- 1Laboratory of Microbiology, Department of Clinical Laboratory, The First Hospital of China Medical University, Shenyang, Liaoning, China
- 2National Clinical Research Center for Laboratory Medicine, Department of Clinical Laboratory, The First Hospital of China Medical University, Shenyang, Liaoning, China
- 3Department of Information Centre, The First Hospital of China Medical University, Shenyang, Liaoning, China
Introduction: Candida auris, a fungal pathogen first reported in 2009, has shown strong resistance to azole antifungal drugs and has caused severe nosocomial outbreaks. It can also form biofilms, which can colonize patients’ skin and transmit to others. Despite numerous reports of C. auris isolation in various countries, many studies have reported contradictory results.
Method: A bibliometric analysis was conducted using VOSviewer to summarize research trends and provide guidance for future research on controlling C. auris infection. The analysis revealed that the United States and the US CDC were the most influential countries and research institutions, respectively. For the researchers, Jacques F. Meis published the highest amount of related articles, and Anastasia P. Litvintseva’s articles with the highest average citation rate. The most cited publications focused on clade classification, accurate identification technologies, nosocomial outbreaks, drug resistance, and biofilm formation. Keyword co-occurrence analysis revealed that the top five highest frequencies were for ‘drug resistance,’ ‘antifungal susceptibility test,’ ‘infection,’ ‘Candida auris,’ and ‘identification.’ The high-frequency keywords clustered into four groups: rapid and precise identification, drug resistance research, pathogenicity, and nosocomial transmission epidemiology studies. These clusters represent different study fields and current research hotspots of C. auris.
Conclusion: The bibliometric analysis identified the most influential country, research institution, and researcher, indicating current research trends and hotspots for controlling C. auris.
1 Introduction
Candida auris was first reported in 2009 and was isolated from the external auditory canal of a Japanese patient (Satoh et al., 2009). Subsequently, there have been a number of reports of C. auris being isolated in several other countries, and a retrospective analysis of preserved strains confirmed that C. auris existed in many countries before 2009; however, its presence was not discovered due to challenges associated with its biological identification (Kim et al., 2009; Shin et al., 2012; Desnos-Ollivier et al., 2021). Owing to its resistance to azole antifungal drugs and its tendency to cause outbreaks in hospitals, it has attracted considerable attention from the medical community and has become a major threat to public health (Lockhart et al., 2017).
The rapid and accurate identification of C. auris is essential for controlling nosocomial outbreaks. At present, traditional identification methods can easily be used to identify C. haemulonii, C. glabrata and other Candida species (Parra-Giraldo et al., 2018). Candida auris is usually resistant to fluconazole, and certain clades of C. auris exhibit a higher MIC against azole antifungals than do other Candida species (Chowdhary et al., 2017). Consequently, the mistaken identification of C. auris not only delays patient treatment but also the optimal timing of nosocomial infection control strategies, resulting in nosocomial outbreaks of C. auris. With the application of new technologies, such as matrix-assisted laser desorption ionization-time of flight mass spectrometry (MALDI-TOF MS), internally transcribed spacer (ITS) sequencing, and other molecular technologies (Borman et al., 2017), the success rate of C. auris identification has also increased.
Studies have shown that C. auris quickly adapts to azoles, as well as other antifungal drugs, and increases its MIC against these drugs (Bravo Ruiz et al., 2019; Carolus et al., 2021), which complicates the treatment of infectious diseases caused by C. auris. In addition, C. auris is able to form biofilms (Eix et al., 2022). Although C. auris grows relatively slowly in Yeast Extract Peptone Dextrose (YEPD or YPD) medium, it can quickly form a dense biofilm on irregular folds of human skin without breaking through the skin tissue, which explains its continuous colonization of patients’ skin and its transmission among patients and between patients and the environment. C. auris does not have as strong of an ability to produce hyphae as does C. albicans; however, it still exhibits considerable virulence (Yue et al., 2018), with different virulence levels exhibited by different strains (Carvajal et al., 2021). Since its discovery, C. auris has become one of the main fungal pathogens due to its high drug resistance, difficulty to treat, and its ability to cause large-scale outbreaks.
Although there are many studies on C. auris, most are based on a small number of strains, and many of these studies have reported contradictory results. Therefore, caution should be exercised when interpreting these data and results, and more in-depth hotspot-based research with a larger number of strains should be conducted to address the challenges associated with C. auris. Bibliometric applies mathematical and statistical methods to analyze changes in the number, distribution, and patterns of published article to describe, compare, and evaluate the hotspots, trends, and contributions of scholars, journals, and countries. To the best of our knowledge, no bibliometric analysis has been conducted on the quantity and quality of C. auris research. Therefore, there is an urgent need to summarize the current research status of C. auris and to help predict promising keywords and trends. This study aimed to evaluate the research status and trends in C. auris to help researchers discover research hotspots in this field and provide a reference for further research.
2 Materials and methods
2.1 Source database and search strategy
In this study, all article data on C. auris was retrieved from the Web of Science (WoS) Core Collection. As the largest comprehensive academic information resource, Web of Science has been utilized as a great tool prior bibliometric studies (Waqas et al., 2020; Ahmad et al., 2021; Saheb et al., 2021; Lin et al., 2022), we chose to use WOS as the data source in this study. The WoS Core Collection database is updated continuously, and it assesses all publications dynamically and rigorously; therefore, it was used for the bibliometric analysis. A WoS-based article analysis was conducted to obtain general information on year of publication, journal, organization, author, and research area distribution. An article search was conducted on the 28 of March 2023. All bibliometric data were downloaded on 28 March 2023 to avoid the impact of database updates.Concerning data collection, the following search strategy was developed: Topic = (‘Candida auris’). The language of publication was set to ‘English’. The document type was set to ‘Article’; and the timespan was not set. In total, 750 studies were identified using these search criteria. The ‘Full Record and Cited References’ for the 750 publications were downloaded in ‘Plain Text’ format. As a result, 750 publications were included in the final dataset.
2.2 Software application and visualization analysis
VOSviewer is a software tool designed to analyze academic records and create maps based on network data. In this study, all valid data downloaded from the WoSCC database were imported into VOSviewer [version 1.6.19] for visualization analysis, including co-authorship analysis of institutions and authors, co-citation analysis of references, and co-occurrence analysis of keywords. In order to avoid any biases, there were two researchers independently proceed bibliometric strategies. Through the comparison of outcomes, these two independent collected data sets displayed highly consistency.
3 Results
3.1 Analysis of publications and citation counts
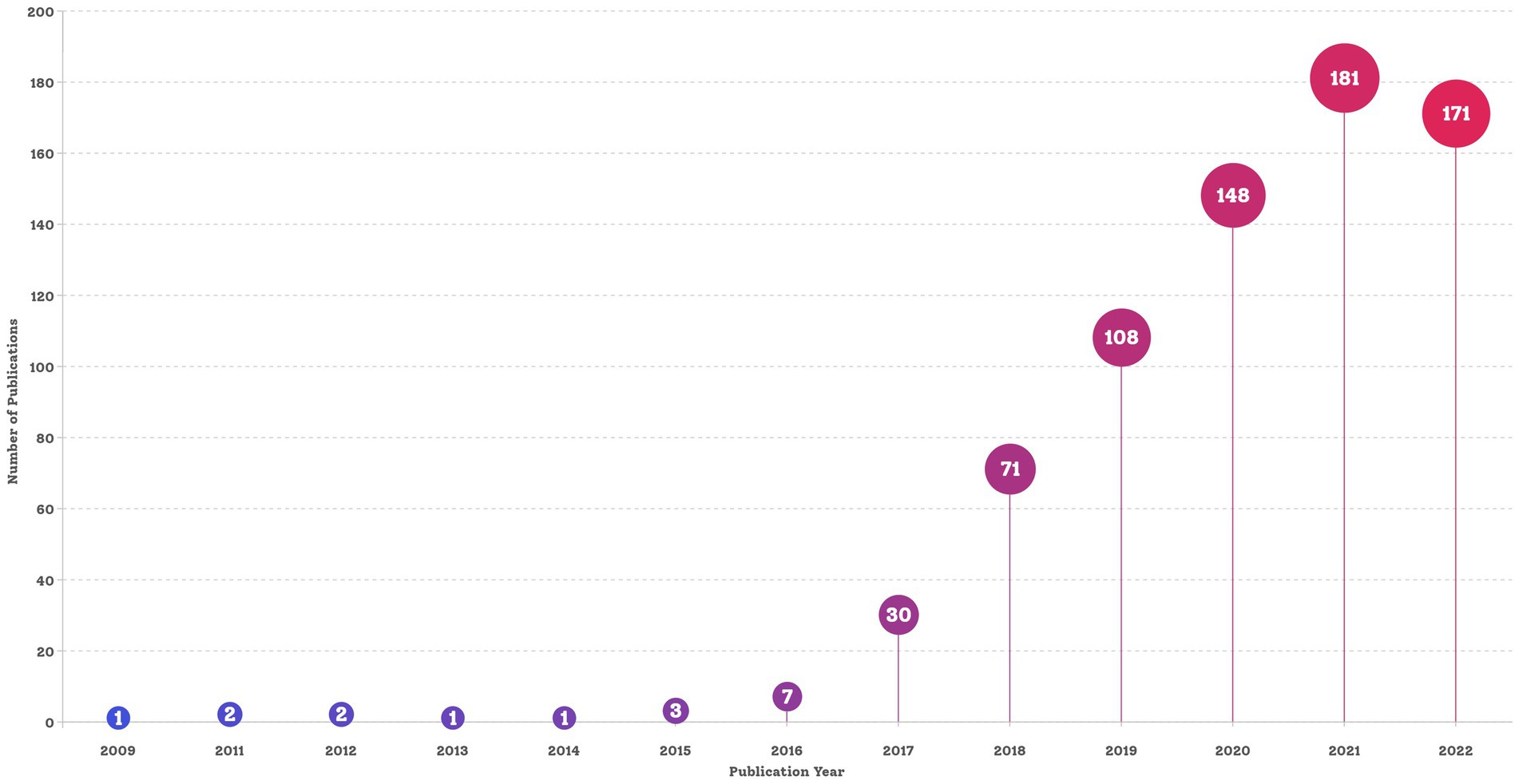
The final dataset contained 750 bibliographic records of English articles reporting on C. auris that were published before 28 March 2023. The search strategy and annual number of publications is shown in Figures 1, 2, respectively. The data for 2023 were not complete annual data; therefore, they are not displayed in Figure 2. The first report on C. auris was published in 2009. Over the past 13 years, the trajectory has exhibited two phases: an initial period (2009–2016), with only a small number of publications and a slow development pace, and a period of growth (2017–2022), with the number of annual publications peaking at 181 in 2021. The number of publications increased from 1 in 2009 to 171 in 2022, with an average of 2.1 in the period of 2009–2016 and 113.2 in the period of 2017–2022. The number of publications in the 3-year period of 2020–2022 reached 66.67% of the total number of publications (500/750). According to the number of citations, we selected 20 highly cited articles (with more than 100 citations) and summarized them in Supplementary Table S1. These articles covered different fields related to C. auris, including biofilm formation, identification methods, multi-drug resistance mechanisms, and nosocomial infections.
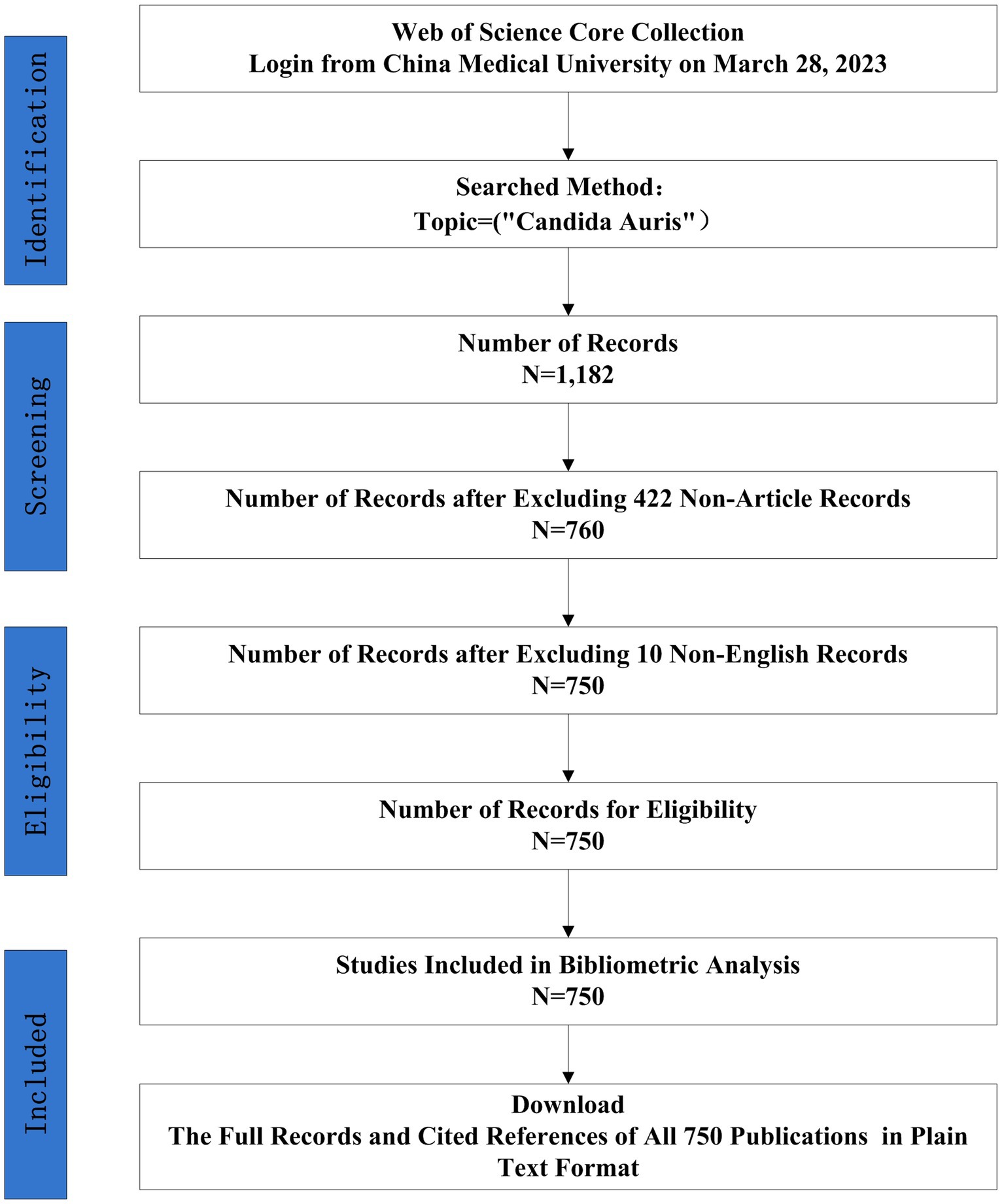
Figure 1. Flow charts of search strategies and article selection methods, followed the PRISMA-ScR guidelines.
3.2 Countries
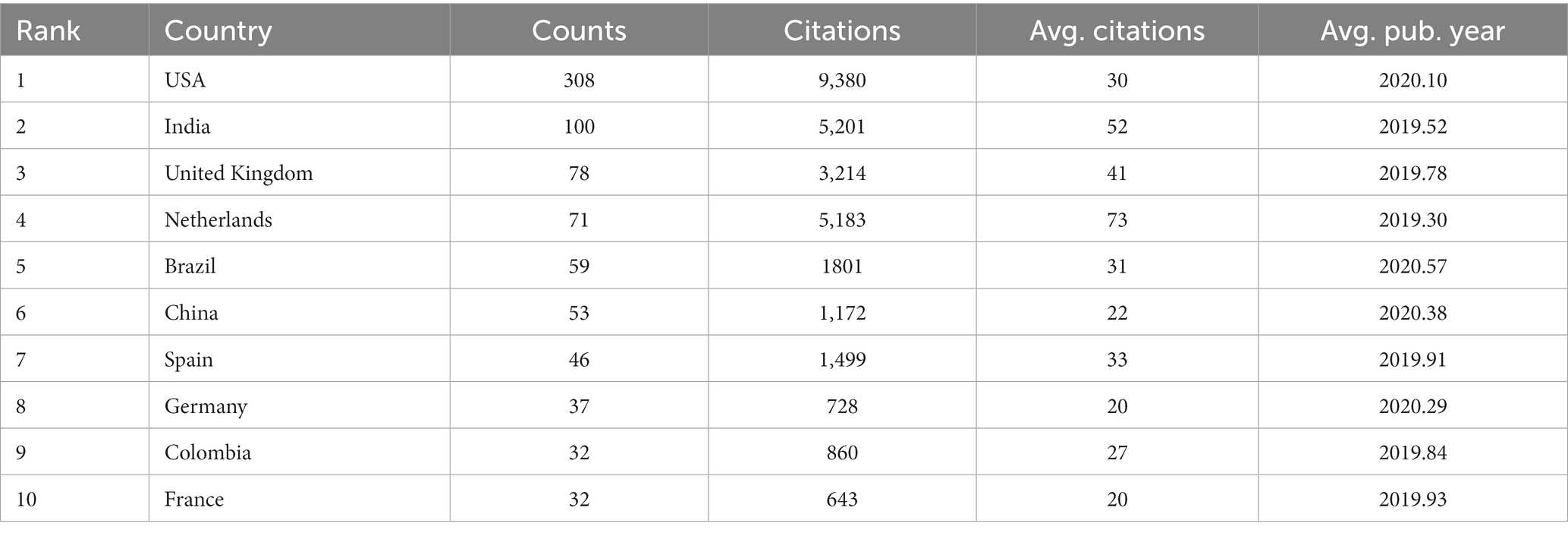
Our data revealed that 74 countries/regions have published relevant articles on C. auris. The global distribution of the countries/regions is shown in Figure 3, with different colors indicating different numbers of publications. The top 10 countries/regions based on the number of publications are listed in Table 1. The United States (308 counts, 9,380 citations) ranked first in terms of the number of publications and citations, followed by India (100 counts, 5,201 citations) and the United Kingdom (78 counts, 3,214 citations). Among the top 10 countries, the Netherlands had the highest average number of citations.
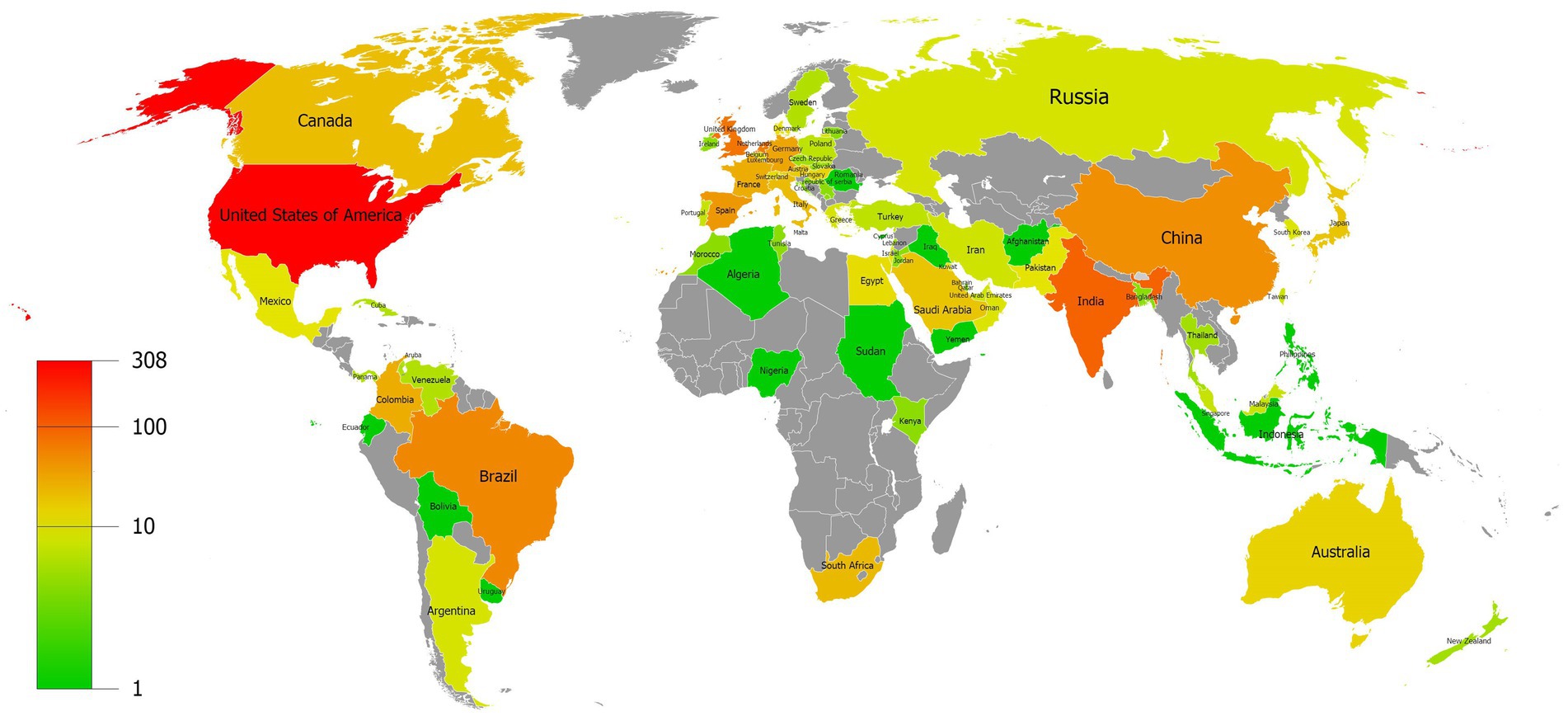
Figure 3. Distribution map of the countries/regions with different colors indicating different numbers of publications.
3.3 Journals
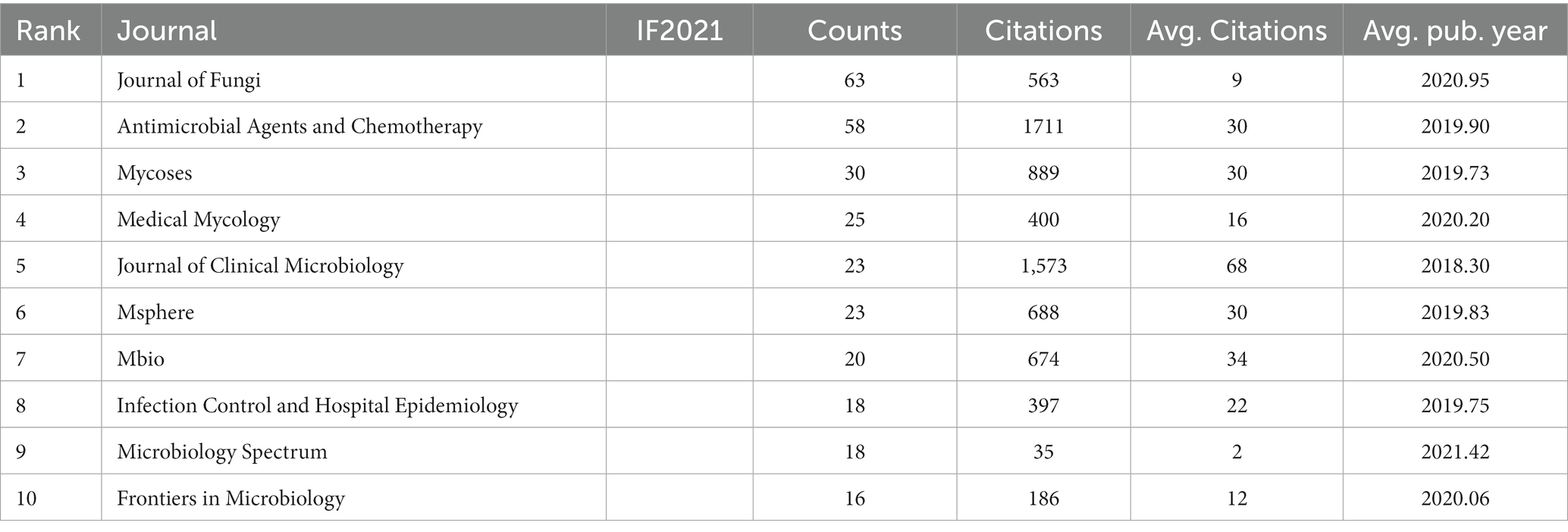
In total, 202 academic journals published articles related to C. auris, with the Journal of Fungi (63 articles) ranking first, followed by Antimicrobial Agents and Chemotherapy (58 articles) and Mycoses (30 articles). Table 2 shows the top 10 journals in terms of the number of publications. The top 10 journals published 294 articles, counting for 39.2% of the total. The Journal of Clinical Microbiology ranked first in terms of average citations, indicating that it was cited most frequently. Other than published amounts and citations, the Microbiology Spectrum had the latest Average Publication Year of 2021.42.
3.4 Institutions
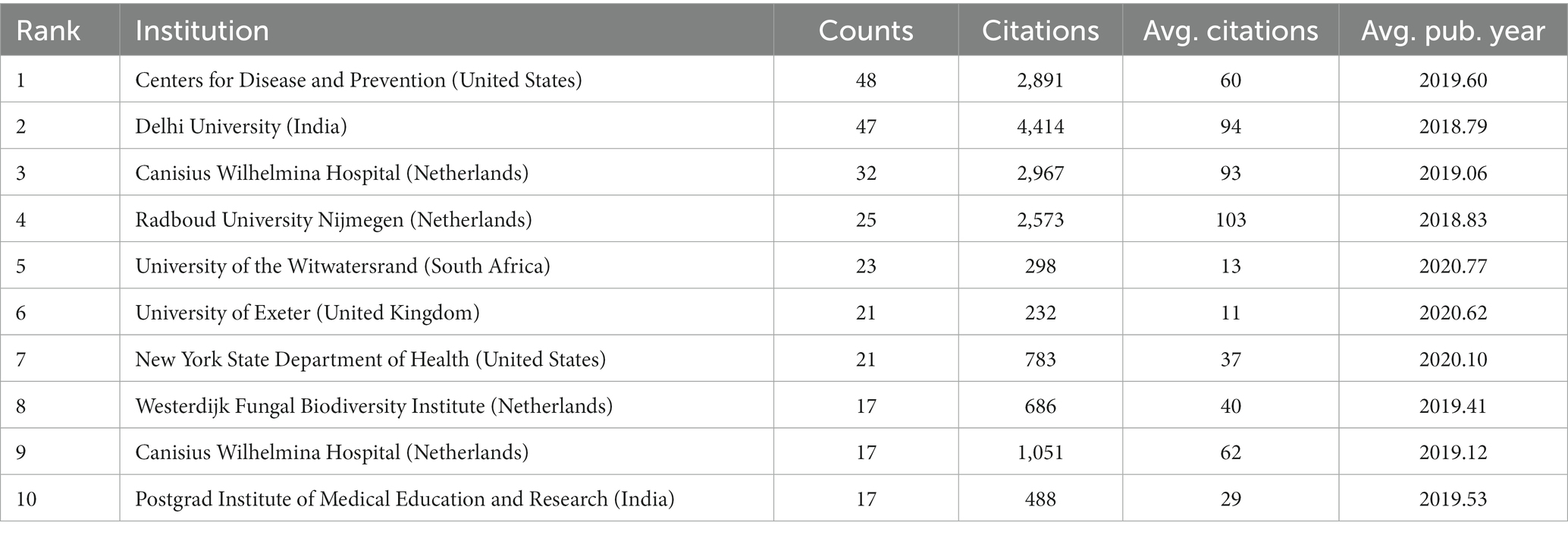
As of 2023, 1,314 institutions have published in this field. Table 3 illustrates the top 10 institutions according to number of publications, 40% of which are in the Netherlands. The top five institutions that contributed the most to the field were the Centers for Disease Control and Prevention in the USA (48 counts), Delhi University (47 counts), Canisius Wilhelmina Hospital (32 counts), Radboud University Nijmegen (25 counts), and the University of the Witwatersrand (23 counts). Among the top 10 institutions, Radboud University Nijmegen had the highest average number of citations.
We set the threshold for the number of publications by institutions at 5 counts and identified 97 highly productive institutions from 1,314 institutions. We used VOSviewer to conduct a co-authorship analysis of 97 highly productive institutions, all of which were in a co-authorship network consisting of nine clusters. The generated institutional network map identifies the nodes and link lines that represent the institutions and their collaborative relationships. As shown in Figure 4, the red cluster was the largest, consisting of 17 institutions. Centres for Disease and Prevention work with 39 highly productive institutions and ranked first. This was closely followed by the Canisius Wilhelmina Hospital, which works with 36 highly productive institutions, and Delhi University and Radboud University Nijmegen, which work with 34 highly productive institutions.
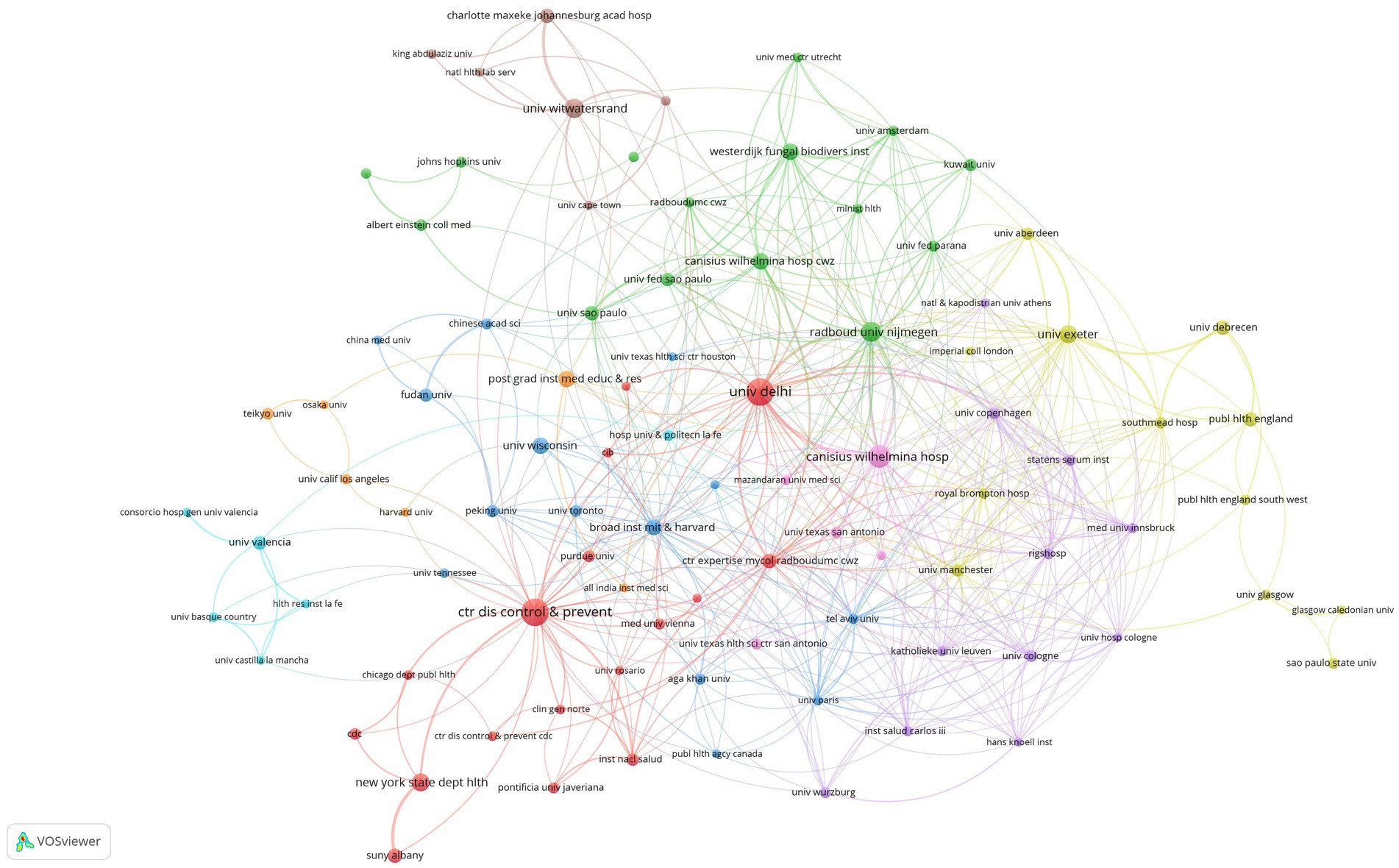
Figure 4. Institutional network map identified nodes and link lines, representing institutions and their collaborative relationships.
3.5 Authors
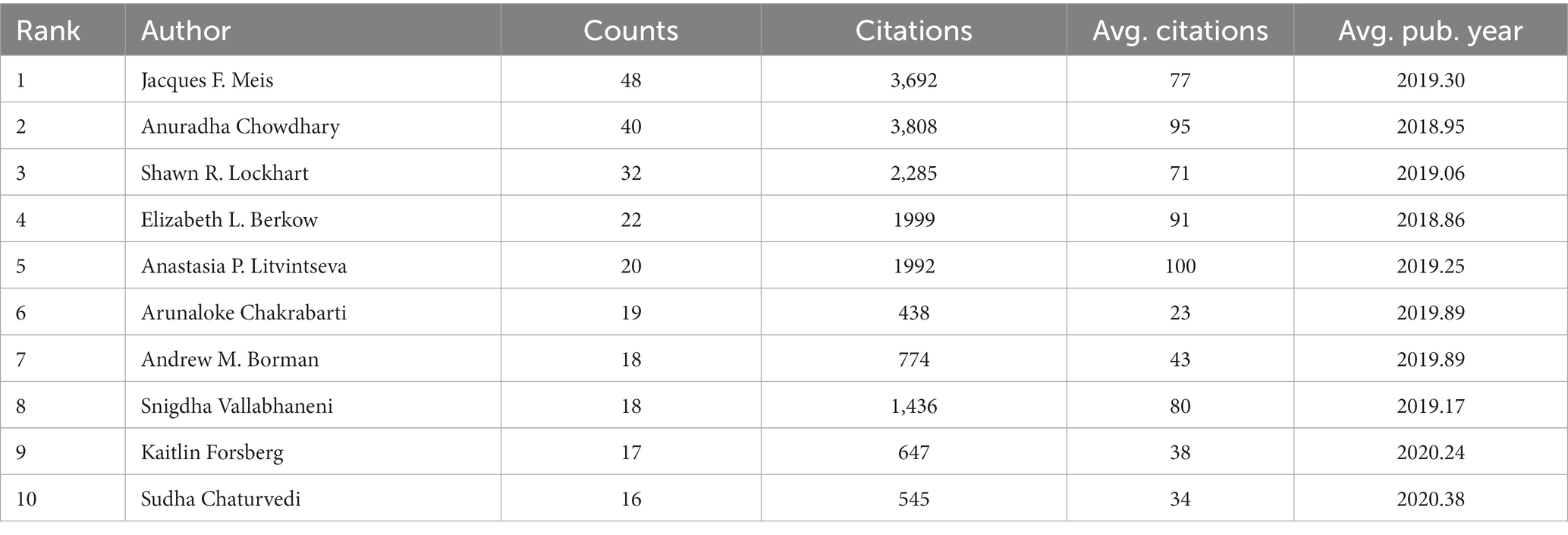
In total, 4,105 authors have published in this field since 2009. Table 4 lists the top 10 authors in terms of number of publications. Jacques F. Meis has published the most articles (48) and is ranked first, followed by Anuradha Chowdhary (40) and Shawn R. Lockhart (32). Anuradha Chowdhary had the highest average citations (3808).
We set the threshold for the number of publications by authors at 5 counts and identified 127 highly productive authors from 4,105 authors. We used VOSviewer to conduct a co-authorship analysis on 127 highly productive authors, 107 of whom formed the largest co-authorship network, consisting of 7 clusters. The generated co-authorship network map identifies nodes and link lines representing authors and their collaborative relationships. Approximately 85% of the highly productive authors are in the co-authorship network, indicating that collaboration among highly productive authors is considerably active. According to the statistics on citation and publication amounts, Jacques F. Meis has published the highest number of related articles, while Anastasia P. Litvintseva’s articles have the highest average citation rate. This result was indicated in Table 4.Furthermore, as shown in Figure 5, Meis worked with 46 highly productive authors and ranked first. He is closely followed by Shawn R. Lockhart, who worked with 44 highly productive authors, and Elizabeth L. Berkow, who worked with 39 highly productive authors. Jacques F. Meis and Anuradha Chowdhary had close partnerships and cooperated in 21 collaborations, ranking first in terms of number of collaborations among highly productive authors. Borman formed a close partnership in the relatively separate blue cluster shown at the bottom left of Figure 6. The authors in this cluster have 11 collaborations with each other, and most articles should have been published together. This may indicate inadequate collaboration between them and other top authors.
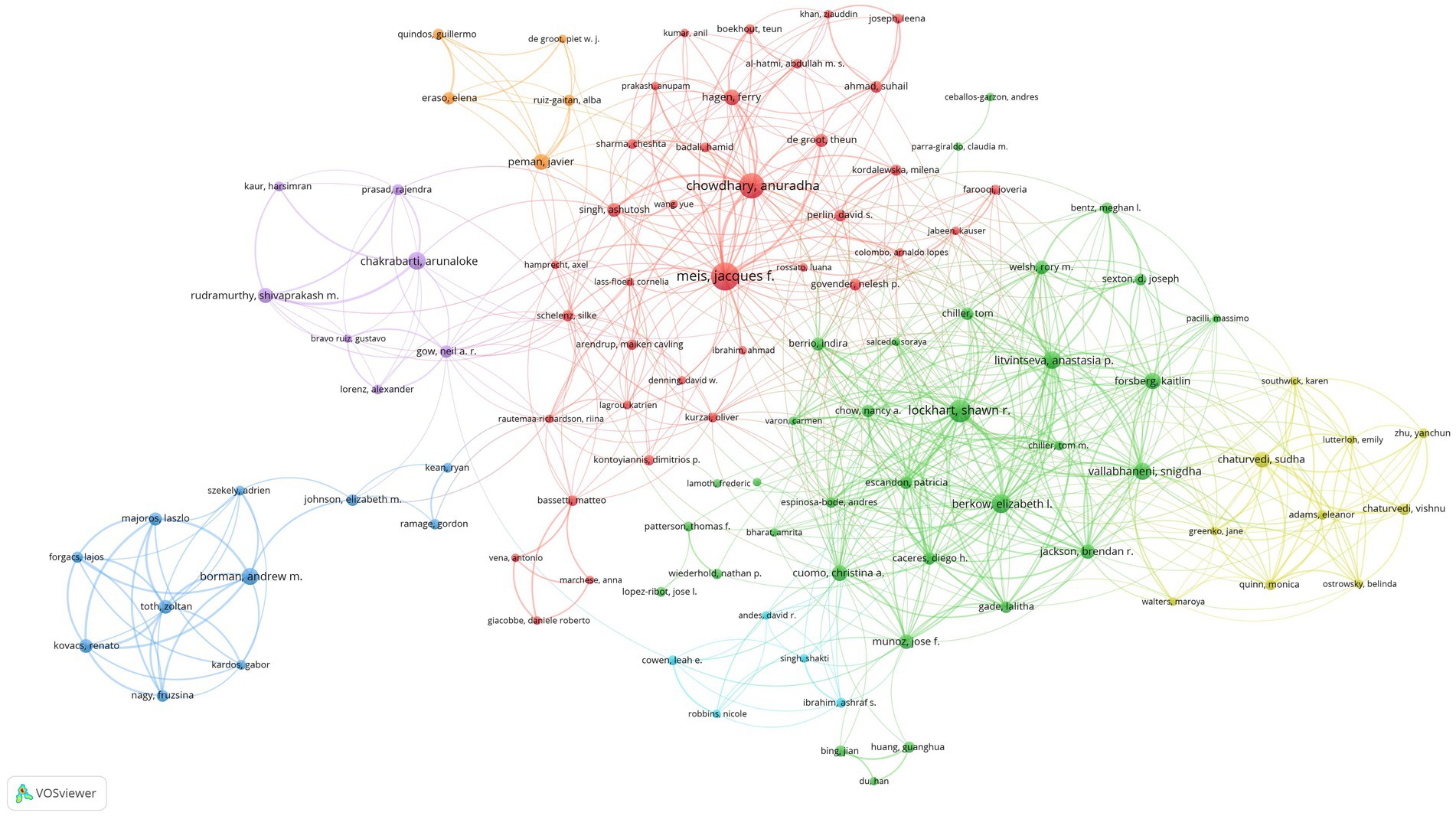
Figure 5. Co-authorship network map identified nodes and link lines, representing authors and their collaborative relationships.
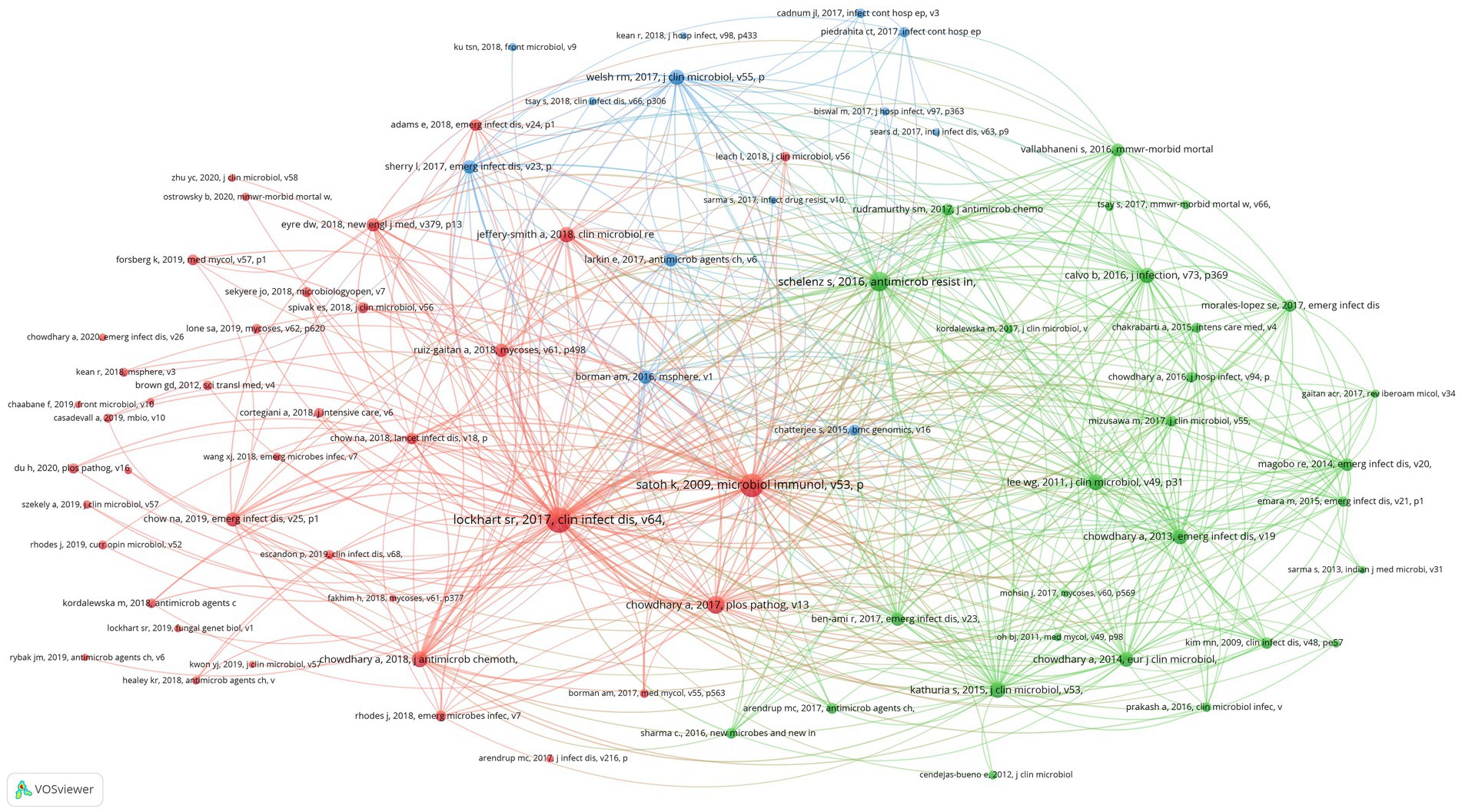
Figure 6. Co-citation network map identified nodes and link lines, representing references and their co-citation relationships.
3.6 References
Scientific studies are based on the research of predecessors, and we can better understand the knowledge base of a certain research field by analyzing references. In this study, 15,892 references were cited in 750 research articles on C. auris, appearing 30,728 times, with an average of 41 references per article. Overall, 80 references have been cited more than 30 times. We used VOSviewer to conduct a co-citation analysis of the 80 references and generate visual maps.
3.7 Keywords
In total, 2,465 keywords occurred in 750 articles. We set the threshold for high-frequency keywords to 10 counts and identified 98 high-frequency keywords. Excluding the search keyword C. auris, the top 5 keywords were resistance (201 counts), C. albicans (151 counts), antifungal susceptibility (102 counts), infections (94 counts), and identification (90 counts).
Co-occurrence analysis was performed on the 98 high-frequency keywords, and a co-contribution network map of the high-frequency keywords was constructed. The co-occurrence analysis of the 98 high-frequency keywords formed 4 clusters, which are represented by different colors in Figure 7. The red cluster comprised 30 keywords and formed the largest cluster. Cluster1 (red), Cluster2 (green), Cluster3 (blue), and Cluster4 (yellow) focus on research topics 1, 2, 3, and 4, respectively.
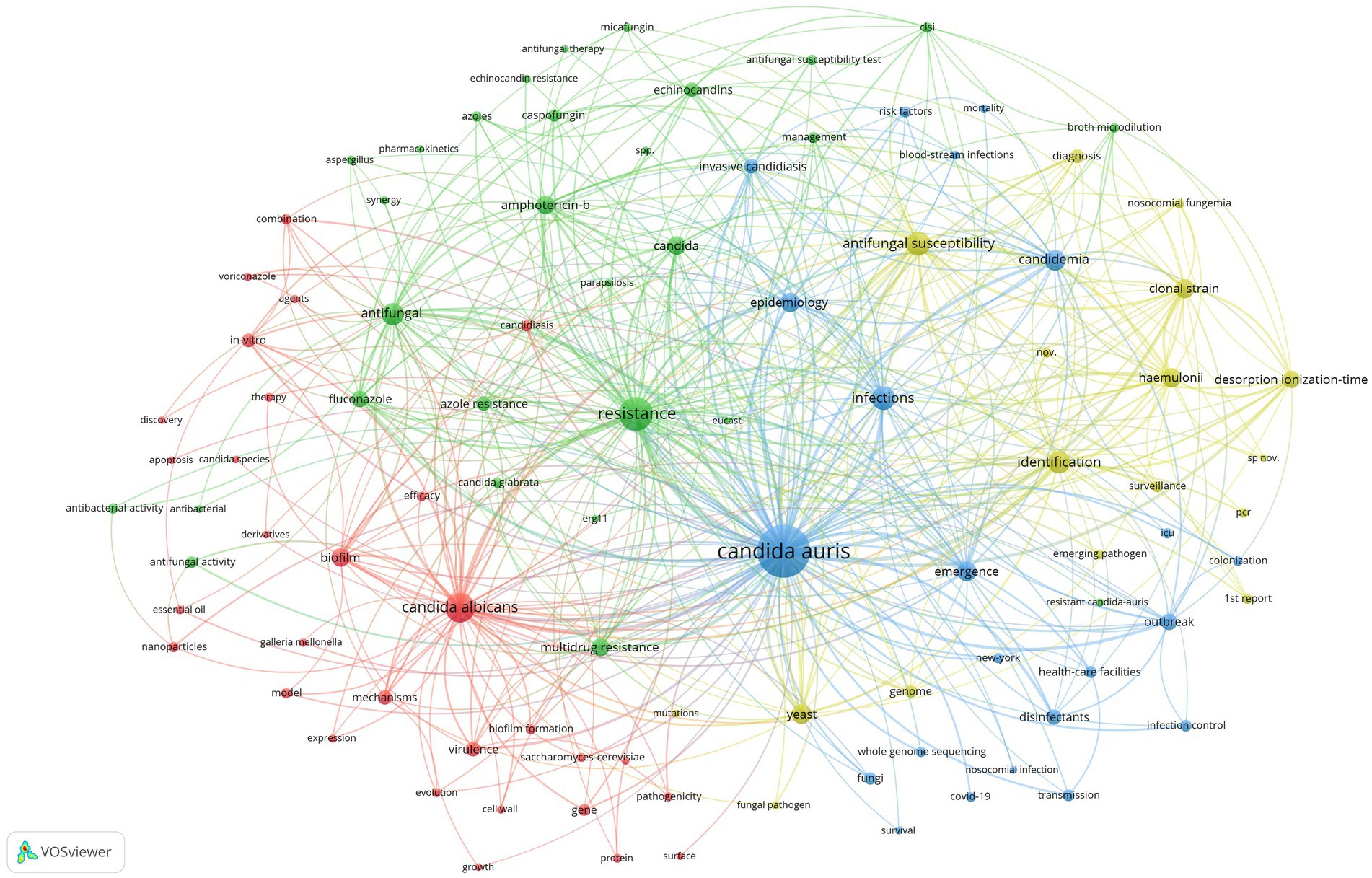
Figure 7. Co-contribution network map identified nodes and link lines, representing keywords and their co-occurrence relationships.
4 Discussion
This is the first bibliometric study of C. auris aimed at analyzing important information, such as the number of publications, publication region, authors, references, and keywords, using VOSviewer (version 1.6.19) software for visualization.
4.1 Trends in publications
Since the discovery of C. auris in 2009, 750 articles on C. auris have been published. Since 2020, the number of studies on C. auris has increased exponentially. Article published during the 2020–2022 period accounted for 66.67% of the total publications, indicating that this particular research field has become more attractive to researchers.
The United States published the most articles on C. auris, followed by India, the United Kingdom, and the Netherlands. According to the average number of publication years, the average value for the United States is higher than that of other countries. Considering that the number of publications in the United States was also ranked first, this indicates that the United States has published more recently with regards to C. auris. According to statistics, the Journal of Clinical Microbiology had the highest average number of citations and a relatively early publication year (average, 2018.3). These articles mainly focused on new progress in the identification of C. auris. This is possibly due to the lack of appropriate laboratory identification technology for C. auris, which is prone to identification errors (Mizusawa et al., 2017; Das et al., 2021). In contrast, Mbio published more recent articles and had a higher single citation rate. The content of the published articles mainly dealt with the evolutionary process of C. auris (Casadevall et al., 2019, 2021; Chow et al., 2020; Yadav et al., 2022) and the mechanism of drug resistance (Kim et al., 2019; Rybak et al., 2020; Carolus et al., 2021), which is in line with current research hotspots.
4.2 International Collaboration
The co-authorship analysis of the research institutions revealed that the United States, the Netherlands, the United Kingdom, and India accounted for the top 10 institutions in terms of publication numbers, while Centers for Disease and Prevention, Canisius Wilhelmina Hospital, Delhi University, and Radboud University Nijmegen were the four institutions that generally worked with other highly productive institutions the most. This shows that these four institutions are currently the centers of cooperation among global institutions in the C. auris research field.
Further analysis of the highly productive authors revealed that half of the top 10 authors, in terms of publication number, were from the Centers for Disease Control and Prevention, with a significant collaboration among these authors. Since the COVID-19 epidemic, C. auris outbreaks have occurred in ICUs and hospitals treating COVID-19 patients and in various environmental settings in hospitals (Sharma and Kadosh, 2023). This has attracted the attention of the Centers for Disease Control and Prevention (CDC) and is consistent with the fact that many of its most productive authors come from the CDC. For instance, Anastasia P. Litvintseva has published 20 articles with approximate 2,000 citations, and her main research interests include the origin of C. auris (Jackson et al., 2019; Chow et al., 2020), nosocomial outbreaks, molecular epidemiology of C. auris (Wilson et al., 2018; Armstrong et al., 2019; Escandon et al., 2019; Proctor et al., 2021), novel identification modes of C. auris (Sexton et al., 2018), and biological characteristics of C. auris (Munoz et al., 2018). Her studies cover the latest research hotspots on C. auris and establish an important position in the field of C. auris research.
4.3 Research hotspots
The high-frequency keywords derived from the bibliometric and visual analysis of C. auris articles can be roughly classified into four research directions: pathogenicity, drug resistance, epidemiological characteristics, and rapid and precise identification technology.
4.3.1 Pathogenicity research
As a critical human pathogen, researchers are still drawing attention to the pathogenicity and virulence of C. auris. Candida auris has a strong biofilm-forming ability, which not only enhances its efflux ability against fungal drugs (Bravo Ruiz and Lorenz, 2021), but also enhances its resistance to antifungal drugs, such as azoles, echinocandins, and polyenes (Kean and Ramage, 2019). In addition, C. auris can cause nosocomial infections through skin colonization or contamination of the surfaces of medical facilities (Kumar et al., 2019), and its ability to form biofilms has resulted in a certain degree of resistance to a variety of hospital disinfectants (Ledwoch and Maillard, 2018). Therefore, the formation mechanisms and elimination methods of C. auris biofilms have emerged as current research hotspots. The ability of C. albicans to form biofilms is much stronger than that of other Candida species under typical experimental conditions; however, Nett et al. found that the biofilm produced by C. auris in a simulated axillary skin model was 10 times higher than that of C. albicans. This also explains the difficulty in eradicating C. auris in hospital settings (Horton et al., 2020). Hu et al. evaluated the killing effects of a variety of chemical disinfectants, including three hand hygiene products, ultraviolet radiation, and ozone, on C. auris (Fu et al., 2020); they found that C. auris has strong resistance to quaternary ammonium salts, ultraviolet radiation, and ozone disinfection and the disinfection time must be extended to effectively inhibit its growth. However, their evaluation process did not consider the killing ability of these sterilization methods on the biofilm of C. auris (Gowri et al., 2020). Therefore, methods for inhibiting or eradicating the biofilms of C. auris formed in skin models will be an important and valuable research topic in the future.
An experiment conducted using mice as an animal model revealed that the virulence of C. auris was comparable to that of C. albicans (Fakhim et al., 2018); when fruit flies were used as the host, the lethality rate of C. auris was even higher than that of C. albicans (Wurster et al., 2019), confirming the strong pathogenicity of C. auris. Compared to C. albicans, C. auris colonizes the skin more deeply, resulting in invasive candidaemia, and IL-17 plays an important role in skin and subcutaneous tissue infections (Huang et al., 2021). Another study on host immune responses to C. auris revealed that C. auris has a strong immune evasion ability (Johnson et al., 2018) and further studies may show that this ability may be derived from PD-1 (Wu et al., 2019), but the specific mechanism must be studied further. In short, C. auris has strong virulence and immune evasion ability, and the study of the underlying mechanisms may have preventive and therapeutic significance.
4.3.2 Drug resistance of Candida auris
The multidrug resistance of Candida (Lockhart et al., 2017) is one of the most important characteristics that has attracted attention, and the resistance mechanism of the Candida species has always been a key focus of research. Recent studies have reported that the resistance of C. auris to echinocandins is related to the hotspot mutation of FKS1 (Chowdhary et al., 2018; Kordalewska et al., 2018); its resistance to azoles is generally related to ERG11 mutations, including K143R of F126L and Y132F (Chowdhary et al., 2018; Chow et al., 2020). Recently, Chaturvedi et al. found that the lack of FUR11 is a novel flucytosine resistance mechanism apart from FCY1, FUR1, and ADE17 mutations; however, in an amphotericin B (AmB) resistance mechanism verification study, they did not find any statistically significant nonsynonymous variants in sensitive or drug-resistant strains. Therefore, the resistance mechanism of amphotericin remains unclear and is also a direction for further research. For multidrug-resistant C. auris, synergistic effects between different antifungal drugs and between antibacterial drugs and antifungal drugs are important research topics for the treatment of C. auris infections, such as voriconazole and micafungin (100%) and flucytosine and amphotericin (100%). Further studies are required to validate these experimental data (Aghaei Gharehbolagh et al., 2021). Novel antifungal drugs, such as rezafungin, ibrexafungerp, olorofim, and fosmanogepix, have demonstrated efficacy against C. auris in in vitro studies (Lamoth et al., 2022; Sobel et al., 2022); however, their clinical efficacy needs to be evaluated in future clinical studies. In addition, several studies have determined the critical roles of nanoparticles in inhibiting the growth and biofilm formation of C. auris. Although these conclusions have not been tested in a clinical trial, nanoparticles have displayed great potential in treating C. auris infection and decolonization (Fayed et al., 2021; Kamli et al., 2021a,b; AlJindan and AlEraky, 2022; Bandara and Samaranayake, 2022).
4.3.3 Epidemiology studies of nosocomial transmission of Candida auris
Since the first strain of C. auris was isolated in 2009, this pathogen has posed enormous threats to humans due to several of its pathogenic properties. In 2011, the first three clinical cases of nosocomial fungaemia caused by C. auris were reported in South Korea; two of these patients died due to septic shock and multiple-organ failure and had received fluconazole therapy. Further, AmB displayed therapeutic failure. Besides the fact that it is multi-drug resistant, its ability to persist, easily transmit, and its highly proliferative clonal lifecycle endows this fungus with an expanding range of nosocomial infections globally (Lee et al., 2011; Schelenz et al., 2016; Vallabhaneni et al., 2017). In the United States, C. auris strains are associated with nosocomial outbreaks, and the excess healthcare costs have increased from $35,000 to $68,000 (Suleyman and Alangaden, 2021). From 2013 to 2018, 18 studies involving more than 200 patients reported nosocomial infections in more than 10 countries (Dahiya et al., 2020). Furthermore, according to several clinical descriptions, invasive candidiasis caused by C. auris is easily misdiagnosed as Saccharomyces cerevisiae, C. famata or other Candida species (Lee et al., 2011; Schelenz et al., 2016). Thus, precise and efficient identification is critical for controlling C. auris-mediated nosocomial transmission.
Since the onset of the COVID-19 epidemic, a large number of patients with severe COVID-19 pneumonia have been admitted to the ICU, and many cases of C. auris coinfection have emerged (Chowdhary et al., 2020; Janniger and Kapila, 2021; Kariyawasam et al., 2022; Thoma et al., 2022). In India, two-thirds of COVID-19 deaths were caused by concomitant C. auris infections (Chowdhary et al., 2020), and similar scenarios were observed in Mexico and Israel. From 2021 to 2022, the C. auris incidence rate increased more than 30-fold, corresponding with the outbreak of the COVID-19 pandemic in Israel. Further, more than 23% of patients suffer from coinfections with these two pathogenic factors; among these co-infected patients, more than 70% require mechanical ventilation (Biran et al., 2023). One study reported 12 patients infected with both C. auris and SARS-CoV-2 in an intensive care unit (ICU) in Mexico in 2020. Fungal strains were isolated from blood or urine samples, or both. The mortality rate is generally >80% (Villanueva-Lozano et al., 2021). Unlike the above-mentioned national studies, one study demonstrated an outbreak of C. auris alongside COVID-19 in a specialty care unit in Florida from July to August 2020. Researchers have indicated that the spread of COVID-19 has significantly enhanced the nosocomial infection of C. auris, in contrast to the situation in 2017. Separate medical care for mono-infections is highly recommended to avoid crossing infection and nosocomial transmission (Prestel et al., 2021).
Although there were many cases indicating a strong correlation between COVID-19 and C. auris infection, as mentioned above, this critical fungal pathogen caused serious nosocomial infection outbreaks prior to the presence of SARS-CoV-2. From the scientific article retrieval, studies have demonstrated that the mortality rate caused by C. auris infection was 30–60% during the pre-COVID-19 era (Lee et al., 2011; Chowdhary et al., 2013, 2014; Lockhart et al., 2017; Ruiz-Gaitan et al., 2018). During the outbreak of nosocomial infections, several predisposing factors of C. auris colonization and candidemia were monitored. Javier Pemán and his team pointed that polytrauma, cardiovascular disease, and cancer were the most common physiological conditions in C. auris infected patients. Moreover, the indwelling central venous catheters (CVC), parenteral nutrition, and mechanical ventilation act as the critical predictors of candidemia. These data were collected from a 12-month prospective, case–control study that included a total of 228 patients (Ruiz-Gaitan et al., 2019). Moreover, administration of immunosuppressors, antifungal therapies and long-term ICU accommodation also increase the risk of C. auris colonization (Eyre et al., 2018; Ruiz-Gaitan et al., 2018, 2019). Furthermore, the risk factors for ICU outbreaks include non-compliance with personal protective equipment (PPE) use, hand hygiene, PPE shortages, and excessive antibiotic use (Thoma et al., 2022). The multi-drug resistance and high fatality rate of C. auris pose great challenges in the treatment of patients. Therefore, the focus of further research will be to prevent nosocomial transmission through early detection of nosocomial epidemics using rapid and effective identification, summary analysis of infection cases, proposals for more effective treatment methods based on new antifungal drugs, and the implementation of more effective combinations of disinfection methods and infection control programs.
4.3.4 Rapid and precise identification technology of Candida auris
Pathogen identification methods should be highly accurate, low-cost and time-efficient. As mentioned above, C. auris is spreading globally and has been identified in various countries. Genetically, C. haemulonii, C. auris, C. pseudohaemulonii, and C. duobushaemulonii are closely related yeast pathogens and display highly multidrug resistant abilities. Thus, low-cost and precise identification methods are required to distinguish these different pathogens.
Since the first identification of C. auris in 2009, it has attracted widespread attention due to the difficulty in distinguishing it from C. haemulonii using traditional methods and its high level of resistance to azoles (Satoh et al., 2009; Kathuria et al., 2015; Kumar et al., 2016). There are five clades of C. auris with tens of thousands of single-nucleotide polymorphisms among clades (Wilson et al., 2018; Spruijtenburg et al., 2022), which result in differences in the drug resistance of each clade (Chowdhary et al., 2018). As these five clades of C. auris are widespread (Waters et al., 2023), it has become important to quickly distinguish between the five clades and develop different response plans according to the characteristics of each clade to control clinical infection. From 2017, there were three studies have demonstrated the Polymerase Chain Reaction (PCR) based method enable to identify the C. auris and phylogenetic relatives (Kordalewska et al., 2017; Arastehfar et al., 2018, 2019). Beside the aforementioned molecular method, Elizabeth M Johnson has established a novel and expeditious method for identifying the C. auris isolates from patient samples, which was named as the CHROMagarTM Candida Plus (Borman et al., 2021). Besides above mentioned methods, the MALDI-TOF MS and ITS sequencing can only be used to identify C. auris species but cannot distinguish between the five clades. Although whole genome sequencing can be used to distinguish between the five clades, it is time-consuming and expensive. A recent study by Jacques F. Meis et al. identified C. auris at the level of different clades through high-resolution mass spectrometry (Jamalian et al., 2023) and this provides us with new tools for faster and more accurate identification for a rapid response to nosocomial C. auris infection and to trace the source of the infection. In addition, YANG et al. applied recombinase-aided amplification combined with lateral flow strips (RAA-LFS) to detect C. auris and successfully developed a rapid and effective method for this purpose (Zhang et al., 2023). In conclusion, C. auris identification methods that are faster, cheaper, and have higher resolutions remain the focus of future research.
4.4 Limitations
Like other bibliometric studies, this study also has some limitations. First, to conduct bibliometric analysis, bibliographic data must be extracted from database such as WOS, Scopus, Dimensions, and PubMed. Due to the different formats of bibliographic data, current bibliometric research can only select one database for analysis. In this study, we only used WOS for data retrieval. WOS is the largest comprehensive academic information resource that provides large coverage of over 20,000 peer-reviewed journals. Consequently, many researchers have used WOS to conduct bibliometric studies on viral diseases with similar objectives to the present study. However, WOS cannot do all the related articles, which may also cause bias in the results. Second, we only used VOSviewer software tool to achieve our analyses. Through article retrieval in WOS database about the bibliometric analyses, both of VOSviewer and CiteSpace software enable to achieve bibliometric analyses, and they share lots of functionalities that include Co-Occurrence-, Co-Citation- and Co-Authorship- analyses. To compare with CiteSpace, VOSviewer is user-friendly and CiteSpace displays advantages in the evaluative analysis of network visualization. Moreover, the statistical analyses are relying on the authorship of researchers and institutions via VOSviewer software tool, the outcome of data may be biased by the unstandardized manner or same name authors.
5 Conclusion
Using bibliometrics and visualization analyses, this study comprehensively evaluated the global research trends and hotspots in the research field of C. auris and predicted future research directions. Close cooperation between institutions has occurred over the past decade, and the number of publications has increased rapidly since 2009. The United States, Netherlands, United Kingdom, and India have contributed the most to this area of research. In addition to the identification, drug resistance, and treatment of C. auris infections, in-hospital outbreak control at the source should be considered an important research focus.
Data availability statement
The raw data supporting the conclusions of this article will be made available by the authors, without undue reservation.
Author contributions
QW: Formal analysis, Funding acquisition, Supervision, Validation, Writing – original draft, Writing – review & editing, Methodology. SC: Conceptualization, Investigation, Supervision, Writing – review & editing. YW: Data curation, Formal analysis, Resources, Visualization, Writing – review & editing. FL: Writing – review & editing. JC: Writing – review & editing. WD: Writing – original draft, Resources, Validation, Visualization, Writing – review & editing. HK: Conceptualization, Supervision, Writing – review & editing. ZW: Formal analysis, Methodology, Supervision, Writing – original draft.
Funding
The author(s) declare financial support was received for the research, authorship, and/or publication of this article. This study was supported by the National Key Research and Development Program of China (2021YFC2300402 to QW).
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2023.1287003/full#supplementary-material
References
Aghaei Gharehbolagh, S., Izadi, A., Talebi, M., Sadeghi, F., Zarrinnia, A., Zarei, F., et al. (2021). New weapons to fight a new enemy: a systematic review of drug combinations against the drug-resistant fungus Candida auris. Mycoses 64, 1308–1316. doi: 10.1111/myc.13277
Ahmad, T., Murad, M. A., Baig, M., and Hui, J. (2021). Research trends in COVID-19 vaccine: a bibliometric analysis. Hum. Vaccin. Immunother. 17, 2367–2372. doi: 10.1080/21645515.2021.1886806
AlJindan, R., and AlEraky, D. M. (2022). Silver nanoparticles: a promising antifungal agent against the growth and biofilm formation of the emergent Candida auris. J Fungi (Basel) 8:744. doi: 10.3390/jof8070744
Arastehfar, A., Fang, W., Badali, H., Vaezi, A., Jiang, W., Liao, W., et al. (2018). Low-cost Tetraplex PCR for the global spreading multi-drug resistant fungus, Candida auris and its phylogenetic relatives. Front. Microbiol. 9:1119. doi: 10.3389/fmicb.2018.01119
Arastehfar, A., Fang, W., Daneshnia, F., Al-Hatmi, A. M., Liao, W., Pan, W., et al. (2019). Novel multiplex real-time quantitative PCR detecting system approach for direct detection of Candida auris and its relatives in spiked serum samples. Future Microbiol. 14, 33–45. doi: 10.2217/fmb-2018-0227
Armstrong, P. A., Rivera, S. M., Escandon, P., Caceres, D. H., Chow, N., Stuckey, M. J., et al. (2019). Hospital-associated multicenter outbreak of emerging fungus Candida auris, Colombia, 2016. Emerg. Infect. Dis. 25, 1339–1346. doi: 10.3201/eid2507.180491
Bandara, N., and Samaranayake, L. (2022). Emerging and future strategies in the management of recalcitrant Candida auris. Med. Mycol. 60:myac008. doi: 10.1093/mmy/myac008
Biran, R., Cohen, R., Finn, T., Brosh-Nissimov, T., Rahav, G., Yahav, D., et al. (2023). Nationwide outbreak of Candida auris infections driven by COVID-19 hospitalizations, Israel, 2021-2022. Emerg. Infect. Dis. 29, 1297–1301. doi: 10.3201/eid2907.221888
Borman, A. M., Fraser, M., and Johnson, E. M. (2021). CHROMagarTM Candida plus: a novel chromogenic agar that permits the rapid identification of Candida auris. Med. Mycol. 59, 253–258. doi: 10.1093/mmy/myaa049
Borman, A. M., Szekely, A., and Johnson, E. M. (2017). Isolates of the emerging pathogen Candida auris present in the UK have several geographic origins. Med. Mycol. 55, 563–567. doi: 10.1093/mmy/myw147
Bravo Ruiz, G., and Lorenz, A. (2021). What do we know about the biology of the emerging fungal pathogen of humans Candida auris? Microbiol. Res. 242:126621. doi: 10.1016/j.micres.2020.126621
Bravo Ruiz, G., Ross, Z. K., Holmes, E., Schelenz, S., Gow, N. A. R., and Lorenz, A. (2019). Rapid and extensive karyotype diversification in haploid clinical Candida auris isolates. Curr. Genet. 65, 1217–1228. doi: 10.1007/s00294-019-00976-w
Carolus, H., Pierson, S., Munoz, J. F., Subotic, A., Cruz, R. B., Cuomo, C. A., et al. (2021). Genome-wide analysis of experimentally evolved Candida auris reveals multiple novel mechanisms of multidrug resistance. MBio 12:e03333–20. doi: 10.1128/mBio.03333-20
Carvajal, S. K., Alvarado, M., Rodriguez, Y. M., Parra-Giraldo, C. M., Varon, C., Morales-Lopez, S. E., et al. (2021). Pathogenicity assessment of Colombian strains of Candida auris in the galleria mellonella invertebrate model. J. Fungi (Basel) 7:401. doi: 10.3390/jof7060401
Casadevall, A., Kontoyiannis, D. P., and Robert, V. (2019). On the emergence of Candida auris: climate change, azoles, swamps, and birds. mBio 10:e01397–19. doi: 10.1128/mBio.01397-19
Casadevall, A., Kontoyiannis, D. P., and Robert, V. (2021). Environmental Candida auris and the global warming emergence hypothesis. MBio 12:e00360–21. doi: 10.1128/mBio.00360-21
Chow, N. A., Munoz, J. F., Gade, L., Berkow, E. L., Li, X., Welsh, R. M., et al. (2020). Tracing the evolutionary history and global expansion of Candida auris using population genomic analyses. MBio 11:e03364–19. doi: 10.1128/mBio.03364-19
Chowdhary, A., Anil Kumar, V., Sharma, C., Prakash, A., Agarwal, K., Babu, R., et al. (2014). Multidrug-resistant endemic clonal strain of Candida auris in India. Eur. J. Clin. Microbiol. Infect. Dis. 33, 919–926. doi: 10.1007/s10096-013-2027-1
Chowdhary, A., Prakash, A., Sharma, C., Kordalewska, M., Kumar, A., Sarma, S., et al. (2018). A multicentre study of antifungal susceptibility patterns among 350 Candida auris isolates (2009-17) in India: role of the ERG11 and FKS1 genes in azole and echinocandin resistance. J. Antimicrob. Chemother. 73, 891–899. doi: 10.1093/jac/dkx480
Chowdhary, A., Sharma, C., Duggal, S., Agarwal, K., Prakash, A., Singh, P. K., et al. (2013). New clonal strain of Candida auris, Delhi. India. Emerg. Infect. Dis. 19, 1670–1673. doi: 10.3201/eid1910.130393
Chowdhary, A., Sharma, C., and Meis, J. F. (2017). Candida auris: a rapidly emerging cause of hospital-acquired multidrug-resistant fungal infections globally. PLoS Pathog. 13:e1006290. doi: 10.1371/journal.ppat.1006290
Chowdhary, A., Tarai, B., Singh, A., and Sharma, A. (2020). Multidrug-resistant Candida auris infections in critically ill coronavirus disease patients, India, April-July 2020. Emerg. Infect. Dis. 26, 2694–2696. doi: 10.3201/eid2611.203504
Dahiya, S., Chhillar, A. K., Sharma, N., Choudhary, P., Punia, A., Balhara, M., et al. (2020). Candida auris and nosocomial infection. Curr. Drug Targets 21, 365–373. doi: 10.2174/1389450120666190924155631
Das, S., Singh, S., Tawde, Y., Chakrabarti, A., Rudramurthy, S. M., Kaur, H., et al. (2021). A selective medium for isolation and detection of Candida auris, an emerging pathogen. J. Clin. Microbiol. 59:e00326–20. doi: 10.1128/JCM.00326-20
Desnos-Ollivier, M., Fekkar, A., and Bretagne, S. (2021). Earliest case of Candida auris infection imported in 2007 in Europe from India prior to the 2009 description in Japan. J. Mycol. Med. 31:101139. doi: 10.1016/j.mycmed.2021.101139
Eix, E. F., Johnson, C. J., Wartman, K. M., Kernien, J. F., Meudt, J. J., Shanmuganayagam, D., et al. (2022). Ex vivo human and porcine skin effectively model Candida auris colonization, differentiating robust and poor fungal colonizers. J. Infect. Dis. 225, 1791–1795. doi: 10.1093/infdis/jiac094
Escandon, P., Chow, N. A., Caceres, D. H., Gade, L., Berkow, E. L., Armstrong, P., et al. (2019). Molecular epidemiology of Candida auris in Colombia reveals a highly related, countrywide colonization with regional patterns in amphotericin B resistance. Clin. Infect. Dis. 68, 15–21. doi: 10.1093/cid/ciy411
Eyre, D. W., Sheppard, A. E., Madder, H., Moir, I., Moroney, R., Quan, T. P., et al. (2018). A Candida auris outbreak and its control in an intensive care setting. N. Engl. J. Med. 379, 1322–1331. doi: 10.1056/NEJMoa1714373
Fakhim, H., Vaezi, A., Dannaoui, E., Chowdhary, A., Nasiry, D., Faeli, L., et al. (2018). Comparative virulence of Candida auris with Candida haemulonii, Candida glabrata and Candida albicans in a murine model. Mycoses 61, 377–382. doi: 10.1111/myc.12754
Fayed, B., Jayakumar, M. N., and Soliman, S. S. M. (2021). Caspofungin-resistance in Candida auris is cell wall-dependent phenotype and potential prevention by zinc oxide nanoparticles. Med. Mycol. 59, 1243–1256. doi: 10.1093/mmy/myab059
Fu, L., Le, T., Liu, Z., Wang, L., Guo, H., Yang, J., et al. (2020). Different efficacies of common disinfection methods against candida auris and other candida species. J. Infect. Public Health 13, 730–736. doi: 10.1016/j.jiph.2020.01.008
Gowri, M., Jayashree, B., Jeyakanthan, J., and Girija, E. K. (2020). Sertraline as a promising antifungal agent: inhibition of growth and biofilm of Candida auris with special focus on the mechanism of action in vitro. J. Appl. Microbiol. 128, 426–437. doi: 10.1111/jam.14490
Horton, M. V., Johnson, C. J., Kernien, J. F., Patel, T. D., Lam, B. C., Cheong, J. Z. A., et al. (2020). Candida auris forms high-burden biofilms in skin niche conditions and on porcine skin. mSphere 5:e00910–19. doi: 10.1128/mSphere.00910-19
Huang, X., Hurabielle, C., Drummond, R. A., Bouladoux, N., Desai, J. V., Sim, C. K., et al. (2021). Murine model of colonization with fungal pathogen Candida auris to explore skin tropism, host risk factors and therapeutic strategies. Cell Host Microbe 29, 210–221.e6. doi: 10.1016/j.chom.2020.12.002
Jackson, B. R., Chow, N., Forsberg, K., Litvintseva, A. P., Lockhart, S. R., Welsh, R., et al. (2019). On the origins of a species: what might explain the rise of Candida auris? J. Fungi (Basel) 5:58. doi: 10.3390/jof5030058
Jamalian, A., Freeke, J., Chowdhary, A., de Hoog, G. S., Stielow, J. B., and Meis, J. F. (2023). Fast and accurate identification of Candida auris by high resolution mass spectrometry. J. Fungi (Basel) 9:267. doi: 10.3390/jof9020267
Janniger, E. J., and Kapila, R. (2021). Public health issues with Candida auris in COVID-19 patients. World Med Health Policy 13, 766–772. doi: 10.1002/wmh3.472
Johnson, C. J., Davis, J. M., Huttenlocher, A., Kernien, J. F., and Nett, J. E. (2018). Emerging fungal pathogen Candida auris evades neutrophil attack. MBio 9:e01403–18. doi: 10.1128/mBio.01403-18
Kamli, M. R., Srivastava, V., Hajrah, N. H., Sabir, J. S. M., Ali, A., Malik, M. A., et al. (2021a). Phytogenic fabrication of ag-Fe bimetallic nanoparticles for cell cycle arrest and apoptosis signaling pathways in Candida auris by generating oxidative stress. Antioxidants (Basel) 10:182. doi: 10.3390/antiox10020182
Kamli, M. R., Srivastava, V., Hajrah, N. H., Sabir, J. S. M., Hakeem, K. R., Ahmad, A., et al. (2021b). Facile bio-fabrication of ag-cu-co Trimetallic nanoparticles and its fungicidal activity against Candida auris. J Fungi (Basel) 7:62. doi: 10.3390/jof7010062
Kariyawasam, R. M., Julien, D. A., Jelinski, D. C., Larose, S. L., Rennert-May, E., Conly, J. M., et al. (2022). Antimicrobial resistance (AMR) in COVID-19 patients: a systematic review and meta-analysis (November 2019-June 2021). Antimicrob. Resist. Infect. Control 11:45. doi: 10.1186/s13756-022-01085-z
Kathuria, S., Singh, P. K., Sharma, C., Prakash, A., Masih, A., Kumar, A., et al. (2015). Multidrug-resistant Candida auris misidentified as Candida haemulonii: characterization by matrix-assisted laser desorption ionization-time of flight mass spectrometry and DNA sequencing and its antifungal susceptibility profile variability by Vitek 2, CLSI broth microdilution, and Etest method. J. Clin. Microbiol. 53, 1823–1830. doi: 10.1128/JCM.00367-15
Kean, R., and Ramage, G. (2019). Combined antifungal resistance and biofilm tolerance: the global threat of Candida auris. mSphere 4:e00458–19. doi: 10.1128/mSphere.00458-19
Kim, S. H., Iyer, K. R., Pardeshi, L., Munoz, J. F., Robbins, N., Cuomo, C. A., et al. (2019). Genetic analysis of Candida auris implicates Hsp90 in morphogenesis and azole tolerance and Cdr1 in azole resistance. MBio 10:e02529–18. doi: 10.1128/mBio.02529-18
Kim, M. N., Shin, J. H., Sung, H., Lee, K., Kim, E. C., Ryoo, N., et al. (2009). Candida haemulonii and closely related species at 5 university hospitals in Korea: identification, antifungal susceptibility, and clinical features. Clin. Infect. Dis. 48, e57–e61. doi: 10.1086/597108
Kordalewska, M., Lee, A., Park, S., Berrio, I., Chowdhary, A., Zhao, Y., et al. (2018). Understanding Echinocandin resistance in the emerging pathogen Candida auris. Antimicrob. Agents Chemother. 62:e00238–18. doi: 10.1128/AAC.00238-18
Kordalewska, M., Zhao, Y., Lockhart, S. R., Chowdhary, A., Berrio, I., and Perlin, D. S. (2017). Rapid and accurate molecular identification of the emerging multidrug-resistant pathogen Candida auris. J. Clin. Microbiol. 55, 2445–2452. doi: 10.1128/JCM.00630-17
Kumar, J., Eilertson, B., Cadnum, J. L., Whitlow, C. S., Jencson, A. L., Safdar, N., et al. (2019). Environmental contamination with Candida species in multiple hospitals including a tertiary care hospital with a Candida auris outbreak. Pathog. Immun. 4, 260–270. doi: 10.20411/pai.v4i2.291
Kumar, A., Prakash, A., Singh, A., Kumar, H., Hagen, F., Meis, J. F., et al. (2016). Candida haemulonii species complex: an emerging species in India and its genetic diversity assessed with multilocus sequence and amplified fragment-length polymorphism analyses. Emerg. Microbes Infect. 5:e49. doi: 10.1038/emi.2016.49
Lamoth, F., Lewis, R. E., and Kontoyiannis, D. P. (2022). Investigational antifungal agents for invasive mycoses: a clinical perspective. Clin. Infect. Dis. 75, 534–544. doi: 10.1093/cid/ciab1070
Ledwoch, K., and Maillard, J. Y. (2018). Candida auris dry surface biofilm (DSB) for disinfectant efficacy testing. Materials (Basel) 12:18. doi: 10.3390/ma12010018
Lee, W. G., Shin, J. H., Uh, Y., Kang, M. G., Kim, S. H., Park, K. H., et al. (2011). First three reported cases of nosocomial fungemia caused by Candida auris. J. Clin. Microbiol. 49, 3139–3142. doi: 10.1128/JCM.00319-11
Lin, J., Li, G., Zhong, P., Zeng, Q., Liu, L., and Chen, L. (2022). Bibliometric analysis of human monkeypox research from 1975 to 2022 and novel prevention and control strategies. Front. Public Health 10:995965. doi: 10.3389/fpubh.2022.995965
Lockhart, S. R., Etienne, K. A., Vallabhaneni, S., Farooqi, J., Chowdhary, A., Govender, N. P., et al. (2017). Simultaneous emergence of multidrug-resistant Candida auris on 3 continents confirmed by whole-genome sequencing and epidemiological analyses. Clin. Infect. Dis. 64, 134–140. doi: 10.1093/cid/ciw691
Mizusawa, M., Miller, H., Green, R., Lee, R., Durante, M., Perkins, R., et al. (2017). Can multidrug-resistant Candida auris be reliably identified in clinical microbiology laboratories? J. Clin. Microbiol. 55, 638–640. doi: 10.1128/JCM.02202-16
Munoz, J. F., Gade, L., Chow, N. A., Loparev, V. N., Juieng, P., Berkow, E. L., et al. (2018). Genomic insights into multidrug-resistance, mating and virulence in Candida auris and related emerging species. Nat. Commun. 9:5346. doi: 10.1038/s41467-018-07779-6
Parra-Giraldo, C. M., Valderrama, S. L., Cortes-Fraile, G., Garzon, J. R., Ariza, B. E., Morio, F., et al. (2018). First report of sporadic cases of Candida auris in Colombia. Int. J. Infect. Dis. 69, 63–67. doi: 10.1016/j.ijid.2018.01.034
Prestel, C., Anderson, E., Forsberg, K., Lyman, M., de Perio, M. A., Kuhar, D., et al. (2021). Candida auris outbreak in a COVID-19 specialty care unit - Florida, July-august 2020. MMWR Morb. Mortal. Wkly Rep. 70, 56–57. doi: 10.15585/mmwr.mm7002e3
Proctor, D. M., Dangana, T., Sexton, D. J., Fukuda, C., Yelin, R. D., Stanley, M., et al. (2021). Integrated genomic, epidemiologic investigation of Candida auris skin colonization in a skilled nursing facility. Nat. Med. 27, 1401–1409. doi: 10.1038/s41591-021-01383-w
Ruiz-Gaitan, A., Martinez, H., Moret, A. M., Calabuig, E., Tasias, M., Alastruey-Izquierdo, A., et al. (2019). Detection and treatment of Candida auris in an outbreak situation: risk factors for developing colonization and candidemia by this new species in critically ill patients. Expert Rev. Anti-Infect. Ther. 17, 295–305. doi: 10.1080/14787210.2019.1592675
Ruiz-Gaitan, A., Moret, A. M., Tasias-Pitarch, M., Aleixandre-Lopez, A. I., Martinez-Morel, H., Calabuig, E., et al. (2018). An outbreak due to Candida auris with prolonged colonisation and candidaemia in a tertiary care European hospital. Mycoses 61, 498–505. doi: 10.1111/myc.12781
Rybak, J. M., Munoz, J. F., Barker, K. S., Parker, J. E., Esquivel, B. D., Berkow, E. L., et al. (2020). Mutations in TAC1B: a novel genetic determinant of clinical fluconazole resistance in Candida auris. MBio 11:e00365–20. doi: 10.1128/mBio.00365-20
Saheb, T., Saheb, T., and Carpenter, D. O. (2021). Mapping research strands of ethics of artificial intelligence in healthcare: a bibliometric and content analysis. Comput. Biol. Med. 135:104660. doi: 10.1016/j.compbiomed.2021.104660
Satoh, K., Makimura, K., Hasumi, Y., Nishiyama, Y., Uchida, K., and Yamaguchi, H. (2009). Candida auris sp. nov., a novel ascomycetous yeast isolated from the external ear canal of an inpatient in a Japanese hospital. Microbiol. Immunol. 53, 41–44. doi: 10.1111/j.1348-0421.2008.00083.x
Schelenz, S., Hagen, F., Rhodes, J. L., Abdolrasouli, A., Chowdhary, A., Hall, A., et al. (2016). First hospital outbreak of the globally emerging Candida auris in a European hospital. Antimicrob. Resist. Infect. Control 5:35. doi: 10.1186/s13756-016-0132-5
Sexton, D. J., Kordalewska, M., Bentz, M. L., Welsh, R. M., Perlin, D. S., and Litvintseva, A. P. (2018). Direct detection of emergent fungal pathogen Candida auris in clinical skin swabs by SYBR Green-based quantitative PCR assay. J. Clin. Microbiol. 56:e01337–18. doi: 10.1128/JCM.01337-18
Sharma, C., and Kadosh, D. (2023). Perspective on the origin, resistance, and spread of the emerging human fungal pathogen Candida auris. PLoS Pathog. 19:e1011190. doi: 10.1371/journal.ppat.1011190
Shin, J. H., Kim, M. N., Jang, S. J., Ju, M. Y., Kim, S. H., Shin, M. G., et al. (2012). Detection of amphotericin B resistance in Candida haemulonii and closely related species by use of the Etest, Vitek-2 yeast susceptibility system, and CLSI and EUCAST broth microdilution methods. J. Clin. Microbiol. 50, 1852–1855. doi: 10.1128/JCM.06440-11
Sobel, R., Nyirjesy, P., Ghannoum, M. A., Delchev, D. A., Azie, N. E., Angulo, D., et al. (2022). Efficacy and safety of oral ibrexafungerp for the treatment of acute vulvovaginal candidiasis: a global phase 3, randomised, placebo-controlled superiority study (VANISH 306). BJOG 129, 412–420. doi: 10.1111/1471-0528.16972
Spruijtenburg, B., Badali, H., Abastabar, M., Mirhendi, H., Khodavaisy, S., Sharifisooraki, J., et al. (2022). Confirmation of fifth Candida auris clade by whole genome sequencing. Emerg. Microbes Infect. 11, 2405–2411. doi: 10.1080/22221751.2022.2125349
Suleyman, G., and Alangaden, G. J. (2021). Nosocomial fungal infections: epidemiology, infection control, and prevention. Infect. Dis. Clin. N. Am. 35, 1027–1053. doi: 10.1016/j.idc.2021.08.002
Thoma, R., Seneghini, M., Seiffert, S. N., Vuichard Gysin, D., Scanferla, G., Haller, S., et al. (2022). The challenge of preventing and containing outbreaks of multidrug-resistant organisms and Candida auris during the coronavirus disease 2019 pandemic: report of a carbapenem-resistant Acinetobacter baumannii outbreak and a systematic review of the literature. Antimicrob. Resist. Infect. Control 11:12. doi: 10.1186/s13756-022-01052-8
Vallabhaneni, S., Kallen, A., Tsay, S., Chow, N., Welsh, R., Kerins, J., et al. (2017). Investigation of the first seven reported cases of Candida auris, a globally emerging invasive, multidrug-resistant fungus-United States, May 2013-august 2016. Am. J. Transplant. 17, 296–299. doi: 10.1111/ajt.14121
Villanueva-Lozano, H., Trevino-Rangel, R. J., Gonzalez, G. M., Ramirez-Elizondo, M. T., Lara-Medrano, R., Aleman-Bocanegra, M. C., et al. (2021). Outbreak of Candida auris infection in a COVID-19 hospital in Mexico. Clin. Microbiol. Infect. 27, 813–816. doi: 10.1016/j.cmi.2020.12.030
Waqas, A., Teoh, S. H., Lapao, L. V., Messina, L. A., and Correia, J. C. (2020). Harnessing telemedicine for the provision of health care: bibliometric and Scientometric analysis. J. Med. Internet Res. 22:e18835. doi: 10.2196/18835
Waters, A., Chommanard, C., Baltozer, S., Angel, L. C., Abdelfattah, R., Lyman, M., et al. (2023). Investigation of a Candida auris outbreak in a skilled nursing facility - Virginia, United States, October 2020-June 2021. Am. J. Infect. Control 51, 472–474. doi: 10.1016/j.ajic.2022.12.003
Wilson, M. R., O'Donovan, B. D., Gelfand, J. M., Sample, H. A., Chow, F. C., Betjemann, J. P., et al. (2018). Chronic meningitis investigated via metagenomic next-generation sequencing. JAMA Neurol. 75, 947–955. doi: 10.1001/jamaneurol.2018.0463
Wu, X., Gu, Z., Chen, Y., Chen, B., Chen, W., Weng, L., et al. (2019). Application of PD-1 blockade in Cancer immunotherapy. Comput. Struct. Biotechnol. J. 17, 661–674. doi: 10.1016/j.csbj.2019.03.006
Wurster, S., Bandi, A., Beyda, N. D., Albert, N. D., Raman, N. M., Raad, I. I., et al. (2019). Drosophila melanogaster as a model to study virulence and azole treatment of the emerging pathogen Candida auris. J. Antimicrob. Chemother. 74, 1904–1910. doi: 10.1093/jac/dkz100
Yadav, A., Jain, K., Wang, Y., Pawar, K., Kaur, H., Sharma, K. K., et al. (2022). Candida auris on apples: diversity and clinical significance. MBio 13:e0051822. doi: 10.1128/mbio.00518-22
Yue, H., Bing, J., Zheng, Q., Zhang, Y., Hu, T., Du, H., et al. (2018). Filamentation in Candida auris, an emerging fungal pathogen of humans: passage through the mammalian body induces a heritable phenotypic switch. Emerg. Microbes Infect. 7, 1–13. doi: 10.1038/s41426-018-0187-x
Keywords: Candida auris , nosocomial outbreaks, drug resistance, bibliometric analysis, research trends
Citation: Wang Q, Cheng S, Wang Y, Li F, Chen J, Du W, Kang H and Wang Z (2023) Global characteristics and trends in research on Candida auris. Front. Microbiol. 14:1287003. doi: 10.3389/fmicb.2023.1287003
Edited by:
Sadegh Khodavaisy, Tehran University of Medical Sciences, IranReviewed by:
Kazem Ahmadikia, Tehran University of Medical Sciences, IranMohammad Kord, Tehran University of Medical Sciences, Iran
Copyright © 2023 Wang, Cheng, Wang, Li, Chen, Du, Kang and Wang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Wei Du, d2VpZHUyMzVAMTYzLmNvbQ==: Hui Kang, a2FuZ2h1aTY1QHNpbmEuY29t: Zhongqing Wang, d2FuZ3pob25ncWluZ0BjbXUuZWR1LmNu
†These authors have contributed equally to this work and share first authorship
 Qihui Wang
Qihui Wang Shitong Cheng1†
Shitong Cheng1† Wei Du
Wei Du Zhongqing Wang
Zhongqing Wang