- ICAR-Indian Institute of Wheat and Barley Research (IIWBR), Karnal, India
The current study describes a new diagnostic method for the rapid and accurate detection of Tilletia indica, the pathogen accountable for causing Karnal bunt (KB) disease in wheat. This method uses quantitative real-time polymerase chain reaction (qPCR) and a primer set derived from glyceraldehyde 3-phosphate dehydrogenase (GAPDH) gene of T. indica to identify the presence of the pathogen. The qPCR assay using this primer set was found highly sensitive, with a limit of detection (LOD) value of 4 pg of T. indica DNA. This level of sensitivity allows for the detection of the pathogen even in cases of different growth stages of wheat, where no visible symptoms of infection on the wheat plants can be seen by naked eyes. The study also validated the qPCR assay on ten different wheat cultivars. Overall, this study presents a valuable molecular tool for rapid, specific and sensitive detection of KB fungus in wheat host. This method has practical applications in disease management, screening of wheat genotypes against KB and can aid in the development of strategies to mitigate the impact of Karnal bunt disease on wheat production.
Introduction
Wheat is recognized as the prime cereal crop globally due to its widespread cultivation and essential role in human nutrition (Singh et al., 2023a). Fungal diseases in wheat are responsible for substantial yield losses, accounting for approximately 22% of total losses (Kayim et al., 2022). Karnal bunt (KB), provoked by fungal pathogen T. indica, is a significant quarantine disease that influences global wheat cultivation (Kashyap et al., 2023). KB was first identified in 1931 and was primarily confined to North–West India during its early years. Over the following decades, KB was officially reported in several other countries, including Afghanistan, Iraq, Nepal, Pakistan (specifically in Punjab and North–West Frontier Provinces), and Iran. It also appeared in Mexico (Sonora, Sinaloa, and Baja California Sur), Brazil (Rio Grande do Sul), the USA (New Mexico, Arizona, Texas, and California), and eventually South Africa (Northern Cape Province).1 The reason for the rapid spread of KB is the wheat seed movement, especially during and after the Green Revolution. This movement likely facilitated the dissemination of KB from its initial localized region to various other countries (Bishnoi et al., 2020). KB fungus infects the ovaries of growing wheat heads, resulting in the conversion of seeds into a black powder of teliospores (Fuentes-Davila and Duran, 1986; Kashyap et al., 2018). This severely impacts the quality and marketability of the grain (Sharma et al., 1998; Singh and Gogoi, 2011). Detecting Karnal bunt in wheat is challenging because not all seeds on an ear of wheat are infected, and infected seeds do not appear different from healthy ones. Additionally, symptoms are often not visible until harvest (Brar et al., 2018). It is worth to mention that T. indica is capable of being transmitted through seeds, soil, and air, making it particularly challenging to control and manage (Kaur and Kaur, 2005; Carris et al., 2006; Kashyap et al., 2019; Bishnoi et al., 2020). Several disease management options are available, including prophylactic measures, fungicide sprays, and deploying partially resistant wheat cultivars (Emebiri et al., 2021; Kashyap et al., 2022a; Singh et al., 2023b). However, these measures may not be entirely effective, especially under high disease pressure (Goates and Jackson, 2006; Bala et al., 2022). It is pertinent to mention that KB disease is sporadic and tends to invade during the late stages of host plant growth. Moreover, a single infected kernel can contain over 100,000 teliospores which are buried in the soil and are well-protected from both physical and chemical adverse conditions (Fuentes-Dávila et al., 2018; Thapa et al., 2022). Teliospores can persist in the soil for a long duration (Smilanick et al., 1989; Babadoost et al., 2004), making complete eradication of the disease through cultural practices and crop rotation complicated. Further, the localized mode of KB infection adds complexity to its management compared to other systemic smut diseases in plants (Fuentes-Davila, 1996; Riccioni et al., 2008). Conspicuously, T. indica infects wheat crops at a partial level, often not reaching a threshold level where it leads to significant economic losses. As a result, the chemical measures are generally neither applied nor completely effective. Additionally, the intricate nature of T. indica infection process makes it challenging to manage KB disease using both cultural practices and fungicides (Emebiri et al., 2019). Chemical fungicides such as Carbendazim, Triadimefon, and Propiconazole, when applied as foliar sprays during flowering or late booting stage have been effective against KB disease. However, concerns about their economic and environmental impact persist (Duveiller and Mezzalama, 2009; Kashyap et al., 2018). The efficacy of seed treatment with fungicides, such as Chlorothalonil and Carboxin + Thiram, is limited because teliospores are protected by the pericarp of infected kernels (Kumar et al., 2021). To address the challenges associated with Karnal bunt management, there is a prerequisite of a precise, responsive, and quick tool to detect and recognize T. indica fungus in wheat during its growth. Furthermore, it is crucial to have effective molecular markers that can differentiate T. indica from other pathogens that affect the wheat phyllosphere under field conditions as well as for large screening of KB resistant genotypes.
Traditional methods for diagnosing T. indica infection in wheat crops rely on morphological identification of symptoms in seeds and spore load (teliospore) counts after harvesting (Sharma et al., 2022). These morphological methods are tedious, time-consuming, and may not be very precise (Gurjar et al., 2021; Iquebal et al., 2021). Moreover, several smut-causing fungi, including T. controversa, T. tritici, T. laevis, T. barclayana, T. ehrhartae, and T. walkeri, share morphological similarities with T. indica and results in the misidentification when relying solely on the morphology or microscopic examination (Ferreira et al., 1996; Pascoe et al., 2005). Other technique such as isozyme analysis has been reported for KB detection (Luster et al., 1998). The prime limitation of this assay is that it requires at least 10 days duration for spore germination. Additionally, it demands specialized knowledge and expertise to perform accurately (Kutilek et al., 2001). Currently, polymerase chain reaction technology is considered a promising alternative for diagnosing T. indica. It is simple, rapid, accurate, and sensitive, making it suitable for detecting and identifying plant pathogens. Many of the PCR assays target the internal transcribed spacer (ITS) region (Levy et al., 2001; Kashyap et al., 2016; Gurjar et al., 2017; Gupta et al., 2022; Ren et al., 2022a), which has limited variation between T. indica and its closest relative, T. walkeri that differ by one nucleotide only (Bonde et al., 1997; Levy et al., 2001; Tremblay et al., 2021). This can affect sensitivity, a critical factor when dealing with diseases like Karnal bunt. Some assays require teliospore germination before molecular analysis, which can slow down the diagnostic process (Tan et al., 2010). Additionally, many previously developed assays are low-throughput, and international protocols involve time-consuming processes like morphological observations and spore isolation (Valente et al., 2019). A series of DNA-based methods have attempted to distinguish teliospores of Tilletia species from seeds or soil samples, which serve as the primary source of pathogen spread (Eibel et al., 2005; Chen et al., 2016; Valente et al., 2023). In earlier studies, PCR based techniques such as RAPD primer-mediated asymmetric polymerase chain reaction (Yuan et al., 2009), sequence-characterized amplified region (Liu et al., 2009; Gao et al., 2010), repetitive element polymerase chain reaction (Vesna et al., 2011), inter-simple sequence repeat (Gao et al., 2011), amplified fragment length polymorphism (Zhang et al., 2012), and sequence characterized amplified region (Ren et al., 2022a; Xu et al., 2022) have been successfully demonstrated for the identification of Tilletia species. However, real time qPCR based assays are more sensitive, precise, reliable and time saving than conventional PCR method (Zhang et al., 2012; Fang and Ramasamy, 2015; Yao et al., 2019). Moreover, real-time PCR (qPCR) eliminates the need to run gels after the reaction, reducing the risk of cross-contamination. At present, there is limited information available regarding the qPCR based assay for the diagnosis of T. indica infection in wheat host under field conditions.
Selecting the right target region is crucial for detecting T. indica infection in plants. Since the whole genomes of T. indica are publicly available, researchers have the opportunity to identify new target loci for molecular detection. This can be achieved through bioinformatic analysis of T. indica genomes and comparing them with other pathogens causing wheat diseases. Pandey et al. (2018) conducted a study comparing highly virulent (TiK) and low virulent (TiP) isolates of T. indica. They found that glyceraldehyde 3-phosphate dehydrogenase (GAPDH) plays a crucial role in determining virulence in T. indica. GAPDH is highly abundant in T. indica mycelium and serves as a virulence factor, similar to its role in other pathogenic fungi. Interestingly, GAPDH is not only present in the cytosol but also on the surface of virulent T. indica isolates. This surface presence of GAPDH contributes to fungal incursion and colonization of wheat tissues.
Real-time PCR has become a significant tool for the diagnosis and detection of phytopathogenic fungi over the last few decades (Kumar et al., 2013; Kashyap et al., 2020a). It has several advantages over conventional PCR methods in terms of sensitivity, reducing the peril of pseudo-positive results, and enabling quantitative analysis (Schena et al., 2006; Choudhary et al., 2020). Abdullah et al. (2018) reported real time PCR assay that concurrently differentiate and enumerate co-infections by Parastagonospora nodorum and Pyrenophora tritici-repentis in wheat. Loop-mediated isothermal amplification (LAMP) method has been reported for the diagnosis of Tilletia spp. (Gao et al., 2016; Pieczul et al., 2018). There are several studies that reported lower sensitivity of LAMP assay than qPCR (Ou et al., 2012; Yang et al., 2013; Nixon et al., 2014; Naveen and Bhat, 2020). Khan et al. (2018) also mentioned that LAMP assay was less sensitive than nested PCR and qPCR in diagnosis Alternaria solani. Similarly, qPCR assay developed by Frederick et al. (2000) was reported to detect 5 pg of DNA, showing relatively more sensitivity than LAMP assays by Gao et al. (2016) at ≥10 pg of DNA and Tan et al. (2016) at 10 pg of fungal DNA. Harper et al. (2010) highlighted that LAMP and real-time assays are highly specific, however, qPCR showed greater sensitivity for rapid detection of Xylella fastidiosa than LAMP assay. The most common drawback of the LAMP assay is the misamplifications that occur from redundant secondary structures. Even with the careful primer design and the availability of programs that confirm dimer and hairpin structure formation, there is no assurance that these structures will not be produced practically (Alhamid et al., 2023). Recently, droplet digital PCR (ddPCR) technique has been explored for the precise and sensitive diagnosis of Tilletia caries fungus in wheat (Ren et al., 2022b). ddPCR’s ability to provide highly sensitive and specific absolute quantification, combined with its reproducibility and tolerance to inhibitors, makes it a valuable emerging tools in the field of diagnostics (Xu et al., 2020). However, the prime demerits and concerns associated with ddPCR include high cost, technical complexity and a reduced dynamic range in comparison to qPCR (Jones et al., 2016). Another major concern with ddPCR is false-positive partitions in no template control (NTC) well (Kojabad et al., 2021). Most importantly, a droplet digital PCR system consists of multiple instruments (droplet generator, thermocycler, and droplet reader) that take up valuable lab space and require trained personnel for operation. Keeping aforementioned points in mind, the current study was planned for developing qPCR assay to differentiate and quantify T. indica in wheat under field conditions and for screening wheat genotypes against KB. To achieve this, research has been performed to design species specific oligonucleotide primers derived from the GAPDH gene and conducted validation tests to assess the performance of these primers in detecting T. indica in pure cultures as well as in artificially inoculated wheat seedlings at different growth stages of wheat under field conditions.
Materials and methods
Fungal isolates and DNA extraction
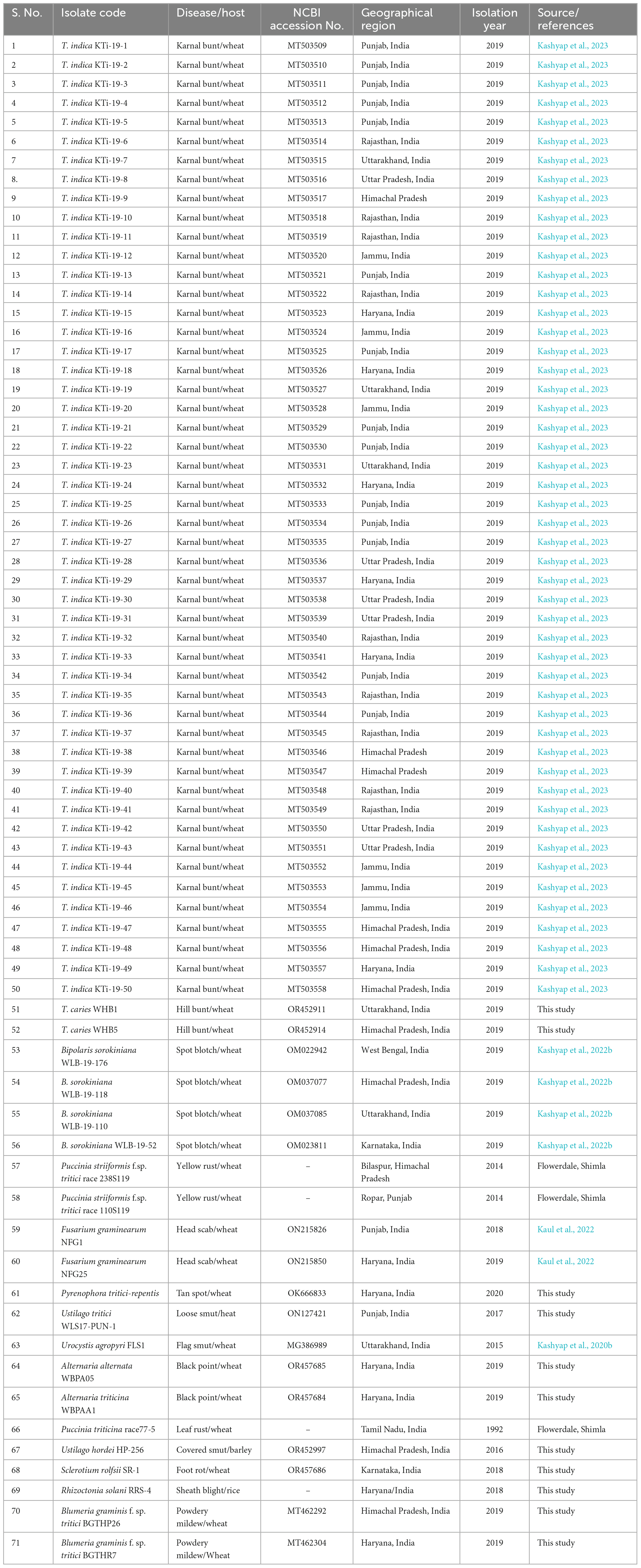
The study involves a total of 71 fungal isolates. A detailed description pertaining to host, pathogen identification, geographical region, year of isolation of the fungal isolates used in the current study is mentioned in Table 1. Usually, an annual crop health field survey is conducted every year to assess the wheat crop situation in the country. During the surveys at farmer’s field and grain mandies from 2017 to 2020, diseased samples (either plant parts or harvested seed) were collected. Among them, 50 were isolates of T. indica, and the remaining 21 isolates were from other unrelated fungal pathogens, majority of them were potential wheat pathogens (Table 1). These pathogens include Tilletia caries, Blumeria graminis, Bipolaris sorokiniana, Alternaria alternata, Alternaria triticina, Fusarium graminearum, P. striiformis, Sclerotium rolfsii, P. triticina, Pyrenophora tritici-repentis, Rhizoctonia solani, Urocystis tritici, Ustilago tritici, and Ustilago hordei. Ungerminated teliospores of U. agropyri, Tilletia indica, Tilletia caries, Ustilago tritici, and Ustilago nuda f. sp. hordei collected from various regions were directly processed for genomic DNA isolation. The mycelia of fungal isolates (Fusarium graminearum, Alternaria triticina, Bipolaris sorokiniana, Pyrenophora tritici-repentis, Alternaria alternata, Sclerotium rolfsii, and Rhizoctonia solani) were cultivated on potato dextrose broth (Hi Media, India) following single spore methodology for 7 days at 25 ± 2°C. The mycelial mat of each isolate was separated through sterile Whatman filter paper and ground to fine powder with mortar and pestle by incorporating liquid nitrogen. Total genomic DNA from these fungal isolates was extracted by following a protocol reported by Kumar et al. (2013). The purity and concentration of the isolated genomic DNA were assessed using a ScanDrop2 instrument from Analytik Jena, Germany. The isolated DNA was stored at a temperature of −20°C for future use.
Development of T. indica specific primers
For designing the species-specific primers for T. indica, GAPDH gene sequences of T. indica from the NCBI GenBank were downloaded. Clustal W software (Thompson et al., 1997) was used to align multiple sequences (Figure 1). Sequence alignment was performed to identify conserved regions and polymorphic sites, which later used to design specific primers. Using the Primer 3 plus software,2 species-specific primers (Table 2) were designed to amplify only the target species, i.e., Tilletia indica and not other related organisms. Basic Local Alignment Search Tool (BLAST) tool3 was used to corroborate the specificity of identified primers and to ensure that the primers did not amplify similar sequences in other microorganisms present in the NCBI GenBank database.4 The primer quality was assessed using the IDT oligo analyzer.5 This involves checking for potential issues like secondary structure formation, self-complementarity, and dimer formation, as these can affect the efficiency and specificity of PCR reactions.
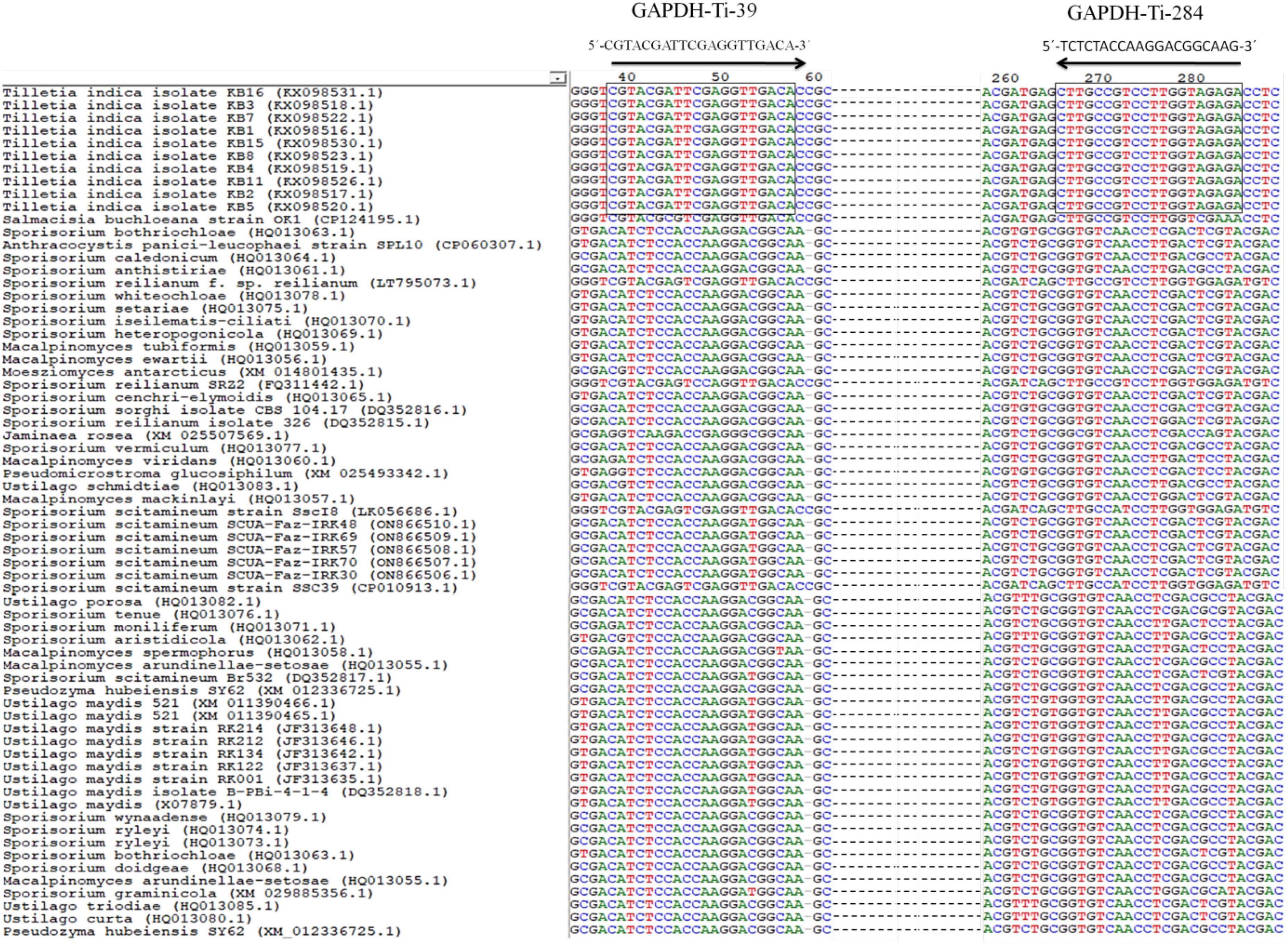
Figure 1. Alignment of GAPDH gene sequences from T. indica showing the position and orientation of unique region of T. indica. The GAPDH-Ti-39 (Forward) and GAPDH-Ti-284 (reverse) primers used for the PCR and qPCR assay that are identical between the species are shown. Consensus nucleotides are indicated with a rectangle box.
PCR amplification assay
Polymerase chain reaction (PCR) experiment for the amplification of a DNA target using specific primers was performed. The total reaction volume was 10 μl and encompassed 1 μl of template DNA with a concentration of 40 ng μl–1. A total
of 5 μl of GoTaq Green master mix by Promega Biotech, India was used. A total of 1 μl of each forward and reverse primer was added, and these primers had a concentration of 10 μM. Nuclease-free water was incorporated to make up the remaining volume to 10 μl. A control check without genomic DNA (NC) was also included. The thermal cycler used was the Sure Cycler 8800 (Agilent Technologies). The PCR program composed of initial denaturation at 94°C for 5 min followed by 35 cycles of: denaturation at 94°C for 60 s, annealing at 55°C for 60 s and extension at 72°C for 60 s. A final extension step at 72°C for 10 min was maintained with holding time at 4°C. The results of PCR amplicon generation were analyzed through electrophoresis. A 1.2% agarose gel in 1 × TBE buffer was used for this purpose. Electrophoresis was run for 1 h at 90 V. To estimate the size of the amplicon (the DNA fragment that was amplified), a 100 bp DNA ladder by Promega Biotech India Pvt. Ltd., was used as a reference.
In vivo validation of the assay
The field experiment was performed at ICAR-Indian Institute of Wheat and Barley Research (IIWBR) in Karnal, India (29°42′19.5″N 76°59′24.8″E), during the rabi cropping season of 2020–2021. Two wheat cultivars, PBW343 and HD2967, were used in the experiment for preliminary validation of the developed markers under field conditions. The soil texture of experimental field was sandy loam (62.1% sand, 26.9% silt, and 11.0% clay) with pH 7.8 and electrical conductivity 0.25 dS/m. The soil was having 0.42% organic carbon, 193 Kg ha–1 available N, 17.9 Kg ha–1 available P, and 241 Kg ha–1 available K. Plantation of the viable (95% seed germination) foundation wheat seeds (99% seed purity) was done in experimental plot (2 m2) with 1 m long rows with a space of 22 cm between rows. Each cultivar was sown in three replicates. Sowing operations of the wheat seeds were performed in the second week of November, which is the optimal time for wheat sowing in North India. T. indica isolates (Table 1) were grown on Petri plates amended with potato dextrose agar (Hi Media, India) for 7 days. The load of the liquid suspension of secondary sporidia (5 × 106 ml–1) in sterilized distilled water was optimized with the help of a hemocytometer. During evening hours, two milliliter of standardized liquid suspension was injected into tillers of each wheat cultivar at Zadok’s growth stage 49 (ZGS49; boot leaf stage) using a hypodermal syringe (Aujla et al., 1989). High humidity (>70%) was sustained by regular mist sprays. At the stage of crop maturity, ears heads of the inoculated tillers were harvested. Each seed from the inoculated tillers was inspected for the presence of KB teliospores. The severity of infection was recorded using numerical values (0, 0.25, 0.50, 0.75, and 1.0) to indicate infection grade. The percent coefficient of infection (CI) was calculated using a specific formula mentioned by Kashyap et al. (2017). Genomic DNA was extracted from healthy and T. indica-inoculated wheat spikes at various growth stages [ZGS 59 (ear emergence stage), ZGS 69 (flowering stage), ZGS77 (milking stage) ZGS87 (dough stage) and ZGS92 (ripening stage)]. PCR assays were performed using species-specific primers, and gel electrophoresis by employing the procedure mentioned earlier.
Real-time PCR assay and melting curve analysis
A quantitative real-time polymerase chain reaction (qRT-PCR) was performed by employing SYBR Green-I dye (Thermo Scientific, USA) on the AriaMx Real-Time PCR machine (Agilent Technologies, USA). The qPCR cocktail was set-up by combining the following components in a 20 μl reaction volume: 10 μl SYBR Green qPCR master mix (Thermo Scientific, USA), 10 picomoles of each primer (forward and reverse) and 1 μl of genomic DNA, with a concentration ranged from 40 to 0.004 ng μl–1. qPCR cycling conditions included initial heat activation at 94°C for 5 min followed by amplification with 35 cycles using a 2-step cycling method. This included denaturation at 94°C for 30 s and annealing at 55°C for 60 s. Melting curve analysis was performed after the amplification cycles to check the specificity of the PCR product. The plate was heated to 95°C for 30 s to denature the DNA. It is then incubated at 65°C for 30 s. Finally, it was heated again to 95°C for 30 s. Fluorescence is measured once per cycle at the completion of the extension step. The machine automatically computed the cycle quantification (Cq) values and were used to estimate the initial amount of target genomic DNA in the sample.
Standard curves construction for absolute quantification of T. indica
An experiment was performed for absolute quantification of T. indica by constructing a standard curve for the GAPDH-Ti-39/GAPDH-Ti-284 primer. Genomic DNA was serially diluted with nuclease-free water to create a range of concentrations from 40 to 0.004 ng reaction–1. Each concentration was tested in three replicates independently. The entire experiment performed twice in an independent manner for confirming the consistency of results and ruling out any experimental errors. A no template control (NC) was also used to ensure the specificity of the reaction and avoid cross contamination.
qPCR based detection of T. indica fungus load in wheat cultivars
Ten different wheat cultivars (DBW173, DBW187, DBW303, HD2967, HD3086, DBW252, PBW343, RAJ4083, WH542, and WL711) were used in the experiment to validate the applicability of the developed assay to check the genotype influence at wheat KB susceptible stage, i.e., boot leaf stage (ZGS-49 stage). Healthy seeds of these wheat cultivars were planted and grown under field conditions during the rabi season of 2021–2022. At the boot leaf stage (ZGS-49 stage), the wheat plants were artificially inoculated with T. indica at a concentration of 5 × 106 sporidia ml–1. Wheat samples from each cultivar (1 gram each) were harvested at ZGS-77. Total genomic DNA was extracted from the collected wheat samples. The methodology used for DNA isolation was similar to a previous section. Real-time PCR assays were performed on the extracted genomic DNA. The qPCR cocktail (20 μl) was prepared by incorporating 10 μl SYBR Green qPCR master mix (Thermo Scientific, USA), 10 picomoles of each primer (forward and reverse) and 1 μl of genomic DNA. A negative control (NC) was included in the experiment. In this control, the DNA template was replaced by nuclease free water. qPCR cycling conditions were as follows: initial heat activation at 94°C for 5 min followed by amplification with 35 cycles using a 2-step cycling of denaturation at 94°C for 30 s and annealing at 55°C for 60 s. Melting curve analysis was performed after the amplification cycles to check the specificity of the PCR product. The plate was heated to 95°C for 30 s to denature the DNA. It is then incubated at 65°C for 30 s. Finally, it was heated again to 95°C for 30 s. Fluorescence is measured once per cycle at the end of extension step. Cycle quantification (Cq) values were used to estimate the initial amount of target genomic DNA in the sample. The assay was performed twice.
Statistical analysis
The significance of the data generated for KB disease was statistically analyzed by conducting an analysis of variance (ANOVA). DMRT (Duncan’s Multiple Range Test) was carried out for post hoc comparative analysis of the mean data. Genetic analysis and sequence alignments were carried out using MEGA 10 software (Tamura et al., 2021). The R2 value was calculated according to the method described by Bustin et al. (2009). The criteria outlined by Broeders et al. (2014) were followed to determine the efficiency (which should be in the range of 90–110%), repeatability (with a relative standard deviation ≤25%), and linearity (with R2 ≥ 0.98) of the qPCR assay.
Results
Design of T. indica-specific primers
The result of the in silico analysis to monitor the homogeneity or genetic consistency of a specific genomic region (GAPDH) within isolates of T. indica was illustrated in Figure 1. Primers named GAPDH-Ti-39 and GAPDH-Ti-284 were designed on the basis of comparison of T. indica regions of divergence with other related species (Table 2). The amplification product generated by these primers had a size of 243 bp. The melting temperature of the DNA fragment was determined as 57°C. The BLAST analysis against the NCBI database was performed using BLAST 2.2.14.6 The designed primer pair was found specific for T. indica (Figure 1). Besides this, in silico results obtained after multiple alignments of GAPDH amino acid sequences of T. indica with animal and plant GAPDH amino acid sequences indicated that T. indica GAPDH region is unique and not matched with plant and animal GAPDH amino acid sequences (Supplementary Figure 1).
Determination of primers specificity
The PCR reaction successfully generated a 243 bp amplicon from all 50 isolates of T. indica (Figure 2). This suggests that the primer pair used in your PCR assay was specific to T. indica and that all the fifty isolates of T. indica share the same DNA sequence at the target region, resulting in the production of the same 243 bp product. For the remaining 21 isolates representing different fungal species, including T. caries, B. graminis, B. sorokiniana, A. alternata, A. triticina, F. graminearum, P. striiformis, S. rolfsii, P. triticina, P. tritici-repentis, R. solani, U. tritici, U. agropyri, and U. hordei, no amplified products were generated (Figure 2). This indicates that the primer pair used for PCR did not bind or amplify DNA from these fungal species.
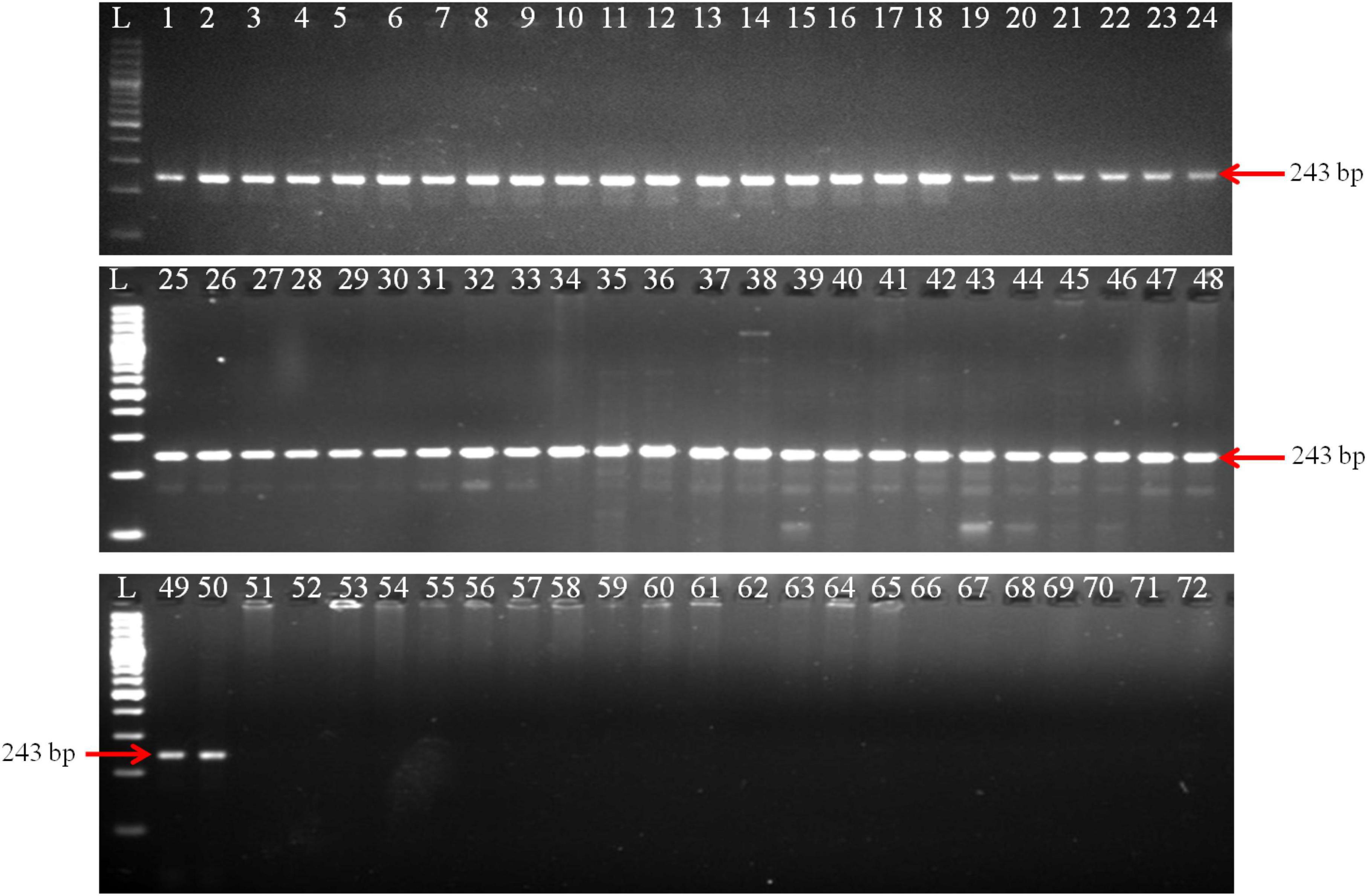
Figure 2. Polymerase chain reaction (PCR) amplification of GAPDH gene fragments from T. indica isolates using GAPDH-Ti-39 (Forward) and GAPDH-Ti-284 (reverse) primers. Lane 1–71 indicates the isolates as per Table 1. Lane72: NC (No template control); L: 100 bp DNA Ladder.
Detection of T. indica in infected plants at different growth stages
The PCR assay utilized the GAPDH-Ti-39/GAPDH-Ti-284 primer pair and successfully generated the specific amplicon (243 bp) from infected wheat varieties, namely PBW343 and HD2967. The specific amplicon was detected in infected wheat samples at all of these growth stages (i.e., ZGS 59, ZGS 69, ZGS 77, ZGS 87, and ZGS 92). Importantly, there was no amplification of the specific amplicon detected in healthy tissue from any of the growth stages (Figure 3). The observation pertaining to the occurrence of T. indica was made in the harvested seeds from the T. indica-inoculated wheat spikes (Table 3). During 2020–2021, the coefficient of infection (CI) were reported as 44.50% for PBW343 and 33.46% for HD2967 (Table 3), indicating the extent of disease infection in these wheat varieties.
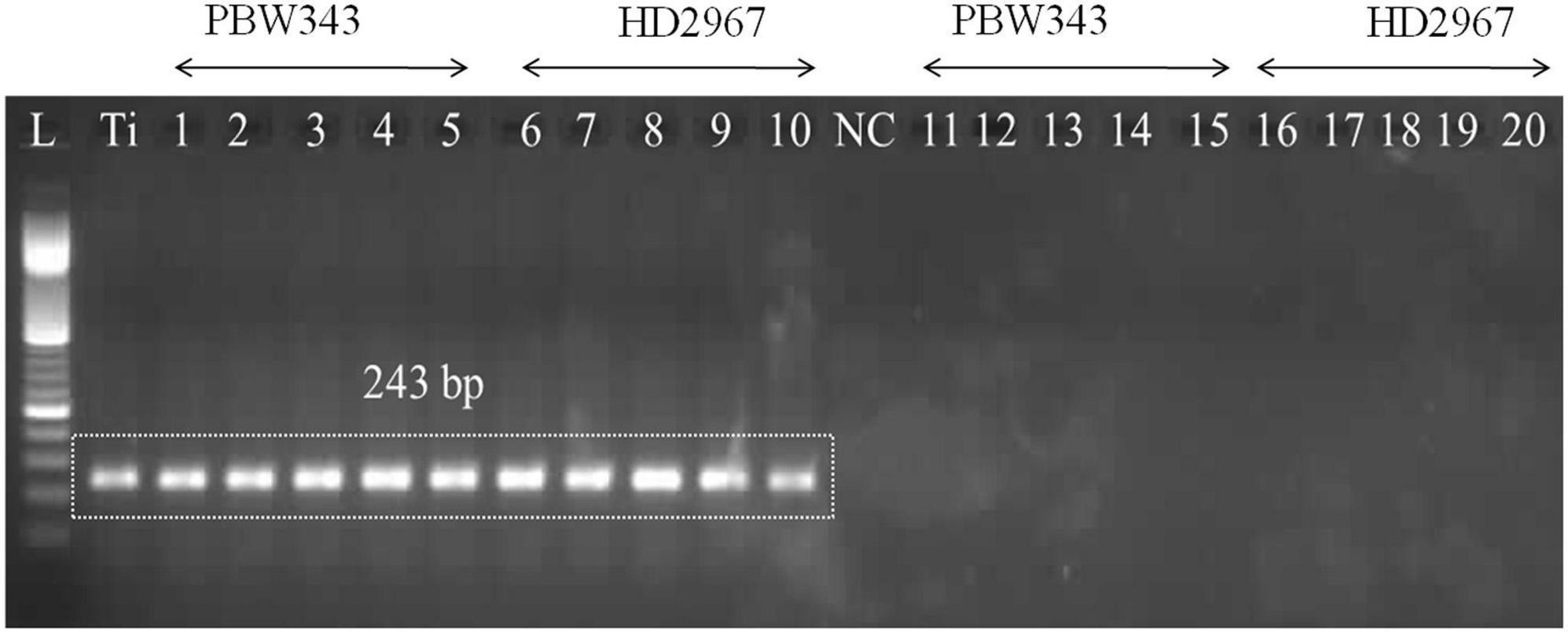
Figure 3. Polymerase chain reaction (PCR) amplification using DNA extracted from wheat seedlings of two different cultivars (PBW343 and HD2967) at five different crop stages, i.e., ZGS 59 (ear emergence stage), ZGS 69 (Flowering stage), ZGS77 (Milking stage) ZGS87 (Dough stage) and ZGS92 (ripening stage) using GAPDH-Ti-39/GAPDH-Ti-284 primer set. L: DNA ladder 100 bp; Ti, positive control (T. indica KTi-19-1); NC: Negative control (without KTi-19-1DNA template), 1–5: Zadoks growth stage DNA obtained from inoculated wheat seedlings at different growth stages; Lane 11–20: negative healthy control (DNA obtained from wheat seedlings, where only water injected in place of T. indica inoculation).
Real-time PCR for absolute quantification of T. indica
The qPCR experiment using GAPDH-Ti-39/GAPDH-Ti-284 primers resulted in Ct (Cycle threshold) values ranging from 19.42 ± 0.03 to 33.53 ± 0.03 when analyzing different dilutions of T. indica DNA. The assay was able to detect T. indica DNA down to a concentration as low as 0.004 ng per microliter (Figure 4A). There is a linear correlation between the Ct values obtained in the qPCR assay and the concentration of the target T. indica DNA. The coefficient of determination (R2) was greater than 0.989, indicating a strong and positive correlation between the Ct values and DNA concentration. The linear equation y = −3.395x + 36.82 describes this relationship, where “y” represents Ct values, and “x” represents the target DNA concentration (Figure 4A). The melting temperature (Tm) for specific amplicon generation was determined to be 86.50 ± 0.50°C, indicating the specificity of the amplicon (Figure 4B).
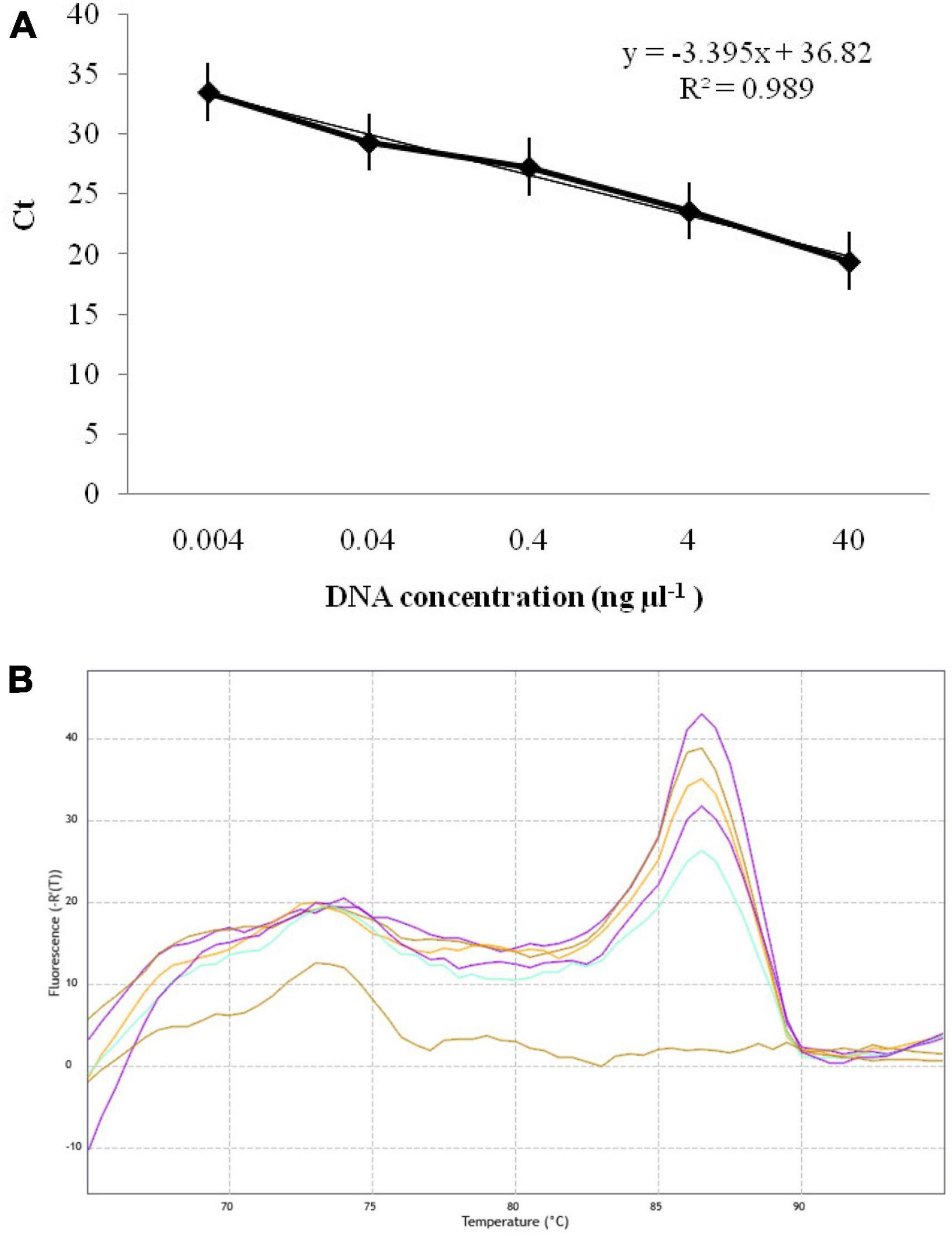
Figure 4. (A) Standard curves displaying the regression between DNA log quantities (ng, X-axis) and cycle thresholds (Ct, y-axis) for qPCR assay using GAPDH-Ti-39 (Forward) and GAPDH-Ti-284 (reverse) primers. The standard curve was developed with serial dilutions (40 to 0.004 ng μl–1) of T. indica KTi-19-1; (B) melting curve showing melting temperatures (86.50) required for 243 bp qPCR product amplification with GAPDH-Ti-39/GAPDH-Ti-284 primers.
qPCR quantification of T. indica DNA in wheat varieties
The GAPDH-Ti-39 and GAPDH-Ti-284 primers were used in the qPCR analysis, and they produced distinct fluorescent signals from all the inoculated wheat samples. During 2021–2022, the mean Ct (Cycle threshold) values for T. indica inoculated wheat cultivars at ZGS87 (Dough stage) were measured and were ranged between 19.54 (cv. PBW343)–31.45 (cv. DBW187) (Table 4). The corresponding genomic concentration of T. indica in the wheat plant ranged from 0.001 (cv. DBW187) to 33.60 ng μl–1 (cv. PBW343). Maximum CI value was observed in cultivar PBW343 (44.50 ± 1.24%) followed by WL711 (38.62 ± 1.99%), HD2967 (35.46 ± 1.77%), WH542 (34.78 ± 1.45%), DBW303 (32.10 ± 1.65%), DBW173 (19.30 ± 0.58%), DBW252 (18.80 ± 1.32%), RAJ4083 (16.30 ± 0.95%), HD3086 (15.70 ± 1.24%), DBW187 (13.50 ± 0.42%) (Table 4). This suggests varying degrees of infection among the different wheat cultivars.
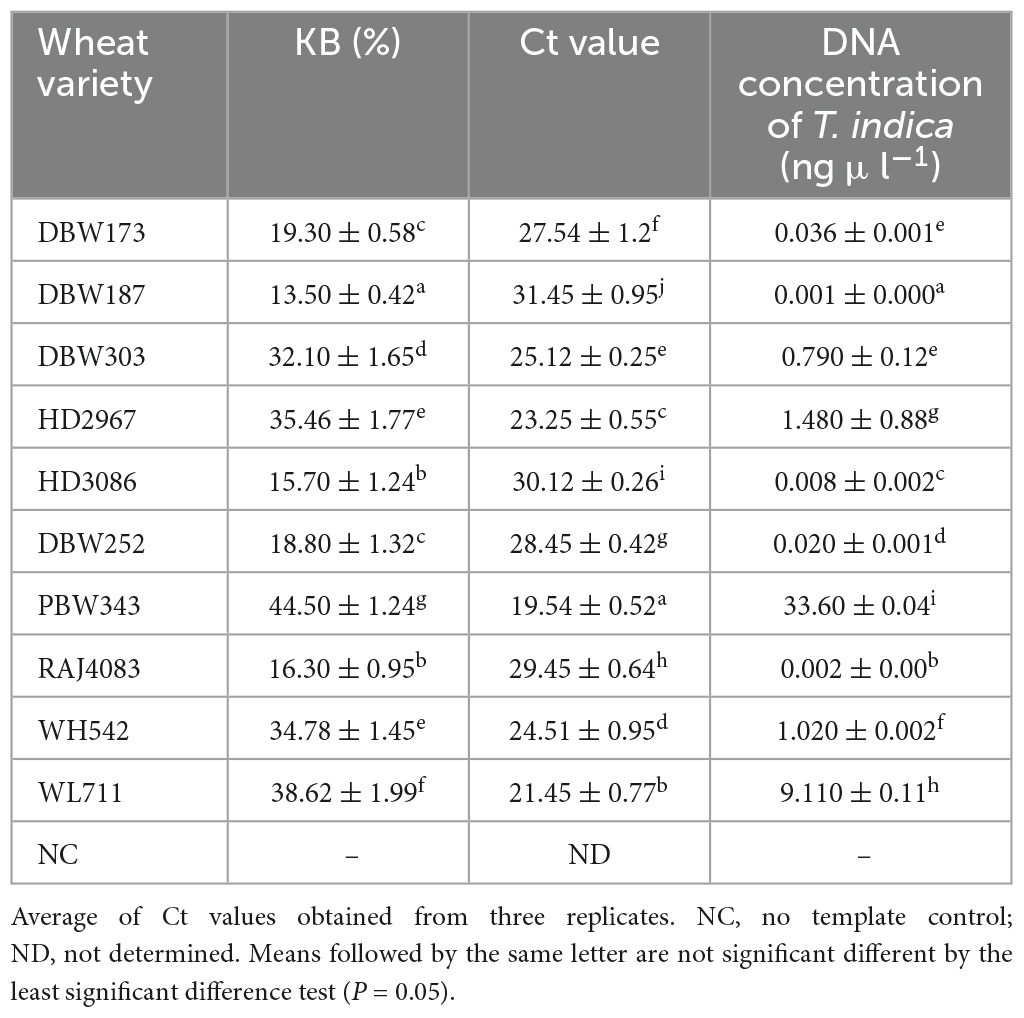
Table 4. Field evaluation of the specificity of primers to quantify T. indica load in artificially inoculated wheat varieties by qPCR assay during 2021–2022.
Discussion
Karnal bunt is a significant quarantine fungus, and its detection in wheat crops is crucial for wheat researchers and implementing regulatory procedures related to global surveillance and quarantine (IPPC, 2016; Singh et al., 2023b). Traditional methods such as microscopic analysis, isozyme analysis, and pathogenicity tests have been used for seed testing and confirming the presence of T. indica (Kashyap et al., 2011; Iquebal et al., 2021). However, these methods have limitations, including subjectivity in observation and examination. Spore morphology analysis, in particular, is mentioned as being unable to distinguish T. indica from related Tilletia species and other wheat pathogens, leading to incorrect identifications (Ferreira et al., 1996; Smith et al., 1996; Pimentel et al., 1998; Castlebury and Carris, 1999). PCR methods have been developed for diagnosing various plant pathogens, including T. indica. These methods are favored in plant disease diagnostic laboratories due to their sensitivity, specificity, ease of execution, and rapid reporting (Kumar et al., 2013; Singh et al., 2014; Kashyap et al., 2016; Sharma et al., 2017; Chakdar et al., 2019; Kaur et al., 2020). Additionally, they require only a small amount of plant tissue for testing. At present, there is no established PCR-based diagnostic assay for confirming the presence of T. indica in different growth stages of wheat under field conditions. Such assays could be valuable for rapid field screening of wheat germplasm to identify Karnal bunt disease in breeding programs. In present research, efforts have been made to identify specific genomic regions that can be used to design T. indica-specific markers for accurate and precise detection of KB disease in wheat. These markers could potentially improve the accuracy and reliability of diagnostic assays for T. indica.
Previous research reports have explored the potential of using specific genetic regions, such as the internal transcribed spacer (ITS) and mitochondrial regions, to develop species-specific genetic markers (Levy et al., 2001; Tan and Murray, 2006; Thirumalaisamy et al., 2011; Gao et al., 2016; Kuzdraliński et al., 2017; Kashyap et al., 2020b; Gupta et al., 2022). However, the current study is the first of its kind to develop novel T. indica-specific markers that are derived from the GAPDH gene of T. indica. The purpose of these markers is to accurately identify the presence of T. indica infection in different wheat growth stages under field conditions.
The selection of an appropriate target region is a critical step in molecular detection of T. indica in wheat. In this study, GAPDH gene region was chosen for analysis because it is implicated in determining virulence in T. indica, making it a promising target (Pandey et al., 2018). It is worth to mention here that GAPDH exists in nearly all organisms (Henry et al., 2015). For instance, wheat has 22 GAPDH genes dispersed throughout the genome. As a result, it was decided to conduct bioinformatic analysis of the GAPDH region from various fungi, animals and wheat available in the NCBI database. This analysis likely involved comparing and identifying unique sequences or regions specific to T. indica. Multiple alignments of animal, fungi, wheat, and T. indica GAPDH amino acid sequences revealed that T. indica GAPDH identified for the development of species-specific marker was distinct and did not match with wheat, fungi, and animal amino acid sequences studied by Zeng et al. (2016). This clearly revealed that the GAPDH region used for the development of T. indica specific markers is unique and suitable for T. indica detection in wheat. As a consequence, T. indica specific primer set was developed by targeting GAPDH region that produces a single, specific DNA band of 243 bp during PCR assay. The designed primers were shown to be effective in generating the desired amplicons from the genomic DNA of various fungal pathogens responsible for diseases like head scab, yellow rust, powdery mildew, and others in wheat. Analogous report regarding the development of GAPDH based genetic markers for the detection of Colletotrichum camelliae fungus in tea host has been reported by He et al. (2020). A qPCR assay based on GAPDH gene with LOD of 80 fg μl–1 C. kahawae DNA in Coffea arabica has been demonstrated by Tao et al. (2013).
At present, for both culture dependent and PCR based procedures, the severity of infection and LOD (i.e., detection sensitivity) of each infected plants is still unclear in case of wheat-T. indica system. Therefore, a PCR and qPCR assay were reported in current study for species specific identification of T. indica from pure cultures as well as for the confirmation of T. indica presence in wheat plants at five different growth stages [i.e., ZGS 59 (ear emergence stage), ZGS 69 (flowering stage), ZGS77 (milking stage), ZGS87 (dough stage), and ZGS92 (ripening stage)] of two different cultivars. The study confirms that the developed assay was able to diagnose T. indica infection in all the growth stages of asymptomatic wheat tissues, which otherwise seen with naked eye only after crop harvesting. In terms of sensitivity, LOD of 1 pg of T. indica DNA in DBW187 has been recorded in present research by employing new primers in qPCR assay and found more sensitive than earlier reported qPCR assays in case of T. indica fungus (Frederick et al., 2000; Gao et al., 2016; Tan et al., 2016). A series of real-time PCR assays for detecting fungal species in plants have been developed by earlier workers. For instance, a qPCR assay targeting the GAPDH gene was reported by Ciampi-Guillardi et al. (2020) which have the potential to detect 3 pg of S. sclerotiorum DNA and 300 fg of both C. truncatum and C. cassiicola DNA in soybean seeds. Besides this, LOD of 4 pg of C. cassiicola DNA in soybean seeds (Guimarães et al., 2017) and 191.31 fg μl–1 of Ilyonectria robusta DNA in ginseng roots have been obtained using SYBR Green I based qPCR assay (Jiang et al., 2023). Similarly, a SYBR Green I real-time PCR assay for detecting T. laevis with a 100 fg μl–1 detection limit has also been reported by Xu et al. (2020). However, it is worth to mention that our results are comparable with the study of Tremblay et al. (2021), who reported LOD as low as 0.1 pg T. indica DNA in wheat seeds using qPCR method. Similarly, qPCR assay developed by Frederick et al. (2000) and Tan et al. (2016) was documented to detect as low as 5 and 10 pg of fungal DNA, respectively.
It is important to mention here that the detection limit of the qPCR assay was noticed to be 4 pg of total DNA isolated from pure fungal cultures of T. indica. This is more to the sensitivity of the LAMP assay for T. controversa with the LOD of 5 pg total DNA of pure fungal cultures (Sedaghatjoo et al., 2021). Similar reports of lower sensitivity of the LAMP assay for T. indica with the LOD of 10 pg reported by Gao et al. (2016) and Tan et al. (2016). However, it is less sensitive than the reported LOD of 0.001 pg for the LAMP assay not differentiating among T. caries, T. controversa, and T. laevis (Pieczul et al., 2018). Therefore, further comparative investigation with respect to qPCR, LAMP and ddPCR to determine the detection limit and their correlation with fungal biomass per wheat spike is highly warranted. But irrespective of this, the unambiguous results obtained in the present study by using qPCR to detect T. indica biomass in different wheat varieties advocate that it might play an essential role in the rapid and effective screening of genotypes against KB disease. Nevertheless, there is no specific DNA microarray available for the detection of fungal pathogens of wheat pathogens. The assay described here is an initial study of identifying GAPDH as a potential region for the identification of T. indica. Therefore, for developing large-scale and robust systems for screening wheat materials against KB, efforts should be made to explore, develop, and validate T. indica-specific probes targeting the entire genomes of T. indica to improve testing efficiency and reduce costs.
Conclusion
The study presents a valuable molecular tool for rapid, specific and sensitive detection of KB fungus in wheat host. Identification and quantification of KB based on qPCR can be applied in the screening process for KB resistance in wheat as well as in certification and breeding processes at early stages of plant development. It could also assist the selection of potential resistance donors for breeding. However, there is room for further improvement through the exploration of combined post-harvest disease severity assessments and large-scale qPCR or GADPH based array, aiming to enhance sensitivity and optimization. Additionally, we accentuated the need for further investigation into the relationship between T. indica DNA concentrations and post-harvest disease severity across different locations and years to unlock the full potential of these new assays for high-throughput screening of wheat germplasm against KB.
Data availability statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/Supplementary material.
Author contributions
PK: Conceptualization, Funding acquisition, Investigation, Methodology, Project administration, Software, Validation, Writing – original draft, Writing – review and editing. SK: Funding acquisition, Investigation, Methodology, Project administration, Supervision, Validation, Visualization, Writing – review and editing. RK: Data curation, Investigation, Methodology, Validation, Visualization, Writing – review and editing. AS: Data curation, Formal analysis, Investigation, Methodology, Software, Validation, Visualization, Writing – review and editing. AK: Formal analysis, Investigation, Methodology, Software, Validation, Visualization, Writing – review and editing. SR: Investigation, Methodology, Validation, Writing – review and editing. PJ: Project administration, Resources, Software, Supervision, Writing – review and editing. GS: Funding acquisition, Supervision, Writing – review and editing.
Funding
The author(s) that no declare financial support was received for the research, authorship, and/or publication of this article.
Acknowledgments
We are grateful to ICAR-National Bureau of Agriculturally Important Microorganisms (NBAIM) for providing necessary financial support under AMAAS project on “Development of diagnostic tools for detection of Karnal bunt and loose smut of wheat” (Project Code 1002588).
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
The author(s) declared that they were an editorial board member of Frontiers, at the time of submission. This had no impact on the peer review process and the final decision.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2023.1291000/full#supplementary-material
Footnotes
- ^ https://www.cabidigitallibrary.org/doi/10.1079/cabicompendium.36168
- ^ https://www.primer3plus.com/
- ^ https://www.ncbi.nlm.nih.gov/tools/primer-blast/
- ^ https://www.ncbi.nlm.nih.gov/genbank/
- ^ https://www.idtdna.com
- ^ http://www.ncbi.nlm.nih.gov
References
Abdullah, A. S., Turo, C., Moffat, C. S., Lopez-Ruiz, F. J., Gibberd, M. R., Hamblin, J., et al. (2018). Real-time PCR for diagnosing and quantifying co-infection by two globally distributed fungal pathogens of wheat. Front. Plant Sci. 9:1086. doi: 10.3389/fpls.2018.01086
Alhamid, G., Tombuloglu, H., and Al-Suhaimi, E. (2023). Development of loop-mediated isothermal amplification (LAMP) assays using five primers reduces the false-positive rate in COVID-19 diagnosis. Sci. Rep. 13:5066. doi: 10.1038/s41598-023-31760-z
Aujla, S. S., Sharma, I., and Singh, V. B. (1989). Rating scale for identifying wheat varieties resistant to Neovossia indica. Short communication. Ind. Phytopathol. 42:161.
Babadoost, M., Mathre, D. E., Johnston, R. H., and Bonde, M. R. (2004). Survival of teliospores of Tilletia indica in soil. Plant Dis. 88, 56–62.
Bala, R., Kaur, J., Tak, P. S., Sandhu, S. K., and Pannu, P. P. S. (2022). A model for Tilletia indica (Karnal bunt)—Triticum aestivum (Wheat) system under changing environmental conditions. Ind. Phytopathol. 75, 723–730. doi: 10.1007/s42360-022-00520-w
Bishnoi, S. K., He, X., Phuke, R. M., Kashyap, P. L., Alakonya, A., Chhokar, V., et al. (2020). Karnal bunt: a re-emerging old foe of wheat. Front. Plant Sci. 11:569057. doi: 10.3389/fpls.2020.569057
Bonde, M. R., Peterson, G. L., Schaad, N. W., and Smilanick, J. L. (1997). Karnal bunt of wheat. Plant Dis. 81, 1370–1377. doi: 10.1094/PDIS.1997.81.12.1370
Brar, G. S., Fuentes-Dávila, G., He, X., Sansaloni, C. P., Singh, R. P., and Singh, P. K. (2018). Genetic mapping of resistance in hexaploid wheat for a quarantine disease: Karnal bunt. Front. Plant Sci. 9:1497. doi: 10.3389/fpls.2018.01497.0
Broeders, S., Huber, I., Grohmann, L., Berben, G., Taverniers, I., Mazzara, M., et al. (2014). Guidelines for validation of qualitative real-time PCR methods. Trends Food Sci. Technol. 37, 115–126. doi: 10.1016/j.tifs.2014.03.008
Bustin, S. A., Benes, V., Garson, J. A., Hellemans, J., Huggett, J., Kubista, M., et al. (2009). The MIQE guidelines: minimum information for publication of quantitative real-time PCR experiments. Clin. Chem. 55, 611–622. doi: 10.1373/clinchem.2008.112797
Carris, L., Castlebury, L., and Goates, B. (2006). Nonsystemic bunt fungi-Tilletia indica and T. horrida: A review of history, systematics, and biology. Annu. Rev. Phytopathol. 44, 113–133. doi: 10.1146/annurev.phyto.44.070505.143402
Castlebury, L. A., and Carris, L. M. (1999). Tilletia walkeri, a new species on Lolium multiflorum and L. perenne. Mycologia 91, 121–131.
Chakdar, H., Goswami, S. K., Singh, E., Choudhary, P., Yadav, J., Kashyap, P. L., et al. (2019). noxB-based marker for Alternaria spp.: a new diagnostic marker for specific and early detection in crop plants. 3 Biotech. 9, 1–9. doi: 10.1007/s13205-019-1779-4
Chen, Y., Yang, X., Yao, J., Kyaw, E. P., Zhang, A. F., Li, Y. F., et al. (2016). Simple and rapid detection of Tilletia horrida causing rice kernel smut in rice seeds. Sci Rep. 6:33258. doi: 10.1038/srep33258
Choudhary, P., Rai, P., Yadav, J., Verma, S., Chakdar, H., Goswami, S. K., et al. (2020). A rapid colorimetric LAMP assay for detection of Rhizoctonia solani AG-1 IA causing sheath blight of rice. Sci Rep. 10:22022. doi: 10.1038/s41598-020-79117-0
Ciampi-Guillardi, M., Ramiro, J., Moraes, M. H. D., Barbieri, M. C. G., and Massola, N. S. Jr. (2020). Multiplex qPCR assay for direct detection and quantification of Colletotrichum truncatum, Corynespora cassiicola, and Sclerotinia sclerotiorum in soybean seeds. Plant Dis. 104, 3002–3009. doi: 10.1094/PDIS-02-20-0231-RE
Duveiller, E., and Mezzalama, M. (2009). Karnal bunt screening for resistance and distributing KB free seed. Available online at: https://repository.cimmyt.org/xmlui/bitstream/handle/10883/1289/96166.pdf
Eibel, P., Wolf, G. A., and Koch, E. (2005). Detection of Tilletia caries, causal agent of common bunt of wheat, by ELISA and PCR. J. Phytopathol. 153, 297–306. doi: 10.1111/j.1439-0434.2005.00973.x
Emebiri, L., Hildebrand, S., Tan, M., Juliana, P., Singh, P. K., Fuentes-Davila, G., et al. (2021). Pre-emptive Breeding against Karnal Bunt Infection in Common Wheat: Combining Genomic and Agronomic Information to Identify Suitable Parents. Front. Plant Sci. 12:675859. doi: 10.3389/fpls.2021.675859
Emebiri, L., Singh, P. K., Tan, M. K., Fuentes-Davila, G., He, X., and Singh, R. P. (2019). Reaction of Australian durum, common wheat and triticale genotypes to Karnal bunt (Tilletia indica) infection under artificial inoculation in the field. Crop Past. Sci. 70, 107–112.
Fang, Y., and Ramasamy, R. P. (2015). Current and prospective methods for plant disease detection. Biosensors 5, 537–561.
Ferreira, M. A., Tooley, P. W., Hatziloukas, E., Castro, C., and Schaad, N. W. (1996). Isolation of a species-specific mitochondrial DNA sequence for identification of Tilletia indica, the Karnal bunt of wheat fungus. Appl. Environ. Microbiol. 62, 87–93. doi: 10.1128/aem.62.1.87-93.1996
Frederick, R., Snyder, K., Tooley, P., Berthier-Schaad, Y., Peterson, G., Bonde, M., et al. (2000). Identification and differentiation of Tilletia indica and T. walkeri using the polymerase chain reaction. Phytopathology 90, 951–960. doi: 10.1094/PHYTO.2000.90.9.951
Fuentes-Davila, G., and Duran, R. (1986). Tilletia indica: Cytology and teliospore formation in vitro and in immature kernels. Canad. J. Bot. 8, 1712–1719.
Fuentes-Davila, G. (1996). “Karnal bunt,” in Bunt and smut diseases of wheat: Concepts and methods of disease management, eds R. D. Wilcoxson and E. E. Saari (Mexico: CIMMYT), 26–32.
Fuentes-Dávila, G., Rosas-Jáuregui, I. A., Ayón-Ibarra, C. A., Álvarez-Amado, K. D., Félix-Valencia, P., and Félix-Fuentes, J. L. (2018). Biological effectiveness of Opus, Folicur, Juwel, and Bemistop for control of Karnal bunt (Tilletia indica) of wheat in the field. Annu. Wheat Newsl. 64, 30–33.
Gao, L., Chen, W., and Liu, T. (2011). An ISSR-based approach for the molecular detection and diagnosis of dwarf bunt of wheat, caused by Tilletia controversa Kühn. J. Phytopathol. 159, 155–158.
Gao, L., Chen, W. Q., and Liu, T. G. (2010). Development of a SCAR marker by inter-simple sequence repeat for diagnosis of dwarf bunt of wheat and detection of Tilletia controversa Kühn. Folia Microbiol. 55, 258–264.
Gao, Y., Tan, M. K., and Zhu, Y. G. (2016). Rapid and specific detection of Tilletia indica using loop-mediated isothermal DNA amplification. Australas. Plant Pathol. 45, 361–367. doi: 10.1007/s13313-016-0422-7
Goates, B. J., and Jackson, E. W. (2006). Susceptibility of wheat to Tilletia indica during stages of spike development. Phytopathology 96, 962–966. doi: 10.1094/PHYTO-96-0962
Guimarães, M. R. F., Siqueira, C. S., Machado, J. C., França, S. K. S., and Guimarães, G. C. (2017). Evaluation of inoculum potential of pathogens in seeds: Relation to physiological quality and DNA quantification by qPCR. J. Seed Sci. 39, 224–233. doi: 10.1590/2317-1545v39n3163901
Gupta, S., Aggarwal, R., Sharma, S., Gurjar, M. S., Bashyal, B. M., Saharan, M. S., et al. (2022). Multiple sequence alignment and phylogenetic analysis of wheat pathogens using conserved genes for identification and development of diagnostic markers. Cereal Res. Comm. 2022, 463–472. doi: 10.1007/s42976-021-00193-7
Gurjar, M. S., Aggarwal, R., Sharma, S., Kulshreshtha, D., Gupta, A., Gogoi, R., et al. (2017). Development of real time PCR assay for the detection and quantification of teliospores of Tilletia indica causing wheat Karnal bunt in soil. Ind. J. Exp. Biol. 55, 549–554.
Gurjar, M. S., Mohan, M. H., Singh, J., Saharan, M. S., and Aggarwal, R. (2021). Tilletia indica: biology, variability, detection, genomics and future perspective. Ind. Phytopathol. 74, 21–31. doi: 10.1007/s42360-021-00319-1
Harper, S. J., Ward, L. I., and Clover, G. R. G. (2010). Development of LAMP and real-time PCR methods for the rapid detection of Xylella fastidiosa for quarantine and field applications. Phytopathology 100, 1282–1288. doi: 10.1094/PHYTO-06-10-0168
He, S., Chen, H., Wei, Y., An, T., and Liu, S. (2020). Development of a DNA-based real-time PCR assay for the quantification of Colletotrichum camelliae growth in tea (Camellia sinensis). Plant Methods 16, 1–11. doi: 10.1186/s13007-020-00564-x
Henry, E., Fung, N., Liu, J., Drakakaki, G., and Coaker, G. (2015). Beyond Glycolysis: GAPDHs are multi-functional enzymes involved in regulation of ROS, autophagy, and plant immune responses. PLoS Genet 11:e1005199. doi: 10.1371/journal.pgen.1005199
IPPC (2016). International Standard for Phytosanitary Measures 27. In Annex 04 DP 04: Tilletia indica Mitra. Rome: International Plant Protection Convention (IPPC).
Iquebal, M. A., Mishra, P., Maurya, R., Jaiswal, S., Rai, A., and Kumar, D. (2021). Centenary of soil and air borne wheat Karnal bunt disease research: a review. Biology 10:1152. doi: 10.3390/biology10111152
Jiang, Y., Jin, H., Li, X., Yan, D., Zhang, Y., Li, M., et al. (2023). Development of a novel real-time quantitative PCR method for detection of Ilyonectria robusta, the predominant species causing ginseng rusty root rot. Plant Dis. 107, 1680–1689. doi: 10.1094/PDIS-06-22-1471-RE
Jones, G. M., Busby, E., Garson, J. A., Grant, P. R., Nastouli, E., Devonshire, A. S., et al. (2016). Digital PCR dynamic range is approaching that of real-time quantitative PCR. Biomol. Detect. Quantif. 10, 31–33. doi: 10.1016/j.bdq.2016.10.001
Kashyap, P. L., Kaur, S., and Pannu, P. P. S. (2018). Induction of systemic tolerance to Tilletia indica in wheat by plant defence activators. Arch. Phytopathol. Plant Prot. 51, 1–13. doi: 10.1080/03235408.2018.1438778
Kashyap, P. L., Kaur, S., and Pannu, P. P. S. (2019). Expression analysis of pathogenesis related proteins induced by compatible and incompatible interactions of Tilletia indica in wheat plants. J. Mycol. Pl. Pathol. 49, 39–47.
Kashyap, P. L., Kaur, S., Sanghera, G. S., Kang, S. S., and Pannu, P. P. S. (2011). Novel methods for quarantine detection of Karnal bunt (Tilletia indica) of wheat. Elixir Agri. 31, 1873–1876.
Kashyap, P. L., Kumar, S., Jasrotia, P., Singh, D. P., and Singh, G. P. (2020a). “Nanotechnology in Wheat Production and Protection,” in Environmental Nanotechnology Volume 4. Environmental Chemistry for a Sustainable World, eds N. Dasgupta, S. Ranjan, and E. Lichtfouse (Cham: Springer), doi: 10.1007/978-3-030-26668-4_5
Kashyap, P. L., Kumar, S., Kumar, R. S., Sharma, A., Jasrotia, P., Singh, D. P., et al. (2020b). Molecular diagnostic assay for rapid detection of flag smut fungus (Urocystis agropyri) in wheat plants and field soil. Front. Plant Sci. 11:1039. doi: 10.3389/fpls.2020.01039
Kashyap, P. L., Kumar, S., Kaul, N., Aggarwal, S. K., Jasrotia, P., Bhardwaj, A. K., et al. (2022a). “Nanotechnology for Wheat and Barley Health Management: Current Scenario and Future Prospectus,” in New Horizons in Wheat and Barley Research: Crop Protection and Resource Management, ed. P. Kashyap (Singapore: Springer Nature Singapore), doi: 10.1007/978-981-16-4134-3_12
Kashyap, P. L., Kumar, S., Sharma, A., Kumar, R. S., Mahapatra, S., Kaul, N., et al. (2022b). Molecular diversity, haplotypes distribution and genetic variation flow of Bipolaris sorokiniana fungus causing spot blotch disease in different wheat growing zones. J. Appl. Genet. 63, 793–803. doi: 10.1007/s13353-022-00716-w
Kashyap, P. L., Kumar, S., Kumar, R. S., Sharma, A., Khanna, A., and Kajal, et al. (2023). Comparative analysis of nine Tilletia indica genomes for the development of novel microsatellite markers for genetic diversity and population structure analysis. Front. Microbiol. 14:1227750. doi: 10.3389/fmicb.2023.1227750
Kashyap, P. L., Rai, P., Kumar, S., Chakdar, H., and Srivastava, A. K. (2017). “DNA Barcoding for Diagnosis and Monitoring of Fungal Plant Pathogens,” in Molecular Markers in Mycology. Fungal Biology, eds B. P. Singh and V. K. Gupta (Cham: Springer), doi: 10.1007/978-3-319-34106-4_5
Kashyap, P. L., Rai, P., Sharma, S., Chakdar, H., Kumar, S., Pandiyan, K., et al. (2016). Nanotechnology for the detection and diagnosis of plant pathogens. Nanosci. Food Agric. 2, 253–276. doi: 10.1007/978-3-319-39306-3_8
Kaul, N., Kashyap, P. L., Kumar, S., Singh, D., and Singh, G. P. (2022). Genetic diversity and population structure of head blight disease causing Fusarium graminearum in Northern Wheat belt of India. J. Fungi 8, 820. doi: 10.3390/jof8080820
Kaur, G., and Kaur, S. (2005). Simulating natural infection of wheat ovaries with Tilletia indica and interaction with date of sowing and varieties. Crop Improv. 32, 26–34.
Kaur, S. I., Kashyap, P. L., Kang, S. S., and Sharma, A. (2020). “Detection and Diagnosis of Seed-Borne Viruses and Virus-Like Pathogens,” in Seed-Borne Diseases of Agricultural Crops: Detection, Diagnosis & Management, eds R. Kumar and A. Gupta (Singapore: Springer), 169–199. doi: 10.1007/978-981-32-9046-4_7
Kayim, M., Nawaz, H., and Alsalmo, A. (2022). Fungal Diseases of Wheat. London: IntechOpen, doi: 10.5772/intechopen.102661
Khan, M., Wang, R., Li, B., Liu, P., Weng, Q., and Chen, Q. (2018). Comparative Evaluation of the LAMP Assay and PCR-Based Assays for the Rapid Detection of Alternaria solani. Front. Microbiol. 9:2089. doi: 10.3389/fmicb.2018.02089
Kojabad, A. A., Farzanehpour, M., Galeh, H. E. G., Dorostkar, R., Jafarpour, A., Bolandian, M., et al. (2021). Droplet digital PCR of viral DNA/RNA, current progress, challenges, and future perspectives. J. Med. Virol. 93, 4182–4197. doi: 10.1002/jmv.26846
Kumar, S., Singh, R., Kashyap, P. L., and Srivastava, A. K. (2013). Rapid detection and quantification of Alternaria solani in tomato. Sci. Hortic. 151, 184–189.
Kumar, S., Singroha, G., Singh, G. P., and Sharma, P. (2021). Karnal bunt of wheat: Etiology, breeding and integrated Management. Crop Protect. 139:105376.
Kutilek, V., Lee, R., and Kitto, G. B. (2001). Development of immunochemical techniques for detecting karnal bunt in wheat. Food Agric. Immunol. 13, 103–114. doi: 10.1080/09540100120055675
Kuzdraliński, A., Kot, A., Szczerba, H., Nowak, M., and Muszyńska, M. (2017). A Review of Conventional PCR Assays for the Detection of Selected Phytopathogens of Wheat. J. Mol. Microbiol. Biotechnol. 27, 175–189. doi: 10.1159/000477544
Levy, L., Castlebury, L., Carris, L., Meyer, R., and Pimentel, G. (2001). Internal transcribed spacer sequence-based phylogeny and polymerase chain reaction-restriction fragment length polymorphism differentiation of Tilletia walkeri and T. indica. Phytopathology 91, 935–940. doi: 10.1094/PHYTO.2001.91.10.935
Liu, J. H., Gao, L., Liu, T. G., and Chen, W. Q. (2009). Development of a sequence-characterized amplified region marker for diagnosis of dwarf bunt of wheat and detection of Tilletia controversa Kühn. Lett. Appl. Microbiol. 2009, 235–240. doi: 10.1111/j.1472-765X.2009.02645.x
Luster, D. G., Bonde, M. R., Peterson, G. L., Hack, M. A., and Schaad, N. W. (1998). Antigenic glycoproteins in the teliospore wall of Tilletia indica. Mycologia 90, 180–188.
Naveen, K. P., and Bhat, A. I. (2020). Development of reverse transcription loop-mediated isothermal amplification (RT-LAMP) and reverse transcription recombinase polymerase amplification (RT-RPA) assays for the detection of two novel viruses infecting ginger. J. Virol. Methods 282, 113884. doi: 10.1016/j.jviromet.2020.113884
Nixon, G., Garson, J. A., Grant, P., Nastouli, E., Foy, C. A., and Huggett, J. F. (2014). Comparative study of sensitivity, linearity, and resistance to inhibition of digital and Nondigital polymerase chain reaction and loop mediated isothermal amplification assays for quantification of human cytomegalovirus. Anal. Chem. 86, 4387–4394. doi: 10.1021/ac500208w
Ou, S. C., Giambrone, J. J., and Macklin, K. S. (2012). Comparison of a TaqMan real-time polymerase chain reaction assay with a loop-mediated isothermal amplification assay for detection of Gallid herpesvirus 1. J. Vet. Diagn. Investig. 24, 138–141. doi: 10.1177/1040638711427578
Pandey, V., Singh, M., Pandey, D., and Kumar, A. (2018). Integrated proteomics, genomics, metabolomics approaches reveal oxalic acid as pathogenicity factor in Tilletia indica inciting Karnal bunt disease of wheat. Sci. Rep. 8:7826. doi: 10.1038/s41598-018-26257-z
Pascoe, I., Priest, M., Shivas, R., and Cunnington, J. (2005). Ustilospores of Tilletia ehrhartae, a smut of Ehrharta calycina, are common contaminants of Australian wheat grain, and a potential source of confusion with Tilletia indica, the cause of Karnal bunt of wheat. Plant Pathol. 54, 161–168. doi: 10.1111/j.1365-3059.2005.01145.x
Pieczul, K., Perek, A., and Kubiak, K. (2018). Detection of Tilletia caries. J. Microbiol. Methods 154, 141–146. doi: 10.1016/j.mimet.2018.10.018
Pimentel, G., Carris, L., Levy, L., and Meyer, R. (1998). Genetic variability among isolates of Tilletia barclayana, T. indica and allied species. Mycologia 90, 1017–1027.
Ren, Y., Wang, Y., Wang, L. Y., Lv, X. L., Liu, T., Nie, J. Y., et al. (2022a). Development of diagnostic molecular markers to differentiate sharp eyespot and Fusarium crown rot isolates collected from common wheat fields in China. Phytoparasitica 50, 1097–1105. doi: 10.1007/s12600-022-01027-w
Ren, Z., Chen, R., Muhae-Ud-Din, G., Fang, M., Li, T., Yang, Y., et al. (2022b). Development of real-time PCR and droplet digital PCR based marker for the detection of Tilletia caries inciting common bunt of wheat. Front. Plant Sci. 13:4467. doi: 10.3389/fpls.2022.1031611
Riccioni, L. U. C. A., Inman, A. L. A. N., Magnus, H. A., Valvassori, M. A. R. C. O., Porta-Puglia, A., Conca, G., et al. (2008). Susceptibility of European bread and durum wheat cultivars to Tilletia indica. Plant Pathol. 57, 612–622.
Schena, L., Hughes, K. J., and Cooke, D. E. (2006). Detection and quantification of Phytophthora ramorum, P. kernoviae, P. citricola and P. quercina in symptomatic leaves by multiplex real-time PCR. Mol. Plant Pathol. 7, 365–379. doi: 10.1111/j.1364-3703.2006.00345.x
Sedaghatjoo, S., Forster, M. K., Niessen, L., Karlovsky, P., Killermann, B., and Maier, W. (2021). Development of a loop-mediated isothermal amplification assay for the detection of Tilletia controversa based on genome comparison. Sci. Rep. 11:11611. doi: 10.1038/s41598-021-91098-2
Sharma, A. K., Kumar, J., and Nagarajan, S. S. (1998). “Worldwide movement of smuts and bunts,” in Bunts and Smuts of Wheat: An International Symposium, eds V. S. Malik and D. E. Mathre (Ottawa, ON: North American Plant Protection Organization (NAPPO)).
Sharma, P., Chauhan, R., Pande, V., Basu, T., and Kumar, A. (2022). Rapid sensing of Tilletia indica-Teliospore in wheat extract by a piezoelectric label free immunosensor. Bioelectrochemistry 147:108175. doi: 10.1016/j.bioelechem.2022.108175
Sharma, S., Rai, P., Rai, S., Srivastava, M., Kashyap, P. L., Sharma, A., et al. (2017). “Genomic revolution in crop disease diagnosis: a review,” in Plants and microbes in an ever changing environment, ed. S. Singh (Hauppauge, NY: Nova Science Publishers), pp 257–293.
Singh, D. V., and Gogoi, R. (2011). Karnal bunt of wheat (Triticum spp.)—A global scenario. Indian J. Agric. Sci. 81, 3–14.
Singh, J., Chhabra, B., Raza, A., Yang, S. H., and Sandhu, K. S. (2023b). Important wheat diseases in the US and their management in the 21st century. Front. Plant Sci. 13:1010191. doi: 10.3389/fpls.2022.1010191
Singh, S. K., Kumar, S., Kashyap, P. L., Sendhil, R., and Gupta, O. P. (2023a). “Wheat,” in Trajectory of 75 years of Indian agriculture after independence, eds P. K. Ghosh, A. Das, R. Saxena, K. Banerjee, G. Kar, and D. Vijay (Singapore: Springer), 137–162. doi: 10.1007/978-981-19-7997-2_7
Singh, R., Kumar, S., Kashyap, P. L., Srivastava, A. K., Mishra, S., and Sharma, A. K. (2014). Identification and Characterization of Microsatellite from Alternaria brassicicola to Assess Cross-Species Transferability and Utility as a Diagnostic Marker. Mol. Biotechnol. 56, 1049–1059. doi: 10.1007/s12033-014-9784-7
Smilanick, J. L., Prescott, J. M., Hoffmann, J. A., Secrest, L. R., and Weise, K. (1989). Environmental effects on survival and growth of secondary sporidia and teliospores of Tilletia indica. Crop Prot. 8, 86–90.
Smith, O. P., Peterson, G. L., Beck, R. J., Schaad, N. W., and Bonde, M. R. (1996). Development of a PCR-based method for identification of Tilletia indica, causal agent of Karnal bunt of wheat. Phytopathology 86, 115–122.
Tamura, K., Stecher, G., and Kumar, S. (2021). MEGA11: Molecular Evolutionary Genetics Analysis Version 11. Mol. Biol. Evol. 38, 3022–3027. doi: 10.1093/molbev/msab120
Tan, M., Brennan, J. P., Wright, D., and Murray, G. M. (2010). An enhanced protocol for the quarantine detection of Tilletia indica and economic comparison with the current standard. Australasian Plant Pathol. 39, 334–342. doi: 10.1071/AP10003
Tan, M. K., and Murray, G. M. (2006). A molecular protocol using quenched FRET probes for the quarantine surveillance of Tilletia indica, the causal agent of Karnal bunt of wheat. Mycol. Res. 110, 203–210. doi: 10.1016/j.mycres.2005.08.006
Tan, M. K., Raman, H., Chambers, G., Sharma, I., Chen, Z., Deshpande, N., et al. (2016). Characterization of SNP and structural variations in the mitochondrial genomes of Tilletia indica and its closely related species formed basis for a simple diagnostic assay. PLoS One 11:e0166086. doi: 10.1371/journal.pone.0166086
Tao, G., Hyde, K. D., and Cai, L. (2013). Species-specific real-time PCR detection of Colletotrichum kahawae. J. Appl. Microbiol. 114, 828–835. doi: 10.1111/jam.12068
Thapa, S., Bala, R., Kumar Sharma, V., Srivastava, P., Kaur, J., Bromand, F., et al. (2022). Basis of Karnal bunt resistance in diploid and tetraploid Triticeae species. Indian Phytopathol. 75, 251–257.
Thirumalaisamy, P., Singh, D., Aggarwal, R., Gogoi, R., Kumar Gupta, P., and Kumar Singh, P. (2011). Development of specific primers for detection of Karnal bunt pathogen of wheat. Ind. Phytopathol. 64, 164–172.
Thompson, J. D., Gibson, T. J., Plewniak, F., Jeanmougin, F., and Higgins, D. G. (1997). The CLUSTAL_X windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res. 25, 4876–4882. doi: 10.1093/nar/25.24.4876
Tremblay, ÉD., Carey, J., Bilodeau, G. J., and Hambleton, S. (2021). Four in silico designed and validated qPCR assays to detect and discriminate Tilletia indica and T. walkeri, individually or as a complex. Biology 10:1295. doi: 10.3390/biology10121295
Valente, M., Bragaloni, M., Di Giambattista, G., and Riccioni, L. (2019). Test validation for the detection of Tilletia indica Mitra by multiplex real-time PCR. EPPO Bull. 49, 104–110. doi: 10.1111/epp.12556
Valente, M. T., Orzali, L., Manetti, G., Magnanimi, F., Matere, A., Bergamaschi, V., et al. (2023). Rapid molecular assay for the evaluation of clove essential oil antifungal activity against wheat common bunt. Front. Plant Sci. 14:1130793. doi: 10.3389/fpls.2023.1130793
Vesna, U., Dragana, I. M., Ana, N., and Slavica, S. I. (2011). Identification of Tilletia species using rep-PCR fingerprinting technique. Genetika 43, 183–195.
Xu, T., Yao, Z., Liu, J., Zhang, H., Din, G. M. U., Zhao, S., et al. (2020). Development of droplet digital PCR for the detection of Tilletia laevis, which causes common bunt of wheat, based on the SCAR marker derived from ISSR and real-time PCR. Sci. Rep. 10:16106. doi: 10.1038/s41598-020-72976-7
Xu, T., Yao, Z., Liu, J., Zhang, H., Din, G. M. U., Zhao, S., et al. (2022). Development of droplet digital PCR for the detection of Tilletia laevis, which causes common bunt of wheat, based on the SCAR marker derived from ISSR and real-time PCR. Sci. Rep. 10:16106.
Yang, B. C., Wang, F. X., Zhang, S. Q., Song, N., Li, J. X., Yang, Z. Q., et al. (2013). Comparative evaluation of conventional polymerase chain reaction (PCR), with loop-mediated isothermal amplification and SYBR green I-based real-time PCR for the quantitation of porcine circovirus-1 DNA in contaminated samples destined for vaccine production. J. Virol. Methods 191, 1–8. doi: 10.1016/j.jviromet.2013.03.014
Yao, Z., Qin, D., Chen, D., Liu, C., Chen, W., Liu, T., et al. (2019). Development of ISSR-derived SCAR marker and SYBR Green I Real-time PCR method for detection of teliospores of Tilletia laevis Kühn. Sci. Rep. 9:17651. doi: 10.1038/s41598-019-54163-5
Yuan, Q., Nian, S., Yin, Y., Li, M., Cai, J., and Wang, Z. (2009). Development of a PCR-based diagnostic tool specific to wheat dwarf bunt, caused by Tilletia controversa. Eur J Plant Pathol. 124, 585–594. doi: 10.1007/s10658-009-9445-z
Zeng, L., Deng, R., Guo, Z., Yang, S., and Deng, X. (2016). Genome-wide identification and characterization of Glyceraldehyde-3-phosphate dehydrogenase genes family in wheat (Triticum aestivum). BMC Genomics 17:240. doi: 10.1186/s12864-016-2527-3
Keywords: Diagnostics, GAPDH, Karnal bunt, real time PCR, Tilletia, Wheat
Citation: Kashyap PL, Kumar S, Kumar RS, Sharma A, Khanna A, Raj S, Jasrotia P and Singh G (2023) Molecular diagnostic assay for pre-harvest detection of Tilletia indica infection in wheat plants. Front. Microbiol. 14:1291000. doi: 10.3389/fmicb.2023.1291000
Received: 08 September 2023; Accepted: 13 October 2023;
Published: 01 November 2023.
Edited by:
Ravinder Kumar, Central Potato Research Institute (ICAR), IndiaReviewed by:
Nagaraju Yalavarthi, National Bureau of Agriculturally Important Microorganisms (ICAR), IndiaSumit Jangra, University of Florida, United States
Copyright © 2023 Kashyap, Kumar, Kumar, Sharma, Khanna, Raj, Jasrotia and Singh. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Prem Lal Kashyap, cHJlbS5rYXNoeWFwQGljYXIuZ292Lmlu, cGxrYXNoeWFwQGdtYWlsLmNvbQ==; Sudheer Kumar, c3VkaGVlci5rdW1hckBpY2FyLmdvdi5pbg==, c3VkaGVlci5pY2FyQGdtYWlsLmNvbQ==; Annie Khanna, YW5uaWVraGFubmFhbm5pZUBnbWFpbC5jb20=
 Prem Lal Kashyap
Prem Lal Kashyap Sudheer Kumar
Sudheer Kumar Ravi Shekhar Kumar
Ravi Shekhar Kumar Anju Sharma
Anju Sharma Annie Khanna
Annie Khanna Shubham Raj
Shubham Raj Poonam Jasrotia
Poonam Jasrotia Gyanendra Singh
Gyanendra Singh