- 1Department of Molecular Microbiology and Immunology, USDA ARS Meat Animal Research Center, Clay Center, NE, United States
- 2South Texas Center for Emerging Infectious Diseases, San Antonio, TX, United States
Shiga toxin-producing Escherichia coli (STEC) O157:H7 strains with the T allele in the translocated intimin receptor polymorphism (tir) 255 A > T gene associate with human disease more than strains with an A allele; however, the allele is not thought to be the direct cause of this difference. We sequenced a diverse set of STEC O157:H7 strains (26% A allele, 74% T allele) to identify linked differences that might underlie disease association. The average chromosome and pO157 plasmid size and gene content were significantly greater within the tir 255 A allele strains. Eighteen coding sequences were unique to tir 255 A allele chromosomes, and three were unique to tir 255 T allele chromosomes. There also were non-pO157 plasmids that were unique to each tir 255 allele variant. The overall average number of prophages did not differ between tir 255 allele strains; however, there were different types between the strains. Genomic and mobile element variation linked to the tir 255 polymorphism may account for the increased frequency of the T allele isolates in human disease.
Introduction
Shiga toxin-producing Escherichia coli (STEC) O157:H7 is a foodborne pathogen that causes illnesses ranging from mild self-limiting cases to hemolytic uremic syndrome (Slutsker et al., 1997). A 2011 assessment estimated that it causes 63,153 illnesses, 2,138 hospitalizations, and 20 deaths annually in the United States (Scallan et al., 2011). Cattle are recognized as a major reservoir for this pathogen (Smith et al., 2001), although it has been isolated from other animals such as wildlife (deer and birds), other domestic livestock (sheep and goats), and companion animals (dogs), as well as from non-animal sources such as vegetables and environmental samples (Mead and Griffin, 1998).
STEC O157:H7 is a relatively recently emerged pathogen with the first reported case linked to it in the 1980s (Mead and Griffin, 1998; Karch et al., 1999). Evolutionary models have described the emergence of this serotype via a stepwise evolution from O55:H7 chromosomes (Wick et al., 2005). STEC O157:H7 strains harbor a highly conserved non-conjugative F-like plasmid known as pO157, ranging between 92 and 104 kilobases (kb) in length (Lim et al., 2010) that has been linked to chromosome evolution (Nyong et al., 2020). While several proteins encoded on it have been associated with virulence, there is still uncertainty around the role pO157 plays in infection and overall virulence (Lim et al., 2010).
STEC O157:H7 genomes may carry many different mobile genetic elements (MGEs) in addition to pO157. Other plasmids include pEC4115, which is a 37 kb conjugal transfer plasmid (Eppinger et al., 2011), pColD157, which is a 6,675 bp colicin-containing plasmid (Hofinger et al., 1998), and pOSAK1, which is a 3,306 kb plasmid-containing ColE1-like replication system (Makino et al., 1998). Another major type of MGE found in the STEC O157:H7 genome is integrated bacteriophages (prophages). The genomes of STEC O157:H7 contain an average of 11.9% prophages that are classified as lambda-like, P4-like, and unclassified (Bobay et al., 2013). Some prophages contain Shiga toxin genes which encode for a significant virulence factor of STEC O157:H7 and have integrated into several known integration sites in the genome (Recktenwald and Schmidt, 2002; Bielaszewska et al., 2006; Serra-Moreno et al., 2007; González-Escalona et al., 2019). Insertion sequence (IS) element IS629 is the most abundant IS element in STEC O157:H7 and has been shown to be responsible for genome diversification (Ooka et al., 2009a; Rump et al., 2011b). IS629 has also been used to develop typing methods for STEC O157:H7 (Ooka et al., 2009b; Yokoyama et al., 2011; Stanton et al., 2014; Kameyama et al., 2015; Toro et al., 2015). MGEs are also a source of antibiotic resistance genes (ARGs) in STEC O157:H7. The prevalence of ARGs in STEC O157:H7 is generally low and ranges from 10% to 14.7% (Schroeder et al., 2002; Greig et al., 2023). Interestingly, ARGs can be located on both chromosomes and plasmids in STEC O157:H7 (Greig et al., 2023).
Several studies have found that not all STEC O157:H7 subtypes are equally likely to cause disease in humans (Kim et al., 1999; Bono et al., 2007; Manning et al., 2008; Whitworth et al., 2010). Octamer-based genome scanning of STEC O157 strains showed a biased distribution of isolates based on clade distributions with most human isolates being found in clade I compared to clade II that had more non-human isolates (Kim et al., 1999). Genotyping of enterohemorrhagic E. coli strains identified 96 SNPs that cluster isolates into nine clades (Manning et al., 2008). The Manning clade eight strains were responsible for more severe disease in humans and were seven times more likely to cause hemolytic uremic syndrome.
The ability of STEC O157:H7 to cause disease is driven by its attachment to intestinal epithelium via the products of a pathogenicity island in the genome known as the locus of enterocyte effacement (LEE) and the production of Shiga toxins (Kaper et al., 2004). The translocated intimin receptor gene tir, found in the LEE, has a single-nucleotide polymorphism (SNP; tir 255 T > A), with strains having the T allele being 34 times more likely to cause disease in humans (Bono et al., 2007). However, strains isolated from diseased humans with the tir 255 A allele suggest that the non-synonymous SNP, while valuable as a marker for disease association, is not itself responsible for the lower propensity to cause disease. In fact, the SNP does not appear to affect Tir protein activity, and tir 255 A allele strains are able to cause attaching and effacing lesions (Besser et al., 2007). Thus, genetic differences responsible for the association of the tir 255 T allele with human disease have not as yet been identified.
Whole-genome sequencing (WGS) of bacterial genomes has allowed a high level of discernment between strains in research and public health settings (Didelot et al., 2012; Rusconi et al., 2016; Nadon et al., 2017; Tagini and Greub, 2017; Jenkins et al., 2019). Bacterial genomes can be comprehensively analyzed for SNPs within genes of interest and macro-differences with WGS. Short-read sequencing technologies have limited ability to resolve complex genomes with large inversions and repeated phage regions such as STEC O157:H7 (Koren et al., 2013; Koren and Phillippy, 2015; De Maio et al., 2019). In contrast, long-read sequencing technologies can bridge repetitive regions, providing a better and more accurate understanding of genome structure. Integrating long-read sequencing as scaffolding with short reads to polish the assembly and find smaller plasmids can produce a higher quality genome (De Maio et al., 2019). This level of assembly gives the best chance of finding genetic determinants that may be responsible for tir 255 T allele strains associating with human disease more than A allele strains. The objectives of this study were to use the hybrid short- and long-read sequencing strategy on a collection of STEC O157:H7 strains to characterize the genetic diversity and compare the genomes of strains with either the T or A allele at tir 255. These comparisons provide a more complete understanding of this important public health pathogen and new insight into the genomic features that contribute to the enhanced association of tir 255 T allele strains with human disease.
Materials and methods
Bacterial strain selection
A total of 58 E. coli O157:H7 strains [isolated from cattle (n = 28), deer (n = 1)], ground beef (n = 6), and humans (n = 23) were included in this study for phylogenetic and comparative genome analyses (Supplementary Table 1). The strains were mainly isolated in the United States (n = 50), but some were included from Japan (n = 3), Scotland (n = 2), Germany (n = 1), and Denmark (n = 1), with one of unknown origin. Forty-eight strains were sequenced based on their tir 255 genotype and their diverse distribution across a previously described phylogenetic classification method based on polymorphism-derived genotypes as described by Bono et al. (2012). Within the 48 sequenced strains, approximately the same relative proportion of tir 255 A genotype (13/48 = 31%) and tir 255 T genotype (33/48 = 61%) strains were used as described from the polymorphism-derived genotyping system [tir 255 A strains (56/175 = 32%) and genotype tir 255 T strains (119/175 = 68%); Bono et al., 2012]. Ten complete closed genomes with the tir 255 T genotypes were downloaded from the National Center for Biotechnology Information (NCBI) database and were used for all analyses except those on the distribution of transposable insertion sequence.
Genome sequencing, genotyping, and annotation
Strains frozen in glycerol were struck onto CHROMagar O157 plates (DRG International, Mountainside, NJ) and incubated overnight at 37°C. The following day strains were inoculated into 1 mL of LB and grown overnight at 37°C with shaking. The next morning, 250 ul of the overnight culture was added to 10 mL of LB and incubated at 37°C with shaking for 3.5 h before undergoing DNA extraction. DNA was extracted as described (Clawson et al., 2009) using Qiagen Genomic-tip 100/G columns (Valencia, CA, United States) and quantified using a nanodrop and a Quantus Fluorometer with QuantiFluor dye (Promega, Madison, WI, United States). An aliquot of DNA (10 μg) was sheared to a 30 kb target fragment length using g-TUBEs (Covaris, Woburn, MA, United States) and concentrated with 0.45x volume AMPure PB beads (Pacific Biosciences, Menlo Park, CA, United States), as per manufacturer’s instruction. Sheared and concentrated DNA (5 μg) was used to make sequencing libraries using the SMRTbell Template Prep Kit 1.0 according to the manufacturer’s protocol. Fragments 15 kb were selected using the BluePippin (Sage Science, Beverly, MA, United States). The library was bound with polymerase P6 for sequencing on a Pacific BioSciences (Pacific Biosciences) RS II sequencing platform with chemistry C4 and the 360-min data collection protocol.
Another aliquot of the same DNA preparation used for long-read sequencing was used for short-read library construction. One microgram of DNA was sheared to 350 bp fragment size using a microTUBE AFA Fiber Pre-Slit Snap-Cap 6x16mm (Corvaris, Woburn, MA). Libraries were constructed using the TruSeq DNA PCR-Free HT Library Preparation kit (Illumina, Inc., San Diego, CA) and quantitated using the KAPA Library Quantification Kit (F. Hoffmann-La Roche Ltd., Basel, Switzerland). Libraries were pooled into groups of 24 and run on an Illumina MiSeq with the MiSeq Reagent Kit v3 (600 cycles).
Raw PacBio reads were assembled using HGAP (Chin et al., 2013) in SMRT analysis v8.0 and contigs imported into Geneious v2019.1(Biomatters Ltd., Auckland, New Zealand). If present, overlapping sequence on the ends of the contigs was removed from the 5′ and 3′ ends to generate circularized chromosomes and plasmids. Closed chromosomes were reoriented after finding a putative origin of replication with Ori-Finder 2 (Luo et al., 2014) for the chromosome of one strain and then orienting all subsequent chromosomes to that same starting position. The closed chromosomes and plasmids were polished twice for accuracy using the RS_Resequencing v1.0 protocol in SMRT analysis by mapping PacBio reads to the chromosomes and plasmids. Further error correction was performed using the short-read data via Pilon v1.23 (Walker et al., 2014). Finally, short reads were mapped to the chromosome and assembled plasmids using Geneious Mapper (Biomatters, Ltd.), and unused reads were de novo assembled using the Geneious assembler (Biomatters, Ltd.) for the identification of additional plasmids. All genomes and plasmids were annotated with the NCBI Prokaryotic Genome Annotation Pipeline v4.8 (Tatusova et al., 2016). The genomes were previously genotyped for the tir 255 allele (Bono et al., 2007), a 175 polymorphism-derived genotype (Bono et al., 2012), and the Shiga toxin-encoding bacteriophage insertion site (Besser et al., 2007; Whitworth et al., 2008). Clade typing was performed according to Manning et al. (2008). Clades and subgroups were assigned by in silico interrogation of the allelic status of 89 core genome SNPs in the assembled genomes using a custom workflow on Galaxy (Giardine et al., 2005; Goecks et al., 2010), which was informed by eight definitive polymorphic positions (Riordan et al., 2008; Yokoyama et al., 2012). VirulenceFinder v2.0.3 (Camacho et al., 2009; Joensen et al., 2014; Malberg Tetzschner et al., 2020) was used to type the Shiga toxin variants.
Phylogenetic tree construction and linear fragment visualization.
A phylogenetic tree was constructed from all O157 chromosomes with Parsnp v1.6.2 (Treangen et al., 2014) with the “-c” flag to include all files with strain 493/89 as the root because it is proposed to be the oldest E. coli O157:H7 ancestor. FigTree v1.4.4 (Rambaut, 2009) was used for tree visualization, and Evolview v3 (Subramanian et al., 2019) was used to annotate tree branches and leaves, as well as the origin of the isolates. EasyFig v2.2.2 (Sullivan et al., 2011) was used to visualize linear comparisons of multiple genomic loci insertions, while individual gene annotations were visualized in SnapGene Viewer v5.2 (GSL Biotech, LLC.).
Core and pangenome
The fasta files from all isolates were exported from Geneious (Biomatters, Ltd.) and annotated with Prokka v1.14.5 (Seemann, 2014) using the—proteins option with the annotated GenBank file from STEC O157:H7 isolate TX 376-2 (GenBank CP038287–CP038289) for annotation. The.gff files from the Prokka annotation were used with Roary v3.13.0 (Page et al., 2015) to define the core and pangenome of the 58 isolates. The gene_presence_absence.csv from the Roary analysis and the tir 255 A vs. T trait.csv file were used as input into Scoary v1.6.16 to look for associations of components of the pangenome with the tir 255 alleles. The.grr file from the Parsnp analysis of the STEC O157 genomes containing all variant sites was converted to a.vcf file using Harvest tools (Treangen et al., 2014). The.vcf file was imported into PLINK v1.90 (Chang et al., 2015) and the linkage disequilibrium based variant pruner option “--indep-pairwise” with a window size the entire length of all SNPs identified with an r2 = 0.99 (and visually inspected to assure r2 = 1) was used to reduce all SNPs to just those that were informative. Linkage disequilibrium SNP associations were visualized with the LinkageMapView v2.1.2 (Ouellette et al., 2018) in R v3.6.
Identification of prophage insertion sites
Prophages were identified using the PHASTER API (Arndt et al., 2016). The number of phages present (including complete, intact, and questionable) and percentage of the chromosome associated with prophages (computed by dividing the total region length of all prophages associated with a chromosome by the total chromosome size) were compared between strains, source of isolate, and tir 255 allele using the SAS Studio v3.8 PROC MIXED procedure with the LSMeans statement with pdiff and cl options at an alpha = 0.05.
The stx-containing prophages were extracted with the flanking core chromosomal genes using Geneious (Biomatters, Ltd.). The extracted prophages were grouped according to their insertion sites and aligned using the Mauve (Darling et al., 2004) plugin in Geneious (Biomatters, Ltd.). Shiga toxin genes were extracted from the prophage sequences, grouped according to subtype, and aligned using the Geneious alignment tool in Geneious (Biomatters, Ltd.).
Evaluation of insertion sequence elements
The insertion sequence (IS) elements from the chromosome and their corresponding pO157 plasmids were extracted from their GenBank annotations using Geneious. The genomes from the 10 closed genomes downloaded from GenBank were not used in this study because they were annotated with a different version of the NCBI prokaryotic genome annotation pipeline. The IS elements were grouped into families, and the number of transposases per family was used in the analysis.
Antibiotic resistance gene identification
Antibiotic resistance genes were identified using the Comprehensive Antibiotic Resistance Database (CARD; Alcock et al., 2020) resistance gene identifier using perfect (100% sequence similarity) and strict (≥80% sequence similarity) hits only and excluding nudges.
pO157 plasmid analysis
The relatedness of 56 pO157 plasmids was determined using Parsnp v1.2 (Treangen et al., 2014) with the Sakai pO157 plasmid as the reference and visualized with Gingr v1.3 (Treangen et al., 2014). pO157 plasmids from strains LSU61 and 493/89 were excluded from this analysis because their pO157 plasmids are approximately 30 kb larger and are missing the known virulence gene toxB (Nyong et al., 2020). Comparison of the phylogenetic trees of the pO157 plasmids to their respective chromosomes was done with the.tree files from Parsnp using the tanglegram option from Dendroscope v3.7.1 (Scornavacca et al., 2011; Huson and Scornavacca, 2012). Plasmids were grouped based on similarities using the Mauve (Darling et al., 2004) plugin in Geneious (Biomatters, Ltd.).
Description of other plasmids identified in the genome
Non-pO157 plasmids were grouped according to size in Geneious (Biomatters, Ltd.). Visual inspection of the gene annotations from each size group was used to further refine the grouping into those with similar gene annotations. The plasmids grouped by size and annotation were compared using the Mauve (Darling et al., 2004) plugin in Geneious (Biomatters, Ltd.).
Results and discussion
Sequencing and assembly
The chromosome from MB9-1 was the only example of the 48 strains sequenced that could not be closed due to the presence of two ~60 kb duplicated regions that could not be resolved with the long reads produced by the RSII. Thirty-one plasmids smaller than 30 kb were identified by assembling Illumina reads that failed to map to the long-read-derived genomes. Information about each genome is available in Supplementary Table 1. The isolates had an average chromosome length of 5,507,515 bp with a range from 5,362,528 bp to 5,643,409 bp and an average of 1.7 plasmids (ranging from 1 to 4, with size ranging from 1.5 to 145.3 kb). The average number of protein-coding sequences (CDS) within strains was 5,633 and ranged from 5,393 to 5,868.
Genome comparisons of tir 255 allele strains
Genomes were compared between strains carrying either the tir 255 T or tir 255 A allele. Strains with the tir A allele chromosomes were found to have larger chromosomes (p = 0.015) than tir T chromosomes (means 5,539,556 vs. 5,496,221 bp, respectively [95% C.I. 5,509,981 bp to 5,569,131 bp and 5,478,332 bp to 5,514,109 bp]); the pO157 plasmid size was also larger on average (p < 0.001, tir A mean = 95,890 bp, [95% C.I. 95,019 bp to 96,761 bp] tir T mean = 93,013 bp [95% C.I. 92,486 bp to 93,539 bp]). Similarly, strains with the tir A allele genomes had a greater number of CDS when compared to strains with the tir T genomes (means 5,701 vs. 5,601, respectively).
These results are consistent with previous studies that show an association of a smaller genome size in pathogenic bacteria than their nearest neighbors that are non- or less-pathogenic (Ochman and Moran, 2001; Moran, 2002; Toft and Andersson, 2010; Weinert and Welch, 2017). This reduction in genome size of pathogenic strains of bacteria is mainly seen between bacteria species and is rarely seen within a single bacterial species (Weinert and Welch, 2017). However, Streptococcus suis, an emerging pathogen in swine, shows a reduction in genome size that is associated with its ability to cause disease more often than expected by chance (Murray et al., 2021). The loss of mobile genetic elements was responsible for a large portion of the reduced genomes with the remaining reductions located in the other genomic elements (Murray et al., 2021). Here, the increased size of the tir A allele genomes and increased number of genes could be from the gain of mobile genetic elements, but prophages do not appear to be responsible for this as there is not a significant difference in the number of integrated prophages between tir A and tir T allele genomes. The most parsimonious explanation for the reduced association of tir A allele strains with human disease is that their predecessor acquired genetic material that reduced fitness or propensity to cause disease in humans. Alternatively, a less parsimonious explanation is that the tir T allele strains may have reduced genome size in multiple events, and not as a part of continuous evolution because the tir A allele strains are flanked by tir T allele genomes on both the ancestral and progeny branches of the phylogenetic tree (Figure 1).
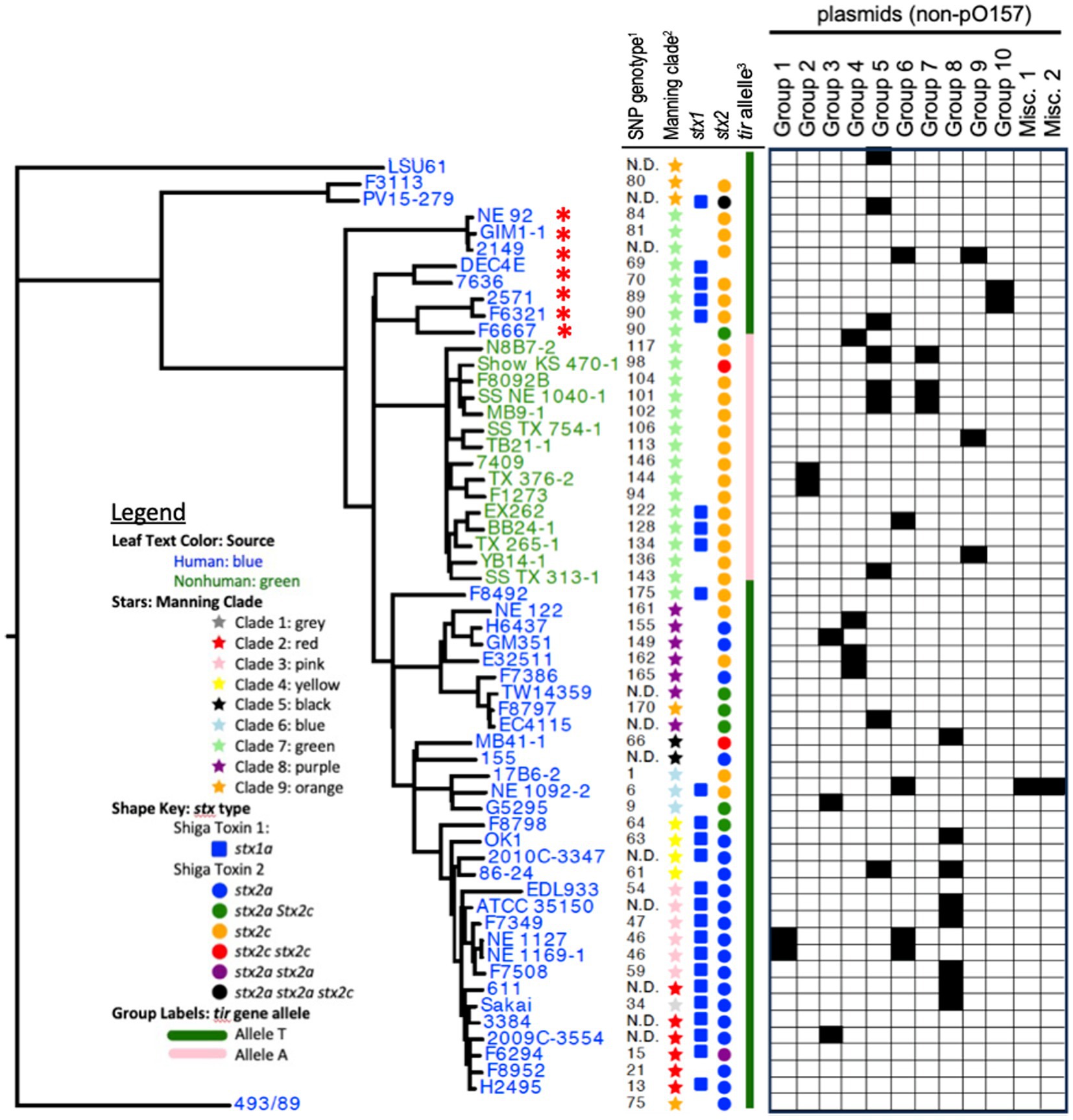
Figure 1. Phylogenetic tree of all STEC O157:H7 chromosomes included in the study. Leaf text colors indicate the source of the isolate as human (blue) or non-human (green). Leaf decorations and labels are based on previously published SNP-based phylogeny (1Manning et al., 2008, 2Bono et al., 2012), tir 255 allele genotype (3Bono et al., 2007), and Shiga toxin subtype and variant as determined by VirulenceFinder (Camacho et al., 2009; Joensen et al., 2014; Malberg Tetzschner et al., 2020). N.D. abbreviation is for not determined. The non-pO157 plasmids are separated into plasmid groups across the top of the columns. The black boxes beneath the group name indicate that the plasmid was found in the corresponding strain. The red asterisk indicates strains that were not found to contain the mdfA gene by CARD because of a missense mutation resulting in a truncated and presumably inactive protein.
Phylogenetic tree clusters with Shiga toxin content from tir 255 T allele and tir 255 A allele strains
The phylogenetic tree of core genomes from the 58 strains resembled the trees from which the strains were chosen according to their polymorphism-derived genotypes (Bono et al., 2012) and the Manning clades (Manning et al., 2008; Figure 1). As seen previously, the tir 255 A and tir 255 T allele strains group separately on the tree as previously described, with only one of the tir 255 A allele strains isolated from a human source while the rest were from a non-human source. The human and non-human sources of tir 255 T allele strains were disbursed throughout the tree with no association with clusters.
Shiga toxin genes were also found both on their own and in combination. However, LSU 61, a proposed evolutionary intermediate strain, was the only strain to not contain a Shiga toxin gene in its genome (Nyong et al., 2020). In addition, DEC 4E was the only strain with no stx2 gene in its genome. The stx1a gene was found in 25 of the strains, while 28 had the stx2a gene and 35 had the stx2c gene. Thirty-five strains contained multiple stx genes with the most common combination being stx1a and stx2a (n = 13), followed by stx1a and stx2c (n = 8), stx2a and stx2c (n = 5), stx2c and stx2c (n = 2), stx1a, stx2a, and stx2a (n = 1), stx1a, stx2a, and stx2c (n = 1), and lastly stx1a, stx2a, stx2a, and stx2c (n = 1; Figure 1). The stx2c gene in STEC O157:H7 strain MB41-1 had an IS629 element inserted into the Shiga toxin subunit b gene presumably causing the Shiga toxin protein to be inactivated. Disruption of stx2 genes by IS element has been previously reported. The stx2c subunit A (Jinneman et al., 2000; Loftsdóttir et al., 2017; Nyong et al., 2020) and subunit B (Kusumoto et al., 1999) genes and the stx2a subunit A gene (Fitzgerald et al., 2019) were disrupted by different IS elements including IS1203, IS629, and IS66. Disruption of the stx2 gene has been identified in different countries and different regions of the genes indicating the potential for IS elements to integrate into any strain, and there does not appear to be a targeted sequence for IS element integration into the stx genes (Kusumoto et al., 1999; Loftsdóttir et al., 2017; Fitzgerald et al., 2019). Strains with a stx gene disrupted by an IS element are a potential risk to human health as IS elements can excise from the gene leaving behind an intact stx gene capable of making toxins (Kusumoto et al., 2001; Loftsdóttir et al., 2017). The tir 255 A allele strains only contained the stx2c gene variant, while the tir 255 T allele contained both the stx2a and stx2c variants. Both tir 255 A and tir 255 T allele strains contained stx1a although tir 255 T allele strains had a higher proportion (22/43 = 51%) than the tir 255 A allele strains (3/15 = 20%; Figure 1; Supplementary Table 1).
Some CDS and SNP-linked alleles associated with only one tir 255 SNP allele
The core STEC O157:H7 chromosome was composed of 4,091 coding sequences (CDS) while the pangenome encompassed 10,079 CDS, with 1,789 CDS being singletons. Chromosomes had on average 5,392 identifiable CDS (range 5,153 to 5,612), or 76% (4,091/5,392) of the chromosome being conserved among the STEC O157:H7 analyzed in this study. Of the singletons identified, the chromosomes contained an average of 30 (range 2 to 232) unique genes. Three strains LSU61, 493/89, and PV15-279 accounted for 25.6% (n = 458/1,789) of the singletons (Supplementary Table 2).
There were 18 CDS unique to the tir 255 A allele chromosomes and three CDS unique to the tir 255 T allele chromosomes (Supplementary Table 3). Specific genes in tir 255 T allele chromosomes encode sel1 repeat family protein (n = 1), RHS repeat protein (n = 1), and phage tail protein (n = 1). Sel1 repeat proteins have been associated with virulence in Legionella pneumophila though when investigated in the more closely related Klebsiella pneumoniae, their function was not characterized (Lery et al., 2014). The tir 255 A allele-specific genes encode AlpA family transcriptional regulator (n = 1), Clp protease ClpB (n = 1), DNA cytosine methyltransferase (n = 1), glycerophosphodiester phosphodiesterase (n = 1), host cell division inhibitor Icd-like protein (n = 1), hypothetical proteins (n = 6), IS3 family transposase (n = 2), IS66 family insertion sequence hypothetical protein (n = 1), phage polarity suppression protein (n = 1), phage receptor (n = 1), phage tail protein (n = 1), and a DNA primase (n = 1).
Analysis for SNPs in linkage disequilibrium with the tir 255 alleles that could contribute to the ability of a strain to cause human disease revealed 204 SNPs on the chromosome and two on the pO157 plasmid. In total, 101 were non-synonymous and 62 were synonymous, with seven resulting in amber mutations and 32 residing in intergenic regions (Supplementary Table 4). The amber mutations were in the glycerophosphodiester phosphodiesterase gene (glpQ), ethanolamine ammonia-lyase reactivase gene (eutA), aquaporin water channel gene (apqZ), a fimbrial protein precursor CDS (lpfE), two hypothetical genes and a bacteriophage N4 receptor, and outer membrane subunit gene. While it is difficult to speculate on the function of the truncation of the hypothetical genes and bacteriophage to reduce the ability of tir 255 A alleles to cause disease in humans, the remaining four truncated genes have metabolic significance. glpQ is periplasmic and can catalyze the hydrolysis of glycerophosphodiesters. This enzyme allows the cell to use a variety of glycerophosphodiesters in the glp system (Larson et al., 1983). eutA is the sole enzyme responsible for reactivating ethanolamine ammonia-lyase (EAL). EAL is induced in the presence of both ethanolamine and vitamin B12 and is involved with both anaerobic fermentation and catabolism (Mori et al., 2004). apqZ prompts changes to the environmental osmolality by allowing the flux of water and is required for rapidly growing cells (Calamita, 2000). lpfE is a predicted fimbriae subunit protein and is found in the lpf1 operon in STEC O157:H7. Its proposed ligands are fibronectin, collagen IV, and laminin (McWilliams and Torres, 2014).
One of the 32 linked SNPs in the intergenic regions was found in the disrupted coding sequence for the propionyl-CoA:succinate CoA transferase (scpC) gene that was truncated previously by a 28 or 56 bp indel. The scpC indel was found in 13 of the 15 tir 255 A allele strains but was not found in any tir 255 T allele strains. Upon further investigation, the scpB gene just upstream of scpC also contained an indel that is 10 bp in length. All 15 tir 255 A allele strains have this indel, while it was not found in any tir 255 T allele strains. scpB and scpC are two of three genes in the succinate to propionate conversion pathway, one of three known pathways for propionate utilization and the only one found in STEC O157:H7. Propionate is an abundant volatile fatty acid that is produced by gut microbiota and found in high concentrations in the mammalian gastrointestinal tract. It has an antimicrobial activity (Salmond et al., 1984) and can block a number of metabolic pathways in Salmonella enterica (Horswill et al., 2001). To combat the negative effects of propionate, several pathways are used by bacteria to catabolize this compound into a carbon and energy source. The indels in the scbB and scbC genes appear to inactivate the propionate to succinate conversion and remove the beneficial properties of this pathway to STEC O157:H7 in the mammalian gut.
Prophage content of tir 255 allele chromosomes
There were 1,144 prophage regions identified in the 58 chromosomes analyzed that could be categorized into 33 different types (Table 1). Chromosomes had an average of 20 prophages (range = 16 to 36). There was no statistical difference (p = 0.07) between tir 255 allele strains when comparing the number of prophages. Seven of the 33 different phage sequences were found in similar percentages between the tir 255 A or T allele strains, while 16 were found more abundant in the tir 255 T allele strains, and 10 were commonly observed in the tir 255 A allele strains (Supplementary Table 5). There were nine prophages that were identified in all the tir 255 A allele strains, while only one was observed in all the tir 255 T allele strains, although this number is likely somewhat impacted by differing strain number representation. Three other prophages were only found in the tir 255 A allele strains, while 11 were found only in the tir 255 T allele strains. Twelve of the fourteen tir 255 strains contained less than five allele-specific prophages with most of them clustering with related strains. One tir 255 T allele-specific prophage, PHAGE_Entero_SfI_NC_027339, was found in 67% (29/43) strains. PHAGE_Entero_SfV_NC_003444 was the most specific prophage for tir 255 A allele strains with 87% of the strains containing this prophage compared to only 7% of the tir 255 T allele strains. Enterobacteria phage BP-4795 was the only prophage that was common to all strains; it has been characterized to contain two IS629 elements genes associated with type III secretion (Creuzburg et al., 2005). Additionally, two prophages (Stx2-converting phage 1717 and Shigella phage POCJ13) were found in 96.6% (56/58) of the strains. The human isolates did show statistical differences in phage content, and the small magnitude of these differences may not be of biological interest, a conclusion that is further supported by the fact that there is not a specific phage associated with only human-associated strains.
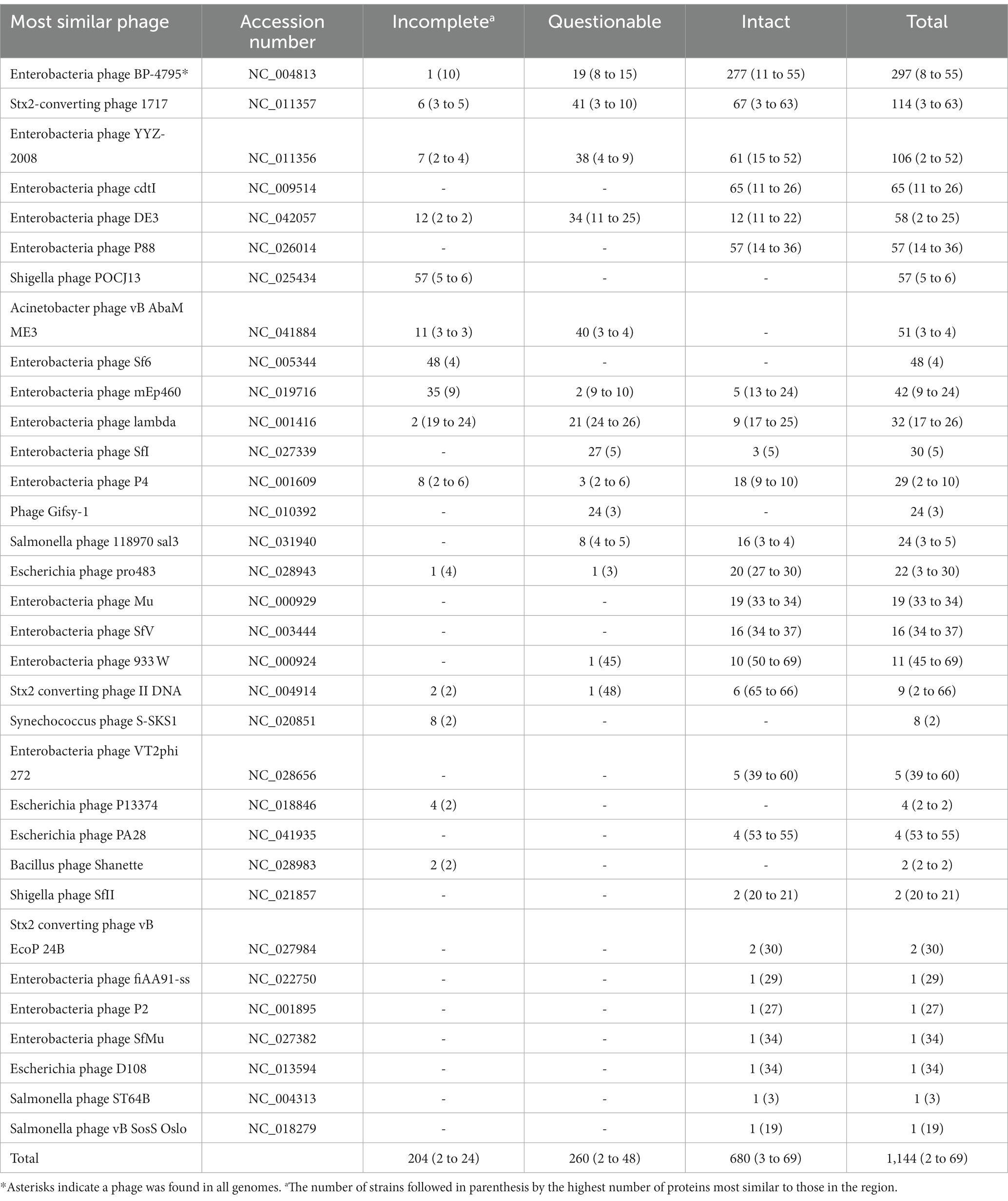
Table 1. Prophages identified within all Escherichia coli O157:H7 genomes and the levels of their completeness.
An average of 817 kb of each chromosome was comprised of prophage regions, which corresponds to an average of 14.8% (range 11.4% to 27.7%) of the chromosome. There was no difference when prophage percentages were compared between tir 255 alleles (p = 0.248). Bobay et al. previously observed the chromosome of STEC O157:H7 strain EC4115 to be composed of 13.5% prophage with the average chromosomal content of prophage in the five compared STEC O157:H7 strains to be 11.9% (Bobay et al., 2013). A similar study in E. coli O26:H11 found that the prophage content of those genomes ranged between 8.7% and 16.7%. While a review of the genes within these prophage regions was mostly non-specific (with the majority of annotations attributed to hypothetical genes), past study has suggested that these regions contribute to survivability and virulence (Delannoy et al., 2017).
Prophages that harbored stx genes in STEC O157:H7 strains were further scrutinized. Of the 33 prophages identified in the STEC O157:H7 strains, 10 were found to harbor at least one stx gene in one genome, with 66% residing in one of the three most common: Stx2-converting phage 1717 (34%), Enterobacteria phage BP-4795 (19%), or Enterobacteria phage 933 W (12%). Further characterization of these three prophages found that the phages differed substantially. Enterobacteria 933 W was only present while carrying stx genes, and only one copy of the phage per genome was present. On the other hand, both Stx2-converting 1717 and Enterobacteria BP-4795 were found in multiple copies within genomes (an average of 1.6 intact copies [range 1 to 2] and an average of 4.8 intact copies [range 2 to 6], respectively) and both with and without stx genes. From a composition standpoint, Enterobacteria phage BP-4795 was diverse within the population and even within a genome. Finally, Stx2-converting phage 1717 was observed in two forms (at least in some genomes), one in the form of a smaller ~20 kb prophage that does not harbor stx genes and the other contained in a larger ~60 kb prophage that does harbor stx.
Shiga toxin-containing prophage insertion sites are more conserved in tir 255 A allele strains
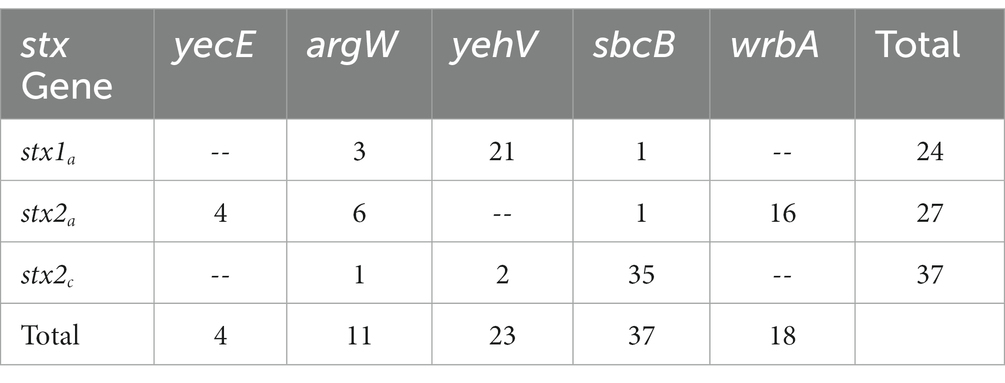
Prophages that contained stx genes were found to integrate into five chromosomally encoded genes (Table 2) that were previously identified as sites for stx-containing prophages. All 15 tir 255 A allele strains had stx2c containing prophage integrated at sbcB with two strains harboring two stx2c containing prophage integrated into tandem at the sbcB integration site (Supplementary Table 6). Three of the 15 tir 255 A allele strains had a stx1a, stx2a, and stx2c prophage integrated at the yehV integration site. The stx1a containing prophage identified in tir 255 T allele strains was integrated into three sites on the chromosome, yehV, argW, and sbcB, while stx2a containing prophage was integrated into four sites on the chromosome, argW, sbcB, wbdR, and yecE. stx2c containing prophage was integrated into sbcB and yehV. Past study has shown that the stx1 containing prophage integrates into yehV while stx2 containing prophage integrates into wrbA or sbcB (Bielaszewska et al., 2006) and argW is associated with stx1 integration (González-Escalona et al., 2019). The yecE gene has been described as an integration site in E. coli for prophage containing Stx2e in a ONT:H− (Recktenwald and Schmidt, 2002) and stx2 from both an E. coli O157:H7 (De Greve et al., 2002) and an E. coli O157:NM (Bielaszewska et al., 2006).
While most Stx prophages were found integrated into previously described genes, this was not always the case. Only prophages integrated at YecE were found to harbor a single type of stx gene; the rest were found to have one to three types of stx-containing prophage integrated into specific sites (Supplementary Table 6). This is in agreement with Serra-Moreno et al. (2007) who found that while these phages have a preferred primary insertion site, a secondary site is used if the primary site is occupied. With this in mind, strains described here that fall outside of insertion site norms may provide useful insight into variation in strain evolution.
Prophages can excise from genomes and integrate. When a prophage is completely excised from a genome, there typically are no remnant genes left behind. However, prophage excision can leave scars (Feiner et al., 2015), which allows for a more complete understanding of the strain’s evolutionary history. Here, prophage scarring was observed at the sbcB insertion site (Figure 2) when compared to an empty interion site from an O55 isolate. The scaring provides evidence of a prophage containing stx2c excising from the genome. This is interesting as a previous study (Asadulghani et al., 2009) has noted that most O157 phages contain one or more genetic defects and have low potential activity as mobile genetic elements (with their role being more evolutionary in STEC emergence).
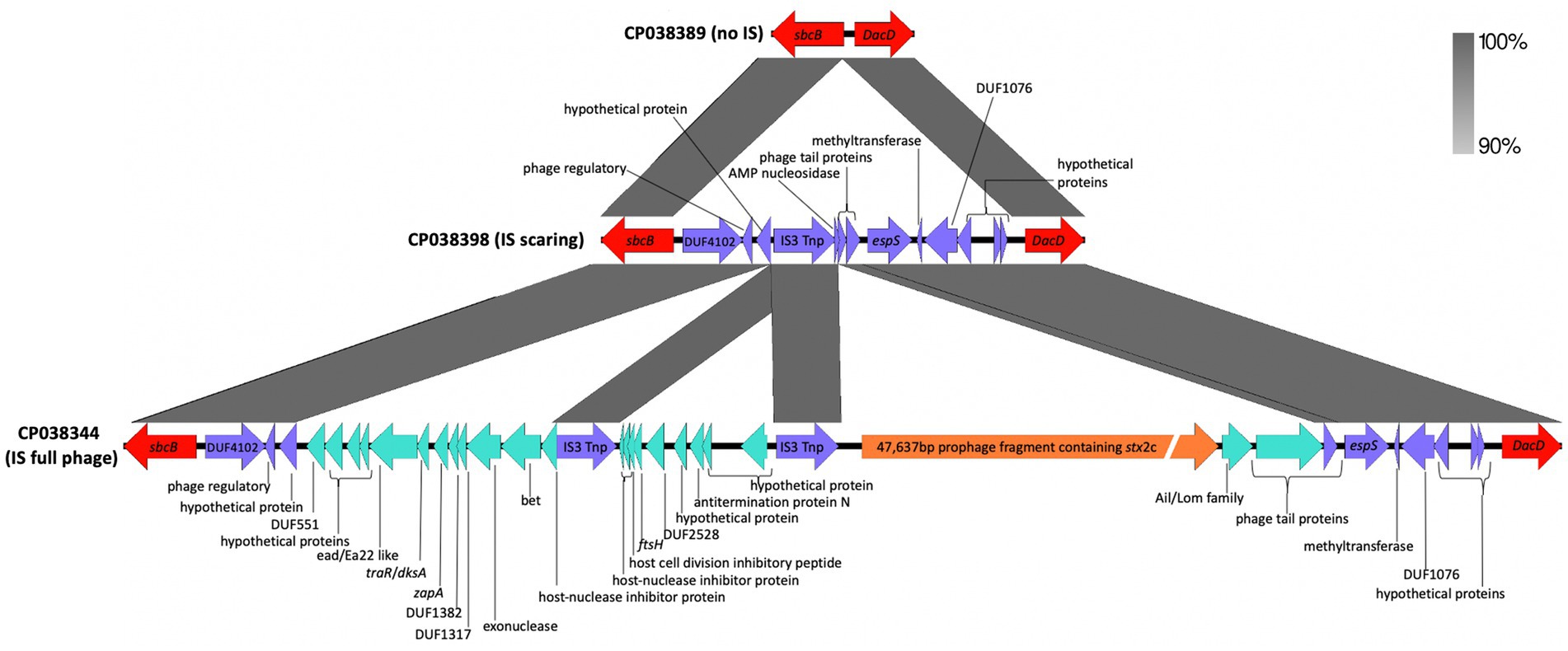
Figure 2. Genetic map showing the apparent scarring left by excision of a phage. The region from an ancestral Escherichia coli O55:H7 chromosome (CP038389) (top) did not have an insertion of a stx2c containing prophage at the sbcB gene. The intact stx2c contains prophage from strain Gim1-1 (CP038344) (bottom), and the genes left behind from the excision of the prophage containing the stx2c gene from strain DEC4E (CP038398) (middle) has some scarring associated with a prophage insertion and subsequent excision. The genes are represented by arrows with the red arrow representing the flanking chromosome genes and the purple arrow representing the genes shared by all three chromosomes. The green and orange arrows represent prophage genes. The shaded areas between the genetic map represent the percent similarly of the regions to each other.
Number of insertion sequence CDS was not found to differ between tir 255 allele strains
The insertion sequence (IS) elements from the chromosome of 48 strains and their corresponding pO157 plasmids were extracted from their GenBank annotations using Geneious. The IS elements were grouped into families, and the number of transposases per family was used in the analysis. There was no difference in the number of IS elements in the chromosome between the tir 255 allele strains. The tir 255 T allele strains contained an average of 99 IS elements (range 54 to 123), while the tir 255 A allele strains contained an average of 101 (range 84 to 118). On the other hand, the tir 255 A allele pO157 plasmids contained an average of 18 IS elements per plasmid (range 14 to 24), while the tir 255 T allele strains contained an average of 14 IS elements per plasmid (range 10 to 19).
The total number of IS elements identified across the 48 chromosomes and pO157 plasmids was 5,531 with 4,793 identified in the chromosomes and 738 in pO157 plasmids (Supplementary Table 7). This averages to about 115 transposases per genome with 100 per chromosome and 15 per pO157 plasmid. The most common IS element identified on both the chromosome and pO157 was the IS3 family transposase CDS (n = 1,222). The second most common, IS66 family insertion sequence hypothetical protein CDS had only two fewer IS elements (n = 1,220), with the third most common IS element being the IS66-like element ISEc8 family transposase CDS (n = 494). While 46 of the 48 strains had the same top three chromosomal IS elements, IS30-like element IS30 family transposase CDS was the most common IS element found in two ancestral strains 493–89 and LSU61. The top two most common IS elements identified on pO157 were the same as the chromosome, IS3 family transposase CDS (n = 295) and IS66 family insertion sequence hypothetical protein CDS (n = 104), respectively. The third most common IS element on pO157 was IS1 family transposase CDS (n = 95). Overall, IS3 family transposase CDS (n = 1,516) and IS66 family insertion sequence hypothetical protein CDS (n = 1,324) were the most abundant IS elements in the 48 strains in this study representing 62% of all IS elements identified.
IS elements have been previously studied in STEC O157:H7, but the main focus has been on IS629, the most abundant IS element in STEC O157 strains. IS629 is a non-replicative 1.31 kb region made up of orfA and orfB and belongs to the IS3 family, and subgroup IS51. IS629 has played a role in the insertional inactivation of genes including the stx gene (Kusumoto et al., 2004; Loftsdóttir et al., 2017; Nyong et al., 2020). We also observed the integration of an IS629 transposase into the stx2 beta subunit gene in strain MB41-1. The inactivation of stx genes by IS629 has no apparent effect in cattle but could excise itself from the stx gene causing human disease with a negative PCR result (Loftsdóttir et al., 2017). Several groups have used IS629 distribution as a way to type strains for epidemiological investigation, given the biased distribution associated with major phenotypic lineages (Ooka et al., 2009a; Yokoyama et al., 2011; Rump et al., 2011a; Stanton et al., 2014; Toro et al., 2015). The association was further enhanced by the inclusion of other STEC O157:H7 genes including stx and norV in genotyping assays (Stanton et al., 2014; Kameyama et al., 2015).
Antimicrobial resistance genes on the chromosome were not found to differ between tir 255 allele strains
The average number of antimicrobial resistance genes (ARGs) was not different between tir 255 A allele strains (avg = 53.9, range 53 to 58) and tir 255 T allele strains (avg = 52.8, range 51 to 55). An average of 53 (range 51 to 58) resistance genes were identified within all chromosomes, and the majority of these (44 genes with elfamycin-resistant EF-Tu identified in all genomes) were conserved across all chromosomes Supplementary Table 8. This result is not surprising because many species of bacteria, including E. coli, are known to be intrinsically resistant to certain antibiotics (Duo et al., 2008; Cox and Wright, 2013). Efflux pumps were the most common mechanisms of resistance followed by target alteration. Nine of the 16 ARGs that were not found in all chromosomes were present in more than 86% ( 50/58) of them. One of the nine genes was the putative tetracycline antibiotic efflux pump, multidrug transporter mdfA. This gene was found in 50 of the 58 strains. Eight strains (2149, 2571, 7636, DEC4E, F6321, F6667, Gim1-1, and NE92) were not found to contain the mdfA gene by CARD because of a missense mutation resulting in a truncated and presumably inactive protein. These eight strains were also found grouped next to each other on the phylogenetic tree (Figure 1). A set of five ARGs occurred in three chromosomes (two aminoglycoside resistance genes [aph(6)-Id and aph(3″)-Ib], two major facilitator superfamily efflux pumps that confer resistance to tetracycline (tetB and tetC), and a target replacement gene associated with sulfonamide resistance (sul2; Figure 3)). The resistance genes were divided into two cassettes, one consisting of aph(6)-Id, aph(3″)-Ib, and sul2 and the other consisting of the tetB, tetC, and tetR. The cassettes were part of an 80–84 kb genomic island inserted between cycD and clpA. Of the 91 genes in this genomic island, there were 22 hypothetical proteins and 11 insertion sequence/transposase genes. The three genomes with the integrated antibiotic cassettes were highly related (all tir 255 A, Manning clade 7, and containing stx2c). This cassette has been previously reported in STEC O157:H7 tir 255 A allele strains FREK944 (CP016625.1) and strain 446,541 (Greig et al., 2023), a STEC O145:H28 strain (FWSEC0002; Tyson et al., 2019), and an E. coli O22:H8 strain (154; Da Silva et al., 2022) that can interfere with STEC O157:H7 in vivo and in vitro colonization. The cassettes in strains FRIK944, FWSEC0002, and 446,541 are inserted between cycD and clpA and are more similar to each other than the STEC cassettes reported here. There were 14 other strains from England in addition to strain 446,541 isolated over a 4-year period that had the same ARG content. However, placement on their chromosome could not be determined because the genomes were sequenced using short-read sequencing technology. These cassettes appeared in both tir 255 allele strains, indicating no preference for one allele over the other, and on multiple continents, indicating that these cassettes are widely disseminated. The cassette from strain 154 is integrated between the yagQ and proA genes and has sequence similarity with ~60 kb of the other cassettes. STEC O157:H7 strain 1,090–2, tir 255 T allele strain, has two aminoglycoside resistance genes [aph(6)-Id and aph(3″)-Ib] flanked by a transposase and IS elements on one end and a transporter gene on the other end. This cassette was integrated into the chromosome between the lsrK autoinducer-2 kinase gene and an autotransporter barrel domain-containing lipoprotein gene. Tadesse et al. reported that an almost identical cassette minus the transporter gene was found on a 54 kb (CP018646 and CP018649) and 118 kb (CP018643) plasmid from Salmonella enterica serovar Enteritidis (Tadesse et al., 2018). These strains are historical S. enteritidis isolates isolated from humans in 1974 (CP018643), 1979 (CP018646), and 1981 (CP018649). STEC O157:H7 strain 1,090–2 was isolated from a steer at harvest in Nebraska in September 2000. This cassette appears to be ubiquitous and promiscuous, in nature as it is found across multiple decades and genera.
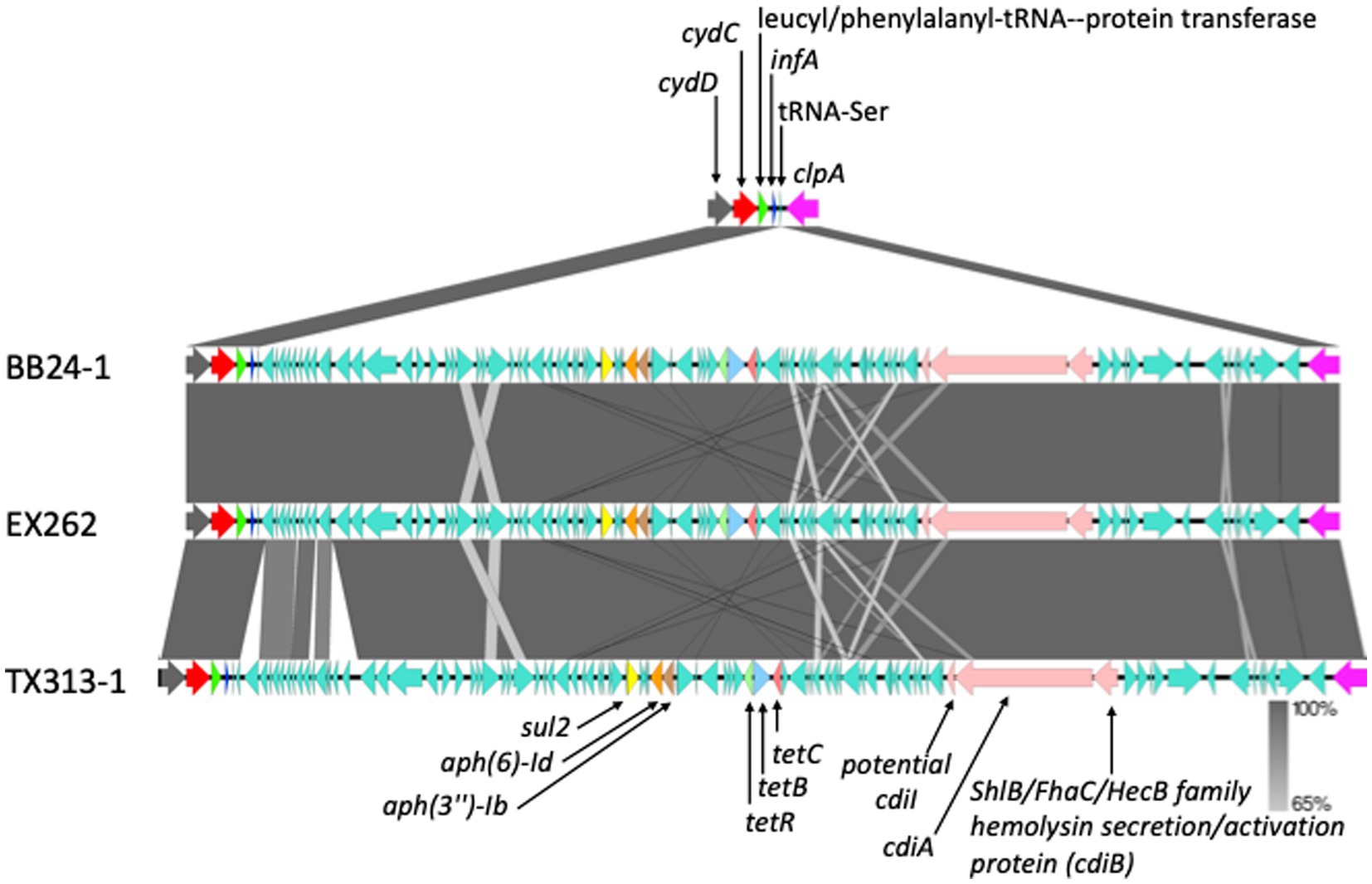
Figure 3. Genetic map showing an 80 kb integrated region in the chromosome from three STEC O157:H7 strains contains antibiotic resistance genes. Each colored arrow represents a different gene, except the green color which represents different genes in the inserted region. Gene names are located either above or below their respective gene. The shaded areas between the genetic map represent the percent similarly of the insertion regions to each other.
Most ARGs identified were present in all or most genomes, indicating that the resistome of E. coli O157:H7 seems to be stable and not to vary widely among this population. Some of the ARGs observed in the present study were associated with an insertion region, but there is little evidence to suggest that there is a diverse movement of ARGs within the STEC population. Nonetheless, this finding places no measure of likelihood on the possibility of a strain’s acquisition of ARGs within niche environments.
Plasmid pO157 has a strong evolutionary link to the host chromosome and the tir 255 allele
A tanglegram constructed between 56 STEC O157:H7 chromosomes and their associated pO157 plasmids showed a clear association of groupings between the two, regardless of their tir 255 allele variant (Figure 4). Chromosomes and O157 plasmids fell into the same phylogenetic tree structure in nearly every case, with very few incongruences between the two trees. This is in agreement with Nyong et al. (2020) who found a stable evolutionary relationship between the host chromosome and pO157 plasmids. pO157 plasmids from strains PV15-279 and F3113 were distinct from the rest of the pO157 tree (Figure 4), and upon further inspection, this was due to a 171 bp region containing 20 SNPs. The SNPs lie outside of any coding regions and were flanked by a replication initiation protein upstream and a transposase downstream. These two strains were from disparate geographically and sample source, with one coming from a Japanese human sample (PV15-279) and a United States FSIS sample (F3113). Further inspection of this region did not identify annotations that might have provided a biological explanation for variation from the other plasmids.
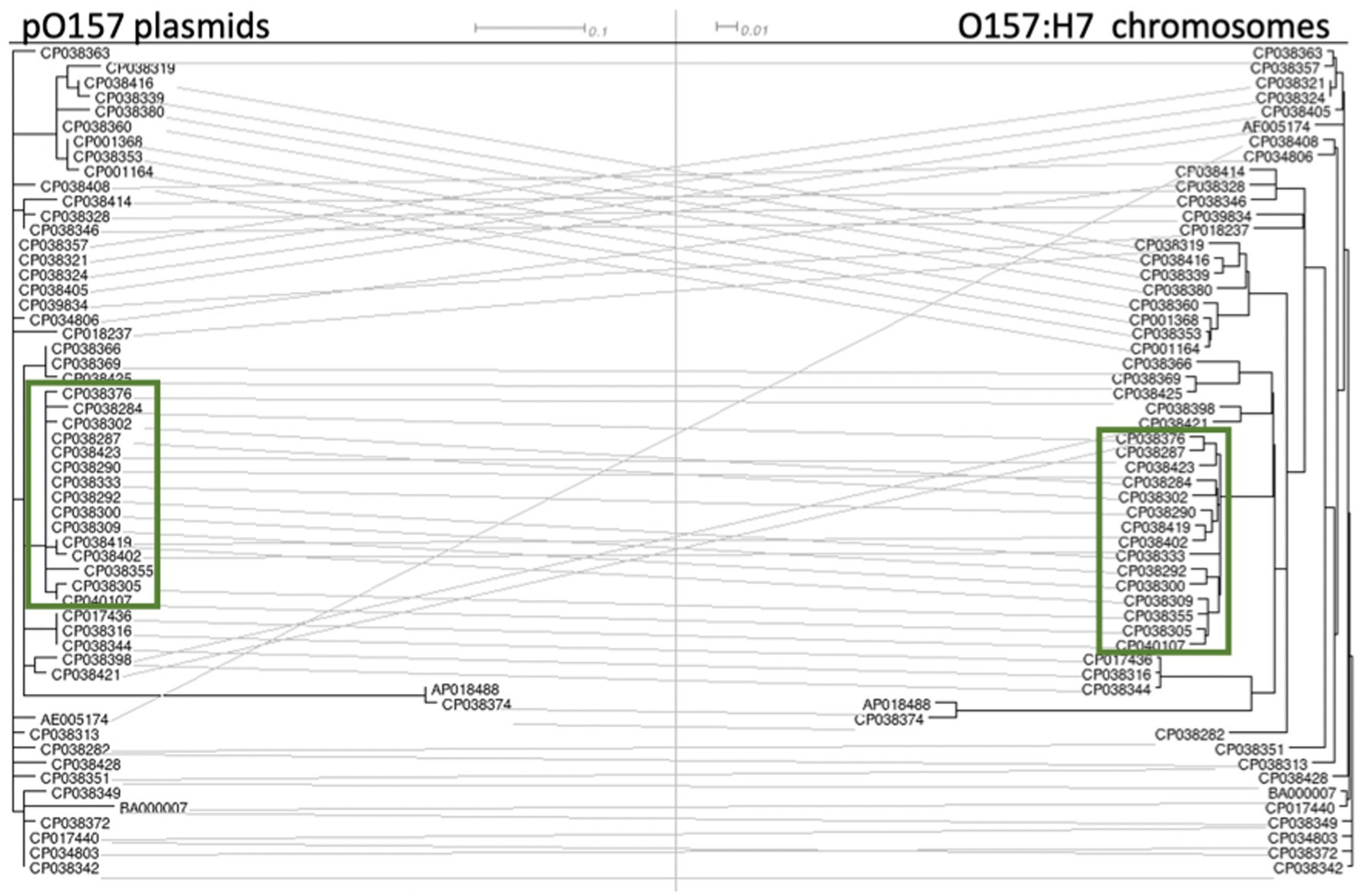
Figure 4. Tanglegram of pO157 plasmids (numbered with their chromosome’s GenBank accession number for ease of reading) and their STEC O157:H7 chromosome. The accession numbers boxed in green are tir 255 A allele strains. The accession numbers from the remaining non-highlighted strains are tir 255 T allele strains. The intermediate ancestral strains 493/89 and LSU 61 were not used in this analysis due to their pO157 plasmid being approximately 20 kb larger than the pO157 plasmids in other STEC O157:H7 strains.
There were 42 informative SNPs on the pO157 plasmid (pruned from 107 SNPs total). Two of the 42 SNPs were in linkage disequilibrium with the tir 255 allele. Both SNPs resulted in non-synonymous amino acid changes. The first was identified within the tagA gene 1,438 G > A (GenBank reference: AB011549, gene BAA31757.3) with the G allele occurring in tir 255 T allele strains and the A occurring with the tir 255 A allele strains. The second SNP was found in the toxB gene 7453 A > G (GenBank reference: AB011549, gene BAA31815.1) with the G allele occurring with the tir 255 T allele strains and the A occurring with the tir 255 A allele strains (Supplementary Table 4).
These two SNPs are noteworthy from both an evolutionary perspective and in understanding the likelihood of an isolate to cause human disease. In terms of evolution, these two SNPs lend further support to the growing understanding of the evolutionary link between the chromosome and the plasmid. Both toxB and tagA are associated with the ability to cause human disease. The toxB gene promotes the production of type III secreted proteins which contributes to the adherence on epithelial cells (Tatsuno et al., 2001), while tagA [later renamed StcE (Lathem et al., 2002)] expression has been found to elicit a host immune response during human infection with STEC O157:H7 (Paton and Paton, 2002). This response is due to tagA/StcE’s ability to cleave C1 esterase inhibitor (C1-INH), which has been suggested as the cause of localized pro-inflammatory and coagulation responses resulting in tissue damage (Lathem et al., 2002). The SNP profile of toxB 7,453 G allele and the tagA/stcE 1,438 G allele could be an indicator of a group of strains more likely to cause disease in humans, much like the tir 255 T found on the chromosome (Bono et al., 2007). The finding of the tir-linked SNPs in toxB and tagA further verifies that the pO157 plasmid plays an important role in the ability of a strain to cause disease in humans.
Additional plasmids identified in tir 255 T allele and tir 255 A allele strains
The remaining non-pO157 plasmids identified were placed into one of 10 groups by nucleotide sequence and gene content similarity. Of the 10 groups, four plasmid groups were found in both tir 255 T allele and tir 255 A allele strains, while four plasmid groups were only found in tir 255 T allele strains and two plasmid groups in tir 255 A allele strains (Supplementary Table 9). The two 145 kb IncA/C plasmids in group 1 were identical in sequence and belonged to tir 255 T allele strains. These plasmids contained ARGs encoding chloramphenicol/florfenicol, tetracycline, trimethoprim, aminoglycoside, and sulfonamide resistance. They also contained the QacE delta 1 gene for quaternary ammonium compound resistance. These plasmids are also related to plasmids in Klebsiella pneumoniae (CP110604), Shewanella algae (CP032414), and Salmonella enterica subspecies enterica serovars Diarizonae (CP117189) and Typhimurium (CP100733).
The two 99 kb plasmids in group 2 differ by 4 SNPs and the location of an IS3 family transposase. They belong to tir 255 A allele strains and the pO111 incompatibility type. These plasmids shared homology with bacteriophages that are chromosomally encoded (accession numbers NC_050153 and LM996300). Phaster identified the phage to be complete with a score greater than 90 with phage_Escher_RCS47_NC_042128 as the phage with the highest number of proteins most similar between the two.
The three plasmids in group 3 are 89 kb or 83 kb in size. The plasmids were found in tir 255 T allele strains and were the IncI1_I(Alpha) incompatibility type. The difference in size is due to deletions that include a colicin operon from the 83 kb plasmid. These plasmids contain a shufflon operon, an incomplete tra system, pilus relate genes, and colicin 1A. These plasmids are highly related to an unnamed plasmid from STEC O157:H7 strain ECP19-598 (CP0066754). They are also related to plasmids found in Salmonella enterica subspecies enterica serovars Agona (CP082483), Typhimurium (MW655523), and Derby (CP074320).
Group 4 plasmids belonging to the IncI2(Delta) incompatibility type are between 56 kb and 58 kb in size and were found one tir 255 A allele and three tir 255 T strains. The gene sequence order is conserved in this group of plasmids with the size differences related to deletions. The defining features of this plasmid group are pilus genes, incomplete tra operon, and a shufflon operon. The related plasmids from this group can also be found in other E. coli including strains 588888_1 (CP073614) and AR_0011 (CP024858).
Plasmids in group 5 ranging from 31 kb to 43 kb in size were identified in nine strains with four being tir 255 A allele strains and five tir 255 T allele strains. Two strains, EC4115 and pMB9_3, belonged to the pEC4115 incompatibility type, while the remaining seven were unclassified. The size difference between these plasmids can be associated with IS elements and deletions. The plasmids of group 4 contain some of the tra genes that are associated with transferring plasmids from one strain to another. Plasmids belonging to this group are not only found in different E. coli but are also found in Salmonella enterica strains.
Group 6 plasmids range in size from 6,675 bp to 7,324 kb and occurred in three tir 255 T alleles and one tir 255 A allele strain. The incompatibility type of these plasmids is unclassified. There are 51 SNP differences between this group of plasmids with most found in a hypothetical gene and its upstream and downstream untranslated region. This group of plasmids is similar to the pColD157 plasmid isolated from EHEC O157:H7 strain CL40cu (Y10412) with the defining feature being the colicin D gene with its immunity and lysis genes.
Group 7 was comprised of a single plasmid sequence of 6,402 bp found in three tir 255 A allele strains and an unclassified incompatibility type. These plasmids contained five hypothetical proteins, type III toxin-antitoxin system, and several genes associated with plasmid mobilization. The plasmids share the same gene arrangement as plasmid pEH09-18-41_4 from E. coli EH09-18-41 (CP063507) but differ by 37 SNPs located mainly in a nuclease gene.
The 3,306 bp plasmids in group 8 were first identified in the Sakai strain and named pOSAK1. pOSAK1 has an unclassified incompatibility type and was only found in tir 255 T allele strains. The plasmids are identical except for one plasmid having a SNP difference from the other plasmids.
The three plasmids in group 9 vary in size between 2,679 bp and 4,634 bp in size and have an unclassified incompatibility. Two were found in tir 255 A allele strains and one in a tir 255 T allele strain. Group 10 contains two 2.7 kb plasmids found in tir 255 A allele strains. These plasmids contain three genes annotated as encoding two hypothetical genes and a plasmid recombination enzyme gene. There were no plasmids in the GenBank non-redundant nucleotide database that had a similar sequence to these plasmids.
There were two miscellaneous plasmids that were not placed in any of the 10 groups. Both plasmids, 1,538 bp and 93,970 bp in size, were found in STEC strain NE1092-2. The 1,538 bp plasmid belongs to incompatibility type IncC and is closely related to plasmids found in E. coli strain Iso00041 (CP095159) and Shigella flexneri 1b strain A-10383 (CP130069). The 93 kb plasmid was found in a tir 255 T allele strain with the incompatibility type Col(MG828). This plasmid contains two aminoglycoside resistance genes along with genes encoding for chloramphenicol/florfenicol, tetracycline, and sulfonamide resistance. This plasmid also contains a mercury resistance operon. This plasmid has regions of homology to plasmids in Klebsiella pneumoniae (CP129795, CP124165, and CP124154) and Phytobacter diazotrophicus (AP028052).
Conclusion
The goal of this study was to identify genomic differences between tir 255 T allele and tir 255 A allele strains and determinants that could be responsible for the different tir 255 allele associations with human disease. The most accurate way to achieve that was with complete or nearly complete closed genomes. We used 57 completely closed genomes and another that was complete except for the chromosome being in two contigs for this study. STEC O157:H7 genomes are constantly being sequenced and added to the public databases; therefore, new unique genes or mobile elements may be discovered in future as part of the STEC O157:H7 pangenome. However, the differences identified between tir 255 T allele and tir 255 A allele core genomes in this study were established with strains that were from five different countries and that were well separated in time and space. Consequently, the core tir 255 T and A allele genomes will likely not fluctuate as much as the pangenome in future with the addition of new genomes.
The chromosome and pO157 plasmid from tir 255 T allele and tir 255 A allele strains differed in size and the average number of CDS from each other. The size differences in the chromosome were not due to additional mobile elements such as prophage or IS elements. However, there were differences between the types of annotated prophage between tir 255 T and A allele strains, including allele-specific prophages that were grouped by strain relatedness. This suggests that prophages have remained integrated into the chromosome long enough to become part of a clonal lineage. The pO157 plasmid size differences could be attributed to IS elements with the tir 255 A allele strains having on average four more IS elements than the tir 255 T allele strains. There were also compelling SNPs on pO157 that were in linkage disequilibrium with the tir 255 alleles.
The overall percentage of STEC O157 strains that carried ARGs was quite low, with resistance genes observed on the chromosome in tir 255 A allele strains but three ARGs carrying plasmids in tir 255 T allele strains. Certain plasmid sequences were unique to either the tir 255 A or T allele strains. However, the allele-specific plasmids were not found in all allele strains suggesting additional plasmids are not likely responsible for the association of tir 255 T variants having a greater probability of causing disease in humans.
Within the core chromosomes of tir 255 A and T strains, 18 CDS were unique to tir 255A and 3 were unique to tir 255 T. SNP alleles consistently observed in the tir 255A lineage resulted in amber mutations in four genes (glpQ, eutA, apqZ, and lpfE). Out-of-frame indel alleles linked to the tir 255A lineage were also identified in scpB and scpC genes. While the inactivated genes were predicted to provide a growth advantage, none necessarily explain why tir 255 A allele strains are less likely to cause disease in humans than tir 255 T allele strains without additional research. The results presented provide a more complete understanding of the genomics of this important public health pathogen and new insight into genomic features that may contribute to decreased association with human disease. Further investigation using transcriptomic and metabolomics methods, phenotypic antimicrobial resistance profiles, and disease models to assess the role of the variants may provide a greater explanation for the tir 255 allele differences in association with human disease.
Data availability statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/Supplementary material.
Author contributions
MW: Conceptualization, Data curation, Formal Analysis, Writing – original draft. MC: Formal Analysis, Writing – original draft. GH: Formal Analysis, Writing – review & editing. ME: Formal Analysis, Writing – review & editing. DH: Formal Analysis, Writing – review & editing. TS: Formal Analysis, Writing – review & editing. JB: Conceptualization, Data curation, Formal Analysis, Project administration, Supervision, Writing – original draft.
Funding
The author(s) declare financial support was received for the research, authorship, and/or publication of this article. This research was funded by the USDA Agricultural Research Service: 3040-42000-017-00D. The contribution of ME was supported by the National Institutes of Health under Award Number (SC1GM135110).
Acknowledgments
The authors would like to thank Sandy Fryda-Bradley and Kerry Brader for technical assistance; Bob Lee, Kelsey McClure, and Kristen Kuhn for sequencing support; Aaron Rogge and Randy Bradley for IT support; and Jody Gallagher for secretarial assistance. The use of product and company names is necessary to accurately report the methods and results; however, the United States Department of Agriculture (USDA) neither guarantees nor warrants the standard of the products, and the use of names by the USDA implies no approval of the product to the exclusion of others that may also be suitable. The USDA is an equal opportunity provider and employer.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
The author(s) declared that they were an editorial board member of Frontiers, at the time of submission. This had no impact on the peer review process and the final decision.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2023.1303387/full#supplementary-material
References
Alcock, B. P., Raphenya, A. R., Lau, T. T. Y., Tsang, K. K., Bouchard, M., Edalatmand, A., et al. (2020). CARD 2020: antibiotic resistome surveillance with the comprehensive antibiotic resistance database. Nucleic Acids Res. 48, D517–D525. doi: 10.1093/nar/gkz935
Arndt, D., Grant, J. R., Marcu, A., Sajed, T., Pon, A., Liang, Y., et al. (2016). PHASTER: a better, faster version of the PHAST phage search tool. Nucleic Acids Res. 44, W16–W21. doi: 10.1093/nar/gkw387
Asadulghani, M., Ogura, Y., Ooka, T., Itoh, T., Sawaguchi, A., Iguchi, A., et al. (2009). The defective prophage pool of Escherichia coli O157: prophage–prophage interactions potentiate horizontal transfer of virulence determinants. PLoS Pathog. 5:e1000408. doi: 10.1371/journal.ppat.1000408
Besser, T. E., Shaikh, N., Holt, N. J., Tarr, P. I., Konkel, M. E., Malik-Kale, P., et al. (2007). Greater diversity of Shiga toxin-encoding bacteriophage insertion sites among Escherichia coli O157:H7 isolates from cattle than in those from humans. Appl. Environ. Microbiol. 74:554. doi: 10.1128/AEM.02632-07
Bielaszewska, M., Prager, R., Zhang, W., Friedrich, A. W., Mellmann, A., Tschäpe, H., et al. (2006). Chromosomal dynamism in progeny of outbreak-related sorbitol-fermenting enterohemorrhagic Escherichia coli O157:NM. Appl. Environ. Microbiol. 72, 1900–1909. doi: 10.1128/AEM.72.3.1900-1909.2006
Bobay, L.-M., Rocha, E. P. C., and Touchon, M. (2013). The adaptation of temperate bacteriophages to their host genomes. Mol. Biol. Evol. 30, 737–751. doi: 10.1093/molbev/mss279
Bono, J. L., Keen, J. E., Clawson, M. L., Durso, L. M., Heaton, M. P., and Laegreid, W. W. (2007). Association of Escherichia coli O157:H7 tir polymorphisms with human infection. BMC Infect. Dis. 7:98. doi: 10.1186/1471-2334-7-98
Bono, J. L., Smith, T. P. L., Keen, J. E., Harhay, G. P., McDaneld, T. G., Mandrell, R. E., et al. (2012). Phylogeny of Shiga toxin-producing Escherichia coli O157 isolated from cattle and clinically ill humans. Mol. Biol. Evol. 29, 2047–2062. doi: 10.1093/molbev/mss072
Calamita, G. (2000). The Escherichia coli aquaporin-Z water channel. Mol. Microbiol. 37, 254–262. doi: 10.1046/j.1365-2958.2000.02016.x
Camacho, C., Coulouris, G., Avagyan, V., Ma, N., Papadopoulos, J., Bealer, K., et al. (2009). BLAST+: architecture and applications. BMC Bioinformatics 10:421. doi: 10.1186/1471-2105-10-421
Chang, C. C., Chow, C. C., Tellier, L. C., Vattikuti, S., Purcell, S. M., and Lee, J. J. (2015). Second-generation PLINK: rising to the challenge of larger and richer datasets. GigaScience 4:7. doi: 10.1186/s13742-015-0047-8
Chin, C.-S., Alexander, D. H., Marks, P., Klammer, A. A., Drake, J., Heiner, C., et al. (2013). Nonhybrid, finished microbial genome assemblies from long-read SMRT sequencing data. Nat. Methods 10, 563–569. doi: 10.1038/nmeth.2474
Clawson, M. L., Keen, J. E., Smith, T. P., Durso, L. M., McDaneld, T. G., Mandrell, R. E., et al. (2009). Phylogenetic classification of Escherichia coli O157:H7 strains of human and bovine origin using a novel set of nucleotide polymorphisms. Genome Biol. 10:R56. doi: 10.1186/gb-2009-10-5-r56
Cox, G., and Wright, G. D. (2013). Intrinsic antibiotic resistance: mechanisms, origins, challenges and solutions. Int. J. Med. Microbiol. 303, 287–292. doi: 10.1016/j.ijmm.2013.02.009
Creuzburg, K., Recktenwald, J., Kuhle, V., Herold, S., Hensel, M., and Schmidt, H. (2005). The Shiga toxin 1-converting bacteriophage BP-4795 encodes an NleA-like type III effector protein. J. Bacteriol. 187, 8494–8498. doi: 10.1128/JB.187.24.8494-8498.2005
Da Silva, W. M., Larzabal, M., Aburjaile, F. F., Riviere, N., Martorelli, L., Bono, J., et al. (2022). Whole-genome sequencing analysis of Shiga toxin-producing Escherichia coli O22:H8 isolated from cattle prediction pathogenesis and colonization factors and position in STEC universe phylogeny. J. Microbiol. 60, 689–704. doi: 10.1007/s12275-022-1616-z
Darling, A. C. E., Mau, B., Blattner, F. R., and Perna, N. T. (2004). Mauve: multiple alignment of conserved genomic sequence with rearrangements. Genome Res. 14, 1394–1403. doi: 10.1101/gr.2289704
De Greve, H., Qizhi, C., Deboeck, F., and Hernalsteens, J.-P. (2002). The Shiga-toxin VT2-encoding bacteriophage ϕ297 integrates at a distinct position in the Escherichia coli genome. Biochim Biophys 1579, 196–202. doi: 10.1016/S0167-4781(02)00539-0
De Maio, N., Shaw, L. P., Hubbard, A., George, S., Sanderson, N. D., Swann, J., et al. (2019). Comparison of long-read sequencing technologies in the hybrid assembly of complex bacterial genomes. Microb. Genomics 5:e000294. doi: 10.1099/mgen.0.000294
Delannoy, S., Mariani-Kurkdjian, P., Webb, H. E., Bonacorsi, S., and Fach, P. (2017). The Mobilome; a major contributor to Escherichia coli stx2-positive O26:H11 strains intra-serotype diversity. Front. Microbiol. 8:1625. doi: 10.3389/fmicb.2017.01625
Didelot, X., Bowden, R., Wilson, D. J., Peto, T. E. A., and Crook, D. W. (2012). Transforming clinical microbiology with bacterial genome sequencing. Nat. Rev. Genet. 13, 601–612. doi: 10.1038/nrg3226
Duo, M., Hou, S., and Ren, D. (2008). Identifying Escherichia coli genes involved in intrinsic multidrug resistance. Appl. Microbiol. Biotechnol. 81, 731–741. doi: 10.1007/s00253-008-1709-6
Eppinger, M., Mammel, M. K., Leclerc, J. E., Ravel, J., and Cebula, T. A. (2011). Genomic anatomy of Escherichia coli O157:H7 outbreaks. Proc. Natl. Acad. Sci. 108, 20142–20147. doi: 10.1073/pnas.1107176108
Feiner, R., Argov, T., Rabinovich, L., Sigal, N., Borovok, I., and Herskovits, A. A. (2015). A new perspective on lysogeny: prophages as active regulatory switches of bacteria. Nat. Rev. Microbiol. 13, 641–650. doi: 10.1038/nrmicro3527
Fitzgerald, S. F., Beckett, A. E., Palarea-Albaladejo, J., McAteer, S., Shaaban, S., Morgan, J., et al. (2019). Shiga toxin sub-type 2a increases the efficiency of Escherichia coli O157 transmission between animals and restricts epithelial regeneration in bovine enteroids. PLoS Pathog. 15:e1008003. doi: 10.1371/journal.ppat.1008003
Giardine, B., Riemer, C., Hardison, R. C., Burhans, R., Elnitski, L., Shah, P., et al. (2005). Galaxy: a platform for interactive large-scale genome analysis. Genome Res. 15, 1451–1455. doi: 10.1101/gr.4086505
Goecks, J., Nekrutenko, A., and Taylor, J., The Galaxy Team (2010). Galaxy: a comprehensive approach for supporting accessible, reproducible, and transparent computational research in the life sciences. Genome Biol. 11:R86. doi: 10.1186/gb-2010-11-8-r86
González-Escalona, N., Allard, M. A., Brown, E. W., Sharma, S., and Hoffmann, M. (2019). Nanopore sequencing for fast determination of plasmids, phages, virulence markers, and antimicrobial resistance genes in Shiga toxin-producing Escherichia coli. PloS One 14:e0220494. doi: 10.1371/journal.pone.0220494
Greig, D. R., Do Nascimento, V., Olonade, I., Swift, C., Nair, S., and Jenkins, C. (2023). Surveillance of antimicrobial resistant Shiga toxin-producing E. coli O157:H7 in England, 2016–2020. J. Antimicrob. Chemother. 78:dkad231. doi: 10.1093/jac/dkad231
Hofinger, C., Karch, H., and Schmidt, H. (1998). Structure and function of plasmid pColD157 of enterohemorrhagic Escherichia coli O157 and its distribution among strains from patients with diarrhea and hemolytic-uremic syndrome. J. Clin. Microbiol. 36, 24–29. doi: 10.1128/JCM.36.1.24-29.1998
Horswill, A. R., Dudding, A. R., and Escalante-Semerena, J. C. (2001). Studies of propionate toxicity in Salmonella enterica identify 2-methylcitrate as a potent inhibitor of cell growth. J. Biol. Chem. 276, 19094–19101. doi: 10.1074/jbc.M100244200
Huson, D. H., and Scornavacca, C. (2012). Dendroscope 3: an interactive tool for rooted phylogenetic trees and networks. Syst. Biol. 61, 1061–1067. doi: 10.1093/sysbio/sys062
Jenkins, C., Dallman, T. J., and Grant, K. A. (2019). Impact of whole genome sequencing on the investigation of food-borne outbreaks of Shiga toxin-producing Escherichia coliserogroup O157:H7, England, 2013 to 2017. Eur. Secur. 24:346. doi: 10.2807/1560-7917.ES.2019.24.4.1800346
Jinneman, K. C., Weagant, S. D., Johnson, J. M., Abbott, S. L., Hill, W. E., Tenge, B. J., et al. (2000). A large insertion in the Shiga-like toxin 2 gene (stx2) of an Escherichia coli O157:H7 clinical isolate. Int. J. Food Microbiol. 57, 115–124. doi: 10.1016/S0168-1605(00)00237-3
Joensen, K. G., Scheutz, F., Lund, O., Hasman, H., Kaas, R. S., Nielsen, E. M., et al. (2014). Real-time whole-genome sequencing for routine typing, surveillance, and outbreak detection of verotoxigenic Escherichia coli. J. Clin. Microbiol. 52, 1501–1510. doi: 10.1128/JCM.03617-13
Kameyama, M., Tominaga, K., Yabata, J., and Nomura, Y. (2015). Distribution of IS629 and stx genotypes among enterohemorrhagic Escherichia coli O157 isolates in Yamaguchi prefecture, Japan, 2004-2013. J. Vet. Med. Sci. 77, 1437–1441. doi: 10.1292/jvms.15-0166
Kaper, J. B., Nataro, J. P., and Mobley, H. L. T. (2004). Pathogenic Escherichia coli. Nat. Rev. Microbiol. 2, 123–140. doi: 10.1038/nrmicro818
Karch, H., Bielaszewska, M., Bitzan, M., and Schmidt, H. (1999). Epidemiology and diagnosis of Shiga toxin-producing Escherichia coli infections. Diagn. Microbiol. Infect. Dis. 34, 229–243. doi: 10.1016/S0732-8893(99)00031-0
Kim, J., Nietfeldt, J., and Benson, A. K. (1999). Octamer-based genome scanning distinguishes a unique subpopulation of Escherichia coli O157:H7 strains in cattle. Proc. Natl. Acad. Sci. U. S. A. 96, 13288–13293. doi: 10.1073/pnas.96.23.13288
Koren, S., Harhay, G. P., Smith, T. P., Bono, J. L., Harhay, D. M., Mcvey, S. D., et al. (2013). Reducing assembly complexity of microbial genomes with single-molecule sequencing. Genome Biol. 14:R101. doi: 10.1186/gb-2013-14-9-r101
Koren, S., and Phillippy, A. M. (2015). One chromosome, one contig: complete microbial genomes from long-read sequencing and assembly. Curr. Opin. Microbiol. 23, 110–120. doi: 10.1016/j.mib.2014.11.014
Kusumoto, M., Nishiya, Y., Kawamura, Y., and Shinagawa, K. (1999). Identification of an insertion sequence, IS1203 variant, in a Shiga toxin 2 gene of Escherichia coli O157:H7. J. Biosci. Bioeng. 87, 93–96. doi: 10.1016/s1389-1723(99)80014-0
Kusumoto, M., Okitsu, T., Nishiya, Y., Suzuki, R., Yamai, S., and Kawamura, Y. (2001). Spontaneous reactivation of Shiga toxins in Escherichia coli O157:H7 cells caused by transposon excision. J. Biosci. Bioeng. 92, 114–120. doi: 10.1016/S1389-1723(01)80210-3
Kusumoto, M., Suzuki, R., Nishiya, Y., Okitsu, T., and Oka, M. (2004). Host-dependent activation of IS1203v excision in Shiga toxin-producing Escherichia coli. J. Biosci. Bioeng. 97, 406–411. doi: 10.1016/S1389-1723(04)70227-3
Larson, T. J., Ehrmann, M., and Boos, W. (1983). Periplasmic glycerophosphodiester phosphodiesterase of Escherichia coli, a new enzyme of the glp regulon. J. Biol. Chem. 258, 5428–5432. doi: 10.1016/S0021-9258(20)81908-5
Lathem, W. W., Grys, T. E., Witowski, S. E., Torres, A. G., Kaper, J. B., Tarr, P. I., et al. (2002). StcE, a metalloprotease secreted by Escherichia coli coli O157:H7, specifically cleaves C1 esterase inhibitor. Mol. Microbiol. 45, 277–288. doi: 10.1046/j.1365-2958.2002.02997.x
Lery, L. M., Frangeul, L., Tomas, A., Passet, V., Almeida, A. S., Bialek-Davenet, S., et al. (2014). Comparative analysis of Klebsiella pneumoniae genomes identifies a phospholipase D family protein as a novel virulence factor. BMC Biol. 12:41. doi: 10.1186/1741-7007-12-41
Lim, J. Y., Yoon, J. W., and Hovde, C. J. (2010). A brief overview of Escherichia coli O157:H7 and its plasmid O157. J. Microbiol. Biotechnol. 20, 5–14. doi: 10.4014/jmb.0908.08007
Loftsdóttir, H., Söderlund, R., Jinnerot, T., Eriksson, E., Bongcam-Rudloff, E., and Aspán, A. (2017). Dynamics of insertion sequence element IS629 inactivation of verotoxin 2 genes in Escherichia coli O157:H7. FEMS Microbiol. Lett. 364:fnx074. doi: 10.1093/femsle/fnx074
Luo, H., Zhang, C.-T., and Gao, F. (2014). Ori-finder 2, an integrated tool to predict replication origins in the archaeal genomes. Front. Microbiol. 5:482. doi: 10.3389/fmicb.2014.00482
Makino, K., Ishii, K., Yasunaga, T., Hattori, M., Yokoyama, K., Yutsudo, C. H., et al. (1998). Complete nucleotide sequences of 93-kb and 3.3-kb plasmids of an enterohemorrhagic Escherichia coli O157:H7 derived from Sakai outbreak. DNA Res. 5, 1–9. doi: 10.1093/dnares/5.1.1
Malberg Tetzschner, A. M., Johnson, J. R., Johnston, B. D., Lund, O., and Scheutz, F. (2020). In silico genotyping of Escherichia coli isolates for extraintestinal virulence genes by use of whole-genome sequencing data. J. Clin. Microbiol. 58:20. doi: 10.1128/jcm.01269-20
Manning, S., Motiwala, A., Springman, A., Qi, W., Lacher, D., Ouellette, L., et al. (2008). Variation in virulence among clades of Escherichia coli O157:H7 associated with disease outbreaks. Proc. Natl. Acad. Sci. U. S. A. 105, 4868–4873. doi: 10.1073/pnas.0710834105
McWilliams, B. D., and Torres, A. G. (2014). Enterohemorrhagic Escherichia coli Adhesins. Microbiol. Spectr. 2:EHEC00032013. doi: 10.1128/microbiolspec.EHEC-0003-2013
Mead, P. S., and Griffin, P. M. (1998). Escherichia coli O157:H7. Lancet 352, 1207–1212. doi: 10.1016/S0140-6736(98)01267-7
Moran, N. A. (2002). Microbial minimalism: genome reduction in bacterial pathogens. Cells 108, 583–586. doi: 10.1016/S0092-8674(02)00665-7
Mori, K., Bando, R., Hieda, N., and Toraya, T. (2004). Identification of a reactivating factor for adenosylcobalamin-dependent ethanolamine ammonia lyase. J. Bacteriol. 186, 6845–6854. doi: 10.1128/JB.186.20.6845-6854.2004
Murray, G. G. R., Charlesworth, J., Miller, E. L., Casey, M. J., Lloyd, C. T., Gottschalk, M., et al. (2021). Genome reduction is associated with bacterial pathogenicity across different scales of temporal and ecological divergence. Mol. Biol. Evol. 38, 1570–1579. doi: 10.1093/molbev/msaa323
Nadon, C., Van Walle, I., Gerner-Smidt, P., Campos, J., Chinen, I., Concepcion-Acevedo, J., et al. (2017). PulseNet international: vision for the implementation of whole genome sequencing (WGS) for global food-borne disease surveillance. Eur. Secur. 22:544. doi: 10.2807/1560-7917.ES.2017.22.23.30544
Nyong, E. C., Zaia, S. R., Allué-Guardia, A., Rodriguez, A. L., Irion-Byrd, Z., Koenig, S. S. K., et al. (2020). Pathogenomes of atypical non-Shiga-toxigenic Escherichia coli NSF/SF O157:H7/NM: comprehensive phylogenomic analysis using closed genomes. Front. Microbiol. 11:619. doi: 10.3389/fmicb.2020.00619
Ochman, H., and Moran, N. A. (2001). Genes lost and genes found: evolution of bacterial pathogenesis and symbiosis. Science 292, 1096–1099. doi: 10.1126/science.1058543
Ooka, T., Ogura, Y., Asadulghani, M., Ohnishi, M., Nakayama, K., Terajima, J., et al. (2009a). Inference of the impact of insertion sequence (IS) elements on bacterial genome diversification through analysis of small-size structural polymorphisms in Escherichia coli O157 genomes. Genome Res. 19, 1809–1816. doi: 10.1101/gr.089615.108
Ooka, T., Terajima, J., Kusumoto, M., Iguchi, A., Kurokawa, K., Ogura, Y., et al. (2009b). Development of a multiplex PCR-based rapid typing method for enterohemorrhagic Escherichia coli O157 strains. J. Clin. Microbiol. 47, 2888–2894. doi: 10.1128/JCM.00792-09
Ouellette, L. A., Reid, R. W., Blanchard, S. G., and Brouwer, C. R. (2018). LinkageMapView—rendering high-resolution linkage and QTL maps. Bioinformatics 34, 306–307. doi: 10.1093/bioinformatics/btx576
Page, A. J., Cummins, C. A., Hunt, M., Wong, V. K., Reuter, S., Holden, M. T. G., et al. (2015). Roary: rapid large-scale prokaryote pan genome analysis. Bioinformatics 31, 3691–3693. doi: 10.1093/bioinformatics/btv421
Paton, A. W., and Paton, J. C. (2002). Reactivity of convalescent-phase hemolytic-uremic syndrome patient sera with the megaplasmid-encoded TagA protein of Shiga toxigenic Escherichia coli O157. J. Clin. Microbiol. 40, 1395–1399. doi: 10.1128/JCM.40.4.1395-1399.2002
Rambaut, A. (2009). FigTree. Available at: https://github.com/rambaut/figtree/.
Recktenwald, J., and Schmidt, H. (2002). The nucleotide sequence of Shiga toxin (Stx) 2e-encoding phage φP27 is not related to other Stx phage genomes, but the modular genetic structure is conserved. Infect. Immun. 70, 1896–1908. doi: 10.1128/IAI.70.4.1896-1908.2002
Riordan, J. T., Viswanath, S. B., Manning, S. D., and Whittam, T. S. (2008). Genetic differentiation of Escherichia coli O157:H7 clades associated with human disease by real-time PCR. J. Clin. Microbiol. 46, 2070–2073. doi: 10.1128/JCM.00203-08
Rump, L. V., Fischer, M., and González-Escalona, N. (2011a). Different IS629 transposition frequencies exhibited by Escherichia coli O157:H7 strains in the stepwise evolutionary model. Appl. Environ. Microbiol. 77, 5030–5033. doi: 10.1128/AEM.00249-11
Rump, L. V., Fischer, M., and Gonzalez-Escalona, N. (2011b). Prevalence, distribution and evolutionary significance of the IS629 insertion element in the stepwise emergence of Escherichia coli O157:H7. BMC Microbiol. 11:133. doi: 10.1186/1471-2180-11-133
Rusconi, B., Sanjar, F., Koenig, S. S. K., Mammel, M. K., Tarr, P. I., and Eppinger, M. (2016). Whole genome sequencing for genomics-guided investigations of Escherichia coli O157:H7 outbreaks. Front. Microbiol. 7:985. doi: 10.3389/fmicb.2016.00985
Salmond, C. V., Kroll, R. G., and Booth, I. R. (1984). The effect of food preservatives on pH homeostasis in Escherichia coli. Microbiology 130, 2845–2850. doi: 10.1099/00221287-130-11-2845
Scallan, E., Hoekstra, R. M., Angulo, F. J., Tauxe, R. V., Widdowson, M.-A., Roy, S. L., et al. (2011). Foodborne illness acquired in the United States—major pathogens. Emerg. Infect. Dis. 17, 7–15. doi: 10.3201/eid1701.P11101
Schroeder, C. M., Zhao, C., DebRoy, C., Torcolini, J., Zhao, S., White, D. G., et al. (2002). Antimicrobial resistance of Escherichia coli O157 isolated from humans, cattle, swine, and food. Appl. Environ. Microbiol. 68, 576–581. doi: 10.1128/AEM.68.2.576-581.2002
Scornavacca, C., Zickmann, F., and Huson, D. H. (2011). Tanglegrams for rooted phylogenetic trees and networks. Bioinformatics 27, i248–i256. doi: 10.1093/bioinformatics/btr210
Seemann, T. (2014). Prokka: rapid prokaryotic genome annotation. Bioinformatics 30, 2068–2069. doi: 10.1093/bioinformatics/btu153
Serra-Moreno, R., Jofre, J., and Muniesa, M. (2007). Insertion site occupancy by stx2 bacteriophages depends on the locus availability of the host strain chromosome. J. Bacteriol. 189, 6645–6654. doi: 10.1128/JB.00466-07
Slutsker, L., Ries, A. A., Greene, K. D., Hutwagner, L., and Griffin, P. M. (1997). Escherichia coli O157:H7 diarrhea in the United States: clinical and epidemiologic features. Ann. Intern. Med. 126, 505–513. doi: 10.7326/0003-4819-126-7-199704010-00002
Smith, D., Blackford, M., Younts, S., Moxley, R., Gray, J., Hungerford, L., et al. (2001). Ecological relationships between the prevalence of cattle shedding Escherichia coli O157:H7 and characteristics of the cattle or conditions of the feedlot pen. J. Food Prot. 64, 1899–1903. doi: 10.4315/0362-028X-64.12.1899
Stanton, E., Park, D., Döpfer, D., Ivanek, R., and Kaspar, C. W. (2014). Phylogenetic characterization of Escherichia coli O157:H7 based on IS629 distribution and Shiga toxin genotype. Microbiology 160, 502–513. doi: 10.1099/mic.0.073437-0
Subramanian, B., Gao, S., Lercher, M. J., Hu, S., and Chen, W.-H. (2019). Evolview v3: a webserver for visualization, annotation, and management of phylogenetic trees. Nucleic Acids Res. 47, W270–W275. doi: 10.1093/nar/gkz357
Sullivan, M. J., Petty, N. K., and Beatson, S. A. (2011). Easyfig: a genome comparison visualizer. Bioinformatics 27, 1009–1010. doi: 10.1093/bioinformatics/btr039
Tadesse, D. A., Hoffmann, M., Sarria, S., Lam, C., Brown, E., Allard, M., et al. (2018). Complete genome sequences of 14 Salmonella enterica serovar Enteritidis strains recovered from human clinical cases between 1949 and 1995 in the United States. Genome Announc. 6, e01406–e01417. doi: 10.1128/genomeA.01406-17
Tagini, F., and Greub, G. (2017). Bacterial genome sequencing in clinical microbiology: a pathogen-oriented review. Eur. J. Clin. Microbiol. Infect. Dis. 36, 2007–2020. doi: 10.1007/s10096-017-3024-6
Tatsuno, I., Horie, M., Abe, H., Miki, T., Makino, K., Shinagawa, H., et al. (2001). toxB gene on pO157 of enterohemorrhagic Escherichia coli O157:H7 is required for full epithelial cell adherence phenotype. Infect. Immun. 69, 6660–6669. doi: 10.1128/IAI.69.11.6660-6669.2001
Tatusova, T., DiCuccio, M., Badretdin, A., Chetvernin, V., Nawrocki, E. P., Zaslavsky, L., et al. (2016). NCBI prokaryotic genome annotation pipeline. Nucleic Acids Res. 44, 6614–6624. doi: 10.1093/nar/gkw569
Toft, C., and Andersson, S. G. E. (2010). Evolutionary microbial genomics: insights into bacterial host adaptation. Nat. Rev. Genet. 11, 465–475. doi: 10.1038/nrg2798
Toro, M., Rump, L. V., Cao, G., Meng, J., Brown, E. W., and Gonzalez-Escalona, N. (2015). Simultaneous presence of insertion sequence excision enhancer and insertion sequence IS629 correlates with increased diversity and virulence in Shiga toxin-producing Escherichia coli. J. Clin. Microbiol. 53, 3466–3473. doi: 10.1128/jcm.01349-15
Treangen, T. J., Ondov, B. D., Koren, S., and Phillippy, A. M. (2014). The harvest suite for rapid core-genome alignment and visualization of thousands of intraspecific microbial genomes. Genome Biol. 15:524. doi: 10.1186/s13059-014-0524-x
Tyson, S., Peterson, C.-L., Olson, A., Tyler, S., Knox, N., Griffiths, E., et al. (2019). Eleven high-quality reference genome sequences and 360 draft assemblies of Shiga toxin-producing Escherichia coli isolates from human, food, animal, and environmental sources in Canada. Microbiol. Resour. Announc. 8, e00625–e00619. doi: 10.1128/MRA.00625-19
Walker, B. J., Abeel, T., Shea, T., Priest, M., Abouelliel, A., Sakthikumar, S., et al. (2014). Pilon: an integrated tool for comprehensive microbial variant detection and genome assembly improvement. PloS One 9:e112963. doi: 10.1371/journal.pone.0112963
Weinert, L. A., and Welch, J. J. (2017). Why might bacterial pathogens have small genomes? Trends Ecol. Evolution 32, 936–947. doi: 10.1016/j.tree.2017.09.006
Whitworth, J. H., Fegan, N., Keller, J., Gobius, K. S., Bono, J. L., Call, D. R., et al. (2008). International comparison of clinical, bovine, and environmental Escherichia coli O157 isolates on the basis of Shiga toxin-encoding bacteriophage insertion site genotypes. Appl. Environ. Microbiol. 74, 7447–7450. doi: 10.1128/AEM.01190-08
Whitworth, J., Zhang, Y., Bono, J., Pleydell, E., French, N., and Besser, T. (2010). Diverse genetic markers concordantly identify bovine origin Escherichia coli O157 genotypes underrepresented in human disease. Appl. Environ. Microbiol. 76, 361–365. doi: 10.1128/AEM.01761-09
Wick, L. M., Qi, W., Lacher, D. W., and Whittam, T. S. (2005). Evolution of genomic content in the stepwise emergence of Escherichia coli O157:H7. J. Bacteriol. 187, 1783–1791. doi: 10.1128/JB.187.5.1783-1791.2005
Yokoyama, E., Hashimoto, R., Etoh, Y., Ichihara, S., Horikawa, K., and Uchimura, M. (2011). Biased distribution of IS629 among strains in different lineages of enterohemorrhagic Escherichia coli serovar O157. Infect. Genet. Evol. 11, 78–82. doi: 10.1016/j.meegid.2010.10.007
Yokoyama, E., Hirai, S., Hashimoto, R., and Uchimura, M. (2012). Clade analysis of enterohemorrhagic Escherichia coli serotype O157:H7/H-strains and hierarchy of their phylogenetic relationships. Infect. Genet. Evol. J. Mol. Epidemiol. Evol. Genet. Infect. Dis. 12, 1724–1728. doi: 10.1016/j.meegid.2012.07.003
Keywords: whole genome sequencing (WGS), comparative genomics, bacterial genomics, STEC, Plasmids, Bacteriophage, Translocated Intimin Receptor
Citation: Weinroth MD, Clawson ML, Harhay GP, Eppinger M, Harhay DM, Smith TPL and Bono JL (2023) Escherichia coli O157:H7 tir 255 T > A allele strains differ in chromosomal and plasmid composition. Front. Microbiol. 14:1303387. doi: 10.3389/fmicb.2023.1303387
Edited by:
Ben Pascoe, University of Oxford, United KingdomReviewed by:
Shaimaa Mouftah, Zewail City of Science and Technology, EgyptYujie Hu, China National Center for Food Safety Risk Assessment, China
Copyright © 2023 Weinroth, Clawson, Harhay, Eppinger, Harhay, Smith and Bono. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: James L. Bono, amltLmJvbm9AdXNkYS5nb3Y=
 Margaret D. Weinroth
Margaret D. Weinroth Michael L. Clawson
Michael L. Clawson Gregory P. Harhay1
Gregory P. Harhay1 Mark Eppinger
Mark Eppinger Dayna M. Harhay
Dayna M. Harhay Timothy P. L. Smith
Timothy P. L. Smith James L. Bono
James L. Bono