- 1School of Biotechnology and Biomolecular Sciences, University of New South Wales, Sydney, NSW, Australia
- 2Helicobacter Research Laboratory, School of Pathology and Laboratory Medicine, Marshall Centre for Infectious Diseases Research and Training, University of Western Australia, Perth, WA, Australia
- 3Laboratory Medicine, Guangdong Provincial People’s Hospital (Guangdong Academy of Medical Sciences), Southern Medical University, Guangzhou, Guangdong, China
- 4The Center for Precision Health, School of Medical and Health Sciences, Edith Cowan University, Perth, WA, Australia
- 5Department of Medical Informatics, School of Medical Informatics and Engineering, Xuzhou Medical University, Xuzhou, Jiangsu, China
- 6Gastrointestinal and Liver Unit, Prince of Wales Hospital, University of New South Wales, Sydney, NSW, Australia
Campylobacter concisus is an oral bacterium. Recent studies suggest that C. concisus may be involved in human gastric diseases. The mechanisms, however, by which C. concisus causes human gastric diseases have not been investigated. Here we examined the gastric epithelial pathogenicity of C. concisus using a cell culture model. Six C. concisus strains and the human gastric epithelial cell line AGS cells were used. IL-8 produced by AGS cells after incubation with C. concisus was measured using enzyme-linked immunosorbent assay (ELISA), and AGS cell apoptosis was determined by caspase 3/7 activities. The effects of C. concisus on actin arrangement in AGS cells was determined using fluorescence staining. The effects of C. concisus on global gene expression in AGS cells was determined by transcriptomic analysis and quantitative real-time PCR (qRT-PCR). The role of the upregulated CYP1A1 gene in gastric cancer survival was assessed using the Kaplan-Meier method. C. concisus induced production of IL-8 by AGS cells with strain variation. Significantly increased caspase 3/7 activities were observed in AGS cells incubated with C. concisus strains when compared to AGS cells without bacteria. C. concisus induced actin re-arrangement in AGS cells. C. concisus upregulated 30 genes in AGS cells and the upregulation of CYP1A1 gene was confirmed by qRT-PCR. The Kaplan-Meier analysis showed that upregulation of CYP1A1 gene is associated with worse survival in gastric cancer patients. Our findings suggest that C. concisus may play a role in gastric inflammation and the progression of gastric cancer. Further investigation in clinical studies is warranted.
Introduction
Campylobacter concisus is a gram-negative bacterium that is motile, with a curved or spiral shape. The bacterium can grow under both anaerobic and microaerophilic conditions, with hydrogen gas being crucial for its growth (Lee et al., 2014). C. concisus is further classified into two genomospecies (GS): GS1 and GS2, distinguished by the core-genome, 23 rRNA gene, and GS-specific genes (Miller et al., 2012; Chung et al., 2016; Huq et al., 2017; Liu et al., 2018; Aagaard et al., 2021; Cornelius et al., 2021). Previous studies have reported that C. concisus GS2 strains exhibit better adaptation in the gastrointestinal tract (Wang et al., 2017) and a better ability to invade intestinal epithelial cells compared to GS1 strains (Kalischuk and Inglis, 2011; Ismail et al., 2012; Mahendran et al., 2015). These findings align with the observation that GS2 strains are more frequently detected in mucosal biopsy samples and fecal samples from patients with gastrointestinal diseases (Kirk et al., 2018). Virulence factors of C. concisus, such as zonula occludens toxin, phospholipase A, as well as the functional protein BisA, have been characterized (Istivan et al., 2004; Mahendran et al., 2016; Benoit and Maier, 2023).
While commonly present in the human oral cavity as a commensal bacterium (Zhang et al., 2010), C. concisus is associated with inflammatory conditions of extraoral diseases such as inflammatory bowel disease (IBD), including Crohn’s disease (CD) and ulcerative colitis (UC) (Zhang L. et al., 2009; Man et al., 2010; Mukhopadhya et al., 2011), microscopic colitis, and Barrett’s esophagus (Macfarlane et al., 2007; Yang et al., 2009; Nielsen et al., 2020). Previous studies have also investigated the pathogenic mechanisms by which C. concisus may contribute to the development of the associated intestinal and esophageal diseases, including the induction of proinflammatory cytokines such as interleukin 8 (IL-8) and tumor necrosis factor alpha (TNF-α), epithelial cell death, immunmodulators MD-2 and programmed death-ligand 1 (PD-L1), as well as enhancing the responses of epithelial cells and macrophages to commensal bacterial species (Lee et al., 2021).
Recent studies suggest that C. concisus may also be involved in human gastric diseases. A study by Ferreira et al. (2022) examined the cultivation of Helicobacter pylori and C. concisus from 2,191 gastric biopsies. They reported that C. concisus was cultured from 50 gastric biopsies (50/2191, 2.3%) and H. pylori cultured from 168 gastric biopsies (168/2191, 7.7%). In twenty-eight cases with concurrent histology, C. concisus was found to be H. pylori immunoreactive positive (Ferreira et al., 2022). A study by Cui et al. (2019) examined the tongue coating microbiome of 78 patients with gastritis and 50 healthy controls. This study found that the abundance of C. concisus in tongue coating microbiome was associated with the gastric precancerous cascade. They also detected C. concisus in gastric fluids of patients with gastritis (Cui et al., 2019).
Despite being suggested to play a role in human gastric diseases, no studies have examined the mechanisms by which C. concisus may contribute to the pathogenesis of gastric diseases. Considering that gastric epithelial cells are the first line of human cells to encounter pathogens in the stomach, this study aimed to examine the pathogenic effects of C. concisus strains on human gastric epithelial cells using a cell culture model. Our data provide novel insights into understanding C. concisus gastric pathogenicity.
Materials and methods
Bacterial strains used in this study
For this study, we randomly selected six oral C. concisus strains with complete genomes sequenced from C. concisus strains we previously isolated from human saliva samples (Liu et al., 2020). P10CDO-S2, P3UCO1, and H1O1 are GS1 strains and P2CDO4, P15UCO-S2, and H16O-S1 are GS2 strains. The details of the six C. concisus strains were provided in Supplementary Table 1. The C. concisus strains were cultured on horse blood agar (HBA) plates, using blood agar base No. 2 (Thermo Fisher Scientific, CA, USA), supplemented with 6% defibrinated horse blood. The cultures were incubated at 37oC under anaerobic conditions with 5% hydrogen for 48 h, as previously described (Lee et al., 2014).
H. pylori strain 26695, a human gastric pathogen, was used as a positive control in this study (Ashktorab et al., 2008; Fazeli et al., 2016). H. pylori strain 26695 was cultured on HBA plates at 37oC under microaerobic conditions generated using CampyGen 2.5L Atmosphere Generation System (Thermo Fisher Scientific) for 48 h before being used in experiments.
Maintenance of AGS cells
The human gastric adenocarcinoma cell line AGS (ATCC No. CRL-1739) was used as a model for human gastric epithelium. AGS cells were maintained in F-12K medium (Thermo Fisher Scientific) supplemented with 10% fetal bovine serum (FBS) (Cytiva, MA, USA), 100 U/mL penicillin, and 100 μg/mL streptomycin (Thermo Fisher Scientific), which was referred to as F-12K/FBS/Antibiotics medium in this study. The AGS cells were incubated in a humidified incubator at 37°C with 5% CO2, following recommended maintenance procedures by ATCC.
Measurement of IL-8 by enzyme-linked immunosorbent assay (ELISA)
Enzyme-linked immunosorbent assay was used to measure IL-8 production by AGS cells in response to C. concisus. AGS cells were seeded on a 96-well plate at a concentration of 1 × 105 cells/well in F-12K/FBS/Antibiotics medium. Following 24 h incubation, the cell culture medium was replaced with F-12K medium supplemented with FBS but without penicillin and streptomycin, which was referred to as F-12K/FBS medium. The AGS cells were then incubated with the six C. concisus strains (in triplicates) described above at a multiplicity of infection (MOI) of 100 for 24 h. As C. concisus at MOI 100 induced the production of IL-8 by other epithelial cells of the gastrointestinal tract in a previous study, this condition was therefore used in the current study (Lee et al., 2021). AGS cells without bacterial infection served as the negative control. H. pylori strain 26695 was also introduced at MOI 10 and 100, which served as a positive control (O’Hara et al., 2006; Zhang Y. et al., 2009). To investigate the combined effects of C. concisus and H. pylori on IL-8 production by AGS cells, AGS cells were incubated with C. concisus strains P2CDO4 and P3UCO1 at MOI 100 along with H. pylori strain 26695 at MOI 10 and 100 for 24 h, respectively. The AGS cell culture supernatants were then collected to measure the concentration of IL-8 using commercially available ELISA kits (Invitrogen, CA, USA) in triplicates, following the manufacturer’s instructions.
Caspase 3/7 assay
Measurement of caspase 3/7 activity was used to assess apoptotic activity in AGS cells induced by C. concisus, as described previously (Lee et al., 2021). In brief, AGS cells were seeded on a black-walled 96-well plate with transparent bottoms at a concentration of 1 × 105 cells/well in F-12K/FBS/Antibiotics medium. After 24 h incubation, the cell culture medium was replaced with F-12K/FBS medium. The AGS cells were then incubated with the six C. concisus strains described above at MOI 100 for 24 h, with untreated AGS cells serving as the negative control. H. pylori strain 26695 was included at MOI 10 and 100 as the bacterial control. To examine the combined effects of C. concisus and H. pylori on the apoptotic activity of AGS cells, AGS cells were incubated with C. concisus strains P2CDO4 and P3UCO1 at MOI 100 along with H. pylori strain 26695 at MOI 10 and 100 for 24 h, respectively. AGS cells were washed three times with Dulbecco’s phosphate-buffered saline (DPBS) before being stained with CellEvent Caspase-3/7 Green ReadyProbes reagent (Invitrogen), following manufacturer’s instructions. The fluorescence readings of caspase 3/7 activity were measured in triplicates and expressed in fold change relative to the untreated control.
Examination of the effects of C. concisus on F-actin arrangement in AGS cells by fluorescence staining
AGS cells were seeded on coverslips in a 24-well plate at a concentration of 1 × 106 cells/well in F-12K/FBS/Antibiotics medium. After 24 h incubation, the cell culture medium was replaced with F-12K/FBS medium. AGS cells were then incubated with C. concisus strains P2CDO4, P3UCO1, or H. pylori strain 26695 at MOI 100 for 24 h, with untreated AGS cells serving as the negative control. The AGS cells were fixed with 3.6% paraformaldehyde for 15 min, permeabilized with 0.1% triton for 10 min, and blocked with 1% bovine serum albumin (BSA) for 1 h. The filamentous actin (F-actin) and nuclei were then stained with Alexa Fluor 488 phalloidin (8878S, Cell Signaling Technology, MA, USA) and Hoechst 33342 (Invitrogen) respectively. The cells were mounted onto glass slides with 50% glycerol in water and examined using a fluorescent microscope (Olympus BX61; Olympus, Tokyo, Japan) with FITC (Excitation wavelength: 480 nm; Emission wavelength: 520 nm) and DAPI (Excitation wavelength: 365 nm; Emission wavelength: 430 nm) filters under the 100X objective. AGS cells without bacteria served as the negative control, and AGS cells incubated with H. pylori served as the positive control (Chang et al., 2016).
Examination of the global gene responses induced by C. concisus in AGS cells by transcriptomic analysis
AGS cells were seeded in triplicates on 6-well cell culture plates at a concentration of 2 × 106 cells/well in F-12K/FBS/Antibiotics medium. After 24 h incubation, the medium was replaced with F-12K/FBS medium. The AGS cells were then incubated with C. concisus strain P2CDO4, which was randomly selected.
Supernatants from AGS cells incubated with C. concisus strain P2CDO4 at MOI 50 for 4 h and without bacterial infection were collected for IL-8 measurement, as described above. The AGS cells were then washed three times with DPBS before being collected for RNA extraction. The total RNA of AGS cells was extracted using the ISOLATE II RNA Mini Kit (cat. no. BIO-52072; Bioline, NSW, Australia), following the manufacturer’s instructions. The purity and concentration of the extracted total RNA were measured using a NanoDrop spectrophotometer. The extracted total RNA was then submitted to the Ramaciotti Centre for Genomics, University of New South Wales, for RNA sequencing. The library preparation was conducted as previously described (Lee et al., 2023).
For RNA-seq data analysis, the raw RNA-seq reads were first checked for quality using FastQC (version 0.11.8). Adapters and low-quality reads were then trimmed using Trimmomatic (version 0.38) with the leading and trailing filters set to a minimum of Phred score 3 and a sliding window of 4:15, filtered reads with length less than 30 bp were also removed (Bolger et al., 2014). The trimmed reads were mapped against the human reference genome GRCh38.p14 using HISAT2 (version 2.1.0) under default settings (Kim et al., 2019). The mapped read counts SAM files generated from HISAT2 were then converted into BAM files using SAMtools (version 1.11) (Danecek et al., 2021). The mapped read counts were then quantified using featureCounts under the Subread package (version 2.0.1) for DEG analysis (Liao et al., 2014). Significantly differentially expressed genes between AGS cells with and without C. concisus infection were identified using the BioConductor package DESeq2 (version 1.36.0) in the R programming environment under default normalization methods, with adjusted P < 0.05 and log2 fold change < -1 and >1 being considered significant (Love et al., 2014).
Gene ontology (GO) enrichment analysis
The list of differentially expressed genes (DEG) was uploaded to Metascape for GO enrichment analysis under default settings (Zhou et al., 2019). Enriched clusters of cellular biological processes were sorted according to P-value.
Quantitative real-time PCR (qRT-pCR)
A literature search in the PubMed database was conducted to examine whether the upregulated genes in AGS cells, as revealed by transcriptomic analysis, were associated with the development of gastric cancer, using the gene name and gastric cancer as keywords. The upregulation of the gastric cancer-associated gene, CYP1A1, was further confirmed using qRT-PCR. For qRT-PCR, the total RNA (2 μg/sample) extracted from AGS cells with or without C. concisus infection was subjected to cDNA synthesis using the Tetro cDNA Synthesis kit (Bioline, NSW, Australia), following manufacturer’s instructions. SensiFAST SYBR No-ROX Mix (Bioline, NSW, Australia) was used for quantifying the synthesized cDNA in qRT-PCR in triplicates. The mRNA expression levels were normalized to the levels of the housekeeping gene glyceraldehyde 3-phosphate dehydrogenase (GAPDH) and expressed as fold changes relative to untreated cells, using the comparative threshold cycle CT (2–ΔΔCT) method (Livak and Schmittgen, 2001). The sequences of PCR primers for quantification of CYP1A1 and qRT-PCR conditions are in Supplementary Table 2.
Analysis of the role of CYP1A1 in gastric cancer patient survival
The gene encoding CYP1A1 was subjected to survival analysis in gastric cancer patients using the Kaplan–Meier Plotter website (Lánczky and Gyõrffy, 2021). The survival plot was generated using JetSet best probe set for the submitted genes, with a database of 631 gastric cancer patients which are classified as low or high expression cohorts. The overall survival differences between low and high expression cohorts were analyzed using the Kaplan-Meier method and log-rank test, P < 0.05 was considered as significant and other options remained default.
Summary of experimental workflow
The experimental workflow of this study is summarized in Figure 1, outlining the key experiments conducted. These include the examination of C. concisus pathogenicity to AGS cells, transcriptomic analysis of gene expression changes, and the subsequent validation of CYP1A1 gene expression through qRT-PCR.
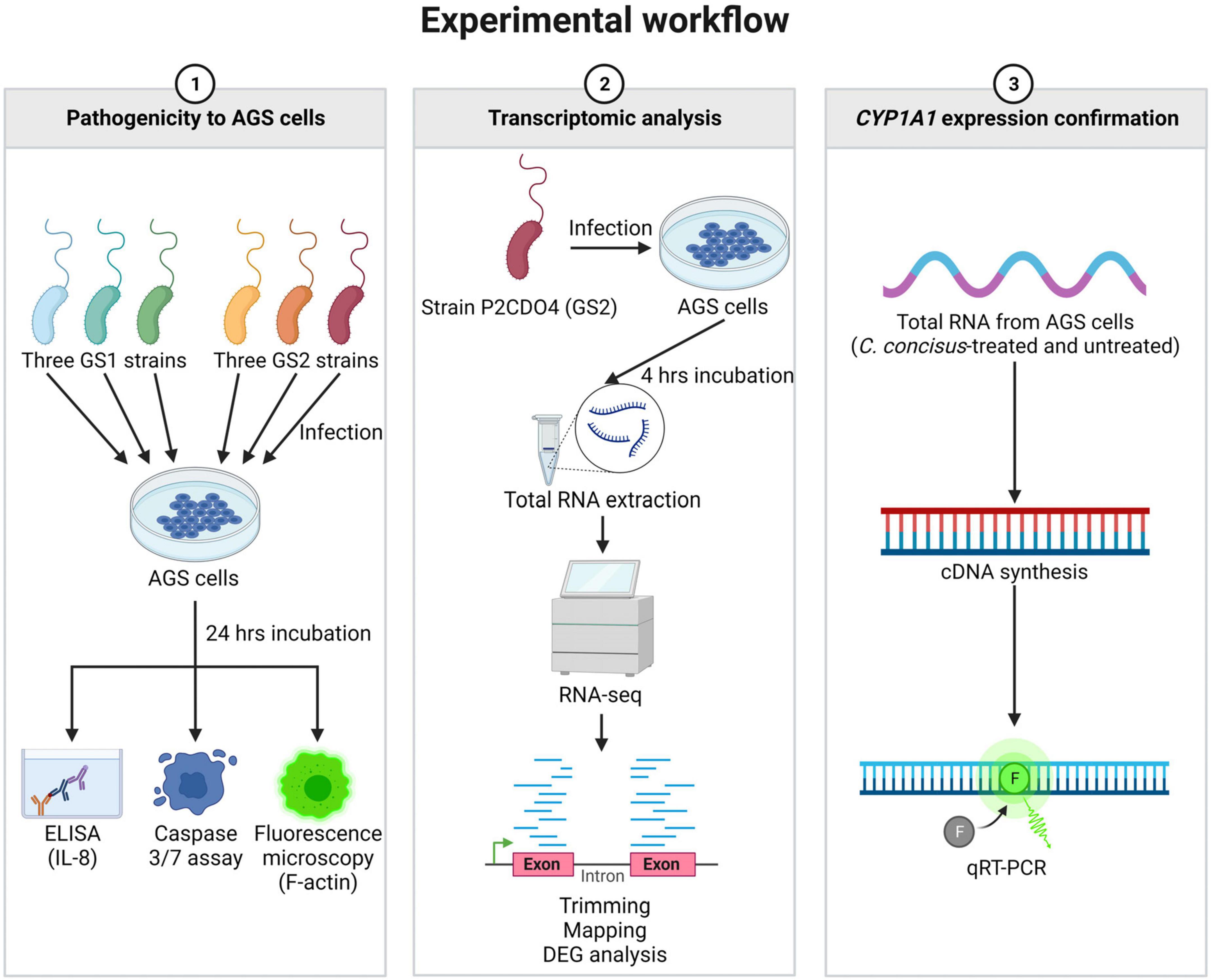
Figure 1. Summary of experimental workflow of this study. Experiments conducted in this study were classified into three categories. The first category involved examining the pathogenicity of C. concisus in AGS cells using ELISA, caspase 3/7 assay, and fluorescence microscopy. The second category focused on transcriptomic analysis of global gene response in AGS cells incubated with C. concisus through RNA-seq. The third category aimed to validate the gene expression level of CYP1A1 using qRT-PCR. Figure created with BioRender.com.
Statistical analysis
P-values for different samples in ELISA and caspase 3/7 assay were calculated using one-way analysis of variance (ANOVA) with Dunnett’s test, while P-value for samples in qRT-PCR were calculated using two-tailed unpaired t-test. P < 0.05 was considered statistically significant. All statistical analyses were conducted using GraphPad Prism (version 9.5.1).
Results
C. concisus induced IL-8 production in AGS cells
All six C. concisus strains examined at MOI 100 induced the production of IL-8 by AGS cells after a 24-h incubation period. Notably, the levels of IL-8 production in AGS cells incubated with C. concisus strains P2CDO4 and H1O1 were 286.64 ± 4.75 and 295.29 ± 11.5 pg/ml, respectively (P < 0.01). These values were statistically significant, indicating a higher induction of IL-8 production compared to AGS cells without bacterial infection (114.63 ± 0.68 pg/ml) (Figure 2). IL-8 production by AGS cells incubated with C. concisus strains P15UCO-S2, H16O-S1, P10CDO-S2, and P3UCO1 were 244.9 ± 1.22, 135.19 ± 2.41, 237.2 ± 3.12, and 224.54 ± 3.92 pg/ml, respectively. While these values were higher than the untreated sample, they were not considered statistically significant (P > 0.05) (Figure 2). Furthermore, H. pylori strain 26695 induced higher IL-8 production in AGS cells compared to C. concisus strains. The levels of IL-8 production by AGS cells incubated with H. pylori strain 26695 at MOI 10 and 100 were 1099.58 ± 29.18 and 1557.94 ± 36.05 pg/ml, respectively, which were significantly higher than the IL-8 production induced by the C. concisus strains (P < 0.0001) (Figure 2).
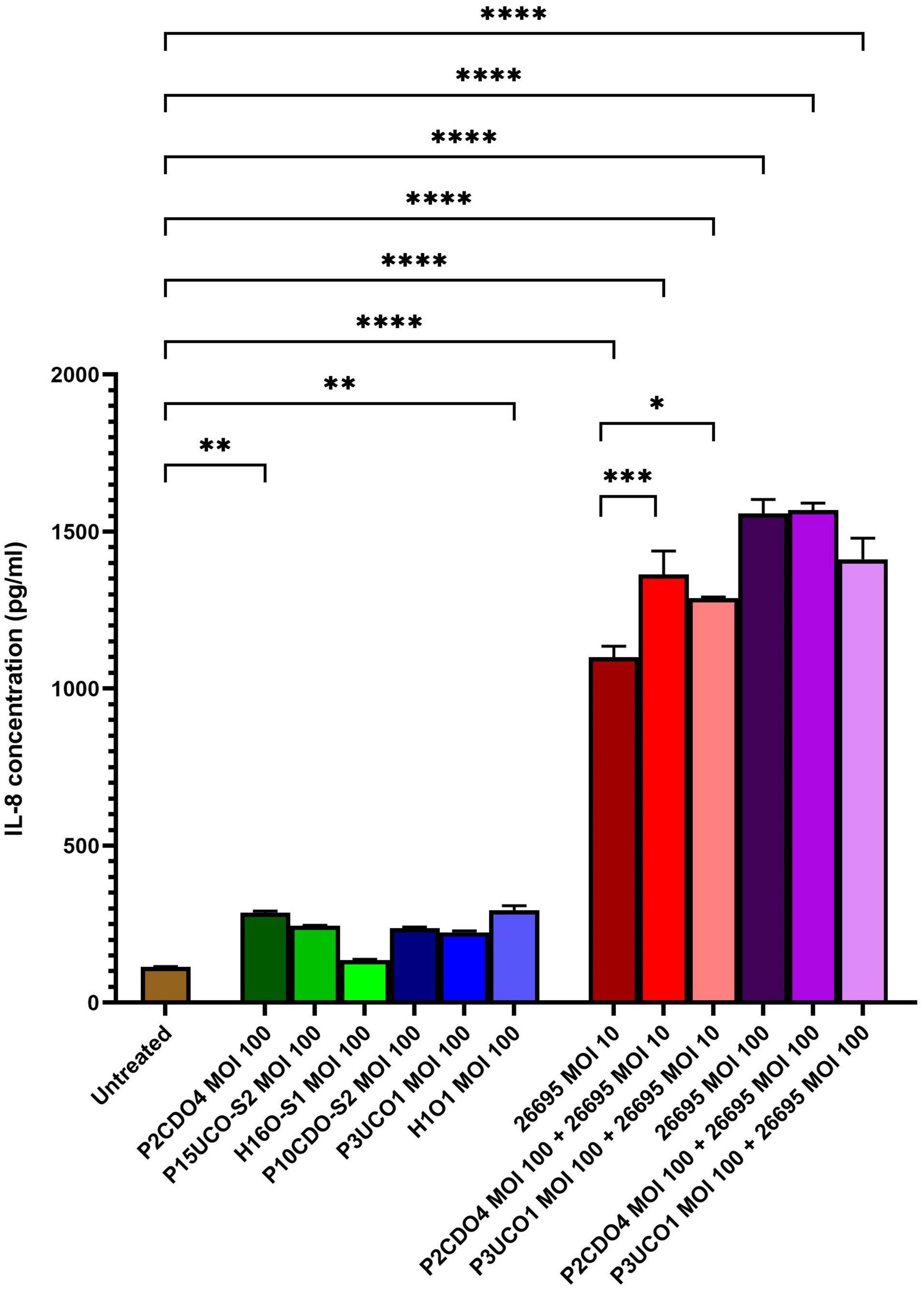
Figure 2. IL-8 production by AGS cells induced by C. concisus strains and H. pylori strain 26695 after 24 h incubation. IL-8 concentrations were measured by ELISA using supernatants of AGS cells after incubating with C. concisus strains and H. pylori strain 26695 for 24 h. C. concisus strains P2CDO4 and H1O1 at MOI 100 induced significantly higher IL-8 productions in AGS cells as compared to the untreated control (P < 0.01). H. pylori strain 26695 at both MOI 10 and 100 induced significantly higher IL-8 productions as compared to C. concisus at MOI 100. Co-incubation of H. pylori at MOI 10 with C. concisus strains (P2CDO4 and P3UCO1) induced significantly higher levels of IL-8 production as compared to that incubated by H. pylori alone (P < 0.001 and P < 0.05 respectively), however this increase was not observed when H. pylori was incubated at MOI 100. One-way analysis of variance (ANOVA) with Dunnett’s test was performed to test for statistical significance between untreated and infected AGS cells. Graph columns represent averages of triplicate experiments ± standard error (****P < 0.0001, ***P < 0.001, **P < 0.01, *P < 0.05; MOI, multiplicity of infection).
In AGS cells incubated with H. pylori strain 26695 at MOI 10, the presence of C. concisus increased the production of IL-8. Co-incubation of H. pylori strain 26695 with P2CDO4 and P3UCO1 resulted in significantly higher levels of IL-8 compared to AGS cell incubated with H. pylori alone, with concentrations of 1363.63 ± 60.97 (P < 0.001) and 1287.99 ± 3.47 pg/ml (P < 0.05), respectively (Figure 2). However, in AGS cells incubated with H. pylori strain 26695 at MOI 100, co-incubation with C. concisus strains P2CDO4 and P3UCO1 induced IL-8 production at 1568.5 ± 18.48 and 1412.19 ± 55 pg/ml, respectively, which was not significantly different from that induced by H. pylori alone (P > 0.05) (Figure 2).
C. concisus induced apoptosis in AGS cells
C. concisus induced apoptosis in AGS cells when incubated with C. concisus strains at MOI 100 for 24 h, as indicated by increased caspase 3/7 activity. The levels of caspase 3/7 activity (expressed as fold change relative to the untreated AGS cells) in AGS cells incubated with C. concisus strains P2CDO4, P15UCO-S2, H16O-S1, P10CDO-S2, P3UCO1, and H1O1 at MOI 100 were 1.47 ± 0.01, 1.67 ± 0.03, 1.48 ± 0.02, 1.58 ± 0.04, 1.56 ± 0.06, and 1.48 ± 0.06 fold, respectively (P < 0.0001) (Figure 3). These values were all significantly higher than that of the untreated AGS cells. The caspase 3/7 activity in AGS cells incubated with H. pylori strain 26695 at MOI 10 and 100 for 24 h was 2.03 ± 0.07 and 2.44 ± 0.06 fold, respectively (P < 0.0001), indicating a higher apoptotic activity compared to that induced by C. concisus strains. Moreover, AGS cells incubated with C. concisus strain P2CDO4 at MOI 100, and P3UCO1 at MOI 100 along with H. pylori strain 26695 at MOI 10 and 100 respectively, also exhibited significantly higher apoptotic activity at 2.4 ± 0.02, 2.57 ± 0.07, 2.71 ± 0.02, and 2.51 ± 0.02 fold, respectively (P < 0.0001).
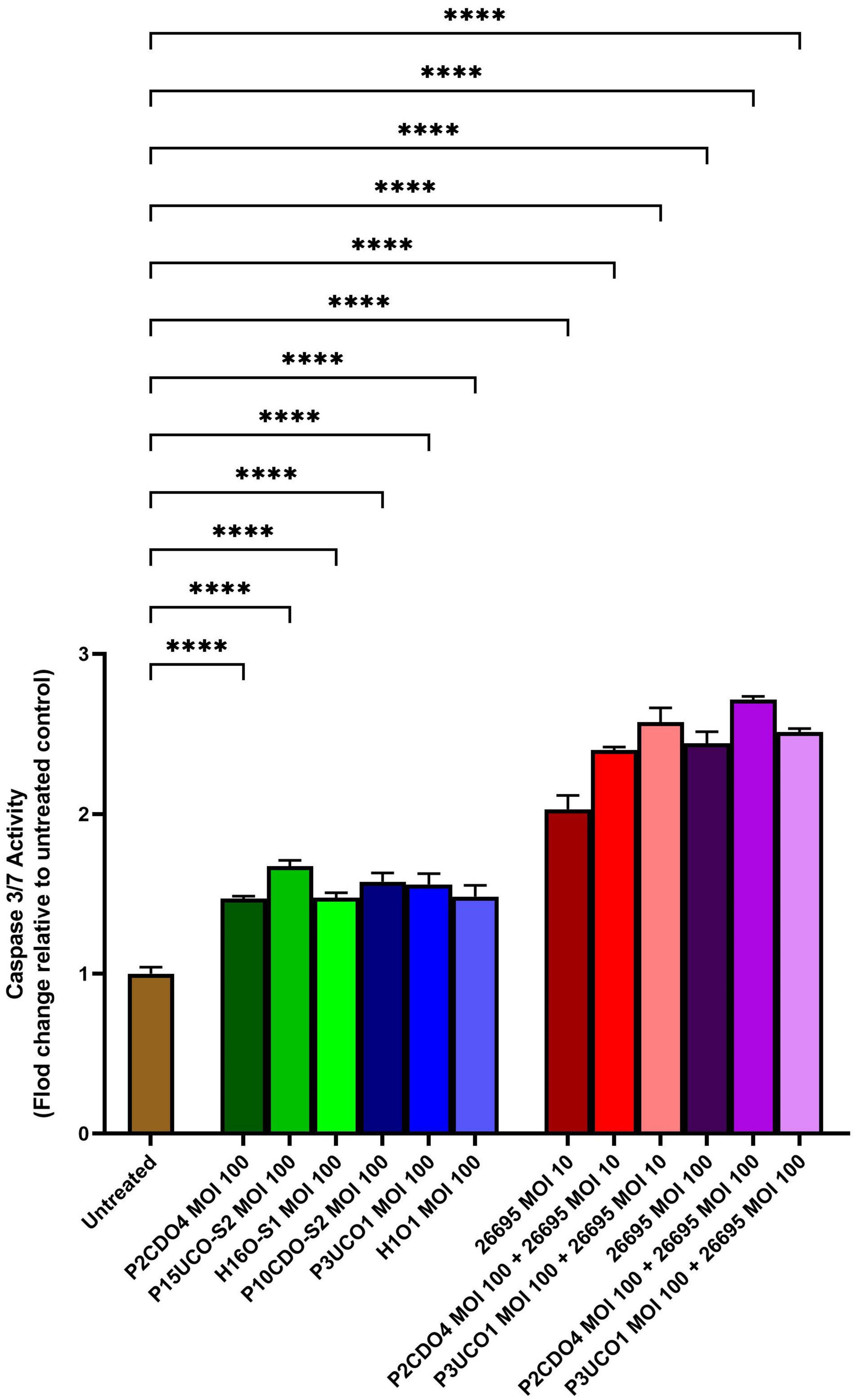
Figure 3. Caspase 3/7 activity of AGS cells infected with C. concisus strains and H. pylori strain 26695 after 24 h. Caspase 3/7 activity was measured using CellEvent caspase 3/7 green detection reagent. All six C. concisus strains at MOI 100, as well as H. pylori strain 26695 at both MOI 10 and 100, induced significantly higher levels of caspase 3/7 activity as compared to the untreated control (P < 0.0001). Co-incubation of C. concisus with H. pylori did not induce higher levels of caspase 3/7 activity as compared to that incubated with H. pylori alone. One-way analysis of variance (ANOVA) with Dunnett’s test was performed to test for statistical significance between untreated and infected AGS cells. Graph columns represent averages of triplicate experiments ± standard error (****P < 0.0001; MOI, multiplicity of infection).
C. concisus induced F-actin aggregation in AGS cells
Both C. concisus strains examined caused F-actin aggregation in comparison to AGS cells without bacterial infection (Figure 4). AGS cells incubated with H. pylori strain 26695 showed sign of elongation, indicative of the hummingbird phenotype caused by the protein encoded by cytotoxin-associated gene A (CagA) in H. pylori (Moese et al., 2004; Chang et al., 2016).
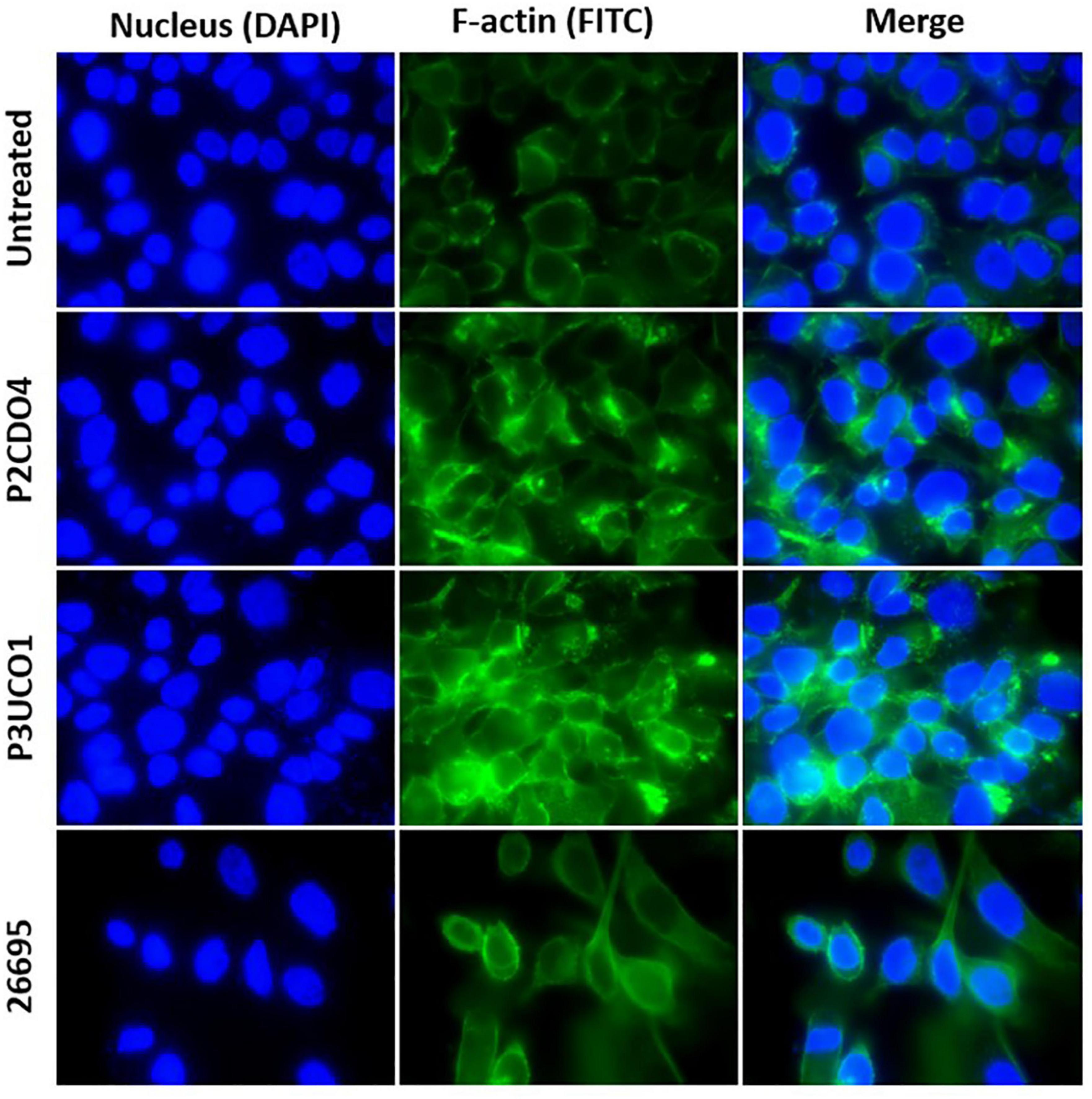
Figure 4. F-actin arrangement in AGS cells incubated with C. concisus strain P2CDO4, P3UCO1, and H. pylori strain 26695 for 24 h. Cell nucleus and F-actin were stained with Hoechst 33342 and Alexa Fluor 488 phalloidin and visualized using DAPI and FITC filters, respectively. Infection of AGS cells with the C. concisus strains P2CDO4 and P3UCO1 showed F-actin aggregation. H. pylori infection caused an elongated cell phenotype of AGS cells.
C. concisus upregulated the expression of gastric cancer associated CYP1A1 gene in AGS cells
Analysis of RNA-seq data revealed notable alterations in global gene response in AGS cells incubated with C. concisus strain P2CDO4 for 4 h, compared to untreated AGS cells (Supplementary Figure 1). From the global gene response, 40 genes were considered significantly altered in expression (adjusted P-value < 0.05, log2 fold change < -1 and >1), of which 30 genes were upregulated and 10 genes downregulated (Figures 5A, B and Supplementary Table 3). The upregulated genes are predicted to predominately affect organelle and mitochondrion organization, and the down regulated genes mainly affect the regulation of cell-cell adhesion (Figures 5C, D). Of the 30 upregulated genes by C. concisus, four genes were reported to be associated with gastric cancer including HSD17B8_2, TRIM27_2, TC2N_1, and CYP1A1. However, three of these genes had an underscore following their gene names, showing that they are on alternate reference locus (Bruford et al., 2020). We therefore decided to further characterize only CYP1A1 gene using qRT-PCR.
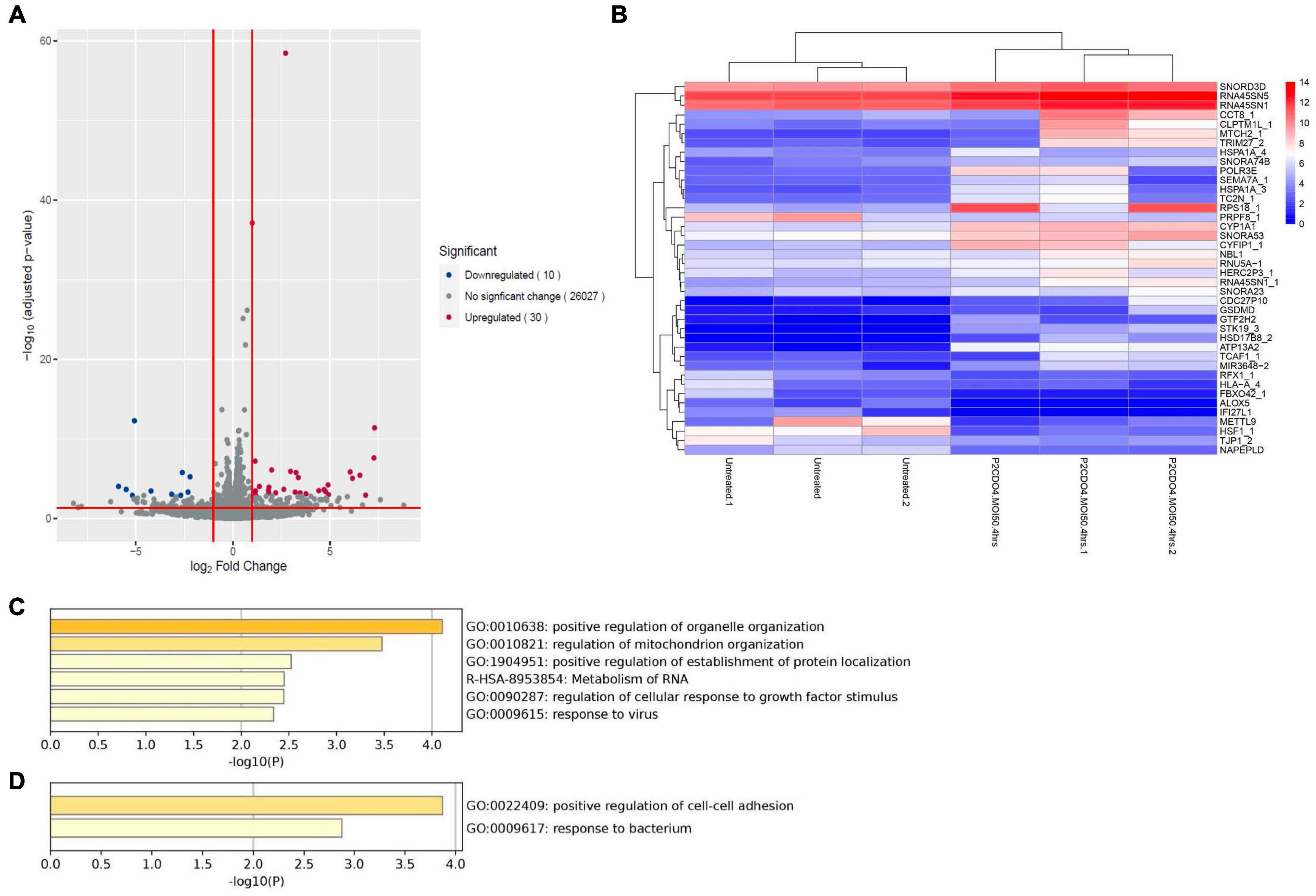
Figure 5. Transcriptomic analysis on global gene expression in AGS cells induced by C. concisus strain P2CDO4 after 4 h incubation. (A) The volcano plot illustrates a total of 26,067 transcripts from RNA-seq reads, with 30 transcripts significantly upregulated (red) and 10 transcripts significantly downregulated (blue). Genes with adjusted P-value < 0.05 and log2 fold change < -1 and >1 were considered statistically significant. Volcano plot generated using the EnhancedVolcano package on the R platform. (B) The heatmap shows the 40 differentially expressed genes based on log2 gene counts. Heatmap generated using the pheatmap package on the R platform. (C) The Gene ontology (GO) enrichment analysis revealed that upregulated genes mainly affect the regulation of organelle and mitochondria organization. (D) The GO enrichment analysis revealed that downregulated genes mainly affect the regulation of cell-cell adhesion.
Transcriptomic analysis revealed that the expression of the upregulated gene CYPIA1 was 6.57 fold in AGS cells incubated with C. concisus P2CDO4 at MOI 50 for 4 h, with a read count of 342.7 ± 35.38, compared to 62.3 ± 5.17 reads in untreated AGS cells (Figure 6A). This was confirmed using by qRT-PCR, which showed a 4.2 ± 0.64 fold increase relative to AGS cells without C. concisus infection (Figure 6B). According to the Kaplan-Meier Plotter survival analysis, a higher level expression of CYP1A1 is associated with poor prognosis in gastric cancer patients, with a hazard ration of 1.86 and median survival period of 20.4 months as opposed to 56.9 months in low expression cohort (Figure 6C).
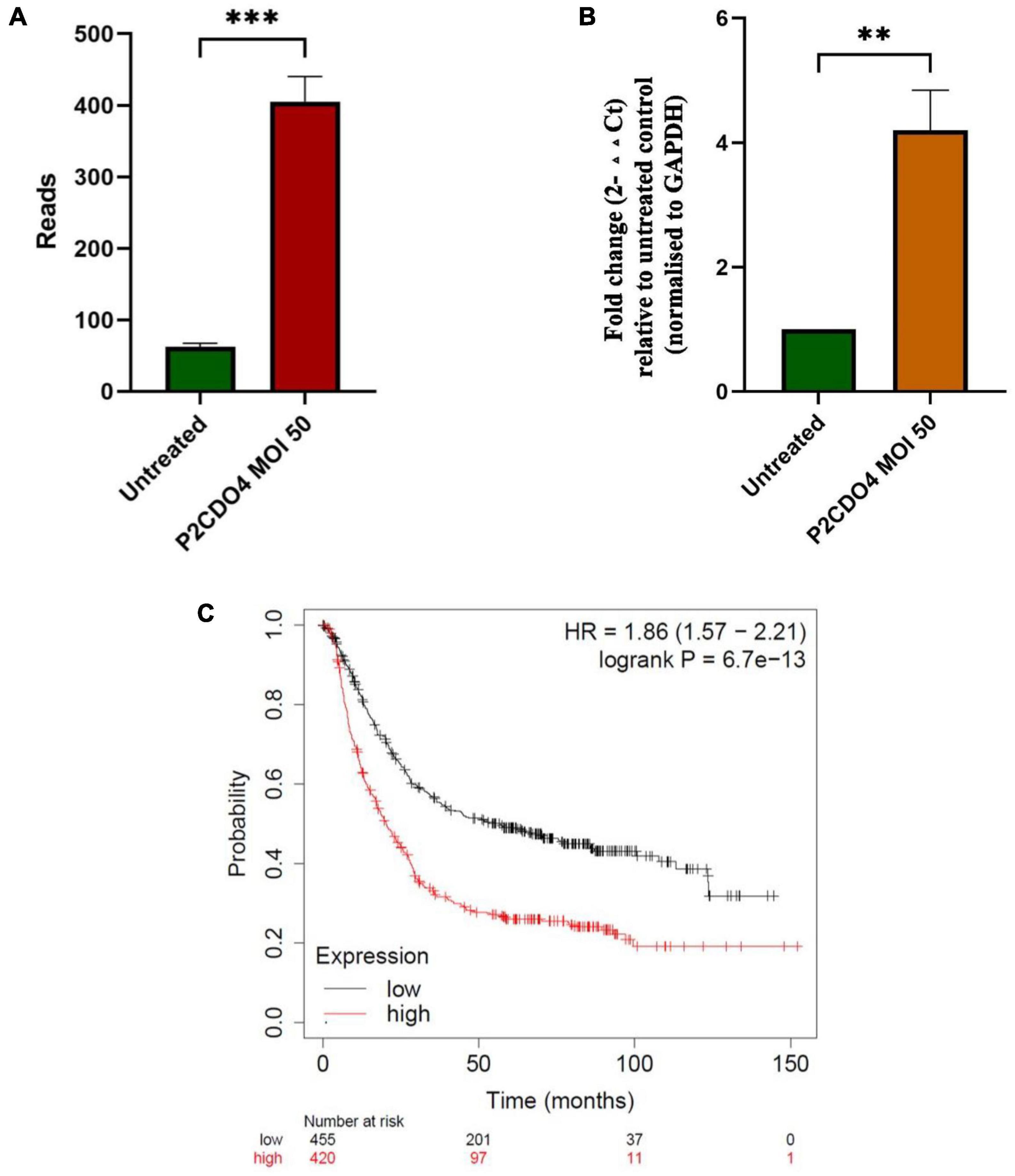
Figure 6. C. concisus strain P2CDO4 upregulated expression of gastric cancer associated CYP1A1 gene in AGS cells. (A) The upregulation of the CYP1A1 gene induced by C. concisus P2CDO4 was shown by gene reads from RNA-seq (***P < 0.001). (B) The upregulation of the CYP1A1 gene was confirmed by qRT-PCR, with the experimental samples being AGS cells incubated with C. concisus strain P2CDO4 at MOI 50 for 4 h, and the untreated AGS cells as negative control. Fold change is calculated using the comparative threshold cycle CT (2–ΔΔCT) method. Target gene fold change is relative to the untreated control, normalized to housekeeping gene GAPDH. Graph columns represent averages of triplicate experiments ± standard error (**P < 0.01; MOI, multiplicity of infection). (C) The survival plot shows that a higher level expression of CYP1A1 is associated with poor prognosis in gastric cancer patients, with a hazard ratio (HR) of 1.86 and median survival period of 20.4 months as opposed to 56.9 months in low expression cohort.
The IL-8 level was 199.96 ± 1.58 pg/ml in the supernatant of AGS cells incubated with C. concisus strain P2CDO4 for 4 h, and 179.83 ± 6.02 pg/ml in the supernatant of untreated AGS cells (P < 0.005).
The RNA-seq data have been deposited in NCBI’s Gene Expression Omnibus and are accessible through GEO Series accession number GSE242316.1
Discussion
This study aimed to investigate the pathogenic effects of C. concisus strains on human gastric epithelial cells using AGS cells as a model for gastric epithelium.
C. concisus strains were found to induce the production of proinflammatory cytokine IL-8 in AGS cells following 24 h of incubation. IL-8, a chemokine known for inducing chemotaxis of neutrophils and other immune cells (Harada et al., 1994). In our study, all C. concisus strains induced the production of IL-8 in AGS cells, suggesting that C. concisus has the potential to induce gastric inflammation. The significant genomic diversity observed among C. concisus strains aligns with their varying capacities to induce IL-8 (Figure 2; Gemmell et al., 2018; Aagaard et al., 2021). C. concisus strains induced much less gastric epithelial production of IL-8, as compared to H. pylori strain 26695 (Figure 2). This finding suggests that C. concisus bacteria are more inclined to cause mild gastric inflammation in comparison to the more robust inflammatory response triggered by H. pylori.
In a study by Ma et al. (2015) it was observed that 4 out of 20 C. concisus strains were able to survive a 30-min exposure to low pH in a test tube. This suggests that certain C. concisus bacteria may endure the gastric environment for a limited period, potentially allowing them to relocate to a safer zone (Ma et al., 2015). In the human stomach, the mucus layer proximal to the epithelial cells maintains a higher pH. Given the motility and spiral to curved shape of C. concisus bacteria, it is conceivable that they could swim toward a more secure region closer to the gastric epithelium, similar to H. pylori. This ability to survive in the gastric environment aligns with findings by Ferreira et al. (2022) who cultured C. concisus from gastric biopsies, providing further evidence for the potential colonization of the human stomach by C. concisus bacteria. However, as C. concisus does not possess urease, their ability in resisting gastric acid would be lower than H. pylori.
Interestingly, we found that co-infection of C. concisus and H. pylori increased the production of IL-8 in AGS cells, suggesting a potential synergistic effect between these bacteria in inducing inflammation. This could have implications for individuals with gastric H. pylori colonization, where the presence of C. concisus might enhance the inflammatory response in gastric epithelial cells. On the other hand, C. concisus did not increase IL-8 production in AGS cells incubated with H. pylori at MOI 100, most likely due to that the AGS cells have reached its maximum capability of producing IL-8 under the stimulation of higher numbers of H. pylori bacteria (Figure 2).
The increase in caspase 3/7 activities in AGS cells following 24 h incubation with C. concisus strains were significant, although H. pylori strain 26695 induced a significantly higher level of caspase 3/7 activity than C. concisus (Figure 3). This suggests that H. pylori has caused more damage to gastric epithelial cells than C. concisus under the same MOI, which apoptotic activity in gastric epithelial cells was known to increase in H. pylori-induced gastritis (Dang et al., 2020). The co-infection of C. concisus and H. pylori further increased the level of caspase 3/7 activity in AGS cells, suggesting a potential synergistic effect on apoptosis (Figure 3). Our findings suggest that induction of apoptosis is a potential mechanism by which C. concisus may contribute to gastric diseases, especially when coexisting with H. pylori.
C. concisus induced F-actin aggregation in AGS cells. The actin cytoskeleton regulates various cellular processes in eukaryotic cells, providing structural integrity at cell-cell junctions and promote membrane extensions (Hartsock and Nelson, 2008; Svitkina, 2018). Actin filaments are frequently targeted by bacterial pathogens, including H. pylori. H. pylori adheres to gastric epithelial cells via adhesins BabA and SabA and delivers CagA and other bacterial factors into the host epithelial cytosol. This process results in the phosphorylation and dephosphorylation of proteins in different pathways, rearrangement of actin and ZO-1, and disruption of the gastric epithelial barrier. Previous studies have reported that H. pylori CagA can induce a “hummingbird” phenotype change in gastric epithelial cells, which was also observed in our experiment (Figure 4). Interestingly, we found that C. concisus caused a different type of actin rearrangement, specifically actin aggregations (Figure 4). The virulence factors of C. concisus that affect the epithelial actin arrangement and their mechanisms are not clear, warranting further investigation in future studies.
C. concisus bacteria transported from the oral cavity to the stomach through swallowed saliva may not establish long-term gastric colonization. To investigate potential changes in gene expression after short-term contact with gastric epithelial cells, we investigated the global gastric epithelial gene responses to C. concisus using transcriptomic analysis following a 4-h incubation at a lower dose (MOI 50). Global gene response in AGS cells incubated with C. concisus strain P2CDO4 for 4 h was altered when compared to the untreated AGS cells (Supplementary Figure 1), of which a total of 30 genes and 10 genes were significantly upregulated and downregulated, respectively, which were predicted to affect several cellular functional pathways (Figure 5 and Supplementary Table 3). These findings suggests that C. concisus bacteria passing through the stomach from the oral cavity may exert regulatory effects on gastric epithelial cell gene expression. We further confirmed the upregulation of expression of the gene coding CYP1A1 through qRT-PCR. CYP1A1, a cytochrome P450 enzyme involved in metabolism of xenobiotics (Androutsopoulos et al., 2009), has been associated with gastric cancer and precancerous gastric cancer as well as lung cancer (Kouri et al., 1982; Zhang et al., 2004; Hidaka et al., 2016; Huang et al., 2021; Sadeghi-Amiri et al., 2021). Previous research by Cui et al. (2019) linked the abundance of C. concisus in tongue coating microbiome to gastric precancerous cascade, which aligns with our finding of C. concisus upregulating CYP1A1 expression. This provides additional evidence that oral C. concisus may play a role in facilitating gastric cancer development or progression. This is further supported by the survival analysis of CYP1A1 in gastric cancer patients (Figure 6).
In summary, our study reveals that incubation of C. concisus with human gastric epithelial cells (AGS cells) induces the production of IL-8, apoptosis, and upregulation of CYP1A1 gene expression in gastric epithelial cells. These findings suggest that C. concisus may play a role in gastric inflammation and the progression of gastric cancer. Further investigation in clinical studies is warranted.
Data availability statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in this article/Supplementary material.
Ethics statement
Ethical approval was not required for the studies on humans in accordance with the local legislation and institutional requirements because only commercially available established cell lines were used.
Author contributions
CL: Formal analysis, Writing – original draft, Investigation. SL: Methodology, Writing – review and editing. NN: Writing – review and editing, Investigation. FL: Methodology, Writing – review and editing. AT: Funding acquisition, Writing – review and editing. LW: Writing – review and editing, Conceptualization, Funding acquisition. SR: Writing – review and editing, Conceptualization. LZ: Writing – review and editing, Conceptualization, Funding acquisition, Supervision.
Funding
The author(s) declare financial support was received for the research, authorship, and/or publication of this article. This project was supported by the Xuzhou Medical University Open Research Grant (XYKF202102) and the Faculty Research Grant (Grant no. PS46772) of the University of New South Wales awarded to Associate Professor LZ.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
The author(s) declared that they were an editorial board member of Frontiers, at the time of submission. This had no impact on the peer review process and the final decision.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2023.1289549/full#supplementary-material
Supplementary Figure 1 | Principal component analysis (PCA) plot of RNA-seq data. A total of 6 samples were included in the analysis, red dots for treatment with C. concisus strain P2CDO4 and blue dots for untreated control. The plot indicates separation between control and treatment groups, suggesting significant changes in overall gene response in AGS cells upon C. concisus infection. Gene counts were normalized using log2 transformation. PCA plot generated using the ggplot2 package on the R platform.
Footnotes
References
Aagaard, M. E. Y., Kirk, K. F., Nielsen, H., and Nielsen, H. L. (2021). High genetic diversity in Campylobacter concisus isolates from patients with microscopic colitis. Gut Pathog. 13, 1–5. doi: 10.1186/s13099-020-00397-y
Androutsopoulos, V. P., Tsatsakis, A. M., and Spandidos, D. A. (2009). Cytochrome P450 CYP1A1: Wider roles in cancer progression and prevention. BMC Cancer 9:187. doi: 10.1186/1471-2407-9-187
Ashktorab, H., Dashwood, R. H., Dashwood, M. M., Zaidi, S. I., Hewitt, S. M., Green, W. R., et al. (2008). H. pylori-induced apoptosis in human gastric cancer cells mediated via the release of apoptosis-inducing factor from mitochondria. Helicobacter 13, 506–517. doi: 10.1111/j.1523-5378.2008.00646.x
Benoit, S. L., and Maier, R. J. (2023). The Campylobacter concisus BisA protein plays a dual role: Oxide-dependent anaerobic respiration and periplasmic methionine sulfoxide repair. mBio 14:e01475-23. doi: 10.1128/mbio.01475-23
Bolger, A. M., Lohse, M., and Usadel, B. (2014). Trimmomatic: A flexible trimmer for Illumina sequence data. Bioinformatics 30, 2114–2120.
Bruford, E. A., Braschi, B., Denny, P., Jones, T. E., Seal, R. L., and Tweedie, S. (2020). Guidelines for human gene nomenclature. Nat. Genet. 52, 754–758.
Chang, C.-C., Kuo, W.-S., Chen, Y.-C., Perng, C.-L., Lin, H.-J., and Ou, Y.-H. (2016). Fragmentation of CagA reduces hummingbird phenotype induction by Helicobactor pylori. PLoS One 11:e0150061. doi: 10.1371/journal.pone.0150061
Chung, H. K. L., Tay, A., Octavia, S., Chen, J., Liu, F., Ma, R., et al. (2016). Genome analysis of Campylobacter concisus strains from patients with inflammatory bowel disease and gastroenteritis provides new insights into pathogenicity. Sci. Rep. 6, 1–14. doi: 10.1038/srep38442
Cornelius, A. J., Huq, M., On, S. L., French, N. P., Vandenberg, O., Miller, W. G., et al. (2021). Genetic characterisation of Campylobacter concisus: Strategies for improved genomospecies discrimination. Syst. Appl. Microbiol. 44:126187. doi: 10.1016/j.syapm.2021.126187
Cui, J., Cui, H., Yang, M., Du, S., Li, J., Li, Y., et al. (2019). Tongue coating microbiome as a potential biomarker for gastritis including precancerous cascade. Protein Cell 10, 496–509. doi: 10.1007/s13238-018-0596-6
Danecek, P., Bonfield, J. K., Liddle, J., Marshall, J., Ohan, V., Pollard, M. O., et al. (2021). Twelve years of SAMtools and BCFtools. Gigascience 10:giab008. doi: 10.1093/gigascience/giab008
Dang, Y., Zhang, Y., Xu, L., Zhou, X., Gu, Y., Yu, J., et al. (2020). PUMA-mediated epithelial cell apoptosis promotes Helicobacter pylori infection-mediated gastritis. Cell Death Dis. 11:139. doi: 10.1038/s41419-020-2339-x
Fazeli, Z., Alebouyeh, M., Tavirani, M. R., Azimirad, M., and Yadegar, A. (2016). Helicobacter pylori CagA induced interleukin-8 secretion in gastric epithelial cells. Gastroenterol. Hepatol. Bed Bench 9(Suppl.1), S42–S46.
Ferreira, E. O., Lagacé-Wiens, P., and Klein, J. (2022). Campylobacter concisus gastritis masquerading as Helicobacter pylori on gastric biopsy. Helicobacter 27:e12864. doi: 10.1111/hel.12864
Gemmell, M. R., Berry, S., Mukhopadhya, I., Hansen, R., Nielsen, H. L., Bajaj-Elliott, M., et al. (2018). Comparative genomics of Campylobacter concisus: Analysis of clinical strains reveals genome diversity and pathogenic potential. Emerg. Microbes Infect. 7, 1–17. doi: 10.1038/s41426-018-0118-x
Harada, A., Sekido, N., Akahoshi, T., Wada, T., Mukaida, N., and Matsushima, K. (1994). Essential involvement of interleukin-8 (IL-8) in acute inflammation. J. Leukoc. Biol. 56, 559–564.
Hartsock, A., and Nelson, W. J. (2008). Adherens and tight junctions: Structure, function and connections to the actin cytoskeleton. Biochim. Biophys. Acta Biomembr. 1778, 660–669. doi: 10.1016/j.bbamem.2007.07.012
Hidaka, A., Sasazuki, S., Matsuo, K., Ito, H., Charvat, H., Sawada, N., et al. (2016). CYP1A1, GSTM1 and GSTT1 genetic polymorphisms and gastric cancer risk among Japanese: A nested case–control study within a large-scale population-based prospective study. Int. J. Cancer 139, 759–768. doi: 10.1002/ijc.30130
Huang, L., He, R., Zhang, Y., Yan, Q., and Veronica, C. (2021). “The role of the aryl hydrocarbon receptor (AhR) in the immune response against microbial infections,” in Antimicrobial Immune Response, ed. O. Maria del Mar (London: IntechOpen).
Huq, M., Van, T. T. H., Gurtler, V., Elshagmani, E., Allemailem, K. S., Smooker, P. M., et al. (2017). The ribosomal RNA operon (rrn) of Campylobacter concisus supports molecular typing to genomospecies level. Gene Rep. 6, 8–14.
Ismail, Y., Mahendran, V., Octavia, S., Day, A. S., Riordan, S. M., Grimm, M. C., et al. (2012). Investigation of the enteric pathogenic potential of oral Campylobacter concisus strains isolated from patients with inflammatory bowel disease. PLoS One 7:e38217. doi: 10.1371/journal.pone.0038217
Istivan, T. S., Coloe, P. J., Fry, B. N., Ward, P., and Smith, S. C. (2004). Characterization of a haemolytic phospholipase A2 activity in clinical isolates of Campylobacter concisus. J. Med. Microbiol. 53, 483–493. doi: 10.1099/jmm.0.45554-0
Kalischuk, L. D., and Inglis, G. D. (2011). Comparative genotypic and pathogenic examination of Campylobacter concisus isolates from diarrheic and non-diarrheic humans. BMC Microbiol. 11:53. doi: 10.1186/1471-2180-11-53
Kim, D., Paggi, J. M., Park, C., Bennett, C., and Salzberg, S. L. (2019). Graph-based genome alignment and genotyping with HISAT2 and HISAT-genotype. Nat. Biotechnol. 37, 907–915. doi: 10.1038/s41587-019-0201-4
Kirk, K. F., Méric, G., Nielsen, H. L., Pascoe, B., Sheppard, S. K., Thorlacius-Ussing, O., et al. (2018). Molecular epidemiology and comparative genomics of Campylobacter concisus strains from saliva, faeces and gut mucosal biopsies in inflammatory bowel disease. Sci. Rep. 8, 1–8. doi: 10.1038/s41598-018-20135-4
Kouri, R., McKinney, C., Slomiany, D., Snodgrass, D., Wray, N., and McLemore, T. (1982). Positive correlation between high aryl hydrocarbon hydroxylase activity and primary lung cancer as analyzed in cryopreserved lymphocytes. Cancer Res. 42, 5030–5037.
Lánczky, A., and Gyõrffy, B. (2021). Web-based survival analysis tool tailored for medical research (KMplot): Development and implementation. J. Med. Internet Res. 23:e27633. doi: 10.2196/27633
Lee, H., Ma, R., Grimm, M. C., Riordan, S. M., Lan, R., Zhong, L., et al. (2014). Examination of the anaerobic growth of Campylobacter concisus strains. Int. J. Microbiol 2014, 476047. doi: 10.1155/2014/476047
Lee, S. A., Liu, F., Yun, D. Y., Riordan, S. M., Tay, A. C. Y., Liu, L., et al. (2021). Campylobacter concisus upregulates PD-L1 mRNA expression in IFN-γ sensitized intestinal epithelial cells and induces cell death in esophageal epithelial cells. J. Oral Microbiol. 13:1978732.
Lee, S. A., Liu, F., Yuwono, C., Phan, M., Chong, S., Biazik, J., et al. (2023). Emerging Aeromonas enteric infections: Their association with inflammatory bowel disease and novel pathogenic mechanisms. Microbiol. Spectr. 11:e01088-23 doi: 10.1128/spectrum.01088-23
Liao, Y., Smyth, G. K., and Shi, W. (2014). featureCounts: An efficient general purpose program for assigning sequence reads to genomic features. Bioinformatics 30, 923–930. doi: 10.1093/bioinformatics/btt656
Liu, F., Chen, S., Luu, L. D. W., Lee, S. A., Tay, A. C. Y., Wu, R., et al. (2020). Analysis of complete Campylobacter concisus genomes identifies genomospecies features, secretion systems and novel plasmids and their association with severe ulcerative colitis. Microb. Genom 6:mgen000457. doi: 10.1099/mgen.0.000457
Liu, F., Ma, R., Tay, C. Y. A., Octavia, S., Lan, R., Chung, H. K. L., et al. (2018). Genomic analysis of oral Campylobacter concisus strains identified a potential bacterial molecular marker associated with active Crohn’s disease. Emerg. Microbes Infect. 7, 1–14. doi: 10.1038/s41426-018-0065-6
Livak, K. J., and Schmittgen, T. D. (2001). Analysis of relative gene expression data using real-time quantitative PCR and the 2- ΔΔCT method. Methods 25, 402–408.
Love, M. I., Huber, W., and Anders, S. (2014). Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 15, 1–21. doi: 10.1186/s13059-014-0550-8
Ma, R., Sapwell, N., Chung, H. K. L., Lee, H., Mahendran, V., Leong, R. W., et al. (2015). Investigation of the effects of pH and bile on the growth of oral Campylobacter concisus strains isolated from patients with inflammatory bowel disease and controls. J. Med. Microbiol. 64, 438–445. doi: 10.1099/jmm.0.000013
Macfarlane, S., Furrie, E., Macfarlane, G. T., and Dillon, J. F. (2007). Microbial colonization of the upper gastrointestinal tract in patients with Barrett’s esophagus. Clin. Infect. Dis. 45, 29–38.
Mahendran, V., Liu, F., Riordan, S. M., Grimm, M. C., Tanaka, M. M., and Zhang, L. (2016). Examination of the effects of Campylobacter concisus zonula occludens toxin on intestinal epithelial cells and macrophages. Gut Pathog. 8, 1–10. doi: 10.1186/s13099-016-0101-9
Mahendran, V., Octavia, S., Demirbas, O. F., Sabrina, S., Ma, R., Lan, R., et al. (2015). Delineation of genetic relatedness and population structure of oral and enteric Campylobacter concisus strains by analysis of housekeeping genes. Microbiol. 161, 1600–1612. doi: 10.1099/mic.0.000112
Man, S. M., Zhang, L., Day, A. S., Leach, S. T., Lemberg, D. A., and Mitchell, H. (2010). Campylobacter concisus and other Campylobacter species in children with newly diagnosed Crohn’s disease. Inflamm. Bowel Dis. 16, 1008–1016. doi: 10.1002/ibd.21157
Miller, W. G., Chapman, M. H., Yee, E., On, S. L., McNulty, D. K., Lastovica, A. J., et al. (2012). Multilocus sequence typing methods for the emerging Campylobacter species C. hyointestinalis, C. lanienae, C. sputorum, C. concisus, and C. curvus. Front. Cell. Infect. Microbiol. 2:45. doi: 10.3389/fcimb.2012.00045
Moese, S., Selbach, M., Kwok, T., Brinkmann, V., König, W., Meyer, T. F., et al. (2004). Helicobacter pylori induces AGS cell motility and elongation via independent signaling pathways. Infect. Immun. 72, 3646–3649. doi: 10.1128/IAI.72.6.3646-3649.2004
Mukhopadhya, I., Thomson, J. M., Hansen, R., Berry, S. H., El-Omar, E. M., and Hold, G. L. (2011). Detection of Campylobacter concisus and other Campylobacter species in colonic biopsies from adults with ulcerative colitis. PLoS One 6:e21490. doi: 10.1371/journal.pone.0021490
Nielsen, H. L., Dalager-Pedersen, M., and Nielsen, H. (2020). High risk of microscopic colitis after Campylobacter concisus infection: Population-based cohort study. Gut 69, 1952–1958. doi: 10.1136/gutjnl-2019-319771
O’Hara, A. M., Bhattacharyya, A., Mifflin, R. C., Smith, M. F., Ryan, K. A., Scott, K. G.-E., et al. (2006). Interleukin-8 induction by Helicobacter pylori in gastric epithelial cells is dependent on apurinic/apyrimidinic endonuclease-1/redox factor-1. J. Immunol. 177, 7990–7999. doi: 10.4049/jimmunol.177.11.7990
Sadeghi-Amiri, L., Barzegar, A., Nikbakhsh-Zati, N., and Mehraban, P. (2021). Hypomethylation of the XRE- 1383 site is associated with the upregulation of CYP1A1 in gastric adenocarcinoma. Gene 769:145216. doi: 10.1016/j.gene.2020.145216
Svitkina, T. (2018). The actin cytoskeleton and actin-based motility. Cold Spring Harb. Perspect. Biol. 10:a018267.
Wang, Y., Liu, F., Zhang, X., Chung, H. K. L., Riordan, S. M., Grimm, M. C., et al. (2017). Campylobacter concisus genomospecies 2 is better adapted to the human gastrointestinal tract as compared with Campylobacter concisus genomospecies 1. Front. Physiol. 8:543. doi: 10.3389/fphys.2017.00543
Yang, L., Lu, X., Nossa, C. W., Francois, F., Peek, R. M., and Pei, Z. (2009). Inflammation and intestinal metaplasia of the distal esophagus are associated with alterations in the microbiome. Gastroenterology 137, 588–597. doi: 10.1053/j.gastro.2009.04.046
Zhang, K.-L., Ma, J.-X., Chen, X.-Y., Sun, Y., Kong, Q.-Y., Liu, J., et al. (2004). Frequent CYP1A1 expression in gastric cancers and their related lesions. Oncol. Rep. 12, 1335–1340.
Zhang, L., Budiman, V., Day, A. S., Mitchell, H., Lemberg, D. A., Riordan, S. M., et al. (2010). Isolation and detection of Campylobacter concisus from saliva of healthy individuals and patients with inflammatory bowel disease. J. Clin. Microbiol. 48, 2965–2967. doi: 10.1128/JCM.02391-09
Zhang, L., Man, S. M., Day, A. S., Leach, S. T., Lemberg, D. A., Dutt, S., et al. (2009). Detection and isolation of Campylobacter species other than C. jejuni from children with Crohn’s disease. J. Clin. Microbiol. 47, 453–455.
Zhang, Y., Takeuchi, H., Nishioka, M., Morimoto, N., Kamioka, M., Kumon, Y., et al. (2009). Relationship of IL-8 production and the CagA status in AGS cells infected with Helicobacter pylori exposed to low pH and activating transcription factor 3 (ATF3). Microbiol. Res. 164, 180–190. doi: 10.1016/j.micres.2006.10.010
Keywords: Campylobacter concisus, Campylobacter, gastritis, oral Campylobacter, CYP1A1
Citation: Luk CYM, Lee SA, Naidovski N, Liu F, Tay ACY, Wang L, Riordan S and Zhang L (2024) Investigation of Campylobacter concisus gastric epithelial pathogenicity using AGS cells. Front. Microbiol. 14:1289549. doi: 10.3389/fmicb.2023.1289549
Received: 06 September 2023; Accepted: 19 December 2023;
Published: 11 January 2024.
Edited by:
Stuart A. Thompson, Augusta University, United StatesReviewed by:
Sankarasubramanian Jagadesan, University of Nebraska Medical Center, United StatesMohsina Huq, Qassim University, Saudi Arabia
Copyright © 2024 Luk, Lee, Naidovski, Liu, Tay, Wang, Riordan and Zhang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Li Zhang, TC5aaGFuZ0B1bnN3LmVkdS5hdQ==; Liang Wang, d2FuZ2xpYW5nQGdkcGgub3JnLmNu
 Christopher Yau Man Luk
Christopher Yau Man Luk Seul A. Lee1
Seul A. Lee1 Fang Liu
Fang Liu Alfred Chin Yen Tay
Alfred Chin Yen Tay Liang Wang
Liang Wang Stephen Riordan
Stephen Riordan Li Zhang
Li Zhang