- 1Department of Animal Science, The University of Tennessee, Knoxville, TN, United States
- 2Department of Genome Science and Technology, The University of Tennessee, Knoxville, TN, United States
Introduction: The rise in extended-spectrum beta-lactamase (ESBL)-producing Enterobacteriaceae in dairy cattle farms poses a risk to human health as they can spread to humans through the food chain, including raw milk. This study was designed to determine the status, antimicrobial resistance, and pathogenic potential of ESBL-producing -E. coli and -Klebsiella spp. isolates from bulk tank milk (BTM).
Methods: Thirty-three BTM samples were collected from 17 dairy farms and screened for ESBL-E. coli and -Klebsiella spp. on CHROMagar ESBL plates. All isolates were confirmed by matrix-assisted laser desorption ionization time-of-flight mass spectrometry (MALDI-TOF MS) and subjected to antimicrobial susceptibility testing and whole genome sequencing (WGS).
Results: Ten presumptive ESBL-producing bacteria, eight E. coli, and two K. pneumoniae were isolated. The prevalence of ESBL-E. coli and -K. pneumoniae in BTM was 21.2% and 6.1%, respectively. ESBL-E. coli were detected in 41.2% of the study farms. Seven of the ESBL-E. coli isolates were multidrug resistant (MDR). The two ESBL-producing K. pneumoniae isolates were resistant to ceftriaxone. Seven ESBL-E. coli strains carry the blaCTX-M gene, and five of them co-harbored blaTEM-1. ESBL-E. coli co-harbored blaCTX-M with other resistance genes, including qnrB19, tet(A), aadA1, aph(3’’)-Ib, aph(6)-Id), floR, sul2, and chromosomal mutations (gyrA, gyrB, parC, parE, and pmrB). Most E. coli resistance genes were associated with mobile genetic elements, mainly plasmids. Six sequence types (STs) of E. coli were detected. All ESBL-E. coli were predicted to be pathogenic to humans. Four STs (three ST10 and ST69) were high-risk clones of E. coli. Up to 40 virulence markers were detected in all E. coli isolates. One of the K. pneumoniae was ST867; the other was novel strain. K. pneumoniae isolates carried three types of beta-lactamase genes (blaCTX-M, blaTEM-1 and blaSHV). The novel K. pneumoniae ST also carried a novel IncFII(K) plasmid ST.
Conclusion: Detection of high-risk clones of MDR ESBL-E. coli and ESBL-K. pneumoniae in BTM indicates that raw milk could be a reservoir of potentially zoonotic ESBL-E. coli and -K. pneumoniae.
1. Introduction
In the United States of America (U.S.), beta-lactam antibiotics, specifically ceftiofur, are frequently used for prophylactic and therapeutic purposes in dairy cattle farms (Food and Drug Administration (FDA), 2016; Redding et al., 2019; Gelalcha et al., 2021; Gelalcha and Kerro Dego, 2022). This heavy use can drive the emergence of extended-spectrum beta-lactamases (ESBL) producing bacteria, which confers resistance to beta-lactam antibiotics, including third-generation cephalosporins (3GC) (Afema et al., 2018). As a result, multi-drug resistant (MDR) ESBL-producing bacteria such as E. coli and Klebsiella spp., the main trafficators of ESBL genes (blaCTX-M, blaTEM, and blaSHV), are becoming increasingly prevalent in the U.S. dairy farms (Afema et al., 2018; Gelalcha et al., 2022; Gelalcha and Kerro Dego, 2022). These ESBL-E. coli and Klebsiella spp. can enter the bulk tank milk from subclinically infected mammary glands, contaminated udders, milking machines, and the dairy farm environment (Straley et al., 2006). The ESBL-E. coli and Klebsiella spp. in dairy farms may spill over to humans indirectly by consuming contaminated, unprocessed dairy products such as raw milk (Christy et al., 2018).
In recent years, there has been a growing trend toward more natural products and the promotion of the “probiotic effects” (i.e., naturally occurring beneficial bacteria or microorganisms in milk that promote or improve health when consumed) of raw milk, leading to an increase in raw milk consumption in the U.S. Some reports estimated that more than 3% of the population in the U.S. consumes raw milk, and at least 30 states in the U.S. legally accepted the sale of unpasteurized milk for human consumption (Whitehead and Lake, 2018). This highlights the increased risk of infection with MDR bacteria that are difficult to treat.
Consuming raw dairy products can increase the risk of ESBL-producing bacteria, such as E. coli and Klebsiella spp., spreading to humans through the food chain, especially milk, and colonizing the gastrointestinal tract (Liu et al., 2020). These bacteria can spread ESBL genes commonly associated with the mobile genetic element (MGEs) to other more harmful human pathogenic bacteria via horizontal gene transfer, creating a reservoir of antibiotic-resistant bacteria that threatens public health (Afema et al., 2018; Kuehn, 2018). In addition to transferring ESBL genes, some E. coli and Klebsiella pneumoniae strains are highly pathogenic and can cause various diseases in humans on their own (Decano et al., 2021).
Some studies reported a possible co-occurrence of ESBL genes, other multiple resistance genes, and virulence in E. coli and K. pneumoniae isolates from dairy cattle and other farm environmental samples (Hennequin and Robin, 2016; Ferreira et al., 2019; Lam et al., 2019; Yang et al., 2019). This further increases public health risks if these bacteria end up in raw milk meant for human consumption.
Although there is limited evidence linking dairy products as a source of ESBL producing-Enterobacteriaceae for humans, the high prevalence of ESBL-Enterobacteriaceae in dairy farms combined with the growing demand for unpasteurized milk suggests that raw milk may serve as a potential source and exit route for ESBL-E. coli and -Klebsiella spp. (Nobrega et al., 2018) from dairy farms to humans. This could be a potential source of the rising incidence of community-associated ESBL-Enterobacteriaceae infections in the U.S. (CDC, 2019).
Previous studies on raw milk in the U.S. were focused on microbiological identification of the commonly reported milk borne pathogens, such as Staphylococcus aureus, Salmonella spp., Listeria monocytogenes, and E. coli O157:H7 (Karns et al., 2007; Langer et al., 2012; Mungai and Gould, 2015; Liu et al., 2020; Gelalcha and Kerro Dego, 2022). Few other studies in Asia (Tark et al., 2017; Liu et al., 2020) and Europe (Filioussis et al., 2020) reported the detection of ESBL-E. coli from mastitic milk samples. A 12-year retrospective study on mastitic milk in Wisconsin also identified a significant number of Klebsiella spp. resistant to ceftiofur, a 3GC whose resistance is mainly mediated by the production of ESBL genes (Fuenzalida et al., 2021). This study was solely based on phenotypic detection of resistance and did not examine the genetic characteristic of the isolates. Another recent research on Klebsiella isolates from mastitic milk reported several resistance genes that mediate resistance to critically important antibiotics (CIAs), including ESBL genes (Zheng et al., 2022). Milk from cows with mastitis is usually discarded and not used for human consumption. Thus, detecting ESBL-producing bacteria in such milk may not entirely indicate public health risks posed by raw milk consumption.
In our recent study, we detected ESBL-E. coli co-carrying resistance genes to multiple classes of antibiotics in bulk tank milk obtained from dairy farms (Gelalcha et al., 2022). In that study, we tested a limited number of farms and detected ESBL-E. coli using PCR. However, PCR is less informative (i.e., it does not offer a comprehensive view of the entire genome) only provides information about the specific gene(s); it cannot identify novel variants of ESBL genes, and it does not provide information about the genetic context or neighboring genes. WGS provides a complete genomic profile allowing the detection of not only ESBL genes and their variants but also other resistance genes, novel or unknown variants of resistance genes, mutations, mobile genetic elements and more complete understanding of genetic diversity. It also helps to identify genetic similarities and differences between bacterial strains and possible sources of milk contamination. Furthermore, WGS provides information about the genomic context of the ESBL genes (e.g., their association with mobile genetic elements), which is crucial in understanding how they are disseminated and how they might be linked to other genetic elements.
To the best of our knowledge, published reports on the detection of ESBL-K. pneumoniae from BTM in the U.S. and elsewhere are unavailable and that of ESBL-E. coli is limited. Thus, further studies using high-resolution molecular tools such as WGS are necessary to understand the status and distribution of ESBL-E. coli and -Klebsiella spp. in dairy farms and the risk for human health. This information can help increase public awareness about the potential health risks of raw milk consumption. Therefore, the objective of this study was to determine the prevalence of ESBL-E. coli and -Klebsiella spp. in bulk tank milk and characterize their antimicrobial resistance phenotypes, genotypes, virulence genes, and potential pathogenicity for humans.
2. Materials and methods
2.1. Isolation and identification of ESBL-Escherichia coli and -Klebsiella pneumoniae from bulk tank milk
Approximately 40 mL of bulk tank milk samples were collected into sterile 50 mL falcon tubes (mostly in duplicates to increase chance of detection of the bacteria) from 17 farms. The samples were transported to the laboratory in a cooler. A 20 mL of milk samples were suspended in 80 mL TSB-PO4 (MG Scientific, Pleasant Prairie, WI, United States). A 100 μL of the suspension was streaked on CHROMagar ESBL plates (a differential and selective media; DRG International, Inc., Springfield, NJ, United States) to isolate ESBL-E. coli and -Klebsiella spp. Plates were aerobically incubated for 24 h at 37°C. From each plate with visible growth, two presumptive ESBL-E. coli (dark pink to reddish) and Klebsiella spp. (metallic blue) colonies were subcultured into new CHROMagar ESBL plates (DRG International Inc.) and incubated for 24 h at 37°C to obtain a pure culture. Up to two well-isolated colonies were selected and inoculated into 5 mL of Luria Bertani broth (LBB; Thermo Fischer Scientific, Waltham, MA, United States) and grown overnight at 37°C while shaking. A 0.5 mL overnight culture was added to a 2 mL serum block, mixed with an equal volume of 80% sterile glycerol, and stored at −80°C for further tests.
Presumptive E. coli and Klebsiella isolates stored at -80oc were thawed and plated on CHROMagar ESBL plates and incubated at 37°C for about 18 h. A single colony of E. coli and Klebsiella spp. was picked and subcultured on blood agar (Thermo Fischer Scientific) at 37°C for about 18 h. Subsequently, E. coli and Klebsiella spp. was identified using matrix-assisted laser desorption ionization time-of-flight mass spectrometry (MALDI-TOF MS) as described by the manufacturer (Bruker Daltonics, Billerica, MA, United States) at the University of Tennessee, College of Veterinary Medicine, Diagnostic Bacteriology and Mycology Lab. The samples were prepared using a formic acid (FA) extraction method (Doern and Butler-Wu, 2016). The MALDI Biotyper library version 11.0 and MALDI Biotyper software version 4.1.100 (Bruker Daltonics) were used for this analysis. For identifying E. coli and Klebsiella spp, the manufacturer-recommended cutoff score values of 2.0 to 3.00, which is considered high-confidence identification, were used.
2.2. Antimicrobial susceptibility testing
Antimicrobial susceptibility testing (AST) was performed on all MALDI-TOF MS-confirmed E. coli and Klebsiella spp. isolates against 14 antimicrobials representing 10 classes of antimicrobials using the broth microdilution method. Commercially available 96-well microtiter plates containing the 14 antimicrobial panels (Sensitire™ CMV4AGNF; Thermo Fisher Scientific) were used. The 14-antimicrobials include β-lactams (ampicillin, ceftriaxone, cefoxitin, amoxicillin-clavulanic acid, meropenem), Aminoglycosides (gentamicin and streptomycin), sulfonamides (sulfisoxazole, trimethoprim/sulfamethoxazole), macrolides (azithromycin), phenicols (chloramphenicol), quinolones (ciprofloxacin and nalidixic acid), and tetracyclines. The minimum inhibitory concentrations (MIC) of the 14 antimicrobials were determined following the manufacturer’s recommended protocol and following CLSI M100 Clinical Laboratory Standards Institute guidelines (CLSI M100: Enterobacteriaceae; Clinical and Laboratory Standards Institute (CLSI), 2022).
2.3. DNA extraction
The MALDI-TOF MS confirmed E. coli and Klebsiella isolates stored at -80°C were thawed and plated on CHROMagar ESBL plates (DRG International Inc.). After overnight incubation, 2–3 colonies were picked with a sterile inoculating wire loop, subcultured into 5 mL LBB (Thermo Fischer Scientific) and incubated at 37°C for 18–24 h. Bacterial DNA was extracted using the Qiagen MagAttract HMW DNA kit (Qiagen, Germantown, MD, United States) following the manufacturer’s procedure with some modifications. Briefly, 2–3 bacterial colonies were taken from a pure overnight culture using a sterile wire loop and dissolved into 5 mL LBB (Thermo Fisher Scientific). The broth was incubated overnight at 37°C, and 2 mL was transferred into the 2 mL microcentrifuge tube. The tube was placed in a centrifuge (Thermo Fisher Scientific) and centrifuged at 5,000 × g for 10 min. Then, the supernatant was discarded, and the pellet was suspended in a lysis solution, placed in shaker-incubator series (VWR International, Philadelphia, PA, United States), and shaken at 500 rpm for 1.25 h at 57°C. After adding 4 μL RNase and incubating for 2 min at room temperature, 15 μL magnetic beads and 280 μL binding buffer (Buffer MB) were added to the lysate solution and pulse vortexed and placed in a MagAttract magnetic rack (Qiagen) and shaken in a shaker-incubator serious (VWR International) at 500 rpm for 15 min. The Magnetic Rack was placed on the magnetic base so that the magnetic beads, along with the DNA, would get separated by sticking to the side of the tube toward the magnetic base, and the supernatant was removed. The magnetic beads (together with the DNA samples attached) were washed twice each by Buffer MW1 and Buffer PE (Qiagen). The magnetic beads were rinsed twice with distilled water while the tube holder was on the magnetic base, and the beads were fixed to the walls of the microcentrifuge tube. Then, the tube holder of the Magnetic Rack was removed from its magnetic base, and 150 μL Buffer AE (elution buffer; Qiagen) was added. The tube holder of the Magnetic Rack was placed in incubator-shaker series (VWR International) and incubated at room temperature for 15 min at 500 rpm. The tube holder of the Magnetic Rack was placed on its magnetic base to separate the beads from the supernatant. Finally, the supernatant (elute) that was expected to contain high-molecular-weight DNA was transferred into a new sample tube. DNA concentration and purity were measured by a Nanodrop 2000 spectrophotometer (Thermo Fisher Scientific).
2.4. Library preparation and whole-genome sequencing of ESBL-Escherichia coli and -Klebsiella pneumoniae
Genomic libraries were prepared using the Nextera XT DNA library prep kit (Illumina Corporation, San Diego, CA, United States) following the manufacturer’s instructions. Final concentrations were confirmed on a Qubit fluorometer (Thermo Fischer Scientific). Some libraries checked for size and concentration on a high-sensitivity Bioanalyzer (Agilent Technologies, Santa Clara, CA, United States). Samples were normalized and pooled, and a final quality confirmation analysis was performed on a Bioanalyzer (Agilent Technologies). The pooled samples were diluted to a final loading concentration of 0.5 nM and combined with 5% PhiX control (Illumina Corporation). The sample pool and PhiX were then loaded on one lane of an SP flow cell reading 250 nucleotides paired-end (2× 250) on the Illumina NovaSeq (Illumina Corporation). The DNA library was sequenced at the University of Tennessee Genomics Core (Knoxville, TN, United States).
2.5. Quality control, assembly, and analyses of whole genome sequence data
Samples were demultiplexed, and quality assessment was done via FastQC v0.11.9 (FastQC, 2010) and multiQC v1.14 (Ewels et al., 2016). Adapter sequence removal, filtering, and trimming of low-quality raw reads were done using Trimmomatic v0.39 (Bolger et al., 2014; LEADING:5 TRAILING:5 SLIDINGWINDOW:5:20 MINLEN:30). Preprocessed reads of each isolate were de novo assembled using SPAdes genome assembler v3.15.5 (Bankevich et al., 2012) with default parameters. PlasmidSPAdes modes were used for the de novo assembly of the “plasmids only” for downstream analysis.
To further confirm the species of bacteria identified by MALDI-TOF MS, KmerFinder 3.21 was used to predict bacterial species using a fast K-mer algorithm from the assembled genome (Hasman et al., 2014; Larsen et al., 2014; Clausen et al., 2018). The WGS data were used to identify variants of ESBL genes and detect virtually all known antibiotic resistance genes (ARGs) or mutations co-harbored with ESBL genes. To determine the presence of acquired antibiotic resistance genes and chromosomal point mutations in assembled genomes of the isolates, ResFinder v4, online bioinformatics tools at the Center for Genomic Epidemiology2 (Zankari et al., 2017; Bortolaia et al., 2020) and Comprehensive Antimicrobial Resistance Database (CARD3; Alcock et al., 2020) were employed using E. coli and Klebsiella spp. specific panels. ESBL-encoding genes and other resistance determinants were considered present when at least 60% of the length of the best-matching gene in the ResFinder database was covered with sequence identity greater than or equal to 95% (Feldgarden et al., 2019).
To identify plasmids and other mobile genetic elements (MGE) associated with ARGs, draft assemblies of ESBL-E. coli and Klebsiella spp. genome was analyzed using PlasmidFinder 2.14 and MGE Finder v1.0.3.5 The PlasmidFinder detects plasmids and assigns them to incompatibility groups (Camacho et al., 2009; Carattoli et al., 2014). At the same time, MGE Finder was used to identify plasmids and other MGEs, such as transposon (Tn) and Insertion Sequences (IS) associated with ARGs and virulence genes (Camacho et al., 2009; Zankari et al., 2012; Joensen et al., 2014). The MGEs were identified with a minimum 98% sequence identity threshold with the closest-matching hits and minimum 90% coverage (Carattoli et al., 2014). The virulence gene associated with MGE in each genome was identified with 98% sequence identity and a minimum of 95% coverage using virulenceFinder. The virulence markers in the isolates were identified using virulenceFinder6 as described before (Joensen et al., 2014; Malberg Tetzschner et al., 2020).
To investigate genetic relatedness, the WGS of isolates were analyzed using in silico sequence serotyping (E. coli only), multilocus sequence types (MLST), and Core genome Multi Locus Sequence Type (cgMLST) for both E. coli and Klebsiella pneumoniae. Multilocus sequence types (MLST) of E. coli were identified according to the Achtman seven-locus MLST approach, which is based on sequences of seven housekeeping genes (adk, fumC, gyrB, icd, mdh, purA, and recA) which are known to present in all strains of E. coli and are relatively stable in terms of their sequence (Wirth et al., 2006; Larsen et al., 2012; Clermont et al., 2015). For Klebsiella spp., MLST was identified based on seven housekeeping genes (gapA, InfB, mdh, pgi, phoE, rpoB, and tonB) described in other studies (Diancourt et al., 2005; Larsen et al., 2012). The serotype of E. coli isolates was predicted from the draft assembly using SerotypeFinder 2.0.17 with 95% threshold identity and 80% minimum length (Camacho et al., 2009; Clausen et al., 2018). The MLST of both species of bacteria was predicted from the assembled draft genome using MLST 2.0 2.08 as described in Bartual et al. (2005), Lemee and Pons (2010), and Larsen et al. (2012). The assignment of cgMLST of E. coli was done using cgMLSTFinder 1.29 as described in previous studies (Clausen et al., 2018; Zhou et al., 2020). The pathogenicity of both species of bacteria was predicted from their genome based on gene families that correlate with pathogenicity using PathogenFinder 1.110 as described in Cosentino et al. (2013). The pathogen finder tool evaluates the disease-causing potential of a bacterial strain. We begin by uploading the draft sequence of the E. coli genomes onto the web-based platform. The tool then compares the proteins encoded by the E. coli strains to a database containing groups of proteins or protein families that were associated with pathogenic or non-pathogenic organisms. Initially, the tool clusters these proteins, identifying significant clusters where most proteins originate from either pathogens or non-pathogens. Subsequently, protein families are labeled as pathogenic or non-pathogenic, and a weight (Z-score) is calculated for each based on matched protein families. Finally, the tool predicts whether the input organism is human pathogenic or not based on the associated probability value from the prediction (Cosentino et al., 2013). Plasmid Multilocus Sequence Typing (pMLST) was carried out on draft assembled plasmids on incompatibility groups for which a pMLST scheme is available using11 as described in Carattoli et al. (2014).
3. Results
3.1. Prevalence and antimicrobial resistance profiles of ESBL-Escherichia coli and -Klebsiella pneumoniae in bulk tank milk
A total of eight ESBL-E. coli and two ESBL-K. pneumoniae strains were isolated from 33 milk samples collected from 17 farms. The isolates were detected from nine farms; one E. coli and K. pneumoniae were isolated from the same farm, whereas the others were detected from 8 different farms. From the eight E. coli isolates identified with ESBL phenotype, one isolate, CD-M, was later confirmed to be susceptible to all tested antibiotics. Thus, the sample level prevalence of ESBL-E. coli was 21.2% (7/33), whereas that of Klebsiella pneumoniae was 6.1% (2/33). At the farm level, ESBL-E. coli and -K. pneumoniae prevalence was 41.2% (7/17) and 11.8% (2/17), respectively.
All eight E. coli isolates identified in this study were tested for antimicrobial susceptibility against 14 antimicrobial agents. All E. coli isolates with ESBL phenotype (n = 7) were co-resistant to ampicillin, ceftriaxone, and tetracycline, and all of them were multidrug resistant (resistant to three or more antimicrobial classes). More than half of the ESBL-E. coli isolates (n = 4) were phenotypically resistant to five or more antimicrobials, including those considered the highest priority critically important classes (HPCI) such as third generation cephalosporins and fluoroquinolones. The ESBL phenotype co-existed with other resistant phenotypes to CIAs, such as ceftriaxone, streptomycin, and fluoroquinolones (nalidixic acid or ciprofloxacin) in four E. coli strains. One E. coli isolates (AG-M) was resistant to nine antimicrobial agents belonging to 7 classes. All E. coli isolates were susceptible to meropenem, cefoxitin, and amoxicillin-clavulanic acid (Table 1).
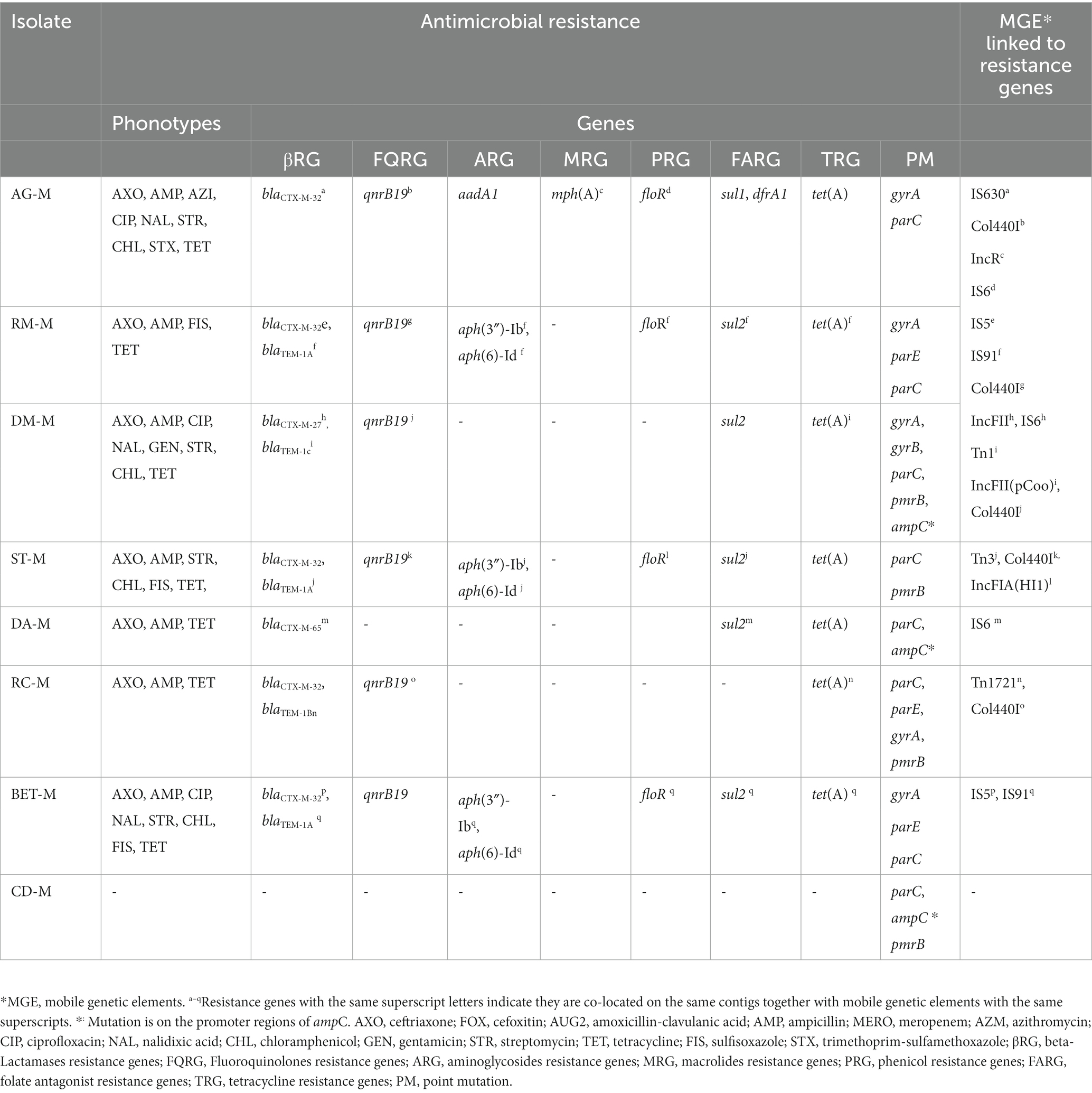
Table 1. Antimicrobial resistance phenotypes and mobile genetic elements associated with resistance genes in ESBL-Escherichia coli.
3.2. Resistance genes and their associations with mobile genetic elements in Escherichia coli
The eight bacterial strains initially identified as E. coli by MALDI-TOF MS were indeed confirmed to be E. coli through WGS. One of these is ESBL-E. coli isolates (CD-M) were found to have no resistance genes to beta-lactam antibiotics or many other classes of antibiotics. Genome sequence analysis did not find the presence of ESBL or other ARGs in E. coli isolates that were phenotypically susceptible to all tested antibiotics.
All seven ESBL-E. coli strains carry the blaCTX-M variant and five isolates co-carry blaTEM-1. Among ESBL genes, blaCTX-M-32 was a common variant and was detected in five of the isolates. All ESBL-E. coli strains co-harbored blaCTX-M with plasmid-mediated quinolone resistance gene (qnrB19), and a tetracycline resistance gene, tet(A).
Resistance genes to aminoglycosides [aadA1 or aph(3″)-Ib, aph (6)-Id], quinolones (plasmid-mediated and point mutations), phenicol (floR), and sulphonamides (sul2) were concurrently detected in four ESBL-E. coli strains that harbored the blaCTX-M-32 variant. The macrolide phosphotransferases [mph(A)], aminoglycoside nucleotidyltransferase (aadA1), dihydrofolate reductase (dfrA), and dihydropteroate synthase (sul1) resistance genes were detected in a single isolate, AG-M. As expected, blaAmpCs and carbapenemase-associated genes were not identified.
Mutation in the ampC gene promoter region was detected in three isolates (CD-M, DA-M, and DC-M) but was not expressed. Double or triple mutations in Quinolone-Resistance Determining Regions (QRDRs) of gyrA, gyrB, parC, and parE were detected in six isolates. These multiple chromosomal mutations were (gyrA-S83L, D87Y, D741E, D87G, D87N, D678E, Q454H, and D741E; parC-E62K or S80I; parE-L416F and gyrB-Y184D). A point mutation that potentially confers resistance to colistin, pmrB, was detected in four of the isolates. For one isolate (DM-M), the genotypes that mediate the phenotypic resistance, such as aminoglycosides and chloramphenicol, were not detected in the Center of Genomic Epidemiology database (see text footnote 2). But we identified marA and evgA, genes that encode efflux pumps known to mediate resistance to several classes of antibiotics, including aminoglycosides and chloramphenicol in comprehensive Antibiotic Resistance Database (CARD) (see text footnote 3).
A quinolone resistance gene, qnrB19, was consistently associated with Col440I plasmid replicon types in six ESBL-E. coli isolates. All except one blaCTX-M gene were associated with MGEs alone or with other resistance genes. In some of the ESBL-E. coli isolates, multiple resistance genes are co-located on the same contigs associated with the same or different MGEs (Table 1). In two of the ESBL-E. coli strains, BET-M, and RM-M, six other resistance genes (aph(3″)-Ib, aph(6)-Id, tet(A), floR, sul2, blaTEM-1A), mediating resistance to five different classes of antibiotics were all found on the same contigs associated with IS91. All except one blaCTX-M and almost all other co-harbored acquired ARGs were associated with MGEs such as plasmids, Insertion sequences (IS), or transposons (Tn) on the same genetic fragment.
3.3. Genetic diversity, plasmid types, and pathogenomics of ESBL-Escherichia coli isolates
The E. coli isolates were assigned to seven distinct serotypes, six multilocus sequence types (STs), and eight-core genome sequence types (cgSTs) based on in silico sequence typing of the WGS data. Two isolates from different farms, RM-M and BET-M, had the same serotype (O101:H9) and sequence type (ST10). All eight strains were distinct according to their core genome sequences, based on comparing nearly all the genes shared by E. coli. The ST10 type was relatively common and found in three farms. Three isolates, AG-M, ST-M, and DA-M had identical genotypes (ST and resistance gene pattern) as E. coli isolates from fecal samples collected from dairy cattle in the same farms where BTM was also obtained (unpublished data).
PlasmidFinder identified 13 different plasmid replicon types from the WGS data of the seven ESBL-E. coli isolates. Each ESBL-E. coli isolate had at least two plasmid replicon types. The IncF family plasmids (FII, FIA, and FIB replicon types) were common and found in four ESBL-E. coli isolates. Colcinogenic plasmids (Col440I) were the most frequent and detected in all seven ESBL-E. coli isolates. The ST10 type carried the largest number (6 to 8) and types of plasmids. Two ST10 and ST69 had epidemic resistance plasmids of the IncF family. No plasmid was detected in the WGS of one E. coli isolate, CD-M, which was susceptible to all tested antibiotics.
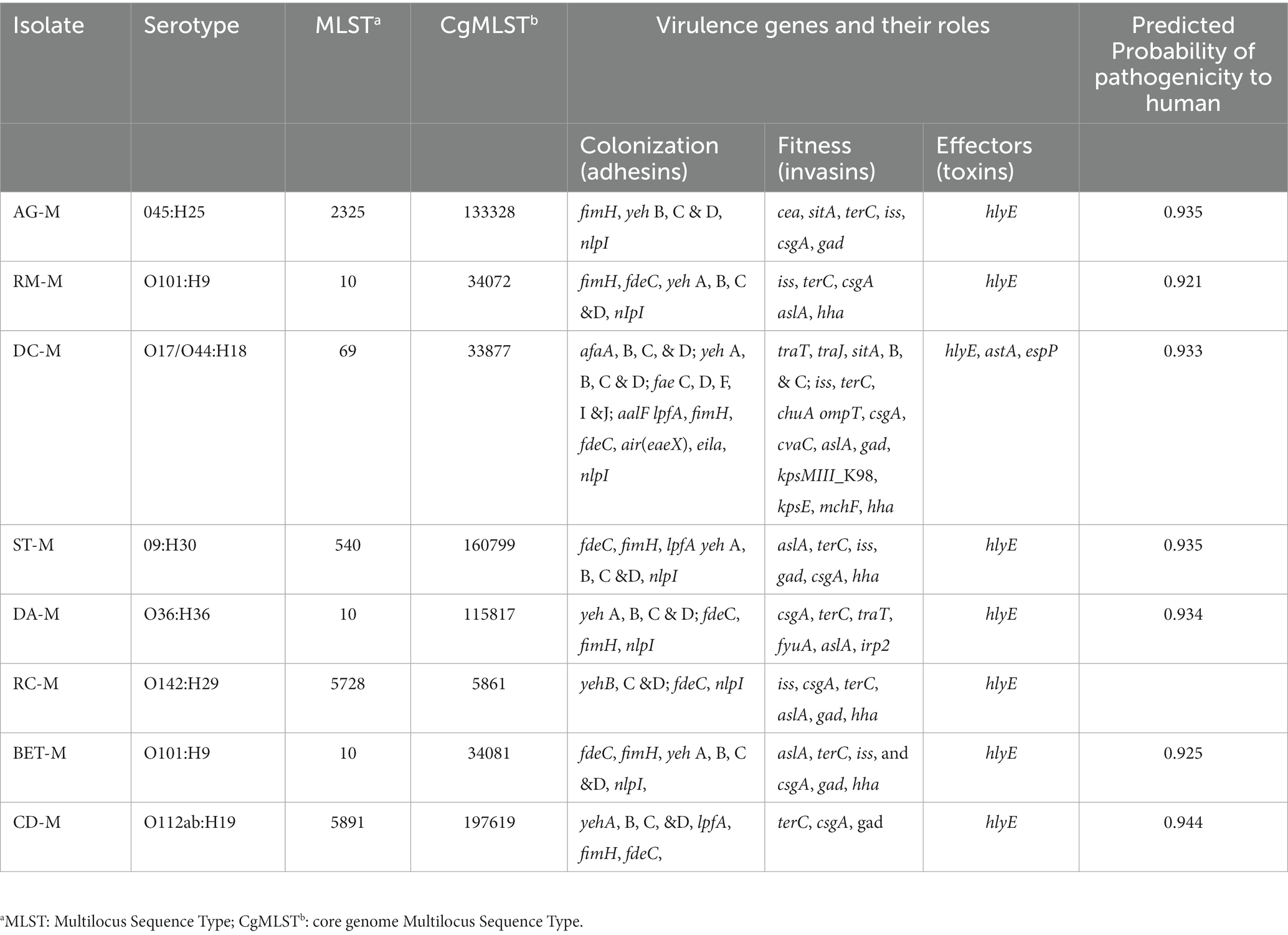
Several virulence genes were detected in all eight E. coli isolates. Each isolate had multiple virulence genes involved in adhesion (adhesin), invasion (invasins for bacterial fitness), and effectors or toxins (Table 2). The highest number of virulence genes (Zankari et al., 2017) were detected in the ST69 isolate, and the lowest number (Christy et al., 2018) in the isolate ST5891 susceptible to all tested antibiotics. Widespread virulence genes, such as aslA (arylsulfatase-like gene), fimH (type1 fimbriae), yeh, treC (tellurium ion resistance protein encoding gene), gad (glutamate decarboxylase encoding gene), csgA (curli fimbriae gene), terM, hylF (hemolysin), iss (increased serum survival protein encoding gene), and traT (outer membrane protein responsible for complement resistance), were found in the E. coli genomes. No hits for the Shiga toxin-producing gene were detected in the WGS of the isolates. The draft sequences of all E. coli isolates were submitted to PathogenFinder, (see text footnote 10) and all were predicted as human pathogens with a probability greater than 0.92.
3.4. Co-localization of virulence genes and mobile genetic elements
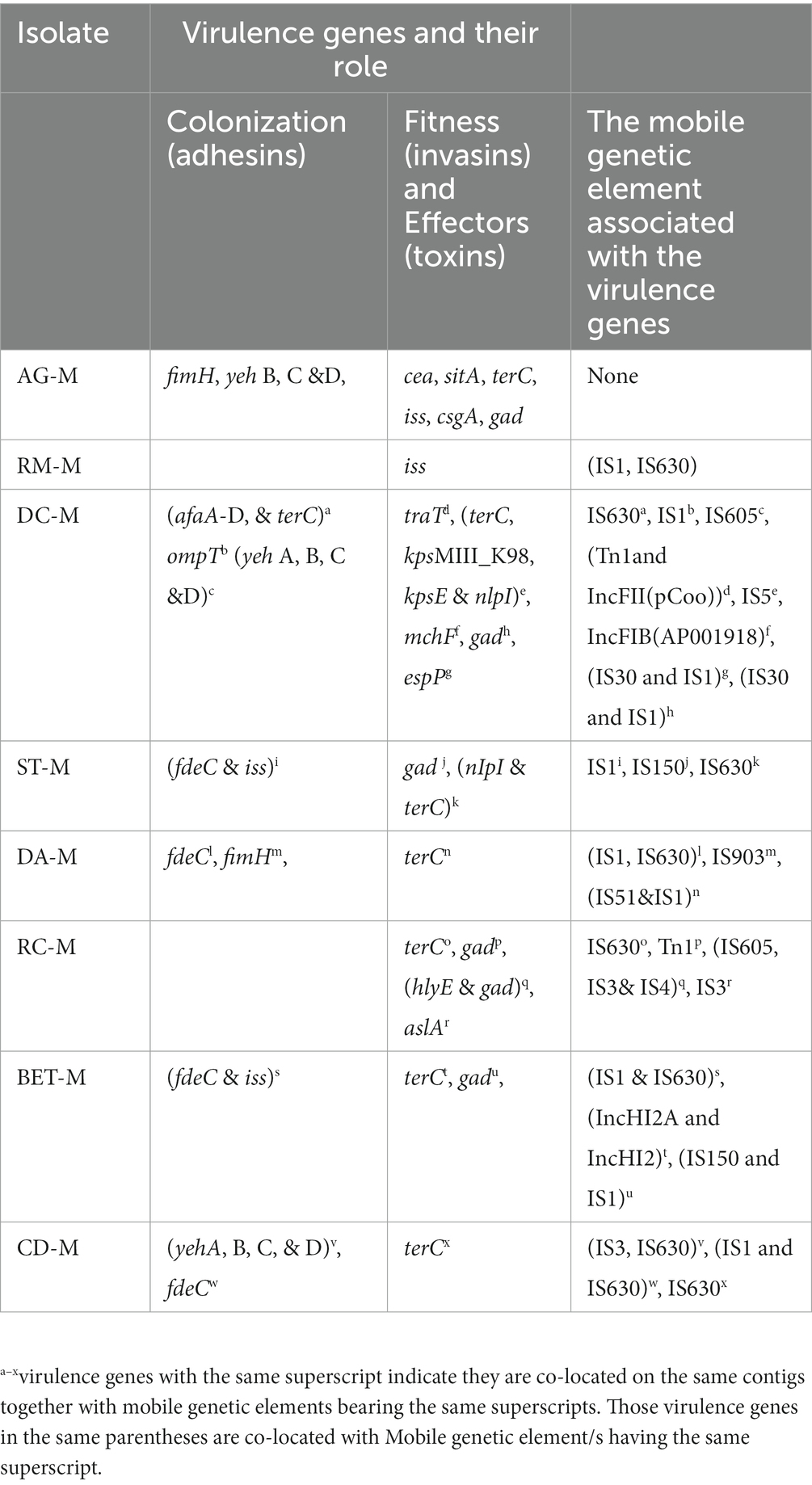
Several virulence genes were detected on ESBL-E. coli were co-located on the same contigs with the MGEs (plasmids, insertion sequences, and transposons). In addition, multiple virulence genes were often co-located with or on the same MGEs. In some instances, copies of the same virulence genes were associated with different MGEs (e.g., plasmids and transposon). Most virulence genes related to MGE are those involved in the invasion and adhesion of mammalian cells. Only two isolates have effector (protease) encoding genes associated with MGEs. Table 3 showed several virulence genes associated with one or more MGEs.
3.5. Resistance phenotypes and genotypes in ESBL-Klebsiella pneumoniae
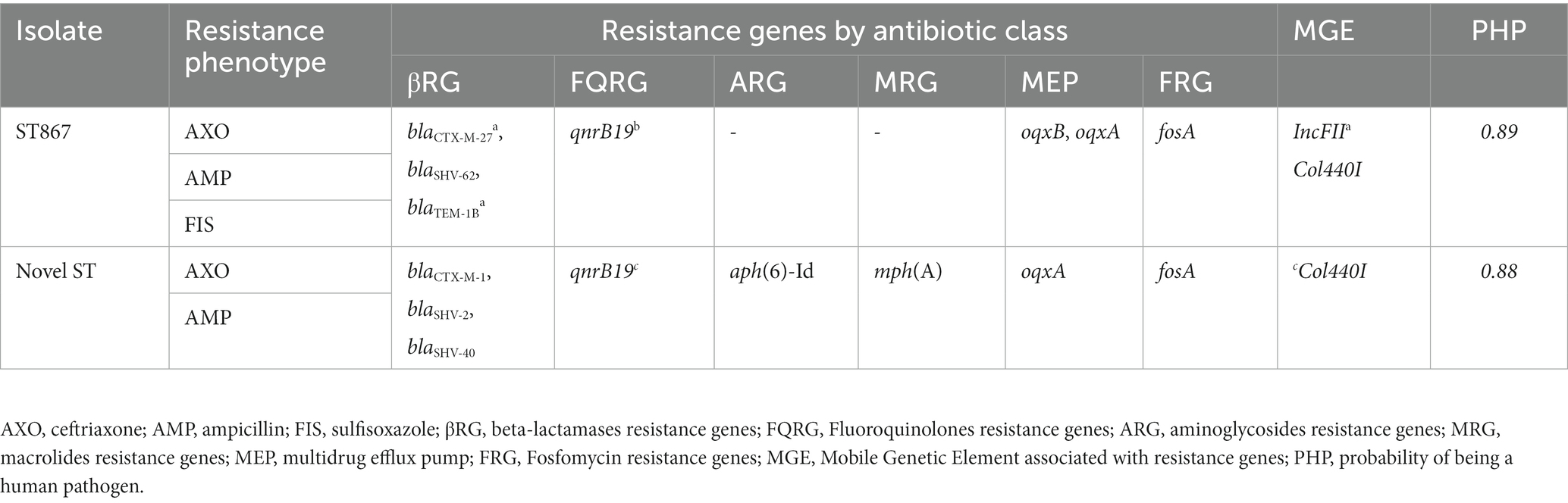
The two K. pneumoniae isolates were resistant to ceftriaxone and ampicillin. In addition to the two antibiotics, one of the isolates, DM-MK, was resistant to sufisoxazole. Each K. pneumoniae isolate carried two distinct types and variants of ESBL-genes, the blaCTX-M, and blaSHV.
The K. pneumoniae isolates contain a multidrug efflux pump encoding gene, oqxAB. Both oqxB and oqxA genes were found in one of the isolates, and this isolate was resistant to sufisoxazole. The other isolate only had the oqxA gene and was susceptible to the same antibiotic. Both isolates are considered genotypically multidrug resistant, carrying at least eight resistant determinants against multiple classes of antibiotics. However, the observed phenotypic resistance was low (Table 4).
3.6. Genetic diversity, plasmid types, and pathogenomics of Klebsiella pneumoniae
The WGS data were used for in silico sequence typing on the two K. pneumoniae isolates. One of the isolates (BM-MK) was assigned as ST867, while the remaining one was novel and could not be assigned. The ST867 is identical to three other STs identified from fecal samples from the same farm where BTM was obtained. Both K. pneumoniae obtained from BTM were identified as new serotypes of K. pneumoniae. Both strains contain IncF family plasmids with replicon types FII. The ST867 strain only had the FII replicon. The novel strain had seven plasmids, including two Col plasmids (Col440I and II), four epidemic resistance plasmids with IncF plasmid families [FII, FII(K), FIA, and FIB replicons], and one IncN plasmid, which is a broad host range plasmid. This study identified a new allele type for IncFIA (HI1) plasmid using MLST. The WGS data also revealed the presence of multiple virulence genes such as fimH (type 1fimbriae), fyuA (siderophore receptor), ituA (ferric aerobactin receptor), traT (outer membrane protein complement resistance), and mrkA (type 3 Fimbrial Shaft) in ST867. More number of virulence genes were detected in the novel K. pneumoniae ST. The virulence genes detected in this ST include fimH, traT, fyuA, iutA, mrkA, Irp2 (non-ribosomal peptide synthetase), ccI (Cloacin), clpK1(thermal stress survival ATPase). Both K. pneumoniae were predicted to be human pathogens with a probability greater than 0.82.
4. Discussion
This study found a high prevalence of ESBL-producing E. coli in bulk tank milk (BTM) from dairy farms showing the significant public health associated with raw milk consumption. ESBL-E. coli was found in the BTM of 41% of the farms, and 21.2% of the BTM samples tested positive for these bacteria. Additionally, ESBL-producing K. pneumoniae was detected in two BTM samples from two farms. Since we did not come across similar studies conducted in the U.S. or elsewhere, a comparison of prevalence was not possible in the current study. However, in our earlier study that used different screening techniques, ESBL-E. coli was detected in at least one of the BTM samples collected from four farms (Gelalcha et al., 2022). Similarly, a study conducted in Turkey on bulk tank milk, which involved an initial enrichment step, reported a comparable ESBL-E. coli prevalence of 22.6% (Kürekci et al., 2019).
Similarly, studies conducted on milk from cows with mastitis in New York (Yang et al., 2019), South Korea (Tark et al., 2017), Greece (Filioussis et al., 2020), and China (Liu et al., 2020) detected ESBL-E. coli. A comparison of the prevalence of ESBL-E. coli between the current study and these studies are not plausible as the sample source, and the initial screening methods used were different. This suggests the prevalence of ESBL-E. coli in BTM may be higher than previously thought, highlighting the need for continuous monitoring and evaluation of associated risk and developing innovative control tools at farm levels.
The study also indicated that all ESBL-E. coli were MDR, with most of them displaying concurrent resistance to five or more antibiotic classes. These findings are consistent with previous studies in animals and humans that showed ESBL-Enterobacteriaceae frequently showed co-resistance to multiple classes of antibiotics (Masse et al., 2021; Badr et al., 2022; Gelalcha et al., 2022; Gelalcha and Kerro Dego, 2022). Four ESBL-E. coli also showed concurrent resistance to CIAs, such as ceftriaxone, fluoroquinolone (ciprofloxacin and/or nalidixic acid), and aminoglycosides (streptomycin and/or gentamycin). This result also agrees with our recent study and several other studies that reported ESBL-producers frequently exhibit co-resistance to fluoroquinolones, aminoglycosides, chloramphenicol, and tetracycline (Bradford, 2001; Canton and Ruiz-Garbajosa, 2011; Timofte et al., 2014; Afema et al., 2018; Agga et al., 2022; Gelalcha et al., 2022; Gelalcha and Kerro Dego, 2022). However, the study farms did not report aminoglycosides, tetracyclines, and fluoroquinolones as frequently used antibiotics. In addition, fluoroquinolones and macrolide are not allowed to use in the U.S. dairy cattle of more than 20 months of age (FDA, 2023). Thus, the detection of fluoroquinolone resistance in half of the isolates and azithromycin resistance in one of the isolates from BTM pooled from adult cows may not be related to antibiotic use. Further investigation is needed to identify if the use of these antibiotics at a young age on calves has an impact during adult age or if ceftiofur uses in dairy cattle co-select for resistance determinants against these two CIAs, as previously hypothesized (Taylor et al., 2020). In addition, it is also vital to examine if resistance genes against these two classes of antibiotics are maintained through bacterial generation via MGEs that confer a fitness advantage to the host.
In line with this thought, WGS analysis of the isolates identified that qnrB19 (a gene mediating resistance to ciprofloxacin) was consistently associated with Col440I plasmids. A previous study showed that ColE1-derived plasmids, including Col440I, contain genes that encode for bacteriocins, proteins that can kill other bacteria and provide a competitive advantage to the host bacterium (Riley and Gordon, 1992). This suggests that the association of qnrB19 with Col440I might be responsible for its maintenance in the absence of selection pressure. A recent study in Germany also reported frequent association of qnrB19 with Col440I plasmids in ESBL-producing E. coli from livestock (Juraschek et al., 2022).
All isolates were susceptible to meropenem, one of the last resort antibacterial agents whose use is typically reserved for severe infections caused by pathogenic members of ESBL-Enterobacteriaceae (Karaiskos and Giamarellou, 2020). This finding is not unexpected, as carbapenems are not approved for animal use in the U.S. due to the risk of promoting the emergence of carbapenemase-producing Enterobacteriaceae. Detection of ESBL-E. coli with phenotypic resistance to multiple classes of antibiotics, particularly to those CIAs, makes consuming raw milk a dangerous practice for public health as only limited therapeutic options are available to cure the infection caused by the bacteria with these phenotypes (WHO, 2019).
The WGS analysis showed that the observed phenotypic resistance patterns are generally congruent with genotypic results, as reported in a previous study suggesting that the genotypes of the bacteria are a reliable indicator of their antibiotic resistance (Tyson et al., 2015). The blaCTX-M was the only ESBL-variants observed from all ESBL-E. coli isolates phenotypically resistant to ceftriaxone, a 3GC. This finding concurs with our previous study, where we reported blaCTX-M as the only ESBL variant from E. coli isolates retrieved from a dairy farm environment with phenotypic resistance to cefotaxime, another 3GC (Gelalcha et al., 2022). Similarly, other studies in the U.S. and elsewhere also showed the predominance of the blaCTX-M variant of ESBL over the others among the Enterobacteriaceae in humans and food animals, including dairy cattle (Canton et al., 2012; Peirano et al., 2012; Weissman et al., 2013; Davis et al., 2015; Bevan et al., 2017; Afema et al., 2018; Collis et al., 2019; Gelalcha and Kerro Dego, 2022). It is hypothesized that the frequent occurrence and the dominance of the blaCTX-M variant over the others in ESBL-E. coli may be related to its low fitness cost to the host, successful dissemination through MGEs, and rapid response to beta-lactam antibiotics selection pressure (influence or changes in bacteria exerted by the use of beta-lactam antibiotics that enable bacteria to became resistant; D’Andrea and Pallecchi, 2013). As we already discussed in our published review (Gelalcha and Kerro Dego, 2022), further controlled study is needed to identify the cause of the spread and dominance of this variant of ESBL genes.
The blaCTX-M-32 was detected in five farms, whereas the remaining two, blaCTX-M-27 and blaCTX-M − 65, were obtained from ESBL-E. coli retrieved from two separate farms. These blaCTX-M − 32 variants were previously reported from ESBL-E. coli isolated from fecal samples in the U.S. dairy farms (Afema et al., 2018; Taylor et al., 2020; Carey et al., 2022). The frequent presence of the non-ESBL beta-lactamase gene (blaTEM-1) along with the ESBL gene in E. coli may aid in maintaining and transmitting one or both genes to future generations clonally and to other bacteria horizontally via MGEs.
The fact that all ESBL-E. coli strains co-harbored blaCTX-M with qnrB19 (plasmid-mediated quinolone resistance gene), and tet(A) concurs with the phenotypic observation. But despite the presence of qnrB19, no corresponding resistance to ciprofloxacin was observed in some isolates. This discrepancy may be due to a low or lack of gene expression in that isolate due to various factors, including lack of activation of regulatory genes as we did not find mutation in the genes. Four ESBL-E. coli strains, co-harbored blaCTX-M-32 with resistance genes to aminoglycosides (aadA1 or aph(3″)-Ib, aph(6)-Id), quinolones (qnrB19), and mutation in one or more of fluoroquinolone resistance determining regions (QRDRs), phenicol (floR), and sulfonamides (sul2). This observation is also consistent with the phenotypic result and previous molecular studies that detect frequent co-occurrence of ESBL genes with multiple other ARGs (Afema et al., 2018; Peirano and Pitout, 2019; Gelalcha et al., 2022).
It is noteworthy that strains of ESBL-E. coli from two farms have the same six antibiotic resistance genes co-located on the same contigs with IS91, a mobile genetic element. Multilocus Sequence Typing (MLST) results showed that both strains belong to the same Sequence Type (ST10). However, the cgMLST typing identified both isolates as distinct strains. The two strains carry the same mutation in QRDRs (gyrA, parC, and parE) but differ in the pmrB gene presence and their plasmid replicon types. The detection of the same ARG with a similar genetic arrangement on the identical STs of E. coli needs further investigation. Still, a possible explanation could be that the strains may have a common ancestry and may have spread from one farm to another, parallel evolution between the two strains with the ARGs they carry, or contamination in the laboratory. However, contamination is less likely as the samples were collected and processed at different times.
All but one of the blaCTX-M genes and most other ARGs co-harbored by the ESBL-E. coli were associated with mobile genetic elements (MGEs), such as plasmids, insertion sequences (IS), or transposons (Tn). The presence of ESBL and non-ESBL beta-lactamase genes, along with multiple ARGs on MGEs, is consistent with findings from previous studies on ESBL-E. coli isolates from dairy cattle and humans (Poirel et al., 2005; Afema et al., 2018; Filioussis et al., 2020; Liu et al., 2020; Toth et al., 2020; Gelalcha and Kerro Dego, 2022). This suggests that MGEs may facilitate the spread of these ARGs. The presence of multiple ARGs located on the same MGEs could make the transfer of these genes to other bacteria of the same or different species easier. This could lead to the development of extensively drug-resistant bacteria, which may risk human health through indirect transmission (Long et al., 2019; Toth et al., 2020).
Some of the ESBL-E. coli strains, ST10 and ST69, reported in this study had the same STs as previously reported as “high-risk” clones. These two sequence types were identified globally as the most prevalent extra-intestinal pathogenic E. coli (ExPEc) lineages after ST131 (Manges et al., 2019). A study on clinical E. coli isolates from 1999 to 2017 in California also identified these two STs as the most common causes of urinary tract infection (Yamaji et al., 2018). The ST10 and ST69 carry IncFII and/or IncI1, plasmid families. This finding is consistent with previous studies that reported high-risk clones of E. coli are frequently harbored by easily transmissible narrow host range or epidemic plasmids such as IncF and/or IncI1 (Johnson et al., 2007; Carattoli, 2009; Afema et al., 2018).
The detection of triple mutations in the QRDR region in two of the STs, ST10, and ST69, along with other ARGs (qnrB19, floR, tet(A), and sul2), indicated a public health risk associated with these bacterial clones. The public health risk or hazard is the potential that multidrug-resistant pathogenic ESBL-E. coli and Klebsiella species may reach the public indirectly through food chain (consumption of raw milk) and cause life threatening infections. These STs have been previously reported in fecal samples from dairy cattle and other food-producing animals in the U.S. (Afema et al., 2018; Hayer et al., 2020). The ST69 carried blaCTX-M-27 and blaTEM-1c on IncFII (F2:A-:B-), an epidemic resistance plasmids known by its ability to acquire resistance genes and easily and rapidly spreading within the same or related species of bacteria (Carattoli, 2009). This ability to acquire and spread resistance genes quickly increases the prevalence of ESBL-producing bacteria and the difficulty of treating infections and is a significant public health concern.
In this study, the pmrB gene, which is commonly associated with colistin resistance (Delannoy et al., 2017), was detected in four E. coli isolates, including in two pandemic clones. Detecting a gene associated with colistin resistance is a public health concern as colistin is considered a last-resort antibiotic for treating infections caused by ESBL- and carbapenemase-producing Enterobacteriaceae (Falagas et al., 2011; Bialvaei and Samadi, 2015). The current study did not test for phenotypic colistin resistance because the panel of antibiotics used did not include colistin. Thus, the colistin gene’s impact on colistin resistance needs further study, but its presence still raises significant health risks. Furthermore, detecting colistin resistance genes in ST10 and ST69, epidemic strains, highlights the high risk associated with consuming raw milk. The plasmid-mediated colistin resistance gene, mcr-1, was previously detected in E. coli at a very low level (0.1%–0.02%) in the U.S. (Meinersmann et al., 2017; Wang et al., 2020) but no previous report on Enterobacteriaceae with any colistin resistance gene from BTM. The MDR ESBL-producing strains are already challenging to treat, and the additional presence of colistin-resistance genes increases the risk of treatment failure (Binsker et al., 2022). Detection of colistin resistance determinant in BTM was unexpected. This is because colistin has never been marketed for use in the U.S. dairy farms due to concerns about potential antibiotic resistance and to preserve it for human medicine as it is considered a last-resort antibiotic.15
All ESBL-E. coli isolates in this study carried several virulence genes exhibiting pathogenic properties (adhesion, invasion, and toxin production) in their mammalian host. The most frequently detected virulence genes in ESBL-E. coli were fimH, csgA, yehA-D, and fdeC for adhesion; iss, aslA, gad, and traT and sitA for bacterial replication and invasion; and hylF for toxin secretion (Kaper et al., 2004; Paniagua-Contreras et al., 2017; Najafi et al., 2019; Abd El-Baky et al., 2020; Bakhshi et al., 2020). The virulence genes belong to various pathotypes of E. coli, but most are related to extra-intestinal pathogenic E. coli (ExPEC; Paniagua-Contreras et al., 2017; Najafi et al., 2019; Abd El-Baky et al., 2020). Among the E. coli strains, the ST69 has the most abundant and diverse virulence genes, including three toxin genes, hlyE (hemolysin), astA (heat-stable enterotoxin EAST-1), espP (serine protease autotransporter), suggesting the public health hazard associated with this strain. The ST69 E. coli is reported as the second most common cause of extraintestinal infection across the globe (Manges et al., 2019).
In some isolates, virulence genes and ARGs were co-located on the same plasmids or associated with other MGEs. For example, in ST69, traT (a virulence gene that helps the bacteria to survive the host serum bactericidal effect of the complement system) is located on IncFII (F2:A: B) plasmids together with tet(A) and blaTEM-1. Hayer et al. (2020) also reported an association between the traT virulence gene and IncFI plasmids from E. coli isolates obtained from pigs in the U.S. The virulence gene, traT, was indicated to be frequently associated with sepsis and urinary tract infections in humans (Nojoomi and Ghasemian, 2019; Abd El-Baky et al., 2020). Thus, co-location favors the clonal and horizontal spread of the virulence and resistance genes and may cause a significant clinical impact on people consuming raw milk. In addition to detecting the virulence gene in each strain, in silico assessment of the pathogenicity of the bacteria to the human host predicted that all the ESBL-E. coli are highly likely (probability > 0.92) to be a human pathogen (Cosentino et al., 2013). Escherichia coli is one of the most important milk borne human pathogens (WHO, 2018). Escherichia coli causes a wide range of severe infections in humans (Lim et al., 2010) and is also among the frequent cause of environmental mastitis in dairy cattle (Herry et al., 2017), along with colibacillosis in calves (Gelalcha et al., 2022). Thus, the presence of multiple ARGs, combined with the presence of virulence factors, make ESBL-E. coli, a greater threat to human health. Furthermore, the detection of potential pathogenic and MDR ESBL-E. coli isolates in this study support the argument about the possible convergence between ESBL-production and virulence (De Koster et al., 2022).
The ESBL-E. coli isolate that showed resistance to nine antibiotics was found to have the same resistant pattern as most E. coli isolates retrieved from fecal samples from the same farm (Farm A), suggesting possible fecal contamination of the bulk tank milk from the cow (unpublished data). Similarly, WGS analysis of this isolate identified 11 different ARGs, including mph(A; macrolide phosphotransferases), aadA1 (aminoglycoside nucleotidyltransferase), dfrA (dihydrofolate reductase), and sul1(dihydropteroate synthase) which was detected only in this isolate. Multiple E. coli isolates with the same serotype (045, H25) and sequence types (ST2325) were isolated from a fecal sample obtained from the same farm. Detection of the same serotypes and sequence of E. coli isolates from fecal samples strongly indicates fecal contamination of the BTM. This highlights the importance of implementing hygiene and sanitation measures to prevent contamination and reduce the risk of transmission of pathogenic or commensal MDR bacteria to humans.
Unlike ESBL-E. coli, the two K. pneumoniae isolates did not exhibit acquired multi-drug resistance phenotypes. However, the detected resistance genotypes of the K. pneumoniae isolates were consistent with their expressed phenotypes. Even without MDR phenotypes, the presence of ESBL-producing K. pneumoniae is still a concern, as it can contribute to the spread of antibiotic resistance (Navon-Venezia et al., 2017). Both Klebsiella strains harbored three beta-lactamase encoding genes, two ESBL genotypes, and one non-ESBL genotype. The detection of non-ESBL or narrow spectrum beta-lactamase, blaTEM-1B in one and blaSHV-2 in the other strain is consistent with the established literature as Klebsiella spp. are inherently resistant to penicillin (Paterson, 2006). Detection of multiple resistance genes to beta-lactam antibiotics suggests that these isolates have accumulated multiple mechanisms for resistance to beta-lactam antibiotics. The observed ESBL phenotypes might be mediated by blaCTX-M-27 and blaSHV-62 in ST867 and blaCTX-M-1 and blaSHV-40 in the novel strain (Bush and Bradford, 2020).
Sequence typing assigned one of the Klebsiella isolates, BM-MK, to ST867, but its serotype was new and could not be assigned. The other isolate, DM-MK, was also not assigned to a particular sequence type or serotype, i.e., its genetic makeup or antigenic characteristics did not match any known K. pneumoniae subtypes based on its DNA sequence. This K. pneumoniae isolate was identified as a novel serotype and sequence type; it was not previously reported in the scientific literature. This finding highlights the importance of continuous monitoring and surveillance for the emergence of new strains of ESBL-producing bacteria to detect them early before their spread and inform public health efforts to control them.
In the novel strain of K. pneumoniae, the ESBL genotypes co-existed with resistance genotypes that mediate resistance to three CIAs, including aminoglycosides, fluoroquinolone, and macrolide. However, the corresponding resistance phenotypes were not observed against these antibiotics during AST, suggesting a lack of proper expression of the respective genes. The resistance to sulfisoxazole observed in the ST867 Klebsiella strain may be due to the oqxAB operon, a plasmid-borne multi-drug efflux pump (Li et al., 2019). The absence of known genes related to folate antagonists does not rule out the possibility of efflux pump-mediated resistance. Detection of multiple resistance genes in K. pneumoniae is consistent with the previous studies and reviews that identified K. pneumoniae as a” key trafficker” of multi-drug resistance genes (Holt et al., 2015; Navon-Venezia et al., 2017; Wyres and Holt, 2018).
The predominance of epidemic resistance plasmids (formerly known as narrow host plasmids), IncF, in these two strains of K. pneumoniae is consistent with the previous findings that reported a frequent association between ESBL-producing Klebsiella spp. and InF plasmid families (Carattoli, 2009). The IncFII(K) and IncFIB(K; both IncF plasmid families) identified in the novel strain are highly similar to (96%–99% nucleotide identity) those plasmids identified from MDR carbapenemase-producing ST258 K. pneumoniae isolate from a human in Italy (Garcia-Fernandez et al., 2012). This suggests closely related epidemic-resistance plasmids are circulating in both animals and humans across the globe. The col440I plasmids that carried qnrB19 in the novel strain were similar (91% nucleotide identity) to the plasmid identified from K. pneumoniae isolates from human blood samples in the U.S. (Accession: CP023920.1; the data not published).
The ESBL-K. pneumoniae were also enriched with virulence determinants that may enhance their pathogenic potential. The siderophore receptor gene (fyuA), the Ferric aerobactin receptor gene (iutA), and the iron regulatory protein (Irp2) encode proteins associated with iron uptake and metabolism and thereby enhance their growth, survival and pathogenicity. Type 1 fimbriae (FimH) help the bacteria adhere to the mammalian host cell. The genes ccI (encodes for cloacin that help the bacteria establish a niche in the host environment without as much competition), clpK1(encodes heat shock survival AAA family ATPase that allow the bacteria adapt to elevated host body temperatures and also degrade host proteins), and traT (encodes outer membrane protein responsible for complement resistance) help evade the host defense mechanism and contribute to the pathogenicity (Tu et al., 2016; Sarowska et al., 2019; Juraschek et al., 2022). In addition, further analysis of the Klebsiella genome predicted both strains as possible human pathogens with a high probability (probability >0.87). The presence of ESBL and multiple virulence genes in K. pneumoniae, a frequent cause of severe infections in humans and mastitis in dairy cattle, highlights the significant public health and veterinary risk associated with these bacteria (Fuenzalida et al., 2021; Zheng et al., 2022). Monitoring and controlling the ESBL-producing Enterobacteriaceae, particularly those with increased virulence, is crucial to reduce infection risk and to improve public and animal health.
5. Conclusion
The results of this study revealed a high prevalence of multidrug resistant ESBL-E. coli and presence of ESBL-K. pneumoniae in bulk tank milk in East Tennessee dairy farms. These bacteria carry several resistance and virulence genes often linked to mobile genetic elements, making it easy for them to spread to other bacteria. Half of ESBL-E. coli isolates from this study are considered high-risk clones due to their ability to cause disease and acquire genetic traits that give them an advantage over other bacterial clones, including virulence factors, epidemic plasmids, and ARGs. Detection of these highly resistant and virulent bacteria in raw bulk tank milk threatens public health. The bacteria can cause clinical infection or transfer their resistance and virulence gene to a more clinically relevant strain. Feces were also found to be a potential source of ESBL-E. coli, which might be a source of milk contamination, suggesting the need for hygienic milking practices on farms. The WGS data of E. coli isolates revealed a diverse population of bacteria with seven distinct serotypes, six STs, and eight different cgSTs. The discovery of a new allele type for the IncFIA (HI1) plasmid in the novel ST of K. pneumoniae can help to further our understanding of the diversity of strains of this bacterium and its plasmids. This study indicates that raw milk is a reservoir of potentially zoonotic MDR E. coli and K. pneumoniae, carrying readily transferable resistance and virulence genes. Thus, the consumption of raw milk and milk products has a public health risk with potentially MDR infections.
Data availability statement
The sequence data are deposited in the NCBI database under BioProject PRJNA1011909.
Author contributions
BG: Conceptualization, Writing – original draft, Methodology, Investigation, Formal analysis, Writing – review & editing. RM: Writing – review & editing, Formal analysis. AG: Investigation, Writing – review & editing. OK: Supervision, Resources, Project administration, Funding–acquisition, Conceptualization, Writing – review & editing.
Funding
The author(s) declare that no financial support was received for the research, authorship, and/or publication of this article.
Acknowledgments
The authors would like to thank East Tennessee dairy farms for volunteering to participate in this study.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Footnotes
1. ^https://cge.food.dtu.dk/services/KmerFinder/
2. ^https://cge.food.dtu.dk/services/ResFinder/
3. ^https://card.mcmaster.ca/analyze/rgi
4. ^https://cge.food.dtu.dk/services/PlasmidFinder/
5. ^https://cge.food.dtu.dk/services/MobileElementFinder/
6. ^https://cge.food.dtu.dk/services/VirulenceFinder/
7. ^https://cge.food.dtu.dk/services/SerotypeFinder/
8. ^https://cge.food.dtu.dk/services/MLST/
9. ^https://cge.food.dtu.dk/services/cgMLSTFinder/
10. ^https://cge.food.dtu.dk/services/PathogenFinder/
References
Abd El-Baky, R. M., Ibrahim, R. A., Mohamed, D. S., Ahmed, E. F., and Hashem, Z. S. (2020). Prevalence of virulence genes and their association with antimicrobial resistance among Pathogenic E. coli isolated from Egyptian patients with different clinical infections. Infect Drug Resist. 13, 1221–1236. doi: 10.2147/IDR.S241073
Afema, J. A., Ahmed, S., Besser, T. E., Jones, L. P., Sischo, W. M., and Davis, M. A. (2018). Molecular epidemiology of dairy cattle-associated Escherichia coli carrying blaCTX-M genes in Washington state. Appl. Environ. Microbiol. 84:e02430. doi: 10.1128/AEM.02430-17
Agga, G. E., Galloway, H. O., Netthisinghe, A. M. P., Schmidt, J. W., and Arthur, T. M. (2022). Tetracycline-resistant, third-generation cephalosporin-resistant, and extended-Spectrum beta-lactamase-producing Escherichia coli in a beef cow-calf production system. J. Food Prot. 85, 1522–1530. doi: 10.4315/JFP-22-178
Alcock, B. P., Raphenya, A. R., Lau, T. T. Y., Tsang, K. K., Bouchard, M., Edalatmand, A., et al. (2020). CARD 2020: antibiotic resistome surveillance with the comprehensive antibiotic resistance database. Nucleic Acids Res. 48, D517–D525. doi: 10.1093/nar/gkz935
Badr, H., Reda, R. M., Hagag, N. M., Kamel, E., Elnomrosy, S. M., Mansour, A. I., et al. (2022). Multidrug-resistant and genetic characterization of extended-Spectrum Beta-lactamase-Producing E. coli recovered from chickens and humans in Egypt. Animals 12:346. doi: 10.3390/ani12030346
Bakhshi, M., Zandi, H., Bafghi, M. F., Astani, A., Ranjbar, V. R., and Vakili, M. (2020). A survey for phylogenetic relationship; presence of virulence genes and antibiotic resistance patterns of avian pathogenic and uropathogenic Escherichia coli isolated from poultry and humans in Yazd. Iran. Gene Rep. 20:100725. doi: 10.1016/j.genrep.2020.100725
Bankevich, A., Nurk, S., Antipov, D., Gurevich, A. A., Dvorkin, M., Kulikov, A. S., et al. (2012). SPAdes: a new genome assembly algorithm and its applications to single-cell sequencing. J. Comput. Biol. 19, 455–477. doi: 10.1089/cmb.2012.0021
Bartual, S. G., Seifert, H., Hippler, C., Luzon, M. A., Wisplinghoff, H., and Rodriguez-Valera, F. (2005). Development of a multilocus sequence typing scheme for characterization of clinical isolates of Acinetobacter baumannii. J. Clin. Microbiol. 43, 4382–4390. doi: 10.1128/JCM.43.9.4382-4390.2005
Bevan, E. R., Jones, A. M., and Hawkey, P. M. (2017). Global epidemiology of CTX-M beta-lactamases: temporal and geographical shifts in genotype. J. Antimicrob. Chemother. 72, 2145–2155. doi: 10.1093/jac/dkx146
Bialvaei, A. Z., and Samadi, K. H. (2015). Colistin, mechanisms and prevalence of resistance. Curr. Med. Res. Opin. 31, 707–721. doi: 10.1185/03007995.2015.1018989
Binsker, U., Kasbohrer, A., and Hammerl, J. A. (2022). Global colistin use: a review of the emergence of resistant Enterobacterales and the impact on their genetic basis. FEMS Microbiol. Rev. 46:fuab049. doi: 10.1093/femsre/fuab049
Bolger, A. M., Lohse, M., and Usadel, B. (2014). Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics 30, 2114–2120. doi: 10.1093/bioinformatics/btu170
Bortolaia, V., Kaas, R. S., Ruppe, E., Roberts, M. C., Schwarz, S., Cattoir, V., et al. (2020). ResFinder 4.0 for predictions of phenotypes from genotypes. J. Antimicrob. Chemother. 75, 3491–3500. doi: 10.1093/jac/dkaa345
Bradford, P. A. (2001). Extended-spectrum beta-lactamases in the 21st century: characterization, epidemiology, and detection of this important resistance threat. Clin. Microbiol. Rev. 14, 933–951. doi: 10.1128/CMR.14.4.933-951.2001
Bush, K., and Bradford, P. A. (2020). Epidemiology of beta-lactamase-producing pathogens. Clin. Microbiol. Rev. 33. doi: 10.1128/CMR.00047-19
Camacho, C., Coulouris, G., Avagyan, V., Ma, N., Papadopoulos, J., Bealer, K., et al. (2009). BLAST+: architecture and applications. BMC Bioinformatics. 10:421. doi: 10.1186/1471-2105-10-421
Canton, R., Gonzalez-Alba, J. M., and Galan, J. C. (2012). CTX-M enzymes: origin and diffusion. Front. Microbiol. 3:110. doi: 10.3389/fmicb.2012.00110
Canton, R., and Ruiz-Garbajosa, P. (2011). Co-resistance: an opportunity for the bacteria and resistance genes. Curr. Opin. Pharmacol. 11, 477–485. doi: 10.1016/j.coph.2011.07.007
Carattoli, A. (2009). Resistance plasmid families in Enterobacteriaceae. Antimicrob. Agents Chemother. 53, 2227–2238. doi: 10.1128/AAC.01707-08
Carattoli, A., Zankari, E., Garcia-Fernandez, A., Larsen, M. V., Lund, O., Villa, L., et al. (2014). In silico detection and typing of plasmids using PlasmidFinder and plasmid multilocus sequence typing. Antimicrob. Agents Chemother. 58, 3895–3903. doi: 10.1128/AAC.02412-14
Carey, A. M., Capik, S. F., Giebel, S., Nickodem, C., Pineiro, J. M., Scott, H. M., et al. (2022). Prevalence and profiles of antibiotic resistance genes mph(a) and qnrB in extended-Spectrum Beta-lactamase (ESBL)-producing Escherichia coli isolated from dairy calf feces. Microorganisms. 10:411. doi: 10.3390/microorganisms10020411
CDC. (2019). Antimicrobial resistance threats report. Available at: https://www.cdc.gov/DrugResistance/Biggest-Threats.html#cam (Accessed 25 July 2023).
Christy, M. L. S., Meyer, E., and Okoh, A. (2018). Antibiotic use in agriculture and its consequential resistance in environmental sources: potential public health implications. Molecules 23:795. doi: 10.3390/molecules23040795
Clausen, P., Aarestrup, F. M., and Lund, O. (2018). Rapid and precise alignment of raw reads against redundant databases with KMA. BMC Bioinformatics. 19:307. doi: 10.1186/s12859-018-2336-6
Clermont, O., Gordon, D., and Denamur, E. (2015). Guide to the various phylogenetic classification schemes for Escherichia coli and the correspondence among schemes. Microbiology 161, 980–988. doi: 10.1099/mic.0.000063
Clinical and Laboratory Standards Institute (CLSI) (2022). Available at: https://clsi.org/
Collis, R. M., Burgess, S. A., Biggs, P. J., Midwinter, A. C., French, N. P., Toombs-Ruane, L., et al. (2019). Extended-Spectrum Beta-lactamase-producing Enterobacteriaceae in dairy farm environments: a New Zealand perspective. Foodborne Pathog. Dis. 16, 5–22. doi: 10.1089/fpd.2018.2524
Cosentino, S., Voldby Larsen, M., Moller Aarestrup, F., and Lund, O. (2013). PathogenFinder—distinguishing friend from foe using bacterial whole genome sequence data. PloS One 8:e77302. doi: 10.1371/journal.pone.0077302
D’Andrea, M. M. A. F., and Pallecchi, L. (2013). Rossolini GM CTX-M-type-lactamases: a successful story of antibiotic resistance. Int. J. Med. Microbiol. 303, 305–317. doi: 10.1016/j.ijmm.2013.02.008
Davis, M. A., Sischo, W. M., Jones, L. P., Moore, D. A., Ahmed, S., Short, D. M., et al. (2015). Recent emergence of Escherichia coli with cephalosporin resistance conferred by Bla(CTX-M) on Washington state dairy farms. Appl. Environ. Microbiol. 81, 4403–4410. doi: 10.1128/AEM.00463-15
De Koster, S., Rodriguez Ruiz, J. P., Rajakani, S. G., Lammens, C., Glupczynski, Y., Goossens, H., et al. (2022). Diversity in the characteristics of Klebsiella pneumoniae ST101 of human, environmental, and animal origin. Front. Microbiol. 13:838207. doi: 10.3389/fmicb.2022.838207
Decano, A. G., Pettigrew, K., Sabiiti, W., Sloan, D. J., Neema, S., Bazira, J., et al. (2021). Pan-Resistome characterization of Uropathogenic Escherichia coli and Klebsiella pneumoniae strains circulating in Uganda and Kenya, isolated from 2017-2018. Antibiotics 10:1547. doi: 10.3390/antibiotics10121547
Delannoy, S., Le Devendec, L., Jouy, E., Fach, P., Drider, D., and Kempf, I. (2017). Characterization of Colistin-resistant Escherichia coli isolated from diseased pigs in France. Front. Microbiol. 8:2278. doi: 10.3389/fmicb.2017.02278
Diancourt, L., Passet, V., Verhoef, J., Grimont, P. A., and Brisse, S. (2005). Multilocus sequence typing of Klebsiella pneumoniae nosocomial isolates. J. Clin. Microbiol. 43, 4178–4182. doi: 10.1128/JCM.43.8.4178-4182.2005
Doern, C. D., and Butler-Wu, S. M. (2016). Emerging and future applications of matrix-assisted laser desorption ionization time-of-flight (MALDI-TOF) mass spectrometry in the clinical microbiology laboratory: a report of the Association for Molecular Pathology. J. Mol. Diagn. 18, 789–802. doi: 10.1016/j.jmoldx.2016.07.007
Ewels, P., Magnusson, M., Lundin, S., and Kaller, M. (2016). MultiQC: summarize analysis results for multiple tools and samples in a single report. Bioinformatics 32, 3047–3048. doi: 10.1093/bioinformatics/btw354
Falagas, M. E., Karageorgopoulos, D. E., and Nordmann, P. (2011). Therapeutic options for infections with Enterobacteriaceae producing carbapenem-hydrolyzing enzymes. Future Microbiol. 6, 653–666. doi: 10.2217/fmb.11.49
FastQC. (2010). A Quality Control Tool for High Throughput Sequence Data. Available at: http://www.bioinformatics.babraham.ac.uk/projects/fastqc/.
Food and Drug Administration (FDA) (2016). Antimicrobial use and resistance in animal agriculture in the United States. Available at: https://www.fda.gov/media/159544/download
FDA. (2023) Extralabel use and antimicrobials. Available at: https://www.fda.gov/animal-veterinary/antimicrobial-resistance/extralabel-use-and-antimicrobials (Accessed 6 August 2023).
Feldgarden, M., Brover, V., Haft, D. H., Prasad, A. B., Slotta, D. J., Tolstoy, I., et al. (2019). Validating the AMRFinder tool and resistance gene database by using antimicrobial resistance genotype-phenotype correlations in a collection of isolates. Antimicrob. Agents Chemother. 63. doi: 10.1128/AAC.00483-19
Ferreira, R. L., da Silva, B. C. M., Rezende, G. S., Nakamura-Silva, R., Pitondo-Silva, A., Campanini, E. B., et al. (2019). High prevalence of multidrug-resistant Klebsiella pneumoniae harboring several virulence and β-lactamase encoding genes in a Brazilian intensive care unit. Front. Microbiol. 9:3198. doi: 10.3389/fmicb.2018.03198
Filioussis, G., Kachrimanidou, M., Christodoulopoulos, G., Kyritsi, M., Hadjichristodoulou, C., Adamopoulou, M., et al. (2020). Short communication: bovine mastitis caused by a multidrug-resistant, mcr-1-positive (colistin-resistant), extended-spectrum beta-lactamase-producing Escherichia coli clone on a Greek dairy farm. J. Dairy Sci. 103, 852–857. doi: 10.3168/jds.2019-17320
Fuenzalida, M., Furmaga, E., and Aulik, N. (2021). Antimicrobial resistance in Klebsiella species from milk specimens submitted for bovine mastitis testing at the Wisconsin veterinary diagnostic laboratory, 2008–2019. JDS. Communications 2, 148–152. doi: 10.3168/jdsc.2020-0031
Garcia-Fernandez, A., Villa, L., Carta, C., Venditti, C., Giordano, A., Venditti, M., et al. (2012). Klebsiella pneumoniae ST258 producing KPC-3 identified in Italy carries novel plasmids and OmpK36/OmpK35 porin variants. Antimicrob. Agents Chemother. 56, 2143–2145. doi: 10.1128/AAC.05308-11
Gelalcha, B. D., Agga, G. E., and Dego, O. K. (2021). “Antimicrobial usage for the Management of Mastitis in the USA: impacts on antimicrobial resistance and potential alternative approaches” in Mastitis in dairy cattle, Sheep and Goats. ed. O. K. Dego (Intech Open), 1–21.
Gelalcha, B. D., Ensermu, D. B., Agga, G. E., Vancuren, M., Gillespie, B. E., D'Souza, D. H., et al. (2022). Prevalence of antimicrobial resistant and extended-Spectrum Beta-lactamase-producing Escherichia coli in dairy cattle farms in East Tennessee. Foodborne Pathog. Dis. 19, 408–416. doi: 10.1089/fpd.2021.0101
Gelalcha, B., and Kerro Dego, O. (2022). Extended-Spectrum Beta-lactamases producing Enterobacteriaceae in the USA dairy cattle farms and implications for public health. Antibiotics 11:1313. doi: 10.3390/antibiotics11101313
Hasman, H., Saputra, D., Sicheritz-Ponten, T., Lund, O., Svendsen, C. A., Frimodt-Moller, N., et al. (2014). Rapid whole-genome sequencing for detection and characterization of microorganisms directly from clinical samples. J. Clin. Microbiol. 52, 139–146. doi: 10.1128/JCM.02452-13
Hayer, S. S., Lim, S., Hong, S., Elnekave, E., Johnson, T., Rovira, A., et al. (2020). Genetic determinants of resistance to extended-Spectrum cephalosporin and fluoroquinolone in Escherichia coli isolated from diseased pigs in the United States. mSphere 5:e00990-20. doi: 10.1128/mSphere.00990-20
Hennequin, C., and Robin, F. (2016). Correlation between antimicrobial resistance and virulence in Klebsiella pneumoniae. Eur. J. Clin. Microbiol. 35, 333–341. doi: 10.1007/s10096-015-2559-7
Herry, V., Gitton, C., Tabouret, G., Répérant, M., Forge, L., Tasca, C., et al. (2017). Local immunization impacts the response of dairy cows to Escherichia coli mastitis. Local immunization impacts the response of dairy cows to Escherichia coli mastitis. Sci. Rep. 7:3441. doi: 10.1038/s41598-017-03724-7
Holt, K. E., Wertheim, H., Zadoks, R. N., Baker, S., Whitehouse, C. A., Dance, D., et al. (2015). Genomic analysis of diversity, population structure, virulence, and antimicrobial resistance in Klebsiella pneumoniae, an urgent threat to public health. Proc. Natl. Acad. Sci. U. S. A. 112, E3574–E3581. doi: 10.1073/pnas.1501049112
Joensen, K. G., Scheutz, F., Lund, O., Hasman, H., Kaas, R. S., Nielsen, E. M., et al. (2014). Real-time whole-genome sequencing for routine typing, surveillance, and outbreak detection of verotoxigenic Escherichia coli. J. Clin. Microbiol. 52, 1501–1510. doi: 10.1128/JCM.03617-13
Johnson, T. J., Wannemuehler, Y. M., Johnson, S. J., Logue, C. M., White, D. G., Doetkott, C., et al. (2007). Plasmid replicon typing of commensal and pathogenic Escherichia coli isolates. Appl. Environ. Microbiol. 73, 1976–1983. doi: 10.1128/AEM.02171-06
Juraschek, K., Malekzadah, J., Malorny, B., Kasbohrer, A., Schwarz, S., Meemken, D., et al. (2022). Characterization of qnrB-carrying plasmids from ESBL- and non-ESBL-producing Escherichia coli. BMC Genomics 23:365. doi: 10.1186/s12864-022-08564-y
Kaper, J. B., Nataro, J. P., and Mobley, H. L. (2004). Pathogenic Escherichia coli. Nat. Rev. Microbiol. 2, 123–140. doi: 10.1038/nrmicro818
Karaiskos, I., and Giamarellou, H. (2020). Carbapenem-sparing strategies for ESBL producers: when and how. Antibiotics. 9:61. doi: 10.3390/antibiotics9020061
Karns, J. S., Van Kessel, J. S., McClusky, B. J., and Perdue, M. L. (2007). Incidence of Escherichia coli O157: H7 and E-coli virulence factors in US bulk tank milk as determined by polymerase chain reaction. J. Dairy Sci. 90, 3212–3219. doi: 10.3168/jds.2006-009
Kuehn, B. (2018). Drug-resistant infections from raw Milk. JAMA 319:1191. doi: 10.1001/jama.2018.2595
Kürekci, C., Osek, J., Aydın, M., Tekeli, İ. O., Kurpas, M., Wieczorek, K., et al. (2019). Evaluation of bulk tank raw milk and raw chicken meat samples as source of ESBL producing Escherichia coli in Turkey: recent insights. J. Food Saf. 39:e12605. doi: 10.1111/jfs.12605
Lam, M. M. C., Wyres, K. L., Wick, R. R., Judd, L. M., Fostervold, A., Holt, K. E., et al. (2019). Convergence of virulence and MDR in a single plasmid vector in MDR Klebsiella pneumoniae ST15. J. Antimicrob. Chemother. 74, 1218–1222. doi: 10.1093/jac/dkz028
Langer, A. J. A. T., Grass, J., Lynch, M., Angulo, F. J., and Mahon, B. E. (2012). Nonpasteurized dairy products, disease outbreaks, and state laws—United States, 1993–2006. Emerg. Infect. Dis. 18, 385–391. doi: 10.3201/eid1803.111370
Larsen, M. V., Cosentino, S., Lukjancenko, O., Saputra, D., Rasmussen, S., Hasman, H., et al. (2014). Benchmarking of methods for genomic taxonomy. J. Clin. Microbiol. 52, 1529–1539. doi: 10.1128/JCM.02981-13
Larsen, M. V., Cosentino, S., Rasmussen, S., Friis, C., Hasman, H., Marvig, R. L., et al. (2012). Multilocus sequence typing of Total-genome-sequenced Bacteria. J. Clin. Microbiol. 50, 1355–1361. doi: 10.1128/JCM.06094-11
Lemee, L., and Pons, J. L. (2010). Multilocus sequence typing for Clostridium difficile. Methods Mol. Biol. 646, 77–90. doi: 10.1007/978-1-60327-365-7_6
Li, J., Zhang, H., Ning, J., Sajid, A., Cheng, G., Yuan, Z., et al. (2019). The nature and epidemiology of OqxAB, a multidrug efflux pump. Antimicrob. Resist. Infect. Control 8:44. doi: 10.1186/s13756-019-0489-3
Lim, J. Y., Yoon, J., and Hovde, C. J. (2010). A brief overview of Escherichia coli O157:H7 and its plasmid O157. J. Microbiol. Biotechnol. 20, 5–14. doi: 10.4014/jmb.0908.08007
Liu, G., Ali, T., Gao, J., Ur Rahman, S., Yu, D., Barkema, H. W., et al. (2020). Co-occurrence of plasmid-mediated Colistin resistance (mcr-1) and extended-Spectrum beta-lactamase encoding genes in Escherichia coli from bovine Mastitic Milk in China. Microb. Drug Resist. 26, 685–696. doi: 10.1089/mdr.2019.0333
Liu, J. Z., Zhu, Y., Jay-Russell, M., Lemay, D. G., and Mills, D. A. (2020). Reservoirs of antimicrobial resistance genes in retail raw milk. Microbiome. 8:99. doi: 10.1186/s40168-020-00861-6
Long, H. F. Y., Feng, Y., Ma, K., Liu, L., McNally, A., and Zong, Z. (2019). The co-transfer of plasmid-borne colistin-resistant genes mcr-1 and mcr-3.5, the carbapenemase gene blaNDM-5 and the 16S methylase gene rmtB from Escherichia coli. Sci. Rep. 9:696. doi: 10.1038/s41598-018-37125-1
Malberg Tetzschner, A. M., Johnson, J. R., Johnston, B. D., Lund, O., and Scheutz, F. (2020). In silico genotyping of Escherichia coli isolates for Extraintestinal virulence genes by use of whole-genome sequencing data. J. Clin. Microbiol. 58:e01269. doi: 10.1128/JCM.01269-20
Manges, A. R., Geum, H. M., Guo, A., Edens, T. J., Fibke, C. D., and Pitout, J. D. D. (2019). Global Extraintestinal pathogenic Escherichia coli (ExPEC) lineages. Clin. Microbiol. Rev. 32:e00135. doi: 10.1128/CMR.00135-18
Masse, J., Larde, H., Fairbrother, J. M., Roy, J. P., Francoz, D., Dufour, S., et al. (2021). Prevalence of antimicrobial resistance and characteristics of Escherichia coli isolates from fecal and manure pit samples on dairy farms in the province of Québec, Canada. Front Vet Sci 8:654125. doi: 10.3389/fvets.2021.654125
Meinersmann, R. J., Ladely, S. R., Plumblee, J. R., Cook, K. L., and Thacker, E. (2017). Prevalence of mcr-1 in the Cecal contents of food animals in the United States. Antimicrob. Agents Chemother. 61:e02244-16. doi: 10.1128/AAC.02244-16
Mungai, E. A. B. C., and Gould, L. H. (2015). Increased outbreaks associated with nonpasteurized milk, United States, 2007–2012. Emerg. Infect. Dis. 21, 119–122. doi: 10.3201/eid2101.140447
Najafi, S., Rahimi, M., and Nikousefat, Z. (2019). Extra-intestinal pathogenic Escherichia coli from human and avian origin: detection of the most common virulence-encoding genes. Vet Res Forum. 10, 43–49. doi: 10.30466/vrf.2019.34307
Navon-Venezia, S., Kondratyeva, K., and Carattoli, A. (2017). Klebsiella pneumoniae: a major worldwide source and shuttle for antibiotic resistance. FEMS Microbiol. Rev. 41, 252–275. doi: 10.1093/femsre/fux013
Nobrega, D. B. D. B. J., de Buck, J., and Barkema, H. W. (2018). Antimicrobial resistance in non-aureus staphylococci isolated from milk is associated with systemic but not intramammary administration of antimicrobials in dairy cattle. J. Dairy Sci. 101, 7425–7436. doi: 10.3168/jds.2018-14540
Nojoomi, F., and Ghasemian, A. (2019). The relation of phylogroups, serogroups, virulence factors and resistance pattern of Escherichia coli isolated from children with septicemia. New Microbes New Infect. 29:100517. doi: 10.1016/j.nmni.2019.100517
Paniagua-Contreras, G. L., Monroy-Perez, E., Rodriguez-Moctezuma, J. R., Dominguez-Trejo, P., Vaca-Paniagua, F., and Vaca, S. (2017). Virulence factors, antibiotic resistance phenotypes and O-serogroups of Escherichia coli strains isolated from community-acquired urinary tract infection patients in Mexico. J. Microbiol. Immunol. Infect. 50, 478–485. doi: 10.1016/j.jmii.2015.08.005
Paterson, D. L. (2006). Resistance in gram-negative bacteria: Enterobacteriaceae. Am. J. Infect. Control 34, S20–S28. doi: 10.1016/j.ajic.2006.05.238
Peirano, G., and Pitout, J. (2019). Extended-Spectrum β-lactamase-producing Enterobacteriaceae: update on molecular epidemiology and treatment options. Drugs 79, 1529–1541. doi: 10.1007/s40265-019-01180-3
Peirano, G., Sang, J. H., Pitondo-Silva, A., Laupland, K. B., and Pitout, J. D. (2012). Molecular epidemiology of extended-spectrum-beta-lactamase-producing Klebsiella pneumoniae over a 10 year period in Calgary, Canada. J. Antimicrob. Chemother. 67, 1114–1120. doi: 10.1093/jac/dks026
Poirel, L., Lartigue, M. F., Decousser, J. W., and Nordmann, P. (2005). ISEcp1B-mediated transposition of blaCTX-M in Escherichia coli. Antimicrob. Agents Chemother. 49, 447–450. doi: 10.1128/AAC.49.1.447-450.2005
Redding, L. E. B., Bender, J., and Baker, L. (2019). Quantification of antibiotic use on dairy farms in Pennsylvania. J. Dairy Sci. 102, 1494–1507. doi: 10.3168/jds.2018-15224
Riley, M. A., and Gordon, D. M. (1992). A survey of col plasmids in natural isolates of Escherichia-Coli and an investigation into the stability of col-plasmid lineages. J. Gen. Microbiol. 138, 1345–1352. doi: 10.1099/00221287-138-7-1345
Sarowska, J., Futoma-Koloch, B., Jama-Kmiecik, A., Frej-Madrzak, M., Ksiazczyk, M., Bugla-Ploskonska, G., et al. (2019). Virulence factors, prevalence and potential transmission of extraintestinal pathogenic Escherichia coli isolated from different sources: recent reports. Gut Pathog. 11:10. doi: 10.1186/s13099-019-0290-0
Straley, B. A., Donaldson, S. C., Hedge, N. V., Sawant, A. A., Srinivasan, V., Oliver, S. P., et al. (2006). Public health significance of antimicrobial-resistant gram-negative Bacteria in raw bulk tank Milk. Foodborne Pathog. Dis. 3, 222–233. doi: 10.1089/fpd.2006.3.222
Tark, D. S., Moon, D. C., Kang, H. Y., Kim, S. R., Nam, H. M., Lee, H. S., et al. (2017). Antimicrobial susceptibility and characterization of extended-spectrum beta-lactamases in Escherichia coli isolated from bovine mastitic milk in South Korea from 2012 to 2015. J. Dairy Sci. 100, 3463–3469. doi: 10.3168/jds.2016-12276
Taylor, E. A., Ossa-Trujillo, C., Vinasco, J., Jordan, E. R., García Buitrago, J. A., Hagevoort, R., et al. (2020). Use of critically important antimicrobial classes early in life may adversely impact bacterial resistance profiles during adult years: potential co-selection for plasmid-borne fluoroquinolone and macrolide resistance via extendedspectrum beta-lactam use in dairy cattle. Lett. Appl. Microbiol. 72, 220–224. doi: 10.1111/lam.13419
Timofte, D., Maciuca, I. E., Evans, N. J., Williams, H., Wattret, A., Fick, J. C., et al. (2014). Detection and molecular characterization of Escherichia coli CTX-M-15 and Klebsiella pneumoniae SHV-12 beta-lactamases from bovine mastitis isolates in the United Kingdom. Antimicrob. Agents Chemother. 58, 789–794. doi: 10.1128/AAC.00752-13
Toth, A. G., Csabai, I., Kriko, E., Tozser, D., Maroti, G., Patai, A. V., et al. (2020). Antimicrobial resistance genes in raw milk for human consumption. Sci. Rep. 10:7464. doi: 10.1038/s41598-020-63675-4
Tu, J., Xue, T., Qi, K., Shao, Y., Huang, B., Wang, X., et al. (2016). The irp2 and fyuA genes in high Pathogenicity Islands are involved in the pathogenesis of infections caused by avian pathogenic Escherichia coli (APEC). Pol. J. Vet. Sci. 19, 21–29. doi: 10.1515/pjvs-2016-0004
Tyson, G. H., McDermott, P. F., Li, C., Chen, Y., Tadesse, D. A., Mukherjee, S., et al. (2015). WGS accurately predicts antimicrobial resistance in Escherichia coli. J. Antimicrob. Chemother. 70, 2763–2769. doi: 10.1093/jac/dkv186
Wang, Y., Hou, N. X., Johnston, J., Sarreal, C., Jarosh, J., Hughes, A. C., et al. (2020). Low prevalence of mobile colistin-resistance in US meat, catfish, poultry and genomic characterization of a mcr-1 positive Escherichia coli strain. Food Control 118:107434. doi: 10.1016/j.foodcont.2020.107434
Weissman, S. J. A. A., Qin, X., and Zerr, D. M. (2013). Emergence of extended-spectrum β-lactam resistance among Escherichia coli at a US academic children's hospital is clonal at the sequence type level for CTX-M-15 but not for CMY-2. Emergence of extended-spectrum β-lactam resistance among Escherichia coli at a US academic children's hospital is clonal at the sequence type level for CTX-M-15, but not for CMY-2. Int. J. Antimicrob. Agents 41, 414–420. doi: 10.1016/j.ijantimicag.2013.01.006
Whitehead, J., and Lake, B. (2018). Recent trends in unpasteurized fluid Milk outbreaks, legalization, and consumption in the United States. PLoS Curr. 10. doi: 10.1371/currents.outbreaks.bae5a0fd685616839c9cf857792730d1
WHO. (2018). E. coli fact sheet. Avaialble at: https://www.who.int/news-room/fact-sheets/detail/e-coli
WHO (2019). “Critically important antimicrobials for human medicine 6th revision 2018” in Ranking of medically important antimicrobials for risk management of antimicrobial resistance due to non-human use. WHO Advisory Group on Integrated Surveillance of Antimicrobial Resistance.
Wirth, T., Falush, D., Lan, R., Colles, F., Mensa, P., Wieler, L. H., et al. (2006). Sex and virulence in Escherichia coli: an evolutionary perspective. Mol. Microbiol. 60, 1136–1151. doi: 10.1111/j.1365-2958.2006.05172.x
Wyres, K. L., and Holt, K. E. (2018). Klebsiella pneumoniae as a key trafficker of drug resistance genes from environmental to clinically important bacteria. Curr. Opin. Microbiol. 45, 131–139. doi: 10.1016/j.mib.2018.04.004
Yamaji, R., Rubin, J., Thys, E., Friedman, C. R., and Riley, L. W. (2018). Persistent pandemic lineages of Uropathogenic Escherichia coli in a college community from 1999 to 2017. J. Clin. Microbiol. 56:e01834. doi: 10.1128/JCM.01834-17
Yang, Y., Higgins, C. H., Rehman, I., Galvao, K. N., Brito, I. L., Bicalho, M. L., et al. (2019). Genomic diversity, virulence, and antimicrobial resistance of Klebsiella pneumoniae strains from cows and humans. Appl. Environ. Microbiol. 85:e02654. doi: 10.1128/AEM.02654-18
Zankari, E., Allesoe, R., Joensen, K. G., Cavaco, L. M., Lund, O., and Aarestrup, F. M. (2017). PointFinder: a novel web tool for WGS-based detection of antimicrobial resistance associated with chromosomal point mutations in bacterial pathogens. J. Antimicrob. Chemother. 72, 2764–2768. doi: 10.1093/jac/dkx217
Zankari, E., Hasman, H., Cosentino, S., Vestergaard, M., Rasmussen, S., Lund, O., et al. (2012). Identification of acquired antimicrobial resistance genes. J. Antimicrob. Chemother. 67, 2640–2644. doi: 10.1093/jac/dks261
Zheng, Z., Gorden, P. J., Xia, X., Zheng, Y., and Li, G. (2022). Whole-genome analysis of Klebsiella pneumoniae from bovine mastitis milk in the U.S. Environ. Microbiol. 24, 1183–1199. doi: 10.1111/1462-2920.15721
Keywords: ESBL, Escherichia coli , Klebsiella pneumoniae , sequence types, virulence gene, high-risk clones, bulk tank milk
Citation: Gelalcha BD, Mohammed RI, Gelgie AE and Kerro Dego O (2023) Molecular epidemiology and pathogenomics of extended-spectrum beta-lactamase producing- Escherichia coli and - Klebsiella pneumoniae isolates from bulk tank milk in Tennessee, USA. Front. Microbiol. 14:1283165. doi: 10.3389/fmicb.2023.1283165
Edited by:
Armin Tarrah, University of Guelph, CanadaReviewed by:
Manoj Kumar Solanki, University of Silesia in Katowice, PolandZahid Rehman, University of Guelph, Canada
Opeyemi U. Lawal, University of Guelph, Canada
Copyright © 2023 Gelalcha, Mohammed, Gelgie and Kerro Dego. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Oudessa Kerro Dego, b2tlcnJvZGVAdXRrLmVkdQ==
 Benti D. Gelalcha
Benti D. Gelalcha Ruwaa I. Mohammed
Ruwaa I. Mohammed Aga E. Gelgie
Aga E. Gelgie Oudessa Kerro Dego
Oudessa Kerro Dego