
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Microbiol. , 22 November 2023
Sec. Microbial Physiology and Metabolism
Volume 14 - 2023 | https://doi.org/10.3389/fmicb.2023.1292937
Rhodopseudomonas palustris CGA009 is a Gram-negative, purple non-sulfur, metabolically diverse bacterium with wide-ranging habitats. The extraordinary ability of R. palustris to decompose a variety of raw materials and convert them into high-value products makes it an attractive host for biotechnology and industrial applications. However, being a freshwater bacterium R. palustris has limited application in highly-saline environments. Therefore, it is of great significance to obtain the salt-tolerant strain of R. palustris and understand its tolerance mechanism. In this study, R. palustris CGA009 was successfully evolved into eight salt-tolerant strains using an adaptive laboratory evolution technique. RPAS-11 (R. palustris anti-salt strain 11) was selected as the best salt-tolerant strain and was used in further studies to explore the salt-tolerance mechanism. The expression of most genes associated with the carotenoid synthesis in RPAS-11 increased significantly under high concentration of salt stress, suggesting that carotenoid synthesis is one of the reasons for the salt tolerance of RPAS-11. Gene overexpression and knockout experiments were performed to get clear about the role of carotenoids in salt stress tolerance. RPAS-11-IDI, the mutant with overexpression of IDI (Isopentenyl diphosphate isomerase) exhibited enhanced salt tolerance, whereas the knockout mutant CGA009-∆crtI showed a decline in salt tolerance. In addition, the results indicated that rhodopin, a carotenoid compound, was the key pigment responsible for the salt tolerance in R. palustris. Furthermore, the production of lycopene, a widely-used carotenoid, was also increased. Taken together, our research helps to deepen the understanding of the salt tolerance mechanism of R. palustris and also widens the application of R. palustris in highly-saline environments.
Rhodopseudomonas palustris is a Gram-negative, purple non-sulfur bacterium (PNSB). Its ability to utilize versatile carbon sources and to flexibly switch between photoautotrophic, photoheterotrophic, chemoheterotrophic, and chemoautotrophic modes of metabolism, makes it a metabolically diverse and environmentally widespread bacterium. It has been isolated from different anaerobic water environments such as lakes, soils, swamps, and oceans (Larimer et al., 2004; Guan et al., 2017). Rhodopseudomonas palustris has excellent degradation and detoxification ability, which enables it to degrade lignin, aromatic compounds, insecticides, herbicides and assimilate heavy metals (Li et al., 2022a). Numerous studies are available on the application of R. palustris in wastewater treatment, environmental remediation, biofuel production, agricultural biological stimulation, and bioelectricity production (Harwood et al., 1998; Sakpirom et al., 2017; Wu et al., 2019; Li et al., 2022a). Most of these wastes, such as effluents from organic peroxides production, pharmacy, tanneries (1–10%), textiles (3–10%), seafood processing (1.3–3.9%), and petroleum refineries (sometimes higher than 10.5%), exhibit high salt (mainly NaCl) concentrations (Woolard and Irvine, 1995; Díaz et al., 2002; Lefebvre et al., 2005; Lefebvre and Moletta, 2006; Lyles et al., 2008; Xiao and Roberts, 2010; Ogugbue et al., 2011; Amin et al., 2014). However, the low salt tolerance of freshwater R. palustris makes it difficult to grow in salt-containing media, which will limit its practical application in high-salinity wastewater treatment. The ability to utilize diverse carbon sources and produce multiple organic compounds makes R. palustris an excellent chassis organism for biotechnological research. The bacterium produces not only different biofuels like hydrogen (Zhu X. et al., 2011), methane (Fixen et al., 2016), and butanol (Doud et al., 2017), but also produces high-value compounds, such as terpenoids (Xu et al., 2016), poly-β-hydroxybutyrate (PHB; Wu et al., 2012), and 5-aminolevulinic acid (ALA; Nunkaew et al., 2018). Almost all fermentation processes require sterilization to reduce microbial contamination, which consumes a lot of energy and money. Seawater (contains about 3.5% NaCl; Pernetti and Di Palma, 2005) has a high osmotic pressure, and proper adjustment of its pH can greatly reduce the microbial contamination, which makes it possible to construct a non-sterile open system (Li et al., 2014). A variety of salts in seawater have been shown to facilitate the decomposition of some refractory organics, and a large number of nutrients in polluted seawater can also provide carbon sources for bacteria (Fang et al., 2015; Adessi et al., 2016). At present, there have been many examples of successful fermentation using seawater (Li et al., 2014; Yue et al., 2014; Yu et al., 2019; Meng et al., 2022). Therefore, the utilization of seawater as the substrate for fermentation can be a promising substitute (Pernetti and Di Palma, 2005).
There are two researching directions to realize fermentation in culture media with high concentrations of salt to avoid strict sterile conditions. One researching direction is to engineer the halophilic archaea which can grow optimally with high salt concentrations to produce valuable compounds such as carotenoids, PHBs, and proteins(Giani et al., 2021; Simó-Cabrera et al., 2021). The other researching direction is to evolve the freshwater strains artificially, like Escherichia coli, which has been widely utilized as a microbial cell factory (Wu et al., 2014; Adams et al., 2023). Rhodopseudomonas palustris, with the properties of extraordinary metabolic versatility, carbon source diversity and metabolite diversity, has been widely applied in wastewater treatment and bioremediation, and it also showed high potential in valuable chemical production, such as lycopene (Li et al., 2022a,b). Our group has focused on valuable chemical production by engineered R. palustris through photosynthetic pathways using CO2 as the carbon source for 6 years. To widen the application of R. palustris, to obtain the salt-tolerant strain and study the mechanism are required.
Most of the reported R. palustris strains are not salt-tolerant, with R. palustris 42OL as one of the exceptions. Adessi et al. reported that the accumulation of trehalose as a compatible solute protected the bacterial enzyme functions against salt stress, enabling the growth of R. palustris 42OL in the salt-amended medium (4.5% salt) without the addition of any exogenous osmoprotectants. Additionally, the production of hydrogen was noticed even with a 3% salt concentration (Adessi et al., 2016). Moreover, the nutrient starvation greatly improved the tolerance of R. palustris CGA009 to high-concentration salt treatment (2.5 M NaCl for 1 h), indicating that ATP-dependent rearrangement of cell components can induce salt tolerance (Wasai et al., 2018). These findings build up the understanding of the salt tolerance mechanism of R. palustris. However, salt-tolerant R. palustris strain has rarely been obtained, and little is known about its salt tolerance mechanisms.
In the present study, salinity evolved R. palustris strains were obtained using the adaptive laboratory evolution technique. Based on the salt tolerance analysis RPAS-11 was selected to further explore the salt-tolerant mechanism. In comparison with the wild-type R. palustris CGA009, significant color variation was evident in RPAS-11. To understand the color change, the relationship between carotenoid production and salt tolerance in R. palustris was evaluated. In addition, the effect of salt stress on lycopene production was also investigated.
Rhodopseudomonas palustris CGA009 (Kim and Harwood, 1991) was chosen as the original strain for adaptive laboratory evolution (ALE) and the construction of knockout/overexpression strains. E. coli strain DH5α was used for gene cloning and strain S17-1 for conjugation. All E. coli strains were cultivated in LB (Luria-Bertani) medium supplemented with appropriate antibiotics at 37°C, providing 220 rpm constant shaking. The defined mineral medium (photosynthetic medium, PM) was prepared in accordance with the published article (Brown et al., 2020) for the growth of R. palustris strains. A series of R. palustris strains were grown anaerobically in PM medium at 30°C, using a 60 W incandescent lamp (General Electric) which was kept on for 24 h a day. Appropriate carbon sources and antibiotics were provided. The salinity of the medium was represented by % w/v, simplified as %, which was the measure of NaCl weight (g) per 100 mL solution. The calculation formula of generation time is: G = lg2* (t2-t1)/lg (x2)-lg (x1). G, the generation time (h); t2 and t1, culture time (h); x2 and x1, OD660 corresponding to culture time t2 and t1 (Anderson et al., 2021).
To obtain strains with better salt tolerance, ALE was performed. PM liquid medium supplied with 20 mM NaAc (sodium acetate) was used as the basic medium. At first, R. palustris CGA009 was cultured in 10 mL basic medium. Then the bacterial medium in the logarithmic phase was transferred into the basic medium supplied with 1.5% salt (NaCl), and two generations of cultivation at the same salt concentration were performed. Then, the salt concentration of the medium was gradually increased by 0.2% and the salt concentration was increased to the final 4%. To isolate single colonies, the bacterial culture mixture was spread on plates with 2 × PM solid medium supplemented with 4% salt, 20 mM NaHCO3 and 4 mM Na2S2O3·5H2O, and the plates were cultivated under anaerobic conditions. Then, the cell growth of the single colonies under different salt concentrations was tested to screen the colonies with increased salt tolerance. At first, the single colonies on the plates were transferred into PM medium supplied with 20 mM NaAc under 1.5% salt concentration, separately. Then, the bacterial medium was transferred into PM medium supplied with 20 mM NaAc under high salt concentrations, including 0, 2.0, 2.5 and 3.0%. The cell growth was monitored and a single colony named RPAS-11 which showed higher cell growth was selected for further research.
Plasmids and strains used in this work were listed in Table 1. crtC was amplified from the genome of R. palustris CGA009 using primers crtC-F/R, and the vector was amplified using primers pBBRzt-F/R from pBBRMCS-5, and Gibson assembly was applied to construct pBBR-crtC. pBBR-IDIsc (Li et al., 2022b) and pBBRMCS-5 (Kovach et al., 1995) was obtained before. Through conjugation, pBBR-IDIsc was transformed into CGA009 and RPAS-11, obtaining CGA009-IDI and RPAS-11-IDI, respectively, (Wang et al., 2020). Using the same method, the empty plasmid pBBRMCS-5 was transformed into CGA009 and RPAS-11, constructing CGA009-pBBRkz and RPAS-11-pBBRkz, respectively. Also, pBBR-crtC was transformed into CGA009, obtaining CGA009-CrtC. In accordance with the method previously described (Quandt and Hynes, 1993; Egland et al., 1995; Jiao and Newman, 2007), the gene knockout in R. palustris CGA009 was carried out. All primers used for construction of suicide plasmid and validation of recombinant strain are listed in Supplementary Table S2. Through conjugation, the constructed plasmid was transformed from E. coli S17-1 to target R. palustris strain (Egland et al., 1995). Constructed suicide plasmid and recombinant strain are listed in Table 1.
Rhodopseudomonas palustris CGA009 and RPAS-11 were cultured at 30°C in PM, supplemented with 20 mM NaAc and various concentrations of salt (0, 1.0 and 2.0%). The cells were harvested in the logarithmic phase with a OD660 at 0.8. RNA extraction, reverse transcription, and real-time quantitative PCR were performed according to the procedures reported before (Wang et al., 2020). All primers used for real time qPCR analysis are listed in Supplementary Tables S1, S2.
Rhodopseudomonas palustris CGA009 and RPAS-11 were cultured at 30°C in PM, supplemented with 20 mM NaAc and various concentrations of salt (0, 1.0 and 2.0%). The cells were harvested in the logarithmic phase and washed with PBS (phosphate buffer saline). ROS was detected using fluorometric probe 2′, 7′-dichlorodihydrofluoresceine diacetate (DCFH-DA), which was dissolved with ethanol and stored at −20°C in dark (Sigma-Aldrich, United States; He and Häder, 2002). The collected was resuspend by DCFH-DA with the final concentration of 10 μM, and kept for 30 min in dark. Excess DCFH-DA was removed by three washes with PBS. The fluorescence of samples was analyzed with an excitation wavelength of 485 nm and an emission band between 500 and 600 nm using an Ultra-sensitive multi-function microchannel plate detector (Biotek, United States; Verma et al., 2019).
200 μL cells were collected by centrifugation at 10,000 × g for 2 min at 4°C and rinsed twice with deionized H2O to wash the cells. 1 mL of acetone: methanol (1:1 v/v) solution was added into the bacterial pellet to extract the pigment. The suspension was shaking at 30°C for 10 min in the dark, centrifuged at 4°C with 10,000 × g for 2 min. The shaking was repeated until the bacterial pellet was colorless, and the acetone: methanol layer containing the pigment was transferred to a new test tube. Absorption spectrum of the extracted pigments was measured in the range of 100 to 900 nm by a UV/VIS spectrophotometer (Cary 60 UV–Vis, Agilent Technologies, United States).
All steps were carried out under dim light. Carotenoids were transferred into hexane by three successive extractions of the acetone: methanol layer. Quantification of total carotenoids (C, mg/g DCW, Dry cell weight) was determined by the following formula (Abidin et al., 2014): C = (D·V·f × (10/2500))/DCW (g), where, D, absorbance at 480 nm; V, total volume of sample used (mL); f, dilution factor of sample. 2,500 is the E1cm1%, namely average extinction co-efficient for carotenoids, and 10 is the factor to convert % to mg/mL. DCW was determined according to the reference (Li et al., 2022b).
The content of lycopene was analyzed by HPLC (Agilent Technologies Series 1,200 system, Agilent, United States). The detection wavelength of UV detector was set to 480 nm, and the column used was a symmetrical C18 column (250 mm × 4.6 mm, 5 μm, Waters, Ireland). Methanol/acetonitrile/dichloromethane (21:21:8, volume ratio) was selected as the mobile phase. The flow rate was set at 1 mL/min and the chromatographic column was kept at 30°C. Lycopene standard was purchased from Sigma-Aldrich and dissolved with methylene dichloride. Standard curve of lycopene was obtained by stepwise dilution of the standard and injection analysis, which was used for subsequent yield calculation.
To obtain R. palustris strains with salt-tolerant properties, wild-type R. palustris CGA009 was transferred into 10 mL liquid PM medium using 20 mM NaAc as a carbon source with increasing salt concentration. Starting with 1.5%, the salt concentration was gradually increased by 0.2% and the salt concentration was increased to the final 4%. After ALE, the bacterial culture was spread plated on a 2 × PM solid medium supplemented with 20 mM NaAc and 4% salt. Single colonies were isolated and inoculated into PM liquid media with 1.5% salt concentration for activation. Bacterial cultures in the logarithmic phase were transferred into PM media supplied with 20 mM NaAc under high concentrations (0, 2.0, 2.5, and 3.0%) of salt. Growth curves were measured to test the salt tolerance of strains and obtain the final salt-tolerant strain (Figure 1).
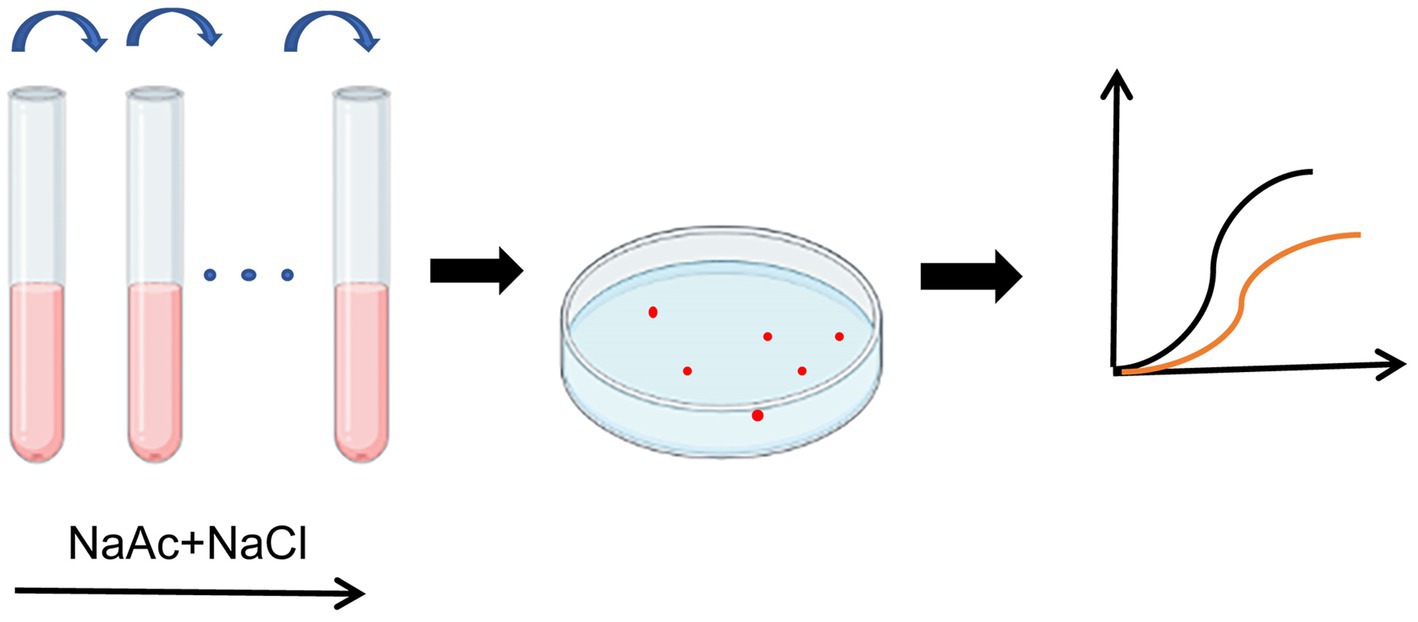
Figure 1. Illustration of the ALE process. Rhodopseudomonas palustris CGA009 was anaerobically cultured in 10 mL PM medium at 30°C under the incandescent lamp, using 20 mM NaAc as a carbon source and 1.5% salt concentration. The culture in the logarithmic phase was further transferred to a fresh medium. After every three generations, the salt concentration in the medium was increased by 0.2%. After 8 months of culture, the salt concentration was increased to the final 4% and stabilized for about 10 generations. The single colonies were isolated and screened for salt tolerance by growth curve.
Eight salt-tolerant R. palustris strains were screened for their growth at different salt concentrations, in comparison to the growth rate of ancestral wild-type CGA009, as represented in Figure 2. Out of eight, RPAS-11 exhibited salt tolerance at all salt concentrations, as indicated by the reduced generation time and optimized growth curve (Figure 2; Table 2). Besides, RPAS-11 also showed growth in a medium containing 4.5% salt concentration, while wild-type CGA009 could hardly grow (Supplementary Figure S1). Seawater contains about 3.5% NaCl, in which many microorganisms are difficult to survive (Pernetti and Di Palma, 2005). The strain we obtained can tolerate up to 4.5% salt concentration, so it is entirely possible to apply it to seawater fermentation. Further, the growth of RPAS-11 and CGA009 was evaluated using four different carbon sources at 1.5% salt concentration (Figure 3). RPAS-11 showed comparatively better growth than that of wild-type CGA009 under salt stress using different carbon sources (Figure 3), which further demonstrated the salt tolerance of RPAS-11 irrespective of carbon source.
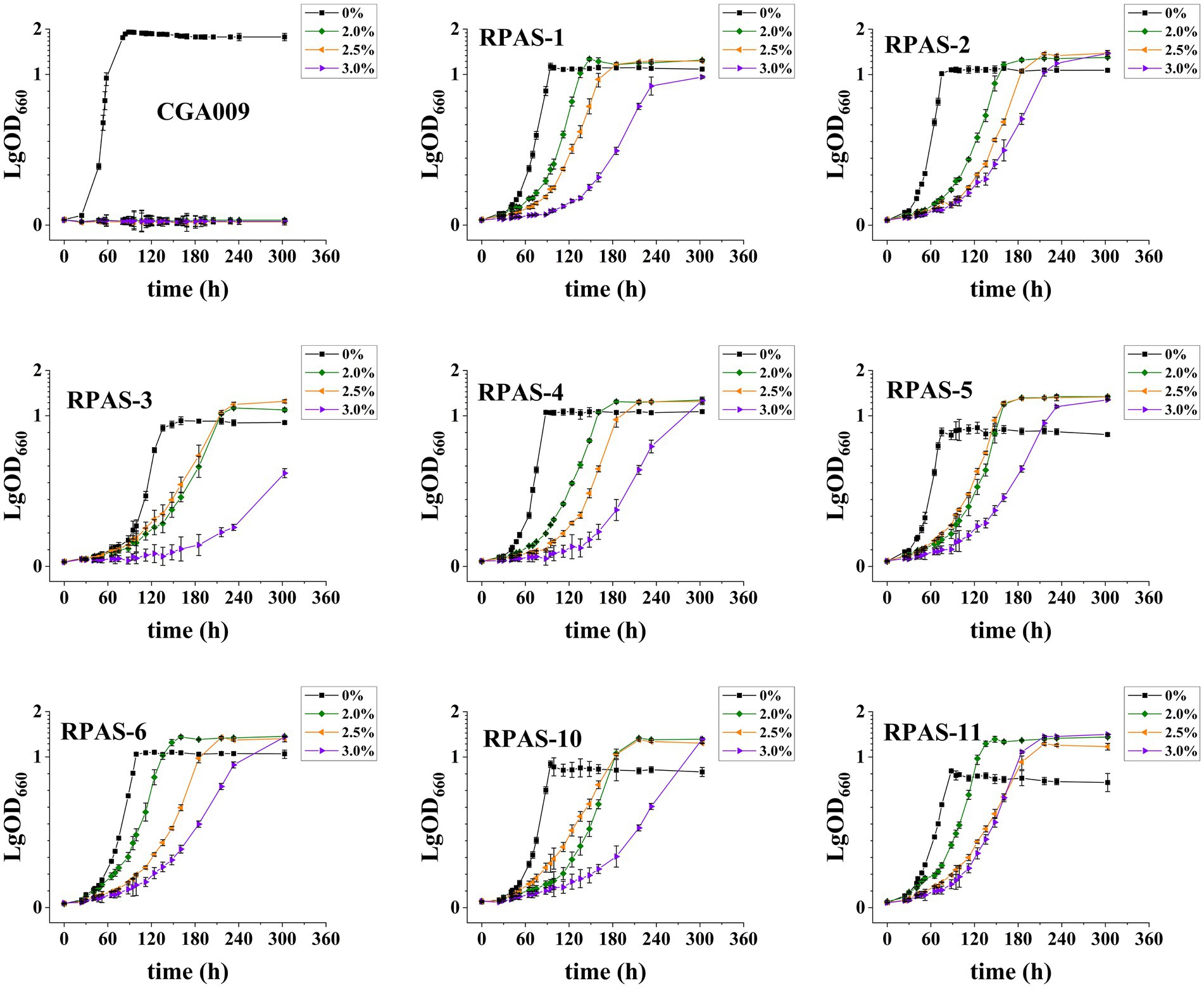
Figure 2. Growth curves of eight screened salt-tolerant R. palustris strains. Strains were cultivated in PM medium with 20 mM acetate as the carbon source at different salt concentrations (0, 2.0, 2.5, and 3.0%). Three biological replicates were performed for each treatment.
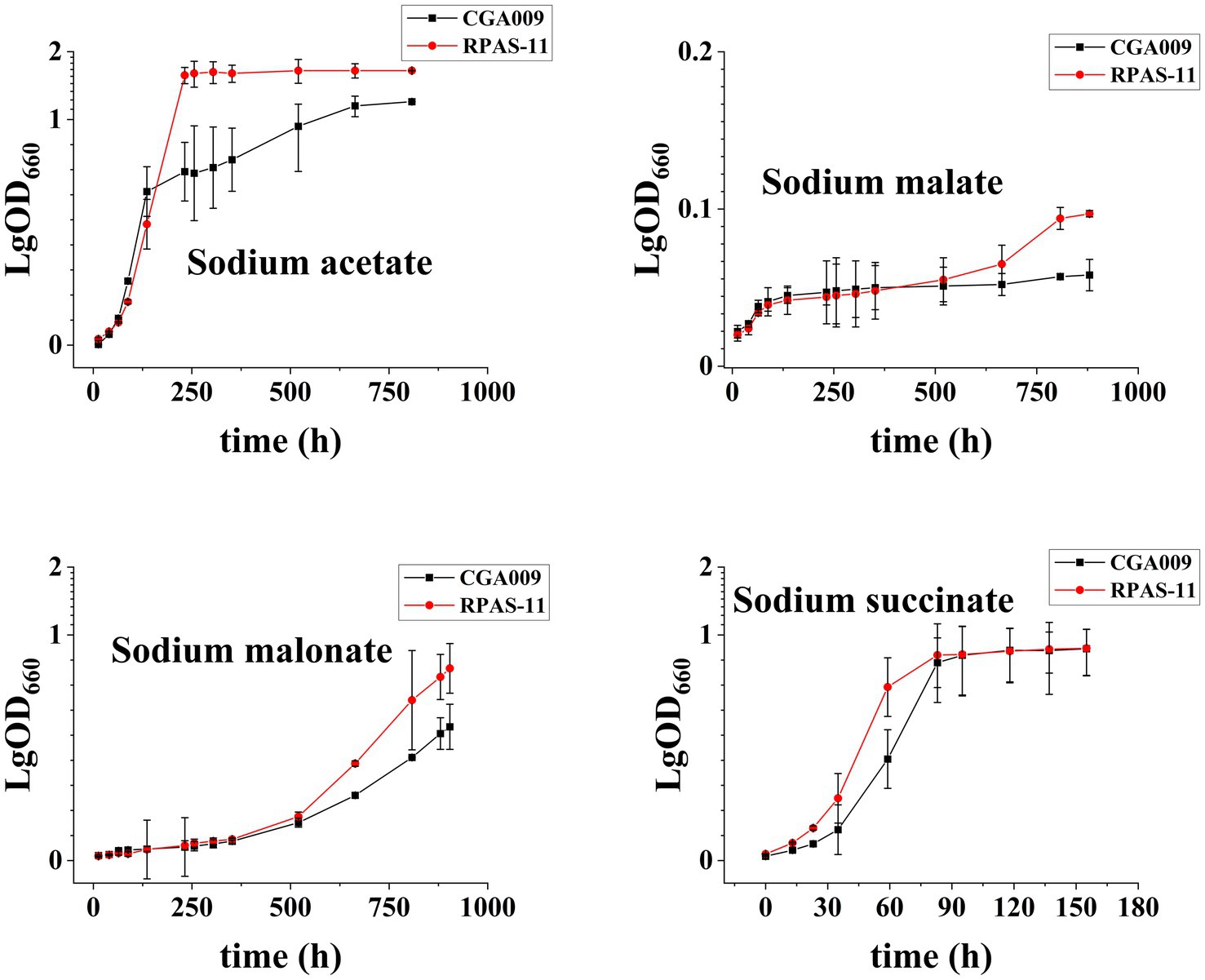
Figure 3. Growth curves of CGA009 and RPAS-11 using different carbon sources. Four carbon sources, including acetate, succinate, malate, and malonate, were selected for CGA009 and RPAS-11 growth with a 1.5% salt concentration. Three biological replicates were performed for each treatment.
In comparison to wild-type CGA009, the screened strains displayed variation in colors at different salt concentrations (Supplementary Figure S2). This change in color can be attributed to the change in carotenoid synthesis in the cell membranes of R. palustris. CGA009 showed a decrease in carotenoids production with increasing salt concentrations, where 16.15 mg/g DCW and 2.61 mg/g DCW carotenoids were produced at 0 and 2% salt concentrations, respectively (Figure 4B). This result was in concordance with the color change of CGA009 observed under salt stress (Supplementary Figure S2). However, the carotenoids production was increased in RPAS-11 from 15.58 mg/g DCW to 32.40 mg/g DCW with an increase in salt concentration from 0 to 2%, respectively (Figure 4B). It was reasonable to say that the increase in carotenoids production under salt stress might be related to the salt tolerance mechanism of RPAS-11. To further elucidate the mechanism of carotenoids production under salt stress, reactive oxygen species (ROS) levels were measured in CGA009 and RPAS-11 under salt stress (1 and 2%; Figure 4D; Ren et al., 2021). Salt stress increases the production of ROS, which can be eliminated by antioxidants, such as carotenoids. Therefore, under the salt stress, ROS level is directly proportional to carotenoids accumulation. In RPAS-11, increased ROS level and carotenoids production under salt stress were detected (Figure 4D). In CGA009, the ROS value was decreased under salt stress, just as the reduction of carotenoids production under salt stress (Figure 4D). It was speculated that, in wild-type CGA009, salt stress (1 and 2%) might damage the cells metabolism to produce ROS and carotenoids.
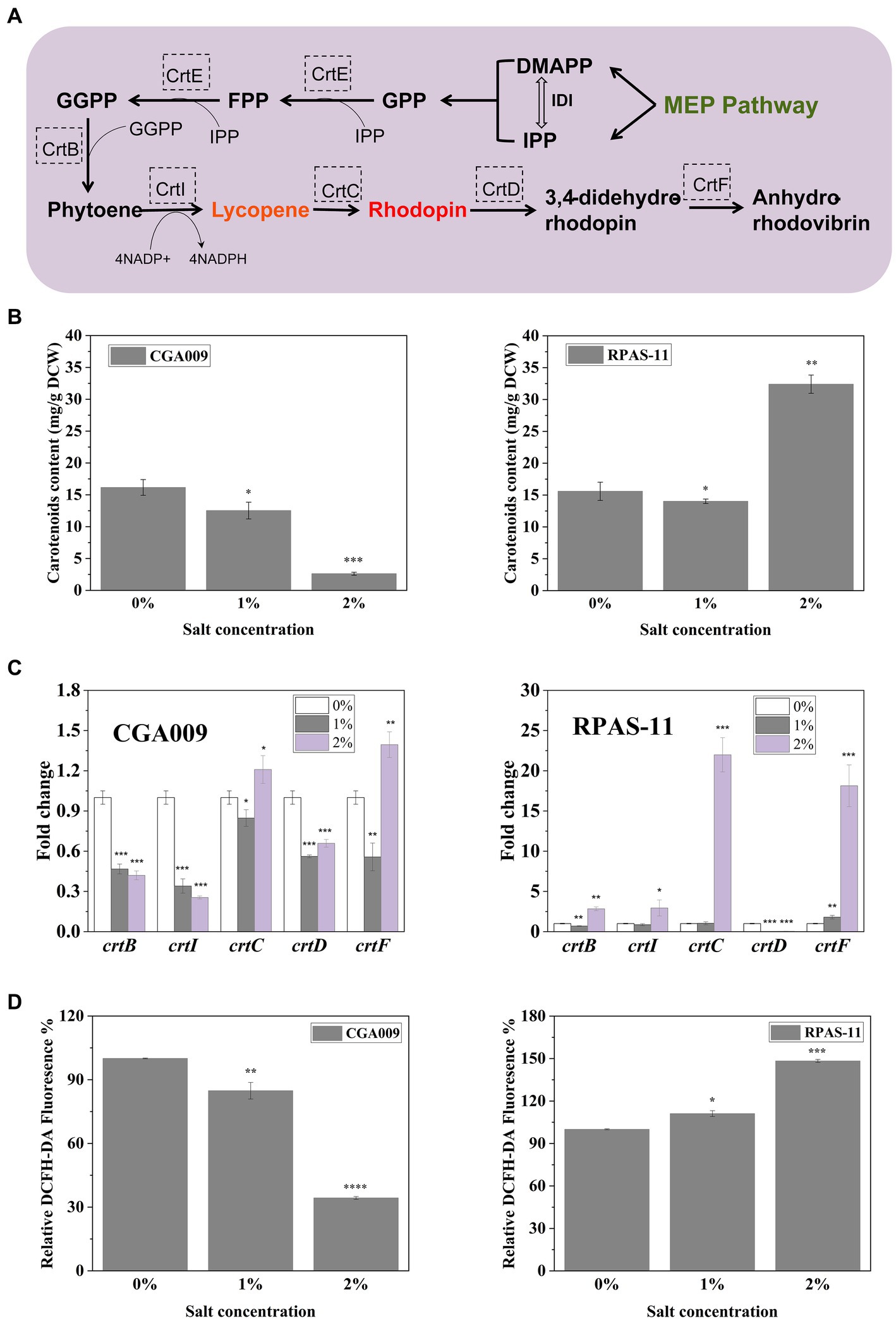
Figure 4. Transcription of carotenoid biosynthesis genes in RPAS-11 and CGA009. (A) Illustration of the related carotenoid biosynthetic pathway in R. palustris. The endogenous MEP pathway is responsible for the synthesis of precursors DMAPP and IPP. MEP Pathway, Methylerythritol phosphate pathway; IPP, Isopentenyl diphosphate; DMAPP, Dimethylallyl diphosphate; GPP, Geranyl diphosphate; FPP, Farnesyl diphosphate; GGPP, Geranylgeranyl diphosphate; IDI, Isopentenyl-diphosphate isomerase; CrtE, FPP/GGPP synthase; CrtB, Phytoene synthase; CrtI, Phytoene desaturase; CrtC, Hydroxyneurosporene synthase; CrtD, FAD dependent oxidoreductase; CrtF, O-Methyltransferase family (B) Carotenoids content of CGA009 and RPAS-11 under different salt concentrations (0, 1, and 2%). (C) Real-time quantitative analysis of genes related to carotenoid synthesis in CGA009 and RPAS-11. The transcription level of genes related to carotenoid synthesis at 0% salt concentration was used as the control. (D) Relative DCFH-DA fluorescence (%) of ROS produced after NaCl stress. Three biological replicates were performed for each treatment. Error bars: mean ± standard deviation (n = 3). *p < 0.05 was considered to be statistically significant. **p < 0.01, ***p < 0.001. p value was calculated by Excel.
No significant difference was evident in the carotenoids content of CGA009 (16.15 mg/g DCW) and RPAS-11 (15.58 mg/g DCW) at 0% salt concentration; however, both the strains displayed different colors (Supplementary Figure S2). In R. palustris, carotenoids, including lycopene, anhydro-rhodovibrin, rhodovibrin, rhodopin, and spirilloxanthin are synthesized through the MEP pathway, and the carotenoids synthetic pathway (Figure 4A; Li et al., 2022b). It was indicated that the expression of carotenoid synthetic genes (crtB, crtI, crtC, crtD, and crtF) might also change under salt stress. To test this speculation, RT-qPCR was performed to determine the effect of different salt concentrations on the transcription of genes associated with the carotenoid synthesis in CGA009 and RPAS-11 (Figure 4C).
In CGA009, the transcriptional expressions of all five genes (crtB, crtI, crtC, crtD, and crtF) were decreased under 1% salt stress (Figure 4C). Under salt stress, cell metabolism was inhibited, which might weaken the synthesis of carotenoids, showing a decrease in the expression of related genes in the experimental results. However, the expression levels of crtB, crtI and crtD were reduced, whereas crtC and crtF showed elevated expression at 2% salt stress in comparison to expression without salt stress. In the carotenoids synthetic pathway, enzymes CrtC and CrtF were responsible for rhodopin, anhydrorhodovibrin, rhodovibrin, and spirilloxanthin production (Giraud et al., 2018). Based on this, we speculated that 2% salt stress might stimulate the synthesis of these pigments, helping R. palustris to adapt to salt stress in turn.
In the salt-tolerant strain RPAS-11, expression levels of most carotenoid synthesis genes remained unchanged or increased under salt stress (1 and 2%), except crtD (Figure 4C). Further, the transcription of crtB, crtI, and crtC was up-regulated by 3-, 3-, and 22-fold, respectively, at 2% salt concentration, in comparison to no salt stress (Figure 4C). Our results indicated that the expression of carotenoid synthesis genes in RPAS-11 was up-regulated under salt stress. Therefore, we speculated that RPAS-11 acquired mutations that increased carotenoid production to combat salt stress. However, the results of RT-qPCR analysis indicated that crtD was hardly expressed under salt stress. The enzyme CrtD is responsible for catalyzing the synthesis of 3,4-didehydro-rhodopin from rhodopin (Figure 4A). Therefore, the reduced expression of crtD allows RPAS-11 to accumulate rhodopin. These results suggested rhodopin accumulation in salt-tolerant RPAS-11 cells. The expression of crtF was also increased under salt stress. Enzyme CrtF is responsible for catalyzing the synthesis of anhydrorhodovibrin from 3,4-didehydro-rhodopin (Figure 4A). If crtD was hardly expressed, the expression of crtF could not function as usual. The results suggested that the salt-tolerant RPAS-11 could accumulate carotenoids to neutralize the salt stress, and rhodopin might play a crucial role among these carotenoids.
Our results demonstrated that RPAS-11 increases carotenoid production during salt stress, and that rhodopin may be the primary carotenoid that is overproduced. In order to establish the role of carotenoids in the salt tolerance of R. palustris, a series of mutant strains with knockout or overexpression of genes related to carotenoid synthesis were constructed, and their salt tolerance was evaluated.
First of all, the effect of increased carotenoid accumulation on salt tolerance was determined. In R. palustris, the precursors of the carotenoids synthetic pathway, isopentenyl diphosphate (IPP) and dimethylallyl diphosphate (DMAPP), are synthesized through the methylerythritol phosphate (MEP) pathway (Figure 4A; Li et al., 2022b). The overexpression of IDI is responsible for catalyzing the reversible conversion of IPP and DMAPP in the MEP pathway (Figure 4A), resulting in increased production of carotenoids, especially lycopene (Li et al., 2022b). Therefore, strains CGA009-IDI and RPAS-11-IDI exhibiting overexpression of IDI were constructed to improve carotenoid production in CGA009 and RPAS-11 (Table 1). Strains CGA009-pBBRkz and RPAS-11-pBBRkz carrying an empty vector pBBRMCS-5 were constructed and used as the controls (Table 1). The IDI overexpression resulted in a slight increase in the total carotenoids content in both CGA009 and RPAS-11 (Figure 5A). Further, the growth of the bacterial strains was analyzed at different salt concentrations (1.5, 2.0, and 2.5%) to determine the salt tolerance. The resultant decline in the growth rate of CGA009-IDI at 1.5 and 2.0% salt concentrations might be due to the consumption of cell energy in IDI overexpression (Figure 5B; Table 3). On the contrary, strain RPAS-11-IDI exhibited a higher value of final OD660 irrespective of the IDI overexpression, as compared to RPAS-11-pBBRkz, under different salt concentrations, especially under 2.0% salt concentration, indicating higher tolerance of RPAS-11-IDI (Figure 5C). These results indicated that the increased carotenoids content by overexpression of IDI might be a reason for the improved salt tolerance in strain RPAS-11-IDI.
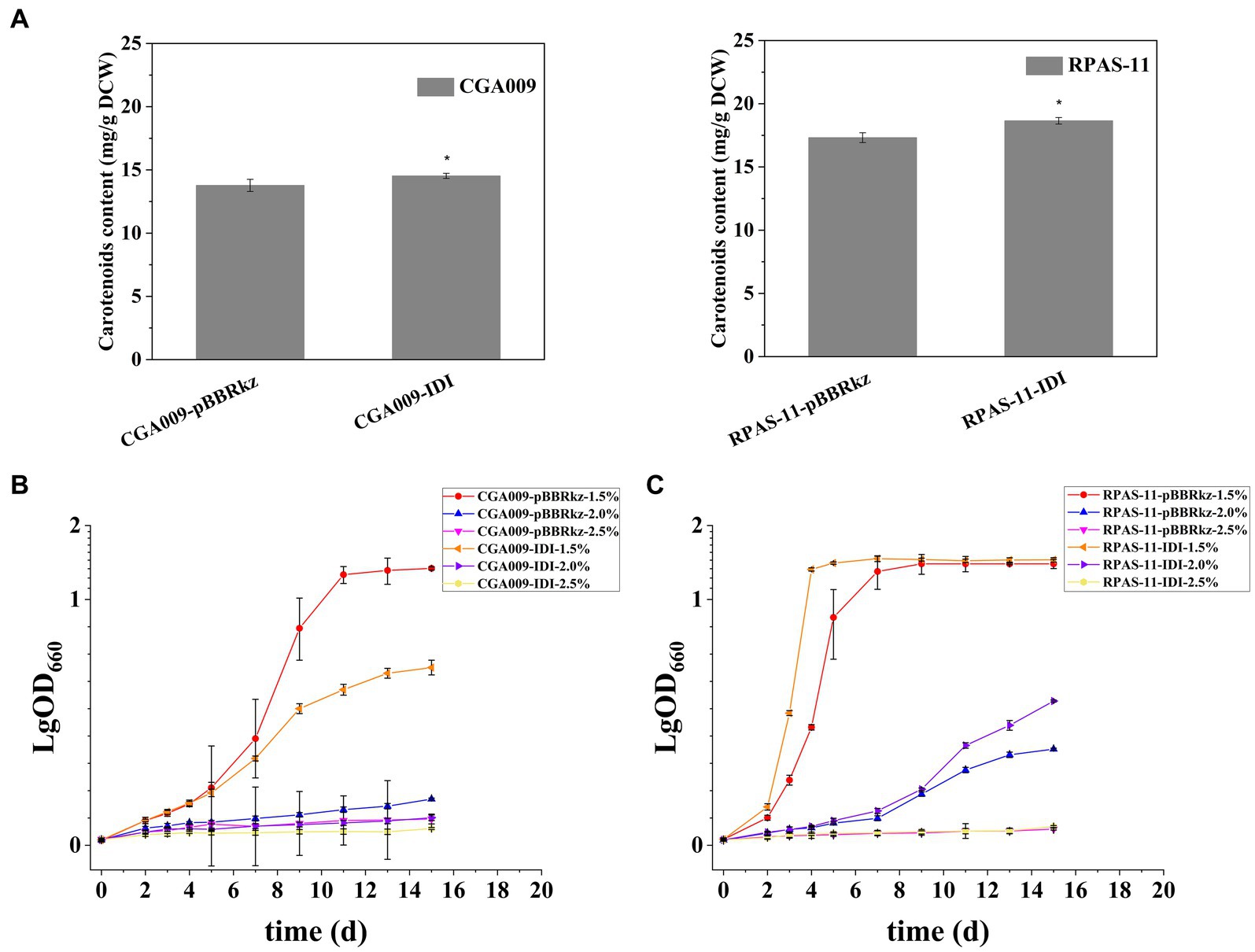
Figure 5. Carotenoids and growth curve after overexpression of IDI in CGA009 and RPAS-11. (A) Carotenoids content of CGA009-pBBRkz, CGA009-IDI, RPAS-11-pBBRkz, and RPAS-11-IDI without salt stress. Error bars: mean ± standard deviation (n = 3). *p < 0.05 was considered to be statistically significant. p value was calculated by Excel. (B) Growth curve of CGA009-pBBRkz and CGA009-IDI at different salt concentrations (1.5, 2.0, and 2.5%). (C) Growth curve of RPAS-11-pBBRkz and RPAS-11-IDI with different salt concentrations (1.5, 2.0, and 2.5%). All these strains were cultured in PM medium with 20 mM acetate as the carbon source. Three biological replicates were performed for each treatment.
In order to reverse verify the effect of carotenoids on the salt tolerance of R. palustris, we studied the salt tolerance of R. palustris without carotenoids. In the carotenoid synthetic pathway, enzyme CrtI is known to catalyze the conversion of phytoene to lycopene; therefore, the deletion of gene crtI (strain CGA009-ΔcrtI) would not only inhibit the lycopene production but also block the formation of other carotenoids (Zhu L. et al., 2011; Giraud et al., 2018). The crtI knockout strain CGA009-ΔcrtI displayed green color, which can be attributed to bacteriochlorophyll (BChl; Figure 6A), another light-absorbing pigment present in R. palustris. The results of absorption spectra (400–850 nm) of extracted pigments from CGA009 and CGA009-ΔcrtI indicated that no carotenoids were synthesized in CGA009-ΔcrtI (Figure 6B). Further, the growth of CGA009-ΔcrtI and CGA009 was analyzed at different salt concentrations (Figure 6C). Under salt-free condition, the final OD660 value of CGA009-ΔcrtI was similar to CGA009, and the generation time, 11.97 ± 0.18, was slightly higher than that of CGA009, 10.27 ± 0.10 (Figure 6C; Table 4). This result indicated that deletion of crtI slightly affected the cell growth. Deletion of crtI inhibited the synthesis of carotenoids, which can capture light of the visible spectrum and, then, transfer the excitation energy to BChl (Adessi and De Philippis, 2014). At higher salt concentrations (1.5, 2, and 2.5%), the final OD660 values of CGA009-ΔcrtI were lower than that of wild-type CGA009, and the growth rate was also affected (Figure 6C; Table 4). Under salt stress, the extent of the effect on cell growth was more serious than that under salt-free condition, indicating the weakening salt tolerance of strain CGA009-ΔcrtI. These results also established the role of carotenoid production in salt stress tolerance. Therefore, it was reasonable to say that the deletion of crtI not only disrupted the carotenoid production but also weakened the salt tolerance, thereby confirming the positive effect of carotenoids on the salt tolerance of R. palustris.
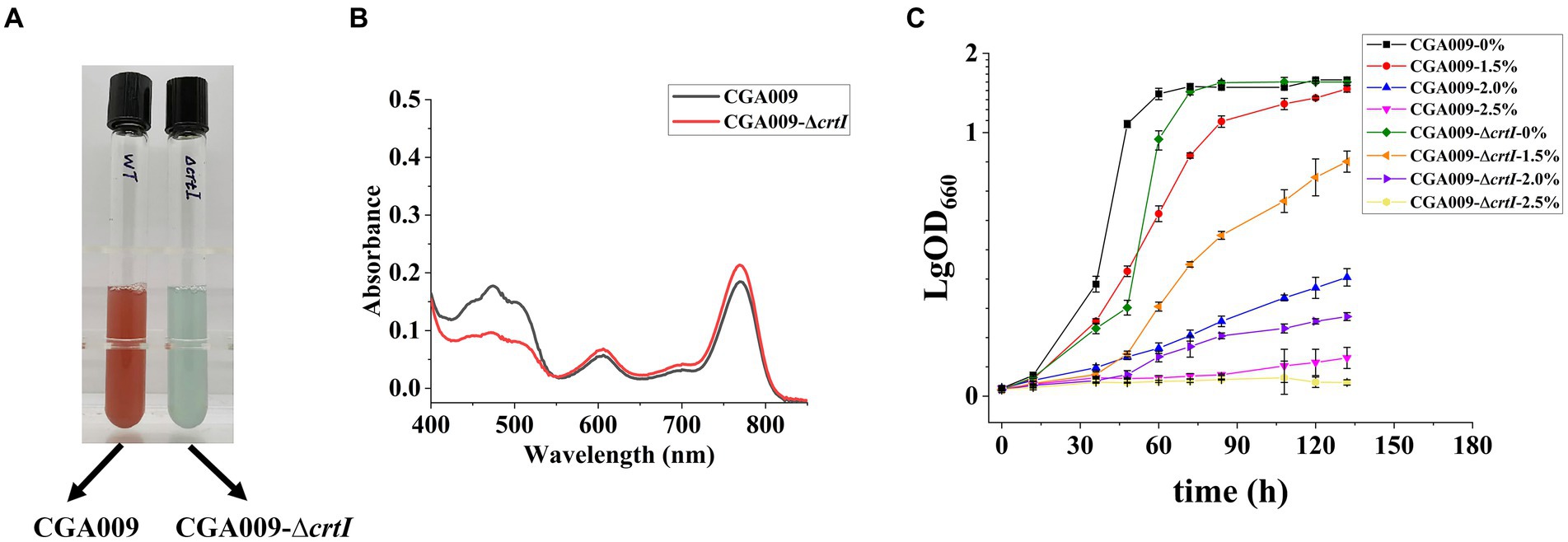
Figure 6. Growth curve and UV–visible absorption spectrum for pigments of CGA009 strains with and without crtI deletion. (A) The color of strain CGA009 and CGA009-ΔcrtI cultured in a salt-free environment. (B) UV–visible absorption spectra (400–850 nm) of extracted pigments of CGA009 and CGA009-ΔcrtI. (C) Growth curve of CGA009 and CGA009-ΔcrtI at different salt concentrations (0, 1.5, 2.0, and 2.5%). Strains were cultivated in a PM medium with 20 mM acetate as the carbon source. Three biological replicates were performed for each treatment.
After demonstrating the effect of carotenoids on salt tolerance, the key pigment among the carotenoids was identified. To determine the key pigment, three carotenoid biosynthetic genes knockout strains, namely CGA009-ΔcrtC, CGA009-ΔcrtD, and CGA009-ΔcrtF, were constructed and evaluated for their growth at different salt concentrations to assess their salt tolerance (Figure 7; Table 5). The results indicated that gene knockout significantly affected the growth of R. palustris under different salt concentrations. At higher salt concentrations (2.0 and 2.5%), the strains CGA009-ΔcrtC and CGA009-ΔcrtF barely showed any growth, whereas strain CGA009-ΔcrtD showed a higher growth rate than CGA009. These results suggested that strain CGA009-ΔcrtD exhibited higher salt tolerance, while strains CGA009-ΔcrtC and CGA009-ΔcrtF showed lower tolerance than that of wild-type CGA009. Lycopene desaturase (CrtI) converts phytoene to lycopene through four desaturation steps. CrtC catalyzes the hydration reaction to convert lycopene into rhodopin, which in turn saturates to 3,4-dihydrorhodopin under the catalysis of CrtD. 3,4-dihydrorhodopin can subsequently undergo methylation reaction under the catalysis of crtF and become anhydrohodovibrin (Figure 4A). Theoretically, strain CGA009-ΔcrtC, CGA009-∆crtD, and CGA009-ΔcrtF can accumulate lycopene, rhodopin and 3,4-dihydrorodopine, respectively (Takaichi, 1999). Therefore, it was reasonable to say that, among the carotenoids, rhodopin played a key role in salt tolerance. These results were backed by the findings of RT-qPCR, where a reduction in crtD expression was detected in RPAS-11, in response to salt stress. To further illustrate the effect of rhodopin on salt tolerance, crtC was overexpressed in CGA009, obtaining strain CGA009-CrtC. However, at higher salt concentrations (2.0 and 2.5%), the growth of strain CGA009-CrtC has no significant difference with the control strain CGA009-pBBRkz (Supplementary Figure S3). It seems that the wild-type CGA009 has different salt-tolerant mechanism with RPAS-11. In the strain RPAS-11, high salt stress would cause ROS production and carotenoid accumulation. And accumulation of carotenoids, especially rhodopin, was speculated to help RPAS-11 to tolerate salt stress. The wild-type CGA009 lacks the potential mutations like RPAS-11 that cause salt tolerance mechanisms. Therefore, overexpression of crtC did not lead to better salt tolerance in CGA009 (Supplementary Figure S3). The other possible reason was that single overexpression of crtC might not lead to increase in rhodopin content, and overexpression of crtC would consume the cell energy for cell growth. In conclusion, carotenoid synthesis, especially rhodopin, plays a role in the salt tolerance ability of R. palustris.
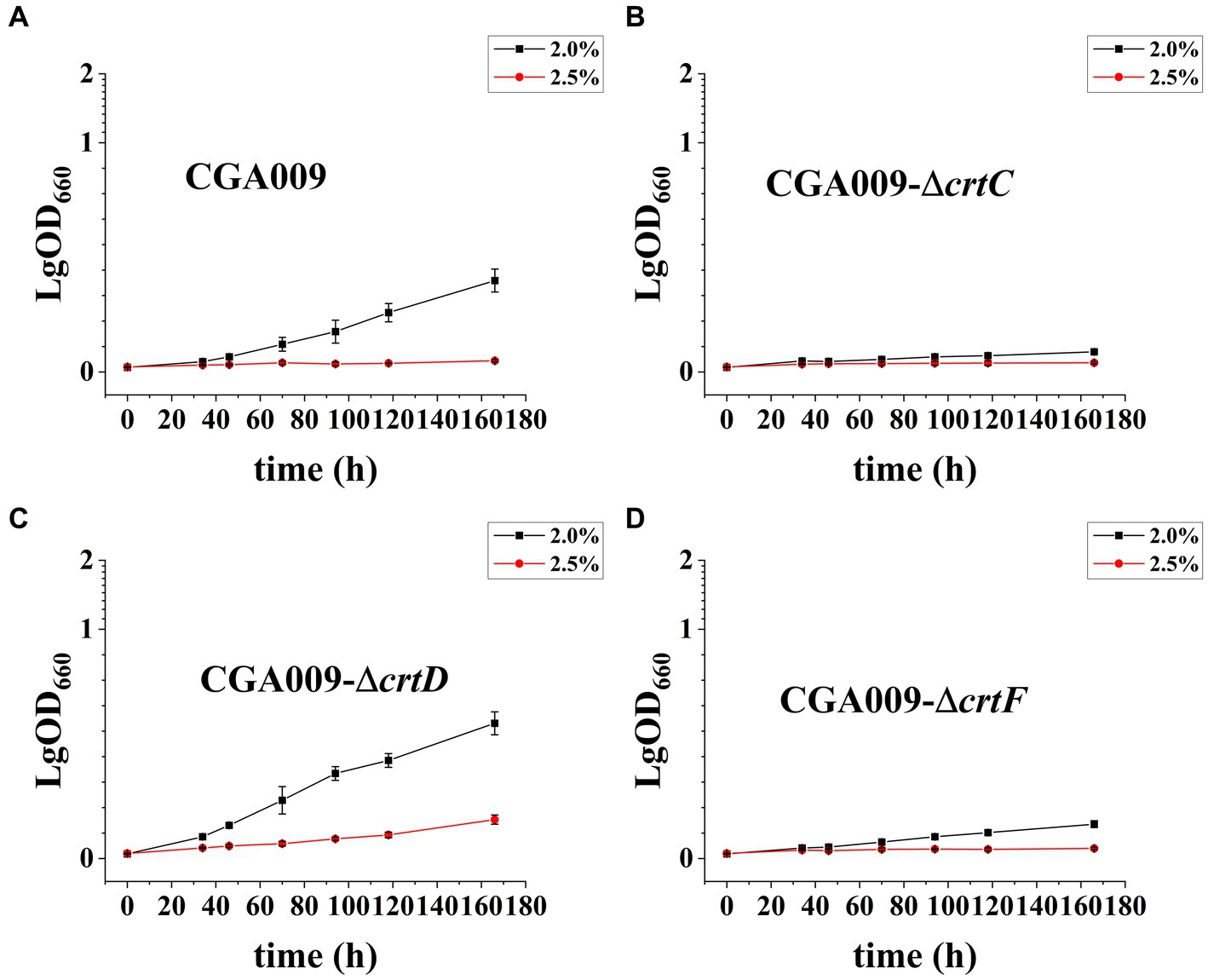
Figure 7. Growth curve of R. palustris strains with crtC, crtD, or crtF deletion under salt stress. (A) Growth curve of CGA009 under salt stress. (B) Growth curve of CGA009-ΔcrtC under salt stress. (C) Growth curve of CGA009-ΔcrtD under salt stress. (D) Growth curve of CGA009-ΔcrtF under salt stress. Strains were cultivated in a PM medium containing different salt concentrations (2.0 and 2.5%) with 20 mM acetate as the carbon source. Three biological replicates were performed for each treatment.
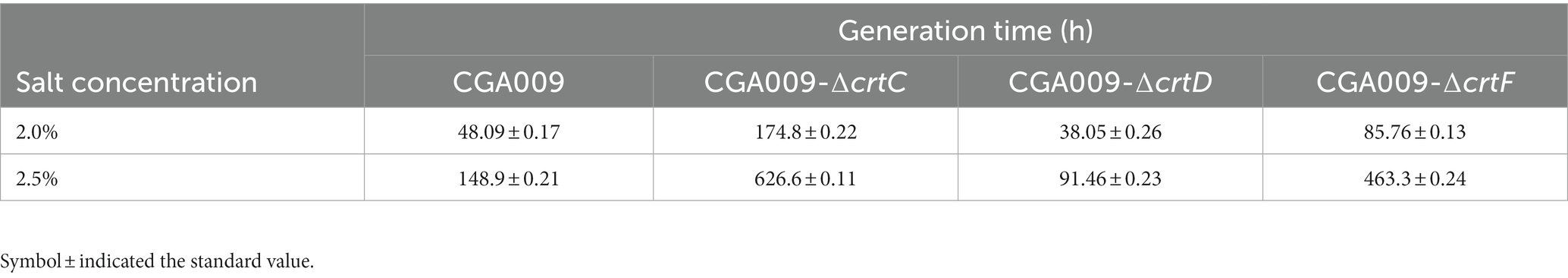
Table 5. Generation time of R. palustris strains with crtC, crtD, or crtF deletion under salt stress.
In the present study, the effect of salt stress on lycopene accumulation in R. palustris (Li et al., 2022b). Lycopene production in wild-type CGA009 and CGA009-ΔcrtC knockout palustris was investigated. To improve lycopene production, CGA009-ΔcrtC knockout was constructed to block the carotenoids synthetic pathway, consequently accumulating lycopene strains grown at different salt concentrations (0, 0.5, 1, 1.5, and 2%) was measured (Figure 8). In wild-type CGA009, the lycopene production was increased from 3.24 mg/g DCW to 5.29 mg/g DCW with an increase in salt concentration from 0 to 1.5% (Figure 8A). In crtC knockout, CGA009-ΔcrtC, the lycopene production was increased from 32.26 mg/g DCW to 63.53 mg/g DCW with an increase in salt concentration from 0 to 1.5% (Figure 8B). These results indicated that salt stress stimulated production of lycopene in R. palustris. However, at the higher salt concentration (2%), the lycopene production in CGA009 and CGA009-ΔcrtC was 2.49 mg/g DCW and 32.67 mg/g DCW, respectively. Based on this, it was speculated that higher salt concentration (2%) might have harmed the normal metabolism in R. palustris, leading to decreased lycopene production. As a result, salt stress could stimulate lycopene production in R. palustris under salt concentrations lower than 2%.
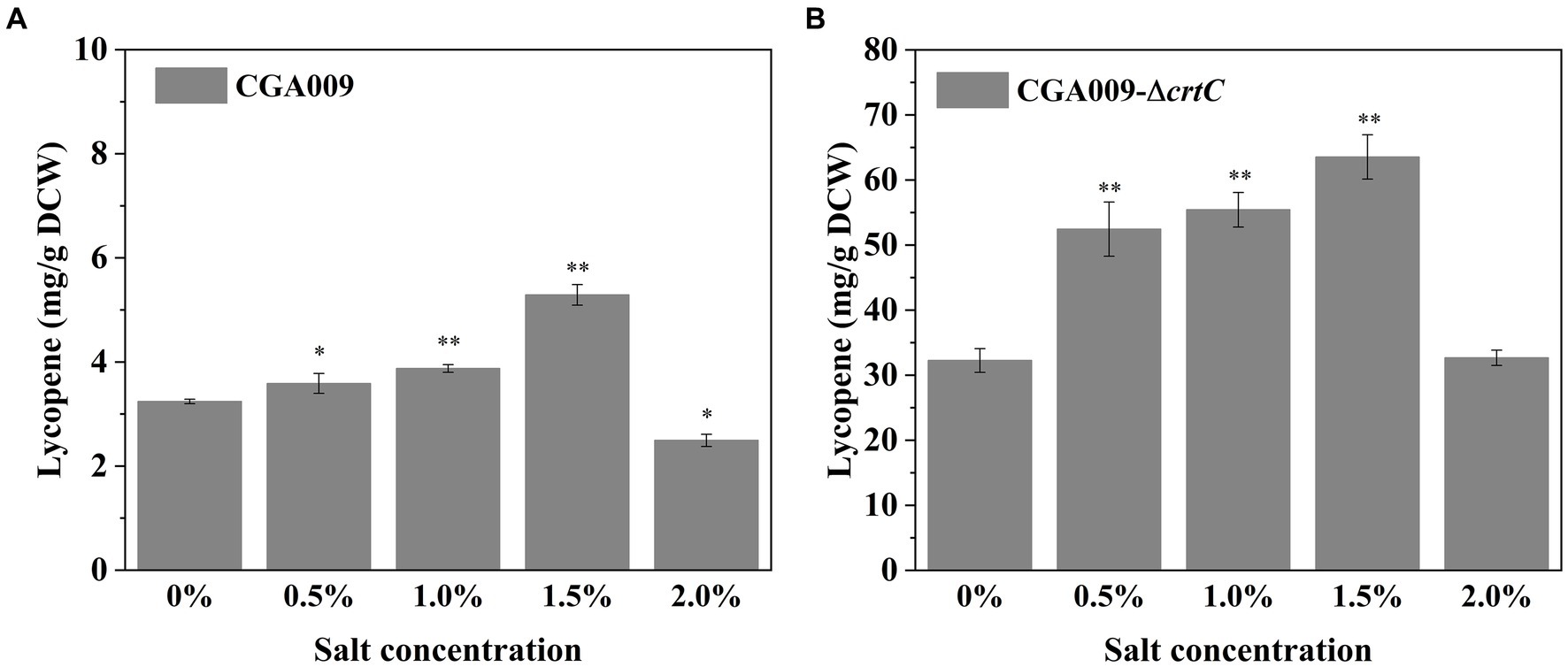
Figure 8. Lycopene content of CGA009 and CGA009-ΔcrtC under salt stress. (A) Lycopene content of CGA009 under salt stress. (B) Lycopene content of CGA009-ΔcrtC under salt stress. Strains were cultivated in a PM medium containing 20 mM acetate as the carbon source and different salt concentrations (0, 0.5, 1.0, 1.5, and 2.0%). Three biological replicates were performed for each treatment. Error bars: mean ± standard deviation (n = 3). *p < 0.05 was considered to be statistically significant. **p < 0.01. p value was calculated by Excel.
Due to its metabolic versatility and carbon source diversity, R. palustris has great potential in wastewater treatment and the production of valuable compounds. However, most kinds of R. palustris belong to freshwater bacteria, whose application in high salinity conditions is limited. In this study, salinity-evolved strains were screened, and the relationship between salt tolerance and carotenoid production was investigated.
Adaptive laboratory evolution was used to obtain RPAS-11 strain with an ability to grow at 4.5% salt concentration which is higher than the average seawater salinity of 3.5%. The selected RPAS-11 exhibited relatively higher salt tolerance than that of its ancestral strain CGA009. Further, the selected RPAS-11 showed increased salt tolerance with different carbon sources, including sodium succinate, sodium malate, sodium acetate, and sodium malonate, which mainly exist in industrial and agricultural wastewater (Li et al., 2022a). Our results displayed the potential application of salt-tolerant RPAS-11 in highly-saline wastewater treatment.
In comparison to CGA009, RPAS-11 displayed a significant change in color, indicating a change in carotenoid synthesis under salt stress. Therefore, the possible association between carotenoid production and salt tolerance was further analyzed. With an increase in salt concentration, a reduction in carotenoid production was recorded in wild-type CGA009, whereas an increase was noticed in the case of salt-tolerant RPAS-11. In turn, the positive effect of carotenoid accumulation on salt tolerance was further demonstrated. In strain RPAS-11-IDI, an overexpression of IDI resulted in an increased carotenoid accumulation and high salt tolerance. On the contrary, the knockout strain CGA009-ΔcrtI showed decreased salt tolerance in the absence of carotenoid synthesis. Likewise, the salt and drought stress in Nicotiana tabacum induced the tobacco lycopene β-cyclase gene Ntβ-LCY1, further enhancing the expression of carotenoid synthetic genes and carotenoid content (Shi et al., 2015).
Likewise, the relationship between carotenoids and salt tolerance has been studied in some other organisms. Carotenoids are considered as strong antioxidant which can improve the fluidity of the cell membrane and protect the cells from osmotic stress and oxidative damages (D'Souza et al., 1997; Camacho-Córdova et al., 2014). Salt stress was usually considered to stimulate carotenoids production. In the salt-tolerant RPAS-11, increase in carotenoid production was obtained under salt stress (2%). In the case of certain microalgae, moderate salt stress was noticed to induce the accumulation of carotenoids, which further acted as antioxidants and cell protectants, thereby increasing the likelihood of microalgae survival (Giani et al., 2021; Matarredona et al., 2021; Ren et al., 2021). However, for Haloferax mediterranei, known for its tolerance to extreme salt levels, the optimal salt concentration for pigmentation accumulation was observed at 12.5% w/v, and decreased carotenoids production was observed with increased salt concentrations (>12.5% w/v), which might due to that for hyper-halophilic strain, low concentration might also be a stress (Thombre et al., 2016). For another kind of hyper-halophilic archaeon Haloarcula marismortui RR12, similar relationship between carotenoids production and salt concentration was obtained, and the optimal salt concentration for red carotenoid production was 25% w/v. Furthermore, high salt stress which exceeds the tolerance range would affect the cell growth and carotenoids. In non-salt resistant Bixa orellana L., salt stress greatly affected the photosynthetic machinery by reducing the accumulation of chlorophyll pigments and carotenoids (Sankari et al., 2019). In the wild-type CGA009, the carotenoids production was decreased when the salt concentration was increased to 1 and 2%. The osmoprotective effect of another kind of isoprenoid, ubiquinones, have also been researched. In E. coli, isoprenoid ubiquinone-8 accumulation improves osmotic-stress tolerance by stabilizing the cell membrane (Sévin and Sauer, 2014). Ubiquinone-10 can modulate the mechanical strength and permeability of lipid membranes, and it is considered as a powerful antioxidant (Agmo Hernández et al., 2015). Of course, the salt tolerance of R. palustris may not be solely related to its carotenoid synthesis. It is well known that the regulation of cellular metabolism is a complex process, and many factors may jointly affect its salt tolerance, such as sodium efflux pump, accumulation of compatible solutes, the composition of the cell wall and membrane and so on (Hagemann, 2011; Dakal et al., 2014; Verma et al., 2019). However, it is undeniable that the synthesis of carotenoids is important for the salt tolerance of R. palustris.
In R. palustris, several pigments, including lycopene, anhydro rhodovibrin, rhodovibrin, rhodopin, and spirilloxanthin were produced, and the key pigment related to salt tolerance was further identified. The RT-qPCR analysis of RPAS-11 revealed that crtD was barely expressed under 2% salt stress, which might block the subsequent reactions of carotenoid synthesis and lead to the accumulation of rhodopin. Besides, the knockout strain CGA009-ΔcrtD not only exhibited relatively better salt tolerance than CGA009, but also displayed salt tolerance at higher salt concentrations (2.0 and 2.5%). These results suggested that the acquired salt tolerance in RPAS-11 might be related to the accumulation of carotenoids, particularly rhodopin. To the best of our knowledge, the promoting effect of rhodopin on salt tolerance was speculated for the first time in the present study. Besides, the effect of high light irradiance on rhodopin content was also reported in R. palustris 42OL, where an increase in the percent of rhodopin among the total carotenoids was noticed under aerobic and H2-producing conditions (Muzziotti et al., 2017). Moreover, the antioxidant activity of rhodopin in marine seaweeds has already been established (Mohy and Elahwany, 2015; Ashour et al., 2020). These results indicated that antioxidant rhodopin confers tolerance to salt and high light.
Lycopene is a carotenoid with antioxidant, anticancer, and anti-inflammatory properties with numerous applications in the pharmaceutical, cosmetic, and food industries (Li et al., 2020). The production of lycopene using R. palustris has already been reported (Niedzwiedzki et al., 2007; Muzziotti et al., 2017). Lycopene generation is one of the physiological responses to stress conditions. Both CGA009 and CGA009-ΔcrtC showed an increase in lycopene production at low salt concentrations, 0.5, 1.0, and 1.5% (Figure 8). However, the overall carotenoids content in CGA009 registered a decline at 1.0% salt concentration (Figure 4B). This phenomenon indicated a change in the percentage distribution of carotenoids, at 1.0% salt concentration. A previous study has also reported an enhancement in lycopene production in Blakeslea trispora, under cold stress conditions (Lingran et al., 2019). Another study reported enhanced synthesis of β-carotene (another carotenoid compound) in Dunaliella salina, a halophyilic bacteria that grows well in higher salinity environments, in an environment with salt concentrations lower than optimal for growth, where the highest amounts of β-carotene were obtained at 2.5 mol/L salinity (Hashemi et al., 2020).
In the present study, a salt-tolerant R. palustris strain RPAS-11 was obtained through the adaptive laboratory evolution method. The improvement in salt tolerance was evaluated in relation to the carotenoid production in the RPAS-11. The results suggested that salt stress could induce carotenoid production in salt-tolerant strains, and in turn, carotenoid accumulation could help the cells to tolerate salt stress. Further, rhodopin was presumed to be the key pigment related to salt tolerance in R. palustris for the first time. The enhancement of salt stress resulted in increased production of another carotenoid compound, lycopene. To sum up, our research provides insight into the salt tolerance mechanism of R. palustris and also provides the theoretical basis for the application of R. palustris in highly saline wastewater treatment and valuable chemicals production.
The original contributions presented in the study are included in the article/Supplementary material, further inquiries can be directed to the corresponding authors.
MEL: Writing – original draft, Writing – review & editing. TZ: Writing – original draft, Writing – review & editing. RY: Writing – review & editing. ZW: Writing – review & editing. MIL: Writing – review & editing. JY: Writing – review & editing.
The author(s) declare financial support was received for the research, authorship, and/or publication of this article. This work was supported by grants from the “First class grassland science discipline” program in Shandong Province, the National Natural Science Foundation of China (31860011), the Talents of High Level Scientific Research Foundation (grants 6651120032, 6651117005 and 6651121004) of Qingdao Agricultural University, Supported by State Key Laboratory of Microbial Technology Open Projects Fund (M2022-07), CAS Key Laboratory of Biobased Materials, Qingdao Institute of Bioenergy and Bioprocess Technology, Chinese Academy of Sciences (BMF-2020-01), and Key Laboratory of Biofuels, Qingdao Institute of Bioenergy and Bioprocess Technology, Chinese Academy of Sciences (CASKLB201805).
The authors would also like to thank the “Laboratory for Agricultural Molecular Biology” for providing laboratory apparatus.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2023.1292937/full#supplementary-material
Abidin, K. C. A. J., Zakaria, Z., Yunus, N. H. Z. M., and Faizul, K. (2014). Carotenoid contents in anoxygenic phototrophic purple bacteria, Marichromatium sp. and Rhodopseudomonas sp. of tropical aquatic environment. Malaysia. Orient. J. Chem. 30, 607–613. doi: 10.13005/ojc/300228
Adams, J. D., Sander, K. B., Criddle, C. S., Arkin, A. P., and Clark, D. S. (2023). Engineering osmolysis susceptibility in Cupriavidus necator and Escherichia coli for recovery of intracellular products. Microb. Cell Factories 22:69. doi: 10.1186/s12934-023-02064-8
Adessi, A., Concato, M., Sanchini, A., Rossi, F., and De Philippis, R. (2016). Hydrogen production under salt stress conditions by a freshwater Rhodopseudomonas palustris strain. Appl. Microbiol. Biotechnol. 100, 2917–2926. doi: 10.1007/s00253-016-7291-4
Adessi, A., and De Philippis, R. (2014.) Photosynthesis and hydrogen production in purple non sulfur bacteria: Fundamental and applied aspects. Microbial BioEnergy: Hydrogen production. Dordrecht: Springer Netherlands. p. 269–290.
Agmo Hernández, V., Eriksson, E. K., and Edwards, K. (2015). Ubiquinone-10 alters mechanical properties and increases stability of phospholipid membranes. Biochim. Biophys. Acta 1848, 2233–2243. doi: 10.1016/j.bbamem.2015.05.002
Amin, M. M., Khiadani, M. H., Fatehizadeh, A., and Taheri, E. J. D. (2014). Validation of linear and non-linear kinetic modeling of saline wastewater treatment by sequencing batch reactor with adapted and non-adapted consortiums. Desalination 344, 228–235. doi: 10.1016/j.desal.2014.03.032
Anderson, M. T., Brown, A. N., Pirani, A., Smith, S. N., Photenhauer, A. L., Sun, Y., et al. (2021). Replication dynamics for six gram-negative bacterial species during bloodstream infection. MBio 12:e0111421. doi: 10.1128/mBio.01114-21
Ashour, M., Mabrouk, M. M., Ayoub, H. F., el-Feky, M. M. M. M., Zaki, S. Z., Hoseinifar, S. H., et al. (2020). Effect of dietary seaweed extract supplementation on growth, feed utilization, hematological indices, and non-specific immunity of Nile Tilapia, Oreochromis niloticus challenged with Aeromonas hydrophila. J. Appl. Phycol. 32, 3467–3479. doi: 10.1007/s10811-020-02178-1
Brown, B., Immethun, C., Wilkins, M., and Saha, R. (2020). Rhodopseudomonas palustris CGA009 polyhydroxybutyrate production from a lignin aromatic and quantification via flow cytometry. Bioresource Technol. Reports. 11:100474. doi: 10.1016/j.biteb.2020.100474
Camacho-Córdova, D. I., Camacho-Ruíz, R. M., Córdova-López, J. A., and Cervantes-Martínez, J. (2014). Estimation of bacterioruberin by Raman spectroscopy during the growth of halophilic archaeon Haloarcula marismortui. Appl. Opt. 53, 7470–7475. doi: 10.1364/ao.53.007470
Dakal, T. C., Solieri, L., and Giudici, P. (2014). Adaptive response and tolerance to sugar and salt stress in the food yeast Zygosaccharomyces rouxii. Int. J. Food Microbiol. 185, 140–157. doi: 10.1016/j.ijfoodmicro.2014.05.015
Díaz, M. P., Boyd, K. G., Grigson, S. J., and Burgess, J. G. (2002). Biodegradation of crude oil across a wide range of salinities by an extremely halotolerant bacterial consortium MPD-M, immobilized onto polypropylene fibers. Biotechnol. Bioeng. 79, 145–153. doi: 10.1002/bit.10318
Doud, D. F. R., Holmes, E. C., Richter, H., Molitor, B., Jander, G., and Angenent, L. T. (2017). Metabolic engineering of Rhodopseudomonas palustris for the obligate reduction of n-butyrate to n-butanol. Biotechnol. Biofuels 10:178. doi: 10.1186/s13068-017-0864-3
D'Souza, S. E., Altekar, W., and D'Souza, S. F. (1997). Adaptive response of Haloferax mediterranei to low concentrations of NaCl (< 20%) in the growth medium. Arch. Microbiol. 168, 68–71. doi: 10.1007/s002030050471
Egland, P. G., Gibson, J., and Harwood, C. S. (1995). Benzoate-coenzyme a ligase, encoded by badA, is one of three ligases able to catalyze benzoyl-coenzyme a formation during anaerobic growth of Rhodopseudomonas palustris on benzoate. J. Bacteriol. 177, 6545–6551. doi: 10.1128/jb.177.22.6545-6551.1995
Fang, C., Thomsen, M. H., Brudecki, G. P., Cybulska, I., Frankær, C. G., Bastidas-Oyanedel, J. R., et al. (2015). Seawater as alternative to freshwater in pretreatment of date palm residues for bioethanol production in coastal and/or arid areas. ChemSusChem 8, 3823–3831. doi: 10.1002/cssc.201501116
Fixen, K. R., Zheng, Y., Harris, D. F., Shaw, S., Yang, Z. Y., Dean, D. R., et al. (2016). Light-driven carbon dioxide reduction to methane by nitrogenase in a photosynthetic bacterium. Proc. Natl. Acad. Sci. U. S. A. 113, 10163–10167. doi: 10.1073/pnas.1611043113
Giani, M., Montero-Lobato, Z., Garbayo, I., Vílchez, C., Vega, J. M., and Martínez-Espinosa, R. M. (2021). Haloferax mediterranei cells as C50 carotenoid factories. Mar. Drugs 19:100. doi: 10.3390/md19020100
Giraud, E., Hannibal, L., Chaintreuil, C., Fardoux, J., and Verméglio, A. (2018). Synthesis of carotenoids of industrial interest in the photosynthetic bacterium Rhodopseudomonas palustris: bioengineering and growth conditions. Methods Mol. Biol. 1852, 211–220. doi: 10.1007/978-1-4939-8742-9_12
Guan, C. J., Ji, Y. J., Hu, J. L., Hu, C. N., Yang, F., and Yang, G. E. (2017). Biotransformation of rutin using crude enzyme from Rhodopseudomonas palustris. Curr. Microbiol. 74, 431–436. doi: 10.1007/s00284-017-1204-3
Hagemann, M. (2011). Molecular biology of cyanobacterial salt acclimation. FEMS Microbiol. Rev. 35, 87–123. doi: 10.1111/j.1574-6976.2010.00234.x
Harwood, C. S., Burchhardt, G., Herrmann, H., and Fuchs, G. (1998). Anaerobic metabolism of aromatic compounds via the benzoyl-CoA pathway. FEMS Microbiol. Rev. 22, 439–458. doi: 10.1016/S0168-6445(98)00026-6
Hashemi, A., Moslemi, M., Pajoum Shariati, F., and Delavari Amrei, H. (2020). Beta-carotene production within Dunaliella salina cells under salt stress condition in an indoor hybrid helical-tubular photobioreactor. Can. J. Chem. Eng. 98, 69–74. doi: 10.1002/cjce.23577
He, Y. Y., and Häder, D. P. (2002). UV-B-induced formation of reactive oxygen species and oxidative damage of the cyanobacterium Anabaena sp.: protective effects of ascorbic acid and N-acetyl-L-cysteine. J. Photochem. Photobiol. B 66, 115–124. doi: 10.1016/s1011-1344(02)00231-2
Jiao, Y., and Newman, D. K. (2007). The pio operon is essential for phototrophic Fe(II) oxidation in Rhodopseudomonas palustris TIE-1. J. Bacteriol. 189, 1765–1773. doi: 10.1128/jb.00776-06
Kim, M.-K., and Harwood, C. S. (1991). Regulation of benzoate-CoA ligase in Rhodopseudomonas palustris. FEMS Microbiol. Lett. 83, 199–203. doi: 10.1111/j.1574-6968.1991.tb04440.x-i1
Kovach, M. E., Elzer, P. H., Steven Hill, D., Robertson, G. T., Farris, M. A., Roop, R. M., et al. (1995). Four new derivatives of the broad-host-range cloning vector pBBR1MCS, carrying different antibiotic-resistance cassettes. Gene 166, 175–176. doi: 10.1016/0378-1119(95)00584-1
Larimer, F. W., Chain, P., Hauser, L., Lamerdin, J., Malfatti, S., do, L., et al. (2004). Complete genome sequence of the metabolically versatile photosynthetic bacterium Rhodopseudomonas palustris. Nat. Biotechnol. 22, 55–61. doi: 10.1038/nbt923
Lefebvre, O., and Moletta, R. (2006). Treatment of organic pollution in industrial saline wastewater: a literature review. Water Res. 40, 3671–3682. doi: 10.1016/j.watres.2006.08.027
Lefebvre, O., Vasudevan, N., Torrijos, M., Thanasekaran, K., and Moletta, R. (2005). Halophilic biological treatment of tannery soak liquor in a sequencing batch reactor. Water Res. 39, 1471–1480. doi: 10.1016/j.watres.2004.12.038
Li, T., Chen, X. B., Chen, J. C., Wu, Q., and Chen, G. Q. (2014). Open and continuous fermentation: products, conditions and bioprocess economy. Biotechnol. J. 9, 1503–1511. doi: 10.1002/biot.201400084
Li, M., Ning, P., Sun, Y., Luo, J., and Yang, J. (2022a). Characteristics and application of Rhodopseudomonas palustris as a microbial cell factory. Front. Bioeng. Biotechnol. 10:897003. doi: 10.3389/fbioe.2022.897003
Li, M., Xia, Q., Lv, S., Tong, J., Wang, Z., Nie, Q., et al. (2022b). Enhanced CO2 capture for photosynthetic lycopene production in engineered Rhodopseudomonas palustris, a purple nonsulfur bacterium. Green Chem. 24, 7500–7518. doi: 10.1039/D2GC02467E
Li, M., Xia, Q., Zhang, H., Zhang, R., and Yang, J. (2020). Metabolic engineering of different microbial hosts for lycopene production. J. Agric. Food Chem. 68, 14104–14122. doi: 10.1021/acs.jafc.0c06020
Lingran, F., Qiang, W., Xiaobin, Y., and Kwame, F. (2019). Effects of exogenous lipids and cold acclimation on lycopene production and fatty acid composition in Blakeslea trispora. AMB Express 9:162. doi: 10.1186/s13568-019-0891-5
Lyles, C., Boopathy, R., Fontenot, Q., and Kilgen, M. (2008). Biological treatment of shrimp aquaculture wastewater using a sequencing batch reactor. Appl. Biochem. Biotechnol. 151, 474–479. doi: 10.1007/s12010-008-8216-1
Matarredona, L., Camacho, M., Zafrilla, B., Bravo-Barrales, G., Esclapez, J., and Bonete, M. J. (2021). The survival of Haloferax mediterranei under stressful conditions. Microorganisms. 9:336. doi: 10.3390/microorganisms9020336
Meng, W., Zhang, Y., Ma, L., Lü, C., Xu, P., Ma, C., et al. (2022). Non-sterilized fermentation of 2,3-butanediol with seawater by metabolic engineered fast-growing Vibrio natriegens. Front. Bioeng. Biotechnol. 10:955097. doi: 10.3389/fbioe.2022.955097
Mohy, E. D. S., and Elahwany, A. (2015). Bioactivity and phytochemical constituents of marine red seaweeds (Jania rubens, Corallina mediterranea and Pterocladia capillacea). J. Taibah Univ. Sci. 10, 471–484. doi: 10.1016/j.jtusci.2015.06.004
Muzziotti, D., Adessi, A., Faraloni, C., Torzillo, G., and De Philippis, R. (2017). Acclimation strategy of Rhodopseudomonas palustris to high light irradiance. Microbiol. Res. 197, 49–55. doi: 10.1016/j.micres.2017.01.007
Niedzwiedzki, D., Koscielecki, J. F., Cong, H., Sullivan, J. O., Gibson, G. N., Birge, R. R., et al. (2007). Ultrafast dynamics and excited state spectra of open-chain carotenoids at room and low temperatures. J. Phys. Chem. B 111, 5984–5998. doi: 10.1021/jp070500f
Nunkaew, T., Kantachote, D., Chaiprapat, S., Nitoda, T., and Kanzaki, H. (2018). Use of wood vinegar to enhance 5-aminolevulinic acid production by selected Rhodopseudomonas palustris in rubber sheet wastewater for agricultural use. Saudi J. Biol. Sci. 25, 642–650. doi: 10.1016/j.sjbs.2016.01.028
Ogugbue, C. J., Sawidis, T., and Oranusi, N. A. (2011). Evaluation of colour removal in synthetic saline wastewater containing azo dyes using an immobilized halotolerant cell system. Ecol. Eng. 37, 2056–2060. doi: 10.1016/j.ecoleng.2011.09.003
Pernetti, M., and Di Palma, L. (2005). Experimental evaluation of inhibition effects of saline wastewater on activated sludge. Environ. Technol. 26, 695–704. doi: 10.1080/09593330.2001.9619509
Quandt, J., and Hynes, M. F. (1993). Versatile suicide vectors which allow direct selection for gene replacement in gram-negative bacteria. Gene 127, 15–21. doi: 10.1016/0378-1119(93)90611-6
Ren, Y., Sun, H., Deng, J., Huang, J., and Chen, F. J. M. D. (2021). Carotenoid production from microalgae: biosynthesis, salinity responses and novel biotechnologies. Mar. Drugs 19:713. doi: 10.3390/md19120713
Sakpirom, J., Kantachote, D., Nunkaew, T., and Khan, E. (2017). Characterizations of purple non-sulfur bacteria isolated from paddy fields, and identification of strains with potential for plant growth-promotion, greenhouse gas mitigation and heavy metal bioremediation. Res. Microbiol. 168, 266–275. doi: 10.1016/j.resmic.2016.12.001
Sankari, M., Hridya, H., Sneha, P., Doss, C. G. P., Christopher, J. G., Mathew, J., et al. (2019). Implication of salt stress induces changes in pigment production, antioxidant enzyme activity, and qRT-PCR expression of genes involved in the biosynthetic pathway of Bixa orellana L. Funct. Integr. Genomics 19, 565–574. doi: 10.1007/s10142-019-00654-7
Sévin, D. C., and Sauer, U. (2014). Ubiquinone accumulation improves osmotic-stress tolerance in Escherichia coli. Nat. Chem. Biol. 10, 266–272. doi: 10.1038/nchembio.1437
Shi, Y., Guo, J., Zhang, W., Jin, L., Liu, P., Chen, X., et al. (2015). Cloning of the lycopene β-cyclase gene in Nicotiana tabacum and its overexpression confers salt and drought tolerance. Int. J. Mol. Sci. 16, 30438–30457. doi: 10.3390/ijms161226243
Simó-Cabrera, L., García-Chumillas, S., Hagagy, N., Saddiq, A., Tag, H., Selim, S., et al. (2021). Haloarchaea as cell factories to produce bioplastics. Mar. Drugs 19:159. doi: 10.3390/md19030159
Takaichi, S. (1999). Carotenoids and carotenogenesis in anoxygenic photosynthetic bacteria. Springer: Netherlands 8:39–69.
Thombre, R. S., Shinde, V. D., Oke, R. S., Dhar, S. K., and Shouche, Y. S. (2016). Biology and survival of extremely halophilic archaeon Haloarcula marismortui RR12 isolated from Mumbai salterns, India in response to salinity stress. Sci. Rep. 6:25642. doi: 10.1038/srep25642
Verma, E., Singh, S. N., and Mishra, A. K. (2019). Salinity-induced oxidative stress-mediated change in fatty acids composition of cyanobacterium Synechococcus sp. PCC7942. Int. J. Environ. Sci. Te. 16, 875–886. doi: 10.1007/s13762-018-1720-0
Wang, Z., Wen, Q., Harwood, C. S., Liang, B., and Yang, J. (2020). A disjointed pathway for malonate degradation by Rhodopseudomonas palustris. Appl. Environ. Microbiol. 86, e00631–e00620. doi: 10.1128/aem.00631-20
Wasai, S., Kanno, N., Matsuura, K., and Haruta, S. J. M. (2018). Increase of salt tolerance in carbon-starved cells of Rhodopseudomonas palustris depending on photosynthesis or respiration. Microorganisms 6:4. doi: 10.3390/microorganisms6010004
Woolard, C. R., and Irvine, R. L. (1995). Treatment of hypersaline wastewater in the sequencing batch reactor. Water Res. 29, 1159–1168. doi: 10.1016/0043-1354(94)00239-4
Wu, X., Altman, R., Eiteman, M. A., and Altman, E. (2014). Adaptation of Escherichia coli to elevated sodium concentrations increases cation tolerance and enables greater lactic acid production. Appl. Environ. Microbiol. 80, 2880–2888. doi: 10.1128/aem.03804-13
Wu, P., Chen, Z., Zhang, Y., Wang, Y., Zhu, F., Cao, B., et al. (2019). Rhodopseudomonas palustris wastewater treatment: Cyhalofop-butyl removal, biochemicals production and mathematical model establishment. Bioresour. Technol. 282, 390–397. doi: 10.1016/j.biortech.2018.11.087
Wu, S. C., Liou, S. Z., and Lee, C. M. (2012). Correlation between bio-hydrogen production and polyhydroxybutyrate (PHB) synthesis by Rhodopseudomonas palustris WP3-5. Bioresour. Technol. 113, 44–50. doi: 10.1016/j.biortech.2012.01.090
Xiao, Y., and Roberts, D. J. (2010). A review of anaerobic treatment of saline wastewater. Environ. Technol. 31, 1025–1043. doi: 10.1080/09593331003734202
Xu, W., Chai, C., Shao, L., Yao, J., and Wang, Y. (2016). Metabolic engineering of Rhodopseudomonas palustris for squalene production. J. Ind. Microbiol. Biotechnol. 43, 719–725. doi: 10.1007/s10295-016-1745-7
Yu, L. P., Wu, F. Q., and Chen, G. Q. (2019). Next-generation industrial biotechnology-transforming the current industrial biotechnology into competitive processes. Biotechnol. J. 14:e1800437. doi: 10.1002/biot.201800437
Yue, H., Ling, C., Yang, T., Chen, X., Chen, Y., Deng, H., et al. (2014). A seawater-based open and continuous process for polyhydroxyalkanoates production by recombinant Halomonas campaniensis LS21 grown in mixed substrates. Biotechnol. Biofuels 7, 1–12. doi: 10.1186/1754-6834-7-108
Zhu, L., Wu, X., Li, O., Chen, Y., Qian, C., Teng, Y., et al. (2011). Cloning and knockout of phytoene desaturase gene in Sphingomonas elodea ATCC 31461 for economic recovery of gellan gum. J. Ind. Microbiol. Biotechnol. 38, 1507–1513. doi: 10.1007/s10295-010-0937-9
Keywords: salt-tolerant, Rhodopseudomonas palustris, carotenoid, lycopene, rhodopin
Citation: Li M, Zhu T, Yang R, Wang Z, Liu M and Yang J (2023) Carotenoids synthesis affects the salt tolerance mechanism of Rhodopseudomonas palustris. Front. Microbiol. 14:1292937. doi: 10.3389/fmicb.2023.1292937
Received: 12 September 2023; Accepted: 08 November 2023;
Published: 22 November 2023.
Edited by:
Harold J. Schreier, University of Maryland, Baltimore County, United StatesReviewed by:
Santosh Kumar, University of Wisconsin-Madison, United StatesCopyright © 2023 Li, Zhu, Yang, Wang, Liu and Yang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Jianming Yang, eWptaW5nODg4QDEyNi5jb20=
†These authors have contributed equally to this work and share first authorship
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.