
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
REVIEW article
Front. Microbiol., 30 November 2023
Sec. Infectious Agents and Disease
Volume 14 - 2023 | https://doi.org/10.3389/fmicb.2023.1279159
 C. Lalbiaktluangi1
C. Lalbiaktluangi1 Mukesh Kumar Yadav1*
Mukesh Kumar Yadav1* Prashant Kumar Singh2
Prashant Kumar Singh2 Amit Singh1
Amit Singh1 Mahalaxmi Iyer3
Mahalaxmi Iyer3 Balachandar Vellingiri3
Balachandar Vellingiri3 Ruth Zomuansangi1
Ruth Zomuansangi1 Zothanpuia2
Zothanpuia2 Heera Ram4
Heera Ram4Respiratory tract infections remain the leading cause of morbidity and mortality worldwide. The burden is further increased by polymicrobial infection or viral and bacterial co-infection, often exacerbating the existing condition. Way back in 1918, high morbidity due to secondary pneumonia caused by bacterial infection was known, and a similar phenomenon was observed during the recent COVID-19 pandemic in which secondary bacterial infection worsens the Severe Acute Respiratory Syndrome Coronavirus 2 (SARS-CoV-2) condition. It has been observed that viruses paved the way for subsequent bacterial infection; similarly, bacteria have also been found to aid in viral infection. Viruses elevate bacterial infection by impairing the host’s immune response, disrupting epithelial barrier integrity, expression of surface receptors and adhesion proteins, direct binding of virus to bacteria, altering nutritional immunity, and effecting the bacterial biofilm. Similarly, the bacteria enhance viral infection by altering the host’s immune response, up-regulation of adhesion proteins, and activation of viral proteins. During co-infection, respiratory bacterial and viral pathogens were found to adapt and co-exist in the airways of their survival and to benefit from each other, i.e., there is a cooperative existence between the two. This review comprehensively reviews the mechanisms involved in the synergistic/cooperativity relationship between viruses and bacteria and their interaction in clinically relevant respiratory infections.
Respiratory tract infections are among the most prevalent human infections and are a major health and economic burden worldwide. They include upper respiratory tract infections (URTI) like pharyngitis, nasopharyngitis, otitis media, and tonsillitis and lower respiratory tract infections (LRTI) like pneumonia, tuberculosis, bronchitis, and bronchiolitis (Dasaraju and Liu, 1996). World Health Organization (WHO) estimated that 4 million people die each year due to acute respiratory infections (ARI), out of which the majority are infants, children, and geriatric patients (World Health Organization, 2020). In developed countries, it was estimated that respiratory infections account for 60% of antibiotic prescriptions (Lindbaek, 2006) and more than 30% of sick leave. The burden is much higher in developing countries where respiratory infections accounted for the majority of deaths in children below the age of 5 years (Bryce et al., 2005). Both bacteria and viruses have been found to cause respiratory infections in humans. Streptococcus pneumoniae, Staphylococcus aureus, Haemophilus influenzae, Moraxella catarrhalis, Klebsiella pneumoniae, and Pseudomonas aeruginosa are the most common bacterial pathogens detected in respiratory tract infections (Cappelletty, 1998; Burillo and Bouza, 2010; Gadsby et al., 2015). Viral pathogens like Influenza A (IAV) and B, Respiratory Syncytial Virus (RSV), Adenovirus, Parainfluenza viruses, and Human Bocavirus account for 30–40% of respiratory infections (Huang et al., 2013; Wen et al., 2019).
Recent technological advances have detected the presence of both viruses and bacteria in different respiratory diseases such as pneumonia, otitis media (OM), cystic fibrosis (CF), Coronavirus – 19 (COVID-19), etc. Research has suggested that high morbidity during the 1918 pandemic may be due to secondary pneumonia caused by bacterial infection (Morens et al., 2008). A similar phenomenon was observed during the recent COVID-19 pandemic in which secondary bacterial infection worsens the SARS-CoV-2 condition (Lansbury et al., 2020). De Bruyn et al. (2022) found that 68% of the COVID-19 patients admitted to the ICU developed secondary bacterial infections like pneumonia, bacteremia, etc. In a study conducted in Saudi Arabia, higher mortality rates were observed in COVID-19 patients co-infected with bacteria (50%) as compared to the SARS-CoV-2- only infected group (18.7%) (Alqahtani et al., 2022). Different studies have shown that there is a symbiotic interaction between viruses and bacteria to establish an infection. Bacteria have been found to aid viral infection by altering the host’s immune response (Bellinghausen et al., 2016), upregulating the surface adhesion proteins (Gulraiz et al., 2015), by activation of viral proteins (Tashiro et al., 1987a,b), etc. Similarly, viruses have been found to aid bacteria in establishing secondary infection through several mechanisms, including impairment of the host’s immune response (Ghoneim et al., 2013), disruption of epithelial barrier integrity (Short et al., 2016), expression of surface receptors and adhesion proteins (Avadhanula et al., 2006), direct binding of virus to bacteria (Rowe et al., 2019), alteration of nutritional immunity (Siegel et al., 2014), etc. For better management of infectious diseases, it is essential to understand the underlying mechanism by which bacteria and viruses interact and aggregate, infect, or establish infection. This review provides a comprehensive look into the mechanisms involved in viral and bacterial co-infections and how this interaction played out in different respiratory infections in human.
The review was conducted through searches of the PubMed database using different search terms. The search term “viral bacterial co-infection” yielded 3,289 results; the results were filtered to focus on co-infection in the respiratory tract and to eliminate co-infections in other systems like urinary tract infections and intestinal and gut infections. For the keyword “viral, bacterial co-infection,” the number of published articles increased significantly from 84 papers in 2010 to 315 papers in 2022. Other specific keywords were employed to study the co-infection patterns in different diseases. In total, 147 papers were reviewed in this study.
Viruses and bacteria can interact directly or indirectly in human infection, and interaction between these two could be either positive or negative (Almand et al., 2017). Mutualism, symbiosis, or aiding in the evasion of the host’s immune system are all examples of positive associations. At the same time, negative interactions occur through ammensalism, predation, or when the host immune system affects one organism over the other. The processes driving the synergy between virus and bacteria are complicated and multifaceted, with bacterial, viral, and host immunological components contributing to increased predisposition of infection. Many bacteria and viruses interact with each other and establish successful respiratory infections (Table 1). The respiratory viruses and bacterial interactions predispose bacterial superinfections, resulting in severe illness, and these interactions can influence microbial pathogenesis, including increased bacterial adhesion, enhanced virion stability, and modulation of the immune response by one microbe that benefits the other (Håkansson et al., 1994; El Ahmer et al., 1999). Similarly, bacterial infection has also been found to modulate viral infections exacerbating the disease (Verkaik et al., 2011; Smith et al., 2013; Nguyen et al., 2015).
It has been established that prior viral infection augmented bacterial adherence to host cells. For example, it has been observed that in children with acute otitis media, prior infection by adenovirus type 1, 2, 3, and 5 significantly enhances pneumococcal colonization (Håkansson et al., 1994). Another study by El Ahmer et al. (1999) also found that influenza A infection of Hep-2 cells increased the adherence of S. pneumonia, H. influenza, and M. catarrhalis to virus-infected Hep-2 cells. Virus-induced bacterial adherence is achieved through several mechanisms, which are briefly discussed in this section. The mechanisms are represented and summarized in Figure 1 and Table 1, respectively.
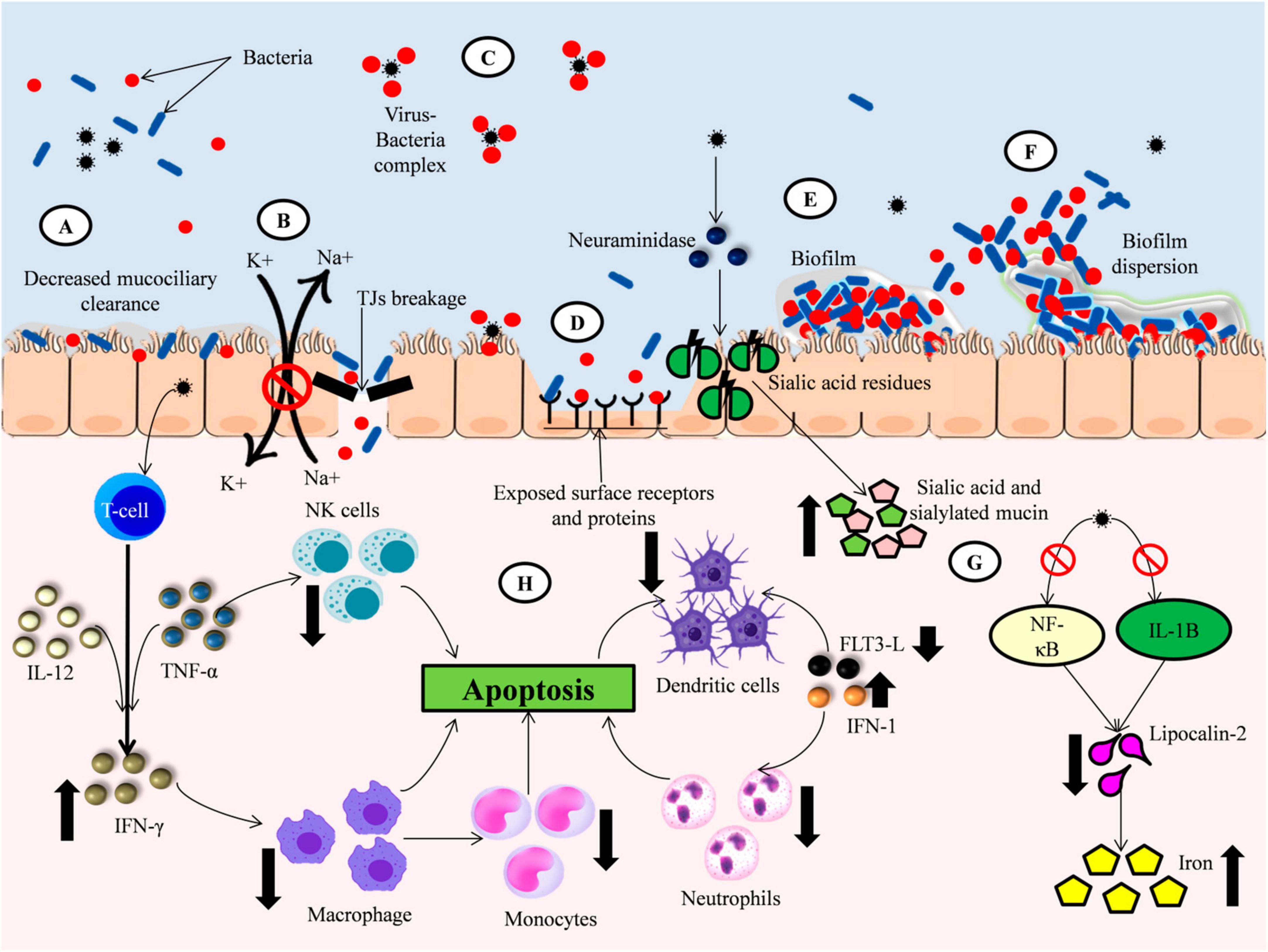
Figure 1. Elevation of bacterial colonization by viral infection. (A) Virus infection decrease the mucociliary clearance action resulting in increased bacterial colonization. (B) Viral infection impaired the epithelial barrier integrity by breaking Tight Junctions (TJs) and inhibiting Na+/K + ATPase pump promoting bacterial entry to the host cells. (C) Viruses directly binds to bacterial surface proteins forming a viral-bacterial complex that can infect uninfected cells as well as infected cells. (D) Viral proteins like Neuraminidase (NA) cleaves the sialic acid residues on the epithelial cell membrane exposing surface receptors and adhesion proteins aiding bacterial colonization. (E) Viral infection promotes bacterial biofilm formation and growth by providing increased nutrients. (F) Viruses induced changes in host environment promotes dispersion of bacterial biofilm leading to increased translocation of more virulent bacteria to other sites. (G) Viral infection induce change in nutritional immunity of the host resulting in elevated levels of nutrients which are utilized by bacteria for their growth. (H) Viral infection altered the host’s immune response to bacteria by decreasing the production and function of important immune cells like macrophages, NK cells, Dendritic cells and Neutrophils.
(i) Impairment of the host’s immune response:
Respiratory viruses can disrupt the host’s immune system through the malfunction and depletion of innate immune cells. One of the most well-studied cases is the influenza virus-induced dysfunction and depletion of alveolar macrophages, promoting subsequent bacterial infection (Ghoneim et al., 2013; Short et al., 2016). RSV infection has been found to cause neutrophil dysfunction by lowering oxidative burst and promoting apoptosis of neutrophils, elevating the adherence of S. aureus or P. aeruginosa (Stark et al., 2006). Influenza infection also modifies monocyte functions, inhibiting proper immune response and reducing cytokine activity and synthesis, resulting in increased bacterial colonization and mortality risk (Abramson et al., 1982). Another innate immune cell affected by viral infection is Natural Killer (NK) cells, which are the critical components of the innate immune response. Small et al. (2010) found that influenza infection increased susceptibility to S. aureus by reducing TNF-α induced NK cell production and impairing the activation of NK cells. Viral infection also affects the adaptive immune response like T CD8 + and T CD4 + cell and antibody responses. RSV has been found to facilitate reinfection by reducing immune memory. This is achieved by impairing the interaction between dendritic cells and T-cells, reducing activation of naïve antigen-specific T-cells (González et al., 2008). In influenza-infected mice, a decrease in the production of FMS-like tyrosine kinase 3 ligand (FLT3-L), a dendritic cell differentiation factor was observed, resulting in decreased dendritic cell population in the lungs contributing to secondary bacterial infection (Beshara et al., 2018).
Depletion and dysfunction of innate and adaptive immune cells significantly affect the production of cytokines and chemokines. Influenza infection has been found to decrease the production of pro-inflammatory cytokines like IL-1β, IL-6, and TNF-α, decreasing bacterial clearance. Influenza infection has also been found to affect the adaptive Type 17 response. Nakamura et al. (2011) found that influenza inhibited the production of chemokine CCL2 responsible for bacterial clearance through the enhanced expression of type I IFN, thus promoting pneumococcal colonization. Robinson et al. (2013) also found that Influenza infection exacerbates S. aureus infection by decreasing IL-1β production, which attenuates Type 17 immunity. Lastly, Kudva et al. (2011) found that in mice infected with influenza virus and Staphylococcus aureus, a significant decrease in production of IL-17 and IL-22 cytokines was observed. Interestingly, cytokine release during the acute inflammation stage has been found to be capable of enhancing the growth of bacteria and prolonging bacterial infection (dos Santos et al., 2002). Kanangat et al. (1999) and Meduri et al. (1999) found that high concentrations of cytokines like IL-1b, IL-6, and TNF-α enhance the in vitro growth of bacteria, including S. aureus, P. aeruginosa, and Actinobacteria sp.
(ii) Disruption of epithelial barrier integrity
Viral infection has been found to cause extensive damage to the human epithelial and endothelial cells. Epithelial barrier damage may occur due to direct cleavage of tight junctions. Downregulation of junction proteins through the host’s pro-inflammatory response against the virus results in impaired barrier integrity, increased membrane permeability, and decreased trans-barrier electrical resistance (Rossi et al., 2020). Tight Junctions (TJ) act as a sealant between neighboring cells and maintain the integrity of the airway epithelial cells (Steed et al., 2010). Viral infections induce the breakage of tight junctions (TJs), which increases the permeability of molecules in the airway epithelial cells, facilitating the entry and transmigration of bacteria in the airway (Short et al., 2016). ZO-1 protein in TJ established a link between the transmembrane protein occludin and the actin cytoskeleton and plays an essential role in maintaining the structural integrity of TJs (Itoh et al., 1997). When infected by Rhinovirus, loss of ZO-1 protein was observed, which facilitates the entry of Non-typeable Haemophilus influenzae (NTHi) (Sajjan et al., 2008). Infection by human bocavirus has also been found to reduce transbarrier electrical resistance via the decreased expression of occludin and reduced function of the Na + /K + ATPase pump (Chiu et al., 2014; Balasubramaniam et al., 2015). Viral infection may also cause denudation of the epithelial layer by killing the epithelial cells through direct lysis or metabolic exhaustion (Bosch et al., 2013).
(iii) Expression of surface receptors and adhesion proteins
Viruses have been found to facilitate bacterial adherence by exposing cell surface receptors on the epithelial cells. Influenza A and B exposed the carbohydrate ligands on the epithelial cells by cleaving the terminal sialic acid residues using neuraminidase. The exposed carbohydrate ligands then act as a binding site for S. pneumoniae (McCullers and Bartmess, 2003a; Li et al., 2015). Inflammatory response to influenza, parainfluenza, and RSV infection caused epithelial injury resulting in increased expression of cell surface receptors such as platelet-activating factor receptor (PAFr), intercellular adhesion molecule-1 (ICAM-1), which serves as bacterial receptors (Ishizuka et al., 2003; McCullers and Bartmess, 2003b; Avadhanula et al., 2006). Upregulation of other surface receptors, such as carcinoembryonic adhesion molecule-1 (CEACAM-1), and P5-homologous fimbriae (P5 fimbriae) were also observed (Jiang et al., 1999). Studies showed that pathogenic bacteria like S. pneumoniae, S. aureus, and M.catarhallis could strongly bind to fibronectin and α5 integrin, which were exposed after viral-induced cell injury (Sajjan et al., 2006; Wang et al., 2009). Viruses also induce bacterial adherence through the increased expression of viral surface proteins, which are known receptors for bacteria.
(iv) Direct binding of the virus to bacteria
The role played by viral glycoproteins and surface receptors on enhancing bacterial adherence is well established. This can explain the increased adherence to virus-infected cells but doesn’t fully explain the increased bacterial adherence to uninfected cells. An alternative mechanism has been introduced where the virus directly binds to the incoming bacteria, thus explaining the bacterial adherence not only to infected cells but also to uninfected cells. The hypothesis is that virus binding to bacteria acts as a coupling agent between the uninfected epithelial cells and bacteria, leading to increased bacterial adherence. Hament et al. (2005) found that G-protein produced by RSV facilitates the binding to S. pneumoniae, after which the pneumococcal adherence to uninfected epithelial cells is significantly increased. Smith et al. (2014) found that the G-protein of RSV binds to penicillin-binding protein 1a of S. pneumoniae, increasing the virulence of S. pneumoniae. Viruses like influenza and parainfluenza viruses recognize the capsular polysaccharides expressed by Group B streptococci (GBS) and bind to the bacteria, enhancing bacterial adherence (Tong et al., 2018a,b). Okamoto et al. (2004) found that the influenza virus can bind to encapsulated Group A Streptococci (GAS), facilitating bacterial adherence to virus-infected alveolar epithelial cells. Similarly, Rowe et al. (2019) studied the direct interaction between the influenza virus and Gram-positive bacteria like S. pneumoniae and S. aureus, as well as Gram-negative bacteria, M. catarrhalis, and non-typeable Haemophilus. They incubated the influenza virus with bacteria, and increased bacterial adherence was observed even after the removal of unbound viruses. The same phenomenon was also observed in in vivo, where higher bacterial adherence was observed in murine tissues infected with bacteria pre-incubated with viruses compared to normal bacteria (Rowe et al., 2019).
(v) Increased availability of nutrients
In healthy cells, the minerals, growth factors, and other nutrients needed by the bacteria are sequestered by the host to prevent pathogenicity in a phenomenon called nutritional immunity (Hennigar and McClung, 2016). This phenomenon is altered when viruses infect the host cells, promoting further infection by bacteria. Influenza virus infection has been found to reduce the production of an iron sequester, Lipocalin -2, by suppressing the activation of NF-kB and expression of IL-1B. This increased iron availability in the host, exacerbating S. aureus acute pneumonia in mice (Robinson et al., 2013). Influenza virus infection also increases the availability of sialic acid and sialylated mucin in the host, stimulating pneumococcal growth in the airway (Siegel et al., 2014). RSV infection has also been found to increase iron concentration in the host, promoting P. aeruginosa biofilm formation (Hendricks et al., 2016). In another study, Hendricks et al. (2016) also found that the extracellular vesicles released from the virally infected cells act as a transferrin transport vehicle between cells, enhancing the growth of P. aeruginosa biofilm. The concentration of other nutrient sources like mucins and surfactant proteins in the host was also altered during virus co-infection (Kiedrowski and Bomberger, 2018).
(vi) Effect on bacterial biofilm
It has been observed that viruses induce the transition of bacterial growth to a biofilm mode by altering nutrient immunity (Hendricks et al., 2016; Kiedrowski et al., 2018). Using a co-culture model, Hendricks et al. (2016) found that RSV infection-induced IFN signaling stimulates the release of iron-bound transferrin protein, which promotes thriving biofilm formation by P. aeruginosa. Similarly, Kiedrowski et al. (2018) found that RSV infection enhanced S. aureus biofilm formation due to the increased release of nutrients by the host. They also found that S. aureus genes involved in protein translation and transport, growth, and amino acid metabolism were upregulated during co-infection.
Biofilm is the reservoir of the bacteria, and the dispersion plays a vital role in the pathogenesis and progression of a disease. Viral infections have been found to influence bacterial biofilm dispersion and its transition from asymptomatic colonization to invasive disease. In Cystic Fibrosis patients, Rhinovirus induces the release of H2O2, which facilitates the dispersion of P. aeruginosa biofilms (Chattoraj et al., 2011a). Reddinger et al. (2016) found that S. aureus biofilms on the upper respiratory tract dispersed due to changes in the host induced by influenza infection. The biofilm dispersion induces the translocation of cells from the nasal tissue to the lungs in a mice model, elevating the progression of pneumonia. Marks et al. (2013) observed the same phenomenon in which influenza A infection induced the dispersion of S. pneumoniae biofilm. The dispersed cells were found to have different phenotypes with upregulation of different virulence genes, exacerbating disease progression (Marks et al., 2013; Pettigrew et al., 2014).
Multiple studies have been carried out to study the role of bacteria on viral pathogenesis with varying results. Some studies found that bacterial infection negatively impacts viral survival and growth (Wolf et al., 2014); however, most studies showed a positive correlation between bacteria and viruses (Blevins et al., 2014; Wu et al., 2015). Verkaik et al. (2011) found that prior infection of human bronchial epithelial cells by S. pneumoniae renders the cells more susceptible to hMPV infections. Nguyen et al. (2015) carried out in vitro and in vivo studies to observe the effect of S. pneumoniae on RSV infection and found that S. pneumoniae enhances RSV infection in both in vitro and in vivo studies. Similarly, Smith et al. (2013) also observed increased Influenza load in the presence of S. pneumoniae, which suggests that bacteria aids in the release of Influenza virus from airway cells as well as the release of virions. Bacteria-mediated modulation of viral infections is achieved mainly through the bacterial influence on the host and the host’s immune system, as well as its direct effect on the virus itself (Figure 2 and Table 2), which are briefly discussed below:
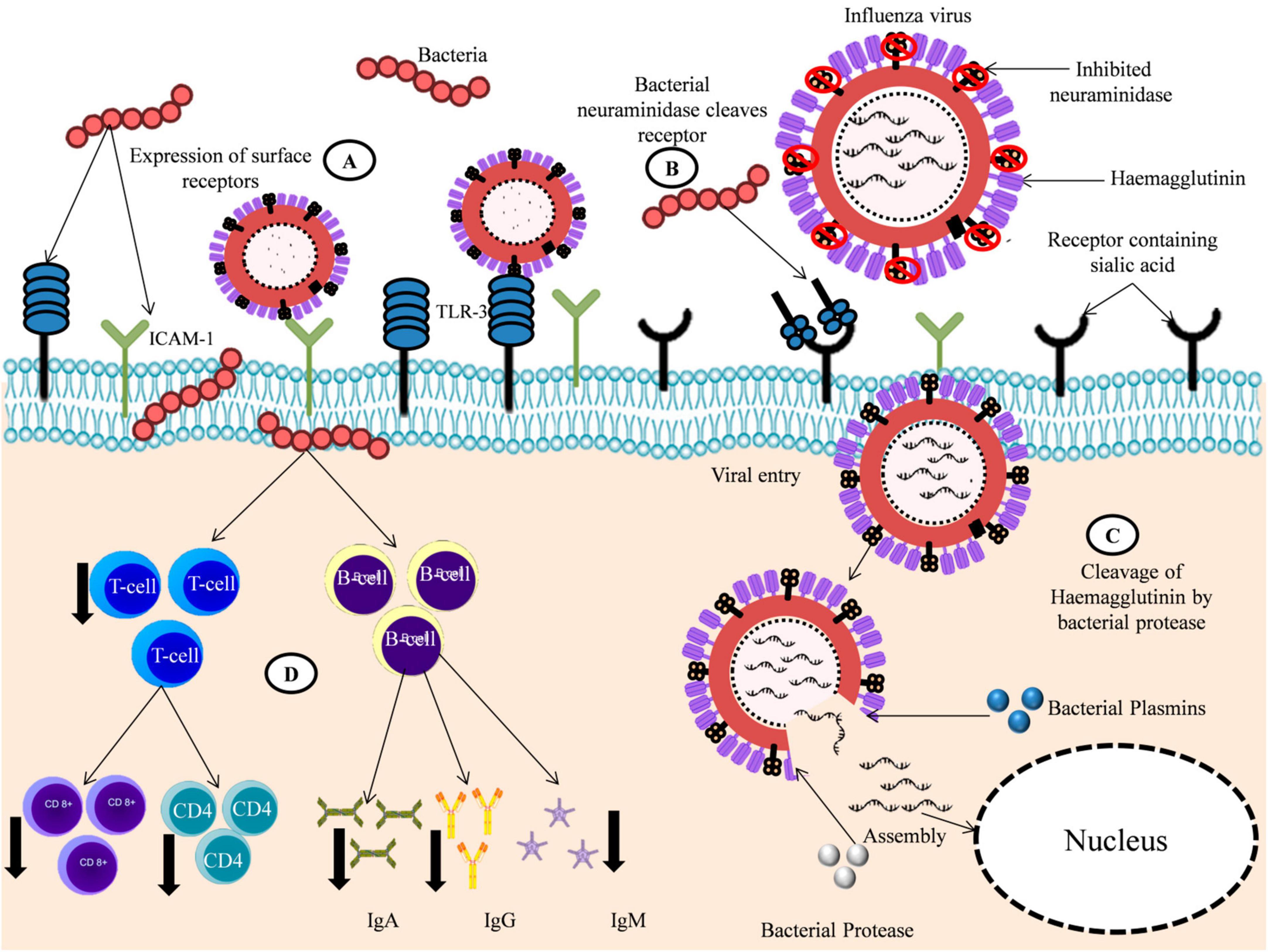
Figure 2. Bacteria aids in viral infection. (A) Bacterial infection increased the expression of surface proteins like ICAM-1 and TLR-3 aiding the attachment of viruses to host cells. (B) Bacterial Neuraminidase (NA) aids in the viral entry to host cells by cleaving sialic acid receptors when viral Neuraminidase (NA) are inhibited. (C) Bacterial proteases and plasmins aids in the release of viral RNA by cleaving the viral protein Haemaglutinin (HA). (D) Bacteria alters the immune response to virus by decreasing the production of viral-specific immune cells CD8+, CD4+, and antibodies like IgG, IgA, and IgM.
(i) Alteration of the host’s immune response
Bacterial co-infection has been found to significantly influence a host’s immune response against viral infection. In most cases, these bacteria-modulated immune changes support viral infection, but some studies found adverse effects on viruses. Wolf et al. (2014) found that co-infection with S. pneumoniae elevates the B-cell response to the Influenza virus. Bellinghausen et al. (2016) also observed that although bacteria altered the airway immune response, it does not necessarily affect the subsequent viral infection. However, the contrasting results were reported by Blevins et al. (2014), which showed that prior infection by S. pneumoniae markedly decreased the lung’s anti-viral CD8 + T cell population, resulting in increased survival of the Influenza A virus and increased mortality. Using a murine mouse model, Wu et al. (2015) also found that secondary infection by S. pneumoniae significantly increased viral titers and decreased the concentration of IgA, IgM, and IgG, which are viral specific antibodies. They also detected a decrease in the population of immune cells, such as CD4 T cells, B cells, and plasma cells, facilitating viral survival in the host.
(ii) Up-regulation of adhesion proteins
Bacteria are also involved in up-regulation of the surface protein through altered immune response. Sajjan et al. (2006) found that H. influenzae infection increased the expression of intracellular adhesion molecules like ICAM-1 and TLR3, leading to increased infection by RV and subsequent RV-induced increased cytokine release (Sajjan et al., 2006). Similar results were obtained by Gulraiz et al. (2015), who found that prior non-typable H. influenzae infection dramatically increases the binding of HRV16 to two bronchial epithelial cell lines pBECs and BEAS-2B, most probably through the upregulation of ICAM-1. However, no such increase in binding was detected in HRV1B and RSV infection.
(iii) Activation of viral proteins
Various components of bacteria have been found to aid viral attachment and infection. Proteases produced by S. aureus have been found to increase the replication of various influenza strains in the lungs by cleaving and activating the viral haemagglutinin (HA) (Tashiro et al., 1987a,b). The activation of hemagglutinin was found to be strain-dependent, i.e., proteases produced by one bacterium work only for specific viral strains. Similarly, Scheiblauer et al. (1992) also found that protease secreted by a pathogen Aerococcus viridans could cleave the HA of various influenza A virus isolates, causing severe pneumonia in mice and leading to mortality. Other bacterial proteases that cannot directly cleave viral haemagglutinin (HA) have also been found to increase the viral load. Simultaneous administration of Influenza A virus and non-cleaving proteases secreted by P. aeruginosa has markedly increased the viral load (Scheiblauer et al., 1992). Although the specific proteases that can cleave viral HA from bacteria are yet to be identified, these proteases might provide a novel drug target for treating viral and bacterial co-infection. Plasminogen activators of S. aureus and S. pneumoniae, like staphylokinase and streptokinase, could generate plasmins, an important fibrinolytic protease. These plasminogens could cleave the hemagglutinin of H1N1 Influenza virus that has Ser-Tyr substitution in their cleavage site (Tse et al., 2013). The commensal bacteria of the upper and lower respiratory tract have been found to secrete neuraminidase, which can aid in viral infection. For example, Nishikawa et al. (2012) found that the neuraminidase secreted by S. pneumoniae can facilitate the successful release of virions even after the inhibition of viral neuraminidase. This can potentially hinder the efficacy of virus neuraminidase drugs like oseltamivir and zanamivir used during influenza infection.
Apart from the cleavage of viral proteins, bacterial peptides have been found to enhance viral infection by directly binding to the virus. A synthetic bacterial lipopeptide Pam3CSK4 has been found to enhance the infection of primary epithelial, lymphoid, and myeloid cells by RSV, measles virus, HIV-1, and human metapneumovirus independent of TLR activation. Pam3CSK4 contains two important cationic structures, N-palmitoylated cysteine and cationic SK4 sequence, which may interact directly with the negatively charged structures of the viral envelope or the host cells, enhancing the viral infection (Nguyen et al., 2010).
The use of molecular assays like PCR to assess clinical samples has provided a better picture of the synergy between bacteria and viruses in the pathogenesis of various infectious diseases (Ruuskanen et al., 2011). Co-infection has been observed in various infectious diseases with mostly adverse patient outcomes. Johansson et al. (2011) found that co-infection in CAP often increases the severity of CAP and results in more extended hospitalization. It has been suggested that a majority of deaths during the “Spanish Flu” pandemic may be due to the development of secondary pneumonia (Morens et al., 2008). Similarly, during the 2009 H1N1 pandemic, 29–55% of mortalities were attributed to secondary pneumonia (Center for Disease Control and Prevention [CDC], 2012). This is again observed during the most recent global COVID-19 pandemic, where secondary infection by bacteria worsens the severity of the disease (Langford et al., 2020). Interestingly, many epidemiological studies detected no pathogens in some patients (Huijskens et al., 2013; Luchsinger et al., 2013; Jain et al., 2015; Voiriot et al., 2016). For example, an epidemiological study on CAP adult patients requiring hospitalization in the US detected no pathogens in the majority of the patients. However, they detected co-infection in 38% of the patients (Jain et al., 2015). Another study by Luchsinger et al. (2013) also detected no pathogenic agents in 35% of the patients tested. The viral and bacterial pathogens co-infections detected in various respiratory diseases are summarized in Table 3.
Multiple tests have been used to detect the type of bacterial and viral species involved in various respiratory infectious diseases, including bacterial cultures, PCR, etc. After the first US FDA approval of respiratory syndromic panel in 2011, syndromic panels emerged as a quick and efficient way of detecting bacteria and viruses in co-infection patients. Currently, eight multiplex upper respiratory panels and two multiplex lower respiratory panels have been approved by the FDA for the diagnosis of upper and lower respiratory tract infections. An example of a URT disease syndromic panel is Applied BioCode Respiratory Pathogen Panel (RPP), which can detect three bacteria: Bordetella pertussis, Chlamydia pneumoniae and Mycoplasma pneumoniae and at least five viral species including Influenza A and B, Human metapneumovirus, RSV A and B, Parainfluenza virus 1, 2, 3, and 4, Human Rhinovirus in 4 h. Another syndromic panel, BioFire FilmArray Respiratory Panel EZ (CLIA-waived), can detect three bacterial species and eight viral species, including Coronavirus, in just 1 h. Multiplex lower respiratory panels BioFire FilmArray Pneumonia plus Panel and Curetis Unyvero Lower Respiratory Tract Panel can detect many bacterial and viral pathogens. The bacterial pathogens that these panels can detect include S. aureus, Stenotrophomonas maltophilia, S. pneumoniae, P. aeruginosa. These lower respiratory panels can detect viruses like Influenza A and B, Coronavirus, RSV, Adenovirus, and Parainfluenza virus. In addition to detecting the pathogens, multiplex lower respiratory panels can also be used for the detection of antibiotic resistance genes like Methicillin resistance genes: mecA/mecC and MREJ, Penicillin resistance gene; TEM (Bard and McElvania, 2020; Dumkow et al., 2021). False positive cultures can be a significant problem in clinical settings. Using strict, standardized criteria for sample collection, culture, and immunological procedures is required to determine whether the collected bacterial or viral pathogens are true pathogens, not contaminants or innocent bystanders.
Pneumonia, a severe inflammation of the lungs, is one of the most commonly known respiratory tract infections responsible for the loss of thousands of lives each year (World Health Organization, 2013). It is the leading cause of mortality in children under 5 years old and attributed to total mortality of 2.5 million people in 2019 (Liu et al., 2012). Either viruses or bacteria cause pneumonia and sometimes fungi; however, bacteria is the primary causative agent in most cases. S. pneumonia and RSV are the most common bacterial and viral causes of pneumonia (Musher et al., 2017).
Co-infection by viruses and bacteria is highly prevalent in community-acquired and hospital-acquired pneumonia (CAP). It has been reported that 45% of CAP cases in children are due to viral and bacterial co-infection, while reported cases of mixed infection in CAP cases in adults are lower (Ruuskanen et al., 2011). Cillóniz et al. (2012) studied the incidence of co-infection in CAP patients and detected bacterial co-infection in 33% of the patients.
Most reported cases of viral-bacterial pneumonia involved S. pneumoniae with different viruses. Other bacterial species like S. aureus, P. aeruginosa, Neisseria meningitides, H. influenza, and K. pneumoniae were also detected in secondary infection. A combination of the influenza virus and S. pneumoniae is the most frequent combination of co-infection (Handel et al., 2010; Honkinen et al., 2012). Before the prevalence of COVID-19, the most commonly found viral pathogens in community-acquired pneumonia patients were RSV, metapneumovirus, and influenza virus (Mandell, 2015). However, after and during the COVID-19 pandemic, pneumonia patients were detected with respiratory pathogenic bacteria and viruses, indicating that the interplay of respiratory bacteria and viruses is a major risk in pneumonia (Mirzaei et al., 2020; Ladas et al., 2023; Zhang et al., 2023).
The synergism between Influenza virus and S. pneumoniae in pneumonia involved several mechanisms, including altered immune response, epithelial barrier damage (Nita-Lazar et al., 2015), expression of surface receptors (McCullers and Rehg, 2002), impaired mucociliary clearance, dispersion of biofilm to sterile sites (Marks et al., 2013; Pettigrew et al., 2014), etc. which are explained in detail in Section 3. However, in addition to facilitating pneumococcal colonization/disease, recent studies have found that Influenza virus and other viruses play an important role in the transmission of pneumococci. In animal studies involving mice and ferrets, IAV infection has been found to facilitate intra- and inter-cage transmission of pneumococci (Diavatopoulos et al., 2010; McCullers et al., 2010). In mice, Diavatopoulos et al. (2010) found that pneumococci transmission occurred when all of the mice were infected with IAV, and transmission was prevented when IAV replication was inhibited in contact (uncolonized) or index (those colonized with S. pneumoniae) mice. In ferrets, McCullers et al. (2010) found that contact ferrets have a higher chance of acquiring pneumococcal infection when they are infected with IAV. Short et al. (2012a,b) investigated the factors determining the virus-mediated transmission of S. pneumoniae and found that elevation of the nasopharyngeal bacterial load in the index mouse and induction of inflammation in the contact mice facilitated pneumococcal colonization. Epidemiological studies on the role of viruses in the transmission of pneumococci found polarizing results. While some studies (Kuster et al., 2011) found no relationship between influenza seasonality and pneumococcal transmission, other studies (Grijalva et al., 2014; Karppinen et al., 2017) found that viruses like Influenza, Rhinovirus, and Parainfluenza infection facilitate the acquisition and transmission of Streptococcus pneumoniae.
Otitis media (OM) is a highly prevalent pediatric disease mainly affecting children under five. A prospective cohort study showed that 80% of children experience OM in childhood, one of the most common reasons for a doctor visit (Chonmaitree et al., 2008). It occurs due to impaired fluid drainage from the middle ear chamber, which facilitates the colonization of the middle ear by commensal bacteria residing in the nasopharynx (Vergison et al., 2010).
Concurrent infection by bacteria and viruses was observed in most OM cases (Vesa et al., 2001; Ruohola et al., 2006; Pettigrew et al., 2011). A cohort study on children aged 2–24 months detected with acute OM showed that most children had viral upper respiratory infections before being diagnosed with acute OM (Vesa et al., 2001). Ruohola et al. (2006) conducted a study to determine the microbiology of acute OM in children and detected viral bacterial co-infection in most (66%) patients. OM is a polymicrobial disease caused by a wide variety of viruses and bacteria, including S. pneumonia, H. influenza, and M. catarrhalis, and viruses like Adenovirus, Rhinovirus, and respiratory syncytial viruses (Chonmaitree et al., 2008). Among bacteria, non-typeable H. influenza was the most common among those pathogens (Vergison et al., 2010).
Several mechanisms involved in the cooperative relationship between bacterial pathogens of otitis media and viruses have been identified, including expression of adhesion molecules and receptors in the cells (Nita-Lazar et al., 2015), inflammatory injury, etc. (McNamee and Harmsen, 2006; Spelmink et al., 2016). A mechanism of viral, bacterial synergism unique to otitis media is the virus-induced dysfunction of the Eustachian tubes in the middle ear. Giebink et al. (1987) studied the effect of influenza virus infection on Eustachian tubes. They found that the viral infection induced inflammation of the tympanic membrane and the epithelium of the Eustachian tube. This resulted in an under-pressured middle ear that facilitates the entry of pathogens into the middle ear cavity (Bakaletz, 2017). Virus infections also affect the mucociliary clearance of middle ear fluid, making the middle ear much more susceptible to secondary bacterial infection (Park et al., 1993).
Bronchiolitis is an infection of the lower respiratory tract commonly affecting children below 2 years. It is considered one of the most frequent causes of infant hospitalization and is often associated with high infant morbidity. Respiratory Syncytial Virus (RSV) was the primary cause of Bronchiolitis and was detected in more than 70% of the patients (Smyth and Openshaw, 2006). Other viruses, such as Parainfluenza virus (PIV), Influenza virus, Rhinovirus (RV), and Adenovirus, were also often detected in bronchiolitis patients (Nascimento et al., 2010). Although bronchiolitis is frequently associated with viruses, recent etiological studies also reported the presence of bacterial pathogens in patients (Thorburn et al., 2006; Wiegers et al., 2019). Wiegers et al. (2019) conducted a study on ventilated bronchiolitis patients and detected bacterial co-infection in more than one-third (37.7%) of the patients. They also found that patients with bacterial co-infection require more prolonged mechanical ventilation and PICU stay (Wiegers et al., 2019). Similarly, Thorburn et al. (2006) concluded that patients with bacterial co-infection require more extended hospital stays and detected bacterial co-infection in almost half (40%) of the patients. The most frequently isolated pathogens are H. influenzae, S. pneumoniae, M.catarrhalis, and S. aureus. However, other bacterial pathogens like P. aeruginosa, K. pneumoniae, E. coli, etc., were also isolated from some patients’ samples (Thorburn et al., 2006; Fares et al., 2011; Wiegers et al., 2019).
Co-infection, especially bacterial co-infection, is commonly found in Cystic Fibrosis (CF), a fatal genetic disease caused by Fibrosis Transmembrane Conductance Regulator (CFTR) gene mutations. CF is characterized by a progressive decline of pulmonary function due to the accumulation of thick mucus in the lungs and persistent chronic infection (Nichols et al., 2008; Scott et al., 2020). A wide variety of pathogenic bacteria, viruses, fungi, and sometimes yeast is involved in chronic infection of the lungs (Skov et al., 2005; Pihet et al., 2009; Sudfeld et al., 2010; Sherrard et al., 2014). Among these pathogens, P.aeruginosa is the most common cause of death in CF (Kiedrowski et al., 2018). The most frequently isolated pathogen from pediatric CF patients is S. aureus, while P. aeruginosa is most frequently isolated from adult CF patients (Cystic Fibrosis Foundation, 2014). It has been established that different species of microbes interact in the airways of CF patients, resulting in a pathogenesis different from a single-species infection. S. aureus and P.aeruginosa is the predominant combination found in CF patients. Interspecies as well as intraspecies interaction has been observed in CF patients.
Rhinovirus, RSV, Influenza A and B are the most common viruses detected in CF patients, while S. aureus and P. aeruginosa are the most common bacterial pathogens. RSV-induced elevated bacterial adherence in CF is achieved through the direct binding of RSV to P. aeruginosa via RSV glycoprotein G (Van Ewijk et al., 2007). Other mechanisms of interaction between RSV and P.aeruginosa include viral-induced increased biofilm growth (Hendricks et al., 2016; Kiedrowski et al., 2018), viral-induced biofilm dispersion by production of H2O2 (Chattoraj et al., 2011a), altered immune response, etc.
Conversely, prior P. aeruginosa infection has been found to modulate antiviral response significantly. Chattoraj et al. (2011b) found higher viral load in co-infected cells compared to RV infection alone. Sörensen et al. (2020) found that P. aeruginosa blocks the antiviral IFN signaling via LasR-dependent degradation of IFNλ by protease AprA. Endres et al. (2022) also found that non-mucoid P. aeruginosa significantly controls the innate antiviral response, creating a favorable environment for RV infection. For example, IL-6 was almost completely degraded by bacterial protease, and the epithelial barrier function was also compromised. Bomberger et al. (2014) found that the P. aeruginosa protein, Cif, enhanced viral infection of CF epithelial cells by preventing MHC class I antigen presentation and CD8 T cell-mediated removal of influenza A-infected cells.
Various studies suggested that the percentage of COVID-19 patients with co-infections varies greatly, from 0 to 100 percent in those who die, and antimicrobial use varies widely by the severity of the disease, ranging from 20 to 100 percent for antibiotics (Langford et al., 2020). Before COVID-19, Zahariadis et al. (2006) had identified co-infections of pulmonary microbes in patients diagnosed with SARS; 9% were caused by M. pneumonia, while 30% were by C. pneumonia. In a study carried out at a hospital in Wuhan, China, Zhang et al. (2020) found that out of all the 221 patients with COVID-19 pneumonia that were admitted, co-infection occurred in 57 of them and again among these 57 patients, 17 of them were confirmed to be caused by bacterial-co-infection. A systemic review of thirty studies involving more than 3,000 patients found that 7% have bacterial co-infection (Lansbury et al., 2020). Another systemic review by Langford et al. (2020) identified secondary bacterial co-infection in 14.3% of the patients. The important bacteria detected in COVID-19 co-infection include S. pneumoniae, S. aureus, Legionella pneumophila, M. pneumonia, A. baumanii, Enterobacter cloaca, K. pneumonia (Chen et al., 2020; Duployez et al., 2020; Wang et al., 2020; Yu et al., 2020). Secondary bacterial infection in COVID-19 patients often exacerbates the disease, resulting in more extended hospitalization and even mortality in many cases. Zhou et al. (2020) suggested that 50% of COVID-19 deaths were due to bacterial co-infections.
The interaction mechanism between COVID-19 and respiratory tract bacteria is similar to bacterial interactions with viruses like RSV, Influenza, etc. One mechanism by which virus-bacteria co-infection occurs is due to the host’s immune system dysfunction, which can be weakened due to viral infection, thus promoting viral-bacterial co-infection. COVID-19, in this case, is a good example in which bacterial co-infection is identified in most hospitalized COVID-19 patients. The COVID-19 patients were found to acquire increased levels of inflammatory cytokines and biomarkers, indicating that the dysregulation of the immune system paved the way for secondary bacterial infection (Tetro, 2020). Another common interaction mechanism between bacteria and viruses is the enhancement of bacterial adhesion to epithelial cells via the upregulation of surface receptors. Golda et al. (2011) found that prior human coronavirus NL63 infection increases the expression of surface receptors like PAFR in the human epithelial cells. This enhances the adherence of S. pneumoniae to human epithelial cells. It has been shown that SARS-CoV-2 infection damages the lung epithelial cells and the lung infrastructure, recruiting a number of immune cells, such as macrophages and neutrophils, which cause inflammation and eventually facilitate bacterial invasion and adherence (MacIntyre et al., 2018; Vellingiri et al., 2020). Feldman and Anderson (2021) suggested that severe immunosuppression by SARS-CoV-2 infection may also activate the quiescent airway pathogens like S. pneumoniae, S. aureus, and H. influenzae enclosed in biofilms. Increased cytokine production by SARS-CoV-2 may reactivate latent or facilitate active TB development (Al Lawati et al., 2021). The prevalence of co-infection can also be attributed to the fact that hospital-associated bacteria can quickly adapt to the host environment of immunocompromised patients (Mirzaei et al., 2020). It has also been hypothesized that the accumulation of H2O2 in the lungs due to SARS-CoV-2 infection may suppress the innate immune response, promoting secondary bacterial infection (Bayindir and Bayindir, 2020). However, experimental studies have yet to be performed to prove this hypothesis.
The human respiratory tract inhabits a complex microbial community composed of commensal and opportunistic pathogens, the most common being bacteria such as S. aureus, S. pneumoniae, and M. catarrhalis, along with Influenza virus, Rhinovirus, SARS-CoV-2, etc. There is a synergistic and competitive interaction among these pathogens. Typically, respiratory tract microbiota causes no harm to the body, and homeostasis exists between formal micro-flora and commensal and opportunistic pathogens. However, alterations or dysbiosis in this community may result in the attainment of bacterial or viral pathogens and displacement of opportunistic pathogens to harmful pathogens, resulting in pathogenic invasion. Respiratory viruses such as influenza, Rhinovirus, and SARS-CoV-2 could cause bacterial superinfections by triggering opportunistic bacteria in the respiratory tract. As a result of bacterial co-infections, there is a rise in the morbidity rate, and subsequently, the mortality rate has significantly increased in patients with viral infections. This interplay between virus and bacteria is complex, and in order to predispose co-infections, many components, including the bacteria, virus, and the host’s immune system, are involved. The mechanism by which viruses can trigger bacterial co-infection is by damaging the respiratory epithelial cells, exposing the bacterial receptors, damaging the cilia, altering the mucosal environment, dysregulation of the immune system, changing the function of phagocytes, promoting the release of inflammatory mediators such as chemokines, cytokines release, overexpression of antimicrobial peptides and by enhancing bacterial adherence. With the onset of the current COVID-19 pandemic comes a new surge of interest in understanding the relationship between respiratory bacteria and the virus. From several experiments and observations, it is clear that the interaction between viruses and bacteria could aggravate infections and even enhance secondary infections before existing diagnosis. Since the co-infection included bacteria and viruses, an effective strategy is required to manage the infection.
CL: Conceptualization, Investigation, Writing – original draft, Writing – review and editing, Methodology. MY: Conceptualization, Supervision, Writing – review and editing, Investigation, Writing – original draft. PS: Conceptualization, Supervision, Writing – review and editing. AS: Conceptualization, Writing – review and editing. MI: Writing – review and editing, Supervision. BV: Conceptualization, Methodology, Supervision, Writing – review and editing. RZ: Writing – review and editing. ZP: Writing – review and editing, Supervision. HR: Writing – review and editing, Supervision, Methods.
The author(s) declare financial support was received for the research, authorship, and/or publication of this article. This study was supported by funding to MY from the Science and Engineering Research Board (SERB), Govt. of India, vide Project Sanction No: SRG/2021/000371.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Abramson, J. S., Giebink, G. S., and Quie, G. (1982). Influenza A virus-induced polymorphonuclear leukocyte dysfunction in the pathogenesis of experimental pneumococcal otitis media. Infect. Immun. 36, 289–296. doi: 10.1128/iai.36.1.289-296.1982
Al Lawati, R., Al Busaidi, N., Al Umairi, R., Al Busaidy, M., Al Naabi, H. H., and Khamis, F. (2021). COVID-19 and pulmonary Mycobacterium tuberculosis co-infection. Oman Med. J. 36, e298.
Almand, E. A., Moore, M. D., and Jaykus, L. A. (2017). Virus-bacteria interactions: An emerging topic in human infection. Viruses 9:58.
Alqahtani, A., Alamer, E., Mir, M., Alasmari, A., Alshahrani, M. M., Asiri, M., et al. (2022). Bacterial co-infections increase mortality of severely ill COVID-19 patients in Saudi Arabia. Int. J. Environ. Res. Public Health 19:2424.
Avadhanula, V., Rodriguez, C. A., DeVincenzo, J. P., Wang, Y., Webby, R. J., Ulett, G. C., et al. (2006). Respiratory viruses augment the adhesion of bacterial pathogens to respiratory epithelium in a viral species-and cell type-dependent manner. J. Virol. 80, 1629–1636. doi: 10.1128/JVI.80.4.1629-1636.2006
Bakaletz, L. O. (2017). Viral–bacterial co-infections in the respiratory tract. Curr. Opin. Microbiol. 35, 30–35.
Balasubramaniam, S. L., Gopalakrishnapillai, A., and Barwe, S. P. (2015). Ion dependence of Na-K-ATPase-mediated epithelial cell adhesion and migration. Am. J. Physiol. Cell Physiol. 309, C437–C441. doi: 10.1152/ajpcell.00140.2015
Bard, J. D., and McElvania, E. (2020). Panels and syndromic testing in clinical microbiology. Clin. Lab. Med. 40, 393–420.
Bayindir, M., and Bayindir, E. E. (2020). Synergic viral-bacterial co-infection in catalase-deficient COVID-19 patients causes suppressed innate immunity and lung damages due to detrimental elevation of hydrogen peroxide concentration. Amsterdam: SSRN.
Bellinghausen, C., Gulraiz, F., Heinzmann, A. C., Dentener, M. A., Savelkoul, H., Wouters, E. F., et al. (2016). Exposure to common respiratory bacteria alters the airway epithelial response to subsequent viral infection. Respir. Res. 17, 1–12. doi: 10.1186/s12931-016-0382-z
Beshara, R., Sencio, V., Soulard, D., Barthélémy, A., Fontaine, J., Pinteau, T., et al. (2018). Alteration of Flt3-Ligand-dependent de novo generation of conventional dendritic cells during influenza infection contributes to respiratory bacterial superinfection. PLoS Pathog. 14:e1007360. doi: 10.1371/journal.ppat.1007360
Blevins, L. K., Wren, J. T., Holbrook, B. C., Hayward, S. L., Swords, W. E., Parks, G. D., et al. (2014). Co-infection with Streptococcus pneumoniae negatively modulates the size and composition of the ongoing influenza-specific CD8+ T cell response. J. Immunol. 193, 5076–5087. doi: 10.4049/jimmunol.1400529
Bomberger, J. M., Ely, K. H., Bangia, N., Ye, S., Green, K. A., Green, W. R., et al. (2014). Pseudomonas aeruginosa Cif protein enhances the ubiquitination and proteasomal degradation of the transporter associated with antigen processing (TAP) and reduces major histocompatibility complex (MHC) class I antigen presentation. J. Biol. Chem. 289, 152–162. doi: 10.1074/jbc.M113.459271
Bosch, A. A., Biesbroek, G., Trzcinski, K., Sanders, E. A., and Bogaert, D. (2013). Viral and bacterial interactions in the upper respiratory tract. PLoS Pathog. 9:e1003057. doi: 10.1371/journal.ppat.1003057
Bryce, J., Boschi-Pinto, C., Shibuya, K., and Black, R. E. (2005). WHO estimates of the causes of death in children. Lancet 365, 1147–1152.
Cappelletty, D. (1998). Microbiology of bacterial respiratory infections. Pediatr. Infect. Dis. J. 17, S55–S61.
Center for Disease Control and Prevention [CDC] (2012). First Global Estimates of 2009 H1N1 Pandemic Mortality Released by CDC-Led Collaboration. Atlanta, GA: Center for Disease Control and Prevention.
Chattoraj, S. S., Ganesan, S., Faris, A., Comstock, A., Lee, W. M., and Sajjan, U. S. (2011a). Pseudomonas aeruginosa suppresses interferon response to rhinovirus infection in cystic fibrosis but not in normal bronchial epithelial cells. Infect. Immun. 79, 4131–4145. doi: 10.1128/IAI.05120-11
Chattoraj, S. S., Ganesan, S., Jones, A. M., Helm, J. M., Comstock, A. T., Bright-Thomas, R., et al. (2011b). Rhinovirus infection liberates planktonic bacteria from biofilm and increases chemokine responses in cystic fibrosis airway epithelial cells. Thorax 66, 333–339. doi: 10.1136/thx.2010.151431
Chen, N., Zhou, M., Dong, X., Qu, J., Gong, F., Han, Y., et al. (2020). Epidemiological and clinical characteristics of 99 cases of 2019 novel coronavirus pneumonia in Wuhan, China: A descriptive study. Lancet 395, 507–513. doi: 10.1016/S0140-6736(20)30211-7
Chiu, C. C., Shi, Y. F., Yang, J. J., Hsiao, Y. C., Tzang, B. S., and Hsu, T. C. (2014). Effects of human parvovirus B19 and bocavirus VP1 unique region on tight junction of human airway epithelial A549 cells. PLoS One 9:e107970. doi: 10.1371/journal.pone.0107970
Chonmaitree, T., Revai, K., Grady, J. J., Clos, A., Patel, J. A., Nair, S., et al. (2008). Viral upper respiratory tract infection and otitis media complication in young children. Clin. Infect. Dis. 46, 815–823.
Cillóniz, C., Ewig, S., Menéndez, R., Ferrer, M., Polverino, E., Reyes, S., et al. (2012). Bacterial co-infection with H1N1 infection in patients admitted with community-acquired pneumonia. J. Infect. 65, 223–230.
Cystic Fibrosis Foundation (2014). Patient registry annual data report. Washington, DC: Cystic Fibrosis Foundation.
Dasaraju, V., and Liu, C. (1996). “Infections of the respiratory system,” in Medical Microbiology, ed. S. Baron (Galveston, TX: University of Texas Medical Branch at Galveston).
De Bruyn, A., Verellen, S., Bruckers, L., Geebelen, L., Callebaut, I., De Pauw, I., et al. (2022). Secondary infection in COVID-19 critically ill patients: a retrospective single-center evaluation. BMC Infect. Dis. 22:207. doi: 10.1186/s12879-022-07192-x
Diavatopoulos, D. A., Short, K. R., Price, J. T., Wilksch, J. J., Brown, L. E., Briles, D. E., et al. (2010). Influenza A virus facilitates Streptococcus pneumoniae transmission and disease. FASEB J. 24, 1789–1798.
dos Santos, C. C., Zhang, H., and Slutsky, A. S. (2002). From bench to bedside: bacterial growth and cytokines. Crit. Care 6, 1–3.
Dumkow, L. E., Worden, L. J., and Rao, S. N. (2021). Syndromic diagnostic testing: a new way to approach patient care in the treatment of infectious diseases. J. Antimicrob. Chemother. 76, 4–11. doi: 10.1093/jac/dkab245
Duployez, C., Le Guern, R., Tinez, C., Lejeune, A. L., Robriquet, L., Six, S., et al. (2020). Panton-valentine leukocidin–secreting Staphylococcus aureus pneumonia complicating COVID-19. Emerg. Infect. Dis. 26:1939. doi: 10.3201/eid2608.201413
El Ahmer, O. R., Raza, M. W., Ogilvie, M. M., Weir, D. M., and Blackwell, C. C. (1999). Binding of bacteria to HEp-2 cells infected with influenza A virus. FEMS Immunol. Med. Microbiol. 23, 331–341. doi: 10.1111/j.1574-695X.1999.tb01255.x
Endres, A., Hügel, C., Boland, H., Hogardt, M., Schubert, R., Jonigk, D., et al. (2022). Pseudomonas aeruginosa affects airway epithelial response and barrier function during rhinovirus infection. Front. Cell. Infect. Microbiol. 12:846828. doi: 10.3389/fcimb.2022.846828
Fares, M., Mourad, S., Rajab, M., and Rifai, N. (2011). The use of C-reactive protein in predicting bacterial co-infection in children with bronchiolitis. North Am. J. Med. Sci. 3:152. doi: 10.4297/najms.2011.3152
Feldman, C., and Anderson, R. (2021). The role of co-infections and secondary infections in patients with COVID-19. Pneumonia 13, 1–15.
Gadsby, N. J., McHugh, M. P., Russell, C. D., Mark, H., Morris, A. C., Laurenson, I. F., et al. (2015). Development of two real-time multiplex PCR assays for the detection and quantification of eight key bacterial pathogens in lower respiratory tract infections. Clin. Microbiol. Infect. 21, 788–e1. doi: 10.1016/j.cmi.2015.05.004
Ghoneim, H. E., Thomas, G., and McCullers, J. A. (2013). Depletion of alveolar macrophages during influenza infection facilitates bacterial superinfections. J. Immunol. 191, 1250–1259.
Giebink, G. S., Ripley, M. L., and Wright, F. (1987). Eustachian tube histopathology during experimental influenza A virus infection in the chinchilla. Ann. Otol. Rhinol. Laryngol. 96, 199–206.
Golda, A., Malek, N., Dudek, B., Zeglen, S., Wojarski, J., Ochman, M., et al. (2011). Infection with human coronavirus NL63 enhances streptococcal adherence to epithelial cells. J. Gen. Virol. 92:1358. doi: 10.1099/vir.0.028381-0
González, A., Prado, C. E., Leiva, E. D., Carreño, L. J., Bueno, S. M., Riedel, C. A., et al. (2008). Respiratory syncytial virus impairs T cell activation by preventing synapse assembly with dendritic cells. Proc. Natl. Acad. Sci. U.S.A. 105, 14999–15004.
Grijalva, C. G., Griffin, M. R., Edwards, K. M., Williams, J. V., Gil, A. I., Verastegui, H., et al. (2014). The role of influenza and parainfluenza infections in nasopharyngeal pneumococcal acquisition among young children. Clin. Infect. Dis. 58, 1369–1376. doi: 10.1093/cid/ciu148
Gulraiz, F., Rellinghausen, C., Bruggeman, C. A., and Stassen, F. R. (2015). Haemophilus influenzae increases the susceptibility and inflammatory response of airway epithelial cells to viral infections. FASEB J. 29, 849–858. doi: 10.1096/fj.14-254359
Håkansson, A., Kidd, A., Wadell, G., Sabharwal, H., and Svanborg, C. (1994). Adenovirus infection enhances in vitro adherence of Streptococcus pneumoniae. Infect. Immun. 62, 2707–2714. doi: 10.1128/iai.62.7.2707-2714.1994
Hament, J. M., Aerts, C., Fleer, A., van Dijk, H., Harmsen, T., Kimpen, J. L., et al. (2005). Direct binding of respiratory syncytial virus to pneumococci: a phenomenon that enhances both pneumococcal adherence to human epithelial cells and pneumococcal invasiveness in a murine model. Pediatr. Res. 58, 1198–1203. doi: 10.1203/01.pdr.0000188699.55279.1b
Handel, A., Longini, I. M. Jr., and Antia, R. (2010). Towards a quantitative understanding of the within-host dynamics of influenza A infections. J. R. Soc. Interf. 7, 35–47.
Hendricks, M. R., Lashua, L. P., Fischer, D. K., Flitter, B. A., Eichinger, K. M., Durbin, J. E., et al. (2016). Respiratory syncytial virus infection enhances Pseudomonas aeruginosa biofilm growth through dysregulation of nutritional immunity. Proc. Natl. Acad. Sci. U.S.A. 113, 1642–1647. doi: 10.1073/pnas.1516979113
Hennigar, S. R., and McClung, J. P. (2016). Nutritional immunity: starving pathogens of trace minerals. Am. J. Lifestyle Med. 10, 170–173.
Honkinen, M., Lahti, E., Österback, R., Ruuskanen, O., and Waris, M. (2012). Viruses and bacteria in sputum samples of children with community-acquired pneumonia. Clin. Microbiol. Infect. 18, 300–307.
Huang, G., Yu, D., Mao, N., Zhu, Z., Zhang, H., Jiang, Z., et al. (2013). Viral etiology of acute respiratory infection in Gansu Province, China, 2011. PLoS One 8, e64254. doi: 10.1186/s12985-015-0388-z
Huijskens, E. G., van Erkel, A. J., Palmen, F. M., Buiting, A. G., Kluytmans, J. A., and Rossen, J. W. (2013). Viral and bacterial aetiology of community-acquired pneumonia in adults. Influenza Other Respir. Vir. 7, 567–573.
Ishizuka, S., Yamaya, M., Suzuki, T., Takahashi, H., Ida, S., Sasaki, T., et al. (2003). Effects of rhinovirus infection on the adherence of Streptococcus pneumoniae to cultured human airway epithelial cells. J. Infect. Dis. 188, 1928–1939. doi: 10.1086/379833
Itoh, M., Nagafuchi, A., Moroi, S., and Tsukita, S. (1997). Involvement of ZO-1 in cadherin-based cell adhesion through its direct binding to α catenin and actin filaments. J. Cell Biol. 138, 181–192. doi: 10.1083/jcb.138.1.181
Jain, S., Self, W. H., Wunderink, R. G., Fakhran, S., Balk, R., Bramley, A. M., et al. (2015). Community-acquired pneumonia requiring hospitalization among US adults. N. Engl. J. Med. 373, 415–427.
Jiang, Z., Nagata, N., Molina, E., Bakaletz, L. O., Hawkins, H., and Patel, J. A. (1999). Fimbria-mediated enhanced attachment of nontypeable Haemophilus influenzae to respiratory syncytial virus-infected respiratory epithelial cells. Infect. Immun. 67, 187–192. doi: 10.1128/IAI.67.1.187-192.1999
Johansson, N., Kalin, M., and Hedlund, J. (2011). Clinical impact of combined viral and bacterial infection in patients with community-acquired pneumonia. Scand. J. Infect. Dis. 43, 609–615.
Kanangat, S., Meduri, G. U., Tolley, E. A., Patterson, D. R., Meduri, C. U., Pak, C., et al. (1999). Effects of cytokines and endotoxin on the intracellular growth of bacteria. Infect. Immun. 67, 2834–2840.
Karppinen, S., Teräsjärvi, J., Auranen, K., Schuez-Havupalo, L., Siira, L., He, Q., et al. (2017). Acquisition and transmission of Streptococcus pneumoniae are facilitated during rhinovirus infection in families with children. Am. J. Respir. Crit. Med. 196, 1172–1180. doi: 10.1164/rccm.201702-0357OC
Kiedrowski, M. R., and Bomberger, J. M. (2018). Viral-bacterial co-infections in the cystic fibrosis respiratory tract. Front. Immunol. 9:3067. doi: 10.3389/fimmu.2018.03067
Kiedrowski, M. R., Gaston, J. R., Kocak, B. R., Coburn, S. L., Lee, S., Pilewski, J. M., et al. (2018). Staphylococcus aureus biofilm growth on cystic fibrosis airway epithelial cells is enhanced during respiratory syncytial virus co-infection. MSphere 3, 10–1128. doi: 10.1128/mSphere.00341-18
Kreitmann, L., Monard, C., Dauwalder, O., Simon, M., and Argaud, L. (2020). Early bacterial co-infection in ARDS related to COVID-19. Intens. Care Med. 46, 1787–1789.
Kudva, A., Scheller, E. V., Robinson, K. M., Crowe, C. R., Choi, S. M., Slight, S. R., et al. (2011). Influenza A inhibits Th17-mediated host defense against bacterial pneumonia in mice. The Journal of Immunology 186, 1666–1674. doi: 10.4049/jimmunol.1002194
Kuster, S. P., Tuite, A. R., Kwong, J. C., McGeer, A., Toronto Invasive Bacterial Diseases Network, et al. (2011). Evaluation of coseasonality of influenza and invasive pneumococcal disease: results from prospective surveillance. PLoS Med. 8:e1001042. doi: 10.1371/journal.pmed.1001042
Ladas, P., Porfyridis, I., Tryfonos, C., Ioannou, A., Adamide, T., Christodoulou, C., et al. (2023). Aetiology of Community-Acquired Pneumonia and the Role of Genetic Host Factors in Hospitalized Patients in Cyprus. Microorganisms 11:2051. doi: 10.3390/microorganisms11082051
Langford, B. J., So, M., Raybardhan, S., Leung, V., Westwood, D., MacFadden, D. R., et al. (2020). Bacterial co-infection and secondary infection in patients with COVID-19: a living rapid review and meta-analysis. Clin. Microbiol. Infect. 26, 1622–1629. doi: 10.1016/j.cmi.2020.07.016
Lansbury, L., Lim, B., Baskaran, V., and Lim, W. S. (2020). Co-infections in people with COVID-19: a systematic review and meta-analysis. J. Infect. 81, 266–275.
Li, N., Ren, A., Wang, X., Fan, X., Zhao, Y., Gao, G. F., et al. (2015). Influenza viral neuraminidase primes bacterial co-infection through TGF-β–mediated expression of host cell receptors. Proc. Natl. Acad. Sci. U.S.A. 112, 238–243.
Lindbaek, M. (2006). Prescribing antibiotics to patients with acute cough and otitis media. Br. J. Gen. Pract. 56, 164–165.
Liu, L., Johnson, H. L., Cousens, S., Perin, J., Scott, S., Lawn, J. E., et al. (2012). Global, regional, and national causes of child mortality: an updated systematic analysis for 2010 with time trends since 2000. Lancet 379, 2151–2161. doi: 10.1016/S0140-6736(12)60560-1
Luchsinger, V., Ruiz, M., Zunino, E., Martínez, M. A., Machado, C., Piedra, A., et al. (2013). Community-acquired pneumonia in Chile: the clinical relevance in the detection of viruses and atypical bacteria. Thorax 68, 1000–1006. doi: 10.1136/thoraxjnl-2013-203551
MacIntyre, C. R., Chughtai, A. A., Barnes, M., Ridda, I., Seale, H., Toms, R., et al. (2018). The role of pneumonia and secondary bacterial infection in fatal and serious outcomes of pandemic influenza a (H1N1) pdm09. BMC Infect. Dis. 18:637. doi: 10.1186/s12879-018-3548-0
Marks, L. R., Davidson, B. A., Knight, R., and Hakansson, A. P. (2013). Interkingdom signaling induces Streptococcus pneumoniae biofilm dispersion and transition from asymptomatic colonization to disease. MBio 4, 10–1128. doi: 10.1128/mBio.00438-13
McCullers, J. A., and Bartmess, K. C. (2003a). Lethal synergism between influenza virus and Streptococcus pneumoniae. J Infect. Dis. 187, 1674–1675.
McCullers, J. A., and Bartmess, K. C. (2003b). Role of neuraminidase in lethal synergism between influenza virus and Streptococcus pneumoniae. J. Infect. Dis. 187, 1000–1009.
McCullers, J. A., McAuley, J. L., Browall, S., Iverson, A. R., Boyd, K. L., and Henriques Normark, B. (2010). Influenza enhances susceptibility to natural acquisition of and disease due to Streptococcus pneumoniae in ferrets. J. Infect. Dis. 202, 1287–1295. doi: 10.1086/656333
McCullers, J. A., and Rehg, J. E. (2002). Lethal synergism between influenza virus and Streptococcus pneumoniae: characterization of a mouse model and the role of platelet-activating factor receptor. J. Infect. Dis. 186, 341–350. doi: 10.1086/341462
McNamee, L. A., and Harmsen, A. G. (2006). Both influenza-induced neutrophil dysfunction and neutrophil-independent mechanisms contribute to increased susceptibility to a secondary Streptococcus pneumoniae infection. Infect. Immun. 74, 6707–6721. doi: 10.1128/IAI.00789-06
Meduri, G. U., Kanangat, S., Stefan, J., Tolley, E., and Schaberg, D. (1999). Cytokines IL-1 β, IL-6, and TNF-α enhance in vitro growth of bacteria. Am. J. Respir. Crit. Care Med. 160, 961–967.
Miró-Cañís, S., Capilla-Rubio, S., Marzo-Checa, L., Fontanals-Aymerich, D., Sanfeliu-Sala, I., Espasa-Soley, M., et al. (2017). Multiplex PCR reveals that viruses are more frequent than bacteria in children with cystic fibrosis. J. Clin. Virol. 86, 1–4. doi: 10.1016/j.jcv.2016.11.004
Mirzaei, R., Goodarzi, P., Asadi, M., Soltani, A., Aljanabi, H. A. A., Jeda, A. S., et al. (2020). Bacterial co-infections with SARS-CoV-2. IUBMB Life 72, 2097–2111.
Morens, D. M., Taubenberger, J. K., and Fauci, A. S. (2008). Predominant role of bacterial pneumonia as a cause of death in pandemic influenza: implications for pandemic influenza preparedness. J. Infect. Dis. 198, 962–970.
Musher, D. M., Abers, M. S., and Bartlett, J. G. (2017). Evolving understanding of the causes of pneumonia in adults, with special attention to the role of pneumococcus. Clin. Infect. Dis. 65, 1736–1744. doi: 10.1093/cid/cix549
Nakamura, S., Davis, K. M., and Weiser, J. N. (2011). Synergistic stimulation of type I interferons during influenza virus co-infection promotes Streptococcus pneumoniae colonization in mice. J. Clin. Investig. 121, 3657–3665.
Nascimento, M. S., de Souza, A. V., de Souza Ferreira, A. V., Rodrigues, J. C., Abramovici, S., and da Silva Filho, L. V. F. (2010). High rate of viral identification and co-infections in infants with acute bronchiolitis. Clinics 65, 1133–1137. doi: 10.1590/s1807-59322010001100014
Nguyen, D. T., de Witte, L., Ludlow, M., Yüksel, S., Wiesmüller, K. H., Geijtenbeek, T. B., et al. (2010). The synthetic bacterial lipopeptide Pam3CSK4 modulates respiratory syncytial virus infection independent of TLR activation. PLoS Pathog. 6:e1001049. doi: 10.1371/journal.ppat.1001049
Nguyen, D. T., Louwen, R., Elberse, K., van Amerongen, G., Yüksel, S., Luijendijk, A., et al. (2015). Streptococcus pneumoniae enhances human respiratory syncytial virus infection in vitro and in vivo. PLoS One 10:e0127098. doi: 10.1371/journal.pone.0127098
Nichols, D., Chmiel, J., and Berger, M. (2008). Chronic inflammation in the cystic fibrosis lung: alterations in inter-and intracellular signaling. Clin. Rev. Allergy Immunol. 34, 146–162. doi: 10.1007/s12016-007-8039-9
Nishikawa, T., Shimizu, K., Tanaka, T., Kuroda, K., Takayama, T., Yamamoto, T., et al. (2012). Bacterial neuraminidase rescues influenza virus replication from inhibition by a neuraminidase inhibitor. PLoS One 7:e45371. doi: 10.1371/journal.pone.0045371
Nita-Lazar, M., Banerjee, A., Feng, C., Amin, M. N., Frieman, M. B., Chen, W. H., et al. (2015). Desialylation of airway epithelial cells during influenza virus infection enhances pneumococcal adhesion via galectin binding. Mol. Immunol. 65, 1–16. doi: 10.1016/j.molimm.2014.12.010
Okamoto, S., Kawabata, S., Terao, Y., Fujitaka, H., Okuno, Y., and Hamada, S. (2004). The Streptococcus pyogenes capsule is required for adhesion of bacteria to virus-infected alveolar epithelial cells and lethal bacterial-viral superinfection. Infect. Immun. 72, 6068–6075. doi: 10.1128/IAI.72.10.6068-6075.2004
Park, K., Coticchia, J. M., Bakaletz, L. O., and Lim, D. J. (1993). Effect of influenza A virus on ciliary activity and dye transport function in the chinchilla eustachian tube. Ann. Otol. Rhinol. Laryngol. 102, 551–558. doi: 10.1177/000348949310200711
Pettigrew, M. M., Gent, J. F., Pyles, R. B., Miller, A. L., Nokso-Koivisto, J., and Chonmaitree, T. (2011). Viral-bacterial interactions and risk of acute otitis media complicating upper respiratory tract infection. J. Clin. Microbiol. 49, 3750–3755.
Pettigrew, M. M., Marks, L. R., Kong, Y., Gent, J. F., Roche-Hakansson, H., and Hakansson, A. P. (2014). Dynamic changes in the Streptococcus pneumoniae transcriptome during transition from biofilm formation to invasive disease upon influenza A virus infection. Infect. Immun. 82, 4607–4619. doi: 10.1128/IAI.02225-14
Pihet, M., Carrere, J., Cimon, B., Chabasse, D., Delhaes, L., Symoens, F., et al. (2009). Occurrence and relevance of filamentous fungi in respiratory secretions of patients with cystic fibrosis—a review. Sabouraudia 47, 387–397. doi: 10.1080/13693780802609604
Reddinger, R. M., Luke-Marshall, N. R., Hakansson, A. P., and Campagnari, A. A. (2016). Host physiologic changes induced by influenza A virus lead to Staphylococcus aureus biofilm dispersion and transition from asymptomatic colonization to invasive disease. MBio 7, 10–1128. doi: 10.1128/mBio.01235-16
Robinson, K. M., Choi, S. M., McHugh, K. J., Mandalapu, S., Enelow, R. I., Kolls, J. K., et al. (2013). Influenza A exacerbates Staphylococcus aureus pneumonia by attenuating IL-1β production in mice. J. Immunol. 191, 5153–5159.
Rossi, G. A., Fanous, H., and Colin, A. A. (2020). Viral strategies predisposing to respiratory bacterial superinfections. Pediatr. Pulmonol. 55, 1061–1073.
Rowe, H. M., Meliopoulos, V. A., Iverson, A., Bomme, Schultz-Cherry, S., and Rosch, J. W. (2019). Direct interactions with influenza promote bacterial adherence during respiratory infections. Nat. Microbiol. 4, 1328–1336. doi: 10.1038/s41564-019-0447-0
Ruohola, A., Meurman, O., Nikkari, S., Skottman, T., Salmi, A., Waris, M., et al. (2006). Microbiology of acute otitis media in children with tympanostomy tubes: prevalences of bacteria and viruses. Clin. Infect. Dis. 43, 1417–1422.
Ruuskanen, O., Lahti, E., Jennings, L. C., and Murdoch, D. R. (2011). Viral pneumonia. Lancet 377, 1264–1275.
Sajjan, U., Wang, Q., Zhao, Y., Gruenert, D. C., and Hershenson, M. B. (2008). Rhinovirus disrupts the barrier function of polarized airway epithelial cells. Am. J. Respir. Crit. Care Med. 178, 1271–1281.
Sajjan, U. S., Jia, Y., Newcomb, D. C., Bentley, J. K., Lukacs, N. W., LiPuma, J. J., et al. (2006). H. influenzae potentiates airway epithelial cell responses to Rhinovirus by increasing ICAM-1 and TLR3 expression. FASEB J. 20, 2121–2123. doi: 10.1096/fj.06-5806fje
Scheiblauer, H., Reinacher, M., Tashiro, M., and Rott, R. (1992). Interactions between bacteria and influenza A virus in the development of influenza pneumonia. J. Infect. Dis. 166, 783–791.
Scott, P., Anderson, K., Singhania, M., and Cormier, R. (2020). Cystic fibrosis, CFTR, and colorectal cancer. Int. J. Mol. Sci. 21:2891.
Sherrard, L. J., Tunney, M. M., and Elborn, J. S. (2014). Antimicrobial resistance in the respiratory microbiota of people with cystic fibrosis. Lancet 384, 703–713.
Short, K. R., Brooks, A. G., Reading, C., and Londrigan, S. L. (2012a). The fate of influenza A virus after infection of human macrophages and dendritic cells. J. Gen. Virol. 93, 2315–2325. doi: 10.1099/vir.0.045021-0
Short, K. R., Kasper, J., Van Der, Aa, S., Andeweg, A. C., Zaaraoui-Boutahar, F., et al. (2016). Influenza virus damages the alveolar barrier by disrupting epithelial cell tight junctions. Eur. Respir. J. 47, 954–966. doi: 10.1183/13993003.01282-2015
Short, K. R., Reading, C., Wang, N., Diavatopoulos, D. A., and Wijburg, O. L. (2012b). Increased nasopharyngeal bacterial titers and local inflammation facilitate transmission of Streptococcus pneumoniae. MBio 3, 10–1128. doi: 10.1128/mBio.00255-12
Siegel, S. J., Roche, A. M., and Weiser, J. N. (2014). Influenza promotes pneumococcal growth during co-infection by providing host sialylated substrates as a nutrient source. Cell Host Microbe 16, 55–67. doi: 10.1016/j.chom.2014.06.005
Skov, M., McKay, K., Koch, C., and Cooper, J. (2005). Prevalence of allergic bronchopulmonary aspergillosis in cystic fibrosis in an area with a high frequency of atopy. Respir. Med. 99, 887–893. doi: 10.1016/j.rmed.2004.11.018
Small, C. L., Shaler, C. R., McCormick, S., Jeyanathan, M., Damjanovic, D., Brown, E. G., et al. (2010). Influenza infection leads to increased susceptibility to subsequent bacterial superinfection by impairing NK cell responses in the lung. J. Immunol. 184, 2048–2056. doi: 10.4049/jimmunol.0902772
Smith, A. M., Adler, F. R., Ribeiro, R. M., Gutenkunst, R. N., McAuley, J. L., McCullers, J. A., et al. (2013). Kinetics of co-infection with influenza A virus and Streptococcus pneumoniae. PLoS Pathog. 9:e1003238. doi: 10.1371/journal.ppat.1003238
Smith, C. M., Sandrini, S., Datta, S., Freestone, Shafeeq, S., and Radhakrishnan, et al. (2014). Respiratory syncytial virus increases the virulence of Streptococcus pneumoniae by binding to penicillin binding protein 1a. A new paradigm in respiratory infection. Am. J. Respir. Crit. Care Med. 190, 196–207. doi: 10.1164/rccm.201311-2110OC
Sörensen, M., Kantorek, J., Byrnes, L., Boutin, S., Mall, M. A., Lasitschka, F., et al. (2020). Pseudomonas aeruginosa modulates the antiviral response of bronchial epithelial cells. Front. Immunol. 11:96. doi: 10.3389/fimmu.2020.00096
Spelmink, L., Sender, V., Hentrich, K., Kuri, T., Plant, L., and Henriques-Normark, B. (2016). Toll-like receptor 3/TRIF-dependent IL-12p70 secretion mediated by Streptococcus pneumoniae RNA and its priming by influenza A virus co-infection in human dendritic cells. MBio 7, 10–1128. doi: 10.1128/mBio.00168-16
Stark, J. M., Stark, M. A., Colasurdo, G. N., and LeVine, A. M. (2006). Decreased bacterial clearance from the lungs of mice following primary respiratory syncytial virus infection. J. Med. Virol. 78, 829–838. doi: 10.1002/jmv.20631
Steed, E., Balda, M. S., and Matter, K. (2010). Dynamics and functions of tight junctions. Trends Cell Biol. 20, 142–149.
Stelzer-Braid, S., Johal, H., Skilbeck, K., Steller, A., Alsubie, H., Tovey, E., et al. (2012). Detection of viral and bacterial respiratory pathogens in patients with cystic fibrosis. J. Virol. Methods 186, 109–112.
Sudfeld, C. R., Dasenbrook, E. C., Merz, W. G., Carroll, K. C., and Boyle, M. P. (2010). Prevalence and risk factors for recovery of filamentous fungi in individuals with cystic fibrosis. J. Cystic Fibr. 9, 110–116.
Tashiro, M., Ciborowski, P., Klenk, H. D., Pulverer, G., and Rott, R. (1987a). Role of Staphylococcus protease in the development of influenza pneumonia. Nature 325, 536–537.
Tashiro, M., Ciborowski, P., Reinacher, M., Pulverer, G., Klenk, H. D., and Rott, R. (1987b). Synergistic role of staphylococcal proteases in the induction of influenza virus pathogenicity. Virology 157, 421–430. doi: 10.1016/0042-6822(87)90284-4
Thorburn, K., Harigopal, S., Reddy, V., Taylor, N., and Van Saene, H. K. F. (2006). High incidence of pulmonary bacterial co-infection in children with severe respiratory syncytial virus (RSV) bronchiolitis. Thorax 61, 611–615.
Tong, J., Fu, Y., Meng, F., Krüger, N., Valentin-Weigand, P., and Herrler, G. (2018a). The sialic acid binding activity of human parainfluenza virus 3 and mumps virus glycoproteins enhances the adherence of group B streptococci to HEp-2 cells. Front. Cell. Infect. Microbiol. 8:280. doi: 10.3389/fcimb.2018.00280
Tong, J., Fu, Y., Wu, N. H., Rohde, M., Meng, F., Valentin-Weigand, P., et al. (2018b). Sialic acid-dependent interaction of group B streptococci with influenza virus-infected cells reveals a novel adherence and invasion mechanism. Cell. Microbiol. 20, e12818. doi: 10.1111/cmi.12818
Tse, L. V., Marcano, V. C., Huang, W., Pocwierz, M. S., and Whittaker, G. R. (2013). Plasmin-mediated activation of pandemic H1N1 influenza virus hemagglutinin is independent of the viral neuraminidase. J. Virol. 87, 5161–5169. doi: 10.1128/JVI.00210-13
Van Ewijk, B. E., Wolfs, T. F., Aerts, C., Van Kessel, K. P., Fleer, A., Kimpen, J. L., et al. (2007). RSV mediates Pseudomonas aeruginosa binding to cystic fibrosis and normal epithelial cells. Pediatr. Res. 61, 398–403. doi: 10.1203/pdr.0b013e3180332d1c
Vellingiri, B., Jayaramayya, K., Iyer, M., Narayanasamy, A., Govindasamy, V., Giridharan, B., et al. (2020). COVID-19: A promising cure for the global panic. Sci. Total Environ. 725:138277. doi: 10.1016/j.scitotenv.2020.138277
Vergison, A., Dagan, R., Arguedas, A., Bonhoeffer, J., Cohen, R., DHooge, I., et al. (2010). Otitis media and its consequences: beyond the earache. Lancet Infect. Dis. 10, 195–203. doi: 10.1016/S1473-3099(10)70012-8
Verkaik, N. J., Nguyen, D. T., De Vogel, C. P., Moll, H. A., Verbrugh, H. A., Jaddoe, V. W. V., et al. (2011). Streptococcus pneumoniae exposure is associated with human metapneumovirus seroconversion and increased susceptibility to in vitro HMPV infection. Clin. Microbiol. Infect. 17, 1840–1844. doi: 10.1111/j.1469-0691.2011.03480.x
Vesa, S., Kleemola, M., Blomqvist, S., Takala, A., Kilpi, T., and Hovi, T. (2001). Epidemiology of documented viral respiratory infections and acute otitis media in a cohort of children followed from two to twenty-four months of age. Pediatr. Infect. Dis. J. 20, 574–581. doi: 10.1097/00006454-200106000-00006
Voiriot, G., Visseaux, B., Cohen, J., Nguyen, L. B. L., Neuville, M., Morbieu, C., et al. (2016). Viral-bacterial co-infection affects the presentation and alters the prognosis of severe community-acquired pneumonia. Crit. Care 20, 1–9. doi: 10.1186/s13054-016-1517-9
Wang, J. H., Kwon, H. J., and Jang, Y. J. (2009). Rhinovirus enhances various bacterial adhesions to nasal epithelial cells simultaneously. Laryngoscope 119, 1406–1411. doi: 10.1002/lary.20498
Wang, Z., Yang, B., Li, Q., Wen, L., and Zhang, R. (2020). Clinical features of 69 cases with coronavirus disease 2019 in Wuhan, China. Clin. Infect. Dis. 71, 769–777.
Wen, X., Huang, Q., Tao, H., Zou, W., Gao, M., Guo, H., et al. (2019). Clinical characteristics and viral etiologies of outpatients with acute respiratory infections in Huzhou of China: a retrospective study. BMC Infect. Dis. 19:32. doi: 10.1186/s12879-018-3668-6
Wiegers, H. M., van Nijen, L., van Woensel, J. B., Bem, R. A., de Jong, M. D., and Calis, J. C. (2019). Bacterial co-infection of the respiratory tract in ventilated children with bronchiolitis; a retrospective cohort study. BMC Infect. Dis. 19:938. doi: 10.1186/s12879-019-4468-3
Williams, D. J., Hall, M., Brogan, T. V., Farris, R. W., Myers, A. L., Newland, J. G., et al. (2011). Influenza co-infection and outcomes in children with complicated pneumonia. Arch. Pediatr. Adolesc. Med. 165, 506–512.
Wolf, A. I., Strauman, M. C., Mozdzanowska, K., Whittle, J. R., Williams, K. L., Sharpe, A. H., et al. (2014). Co-infection with Streptococcus pneumoniae modulates the B cell response to influenza virus. J. Virol. 88, 11995–12005. doi: 10.1128/JVI.01833-14
World Health Organization (2020). Severe acute respiratory infections treatment centre: practical manual to set up and manage a SARI treatment centre and a SARI screening facility in health care facilities (No. WHO/2019-nCoV/SARI_treatment_center/2020.1). Geneva: World Health Organization.
Wu, Y., Tu, W., Lam, K. T., Chow, K. H., Ho, L., Guan, Y., et al. (2015). Lethal co-infection of influenza virus and Streptococcus pneumoniae lowers antibody response to influenza virus in lung and reduces numbers of germinal center B cells, T follicular helper cells, and plasma cells in mediastinal lymph node. J. Virol. 89, 2013–2023. doi: 10.1128/JVI.02455-14
Yu, N., Li, W., Kang, Q., Xiong, Z., Wang, S., Lin, X., et al. (2020). Clinical features and obstetric and neonatal outcomes of pregnant patients with COVID-19 in Wuhan, China: a retrospective, single-centre, descriptive study. Lancet Infect. Dis. 20, 559–564. doi: 10.1016/S1473-3099(20)30176-6
Zahariadis, G., Gooley, T. A., Ryall, P., Hutchinson, C., Latchford, M. I., Fearon, M. A., et al. (2006). Risk of ruling out severe acute respiratory syndrome by ruling in another diagnosis: variable incidence of atypical bacteria co-infection based on diagnostic assays. Can. Respir. J. 13, 17–22. doi: 10.1155/2006/862797
Zhang, G., Hu, C., Luo, L., Fang, F., Chen, Y., Li, J., et al. (2020). Clinical features and short-term outcomes of 221 patients with COVID-19 in Wuhan, China. J. Clin. Virol. 127:104364. doi: 10.1016/j.jcv.2020.104364
Zhang, L., Xiao, Y., Zhang, G., Li, H., Zhao, J., Chen, M., et al. (2023). Identification of priority pathogens for aetiological diagnosis in adults with community-acquired pneumonia in China: a multicentre prospective study. BMC Infect. Dis. 23:231. doi: 10.1186/s12879-023-08166-3
Keywords: viral infection, respiratory diseases, COVID-19, bacterial contamination, cooperative relation
Citation: Lalbiaktluangi C, Yadav MK, Singh PK, Singh A, Iyer M, Vellingiri B, Zomuansangi R, Zothanpuia and Ram H (2023) A cooperativity between virus and bacteria during respiratory infections. Front. Microbiol. 14:1279159. doi: 10.3389/fmicb.2023.1279159
Received: 17 August 2023; Accepted: 27 October 2023;
Published: 30 November 2023.
Edited by:
Efthymia Giannitsioti, University General Hospital Attikon, GreeceReviewed by:
Evdoxia Kyriazopoulou, National and Kapodistrian University of Athens, GreeceCopyright © 2023 Lalbiaktluangi, Yadav, Singh, Singh, Iyer, Vellingiri, Zomuansangi, Zothanpuia and Ram. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Mukesh Kumar Yadav, bXVraXlhZGF2QGdtYWlsLmNvbQ==
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.