
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Microbiol. , 10 November 2023
Sec. Antimicrobials, Resistance and Chemotherapy
Volume 14 - 2023 | https://doi.org/10.3389/fmicb.2023.1278821
This article is part of the Research Topic Antimicrobial Resistance in Zoonotic Bacteria View all 12 articles
Salmonella enterica serovar Enteritidis (SE) remains a frequent cause of foodborne illnesses associated with the consumption of contaminated hen eggs. Such a food–pathogen association has been demonstrated epidemiologically, but the molecular basis for this association has not been explored. Comparative genomic analysis was implemented to decipher the phylogenomic characteristics, antimicrobial resistance, and virulence potential of eggs-associated SE. Analyzing 1,002 genomes belonging to 841 sequence types of food-isolated SE strains suggests a high genomic similarity within the egg-related lineage, which is phylogenetically close to SE strains isolated from poultry but is different from those isolated from beef. Core genome- and single nucleotide polymorphism (SNP)-based phylogeny of 74 SE strains of egg origin showcased two distinct sublineages. Time-scaled phylogeny supported the possibility of a common ancestor of egg-related SE lineages. Additionally, genome mining revealed frequent antibiotic resistance due to the presence of aac(6’)-Iaa and mdsAB encoded on the genomes of egg-associated SE strains. For virulence gene profiling, 103–113 virulence determinants were identified in the egg-associated SE, which were comparable to 112 determinants found in human-associated SE, emphasizing the capacity of egg-associated strains to infect humans and cause diseases. The findings of this study proved the genomic similarity of egg-associated SE strains, and these were closely related to poultry strains. The egg-associated strains also harbor virulence genes equivalent to those found in human-associated SE strains. The analysis provided critical insights into the genetic structure, phylogenomics, dynamics of virulence, and antibiotic resistance of Salmonella Enteritidis, circulating in eggs and emphasizing the necessity of implementing anti-Salmonella intervention strategies, starting at the production stage of the poultry supply chain.
Non-typhoidal Salmonella enterica is estimated to cause 153 million illnesses and 57,000 deaths annually worldwide (Healy and Bruce, 2019). According to the European Food Safety Authority (EFSA) and European Center for Disease Prevention and Control (ECDC), Salmonella was involved in 30.7% of foodborne disease outbreaks in 2018, and salmonellosis is the second most reported disease and the second cause of death due to the consumption of contaminated food (EFSA and ECDC, 2019). In the United States, it was estimated that 1 million cases of non-typhoidal salmonellosis occurred in 2013; this infection level accounted for 24% of the economic burden of all foodborne diseases (USDA, 2019). Certain foods are disproportionately linked to salmonellosis outbreaks. Eggs and egg products accounted for 45.6% of salmonellosis outbreaks in the European Union (EFSA and ECDC, 2019). In a recent report by EFSA and ECDC, salmonellosis was the second most reported zoonoses in humans, Salmonella Enteritidis remained the most frequent causative agent of foodborne outbreaks, and Salmonella serovars in eggs and egg products were of the most concern (EFSA and ECDC, 2022). Similarly, the Salmonella–egg combination caused most of the salmonellosis outbreaks in the United States (Dewey-Mattia et al., 2018). In fact, Salmonella Enteritidis has been the main serovar associated with human salmonellosis (Ferrari et al., 2019), including those associated with eggs, and the serovar has been associated with disease outbreaks in several countries (Cardoso et al., 2021). The terms “egg” or “eggs,” used in the current study, refer to hen whole shell eggs, egg components such as white and yolk, or raw whole liquid egg.
Despite the obvious Salmonella Enteritidis–egg epidemiological association, there is a knowledge gap about the genetic basis for the colonization and survival of Salmonella Enteritidis in eggs. Several researchers attempted to determine the underlying genetic mechanisms governing the ability of this serovar to colonize internal egg contents. Previous research showed that non-motile mutants (ΔfliC, ΔfljB, and ΔfliH), compared with the wild-type strain, were impaired in the survival of egg albumin (Cogan et al., 2004; Shah et al., 2012). In addition to motility, genes involved in curli fimbriae production (agfA), DNA replication, repair, and recombination (e.g., yafD) were significantly important for the survival of the pathogen in egg albumin (Lu et al., 2003; Cogan et al., 2004). In a previous study (Clavijo et al., 2006), screening 2,850 libraries of Salmonella Enteritidis mutants revealed the importance of 32 genes, broadly involved in cell wall structure and nucleic acid and amino acid metabolism, for survivability of the serovar in egg albumin. Adaptation of Salmonella Enteritidis to egg yolk has been investigated. Passage of Salmonella Enteritidis in egg yolk increased intestinal colonization in a colitis mouse model, but the infection dose of the pathogen and the mode of action were not reported in the study (Moreau et al., 2016). In a recent research, the growth of Salmonella Enteritidis in egg yolk, in comparison with growth in a synthetic microbiological medium, increased virulence of the pathogen in mice, and a dose of only 280 CFU in yolk was sufficient to kill 50% of the mice (Xu et al., 2022). However, it is still unknown whether Salmonella Enteritidis strains isolated from eggs share genomic similarities or can be phylogenomically distinguishable from those isolated from other sources. It is not known whether egg-associated Salmonella Enteritidis strains possess important genetic traits (e.g., virulence and antimicrobial resistance) in a similar manner to strains associated with human illnesses. To reduce this knowledge gap, the current study was conducted to provide insight into phylogenetic characteristics, antimicrobial resistance, virulence, and genetic signatures of Salmonella Enteritidis associated with eggs.
To evaluate the distinct global genomic characteristics of Salmonella Enteritidis, compared with other serovars isolated from food sources, a dataset consisting of 10,869 strains characterized by 9,235 core-genome multilocus sequence types was extracted from the EnteroBase database. This dataset was subsequently utilized to construct a phylogenetic tree using the rapid neighbor-joining model integrated in EnteroBase, thereby elucidating potential genetic distinctions. Additionally, to further underscore the genetic distinctiveness of Salmonella Enteritidis strains originated from eggs compared with those from other food sources, a subset of 1,002 strains represented by 841 core-genome multilocus sequence types was extracted from the EnteroBase database. A minimum spanning tree model, integrated in EnteroBase, was then employed to construct a phylogenetic tree.
For a focused assessment of the phylogenetic attributes, antimicrobial resistance genetic determinants, and virulence traits of Salmonella Enteritidis strains associated with eggs, we selected strains from the National Center for Biotechnology Information (NCBI) due to their adequate metadata and the comprehensive genomic sequence information. The selection of Salmonella Enteritidis strains was based on a defined set of criteria, ensuring that the strains are representative and suitable for analysis. A total of 94 Salmonella Enteritidis strains submitted to NCBI between 2015 and 2021 were used to represent the following two groups, egg-related and non-related groups. The quality of the selected genomes was also performed by assessing the N50 and coverage values of the genomes. The ranges of coverage and N50 for the selected genomes were 29–301x and 32 kb–4.7 Mb, respectively (Supplementary File 1). The egg-related group included 74 strains isolated from 23 whole raw liquid egg samples, 20 shell eggs, and 31 raw egg yolk samples. These were all the strains that met the selection criteria below and retrieved from the NCBI pathogen isolates database.1 The isolates, which were (a) associated with “egg” in their “isolation source,” (b) originated from different single nucleotide polymorphism (SNP) clusters, and (c) not belonging to the same outbreak, were retrieved from NCBI in November 2022. Among the selected egg-related strains, three shell egg isolates (Salmonella Enteritidis PT8, PT13, and ODA 99-30581-13) were sequenced in our laboratory at The Ohio State University, Columbus, United States, as described in a previous study (Xu et al., 2021). The second group was a control group (non-egg-related) integrated into the analysis to provide a comparative baseline against egg-related strains. The control group consisted of 19 clinical strains originating from human feces and one strain originating from a farm environment. These strains were selected from geographical regions that were different from those of the egg-related group, and their isolation source metadata explicitly indicated no association with eggs. Detailed information about the investigated strains is presented in Supplementary File 1.
For SNP calling, genomes of the 94 Salmonella Enteritidis strains were uploaded as Fasta files into CSI Phylogeny 4.1.0 tool (available at the Center for Genomic Epidemiology; https://cge.cbs.dtu.dk/services/CSIPhylogeny/; Kaas et al., 2014) and mapped against a reference genome of Salmonella Enteritidis strain P125109 (accession number GCA_015240635.1). SNPs were called at default settings (minimum SNP quality of 30, and minimum map quality of 25). CSI phylogeny yields a Newick maximum likelihood phylogenetic tree inferred by FastTree 2.1.7, and the tree was visualized using the interactive Tree Of Life tool (iTOL; https://itol.embl.de/; Letunic and Bork, 2019).
Pangenomic analysis was performed utilizing the 94 Salmonella Enteritidis strains in addition to the reference genome of the P125109 strain using the Roary pipeline (Page et al., 2015). The whole genomes of all strains were annotated using Prokka program, version 1.13 (Seemann, 2014), which yields annotated genomes in the GFF3 format; these genomes were used as an input for Roary that created a multi-FASTA alignment of the core genes with MAFFT (version 7.477; Katoh et al., 2002) at a minimum BLAST percentage identity of 95. The alignment was used to infer a maximum likelihood phylogeny using IQ-TREE (version 1.5.4; Nguyen et al., 2015), and the phylogenetic tree was visualized and annotated with iTOL.
The SNP-based maximum likelihood phylogenetic tree produced by CSI phylogeny tool was time-scaled and rooted by LSD2 (version 1.4.2.2.; To et al., 2016), as described previously (Carroll et al., 2021) using the default settings with modifications as follows: (i) a substation rate of 2.79 × 10−7 substitutions/site/year, a rate that corresponds to the estimate determined for Salmonella Typhimurium DT104 in a previous study (Leekitcharoenphon et al., 2016); (ii) tip dates corresponding to the collection year for each strain; (iii) genome sequence length of the reference strain Salmonella Enteritidis P125109 (4,685,848 bp); and (iv) variances of input branch lengths. The resulting time-scaled phylogenetic tree was visualized and annotated using iTOL.
For AMR determinants, two different genome-mining analyses were applied to Salmonella Enteritidis genomes. First, ABRicate (version 1.0.1) in GalaxyTrakr pipeline (Gangiredla et al., 2021) was used to identify the AMR determinants in the 94 Salmonella Enteritidis genomes, in addition to the reference genome, Salmonella Enteritidis P125109, using the Resfinder database with 80% minimum DNA identity and coverage for each gene. Second, AMRFinderplus (version 3.8.28; Feldgarden et al., 2019) was used to detect AMR and stress response genes using the following settings: AMRFinderplus database (version 2020-09-30.1), Salmonella organism option, minimum coverage percentage of 50, and minimum identity threshold of 75. In addition, plasmid replicons, in Salmonella Enteritidis genomes, were screened using ABRicate (version 1.0.1) and PlasmidFinder databases with an identity and coverage percentage of 80. For virulence factor determination in the Salmonella Enteritidis genomes, ABRicate (version 1.0.1) was implemented against the Virulence Factor Database (VFDB) with a minimum coverage and identity percentage of 80. The presence or absence of AMR and virulence genes were represented as heatmaps using GraphPad Prism software (version 9.4.1; GraphPad Software, San Diego, CA, United States).
To gain a global overview of the genetic diversity of S. enterica isolated from food, we investigated the population structure and genomic diversity of non-typhoidal S. enterica isolated from “food” as a “source niche” according to the EnteroBase database. A neighbor-joining phylogenetic tree (Figure 1) was constructed based on cgMLST and allelic differences; this tree comprised 10,869 strains belonging to 9,235 sequence types (STs) of more than 30 Salmonella serovars. The S. enterica strains, included in this analysis, were geographically dispersed and originated from more than 30 countries located in Europe, North America, South America, Asia, Africa, and Oceania. Food-associated S. enterica strains formed separate clusters when based on the serovar. It was obvious that “Enteritidis,” “Heidelberg,” and “Typhimurium” were the most common serovars in food, each of which formed a distinct lineage in this cgMLST-based phylogenetic tree (Figure 1). The emphasis in this manuscript will be on serovar “Enteritidis” strains from food. By analyzing 1,002 Salmonella Enteritidis strains belonging to 841 STs and isolated from more than 20 food sources, the strains were grouped into different lineages (Figure 2), among which one single lineage contained all strains isolated from eggs or egg products (Figure 2, red-colored clusters). This observation indicates relatively small genomic diversity within the egg-related strains. Nevertheless, the egg-related group clustered closely with the poultry-associated strains and distantly from the beef-associated strains (Figure 2).
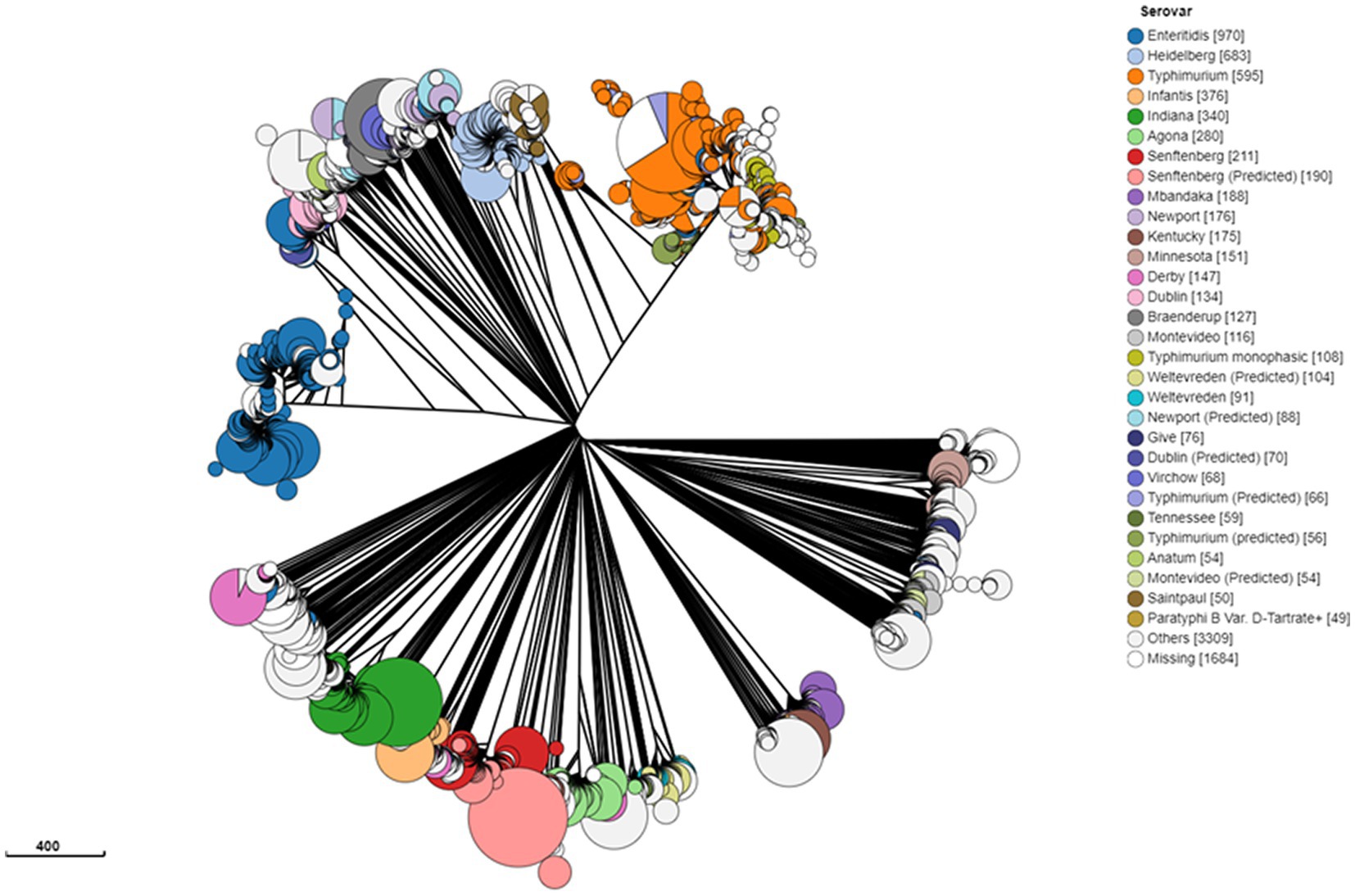
Figure 1. Genomic diversity of Salmonella enterica isolated from food worldwide, as determined by a neighbor-joining tree of 10,869 strains of 9,235 core-genome multilocus sequence types, according to EnteroBase. Each node represents a unique sequence type, and the size of each node corresponds to the number of strains within that node. Salmonella strains are color-coded by serovar, and the number of isolates is presented between brackets. The scale bar indicates 400 core-genome multilocus sequence-type alleles.
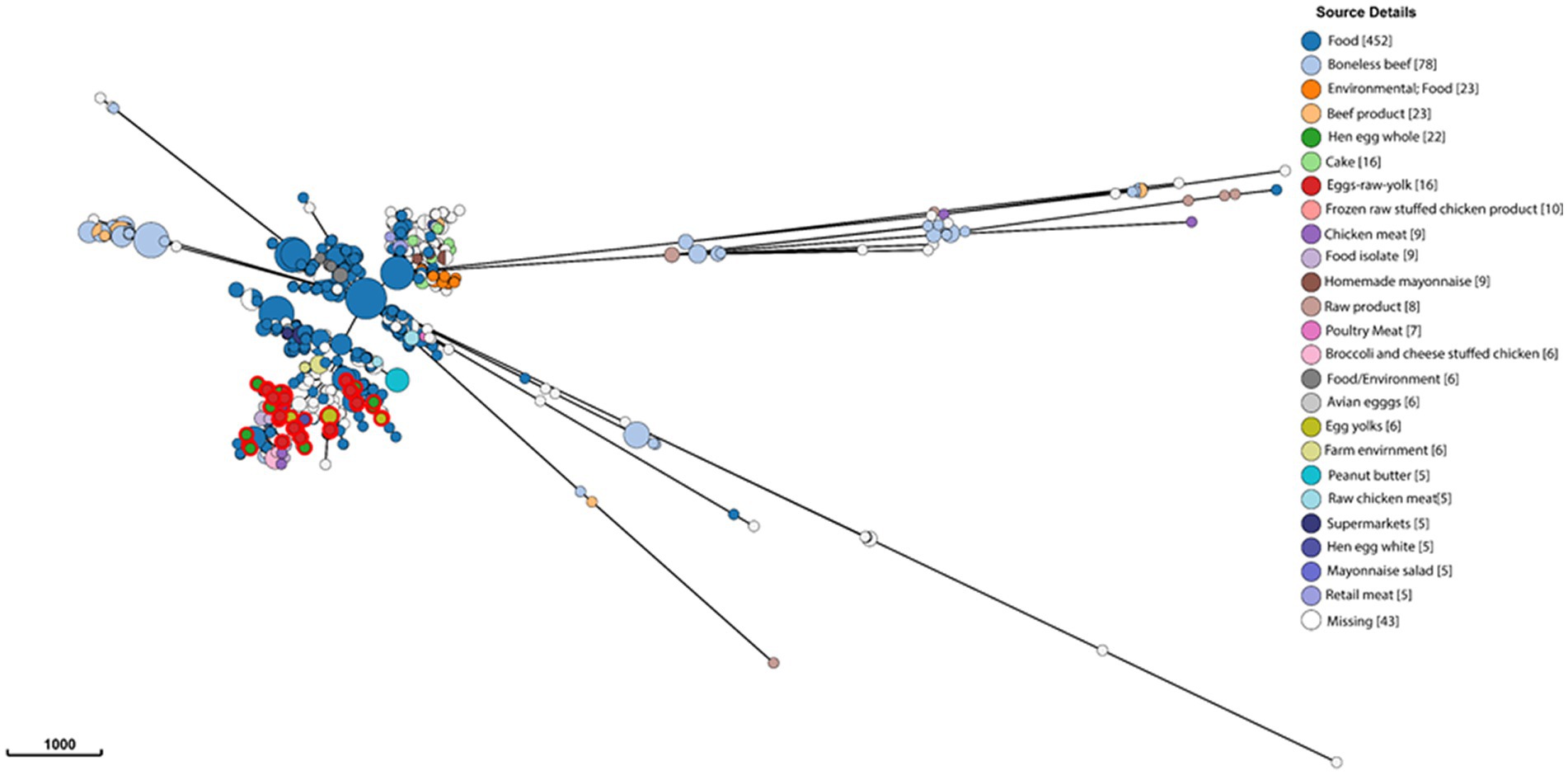
Figure 2. Genomic diversity of Salmonella Enteritidis isolated from food, according to the EnteroBase database, and determined by the minimum spanning tree of 1,002 strains of 841 core-genome multilocus sequence types. Each node represents a unique sequence type, and the node size corresponds to the number of Salmonella Enteritidis strains within that node. Salmonella strains are color-coded by EnteroBase-defined isolation source details (the number of isolates is presented between brackets), and the scale bar indicates 1,000 core-genome multilocus sequence type alleles. Reference to “food” in the legend indicates that the food types were not specified in the EnteroBase.
A maximum likelihood phylogenetic tree (Figure 3) was constructed based on 2,164 SNPs detected in the core genome regions of 94 Salmonella Enteritidis strains (74 from eggs, 19 from human feces, and one from farm environment for comparison) against Salmonella Enteritidis P125109 reference strain. Pairwise comparisons between strains showed a range of SNPs varying from 2 to 542. The SNP-based phylogeny demonstrated that most of the egg-associated strains formed a distinct sublineage from the human-associated strains (Figure 3). These findings were confirmed by constructing a phylogeny (Figure 4A), based on the core genome sequences, in all 94 Enteritidis strains, in addition to the reference P125109 strain. The phylogenetic tree comprised two major clades, and the egg-associated strains formed unique sublineages separated from the human and farm-associated strains (Figure 4A).
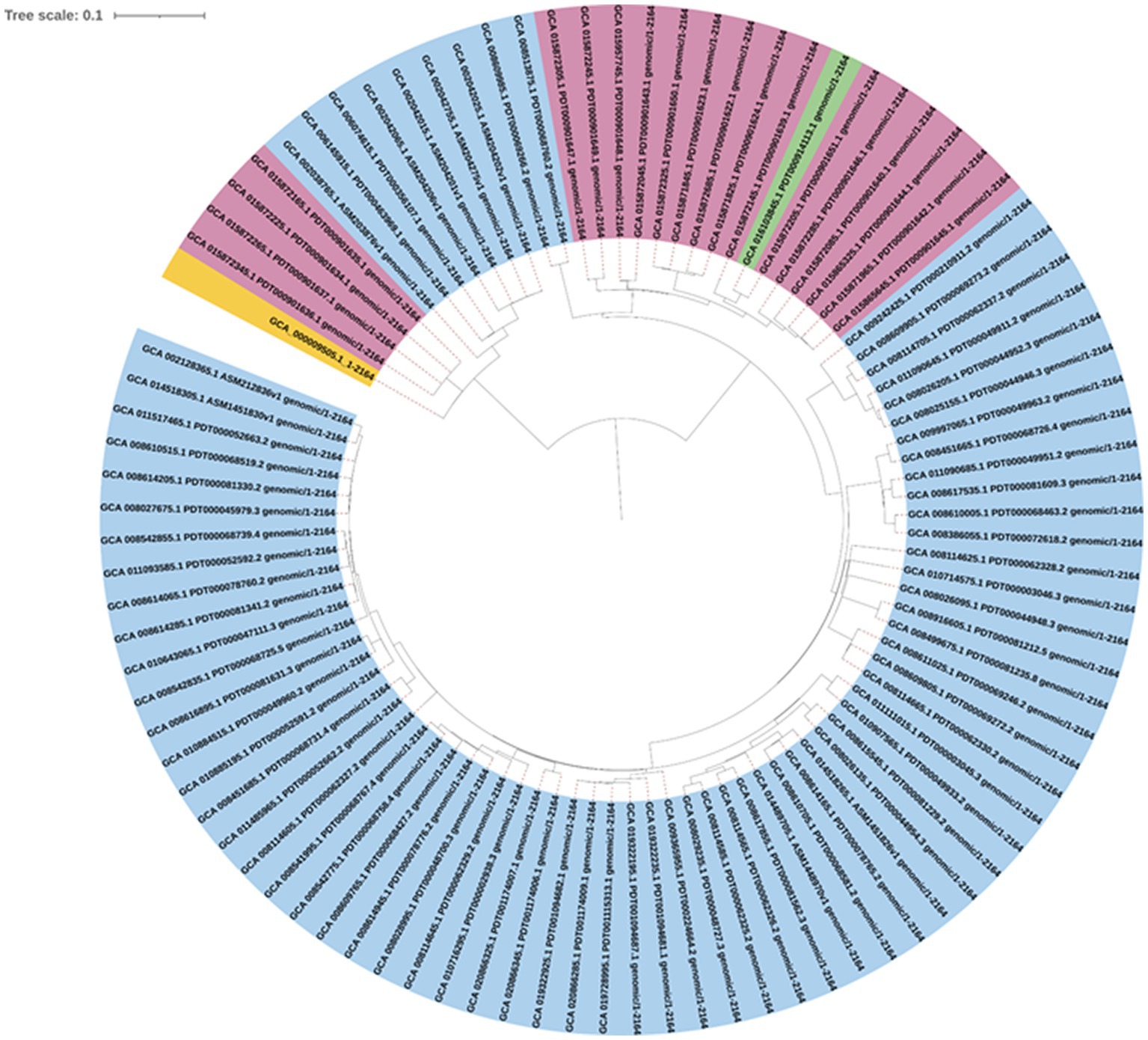
Figure 3. Maximum likelihood phylogeny constructed based on core single nucleotide polymorphism (SNPs; n = 2,164) among 94 Salmonella Enteritidis genomes. The strains were color-shaded according to their source as follows: eggs (blue), pink (human feces), green (farm environment), and yellow (reference genome of Salmonella Enteritidis P125109). Core SNPs were determined by CSI Phylogeny v.4.1.0, whereas the phylogenetic tree was inferred using FastTree v.2.1.7. The scale bar denotes substitutions per site.
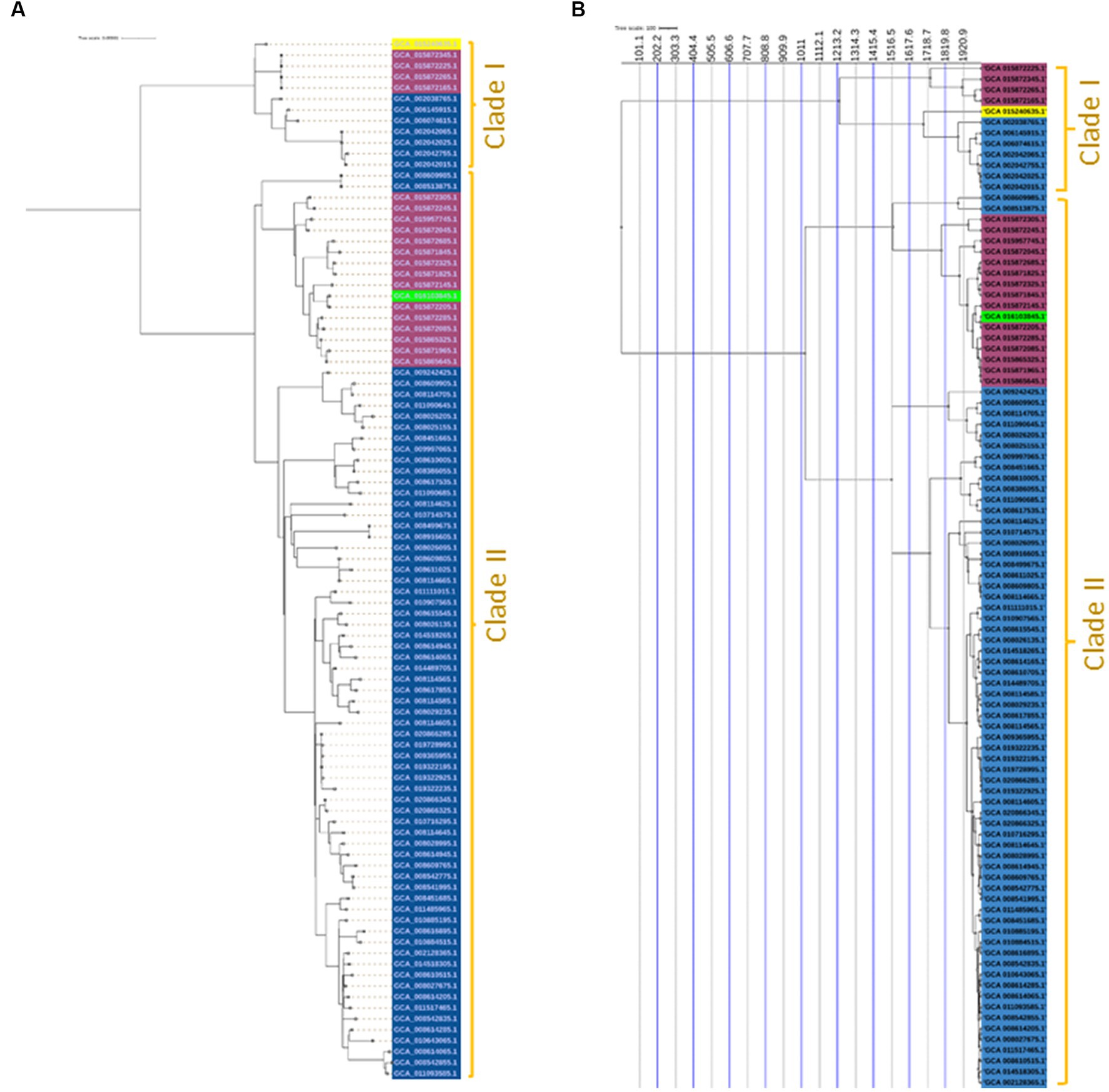
Figure 4. Core-genome and time-scaled phylogeny of Salmonella Enteritidis from eggs. (A) Maximum likelihood phylogeny of 94 Salmonella Enteritidis strains, constructed using the core-genes sequence alignment determined by the Roary pipeline and inferred by IQ-TREE v.1.5.4; (B) Time-scaled maximum likelihood phylogeny constructed using core SNPs identified among the 94 Salmonella Enteritidis strains; the phylogeny was rooted and scaled using LSD2 with scale bar reported in years. The strains were color-shaded according to their source as follows: eggs (blue), pink (human feces), green (farm environment), and yellow (reference genome of Salmonella Enteritidis P125109).
A time-scaled phylogeny was built to infer the possible timing of the emergence of the egg-associated Salmonella Enteritidis (Figure 4B). The SNP-based time-scaled phylogenetic tree demonstrated the presence of two major clades containing the egg-related strains. A small clade “Clade I” contained a sublineage of seven egg-associated strains that shared a common most recent ancestor dated to approximately 1,920 as determined by the LSD v.2.0.0 tool. The large clade “Clade II” encompassed most egg-associated strains, which formed a distinct sublineage that shared a common most recent ancestor dated approximately to 1,516. Additionally, these findings illustrate the genetic distinctiveness of egg-associated Salmonella Enteritidis since most of these strains were clustered in Clade II. Genetic distinction of egg-associated Salmonella Enteritidis would become more apparent when more genome sequences from eggs become available in public databases.
Two genome mining approaches, Resfinder and AMRFinderplus, identified AMR determinants in the genomes of egg-associated Salmonella Enteritidis while selected human-associated strains (n = 19) were used for comparison. AMR results from Resfinder showed that egg-associated Salmonella Enteritidis showcased two AMR genes, aac(6′)-Iaa and mdsAB (Figure 5A), which confer resistance to aminoglycosides and other antibiotics (e.g., chloramphenicol, cephalosporin, and monobactam). These AMR genes were also conserved in the investigated human-related strains. Exceptionally, the egg isolate Salmonella Enteritidis PT13 (GCA_014518265.1) harbored genes conferring resistance to aminoglycosides (aph(3″)-Ib and aph(6)-Id), beta lactams (blaTEM-1b), sulfonamide (sul2), and tetracycline (tetA). With AMRFinderplus analysis, the results (Figure 5B) revealed the presence of a colonization factor (sinH), and some stress response genes (golS and golT) that confer resistance to metal ions (i.e., gold and copper). These genes were similarly found in egg- and human-related strains. Moreover, two gyrA mutations (D87Y and S83F) were found in a few Salmonella Enteritidis strains isolated from eggs and human feces, and these gene mutations mediate quinolone resistance.
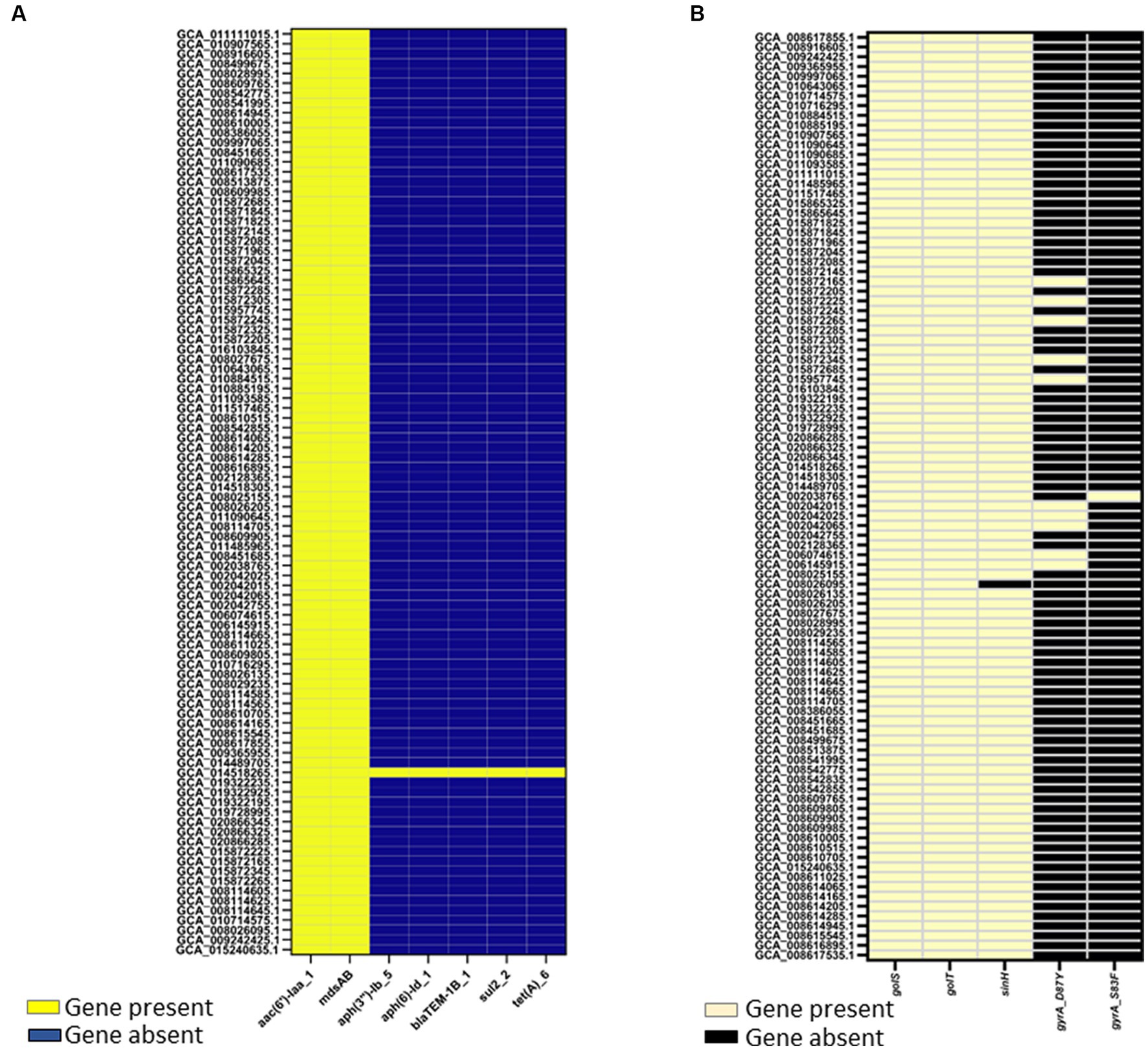
Figure 5. Antimicrobial resistance (AMR) determinants identified in 94 Salmonella Enteritidis strains including the 74 strains from eggs. AMR determinants were identified as “present” using ABRicate with the Resfinder database (A) at a minimum 80% identity and coverage, or (B) AMRFinderplus v.3.8.28 with the AMRFinderplus database at a minimum threshold of 50 and 75% for coverage and identity, respectively. Genes present in Salmonella Enteritidis genomes were colored “yellow” in the heatmap illustrations.
Genomic screening was conducted for plasmid replicons known to harbor antibiotic-resistant genes in Salmonella genomes. Out of eight different plasmid replicons identified in Salmonella Enteritidis genomes, IncFIB(S)_1 and IncFII(S)_1 were detected in 86 genomes (Supplementary File 2). The plasmid replicons IncN_1, Col156_1, ColRNAI_1, and ColpVC_1 were detected in 2–3 genomes while IncX1_1 and IncX4_2 were found in 10–11 genomes. Most of the egg-associated Salmonella Enteritidis genomes harbored IncFIB(S)_1 and IncFII(S)_1, and 9–10 of the genomes harbored IncX1_1 and IncX4_2 plasmid replicons (Supplementary File 2). The presence of such plasmid replicons indicates a potential antibiotic resistance capability of egg-associated Salmonella Enteritidis.
Profiling of virulence genes using the ABRicate tool against the virulence factor database revealed that the egg-associated Salmonella Enteritidis possessed 103–113 virulence factors in their genomes, mainly encoded on the Salmonella pathogenicity islands (SPIs), such as SPI-1, SPI-2, SPI-3, SPI-4, and SPI-5; these SPIs are indispensable for disease onset in the host. Comparably, the human-associated strains harbored 112 virulence genes (Figure 6). These results augment the notion that egg-associated Salmonella Enteritidis strains possess putative AMR determinants and the essential virulence factors required for causing human diseases.

Figure 6. Virulence gene profiling in 94 Salmonella Enteritidis strains including the 74 strains from eggs. Up to 113 virulence determinants were identified as “present” using ABRicate with the Virulence Factor database at a minimum of 80% identity and coverage. Virulence genes reported as “present” were colored “yellow” in the heatmap illustration.
The annotated genomes obtained with the Prokka tool were used as an input for the Roary pipeline to infer the pangenome, which is the entire set of genes within the investigated Salmonella Enteritidis strains. The pangenome results produced a gene presence/absence matrix by comparing the presence of 6,148 genes with the 94 Salmonella Enteritidis genomes (74 for the egg-, 19 for the human-, and one for the farm-associated strains), in addition to the reference Salmonella Enteritidis P125109 genome. Our findings (Table 1) showed that two genes (oadA2 and oadB2), involved in sodium ion transport and anaerobic growth, were identified in 30 genomes of the egg-associated strains but were absent in the human-associated group. Additional genes (Table 1) involved in cellular processes, such as DNA-dependent transcription, DNA repair, and conjugal transfer, were identified only in 12–13 of the egg-associated Salmonella Enteritidis group but were absent in the human-associated group.
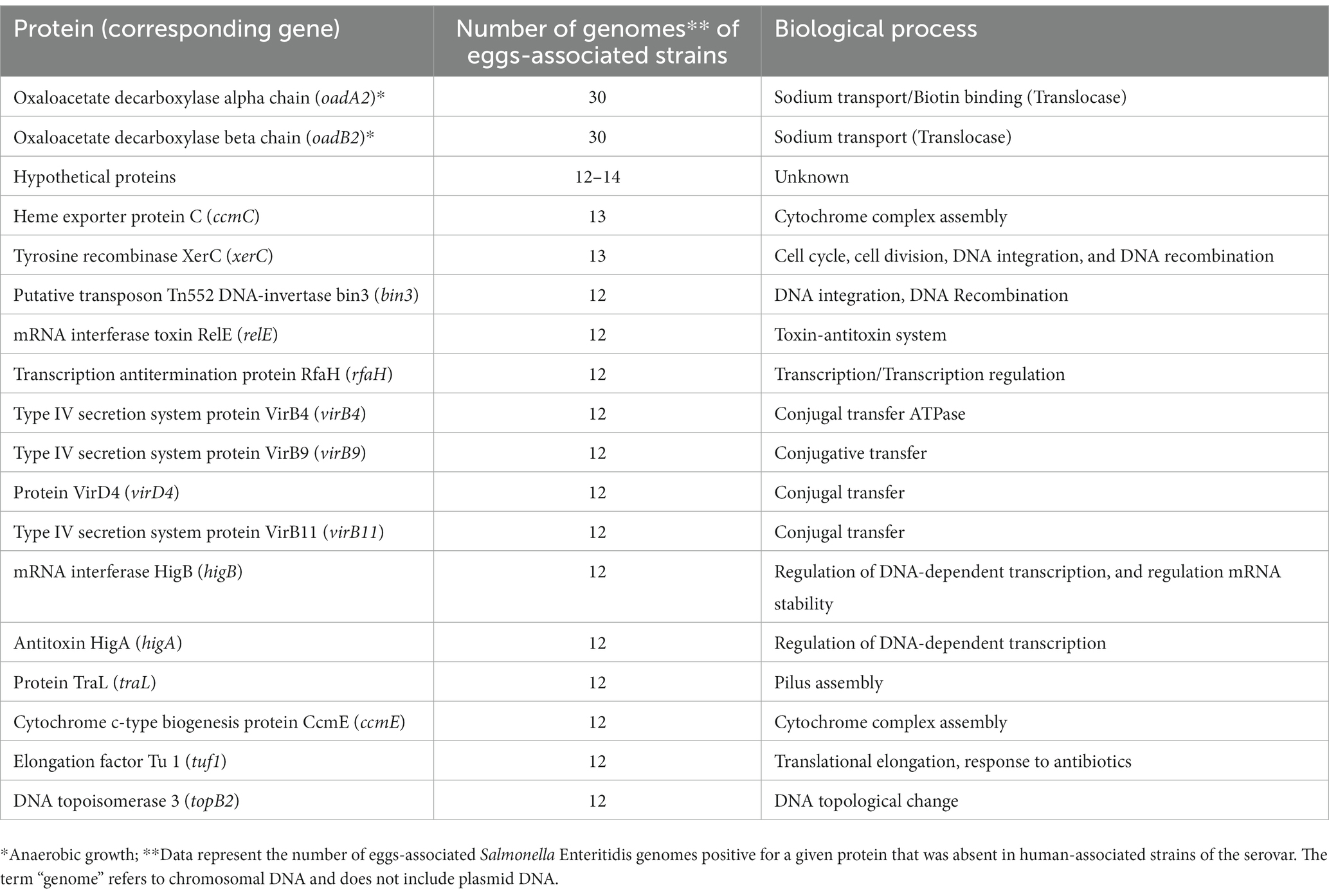
Table 1. Proteins and corresponding genes uniquely found in genomes of Salmonella enterica serovar Enteritidis from eggs.
Salmonella Enteritidis is the main S. enterica serovar that causes frequent disease outbreaks linked to food, particularly eggs and other poultry products (Rodrigue et al., 1990). The historical dilemma of Salmonella Enteritidis–egg association remains unresolved, and understanding the biological mechanisms behind the serovar fitness in such a food matrix is still insufficiently explored. It is unknown whether Salmonella Enteritidis strains associated with eggs are genetically identical. Analysis of the global lineages of non-typhoidal Salmonella isolated from food (Figure 1) suggests that Salmonella Enteritidis is genetically different from the rest of the serovars, and the genomes of the serovar formed a unique cluster. The analysis also showed that Salmonella Enteritidis strains from eggs and poultry are closely related genomically (Figure 2), and both are relatively different from the beef-related strains of that serovar. This obvious genetic relatedness emphasizes the importance of the Salmonella Enteritidis vertical transmission route, i.e., from breeding chicken to hatching eggs (Gantois et al., 2009). According to a recent study, poultry breeding stocks drove the dispersal of Salmonella Enteritidis globally, and that hatching eggs were the key driver for the global spread of this pathogen (Li et al., 2021). The latter study provided evidence that the intercontinental spread of Salmonella Enteritidis arose from centralized origins because the global lineages of this pathogen were similar to the strains that were isolated originally from poultry products in the United States and Europe, where the first industrialized poultry production appeared. Moreover, surveys conducted in the United States in 1991 highlighted the presence of Salmonella on eggshells from breeder hatcheries (Cox et al., 1991). Together, these findings explain the genomic similarity between Salmonella Enteritidis strains from eggs and poultry sources. Intriguingly, Salmonella Enteritidis strains from eggs were genetically distinct from those isolated from beef, indicating that beef is an unlikely transmission vehicle of this serovar to eggs.
In the current study, the egg-associated Salmonella Enteritidis strains found in the NCBI database (as of November 2022) were included in our analyses. Most of these strains were isolated in the United States; this coincides with the fact that the United States was one of the earliest exporters of chicken breeding stocks to the world (Li et al., 2021). Our SNP- and core-genome-based phylogeny demonstrated that Salmonella Enteritidis strains from eggs displayed limited intra-genomic diversity, and these strains were closely related phylogenetically, indicating that they may have a common ancestor. The time-scaled phylogeny constructed in the current study (Figure 4) suggests that the egg-related strains may have naturally evolved from a common ancestor. Comparably, a recent study found that Salmonella Enteritidis ST11 lineage from Africa shared a common ancestor dated to 1,551–1,801 (Carroll et al., 2021).
Our findings showcased Salmonella Enteritidis strains from eggs to be a reservoir of antibiotic resistance genes, mainly aac(6′)-Iaa and mdsAB, which encode aminoglycoside acetyl transferase and efflux pump complex, respectively; these genetic traits confer resistance to aminoglycosides and other antibiotics (Alcaine et al., 2007; Song et al., 2014; Nair et al., 2018). The identification of such antibiotic-resistant determinants (Figure 5), in addition to several plasmid replicons (Supplementary File 2), including IncFIB(S)_1, IncFII(S)_1, IncX1_1, and IncX4_1, which have been associated with harboring antibiotic resistance genes (van den Berg et al., 2019; Hull et al., 2022), suggests that the egg-associated Salmonella Enteritidis could harbor and possibly disseminate antibiotic resistance genes via plasmid-mediated gene transfer. Antibiotic resistance among Salmonella Enteritidis strains from eggs can be explained by the intense use of antibiotics during poultry production (Sapkota et al., 2014). This is consistent with a previous study, which concluded that the prevalence of antibiotic-resistant Salmonella was reduced when a conventional farm was converted to organic farming (Sapkota et al., 2014). In addition to that assumption, antibiotic resistance genes can spread among Salmonella Enteritidis strains via horizontal transfer of antibiotic-resistant plasmids or chromosomal acquisition of transposons that carry antibiotic-resistant genes (Domingues et al., 2012). The prevalence of golS and golT in Salmonella Enteritidis genomes from eggs highlights the strains’ ability to confer metal ion resistance; the two genes were also implicated in regulating antibiotic resistance efflux pumps (Pontel et al., 2007). These findings alert the egg industry sector to revise the egg/poultry production regimes to reduce the spread of antibiotic resistance among associated pathogens such as Salmonella Enteritidis. Our genome (Chromosomal DNA) mining analysis also uncovered a comprehensive virulence machinery in Salmonella Enteritidis strains from eggs, which implies the capability of these strains to infect humans and cause diseases.
From a pathogen–food adaptive perspective, the pangenome analysis revealed the abundance of oadA and oadB and other genes (e.g., xerC and higB; Table 1) in egg-associated Salmonella Enteritidis. These genes enhance the capacity of Salmonella to grow anaerobically (similar to the eggshell environment) and in assisting nucleic acid-related processes such as DNA repair in response to damage encountered in environments such as egg white (Woehlke and Dimroth, 1994; Huang et al., 2019). These hypotheses have been overlooked and are now worth further investigation.
Salmonella Enteritidis is a predominant causative agent of egg-associated foodborne disease outbreaks and leads to a multitude of human illnesses and economic losses. This study utilized comparative genomic analysis for an understanding of the phylogenomic links, genomically encoded virulence and antimicrobial resistance traits, and genomic landscape of Salmonella Enteritidis strains that were historically associated with disease outbreaks in eggs or egg products. The genetic homology within the egg-associated Salmonella Enteritidis indicates a possible common ancestry origin for the circulating strains in egg products, presumably due to the trading of poultry breeding stocks. A whole genome-based comparison gave a glimpse of the virulence aspect of Salmonella Enteritidis associated with poultry eggs, and such assessment confirmed the need for advancing the anti-Salmonella intervention strategies, starting at the poultry production chain. Further investigations utilizing genome-wide association studies could underpin specific genetic targets characteristics of Salmonella Enteritidis associated with eggs.
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/Supplementary material.
AA: Conceptualization, Data curation, Formal analysis, Investigation, Methodology, Writing – original draft, Writing – review & editing. AY: Conceptualization, Funding acquisition, Project administration, Supervision, Writing – review & editing.
The author(s) declare financial support was received for the research, authorship, and/or publication of this article. This study is supported by the USDA National Institute of Food and Agriculture, AFRI project 2020-67017-30794.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2023.1278821/full#supplementary-material
Alcaine, S. D., Warnick, L. D., and Wiedmann, M. (2007). Antimicrobial resistance in nontyphoidal Salmonella. J. Food Prot. 70, 780–790. doi: 10.4315/0362-028X-70.3.780
Cardoso, M. J., Nicolau, A. I., Borda, D., Nielsen, L., Maia, R. L., Møretrø, T., et al. (2021). Salmonella in eggs: from shopping to consumption—a review providing an evidence-based analysis of risk factors. Compr. Rev. Food Sci. Food Saf. 20, 2716–2741. doi: 10.1111/1541-4337.12753
Carroll, L. M., Pierneef, R., Mathole, M., and Matle, I. (2021). Genomic characterization of endemic and ecdemic non-typhoidal Salmonella enterica lineages circulating among animals and animal products in South Africa. Front. Microbiol. 12:748611. doi: 10.3389/fmicb.2021.748611
Clavijo, R. I., Loui, C., Andersen, G. L., Riley, L. W., and Lu, S. (2006). Identification of genes associated with survival of Salmonella enterica Serovar Enteritidis in chicken egg albumen. Appl. Environ. Microbiol. 72, 1055–1064. doi: 10.1128/AEM.72.2.1055-1064.2006
Cogan, T. A., Jørgensen, F., Lappin-Scott, H. M., Benson, C. E., Woodward, M. J., and Humphrey, T. J. (2004). Flagella and curli fimbriae are important for the growth of Salmonella enterica serovars in hen eggs. Microbiology 150, 1063–1071. doi: 10.1099/MIC.0.26791-0
Cox, N. A., Bailey, J. S., Mauldin, J. M., Blankenship, L. C., and Wilson, J. L. (1991). Extent of salmonellae contamination in breeder hatcheries. Poult. Sci. 70, 416–418. doi: 10.3382/PS.0700416
Dewey-Mattia, D., Manikonda, K., Hall, A. J., Wise, M. E., and Crowe, S. J. (2018). Surveillance for foodborne disease outbreaks—United States, 2009–2015. MMWR Surveill. Summ. 67, 1–11. doi: 10.15585/mmwr.ss6710a1
Domingues, S., da Silva, G. J., and Nielsen, K. M. (2012). Integrons: vehicles and pathways for horizontal dissemination in bacteria. Mob. Genet. Elem. 2, 211–223. doi: 10.4161/MGE.22967
EFSA and ECDC (2019). The European Union one health 2018 zoonoses report. EFSA J. 17:e05926. doi: 10.2903/j.efsa.2019.5926
EFSA and ECDC (2022). The European Union one health 2021 zoonoses report. EFSA J. 20:e07666. doi: 10.2903/j.efsa.2022.7666
Feldgarden, M., Brover, V., Haft, D. H., Prasad, A. B., Slotta, D. J., Tolstoy, I., et al. (2019). Validating the AMRFINder tool and resistance gene database by using antimicrobial resistance genotype-phenotype correlations in a collection of isolates. Antimicrob. Agents Chemother. 63, e00483–19. doi: 10.1128/AAC.00483-19/SUPPL_FILE/AAC.00483-19-S0004.XLSX
Ferrari, R. G., Rosario, D. K. A., Cunha-Neto, A., Mano, S. B., Figueiredo, E. E. S., and Conte-Juniora, C. A. (2019). Worldwide epidemiology of Salmonella serovars in animal-based foods: a meta-analysis. Appl. Environ. Microbiol. 85:e00591. doi: 10.1128/AEM.00591-19
Gangiredla, J., Rand, H., Benisatto, D., Payne, J., Strittmatter, C., Sanders, J., et al. (2021). GalaxyTrakr: a distributed analysis tool for public health whole genome sequence data accessible to non-bioinformaticians. BMC Genomics 22, 1–11. doi: 10.1186/S12864-021-07405-8/TABLES/2
Gantois, I., Ducatelle, R., Pasmans, F., Haesebrouck, F., Gast, R., Humphrey, T. J., et al. (2009). Mechanisms of egg contamination by Salmonella Enteritidis. FEMS Microbiol. Rev. 33, 718–738. doi: 10.1111/j.1574-6976.2008.00161.x
Healy, J., and Bruce, B. (2019). “Salmonellosis (Nontyphoidal)” in Chapter 4—CDC yellow book travel health 2019. Available at https://wwwnc.cdc.gov/travel/yellowbook/2020/travel-related-infectious-diseases/salmonellosis-nontyphoidal (Accessed November 9, 2022).
Huang, X., Zhou, X., Jia, B., Li, N., Jia, J., He, M., et al. (2019). Transcriptional sequencing uncovers survival mechanisms of Salmonella enterica serovar Enteritidis in antibacterial egg white. mSphere 4, e00700–e00718. doi: 10.1128/mSphere.00700-18
Hull, D. M., Harrell, E., Harden, L., and Thakur, S. (2022). Multidrug resistance and virulence genes carried by mobile genomic elements in Salmonella enterica isolated from live food animals, processed, and retail meat in North Carolina, 2018–2019. Int. J. Food Microbiol. 378:109821. doi: 10.1016/j.ijfoodmicro.2022.109821
Kaas, R. S., Leekitcharoenphon, P., Aarestrup, F. M., and Lund, O. (2014). Solving the problem of comparing whole bacterial genomes across different sequencing platforms. PLoS One 9:e104984. doi: 10.1371/JOURNAL.PONE.0104984
Katoh, K., Misawa, K., Kuma, K. I., and Miyata, T. (2002). MAFFT: a novel method for rapid multiple sequence alignment based on fast Fourier transform. Nucleic Acids Res. 30, 3059–3066. doi: 10.1093/NAR/GKF436
Leekitcharoenphon, P., Hendriksen, R. S., Le Hello, S., Weill, F. X., Baggesen, D. L., Jun, S. R., et al. (2016). Global genomic epidemiology of Salmonella enterica serovar typhimurium DT104. Appl. Environ. Microbiol. 82, 2516–2526. doi: 10.1128/AEM.03821-15
Letunic, I., and Bork, P. (2019). Interactive tree of life (iTOL) v4: recent updates and new developments. Nucleic Acids Res. 47, W256–W259. doi: 10.1093/NAR/GKZ239
Li, S., He, Y., Mann, D. A., and Deng, X. (2021). Global spread of Salmonella Enteritidis via centralized sourcing and international trade of poultry breeding stocks. Nat. Commun. 12:5109. doi: 10.1038/s41467-021-25319-7
Lu, S., Killoran, P. B., and Riley, L. W. (2003). Association of Salmonella enterica Serovar Enteritidis YafD with resistance to chicken egg albumen. Infect. Immun. 71, 6734–6741. doi: 10.1128/IAI.71.12.6734-6741.2003
Moreau, M. R., Wijetunge, D. S. S., Bailey, M. L., Gongati, S. R., Goodfield, L. L., Hewage, E. M. K. K., et al. (2016). Growth in egg yolk enhances Salmonella Enteritidis colonization and virulence in a mouse model of human colitis. PLoS One 11:e0150258. doi: 10.1371/journal.pone.0150258
Nair, D. V. T., Venkitanarayanan, K., and Johny, A. K. (2018). Antibiotic-resistant Salmonella in the food supply and the potential role of antibiotic alternatives for control. Foods 7:167. doi: 10.3390/FOODS7100167
Nguyen, L. T., Schmidt, H. A., Von Haeseler, A., and Minh, B. Q. (2015). IQ-TREE: a fast and effective stochastic algorithm for estimating maximum-likelihood phylogenies. Mol. Biol. Evol. 32, 268–274. doi: 10.1093/MOLBEV/MSU300
Page, A. J., Cummins, C. A., Hunt, M., Wong, V. K., Reuter, S., Holden, M. T. G., et al. (2015). Roary: rapid large-scale prokaryote pan genome analysis. Bioinformatics 31, 3691–3693. doi: 10.1093/BIOINFORMATICS/BTV421
Pontel, L. B., Audero, M. E. P., Espariz, M., Checa, S. K., and Soncini, F. C. (2007). GolS controls the response to gold by the hierarchical induction of Salmonella-specific genes that include a CBA efflux-coding operon. Mol. Microbiol. 66, 814–825. doi: 10.1111/J.1365-2958.2007.05963.X
Rodrigue, D. C., Tauxe, R. V., and Rowe, B. (1990). International increase in Salmonella Enteritidis: a new pandemic? Epidemiol. Infect. 105, 21–27. doi: 10.1017/S0950268800047609
Sapkota, A. R., Kinney, E. L., George, A., Hulet, R. M., Cruz-Cano, R., Schwab, K. J., et al. (2014). Lower prevalence of antibiotic-resistant Salmonella on large-scale U.S. conventional poultry farms that transitioned to organic practices. Sci. Total Environ. 476-477, 387–392. doi: 10.1016/J.SCITOTENV.2013.12.005
Seemann, T. (2014). Prokka: rapid prokaryotic genome annotation. Bioinformatics 30, 2068–2069. doi: 10.1093/BIOINFORMATICS/BTU153
Shah, D. H., Zhou, X., Kim, H. Y., Call, D. R., and Guard, J. (2012). Transposon mutagenesis of Salmonella enterica serovar Enteritidis identifies genes that contribute to invasiveness in human and chicken cells and survival in egg albumen. Infect. Immun. 80, 4203–4215. doi: 10.1128/IAI.00790-12
Song, S., Hwang, S., Lee, S., Ha, N. C., and Lee, K. (2014). Interaction mediated by the putative tip regions of Mdsa and Mdsc in the formation of a Salmonella-specific tripartite efflux pump. PLoS One 9:e100881. doi: 10.1371/JOURNAL.PONE.0100881
To, T. H., Jung, M., Lycett, S., and Gascuel, O. (2016). Fast dating using least-squares criteria and algorithms. Syst. Biol. 65, 82–97. doi: 10.1093/SYSBIO/SYV068
USDA (2019). Cost estimates of foodborne illnesses. Avialable at: https://www.ers.usda.gov/data-products/cost-estimates-of-foodborne-illnesses/cost-estimates-of-foodborne-illnesses/#Pathogen (Accessed November 9, 2022).
van den Berg, R. R., Dissel, S., Rapallini, M. L., van Der Weijden, C. C., Wit, B., and Heymans, R. (2019). Characterization and whole genome sequencing of closely related multidrug-resistant Salmonella enterica serovar Heidelberg isolates from imported poultry meat in the Netherlands. PLoS One 14:e0219795. doi: 10.1371/journal.pone.0219795
Woehlke, G., and Dimroth, P. (1994). Anaerobic growth of Salmonella Typhimurium on l L(+)- and D(−)-tartrate involves an oxaloacetate decarboxylase Na+ pump. Arch. Microbiol. 162, 233–237. doi: 10.1007/BF00301843
Xu, Y., Abdelhamid, A. G., Sabag-Daigle, A., Sovic, M. G., Ahmer, B. M. M., and Yousef, A. E. (2022). The role of egg yolk in modulating the virulence of Salmonella enterica serovar Enteritidis. Front. Cell. Infect. Microbiol. 12:572. doi: 10.3389/fcimb.2022.903979
Keywords: whole genome sequencing, Salmonella Enteritidis, salmonellosis, foodborne disease outbreaks, eggs, egg safety, comparative genomics
Citation: Abdelhamid AG and Yousef AE (2023) Egg-associated Salmonella enterica serovar Enteritidis: comparative genomics unveils phylogenetic links, virulence potential, and antimicrobial resistance traits. Front. Microbiol. 14:1278821. doi: 10.3389/fmicb.2023.1278821
Received: 16 August 2023; Accepted: 18 October 2023;
Published: 10 November 2023.
Edited by:
Guyue Cheng, Huazhong Agricultural University, ChinaReviewed by:
Yolande Proroga, Experimental Zooprophylactic Institute of Southern Italy (IZSM), ItalyCopyright © 2023 Abdelhamid and Yousef. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Ahmed E. Yousef, eW91c2VmLjFAb3N1LmVkdQ==
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.