- Institute for Research in Molecular Medicine (INFORMM), University Sains Malaysia, Penang, Malaysia
Background: S. Typhi is a Gram-negative bacterium that causes typhoid fever in humans. Its virulence depends on the TolC outer membrane pump, which expels toxic compounds and antibiotics. However, the role of TolC in the host cell adhesion and invasion by S. Typhi is unclear.
Objective: We aimed to investigate how deleting the tolC affects the adhesion and invasion of HT-29 epithelial and THP-1 macrophage cells by S. Typhi in vitro.
Methods: We compared the adhesion and invasion rates of the wild-type and the tolC mutant strains of S. Typhi using in vitro adhesion and invasion assays. We also measured the expression levels of SPI-1 genes (invF, sipA, sipC, and sipD) using quantitative PCR.
Results: We found that the tolC mutant showed a significant reduction in adhesion and invasion compared to the wild-type strain in both cell types. We also observed that the expression of SPI-1 genes was downregulated in the tolC mutant.
Discussion: Our results suggest that TolC modulates the expression of SPI-1 genes and facilitates the adhesion and invasion of host cells by S. Typhi. Our study provides new insights into the molecular mechanisms of S. Typhi pathogenesis and antibiotic resistance. However, our study is limited by the use of in vitro models and does not reflect the complex interactions between S. Typhi and host cells in vivo.
1. Introduction
Efflux pumps are present in all major pathogenic bacterial species lineages (Piddock, 2006a,b). In Gram-negative bacteria, this active efflux mainly contributes to the intrinsic resistance to several classes of antibiotics, dyes, and detergents (Li and Nikaido, 2004; Piddock, 2006a,b; Sun et al., 2014). In Gram-negative bacterial pathogens, TolC is an outer membrane efflux pump protein that facilitates efflux and contributes to virulence and pathogenesis (Piddock, 2006a,b). The expression of efflux pumps was observed along with the infection process of Gram-negative pathogens (Fernando and Kumar, 2013). Outer membrane efflux pump proteins (OMPs) have essential functions in the physiology of bacteria, such as adhesion and invasion of the host cell, resistance to host serum, maintenance of the membrane integrity, and passive and active transfer of substances (Tokuda, 2009). Although the biological functions of TolC homologs in several Gram-negative bacteria have already been reported, the role of TolC in Salmonella enterica serovar Typhi has not been investigated, especially its role in host cell adhesion and invasion. S. Typhi is a human-restricted pathogen that causes typhoid fever.
In this study, the role of S. Typhi TolC in the adhesion and invasion of host cells were first investigated using a tolC mutant. As the expression of Salmonella pathogenicity island 1 (SPI-1) genes for invasion (sipA, sipC, sipD, and invF) is required for penetration into host cells (Darwin and Miller, 1999a,b), we also investigated the expression of the invasion genes under SPI-1-inducing conditions.
2. Materials and methods
2.1. Bacterial strains and plasmids
A Salmonella enterica serovar Typhi (S. Typhi) strain (a clinical isolate from an acute typhoid fever patient), denoted as ST-WT was used as the wild-type strain in this study.
This study was conducted in accordance with the ethical principles of the Declaration of Helsinki and the EEC directive of 1986. The Salmonella enterica serovar Typhi strain was obtained from patients with acute typhoid fever subjects in Hospital Universiti Sains Malaysia (HUSM). The strain was deposited in the Bank of the Institute for Research in Molecular Medicine (INFORMM), Kubang Kerian, Kelantan, Malaysia. The collection and use of the strain were approved by the Committee for Human Ethical Clearance of Universiti Sains Malaysia, Kubang Kerian, Malaysia under ethical clearance number USMKK/PPP/JEPeM [229.3. (03)]. All participants gave their informed consent before enrollment in the study. Bacterial strains and plasmids used in this study are summarized in Table 1. All strains were routinely grown in Luria-Bertani (LB) agar and broth (Hi-media) at 37°C with antibiotics for selection when required.
2.2. Construction of tolC mutant
A tolC deletion mutant (ST-ΔtolC) was constructed using the one-step inactivation of the chromosomal gene method (Baba et al., 2006). The tolC gene of ST-WT strain was replaced by inserting the kanamycin resistance gene aph (3′)-II genes that confers kanamycin resistance to generate strain ST-ΔtolC. A complementation mutant (ST-∆tolC+) was also constructed by cloning the tolC gene, including its native promoter, into the pKK223-3 plasmid and transformed into ST-ΔtolC. Primers for construction of tolC mutant summarized in Table 2.
2.3. Growth of Salmonella enterica serovar Typhi strains
The growth of all S. Typhi strains was determined by measuring the optical density (OD 595 nm) of bacterial culture using LB broth in a microtiter plate at 37°C as previously described (Sheridan et al., 2013). The sample was placed in a microtiter plate, and bacterial growth was recorded at 2-h time intervals (Multiskan Spectrum, Thermo Scientific). All growth experiments were performed three times, and results were analyzed by plotting average values for each strain on a logarithmic graph with calculated standard deviations to use as error bars. Generation times were also calculated using the logarithmic growth phase of the line graph by the following equation;
(Where g is generation [doubling] time and T is time, ln is Natural log of given value). Values which appeared different to the parent strain were evaluated for significance at defined time points using a student’s ‘t’ test. OD end is the optical density at the end of the exponential growth phase, OD start is the optical density at the start of the exponential growth phase.
2.4. Efflux activity assay
Efflux activity was evaluated with the ethidium bromide agar screening method (Martins et al., 2006). Overnight cultures of all strains were swabbed onto LB agar plates containing 0.5 and 1 mg/L of ethidium bromide. The plates were incubated at 37°C for 16 h, and the fluorescence intensity associated with efflux pump function in the bacterial mass was photographed.
2.5. Adhesion and invasion assays
The adhesion and invasion of the S. Typhi strains were tested using in vitro adhesion and invasion assays with THP-1-derived human macrophages and HT-29 human epithelial cells according to the methods of Dibb-Fuller et al. (1999) and Buckley et al. (2006) with some modification. S. Typhi cells were grown overnight in 10 mL LB broth at 37°C. Cell monolayers were grown to confluence in 6-well plates, and the cells were then infected for 2 h at an infection multiplicity of 50. For the adhesion assay, cells were gently washed six times with phosphate-buffered saline (PBS) (pH 7.3) and then disrupted with 1 mL distilled water (Roche et al., 2005). First, all viable bacteria (intra- and extracellular) were counted as colony-forming units after plating serial dilutions with PBS. Then, the entry of S. Typhi into HT-29 cells and THP-1 macrophages was quantified by the invasion assay (also called gentamicin protection assay) as previously described (Amy et al., 2004) to quantify intracellular bacteria. All the bacterial strains tested have similar gentamicin susceptibility (data not shown). To calculate the bacterial adhesion, we subtracted the cfu/ml of the invaded bacteria from the cfu/ml of the total viable bacteria that were either adherent to the cell surface or inside the cell after washing step. Results are expressed as cfu/ml of adherent and invasive bacteria compared to the ST-WT strain.
All quantitative invasion assays were performed separately for each strain in triplicates. ST-∆tolC and ST-∆tolC+ were compared with the ST-WT reference strain using Student’s t-test. Each strain’s overall mean cfu/ml was calculated for each biological replicate.
2.6. Reverse transcription PCR of SPI-1 gene expression
Reverse transcription polymerase chain reaction (RT-PCR) was performed to measure the transcription of the invasion-related genes of S. Typhi, according to the study of Webber et al. (2009). Bacteria were grown in LB broth until the mid-log phase (OD600 of 0.6) containing 0.3 M NaCl for SPI-1-inducing conditions (Arricau et al., 1998). Total bacterial RNA was obtained from the bacteria by using the RNeasy mini kit (Qiagen) according to the manufacturer’s recommendation. Possible DNA contamination was removed by treating with DNase I (Sigma). The absence of DNA contamination was tested by PCR amplification using total RNA as a template and primers specific for the recA housekeeping gene (Wong et al., 2013). The purity and concentration of RNA were determined by measuring the optical density at 230, 260, and 280 nm before use (A260/280 ranged from 1.8 to 2.0). The quality of the RNA was assessed by gel electrophoresis and ethidium bromide staining.
Real-time RT-PCR was performed to measure the transcriptional level of the invF, sipA, sipC, and sipD, and the housekeeping gene recA was used as a control (Wong et al., 2013). Specific primer sequences were designed for each gene using the Primer3 software program (Table 3). First, 1 μg of DNase-treated total RNA from at least three independent cultures were reverse transcribed using random hexamers and Superscript III 1st Strand Kit (Invitrogen, Cat #18080-051). Then, the amplification was performed using the QuantiFast SYBR Green PCR Kit (Qiagen) on an Applied Biosystems™ 7500 Real-Time PCR System according to the manufacturer’s instructions. The amplification of recA for target gene expression normalization was conducted simultaneously with the amplification of targeted genes. Fold-change of expression level for a target gene was determined using the comparative Ct approach, whereby the Ct value of the target gene in each sample was normalized to recA, and the relative expression level of the target gene and the fold-change in gene expression were calculated (Livak and Schmittgen, 2001). The expression of a target gene was presented as the fold change relative to the ST-WT strain. Data were obtained in three separate experiments with three technical replicates. All results were analyzed by the Student’s t-test, and p values of <0.05 indicate significance.
3. Results
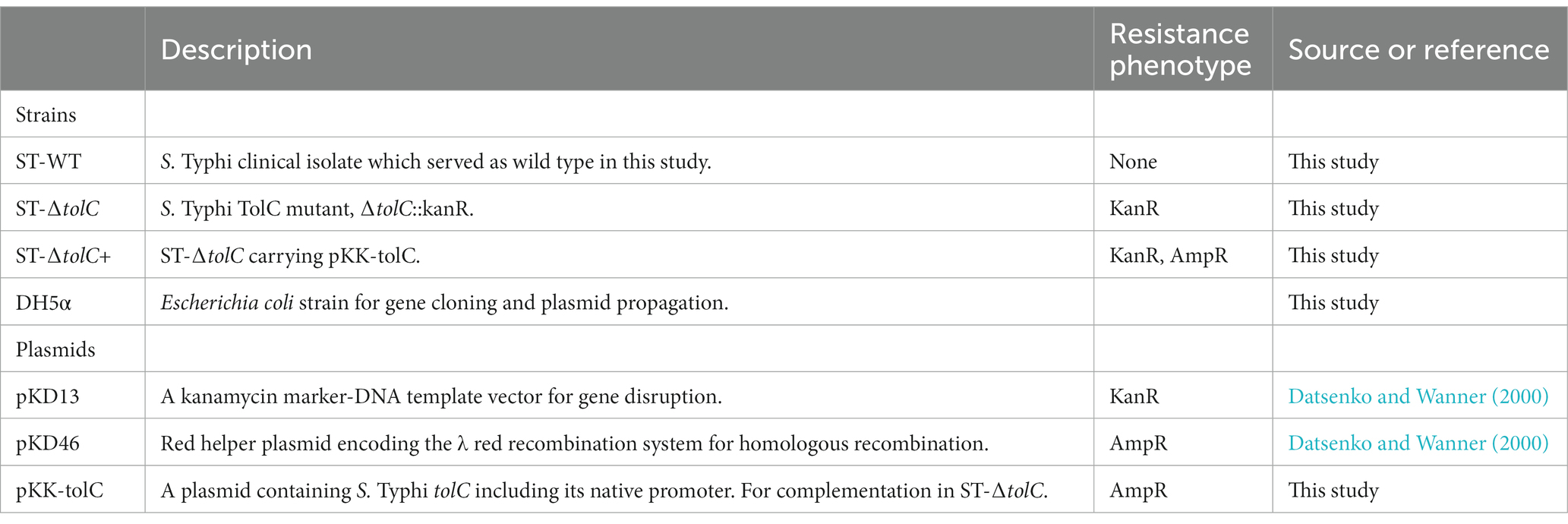
3.1. The growth of the tolC mutant was defective
To evaluate the fitness of the strains, we observed the growth of the strains by OD measurement. The ST-WT and ST-ΔtolC had similar doubling times of 112 and 116 min (p = 0.55), respectively (Figure 1A), while the ST-∆tolC+ had a shorter doubling time of ~80 min (p = 0.004). The ST-ΔtolC and ST-∆tolC+ strains arrived at the stationary stage at 8 h; however, ST-WT continued growing at a slower rate with an OD595 reading of ~0.63, ~0.70, and ~ 0.55, respectively. The growth of the bacteria reached a stationary phase after 18 h of incubation. The OD595 of the ST-WT and the ST-∆tolC+ strain was similar, with mean values of ~ 0.76 and ~ 0.74, respectively (p = 0.20). However, the OD595 of the ST-ΔtolC strain was significantly lower, with a mean value of ~0.56 (p < 0.05). Based on the growth curves (Figure 1A), it is deduced that the ST-ΔtolC cell growth defect may be due to the lack of the TolC efflux pump. In contrast, the growth defect of the mutant was rescued by the presence of the plasmid carrying tolC (ST-∆tolC+) complement strain.
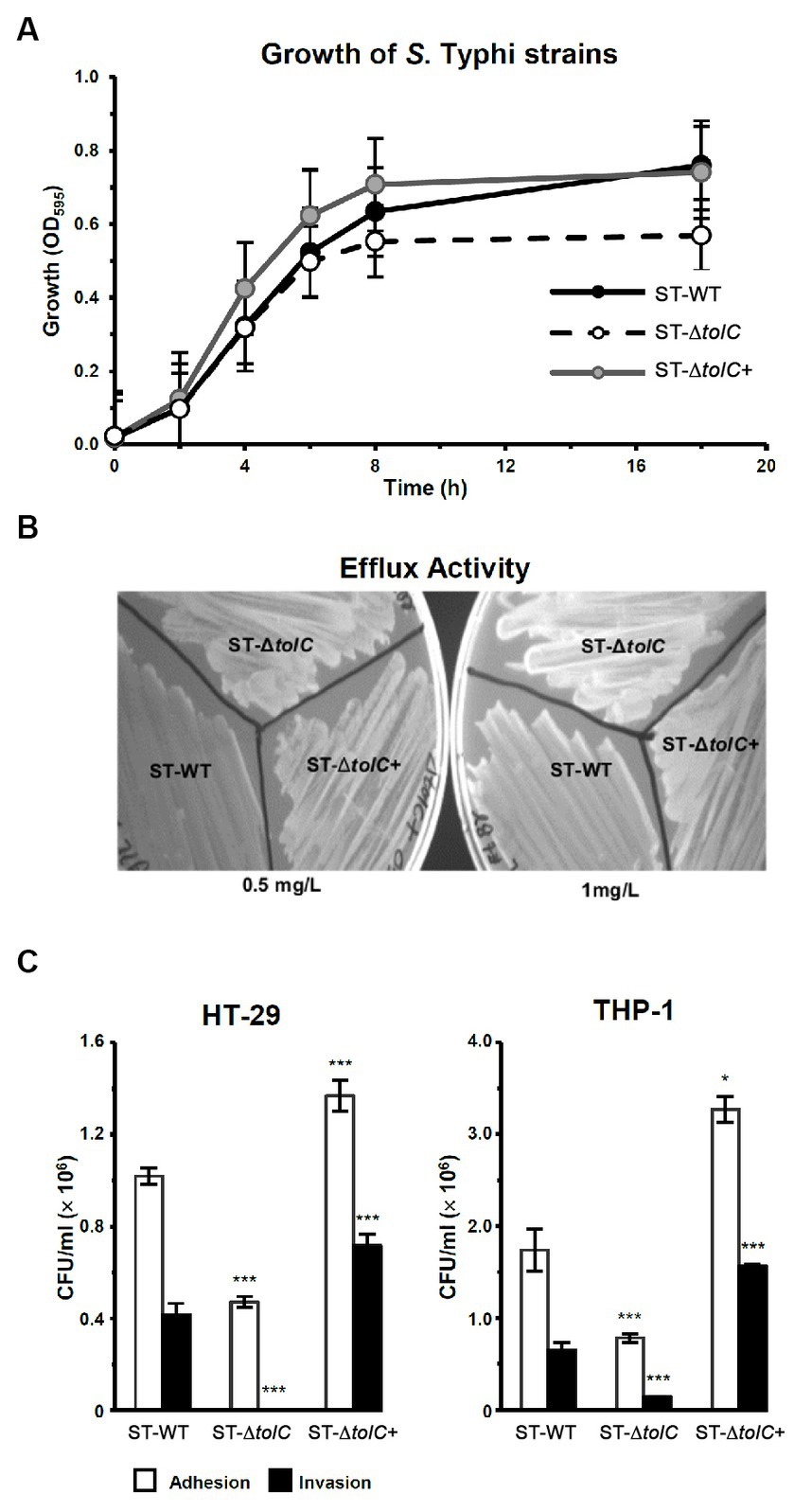
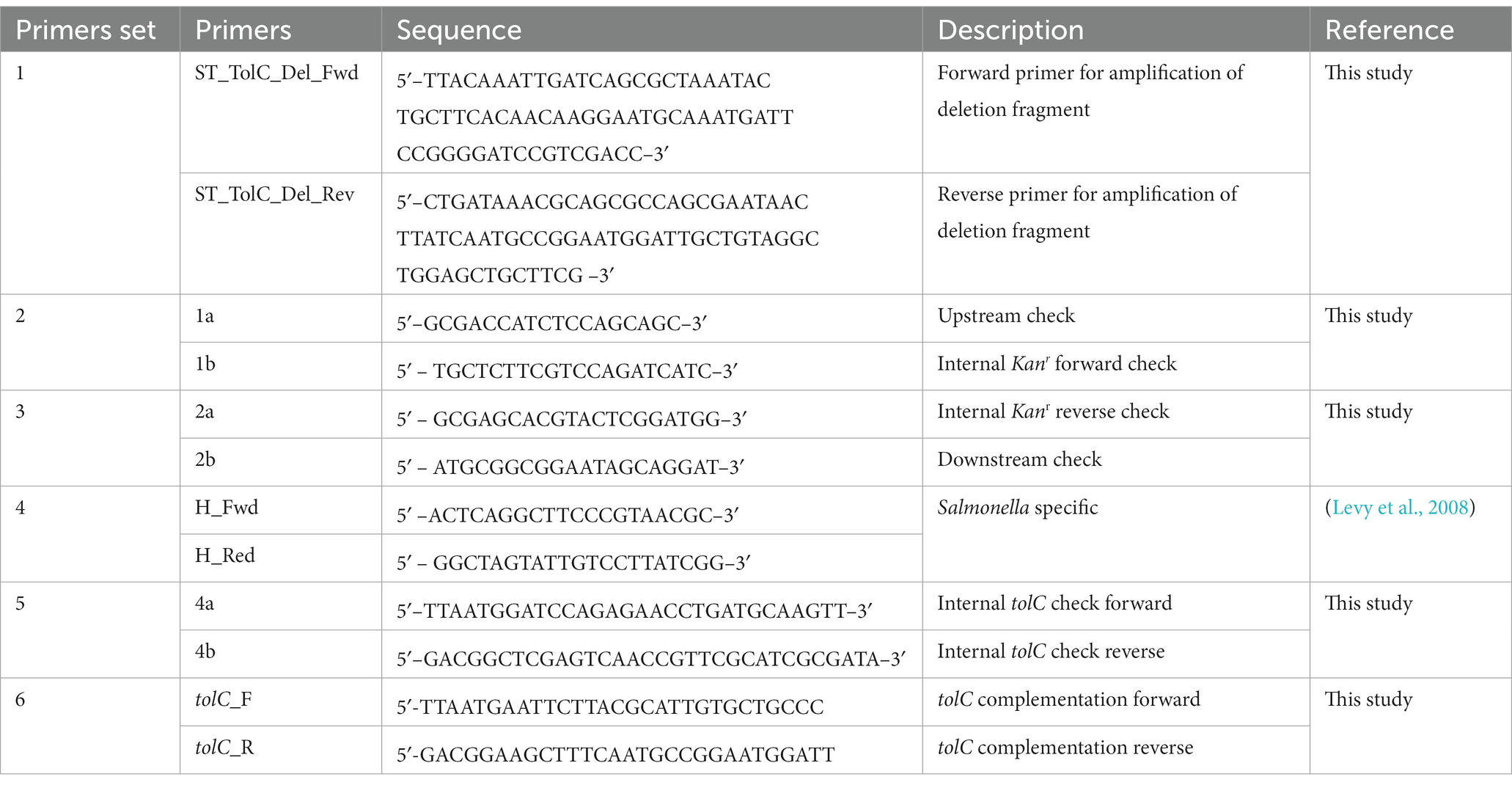
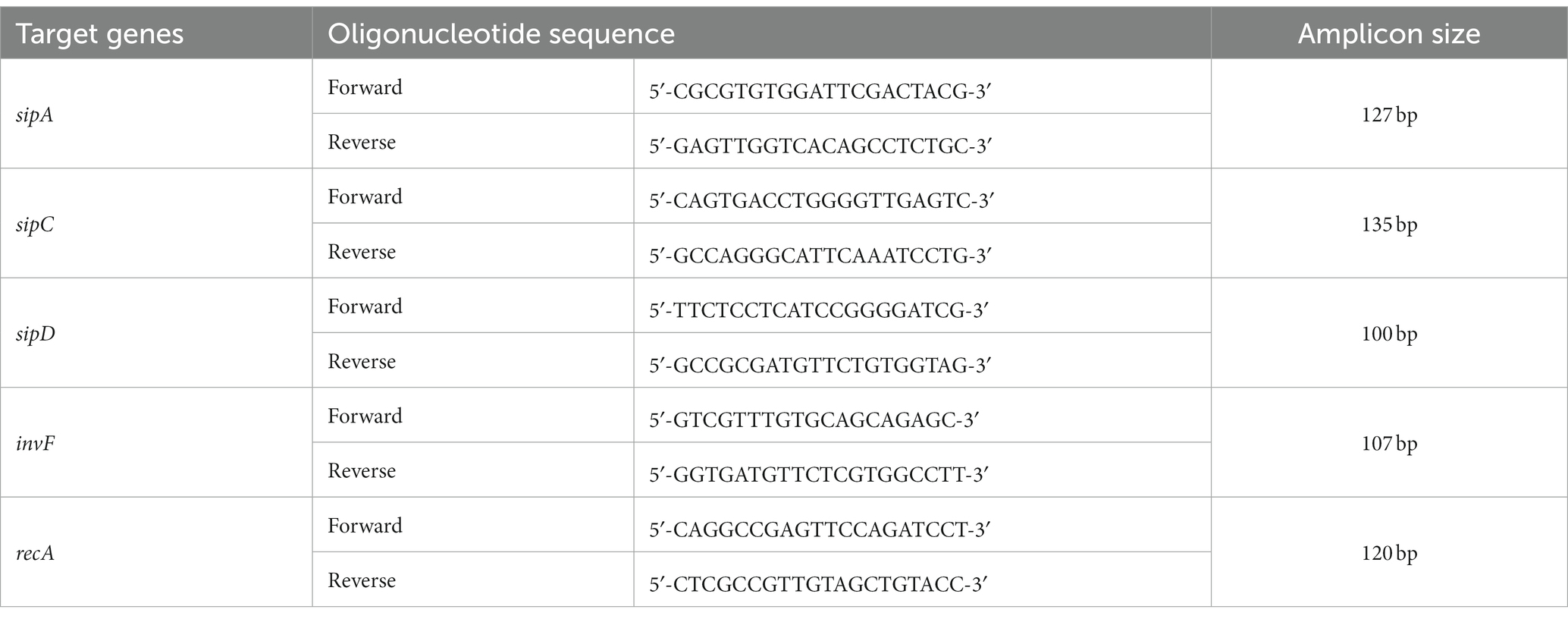
Figure 1. Deletion of S. Typhi tolc affects bacterial growth, efflux, adhesion, and invasion. (A) S. Typhi strains ST-WT, ST-∆tolC, and ST-∆tolC+ were grown at 37°C, and their OD was recorded for up to 18 h. (B) Evaluation of the efflux activity of S. Typhi strains. Strains were cultured on an LB plate containing 0.5 and 1.0 mg/L ethidium bromide overnight at 37°C and observed under UV light. (C) Evaluation of the adhesion and invasion activity of S. Typhi strains on HT-29 and THP-1 cells. Data are displayed as the mean of at least three separate experiments performed in triplicate ± standard deviation. Values returning a p value of ≤0.001 from a Student’s t-test comparing ST-∆tolC and ST-∆tolC+ strains to the ST-WT strain. The asterisks above the bars represent the significance of the t-test, *** < 0.001.
3.2. The efflux activity of the tolC mutant was impaired
To evaluate the efflux activity of the ST-ΔtolC strain, all strains (ST-WT and ST-ΔtolC+ as controls) were streaked on media containing ethidium bromide. After incubation, the plates were observed under UV light. The ST-WT appeared to fluoresce the least, followed by ST-ΔtolC+ and ST-ΔtolC (Figure 1B). Fluorescence intensity from the cells is inversely proportional to efflux pump function, indicating pump functionality in ST-WT and impairment in ST-ΔtolC (at both lower and higher ethidium bromide concentrations). Even though ST-ΔtolC+ was expected to have efflux activity similar to ST-WT, the strain’s efflux activity was affected (Figure 1B), suggesting that the TolC expressed from the plasmid could be different from the chromosome expressed protein in ST-WT.
3.3. The adhesion and invasion of tolC mutant were reduced
To investigate the adhesion and invasion abilities of the strains, the bacterial cells were added to the human intestine epithelial HT-29 and macrophage THP-1 cells. Declines in the adhesion and invasion abilities of ST-∆tolC were observed when the bacterial cell interacted with both host cell types in vitro (Figure 1C). The adhesion efficiency of ST-∆tolC was reduced by ~55% in both host cell types compared to ST-WT. The significant loss of invasion is most likely due to the loss of adhesion capability in ST-∆tolC because adhesion is a crucial step preceding invasion. Interestingly, the ST-∆tolC+ strain showed increased adhesion and invasion in both host cell types. We speculate that the presence of multiple copies of tolC on the plasmids likely contributed to this phenomenon.
3.4. Invasion-related genes were downregulated in the tolC mutant
To evaluate the direct efflux function of TolC and the indirect role of the presence of tolC on the expression of SPI-1 TTSS-1 genes, we cultured the strains in the SPI-1-inducing condition (0.3 M NaCl). We performed RT-PCR to quantify relative gene expression. The mRNA expression of invasion-related genes such as sipA, sipC, sipD, and invF was significantly reduced in ST-ΔtolC when compared with the ST-WT reference strain that was used as control; indeed, the transcripts of these genes revealed that transcriptions of invF, sipA, sipC, and sipD genes were decreased, 15–fold, 1.6–fold, 9.6–fold, and 2.5–fold, respectively, in the ST-ΔtolC when compared with the ST-WT reference strain. Although the complementation ST-∆tolC+ strain significantly increased the transcriptions of these genes, sipA (2.8-fold), invF (2.4-fold), sipC (1.2 -fold), and sipD (1.8-fold), when compared with ST-WT (Figure 2).
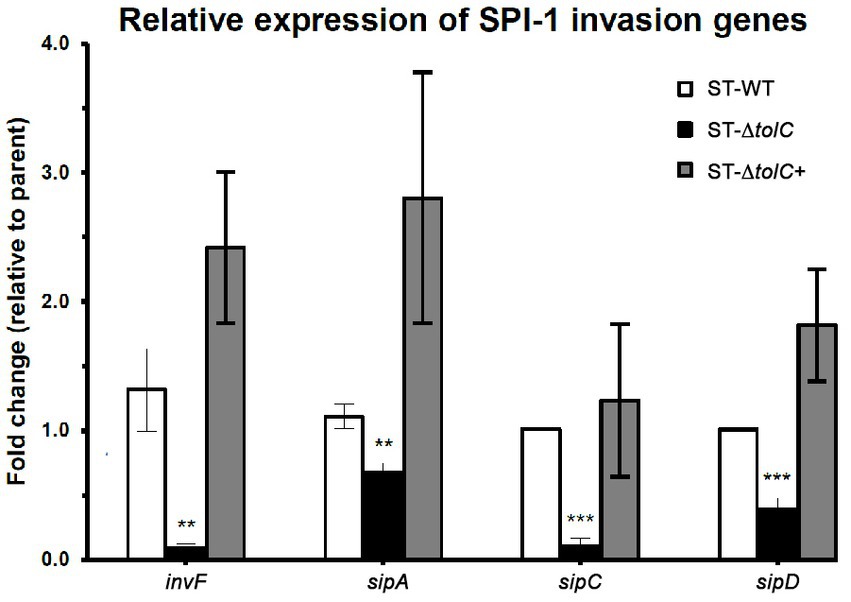
Figure 2. The relative expression of selected invasion-related SPI-1 genes in S. Typhi strains. The strains were cultured in SPI-1-inducing condition (0.3 M NaCl), and RT-PCR was performed to quantify relative gene expression. The expression of a target gene was presented as the fold change relative to the ST-WT strain used as a control in this study. White bars indicate ST-WT, black bars ST-∆tolC, and gray bars for ST-∆tolC+. Bars indicate the messenger RNA fold-changes observed in ST-∆tolC, and ST-∆tolC+ compared to their ST-WT reference strain with +/− standard deviation and the mean of three independent experiments. The asterisks above bars represent significance from a Student’s t-test, ** < 0.01, *** < 0.001.
4. Discussion
The molecular mechanisms of Salmonella host cell entry and its intracellular survival have been widely investigated in the past few decades, and key bacterial invasion factors (e.g., SPI-1 TTSS-1) have been identified (Kaufmann et al., 2001; Ribet and Cossart, 2015). While previous studies have reported TolC-related functions in S. Typhimurium virulence (i.e., colonization, persistence, adhesion, and invasion) (Buckley et al., 2006; Nishino et al., 2006), no studies have been reported for the role of TolC in S. Typhi. In this study, we hypothesized that the S. Typhi TolC would similarly play an essential role in bacterial adhesion and invasion during the infection of human cells and also investigated whether the lack of TolC will affect SPI-1 gene expression known to be upregulated during the bacterial invasion.
We first constructed a tolC mutant, and the analysis of its growth showed that during the log phase, the growth profiles of ST-ΔtolC were similar to the ST-WT strain; however, it reached the stationary phase earlier and at a lower OD (Figure 1A). One plausible explanation is that intracellular waste and metabolites accumulated due to impaired efflux, and this internal toxicity affects cell viability, thus leading to an earlier stationary phase and lower cell density. Alternatively, it may be due to the altered expression of genes involved in stress response, virulence, or metabolism that are regulated by tolC or its associated operons. Further studies are needed to elucidate the molecular mechanisms underlying the growth defect of ST-ΔtolC in the stationary phase and its implications for pathogenesis.
Our results are consistent with some previous reports on tolC mutants of other bacterial species. For example, Virlogeux-Payant et al. (2008) showed that a tolC mutant of Salmonella Typhimurium had a comparable growth rate to the wild-type strain based on the growth curve, but they did not report the growth characteristics of the mutant in the stationary phase. Webber et al. (2009) reported that a tolC mutant of Salmonella Typhimurium did not show a growth defect in the log phase compared to the wild-type strain. However, Santos et al. (2010) reported that a tolC mutant of Sinorhizobium meliloti had a similar growth rate to the wild-type strain for the first 8 h of growth, but then showed a reduced growth rate and decreased biomass formation (Santos et al., 2010). Similarly, in our study, ST-ΔtolC had reduced growth after 8 h compared to ST-WT.
The ST-ΔtolC also showed hyper-susceptibility to detergents and antibiotics (results shown in Supplementary material) as demonstrated by its weaker ability to efflux ethidium bromide, unlike the ST-WT strain (Figure 1B).
In the cell adhesion and invasion assay, the ST-∆tolC was significantly less invasive than the ST-WT reference strain in both the epithelium (HT-29) and macrophage (THP-1) cells. The invasion of ST-∆tolC was removed entirely in HT-29 cells and was approximately 20% in THP-1 cells, possibly due to the phagocytic activity of the macrophages. As predicted, the adhesion and invasion activity were restored to a higher level in the ST-∆tolC+ strain in both HT-29 and THP-1 (Figure 1C). These findings are consistent with the study on an S. Typhimurium tolC mutant where it was also reported to be less adherent to epithelial cells than its WT parent and that TolC is crucial for virulence-related phenotypes such as adhesion and invasion to the host cells (Buckley et al., 2006; Virlogeux-Payant et al., 2008).
The growth curves and invasion assays resulting from this report are consistent with our previous preliminary study that was done with other S. Typhi strains (Hussain et al., 2016).
Next, we wanted to see if the suppressed invasion activity of ST-ΔtolC was also linked to the TTSS-1 system, which is primarily associated with invasion (Galan, 2001). Using RT-PCR, we found lower expression of invasion-related genes of the TTSS-1 (invF, sipA, sipC, and sipD) (Figure 2). The transcription of invF, a transcriptional regulator which activates the transcription of other SPI-1 genes (Darwin and Miller, 1999a,b), was downregulated by fifteen-fold in the ST-ΔtolC compared to the ST-WT. When the tolC was complemented, the expressions of the SPI-1 genes in the ST-ΔtolC+ strain were higher than ST-WT. The higher gene expression levels of ST-∆tolC+ also reflect the strain’s higher adhesion and invasion activities (Figure 1C). In S. Typhimurium, deletion of another gene invH, located downstream of invF, was shown to partially impair the secretion of Sip effector proteins (SipABCD) (Pati et al., 2013).
The reduced expressions of the invasion-related genes are likely mediated by the lowered expression of invF in the ST-ΔtolC. This downregulation is in agreement with the observation in S. Typhimurium (tolC::aph), where the invF was significantly downregulated in the tolC mutant (Webber et al., 2009). The invF is a positive regulator of SPI-1 that has been confirmed to be essential for the expression of several SPI-1 genes (Darwin and Miller, 1999a,b).
5. Conclusion
In this study, we demonstrated that TolC-dependent efflux systems play a vital role in the adhesion and invasion of S. Typhi to host cells, which are key steps in the pathogenesis of typhoid fever. We also revealed that TolC influences the expression of SPI-1 genes, which encode the type III secretion system (TTSS-1) that mediates the invasion process. Our findings suggest that TolC is a multifunctional protein that modulates both the efflux activity and the virulence gene expression of S. Typhi. However, the exact mechanism by which TolC regulates SPI-1 genes remains unknown and requires further investigation. Moreover, the potential of TolC as a target for developing novel anti-virulence strategies against S. Typhi needs to be explored in future studies.
Data availability statement
The original contributions presented in the study are included in the article/Supplementary material, further inquiries can be directed to the corresponding authors.
Ethics statement
The studies involving humans were approved by ethical clearance number USMKK/PPP/JEPeM [229.3. (03)]. The studies were conducted in accordance with the local legislation and institutional requirements. Written informed consent for participation in this study was provided by the participants’ legal guardians/next of kin.
Author contributions
AH: Conceptualization, Data curation, Formal analysis, Investigation, Writing – original draft, Writing – review & editing. EO: Funding acquisition, Project administration, Resources, Supervision, Validation, Visualization, Writing – review & editing. PB: Conceptualization, Formal analysis, Funding acquisition, Project administration, Resources, Supervision, Validation, Visualization, Writing – review & editing. AI: Funding acquisition, Project administration, Resources, Supervision, Writing – review & editing. PK: Funding acquisition, Project administration, Resources, Supervision, Validation, Visualization, Writing – review & editing.
Funding
The author(s) declare financial support was received for the research, authorship, and/or publication of this article. This study was supported by the University Sains Malaysia Enteric Diseases Research Cluster Grants (1001/PSKBP/8630011 and PSKBP/86300111) and Postgraduate Research Grant (1001/CIPPM/846046). AH was financially supported by the USM Fellowship during his study.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2023.1301478/full#supplementary-material
References
Amy, M., Velge, P., Senocq, D., Bottreau, E., Mompart, F., and Virlogeux-Payant, I. (2004). Identification of a new Salmonella enterica serovar Enteritidis locus involved in cell invasion and in the colonisation of chicks. Res. Microbiol. 155, 543–552. doi: 10.1016/j.resmic.2004.03.005
Arricau, N., Hermant, D., Waxin, H., Ecobichon, C., Duffey, P. S., and Popoff, M. Y. (1998). The RcsB-RcsC regulatory system of Salmonella typhi differentially modulates the expression of invasion proteins, flagellin and Vi antigen in response to osmolarity. Mol. Microbiol. 29, 835–850. doi: 10.1046/j.1365-2958.1998.00976.x
Baba, T., Ara, T., Hasegawa, M., Takai, Y., Okumura, Y., Baba, M., et al. (2006). Construction of Escherichia coli K-12 in-frame, single-gene knockout mutants: the Keio collection. Mol. Syst. Biol. 2:2006.0008. doi: 10.1038/msb4100050
Buckley, A. M., Webber, M. A., Cooles, S., Randall, L. P., La Ragione, R. M., Woodward, M. J., et al. (2006). The AcrAB-TolC efflux system of Salmonella enterica serovar Typhimurium plays a role in pathogenesis. Cell. Microbiol. 8, 847–856. doi: 10.1111/j.1462-5822.2005.00671.x
Darwin, K. H., and Miller, V. L. (1999a). InvF is required for expression of genes encoding proteins secreted by the SPI1 type III secretion apparatus in Salmonella typhimurium. J. Bacteriol. 181, 4949–4954. doi: 10.1128/JB.181.16.4949-4954.1999
Darwin, K. H., and Miller, V. L. (1999b). Molecular basis of the interaction of Salmonella with the intestinal mucosa. Clin. Microbiol. Rev. 12, 405–428. doi: 10.1128/CMR.12.3.405
Datsenko, K. A., and Wanner, B. L. (2000). One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc. Natl. Acad. Sci. U S A, 97, 640–6645. doi: 10.1073/pnas.120163297
Dibb-Fuller, M. P., Allen-Vercoe, E., Thorns, C. J., and Woodward, M. J. (1999). Fimbriae- and flagella-mediated association with and invasion of cultured epithelial cells by Salmonella enteritidis. Microbiology 145, 1023–1031.
Fernando, D. M., and Kumar, A. (2013). Resistance-nodulation-division multidrug efflux pumps in gram-negative bacteria: role in virulence. Antibiotics 2, 163–181. doi: 10.3390/antibiotics2010163
Galan, J. E. (2001). Salmonella interactions with host cells: type III secretion at work. Annu. Rev. Cell Dev. Biol. 17, 53–86. doi: 10.1146/annurev.cellbio.17.1.53
Hussain, A., Ong, E. B. B., Phei, P. S. C., Hossain, K., Balaram, P., Ismail, A., et al. (2016). Role of TolC in virulence of salmonella enterica serovar Typhi. J. Pure Appl. Microbiol. 10:887.
Kaufmann, S. H., Raupach, B., and Finlay, B. B. (2001). Introduction: microbiology and immunology: lessons learned from Salmonella. Microbes Infect. 3, 1177–1181. doi: 10.1016/S1286-4579(01)01498-8
Levy, H., Diallo, S., Tennant, S. M., Livio, S., Sow, S. O., Tapia, M., et al. (2008). PCR method to identify Salmonella enterica serovars Typhi, Paratyphi A, and Paratyphi B among Salmonella Isolates from the blood of patients with clinical enteric fever. J. Clin. Microbiol. 46, 1861–1866. doi: 10.1128/jcm.00109-08
Li, X. Z., and Nikaido, H. (2004). Efflux-mediated drug resistance in bacteria. Drugs 64, 159–204. doi: 10.2165/00003495-200464020-00004
Livak, K. J., and Schmittgen, T. D. (2001). Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods 25, 402–408. doi: 10.1006/meth.2001.1262
Martins, M., Santos, B., Martins, A., Viveiros, M., Couto, I., Cruz, A., et al. (2006). An instrument-free method for the demonstration of efflux pump activity of bacteria. In Vivo 20, 657–664.
Nishino, K., Latifi, T., and Groisman, E. A. (2006). Virulence and drug resistance roles of multidrug efflux systems of Salmonella enterica serovar Typhimurium. Mol. Microbiol. 59, 126–141. doi: 10.1111/j.1365-2958.2005.04940.x
Pati, N. B., Vishwakarma, V., Jaiswal, S., Periaswamy, B., Hardt, W. D., and Suar, M. (2013). Deletion of invH gene in Salmonella enterica serovar Typhimurium limits the secretion of Sip effector proteins. Microbes Infect. 15, 66–73. doi: 10.1016/j.micinf.2012.10.014
Piddock, L. J. (2006a). Clinically relevant chromosomally encoded multidrug resistance efflux pumps in bacteria. Clin. Microbiol. Rev. 19, 382–402. doi: 10.1128/CMR.19.2.382-402.2006
Piddock, L. J. (2006b). Multidrug-resistance efflux pumps - not just for resistance. Nat. Rev. Microbiol. 4, 629–636. doi: 10.1038/nrmicro1464
Ribet, D., and Cossart, P. (2015). How bacterial pathogens colonize their hosts and invade deeper tissues. Microbes Infect. 17, 173–183. doi: 10.1016/j.micinf.2015.01.004
Roche, S. M., Gracieux, P., Milohanic, E., Albert, I., Virlogeux-Payant, I., Temoin, S., et al. (2005). Investigation of specific substitutions in virulence genes characterizing phenotypic groups of low-virulence field strains of Listeria monocytogenes. Appl. Environ. Microbiol. 71, 6039–6048. doi: 10.1128/AEM.71.10.6039-6048.2005
Santos, M. R., Cosme, A. M., Becker, J. D., Medeiros, J. M., Mata, M. F., and Moreira, L. M. (2010). Absence of functional TolC protein causes increased stress response gene expression in Sinorhizobium meliloti. BMC Microbiol. 10:180. doi: 10.1186/1471-2180-10-180
Sheridan, A., Lenahan, M., Condell, O., Bonilla-Santiago, R., Sergeant, K., Renaut, J., et al. (2013). Proteomic and phenotypic analysis of triclosan tolerant verocytotoxigenic Escherichia coli O157: H19. J. Proteome 80, 78–90. doi: 10.1016/j.jprot.2012.12.025
Sun, J., Deng, Z., and Yan, A. (2014). Bacterial multidrug efflux pumps: mechanisms, physiology and pharmacological exploitations. Biochem. Biophys. Res. Commun. 453, 254–267. doi: 10.1016/j.bbrc.2014.05.090
Tokuda, H. (2009). Biogenesis of outer membranes in gram-negative bacteria. Biosci. Biotechnol. Biochem. 73, 465–473. doi: 10.1271/bbb.80778
Virlogeux-Payant, I., Baucheron, S., Pelet, J., Trotereau, J., Bottreau, E., Velge, P., et al. (2008). TolC, but not AcrB, is involved in the invasiveness of multidrug-resistant Salmonella enterica serovar typhimurium by increasing type III secretion system-1 expression. Int. J. Med. Microbiol. 298, 561–569. doi: 10.1016/j.ijmm.2007.12.006
Webber, M. A., Bailey, A. M., Blair, J. M. A., Morgan, E., Stevens, M. P., Hinton, J. C. D., et al. (2009). The global consequence of disruption of the AcrAB-TolC efflux pump in Salmonella enterica includes reduced expression of SPI-1 and other attributes required to infect the host. J. Bacteriol. 191, 4276–4285. doi: 10.1128/JB.00363-09
Keywords: Salmonella Typhi, TolC, Salmonella pathogenicity island 1, invasion, efflux pump protein, adhesion, pathogenesis, antibiotic resistance
Citation: Hussain A, Ong EBB, Balaram P, Ismail A and Kien PK (2023) Deletion of Salmonella enterica serovar Typhi tolC reduces bacterial adhesion and invasion toward host cells. Front. Microbiol. 14:1301478. doi: 10.3389/fmicb.2023.1301478
Edited by:
George Grant, University of Aberdeen, United KingdomReviewed by:
France Daigle, University of Montreal, CanadaTimothy James Wells, The University of Queensland, Australia
Derek Pickard, University of Cambridge, United Kingdom
Copyright © 2023 Hussain, Ong, Balaram, Ismail and Kien. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Ashraf Hussain, YXhoMTIyNEBtaWFtaS5lZHU=; Eugene Boon Beng Ong, ZXVnZW5lQHVzbS5teQ==
†Present address: Ashraf Hussain, John P. Hussman Institute for Human Genomics, University of Miami Miller School of Medicine, Miami, FL, United States
 Ashraf Hussain
Ashraf Hussain Eugene Boon Beng Ong
Eugene Boon Beng Ong Prabha Balaram
Prabha Balaram