
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Microbiol. , 15 November 2023
Sec. Antimicrobials, Resistance and Chemotherapy
Volume 14 - 2023 | https://doi.org/10.3389/fmicb.2023.1293363
Background: The increase in antibiotic resistance of bacteria has become a major concern in clinical treatment. Silver nanoparticles (AgNPs) have significant antibacterial effects against Streptococcus suis. Therefore, this study aimed to investigate the antibacterial activity and mechanism of action of AgNPs against multidrug-resistant S. suis.
Methods: The effect of AgNPs on the morphology of multidrug-resistant S. suis was observed using scanning electron microscopy (SEM). Differentially expressed proteins were analyzed by iTRAQ quantitative proteomics, and the production of reactive oxygen species (ROS) was assayed by H2DCF-DA staining.
Results: SEM showed that AgNPs disrupted the normal morphology of multidrug-resistant S. suis and the integrity of the biofilm structure. Quantitative proteomic analysis revealed that a large number of cell wall synthesis-related proteins, such as penicillin-binding protein and some cell cycle proteins, such as the cell division protein FtsZ and chromosomal replication initiator protein DnaA, were downregulated after treatment with 25 μg/mL AgNPs. Significant changes were also observed in the expression of the antioxidant enzymes glutathione reductase, alkyl hydroperoxides-like protein, α/β superfamily hydrolases/acyltransferases, and glutathione disulfide reductases. ROS production in S. suis positively correlated with AgNP concentration.
Conclusion: The potential antibacterial mechanism of AgNPs may involve disrupting the normal morphology of bacteria by inhibiting the synthesis of cell wall peptidoglycans and inhibiting the growth of bacteria by inhibiting the cell division protein FtsZ and Chromosomal replication initiator protein DnaA. High oxidative stress may be a significant cause of bacterial death. The potential mechanism by which AgNPs inhibit S. suis biofilm formation may involve affecting bacterial adhesion and interfering with the quorum sensing system.
Streptococcus suis is an important zoonotic pathogen with worldwide prevalence and is considered to be one of the most important bacterial pathogens causing significant economic losses in the swine industry (Segura, 2020). As with most pathogens, the ability of S. suis to form biofilms plays a significant role in its virulence and drug resistance (Wang et al., 2018). Currently, the treatment of S. suis infections relies on antibiotics; however, drug resistance is a concern. Data suggest that available veterinary drugs, such as ampicillin, cefepime, cefotaxime, ceftiofur, ceftriaxone, chloramphenicol, florfenicol, gentamicin, penicillin, and tiamulin, tend to be less effective in treating S. suis infections (Lunha et al., 2022). S. suis has an extremely high rate of resistance to tetracyclines, lincosamides, and macrolides, and resistance has spread globally (Uruen et al., 2022). Therefore, there is an urgent need to develop efficient alternatives to antibiotics.
Silver has strong antimicrobial potential and has been used since ancient times (Rai et al., 2012). AgNPs are now considered a viable alternative to antibiotics and appear to have appreciable potential to address the concern of bacterial multidrug resistance (Franci et al., 2015). AgNPs show distinct antibacterial and anti-biofilm formation effects on bacteria. For example, it has been proven that AgNPs could play antimicrobial roles in the multidrug-resistant (MDR) Pseudomonas aeruginosa and the main mechanism involves the disequilibrium of oxidation and antioxidation processes and the failure to eliminate the excessive ROS (Liao et al., 2019). Siddique et al. provided evidence of AgNPs being safe antibacterial and antibiofilm compounds against MDR Klebsiella pneumoniae (Siddique et al., 2020). Farouk et al. demonstrated the effective ability of AgNPs to fight MDR Salmonella spp. in vitro and in vivo without adverse effects (Farouk et al., 2020). AgNPs have antibacterial activity against multidrug-resistant bacteria pathogens, such as Vibrio cholerae, Staphylococcus aureus, Streptococcus pyogenes, Escherichia coli, and Klebsiella pneumoniae (Chinnathambi et al., 2023). The mechanism of antimicrobial activity of AgNPs involves four steps: (i) adhesion of AgNPs to cell wall/membrane and their disruption; (ii) intracellular penetration and damage; (iii) oxidative stress; and (iv) modulation of signal transduction pathways (Wahab et al., 2021; Tripathi and Goshisht, 2022). In our previous study, AgNPs showed significant activity against S. suis in vitro (Liu et al., 2022), however, the mechanism of AgNPs against S. suis remains unclear.
The occurrence of disease and the therapeutic effect of drugs are always accompanied by fluctuations and changes in numerous proteins. The application of quantitative proteomics can enable visualization of the up-regulation and down-regulation of differential proteins through charts, and provide functional annotations to intuitively analyze the possible mechanism of action of drugs (Saleh et al., 2019). In particular, iTRAQ technology has good applicability in the study of antibacterial mechanisms.
In this study, ultrastructural observations using scanning electron microscopy (SEM), fluorescence microscopy, and iTRAQ-based quantitative proteomics were used to investigate the antibacterial mechanism of action of AgNPs against multidrug-resistant S. suis. The findings can provide significant insights into the molecular mechanism of AgNPs against S.suis.
The ready-to-use AgNP stock solution (concentration: 30000 μg/mL, size: 5–7 nm, density: 1.01 g/cm3) was provided by Guangdong Shunde Zhengshanchuan Biotechnology Co., Ltd. (China).
A S. suis type 2 strain isolated from a diseased pig was employed and preserved in our laboratory. In our previous study, we identified it as an MDR bacterium that exhibits resistance to various antibiotics, such as tetracycline, doxycycline, penicillin, florfenicol, cefotaxime, kanamycin, and lincomycin, and that the minimum inhibitory concentration (MIC) of AgNPs against it is 25 μg/mL (Liu et al., 2022). S. suis was cultured in trypticase soy broth (TSB) or maintained on trypticase soy agar (TSA) supplemented with 5% bovine serum (Gibco, Auckland, New Zealand) at 37°C.
To observe the impact of AgNPs on the morphology of S. suis, S. suis bacteria were proliferated to 1 × 108 CFU/mL. After being exposed to 6.25, 12.5, 25, 50, and 100 μg/mL AgNPs for 12 h, the culture was precipitated by centrifugation and washed with PBS and then centrifuged; the precipitates were fixed in 2.5% glutaraldehyde for 2 h at 4. Subsequently, the precipitates were centrifuged, and washed with sterile PBS. The precipitates were then sequentially dehydrated through a series of alcohols (30, 50, 70, 90, and 100%) to the critical point. After coating with gold, the samples were examined using SEM (Zeiss Sigma 300).
To observe the effect of AgNPs on the biofilms of S. suis, 1 cm × 1 cm coverslips were ultrasonically cleaned for 1 h, and surface impurities and grease were removed and sterilized with high-pressure steam. A 6-well plate was prepared by placing a sterile coverslip in each hole, and then adding 1 mL of bacterial solution to cover the surface of the coverslips. The culture plate was cultured in an incubator at 37°C for 24 h, and the supernatant was discarded. The experimental group was added with TSB containing 25 μg/mL AgNPs and the control group was added with blank TSB. The culture plate was again cultured in an incubator at 37°C for 24 h and the supernatant was discarded. The plates were immersed in 2.5% glutaraldehyde for 3 h, then dehydrated with a series of concentrations of ethanol (30, 50, 70, 90, and 100%), dried, sprayed with gold, and observed with a scanning electron microscope.
The bacterial cells were treated with 25 μg/mL AgNPs and harvested by centrifugation. Proteins were extracted as follows:
The appropriate amount of sample was weighed and transferred into a 2 mL centrifuge tube, two steel beads and 1XCocktail with an appropriate amount of SDS L3 and EDTA were added, placed on ice for 5 min, and DTT was added at a final concentration of 10 mM. A grinder (frequency 60 HZ, duration 2 min) was used to crush the tissue, which was then centrifuged at 25,000 g*4°C for 15 min, and the supernatant was collected. DTT was added at a final concentration of 10 mM, and the mixture was placed in a water bath at 56°C for 1 h. IAM was added at a final concentration of 55 mM and the mixture was placed in a dark room for 45 min. Cold acetone was added to the protein solution at a ratio of 1:5, placed in a refrigerator at −20°C for 30 min, centrifuged at 25,000 g*4°C for 15 min, and the supernatant was discarded. The precipitate was air-dried, lysis buffer without SDS L3 was added, and a grinder (frequency 60HZ, duration 2 min) was used to promote protein solubilization. This was centrifuged for 15 min at 25,000 g*4°C to collect the supernatant; the supernatant is the protein solution. The proteins were digested, and the resultant peptides were labeled using iTRAQ 8-plex kits (AB Sciex). The untreated samples were labeled as 118, 119, and 121, and the samples treated with AgNPs were labeled as 114, 116, and 118.
The dried peptide samples were reconstituted with mobile phase A (2% ACN, 0.1% FA), centrifuged at 20,000 g for 10 min, and the supernatant was collected for injection. The separation was performed using a Thermo UltiMate 3,000 UHPLC system. The sample was first enriched in a trap column and desalted, and then separated on a self-packed C18 column (75 μm internal diameter, 3 μm column size, 25 cm column length) at a flow rate of 300 nL/min by the following effective gradient: 0 ~ 5 min, 5% mobile phase B (98% ACN, 0.1% FA); 5 ~ 45 min, mobile phase B linearly increased from 5 to 25%; 45 ~ 50 min, mobile phase B increased from 25 to 35%; 50 ~ 52 min, mobile phase B rose from 35 to 80%; 52 ~ 54 min, 80% mobile phase B; 54 ~ 60 min, 5% mobile phase B. The nanolitre liquid-phase separation end was directly connected to a mass spectrometer.
The peptides separated by liquid-phase chromatography were ionized using a Nano ESI source and then passed to a tandem mass spectrometer Q-Exactive HF X (Thermo Fisher Scientific, San Jose, CA) for data-dependent acquisition (DDA) mode detection. The main parameters were set as follows: ion source voltage, 1.9 kV; MS1 scanning range, 350 ~ 1,500 m/z; resolution, 60,000; MS2 starting m/z, 100; and resolution, 15,000. The ion screening conditions for MS2 fragmentation were as follows: charge 2+ to 6+ and the top 20 parent ions with a peak intensity exceeding 10,000. The ion fragmentation mode was HCD and fragment ions were detected using the Orbitrap. The dynamic exclusion time was set to 30 s. The AGC was set to 3E6 in MS1 and 1E5 in MS2.
The bacteria of S.suis proliferated to 1 × 108 CFU/mL after exposure to 6.25, 12.5, 25, 50, and 100 μg/mL AgNPs for 6 h. ROS was measured by 2′,7′-dichloro fluorescein diacetate (H2DCF-DA) based on the method of Liao et al. (2019). Initially, a 10 mM H2DCF-DA stock solution in dimethyl sulfoxide was diluted to 1 mM working solution with a TSB medium. The collected bacteria were washed with PBS and suspended in 1.8 mL of PBS. Then the samples were incubated with 200 μL of working solution at 37°C for 30 min in darkness. Subsequently, the cells were harvested, washed, and resuspended in PBS. This bacterial suspension was dropped on a slide and naturally dried in the darkness at room temperature before fluorescence microscopy (ZEISS, Axio vert. A1) detection. The cultured bacteria were lysed using an alkaline lysis buffer and centrifuged at 3,000 rpm for 5 min. Subsequently, 1 mL of lysate supernatant was prepared for fluorescence detection (Multifunctional microplate reader, Thermo Scientific varios) at excitation and emission wavelengths of 470 and 529 nm, respectively.
All data are expressed as means ± standard deviation with three biological replicates. GraphPad Prism 8.0 was used to perform one-way ANOVA analysis at p ≤ 0.05 and create graphs. Quantification of iTRAQ data was performed using the IQuant software (2.4.0), and the Mascot search engine (v2.3.02, Matrix Sciences, London, United Kingdom) was used to search the UniProt database.
After treatment with AgNP for 12 h, the bacteria of each multidrug-resistant S. suis strain were collected for morphological examination using SEM. Compared to the control, pits appeared on the surface of the bacterial cells after AgNP treatment, and the cell morphology was distorted (Figure 1). The analysis showed that AgNPs disrupted the morphology of S. suis, and the destruction of cells was aggravated with an increase in AgNP concentration. Further, the structure of the biofilm was destroyed by AgNPs (Figure 2).
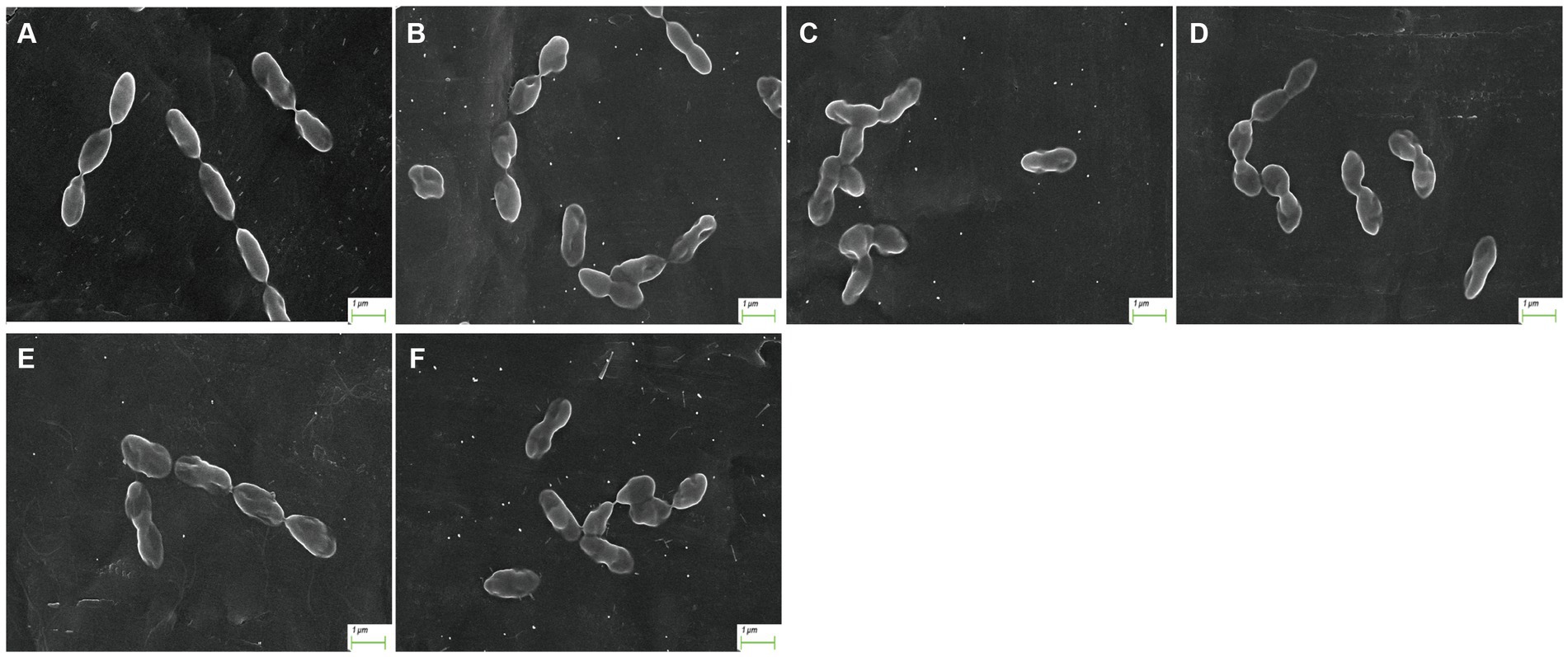
Figure 1. Morphology of S. suis after AgNP treatment. (A) control, (B) 6.25 μg/mL, (C) 12.5 μg/mL, (D) 25 μg/mL, (E) 50 μg/mL, (F) 100 μg/mL.
Quantitative proteomic analysis revealed 1,268 bacterial proteins, and a volcano map of the differentially expressed proteins (DEPs) in S. suis was analyzed (X-axis is log2 fold change) (Figure 3A). In total, 633 upregulated and 635 downregulated genes were significantly altered in response to AgNP exposure at 25 μg/mL. This depicts a volcano plot of log2 fold-change (x-axis) versus the -log10 Q-value (y-axis, representing the probability that the protein is differentially expressed). Q-value <0.05 and fold change >1.2 are considered significant differential expression thresholds. Quantitative repeatability was assessed using CV (CV = SD/mean). The lower the CV value, the better the repeatability. The CV value in this experiment was 0.12, indicating good repeatability (Figure 3B).
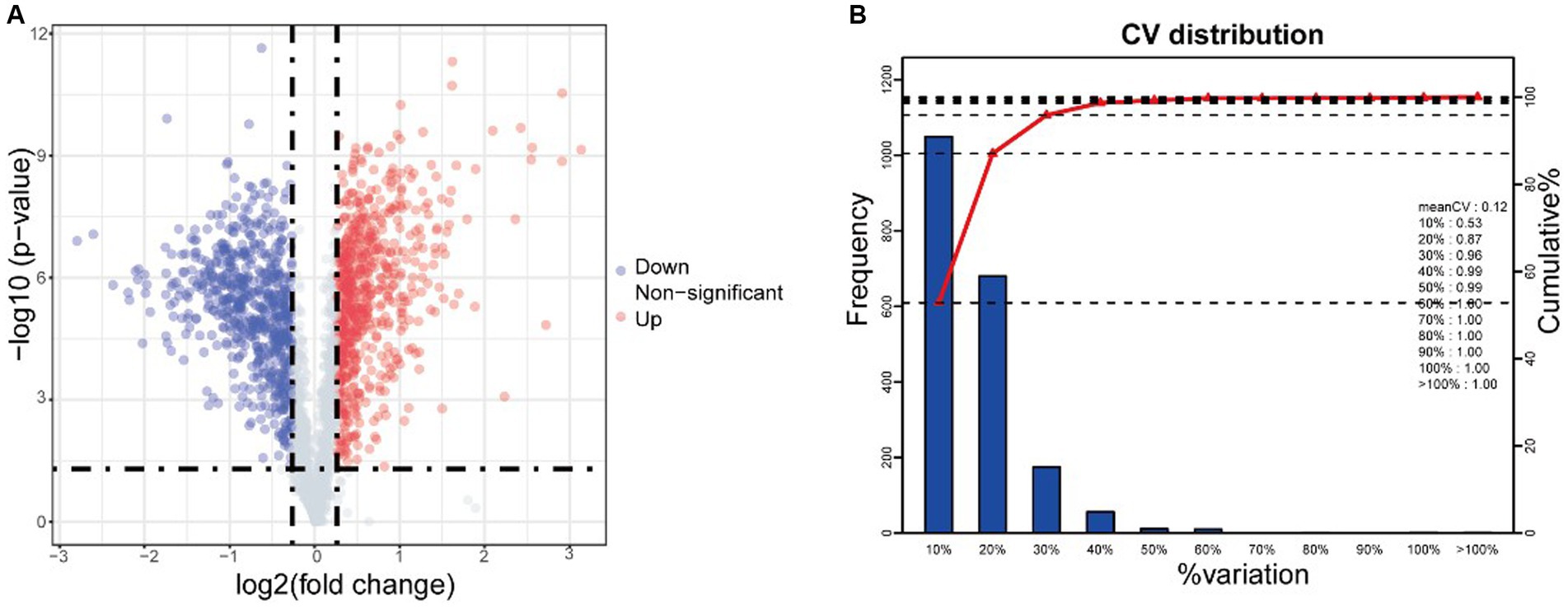
Figure 3. iTRAQ analysis reveals differentially expressed proteins (DEPs) after AgNPs treatment. (A) Volcano of differentially expressed proteins, (B) CV distribution in replicate. Red and green dots indicate points of interest that display both large- and high-magnitude fold changes, respectively. Red dots indicate significantly upregulated proteins that passed the screening threshold. Blue dots indicate significantly downregulated proteins that passed the screening threshold. Gray dots represent non-significantly differentially expressed proteins.
Gene Ontology (GO) analysis classified all the identified proteins and DEPs into three broad categories: molecular function, cellular components, and biological processes (Figure 4A). Molecular functional analysis showed that AgNP treatment significantly affected catalytic activity, binding, transporter activity, structural molecule activity, and transcription regulator activity. Partial GO enrichment analysis of DEPs is shown in Table 1.
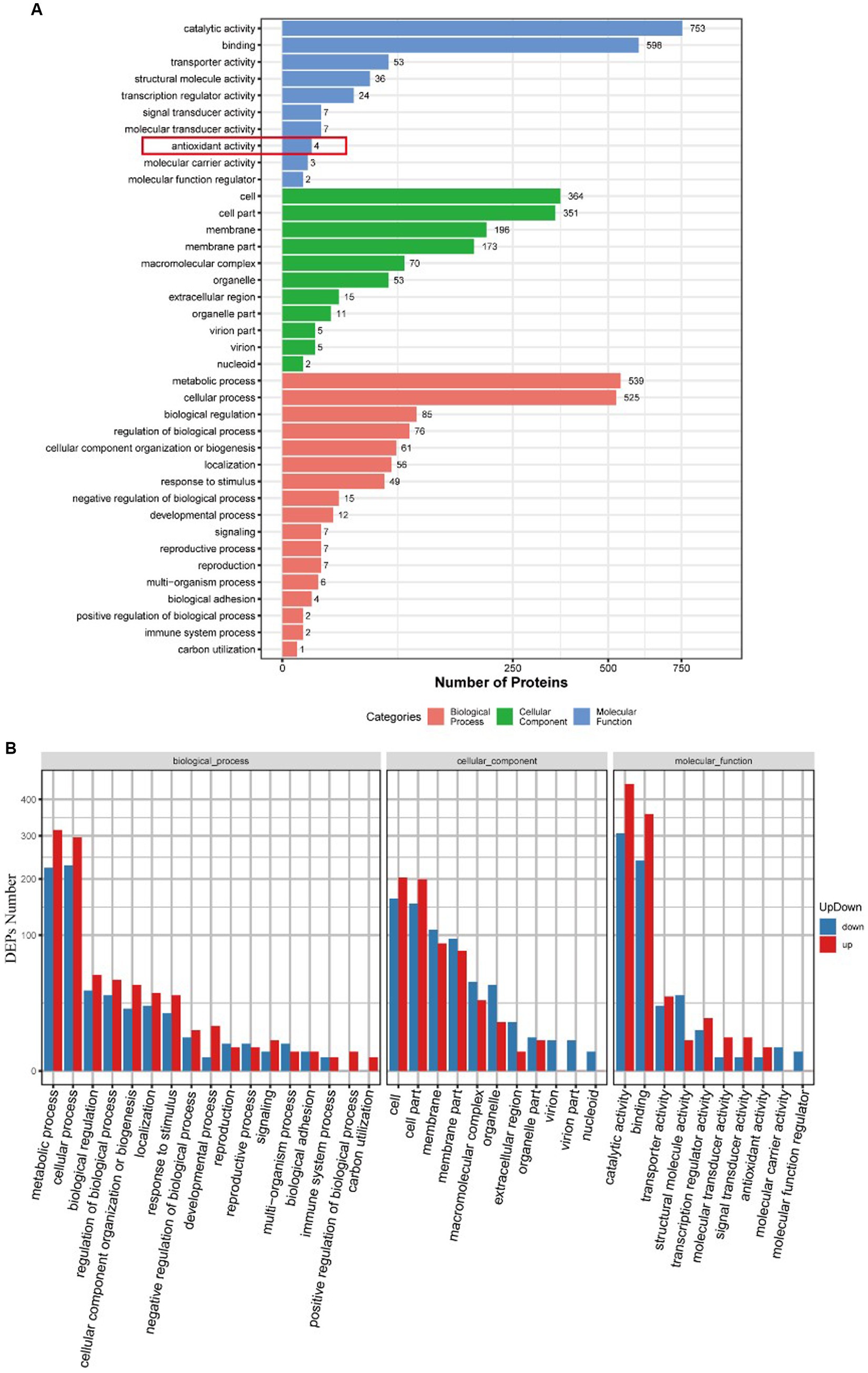
Figure 4. GO functional annotation and enrichment analysis of DEPs between the control and AgNPs-treated group. (A) GO functional annotation. (B) GO enrichment analysis; red: Up-regulation, blue: down-regulation.
Figure 4B illustrates the upregulation and downregulation of differentially expressed proteins in each classification. There were 196 proteins enriched in the membrane, of which 88 were upregulated, and 108 were downregulated.
The heatmap of antioxidant activity proteins between the AgNPs-treated group and the control group Most of these antioxidant proteins exhibited a trend of upregulated expression, except the hydrolases/acyltransferases of the α/β superfamily (Figure 5).
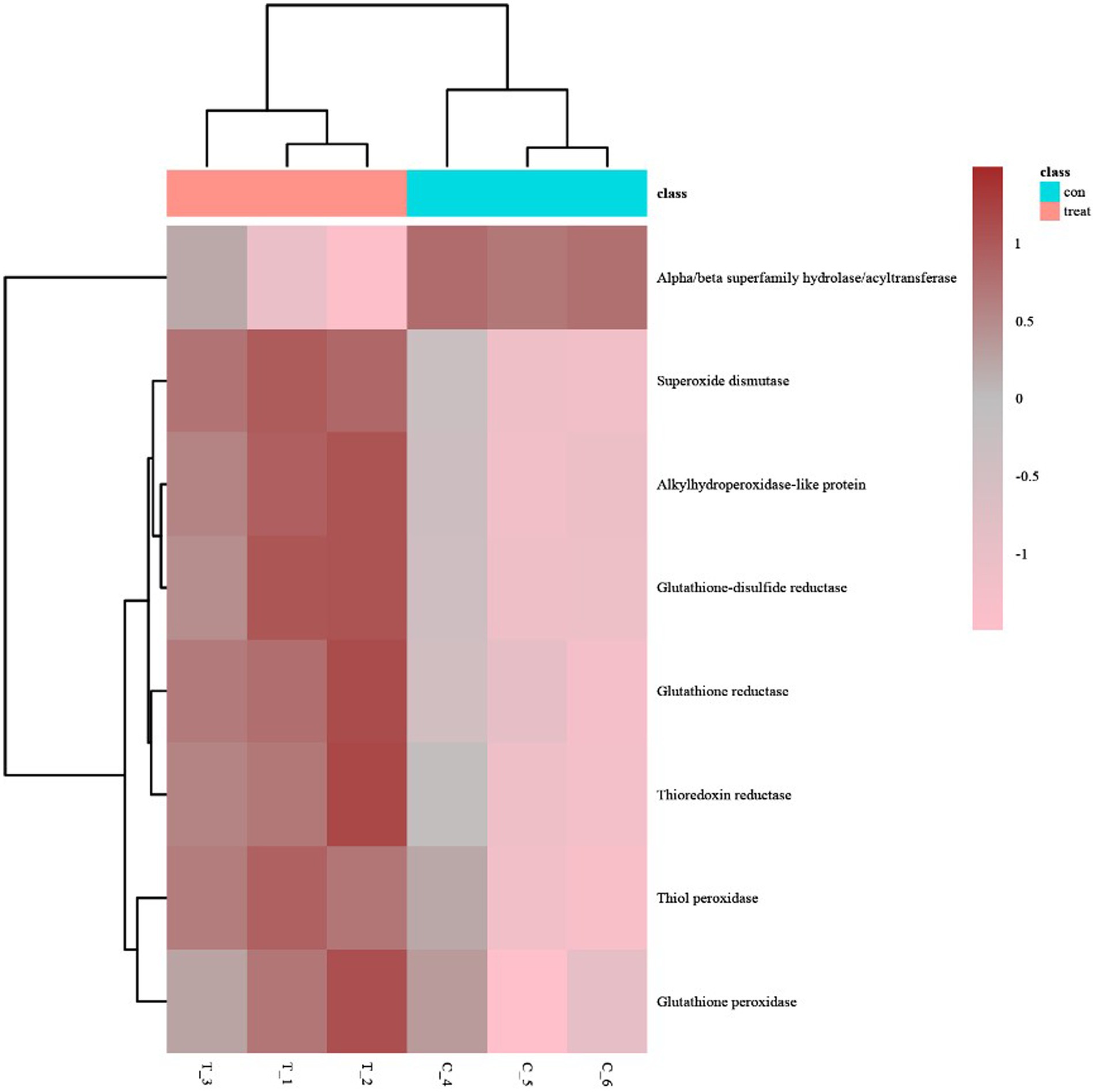
Figure 5. The heatmap of antioxidant activity proteins between AgNP-treated and control samples; pink indicates down-regulation, brown indicates up-regulation, and gray indicates no detectable expression change.
GO analysis of differentially expressed proteins revealed four differentially expressed proteins enriched in antioxidant activity (Figure 4A): glutathione reductase, alkyl hydroperoxides-like protein, α/β superfamily hydrolases/acyltransferases, and glutathione disulfide reductases, of which the α/β superfamily hydrolases/acyltransferases were significantly downregulated. The remaining three proteins were significantly upregulated (Table 2).
Clusters of Orthologous Groups of proteins (COGs) representing major phylogenetic lineages were delineated by comparing protein sequences encoded in complete genomes. The COG annotation of DEPs is shown in Figure 6, where among all the differentially expressed proteins, we focused on 111 proteins that were related to cell wall/membrane/environment biogenesis.
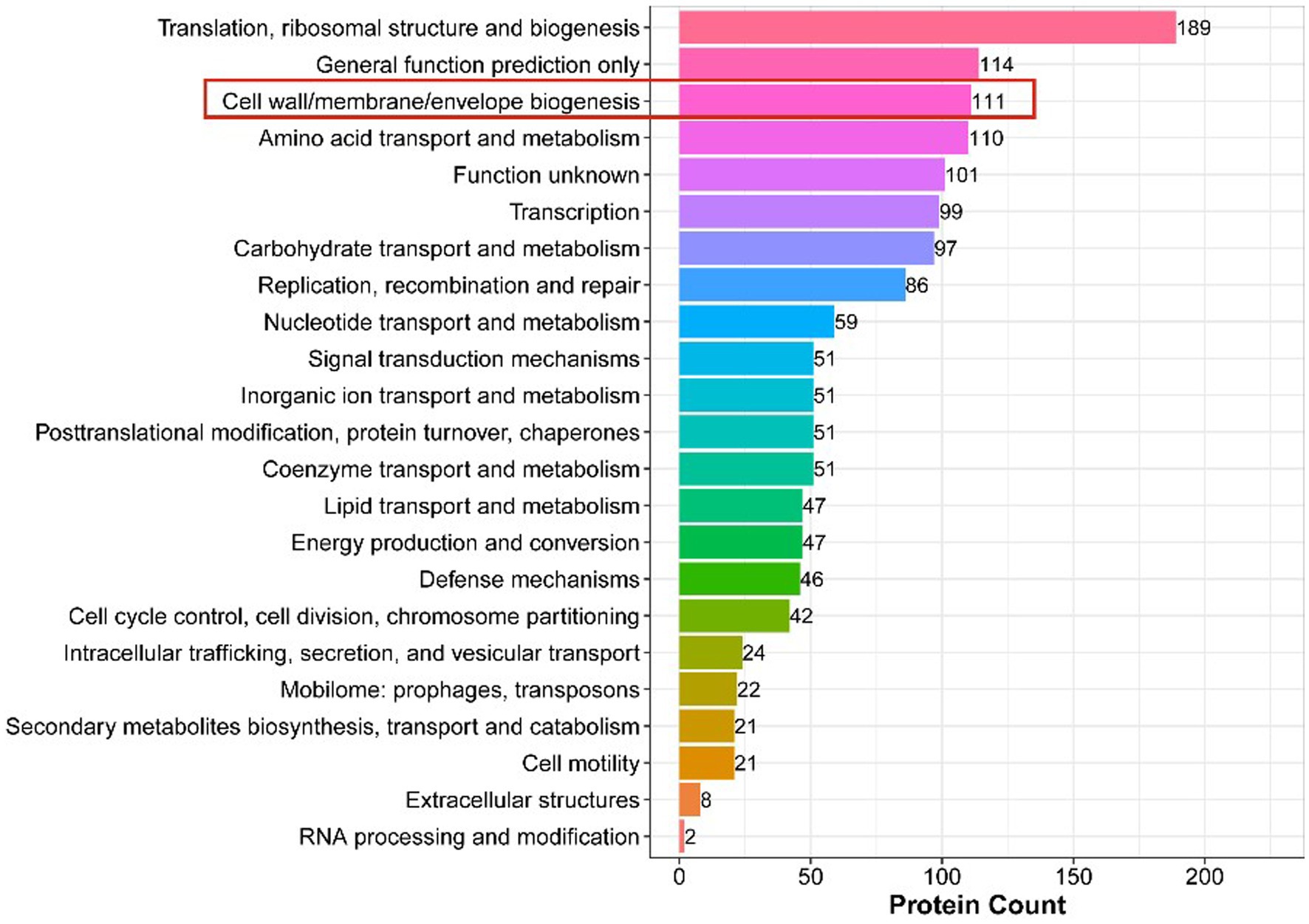
Figure 6. COG Annotation of DEPs. The x-axis displays the COG term, y-axis displays the corresponding protein count illustrating the protein number of different functions.
Capsular polysaccharides (CPS) are the main components of the outer capsule of the bacterial cell wall. CPS is an essential virulence factor in the pathogenesis of S. suis 2, and the synthesis of CPS repeating units involves multiple glycosyltransferases (Zhang et al., 2016). This analysis revealed differential expression of many CPS-related proteins (Table 3).
Peptidoglycan is an important component of the cell wall of Gram-positive bacteria, necessary for maintenance of cell morphology, size, osmotic pressure, and survival (Egan et al., 2020). Among the 111 DEPs annotated for the biosynthesis of the cell wall, cell membrane, and envelope, we found that 14 peptidoglycan synthesis-related proteins were downregulated (Table 4). Therefore, we speculated that AgNPs inhibited the synthesis of peptidoglycan, thus, affecting the normal structure of the cell wall and destroying bacterial morphology.
There were 34 differentially expressed proteins enriched in the quorum sensing pathway, of which 23 were upregulated and 11 were downregulated (Figure 7). The quorum sensing system had a regulatory effect on various life activities of S. suis, and AgNPs interfered with the expression of related proteins in the quorum sensing system, which would affect the regulation of bacterial density of fine communities. This could potentially be one of the mechanisms through which AgNPs inhibit biofilm formation or eliminate the formed biofilms of S. suis.
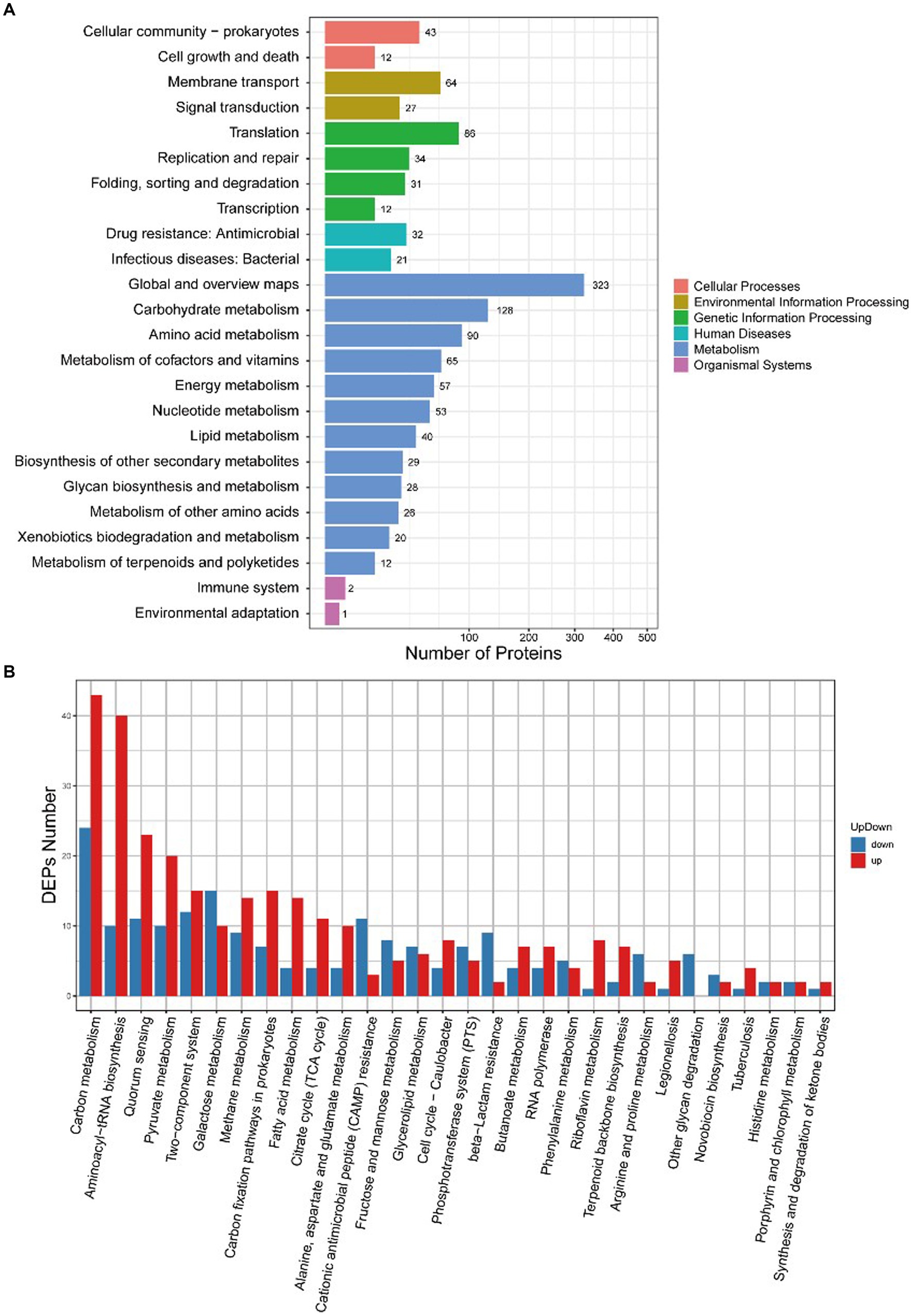
Figure 7. KEGG enrichment analysis of DEPs between the control group and the AgNP-treated group. (A) Enrichment at levels 1 and 2, (B) Specific enrichment pathways of DEPs.
DEPs enriched in the cell cycle are shown in Figure 8. Table 5 describes the significantly downregulated cell division-related proteins. This includes the chromosome replication initiator protein DnaA, cell division proteins FtsZ and DivIB, and the ATP-dependent Clp protease ATP-binding subunit. The results showed that AgNPs affected the expression of cell division protein-related proteins and inhibited the division of S. suis at the initial stage, which may be an important factor affecting bacterial proliferation.
H2DCF-DA staining and fluorescence microscopy revealed that, compared to the weak fluorescence of untreated S. suis, the fluorescence intensity of the AgNP-treated bacteria increased with an increase in AgNP concentration within 6 h (Figure 9). The bacteria displayed increased fluorescence intensity when the AgNPs were added (Figure 10), indicating that the AgNPs induced ROS production in a dose-dependent manner.
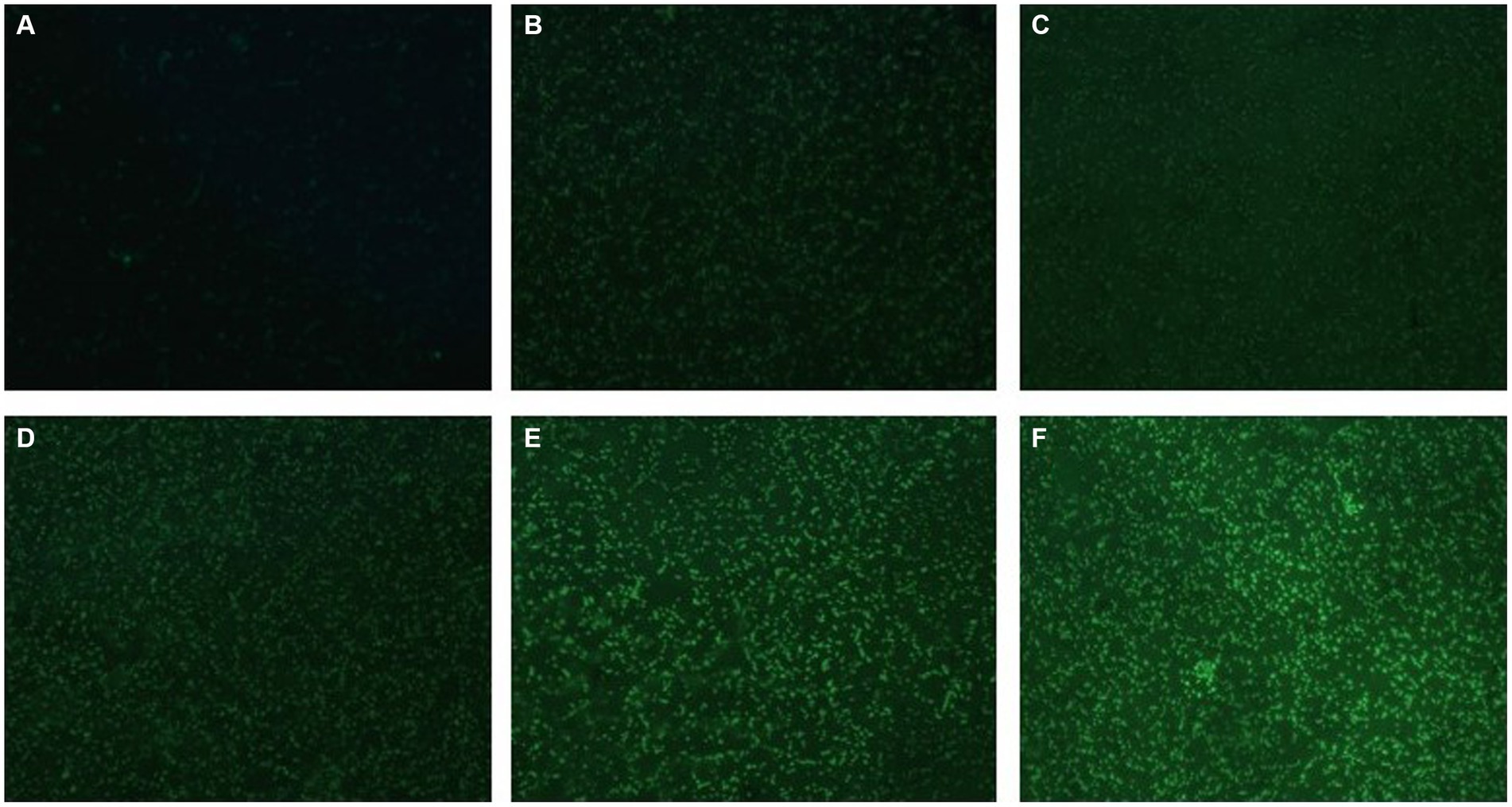
Figure 9. Changes in ROS production in AgNP-treated multidrug-resistant S. suis at different concentrations under fluorescence microscopy with ×400 magnification. (A) The untreated S.suis without observable fluorescence. (B–F) Fluorescence observation of the bacteria treated with AgNPs at different concentrations of 6.25, 12.5, 25, 50, and 100 μg/mL.
The lack of an effective vaccine to prevent S. suis disease has led to the widespread use of antibiotics worldwide, with the attendant problem of bacterial resistance. AgNPs exhibit antibacterial, antifungal, antiviral, anti-inflammatory, and antiangiogenic properties owing to their unique physical, chemical, and biological properties (Gurunathan et al., 2014). The antibacterial activity of AgNPs is reportedly due to the production of ROS, malondialdehyde, and leakage of proteins and sugars from bacterial cells (Yuan et al., 2017). It is well known that excessive ROS can lead to oxidative stress in cells; relevant evidence indicates that intensified oxidative stress disturbs energy metabolism and protein metabolism of bacteria (Chen et al., 2022; Zhou et al., 2023a).
In our previous study, AgNPs showed significant antibacterial and anti-biofilm effects against S. suis (Liu et al., 2022). In the present study, the mechanism of action of AgNPs against S. suis was explored based on their previously established antibacterial effects. Our results indicate that AgNPs may damage the morphology of multidrug-resistant S. suis and the structure of its biofilm (Figures 1, 2). The cell wall and membrane are important structures for maintenance of normal bacteria morphology. We found that many proteins related to peptidoglycan synthesis were downregulated, including penicillin-binding proteins (PBPs), glycosyltransferases, and LytR family transcriptional regulators (Table 4). The normal presence of PBPs is necessary for maintenance of normal bacteria morphology and function (Wei et al., 2003). Therefore, we inferred that the inhibition of PBP expression by AgNPs is an important factor in their antibacterial effect.
Quorum sensing (QS) and capsular polysaccharides have important influences on biofilm formation in S.suis biofilms. QS is a microbial cell-to-cell communication process that dynamically regulates various metabolic and physiological activities (Wu et al., 2020). LuxS/AI-2-mediated QS is a key system involved in the formation of biofilms (Wang et al., 2018). Our results identified 34 differentially expressed proteins enriched in the quorum sensing pathway, of which the expression of S-ribosylhomocysteine lyase was significantly up-regulated. S-ribosylhomocysteine lyase is regulated by LuxS, which is involved in the synthesis of autoinducer 2 (AI-2) which is secreted by bacteria and is used to communicate both the cell density and the metabolic potential of the environment. Moreover, upregulation of cps2J may promote the synthesis of capsular polysaccharides (Wang S. et al., 2017), thereby decreasing the formation of S. suis biofilm. These results suggest that AgNPs inhibit the formation of S. suis biofilms by affecting QS and capsular polysaccharide synthesis.
Cell division is an important core process in almost all organisms and is regulated by multiple genes and proteins. Bacterial cell division is coordinated by macromolecular protein complexes. DnaA is the most conserved DNA replication initiation protein, which can initiate chromosome replication and acts as a transcription factor (Menikpurage et al., 2021). FtsZ is the initiation structure of division formation and cytokinesis (Silber et al., 2020). It is an essential cell division protein that forms a contraction ring structure (Z ring) at the site of cell division. It is also an important target of antibacterial drugs (Ur et al., 2020). This study found that the expression of the chromosome replication initiation protein DnaA, cell division initiation proteins FtsZ and DivIB, and the ATP-dependent Clp protease proteolytic subunit was downregulated after AgNP treatment. These proteins are essential for bacterial replication and division; therefore, they could be important factors for the effect of AgNPs on bacterial proliferation.
ROS is an umbrella term for an array of molecular oxygen derivatives that occur as a normal attribute of aerobic life (Sies and Jones, 2020). Most living organisms possess enzymatic defenses (superoxide dismutase [SOD], glutathione peroxidase [GPx], and glutathione reductase [GR]), non-enzymatic antioxidant defenses (glutathione, thioredoxin, Vitamin C, Vitamin E), and repair systems that protect them against oxidative stress (Wang Y. et al., 2017). However, excessive ROS causes an imbalance between oxidation and antioxidation, resulting in oxidative stress that damages various cellular components (including proteins, lipids, and DNA) (Schieber and Chandel, 2014; Huang et al., 2021) and ultimately induces bacterial death. Van Acker and Coenye (2017) demonstrated that ROS mediates the bactericidal mechanisms of some antibiotics. ROS levels that exceed the capacity of the cellular antioxidant defense system induce oxidative stress (Niki, 2016). The intensified oxidative stress may also affect the structure and permeability of the cell membrane, subsequently leading to cell death (Zhou et al., 2023b).
Our results revealed that the production amount of bacterial ROS was positively correlated with the concentration of added AgNPs to some extent. Based on iTRAQ quantitative proteomic analysis, our results indicate that AgNPs significantly affected the expression of a large number of bacterial proteins, including oxidoreductases. We found that the expression of some oxidoreductases, such as GR, increased significantly, which may have been due to an increase in ROS. We speculate that the action of AgNPs leads to the enhancement of the metabolic activity of cells, which then produces excessive ROS, and the increasing ROS induces the continuous expression of antioxidant enzymes to eliminate excessive ROS. While four antioxidant enzymes were significantly differentially expressed, our results suggest that this is insufficient to effectively eliminate excessive ROS in the bacterial cells. As a result, the cells experience oxidative stress, leading to oxidative damage and, ultimately, death. These findings are consistent with previous research by Liao et al. (2019) and suggest that excessive oxidative stress is a key factor in bacterial mortality.
In summary, our findings indicate that AgNPs disturb the natural morphology of S. suis and its biofilm. The results obtained through proteomic analysis suggest that AgNPs may negatively impact the cell wall structure by inhibiting the synthesis of peptidoglycan, thus, leading to bacterial morphology disruption. Additionally, AgNPs can impede bacterial adhesion, interfere with QS system, and inhibit bacterial growth by targeting the cell division protein FtsZ and the chromosomal replication initiator protein DnaA. AgNPs’ induction of considerable oxidative stress is a significant contributing factor to bacterial death.
These findings contribute to a better understanding of the molecular basis of AgNPs’ antibacterial activity, highlighting potential targets for the development of new antimicrobial agents against S. suis infections.
The datasets for this study can be found in iProX, https://www.iprox.cn/page/project.html?id=IPX0007482000. The mass spectrometry proteomics data have been deposited to the ProteomeXchange Consortium, https://proteomecentral.proteomexchange.org, via the iProX partner repository with the dataset identifier PXD046681.
BL: Writing – original draft, Writing – review & editing. DL: Writing – review & editing. TC: Writing – review & editing. XW: Writing – review & editing. HX: Writing – review & editing. GW: Writing – review & editing. RC: Writing – review & editing.
The author(s) declare financial support was received for the research, authorship, and/or publication of this article. This study was supported by the Guangdong Modern Agro-industry Technology Research System (2022KJ114), Science and Technology Plan Projects of Guangdong Province (Grant No. 2021B1212050021), Prevention and control of African swine fever and construction of African swine fever-free zone (20212100015), and scientific and technological support and technical demonstration of pig industrial park in Yun’an District, Yunfu City (K06-005, 2022).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Chen, H., Ji, P. C., Qi, Y. H., Chen, S. J., Wang, C. Y., Yang, Y. J., et al. (2022). Inactivation of Pseudomonas aeruginosa biofilms by thymoquinone in combination with nisin. Front. Microbiol. 13:1029412. doi: 10.3389/fmicb.2022.1029412
Chinnathambi, A., Alharbi, S. A., Joshi, D., Jhanani, G. K., On-uma, R., Jutamas, K., et al. (2023). Synthesis of AgNPs from leaf extract of Naringi crenulata and evaluation of its antibacterial activity against multidrug resistant bacteria. Environ. Res. 216:114455. doi: 10.1016/j.envres.2022.114455
Egan, A., Errington, J., and Vollmer, W. (2020). Regulation of peptidoglycan synthesis and remodelling. Nat. Rev. Microbiol. 18, 446–460. doi: 10.1038/s41579-020-0366-3
Farouk, M. M., El-Molla, A., Salib, F. A., Soliman, Y. A., and Shaalan, M. (2020). The role of silver nanoparticles in a treatment approach for multidrug-resistant salmonella species isolates. Int. J. Nanomed. 15, 6993–7011. doi: 10.2147/IJN.S270204
Franci, G., Falanga, A., Galdiero, S., Palomba, L., Rai, M., Morelli, G., et al. (2015). Silver nanoparticles as potential antibacterial agents. Molecules 20, 8856–8874. doi: 10.3390/molecules20058856
Gurunathan, S., Han, J. W., Kwon, D. N., and Kim, J. H. (2014). Enhanced antibacterial and anti-biofilm activities of silver nanoparticles against gram-negative and gram-positive bacteria. Nanoscale Res. Lett. 9:373. doi: 10.1186/1556-276X-9-373
Huang, X., He, D., Pan, Z., Luo, G., and Deng, J. (2021). Reactive-oxygen-species-scavenging nanomaterials for resolving inflammation. Mater Today Bio. 11:100124. doi: 10.1016/j.mtbio.2021.100124
Liao, S., Zhang, Y., Pan, X., Zhu, F., Jiang, C., Liu, Q., et al. (2019). Antibacterial activity and mechanism of silver nanoparticles against multidrug-resistant Pseudomonas aeruginosa. Int. J. Nanomed. 14, 1469–1487. doi: 10.2147/IJN.S191340
Liu, B., Li, C., Jiang, Z., Yang, D., Zhang, K., Chu, P., et al. (2022). The effect of silver Nanparticles on the activity of Streptococcus suis and its biofilm formation in vitro. China Anim. Husband. Vet. Med. 49, 1172–1180. doi: 10.16431/j.cnki.1671-7236.2022.03.040 (in Chinese)
Lunha, K., Chumpol, W., Samngamnim, S., Jiemsup, S., Assavacheep, P., and Yongkiettrakul, S. (2022). Antimicrobial susceptibility of Streptococcus suis isolated from diseased pigs in Thailand, 2018-2020. Antibiotics (Basel). 11:410. doi: 10.3390/antibiotics11030410
Menikpurage, I. P., Woo, K., and Mera, P. E. (2021). Transcriptional activity of the bacterial replication initiator Dna a. Front. Microbiol. 12:662317. doi: 10.3389/fmicb.2021.662317
Niki, E. (2016). Oxidative stress and antioxidants: distress or eustress? Arch. Biochem. Biophys. 595, 19–24. doi: 10.1016/j.abb.2015.11.017
Rai, M. K., Deshmukh, S. D., Ingle, A. P., and Gade, A. K. (2012). Silver nanoparticles: the powerful nanoweapon against multidrug-resistant bacteria. J. Appl. Microbiol. 112, 841–852. doi: 10.1111/j.1365-2672.2012.05253.x
Saleh, S., Staes, A., Deborggraeve, S., and Gevaert, K. (2019). Targeted proteomics for studying pathogenic bacteria. Proteomics 19:e1800435. doi: 10.1002/pmic.201800435
Schieber, M., and Chandel, N. S. (2014). ROS function in redox signaling and oxidative stress. Curr. Biol. 24, R453–R462. doi: 10.1016/j.cub.2014.03.034
Segura, M. (2020). Streptococcus suis research: Progress and challenges. Pathogens 9:707. doi: 10.3390/pathogens9090707
Siddique, M. H., Aslam, B., Imran, M., Ashraf, A., Nadeem, H., Hayat, S., et al. (2020). Effect of silver nanoparticles on biofilm formation and EPS production of multidrug-resistant Klebsiella pneumoniae. Biomed. Res. Int. 2020, 1–9. doi: 10.1155/2020/6398165
Sies, H., and Jones, D. P. (2020). Reactive oxygen species (ROS) as pleiotropic physiological signalling agents. Nat. Rev. Mol. Cell Biol. 21, 363–383. doi: 10.1038/s41580-020-0230-3
Silber, N., Matos, D. O. C., Mayer, C., and Sass, P. (2020). Cell division protein Fts Z: from structure and mechanism to antibiotic target. Future Microbiol. 15, 801–831. doi: 10.2217/fmb-2019-0348
Tripathi, N., and Goshisht, M. K. (2022). Recent advances and mechanistic insights into antibacterial activity, antibiofilm activity, and cytotoxicity of silver nanoparticles. ACS Appl. Biol. Mater. 5, 1391–1463. doi: 10.1021/acsabm.2c00014
Ur, R. M., Wang, P., Wang, N., and Chen, Y. (2020). A key bacterial cytoskeletal cell division protein Fts Z as a novel therapeutic antibacterial drug target. Bosn. J. Basic Med. Sci. 20, 310–318. doi: 10.17305/bjbms.2020.4597
Uruen, C., Garcia, C., Fraile, L., Tommassen, J., and Arenas, J. (2022). How Streptococcus suis escapes antibiotic treatments. Vet. Res. 53:91. doi: 10.1186/s13567-022-01111-3
Van Acker, H., and Coenye, T. (2017). The role of reactive oxygen species in antibiotic-mediated killing of bacteria. Trends Microbiol. 25, 456–466. doi: 10.1016/j.tim.2016.12.008
Wahab, S., Khan, T., Adil, M., and Khan, A. (2021). Mechanistic aspects of plant-based silver nanoparticles against multi-drug resistant bacteria. Heliyon 7:e07448. doi: 10.1016/j.heliyon.2021.e07448
Wang, S., Wang, C., Gao, L., Cai, H., Zhou, Y., Yang, Y., et al. (2017). Rutin inhibits Streptococcus suis biofilm formation by affecting CPS biosynthesis. Front. Pharmacol. 8:379. doi: 10.3389/fphar.2017.00379
Wang, Y., Wang, Y., Sun, L., Grenier, D., and Yi, L. (2018). Streptococcus suis biofilm: regulation, drug-resistance mechanisms, and disinfection strategies. Appl. Microbiol. Biotechnol. 102, 9121–9129. doi: 10.1007/s00253-018-9356-z
Wang, Y., Wu, Y., Wang, Y., Xu, H., Mei, X., Yu, D., et al. (2017). Antioxidant properties of probiotic bacteria. Nutrients 9:521. doi: 10.3390/nu9050521
Wei, Y., Havasy, T., McPherson, D. C., and Popham, D. L. (2003). Rod shape determination by the Bacillus subtilis class B penicillin-binding proteins encoded by pbp a and pbp H. J. Bacteriol. 185, 4717–4726. doi: 10.1128/JB.185.16.4717-4726.2003
Wu, S., Liu, J., Liu, C., Yang, A., and Qiao, J. (2020). Quorum sensing for population-level control of bacteria and potential therapeutic applications. Cell. Mol. Life Sci. 77, 1319–1343. doi: 10.1007/s00018-019-03326-8
Yuan, Y., Peng, Q., and Gurunathan, S. (2017). Effects of silver nanoparticles on multiple drug-resistant strains of Staphylococcus aureus and Pseudomonas aeruginosa from mastitis-infected goats: an alternative approach for antimicrobial therapy. Int. J. Mol. Sci. 18:569. doi: 10.3390/ijms18030569
Zhang, Y., Ding, D., Liu, M., Yang, X., Zong, B., Wang, X., et al. (2016). Effect of the glycosyltransferases on the capsular polysaccharide synthesis of Streptococcus suis serotype 2. Microbiol. Res. 185, 45–54. doi: 10.1016/j.micres.2016.02.002
Zhou, J., Ji, P., Wang, C., Yang, Y., Zhao, X., and Tang, H. (2023a). Anti-virulence activity of dihydrocuminyl aldehyde and nisin against spoilage bacterium Pseudomonas aeruginosa XZ01. LWT-Food Sci. Technol. 177:114573. doi: 10.1016/j.lwt.2023.114573
Keywords: AgNPs, iTRAQ, Streptococcus suis , antibacterial mechanism, Biofilm
Citation: Liu B, Liu D, Chen T, Wang X, Xiang H, Wang G and Cai R (2023) iTRAQ-based quantitative proteomic analysis of the antibacterial mechanism of silver nanoparticles against multidrug-resistant Streptococcus suis. Front. Microbiol. 14:1293363. doi: 10.3389/fmicb.2023.1293363
Received: 13 September 2023; Accepted: 24 October 2023;
Published: 15 November 2023.
Edited by:
Geelsu Hwang, University of Pennsylvania, United StatesReviewed by:
Zhenting Xiang, University of Pennsylvania, United StatesCopyright © 2023 Liu, Liu, Chen, Wang, Xiang, Wang and Cai. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Rujian Cai, NDY2ODY2NTY5QHFxLmNvbQ==
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.