
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
REVIEW article
Front. Microbiol. , 11 January 2024
Sec. Microbial Physiology and Metabolism
Volume 14 - 2023 | https://doi.org/10.3389/fmicb.2023.1294182
Saccharomyces cerevisiae is a eukaryotic organism with a clear genetic background and mature gene operating system; in addition, it exhibits environmental tolerance. Therefore, S. cerevisiae is one of the most commonly used organisms for the synthesis of biological chemicals. The investigation of fatty acid catabolism in S. cerevisiae is crucial for the synthesis and accumulation of fatty acids and their derivatives, with β-oxidation being the predominant pathway responsible for fatty acid metabolism in this organism, occurring primarily within peroxisomes. The latest research has revealed distinct variations in β-oxidation among different fatty acids, primarily attributed to substrate preferences and disparities in the metabolic regulation of key enzymes involved in the S. cerevisiae fatty acid metabolic pathway. The synthesis of lipids, on the other hand, represents another crucial metabolic pathway for fatty acids. The present paper provides a comprehensive review of recent research on the key factors influencing the efficiency of fatty acid utilization, encompassing β-oxidation and lipid synthesis pathways. Additionally, we discuss various approaches for modifying β-oxidation to enhance the synthesis of fatty acids and their derivatives in S. cerevisiae, aiming to offer theoretical support and serve as a valuable reference for future studies.
Fatty acids, a class of alkane compounds with carboxyl groups, are widely used in the chemical, medicine, food, and agricultural sectors. Conventional fatty acids are primarily prepared by cracking animal and vegetable oils and petroleum extracts, and by olefin oxidation. In recent years, owing to the shortage of petroleum resources and the rising prices of petrochemical products (Röttig et al., 2010), the synthesis of fatty acids and their derivatives using microorganisms has gained significant interest (Peralta-Yahya et al., 2012).
The whole genome sequencing and phenotypic analysis of S. cerevisiae are more comprehensive compared to those of oil-producing yeast strains. Moreover, as a food-safe strain (Son et al., 2023), it plays a pivotal role as a chassis organism for the efficient production of fatty acids and their derivatives.
The synthesis of fatty acids occurs when glucose is sufficient, and acetyl-CoA synthesizes fatty acids under the action of fatty acid synthase, forming related lipid derivatives in the endoplasmic reticulum and cytoplasm (Gajewski et al., 2017). Under the condition of a low-carbon source, β-oxidation is the only fatty acid degradation pathway can provide energy in S. cerevisiae. Long-chain fatty acids (LCFA) are activated by the fatty acyl-CoA synthetase of S. cerevisiae, then transported to the peroxisomes through the ABC transporter for β-oxidation (Ferreira et al., 2018). Medium-chain fatty acids are activated by Faa2p and pex11p, located on peroxisome membranes, as medium-chain lipoyl-CoA, and transported into peroxisomes for β oxidation.
Saturated fatty acids can be directly metabolized via β-oxidation, whereas unsaturated fatty acids must first be converted to trans-dienoyl-CoA by auxiliary enzymes before entering the β-oxidation pathway (Yocum et al., 2022). A β-oxidation cycle is the process of releasing an acetyl-CoA molecule from the carboxyl terminus of a fatty acid. Fatty acids are first activated by fatty acyl-CoA synthetase to form fatty acyl-CoA, and then dehydrogenated by fatty acyl-CoA oxidase to form trans-2-enoyl-CoA. Trans-2-enoyl-CoA is hydrated and dehydrogenated by the multifunctional enzyme Fox2 to form 3-ketoyl-CoA. Finally, 3-ketoacyl-CoA removes one molecule of acetyl-CoA under the action of 3-ketoyl-CoA thiolase Pot1p, and then proceeds to the next β-oxidation cycle. With the release of acetyl-CoA, fatty acids are eventually degraded into acetyl-CoA, which is then transported to the cytoplasm or mitochondria to be used in other biological pathways. However, in addition to the key enzymes mentioned above, the β-oxidation process requires cofactors such as CoA, NADPH, NADH, and ATP.
Fatty acyl-CoA is a key intermediate in fatty acid metabolism and transformation in S. cerevisiae. In addition to being degraded into acetyl-CoA during β-oxidation, it is an essential intermediate for the synthesis of fatty acid derivatives, such as storage lipids and membrane esters (Figure 1).
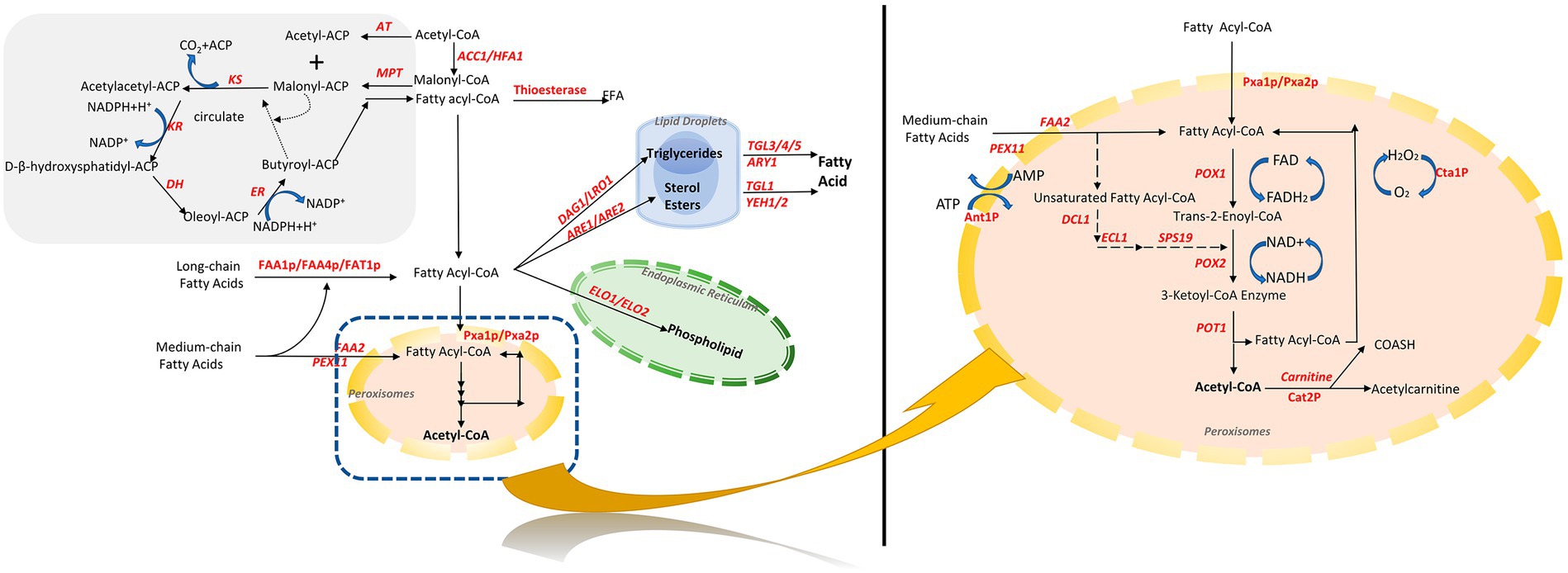
Figure 1. Fatty acid synthesis and metabolic pathways in S. cerevisiae. Orange is peroxisomes, green is endoplasmic reticulum, and blue is lipid droplets. Acetyl-CoA and malonic acid monoacyl-CoA synthesize fatty acids in the cytosol through fatty acid synthase. Fatty acids are activated in the cytosol as fatty acyl-CoA, followed by the synthesis of phospholipids by elongation enzymes in the endoplasmic reticulum, and triglycerides and sterol esters in the cytosol, which are located inside the sterol esters, which are in the center of the lipid droplets. Exogenous and endogenous fatty acids are finally degraded to acetyl-CoA by a unique β-oxidation pathway within S. cerevisiae. PGI, phosphoglucose isomerase; PDC, pyruvate decarboxylase; ALD, acetaldehyde dehydrogenase; Acs, acetyl-CoA synthetase; ACC1, acetyl CoA carboxylase; FAS, fatty acid synthase.
The intracellular metabolism of fatty acids plays a crucial role in the accumulation of fatty acids, and investigating the functionality and regulatory expression of enzymes involved in key metabolic pathways is beneficial for controlling fatty acid production in engineered strains. The construction of the reverse pathway for fatty acid β-oxidation is currently a prominent area of research in synthetic fatty acid pathways, and studying the substrate preferences and functional properties of key enzyme catalysis within the β-oxidation pathway is helpful to enrich the catalytic element library of the β-reversal pathway.
Studies have shown that a variety of ester acyl-CoA synthases (EC 6.2.1.3) are present in S. cerevisiae, including Faa1-4p and Fat1-2p (Watkins and Ellis, 2012), which are localized in the endoplasmic reticulum, lipid granules, plasma membrane, vacuolar membrane, cell periphery, and peroxisomes (Black and DiRusso, 2007). They play a key role in the transportation and activation of fatty acids and have certain preferences for fatty acids of different lengths (Ullah et al., 2021), there are similarities between oil-producing yeast and S. cerevisiae in fatty acid metabolism, as shown in Table 1.
Faa1p is an important protein involved in the transport and activation of fatty acids (Li et al., 2007) and is mainly located in lipid granules, the plasma membrane, the endoplasmic reticulum, and mitochondria (Hu et al., 2019; Zhu et al., 2019). Studies have shown that Faa1p plays a key role in the metabolism of long-chain fatty acids (Zhou et al., 2016). Three fatty acyl-CoA (Faa1p-6xHis, Faa2p-6xHis, and Faa3p-6xhis) were isolated and purified from S. cerevisiae at 25°C and pH 7.1 by affinity chromatography. Analysis of the activity of the three ester acyl-CoA synthetases on different saturated fatty acid carbon chains (C3–C24) showed that Faa1p has a wide range of substrate catalytic activities on C10–C18 fatty acids, with its most suitable substrate being pentadecanoic acid (Knoll et al., 1994).
Faa4p, which is localized in the endoplasmic reticulum and lipid droplets (Gaspar et al., 2022), acts on long-chain fatty acids (Black and DiRusso, 2007) and has high catalytic effects on C14–C18 fatty acids (Melton et al., 2013). In another study, after using fluorescence-labeled oleate as substrate at a final concentration of 100 μM for 10 min, the intracellular oleyl-CoA concentration in wild type S. cerevisiae increased by almost six fold, while that in the ΔFAA4 strain decreased to approximately 50% of that in the wild type strain (Færgeman et al., 2001). Faa1p and Faa4p play major roles in activating intracellular C14 and C16 saturated fatty acids in S. cerevisiae (Knoll et al., 1995). Knockout of FAA1 or FAA4 alone did not effectively reduce the activation and consumption of intracellular fatty acids, whereas their combined knockout significantly reduced the activation of cytoplasmic fatty acids (Scharnewski et al., 2008).
Fat1p, an ultra-long-chain fatty acyl-CoA synthetase located in the endoplasmic reticulum, is mainly responsible for the transport and activation of C20–C26 ultra long-chain fatty acids. The activation of 90% of C24 fatty acids in S. cerevisiae cells is completed by Fat1p, and FAT1 knockdown hinders the consumption of ultra long-chain fatty acids (Choi and Martin, 1999). Fat1p and Faa1p or Faa4p are required for the transport of long-chain fatty acid in S. cerevisiae (Zou et al., 2003). When FAA1, FAA4, and FAT1, the three synthase genes that are mainly responsible for activating fatty acids, are knocked out at the same time, most of the free fatty acids in the cytosol cannot be activated into fatty acyl-CoA, which means that they cannot be transported to the peroxisomes for β-oxidative degradation, and the fatty acids in the cytosol are accumulated, reaching 490 mg/L (Leber et al., 2015).
Faa2p is located in the peroxisome membrane (Poudyal and Paul, 2022) and its catalytic center is situated at its C-end site. It belongs to a class of fatty acyl-CoA synthetases (Van Roermund et al., 2001) reported to have a predilection for medium-chain fatty acids (C6–C12 or C9–C13; Knoll et al., 1994). S. cerevisiae lacking Faa2p cannot effectively utilize medium-chain fatty acids, such as caprylic acid, decanoic acid, and dodecanoic acid, via β-oxidation (Zou et al., 2003). It has been reported that Faa2p acts synergistically with Fat1p in the activation of ultra long-chain fatty acids (Falcon et al., 2010). After single knockout of FAT1, oleic acid induces Faa2p expression in peroxisomes, which leads to ultra-long chain synthase activity. After continuing to knock out the FAA2 gene, the residual ultra-long chain acyl-CoA activity in strains that double-knocked out FAT1 and FAA2 genes was less than 5% of wild-type strains and less than 20% of single-knockout FAT1 strains (Choi and Martin, 1999).
Only a few studies have been published on the function of Faa3p. Through the induction of FAA3 overexpression in Escherichia coli, Faa3p was found to have catalytic activity on 9-hexadecanoic acid and 9-ocadecanoic acid (Knoll et al., 1994). The activation of fatty acid transport in S. cerevisiae, as well as the activation of the five ester acyl CoA synthetase genes, FAA1, FAA2, FAA4, FAT1, and FAT2, involve some unidentified proteins (Van Roermund et al., 2000). In a previous study, the acyl-CoA synthetases genes FAA1 and FAA4 were completely knocked out, radiolabeled oleic acid was added to the medium, and gas chromatography analysis revealed that Faa1p and Faa4p were mainly responsible for the utilization of intracellular fatty acids (Scharnewski et al., 2008). Compared with S. cerevisiae, Yarrowia lipolytica fatty acyl-CoA synthetase has a transport function in addition to the role of activating fatty acids and the intracellular Long-chain fatty acids is activated to fatty acyl-CoA by fatty acyl-CoA synthetase ACS I. Long chain fatty acyl-coA is degraded by the transfer protein into the peroxisome, while medium and short chain fatty acids enter the peroxisome directly to be activated by ACSII and then undergo β-oxidation (Zhang et al., 2022).
Fox1p (EC 1.3.3.6) is the second key enzyme in the S. cerevisiae β-oxidation pathway; it is located in the peroxisome matrix and oxidizes acyl-CoA to trans-2,3-enoyl-CoA (Han et al., 2017). Following FOX1 knockout, S. cerevisiae was found not to grow normally on a medium with oleic acid as the sole carbon source (Lefevre et al., 2013), suggesting that Fox1p is the only fatty acyl-CoA oxidase in S. cerevisiae. FOX1 transcription is regulated by intracellular glucose, ethanol, and long-chain fatty acid levels (Wang et al., 1994). No FOX1 transcription was detected when the glucose concentration in the medium was >0.1% (Wang et al., 1992). However, when ethanol was used as the sole carbon source for culturing, a small amount of FOX1 mRNA in the stable state was detected. Furthermore, when the medium contained only oleic acid, higher FOX1 transcription levels were detected, with its mRNA levels being approximately 25 and 10 times higher than that in the glucose- and ethanol-containing media, respectively. Moreover, it has been reported that FOX1 transcription is strictly regulated by the carbon source (Stanway et al., 1995).
Oleogenic yeast cells often contain multiple fatty acyl-CoA oxidase genes. For example, Yarrowia lipolytica contains six fatty acyl-CoA oxidase genes (ACOX1–6). Acox1p is inactive, and in its presence alone, the strain cannot grow on a medium with oleic acid as the sole carbon source (Luo et al., 2002). Acox2p oxidizes medium- and long-chain acyl-CoAs and has good catalytic activity on C10–C14 straight chain acyl-CoAs (Kim and Kim, 2018). Acox3p preferentially oxidizes short-chain acyl-CoA, and Yarrowia lipolytica cells lacking Acox2p and Acox3p exhibited normal growth on a medium with glucose; however, the activity of intracellular fatty acyl-CoA oxidase was only one-fifth the total oxidase activity. This suggests that Acox2p and Acox3p are important acyl-CoA oxidases in Y. lipolytica (Mlíčková et al., 2004). Acox4p exhibits catalytic activity on both medium- and long-chain fatty acids (C6–C16), but has a predilection for long-chain fatty acids; in addition, Acox5p exhibits significant catalytic activity on fatty acids longer than 12 carbon atoms (Hilmi Ibrahim et al., 2020). Three fatty acyl-CoA oxidases, Pox2p, Pox4p, and Pox5p, have been identified in Candida tropicalis; Pox4p is a long-chain fatty acyl-CoA oxidase (Cao et al., 2017). Plant cells also typically contain several acyl-CoA oxidase isoenzymes, which are specific to long, medium, and short-chain fatty acids depending on the structural function of the enzyme (Zhang et al., 2017).
Fox2p [Includes: D-3-hydroxyacyl-CoA dehydratase (EC 4.2.1.-); 3-hydroxyacyl-CoA dehydrogenase (EC 1.1.1.35)] is a multifunctional S. cerevisiae β-oxidation enzyme with both enoyl-CoA hydratase and hydroxyl-CoA dehydrogenase activity; it catalyzes the transformation of trans-2,3-enoyl-CoA to 3-ketoacyl-CoA (Knoblach and Rachubinski, 2018). Researchers have attempted to induce FOX2 expression in the cytoplasm of S. cerevisiae; however, no enzyme activity was detecte, probably because FOX2 cannot fold into the correct conformation in the cytoplasm of S. cerevisiae (Lian and Zhao, 2015).
FOX2 transcription also requires oleic acid induction (Fickers et al., 2011; Darvishi, 2012) and is regulated by the Pip2-Oaf1 complex (Bergenholm et al., 2018). The oleic acid-activated transcription factor, Pip2p, which is a subunit of the Pip2-Oaf1 complex, contains a typical Zn (2)-Cys (6) cluster domain. In the absence of glucose in the culture medium but with sufficient oleic acid, both Pip2 and Oaf1 bind to OREs in the promoter region of the gene that encodes the peroxisome protein (Otzen et al., 2013) to form a heterodimer (Han et al., 2017) that positively regulates the transcription of genes involved in fatty acid β-oxidation and that of RNA polymerase II (Gurvitz et al., 1999). Rottensteiner et al. (1996) found that strains with PIP2 deletion exhibited impaired growth, could not transcribe β-oxidation-related enzymes, and had reduced peroxisome number and size in a medium with oleate as the sole carbon source. These results indicate that PIP2 and OAF1 are essential genes for fatty acid-induced peroxisome proliferation and the regulation of fatty acid oxidation; these findings also indicate that fatty acids can activate PIP2, thereby improving the transcription of β-oxidation-related genes.
Only one thiolase gene, POT1, has been identified in the S. cerevisiae genome (Huang et al., 2022). Mercaptase, which plays an important role in lipid metabolism, is localized in the mitochondrial membrane space and peroxisomal matrix as a homologous dimer. The sequences of the first 16 amino acids at its N-terminus are the same as the localization sequences of thiolases (Huang et al., 2022).
Studies on the crystal structure of S. cerevisiae peroxisome thiolases have revealed their binding pattern to substrate molecules and their reaction mechanisms (Mathieu et al., 1997). Pot1p (EC 2.3.1.16) catalyzes linear fatty acid carbon chain shortening during β-oxidation and together with CoA catabolizes 3-ketoacyl-CoA into acetyl-CoA and acyl-CoA (Lee et al., 2009). Each oxidation cycle was found to release one acetyl-CoA molecule until the fatty acid was completely converted into acetyl-CoA (Infant et al., 2021). POT1 deletion was found to induce abnormal growth morphology (van Roermund et al., 2022) and a significant decrease in intracellular ATP content in S. cerevisiae, confirming that its deletion had an effect on β-oxidation blockade (Weber et al., 2020).
Three 3-ketoacyl-CoA thiolases have been identified in Candida albicans: Pot1p, Fox3p, and Pot13p. Pot1p and Fox3p share high homology with the S. cerevisiae Pot1p (Otzen et al., 2013).
In addition to the key enzymes directly involved in β-oxidation in S. cerevisiae, fatty acid metabolism in the organism also involves peroxisome regulatory proteins. Pxa1p/Pxa2p is a protein complex located on the peroxisome membrane, the primary function of which is to transport long-chain ester acyl-CoA activated in the cytoplasm to peroxisomes for β-oxidation (Baker et al., 2015). Pxa1p/Pxa2p controls the balance between internal and external acyl-CoA and fatty acid levels of peroxidase, and regulates acetyl-CoA transport according to cellular demand for acetyl-CoA (van Roermund et al., 2021).
The PEX gene family encodes a variety of specific peroxisome-associated proteins (Nuttall et al., 2011) and regulates various biochemical reactions in peroxisomes (Table 1). Pex11p, located on the peroxisome membrane, participates in the transportation of medium-chain fatty acids from the cytoplasmic matrix to peroxisomes. During the process of β-oxidation transport, medium-chain fatty acids are first transported to peroxisome by Pex11p, are activated by Faa2p to medium-chain fatty acyl-CoA, and then undergo β-oxidation.
The number, size, and function of peroxisomes are closely related, and the larger the peroxisomes, the stronger the β-oxidation function, but the number will be smaller. The Pex11p family is involved in the regulation of peroxisome proliferation and size. However, the exact function of Pex11p is unclear. Some researchers believe that the deletion of PEX11 may induce decreased peroxisome function and the inability to use fatty acids as carbon source in S. cerevisiae, and that its overexpression may induce the formation of numerous peroxisomes with significantly reduced volumes (Huber et al., 2012). Other researchers hold a different view, suggesting that PEX11 deletion induces the formation of large peroxisomes in S. cerevisiae (Van Roermund et al., 2000). Through extensive research on the regulation of peroxisome proliferation, in addition to Pex11p, several PEX11 isoenzymes, such as Pex25p, Pex27p, Pex28p, Pex29p, Pex23p, Pex31p, and Pex32p, have been identified (Mast et al., 2016).
Aside from their involvement in the regulation of peroxisome body proliferation in S. cerevisiae, multiple PEX genes (PEX3, PEX19 PEX5, PEX22, PEX1, and PEX6) have been found to participate in protein transport and positioning in the peroxisome enzyme body; their specific functions are shown in Table 2.
Peroxisome-related proteins (Glover et al., 1994), which include matrix and membrane proteins (Neuberger et al., 2003), have a peroxisome localization signal (PTS sequence S/A-K/R-L/M, R-L/I-X(5)-HL). These PTS-targeted sequences include PTS1, PTS2, and PEX19BS. Matrix proteins contain the PTS1 or PTS2 sequences, which are transferred to the peroxisome matrix through binding to the cytoplasmic receptors, PEX5 or PEX7. Peroxisome membrane proteins, which have the PEX19BS signal, bind to PEX19 membrane protein receptors and are transferred to the peroxisome membrane.
The acyl-CoA thioesterase, Pte1p, which is the only acyl-CoA hydrolase identified in S. cerevisiae (Ntamack et al., 2009), is localized in the peroxisome matrix and participates in the hydrolysis of acyl-CoA, with free fatty acid release (Caswell et al., 2022). PTE1 is expressed exogenously in E. coli; a wide range of substrates have been identified for the enzyme, including long-chain (C14–C18), medium-chain (C8–C12), and short-chain (C4–C6) ester acyl-CoAs (Ntamack et al., 2009). Pte1p is more active on short- and medium-chain acyl-CoA than on long-chain acyl-CoA; these include C4, C6, C10, and C12 fatty acids. Furthermore, it has a predilection for unsaturated enoyl-CoA (Kawaguchi et al., 2021).
Four acyl-CoA thioesterases were identified in mice (Table 3). ACOT3 and ACOT5 are long- and medium-chain acyl-CoA thioesterases, respectively (Chinopoulos, 2021). ACOT8 has broad substrate specificity and hydrolyzes straight and branched ester acyl-CoAs. It has high homology with S. cerevisiae PTE1. Acot4p is a succinyl-CoA thioesterase (Tillander et al., 2017) (Table 3).
Thioesterase is a key enzyme required for the β-oxidation cycle and plays an important role in maintaining the balance of free fatty acid and coenzyme A levels. It requires five essential enzymes (CAB1–CAB5) to catalyze a five-step reaction synthesis, which can be used to synthesize acetyl-CoA or enter other reaction pathways (Nielsen, 2014). The coenzyme A released by thioesterase can enter the next round of the β-oxidation cycle, and acetyl-CoA can also be synthesized into other reaction pathways, which is conducive to the intracellular circulation of coenzyme A (Zhang et al., 2021).
In addition to the aforementioned key enzymes and regulatory genes involved in β-oxidation, some unsaturated fatty acids can enter the β-oxidation pathway only by means of auxiliary enzymes (Baker et al., 2015). When acyl-CoA substrates cannot be recognized by the key β-oxidation enzyme, Pox2p, a series of auxiliary enzymes are required to transform the substrates into suitable forms for β-oxidation. SPS19 encodes peroxisome 2,4-dienoyl-CoA reductase, which metabolizes 2,4-dienoyl-CoA in the peroxisome to 3-enoyl-CoA in a NADPH-dependent process (Robert et al., 2005). The dodecenoyl-CoA isomerase, Eci1p, also known as ∆3(cis), ∆2(trans)-enoyl-CoA isomerase, which exhibits elevated transcription levels in oleic acid-containing media, can convert 3-enoyl-CoA into trans-2-enoyl-CoA. It is a member of the hydratase/isomerase protein family and a hexameric enzyme comprising six identical 32 kDa subunits with 280 residues each (Mursula et al., 2000; Ntamack et al., 2009).
The beta-oxidation process also requires the participation of cofactors, such as ATP and CoA, necessitating the involvement of a series of key cofactor transport-associated enzymes. (Ant1p) YPR128cp is a peroxisome ATP/AMP transporter and is necessary for peroxisome proliferation. This protein is localized in the peroxisome membrane and belongs to a subgroup of S. cerevisiae medium-chain fatty acid ATP/AMP transporters (El Moualij et al., 1997). YPR128cp was found to show the highest homology with C. albicans PMP47, Plasmodium falciparum adenine nucleotide translocase, and homo PMP34 (Van Roermund et al., 2001).
During the process of fatty acid degradation, H2O2 produced is degraded by catalase Cta1p. When the acyl-CoA oxidase, Fox1p, oxidizes acyl-CoA to trans-2-enoyl-CoA, H2O2 is degraded by Cta1p, thereby preventing cellular H2O2 toxicity (Lefevre et al., 2013; Klug and Daum, 2014).
In S. cerevisiae, aside from participating in the β-oxidation metabolic pathway, fatty acids can also be used as precursors for the synthesis of lipid substances (Yadav et al., 2018), providing material for cell membrane synthesis and allowing for the accumulation of intracellular fats (Gossing et al., 2018). When it comes to synthesizing products such as ceramides or other fatty acid derivatives, Yarrowia lipolytica has a great advantage over S. cerevisiae in terms of yield. However, Saccharomyces cerevisiae, as a gene model strain, is a good research object, which can provide metabolic pathway modification ideas for other oil-producing yeasts (Su et al., 2023).
There are three main fatty acid derivatives of S. cerevisiae, which are phospholipids, sphingolipids, and neutral lipids. Neutral lipids are mainly composed of sterols and triacylglycerol. Among them, phospholipids, sterols and sphingolipids are important components of cell membranes (Lv et al., 2023). When the supply of carbon sources in S. cerevisiae is insufficient, neutral lipids stored in the cell are degraded to produce FFA (Free long-chain fatty acids). FFA is activated to form fatty acyl-CoA, which enters β-oxidation to provide energy to cells (Figure 2).
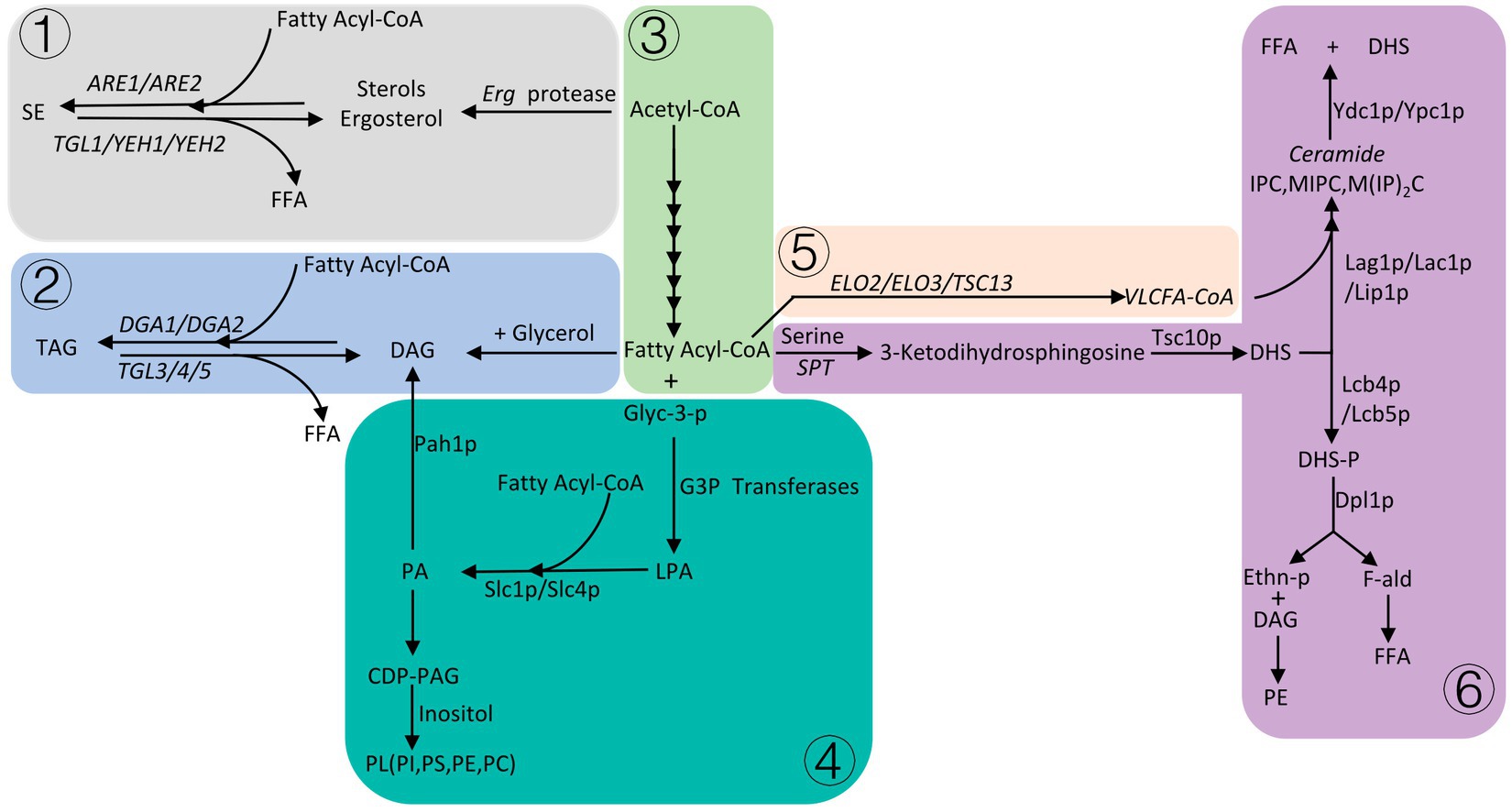
Figure 2. Please refer to Section 5 for details, the gray area ① is the formation process of sterol esters, the light blue area ② is the process of triglyceride formation, the light green area ③ is the abbreviation of fatty acid synthesis, the dark green area ④ is the phospholipid formation process, the light orange area ⑤ is the process of fatty acid elongation, and the purple area ⑥ is the sphingolipid anabolic process.
In S. cerevisiae, sterols are necessary for the integrity of cell membranes, and maintain the integrity, fluidity, and permeability of the membrane structure (Jordá and Puig, 2020). The synthesis of sterols occurs mainly in the endoplasmic reticulum, and the end product of the synthesis pathway is ergosterol (Khatri and Choudhary, 2023). Ergosterol starts from acetyl-CoA and is synthesized by nearly 30 proteases, ergosterol esters/sterol esters (SE) are formed by the action of fatty acyl-CoA and ergosterol/sterols under the action of sterol acyltransferases (ARE1/ARE2) (Koch et al., 2014). The balanced regulation of sterol synthesis pathways in yeast is very important, and excess sterols can cause toxicity to cells. At this point, cells induce proteasomal degradation of HMG-CoA reductase (HMGR), resulting in reduced sterol synthesis (Wang et al., 2023). The amount of sterols in yeast is very low, the rate of sterolipid synthesis is relatively stable during the logarithmic phase of yeast growth, and sterolipid synthesis significantly increases when yeast growth enters the stationary phase (Alvarez-Vasquez et al., 2011), under aerobic conditions, yeast is prone to produce excess sterols. In order to reduce the toxicity caused by excess sterols, sterols can be transferred to the plasma membrane in the form of sterol acetate and then secreted into culture media, or sterols can be esterified and stored in lipid droplets (LD) inside the cell (Pagac et al., 2011).
Triacylglycerol (TAG) is formed by the glycerol backbone and fatty acyl-CoA under the action of diacylglycerol O-acyltransferase (DGA1/LRO1), which can be used as energy storage and as a component of cell membranes (Khatri and Choudhary, 2023). When cells are starved, sterol esterases (TGL1/YEH1/YEH2) degrade sterol esters stored in LD to free fatty acids, and difunctional triglyceride lipase/lysophosphatidyletha nolamine acyltransferase (TGL3/4/5, ARY1) degrades triacylglycerol to FFA, which participates in β-oxidative degradation to acetyl-CoA for cellular energy (Ke et al., 2023).
The phospholipids (PL) in S. cerevisiae mainly include phosphatidylcholine (PC), phosphatidylethanolamine (PE), phosphatidylinositol (PI), and phosphatidylserine (PS). Phosphatidic acid (PA) is a central metabolite in the synthesis of PL (Engelhardt et al., 2023), and a series of synthesis occurs in the endoplasmic reticulum. First, fatty acyl-CoA and glycerol-3-phosphate (Glyc-3-P) synthesized lysophosphatidic acid (LPA) through G3P acyltransferases encoded by SCT1 (GAT2) and GPT2 (GAT1) in the endoplasmic reticulum (Yasukawa et al., 2023), and LPA and fatty acyl-CoA formed PA through the LPA acyltransferase Slc1p/Slc4p in the endoplasmic reticulum, and then formed cytidine diphosphate diacylglycerol (CDP-DAG). CDP-DAG is a precursor for the synthesis of PL, which forms PI, PS, PE, PC under the action of celluloalcohol (Matos et al., 2023). PA can be cleaved into diacylglycerol (DAG) by phospholipidase phosphatase Pah1p, which is then combined with fatty acyl-CoA to act as a storage lipid in cells (Speer et al., 2024). Although the composition of organelle membranes varies, film-forming PL is necessary for the formation of various organelles. The expression of synthetic PL genes was affected by various growth conditions such as temperature, growth medium, pH and nutrients, but PA, as the central metabolite of synthetic PL, had the greatest impact on PL. PA is regulated by a variety of enzymes, the most important being PA phosphatase Pah1p and DG kinase Dgk1p, overexpression of DGK1 or knockout of PAH1 can enhance PA synthesis (Khaddaj et al., 2023).
Sphingolipids (SL) are mainly synthesized in the endoplasmic reticulum and are transported to the plasma membrane or other organelles after synthesis (Klug and Daum, 2014). The synthesis of complex ceramides begins with fatty acyl-CoA (mainly palmitoyl-CoA or stearoyl-CoA) and condenses with serine in a serine palmitoyltransferase (SPT) complex to synthesize 3-ketodihydrosphingosine. 3-Ketodihydrosphingosine can be transformed into dihydrosphingosine (DHS) by the 3-ketosphingosine reductase (Tsc10p) (Schäfer et al., 2023), and one of the pathways for DHS is to bind to VLCFA (C20–C26) (Very-long-chain fatty acids) acyl-CoA (extended by the enzymes encoded by the three genes ELO2, ELO3, and TSC13) in response to the ceramide synthase complex (Lag1p, Lac1p, and Lip1p), and then undergo a series of reactions to produce ceramides, including inositol phosphoceramide (IPC)/Mannositide phosphoceramide (MIPC)/mannosyldiinositide phosphoceramide (M(IP)2C) (Willet et al., 2023). The alkaline dihydrofibrinoenzymes Ydc1p and Ypc1p are responsible for catalyzing ceramides into FFA and DHS. The other route is phosphorylation catalyzed by Lcb4p and Lcb5p to form dihydrosphingosine phosphate (DHS-P), which is cleaved by the DHS phospholyase Dpl1p to synthesis ethanolamine. Phosphate (Ethn-P) and fatty aldehyde (F-ald). Ethn-P can synthesize PE with DAG through a series of reactions, F-ald is metabolized to FFA. FFA is activated in the cytosol and then enters the β-oxidative degradation for energy.
As a single cell-type strain, S. cerevisiae exhibits good biosafety and robustness. Compared with strains that are harmful to humans, S. cerevisiae is certified as a GRAS (Generally Recognized as Safe) strain (Lian et al., 2018), produces fatty acids and products that are safer, the comparison of oil-producing yeast to S. cerevisiae is shown in Table 4. Modifying the fatty acid metabolic pathway is of great value in improving the synthesis of fatty acids and their derivatives.
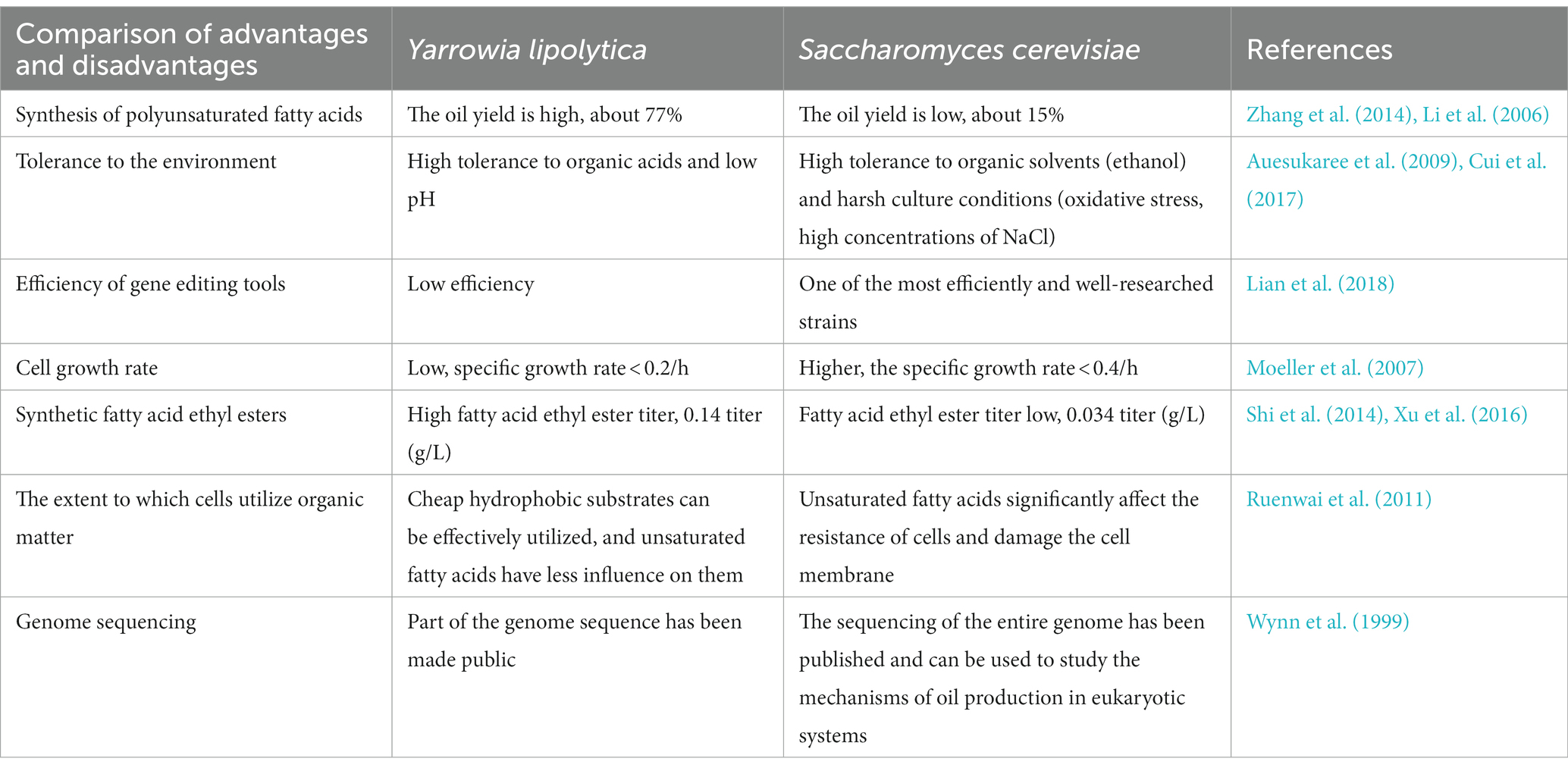
Table 4. Comparison of the advantages and disadvantages of Saccharomyces cerevisiae and Yarrowia lipolytica.
Medium-chain fatty acids contain 8–12 carbon atoms. Compared with long-chain fatty acids, esters related to medium-chain fatty acids have an aroma and can be combined with other substances to form fuels; polymers can also be synthesized in materials, and are used in fields such as health care drugs, cosmetics, and foods to speed up metabolism (Garces Daza et al., 2023). The accumulation of saturated long-chain fatty acids under fermentation conditions induces the release of medium-chain acyl-CoA by fatty acid synthase (FAS) complexes (Mbuyane et al., 2022). To induce the production of more medium-chain fatty acids in S. cerevisiae, Pox1p was knocked down, while Pox2p (from Y. lipolytica) and carnitine O-octanoyl transferase Crotp overexpression was induced; this resulted in the secretion and accumulation of medium-chain fatty acids in the medium, with a corresponding increase in medium-chain fatty content. The intracellular medium-chain fatty acid (C8:0, C10:0, and C12:0) titers in Δ pox1 [pox2+] 和 Δpox1 [pox2+,crot+] train were 2.26 and 1.87 fold higher than those in WT strain, and their extracellular titers in the modified strain were 3.29 and 3.34 fold higher than those in the WT strain (Chen et al., 2014a).
Long-chain fatty acids are components of cell membranes, which have strong inhibitory properties against foodborne pathogens such as Salmonella, can effectively regulate intestinal health after use, and are widely used in medical and food fields (Borreby et al., 2023). Currently, they are typically obtained from plants, animals, and petroleum. Producing them using microorganisms is a renewable approach. Fatty acids (16C–18C) are synthesized in yeast from acetyl-CoA by the action of type I FAS (Nancolas et al., 2017). By inducing the overexpression of the acetyl-CoA carboxylase, Acc1p, the fatty acid synthase, RtFAS (Rhodosporidium toruloides FAS), and E. coli thioesterase, TesA, in combination with the knock out of the two main acyl-CoA synthetase genes, FAA1 and FAA4, as well as that of the torulosis acyl-CoA oxidase gene, POX1, fatty acid production was significantly improved to 10.4 g/L in S. cerevisiae (Zhou et al., 2016).
Genetic engineering is used to regulate cell metabolic pathways to increase the accumulation of FFA in yeast cells (Zhang et al., 2020). Firstly, the conversion of FFA to acyl-CoA was blocked by knocking out FAA1 and FAA4 genes (Chen et al., 2014b). A truncated version of acyl-CoA thioesterase ACOT5 (Acot5s), which encodes Mus musculus peroxisomal acyl-CoA thioesterase 5, is then expressed in the cytoplasm, enhancing the conversion of Acyl-CoA to FFA. Strains with double deletion of FAA1 and FAA4 genes and overexpression of thioesterase ACOT5s increased FFA levels nearly 6.43 times in the medium, and the proportion of unsaturated fatty acids (UFA) was higher. In addition, the expression levels of genes associated with fatty acid synthesis, ACC1, FAS1, FAS2, and OLE1, were also increased in RT-PCR analysis. These results suggest that FFA accumulation in yeast cells can be increased by regulating acyl-coA metabolism.
To address the imbalance of redox factors in FFA production (acetyl-CoA supply, low NADH/NAD ratio), FFA-producing strains with four gene deletions (faa1, Δfaa4, Δpox1, Δfaa4) were used. ΔHFD1 as a chassis cell (Zhou et al., 2016) knocked out two endogenous NAD-dependent glycerol-3-phosphate dehydrogenase genes GPD1 and GPD2 (to reduce redox imbalance during glucose synthesis of fatty acids). Overexpression of NADP-dependent glyceraldehyde-3-phosphate dehydrogenase (GapN) in heterostreptococcus mutants (Zhang et al., 2016) balanced the redox factors of NAD and NADH in the cytoplasm. Overexpression of the bacterial pyruvate dehydrogenase PDH complex (increased supply of acetyl-CoA) significantly increased cell growth, decreased ethanol consumption, and decreased glycerol accumulation. The PDH7 strain produced 840.5 mg/L FFA at 72 h, and the final experimental results showed that the optimal fatty acid producing strain PDH7 produced 840.5 mg/L FFAs in the shaker, which was 2.08% higher than that of the control strain. At the same time, the distribution ratios of saturated and unsaturated FFA (C16 and C18) also increased (Zhang et al., 2020).
The synthesis of fatty alcohols using microorganisms has several applications (Hu et al., 2020). Biosynthetic natural higher fatty alcohols (medium-chain fatty alcohols) can be used as the basic raw materials for fine chemical products such as cosmetics, detergents, surfactants, plastic plasticization, etc., and medium-chain fatty alcohols have high energy density properties and can be used as alternative fuels to traditional diesel. Most of the fatty alcohols currently used are extracted from petroleum or produced from fatty acids extracted from vegetable oils (oilseeds). Biosynthetic fatty alcohols are sustainable. Fatty acids and fatty aldehyde intermediates are usually used for synthesis reactions (Yaguchi et al., 2018). Fatty alcohol synthesis in wild-type S. cerevisiae is completed in the cytoplasm. After fatty acids are activated by fatty acyl-CoA synthetase in the cytoplasm, a portion of the activated fatty acids is reduced to form fatty aldehydes and alcohols. By knocking out the fatty alcohol oxidation gene, HFD1, and inducing the overexpression of alcohol dehydrogenase and aldehyde reductase, the fatty alcohol yield can reach 1.5 g/L (Zhou et al., 2016). To produce more medium-chain fatty alcohols in peroxisomes, the overexpression of fatty acyl-CoA reductase (TaFAR), which utilizes medium-chain fatty acyl-CoA produced in peroxisomes to produce medium-chain fatty alcohols, but no 1-tetradecanool production was found in the assay results. On this basis, acetyl-CoA carboxylase (Acc1p) and ATP-dependent citrate lyase (Aclp) overexpression was induced in S. cerevisiae to increase fatty acid titers in the cytoplasm; finally, the medium-chain fatty alcohol titer in the modified strain was increased to 1.3 g/L (Sheng et al., 2016).
The formation of fatty acids and their derivatives in S. cerevisiae using acetyl-CoA is catalyzed by fatty acid synthetase, and this requires the use of ATP and ACP (acyl carrier protein), as well as the reducing power of NAPDH. Compared with this pathway, reversing β-oxidation requires less energy and has greater potential in fatty acid synthesis.
Thus, induction in the cytoplasm can reverse the overexpression of core enzymes of β-oxidative function (Lian and Zhao, 2015), including acetyl-CoA synthase (ACS), β-ketoacyl-CoA synthase (KS), β-ketoacyl-CoA reductase (KR), β-hydroxyacyl-CoA dehydrase (HTD), trans-2-ene-CoA reductase (TER), and thioesterase (CpFatB1), and can increase the production of short- and medium-chain fatty acids. “N-butanol” accumulation was induced through the overexpression of CoA-acylated aldehyde dehydrogenase (EcEutE) (Eriksen et al., 2014) and clostridium acetylbutanol dehydrogenase (CaBdhB). In addition, acyl-CoA:ethanol O-acyltransferase (EEB1/EHT1) overexpression was induced to successfully synthesize fatty acid ethyl esters (Strijbis and Distel, 2010).
To carry out the β-oxidation reversal reaction more efficiently, the supply mechanism of redox cofactors in the β-oxidation process was explored, and MCFA was used as the product to optimize the conditions. Stoichiometric analysis of the β-oxidation pathway showed that NADH was the key factor affecting the oxidation efficiency of β, and the highest titer of MCFA reached 4.7 g/L after strengthening the supply of acetyl-CoA and NADH, overexpressing the formate dehydrogenase of Candida tropicalis, and optimizing the acetate reassimilation pathway (Wu et al., 2017). (This section refers to Figure 3).
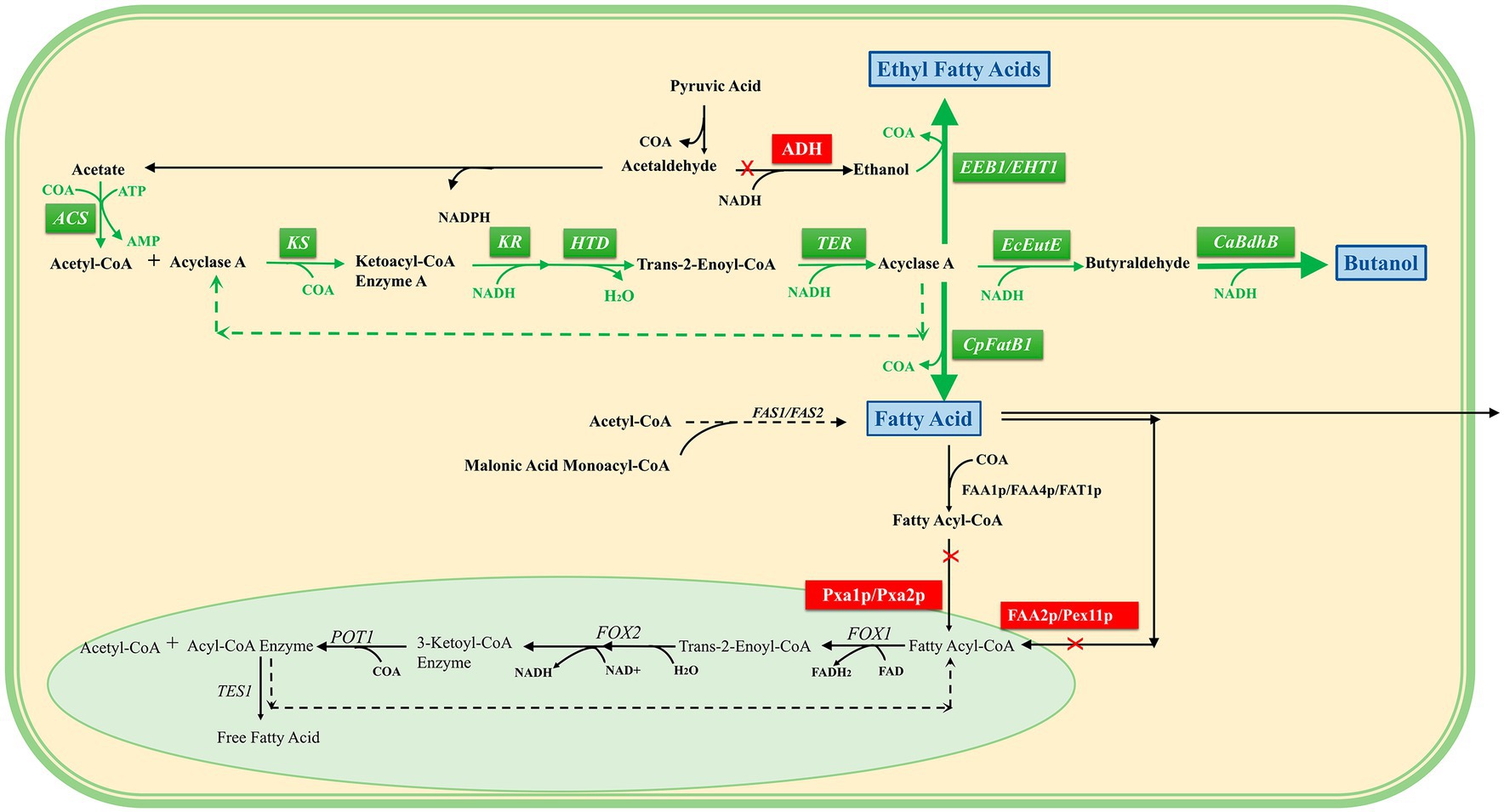
Figure 3. Reversal of the β-oxidation pathway in the cytosol. A pale green background indicates peroxisomes, and a pale yellow background indicates cytoplasm. The green box is the overexpressed reverse β-oxidation-related enzyme, the blue box is the product, the red box is the blocking pathway, the black arrow is the direction of the intracellular original reaction, and the green arrow is the direction of the overexpression construction pathway. It can be seen that the intermediate products of the reversed β-oxidation pathway constructed in the cytosol are consistent with the β-oxidation intermediate products in the peroxisome, and finally three target products can be obtained.
In recent years, the demand for fatty acids and their derivatives in the medical, health, cosmetic, and food sectors has increased. The efficient production of fatty acids by microorganisms is a promising approach (Saravanan et al., 2022). Saccharomyces cerevisiae is a preferred host for the synthesis of high-value products. By controlling the β-oxidation process, as well as other metabolic pathways, S. cerevisiae can be modified to be a novel platform for the production and accumulation of fatty acids and their derivatives (Akşit and Van Der Klei, 2018).
Taking advantage of the similarity of fatty acid synthesis pathways in oil-producing yeast and non-oil-producing yeast, the content of specific fatty acids is increased by genetic engineering of S. cerevisiae to overexpress heterologous genes, thereby enriching the potential products of S.cerevisiae (Biernat et al., 2023).
At present, the fatty acid synthesis of S. cerevisiae is dominated by long-chain fatty acids C16:0, C16:1, C18:0 and C18:1, because the content of C16/C18 in the cell membrane of S. cerevisiae is higher (Hashem et al., 2021), which is more conducive to the synthesis of fatty acid derivatives. Saccharomyces cerevisiae has many potential products of fatty acids, aldehydes, alcohols and alkanes (Kim et al., 2021), but the tolerance of these potential products needs to be improved. For example, lipophilic medium-chain fatty acids tend to damage the plasma membrane of S. cerevisiae, reducing the plasma membrane’s ability to resist the external environment (Liu et al., 2022). In a study, it was found that the production of medium-chain fatty acids in wild-type S. cerevisiae was low, and exogenous medium-chain fatty acids had an inhibitory effect on the growth of S. cerevisiae; The accumulation of alkanes through overexpression of thioesterase can alleviate the damage of medium-chain fatty acids to cell membranes (Zhu et al., 2017). Medium chain fatty acids and their derivatives can be used as potential products of Saccharomyces cerevisiae and have great room for improvement. In the future, transcriptome analysis can be used to screen key genes for medium-chain fatty acid tolerance in Saccharomyces cerevisiae, and then use the advanced gene editing technology CRISPR-Cas9 to modify related metabolic pathways in Saccharomyces cerevisiae. In future fermentation production, fed-batch strategies such as in-situ adsorption, substrate product separation, and substrate embedding techniques can be carried out according to different substrates to minimize the damage of the product to cells.
Peroxisomes are a promising compartment for regulating fatty acid degradation. Knockdown of peroxisome regulatory genes PEX11, PEX25, and PEX27 showed that changes in the number of peroxisomes had an impact on the longevity of yeast (Deb and Nagotu, 2023). During β-oxidation, energy is required for fatty acid degradation, and the energy provided by mitochondria to peroxisomes passes through the cytoplasmic matrix. How to significantly improve the β-oxidative energy supply of peroxisomes and effectively regulate the metabolic flux of NADP/NADPH and ATP/AMP transport rate is a difficult task to improve the metabolic efficiency of peroxisome, In the future, the pathway of coenzyme A in peroxisome can be explored by isotope labeling, and the regulation process of key genes or protein families on cofactor delivery can be reasoned by transcriptome analysis.
ZW: Writing – original draft. CS: Writing – original draft. RW: Writing – review & editing. JS: Writing – review & editing. YZ: Writing – review & editing and software. SS: Writing – review & editing.
The author(s) declare financial support was received for the research, authorship, and/or publication of this article. This study was funded by the Key Innovation Project of Qilu University of Technology [Shandong Academy of Sciences] (Grant No. 2022JBZ01-06), Science Foundation of ShanDong Province (Grant No. ZR2023MC089), and Focus on Research and Development Plan in Shandong Province (Grant No. 2023CXGC010714).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Akşit, A., and Van Der Klei, I. J. (2018). Yeast peroxisomes: how are they formed and how do they grow? Int. J. Biochem. Cell Biol. 105, 24–34. doi: 10.1016/j.biocel.2018.09.019
Alvarez-Vasquez, F., Riezman, H., Hannun, Y. A., and Voit, E. O. (2011). Mathematical modeling and validation of the Ergosterol pathway in Saccharomyces cerevisiae. PLoS One 6:e28344. doi: 10.1371/journal.pone.0028344
Auesukaree, C., Damnernsawad, A., Kruatrachue, M., Pokethitiyook, P., and Boonchird, C. (2009). Genome-wide identification of genes involved in tolerance to various environmental stresses Insaccharomyces cerevisiae. J. Appl. Genet. 50, 301–310. doi: 10.1007/BF03195688
Baker, A., Carrier, D. J., Schaedler, T., Waterham, H. R., van Roermund, C. W., and Theodoulou, F. L. (2015). Peroxisomal ABC transporters: functions and mechanism. Biochem. Soc. Trans. 43, 959–965. doi: 10.1042/BST20150127
Beopoulos, A., Cescut, J., Haddouche, R., Uribelarrea, J.-L., and Molina-Jouve, C. (2009). Yarrowia lipolytica as a model for bio-oil production. Prog. Lipid Res. 48, 375–387. doi: 10.1016/j.plipres.2009.08.005
Bergenholm, D., Liu, G., Holland, P., and Nielsen, J. (2018). Reconstruction of a global transcriptional regulatory network for control of lipid metabolism in yeast by using chromatin immunoprecipitation with lambda exonuclease digestion. mSystem 3, e00215–e00217. doi: 10.1128/mSystems.00215-17
Biernat, E., Khan, U., and Govind, C. K. (2023). Measuring occupancies of the nucleosome and nucleosome-interacting factors in vivo in Saccharomyces cerevisiae genome-wide. Methods 218, 167–175. doi: 10.1016/j.ymeth.2023.08.007
Black, P. N., and DiRusso, C. C. (2007). Yeast acyl-CoA synthetases at the crossroads of fatty acid metabolism and regulation. Biochim. Biophys. Acta 1771, 286–298. doi: 10.1016/j.bbalip.2006.05.003
Borreby, C., Lillebæk, E. M. S., and Kallipolitis, B. H. (2023). Anti-infective activities of long-chain fatty acids against foodborne pathogens. FEMS Microbiol. Rev. 47:fuad037. doi: 10.1093/femsre/fuad037
Cao, W., Li, H., Luo, J., Yin, J., and Wan, Y. (2017). High-level productivity of α,ω-dodecanedioic acid with a newly isolated Candida viswanathii strain. Regul. Phosph. Syn. 44, 1191–1202. doi: 10.1007/s10295-017-1948-6
Carrier, D. J., van Roermund, C. W. T., Schaedler, T. A., Rong, H. L., IJlst, L., and Wanders, R. J. A. (2019). Mutagenesis separates ATPase and thioesterase activities of the peroxisomal ABC transporter, comatose. Sci. Rep. 9:10502. doi: 10.1038/s41598-019-46685-9
Caswell, B. T., De Carvalho, C. C., Nguyen, H., Roy, M., Nguyen, T., and Cantu, D. C. (2022). Thioesterase enzyme families: functions, structures, and mechanisms. Protein Sci. 31, 652–676. doi: 10.1002/pro.4263
Chen, L., Zhang, J., and Chen, W. N. (2014a). Engineering the Saccharomyces cerevisiae β-oxidation pathway to increase medium chain fatty acid production as potential biofuel. PLoS One 9:e84853. doi: 10.1371/journal.pone.0084853
Chen, L., Zhang, J., Lee, J., and Chen, W. N. (2014b). Enhancement of free fatty acid production in Saccharomyces cerevisiae by control of fatty acyl-CoA metabolism. Appl. Microbiol. Biotechnol. 98, 6739–6750. doi: 10.1007/s00253-014-5758-8
Chinopoulos, C. (2021). The mystery of extramitochondrial proteins lysine Succinylation. IJMS 22:6085. doi: 10.3390/ijms22116085
Choi, J.-Y., and Martin, C. E. (1999). The Saccharomyces cerevisiae FAT1 gene encodes an acyl-CoA synthetase that is required for maintenance of very long chain fatty acid levels. J. Biol. Chem. 274, 4671–4683. doi: 10.1074/jbc.274.8.4671
Cui, Z., Gao, C., Li, J., Hou, J., and Lin, C. S. K. (2017). Engineering of unconventional yeast Yarrowia lipolytica for efficient succinic acid production from glycerol at low pH. Metab. Eng. 42, 126–133. doi: 10.1016/j.ymben.2017.06.007
Darvishi, F. (2012). Expression of native and mutant extracellular lipases from Yarrowia lipolytica in Saccharomyces cerevisiae: expression Y. Lipolytica lipase in S. cerevisiae. Microb. Biotechnol. 5, 634–641. doi: 10.1111/j.1751-7915.2012.00354.x
Deb, R., and Nagotu, S. (2023). The nexus between peroxisome abundance and chronological ageing in Saccharomyces cerevisiae. Biogerontology 24, 81–97. doi: 10.1007/s10522-022-09992-9
DeLoache, W. C., Russ, Z. N., and Dueber, J. E. (2016). Towards repurposing the yeast peroxisome for compartmentalizing heterologous metabolic pathways. Nat. Commun. 7:11152. doi: 10.1038/ncomms11152
El Moualij, B., Duyckaerts, C., Lamotte-Brasseur, J., and Sluse, F. E. (1997). Phylogenetic classification of the mitochondrial carrier family of saccharomyces cerevisiae. Yeast 13, 573–581. doi: 10.1002/(SICI)1097-0061(199705)13:6<573::AID-YEA107>3.0.CO;2-I
Engelhardt, M., Hintze, S., Wendegatz, E.-C., Lettow, J., and Schüller, H.-J. (2023). Ino2, activator of yeast phospholipid biosynthetic genes, interacts with basal transcription factors TFIIA and Bdf1. Curr. Genet. 69, 289–300. doi: 10.1007/s00294-023-01277-z
Eriksen, D. T., Lian, J., and Zhao, H. (2014). Protein design for pathway engineering. J. Struct. Biol. 185, 234–242. doi: 10.1016/j.jsb.2013.03.011
Færgeman, N. J., Black, P. N., Zhao, X. D., Knudsen, J., and DiRusso, C. C. (2001). The acyl-CoA synthetases encoded within FAA1 and FAA4 in Saccharomyces cerevisiae function as components of the fatty acid transport system linking import, activation, and intracellular utilization. J. Biol. Chem. 276, 37051–37059. doi: 10.1074/jbc.M100884200
Falcon, A., Doege, H., Fluitt, A., Tsang, B., Watson, N., and Kay, M. A. (2010). FATP2 is a hepatic fatty acid transporter and peroxisomal very long-chain acyl-CoA synthetase. Am. J. Physiol. Endocrinol. Metab. 299, E384–E393. doi: 10.1152/ajpendo.00226.2010
Farré, J.-C., Carolino, K., Stasyk, O. V., Stasyk, O. G., Hodzic, Z., Agrawal, G., et al. (2017). A new yeast Peroxin, Pex36, a functional homolog of mammalian PEX16, functions in the ER-to-peroxisome traffic of Peroxisomal membrane proteins. J. Mol. Biol. 429, 3743–3762. doi: 10.1016/j.jmb.2017.10.009
Ferreira, R., Teixeira, P. G., Gossing, M., David, F., Siewers, V., and Nielsen, J. (2018). Metabolic engineering of Saccharomyces cerevisiae for overproduction of triacylglycerols. Metab. Eng. Commun. 6, 22–27. doi: 10.1016/j.meteno.2018.01.002
Fickers, P., Marty, A., and Nicaud, J. M. (2011). The lipases from Yarrowia lipolytica: genetics, production, regulation, biochemical characterization and biotechnological applications. Biotechnol. Adv. 29, 632–644. doi: 10.1016/j.biotechadv.2011.04.005
Gajewski, J., Pavlovic, R., Fischer, M., Boles, E., and Grininger, M. (2017). Engineering fungal de novo fatty acid synthesis for short chain fatty acid production. Nat. Commun. 8:14650. doi: 10.1038/ncomms14650
Garces Daza, F., Haitz, F., Born, A., and Boles, E. (2023). An optimized reverse β-oxidation pathway to produce selected medium-chain fatty acids in Saccharomyces cerevisiae. Biotechnol Biofuels Bioprod 16:71. doi: 10.1186/s13068-023-02317-z
Gaspar, M. L., Aregullin, M. A., Chang, Y.-F., Jesch, S. A., and Henry, S. A. (2022). Phosphatidic acid species 34:1 mediates expression of the myo-inositol 3-phosphate synthase gene INO1 for lipid synthesis in yeast. J. Biol. Chem. 298:102148. doi: 10.1016/j.jbc.2022.102148
Gaussmann, S., Gopalswamy, M., Eberhardt, M., Reuter, M., Zou, P., and Schliebs, W. (2021). Membrane interactions of the Peroxisomal proteins PEX5 and PEX14. Front. Cell Dev. Biol. 9:651449. doi: 10.3389/fcell.2021.651449
Glover, J. R., Andrews, D. W., Subramani, S., and Rachubinski, R. A. (1994). Mutagenesis of the amino targeting signal of Saccharomyces cerevisiae 3-ketoacyl-CoA thiolase reveals conserved amino acids required for import into peroxisomes in vivo. J. Biol. Chem. 269, 7558–7563. doi: 10.1016/S0021-9258(17)37323-4
Gossing, M., Smialowska, A., and Nielsen, J. (2018). Impact of forced fatty acid synthesis on metabolism and physiology of Saccharomyces cerevisiae. FEMS Yeast Res. 18:e96. doi: 10.1093/femsyr/foy096
Gurvitz, A., Hamilton, B., Hartig, A., Ruis, H., Dawes, I. W., and Rottensteiner, H. (1999). A novel element in the promoter of the Saccharomyces cerevisiae gene SPS19 enhances ORE-dependent up-regulation in oleic acid and is essential for de-repression. Mol. Gen. Genet. 262, 481–492. doi: 10.1007/s004380051109
Han, L., Peng, Y., Zhang, Y., Chen, W., Lin, Y., and Wang, Q. (2017). Designing and creating a synthetic omega oxidation pathway in Saccharomyces cerevisiae enables production of medium-chain α, ω-dicarboxylic acids. Front. Microbiol. 8:2184. doi: 10.3389/fmicb.2017.02184
Hanko, E. K. R., Denby, C. M., Sànchez i Nogué, V., Lin, W., and Ramirez, K. J. (2018). Engineering β-oxidation in Yarrowia lipolytica for methyl ketone production. Metab. Eng. 48, 52–62. doi: 10.1016/j.ymben.2018.05.018
Hashem, M., Alamri, S. A., Asseri, T. A. Y., Mostafa, Y. S., and Lyberatos, G. (2021). On the optimization of fermentation conditions for enhanced bioethanol yields from starchy biowaste via yeast co-cultures. Sustainability 13:1890. doi: 10.3390/su13041890
Henritzi, S., Fischer, M., Grininger, M., Oreb, M., and Boles, E. (2018). An engineered fatty acid synthase combined with a carboxylic acid reductase enables de novo production of 1-octanol in Saccharomyces cerevisiae. Biotechnol. Biofuels 11:150. doi: 10.1186/s13068-018-1149-1
Hilmi Ibrahim, Z., Bae, J.-H., Lee, S.-H., Sung, B. H., and Ab Rashid, A. H. (2020). Genetic manipulation of a Lipolytic yeast Candida aaseri SH14 using CRISPR-Cas9 system. Microorganisms 8:526. doi: 10.3390/microorganisms8040526
Hu, Y., Zhu, Z., Gradischnig, D., Winkler, M., Nielsen, J., and Siewers, V. (2020). Engineering carboxylic acid reductase for selective synthesis of medium-chain fatty alcohols in yeast. Proc. Natl. Acad. Sci. U. S. A. 117, 22974–22983. doi: 10.1073/pnas.2010521117
Hu, Y., Zhu, Z., Nielsen, J., and Siewers, V. (2019). Engineering Saccharomyces cerevisiae cells for production of fatty acid-derived biofuels and chemicals. Open Biol. 9:190049. doi: 10.1098/rsob.190049
Huang, H., Niu, Y., Jin, Q., Qin, K., Wang, L., and Shang, Y. (2022). Identification of six Thiolases and their effects on fatty acid and Ergosterol biosynthesis in aspergillus oryzae. Appl. Environ. Microbiol. 88:e0237221. doi: 10.1128/aem.02372-21
Huber, A., Koch, J., Kragler, F., Brocard, C., and Hartig, A. (2012). A subtle interplay between three Pex11 proteins shapes De novo formation and fission of peroxisomes. Traffic 13, 157–167. doi: 10.1111/j.1600-0854.2011.01290.x
Ibrahim, Z. H., Bae, J.-H., Sung, B. H., Kim, M.-J., and Rashid, A. H. A. (2022). Characterization of acyl-CoA oxidases from the Lipolytic yeast Candida aaseri SH14. J. Microbiol. Biotechnol. 32, 949–954. doi: 10.4014/jmb.2205.05029
Infant, T., Deb, R., Ghose, S., and Nagotu, S. (2021). Post-translational modifications of proteins associated with yeast peroxisome membrane: an essential mode of regulatory mechanism. Genes Cells 26, 843–860. doi: 10.1111/gtc.12892
Jordá, T., and Puig, S. (2020). Regulation of Ergosterol biosynthesis in Saccharomyces cerevisiae. Genes 11:795. doi: 10.3390/genes11070795
Judy, R. M., Sheedy, C. J., and Gardner, B. M. (2022). Insights into the structure and function of the Pex1/Pex6 AAA-ATPase in peroxisome homeostasis. Cells 11:2067. doi: 10.3390/cells11132067
Kawaguchi, K., Mukai, E., Watanabe, S., Yamashita, A., Morita, M., and So, T. (2021). Acyl-CoA thioesterase activity of peroxisomal ABC protein ABCD1 is required for the transport of very long-chain acyl-CoA into peroxisomes. Sci. Rep. 11:2192. doi: 10.1038/s41598-021-81949-3
Ke, X., Pan, Z., Du, H., Shen, Y., and Shen, J. (2023). Secretory production of 7-dehydrocholesterol by engineered Saccharomyces cerevisiae. Biotechnol. J. 18:e2300056. doi: 10.1002/biot.202300056
Khaddaj, R., Stribny, J., Cottier, S., and Schneiter, R. (2023). Perilipin 3 promotes the formation of membrane domains enriched in diacylglycerol and lipid droplet biogenesis proteins. Front. Cell Dev. Biol. 11:1116491. doi: 10.3389/fcell.2023.1116491
Khatri, A., and Choudhary, V. (2023). Protocol to induce de novo lipid droplets in Saccharomyces cerevisiae. STAR Protocols 4:102732. doi: 10.1016/j.xpro.2023.102732
Kiel, J. A. K. W., Veenhuis, M., and Van Der Klei, I. J. (2006). PEX genes in fungal genomes: common, rare or redundant. Traffic 7, 1291–1303. doi: 10.1111/j.1600-0854.2006.00479.x
Kim, J., Baidoo, E. E. K., Amer, B., Mukhopadhyay, A., Adams, P. D., and Simmons, B. A. (2021). Engineering Saccharomyces cerevisiae for isoprenol production. Metab. Eng. 64, 154–166. doi: 10.1016/j.ymben.2021.02.002
Kim, S., and Kim, K.-J. (2018). Structural insight into the substrate specificity of acyl-CoA oxidase1 from Yarrowia lipolytica for short-chain dicarboxylyl-CoAs. Biochem. Biophys. Res. Commun. 495, 1628–1634. doi: 10.1016/j.bbrc.2017.11.191
Klug, L., and Daum, G. (2014). Yeast lipid metabolism at a glance. FEMS Yeast Res. 14, 369–388. doi: 10.1111/1567-1364.12141
Knoblach, B., and Rachubinski, R. A. (2018). Reconstitution of human peroxisomal β-oxidation in yeast. FEMS Yeast Res. 18:e92. doi: 10.1093/femsyr/foy092
Knoll, L. J., Johnson, D. R., and Gordon, J. I. (1994). Biochemical studies of three Saccharomyces cerevisiae acyl-CoA synthetases, Faa1p, Faa2p, and Faa3p. J. Biol. Chem. 269, 16348–16356. doi: 10.1016/S0021-9258(17)34014-0
Knoll, L. J., Johnson, D. R., and Gordon, J. I. (1995). Complementation of Saccharomycescerevisiae strains containing fatty acid activation gene (FAA) deletions with a mammalian acyl-CoA Synthetase. J. Biol. Chem. 270, 10861–10867. doi: 10.1074/jbc.270.18.10861
Koch, B., Schmidt, C., and Daum, G. (2014). Storage lipids of yeasts: a survey of nonpolar lipid metabolism in Saccharomyces cerevisiae, Pichia pastoris, and Yarrowia lipolytica. FEMS Microbiol. Rev. 38, 892–915. doi: 10.1111/1574-6976.12069
Leber, C., Polson, B., Fernandez-Moya, R., and Da Silva, N. A. (2015). Overproduction and secretion of free fatty acids through disrupted neutral lipid recycle in Saccharomyces cerevisiae. Metab. Eng. 28, 54–62. doi: 10.1016/j.ymben.2014.11.006
Lee, J.-R., Kim, S.-Y., Chae, H.-B., Jung, J.-H., and Lee, S.-Y. (2009). Antifungal activity of Saccharomyces cerevisiae peroxisomal 3-ketoacyl-CoA thiolase. BMB Rep. 42, 281–285. doi: 10.5483/BMBRep.2009.42.5.281
Lefevre, S. D., Roermund, C. W., Wanders, R. J. A., Veenhuis, M., and Klei, I. J. (2013). The significance of peroxisome function in chronological aging of Saccharomyces cerevisiae. Aging Cell 12, 784–793. doi: 10.1111/acel.12113
Li, Y.-H., Liu, B., Zhao, Z.-B., and Bai, F.-W. (2006). Optimized culture medium and fermentation conditions for lipid production by Rhodosporidium toruloides. Sheng Wu Gong Cheng Xue Bao 22, 650–656. doi: 10.1016/S1872-2075(06)60050-2
Li, H., Melton, E. M., Quackenbush, S., DiRusso, C. C., and Black, P. N. (2007). Mechanistic studies of the long chain acyl-CoA synthetase Faa1p from Saccharomyces cerevisiae. Biochimica et Biophysica Acta (BBA) - molecular and cell biology of. Lipids 1771, 1246–1253. doi: 10.1016/j.bbalip.2007.05.009
Lian, J., Mishra, S., and Zhao, H. (2018). Recent advances in metabolic engineering of Saccharomyces cerevisiae: new tools and their applications. Metab. Eng. 50, 85–108. doi: 10.1016/j.ymben.2018.04.011
Lian, J., and Zhao, H. (2015). Reversal of the β-oxidation cycle in Saccharomyces cerevisiae for production of fuels and chemicals. ACS Synth. Biol. 4, 332–341. doi: 10.1021/sb500243c
Liu, H.-H., Ji, X.-J., and Huang, H. (2015). Biotechnological applications of Yarrowia lipolytica: past, present and future. Biotechnol. Adv. 33, 1522–1546. doi: 10.1016/j.biotechadv.2015.07.010
Liu, H., Yuan, W., Zhou, P., Liang, G., Gao, C., and Guo, L. (2022). Engineering membrane asymmetry to increase medium-chain fatty acid tolerance in Saccharomyces cerevisiae. Biotech Bioeng 119, 277–286. doi: 10.1002/bit.27973
Luo, Y.-S., Nicaud, J.-M., Van Veldhoven, P. P., and Chardot, T. (2002). The acyl–CoA oxidases from the yeast Yarrowia lipolytica: characterization of Aox2p. Arch. Biochem. Biophys. 407, 32–38. doi: 10.1016/S0003-9861(02)00466-6
Lv, X., Jin, K., Yi, Y., Song, L., Xiu, X., and Liu, Y. (2023). Analysis of acid-tolerance mechanism based on membrane microdomains in Saccharomyces cerevisiae. Microb. Cell Factories 22:180. doi: 10.1186/s12934-023-02195-y
Mast, F. D., Jamakhandi, A., Saleem, R. A., Dilworth, D. J., Rogers, R. S., and Rachubinski, R. A. (2016). Peroxins Pex30 and Pex29 dynamically associate with Reticulons to regulate peroxisome biogenesis from the endoplasmic reticulum. J. Biol. Chem. 291, 15408–15427. doi: 10.1074/jbc.M116.728154
Mathieu, M., Modis, Y., Zeelen, J. P., Engel, C. K., Abagyan, R. A., and Ahlberg, A. (1997). The 1.8 Å crystal structure of the dimeric peroxisomal 3-ketoacyl-CoA thiolase of Saccharomyces cerevisiae: implications for substrate binding and reaction mechanism. J. Mol. Biol. 273, 714–728. doi: 10.1006/jmbi.1997.1331
Matos, G. S., Vogt, L., Santos, R. S., Devillars, A., Yoshinaga, M. Y., and Miyamoto, S. (2023). Lipidome remodeling in response to nutrient replenishment requires the TRNA modifier Deg1/Pus3 in yeast. Mol. Microbiol. 120, 893–905. doi: 10.1111/mmi.15185
Mbuyane, L. L., Bauer, F. F., Bloem, A., Camarasa, C., and Ortiz-Julien, A. (2022). Species-dependent metabolic response to lipid mixtures in wine yeasts. Front. Microbiol. 13:823581. doi: 10.3389/fmicb.2022.823581
Melton, E. M., Cerny, R. L., DiRusso, C. C., and Black, P. N. (2013). Overexpression of human fatty acid transport protein 2/very long chain acyl-CoA synthetase 1 (FATP2/Acsvl1) reveals distinct patterns of trafficking of exogenous fatty acids. Biochem. Biophys. Res. Commun. 440, 743–748. doi: 10.1016/j.bbrc.2013.09.137
Mlíčková, K., Roux, E., Athenstaedt, K., d’Andrea, S., and Daum, G. (2004). Lipid accumulation, lipid body formation, and acyl coenzyme a oxidases of the yeast Yarrowia lipolytica. Appl. Environ. Microbiol. 70, 3918–3924. doi: 10.1128/AEM.70.7.3918-3924.2004
Moeller, L., Strehlitz, B., Aurich, A., Zehnsdorf, A., and Bley, T. (2007). Optimization of citric acid production from glucose by Yarrowia lipolytica. Eng. Life Sci. 7, 504–511. doi: 10.1002/elsc.200620207
Mursula, A. M., Hiltunen, J. K., and Wierenga, R. K. (2004). Structural studies on Δ3-Δ2-enoyl-CoA isomerase: the variable mode of assembly of the trimeric disks of the crotonase superfamily. FEBS Lett. 557, 81–87. doi: 10.1016/S0014-5793(03)01450-9
Mursula, A. M., Van Aalten, D. M. F., Modis, Y., Hiltunen, J. K., and Wierenga, R. K. (2000). Crystallization and X-ray diffraction analysis of peroxisomal Δ3-Δ2-enoyl-CoA isomerase from Saccharomyces cerevisiae. Acta Crystallogr. D Biol. Crystallogr. 56, 1020–1023. doi: 10.1107/S0907444900006533
Nancolas, B., Bull, I. D., Stenner, R., Dufour, V., and Curnow, P. (2017). Saccharomyces cerevisiae Atf1p is an alcohol acetyltransferase and a thioesterase in vitro: characterization of Atf1p. Yeast 34, 239–251. doi: 10.1002/yea.3229
Neuberger, G., Maurer-Stroh, S., Eisenhaber, B., Hartig, A., and Eisenhaber, F. (2003). Motif refinement of the Peroxisomal targeting signal 1 and evaluation of taxon-specific differences. J. Mol. Biol. 328, 567–579. doi: 10.1016/S0022-2836(03)00318-8
Nielsen, J. (2014). Synthetic biology for engineering acetyl coenzyme a metabolism in yeast. MBio 5:e02153. doi: 10.1128/mBio.02153-14
Ntamack, A. G., Karpichev, I. V., Gould, S. J., Small, G. M., and Schulz, H. (2009). Oleate β-oxidation in yeast involves thioesterase but not Yor180c protein that is not a dienoyl-CoA isomerase. Biochimica et Biophysica Acta (BBA) 1791, 371–378. doi: 10.1016/j.bbalip.2009.01.026
Nuttall, J. M., Motley, A., and Hettema, E. H. (2011). Peroxisome biogenesis: recent advances. Curr. Opin. Cell Biol. 23, 421–426. doi: 10.1016/j.ceb.2011.05.005
Otzen, C., Müller, S., Jacobsen, I. D., and Brock, M. (2013). Phylogenetic and phenotypic characterisation of the 3-ketoacyl-CoA thiolase gene family from the opportunistic human pathogenic fungus Candida albicans. FEMS Yeast Res. 13, 553–564. doi: 10.1111/1567-1364.12057
Pagac, M., De La Mora, H. V., Duperrex, C., Roubaty, C., and Vionnet, C. (2011). Topology of 1-acyl-sn-glycerol-3-phosphate acyltransferases SLC1 and ALE1 and related membrane-bound O-acyltransferases (MBOATs) of Saccharomyces cerevisiae. J. Biol. Chem. 286, 36438–36447. doi: 10.1074/jbc.M111.256511
Peralta-Yahya, P. P., Zhang, F., Del Cardayre, S. B., and Keasling, J. D. (2012). Microbial engineering for the production of advanced biofuels. Nature 488, 320–328. doi: 10.1038/nature11478
Plett, A., Charton, L., and Linka, N. (2020). Peroxisomal cofactor transport. Biomol. Ther. 10:1174. doi: 10.3390/biom10081174
Poudyal, N. R., and Paul, K. S. (2022). Fatty acid uptake in Trypanosoma brucei: host resources and possible mechanisms. Front. Cell. Infect. Microbiol. 12:949409. doi: 10.3389/fcimb.2022.949409
Pozdniakova, T. A., Cruz, J. P., Silva, P. C., Azevedo, F., Parpot, P., and Domingues, M. R. (2023). Optimization of a hybrid bacterial/Arabidopsis thaliana fatty acid synthase system II in Saccharomyces cerevisiae. Metab Eng Commun 17:e00224. doi: 10.1016/j.mec.2023.e00224
Robert, J., Marchesini, S., Delessert, S., and Poirier, Y. (2005). Analysis of the β-oxidation of trans-unsaturated fatty acid in recombinant Saccharomyces cerevisiae expressing a peroxisomal PHA synthase reveals the involvement of a reductase-dependent pathway. Biochimica et Biophysica Acta (BBA) 1734, 169–177. doi: 10.1016/j.bbalip.2005.02.010
Rottensteiner, H., Kal, A. J., Filipits, M., Binder, M., and Hamilton, B. (1996). Pip2p: a transcriptional regulator of peroxisome proliferation in the yeast Saccharomyces cerevisiae. EMBO J. 15, 2924–2934. doi: 10.1002/j.1460-2075.1996.tb00655.x
Röttig, A., Wenning, L., Bröker, D., and Steinbüchel, A. (2010). Fatty acid alkyl esters: perspectives for production of alternative biofuels. Appl. Microbiol. Biotechnol. 85, 1713–1733. doi: 10.1007/s00253-009-2383-z
Ruenwai, R., Neiss, A., Laoteng, K., Vongsangnak, W., and Dalfard, A. B. (2011). Heterologous production of polyunsaturated fatty acids in Saccharomyces cerevisiae causes a global transcriptional response resulting in reduced proteasomal activity and increased oxidative stress. Biotechnol. J. 6, 343–356. doi: 10.1002/biot.201000316
Saravanan, A., Senthil Kumar, P., Jeevanantham, S., Karishma, S., and Vo, D.-V. N. (2022). Recent advances and sustainable development of biofuels production from lignocellulosic biomass. Bioresour. Technol. 344:126203. doi: 10.1016/j.biortech.2021.126203
Sartaj, K., Prasad, R., Matsakas, L., and Patel, A. (2023). Transforming recalcitrant wastes into biodiesel by oleaginous yeast: an insight into the metabolic pathways and multi-omics landscape. Chem. Eng. J. 474:145625. doi: 10.1016/j.cej.2023.145625
Schäfer, A., Kerssen, D., Veenhuis, M., Kunau, W.-H., and Schliebs, W. (2004). Functional similarity between the peroxisomal PTS2 receptor binding protein Pex18p and the N-terminal half of the PTS1 receptor Pex5p. Mol. Cell. Biol. 24, 8895–8906. doi: 10.1128/MCB.24.20.8895-8906.2004
Schäfer, J.-H., Körner, C., Esch, B. M., Limar, S., Parey, K., and Walter, S. (2023). Structure of the ceramide-bound SPOTS complex. Nat. Commun. 14:6196. doi: 10.1038/s41467-023-41747-z
Scharnewski, M., Pongdontri, P., Mora, G., Hoppert, M., and Fulda, M. (2008). Mutants of Saccharomyces cerevisiae deficient in acyl-CoA synthetases secrete fatty acids due to interrupted fatty acid recycling: fatty acid secretion in mutants of yeast. FEBS J. 275, 2765–2778. doi: 10.1111/j.1742-4658.2008.06417.x
Sheng, J., Stevens, J., and Feng, X. (2016). Pathway compartmentalization in peroxisome of Saccharomyces cerevisiae to produce versatile medium chain fatty alcohols. Sci. Rep. 6:26884. doi: 10.1038/srep26884
Shi, S., Valle-Rodríguez, J. O., Siewers, V., and Nielsen, J. (2014). Engineering of chromosomal wax ester synthase integrated Saccharomyces cerevisiae mutants for improved biosynthesis of fatty acid ethyl esters. Biotech Bioeng 111, 1740–1747. doi: 10.1002/bit.25234
Son, Y. J., Jeon, M.-S., Moon, H. Y., Kang, J., Jeong, D. M., and Lee, D. W. (2023). Integrated genomics and phenotype microarray analysis of Saccharomyces cerevisiae industrial strains for rice wine fermentation and recombinant protein production. Microb. Biotechnol. 16, 2161–2180. doi: 10.1111/1751-7915.14354
Speer, N. O., Braun, R. J., Reynolds, E. G., Brudnicka, A., and Swanson, J. M. J. (2024). Tld1 is a regulator of triglyceride lipolysis that demarcates a lipid droplet subpopulation. J. Cell Biol. 223:e202303026. doi: 10.1083/jcb.202303026
Stanway, C. A., Gibbs, J. M., and Berardi, E. (1995). Expression of the FOX1 gene of Saccharomyces cerevisiae is regulated by carbon source, but not by the known glucose repression genes. Curr. Genet. 27, 404–408. doi: 10.1007/BF00311208
Strijbis, K., and Distel, B. (2010). Intracellular acetyl unit transport in fungal carbon metabolism. Eukaryot. Cell 9, 1809–1815. doi: 10.1128/EC.00172-10
Su, H., Shi, P., Shen, Z., Meng, H., Meng, Z., Han, X., et al. (2023). High-level production of nervonic acid in the oleaginous yeast Yarrowia lipolytica by systematic metabolic engineering. Commun Biol 6:1125. doi: 10.1038/s42003-023-05502-w
Thevenieau, F., Le Dall, M.-T., Nthangeni, B., Mauersberger, S., and Marchal, R. (2007). Characterization of Yarrowia lipolytica mutants affected in hydrophobic substrate utilization. Fungal Genet. Biol. 44, 531–542. doi: 10.1016/j.fgb.2006.09.001
Tillander, V., Alexson, S. E. H., and Cohen, D. E. (2017). Deactivating fatty acids: acyl-CoA Thioesterase-mediated control of lipid metabolism. Trends Endocrinol. Metab. 28, 473–484. doi: 10.1016/j.tem.2017.03.001
Ullah, N., Shahzad, K., and Wang, M. (2021). The role of metabolic engineering technologies for the production of fatty acids in yeast. Biology 10:632. doi: 10.3390/biology10070632
Van Roermund, C. W. T., Drissen, R., Van Den Berg, M., Ijlst, L., and Hettema, E. H. (2001). Identification of a Peroxisomal ATP Carrier required for medium-chain fatty acid β-oxidation and Normal peroxisome proliferation in Saccharomyces cerevisiae. Mol. Cell. Biol. 21, 4321–4329. doi: 10.1128/MCB.21.13.4321-4329.2001
van Roermund, C. W. T., IJlst, L., Baker, A., Wanders, R. J. A., and Theodoulou, F. L. (2021). The Saccharomyces cerevisiae ABC subfamily D transporter Pxa1/Pxa2p co-imports CoASH into the peroxisome. FEBS Lett. 595, 763–772. doi: 10.1002/1873-3468.13974
van Roermund, C. W. T., IJlst, L., Linka, N., Wanders, R. J. A., and Waterham, H. R. (2022). Peroxisomal ATP uptake is provided by two adenine nucleotide transporters and the ABCD transporters. Front. Cell Dev. Biol. 9:788921. doi: 10.3389/fcell.2021.788921
Van Roermund, C. W. T., Tabak, H. F., Van Den Berg, M., and Wanders, R. J. A. (2000). Pex11p plays a primary role in medium-chain fatty acid oxidation, a process that affects peroxisome number and size in Saccharomyces cerevisiae. J. Cell Biol. 150, 489–498. doi: 10.1083/jcb.150.3.489
Veenhuis, M., and Van Der Klei, I. J. (2014). A critical reflection on the principles of peroxisome formation in yeast. Front. Physiol. 5:e110. doi: 10.3389/fphys.2014.00110
Wang, H., Le Clainche, A., Le Dall, M.-T., Wache, Y., and Pagot, Y. (1998). Cloning and characterization of the peroxisomal acyl CoA oxidaseACO3 gene from the alkane-utilizing yeastYarrowia lipolytica. Yeast 14, 1373–1386. doi: 10.1002/(SICI)1097-0061(199811)14:15<1373::AID-YEA332>3.0.CO;2-1
Wang, T. W., Lewin, A. S., and Small, G. M. (1992). A negative regulating element controlling transcription of the gene encoding acyl-CoA oxidase in Saccharomyces cerevisiae. Nucl Acids Res 20, 3495–3500. doi: 10.1093/nar/20.13.3495
Wang, T., Luo, Y., and Small, G. M. (1994). The POX1 gene encoding peroxisomal acyl-CoA oxidase in Saccharomyces cerevisiae is under the control of multiple regulatory elements. J. Biol. Chem. 269, 24480–24485. doi: 10.1016/S0021-9258(19)51109-7
Wang, Y., Wu, T., Fan, X., Ruan, H., Fan, F., and Zhang, X. (2023). Sterol transport proteins in yeast: a review. Sheng Wu Gong Cheng Xue Bao 39, 3204–3218. doi: 10.13345/j.cjb.230042
Watkins, P. A., and Ellis, J. M. (2012). Peroxisomal acyl-CoA synthetases. Biochim. Biophys. Acta (BBA) 1822, 1411–1420. doi: 10.1016/j.bbadis.2012.02.010
Weber, C. A., Sekar, K., Tang, J. H., Warmer, P., Sauer, U., and Weis, K. (2020). β-Oxidation and autophagy are critical energy providers during acute glucose depletion in S accharomyces cerevisiae. Proc. Natl. Acad. Sci. U. S. A. 117, 12239–12248. doi: 10.1073/pnas.1913370117
Willet, A. H., Wos, M., Igarashi, M. G., Ren, L., and Turner, L. A. (2023). Elevated levels of sphingolipid MIPC in the plasma membrane disrupt the coordination of cell growth with cell wall formation in fission yeast. PLoS Genet. 19:e1010987. doi: 10.1371/journal.pgen.1010987
Wu, J., Zhang, X., Zhou, P., Huang, J., Xia, X., Li, W., et al. (2017). Improving metabolic efficiency of the reverse beta-oxidation cycle by balancing redox cofactor requirement. Metab. Eng. 44, 313–324. doi: 10.1016/j.ymben.2017.11.001
Wynn, J. P., Hamid, A., and Ratledge, C. (1999). The role of malic enzyme in the regulation of lipid accumulation in filamentous fungi. Microbiology 145, 1911–1917. doi: 10.1099/13500872-145-8-1911
Xu, P., Qiao, K., Ahn, W. S., and Stephanopoulos, G. (2016). Engineering Yarrowia lipolytica as a platform for synthesis of drop-in transportation fuels and oleochemicals. Proc. Natl. Acad. Sci. U. S. A. 113, 10848–10853. doi: 10.1073/pnas.1607295113
Yadav, P. K., Rajvanshi, P. K., and Rajasekharan, R. (2018). The role of yeast m6A methyltransferase in peroxisomal fatty acid oxidation. Curr. Genet. 64, 417–422. doi: 10.1007/s00294-017-0769-5
Yaguchi, A., Spagnuolo, M., and Blenner, M. (2018). Engineering yeast for utilization of alternative feedstocks. Curr. Opin. Biotechnol. 53, 122–129. doi: 10.1016/j.copbio.2017.12.003
Yasukawa, T., Iwama, R., Yamasaki, Y., Masuo, N., and Noda, Y. (2023). Yeast Rim11 kinase responds to glutathione-induced stress by regulating the transcription of phospholipid biosynthetic genes. MBoC 35:ar8. doi: 10.1091/mbc.E23-03-0116
Yocum, H. C., Bassett, S., and Da Silva, N. A. (2022). Enhanced production of acetyl-CoA-based products via peroxisomal surface display in Saccharomyces cerevisiae. Proc. Natl. Acad. Sci. U. S. A. 119:e2214941119. doi: 10.1073/pnas.2214941119
Zhang, L., Li, H., Gao, L., Qi, Y., Fu, W., and Li, X. (2017). Acyl-CoA oxidase 1 is involved in γ-decalactone release from peach (Prunus persica) fruit. Plant Cell Rep. 36, 829–842. doi: 10.1007/s00299-017-2113-4
Zhang, B., Ni, L., Tang, X., Chen, X., and Hu, B. (2022). Engineering the β-oxidation pathway in Yarrowia lipolytica for the production of trans -10, cis -12-conjugated linoleic acid. J. Agric. Food Chem. 70, 8377–8384. doi: 10.1021/acs.jafc.2c02242
Zhang, J., Sonnenschein, N., Pihl, T. P. B., Pedersen, K. R., and Jensen, M. K. (2016). Engineering an NADPH/NADP + redox biosensor in yeast. ACS Synth. Biol. 5, 1546–1556. doi: 10.1021/acssynbio.6b00135
Zhang, Y., Su, M., Qin, N., Nielsen, J., and Liu, Z. (2020). Expressing a cytosolic pyruvate dehydrogenase complex to increase free fatty acid production in Saccharomyces cerevisiae. Microb. Cell Factories 19:226. doi: 10.1186/s12934-020-01493-z
Zhang, Q., Zeng, W., Xu, S., and Zhou, J. (2021). Metabolism and strategies for enhanced supply of acetyl-CoA in Saccharomyces cerevisiae. Bioresour. Technol. 342:125978. doi: 10.1016/j.biortech.2021.125978
Zhang, H., Zhang, L., Chen, H., Chen, Y. Q., and Chen, W. (2014). Enhanced lipid accumulation in the yeast Yarrowia lipolytica by over-expression of ATP:citrate lyase from Mus musculus. J. Biotechnol. 192, 78–84. doi: 10.1016/j.jbiotec.2014.10.004
Zhou, Y. J., Buijs, N. A., Zhu, Z., Qin, J., Siewers, V., and Nielsen, J. (2016). Production of fatty acid-derived oleochemicals and biofuels by synthetic yeast cell factories. Nat. Commun. 7:11709. doi: 10.1038/ncomms11709
Zhu, J., Zhang, Z.-T., Tang, S.-W., Zhao, B.-S., and Li, H. (2019). A validated set of fluorescent-protein-based markers for major organelles in yeast (Saccharomyces cerevisiae). MBio 10:e01691. doi: 10.1128/mBio.01691-19
Zhu, Z., Zhou, Y. J., Kang, M.-K., Krivoruchko, A., Buijs, N. A., and Nielsen, J. (2017). Enabling the synthesis of medium chain alkanes and 1-alkenes in yeast. Metab. Eng. 44, 81–88. doi: 10.1016/j.ymben.2017.09.007
Keywords: β-oxidation, lipid metabolism, Saccharomyces cerevisiae , fatty acids, fatty acyl-CoA, yeast
Citation: Wang Z, Su C, Zhang Y, Shangguan S, Wang R and Su J (2024) Key enzymes involved in the utilization of fatty acids by Saccharomyces cerevisiae: a review. Front. Microbiol. 14:1294182. doi: 10.3389/fmicb.2023.1294182
Received: 14 September 2023; Accepted: 26 December 2023;
Published: 11 January 2024.
Edited by:
Gennaro Agrimi, University of Bari Aldo Moro, ItalyReviewed by:
Dongming Xie, University of Massachusetts Lowell, United StatesCopyright © 2024 Wang, Su, Zhang, Shangguan, Wang and Su. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Jing Su, c2pfMTIzNDZAMTYzLmNvbQ==
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.