- 1Facultad de Ingeniería Zootecnista, Agronegocios y Biotecnología, Instituto de Investigación en Ganadería y Biotecnología, Universidad Nacional Toribio Rodríguez de Mendoza de Amazonas, Chachapoyas, Peru
- 2Facultad de Ingeniería Zootecnista, Agronegocios y Biotecnología, Universidad Nacional Toribio Rodríguez de Mendoza de Amazonas, Chachapoyas, Peru
- 3Laboratorio de Fisiología Molecular, Facultad de Ingeniería Zootecnista, Agronegocios y Biotecnología, Instituto de Investigación en Ganadería y Biotecnología, Universidad Nacional Toribio Rodríguez de Mendoza de Amazonas, Chachapoyas, Peru
- 4Laboratorio de Enfermedades Infecciosas y Parasitarias, Facultad de Ingeniería Zootecnista, Agronegocios y Biotecnología, Instituto de Investigación en Ganadería y Biotecnología, Universidad Nacional Toribio Rodríguez de Mendoza de Amazonas, Chachapoyas, Peru
- 5Laboratory of Bacterial Diseases, Sector of Preventive Veterinary Medicine and Public Health, Department of Veterinary, Universidade Federal de Viçosa, Viçosa, MG, Brazil
- 6Department of Basic Medical Sciences, College of Veterinary Medicine, Purdue University, West Lafayette, IN, United States
- 7Chemistry Department, Institute for Drug Discovery, Purdue University, West Lafayette, IN, United States
- 8Programa de Pós-Graduação em Ciência Animal, Departamento de Zootecnia, Centro de Ciências Agrárias, Universidade Federal da Paraíba, Areia, Brazil
- 9Facultad de Medicina, Universidad Nacional Toribio Rodríguez de Mendoza de Amazonas, Chachapoyas, Peru
Guinea pigs have historically been used as a food source and are also an important model for studying the human intestines. Fasting is the act of temporarily stopping the intake of food. This process can alter the microbiota of various animals. This study is the first to investigate the impact of fasting on the cecum microbiome of three guinea pig breeds. We investigated the impact of fasting on the microbiome population structure in the cecum of three guinea pig breeds. This was done by sequencing and analyzing the V4 hypervariable region of the 16S rRNA gene in bacterial communities found in cecum mucosa samples. To achieve this, we established two treatment groups (fasting and fed), for each of the three guinea pig breeds: Andina, Inti, and Peru. The study involved twenty-eight guinea pigs, which were divided into the following groups: Andina-fed (five), Andina-fasting (five), Inti-fed (four), Inti-fasting (five), Peru-fed (five), and Peru-fasting (four). The results indicated a significant difference in beta diversity between the treatment groups for the Peru breed (P-value = 0.049), but not for the treatment groups of the Andina and Inti breeds. The dominant phyla across all groups were Firmicutes and Bacteroidetes. We observed variations in the abundance of different taxa in the cecum microbiota when comparing the treatment groups for each breed. Additionally, there was a higher number of unique taxa observed in the fasting groups compared to the fed groups. We discovered that the genus Victivallis was the only one present in all fasting groups across all breeds. Despite the findings, the resilience of the gut microbiome was not challenged in all three breeds, which can lead to disruptive changes that may affect the overall maintenance of the cecum microbiome. Based on the observed differences in the treatment groups of the Peru breed, it can be suggested that fasting has a greater impact on this particular breed.
1 Introduction
Fasting is a process that can generate several changes in the human and animal body. Since the 1920s, it has been known that fasting has an impact on the composition of the microbiome (Benedict, 1915); however, the direct relationship is still uncertain. The process of fasting can cause a reduction in nutrient availability since the microbiome can be affected (McCue et al., 2017). During fasting, bacteria that use host-derived substrates, such as mucins, shed epithelial cells and proliferate. Still, bacteria that depend on food substrates decrease their abundance because of the food intake irruption (Ducarmon et al., 2023). Another factor associated with the microbiome change during fasting is intestinal tissue remodeling. This process is known as transient atrophy of the intestinal tissue. It is characterized by a decrease in mitosis in crypts and an increment in the rate of apoptosis and autophagy (Habold et al., 2004). For example, in rats, the mucosal mass decreases by 50% during fasting (Dunel-Erb et al., 2001).
Guinea pigs (Cavia porcellus) have been used for different motives such as a food source and popular pets by humans (Buela et al., 2022). Another human utility of this animal is human intestinal research (Hildebrand et al., 2012). As a food resource, its meat has a big amount of protein and a reduced percentage of fat content (Avilés et al., 2014). Over the years, the National Institute for Agricultural Innovation (INIA) of Peru developed three breeds of guinea pigs (Andina, Inti, and Peru). These breeds are consumed commonly in the Peruvian gastronomy (Rubio Arias, 2018; Chauca Francia, 2023). Andina breed stands out for its remarkable prolificity (litter size = 3.4), boasting a fertility rate of 98.5% (Rubio Arias, 2018). Inti breed achieves a weight of 0.9 kg at 56 days and a meat carcass yield of 71.1%. Lastly, Peru breed has a great ability to gain weight and meat carcass yield compared to the Andina and Inti breeds (Reynaga Rojas et al., 2020). Peru breed have meat-oriented characteristics, boasting high precocity, fast growth, and prolificity (litter size = 2.8) (Rubio Arias, 2018).
In animal production, the “control” of food intake can influence general aspects of meat and milk composition, quality, and characteristics. Diet alterations in animals will affect muscle composition, fat development, and protein and lipid quality. On the other hand, the health of production animals can be altered by the microbiome of the gastrointestinal tract (GIT), besides is a key component of the development, nutritional absorption, metabolism, and productivity of these animals (Yeoman and White, 2014; Zhang et al., 2021). Therefore, the study of GIT microbiome can generate information and knowledge that allows the improvement of meat. This is one of the reasons for the increasing number of studies about GIT microbiome of production animals, including different guinea pig breeds (Hildebrand et al., 2012; Phillips Campbell et al., 2016; Al et al., 2017; Crowley et al., 2017; Lucking et al., 2018; Palakawong Na Ayudthaya et al., 2019; Shin et al., 2021; Tang et al., 2022; Wada et al., 2022; Frias et al., 2023).
The gut microbiome can be influenced by various factors. For instance, factors such as the host's genetics, diet, fasting, age, and use of antibiotics can all play a role (Francino, 2015; Kurilshikov et al., 2017; Angoorani et al., 2021; Maifeld et al., 2021; Ducarmon et al., 2023; Frias et al., 2023). On the other hand, fasting can impact the gut microbiome in rodents (Angoorani et al., 2021). Additionally, research has shown that host genetics can also influence the effect of fasting on the gut microbiome within the same species (Yan et al., 2021).
The investigation into the effects of fasting on the gut microbiome of guinea pigs remains unexplored thus far. It has been demonstrated by Turley and West (1976) that a 24-h fasting period can lead to a reduction in sterol synthesis in the ileum of guinea pigs at the age of 4 months. Numerous studies have highlighted the potential of sterols in modulating the composition of gut microbiota, resulting in a range of beneficial health effects (Le et al., 2022; Manoppo et al., 2022). Additionally, Langley and Kelly (1992) found that adult guinea pigs, aged 6 months or older, experienced a 9% reduction in body weight following a 48-h fasting period. Weight loss has been found to be linked to alterations in the diversity of the gut microbiota (Jian et al., 2022; Koutoukidis et al., 2022). Based on the aforementioned information, it can be suggested that a 24-h period has the potential to impact the gut microbiome of guinea pigs. The benefits of fasting has been demonstrated in the dietary practices of numerous livestock animal species. The utilization of intermittent fasting as a feeding strategy for chickens is widely acknowledged for its capacity to improve flock uniformity by increasing portion sizes, thereby prolonging feeding periods and reducing feed competition (Lindholm, 2019; Lindholm and Altimiras, 2023). The incorporation of fasting as a pre-slaughter practice for pigs has the potential to enhance carcass hygiene (Faucitano et al., 2010), pork quality, and animal welfare (Driessen et al., 2020). In the context of rabbits, fasting offers several benefits, including enhanced digestive function, a shift in the body's energy allocation from fat to protein, and a decrease in mortality and morbidity associated with digestive issues (Abou-Hashim et al., 2023). Based on the aforementioned information, providing additional information about the impact of fasting on the gut microbiome of guinea pigs can deepen our comprehension and assist in making well-informed decisions regarding the implementation of enhanced feeding strategies that incorporate fasting periods for guinea pigs.
The impact of fasting on the microbiome of livestock animals, specifically guinea pigs, has not been extensively investigated in comparison to studies conducted on humans or mice. The impact of these animals on meat production is currently unknown. Therefore, the present study aims to characterize the microbiome of the cecum in three distinct breeds of guinea pigs (Andina, Inti, and Peru) belonging to two separate groups: a control group (fed group) and a group subjected to a time-restricted fasting regimen (fasting group). The objective is to compare the impact of fasting on the composition of their microbiome. The understanding of these variations and similarities in the circumstances of food limitation can contribute to the advancement of more effective feeding strategies for guinea pigs and enhance the exploration of particular taxa that may be associated with fasting.
2 Materials and methods
2.1 Ethics statement
The experimental protocol (CIEI-N°005) has received approval from the Institutional Committee on Research Ethics (CIEI) of the National University Toribio Rodriguez de Mendoza (UNTRM).
2.2 Criteria for selection, treatment, and sampling of animals
The facilities of the small animal shed of the experimental research center of the Institute of Livestock and Biotechnology (IGBI) of the UNTRM were used for the breeding of the guinea pigs, and the Molecular Physiology Laboratory was used for the molecular procedures.
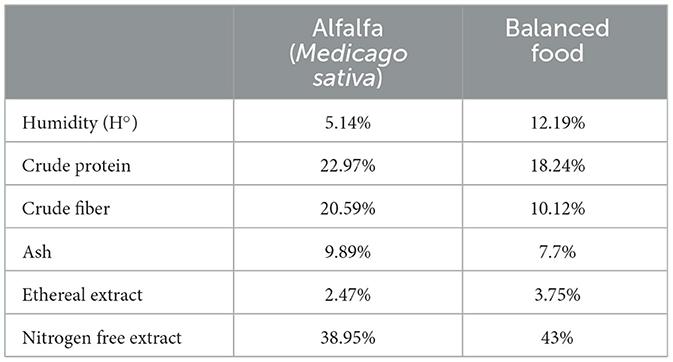
This study explored whether fasting could modify the population structure of the microbiome of the cecum of three breeds of guinea pigs, To achieve this, we established two treatment groups: fasting and fed, for each of the three guinea pig breeds: Andina, Inti, and Peru. Thirty samples of guinea pig cecum mucosa were used. Still, one of the samples from the Inti breed and one of the samples from the Peru breed did not show enough sequences during the filtering performed and were removed from the analysis, determining 28 samples downstream. The global sample size was 28 male guinea pigs, divided into 10 specimens for the Andina breed (five of the fed group; five of the fasting group), nine specimens for the Inti breed (four of the fed group; five of the fasting group) and nine specimens for the Peru breed (five of the fed group; four of the fasting group). The study involved 70–90-day-old guinea pigs fed with 80% alfalfa and 20% balanced food and a fasting treatment with 24-h fasting before his euthanasia. The bromatological analysis of the alfalfa (Medicago sativa) and the balanced food that has been used to feed the guinea pigs was carried out in the Laboratory of Animal Nutrition and Food Bromatology of the UNTRM, where the parameters of humidity (H°), crude protein, ashes, crude fiber, ethereal extract, nitrogen-free extract, according to the protocols established by the same laboratory (Table 1). Guinea pigs were euthanized by cervical dislocation, and microbial samples were obtained from mucosal samples of the cecum.
The protocol for collecting samples was modified from Hu et al. (2021). Using sterile swabs, we extracted microbial samples from the cecum. Flushing sterile PBS buffer was used to obtain these samples. After carefully locating the respective body site for each sample, we cut a 1–1.5 cm hole there, collected the contents with sterilized spoons, and then put the contents into sterile microcentrifuge tubes. For each gastrointestinal site (cecum) sample, we used a new, sterilized spoon. We repeatedly gently kneaded 1–2 ml PBS buffer samples for 2 min after flushing them with a sterile syringe. Fifteen milliliter sterile centrifuge tubes were used to collect the lavage fluid, which was then chilled to 4°C. Each sample needed between 5 and 10 ml of lavage fluid, which was then gathered, centrifuged at 4,000 × g for 30 min at 4°C to form a pellet, and then transferred into a 2 ml sterile centrifuge tube. The pellet was kept at −80°C until DNA extraction.
2.3 DNA extraction, amplification, and sequencing of the 16S rRNA gene
DNA was extracted using the PureLink Genomic DNA Extraction MiniKit (Invitrogen) according to the manufacturer's instructions for Gram-Positive Bacterial Cell Lysate, with a few minor modifications. The “DNA Clean and Concentrator ®-5” kit (Zymo Research) was used to purify the extracted genomic DNA. DNA concentration and purity were evaluated using a NanoDrop® Thermo Fisher Scientific Spectrophotometer (Waltham, Massachusetts, USA). Agar gel electrophoresis was used to verify the results.
The DNA samples were sent to the Argonne Laboratory for the amplification and sequencing of the V4 hypervariable region of the bacterial 16S rRNA. The Argonne Laboratory (Argonne, IL, USA) used the MiSeq Reagent Kit V2 with primers 515 F and 806 R created for the Illumina MiSeq platform (Illumina Inc., San Diego, CA) to amplify the V4 hypervariable region of the bacterial 16S rRNA gene from genomic DNA (Caporaso et al., 2011). Degeneracy was added to both the forward and reverse primers to correct known biases against the marine and freshwater Alphaproteobacterial clade SAR11 [806R, (Apprill et al., 2015)] and Crenarchaeota/Thaumarchaeota [515F, also known as 515F-Y, (Parada et al., 2016)].
2.4 Sequence and bioinformatics analyses
The Quantitative Insights Into Microbial Ecology 2 (QIIME2) software (v. 2022.11) was used to analyze the microbiome of the cecum of the guinea pigs with the evaluation of the sequences of the V4 hypervariable region of the 16S rRNA gene (Bolyen et al., 2019), on the bioinformatics server of the Molecular Physiology Laboratory of the National University Toribio Rodrguez de Mendoza using Python programming. We used the “DADA2” plugin (v. 1.26.0) (Callahan et al., 2016) to follow the QIIME2 pipeline to execute the demultiplexing of the reads, the trimming process of the sequence adapters, and the deletion of ambiguous, duplicate, low-quality, chimeric, and other sequences. From then, we were only able to continue the analysis up to positions 225 and 193 of the forward and reverse reads, respectively. Additionally, sequences with insufficient amplicon sequence variants (ASVs) per sample were eliminated using alpha rarefaction.
The taxonomic categorization using the SILVA v. 138 database (Quast et al., 2013) and the sklearn classifier was applied to the representative and high-quality sequences to produce the taxonomy tables and ASVs. The data was filtered using the software phyloseq (McMurdie and Holmes, 2013) in R (R Core Team, 2022) to eliminate any ASVs that were unassigned, assigned as being of Archaea, Chloroplast, or Mitochondrial origin, or had no assigned bacterial phylum.
The software R version 4.2.2 (R Core Team, 2022) was used to do all statistical analyses, and a number of packages and techniques were used. Plotting alpha rarefaction curves was done using the vegan package (Oksanen et al., 2022). Utilizing metrics from the indices Shannon diversity (Shannon, 1948), Chao1 richness (Chao, 1984), Abundance-based Coverage Estimator (ACE) of species richness (Chao and Lee, 1992), and Observed operational taxonomic units (OTUs) in the R statistical program, the alpha diversity indices were assessed in the phyloseq package (McMurdie and Holmes, 2013) to compute bacterial diversity. The box plots were created using the MicrobiotaProcess package, and the Alpha diversity box-and-whisker plots were created using the same package (Xu et al., 2023). Analysis of variance (ANOVA; α < 0.05) and the Tukey's honestly-significant-difference (HSD) post hoc test were used to compare the index values for the three breed types (Andina, Inti, and Peru) (R Core Team, 2022).
Beta diversity was examined for variations in community structure between different treatments for each breed using the canonical analysis of principal coordinates [CAP; (Anderson and Willis, 2003)] and non-metric multidimensional scaling [NMDS; (44)] methods. These methodologies were used, respectively, in the packages phyloseq (McMurdie and Holmes, 2013) and vegan (Oksanen et al., 2022). A deeper analysis using Permutational Multivariate Analysis of Variance (PERMANOVA) was conducted to assess the differences of the communities among different treatments for each breed with the aid of the function adonis2 from the vegan package (Oksanen et al., 2022) and all the metrics mentioned above over 1,000 permutations. An analysis of similarities (ANOSIM) and an analysis of the multivariate homogeneity of group dispersions were also carried out using the functions anosim and betadisper, respectively. Post hoc tests were run in pairs using the pairwise function. Additionally, we used the Euclidean method and Bonferroni correction with the pairwiseAdonis package's adonis to determine the statistical significance of these tests (Martinez Arbizu, 2020).
To compare taxonomic bar plots with relative and absolute abundance at the phylum and genus levels, the microbial composition in the stacked bar plots was analyzed using the R packages qiime2R (Bisanz, 2018) and ggplot2 (Wickham, 2009). The Firmicutes to Bacteroidetes ratio (F/B ratio) was calculated by dividing the relative abundance of Firmicutes by the relative abundance of Bacteroidetes, and then the Mann-Whitney U test was used to identify significant statistical differences between the treatment groups for each breed. Through the use of the software microeco (Liu C. et al., 2021), a linear discriminant analysis (LDA) effect size (LEfSe) analysis was conducted to identify the taxa with an LDA of ±2 for effect size among the treatment groups for each breed and their relative abundances. Taxonomic abundance was represented as a differential heat tree using the R package metacoder (Foster et al., 2017), with a Wilcox rank sum test and Benjamin-Hochberg (False discovery rate: FDR) correction for multiple comparisons. Furthermore, lists of the distinct and shared taxa between the treatment groups (for each breed) were created using the packages MicrobiotaProcess (Xu et al., 2023), zoo (Zeileis and Grothendieck, 2005), and VennDiagram (Chen and Boutros, 2011) as well as a Venn diagram showing the different treatments for each breed. The methodology of the present article was based on the methodology described previously in one of our previous articles: Frias et al. (2023).
2.5 Data availability
The DNA sequences of the samples used in this investigation can be found in the NCBI SRA repository under the project names BioProject PRJNA956576 (for samples from guinea pigs in the fed groups) and BioProject PRJNA982863 (for samples from guinea pigs in the fasting groups).
3 Results
3.1 Summary of breeds and sequencing
We used 28 samples of guinea pig cecum's mucosa, 10 samples of the Andina breed (five for the fed group and five for the fasting group), nine samples of the Inti breed (four for the fed group and five for the fasting group), and nine samples of the Peru breed (five for the fed group and four for the fasting group).
A total of 785,617 sequences (Andina), 640,893 sequences (Inti), and 784,554 sequences (Peru) were obtained from sequencing the guinea pig cecum mucosa samples in the V4 region of the 16S rRNA gene. These sequences were used for downstream analyses relevant to the study of the structure and composition of the cecum microbiota of guinea pig breeds treatment groups. On the other hand, the number of reads per sample was: 78,561.700 ± 22,020.000 (mean ± SD) reads/sample (Andina); 71,210.333 ± 25,540.382 (mean ± SD) reads/sample (Inti), and 87,172.667 ± 17,509.425 (mean ± SD) reads/sample (Peru). The median length for all reads was 252.85 bp (Andina), 252.82 bp (Inti), and 252.85 bp (Peru). Overall, 1,716 (Andina), 1,466 (Inti), and 1,680 (Peru) taxa identified were used in the analyses.
3.2 Alpha diversity of the cecum microbiota of the guinea pig breeds treatment groups
Rarefaction curves showed that all of the samples had reached the saturation plateau, demonstrating that the volume of sequencing data was sufficient and could accurately represent the vast majority of the microorganisms in the samples (Supplementary Figure S1 in Supplementary Presentation 1). To determine if there were any differences among the sample groups, richness (Chao1 index) and diversity (Shannon index) were examined. The Shannon and Chao1 indices did not differ statistically by treatment group within samples (Tables S1–S9 in Supplementary Data Sheet 1). There was a tendency for a higher Chao1 index and the number of Observed OTUs of the fed group in comparison to the fasting group in the Inti and Peru breeds, but not in the Andina breed. On the other hand, there was a tendency for a higher Shannon index of the fasting group in comparison to the fed group in the Inti and Peru breeds, but not in the Andina breed.
Interestingly, we observed in the figures that there were slight changes in the structure of alpha diversity among samples of all breeds. The Andina breed increased the richness index (Chao1) and decreased sample diversity (Shannon). On the other hand, the Inti and Peru breeds had a slight decrease in richness and an increase in diversity. This demonstrates that statistically fasting does not change alpha diversity, but the graphical data show a slight change (Figure 1).
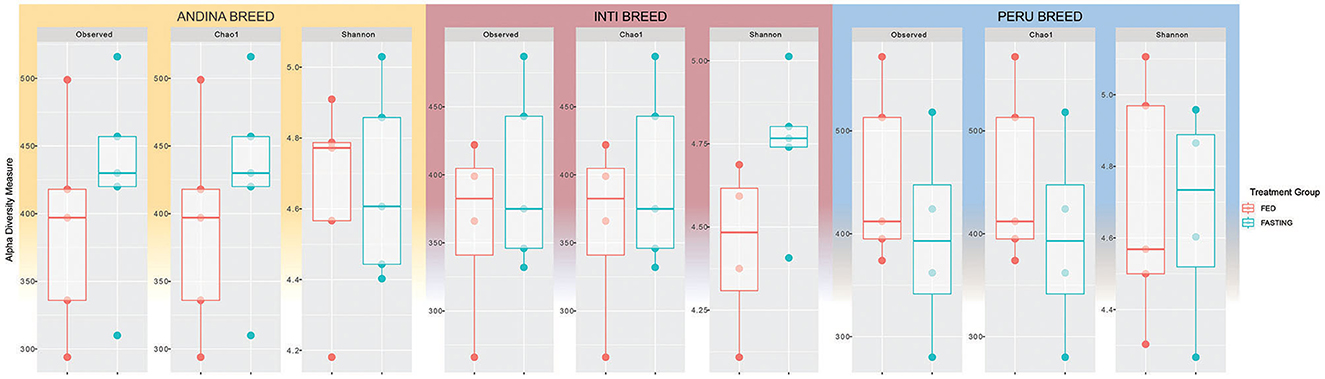
Figure 1. Box plots representing alpha diversity indices for the comparison of guinea pig breeds treatment groups. Different colors indicate different treatments (red: fed and green: fasting) for each breed: Andina (left), Inti (center), and Peru (right). The horizontal line inside the boxes represents the median, the box indicates the interquartile range, and the thin vertical black line represents the rest of the distribution.
3.3 Differences in microbial composition among groups based on beta diversity
CAP and NMDS were used to investigate the beta diversity of microbial communities. The CAP in Figure 2 illustrates how the treatment groups differ from one another in each breed (Andina, Inti, and Peru). A clear separation of the treatment groups was found in the Andina and Peru breeds, but a slight separation for Inti.
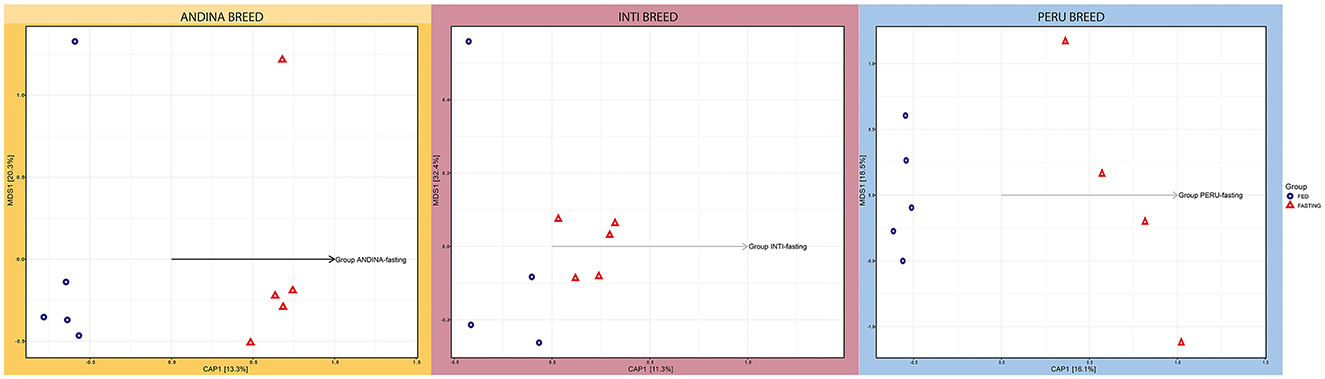
Figure 2. Canonical Principal Coordinate Analysis (CAP) of the guinea pig breeds treatment groups. The forms indicate the types of treatment: a triangle for the fasting group and a circle for the fed group in each breed: Andina (left), Inti (center), and Peru (right). The arrangement of the arrows illustrates the formation of groups of individuals selected in different coordinates, indicating the dissimilarity and similarity of microbiota composition among the independent samples and groups of treatments within breed groups. The CAP was built on an unweighted UniFrac distance. CAP, Canonical Principal Analysis; NMDS, Non-metric MultiDimensional Scaling.
However, we only found a significant difference between the treatment groups of the Peru breed (P-value < 0.05), but not for the Andina and Inti breeds (Supplementary Tables S10–S12 in Supplementary Data Sheet 1). Following qualitative ecological data of the various bacterial species grouped, the intestinal microbiota of the Peru breed varies between the treatment groups. The treatment groups of each breed were separated based on unique fraction metric (Unifrac) unweighted distances (which take into account the presence or absence of a species) and Unifrac weighted distances (which take into account information about species abundance), as shown in the Supplementary Figures S2, S3. These distances are based on phylogenetic distance measurements and were used in the NMDS plots. The NMDS plots showed no evident clustering of treatment groups for each breed. Based on unweighted Unifrac distances, only the treatment groups of the Peru breed displayed significant similarity, with an even distribution of high and low ranks within and between groups, in the NMDS plot (ANOSIM: R2 = 0.3563; P-value = 0.04).
3.4 Differences in the composition of the cecum microbiota of the guinea pig breeds treatment groups
The microbial compositions present in the cecum microbiota of guinea pigs from different treatment groups within breeds are shown in Figures 3–6. The figures of the absolute microbiota composition are displayed in Supplementary Figures S4–S6.
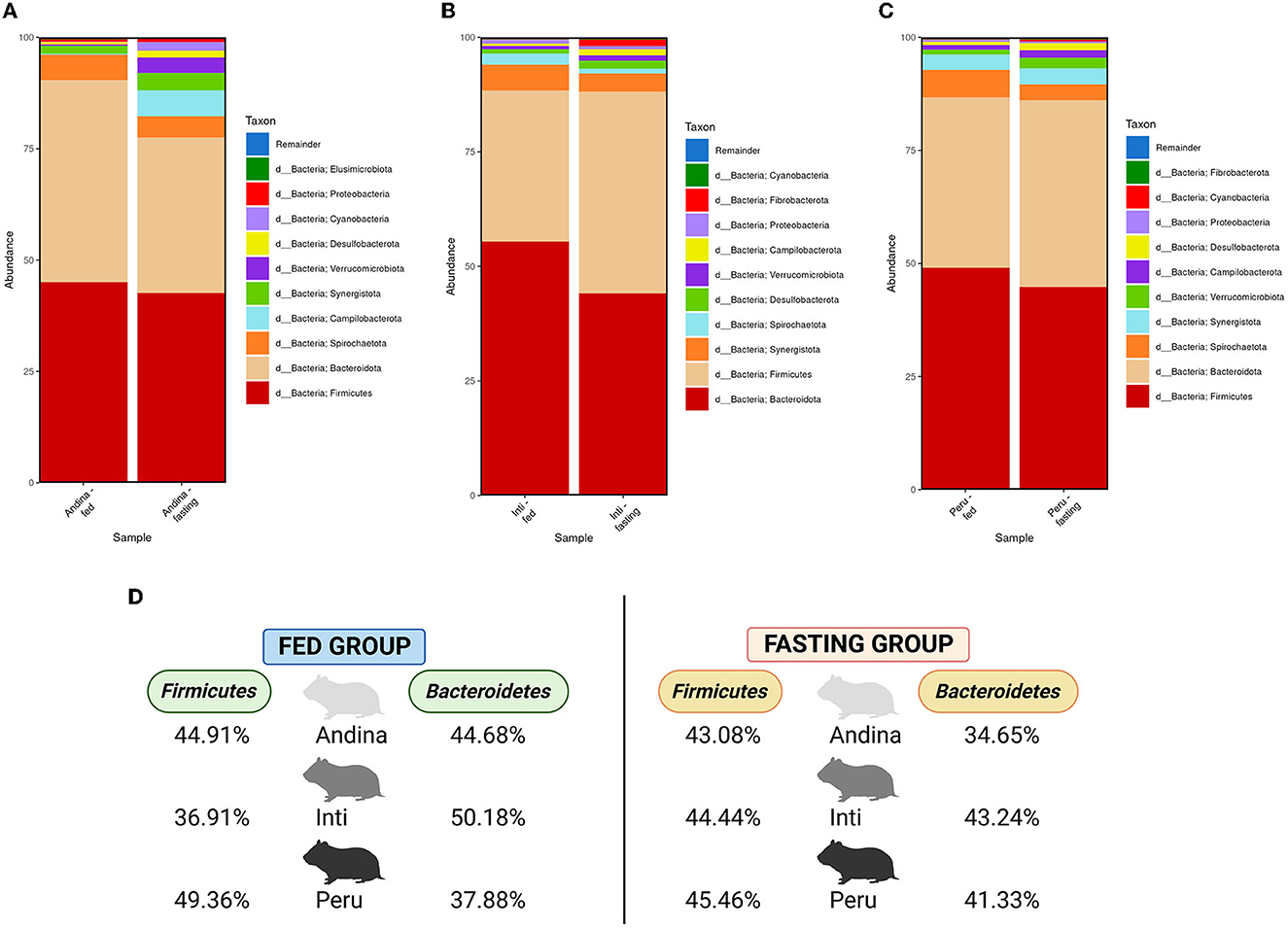
Figure 3. Relative abundance of the main phyla in the guinea pig breeds treatment groups. The 10 main phyla of the cecum microbiota present in the treatment groups (fed and fasting groups) within guinea pig breed groups: Andina (A), Inti (B), and Peru (C). Representation of the relative abundance percentages of the two main phyla present in all the treatment breed groups (D). d, domain. The subfigure (D) was created with BioRender.com.
In Figure 3, among the 10 main phyla that we found in the cecum microbiota of the guinea pig breeds treatment groups, we found that Firmicutes and Bacteroidetes were the 2 most abundant phyla for all the guinea pig breeds treatment groups.
In Figure 4, we found an increase in the F/B ratio in the fasting groups of the Andina and Inti breeds but not for the Peru breed, in this case, we found an opposite effect. Denote, that the animals were maintained in the same environment and received the same food/fasting procedures.
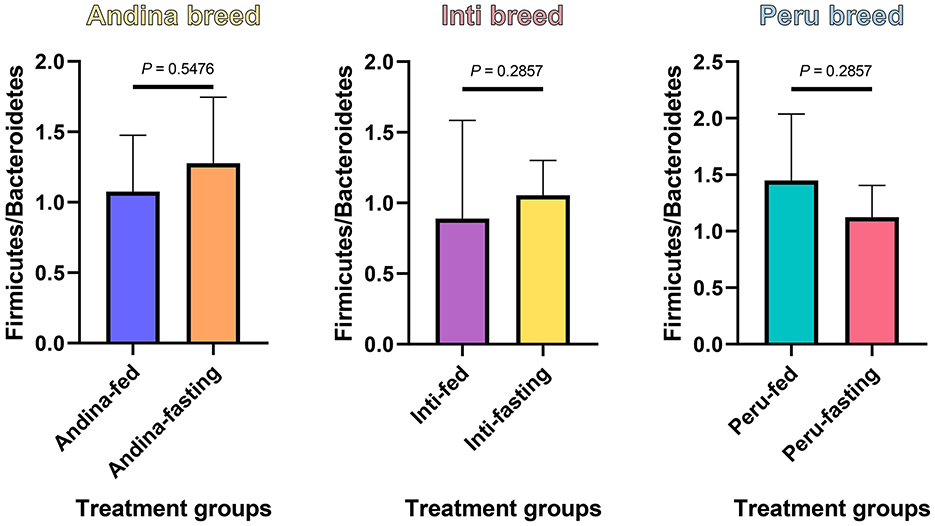
Figure 4. The ratio of Firmicutes to Bacteroidetes in the guinea pig breed treatment groups. The P-values were obtained from the Mann–Whitney U-test. The box and whisker represent the mean ± standard deviation (SD).
We found that the most dominant genus in all groups was Muribaculaceae (Muribaculaceae), with variable rates of 15.85% and 9.24% for the Andina-fed and Andina-fasting groups; 14.53% and 17.06% for Inti-fed and Inti-fasting groups; 18.78% and 13.65% for the Peru-fed and Peru-fasting groups, respectively (Figure 5).

Figure 5. Relative abundance of the 35 main genera present in the guinea pig treatment breed groups. The relative abundance of the 35 main genera present in the cecum microbiota of the treatment groups (fed and fasting groups) within guinea pig breed groups: Andina (A), Inti (B), and Peru (C). d, domain.
Also, in the majority of the samples of all groups, the most dominant genus was Muribaculaceae (Muribaculaceae; Figure 6). The other two predominant genera after Muribaculaceae were Prevotella (7.99%) and Oscillospiraceae (UCG-005) for the Andina-fed group, Ruminococcus (7.36%) and Prevotellaceae (Prevotellaceae UCG-004; 5.69%) for the Andina-fasting group, Prevotella (11.93%) and Prevotellaceae (Prevotellaceae UCG-004; 8.26%) for the Inti-fed group, Prevotellaceae (Prevotellaceae UCG-001; 8.17%) and Ruminococcus (7.70%) for the Inti-fasting, Prevotella (8.62%) and Ruminococcus (7.55%) for the Peru-fed group, Ruminococcus (10.03%) and Oscillospiraceae (UCG-005; 6.75%) for the Peru-fasting group. The values of the whole taxa relative abundance have been stored in the Supplementary Data Sheet 2.

Figure 6. Relative abundance of the 45 main genera of the cecum microbiota present in the samples. The relative abundance of the 45 main genera present in the cecum microbiota of the treatment groups (fed and fasting groups) within guinea pig breed groups: Andina (A), Inti (B), and Peru (C). d, domain; INT, Inti breed; AND, Andina breed; P, Peru breed; AL, fed treatment; AY, fasting treatment.
We identified an enrichment of 59 taxa in the Andina breed, 43 taxa in the Inti breed, and 57 taxa in the Peru breed (Figure 7). Four taxa were enriched in all the fasting groups: Clostridia (genus Clostridia vadinBB60 group), Rikenellaceae (dgA-11 gut group), Helicobacter, and Oscillospirales (genus UCG-010). Also, we found 28 taxa that have a significant enrichment (P < 0.05; LDA score >2; LDA score < -2) with the LefSe comparison in some of the treatment groups. For the Andina-fed group, the significantly enriched taxa were Muribaculaceae (Muribaculaceae), Prevotellaceae (Prevotellaceae UCG-001), Blautia, Ruminobacter, Streptococcus, Butyricicoccaceae (UCG-008), Veillonella, and Caproiciproducens. For the Andina-fasting group, the significantly enriched taxa were Gastranaerophilales (genus Gastranaerophilales), Victivallales (genus vadinBE97), Helicobacter, Kiritimatiellae (genus WCHB1-41), Oscillospirales (genus UCG-010), Izemoplasmatales (genus Izemoplasmatales), Peptococcus, and Erysipelatoclostridiaceae (UCG-004) were the enriched taxa. For the Inti-fasting group, the significantly enriched taxa were Ruminococcus. For the Peru-fed group, the significantly enriched taxa were Clostridia (genus Clostridia UCG-014) and Oscillospiraceae (NK4A214 group). For the Peru-fasting group, the significantly enriched taxa were Prevotellaceae (Prevotellaceae UCG-004), Kiritimatiellae (genus WCHB1-41), Monoglobus, Elusimicrobium, Bacteroides, Butyricicoccaceae (UCG-009), Frisingicoccus, and Victivallales (genus vadinBE97). The complete data of the LefSe comparison has been stored in the Supplementary Data Sheet 3.
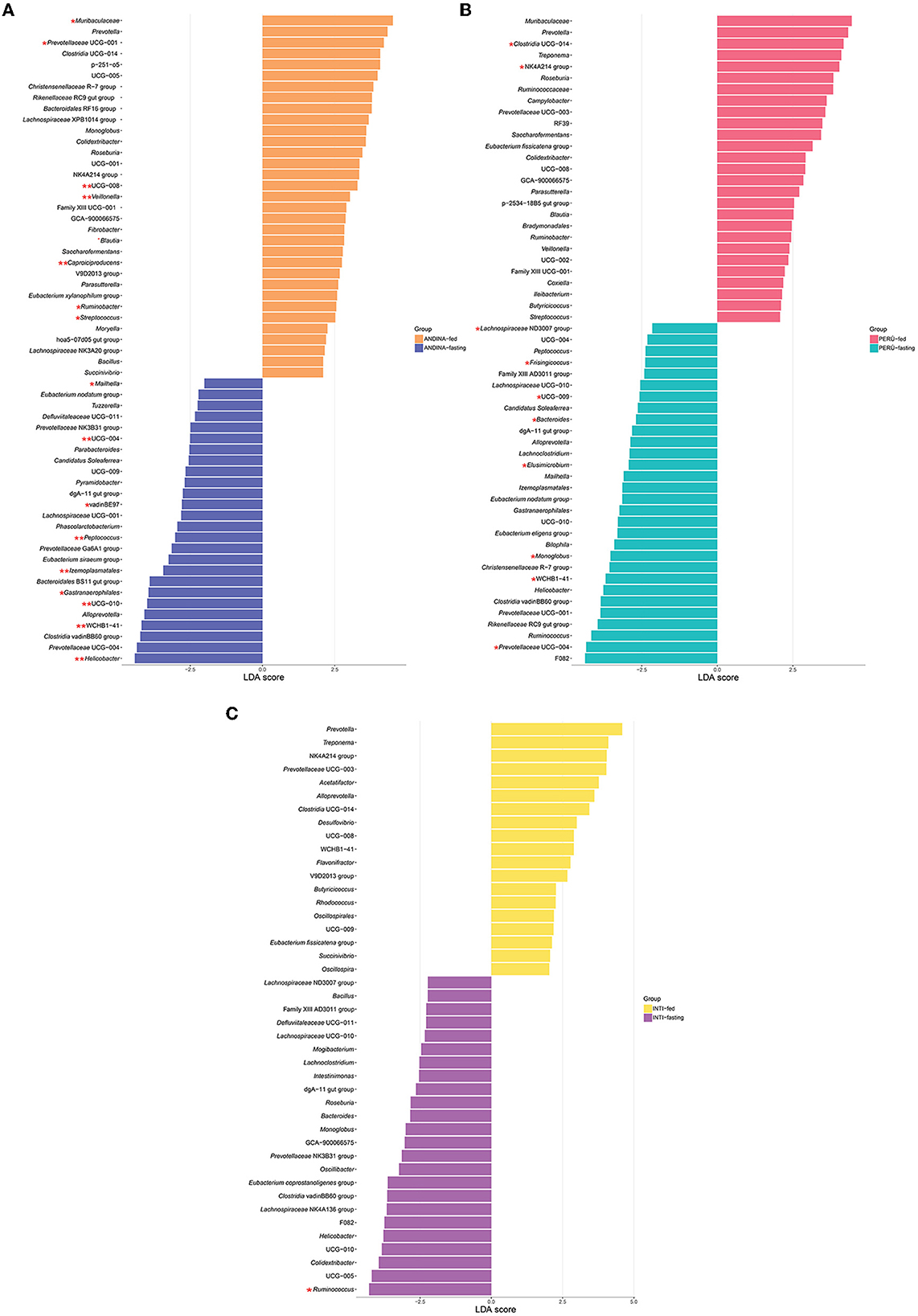
Figure 7. Linear discriminant analysis (LDA) effect size (LEfSe) comparison of the guinea pig breeds treatment groups. LefSe comparison of differentially abundant bacterial taxa between the treatment groups (fed and fasting groups) within the breeds of the guinea pig: Andina (A), Peru (B), and Inti (C). Horizontal bars represent the effect size for each taxon. LDA score cutoff of 2.0 was used to determine an enrichment in a bacterial taxon. One red asterisk (P < 0.05) and two red asterisks (P < 0.01) denote taxa with statistically significant differences between the abundances of treatment groups within breed groups.
The heat tree analysis showed 45 taxa with a significant difference between the treatment groups in the Andina breed, six taxa between the treatment groups of the Inti breed, 23 taxa between the treatment groups of the Peru breed (Figure 8, Supplementary Data Sheet 4).
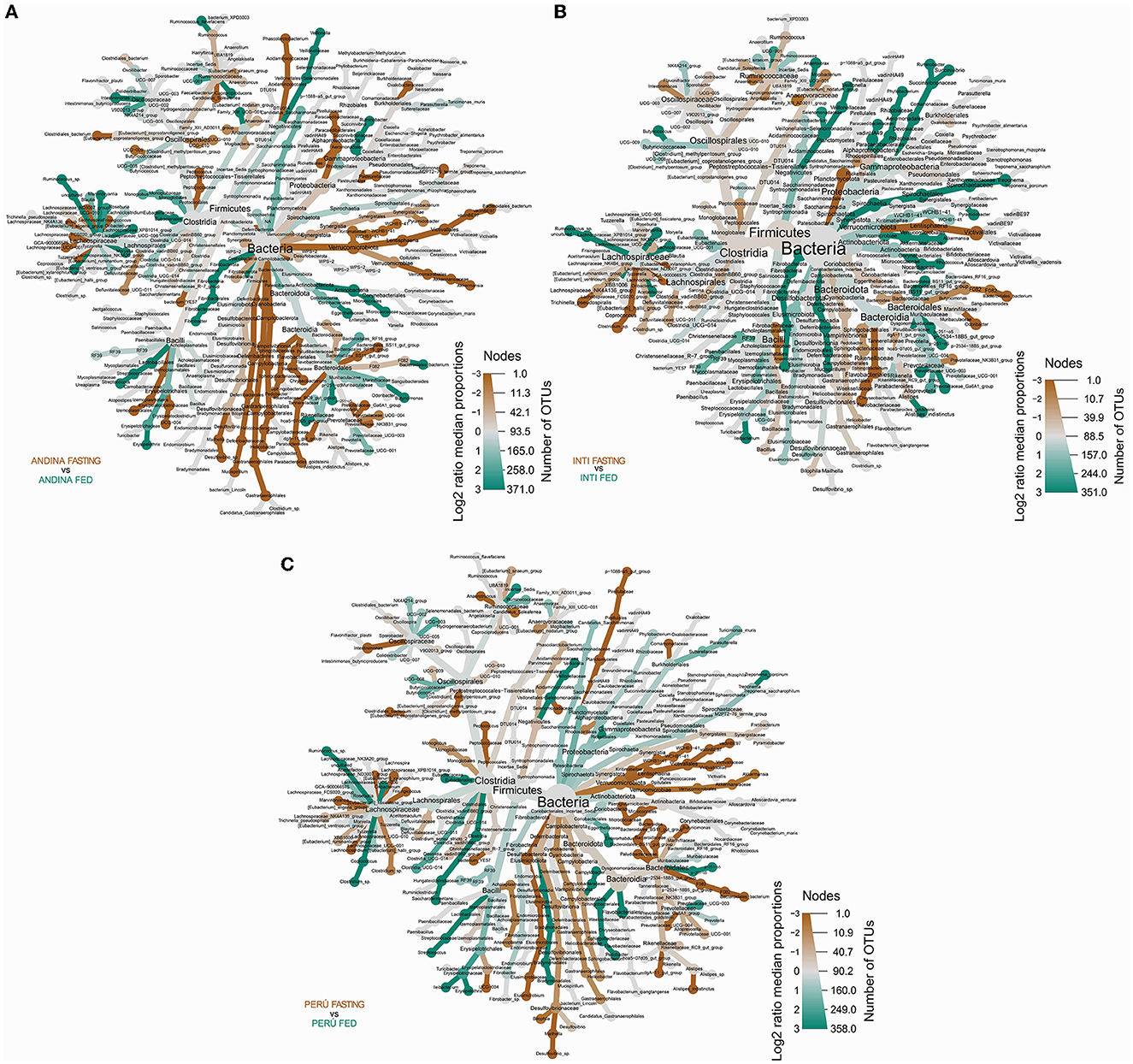
Figure 8. Differential heat tree comparison between the guinea pig breeds treatment groups. The differential heat trees show the taxonomy and the comparison between the taxa of the treatment groups (fed and fasting groups) within the breed groups: Andina (A), Inti (B), and Peru (C). The color and the size of the nodes and edges correlate with the richness or number of operational taxonomic units (OTUs) of organisms within the community in which they are found. Color intensity is related to the log2 ratio of the difference in median proportions and the Wilcoxon test applied to the readings in each treatment group (fed and fasting groups) for each breed group. The brown taxa show an enhancement in the fasting group, and the green taxa show the opposite in the other comparative group (fed group) within the breed groups: Andina (A), Inti (B), and Peru (C). In gray, the nodes are equally present in both compartments. OTUs, operational taxonomic units.
We found several differences between the treatment groups in the Andina breed: in the phylum level: Campylobacterota: Campylobacteria (Campylobacterales); Cyanobacteria: Vampirivibrionia [Gastranaerophilales (family and genus Gastranaerophilales)]; and Verrumicrobiota: Kiritimatiellae (order, family, and genus WCHB1-41) and Lentisphaeria [Victivallales (family and genus vadinBE97)]. In the order level: Izemoplasmatales (family and genus Izemoplasmatales); Oscillospirales [family and genus UCG-010, Butyricicoccaceae (UCG-008)]; Peptococcales: Peptococcaceae (Peptococcus); and Eubacteriales (Eubacteriaceae). In the family level: Veillonellaceae (Veillonella); Helicobacteraceae (Helicobacter); Erysipelatoclostridiaceae (UCG-004); Streptococcaceae (Streptococcus); Muribaculaceae (Muribaculaceae); and Prevotellaceae (Prevotellaceae UCG-001). In the genus level: Desulfovibrio, Mailhella, Ruminococcus, Caproiciproducens, and Blautia.
We found several differences between the treatment groups in the Inti breed: in the phylum level: Spirochaetota (Spirochaetia). In the family level: Weeksellaceae (Chryseobacterium). In the genus level: Ruminococcus. In the species level: Trichinella pseudospiralis.
We found several differences between the treatment groups in the Peru breed: in the class level: Clostridia (order, family, and genus Clostridia UCG-014) and Kiritimatiellae (order, family, and genus WCHB1-41). In the order level: Eubacteriales (Eubacteriaceae); Peptococcales (Peptococcaceae); Monoglobales: Monoglobaceae (Monoglobus). In the family level: Butyricicoccaceae (UCG-009); Ruminococcaceae (Incertae Sedis); Oscillospiraceae (NK4A214 group); Lachnospiraceae (Lachnospiraceae ND3007 group and Frisingicoccus); Bacteroidaceae (Bacteroides); and Prevotellaceae (Prevotellaceae UCG-004).
The Venn diagram shows that most of the taxa were shared between the treatment groups within the breed groups (Andina: 116 taxa, Inti: 112 taxa, and Peru: 111 taxa). Furthermore, we found a greater number of unique taxa in the fasting groups in comparison with the fed groups of the different breeds. Also, the only unique genus that the fasting groups of the three breeds have in common is Victivallis (Figure 9). The shared taxa between treatment groups of each breed group were specified in the Supplementary Data Sheet 5.
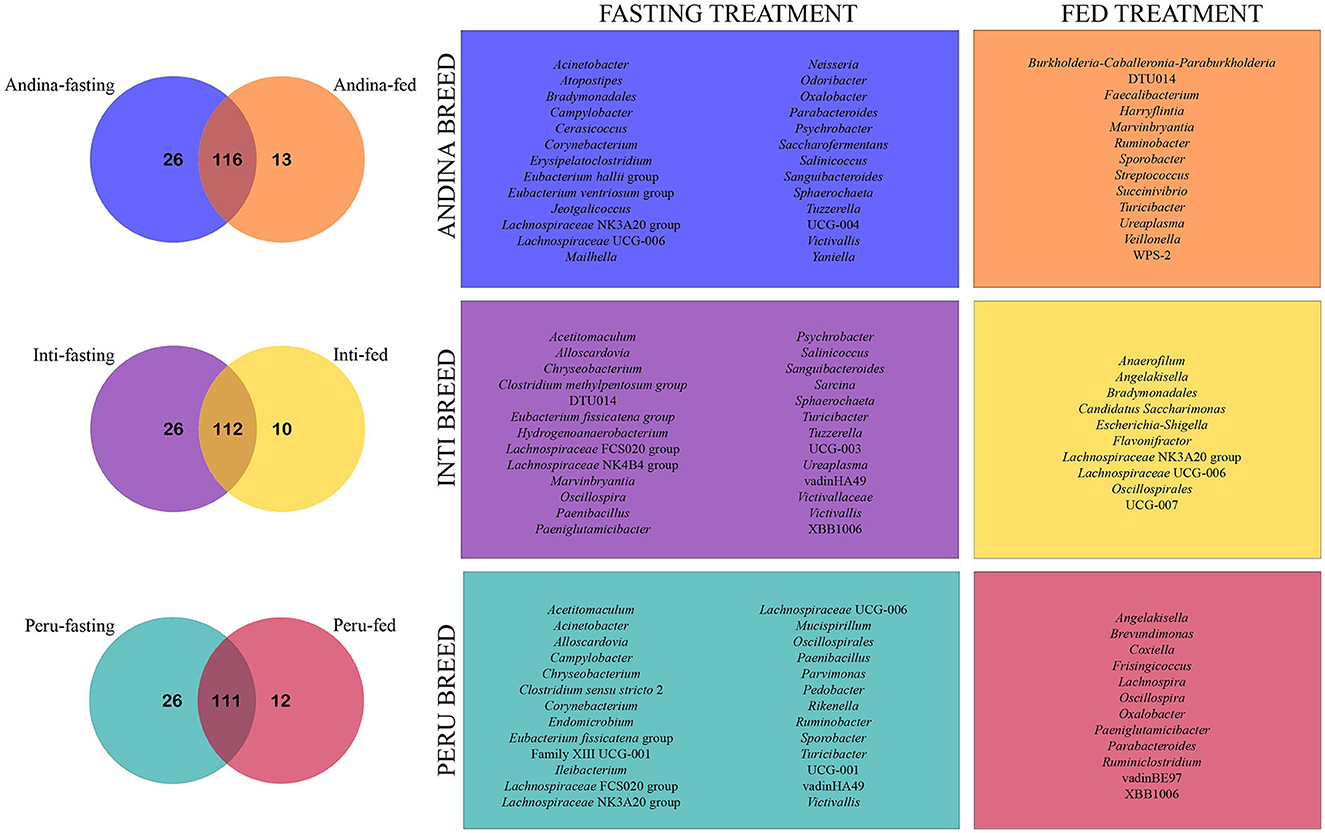
Figure 9. Venn chart illustrating unique and shared genera among different guinea pig breeds treatment groups. The Venn charts illustrate the comparison of the unique and shared genera present in the treatment groups (fed and fasting) for each breed group (Andina, Inti, and Peru). The numbers inside the subsets on the Venn diagrams represent the number of genera identified for each subset (unique breed-fasting subset, unique breed-fed subset, and shared subset). On the right side of the Venn diagrams, the frames are labeled with the colors of their respective subsets and show each unique genus that has been cataloged for their subsets. Some genera may have the same name but different amplicon sequence variants (ASVs).
4 Discussion
The cecum microbiota of the guinea pig plays a key role in the fermentation of the vegetal material eaten by the animal (Tang et al., 2022; Frias et al., 2023). To the best of our knowledge, this is the first study that explores the effect of fasting on the cecum microbiome of the guinea pig Andina, Inti, and Peru breeds. The alpha diversity between the samples of the fasting and fed groups showed an observational difference between the samples and beta diversity demonstrated that the Peru breed may be the most affected by the fasting period. For the composition of the microbiota, we identified notable changes or fluctuations in the taxonomy, with the Firmicutes to Bacteroidetes relationship possibly being the most affected. However, the family Muribaculaceae, being the most abundant, was the most present among the breeds and the most resilient to the fasting period. We also found a higher number of unique genera in the fasting groups of the breeds.
In the analysis of the alpha diversity, we found that there was an increase in the richness for the fasting group in the Andina and Inti breeds, but not in the Peru breed. Also, there was an increase in the diversity for the fasting group in the Inti and Peru breeds, but not for the Andina breed. Some studies have found that intermittent fasting can increase the gut microbiome richness (Observed OTUs) and the gut microbiome diversity (Simpson index) in a diabetic model and diet-induced obese mice, respectively (Deng et al., 2020; Liu et al., 2020). In the analysis of the beta diversity, we found a significant difference between the bacterial communities of the treatment groups of the Peru breed, but not for the other breeds. Based on these findings we could argue that the fasting treatment can have a significant effect on the bacterial communities of the cecum microbiota of the Peru breed, but not in the other breeds. These findings are related to other studies in humans and mice: Ali et al. (2021) identified that fasting can significantly change the beta diversity in the gut microbiome of humans. Also, Daas and de Roos (2021) report that multiple studies that investigated the effects of fasting on the gut microbiome of mice have similar results regarding the increase of beta diversity.
There are several phenotypic differences between the guinea pig breeds: Andina, Inti, and Peru. For example, the Peru breed has a major meat carcass yield and weight gain in comparison with Andina and Inti, when fed with the same feeding system (Reynaga Rojas et al., 2020). These phenotypic differences between the Peru breed with the Andina and Inti breed could be associated with the compositional differences in the microbiota of the cecum that the Peru breed presents in his fasting state and fed state, as detailed in the present study.
The phyla that dominated all guinea pig breeds treatment groups were Firmicutes and Bacteroidetes, normally found in the cecum microbiota of guinea pigs (Tang et al., 2022; Frias et al., 2023), and several studies detailed their importance for fatty acid production through the fermentation of different substrates (Thomas et al., 2011; Rowland et al., 2018; Parada Venegas et al., 2019; Stojanov et al., 2020). These two phyla are important for fatty acid production through fermentation. However, these phyla are the ones that may have fluctuated the most during the fasting period, this was assessed in this study with the analysis of the F/B ratio. The F/B ratio is associated with intestinal homeostasis. The balance of the intestinal ecosystem is critical for maintaining normal body function and significant changes in the ratio F/B can be meaningful to the health aspect of the organism (Stojanov et al., 2020). The F/B ratio was calculated by dividing the relative abundances of Firmicutes by the relative abundance of the Bacteroidetes, like in previous studies (Houtman et al., 2022; Tang et al., 2022). We found a reduction of the F/B ratio in the fasting group of the Peru breed, and in the long term, this could influence the transformation of nutrients into the necessary fat (Deng et al., 2020; Angoorani et al., 2021). Based on the aforementioned, we could suggest that the Peru breed has a higher susceptibility to the fasting treatment.
Furthermore, we observed an increase in the relative abundance of the genera Akkermansia and Bacteroides in the fasting groups of all three breeds. Members of the genus Akkermansia are mucolytic bacteria and do mucin forage and use host-derived substances, similar to members of Bacteroides that have a growth advantage over the other populations of bacteria that rely strictly on dietary substrates (Sonoyama et al., 2009; Ducarmon et al., 2023). Furthermore, we also observed an increase in the relative abundance of the genera from the family Desulfovibrionaceae. The majority of the members of this family are sulfate-reducing bacteria (Spring et al., 2019), and the increase in this family could be related to the fact that the increased mucin foraging during the fasting treatment can cause the proliferation of sulfur-reducing bacteria (Ducarmon et al., 2023).
The LefSe analysis contributed to the identification of four taxa that were enriched in all the fasting groups of the three breeds: Clostridia (genus Clostridia vadinBB60 group), Rikenellaceae (dgA-11 gut group), Helicobacter, and Oscillospirales (genus UCG-010). Furthermore, Maifeld et al. (2021) found that members of Clostridia showed an opposite effect with the fasting treatment in the gut microbiome of humans. An enrichment in the family Rikenellaceae was already found in the gut microbiome of rodents after a fasting treatment: Zhang et al. (2020) determined an increase of Rikenellaceae in the intermittent energy fasting group in the gut microbiome of a colitis mouse model and Su et al. (2022) found similar results in the gut microbiome of BALB/c mice after Ramadan fasting. We could argue that the increase in the abundance of the members of Helicobacter in the fasting groups could be related to a reduction in the microbiome-mediated colonization resistance against potential pathogens of the gut microbiota of the guinea pig (Ducarmon et al., 2023). On the other hand, Liu J. et al. (2021) found that an intermittent fasting treatment in mice can cause a significant reduction in the abundance of Helicobacter. Some members of the order Oscillospirales can metabolize sugars and produce short-chain fatty acids (SCFAs) as fermentation products (Yang et al., 2021).
We found several abundance differences between the treatment groups in different taxonomic ranks with the Heat tree analysis. The fasting treatment significantly increased the abundance of Bacteroidetes, Bacteroidia, Lachnospirales, and Lachnospiraceae in the Inti and Peru breeds, but not in the Andina breed, in this breed decreased. Mesnage et al. (2019) found that the Buchinger fasting treatment led to an increase in the abundance of Bacteroidetes, which uses derived energy substrates. Also, Su et al. (2021) discovered that the Ramadan fasting treatment increased the amount of Lachnospiraceae, a family of bacteria that break down dietary polysaccharides (Angoorani et al., 2021).
The fasting treatment significantly increased the abundance of Firmicutes, Clostridia, and Oscillospirales in the Inti breed, but not in the Andina and Peru breeds, in these breeds, some inferior taxonomic ranks showed an increased or decreased abundance. Angoorani et al. (2021) reported that the Ramadan fasting treatment led to a decreasing trend in the abundance of Firmicutes. Sonoyama et al. (2009) found that the fed active group led to an increase in the abundance of Clostridia (compared with a fasting active group, which fasted for 96 hours) in the cecum microbiome of hamsters. Kohl et al. (2014) found that the fasting treatment led to an increase in the abundance of Oscillospirales in the cecum microbiome of mice, tilapia, and quail.
The fasting treatment significantly increased the abundance of Ruminococcaceae in the Inti breed, but not in the Andina and Peru breeds, in these breeds decreased. Mesnage et al. (2019) found that a Buchinger fasting treatment reduced the abundance of Ruminococcaceae. On the other hand, Su et al. (2021) found the opposite effect with a Ramadan fasting treatment.
There are several reports that host genetics can influence the differences in the abundance of the microorganisms present in the gut microbiota of guinea pig breeds (Frias et al., 2023), pig breeds (Bergamaschi et al., 2020), chicken breeds (Sun et al., 2018; Yan et al., 2021), dog breeds (Morelli et al., 2022), and mice breeds (Campbell et al., 2012). These differences then could have an impact on the effect of a fasting treatment or a feed restriction treatment such as that observed in the cecal microbiome of different chicken breeds (Yan et al., 2021). Furthermore, fasting has been shown to elicit weight loss and inhibit sterol synthesis in the ileum of guinea pigs (Turley and West, 1976; Langley and Kelly, 1992). These two effects have also been observed in conjunction with alterations in the gut microbiome of other animal species (Jian et al., 2022; Koutoukidis et al., 2022; Le et al., 2022; Manoppo et al., 2022). Hence, based on the aforementioned evidence, it can be argued that the differences in the impact of fasting among guinea pig breeds at different taxonomic levels may be attributed to a potential genetic influence of the host on the cecum microbiota.
Finally, we found unique genera in all the treatment groups within the three breeds of guinea pigs. The fasting group has more unique genera than the fed group in all three breeds, this could be related to the finding made by another study that identified an increase in the richness of the fasting group in the gut microbiome of humans (Ozkul et al., 2020). The only unique genus that the fasting groups of the three breeds shared was Victivallis. This genus has members that are cellobiose-degrading, produce SCFAs, and are positively associated with fat-derived energy from dietary intake in humans (Zoetendal et al., 2003; Ali et al., 2021).
As we explained, fasting seems to direct to a more homogeneous group of microbes. However, there are lots of different strategies for fasting that could lead to different changes in the composition of the microbiome, for example, the most representative are Buchinger fasting (calorie-restricted regimen) and Ramadan fasting (time-restricted regimen) that have some different effects in the gut microbiota of several animals and humans (Angoorani et al., 2021). However, there are no reports of the effect of fasting on the cecum microbiota of guinea pigs. Therefore, the interaction between fasting and the cecum microbiome from guinea pigs can be further clarified by our research. But understanding more of the effects of prolonged fasting or intermittent fasting should be applied, expecting a long-term reflection of the microbiome and a more complex understanding. Intermittent fasting promotes microbial fermentation, forming several bioproducts with beneficial effects on metabolic disorders (Li et al., 2017). This could be one future approach led by this research since the microbiome, as it is known, can influence several biological aspects in humans and animals. Additionally, future research endeavors may contemplate the inclusion of extended fasting durations preceding euthanasia or delve into the examination of feeding strategies that encompass intermittent fasting.
5 Conclusions
Ceasing food intake can modify the structure of the microbiome. The current study discovered a different effect of fasting on the cecum microbiome of the guinea pigs: Andina, Inti, and Peru breeds. The analysis of the beta diversity shows significant differences only between the treatment groups of the breed Peru, but not for the other breeds. We found that two main phyla were shared between all the guinea pig breeds treatment groups: Bacteroidetes and Firmicutes, with fluctuations in the bacterial count after fasting. Additionally, we discovered that the Peru breed was the only breed that demonstrated that the fasting treatment reduced the F/B ratio. Although we found unique genera in all the guinea pig breeds treatment groups, the only unique genus that the fasting groups of the three breeds shared was Victivallis. Finally, this study is the first to elucidate how fasting can affect the cecum microbiome of different breeds of guinea pigs. Despite the results found, the resilience of the gut microbiome was not challenged, causing disruptive changes that can influence the general maintenance of the cecum microbiome. Although, based on the several differences found in the treatment groups of the Peru breed, we could suggest that the fasting treatment has a bigger effect on this breed.
Data availability statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found at: https://www.ncbi.nlm.nih.gov/bioproject/PRJNA956576, PRJNA956576; https://dataview.ncbi.nlm.nih.gov/object/PRJNA982863?reviewer=il3qiric5go0ur1n6uln77u5jv, PRJNA982863.
Ethics statement
The experimental protocol (CIEI-N°005) has received approval from the Institutional Committee on Research Ethics (CIEI) of the National University Toribio Rodriguez de Mendoza (UNTRM). The study was conducted in accordance with the local legislation and institutional requirements.
Author contributions
HF: Conceptualization, Investigation, Writing – original draft, Writing – review & editing. NM: Conceptualization, Investigation, Writing – original draft, Writing – review & editing. GF: Conceptualization, Investigation, Writing – original draft, Writing – review & editing. VC: Data curation, Formal Analysis, Methodology, Software, Writing – original draft. AR: Investigation, Writing – review & editing. WB: Writing – review & editing. GS: Writing – review & editing. RP: Investigation, Methodology, Writing – review & editing. DV: Investigation, Writing – original draft. ER: Writing – review & editing. RL: Project administration, Supervision, Writing – review & editing. JM: Project administration, Supervision, Writing – review & editing.
Funding
The author(s) declare financial support was received for the research, authorship, and/or publication of this article. This research was funded, in whole, by the project CUI No 2254946 from the National University Toribio Rodríguez de Mendoza, Amazonas, Peru.
Acknowledgments
The authors acknowledge the assistance of the practitioners of the different Livestock and Biotechnology Research Institute laboratories from the National University Toribio Rodríguez de Mendoza.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher's note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2023.1283738/full#supplementary-material
Abbreviations
16S rRNA, 16S ribosomal ribonucleic acid; ACE, abundance-based coverage estimator; ANOSIM, analysis of similarities; ANOVA, analysis of variance; ASV, amplicon sequence variant; CAP, canonical analysis of principal coordinates; CIEI, Institutional Committee on Research Ethics; DNA, deoxyribonucleic acid; F/B, firmicutes to bacteroidetes; FDR, false discovery rate; GIT, gastrointestinal tract; HSD, honestly significant difference; IGBI, Institute of Livestock and Biotechnology; INIA, National Institute for Agricultural Innovation; LDA, linear discriminant analysis; LEfSe, linear discriminant analysis effect size; NCBI, National Center for Biotechnology Information; NMDS, non-metric multidimensional Scaling; OTU, operational taxonomic unit; PBS, phosphate buffer saline; PERMANOVA, permutational multivariate analysis of variance; QIIME2, Quantitative Insights Into Microbial Ecology 2; R2, R squared; SCFA, short-chain fatty acid; SD, standard deviation; SRA, sequence read archive; UCG, uncultured genus-level group; UniFrac, Unique FRACTION metric; UNTRM, National University Toribio Rodriguez de Mendoza; v, version; × g, times gravity.
References
Abou-Hashim, F., Khalifa, W. H., Shalaby, M. B., Kassem, S. M., and Khalil, W. K. B. (2023). Evaluation of fasting and probiotics in reducing postweaning stress in rabbits: Study of their effects on biochemical and gene expression patterns. Appl. Biochem. Biotechnol. doi: 10.1007/s12010-023-04479-w
Al, K., Sarr, O., Dunlop, K., Gloor, G. B., Reid, G., Burton, J., et al. (2017). Impact of birth weight and postnatal diet on the gut microbiota of young adult guinea pigs. PeerJ 5, e2840. doi: 10.7717/peerj.2840
Ali, I., Liu, K., Long, D., Faisal, S., Hilal, M. G., Ali, I., et al. (2021). Ramadan fasting leads to shifts in human gut Microbiota structured by dietary composition. Front. Microbiol. 12, 642999. doi: 10.3389/fmicb.2021.642999
Anderson, M. J., and Willis, T. J. (2003). Canonical analysis of principal coordinates: a useful method of constrained ordination for ecology. Ecology 84, 511–525. doi: 10.1890/0012-9658(2003)084(0511:CAOPCA)2.0.CO;2
Angoorani, P., Ejtahed, H.-S., Hasani-Ranjbar, S., Siadat, S. D., Soroush, A. R., Larijani, B., et al. (2021). Gut microbiota modulation as a possible mediating mechanism for fasting-induced alleviation of metabolic complications: a systematic review. Nutr. Metab. 18, 105. doi: 10.1186/s12986-021-00635-3
Apprill, A., Mcnally, S., Parsons, R., and Weber, L. (2015). Minor revision to V4 region SSU rRNA 806R gene primer greatly increases detection of SAR11 bacterioplankton. Aquat. Microb. Ecol. 75, 129–137. doi: 10.3354/ame01753
Avilés, D. F., Martínez, A. M., Landi, V., and Delgado, J. V. (2014). El cuy (Cavia porcellus): un recurso andino de interés agroalimentario the guinea pig (Cavia porcellus): an Andean resource of interest as an agricultural food source. Anim. Genet. Resour. 55, 87–91. doi: 10.1017/S2078633614000368
Benedict, F. G. (1915). Investigations at the nutrition laboratory of the Carnegie institution of Washington, Boston, Massachusetts. Science 42, 75–84. doi: 10.1126/science.42.1072.75
Bergamaschi, M., Tiezzi, F., Howard, J., Huang, Y. J., Gray, K. A., Schillebeeckx, C., et al. (2020). Gut microbiome composition differences among breeds impact feed efficiency in swine. Microbiome 8, 110. doi: 10.1186/s40168-020-00888-9
Bisanz, J. E. (2018). qiime2R: Importing QIIME2 Artifacts and Associated Data into R Sessions. R package version 0.99. Available online at: https://github.com/jbisanz/qiime2R
Bolyen, E., Rideout, J. R., Dillon, M. R., Bokulich, N. A., Abnet, C. C., Al-Ghalith, G. A., et al. (2019). Reproducible, interactive, scalable and extensible microbiome data science using QIIME 2. Nat. Biotechnol. 37, 852–857. doi: 10.1038/s41587-019-0209-9
Buela, L., Cuenca, M., Sarmiento, J., Peláez, D., Mendoza, A. Y., Cabrera, E. J., et al. (2022). Role of guinea pigs (Cavia porcellus) raised as livestock in Ecuadorian Andes as reservoirs of zoonotic yeasts. Animals 12, 3449. doi: 10.3390/ani12243449
Callahan, B. J., McMurdie, P. J., Rosen, M. J., Han, A. W., Johnson, A. J. A., Holmes, S. P., et al. (2016). DADA2: high-resolution sample inference from illumina amplicon data. Nat. Methods 13, 581–583. doi: 10.1038/nmeth.3869
Campbell, J. H., Foster, C. M., Vishnivetskaya, T., Campbell, A. G., Yang, Z. K., Wymore, A., et al. (2012). Host genetic and environmental effects on mouse intestinal microbiota. ISME J. 6, 2033–2044. doi: 10.1038/ismej.2012.54
Caporaso, J. G., Lauber, C. L., Walters, W. A., Berg-Lyons, D., Lozupone, C. A., Turnbaugh, P. J., et al. (2011). Global patterns of 16S rRNA diversity at a depth of millions of sequences per sample. Proc. Natl. Acad. Sci. USA. 108(Suppl 1), 4516–4522. doi: 10.1073/pnas.1000080107
Chao, A. (1984). Nonparametric estimation of the number of classes in a population. Scand. J. Stat. 11, 265–270.
Chao, A., and Lee, S.-M. (1992). Estimating the number of classes via sample coverage. J. Am. Stat. Assoc. 87, 210. doi: 10.1080/01621459.1992.10475194
Chauca Francia, L. (2023). Desarrollo del mejoramiento genetico en cuyes en el peru: formación de nuevas razas. An. Cientí. 83, 109–125. doi: 10.21704/ac.v83i2.1879
Chen, H., and Boutros, P. C. (2011). VennDiagram: a package for the generation of highly-customizable Venn and Euler diagrams in R. BMC Bioinformatics 12, 35. doi: 10.1186/1471-2105-12-35
Crowley, E. J., King, J. M., Wilkinson, T., Worgan, H. J., Huson, K. M., Rose, M. T., et al. (2017). Comparison of the microbial population in rabbits and guinea pigs by next generation sequencing. PLoS ONE 12, e0165779. doi: 10.1371/journal.pone.0165779
Daas, M. C., and de Roos, N. M. (2021). Intermittent fasting contributes to aligned circadian rhythms through interactions with the gut microbiome. Benef. Microbes 12, 147–161. doi: 10.3920/BM2020.0149
Deng, Y., Liu, W., Wang, J., Yu, J., and Yang, L.-Q. (2020). Intermittent fasting improves lipid metabolism through changes in gut microbiota in diet-induced obese mice. Med. Sci. Monit. 26, e926789. doi: 10.12659/MSM.926789
Driessen, B., Freson, L., and Buyse, J. (2020). Fasting finisher pigs before slaughter influences pork safety, pork quality and animal welfare. Animals 10, 2206. doi: 10.3390/ani10122206
Ducarmon, Q. R., Grundler, F., Le Maho, Y., Wilhelmi De Toledo, F., Zeller, G., Habold, C., et al. (2023). Remodelling of the intestinal ecosystem during caloric restriction and fasting. Trends Microbiol. 31, 832–844. doi: 10.1016/j.tim.2023.02.009
Dunel-Erb, S., Chevalier, C., Laurent, P., Bach, A., Decrock, F., Le Maho, Y., et al. (2001). Restoration of the jejunal mucosa in rats refed after prolonged fasting. Comp. Biochem. Physiol,. Part A Mol. Integr. Physiol. 129, 933–947. doi: 10.1016/S1095-6433(01)00360-9
Faucitano, L., Chevillon, P., and Ellis, M. (2010). Effects of feed withdrawal prior to slaughter and nutrition on stomach weight, and carcass and meat quality in pigs. Livest. Sci. 127, 110–114. doi: 10.1016/j.livsci.2009.10.002
Foster, Z. S. L., Sharpton, T. J., and Grünwald, N. J. (2017). Metacoder: an R package for visualization and manipulation of community taxonomic diversity data. PLoS Comput. Biol. 13, e1005404. doi: 10.1371/journal.pcbi.1005404
Francino, M. P. (2015). Antibiotics and the human gut microbiome: dysbioses and accumulation of resistances. Front. Microbiol. 6, 1543. doi: 10.3389/fmicb.2015.01543
Frias, H., Murga Valderrama, N. L., Flores, G. J., Cornejo, V. G., Del Solar, J. C., Romani, A. C., et al. (2023). An analysis of the cecum microbiome of three breeds of the guinea pig: Andina, Inti, and Peru. Res. Vet. Sci. 161, 50–61. doi: 10.1016/j.rvsc.2023.06.005
Habold, C., Chevalier, C., Dunel-Erb, S., Foltzer-Jourdainne, C., Le Maho, Y., Lignot, J. H., et al. (2004). Effects of fasting and refeeding on jejunal morphology and cellular activity in rats in relation to depletion of body stores. Scand. J. Gastroenterol. 39, 531–539. doi: 10.1080/00365520410004514
Hildebrand, F., Ebersbach, T., Nielsen, H. B., Li, X., Sonne, S. B., Bertalan, M., et al. (2012). A comparative analysis of the intestinal metagenomes present in guinea pigs (Cavia porcellus) and humans (Homo sapiens). BMC Genomics 13, 514. doi: 10.1186/1471-2164-13-514
Houtman, T. A., Eckermann, H. A., Smidt, H., and De Weerth, C. (2022). Gut microbiota and BMI throughout childhood: the role of firmicutes, bacteroidetes, and short-chain fatty acid producers. Sci. Rep. 12, 3140. doi: 10.1038/s41598-022-07176-6
Hu, X., Wang, F., Yang, S., Yuan, X., Yang, T., Zhou, Y., et al. (2021). Rabbit microbiota across the whole body revealed by 16S rRNA gene amplicon sequencing. BMC Microbiol. 21, 312. doi: 10.1186/s12866-021-02377-x
Jian, C., Silvestre, M. P., Middleton, D., Korpela, K., Jalo, E., Broderick, D., et al. (2022). Gut microbiota predicts body fat change following a low-energy diet: a PREVIEW intervention study. Genome Med. 14, 54. doi: 10.1186/s13073-022-01053-7
Kohl, K. D., Amaya, J., Passement, C. A., Dearing, M. D., and Mccue, M. D. (2014). Unique and shared responses of the gut microbiota to prolonged fasting: a comparative study across five classes of vertebrate hosts. FEMS Microbiol. Ecol. 90, 883–894. doi: 10.1111/1574-6941.12442
Koutoukidis, D. A., Jebb, S. A., Zimmerman, M., Otunla, A., Henry, J. A., Ferrey, A., et al. (2022). The association of weight loss with changes in the gut microbiota diversity, composition, and intestinal permeability: a systematic review and meta-analysis. Gut Microbes 14, 2020068. doi: 10.1080/19490976.2021.2020068
Kurilshikov, A., Wijmenga, C., Fu, J., and Zhernakova, A. (2017). Host genetics and gut microbiome: challenges and perspectives. Trends Immunol. 38, 633–647. doi: 10.1016/j.it.2017.06.003
Langley, S. C., and Kelly, F. J. (1992). Differing response of the glutathione system to fasting in neonatal and adult guinea pigs. Biochem. Pharmacol. 44, 1489–1494. doi: 10.1016/0006-2952(92)90462-R
Le, H. H., Lee, M.-T., Besler, K. R., Comrie, J. M. C., and Johnson, E. L. (2022). Characterization of interactions of dietary cholesterol with the murine and human gut microbiome. Nat. Microbiol. 7, 1390–1403. doi: 10.1038/s41564-022-01195-9
Li, Z., Wright, A.-D. G., Yang, Y., Si, H., and Li, G. (2017). Unique bacteria community composition and co-occurrence in the milk of different ruminants. Sci. Rep. 7, 40950. doi: 10.1038/srep40950
Lindholm, C. (2019). Intermittent Fasting in Chickens: Physiological Mechanisms and Welfare Implications for Broiler Breeders. Linköping: Linköping University Electronic Press. doi: 10.3384/diss.diva-160814
Lindholm, C., and Altimiras, J. (2023). Physiological and behavioural effects of intermittent fasting vs daily caloric restriction in meat-type poultry. Animal 17, 100849. doi: 10.1016/j.animal.2023.100849
Liu, C., Cui, Y., Li, X., and Yao, M. (2021). microeco: an R package for data mining in microbial community ecology. FEMS Microbiol. Ecol. 97, fiaa255. doi: 10.1093/femsec/fiaa255
Liu, J., Zhong, Y., Luo, X. M., Ma, Y., Liu, J., Wang, H., et al. (2021). Intermittent fasting reshapes the gut Microbiota and metabolome and reduces weight gain more effectively than melatonin in mice. Front. Nutr. 8, 784681. doi: 10.3389/fnut.2021.784681
Liu, Z., Dai, X., Zhang, H., Shi, R., Hui, Y., Jin, X., et al. (2020). Gut microbiota mediates intermittent-fasting alleviation of diabetes-induced cognitive impairment. Nat. Commun. 11, 855. doi: 10.1038/s41467-020-14676-4
Lucking, E. F., O'Connor, K. M., Strain, C. R., Fouhy, F., Bastiaanssen, T. F. S., Burns, D. P., et al. (2018). Chronic intermittent hypoxia disrupts cardiorespiratory homeostasis and gut microbiota composition in adult male guinea-pigs. EBioMed. 38, 191–205. doi: 10.1016/j.ebiom.2018.11.010
Maifeld, A., Bartolomaeus, H., Löber, U., Avery, E. G., Steckhan, N., Markó, L., et al. (2021). Fasting alters the gut microbiome reducing blood pressure and body weight in metabolic syndrome patients. Nat. Commun. 12, 1970. doi: 10.1038/s41467-021-22097-0
Manoppo, J. I. C., Nurkolis, F., Gunawan, W. B., Limen, G. A., Rompies, R., Heroanto, J. P., et al. (2022). Functional sterol improves breast milk quality by modulating the gut microbiota: a proposed opinion for breastfeeding mothers. Front. Nutr. 9, 1018153. doi: 10.3389/fnut.2022.1018153
Martinez Arbizu, P. (2020). pairwiseAdonis: Pairwise Multilevel Comparison using Adonis. R package version 0.4. Available online at: https://github.com/pmartinezarbizu/pairwiseAdonis
McCue, M. D., Passement, C. A., and Meyerholz, D. K. (2017). Maintenance of distal intestinal structure in the face of prolonged fasting: a comparative examination of species from five vertebrate classes. Anat. Rec. 300, 2208–2219. doi: 10.1002/ar.23691
McMurdie, P. J., and Holmes, S. (2013). phyloseq: an R package for reproducible interactive analysis and graphics of microbiome census data. PLoS ONE 8, e61217. doi: 10.1371/journal.pone.0061217
Mesnage, R., Grundler, F., Schwiertz, A., Le Maho, Y., and Wilhelmi De Toledo, F. (2019). Changes in human gut microbiota composition are linked to the energy metabolic switch during 10 d of Buchinger fasting. J. Nutr. Sci. 8, e36. doi: 10.1017/jns.2019.33
Morelli, G., Patuzzi, I., Losasso, C., Ricci, A., Contiero, B., Andrighetto, I., et al. (2022). Characterization of intestinal microbiota in normal weight and overweight Border Collie and Labrador Retriever dogs. Sci. Rep. 12, 9199. doi: 10.1038/s41598-022-13270-6
Oksanen, J., Simpson, G., Blanchet, F. G., Kindt, R., Legendre, P., Minchin, P., et al. (2022). Vegan: Community Ecology Package. R package version 2.6-4. Available online at: https://github.com/vegandevs/vegan
Ozkul, C., Yalinay, M., and Karakan, T. (2020). Structural changes in gut microbiome after Ramadan fasting: a pilot study. Benef. Microbes 11, 227–233. doi: 10.3920/BM2019.0039
Palakawong Na Ayudthaya, S., Van Der Oost, H., Van Der Oost, J., Van Vliet, D. M., and Plugge, C. M. (2019). Microbial diversity and organic acid production of guinea pig faecal samples. Curr. Microbiol. 76, 425–434. doi: 10.1007/s00284-019-01630-x
Parada Venegas, D., De La Fuente, M. K., Landskron, G., González, M. J., Quera, R., Dijkstra, G., et al. (2019). Short chain fatty acids (SCFAs)-mediated gut epithelial and immune regulation and its relevance for inflammatory bowel diseases. Front. Immunol. 10, 277. doi: 10.3389/fimmu.2019.01486
Parada, A. E., Needham, D. M., and Fuhrman, J. A. (2016). Every base matters: assessing small subunit rRNA primers for marine microbiomes with mock communities, time series and global field samples. Environ. Microbiol. 18, 1403–1414. doi: 10.1111/1462-2920.13023
Phillips Campbell, R. B., Duffourc, M. M., Schoborg, R. V., Xu, Y., Liu, X., Kenknight, B. H., et al. (2016). Aberrant fecal flora observed in guinea pigs with pressure overload is mitigated in animals receiving vagus nerve stimulation therapy. Am. J. Physiol. Gastrointest. Liver Physiol. 311, G754–G762. doi: 10.1152/ajpgi.00218.2016
Quast, C., Pruesse, E., Yilmaz, P., Gerken, J., Schweer, T., Yarza, P., et al. (2013). The SILVA ribosomal RNA gene database project: improved data processing and web-based tools. Nucleic Acids Res. 41, D590–D596. doi: 10.1093/nar/gks1219
R Core Team (2022). R: A Language and Environment for Statistical Computing. Vienna: R Foundation for Statistical Computing.
Reynaga Rojas, M. F., Vergara Rubín, V., Chauca Francia, L., Muscari Greco, J., and Higaonna Oshiro, R. (2020). Sistemas de alimentación mixta e integral en la etapa de crecimiento de cuyes (Cavia porcellus) de las razas Perú, Andina e Inti. Rev. Investig. Vet. Peru 31, e18173. doi: 10.15381/rivep.v31i3.18173
Rowland, I., Gibson, G., Heinken, A., Scott, K., Swann, J., Thiele, I., et al. (2018). Gut microbiota functions: metabolism of nutrients and other food components. Eur. J. Nutr. 57, 1–24. doi: 10.1007/s00394-017-1445-8
Rubio Arias, P. G. (2018). Estimación de parámetros fenotípicos y genéticos para medidas de carcasa en cuyes (Cavia porcellus) del genotipo Cieneguilla [Doctor Degree Thesis Dissertation]. Lima: Universidad Nacional Agraria La Molina.
Shannon, C. E. (1948). A mathematical theory of communication. Bell Syst. Tech. J. 27, 379–423. doi: 10.1002/j.1538-7305.1948.tb01338.x
Shin, S. Y., Hussain, Z., Lee, Y. J., and Park, H. (2021). An altered composition of fecal microbiota, organic acids, and the effect of probiotics in the guinea pig model of postoperative ileus. Neurogastroenterol. Motil. 33, e13966. doi: 10.1111/nmo.13966
Sonoyama, K., Fujiwara, R., Takemura, N., Ogasawara, T., Watanabe, J., Ito, H., et al. (2009). Response of gut microbiota to fasting and hibernation in Syrian hamsters. Appl. Environ. Microbiol. 75, 6451–6456. doi: 10.1128/AEM.00692-09
Spring, S., Sorokin, D. Y., Verbarg, S., Rohde, M., Woyke, T., Kyrpides, N. C., et al. (2019). Sulfate-reducing bacteria that produce exopolymers thrive in the calcifying zone of a hypersaline cyanobacterial mat. Front. Microbiol. 10, 862. doi: 10.3389/fmicb.2019.00862
Stojanov, S., Berlec, A., and Štrukelj, B. (2020). The influence of probiotics on the Firmicutes/Bacteroidetes ratio in the treatment of obesity and inflammatory bowel disease. Microorganisms 8, 1715. doi: 10.3390/microorganisms8111715
Su, J., Li, F., Wang, Y., Su, Y., Verhaar, A., Ma, Z., et al. (2022). Investigating Ramadan like fasting effects on the gut microbiome in BALB/c mice. Front. Nutr. 9, 832757. doi: 10.3389/fnut.2022.832757
Su, J., Wang, Y., Zhang, X., Ma, M., Xie, Z., Pan, Q., et al. (2021). Remodeling of the gut microbiome during Ramadan-associated intermittent fasting. Am. J. Clin. Nutr. 113, 1332–1342. doi: 10.1093/ajcn/nqaa388
Sun, J., Wang, Y., Li, N., Zhong, H., Xu, H., Zhu, Q., et al. (2018). Comparative analysis of the gut microbial composition and meat flavor of two chicken breeds in different Rearing Patterns. Biomed Res. Int. 2018, 4343196. doi: 10.1155/2018/4343196
Tang, C., Ma, J., Kong, F., Li, B., Du, Q., Zhang, Y., et al. (2022). The analysis of transcriptomes and microorganisms reveals differences between the intestinal segments of guinea pigs. Animals 12, 2925. doi: 10.3390/ani12212925
Thomas, F., Hehemann, J.-H., Rebuffet, E., Czjzek, M., and Michel, G. (2011). Environmental and gut bacteroidetes: the food connection. Front. Microbiol. 2, 93. doi: 10.3389/fmicb.2011.00093
Turley, S. D., and West, C. E. (1976). Effect of cholesterol and cholestyramine feeding and of fasting on sterol synthesis in the liver, ileum, and lung of the guinea pig. Lipids 11, 571–577. doi: 10.1007/BF02532904
Wada, Y., Nishiyama, M., Uehara, H., Sato, K., Hamamoto, Y., Ogihara, H., et al. (2022). Microbiome biomarkers associated with the gut contraction response elicited by the Japanese traditional medicine daikenchuto. Gene 826, 146262. doi: 10.1016/j.gene.2022.146262
Xu, S., Zhan, L., Tang, W., Wang, Q., Dai, Z., Zhou, L., et al. (2023). MicrobiotaProcess: a comprehensive R package for deep mining microbiome. Innovation 4, 100388. doi: 10.1016/j.xinn.2023.100388
Yan, C., Xiao, J., Chen, D., Turner, S. P., Li, Z., Liu, H., et al. (2021). Feed restriction induced changes in behavior, corticosterone, and microbial programming in slow- and fast-growing chicken breeds. Animals 11, 141. doi: 10.3390/ani11010141
Yang, J., Li, Y., Wen, Z., Liu, W., Meng, L., Huang, H., et al. (2021). Oscillospira - a candidate for the next-generation probiotics. Gut Microbes 13, 1987783. doi: 10.1080/19490976.2021.1987783
Yeoman, C. J., and White, B. A. (2014). Gastrointestinal tract microbiota and probiotics in production animals. Annu. Rev. Anim. Biosci. 2, 469–486. doi: 10.1146/annurev-animal-022513-114149
Zeileis, A., and Grothendieck, G. (2005). zoo: S3 Infrastructure for regular and irregular time series. J. Stat. Softw. 14, 1–27. doi: 10.18637/jss.v014.i06
Zhang, X., Zou, Q., Zhao, B., Zhang, J., Zhao, W., Li, Y., et al. (2020). Effects of alternate-day fasting, time-restricted fasting and intermittent energy restriction DSS-induced on colitis and behavioral disorders. Redox Biol. 32, 101535. doi: 10.1016/j.redox.2020.101535
Zhang, Y., Choi, S. H., Nogoy, K. M., and Liang, S. (2021). Review: The development of the gastrointestinal tract microbiota and intervention in neonatal ruminants. Animal 15, 100316. doi: 10.1016/j.animal.2021.100316
Keywords: cecum microbiome, Guinea pig, Andina breed, Inti breed, Peru breed, fasting
Citation: Frias H, Murga Valderrama NL, Flores Durand GJ, Cornejo VG, Romani AC, Bardales W, Segura GT, Polveiro RC, Vieira DdS, Ramos Sanchez EM, Lopez Lapa RM and Maicelo Quintana JL (2023) Comparative analysis of fasting effects on the cecum microbiome in three guinea pig breeds: Andina, Inti, and Peru. Front. Microbiol. 14:1283738. doi: 10.3389/fmicb.2023.1283738
Received: 27 August 2023; Accepted: 04 December 2023;
Published: 20 December 2023.
Edited by:
Eugenia Bezirtzoglou, Democritus University of Thrace, GreeceReviewed by:
Hui Jiang, Jiangxi Agricultural University, ChinaChrysa Voidarou, University of Ioannina, Greece
Copyright © 2023 Frias, Murga Valderrama, Flores Durand, Cornejo, Romani, Bardales, Segura, Polveiro, Vieira, Ramos Sanchez, Lopez Lapa and Maicelo Quintana. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Rainer M. Lopez Lapa, cmFpbmVyLmxvcGV6QHVudHJtLmVkdS5wZQ==
†These authors have contributed equally to this work and share first authorship
 Hugo Frias
Hugo Frias Nilton Luis Murga Valderrama1,2†
Nilton Luis Murga Valderrama1,2† Gary J. Flores Durand
Gary J. Flores Durand Victor G. Cornejo
Victor G. Cornejo Ana C. Romani
Ana C. Romani William Bardales
William Bardales Richard C. Polveiro
Richard C. Polveiro Dielson da S. Vieira
Dielson da S. Vieira Eduardo M. Ramos Sanchez
Eduardo M. Ramos Sanchez Rainer M. Lopez Lapa
Rainer M. Lopez Lapa Jorge Luis Maicelo Quintana
Jorge Luis Maicelo Quintana