
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Microbiol. , 06 October 2023
Sec. Antimicrobials, Resistance and Chemotherapy
Volume 14 - 2023 | https://doi.org/10.3389/fmicb.2023.1271733
This article is part of the Research Topic The role of regulatory networks in virulence and antimicrobial resistance of microbial pathogens View all 7 articles
Introduction: Although carbapenemases are frequently reported in resistant A. baumannii clinical isolates, other chromosomally mediated elements of resistance that are considered essential are frequently underestimated. Having a wide substrate range, multidrug efflux pumps frequently underlie antibiotic treatment failure. Recognizing and exploiting variations in multidrug efflux pumps and penicillin-binding proteins (PBPs) is an essential approach in new antibiotic drug discovery and engineering to meet the growing challenge of multidrug-resistant Gram-negative bacteria.
Methods: A total of 980 whole genome sequences of A. baumannii were analyzed. Nucleotide sequences for the genes studied were queried against a custom database of FASTA sequences using the Bacterial and Viral Bioinformatics Resource Center (BV-BRC) system. The correlation between different variants and carbapenem Minimum Inhibitory Concentrations (MICs) was studied. PROVEAN and I-Mutant predictor suites were used to predict the effect of the studied amino acid substitutions on protein function and protein stability. Both PsiPred and FUpred were used for domain and secondary structure prediction. Phylogenetic reconstruction was performed using SANS serif and then visualized using iTOL and Phandango.
Results: Exhibiting the highest detection rate, AdeB codes for an important efflux-pump structural protein. T48V, T584I, and P660Q were important variants identified in the AdeB-predicted multidrug efflux transporter pore domains. These can act as probable targets for designing new efflux-pump inhibitors. Each of AdeC Q239L and AdeS D167N can also act as probable targets for restoring carbapenem susceptibility. Membrane proteins appear to have lower predictive potential than efflux pump-related changes. OprB and OprD changes show a greater effect than OmpA, OmpW, Omp33, and CarO changes on carbapenem susceptibility. Functional and statistical evidence make the variants T636A and S382N at PBP1a good markers for imipenem susceptibility and potential important drug targets that can modify imipenem resistance. In addition, PBP3_370, PBP1a_T636A, and PBP1a_S382N may act as potential drug targets that can be exploited to counteract imipenem resistance.
Conclusion: The study presents a comprehensive epidemiologic and statistical analysis of potential membrane proteins and efflux-pump variants related to carbapenem susceptibility in A. baumannii, shedding light on their clinical utility as diagnostic markers and treatment modification targets for more focused studies of candidate elements.
Although initially described as an opportunistic bacterium of low pathogenicity, A. baumannii has emerged as an important pathogen in critical care settings during the last decades. It is known to be widely involved in diseases such as mechanical ventilator-associated pneumonia, osteomyelitis, peritonitis, endocarditis, septicemia, and meningitis, as well as wound, skin, soft tissue, urinary tract, ear, and eye infections (Ayoub Moubareck and Hammoudi Halat, 2020). Additionally, outbreaks caused by A. baumannii are common in intensive care units (ICUs) and burn units (Lima et al., 2019).
Especially concerning is the emergence of multidrug resistant (MDR) A. baumannii and its alarming spread all over the world. Their emergence is usually associated with prolonged hospitalizations and increased morbidity and mortality, particularly in ICU patients. The incidence of MDR A. baumannii has been associated with some underlying risk factors, including burn infections and premature births, as well as the use of mechanical ventilation, indwelling devices, and extensive exposure to antimicrobial therapy (Agyepong et al., 2023).
Carbapenems are extremely important for controlling severe Gram-negative infections, particularly those caused by multidrug-resistant bacteria. Carbapenem antibiotics, such as imipenem, meropenem, and doripenem but not ertapenem, are considered last-resort treatments for MDR A. baumannii infection (Vila and Pachón, 2012; Nguyen and Joshi, 2021). Both imipenem and meropenem are used as empiric therapy for a variety of severe Gram-negative infections. However, since 1991, when the first carbapenem-resistant A. baumannii (CRAB) was recognized, a considerable increase in the number of these strains has been documented worldwide (Peleg et al., 2008; Gales et al., 2019; ECDC, 2022). Although carbapenem is one of the last-resort treatments, hospital outbreaks caused by CRAB isolates have been described earlier worldwide (Durante-Mangoni and Zarrilli, 2011; Migliavacca et al., 2013). This also includes high-resistance levels in burn units (Shoja et al., 2017; Lima et al., 2019). Evidence shows that patients infected with CRAB exhibit significantly higher mortality than those infected with carbapenem-susceptible A. baumannii (Lee et al., 2012). The rate of mortality related to CRAB infection was reported at 45.4%. Ventilator-associated pneumonia and bloodstream infections were the most prevalent infections in these cases (Schimith Bier et al., 2010; Kanafani et al., 2018; Gu et al., 2021). With a strong capacity for clonal transmission and acquirement of antimicrobial resistance determinants, in 2017, the World Health Organization (WHO) announced CRAB as the top-priority pathogen, critically requiring research and development of new antibiotics (Tacconelli and Magrini, 2017).
Mechanisms of carbapenem resistance in A. baumannii can be classified into enzymatic mechanisms, which involve the production of different carbapenemases and non-enzymatic mechanisms. The latter includes genetic alterations of PBPs, overexpression of efflux pumps belonging to the resistance-nodulation-cell division (RND) family, and the loss or modifications of outer membrane porins (Nguyen and Joshi, 2021; Vahhabi et al., 2021).
Efflux pumps such as AdeABC are reportedly involved in multidrug resistance (Vila et al., 2007; Xu et al., 2019). AdeABC, a resistance-nodulation-division (RND) family-type pump, has three component structures, AdeA, AdeB, and AdeC, which form the inner membrane fusion protein, the transmembrane component, and the Outer Membrane Protein (OMP), respectively. The AdeABC operon found in 80% of clinical isolates is chromosomally encoded and normally regulated by a two-component system, including the sensor kinase gene (AdeS) and its related response regulator gene (AdeR). Overexpression of AdeB has been attributed to mutations in the AdeS and AdeR genes and also to the insertion of ISAba1 into the AdeS gene or upstream from the operon, resulting in multidrug resistance in A. baumannii (Marchand et al., 2004; Coyne et al., 2011). Studies have demonstrated that the AdeB gene shows 10–40 times higher levels of expression in resistant isolates than in carbapenem-susceptible isolates, suggesting the association of AdeABC efflux pump with carbapenem resistance (Lee et al., 2010). It has been suggested that the AdeABC genes are common in A. baumannii but may be absent in fully susceptible isolates (Nemec et al., 2007). The AdeB gene, especially when hyperproduced, has been previously suggested as a genetic marker of MDR AB clinical isolates (Huys et al., 2005; Huang et al., 2008). Other efflux pumps include AdeIJK and a third RND efflux pump, AdeFGH, which confers multidrug resistance when overexpressed (Xu et al., 2019). Mutations in the upstream-located transcriptional regulator AdeL have been linked to AdeFGH overexpression (Coyne et al., 2010; Leus et al., 2020).
Next to antibiotic efflux, PBP modifications and changes in outer membrane proteins are among the most important non-enzymatic mechanisms of carbapenem resistance. PBP modifications are considered important mechanisms of chromosomally mediated acquired carbapenem resistance; however, they are less frequently reported (Poirel and Nordmann, 2006; Martínez-Trejo et al., 2022). It has been proposed that the loss or disruption of small outer membrane proteins would cause carbapenem resistance. This suggests their role as influx channels for carbapenem; however, carbapenem-binding sites have not been fully identified in all these types of membrane porins (Vahhabi et al., 2021).
Although both carbapenemase production and outer membrane permeability have been proposed to be jointly responsible for high levels of carbapenem resistance (Poirel and Nordmann, 2006; Nguyen and Joshi, 2021), investigators have associated imipenem-resistant clinical isolates of A. baumannii, which exhibited no carbapenemase activity, with the loss of outer membrane proteins (Limansky et al., 2002; Uppalapati et al., 2020). Other previous studies have also shown that high-level carbapenem resistance could result from the loss of outer membrane porins with the absence of carbapenemase activity (del Mar Tomás et al., 2005). In addition, there is evidence to suggest that clinical resistance to carbapenem develops during treatment in relation to a 4.6-fold increase in AdeB gene expression, while carbapenemase genes remain negative (Yamada and Suwabe, 2013). The same study also reported a loss of CarO resulting in carbapenem resistance with the absence of any carbapenemase activity. Experimental evidence has also shown that increased carbapenem MICs occur solely because of the overexpression of efflux pumps in the absence of carbapenemase activity or the overexpression of structural genes for the enzymes (Yoon et al., 2015). In addition, the overexpression of AdeABC combined with enzymatic resistance has been shown to contribute to carbapenem resistance in more than an additive manner, with a good relationship demonstrated between the level of expression and the level of resistance (Yoon et al., 2015).
The modification of PBPs as a carbapenem-resistance mechanism also appears to be under-reported. Carbapenem-resistant-derived in-vitro mutants exhibited hyperproduction of seven types of PBPs (Gehrlein et al., 1991), while other imipenem-resistant isolates exhibited an absence of 73.2-kDa PBP, besides carbapenemase production (Fernández-Cuenca et al., 2003). Both impermeability of the outer membrane mediated by different membrane porins and efflux-pump activity are important intrinsic determinants of resistance in Gram-negative bacteria, specifically in A. baumannii surviving in harsh hospital environments, and this necessitates a thorough and detailed study of such determinants (Bazyleu and Kumar, 2014).
Although carbapenemase-mediated resistance is studied extensively (Beigverdi et al., 2019; Strateva et al., 2019), its exact contribution to carbapenem resistance remains uncertain (Lee et al., 2012). In addition, the overall contribution of other non-enzymatic mechanisms involved in carbapenem resistance, including reduced outer membrane permeability, penicillin-binding protein alterations, and efflux pumps' overexpression, appears to be less well-studied (Lin, 2017). Carbapenem resistance can be encountered in a wide range of diverse genetic backgrounds. This study focused on determining the genetic structure and examining different membrane-related elements and their role in carbapenem resistance, including alterations in outer membrane porins, PBPs, and efflux-pump changes. This can prove especially valuable in cases of resistance emerging in prolonged treatments in critical settings.
In contrast to carbapenemases, most outer membrane proteins and PBPs are integral parts of the cell and are considered structurally and physiologically indispensable. In addition, it has been suggested that efflux pumps such as AdeABC and AdeIJK are tightly integrated into A. baumannii physiology and play distinct physiological roles related to cellular growth and survival (Leus et al., 2020). Under such circumstances, identifying determinants and markers of susceptibility among such a group of chromosomal cellular components would serve as candidate stable diagnostic markers.
This research aims to study the diversity in membrane genes contributing to carbapenem resistance and to determine the repertoire and phylogeny of resistance determinants among timely and geographically spaced clinical isolates of A. baumannii. A complete understanding of the mechanisms contributing to resistance phenotypes can be used as a target for novel drug development. Consequently, determining the contribution of different resistance mechanisms can aid in screening for new antimicrobial targets by prioritizing the most effective treatment targets.
In this study, we highlight the importance of these chromosomally encoded elements in determining carbapenem resistance. Our aim is to exploit this knowledge to design diagnostic markers that can guide the selection of the best treatment options, ultimately reducing the emergence of resistance. In addition, understanding the mutations that affect efflux-pump function could help design improved active efflux-pump inhibitors.
A total of 980 whole genome sequences of A. baumannii available at the NCBI and the PATRIC databases (Wattam et al., 2017) currently integrated into the Bacterial and Viral Bioinformatics Resource Center (BV-BRC) (Olson et al., 2023) were studied. The studied genome IDs are shown in Supplementary Table S1. A. baumannii genomic FASTA files were downloaded from the PATRIC database with their corresponding imipenem and meropenem MIC data. Nucleotide sequences for the genes were retrieved from the NCBI gene bank and queried using MegaBlast against a custom database of FASTA sequences on the BV-BRC system using BLASTn to search the nucleotide database using a nucleotide query sequence for each of the following entries: multidrug efflux RND transporter periplasmic adaptor subunit AdeA (NCBI Reference Sequence: NZ_CP009257.1: c3978859-3977669), multidrug efflux RND transporter permease subunits AdeB (NZ_CP009257.1: c3977672-3974562) and AdeC (NZ_CP009257.1:c3974485-3973088), efflux system response regulator transcription factor AdeR (NZ_CP009257.1:3979005-3979748), two-component sensor histidine kinase AdeS (NZ_CP009257.1:3979780-3980853), multidrug efflux RND transporter periplasmic adaptor subunit AdeF (NZ_CP009257.1:1450871-1452091), multidrug efflux RND transporter permease subunit AdeG (NZ_CP009257.1:1452098-1455277), multidrug efflux RND transporter outer membrane subunit AdeH (NZ_CP009257.1:1455290-1456738), multidrug efflux transcriptional repressor AdeL (NZ_CP009257.1:c1450673-1449643), multidrug efflux RND transporter periplasmic adaptor subunit AdeI (NZ_CP009257.1:1990739-1991989), multidrug efflux RND transporter permease subunit AdeJ (NZ_CP009257.1:1992002-1995178), multidrug efflux RND transporter outer membrane channel subunit AdeK (NZ_CP009257.1:1995178-1996632), and multidrug efflux transcriptional repressor AdeN (NZ_CP009257.1:4265008-4265661).
tBLASTn was used to search the translated nucleotide database using a protein query sequence for each of the carbapenem-associated resistance proteins, CarO1, CarO2, and CarO3 (with protein data bank accession codes 4RL9, 4RLB, and 4FUV). tBLASTn was also used to query the outer membrane protein A (GenBank: ATG88079.1), OmpW (NCBI Reference Sequence: WP_004712981.1), Omp33-36 (NCBI Reference Sequence: WP_000733003.1), glucose-selective porin OprB (GenBank: BAP65421.1), OprD outer membrane porin (NCBI Reference Sequence: WP_002059892.1), and a group of penicillin-binding proteins, including PBP1a MrcA membrane carboxypeptidase (NCBI Reference Sequence: WP_000736677.1), PBP1b (NCBI Reference Sequence: WP_000667420.1), PBP2_mrdA (NCBI Reference Sequence: WP_000809155.1), PBP3 FtsI (NCBI Reference Sequence: WP_000227939.1), PBP6 dacC (GenBank: AEK25710.1), PBP6b D-alanyl-D-alanine carboxypeptidase (NCBI Reference Sequence: WP_000667917.1), PBP7/8 pbpG D-alanyl-D-alanine endopeptidase (NCBI Reference Sequence: WP_001984577.1), and MtgA monofunctional biosynthetic peptidoglycan transglycosylase (NCBI Reference Sequence: WP_000642942.1).
Genes included in the analysis had >98% sequence identity and >25% query coverage OR > 90% sequence identity and >50% query coverage. The protein BLAST output or the translated protein sequence for each gene locus in each strain was then aligned using the MUSCLE option, following which it was visualized and comprehensively explored for different amino acid substitutions using the MEGA software. Gene variants identified for membrane protein genes were blasted using tBLASTn to blast the variant protein sequences against a custom-built nucleotide database using BV-BRC tools and services (Wattam et al., 2017; Olson et al., 2023).
A literature search was performed on each of the databases of PMC PubMed, ACADEMIC SEARCH COMPLETE (EBSCO host), Scopus, and Web of Science core collection using the following search criteria: “carbapenem resistant Acinetobacter baumannii” [title/abstract/keywords] or [topic] AND “resistance mechanisms” [title/abstract/keywords] or [topic]. A search was also performed to include abbreviations such as “carbapenem resistant A. baumannii” or “CRAB.” Additional, more detailed searches were performed using the following search criteria: “Acinetobacter baumannii” or “A. baumannii” AND “membrane proteins” and also “Acinetobacter baumannii” or “A. baumannii” AND “efflux pumps” to extract all previously identified variants in the studied genes. A summary of output variants previously mentioned in the literature is listed in Supplementary Table S2. These variants, in addition to other variants identified during the current analysis, were studied in further detail. A matrix was built to study the distribution of different variants and their correlation to carbapenem MIC in the agents studied. Predictive values of their performance as predictors for carbapenem resistance and their distribution in relation to genetic background relatedness were studied. This was performed by classifying the sequences under study into a susceptible group and a non-susceptible group based on MIC values at the cutoff value of 2, according to the latest EUCAST breakpoint tables (EUCAST, 2023). The significance of gene variants among the two groups was determined by conducting a chi-squared test for independence, and their predictive values toward the outcome of interest (susceptibility/lack of susceptibility) were determined as previously indicated (Nageeb and Hetta, 2022).
These measures were evaluated in relation to their genetic background distribution and predicted functional effects to filter out the most probable significant phenotype predictors. PROVEAN (Choi and Chan, 2015) and I-Mutant predictor suite (Capriotti et al., 2005) were used to predict the effect of the studied amino acid substitutions on protein function and protein stability, respectively.
Both PsiPred (McGuffin et al., 2000) available at http://bioinf.cs.ucl.ac.uk/psipred/ and FUpred (Zheng et al., 2020) available at https://zhanggroup.org/FUpred/ were used for domain and secondary structure prediction for each of the studied proteins. The CATH/Gene 3D v4.3 (Knudsen and Wiuf, 2010) database available at https://www.cathdb.info/ was searched using FASTA protein sequences to locate the position of structural domains on each efflux pump's protein sequence. UniProt (Consortium, 2019) was also searched for protein features and predicted structures using AlphaFold (Jumper et al., 2021) when available. Variants were mapped and visualized on the Protein 3D structure using BIOVIA Discovery Studio Visualizer v21.1.0.20298, Release 2020, San Diego: Dassault Systèmes and The PyMOL Molecular Graphics System, Version 2, Schrödinger, LLC.
To investigate the genetic relatedness of the studied sequences, whole genome-based phylogenetic reconstruction was performed using SANS serif (Rempel and Wittler, 2021). The length of K-mers was set to the default value of 31, and the assembled genomes' FASTA files were used as the input. SANS serif was used with the option “strict” to generate an output file in the Newick format, which was then visualized using iTOLv6 (Letunic and Bork, 2021). Phandango interactive visualization (Hadfield et al., 2018) was used to plot the variant distribution and other metadata in relation to the genetic relatedness phylogram.
In this study, we report the distribution of the structural and regulatory genes encoding different efflux pump systems among a large group of studied whole genome sequences of A. baumannii and their correlation to carbapenem resistance. The studied sequences included a total of 980 whole genome sequences isolated from different geographic localities, including United States (Alaska, Washington, and Maryland), China (Shandong, Liaoning, Zhejiang, Beijing, Guangdong, Jiangsu, Shanghai, Tianjin, and Hunan), South Korea (Seoul), Thailand, Sweden, Germany, Italy, Iraq, Colombia, United Kingdom, Afghanistan, Honduras, Peru, and India. The studied sequences were isolated from different body sources, including sputum, blood, abdominal fluid, tracheal mucosa, tissue, skin, feces, rectum, urine, and wound, and spanned a range of isolation years between 1982 and 2017. They included 423 imipenem-resistant, 557 imipenem-susceptible, 354 meropenem-resistant, and 208 meropenem-susceptible isolates.
In this study, we examined the molecular epidemiology and the predictive values of different membrane-related elements in relation to carbapenem resistance, including efflux-pump proteins, and we performed a more detailed analysis of the distribution of different variants related to these groups of genes. We aimed to explore the repertoire, predictive capacity, and phylogeny of these studied resistance determinants among a diverse group of clinical isolates.
Figure 1 illustrates the diversity of the studied sequences and shows the distribution of their phenotypes of imipenem and meropenem resistance, their geographic location, and the year of collection in relation to genetic background relatedness. From the figure, it can be observed that the studied sequences are genetically diverse with a heterogeneous distribution of resistance phenotypes in relation to the genetic background, country, and time of collection. Cluster 1 shows imipenem-resistant sequences, which were isolated from South Korea, and this cluster was located between two other carbapenem-sensitive clusters (Clusters 2 and 3) in the phylogram genetic order. Cluster 2 shows both imipenem- and meropenem-susceptible sequences that were collected in Washington, D.C. (USA) between 2003 and 2008. Similarly, Cluster 3 also contained carbapenem-susceptible sequences isolated from Germany and the USA, mostly between 2005 and 2009. Cluster 4 contained sequences that are resistant to both imipenem and meropenem collected from three different localities in China (Shandong, Liaoning, and Beijing), mostly during 2011, and this cluster was related to two other susceptible clusters (Clusters 5 and 6). Cluster 5 contained sequences that are susceptible to both imipenem and meropenem, mostly isolated in the USA between 2003 and 2009, while Cluster 6 contained carbapenem-susceptible isolates that were also isolated in the USA between 2005 and 2006. Both Clusters 7 and 8 showed closely related imipenem and meropenem non-susceptible sequences isolated from different countries, including Thailand, Sweden, USA, and China, with Cluster 7 being isolated between 2011 and 2016 and Cluster 8 between 2003 and 2010. Other examples showing the diverse distribution of the current analyzed set of sequences are Clusters 9 and 10. Both clusters contained imipenem-susceptible isolates that were isolated in the USA between 2006 and 2007 and between 2003 and 2008, respectively. The sequences from Cluster 11 contained imipenem- and meropenem-resistant sequences that were also isolated in the USA between 2003 and 2007.
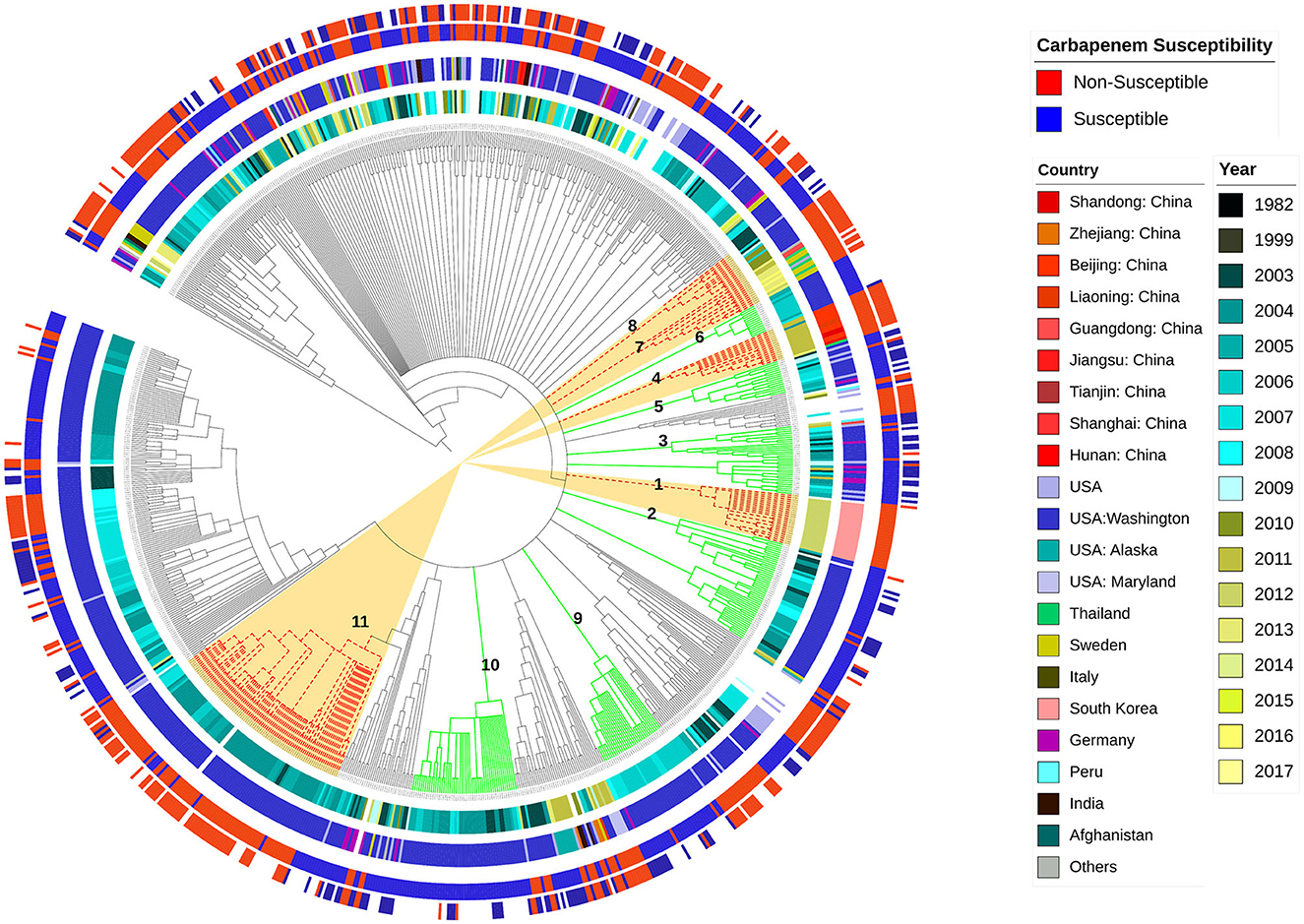
Figure 1. Whole genome-based genetic-relatedness phylogram and a heat map illustrating the correlation of the genetic background with the sequences' phenotype, their genome country of observation, and the year of isolation. Inner circle 1 shows different years of isolation. The second circle shows the country of isolation. The third circle represents the imipenem-susceptibility phenotype, while the fourth circle represents the meropenem-susceptibility phenotype.
AdeABC is known to be regulated by the AdeRS two-component regulatory system, AdeFGH by the LysR-type transcriptional regulator AdeL, and AdeIJK by the TetR-transcriptional regulator AdeN. Exploring these genes in the current analyzed set of sequences showed that AdeA occurred at the frequency of 94.18%, AdeB at the frequency of 95.61%, AdeC at the frequency of 68.57%, AdeR at the frequency of 92.85%, and AdeS at the frequency of 93.46%.
The AdeA gene was absent in 55 sequences, with two other sequences not meeting the identification criteria. The AdeB was absent in 30 sequences, with 13 other sequences not meeting the identification criteria. The AdeC was absent in 305 sequences, with three other sequences not meeting the identification criteria. AdeR was absent in 68 sequences, with two other sequences not meeting the identification criteria. The AdeS gene was absent in 62 sequences, with two other sequences not meeting the identification criteria. Both AdeF and AdeG were absent in six sequences, with one other sequence not meeting the identification criteria in both genes, while AdeH was also absent in six sequences, with two other sequences not meeting the identification criteria. The AdeL was absent in six sequences, while both AdeI and AdeN were absent in five sequences. The AdeJ was absent in four sequences, while AdeK was absent in four sequences, with two other sequences not meeting the identification criteria.
The analyzed structural and regulatory variants, related to the three studied efflux pump systems included a total of 117 variants, including the analysis of variants previously reported in the literature (shown in Supplementary Table S2) and other new variants analyzed in the current study. The percentage distribution of 97 variants occurring at more than 10 sequences among different imipenem MIC levels is shown in Figure 2A. Figure 2A illustrates a frequency of observation of significant efflux-pump gene variants in relation to different imipenem MIC levels. The frequency of significant efflux-pump variants occurring at more than 20 sequences is shown in Figure 2B in the descending order of frequency of occurrence among the entire studied set.
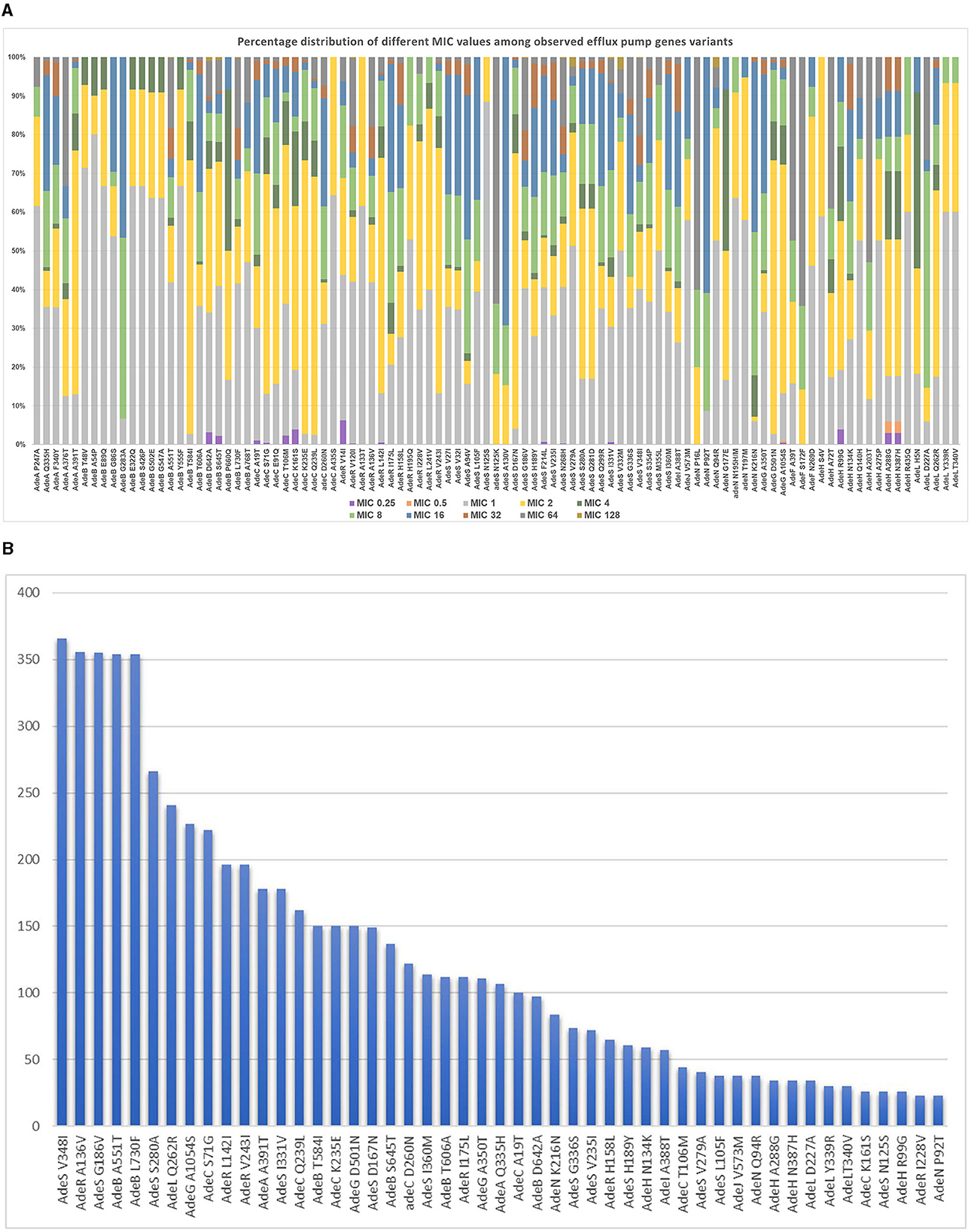
Figure 2. (A) Frequency of observation of significant efflux pump gene variants in relation to different MIC levels. (B) Frequency of observation of the most frequently occurring significant efflux-pump gene variants.
All 117 variants were analyzed for their significant correlation and predictive capacity toward the carbapenem-susceptibility phenotype, and the results of the group of variants showing a significant outcome are summarized in Tables 1, 2. A total of 67 and 76 variants exhibited significant differential distributions in relation to imipenem- and meropenem-susceptibility phenotypes, respectively. Among these, 27 variants were significantly encountered in both the imipenem and meropenem non-susceptible groups, while 40 variants and 49 variants were significantly more distributed in relation to imipenem- and meropenem-susceptible sequences, respectively. The position of variants in relation to each gene structure is diagrammatically illustrated in Figure 3.
Those showing a significant correlation to any of the susceptibility phenotypes were further tested for their predicted effect on gene function. The p-values indicating the significance of both imipenem and meropenem association, as well as the predicted functional effects of these variants, are shown in Tables 1, 2. These variants were also further mapped to the predicted secondary structure of each corresponding protein. Only specificity and PPV for each variant were considered. The sensitivity of the variants was not considered in this study. The variants identified did not occur at a high frequency in the population; therefore, they were not considered suitable candidates for use as screening markers, and as a result, their sensitivity was not assessed.
Regarding the distribution of membrane protein genes, Omp33-36 was completely absent in two sequences, while 292 other sequences showed hits below the threshold used for gene identification. Additionally, for OmpA, 52 sequences showed hits below the threshold used for gene identification. Genes identified below the threshold could be disrupted by insertion sequences and could be non-functional. OmpW was completely absent in nine sequences. OprB was absent in 1 sequence, while OprD was completely absent in 4 sequences, with 79 other sequences showing hits below the threshold used for identification. Three protein variants were identified for Omp33-36. These were further analyzed for their relation to carbapenem susceptibility. For OmpA, most studied sequences showed two variants. OmpW sequence appeared to be highly conserved, with V1 being the most frequently occurring variant and another two variants that were observed at a lower frequency. OmpW T18A was observed and further studied. Three variants were observed in OprD, with V2 and V3 being observed in fewer isolates. Two variants were observed in OprB, with V2 being observed in fewer isolates. Substitutions such as OprB F57L, OprB T304A, OprB S360N, and OprB G403S were observed and further analyzed.
CarO sequence was identified in 71 sequences, with hits below the threshold used for gene identification. CarO1 showed a significant correlation to imipenem resistance, while CarO3 variants showed a correlation to meropenem resistance, although it was non-significant. Omp33_V2 showed a significant correlation to the imipenem-susceptibility phenotype, while Omp33_V3 showed a significant correlation to the meropenem-susceptibility phenotype. OmpA_V1 showed a significant correlation to imipenem and meropenem resistance, while OmpA_V2 showed a significant correlation to imipenem susceptibility. OprD_V2 showed a significant correlation to meropenem susceptibility, while OprD-V3 showed a significant correlation to imipenem susceptibility. OprB G403S showed a significant correlation to both imipenem and meropenem resistance. OprB N360S showed a significant correlation to imipenem susceptibility, while OprB F57L was significantly correlated to meropenem susceptibility.
FASTA sequences of the studied variants are provided in Supplementary Table S3. The significance of the variants in relation to each of the imipenem and meropenem-susceptibility phenotypes along with the predictive values for the observed variants in membrane protein genes are shown in Table 3.
PBPs previously reported in the literature (listed in Supplementary Table S2), in addition to other observed variants, were explored for their correlation to imipenem- and meropenem-susceptibility phenotypes. Variants showing a significant correlation to imipenem- and meropenem-susceptibility/resistance, together with their predictive values and functional effect on protein stability, are shown in Tables 4, 5, respectively.
Figure 4 shows the distribution of different efflux-pump gene variants and carbapenem-resistance phenotypes in relation to the genetic-relatedness phylogram, while Figure 5 shows the distribution of different membrane protein gene variants and carbapenem-resistance phenotypes in relation to the genetic-relatedness phylogram.
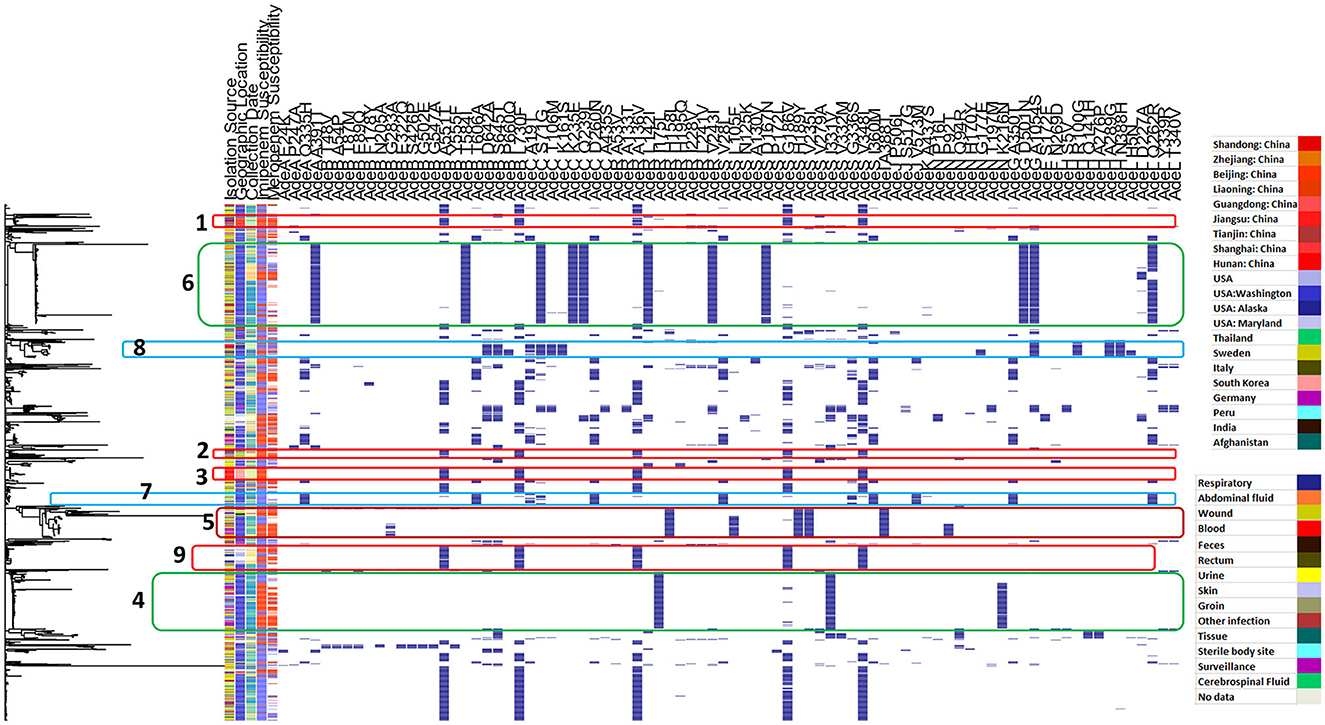
Figure 4. Distribution of different efflux pump gene variants and carbapenem-resistance phenotypes in relation to the genetic-relatedness phylogram. Dark blue color indicates the presence of an efflux-pump gene variant. Blue color indicates carbapenem susceptibility, while red color indicates non-susceptibility to carbapenems.
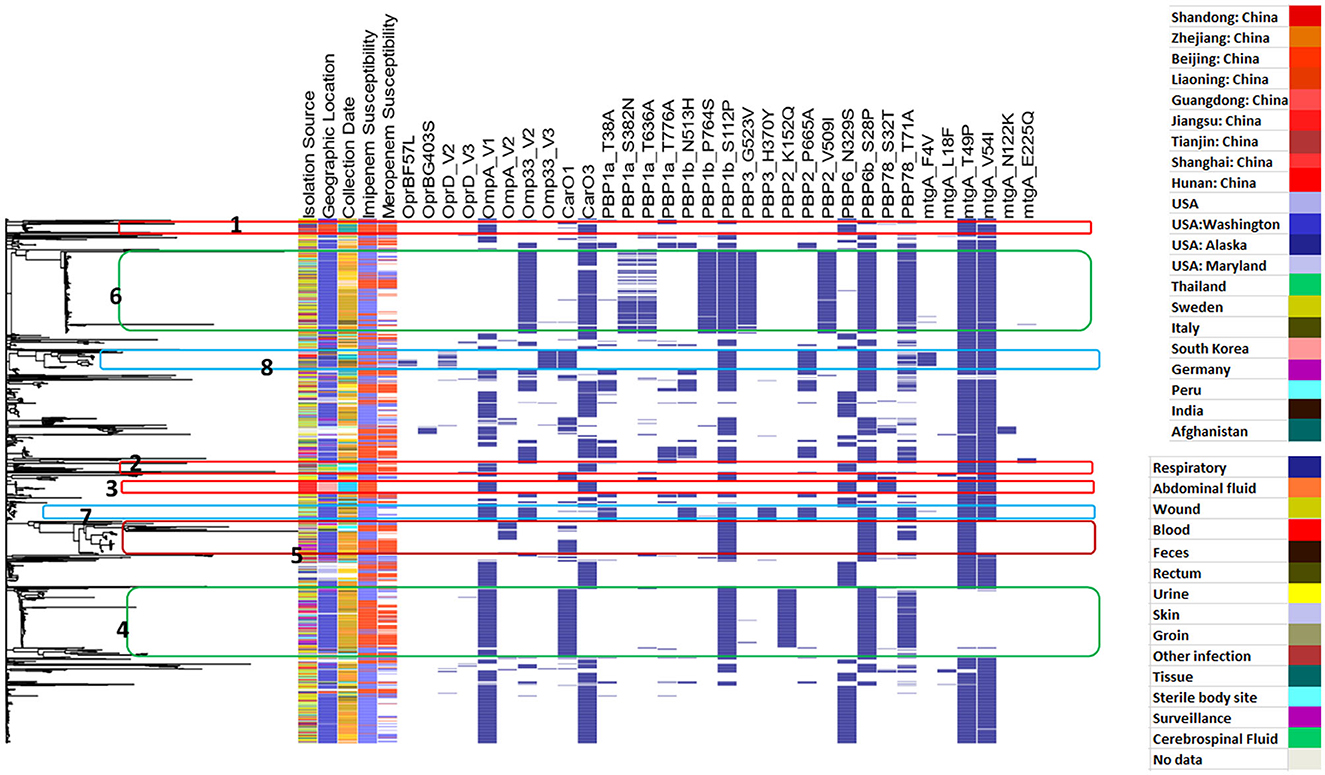
Figure 5. Distribution of different membrane-protein gene variants and carbapenem-resistance phenotypes in relation to the genetic-relatedness phylogram. Dark blue color indicates the presence of a membrane protein variant. Blue color indicates carbapenem susceptibility, while red color indicates non-susceptibility to carbapenems.
AdeABC is a chromosomal RND-type efflux system. It was the first efflux pump described in A. baumannii in 2001, followed by AdeIJK in 2008 and AdeFGH in 2010. AdeABC is the most frequently studied type, and the literature shows evidence that the detection rate of AdeB is the highest in clinical isolates compared to AdeFGH and AdeIJK, although variable among other strains (Xu et al., 2019). Based on MIC changes in different pump mutants, it has been assumed that the three carbapenems could act as substrates for AdeABC, while only meropenem could be a substrate for the AdeIJK pump (Yoon et al., 2015).
Researchers have also shown that overexpression of the AdeABC system could significantly contribute to resistance against meropenem in A. baumannii clinical isolates (Cardoso et al., 2016). The high incidence of AdeABC efflux-pump hyper-expression in MDR A. baumannii has been outlined in several reports and linked to several mutations in the corresponding two-component regulatory system (Yoon et al., 2013). These mutations have been revised and explored in the current study set. Some studies have shown the universal presence of RND-type efflux pumps and their variable expression level, highlighting the complexity of their regulatory expression and the role of expressed pumps in MDR (Pagdepanichkit et al., 2016). AdeABC has been described to be common in imipenem-resistant A. baumannii, leading to higher levels of resistance when combined with other resistance-conferring elements (Xiao et al., 2016). More recently, the contributions of AdeABC and AdeIJK to drug resistance and growth physiology have been analyzed, showing different specificities and non-overlapping changes in both pump systems (Leus et al., 2020).
Different studies have investigated the distribution of different efflux-pump-related genes, their expression, and their correlation to the MDR phenotype (Lin et al., 2017; Chen et al., 2018; Basatian-Tashkan et al., 2020). Although it has been proposed that high resistance levels do not occur because of efflux-pump genes, their variable expression levels cannot be ignored in relation to MDR (Basatian-Tashkan et al., 2020). Furthermore, the presence of the AdeABC genes in sensitive strains is assumed to be low, suggesting the use of AdeABC as a sign of resistance in A. baumannii (Lin et al., 2009; Xu et al., 2019). No direct evidence confirming the link between the presence/absence of porin proteins and efflux systems and carbapenem susceptibility in A. baumannii has been found, with a study concluding that intrinsic resistance mechanisms, including both membrane proteins and efflux pumps, do not play a major role in carbapenem resistance; however, the study may have underestimated this role as it included only 14 isolates from one geographic locality (Wong et al., 2019). This needs to be carefully studied and analyzed in an extensive collection of sequences, especially focusing on different variants and their potential role in modifying the expression or functionality of these genes, which was applied in the current study.
However, the AdeIJK efflux pump has been detected in both sensitive and resistant isolates and is considered a factor of intrinsic resistance with the average expression level of AdeJ being relatively low (Coyne et al., 2011) or entirely non-overexpressed in some studies (Yoon et al., 2013). Its regulatory gene AdeN has been shown to have a high level of amino acid conservation (Touchon et al., 2014). Mutated or truncated inactive AdeN protein has been shown to result in AdeIJK pump overexpression (Rosenfeld et al., 2012), which underlies meropenem and doripenem resistance (Fernando et al., 2014). The overexpression of the AdeIJK efflux pump because of mutations in the negative regulator AdeN has been related to the increase in meropenem MIC (Fernando et al., 2014). No studies have directly compared the effect of AdeABC and AdeIJK efflux pumps, and further studies were recommended to understand the importance of the AdeIJK effect on A. baumannii (Xu et al., 2019).
The AdeABC system is under the regulatory control of the AdeRS two-component regulatory system, which contains a sensor kinase coding gene AdeS and a response regulator coding gene AdeR (Marchand et al., 2004). AdeRS operon is known to express high genetic variability with different alleles and variants being reported, especially the AdeS gene (Sun et al., 2014; Hammerstrom et al., 2015; Montaña et al., 2015). A list of previously reported substitutions in these two genes is shown in Supplementary Table S2.
AdeFGH is another RND efflux system conferring MDR when overexpressed, and its structural genes are under the regulation of the upstream-located AdeL. It is not constitutively expressed in wild-type strains, so it is not believed to contribute to intrinsic resistance (Coyne et al., 2011).
Previous studies have shown AdeAB and AdeRS to be highly distributed among MDR AB clinical isolates, while AdeC was less frequently distributed (Wieczorek et al., 2013). In their study, Wieczorek et al. (2013) showed that AdeB was found throughout the studied strains. In the same study, three strains had no AdeR gene, and a single strain had no AdeA gene or AdeS gene. AdeC was negative in 16 strains among 100 studied randomly selected isolates, making it the least abundant. Other studies have reported the distribution of AdeB at 70% (Chu et al., 2006), 75% (Lin et al., 2009), and 87% (Nemec et al., 2007). The latter study demonstrated the absence of AdeA, AdeB, AdeR, and AdeS in 11 out of 116 studied isolates, while AdeC was detected at a much lower frequency (only in 49 out of 116) (Nemec et al., 2007). These results align with those obtained from the analysis performed in this study; however, our results may be more representative as they include the analysis of a diverse group of sequences that are timely and geographically separated.
A more recent study investigated the distribution of AdeABC and AdeRS genes among 75 A. baumannii environmental isolates isolated from contaminated soil. The study determined the distribution of AdeA at 66.6% (50/75), AdeB at 60% (45/75), AdeC at 30.6% (23/75), AdeR and AdeS at 54.6% (41/75), and 48% (36/75) of isolates, respectively (Al-Tofaily et al., 2020).
Other studies have also shown a high distribution of AdeABC-AdeRS efflux-system genes in CRAB strains, which was reported at 90%, and a significantly increased distribution of the regulator genes AdeR and AdeS in CRAB strains as compared to CSAB strains. CRAB strains also showed a significantly increased expression of AdeB (12.3-fold) but a decreased expression of AdeR (3.3-fold) (Chen et al., 2018). Another study concluded the universal presence of some efflux-pump genes and correlated AdeB and AdeJ upregulation to MDR; however, the study only included 21 isolates (Lin et al., 2017). Similarly, other investigators have reported the high prevalence of AdeA, AdeB, and AdeC genes among clinical isolates of A. baumannii showing imipenem and meropenem resistance. The efflux-pump genes occurred at the frequencies of 96.2, 96.2, and 91.1%, respectively, and were significantly related to imipenem resistance (Ranjbar et al., 2020).
Both AdeA and AdeS were also detected at a high frequency among A.baumannii clinical isolates, where 80% (48/60) of A. baumanii isolates had the AdeA gene and 81.66% (49/60) of isolates had the AdeS gene (Basatian-Tashkan et al., 2020). Other researchers have also identified both genes at similar frequencies of 79.6 and 80.6% for the AdeA and AdeS genes, respectively (Jia et al., 2015). Nemec et al. (2007) identified both AdeA and AdeS at a similar distribution of 81.9%. Contrarily, AdeA was identified at a frequency of only 62.7% in other studies (Kor et al., 2014). In another study, the AdeR and AdeS genes were detected in 36/60 (60%) and 59/60 (98.3%), respectively (Ostad Asadolah-Malayeri et al., 2016). In yet another study, AdeRS was positive in 31/37 (84%) of A. baumannii isolates studied (Montaña et al., 2015). Another study reported the presence of different pump genes at the frequencies of 60 (92%) for AdeA, 40 (61.5%) for AdeB, and 55 (84.6%) for AdeC, while AdeR was detected upstream of the AdeABC operon in 38 (58.4%) of the studied isolates (Modarresi et al., 2015). Another study identified the AdeR, AdeS, and AdeB genes at the percentages of 96.8 (61/63), 63.4 (41/63), and 79.3 (50/63) of isolates, respectively (Hassan et al., 2018).
In addition to AdeABC, two other RND pumps, AdeIJK and AdeFGH, also contribute to resistance in the A. baumannii clinical isolates. Coyne et al. (2010) showed that AdeABC was detected in 80% of clinically isolated resistant strains, while AdeFGH operon was identified in 36 out of 40 A. baumannii clinical isolates. Researchers have also identified AdeL mutations in three AdeFGH-overexpressing mutants in the same study set.
In another study, the genes of the AdeABC, AdeFGH, and AdeIJK efflux pumps existed in all AB-XDR clinical isolates. The transcriptional level of AdeA was significantly higher than the AdeF and AdeI genes. Seventy out of 81 (70/81) isolates (86.4%) overexpressed AdeABC, and only 17 out of 81 isolates (21.0%) overexpressed AdeIJK, while no isolate overexpressed the AdeFGH pump system (Sun et al., 2014).
Positive AdeB and AdeJ were detected in 47 (94%) and 45 (90%) of the 50 studied imipenem-resistant isolates, respectively. Forty-three out of 47 (43/47) AdeB-positive imipenem-resistant A. baumannii (IRAB) carried the AdeRS regulatory system, which was absent in four isolates. Six (12%) isolates of IRAB carried AdeB and AdeE simultaneously. In addition, AdeB and AdeJ showed approximately 4- and 3-fold increases in expression levels in imipenem-resistant isolates as compared to imipenem-susceptible isolates (Hou et al., 2012). Both AdeA and AdeI showed overexpression in MDR strains (Leus et al., 2020).
The reason for some observed differences may be attributed to variations in antibiotic usage patterns in the studied localities, the type of clinical samples isolated, the number of samples studied, the sampling method, environmental factors, or differences in geographical distribution of these genes. However, we believe that the frequencies identified in the current analysis better reflect the general epidemiology of these pump genes due to the inclusive and representative nature of this study.
AdeABC was the first RND-type efflux pump discovered in A. baumannii and has been extensively studied. AdeA is the membrane fusion protein part of the efflux pump acting as a periplasmic adapter with transmembrane transporter activity. It facilitates interaction between transporters and outer membrane channels and links the reactions between the inner and outer membranes together (Zgurskaya et al., 2015). As a membrane fusion protein, AdeA is a linear protein formed of four characteristic domains (Supplementary Figure S1A) that extend from the inner membrane into the periplasm to meet the outer membrane channel (Mikolosko et al., 2006).
In the current analysis, AdeA Q335H was observed in the carbapenem non-susceptible group. The variant was located at the upper end of the membrane-proximal domain at the start of a beta sheet at the upper pocket near the short flexible linker between the membrane-proximal domain and the beta-barrel domain (Supplementary Figure S1B). These linker regions are known to be highly flexible and are capable of making conformational changes due to high structural variation that results in the transmission of energy from the inner membrane to the outer membrane, which is needed for drug efflux (Zgurskaya et al., 2015).
GLN at 335 links through a 3.1 hydrogen bond to ILE 342. When GLN was mutated to HIS, no change in the hydrogen bonding was observed. It appears that the nature of the change of the polar glutamine to histidine with the imidazole side chain may be needed to effectively participate in catalysis. Positively charged residues located at the interface between the β-barrel and the MP domains are important for both binding to the core LPS and the functionality of the pump (Shuo et al., 2013). Researchers have also observed that the protonation/deprotonation of HIS underlies the PH-regulated conformational dynamics of AcrA (Wang et al., 2012), a similar MFP efflux pump. Twisting of the MP domain relative to the beta-barrel domain has been previously observed among the conformational changes needed for the efflux process. This twisting may be necessary for the steps of access and binding of the drug to the efflux transporter (Symmons et al., 2009). Based on this, the change of GLN to HIS at position 335 may contribute to a more stable binding, potentially leading to more effective efflux and contributing to resistance.
Three other variants were identified in the carbapenem-susceptible group: AdeA E24K, AdeA A391T, and AdeA P247A. AdeA P247A was located in the middle of a coil connecting the two groups of beta strands that form the beta-barrel domain, as shown by the predicted structure using AlphaFold (Supplementary Figure S1C). Although the mechanism is not clear, the change of PRO to ALA appears to destabilize drug-binding, thereby compromising the efflux process. Although AdeA E24K was located in an area of very low model confidence, as predicted by the AlphaFold model, position 24 was located just next to the predicted signal peptide found at the start of the efflux RND transporter periplasmic adaptor subunit. Similarly, AdeA A391T was located in a predicted coil at the end of the multidrug resistance protein and also in an area of very low model prediction confidence. Due to their location in an area of low model prediction confidence, the functional effects of these variants could not be hypothesized.
AdeF and AdeI components of the efflux pumps AdeIJK and AdeFGH, respectively, perform a similar periplasmic adapter function as that of AdeA. In this analysis, AdeI A388T and AdeF S172F variants were detected in the carbapenem non-susceptible group. AdeF S172F was located at the lower side of the α-helical hairpin that spans the periplasm (Supplementary Figure S2). The α-helical hairpins are known to oligomerize and assemble a funnel-like structure that interacts with the outer membrane channels to help extrude the drug. It is postulated that the interaction between the MFP transporter and the outer membrane channels is dynamic and results from conformational changes that transmit energy, resulting in open-channel conformations that extrude the drug (Tikhonova et al., 2007). SER at position 172 appears to be tightly bonded to the surrounding residues, forming 5 hydrogen bonds within its containing alpha helix. These include two hydrogen bonds (3A and 3.3A) with ASN 176, a 3.2 A bond with LEU 175, a 3.9 A bond with ALA 169, and a 2.9 A bond with ALA 168. The change of SER to the bulky hydrophobic non-polar PHE may result in chain instability; this may consequently lead to conformational changes because of the hydrophobic interaction with the nearby non-polar amino acids, which is known to stabilize folded protein conformation. AdeI A388T was located in a predicted coil at the end of the proteins in an area of very low model prediction confidence, so its function could not be predicted.
Both the multidrug transporter AdeB and outer membrane channel AdeC are considered structural proteins of the pump that promote drug discharge, where AdeB captures the substrate in the inner membrane of the phospholipid bilayer or cytoplasm, while AdeC transports the substrate outside the cell (Xu et al., 2019). The AdeB shows the highest detection rate in clinical isolates and is considered to be most vitally related to resistance in A. baumannii (Yoon et al., 2016). The three variants AdeB T48V, AdeB T584I, and AdeB P660Q were significantly detected in the carbapenem-susceptible group and were predicted to be deleterious mutations. The mutations were mapped on the PDB available 3D structure found in the RCSB Protein Data Bank with accession number 6OWS.
P660 is one of the important residues for substrate recognition and binding at the periplasmic cleft multidrug-binding sites (Su et al., 2019). AdeB P660Q was located at the apex periplasmic binding cleft between the subdomains PC1 and PC2 at the start of the F loop that connects the cleft entrance to the proximal drug-binding pocket (Su et al., 2019) (Supplementary Figure S3A). On the contrary, both AdeB T48V and AdeB T584I were located at the distal part of the binding cleft (Supplementary Figure S3A). AdeB T48V was located at the last position of the first beta strand located approximately in the middle of the multidrug efflux transporter pore domain. Similarly, AdeB T584I was located in the second predicted multidrug efflux transporter pore domain. It was located at the beginning of an alpha helix just opposite T48V. It appears that these mutations in the periplasmic cleft affect binding to the carbapenem molecule and that destabilization of its structure affects this binding, consequently affecting drug transport and discharge outside the cell. This would consequently make the cell carbapenem susceptible by preventing carbapenem efflux. These positions may act as probable targets for new efflux pump inhibitors.
AdeB G283A was located at the junction of two small beta sheets in close proximity to the multidrug efflux transporter pore domain but on the outer side. The residue was located near the distal drug-binding site (Supplementary Figure S3B). It may also have a role in binding to the carbapenem molecule. It was predicted to be deleterious and was significantly detected in the carbapenem-resistant group.
Similar to AdeB, both AdeG and AdeJ recognize and transport substrates through a drug-proton antiporter mechanism (Ruggerone et al., 2013). AdeG A350T was identified in the carbapenem non-susceptible group, while AdeG D501N, AdeJ P506L, AdeJ S517G, and AdeJ V573M were identified in the carbapenem-susceptible group.
AdeJ P506L, AdeJ S517G, and AdeJ V573M were mapped on the AdeJ efflux-pump structure available at PDB (7M4Q). AdeJ V573M is located near the proximal drug-binding site (Zhang et al., 2021) (Supplementary Figure S4). It has been proposed that non-polar residues at such drug-binding sites are generally involved in the recognition of different antimicrobials through hydrophobic interactions (Zhang et al., 2021). Although both are non-polar hydrophobic amino acids, the change of valine to methionine may affect drug recognition, binding, and, consequently, its extrusion because methionine residue contains sulfur atoms, which are more susceptible to oxidation.
AdeG A350T and AdeG D501N were mapped to the AlphaFold-predicted 3D structure (Supplementary Figure S5). AdeG A350T was linked to carbapenem resistance and was located in the middle of an alpha helix at the drug-access site near the inner membrane. AdeG D501N was also located at the lower end of the molecule in the middle of a predicted loop just below the inner membrane. The functional effect of these mutations needs to be further studied.
AdeC is an outer membrane channel protein that was reported to occur at the lowest frequency, and it is considered not necessary for AdeABC-mediated drug efflux. However, its presence usually correlates with multidrug resistance or pan-resistance (Xu et al., 2019). AdeC A19T was located at the area of the alpha helix identified at very low confidence prediction by AlphaFold and was related to carbapenem resistance. AdeC S71G was located at the center of the third alpha helix, forming a pocket just parallel to the four large alpha helices, while the AdeC Q239L deleterious mutation was located in the opposite alpha helix at the boundaries of the same pocket (Supplementary Figure S6). Both variants were observed in the carbapenem-susceptible group. AdeC T106M was located at the first position of the first beta sheet at the junction between the beta sheets and alpha helices and was also observed in the carbapenem-susceptible group. The mentioned variants appear to affect the function of AdeC in transporting carbapenem molecules outside the cell; however, the dynamics related to this need to be further studied.
Similar to AdeC, both AdeH and AdeK provide channels for substrates to cross the outer membrane. Two variants, AdeH R100G and AdeH A289G, were significantly related to meropenem susceptibility, showing high specificity, and they were predicted to be deleterious (Supplementary Figure S7).
The function of AdeS is to receive the environmental signals causing autophosphorylation and then to transfer the phosphoric acid to the output responder AdeR, which acts as a transcriptional activator (Marchand et al., 2004). The AdeS structure was analyzed in this study, and two domains were predicted using different tools. These domains include the HAMP linker domain and the histidine kinase domain. It has been suggested that the HAMP domain with the alpha-helical regions common to many chemoreceptors plays a role in regulating the phosphorylation of homo-dimeric receptors by transmitting the conformational changes in periplasmic ligand-binding domains to a cytoplasmic signaling kinase (Aravind and Ponting, 1999). It has also been proposed that the two HAMP alpha helices take two different conformational states depending on the ligand-bound state of the chemoreceptor and that a transmembrane signaling helix can switch between these conformational states. Furthermore, it has been proposed that the bihelical HAMP domain of monomeric histidine kinases binds intramolecularly to the bihelical dimerization domain and that ligand-mediated change in HAMP domain conformation leads to the subsequent formation of the four-helix bundle within histidine kinase dimers (Aravind and Ponting, 1999).
Three deleterious mutations were observed in the HAMP linker domain, AdeS L105F, AdeS N125K, and AdeS A130V. Mapping these variants on the AlphaFold-predicted structure showed that L105F was located at the turn between two coils, while A130V was located just opposite to it near the start of the following alpha helix. Similarly, N125K was located in the same area (Supplementary Figure S8A). It appears that L105F may have a role in making bihelical dimerization conformational changes, helping to transmit phosphorylation reactions. Supporting this is the nature of the substitution of leucine into the bulky hydrophobic phenylalanine residue with an aromatic ring that is known to stabilize the folded conformation of proteins and that frequently forms the cores of protein folds.
Three other deleterious variants were observed in the histidine kinase domain, AdeS D167N, AdeS P172L, and AdeS G186V. Previous studies have shown that AdeS inactivation leads to aminoglycoside susceptibility with Thr153 Met mutations downstream from the putative His-149 site of autophosphorylation, probably leading to the loss of phosphorylase activity by the sensor (Marchand et al., 2004). In the current analysis, the two deleterious mutants, AdeS D167N and AdeS P172L, showed a correlation with carbapenem susceptibility. AdeS D167N was located at the last position of an alpha-4 helix and the following coil on which AdeS P172L was located. Contrarily, AdeS G186V was located in the middle of an alpha helix and probably at the boundary of a binding pocket (Supplementary Figure S8B). This substitution was conserved in tigecycline-resistant isolates in other studies (Sun et al., 2014). Although its role was not clear, it was proposed that previous selective pressure resulting from the use of different antibiotic agents, including carbapenem, was related to the emergence of tigecycline resistance.
I175L and H195Q were mapped on the available 3D structure of AdeR from PDB (5X5L) for the C-terminal DNA-binding domain for the positions 139-247. Supplementary Figure S9A shows only chain A and chain E for illustration purposes. I175L was identified in the middle of an alpha helix, probably affecting the DNA-binding pocket, and H195Q was also located near the end of another small alpha helix closely related to the DNA-binding pocket (Supplementary Figure S9A).
AdeR A53V was the only variant identified in the response regulator receiver domain. Mapped on the predicted structure for that domain, this amino acid substitution occurred near the end of the alpha-2 helix. It appears that it is located as a part of the magnesium-binding motif with a role in the phosphorylation of AdeR. Thus, it may be correlated with the expression of the efflux pump AdeABC. ALA 53 forms a polar interaction of 2.2 A with Ile 49 (Supplementary Figure S9B). The change of the ALA 53 to VAL leads to an additional 2.7 A bonding with GLU 50.
A133T and A136V are another two variants located in between the two domains of AdeR. A133T was located at the end of an alpha helix, and A136V was located at the start of the coil connecting the two domains (Supplementary Figure S10A). ALA 133 forms a polar interaction of 2 A with ARG 129, while ALA 136 forms a polar interaction of 2.9 with ASN 138 (Supplementary Figure S10B). Mutating ALA 136 to VAL leads to an additional bond of 2.5 A to ALA 133. Additional mutation of ALA 133 to THR leads to additional 2.2 bonding to VAL 136, probably adding more stability (Supplementary Figure S10B).
Previous studies have demonstrated the contribution of outer membrane channel loss or mutation, including OprD and CarO, to the carbapenem-resistance phenotype in Acinetobacter (Morán-Barrio et al., 2017). Although genetically engineered experimental mutants of A. baumannii with mutated CarO, Omp33, and OprD have shown almost similar MICs (<2-fold difference) compared to parenteral strains, ΔompA mutants were more susceptible to both imipenem and meropenem when compared to parenteral strains (2–3 fold difference) (Tsai et al., 2020). In addition, similar findings showed <2-fold change in imipenem MIC with ΔompA mutant compared to the wild-type strain (Kwon et al., 2017). This slight change in imipenem and meropenem susceptibility resulting from OmpA changes was supported by the finding in the current analysis, which showed lower specificity and PPVs for the OmpA_V1 variant observed (Table 3). This contrasted the significant increase in meropenem resistance, which was observed (>4-fold difference) with single AdeRS mutations. However, a <2-fold increase in resistance was observed with ΔAdeAB. ΔAdeH and ΔAdeL were also shown to have similar results (Tsai et al., 2020).
Large-scale quantitative proteomics analysis demonstrated that membrane proteins, including each of OmpA, CarO, and OmpW, were more abundant in MDR isolates (Chopra et al., 2013). Another study showed that the overexpression of genes encoding RND pumps—especially the AdeFGH pump—and the downregulation of porin-encoding genes are common in clinical CRAB isolates (Fernando et al., 2013). Studies have also shown that imipenem exposure can notably increase the expression levels of different RND efflux pumps and PBPs and decrease the levels of OmpW, OmpA, and CarO to prevent antibiotic import (Yun et al., 2011). In addition, recent proteomic and transcriptional analyses have identified OmpA and OmpW among the major proteins related to extensive drug resistance (Lee et al., 2018). In the current study, OmpW appeared to be highly conserved in the studied set, and although the variant OmpW T18A was observed, it did not show a significant correlation to carbapenem-resistance phenotypes.
Loss of a 43-kDa protein identified as a homolog of the OprD protein in P. aeruginosa has been previously linked to carbapenem resistance (Perez et al., 2007). Contrasting results did not correlate CarO, OprD, and 33-36Kda Omp expression to carbapenem resistance and showed identical outer membrane protein profiles among the studied panel (D'Souza et al., 2019). However, these observations may be deviated by the type of studied isolates. In the current set of studied isolates, two OprD variants correlated significantly with carbapenem susceptibility, and despite being observed in fewer isolates, both OprD_V2 and OprD_V3 showed high specificity to meropenem- and imipenem-susceptibility behavior, with values of 97.7 and 99.5%, respectively (Table 3). However, Omp33_V2 showed a significant correlation to the imipenem-susceptibility phenotype, while Omp33_V3 showed a significant correlation to the meropenem-susceptibility phenotype, with Omp33_V3 showing high specificity (96.6%) (Table 3).
The absence of the 29-kDa OMP (CarO) and 33-kDa outer membrane proteins has been previously associated with carbapenem resistance in A. baumannii (Limansky et al., 2002). In addition, Yang et al. (2015) showed CarO absence in 2 out of 27 studied MDR isolates. From similar findings, we can speculate that the channel would be changed due to mutations in the membrane porin, and this could result in resistance either by totally preventing antibiotic entry or by decreasing its amounts. However, another study from Brazil concluded that OXA-23 carbapenemase activity is the major carbapenem resistance mechanism among 28 studied isolates and that CarO loss plays only a minor role. However, the same study could associate different CarO alleles and their levels of expression with imipenem resistance, highlighting the complexity of understanding the carbapenem-resistance phenotype (Fonseca et al., 2013). Our findings that showed either the absence of a significant correlation or low predictive values of the studied CarO variants toward both meropenem- and imipenem-susceptibility phenotypes support these latter findings.
The glucose-selective porin OprB, the outer membrane protein A precursor, and the 33–36 kDa outer membrane proteins were among the major outer membrane proteins downregulated in imipenem culture conditions (Lee et al., 2014). Two variants located in the carbohydrate-selective porin domain OprB showed significant correlations to carbapenem-susceptibility behavior (Table 3).
In this analysis, OprB F57L was significantly correlated with meropenem susceptibility, showing high specificity and a predicted large decrease in protein stability. The variant is located in the middle of the first beta strand, which has an extracellular location near the protein N-terminal (Supplementary Figure S11). In Pseudomonas, position 56 was described as highly conserved in OprB orthologs and is located at strand S2, lining the porin channel with its side chain (van den Berg, 2012). From the current findings, it appears that the change of the bulky phenylalanine with an unreactive aromatic ring to leucine at position 57 might facilitate meropenem-binding and entry through the porin channel. However, the exact mechanism needs further study.
On the other hand, OprB G403S showed a significant correlation with both imipenem and meropenem resistance with high specificity and PPVs (Table 3). Functional prediction indicated that the variant is associated with a large decrease in protein stability. It is located in a coil preceding the last beta strand in the protein structure and has a cytoplasmic location near the C-terminal, as predicted using Psipred (Supplementary Figure S11). Position 402 was described among the highly conserved residues in OprB orthologs and is located in Loop L8, allowing a tight turn (van den Berg, 2012).
It can be concluded that, among membrane proteins, both OmpA and CarO have a lower contribution to the change in carbapenem resistance, and both OprB and OprD variants show a greater effect.
However, a study by Sen and Joshi (2016) concluded the absence of a correlation between each of OprD, AdeABC, and AdeIJK efflux pumps and carbapenem resistance based on expression data results. Their results may be deviated and non-generalizable because the study only included 16 carbapenem-resistant isolates, which might have greatly underestimated the contribution of these mechanisms. However, other research reports have shown that the combination of carbapenemases, efflux-pump expression, and the absence of outer membrane proteins contribute to carbapenem resistance in A. baumannii (Alcántar-Curiel et al., 2014).
PBPs are an important family of enzymes inactivated by B-lactam drugs, including carbapenems. Recognizing and exploiting variations in these proteins is an essential approach in new antibiotic drug discovery and engineering to meet the growing challenge of multidrug-resistant Gram-negatives. Several studies have reported the role of PBP3 mutations in meropenem resistance (Hawkey et al., 2018; Nelson et al., 2020). The role of other PBPs in imipenem resistance has also been previously demonstrated (Cayô et al., 2011).
Researchers have reported that both PBP1a and PBP3 have unique roles in drug-resistant A. baumannii and could potentially underlie therapeutic failure (Kang and Boll, 2022). The analysis of variants observed in these proteins is, therefore, an extremely vital topic. The essentiality of PBP3 for the survival of A. baumannii has been recently emphasized (Toth et al., 2022). In our study, the variant PBP3_H370Y showed a significant correlation with the imipenem-susceptibility phenotype, showing very high specificity and PPV (Table 4). This position occurs near the drug-binding site, showing hydrophobic interactions, as observed in previous studies (Han et al., 2011; Papp-Wallace et al., 2012). Given the results of previous studies (Toth et al., 2022), in addition to the results of our study, it can be concluded that the amino acid position 370 of PBP3 may act as a potential drug target that can be exploited to counteract imipenem resistance in A. baumannii.
In the same study (Toth et al., 2022), the researchers demonstrated that the inactivation of PBP1a results in bacterial growth retardation and an increase in susceptibility to B-lactam. This was further enhanced by the additional inactivation of PBP2, resulting in a 4-fold increase in B-lactam antibiotic susceptibility. A similar trend can be concluded from the results of this study, where the PBP1a_T636A variant associated with a large decrease in stability can act as a potential drug target together with PBP1a_S382N, both acting as markers of imipenem susceptibility and showing high specificity and positive predictive values (Table 4).
Mapping PBP1a_S382N to the PBP1a 3D structure (ID = 3UDX) showed that the variant occurs at the transpeptidase (TP) domain, whose inactivation was associated with increased B-lactam susceptibility observed in other studies (Toth et al., 2022). Both the statistical and functional evidence reported in this study, as well as additional experimental evidence from other studies (Kang and Boll, 2022; Toth et al., 2022), make these positions potentially important drug targets.
Researchers have recently reported that a decreased affinity of PBP2 to carbapenems is one of the best described target site changes in A. baumannii (Martínez-Trejo et al., 2022). The lack of PBP2 was one of the most common mechanisms reported in CRAB isolates (Fernández-Cuenca et al., 2003), and these facts can be supported by the findings of our study. The results from the current study show that the variant PBP2_K152Q, predicted to cause a large decrease in protein stability, was significantly correlated with meropenem resistance, showing both high specificity and high PPV (Table 4).
Based on the PsiPred output for predicting the 3D structure, MtgA_L18F was located in the middle of the first alpha helix, which is a transmembrane helix showing membrane interaction (Supplementary Figure S12). Mapping the amino acid changes on the AlphaFold predicted 3D structure of MtgA showed that L18 forms three hydrogen bonds of 2.9 A, 2.9 A, and 3.4 A, respectively, within the containing alpha helix. While the F residue contains an unreactive aromatic ring, mutating the hydrophobic residue L into the bulky F residue may affect the protein-membrane interaction. MtgA_N122K is located in the middle of a coil between two alpha helices in the PBP transpeptidase fold that belongs to the biosynthetic peptidoglycan transglycosylase-like superfamily (Supplementary Figure S12). The change of the N residue to the K residue may affect post-translational modification because asparagine side chains are known to be major sites of post-translational modification. MtgA_E225Q was located at the end of a predicted coil at the end of the protein that is located near the center of the folded protein structure (Supplementary Figure S12). The change of glutamic acid to glutamine may affect its metal ion chelating activity.
Figure 4 shows that AdeB A551T, AdeB L730F, AdeR A136V, AdeS G186V, and AdeS V348I co-occurred in Clusters 1, 2, and 3. All of these variants showed a significant correlation with meropenem resistance, while only AdeS G186V showed a significant correlation to both imipenem and meropenem resistance and was expected to be deleterious. The same panel was also observed in Cluster 9.
It is interesting to observe that AdeR I175L was only identified in Cluster 4 across the entire phylogram, and both AdeS I331V and AdeN K216N showed the largest occurrence in this cluster. AdeS I331V showed a significant correlation to imipenem resistance, while both AdeR I175L and AdeN K216N showed a significant correlation to both imipenem and meropenem resistance. Both showed high specificity (Table 1).
Cluster 5 appeared to be more recently acquired, and it included a unique cluster of variants occurring together. These were nearly only identified in this cluster across the entire phylogram. These included AdeR H158L, AdeS L105F, AdeS H189Y, AdeS V235I, and AdeI A388T. In addition, both AdeN P92T and AdeB G283A occurred exclusively at the lower subcluster, showing very high specificity and PPVs to both imipenem and meropenem resistance (Table 1). Each of AdeN P92T, AdeB G283A, AdeR H158L, AdeS H189Y, and AdeI A388T showed a significant correlation to both imipenem and meropenem resistance, while both AdeS L105F and AdeS V235I showed a significant correlation to meropenem resistance.
However, Cluster 6 showed the preserved occurrence of the variants AdeA A391T, AdeB T584I, AdeC S71G, AdeC K235E, AdeC Q239L, AdeR L142I, AdeR V243I, AdeS D167N, AdeG D501N, and AdeL Q262R among mostly carbapenem-susceptible isolates.
Each of AdeA A391T, AdeC S71G, AdeR L142I, AdeR V243I, and AdeS D167N showed a significant correlation to both imipenem and meropenem susceptibility, while each of AdeB T584I, AdeC K235E, AdeC Q239L, AdeG D501N, and AdeL Q262R were significantly correlated to imipenem susceptibility.
AdeJ V573M occurred only in Cluster 7 and showed a significant correlation to imipenem susceptibility with high specificity (Table 2). It occurred together with AdeL Q262R, which showed a significant correlation to imipenem susceptibility. However, other efflux variants from the same cluster did not correlate with susceptibility.
Each of AdeB D642A, AdeB S645T, AdeC S71G, AdeC T106M, AdeC K161S, AdeH R100G, AdeH A289G, and AdeH N388H co-occurred in Cluster 8 and showed a significant correlation with carbapenem susceptibility.
From Figure 5, it was observed that the three most recent clusters regarding the time of isolate collection were Clusters 1–3. Cluster 1 showed the co-occurrence of Omp_V1, CarO3, and PBP6_N329S, in addition to MtgA_T49P and MtgA_V54I. The isolates in this cluster were collected from different parts of China over the period between 2011 and 2013. Most of the clusters included respiratory samples, and the cluster showed both imipenem and meropenem resistance. The isolates in Cluster 2 were collected from Sweden and Thailand between 2011 and 2016 and similarly showed the co-occurrence of Omp_V1, CarO3, and PBP6_N329S, in addition to MtgA_T49P and MtgA_V54I. This cluster also showed imipenem and meropenem resistance; however, the isolates were isolated from different types of samples, including respiratory, wound, sterile body sites, and other infections. Imipenem-resistant Cluster 3 was observed in South Korea in blood isolates obtained in 2012 and showed the occurrence of the same panel listed in Clusters 1 and 2, in addition to PBP78_S32T. Out of these variants, Omp_V1 and PBP6_N329S showed a significant correlation to meropenem resistance, while both Omp_V1 and PBP78_S32T showed a significant correlation to imipenem resistance.
It can be observed that MtgA_T49P and MtgA_V54I are the most commonly occurring variants among the presented membrane-protein variants; however, both were nearly co-absent in a large cluster of related isolates, with nearly all isolates collected from Washington between 2003 and 2010 (Cluster 4). The cluster showed the co-occurrence of Omp_V1, CarO1, PBP1b_S112P, PBP2_K152Q, PBP6b_S28P, and PBP78_T71A. It is interesting to observe that PBP2_K152Q was only identified in this cluster and showed a significant correlation to both imipenem and meropenem resistance with high specificity (Table 5). Omp_V1 showed a significant correlation to both imipenem and meropenem resistance, while CarO1 showed a correlation to both imipenem and meropenem resistance (only significant with imipenem). Additionally, each of PBP1b_S112P, PBP6b_S28P, and PBP78_T71A showed a significant correlation to meropenem resistance.
MtgA_V54I was also not observed in another two related clusters from the USA (2003-2007) and from Sweden, Thailand, and Germany (2008-2013) (Cluster 5).
Another large cluster isolated from Washington (2003–2008) showed the co-occurrence of Omp33_V2, CarO3, PBP1a_S382N, PBP1a_T363A, PBP1b_P764S, PBP1b_S112P, PBP3_G523V, PBP2_V509I, PBP6b_S28P, and PBP78_T71A, in addition to MtgA_T49P and MtgA_V54I (Cluster 6). Among these, the variants Omp33_V2, PBP1a_S382N, PBP1a_T636A, PBP1b_P764S, PBP3_G523V, and PBP2_V509I occurred only in this cluster across the entire phylogram and were significantly related to carbapenem susceptibility.
The variant PBP3_H370Y was only observed across the whole phylogramin in Cluster 7 (shown in blue). It showed a significant correlation to imipenem susceptibility with very high specificity and PPV (Table 5). Isolates from this cluster were isolated from either wounds or sterile body sites and were isolated in Washington between 2003 and 2007. Most of these isolates were imipenem susceptible. Among the variants occurring in the same cluster, each of MtgA_T49P, MtgA_V54I, and Omp33_V2 showed a significant correlation to imipenem susceptibility.
Cluster 8 showed an interesting observation. The four variants Omp33_V3, OprD_V2, OprB_F57L, and MtgA_F4V were observed almost exclusively in cluster 8 across the entire phylogram. These four variants showed a significant correlation to meropenem susceptibility. Although genetically related, the cluster contained isolates collected from different parts of the USA, China, and India. It also spanned 10 years between 2003 and 2013 and was collected from different types of specimens, including respiratory infection, wound infection, abdominal fluid, blood, and groin, in addition to sterile body sites, which might indicate the more informative and predictive value of these variants.
Considerably more is known about efflux systems in Ps. aeruginosa and E. coli than about those in Acinetobacter spp. Most efflux pump systems possess physiological functions related to the homeostasis of the bacterial cell. An improved understanding of these systems, including the comprehension of their regulation and, particularly, of the RND family, can help us decode their natural functions and offer approaches to block or alter these pieces of machinery. It is also obvious that a complex network of regulations directs the efflux capacity of bacteria and that interplay or compensatory mechanisms between efflux systems could exist. It is worth noting that both membrane proteins and efflux-pump gene changes, when compared to other carbapenem-related elements of resistance, were not linked to phenotypic heteroresistance as in the case of other mobile elements (Fernández Cuenca et al., 2012), which may enforce their more essential and stable role in relation to carbapenem resistance.
From the predictive capacity and genetic relatedness analysis, it can be concluded that the following combinations occur together to predict carbapenem resistance:
(1) The combination of AdeN P92T, AdeB G283A, AdeR H158L, AdeI A388T, AdeS H189Y, AdeS L105F, and AdeS V235I.
(2) The combination of AdeR I175L, AdeS I331V, AdeN K216N, Omp_V1, CarO1, PBP1b_S112P, PBP2_K152Q, PBP6b_S28P, and PBP78_T71A, with higher predictive values especially for efflux pumps.
(3) The combination of AdeB A551T, AdeB L730F, AdeR A136V, AdeS G186V, AdeS V348I, CarO3, Omp_V1, and PBP6_N329S co-occurs and is correlated with carbapenem resistance (significantly with meropenem resistance), with lower predictive values.
However, other combinations occur together and can be used to predict carbapenem susceptibility.
(1) The combination of AdeA A391T, AdeB T584I, AdeC S71G, AdeC K235E, AdeC Q239L, AdeR L142I, AdeR V243I, AdeS D167N, AdeG D501N, and AdeL Q262R from efflux-pump variants and Omp33_V2, PBP1a_S382N, PBP1a_T636A, PBP1b_P764S, PBP3_G523V, and PBP2_V509I from membrane-protein variants.
(2) The combination of AdeB D642A, AdeB S645T, AdeC S71G, AdeC T106M, AdeC K161S, AdeH R100G, AdeH A289G, and AdeH N388H from efflux-pump variants and Omp33_V3, OprD_V2, OprB_F57L, and MtgA_F4V from membrane-protein variants.
Carbapenem resistance is currently widespread worldwide in ICUs in different parts of the world; however, the currently available rapid diagnostic panels often depend on the detection of carbapenemases to predict carbapenem resistance and usually do not differentiate antibiotic agents from the same antimicrobial class. This approach is apparently lacking and incomplete as it disregards important chromosomal resistance predictors. Based on the results we presented in this study, the identified sets of markers can act as useful diagnostic predictor panels for inclusion in sequence-based diagnostic workflows.
The variants identified in this study can act as potential diagnostic markers that can be used to guide the clinical prescription of carbapenems, which would preserve the utility and effectiveness of such an antimicrobial class, especially in high-risk settings. The elements identified can also provide potential valuable targets for designing agents that might restore carbapenem susceptibility.
The original contributions presented in the study are included in the article/Supplementary material, further inquiries can be directed to the corresponding author.
WN: Conceptualization, Formal analysis, Investigation, Methodology, Resources, Visualization, Writing—original draft, Writing—review and editing, Supervision. NA: Funding acquisition, Investigation, Writing—original draft, Methodology, Resources, Validation. AA: Funding acquisition, Investigation, Writing—original draft, Resources, Validation, Visualization. SZ: Funding acquisition, Investigation, Writing—review and editing, Methodology, Resources, Validation. AE: Writing—original draft, Visualization. HH: Formal analysis, Investigation, Resources, Supervision, Writing—original draft, Writing—review and editing, Validation.
The author(s) declare no financial support was received for the research, authorship, and/or publication of this article.
We extend our thanks to Princess Nourah bint Abdulrahman University Researchers Supporting Project number (PNURSP2023R153), Princess Nourah bint Abdulrahman University, Riyadh, Saudi Arabia.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2023.1271733/full#supplementary-material
Agyepong, N., Fordjour, F., and Owusu-Ofori, A. (2023). Multidrug-resistant Acinetobacter baumannii in healthcare settings in Africa. Front. Trop. Dis. 4, 1110125. doi: 10.3389/fitd.2023.1110125
Alcántar-Curiel, M. D., García-Torres, L. F., González-Chávez, M. I., Morfín-Otero, R., Gayosso-Vázquez, C., Jarillo-Quijada, M. D., et al. (2014). Molecular mechanisms associated with nosocomial carbapenem-resistant Acinetobacter baumannii in Mexico. Arch. Med. Res. 45, 553–560. doi: 10.1016/j.arcmed.2014.10.006
Al-Tofaily, A. H., Kadhum, H. F., Ali, A. A., and Al-Yasseen, A. K. (2020). Distribution of some efflux pump system genes among environmental isolates of Acinetobacter baumannii. EurAsian J. Biosci. 14, 2101–2106.
Aravind, L., and Ponting, C. P. (1999). The cytoplasmic helical linker domain of receptor histidine kinase and methyl-accepting proteins is common to many prokaryotic signalling proteins. FEMS Microbiol. Lett. 176, 111–116. doi: 10.1111/j.1574-6968.1999.tb13650.x
Ayoub Moubareck, C., and Hammoudi Halat, D. (2020). Insights into Acinetobacter baumannii: a review of microbiological, virulence, and resistance traits in a threatening nosocomial pathogen. Antibiotics 9, 119. doi: 10.3390/antibiotics9030119
Basatian-Tashkan, B., Niakan, M., Khaledi, M., Afkhami, H., Sameni, F., Bakhti, S., et al. (2020). Antibiotic resistance assessment of Acinetobacter baumannii isolates from Tehran hospitals due to the presence of efflux pumps encoding genes (adeA and adeS genes) by molecular method. BMC Res. Notes 13, 543. doi: 10.1186/s13104-020-05387-6
Bazyleu, A., and Kumar, A. (2014). Incubation temperature, osmolarity, and salicylate affect the expression of resistance–nodulation–division efflux pumps and outer membrane porins in Acinetobacter baumanniiATCC19606T. FEMS Microbiol. Lett. 357, 136–143. doi: 10.1111/1574-6968.12530
Beigverdi, R., Sattari-Maraji, A., Emaneini, M., and Jabalameli, F. (2019). Status of carbapenem-resistant Acinetobacter baumannii harboring carbapenemase: First systematic review and meta-analysis from Iran. Infect. Genet. Evol. 73, 433–443. doi: 10.1016/j.meegid.2019.06.008
Capriotti, E., Fariselli, P., and Casadio, R. (2005). I-Mutant2.0: predicting stability changes upon mutation from the protein sequence or structure. Nucleic. Acids Res. 33, W306–W310. doi: 10.1093/nar/gki375
Cardoso, J. P., Cayô, R., Girardello, R., and Gales, A. C. (2016). Diversity of mechanisms conferring resistance to β-lactams among OXA-23-producing Acinetobacter baumannii clones. Diagn. Microbiol. Infect. Dis. 85, 90–97. doi: 10.1016/j.diagmicrobio.2016.01.018
Cayô, R., Rodríguez, M.-C., Espinal, P., Fernández-Cuenca, F., Ocampo-Sosa, A. A., Pascual, A., et al. (2011). Analysis of genes encoding penicillin-binding proteins in clinical isolates of Acinetobacter baumannii. Antimicrob. Agents Chemother. 55, 5907–5913. doi: 10.1128/AAC.00459-11
Chen, Y., Ai, L., Guo, P., Huang, H., Wu, Z., Liang, X., et al. (2018). Molecular characterization of multidrug resistant strains of Acinetobacter baumannii isolated from pediatric intensive care unit in a Chinese tertiary hospital. BMC Infect. Dis. 18, N.PAG-N.PAG. doi: 10.1186/s12879-018-3511-0
Choi, Y., and Chan, A. P. (2015). PROVEAN web server: a tool to predict the functional effect of amino acid substitutions and indels. Bioinformatics 31, 2745–2747. doi: 10.1093/bioinformatics/btv195
Chopra, S., Ramkissoon, K., and Anderson, D. C. (2013). A systematic quantitative proteomic examination of multidrug resistance in Acinetobacter baumannii. J. Proteomics 84, 17–39. doi: 10.1016/j.jprot.2013.03.008
Chu, Y. W., Chau, S. L., and Houang, E. T. S. (2006). Presence of active efflux systems AdeABC, AdeDE and AdeXYZ in different Acinetobacter genomic DNA groups. J. Med. Microbiol. 55, 477–478. doi: 10.1099/jmm.0.46433-0
Consortium, T. U. (2019). UniProt: a worldwide hub of protein knowledge. Nucleic Acids Res. 47, D506–D515. doi: 10.1093/nar/gky1049
Coyne, S., Courvalin, P., and Périchon, B. (2011). Efflux-mediated antibiotic resistance in Acinetobacter spp. Antimicrob. Agents Chemother. 55, 947–953. doi: 10.1128/AAC.01388-10
Coyne, S., Rosenfeld, N., Lambert, T., Courvalin, P., and Périchon, B. (2010). Overexpression of resistance-nodulation-cell division pump AdeFGH confers multidrug resistance in Acinetobacter baumannii. Antimicrob. Agents Chemother. 54, 4389–4393. doi: 10.1128/AAC.00155-10
del Mar Tomás, M., Beceiro, A., Pérez, A., Velasco, D., Moure, R., Villanueva, R., et al. (2005). Cloning and functional analysis of the gene encoding the 33- to 36-kilodalton outer membrane protein associated with carbapenem resistance in Acinetobacter baumannii. Antimicrob. Agents Chemother. 49, 5172–5175. doi: 10.1128/AAC.49.12.5172-5175.2005
D'Souza, R., Pinto, N. A., Phuong, N., Le Higgins, P. G., Vu, T. N., Byun, J.-H., et al. (2019). Phenotypic and genotypic characterization of acinetobacter spp. panel strains: a cornerstone to facilitate antimicrobial development. Front. Microbiol. 10, 559. doi: 10.3389/fmicb.2019.00559
Durante-Mangoni, E., and Zarrilli, R. (2011). Global spread of drug-resistant Acinetobacter baumannii: molecular epidemiology and management of antimicrobial resistance. Future Microbiol. 6, 407–422. doi: 10.2217/fmb.11.23
ECDC (2022). European Centre for Disease Prevention and Control. WHO Regional Office for Europe/European Centre for Disease Prevention and Control. Antimicrobial resistance surveillance in Europe 2022–2020 data 2022. Technical Report.
EUCAST (2023). The European Committee on Antimicrobial Susceptibility Testing. Breakpoint tables for interpretation of MICs and zone diameters. Version 13.0 2023.
Fernández Cuenca, F., Sánchez, M., del, C. G., Caballero-Moyano, F. J., Vila, J., Martínez-Martínez, L., et al. (2012). Prevalence and analysis of microbiological factors associated with phenotypic heterogeneous resistance to carbapenems in Acinetobacter baumannii. Int. J. Antimicrob. Agents 39, 472–477. doi: 10.1016/j.ijantimicag.2012.01.015
Fernández-Cuenca, F., Martínez-Martínez, L., Conejo, M. C., Ayala, J. A., Perea, E. J., and Pascual, A. (2003). Relationship between beta-lactamase production, outer membrane protein and penicillin-binding protein profiles on the activity of carbapenems against clinical isolates of Acinetobacter baumannii. J. Antimicrob. Chemother. 51, 565–574. doi: 10.1093/jac/dkg097
Fernando, D., Zhanel, G., and Kumar, A. (2013). Antibiotic resistance and expression of resistance-nodulation-division pump- and outer membrane porin-encoding genes in Acinetobacter species isolated from Canadian hospitals. Can. J. Infect. Dis. Med. Microbiol. 24, 17–21. doi: 10.1155/2013/696043
Fernando, D. M., Xu, W., Loewen, P. C., Zhanel, G. G., and Kumar, A. (2014). Triclosan can select for an AdeIJK-overexpressing mutant of Acinetobacter baumannii ATCC 17978 that displays reduced susceptibility to multiple antibiotics. Antimicrob. Agents Chemother. 58, 6424–6431. doi: 10.1128/AAC.03074-14
Fonseca, E. L., Scheidegger, E., Freitas, F. S., Cipriano, R., and Vicente, A. C. P. (2013). Carbapenem-resistant Acinetobacter baumannii from Brazil: role of carO alleles expression and blaOXA-23 gene. BMC Microbiol. 13, 245. doi: 10.1186/1471-2180-13-245
Gales, A. C., Seifert, H., Gur, D., Castanheira, M., Jones, R. N., and Sader, H. S. (2019). Antimicrobial susceptibility of acinetobacter calcoaceticus-Acinetobacter baumannii complex and stenotrophomonas maltophilia clinical isolates: results from the SENTRY antimicrobial surveillance program (1997-2016). Open Forum Infect. Dis. 6, S34–S46. doi: 10.1093/ofid/ofy293
Gehrlein, M., Leying, H., Cullmann, W., Wendt, S., and Opferkuch, W. (1991). Imipenem resistance in Acinetobacter baumanii is due to altered penicillin-binding proteins. Chemotherapy 37, 405–412. doi: 10.1159/000238887
Gu, Y., Jiang, Y., Zhang, W., Yu, Y., He, X., Tao, J., et al. (2021). Risk factors and outcomes of bloodstream infections caused by Acinetobacter baumannii: a case–control study. Diagn. Microbiol. Infect. Dis. 99, 115229. doi: 10.1016/j.diagmicrobio.2020.115229
Hadfield, J., Croucher, N. J., Goater, R. J., Abudahab, K., Aanensen, D. M., and Harris, S. R. (2018). Phandango: an interactive viewer for bacterial population genomics. Bioinformatics 34, 292–293. doi: 10.1093/bioinformatics/btx610
Hammerstrom, T. G., Beabout, K., Clements, T. P., Saxer, G., and Shamoo, Y. (2015). Acinetobacter baumannii repeatedly evolves a hypermutator phenotype in response to tigecycline that effectively surveys evolutionary trajectories to resistance. PLoS ONE 10, e0140489. doi: 10.1371/journal.pone.0140489
Han, S., Caspers, N., Zaniewski, R. P., Lacey, B. M., Tomaras, A. P., Feng, X., et al. (2011). Distinctive attributes of β-Lactam target proteins in Acinetobacter baumannii relevant to development of new antibiotics. J. Am. Chem. Soc. 133, 20536–20545. doi: 10.1021/ja208835z
Hassan, R., Mukhtar, A., Hasanin, A., and Ghaith, D. (2018). Role of insertion sequence Aba-1 and AdeS in reduced tigecycline susceptibility in MDR-Acinetobacter baumannii clinical isolates from Cairo, Egypt. J. Chemother. 30, 89–94. doi: 10.1080/1120009X.2017.1396057
Hawkey, J., Ascher, D. B., Judd, L. M., Wick, R. R., Kostoulias, X., Cleland, H., et al. (2018). Evolution of carbapenem resistance in Acinetobacter baumannii during a prolonged infection. Microb. genomics 4, e000165. doi: 10.1099/mgen.0.000165
Hou, P. F., Chen, X. Y., Yan, G. F., Wang, Y. P., and Ying, C. M. (2012). Study of the correlation of imipenem resistance with efflux pumps AdeABC, AdeIJK, AdeDE and AbeM in clinical isolates of Acinetobacter baumannii. Chemotherapy 58, 152–158. doi: 10.1159/000335599
Huang, L., Sun, L., Xu, G., and Xia, T. (2008). Differential susceptibility to carbapenems due to the AdeABC efflux pump among nosocomial outbreak isolates of Acinetobacter baumannii in a Chinese hospital. Diagn. Microbiol. Infect. Dis. 62, 326–332. doi: 10.1016/j.diagmicrobio.2008.06.008
Huys, G., Cnockaert, M., Nemec, A., and Swings, J. (2005). Sequence-based typing of ade B as a potential tool to identify intraspecific groups among clinical strains of multidrug-resistant Acinetobacter baumannii. J. Clin. Microbiol. 43, 5327–5331. doi: 10.1128/JCM.43.10.5327-5331.2005
Jia, W., Li, C., Zhang, H., Li, G., Liu, X., and Wei, J. (2015). Prevalence of Genes of OXA-23 Carbapenemase and AdeABC Efflux Pump Associated with Multidrug Resistance of Acinetobacter baumannii Isolates in the ICU of a Comprehensive Hospital of Northwestern China. Int. J. Environ. Res. Public Health 12, 10079–10092. doi: 10.3390/ijerph120810079
Jumper, J., Evans, R., Pritzel, A., Green, T., Figurnov, M., Ronneberger, O., et al. (2021). Highly accurate protein structure prediction with AlphaFold. Nature 596, 583–589. doi: 10.1038/s41586-021-03819-2
Kanafani, Z. A., Zahreddine, N., Tayyar, R., Sfeir, J., Araj, G. F., Matar, G. M., et al. (2018). Multi-drug resistant Acinetobacter species: a seven-year experience from a tertiary care center in Lebanon. Antimicrob. Resist. Infect. Control 7, 9. doi: 10.1186/s13756-017-0297-6
Kang, K. N., and Boll, J. M. (2022). PBP1A Directly Interacts with the Divisome Complex to Promote Septal Peptidoglycan Synthesis in Acinetobacter baumannii. J. Bacteriol. 204, e0023922. doi: 10.1128/jb.00239-22
Knudsen, M., and Wiuf, C. (2010). The CATH database. Hum. Genomics 4, 207–212. doi: 10.1186/1479-7364-4-3-207
Kor, S., Tou, B., Chieng, C., and Hiew, M. (2014). Distribution of the multidrug efflux pump genes adeA, adeI, adeJ, adeY and integrons in clinical isolates of Acinetobacter baumannii from Malaysian hospitals. Biomed. Res. 25, 143–148.
Kwon, H., Il, K.im, S., Oh, M. H., Na, S. H., Kim, Y. J., Jeon, Y. H., et al. (2017). Outer membrane protein A contributes to antimicrobial resistance of Acinetobacter baumannii through the OmpA-like domain. J. Antimicrob. Chemother. 72, 3012–3015. doi: 10.1093/jac/dkx257
Lee, H.-Y., Chang, R.-C., Su, L.-H., Liu, S.-Y., Wu, S.-R., Chuang, C.-H., et al. (2012). Wide spread of Tn2006 in an AbaR4-type resistance island among carbapenem-resistant Acinetobacter baumannii clinical isolates in Taiwan. Int. J. Antimicrob. Agents 40, 163–167. doi: 10.1016/j.ijantimicag.2012.04.018
Lee, S.-Y., Oh, M. H., Yun, S. H., Choi, C.-W., Park, E. C., Song, H. S., et al. (2018). Genomic characterization of extensively drug-resistant Acinetobacter baumannii strain, KAB03 belonging to ST451 from Korea. Infect. Genet. Evol. J. Mol. Epidemiol. Evol. Genet. Infect. Dis. 65, 150–158. doi: 10.1016/j.meegid.2018.07.030
Lee, S. Y., Yun, S. H., Lee, Y. G., Choi, C. W., Leem, S. H., Park, E. C., et al. (2014). Proteogenomic characterization of antimicrobial resistance in extensively drug-resistant Acinetobacter baumannii DU202. J. Antimicrob. Chemother. 69, 1483–1491. doi: 10.1093/jac/dku008
Lee, Y., Yum, J. H., Kim, C.-K., Yong, D., Jeon, E. H., Jeong, S. H., et al. (2010). Role of OXA-23 and AdeABC efflux pump for acquiring carbapenem resistance in an Acinetobacter baumannii strain carrying the blaOXA-66 gene. Ann. Clin. Lab. Sci. 40, 43–48.
Letunic, I., and Bork, P. (2021). Interactive Tree Of Life (iTOL) v5: an online tool for phylogenetic tree display and annotation. Nucleic Acids Res. 49, W293–W296. doi: 10.1093/nar/gkab301
Leus, I. V., Adamiak, J., Trinh, A. N., Smith, R. D., Smith, L., Richardson, S., et al. (2020). Inactivation of AdeABC and AdeIJK efflux pumps elicits specific nonoverlapping transcriptional and phenotypic responses in Acinetobacter baumannii. Mol. Microbiol. 114, 1049–1065. doi: 10.1111/mmi.14594
Lima, W. G., Silva Alves, G. C., Sanches, C., Antunes Fernandes, S. O., and de Paiva, M. C. (2019). Carbapenem-resistant Acinetobacter baumannii in patients with burn injury: A systematic review and meta-analysis. Burns 45, 1495–1508. doi: 10.1016/j.burns.2019.07.006
Limansky, A. S., Mussi, M. A., and Viale, A. M. (2002). Loss of a 29-kilodalton outer membrane protein in Acinetobacter baumannii is associated with imipenem resistance. J. Clin. Microbiol. 40, 4776–4778. doi: 10.1128/JCM.40.12.4776-4778.2002
Lin, L., Ling, B.-D., and Li, X.-Z. (2009). Distribution of the multidrug efflux pump genes, adeABC, adeDE and adeIJK, and class 1 integron genes in multiple-antimicrobial-resistant clinical isolates of Acinetobacter baumannii-Acinetobacter calcoaceticus complex. Int. J. Antimicrob. Agents 33, 27–32. doi: 10.1016/j.ijantimicag.2008.06.027
Lin, M.-F., Lin, Y.-Y., Tu, C.-C., and Lan, C.-Y. (2017). Distribution of different efflux pump genes in clinical isolates of multidrug-resistant Acinetobacter baumannii and their correlation with antimicrobial resistance. J. Microbiol. Immunol. Infect. 50, 224–231. doi: 10.1016/j.jmii.2015.04.004
Lin, Y.-T. (2017). What can we learn from the dissemination of carbapenem-resistant Acinetobacter baumannii in patients with burn injury? J. Chin. Med. Assoc. 80, 189–190. doi: 10.1016/j.jcma.2016.12.004
Marchand, I., Damier-Piolle, L., Courvalin, P., and Lambert, T. (2004). Expression of the RND-type efflux pump AdeABC in Acinetobacter baumannii is regulated by the AdeRS two-component system. Antimicrob. Agents Chemother. 48, 3298–3304. doi: 10.1128/AAC.48.9.3298-3304.2004
Martínez-Trejo, A., Ruiz-Ruiz, J. M., Gonzalez-Avila, L. U., Saldaña-Padilla, A., Hernández-Cortez, C., Loyola-Cruz, M. A., et al. (2022). Evasion of antimicrobial activity in acinetobacter baumannii by target site modifications: an effective resistance mechanism. Int. J. Mol. Sci. 23, 6582. doi: 10.3390/ijms23126582
McGuffin, L. J., Bryson, K., and Jones, D. T. (2000). The PSIPRED protein structure prediction server. Bioinformatics 16, 404–405. doi: 10.1093/bioinformatics/16.4.404
Migliavacca, R., Espinal, P., Principe, L., Drago, M., Fugazza, G., Roca, I., et al. (2013). Characterization of resistance mechanisms and genetic relatedness of carbapenem-resistant Acinetobacter baumannii isolated from blood, Italy. Diagn. Microbiol. Infect. Dis. 75, 180–186. doi: 10.1016/j.diagmicrobio.2012.11.002
Mikolosko, J., Bobyk, K., Zgurskaya, H. I., and Ghosh, P. (2006). Conformational flexibility in the multidrug efflux system protein AcrA. Structure 14, 577–587. doi: 10.1016/j.str.2005.11.015
Modarresi, F., Azizi, O., Shakibaie, M. R., Motamedifar, M., Valibeigi, B., and Mansouri, S. (2015). Effect of iron on expression of efflux pump (adeABC) and quorum sensing (luxI, luxR) genes in clinical isolates of Acinetobacter baumannii. APMIS 123, 959–968. doi: 10.1111/apm.12455
Montaña, S., Vilacoba, E., Traglia, G. M., Almuzara, M., Pennini, M., Fernández, A., et al. (2015). Genetic variability of AdeRS two-component system associated with tigecycline resistance in XDR-Acinetobacter baumannii isolates. Curr. Microbiol. 71, 76–82. doi: 10.1007/s00284-015-0829-3
Morán-Barrio, J., Cameranesi, M. M., Relling, V., Limansky, A. S., Brambilla, L., and Viale, A. M. (2017). The Acinetobacter outer membrane contains multiple specific channels for carbapenem β-lactams as revealed by kinetic characterization analyses of imipenem permeation into Acinetobacter baylyi cells. Antimicrob. Agents Chemother. 61, 1–15. doi: 10.1128/AAC.01737-16
Nageeb, W. M., and Hetta, H. F. (2022). The predictive potential of different molecular markers linked to amikacin susceptibility phenotypes in Pseudomonas aeruginosa. PLoS ONE 17, e0267396. doi: 10.1371/journal.pone.0267396
Nelson, K., Rubio-Aparicio, D., Tsivkovski, R., Sun, D., Totrov, M., Dudley, M., et al. (2020). In Vitro Activity of the Ultra-Broad-Spectrum Beta-Lactamase Inhibitor QPX7728 in Combination with Meropenem against Clinical Isolates of Carbapenem-Resistant Acinetobacter baumannii. Antimicrob. Agents Chemother. 64, e01406–20. doi: 10.1128/AAC.01406-20
Nemec, A., Maixnerová, M., van der Reijden, T. J. K., van den Broek, P. J., and Dijkshoorn, L. (2007). Relationship between the AdeABC efflux system gene content, netilmicin susceptibility and multidrug resistance in a genotypically diverse collection of Acinetobacter baumannii strains. J. Antimicrob. Chemother. 60, 483–489. doi: 10.1093/jac/dkm231
Nguyen, M., and Joshi, S. G. (2021). Carbapenem resistance in Acinetobacter baumannii, and their importance in hospital-acquired infections: a scientific review. J. Appl. Microbiol. 131, 2715–2738. doi: 10.1111/jam.15130
Olson, R. D., Assaf, R., Brettin, T., Conrad, N., Cucinell, C., Davis, J. J., et al. (2023). Introducing the Bacterial and Viral Bioinformatics Resource Center (BV-BRC): a resource combining PATRIC, IRD and ViPR. Nucleic Acids Res. 51, D678–D689. doi: 10.1093/nar/gkac1003
Ostad Asadolah-Malayeri, H., Hakemi-Vala, M., and Davari, K. (2016). Role of aders and OXA23 genes among imipenem resistant Acinetobacter baumannii isolates from two hospitals of Tehran, Iran. Iran. J. Pathol. 11, 345–353.
Pagdepanichkit, S., Tribuddharat, C., and Chuanchuen, R. (2016). Distribution and expression of the Ade multidrug efflux systems in Acinetobacter baumannii clinical isolates. Can. J. Microbiol. 62, 794–801. doi: 10.1139/cjm-2015-0730
Papp-Wallace, K. M., Senkfor, B., Gatta, J., Chai, W., Taracila, M. A., Shanmugasundaram, V., et al. (2012). Early insights into the interactions of different β-lactam antibiotics and β-lactamase inhibitors against soluble forms of Acinetobacter baumannii PBP1a and Acinetobacter sp. PBP3. Antimicrob. Agents Chemother. 56, 5687–5692. doi: 10.1128/AAC.01027-12
Peleg, A. Y., Seifert, H., and Paterson, D. L. (2008). Acinetobacter baumannii: emergence of a successful pathogen. Clin. Microbiol. Rev. 21, 538–582. doi: 10.1128/CMR.00058-07
Perez, F., Hujer, A. M., Hujer, K. M., Decker, B. K., Rather, P. N., and Bonomo, R. A. (2007). Global challenge of multidrug-resistant Acinetobacter baumannii. Antimicrob. Agents Chemother. 51, 3471–3484. doi: 10.1128/AAC.01464-06
Poirel, L., and Nordmann, P. (2006). Carbapenem resistance in Acinetobacter baumannii: mechanisms and epidemiology. Clin. Microbiol. Infect. 12, 826–836. doi: 10.1111/j.1469-0691.2006.01456.x
Ranjbar, R., Zayeri, S., and Afshar, D. (2020). High frequency of AdeA, AdeB and AdeC genes among Acinetobacter baumannii isolates. Iran. J. Public Health 49, 1539–1545. doi: 10.18502/ijph.v49i8.3898
Rempel, A., and Wittler, R. (2021). SANS serif: alignment-free, whole-genome based phylogenetic reconstruction. Bioinformatics 37, 4868–4870. doi: 10.1093/bioinformatics/btab444
Rosenfeld, N., Bouchier, C., Courvalin, P., and Périchon, B. (2012). Expression of the resistance-nodulation-cell division pump AdeIJK in Acinetobacter baumannii is regulated by AdeN, a TetR-type regulator. Antimicrob. Agents Chemother. 56, 2504–2510. doi: 10.1128/AAC.06422-11
Ruggerone, P., Murakami, S., Pos, K. M., and Vargiu, A. V. (2013). RND efflux pumps: structural information translated into function and inhibition mechanisms. Curr. Top. Med. Chem. 13, 3079–3100. doi: 10.2174/15680266113136660220
Schimith Bier, K. E., Luiz, S. O., Scheffer, M. C., Gales, A. C., Paganini, M. C., do Nascimento, A. J., et al. (2010). Temporal evolution of carbapenem-resistant Acinetobacter baumannii in Curitiba, southern Brazil. Am. J. Infect. Control 38, 308–314. doi: 10.1016/j.ajic.2009.09.012
Sen, B., and Joshi, S. G. (2016). Studies on Acinetobacter baumannii involving multiple mechanisms of carbapenem resistance. J. Appl. Microbiol. 120, 619–629. doi: 10.1111/jam.13037
Shoja, S., Moosavian, M., Rostami, S., Farahani, A., Peymani, A., Ahmadi, K., et al. (2017). Dissemination of carbapenem-resistant Acinetobacter baumannii in patients with burn injuries. J. Chinese Med. Assoc. 80, 245–252. doi: 10.1016/j.jcma.2016.10.013
Shuo, L.I Z. H. (2013). MacA, a periplasmic membrane fusion protein of the macrolide transporter MacAB-TolC, binds lipopolysaccharide core specifically and with high affinity. J. Bacteriol. 195, 4865–4872. doi: 10.1128/JB.00756-13
Strateva, T., Sirakov, I., Stoeva, T., Stratev, A., Dimov, S., Savov, E., et al. (2019). Carbapenem-resistant Acinetobacter baumannii: Current status of the problem in four Bulgarian university hospitals (2014-2016). J. Glob. Antimicrob. Resist. 16, 266–273. doi: 10.1016/j.jgar.2018.10.027
Su, C.-C., Morgan, C. E., Kambakam, S., Rajavel, M., Scott, H., Huang, W., et al. (2019). Cryo-electron microscopy structure of an Acinetobacter baumannii multidrug efflux pump. MBio 10, e01295–19. doi: 10.1128/mBio.01295-19
Sun, J.-R., Perng, C.-L., Lin, J.-C., Yang, Y.-S., Chan, M.-C., Chang, T.-Y., et al. (2014). AdeRS combination codes differentiate the response to efflux pump inhibitors in tigecycline-resistant isolates of extensively drug-resistant Acinetobacter baumannii. Eur. J. Clin. Microbiol. Infect. Dis. 33, 2141–2147. doi: 10.1007/s10096-014-2179-7
Symmons, M. F., Bokma, E., Koronakis, E., Hughes, C., and Koronakis, V. (2009). The assembled structure of a complete tripartite bacterial multidrug efflux pump. Proc. Natl. Acad. Sci. 106, 7173–7178. doi: 10.1073/pnas.0900693106
Tacconelli, E., and Magrini, N. (2017). Global priority list of antibiotic-resistant bacteria to guide research, discovery, and development of new antibiotics. Organ. Mund. la Salud, 1–7. Available online at: https://www.who.int/medicines/publications/global-priority-list-antibiotic-resistant-bacteria/en/ (accessed March 15, 2021).
Tikhonova, E. B., Devroy, V. K., Lau, S. Y., and Zgurskaya, H. I. (2007). Reconstitution of the Escherichia coli macrolide transporter: the periplasmic membrane fusion protein MacA stimulates the ATPase activity of MacB. Mol. Microbiol. 63, 895–910. doi: 10.1111/j.1365-2958.2006.05549.x
Toth, M., Lee, M., Stewart, N. K., and Vakulenko, S. B. (2022). Effects of Inactivation of d,d-Transpeptidases of Acinetobacter baumannii on bacterial growth and susceptibility to β-lactam antibiotics. Antimicrob. Agents Chemother. 66, e0172921. doi: 10.1128/AAC.01729-21
Touchon, M., Cury, J., Yoon, E.-J., Krizova, L., Cerqueira, G. C., Murphy, C., et al. (2014). The genomic diversification of the whole acinetobacter genus: origins, mechanisms, and consequences. Genome Biol. Evol. 6, 2866–2882. doi: 10.1093/gbe/evu225
Tsai, Y.-K., Liou, C.-H., Lin, J.-C., Fung, C.-P., Chang, F.-Y., and Siu, L. K. (2020). Effects of different resistance mechanisms on antimicrobial resistance in Acinetobacter baumannii: a strategic system for screening and activity testing of new antibiotics. Int. J. Antimicrob. Agents 55, 105918. doi: 10.1016/j.ijantimicag.2020.105918
Uppalapati, S. R., Sett, A., and Pathania, R. (2020). The outer membrane proteins OmpA, CarO, and OprD of Acinetobacter baumannii confer a two-pronged defense in facilitating its success as a potent human pathogen. Front. Microbiol. 11, 589234. doi: 10.3389/fmicb.2020.589234
Vahhabi, A., Hasani, A., Rezaee, M. A., Baradaran, B., Hasani, A., Samadi Kafil, H., et al. (2021). A plethora of carbapenem resistance in Acinetobacter baumannii: no end to a long insidious genetic journey. J. Chemother. 33, 137–155. doi: 10.1080/1120009X.2020.1847421
van den Berg, B. (2012). Structural basis for outer membrane sugar uptake in pseudomonads. J. Biol. Chem. 287, 41044–41052. doi: 10.1074/jbc.M112.408518
Vila, J., Mart,í, S., and Sánchez-Céspedes, J. (2007). Porins, efflux pumps and multidrug resistance in Acinetobacter baumannii. J. Antimicrob. Chemother. 59, 1210–1215. doi: 10.1093/jac/dkl509
Vila, J., and Pachón, J. (2012). Therapeutic options for Acinetobacter baumannii infections: an update. Expert Opin. Pharmacother. 13, 2319–2336. doi: 10.1517/14656566.2012.729820
Wang, B., Weng, J., Fan, K., and Wang, W. (2012). Interdomain flexibility and pH-induced conformational changes of AcrA revealed by molecular dynamics simulations. J. Phys. Chem. B 116, 3411–3420. doi: 10.1021/jp212221v
Wattam, A. R., Davis, J. J., Assaf, R., Boisvert, S., Brettin, T., Bun, C., et al. (2017). Improvements to PATRIC, the all-bacterial bioinformatics database and analysis resource center. Nucleic Acids Res. 45, D535–D542. doi: 10.1093/nar/gkw1017
Wieczorek, P., Sacha, P., Czaban, S., Hauschild, T., Ojdana, D., Kowalczuk, O., et al. (2013). Distribution of AdeABC efflux system genes in genotypically diverse strains of clinical Acinetobacter baumannii. Diagn. Microbiol. Infect. Dis. 77, 106–109. doi: 10.1016/j.diagmicrobio.2013.06.017
Wong, M. H., yin, Chan, B. K., wai Chan, E. W., and chi Chen, S. (2019). Over-expression of ISAba1-linked intrinsic and exogenously acquired OXA type carbapenem-hydrolyzing-class D-ß-lactamase-encoding genes is key mechanism underlying carbapenem resistance in Acinetobacter baumannii. Front. Microbiol. 10, 2809. doi: 10.3389/fmicb.2019.02809
Xiao, S.-Z., Chu, H.-Q., Han, L.-Z., Zhang, Z.-M., Li, B., Zhao, L., et al. (2016). Resistant mechanisms and molecular epidemiology of imipenem-resistant Acinetobacter baumannii. Mol. Med. Rep. 14, 2483–2488. doi: 10.3892/mmr.2016.5538
Xu, C., Bilya, S. R., and Xu, W. (2019). adeABC efflux gene in Acinetobacter baumannii. New Microb. Infect. 30, 100549. doi: 10.1016/j.nmni.2019.100549
Yamada, Y., and Suwabe, A. (2013). Diverse carbapenem-resistance mechanisms in 16S rRNA methylase-producing Acinetobacter baumannii. J. Med. Microbiol. 62, 618–622. doi: 10.1099/jmm.0.048991-0
Yang, H., Huang, L., Barnie, P. A., Su, Z., Mi, Z., Chen, J., et al. (2015). Characterization and distribution of drug resistance associated β-lactamase, membrane porin and efflux pump genes in MDR A. baumannii isolated from Zhenjiang, China. Int. J. Clin. Exp. Med. 8, 15393–15402.
Yoon, E.-J., Balloy, V., Fiette, L., Chignard, M., Courvalin, P., and Grillot-Courvalin, C. (2016). Contribution of the Ade Resistance-Nodulation-Cell Division-Type Efflux Pumps to Fitness and Pathogenesis of Acinetobacter baumannii. MBio 7, e00697–16. doi: 10.1128/mBio.00697-16
Yoon, E.-J., Chabane, Y. N., Goussard, S., Snesrud, E., Courvalin, P., Dé, E., et al. (2015). Contribution of resistance-nodulation-cell division efflux systems to antibiotic resistance and biofilm formation in Acinetobacter baumannii. MBio 6, e00309–15. doi: 10.1128/mBio.00309-15
Yoon, E.-J., Courvalin, P., and Grillot-Courvalin, C. (2013). RND-type efflux pumps in multidrug-resistant clinical isolates of Acinetobacter baumannii: major role for AdeABC overexpression and AdeRS mutations. Antimicrob. Agents Chemother. 57, 2989–2995. doi: 10.1128/AAC.02556-12
Yun, S.-H., Choi, C.-W., Kwon, S.-O., Park, G. W., Cho, K., Kwon, K.-H., et al. (2011). Quantitative proteomic analysis of cell wall and plasma membrane fractions from multidrug-resistant Acinetobacter baumannii. J. Proteome Res. 10, 459–469. doi: 10.1021/pr101012s
Zgurskaya, H. I., Weeks, J. W., Ntreh, A. T., Nickels, L. M., and Wolloscheck, D. (2015). Mechanism of coupling drug transport reactions located in two different membranes. Front. Microbiol. 6, 100. doi: 10.3389/fmicb.2015.00100
Zhang, Z., Morgan, C. E., Bonomo, R. A., and Yu, E. W. (2021). Cryo-EM determination of eravacycline-bound structures of the ribosome and the multidrug efflux pump AdeJ of Acinetobacter baumannii. MBio 12, e0103121. doi: 10.1128/mBio.01031-21
Keywords: Acinetobacter baumannii, carbapenem resistance, multidrug efflux pump, penicillin binding proteins, new drug targets, molecular predictors, diagnostic markers
Citation: Nageeb WM, AlHarbi N, Alrehaili AA, Zakai SA, Elfadadny A and Hetta HF (2023) Global genomic epidemiology of chromosomally mediated non-enzymatic carbapenem resistance in Acinetobacter baumannii: on the way to predict and modify resistance. Front. Microbiol. 14:1271733. doi: 10.3389/fmicb.2023.1271733
Received: 02 August 2023; Accepted: 05 September 2023;
Published: 06 October 2023.
Edited by:
Rami M. Elshazli, Horus University, EgyptReviewed by:
Mansoor Khaledi, Shahed University, IranCopyright © 2023 Nageeb, AlHarbi, Alrehaili, Zakai, Elfadadny and Hetta. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Wedad M. Nageeb, d2VkYWRfc2FsZWhAbWVkLnN1ZXouZWR1LmVn
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.