
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Microbiol. , 03 October 2023
Sec. Infectious Agents and Disease
Volume 14 - 2023 | https://doi.org/10.3389/fmicb.2023.1273095
In the human pathogen Vibrio vulnificus, the quorum-sensing (QS) signal molecule cyclo-(L-phenylalanine-L-proline) (cFP) plays a critical role in triggering a signaling pathway involving the components LeuO-vHUαβ-RpoS-KatG via the membrane signal receptor ToxR. In this study, we investigated the impact of iron on the expression of these signaling components. We found that the transcription of the membrane sensor protein ToxR was not significantly affected by Fur-iron. However, Fur-iron repressed the transcription of genes encoding all the downstream cytoplasmic components in this pathway by binding to the upstream regions of these genes. Consequently, the expression of genes regulated by the alternative sigma factor RpoS, as well as the resistance to hydrogen peroxide conferred by KatG, were repressed. Additionally, we observed that in Vibrio cholerae, genes dependent on ToxR showed higher expression levels in a fur-deletion mutant compared to the wild type. These findings indicate that iron, in association with Fur, represses virtually all the cytoplasmic components responsible for the ToxR-dependent cFP-signaling pathways in these two pathogenic Vibrio species. This study, along with our previous reports demonstrating the repression of components involved in AI-2 dependent QS signaling by Fur-iron, highlights the crucial role of iron in quorum-sensing regulation, which is closely associated with the pathogenicity of this human pathogen.
The opportunistic human pathogen Vibrio vulnificus is a halophilic Gram-negative bacterium that and causes primary sepsis in a certain high-risk human population (Kumamoto and Vukich, 1998). Like many other pathogenic bacteria, V. vulnificus employs complex signal transduction systems to sense various environmental factors and adjust corresponding functions accordingly for survival and pathogenicity (Kim et al., 2013b). A typical example is the quorum-sensing (QS) regulation, which is triggered by diffusible signal molecules produced by cognate bacterial cells and plays a significant role in regulation of sets of virulence factors.
A QS signaling pathway initiated by a diketopiperazine (DKP) signal molecule cyclo-(L-phenylalanine-L-proline) (cFP) has been identified in Vibrio spp., including pathogens V. vulnificus, V. cholerae and Vibrio parahaemolyticus as well as a non-pathogen Vibrio harveyi (Park et al., 2006; Bina and Bina, 2010). This signal is released from cells into environment through simple diffusion across the bacterial membrane (Park et al., 2020a) and the cFP signal is recognized in neighboring cells by the inner membrane receptor protein ToxR (Park et al., 2006; Bina et al., 2013), which is known to form either homodimers or heterodimers with ToxS (DiRita and Mekalanos, 1991). The transduction of the cFP signal from ToxR leads to induction of the expression of LeuO, the master regulator in the cFP-dependent signal pathway (Bina et al., 2013; Park et al., 2019). However, when the signal is not transduced, H-NS, a histone-like nucleoid structural protein, plays as a basal stopper role to suppress the expression of LeuO (Ghosh et al., 2006; Picker and Wing, 2016; Park et al., 2020b). LeuO, when over-expressed, exhibits a feedback control mechanism by inhibiting its own transcription (Park et al., 2019).
The LeuO master regulator subsequently activates the expression of several genes. This includes the porin OmpU (Park et al., 2006), as well as the histone-like proteins vHUα and vHUβ which enhance the post-transcriptional stability of the rpoS mRNA. The rpoS gene encodes an alternate sigma factor (Kim et al., 2018; Park et al., 2020b) that directs the transcription of a series of genes including katG encoding a catalase that confers resistance to hydrogen peroxide in the pathogen (Kim et al., 2018). Furthermore, transcriptomic analysis has revealed that more than 950 genes in V. vulnificus are modulated by cFP (Kim et al., 2013a), indicating that the cognate signal transductions exert a profound influence on physiology of the pathogen.
Iron is essential element for most living organisms including animals, plants, and bacteria (Schaible and Kaufmann, 2004; Waldron and Robinson, 2009). However, high concentrations of ferrous ion (Fe2+) can be toxic as they can generate highly reactive radicals through the Fenton reaction (Massé and Arguin, 2005). To survive, cells have developed strategies to maintain iron homeostasis in the cytoplasm. One common strategy employed by bacteria is the production of low-molecular weight compounds called siderophores, which have a high affinity for ferric iron (Fe3+) (Troxell and Hassan, 2013). In most bacteria, iron homeostasis is mainly regulated by the ferric uptake regulator (Fur) (Deng et al., 2015). Fur is crucial in host–parasite interactions as it controls the expression of various proteins involved in iron removal and uptake systems, which allow bacteria to acquire iron from heme or specifically internalize host iron-binding proteins (Vasil and Ochsner, 1999). V. vulnificus possesses a 143-amino acid Fur protein, which shares 79% homology with Escherichia coli Fur and 93% homologous to V. cholerae Fur (Litwin and Calderwood, 1993). Under iron-rich conditions, Fur-iron complexes typically recognize a DNA sequence called Fur box (5′-GATAATGATAATCATTATC-3′) which is present in the promoter region of target genes and affect RNA polymerase binding to modulate the expression of the genes (Wen et al., 2016).
We have demonstrated that the Fur-iron complex regulates the autoinducer-2 (AI-2) signaling QS pathway in V. vulnificus. The QS, along with Fur-iron complex, controls the production vulnibactin encoded by vvsAB, thereby maintaining the intracellular iron concentration at an appropriate level. Under conditions of iron limitation, the transcription level of vvsAB is low at low cell density but induced at high cell density. However, in the presence of iron, the Fur-iron complex represses the transcription of the genes regardless of cell density (Wen et al., 2012). We also have found that the Fur-iron complex also regulates virulence factors by modulate the expression of smcR, which encodes the master regulator of the AI-2 QS signaling system. This repression is achieved through the direct binding of the Fur-iron complex to the cis-acting element in the upstream region of smcR (Kim et al., 2013b). Five small RNA molecules called Qrrs1-5, which are involved in the QS regulation, are also regulated by the Fur-iron complex (Wen et al., 2016). We also have shown that the small RNA RyhB, which enhances the stability and translation of the LuxS mRNA responsible for produces AI-2, is inhibited by the Fur-iron complex (Lee et al., 2022).
Our findings suggest that iron antagonizes QS signaling, and the regulation is primarily mediated by the Fur protein. On the basis, we further investigated the impact of the Fur-iron complex on another QS system mediated by cFP signaling in V. vulnificus. In this study, we demonstrate that the Fur-iron complex exerts control over this signaling system as well, by repressing the expression of virtually all known components associated with the cFP signaling pathway. The results presented here underscore the significance of iron in the signaling mechanisms of V. vulnificus and, consequently, in the pathogenicity of this virulent species.
The bacterial strains and plasmids used in this study are listed in Supplementary Table S1. Escherichia coli and V. cholerae strains were cultured at 37°C in Luria-Bertani (LB) broth supplemented with appropriate antibiotics. V. vulnificus strains were cultured in LB broth or thiosulfate citrate bile salt sucrose (TCBS) agar at 30°C. When necessary, either ferrous sulfate (25 μM) as an iron source or 2,2′-dipyridyl (100 μM) as an iron chelator was added exogenously to the LB broth when the A600 value of the culture reached approximately 0.1. Antibiotics were used at the following concentration: For E. coli, ampicillin 50 μg/mL, kanamycin 25 μg/mL, tetracycline 10 μg/mL, chloramphenicol 25 μg/mL; for V. vulnificus and V. cholerae, kanamycin 100 μg/mL, tetracycline 2 μg/mL, chloramphenicol 2 μg/mL. All media used in this study were purchased from Difco (MI, United States). All reagents and antibiotics were purchased from Sigma Aldrich (MO, United States).
To construct a fur deletion derivative, Δfur, the primers Δfur_FF_xbaI and Δfur_FR_speI (Supplementary Table S2) were used for amplification of the upstream region of fur, and Δfur_BF_speI and Δfur_BR_xhoI for the downstream region of fur. The PCR products were cloned to the predigested suicide vector pDM4. The resulting plasmid was mobilized from S17-1 λpir to the wild type V. vulnificus MO6-24/O strain by conjugation. A double crossover was selected in LB plated with 10% sucrose. Colonies that grew on sucrose plate but sensitive to chloramphenicol were selected. The mutation was confirmed through PCR and DNA nucleotide sequencing.
Derivatives of V. vulnificus MO6-24/O or its fur-deletion isotype, Δfur, harboring the luxAB reporter transcriptionally fused to each of leuO, vhuα, vhuβ, rpoS, and katG were described previously (Kim et al., 2018). Overnight cultures of tested strains harboring the luxAB reporter fusions grown in LB were inoculated into fresh LB medium. To make an iron-limiting condition, 100 μM 2,2′-dipyridyl was added exogenously to the LB broth when the A600 value of the culture reached approximately 0.1, and samples were diluted 125-fold with LB broth. At various growth stages, 0.006% (v/v) n-decylaldehyde (in 50% ethanol) was added and luminescence was measured using a microplate reader (Mithras LB 940; Berthold, Bad Wildbach, Germany) as previously described (Wen et al., 2012). The specific transcription level was expressed as relative light units (RLU) normalized to cell density.
The 1,540-bp DNA fragment of the leuO upstream region (−975 to +565 with respect to the translation start site) was amplified by PCR using the primers leuO_DCO_F and leuO_DCO_R. The resulting product was ligated to the pGEM-T Easy vector to construct pGEM-LeuO. To introduce mutations into each of the four regions (SM1 ~ 4) containing putative Fur boxes, mutagenesis was performed using primer sets leuO_SDM_1_F and leuO_SDM_1_R for SM1, leuO_SDM_2_F and leuO_SDM_2_R for SM2, leuO_SDM_3_F and leuO_SDM_3_R for SM3, and leuO_SDM_4_F and leuO_SDM_4_R for SM4. The resulting four plasmids were named pGEM-SM1 through pGEM-SM4, respectively.
The Strep-tagged Fur was expressed in E. coli BL21(DE3) cells harboring the fur clone pASK-IBA7-Fur (Kim et al., 2013b) by induction with 0.2 μg/mL anhydrotetracycline. After centrifugation, bacterial pellets were resuspended in a buffer (100 mM Tris-Cl, 150 mM NaCl, and 1 mM EDTA, pH 7.5). The cells were then sonicated and centrifuged at 4,585 × g for 10 min. The resulting supernatant was subjected to purification using Strep-Tactin affinity resin (IBA BioTAGnology, Göttingen, Germany), and specifically bound protein was eluted with E buffer (100 mM Tris-Cl, 150 mM NaCl, and 1 mM EDTA, and 2.5 mM desthiobiotin, pH 7.5) according to the manufacturer’s instructions. The eluted protein was separated on a 12% SDS-PAGE to assess the purity. The purified Fur protein was dialyzed using Spectra/Por molecular porous membrane tubing (molecular weight cutoff of 10,000; Spectrum Laboratoried Inc., Rancho Dominguez, CA) with A buffer (50 mM Tris-Cl, 100 mM NaCl, 1 mM MgCl2, and 2 mM dithiothreitol, pH 8.0). The protein was concentrated using the Vivaspin 6 instrument (Vivagen, Seoul, Korea). The protein concentration was determined by the Bradford method (Bradford, 1976).
To assess the binding of Fur to upstream region of leuO, the 299-bp regions (−288 to +11 with respect to the translation start site) of the gene containing wild type and mutated bases at each of Fur box candidates (SM1 ~ 4) were amplified by PCR with the primers leuO_EMSA_F and 32P-labeled leuO_EMSA_R using pGEM-leuO and pGEM-SM1 ~ 4 as template DNAs. Similarly, to assess the binding of Fur on the region upstream to vhuα, the 303-bp regions (−261 to +42 with respect to the translation start site) were amplified by PCR with the primers HU_alpha_EMSAF and 32P-labeled HU_alpha_EMSAB. In the same way, for vhuβ used the 257-bp regions (−212 to +45 with respect to the translation start site); for rpoS, the 650-bp regions (−556 to +94 with respect to the translation start site); and for katG, the 495-bp regions (−401 to +94 with respect to the translation start site) were amplified by PCR with the each primer set HU_beta_EMSAF and 32P-labeled HU_beta_EMSAB, rpoS_EMSA_longF and 32P-labeled rpoS_EMSA_R, katG_EMSA_F and 32P-labeled katG_EMSA_R, respectively.
For gel shift assays, 10 ng of the labeled probe was incubated with increasing amounts of purified Fur protein in a 20 μL reaction mixture in binding buffer (Alice et al., 2008) containing 10 mM Tris-borate (pH 7.5), 100 μg/mL bovine serum albumin, 5% (v/v) glycerol, 40 mM KCl, 1 mM MgCl2, and 1 μg poly(dI-dC). The reaction mixture was supplemented with either 1 mM MnCl2 or 1 mM EDTA for 30 min at 30°C. The resulting mixtures were resolved in a 6% neutral polyacrylamide gel. Each of the labeled probes (10 ng) was incubated with increasing amounts of purified Fur protein, and gel shift assays were performed as described above. The gels were exposed to a BAS_MP 2040s imaging plate (Fujifilm, Tokyo, Japan) and scanned using a BAS-1500 instrument (Fujifilm).
An end-labeled 405-bp DNA fragments of the leuO upstream region (−376 to +29 with respect to the translation start site) was amplified using the primers leuO_fp_F2 and 6-FAM labeled leuO_fp2_R_FAM. To determine the Fur binding site, 200 ng of the amplified leuO upstream region was incubated with purified Fur (2 μM) in 50 μL of binding buffer containing 10 mM Tris-borate (pH 7.5), 100 μg/mL bovine serum albumin, 5% (v/v) glycerol, 40 mM KCl, 1 mM MgCl2, 1 μg poly(dI-dC) for 30 min at 30°C. After incubation, 0.01 unit of DNase I (Promega, Madison, WI) was added, and the reaction mixture was incubated at 37°C for 1 min. The reaction was terminated by the addition of 5 μL of RQ1 DNase stop solution (Promega, Madison, WI), and inactivated at 65°C for 10 min. To precipitate the samples, 55 μL phenol-chloroform was added, and the mixture was precipitated at room temperature for 5 min. After centrifugation at 11,323 × g at 4°C for 5 min sodium acetate (NaCOOH, pH 5.2) was added to 50 μL of supernatant. Following the addition of 100 μL of 100% ethanol, the mixture was incubated for 1 h at −80°C and then centrifuged in 11,323 × g at 4°C for 15 min. The sample was then washed with 70% ethanol, dried in water bath at 60°C, and dissolved in 10 μL of distilled water. DNA sequencing of the sample was carried out in GBST (Green-Bio Science and Technology, Seoul National University). Raw data obtained through ABI 3730xl were analyzed using the Peak Scanner software (Applied Biosystems, Waltham, MA, United States).
β-galactosidase activity was measured as described previously (Miller, 1972). Briefly, V. vulnificus strains were cultured overnight in LB medium, harvested, and diluted to an A600 of 0.005, and β-galactosidase activities from cells harboring the genes transcriptionally fused with lacZ as described above was measured.
RNA was isolated from V. vulnificus using the easy-BLUE™ total RNA extraction Kit (iNtRON Biotechnology, Seongnam, Korea) and treated with the RNase-free DNase set (Promega, Madison, WI, United States) to remove any residual DNA. The purified RNA was quantified using a Biophotometer (Eppendorf, Hamburg, Germany). Subsequently, cDNA was synthesized from 500 ng of RNA using the CellScript™ All-in-One cDNA Master Mix (Cellsafe, Yongin, Korea) following the manufacturer’s instructions. One microliter of cDNA was used for RT-PCR analysis on a Stratagene Mx3000p qPCR machine (Agilent Technologies, Santa Clara, CA, United States) using QGreenBlue 2 × Green qPCR Master Mix (Cellsafe, Yongin, Korea). The RT-PCR reactions were performed in triplicate in a 96-well plate using primer shown in Supplementary Table S2. The PCR conditions used to amplify all genes were: 10 min at 95°C and 40 cycles of 95°C for 15 s and 64°C for 40 s. The genes encoding type I glyceraldehyde-3-phosphate dehydrogenase (RS_10395) and DNA-directed RNA polymerase subunit alpha (RS_13660) of V. vulnificus were used as endogenous loading controls. Quantification was carried out using the Light Cycle 480 II real-time PCR system software program.
V. vulnificus strains (MO6-24/O, Δfur, Δfur complemented with pRK415-fur, ΔkatG) were cultured overnight in LB medium, washed, and sub-cultured in fresh LB medium. All cells were harvested at exponential phase (A600 value of approximately 1.0) and sonicated using Ultrasonic Homogenizer (KUS-650, KBT, Seongnam, Korea) in 10 cycles with 1 s of sonication followed by 2 s of rest for each cycle. After centrifugation, the supernatant was concentrated using Amicon Ultra-0.5 mL Centrifugal Filter Units (10,000 NMWL, UFC501024, Merck-Millipore, Germany). Each of 50 μL sample was mixed with 10 mM hydrogen peroxide (Duksan, Ansan, Korea). The reaction tubes were vortexed, incubated for 2 min at 37°C, and 600 μL of working solution was added. The working solution consisted of 100 mL cobalt (II) solution (20.3 g / 1 L DIW), 100 mL sodium hexametaphosphate solution (10 g / 1 L DIW), 800 mL sodium bicarbonate solution (180 g/ 2 L DIW) (Hadwan, 2018). The tubes were vortexed for 5 s and then kept at room temperature for 10 min in the dark. After 10 min, the catalase activities were measured at 440 nm using a Multimode Plate Reader (PerkinElmer, Waltham, MA, United States).
The flow rate was set to 1.0 mL/min, and the cFP peak was detected at 256 nm. As a reference, a 1 mM cFP solution (from a company) dissolved in 30% methanol was also prepared and analyzed using the same HPLC method. The amount of cFP in each sample was estimated based on the area of the cFP peak observed in the 1 mM cFP reference sample.
After culturing cells in LB broth for 24 h, a 20 mL supernatant of each sample was collected and mixed with an equal volume of ethyl acetate, and then finally dissolved in 150 μL of 30% methanol. The cFP in each sample was resolved using high-performance liquid chromatography (HPLC) using a C18-reverse phase column Mightysil RP-18 GP (Kanto, Tokyo, Japan) with 30% methanol as the mobile phase. The flow rate was set to 1.0 mL/min, and the cFP peak was detected at 256 nm. As a reference, 1 mM of cFP (Bachem, Bubendorf, Switzerland) dissolved in 30% methanol were also prepared in the same way, and analyzed using the same HPLC methods. The amount of cFP in each sample was estimated based on the area of the cFP peak observed in the 1 mM cFP reference sample.
We initially examined the effect of Fur-iron on the transcription of toxR encoding inner membrane porin, which serves as receptor for cFP (Park et al., 2006, 2019). Using a lacZ-fusion, we quantitatively compared the transcription level of ToxR in wild type and Δfur (fur-deletion isotype) strains in the presence or absence of iron. However, no significant difference was observed between two groups of cells (data not shown). Furthermore, through a gel-shift assay using 363-bp region upstream to toxR as a probe (Supplementary Figure S1A) and purified ToxR, we determined that Fur does not bind to the upstream region, regardless of iron availability (Supplementary Figure S1B). These results indicate that the expression of ToxR is not influenced by iron.
In our previous study (Park et al., 2019), we investigated the regulatory elements in the region upstream to leuO, which encodes the master regulator for the cFP-signaling pathway in V. vulnificus. Within this region, we identified a putative Fur-binding sequence, suggesting that Fur may be involved in the regulation of leuO expression. To examine this possibility, we constructed the luxAB reporter gene fusions with leuO in both wild-type MO6-24/O strain and the Δfur, and quantitatively measured luciferase activities in the presence or absence of iron. When iron was supplied, the RLU values in the Δfur mutant were approximately three times higher than these in the wild-type strain (Figure 1). Under iron-limited conditions, there was no significant difference in leuO expression was observed between the wild-type strain and Δfur mutant. It is noteworthy that, even in Δfur cells, the expression of leuO was still repressed to some extent by iron, albeit significantly less compared to the wild type cells. This suggests the possible involvement of unidentified factor(s) in the iron-dependent regulation of leuO.
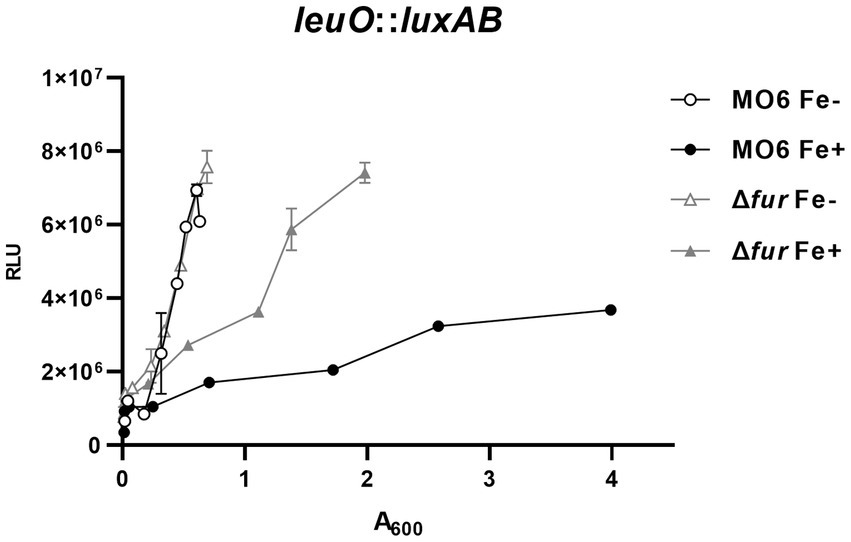
Figure 1. Fur-iron represses the transcription of leuO. The transcriptional activities of leuO were assessed using the leuO-luxAB reporter in wild-type V. vulnificus strain MO6-24/O (represented by circles) and the fur deletion mutant (Δfur) (represented by triangles) under conditions of iron limitation (blank) or iron abundance (solid). Relative light units (RLU) were measured and normalized to cell density (A600). To induce iron limitation, 2,2′-dipyridyl (100 μM), an iron chelator, was added when the A600 reached 0.1.
The leuO gene encodes a key regulatory component in cFP – signaling pathway which is closely associated with virulence of pathogenic Vibrio species, and hence its regulation is important for the pathogenicity. Therefore, we defined in detail the cis-acting elements for Fur in the gene. There exist four regions with nucleotide sequences homologous to the Fur box (Wen et al., 2016) in the leuO upstream region, named PF (putative Fur box) sites 1 ~ 4 (Figure 2A). Meanwhile, gel shift assay demonstrated that purified Fur binds to the upstream region of leuO and it appears that at least two shifts occurred (Supplementary Figure S2A). To determine which sites of these putative Fur-binding sites are actually bound by Fur, we performed competition gel-shift assay using a radiolabeled 299-bp oligomer, as described in Materials and Methods, as a probe and purified Fur. The probe was synthesized by PCR using leuO_EMSA_F and leuO_EMSA_R as primers (Supplementary Table S2). We then prepared four DNA oligomers of the same size with mutations in each of those four PF sites (named SM1 ~ 4) as shown in Figure 2A. Each of these mutated oligomers, without radiolabeling, was added as a competitor at about 10-fold higher concentration than the radiolabeled probe oligomer. As shown in the lane 3 of Figure 2B, the competitor without any mutation completely outcompeted the binding of Fur for the probe. The competitor without any mutations in each of the PF1 and PF3 sites also hindered the binding to Fur to the probe (lanes 4 and 6). In contrast, the competitor oligomer with mutations in each of PF2 or PF4 sites did not significantly interfere in the binding of Fur onto the probe oligomer, while the competitor with mutations in PF2 slightly interfered in the binding, but much less than the one with mutation in PF4. In addition, footprinting experiments also show high binding affinity to the PF2 and PF4 regions (Supplementary Figure S2B). These results suggested that Fur strongly binds to the PF4 region, and weakly to the PF2 region.
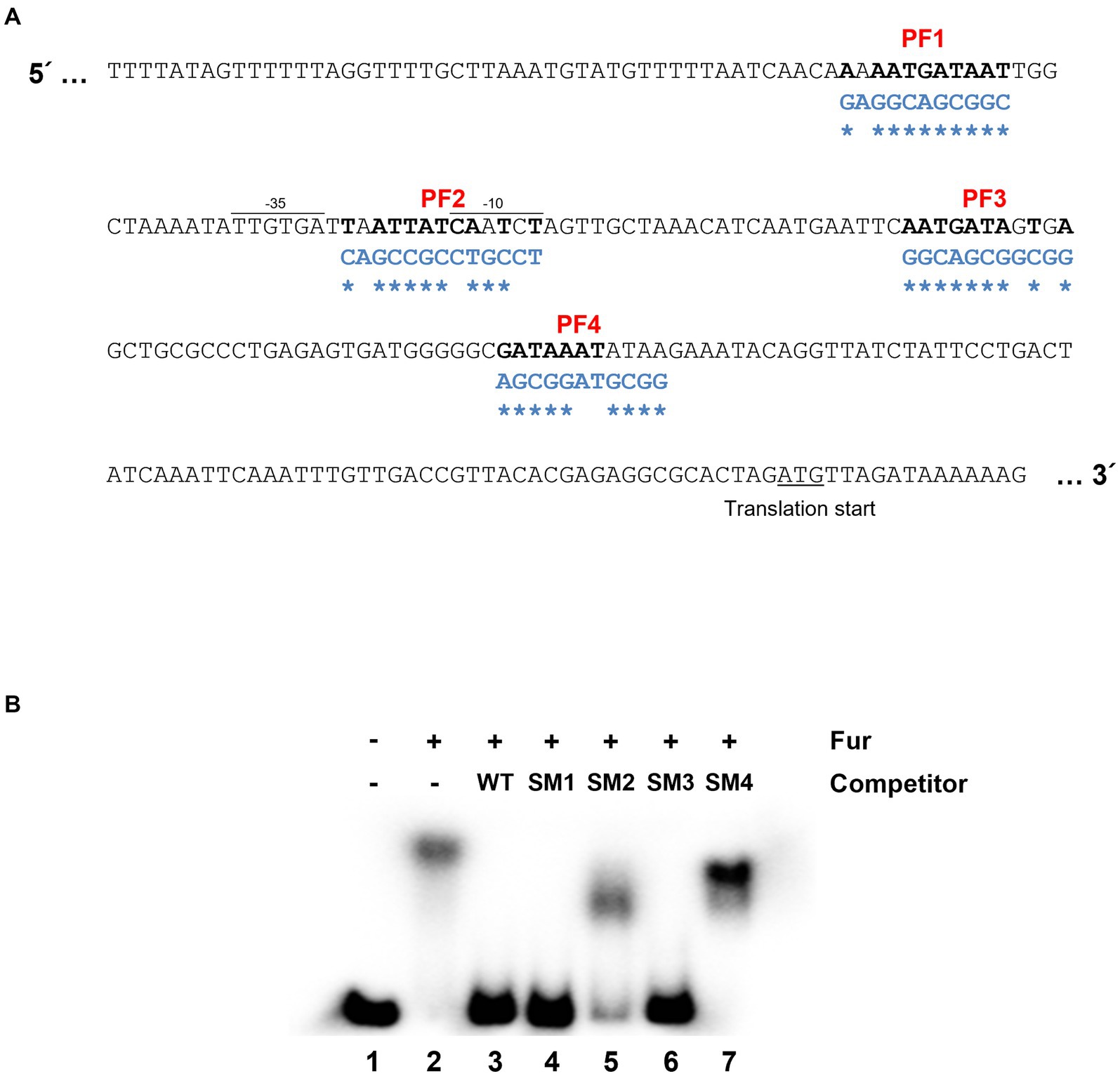
Figure 2. Fur-iron binding sites on the region upstream to leuO. (A) Nucleotide sequences upstream to leuO. The promoter region (−35 and −10 sites) and translation start site are indicated. Nucleotide sequences homologous to the Fur box (5′-GATAATGATAATCATTATC-3′) are named PF (putative Fur box) 1 through 4, and are denoted in bold. Nucleotide sequences of oligomers with site-directed mutations are denoted in blue letters, and the mutated sites are indicated with asterisks. (B) Gel shift assay indicates that Fur-iron binds to PF2 and PF4 regions. Shown here are the results of gel shift assays using the radiolabeled 299-bp fragment probe with the sequences of the region upstream to leuO and purified 200 nM Fur with 1 mM MnCl2. Lane 1 represents a negative control, which does not have any competitor or Fur, but only has 10 ng of the wild-type probe. Lane 2 has no competitor, but 200 nM Fur and 10 ng of the wild-type probe. Lanes 3 to 7 contain 200 nM Fur with 100 ng of the wild-type probe, and each of competitor SM1, SM2, SM3, and SM4, respectively, which are the 299-bp fragments with mutated sequences in PF1 through 4 regions.
In a previous study, it was demonstrated that LeuO has a negative regulatory effect on two genes, vhuα and vhuβ, which encode histone-like proteins that are involved in the regulation of numerous genes (Balandina et al., 2002; Grove, 2011; Kim et al., 2018). To investigate the potential regulation exerted by Fur-iron on these genes, we examined their transcriptional levels using lacZ transcriptional fusions in both wild-type and Δfur cells under iron-rich and iron-depleted conditions (Figure 3). In wild-type cells, the transcription of both vhuα and vhuβ genes was repressed in the presence of iron. However, in the fur-deletion mutant cells, the expression of these genes was derepressed under iron-depleted conditions. In the presence of iron, the expression was still repressed, but to a significantly lesser extent than in wild-type cells. To further investigate the binding of Fur to the upstream regions of these two genes, we performed gel-shift assays using purified Fur in the presence or absence of iron. The results showed that Fur only binds to the upstream regions when iron is present (Supplementary Figure S3). These findings suggest that Fur-iron complex plays a role in the regulation of vhuα and vhuβ genes, and its binding is dependent on the availability of iron.
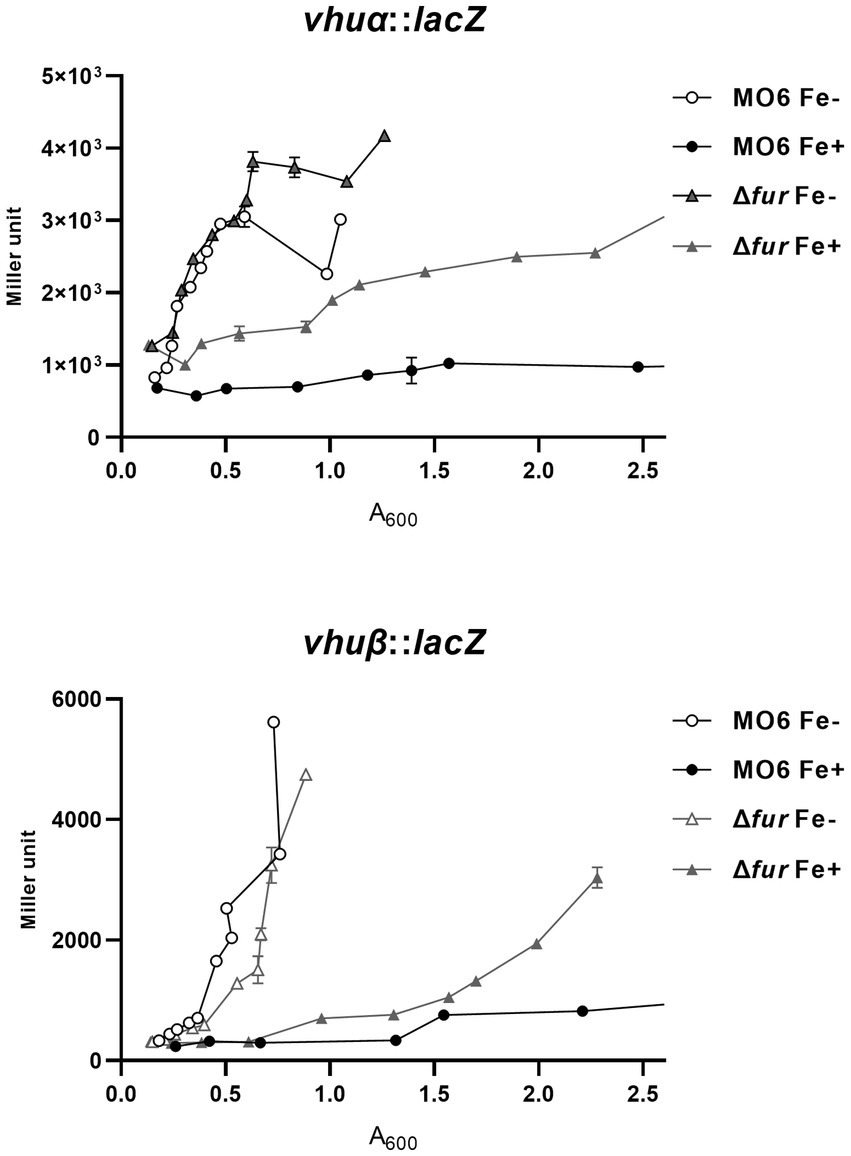
Figure 3. Transcriptional expression of vhuα and vhuβ also is repressed by Fur and iron. The transcription levels of vhuα and vhuβ were measured by β-galactosidase activities using vhuα-lacZ and vhuβ-lacZ transcription reporter in wild-type V. vulnificus, MO6-24/O (circles) and Δfur (fur deletion mutant) (triangles) under iron-limiting (white symbols) or iron-rich (black symbols) conditions. When A600 reached 0.1, 100 μM 2,2′-dipyridyl was added as a chelator. Miller units represent the production of β-galactosidase values normalized to cell density. The error bars denote standard deviations of the results of three independent experiments.
We further investigated the impact of iron on the expression of rpoS, which encodes an alternative sigma factor and is involved in the cFP pathway. The activity of β-galactosidase, measured from a lacZ fusion to rpoS, was found to be strongly repressed by iron in wild-type cells. However, in the fur-deletion mutant, the activity was derepressed regardless of iron availability (Figure 4). To confirm the direct binding of Fur to the upstream region of rpoS, we performed a gel-shift assay, and the results are shown in Supplementary Figure S4.
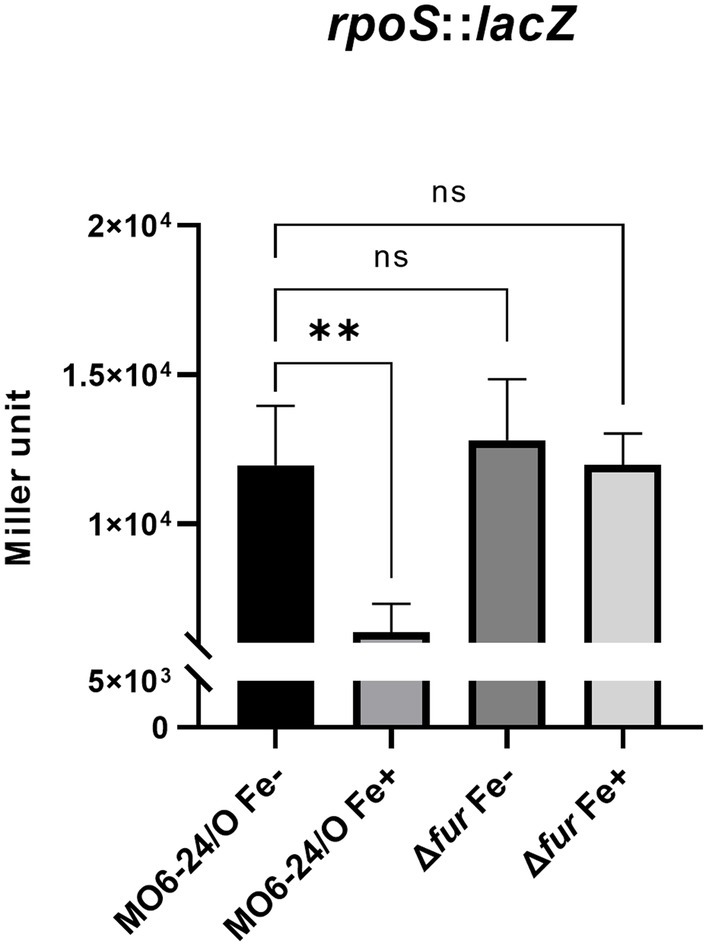
Figure 4. Fur-iron that represses transcription level of rpoS. The transcription level of rpoS were measured by β-galactosidase activities from rpoS-lacZ transcription reporter in wild-type V. vulnificus, MO6-24/O and Δfur under iron-limiting (Fe-) or iron-rich (Fe+) conditions. Miller units represent the production of β-galactosidase values normalized to cell density. The data are average values from three independent experiments, and error bars denote the standard deviations indicated (Student’s t-test; **, 0.005 ≤ p < 0.05; ns, not significant).
Our previous study demonstrated that katG, which encodes a catalase, is a member of the cFP-signaling regulon, and its transcription is regulated by RpoS (Kumamoto and Vukich, 1998; Kim et al., 2013b). Consequently, we investigated the direct regulation of the katG gene by Fur. To assess the impact of Fur on katG transcription, we utilized a katG-lacZ transcription fusion. The β-galactosidase activity from the reporter fusion in the Δfur mutant was higher than that in the wild-type strain under iron-depleted conditions (Figure 5). In the fur-deletion mutant, the expression of katG was derepressed in the absence of iron; however, in the presence of iron, it remained repressed. Fur-iron repression occurs through direct binding to the upstream region of the gene (Supplementary Figure S5).
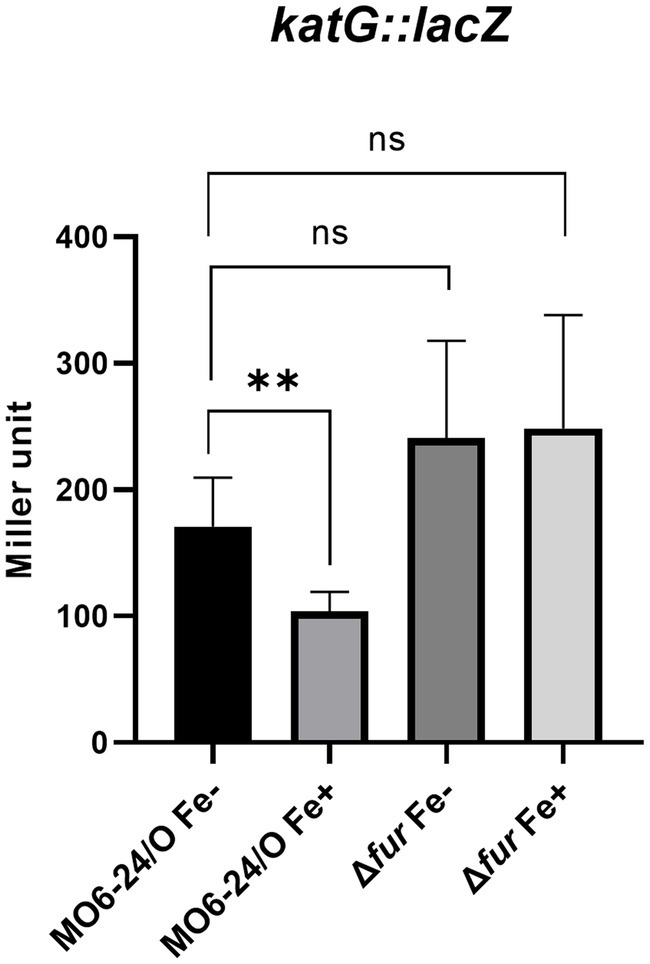
Figure 5. Fur-iron represses the catalase activity of V. vulnificus via regulating the transcription level of katG. The transcription level of katG were measured by β-galactosidase activities from katG-lacZ transcription reporter in wild-type V. vulnificus, MO6-24/O and Δfur (fur deletion mutant) under iron-limiting or iron-rich conditions.
RpoS is an alternative sigma factor known to initiate the transcription of numerous genes, forming a regulon. This suggests that these downstream genes may also be regulated by Fur-iron through modulation of RpoS expression. To investigate this, we measured the expression of three representative genes, aldA (encoding an aldehyde dehydrogenase), vvpE (encoding a metalloprotease), and gabD (encoding a succinate-semialdehyde dehydrogenase), known to be transcribed by RpoS in V. vulnificus (Jeong et al., 2001; Vijayakumar et al., 2004; Kim et al., 2018). The expression levels of these genes were semi-quantitatively measured using qRT-PCR. As we predicted, the results showed that all three genes were repressed by Fur in the presence of iron. In Δfur isotype cells, the repression was relieved, and the introduction of exogenous fur in plasmid restored the repression (Figure 6). The nucleotide sequences in the regions upstream of these three genes did not show any apparent Fur box (Supplementary Figure S7), suggesting that the Fur-dependent repression of these genes was exerted via RpoS.
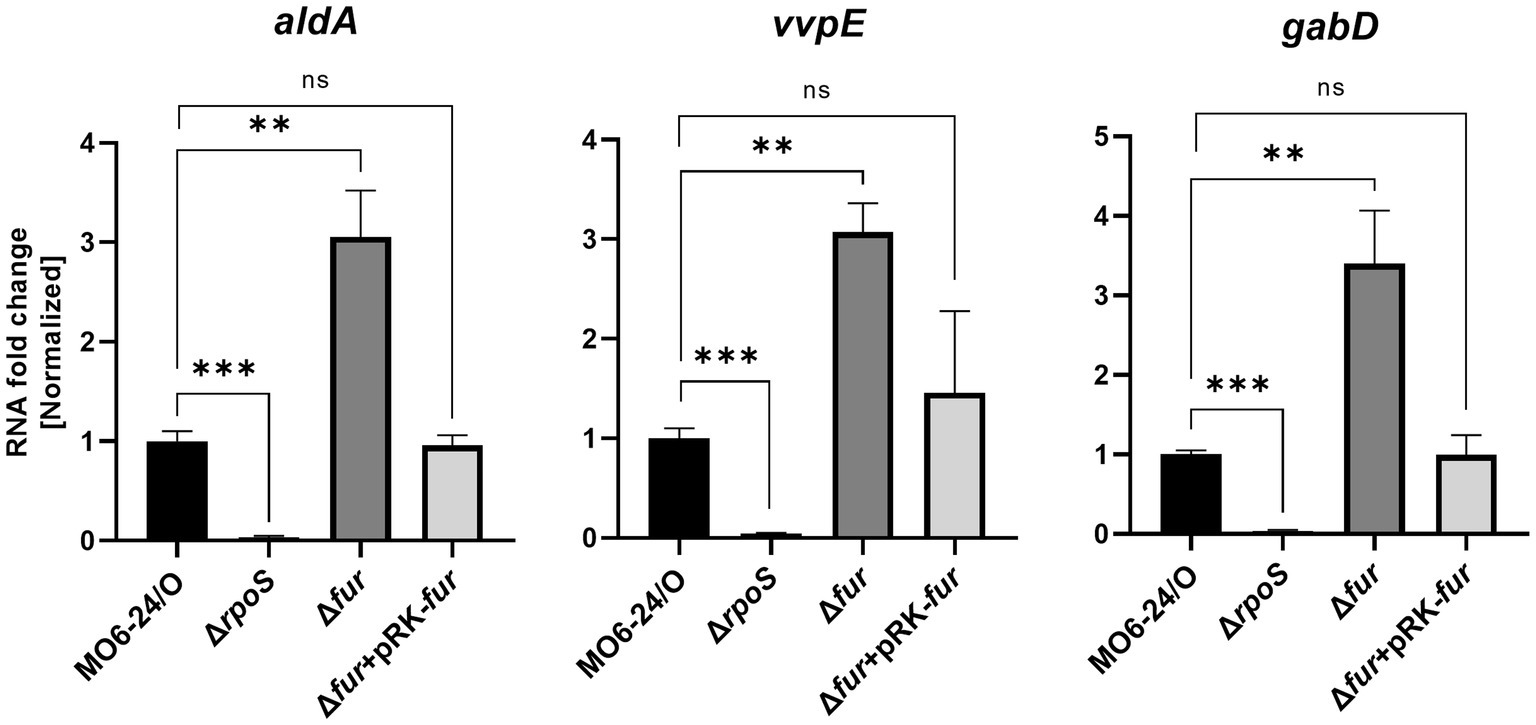
Figure 6. Transcription of the RpoS-inducing genes aldA, vvpE, and gabD is also repressed by Fur. The transcription levels of aldA, vvpE, and gabD in wild-type V. vulnificus MO6-24/O (pRK415), ΔrpoS (pRK415), Δfur (pRK415), and Δfur (pRK-fur) cultured in LB broth as measured by qRT-PCR are shown. RNA levels were quantified using the comparative threshold cycle (ΔCt) method, and RNA-fold change was normalized to the value for MO6-24/O harboring pRK415. Values are averages from three independent experiments, and error bars denote standard deviations. The p-values for comparison with MO6-24/O are indicated (Student’s t-test; ***, 0.001 ≤ p < 0.005; **, 0.005 ≤ p < 0.05; ns, no significant).
We expanded our investigation to examine the production of cFP, which serves as the signal molecule of the ToxR-LeuO-dependent QS system (Kim et al., 2018). To compare the levels of cFP in the culture supernatants, we employed HPLC. We found that the amounts of cFP detected in both wild-type cells and fur-deletion cells did not exhibit a significant difference (Supplementary Figure S6).
The katG gene is a target virulence factor regulated by the cFP-mediated QS pathway. As shown in Figure 6, the transcription of katG, which encodes a catalase, is negatively controlled by Fur-iron. To quantitatively measure catalase activity, we performed assays on MO6-24/O, Δfur, and Δfur(pRK-fur) cells following the procedure outlined in the Materials and methods section. In Δfur cells, a higher level of catalase activity was observed compared to wild-type cells. However, the introduction of the fur gene on a plasmid reduced the activity to the wild-type level (Figure 7). This finding is consistent with the results presented in Figure 5, suggesting that Fur represses the catalase activity of V. vulnificus by regulating katG expression.
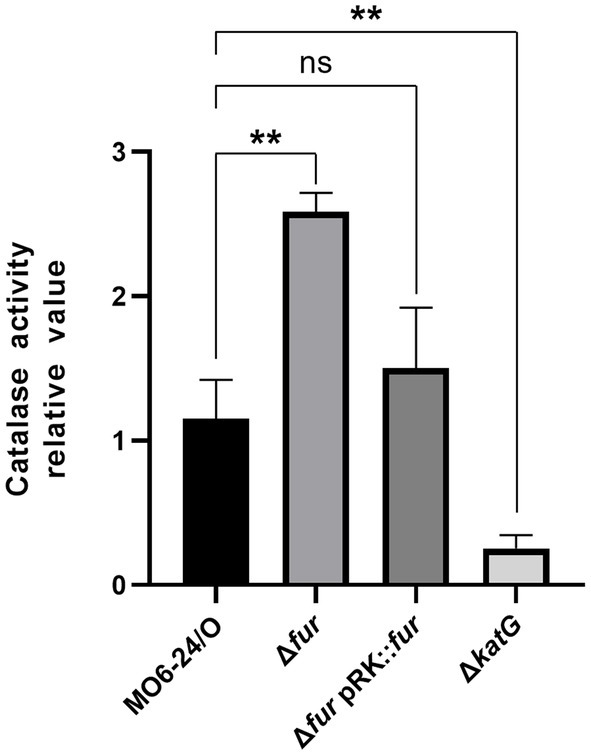
Figure 7. Fur-iron complex inhibits the catalase activity of V. vulnificus. Catalase activity was measured at 440 nm as described in the Materials and Methods section. Values are averages obtained from three independent experiments, and error bars denote standard deviations. The p-values for comparison with MO6-24/O under iron-limiting condition are indicated (Student’s t-test; **, 0.005 ≤ p < 0.05; ns, no significant).
The cFP-mediated QS pathway is also present in the human pathogen V. cholerae (Park et al., 2006; Bina and Bina, 2010; Bina et al., 2013). In this pathogen, LeuO, which receives the cFP signal through ToxR similar to V. vulnificus, positively regulates the expression of ctxAB, the genes encoding the exotoxin cholera toxin, as well as ToxT, which induces the expression of various virulence-associated genes (Silva and Benitez, 2016; Bhandari et al., 2021; Bina and Bina, 2023). To explore whether Fur-iron also regulates these genes involved in the cFP pathway in V. cholerae, we compared the transcription levels of leuO, toxT, and ctxAB in the V. cholerae O1 El Tor strain N16961 and its fur-deletion derivative using qRT-PCR. As shown in Figure 8A, the expression levels of these four genes were significantly higher in the fur-deletion mutant compared to wild-type cells. Additionally, the transcription level of ctxA, which encodes the effector subunit of the cholera toxin, was quantitatively assessed using the luxAB reporter fusion (Figure 8B). Deletion of fur significantly enhanced ctxA transcription, and the introduction of the fur gene in trans complemented the mutant phenotype, confirming the repression of ctx by Fur.
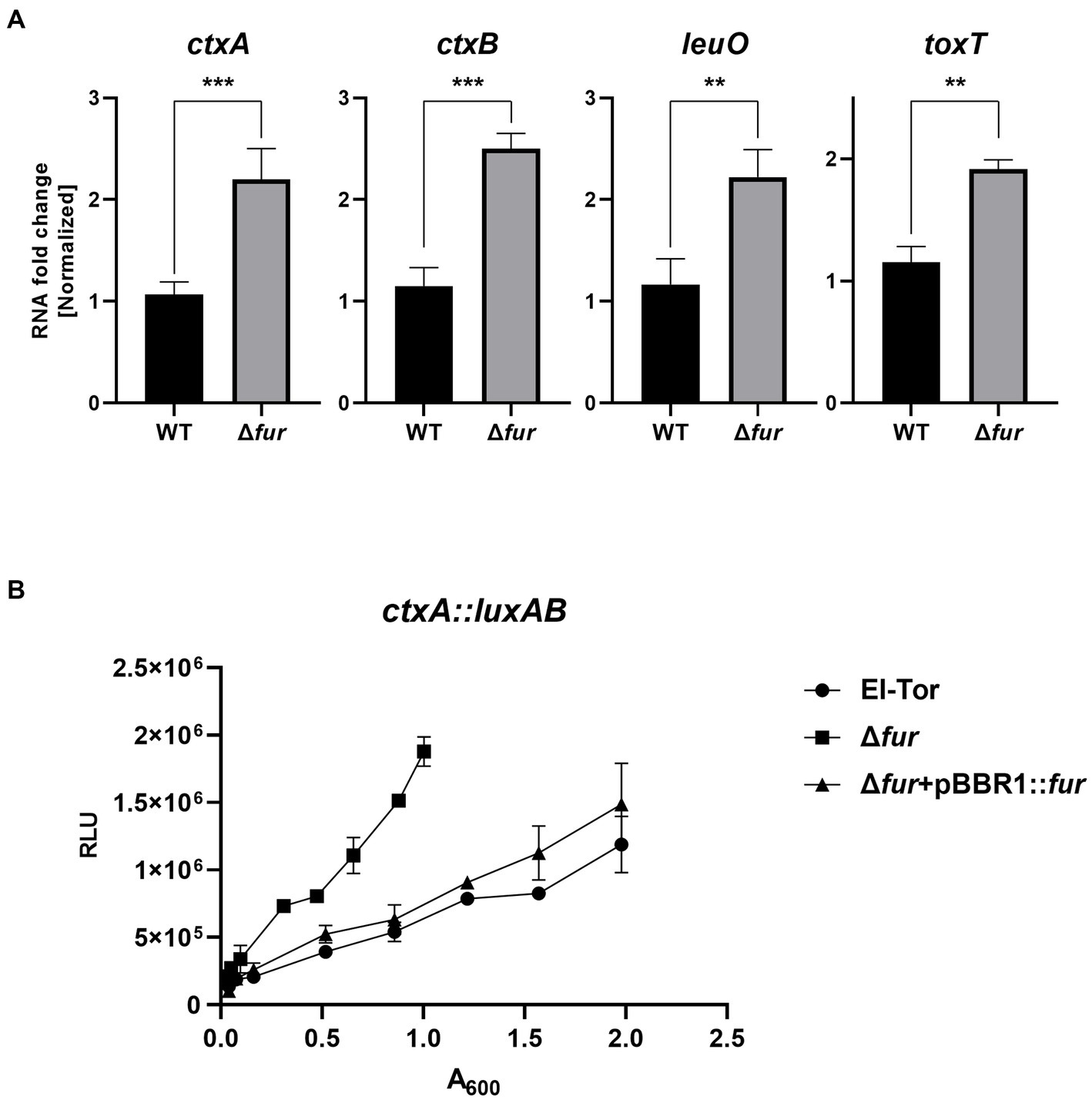
Figure 8. Fur-iron also inhibits leuO and virulence factors in V. cholerae. (A) The transcription levels of ctxAB, leuO, and toxT in V. cholerae (El Tor), and Δfur cultured in LB broth as measured by qRT-PCR are shown. RNA samples obtained during the exponential phase were subjected to qRT-PCR analysis using the primers shown in Supplementary Table S2. The RNA levels were quantified using the comparative threshold cycle (ΔCt) method, and RNA-fold change was normalized to the value for El Tor. The p-values for comparison with El Tor are indicated (Student’s t-test; ***, 0.001 ≤ p < 0.005; **, 0.005 ≤ p < 0.05). (B) The transcriptional activities of ctxA1 were assessed using the ctxA1-luxAB reporter in wild-type V. cholerae, El Tor (pBBR1-mcs2) (circles), Δfur (pBBR1-mcs2) (squares) and Δfur (pBBR1-fur) (triangles). Relative light units (RLU) represent the luminescence values, which were normalized to cell density (A600).
Availability of iron ion is an important environmental factor affecting virulence of pathogenic microorganisms. We have studied QS signal transductions, which play important roles in regulating virulence factors, and various factors affecting the pathways, using pathogenic Vibrio species as model systems. Previous studies demonstrated that iron affects the AI-2-mediated QS signal pathway by regulating the expressions of components associated with the pathway (Wen et al., 2012, 2016; Lee et al., 2022). Iron is related to oxidative stress, and one of the target genes in the cFP-mediated QS signal pathway is katG encoding a catalase. Therefore, we extended our study to the cFP-mediated QS pathway. In this study, we found that iron also affects the expression of regulatory components in the cFP-mediated quorum-sensing pathway, and Fur is a major regulator responsible for the regulation, as is in the case of AI-2 QS.
All the cytoplasmic regulatory components of the cFP QS pathway including the master regulator LeuO, vHUα and β, and RpoS are repressed by Fur-iron, which directly binds to Fur boxes in the upstream region of each of those coding genes. Each of these cytoplasmic component also functions as a regulator forming its own regulon. Therefore, it was expected that the target genes of these regulators would also be indirectly regulated by Fur-iron. The role as a regulator of the alternative sigma factor RpoS has been well described. As such, we examined three target genes known to be downstream genes of RpoS; aldA (aldehyde dehydrogenase), vvpE (metalloprotease), and gabD (succinate-semialdehyde dehydrogenase). These genes, as expected, were regulated by iron in Fur-dependent manner, and the regulatory patterns were similar to that of RpoS. However, none of these three genes has a distinct Fur box near the promotes (data not shown), indicating that the Fur-dependent iron regulation is elicited via RpoS. Among these genes, vvpE has been well studied for its roles as virulence factor (Lee et al., 2016). This protease functions as an elastase that disrupts the tight junctions of human intestinal cells. The expression of vvpE is activated by SmcR, which is the master regulator of AI-2 QS system. Our previous study showed that expression of smcR is also directly repressed by Fur-iron (Kim et al., 2013b). Therefore, it is reasonable to postulate that VvpE, an elastase, contributes to release of iron from host human cells, and the expression of this enzyme is not necessary when iron is available. Unlike above three genes, which are transcribed by RpoS and their expressions are repressed by Fur-iron rather indirectly through RpoS, katG is directly repressed by Fur-iron. As shown in this study, the expression of this gene is also repressed by Fur-iron. The catalase activities from wild-type and fur-deletion isotype cells of V. vulnificus (Figure 7) also confirm this regulation. At the moment, we do not understand biological meaning of the tight repression of katG caused by Fur-iron. One possible scenario is that KatG may be defense mechanism required for the pathogen to protect itself against ROS produced by host cells. By the time infected host cells erupt and release iron, they may have lost the ability to produce ROS, and pathogen would not need KatG anymore. Therefore, the pathogen may tightly regulate the katG expression to save energy. Meanwhile three other RpoS-directed genes may have some other functions regardless of the presence of iron. Further study on biological functions of KatG would enable a more precise interpretation.
We observed that the signal molecule cFP is produced independently of iron. Functions responsible for the biosynthesis of cFP have not yet been elucidated in Vibrio spp., and therefore, we could not confirm this in a molecular genetic level, but HPLC analysis of cFP production clearly indicated that the product of cFP is not affected by either iron or Fur. Our previous study showed that the production of AI-2, which is a signal molecule for another QS pathway in Vibrio spp. is negatively controlled by the master regulator SmcR via repressing the expression of the small RNA RyhB, which is necessary for the full expression of LuxS, the AI-2 biosynthase (Lee et al., 2022). The expression of this small RNA is negatively controlled by Fur-iron. In other words, the expression of AI-2 signal molecule is negatively controlled by Fur-iron as is high cell density. This mechanism enables the feedback regulation of the AI-2-mediated QS pathway, and hence it is promptly shut off when cell density decreases, or when the environmental iron level is too high. Supplementary Figure S8 provides a schematic summary of the Fur-iron-mediated regulatory networks in QS pathways in V. vulnificus. These differences in regulation patterns between biosynthesis of AI-2 and cFP suggest that these two cognate signal pathways may have some different biological functions. It is possible that the AI-2 QS may be more closely related with cell density or iron level than the cFP QS. A clear explanation awaits identification of the functions of cFP biosynthesis and its regulatory aspects.
It is noteworthy that, in the fur-deletion mutant, expression levels of leuO and vhuαβ still appear to be modulated by iron. Expression levels of these genes in the fur-deletion mutant are much higher than in wild-type cells. Nevertheless, in the fur-deletion mutant, the expression is repressed significantly, even though it is still higher than in the wild-type cells (Figures 1, 3). This suggests that there is some unknown factor other than Fur involved in an iron-dependent regulation, and its effect is not as strong as that of Fur. One possible candidate responsible for the iron-dependent repression is the Csr pathway, which is reported to be involved in iron-dependent regulation of toxR in the related species V. cholerae (Mey et al., 2015). We recently found that this pathway is also functional in V. vulnificus, and that the expression is repressed by Fur-iron (unpublished data). Further study would clarify the connection between Csr and cFP QS pathways in terms of iron-dependent regulation.
Cells could repress the expression of all the downstream components just by repressing upstream regulatory components. However, our result apparently showed that Fur-iron directly represses each of the individual components of the downstream genes in the ToxR-dependent QS pathway to achieve a tight regulation in V. vulnificus. We cannot clearly rationalize this phenotype and it is possible that some ‘leaky’ expression of downstream genes may have inadvertent or disadvantageous effects on cells in the presence of iron. Alternatively, the main biological role of the cFP-ToxR dependent regulatory pathway of V. vulnificus may be closely associated with the availability of iron in host body, which is absolutely necessary for the survival of the pathogen. Hence, with iron available, the pathogenic cells may want to save energy by tightly shutting down those components. The pathogenic bacterium may have evolved such a modulation strategy to optimize its own physiology in response to iron levels in a host in various stages of infection. This study together with previous studies once again highlights the importance of iron in pathogenicity of virulent bacteria and the co-relationship between iron and QS regulation.
The original contributions presented in the study are included in the article/Supplementary material, further inquiries can be directed to the corresponding author.
K-WL: Investigation, Methodology, Visualization, Writing – original draft. SK: Investigation, Writing – original draft. SL: Investigation, Writing – original draft. MK: Investigation, Writing – original draft. SS: Investigation, Writing – review & editing. K-SK: Conceptualization, Formal analysis, Funding acquisition, Project administration, Supervision, Validation, Writing – original draft, Writing – review & editing.
The author(s) declare financial support was received for the research, authorship, and/or publication of this article. This work was supported by grants from the National Research Foundation (NRF) of Korea, funded by the Ministry of Science and ICT (2022R1A2C1008958), Republic of Korea.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2023.1273095/full#supplementary-material
cFP, cyclic-phenylalanine-proline; QS, quorum sensing; AI-2, autoinducer-2.
Alice, A. F., Naka, H., and Crosa, J. H. (2008). Global gene expression as a function of the iron status of the bacterial cell: influence of differentially expressed genes in the virulence of the human pathogen Vibrio vulnificus. Infect. Immun. 76, 4019–4037. doi: 10.1128/IAI.00208-08
Balandina, A., Kamashev, D., and Rouviere-Yaniv, J. (2002). The bacterial histone-like protein HU specifically recognizes similar structures in all nucleic acids. DNA, RNA, and their hybrids. J. Biol. Chem. 277, 27622–27628. doi: 10.1074/jbc.M201978200
Bhandari, M., Jennison, A. V., Rathnayake, I. U., and Huygens, F. (2021). Evolution, distribution and genetics of atypical Vibrio cholerae –a review. Infect. Genet. Evol. 89:104726. doi: 10.1016/j.meegid.2021.104726
Bina, X. R., and Bina, J. E. (2010). The cyclic dipeptide cyclo(Phe-pro) inhibits cholera toxin and toxin-coregulated pilus production in O1 El Tor Vibrio cholerae. J. Bacteriol. 192, 3829–3832. doi: 10.1128/JB.00191-10
Bina, X. R., and Bina, J. E. (2023). Vibrio cholerae RND efflux systems: mediators of stress responses, colonization and pathogenesis. Front. Cell. Infect. Microbiol. 13:1203487. doi: 10.3389/fcimb.2023.1203487
Bina, X. R., Taylor, D. L., Vikram, A., Ante, V. M., and Bina, J. E. (2013). Vibrio cholerae ToxR downregulates virulence factor production in response to cyclo(Phe-pro). MBio 4, e00366–e00313. doi: 10.1128/mBio.00366-13
Bradford, M. M. (1976). A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 72, 248–254. doi: 10.1006/abio.1976.9999
Deng, Z., Wang, Q., Liu, Z., Zhang, M., Machado, A. C., Chiu, T. P., et al. (2015). Mechanistic insights into metal ion activation and operator recognition by the ferric uptake regulator. Nat. Commun. 6:7642. doi: 10.1038/ncomms8642
DiRita, V. J., and Mekalanos, J. J. (1991). Periplasmic interaction between two membrane regulatory proteins, ToxR and ToxS, results in signal transduction and transcriptional activation. Cells 64, 29–37. doi: 10.1016/0092-8674(91)90206-E
Ghosh, A., Paul, K., and Chowdhury, R. (2006). Role of the histone-like nucleoid structuring protein in colonization, motility, and bile-dependent repression of virulence gene expression in Vibrio cholerae. Infect. Immun. 74, 3060–3064. doi: 10.1128/IAI.74.5.3060-3064.2006
Grove, A. (2011). Functional evolution of bacterial histone-like HU proteins. Curr. Issues Mol. Biol. 13, 1–12. doi: 10.21775/cimb.013.001
Hadwan, M. H. (2018). Simple spectrophotometric assay for measuring catalase activity in biological tissues. BMC Biochem. 19:7. doi: 10.1186/s12858-018-0097-5
Jeong, H. S., Jeong, K. C., Choi, H. K., Park, K.-J., Lee, K.-H., Rhee, J. H., et al. (2001). Differential expression of Vibrio vulnificus elastase gene in a growth phase-dependent manner by two different types of promoters. J. Biol. Chem. 276, 13875–13880. doi: 10.1074/jbc.M010567200
Kim, I. H., Kim, S.-Y., Park, N.-Y., Wen, Y., Lee, K.-W., Yoon, S.-Y., et al. (2018). Cyclo-(L-Phe-L-pro), a quorum-sensing signal of Vibrio vulnificus, induces expression of hydroperoxidase through a ToxR-LeuO-HU-RpoS signaling pathway to confer resistance against oxidative stress. Infect. Immun. 86, e00932–e01017. doi: 10.1128/IAI.00932-17
Kim, I. H., Son, J.-S., Wen, Y., Jeong, S.-M., Min, G.-Y., Park, N.-Y., et al. (2013a). Transcriptomic analysis of genes modulated by cyclo(L-phenylalanine-L-proline) in Vibrio vulnificus. J. Microbiol. Biotechnol. 23, 1791–1801. doi: 10.4014/jmb.1308.08068
Kim, I. H., Wen, Y., Son, J.-S., Lee, K.-H., and Kim, K.-S. (2013b). The Fur-iron complex modulates expression of the quorum-sensing master regulator, SmcR, to control expression of virulence factors in Vibrio vulnificus. Infect. Immun. 81, 2888–2898. doi: 10.1128/IAI.00375-13
Kumamoto, K. S., and Vukich, D. J. (1998). Clinical infection of Vibrio vulnificus: a case report and review of the literature. J. Emerg. Med. 16, 61–66. doi: 10.1016/s0736-4679(97)00230-8
Lee, S.-J., Jung, Y. H., Ryu, J. M., Jang, K. K., Choi, S. H., and Han, H. J. (2016). VvpE mediates the intestinal colonization of Vibrio vulnificus by the disruption of tight junctions. Int. J. Med. Microbiol. 306, 10–19. doi: 10.1016/j.ijmm.2015.10.006
Lee, K.-W., Wen, Y., Park, N.-Y., and Kim, K.-S. (2022). Quorum sensing and iron-dependent coordinated control of autoinducer-2 production via small RNA RyhB in Vibrio vulnificus. Sci. Rep. 12:831. doi: 10.1038/s41598-021-04757-9
Litwin, C. M., and Calderwood, S. B. (1993). Role of iron in regulation of virulence genes. Clin. Microbiol. Rev. 6, 137–149. doi: 10.1128/Cmr.6.2.137-149.1993
Massé, E., and Arguin, M. (2005). Ironing out the problem: new mechanisms of iron homeostasis. Trends Biochem. Sci. 30, 462–468. doi: 10.1016/j.tibs.2005.06.005
Mey, A. R., Butz, H. A., and Payne, S. M. (2015). Vibrio cholerae CsrA regulates ToxR levels in response to amino acids and is essential for virulence. MBio 6, e01064–e01115. doi: 10.1128/mBio.01064-15
Miller, J. (1972). Experiments in molecular genetics: assay of β-galactosidase. Cold Spring Harb. Lab., 352–355.
Park, N.-Y., Cho, Y. B., Kim, O. B., and Kim, K.-S. (2020a). Cyclo(Phe-pro) produced by Vibrio species passes through biological membranes by simple diffusion. Appl. Microbiol. Biotechnol. 104, 6791–6798. doi: 10.1007/s00253-020-10646-4
Park, N.-Y., Kim, I. H., Wen, Y., Lee, K.-W., Lee, S., Kim, J. A., et al. (2019). Multi-factor regulation of the master modulator LeuO for the cyclic-(Phe-pro) signaling pathway in Vibrio vulnificus. Sci. Rep. 9:20135. doi: 10.1038/s41598-019-56855-4
Park, D.-K., Lee, K.-E., Baek, C.-H., Kim, I. H., Kwon, J.-H., Lee, W. K., et al. (2006). Cyclo(Phe-pro) modulates the expression of ompU in Vibrio spp. J. Bacteriol. 188, 2214–2221. doi: 10.1128/JB.188.6.2214-2221.2006
Park, N.-Y., Lee, K.-W., and Kim, K.-S. (2020b). H-NS silences gene expression of LeuO, the master regulator of the cyclic(Phe-pro)-dependent signal pathway, in Vibrio vulnificus. J. Microbiol. Biotechnol. 30, 830–838. doi: 10.4014/jmb.2001.01021
Picker, M. A., and Wing, H. J. (2016). H-NS, its family members and their regulation of virulence genes in Shigella species. Genes (Basel) 7:112. doi: 10.3390/genes7120112
Schaible, U. E., and Kaufmann, S. H. E. (2004). Iron and microbial infection. Nat. Rev. Microbiol. 2, 946–953. doi: 10.1038/nrmicro1046
Silva, A. J., and Benitez, J. A. (2016). Vibrio cholerae biofilms and cholera pathogenesis. PLoS Negl. Trop. Dis. 10:e0004330. doi: 10.1371/journal.pntd.0004330
Troxell, B., and Hassan, H. M. (2013). Transcriptional regulation by ferric uptake regulator (Fur) in pathogenic bacteria. Front. Cell. Infect. Microbiol. 3:59. doi: 10.3389/fcimb.2013.00059
Vasil, M. L., and Ochsner, U. A. (1999). The response of Pseudomonas aeruginosa to iron: genetics, biochemistry and virulence. Mol. Microbiol. 34, 399–413. doi: 10.1046/j.1365-2958.1999.01586.x
Vijayakumar, S. R. V., Kirchhof, M. G., Patten, C. L., and Schellhorn, H. E. (2004). RpoS-regulated genes of Escherichia coli identified by random lacZ fusion mutagenesis. J. Bacteriol. 186, 8499–8507. doi: 10.1128/JB.186.24.8499-8507.2004
Waldron, K. J., and Robinson, N. J. (2009). How do bacterial cells ensure that metalloproteins get the correct metal? Nat. Rev. Microbiol. 7, 25–35. doi: 10.1038/nrmicro2057
Wen, Y., Kim, I. H., and Kim, K.-S. (2016). Iron- and quorum-sensing signals converge on small quorum-regulatory RNAs for coordinated regulation of virulence factors in Vibrio vulnificus. J. Biol. Chem. 291, 14213–14230. doi: 10.1074/jbc.M116.714063
Keywords: Vibrio vulnificus, cyclo-(L-Phe-L-Pro), quorum-sensing, iron, Fur, virulence factors
Citation: Lee K-W, Kim S, Lee S, Kim M, Song S and Kim K-S (2023) Iron-Fur complex suppresses the expression of components of the cyclo-(Phe-Pro)-signaling regulatory pathway in Vibrio vulnificus. Front. Microbiol. 14:1273095. doi: 10.3389/fmicb.2023.1273095
Received: 05 August 2023; Accepted: 18 September 2023;
Published: 03 October 2023.
Edited by:
Stefan Schild, University of Graz, AustriaReviewed by:
Ryan Kenton, University of Portland, United StatesCopyright © 2023 Lee, Kim, Lee, Kim, Song and Kim. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Kun-Soo Kim, a3NraW1Ac29nYW5nLmFjLmty
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.