- Department of Natural Sciences, University of Michigan-Dearborn, Dearborn, MI, United States
Rapid industrialization and urbanization have led to widespread metal contamination in aquatic ecosystems. This study explores the metal tolerance and biosorption characteristics of four bacterial strains (Serratia sp. L2, Raoultella sp. L30, Klebsiella sp. R3, and Klebsiella sp. R19) isolated from Saint Clair River sediments. These strains effectively removed various metal cations (As3+, Pb2+, Cu2+, Mn2+, Zn2+, Cd2+, Cr6+, and Ni2+) in single and multi-metal solutions. Minimum inhibitory concentration (MIC) assays revealed strain-specific variations in metal tolerance, with L2 and L30 exhibiting higher tolerance. Surprisingly, R3 and R19, despite lower tolerance, demonstrated superior metal removal efficiency, challenging the notion that tolerance dictates removal efficacy. In single-metal solutions, R3 and R19 excelled at extracting various metal ions, while competitive binding in multi-metal solutions hindered removal. However, R3 and R19 retained higher removal efficiencies, possibly due to enhanced flocculation activities facilitating metal-ion contact. Comprehensive Fourier-transform infrared (FTIR) analysis highlighted the strains’ metal-binding capabilities, with novel peaks emerging after metal exposure, indicative of extracellular polymeric substance (EPS) production. Scanning electron microscopy (SEM) and energy-dispersive X-ray spectroscopy (EDX) confirmed metal accumulation on bacterial surfaces and within cytoplasmic regions and revealed morphological changes and metal adsorption patterns, emphasizing the strains’ ability to adapt to metal stress. Scanning transmission microscopy (STEM) and EDX analysis uncovered metal accumulation within bacterial cells, underscoring the complexity of microbial-metal interactions. This study also confirms that the simultaneous presence of an aqueous solution may cause a mutual inhibition in the adsorption of each metal to the EPS resulting in reduced metal uptake, which emphasizes the need to select specific bacterial strains for a given metal-containing effluent. The differences in metal distribution patterns between Klebsiella sp. R19 and Raoultella sp. L30 suggest species-specific metal accumulation strategies driven by environmental conditions and metal availability. The heavy metal-removing capabilities and the ability to grow over a wide range of metal concentrations of the strains used in this study may offer an advantage to employ these organisms for metal remediation in bioreactors or in situ.
1. Introduction
Wastewater contamination has emerged as a critical environmental concern in recent years. The escalating processes of industrialization and urbanization have contributed to the significant release of heavy metals into aquatic ecosystems (Boening, 2000; Zamora-Ledezma et al., 2021; Goswami and Neog, 2023). The release of heavy metals into aquatic environments arises from a variety of sources, including industrial discharges, urban runoff, and agricultural activities (Tiquia, 2010; Oest et al., 2018; Patel et al., 2019; Narwal et al., 2023). These pollutants pose a substantial threat to both aquatic ecosystems and human health due to their persistence and potential toxicity (Goswami and Neog, 2023). As heavy metals accumulate in aquatic environments, they can disrupt the delicate balance of these ecosystems, causing harm to aquatic life and endangering the health of humans who rely on these water bodies for various purposes, including drinking water and recreational activities (Tiquia, 2011; Sharma et al., 2023). Additionally, heavy metal contamination can have far-reaching ecological implications, affecting the food chain and ultimately impacting terrestrial ecosystems as well (Ledin, 2000; Ahmad et al., 2021). Among the heavy metals under rigorous investigation are chromium (Cr), cadmium (Cd), copper (Cu), zinc (Zn), mercury (Hg), lead (Pb), nickel (Ni), manganese (Mn), and arsenic (As), primarily due to their pronounced threats to both public health and the environment (Boulanger and Nikolaidis, 2003; Tchounwou et al., 2012; Queiroz et al., 2021). These heavy metals find their way into the environment through human-driven activities like mining and agriculture (Goswami and Neog, 2023). Traditional techniques such as reverse osmosis, ion exchange, precipitation, and solvent extraction have been utilized to treat wastewater before discharge into natural ecosystems. However, these methods either incur substantial costs or fall short in terms of efficacy (Volesky, 1990; Qasem et al., 2021). In response, the employment of microorganisms for heavy metal removal has emerged as a highly promising alternative. This approach holds advantages by preventing secondary pollution and offering cost-effectiveness through the avoidance of sludge disposal requirements (Kim et al., 1996; Bruins et al., 2000; Lakherwal, 2014).
Bioremediation processes using microorganisms are cost effective and are highly efficient as compared to physicochemical methods mentioned above; therefore, over the last several decades attention has focused toward exploiting microbes for heavy metal bioremediation (Tiquia-Arashiro, 2018). The bioremediation of heavy metals can be categorized into two main approaches: bioaccumulation, which is an actively controlled process necessitating the uptake of heavy metal ions by living biomass, and biosorption, a passive process where metal cations are adhered to non-living biomass (Razzak et al., 2022; Jeyakumari et al., 2023). In the context of bioaccumulation, energy is expended for the absorption of metal ions, usually achieved through interactions with the cell wall. Following this, intracellular uptake takes place, with metal ions permeating the cell membrane and binding to active sites provided by polysaccharides and proteins. In contrast, biosorption capitalizes on the presence of functional groups in the cell wall or exported metabolites within the external environment. The mechanisms behind biosorption include ion exchange, complexation, precipitation, reduction, and chelation (Vishan et al., 2017; Priya et al., 2022; Sreedevi et al., 2022).
Accordingly, understanding the mechanisms by which microorganisms sequester toxic heavy metals is crucial to the development of microbial processes that will concentrate, remove, and recover metals from industrial effluents. The bioavailability of metals in the environment is an important factor for metal toxicity since soluble metals can readily penetrate cell membranes (Roane, 1999; Tiquia-Arashiro, 2018; Jeong et al., 2023). To counteract this, bacteria employ immobilization strategies to counteract the toxic effect of heavy metals, which includes precipitation, intracellular accumulation, and extracellular sequestration in exopolysaccharides (EPSs) (Higham and Sadler, 1984; Roane, 1999; van Hullebusch et al., 2003; Tiquia-Arashiro, 2018; Vandana et al., 2023). EPSs typically consist of polysaccharides featuring ionizable functional groups that can bind to anionic species. This transformation facilitates electrostatic interactions with cationic metal ions, ultimately leading to the immobilization of heavy metals within the EPS matrix (Saba et al., 2019). The effectiveness of EPS-mediated biosorption in efficiently sequestering toxic metals has led to its extensive investigation (Shameer, 2016; Vishan et al., 2017; Saba et al., 2019; Sharma and Saraf, 2023). As a result, biosorption via EPS has garnered substantial research attention. Heavy metals do not only need to be remediated from the ecosystem but are also required to be recovered from every possible source given their importance in commercial and industrial applications. The interaction between several metal cations and the EPS-producing microorganisms combined with their potential for removing heavy metals from polluted waters has stimulated scientific interests due to their ecological importance and practical implications.
Despite the isolation of diverse bacterial strains, the significance of isolating and characterizing indigenous bacteria remains paramount. This approach is not only environmentally sound but also preserves the ecological balance within their specific habitats. This assertion is corroborated by studies that have harnessed metal-tolerant bacteria sourced from industrial effluents (Mishra et al., 2013; Bhatt et al., 2023). In our previous study (Bowman et al., 2018), we isolated Pb-resistant bacterial strains, Klebsiella sp. R3, Klebsiella sp. R19, Serratia sp. L2, and Raoultella sp. L30 from sediments of the Saint Clair River. These bacterial strains remove large amounts of Pb2+ from solution and produce a high rate of flocculation activity (Bowman et al., 2018). Furthermore, these bacterial strains are well adapted to unfavorable conditions due to their resistance to metals (e.g., Pb) and antibiotics and can grow in a wide range of temperatures. These characteristics may help in developing an effective process for wastewater treatment. However, the metal removal of these strains in aqueous multi-metal solutions has not been investigated in detail, which is particularly useful for building up processes aimed at recovering metals from industrial wastewaters. This study aims to (1) assess the selective metal removal abilities of the bacterial strains in the presence of multiple heavy metals (As3+, Pb2+, Cu2+, Mn2+, Zn2+, Cd2+, Cr6+), (2) investigate the interactions between these metals during the sorption process, and (3) uncover mechanisms of the biosorption process using FT-IR, SEM–EDX, and STEM-EDX techniques.
2. Materials and methods
2.1. Bacterial strains
In this study, the strains were grown on M9 minimal media (M9 salts [BD Difco, Franklin Lakes, NJ], 20% glucose, 1 M MgSO4, 1 M CaCl2). The biochemical properties of bacterial strains were characterized using the API 20E system (Bio-Mérieux, Marcy-l’Étoile, France). Furthermore, the 16S rRNA sequence of the bacterial strains were determined by direct sequencing of the PCR product. DNA extracts from each strain were prepared using the DNEasy kit (Qiagen, Inc., Valencia, CA). Genomic DNA from these isolates were extracted using the DNEasy kit. Identification of isolates was performed by amplifying the full-length 16S rRNA genes using bacteria-specific primers sequence FD1 (5′ AGA GTT TGA TCC TGG CTC AG 3′) and 1540r (5′ > AAG GAG GTG ATC CAG CC < 3′) (Weisburg et al., 1991) with cycling conditions described previously (Tiquia et al., 2007, 2008; Cho et al., 2012; Nguyen et al., 2013; Plecha et al., 2013). The purified PCR products were sequenced and analyzed directly without cloning. Sequence fragments generated from a given template were edited against electropherograms and then assembled into contigs using SeqMan (Lasergene DNASTAR, Inc., Madison, WI). Two to four overlapping fragments (from both coding and noncoding strand) were used to assemble the contigs. Chimeric sequences were checked by the Check_Chimera program available at the Ribosomal Database Project (RDP II) (Hugenholtz and Huber, 2003). Complete 16S rRNA gene sequences were compared with other reference sequences as available in the NCBI database using the Basic Local Alignment Search Tool (BLAST) algorithm. Closely related sequences were retrieved from Genbank and were aligned along with the 16S rRNA gene sequences of the bacterial strains using the ClustalW program. Phylogenetic trees (Supplementary Figure S1) were constructed from the evolutionary distance matrix calculated through the neighbor-joining method (Saitou and Nei, 1987). Neighbor joining analysis was performed using MEGA 11 (Tamura et al., 2021). Based on the morphological, physiological, biochemical characteristics (Supplementary Table S1) and comparative analysis of the sequence to references retrieved from the NCBI database, the strains were found to be similar to the 16S rRNA sequences of Klebsiella sp. (strain R2), Klebsiella sp. (strain R19), Serratia sp. (strain L2) and Raoultella sp. (strain L30).
2.2. Determination of minimum inhibitory concentrations to metals
The bacterial strains used in this study are resistant to Pb and grew in enrichment cultures containing high concentrations (1.25 or 1.5 g L−1) of Pb (NO3)2 (Bowman et al., 2018). To determine their resistance to other metals (As3+, Pb2+, Cu2+, Mn2+, Zn2+, Cd, 2+ Cr6+, or Ni2+), the MIC on the strains to these metals was carried out. Stock solutions of As (NaAsO2), Cr (K2Cr2O7), Cd (CdCl2), Cu (CuCl2.2H20), Pb (Pb [NO3]2), Mn (MnCl2.4H2O), Ni (NiCl2.6H2O), and Zn (ZnSO4.7H2O) were prepared to achieve the final concentration of 1,000 and 10,000 mg L−1 with distilled water. The metal solutions were filter sterilized using 0.2 μm Nalgene vacuum filtration system (Thermo Scientific, Waltham, MA). Polycarbonate filters were required for the Cu study. For other metals, polysulfone filters were used to prevent sorption of metals to the filter apparatus (Shuttleworth and Unz, 1993; Wu et al., 2005). Susceptibility testing of the strains to eight different metals was conducted in M9 minimal media amended with metals. Metal-amended media were prepared by adding increasing amounts of metal stock solutions to the autoclaved media. The broth media (200 μL) were poured into 96-well plates and inoculated with mid-logarithmic-phase cultures (20 μL). The plates were incubated at room temperature with shaking (SBT1500 Microplate Shaker, Southwest Science, Hamilton, NJ) at 150 rpm. The MICs (minimal inhibitory concentrations) reported in this study were the lowest concentration of the metal that inhibited bacterial growth after an incubation of 24 h at room temperature in the dark. Growth media inoculated with bacterial strains without metals were used as positive controls. Growth of the strains was monitored by optical density at 595 nm with Sunrise microwell plate reader (Tecan, Research Triangle Park, NC). The absence of growth was determined by the detection threshold of the plate reader.
2.3. Metal removal assay in aqueous solutions
Cells of each culture (100 mL) were confined in dialysis tubing (12–14 kDa of molecular weight cut-off; Spektra/por 5, Spectrum Laboratories, CA) and pretreated with 0.1 N HCl to remove the metal ions possibly bound to the negatively charged groups of the cell envelopes. After 30 min of soaking in 0.1 N HCl, the cell cultures were dialyzed against deionized water for 24 h with continuous stirring at 100 rpm, to remove residual HCl from the treated cultures. In the mono-metal systems, dialyzed cultures were subsequently transferred in 1 L aqueous solutions containing 10 mg L−1 of As3+, Pb2+, Cu2+, Mn2+, Zn2+, Cd2+, Cr6+, or Ni2+ for 24 h. This metal concentration (10 mg L−1) is like that used in other dialysis assays to investigate the binding capacity of bacterial cells or bacterial EPS with metals (Perez et al., 2008; Pereira et al., 2011; Wang et al., 2014; Bowman et al., 2018). To avoid any possible hydroxide precipitation, the pH of the system (biosorbent + metal solution) was adjusted to 4.5–5.5 by addition of either 1 N HCl or 1 N NaOH. The experiment was performed at room temperature. For the multi-metal systems, 10 mg L−1 of each metal was used. Such concentration was used for each metal to be consistent with the concentrations used for the single-metal assays.
Ten mL of the aqueous solutions were withdrawn at different time intervals over 24 h and filtered through 0.2 μm Acrodisc syringe filters (Sigma-Aldrich, Saint Louis, MO). The metal uptake by the bacterial strains was determined from the difference between the concentration of the metals in solution using an atomic absorption spectrometer (PinAAcle 900 T, Perkin Elmer, MA). The As3+, Pb2+, Cu2+, Mn2+, Zn2+, Cd2+, Cr6+, and Ni2+ were determined at 193.70, 283.31, 324.75, 279.48, 213.86, 228.00, 357.87, and 232.0 nm, respectively. The amount of metal uptake in blanks (carried out in parallel with just 100 mL of culture media without cells) was also monitored to confirm that negligible metal was lost from solution. The amount of metals removed in blanks, which ranged from 0.1 to 0.2 mg L−1, was subtracted from the experimental values obtained in the tests with the bacterial cultures.
Specific metal uptake (q) was expressed as mg metal removed per g of dry biomass. The dry weight (g L−1) was determined by centrifugation 10 mL of the dialyzed cultures, followed by drying at 50°C until a constant weight was reached (Volesky and May-Philips, 1995). The experiments were performed in triplicates and the data were reported as mean values ± standard deviation. As metal sorption was followed, cell viability was also determined at the beginning and at 24 h using the plate count method (Gerhardt et al., 1994).
2.4. FT-IR analysis of bacterial biomass
Fourier transform infrared spectroscopy (FT-IR) was performed to identify the functional groups in the bacterial strains that might be involved in metal uptake during the biosorption process. This technique has proven to be effective in providing structural information on metal cation binding in microbes (Gupta et al., 2020). FT-IR analysis was carried out on cells before and after metal uptake in aqueous solution containing the 8 metals (As3+, Pb2+, Cu2+, Mn2+, Zn2+, Cd2+, Cr6+, and Ni2+), each had a concentration of 10 mg/L. The infrared spectra of the control before and after metal uptake were analyzed on lyophilized cells using Attenuated Total Reflection (ATR).
2.5. SEM–EDX analysis
Field emission scanning electron microscopy (Tescan Mira3 FEG SEM, Tescan USA, Inc., Pleasanton, CA 94588, United States) coupled with energy dispersive X-ray spectroscopy was carried out to examine the outer morphology of the bacterial cells (Raoultella sp. L30 and Klebsiella R19) grown without metal stress and bacterial cells grown with metals. Samples were fixed in 2.5% glutaraldehyde in Sorensen phosphate buffer (pH 7.2), dehydrated with increasing solutions of ethanol (30, 50, 70, 80, 90, 95, and 100%) and HDMS. Prepared samples were placed on the sample holder (stub) with carbon tape. To improve electron conduction, the samples were sputter coated with carbon.
2.6. STEM-EDX analysis
Scanning Transmission electron microscopy with energy dispersive X-ray (EDS) analysis (Thermo Fisher Talos F200X G2, Waltham, Massachusetts, United States) was used to determine the location of the metals in the cell. Strains R19 and L30 were washed with phosphate buffer saline (pH 6.8). After washing, the samples were embedded in agarose and then fixed in 3% Glutaraldehyde +3% paraformaldehyde solutions in 0.1 M Cacodylate buffer (pH 7.2). After 24 h, the cells were fixed with osmium tetroxide (2%) and potassium ferrocyanide (1%) 0.1 M carbonate buffer for 1 h and then washed with 0.1 M carbonate buffer and 0.1 M Na2 + acetate buffer (pH 5.2) before en-bloc staining for 1 h. Using an automated tissue processor (Leica EM TP, Leica Biosystems, Deerfield, Illinois, USA) the samples were washed with 0.1 M carbonate buffer and 0.1 M Na2 + acetate buffer (pH 5.2) and then dehydrated with increasing concentrations of ethanol before infiltrated with acetone and Spurr’s resin. Thin sections of 60–80 nm thick were obtained using an ultramicrotome (Leica EM UC7, Leica Biosystems, Deerfield, Illinois, United States) and stained with uranyl acetate. The section was fixed on a carbon grid and examined TEM.
2.7. Nucleotide sequence accession numbers and taxon numbers
The nucleotide sequence coding for 16S rRNA genes of Klebsiella sp. R3, Klebsiella sp. R19, Serratia sp. L2, and Raoultella sp. L30 were submitted to the GenBank database under the accession numbers OR143353, MG022653, OR143351, and OR143352, respectively. The respective digital protologue database (DPD) Taxon Number were: TA00308 (Klebsiella sp. R3), TA00308 (Klebsiella sp. R19), TA00309 (Serratia sp. L2), and TA00310 (Raoultella sp. L30).
3. Results
3.1. Tolerance to metals of the bacterial strains
Table 1 provides the minimum inhibitory concentrations (MICs) of the eight different metals for the respective bacterial strains. Overall, strains L2 and L30 (Serratia sp. strain L2 and Raoultella sp. strain L30) exhibited higher metal tolerance compared to strains R3 (Klebsiella sp.) and R19 (Klebsiella sp.). In a broader perspective, the strains demonstrated greater resilience against As3+ (ranging from 250 to 450 mg L−1), Pb2+ (ranging from 700 to 800 g mL−1), Mn2+ (>2,000 mg L−1), and Zn2+ (ranging from 500 to 1,100 mg L−1), in contrast to Cd2+ (ranging from 20 to 75 mg L−1), Cr6+ (ranging from 5 to 10 mg L−1), Cu2+ (ranging from 20 to 50 mg L−1), and Ni2+ (ranging from 50 to 75 mg L−1).
The hierarchy of metal tolerance for each bacterial strain was as follows: for Klebsiella sp. R3, it was Mn2+ > Zn2+ > Pb2+ > As3+ > Cu2+ = Ni2+ > Cd2+ > Cr6+; for Klebsiella sp. R19, it was Mn2+ > Pb2+ > Zn2+ > As3+ > Cu2+ = Ni2+ > Cd2+ > Cr6+; for Serratia sp. L2, it was Mn2+ > Zn2+ > Pb2+ > As3+ > Ni2+ = Cd2+ > Cu2+ > Cr6+; and for Raoultella sp. L30, it was Mn > Zn > Pb > As > Cd > Ni = Cu > Cr (Table 1).
3.2. Metal removal in single-metal and multi-metal systems
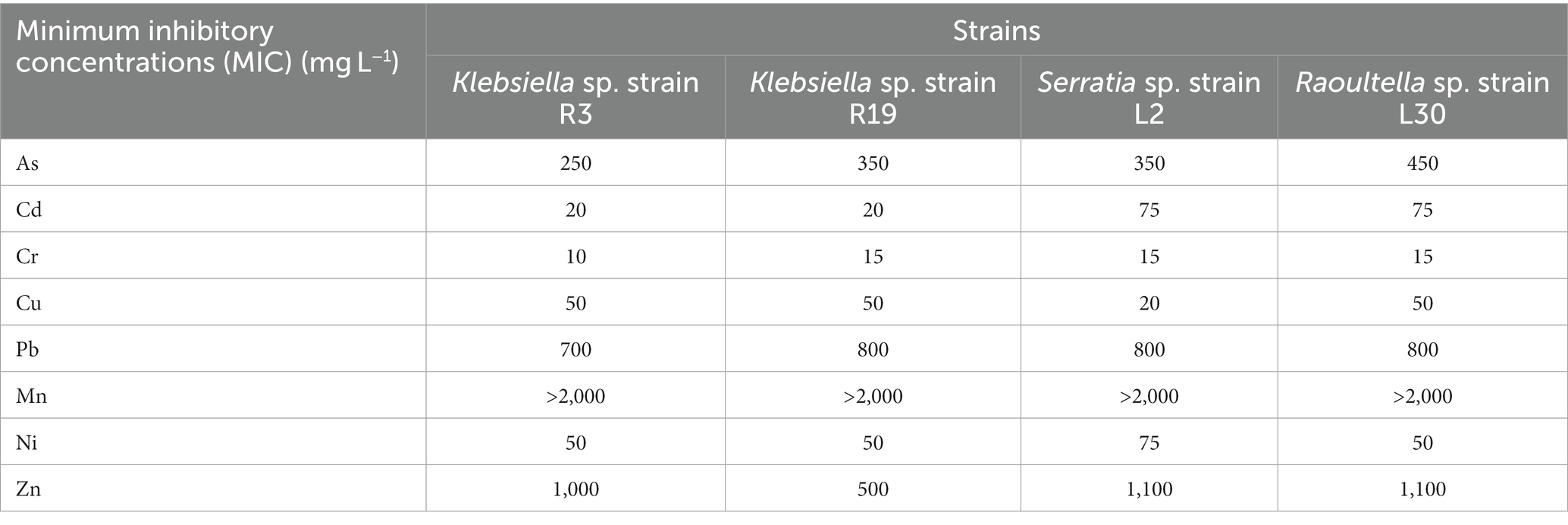
The investigation into the specific metal removal capabilities of the bacterial strains was conducted using individual single-metal solutions, each containing 10 mg L−1 of As3+, Pb2+, Cu2+, Mn2+, Zn2+, Cd2+, Cr6+, or Ni2+. The study spanned various time intervals over a 24-h period (as demonstrated in Supplementary Figure S2). Over time, the metals were gradually extracted from the solutions, with saturation of the metal removal capacity achieved within approximately 4 to 5 h (Supplementary Figure S2), except for As3+ (notably for strains L2 and L30) and Cr6+, for which minimal or negligible removal occurred. Generally, the metal removal efficiency was more pronounced for Klebsiella sp. R3 and Klebsiella sp. R19 (ranging from 4.4 to 318 mg g−1 dry mass) compared to Serratia sp. L2 and Raoultella sp. L30 (ranging from 0 to 155.42 mg g−1 dry mass) (Supplementary Figure S2).
Comparatively, the efficiency of metal removal in single-metal solutions was notably higher than that in mixed solutions containing all eight metal cations (as depicted in Figure 1). Notably, in single-metal solutions, Klebsiella sp. R3 exhibited removal efficiencies of ≥150 mg g−1 dry mass for Pb2+, Cu2+, and Zn2+ (Figure 1A). Similarly, Klebsiella sp. R19 displayed high removal efficiency for Pb2+, Cd2+, Cu2+, Mn2+, and Zn2+ (Figure 1B), while Raoultella sp. L30 showed effective removal of Pb2+ (Figure 1D). In the presence of all eight metal cations, the binding process was primarily inhibitory rather than stimulatory, as the cations competed for binding sites on the bacterial cells. Unexpectedly, a stimulatory effect on the removal efficiency of Pb2+ and Cr6+ was observed in strain R19 (Figure 1B) in the presence of multiple metal cations. Specifically, Pb2+ removal in the multi-metal solution was 31% higher compared to the solution with only Pb2+, and 91% higher compared to the solution with only Cr6+. A lesser stimulatory effect was observed for Cr6+ removal in strains L2 and L30 (Figures 1C,D).
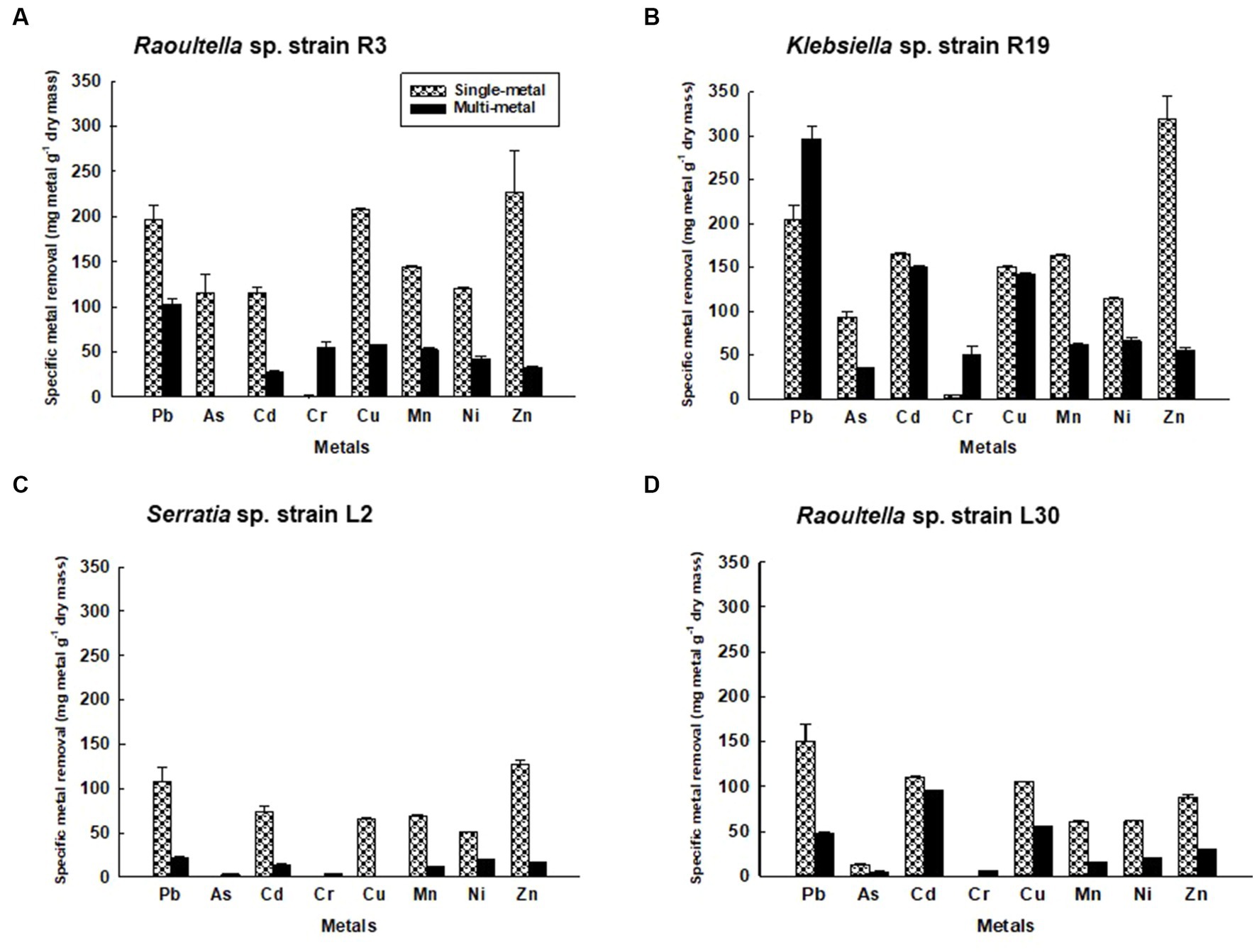
Figure 1. Metal removal by (A) Klebsiella sp. R3; (B) Klebsiella sp. R19; (C) Serratia sp. L2; and (D) Raoultella sp. L30 in single- (dotted bars) and multiple- (black bars) metal solutions. All the values are the mean of three replicates ± standard deviation.
Notably, the kinetics of metal uptake in the eight-metal solutions did not significantly deviate from those observed in the single-metal solutions. The saturation of metal removal capacity occurred within 4–5 h in the presence of all eight metals (Supplementary Figure S3). Moreover, the viability of the strains remained unaffected by the presence of metals in both single-metal and multi-metal solutions. The number of viable cells was consistent, ranging from 7.3 to 8.9 Log10 CFU g−1 of dry cell mass for strains R3 and R19, and from 8.0 to 9.7 Log10 CFU g−1 of dry cell mass for strains L2 and L30 throughout the 24-h metal binding assay (Supplementary Figure S4). There was no statistically significant difference (p ≤ 0.05) in the number of viable cells between the single-metal and multi-metal solutions.
3.3. FT-IR
FTIR spectra were acquired for bacterial strains before and after the uptake of a combination of eight metals, spanning the range of 4,000 to 400 cm−1 (depicted in Figure 2). The FT-IR profiles of metal-free bacterial strains exhibited diverse peaks, revealing the intricate nature of the bacterial cell surface. Common bands were evident in the strains prior to metal uptake across all four bacterial types, albeit with fewer IR peaks observed in the Serratia sp. L2 and Raoultella sp. L30 strains (illustrated in Figures 2A,B) compared to the Klebsiella sp. R3 and Klebsiella sp. R19 strains (shown in Figures 2C,D). These IR bands corresponded to functional groups including amino (N-H, NH2), alkyne (C ≡ C), carbonyl (C=O), carboxylic (C-O), hydroxyl (-OH), and phosphate (P=O) groups (depicted in Figure 2). The assignments of these bands and the specific functional groups for each of the four strains are detailed in Tables 2–5. Upon exposure to metals, alterations in the intensity of bands became apparent, accompanied by shifts in absorption bands and the emergence of new peaks. The number of IR bands expanded as the strains encountered the multi-metal environment. Notably, the occurrences of IR shifts and new peaks were more pronounced in Klebsiella sp. R3 and Klebsiella sp. R19 than in Serratia sp. L2 and Raoultella sp. L30 strains. In the metal-loaded L2 and L30 strains, IR spectra shifts indicated the involvement of functional groups linked to aromatic organics, alkynes (C ≡ C), and alkanes (C-H). Conversely, the R3 and R19 strains demonstrated shifts associated with aromatic organics, alkynes (C ≡ C), alkanes (C-H), as well as hydroxyl, amine, and aldehyde functional groups. Moreover, new peaks emerged in the spectra of the R3 and R19 metal-loaded strains, indicating the presence of aromatic compounds, alkanes (C-H), carboxyl (C-C), alkynes (C ≡ C), alcohol (R-CHO), amine (P-NH, NH2), and hydroxyl (O-H) functional groups. These observations underscored the intricate alterations in the bacterial cell surfaces induced by the uptake of multiple metals, with distinct variations based on bacterial strain type.
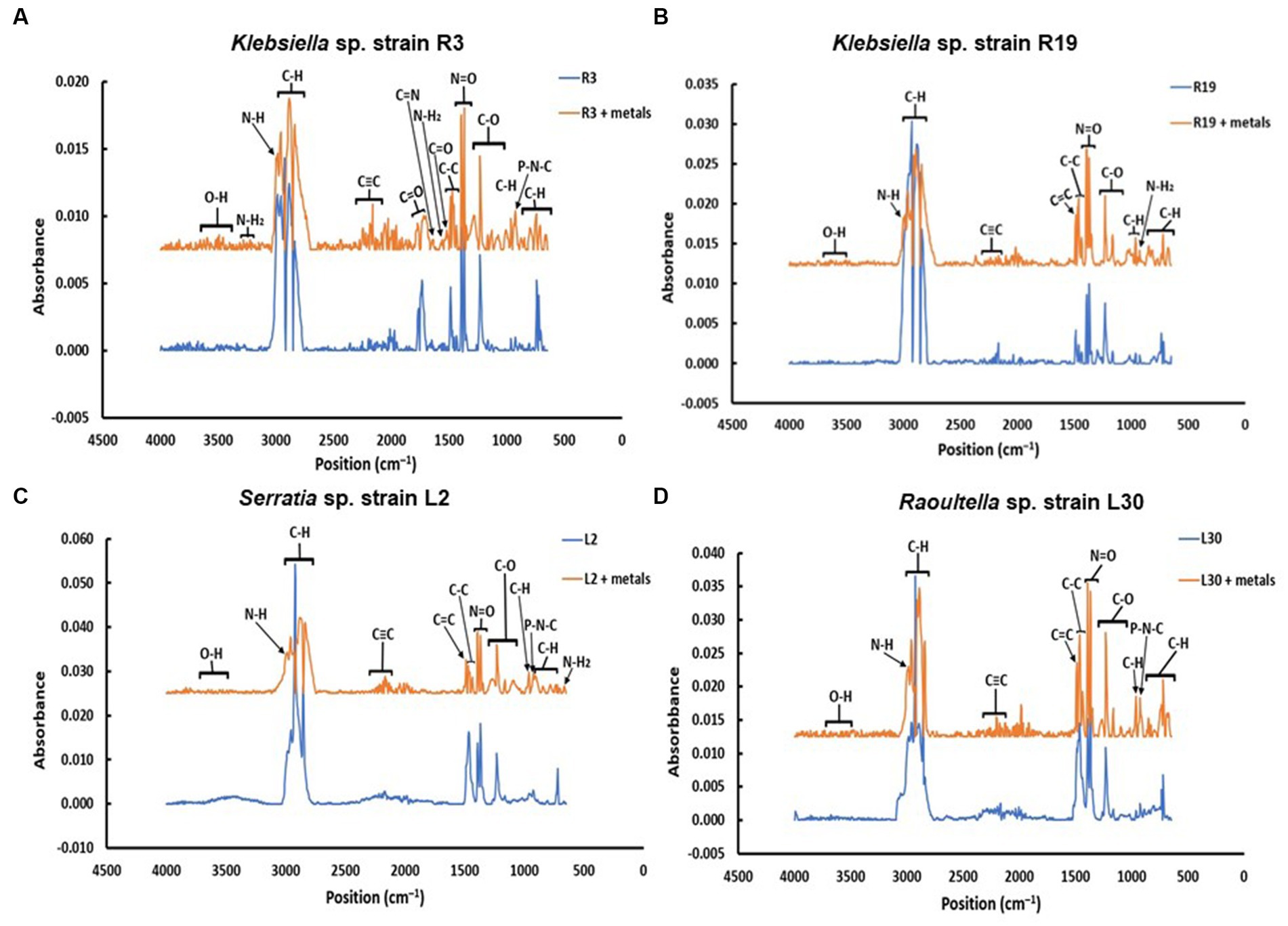
Figure 2. Comparison of the IR spectra of (A) Klebsiella sp. R19; (B) Klebsiella sp. R19; (C) Serratia sp. L2; and (D) Raoultella sp. L30 before and after absorption of mixed metals.
Table 2. IR absorption band changes and possible assignment for the metal-free and metal-loaded Klebsiella strain sp. R3.
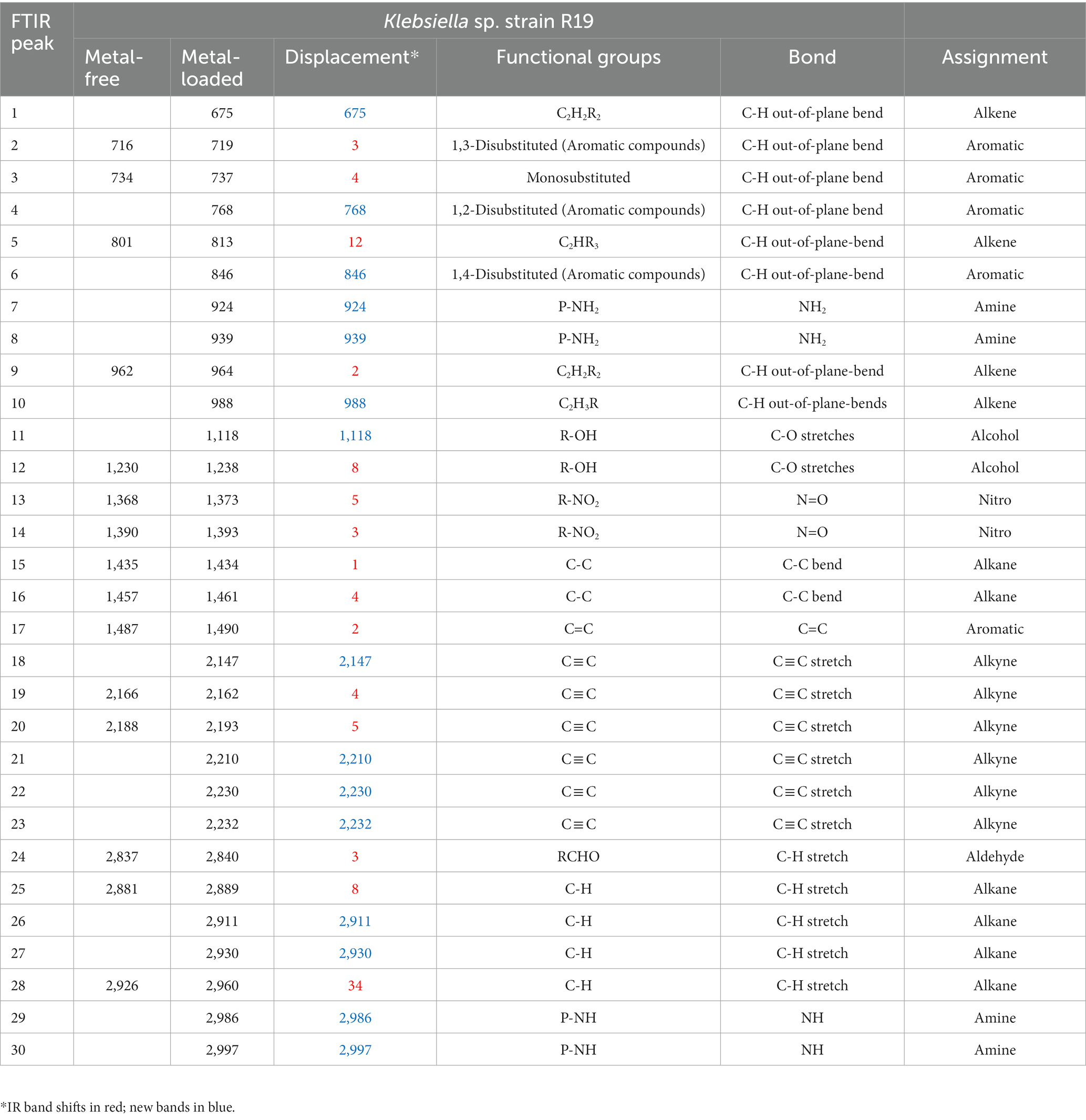
Table 3. IR absorption band changes and possible assignment for the metal-free and metal-loaded Klebsiella sp. strain R19.
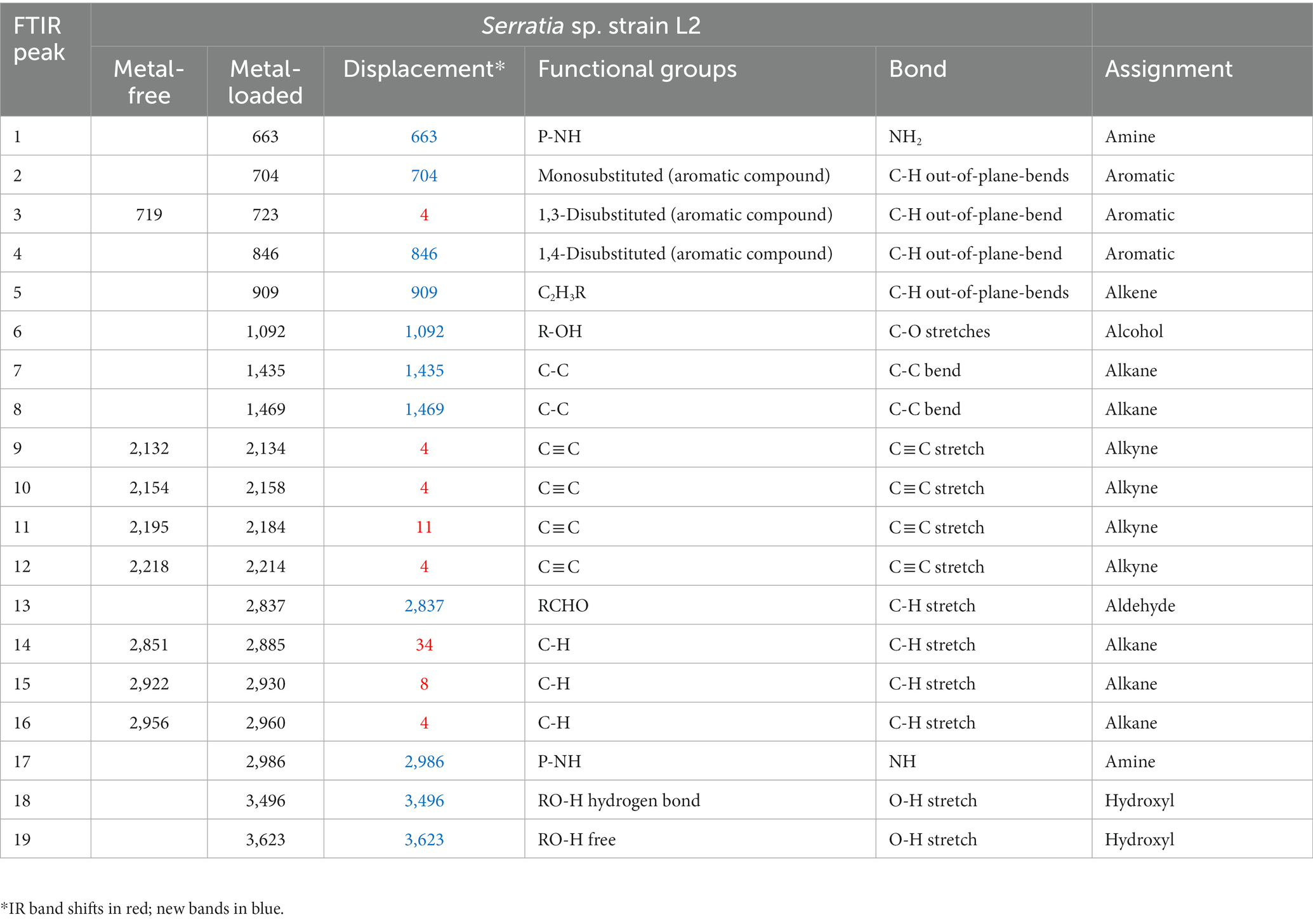
Table 4. IR absorption band changes and possible assignment for the metal-free and metal-loaded Serratia sp. strain L2.
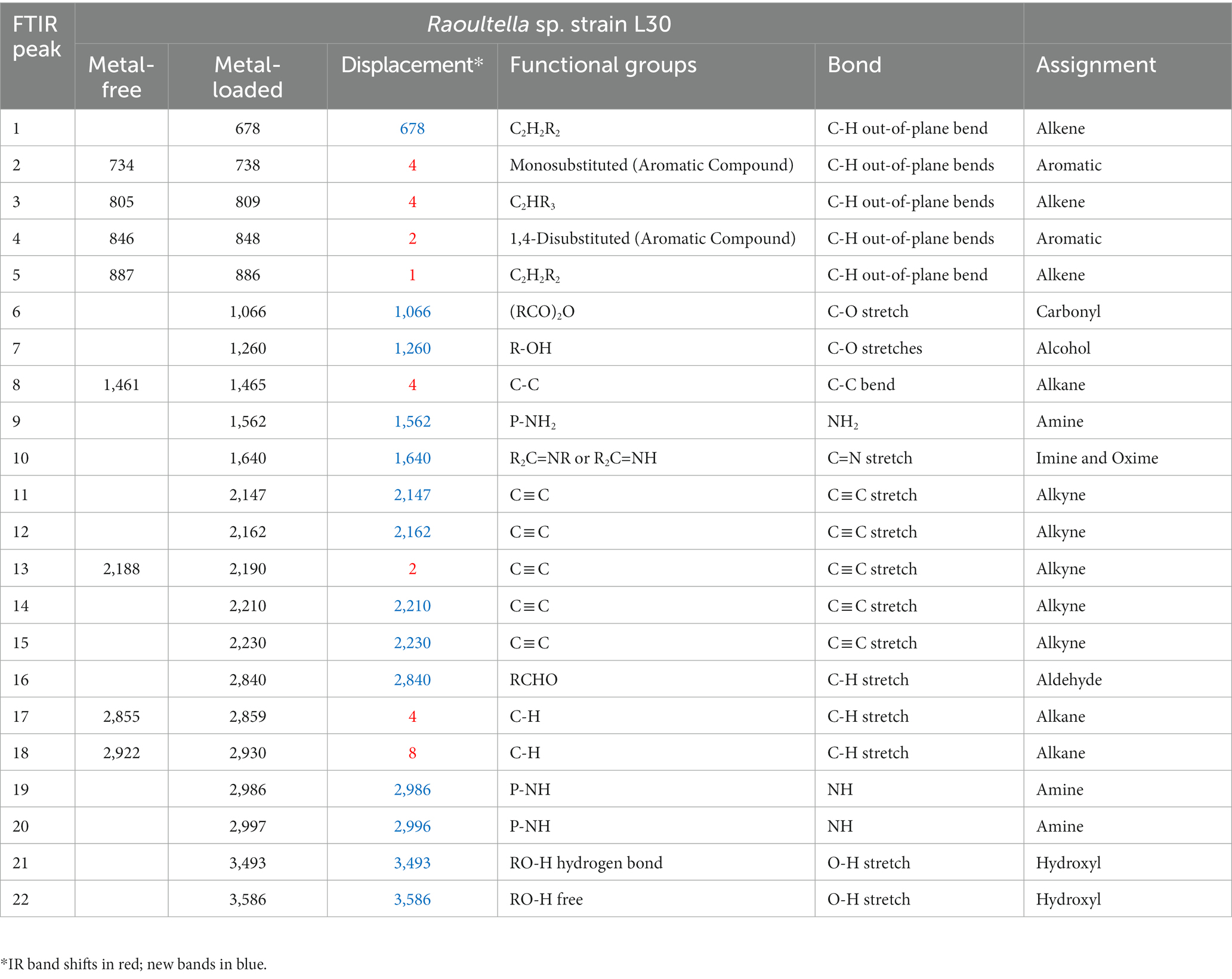
Table 5. IR absorption band changes and possible assignment for the metal-free and metal-loaded Raoultella sp. strain L30 strain.
3.4. SEM–EDX
Examination of SEM micrographs revealed alterations in both size and morphology of Klebsiella sp. R19 (depicted in Figure 3) and Raoultella sp. L30 (depicted in Figure 4) before and after metal adsorption. Initially, Klebsiella sp. R19 exhibited an average length of 1.78 ± 0.40 μm and an average width of 0.71 ± 0.12 μm (Figure 3A). Subsequent to the biosorption process in an aqueous solution containing eight metals, its dimensions were diminished to 1.32 ± 0.27 μm x 0.70 ± 0.11 μm (Figure 3B), reflecting reductions of 26% in length and 3% in width. A parallel outcome was observed for Raoultella sp. L30, wherein its size underwent a decrease from 1.53 ± 0.34 μm x 0.65 ± 0.09 μm (Figure 4A) to 1.17 ± 0.27 μm x 0.70 ± 0.07 μm (Figure 4B). SEM analysis also provided insights into notable modifications observed on the cell surfaces of both Klebsiella sp. R19 and Raoultella sp. L30 bacteria Following the process of metal biosorption. The cell surfaces of Klebsiella sp. R19 exhibited a distinctive texture characterized by irregularities, including dents, wrinkles, and elongations (Figures 3C,D). These surface features appeared to be a direct consequence of the interaction between the bacterial cells and the adsorbed metals. The metals’ toxic influence appeared to induce these alterations in the cell surface structure. Additionally, a considerable amount of flocculation was evident on the cell surfaces. This phenomenon could be attributed to the secretion of extracellular polysaccharides prompted by the presence of metals. The extracellular polysaccharides might have contributed to the aggregation of bacterial cells, leading to the formation of these flocculent clusters (Figure 3C). The cell surfaces of Raoultella sp. exhibited varying degrees of deformations, creases, or elongations due to the adverse impact of the metals. Additionally, a notable amount of flocculation emerged on the cell surfaces, prompting cellular aggregation (Figures 4C,D). This phenomenon might be attributed to the metal-induced secretion of extracellular polysaccharides (Gupta and Diwan, 2017).
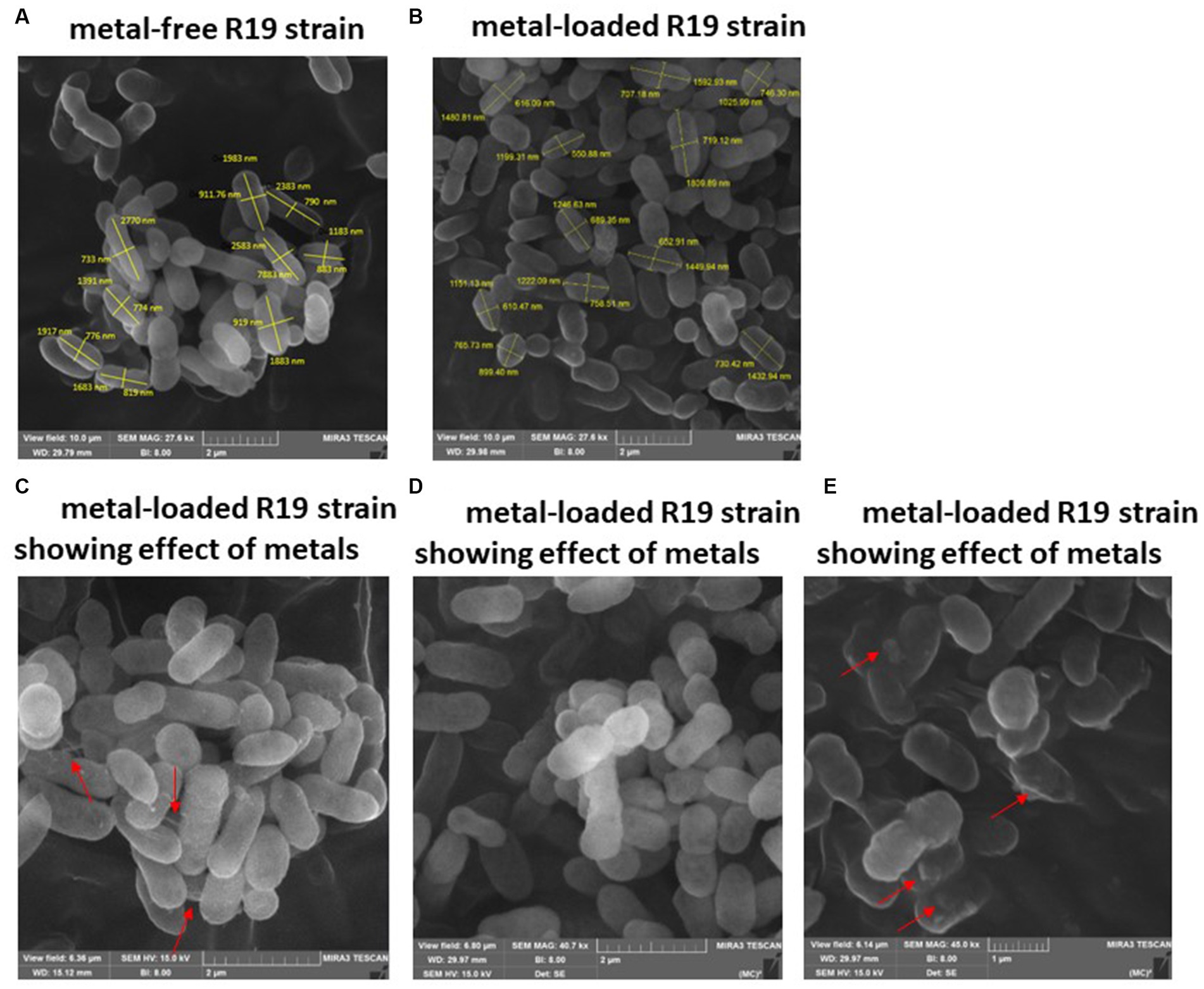
Figure 3. SEM images of Klebsiella sp. R19 before (A) and after absorption of mixed metals (B) and morphological changes when exposed to the eight metals (C–E). The size (length and width) and cells are shown in nm. The red arrows point to the presence of flocs on the cell surface.
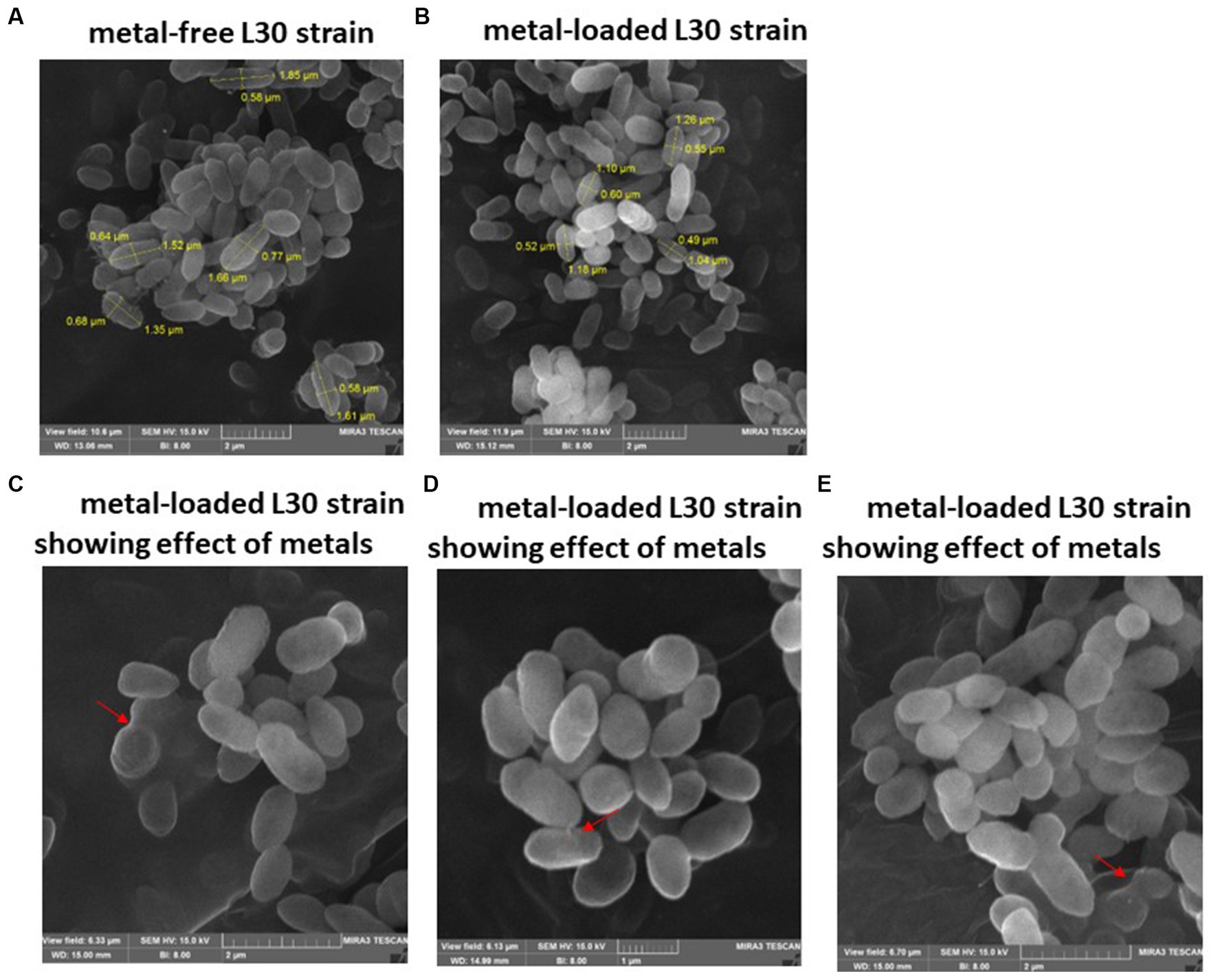
Figure 4. SEM images of Raoultella sp. L30 L30 before (A) and after absorption of mixed metals (B) and morphological changes when exposed to the eight metals (C–E). The size (length and width) and cells are shown in μm. The red arrows point to the presence of flocs on the cell surface.
The energy-dispersive X-ray spectroscopy (EDX) analysis of Klebsiella sp. R19 and Serratia sp. L30 (Figure 5) affirmed the presence of adsorbed metals on the cell surfaces. The quantified outcomes, illustrating the distribution of the eight metals on the cell surfaces of Klebsiella sp. R19 and Raoultella sp. L30, are presented in Figure 5. Among these eight metals, Cd2+ predominated the cell surface of Klebsiella sp. followed by Cu2+, Zn2+ and Pb2+ (Figure 5A), while Cu2+ stood out as the foremost element, followed by Zn2+, Cd2+, and As3+ (Figure 5B).
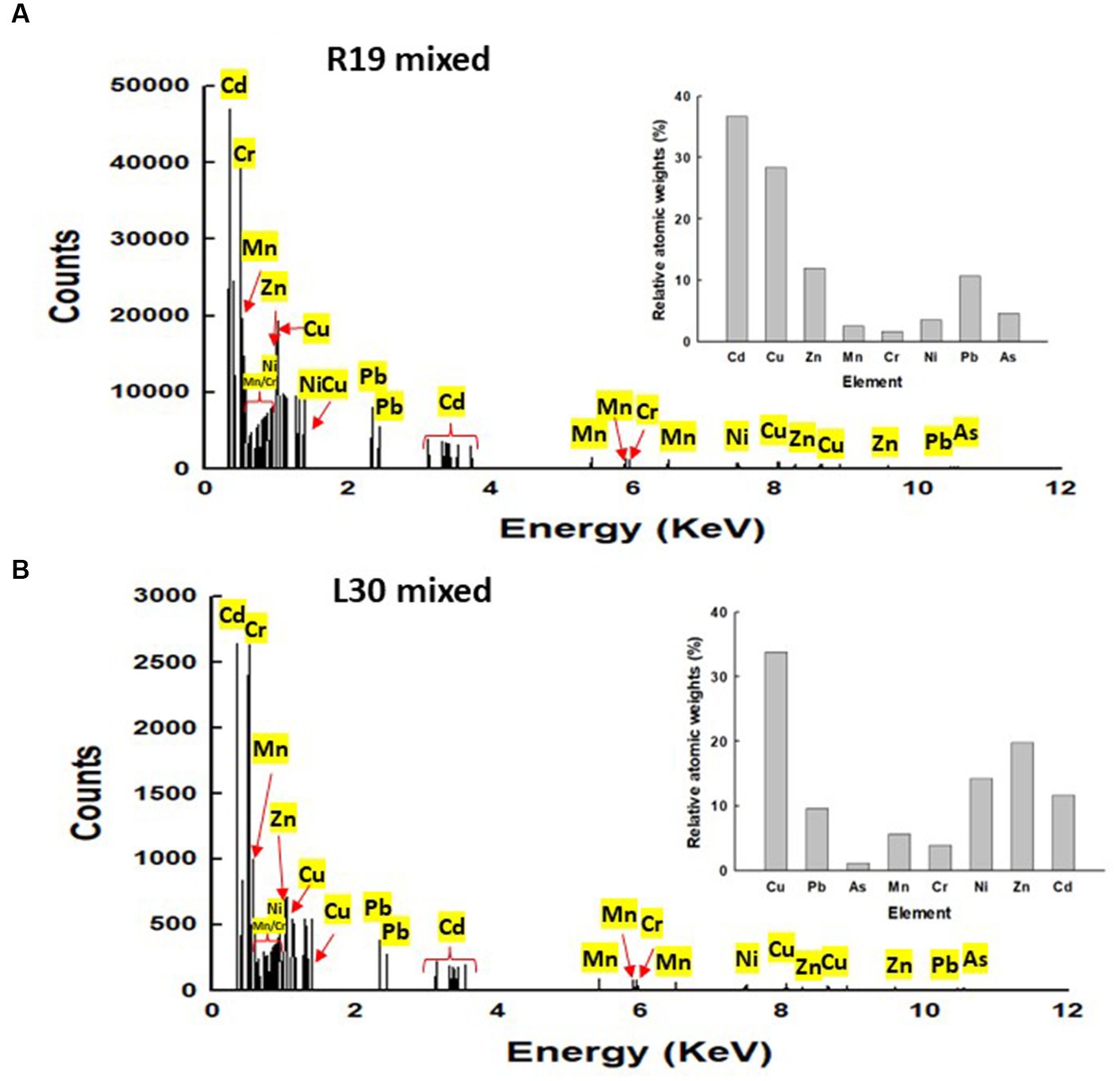
Figure 5. Energy dispersive X-ray spectroscopy (EDX) results for (A) Klebsiella sp. R19 and Serratia sp. L30 after absorption of mixed metals (B). Quantification results of Pb, As, Cd, Cr, Cu, Mn, Ni or Zn in Klebsiella sp. R19 and Serratia sp. L30 strains at 15 kV.
3.5. STEM-EDX
STEM images unveiled the accumulation of metals within both the cytoplasm and cell walls of Klebsiella sp. R19 (depicted in Figures 6, 7) and Raoultella sp. L30 (illustrated in Figures 8, 9). Numerous electron-dense granules were discernible, situated across cell walls, membrane fractions, and within the cytoplasm. These electron-dense granules appeared vividly in the STEM-HAADF image. The granules exhibited three distinct types: sizable and compact granules (indicated by blue arrows), smaller granules (marked by green arrows), and saturation clustering around storage granules (designated by purple arrow) (depicted in Figure 7). Interestingly, Raoultella sp. L30 did not exhibit evident storage granules. The verification via EDX analysis affirmed the presence of metal accumulation on the cell wall, cell membrane, and cytoplasm. The position beam spectra clearly indicated the detection of all eight metals within the electron-dense granules for both strains (portrayed in Figures 6–9). These electron-dense granules encapsulate metal complexes, effectively binding the metals. To determine the relative distribution of the eight metals within the electron-dense granules, the atomic percentages (atm %) of As3+, Pb2+, Cu2+, Mn2+, Zn2+, Cd2+, Cr6+, and Ni2+ were normalized to 100%, after excluding contaminants (such as C, O, etc.).
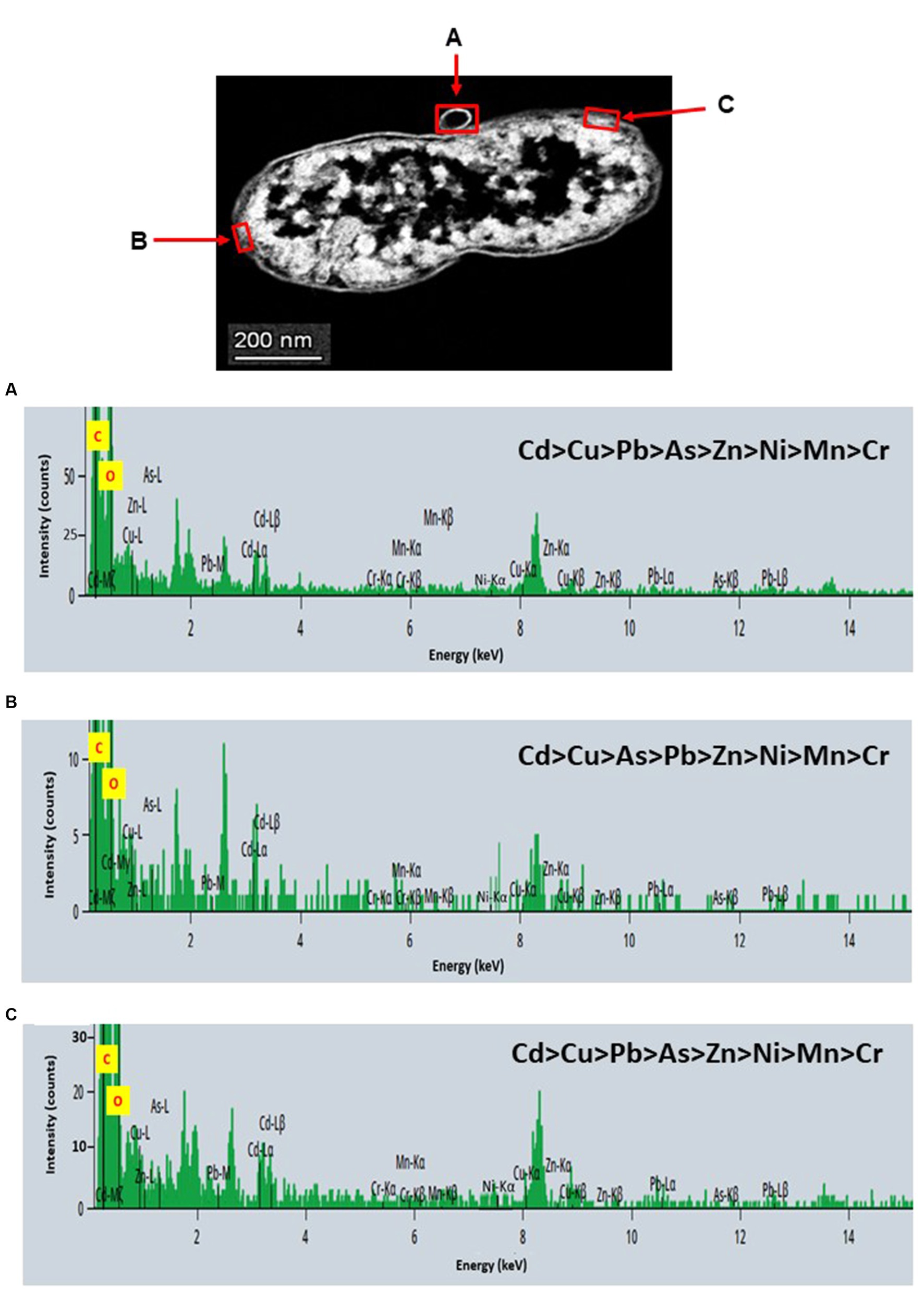
Figure 6. Scanning transmission electron micrographs (STEM) off a thin section of metal-loaded Klebsiella sp. R19 and the location of metals. (A–C) Energy dispersive X-ray spectra of the surface biosorption of metals acquired from the region indicated by arrow A, B, and C. The electron-dense granules appear bright on the STEM-HAADF image.
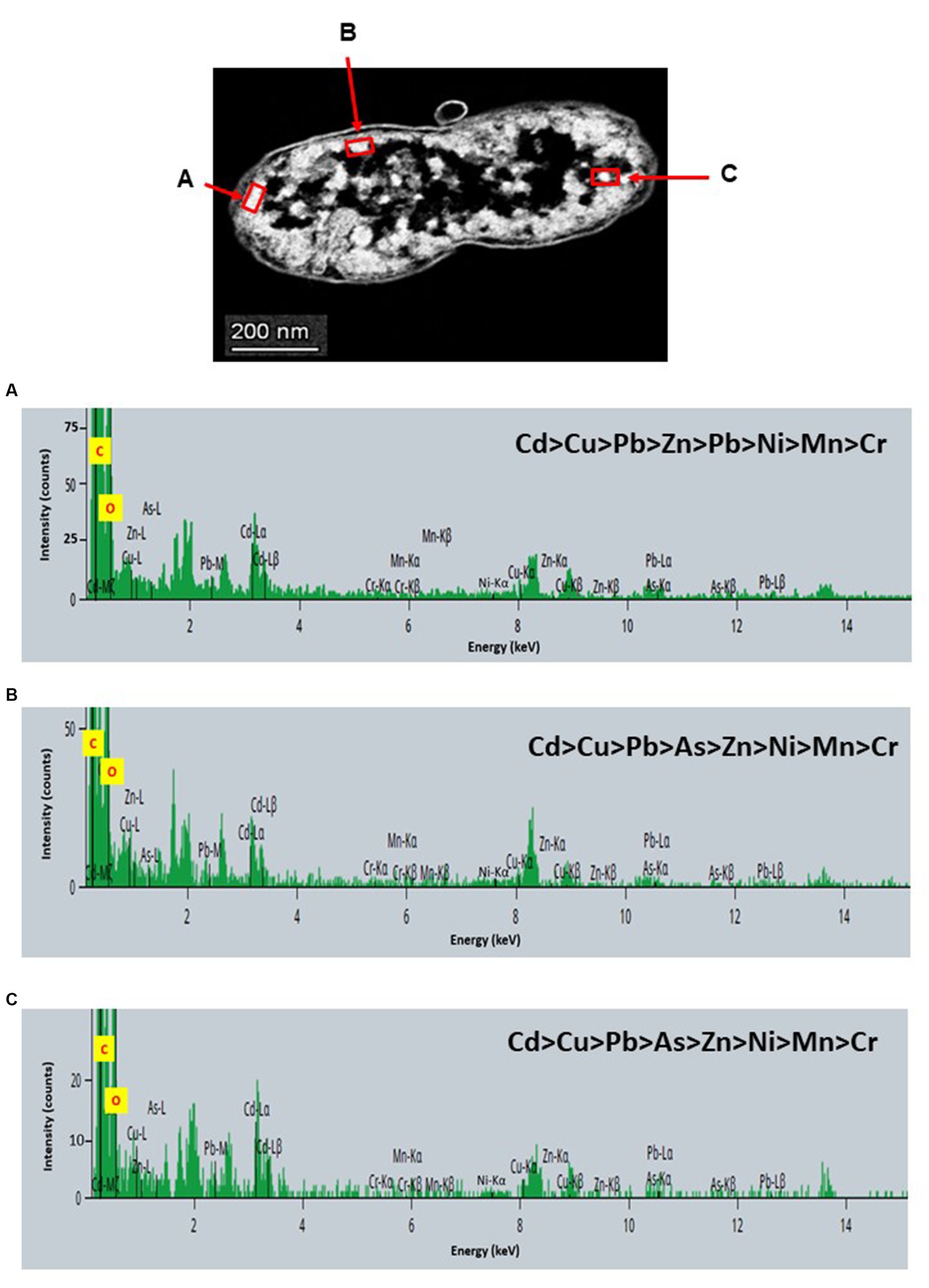
Figure 7. Scanning transmission electron micrographs (STEM) off a thin section of metal-loaded Klebsiella sp. R19 and the location of metals. (A–C) Energy dispersive X-ray spectra of the intracellular accumulation of metals acquired from the region indicated by arrow A, B, and C. The electron-dense granules appear bright on the STEM-HAADF image. Granules are of three types: large and compact (blue arrows), small (green arrows), and saturated around storage granules (purple arrow).
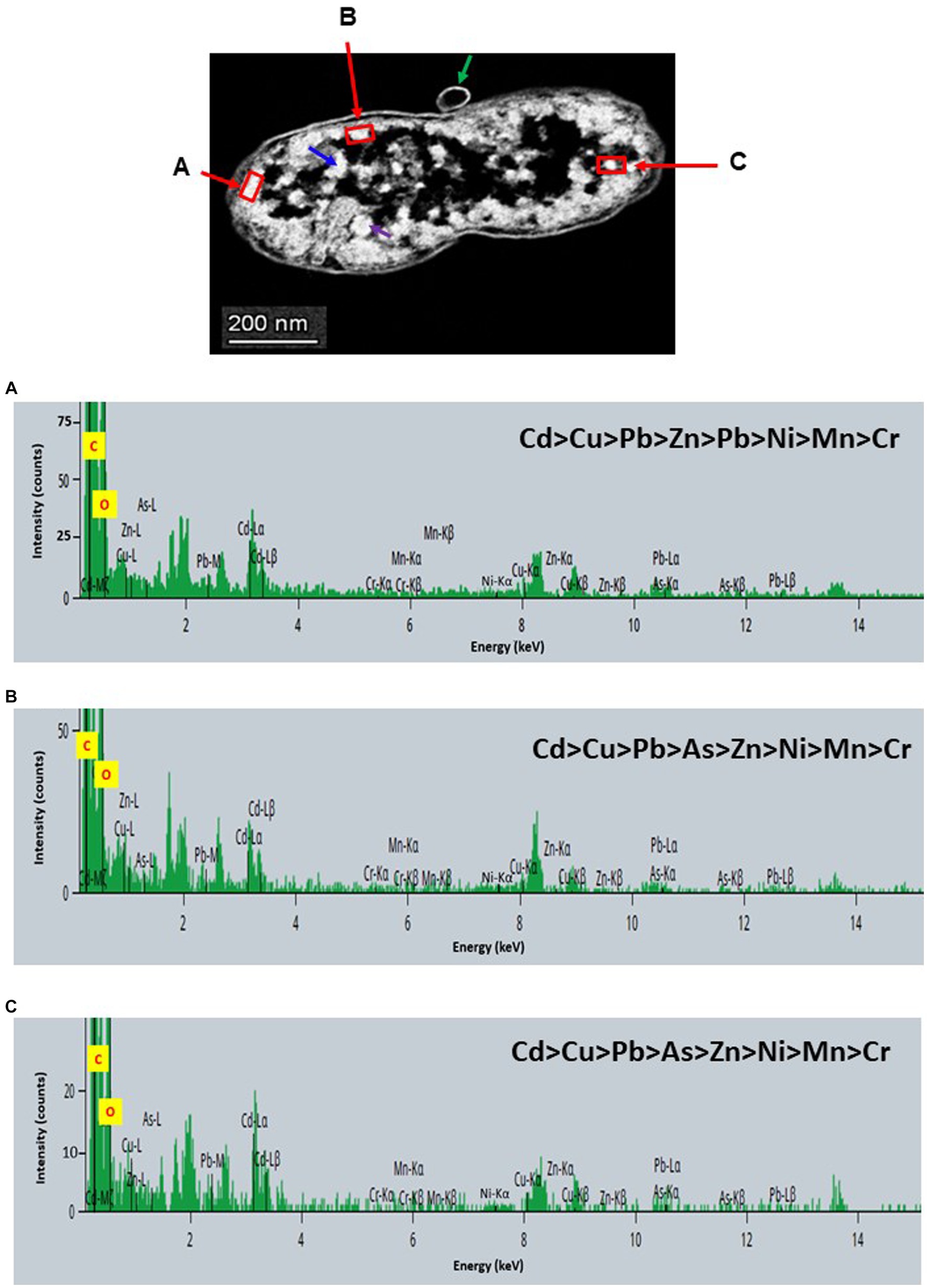
Figure 8. Scanning transmission electron micrographs (STEM) off a thin section of metal-loaded Raoultella sp. L30 and the location of metals. (A–C) Energy dispersive X-ray spectra of the surface biosorption of metals acquired from the region indicated by arrow A, B, and C. The electron-dense granules appear bright on the STEM-HAADF image.
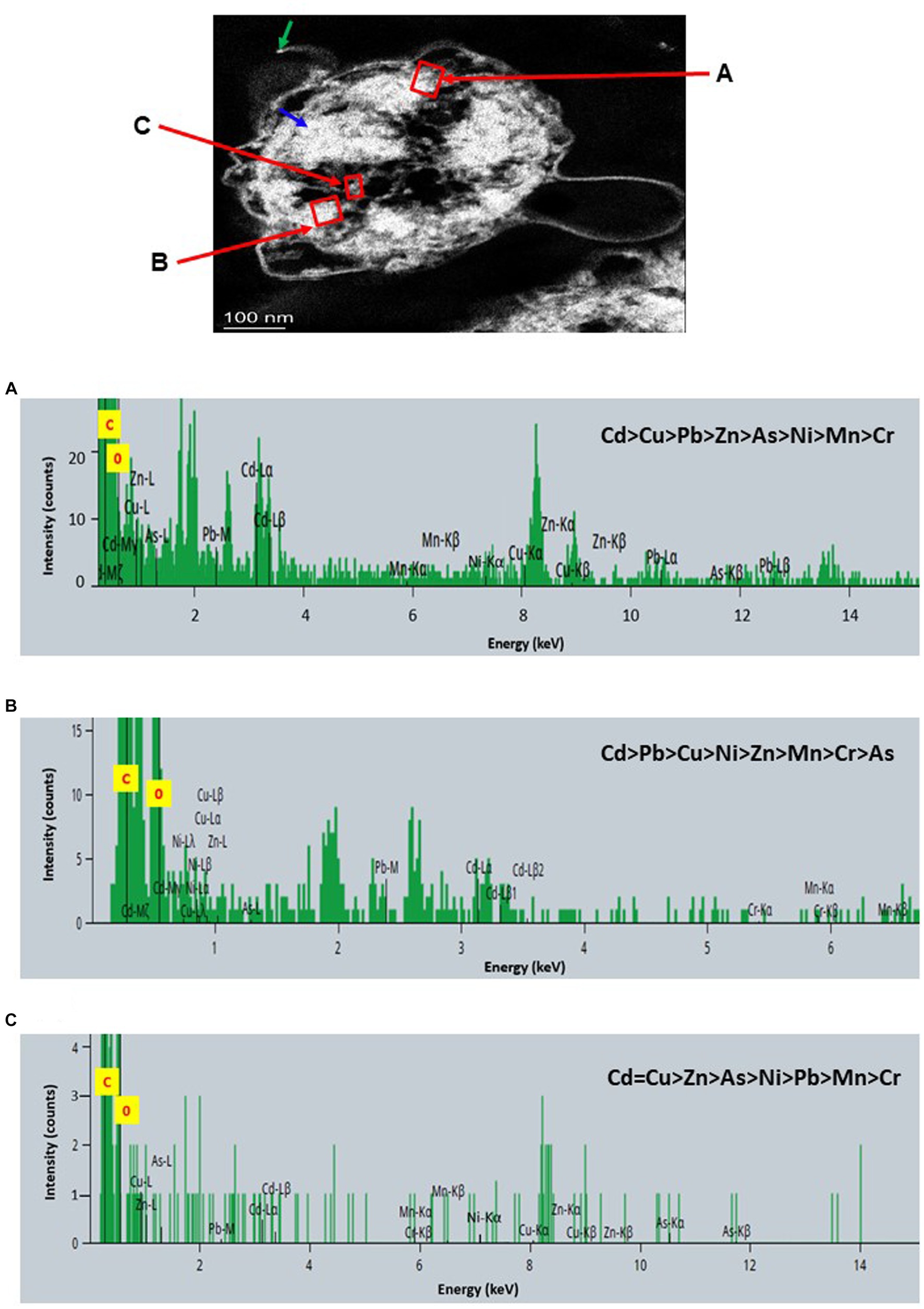
Figure 9. Scanning transmission electron micrographs (STEM) off a thin section of metal-loaded Raoultella sp. L30 and the location of metals. (A–C) Energy dispersive X-ray spectra of the intracellular accumulation of metals acquired from the region indicated by arrow A, B, and C. The electron-dense granules appear bright on the STEM-HAADF image. Granules are of three types: large and compact (blue arrows), small (green arrows), and saturated around storage granules (purple arrow).
For Klebsiella sp. R19, Cd overwhelmingly dominated the electron-dense granules within the cell wall and cell membrane. The order of metal distribution was as follows: Cd2+ > Cu2+ > Pb2+ > As3+ > Zn2+ > Ni2+ > Mn2+ > Cr6+ (depicted in Figure 6). A similar pattern emerged for the electron-dense granules within the cytoplasm (illustrated in Figure 4). Meanwhile, in the case of Raoultella sp. L30, the electron-dense granules in the cell wall [Cu2+ > Cd2+ > Pb2+ = Zn2+ > Cr6+ > Ni2+ > Mn2+ > As3+] and cell membrane [Cu2+ > Pb2+ > Cd2+ > Zn2+ > Cr6+ > Ni2+ > Mn2+ > As3+] were notably enriched in Cu2+ (depicted in Figure 8). While the relative metal distribution in the electron-dense granules within the cytoplasm of Raoultella sp. L30 exhibited some variability (shown in Figure 9), the dominant metal consistently observed in these granules was Cu2+ (as shown in Figure 9).
4. Discussion
Metal tolerance in bacteria is an essential trait and may help in developing an effective process for the treatment of heavy metal contaminated wastewaters. The results of the MIC assay provided insights into the range of metal concentrations at which these strains could withstand metal exposure. The strains were adapted to high concentrations of not only Pb but also As3+, Mn2+, and Zn2+ (Table 1). From a broader perspective, it is notable that the bacterial strains exhibited considerable resistance against certain metals. Specifically, they demonstrated robust resilience against As3+, with MICs ranging from 250 to 450 mg L−1. Similarly, they showcased noteworthy tolerance against Pb2+, spanning from 700 to 800 g L−1. Moreover, the strains exhibited impressive resilience against Mn concentrations exceeding 2,000 mg L−1, as well as Zn2+ concentrations ranging from 500 to 1,100 mg L−1. These findings suggest that these bacterial strains have evolved mechanisms to withstand and adapt to these specific heavy metals. Conversely, the strains exhibited comparatively lower tolerance toward other heavy metals. Cd2+, Cr6+, Cu2+, and Ni2+ elicited relatively lower MIC values, indicating that the bacterial strains are more sensitive to these metals. This variability in sensitivity across different metals underscores the complexity of bacterial responses to heavy metal exposure. it becomes evident that strains L2 and L30 (Serratia sp. strain L2 and Raoultella sp. strain L30) displayed heightened levels of metal tolerance compared to strains R3 (Klebsiella sp.) and R19 (Klebsiella sp.). This disparity in metal tolerance could stem from a multitude of factors, such as variations in the strains’ genetic makeup, metabolic pathways, and the presence of specific metal detoxification mechanisms (Rosen, 1995; Silver, 1996; Nies, 1999; Rensing and Grass, 2003; Silver and Phung, 2005; Helbig and Grass, 2017).
The biosorption abilities of bacterial isolates in both single metal and multi-metal solutions, shedding light on their potential effectiveness in metal removal and the intricate dynamics of simultaneous metal exposure. In the context of single metal solutions, Within the realm of single metal solutions, the capabilities of four metal-tolerant strains emerge prominently. These strains demonstrated impressive efficacy in extracting substantial concentrations of diverse metal ions, including Pb2+, As2+, Zn2+, Cd2+, Cr6+, and Mn2+—each exceeding 50 mg of metal per gram of cell dry mass. Notably, strains R3 and R19 exhibited heightened metal cation removal compared to L2 and L30. These findings underscore the strains’ adaptability and potential applicability in targeted scenarios where specific metal pollutants predominate. Transitioning into solutions encompassing eight metals, a more intricate picture unfolds. The co-presence of multiple metal cations elicited inhibitory effects in the cation binding process, stemming from the competition for binding sites on both bacterial cell surfaces and extracellular polymeric substances (EPS). It has been reported that in solutions containing multiple metals, some of the metal cations already bound on the EPS exert a strong hindrance to the access of other cations at the adjacent adsorption sites (Sag et al., 2000; Micheletti et al., 2008; Pradhan and Rai, 2011; Lu et al., 2021). This steric effect results in EPS conformational change (Huang et al., 2022) that leads to the general reduction in metal removal. Klebsiella sp. R19 showed the highest affinity to Pb2+ in mixed metal solutions with q value even higher than in single-metal solutions (Supplementary Figure S2). In contrast to Pb2+ the other seven metals performed poorer in the multiple-metal tests. The reduction in metal removal could be because the presence of many metals may have overwhelmed the cells and EPS to a point that they cannot bind to as many metal cations as in the single-metal tests. Nonetheless, Klebsiella sp. R19 was the strain that removed the most metal cations of all the four bacterial strains tested. In the mixed metal solutions, Klebsiella sp. R3, Klebsiella sp. R19 showed higher metal cation removal efficiencies than Serratia sp. L2 and Raoultella sp. L30. Insights from the study align with Bowman et al.’s (2018) observations, linking higher flocculation activities of bacterial strains to enhanced metal cation removal capacity. Both Klebsiella sp. R3 and Klebsiella sp. R19 exhibited superior flocculation activities (79 and 71%, respectively) compared to Serratia sp. L2 and Raoultella sp. L30 (L2 = 56%; L30 = 67%). This correlation underscores that flocculation activity directly influences the EPS’s ability to bind metal ions, thus strengthening the strains’ metal removal potential.
Surprisingly, despite R3 and R19 strains showcasing lower metal tolerance than L2 and L30 strains (Table 1), they exhibit a remarkable ability to efficiently remove a greater quantity of metals (Figure 10). Overall, Klebsiella sp. R3, Klebsiella sp. R19 had higher metal cation removal efficiencies than Serratia sp. L2 and Raoultella sp. L30 in single- and multi-metal solutions. These findings challenge the conventional belief that metal tolerance dictates the efficacy of metal removal by bacterial strains. The results underline that metal removal efficiency is a complex phenomenon, shaped not only by a strain’s ability to endure metal stress but also by its capacity to effectively bind metal cations. The high flocculant-production ability of R3 and R19 strains (Bowman et al., 2018) emerges as a crucial factor, enabling them to interact with a higher number of metal cations in aqueous solutions. Bowman et al. (2018) suggest that the efficiency of metal removal is linked to the strains’ capability to create contact with metal cations, a vital aspect often overshadowed by metal tolerance considerations. A pertinent facet of metal resistance is its association with decreased metal uptake or impermeability, as discussed by Gadd (1990). The lower overall metal uptake observed in the L2 and L30 strains may be attributed to their metal resistance mechanisms, which inadvertently diminish their metal uptake capacity. Conversely, R3 and R19 strains’ ability to effectively remove metals could be attributed to their dynamic interaction with metal cations, potentially due to a different balance between metal resistance and efficient metal removal strategies. R3 and R19 strains might possess a higher number or more accessible functional groups, recognized as crucial metal binding sites.
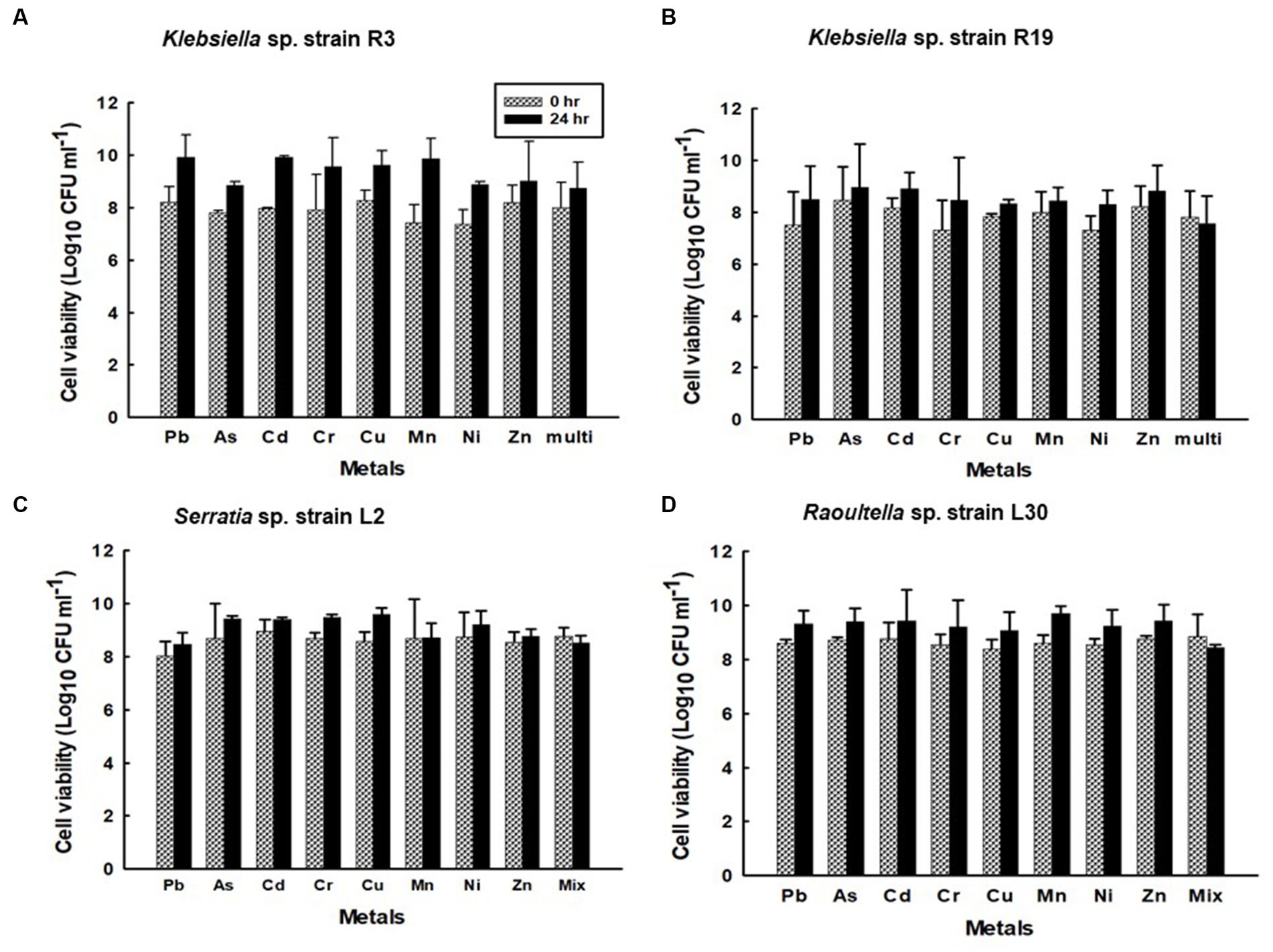
Figure 10. Viability of the confined (A) Klebsiella sp. R3; (B) Klebsiella sp. R19; (C) Serratia sp. L2; and (D) Raoultella sp. L30 strains during sorption in aqueous solutions single metal solutions containing Pb, Cu, Cd, Ni, As, Zn, Cr, Mn, or in mixed metal solutions containing eight metals.
To understand the interactions between bacterial cell surfaces and multiple metal ions, the four strains were subjected to a comprehensive FTIR analysis, both before and after exposure to a complex blend of eight metal ions. When metals first interact with the cell surface, they get sequestered on the surface via adsorption. This could be a result of surface complexation of the metals with the functional groups present on the cell wall (Wang et al., 2021). As a result, changes in the absorption peaks of the spectrum were witnessed in this study (Tables 2–5). The bacterial strains devoid of metal ions revealed a nuanced spectrum, marked by a multitude of absorption peaks. This intricate profile reflects the complex composition of the cellular biomass. A robust and expansive FTIR region, spanning from 3,600 to 2,500 cm−1, is emblematic of the vibrational stretches encompassing C-H (alkane), OH (hydroxyl), and N-H (amine) stretches (Panda et al., 2007). The distinctive C ≡ C stretch between 2,260 and 2,100 cm−1 aligns with alkynes, while the span from, while the span from 1,500 to 900 cm−1 encapsulates the overlap of C-C, C-O, and C-O-P stretches, characteristic of cellular polysaccharides (Wolkers et al., 2004). Notably, bands resonating within the range of 860 to 700 cm−1 correspond to aromatic organic components, further mirroring the intricate nature of cellular constitution (Grube et al., 2008). It is important to note that the appearance of a new peak at 3,496 cm−1 for Serratia sp. L2 and 3,493 for Raoultella sp. L30 appeared after contact with metals and these shifts may be due to Cd+2, Cr2+, Cu2+, or Ni2+ cationic interaction with the hydroxyl group for metal oxygen binding (Panda et al., 2007). The appearance of new peaks at 3,224 and 3,280 cm−1 for Klebsiella sp. R3, and 1,118, 2,986, and 2,997 cm−1 for Klebsiella sp. R19 can be attributed to the presence of phosphate and amino groups and are linked to the presence of phosphate and amino groups, playing a pivotal role in the biosorption of metals (Sodhi et al., 2020).
The appearance of new IR peaks within the spectra can be attributed to the dynamic response of bacterial strains to metal exposure, resulting in the production of extracellular polymeric substances (EPS). This phenomenon has garnered the attention of numerous researchers who have noted enhanced EPS production in bacterial cells under metal-amended conditions (Redmile-Gordon and Chen, 2017; Rizvi et al., 2019a). This observation underscores the notion that metal-induced stress triggers heightened EPS production, potentially acting as a protective layer against the deleterious effects of metal toxicity (Mathivanan et al., 2021a). EPS are such complex blends of high molecular weight polyanionic polymers, such as proteins, humic acids, polysaccharides, and nucleic acids that bind cationic metals with different degrees of specificity and affinity (Bhaskar and Bhosle, 2006; Pal and Paul, 2008; Tiquia-Arashiro, 2018; Pagliaccia et al., 2022). The multifaceted functions of EPS span beyond metal binding, extending to the safeguarding of cells against environmental stressors and the augmentation of metal biosorption capabilities (Planchon et al., 2013; Rajaram et al., 2013; Deepika et al., 2016). Notably, Mathivanan et al. (2021b) conducted FT-IR analysis on EPS produced by Bacillus cereus KMS3-1, revealing the presence of hydroxyl, carboxyl, or carbonyl groups and glycosidic bonds—a testament to the diversity of functional groups contributing to metal binding. Insightful research by Shuhong et al. (2014) shed light on the metal-binding mechanism, emphasizing that this process occurs on the EPS surface. Their findings indicated the pivotal roles of functional groups such as O–H, CH, C=O, C–O, and C–C=O in the binding of metal ions like Cd+2, Cu2+, and Pb2. Notably, these very functional groups were evident in the metal-loaded bacterial cells in the current study, as exemplified in Tables 2–5. The distinctive spectrum alterations and emergence of new bands, particularly prominent in Klebsiella sp. R3 and Klebsiella sp. R19 (Tables 2, 3), allude to the abundance of binding sites on the cell surface for metal interactions. Consequently, this abundance of interaction sites aligns with the higher observed metal removal capacity in these strains compared to Serratia sp. L2 and Raoultella sp. L30 (Figure 1). In essence, the appearance of novel IR peaks underscores the dynamic interplay between bacterial strains and metal ions, triggering EPS production and instigating a repertoire of metal-binding functional groups. This mechanism not only shields cells against metal toxicity but also enhances their metal removal prowess, accentuating the multifaceted nature of microbial-metal interactions.
The investigation into the effects of metal exposure on bacterial strains was enriched by employing SEM and EDX analyses. These techniques provided valuable insights into the morphological changes and metal adsorption patterns exhibited by the bacterial strains Klebsiella sp. R19 and Raoultella sp. L30, both before and after exposure to a complex mixture of eight metal ions. The SEM micrographs unveiled significant alterations in the size and morphology of the bacterial strains following metal adsorption. The initial dimensions of Klebsiella sp. R19 and Raoultella sp. L30 were reduced after the biosorption process, indicating a potential interaction between the bacterial cells and the adsorbed metals. These changes in size could be attributed to the toxic effects of metals on the cells, leading to structural modifications on the cell surface (Rizvi et al., 2019b). Particularly noteworthy were the distinct textures observed on the cell surfaces of both strains. The irregularities such as dents, wrinkles, and elongations reflected the complex interplay between bacterial cells and metal ions. The appearance of these surface features was likely driven by the binding of metals to specific functional groups on the bacterial cell surfaces. Dadrasnia et al. (2015) noted that the roughness and wrinkling of the cells could also be due to sequestration and precipitation of functional groups on the cell wall. The EDX analysis confirmed the presence of metals on the surface of the cells. The observed phenomenon of flocculation on the cell surfaces of both strains further underscored the intricate response to metal exposure. This flocculation could be linked to the secretion of extracellular polysaccharides as a protective mechanism against metal toxicity. The aggregation of bacterial cells resulting from this flocculation indicated a dynamic adaptation to the metal stress environment. The SEM analysis thus provided a visual representation of the cellular changes induced by metal exposure, shedding light on the intricate dynamics of bacterial response to multi-metal conditions. Moreover, EDX analysis validated the presence of adsorbed metals on the cell surfaces. The quantification of metal distribution further illuminated the preference of certain metals for binding to the cell surfaces. The dominance of specific elements on the cell surface, such as Cd2+ and Cu2+, highlighted the varying degrees of metal binding affinity exhibited by the strains. These findings corroborated the importance of examining the interaction between bacterial cell surfaces and metal ions in multi-metal environments.
The comprehensive understanding of metal uptake and accumulation within bacterial cells has been facilitated by STEM EDX analysis. This advanced analytical approach not only confirmed the presence of metals on the cell surface but also provided valuable insights into the intracellular accumulation of these metals. The coexistence of metal ions on both the cell wall and within the cytoplasm suggests intricate interactions and potential bioaccumulation mechanisms at play. The identification of metal accumulation within the cytoplasm holds significance as it hints at the presence of internalized mechanisms for metal uptake. This phenomenon has been reported in various microorganisms and underpins the complexity of microbial-metal interactions. This is particularly evident from studies conducted by different researchers (Perdrial et al., 2008; Ozdemir et al., 2012; Diba et al., 2021; Li et al., 2021), indicating the universality of intracellular metal accumulation mechanisms. However, the process of intracellular absorption is considered slower and more intricate compared to the relatively straightforward cell surface absorption (Ma et al., 2022). The initial binding of metal ions to the cell surface is often a precursor to their subsequent transport into the cell. Many established mechanisms of metal transport hinge upon the electrochemical proton gradient existing across the cell membrane. This gradient, comprising a chemical component in the form of the pH gradient and a membrane potential, actively facilitates the transport of ionized solutes across cellular membranes. This process has been explored in prior research (Borst-Pauwels, 1981; Ma et al., 2022), underscoring the critical role of membrane electrochemistry in mediating metal movement within cells. It’s noteworthy that intracellular metal uptake may also transpire via diffusion, especially in scenarios where the toxic influence of the metal leads to alterations in membrane permeability. These adaptive changes in cellular structure may inadvertently facilitate metal penetration, a phenomenon explored in studies by Gadd (1988). This underscores the multifaceted nature of microbial responses to metal exposure, wherein cells dynamically adjust their mechanisms to counteract toxicity. Comparing the metal distribution patterns between Klebsiella sp. R19 and Raoultella sp. L30 reveals intriguing differences. While both species exhibit a preference for Cu2+ accumulation, Klebsiella sp. R19 displays a unique preference for Cd2+, whereas Raoultella sp. L30 shows a more varied distribution in the cytoplasmic granules. These patterns might reflect the adaptation of each species to its specific environmental conditions, suggesting that metal availability and toxicity play crucial roles in shaping their metal accumulation strategies. The observed metal distribution patterns could be attributed to the presence of specialized metal transporters or binding proteins within these bacteria. The differences in distribution might also arise from variations in the metal-binding affinities of their cellular components.
5. Conclusion
The bacterial strains in the study effectively removed various metal cations, both in single- and multi-metal solutions. They exhibited both heavy metal-removing capabilities and the ability to thrive over a wide range of metal concentrations, making them suitable for potential use in metal remediation in bioreactors or in situ applications. Strains R3 and R19, despite being less metal-tolerant than L2 and L30 strains, demonstrated efficient metal removal because possess a higher number and more accessible carboxyl and amide functional groups, which are crucial for metal binding. This study confirmed that the simultaneous presence of multiple metals in an aqueous solution can lead to mutual inhibition in metal adsorption by the extracellular polymeric substance (EPS), resulting in reduced overall metal uptake. FTIR and SEM–EDX techniques confirmed the interactions between metal ions and functional groups on the surface of the strains. Moreover, TEM-EDX analysis showed the presence of metals on the cell surface and the cytoplasm.
Collectively, these findings advance our understanding of the metal removal capabilities and adaptive mechanisms of bacterial strains. They underscore the potential of these strains for applications in environmental remediation and the treatment of metal-contaminated effluents. Furthermore, the study emphasizes the critical role of tailoring strain selection to suit the specific metal composition of effluent environments, thus enhancing the efficiency and effectiveness of metal removal strategies.
6. Future studies
Future research should explore the practical applications of the highly efficient metal-resistant bacterial strains identified in this study by conducting pilot-scale studies and field trials to assess their real-world performance in diverse contaminated environments. Additionally, efforts should be directed toward optimizing bioremediation techniques and strategies that harness these strains, including the design of specialized bioreactor systems and the evaluation of scalability. Finally, investigating genetic and metabolic engineering approaches to enhance the metal-binding capabilities of these strains could lead to the development of even more specialized and efficient bioremediation tools.
Data availability statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/Supplementary material.
Author contributions
GP: Data curation, Formal analysis, Investigation, Software, Validation, Writing – original draft. DO: Data curation, Formal analysis, Investigation, Methodology, Writing – review & editing. YC: Data curation, Formal analysis, Investigation, Methodology, Validation, Writing – review & editing. AF: Formal analysis, Investigation, Methodology, Validation, Writing – review & editing. FS: Data curation, Formal analysis, Investigation, Methodology, Validation, Writing – review & editing. JF: Data curation, Formal analysis, Investigation, Methodology, Software, Writing – review & editing. RS: Data curation, Formal analysis, Methodology, Writing – review & editing. CS: Data curation, Formal analysis, Investigation, Methodology, Software, Validation, Writing – review & editing. AK: Data curation, Formal analysis, Investigation, Methodology, Software, Validation, Writing – review & editing. SO: Data curation, Formal analysis, Investigation, Methodology, Software, Validation, Writing – review & editing. HN: Data curation, Formal analysis, Investigation, Methodology, Validation, Writing – review & editing. LA: Data curation, Formal analysis, Investigation, Methodology, Validation, Writing – review & editing. OC: Formal analysis, Investigation, Methodology, Writing – review & editing, Data curation, Software. BC: Data curation, Formal analysis, Investigation, Methodology, Validation, Writing – review & editing. XL: Investigation, Resources, Supervision, Validation, Writing – review & editing. ST-A: Conceptualization, Formal analysis, Funding acquisition, Investigation, Methodology, Project administration, Resources, Supervision, Validation, Visualization, Writing – original draft, Writing – review & editing.
Funding
The author(s) declare financial support was received for the research, authorship, and/or publication of this article. This work was financially supported by the University of Michigan-Dearborn. Office of Research and Sponsored Programs (Research Initiation and Development Grant). GP, YC, and BC were supported by the Summer Undergraduate Research Experience (SURE) Program at the University of Michigan-Dearborn.
Acknowledgments
The authors like to express their sincere thanks and deep gratitude to Haiping Sun of Michigan Center for Materials Characterization and the staff at the Microscopy Core at University of Michigan-Ann Arbor for SEM and TEM training to ST-A’s undergraduate students.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2023.1278886/full#supplementary-material
References
Ahmad, W., Alharthy, R. D., Zubair, M., Ahmed, M., Hameed, A., and Rafique, S. (2021). Toxic and heavy metals contamination assessment in soil and water to evaluate human health risk. Sci. Rep. 11:17006. doi: 10.1038/s41598-021-94616-4
Bhaskar, P. V., and Bhosle, N. B. (2006). Bacterial extracellular polymeric substances (EPS) a carrier of heavy metals in the marine food-chain. Environ. Int. 32, 191–198. doi: 10.1016/J.ENVINT.2005.08.010
Bhatt, J., Desai, S., Wagh, N. S., and Lakkakula, J. (2023). “New bioremediation technologies to remove heavy metals and radionuclides” in Industrial wastewater reuse. ed. M. P. Shah (Singapore: Springer) doi: 10.1007/978-981-99-2489-9_14
Boening, D. W. (2000). Ecological effects, transport, and fate of mercury: a general review. Chemosphere 40, 1335–1351. doi: 10.1016/S0045-6535(99)00283-0
Borst-Pauwels, G. (1981). Ion transport in yeast. Biochim. Biophys. Acta 650, 88–127. doi: 10.1016/0304-4157(81)90002-2
Boulanger, B., and Nikolaidis, N. P. (2003). Mobility and aquatic toxicity of copper in an urban watershed. J. Am. Water Resour. Assoc. 39, 325–336. doi: 10.1111/j.1752-1688.2003.tb04387.x
Bowman, N., Patel, P., Xu, W., Alsaffar, A., and Tiquia-Arashiro, S. M. (2018). Enrichment and isolation of Pb-resistant bacteria from Saint Clair River sediments and their potential for Pb removal in aqueous solutions. Appl. Microbiol. Biotechnol. 102, 2391–2398. doi: 10.1007/s00253-018-8772-4
Bruins, M. R., Kapil, S., and Oehme, F. W. (2000). Microbial resistance to metals in the environment. Ecotoxicol. Environ. Saf. 45, 198–207. doi: 10.1006/eesa.1999.1860
Cho, K., Zholi, A., Frabutt, D., Flood, M., Floyd, D., and Tiquia, S. M. (2012). Linking bacterial diversity and geochemistry of uranium-contaminated groundwater. Environ. Technol. 33, 1629–1640. doi: 10.1080/09593330.2011.641036
Dadrasnia, A., Chuan Wei, K. S., Shahsavari, N., Azirun, M. S., and Ismail, S. (2015). Biosorption potential of Bacillus salmalaya strain 139SI for removal of Cr(VI) from aqueous solution. Int. J. Environ. Res. Public Health 12, 15321–15338. doi: 10.3390/ijerph121214985
Deepika, K. V., Raghuram, M., Kariali, E., and Bramhachari, P. V. (2016). Biological responses of symbiotic Rhizobium radiobacter strain VBCK1062 to the arsenic contaminated rhizosphere soils of mung bean. Ecotoxicol. Environ. Saf. 134, 1–10. doi: 10.1016/j.ecoenv.2016.08.008
Diba, F., Khan, M. Z. H., Uddin, S. Z., Istiaq, A., Shuvo, M. S. R., Ul Alam, A. S. M. R., et al. (2021). Bioaccumulation and detoxification of trivalent arsenic by Achromobacter xylosoxidans BHW-15 and electrochemical detection of its transformation efficiency. Sci. Rep. 11:21312. doi: 10.1038/s41598-021-00745-1
Gadd, G. M. (1988). “Accumulation of metals by microorganisms and algae” in Biotechnology a comprehensive treatise, vol. 6 (Weinheim: VCH Yerlagsgesellschaft), 401–433.
Gadd, G. M. (1990). Heavy metal accumulation by bacteria and other microorganisms. Experientia 46, 834–840. doi: 10.1007/BF01935534
Gerhardt, P., Murray, R. G. E., Costilow, R. N., Nester, E. W., Wood, W. A., Kreig, N. R., et al. Manual of methods for general bacteriology. Washington, DC: American Society for Microbiology. (1994); 40: 791.
Goswami, R., and Neog, N. (2023). “Heavy metal pollution in the environment: impact on air quality and human health implications” in Heavy metal toxicity: Environmental concerns, remediation and opportunities. eds. R. P. Singh, P. Singh, and A. Srivastava (Singapore: Springer) doi: 10.1007/978-981-99-0397-9_4
Grube, M., Muter, O., Strikauska, S., Gavare, M., and Limane, B. (2008). Application of FT-IR spectroscopy for control of the medium composition during the biodegradation of nitro aromatic compounds. J. Ind. Microbiol. Biotechnol. 35, 1545–1549. doi: 10.1007/s10295-008-0456-0
Gupta, P., and Diwan, B. (2017). Bacterial exopolysaccharide mediated heavy metal removal: a review on biosynthesis, mechanism and remediation strategies. Biotechnol. Rep. 13, 58–71. doi: 10.1016/j.btre.2016.12.006
Gupta, A. D., Kavitha, E., Singh, S., and Karthikeyan, S. (2020). Toxicity mechanism of Cu2+ ion individually and in combination with Zn2+ ion in characterizing the molecular changes of Staphylococcus aureus studied using FTIR coupled with chemometric analysis. J. Biol. Phys. 46, 395–414. doi: 10.1007/s10867-020-09560-7
Helbig, K., and Grass, G. (2017). Mechanisms of copper homeostasis in bacteria. Front. Cell. Infect. Microbiol. 287, 13549–13555. doi: 10.1074/jbc.R111.316406
Higham, D. P., and Sadler, P. J. (1984). Scawen MO. Cadmium resistant Pseudomas putida synthesizes novel cadmium binding proteins. Science 225, 1043–1046. doi: 10.1126/science.225.4666.1043
Huang, L., Jin, Y., Zhou, D., Liu, L., Huang, S., Zhao, Y., et al. (2022). A review of the role of extracellular polymeric substances (EPS) in wastewater treatment systems. Int. J. Environ. Res. Public Health 19:12191. doi: 10.3390/ijerph191912191
Hugenholtz, P., and Huber, T. (2003). Chimeric 16S rDNA sequences of diverse origin are accumulating in public databases. Int. J. Syst. Evol. Microbiol. 53, 289–293. doi: 10.1099/ijs.0.02441-0
Jeong, H., Byeon, E., Kim, D. H., Maszczyk, P., and Lee, J. S. (2023). Heavy metals and metalloid in aquatic invertebrates: a review of single/mixed forms, combination with other pollutants, and environmental factors. Mar. Pollut. Bull. 191:114959. doi: 10.1016/j.marpolbul.2023.114959
Jeyakumari, P., Debnath, C., Vijayaaraghavan, R., and Muthura, M. (2023). Trends in bioremediation of heavy metal contaminations. Environ. Eng. Res. 28:220631, –220630. doi: 10.4491/eer.2021.631
Kim, S. Y., Kim, J. H., Kim, C. I., and Oh, O. (1996). Metal adsorption of the polysaccharide produced from Methylobacterium organophilum. Biotechnol. Lett. 18, 1161–1164. doi: 10.1007/BF00128585
Lakherwal, D. (2014). Adsorption of heavy metals: a review. Int. J. Environ. Res. Dev. 18, 4745–4750. doi: 10.1016/j.matpr.2019.07.462
Ledin, M. (2000). Accumulation of metals by microorganisms: processes and importance for soil systems. Earth Sci. Rev. 51, 1–31. doi: 10.1016/S0012-8252(00)00008-8
Li, N., Qin, L., Jin, M., Zhang, L., Geng, W., and Xiao, X. (2021). Extracellular adsorption, intracellular accumulation and tolerance mechanisms of Cyclotella sp. to Cr(VI) stress. Chemosphere 270:128662. doi: 10.1016/j.chemosphere.2020.128662
Lu, S., Li, X., Xi, Y., Liu, H., Zhang, Z., Huang, Y., et al. (2021). Insight the roles of loosely bound and tightly-bound extracellular polymeric substances on Cu2+, Zn2+ and Pb2+ biosorption process with Desulfovibrio vulgaris. J. Colloid Interface Sci. 596, 408–419. doi: 10.1016/j.jcis.2021.03.152
Ma, Y., Bantec, T. N., Oliveira, R. S., Coutinho, A., Zhang, C., and Freitas, H. (2022). “The role of bacteria in metal bioaccumulation and biosorption” in Advances in microbe-assisted phytoremediation of polluted sites. eds. K. Bauddh and Y. Ma (New York, USA: Elsevier), 103–112.
Mathivanan, K., Chandirika, J. U., Mathimani, T., Rajaram, R., Annadurai, G., and Yin, H. (2021b). Production and functionality of exopolysaccharides in bacteria exposed to a toxic metal environment. Ecotoxicol. Environ. Safety 208:111567. doi: 10.1016/j.ecoenv.2020.111567
Mathivanan, K., Chandirika, J. U., Vinothkanna, A., Yin, H., Liu, X., and Meng, D. (2021a). Bacterial adaptive strategies to cope with metal toxicity in the contaminated environment-a review. Ecotoxicol. Environ. Saf. 226:112863. doi: 10.1016/j.ecoenv.2021.112863
Micheletti, E., Colica, G., Viti, C., Tamagnini, P., and De Philippis, R. (2008). Selectivity in the heavy metal removal by exopolysaccharide-producing cyanobacteria. J. Appl. Microbiol. 105, 88–94. doi: 10.1111/j.1365-2672.2008.03728.x
Mishra, V., Balomajumder, C., and Agarwal, V. K. (2013). Biological removal of heavy metal zinc from industrial effluent by zinc sequestering bacterium VMSDCM. Clean Techn. Environ. Policy 16, 555–568. doi: 10.1007/s10098-013-0655-x
Narwal, N., Katyal, D., Kataria, N., Rose, P. K., Warkar, S. G., Pugazhendhi, A., et al. (2023). (2023) emerging micropollutants in aquatic ecosystems and nanotechnology-based removal alternatives: a review. Chemosphere 341:139945. doi: 10.1016/j.chemosphere.2023.139945
Nguyen, S., Ala, F., Cardwell, C., Cai, D., McKindles, K. M., Lotvola, A., et al. (2013). Isolation and screening of carboxydotrophs isolated from composts and their potential for butanol synthesis. Environ. Technol. 34, 1995–2007. doi: 10.1080/09593330.2013.795987
Nies, D. H. (1999). Microbial heavy-metal resistance. Appl. Microbiol. Biotechnol. 51, 730–750. doi: 10.1007/s002530051457
Oest, A., Alsaffar, A., Fenner, M., Azzopardi, D., and Tiquia-Arashiro, S. M. (2018). Patterns of change in metabolic capabilities of sediment microbial communities in river and lake ecosystems. Int. J. Microbiol. 2018, 1–15. doi: 10.1155/2018/6234931
Ozdemir, S., Kilinc, E., Poli, A., Nicolaus, B., and Güven, K. (2012). Cd, cu, Ni, Mn and Zn resistance and bioaccumulation by thermophilic bacteria, Geobacillus toebii subsp. decanicus and Geobacillus thermoleovorans subsp. stromboliensis. World J. Microbiol. Biotechnol. 28, 155–163. doi: 10.1007/s11274-011-0804-5
Pagliaccia, B., Carretti, E., Severi, M., Berti, D., Lubello, C., and Lotti, T. (2022). Heavy metal biosorption by extracellular polymeric substances (EPS) recovered from anammox granular sludge. J. Haz. Mat. 424:126661. doi: 10.1016/j.jhazmat.2021.126661
Pal, A., and Paul, A. K. (2008). Microbial extracellular polymeric substances: central elements in heavy metal bioremediation. Indian J. Microbiol. 48, 49–64. doi: 10.1007/s12088-008-0006-5
Panda, G., Das, S., Bandopadhyay, T., and Guha, A. (2007). Adsorption of nickel on husk of Lathyrus sativus: behavior and binding mechanism. Colloids Surf. B Biointerfaces 57, 135–142. doi: 10.1016/j.colsurfb.2007.01.022
Patel, D., Gismondi, R., Alsaffar, A., and Tiquia-Arashiro, S. M. (2019). (2019) applicability of API ZYM to capture seasonal and spatial variabilities in lake and river sediments. Environ. Technol. 40, 3227–3239. doi: 10.1080/09593330.2018.1468492
Perdrial, N., Liewig, N., Delphin, J. E., and Elsass, F. (2008). TEM evidence for intracellular accumulation of lead by bacteria in subsurface environments. Chem. Geol. 253, 196–204. doi: 10.1016/j.chemgeo.2008.05.008
Pereira, S., Micheletti, E., Zille, A., Santos, A., Moradas-Ferreira, P., Tamagnini, P., et al. (2011). Using extracellular polymeric substances (EPS)- producing cyanobacteria for the bioremediation of heavy metals: do cations compete for the EPS functional groups and also accumulate inside the cell? Microbiology 157, 451–458. doi: 10.1099/mic.0.041038-0
Perez, J. A. M., Garcia-Ribera, R., Quesada, T., Aguilera, M., Ramos-Cormenzana, A., and Monteoliva-Sanchez, M. (2008). Biosorption of heavy metals by exopolysaccharide produced by Paenibacillus jamilae. World J. Microbiol. Biotechnol. 24, 2699–2704. doi: 10.1007/s11274-008-9800-9
Planchon, M., Jittawuttipoka, T., Cassier-Chauvat, C., Guyot, F., Gelabert, A., Benedetti, M. F., et al. (2013). Exopolysaccharides protect Synechocystis against the deleterious effects of titanium dioxide nanoparticles in natural and artificial waters. J. Colloid Interface Sci. 405, 35–43. doi: 10.1016/j.jcis.2013.05.061
Plecha, S., Hall, D., and Tiquia-Arashiro, S. M. (2013). Screening for novel bacteria from the bioenergy feedstock switchgrass (Panicum virgatum L.). Environ. Technol. 34, 1895–1904. doi: 10.1080/09593330.2013.818701
Pradhan, S., and Rai, L. C. (2011). Biotechnological potential of Microcystis sp. in cu, Zn and cd biosorption from single and multimetallic systems. Biometals 14, 67–74. doi: 10.1023/a:1016607729691
Priya, A. K., Gnanasekaran, L., Dutta, K., Rajendran, S., Balakrishnan, D., and Soto-Moscoso, M. (2022). Biosorption of heavy metals by microorganisms: evaluation of different underlying mechanisms. Chemosphere 307:135957. doi: 10.1016/j.chemosphere.2022.135957
Qasem, N. A. A., Mohammed, R. H., and Lawal, D. U. (2021). Removal of heavy metal ions from wastewater: a comprehensive and critical review. NPI Clean Water 4:36. doi: 10.1038/s41545-021-00127-0
Queiroz, H. M., Ying, S. C., Abernathy, M., Barcellos, D., Gabriel, F. A., Otero, X. L., et al. (2021). Manganese: the overlooked contaminant in the world largest mine tailings dam collapse. Environ. Int. 146, 146:106284. doi: 10.1016/j.envint.2020.106284
Rajaram, R., Banu, J. S., and Mathivanan, K. (2013). Biosorption of cu (II) ions by indigenous copper-resistant bacteria isolated from polluted coastal environments. Toxicol. Environ. Chem. 95, 590–604. doi: 10.1080/02772248.2013.801979
Razzak, S. A., Faruque, M. O., Alsheikh, Z., Alsheikhmohamad, L., Alkuroud, D., Alfayez, A., et al. (2022). A comprehensive review on conventional and biological-driven heavy metals removal from industrial wastewater. Environ. Adv. 7:100168. doi: 10.1016/j.envadv.2022.100168
Redmile-Gordon, M., and Chen, L. (2017). Zinc toxicity stimulates microbial production of extracellular polymers in a copiotrophic acid soil. Int. Biodeterior. Biodegrad. 119, 413–418. doi: 10.1016/j.ibiod.2016.10.004
Rensing, C., and Grass, G. (2003). Escherichia coli mechanisms of copper homeostasis in a changing environment. FEMS Microbiol. Rev. 27, 197–213. doi: 10.1016/S0168-6445(03)00049-4
Rizvi, A., Ahmed, B., Zaidi, A., and Khan, M. S. (2019a). Heavy metal mediated phytotoxic impact on winter wheat: oxidative stress and microbial management of toxicity by Bacillus subtilis BM2. RSC Adv. 9, 6125–6142. doi: 10.1039/c9ra00333a
Rizvi, A., Ahmed, B., Zaidi, A., and Khan, M. S. (2019b). Bioreduction of toxicity influenced by bioactive molecules secreted under metal stress by Azotobacter chroococcum. Ecotoxicology 28, 302–322. doi: 10.1007/s10646-019-02023-3
Roane, T. M. (1999). Lead resistance in two bacterial isolates from heavy metal-contaminated soils. Microb. Ecol. 37, 218–224. doi: 10.1007/s002489900145
Rosen, B. P. (1995). Resistance mechanisms to arsenicals and antimonials. J. Basic Microbiol. 35, 453–460.
Saba, Y. R., Ahmed, M., and Sabri, A. N. (2019). Potential role of bacterial extracellular polymeric substances as biosorbent material for arsenic bioremediation. Biorem. J. 23, 72–81. doi: 10.1080/10889868.2019.1602107
Sag, Y., Kaya, A., and Kutsal, T. (2000). (2000) Lead, copper and zinc biosorption from bicomponent systems modelled by empirical Freundlich isotherm. Appl. Microbiol. Biotechnol. 53, 338–341. doi: 10.1007/s002530050031
Saitou, N., and Nei, M. (1987). The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol. Biol. Evol. 4, 406–425. doi: 10.1093/oxfordjournals.molbev.a040454
Shameer, S. (2016). Biosorption of lead, copper and cadmium using the extracellular polysaccharides (EPS) of Bacillus sp., from solar salterns. Biotech 6, 194–110. doi: 10.1007/s13205-016-0498-3
Sharma, A., Grewal, A. S., Sharma, D., and Srivastav, A. L. (2023). “Chapter 3- heavy metal contamination in water: Consequences on human health and environment” in Advances in environmental pollution research, metals in water. eds. S. K. Shukla, S. Kumar, S. Madhav, and P. K. Mishra (New York, USA: Elsevier), 39–52.
Sharma, S., and Saraf, M. (2023). Enhanced exopolysaccharide production by multi metal tolerant Klebsiella variicola SMHMZ46 isolated from mines area and application in metal bioremediation. Int. Microbiol. doi: 10.1007/s10123-023-00366-w
Shuhong, Y., Meiping, Z., Hong, Y., Han, W., Shan, X., Yan, L., et al. (2014). Biosorption of Cu2+, Pb2+ and Cr6+ by a novel exopolysaccharide from Arthrobacter ps-5. Carbohydr. Polym. 101, 50–56. doi: 10.1016/j.carbpol.2013.09.021
Shuttleworth, K. L., and Unz, R. F. (1993). Sorption of heavy metal to the filamentous bacterium Thiothrix strain A1. Appl. Environ. Microbiol. 59, 1274–1282. doi: 10.1128/aem.59.5.1274-1282.1993
Silver, S. (1996). Bacterial resistances to toxic metal ions—a review. Gene 179, 9–19. doi: 10.1016/S0378-1119(96)00323-X
Silver, S., and Phung, L. T. (2005). Bacterial heavy metal resistance: new surprises. Annu. Rev. Microbiol. 59, 273–298. doi: 10.1146/annurev.micro.50.1.753
Sodhi, K. K., Kumar, M., and Singh, D. K. (2020). Multi-metal resistance and potential of Alcaligenes sp. MMA for the removal of heavy metals. SN. Appl. Sci. 2, 1–13. doi: 10.1007/s42452-020-03583-4
Sreedevi, P. R., Suresh, K., and Jiang, G. (2022). Bacterial bioremediation of heavy metals in wastewater: a review of processes and applications. J. Wat. Process Eng. 48:102884. doi: 10.1016/j.jwpe.2022.102884
Tamura, K., Stecher, G., and Kumar, S. (2021). MEGA11: molecular evolutionary genetics analysis. Mol. Biol. Evol. 38, 3022–3027. doi: 10.1093/molbev/msab120
Tchounwou, P. B., Yedjou, C. G., Patlolla, A. K., and Sutton, D. J. (2012). Heavy metal toxicity and the environment. Exp. Suppl. 101, 133–164. doi: 10.1007/978-3-7643-8340-4_6
Tiquia, S. M. (2010). Metabolic diversity of the heterotrophic microorganisms and potential link to pollution of the Rouge River. Environ. Pollut. 158, 1435–1443. doi: 10.1016/j.envpol.2009.12.035
Tiquia, S. M. (2011). Extracellular hydrolytic enzyme activities of the heterotrophic microbial communities of the Rouge River: an approach to evaluate ecosystem response to urbanization. Microb. Ecol. 62, 679–689. doi: 10.1007/s00248-011-9871-2
Tiquia, S. M., Davis, D., Hadid, H., Kasparian, S., Ismail, M., and Murray, K. S. (2007). Halotolerant bacteria from river waters and shallow groundwater along the Rouge River of southeastern Michigan. Environ. Technol. 28, 297–307. doi: 10.1080/09593332808618789
Tiquia, S. M., Schleibak, M., Schlaff, J., Floyd, J. C., Benipal, B., Zakhem, E., et al. (2008). Microbial community profiling and characterization of some heterotrophic bacterial isolates from river waters and shallow groundwater wells along the Rouge River, Southeast Michigan. Environ. Technol. 29, 651–663. doi: 10.1080/09593330801986998
Tiquia-Arashiro, S. M. (2018). Lead absorption mechanisms in bacteria as strategies for lead bioremediation. Appl. Microbiol. Biotechnol. 102, 5437–5444. doi: 10.1007/s00253-018-8969-6
van Hullebusch, E. D., Zandvoort, M. H., and Lens, P. N. L. (2003). Metal immobilization by biofilms: mechanisms and analytical tools. Rev. Environ. Sci. Biotechnol. 2, 9–33. doi: 10.1023/B:RESB.0000022995.48330.55
Vandana, M., Priyadarshanee, M., and Das, S. (2023). Bacterial extracellular polymeric substances: biosynthesis and interaction with environmental pollutants. Chemosphere 332:138876. doi: 10.1016/j.chemosphere.2023.138876
Vishan, I., Laha, A., and Kalamdhad, A. (2017). Biosorption of Pb(II) by Bacillus badius AK strain originating from rotary drum compost of water hyacinth. Water Sci. Technol. 75, 1071–1083. doi: 10.2166/wst.2016.590
Volesky, B., and May-Philips, H. A. (1995). Biosorption of heavy metals by Saccharomyces cerevisiae. Appl. Microbiol. Biotechnol. 42, 797–806. doi: 10.1007/bf00171964
Wang, J., Li, Q., Li, M. M., Chen, T. H., Zhou, Y. F., and Yue, Z. B. (2014). Competitive adsorption of heavy metal by extracellular polymeric substances (EPS) extracted from sulfate reducing bacteria. Bioresour. Technol. 163, 374–376. doi: 10.1016/j.biortech.2014.04.073
Wang, Z., Xu, W., Jie, F., Zhao, Z., Zhou, K., and Liu, H. (2021). The selective adsorption performance and mechanism of multiwall magnetic carbon nanotubes for heavy metals in wastewater. Sci. Rep. 11:16878. doi: 10.1038/s41598-021-96465-7
Weisburg, W. W., Barns, S. M., Pelletier, D. A., and Lane, D. J. (1991). SSU ribosomal DNA amplification for phylogenetic study. J. Bacteriol. 173, 697–703. doi: 10.1128/jb.173.2.697-703.1991
Wolkers, W. F., Oliver, A. E., Tablin, F., and Crowe, J. H. (2004). A Fourier-transform infrared spectroscopy study of sugar glasses. Carbohydr. Res. 339, 1077–1085. doi: 10.1016/j.carres.2004.01.016
Wu, W., Gu, B., Fields, M. W., Gentile, M., Ku, Y.-K., Yan, H., et al. (2005). Uranium (VI) reduction by denitrifying biomass. Biorem. J. 9, 49–61. doi: 10.1080/10889860590929628
Keywords: biosorption, metal-resistant microbes, metal selectivity, exopolysaccharide-producing bacteria, heavy metal removal, Fourier-transform infrared analysis, scanning electron microscopy, transmission electron microscopy
Citation: Pagnucco G, Overfield D, Chamlee Y, Shuler C, Kassem A, Opara S, Najaf H, Abbas L, Coutinho O, Fortuna A, Sulaiman F, Farinas J, Schittenhelm R, Catalfano B, Li X and Tiquia-Arashiro SM (2023) Metal tolerance and biosorption capacities of bacterial strains isolated from an urban watershed. Front. Microbiol. 14:1278886. doi: 10.3389/fmicb.2023.1278886
Edited by:
Ilana Kolodkin-Gal, Reichman University, IsraelReviewed by:
Edwin Hualpa Cutipa, National University of San Marcos, PeruShafinaz Shahir, University of Technology Malaysia, Malaysia
Copyright © 2023 Pagnucco, Overfield, Chamlee, Shuler, Kassem, Opara, Najaf, Abbas, Coutinho, Fortuna, Sulaiman, Farinas, Schittenhelm, Catalfano, Li and Tiquia-Arashiro. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Sonia M. Tiquia-Arashiro, c210aXF1aWFAdW1pY2guZWR1
 Grace Pagnucco
Grace Pagnucco Dustin Overfield
Dustin Overfield Yanesa Chamlee
Yanesa Chamlee Claudia Shuler
Claudia Shuler Somie Opara
Somie Opara Hawraa Najaf
Hawraa Najaf Oliver Coutinho
Oliver Coutinho Xiaohua Li
Xiaohua Li Sonia M. Tiquia-Arashiro
Sonia M. Tiquia-Arashiro