
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Microbiol. , 04 January 2024
Sec. Food Microbiology
Volume 14 - 2023 | https://doi.org/10.3389/fmicb.2023.1272892
This article is part of the Research Topic Microbial Food Safety in Retail Stores and Restaurants View all 8 articles
Introduction: Salmonella infections have been intensely increasing and becoming a universal public health crisis. This study investigated the prevalence of Salmonella in organic and non-organic chickens and the antimicrobial resistance profiles and virulence genes (invA, pagC, and spvC) in recovered Salmonella isolates.
Methods: Whole chicken carcasses [organic (n = 240) and non-organic (n = 240)] were obtained monthly for 1 year (n = 480) from a retail store on the Eastern Shore of Maryland. Salmonella isolation and identification were conducted by following the whole carcass enrichment method recommended by USDA-FSIS. Confirmed Salmonella isolates (organic n = 76; non-organic n = 137) were serotyped and tested for antibiotic susceptibility and virulence genes using standard methods.
Results: Forty-nine percent (237/480) of the carcasses were positive for Salmonella. Organic and non-organic positivity rates were 37.1 and 61.8%, respectively. A significantly higher Salmonella contamination was observed in non-organic chickens (p < 0.05). The most common serovars were Salmonella Kentucky (47%), S. Infantis (35%), S. Enteritidis (6%), S. Typhimurium (5%), and S. Blockley (4%). Isolates were frequently resistant to at least one antibiotic (91.24%) or multidrug resistant (45.54%). Resistance was observed to tetracycline (82.8%), minocycline (42.3%), nitrofurantoin (40.3%), cefazolin (38.3%), ampicillin (32.1%), and ceftriaxone (26%). All isolates were susceptible to fluoroquinolone, carbapenem, and glycylcycline. The majority of isolates (99.1%) possessed at least one of three virulence genes of concern and 4.2% tested positive for all three. Ninety-five, 89, and 6.6% of isolates contained invA, pagC, and spvC genes, respectively. The spvC gene was not detected in serovars recovered from organic chickens though 92% and 82% of isolates were positive for invA and pagC. The frequency of Salmonella recovered from non-organic chickens possessing invA, pagC, and spvC genes were 97.1, 89.8, and 10.2%, respectively. Detection of invA and pagC genes showed no significant difference (p > 0.05) between organic and non-organic chickens but a significantly higher spvC gene (p < 0.05) was detected in non-organic chickens due to the majority of S. Enteritidis (92.3%) exclusively recovered from non-organic chicken carried spvC gene.
Discussion: This study reveals a high prevalence of Salmonella in both organic and non-organic chickens, which exhibit resistance to vital antibiotics and carry virulence genes, thereby creating a potential risk of salmonellosis.
Among foodborne pathogens, Salmonella is recognized as a prominent and hazardous foodborne pathogen which is often associated with chickens and the causative agent of the zoonotic disease salmonellosis. Salmonellosis is the second leading foodborne illness in the United States, after norovirus infection (FDA, 2020). In the United States, Salmonella is responsible for approximately 1.35 million illnesses, 26,500 hospitalizations, and 420 deaths per annum (CDC, 2023). People get sick by consuming undercooked chicken/poultry products and any other foods that are contaminated by raw chicken or its juices (CDC, 2022). Pathogenesis of Salmonella requires the action of multiple virulence factors including invA, pagC, and spvC to invade, survive, and proliferate in the host. Salmonella containing these virulence genes have the potential to cause human illness (Olah et al., 2005; Mohamed et al., 2014). Therefore, investigating virulence factors is crucial for understanding and preventing Salmonella-induced foodborne illnesses.
Several poultry-related Salmonella outbreaks have been occurring every year in the United States (Punchihewage-Don et al., 2022). Antimicrobial resistance of Salmonella is becoming a significant concern for public health in the United States (CDC, 2019; Marchello et al., 2020). Antimicrobials can mitigate the outcome of an infection by destroying or suppressing the growth of pathogens (bacteria, parasites, viruses, and fungi). Antibiotics are medicines that can be used to treat bacterial infections by killing the bacteria (bactericidal) or preventing their multiplication (bacteriostatic) (Punchihewage-Don et al., 2022; World Health Organization [WHO], 2023). Patients with severe Salmonella infections are treated with fluoroquinolones (ciprofloxacin), third-generation cephalosporins (ceftriaxone), and macrolides (azithromycin); however, fewer antibiotics are available to treat these severe cases because of increasing AMR. During the past decade, an increasing trend of resistance to medically important antibiotics such as ceftriaxone, ciprofloxacin (non-susceptible), and azithromycin (decreased susceptibility) in non-typhoid Salmonella was observed (CDC, 2018, 2019).
Despite the risk of Salmonella contamination, chicken is the most frequently consumed meat in the United States (CDC, 2022). According to the National Organic Program (NOP), organically raised chickens must be fed 100% organic feed and no usage of antibiotics, added growth hormones, mammalian or avian byproducts, or other prohibited feed ingredients. Organically raised chickens should also have year around access to the outdoors except during inclement weather conditions (eCFR, 2000; Bailey and Cosby, 2005; USDA-Agricultural Marketing Service, 2013, 2015). In addition, chickens that are treated with antibiotics are prohibited from being sold under the “organic” label (eCFR, 2000). On the other hand, non-organic (conventional) chicken production is a more common chicken farming practice than organic farming and the majority (99%) of the total poultry production in the U.S. comes from non-organic farms (University of Georgia Extension, 2022). Non-organic chickens are fed commercial feed that may contain antimicrobials and dietary supplements. According to the poultry production and value summary, in 2021 more than five billion pounds of broiler chickens were produced in Delaware, Maryland, and Virginia (Delmarva). This is approximately 9% of the total chicken production in the United States and worth over $2.7 billion (USDA-National Agricultural Statistics Service, 2022). The Delmarva peninsula is recognized as the pioneer of chicken processing because the first broiler processing plant in the United States was established in Delaware in 1937 (Constance, 2008).
In order to mitigate foodborne illnesses caused by Salmonella, reducing Salmonella in food and monitoring the prevalence of resistant strains are important. A few studies were conducted on the prevalence and antimicrobial resistance of Salmonella in chicken a few years ago (Parveen et al., 2007; Mazengia et al., 2014; Nguyen et al., 2016). However, little information is available on the prevalence and antimicrobial resistance of Salmonella in organic and non-organic chickens (Mak et al., 2022) on the Eastern Shore of Maryland. Therefore, this study was undertaken to determine the prevalence of Salmonella in organic and non-organic chickens and investigate antimicrobial resistance profiles and virulence properties of recovered isolates.
The sample collection and processing were carried out according to USDA-FSIS (2019) from March 2019 to February 2020. Whole chicken carcasses (organic and non-organic) were obtained monthly from a retail store in the Eastern Shore area, Maryland. A retail store was selected based on the affordability and availability of whole chicken carcasses. During each sampling, 20 organic and 20 non-organic carcasses were collected. These organic and non-organic chickens belonged to two different brands based on their farming practices. All samples were placed in coolers with ice and transported to the food microbiology laboratory at the University of Maryland Eastern Shore within an hour of collection. All the bacterial media utilized in this study were purchased from Becton Dickinson, Sparks, MD, USA unless otherwise specified.
In brief, each carcass was placed in a 4 L sterile plastic stomacher bag (Thermo Fisher Scientific, Hampton, NH, USA). Then sterile buffered peptone water (500 mL) (BPW) was added to the interior and exterior surfaces of each carcass, and the carcass in the bag was shaken vigorously for 60 s. The bag containing the whole carcass and rinse solution were incubated at 37°C for 24 h. Salmonella enterica serovar Typhimurium (H2S positive) was used as a positive control and sterile BPW was used as a negative control with each batch of samples. After incubation, the sample was screened for Salmonella using the BAX system, a commercial PCR-based system (Qualicon Diagnostic, Camarillo, CA, USA). The BAX testing was carried out according to the manufacturer’s guidelines. Samples positive for Salmonella were added (0.1 mL) into 10 mL of Rappaport Vassiliadis (RV) broth tubes and the tubes were incubated at 42°C for 24 h. Enriched samples were streaked onto Xylose Lysine agar supplemented with Tergitol 4 (XLT4) and incubated at 37°C for 24 h. After incubation, isolated presumptive Salmonella colonies (black centered) were randomly selected and inoculated into Tryptic Soy Broth (TSB) and incubated at 37°C for 24 h to preserve them for further analysis. The incubated samples were centrifuged at 5,000 rpm for 5 min. The remaining pellet was resuspended in TSB with 25% glycerol and stored at −80°C for further analysis (Parveen et al., 2007). Presumptive Salmonella isolates were biochemically confirmed using triple sugar iron agar (TSI) and lysine iron agar (LIA) slants (Andrews et al., 2023).
All Salmonella isolates were serotyped using standard methods at the USDA National Veterinary Services Laboratories (NVSL). Briefly, the isolates were subjected to molecular typing using the xMAP Salmonella serotyping assay and classical serotyping using standardized animal antisera to test for the lipopolysaccharide (O antigen) and the flagellar proteins (H antigens) in accordance with the methods described by Edwards and Ewing (Ewing, 1986). Then, the serotypes were designated according to the Kauffmann-White Scheme (Grimont and Weill, 2007).
Salmonella isolates were tested for antimicrobial susceptibilities to a panel of 24 antimicrobials of veterinary and human health importance using Sensititre® antimicrobial susceptibility plates following the manufacturer’s instructions (Thermo Fisher Scientific, Hampton, NH, USA). Escherichia coli ATCC 25922, and Pseudomonas aeruginosa ATCC 27853 were used as controls. The Minimum Inhibitory Concentrations (MICs) were determined as the lowest concentration of an antimicrobial that completely inhibits the growth of bacteria according to the Clinical and Laboratory Standards Institute [CLSI] (2016). The types and ranges of concentrations of the antibiotics are shown in Table 1.
DNA extraction of the preserved isolates was done using the InstaGene matrix DNA kit (Bio-Rad, PA, USA) following the manufacturer’s instructions. Briefly, 1–3 confirmed isolated Salmonella colonies were suspended with 200 μl of InstaGene matrix and incubated for 30 min at 56°C. The incubated mixture was vortexed for 30 s and proceeded to another incubation at 100°C for 8 min on a heated block. Then, the DNA was separated via centrifugation at 13,200 rpm for 3 min. The supernatant was stored at −20°C for further experiment after measuring DNA concentrations using the Qubit dsDNA HS Assay Kit on a Qubit 3.0 fluorometer (Fisher Scientific, Hampton, NH, USA).
To determine the presence of the invA, pagC, and spvC genes a set of primers, shown in Table 2, was used according to Pulkkinen and Miller (1991), Rahn et al. (1992) and Suzuki et al. (1994), respectively. The 50 μL PCR master mix consists of a DNA template (2 μL), deoxynucleotide triphosphates (dATP, dGTP, dCTP, dTTP) at a concentration of 0.25 mM each, MgCl2 (2.5 mM), primer (50 pmol/μL), Taq DNA polymerase (1 U), 1 X PCR buffer and distilled water. The amplification parameters were carried out as shown in Table 2 using a PCR system (Applied Biosystems, CA, USA). Thirty cycles of 94°C for 1 min were run to complete the amplification. The amplicon sizes of invA, pagC, and spvC genes were 284, 318, and 400 base pairs, respectively. For all reactions, S. Typhimurium strain LT2 × 3324 containing a recombinant plasmid with invA, E. coli DH5-α containing a recombinant plasmid with spvC, and S. Typhimurium SR 11 × 3337 containing a recombinant plasmid with pagC were used as positive controls, while Escherichia coli DH5-α (Invitrogen, Carlsbad, CA, USA) was used as a negative control (Olah et al., 2005; Mohamed et al., 2014). The PCR products were separated by electrophoresis in a 1% agarose gel and the gels were stained with GelRed™ (Biotium, Fremont, CA, USA) and viewed with UV light to determine the existence of the PCR products.
A Supplementary table, which displays the collection month, serovar, AMR profiles and presence of virulence genes for each isolate, can be found in Supplementary Table 1.
The statistical significance of differences in the prevalence of Salmonella in organic and non-organic chickens, differences in prevalence of invA, pagC, and spvC genes, and differences in resistance rate between Salmonella serovars and chicken types for each antimicrobial agent tested were determined using Fisher’s exact test. An alpha level of 0.05 was considered the minimum level for statistical significance and, consistent with a per-comparison error rate control approach, p-values where unadjusted for the total number of pairwise comparisons. All statistical analyses were performed using R version 4.0.2 (R Core Team, 2021).
A total of 480 whole chicken carcasses (240 organic and 240 non-organic) were collected during the sampling period (March 2019 to February 2020). Out of 480 whole chicken carcasses, 237 carcasses (49.38%) tested positive for Salmonella through molecular screening of primary enrichment. Subsequently, 213 Salmonella isolates were cultured, confirmed, and subjected to further testing. Twenty-four Salmonella isolates that did not produce typical black-centered colonies during culture confirmation were not chosen for further experiments. According to the present results, 89 (37.08%) and 148 (61.67%) of organic and non-organic chicken carcasses were positive for Salmonella, respectively. The results indicated a significantly higher Salmonella contamination among non-organic chickens compared to the organic chickens (p < 0.05). Contrary to the present results other investigators reported a higher rate of Salmonella prevalence in organic chickens (Bailey and Cosby, 2005; Cui et al., 2005). In addition, Lestari et al. (2009) did not find a significant difference between the prevalence of Salmonella in organic and non-organic chickens isolated from Louisiana retail stores. Compared to Lestari et al. (2009), the present study has a higher detection rate of Salmonella. This might be due to the use of the USDA-FSIS recommended Whole Carcass Enrichment method (WCE). In this method, the entire carcass is subjected to incubation for 24 h at 37°C after vigorously mixing with primary enrichment buffer (BPW) and it helps to proliferate loosely and firmly attached Salmonella and increase detection rate (Cox et al., 2014; USDA-FSIS, 2019). A previous study that used the WCE method for the detection of Salmonella also showed a high prevalence of Salmonella in chickens (Parveen et al., 2007).
Figure 1 shows the prevalence of Salmonella in organic and non-organic chicken carcasses during the sampling period. Throughout the 1-year survey period, the prevalence of Salmonella fluctuated widely. Comparatively higher rates were observed in the months of March, April and May 2019 in both types of chickens. Thereafter, the prevalence of Salmonella in organic chickens was significantly lower and fluctuated during the rest of the sampling period. In the months of July and October, no Salmonella was detected in organic chickens and in the months of August, November 2019 and January 2020, only one sample was positive for Salmonella in each month. In the case of non-organic chickens, significantly lower Salmonella prevalence was observed in the months of June to October 2019 and then significantly higher Salmonella prevalence was observed in the months of November 2019 to January 2020. In addition, the month of July was the lowest Salmonella prevalence (four positive carcasses) recorded in the entire sampling period among non-organic chickens. However, no seasonal effects with regard to Salmonella prevalence on chicken carcasses were observed during the study period. These results are consistent with findings reported by Parveen et al. (2007) and Lestari et al. (2009) who did not find a correlation between Salmonella prevalence and the season/month of the year.
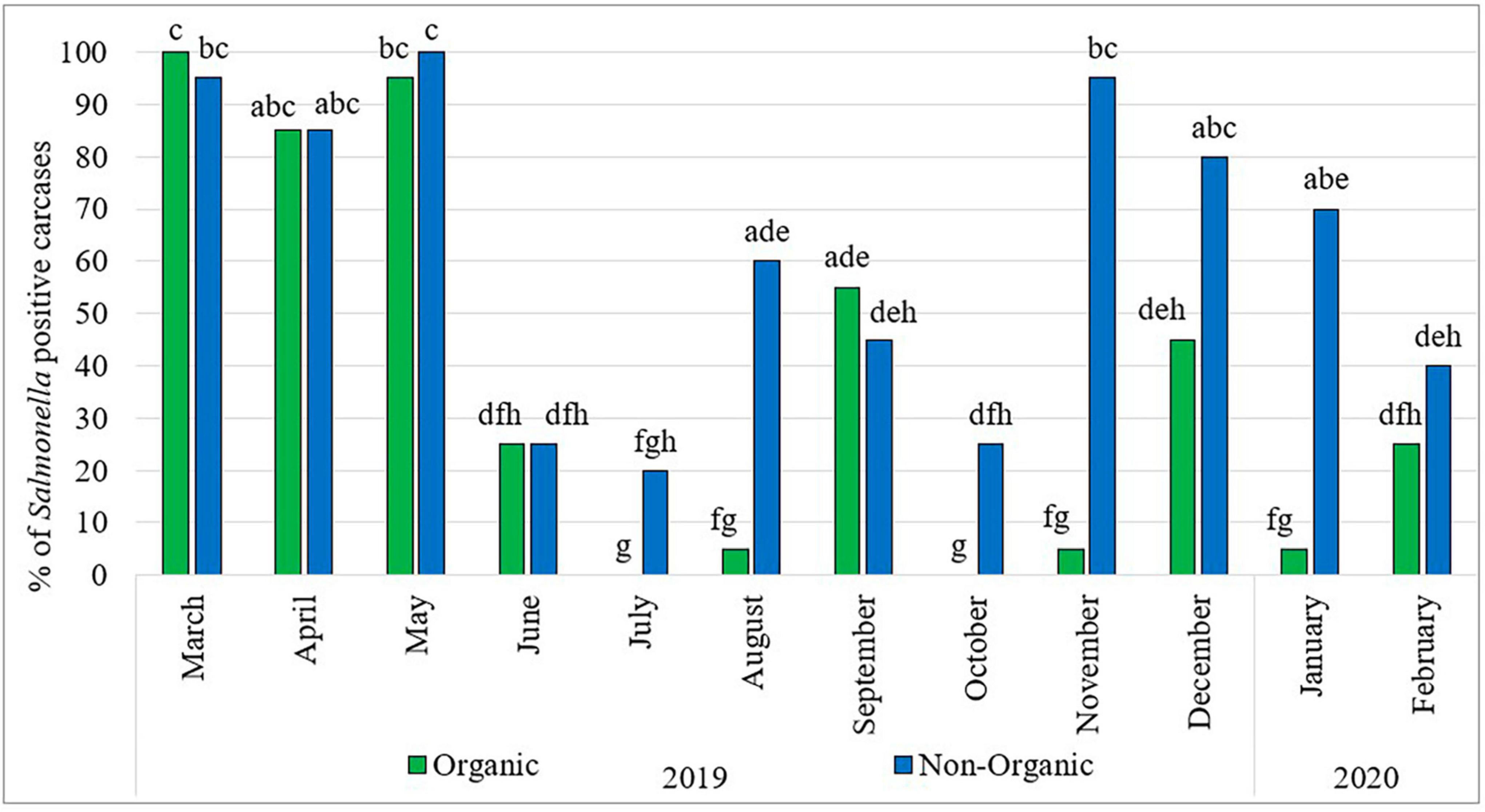
Figure 1. Prevalence of Salmonella in organic and non-organic chicken carcasses from March 2019 to February 2020. The Percentage values with different lowercase letters (a-h) are significantly different (p < 0.05) amongst comparisons between different months and types of chicken.
According to the serotyping results of 213 isolates (Table 3), the top five Salmonella serovars were S. Kentucky (46.95%), S. Infantis (34.27%), S. Enteritidis (6.1%), S. Typhimurium (5.1%), and S. Blockley (5.1%). Table 3 also shows the distribution of Salmonella serovars recovered from organic and non-organic chicken carcasses. Similar to the present results, Cui et al. (2005) and Lestari et al. (2009) also reported that S. Kentucky was the dominant serovar. A previous study conducted by our lab in 2007 to observe Salmonella prevalence in pre- and post-chill broiler carcasses also reported that the predominant serovar was S. Kentucky followed by S. Typhimurium (Parveen et al., 2007). Furthermore, S. Enteritidis was associated only with non-organic chickens while S. Blockley was recovered only from organic chickens. In addition, S. Typhimurium was more prevalent in organic chickens (10.53%) than in non-organic chickens (2.19%). One of the reasons for the higher S. Kentucky and S. Infantis observation throughout the year may be the cross contaminations that occurred during the processing of chicken carcasses (Parveen et al., 2007). According to the USDA-FSIS (2023), S. Infantis has shown an increasing trend in chicken and has emerged as one of the top serotypes in both cecal and product samples. In addition, Siceloff et al. (2022) reported that the prevalence of S. Infantis exceeds that of S. Kentucky and becoming a predominant Salmonella serovar in this region since 2019. Siceloff et al. (2022) also hypothesized that somehow climate or environmental factors are promoting the colonization of poultry by S. Infantis or suppressing the growth of other serovars such as S. Kentucky. This explains the higher prevalence of S. Infantis occurrence in the present study, even though S. Kentucky was dominant among the Salmonella collection. However, the association of specific serovars with poultry is not fully understood yet.
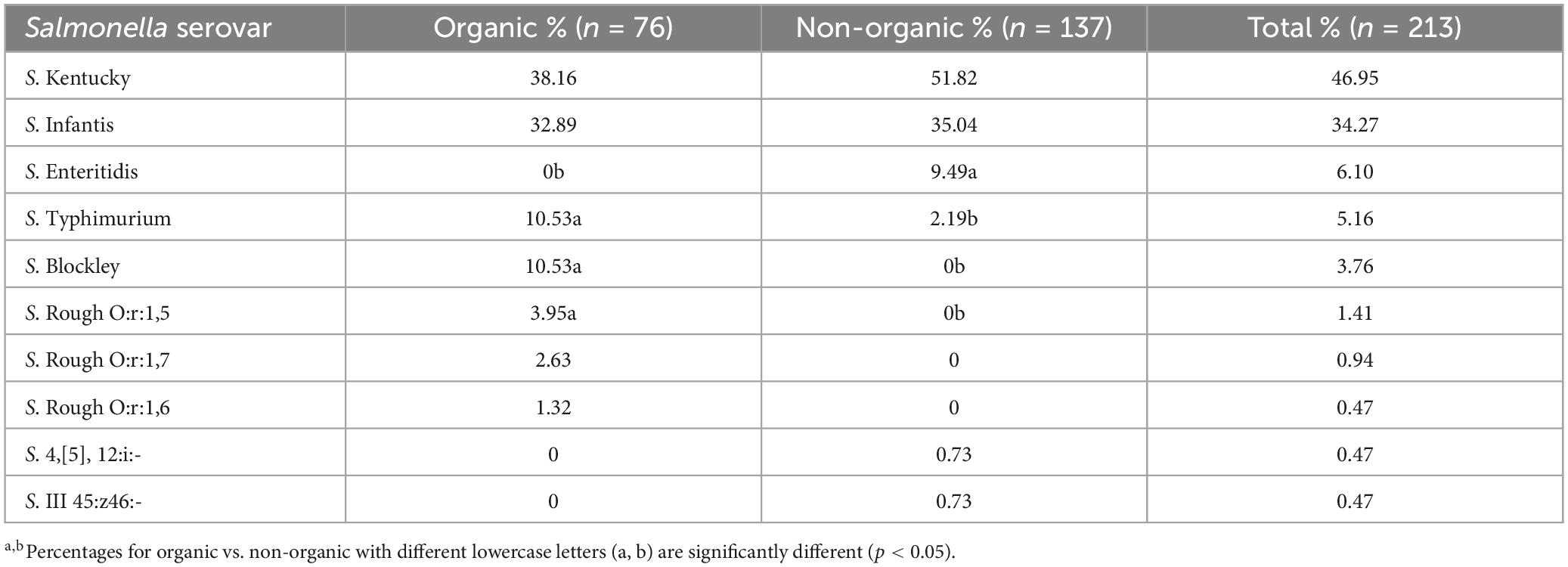
Table 3. Distribution of Salmonella serovars recovered from organic and non-organic chicken carcasses.
Out of 213 Salmonella isolates, 91.24% were resistant to at least one antibiotic and 8.76% of Salmonella isolates were susceptible to all tested antibiotics (Figure 2). Five isolates in each type of chicken showed dose-dependent susceptibility (SDD) to cefepime. Intermediate resistance was observed in 15.78% of isolates recovered in organic chickens [tobramycin (n = 1), cefazolin (n = 1), and aztreonam (n = 10)] and 15.32% of isolates recovered in non-organic chickens [piperacillin/tazobactam constant (n = 1), ceftazidime (n = 6), ceftriaxone (n = 2), cefazolin (n = 3), and aztreonam (n = 9)]. Salmonella isolates were tested for susceptibility to 24 antimicrobial agents belonging to 10 antimicrobial classes that are often prescribed in veterinary and human health. The resistance was often observed to tetracycline (82.8%), minocycline (42.3%), nitrofurantoin (40.3%), cefazolin (38.3%), and ampicillin (32.1%). In this study, ceftriaxone resistant isolates were observed (26%). But all the isolates were susceptible to ciprofloxacin. The frequency of resistant to ceftriaxone in Salmonella isolates recovered from the organic and non-organic chickens was 31.6 and 24.1%, respectively. In addition, the frequency of resistance to tobramycin, ampicillin-sulbactam, trimethoprim/sulfamethoxazole, gentamicin, aztreonam, and ceftazidime was 23.5, 11.9, 11.5, 11.0, 10.2, and 3.4%, respectively. Eight percent of Salmonella isolates recovered from non-organic chickens were susceptible to all tested antibiotics compared to 10.5% of Salmonella isolates recovered from organic chickens. All isolates were susceptible to antibiotic classes of fluoroquinolone, carbapenem, and glycylcycline regardless of the types of chickens (Table 4).
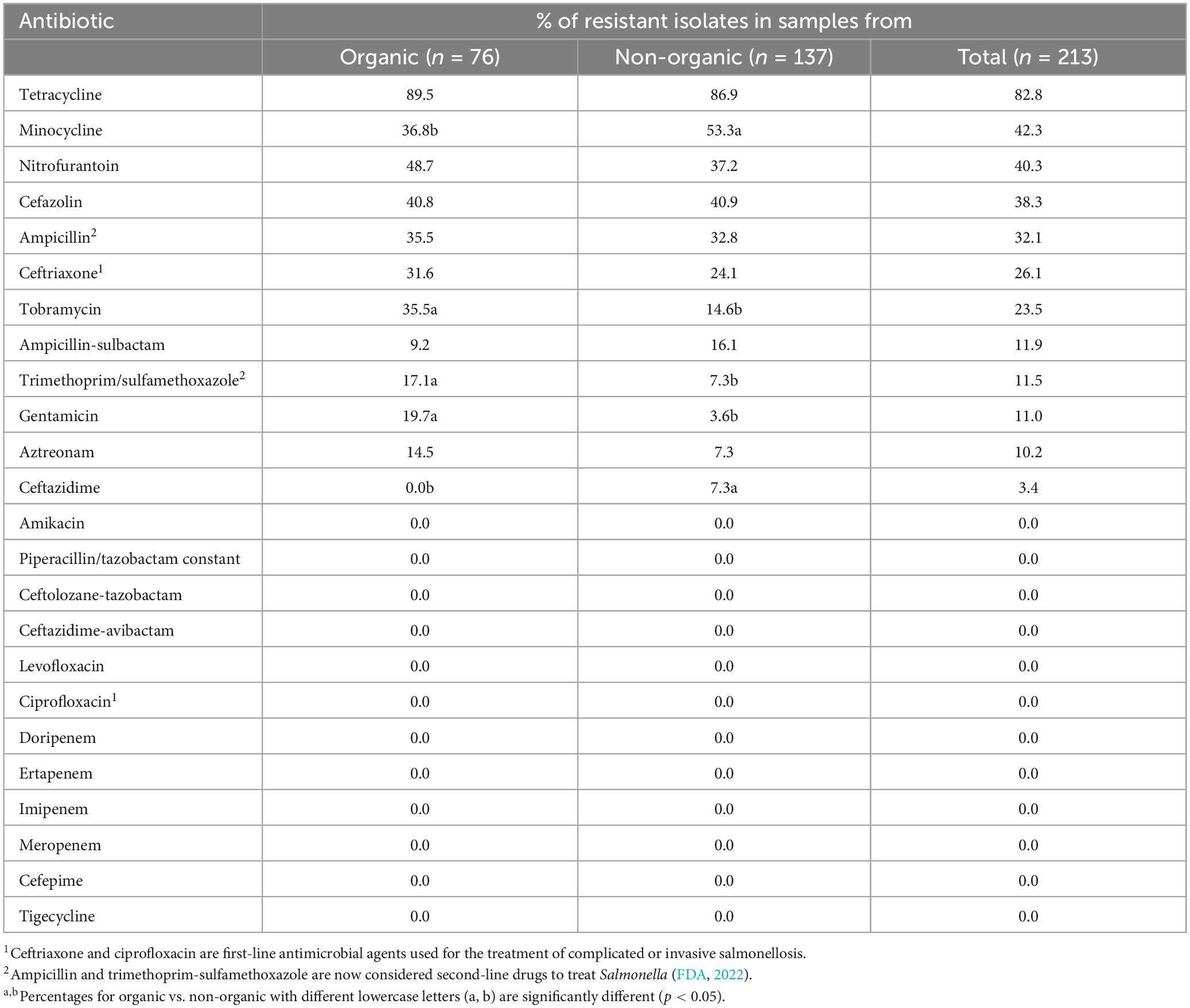
Table 4. Antimicrobial resistance phenotypes of Salmonella isolates recovered from organic and non-organic chickens.
Salmonella has the capability to adapt to undesirable external environment conditions and it can develop mechanisms to overcome the burden given by the external environment such as antimicrobial drugs (Tomičić et al., 2018). This process is known as antimicrobial resistance. Multiple studies have indicated the prevalence of tetracycline resistance in Salmonella (Parveen et al., 2007; Liljebjelke et al., 2017; Velasquez et al., 2018). Previous studies conducted in the United States have similarly documented the resistance of Salmonella isolates associated with chickens to cefazolin (Bythwood et al., 2019), ampicillin (Velasquez et al., 2018; Bythwood et al., 2019), and ceftriaxone (Velasquez et al., 2018).
Table 5 shows the distribution of resistant isolates by antibiotic classes and serovars. A significantly higher number (p < 0.05) of S. Kentucky recovered from non-organic chickens were resistant to ampicillin, ampicillin-sulbactam, and ceftriaxone compared to the S. Kentucky isolated from organic chickens. In contrast, a significantly higher number (p < 0.05) of S. Infantis recovered from organic chickens were resistant to tobramycin, ampicillin, and ceftriaxone. Among S. Typhimurium, significantly higher (p < 0.05) gentamicin resistance was observed in those recovered from organic chickens. All S. Typhimurium recovered from non-organic chickens were resistant to both cefazolin and ampicillin and significantly different from the frequency of resistance of those recovered from organic chickens (p < 0.05).
Unexpectedly, a higher rate of Salmonella isolates recovered from organic chickens showed resistance to some antibiotics compared to Salmonella from non-organic chickens (Tables 4, 5). Bailey et al. (2020) also reported such a noticeable antimicrobial resistance in Salmonella recovered from organic chickens compared to non-organic chickens. A possible reason for this higher rate of prevalence of antibiotic resistance Salmonella among organic chickens could be the farming practice; the organic chickens have access to the outdoors where proper biosecurity measures are difficult to provide. Therefore, there is a higher probability to interact with other avian species, insects, and other wild animals. Avian species such as migratory birds, waterfowl, and other animals that have already been exposed to sources of AMR genes (aquatic environments and other anthropogenic sources) may potentially carry and contaminate the organic chickens with the AMR genes over great distances (Allen et al., 2010; Bailey et al., 2020). Another possible reason for this higher rate of AMR Salmonella isolates may be contaminants or antibiotic residues which have been on site before the conversion of a conventional farm to an organic farm (Pesciaroli et al., 2020). One of the requirements for poultry products, that are to be labeled as organic, the poultry must be subjected to organic management from at least the second day of life (eCFR, 2000). But, Pesciaroli et al. (2020) reported that AMR bacteria can be present in 1-day-old chickens due to vertical transmission from the parent and/or contamination at the hatchery.
Sixty resistance profiles were observed in Salmonella isolates recovered from both organic and non-organic chickens (Table 6). Twenty-five resistance profiles were observed in Salmonella isolates recovered from organic chickens while 50 resistance profiles were detected in Salmonella isolates recovered from non-organic chickens. The most common resistance profile was tetracycline-minocycline (27.2%) followed by tetracycline-nitrofurantoin (8.5%) in both types of chickens. The prominent profile among the isolates recovered from the organic chickens was TET-MIN (25.0%) followed by TET-FAZ-NIT-AMP-AXO-TOB (9.2%). The prevalence of the TET-FAZ-NIT-AMP-AXO-TOB-AZT-SXT resistance profile was significantly higher (p < 0.05) in Salmonella from organic chickens (6.6%) compared to non-organic chickens. The prevalence of the TET-NIT-TOB resistance profile was also significantly higher (p < 0.05) in Salmonella recovered from organic chickens than in non-organic chickens. One S. Infantis isolated from organic chickens showed resistance to 11 antibiotics (TET-FAZ-NIT-MIN-AMP-AXO-AS2-TOB-AZT-SXT-GEN). In addition, two S. Infantis and an S. Rough_O:1:1,5 were resistant to the profile containing ten antibiotics (TET-FAZ-NIT-AMP-AXO-AS2-TOB-AZT-SXT-GEN). Regarding non-organic chickens, the most common antibiotic resistance profile was TET-MIN (28.5%). The prevalence of the profile containing tetracycline and nitrofurantoin (11.7%) was significantly higher in the isolates recovered from non-organic compared to organic chickens. Five percent of isolates (S. Kentucky) were resistant to TET-MIN-FAZ-AMP-AXO-AS2-TAZ. Among the isolates, 3.6% exhibited resistance to TET-MIN-FAZ-AMP profile. One S. Infantis isolate (0.7%) showed resistance to ten antimicrobials (TET-FAZ-NIT-AMP-AXO-AS2-TOB-AZT-SXT-GEN) and two S. Infantis isolates (1.5%) showed resistance to eight antimicrobials (TET-FAZ-NIT-AMP-AXO-TOB-AZT-GEN). Another S. Infantis isolate (0.7%) showed resistance to eight antimicrobials (TET-FAZ-NIT-AMP-AXO-TOB-AZT-SXT). Our results demonstrated a large number of recovered Salmonella isolates were resistant to multiple antimicrobials including third-generation cephalosporins (ceftriaxone and ceftazidime). Also, we observed antibiotic resistance profiles with various combinations of antibiotics. Using a higher number of antibiotics (twenty-four different antibiotics) in the AST, we were able to test a large spectrum of antibiotic phenotypes and 60 antibiotic resistance profiles. This is one of the reasons we observed a higher number of antibiotic resistance profiles in our isolates compared to the other studies (Parveen et al., 2007; Lestari et al., 2009).
After exposure to misused/over-used antibiotics residuals for extended periods, Salmonella gains resistance to an antibiotic, it eventually suppresses the actions of the combinations of drugs, leading to the development of Multi Drug Resistance (MDR) (Hetta et al., 2023). In this study, we considered MDR as resistance to at least one agent in three or more antimicrobial classes (Magiorakos et al., 2012; Sapkota et al., 2014). In the present study, we observed MDR in 45.54% of recovered Salmonella isolates (Figure 2). Sixty isolates in non-organic chickens (43.80%) and 37 isolates in organic chickens (48.68%) were shown MDR for tested antimicrobial classes. Moreover, a lower prevalence of MDR was observed in S. Kentucky (10.35%) isolated from organic chickens compared to S. Kentucky (33.8%) in non-organic chickens. In contrast, S. Infantis in organic chickens showed a higher prevalence of MDR (96%) compared to S. Infantis in non-organic chickens (64.6%). All three isolates of S. Typhimurium showed MDR in non-organic chickens and 50% of S. Typhimurium isolates showed MDR in organic chickens. In addition, 15.39% of S. Enteritidis were MDR. In the present study, we did not observe Extensively Drug Resistant (XDR; resistant to at least one agent in all antimicrobial classes, but susceptible to 1–2 antimicrobial classes) and Pan Drug Resistant (PDR; resistant to all agents in all antimicrobial classes) Salmonella isolates among both types of chickens. If Salmonella develops its resistance to the multiple drugs that are used to treat severe bacterial diseases in both animals and humans, new effective drugs must be invented to control Salmonella infections.
The majority of isolates (99.1%) possessed at least one of the invA, pagC, and spvC genes. Only two isolates did not possess any of these three genes (S. III 45:z46:- and an S. Infantis). Among tested isolates, 4.2% were positive for all three virulence genes. Regardless of the type of chickens, the prevalence of invA, pagC, and spvC genes in Salmonella isolates was 95.3, 89.2, and 6.6%, respectively. The prevalence of the invA gene in the Salmonella isolates recovered from organic chickens was 92% and the prevalence of pagC was 88%. The spvC gene was not detected in the Salmonella isolates recovered from organic chickens. On the other hand, invA, pagC, and spvC genes were detected in Salmonella recovered from non-organic chickens with a prevalence of 97, 89, and 10%, respectively. A significantly higher prevalence of the spvC gene (p < 0.05) was observed in non-organic chickens compared to organic chickens (p < 0.05). The majority of spvC genes were carried by S. Enteritidis (92.3%) which was exclusively recovered from non-organic chicken samples. Furthermore, one S. 4,[5],12:i:- and one S. Kentucky were recovered from non-organic chickens carried the spvC gene as well as invA and pagC genes (as shown in Table 7). Importantly, 53.8% of S. Enteritidis isolates were found to possess all three genes.
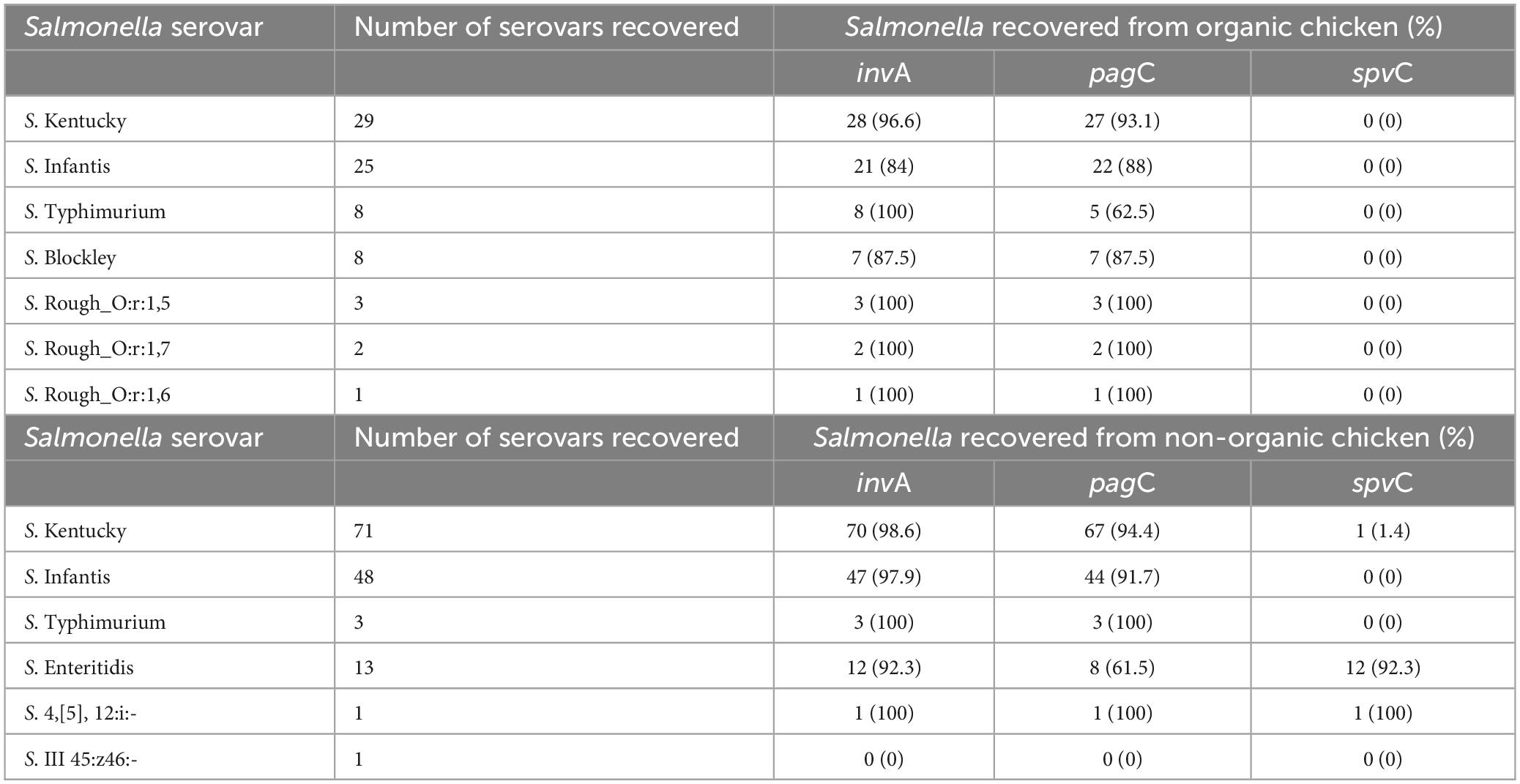
Table 7. The distribution of invA, pagC, and spvC genes in Salmonella isolates recovered from organic and non-organic chicken.
These invA, pagC, and spvC genes are playing a major role in Salmonella virulence (Nolan et al., 1995; Olah et al., 2005). The invA gene which is located in the chromosome of Salmonella promotes the invasion of the host cell by stimulating the formation of inner and outer membrane proteins (Olah et al., 2005; Mohamed et al., 2014). PagC is another essential chromosomal virulence gene that encodes an outer membrane/envelope protein that promotes survival within macrophages by suppressing bacterial cell division and prolonging the cell cycle (Olah et al., 2005; Xu et al., 2018). The expression of pagC gene is activated by Salmonella cells as a defensive mechanism against stress conditions inside the host. In unfavorable environmental conditions, pagC gene promotes growth under low Mg2+ concentrations, resistance to low pH, bile salts, and cationic antimicrobial proteins (Xu et al., 2018). Therefore, pagC plays an important role in Salmonella virulence, inducing Salmonella cells to enter a Viable but Non-Culturable (VBNC) state under unfavorable environmental conditions (Xu et al., 2018). The spvC gene which is located on Salmonella Typhimurium virulence plasmid, promotes the prolific growth of Salmonella in host reticuloendothelial tissues (Olah et al., 2005). Our results indicated a high prevalence of invA and pagC among the Salmonella isolates recovered from both organic and non-organic chickens compared to spvC gene. Olah et al. (2005) and Mohamed et al. (2014) observed that all the tested isolates were positive for the invA and pagC but observed low detection of spvC gene. It has been suggested that the invA gene is exclusive to the Salmonella genome and serves as a distinctive marker, allowing for molecular detection of Salmonella (Rahn et al., 1992; Chiu and Ou, 1996; Wolffs et al., 2006; Andrews et al., 2023). However, mutations occurred in the invA gene may not be identifiable through traditional PCR based methods. In addition, the Salmonella carrying mutant invA gene showed less virulent (Galán et al., 1992; Ginocchio and Galán, 1995; Darwin and Miller, 1999). According to previous studies, the detection of spvC gene is low compared to invA and pagC genes (Nolan et al., 1995; Mohamed et al., 2014; Sharma et al., 2019; Tasmin et al., 2019). It has been reported that spvC gene is frequently found in a limited number of serovars including S. Enteritidis, S. Typhimurium, S. Choleraesuis, and S. Dublin (mostly from infected animals and birds) (Krzyzanowski et al., 2014; Tasmin et al., 2019) and its presence or absence seems to be correlated with the possession of virulence plasmid (Olah et al., 2005). In the present study, the majority of S. Enteritidis (n = 12), S. 4,[5],12:i:- (n = 1), and S. Kentucky (n = 1) had spvC gene in their genome. Apparently, in our Salmonella collection, S. Typhimurium recovered from both types of chickens did not possess spv operon. In the present study, we observed a comparatively higher rate of spvC gene in the non-organic chickens because the S. Enteritidis recovered only from the non-organic chickens and the majority (92.3%) of them carried spvC gene. Another possible reason for the lower spvC gene detection is the method of DNA extraction. In the present study, we used a DNA extraction method that focused on chromosomal DNA extraction but not specifically designed for plasmids. However, it has been suggested that further research is needed to understand the factors that affect the presence and expression of spvC gene in Salmonella (Mohamed et al., 2014).
Salmonella which carries the invA, pagC, and spvC genes has the potential to cause foodborne illnesses when ingested with contaminated food (Olah et al., 2005; Mohamed et al., 2014). Four isolates identified in our study that carried all three of these genes showed resistance to multiple drugs. The S. Kentucky isolate which had all three genes showed resistance to cefazolin, ampicillin, tetracycline, and minocycline. In addition, S. Enteritidis isolates which carried all three genes were resistant to multiple drugs in classes of cephem (ceftriaxone and cefazolin), tetracycline, nitrofurantoin, and penicillin. Moreover, an S. Enteritidis isolate was resistant to gentamicin which belongs to the class of aminoglycoside in addition to the above-mentioned antibiotics. Ceftriaxone is one of the important antibiotics that can be used to treat severe Salmonella infection (CDC, 2018). If salmonellosis occurs due to ingestion of an MDR Salmonella serovar which carries these virulence genes, it would be a challenge to select appropriate antibiotics to control the disease.
The results of this study demonstrate a high prevalence of Salmonella contamination in organic and non-organic chickens and a significant number of these isolates were resistant to commonly used antibiotics. The Salmonella isolates recovered from both types of chickens possessed virulence genes and thus have the potential to cause salmonellosis. It will be a challenge to treat a patient who has a severe Salmonella infection caused by an MDR Salmonella strain. Conscious action must be taken to reduce Salmonella contamination throughout the food chain. In order to have a broader knowledge of the topic a large-scale and multi-state-wide study is highly recommended. The result of our study highlighted the importance of educating consumers on safe food handling practices in the home to improve their self-hygiene practices, eliminate cross-contamination and avoid consuming undercooked food.
The raw data supporting the conclusions of this article will be made available by the authors, without undue reservation.
AJP-D: Data curation, Formal analysis, Investigation, Methodology, Software, Visualization, Writing—original draft, Writing—review and editing. JS: Conceptualization, Writing—review and editing, Funding acquisition. AD: Methodology, Writing—review and editing. JB: Formal analysis, Software, Writing—review and editing. SP: Conceptualization, Funding acquisition, Investigation, Project administration, Resources, Supervision, Writing—review and editing, Data curation, Visualization.
The author(s) declare financial support was received for the research, authorship, and/or publication of this article. This research was supported by USDA Evans-Allen funds (Grant No: 4-451621) and from the School of Graduate Studies of the University of Maryland Eastern Shore (UMES).
We would like to express our gratitude to Ms. Joan Meredith for her excellent support.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
The author(s) declared that they were an editorial board member of Frontiers, at the time of submission. This had no impact on the peer review process and the final decision.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2023.1272892/full#supplementary-material
Allen, H. K., Donato, J., Wang, H. H., Cloud-Hansen, K. A., Davies, J., and Handelsman, J. (2010). Call of the wild: Antibiotic resistance genes in natural environments. Nat. Rev. Microbiol. 8, 251–259. doi: 10.1038/nrmicro2312
Andrews, W. H., Wang, H., Jacobson, A., Ge, B., Zhang, G., and Hammack, T. (2023). Bacteriological analytical manual (BAM) chapter 5: Salmonella. Available online at: https://www.fda.gov/food/laboratory-methods-food/bam-chapter-5-salmonella (Accessed August 19, 2023).
Bailey, J. S., and Cosby, D. E. (2005). Salmonella prevalence in free-range and certified organic chickens. J. Food Prot. 68, 2451–2453. doi: 10.4315/0362-028x-68.11.2451
Bailey, M., Taylor, R., Brar, J., Corkran, S., Velásquez, C., Novoa-Rama, E., et al. (2020). Prevalence and antimicrobial resistance of Salmonella from antibiotic-free broilers during organic and conventional processing. J. Food Prot. 83, 491–496. doi: 10.4315/0362-028X.JFP-19-269
Bythwood, T. N., Soni, V., Lyons, K., Hurley-Bacon, A., Lee, M. D., Hofacre, C., et al. (2019). Antimicrobial resistant Salmonella enterica Typhimurium colonizing chickens: The impact of plasmids, genotype, bacterial communities, and antibiotic administration on resistance. Front. Sust. Food Syst. 3:20. doi: 10.3389/fsufs.2019.00020
CDC (2018). Advice to clinicians. Available online at: https://www.cdc.gov/salmonella/infantis-10-18/advice.html (accessed January 17).
CDC (2019). Antibiotic resistance threats in the United States. Available online at: https://www.cdc.gov/drugresistance/pdf/threats-report/2019-ar-threats-report-508.pdf (accessed January 17, 2023).
CDC (2022). Chicken and food poisoning. Available online at: https://www.cdc.gov/foodsafety/chicken.html (accessed January 17, 2023).
CDC (2023). Salmonella. Available online at: https://www.cdc.gov/salmonella/index.html (accessed June 19, 2023).
Chiu, C. H., and Ou, J. T. (1996). Rapid identification of Salmonella serovars in feces by specific detection of virulence genes, invA and spvC, by an enrichment broth culture-multiplex PCR combination assay. J. Clin. Microbiol. 34, 2619–2622. doi: 10.1128/jcm.34.10.2619-2622.1996
Clinical and Laboratory Standards Institute [CLSI] (2016). Performance standards for antimicrobial susceptibility testing. Pennsylvania: Clinical and Laboratory Standards Institute.
Constance, D. H. (2008). The southern model of broiler production and its global implications. Cult. Agricul. 30, 17–31.
Cox, N. A., Buhr, R. J., Smith, D. P., Cason, J. A., Rigsby, L. L., Bourassa, D. V., et al. (2014). Sampling naturally contaminated broiler carcasses for Salmonella by three different methods. J. Food Protect. 77, 493–495. doi: 10.4315/0362-028x.Jfp-13-320
Cui, S., Ge, B., Zheng, J., and Meng, J. (2005). Prevalence and antimicrobial resistance of Campylobacter spp. and Salmonella serovars in organic chickens from Maryland retail stores. Appl. Environ. Microbiol. 71, 4108–4111. doi: 10.1128/AEM.71.7.4108-4111.2005
Darwin, K. H., and Miller, V. L. (1999). Molecular basis of the interaction of Salmonella with the intestinal mucosa. Clin. Microbiol. Rev. 12, 405–428. doi: 10.1128/cmr.12.3.405
eCFR (2000). National organic program. 7 CFR §205. Available online at: https://www.federalregister.gov/documents/2000/12/21/00-32257/national-organic-program (accessed August 26, 2023).
Ewing, W. H. (1986). Edwards and Ewing’s identification of Enterobacteriaceae. New York, NY: Elsevier Science Publishing Co., Inc.
FDA (2020). Get the facts about Salmonella. Available online at: https://www.fda.gov/animal-veterinary/animal-health-literacy/get-facts-about-salmonella (accessed January 17, 2023).
FDA (2022). 2019 NARMS Update: Integrated Report Summary Interactive Version. Available online at: https://www.fda.gov/animal-veterinary/national-antimicrobial-resistance-monitoring-system/2019-narms-update-integrated-report-summary-interactive-version (accessed January 31, 2023).
Galán, J. E., Ginocchio, C., and Costeas, P. (1992). Molecular and functional characterization of the Salmonella invasion gene invA: Homology of InvA to members of a new protein family. J. Bacteriol. 174, 4338–4349. doi: 10.1128/jb.174.13.4338-4349.1992
Ginocchio, C. C., and Galán, J. E. (1995). Functional conservation among members of the Salmonella Typhimurium InvA family of proteins. Infect. Immun. 63, 729–732. doi: 10.1128/iai.63.2.729-732.1995
Grimont, P. A., and Weill, F.-X. (2007). “Antigenic formulae of the Salmonella serovars,” in WHO collaborating center for reference and research on Salmonella, Vol. 9, (Paris: Institut Pasteur), 1–166.
Hetta, H. F., Ramadan, Y. N., Al-Harbi, A. I., Ahmed, A., Battah, B., Abd Ellah, N. H., et al. (2023). Nanotechnology as a promising approach to combat multidrug resistant bacteria: A comprehensive review and future perspectives. Biomedicines 11:413. doi: 10.3390/biomedicines11020413
Krzyzanowski, F., Zappelini, L., Martone-Rocha, S., Dropa, M., Matté, M. H., Nacache, F., et al. (2014). Quantification and characterization of Salmonella spp. isolates in sewage sludge with potential usage in agriculture. BMC Microbiol. 14:263. doi: 10.1186/s12866-014-0263-x
Lestari, S. I., Han, F., Wang, F., and Ge, B. (2009). Prevalence and antimicrobial resistance of Salmonella serovars in conventional and organic chickens from Louisiana retail stores. J. Food. Prot. 72, 1165–1172. doi: 10.4315/0362-028x-72.6.1165
Liljebjelke, K. A., Hofacre, C. L., White, D. G., Ayers, S., Lee, M. D., and Maurer, J. J. (2017). Diversity of antimicrobial resistance phenotypes in Salmonella isolated from commercial poultry farms. Front. Vet. Sci. 4:96. doi: 10.3389/fvets.2017.00096
Magiorakos, A. P., Srinivasan, A., Carey, R. B., Carmeli, Y., Falagas, M. E., Giske, C. G., et al. (2012). Multidrug-resistant, extensively drug-resistant and pandrug-resistant bacteria: An international expert proposal for interim standard definitions for acquired resistance. Clin. Microbiol. Infect. 18, 268–281.
Mak, P. H. W., Rehman, M. A., Kiarie, E. G., Topp, E., and Diarra, M. S. (2022). Production systems and important antimicrobial resistant-pathogenic bacteria in poultry: A review. J. Anim. Sci. Biotechnol. 13:148. doi: 10.1186/s40104-022-00786-0
Marchello, C. S., Carr, S. D., and Crump, J. A. (2020). A systematic review on antimicrobial resistance among Salmonella Typhi worldwide. Am. J. Trop Med. Hyg. 103, 2518–2527. doi: 10.4269/ajtmh.20-0258
Mazengia, E., Samadpour, M., Hill, H. W., Greeson, K., Tenney, K., Liao, G., et al. (2014). Prevalence, concentrations, and antibiotic sensitivities of Salmonella serovars in poultry from retail establishments in Seattle. Washington. J. Food Prot. 77, 885–893. doi: 10.4315/0362-028x.Jfp-13-394
Mohamed, T., Zhao, S., White, D. G., and Parveen, S. (2014). Molecular characterization of antibiotic resistant Salmonella Typhimurium and Salmonella Kentucky isolated from pre- and post-chill whole broilers carcasses. Food Microbiol. 38, 6–15. doi: 10.1016/j.fm.2013.08.002
Nguyen, D. T. A., Kanki, M., Nguyen, P. D., Le, H. T., Ngo, P. T., Tran, D. N. M., et al. (2016). Prevalence, antibiotic resistance, and extended-spectrum and AmpC β-lactamase productivity of Salmonella isolates from raw meat and seafood samples in Ho Chi Minh City, Vietnam. Int. J. Food Microbiol. 236, 115–122. doi: 10.1016/j.ijfoodmicro.2016.07.017
Nolan, L. K., Giddings, C. W., and Brown, J. (1995). The distribution of invA, pagC and spvC genes among Salmonella isolates from animals. Vet. Res. Commun. 19, 167–177. doi: 10.1007/bf01839295
Olah, P. A., Sherwood, J. S., and Logue, C. M. (2005). Molecular analysis of Salmonella isolates recovered from processed Turkey carcasses. J. Food Prot. 68, 845–849. doi: 10.4315/0362-028x-68.4.845
Parveen, S., Taabodi, M., Schwarz, J. G., Oscar, T. P., Harter-Dennis, J., and White, D. G. (2007). Prevalence and antimicrobial resistance of Salmonella recovered from processed poultry. J. Food Prot. 70, 2466–2472. doi: 10.4315/0362-028x-70.11.2466
Pesciaroli, M., Magistrali, C. F., Filippini, G., Epifanio, E. M., Lovito, C., Marchi, L., et al. (2020). Antibiotic-resistant commensal Escherichia coli are less frequently isolated from poultry raised using non-conventional management systems than from conventional broiler. Int. J. Food Microbiol. 314:108391. doi: 10.1016/j.ijfoodmicro.2019.108391
Pulkkinen, W. S., and Miller, S. I. (1991). A Salmonella Typhimurium virulence protein is similar to a Yersinia enterocolitica invasion protein and a bacteriophage lambda outer membrane protein. J. Bacteriol. 173, 86–93. doi: 10.1128/jb.173.1.86-93.1991
Punchihewage-Don, A. J., Hawkins, J., Adnan, A. M., Hashem, F., and Parveen, S. (2022). The outbreaks and prevalence of antimicrobial resistant Salmonella in poultry in the United States: An overview. Heliyon 8:e11571. doi: 10.1016/j.heliyon.2022.e11571
R Core Team (2021). R: A language and environment for statistical computing. Vienna: R Foundation for Statistical Computing.
Rahn, K., De Grandis, S. A., Clarke, R. C., McEwen, S. A., Galán, J. E., Ginocchio, C., et al. (1992). Amplification of an invA gene sequence of Salmonella Typhimurium by polymerase chain reaction as a specific method of detection of Salmonella. Mol. Cell. Probes. 6, 271–279. doi: 10.1016/0890-8508(92)90002-f
Sapkota, A. R., Kinney, E. L., George, A., Hulet, R. M., Cruz-Cano, R., Schwab, K. J., et al. (2014). Lower prevalence of antibiotic-resistant Salmonella on large-scale U.S. conventional poultry farms that transitioned to organic practices. Sci. Total Environ. 476-477, 387–392. doi: 10.1016/j.scitotenv.2013.12.005
Sharma, J., Kumar, D., Hussain, S., Pathak, A., Shukla, M., Prasanna Kumar, V., et al. (2019). Prevalence, antimicrobial resistance and virulence genes characterization of nontyphoidal Salmonella isolated from retail chicken meat shops in Northern India. Food Control 102, 104–111.
Siceloff, A. T., Waltman, D., and Shariat, N. W. (2022). Regional Salmonella differences in United States broiler production from 2016 to 2020 and the contribution of multiserovar populations to Salmonella Surveillance. J. Appl. Environ. Microbiol. 88:e00204-22. doi: 10.1128/aem.00204-22
Suzuki, S., Komase, K., Matsui, H., Abe, A., Kawahara, K., Tamura, Y., et al. (1994). Virulence region of plasmid pNL2001 of Salmonella enteritidis. Microbiology 140(Pt 6), 1307–1318. doi: 10.1099/00221287-140-6-1307
Tasmin, R., Gulig, P. A., and Parveen, S. (2019). Detection of virulence plasmid-encoded genes in Salmonella Typhimurium and Salmonella Kentucky isolates recovered from commercially processed chicken carcasses. J. Food Prot. 82, 1364–1368. doi: 10.4315/0362-028x.Jfp-18-552
Tomičić, Z., Čabarkapa, I., Čolović, R., Ðuragić, O., and Tomičić, R. (2018). Salmonella in the feed industry: Problems and potential solutions. J. Agron. 22:2019.
University of Georgia Extension (2022). Organic poultry production vs. other systems. Available online at: https://extension.uga.edu/publications/detail.html?number=C1139&title=organic-poultry-production-vs-other-systems (accessed August 25, 2023).
USDA-Agricultural Marketing Service (2013). Organic livestock requirements. Available online at: https://www.ams.usda.gov/publications/content/organic-livestock-requirements (accessed January 18, 2023).
USDA-Agricultural Marketing Service (2015). Fact sheet: Introduction to organic practices. Available: https://www.ams.usda.gov/publications/content/fact-sheet-introduction-organic-practices (accessed January 18, 2023).
USDA-FSIS (2019). Isolation and identification of Salmonella from meat, poultry, pasteurized egg, siluriformes (fish) products and carcass and environmental sponges. Available online at: https://s27415.pcdn.co/wp-content/uploads/2020/01/64ER20-7/Microbial/5-USDA-FSIS-4.10-Isolation-and-Identification-of-Salmonella.pdf (accessed January 19, 2023).
USDA-FSIS (2023). A snapshot of multi-year trends for salmonella antimicrobial resistance. Available: https://www.fsis.usda.gov/news-events/news-press-releases/a-snapshot-multi-year-trends-salmonella-antimicrobial-resistance-0 (accessed October 27, 2023).
USDA-National Agricultural Statistics Service (2022). Poultry-production and value 2021 summary. Available online at: https://www.uspoultry.org/economic-data/docs/broiler-production-and-value-2021.pdf (accessed Januaray 18, 2023).
Velasquez, C. G., Macklin, K. S., Kumar, S., Bailey, M., Ebner, P. E., Oliver, H. F., et al. (2018). Prevalence and antimicrobial resistance patterns of Salmonella isolated from poultry farms in Southeastern United States. Poul. Sci. 97, 2144–2152. doi: 10.3382/ps/pex449
World Health Organization [WHO] (2023). What is the difference between antibiotic and antimicrobial resistance? Available online at: https://www.emro.who.int/health-topics/drug-resistance/what-is-the-difference-between-antibiotic-and-antimicrobial-resistance.html (accessed January 31, 2023).
Wolffs, P. F., Glencross, K., Thibaudeau, R., and Griffiths, M. W. (2006). Direct quantitation and detection of salmonellae in biological samples without enrichment, using two-step filtration and real-time PCR. Appl. Environ. Microbiol. 72, 3896–3900. doi: 10.1128/aem.02112-05
Keywords: Salmonella, prevalence, antibiotic, resistance, virulence genes
Citation: Punchihewage-Don AJ, Schwarz J, Diria A, Bowers J and Parveen S (2024) Prevalence and antibiotic resistance of Salmonella in organic and non-organic chickens on the Eastern Shore of Maryland, USA. Front. Microbiol. 14:1272892. doi: 10.3389/fmicb.2023.1272892
Received: 04 August 2023; Accepted: 08 December 2023;
Published: 04 January 2024.
Edited by:
Lucilla Iacumin, University of Udine, ItalyReviewed by:
Adriana Morar, Banat University of Agricultural Sciences and Veterinary Medicine, RomaniaCopyright © 2024 Punchihewage-Don, Schwarz, Diria, Bowers and Parveen. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Salina Parveen, c3BhcnZlZW5AdW1lcy5lZHU=
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.