- 1State Key Laboratory for Animal Disease Control and Prevention, Guangdong Laboratory for Lingnan Modern Agriculture, College of Veterinary Medicine, South China Agricultural University, Guangzhou, China
- 2Key Laboratory of Zoonosis of Ministry of Agricultural and Rural Affairs, Guangdong Provincial Key Laboratory of Veterinary Pharmaceutics Development and Safety Evaluation, Guangzhou, Guangdong, China
The New Delhi Metallo-β-lactamase (NDM) producing Enterobacterales has been detected from diverse sources but has rarely been reported in retail eggs. In this study, 144 eggshell and 96 egg content samples were collected in 2022 from Guangdong province and were screened for NDM-producing strains. Four Escherichia coli strains (ST3014, ST10, ST1485, and ST14747) recovered from two (1.39%, 2 of 144) eggshells and two (2.08%, 2 of 96) egg content samples were identified as blaNDM−5-positive strains. Oxford Nanopore MinION sequencing and conjugation assays revealed that the blaNDM−5 gene was carried by IncX3 (n = 1), IncI1 (n = 1), and IncHI2 (n = 2). The IncI1-plasmid-carrying blaNDM−5 displayed high homology with one plasmid pEC6563-NDM5 from the human clinic, while the IncHI2 plasmid harboring blaNDM−5 shared highly similar structures with plasmids of animal origin. To the best of our knowledge, this is the first report on the identification of blaNDM−5-positive bacteria in retail eggs. NDM-producing E. coli could be transmitted to humans by the consumption of eggs or direct contact, which could pose a potential threat to human health.
1 Introduction
Carbapenemase-resistant Enterobacterales (CRE) have increased rapidly over the last decades and have become an urgent public health threat (El-Gamal et al., 2017). Carbapenem resistance in Enterobacteriaceae is attributed to three dominant carbapenemase enzymes, including New Delhi metallo-beta-lactamases (NDM), Klebsiella pneumoniae carbapenemases (KPC), and carbapenem-hydrolyzing oxacillinase-48-type β-lactamases (OXA-48) (Iovleva and Doi, 2017). Among these CRE, NDM-producing strains are highly prevalent around the world, especially in China and South Asia (Wu et al., 2019). At present, 47 variants of NDM have been identified (https://www.ncbi.nlm.nih.gov/pathogens/refgene/#NDM); of these, NDM-1 and NDM-5 remain the most prevalent carbapenemases (Shen et al., 2022; Saravanan et al., 2023). The NDM-5 possesses higher carbapenemase activity than NDM-1 (Hornsey et al., 2011). Currently, the blaNDM−5 gene has been disseminated to various bacterial species (e.g., Escherichia coli, K. pneumoniae, and Klebsiella aerogenes), with E. coli as the main bacterial host (Nordmann and Poirel, 2019; Jean et al., 2022; Ma et al., 2023).
Plasmid-mediated transmission has facilitated the widespread distribution of the blaNDM−5 gene among bacteria from various environmental sources and geographical regions. The blaNDM−5 gene has been found in an array of plasmid replicon types, such as IncX3, IncFII, IncF, IncN, and IncHI2 (Nordmann and Poirel, 2019; Jean et al., 2022; Lv et al., 2022). The IncX3 has long been recognized as the primary carrier for transmission of the blaNDM−5 gene (Shen et al., 2022); however, recently, there is an increase in IncHI2 plasmid as a carrier of blaNDM−5 in China (Ma et al., 2021; Zhao Q. et al., 2021; Lv et al., 2022; Wang et al., 2022; He et al., 2023). Alarmingly, IncHI2 has also been found to carry multiple antibiotic resistance genes, including colistin resistance gene (mcr), extended-spectrum beta-lactamase, and quinolone resistance genes (Webb et al., 2016; Mmatli et al., 2022).
Although carbapenems have not been approved for food animals, CRE has been continuously detected in pigs, poultry, and animal-derived foods, especially from chickens and poultry products. Eggs are important poultry products that play an essential role in the daily healthy diet of human beings and are the most consumed food all over the world [https://www.who.int/zh/news-room/fact-sheets/detail/salmonella-(non-typhoidal)]. Poultry eggs are also considered as reservoirs and transmission vectors of resistance genes, such as mcr-1, fosA, qnrS1, blaCTX−M−1, blaIMP, and blaOXA−48−like (Benameur et al., 2018; Kapena et al., 2020; Zhang et al., 2021, 2022b; Kanaan et al., 2022; Li et al., 2022). Resistant bacteria and genes in eggs have the risk of spreading to humans through various ways, such as hand-to-egg contact and storing unwashed eggs in fridge. However, the occurrence of clinically important resistant bacteria, NDM-producing Enterobacterales, in eggs has rarely been studied. Hence, we investigated the prevalence of NDM-producing Enterobacterales among egg samples recovered from markets in Guangzhou and characterized the molecular traits of blaNDM-positive isolates.
2 Methods
2.1 Sampling
From June to September 2022, 144 non-repetitive egg samples were randomly collected from 29 farmer markets located in four districts (Tianhe, Baiyun, Yuexiu, and Haizhu) of Guangzhou. To ensure diversity in the sampling and prevent repeated sampling from a singular supplier, we selected different stalls within each market, with a maximum of three eggs procured from any single stall. Each sample was placed in a separate sterile sample bag, and all samples were transported to the laboratory in a cool box within 8 h.
2.2 Bacterial isolation and detection of carbapenemase-encoding genes
For the isolation of bacteria from eggshells, the surface of eggs was wiped with a sterile swab, and then, the swab was placed into 4 ml sterilized Luria–Bertani (LB) broth medium for enrichment cultivation at 37°C overnight. For the isolation of bacteria from egg content, the eggshell was wiped with gauze with 70% ethanol, followed by being homogenized. During the processing of the first batch, 48 cracked eggs were collided and discarded to avoid cross-contamination (detailed information about all the samples is shown in Supplementary Table S1), and then, the remaining eggs were opened to extract the whole egg content. In total, 1 ml of egg content was dispensed into 4 ml sterilized Luria–Bertani (LB) broth medium and enriched at 37°C overnight with shaking. The overnight cultures of each sample were incubated on MacConkey agar plates supplemented with 0.5 mg/L meropenem, and the plates were incubated at 37°C for 16 h. One to three colonies with different morphologies in each plate were selected for the detection of carbapenemase-encoding genes, blaNDM, using PCR and DNA sequencing (primers are shown in Supplementary Table S2). All blaNDM-positive isolates were collected for species identification by direct smear and matrix-assisted laser desorption/ionization–time-of-flight mass spectrometry (MALDI-TOF MS; Bruker Daltonik GmbH, Bremen, Germany).
2.3 Antimicrobial susceptibility testing
According to the recommendations of the Clinical and Laboratory Standards Institute, the minimal inhibitory concentrations (MICs) of 19 antimicrobials against NDM-positive isolates were determined using the agar dilution method or broth microdilution (colistin and tigecycline) method. E. coli ATCC 25922 was used as a quality control strain. The results of MICs were interpreted according to CLSI (M100-S30) criteria and EUCAST (http://www.eucast.org/clinical_breakpoints/).
2.4 Plasmid transferability and stability
Conjugation experiments were performed by broth mating using blaNDM-positive strains as the donor and a sodium azide-resistant (MIC > 2,000 μg/ml) E. coli J53 strain as the recipient. In detail, the donor and recipient strains were incubated separately in LB broth for 4 h, followed by mating the bacterial cultures with a ratio of 1:1 and incubating at 37°C without shaking overnight. A 50-μL overnight mixture was plated onto MacConkey agar plates containing 0.5 mg/L meropenem and 150 mg/L sodium azide, and incubated for 18 h to count and select transconjugants. Conjugation frequency was calculated as the number of transconjugants per recipient. Chemical transformation experiments were performed in those cases that blaNDM-positive strains failed to conjugate. All transconjugants and transformants were confirmed by PCR (primers are shown in Supplementary Table S2) and antimicrobial susceptibility testing.
The stability of blaNDM−5-carrying plasmids in host bacteria was performed by a passage in the absence of antibiotic Luria broth (LB). Three single clones of each blaNDM−5-positive strain were grown in 3 ml LB without antibiotic treatment overnight at 37°C. The overnight culture was daily diluted 1:100 in fresh LB broth for 15 days. Cultures were collected at the end of each of 3 days for streaking on antibiotic-free MacConkey agar plates. Then, 100 colonies were selected, and the presence of blaNDM−5 and the corresponding plasmids was verified by PCR amplification of blaNDM−5 and repA (primers are shown in Supplementary Table S2). Plasmid retention was calculated as the ratio of strains with blaNDM−5 and repA and over 100 colonies.
2.5 Whole-genome sequencing and bioinformatics analysis
Genomic DNAs of four NDM-positive isolates were extracted by using HiPure Bacterial DNA Kit (Magen, Beijing, China), according to the manufacturer's instructions. Whole genomic DNA was sequenced using the Illumina NovaSeq 6000 and MinION platform (Nanopore, UK). Hybrid assembly of complete genomes was carried out using the Unicycler version 0.4.8 (Wick et al., 2017). MLST v2.19 (https://github.com/tseemann/mlst) was applied to the verified sequence type (ST). Center for Genomic Epidemiology (CGE) (http://genomicepidemiology.org/services/) and PubMLST were used to identify antimicrobial resistance genes (ARGs) and plasmid replication types. Prokka software was used to annotate the draft genome (Seemann, 2014). EasyFig tool (http://mjsull.github.io/Easyfig/) was used to draw the genetic context of blaNDM and plasmid alignments and comparisons (Sullivan et al., 2011).
2.6 Accession numbers
The complete sequences of NDM-positive isolates have been deposited in the GenBank database under accession numbers PRJNA983957.
3 Results
3.1 Prevalence of NDM-producing enterobacterales in egg samples
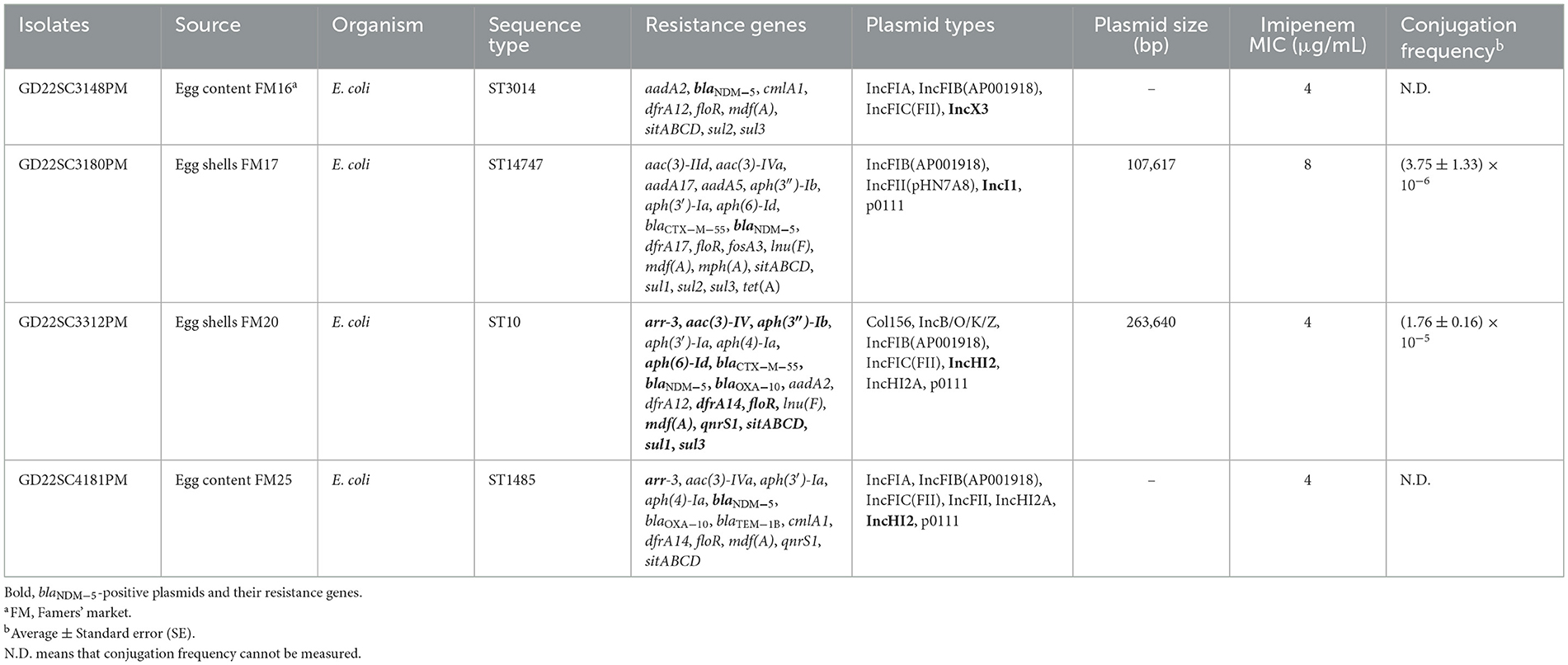
From the 144 eggs, 144 eggshell and 96 egg content samples were collected, and 4 blaNDM-positive isolates were recovered from 2 (1.39%, 2/144) eggshell samples (GD22SC3180PM and GD22SC3312PM) and 2 (2.08%, 2/96) egg content samples (GD22SC3148PM and GD22SC4181PM). These strains were further identified as E. coli by MALDI-TOF MS. All the blaNDM genes were identified as blaNDM−5.
3.2 Antimicrobial resistance patterns of blaNDM-5-positive E. coli strains
Antimicrobial susceptibility testing showed that all four isolates were resistant to most of the antimicrobials, including β-lactams (ampicillin, cefoxitin, cefotaxime, ceftazidime, cefquinome, and imipenem), aminoglycosides (apramycin, neomycin, and streptomycin), fosfomycin, florfenicol, tetracycline, and sulfamethoxazole. However, all isolates remained susceptible to amikacin, colistin, and tigecycline (Supplementary Table S3).
3.3 Genotyping and genetic background of blaNDM-5-positive E. coli strains
The four blaNDM−5-positive E. coli strains were subjected to short- and long-read sequencing to acquire complete genomes. Sequence analysis revealed that GD22SC3180PM, GD22SC3312PM, GD22SC3148PM, and GD22SC4181PM belonged to ST14747, ST10, ST3014, and ST1485, respectively (Table 1). All blaNDM-positive isolates harbored multiple antimicrobial resistance genes (ARGs) and plasmid replicon types (Table 1). Aminoglycoside resistance gene, β-lactam resistance gene, florfenicol resistance gene floR, macrolide resistance gene mdf(A), and sulfonamide resistance gene dfrA were detected in all the blaNDM−5-positive E. coli strains. In addition, the fluoroquinolone resistance gene qnrS and rifampicin resistance gene arr-3 were identified in two isolates (GD22SC3312PM and GD22SC4181PM). GD22SC3180PM additionally harbored tetracycline resistance gene tet(A), fosfomycin resistance gene fosA3, and lincomycin resistance gene lnu(F).
3.4 Characterization of blaNDM-5-carrying plasmids
Bioinformatics analysis revealed that there were three types of replicons carrying the blaNDM−5 gene, namely, IncX3 (pHN22SC3148), IncI1 (pHN22SC3180), and IncHI2 (pHN22SC3312 and pHN22SC4181) (Table 1). The complete sequence of IncI1 plasmid pHN22SC3180 is a 107,617 circular molecule with a GC content of 47% (Figure 1A). The backbone regions of the pHN22SC3180 displayed the highest similarity to plasmid pEC6563-NDM5 (CP095858.1, urine, Homo sapiens, E. coli, Zhejiang, China), with 100% identity and 99.0% coverage (Figure 1A) (Zhang et al., 2022a), but had low similarity to the other four blaNDM−5-bearing IncI1 plasmids deposited in the NCBI database (Supplementary Table S4). The plasmid pHN22SC3312 (IncHI2) was 263,640 bp in size with a GC content of 54% (Figure 1B) and had a high degree of homology with pHNGD64-NDM (MW296099.1, pig, E. coli, Guangdong, China) with 100% identity and 98.0% coverage, pNDM33-1 (CP076648.1, duck, E. coli, Guangdong, China) with 99.99% identity and 99.0% coverage, and pHNBYF33-1 (CP101733.1, fish, E. coli, Guangdong, China) with 99.99% identity and 98.0% coverage (Figure 1B). Furthermore, in addition to blaNDM−5, the variable region of pHN22SC3312 contained several antibiotic resistance genes [e.g., blaOXA−10, tet(A), qnrS1, floR, sul3, aadA2, and aph(4)-Ia].
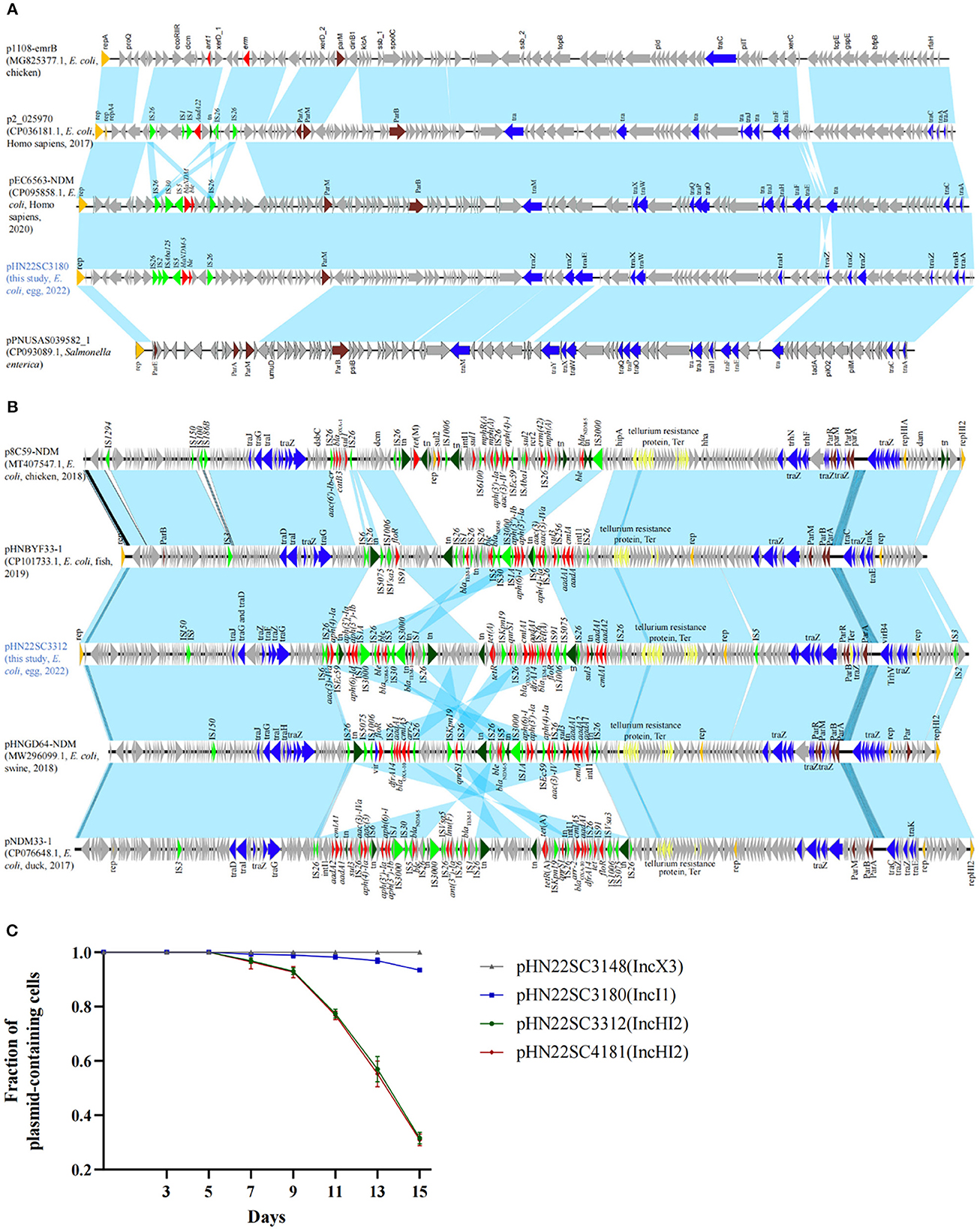
Figure 1. Comparison and stability of blaNDM−5-carrying plasmids. (A) blaNDM−5-harboring IncI1 plasmids in this study with other similar plasmids. p1108-emrB (NZ_MG825377.1), p2_025970 (CP036181.1), pPNUSAS039582_1 (CP093089.1), and pEC6563-NDM5 (CP095858.1). (B) blaNDM−5-harboring IncHI2 plasmids in this study with other similar plasmids. p8C59-NDM (MT407547.1), pHNBYF33-1 (CP101733.1), pHNGD64-NDM (MW296099.1), and pNDM33-1 (CP076648.1). (C) Stability of blaNDM−5-carrying plasmids in their corresponding host bacteria. Error bars represent standard deviations (n = 3).
3.5 The biological features of blaNDM-5-carrying plasmids
To evaluate the transferability of the blaNDM−5 gene, all four blaNDM−5 positive strains were conducted on a conjugation assay. The blaNDM−5-carrying plasmids were successfully transferred to recipients E. coli J53 at a frequency of 10−5-10−6. The imipenem MICs of the transconjugants were 2–4 μg/ml, which were 32–64-fold the MICs of the recipient (Supplementary Table S3). To evaluate the stability of blaNDM−5-carrying plasmids, we performed passage with the four blaNDM−5-positive strains in antibiotic-free Luria broth. The stability of the IncX3 plasmid pHN22SC3148 was 100% in the absence of antibiotic after 15 days (i.e., ~150 generations) in the natural host GD22SC3148PM, while IncI1 plasmid pHN22SC3180 and IncHI2 plasmids, pHN22SC3312 and pHN22SC4181, were gradually lost from their corresponding host strains after 7 days of passage, with 93.40, 31.60, and 30.90% retention after 15 days, respectively (Figure 1C). Thus, in the absence of antibiotic selection, IncX3 plasmid pHN22SC3148 is stable in the original isolate, and the other three plasmids (IncHI2 and IncI1) are less stable in the host strains.
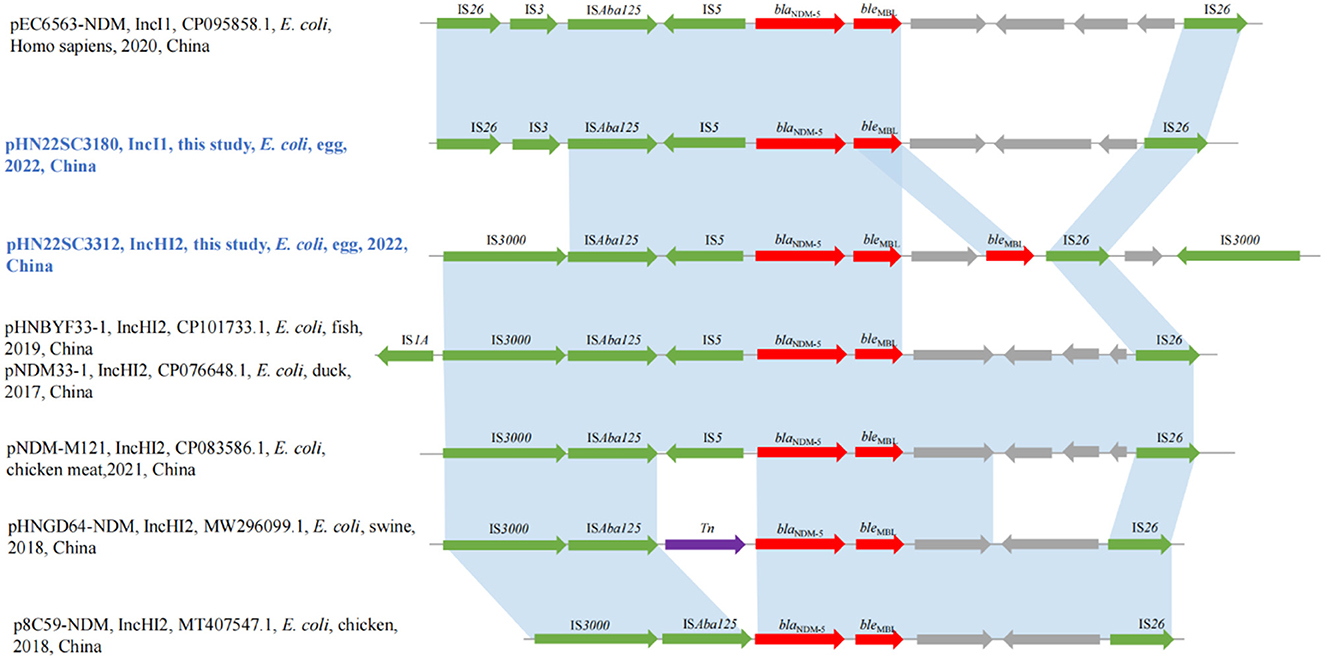
3.6 Genetic environments of blaNDM-5 genes in IncI1 and IncHI2
The genetic context of the blaNDM−5 gene in IncI1 plasmid pHN22SC3180 was IS26-IS3-ISAba125-IS5-blaNDM−5-bleMBL-trpF-dsbD-IS26, which was similar to the genetic environment recently discovered in IncI1 plasmid pEC6563-NDM5-carrying blaNDM−5 (GenBank accession no. CP095858.1) (Figure 2). Although the blaNDM−5 region (IS26-IS3-ISAba125-IS5-blaNDM−5-bleMBL-trpF-dsbD-IS26) was surrounded by two copies of IS26 with the same direction, no circular intermediate was obtained in this study, similar to previous report (Zhang et al., 2022a). Moreover, the genetic contexts of blaNDM−5 in IncHI2 plasmid pHN22SC3312, IS3000-ISAba125-IS5-blaNDM−5-bleMBL-trpF-bleMBL-IS26-dsbD-IS3000, were highly similar to other blaNDM−5-harboring IncHI2 plasmids, pNDM-M121 (GenBank accession no. CP083586.1), pHNBYF33-1 (GenBank accession no. CP101733.1), and pNDM33-1 (GenBank accession no. CP076648.1), except for the presence of two copies of bleMBL downstream of blaNDM−5 in pHN22SC3312 in this study (the information of all egg samples is shown in Supplementary Table S1).
4 Discussion
To date, blaNDM-positive Enterobacterales have been identified in various sources, including food animals, pets, human beings, animal foods, vegetables, and the environment (Zhai et al., 2020; Huang et al., 2023; Ma et al., 2023). However, there are few reports of NDM-producing bacteria in egg sources, except for one study, which reported the presence of blaNDM-positive Salmonella enterica in eggs from Iraq (Kanaan et al., 2022). To the best of our knowledge, this is the first report of NDM-5-producing Enterobacterales in retail egg samples from China. As eggs are an important food in the human diet and its consumption continues to increase, the NDM-positive Enterobacterales in eggs have the risk of spreading to humans via the food chain and even hand–egg contact.
Previous studies revealed that IncX3 is the most epidemiologically successful vehicle for spreading blaNDM-5 (Zhang et al., 2019; Zhao Q. Y. et al., 2021; Ma et al., 2023). blaNDM−5-bearing IncX3 plasmids are widely distributed in animals, human beings, and environments worldwide (Lv et al., 2022; Ma et al., 2023). The IncX3 plasmids carrying blaNDM−5 in this study further confirmed the importance of the IncX3 plasmid by acting as a vehicle for blaNDM−5 transfer. The blaNDM−5-positive IncX3 plasmids can be stably inherited in the original isolate (Figure 1C), which may partly explain the rapid global dissemination of blaNDM−5-bearing IncX3 plasmids (Ma et al., 2020).
In this study, we also detected the IncI1 plasmid (pHN22SC3180) and IncHI2 plasmid (pHN22SC3312) carrying the blaNDM−5 gene. IncI1 is an epidemic plasmid and can carry many resistance genes, especially the extended-spectrum beta-lactamase gene blaCTX, which has widely spread in patients and animals (Yang et al., 2014; Chong et al., 2018; Carattoli et al., 2021; Liu et al., 2021). However, the reports of the blaNDM−5-bearing IncI1 plasmid are few, and blaNDM−5-bearing IncI1 plasmid has just been detected in isolates from clinical and duck samples in China (Zhao Q. Y. et al., 2021; Dong et al., 2022; Zhang et al., 2022a). Of note, by searching through the NCBI database, we found only five blaNDM−5-bearing IncI1 plasmids, four of which were clinical samples isolated from China in recent years, implying that the prevalence and risk of blaNDM−5-bearing IncI1 in the clinic might be underestimated and need further investigation.
IncHI2 is a wide host plasmid and acts as an important vector for the dissemination of multiple ARGs, especially mcr-1 (Webb et al., 2016; Liu and Liu, 2018; Wu et al., 2018; Cao et al., 2020). To date, IncHI2-type plasmids carrying blaNDM−5 have only been detected in strains recovered from chicken, duck, pig feces, and freshwater fish (Ma et al., 2021; Zhao Q. Y. et al., 2021; Lv et al., 2022). These IncHI2-blaNDM−5 plasmids mainly spread regionally in Guangdong province in China but have also spread to other regions (Wang et al., 2022; He et al., 2023). The high similarity of the blaNDM−5-bearing IncHI2 plasmids in eggs and other origins suggested that these plasmids are spreading. However, IncHI2-type plasmids carrying blaNDM−5 are not stable in bacterial hosts (Figure 1C). The increasing occurrence of IncHI2-type plasmids carrying blaNDM−5 in China might be associated with the co-selection by other antimicrobials as IncHI2 plasmids usually carry various antimicrobial resistance genes.
While our findings indicate a slightly higher prevalence of blaNDM-positive isolates in egg contents (2.08%, 2/96) compared with eggshells (1.39%, 2/144), this study is not without limitations. The exclusion of 48 egg content samples may influence the overall contamination rates. Furthermore, the sample size, hovering around a hundred, does not offer a comprehensive representation of the prevalence of blaNDM in egg samples, highlighting the need for continuous surveillance.
5 Conclusion
In summary, to the best of our knowledge, we report the first case of Enterobacterales carrying blaNDM−5 of retail eggs in China. The blaNDM−5-bearing plasmids displayed high homology with those of plasmids from other sources. Of note, the IncHI2 plasmids carrying both carbapenem and multiple resistance genes showed an increasing trend that pose another threat to human health. Considering the clinical importance of carbapenem together with the fact that the consumption of eggs is substantial in our diet, the carbapenem resistance in eggs has the risk to spread to humans through the food chain or contact with the contaminant. Continued monitoring of carbapenem resistance in eggs is urgently needed.
Data availability statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/Supplementary material.
Author contributions
Y-YL: Investigation, Writing—original draft, Writing—review & editing. TL: Formal analysis, Investigation, Writing—original draft. HY: Investigation, Writing—original draft. CY: Investigation, Writing—original draft. LL: Formal analysis, Writing—original draft. JC: Formal analysis, Writing—original draft. HD: Formal analysis, Writing—original draft. XG: Formal analysis, Writing—original draft. J-HL: Conceptualization, Funding acquisition, Writing—review & editing.
Funding
The author(s) declare financial support was received for the research, authorship, and/or publication of this article. This study was supported by the National Key Research and Development Program of China (No. 2022YFC2303900), the National Natural Science Foundation of China (grants no. 32141002), and the Local Innovative and Research Teams Project of Guangdong Pearl River Talents Program (2019BT02N054).
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher's note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2023.1281838/full#supplementary-material
References
Benameur, Q., Tali-Maamar, H., Assaous, F., Guettou, B., Tahrat, N., and Aggoune, N., et al. (2018). Isolation of Escherichia coli carrying the bla(CTX-M-1) and qnrS1 genes from reproductive organs of broiler breeders and internal contents of hatching eggs. J. Vet. Med. Sci. 80, 1540–1543. doi: 10.1292/jvms.18-0283
Cao, Y. P., Lin, Q. Q., He, W. Y., Wang, J., Yi, M. Y., and Lv, L. C., et al. (2020). Co-selection may explain the unexpectedly high prevalence of plasmid-mediated colistin resistance gene mcr-1 in a chinese broiler farm. Zool. Res. 41, 569–575. doi: 10.24272/j.issn.2095-8137.2020.131
Carattoli, A., Villa, L., Fortini, D., and Garcia-Fernandez, A. (2021). Contemporary IncI1 plasmids involved in the transmission and spread of antimicrobial resistance in Enterobacteriaceae. Plasmid 118, 102392. doi: 10.1016/j.plasmid.2018.12.001
Chong, Y., Shimoda, S., and Shimono, N. (2018). Current epidemiology, genetic evolution and clinical impact of extended-spectrum beta-lactamase-producing Escherichia coli and Klebsiella pneumoniae. Infect. Genet. Evol. 61, 185–188. doi: 10.1016/j.meegid.2018.04.005
Dong, H., Li, Y., Cheng, J., Xia, Z., Liu, W., and Yan, T., et al. (2022). Genomic epidemiology insights on ndm-producing pathogens revealed the pivotal role of plasmids on bla(NDM) transmission. Microbiol. Spectr. 10, e215621. doi: 10.1128/spectrum.02156-21
El-Gamal, M. I., Brahim, I., Hisham, N., Aladdin, R., Mohammed, H., and Bahaaeldin, A. (2017). Recent updates of carbapenem antibiotics. Eur. J. Med. Chem. 131, 185–195. doi: 10.1016/j.ejmech.2017.03.022
He, W., Gao, M., Lv, L., Wang, J., Cai, Z., and Bai, Y., et al. (2023). Persistence and molecular epidemiology of bla(NDM)-positive gram-negative bacteria in three broiler farms: a longitudinal study (2015-2021). J. Hazard. Mater. 446, 130725. doi: 10.1016/j.jhazmat.2023.130725
Hornsey, M., Phee, L., and Wareham, D. W. (2011). A novel variant, NDM-5, of the new delhi metallo-beta-lactamase in a multidrug-resistant Escherichia coli ST648 isolate recovered from a patient in the United Kingdom. Antimicrob. Agents Chemother. 55, 5952–5954. doi: 10.1128/AAC.05108-11
Huang, E., Yang, X., Leighton, E., and Li, X. (2023). Carbapenem resistance in the food supply chain. J. Food Prot. 86, 100108. doi: 10.1016/j.jfp.2023.100108
Iovleva, A., and Doi, Y. (2017). Carbapenem-resistant Enterobacteriaceae. Clin. Lab. Med. 37, 303–315. doi: 10.1016/j.cll.2017.01.005
Jean, S. S., Harnod, D., and Hsueh, P. R. (2022). Global threat of carbapenem-resistant gram-negative bacteria. Front. Cell. Infect. Microbiol. 12, 823684. doi: 10.3389/fcimb.2022.823684
Kanaan, M., Khalil, Z. K., Khashan, H. T., and Ghasemian, A. (2022). Occurrence of virulence factors and carbapenemase genes in salmonella enterica serovar enteritidis isolated from chicken meat and egg samples in Iraq. BMC Microbiol. 22, 279. doi: 10.1186/s12866-022-02696-7
Kapena, M. S., Muma, J. B., Mubita, C. M., and Munyeme, M. (2020). Antimicrobial resistance of Escherichia coli and salmonella in raw retail table eggs in Lusaka, Zambia. Vet. World 13, 2528–2533. doi: 10.14202/vetworld.2020.2528-2533
Li, C., Gu, X., Zhang, L., Liu, Y., Li, Y., and Zou, M., et al. (2022). The occurrence and genomic characteristics of mcr-1-harboring salmonella from retail meats and eggs in qingdao, China. Foods 11, 1–12. doi: 10.3390/foods11233854
Liu, Y., and Liu, J. H. (2018). Monitoring colistin resistance in food animals, an urgent threat. Expert Rev. Anti Infect. Ther. 16, 443–446. doi: 10.1080/14787210.2018.1481749
Liu, Y. Y., Chen, S., Burrus, V., and Liu, J. H. (2021). Editorial: globally or regionally spread of epidemic plasmids carrying clinically important resistance genes: epidemiology, molecular mechanism, and drivers. Front. Microbiol. 12, 822802. doi: 10.3389/fmicb.2021.822802
Lv, L. C., Lu, Y. Y., Gao, X., He, W. Y., Gao, M. Y., and Mo, K. B., et al. (2022). Characterization of ndm-5-producing Enterobacteriaceae isolates from retail grass carp (ctenopharyngodon idella) and evidence of bla(ndm-5)-bearing IncHI2 plasmid transfer between ducks and fish. Zool. Res. 43, 255–264. doi: 10.24272/j.issn.2095-8137.2021.426
Ma, J., Song, X., Li, M., Yu, Z., Cheng, W., and Yu, Z., et al. (2023). Global spread of carbapenem-resistant Enterobacteriaceae: epidemiological features, resistance mechanisms, detection and therapy. Microbiol. Res. 266, 127249. doi: 10.1016/j.micres.2022.127249
Ma, T., Fu, J., Xie, N., Ma, S., Lei, L., and Zhai, W., et al. (2020). Fitness cost of bla(NDM-5)-carrying p3r-IncX3 plasmids in wild-type ndm-free Enterobacteriaceae. Microorganisms 8, 1–11. doi: 10.3390/microorganisms8030377
Ma, Z., Zeng, Z., Liu, J., Liu, C., Pan, Y., and Zhang, Y., et al. (2021). Emergence of IncHI2 plasmid-harboring blaNDM-5 from porcine Escherichia coli isolates in guangdong, China. Pathogens 10, 1–7. doi: 10.3390/pathogens10080954
Mmatli, M., Mbelle, N. M., and Osei, S. J. (2022). Global epidemiology, genetic environment, risk factors and therapeutic prospects of mcr genes: a current and emerging update. Front. Cell. Infect. Microbiol. 12, 941358. doi: 10.3389/fcimb.2022.941358
Nordmann, P., and Poirel, L. (2019). Epidemiology and diagnostics of carbapenem resistance in gram-negative bacteria. Clin. Infect. Dis. 69, S521–S528. doi: 10.1093/cid/ciz824
Saravanan, B. S., Kumar, D. J. M., and Venkataramaniah, C. (2023). A review on molecular description of carbapenem resistant gram-negative Bacilli. Clin. Med. 10.
Seemann, T. (2014). Prokka: rapid prokaryotic genome annotation. Bioinformatics 30, 2068–2069. doi: 10.1093/bioinformatics/btu153
Shen, Y., Hu, F., Wang, Y., Yin, D., Yang, L., and Chen, Y., et al. (2022). Transmission of carbapenem resistance between human and animal NDM-positive Escherichia coli Strains. Engineering 15, 24–33. doi: 10.1016/j.eng.2021.07.030
Sullivan, M. J., Petty, N. K., and Beatson, S. A. (2011). Easyfig: a genome comparison visualizer. Bioinformatics 27, 1009–1010. doi: 10.1093/bioinformatics/btr039
Wang, J., Tang, B., Lin, R., Zheng, X., Ma, J., and Xiong, X., et al. (2022). Emergence of mcr-1- and bla(NDM-5)-harbouring IncHI2 plasmids in Escherichia coli strains isolated from meat in zhejiang, China. J. Glob. Antimicrob. Resist. 30, 103–106. doi: 10.1016/j.jgar.2022.06.002
Webb, H. E., Granier, S. A., Marault, M., Millemann, Y., den Bakker, H. C., and Nightingale, K. K., et al. (2016). Dissemination of the mcr-1 colistin resistance gene. Lancet Infect. Dis. 16, 144–145. doi: 10.1016/S1473-3099(15)00538-1
Wick, R. R., Judd, L. M., Gorrie, C. L., and Holt, K. E. (2017). Unicycler: resolving bacterial genome assemblies from short and long sequencing reads. PLoS Comput. Biol. 13, e1005595. doi: 10.1371/journal.pcbi.1005595
Wu, R., Yi, L. X., Yu, L. F., Wang, J., Liu, Y., and Chen, X., et al. (2018). Fitness advantage of mcr-1-bearing IncI2 and IncX4 plasmids in vitro. Front. Microbiol. 9, 331. doi: 10.3389/fmicb.2018.00331
Wu, W., Feng, Y., Tang, G., Qiao, F., McNally, A., and Zong, Z. (2019). Ndm metallo-beta-lactamases and their bacterial producers in health care settings. Clin. Microbiol. Rev. 32, 2–30. doi: 10.1128/CMR.00115-18
Yang, X., Liu, W., Liu, Y., Wang, J., Lv, L., and Chen, X., et al. (2014). F33: A-: B-, IncHI2/ST3, and IncI1/ST71 plasmids drive the dissemination of fosA3 and bla CTX-M-55/-14/-65 in Escherichia coli from chickens in China. Front. Microbiol. 5, 688. doi: 10.3389/fmicb.2014.00688
Zhai, R., Fu, B., Shi, X., Sun, C., Liu, Z., and Wang, S., et al. (2020). Contaminated in-house environment contributes to the persistence and transmission of ndm-producing bacteria in a Chinese poultry farm. Environ. Int. 139, 105715. doi: 10.1016/j.envint.2020.105715
Zhang, Q., Lv, L., Huang, X., Huang, Y., Zhuang, Z., and Lu, J., et al. (2019). Rapid increase in carbapenemase-producing Enterobacteriaceae in retail meat driven by the spread of the bla(NDM-5)-carrying IncX3 plasmid in China from 2016 to 2018. Antimicrob. Agents Chemother. 63, 1–5. doi: 10.1128/AAC.00573-19
Zhang, X., Chen, L., Zhang, X., Wang, Q., Quan, J. and He, J., et al. (2022a). Emergence of coexistence of a novel bla(NDM-5)-harbouring IncI1-i plasmid and an mcr-1.1-harbouring IncHI2 plasmid in a clinical Escherichia coli isolate in China. J. Infect. Public Health 15, 1363–1369. doi: 10.1016/j.jiph.2022.10.020
Zhang, X., Ma, M., Cheng, Y., Huang, Y., Tan, Y., and Yang, Y., et al. (2022b). Spread and molecular characteristics of Enterobacteriaceae carrying fosA-like genes from farms in China. Microbiol. Spectr. 10, e54522. doi: 10.1128/spectrum.00545-22
Zhang, Y., Liu, K., Zhang, Z., Tian, S., Liu, M., and Li, X., et al. (2021). A severe gastroenteritis outbreak of Salmonella enterica serovar enteritidis linked to contaminated egg fried rice, China, 2021. Front. Microbiol. 12, 779749. doi: 10.3389/fmicb.2021.779749
Zhao, Q., Berglund, B., Zou, H., Zhou, Z., Xia, H., and Zhao, L., et al. (2021). Dissemination of bla(NDM-5) via IncX3 plasmids in carbapenem-resistant Enterobacteriaceae among humans and in the environment in an intensive vegetable cultivation area in eastern China. Environ. Pollut. 273, 116370. doi: 10.1016/j.envpol.2020.116370
Keywords: resistance, food, carbapenemase, egg, plasmid
Citation: Liu Y-Y, Li T, Yue H, Yue C, Lu L, Chen J, Deng H, Gao X and Liu J-H (2023) Occurrence and characterization of NDM-5-producing Escherichia coli from retail eggs. Front. Microbiol. 14:1281838. doi: 10.3389/fmicb.2023.1281838
Received: 23 August 2023; Accepted: 24 October 2023;
Published: 23 November 2023.
Edited by:
Yujie Hu, China National Center for Food Safety Risk Assessment, ChinaReviewed by:
Fengqin Li, China National Center for Food Safety Risk Assessment, ChinaThomas Gronthal, Finnish Food Safety Authority Evira, Finland
Copyright © 2023 Liu, Li, Yue, Yue, Lu, Chen, Deng, Gao and Liu. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Jian-Hua Liu, amhsaXUyMUAxNjMuY29t
†These authors have contributed equally to this work
‡ORCID: Yi-Yun Liu orcid.org/0000-0002-1269-2441
 Yi-Yun Liu
Yi-Yun Liu Tong Li1†
Tong Li1† Jian-Hua Liu
Jian-Hua Liu