control. The samples were fixed with 2.5% glutaraldehyde at 4°C overnight and then treated with 1% osmic acid at 4°C for 2 h, and the samples were dehydrated with a 50, 70, 90, and 100% (v/v) alcohol series. Then, the samples were embedded into epoxy resin overnight at 25°C, and polymerization was performed at 70°C for 48 h. Ultrathin sections (thicknesses ranging from 65 to 70 nm) were sliced and deposited on copper grids. After staining with uranyl acetate and lead citrate, morphology was observed under a transmission electron microscope (Hitachi, Tokyo, Japan).
2.7 Effects of temperature, salinity, and pH on Ply113 activity
To assess the effect of temperature on Ply113 lytic activity, a bacterial suspension of E. faecalis ATCC29212 (5.0 × 106 CFU/mL) in PBS was treated with 4 μg/mL Ply113 and incubated at different temperatures ranging from 4°C to 45°C. The effect of salinity on Ply113 lytic activity was investigated by incubating the bacterial suspension in 20 mM Tris–HCl buffer (pH 7.0) with different NaCl concentrations (0–400 mM) at 37°C. For the pH stability assay, the bacterial suspension was treated with 4 μg/mL Ply113 in buffers with different pH ranges (50 mM sodium acetate for pH 5.0–6.0 and 20 mM Tris–HCl for pH 7.0–9.0). After 1 h of incubation, each sample was diluted and spread onto BHI or TSB agar plates to count CFU. The lytic activity was expressed as the relative reduction in bacterial CFU. The relative activity was determined by setting the maximum activity of Ply113 under various test conditions at 100%. The experiment was performed in triplicate.
2.8 Biofilm elimination assay in 24-well polystyrene plates
The effect of Ply113 against established biofilms of E. faecium, E. faecalis, and S. aureus was determined as previously described (Duarte et al., 2021). E. faecium D73, E. faecalis E141, and S. aureus YD2 were grown in TSB medium to the logarithmic growth phase. Then, 100 μL of bacterial culture was added to 900 μL of TSB medium in the wells of a 24-well polypropylene microtiter plate, which was incubated at 37°C for 24 h to induce the formation of mono-species biofilms. For the establishment of dual-species biofilms, E. faecium or E. faecalis culture was mixed with S. aureus culture at a 1:1 ratio, and 100 μL of the E. faecium–S. aureus mixture or E. faecalis–S. aureus mixture was added to 900 μL of TSB medium in 24-well plates to allow dual-species biofilm formation. For the biofilm elimination assay, the preformed biofilms were incubated with various concentrations of Ply113 (0–64 μg/mL) at 37°C for 2 h and washed with PBS. The remaining biofilms were stained with 0.1% crystal violet solution and dissolved in 33% acetic acid. The absorbance of the obtained solution in each well was measured at 570 nm using an EnVision Multimode Plate Reader (PerkinElmer, Waltham, United States). The number of culturable bacteria in the biofilm following Ply113 treatment was also quantified by counting CFU to evaluate the biofilm elimination efficiency of Ply113. All assays were performed in triplicate.
2.9 Confocal laser scanning microscopy
Cultures of E. faecium D73 or E. faecalis E141 in the logarithmic growth phase were mixed with S. aureus YD2 culture in a 1:1 ratio. Next, 200 μL of the E. faecium–S. aureus mixture or the E. faecalis–S. aureus mixture was added to 1.8 mL of TSB medium in a glass-bottom culture dish, which was incubated for 24 h at 37°C to induce the formation of dual-species biofilms. The established biofilms were washed twice with PBS and exposed to 8 μg/mL Ply113 for 2 h. Biofilms treated with PBS were the control. After Ply113 treatment, the biofilms were stained with the Live/Read® BacLight™ kit (Invitrogen, Eugene, OR, United States) and observed using a confocal scanning laser microscope (Zeiss LSM 800, Jena, Germany).
2.10 Hemolysis assay
The in vitro hemolytic activity of Ply113 was determined as previously described (Tucker et al., 2018). First, 4% (w/v) red blood cells (RBCs) of pig and rabbit in PBS were incubated with various concentrations of Ply113 (ranging from 0 to 256 μg/mL) in 96-well polystyrene plates for 1 h at 37°C. RBC suspensions treated with 0.5% Triton X-100 and PBS were, respectively, used as positive and negative controls. The samples were centrifuged at 1,000 × g for 5 min, and the supernatants were transferred to new 96-well microplates. The absorbance was measured at 545 nm. The hemolysis rate was calculated using the following equation: hemolysis (%) = [(Asample - APBS)/(Atriton X-100 - APBS)] × 100. The experiment was performed in triplicate.
2.11 Cytotoxicity assay
Human skin keratinocyte HaCaT cells and mouse bone marrow-derived macrophages (BMDMs) were used to evaluate the cytotoxicity of Ply113. Cells were seeded in 96-well plates at 1 × 105 cells per well and cultured for 12 h. Then the cells were exposed to different concentrations of Ply113 (ranging from 0 to 256 μg/mL) for 24 h. PBS buffer was used as a control. Cell viability was measured using the Cell Counting Kit-8 (CCK-8; Beyotime, Beijing, China), and absorbance was measured at 450 nm. The experiment was performed in triplicate.
2.12 Murine model of peritoneal septicemia
The procedure for the mouse experiment was approved by the Institutional Animal Care and Use Committee of the Harbin Veterinary Research Institute (Permission No. HVRI-IACUC-23110803GR). Peritoneal septicemia of mice was induced with E. faecium, E. faecalis, and S. aureus as previously described, with some modifications (Schuch et al., 2014; Brown Gandt et al., 2018). 6-week-old BALB/c mice were randomly assigned to five groups (n = 10 per group). The five groups were, respectively, inoculated intraperitoneally with 0.2 mL of E. faecium D73 (8 × 107 CFU), E. faecalis E141 (7 × 107 CFU), S. aureus YD2 (2 × 107 CFU), E. faecium–S. aureus mixture (1:1 ratio), or E. faecalis–S. aureus mixture (1:1 ratio). After 1 h post-infection, five mice in each group were administered intraperitoneally with either 0.2 mL of Ply113 (10 mg/kg) or PBS. The mice were humanely euthanized after 24 h of infection, and the organs were homogenized with sterile PBS, and the bacterial loads in these organs were calculated by plate counting.
2.13 Statistical analysis
Statistical analysis was conducted and the graphs were created with GraphPad Prism 9.0 (GraphPad Software, Inc., La Jolla, CA, United States). All the data were assessed for normality using the Shapiro–Wilk test. The equality of variance was assessed using the F test or Brown-Forsythe test. If the data were normally distributed and had equal variance, an unpaired t-test or one-way analysis of variance (ANOVA) was used to determine statistical significance. A p-value of <0.05 was considered to be statistically significant.
3 Results
3.1 Lytic activity of Ply113 against Enterococcus faecium and Enterococcus faecalis
The bacteriophage endolysin Ply113 consists of 314 amino acids with an expected molecular weight of 34.1 kDa. According to SMART predictions, Ply113 has an N-terminal NlpC/P60 domain for enzymatic catalysis and a C-terminal SH3b domain for cell wall binding (Supplementary Figure S1A). The 3D structure of Ply113 was obtained by homology modeling using the crystal structure of the Streptococcus pyogenes bacteriophage endolysin PlyPy (PDB code: 5UDM) as a template. Molecular models and 3D structure analysis revealed that Ply113 has three α-helices and five β-sheets in the NlpC/P60 domain and seven β-sheets in the SH3 domain (Supplementary Figure S1B).
The in vitro antibacterial activities of Ply113 against E. faecium and E. faecalis strains were examined. The MICs of Ply113 against these strains ranged from 4 to 8 μg/mL, as shown in Table 1. The lytic activity of Ply113 and antibiotics at their 1 × MIC were tested against E. faecium and E. faecalis strains. When vancomycin-resistant E. faecium (VREfm) strain ATCC700221 was treated with 4 μg/mL (1 × MIC) Ply113 for 10 min, log10 (CFU/mL) decreased by approximately 4.1; after 5 h, log10 (CFU/mL) decreased by nearly 6.4 (Figure 1A). Similarly, when vancomycin-resistant E. faecalis (VREfs) strain ATCC51299 was treated with 8 μg/mL (1 × MIC) Ply113 for 10 min, log10 (CFU/mL) decreased by 3.5, and after 5 h, log10 (CFU/mL) decreased by 5.8 (Figure 1B). Compared to Ply113, linezolid and daptomycin exhibited slower bactericidal activity. When VREfm and VREfs were treated with these antibiotics for 5 h, log10 (CFU/mL) reduced by no more than 2.5 (Figures 1A,B). The bacteriolytic spectrum of Ply113 against different Enterococcus isolates was also determined. As shown in Figures 1C,D, Ply113 was effective against all E. faecium and E. faecalis isolates tested, including VRE and linezolid-resistant Enterococcus (LRE).
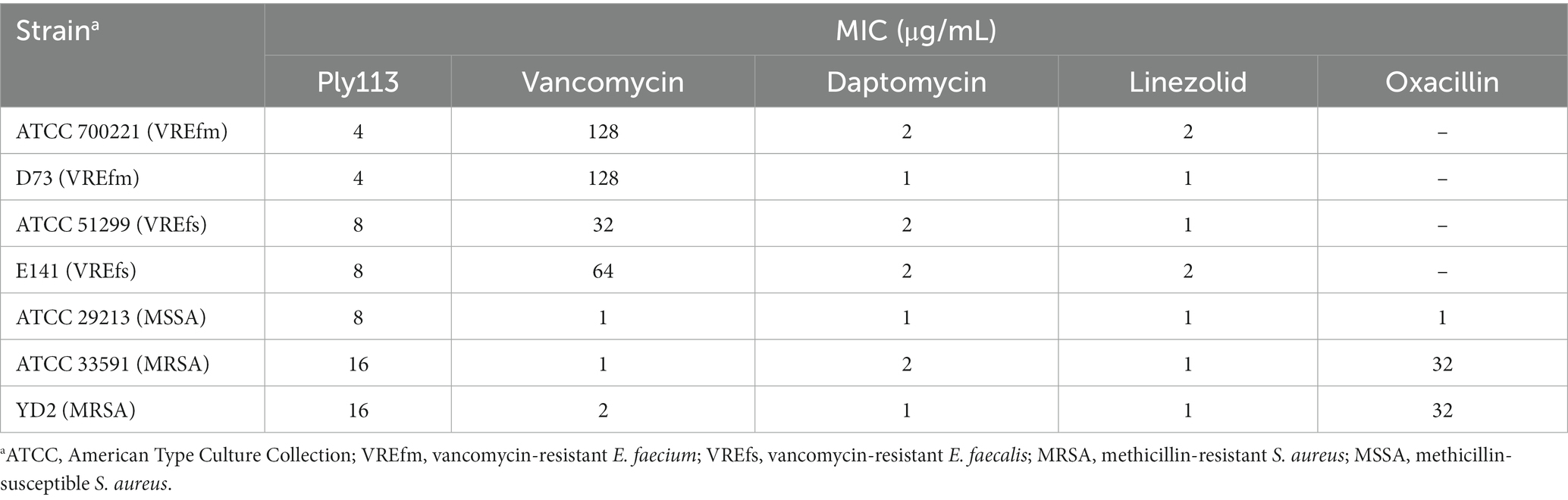
Table 1. MICs of Ply113 and antibiotics against Enterococcus faecium, Enterococcus faecalis, and Staphylococcus aureus.
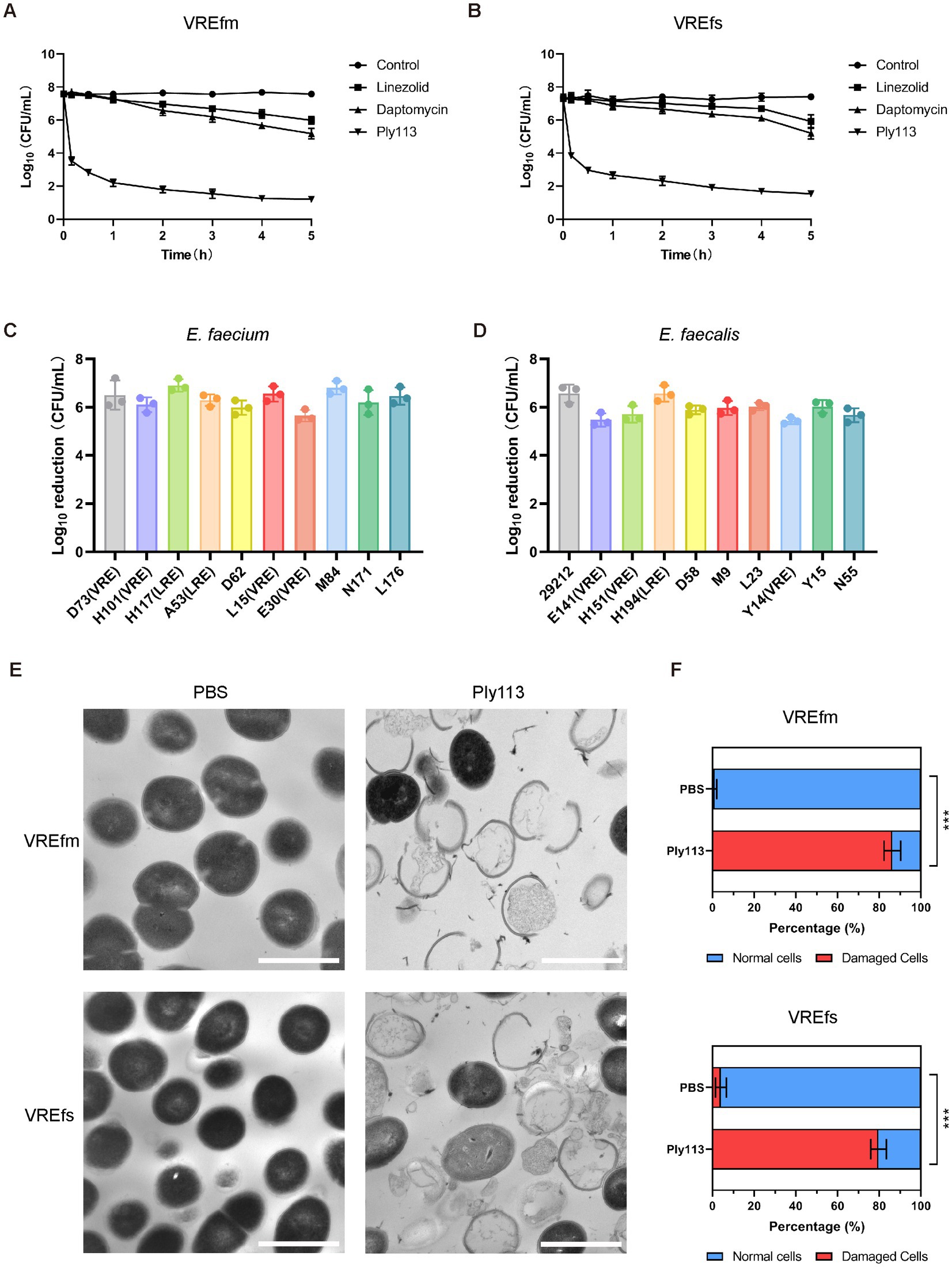
Figure 1. Lytic activity of Ply113 against E. faecium and E. faecalis. (A,B) Time-dependent killing curves of Ply113 and antibiotics against (A) VREfm strain ATCC700221 and (B) VREfs strain ATCC51299. Bacterial cells were incubated with Ply113 or antibiotics for different periods of time. The drug concentrations correspond to strain-specific 1 × MIC values. Each sample was diluted and spread onto BHI agar plates to count CFU. (C,D) Susceptibility of multiple isolates of (C) E. faecium and (D) E. faecalis to Ply113. An inoculum of approximately 108 CFU/mL of different isolates was incubated with 8 μg/mL Ply113 at 37°C for 1 h. The reduction in bacterial CFU after Ply113 treatment indicates the bactericidal activity. (E) Morphological changes of VREfm and VREfs after exposure to Ply113. Scale bar, 1 μm. Cultures of VREfm and VREfs in the logarithmic growth phase were treated with 4 μg/mL Ply113 at 37°C for 1 h and observed using TEM. (F) The percentages of morphologically damaged cells of VREfm and VREfs after exposure to Ply113. Averages and standard deviations (SD) of three independent counts are shown. The number of cells for each count is 100 (n = 100). Statistical significance was calculated using unpaired t tests, ***p < 0.001.
The morphological changes of VREfm and VREfs after treatment with Ply113 were observed using transmission electron microscopy (TEM). As shown in Figure 1E, VREfm and VREfs in PBS displayed a regular spherical shape, with intact cell walls and a homogeneous electron density in the cytoplasm. However, Ply113 treatment led to marked morphological damage in VREfm and VREfs. A majority of treated cells exhibited a fractured peptidoglycan structure and cytoplasmic membrane damage, along with the leakage of intracellular content from the rupture site. After Ply113 treatment, approximately 86.3% of VREfm cells were destroyed, whereas approximately 79.7% of VREfs cells were abnormal (Figure 1F). The percentage of damaged cells in treatment groups and untreated controls showed statistically significant differences (p < 0.001).
3.2 Lytic activity of Ply113 against Staphylococcus aureus
The MICs of Ply113 against S. aureus strains ranged from 8 to 16 μg/mL, as shown in Table 1. The lytic activities of Ply113 against S. aureus strains were further tested in a time-killing assay. As shown in Figure 2A, Ply113 was highly active against MRSA ATCC33591. After treatment with 16 μg/mL (1 × MIC) Ply113 for 10 min, log10 (CFU/mL) decreased by 3.3. After treatment for 5 h, log10 (CFU/mL) decreased by 6.0 (Figure 2A). Log10 (CFU/mL) dropped by no more than 2.6 when MRSA was treated with linezolid, daptomycin, and vancomycin for 5 h. Ply113 was also shown to be active against a variety of S. aureus isolates, including MRSA isolates, with log10 (CFU/mL) decreasing by 4.7–5.6 (Figure 2B). TEM observation revealed that MRSA cells exposed to Ply113 exhibited substantial morphological alterations (Figure 2C). Untreated MRSA cells had a smooth outer layer of normal cells with a typical ball shape. Ply113 treatment caused cell wall detachment in MRSA, exhibiting a deep distortion and rupture of the cytoplasmic membrane. The electron density in the cytoplasm was drastically reduced, and many cells were lysed like “ghost cells.” Nearly 71.7% of cells were damaged after Ply113 treatment (Figure 2D). The percentage of damaged cells in the treatment and control groups differed statistically significantly (p < 0.001).
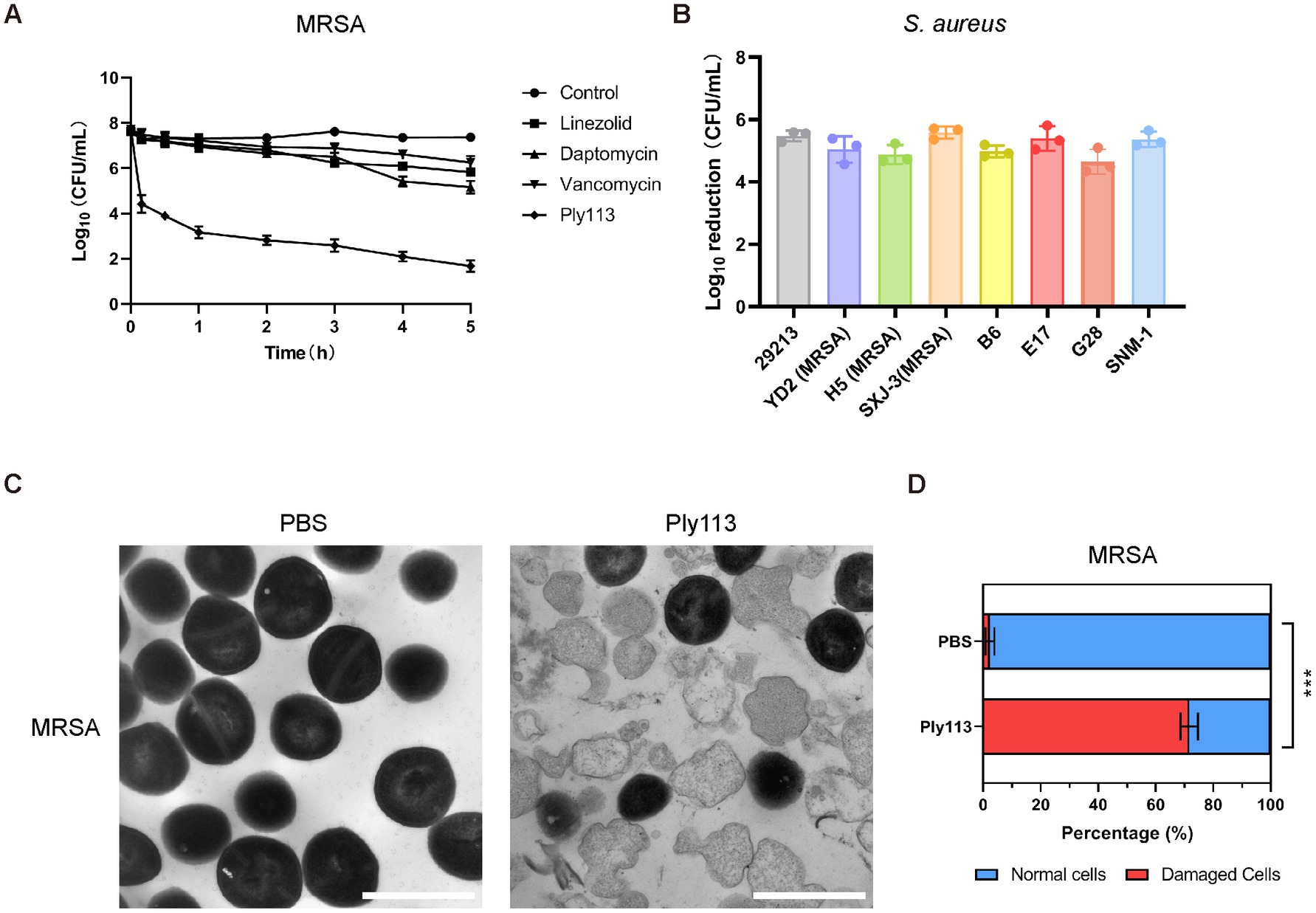
Figure 2. Lytic activity of Ply113 against S. aureus. (A) Time-dependent killing curves of Ply113 and antibiotics against MRSA strain ATCC33591. Bacterial cells were incubated with Ply113 or antibiotics for different periods of time. The drug concentrations correspond to strain-specific 1 × MIC values. Each sample was diluted and spread onto TSB agar plates to count CFU. (B) Susceptibility of multiple isolates of S. aureus to Ply113. An inoculum of approximately 108 CFU/mL of different isolates was incubated with 8 μg/mL Ply113 at 37°C for 1 h. The reduction in bacterial CFU after Ply113 treatment indicates the bactericidal activity. (C) Morphological changes of MRSA after exposure to Ply113. Scale bar, 1 μm. Cultures of MRSA in the logarithmic growth phase were treated with 4 μg/mL Ply113 at 37°C for 1 h and observed using TEM. (D) The percentages of morphologically damaged cells of MRSA after exposure to Ply113. Averages ± SD of three independent counts are shown. The number of cells for each count is 100 (n = 100). Statistical significance was calculated using unpaired t tests, ***p < 0.001.
3.3 Biochemical characterization and stability of Ply113
The influence of temperature, salinity, and pH on the lytic activity of Ply113 was investigated by assessing CFU. Ply113 remained relatively stable, with high activity at temperatures ranging from 4°C to 45°C (Figure 3A). The effect of salt concentration on the lytic activity of Ply113 was also minimal. Ply113 exhibited the highest lytic activity when the salt concentration was 100 mM. As the concentration was increased to 400 mM, the lytic activity of Ply113 decreased marginally (Figure 3B). Although the lytic activity of Ply113 declined to 36.2% at pH 9.0, it maintained a high lytic activity over a wide pH range of 5.0–8.0 (Figure 3C).
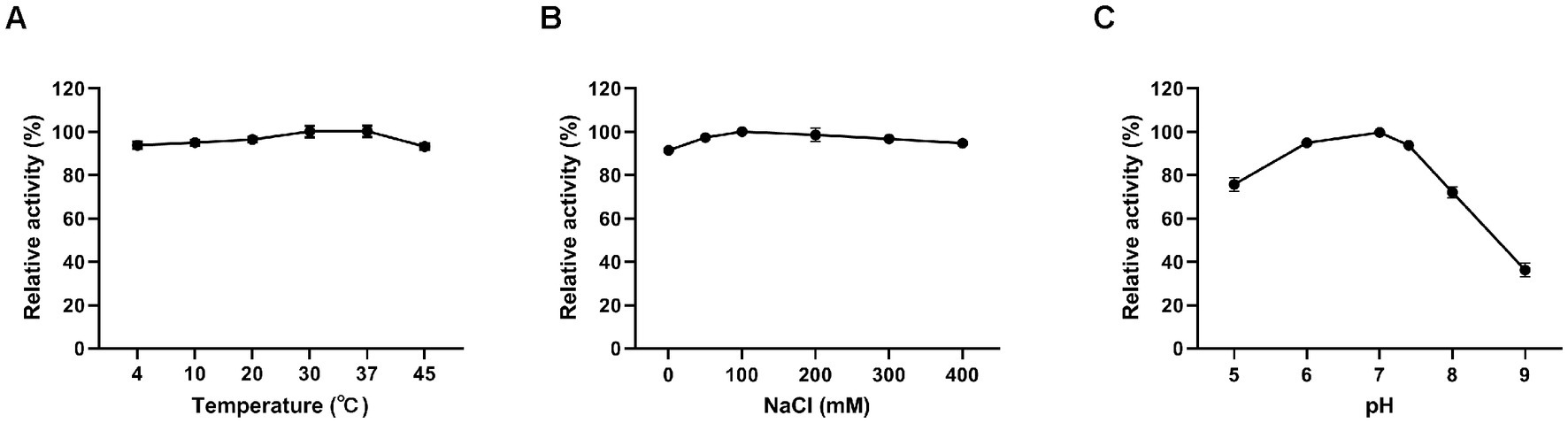
Figure 3. Effects of different conditions on the lytic activity of Ply113. The effects of (A) temperature, (B) NaCl, and (C) pH on the lytic activity of Ply113 were examined by incubating bacterial suspensions of E. faecalis with 4 μg/mL Ply113 under different conditions for 1 h. The maximum activity of Ply113 under each test condition was set as 100%. Data are presented as the mean ± SD from three independent tests.
3.4 Antibiofilm activity of Ply113 against the mono-species biofilms of Enterococcus faecium, Enterococcus faecalis, and Staphylococcus aureus
To determine the antibiofilm activity of Ply113, the mono-species biofilms of E. faecium, E. faecalis, and S. aureus were formed in 24-well polystyrene microplates, and each well was exposed to Ply113 for 2 h. The VREfm isolate D73, VREfs isolate E141, and MRSA isolate YD2 were chosen for the biofilm elimination assay on the basis of their capacity to form strong biofilms. The crystal violet staining results showed that Ply113 could significantly eliminate the biofilms of E. faecium, E. faecalis, and S. aureus in a dose-dependent manner (Figures 4A–C). Following treatment with 64 μg/mL Ply113, approximately 91.7, 95.8, and 78.7% of the biofilms formed by E. faecium, E. faecalis, and S. aureus, respectively, were eliminated. The number of culturable bacteria in biofilms after exposure to Ply113 was also evaluated. As shown in Figures 4D–F, the 24-h biofilms contained 107.2 CFU/mL of E. faecium, 107.5 CFU/mL of E. faecalis, and 107.8 CFU/mL of S. aureus. After 2 h of Ply113 treatment, the numbers of culturable bacteria in mono-species biofilms decreased with increasing Ply113 concentrations. E. faecalis was the most sensitive to Ply113; log10 (CFU/mL) was reduced by 4.5 when 64 μg/mL of Ply113 was applied. Under the same concentration of Ply113, log10 (CFU/mL) was reduced by 4.1 in E. faecium biofilms and by 3.7 in S. aureus biofilms. These results suggest that Ply113 can disrupt the mono-species biofilms of E. faecium, E. faecalis, and S. aureus.
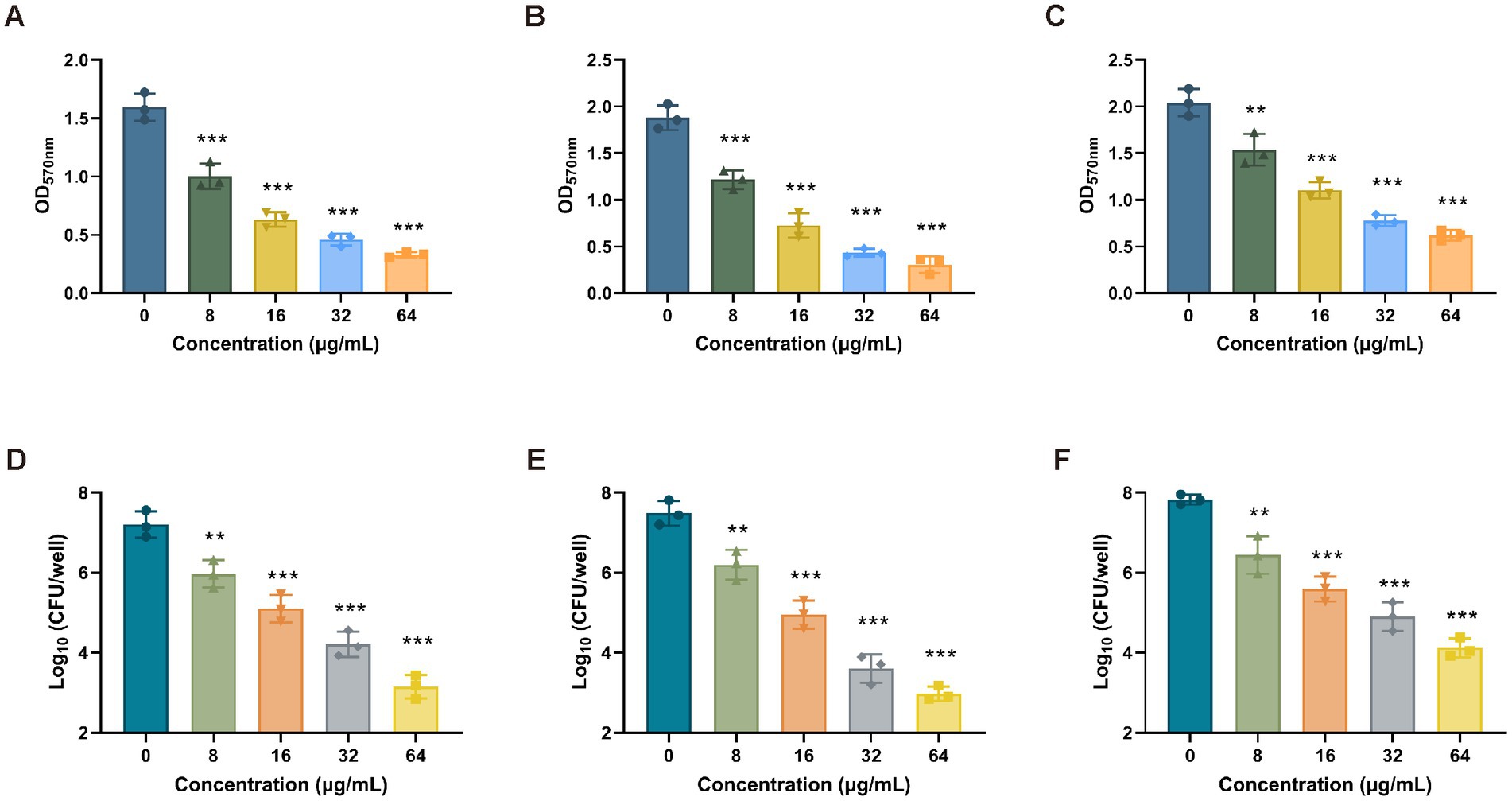
Figure 4. Biofilm removal efficacy of Ply113 against the mono-species biofilms. (A–C) Effectiveness of Ply113 on the mono-species biofilms of (A) E. faecium, (B) E. faecalis, and (C) S. aureus preformed in 24-well plates. The preformed mono-species biofilms were treated with different concentrations of Ply113 at 37°C for 2 h and quantified using crystal violet staining. (D–F) Quantifications of (D) E. faecium, (E) E. faecalis, and (F) S. aureus by CFU counting after Ply113 treatment of preformed mono-species biofilms. Data are presented as the mean ± SD from three independent tests. Statistical significance was calculated using one-way ANOVA. **p < 0.01, ***p < 0.001.
3.5 Capability of Ply113 to eliminate dual-species biofilms produced by Enterococcus species and Staphylococcus aureus
To produce dual-species biofilms, E. faecium or E. faecalis was mixed with S. aureus in polystyrene microplates. When E. faecium and S. aureus were cultured together for 24 h, greater biofilm production was observed compared with each mono-species biofilm (Figure 5A). The biofilm production between the dual-species biofilms and mono-species biofilms differed statistically significantly (p < 0.05). A similar observation was made in the dual-species biofilms produced by E. faecalis and S. aureus (Figure 5B). Preformed dual-species biofilms were treated with Ply113 for 2 h and analyzed using crystal violet staining. Ply113 eliminated the dual-species biofilms of E. faecium–S. aureus and E. faecalis–S. aureus in a dose-dependent manner (Figures 5C,D). Ply113 at a concentration of 64 μg/mL eliminated approximately 78.3% of the E. faecium–S. aureus biofilms and 79% of the E. faecalis–S. aureus biofilms. The number of culturable bacteria in dual-species biofilms reduced as Ply113 concentrations increased (Figures 5E–H). To visualize the efficacy of Ply113 in disrupting the dual-species biofilms, SYTO 9–PI staining was performed on 24-h-old dual-species biofilms following exposure to 8 μg/mL Ply113. Ply113 treatment of E. faecium–S. aureus or E. faecalis–S. aureus dual-species biofilms resulted in a considerable decrease in SYTO 9-stained green bacterial cells and a significant increase in PI-stained red dead cells (Figure 5I). These results suggest that Ply113 has a high biofilm-disrupting potential not only against mono-species but also against polymicrobial biofilms.
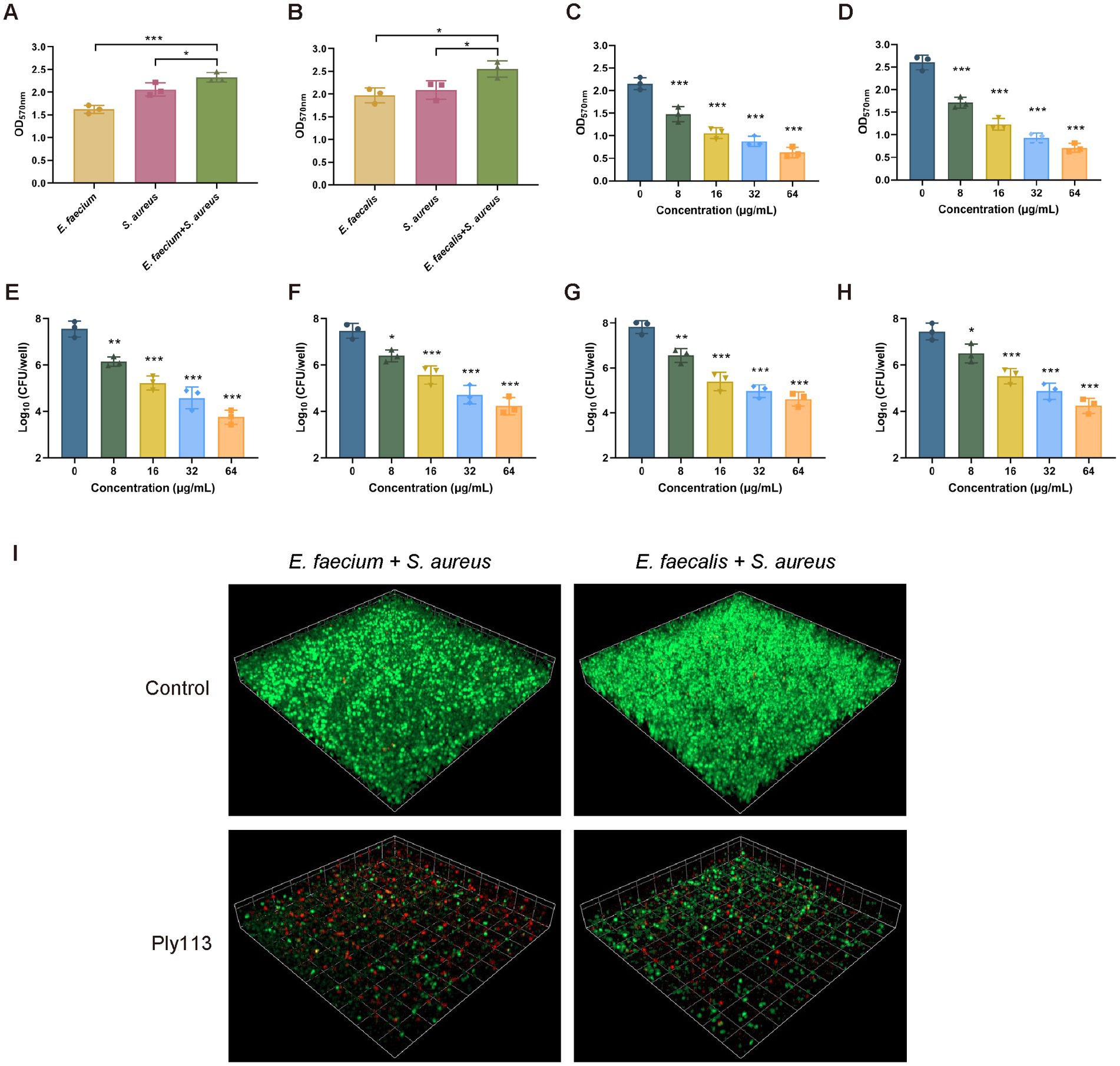
Figure 5. Biofilm elimination efficacy of Ply113 against dual-species biofilms. (A,B) Dual-species biofilms formed in 24 h by either (A) E. faecium–S. aureus or (B) E. faecalis–S. aureus at a 1:1 ratio in 24-well plates. (C,D) Effects of Ply113 on the dual-species biofilms of (C) E. faecium–S. aureus and (D) E. faecalis–S. aureus. The preformed dual-species biofilms were incubated with various concentrations of Ply113 at 37°C for 2 h and quantified using crystal violet staining. (E–H) Quantifications of (E) E. faecium and (F) S. aureus in the E. faecium–S. aureus biofilms and (G) E. faecalis and (H) S. aureus in the E. faecalis–S. aureus biofilms by CFU counting after Ply113 treatment. (I) Confocal micrographs of SYTO 9–PI-stained dual-species biofilms of E. faecium–S. aureus and E. faecalis–S. aureus after Ply113 treatment for 2 h. Data are presented as the mean ± SD from three independent tests. Statistical significance was calculated using one-way ANOVA. *p < 0.05, **p < 0.01, ***p < 0.001.
3.6 Hemolytic activity and cytotoxicity of Ply113
To determine the hemolytic activity of Ply113, hemolytic assays were conducted by incubating RBCs from pigs and rabbits with Ply113. As shown in Figures 6A,B, Ply113 was found to have negligible hemolytic activity (<2%) in either pig or rabbit RBCs, even at concentrations of up to 256 μg/mL. Hemolysis in the treatment and positive control groups differed statistically significantly (p < 0.001). The potential cytotoxicity of Ply113 was also evaluated by performing the CCK-8 assay using HaCaT cells and BMDMs. No significant reduction (p > 0.05) in cell viability was detected after 24 h of Ply113 treatment at concentrations ranging from 16 to 256 μg/mL, but a 10–14.3% drop in the cell viability of mouse BMDMs was observed (Figures 6C,D).
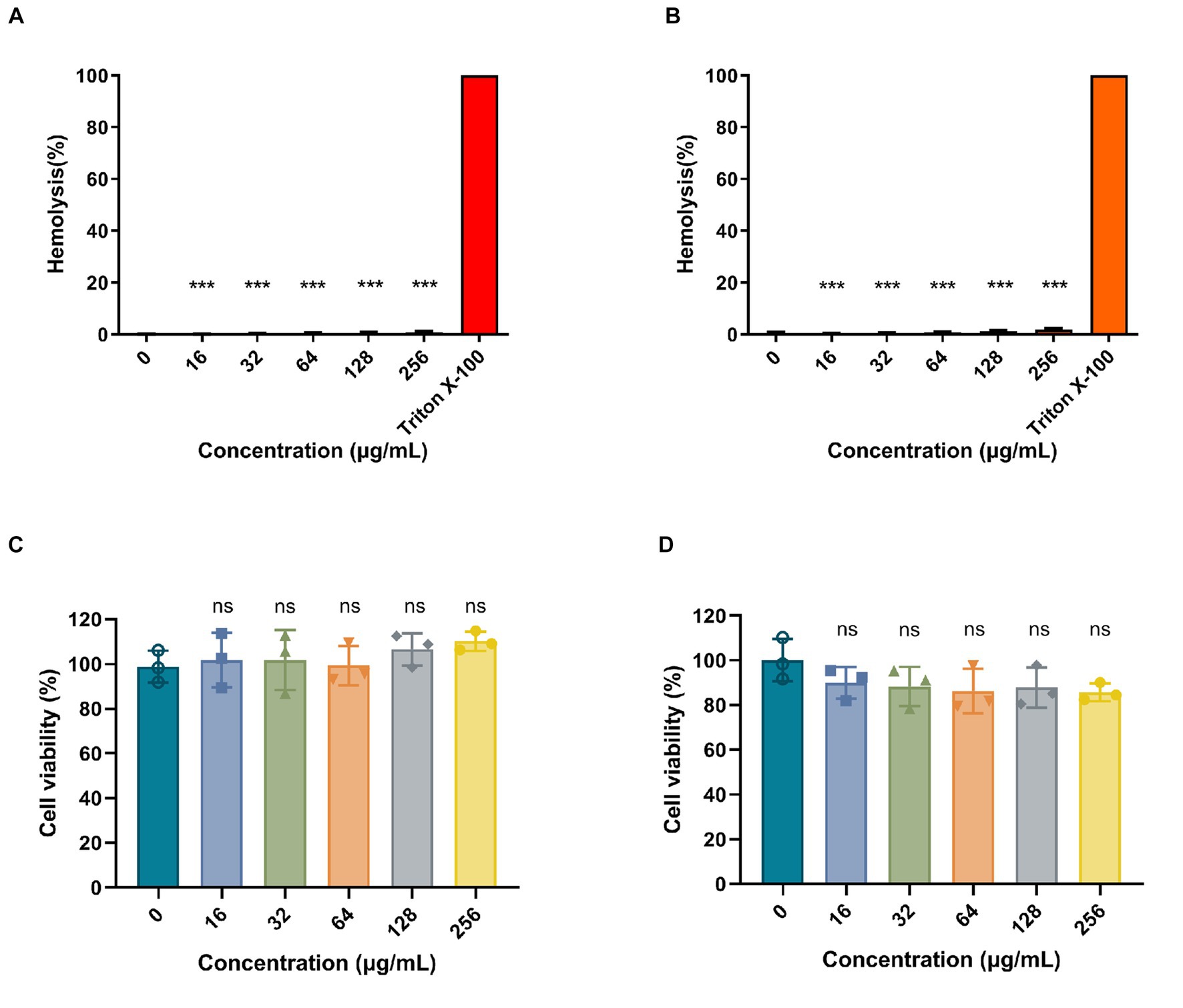
Figure 6. Hemolytic activity and cytotoxicity of Ply113. (A,B) Hemolytic activities of Ply113 on (A) pig red blood cells (RBCs) and (B) rabbit RBCs. RBCs were incubated with different concentrations of Ply113 at 37°C for 1 h. Hemolysis was determined by measuring the absorbance at 545 nm. PBS served as a negative control, Triton X-100 served as a positive control. (C,D) Cytotoxicity of Ply113 on (C) HaCaT cells and (D) BMDMs. Cell viability was examined after incubating cell suspensions with different concentrations of Ply113 for 24 h. Data are presented as the mean ± SD from three independent tests. Statistical significance was calculated using one-way ANOVA. ***p < 0.001, ns, not significant.
3.7 In vivo antibacterial efficacy of Ply113
The in vivo efficacy of Ply113 was evaluated in a murine model of peritoneal septicemia. Mice were, respectively, infected with E. faecium, E. faecalis, S. aureus, the E. faecium–S. aureus mixture, or the E. faecalis–S. aureus mixture, and then treated with either Ply113 or PBS 1 h later. As shown in Figures 7A–C, the bacterial loads in the livers and kidneys of Ply113-treated mice decreased by 3.0 and 2.5 log units in the E. faecium-infected group, 2.7 and 2.3 log units in the E. faecalis-infected group, and 2.4 and 1.8 log units in the S. aureus-infected group, respectively. In addition, Ply113 treatment was also found to reduce the bacterial loads (>2.5 log units) in the livers and kidneys of the E. faecium–S. aureus and E. faecalis–S. aureus infected groups using this single dose (Figures 7D,E). The bacterial loads in the organs between the treatment and control groups differed statistically significantly (p < 0.001).
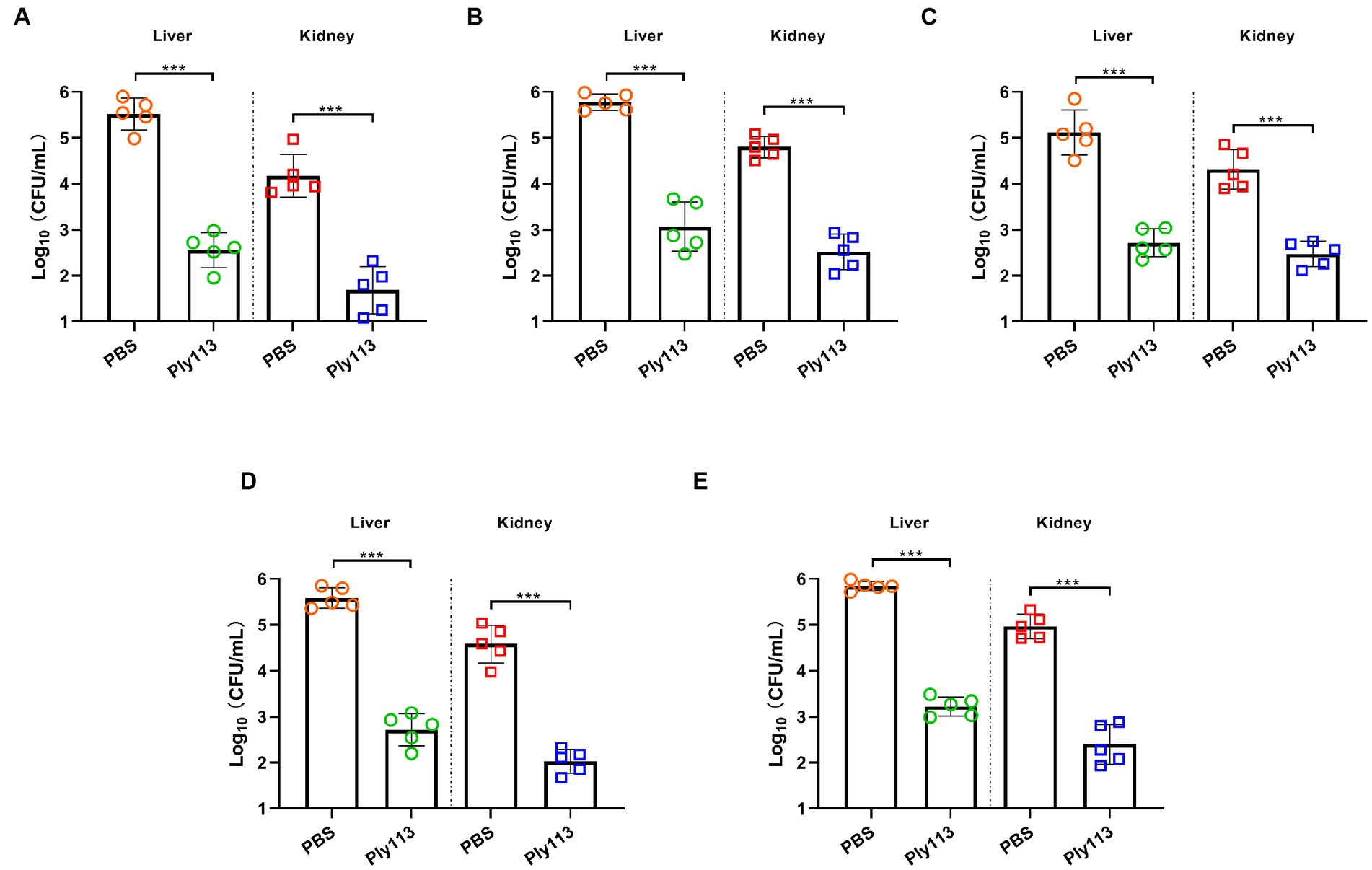
Figure 7. In vivo antibacterial efficacy of Ply113 in a murine model of peritoneal septicemia. BALB/c mice were inoculated intraperitoneally with (A) E. faecium, (B) E. faecalis, (C) S. aureus, (D) the E. faecium–S. aureus mixture, or (E) the E. faecalis–S. aureus mixture, and then treated with Ply113 1 h later. After 24 h of treatment, samples were collected from liver and kidney for bacterial quantification. Values represent the mean ± SD from five mice. Statistical significance was calculated using unpaired t tests. ***p < 0.001.
4 Discussion
The Gram-positive pathogenic bacteria E. faecium, E. faecalis, and S. aureus are internationally recognized as the three most concerning antibiotic-resistant pathogens causing hospital and community-related infections (Huang et al., 2019; Weiner-Lastinger et al., 2020). These three pathogenic bacteria can infect both humans and animals and cause sepsis, mastitis, and other diseases, threatening public health globally (Fitzgerald, 2012; Abat et al., 2016; Argudin et al., 2017). In recent years, the evolution of antibiotic resistance has become increasingly serious. The prevalence of MRSA and VRE is still steadily increasing, while vancomycin-resistant S. aureus and daptomycin-resistant Enterococcus are emerging (Munoz-Price et al., 2005; Cong et al., 2020). The antibiotic resistance crisis has highlighted the urgent need for innovation in therapeutics and treatment strategies.
Bacteriophage-derived endolysins are considered to have significant potential as novel therapeutic agents for effectively combating antibiotic-resistant bacterial infections. In the present study, the endolysin Ply113 from an E. faecium bacteriophage was identified and expressed, and its antibacterial activity was evaluated. Ply113 endolysin was chosen after searching for endolysin domains in the National Center for Biotechnology Information database (Oliveira et al., 2013), and we discovered that it had outstanding antibacterial activity against some Gram-positive bacteria. Ply113 showed high lytic activity against a broad range of E. faecium and E. faecalis strains, including clinical isolates of VRE and LRE. Ply113 also exhibited cross-species bactericidal efficacy against S. aureus strains, including MRSA isolates. Bacteriophages typically have species specificity, but several phage endolysins exhibit cross-species lytic activity. For example, the endolysin PlySs2 was found to have lytic activity against MRSA as well as a variety of Streptococcus species (Gilmer et al., 2013). Similarly, the LysSS endolysin has been shown to have activity against Acinetobacter baumannii, Klebsiella pneumoniae, Pseudomonas aeruginosa, and other bacterial species (Kim et al., 2020). Similar to PlySs2 and LysSS, Ply113 has cross-species lytic activity and is highly effective against both MRSA and VRE, which are notorious antibiotic-resistant pathogens. This relatively wide lytic spectrum of Ply113 also enhanced the possibility of its therapeutic application for the treatment of infections caused by Gram-positive pathogens.
An issue that cannot be ignored is that biofilms play an important role in bacterial infections, accounting for nearly 80% of human microbial infections (Romling and Balsalobre, 2012). Thus, the choice of antibacterial therapies also needs to consider the effectiveness of biofilm eradication. Endolysins have been recommended as a promising antimicrobial alternative for controlling bacterial biofilms (Liu et al., 2023). LysSA52, an endolysin derived from an S. aureus bacteriophage, was reported to remove 60% of biofilms after 12 h of treatment at a concentration of 500 μg/mL (Abdurahman et al., 2023). Another study showed that the Lys84 endolysin at concentrations greater than 10 μM could effectively eliminate 90% of the biofilms of S. aureus in 2 h (Ning et al., 2021). According to the present findings, Ply113 at concentrations of more than 8 μg/mL (0.24 μM) was capable of significantly eliminating biofilms of E. faecium, E. faecalis, and S. aureus after 2 h of treatment. At a Ply113 concentration of 64 μg/mL (1.9 μM), up to 95.8% of the biofilm could be eliminated, demonstrating the high efficiency of Ply113 in biofilm removal.
In clinical settings, it is common for multiple bacterial species to co-infect and invade the same tissue in a host (Pammi et al., 2014; Azevedo et al., 2017). The formation of polymicrobial biofilms involves two or more bacterial species interacting with each other, enhancing their antibiotic resistance and pathogenicity through a variety of mechanisms (Sadiq et al., 2022). Here, we observed a significant increase in biofilm formation when E. faecalis was co-cultured with S. aureus, in comparison to individual cultures. A previous study has shown that interactions between E. faecalis and S. aureus could promote the formation of dual-species biofilms (Ch'ng et al., 2022), which is consistent with our findings. In addition, we observed that dual-species biofilm formation was also enhanced in the combination of E. faecium–S. aureus. Several phage endolysins have been demonstrated to be efficient against bacterial biofilms (Poonacha et al., 2017; Ning et al., 2021; Abdurahman et al., 2023; Liu et al., 2023); however, these antibiofilm effects have largely been verified on mono-species biofilms. Polymicrobial biofilms are more tolerant to antimicrobial agents than mono-species biofilms; for example, ciprofloxacin concentrations must be higher against polymicrobial biofilms formed by S. aureus and P. aeruginosa compared with mono-species biofilms (Tkhilaishvili et al., 2020). Therefore, the original therapy dose may be ineffective in eliminating polymicrobial biofilms, which is also the main reason why polymicrobial infections often result in treatment failure and even death (Pammi et al., 2014). Several antibacterial agents have recently been reported for their action on polymicrobial biofilms formed by enterococci and S. aureus. It has been discovered that the bacteriophage phiIPLA-RODI can kill S. aureus but not E. faecium in polymicrobial biofilms (Gonzalez et al., 2017). Additionally, the cationic peptide Brevinin2 HYba5 was found to inhibit the biofilm formation of S. aureus and E. faecalis individually as well as in polymicrobial biofilm; however, its concentration is up to 75 μM (Radhakrishnan et al., 2023), which may be too high to reduce its therapeutic value. In the present study, we found that Ply113 at a concentration of 64 μg/mL (1.9 μM) was able to remove the majority of the mono-species biofilms of E. faecium, E. faecalis, and S. aureus. When Ply113 was applied to dual-species biofilms at the same concentration, it eliminated the biofilms with equivalent efficiency. Our findings imply that Ply113 has the potential to be a valuable antimicrobial agent against polymicrobial infections (Figure 8).
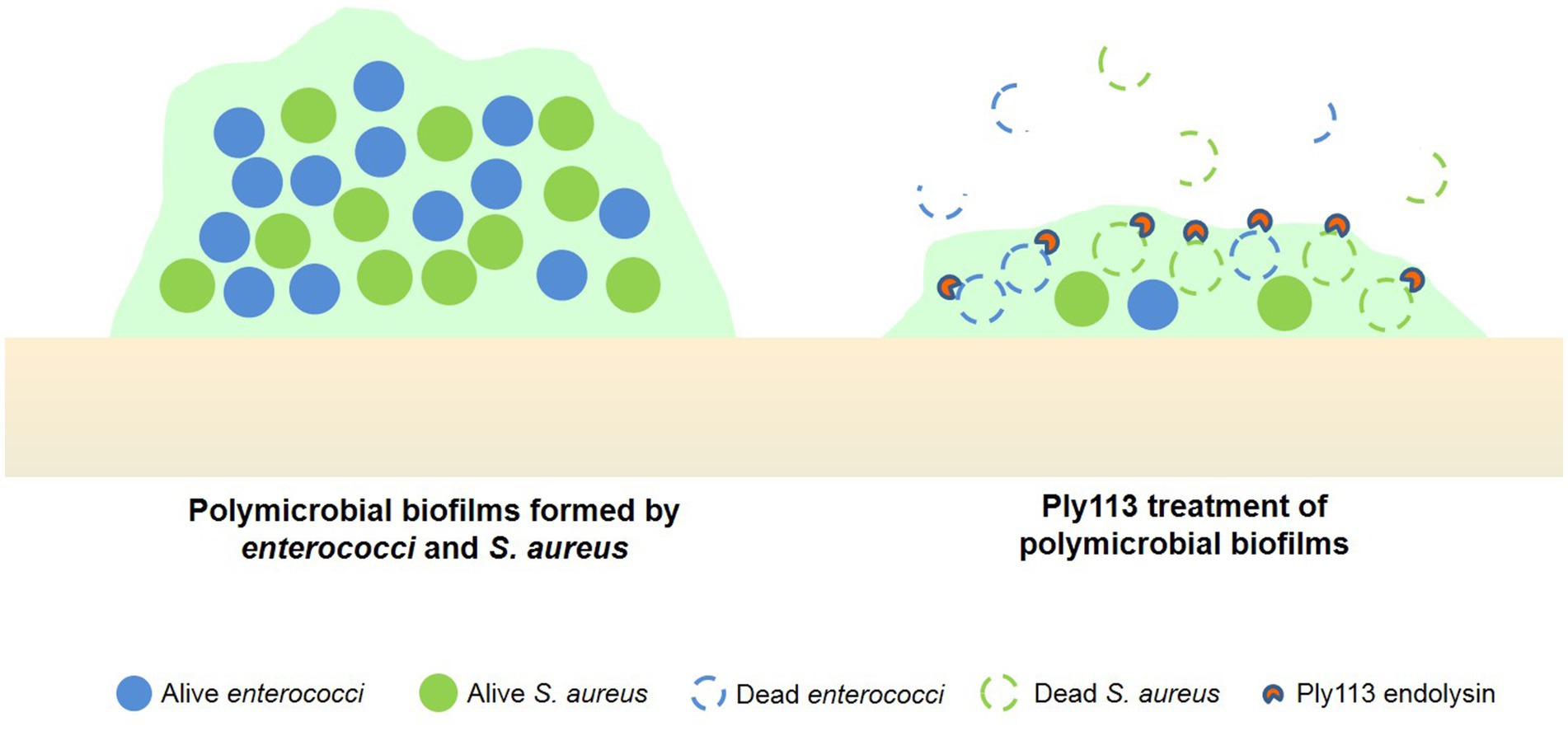
Figure 8. Ply113 treatment of dual-species biofilms formed by enterococci and S. aureus. Ply113 exerts rapid and high lytic activity against strains of E. faecium, E. faecalis, and S. aureus, and eliminates the dual-species biofilms of E. faecium–S. aureus and E. faecalis–S. aureus.
In summary, our findings show that bacteriophage endolysin Ply113 can be applied to effectively decrease the bacterial burden of E. faecium, E. faecalis, and S. aureus in vitro and in vivo. Furthermore, Ply113 exerts the potent bactericidal activity against polymicrobial biofilms formed by enterococci and S. aureus. This makes Ply113 is a promising therapeutic candidate for the treatment of polymicrobial infections and biofilm-associated infections, such as chronic and recurrent infections. We are currently exploring the combination of Ply113 and other drugs, including antibiotics and antimicrobial peptides, to improve their therapeutic efficacy. Although not statistically significant, there was a more than 10% decrease in the cell viability of mouse BMDMs, and the in vivo safety of Ply113 still needs to be further validated. Whether Ply113 therapy can be used in the future also still needs to be further validated using in vivo models, such as skin wound infections and catheter-related biofilm infections.
Data availability statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/Supplementary material.
Ethics statement
The animal study was approved by the Animal Ethics Committee of the Harbin Veterinary Research Institute of the Chinese Academy of Agricultural Sciences. The study was conducted in accordance with the local legislation and institutional requirements.
Author contributions
JW: Conceptualization, Methodology, Writing – original draft. SLia: Methodology, Validation, Writing – original draft. XL: Data curation, Writing – original draft. QX: Investigation, Writing – original draft. YZ: Resources, Writing – original draft. SY: Software, Writing – original draft. WZ: Supervision, Writing – review & editing. SLiu: Project administration, Writing – review & editing. FX: Conceptualization, Funding acquisition, Writing – review & editing.
Funding
The author(s) declare financial support was received for the research, authorship, and/or publication of this article. This research was supported by grants from the National Natural Science Foundation of China (31873016).
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2023.1304932/full#supplementary-material
References
Abat, C., Huart, M., Garcia, V., Dubourg, G., and Raoult, D. (2016). Enterococcus faecalis urinary-tract infections: Do they have a zoonotic origin? J. Infect. 73, 305–313. doi: 10.1016/j.jinf.2016.07.012
Abdurahman, M. A., Durukan, I., Dincer, T., Pektas, S., Karatas, E., and Kilic, A. O. (2023). Staphylococcus aureus Bacteriophage 52 endolysin exhibits anti-biofilm and broad antibacterial activity against gram-positive bacteria. Protein J. 42, 596–606. doi: 10.1007/s10930-023-10145-1
Argudin, M. A., Deplano, A., Meghraoui, A., Dodemont, M., Heinrichs, A., Denis, O., et al. (2017). Bacteria from animals as a pool of antimicrobial resistance genes. Antibiotics (Basel) 6:12. doi: 10.3390/antibiotics6020012
Azevedo, A. S., Almeida, C., Melo, L. F., and Azevedo, N. F. (2017). Impact of polymicrobial biofilms in catheter-associated urinary tract infections. Crit. Rev. Microbiol. 43, 423–439. doi: 10.1080/1040841X.2016.1240656
Bienert, S., Waterhouse, A., De Beer, T. A., Tauriello, G., Studer, G., Bordoli, L., et al. (2017). The SWISS-MODEL repository-new features and functionality. Nucleic Acids Res. 45, D313–D319. doi: 10.1093/nar/gkw1132
Brown Gandt, A., Griffith, E. C., Lister, I. M., Billings, L. L., Han, A., Tangallapally, R., et al. (2018). In vivo and in vitro effects of a ClpP-activating antibiotic against vancomycin-resistant enterococci. Antimicrob. Agents Chemother. 62:e00424-18. doi: 10.1128/AAC.00424-18
Cheung, G. Y. C., Bae, J. S., and Otto, M. (2021). Pathogenicity and virulence of Staphylococcus aureus. Virulence 12, 547–569. doi: 10.1080/21505594.2021.1878688
Ch'ng, J. H., Chong, K. K. L., Lam, L. N., Wong, J. J., and Kline, K. A. (2019). Biofilm-associated infection by enterococci. Nat. Rev. Microbiol. 17, 82–94. doi: 10.1038/s41579-018-0107-z
Ch'ng, J. H., Muthu, M., Chong, K. K. L., Wong, J. J., Tan, C. A. Z., Koh, Z. J. S., et al. (2022). Heme cross-feeding can augment Staphylococcus aureus and Enterococcus faecalis dual species biofilms. ISME J. 16, 2015–2026. doi: 10.1038/s41396-022-01248-1
Clinical and Laboratory Standards Institute [CLSI] (2012). Methods for Dilution Antimicrobial Susceptibility Tests for Bacteria that Grow Aerobically; Approved Standard. 9, CLSI Document M07-A9. Wayne, PA: Clinical and Laboratory Standards Institute [CLSI].
Cong, Y., Yang, S., and Rao, X. (2020). Vancomycin resistant Staphylococcus aureus infections: A review of case updating and clinical features. J. Adv. Res. 21, 169–176. doi: 10.1016/j.jare.2019.10.005
Duarte, A. C., Fernandez, L., De Maesschalck, V., Gutierrez, D., Campelo, A. B., Briers, Y., et al. (2021). Synergistic action of phage phiIPLA-RODI and lytic protein CHAPSH3b: a combination strategy to target Staphylococcus aureus biofilms. NPJ Biofilms Microbiom. 7:39. doi: 10.1038/s41522-021-00208-5
Erez, Z., Steinberger-Levy, I., Shamir, M., Doron, S., Stokar-Avihail, A., Peleg, Y., et al. (2017). Communication between viruses guides lysis-lysogeny decisions. Nature 541, 488–493. doi: 10.1038/nature21049
Fitzgerald, J. R. (2012). Livestock-associated Staphylococcus aureus: origin, evolution and public health threat. Trends Microbiol. 20, 192–198. doi: 10.1016/j.tim.2012.01.006
Furuno, J. P., Perencevich, E. N., Johnson, J. A., Wright, M. O., Mcgregor, J. C., Morris, J. G. Jr., et al. (2005). Methicillin-resistant Staphylococcus aureus and vancomycin-resistant Enterococci co-colonization. Emerg. Infect. Dis. 11, 1539–1544. doi: 10.3201/eid1110.050508
García-Solache, M., and Rice, L. B. (2019). The enterococcus: a model of adaptability to its environment. Clin. Microbiol. Rev. :e00058-18:32. doi: 10.1128/CMR.00058-18
Gilmer, D. B., Schmitz, J. E., Euler, C. W., and Fischetti, V. A. (2013). Novel bacteriophage lysin with broad lytic activity protects against mixed infection by Streptococcus pyogenes and methicillin-resistant Staphylococcus aureus. Antimicrob. Agents Chemother. 57, 2743–2750. doi: 10.1128/AAC.02526-12
Gonzalez, S., Fernandez, L., Campelo, A. B., Gutierrez, D., Martinez, B., Rodriguez, A., et al. (2017). The behavior of Staphylococcus aureus dual-species biofilms treated with bacteriophage phiIPLA-RODI depends on the accompanying microorganism. Appl. Environ. Microbiol. 83, e02821–e02816. doi: 10.1128/AEM.02821-16
Heinze, K., Kabeto, M., Martin, E. T., Cassone, M., Hicks, L., and Mody, L. (2019). Predictors of methicillin-resistant Staphylococcus aureus and vancomycin-resistant enterococci co-colonization among nursing facility patients. Am. J. Infect. Control 47, 415–420. doi: 10.1016/j.ajic.2018.09.026
Huang, L., Zhang, R., Hu, Y., Zhou, H., Cao, J., Lv, H., et al. (2019). Epidemiology and risk factors of methicillin-resistant Staphylococcus aureus and vancomycin-resistant enterococci infections in Zhejiang China from 2015 to 2017. Antimicrob. Resist. Infect. Control 8:90. doi: 10.1186/s13756-019-0539-x
Jaradat, Z. W., Ababneh, Q. O., Sha'aban, S. T., Alkofahi, A. A., Assaleh, D., and Al Shara, A. (2020). Methicillin resistant Staphylococcus aureus and public fomites: a review. Pathog. Glob. Health 114, 426–450. doi: 10.1080/20477724.2020.1824112
Kadariya, J., Smith, T. C., and Thapaliya, D. (2014). Staphylococcus aureus and staphylococcal food-borne disease: an ongoing challenge in public health. Biomed. Res. Int. 2014:827965. doi: 10.1155/2014/827965
Kim, S., Lee, D. W., Jin, J. S., and Kim, J. (2020). Antimicrobial activity of LysSS, a novel phage endolysin, against Acinetobacter baumannii and Pseudomonas aeruginosa. J. Glob. Antimicrob. Resist. 22, 32–39. doi: 10.1016/j.jgar.2020.01.005
Letunic, I., Khedkar, S., and Bork, P. (2021). SMART: recent updates, new developments and status in 2020. Nucleic Acids Res. 49, D458–D460. doi: 10.1093/nar/gkaa937
Liu, B., Guo, Q., Li, Z., Guo, X., and Liu, X. (2023). Bacteriophage endolysin: a powerful weapon to control bacterial biofilms. Protein J. 42, 463–476. doi: 10.1007/s10930-023-10139-z
Long, S. W. (2020). The ongoing threat of methicillin-resistant Staphylococcus aureus. J. Infect. Dis. 222, 1943–1945. doi: 10.1093/infdis/jiaa260
Lusiak-Szelachowska, M., Weber-Dabrowska, B., and Gorski, A. (2020). Bacteriophages and lysins in biofilm control. Virol. Sin. 35, 125–133. doi: 10.1007/s12250-019-00192-3
Ma, X. X., Ito, T., Tiensasitorn, C., Jamklang, M., Chongtrakool, P., Boyle-Vavra, S., et al. (2002). Novel type of staphylococcal cassette chromosome mec identified in community-acquired methicillin-resistant Staphylococcus aureus strains. Antimicrob. Agents Chemother. 46, 1147–1152. doi: 10.1128/AAC.46.4.1147-1152.2002
Moormeier, D. E., and Bayles, K. W. (2017). Staphylococcus aureus biofilm: a complex developmental organism. Mol. Microbiol. 104, 365–376. doi: 10.1111/mmi.13634
Munoz-Price, L. S., Lolans, K., and Quinn, J. P. (2005). Emergence of resistance to daptomycin during treatment of vancomycin-resistant Enterococcus faecalis infection. Clin. Infect. Dis. 41, 565–566. doi: 10.1086/432121
Ning, H., Lin, H., Wang, J., He, X., Lv, X., and Ju, L. (2021). Characterizations of the endolysin Lys84 and its domains from phage qdsa002 with high activities against Staphylococcus aureus and its biofilms. Enzym. Microb. Technol. 148:109809. doi: 10.1016/j.enzmictec.2021.109809
Oliveira, H., Melo, L. D., Santos, S. B., Nobrega, F. L., Ferreira, E. C., Cerca, N., et al. (2013). Molecular aspects and comparative genomics of bacteriophage endolysins. J. Virol. 87, 4558–4570. doi: 10.1128/JVI.03277-12
Pammi, M., Zhong, D., Johnson, Y., Revell, P., and Versalovic, J. (2014). Polymicrobial bloodstream infections in the neonatal intensive care unit are associated with increased mortality: a case-control study. BMC Infect. Dis. 14:390. doi: 10.1186/1471-2334-14-390
Papanicolaou, G. A., Ustun, C., Young, J. H., Chen, M., Kim, S., Woo Ahn, K., et al. (2019). Bloodstream infection due to vancomycin-resistant enterococcus is associated with increased mortality after hematopoietic cell transplantation for acute leukemia and myelodysplastic syndrome: a multicenter, retrospective cohort study. Clin. Infect. Dis. 69, 1771–1779. doi: 10.1093/cid/ciz031
Peters, B. M., Jabra-Rizk, M. A., O'may, G. A., Costerton, J. W., and Shirtliff, M. E. (2012). Polymicrobial interactions: impact on pathogenesis and human disease. Clin. Microbiol. Rev. 25, 193–213. doi: 10.1128/CMR.00013-11
Poonacha, N., Nair, S., Desai, S., Tuppad, D., Hiremath, D., Mohan, T., et al. (2017). Efficient killing of planktonic and biofilm-embedded coagulase-negative staphylococci by bactericidal protein P128. Antimicrob. Agents Chemother. :e00457-17:61. doi: 10.1128/AAC.00457-17
Radhakrishnan, M. P., Suryaletha, K., Joseph, I., George, S., and Thomas, S. (2023). A novel Brevinin2 HYba5 peptide against polymicrobial biofilm of Staphylococcus aureus and Enterococcus faecalis. Protein Pept. Lett. 30, 795–805. doi: 10.2174/0109298665266332231001115508
Roach, D. R., and Donovan, D. M. (2015). Antimicrobial bacteriophage-derived proteins and therapeutic applications. Bacteriophage 5:e1062590. doi: 10.1080/21597081.2015.1062590
Rodriguez-Rubio, L., Gutierrez, D., Donovan, D. M., Martinez, B., Rodriguez, A., and Garcia, P. (2016). Phage lytic proteins: biotechnological applications beyond clinical antimicrobials. Crit. Rev. Biotechnol. 36, 542–552. doi: 10.3109/07388551.2014.993587
Romling, U., and Balsalobre, C. (2012). Biofilm infections, their resilience to therapy and innovative treatment strategies. J. Intern. Med. 272, 541–561. doi: 10.1111/joim.12004
Sadiq, F. A., Hansen, M. F., Burmolle, M., Heyndrickx, M., Flint, S., Lu, W., et al. (2022). Trans-kingdom interactions in mixed biofilm communities. FEMS Microbiol. Rev. :fuac024:46. doi: 10.1093/femsre/fuac024
Sass, P., and Bierbaum, G. (2007). Lytic activity of recombinant bacteriophage phi11 and phi12 endolysins on whole cells and biofilms of Staphylococcus aureus. Appl. Environ. Microbiol. 73, 347–352. doi: 10.1128/AEM.01616-06
Schuch, R., Lee, H. M., Schneider, B. C., Sauve, K. L., Law, C., Khan, B. K., et al. (2014). Combination therapy with lysin CF-301 and antibiotic is superior to antibiotic alone for treating methicillin-resistant Staphylococcus aureus-induced murine bacteremia. J. Infect. Dis. 209, 1469–1478. doi: 10.1093/infdis/jit637
Tacconelli, E., Carrara, E., Savoldi, A., Harbarth, S., Mendelson, M., Monnet, D. L., et al. (2018). Discovery, research, and development of new antibiotics: the WHO priority list of antibiotic-resistant bacteria and tuberculosis. Lancet Infect. Dis. 18, 318–327. doi: 10.1016/S1473-3099(17)30753-3
Tkhilaishvili, T., Wang, L., Perka, C., Trampuz, A., and Gonzalez Moreno, M. (2020). Using bacteriophages as a trojan horse to the killing of dual-species biofilm formed by Pseudomonas aeruginosa and methicillin resistant Staphylococcus aureus. Front. Microbiol. 11:10.3389/fmicb.2020.00695:695.
Tucker, A. T., Leonard, S. P., DuBois, C. D., Knauf, G. A., Cunningham, A. L., Wilke, C. O., et al. (2018). Discovery of next-generation antimicrobials through bacterial self-screening of surface-displayed peptide libraries. Cell 172, 618–628. doi: 10.1016/j.cell.2017.12.009
Van Hal, S. J., Willems, R. J. L., Gouliouris, T., Ballard, S. A., Coque, T. M., Hammerum, A. M., et al. (2022). The interplay between community and hospital Enterococcus faecium clones within health-care settings: a genomic analysis. Lancet Microbe 3, e133–e141. doi: 10.1016/S2666-5247(21)00236-6
Weigel, L. M., Donlan, R. M., Shin, D. H., Jensen, B., Clark, N. C., Mcdougal, L. K., et al. (2007). High-level vancomycin-resistant Staphylococcus aureus isolates associated with a polymicrobial biofilm. Antimicrob. Agents Chemother. 51, 231–238. doi: 10.1128/AAC.00576-06
Weiner-Lastinger, L. M., Abner, S., Edwards, J. R., Kallen, A. J., Karlsson, M., Magill, S. S., et al. (2020). Antimicrobial-resistant pathogens associated with adult healthcare-associated infections: Summary of data reported to the National Healthcare Safety Network, 2015-2017. Infect. Control Hosp. Epidemiol. 41, 1–18. doi: 10.1017/ice.2019.296
Yoon, Y. K., Lee, M. J., Ju, Y., Lee, S. E., Yang, K. S., Sohn, J. W., et al. (2019). Determining the clinical significance of co-colonization of vancomycin-resistant enterococci and methicillin-resistant Staphylococcus aureus in the intestinal tracts of patients in intensive care units: a case-control study. Ann. Clin. Microbiol. Antimicrob. 18:28. doi: 10.1186/s12941-019-0327-8
Keywords: bacteriophage endolysin, Enterococcus faecium , Enterococcus faecalis , Staphylococcus aureus , antibacterial activity, antibiofilm activity, polymicrobial biofilms
Citation: Wang J, Liang S, Lu X, Xu Q, Zhu Y, Yu S, Zhang W, Liu S and Xie F (2023) Bacteriophage endolysin Ply113 as a potent antibacterial agent against polymicrobial biofilms formed by enterococci and Staphylococcus aureus. Front. Microbiol. 14:1304932. doi: 10.3389/fmicb.2023.1304932
Edited by:
Manuela Oliveira, University of Lisbon, PortugalReviewed by:
Junyan Liu, Zhongkai University of Agriculture and Engineering, ChinaEva Cunha, University of Lisbon, Portugal
Catarina Milho, Centro de Valorização e Transferência de Tecnologia da Água (AQUAVALOR), Portugal
Copyright © 2023 Wang, Liang, Lu, Xu, Zhu, Yu, Zhang, Liu and Xie. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Siguo Liu, bGl1c2lndW9AY2Fhcy5jbg==; Fang Xie, eGllZmFuZ0BjYWFzLmNu
†These authors have contributed equally to this work
 Jing Wang
Jing Wang Siyu Liang†
Siyu Liang† Yao Zhu
Yao Zhu Shenye Yu
Shenye Yu Siguo Liu
Siguo Liu Fang Xie
Fang Xie