- 1Department of Microbiology, School of Medicine, Iran University of Medical Sciences, Tehran, Iran
- 2Microbial Biotechnology Research Center, Iran University of Medical Sciences, Tehran, Iran
- 3Department of Medical Laboratory Sciences, Khomein University of Medical Sciences, Khomeyn, Iran
- 4Iran National Tumor Bank, Cancer Institute of Iran, Tehran University of Medical Sciences, Tehran, Iran
Background: Fusobacterium nucleatum has been recognized as an important key bacterium in the cause and spread of colorectal carcinogenesis. Nevertheless, the clinical relevance of F. nucleatum in colorectal cancer (CRC) and its effect on immune factors and the tumor microenvironment have not been fully elucidated.
Materials and methods: The frequency of F. nucleatum was measured in 100 paired tumor and normal tissue specimens by TaqMan quantification Real-Time Polymerase Chain Reaction (qPCR). The mRNA expression levels of cytokines (IL-6, IL-10, IL-12β, IL-17, TNF-α, TLR-2, and TLR-4), and miRNAs (miR-21, miR-31) were examined. Eventually, any potential correlations between the molecular and clinicopathological features of the neoplastic samples and the abundance of F. nucleatum were analyzed.
Results: The relative frequency of F. nucleatum was significantly increased in cancerous tissue compared to adjacent non-tumor tissues. Furthermore, the high level of F. nucleatum was significantly associated with histological grade III and IV CRC tissues (P = 0.027 and P = 0.022, respectively) and perineural invasion-positive patients (P = 0.037). In addition, the expression levels of IL-6, IL-17, TNF-α,IL-12β, TLR-2, and TLR-4 as well as miR-21 and miR-31 showed a significant increase in the cancer group. A notable correlation was also observed between the high status of F. nucleatum and the expression of IL-6, TNF-α and miR-21.
Conclusion: Our results emphasize the importance of F. nucleatum and changes in the expression of genes involved in CRC. Studying the microbial profile and gene expression changes in CRC patients may be a promising approach to improve screening methods and provide therapeutic strategies.
1 Introduction
Colorectal cancer (CRC) is a major global health problem, ranking second in mortality with more than 600,000 deaths and third in incidence with more than 1.8 million new cases annually arising. The International Agency for Research on Cancer (IARC) has predicted that the global number of mortality and new CRC cases will increase to 3 and 1.6 million, respectively, by 2040 (Cheng et al., 2020; Kaźmierczak-Siedlecka et al., 2020; Sung et al., 2021; Stolzer, 2022). The etiology of CRC includes both genetic and environmental factors. CRC with heritability aspects that account for only 12–35% of the risk factors includes multiple associated polyposis (MAP), hereditary non-polyposis colorectal cancer (HNPCC or Lynch syndrome), multiple associated polyposis (MAP), Peutz-Jeghers syndrome and familial adenomatous polyposis (FAP) (Dekker et al., 2019; Stolzer, 2022). Environmental factors have a significant impact on the occurrence and progression of CRC, including sedentary lifestyle and obesity, smoking, heavy alcohol consumption, diabetes mellitus, and bad eating habits such as consumption of a diet containing low fiber, high fat, and food carcinogens (Kaźmierczak-Siedlecka et al., 2020; Stolzer, 2022). Among the aforementioned environmental factors, intestinal microbiota is a crucial component in colorectal carcinogenesis.
Previous research points toward the importance of the human intestinal microbiota in influencing normal physiological activities that contribute to the maturation of the immune system and act as a barrier to pathogens (Rea et al., 2018; Dadgar-Zankbar et al., 2023). Gut microbiota dysbiosis, described as an increase in the levels of pathogens alongside a decrease in the levels of beneficial bacteria, is caused by a variety of factors including lifestyle and dietary changes, widespread use of antibiotics, chronic stress and host genetics (Chen et al., 2019). Bacterial dysbiosis is closely associated with inflammatory gastrointestinal diseases such as ulcerative colitis, inflammatory bowel disease (IBD) and CRC. They lead to carcinoma by inducing inflammatory responses, boosting inflammation, aberrant immune regulation, activation of tumorigenic pathways, production of oncogenic substances, metabolic dysregulation, and damage to host DNA (Chen et al., 2019; Wong and Yu, 2019).
Current studies have identified several specific bacterial species associated with the initiation and progression of CRC through the aforementioned mechanisms, including Fusobacterium nucleatum, Bacteroides fragilis, Salmonella enterica, and Escherichia coli (Zeller et al., 2014; Yachida et al., 2019). Interestingly, considerable evidence has shown that F. nucleatum is more abundant in colorectal tumor tissue compared to adjacent normal mucosa, suggesting its potential involvement in the pathogenesis of CRC (Allali et al., 2015; Thomas et al., 2019; Wong and Yu, 2019; Tran et al., 2022). F. nucleatum is an invasive, pro-tumorigenic, and pro-inflammatory pathogen that adheres to and invades epithelial and endothelial cells by means of several virulence factors and induces inflammatory cytokines in the mucosa surrounding the tumor. This interaction is recognized by Toll-like Receptors (TLRs) and leads to activation of both innate and adaptive immunity (Sakamoto and Maeda, 2010; Huang et al., 2014; Brennan and Garrett, 2016; Henstra et al., 2021). Chemokines and cytokines, which are examples of inflammatory mediators, are extensively produced by inflammatory cells and alter the immune system as well as a wide range of cancers. These cytokines, such as IL-6, IL-17, and TNF-α, directly promote tumorigenesis, tumor cell proliferation, angiogenesis, metastasis, and cell death (Bhat et al., 2022). It is noteworthy that inflammatory cells can also produce cytokines that restrict tumor growth, such as IL-10 and IL-12, which lead to modulating apoptosis and suppressing angiogenesis. However, some studies show their dual role as both tumor promoters and inhibitors (Kabel, 2014).
Numerous genetic factors can serve as molecular markers for CRC diagnosis and prognosis. Among these factors, the family of MicroRNAs (miRNAs) has been identified as a promising candidate (Ghafouri-Fard et al., 2021). miRNAs are small, endogenous, non-coding RNA molecules of 18–23 nucleotides. Aberrant expression patterns and functional abnormalities of miRNAs have been observed in inflammatory processes and several types of human cancer. MiRNAs can act as either tumor inhibitors or oncogenes, depending on their downstream target genes (Chi and Zhou, 2016). miR-21 and miR-31 are considered to be critical miRNAs in CRC and exhibit a statistically significant increase in expression levels in CRC patients compared to the healthy group (Eslamizadeh et al., 2018). miR-21 is a highly prominent miRNAs that is involved in cell proliferation and invasion in CRC via targeting of phosphatase and tensin homolog (PTEN) and Programmed cell death protein 4 (PDCD4) (Li et al., 2014; Shen et al., 2019). Furthermore, miR-31 has been identified as a potent cancer-related miRNA that plays a role in CRC carcinogenesis by targeting tumor suppressor genes (Cottonham et al., 2010).
However, there are few studies regarding the interaction and regulation between the presence of F. nucleatum and inflammatory genes and miRNAs in CRC. Therefore, in the current study, we investigated the amount of F. nucleatum and its relationship with the expression of inflammatory genes (TLR-2, TLR-4, TNF-α, IL-6, IL-10, IL-12β, and IL-17) and miRNAs (miR-21, and miR-31) in tumor and adjacent normal tissue of Iranian CRC patients.
2 Materials and methods
2.1 Sample preparation
A total of 100 pairs of fresh-frozen colorectal adenocarcinoma and matched adjacent non-tumor tissues were provided by the Iran National Tumor Bank, which was founded by the Cancer Institute of Tehran University of Medical Sciences, for Cancer Research, during the 2021–2023 period. After the surgical procedure, the tissue samples were expeditiously conveyed to the pathology unit for expert inspection and assessment by a pathologist. In addition, a segment of the tumor tissues as well as normal adjacent mucosal samples were selected and fixed in RNAlater stabilization buffer (QIAGEN, Hilden, Germany). Samples were preserved at −70°C for further analysis. All clinical histopathological parameters and necessary information were captured from patients’ records. Patients who had colorectal tumors of other types than adenocarcinoma or concomitant malignant tumors originating from other organs were excluded from the study. All patients were new cases and did not use any treatment methods such as antibiotics, probiotics, radiotherapy, or chemotherapy before surgery.
2.2 Acid nucleic extraction and cDNA reverse transcription
Total DNA and RNA were extracted from the frozen CRC and adjacent normal mucosal tissue specimens using a FavorPrepTM DNA Mini Kit and FavorPrep™ Tissue Total RNA Mini Kit (Favorgen, Taiwan, Cat. No: FATGK 001), respectively, according to the manufacturer’s instructions. A nanodrop instrument (WPA Biowave II Nanospectrophotometer, USA) at OD 260 and 280 nm was used to measure the concentration and purity of the extracts. A reverse transcriptase reaction was performed using the cDNA synthesis kit (Yekta Tajhiz Azma, Iran and Cat. No: YT4500). For further analysis, the DNA and synthesized cDNA were stored at −20°C.
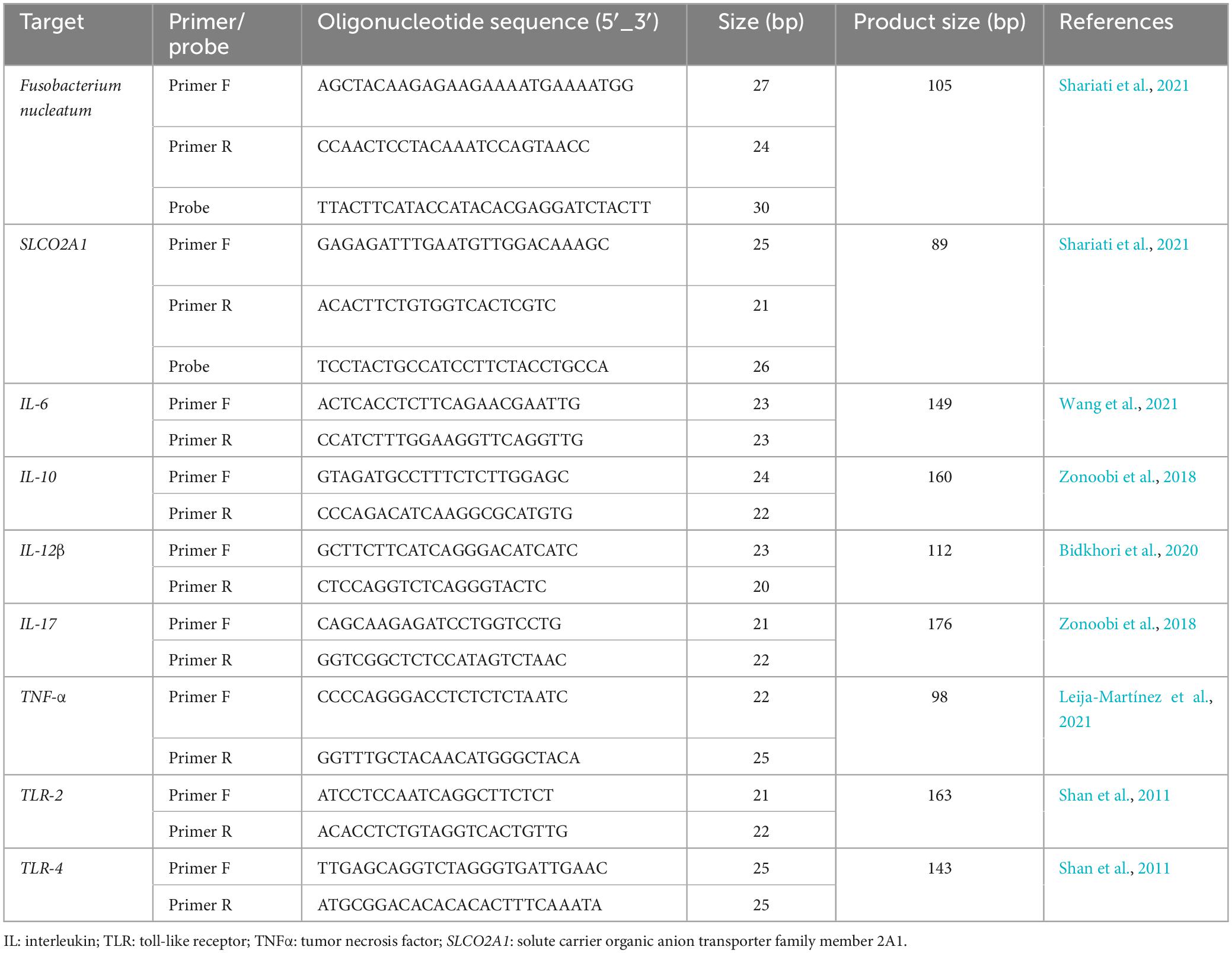
2.3 Relative quantification of F. nucleatum
The levels of F. nucleatum in both cancerous and matched normal tissues were identified by employing the 16S rDNA gene sequence through the utilization of a real-time TaqMan primer/probe on a Rotor-Gene 6000 real-time PCR cycler (Qiagen Corbett, Hilden, Germany). The gene solute carrier organic anion transporter family member 2A1 (SLCO2A1) of the human reference was used to normalize the Cycle quantification (Cq) values, as previously elucidated (Mima et al., 2016). The primer and probe sequences are indicated in Table 1, and their specificities were examined through the use of EMBL-EPI, NCBI BLAST databases, and Allele ID software (v.7.5). Each reaction mixture, having a total volume of 20 μl, contained 20 ng of genomic DNA, 0.5 μM of each primer, 0.25 μM of the probe, 9 μl of Universal Probe Ex Taq PCR Master Mix (Ampliqon, Denmark), and deionized distilled water (6 μL). The qPCR experiment was conducted as follows: an initial incubation at 95°C for 15 min, followed by 40 cycles of denaturation at 95°C for 15 s, and annealing/extension at 62°C for 30 s. Moreover, all assays for each individual sample were carried out in duplicate in a single patch, and the average outcomes of the dual qPCR investigations were utilized for statistical assessment. The negative control in all analyses consisted of all the components of the reaction mixture, excluding bacterial genomic DNA. Following the guidelines on the minimum information for publication of quantitative real-time PCR experiments (MIQE) (Bustin et al., 2009) (Supplementary material). In addition, F. nucleatum ATCC 25586 was used as a positive control. The computation of the fold change (2–ΔΔCq) in F. nucleatum abundance in tumor compared to normal tissue involved subtracting ΔCq tumor from ΔCq normal, where ΔCq represents the average Cq value difference between each F. nucleatum and the reference gene (Mima et al., 2016).
2.4 Inflammatory and anti-inflammatory genes expression
In the present study, relative quantification real-time PCR was used to assess the expression of the interleukin (IL)-6, IL-10, IL-12β, IL-17, TNF-α, TLR-2, and TLR-4 genes. All of the primers utilized in this investigation are presented in Table 1. Real-time PCR was performed using a Rotor-Gene 6000 real-time PCR cycler (Qiagen Corbett, Hilden, Germany) according to the manufacturer’s instructions. A final volume of 12.5 μL was used, consisting of 3 μL cDNA template, 0.5 μM appropriate forward and reverse primers, 5.25 μL Power SYBR Green PCR Master Mix (Bioneer, Korea) and 2.5 μL deionized distilled water. All reactions were performed in duplicate and all experiments had a no-template control. Each amplification protocol included an initial denaturation step of 12 min at 95°C, followed by 40 cycles of 15 s at 95°C, 45 s at 58–61.5°C (depending on the primer temperature), and extension at 72°C for 25 s. The SLCO2A1 was used as an internal control, and mRNA levels were quantified using the 2–ΔΔCq approach as described above.
2.5 microRNA extraction and cDNA synthesis
The FavorPrep™ miRNA Isolation Kit (Favorgen, Taiwan) was used to extract microRNA (miRNA) from frozen tissues. The integrity of the miRNAs was checked using a nanospectrophotometer. Stem-loop primers for specific reverse transcription (RT) of miRNAs and Ana microRNA detection kit (AnaCell Co, Iran) were used according to the manufacturer’s protocol. Briefly, a 20 mL RT reaction master mix was prepared with a 10 ng microRNA sample, 4 μL RT buffer (5X), 1 μL dNTP (10 mM), 0.5 μL RT enzyme (20 U/mL), and 1.5 μL stem-loop RT primers (5×). The reaction conditions were: 37°C for a duration of 60 min, 70°C for 5 min.
2.6 miRNA gene expression
Relative quantification real-time PCR was performed using 2X QPCR Master Mix SYBR Green (AnaCell, Iran) to assess the expression of miR-21, and miR-31 in accordance with the guidelines provided by the manufacturer. Cycle conditions for the aforementioned genes were as follows: initial denaturation at 95°C for 5 min following 40 cycles at 95°C for 30 s, annealing at 60°C for 30 s, and extension at 72°C for 30 s. The mRNA expression levels were analyzed using the 2–ΔΔCq method as previously described. The relative levels of miR-21 and miR-31 were compared to the geometric mean of U6 snRNA (RNU6B) expression (Schee et al., 2012).
2.7 Statistical analyses
Statistical analysis was performed with SPSS version v.26.0 software(SPSS Inc., Chicago, IL, USA) and GraphPad Prism v.8.3.0. A two-tailed P-value < 0.05 was considered statistically significant. Paired samples t-test was used to compare the relative amounts of F. nucleatum and the expression of microRNA and pro-inflammatory genes in the tumor and adjacent normal mucosa of paired samples, while the difference in copy number was analyzed using the rank sum test. Fisher’s exact test and chi-squared test (χ2) were used to assess the relationships between F. nucleatum status and clinicopathological and molecular features. Multivariate logistic regression analysis was performed to estimate odds ratios (ORs) and corresponding 95% confidence intervals (CIs) for associations between high F. nucleatum DNA levels and clinicopathological features.
3 Results
3.1 Clinicopathological characterization of CRC patients
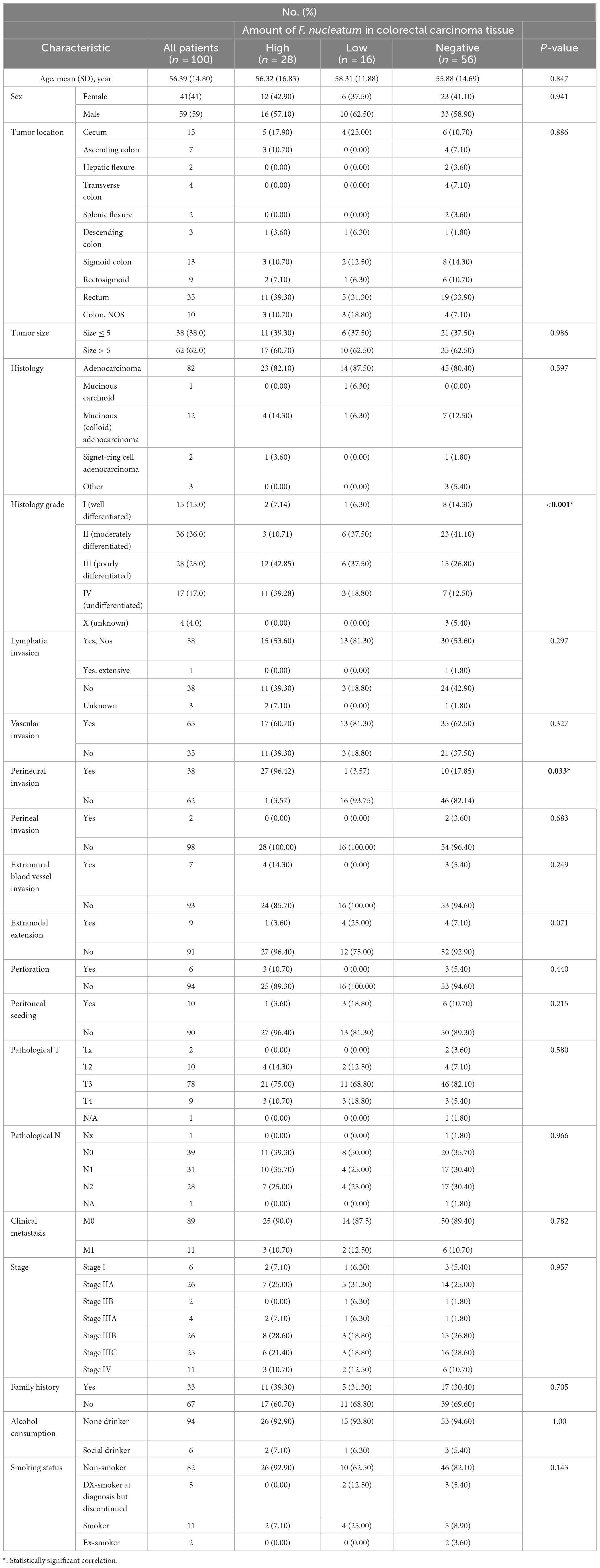
The histopathological and demographic features of the individuals are summarized in Table 2. In brief, the study consisted of a total of 59 men and 41 women with a mean age of 56.39 years (SD ± 14.80). The majority of CRC patients exhibited signs of grade-II cancer (36%), characterized as moderately differentiated. while 28%, 17%, 15%, and 4% of patients demonstrated grade III (poorly differentiated), grade IV (undifferentiated), grade I (well-differentiated), and grade X (unknown) cancers, respectively. Based on the initial examinations conducted by a gastroenterologist and the microscopic examinations performed by a pathologist, the involvement frequency of various sections of the intestine in CRC has been recorded in Table 2. Tumor location mainly involved rectum (35%), cecum (15%), and sigmoid colon (13%). Overall, 82 colorectal carcinoma patients were diagnosed with adenocarcinomas, twelve patients had mucinous (colloid) adenocarcinoma, two patients had Signet-ring Cell Adenocarcinoma and one patient had mucinous carcinoid. Notably, around 3% of CRC patients had other histology of CRC.
3.2 Fusobacterium nucleatum quantification
In this particular research, we quantitated CRC-associated F. nucleatum in both colorectal carcinoma tissue and matched normal mucosal samples through relative quantification Real-time-PCR assay. The mean abundance of F. nucleatum was significantly higher in CRC tissues in comparison to adjacent normal tissues (CRC vs. normal: 28.57 ± 22.82 vs 6.04 ± 10.45, n = 100, P = 0.004, paired t-test) (Figure 1). F. nucleatum was detected in 44% and 25% of cancer tissues and adjacent non-tumor tissue, respectively. The Receiver Operating Characteristic (ROC) curve was utilized to calculate an optimal cutoff value, based on the quantity of F. nucleatum, to categorize F. nucleatum-positive CRCs into low and high groups. Consequently, out of the 44 colorectal carcinoma cases with detectable F. nucleatum, 28 cases were split into high-level groups and 16 cases into low-level groups. None of the patients in the high-level group had previous cancer, with a mean age and tumor sample size of 56.32 years (SD ± 16.83) and 6.13 cm (SD ± 2.15), respectively. However, the statistical analyses revealed no significant correlation (p > 0.05) between F. nucleatum and the aforementioned markers.
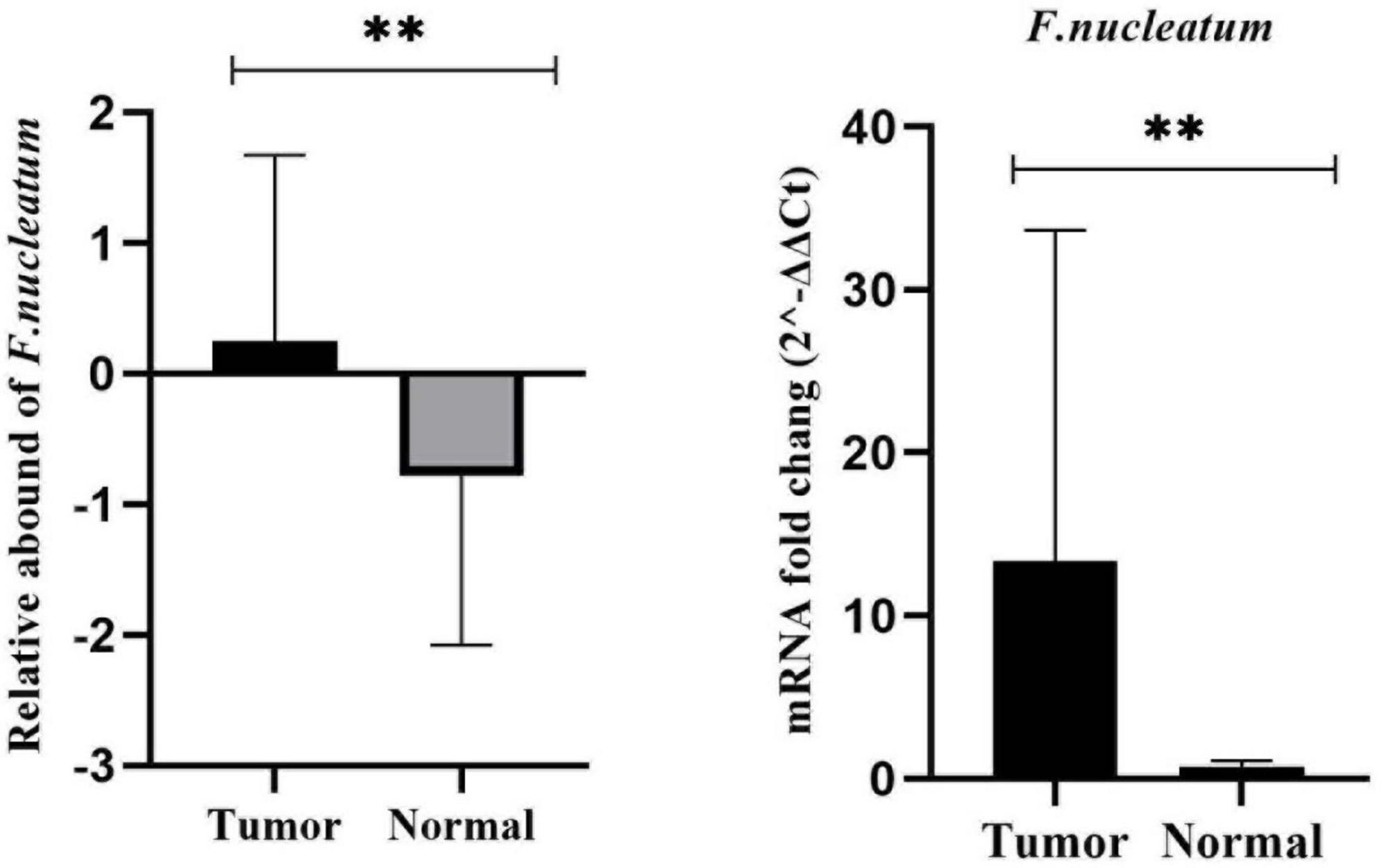
Figure 1. Comparison of the presence of F. nucleatum in cancerous and matched normal tissues (P < 0.004**).
3.3 Inflammatory and anti-inflammatory gene expression
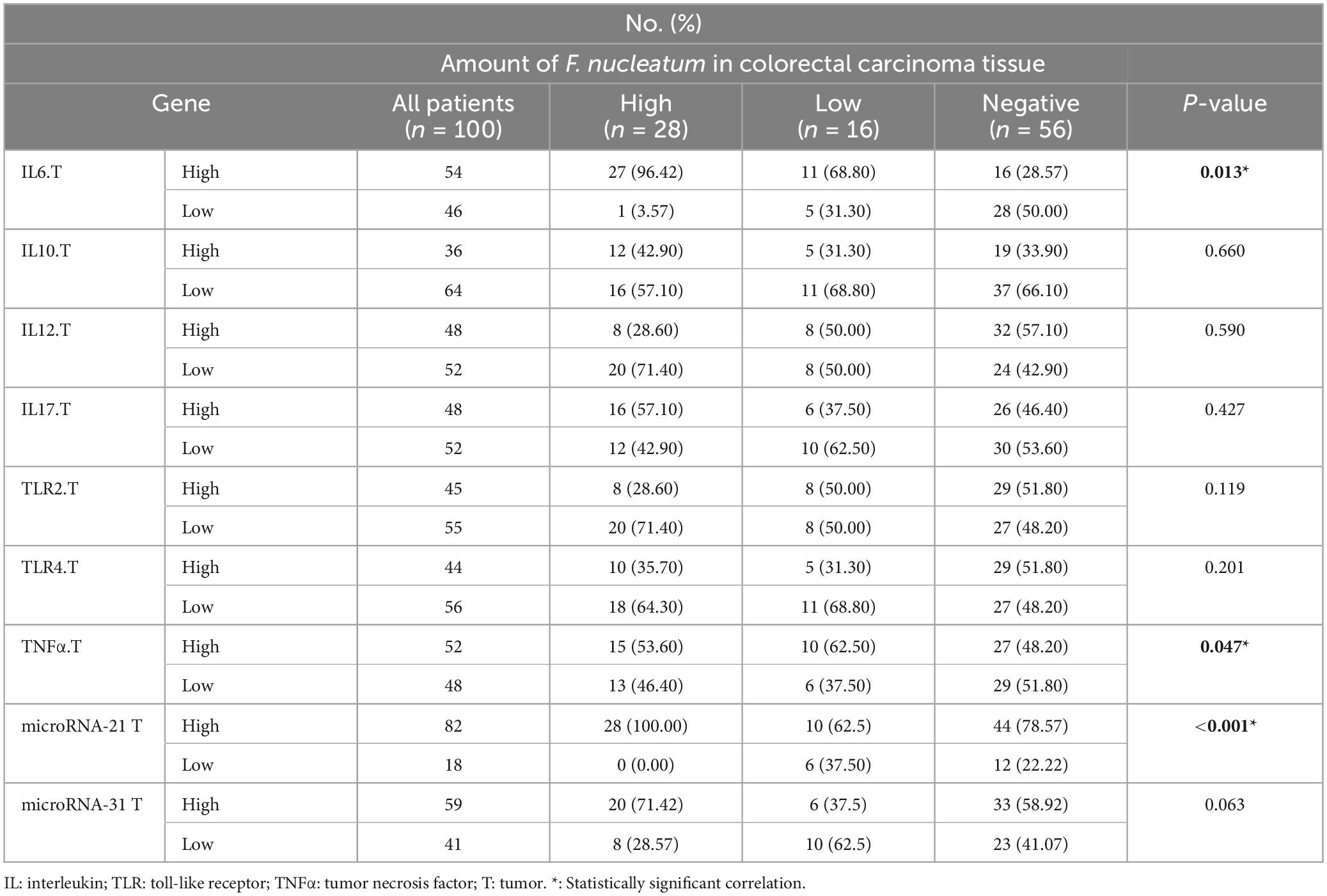
In this study, the expression levels of IL-6, IL-10, IL-12β, IL-17, TNF-α, TLR-2, and TLR-4 were investigated by real-time PCR and 2–ΔΔCq. The ROC curve was used to classify the expression of each gene in CRC into low and high groups. IL-6, IL-17, TNF-α, TLR-2, and TLR-4 were significantly more highly expressed in the cancer tissues (p < 0.05). However, there was no meaningful difference between the tumor and normal samples for IL-10, and IL-12β (p > 0.05) (Figure 2).
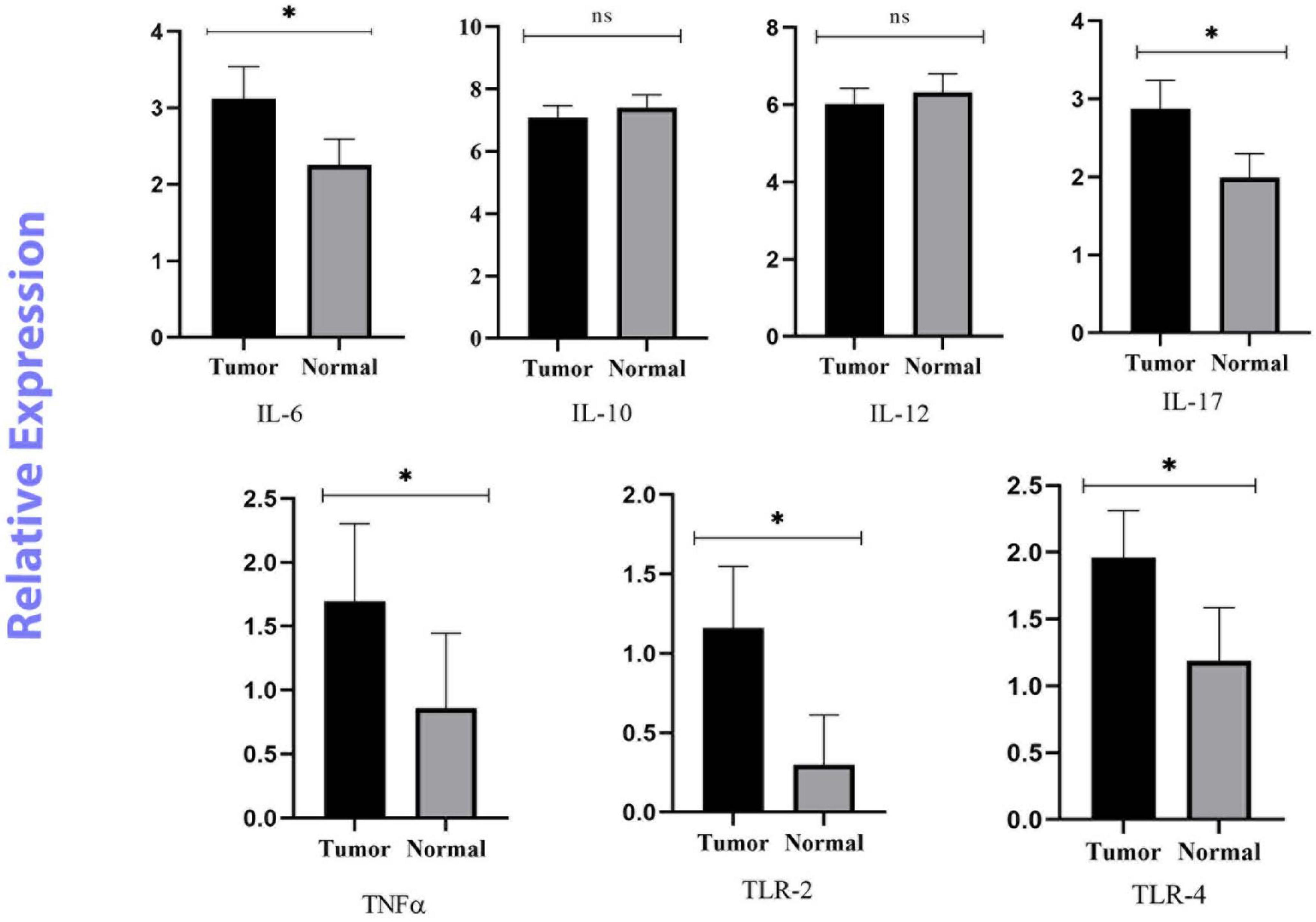
Figure 2. Relative quantification of Inflammatory and anti-inflammatory genes in cancerous and matched normal tissues (P < 0.05* and P > 0.05ns).
3.4 miRNA gene expression
As a control, the U6 gene was used to evaluate the expression levels of miR-21, and miR-31 genes in cancer and adjacent normal tissues (Schee et al., 2012). According to the results, miR-21 and miR-31 gene expression was significantly higher in the cancer tissues compared to the adjacent non-tumor mucosa (P < 0.001) (Figure 3).
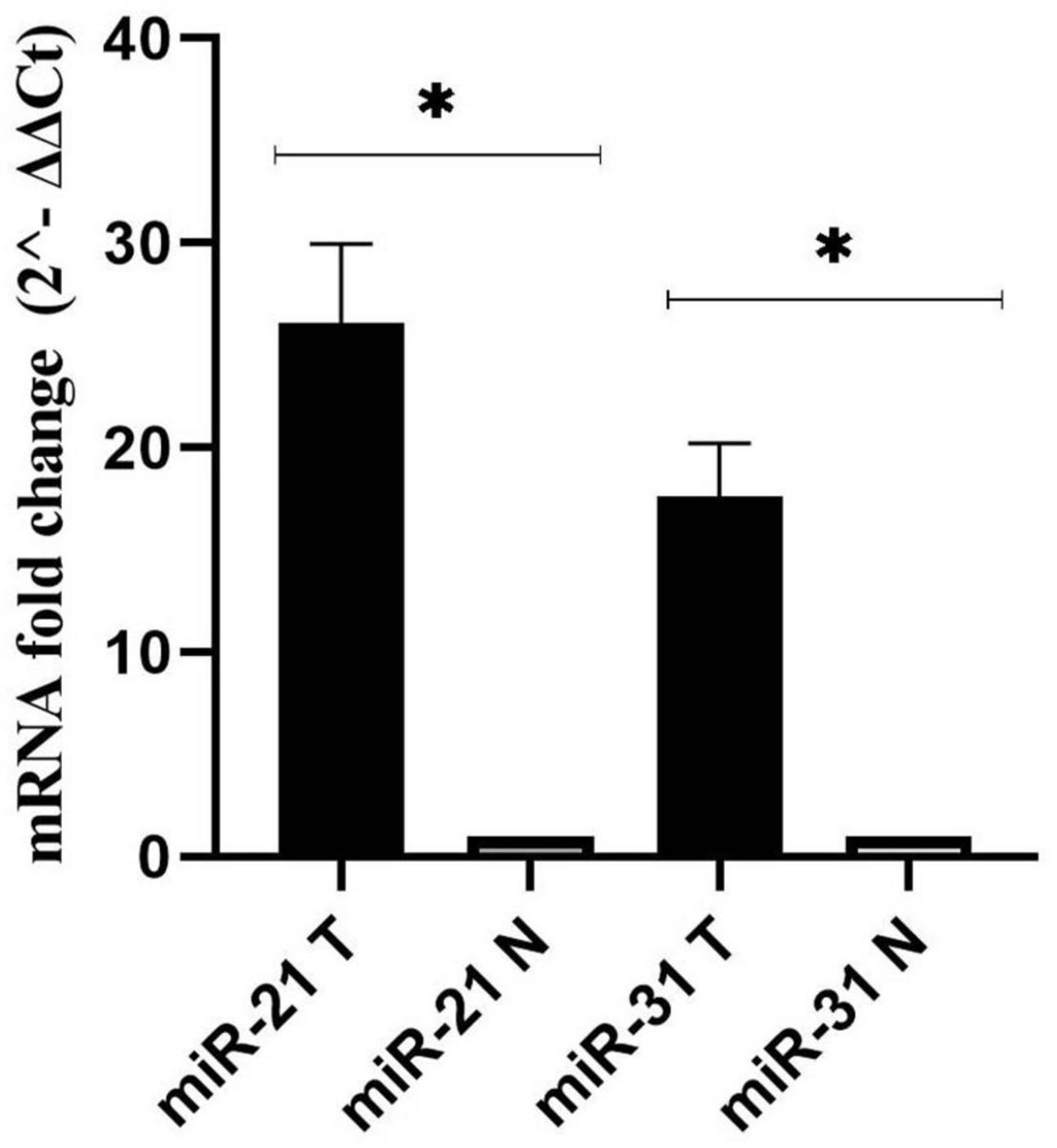
Figure 3. Fold change analysis of miR-21, and miR-31 gene expression in cancerous and matched normal tissues (P < 0.001*).
3.5 Clinicopathological and molecular association of F. nucleatum in CRC
The clinicopathological and molecular characteristics of CRCs according to F. nucleatum status (high expression group vs. low expression group vs. negative group) in CRC tissue are presented in Tables 2, 3.
According to logistic regression analysis, a significant correlation was observed between high levels of F. nucleatum and histology grade III and IV CRC tissues (P = 0.027 and P = 0.022, respectively), in which the levels of F. nucleatum DNA were 5.25-fold and 14-fold higher in grade III and IV CRC tissues, respectively, compared with grade I tissues. Moreover, perineural invasion-positive patients displayed a significant association with high-level F. nucleatum (P = 0.037). Compared to perineural invasion-negative patients, the presence of F. nucleatum DNA was 2.79-fold higher in perineural invasion-positive patients. No significant correlation between F. nucleatum infection with other clinicopathological variables was observed (P > 0.05).
According to the results, significant positive correlations were observed between the greater amount of F. nucleatum and high levels of IL-6 (P = 0.014) and TNF-α (P = 0.047) genes. Logistic regression results suggested that the level of F. nucleatum DNA was 3-fold and 4.4-fold higher in IL-6 and TNF-α high groups compared to low groups. There was a conspicuous correlation between miR-21 gene expression and F. nucleatum-high status in CRC tissues (P = 0.012). Conversely, no significant relation was found between the expression of the miR-31 gene and this group (P = 0.063).
4 Discussion
The dynamic and observable effect of microorganisms involved in carcinogenesis is a key issue in investigating their role in many types of cancer (Collins and Altman, 2012; Guo et al., 2021). F.nucleatum, has been recognized as one of the predisposing factors for CRC and plays a role in the progression of cancer through multiple mechanisms (McIlvanna et al., 2021). Recently, research into the F. nucleatum and its relationship with CRC has been the focus of many studies. However, the mechanism of action and the relationship between F. nucleatum and other microenvironmental factors in the development of CRC are unclear. To address this knowledge gap and gain a more thorough comprehension of the existing relationship, in this study, we investigated the frequency and association of F. nucleatum with the expression of cytokines and miRNAs involved in tumor tissue samples and non-neoplastic mucosa of Iranian patients.
In our current study, F. nucleatum was expressed at significantly higher levels in tumor samples compared to the adjacent normal tissues, which is aligned with previous studies (Castellarin et al., 2012; Viljoen et al., 2015; Rye et al., 2022). The percentage of presence of F. nucleatum in this study is also within the average range reported in previous studies from different countries (8.6–87%) (Mima et al., 2015; Baxter et al., 2016; Suehiro et al., 2017; Wong et al., 2017; Tunsjø et al., 2019; Kim et al., 2020; Shariati et al., 2021). This discrepancy is influenced by a multitude of factors such as the utilization of different diagnostic methods, various biological samples, and sample quality (Li et al., 2016; Suehiro et al., 2017; Leung et al., 2019). In addition, Lee et al. (2018) found that the method of tissue fixation may influence the variable results of F. nucleatum analysis. Shariati et al. (2021) reached similar conclusions by comparing the frequency of F. nucleatum in the colorectal tumor specimens and matched normal tissue by quantitative PCR analysis. They found F. nucleatum DNA in 23% of CRC biopsies (Shariati et al., 2021). On the other hand, in some studies in Iran, approximately 70% of patients with CRC were colonized by F. nucleatum (Kashani et al., 2020; Bostanshirin et al., 2023). The possibility of an excess rate of F. nucleatum in these studies may be due to disparities in technical methodologies employed, including simple PCR, SYBR qPCR, and quantitative PCR. Other factors influencing the inconsistency of reports include the diversity of the gut microbiome within the population, dietary habits, and geographical location. Nishijima et al. (2016) have shown that the gut microbiome of Japanese people is significantly different from that of other populations. Of course, this difference cannot be attributed to differences in geographical location alone (Nishijima et al., 2016). Recent studies have shown that the association between F. nucleatum and CRC is higher in Asian populations than in European or American populations (Huang et al., 2018; Janati et al., 2020). This population-level association may be explained by differences in lifestyle and diet in different communities, or by population-level variations in the human gut microbiome. Studies have shown that a conservative diet that includes more vegetables, fruits and fiber is associated with a reduced risk of F. nucleatum-positive CRC. On the other hand, higher consumption of fats, red/processed meat increases the risk of the F. nucleatum-positive CRC tumor subtype. Therefore, the composition and diversity of the colonic microbiota is influenced by dietary changes, and the balance between beneficial and harmful microbial metabolites is crucial in mediating CRC risk factors (Leng et al., 2018; Zhang et al., 2018). However, these results should be investigated further and different factors should be taken into account.
We also assessed the association of F. nucleatum status with clinicopathological features and found that high levels of F. nucleatum in tumor tissue were related to poorly differentiated and undifferentiated tumors. Previous studies have shown that there is a significant association between elevated levels of this bacterium and poor tumor differentiation (Sun et al., 2016; de Carvalho et al., 2019). Notably, a significant association has been observed between high levels of F. nucleatum and perineural invasion (PNI) positive tumors. However, further studies are needed to prove this association. PNI is the invasion of tumor cells into the perineural space of nerves. PNI is a strong prognostic factor in CRC and is significantly associated with reduced survival and high recurrence rates. Studies have shown a significant improvement in 5-year survival in patients with negative PNI tumors compared with those with positive PNI tumors (Poeschl et al., 2010; Betge and Langner, 2011).
The precise mechanisms by which the gut microbiota influences the development of CRC are not fully understood. However, one of the most encouraging hypotheses is that it occurs via microbe-induced inflammation. In particular, inflammation is a critical and well-known risk factor for the development and progression of CRC (Gerhard Rogler et al., 2010). Dysbiosis of gut bacteria may be the cause of immune dysregulation and pro-inflammatory mediator production (Jm, 2009; Swidsinski et al., 2011). Numerous studies have investigated the potential factors by which F. nucleatum contributes to colorectal tumorigenesis (McCoy et al., 2013; Hashemi Goradel et al., 2019). The presence of F. nucleatum in the gut promotes the expression of tumor-associated cytokines and an inflammatory response through the action of virulence factors.
Fusobacterium nucleatum can activate β-catenin signaling through two pathways. The first pathway is the binding of FadA to E-cadherin which can active the zipper mechanism and transport F. nucleatum into cells. The second pathway is the TLR4/P-PAK1 cascade that leads to the initiation of inflammatory responses, followed by the amplification of transcription of NF-κβ genes and pro-inflammatory cytokines. Also, it can intensify inflammatory responses through the binding of the Fap2 factor to Gal-GalNAc (Kostic et al., 2013; Rubinstein et al., 2013). TLR-2 and TLR-4 are mainly involved in recognizing F. nucleatum and regulating inflammatory factors induced by this bacterium via Tregs (Jia et al., 2017). Both the observations of Kostic et al. (2013) and Rubinstein et al. (2013) support the role of F. nucleatum in stimulating the production of an inflammatory microenvironment, leading to an increase in the oncogenic potential of this microorganism (20, 21). Given the reports of previous studies, we assessed the mRNA expression of mucosal inflammatory cytokines and the association between their expression levels and the abundance of F. nucleatum in CRC tissues compared to adjacent normal tissues. Our results showed that IL-6, IL-17, TNF-α, TLR-2 and TLR-4 genes were overexpressed in tumor tissues compared to adjacent normal tissues. IL-10 and IL-12β genes were downregulated, but the difference was not significant. We also found a significant positive correlation between a high amount of F. nucleatum and high levels of IL-6, and TNF-α expression in the tumor tissues.
IL-6 is a pro-inflammatory cytokine with pro-tumorigenic properties. It regulates multiple signaling pathways including survival, invasion, apoptosis, proliferation, angiogenesis and metastasis. Overexpression of IL-6 has been well-studied in several malignancies including lung, breast, and colon cancer (Sethi et al., 2012; Heichler et al., 2020; Ke et al., 2020). Consistent with our findings, Akhmaltdinova et al. (2020) and Proença et al. (2018) have shown that IL-6 levels are significantly elevated in CRC patients. In contrast, two British studies found no association between IL-6 levels and CRC risk (Heikkilä et al., 2009). The small sample size may be a reason for this conclusion. Another study measured the concentration of IL-6 in serum samples from 208 patients with stage I to IV CRC. Patients with stage III and IV disease had significantly higher serum IL-6 concentrations than those with stage I and II disease (Belluco et al., 2000). TNF-α is another pro-inflammatory cytokine that is produced by tumor or inflammatory cells and is involved in the regulation of a variety of signaling processes. Like IL-6, it is involved in tumor initiation, cell proliferation, promotion of angiogenesis, and metastasis (Lan et al., 2021). Consistent with the study by Akhmaltdinova et al. (2020) our data show that TNF-α levels are significantly elevated in all CRC tissues. Previously published results on colorectal adenomas found that a high abundance of F. nucleatum was positively correlated with the expression of inflammatory cytokine genes, such as IL-6 and TNF-α, which is similar to our findings (McCoy et al., 2013; Velsko et al., 2015; Martinho et al., 2016; Wei et al., 2016). Despite the positive correlation between Fusobacterium species and IL-6, the results were not statistically significant in the study by McCoy et al. (2013). Studies have shown that some markers of inflammatory responses, such as IL-1β, IL-6 and TNF-α, are specific to F. nucleatum infection and their expression is enriched in F. nucleatum-infected CRCs. However, they were not seen in CRC tissue with other bacteria (Wu et al., 2019). As mentioned above, F. nucleatum can shape the inflammatory microenvironment in CRC through multiple mechanisms. It binds to colon epithelium and leads to the activation of NF-κB. Also, it can stimulate IL-6 production by activating both TLR-2 and TLR-4 in bone marrow-derived macrophages (Park et al., 2014). F. nucleatum increases the infiltration of inflammatory cells such as dendritic cells, M2 macrophage polarization, and granulocytes (Chaushu et al., 2012; Noh et al., 2016). Natural killer (NK) cells can directly recognize F. nucleatum through its surface ligand and secrete TNF-α to exacerbate the expression and secretion of IL-6 (Chaushu et al., 2012). Consequently, the concentration of circulating IL-6 and TNF-α as gastrointestinal inflammation may be an indicator of promoting CRC development.
IL-17 is another cytokine investigated in this study, which is mainly synthesized and released by Th-17 lymphocytes and contributes to the development of terminal inflammation. In addition, IL-17 is a potent immunomodulator and can promote angiogenesis and tumor growth (Kuen et al., 2020). Our results showed a significant difference in IL-17 expression levels between CRC patients and similar normal tissues, which seems to be in line with the results of other studies, especially in those with poorly differentiated and well-differentiated tumor tissues (Lin et al., 2015; Feng et al., 2019). Taken together, these results support the role of IL-17 in CRC development and progression. In contrast to our results, Stanilov et al. (2010) found no significant difference in IL-17 levels between plasma samples from CRC patients and healthy subjects. Sample type and method of measurement may explain this difference in results.
IL-12 is an inflammatory cytokine that has been shown to play an anti-tumor and anti-metastatic role in vivo in a number of murine models of colon adenocarcinoma. The anti-tumor activity of IL-l2 is mediated by activation of Th1 adaptive immunity and increased interferon production, which has a direct toxic effect on cancer cells (Trinchieri, 2003; Lan et al., 2021). Our results showed that the expression level of the IL-12β gene was decreased in cancer tissues, suggesting the anti-tumor activity of IL-12β in CRC patients. The findings presented here align with the investigation conducted by O’Hara et al. (1998) in which the authors observed that patients with CRC have decreased IL-12β production. According to the results, the use of IL-12β as a cancer immunotherapy may be beneficial in controlling tumor growth (Briukhovetska et al., 2021).
IL-10 is a cytokine with bidirectional immunomodulatory properties. The immunosuppressive effect of IL-10 on dendritic cells and macrophages results in attenuated antigen presentation, allowing tumor cells to evade immune surveillance and impair cell maturation and differentiation. IL-10 inhibits NF-κB signaling; therefore it can downregulate the expression of proinflammatory cytokines and act as an antitumor cytokine (Li et al., 2020). Abtahi et al. (2017) showed that the serum level of IL-10 was significantly lower in CRC patients than in controls. This finding is consistent with our current research. However, they highlighted the association between the expression of this cytokine and the prognosis of CRC patients, and those with a poor prognosis had high levels of IL-10 (Abtahi et al., 2017). Inconsistent with our results, previous studies have shown overexpression of IL-10 in CRC tissues compared to normal tissues (Li et al., 2019; Cuellar-Gómez et al., 2022). This variation in IL-10 levels is contingent upon the onset and progression of CRC and supports the potential ambivalent function of IL-10. Nevertheless, further investigation is imperative in order to comprehend the underlying processes of IL-10 whether it is a tumor-stimulating or inhibitory factor.
Until recently, a number of studies have suggested aberrant expression of miRNAs and their oncogenic or suppressive functions in the initiation and progression of various malignancies such as CRC (Nosho et al., 2016; Rapado-González et al., 2019). The trend of miR-31, and miR-21 expression in our results was significantly upregulated in CRC patients compared to adjacent normal tissues, which is similar to previous researches (Kanaan et al., 2012; Wu et al., 2012; Nosho et al., 2014; Wang et al., 2017; Sabry et al., 2019; Farouk et al., 2020; Nassar et al., 2021; Zhou et al., 2022). Therefore, with this comparison, we can refer to the role of these genes in the development of CRC. However, Wang et al. (2014) showed that the expression of miR-31 was significantly decreased in serum samples from patients with CRC. This may demonstrate the dual and contradictory function of miRNA in different types of tumors. miR-31 may not only promote the growth and progression of malignancies such as pancreatic, cervical, and CRC, but also suppress carcinogenesis and induce apoptosis in cancers such as ovarian and prostate cancer (Laurila and Kallioniemi, 2013; Braga et al., 2017). Studies suggest that the diverse role of this factor in cancer regulation may be due to spatiotemporal specificity, characteristics of adenocarcinoma tissue and target genes, which have significant interaction with the signaling pathway (Loya et al., 2009; Yu et al., 2018). However, further research is required to understand the underlying mechanism. In general, miR-21 and miR-31 have been reported to be valuable diagnostic biomarkers for CRC (Sur et al., 2022). We identified a significant association between high levels of F. nucleatum and miR-21 expression in CRC tissue, which is consistent with previous studies (Nosho et al., 2016; Yang et al., 2017; Bostanshirin et al., 2023). F. nucleatum can trigger the TLR4/MYD88 signaling pathway by lipopolysaccharide (LPS). Subsequently, hyperactive NF-κB binds to the miR-21 promoter and upregulates its expression in CRC patients (Yang et al., 2017; Yu et al., 2017). This finding partially supports the role of F nucleatum in carcinogenesis through the induction of miR21.
In contrast, no correlation was observed between miR-31 expression and the aforementioned colonization. Similarly, Ito et al. (2015) also showed that there was no significant correlation between miR-31 expression and F. nucleatum status. However, Tang et al. (2023) demonstrated that the upregulation of miR-31 was significantly correlated with the presence of F. nucleatum in CRC tissues and resulted in the promotion of tumorigenesis. Furthermore, they reported that miR-31-mediated inhibition of autophagic flux via suppression of syntaxin-12 (STX12) was linked to enhanced intracellular survival of F. nucleatum infection (Tang et al., 2023). The investigation of the relationship between F. nucleatum and miRNAs expression has been very limited. Understanding this relationship will provide new insights into strategies for cancer control and treatment.
5 Conclusion
The results showed that the abundance of F. nucleatum was significantly greater in cancerous tissue compared to normal tissue. In addition, a significant association was found between F. nucleatum and the expression of miR-21, IL-6 and TNF-α. The current findings provide important insights into the function of F. nucleatum and its potential association with increased gene expression in carcinogenesis, thereby playing a critical role in CRC progression and metastasis. The data presented provide ample evidence for the pathogenic role of F. nucleatum in CRC, thus opening new avenues for targeting the microbiota to accelerate the prognosis of cancer progression and prevent the development of CRC. Furthermore, due to the effect of inflammatory factors and miRNAs and the effect of F. nucleatum on their expression, they may serve as biomarkers for acceptable cancer diagnosis.
Data availability statement
The raw data supporting the conclusions of this article will be made available by the authors, without undue reservation.
Ethics statement
The studies involving humans were approved by the Ethics Committee of Iran University of Medical Sciences “IR.IUMS.FMD.REC.1400.569.” To ensure safety, all participants were informed of the aims of this study and written informed consent was obtained prior to their participation. The studies were conducted in accordance with the local legislation and institutional requirements. The participants provided their written informed consent to participate in this study.
Author contributions
NB: Conceptualization, Data curation, Investigation, Methodology, Resources, Software, Writing – original draft, Writing – review and editing. SR: Project administration, Writing – review and editing. AS: Project administration, Writing – review and editing. MT: Project administration, Writing – review and editing. SM: Project administration, Writing – review and editing. AE: Data curation, Writing – review and editing. DD-S: Data curation, Funding acquisition, Methodology, Project administration, Resources, Software, Supervision, Visualization, Writing – review and editing.
Funding
The author(s) declare financial support was received for the research, authorship, and/or publication of this article. This research was supported by grant no: 1400-2-4-21695 from the Iran University of Medical Sciences.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
The reviewer AD declared a shared affiliation with the authors NB, SR, MT, SM, and DD-S to the handling editor at the time of review.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2023.1302719/full#supplementary-material
References
Abtahi, S., Davani, F., Mojtahedi, Z., Hosseini, S., Bananzadeh, A., and Ghaderi, A. (2017). Dual association of serum interleukin-10 levels with colorectal cancer. J. Cancer Res. Ther. 13, 252–256. doi: 10.4103/0973-1482.199448
Akhmaltdinova, L., Sirota, V., Babenko, D., Zhumaliyeva, V., Kadyrova, I., Maratkyzy, M., et al. (2020). Proinflammatory cytokines and colorectal cancer - the impact of the stage. Contemp. Oncol. 24, 207–210.
Allali, I., Delgado, S., Marron, P., Astudillo, A., Yeh, J., Ghazal, H., et al. (2015). Gut microbiome compositional and functional differences between tumor and non-tumor adjacent tissues from cohorts from the US and Spain. Gut Microbes 6, 161–172. doi: 10.1080/19490976.2015.1039223
Baxter, N., Ruffin, M., Rogers, M., and Schloss, P. (2016). Microbiota-based model improves the sensitivity of fecal immunochemical test for detecting colonic lesions. Genome Med. 8:37. doi: 10.1186/s13073-016-0290-3
Belluco, C., Nitti, D., Frantz, M., Toppan, P., Basso, D., Plebani, M., et al. (2000). Interleukin-6 blood level is associated with circulating carcinoembryonic antigen and prognosis in patients with colorectal cancer. Ann. Surg. Oncol. 7, 133–138.
Betge, J., and Langner, C. (2011). Vascular invasion, perineural invasion, and tumour budding: Predictors of outcome in colorectal cancer. Acta Gastroenterol. Belg. 74, 516–529.
Bhat, A., Nisar, S., Singh, M., Ashraf, B., Masoodi, T., Prasad, C., et al. (2022). Cytokine- and chemokine-induced inflammatory colorectal tumor microenvironment: Emerging avenue for targeted therapy. Cancer Commun. 42, 689–715. doi: 10.1002/cac2.12295
Bidkhori, H., Hedayati-Moghaddam, M., Mosavat, A., Valizadeh, N., Tadayon, M., Ahmadi Ghezeldasht, S., et al. (2020). The IL-18, IL-12, and IFN-gamma expression in HTLV-1-associated myelopathy/tropical spastic paraparesis (HAM/TSP) patients, HTLV-1 carriers, and healthy subjects. J. Neurovirol. 26, 338–346. doi: 10.1007/s13365-020-00832-5
Bostanshirin, N., Hajikhani, B., Vaezi, A., Kermanian, F., Sameni, F., Yaslianifard, S., et al. (2023). Association between colorectal cancer and expression levels of miR-21, miR-17-5P, miR-155 genes and the presence of Fusobacterium nucleatum in biopsy samples obtained from Iranian patients. Infect. Agents Cancer 18, 1–9. doi: 10.1186/s13027-023-00494-y
Braga, E., Fridman, M., and Kushlinskii, N. (2017). Molecular mechanisms of ovarian carcinoma metastasis: Key genes and regulatory MicroRNAs. Biochemistry 82, 529–541.
Brennan, C., and Garrett, W. (2016). Gut microbiota, inflammation, and colorectal cancer. Annu. Rev. Microbiol. 70, 395–411.
Briukhovetska, D., Dörr, J., Endres, S., Libby, P., Dinarello, C., and Kobold, S. (2021). Interleukins in cancer: From biology to therapy. Nat. Rev. Cancer 21, 481–499.
Bustin, S., Benes, V., Garson, J., Hellemans, J., Huggett, J., Kubista, M., et al. (2009). The MIQE Guidelines: M inimum I nformation for Publication of Q uantitative Real-Time PCR E xperiments. Oxford: Oxford University Press.
Castellarin, M., Warren, R., Freeman, J., Dreolini, L., Krzywinski, M., Strauss, J., et al. (2012). Fusobacterium nucleatum infection is prevalent in human colorectal carcinoma. Genome Res. 22, 299–306. doi: 10.1101/gr.126516.111
Chaushu, S., Wilensky, A., Gur, C., Shapira, L., Elboim, M., Halftek, G., et al. (2012). Direct recognition of Fusobacterium nucleatum by the NK cell natural cytotoxicity receptor NKp46 aggravates periodontal disease. PLoS Pathog. 8:e1002601. doi: 10.1371/journal.ppat.1002601
Chen, D., Wu, J., Jin, D., Wang, B., and Cao, H. (2019). Fecal microbiota transplantation in cancer management: Current status and perspectives. Int. J. Cancer 145, 2021–2031. doi: 10.1002/ijc.32003
Cheng, Y., Ling, Z., and Li, L. (2020). The intestinal microbiota and colorectal cancer. Front. Immunol. 11:615056. doi: 10.3389/fimmu.2020.615056
Chi, Y., and Zhou, D. (2016). MicroRNAs in colorectal carcinoma-from pathogenesis to therapy. J. Exp. Clin. Cancer Res. 35, 1–11. doi: 10.1186/s13046-016-0320-4
Collins, G., and Altman, D. (2012). Identifying patients with undetected colorectal cancer: An independent validation of QCancer (Colorectal). Br. J. Cancer 107, 260–265. doi: 10.1038/bjc.2012.266
Cottonham, C., Kaneko, S., and Xu, L. (2010). miR-21 and miR-31 converge on TIAM1 to regulate migration and invasion of colon carcinoma cells. J. Biol. Chem. 285, 35293–35302. doi: 10.1074/jbc.M110.160069
Cuellar-Gómez, H., Ocharán-Hernández, M., Calzada-Mendoza, C., and Comoto-Santacruz, D. (2022). Association of Fusobacterium nucleatum infection and colorectal cancer: A Mexican study. Rev Gastroenterol México. 87, 277–284. doi: 10.1016/j.rgmxen.2021.07.001
Dadgar-Zankbar, L., Shariati, A., Bostanghadiri, N., Elahi, Z., Mirkalantari, S., Razavi, S., et al. (2023). Evaluation of enterotoxigenic Bacteroides fragilis correlation with the expression of cellular signaling pathway genes in Iranian patients with colorectal cancer. Infect. Agents Cancer 18:48. doi: 10.1186/s13027-023-00523-w
de Carvalho, A., de Mattos, P., Datorre, J., Dos Santos, W., Berardinelli, G., and Matsushita, M. (2019). Microbiota profile and impact of Fusobacterium nucleatum in colorectal cancer patients of Barretos cancer hospital. Front. Oncol. 9:813. doi: 10.3389/fonc.2019.00813
Dekker, E., Tanis, P., Vleugels, J., Kasi, P., and Wallace, M. (2019). Risk factors. Lancet 394, 1467–1480.
Eslamizadeh, S., Heidari, M., Agah, S., Faghihloo, E., Ghazi, H., Mirzaei, A., et al. (2018). The role of MicroRNA signature as diagnostic biomarkers in different clinical stages of colorectal cancer. Cell J. 20, 220–230.
Farouk, S., Khairy, A., Salem, A., and Soliman, A. (2020). Bader El Din NG: Differential expression of miR-21, miR-23a, and miR-27a, and their diagnostic significance in Egyptian colorectal cancer patients. Genetic Test. Mol. Biomark. 24, 825–834. doi: 10.1089/gtmb.2020.0184
Feng, H., Ying, R., Chai, T., Chen, H., and Ju, H. (2019). The association between IL-17 gene variants and risk of colorectal cancer in a Chinese population: A case–control study. Biosci. Rep. 39:BSR20190013. doi: 10.1042/BSR20190013
Gerhard Rogler, G., Haller, D., and Jobin, C. (2010). Microbiota in chronic mucosal inflammation. Int. J. Inflam. 2010:395032.
Ghafouri-Fard, S., Hussen, B., Badrlou, E., Abak, A., and Taheri, M. (2021). MicroRNAs as important contributors in the pathogenesis of colorectal cancer. Biomed. Pharmacother. 140:111759.
Guo, Y., Shao, L., Wang, L., Chen, M., Zhang, W., and Huang, W. (2021). Bioconversion variation of ginsenoside CK mediated by human gut microbiota from healthy volunteers and colorectal cancer patients. Chin. Med. 16, 1–10. doi: 10.1186/s13020-021-00436-z
Hashemi Goradel, N., Heidarzadeh, S., Jahangiri, S., Farhood, B., Mortezaee, K., Khanlarkhani, N., et al. (2019). Fusobacterium nucleatum and colorectal cancer: A mechanistic overview. J. Cell. Physiol. 234, 2337–2344. doi: 10.1002/jcp.27250
Heichler, C., Scheibe, K., Schmied, A., Geppert, C., Schmid, B., Wirtz, S., et al. (2020). STAT3 activation through IL-6/IL-11 in cancer-associated fibroblasts promotes colorectal tumour development and correlates with poor prognosis. Gut 69, 1269–1282. doi: 10.1136/gutjnl-2019-319200
Heikkilä, K., Harris, R., Lowe, G., Rumley, A., Yarnell, J., Gallacher, J., et al. (2009). Associations of circulating C-reactive protein and interleukin-6 with cancer risk: Findings from two prospective cohorts and a meta-analysis. Cancer Causes Control 20, 15–26. doi: 10.1007/s10552-008-9212-z
Henstra, C., van Praagh, J., Olinga, P., and Nagelkerke, A. (2021). The gastrointestinal microbiota in colorectal cancer cell migration and invasion. Clin. Exp. Metastasis 38, 495–510.
Huang, H., Zhang, Z., Cao, C., Wang, N., Liu, F., Peng, J., et al. (2014). The TLR4/NF-kappaB signaling pathway mediates the growth of colon cancer. Eur. Rev. Med. Pharmacol. Sci. 18, 3834–3843.
Huang, Q., Peng, Y., and Xie, F. (2018). Fecal fusobacterium nucleatum for detecting colorectal cancer: A systematic review and meta-analysis. Int. J. Biol. Mark. 33, 345–352. doi: 10.1177/1724600818781301
Ito, M., Kanno, S., Nosho, K., Sukawa, Y., Mitsuhashi, K., Kurihara, H., et al. (2015). Association of Fusobacterium nucleatum with clinical and molecular features in colorectal serrated pathway. Int. J. Cancer 137, 1258–1268. doi: 10.1002/ijc.29488
Janati, A., Karp, I., Laprise, C., Sabri, H., and Emami, E. (2020). Detection of Fusobacterium nucleatum in feces and colorectal mucosa as a risk factor for colorectal cancer: A systematic review and meta-analysis. Syst. Rev. 9, 1–15. doi: 10.1186/s13643-020-01526-z
Jia, Y., Wang, K., Zhang, Z., Tong, Y., Han, D., Hu, C., et al. (2017). TLR2/TLR4 activation induces Tregs and suppresses intestinal inflammation caused by Fusobacterium nucleatum in vivo. PLoS One 12:e0186179. doi: 10.1371/journal.pone.0186179
Jm, U. (2009). Modulation of the intestinal microbiota alters colitis-associated colorectal cancer susceptibility. PLoS One 4:e6026. doi: 10.1371/journal.pone.0006026
Kanaan, Z., Rai, S., Eichenberger, M., Roberts, H., Keskey, B., Pan, J., et al. (2012). Plasma miR-21: A potential diagnostic marker of colorectal cancer. Ann. Surg. 256, 544–551.
Kashani, N., Abadi, A., Rahimi, F., and Forootan, M. (2020). FadA-positive Fusobacterium nucleatum is prevalent in biopsy specimens of Iranian patients with colorectal cancer. N. Microbes New Infect. 34:100651. doi: 10.1016/j.nmni.2020.100651
Kaźmierczak-Siedlecka, K., Daca, A., Fic, M., van de Wetering, T., Folwarski, M., and Makarewicz, W. (2020). Therapeutic methods of gut microbiota modification in colorectal cancer management–fecal microbiota transplantation, prebiotics, probiotics, and synbiotics. Gut Microbes 11, 1518–1530. doi: 10.1080/19490976.2020.1764309
Ke, W., Zhang, L., and Dai, Y. (2020). The role of IL-6 in immunotherapy of non-small cell lung cancer (NSCLC) with immune-related adverse events (irAEs). Thoracic Cancer 11, 835–839. doi: 10.1111/1759-7714.13341
Kim, M., Lee, S., Choi, S., Lee, H., Kwon, S., Byun, J., et al. (2020). Fusobacterium nucleatum in biopsied tissues from colorectal cancer patients and alcohol consumption in Korea. Sci. Rep. 10:19915. doi: 10.1038/s41598-020-76467-7
Kostic, A., Chun, E., Robertson, L., Glickman, J., Gallini, C., Michaud, M., et al. (2013). Fusobacterium nucleatum potentiates intestinal tumorigenesis and modulates the tumor-immune microenvironment. Cell Host Microbe 14, 207–215. doi: 10.1016/j.chom.2013.07.007
Kuen, D., Kim, B., and Chung, Y. (2020). IL-17-producing cells in tumor immunity: friends or foes? Immune Netw. 20:e6.
Lan, T., Chen, L., and Wei, X. (2021). Inflammatory cytokines in cancer: Comprehensive understanding and clinical progress in gene therapy. Cells 10:100.
Laurila, E., and Kallioniemi, A. (2013). The diverse role of miR-31 in regulating cancer associated phenotypes. Genes Chromosomes Cancer 52, 1103–1113.
Lee, D., Han, S., Kang, J., Bae, J., Kim, H., Won, J., et al. (2018). Association between Fusobacterium nucleatum, pathway mutation, and patient prognosis in colorectal cancer. Ann. Surg. Oncol. 25, 3389–3395. doi: 10.1245/s10434-018-6681-5
Leija-Martínez, J., Del-Río-Navarro, B., Sanchéz-Muñoz, F., Muñoz-Hernández, O., Hong, E., Giacoman-Martínez, A., et al. (2021). Associations of TNFA, IL17A, and RORC mRNA expression levels in peripheral blood leukocytes with obesity-related asthma in adolescents. Clin. Immunol. 229:108715.
Leng, S., Zhao, A., Li, Q., Pei, L., Zheng, W., Liang, R., et al. (2018). Metabolic status and lifestyle factors associated with gallbladder polyps: A covariance structure analysis. BMC Gastroenterol. 18:159. doi: 10.1186/s12876-018-0882-z
Leung, P., Subramanya, R., Mou, Q., Lee, K., Islam, F., Gopalan, V., et al. (2019). Characterization of mucosa-associated microbiota in matched cancer and non-neoplastic mucosa from patients with colorectal cancer. Front. Microbiol. 10:1317. doi: 10.3389/fmicb.2019.01317
Li, B., Wang, F., Ma, C., Hao, T., Geng, L., and Jiang, H. (2019). Predictive value of IL-18 and IL-10 in the prognosis of patients with colorectal cancer. Oncol. Lett. 18, 713–719.
Li, J., Huang, L., Zhao, H., Yan, Y., and Lu, J. (2020). The role of interleukins in colorectal cancer. Int. J. Biol. Sci. 16:2323.
Li, X., Xin, S., He, Z., Che, X., Wang, J., Xiao, X., et al. (2014). MicroRNA-21 (miR-21) post-transcriptionally downregulates tumor suppressor PDCD4 and promotes cell transformation, proliferation, and metastasis in renal cell carcinoma. Cell. Physiol. Biochem. 33, 1631–1642. doi: 10.1159/000362946
Li, Y., Ge, Q., Cao, J., Zhou, Y., Du, Y., Shen, B., et al. (2016). Association of Fusobacterium nucleatum infection with colorectal cancer in Chinese patients. World J. Gastroenterol. 22:3227. doi: 10.3748/wjg.v22.i11.3227
Lin, Y., Xu, J., Su, H., Zhong, W., Yuan, Y., Yu, Z., et al. (2015). Interleukin-17 is a favorable prognostic marker for colorectal cancer. Clin. Transl. Oncol. 17, 50–56. doi: 10.1007/s12094-014-1197-3
Loya, C., Lu, C., Van Vactor, D., and Fulga, T. (2009). Transgenic microRNA inhibition with spatiotemporal specificity in intact organisms. Nat. Methods 6, 897–903. doi: 10.1038/nmeth.1402
Martinho, F., Leite, F., Nóbrega, L., Endo, M., Nascimento, G., Darveau, R., et al. (2016). Comparison of Fusobacterium nucleatum and Porphyromonas gingivalis lipopolysaccharides clinically isolated from root canal infection in the induction of pro-inflammatory cytokines secretion. Braz. Dental J. 27, 202–207. doi: 10.1590/0103-6440201600572
McCoy, A., Araujo-Perez, F., Azcarate-Peril, A., Yeh, J., Sandler, R., and Keku, T. (2013). Fusobacterium is associated with colorectal adenomas. PLoS One 8:e53653. doi: 10.1371/journal.pone.0053653
McIlvanna, E., Linden, G., Craig, S., Lundy, F., and James, J. (2021). Fusobacterium nucleatum and oral cancer: A critical review. BMC Cancer 21:1212. doi: 10.1186/s12885-021-08903-4
Mima, K., Nishihara, R., Qian, Z., Cao, Y., Sukawa, Y., Nowak, J., et al. (2016). Fusobacterium nucleatum in colorectal carcinoma tissue and patient prognosis. Gut 65, 1973–1980. doi: 10.1136/gutjnl-2015-310101
Mima, K., Sukawa, Y., Nishihara, R., Qian, Z., Yamauchi, M., Inamura, K., et al. (2015). Fusobacterium nucleatum and T cells in colorectal carcinoma. JAMA Oncol. 1, 653–661. doi: 10.1001/jamaoncol.2015.1377
Nassar, F., Msheik, Z., Itani, M., Helou, R., Hadla, R., Kreidieh, F., et al. (2021). Circulating miRNA as biomarkers for colorectal cancer diagnosis and liver metastasis. Diagnostics 11:341.
Nishijima, S., Suda, W., Oshima, K., Kim, S., Hirose, Y., Morita, H., et al. (2016). The gut microbiome of healthy Japanese and its microbial and functional uniqueness. DNA Res. 23, 125–133.
Noh, E., Kang, M., Jeong, Y., Lee, J., Park, J., Choi, H., et al. (2016). Withaferin A inhibits inflammatory responses induced by Fusobacterium nucleatum and Aggregatibacter actinomycetemcomitans in macrophages. Mol. Med. Rep. 14, 983–988.
Nosho, K., Igarashi, H., Nojima, M., Ito, M., Maruyama, R., Yoshii, S., et al. (2014). Association of microRNA-31 with BRAF mutation, colorectal cancer survival and serrated pathway. Carcinogenesis 35, 776–783. doi: 10.1093/carcin/bgt374
Nosho, K., Sukawa, Y., Adachi, Y., Ito, M., Mitsuhashi, K., Kurihara, H., et al. (2016). Association of Fusobacterium nucleatum with immunity and molecular alterations in colorectal cancer. World J. Gastroenterol. 22:557. doi: 10.3748/wjg.v22.i2.557
O’Hara, R., Greenman, J., MacDonald, A., Gaskell, K., Topping, K., Duthie, G., et al. (1998). Advanced colorectal cancer is associated with impaired interleukin 12 and enhanced interleukin 10 production. Clin. Cancer Res 4, 1943–1948.
Park, S., Kim, D., Han, S., Kang, M., Lee, J., Jeong, Y., et al. (2014). Diverse Toll-like receptors mediate cytokine production by Fusobacterium nucleatum and Aggregatibacter actinomycetemcomitans in macrophages. Infect. Immun. 82, 1914–1920. doi: 10.1128/IAI.01226-13
Poeschl, E., Pollheimer, M., Kornprat, P., Lindtner, R., Schlemmer, A., Rehak, P., et al. (2010). Perineural invasion: Correlation with aggressive phenotype and independent prognostic variable in both colon and rectum cancer. J. Clin. Oncol. 28, e358–e360. doi: 10.1200/JCO.2009.27.3581
Proença, M., Biselli, J., Succi, M., Severino, F., Berardinelli, G., Caetano, A., et al. (2018). Relationship between Fusobacterium nucleatum, inflammatory mediators and microRNAs in colorectal carcinogenesis. World J. Gastroenterol. 24:5351. doi: 10.3748/wjg.v24.i47.5351
Rapado-González, Ó, Álvarez-Castro, A., López-López, R., Iglesias-Canle, J., Suárez-Cunqueiro, M., and Muinelo-Romay, L. (2019). Circulating microRNAs as promising biomarkers in colorectal cancer. Cancers 11:898.
Rea, D., Coppola, G., Palma, G., Barbieri, A., Luciano, A., Del Prete, P., et al. (2018). Microbiota effects on cancer: From risks to therapies. Oncotarget 9:17915.
Rubinstein, M., Wang, X., Liu, W., Hao, Y., Cai, G., and Han, Y. (2013). Fusobacterium nucleatum promotes colorectal carcinogenesis by modulating E-cadherin/β-catenin signaling via its FadA adhesin. Cell Host Microbe 14, 195–206.
Rye, M., Garrett, K., Holt, R., Platell, C., and McCoy, M. (2022). Fusobacterium nucleatum and Bacteroides fragilis detection in colorectal tumours: Optimal target site and correlation with total bacterial load. PLoS One 17:e0262416. doi: 10.1371/journal.pone.0262416
Sabry, D., El-Deek, S., Maher, M., El-Baz, M., El-Bader, H., Amer, E., et al. (2019). Role of miRNA-210, miRNA-21 and miRNA-126 as diagnostic biomarkers in colorectal carcinoma: Impact of HIF-1α-VEGF signaling pathway. Mol. Cell. Biochem. 454, 177–189.
Sakamoto, K., and Maeda, S. (2010). Targeting NF-κB for colorectal cancer. Expert Opin. Ther. Targets 14, 593–601.
Schee, K., Boye, K., Abrahamsen, T., Fodstad, Ø, and Flatmark, K. (2012). Clinical relevance of microRNA miR-21, miR-31, miR-92a, miR-101, miR-106a and miR-145 in colorectal cancer. BMC Cancer 12:505. doi: 10.1186/1471-2407-12-505
Sethi, G., Shanmugam, M., Ramachandran, L., Kumar, A., and Tergaonkar, V. (2012). Multifaceted link between cancer and inflammation. Biosci. Rep. 32, 1–15.
Shan, J., Ji, W., Li, H., Tuxun, T., Lin, R., and Wen, H. (2011). TLR2 and TLR4 expression in peripheral blood mononuclear cells of patients with chronic cystic echinococcosis and its relationship with IL-10. Parasite Immunol. 33, 692–696. doi: 10.1111/j.1365-3024.2011.01335.x
Shariati, A., Razavi, S., Ghaznavi-Rad, E., Jahanbin, B., Akbari, A., Norzaee, S., et al. (2021). Association between colorectal cancer and Fusobacterium nucleatum and Bacteroides fragilis bacteria in Iranian patients: A preliminary study. Infect. Agents Cancer 16, 1–10. doi: 10.1186/s13027-021-00381-4
Shen, W., Jin, Z., Tong, X., Wang, H., Zhuang, L., Lu, X., et al. (2019). TRIM14 promotes cell proliferation and inhibits apoptosis by suppressing PTEN in colorectal cancer. Cancer Manage. Res. 11, 5725–5735. doi: 10.2147/CMAR.S210782
Stanilov, N., Miteva, L., Deliysky, T., Jovchev, J., and Stanilova, S. (2010). Advanced colorectal cancer is associated with enhanced IL-23 and IL-10 serum levels. Lab. Med. 41, 159–163.
Stolzer, I. (2022). Role of IFN-STAT Mediated Regulated Cell Death During Gastrointestinal Inflammation and Infection. Erlangen: Friedrich-Alexander-Universität Erlangen-Nürnberg (FAU).
Suehiro, Y., Sakai, K., Nishioka, M., Hashimoto, S., Takami, T., Higaki, S., et al. (2017). Highly sensitive stool DNA testing of Fusobacterium nucleatum as a marker for detection of colorectal tumours in a Japanese population. Ann. Clin. Biochem. 54, 86–91. doi: 10.1177/0004563216643970
Sun, Y., An, Q., Tian, X., Wang, Z., Guan, X., Dong, B., et al. (2016). Fusobacterium nucleatum infection is correlated with tumor metastasis and postoperative survival of colorectal cancer patients in China. Transl. Cancer Res. 5, 579–588.
Sung, H., Ferlay, J., Siegel, R., Laversanne, M., Soerjomataram, I., Jemal, A., et al. (2021). Global cancer statistics 2020: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA 71, 209–249.
Sur, D., Advani, S., and Braithwaite, D. (2022). MicroRNA panels as diagnostic biomarkers for colorectal cancer: A systematic review and meta-analysis. Front. Med. 9:915226. doi: 10.3389/fmed.2022.915226
Swidsinski, A., Dörffel, Y., Loening-Baucke, V., Theissig, F., Rückert, J., Ismail, M., et al. (2011). Acute appendicitis is characterised by local invasion with Fusobacterium nucleatum/necrophorum. Gut 60, 34–40. doi: 10.1136/gut.2009.191320
Tang, B., Lu, X., Tong, Y., Feng, Y., Mao, Y., Dun, G., et al. (2023). MicroRNA-31 induced by Fusobacterium nucleatum infection promotes colorectal cancer tumorigenesis. Iscience 26:106770. doi: 10.1016/j.isci.2023.106770
Thomas, A., Manghi, P., Asnicar, F., Pasolli, E., Armanini, F., Zolfo, M., et al. (2019). Metagenomic analysis of colorectal cancer datasets identifies cross-cohort microbial diagnostic signatures and a link with choline degradation. Nat. Med. 25, 667–678.
Tran, H., Thu, T., Nguyen, P., Vo, C., Doan, K., Nguyen Ngoc Minh, C., et al. (2022). Tumour microbiomes and Fusobacterium genomics in Vietnamese colorectal cancer patients. Npj Biofilms Microb 8:87. doi: 10.1038/s41522-022-00351-7
Trinchieri, G. (2003). Interleukin-12 and the regulation of innate resistance and adaptive immunity. Nat. Rev. Immunol. 3, 133–146.
Tunsjø, H., Gundersen, G., Rangnes, F., Noone, J., Endres, A., and Bemanian, V. (2019). Detection of Fusobacterium nucleatum in stool and colonic tissues from Norwegian colorectal cancer patients. Eur. J. Clin. Microbiol. Infect. Dis. 38, 1367–1376. doi: 10.1007/s10096-019-03562-7
Velsko, I., Chukkapalli, S., Rivera-Kweh, M., Chen, H., Zheng, D., Bhattacharyya, I., et al. (2015). Fusobacterium nucleatum alters atherosclerosis risk factors and enhances inflammatory markers with an atheroprotective immune response in ApoEnull mice. PLoS One 10:e0129795. doi: 10.1371/journal.pone.0129795
Viljoen, K., Dakshinamurthy, A., Goldberg, P., and Blackburn, J. (2015). Quantitative profiling of colorectal cancer-associated bacteria reveals associations between fusobacterium spp., enterotoxigenic Bacteroides fragilis (ETBF) and clinicopathological features of colorectal cancer. PLoS One 10:e0119462. doi: 10.1371/journal.pone.0119462
Wang, J., Huang, S., Zhao, M., Yang, M., Zhong, J., Gu, Y., et al. (2014). Identification of a circulating microRNA signature for colorectal cancer detection. PLoS One 9:e87451. doi: 10.1371/journal.pone.0087451
Wang, J., Liu, Y., Li, Y., Zheng, X., Gan, J., Wan, Z., et al. (2021). Exosomal-miR-10a derived from colorectal cancer cells suppresses migration of human lung fibroblasts, and expression of IL-6, IL-8 and IL-1β. Mol. Med. Rep. 23, 1–1.
Wang, Y., Chen, Z., and Chen, W. (2017). Novel circulating microRNAs expression profile in colon cancer: A pilot study. Eur. J. Med. Res. 22, 1–11. doi: 10.1186/s40001-017-0294-5
Wei, Z., Cao, S., Liu, S., Yao, Z., Sun, T., Li, Y., et al. (2016). Could gut microbiota serve as prognostic biomarker associated with colorectal cancer patients’ survival? A pilot study on relevant mechanism. Oncotarget 7:46158. doi: 10.18632/oncotarget.10064
Wong, S., Kwong, T., Chow, T., Luk, A., Dai, R., Nakatsu, G., et al. (2017). Quantitation of faecal Fusobacterium improves faecal immunochemical test in detecting advanced colorectal neoplasia. Gut 66, 1441–1448.
Wong, S., and Yu, J. (2019). Gut microbiota in colorectal cancer: Mechanisms of action and clinical applications. Nat. Rev. Gastroenterol. Hepatol. 16, 690–704.
Wu, C., Ng, S., Dong, Y., Ng, S., Leung, W., Lee, C., et al. (2012). Detection of miR-92a and miR-21 in stool samples as potential screening biomarkers for colorectal cancer and polyps. Gut 61, 739–745. doi: 10.1136/gut.2011.239236
Wu, J., Li, Q., and Fu, X. (2019). Fusobacterium nucleatum contributes to the carcinogenesis of colorectal cancer by inducing inflammation and suppressing host immunity. Transl. Oncol. 12, 846–851. doi: 10.1016/j.tranon.2019.03.003
Yachida, S., Mizutani, S., Shiroma, H., Shiba, S., Nakajima, T., Sakamoto, T., et al. (2019). Metagenomic and metabolomic analyses reveal distinct stage-specific phenotypes of the gut microbiota in colorectal cancer. Nat. Med. 25, 968–976. doi: 10.1038/s41591-019-0458-7
Yang, Y., Weng, W., Peng, J., Hong, L., Yang, L., Toiyama, Y., et al. (2017). Fusobacterium nucleatum increases proliferation of colorectal cancer cells and tumor development in mice by activating toll-like receptor 4 signaling to nuclear factor- κB, and up-regulating expression of microRNA-21. Gastroenterology 152, 851–866.e24.
Yu, T., Guo, F., Yu, Y., Sun, T., Ma, D., Han, J., et al. (2017). Fusobacterium nucleatum promotes chemoresistance to colorectal cancer by modulating autophagy. Cell 170, 548–563.e16. doi: 10.1016/j.cell.2017.07.008
Yu, T., Ma, P., Wu, D., Shu, Y., and Gao, W. (2018). Functions and mechanisms of microRNA-31 in human cancers. Biomed. Pharmacother. 108, 1162–1169.
Zeller, G., Tap, J., Voigt, A., Sunagawa, S., Kultima, J., Costea, P., et al. (2014). Potential of fecal microbiota for early-stage detection of colorectal cancer. Mol. Syst. Biol. 10:766.
Zhang, S., Cai, S., and Ma, Y. (2018). Association between Fusobacterium nucleatum and colorectal cancer: Progress and future directions. J. Cancer 9:1652. doi: 10.7150/jca.24048
Zhou, P., Yang, D., Sun, D., and Zhou, Y. (2022). Gut microbiome: New biomarkers in early screening of colorectal cancer. J. Clin. Lab. Anal. 36:e24359.
Keywords: Fusobacterium nucleatum, colorectal cancer, cytokines, miR-21, miR-31
Citation: Bostanghadiri N, Razavi S, Shariati A, Talebi M, Mirkalantari S, Emami Razavi A and Darban-Sarokhalil D (2023) Exploring the interplay between Fusobacterium nucleatum with the expression of microRNA, and inflammatory mediators in colorectal cancer. Front. Microbiol. 14:1302719. doi: 10.3389/fmicb.2023.1302719
Received: 26 September 2023; Accepted: 30 October 2023;
Published: 23 November 2023.
Edited by:
Swayam Prakash, University of California, Irvine, United StatesReviewed by:
Atieh Darbandi, Iran University of Medical Sciences, IranAfshana Quadiri, National Institute of Immunology (NII), India
Taniya Mitra, University of California, San Diego, United States
Copyright © 2023 Bostanghadiri, Razavi, Shariati, Talebi, Mirkalantari, Emami Razavi and Darban-Sarokhalil. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Davood Darban-Sarokhalil, ZGF2b29kX2RhcmJhbkB5YWhvby5jb20=; ZGFyYmFuLmRAaXVtcy5hYy5pcg==
 Narjess Bostanghadiri
Narjess Bostanghadiri Shabnam Razavi1,2
Shabnam Razavi1,2 Aref Shariati
Aref Shariati Shiva Mirkalantari
Shiva Mirkalantari Davood Darban-Sarokhalil
Davood Darban-Sarokhalil