- 1National Engineering Laboratory for Applied Technology of Forestry & Ecology in South China, Central South University of Forestry and Technology, Changsha, Hunan, China
- 2Hunan Provincial Key Laboratory of Forestry Biotechnology, Central South University of Forestry and Technology, Changsha, Hunan, China
- 3International Cooperation Base of Science and Technology Innovation on Forest Resource Biotechnology, Central South University of Forestry and Technology, Changsha, Hunan, China
- 4College of Arts and Sciences, Governors State University, University Park, IL, United States
- 5School of Life Sciences, Huizhou University, Huizhou, Guangdong, China
Nitrogen (N) as an essential macronutrient affects the soil nutrient cycle, microbial community abundance, and metabolic function. However, the specific responses of microorganisms and metabolic functions in rhizosphere soil of Phellodendron chinense Schneid seedlings to N addition remain unclear. In this study, four treatments (CK, N5, N10 and N15) were conducted, and the soil physicochemical properties, enzyme activities, microbial community abundances and diversities, metabolism, and gene expressions were investigated in rhizosphere soil of P. chinense Schneid. The results showed that N addition significantly decreased rhizosphere soil pH, among which the effect of N10 treatment was better. N10 treatment significantly increased the contents of available phosphorus (AP), available potassium (AK), ammonium nitrogen (NH4+-N), nitrate nitrogen (NO3−-N) and sucrase (SU) activity, as well as fungal diversity and the relative expression abundances of amoA and phoD genes in rhizosphere soil, but observably decreased the total phosphorus (TP) content, urease (UR) activity and bacterial diversity, among which the pH, soil organic matter (SOM), AP, NH4+-N and NO3−-N were the main environmental factors for affecting rhizosphere soil microbial community structure based on RDA and correlation analyses. Meanwhile, N10 treatment notably enhanced the absolute abundances of the uracil, guanine, indole, prostaglandin F2α and γ-glutamylalanine, while reduced the contents of D-phenylalanine and phenylacetylglycine in rhizosphere soil of P. chinense Schneid seedlings. Furthermore, the soil available nutrients represented a significant correlation with soil metabolites and dominant microorganisms, suggesting that N10 addition effectively regulated microbial community abundance and metabolic functions by enhancing nutrient cycle in the rhizosphere soil of P. chinense Schneid seedlings.
1 Introduction
The continuous increase in nitrogen (N) deposition resulting from various human activities, such as industry, agriculture, and livestock farming, has emerged as a significant driver of global environmental change, impacting both forest and soil ecosystems (Peng et al., 2017; He et al., 2023; Huang et al., 2023). Within the soil ecosystem, N input affects the soil physicochemical property, enzyme activity and the relative abundance of gene expression (Cardinale et al., 2020; Chen et al., 2021; Liu et al., 2022; Zhang X. et al., 2022). Previous studies have demonstrated that N addition significantly raised total nitrogen (TN) and available nitrogen (AN) levels, while reducing pH due to increase hydrogen proton concentration, resulting in soil acidification and promoting soil organic carbon (SOC) degradation in the rhizosphere soil of wheat fields and Cunninghamia lanceolate forests (Liu et al., 2022; Xu et al., 2022). Interestingly, N application shows no evident impact on the TN and total organic carbon (TOC) contents, whereas enhances the activities of urease and acid phosphatase in soil of bamboo and secondary evergreen broad-leaves forests (Tu et al., 2014; Peng et al., 2017). In addition, N addition significantly increases the abundances of soil nirK, nirS, narG and ppx genes, while distinctly decreases the soil nifH abundance in Metasequoia glyptostroboides plantations (Wang et al., 2022). However, N addition does not significantly affect the abundances of soil N and P cycling genes in the Alpine Meadow (Xiao et al., 2022). Therefore, N input plays a crucial role in regulating soil physicochemical properties, enzyme activities and gene expressions, with outcomes depending on factors such as plant types, N concentration and agrotype (Tu et al., 2014; Peng et al., 2017; Xu et al., 2022).
N input not only regulates soil physicochemical property and enzyme activity, but also exerts a profound influence on the proliferation, diversity, community composition, relative abundance and metabolic function of soil microorganisms (Ma et al., 2021; Si et al., 2022; Zhang X. et al., 2022). N addition has been reported to reduce the soil bacterial diversity, with significant correlation with physicochemical properties, and it also leads to increase relative abundances of Proteobacteria and Actinobacteria (Dai et al., 2018; Zhang X. et al., 2022). In addition, N application significantly enhances the fungal diversity and relative abundances of certain fungi, showing significant correlations with soil available nutrients, but it does not appear to impact fungal community composition in tropical/subtropical forests, suggesting that N input primarily influences microbial diversity and relative abundance by modulating soil physicochemical properties (He et al., 2021, 2022; Zhang X. et al., 2022). At the same time, N input influences the contents of proteins, amino acids, nucleic acids, and hormones containing N elements, all of which play pivotal roles in various metabolic processes in soil. These findings underscore the capacity of N addition to affect the metabolic functions of microorganisms in the rhizosphere soil of plants (Cheng et al., 2022).
The P. chinense Schneid is an important traditional medicinal plant which mainly distributed in the elevation range of 800–2000 meters in subtropical evergreen broad-leaved forests of China (He et al., 2020; Zhang et al., 2023). The stem and root barks of this plant are rich in various bioactive compounds such as berberine and phellodendrine, which widely uses for the clinic therapies of cancer, hepatitis, pneumonia, diabete and arthritis (Sun et al., 2019; Rauf et al., 2021; Nematollahi et al., 2022). We have demonstrated that N addition increased the chlorophyll content, photosynthetic efficiency, N metabolic capacity, berberine content and relative expression level of TETRAHYDROPROTOBERBERINE OXIDASE gene, implying that N addition promoted the growth and berberine synthesis of P. chinense Schneid by improving the N metabolic capacity. However, the effects of N addition on physicochemical property, enzyme activity, gene expression, microbial community composition, microbial relative abundance and metabolic function in rhizosphere soil of P. chinense Schneid seedlings are still unclear. Therefore, this study aimed to investigate the response mechanism of microorganisms and metabolic functions to N addition in rhizosphere soil of P. chinense Schneid seedlings.
2 Materials and methods
2.1 Study site
The experiment was conducted at the campus practical field of the Central South University of Forestry and Technology, Changsha City, Hunan Province, China (112.9902°E, 28.1319°N). It is a subtropical monsoon humid climate area with the following average annual values: temperature is 17.2°C, precipitation is 1361.6 mm, frost-free period is 275 days, and sunshine is 1529.3 h. We collected the 10 cm deep soil of experimental field using a completely randomized method before the experiment. The soil samples were sieved through a mesh (2 mm) and used to determine the basic physicochemical properties after drying at room temperature. The experimental soil is laterite which basic physicochemical properties are listed as follows: the pH value is 6.35, the SOM content is 24.28 g kg−1, the TN content is 1.58 g kg−1, and the TP content is 0.68 g kg−1 in the plough layer (0–20 cm).
2.2 Experimental design
The seeds of P. chinense Schneid were collected from a forest farm in Xiangxi, Hunan province, China. The experiment was performed in April 2020. After germination and cultivation, healthy and vital P. chinense Schneid seedlings (plant height ≥ 20 cm) were transplanted to the experimental field, with planting row spacing of 1.0 m × 1.0 m. The seedlings were continuously cultivated in the field and managed by manual weeding and watering. After two-months cultivation, the N (urea, Sinopharm, Shanghai, CHN) addition treatment was performed when the seedlings reached about 40 cm height.
In this study, four N concentration treatments were selected for P. chinense Schneid seedlings which included: 0 g m−2 (CK), 5 g m−2 (N5), 10 g m−2 (N10) and 15 g m−2 (N15). Each treatment contained six seedlings and the experiment were repeated triplicates. N fertilizers were applied around the seedlings in a circular and uniform manner. The experimental fields were subjected to regular management practices, including manual weeding and watering. After the N application treatment 60 days, we performed the destructive sampling of each group. We took out the roots of plant after loosening the soil, shook off the soil tightly combined with the root and brushed them from the root as rhizosphere soil samples. The soil samples were collected from the rhizosphere soils corresponding to each N application treatment and were sieved through a mesh (2 mm) before testing. The treated soil samples were divided into three portions: the first part was used to determine the soil physicochemical properties after drying at room temperature, the second portion was used for analyzing enzyme activities and the third part was saved at −80°C for high throughput sequencing of microorganisms and ultra-high performance liquid chromatography-tandem mass-spectrometry analysis of metabolism.
2.3 Soil physicochemical property measurement
The determination of soil pH was using a pH meter at soil-to-water ratio of 1:2.5 (Zhao et al., 2022). The SOM content was determined by potassium dichromate (Sun et al., 2020). The determination of TN was using semi-micro Kjeldahl method (Ma et al., 2021). The determinations of ammonium N (NH4+-N) and nitrate N (NO3−-N) contents were using the continuous flow analyzer (SAN++, Skalar Analytical B.V, Breda, NED) (Wang X. et al., 2021). The determinations of total phosphorus and available phosphorus were using the molybdenum antimony colorimetric method (Ma et al., 2021). The determination of available potassium (AK) was using an inductively coupled plasma spectrometer (ICP-AES, Thermo Fisher, CA, USA) (Zhao et al., 2022).
2.4 Rhizosphere soil enzyme activity measurement
The determination of soil urease (UR) was using sodium hypochlorite colorimetric method (Sun et al., 2020). The catalase (CAT) activity was determined by potassium dichromate (Li et al., 2022). The determination of sucrase (SU) was using 3, 5-dinitrosalicylic acid (DNSA) method with sucrose used as a substrate and acid phosphatase (ACP) activity was assayed by the p-nitrophenyl phosphate disodium method (PNPP) at pH 6.5 (Sun et al., 2020).
2.5 Microbial community composition and diversity analysis
The DNA from 0.50 g soil samples was extracted using PowerSoil® DNA Isolation Kit (12888-50, MOBio, CA, USA) and stored at −80°C for sequencing analysis. For microbial community and diversity analysis, the V4 + V5 region of bacterial 16S rDNA gene was amplified with primers 515F (5′-GTGCCAGCMGCCGCGGTAA-3′) and 907R (5′-CCGTCAATTCMTTTRAGTTT-3′). The internal transcribed spacer (ITS) region of fungal 18S rDNA gene was amplified with the primers ITS1F (5′-GGAAGTAAAAGTCGTAACAAGG-3′) and ITS2R (5′-GCTGCGTTCTTCATCGATGC-3′) under the following conditions: pre-denaturation at 98°C for 5 min, denaturation at 94°C for 30 s, annealing at 52°C for 30 s, extension at 72°C for 45 s and 30 cycles. The PCR products from different treatment samples were purified and pooled, and then sequenced on the Illumina Novaseq platform (Novaseq PE250, Illumina, CA, USA) by WEHEMO (Shenzhen, Guangdong, China). The raw sequences of different samples were separated using barcodes system, forward and reverse reads of the same sequence were combined using the FLASH tool. The QIIME DATA2 plugin was used for quality filtering, denoising, splicing and chimerism removal of the original data to get the clean data (Ma et al., 2021; Liu et al., 2022). The OTUs were classified using UCLUST software at 97% similarity level and the rarefaction analysis was conducted using the originally detected OTUs. The taxonomic assignment was conducted using the Ribosomal Database Project at minimal 80% confidence estimates (Ma et al., 2021; Song et al., 2023).
2.6 Functional gene abundance analysis
Using the extracted DNA as template, the relative expression abundances of microbial functional genes (nifH, amoA-AOB, nirK and phoD) involved in soil N and P cycling were detected by quantitative real-time PCR (qRT-PCR) which performed on the ABI StepOne instrument (ABI7500, Applied Biosystems, MA, USA) (Lang et al., 2020; Kim et al., 2022; Zhang Z. et al., 2022; Li et al., 2023). All the primer sequences were listed in Supplementary Table 1. The reaction system of qRT-PCR was 20 μL and included PCR reaction mixture contained 10 μL 2 × qPCR mix (2 × ChamQ Universal SYBR, Vazyme, Jiangsu, CHN), 0.8 μL primers (10 μM), 1.5 μL template DNA, 0.4 μL ROXII and 7.3 μL sterile ddH2O. The qRT-PCR procedure was as follows: pre-denaturation at 95°C for 5 min, denaturation at 95°C for 15 s, annealing at 55°C for 30 s, extension at 72°C for 30 s, 40 cycles, extension at 72°C for 10 min. For standard curve generation, the samples were amplified with primers and ligated to the Pgem-T Easy Vector after recycling the target DNA fragments, and then transformed into DH5a competent state. The standard curves were obtained by continuous dilutions of the known copy plasmid DNA inserted with the fragment, and the amplification efficiencies ranged from 87.29 to 93.09% for different genes.
2.7 Soil metabolite analysis
We tested the changes of metabolites in rhizosphere soil of P. chinense Schneid seedlings using untargeted metabolomics (WEHEMO, Shenzhen, Guangdong, China). Soil samples were individually grounded with liquid N and the homogenates were resuspended with prechilled 80% methanol (67-56-1, Thermo Fisher, CA, USA) by well vortex. The samples were incubated on ice for 5 min and then centrifuged at 15,000× g, 4°C for 20 min (D3024R, Scilogex, CT, USA). Some of supernatant was diluted to final concentration containing 53% methanol by LC–MS grade water (7732-12-5, Merck, Darmstadt, Germany). The samples were subsequently transferred to a new Eppendorf tube and centrifuged at 15,000× g, 4°C for 20 min (D3024R, Scilogex, CT, USA). An Orbitrap Q Exactive ™ HF mass spectrometer (Thermo Fisher, CA, USA) coupled with a Vanquish UHPLC system (Thermo Fisher, CA, USA) was used for an ultra-high performance liquid chromatography-tandem mass-spectrometry (UPLC-MS/MS) analysis (Want et al., 2013). Samples were injected onto a Hypesil Goldcolumn (100 mm × 2.1 mm, 1.9 μm) (Thermo Fisher, CA, USA) using a 17-min linear gradient at a flow rate of 0.2 mL min−1. The eluents for the positive polarity mode were eluent A (0.1% FA in Water) and eluent B (Methanol). The eluents for the negative polarity mode were eluent A (5 mM ammonium acetate, pH 9.0) and eluent B (Methanol).The solvent gradient was set as follows: 2% B, 1.5 min; 2–100% B, 12.0 min; 100% B, 14.0 min; 100–2% B, 14.1 min; 2% B, 17 min. Q ExactiveTM HF mass spectrometer was operated in positive/negative polarity mode with spray voltage of 3.5 kV, capillary temperature of 320°C, sheath gas flow rate of 35 psi and aux gas flow rate of 10 L min−1, S-lens RF level of 60, Aux gas heater temperature of 350°C (Cheng et al., 2022).
The raw data were processed using the compound discoverer 3.1 (CD3.1, Thermo Fisher, CA, USA) to perform peak alignment, peak picking and quantitation for each metabolite. We used the normalized data to predict molecular formulas based on additive ions, molecular ion peaks and fragment ions. Then we matched the peaks using the mzCloud, mzVault and MassList database to obtain the accurate quantitative results. We adopted the normalization function to make the data close to normal distribution, and applied a univariate analysis (t-test) to calculate the statistical significance (p < 0.05). The DAMs were screened based on log2(FC) > 1, p < 0.05, then enriched and annotated in the KEGG database.
2.8 Statistical analysis
The data were analyzed using SPSS software (V22.0, IBM, NY, USA) with a significant difference level. The normality of data was tested using the Shapiro–Wilk test and non-normal data was transformed with log10, square root or sine to fit a normal distribution. The significant differences of soil physicochemical properties, enzyme activities, gene expression levels and soil metabolites between N addition treatments were analyzed by the methods of one-way analysis of variance (ANOVA) and Kruskal-Wallis test. A nonmetric multidimensional scaling (NMDS) ordination to illustrate the clustering of bacterial and fungal community composition variation was conducted on the Bray-Curtis distance of the phylum and order. The variation between the groups in NMDS was tested with function betadisper (), and rerified with a permutation test. The redundancy analysis (RDA) was chosen based on the first axis length in DCA analyses of bacteria and fungi (1.65 and 1.70 respectively). RDA were conducted on soil physicochemical properties with microbial diversities. Pearson correlation analysis and the T-test were performed to integrate and visualize the normalized and log-transformed datasets of sequencing and metabolomics. These graphs were conducted using R language (V3.5.1, Lucent, NJ, USA) and Origin software (2021 version, Origin Lab, MA, USA).
3 Results
3.1 Effects of N addition on soil physicochemical property and enzyme activity
Soil pH values significantly decreased by 15.75, 19.21 and 11.81%, respectively, in the N addition groups compared with CK, while they were no significant difference among N5, N10 and N15 treatment groups (Table 1). Under the N addition treatments, no significant changes were observed in the soil TN contents while the soil TP content significantly decreased in N10 treatment. In comparison with CK, the SOM content showed an evident increase under N5 treatment, but no notable difference was found in other two N treatment groups. Under the same condition, the soil AP content was evidently increased with 2.15-fold in N10 treatment group, while no significant changes were found in N5 and N15 treatments compared with CK (Table 1). The soil AK contents showed a distinct increase under N5 and N10 treatment groups, but there was no obvious change in N15 treatment group. Furthermore, the soil NH4+-N was increased with 9.5- and 3.01-fold, concurrently the soil NO3−-N was significantly increased with 1.97- and 0.86-fold in N10 and N15 treatment groups.
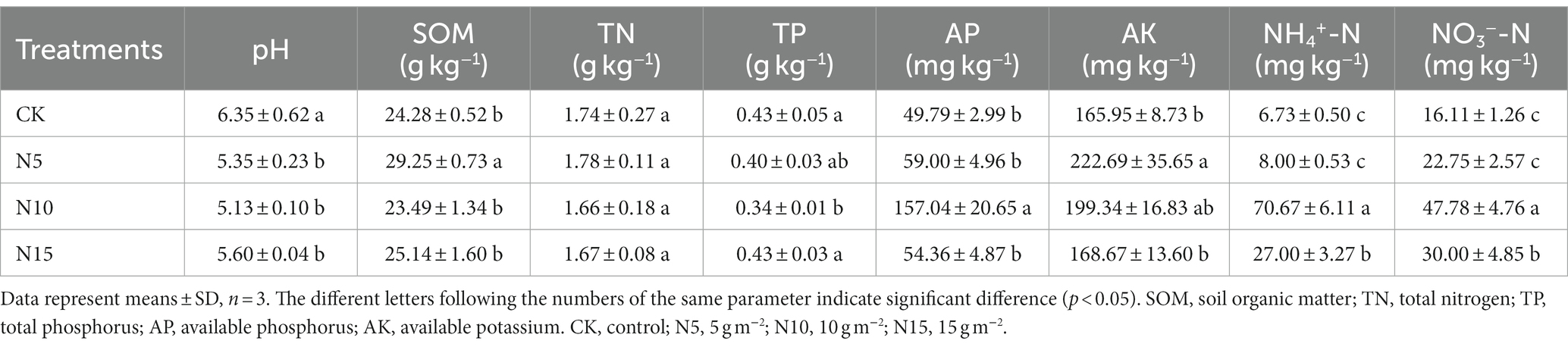
Table 1. Effects of N addition on soil physicochemical properties in rhizosphere soil of P. chinense Schneid seedlings.
The SU activities significantly increased by 59.55 and 21.15% in N5 and N10 treatment groups, but slightly decreased by 20.33% in the N15 group compared to CK (Table 2). Soil CAT activities significantly increased in N5 treatment compared to CK (p < 0.01). Conversely, soil UR activities were showed a notable reduction with 76.00, 30.00 and 60.00%, respectively, in N addition treatments.
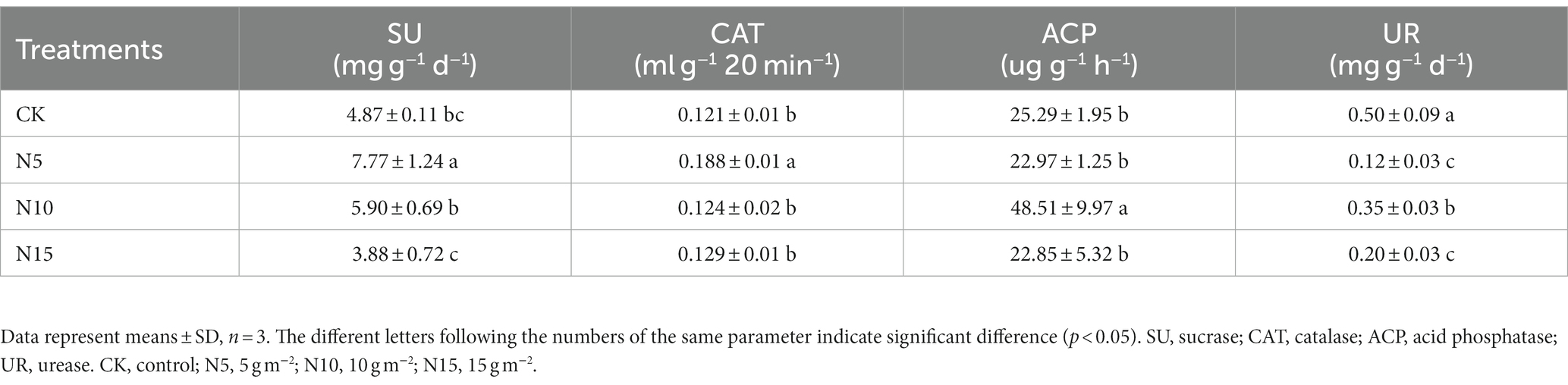
Table 2. Effects of N addition on enzyme activities in rhizosphere soil of P. chinense Schneid seedlings.
3.2 Effects of N addition on microbial diversity
The ACE index of rhizosphere soil bacteria was significantly decreased, and both the indices of Chao1 and Shannon were notably reduced under N addition treatments compared with CK (Figure 1A). Meanwhile, the Simpson index of rhizosphere soil bacteria was evidently increased in N10 treatment, while no notable differences were observed in the N5 and N15 treatment samples compared with CK (Figure 1A).
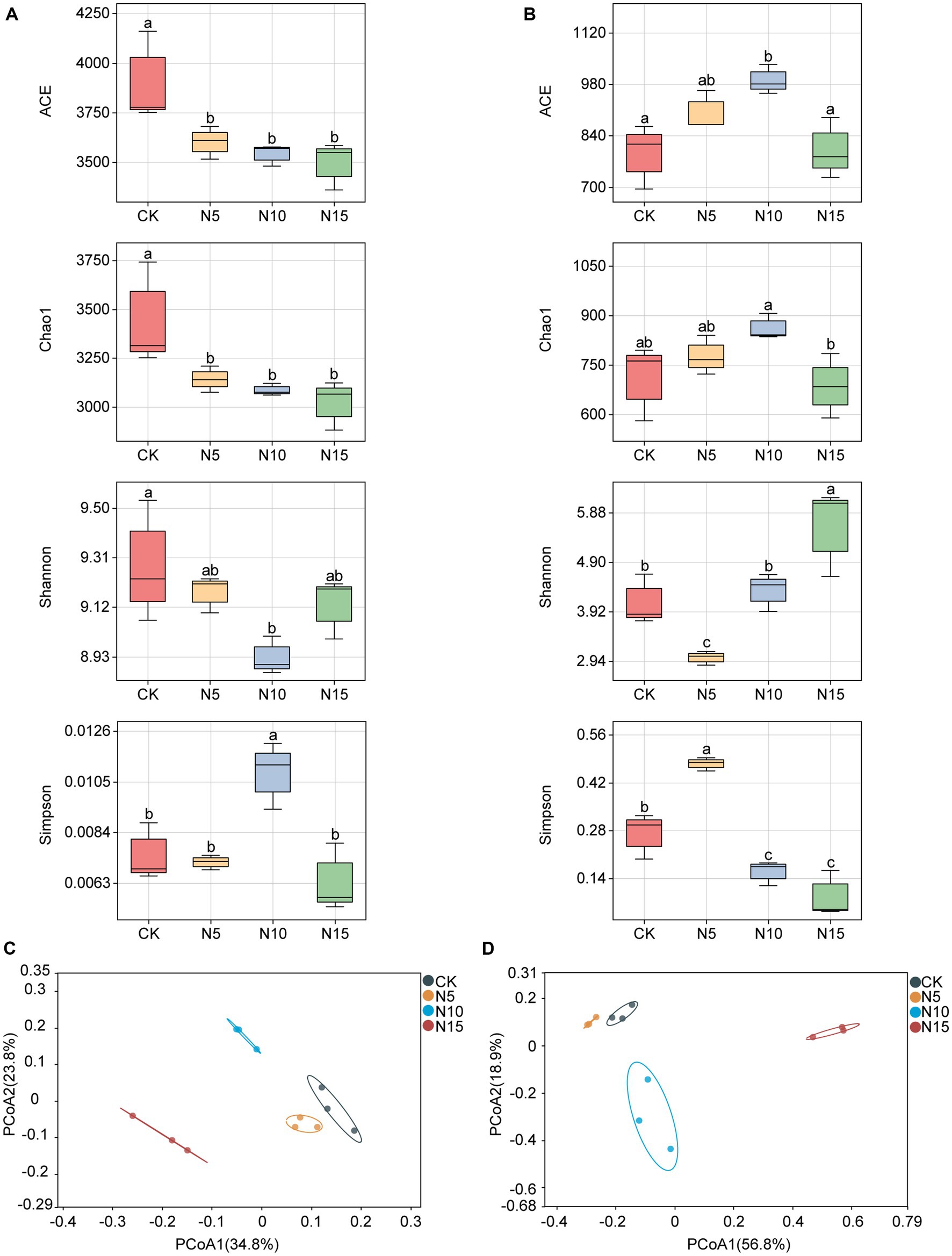
Figure 1. Effects of N addition on microbial diversity in rhizosphere soil of P. chinense Schneid seedlings. (A) Bacterial α diversity. (B) Fungal α diversity. (C) Bacterial β diversity. (D) Fungal β diversity. The different letters on the bar of the same parameter indicate significant difference (p < 0.05). CK, control; N5, 5 g m−2; N10, 10 g m−2; N15, 15 g m−2.
The ACE index of rhizosphere soil fungi exhibited a significant increase under the N5 and N10 treatments, but showed no significant difference in the N15 treatment group compared to the control (CK) (Figure 1B). The Chao1 index showed a notable increase in the N10 treatment but a reduction in the N15 treatment. At the same time, the Shannon indices increased in both the N10 and N15 treatments but decreased in the N5 treatment compared to the CK. Furthermore, the Simpson index of rhizosphere soil fungi showed a substantial increase under the N5 treatment, while it significantly decreased in both the N10 and N15 treatment groups in comparison to the CK (Figure 1B). The PCoA analysis revealed noticeable clustering of bacterial and fungal communities in the N5 and CK treatment samples, while they appeared distinctly separated with the N10 and N15 treatments (Figures 1C,D).
3.3 Effects of N addition on microbial community
Under N addition treatments, the Proteobacteria (43.13% ~ 47.11%), Acidobacteria (18.40% ~ 26.10%), Bacteroidetes (5.18% ~ 8.73%), Actinobacteria (3.32% ~ 5.45%), Chloroflexi (3.43% ~ 5.16%), Gemmatimonadetes (3.54% ~ 4.06%), Planctomycetes (3.34% ~ 4.15%), Rokubacteria (2.29% ~ 2.49%), Nitrospirae (0.98% ~ 2.18%) and Thaumarchaeota (1.10% ~ 1.37%) were the top 10 bacterial phyla in terms of the relative abundance. Notably, the Proteobacteria, Acidobacteria and Bacteroidetes emerged as the dominant bacteria phyla, collectively constituting 72.40% ~ 74.23% of the total bacterial population (Figure 2A). Comparatively, the relative abundances of Proteobacteria, Bacteroidetes and Actinobacteria increased, while the relative abundances of Acidobacteria and Chloroflexi decreased in the N application groups when compared to CK (Supplementary Table 2). At order level, the Betaproteobacteriales (13.98% ~ 15.75%), Rhizobiales (6.79% ~ 10.43%), Chitinophagales (3.60% ~ 5.87%), Xanthomonadales (3.13% ~ 4.90%), Gemmatimonadales (3.17% ~ 3.83%), Sphingomonadales (1.25% ~ 5.94%), Pyrinomonadales (2.17% ~ 3.70%), Acidobacteriales (2.32% ~ 3.04%) and Myxococcales (2.00% ~ 2.69%) were the top 9 bacterial order in terms of the relative abundance, among the Betaproteobacteriales, Rhizobiales and Chitinophagales were the predominant bacteria which accounting for 24.58 ~ 31.76% in bacterial communities under N treatments (Figure 2C).
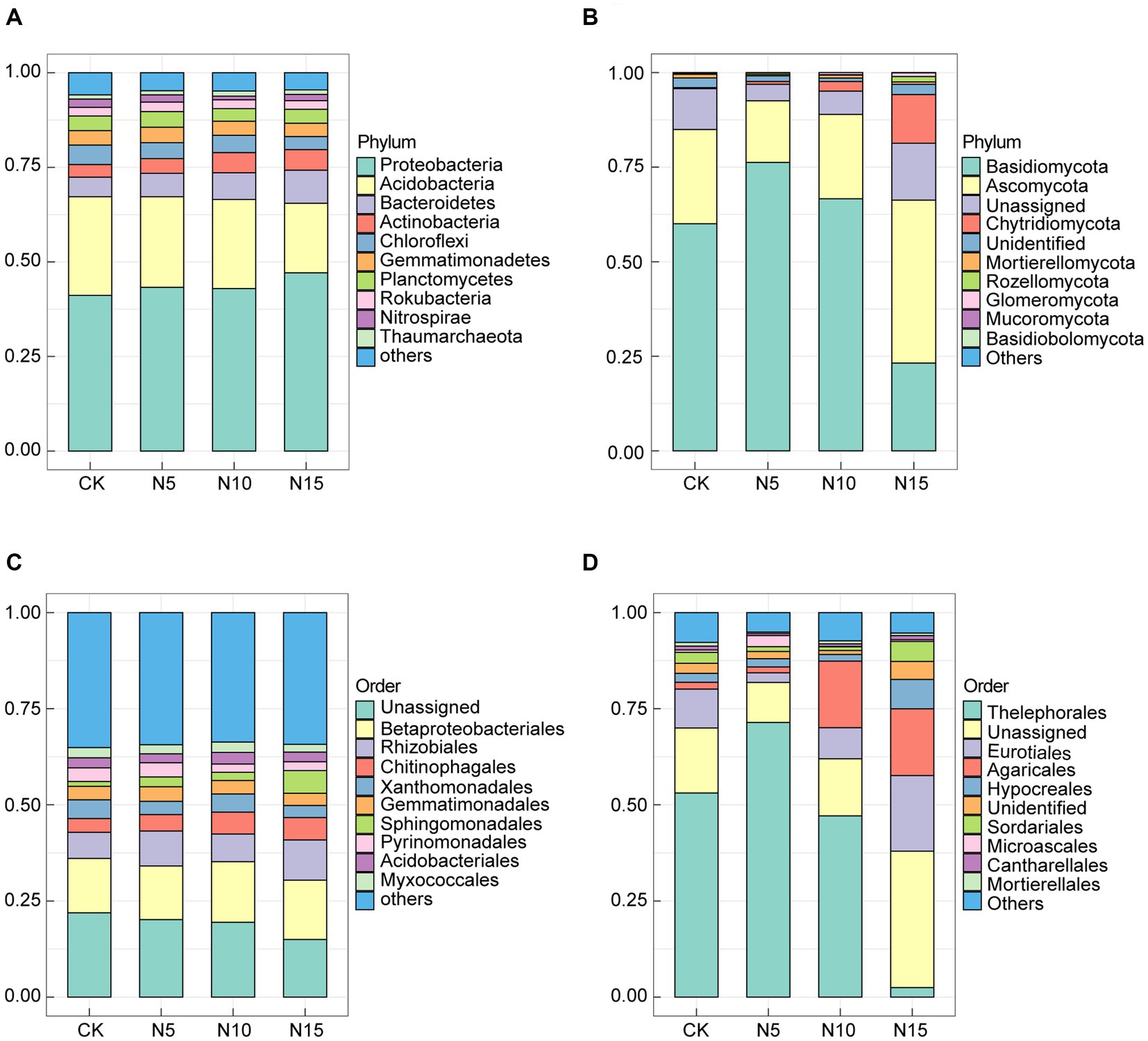
Figure 2. Effects of N addition treatments on microbial community composition in rhizosphere soil of P. chinense Schneid seedlings. (A) Bacterial phylum. (B) Fungal phylum. (C) Bacterial order. (D) Fungal order. CK, control; N5, 5 g m−2; N10, 10 g m−2; N15, 15 g m−2.
In the fungal communities, the Basidiomycota (21.80% ~ 76.65%), Ascomycota (15.98% ~ 43.42%), Unassigned (4.32% ~ 15.06%), Chytridiomycota (0.28% ~ 13.44%), Unidentified (0.90% ~ 2.70%), Mortierellomycota (0.37% ~ 1.07%), Rozellomycota (0.12% ~ 1.51%), Glomeromycota (0.08% ~ 1.04%), Mucoromycota (0.01% ~ 0.04%) and Basidiobolomycota (0.00% ~ 0.06%) were the top 10 fungi phyla in terms of the relative abundance, among the Basidiomycota, Ascomycota and Chytridiomycota emerged as dominant fungi which consisting 78.66 ~ 93.22% of total fungi (Figure 2B). Notably, the relative abundances of Basidiomycota and Chytridiomycota increased in the N10 treatment group, while the relative abundances of Ascomycota, Mortierellomycota and Rozellomycota decreased in N application groups compared to CK (Supplementary Table 3). At order level, the Thelephorales (2.42% ~ 71.58%), Unassigned (10.40% ~ 35.49%), Eurotiales (2.44% ~ 19.84%), Agaricales (1.56% ~ 17.69%), Hypocreales (1.82% ~ 7.93%), Unidentified (1.05% ~ 4.67%), Sordariales (0.94% ~ 4.98%), Microascales (0.09% ~ 2.47%), Cantharellales (0.52% ~ 1.18%) and Mortierellales (0.37% ~ 1.07%) in the rhizosphere soil were the top 10 fungi order in the light of the relative abundance, among the Thelephorales, Eurotiales and Agaricales were the primary fungi which accounting for 38.13 ~ 75.58% in fungal communities under N input treatments (Figure 2D).
3.4 Effects of N addition on gene expression abundance
Under N addition treatments, the relative expression abundances of amoA, nirK and phoD genes related to N and P metabolism in microbe significantly increased when compared with CK (Figures 3A,C,D). However, the relative expression abundances of nifH gene in microbe were no notable difference in comparison with CK (Figure 3B). The amoA gene was negatively correlated with soil UR and soil pH. The phoD gene was positively correlated with soil CAT, soil SOM and AK, but negatively correlated with soil UR (p < 0.05). Furthermore, amoA gene presented a significant positive correlation with nirK gene (p < 0.01) (Figure 3E).
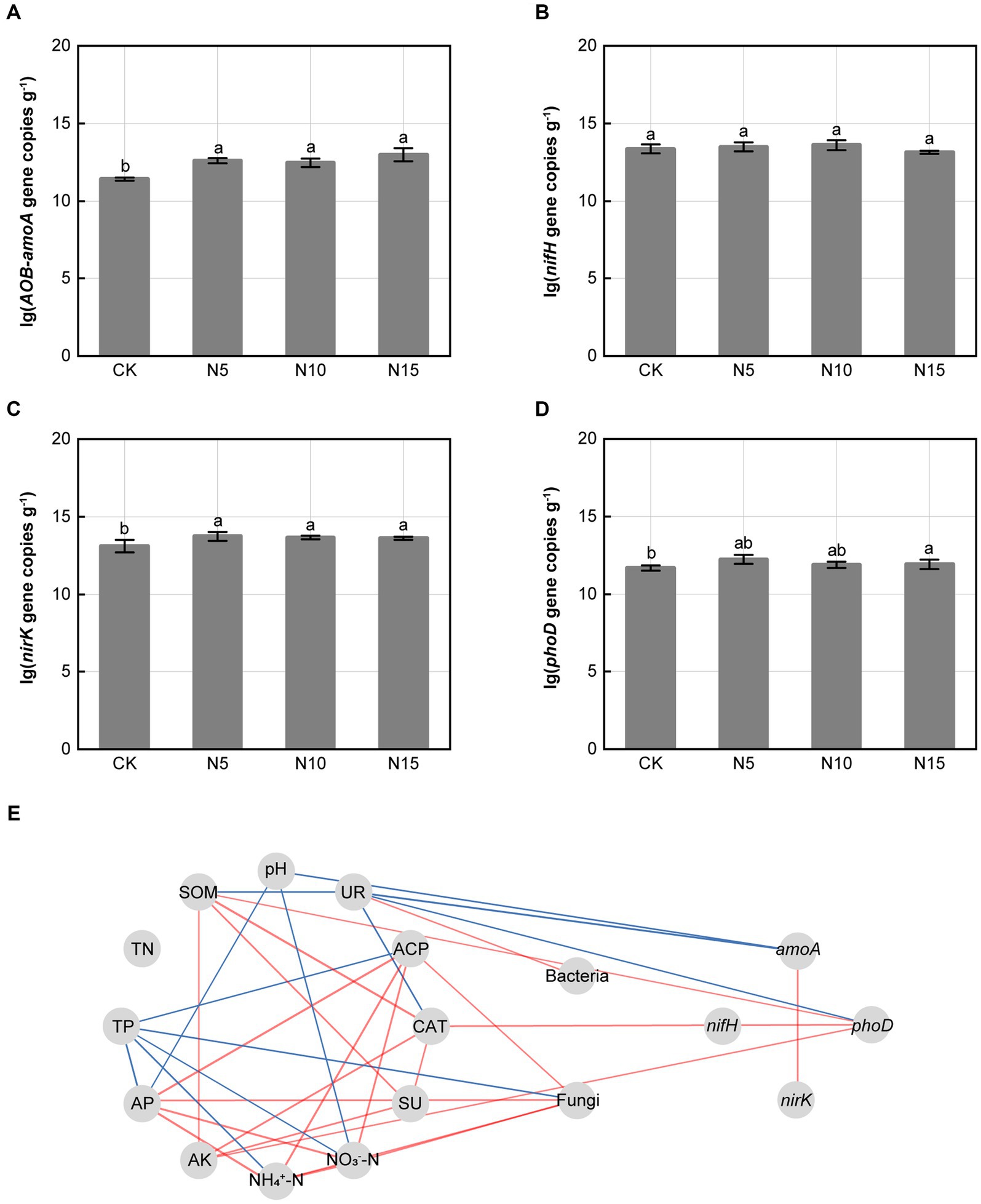
Figure 3. Effects of N addition treatments on the relative expression abundances of functional genes and correlation analysis. (A) AOB-amoA gene, (B) nifH gene, (C) nirK gene, (D) phoD gene. (E) Correlation analysis of soil physicochemical properties, enzyme activities, functional genes and microbial diversity. SOM, soil organic matter; TN, total nitrogen; TP, total phosphorus; AP, available phosphorus; AK, available potassium; SU, sucrase; CAT, catalase; ACP, acid phosphatase; UR, urease. The different letters on the bar of the same parameter indicate significant difference (p < 0.05). The lines marked with red and blue color represent the significant positive and negative correlations, respectively, (p < 0.05). CK, control; N5, 5 g m−2; N10, 10 g m−2; N15, 15 g m−2.
3.5 Correlation and redundancy analysis
The Nitrospirae was negatively correlated with soil NH4+-N, NO3−-N and AP, but positively correlated with soil pH and TN (p < 0.05). The Chloraflexi was positively correlated with soil TN (p < 0.05). The Actinobacteria exhibited a significant positive correlation with soil NH4+-N and NO3−-N (p < 0.01), but displayed a notable negative correlation with soil TN and SOM (p < 0.05). The Bacteroidetes showed a substantial positive correlation with soil NH4+-N and NO3−-N, but while exhibiting a significant negative correlation with soil SOM (p < 0.05). Furthermore, the Acidobacteria showed a obvious positive correlation with soil SOM (p < 0.01) (Figure 4A). Based on RDA analysis, the first two principal components between bacterial community and physicochemical properties explained 18.31 and 14.72%. Among these, the soil pH (r2 = 0.7916, p = 0.0030) and SOM (r2 = 0.7972, p = 0.0015) emerged as the dominant regulating factors in bacterial community composition (Figure 4C; Table 3).
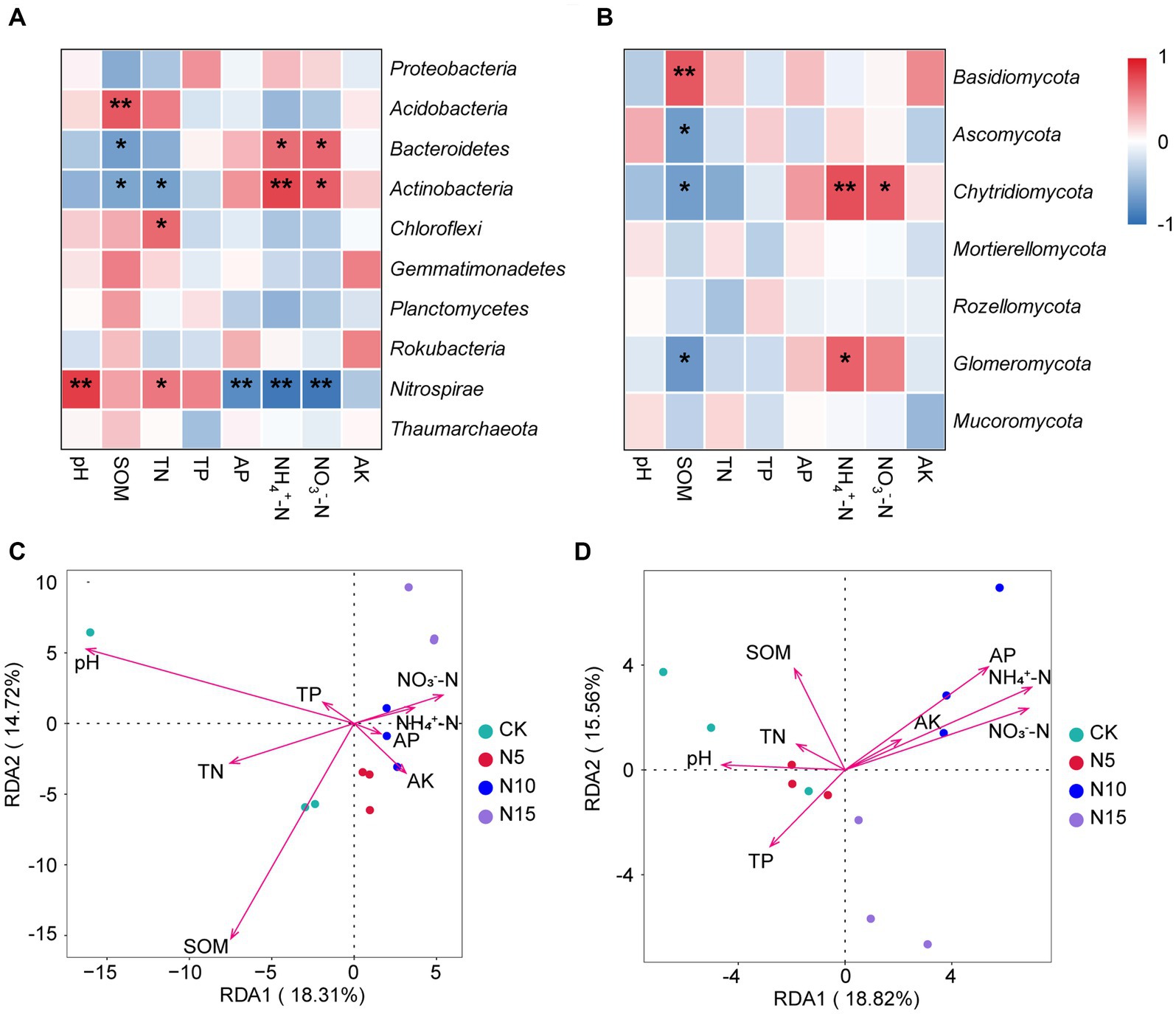
Figure 4. Correlation and RDA analyses of dominant microorganisms at phylum level and microbial communities with soil physicochemical properties under N addition treatments. (A) The heatmap of correlation between Top 10 bacteria and soil properties. (B) The heatmap of correlation between Top 7 fungi and soil properties. (C) RDA analysis between bacterial community and soil physicochemical property. (D) RDA analysis between fungal community and soil physicochemical property. SOM, soil organic matter; TN, total nitrogen; TP, total phosphorus; AP, available phosphorus; AK, available potassium. Significant levels: ** indicates p < 0.01, * indicates p < 0.05. CK, control; N5, 5 g m−2; N10, 10 g m−2; N15, 15 g m−2.
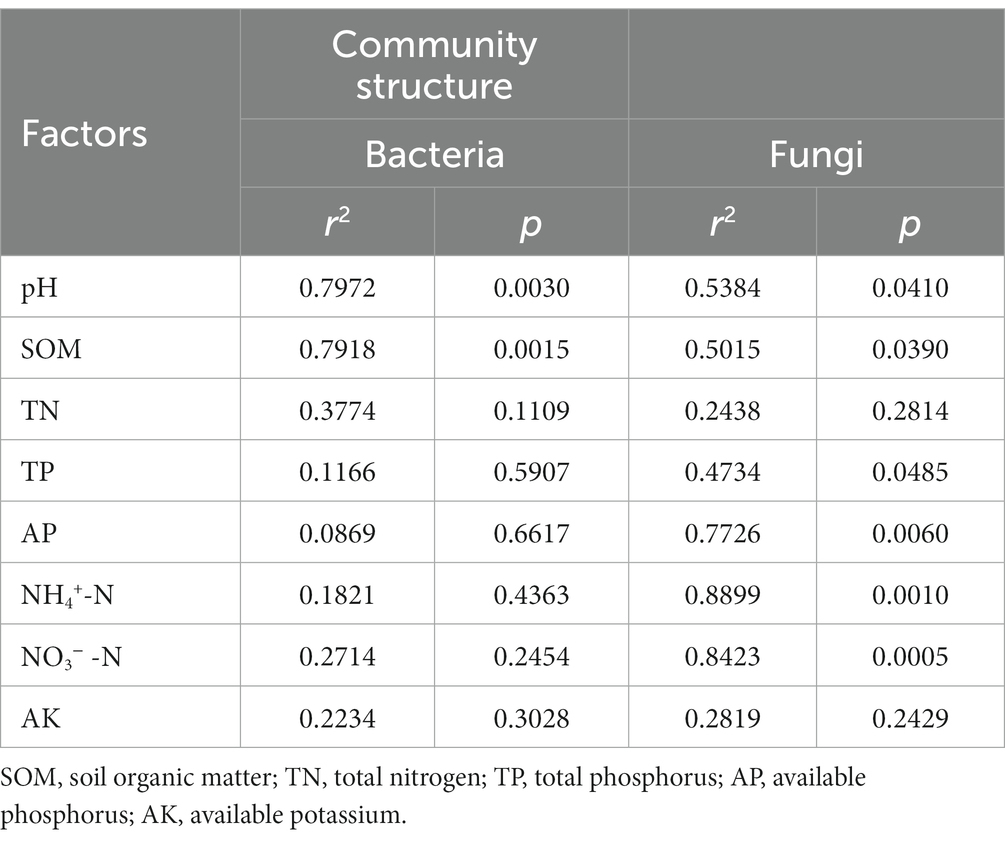
Table 3. Monte Carlo permutation test for influencing factors of microbial community structure in rhizosphere soil of P. chinense Schneid seedlings under N addition.
The Glomeromycota showed a significant positive correlation with soil NH4+-N, but presented a significant negative correlation with soil SOM (p < 0.05). The Chytridiomycota showed a substantial positive correlation with soil NH4+-N and NO3−-N, but had a notable negative correlation with soil SOM (p < 0.05). The Basidiomycete was significant positively correlated with soil SOM (p < 0.01), while the Ascomycota had a obvious negative correlation with soil SOM (p < 0.05) (Figure 4B). Additionally, The RDA analysis results showed that the first two principle components of fungal community explained 18.82 and 15.56%, among which the soil pH (r2 = 0.5384, p = 0.0410), SOM (r2 = 0.5015, p = 0.0390), TP (r2 = 0.4734, p = 0.0485) NH4+-N (r2 = 0.8899, p = 0.0010), NO3−-N (r2 = 0.8423, p = 0.0005) and AP (r2 = 0.7726, p = 0.0060) were the primary regulating factors for fungal community components (Figure 4D; Table 3).
3.6 Effects of N addition on rhizosphere soil metabolite
To further assess the influence of N addition on the microenvironment in rhizosphere soil of P. chinense Schneid seedlings, we performed a metabolomics analysis of both CK and N10 samples using the UPLC-MS/MS platform. The PLS-DA analysis showed that the metabolites were obviously separated under CK and N10 treatments, indicating that N10 treatment exerted a significant effect on rhizosphere soil metabolites (Figure 5A). The cross-validation model of PLS-DA implied that the model was reliable and suitable for the screening of DAMs (R2Y = 0.839, Q2Y = 0.106) (Figure 5B). The volcano map analysis showed that a total of 58 DAMs (31 upregulated and 27 downregulated) were identified between CK and N10 treatment samples (log2(FC) > 1, p < 0.05) (Figure 5C). Notably, seven DAMs, including D-phenylalanine, phenylacetylglycine, uracil, indole, γ-glutamylalanine, prostaglandin F2α and guanine were enriched in various metabolic pathways, such as pantothenate and CoA biosynthesis, β-alanine metabolism, steroid hormone biosynthesis, purine metabolism, arachidonic acid metabolism, pyrimidine metabolism, nucleotide metabolism, phenylalanine tyrosine and tryptophan biosynthesis, glutathione metabolism, tryptophan metabolism and phenylalanine metabolism, as identified in the KEGG database (Figure 5D). Furthermore, under the N10 treatment, the absolute abundance of D-phenylalanine and phenylacetylglycine significantly decreased, while the absolute abundance of uracil, indole, γ-glutamylalanine, prostaglandin F2α and guanine distinctly increased (Supplementary Figure 1).
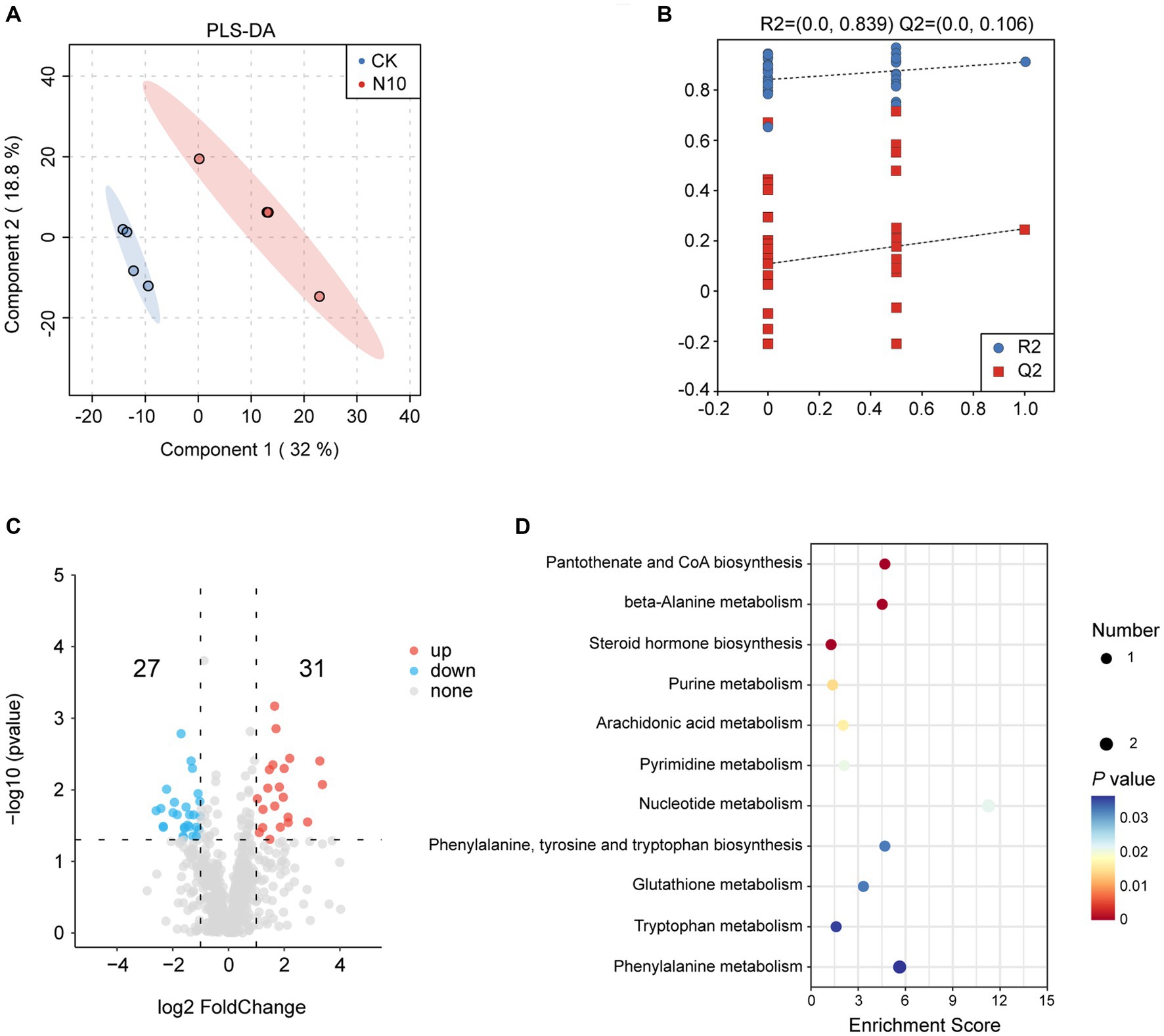
Figure 5. Effects of N addition on metabolites in rhizosphere soil of P. chinense Schneid seedlings. (A) PLS-DA analysis of metabolism between CK and N10. (B) Cross-validation model of PLS-DA. (C) The expression volcano map of DAMs. (D) The enrichment analysis of 7 DAMs in KEGG database. CK, control; N5, 5 g m−2; N10, 10 g m−2; N15, 15 g m−2.
The correlation analysis revealed significant associations between DAMs and dominant microorganisms, which the D-phenylalanine presented a significant positive correlation with Nitrospirae but a notable negative correlation with Actinobacteria and Glomeromycota (p < 0.05). Phenylacetylglycine showed a distinct positive correlation with Chloroflexi and Nitrospirae, while displaying a notable negative correlation with Actinobacteria, Chytridiomycota and Glomeromycota (p < 0.05). Uracil showed a significant positive correlation with Bacteroidetes, Actinobacteria and Chytridiomycota, while having an obvious negative correlation with Nitrospirae (p < 0.05). Indole exhibited a significant positive correlation with Bacteroidetes, Actinobacteria, Chytridiomycota and Glomeromycota, and a notable negative correlation with Nitrospirae (p < 0.05). It was also found that γ-glutamylalanine demonstrated a substantial positive correlation with Bacteroidetes and Chytridiomycota, while showing an obvious negative correlation with Nitrospirae (p < 0.05). Prostaglandin F2α displayed a significant positive correlation with Basidiomycota (p < 0.05). Guanine showed a notable positive correlation with Bacteroidetes, Actinobacteria and Chytridiomycota, while presenting a significant negative correlation with Nitrospirae (p < 0.05) (Figure 6).
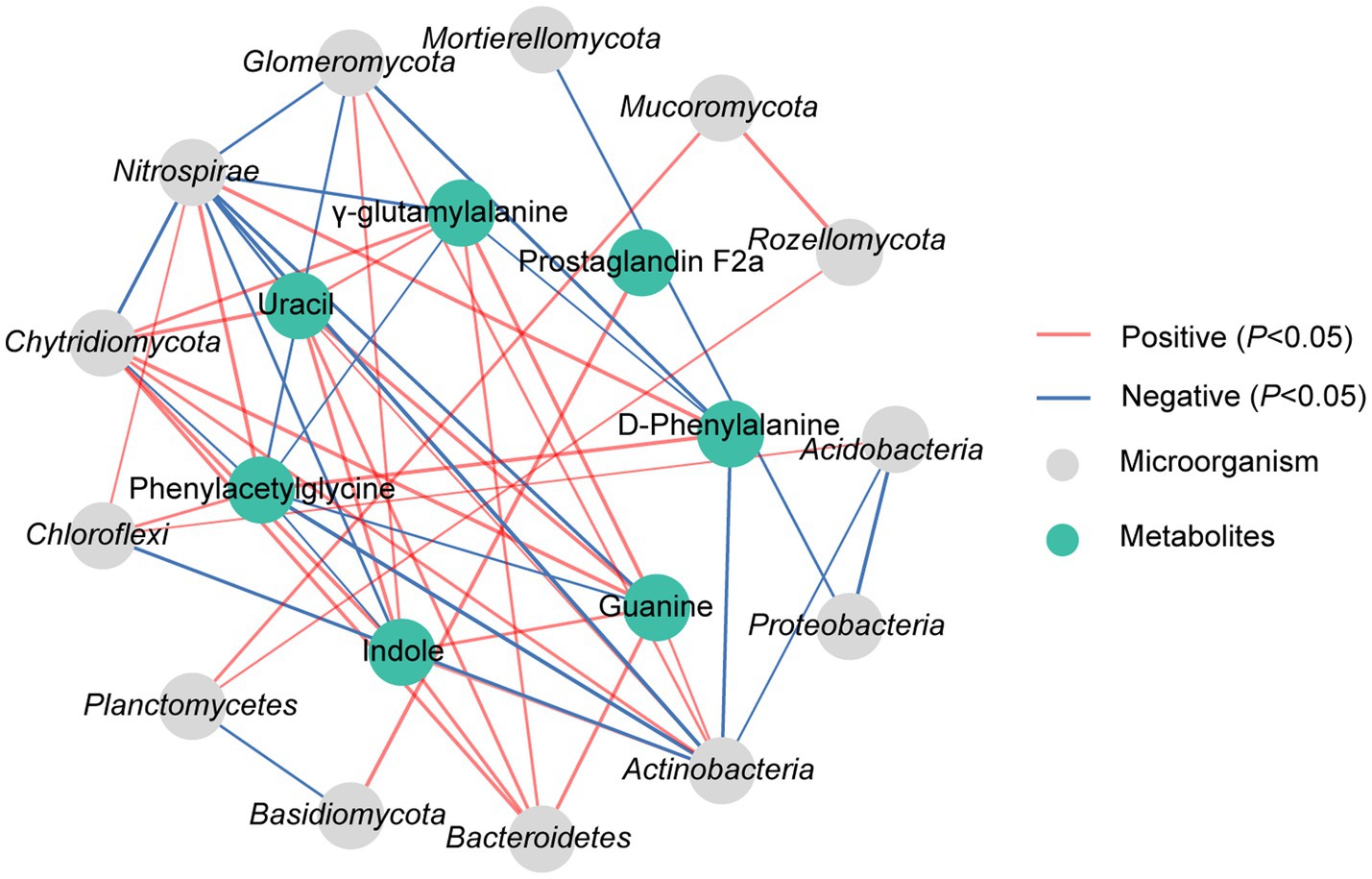
Figure 6. Correlation analysis of dominant microorganisms at phylum with DAMs in rhizosphere soil under N10 treatment. The lines marked with red and blue colors represent the significant positive and negative correlations, respectively, (p < 0.05). Only the indicators that have a significant correlation are shown in the figure.
The correlation analysis revealed significant associations between DAMs and soil physicochemical properties, which the D-phenylalanine presented a significant positive correlation with soil pH, TP and UR, but a notable negative correlation with AP, NH4+-N, NO3−-N, AK and ACP (p < 0.05). Phenylacetylglycine showed a significant positive correlation with soil pH, while having an obvious negative correlation with AP, NH4+-N, NO3−-N, AK and ACP (p < 0.05). The uracil exhibited a significant positive correlation with soil AP and SU, and an evident negative correlation with UR (p < 0.05). It was also found that indole demonstrated a substantial positive correlation with soil AP, NH4+-N, SU and ACP (p < 0.05). The γ-glutamylalanine displayed a significant positive correlation with AP, NH4+-N and NO3−-N (p < 0.05). Guanine showed a notable positive correlation with AP, NH4+-N, NO3−-N and SU (p < 0.05) (Supplementary Figure 2).
4 Discussion
4.1 N addition altered the rhizosphere soil physicochemical property and enzyme activity
N is a crucial element in soil, with substantial effects on physicochemical properties, enzyme activities, and the relative expression abundances of genes related to nutrient element metabolism (Cardinale et al., 2020; Chen et al., 2021; Zhang H. et al., 2022). Previous studies showed that N addition significantly decreased the soil pH value and SOC content, but increased the contents of soil NO3−-N, TN and available N in soil of wheat and Cunninghamia lanceolata forests, suggesting that N input regulated rhizosphere soil physicochemical properties by promoting soil acidification (Liu et al., 2022; Xu et al., 2022). In our present study, N addition led to a noticeable decrease in soil pH while increasing soil NH4+-N and NO3−-N levels, suggesting that N addition also promoted soil acidification and the accumulation of available N in the rhizosphere soil of P. chinense Schneid seedlings. The increase in soil NH4+-N and NO3−-N contents could be attributed to the upregulation of relative expression abundances of amoA and nirK genes, which are involved in nitrification processes. Additionally, our results indicated a significant positive correlation between soil available N and fungal diversity (Supplementary Figure 2), suggesting that the presence of available N supports fungal proliferation. Concurrently, the protons are generated in nitrification processes or secretes from the root following NH4+-N uptake, contributing to soil acidification (Hao et al., 2020). These findings collectively demonstrated that N addition promoted soil acidification and the accumulation of available N in the rhizosphere soil of P. chinense Schneid seedlings.
N input has no effect on the contents of the TN and TOC, but it enhanced the activities of urease and acid phosphatase in soil of bamboo forests (Tu et al., 2014). In our study, the soil TN contents did not exhibit significant changes, but the UR activities were notably reduced after N application. In the early stage after N input, soil N fertilizer was rapidly degraded into available N and were subsequently utilized by both roots and microorganisms. This dynamic process contributed to the maintenance of stable TN levels in the soil. The UR activities were reduced due to the decrease of catalytic substrate during the late period of N application (Xu et al., 2022). Under N10 treatment, the soil TP content was evidently reduced, whereas significantly increased the AP and AK contents, ACP activity and the relative expression abundances of amoA and phoD genes, indicating that appropriate N application could promote the transformation and accumulation of AP in soil through improving ACP activity and gene expression abundance, combining with available N to promote root development of P. chinense Schneid seedlings (data not shown). Furthermore, N input regulated the activities of SU and CAT, implying that they were involved in decomposition of the SOM and hydrogen peroxide in soil of the seedlings.
4.2 N addition regulated microbial diversity and community composition
N input influences the diversity and community composition of microorganism in rhizosphere soils of plants (Cardinale et al., 2020; Liu et al., 2021; Ma et al., 2021; Si et al., 2022). Under long-term N addition, the soil bacterial diversity decreased in agro-ecosystems, which was positively related to both soil pH and SOC, but increased the relative abundances of Proteobacteria and Actinobacteria (Dai et al., 2018). In this study, it was found that N addition significantly reduced the indices of Chao1 and Shannon, and produced obvious separation effect of bacterial communities in rhizosphere soil under N10 and N15 treatments compared with CK and N5 experimental groups, indicating that N input reduced bacterial diversity in rhizosphere soil of P. chinense Schneid seedlings. Under the same condition, the bacterial relative abundance in the rhizosphere soil were changed. At the bacterial phyla level, the Proteobacteria, Acidobacteria and Bacteroidetes were dominant bacteria phyla. The N addition directly and indirectly increased the soil nutrient content, and it could be benefit to the proliferation of eutrophic bacteria, which likely contributed to the increased relative abundance of eutrophic bacteria (Proteobacteria, Acidobacteria and Bacteroidetes) and made them the main bacteria phyla (Chen et al., 2023). There were noticeable increases in the relative abundances of Proteobacteria, Bacteroidetes and Actinobacteria with increasing N addition. Conversely, there were decreases in the relative abundances of Acidobacteria and Chloroflexi, suggesting that N addition could promote the eutrophic rather than oligotrophic and shift bacterial communities to more eutrophic taxa (Liu et al., 2020, 2023). The Bacteroidetes and Actinobacteria showed a significant negative position with SOM, suggesting that N addition could promote the carbon utilization efficiency by increasing the relative abundance of eutrophic bacteria in rhizosphere soil of P. chinense Schneid seedlings (Liu et al., 2020). Importantly, despite these shifts in relative abundances, the overall composition of the bacterial community remained relatively stable under the N addition treatments, suggesting that N input had the effect of the relative abundance of specific bacterial phyla while little influencing the bacterial community in rhizosphere soil of P. chinense Schneid seedlings. Recent studies have shown that nutrients availability was a predictor of bacteria diversity in tropical forest ecosystem, implying that N input directly or indirectly regulated the bacterial diversity and the relative abundance of main bacterial phyla by modulating physicochemical properties (Cui et al., 2021). In the present experiment, N application enhanced the soil available N content, suggesting that N addition decreased bacterial diversity through improving nutrient availability in rhizosphere soil of P. chinense Schneid seedlings. Meanwhile, N input reduced soil pH and regulated SOM content, which had significant correlation with the relative abundances of main bacterial phyla, implying that N addition indirectly affected the relative abundances of domain bacterial phyla by regulating physicochemical properties in rhizosphere soil of P. chinense Schneid seedlings (Du et al., 2019; Li et al., 2019). Additionally, the berberine synthesized in root were secreted into soil, which might inhibit the growth and proliferation of key bacterial phyla in soil, and then reduced its bacterial diversity and relative abundance.
In the subtropical forests, the Shannon and Chao1 indices of fungus in rhizosphere soil were significantly increased, and also enhanced the relative abundances of Mortierellomycota and Rozellomycota under appropriate N input, which had distinctly correlation with the soil NH4+-N and NO3−-N, indicating that N input elevated the diversity and relative abundance of soil fungi in subtropical forests by regulating soil physicochemical properties (Wang J. et al., 2021). In the present study, N input increased the fungal ACE index, and appropriate N addition distinctly elevated the fungal indices of Chao1, Shannon and Simpson, which presented a significant positive correlation with soil AP, NH4+-N and NO3−-N (p < 0.05) (Supplementary Figure 2), suggesting that appropriate N input might significantly enhances the fungal diversity through promoting the N metabolism in rhizosphere soil of P. chinense Schneid seedlings. Our results were in agreement with the findings of previous studies in the subtropical forests (He et al., 2021; Wang J. et al., 2021). Under the same condition, the fungal community compositions were not obviously changed, but the relative abundance of Basidiomycota and Chytridiomycota were enhanced under the N10 application, which the Basidiomycota was eutrophic taxa and Chytridiomycota showed a significant correlation with soil NH4+-N and NO3−-N (p < 0.05), implying that N input elevated the relative abundances of Basidiomycota and Chytridiomycota by promoting the N metabolism in soil, because the soil available N provides available nutrients for the growths and proliferations of Basidiomycota and Chytridiomycota (Digby et al., 2010; Liu et al., 2020). At the same time, N addition reduced the relative abundance of Mortierellomycota, but enhanced the relative abundances of other domain fungal phyla in appropriate N treatment groups, suggesting that different fungal groups adopt various mechanisms to response N fertilizer applications. These results were consistent with the results of RDA analysis.
4.3 N addition regulated soil metabolism
Soil metabolites are mainly derived from microbial metabolism, which change reveals the microbial response to soil nutrients, and they are affected by N addition (Cheng et al., 2022). In this study, a total of 7 DAMs were identified in rhizosphere soil under N10 treatment, they were primarily involved in the metabolism pathways of nucleotide, phenylalanine and hormone. N10 application significantly increased the absolute abundances of guanine and uracil that an important components of nucleotides, which had a significant positive correlation with soil NH4+-N, NO3−-N, AP and SU (Supplementary Figure 2), suggesting that N10 addition promoted nucleotide synthesis by improving the capacity of soil N metabolism in rhizosphere soil of P. chinense Schneid seedlings (Tang et al., 2022). At the same time, guanine and uracil presented a significant positive correlation with Bacteroidetes, Actinobacteria and Chytridiomycota, proposing that N10 addition promoted nucleic acid synthesis by improving the relative abundances of these three fungi.
Indeed, the γ-glutamylalanine is the precursor of L-glutamic acid that related to the N response and urea cycle (Wang et al., 2020; Lian et al., 2021; Navarro-León et al., 2022; Wang et al., 2023). N10 application significantly increased the absolute abundance of γ-glutamylalanine in rhizosphere soil which showed a significant positive correlation with soil NH4+-N, NO3−-N and amoA gene, suggesting that N10 addition promoted the transformation and utilization of N nutrient through improving the capacity of amino acid metabolism in rhizosphere soil of P. chinense Shneid seedlings (Wang et al., 2020; Cheng et al., 2022; Wang et al., 2023). Meanwhile, the γ-glutamylalanine exhibited a significant positive correlation with Bacteroidetes and Chytridiomycota, suggesting that N10 addition improved the γ-glutamylalanine synthesis by promoting microbial proliferation. Furthermore, N10 input evidently enhanced the absolute abundance of indole and prostaglandin F2α. The former is synthesized by Proteobacteria and is the precursor of auxin, the latter regulates the lipid metabolism and Ca2+ signal pathway, these results are consistent with the relative abundance of dominant bacterial phyla and fungal phyla in rhizosphere soil of P. chinense Schneid seedlings.
Under the same condition, N10 input evidently reduced the absolute abundance of D-phenylalanine, and had a significant positive correlation with Nitrospirae, suggesting that N10 addition decreased the D-phenylalanine content through inhibited the Nitrospirae proliferation in rhizosphere soil. The soil phenylalanine is absorbed and utilized by plant root for synthesizing lignin and flavonoids, and promotes root growth and secondary metabolism, while leads to reduce its content, subsequently decreases the nitration and loss of N element in soil (Li et al., 2017; Fu et al., 2023). Furthermore, N10 application decreased the absolute abundance of soil phenylacetylglycine which showed a significant correlation with Nitrospirae, Chloroflexi, Actinobacteria, Chytridiomycota and Glomeromycota, proposing that N10 addition reduced the phenylacetylglycine content by inhibiting some microbial proliferation and metabolism in rhizosphere soil of P. chinense Schneid seedlings. The phenylacetylglycine is used to synthesize antibiotics, while promotes or represses other microbial growth, ultimately improves the efficiency of N utilization of P. chinense Schneid seedlings (Fu et al., 2023).
Based on our results, a conceptual model was developed for describing the response mechanism of microorganisms and metabolic function to N addition in rhizosphere soil of P. chinense Schneid seedlings. The appropriate N addition (N10) decreased the soil pH, TP and UR activity, concurrently enhanced the available nutrient content, SU activity and relative expression abundances of amoA, nirK and phoD genes, subsequently elevated the fungal diversity and the relative abundances of Bacteroidetes, Actinobacteria and Chytridiomycota, simultaneously reduced the bacterial diversity and the relative abundances of Nitrospirae, Chloroflexi and Glomeromycota, ultimately increased the absolute contents of uracil, indole, γ-glutamylalanine, prostaglandin F2α and guanine while reduced the levels of D-phenylalanine and phenylacetylglycine, suggesting that N10 addition regulated the microbial community abundance and metabolic function through improving nutrient cycle in rhizosphere soil of P. chinense Schneid seedlings (Figure 7).
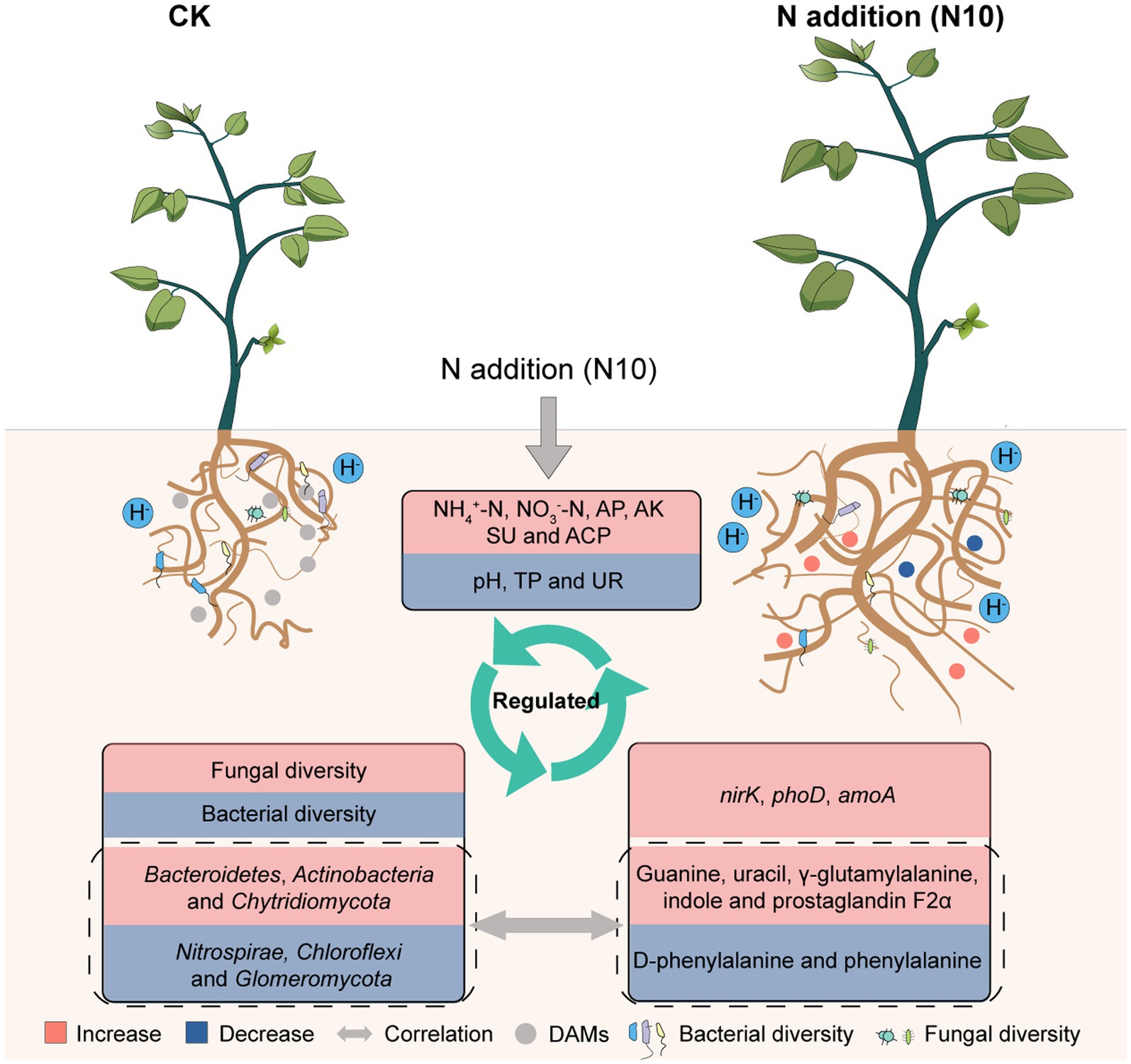
Figure 7. Theoretical mechanism of microorganisms and metabolic function reponse to N addition in rhizosphere soil of P. chinense Schneid seedlings. TP, total phosphorus; AP, available phosphorus; AK, available potassium; ACP, acid phosphatase; UR, urease; CK, control; N5, 5 g m−2; N10, 10 g m−2; N15, 15 g m−2.
5 Conclusion
In summary, N addition significantly increased the contents of AP, AK, NH4+-N, NO3−-N, SU activity, and the relative expression abundances of amoA and phoD genes, concurrently decreased soil pH, TP content and UR activity, demonstrating that N addition promoted acidification and nutrient cycle in rhizosphere. Meanwhile, N addition enhanced the fungal diversity and the relative abundances of Bacteroidetes, Actinobacteria and Chytridiomycota, reduced the bacterial diversity and the relative abundances of Nitrospirae, Chloroflexi and Glomeromycota, which exhibited a significant correlation with soil NH4+-N, NO3−-N, AP and UR, indicating that N addition regulated the microbial diversity and community abundance through modulating the physicochemical properties. Additionally, N10 addition distinctly improved the absolute abundances of the uracil, guanine, indole, prostaglandin F2α and γ-glutamylalanine, whereas reduced the absolute contents of D-phenylalanine and phenylacetylglycine, which showed a significant correlation with soil NH4+-N, NO3−-N, AP, Bacteroidetes, Chytridiomycota, Actinobacteria and Nitrospirae, suggesting that N10 addition impacted the metabolic function through regulating nutrient cycle and microbial community abundance. Therefore, our results indicates that N10 addition regulates microbial community abundance and metabolic function by enhancing nutrient cycling capacity in the rhizosphere soil of P. chinense Schneid seedlings.
Data availability statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found at: NCBI – PRJNA1035961.
Author contributions
YG: Writing – original draft, Data curation, Formal analysis, Investigation, Software, Visualization. XLC: Data curation, Formal analysis, Writing – original draft. YS: Writing – review & editing. XYC: Supervision, Validation, Writing – review & editing. GH: Conceptualization, Funding acquisition, Writing – review & editing. XH: Funding acquisition, Writing – review & editing. GW: Funding acquisition, Writing – review & editing. HH: Conceptualization, Funding acquisition, Investigation, Methodology, Writing – review & editing. ZL: Writing – review & editing, Conceptualization, Investigation, Methodology, Supervision.
Funding
The author(s) declare financial support was received for the research, authorship, and/or publication of this article. This research was financially supported by the National Natural Science Foundation of China (grant no. 32071752), by the Department of Science and Technology of Hunan Province (grant nos. 2020NK2018, 2022NK2018, and 2021RC2083).
Conflict of interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2023.1302775/full#supplementary-material
References
Cardinale, M., Ratering, S., Sadeghi, A., Pokhrel, S., Honermeier, B., and Schnell, S. (2020). The response of the soil microbiota to long-term mineral and organic nitrogen fertilization is stronger in the bulk soil than in the rhizosphere. Genes (Basel) 11:456. doi: 10.3390/genes11040456
Chen, M. Q., Xu, J. S., Li, Z. Q., Li, D. D., Wang, Q. X., Zhou, Y. P., et al. (2023). Long-term nitrogen fertilization-induced enhancements of acid hydrolyzable nitrogen are mainly regulated by the most vital microbial taxa of keystone species and enzyme activities. Sci. Total Environ. 874:162463. doi: 10.1016/j.scitotenv.2023.162463
Chen, Q., Yuan, Y., Hu, Y., Wang, J., Si, G., Xu, R., et al. (2021). Excessive nitrogen addition accelerates N assimilation and P utilization by enhancing organic carbon decomposition in a Tibetan alpine steppe. Sci. Total Environ. 764:142848. doi: 10.1016/j.scitotenv.2020.142848
Cheng, H., Yuan, M., Tang, L., Shen, Y., Yu, Q., and Li, S. (2022). Integrated microbiology and metabolomics analysis reveal responses of soil microorganisms and metabolic functions to phosphorus fertilizer on semiarid farm. Sci. Total Environ. 817:152878. doi: 10.1016/j.scitotenv.2021.152878
Cui, J., Yuan, X., Zhang, Q., Zhou, J., Lin, K., Xu, J., et al. (2021). Nutrient availability is a dominant predictor of soil bacterial and fungal community composition after nitrogen addition in subtropical acidic forests. PLoS One 16:e0246263. doi: 10.1371/journal.pone.0246263
Dai, Z., Su, W., Chen, H., Barberán, A., Zhao, H., Yu, M., et al. (2018). Long-term nitrogen fertilization decreases bacterial diversity and favors the growth of actinobacteria and proteobacteria in agro-ecosystems across the globe. Glob. Chang. Biol. 24, 3452–3461. doi: 10.1111/gcb.14163
Digby, A. L., Gleason, F. H., and McGee, P. A. (2010). Some fungi in the Chytridiomycota can assimilate both inorganic and organic sources of nitrogen. Fungal Ecol. 3, 261–266. doi: 10.1016/j.funeco.2009.11.002
Du, Y., Wang, T., Wang, C., Anane, P. S., Liu, S., and Paz-Ferreiro, J. (2019). Nitrogen fertilizer is a key factor affecting the soil chemical and microbial communities in a Mollisol. Can. J. Microbiol. 65, 510–521. doi: 10.1139/cjm-2018-0683
Fu, Y., Liu, T., Wang, X., Wang, Y., Gong, Q., Li, G., et al. (2023). Untargeted metabolomics reveal rhizosphere metabolites mechanisms on continuous ramie cropping. Front. Plant Sci. 14:1217956. doi: 10.3389/fpls.2023.1217956
Hao, T., Zhu, Q., Zeng, M., Shen, J., Shi, X., Liu, X., et al. (2020). Impacts of nitrogen fertilizer type and application rate on soil acidification rate under a wheat-maize double cropping system. J. Environ. Manag. 270:110888. doi: 10.1016/j.jenvman.2020.110888
He, D., Guo, Z. M., Shen, W. J., Ren, L. J., Sun, D., Yao, Q., et al. (2022). Fungal communities are more sensitive to the simulated environmental changes than bacterial communities in a subtropical forest: the single and interactive effects of nitrogen addition and precipitation seasonality change. Microb. Ecol. 86, 521–535. doi: 10.1007/s00248-022-02092-8
He, J., Jiao, S., Tan, X., Wei, H., Ma, X., Nie, Y., et al. (2021). Adaptation of soil fungal community structure and assembly to long- versus short-term nitrogen addition in a tropical forest. Front. Microbiol. 12:689674. doi: 10.3389/fmicb.2021.689674
He, H., Qin, J., Ma, Z., Sun, W., Yan, W., He, G., et al. (2020). Highly efficient regeneration and medicinal component determination of Phellodendron chinense Schneid. In Vitro Cell. Dev. Biol. Plant 56, 775–783. doi: 10.1007/s11627-020-10080-1
He, J., Tan, X., Nie, Y., Ma, L., Liu, J., Lu, X., et al. (2023). Distinct responses of abundant and rare soil bacteria to nitrogen addition in tropical forest soils. Microbiol. Spectr. 11:e0300322. doi: 10.1128/spectrum.03003-22
Huang, C., Fu, S., Ma, X., Ma, X., Ren, X., Tian, X., et al. (2023). Long-term intensive management reduced the soil quality of a Carya dabieshanensis forest. Sci. Rep. 13:5058. doi: 10.1038/s41598-023-32237-9
Kim, N., Riggins, C. W., Zabaloy, M. C., Rodriguez-Zas, S. L., and Villamil, M. B. (2022). Limited impacts of cover cropping on soil N-cycling microbial communities of long-term corn monocultures. Front. Microbiol. 13:926592. doi: 10.3389/fmicb.2022.926592
Lang, M., Zou, W., Chen, X., Zou, C., Zhang, W., Deng, Y., et al. (2020). Soil microbial composition and phoD gene abundance are sensitive to phosphorus level in a long-term wheat-maize crop system. Front. Microbiol. 11:605955. doi: 10.3389/fmicb.2020.605955
Li, C., Jia, Z., Zhang, S., Li, T., Ma, S., Cheng, X., et al. (2023). The positive effects of mineral-solubilizing microbial inoculants on asymbiotic nitrogen fixation of abandoned mine soils are driven by keystone phylotype. Sci. Total Environ. 882:163663. doi: 10.1016/j.scitotenv.2023.163663
Li, J., Li, Y., Tian, Y., Qu, M., Zhang, W., and Gao, L. (2017). Melatonin has the potential to alleviate cinnamic acid stress in cucumber seedlings. Front. Plant Sci. 8:1193. doi: 10.3389/fpls.2017.01193
Li, Y., Nie, C., Liu, Y., Du, W., and He, P. (2019). Soil microbial community composition closely associates with specific enzyme activities and soil carbon chemistry in a long-term nitrogen fertilized grassland. Sci. Total Environ. 654, 264–274. doi: 10.1016/j.scitotenv.2018.11.031
Li, Q., Yang, J., He, G., Liu, X., and Zhang, D. (2022). Characteristics of soil C:N:P stoichiometry and enzyme activities in different grassland types in Qilian Mountain nature reserve-Tibetan Plateau. PLoS One 17:e0271399. doi: 10.1371/journal.pone.0271399
Lian, L., Lin, Y., Wei, Y., He, W., Cai, Q., Huang, W., et al. (2021). PEPC of sugarcane regulated glutathione S-transferase and altered carbon-nitrogen metabolism under different N source concentrations in Oryza sativa. BMC Plant Biol. 21:287. doi: 10.1186/s12870-021-03071-w
Liu, M., Gan, B., Li, Q., Xiao, W., and Song, X. (2022). Effects of nitrogen and phosphorus addition on soil extracellular enzyme activity and stoichiometry in chinese fir (Cunninghamia lanceolata) forests. Front. Plant Sci. 13:834184. doi: 10.3389/fpls.2022.834184
Liu, W., Jiang, L., Yang, S., Wang, Z., Tian, R., Peng, Z., et al. (2020). Critical transition of soil bacterial diversity and composition triggered by nitrogen enrichment. Ecology 101:e03053. doi: 10.1002/ecy.3053
Liu, H., Wang, R., Lü, X. T., Cai, J., Feng, X., Yang, G., et al. (2021). Effects of nitrogen addition on plant-soil micronutrients vary with nitrogen form and mowing management in a meadow steppe. Environ. Pollut. 289:117969. doi: 10.1016/j.envpol.2021.117969
Liu, M., Wei, Y., Lian, L., Wei, B., Bi, Y., Liu, N., et al. (2023). Macrofungi promote SOC decomposition and weaken sequestration by modulating soil microbial function in temperate steppe. Sci. Total Environ. 899:165556. doi: 10.1016/j.scitotenv.2023.165556
Ma, X., Song, Y., Song, C., Wang, X., Wang, N., Gao, S., et al. (2021). Effect of nitrogen addition on soil microbial functional gene abundance and community diversity in permafrost peatland. Microorganisms 9:2498. doi: 10.3390/microorganisms9122498
Navarro-León, E., López-Moreno, F. J., Borda, E., Marín, C., Sierras, N., Blasco, B., et al. (2022). Effect of l-amino acid-based biostimulants on nitrogen use efficiency (NUE) in lettuce plants. J. Sci. Food Agric. 102, 7098–7106. doi: 10.1002/jsfa.12071
Nematollahi, S., Pishdad, G. R., Zakerkish, M., Namjoyan, F., Ahmadi Angali, K., and Borazjani, F. (2022). The effect of berberine and fenugreek seed co-supplementation on inflammatory factor, lipid and glycemic profile in patients with type 2 diabetes mellitus: a double-blind controlled randomized clinical trial. Diabetol. Metab. Syndr. 14:120. doi: 10.1186/s13098-022-00888-9
Peng, Y., Chen, G., Chen, G., Li, S., Peng, T., Qiu, X., et al. (2017). Soil biochemical responses to nitrogen addition in a secondary evergreen broad-leaved forest ecosystem. Sci. Rep. 7:2783. doi: 10.1038/s41598-017-03044-w
Rauf, A., Abu-Izneid, T., Khalil, A. A., Imran, M., Shah, Z. A., Emran, T. B., et al. (2021). Berberine as a potential anticancer agent: a comprehensive review. Molecules 26:7368. doi: 10.3390/molecules26237368
Si, P., Shao, W., Yu, H., Xu, G., and Du, G. (2022). Differences in microbial communities stimulated by malic acid have the potential to improve nutrient absorption and fruit quality of grapes. Front. Microbiol. 13:850807. doi: 10.3389/fmicb.2022.850807
Song, B., Li, Y., Yang, L., Shi, H., Li, L., Bai, W., et al. (2023). Soil acidification under long-term N addition decreases the diversity of soil bacteria and fungi and changes their community composition in a semiarid grassland. Microb. Ecol. 85, 221–231. doi: 10.1007/s00248-021-01954-x
Sun, Y., Lenon, G. B., and Yang, A. W. H. (2019). Phellodendri cortex: a phytochemical, pharmacological, and pharmacokinetic review. Evid. Based Complement. Alternat. Med. 2019:7621929. doi: 10.1155/2019/7621929
Sun, J., Yang, L., Wei, J., Quan, J., and Yang, X. (2020). The responses of soil bacterial communities and enzyme activities to the edaphic properties of coal mining areas in Central China. PLoS One 15:e0231198. doi: 10.1371/journal.pone.0231198
Tang, X., He, Y., Zhang, Z., Wu, H., He, L., Jiang, J., et al. (2022). Beneficial shift of rhizosphere soil nutrients and metabolites under a sugarcane/peanut intercropping system. Front. Plant Sci. 13:1018727. doi: 10.3389/fpls.2022.1018727
Tu, L. H., Chen, G., Peng, Y., Hu, H. L., Hu, T. X., Zhang, J., et al. (2014). Soil biochemical responses to nitrogen addition in a bamboo forest. PLoS One 9:e102315. doi: 10.1371/journal.pone.0102315
Wang, X., Fang, L., Beiyuan, J., Cui, Y., Peng, Q., Zhu, S., et al. (2021). Improvement of alfalfa resistance against cd stress through rhizobia and arbuscular mycorrhiza fungi co-inoculation in cd-contaminated soil. Environ. Pollut. 277:116758. doi: 10.1016/j.envpol.2021.116758
Wang, J., Shi, X., Zheng, C., Suter, H., and Huang, Z. (2021). Different responses of soil bacterial and fungal communities to nitrogen deposition in a subtropical forest. Sci. Total Environ. 755:142449. doi: 10.1016/j.scitotenv.2020.142449
Wang, Y.-F., Wang, J.-F., Xu, Z.-M., She, S.-H., Yang, J.-Q., and Li, Q.-S. (2020). L-glutamic acid induced the colonization of high-efficiency nitrogen-fixing strain Ac63 (Azotobacter chroococcum) in roots of Amaranthus tricolor. Plant Soil 451, 357–370. doi: 10.1007/s11104-020-04531-2
Wang, G., Wang, J., Yao, L., Li, B., Ma, X., Si, E., et al. (2023). Transcriptome and metabolome reveal the molecular mechanism of barley genotypes underlying the response to low nitrogen and resupply. Int. J. Mol. Sci. 24:4706. doi: 10.3390/ijms24054706
Wang, L., Wen, Y., Tong, R., Zhang, H., Chen, H., Hu, T., et al. (2022). Understanding responses of soil microbiome to the nitrogen and phosphorus addition in metasequoia glyptostroboides plantations of different ages. Microb. Ecol. 84, 565–579. doi: 10.1007/s00248-021-01863-z
Want, E. J., Masson, P., Michopoulos, F., Wilson, I. D., Theodoridis, G., Plumb, R. S., et al. (2013). Global metabolic profiling of animal and human tissues via UPLC-MS. Nat. Protoc. 8, 17–32. doi: 10.1038/nprot.2012.135
Xiao, J., Dong, S., Shen, H., Li, S., Wessell, K., Liu, S., et al. (2022). N addition overwhelmed the effects of P addition on the soil C, N, and P cycling genes in Alpine Meadow of the Qinghai-Tibetan Plateau. Front. Plant Sci. 13:860590. doi: 10.3389/fpls.2022.860590
Xu, A., Li, L., Xie, J., Gopalakrishnan, S., Zhang, R., Luo, Z., et al. (2022). Changes in ammonia-oxidizing archaea and bacterial communities and soil nitrogen dynamics in response to long-term nitrogen fertilization. Int. J. Environ. Res. Public Health 19:2732. doi: 10.3390/ijerph19052732
Zhang, H., Phillip, F. O., Wu, L., Zhao, F., Yu, S., and Yu, K. (2022). Effects of temperature and nitrogen application on carbon and nitrogen accumulation and bacterial community composition in apple rhizosphere soil. Front. Plant Sci. 13:859395. doi: 10.3389/fpls.2022.859395
Zhang, X., Song, X., Wang, T., Huang, L., Ma, H., Wang, M., et al. (2022). The responses to long-term nitrogen addition of soil bacterial, fungal, and archaeal communities in a desert ecosystem. Front. Microbiol. 13:1015588. doi: 10.3389/fmicb.2022.1015588
Zhang, X., Sun, W., Chen, X., Chen, L., Lv, Z., He, H., et al. (2023). Integrated physiological and transcriptomic analysis reveals mechanism of leaf in Phellodendron Chinense Schneid seedlings response to drought stress. Ind. Crop. Prod. 198:116679. doi: 10.1016/j.indcrop.2023.116679
Zhang, Z., Yu, Z., Zhang, Y., and Shi, Y. (2022). Impacts of fertilization optimization on soil nitrogen cycling and wheat nitrogen utilization under water-saving irrigation. Front. Plant Sci. 13:878424. doi: 10.3389/fpls.2022.878424
Zhao, Y., Yan, C., Hu, F., Luo, Z., Zhang, S., Xiao, M., et al. (2022). Intercropping pinto peanut in litchi orchard effectively improved soil available potassium content, optimized soil bacterial community structure, and advanced bacterial community diversity. Front. Microbiol. 13:868312. doi: 10.3389/fmicb.2022.868312
Keywords: Phellodendron chinense Schneid, nitrogen, soil microorganism, soil metabolism, gene expression
Citation: Gu Y, Chen X, Shen Y, Chen X, He G, He X, Wang G, He H and Lv Z (2023) The response of nutrient cycle, microbial community abundance and metabolic function to nitrogen fertilizer in rhizosphere soil of Phellodendron chinense Schneid seedlings. Front. Microbiol. 14:1302775. doi: 10.3389/fmicb.2023.1302775
Edited by:
Amit Kumar, Nanjing University of Information Science and Technology, ChinaReviewed by:
Xiaoli Zhang, Chinese Academy of Sciences (CAS), ChinaFidelis O. Ajibade, Federal University of Technology, Nigeria
Abdulhamid Yusuf, Federal University, Dutsin-Ma, in collaboration with reviewer FA
Copyright © 2023 Gu, Chen, Shen, Chen, He, He, Wang, He and Lv. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Hanjie He, aGVqaWUyMjRAMTYzLmNvbQ==; Zhencheng Lv, c3pza3lAaHp1LmVkdS5jbg==
†These authors have contributed equally to this work and share first authorship
‡ORCID: Hanjie He, https://orcid.org/0000-0002-2771-6151
 Yuanzheng Gu1,2,3†
Yuanzheng Gu1,2,3† Gongxiu He
Gongxiu He Hanjie He
Hanjie He