- Key Laboratory of Biocatalysis and Enzyme Engineering, School of Life Sciences, Hubei University, Wuhan, China
The symbiotic bacterial microbiota of insects has been shown to play essential roles in processes related to physiology, metabolism, and innate immunity. In this study, the symbiotic microbiome of Plagiodera versicolora at different developmental stages was analyzed using 16S rRNA high-throughput sequencing. The result showed that symbiotic bacteria community in P. versicolora was primarily made up of Actinobacteriota, Proteobacteria, Firmicutes, Bacteroidota, and Dependentiae. The bacterial composition among different age individuals were highly diverse, while 65 core genera were distributed in all samples which recommend core bacterial microbiome. The 8 species core bacteria were isolated from all samples, and all of them were classified as Pseudomonas sp. Among them, five species have been proven to promote the vegetable growth of Beauveria bassiana. Moreover, the virulence of B. bassiana against nonaxenic larvae exceeded B. bassiana against axenic larvae, and the introduction of the Pseudomonas sp. to axenic larvae augmented the virulence of fungi. Taken together, our study demonstrates that the symbiotic bacteria of P. versicolora are highly dissimilar, and Pseudomonas sp. core bacteria can promote host infection by entomopathogenic fungus. This result emphasizes the potential for harnessing these findings in the development of effective pest management strategies.
1. Introduction
P. versicolora is a leaf-eating pest that is a member of the Chrysomelidae family and widely distributed in the world, including Asia, Europe, and North America (Yoneya et al., 2014; Liu et al., 2023). Both the adults and larvae of this pest are known for their preference for feeding on the young leaves of poplar and willow plants, which causes huge economic losses for the forestry industry (Ling et al., 2021; Liu et al., 2021). Using chemical insecticides like phoxim and malathion can effectively lower the population density, but their persistent use inevitably results in undesirable and harmful consequences for the environment, plants, and human health (Wade and Breden, 1986). As a result, the entomopathogenic fungi that have low environmental impact are gradually developing into an effective strategy for P. versicolora control (Hou et al., 2023; Tu et al., 2023).
The insect-pathogenic fungus B. bassiana (Hypocreales: Cordycipitaceae) has the broadest host spectrum among all insect pathogens and serves as a main source of fungal pesticides to combat against wide-spectrum arthropod pests (Xiao et al., 2012; Vivekanandhan et al., 2018, 2020; Ding et al., 2023a). The host arthropod’s pathogenesis and mycosis by B. bassiana begins with the adherence of conidia on the epicuticle of the host (Baek et al., 2022; Chavez et al., 2023). Then, infective cells penetrate through the cuticle, proliferate in hemocoel, secrete toxins, and cause the hosts to die (Joop and Vilcinskas, 2016; Perumal et al., 2023a). The fungus undergoes a transformation from pathogenic proliferation to growth on the host cadavers and reproduces conidia to infect new insect hosts (Ding et al., 2023b; Perumal et al., 2023b). During the entire infection cycle, the virulence and growth of B. bassiana can be influenced by symbiotic microbiota of the host (Zhang et al., 2013; Xu et al., 2019; Bai et al., 2023). An example is gut associated bacteria Bacillus and Pseudomonas which, symbiotic with Blattella Germanics, significantly inhibit the germination and growth of B. bassiana (Zhang et al., 2013).
The insect harbors a variety of bacterial symbionts like other animals (Sittipo et al., 2018; Xiong, 2022). The composition of microbiota in insects is highly variable, which is promoted by different drivers including diet, environment, physiology, and development stage (Malacrinò, 2021; Zhao et al., 2022). But not all of them are affected by external or internal factors. Some bacteria persist in the insect host regardless of the environmental or physiological changes, these bacteria constitute the core microbiome (Zhao et al., 2022). The core microbiomes have a close evolutionary relationship with insects that has been retained, enriched, and inherited through natural selection, and plays an important role in the degradation of host defense substances and defense against pathogenic microorganisms (Douglas, 2015; Singh et al., 2021).
In this study, the symbiotic microbiome of different ages P. versicolora was identified through 16 s rRNA high-throughput sequencing and the core microbiome was determined. The phenomenon that symbiotic microbiota affected the fungi virulence was analyzed by removing and reintroducing the core microbiome. Moreover, Pseudomonas sp. of the core microbiome promoted fungal growth and virulence. Overall, this research was described as the intricate relationships between the insects, symbiotic microorganisms, and pathogens, and how these interactions can be harnessed for pest management strategies.
2. Materials and methods
2.1. Strains culture and insect rearing
B.bassiana (Bb476) were obtained from the Institute of Ecology, Shandong Academy of Sciences, Jinan, China, and were maintained on Potato dextrose agar (PDA: 0.6% potato starch, 2% dextrose, and 2% agar) at 25°C. Bacteria isolated from P. versicolora was cultured in Luria-Bertani medium (LB:1% peptone, 0.5% yeast extra, 1% NaCl and 1.5% agar). P. versicolora were collected from Shahu Park in Wuhan, Hubei Province (30.35°N, 114.33°E) following its appearance characteristics, and they were fed the fresh willow leaves and maintained in a room under controlled conditions (temperature 25 ± 2°C, relative humidity 60 ± 5% and 14 h of light) (Wade, 1994). Surface-sterilized eggs were fed on sterile poplar leaves (washed with 75% ethanol for 8 min) to obtain axenic larvae, and axenic larvae were verified to be aseptic through the bacteria isolation experiments (Liu et al., 2023).
2.2. 16S rRNA sequencing
The eggs (E), pupae (P), first instar larva (L1), second instar larva (L1), third instar larva (L1), female adults (BF), and male adults (BM) of P. versicolora were collected from Shahu Park in Wuhan, Hubei Province (30.35°N, 114.33°E). The high-throughput sequencing through a 16S rRNA gene fragment were used to detected symbiotic bacteria of collected samples (Bai et al., 2023). Female adults (BF), and male adults (BM) were divided into the body and gut for testing. Each sample included eight parallel replicates, and the pupae, first instar larva, second instar larva, third instar larva, female adults, and male adults were used at approximately 5 larvae per replicate, and the pupae were used at approximately 15 pupae per replicate.
TIANamp DNA kit (TIANGEN, China) was used to extract total DNA from all samples according to the instructions. PCR amplification of the V3-V4 region of the 16S rRNA gene was performed using primer pairs 338F (5’-ACTCCTACGGGAGGCAGCA-3′) and 806R (5’-GGACTACHVGGGTWTCTAAT-3′). All samples were performed according to formal experimental conditions. PCR products of the same sample were mixed and tested by 2% agarose gel electrophoresis. The PCR products were recovered by cutting glue using an AxyPrepDNA gel recovery kit (AXYGEN). Finally, the PCR products were sequenced at Illumina NovaSeq 6,000 for high throughput sequencing. All raw sequencing reads in this study have been uploaded to the NCBI Sequence Read Archive (SRA) database with accession numbers from SRR25927065 to SRR25927134 (Bio Project: PRJNA1013480). After sequencing, Fastp (v0.19.6) was used to control the quality of the raw sequences, Flash (v1.2.7) was used to splice the double-ended sequences, and DADA2 was used to diagnose the valid tags, to obtain the initial ASV (amplicon sequence variant) for species annotation. Microbial diversity and community composition were analyzed using the vegan package in R (version 3.3.1).
Rarefaction curves constructed using Mothur (version 1.30) based on α diversity indices were used to illustrate whether the amount of sequencing data for the samples was reasonable. The α diversity index of the community (includes Chao, Shannon, Simpson, and Ace) can reflect the sequencing depth, richness, and evenness of the community. The Kruskal-Wallis test was used to compare the α diversity index of different samples. β diversity was evaluated by using dimensionality reduction analysis based on the weighted unifrac distance algorithm. In this study, principal coordinate analysis (PCoA) and non-metric multidimensional scaling (NMDS) were used to compare β diversity among different groups based on Adonis. Hierarchical clustering analysis was performed based on the beta diversity distance matrix, and the UPGMA algorithm was used to construct a tree structure to visualize the different degrees of community distribution in different environmental samples. R (version 3.3.1) and Python (version 2.7) were used to draw heat maps of all samples. The heatmap represents the size of data in tables by color gradients and presents information about community species composition.
2.3. Infection bioassays
To examine fungal virulence, the axenic and nonaxenic larvae of P. versicolora were used as the bioassay hosts, and each assay included three parallel replicates, using approximately 35 larvae per replicate (Liu et al., 2023). Fungal strains were cultured on PDA plates for 10 d at 25°C, and the resultant conidia were used as infectious inocula. The mycelia and conidia were harvested from the plate and suspended in 0.02% Tween 80 solution, followed by violent votexing. The resultant mixture was filtered through the cotton column, and the filtrated conidia was used to infect the hosts. For infection bioassays, the insects were immersed in conidial suspension (1 × 108 cells/mL) for 30 s and the daily mortality was recorded, and then the log-rank test was used to determine the statistical differences between the paired curves.
2.4. Measurement of fungi vegetative growth and virulence
The core bacteria isolated from P. versicolora were cultured in 50 mL liquid Luria-Bertani medium (1% peptone, 0.5% yeast extra and 1% NaCl) at 28°C. After 24 h, the fermentation of core bacteria was collected and filtered through the microporous membrane (0.22 μm in pore size). 2 × PDA culture medium (1.2% potato starch, 4% dextrose, and 4% agar) added to the fermentation suspension in a 1:1 ratio as FB medium was used to measure vegetative growth of fungi, and PDA culture medium as a control. 5 mm hypha colonies were inoculated in different FB mediums at 25°C for 10 days, and the colony diameters were measured.
To investigate how the core microbiome affects the infection virulence of B. bassiana, the axenic larvae reintroduced to the core microbiome were used as the bioassay hosts to test fungi virulence. The core microbiome reintroduction was performed as described previously (Demirci et al., 2013). Axenic larvae were fed with leaves of willow, the surface of which were coated by the core bacteria, and reintroduced larvae were verified through the bacteria isolation experiments. After that, the same pathogenic fungal infection method was used for biometric tests, and mortality was observed and recorded.
2.5. Statistical analysis
Prior to statistical analysis, the Kolmogorov–Smirnov test and Levene test were performed to test the normality and homogeneity of all variances, respectively (Ding et al., 2023a,b). The Kaplan–Meier method was used to analyze the survival curves, and the log-rank test was used to evaluate the significance of differences.
3. Result
3.1. Analysis of the symbiotic bacteria richness and diversity between different age larvae
The symbiotic bacteria richness and diversity between different age P. versicolora were analyzed. A total of 1,592,856 valid sequences of symbiotic bacteria were obtained from 72 samples. Based on a 97% similarity level of classification and an average sequencing coverage of 99.88%, 7,879 bacterial ASVs (Amplicon sequence variants) were found, belonging to 38 phyla and 1,038 genera, respectively (Supplementary Table S1). The rarefaction curve was increased with the increase of sequencing depth and this indicated that the measured data could meet the subsequent experimental analysis (Supplementary Figure S1).
Alpha diversity analysis based on ASV level is shown in Figure 1, The ACE index and Chao index reflected community species richness, and there was no significant difference in microbial community richness among 7 sets of samples (Figures 1A,B). Shannon index and Simpson index indicated community species diversity and showed significant differences among texted samples (Figures 1C,D). β-diversity analysis of all samples based on ASV levels was shown in Figure 2. PCoA (Adonis, p < 0.001) and NMDS (Adonis, stress = 0.138; p < 0.001) analyses based on the weighted UniFrac metric were showing significant differences, this revealed that microbial community composition among the seven groups were significantly different (Figures 2A,B). The Hierarchical clustering analysis based on the bray-Curtis distance metric reflected that seven groups had higher diversity and abundance of the bacterial microbiota (Figure 2C).
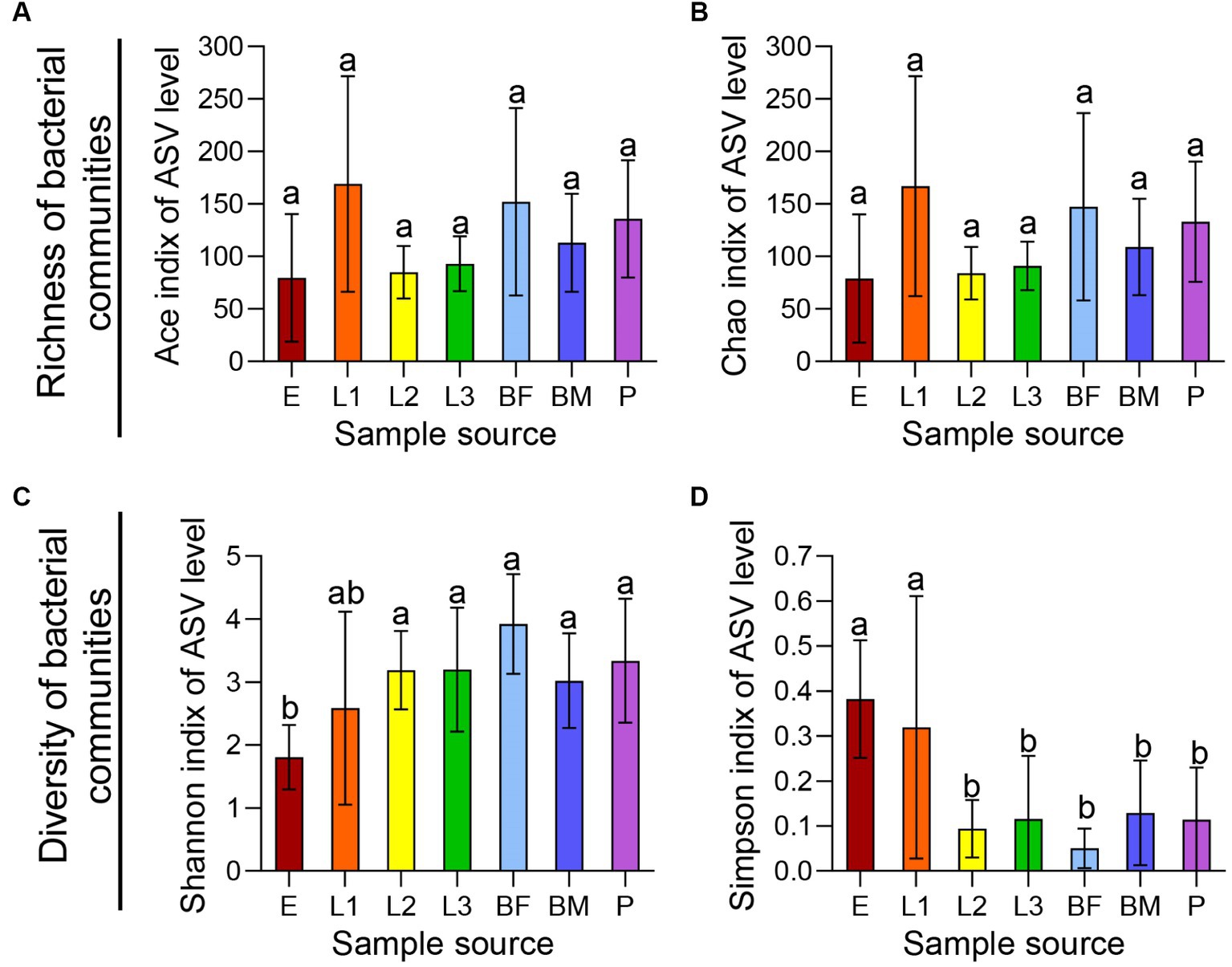
Figure 1. Richness and diversity of symbiotic bacteria in different age P. versicolora. The eggs (E), pupae (P), first instar larva (L1), second instar larva (L2), third instar larva (L3), female adults (BF), and male adults (BM) of P. versicolora were collected from Shahu Park in Wuhan, Hubei Province (30.35°N, 114.33°E). The α diversity of symbiotic bacteria in all samples were shown through alpha diversity analysis. (A) ACE index. (B) Chao index. (C) Shannon index. (D) Simpson index.
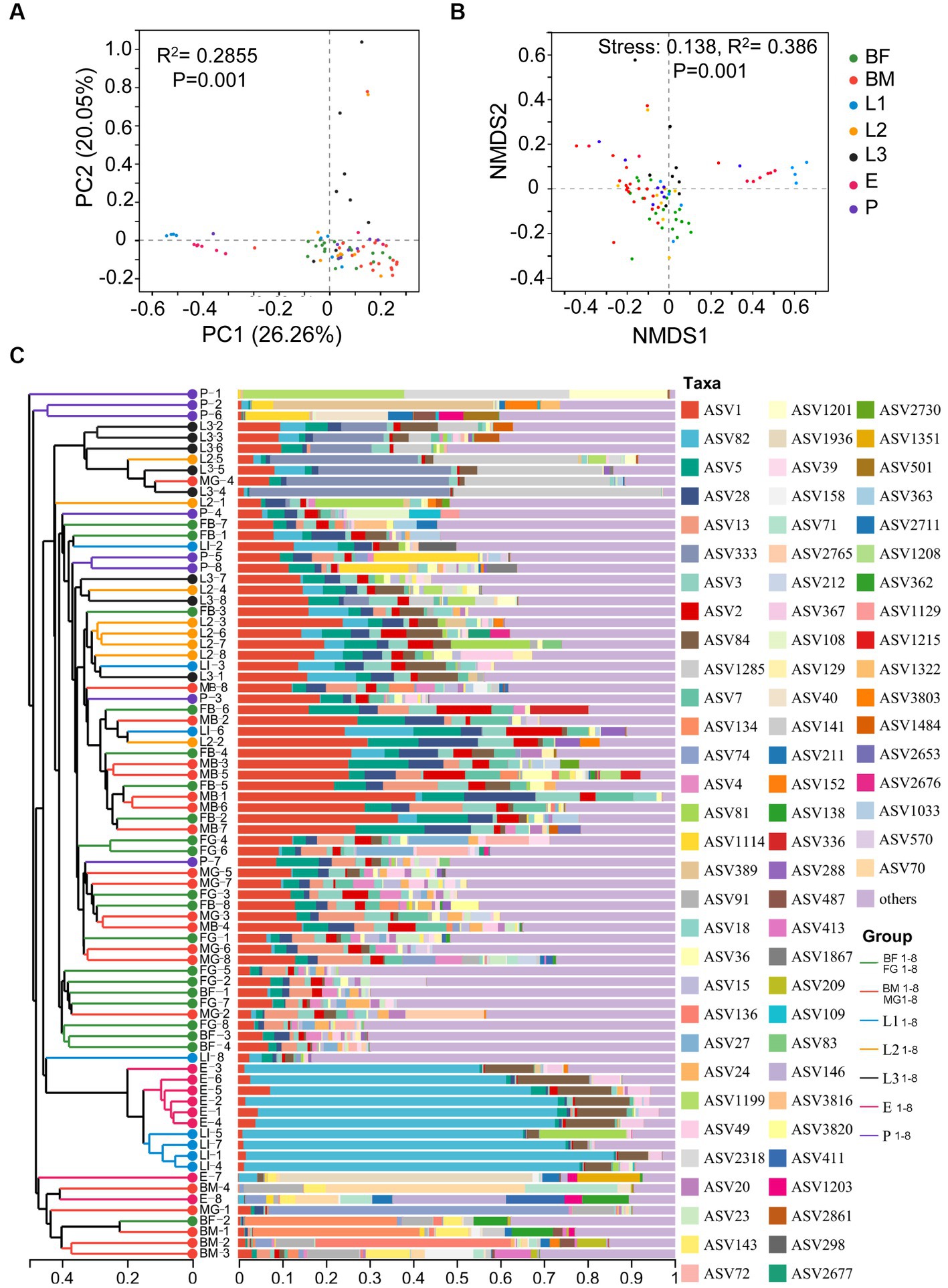
Figure 2. The composition and diversity of bacterial microbiota in different age larvae. (A) Principal Coordinate analysis (PCoA) and (B) Nonmetric Multidimensional Scaling (NMDS) analysis was showed the diversity of symbiotic bacteria in different age larvae based on the weighted UniFrac metric. (C) Community analysis of symbiotic bacteria in different age larvae.
As shown in the Figure 3A, 25 unique genera were found in the BM group, and 163, 76, 11, 14, 20, and 29 unique genera were distributed in BF, L1, L2, L3, E, and P groups respectively, and the unique bacteria species of different sample were listed in Supplementary Table S2. Among those, 65 genera were distributed in all samples, which were named the core microbiome. Out of these 65 genera, 15.84% belong to Pseudomonas, 15.16% belong to Rhodococcus, and 8.74% belong to Delftia (Figure 3B). The community heat map showed that 65 core bacteria belong to 50 genera, with Delftia sp., Pseudomonas sp., Rhodococcus sp., and Enterobacteriaceae sp. as the common genera of all samples (Figure 4).
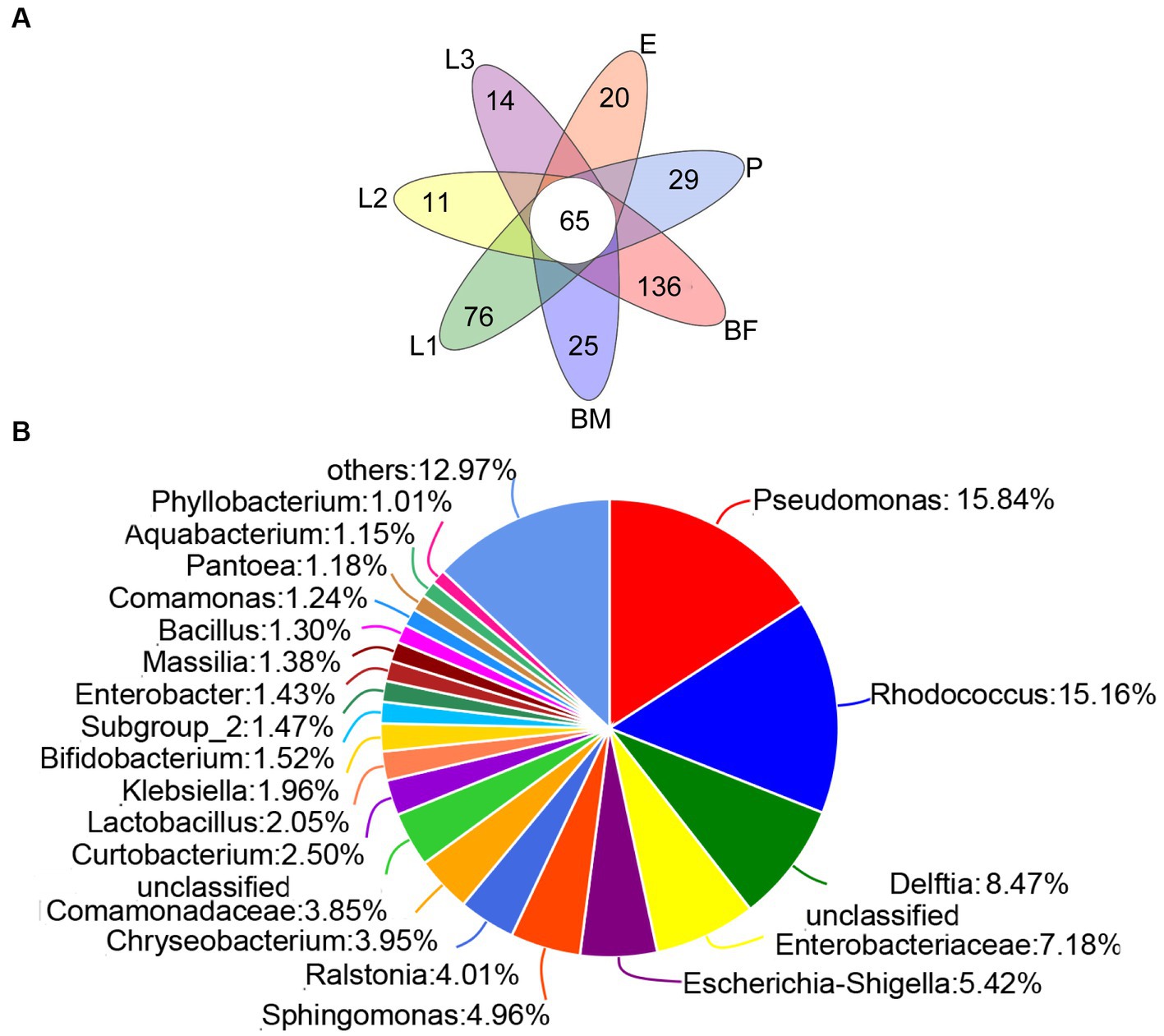
Figure 3. The composition of corn bacterial microbiota in P. versicolora. (A) Venn diagram was displayed the genus number of symbiotic bacteria in different age larvae. (B) A pie chart of common species for corn bacterial microbiota was displayed at the genus level.
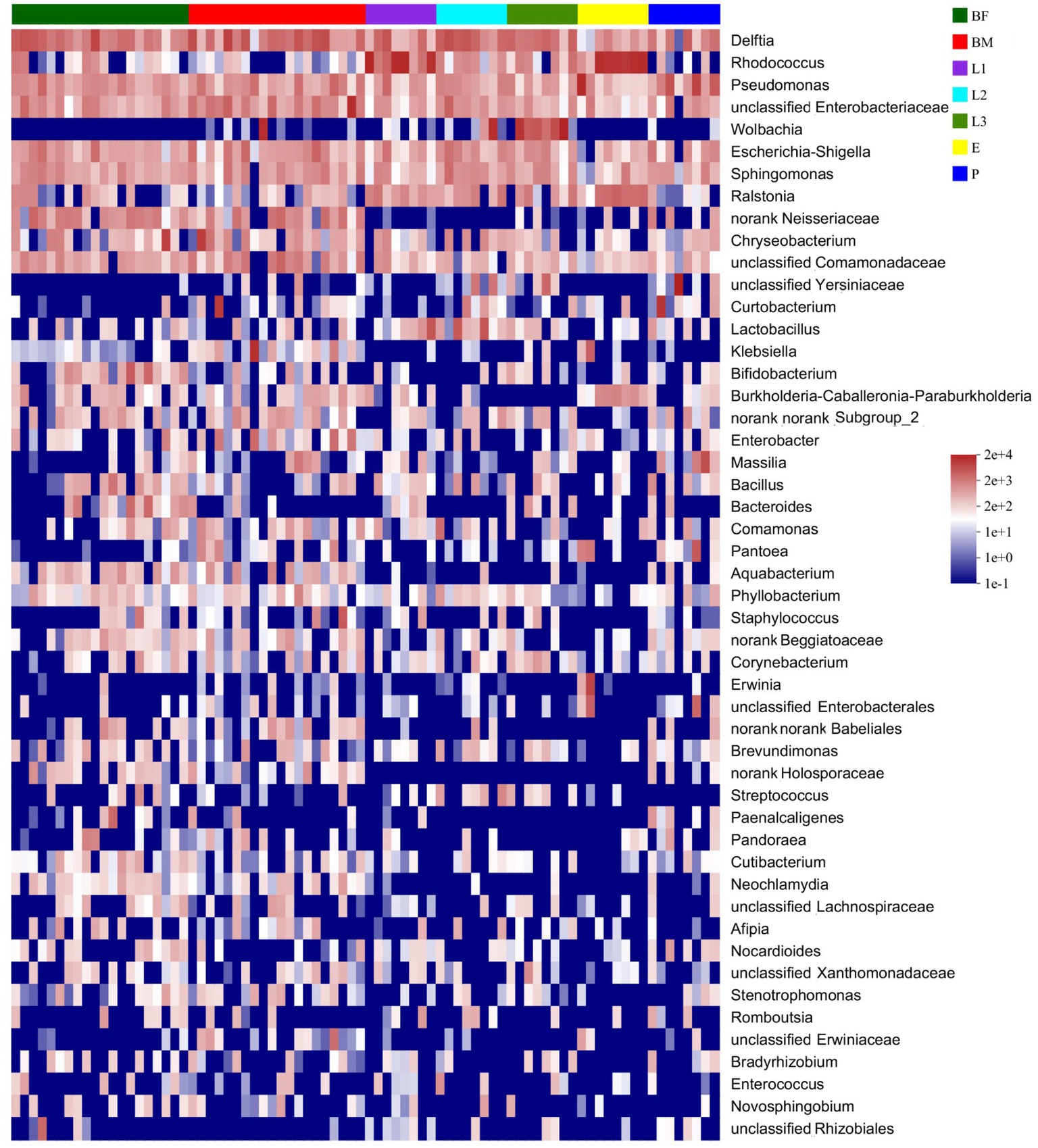
Figure 4. Heatmap of the top 50 genera abundances of corn bacterial microbiota was displayed at the genus classification level.
3.2. Symbiotic bacteria accelerated the mortality of Plagiodera versicolora larvae infected by Beauveria bassiana
To evaluate the role of symbiotic bacteria in the host interplay with pathogenic microorganisms, axenic and nonaxenic larvae were used to test host susceptibility to B. bassiana. Interestingly, at 6 days post-infection, the mortality of nonaxenic larvae infected by B. bassiana was significantly higher than those of axenic larvae (Figure 5, Log-rank test, p < 0.001). These results indicated that symbiotic bacteria accelerated the mortality of P. versicolora larvae infected by B. bassiana.
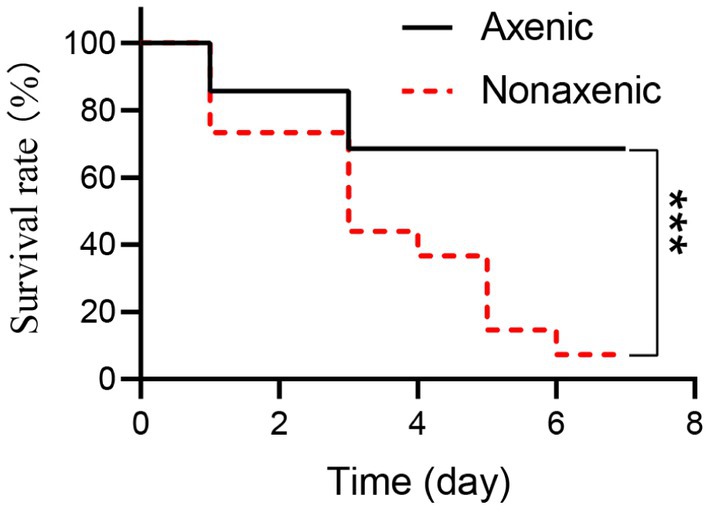
Figure 5. The mortality of axenic and nonaxenic P. versicolora larvae larvae infected by B. bassiana. Fungal strains were incubated on PDA plates for 10 days at 25°C, and the resultant conidia were used as infectious propagules. In bioassay, the insect hosts were immersed in conidial suspension (108 conidia/ml). The resultant insects were reared at 25°C, and accumulative mortality was recorded daily. Survival percentage was plotted against the time post-infection, and statistical difference between the paired curves was analyzed by log rank test.
3.3. Pseudomonas sp. promote fungi vegetative growth
Out of 65 core microbial genera, only 8 species were obtained through the bacteria isolation experiments, and they are Pseudomonas putida, Pseudomonas taiwanensis, Pseudomonas parafulva, Pseudomonas ficuserectae, Pseudomonas fulva, Pseudomonas meliae, Pseudomona aeruginosa, and Pseudomonas punonensis, respectively. To investigate whether the core microbiome plays an important role in P. versicolora susceptibility to B. bassiana, fungi vegetative growth on the fermentation medium was detected. As shown in Figure 6, on the PDA medium, the colony diameter of B.bassiana was 1.25 ± 0.11 cm. On the added P.putida, P. taiwanensis, P. parafulva, P. ficuserectae, and P. fulva fermentation media, the colony diameter increased by approximately 26.46, 54.90, 141.82, 109.96 and 106.26%, respectively. These five strains are highly conserved in evolution and belong to Pseudomonas sp. (Figure 6F). However, there was no significant difference in the colony diameter between the P.meliae, P.aeruginosa, and P.punonensis fermentation media and PDA medium. The results showed that Pseudomonas sp. Played an important role in the growth rate of B. bassiana.
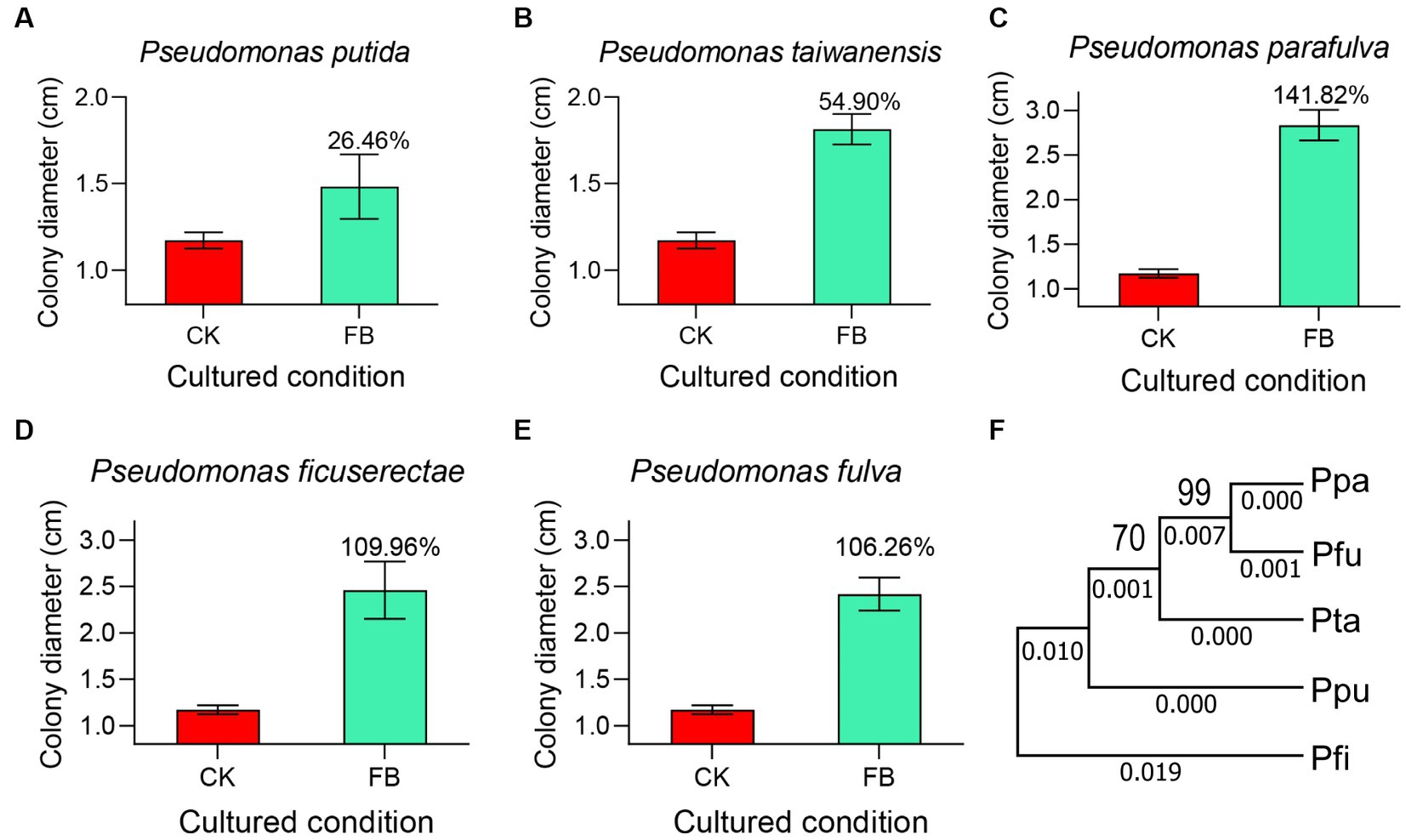
Figure 6. The core microbiome affects the mycelium growth. Fungal strains were cultured on PDA plates and different FB mediums for the condition. After 7 days of incubation at 25°C, colony morphologies were recorded. (A) Fermentation broth medium of Pseudomonas putida. (B) Fermentation broth medium of Pseudomonas taiwanensis. (C) Fermentation broth medium of Pseudomonas parafulva. (D) Fermentation broth medium of Pseudomonas ficuserectae. (E) Fermentation broth medium of Pseudomonas fulva. (F) The evolutionary relationship of five core strains was analyzed through MEGA7. Ppa represents Pseudomonas parafulva; Pfu represents Pseudomonas fulva; Pta represents Pseudomonas taiwanensis; Ppu represents Pseudomonas putida; Pfi represents Pseudomonas ficuserectae.
3.4. The isolated Pseudomonas sp. enhanced the pathogenesis of Beauveria bassiana against Plagiodera versicolora
To refine the specific functions of the core microbiome on B. bassiana pathogenesis to P. versicolora larvae, P.putida, P. taiwanensis, P. parafulva, P. ficuserectae, and P. fulva were reintroduced into axenic larvae. All the bacteria reintroduced into the larvae significantly accelerated the mortality of larvae at 4 to 6 d post-B. bassiana infection. There is a significant difference between the mortality of the reintroduced core microbiome group and the axenic group (Log-rank test, p < 0.001) and no significant difference between the reintroduced core microbiome group and the nonaxenic group (Log-rank test, p = 0.5598). These results suggest that the core microbiome of Pseudomonas sp. promotes B. bassiana pathogenesis to P. versicolora (Figure 7).
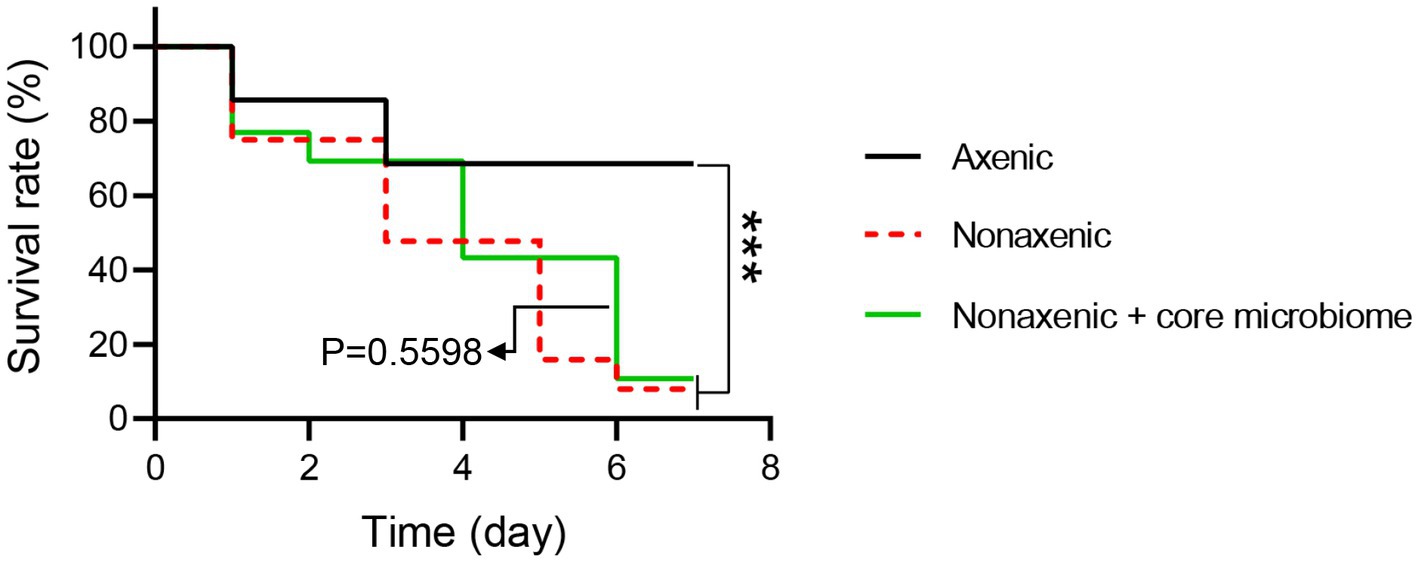
Figure 7. The survival rate of axenic, nonaxenic and bacteria reintroduced P. versicolora larvae infected by B. bassiana. The log-rank test was used to evaluate the significance of differences among groups, ***p < 0.001.
4. Discussion
In this study, 16 s rRNA high-throughput sequencing was used to analyze the diversity and variability of symbiotic bacteria in different age P. versicolora larvae. Taxonomic analysis revealed that the symbiotic bacteria community of P. versicolora was mainly composed of Actinobacteriota, Proteobacteria, Firmicutes, Bacteroidota, and Dependentiae. Similar results were also reported in other Coleoptera, such as Gastrolina depressa, Dendroctonus valens and Dendroctonus rhizophagus (Morales-Jimenez et al., 2012; Xu et al., 2019; Ma et al., 2021). The environment where insects live or the diet that insects feed upon significantly affects the structure of their symbiotic bacteria (Malacrinò, 2021; Zhao et al., 2022). Rahnella sp., Micrococcus luteus and Pantoea agglomerans were present in a large proportion of lab-reared P.versicolora larvae, but in very small quantities in wild-collected larvae (Demirci et al., 2013). Nevertheless, Sphingomonas sp. was detected in all the samples in both wild-collected P. versicolora, which suggested that there was a core community of bacteria (Demirci et al., 2013). Various studies also indicated that the persistence of some core symbiotic bacterial microbiota occurs regardless of the diet and environment in insect species (Turnbaugh et al., 2009; Xu et al., 2019; Ma et al., 2021). Those symbiotic bacteria play an important ecological role in maintaining the population of insects (Turnbaugh et al., 2009). Analysis of symbiotic bacteria of different ages of P. versicolora shows that there are 65 types of symbiotic bacteria present in the all sample, mainly Pseudomonas, Rhodococcus, and Delftia et al. Among them, Pseudomonas is a genus of gram-negative bacteria able to colonize a wide range of niches and is also a core microbiome in humans, soil, and plants (Turnbaugh et al., 2009; Chevalier et al., 2017; Hong et al., 2023).
Bacteria not only act as symbionts but also as pathogens of eukaryotes (mammals, insects, and plants). Their ability to enter and inhabit eukaryotic tissues/cells has given bacteria specialized niches that are stable, nutrient-rich, and enemy-free (Waters and Bassler, 2005). When hosts obtain benefits from bacteria or no negative impact (such as improving the insect’s fitness, or intervening in host-pathogen interactions), the relationship is referred to as symbiosis (Malacrinò, 2021; Zhao et al., 2022). On the other hand, bacteria that cause damage to hosts are called pathogens (Demirci et al., 2013). After the insects were infected by entomopathogenic fungi, the symbiotic bacteria in the host will transform into pathogenic bacteria. An example is Metarhizium rileyi infected insects, in which the gut symbiotic bacteria are translocated to hemolymph, its host humoral antibacterial immunity is activated, and fungi infection is enabled (Wang et al., 2023). Our study showed that symbiotic bacteria significantly enhanced the pathogenesis of B. bassiana against P. versicolora. Notably, the virulence of B. bassiana against nonaxenic larvae exceeded B. bassiana against axenic larvae, and the introduction of the core microbiome to axenic larvae augmented the virulence of fungi. The introduction of core microbiota significantly enhances fungal virulence. A large part of this was caused by the bacterial enzymes, such as proteases (alkaline metalloproteases) and chitinases which were identified as key contributors to tissue degradation and fungal defense system suppression (Ansari et al., 2005; Pham et al., 2007; Cirimotich et al., 2011). The larvae of Dendroctonus Valens LeConte infection by B. bassiana induced gut microbiota dysbiosis, such as Erwinia sp., which expedited entomopathogenic fungal infection (Xu et al., 2019). In other insects, symbiotic bacteria that affect B. bassiana virulence has also been reported. For example, symbiotic bacteria (Pseudomonas sp., Enterobacter sp., and Microbacterium sp.) of Lymantria dispar promoted the infection virulence of B. bassiana (Bai et al., 2023). Analogous outcomes were evident in mosquito research (Davis et al., 2019). Other studies have pointed out that Aphids harboring endosymbiont Regiella insecticola exhibited heightened resistance to entomophthorales fungus (Sanda et al., 2023). This underscores the multifaceted role of symbiotic bacteria of insects in modulating B. bassiana infectivity. The symbiotic microbiome also demonstrated a nutritionally reciprocal relationship with B. bassiana. The fermented liquid of core symbiotic bacteria was enhanced B. bassiana growth rates.
In summary, this study investigated the composition of symbiotic microbiomes of different ages of P. versicolora and identified the core microbiome. In addition, the core microbiome of Pseudomonas sp. was proven to promote the fungal infection process in P. versicolora. These findings provide fundamental insights into the multilateral interactions among insect hosts, communal microbiota, and entomopathogenic fungus, which may ultimately lead to a better strategy for biological control of P. versicolora.
Data availability statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found at: https://www.ncbi.nlm.nih.gov/, PRJNA1013480.
Ethics statement
The manuscript presents research on animals that do not require ethical approval for their study.
Author contributions
MLi: Formal analysis, Investigation, Visualization, Writing – original draft. JD: Formal analysis, Investigation, Writing – review & editing. MLu: Conceptualization, Funding acquisition, Project administration, Supervision, Writing – review & editing.
Funding
The author(s) declare financial support was received for the research, authorship, and/or publication of this article. This project is supported by the Hubei University National talent project (1070017364).
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2023.1290925/full#supplementary-material
References
Ansari, M. A., Tirry, L., and Moens, M. (2005). Antagonism between entomopathogenic fungi and bacterial symbionts of entomopathogenic nematodes. BioControl 50, 465–475. doi: 10.1186/s40064-016-3745-5
Baek, S., Noh, M. Y., Munm, S., Leem, S. J., Arakanem, Y., and Kim, J. S. (2022). Ultrastructural analysis of beetle larva cuticles during infection with the entomopathogenic fungus Beauveria bassiana. Pest Manag Sci. 78, 3356–3364. doi: 10.1002/ps.6962
Bai, J., Xu, Z., Li, L., Zhang, Y., Diao, J., Cao, J., et al. (2023). Gut bacterial microbiota of Lymantria dispar asiatica and its involvement in Beauveria bassiana infection. J. Invertebr. Pathol. 197:107897. doi: 10.1016/j.jip.2023.107897
Chavez, A. V., Duren, E. B., Avery, P. B., Pitino, M., Duncan, R. E., Cruz, L. F., et al. (2023). Evaluation of spore acquisition, spore production, and hostsurvival time for tea shot-hole borer, Euwallacea perbrevis, adults after exposure to four commercial products containing Beauveria bassiana. Insects 14:726. doi: 10.3390/insects14090726
Chevalier, S., Bouffartigues, E., Bodilis, J., Maillot, O., Lesouhaitier, O., Feuilloley, M. G. J., et al. (2017). Structure, function and regulation of Pseudomonas aeruginosa porins. FEMS Microbiol. Rev. 41, 698–722. doi: 10.1093/femsre/fux020
Cirimotich, C. M., Dong, Y., Clayton, A. M., Sandiford, S. L., Souza-Neto, J. A., Mulenga, M., et al. (2011). Natural microbe-mediated refractoriness to Plasmodium infection in Anopheles gambiae. Science 332, 855–8. doi: 10.1126/science.1201618
Davis, T. S., Stewart, J. E., Mann, A., Bradley, C., and Hofstetter, R. W. (2019). Evidence for multiple ecological roles of Leptographium abietinum, a symbiotic fungus associated with the north American spruce beetle. Fungal Ecol. 38, 62–70. doi: 10.1016/j.funeco.2018.04.008
Demirci, M., Sevim, E., Demir, I., and Sevim, A. (2013). Culturable bacterial microbiota of Plagiodera versicolora (L.) (Coleoptera: Chrysomelidae) and virulence of the isolated strains. Folia Microbiol. 58, 201–210. doi: 10.1007/s12223-012-0199-1
Ding, J. L., Lin, H. Y., Hou, J., Feng, M. G., and Ying, S. H. (2023a). The entomopathogenic fungus Beauveria bassiana employs autophagy as a persistence and recovery mechanism during conidial dormancy. MBio 14:e0304922. doi: 10.1128/mbio.03049-22
Ding, J. L., Wei, K., Feng, M. G., and Ying, S. H. (2023b). Homologs of bacterial heat-labile enterotoxin subunit a contribute to development, stress response, and virulence in filamentous entomopathogenic fungus Beauveria bassiana. Front. Immunol. 4:1264560. doi: 10.3389/fimmu.2023
Douglas, A. E. (2015). Multiorganismal insects: diversity and function of resident microorganisms. Annu. Rev. Entomol. 60, 17–34. doi: 10.1146/annurev-ento-010814-020822
Hong, S., Yuan, X., Yang, J., Yang, Y., Jv, H., Li, R., et al. (2023). Selection of rhizosphere communities of diverse rotation crops reveals unique core microbiome associated with reduced banana Fusarium wilt disease. New Phytol. 238, 2194–2209. doi: 10.1111/nph.18816
Hou, J., Zhang, H., Ding, J. L., Feng, M. G., and Ying, S. H. (2023). Transcriptomic investigation reveals a physiological mechanism for Beauveria bassiana to survive under linoleic acid stress. iScience 26:106551. doi: 10.1016/j.isci.2023.106551
Joop, G., and Vilcinskas, A. (2016). Coevolution of parasitic fungi and insect hosts. Zoology 119, 350–358. doi: 10.1016/j.zool.2016.06.005
Ling, J., Li, X., Yang, G., and Yin, T. (2021). Volatile metabolites of willows determining host discrimination by adult Plagiodera versicolora. J. For. Res. 33, 679–687. doi: 10.1007/s11676-021-01349-1
Liu, F., Li, B., Liu, C., Liu, Y., Liu, X., and Lu, M. (2023). Oviposition by Plagiodera versicolora on Salix matsudana cv. 'Zhuliu' alters the leaf transcriptome and impairs larval performance. Front. Plant Sci. 14:1226641. doi: 10.3389/fpls.2023.1226641
Liu, X., Tong, N., Wu, Z., Li, Y., Ma, M., Liu, P., et al. (2021). Identification of chemosensory genes based on the antennal transcriptomic analysis of Plagiodera versicolora. Insects 13:36. doi: 10.3390/insects13010036
Ma, M., Tu, C., Luo, J., Lu, M., Zhang, S., and Xu, L. (2021). Metabolic and immunological effects of gut microbiota in leaf beetles at the local and systemic levels. Integr Zool. 16, 313–323. doi: 10.1111/1749-4877.12528
Malacrinò, A. (2021). Host species identity shapes the diversity and structure of insect microbiota. Mol. Ecol. 31, 723–735. doi: 10.1111/mec.16285
Morales-Jimenez, J., Zuniga, G., Ramirez-Saad, H. C., and Hernandez-Rodriguez, C. (2012). Gut-associated bacteria throughout the life cycle of the bark beetle Dendroctonus rhizophagus Thomas and Bright (Curculionidae: Scolytinae) and their cellulolytic activities. Microb. Ecol. 64, 268–278. doi: 10.1007/s00248-011-9999-0
Perumal, V., Kannan, S., Alford, L., Pittarate, S., Geedi, R., Elangovan, D., et al. (2023a). First report on the enzymatic and immune response of Metarhizium majus bag formulated conidia against Spodoptera frugiperda: an ecofriendly microbial insecticide. Front. Microbiol. 14:1104079. doi: 10.3389/fmicb.2023.1104079
Perumal, V., Kannan, S., Alford, L., Pittarate, S., Mekchay, S., Reddy, G. V. P., et al. (2023b). Biocontrol effect of entomopathogenic fungi Metarhizium anisopliae ethyl acetate-derived chemical molecules: an eco-friendly anti-malarial drug and insecticide. Arch. Insect Biochem. Physiol. 114, 1–19. doi: 10.1002/arch.22037
Pham, L. N., Dionne, M. S., Shirasu-Hiza, M., and Schneider, D. S. (2007). A specific primed immune response in drosophila is dependent on phagocytes. PLoS Pathog. 3:e26. doi: 10.1371/journal.ppat.0030026
Sanda, N. B., Inchauregui, R. A., Tallapragada, K., and Parker, B. J. (2023). Aphid facultative symbionts confer no protection against the fungal entomopathogen Batkoa apiculata. PLoS One 18:e0286095. doi: 10.1371/journal.pone.0286095
Singh, S., Singh, A., Baweja, V., Roy, A., Chakraborty, A., and Singh, I. K. (2021). Molecular rationale of insect-microbes symbiosis-from insect behaviour to mechanism. Microorganisms 9:2422. doi: 10.3390/microorganisms9122422
Sittipo, P., Lobionda, S., Lee, Y. K., and Maynard, C. L. (2018). Intestinal microbiota and the immune system in metabolic diseases. J. Microbiol. 56, 154–162. doi: 10.1007/s12275-018-7548-y
Tu, C., Zhang, Y., Zhu, P., Sun, L., Xu, P., Wang, T., et al. (2023). Enhanced toxicity of entomopathogenic fungi Beauveria bassiana with bacteria expressing immune suppressive dsRNA in a leaf beetle. Pestic. Biochem. Physiol. 193:105431. doi: 10.1016/j.pestbp.2023.105431
Turnbaugh, P. J., Hamady, M., Yatsunenko, T., Cantarel, B. L., Duncan, A., Ley, R. E., et al. (2009). A core gut microbiome in obese and lean twins. Nature 457, 480–484. doi: 10.1038/nature07540
Vivekanandhan, P., Bedini, S., and Shivakumar, M. S. (2020). Isolation and identification of entomopathogenic fungus from eastern Ghats of south Indian forest soil and their efficacy as biopesticide for mosquito control. Parasitol. Int. 76:102099. doi: 10.1016/j.parint.2020.102099
Vivekanandhan, P., Kavitha, T., Karthi, S., Senthil-Nathan, S., and Shivakumar, M. S. (2018). Toxicity of Beauveria bassiana-28 mycelial extracts on larvae of culex quinquefasciatus mosquito (Diptera: Culicidae). Int. J. Environ. Res. Public Health 15:440. doi: 10.3390/ijerph15030440
Wade, M. J. (1994). “The biology of the imported willow leaf beetle, Plagiodera versicolora (Laicharting)” in Novel aspects of the biology of Chrysomelidae. eds. P. H. Jolivet, M. L. Cox, and E. Petitpierre (Dordrecht: Springer) 50
Wade, M. J., and Breden, F. (1986). Life history of natural populations of the imported willow leaf beetle, Plagiodera versicolora (Coleoptera: Chrysomelidae). Ann. Entomol. Soc. Am. 79, 73–79. doi: 10.1093/aesa/79.1.73
Wang, J. L., Sun, J., Song, Y. J., Zheng, H. H., Wang, G. J., Luo, W. X., et al. (2023). An entomopathogenic fungus exploits its host humoral antibacterial immunity to minimize bacterial competition in the hemolymph. Microbiome 11:116. doi: 10.1186/s40168-023-01538-6
Waters, C. M., and Bassler, B. L. (2005). Quorum sensing: cell-to-cell communication in bacteria. Annu. Rev. Cell Dev. Biol. 21, 319–346. doi: 10.1146/annurev.cellbio.21.012704.131001
Xiao, G., Ying, S. H., Zheng, P., Wang, Z. L., Zhang, S., Xie, X. Q., et al. (2012). Genomic perspectives on the evolution of fungal entomopathogenicity in Beauveria bassiana. Sci. Rep. 2:483. doi: 10.1038/srep00483
Xiong, W. (2022). Intestinal microbiota in various animals. Integr Zool. 17, 331–332. doi: 10.1111/1749-4877.12633
Xu, L. T., Deng, J. D., Zhou, F. Y., Cheng, C. H., Zhang, L. W., Zhang, J., et al. (2019). Gut microbiota in an invasive bark beetle infected by a pathogenic fungus accelerates beetle mortality. J. Pest. Sci. 92, 343–351. doi: 10.1007/s10340-018-0999-4
Yoneya, K., Kugimiya, S., and Takabayashi, J. (2014). Leaf beetle larvae, Plagiodera versicolora (Coleoptera: Chrysomelidae), show decreased performance on uninfested host plants exposed to airborne factors from plants infested by conspecific larvae. Appl. Entomol. Zool. 49, 249–253. doi: 10.1007/s13355-013-0243-x
Zhang, F., Huang, Y. H., Liu, S. Z., Zhang, L., Li, B. T., Zhao, X. X., et al. (2013). Pseudomonas reactans, a bacterial strain isolated from the intestinal flora of Blattella germanica with anti-Beauveria bassiana activity. Environ. Entomol. 42, 453–459. doi: 10.1603/EN12347
Keywords: 16S rRNA, Beauveria bassiana, symbiotic microbiota, interactions, Plagiodera versicolora, fungal virulence and pathogenicity
Citation: Liu M, Ding J and Lu M (2023) Influence of symbiotic bacteria on the susceptibility of Plagiodera versicolora to Beauveria bassiana infection. Front. Microbiol. 14:1290925. doi: 10.3389/fmicb.2023.1290925
Edited by:
Vivekanandhan Perumal, Chiang Mai University, ThailandReviewed by:
Nareshkumar Arjunan, Periyar University, IndiaThendralmanikandan Ayyavu, Periyar University, India
Meenambigai K., Vinayaka Missions University, India
Copyright © 2023 Liu, Ding and Lu. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Min Lu, bHVtaW5AaHVidS5lZHUuY24=
 Mei Liu
Mei Liu Jinli Ding
Jinli Ding Min Lu
Min Lu