
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Microbiol. , 15 November 2023
Sec. Food Microbiology
Volume 14 - 2023 | https://doi.org/10.3389/fmicb.2023.1284929
This article is part of the Research Topic Mobile DNA Element-Driven Evolution of Bacterial Pathogens View all 13 articles
 Lee Julia Bartsch1
Lee Julia Bartsch1 Maria Borowiak1
Maria Borowiak1 Carlus Deneke1
Carlus Deneke1 Josephine Gruetzke1
Josephine Gruetzke1 Jens-Andre Hammerl1
Jens-Andre Hammerl1 Burkhard Malorny1
Burkhard Malorny1 Istvan Szabo1
Istvan Szabo1 Thomas Alter2
Thomas Alter2 Kim Katherine Nguyen3
Kim Katherine Nguyen3 Jennie Fischer1*
Jennie Fischer1*Salmonella enterica subsp. enterica serovar Agona has a history of causing food-borne outbreaks and any emergence of multidrug-resistant (MDR) isolates in novel food products is of concern. Particularly, in food products frequently consumed without sufficient heating prior to consumption. Here, we report about the MDR isolate, 18-SA00377, which had been isolated from a dietary supplement in Germany in 2018 and submitted to the German National Reference Laboratory for Salmonella. WGS-based comparative genetic analyses were conducted to find a potential reservoir of the isolate itself or mobile genetic elements associated with MDR. As a phylogenetic analysis did not yield any closely related S. Agona isolates, either globally or from Germany, a detailed analysis of the largest plasmid (295,499 bp) was performed as it is the main carrier of resistances. A combined approach of long-read and short-read sequencing enabled the assembly of the isolate’s chromosome and its four plasmids. Their characterization revealed the presence of 23 different antibiotic resistance genes (ARGs), conferring resistance to 12 different antibiotic drug classes, as well as genes conferring resistance to six different heavy metals. The largest plasmid, pSE18-SA00377-1, belongs to the IncHI2 plasmid family and carries 16 ARGs, that are organized as two distinct clusters, with each ARG associated with putative composite transposons. Through a two-pronged approach, highly similar plasmids to pSE18-SA00377-1 were identified in the NCBI database and a search for Salmonella isolates with a highly similar ARG resistance profile was conducted. Mapping and structural comparisons between pSE18-SA00377-1 and these plasmids and Salmonella isolates showed that both the plasmid backbone and identical or similar ARG clusters can be found not only in Salmonella isolates, originating mostly from a wide variety of livestock, but also in a diverse range of bacterial genera of varying geographical origins and isolation sources. Thus, it can be speculated that the host range of pSE18-SA00377-1 is not restricted to Salmonella and its spread already occurred in different bacterial populations. Overall, this hints at a complex history for pSE18-SA00377-1 and highlights the importance of surveilling multidrug-resistant S. enterica isolates, especially in novel food items that are not yet heavily regulated.
The bacterial genus Salmonella spans two species and more than 2,500 serovars (Grimont and Weill, 2007). Non-typhoidal serovars of one of its constituent subspecies, Salmonella enterica subsp. enterica, are the principal cause of food-borne illness worldwide manifesting as enteric infections (salmonellosis) with symptoms such as diarrhea, fever, abdominal pain and vomiting. Salmonellosis is a worldwide public health issue. In 2021, it was the second most reported zoonosis with over 60,000 reported cases in humans in the European Union (EU), including 71 deaths (European Food Safety Authority and European Centre for Disease Prevention and Control, 2022b). Additionally, the World Health Organization (WHO) estimates that salmonellosis accounts for more than 150,000 deaths worldwide annually (Ao et al., 2015). Despite the large number of serovars, fewer than 100 serovars are the main causative agents of human salmonellosis.
Starting in the 1970s, the serovar Salmonella enterica subsp. enterica serovar (S.) Agona has emerged on the radar of public health agencies, when a major S. Agona outbreak in five different countries was traced to animal feed based on contaminated fish meal from Peru (Clark et al., 1973). S. Agona now ranks among the top 20 most frequent serovars both in the European Economic Area (EEA) and European Union, having caused 784 reported cases of human salmonellosis from 2019 to 2021 (European Food Safety Authority and European Centre for Disease Prevention and Control, 2022a). In the past decades, numerous S. Agona outbreaks have occurred, ranging from smaller, localized outbreaks to larger, transnational and even transcontinental outbreaks. The first recognized transcontinental S. Agona outbreak occurred in 1994/95 in England, Wales, the United States (Killalea et al., 1996), and Israel. Here, the origin was traced to contaminated kosher savory snacks, frequently consumed by children (Shohat et al., 1996). A further outbreak in the United States occurred in a similar isolation source, dry rice and wheat cereal products, in 1998 and re-occurred 10 years later in 2008 (Russo et al., 2013). A similar re-emergence of S. Agona contamination in the same food matrix and linked to the same production facility, but 12 years apart, has also been stipulated with the 2005 (Brouard et al., 2007) and 2017 (Jourdan-da Silva et al., 2018) outbreaks in infant milk formula. Other past S. Agona outbreaks could be traced to air-dried beef products (Taylor et al., 1998), pre-cooked meat products (Nicolay et al., 2011), fresh papaya (Mba-Jonas et al., 2018) and aniseed tea (Koch et al., 2005). Notably, these outbreaks occurred in a diverse range of mostly dried food products that are frequently consumed by children and infants without sufficient prior cooking.
Since salmonellosis symptoms are ordinarily mild, treatments usually do not include antibiotics, except for serious infections or in infections of vulnerable populations such as infants, the elderly and immunocompromised. However, excessive and improper use of antibiotics in the treatment of food-producing animals has led to the emergence of antimicrobial resistance (AMR) as well as multidrug-resistance (MDR) in Salmonella species. Particularly alarming is the occurrence of isolates with increased resistance to ciprofloxacin or combined resistance to fluoroquinolones and third generation cephalosporins (European Food Safety Authority and European Centre for Disease Prevention and Control, 2023). This trend poses a major threat to public health through contamination in the food chain. Salmonella has been shown to harbor antimicrobial resistances genes (ARGs) including those, causing resistance against last-resort antibiotics such as colistin (Borowiak et al., 2017; Fernández et al., 2018) encoded on various plasmid families (Carattoli, 2003; Rozwandowicz et al., 2018). Therefore, monitoring and surveillance of antimicrobial resistance in Salmonella is crucial to prevent further spread and infection. Particular attention has to be given to novel foods such as insect-derived foods or dietary supplements as they are often produced using new methods, with novel ingredients from unfamiliar sources. Especially, the recent increase in the consumption of dietary supplements in the EU (Lordan, 2021) is worrying as similar matrices have caused Salmonella outbreaks before – including S. Agona outbreaks. None of these matrices – dietary supplements, cereals, etc. – are heated or are sufficiently heated immediately prior to consumption. Thus, potential contaminations with Salmonella spp. pose a serious health risk to the consumer, especially since infants and vulnerable or ill populations frequently consume these products. This is evident in the 2021 outbreak of salmonellosis by S. Typhimurium in Denmark, which was linked to contaminated herbal supplement capsules containing psyllium husk (Technical University of Denmark, 2022).
Here, we have investigated a multidrug-resistant S. Agona, isolated from dietary supplements in 2018. We also sequenced and characterized its very large constituent plasmid harboring many antimicrobial and heavy metal resistance genes on mobile genetic elements and tried to reconstruct its origin. Structural comparisons revealed a composite plasmid structure, found in isolates from numerous other genera, geographic origins and isolation matrices highlighting the enormous potential for shuffling resistance determinants within an epidemiologically widely disseminated plasmid backbone.
On average, the National Reference Laboratory (NRL) for Salmonella in Germany receives around 3,000–4,000 Salmonella isolates annually from different sources. Since 2018, whole-genome short-read sequencing is completed on all S. Enteritidis isolates, as well as isolates from national zoonosis monitoring and control programs. Since 2019, these isolates are further supplemented with sequencing of project- and outbreak-specific isolates, rough Salmonella isolates, and since 2020 isolates arising from Phase I of GenoSalmSurv (Uelze et al., 2021). Since 2021, foodborne isolates, isolates from Phase II of GenoSalmSurv and isolates arising from antimicrobial resistance monitoring according to Commission Implementing Decision (EU) 2020/1729 are also sequenced.
Isolate 18-SA00377 was submitted to the NRL for Salmonella in 2018. The sample was isolated from a dietary supplement that had been submitted to the Bavarian Health and Food Safety Authority in 2018, after having been recalled by the distributor in late 2017. According to submission metadata supplied by the Bavarian authority, the dietary supplement consisted of gelatin capsules filled with plant material enriched with vitamins and magnesium, intended for twice daily consumption.
The S. Agona isolates of this study underwent classical serotyping according to the White-Kauffmann-Le Minor scheme (Grimont and Weill, 2007) by slide agglutination with the O- and H-antigen specific sera (Sifin Diagnostics, Berlin, Germany) as part of the routine diagnostics of the NRL for Salmonella.
Antibiotic susceptibility testing was performed by the NRL for Antibiotic Resistance at the BfR, using broth microdilutions or disk diffusion. Following CLSI guidelines (version A7-M11) for broth microdilution procedures, minimum inhibitory concentrations (MIC) were obtained for 37 different antibiotics. These were interpreted following the epidemiological cutoff values provided by EUCAST (v.10.0) (The European Committee on Antimicrobial Susceptibility Testing, 2021).
Preparation of agarose plug molds and subsequent digestion by S1 nuclease (Thermo Fisher Scientific) were performed using a previously described protocol (Rodríguez et al., 2009). A plasmid pattern by PFGE was generated using the CHEF-DR III system (Bio-Rad Laboratories, Madrid, Spain) under standardized run conditions described by PulseNet standardized protocol (available at1). The Salmonella Braenderup strain H9812 (digested with the restriction endonuclease “XbaI” (Thermo Fisher Scientific, Darmstadt, Germany)) was used for size comparisons.
For sequencing of 18-SA00377, genomic DNA was extracted using the PureLink Genomic DNA Mini Kit (Thermo Fisher Scientific, Waltham, MA, USA) and sequenced using MiSeq (Illumina, San Diego, CA, USA) and Oxford Nanopore Technologies (ONT, Oxford, UK) MinION devices.
A short-read sequencing library was prepared using the sparQ DNA Frag & Library Prep Kit (Quantabio Beverly, MA, USA). Paired-end sequencing was performed in 2 × 151 bp cycles on an Illumina MiSeq instrument using the MiSeq Reagent Kit v3 (600 cycle). Trimming of short-reads using fastp v0.19.5 (Chen et al., 2018) resulted in 1.9 million high quality reads (≥ 89% Q30).
An Oxford Nanopore (ONT) sequencing library was prepared according to manufacturer’s protocol using the Rapid Barcoding Kit (SQK-RBK004) and sequenced on an ONT MinION sequencer MK1C using a FLO-MIN106 R9.4.1 flow cell. Basecalling was performed with Albacore v2.3.1 (available at2). Obtained reads were trimmed using Porechop v0.2.3 (available at3), filtered, and quality checked using NanoStat v1.4.0 and NanoFilt v2.7.1 (De Coster et al., 2018), respectively. In total, 123,777 reads with a read length N50 value of 11,288 bp and a mean read quality score of 10.3 were available.
Both data sets were assembled and circularized using Unicycler v0.4.8 (Wick et al., 2017) including Pilon (Walker et al., 2014). Default parameters were used for all software unless otherwise noted.
Characterization of the S. Agona sequences from the NRL for Salmonella, including 18-SA00377 and its four plasmids, was completed using the BakCharak pipeline version 3.0.4 (including -species Salmonella and -bakta options) (available at4). In short, the pipeline relies on the following tools: NCBI AMRFinderPlus (Feldgarden et al., 2021) to find antimicrobial resistance genes, ABRicate (available at5) to classify plasmids using CGE PlasmidFinder (Carattoli et al., 2014) and to find virulence factors using VFDB (Chen et al., 2015), Mash (Ondov et al., 2016) to find a nearest reference and plasmid reference using NCBI RefSeq (O'Leary et al., 2015) and NCBI plasmid database, respectively, Blastn (Camacho et al., 2009) to blast plasmids against the NCBI plasmid database, Bakta (Schwengers et al., 2021) for annotations using the Bakta database (Schwengers, 2021), and lastly, Platon (Schwengers et al., 2020) to predict plasmid contigs utilizing the Platon database (Schwengers, 2020). Further, Salmonella-specific characterization occurred using the SISTR tool with the sistr database and sistr 330 locus scheme for serotyping and cgMLST, respectively (Yoshida et al., 2016).
The annotations were supplemented with NCBI Prokaryotic Genome Annotation Pipeline (PGAP) (Tatusova et al., 2016) and the following tools: ISfinder v. 2–2022-04-06 (Siguier et al., 2006), PhiSpy v.4.2.21 (Akhter et al., 2012), MobileElementFinder v1.0.3 (Johansson et al., 2020) and SPIFinder2 (Roer et al., 2016). For SPIFinder2, thresholds of 80 and 90% of minimum sequence coverage and identity, respectively, were used to identify Salmonella Pathogenicity Islands (SPIs). Visualization occurred with circos v.0.69–6 (Krzywinski et al., 2009).
For a representative selection of the S. Agona isolates received by the strain collection of the German NRL for Salmonella, isolates of different isolation sources, isolation years and geographic origins were sequenced (Supplementary File 6). Genomic DNA was prepared as described above and short-read libraries were prepared using the Nextera DNA Flex Library Prep Kit (Illumina, San Diego, CA, USA Illumina). Sequencing occurred on either a NextSeq 500 or a MiSeq device (Illumina, San Diego, CA, USA). MiSeq sequencing was performed as described for isolate 18-SA00377-0. For the NextSeq, paired-end sequencing was performed in 2 × 151 bp using the NextSeq 500/550 Mid Output Kit v2.5 (300 Cycles).
Short-read data of 18-SA00377 is available in BioProject PRJNA706607 and was compared to genetically similar isolates on NCBI Pathogen Detection database (accessed on 12.12.2022 at6) based on Single Nucleotide Polymorphisms (SNPs). Within the SNP cluster, three closely related isolates were identified and included in the phylogenetic analysis.
Additionally, all available S. Agona sequences (157 isolates, excluding duplicates) from the NRL for Salmonella (BioProject PRJEB31846 and BioProject PRJNA742494) were included for phylogenetic comparisons as well as the following six isolates: one MDR S. Agona sequence isolated from a silver gull and five isolates from recent European S. Agona outbreaks (Supplementary File 6). Where required, read-based sequencing data was assembled with the Assembly-based QUality Assessment for Microbial Isolate Sequencing (AQUAMIS) pipeline (Deneke et al., 2021a). A S. Agona isolate (GCA_011632245.1) with a highly similar resistance profile to 18-SA00377 (more than 80% overlap of ARGs) was also included in the phylogenetic comparison. Thus, a total of 167 isolates were included in the phylogenetic analysis using the cgMLST workflow ChewieSnake (Deneke et al., 2021b). This workflow implements the allele calling software chewBBACA and generates an allele distance matrix, cluster membership, and phylogeny. The resulting allele distance matrix was visualized as a minimum spanning tree using iTOL (Letunic and Bork, 2021). Isolates were classified based on the number of different antimicrobial resistance classes conferred by their ARGs according to the comprehensive antibiotic resistance database (Alcock et al., 2019).
The plasmid pSE18-SA00377-1 was analyzed using two tools in order to find closed plasmid sequences with high levels of similarity: the MOB-cluster tool from the MOB-suite (Robertson and Nash, 2018), which utilizes fast genomic distance estimation using Mash, and the web service COPLA (available at7), a plasmid classifying tool. The resulting plasmids deemed similar to pSE18-SA00377-1 by these two tools were characterized using BakCharak version 3.0.4 and their Average Nucleotide Identity (ANI) scores were calculated using FastANI (Jain et al., 2018). Plasmids with the highest ANI were mapped against pSE18-SA00377-1 using minimap2 version 2.22-r1101 (−asm5 option). The 11 plasmids with the highest mapping coverages against pSE18-SA00377-1 were visualized using circos v. 0.69–6 6 (Krzywinski et al., 2009). Easyfig v. 2.2 (Sullivan et al., 2011) was utilized to compare the structural organization of two subregional clusters of pSE18-SA00377-1 with four of these plasmids. Easyfig visualized the BLAST results from comparing the two clusters (100–146 kb and 220–260 kb) with the four plasmids with the highest mapping coverage (NC_012555, CP011601, CP042552, and NC_012556).
The AMR resistance profile of the 18-SA00377 isolate available on NCBI was compared to all available isolates of S. enterica, E. coli and Shigella sp., Klebsiella pneumoniae, Enterobacter sp., Acinetobacter baumannii on the respective NCBI Pathogen Detection databases (available at https://www.ncbi.nlm.nih.gov/pathogens)8 using a custom R script (Supplementary File 8) (databases download on 31.10.2022). The aim was to identify further Salmonella strains harboring plasmids that are similar to pSE18-SA00377-1, but for which no closed plasmid sequences were available. The draft genomes of all Salmonella genomes with a resistance profile that shared a minimum of 80% of the ARGs of 18-SA00377 were downloaded (Table 1), and mappings to pSE18-SA00377-1 with minimap2 version 2.22-r1101 (−asm5 option) (Li, 2018) were visualized using circos v. 0.69–6 (Krzywinski et al., 2009).
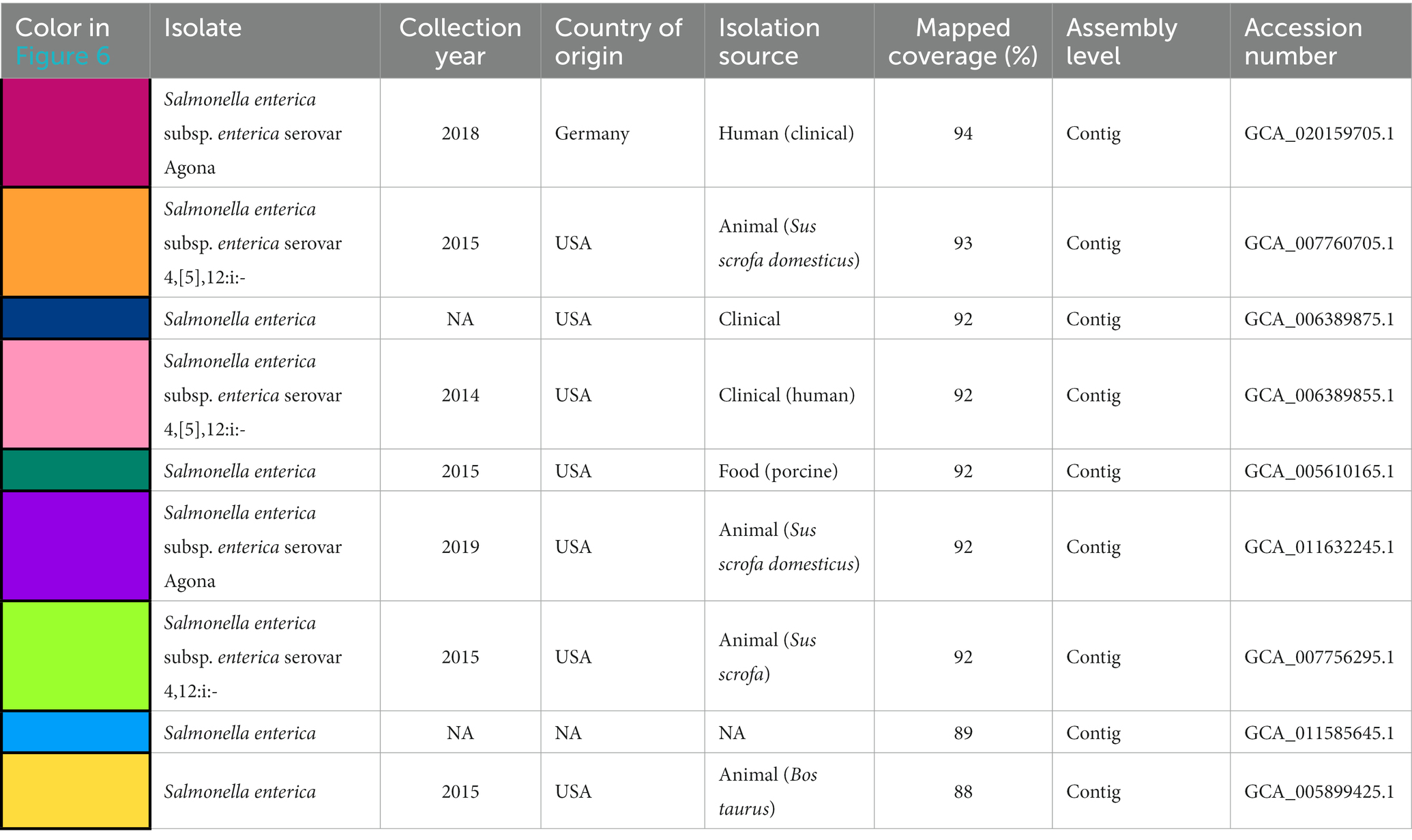
Table 1. Available metadata details of the nine Salmonella isolates mapped against pSE18-SA00377-1, covering their size in bp, date of collection, country of origin, isolation source, mapping coverage (%) to pSE-18-SA0037-1, assembly level and their accession numbers.
To analyze transferability of pSE18-SA00377-1, filter mating conjugation studies using sodium azide-resistant E. coli K-12 J53 recipient cells were conducted. Selective marker for p18-SA00377-1 was the tetD gene, located on the plasmid (see Table 2). Filter mating conjugation was performed as previously described (Hadziabdic et al., 2018). The reaction mixtures were plated on transconjugant selective LBA plates containing 12.5 mg/liter tetracycline (tetracycline, Sigma-Aldrich, Steinheim, Germany) and 100 mg/liter sodium azide (NaN3, Sigma-Aldrich, Darmstadt, Germany) and incubated at 37°C for approximately 42 h. A selection of potential transconjugants were picked and singulated on new selective tet/NaN3 plates. From these plates, single colonies were inoculated in LBL and incubated at 37°C under shaking condition (250 × rpm) for 16 h. Thermal cell lysis preparations were produced as previously described (Borowiak et al., 2017). Transconjugants were confirmed by tetD and J53 K12 screening PCR. The following primer pairs (with primer sequences in 5′-3′ direction and product length in parentheses) were used for the screening PCRs: For the J53 K12 screening K12R (ATCCTGCGCACCAATCAACAA; 1687 bp) and K12L (TTCCCACGGACATGAAGACTACA; 1687 bp) (Bauer et al., 2007), and for the tet(D) screening tet(D)-1 (AAACCATTACGGCATTCTGC; 787 bp) and tet(D)-2 (GACCGGATACACCATCCATC; 787 bp) (Ng et al., 2001).
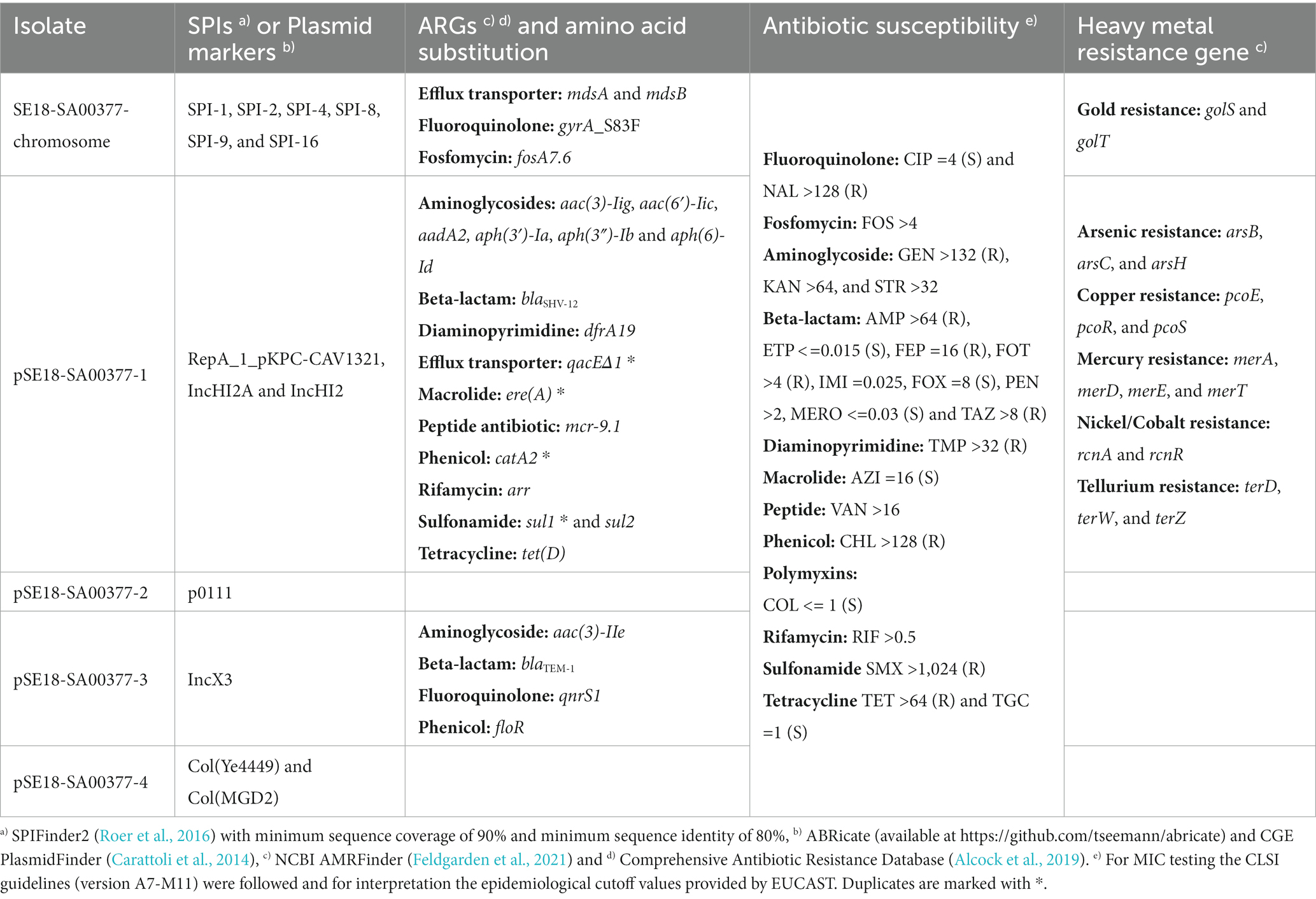
Table 2. Breakdown of SPIs, plasmid markers, antibiotic resistance genes (ARGs), heavy metal resistance genes, and results from antibiotic susceptibility testing of the 18-SA00377 isolate including its four constituent plasmids.
The PCR reactions were prepared in 25 μL including 12.5 μL 2x DreamTaq Green PCR Master Mix (Thermo Scientific, Vilnius, Lithuania), 2.5 μL of each 10 μM primer dilution (s. above for details), 5.5 μL PCR grade water and 2 μL of thermal cell lysis preparation as template DNA and carried out as follows: initial denaturation for 5 min at 95°C, 30 cycles denaturation for 30 s at 95°C, primer annealing for 30 s at 54°C and elongation for 1 min [tet(D)] or 1:40 min (J53 K12) at 72°C followed by a final elongation step for 10 min at 72°C.
Illumina short-read and ONT long-read sequencing resulted in a hybrid assembly of one Salmonella chromosome with 4,856,956 bp and four plasmids: one IncHI2 plasmid (pSE18-SA00377-1) of 295,499 bp, one p0111 plasmid (pSE18-SA00377-2) of 94,574 bp, one IncX3 plasmid (pSE18-SA00377-3) of 50,931 bp and one Col (Ye4449) plasmid (pSE18-SA00377-4) of 5,284 bp. These five constituent assemblies of 18-SA00377 are available online on NCBI, grouped under the GenBank Accession number GCA_021497565.1 and BioProject PRJNA706607 with their individual Genbank Accession numbers as follows: CP071388.1 (chromosome), CP071389.1 (pSE18-SA00377-1), CP071390.1 (pSE18-SA00377-2), CP071391.1 (pSE18-SA00377-3), and CP071392.1 (pSE18-SA00377-4).
The presence of at least three plasmids was confirmed by PFGE using the S1 nuclease (Supplementary File 1). The sizes of these three plasmids were determined to be around 45, 80 and 320 kb. While varying slightly from the whole genome sequencing results this can be explained by the fact that S1-PFGE is more reliable at ascertaining plasmid sizes above 100 kb (Barton et al., 1995; Li et al., 2022) and can be unreliable for smaller plasmid sizes (Zhang et al., 2020; Juraschek et al., 2021).
Following genotypic characterization using the BakCharak pipeline and NCBI PGAP, the isolate 18-SA00377 was found to exhibit one DNA gyrase amino acid substitution at codon 83 (gyrA_S83F) and 23 different antibiotic resistance genes (ARGs). In total, the isolate 18-SA00377 carries 27 ARGs as four ARGs [catA2, qacEΔ1, sul1 and ere(A)] are in duplicate and they confer resistance to 12 different classes of antibiotics resistance (Table 2). This was supported by antimicrobial susceptibility testing against antimicrobials of nearly every drug class, excluding efflux transporters. Moreover, the isolate harbors resistance genes against six heavy metals (gold, tellurium, arsenic, mercury, copper, and nickel/cobalt) as well as containing 131 chromosomal virulence factors belonging to seven classes (adherence, antimicrobial activity/competitive advantage, effector delivery system, immune modulation, invasion, nutritional/metabolic factor, and regulation) (data not shown). The isolate’s chromosome also harbors six Salmonella Pathogenicity Islands (SPIs) – SPI-1, SPI-2, SPI-4, SPI-8, SPI-9, and SPI-16 – with a sequence coverage of minimum 90% and sequence identity of above 80%. The following nine further SPIs were identified having a sequence coverage between 30 and 80%, but sequence identities of above 90%: SPI-3, SPI-12 (two copies), SPI-11, SPI-5, SPI-19, SPI-16 (two copies). Three of these SPIs (SPI-1, SPI-2, and SPI-4) have important roles in the virulence of Salmonella infections and can be found in all serovars of Salmonella enterica. Both SPI-1 and SPI-2 encode a distinct of type III protein secretion system (Jennings et al., 2017; Lou et al., 2019), which are, inter alia, important for the penetration and invasion of epithelial intestinal cells. On the other hand, the role of SPI-4 encodes a type I secretion system, which is crucial for adhesion during Salmonella infections (Gerlach et al., 2007). Two other SPIs – SPI-8 and SPI-9 – have been characterized based on the complete genome sequence of a S. Typhi CT18 strain (Parkhill et al., 2001). The function of SPI-8 is not well understood, but it has been shown to conferring resistance to bacteriocins (van Asten and van Dijk, 2005), while SPI-9, similar to SPI-4, encodes a type I secretion system as well as a RTX toxin-like protein (Velásquez et al., 2016). Lastly, SPI-16 has a role in immune evasion by carrying genes involved in O-antigen variation (Ilyas et al., 2017). Of the nine other SPIs present, the SPI-5 and SPI-3 are of particular importance as they are important mediators in host colonization and intracellular survival (Blanc-Potard et al., 1999), as well as the enteric stage of a Salmonella infections (Marcus et al., 2000), respectively. Overall, the presence of such a wide range of SPIs in a single isolate underlines the pathogenic potential of 18-SA00377. Thus, as previous outbreaks of S. Agona have shown, this serovar can cause human harm and the presence of both multidrug resistance and heavy metal resistances, highlight the importance of finding closely related isolates and establishing phylogeny.
NCBI Pathogen Detection database based on Single Nucleotide Polymorphism (SNP) was searched for closely related isolates. This showed that the isolate 18-SA00377 is, with a minimum SNP distance of 31, only distantly related to three other S. Agona isolates: GCA_011585645.1, GCA_006296875, and GCA_020159705.1) (Supplementary File 6).
Next, all available sequences of S. Agona isolates from the German NRL for Salmonella was supplemented with S. Agona sequences from four other sources. Firstly, from three foodborne outbreaks: the 2017/18 infant formula outbreak in France (Jourdan-da Silva et al., 2018), the 2002/03 herbal tea outbreak (Koch et al., 2005), and the outbreak caused by a Bavarian feed product (Dangel et al., 2019). Secondly, three isolates from the aforementioned NCBI Pathogen Detection database (GCA_011585645.1, GCA_006296875, and GCA_020159705.1) as well as the sequence of a extensively drug-resistant S. Agona isolated from a silver gull (GCA_012169275.1) (Cummins et al., 2020). Lastly, a S. Agona isolate (GCA_011632245.1) with a high similarity ARG resistance profile was also included. A cgMLST analysis of these 167 S. Agona sequences (Supplementary File 6) was completed and visualized as a minimum spanning tree (Figure 1) (Letunic and Bork, 2021).
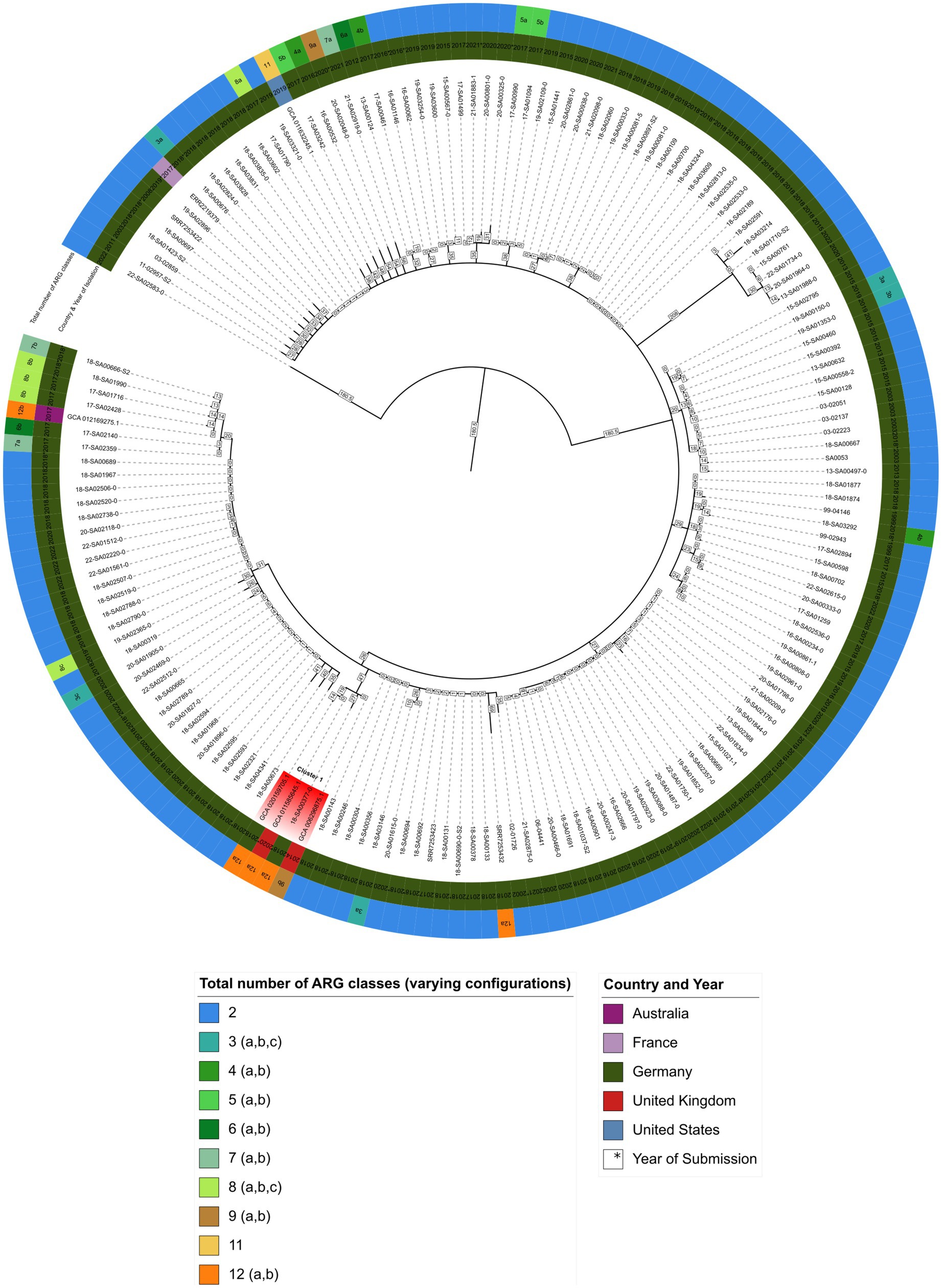
Figure 1. Minimum spanning tree showing hierarchical clustering between 167 S. Agona isolates. Inner circle indicates the country and isolation year, outer circle represents the number of ARG classes for each isolate, respectively (Supplementary File 6). Branch lengths in square boxes indicate minimum allelic distance between isolates. Varying configurations in the total number of different antibiotic resistance drug classes are indicated with a, b, and c – for a detailed breakdown see Supplementary File 7.
As shown in Figure 1, isolate 18-SA00377 is located within a clade, here named cluster A, with three other isolates: GCA_020159705.1, GCA_011585645.1 and GCA_006296875.1 with minimum allelic distances of 60, 19 and 27, respectively. These three isolates are not isolates from the German NRL for Salmonella, but were found via the NCBI Pathogen Detection database search. Two isolates, GCA_020159705.1 and GCA_006296875.1, were isolated from human sources in Germany in 2018 and the United Kingdom in 2014, respectively, while for GCA_011585645.1 metadata was not available. The closest S. Agona isolate to 18-SA00377 from the German NRL Salmonella has a minimum allelic distance of 74. The closest outbreak-associated S. Agona isolate is SRR7253423 from the Bavarian prevalence study, isolated from animal feed in Germany in 2017 (Dangel et al., 2019). As the allelic distances both between isolates within cluster A and to the closest cluster of isolates from the NRL for Salmonella exceed cut-offs previously used for defining distinct Salmonella outbreak clusters (Simon et al., 2018; Meinen et al., 2019), a recent common ancestor cannot be pinpointed by the cgMLST of 167 S. Agona isolates.
The total number of ARG drug classes across the 167 S. Agona isolates varies considerably and is not uniformly distributed. The majority of S. Agona isolates, including all the available S. Agona sequences in the NRL for Salmonella, harbor ARGs against only two classes, fosfomycin (fosA7.2) and efflux transporter subunits (mdsA and mdsB). Only a minority of S. Agona isolates (n = 8) carry resistance genes encoding for nine or more drug classes. Nevertheless, the four isolates in cluster A all carry ARGs against a minimum of nine antibiotic classes. Isolate 18-SA00377 is one of the most resistant isolates with 23 distinct ARGs conferring resistance to 12 different classes. There is an overlap between the antibiotic classes, with all four isolates in cluster A sharing resistances against the aforementioned fosfomycin and efflux transporter subunits, as well as the peptide antibiotic colistin (mcr-9.1), beta-lactams, fluoroquinolones, aminoglycosides, sulfonamides, tetracyclines, and diaminopyrimidine (see Supplementary Files 6, 7). Since the cgMLST analysis did not reveal a recent common ancestor for 18-SA00377, the antibiotic resistance profiles within cluster A and the unique resistance profile of 18-SA00377 highlight the importance of finding another potential source of the resistance properties of 18-SA00377, namely associated mobile genetic elements.
Next, we focused to finding isolates with similar antibacterial resistance profiles in a wider set of genera of Enterobacteriaceae. As the majority of its unique MDR resistance profile is due to the ARGs carried on the isolate’s largest plasmid, pSE18-SA00377-1, the plasmid was characterized. Assembly of the plasmids was possible by combining long-read and short-read sequencing data. Annotation of the assembly revealed that pSE18-SA00377-1 harbors a total of 20 ARGs (Table 2), including two copies of each of catA2, qacEΔ1, sul1, and ere(A). A further four ARGs (qnrS1, blaTEM-1, aac(3)-IIe, and floR) are located on pSE18-SA00377-3 and each associated with a putative composite transposon of the IS6 family (Supplementary File 4). The plasmids, pSE18-SA00377-2 and pSE18-SA00377-4, do not carry any antibiotic resistance genes, but pSE18-SA00377-4 carries two replicons, Col(MGD2) and Col(Ye4449) as well as mobilization genes (mobC, mbeD, mbeB, mbA) (Supplementary File 5).
Similar to the ARG distribution, the distribution of genes conferring heavy metal resistances is skewed toward pSE18-SA00377-1, where genes encoding against six different heavy metal resistances are located (Table 2). While the genome only carries golS and golT genes, encoding for gold resistance, the resistance genes against arsenic (arsB, arsC, and arsH), copper (pcoE, pcoR, pcoS), mercury (merA, merD, merE), nickel/cobalt (rcnA and rcnR), and tellurium (terD, terW, terZ) all are located on pSE18-SA00377-1.
The makeup of pSE18-SA00377-1 is not only limited to a large number of resistance genes, it also carries three plasmid markers: RepA_1_pKPC-CAV1321 as well as IncHI2A and IncHI2. Moreover, it also carries genes for the purpose of conjugational transfer by encoding for numerous conjugal transfer proteins (e.g., traK, traB, traV, etc.) as well as an oriT and repB replication initiator. The presence of these genes supports the assumption that the pSE18-SA00377-1 plasmid is transferrable in vivo.
Transmissibility of pSE18-SA00377-1 by conjugation was confirmed experimentally by in vitro filter mating experiments. Successful transfer of the plasmid to the recipient, E. coli J53 K12, occurred, supporting the bioinformatic analysis that pSE18-SA00377-1 is conjugative. Furthermore, as the recipient E. coli K12 J53 belongs to a different genus of the family Enterobacteriaceae, this supports the subsequent bioinformatic analyses that indicates that large parts of the pSE18-SA00377-1 plasmid backbone can be found in other bacterial genera and that the host range of the pSE18-SA00377-1 plasmid is not limited to Salmonella.
Arrangement of ARGs, heavy metal resistances, conjugation machinery and transposable elements on pSE18-SA00377-1 is concentrated within two distinct clusters, as evident in the visualization of the plasmid’s annotations (Figure 2). The annotations of the remaining plasmids, pSE18-SA00377-2, pSE18-SA00377-3, and pSE18-SA00377-4, were not visualized, but their consensus annotations are available as Supplementary Files 3–5, respectively. The first cluster on pSE18-SA00377-1, ~ 95–140 kb, contains nine ARGs interspersed with transposases. Four ARGs – aadA2, qacEΔ1, sul1, and dfrA19 – are part of a class 1 integron cassette as they are flanked on both sides by class 1 integron integrases (intI1) and have Pc and PintI1 promoters in their vicinity as well as two recombination crossover points (attC and attI). Moreover, all nine ARGs in this region are associated with putative composite transposons. Similarly, six ARGs – aadA2, qacEΔ1, sul1, dfrA19, aph(3″)-Ib and aph(6)-Id – are associated with a single putative composite transposon, cn_14741_IS26, which is flanked by two IS26 elements. A similar picture emerges for heavy metal resistances, with genes encoding for copper, nickel/cobalt, and nickel resistances associated with a putative composite transposon, cn_22931_IS903.
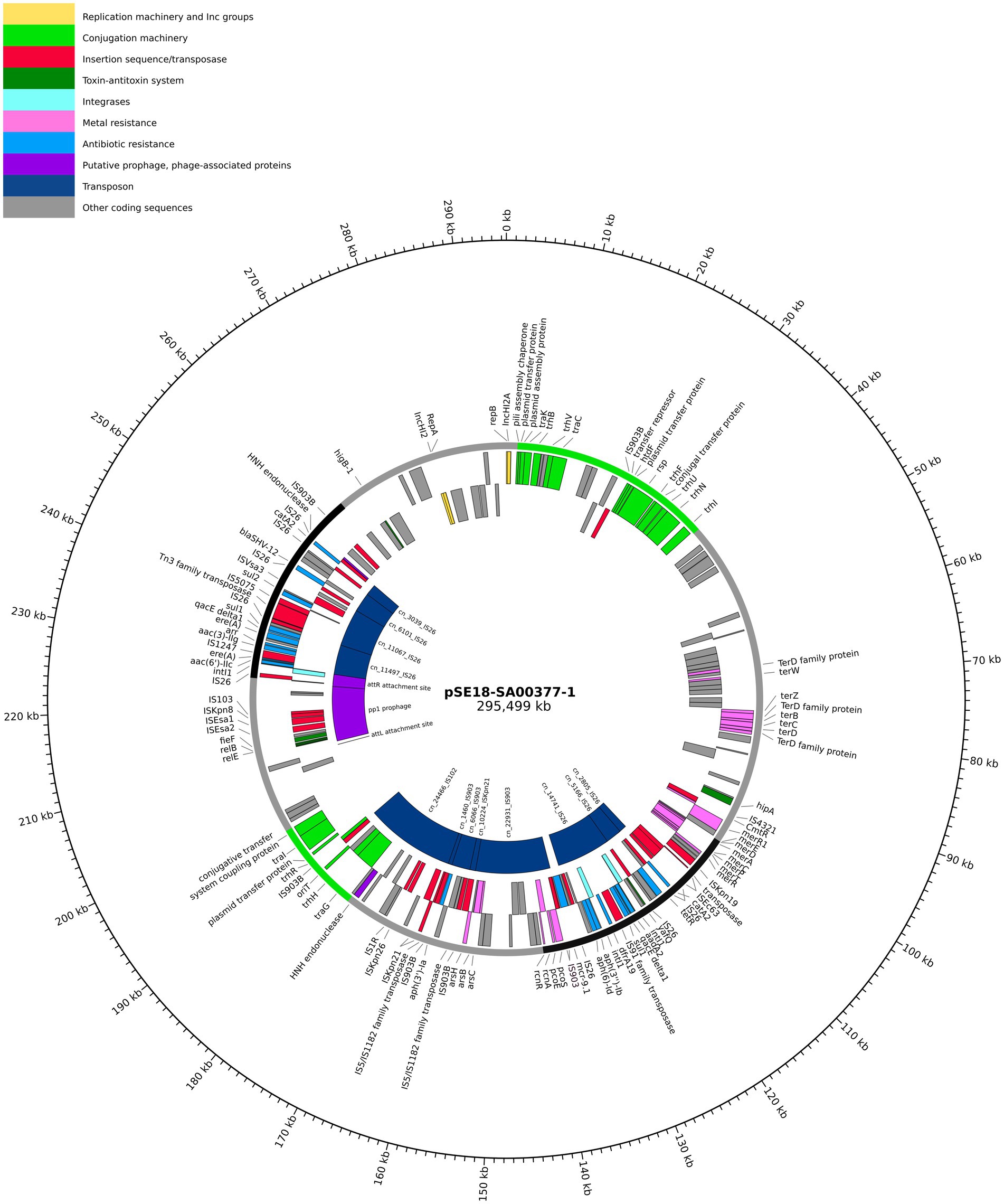
Figure 2. Visualization of the pSE18-SA00377-1 plasmid. Annotations are colored according to nine categories: replication machinery and incompatibility group in yellow, conjugation machinery in light green, insertion sequences and transposases in red, toxin-antitoxin system in dark green, integrases in turquoise, heavy metal resistances in pink, and antibiotic resistances in light blue. On the innermost track, the putative prophage and prophage-associated proteins are shown in purple, while putative transposons are colored dark blue. Coding sequences for other gene products are colored in gray. The outermost ring shows the plasmid backbone in gray and highlights regions of interest: cluster 1 (95–140 kb) and cluster 2 (230–250 kb) in black and regions with conjugation machinery in green (0–40 kb and 180–200 kb). For a complete list of available annotations, see Supplementary File 3.
A second cluster with a high concentration of ARGs spans the range 230–250 kb and contains further 10 ARGs. The aac(6′)-IIc gene is associated with an incomplete class 1 integron as a class 1 integron integrase (intI1), Pc and PintI1 promoters and recombination crossover points (attC and attI) are in its vicinity. Similar to the first cluster, all ARGs are located on putative composite transposons, with the first seven ARGs being associated with the cn_11497_IS26 putative composite transposon, while each of the remaining three ARGs are associated with a composite transposons of the IS6 family. Between these two distinct regions of the plasmid, pSE18-SA00377-1 also encodes components for a type II toxin-antitoxin system (relE, relB) and an iron efflux transporter (fieF).
Closer analysis of the plasmid’s genetic makeup revealed that this large plasmid has two distinct regions of densely clustered ARGs, which are associated with putative composite transposons. This, in conjunction with the presence of numerous copies of highly active IS26 elements, that frequently mediate recombination in Salmonella spp. (Doublet et al., 2009), suggests that this plasmid has a complex evolution with frequent insertions of ARGs.
In order to find closely related plasmids of pSE18-SA00377-1, the outputs of two tools, the MOB-cluster tool and the COPLA web tool, were further analyzed by calculating their ANI scores. Consequent ranking by ANI score and matching antibiotic resistance profiles yielded 11 high-similarity plasmids (Table 3), which were mapped to pSE18-SA00377-1 (Figure 3).
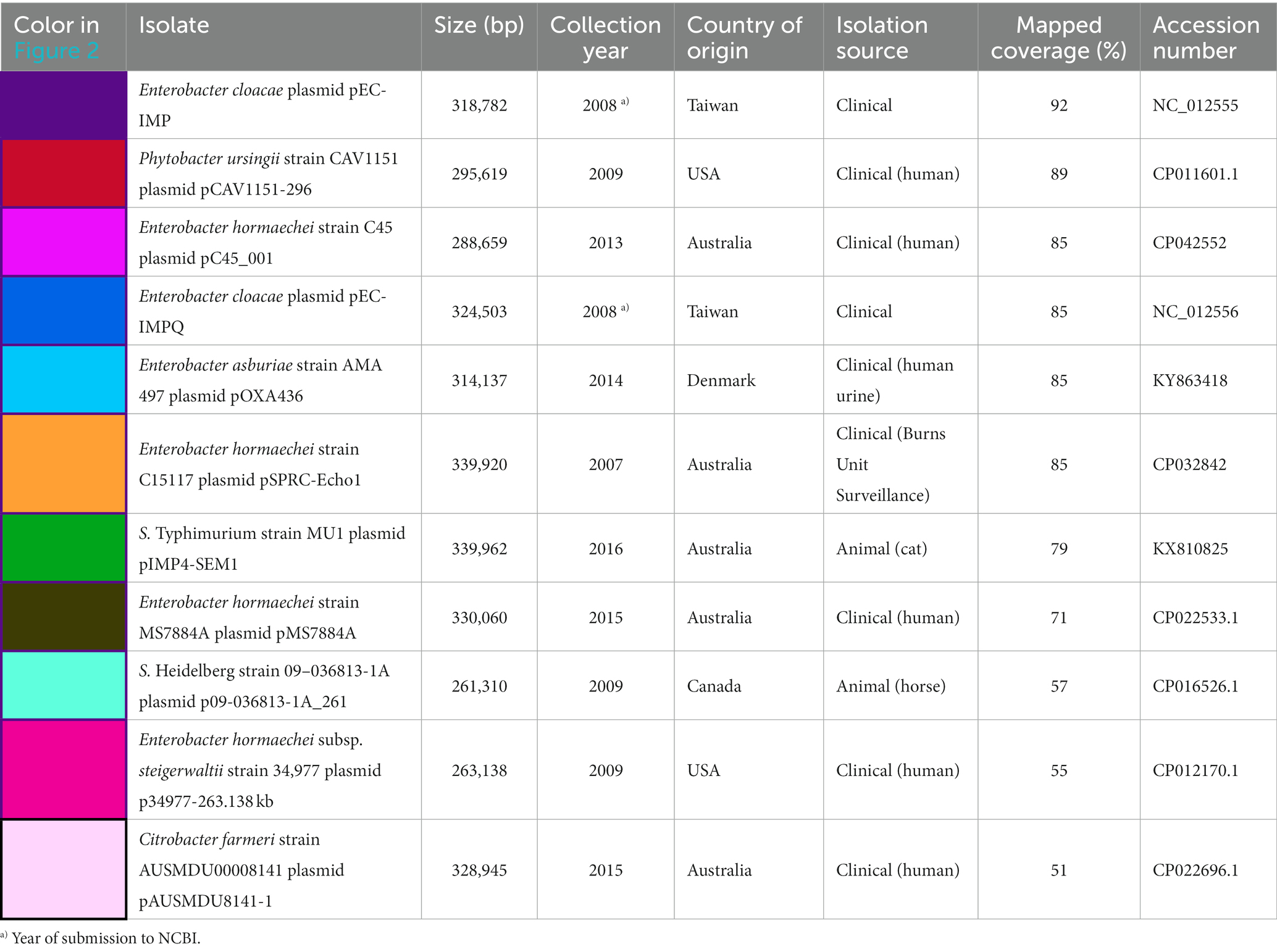
Table 3. Metadata details of the 11 plasmids mapped against pSE18-SA00377-1, covering their size in bp, date of collection or NCBI submission, country of origin, category of isolation source, percentage of coverage when mapped to pSE-18-SA0037-1, and their accession.
When mapped against pSE18-SA00377-1, these 11 plasmids exhibit a nucleotide coverage exceeding 50%. They were isolated from a wide range of bacterial isolates and their geographical origins span Australia (CP022696.1, CP022533.1, CP042552, CP032842, and KX810825), Taiwan (NC_012555 and NC_012556), the United States (CP012170.1 and CP011601.1), Canada (CP016526.1), and Denmark (KY863418). Furthermore, these plasmids were mostly isolated from Enterobacter species in a clinical context, but were also found in Salmonella, Citrobacter, and Phytobacter. The Enterobacter and Citrobacter plasmids were found exclusively in clinical isolates, while the Salmonella plasmids were isolated from animals. Particularly, the occurrence of the Salmonella plasmid, KX810825, is of concern as it was found in a companion animal (Abraham et al., 2016).
Figure 3 shows that the coverages against pSE18-SA00377-1 (E. cloacae pEC-IMP, Phytobacter ursingii, Enterobacter hormaechei, and E. cloacae pEC-IMPQ) all map in the region of the first cluster of ARGs. However, for the second cluster of ARGs at 230–250 kb only the E. cloacae pEC-IMP plasmid (in dark purple) maps against large parts of this cluster, while the other six plasmids do not. All eleven plasmids wholly map against the majority of the pSE18-SA00377-1 plasmid backbone, including the conjugation and replication machinery.
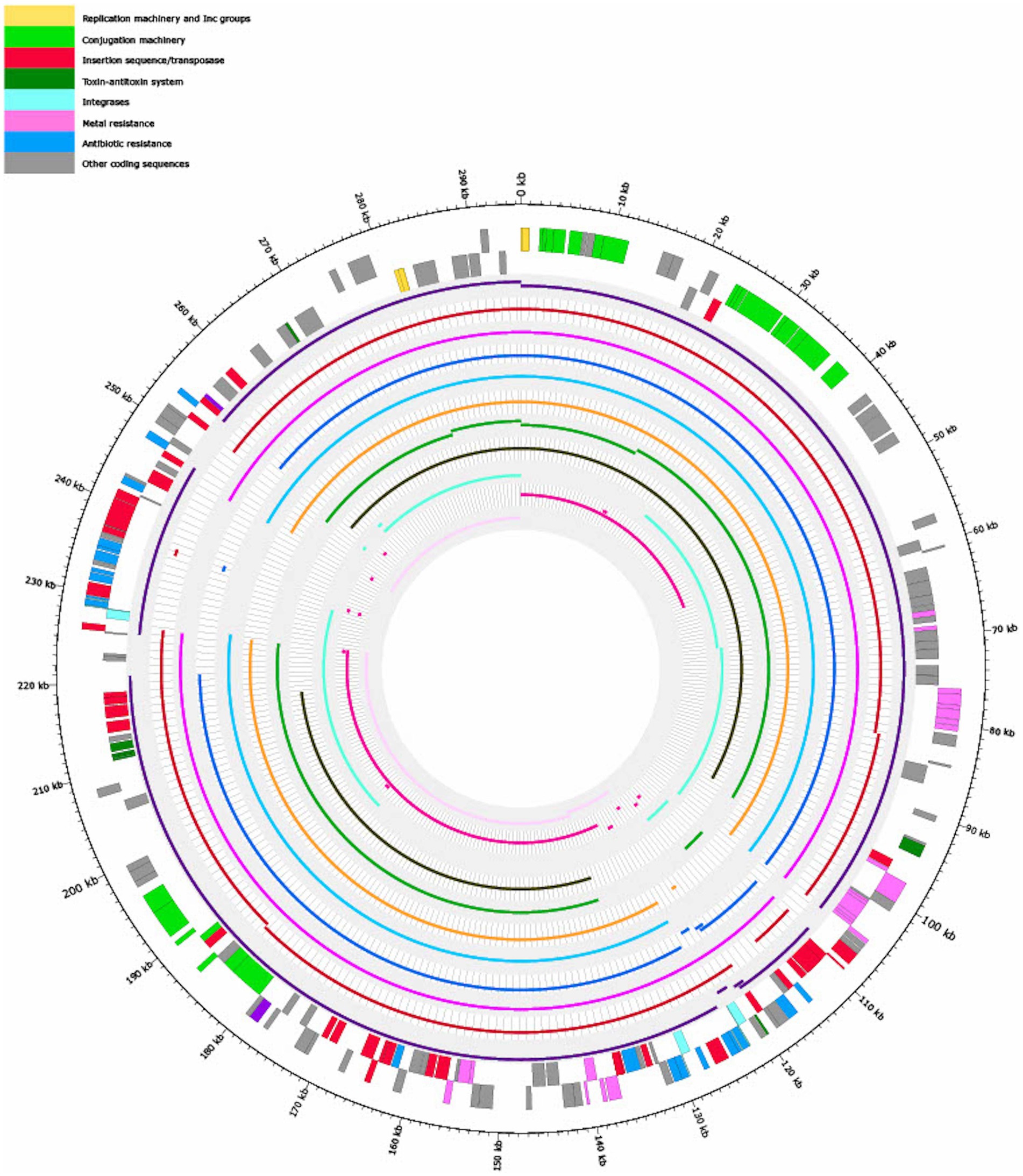
Figure 3. Visualization of mapping results of 11 plasmids against pSE18-SA00377-1 (outermost two tracks), in order of decreasing mapping coverage (%), with lowest coverage of the Citrobacter farmeri plasmid (light pink) and highest coverage of the Enterobacter cloacae plasmid pEC-IMP (dark purple) (see Table 3). Regions of the 11 plasmids that did not map to pSE18-SA00377-1 are not shown. The outermost black circle designates the base positions around the plasmid. Farthest two tracks on the outside represents the pSE18-SA00377-1 plasmid annotations, with replication machinery and incompatability groups colored in yellow, conjugation machinery in light green, insertion sequences and transposases in red, toxin-antitoxin system in dark green, integrases in turquoise, heavy metal resistances in pink, and antibiotic resistances in light blue. Coding sequences for other gene products are colored in gray. For labeling of annotations refer to Figure 2.
For closer comparison of the organization of the ARGs and other features within the two aforementioned ARG clusters of pSE18-SA00377-1, these regions were compared to the 11 high-similarity plasmids using BLAST and visualized using easyfig. This closer inspection revealed that the organization of the first cluster of pSE18-SA00377-1, 100–146 kb, is conserved across four plasmids NC_012555, CP011601, CP042552, and NC_012556 (Figure 4).
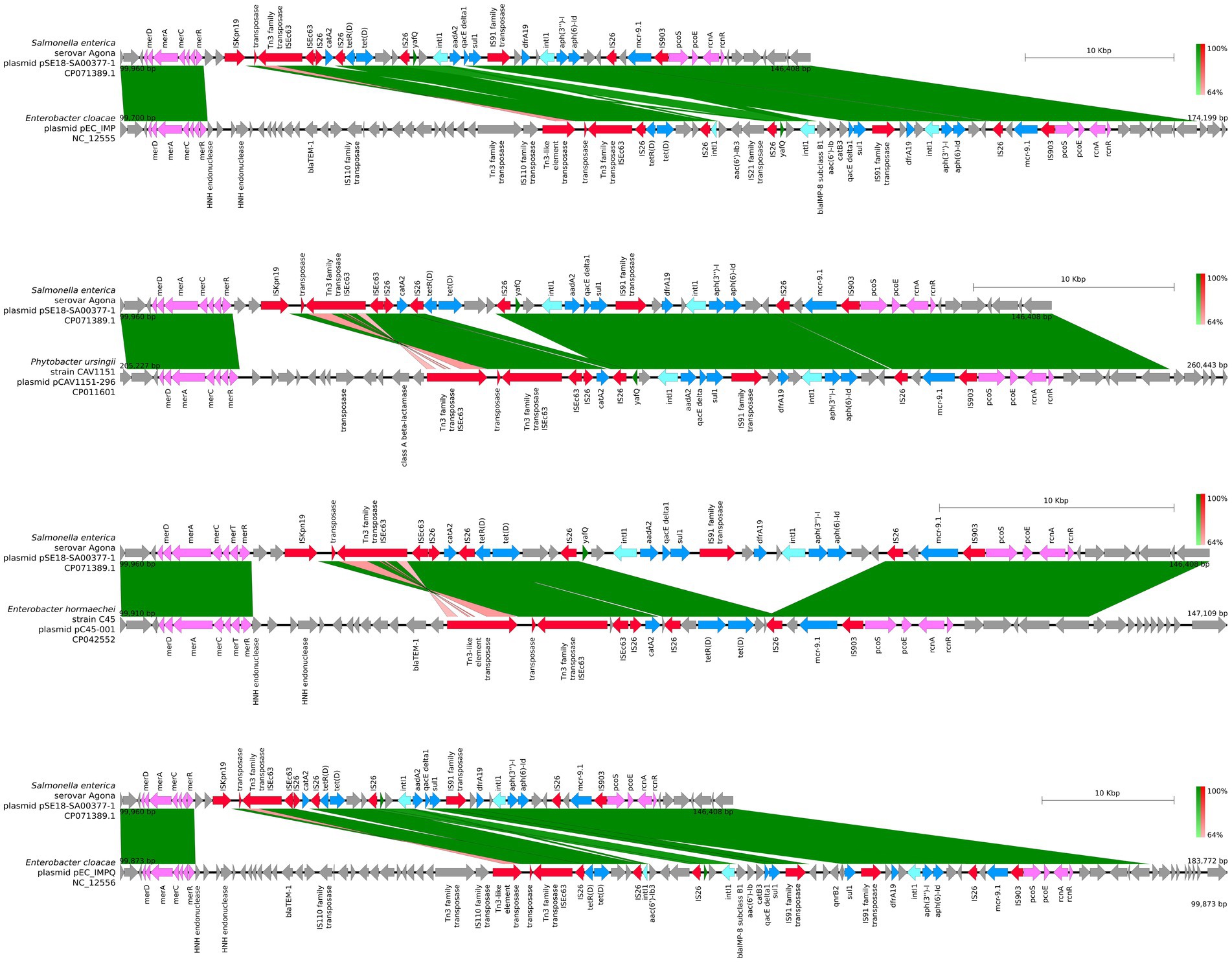
Figure 4. Visualization of comparison of the structural features between the 100–146 kb region of pSE18-SA00377-1 and four plasmids with the highest nucleotide coverages (NC_012555, CP011601, CP042552, and NC_012556). Annotations of coding sequences are only colored if BLAST results indicated homology between pSE18-SA00377-1 and the other plasmid. Annotation coloring is based on the same eight categories as in Figures 2, 3, with insertion sequences and transposases in red, toxin-antitoxin system in dark green, integrases in turquoise, heavy metal resistances in pink, and antibiotic resistances in light blue. Coding sequences present in the plasmids belonging to one of the eight colored annotation categories, but showing no homology by blast to pSE18-SA00377-1, were labeled but left in gray.
All four of these plasmids harbor minimum eight of the nine ARGs constituent of this region, catA2, tetD, aadA2, qacEΔ1, sul1, dfrA19, aph(3″)-Ib, aph(6)-Id, and mcr-9.1. In two plasmids, CP016526 and CP012170, the region with its nine ARGs is present but in an inverted state and only partially. Four ARGs are located toward the end of the region, dfrA19, aph(3″)-Ib, aph(6)-Id, and mcr-9.1. They are present in both plasmids, while the remaining ARGs, catA2, tetD, aadA2, qacEΔ1, sul1 have merged with the second cluster of ARGs (220–260 kb). In the five remaining plasmids, the organization of the ARGs is considerably changed, with the order of the ARGs split or shuffled (CP022696, CP022533, CP042552, CP032842, and KX810825) or merged partially (CP042552) with the second cluster of ARGs.
Similar to the first region, the second cluster of ARGs at 220–260 kb of pSE18-SA00377-1 was compared to four of the 11 aforementioned high-similarity plasmids and visualized using easyfig (Figure 5). For this second cluster, two plasmids (NC_012555 and NC_012556) harbor the same structural features in the same organizational matter as pSE18-SA00377-1, while the remainder of the 11 plasmids only carry a partial ARG load.
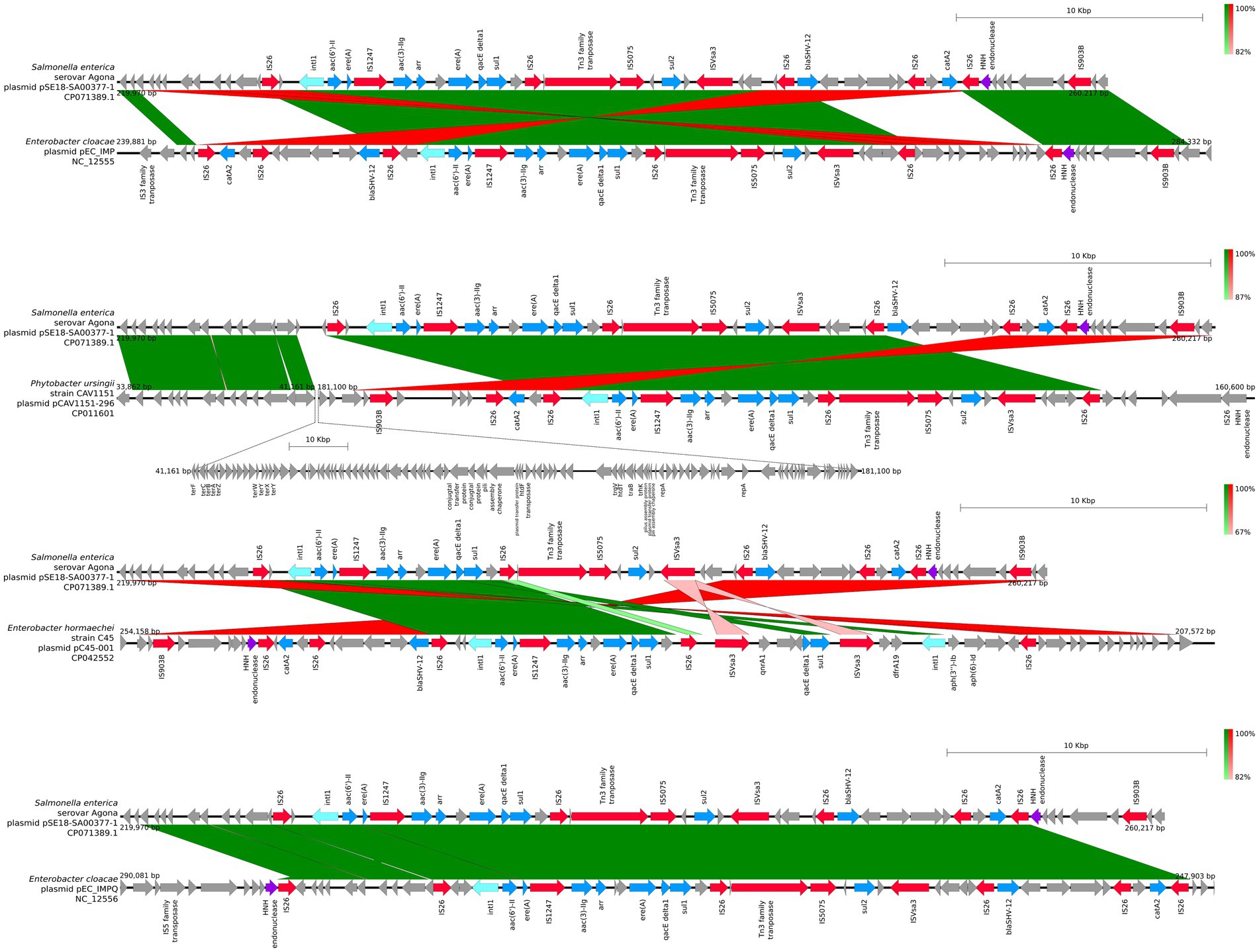
Figure 5. Visualization of comparison of the structural features between the 220–260 kb region of pSE18-SA00377-1 and four plasmids with the highest nucleotide coverages (NC_012555, CP011601, CP042552, and NC_012556). Annotations of coding sequences are only colored if blast results indicated homology between pSE18-SA00377-1 and the other plasmid’s. Annotation coloring is based on the same eight categories as in Figures 2, 3, with insertion sequences and transposases in red, toxin-antitoxin system in dark green, integrases in turquoise, heavy metal resistances in pink, and antibiotic resistances in light blue. Coding sequences present in the plasmids belonging to one of the eight colored annotation categories, but showing no homology by blast to pSE18-SA00377-1, were labeled but left in gray.
The mapping of these 11 plasmids to pSE18-SA00377-1 indicates that the pSE18-SA00377-1 contains a conserved plasmid backbone which frequently occurs in other plasmids of other genera. However, the widespread geographical origin of these 11 plasmids and their occurrence in a wide range of bacterial genera does not allow for ascertaining a potential common origin. Nevertheless, the high number of ARGs as well as their associations with composite transposons indicates that multiple insertion events had occurred and led to this accumulation of resistance genes in two distinct clusters. This accumulation of resistance genes is also present in the other 11 plasmids, although the first cluster of ARGs seems to be more stable, being present in its entirety and same organizational structure in four other plasmids.
This analysis showed that plasmids with high nucleotide similarity to pSE18-SA00377-1 and similar antibiotic resistance profiles can be found. However, no plasmid with a mapping coverage of >80% had been isolated from S. Agona isolates, with the highest mapping coverages all belonging to plasmids isolated either from Enterobacter or Phytobacter isolates. As high-similarity plasmids to pSE18-SA00377-1 seem to be found in a wide variety of genera and different geographical origins, it can be speculated that pSE18-SA00377-1 had been taken up from other sources, potentially in a clinical environment as a majority of the closely-related plasmids were isolated from clinical sources. Alternatively, the pSE18-SA00377-1 plasmid could have been taken up from environmental sources, for example when wastewater is re-used for irrigation in agriculture, as the SE18-SA00377 isolate was isolated from a dietary supplement consisting of plant-based ingredients. These hypotheses are also supported by the potential origins of the other constituent plasmids of the 18-SA00377 isolate. Firstly, the second largest plasmid, pSE18-SA00377-2 (94,574 bp), is a P1-like phage plasmid, carrying the p0111 plasmid replication gene, which was first identified from an enterohemorrhagic E. coli strain (Ogura et al., 2009) and is still frequently found in E. coli isolates, including clinical and food isolates (Balbuena-Alonso et al., 2022). Moreover, it carries two prophage-like elements pp1 and pp2, which were first identified in the core genome of E. faecalis isolates (Matos et al., 2013).
As an IncX3 plasmid and carrier of the blaTEM-1 gene, the second smallest plasmid, pSE18-SA00377-3 (50,931 bp), plays a role in the dissemination of carbapenemase resistance genes. The IncX plasmid family has been reported in a wide variety of Enterobacteriaceae from different sources (Guo et al., 2022) and thus the plasmid is a cause of concern due to its additional ARG load of aac(3)-IIe, qnrS1, and floR. Furthermore, pSE18-SA00377-3 also carries tmrB, a gene encoding the tunicamycin resistance protein which confers resistance to tunicamycin in Bacillus subtilis (Noda et al., 1992).
In order to limit the search to closely related isolates of Salmonella but also other members of the Enterobacteriaceae family, several NCBI Pathogen Detection databases were queried for isolates with highly similar antibiotic resistance profiles (80% overlap in ARGs with 18-SA00377). This resulted in short-read sequences from the following four databases: S. enterica, E. coli and Shigella, Klebsiella, and Citrobacter (Table 4).
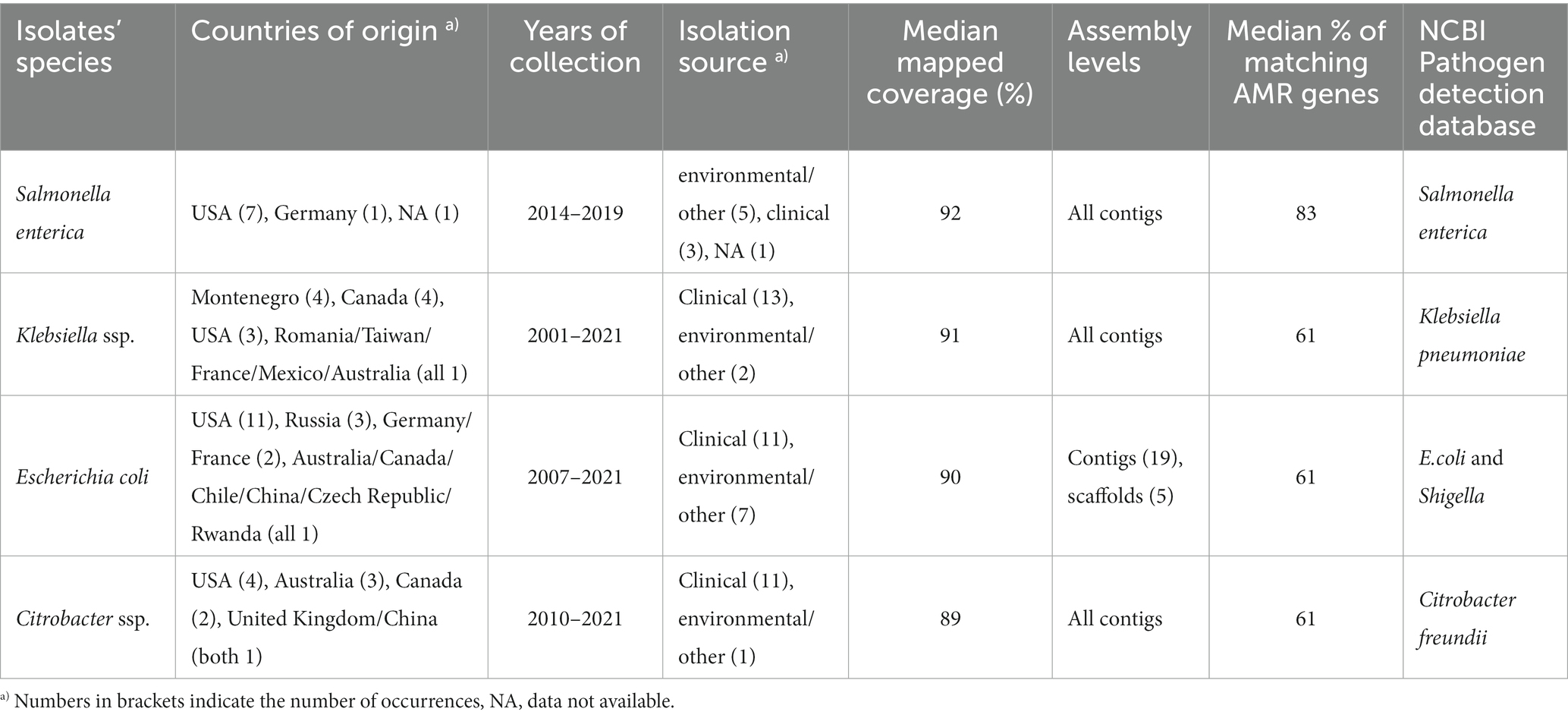
Table 4. Table showing the overview of isolates resulting from querying the respective NCBI Pathogen Detection databases with custom R script.
The nine Salmonella isolates of different serovars, geographical origins, and isolation sources (Table 1) were compared by mapping of contigs to p18-SA00377-1 and mapping results visualized (Figure 6).
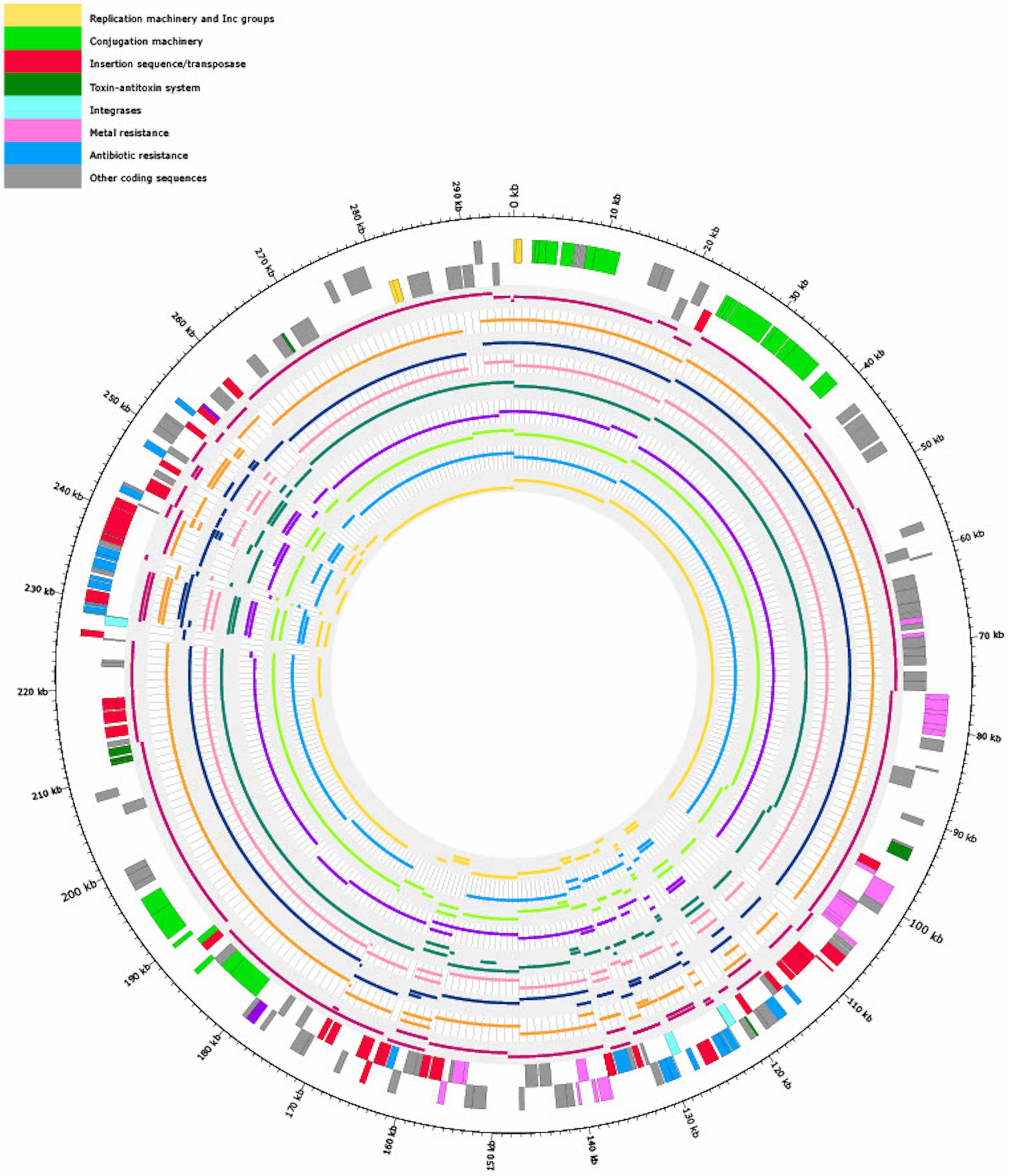
Figure 6. Visualization of mapping the contigs of nine Salmonella isolates (Table 1) with similar antibiotic resistance profiles (80% overlap of ARGs with 18-SA00377) to pSE18-SA00377-1. Regions of the nine Salmonella isolates that did not map to pSE18-SA00377-1 are not shown. The outermost black circle designates the base positions around the plasmid. Farthest two tracks on the outside represents the pSE18-SA00377-1 plasmid annotations, with replication machinery and incompatibility groups colored in yellow, conjugation machinery in light green, insertion sequences and transposases in red, toxin-antitoxin system in dark green, integrases in turquoise, heavy metal resistances in pink, and antibiotic resistances in light blue. Coding sequences for other gene products are colored in gray. For labeling of annotations refer to Figure 2.
Four isolates were isolated from animal sources, three isolates from clinical sources, and one isolate from a porcine food source. The animal samples were all isolated from the United States, but their isolation types range from domestic pigs (Sus scrofa domesticus) to wild boar (Sus scrofa) and cattle (Bos taurus). However, these environmental samples harboring these plasmids were either of serovar Agona or 4,12:i:-, the monophasic variant of S. Typhimurium.
However, based on available isolates’ metadata, it was impossible to infer if the mapping against the pSE18-SA00377-1 occurred within their chromosomes or in constituent plasmids, due to draft character of the used short-read derived assemblies. Nevertheless, based on the numerous breaks in coverage of mapped sections and the large number of very small mapped sections (e.g., IS26), it might be that the ARGs are located in the isolates’ chromosomes and not on a plasmid and the two described ARG clusters. Furthermore, multiple occurrences of these areas could become merged into repetitive regions during assembly of the short-read data.
In conclusion, this SE18-SA00377 isolate belongs to a sublineage of S. enterica serovar Agona that is multidrug-resistant and might be plant-associated. Along with its four plasmids, pSE18-SA00377-1, pSE18-SA00377-2, pSE18-SA00377-3, and pSE18-SA00377-4, the isolate carries a total of 23 different ARGs, conferring resistance to 12 different classes of antibiotics, with its largest plasmid of 295,499 kb in size, pSE18-SA00377-1, conferring the majority of them. Moreover, the pSE18-SA00377-1 plasmid is not only the main carrier of antibiotic resistance genes but also of heavy metal resistances. The structure of this plasmid is striking as its ARGs have accumulated in two distinct regions. This accumulation of ARGs as well as the presence of these clusters and a large part of its backbone in plasmids isolated from a wide range of genera, matrices, years of isolation and geographical origins suggest that this plasmid has a complex history with numerous transmission events.
Further analysis of plasmids from human, veterinary, and environmental sources may provide further insights into the evolution of this plasmid. In particular, due to the highly drug-resistant nature of this plasmid, identifying potential reservoirs of multidrug-resistant isolates is crucial, as they have the capacity to disseminate antibiotic and metal resistance genes.
Here, we present an in-depth characterization of a multidrug-resistant S. Agona, isolated from dietary supplements in 2018. Its phylogeny to other S. Agona isolates from Germany was established and supplemented with available sequences of S. Agona that have been reported globally and are available in the NCBI database. Detailed annotation of its largest constituent plasmid included antimicrobial resistance genes on mobile genetic elements. Closely related plasmids were queried through a two-pronged approach: MOB-typing and taxonomic classification of plasmids. Lastly, structural comparisons with high-similarity plasmids revealed a composite plasmid structure, found in isolates from numerous other genera, geographic origins and isolation matrices. These analyses showed that this plasmid is a potential reservoir for antimicrobial and heavy metal resistance determinants and has the potential to adapt to various hosts and environments. Thus, highlighting the need for continued surveillance to prevent future outbreaks.
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/Supplementary material.
LB: Formal analysis, Investigation, Methodology, Visualization, Writing – review & editing. MB: Formal analysis, Investigation, Methodology, Visualization, Writing – review & editing. CD: Data curation, Formal analysis, Investigation, Software, Writing – review & editing. JG: Conceptualization, Supervision, Writing – review & editing, Software. J-AH: Methodology, Writing – review & editing. BM: Project administration, Resources, Supervision, Writing – review & editing, Funding acquisition. IS: Project administration, Resources, Writing – review & editing. TA: Project administration, Supervision, Writing – review & editing. KN: Resources, Writing – review & editing. JF: Conceptualization, Investigation, Methodology, Project administration, Resources, Supervision, Writing – original draft, Writing – review & editing, Data curation, Formal analysis, Funding acquisition, Visualization.
The author(s) declare financial support was received for the research, authorship, and/or publication of this article. This study was supported by the German Federal Institute for Risk Assessment, Grant no. 60_0103_01.P543. LB received funding by the FARMED project, which is part of the European Union’s Horizon 2020 Research and Innovation Programme under Grant Agreement no. 773830: One Health European Joint Program.
We would like to express our thanks to the whole team at the NRL for Salmonella for serotyping and at the NRL for Antimicrobial Resistance for MIC testing, as well as to Beatrice Baumann, Katharina Thomas, and Angelina Groger for their tireless support in the laboratory, in particular for sequencing.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2023.1284929/full#supplementary-material
1. ^https://www.cdc.gov/pulsenet/pdf/ecoli-shigella-salmonella-pfge-protocol-508c.pdf
2. ^https://community.nanoporetech.com
3. ^https://github.com/rrwick/Porechop
4. ^https://gitlab.com/bfr_bioinformatics/bakcharak
5. ^https://github.com/tseemann/abricate
6. ^https://www.ncbi.nlm.nih.gov/pathogens
Abraham, S., O'Dea, M., Trott, D. J., Abraham, R. J., Hughes, D., Pang, S., et al. (2016). Isolation and plasmid characterization of carbapenemase (IMP-4) producing Salmonella enterica typhimurium from cats. Sci. Rep. 6:35527. doi: 10.1038/srep35527
Akhter, S., Aziz, R. K., and Edwards, R. A. (2012). PhiSpy: a novel algorithm for finding prophages in bacterial genomes that combines similarity- and composition-based strategies. Nucleic Acids Res. 40:e126. doi: 10.1093/nar/gks406
Alcock, B. P., Raphenya, A. R., Lau, T. T. Y., Tsang, K. K., Bouchard, M., Edalatmand, A., et al. (2019). CARD 2020: antibiotic resistome surveillance with the comprehensive antibiotic resistance database. Nucleic Acids Res. 48, D517–D525. doi: 10.1093/nar/gkz935
Ao, T. T., Feasey, N. A., Gordon, M. A., Keddy, K. H., Angulo, F. J., and Crump, J. A. (2015). Global burden of invasive nontyphoidal Salmonella disease, 2010(1). Emerg. Infect. Dis. 21, 941–949. doi: 10.3201/eid2106.140999
Balbuena-Alonso, M. G., Cortés-Cortés, G., Kim, J. W., Lozano-Zarain, P., Camps, M., and del Carmen Rocha-Gracia, R. (2022). Genomic analysis of plasmid content in food isolates of E. coli strongly supports its role as a reservoir for the horizontal transfer of virulence and antibiotic resistance genes. Plasmid 123-124:102650. doi: 10.1016/j.plasmid.2022.102650
Barton, B. M., Harding, G. P., and Zuccarelli, A. J. (1995). A general method for detecting and sizing large plasmids. Anal. Biochem. 226, 235–240. doi: 10.1006/abio.1995.1220
Bauer, A. P., Dieckmann, S. M., Ludwig, W., and Schleifer, K.-H. (2007). Rapid identification of Escherichia coli safety and laboratory strain lineages based on multiplex-PCR. FEMS Microbiol. Lett. 269, 36–40. doi: 10.1111/j.1574-6968.2006.00594.x
Blanc-Potard, A. B., Solomon, F., Kayser, J., and Groisman, E. A. (1999). The SPI-3 pathogenicity island of Salmonella enterica. J. Bacteriol. 181, 998–1004. doi: 10.1128/jb.181.3.998-1004.1999
Borowiak, M., Fischer, J., Hammerl, J. A., Hendriksen, R. S., Szabo, I., and Malorny, B. (2017). Identification of a novel transposon-associated phosphoethanolamine transferase gene, mcr-5, conferring colistin resistance in d-tartrate fermenting Salmonella enterica subsp. enterica serovar Paratyphi B. J. Antimicrob. Chemother. 72, 3317–3324. doi: 10.1093/jac/dkx327
Brouard, C., Espie, E., Weill, F. X., Kerouanton, A., Brisabois, A., Forgue, A. M., et al. (2007). Two consecutive large outbreaks of Salmonella enterica serotype Agona infections in infants linked to the consumption of powdered infant formula. Pediatr. Infect. Dis. J. 26, 148–152. doi: 10.1097/01.inf.0000253219.06258.23
Camacho, C., Coulouris, G., Avagyan, V., Ma, N., Papadopoulos, J., Bealer, K., et al. (2009). BLAST+: architecture and applications. BMC Bioinform. 10:421. doi: 10.1186/1471-2105-10-421
Carattoli, A. (2003). Plasmid-mediated antimicrobial resistance in Salmonella enterica. Curr. Issues Mol. Biol. 5, 113–122. doi: 10.21775/cimb.005.113
Carattoli, A., Zankari, E., García-Fernández, A., Voldby Larsen, M., Lund, O., Villa, L., et al. (2014). In silico detection and typing of plasmids using PlasmidFinder and plasmid multilocus sequence typing. Antimicrob. Agents Chemother. 58, 3895–3903. doi: 10.1128/AAC.02412-14
Chen, L., Zheng, D., Liu, B., Yang, J., and Jin, Q. (2015). VFDB 2016: hierarchical and refined dataset for big data analysis—10 years on. Nucleic Acids Res. 44, D694–D697. doi: 10.1093/nar/gkv1239
Chen, S., Zhou, Y., Chen, Y., and Gu, J. (2018). Fastp: an ultra-fast all-in-one FASTQ preprocessor. Bioinformatics 34, i884–i890. doi: 10.1093/bioinformatics/bty560
Clark, G. M., Kaufmann, A. F., Gangarosa, E. J., and Thompson, M. A. (1973). Epidemiology of an international outbreak of Salmonella Agona. Lancet 2, 490–493. doi: 10.1016/s0140-6736(73)92082-5
Cummins, M. L., Sanderson-Smith, M., Newton, P., Carlile, N., Phalen, D. N., Maute, K., et al. (2020). Whole-genome sequence analysis of an extensively drug-resistant Salmonella enterica serovar Agona isolate from an Australian silver Gull (Chroicocephalus novaehollandiae) reveals the acquisition of multidrug resistance plasmids. mSphere 5:e00743-20. doi: 10.1128/mSphere.00743-20
Dangel, A., Berger, A., Messelhäußer, U., Konrad, R., Hörmansdorfer, S., Ackermann, N., et al. (2019). Genetic diversity and delineation of Salmonella Agona outbreak strains by next generation sequencing, Bavaria, Germany, 1993 to 2018. Euro Surveill. 24:1800303. doi: 10.2807/1560-7917.Es.2019.24.18.1800303
De Coster, W., D'Hert, S., Schultz, D. T., Cruts, M., and Van Broeckhoven, C. (2018). NanoPack: visualizing and processing long-read sequencing data. Bioinformatics 34, 2666–2669. doi: 10.1093/bioinformatics/bty149
Deneke, C., Brendebach, H., Uelze, L., Borowiak, M., Malorny, B., and Tausch, S. H. (2021a). Species-specific quality control, assembly and contamination detection in microbial isolate sequences with AQUAMIS. Genes 12:644. doi: 10.3390/genes12050644
Deneke, C., Uelze, L., Brendebach, H., Tausch, S. H., and Malorny, B. (2021b). Decentralized investigation of bacterial outbreaks based on hashed cgMLST. Front. Microbiol. 12:649517. doi: 10.3389/fmicb.2021.649517
Doublet, B., Praud, K., Weill, F. X., and Cloeckaert, A. (2009). Association of IS26-composite transposons and complex In4-type integrons generates novel multidrug resistance loci in Salmonella genomic island 1. J. Antimicrob. Chemother. 63, 282–289. doi: 10.1093/jac/dkn500
European Food Safety Authority and European Centre for Disease Prevention and Control (2022a). The European union one health 2021 Zoonoses report. EFSA J. 20:e07666. doi: 10.2903/j.efsa.2022.7666
European Food Safety Authority and European Centre for Disease Prevention and Control (2022b). The European union summary report on antimicrobial resistance in zoonotic and indicator bacteria from humans, animals and food in 2019–2020. EFSA J. 20:e07209. doi: 10.2903/j.efsa.2022.7209
European Food Safety Authority and European Centre for Disease Prevention and Control (2023). The European union summary report on antimicrobial resistance in zoonotic and indicator bacteria from humans, animals and food in 2020/2021. EFSA J. 21:e07867. doi: 10.2903/j.efsa.2023.7867
Feldgarden, M., Brover, V., Gonzalez-Escalona, N., Frye, J. G., Haendiges, J., Haft, D. H., et al. (2021). AMRFinderPlus and the reference gene catalog facilitate examination of the genomic links among antimicrobial resistance, stress response, and virulence. Sci. Rep. 11:12728. doi: 10.1038/s41598-021-91456-0
Fernández, J., Guerra, B., and Rodicio, M. R. (2018). Resistance to Carbapenems in non-Typhoidal Salmonella enterica Serovars from humans, animals and food. Vet. Sci. 5:40. doi: 10.3390/vetsci5020040
Gerlach, R. G., Jäckel, D., Geymeier, N., and Hensel, M. (2007). Salmonella pathogenicity island 4-mediated adhesion is coregulated with invasion genes in Salmonella enterica. Infect. Immun. 75, 4697–4709. doi: 10.1128/iai.00228-07
Grimont, P. A. D., and Weill, F.. (2007). Antigenic formulae of the Salmonella serovars. WHO Collaborating Centre for Reference and Research on Salmonella. Paris
Guo, X., Chen, R., Wang, Q., Li, C., Ge, H., Qiao, J., et al. (2022). Global prevalence, characteristics, and future prospects of IncX3 plasmids: a review. Front. Microbiol. 13:979558. doi: 10.3389/fmicb.2022.979558
Hadziabdic, S., Fischer, J., Malorny, B., Borowiak, M., Guerra, B., Kaesbohrer, A., et al. (2018). In vivo transfer and microevolution of avian native IncA/C(2)Bla(NDM-1)-carrying plasmid pRH-1238 during a broiler chicken infection study. Antimicrob. Agents Chemother. 62:e02128-17. doi: 10.1128/AAC.02128-17
Ilyas, B., Tsai, C. N., and Coombes, B. K. (2017). Evolution of Salmonella-host cell interactions through a dynamic bacterial genome. Front. Cell. Infect. Microbiol. 7:428. doi: 10.3389/fcimb.2017.00428
Jain, C., Rodriguez-R, L. M., Phillippy, A. M., Konstantinidis, K. T., and Aluru, S. (2018). High throughput ANI analysis of 90K prokaryotic genomes reveals clear species boundaries. Nat. Commun. 9:5114. doi: 10.1038/s41467-018-07641-9
Jennings, E., Thurston, T. L. M., and Holden, D. W. (2017). Salmonella SPI-2 type III secretion system effectors: molecular mechanisms and physiological consequences. Cell Host Microbe 22, 217–231. doi: 10.1016/j.chom.2017.07.009
Johansson, M. H. K., Bortolaia, V., Tansirichaiya, S., Aarestrup, F. M., Roberts, A. P., and Petersen, T. N. (2020). Detection of mobile genetic elements associated with antibiotic resistance in Salmonella enterica using a newly developed web tool: MobileElementFinder. J. Antimicrob. Chemother. 76, 101–109. doi: 10.1093/jac/dkaa390
Jourdan-da Silva, N., Fabre, L., Robinson, E., Fournet, N., Nisavanh, A., Bruyand, M., et al. (2018). Ongoing nationwide outbreak of Salmonella Agona associated with internationally distributed infant milk products, France, December 2017. Euro Surveill. 23:17-00852. doi: 10.2807/1560-7917.ES.2018.23.2.17-00852
Juraschek, K., Borowiak, M., Tausch, S. H., Malorny, B., Kasbohrer, A., Otani, S., et al. (2021). Outcome of different sequencing and assembly approaches on the detection of plasmids and localization of antimicrobial resistance genes in commensal Escherichia coli. Microorganisms 9:598. doi: 10.3390/microorganisms9030598
Killalea, D., Ward, L. R., Roberts, D., de Louvois, J., Sufi, F., Stuart, J. M., et al. (1996). International epidemiological and microbiological study of outbreak of Salmonella agona infection from a ready to eat savoury snack--I: England and Wales and the United States. BMJ 313, 1105–1107. doi: 10.1136/bmj.313.7065.1105
Koch, J., Schrauder, A., Alpers, K., Werber, D., Frank, C., Prager, R., et al. (2005). Salmonella Agona outbreak from contaminated aniseed, Germany. Emerg. Infect. Dis. 11, 1124–1127. doi: 10.3201/eid1107.041022
Krzywinski, M., Schein, J., Birol, I., Connors, J., Gascoyne, R., Horsman, D., et al. (2009). Circos: an information aesthetic for comparative genomics. Genome Res. 19, 1639–1645. doi: 10.1101/gr.092759.109
Letunic, I., and Bork, P. (2021). Interactive tree of life (iTOL) v5: an online tool for phylogenetic tree display and annotation. Nucleic Acids Res. 49, W293–W296. doi: 10.1093/nar/gkab301
Li, H. (2018). Minimap2: pairwise alignment for nucleotide sequences. Bioinformatics 34, 3094–3100. doi: 10.1093/bioinformatics/bty191
Li, I. C., Yu, G. Y., Huang, J. F., Chen, Z. W., and Chou, C. H. (2022). Comparison of reference-based assembly and De novo assembly for bacterial plasmid reconstruction and AMR gene localization in Salmonella enterica Serovar Schwarzengrund isolates. Microorganisms 10:227. doi: 10.3390/microorganisms10020227
Lordan, R. (2021). Dietary supplements and nutraceuticals market growth during the coronavirus pandemic - implications for consumers and regulatory oversight. PharmaNutrition 18:100282. doi: 10.1016/j.phanu.2021.100282
Lou, L., Zhang, P., Piao, R., and Wang, Y. (2019). Salmonella Pathogenicity Island 1 (SPI-1) and its complex regulatory network. Front. Cell. Infect. Microbiol. 9:270. doi: 10.3389/fcimb.2019.00270
Marcus, S. L., Brumell, J. H., Pfeifer, C. G., and Finlay, B. B. (2000). Salmonella pathogenicity islands: big virulence in small packages. Microbes Infect. 2, 145–156. doi: 10.1016/S1286-4579(00)00273-2
Matos, R. C., Lapaque, N., Rigottier-Gois, L., Debarbieux, L., Meylheuc, T., Gonzalez-Zorn, B., et al. (2013). Enterococcus faecalis prophage dynamics and contributions to pathogenic traits. PLoS Genet. 9:e1003539. doi: 10.1371/journal.pgen.1003539
Mba-Jonas, A., Culpepper, W., Hill, T., Cantu, V., Loera, J., Borders, J., et al. (2018). A multistate outbreak of human Salmonella Agona infections associated with consumption of fresh, whole papayas imported from Mexico-United States, 2011. Clin. Infect. Dis. 66, 1756–1761. doi: 10.1093/cid/cix1094
Meinen, A., Simon, S., Banerji, S., Szabo, I., Malorny, B., Borowiak, M., et al. (2019). Salmonellosis outbreak with novel Salmonella enterica subspecies enterica serotype (11,z41,e,n,z15) attributable to sesame products in five European countries, 2016 to 2017. Euro Surveill. 24:1800543. doi: 10.2807/1560-7917.Es.2019.24.36.1800543
Ng, L. K., Martin, I., Alfa, M., and Mulvey, M. (2001). Multiplex PCR for the detection of tetracycline resistant genes. Mol. Cell. Probes 15, 209–215. doi: 10.1006/mcpr.2001.0363
Nicolay, N., Thornton, L., Cotter, S., Garvey, P., Bannon, O., McKeown, P., et al. (2011). Salmonella enterica serovar Agona European outbreak associated with a food company. Epidemiol. Infect. 139, 1272–1280. doi: 10.1017/S0950268810002360
Noda, Y., Yoda, K., Takatsuki, A., and Yamasaki, M. (1992). TmrB protein, responsible for tunicamycin resistance of Bacillus subtilis, is a novel ATP-binding membrane protein. J. Bacteriol. 174, 4302–4307. doi: 10.1128/jb.174.13.4302-4307.1992
Ogura, Y., Ooka, T., Iguchi, A., Toh, H., Asadulghani, M., Oshima, K., et al. (2009). Comparative genomics reveal the mechanism of the parallel evolution of O157 and non-O157 enterohemorrhagic Escherichia coli. Proc. Natl. Acad. Sci. U. S. A. 106, 17939–17944. doi: 10.1073/pnas.0903585106
O'Leary, N. A., Wright, M. W., Brister, J. R., Ciufo, S., Haddad, D., McVeigh, R., et al. (2015). Reference sequence (RefSeq) database at NCBI: current status, taxonomic expansion, and functional annotation. Nucleic Acids Res. 44, D733–D745. doi: 10.1093/nar/gkv1189
Ondov, B. D., Treangen, T. J., Melsted, P., Mallonee, A. B., Bergman, N. H., Koren, S., et al. (2016). Mash: fast genome and metagenome distance estimation using MinHash. Genome Biol. 17:132. doi: 10.1186/s13059-016-0997-x
Parkhill, J., Dougan, G., James, K. D., Thomson, N. R., Pickard, D., Wain, J., et al. (2001). Complete genome sequence of a multiple drug resistant Salmonella enterica serovar Typhi CT18. Nature 413, 848–852. doi: 10.1038/35101607
Robertson, J., and Nash, J. H. E. (2018). MOB-suite: software tools for clustering, reconstruction and typing of plasmids from draft assemblies. Microb. Genom. 4:e000206. doi: 10.1099/mgen.0.000206
Roer, L., Hendriksen, R. S., Leekitcharoenphon, P., Lukjancenko, O., Kaas, R. S., Hasman, H., et al. (2016). Is the evolution of Salmonella enterica subsp. enterica linked to restriction-modification systems? mSystems 1:e00009-16. doi: 10.1128/mSystems.00009-16
Rodríguez, I. W., Barownick, R., Helmuth, M. C., Mendoza, M. R., Rodicio, A., Schroeter, B., et al. (2009). Extended-spectrum β-lactamases and AmpC β-lactamases in ceftiofur-resistant Salmonella enterica isolates from food and livestock obtained in Germany during 2003-07. J. Antimicrob. Chemother. 64, 301–309. doi: 10.1093/jac/dkp195
Rozwandowicz, M., Brouwer, M. S. M., Fischer, J., Wagenaar, J. A., Gonzalez-Zorn, B., Guerra, B., et al. (2018). Plasmids carrying antimicrobial resistance genes in Enterobacteriaceae. J. Antimicrob. Chemother. 73, 1121–1137. doi: 10.1093/jac/dkx488
Russo, E. T., Biggerstaff, G., Hoekstra, R. M., Meyer, S., Patel, N., Miller, B., et al. (2013). A recurrent, multistate outbreak of Salmonella serotype agona infections associated with dry, unsweetened cereal consumption, United States, 2008. J. Food Prot. 76, 227–230. doi: 10.4315/0362-028X.JFP-12-209
Schwengers, O., Barth, P., Falgenhauer, L., Hain, T., Chakraborty, T., and Goesmann, A. (2020). Platon: identification and characterization of bacterial plasmid contigs in short-read draft assemblies exploiting protein sequence-based replicon distribution scores. Microb. Genom. 6:mgen000398. doi: 10.1099/mgen.0.000398
Schwengers, O., Jelonek, L., Dieckmann, M. A., Beyvers, S., Blom, J., and Goesmann, A. (2021). Bakta: rapid and standardized annotation of bacterial genomes via alignment-free sequence identification. Microb. Genom. 7:000685. doi: 10.1099/mgen.0.000685
Shohat, T., Green, M. S., Merom, D., Gill, O. N., Reisfeld, A., Matas, A., et al. (1996). International epidemiological and microbiological study of outbreak of Salmonella Agona infection from a ready to eat savoury snack--II: Israel. BMJ 313, 1107–1109. doi: 10.1136/bmj.313.7065.1107
Siguier, P., Perochon, J., Lestrade, L., Mahillon, J., and Chandler, M. (2006). ISfinder: the reference centre for bacterial insertion sequences. Nucleic Acids Res. 34, D32–D36. doi: 10.1093/nar/gkj014
Simon, S., Trost, E., Bender, J., Fuchs, S., Malorny, B., Rabsch, W., et al. (2018). Evaluation of WGS based approaches for investigating a food-borne outbreak caused by Salmonella enterica serovar Derby in Germany. Food Microbiol. 71, 46–54. doi: 10.1016/j.fm.2017.08.017
Sullivan, M. J., Petty, N. K., and Beatson, S. A. (2011). Easyfig: a genome comparison visualizer. Bioinform 27, 1009–1010. doi: 10.1093/bioinformatics/btr039
Tatusova, T., DiCuccio, M., Badretdin, A., Chetvernin, V., Nawrocki, E. P., Zaslavsky, L., et al. (2016). NCBI prokaryotic genome annotation pipeline. Nucleic Acids Res. 44, 6614–6624. doi: 10.1093/nar/gkw569
Taylor, J. P., Barnett, B. J., del Rosario, L., Williams, K., and Barth, S. S. (1998). Prospective investigation of cryptic outbreaks of Salmonella agona salmonellosis. J. Clin. Microbiol. 36, 2861–2864. doi: 10.1128/JCM.36.10.2861-2864.1998
Technical University of Denmark. (2022). Annual report on Zoonoses in Denmark 2021. Denmark, Technical University of Denmark.
The European Committee on Antimicrobial Susceptibility Testing. (2021). Breakpoint tables for interpretation of MICs and zone diameters. EUCAST. Denmark
Uelze, L., Becker, N., Borowiak, M., Busch, U., Dangel, A., Deneke, C., et al. (2021). Toward an integrated genome-based surveillance of Salmonella enterica in Germany. Front. Microbiol. 12:626941. doi: 10.3389/fmicb.2021.626941
van Asten, A. J. A. M., and van Dijk, J. E. (2005). Distribution of “classic” virulence factors among Salmonella spp. FEMS Immunol. Med. Microbiol. 44, 251–259. doi: 10.1016/j.femsim.2005.02.002
Velásquez, J. C., Hidalgo, A. A., Villagra, N., Santiviago, C. A., Mora, G., and Fuentes, J. A. (2016). SPI-9 of Salmonella enterica serovar Typhi is constituted by an operon positively regulated by RpoS and contributes to adherence to epithelial cells in culture. Microbiology 162, 1367–1378. doi: 10.1099/mic.0.000319
Walker, B. J., Abeel, T., Shea, T., Priest, M., Abouelliel, A., Sakthikumar, S., et al. (2014). Pilon: an integrated tool for comprehensive microbial variant detection and genome assembly improvement. PLoS One 9:e112963. doi: 10.1371/journal.pone.0112963
Wick, R. R., Judd, L. M., Gorrie, C. L., and Holt, K. E. (2017). Unicycler: resolving bacterial genome assemblies from short and long sequencing reads. PLoS Comput. Biol. 13:e1005595. doi: 10.1371/journal.pcbi.1005595
Yoshida, C. E., Kruczkiewicz, C. R., Laing, E. J., Lingohr, V. P. J., Gannon, V. P., Nash, J. H., et al. (2016). The Salmonella in silico typing resource (SISTR): an open web-accessible tool for rapidly typing and subtyping draft Salmonella genome assemblies. PLoS One 11:e0147101. doi: 10.1371/journal.pone.0147101
Keywords: Salmonella Agona, dietary supplement, whole genome sequencing, antimicrobial resistance, plasmid
Citation: Bartsch LJ, Borowiak M, Deneke C, Gruetzke J, Hammerl J-A, Malorny B, Szabo I, Alter T, Nguyen KK and Fischer J (2023) Genetic characterization of a multidrug-resistant Salmonella enterica serovar Agona isolated from a dietary supplement in Germany. Front. Microbiol. 14:1284929. doi: 10.3389/fmicb.2023.1284929
Received: 29 August 2023; Accepted: 25 October 2023;
Published: 15 November 2023.
Edited by:
Xin Wang, National Institute for Communicable Disease Control and Prevention (China CDC), ChinaReviewed by:
Meiying Yan, National Institute for Communicable Disease Control and Prevention (China CDC), ChinaCopyright © 2023 Bartsch, Borowiak, Deneke, Gruetzke, Hammerl, Malorny, Szabo, Alter, Nguyen and Fischer. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Jennie Fischer, amVubmllLmZpc2NoZXJAYmZyLmJ1bmQuZGU=
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.