- Department of Biochemistry and Biomedical Sciences, McMaster University, Hamilton, ON, Canada
Genus Pseudomonas is a large assemblage of diverse microorganisms, not sharing a common evolutionary history. To clarify their evolutionary relationships and classification, we have conducted comprehensive phylogenomic and comparative analyses on 388 Pseudomonadaceae genomes. In phylogenomic trees, Pseudomonas species formed 12 main clusters, apart from the “Aeruginosa clade” containing its type species, P. aeruginosa. In parallel, our detailed analyses on protein sequences from Pseudomonadaceae genomes have identified 98 novel conserved signature indels (CSIs), which are uniquely shared by the species from different observed clades/groups. Six CSIs, which are exclusively shared by species from the “Aeruginosa clade,” provide reliable demarcation of this clade corresponding to the genus Pseudomonas sensu stricto in molecular terms. The remaining 92 identified CSIs are specific for nine other Pseudomonas species clades and the genera Azomonas and Azotobacter which branch in between them. The identified CSIs provide strong independent evidence of the genetic cohesiveness of these species clades and offer reliable means for their demarcation/circumscription. Based on the robust phylogenetic and molecular evidence presented here supporting the distinctness of the observed Pseudomonas species clades, we are proposing the transfer of species from the following clades into the indicated novel genera: Alcaligenes clade – Aquipseudomonas gen. nov.; Fluvialis clade – Caenipseudomonas gen. nov.; Linyingensis clade – Geopseudomonas gen. nov.; Oleovorans clade – Ectopseudomonas gen. nov.; Resinovorans clade – Metapseudomonas gen. nov.; Straminea clade – Phytopseudomonas gen. nov.; and Thermotolerans clade – Zestomonas gen. nov. In addition, descriptions of the genera Azomonas, Azotobacter, Chryseomonas, Serpens, and Stutzerimonas are emended to include information for the CSIs specific for them. The results presented here should aid in the development of a more reliable classification scheme for Pseudomonas species.
Introduction
Genus Pseudomonas (Migula, 1894) is a large assemblage of motile, rod-shaped, aerobic, non-spore forming, Gram-negative bacteria, generally containing one or more polar flagella that assist in their movement (Palleroni, 2005, 2015). The members of this genus presently contain >300 species with validly published names (Parte et al., 2020), and they span enormous genetic and metabolic diversity, inhabiting diverse niches and environments including soil, water, plants and animal tissues (Peix et al., 2009; Palleroni, 2015). Its members include species which are opportunistic pathogens of humans, animals, and plants, and other species of economic and ecological significance (Palleroni, 2005; Lund-Palau et al., 2016; Winsor et al., 2016; Xin et al., 2018; Rossi et al., 2021). The best studied species from this genus, which is also its nomenclature type (Migula, 1894; Skerman et al., 1980), is Pseudomonas aeruginosa, which is an opportunistic human pathogen capable of causing a wide array of life-threatening acute and chronic diseases (Stover et al., 2000; Planquette et al., 2013). Despite the clinical and environmental importance of Pseudomonas species, evolutionary relationships among the members of this genus are not clearly understood (Anzai et al., 2000; Peix et al., 2009; Palleroni, 2015; García-Valdés and Lalucat, 2016; Jun et al., 2016; Passarelli-Araujo et al., 2022). In different phylogenetic and genomic studies on Pseudomonas species, members of this genus consistently form multiple clades, which are unrelated to each other (i.e., not evolved from a common ancestor) (Peix et al., 2009; Gomila et al., 2015; Jun et al., 2016; Hesse et al., 2018; Peix et al., 2018; Rudra and Gupta, 2021; Saati-Santamaría et al., 2021; Lalucat et al., 2022). Additionally, in these trees, species from several genera including Azomonas, Azotobacter and Chryseomonas branch in between Pseudomonas species, making this genus polyphyletic (Jun et al., 2016; Hesse et al., 2018; Rudra and Gupta, 2021; Saati-Santamaría et al., 2021; Lalucat et al., 2022). In recent work, a large number of Pseudomonas species, which generally branched outside the main cluster of Pseudomonas species, have been reclassified into several novel genera (viz. Atopomonas, Halopseudomonas and Stutzerimonas) (Rudra and Gupta, 2021; Lalucat et al., 2022), or in other existing genera (viz. Chryseomonas, Stenotrophomonas, Thiopseudomonas and Xanthomonas) (Holmes et al., 1987; Rudra and Gupta, 2021; Saati-Santamaría et al., 2021).
Importantly, in all constructed phylogenomic trees, the type species P. aeruginosa, along with a limited number of other species, forms a distinct clade referred to as the “Aeruginosa clade” (Jun et al., 2016; Hesse et al., 2018; Peix et al., 2018; Rudra and Gupta, 2021; Saati-Santamaría et al., 2021; Lalucat et al., 2022; Passarelli-Araujo et al., 2022). The remainder (>95%) of the Pseudomonas species group into 12–18 main clusters, some of which are referred to as the Alcaligenes, Anguilliseptica, Flexibilis, Fluorescens, Kuykendallii, Linyingensis, Lutea, Massiliensis, Oleovorans, Oryzihabitans, Pertucinogena, Putida, Resinovorans, Rhizosphaerae, Straminea, Stutzeri and Syringae clades, named after one of the species from each of these clusters (Palleroni, 2015; Hesse et al., 2018; Peix et al., 2018; Girard et al., 2021; Rudra and Gupta, 2021; Saati-Santamaría et al., 2021; Lalucat et al., 2022). Species from the Pertucinogena and Stutzeri clusters were recently reclassified into the genera Halopseudomonas and Stutzerimonas, respectively (Rudra and Gupta, 2021; Lalucat et al., 2022). Of these species’ clades, according to the Code governing the nomenclature of Prokaryotes (Oren et al., 2023), the “Aeruginosa clade,” which contains the type species P. aeruginosa, constitute the genus Pseudomonas sensu stricto. It is generally recognized that the species from clades other than the “Aeruginosa clade,” should be reclassified into novel genera (Hesse et al., 2018; Peix et al., 2018; Girard et al., 2021; Rudra and Gupta, 2021; Lalucat et al., 2022; Passarelli-Araujo et al., 2022). This task requires that the boundaries of different Pseudomonas species clades, including the “Aeruginosa clade,” are reliably demarcated so that any proposed reclassification is stable. Different Pseudomonas species clades are presently identified primarily based on the clustering of species in phylogenetic trees. However, the numbers of observed species clusters as well as the species grouping within them often vary in different phylogenetic studies (Hesse et al., 2018; Girard et al., 2021; Rudra and Gupta, 2021; Lalucat et al., 2022; Rudra et al., 2022), which makes it difficult to reliably demarcate the boundaries of these clades.
The availability of whole genome sequences is enabling construction of more reliable phylogenetic trees based on large dataset of genes/proteins (Parks et al., 2018). Additionally, the genome sequences also provide an important resource for identification of novel molecular markers, such as conserved signature indels (CSIs), which are uniquely shared characteristics of different monophyletic clades of organisms. Due to their clade specificities, these novel molecular synapomorphies are providing robust means for the demarcation of different observed species clades/taxa in molecular terms (Gupta et al., 2013; Gupta, 2014; Adeolu et al., 2016; Gupta et al., 2020). The use of these markers in conjunction with phylogenomic analyses has recently led to the development of a reliable classification scheme for members of the highly polyphyletic genus Bacillus (Gupta et al., 2020). Genome sequences are now available for >300 Pseudomonas species in the NCBI genome database1 (Sayers et al., 2019). With the objective of clarifying evolutionary relationships and classification of Pseudomonas species, we have conducted comprehensive phylogenomic and molecular marker-based studies on their genome sequences. In two genome scale phylogenetic trees constructed in this study, Pseudomonas species formed approximately 13 main clades, like those seen in earlier work (Hesse et al., 2018; Girard et al., 2021; Lalucat et al., 2022; Passarelli-Araujo et al., 2022). In parallel, our detailed studies on protein sequences from Pseudomonas genomes have identified 98 novel CSIs which are unique characteristics of the species from different observed clades. Based on these CSIs, species from the “Aeruginosa clade” (i.e., genus Pseudomonas sensu stricto), 10 other Pseudomonas species clades, and the genera Azomonas and Azotobacter, can now be reliably demarcated based on multiple uniquely shared molecular characteristics. Based on the strong evidence obtained from our phylogenomic studies and identified molecular markers, we are proposing the reclassification of Pseudomonas species from the following clades, viz. Alcaligenes, Fluvialis, Linyingensis, Oleovorans, Resinovorans, Straminea, and Thermotolerans, into seven novel genera. In addition, we are also emending the descriptions of the genera Azomonas, Azotobacter, Chryseomonas, Serpens and Stutzerimonas to include information for the diagnostic CSIs for these genera.
Methods
Construction of phylogenetic trees
Genome sequences were downloaded from the NCBI for 342 named Pseudomonas species and 46 sequences from other Pseudomonadaceae genera available as of December 16, 2022, in the database. Each species is represented in the tree by a single genomic sequence, which is generally of the type strain, when available. Based on these genome sequences, a rooted phylogenetic tree was constructed based on concatenated sequences of 118 conserved proteins that are a part of the phyloeco set for the class Gammaproteobacteria (Wang and Wu, 2013) (listed in Supplementary Table S1). Genome sequences for Moraxella bovoculi and M. bovis were included in this dataset for rooting purposes. Another comprehensive phylogenetic tree was constructed based on the core proteins from the genomes of Pseudomonadaceae species. This latter tree was based on genome sequences for 174 species, which included most of the species from the other main clades of Pseudomonas species, but only 41 divergent species from the Fluorescens superclade (lineage). Trees were constructed using an internally developed pipeline described in earlier work (Adeolu et al., 2016; Gupta et al., 2020; Rudra and Gupta, 2021; Saini and Gupta, 2021). Briefly, the CD-HIT program (Li and Godzik, 2006; Fu et al., 2012) was used to identify protein families (or homologs of different proteins) where the proteins were present in at least 80% of the genomes in the dataset and they shared at least 50% of sequence length and identity. The Clustal Omega program (Sievers et al., 2011) was then used to generate multiple sequence alignments (MSA) of the proteins. These MSAs were converted into profile Hidden Markov Models (HMMs) using HMMer 3-1b2 (Eddy, 2011), which were then used to search for other members of the protein families in the input genomes. These analyses identified 1,503 protein families meeting the stated criteria (also listed in Supplementary Table S1). The sequence alignments of these proteins were trimmed using TrimAl program (Capella-Gutiérrez et al., 2009) to remove poorly aligned sections prior to their concatenation. The concatenated sequence alignment for the phyloeco set of proteins for Gammaproteobacteria was created similarly using the published profile HMMs for these proteins (Wang and Wu, 2013). The concatenated sequence alignments used for the construction of phyloeco and the core genome trees consisted of 42,362 and 494,143 amino acid (aa) positions, respectively. Using these alignments, maximum likelihood (ML) trees were initially constructed using FastTree 2 (Price et al., 2010) with the Whelan and Goldman (2001) model of protein sequence evolution. The resulting trees were optimized with RAxML 8 (Stamatakis, 2014) and to obtain the Shimodaira-Hasegawa (SH) statistical support values, which are similar to the bootstrap scores, for different nodes. The trees were labeled and formatted using MEGA X (Kumar et al., 2018). The percentage of conserved proteins (POCP) and average amino acid identity (AAI) for different pairs of genomes were calculated as described by Thompson et al. (2013) and Qin et al. (2014).
Identification of conserved signature indels
Identification of CSIs was carried out by similar procedures as described in earlier work (Gupta, 2014, 2016; Gupta et al., 2020). Briefly, local BLASTp searches were carried out on protein sequences from the genomes of several Pseudomonas species representing different clades of interest and other outgroup species. Based on these BLAST searches, sequences of high scoring homologs (E value <1e-20) of different proteins were retrieved for several species (generally between 4 to 12) from the group of interest, and 10–15 species from other Pseudomonas clades or other Pseudomonadaceae genera. Multiple sequence alignments for the proteins were created using Clustal X 2.1 program (Jeanmougin et al., 1998). Alignments were visually examined for insertions or deletions of fixed length that were present in conserved regions (i.e., flanked on both sides by minimally 5–6 conserved aa residues in the neighboring 40–50 aa), and which were only found in the Pseudomonas species from the clade of interest. The indels which were not present in conserved regions were not further considered. The query sequences consisting of the conserved indels and their flanking 30–40 aa on each side were subjected to a second BLASTp search against the NCBI nr database and the top 250–500 hits were evaluated to determine the group specificities of the CSIs. Based on these results, indels which were specific for different clades of Pseudomonas were formatted using the SIG_CREATE and SIG_STYLE programs (Gupta, 2014, 2016). Due to space constraints, sequence information is shown for only a limited number of species in the main figures. However, unless otherwise indicated the CSIs reported here are specifically found in different named Pseudomonas species from the indicated groups. More detailed information for different CSIs is provided in the Supplemental Data files.
Results
Phylogenomic analyses of Pseudomonas and related species
To understand the interspecies relationships among different Pseudomonadaceae species whose genomes were available in the NCBI as of December 16, 2022, two genome-scale phylogenetic trees were constructed. The first of these trees shown in Figure 1 (Supplementary Figure S1), which will be referred to as the phyloeco tree, is based on concatenated sequences for 118 conserved proteins, which comprise the phyloeco set for the class Gammaproteobacteria (Wang and Wu, 2013). Another comprehensive tree constructed is a core genome (protein) tree based on 1,503 proteins which are shared by at least 80% of the input Pseudomonadaceae species. This latter tree included only representative species (41) from the Fluorescens superclade (lineage), which is not the focus of this study. In both constructed trees, most observed nodes are supported with 100% SH values (like bootstrap scores) indicating that the observed evolutionary relationships are reliable.
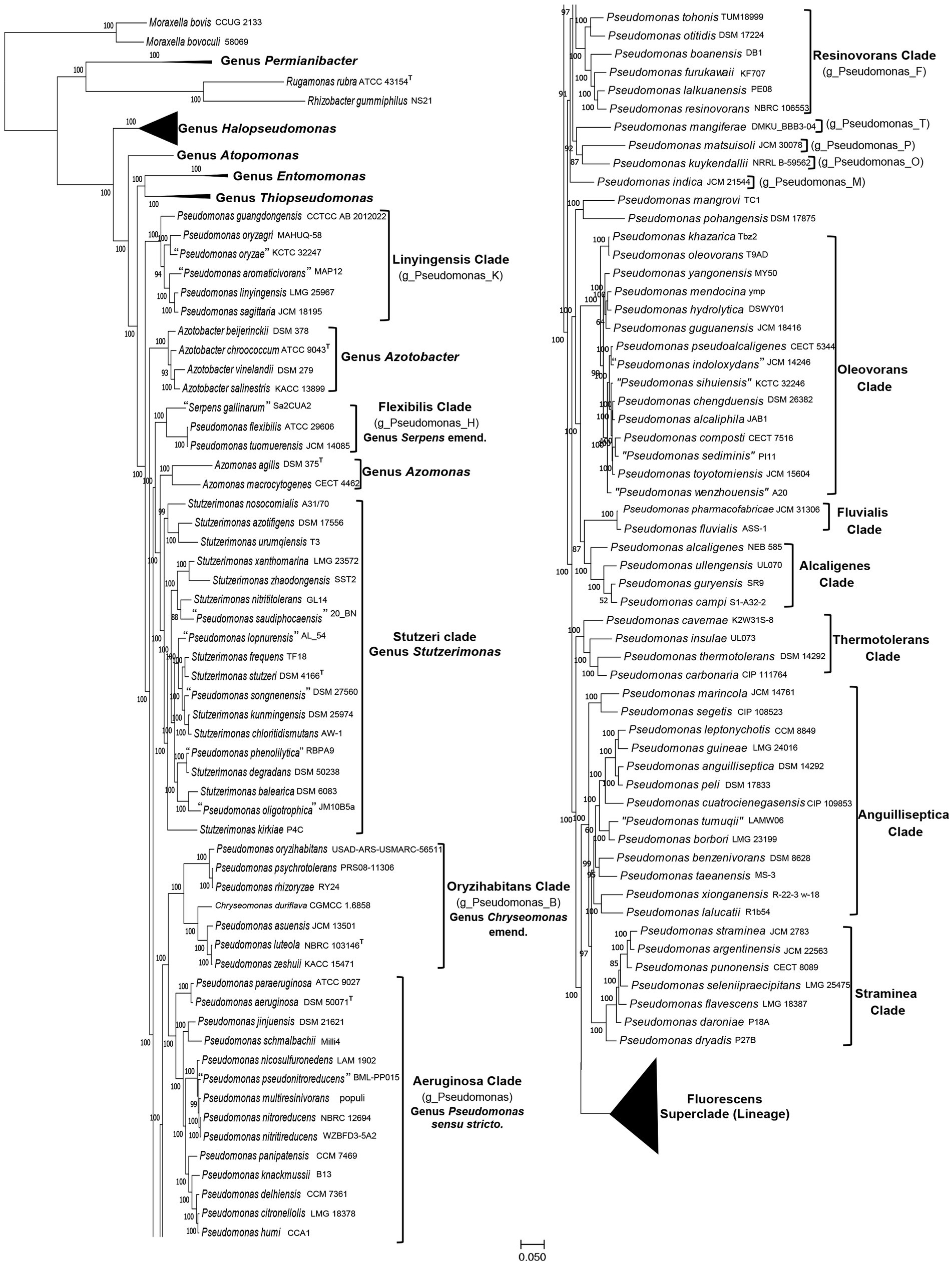
Figure 1. A maximum-likelihood tree for 388 genome-sequenced Pseudomonadaceae species based on concatenated sequences for 118 conserved proteins. The tree is shown into two halves, and species from the Fluorescens superclade (lineage) are compressed, so that the species compositions of other clades of interest can be seen. The species clades of interest are demarcated and labeled with the commonly used names and in some cases with the GTDB taxon assignment for the clade.
The overall branching and grouping of Pseudomonadaceae species in different clusters in both the phyloeco (Figure 1) and the core protein tree (Supplementary Figure S2) is nearly identical, and it is similar to that observed in our earlier work (Rudra and Gupta, 2021), and other phylogenetic studies (Gomila et al., 2015; Hesse et al., 2018; Peix et al., 2018; Lalucat et al., 2020; Girard et al., 2021; Lalucat et al., 2022; Passarelli-Araujo et al., 2022). In both these trees, Pseudomonas species formed several distinct clades/groups, and species from the genera Azomonas and Azotobacter consistently branched between them (Hesse et al., 2018; Rudra and Gupta, 2021; Lalucat et al., 2022; Passarelli-Araujo et al., 2022). Additionally, species from the two recently proposed genera Stutzerimonas and Chryseomonas also branched within other Pseudomonas species, thus further contributing to the polyphyly of this genus. We have labeled different Pseudomonas species clades in Figure 1 and Supplementary Figure S2 by their commonly used clade/group names (Hesse et al., 2018; Girard et al., 2021; Lalucat et al., 2022). One distinct clade observed in all constructed trees is the “Aeruginosa clade,” which contains the type species P. aeruginosa and 13 other Pseudomonas species. As this clade contains the type species of the genus Pseudomonas, we have labeled it as the “Genus Pseudomonas sensu stricto.” Other species’ clades observed and labeled in Figure 1 (Supplementary Figure S2) include: the Alcaligenes, Anguilliseptica, Azomonas, Azotobacter, Flexibilis, Fluvialis, Linyingensis, Oleovorans, Oryzihabitans, Resinovorans, Straminea, Stutzeri (Stutzerimonas), Thermotolerans, and Fluorescens superclade (lineage).The Genome Taxonomy Database (GTDB),2 based on phylogenetic analysis of 120 ubiquitously conserved proteins, now provides an important resource for taxonomic inferences (Parks et al., 2018). The GTDB refers to the “Aeruginosa clade” as the genus Pseudomonas whereas most of the other observed species clades are referred to as distinct genera denoted by designations such as g_Pseudomonas_B, g_Pseudomonas_K, etc., which are also indicated in the tree in Figure 1.
Of these observed clades, the Fluorescens superclade (lineage) is the largest harboring 245 Pseudomonas species. It is separated from all other Pseudomonas species by a long branch in both constructed trees (Figure 1; Supplementary Figure S2). Due to the large number of species present in this clade, it is shown in a compressed form in Figure 1. However, detailed information for species comprising this clade is provided in Supplementary Figure S1. The Fluorescens superclade (lineage) is made up of multiple distinct clades and subclades (see Supplementary Figure S1) (Hesse et al., 2018; Peix et al., 2018; Lalucat et al., 2020; Rudra and Gupta, 2021; Lalucat et al., 2022). However, all species grouping within the Fluorescens superclade (lineage) are part of the GTDB taxon “g_Pseudomonas_E.” Although the Pseudomonas_E cluster in GTDB also encompasses the Alcaligenes, Anguilliseptica, Oleovorans and Thermotolerans clades, these clades in our phylogenomic trees (Figure 1; Supplementary Figure S1), and in several other published studies (Hesse et al., 2018; Girard et al., 2021; Lalucat et al., 2022; Passarelli-Araujo et al., 2022), branch separately from the Fluorescens superclade. This discrepancy in the branching positions of the Alcaligenes, Anguilliseptica, Oleovorans and Thermotolerans clades between the GTDB taxonomy and other phylogenomic trees, was also noted by Lalucat et al. (2022). However, in the present work, we will not be examining the evolutionary relationships of different species within the Fluorescens superclade. Besides the “Aeruginosa clade” and the Fluorescens superclade (lineage), the other clades marked in Figure 1 (Supplementary Figure S2) contain between 2–18 species. Except for the Anguilliseptica clade, which shows poor resolution and weak statistical support, all other clades in our phylogenetic trees are statistically strongly supported. Besides these species’ clades, a limited number of Pseudomonas species (viz. P. indica, P. kuykendallii, P. mangiferae, P. mangrovi, P. matsuisoli and P. pohangensis) are not part of any of the observed clades.
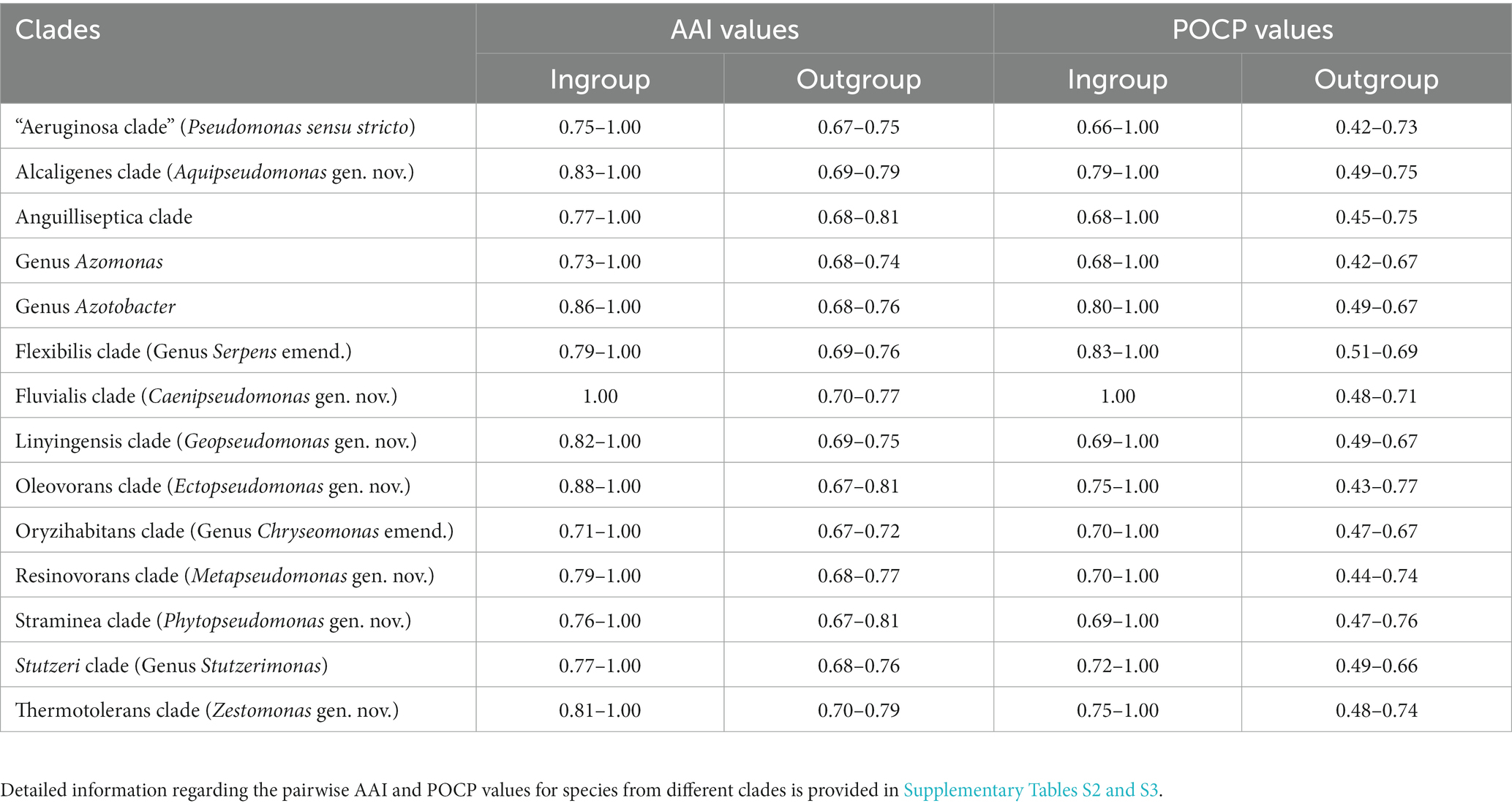
The analyzed genome sequences were also used for determination of percentage of conserved proteins (POCP) and average amino acid identity (AAI) between different pairs of genomes. The results of pairwise AAI and POCP values, for different Pseudomonadaceae genomes are presented in Supplementary Tables S2 and S3, respectively. Genome pairs exhibiting higher AAI or POCP values are shown by a darker shade of green/red, and different clades observed in our phylogenetic trees (Figure 1; Supplementary Figure S2) are outlined. In Table 1, we present a summary of the ranges of the AAI and POCP values for different Pseudomonas species clades for the ingroup and outgroup species. Based on the results in Table 1, the AAI and POCP values for species within different clades are higher (AAI values range: 0.70–1.00; POCP values range: 0.66–1.00) in comparison to these values for species from the other clades (AAI values range: 0.67–0.81; POCP values range: 0.42–0.77), which is an expected result. However, based on the AAI and POCP values (Table 1), only species from the Alcaligenes, Azotobacter, Flexibilis, Fluvialis, Lingyingensis, Oleovorans and Thermotolerans clades show no overlap with species from the other clades. In contrast, these values for several other clades (viz. “Aeruginosa.” Anguiliiseptica, Azomonas, Oryzihabitans, Resinovorans, Straminea, Stutzeri) either show significant overlap or are very close to those from the outgroup species. Thus, based on these genome similarity indices, species from different observed Pseudomonadaceae clades cannot be reliably demarcated. In Table 1, the highest overlap in the AAI and POCP values between the ingroup versus outgroup species is observed for the species from Anguilliseptica clade, which also shows poor resolution and weak statistical support in the phylogenetic trees.
Identification of molecular markers demarcating/distinguishing different Pseudomonas species clades
Although Pseudomonadaceae species form similar clades in different genome scale trees (Hesse et al., 2018; Parks et al., 2018; Girard et al., 2021; Lalucat et al., 2022; Figure 1; Supplementary Figure S2), branching of species in phylogenetic trees is influenced by large numbers of variables (Gupta, 1998; Baldauf, 2003; Felsenstein, 2004). Moreover, in phylogenetic trees for Pseudomonas, species from several clades are separated from each other by short branches (Figure 1; Supplementary Figure S2), which makes it difficult to reliably determine their boundaries. The POCP and AAI values for several clades also overlap or are very close to the other species (Table 1), thus they do not permit reliable determination of the boundaries of these clades. Hence, it was important to discover other reliable means for the demarcation of these clades. Molecular synapomorphies consisting of CSIs in genes/proteins sequences, which are uniquely shared characteristics of species from different clades, provide important means for the demarcation of taxa of different ranks in molecular terms (Gupta, 2014; Adeolu et al., 2016; Gupta et al., 2020; Patel and Gupta, 2020; Rudra and Gupta, 2021). Hence, detailed studies were conducted on protein sequences from Pseudomonadaceae species to identify CSIs which are specific for different observed clades. These analyses have identified 98 novel CSIs which are specific for different Pseudomonadaceae clades, providing independent evidence for the genetic distinctness of these clades and affording reliable means for their demarcation. Brief descriptions of the characteristics of these CSIs are given below.
CSIs specific for the “Aeruginosa clade”
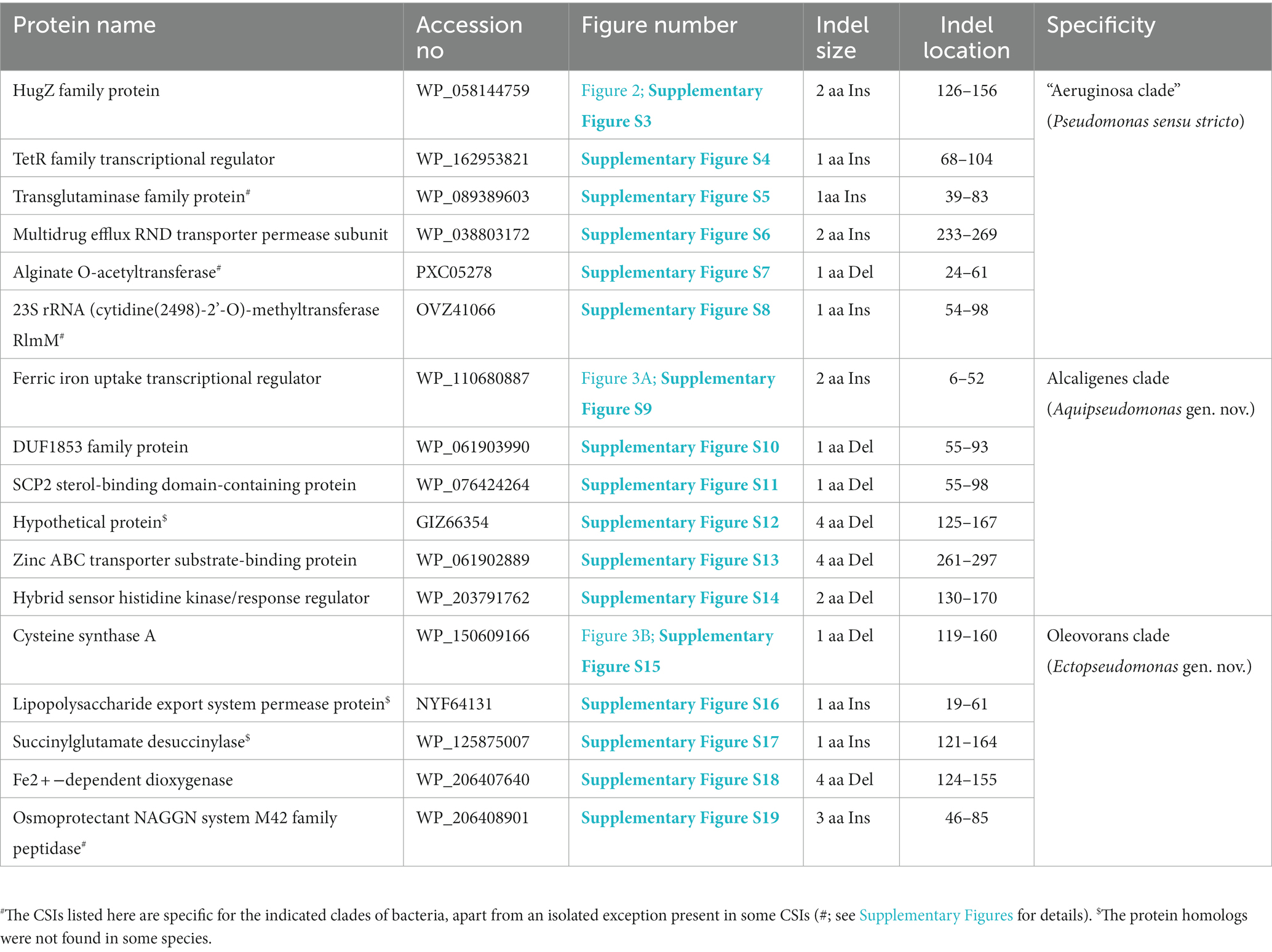
The “Aeruginosa clade” representing the genus Pseudomonas sensu stricto, encompasses 14 named species (viz., P. aeruginosa, P. paraeruginosa, P. citronellolis, P. delhiensis, P. humi, P. jinjuensis, P. knackmussii, P. multiresinivorans, P. nicosulfuronedens, P. nitritireducens, P. nitroreducens, P. panipatensis, “P. pseudonitroreducens” and P. schmalbachii) (Figure 1). Our analyses have identified six CSIs in proteins involved in different functions (Table 2), which are commonly and, in most cases, uniquely shared by different species from the “Aeruginosa clade.” Sequence information for one of these is presented in Figure 2. In the example shown, a two aa insertion (highlighted) in a conserved region of the HugZ family protein is commonly shared by all 14 species from the “Aeruginosa clade” but absent in all other Pseudomonadaceae species. Sequence information is shown in Figure 2 for only a limited number of species. However, more detailed information for this CSI is presented in Supplementary Figure S3. Like the CSI shown in Figure 2, we have identified five additional CSIs in other proteins which, except for an isolated occurrence, are uniquely shared by different species from the “Aeruginosa clade.” Sequence information for these CSIs is provided in Supplementary Figures S4–S8 and some of their characteristics are summarized in Table 2. Due to their unique shared presence in species from the “Aeruginosa clade,” genetic changes responsible for these CSIs likely occurred in a common ancestor of this clade and subsequently inherited by all members. Due to their specificities for the species from the “Aeruginosa clade,” these molecular synapomorphies provide robust means for the demarcation of this clade in molecular terms.
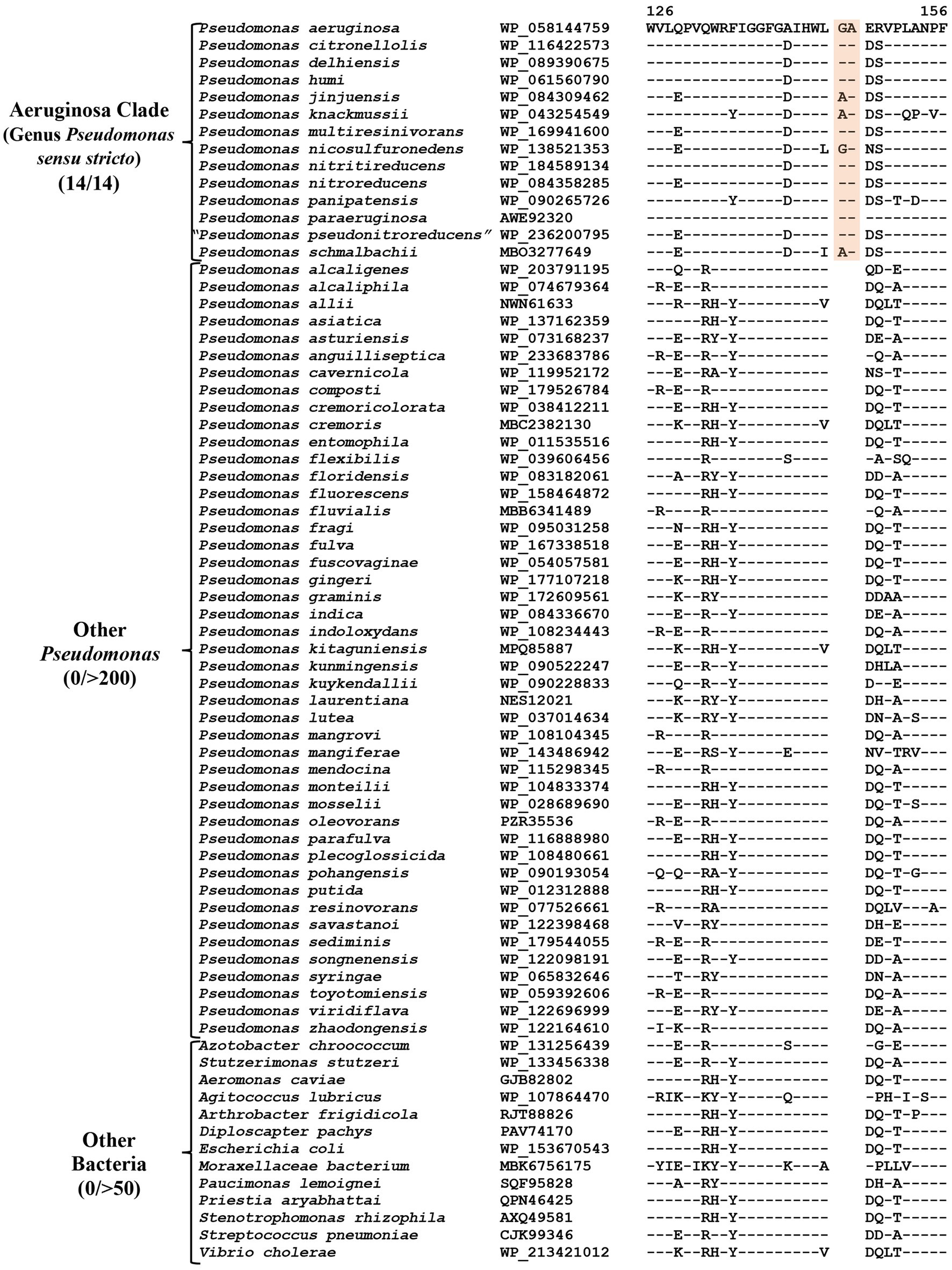
Figure 2. Partial sequence alignment of the HugZ family protein showing a two aa insertion (highlighted) that is exclusively present in all members of the “Aeruginosa clade.” The dashes (−) in this and all other sequence alignments indicate identity with the amino acids on the top line. Accession numbers for different sequences are indicated in the second column and the numbers at the top indicate the position of this sequence in the protein sequences. Detailed sequence information for this CSI and five other CSIs specific for this clade is provided in Supplementary Figures S3–S8.
CSIs specific for the Alcaligenes clade
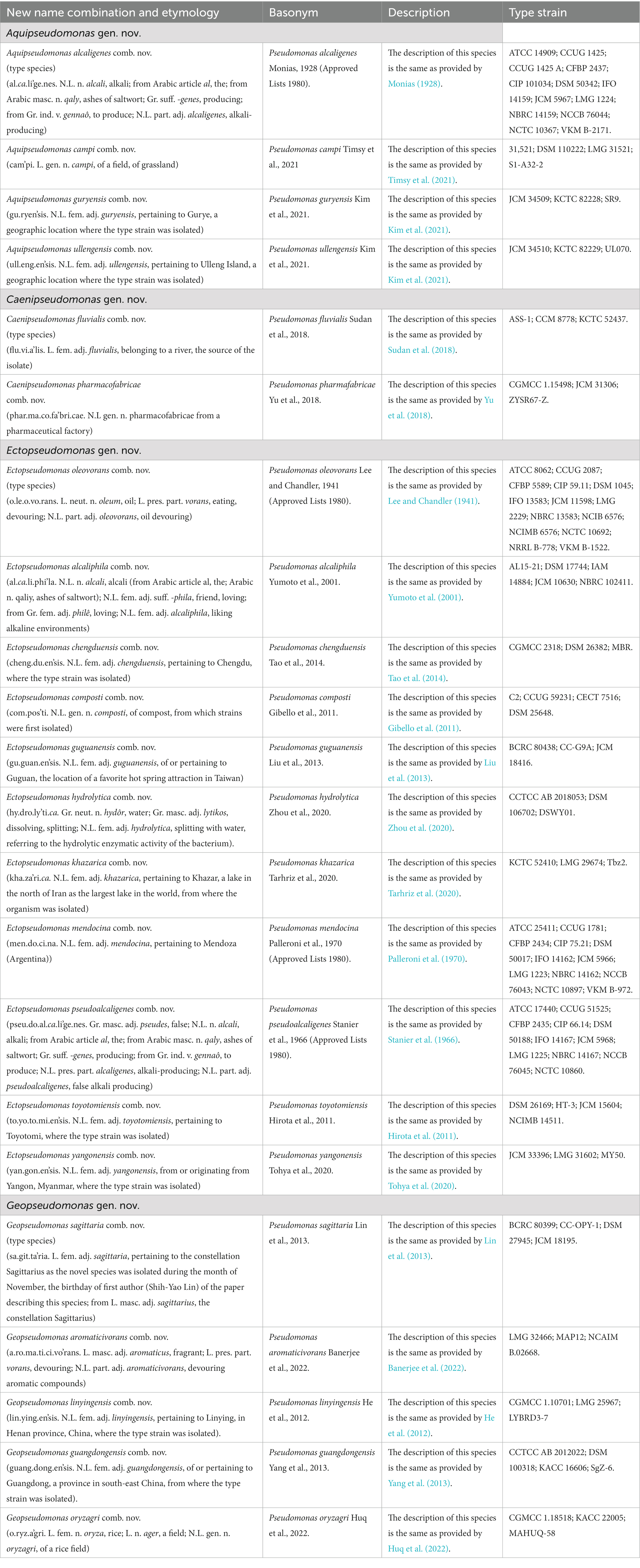
P. alcaligenes was indicated to branch separately from other clades in earlier studies (Hesse et al., 2018; Girard et al., 2021; Lalucat et al., 2022). In our phylogenetic trees (Figure 1; Supplementary Figure S2), three recently identified species (viz., P. campi, P. guryensis, P. ullengensis) also reliably grouped with P. alcaligenes. Our analysis has identified six novel CSIs, which in most cases are exclusively shared by all four species from the Alcaligenes clade. Sequence information for one of these CSIs is presented in Figure 3A, where a two aa insertion in the protein ferric iron uptake transcriptional regulator is exclusively present in all four species from the Alcaligenes clade. Five additional CSIs in other proteins are also generally specific for the species from this clade. Detailed sequence information for these six CSIs is provided in Supplementary Figures S9–S14, and some of their characteristics are listed in Table 2. The identified CSIs provide reliable means for the demarcation of species from the Alcaligenes clade in molecular terms and we are proposing their transfer into Aquipseudomonas gen. nov.
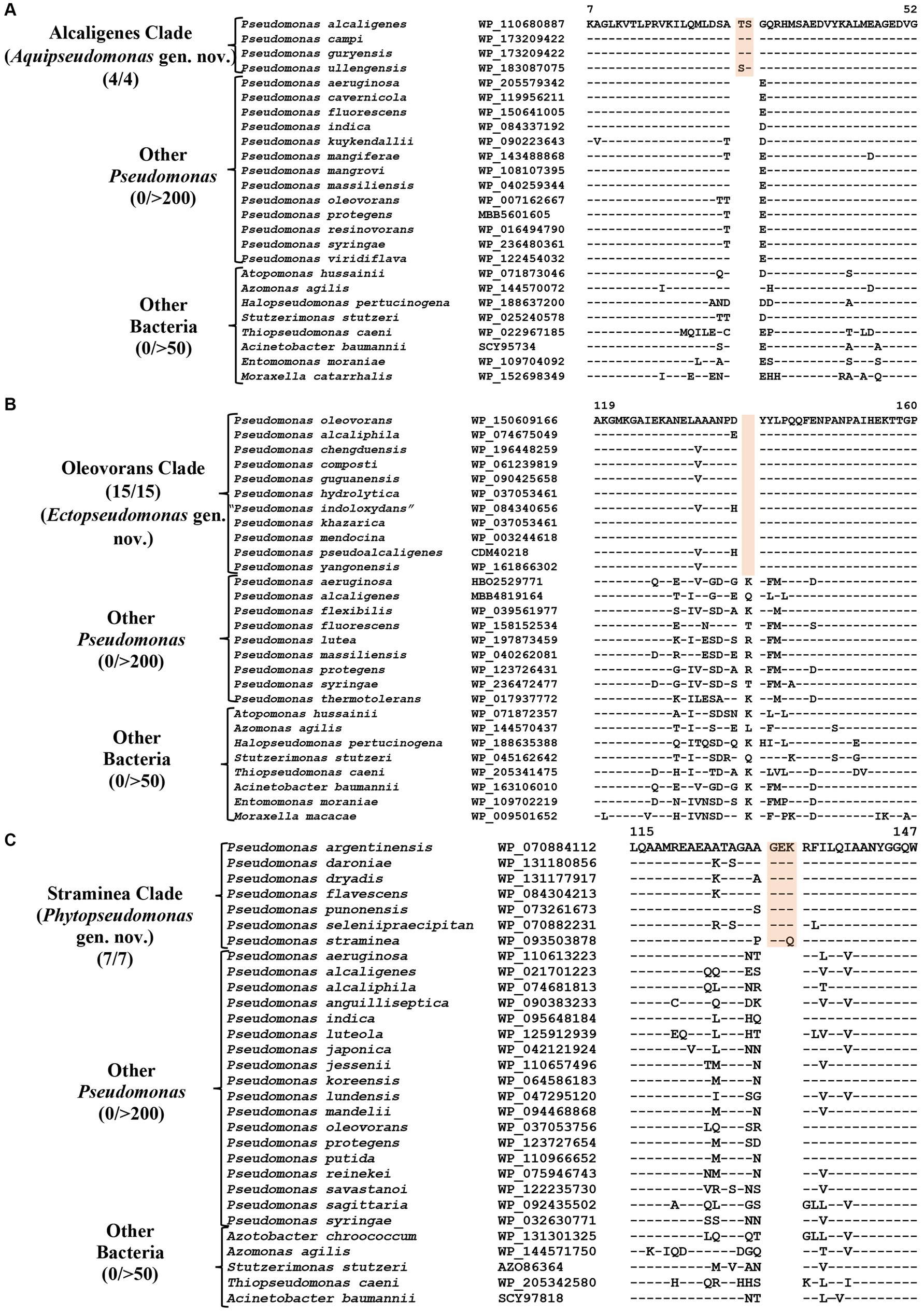
Figure 3. Partial sequence alignments of (A) Ferric iron uptake protein showing a two aa insertion within a conserved region that is a distinctive characteristic of all members of the Alcaligenes clade. (B) A one aa deletion in a conserved region of the protein Cysteine synthase A which is specific for the species from Oleovorans clade. (C) A three aa insertion within a conserved region in the protein Di-trans, poly-cis-decaprenylcistransferase, specific for the species from the Straminea clade. Detailed sequence information for these CSIs along with other CSIs specific for these clades are provided in Supplementary Figures S9–S31.
CSIs specific for the Oleovorans clade
Oleovorans clade is a strongly supported clade consisting of 15 Pseudomonas species (viz., P. alcaliphila, P. chengduensis, P. composti, P. guguanensis, P. hydrolytica, “P. indoloxydans,” P. khazarica, P. mendocina, P. oleovorans, P. pseudoalcaligenes, “P. sediminis,” “P. sihuiensis,” P. toyotomiensis, “P. wenzhouensis,” P. yangonensis), which reliably group together in the constructed phylogenetic trees (Figure 1; Supplementary Figure S2). The genetic distinctness of this clade is also independently supported by five novel identified CSIs which, excepting an isolated occurrence, are uniquely shared by all species from this clade. Sequence information for one of these CSIs is provided in Figure 3B, where a one aa deletion (highlighted), within a conserved region of the protein cysteine synthase A, is exclusively shared by all species from the Oleovorans clade. More detailed sequence information for this CSI and four additional CSIs specific for the Oleovorans clade is provided in Supplementary Figures S15–S19 and some of their characteristics are listed in Table 2. Based on the strong evidence presented here demonstrating the distinctness of species from the Oleovorans clade, we are proposing the transfer of these species into Ectopseudomonas gen. nov.
In addition to the species with validly published names, Oleovorans clade also encompasses four species [viz., “P. indoloxydans” (Manickam et al., 2008), “P. sediminis” (Behera et al., 2018), “P. sihuiensis” (Wu et al., 2014) and “P. wenzhouensis” (Zhang et al., 2021)], whose names have not been validly published. Because of their non-validly published status, new name combinations for these species are not proposed. However, in view of their reliable grouping with the Oleovorans clade, it is suggested that these species should also be recognized as members of the genus Ectopseudomonas with the names “E. indoloxydans,” “E. sediminis,” “E. sihuiensis” and “E. wenzhouensis,” respectively.
CSIs specific for the Straminea clade
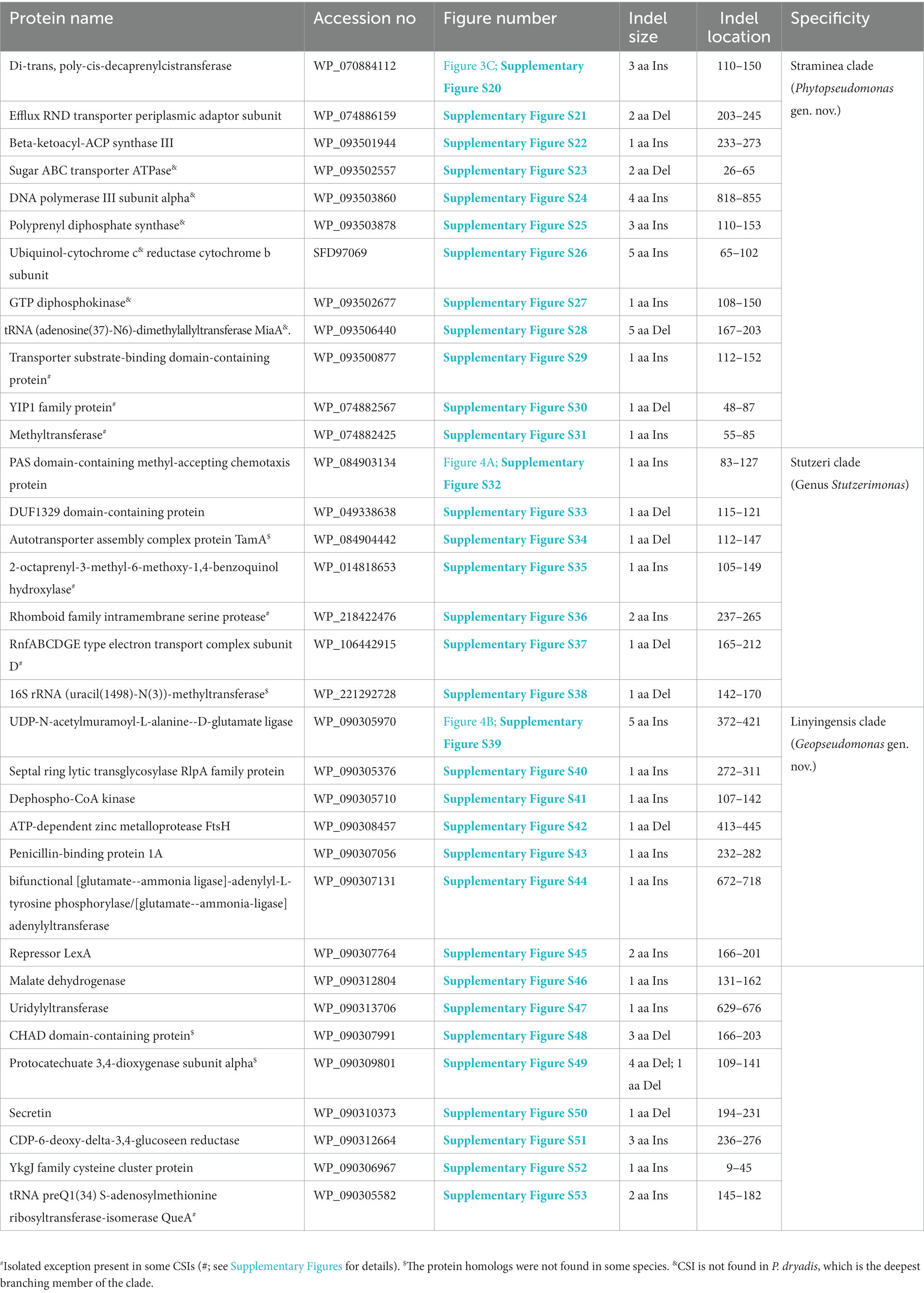
The Straminea clade is a strongly supported cluster encompassing seven Pseudomonas species (P. argentinensis, P. daroniae, P. dryadis, P. flavescens, P. punonensis, P. seleniipraecipitans, P. straminea) (Figure 1; Supplementary Figure S2). Species from this clade have also been found to group together in earlier studies (Hesse et al., 2018; Girard et al., 2021; Lalucat et al., 2022; Passarelli-Araujo et al., 2022). The members of this clade can be reliably distinguished from all other Pseudomonadaceae species by 12 novel CSIs identified in this study, which in most cases are exclusively shared by the species from this clade. Sequence information for one of these CSIs consisting of a three aa insertion in the protein Di-trans, poly-cis-decaprenylcistransferase is presented in Figure 3C. Detailed sequence information for this CSI and the 11 other CSIs specific for this clade are presented in Supplementary Figures S20–S31 and some of their characteristics are listed in Table 3. Based on the presented results showing the distinctness of this clade, we are proposing the transfer of species from this clade into Phytopseudomonas gen. nov.
CSIs specific for the genus Stutzerimonas
The genus Stutzerimonas was recently described by Lalucat et al. (2022) by the transfer of several Pseudomonas species which branched distinctly in their phylogenetic tree. The clade labeled as Stutzerimonas in our phylogenetic tree (Figure 1) encompasses all 13 named Stutzerimonas species, whose genome sequences were available in the NCBI database at the time of analysis, as well as five non-validly published Pseudomonas species. Apart from their clustering in phylogenetic trees, there is no known reliable characteristic which is specific for the members of this genus. Our analyses have identified seven CSIs in different proteins, which in most cases are uniquely shared by all/most species from this clade. Sequence information for one of these CSIs is shown in Figure 4A. In this instance, a one aa insertion in a conserved region of the PAS domain-containing methyl-accepting chemotaxis protein is uniquely shared by all species from the Stutzerimonas clade. Detailed sequence information for this CSI and the six other CSIs specific for this clade/genus is provided in Supplementary Figures S32–S38 and some of their characteristics are summarized in Table 3. The identified CSIs provide reliable means for distinguishing Stutzerimonas species from all other Pseudomonadaceae species. Hence, we are emending the description of this genus to include these diagnostic characteristics.
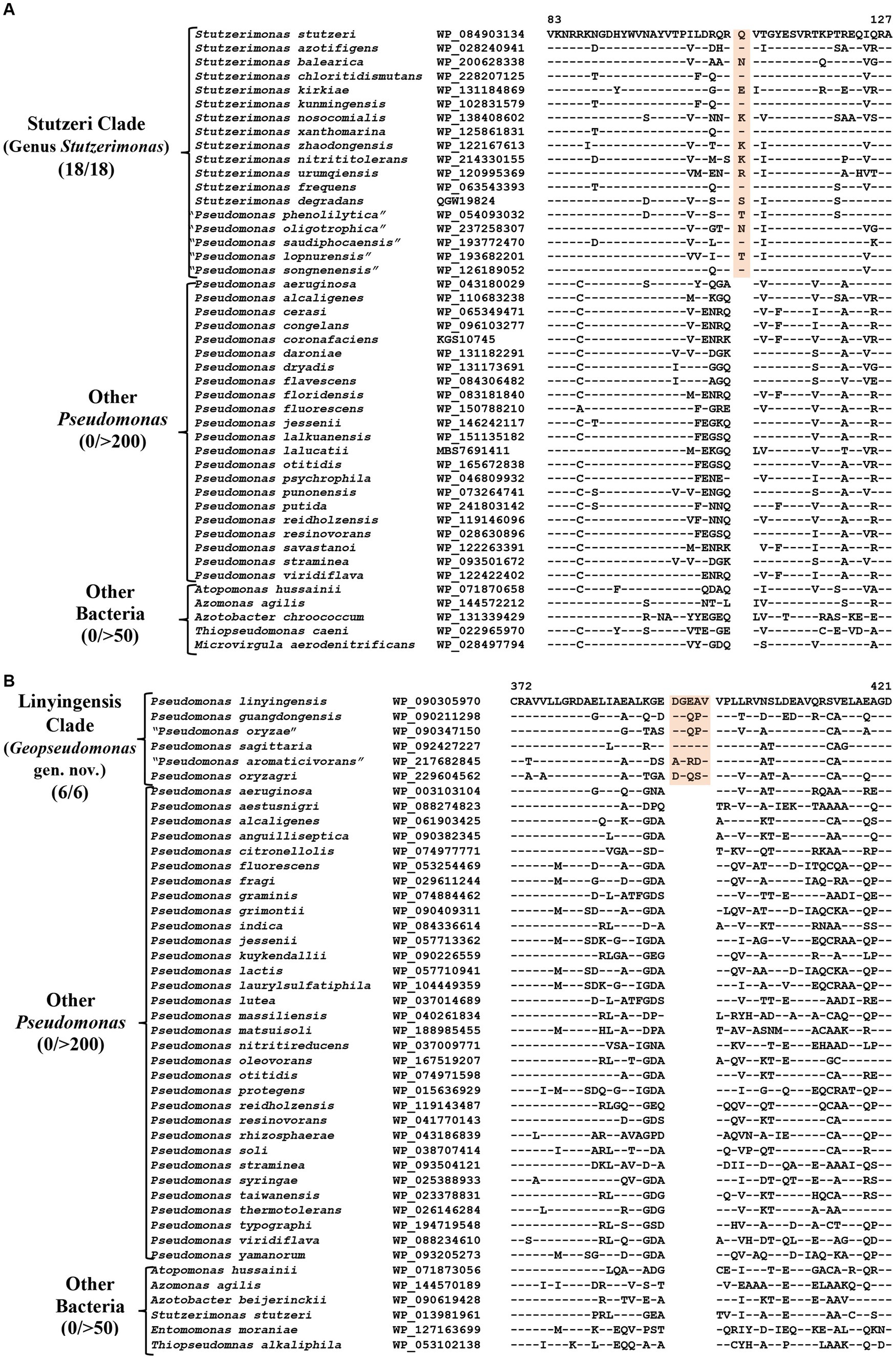
Figure 4. Partial sequence alignment of the protein (A) PAS domain containing Methyl-accepting chemotaxis protein showing a one aa insertion within a conserved region (highlighted) that is uniquely present in all members of the Stutzeri clade. (B) A five aa insertion within a conserved region of the protein UDP-N-acetylmuramoyl-L-alanine, which is specific for the species from Linyingensis clade. Detailed sequence information for these CSIs and other CSIs specific for Stutzeri and Linyingensis clades are provided in Supplementary Figures S32–S53.
Five species with non-validly published names [viz.“P. lopnurensis” (Mamtimin et al., 2021), “P. phenolilytica” (Kujur and Das, 2022), “P. oligotrophica” (Zhang et al., 2022), “P. saudiphocaensis” (Azhar et al., 2017) and “P. songnenensis” (Zhang et al., 2015)], also group reliably within the Stutzerimonas clade and share CSIs specific for this clade. These species should also be recognized as members of this genus with the names “S. lopnurensis,” “S. phenolilytica,” “S. oligotrophica,” “S. saudiphocaensis” and “S. songnenensis” respectively.
CSIs specific for the Linyingensis clade
The Linyingensis clade consists of six Pseudomonas species viz., P. aromaticivorans, P. guangdongensis, P. linyingensis, P. oryzagri, “P. oryzae” and P. sagittaria, which form a strongly supported clade in our phylogenetic trees (Figure 1; Supplementary Figure S2). This clade is also denoted as g_Pseudomonas_K in the GTDB taxonomy (Parks et al., 2018). A specific evolutionary relationship among these species is supported by 15 CSIs (Table 3), which in most cases are uniquely shared by all species from this clade. In Figure 4B, we present one example of a CSI specific for this clade, where a five aa insertion in UDP-N-acetylmuramoyl-L-alanine--D-glutamate ligase protein is uniquely shared by all members of this clade. Detailed sequence information for this CSI and 14 other CSIs specific for this clade is presented in Supplementary Figures S39–S53. Based on these results, which robustly demarcate this species clade, we are proposing the transfer of these species into Geopseudomonas gen. nov.
CSIs specific for the Resinovorans clade
The Resinovorans clade (Figure 1; Supplementary Figure S2), which is denoted as the taxon g_Pseudomonas_F in GTDB taxonomy (Parks et al., 2018), consists of six species viz. P. boanensis, P. furukawaii, P. lalkuanensis, P. otitidis, P. resinovorans and P. tohonis. Species from this clade also formed a distinct clade in earlier studies (Girard et al., 2021; Lalucat et al., 2022; Passarelli-Araujo et al., 2022). The members of this clade can be reliably distinguished from all other Pseudomonadaceae species by five identified CSIs, which in most cases are exclusively shared by all/most species from this clade. One example of a CSI specific for this clade is presented in Figure 5A, where in the Murein L, D-transpeptidase catalytic domain family protein, a two aa insertion is exclusively present in all species from the Resinovorans clade. Detailed sequence information for this CSI and four other identified CSIs, specific for this clade, is presented in Supplementary Figures S54–S58 and some of their characteristics are listed in Table 4. Based on these results, we are proposing the transfer of species from Resinovorans clade into Metapseudomonas gen. nov.
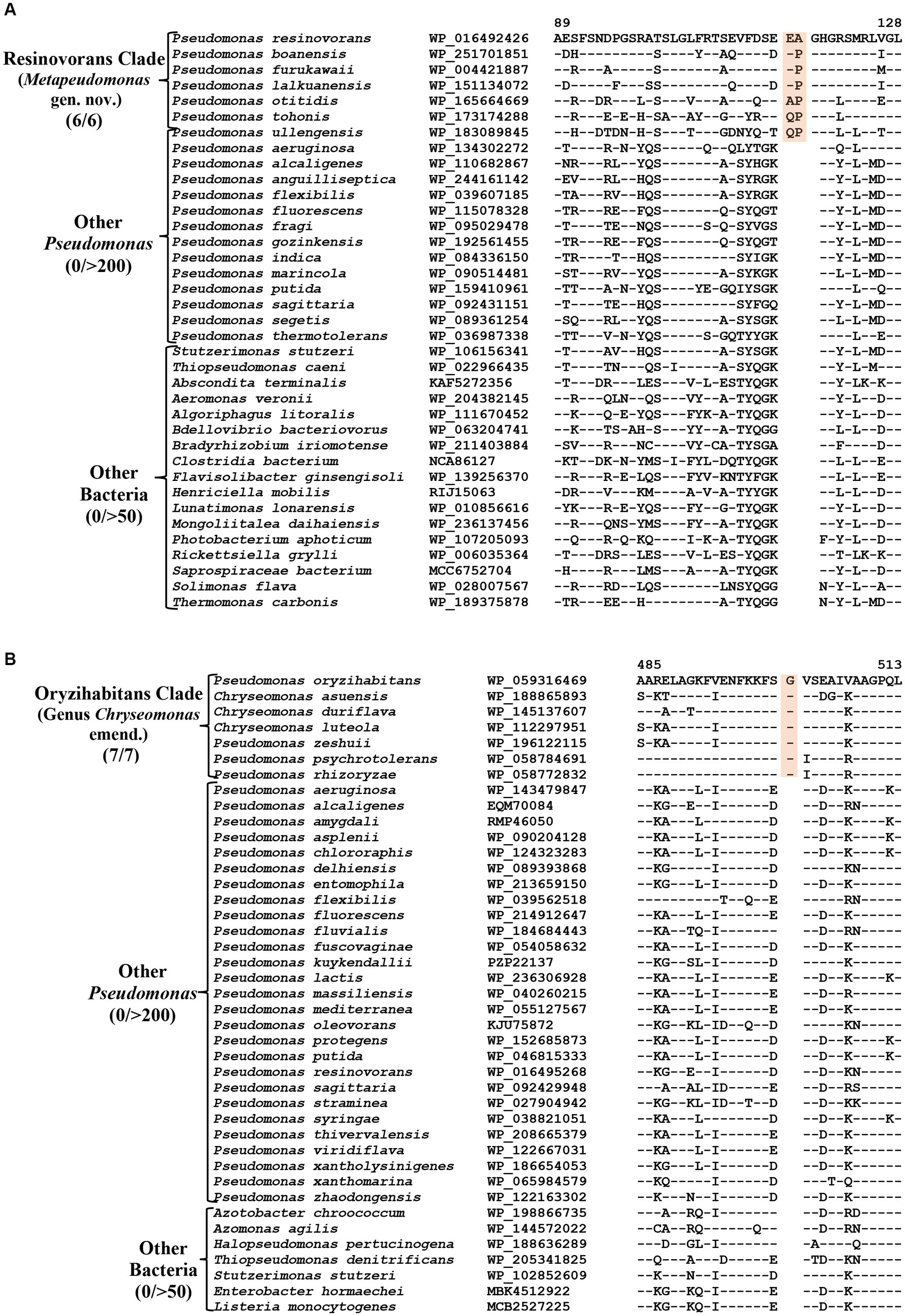
Figure 5. Partial sequence alignment of the protein (A) Murein L, D-transpeptidase catalytic domain family protein showing a two aa insertion within a conserved region that is commonly shared by all members of the Resinovorans clade. (B) A one aa insertion in the protein Cytochrome d ubiquinol oxidase subunit II which is specific for the species from the Oryzihabitans clade. Detailed sequence information for these CSIs and other CSIs specific for Resinovorans and Oryzihabitans clades are provided in Supplementary Figures S54–S69.
Table 4. Summary of CSIs specific for members of the Resinovorans, Oryzihabitans, Thermotolerans, and Flexibilis clades.
CSIs specific for the Oryzihabitans clade (genus Chryseomonas)
Oryzihabitans clade (denoted as the taxon g_Pseudomonas_B in GTDB taxonomy) consists of seven named Pseudomonas species viz. P. asuensis, P. duriflava, P. luteola, P. oryzihabitans, P. psychrotolerans, P. rhizoryzae and P. zeshuii, which form a strongly supported clade in our phylogenetic trees (Figure 1; Supplementary Figure S2). These species also formed a distinct clade in earlier phylogenetic studies (Hesse et al., 2018; Girard et al., 2021; Saati-Santamaría et al., 2021; Passarelli-Araujo et al., 2022). The best-studied species from this clade is P. luteola, which was originally a member of the genus Chryseomonas (Holmes et al., 1986). However, in 1997, based on 16S rRNA gene sequence similarity, this species was transferred into the genus Pseudomonas (Anzai et al., 1997). More recently, based on genomic studies, this species along with two other Pseudomonas species (P. asuensis and P. duriflava) were transferred into the genus Chryseomonas. It should be noted that C. luteola is a synonym of C. polytricha (Holmes et al., 1986), which is the type species of genus Chryseomonas (Parte et al., 2020). The genetic distinctness of the clade formed by these seven species is strongly supported by 11 novel identified CSIs which are uniquely shared by these species. One example of a CSIs specific for this clade is shown in Figure 5B. In this case, a one aa insertion in the protein cytochrome d ubiquinol oxidase subunit II is exclusively shared by all members of this clade. Detailed sequence information for this CSI and 10 other CSIs specific for this clade are presented in Supplementary Figures S59–S69 and some of their characteristics are listed in Table 4. In addition to the three species which are presently assigned to the genus Chryseomonas, four additional Pseudomonas species viz. P. oryzihabitans, P. psychrotolerans, P. rhizoryzae and P. zeshuii reliably group within this clade and share different CSIs specific for this genus. Hence, we are proposing new name combinations of these species to transfer them into the genus Chryseomonas.
CSIs specific for the Thermotolerans clade
The Thermotolerans clade includes the species P. carbonaria, P. cavernae, P. insulae and P. thermotolerans, which form a distinct clade in our phylogenomic trees (Figure 1; Supplementary Figure S2). Species from this clade also formed a distinct cluster in earlier studies (Girard et al., 2021; Lalucat et al., 2022). A specific evolutionary relationship among these species is strongly supported by five CSIs, which are exclusively shared by all members of this clade. One example of a CSI specific for this clade is shown in Figure 6A, where a six aa insertion in the TerB family tellurite resistance protein is exclusively found in all four species from this clade. Detailed sequence information for the five CSIs specific for this clade are presented in Supplementary Figures S70–S74 and some of their characteristics are listed in Table 4. Based on these results, we are proposing the transfer of species from this clade into Zestomonas gen. nov.
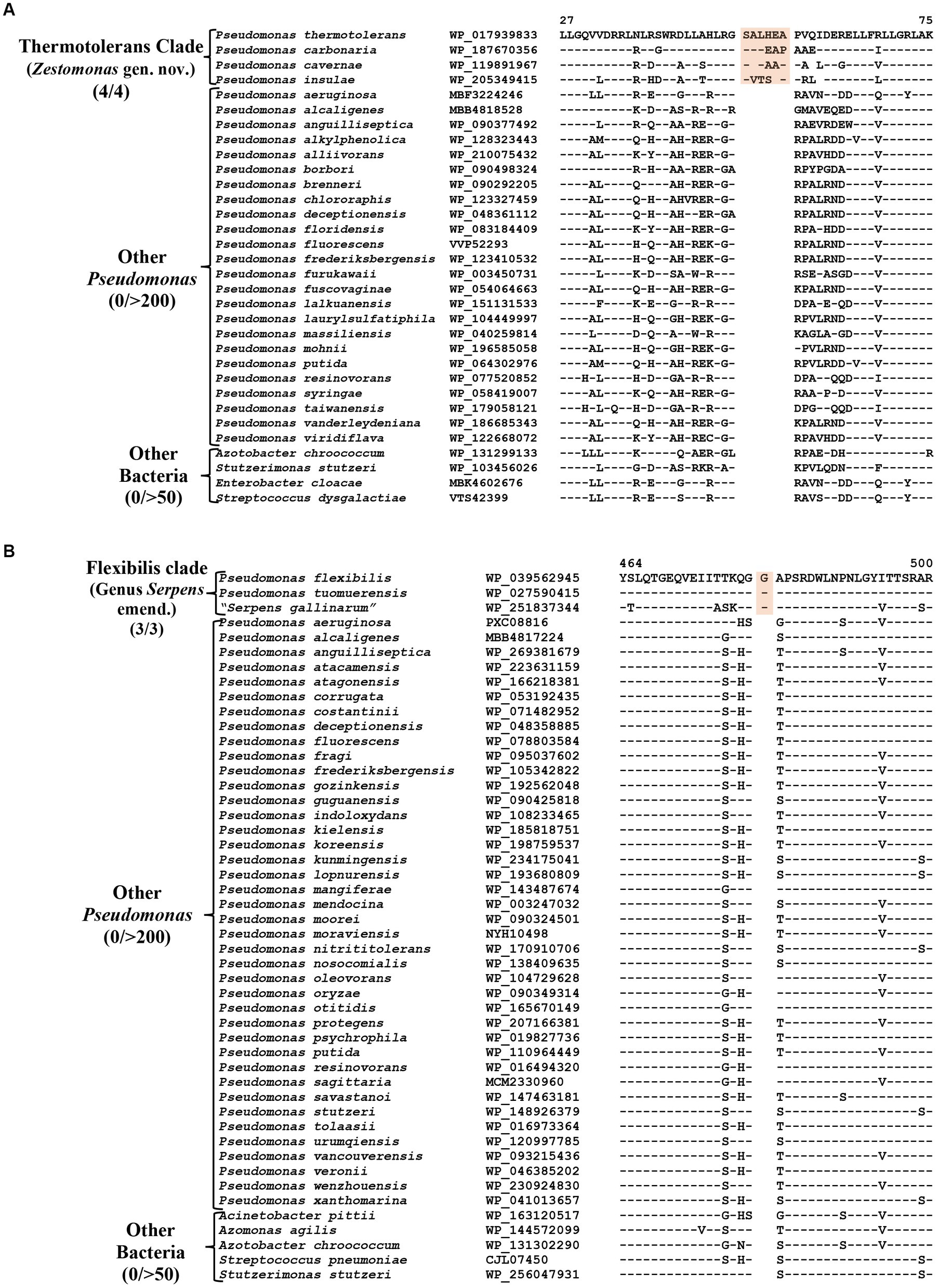
Figure 6. Partial sequence alignment of the protein (A) TerB family tellurite resistance protein showing a six aa insertion within a conserved region (highlighted) that is uniquely shared by members of the Thermotolerans clade. (B) A one aa insertion in a conserved region of the protein GTP diphosphokinase which is specific for the species from Flexibilis clade. Detailed sequence information for these CSIs and other CSIs specific for the Thermotolerans and Flexibilis clades are provided in Supplementary Figures S70–S77.
CSIs specific for the Flexibilis clade (genus Serpens)
Pseudomonas flexibilis, formerly known as Serpens flexibilis (Hespell, 1977) was recently transferred into the genus Pseudomonas based on 16S rRNA similarity with P. pseudoalcaligenes (Shin et al., 2015). In our phylogenomic tree (Figure 1), this species branches separately from other Pseudomonas species and forms a distinct clade together with a newly described non-validly published species “Serpens gallinarum” (Gilroy et al., 2021) and another species P. tuomuerensis, which according to Shin et al. (2015) is a heterotypic synonym of P. flexibilis. This clade is identified as the taxon g_Pseudomonas_H in the GTDB taxonomy (Parks et al., 2018). A close and specific relationship of P. flexibilis (P. tuomuerensis) to “S. gallinarum” is independently supported by three CSIs identified in this study, which are exclusively shared by these species. One example of a CSI specific for this clade is shown in Figure 6B, where a one aa insertion in the protein GTP diphosphokinase is specifically shared by these three species. Detailed sequence information for this CSI and the two other CSIs specific for this clade is presented in Supplementary Figures S75–S77 and some of their characteristics are summarized in Table 4. Based on these results we are presenting an emended description of the genus Serpens with S. flexibilis as its type species.
CSIs specific for the Fluvialis clade
The Fluvialis clade consists of the species P. fluvialis and P. pharmacofabricae, which formed a strongly supported clade in different phylogenetic trees (Figure 1; Supplementary Figure S2). Our analyses have identified eight CSIs in different proteins that are uniquely shared by these two species. Figure 7A depicts an example of a CSI, consisting of a seven aa deletion within a conserved region of an ATP binding protein, which is exclusively shared by these two species. Detailed sequence information for this and the six other CSIs specific for the Fluvalis clade is presented in Supplementary Figures S78–S85 and a summary of some of their sequence characteristics is presented in Table 5. Based on the results presented here, we are proposing the transfer of species from this clade into Caenipseudomonas gen. nov.
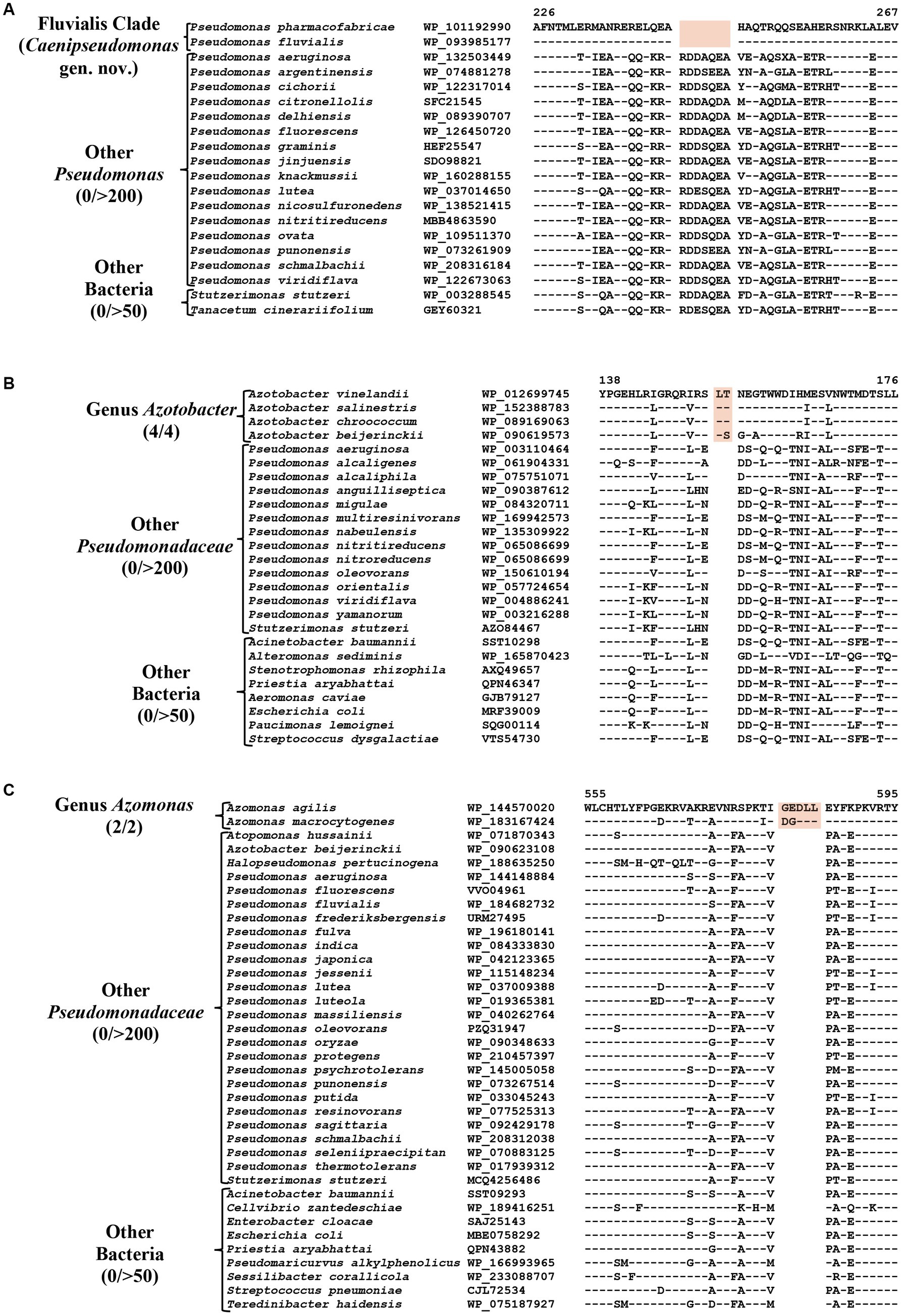
Figure 7. Partial sequence alignment of (A) ATP binding protein showing seven aa deletion within a conserved region (highlighted) that is uniquely shared by species from the Fluvialis clade. (B) A two aa insertion in a conserved region of the Alginate export family protein showing that is exclusively shared by species from the genus Azotobacter. (C) A five aa insertion in the protein Succinate dehydrogenase flavoprotein subunit which is specific for the species from genus Azomonas. Detailed sequence information for these CSIs and other CSIs specific for the Fluvialis clade and the Azotobacter and Azomonas genera are provided in Supplementary Figures S78–S100.
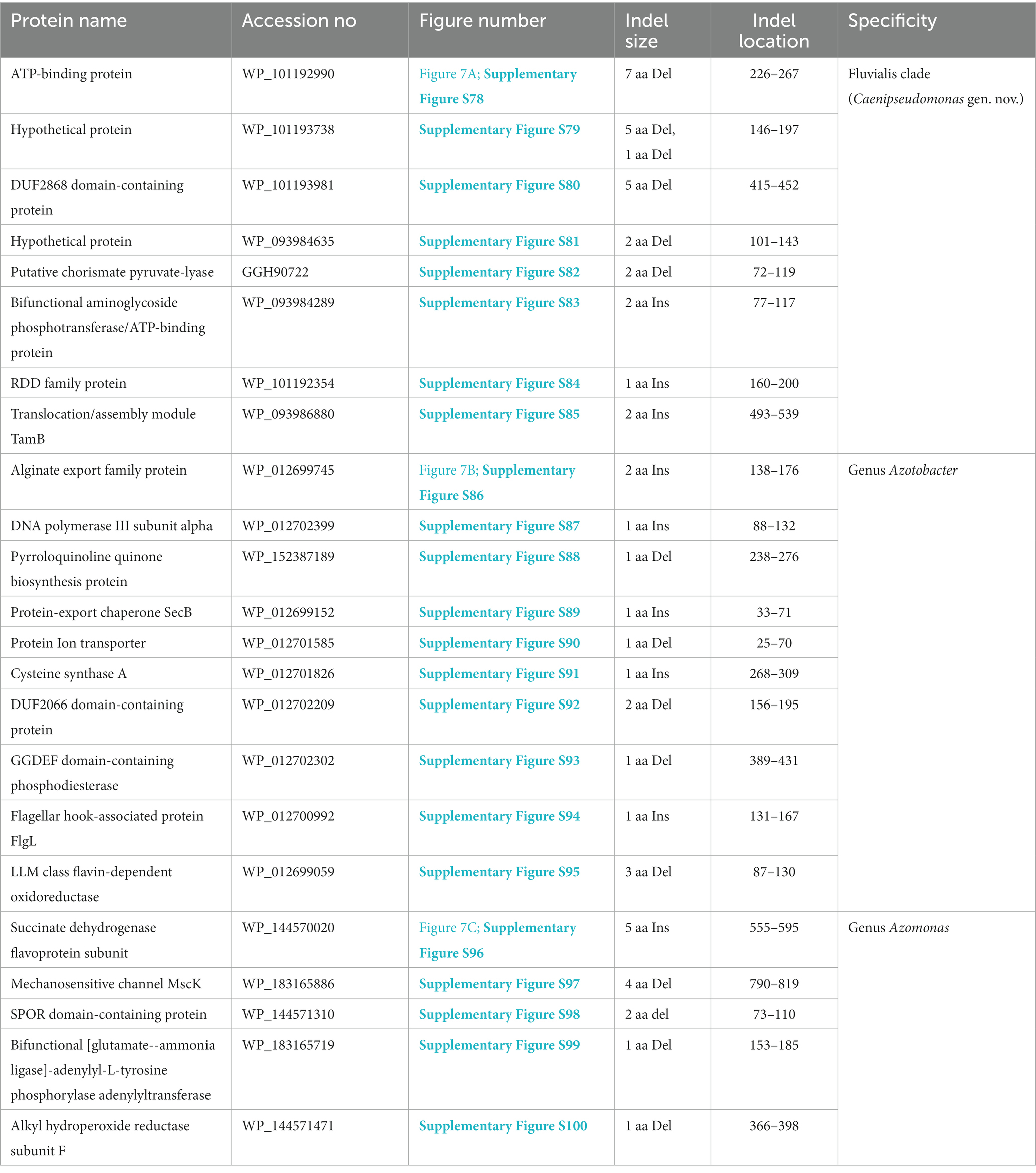
Table 5. Summary of CSIs specific for members of the Fluvialis clade, and the genera Azotobacter and Azomonas.
Identification of CSIs specific for the Azotobacter and Azomonas genera
The genus Azotobacter was described by Beijerinck (1901) and its members are known to branch in between Pseudomonas species (Young and Park, 2007; Özen and Ussery, 2012; Lalucat et al., 2022). Four Azotobacter species whose genome sequences were analyzed in this study (viz. A. beijerinckii, A. chroococcum, A. salinestris, and A. vinelandii), formed a distinct clade branching in the proximity of Stutzeri and Linyingensis clades (Figure 1; Supplementary Figure S2). Similar branching of Azotobacter species has been reported in earlier work (Jun et al., 2016; Hesse et al., 2018; Lalucat et al., 2022). Our analyses have identified 10 CSIs which are exclusively found in all four Azotobacter species providing reliable means for the demarcation of this clade. Partial sequence information for one of the CSIs specific for this genus, found in the alginate export family protein, is shown in Figure 7B. Detailed sequence information for this CSI and nine other CSIs specific for this genus is provided in Supplementary Figures S86–S95, and some of their sequence characteristics are listed in Table 5.
Azomonas is another genus whose members branch in between Pseudomonas species (Figure 1; Supplementary Figure S2; Young and Park, 2007; Kennedy and Rudnick, 2015; Rudra and Gupta, 2021; Lalucat et al., 2022). The two Azomonas species included in our analyses (viz., A. agilis and A. macrocytogenes) formed a distinct cluster in our phylogenomic trees (Figure 1; Supplementary Figure S2). The distinctness of this clade is also supported by five CSIs identified in this work, which are exclusively shared by these two species. Sequence information for one of these CSIs, containing a five aa insertion within the protein succinate dehydrogenase flavoprotein, is shown in Figure 7C. Detailed sequence information for this CSI and the other four CSIs specific for this genus are provided in the Supplementary Figures S96–S100, and a summary of some of their sequence characteristics is listed in Table 5.
Discussion
The genus Pseudomonas is one of the earliest known and largest prokaryotic genera encompassing a large assemblage of organisms exhibiting enormous genetic and metabolic diversity (Palleroni, 2005;Peix et al., 2009; Silby et al., 2011; Palleroni, 2015). The nomenclature type of this genus, P. aeruginosa, is an important human pathogen capable of causing a wide array of life-threatening acute and chronic diseases (Lund-Palau et al., 2016; Rossi et al., 2021). However, this genus also includes some animals and plant pathogenic species, as well as other economically and ecologically significant species (Desnoues et al., 2003; Silby et al., 2011; Xin et al., 2018). According to the LPSN (Parte et al., 2020), the genus Pseudomonas presently contains ≈310 species with validly published names. However, this number is increasing at a rapid pace (Girard et al., 2021), and in 2022 alone, more than 50 novel Pseudomonas species were listed in the LPSN server (Parte et al., 2020). As indicated in the introduction, and reviewed by others (Palleroni, 2010; Peix et al., 2018; Lalucat et al., 2022), evolutionary studies on the genus Pseudomonas have consistently shown that these species form multiple distinct clusters/clades, which are not specifically related to each other (Gomila et al., 2015; Hesse et al., 2018; Girard et al., 2021; Rudra and Gupta, 2021; Saati-Santamaría et al., 2021). Furthermore, it is generally recognized that of these species’ clades, circumscription of the genus Pseudomonas should be limited to the “Aeruginosa clade” harboring its type species, whereas species from the other observed clades should be reclassified into either novel or existing genera. In recent years, although several Pseudomonas species from deep branching clusters have been reclassified into novel genera (viz. Atopomonas, Chryseomonas, Halopseudomonas and Stutzerimonas) (Rudra and Gupta, 2021; Saati-Santamaría et al., 2021; Lalucat et al., 2022), the task of reliably reclassifying majority (>90%) of the Pseudomonas species into well-demarcated genera has proven challenging.
With the aim of reliably demarcating some of the observed Pseudomonas species clades, we have conducted here comprehensive phylogenomic and comparative analyses on the genome sequences of Pseudomonadaceae species. In our phylogenomic trees, Pseudomonas species formed multiple distinct clades (Figure 1; Supplementary Figure S2), which are similar to those reported in earlier studies (Gomila et al., 2015; Peix et al., 2018; Girard et al., 2021; Lalucat et al., 2022) excepting some differences resulting from the inclusion of several new species in our analysis. However, while similar species clusters are observed in different studies, based on their branching in phylogenetic trees (see Figure 1; Supplementary Figure S2), which is dynamic in nature and influenced by multiple variables including addition of new species (Gupta, 1998; Baldauf, 2003; Felsenstein, 2004), it is difficult to reliably demarcate the boundaries of different clades. Thus, a major focus of this study was to identify robust molecular markers, which independent of phylogenetic analyses, can confirm the existence of observed species clades and can provide reliable means for their demarcation.
Although genome sequence based indices such as average nucleotide identity (ANIb) and genome to genome DNA hybridization (GGDC) are now widely used for the delimitation of species level taxa (Goris et al., 2007; Kim et al., 2014; Yarza et al., 2014), such methods including AAI (Konstantinidis and Tiedje, 2007) or POCP (Qin et al., 2014) have shown limited usefulness for the delineation of genus level taxa (Parks et al., 2018; Gupta, 2019; Gupta and Kanter-Eivin, 2023). In the present work, while based on POCP and AAI values, some Pseudomonas species clades appear to be distinct (Table 1 and Supplementary Tables S2 and S3), for most of the observed clades these values generally show some overlap between the ingroup and outgroup species. Thus, based on these indices, it is difficult to reliably demarcate the boundaries of most of the clades. However, genome sequences are also enabling identification of highly specific molecular markers such as CSIs which are uniquely shared by different groups of organisms and provide dependable means for taxonomic and diagnostic studies (Gupta, 2014; Adeolu et al., 2016; Gupta, 2016; Gupta et al., 2020). As the CSIs in genes/proteins sequences result from rare genetic changes, their presence or absence in different species is generally not affected by most factors which can confound inferences from phylogenetic analyses (Baldauf and Palmer, 1993; Gupta, 1998; Rokas and Holland, 2000; Gupta, 2014, 2016). Furthermore, as the CSIs in different genes/proteins result from unrelated genetic changes, each of them provides independent evidence of a close and specific evolutionary relationship among a given group of species. In the present work, detailed analyses conducted on protein sequences from Pseudomonadaceae species, have identified 98 CSIs, which are specific for the species from 13 different Pseudomonadaceae species clades including the genera Azomonas and Azotobacter. Table 6 shows a summary of the CSIs that were identified for different Pseudomonadaceae clades along with the species that currently comprise these clades.
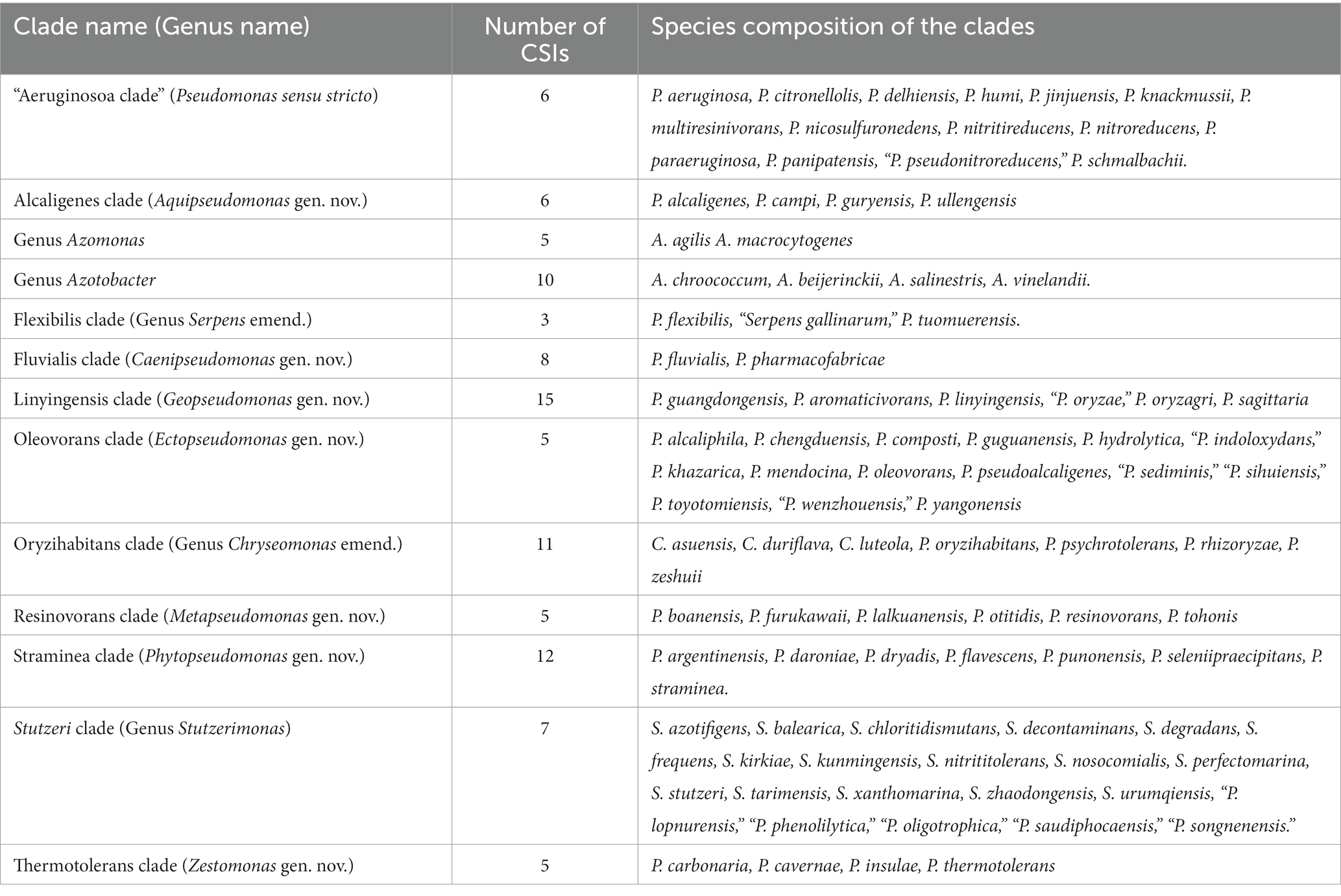
Table 6. Summary of different Pseudomonadaceae species clades reliably demarcated based on phylogenomic analyses and identified CSIs specific for these clades.
The results presented in Table 6 show that most of the Pseudomonas species clades, which are observed in our phylogenomic trees (Figure 1; Supplementary Figure S2), can now be robustly demarcated based on multiple identified CSIs, which are exclusively shared by the species from these clades. The genetic relatedness of the species from several of these clades is also supported by the results from AAI and POCP indices (Table 1). However, one clade for which CSIs were not identified is the Anguilliseptica clade. Species from this clade do not also form a well-resolved and strongly supported lineage in our phylogenetic trees (Figure 1; Supplementary Figure S2), and in earlier studies (Hesse et al., 2018; Busquets et al., 2021; Lalucat et al., 2022). In some phylogenetic trees [Supplementary Figure S2, unpublished results, and (Hesse et al., 2018)], one or more species from this clade (viz. P. cuatrocienegasensis) branch outside this clade. The results from AAI and POCP analyses (Table 1) also do not support the distinctness of this clade. All these observations indicate that the Anguilliseptica clade is not a trustworthy lineage and the cladistic relationships of species from this clade need to be further investigated. Of the CSIs identified by our analysis, six are uniquely shared by different species from the “Aeruginosa clade,” providing reliable molecular means for the demarcation/circumscription of this clade representing the genus Pseudomonas sensu stricto. Our analyses have also identified multiple CSIs reliably demarcating the species from Alcaligenes, Fluvialis, Linyingensis, Oleovorans, Resinovorans, Straminea, and Thermotolerans clades. Based on the strong and consistent evidence provided by phylogenomic analyses and identified molecular signatures supporting the distinctness of these clades, we are proposing that the species from the above noted clades should be reclassified into the following novel genera Aquipseudomonas gen. nov., Caenipseudomonas gen. nov., Geopseudomonas gen. nov., Ectopseudomonas gen. nov., Metapseudomonas gen. nov., Phytopseudomonas gen. nov., and Zestomonas gen. nov., respectively (Table 6). Our work has also identified 11 CSIs which are shared by all species from the Oryzihabitans clade providing robust means for the demarcation of species from this clade. Previously, only three species, which form a subclade of the Oryzihabitans clade, were reclassified into the genus Chryseomonas (Saati-Santamaría et al., 2021). Based on the results presented, we are proposing that the other species from this clade should also be transferred into the emended genus Chryseomonas. Species from the Flexibilis clade containing P. flexibilis are also transferred into the emended genus Serpens. Seven identified CSIs are commonly shared by all 13 species from the Stutzerimonas clade (Lalucat et al., 2022) providing robust molecular means for the demarcation of this genus. Lastly, multiple CSIs identified by our analyses are specific for the genera Azomonas and Azotobacter providing trustworthy means for the demarcation of these genera in molecular terms. As the identified CSIs provide important diagnostic characteristics of the above noted genera, we are also providing emended descriptions of these genera to include this information.
Although the present work represents a significant step toward clarifying the evolutionary relationships and classification scheme for Pseudomonas species, a vast majority of Pseudomonas species representing more than two thirds of the known species (see Supplementary Figure S1), are part of the Fluorescens superclade. As seen from Supplementary Figure S1, this large lineage is comprised of multiple clades and subclades (Palleroni, 2015; Hesse et al., 2018; Peix et al., 2018; Lalucat et al., 2020; Girard et al., 2021). To develop a reliable classification scheme for all Pseudomonas species, it will be necessary to reliably distinguish and demarcate different species clades within the Fluorescens superclade and reclassify them appropriately. In view of this consideration, despite our reliable demarcation of the genus Pseudomonas sensu stricto, an emended description of this genus is not proposed, until most other Pseudomonas species are reliably classified.
All newly proposed genera and other studied genera/clades in this work have been circumscribed based on their harboring multiple uniquely shared CSIs. One notable characteristic of the CSIs, which is of much importance for classification purposes, is that these markers exhibit high degree of predictive ability to be found in other (uncharacterized or unidentified) members of a given group/taxon (Bhandari et al., 2013; Gupta, 2014, 2016; Dobritsa and Samadpour, 2019; Patel and Gupta, 2020; Montecillo and Bae, 2022). Thus, the CSIs specific for the genus Halopseudomonas identified in our earlier work (Rudra and Gupta, 2021) are also present in all newly described species from this genus (Supplementary Figure S2). Similarly, the CSIs specific for the genus Atopomonas were also present in a newly described species from this genus (Li et al., 2023). Due to the demonstrated predictive abilities of the CSIs to be found in other members of specific taxa, we have recently developed a web-based tool/server,3 which can predict taxonomic affiliation based on the presence of known taxon-specific CSIs in a genome sequence (Gupta and Kanter-Eivin, 2023). Therefore, upon the addition of information for these newly identified CSIs to the AppIndels server, it should greatly facilitate the classification of both cultured and uncultured isolates related to the described taxa (Gupta and Patel, 2019). The CSIs specific for different taxa also provide useful means for the development of sensitive and specific diagnostic tests using in silico and experimental methods (Ahmod et al., 2011; Wong et al., 2014). Lastly, the earlier work on CSIs show that these molecular characteristics are functionally important for the group of organisms for which they are specific (Singh and Gupta, 2009; Khadka et al., 2020). Hence, genetic, and biochemical studies on the identified CSIs could lead to the discovery of novel biochemical and/or other characteristics of different groups of organisms.
The descriptions of different novel genera proposed and other emended genera are given below. The new name combinations for different species resulting from the proposed taxonomic changes are provided in Tables 7, 8. The names for the newly proposed genera are generally based on some characteristics of the proposed group of species.
Description of the genus Aquipseudomonas gen. nov.
Aquipseudomonas (A.qui.pseu.do.mo’nas. L. fem. n. aqua, water; N.L. fem. n. Pseudomonas, a bacterial genus; N.L. fem. n. Aquipseudomonas, Pseudomonas-like species isolated from water).
Cells are Gram-stain negative, motile and rod shaped. The species are aerobic in respiration and have been isolated from soil and swimming pool water. Optimum temperature for growth ranges from 30 – 37°C with <2% (w/v) NaCl and pH range from 4–10. Genome sizes for the species vary from 4.3 Mb to 4.6 Mb and the GC content ranges from 63.3 to 65.5%. Of the species from this genus, the type species A. alcaligenes can degrade polycyclic aromatic hydrocarbons and has been proven useful for bioremediation of oil pollution, pesticide substances, and certain chemical substances. Species from this genus form a strongly supported clade in phylogenomic tree based on large datasets of concatenated proteins. Additionally, species from this genus can be reliably distinguished from all other Pseudomonadaceae genera based on six CSIs (Table 2) which are exclusively found in the species from this genus. New name combinations for the species that are part of this genus are provided in Table 7.
The type species of this genus is Aquipseudomonas alcaligenes.
Description of the genus Caenipseudomonas gen. nov.
Caenipseudomonas (Cae.ni.pseu.do.mo’nas. L. neut. n. caenum, mud; N.L. fem. n. Pseudomonas, a bacterial genus; N.L. fem. n. Caenipseudomonas, Pseudomonas-like organism(s) isolated from river sediments).
Cells are strictly aerobic, Gram-stain-negative, non-fluorescent and occur mostly as short rods. Cells are motile and contain a single polar flagellum. Chemoorganotrophic growth. Species have been isolated from river sediment, and wastewater sample from a pharmaceutical company. Growth occurs in the temperature range from 4-22°C with optimum growth occurring between 25-35o C at pH between 7–8 in presence of 0–2% (w/v) NaCl concentration. Genome size range is from 3.3–3.4 Mb and the GC content is 62.6%. Species from this genus form a distinct lineage in phylogenomic trees based on large datasets of proteins, as well as in trees based on rpoD gene, or concatenated partial sequences for the 16S rDNA, gyrB, rpoB, and rpoD genes. In addition, species from this genus can be reliably distinguished based on eight exclusively shared CSIs listed in Table 5. The new name combinations for species from this genus are provided in Table 7.
The type species is Caenipseudomonas fluvialis.
Description of the genus Ectopseudomonas gen. nov.
Ectopseudomonas (Ec.to.pseu.do.mo’nas. Gr. prep. Ecto, outside; N.L. fem. n. Pseudomonas, a bacterial genus; N.L. fem. n. Ectopseudomonas, a genus outside of Pseudomonas).
Cells are Gram-stain negative, motile and rod shaped. Excepting E. chengduensis, all other species from this genus are motile due to the presence of a polar flagellum. Species have been isolated from diverse sources including sea water, soil, hot spring, compost, and lake sediments, etc. Chemoorganotrophic life cycle. Most species grow aerobically; however, some are indicated to be facultatively anerobic. Colonies are generally brownish yellow. Growth can occur from 4o-42°C with optimum growth temperature between 30–37°C, with or without NaCl, in the pH range from 3.0–10.5 (optimum between pH 6–8). Genome sizes for known species vary from 4.5 Mb to 5.6 Mb and the GC content ranges from 62.2 to 65.0%. Of the species from this genus, E. mendocina can degrade toluene and it is indicated to cause opportunistic nosocomial infections. Members of this genus form a monophyletic clade in phylogenetic trees based on concatenated sequences of several large datasets of core genome proteins. Additionally, species from this genus also generally cluster together in phylogenetic trees based on rpoD gene, or concatenated partial sequences for the 16S rDNA, gyrB, rpoB, and rpoD genes. In addition of their distinct branching in phylogenetic trees, members of this genus can be reliably distinguished from other Pseudomonadaceae species based on five CSIs (Table 2) which in most cases are exclusively shared by the members of this genus. The new name combinations for species that are part of this genus are provided in Table 7.
The type species of this genus is Ectopseudomonas oleovorans.
Description of the genus Geopseudomonas gen. nov.
Geopseudomonas (Ge.o.pseu.do.mo’nas. Gr. fem. n. gê, the Earth; N.L. fem. n. Pseudomonas, a bacterial genus; N.L. fem. n. Geopseudomonas, Pseudomonas like organisms isolated from soil).
Strictly aerobic to facultatively anaerobic, rod-shaped bacteria. Motile due to the presence of one or more polar or peritrichous flagella. Chemoorganotrophs, with cells exhibiting Gram-stain negative staining response. Cells generally do not produce fluorescent pigments. Members have been isolated from diverse sources including paddy soil, electroactive biofilm, herbicide applied wheat field and oil contaminated soil. Optimum growth occurs in the range of 30–37°C, between pH 7–8, in medium containing 1–2% NaCl (w/v). Genome lengths of the species vary from 3.2 to 4.7 Mb, and GC contents vary from 66.4 to 68.3%. Members of this genus form a monophyletic clade in phylogenetic tree based on concatenated sequences for several large datasets of proteins. Species from this genus also cluster together in phylogenetic trees based on rpoD gene, or concatenated partial sequences for the 16S rDNA, gyrB, ropB, and rpoD genes. In addition, the members of this genus can be reliably distinguished from all other Pseudomonadaceae genera by the 15 CSIs described in Table 3, which in most cases are exclusively shared by either all or most species from this genus. The new name combinations for species which are part of this genus are provided in Table 7.
The type species is Geopseudomonas sagittaria.
Description of the genus Metapseudomonas gen. nov.
Metapseudomonas (Me.ta.pseu.do.mo’nas. Gr. adv. Meta, besides; N.L. fem. n. Pseudomonas, a bacterial genus; N.L. fem. n. Metapseudomonas, a genus beside Pseudomonas).
Species of this genus are Gram-negative, motile, aerobic and rod shaped. Chemoorganotrophic growth, cells do not produce fluorescent pigments. Members have been isolated from different sources such as clinical samples, soil or oil of wood mills and biphenyl contaminated soil. Optimum growth temperature is in the range of 30-37°C. Genome sizes for known species are in the range of 6.1 Mb to 6.8 Mb and GC content varies from 64.2 to 66.80%. Species from this genus form a strongly supported clade in phylogenomic trees based on large datasets of proteins. In addition, most of the species from this genus also cluster together in phylogenetic trees based on rpoD gene, or concatenated partial sequences for the 16S rDNA, gyrB, ropB, and rpoD genes. Importantly, the species from this genus can also be reliably distinguished from all other Pseudomonadaceae genera by the shared presence of five CSIs listed in Table 4. The new name combinations for the species of this genus are provided in Table 8.
The type species of this genus is Metapseudomonas resinovorans.
Description of the genus Phytopseudomonas gen. nov.
Phytopseudomonas (Phy.to.pseu.do.mo’nas. Gr. neut. n. phyton, plant; N.L. fem. n. Pseudomonas, a bacterial genus; N.L. fem. n. Phytopseudomonas, Pseudomonas-like species isolated from plants).
Cells are Gram-stain negative, motile due to the presence of a polar flagellum, aerobic, and rod shaped. Chemoorganotrophs. Most species have been isolated from different plant sources such as Quercus robur stem tissues, straw grass, rice paddy, walnut blight cankers etc. All species produce a diffusible fluorescent pigment. Optimum temperature for growth is between 25-30°C, with <4% (w/v) or without NaCl in the pH range from 6–8. Genome sizes for the species vary from 4.5 Mb to 5.9 Mb and the GC content ranges from 61.5 to 65.0%. Members of this genus form a monophyletic clade in phylogenetic trees based on concatenated sequences of several large datasets of core genome proteins. Additionally, species from this genus also generally cluster together in phylogenetic trees based on rpoD gene, or concatenated partial sequences for the 16S rDNA, gyrB, ropB, and rpoD genes. Additionally, members of this genus can be reliably distinguished from other Pseudomonadaceae genera based on the presence of 12 CSIs summarized in Table 3. which in most cases are exclusively present in the species from this genus. The new name combinations for species that are part of this genus are provided in Table 8.
The type species of this genus is Phytopseudomonas straminea.
Description of the genus Zestomonas gen. nov.
Zestomonas (Zes.to.mo’nas. Gr. masc. Adj. zestos, hot, boiling; L. fem. n. monas, a unit, monad; N.L. fem. n. Zestomonas, a monad that can grow at high temperature).
Aerobic, motile rods exhibiting Gram-negative staining response. Chemoorganotrophs. Species have been cultivated from different sources such as cooking water, forest soil, charcoal, and cave sediment. Temperature range for growth for species from this genus differs considerably. While the optimum growth of the type species Zestomonas thermotolerans occurs at 47°C (growth range 25–56°C), other species from this genus grow optimally at 28–30°C. Genome length ranges from 3.8 to 5.5 Mb and the GC content varies from 64.5 to 66.8%. Members of this genus form a monophyletic clade in phylogenomic tree based on concatenated sequences for several large datasets of proteins. In addition, members of this genus can be reliably distinguished from other Pseudomonadaceae genera by their uniquely sharing five CSIs listed in Table 4. New name combinations for the species from this genus are provided in Table 8.
The type species is Zestomonas thermotolerans.
Emended description of the genus Azomonas Winogradsky, 1938 (Approved lists 1980)
Azomonas (A.zo.mo.nas. N.L. pref. Azo-, pertaining to nitrogen; L. fem. n. monas, a unit, monad; N.L. fem. n. Azomonas, nitrogen monad).
Description of this genus is in large part based on that provided by Kennedy and Rudnick (2015) in the Bergey’s Manual of Systematics of Archaea and Bacteria. Cells are Gram-stain variable or sometimes Gram-stain negative depending on the culture age, aerobic, ellipsoidal to rod shaped. Species are motile with peritrichous or lophotrichous polar flagella. Cells may occur singly, in pairs, or in clumps. All species fix atmospheric nitrogen under aerobic conditions. Alternative nitrogenases containing vanadium (nitrogenase-2) or iron (nitrogenase-3) may only be synthesized in Mo-deficient media. Cultures can grow both aerobically and microaerobically. Chemoorganotrophic. Sugars, alcohols, and organic acids are used as carbon sources. Ammonium salts and sometimes nitrate (A. insignis only) are used as nitrogen sources; amino acids are not used. Water-soluble and fluorescent pigments are produced by nearly all strains. Species are catalase positive. The optimum pH for nitrogen fixation is close to neutrality, but certain strains can also fix nitrogen at a pH of 4.6–4.8. Species isolated from water or soil. The G + C content of DNA from known species varies from 52.0–58.6% and their genome size ranges from 3.3 to 4.1 MB. Species belonging to this genus form a distinct clade in phylogenomic trees based on concatenated sequences of large number of proteins and in the tree based on 16S rRNA gene sequences. In addition, members of this genus can be reliably distinguished from Azotobacter as well as all other Pseudomonadaceae genera based on their exclusive sharing five CSIs described in this work (Table 5).
Type species is Azomonas agilis (Beijerinck, 1901) Winogradsky, 1938 (Approved Lists 1980).
Emended description of the genus Azotobacter Beijerinck, 1901 (Approved lists 1980)
Azotobacter (A.zo.to.bac.ter. N.L. neut. n. azotum, nitrogen; N.L. masc. n. bacter, a rod; N.L. masc. n. Azotobacter, a nitrogen rod).
Description of this genus is in large part based on that provided by Kennedy et al. (2015) in the 2015 Bergey’s Manual of Systematics of Archaea and Bacteria. Cells range from straight rods with rounded ends to more ellipsoidal or coccoid. Motile with peritrichous flagella or nonmotile. Aerobic, having a strictly respiratory type of metabolism with oxygen as the terminal electron acceptor. Nitrogen is fixed under microaerobic conditions (2% oxygen), under full aerobiosis, or after adaptation in hyperbaric oxygen. N2 fixation uses Mo-, V-, or Fe-containing nitrogenase enzymes, depending on the environmental metal supply. Water-soluble and water-insoluble pigments are produced by some strains. Growth is heterotrophic; sugars, alcohols, and salts of organic acids are used as carbon sources. Ammonium salts, nitrate, and urea are used as sources of fixed nitrogen. The pH range for growth is from 4.8 to 8.5, with optimum pH for diazotrophic growth between 7.0–7.5. Most isolates are from soil, but a few are from water. The GC content of the DNA varies from 65.5–67.5%. Genome size ranges from 4.9–5.4 Mb. Species belonging to this genus group together in phylogenetic trees based on 16S rRNA gene sequences, and in phylogenomic trees based on concatenated sequences of large number of proteins. In addition, members of this genus can be reliably distinguished from all other Pseudomonadaceae genera by 10 uniquely shared CSIs listed in Table 5.
Type species is Azotobacter chroococcum Beijerinck, 1901 (Approved Lists 1980).
Emended description of the genus Chryseomonas Holmes et al., 1986
Chryseomonas (Chry.se.o.mo’nas. Gr. masc. Adj. chryseos, golden; L. fem. n. monas, a unit, monad; N.L. fem. n. Chryseomonas, a yellow unit).
The description of this genus is partially based on that given by Holmes et al. (1986) for the type species (C. polytricha) of this genus. The cells are rod-shaped, Gram-negative, aerobic, and exhibit chemoorganotrophic growth. Except for C. duriflava (and its synonym C. zeshuii), which do not exhibit motility, cells from the other species are motile by either a single or several polar or trichous flagella. Known species have been isolated from diverse sources including rice seeds and paddy, desert soil, herbicide-contaminated soil, grass rhizosphere, clinical specimens, and medical clinic for small animals. C. oryzihabitans has been reported as pathogenic to plants and animals. Some species (C. luteola) can reduce nitrate. Growth can occur in the temperature range from 4–42°C with optimum growth occurring between 30 to 37°C at pH 7.0 (pH range 6–8) in medium supplemented with 1–2% (w/v) NaCl. The cells are catalase positive but oxidase negative. The GC content of species varies from 53.6 to 66.2% and their genome lengths range from 4.3 to 5.4 Mb. Species from this genus form a distinct clade in the phylogenomic trees based on a large number of proteins. Additionally, these species also cluster together in phylogenetic trees based on rpoD gene, or concatenated partial sequences for the 16S rDNA, gyrB, ropB, and rpoD genes. Apart from their grouping together in phylogenetic trees, species from this genus can be reliably distinguished from all other Pseudomonadaceae genera by their 11 CSIs listed in Table 4, which in most cases are exclusively present in the species from this genus. New name combinations for four Pseudomonas species, which are transferred to this genus, are provided in Table 8.
Type species of this genus is Chryseomonas polytrichia (Holmes et al., 1986).
Emended description of the genus Serpens Hespell, 1977 (Approved lists 1980)
Description of this genus is modified from that given by Hespell (1977). Gram-negative, aerobic, rod-shaped, non-spore forming, bacterial cells. Cells from the type species, S. flexibilis, are very flexible, and motile due to containing a flagellum, and exhibit serpentine-like movement in agar gels. Metabolism is respiratory, and molecular oxygen serves as the terminal electron acceptor. S. flexibilis mainly uses lactate as the energy and carbon source. Catalase and oxidase are produced. Temperature range for optimal growth is from 28 to 37°C. The G + C content of DNA ranges from 61.0–65.8 mol% and genome size varies from 3.8–3.9 Mb. Species from this genus form a monophyletic clade in the phylogenetic tree based on large dataset of proteins. The type species also forms a distinct lineage in phylogenetic trees based on rpoD gene, or concatenated partial sequences for the 16S rDNA, gyrB, ropB, and rpoD genes. Additionally, species from this genus can be reliably distinguished from other Pseudomonadaceae genera by the presence of three exclusively shared CSIs (Table 4). New name combinations for the two species which are part of this genus are provided in Table 8.
Type species of this genus is Serpens flexibilis Hespell, 1977 (Approved lists).
Emended description of the genus Stutzerimonas Lalucat et al., 2022
Stutzerimonas (Stut.ze.ri.mo’nas. L. fem. n. monas, a unit, monad; N.L. fem. n. Stutzerimonas, monad of Stutzer, named in honor of Albert Stutzer, who in 1895 described the bacterium today known).
The description of this genus, especially in terms of its morphological, chemotaxonomic and growth characteristics, remains the same as provided by Lalucat et al. (2022). In addition to the genomic characteristics described by Lalucat et al. (2022), members of this genus can be reliably distinguished from other Pseudomonadaceae genera by seven novel CSIs identified in this study (listed in Table 3), which in most cases are exclusively found in the species from this genus. New name combination for P. marianensis (Table 8) is based on its branching in the 16S rRNA gene tree (Yang et al., 2022).
The type species is Stutzerimonas stutzeri (Lehmann and Neumann 1896) Lalucat et al. 2022.
Data availability statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/Supplementary material.
Author contributions
BR: Data curation, Formal analysis, Investigation, Methodology, Writing – original draft, Validation. RG: Conceptualization, Funding acquisition, Project administration, Resources, Software, Supervision, Validation, Writing – review & editing.
Funding
The author(s) declare financial support was received for the research, authorship, and/or publication of this article. This work was supported by a research grant (RGPIN-2019-06397) from the Natural Science and Engineering Research Council of Canada.
Acknowledgments
We thank Dr. Aharon Oren for checking the etymology of the proposed genera.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2023.1273665/full#supplementary-material
Footnotes
References
Adeolu, M., Alnajar, S., Naushad, S., and Gupta, S. (2016). Genome-based phylogeny and taxonomy of the 'Enterobacteriales': proposal for Enterobacterales ord. nov. divided into the families Enterobacteriaceae, Erwiniaceae fam. nov., Pectobacteriaceae fam. nov., Yersiniaceae fam. nov., Hafniaceae fam. Nov., Morganellaceae fam. nov., and Budviciaceae fam. nov. Int. J. Syst. Evol. Microbiol. 66, 5575–5599. doi: 10.1099/ijsem.0.001485
Ahmod, N. Z., Gupta, R. S., and Shah, H. N. (2011). Identification of a Bacillus anthracis specific indel in the yeaC gene and development of a rapid pyrosequencing assay for distinguishing B. anthracis from the B. cereus group. J. Microbiol. Methods 87, 278–285. doi: 10.1016/j.mimet.2011.08.015
Anzai, Y., Kim, H., Park, J. Y., Wakabayashi, H., and Oyaizu, H. (2000). Phylogenetic affiliation of the pseudomonads based on 16S rRNA sequence. Int. J. Syst. Evol. Microbiol. 50, 1563–1589. doi: 10.1099/00207713-50-4-1563
Anzai, Y., Kudo, Y., and Oyaizu, H. (1997). The phylogeny of the genera Chryseomonas, Flavimonas, and Pseudomonas supports synonymy of these three genera. Int. J. Syst. Bacteriol. 47, 249–251. doi: 10.1099/00207713-47-2-249
Azhar, E. I., Papadioti, A., Bibi, F., Ashshi, A. M., Raoult, D., and Angelakis, E. (2017). 'Pseudomonas saudimassiliensis' sp. nov. a new bacterial species isolated from air samples in the urban environment of Makkah, Saudi Arabia. New Microbes New Infect. 16, 43–44. doi: 10.1016/j.nmni.2016.12.021
Baldauf, S. L. (2003). Phylogeny for the faint of heart: a tutorial. Trends Genet. 19, 345–351. doi: 10.1016/S0168-9525(03)00112-4
Baldauf, S. L., and Palmer, J. D. (1993). Animals and fungi are each other's closest relatives: congruent evidence from multiple proteins. Proc. Natl. Acad. Sci. U. S. A. 90, 11558–11562. doi: 10.1073/pnas.90.24.11558
Banerjee, S., Bedics, A., Tóth, E., Kriszt, B., Soares, A. R., Bóka, K., et al. (2022). Isolation of Pseudomonas aromaticivorans sp. nov from a hydrocarbon-contaminated groundwater capable of degrading benzene-, toluene-, m- and p-xylene under microaerobic conditions. Front. Microbiol. 13:929128. doi: 10.3389/fmicb.2022.929128
Behera, P., Mahapatra, M., Seuylemezian, A., Vaishampayan, P., Ramana, V. V., Joseph, N., et al. (2018). Taxonomic description and draft genome of Pseudomonas sediminis sp. nov., isolated from the rhizospheric sediment of Phragmites karka. J. Microbiol. 56, 458–466. doi: 10.1007/s12275-018-7549-x
Beijerinck, M. (1901). Über oligonitrophile Mikroben. Zentralbl. Bakterol. Parasitenkd. Infektionskr. Hyg. Abt. II 7, 561–582.
Bhandari, V., Ahmod, N. Z., Shah, H. N., and Gupta, R. S. (2013). Molecular signatures for Bacillus species: demarcation of the Bacillus subtilis and Bacillus cereus clades in molecular terms and proposal to limit the placement of new species into the genus Bacillus. Int. J. Syst. Evol. Microbiol. 63, 2712–2726. doi: 10.1099/ijs.0.048488-0
Bueno-Gonzalez, V., Brady, C., Denman, S., Plummer, S., Allainguillaume, J., and Arnold, D. (2019). Pseudomonas daroniae sp. nov. and Pseudomonas dryadis sp. nov., isolated from pedunculate oak affected by acute oak decline in the UK. Int. J. Syst. Evol. Microbiol. 69, 3368–3376. doi: 10.1099/ijsem.0.003615
Busquets, A., Mulet, M., Gomila, M., and García-Valdés, E. (2021). Pseudomonas lalucatii sp. nov. isolated from Vallgornera, a karstic cave in Mallorca, Western Mediterranean. Syst. Appl. Microbiol. 44:126205. doi: 10.1016/j.syapm.2021.126205
Capella-Gutiérrez, S., Silla-Martínez, J. M., and Gabaldón, T. (2009). trimAl: a tool for automated alignment trimming in large-scale phylogenetic analyses. Bioinformatics 25, 1972–1973. doi: 10.1093/bioinformatics/btp348
Clark, L. L., Dajcs, J. J., Mclean, C. H., Bartell, J. G., and Stroman, D. W. (2006). Pseudomonas otitidis sp. nov., isolated from patients with otic infections. Int. J. Syst. Evol. Microbiol. 56, 709–714. doi: 10.1099/ijs.0.63753-0
Delaporte, B., Raynaud, M., and Daste, P. (1961). Une bactérie du sol capable d'utiliser, comme source de carbone, la fraction fixe de certaines oléorésines, Pseudomonas resinovorans n. sp. Compte Rendu des Séances de l'Académie des Sci. 252, 1073–1075.
Desnoues, N., Lin, M., Guo, X., Ma, L., Carreño-Lopez, R., and Elmerich, C. (2003). Nitrogen fixation genetics and regulation in a Pseudomonas stutzeri strain associated with rice. Microbiology (Reading) 149, 2251–2262. doi: 10.1099/mic.0.26270-0
Dobritsa, A. P., and Samadpour, M. (2019). Reclassification of Burkholderia insecticola as Caballeronia insecticola comb. nov. and reliability of conserved signature indels as molecular synapomorphies. Int. J. Syst. Evol. Microbiol. 69, 2057–2063. doi: 10.1099/ijsem.0.003431
Eddy, S. R. (2011). Accelerated profile HMM searches. PLoS Comput. Biol. 7:e1002195. doi: 10.1371/journal.pcbi.1002195
Feng, Z., Zhang, J., Huang, X., Zhang, J., Chen, M., and Li, S. (2012). Pseudomonas zeshuii sp. nov., isolated from herbicide-contaminated soil. Int. J. Syst. Evol. Microbiol. 62, 2608–2612. doi: 10.1099/ijs.0.037796-0
Fu, L., Niu, B., Zhu, Z., Wu, S., and Li, W. (2012). CD-HIT: accelerated for clustering the next-generation sequencing data. Bioinformatics 28, 3150–3152. doi: 10.1093/bioinformatics/bts565
García-Valdés, E., and Lalucat, J. (2016). “Pseudomonas: molecular phylogeny and current taxonomy” in Pseudomonas: Molecular and applied biology. ed. R. S. Kahlon (New York: Springer), 1–23.
Gibello, A., Vela, A. I., Martín, M., Mengs, G., Alonso, P. Z., Garbi, C., et al. (2011). Pseudomonas composti sp. nov., isolated from compost samples. Int. J. Syst. Evol. Microbiol. 61, 2962–2966. doi: 10.1099/ijs.0.027086-0
Gilroy, R., Ravi, A., Getino, M., Pursley, I., Horton, D. L., Alikhan, N. F., et al. (2021). Extensive microbial diversity within the chicken gut microbiome revealed by metagenomics and culture. PeerJ 9:e10941. doi: 10.7717/peerj.10941
Girard, L., Lood, C., Höfte, M., Vandamme, P., Rokni-Zadeh, H., Van Noort, V., et al. (2021). The ever-expanding Pseudomonas genus: description of 43 new species and partition of the Pseudomonas putida group. Microorganisms 9:1766. doi: 10.3390/microorganisms9081766
Gomila, M., Peña, A., Mulet, M., Lalucat, J., and García-Valdés, E. (2015). Phylogenomics and systematics in Pseudomonas. Front. Microbiol. 6:214. doi: 10.3389/fmicb.2015.00214
Goris, J., Konstantinidis, K. T., Klappenbach, J. A., Coenye, T., Vandamme, P., and Tiedje, J. M. (2007). DNA-DNA hybridization values and their relationship to whole-genome sequence similarities. Int. J. Syst. Evol. Microbiol. 57, 81–91. doi: 10.1099/ijs.0.64483-0
Gupta, R. S. (1998). Protein phylogenies and signature sequences: a reappraisal of evolutionary relationships among archaebacteria, eubacteria, and eukaryotes. Microbiol. Mol. Biol. Rev. 62, 1435–1491. doi: 10.1128/MMBR.62.4.1435-1491.1998
Gupta, R. S. (2014). Identification of conserved indels that are useful for classification and evolutionary studies. Methods Microbiol. 41, 153–182. doi: 10.1016/bs.mim.2014.05.003
Gupta, R. S. (2016). Impact of genomics on the understanding of microbial evolution and classification: the importance of Darwin's views on classification. FEMS Microbiol. Rev. 40, 520–553. doi: 10.1093/femsre/fuw011
Gupta, R. S. (2019). Distinction between Borrelia and Borreliella is more robustly supported by molecular and phenotypic characteristics than all other neighbouring prokaryotic genera: response to Margos' et al. "the genus Borrelia reloaded" (PLoS One 13: e0208432). PLoS One 14:e0221397. doi: 10.1371/journal.pone.0221397
Gupta, R. S., Chander, P., and George, S. (2013). Phylogenetic framework and molecular signatures for the class Chloroflexi and its different clades; proposal for division of the class Chloroflexia class. nov. [corrected] into the suborder Chloroflexineae subord. nov., consisting of the emended family Oscillochloridaceae and the family Chloroflexaceae fam. nov., and the suborder Roseiflexineae subord. nov., containing the family Roseiflexaceae fam. nov. Antonie Van Leeuwenhoek 103, 99–119. doi: 10.1007/s10482-012-9790-3
Gupta, R. S., and Kanter-Eivin, D. A. (2023). AppIndels.com server: a web based tool for the identification of known taxon-specific conserved signature indels in genome sequences: validation of its usefulness by predicting the taxonomic affiliation of >700 unclassified strains of Bacillus species. Int. J. Syst. Evol. Microbiol. 73:005844. doi: 10.1099/ijsem.0.005844
Gupta, R. S., and Patel, S. (2019). Robust demarcation of the family Caryophanaceae (Planococcaceae) and its different genera including three novel genera based on phylogenomics and highly specific molecular signatures. Front. Microbiol. 10:2821. doi: 10.3389/fmicb.2019.02821
Gupta, R. S., Patel, S., Saini, N., and Chen, S. (2020). Robust demarcation of 17 distinct Bacillus species clades, proposed as novel Bacillaceae genera, by phylogenomics and comparative genomic analyses: description of Robertmurraya kyonggiensis sp. nov. and proposal for an emended genus Bacillus limiting it only to the members of the subtilis and Cereus clades of species. Int. J. Syst. Evol. Microbiol. 70, 5753–5798. doi: 10.1099/ijsem.0.004475
Hauser, E., Kämpfer, P., and Busse, H. J. (2004). Pseudomonas psychrotolerans sp. nov. Int. J. Syst. Evol. Microbiol. 54, 1633–1637. doi: 10.1099/ijs.0.03024-0
He, W.-H., Wang, Y.-N., Du, X., Zhou, Y., Jia, B., Bian, J., et al. (2012). Pseudomonas linyingensis sp. nov.: a novel bacterium isolated from wheat soil subjected to long-term herbicides application. Curr. Microbiol. 65, 595–600. doi: 10.1007/s00284-012-0187-3
Hespell, R. B. (1977). Serpens flexibilis gen. nov., sp. nov., an unusually flexible, lactate-oxidizing bacterium. Int. J. Syst. Evol. Microbiol. 27, 371–381. doi: 10.1099/00207713-27-4-371
Hesse, C., Schulz, F., Bull, C. T., Shaffer, B. T., Yan, Q., Shapiro, N., et al. (2018). Genome-based evolutionary history of Pseudomonas spp. Environ. Microbiol. 20, 2142–2159. doi: 10.1111/1462-2920.14130
Hildebrand, D., Palleroni, N., Hendson, M., Toth, J., and Johnson, J. (1994). Pseudomonas flavescens sp. nov., isolated from walnut blight cankers. Int. J. Syst. Evol. Microbiol. 44, 410–415. doi: 10.1099/00207713-44-3-410
Hirota, K., Yamahira, K., Nakajima, K., Nodasaka, Y., Okuyama, H., and Yumoto, I. (2011). Pseudomonas toyotomiensis sp. nov., a psychrotolerant facultative alkaliphile that utilizes hydrocarbons. Int. J. Syst. Evol. Microbiol. 61, 1842–1848. doi: 10.1099/ijs.0.024612-0
Holmes, B., Steigerwalt, A., Weaver, R., and Brenner, D. J. (1986). Chryseomonas polytricha gen. nov., sp. nov., a Pseudomonas-like organism from human clinical specimens and formerly known as group Ve-1. Int. J. Syst. Evol. Microbiol. 36, 161–165. doi: 10.1099/00207713-36-2-161
Holmes, B., Steigerwalt, A., Weaver, R., and Brenner, D. J. (1987). Chryseomonas luteola comb. nov. and Flavimonas oryzihabitans gen. nov., comb. nov., Pseudomonas-like species from human clinical specimens and formerly known, respectively, as groups Ve-1 and Ve-2. Int. J. Syst. Evol. Microbiol. 37, 245–250. doi: 10.1099/00207713-37-3-245
Hunter, W. J., and Manter, D. K. (2011). Pseudomonas seleniipraecipitatus sp. nov.: a selenite reducing gamma-proteobacteria isolated from soil. Curr. Microbiol. 62, 565–569. doi: 10.1007/s00284-010-9745-8
Huq, M. A., Lee, S. Y., Ma, J., Rahman, M. M., Rahman, M. S., Park, J. H., et al. (2022). Pseudomonas oryzagri sp. nov., isolated from a rice field soil. Int. J. Syst. Evol. Microbiol. 72:005534. doi: 10.1099/ijsem.0.005534
Iizuka, H., and Komagata, K. (1963). New species of Pseudomonas belonging to fluorescent group. (studies on the microorganisms of cereal grains. Part V). Nippon Nogeikagaku Kaishi 37, 137–141. doi: 10.1271/nogeikagaku1924.37.137
Jeanmougin, F., Thompson, J. D., Gouy, M., Higgins, D. G., and Gibson, T. J. (1998). Multiple sequence alignment with Clustal X. Trends Biochem. Sci. 23, 403–405. doi: 10.1016/S0968-0004(98)01285-7
Jun, S. R., Wassenaar, T. M., Nookaew, I., Hauser, L., Wanchai, V., Land, M., et al. (2016). Diversity of Pseudomonas genomes, including populus-associated isolates, as revealed by comparative genome analysis. Appl. Environ. Microbiol. 82, 375–383. doi: 10.1128/AEM.02612-15
Kämpfer, P., Glaeser, S., Mcinroy, J. A., Clermont, D., Criscuolo, A., and Busse, H.-J. (2021). Pseudomonas carbonaria sp. nov., isolated from charcoal. Syst. Appl. Microbiol. 71:004750. doi: 10.1099/ijsem.0.004750
Kennedy, C. R. P., Macdonald, M. L., and Melton, T. (2015). “Azotobacter” in Bergey's Manual of Systematics of Archaea and Bacteria (Hoboken, NJ: John Wiley and Sons)
Kennedy, C., and Rudnick, P. (2015). “Azomonas” in Bergey's manual of systematics of Archaea and Bacteria (Hoboken, NJ: John Wiley and Sons)
Khadka, B., Persaud, D., and Gupta, R. S. (2020). Novel sequence feature of SecA translocase protein unique to the thermophilic bacteria: bioinformatics analyses to investigate their potential roles. Microorganisms 8:59. doi: 10.3390/microorganisms8010059
Kim, C. M., Jeong, J. W., Lee, D. H., and Kim, S. B. (2021). Pseudomonas guryensis sp. nov. and Pseudomonas ullengensis sp. nov., isolated from soil. Int. J. Syst. Evol. Microbiol. 71:005082. doi: 10.1099/ijsem.0.005082
Kim, M., Oh, H. S., Park, S. C., and Chun, J. (2014). Towards a taxonomic coherence between average nucleotide identity and 16S rRNA gene sequence similarity for species demarcation of prokaryotes. Int. J. Syst. Evol. Microbiol. 64, 346–351. doi: 10.1099/ijs.0.059774-0
Kimura, N., Watanabe, T., Suenaga, H., Fujihara, H., Futagami, T., Goto, M., et al. (2018). Pseudomonas furukawaii sp. nov., a polychlorinated biphenyl-degrading bacterium isolated from biphenyl-contaminated soil in Japan. Int. J. Syst. Evol. Microbiol. 68, 1429–1435. doi: 10.1099/ijsem.0.002670
Kodama, K. K., Kimura, N., and Komagata, K. (1985). Two new species of Pseudomonas: P. oryzihabitans isolated from rice paddy and clinical specimens and P. luteola isolated from clinical specimens. Int. J. Syst. Evol. Microbiol. 35, 467–474. doi: 10.1099/00207713-35-4-467
Konstantinidis, K. T., and Tiedje, J. M. (2007). Prokaryotic taxonomy and phylogeny in the genomic era: advancements and challenges ahead. Curr. Opin. Microbiol. 10, 504–509. doi: 10.1016/j.mib.2007.08.006
Kujur, R. R. A., and Das, S. K. (2022). Pseudomonas phenolilytica sp. nov., a novel phenol-degrading bacterium. Arch. Microbiol. 204:320. doi: 10.1007/s00203-022-02912-y
Kumar, S., Stecher, G., Li, M., Knyaz, C., and Tamura, K. (2018). MEGA X: molecular evolutionary genetics analysis across computing platforms. Mol. Biol. Evol. 35, 1547–1549. doi: 10.1093/molbev/msy096
Lalucat, J., Gomila, M., Mulet, M., Zaruma, A., and García-Valdés, E. (2022). Past, present and future of the boundaries of the Pseudomonas genus: proposal of Stutzerimonas gen. nov. Syst. Appl. Microbiol. 45:126289. doi: 10.1016/j.syapm.2021.126289
Lalucat, J., Mulet, M., Gomila, M., and García-Valdés, E. (2020). Genomics in bacterial taxonomy: impact on the genus Pseudomonas. Genes (Basel) 11:139. doi: 10.3390/genes11020139
Lee, M., and Chandler, A. C. (1941). A study of the nature, growth and control of bacteria in cutting compounds. J. Bacteriol. 41, 373–386. doi: 10.1128/jb.41.3.373-386.1941
Lee, D. H., Kim, C. M., and Kim, S. B. (2022). Pseudomonas insulae sp. nov., isolated from island soil. Int. J. Syst. Evol. Microbiol. 72:005363. doi: 10.1099/ijsem.0.005363
Li, W., and Godzik, A. (2006). Cd-hit: a fast program for clustering and comparing large sets of protein or nucleotide sequences. Bioinformatics 22, 1658–1659. doi: 10.1093/bioinformatics/btl158
Li, K., Ye, Z., Chen, G., Zheng, K., Yin, J., Debnath, S. C., et al. (2023). Atopomonas sediminilitoris sp. nov., isolated from beach sediment of Zhairuo Island, China. Antonie Van Leeuwenhoek 116, 97–107. doi: 10.1007/s10482-022-01780-2
Lin, S.-Y., Hameed, A., Liu, Y.-C., Hsu, Y.-H., Lai, W.-A., Chen, W.-M., et al. (2013). Pseudomonas sagittaria sp. nov., a siderophore-producing bacterium isolated from oil-contaminated soil. Int. J. Syst. Evol. Microbiol. 63, 2410–2417. doi: 10.1099/ijs.0.045567-0
Liu, Y.-C., Young, L.-S., Lin, S.-Y., Hameed, A., Hsu, Y.-H., Lai, W.-A., et al. (2013). Pseudomonas guguanensis sp. nov., a gammaproteobacterium isolated from a hot spring. Int. J. Syst. Evol. Microbiol. 63, 4591–4598. doi: 10.1099/ijs.0.047712-0
Lund-Palau, H., Turnbull, A. R., Bush, A., Bardin, E., Cameron, L., Soren, O., et al. (2016). Pseudomonas aeruginosa infection in cystic fibrosis: pathophysiological mechanisms and therapeutic approaches. Expert Rev. Respir. Med. 10, 685–697. doi: 10.1080/17476348.2016.1177460
Mamtimin, T., Anwar, N., Abdurahman, M., Kurban, M., Rozahon, M., Mamtimin, H., et al. (2021). Pseudomonas lopnurensis sp. nov., an endophytic bacterium isolated from Populus euphratica at the ancient Ugan river. Antonie Van Leeuwenhoek 114, 399–410. doi: 10.1007/s10482-021-01524-8
Manaia, C. M., and Moore, E. R. (2002). Pseudomonas thermotolerans sp. nov., a thermotolerant species of the genus Pseudomonas sensu stricto. Int. J. Syst. Evol. Microbiol. 52, 2203–2209. doi: 10.1099/00207713-52-6-2203
Manickam, N., Ghosh, A., Jain, R. K., and Mayilraj, S. (2008). Description of a novel indole-oxidizing bacterium Pseudomonas indoloxydans sp. nov., isolated from a pesticide-contaminated site. Syst. Appl. Microbiol. 31, 101–107. doi: 10.1016/j.syapm.2008.02.002
Monias, B. L. (1928). Classification of bacterium alcaligenes, pyocyaneum, and fluorescens. J. Infect. Dis. 43, 330–334. doi: 10.1093/infdis/43.4.330
Montecillo, J. A. V., and Bae, H. (2022). Reclassification of Brevibacterium frigoritolerans as Peribacillus frigoritolerans comb. nov. based on phylogenomics and multiple molecular synapomorphies. Int. J. Syst. Evol. Microbiol. 72:005389. doi: 10.1099/ijsem.0.005389
Nicklasson, M., Martín-Rodríguez, A. J., Thorell, K., Higdon, S. M., Neves, L., Mussagy, A., et al. (2022). Pseudomonas boanensis sp. nov., a bacterium isolated from river water used for household purposes in Boane District, Mozambique. Int. J. Syst. Evol. Microbiol. 72:005461. doi: 10.1099/ijsem.0.005461
Oren, A., Arahal, D. R., Göker, M., Moore, E. R. B., Rossello-Mora, R., and Sutcliffe, I. C. (2023). International code of nomenclature of prokaryotes. Prokaryotic code (2022 revision). Int. J. Syst. Evol. Microbiol. 73:005585. doi: 10.1099/ijsem.0.005782
Özen, A. I., and Ussery, D. W. (2012). Defining the Pseudomonas genus: where do we draw the line with Azotobacter? Microb. Ecol. 63, 239–248. doi: 10.1007/s00248-011-9914-8
Palleroni, N. J. (2005). “Genus I. Pseudomonas Migula 1894” in Bergey's manual of systematic bacteriology (the Proteobacteria), part B (the Gammaproteobacteria). eds. D. J. Brenner, N. R. Krieg, J. T. Staley, and G. M. Garrity, vol. 2. 2nd ed (New York: Springer), 323–379.
Palleroni, N. J. (2010). The Pseudomonas story. Environ. Microbiol. 12, 1377–1383. doi: 10.1111/j.1462-2920.2009.02041.x
Palleroni, N. J. (2015). “Pseudomonas” in Bergey's Manual of Systematics of Archaea and Bacteria (Hoboken, NJ: John Wiley and Sons)
Palleroni, N. J., Doudoroff, M., Stanier, R. Y., Solanes, R., and Mandel, M. (1970). Taxonomy of the aerobic pseudomonads: the properties of the Pseudomonas stutzeri group. Microbiology 60, 215–231. doi: 10.1099/00221287-60-2-215
Parks, D. H., Chuvochina, M., Waite, D. W., Rinke, C., Skarshewski, A., Chaumeil, P.-A., et al. (2018). A standardized bacterial taxonomy based on genome phylogeny substantially revises the tree of life. Nat. Biotechnol. 36, 996–1004. doi: 10.1038/nbt.4229
Parte, A. C., Carbasse, J. S., Meier-Kolthoff, J. P., Reimer, L. C., and Göker, M. (2020). List of prokaryotic names with standing in nomenclature (LPSN) moves to the DSMZ. Int. J. Syst. Evol. Microbiol. 70, 5607–5612. doi: 10.1099/ijsem.0.004332
Passarelli-Araujo, H., Franco, G. R., and Venancio, T. M. (2022). Network analysis of ten thousand genomes shed light on Pseudomonas diversity and classification. Microbiol. Res. 254:126919. doi: 10.1016/j.micres.2021.126919
Patel, S., and Gupta, R. S. (2020). A phylogenomic and comparative genomic framework for resolving the polyphyly of the genus Bacillus: proposal for six new genera of Bacillus species, Peribacillus gen. nov., Cytobacillus gen. nov., Mesobacillus gen. nov., Neobacillus gen. nov., Metabacillus gen. nov. and Alkalihalobacillus gen. nov. Int. J. Syst. Evol. Microbiol. 70, 406–438. doi: 10.1099/ijsem.0.003775
Peix, A., Berge, O., Rivas, R., Abril, A., and Velázquez, E. (2005). Pseudomonas argentinensis sp. nov., a novel yellow pigment-producing bacterial species, isolated from rhizospheric soil in Cordoba, Argentina. Syst. Appl. Microbiol. 55, 1107–1112. doi: 10.1099/ijs.0.63445-0
Peix, A., Ramírez-Bahena, M.-H., and Velázquez, E. (2009). Historical evolution and current status of the taxonomy of genus Pseudomonas. Infect. Genet. Evol. 9, 1132–1147. doi: 10.1016/j.meegid.2009.08.001
Peix, A., Ramirez-Bahena, M. H., and Velázquez, E. (2018). The current status on the taxonomy of Pseudomonas revisited: an update. Infect. Genet. Evol. 57, 106–116. doi: 10.1016/j.meegid.2017.10.026
Planquette, B., Timsit, J.-F., Misset, B. Y., Schwebel, C., Azoulay, E., Adrie, C., et al. (2013). Pseudomonas aeruginosa ventilator-associated pneumonia. Predictive factors of treatment failure. Am. J. Respir. Crit. Care Med. 188, 69–76. doi: 10.1164/rccm.201210-1897OC
Price, M. N., Dehal, P. S., and Arkin, A. P. (2010). FastTree 2 – approximately maximum-likelihood trees for large alignments. PLoS One 5:e9490. doi: 10.1371/journal.pone.0009490
Qin, Q. L., Xie, B. B., Zhang, X. Y., Chen, X. L., Zhou, B. C., Zhou, J., et al. (2014). A proposed genus boundary for the prokaryotes based on genomic insights. J. Bacteriol. 196, 2210–2215. doi: 10.1128/JB.01688-14
Ramos, E., Ramírez-Bahena, M.-H., Valverde, A., Velázquez, E., Zúñiga, D., Velezmoro, C., et al. (2013). Pseudomonas punonensis sp. nov., isolated from straw. Int. J. Syst. Evol. Microbiol. 63, 1834–1839. doi: 10.1099/ijs.0.042119-0
Rokas, A., and Holland, P. W. (2000). Rare genomic changes as a tool for phylogenetics. Trends Ecol. Evol. 15, 454–459. doi: 10.1016/S0169-5347(00)01967-4
Rossi, E., La Rosa, R., Bartell, J. A., Marvig, R. L., Haagensen, J. A. J., Sommer, L. M., et al. (2021). Pseudomonas aeruginosa adaptation and evolution in patients with cystic fibrosis. Nat. Rev. Microbiol. 19, 331–342. doi: 10.1038/s41579-020-00477-5
Rudra, B., Duncan, L., Shah, A. J., Shah, H. N., and Gupta, R. S. (2022). Phylogenomic and comparative genomic studies robustly demarcate two distinct clades of Pseudomonas aeruginosa strains: proposal to transfer the strains from an outlier clade to a novel species Pseudomonas paraeruginosa sp. nov. Int. J. Syst. Evol. Microbiol. 72:005542. doi: 10.1099/ijsem.0.005542
Rudra, B., and Gupta, R. S. (2021). Phylogenomic and comparative genomic analyses of species of the family Pseudomonadaceae: proposals for the genera Halopseudomonas gen. nov. and Atopomonas gen. nov., merger of the genus Oblitimonas with the genus Thiopseudomonas, and transfer of some misclassified species of the genus Pseudomonas into other genera. Int. J. Syst. Evol. Microbiol. 71:005011. doi: 10.1099/ijsem.0.005011
Saati-Santamaría, Z., Peral-Aranega, E., Velázquez, E., Rivas, R., and García-Fraile, P. (2021). Phylogenomic analyses of the genus Pseudomonas lead to the rearrangement of several species and the definition of new genera. Biology (Basel) 10:782. doi: 10.3390/biology10080782
Saini, N., and Gupta, R. S. (2021). A robust phylogenetic framework for members of the order Legionellales and its main genera (Legionella, Aquicella, Coxiella and Rickettsiella) based on phylogenomic analyses and identification of molecular markers demarcating different clades. Antonie Van Leeuwenhoek 114, 957–982. doi: 10.1007/s10482-021-01569-9
Sayers, E. W., Agarwala, R., Bolton, E. E., Brister, J. R., Canese, K., Clark, K., et al. (2019). Database resources of the National Center for biotechnology information. Nucleic Acids Res. 47, D23–D28. doi: 10.1093/nar/gky1069
Shin, S. K., Hwang, C. Y., Cho, Y. J., Yi, H., and Yi, H. (2015). Reclassification of Serpens flexibilis Hespell 1977 as Pseudomonas flexibilis comb. nov., with Pseudomonas tuomuerensis Xin et al. 2009 as a later heterotypic synonym. Syst. Appl. Microbiol. 38, 563–566. doi: 10.1016/j.syapm.2015.09.007
Sievers, F., Wilm, A., Dineen, D., Gibson, T. J., Karplus, K., Li, W., et al. (2011). Fast, scalable generation of high-quality protein multiple sequence alignments using Clustal omega. Mol. Syst. Biol. 7:539. doi: 10.1038/msb.2011.75
Silby, M. W., Winstanley, C., Godfrey, S. A., Levy, S. B., and Jackson, R. W. (2011). Pseudomonas genomes: diverse and adaptable. FEMS Microbiol. Rev. 35, 652–680. doi: 10.1111/j.1574-6976.2011.00269.x
Singh, B., and Gupta, R. S. (2009). Conserved inserts in the Hsp60 (GroEL) and Hsp70 (DnaK) proteins are essential for cellular growth. Mol. Gen. Genomics. 281, 361–373. doi: 10.1007/s00438-008-0417-3
Skerman, V. B. D., Mcgowan, V., and Sneath, P. H. A. (1980). Approved lists of bacterial names. Int. J. Syst. Bacteriol. 30, 225–420. doi: 10.1099/00207713-30-1-225
Stamatakis, A. (2014). RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics 30, 1312–1313. doi: 10.1093/bioinformatics/btu033
Stanier, R. Y., Palleroni, N. J., and Doudoroff, M. (1966). The aerobic pseudomonads a taxonomic study. Microbiology 43, 159–271. doi: 10.1099/00221287-43-2-159
Stover, C. K., Pham, X. Q., Erwin, A., Mizoguchi, S., Warrener, P., Hickey, M., et al. (2000). Complete genome sequence of Pseudomonas aeruginosa PAO1, an opportunistic pathogen. Nature 406, 959–964. doi: 10.1038/35023079
Sudan, S. K., Pal, D., Bisht, B., Kumar, N., Chaudhry, V., Patil, P., et al. (2018). Pseudomonas fluvialis sp. nov., a novel member of the genus Pseudomonas isolated from the river Ganges, India. Int. J. Syst. Evol. Microbiol. 68, 402–408. doi: 10.1099/ijsem.0.002520
Tao, Y., Zhou, Y., He, X., Hu, X., and Li, D. (2014). Pseudomonas chengduensis sp. nov., isolated from landfill leachate. Int. J. Syst. Evol. Microbiol. 64, 95–100. doi: 10.1099/ijs.0.050294-0
Tarhriz, V., Nouioui, I., Spröer, C., Verbarg, S., Ebrahimi, V., Cortés-Albayay, C., et al. (2020). Pseudomonas khazarica sp. nov., a polycyclic aromatic hydrocarbon-degrading bacterium isolated from Khazar Sea sediments. Antonie Van Leeuwenhoek 113, 521–532. doi: 10.1007/s10482-019-01361-w
Thompson, C. C., Chimetto, L., Edwards, R. A., Swings, J., Stackebrandt, E., and Thompson, F. L. (2013). Microbial genomic taxonomy. BMC Genomics 14:913. doi: 10.1186/1471-2164-14-913
Thorat, V., Kirdat, K., Tiwarekar, B., Dacosta, E., Debbarma, P., Shouche, Y., et al. (2020). Pseudomonas lalkuanensis sp. nov., isolated from a bacterial consortia of contaminated soil enriched for the remediation of e-waste. Int. J. Syst. Evol. Microbiol. 70, 6468–6475. doi: 10.1099/ijsem.0.004559
Timsy, T., Ulrich, A., Kublik, S., Foesel, B. U., Kolb, S., Horn, M. A., et al. (2021). Pseudomonas campi sp. nov., a nitrate-reducing bacterium isolated from grassland soil. Int. J. Syst. Evol. Microbiol. 71:4799. doi: 10.1099/ijsem.0.004799
Tohya, M., Watanabe, S., Teramoto, K., Tada, T., Kuwahara-Arai, K., Mya, S., et al. (2020). Pseudomonas yangonensis sp. nov., isolated from wound samples of patients in a hospital in Myanmar. Int. J. Syst. Evol. Microbiol. 70, 3597–3605. doi: 10.1099/ijsem.0.004181
Wang, X., He, S.-W., Guo, H.-B., Thin, K. K., Gao, J.-S., Wang, Y., et al. (2020). Pseudomonas rhizoryzae sp. nov., isolated from rice. Int. J. Syst. Evol. Microbiol. 70, 944–950. doi: 10.1099/ijsem.0.003852
Wang, Z., and Wu, M. (2013). A phylum-level bacterial phylogenetic marker database. Mol. Biol. Evol. 30, 1258–1262. doi: 10.1093/molbev/mst059
Whelan, S., and Goldman, N. (2001). A general empirical model of protein evolution derived from multiple protein families using a maximum-likelihood approach. Mol. Biol. Evol. 18, 691–699. doi: 10.1093/oxfordjournals.molbev.a003851
Winogradsky, S. (1938). Sur la morphologie et l' oecologie des Azotobacter. Ann. Inst. Pasteur 55, 351–400.
Winsor, G. L., Griffiths, E. J., Lo, R., Dhillon, B. K., Shay, J. A., and Brinkman, F. S. (2016). Enhanced annotations and features for comparing thousands of Pseudomonas genomes in the Pseudomonas genome database. Nucleic Acids Res. 44, D646–D653. doi: 10.1093/nar/gkv1227
Wong, S. Y., Paschos, A., Gupta, R. S., and Schellhorn, H. E. (2014). Insertion/deletion-based approach for the detection of Escherichia coli O157:H7 in freshwater environments. Environ. Sci. Technol. 48, 11462–11470. doi: 10.1021/es502794h
Wu, M., Wen, J., Chang, M., Yang, G., and Zhou, S. (2014). Pseudomonas sihuiensis sp. nov., isolated from a forest soil in South China. Antonie Van Leeuwenhoek 105, 781–790. doi: 10.1007/s10482-014-0134-3
Xin, X. F., Kvitko, B., and He, S. Y. (2018). Pseudomonas syringae: what it takes to be a pathogen. Nat. Rev. Microbiol. 16, 316–328. doi: 10.1038/nrmicro.2018.17
Xin, Y. H., Zhang, D. C., Liu, H. C., Zhou, H. L., and Zhou, Y. G. (2009). Pseudomonas tuomuerensis sp. nov., isolated from a bird's nest. Int. J. Syst. Evol. Microbiol. 59, 139–143. doi: 10.1099/ijs.0.000547-0
Yamada, K., Sasaki, M., Aoki, K., Nagasawa, T., Murakami, H., Ishii, M., et al. (2021). Pseudomonas tohonis sp. nov., isolated from the skin of a patient with burn wounds in Japan. Int. J. Syst. Evol. Microbiol. 71:005115. doi: 10.1099/ijsem.0.005115
Yang, Y., Gao, Y., Liu, Y., Liu, B., Wang, D., Xu, Y., et al. (2022). Pseudomonas marianensis sp. nov., a marine bacterium isolated from deep-sea sediments of the Mariana trench. Arch. Microbiol. 204:638. doi: 10.1007/s00203-022-03250-9
Yang, G., Han, L., Wen, J., and Zhou, S. (2013). Pseudomonas guangdongensis sp. nov., isolated from an electroactive biofilm, and emended description of the genus Pseudomonas Migula 1894. Int. J. Syst. Evol. Microbiol. 63, 4599–4605. doi: 10.1099/ijs.0.054676-0
Yarza, P., Yilmaz, P., Pruesse, E., Glöckner, F. O., Ludwig, W., Schleifer, K.-H., et al. (2014). Uniting the classification of cultured and uncultured bacteria and archaea using 16S rRNA gene sequences. Nat. Rev. Microbiol. 12, 635–645. doi: 10.1038/nrmicro3330
Young, J. M., and Park, D. C. (2007). Probable synonymy of the nitrogen-fixing genus Azotobacter and the genus Pseudomonas. Int. J. Syst. Evol. Microbiol. 57, 2894–2901. doi: 10.1099/ijs.0.64969-0
Yu, X.-Y., Zhai, J.-Y., Wu, C., Zhang, C.-Y., Shi, J.-Y., Ding, L.-X., et al. (2018). Pseudomonas pharmafabricae sp. nov., isolated from pharmaceutical wastewater. Curr. Microbiol. 75, 1119–1125. doi: 10.1007/s00284-018-1495-z
Yumoto, I., Yamazaki, K., Hishinuma, M., Nodasaka, Y., Suemori, A., Nakajima, K., et al. (2001). Pseudomonas alcaliphila sp. nov., a novel facultatively psychrophilic alkaliphile isolated from seawater. Int. J. Syst. Evol. Microbiol. 51, 349–355. doi: 10.1099/00207713-51-2-349
Zhang, P., Dong, X., Zhou, K., Zhu, T., Liang, J., Shi, W., et al. (2021). Characterization of a novel chromosome-encoded AmpC β-lactamase gene, Bla PRC–1, in an isolate of a newly classified Pseudomonas species, Pseudomonas wenzhouensis A20, from animal farm sewage. Front. Microbiol. 12:732932. doi: 10.3389/fmicb.2021.732932
Zhang, M., Li, A., Yao, Q., Xiao, B., and Zhu, H. (2022). Pseudomonas oligotrophica sp. nov., a novel denitrifying bacterium possessing nitrogen removal capability under low carbon–nitrogen ratio condition. Front. Microbiol. 13:882890. doi: 10.3389/fmicb.2022.882890
Zhang, L., Pan, Y., Wang, K., Zhang, X., Zhang, S., Fu, X., et al. (2015). Pseudomonas songnenensis sp. nov., isolated from saline and alkaline soils in Songnen plain, China. Antonie Van Leeuwenhoek 107, 711–721. doi: 10.1007/s10482-014-0365-3
Zhou, S., Wang, Y., Xia, H., Liu, D., Chen, S., and Li, F. (2020). Pseudomonas hydrolytica sp. nov., multiple polymer-degrading bacteria isolated from soil in China. Int. J. Syst. Evol. Microbiol. 70, 3049–3054. doi: 10.1099/ijsem.0.004129
Keywords: Pseudomonas classification, phylogenomic and comparative genomic analyses, conserved signature indels (CSIs), molecular markers specific for Pseudomonas species clades/groups, proposals for reclassification of Pseudomonas species into novel genera
Citation: Rudra B and Gupta RS (2024) Phylogenomics studies and molecular markers reliably demarcate genus Pseudomonas sensu stricto and twelve other Pseudomonadaceae species clades representing novel and emended genera. Front. Microbiol. 14:1273665. doi: 10.3389/fmicb.2023.1273665
Edited by:
Iain Sutcliffe, Northumbria University, United KingdomReviewed by:
Margarita Gomila, University of the Balearic Islands, SpainCarrie Brady, University of the West of England, United Kingdom
Aharon Oren, Hebrew University of Jerusalem, Israel
Copyright © 2024 Rudra and Gupta. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Radhey S. Gupta, Z3VwdGFAbWNtYXN0ZXIuY2E=
 Bashudev Rudra
Bashudev Rudra Radhey S. Gupta
Radhey S. Gupta