- 1Institute of Bioanalysis, Coburg University of Applied Sciences and Arts, Coburg, Germany
- 2Behavioral Physiology and Sociobiology, Biocenter, Julius Maximilian University of Würzburg, Würzburg, Germany
- 3Bayreuth Center of Ecology and Environmental Research (BayCEER), University of Bayreuth, Bayreuth, Germany
Honey bees are crucial for our ecosystems as pollinators, but the intensive use of plant protection products (PPPs) in agriculture poses a risk for them. PPPs do not only affect target organisms but also affect non-targets, such as the honey bee Apis mellifera and their microbiome. This study is the first of its kind, aiming to characterize the effect of PPPs on the microbiome of the cuticle of honey bees. We chose PPPs, which have frequently been detected in bee bread, and studied their effects on the cuticular microbial community and function of the bees. The effects of the fungicide Difcor® (difenoconazole), the insecticide Steward® (indoxacarb), the combination of both (mix A) and the fungicide Cantus® Gold (boscalid and dimoxystrobin), the insecticide Mospilan® (acetamiprid), and the combination of both (mix B) were tested. Bacterial 16S rRNA gene and fungal transcribed spacer region gene-based amplicon sequencing and quantification of gene copy numbers were carried out after nucleic acid extraction from the cuticle of honey bees. The treatment with Steward® significantly affected fungal community composition and function. The fungal gene copy numbers were lower on the cuticle of bees treated with Difcor®, Steward®, and PPP mix A in comparison with the controls. However, bacterial and fungal gene copy numbers were increased in bees treated with Cantus® Gold, Mospilan®, or PPP mix B compared to the controls. The bacterial cuticular community composition of bees treated with Cantus® Gold, Mospilan®, and PPP mix B differed significantly from the control. In addition, Mospilan® on its own significantly changed the bacterial functional community composition. Cantus® Gold significantly affected fungal gene copy numbers, community, and functional composition. Our results demonstrate that PPPs show adverse effects on the cuticular microbiome of honey bees and suggest that PPP mixtures can cause stronger effects on the cuticular community than a PPP alone. The cuticular community composition was more diverse after the PPP mix treatments. This may have far-reaching consequences for the health of honey bees.
1. Introduction
A significant decline in pollinators has been observed in the past decade, despite animal pollination being one of the most important ecosystem services (Potts et al., 2010). One of the main drivers that could lead to this pollinator decline is the intensive use of plant protection products (PPPs) in agriculture (Kluser and Peduzzi, 2007; Goulson et al., 2015). For the investigation of adverse side effects of PPPs on beneficial insects, the honey bee Apis mellifera is an excellent model organism because of its rich behavioral repertoire and the large diversity of methods for investigating their behavior (Scheiner et al., 2013).
Among the different PPP groups, insecticides are the best-studied class as they often show negative effects on beneficial insects (Galvan et al., 2006; El Hassani et al., 2008; Shi et al., 2019). Fungicides are usually not sufficiently investigated since no harmful impact on insects is assumed (Zubrod et al., 2019; Schuhmann et al., 2022). Nevertheless, some fungicides can synergistically interact with insecticides or prolong their undesirable effects on the health of pollinators (Wernecke and Castle, 2020). This study tested combinations of PPPs frequently detected in bee bread (Rosenkranz et al., 2019). The insecticide Steward® was combined with the fungicide Difcor®, and the insecticide Mospilan® was applied together with the fungicide Cantus® Gold (Table 1). Both combinations have been used in agriculture and are consumed by bees as they are applied to mass-flowering crops (Holzschuh et al., 2013).
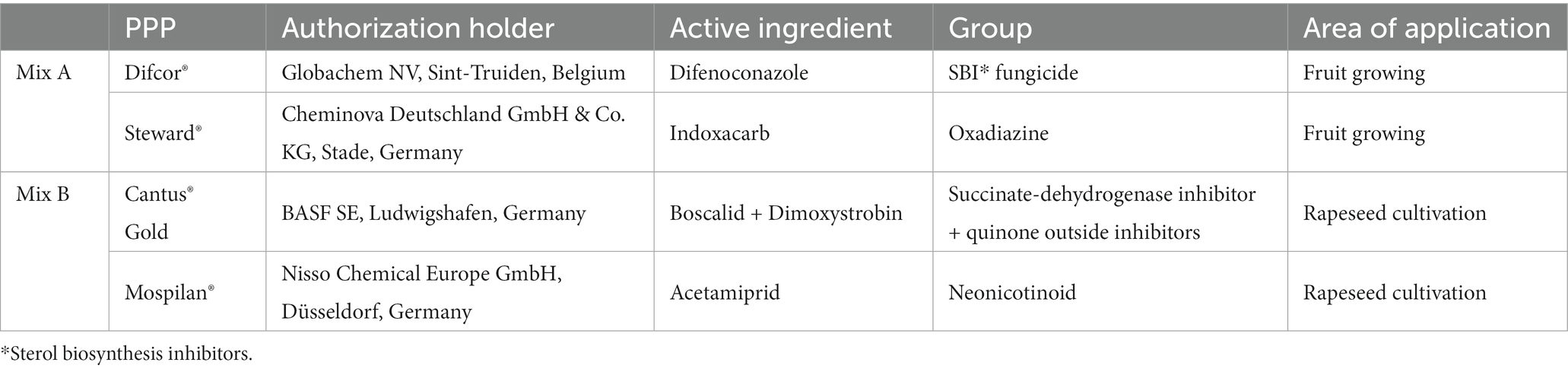
Table 1. Overview of the plant protection products (PPPs) used (FMC Agricultural Solutions, 2020, 2021; BASF SE, 2021; Fungicide Resistance Action Committee, 2021; Insecticide Resistance Action Committee, 2021; PLANTAN GmbH, 2021).
Indoxacarb is the active ingredient of the insecticide Steward® and acts as a sodium channel modulator, leading to a quick inhibition of feeding in pest insects (Börner et al., 2009). Acetamiprid (active ingredient of the insecticide Mospilan®) is a neonicotinoid that acts as an agonist on nicotinic acetylcholine receptors, resulting in constant ion flow and neurotoxic effects (Börner et al., 2009). Several negative side effects of the isolated application of both insecticides on beneficial insects have already been demonstrated (Galvan et al., 2006; Laurino et al., 2011; Shi et al., 2020). In contrast to these insecticides, the fungicides difenoconazole (an active ingredient of Difcor®) and boscalid or dimoxystrobin (active ingredients of Cantus® Gold) appear to have small or no effects on beneficial insects when applied on their own (Wernecke et al., 2019; Almasri et al., 2020).
Bees are often exposed to a mixture of several PPPs. This can result from tank mixtures, spraying sequences, the combined use of seed and spray treatment, or bees foraging sequentially at different flowers (Thompson et al., 2014). Thus, various active substances are present in bee bread (Rosenkranz et al., 2019). It has been shown that certain insecticide–fungicide combinations can lead to synergistic negative effects (Schuhmann et al., 2022), which makes it important to study more intensively the interaction of insecticides and fungicides in insects.
PPPs affect not only pest organisms but also non-targets such as bees and microorganisms in the environment. Honey bees come in contact with a wide variety of microbiomes, namely pollen (Gilliam et al., 1989; Martinson et al., 2011), nectar (Fridman et al., 2012; Alvarez-Pérez and Herrera, 2013), bee bread (Sinpoo et al., 2017), and plants’ surfaces (Keller et al., 2021). Nevertheless, little is known about the cuticular microbiome of bees and its effects on the health and performance of honey bees. In contrast, the gut microbiome is well studied since it plays a central role in metabolism, growth, development, protection against pathogens, and immune defense (Zheng et al., 2017; Kwong et al., 2017a).
The core gut microbiome of honey bees is dominated by nine bacterial species clusters, which account for 95 to 99.9% of the bacteria in almost all individuals (Babendreier et al., 2007; Engel et al., 2012; Moran et al., 2012; Sabree et al., 2012; Corby-Harris et al., 2014). Five core bacterial species clusters and four rarer species clusters form the dominant cluster of bacteria found in honey bees (Kwong et al., 2017b; Raymann and Moran, 2018). The four core bacteria are Snodgrassella alvi, Gilliamella apicola, Lactobacillus Firm-4, Lactobacillus Firm-5, and Bifidobacterium asteroids (Babendreier et al., 2007; Martinson et al., 2011; Kwong and Moran, 2013).
The gut mycobiome of most worker bees is dominated by the members of the genus Saccharomyces, whereas the intestines of forager bees and queens were colonized by various fungal taxa, including Zygosaccharomyces, Candida, and Ascomycota. Katsnelson (2015) pointed out that the gut microbiome is not the only microbial community that is important for bees. For example, bees have close contact with bacteria on the inner walls of the hive (Anderson et al., 2013).
The gut of the honey bee and whole-body extracts appeared to have the same dominant microbiomes but vary in relative abundance and composition (Mattila et al., 2012; Ribière et al., 2019). Aizenberg-Gershtein et al. (2013) analyzed the cuticular bacterial community composition. The cuticular bacterial microbiome is colonized by the bacterial classes Gammaproteobacteria, Actinobacteria, Bacilli, and Alphaproteobacteria. Arsenophonus represented the most dominant genus within Gammaproteobacteria. Arsenophonus represents an extensive cluster of symbiotic bacteria in insects (Nováková et al., 2009), which has already been recovered in the gut of honey bees (Babendreier et al., 2007). Saccà and Lodesani (2020) identified and isolated bacteria (Apilactobacillus kunkeei, Bacillus thuringiensis, and Acetobacteraceae) from the cuticle of honey bees, which might function as a natural antagonist of the external parasitic mite Varroa destructor.
The study herein aimed to analyze (i) the effects of single neonicotinoids and fungicides; (ii) the effects of a mixture of a non-neonicotinoid (Steward®) with an SBI fungicide (Difcor®) (mix A, Table 1), and (iii) the effects of a combination of a neonicotinoid (Mospilan®) and a non-SBI fungicide (Cantus® Gold) (mix B, Table 1) on the bacterial and fungal communities of honey bees. The quantity, composition, and function of the different microbes were investigated. We hypothesize that fungal taxa and, to a lesser extent, bacterial taxa will be reduced in abundance, and some taxa might even be sensitive to PPPs. On the other hand, we expect that other microbes will gain an advantage due to the inactivation of competitors and the addition of PPPs as substrates.
2. Methods
2.1. Treatment of honey bees
Honey bee workers (A. mellifera carnica) were collected randomly from a hive in the departmental apiary at the University of Würzburg. For each treatment, five cages were prepared on five consecutive days. Each cage contained 30 honey bees. The cages were maintained in an incubator (30°C, 50% humidity), and the honey bees received the treatment solutions for 1 week. The feeding solutions were provided via prepared 2 mL cups (Figure 1). The amount of food per cage was adapted to the number of individuals so that the bees could eat ad libitum. The 2 mL cups were replaced every day to guarantee a controlled food supply.
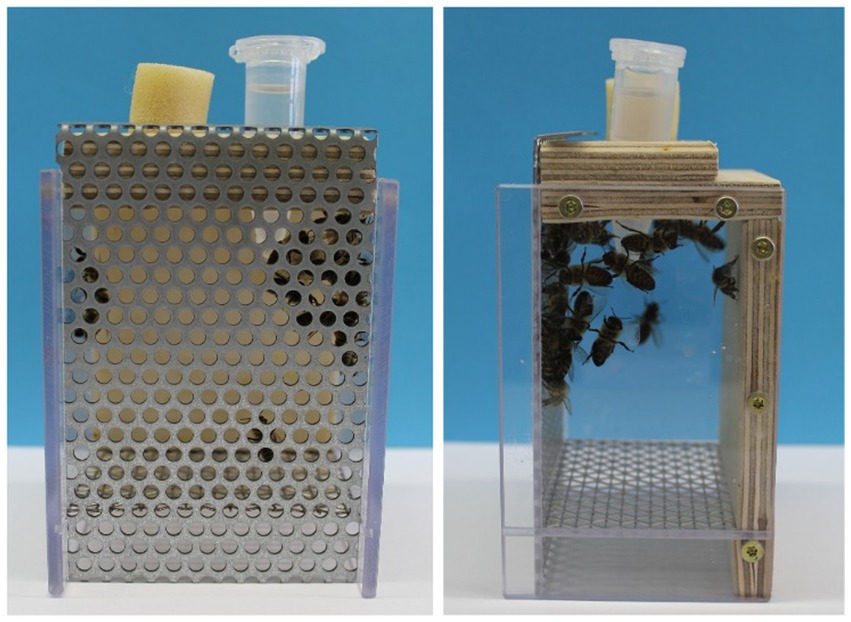
Figure 1. Experimental setup of the bee’s treatment. Experimental setup of Apis mellifera nursing bees in a cage system. The feeding solution is provided via a prepared 2 mL cup with holes at the bottom. Picture A. Schuhmann.
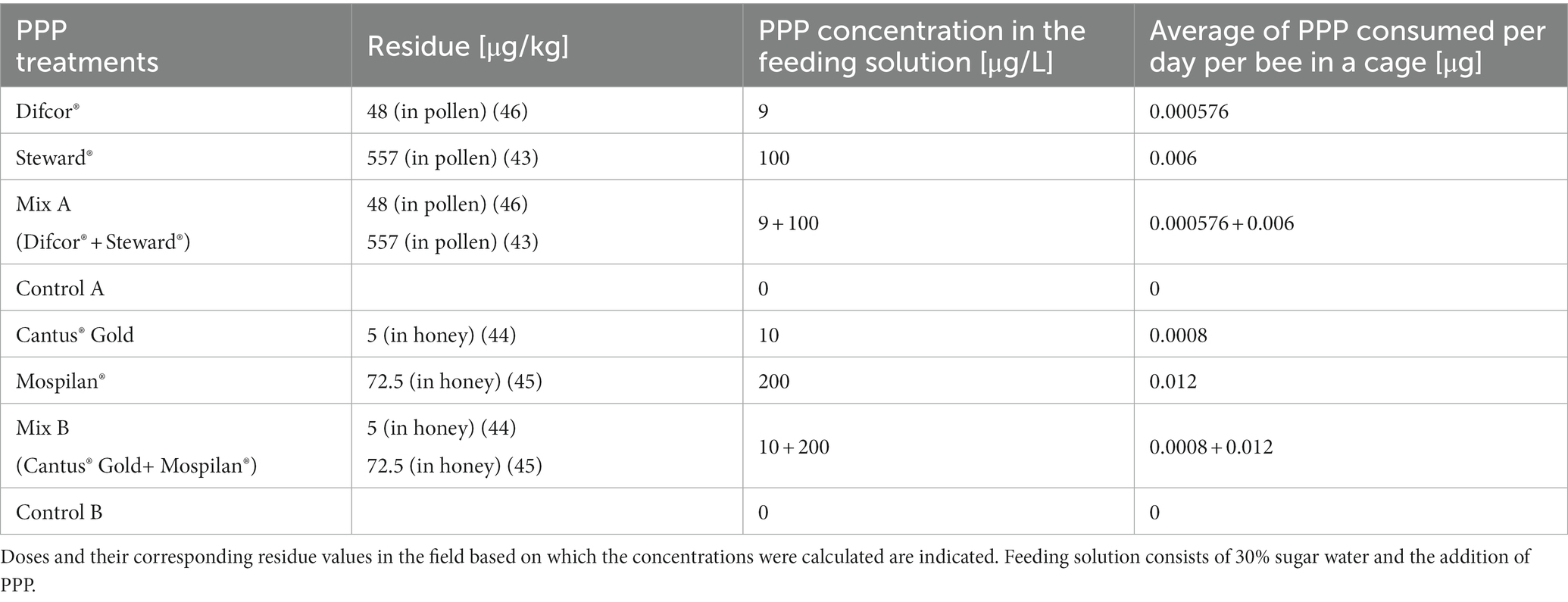
The feeding solutions were calculated based on residue levels found in the field and consisted of 30% sugar water with the addition of PPPs (McArt et al., 2017; Lüken and von der Ohe, 2018; El-Nahhal, 2020; Friedle et al., 2021). For reasons of data availability, we relied on residue values from pollen for the calculation of Steward® and Difcor® and on residue values from honey for Mospilan® and Cantus® Gold.
Using the intake of honey or pollen per bee per day (Rortais et al., 2005), it was calculated how much active ingredient a honey bee would consume per day based on the selected residues. It was assumed that a caged honey bee consumes 60 μL (Hesselbach and Scheiner, 2019) of feeding solution per day. Therefore, the feeding concentration was adjusted so that a honey bee would ingest the corresponding calculated active ingredient concentration in these 60 μL (Table 2).
We used the water-soluble formulation of PPPs to mimic field conditions (Cox and Surgan, 2006). The honey bees were either treated with the insecticide alone, the fungicide alone, a mixture of both, or with a control solution (30% sugar water). All four treatments belonged to one experimental series (Table 2). Each treatment group had its own control (“control A” belongs to Difcor®, Steward®, and PPP mix A treatments; “control B” belongs to Cantus® Gold, Mospilan®, and PPP mix B treatments). The concentrations of the single PPPs were maintained in the mix A and B treatments. After the 1-week treatment, the bees were frozen in liquid nitrogen and stored at −80°C.
2.2. Cuticle preparation and DNA extraction
Dissection and preparation of cuticles were performed under frozen conditions. Antennae, legs, and wings were removed (but not discarded). Afterward, the inner organs (i.e., brain, muscles, gut, and sting apparatus) were carefully removed from the head capsule, the thorax, and the abdomen (Ribière et al., 2019; Subotic et al., 2019). The prepared cuticles and outer body parts underwent DNA isolation. Cuticles from four individuals were pooled for one sample. DNA extraction was performed using the Quick-DNA™ Fecal/Soil Microbe Microprep Kit according to the manufacturer’s protocol (Zymo Research Europe GmbH, Freiburg im Breisgau, Germany). All DNA extracts were stored at −20°C until further use. Five independent bee replicates per treatment were analyzed.
2.3. Quantitative PCR of bees cuticular DNA extracts
The quantitative PCR (qPCR) was performed to quantify the gene copy numbers of the bacterial 16S rRNA gene with the primer sets BAC341f (5′-CCTACGGGNGGCWGCAG-3′) and BAC758R (5′-GACTACHVGGGTATCTAAKCC-3′) (Klindworth et al., 2013) and of the fungal internal transcribed spacer (ITS) DNA regions with the primer sets fITS7 (5′-GTGAATCATCGAATCTTTG-3′) (Ihrmark et al., 2012) and ITS4 (5′-TCCTCCGCTTATTGATATGC-3′) (White et al., 1994). Each independent replicate was quantified in three technical triplicates in 96-well plates using the CFX96™ Real-Time System C1000™ Thermal Cycler (Bio-Rad Laboratories GmbH, Feldkirchen, Germany). Fungal ITS and bacterial 16S rRNA gene-base qPCR were performed in 20 μL reaction mixtures containing 1 μL of DNA template, 0.3 μM each primer, 1x PCR-Enhancer (Biozym Scientific GmbH, Oldendorf, Germany), 1x iTaq Universal SYBR Green Supermix (Bio-Rad, Munich, Germany), and nuclease-free water (Sigma-Aldrich, St. Louis, MO, USA). Nuclease-free master mix blanks were run as negative controls. Reaction conditions for 16S qPCR involved an initial 3-min denaturation at 95°C, followed by 40 cycles of 5 s of denaturation at 95°C, annealing at 52°C for bacteria and 52.7°C for fungi over 30 s, respectively, and elongation at 60°C for 30 s. The final elongation step was at 72°C for 10 min. Gene copy numbers were calculated as previously described by Lasota et al. (2019) by comparing PCR-cycle threshold (CT) values to a standard curve of triplicate 10-fold dilutions of genomic DNA (gDNA) extracted from a known concentration of Escherichia coli K12 (DSM 423) and Fusarium solani (DSM 1164) by employing the Quick-DNA™ Fecal/Soil Microbe Miniprep Kit according to the manufacturer’s instructions (Zymo Research Europe GmbH). The genomic DNA concentration per PCR reaction of E. coli and F. solani standard ranged from 1 × 109 to 5 × 103 and 6.51 × 106 to 65.1 gene copies.
2.4. Amplicon sequencing of the cuticular microbiome
Cuticular DNA samples were further analyzed by amplicon sequencing. The 16S rRNA gene and ITS DNA region were amplified with the same primer sets used for the qPCR analysis to create amplicon sequencing libraries for each of the 40 bees’ cuticular DNA samples. Inline barcodes and Illumina sequencing adapters were added to the amplicon sequence libraries using the Nextera CT Index Kit (Illumina, San Diego, CA, USA) and MiSeq Reagent Kit v3 600 cycles (Illumina) according to the manufacturer’s instructions. PCR products for library preparation were purified by AMPure XP beads (Beckman Coulter, Brea, CA, USA). The sequencing of libraries was performed by 300-bp paired-end sequencing on an Illumina MiSeq platform (Illumina MiSeq V3; Illumina) based on a standard protocol from the manufacturer. Amplicon sequencing library preparation, sequencing, and sequence quality checks were carried out by LGC Genomics GmbH (Berlin, Germany).
2.5. Bioinformatics
Raw data pre-processing with demultiplexing, sorting, adapter trimming, and merging reads was congregated using Illumina bcl2fastq conversion software v2.20 and BBMerge v34.48 (DOE Joint Genome Institute, 2022). The sequence quality of the reads was controlled with the FastQC software, version 0.11.8 (Babraham Bioinformatics, 2020). Sequence pre-processing and Operational Taxonomic Units (OTUs) picking from amplicons were conducted using Mothur 1.35.1 (Schloss et al., 2009). Sequences were aligned against the 16S Mothur-Silva SEED r119 reference alignment depending on their Phred quality score over 33 (Whelan et al., 2019). Filtering short alignments and reducing sequencing errors were conducted by pre-clustering, where a maximum of one base mismatch per 100 bases within a cluster was allowed. Chimeras were eliminated with the UCHIME algorithm (Edgar et al., 2011). Afterward, taxonomical classification of the sequences against the Silva reference classification was conducted, and sequences of other domains of life were removed for OTU picking. OTUs were selected and assigned to a taxonomic level by clustering at the 97% identity level (Edgar, 2018; Nilsson et al., 2019; Kõljalg et al., 2020). Thereby, OTU tables for DNA samples were constituted.
Ecological and metabolic functions of detected bacterial OTUs were predicted using the functional annotation tool of the prokaryotic taxa v.1.1 (FAPROTAX) database (Louca et al., 2016). The functions of each prokaryotic taxon were annotated using the literature on cultivable strains. The FungalTraits database (Põlme et al., 2021), a specific functional prediction tool, was used to taxonomically parse fungal genera by ecological guild independent of the sequencing method. Bacterial and fungal function count tables for each DNA sample were generated. Sequence counts of OTUs for fungi ranged from 919 to 345,516 and from 3,131 to 204,067 for bacteria.
2.6. Statistics
Statistical analyses were performed after OTUs were taxonomically summarized at the genus level. The normal distribution of each dataset was tested via OriginPro 2022 (OriginLab Corporation, Northampton, MA, USA) by the Shapiro–Wilk test (p < 0.05). Rarefaction analysis as well as the estimation of alpha diversity (OTU richness, Shannon index, Simpson index, and Pielou’s Evenness) and OTU richness estimators [bias-corrected Chao1 and an abundance-based coverage estimator (ACE)] were performed for cuticular DNA samples in RStudio (Version 2022.02.1, RStudio, Inc., Boston, MA, USA) and the package vegan 2.5–7 (Oksanen et al., 2022; R: The R Project for Statistical Computing, 2022).
Alpha diversity indices were tested for normal distribution by Shapiro–Wilk test (p < 0.05). Significant effects (p < 0.05) on cuticular alpha diversity indices for each PPP treatment were calculated either by one-way ANOVA with a post-hoc adjusted Tukey test, if data were normally distributed, or Kruskal–Wallis ANOVA with a post-hoc Dunn test, if data were not normally distributed, using OriginPro (Version 2022. OriginLab Corporation). The same statistical procedure was used to analyze the effects of PPP treatments on bacterial and fungal gene copy numbers on the cuticles. Permutation multivariate analysis of variance (NPMANOVA) based on Bray–Curtis similarity was performed using the software PAST 2.17c (Hammer et al., 2001) to analyze the differences between the different PPP treatments on the cuticular microbial communities and functions. The results were visualized by OriginPro (Version 2022. OriginLab Corporation). Noll et al. (2005) explained that relative abundances were calculated for each sample and visualized with OriginPro (Version 2022. OriginLab Corporation). Significantly distinctive cuticular bacterial and fungal genera of the PPP treatments were identified using indicator species analysis conducted using the “multipatt” function in the indicspecies package (de Cáceres and Legendre, 2009), which calculates indicator values with the “r.g.” function.
3. Results
3.1. PPP treatment significantly altered bacterial and fungal gene copy numbers
Our data showed that fungicides can significantly affect the bacterial and fungal gene copy numbers on the cuticle of honey bees. The fungicide Difcor® significantly reduced fungal gene copy numbers compared to control A (p = 0.04246), whereas neither the insecticide Steward® nor the mixture of Difcor® and Steward® affected fungal gene copy numbers (Supplementary Figure 1A).
Bacterial gene copy numbers were not affected by the fungicide Difcor®, the insecticide Steward®, or PPP mix A treatments (Figure 2A). In contrast, the fungicide Cantus® Gold significantly increased the bacterial gene copy numbers compared to control B (p = 0.00167) (Figure 2B). The insecticide Mospilan® and the PPP mix B had no significant effect on bacterial gene copy numbers compared to the control B (Figure 2B). PPP mix B treatment increased fungal gene copy numbers in all treatment groups (Supplementary Figure 1B). The highest fungal gene copy numbers could be found in the fungicide Cantus® Gold treatment, which differed significantly from the control B (p = 0.01528) and the insecticide Mospilan®-treated group (p = 0.01815) (Supplementary Figure 1B).
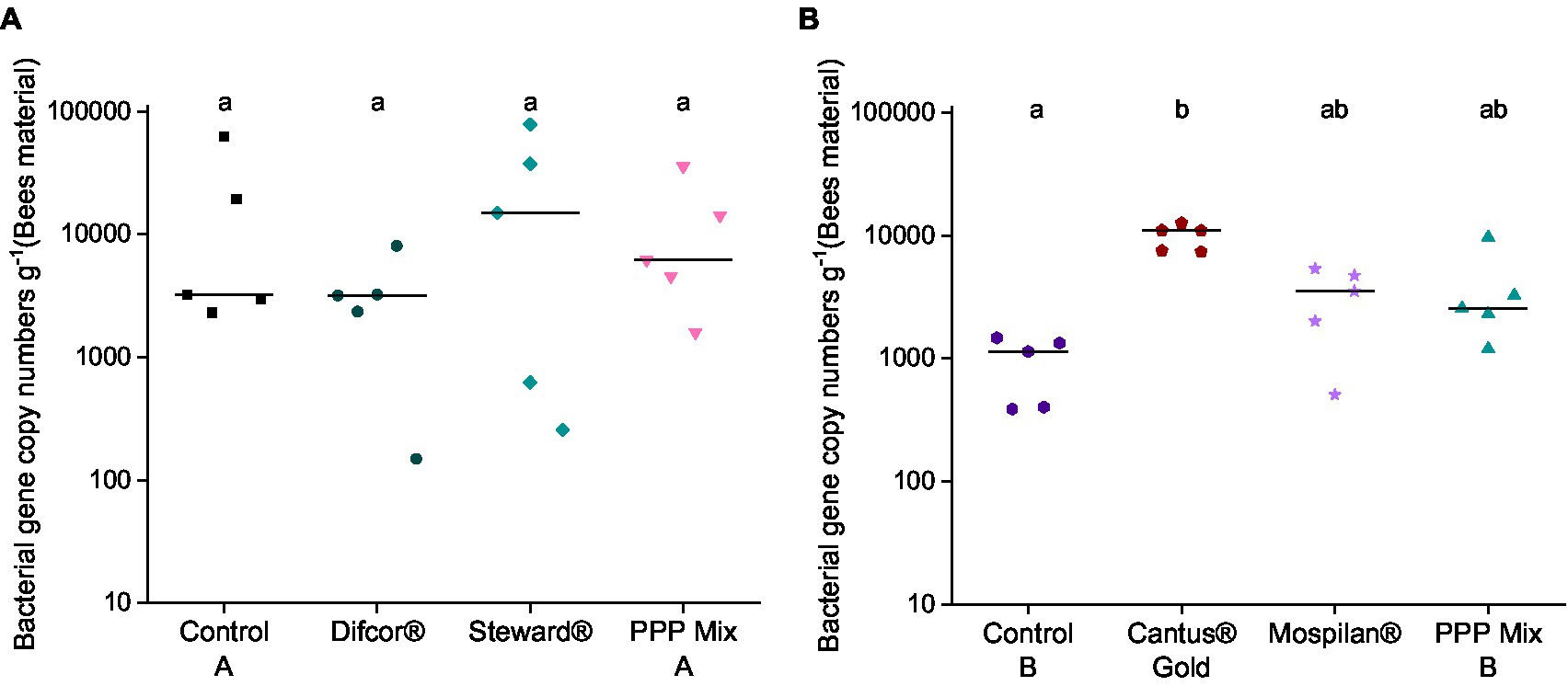
Figure 2. Bacterial gene copy (A,B) numbers after PPP treatment. PPP Difcor®, Steward®, or the combination of both (mix A) (A) and PPP Cantus® Gold, Mospilan® and the combination of both (mix B) (B) treatments (n = 5). Different letters indicate statistically significant differences according to the Dunn test (p = 0.05). If the letters are the same, treatments were not significant to each other, and if they were different, treatments were significantly different. The fungal gene copy numbers are shown in Supplementary Figure 1.
3.2. PPP treatments showed different effects on bacterial and fungal community compositions
The data indicate that the insecticide and fungicide treatments affect the fungal community composition. The fungal community composition was significantly altered by the insecticide Steward® treatment and differed significantly from control A (p = 0.0157). The fungicide Cantus® Gold significantly altered fungal community composition (p = 0.0239) in comparison with control B. Cantus® Gold, Mospilan®, and PPP mix B treatments significantly changed bacterial community composition in all treatments compared to control B (Figure 3; Supplementary Table 1).
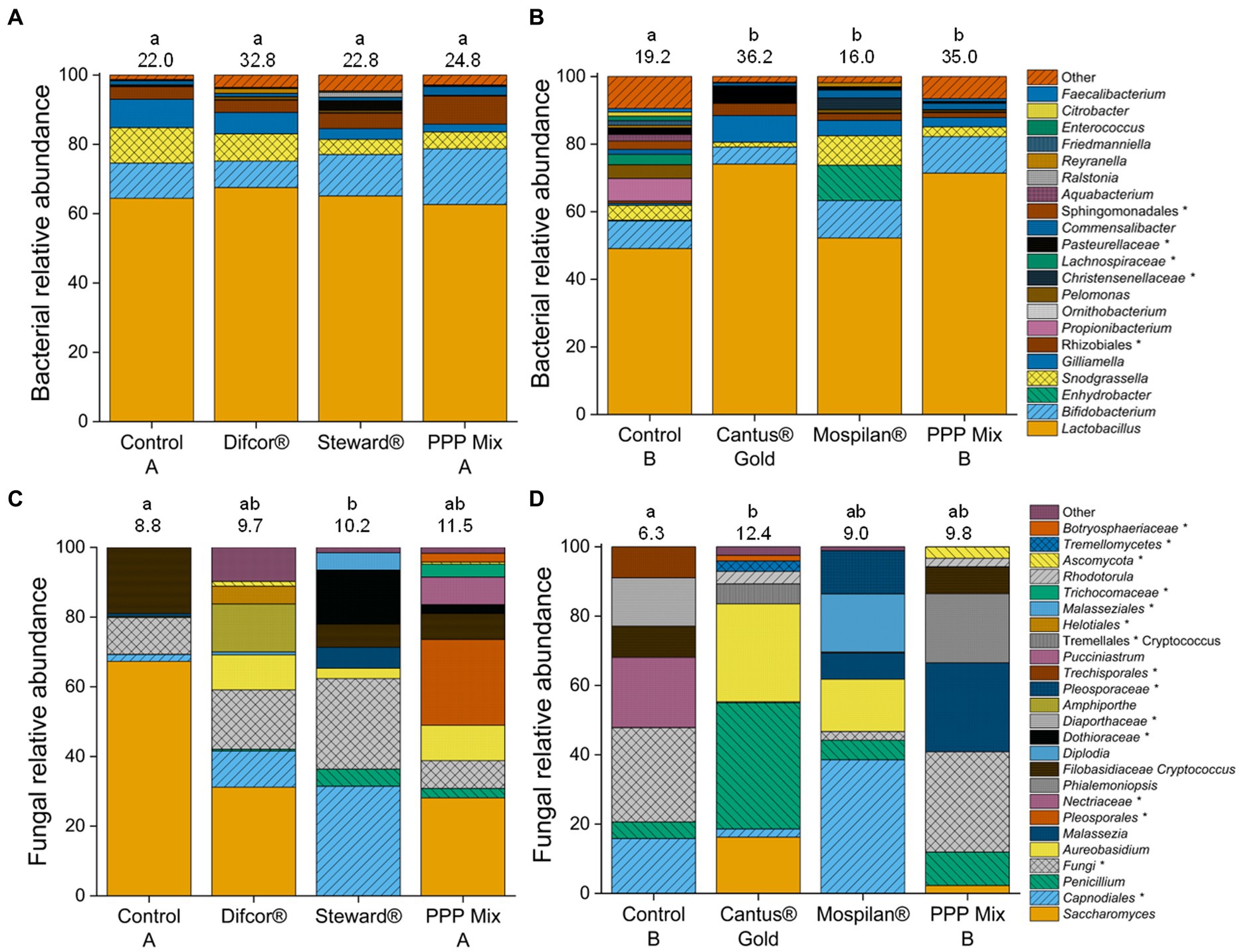
Figure 3. Relative sequence read abundance of the bacteria (A,B) and fungi (C,D) after PPP treatment. PPP Difcor®, Steward®, or the combination of both (mix A) (A,C) and PPP Cantus® Gold, Mospilan® and the combination of both (mix B) (B,D) treatments (n = 5). Bacterial species with a relative abundance of <1% were summarized as other. Complete bacterial abundance graphs without a summary of the low abundance can be found in Supplementary Figure 2. Fungal species with a relative abundance of <2% were summarized as other. Fully fungal abundance graphs without a summary of the low abundance can be found in different letters indicating statistically significant differences according to one-way non-parametric multivariate analysis (p = 0.05). If the letters are the same, treatments were not significantly different to each other, and if they were different, treatments were significantly different. The numbers above the bars reflect the respective OTU richness. Unclassified members of the taxon are marked with *.
In contrast, the insecticide Mospilan® and PPP mix B treatments did not lead to any alterations in the fungal community composition compared to the Cantus® Gold treatment and control B (Supplementary Table 1). The fungicide Difcor®, insecticide Steward®, and PPP mix A treatments did not significantly change the bacterial community composition (Figure 3; Supplementary Table 2). Bacterial and fungal OTU richness were not affected by the Difcor®, Steward®, and PPP mix A treatments [Dunn’s test (p = 0.05)] or Cantus® Gold, Mospilan®, and PPP mix B treatments [Tukey test (p = 0.05); Figure 3].
3.3. PPP treatments showed different effects on bacterial and fungal community functions
According to our data, insecticides and fungicides had an impact on fungal and bacterial functional composition. The insecticide Steward® treatment significantly impacted the fungal functional composition compared to control A (p = 0.0154) (Figure 4A). Furthermore, the fungicide Cantus® Gold treatment significantly altered fungal functional community composition in comparison with control B (p = 0.0255) (Figure 4B).
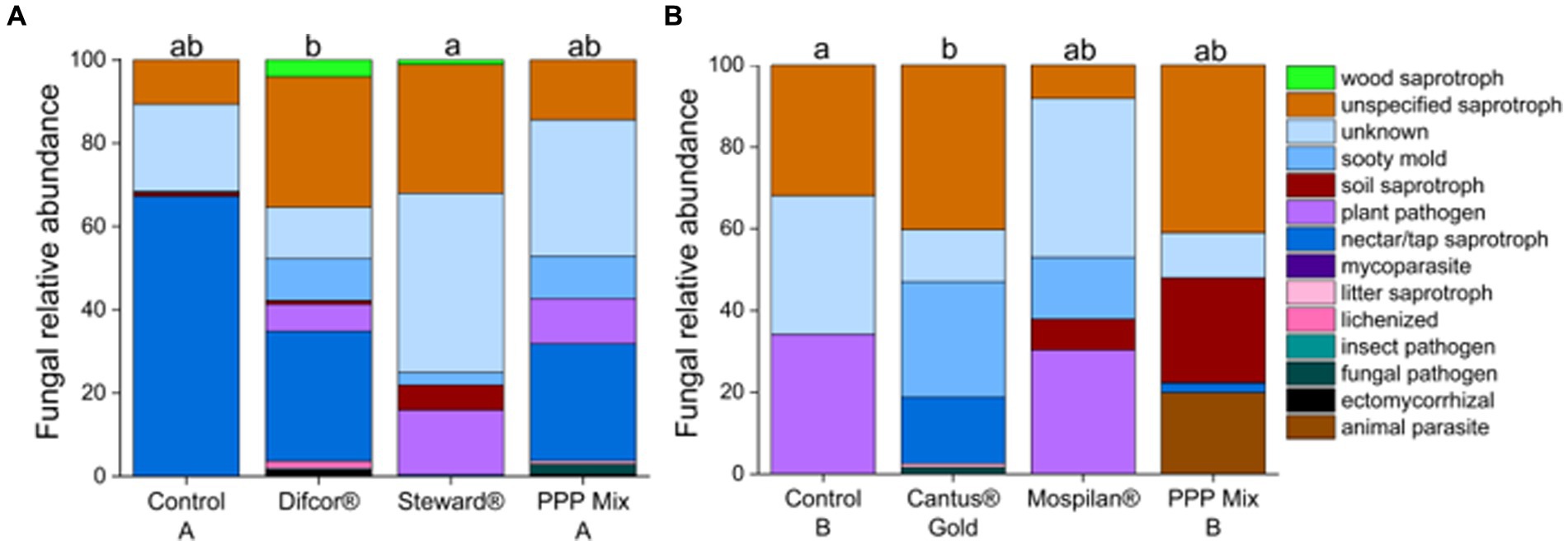
Figure 4. Fungal functional composition on genus level after PPP treatment. PPP Difcor®, Steward®, or the combination of both (mix A) (A) and PPP Cantus® Gold, Mospilan® and the combination of both (mix B) (B) treatments (n = 5). Different letters indicate statistically significant differences according to one-way non-parametric multivariate analysis (p = 0.05). If the letters are the same, treatments were not significant to each other, and if they were different, treatments were significantly different. Supplementary Tables 3, 4 provide which fungal genera were assigned to which functional traits.
Plant pathogenic fungi had increased sequence read abundance in the fungicide Difcor® (19%) and insecticide Steward® (22%)-treated bees in comparison with control A (<1%) (Figure 4A). Whereas sequence read abundance of plant pathogenic fungi was reduced in the treatment’s fungicide Cantus® Gold (2%), and PPP mix B (20%) or not altered in the insecticide Mospilan® (30%) treatment in comparison with control B (34%) (Figure 4B). The fungicide Difcor®-treated bees’ cuticular fungal microbiome was highly associated with the fungal genus Amphiporthe (Table 3), and the fungicide Cantus® Gold-treated bees’ cuticular fungal microbiome was associated with the genus Saccharomyces. The insecticides Steward® and PPP mix A, and control A, as well as the insecticides Mospilan®, PPP mix B, and control B, were not associated with any fungal indicator species. Fungicide Difcor® and PPP mix A did not differ in the fungal functional community composition compared to control A and the insecticide Steward® treatment (Figure 4A). The same could be observed for the insecticide Mospilan® and PPP mix B in comparison with control B and the Cantus® Gold treatment (Figure 4A).
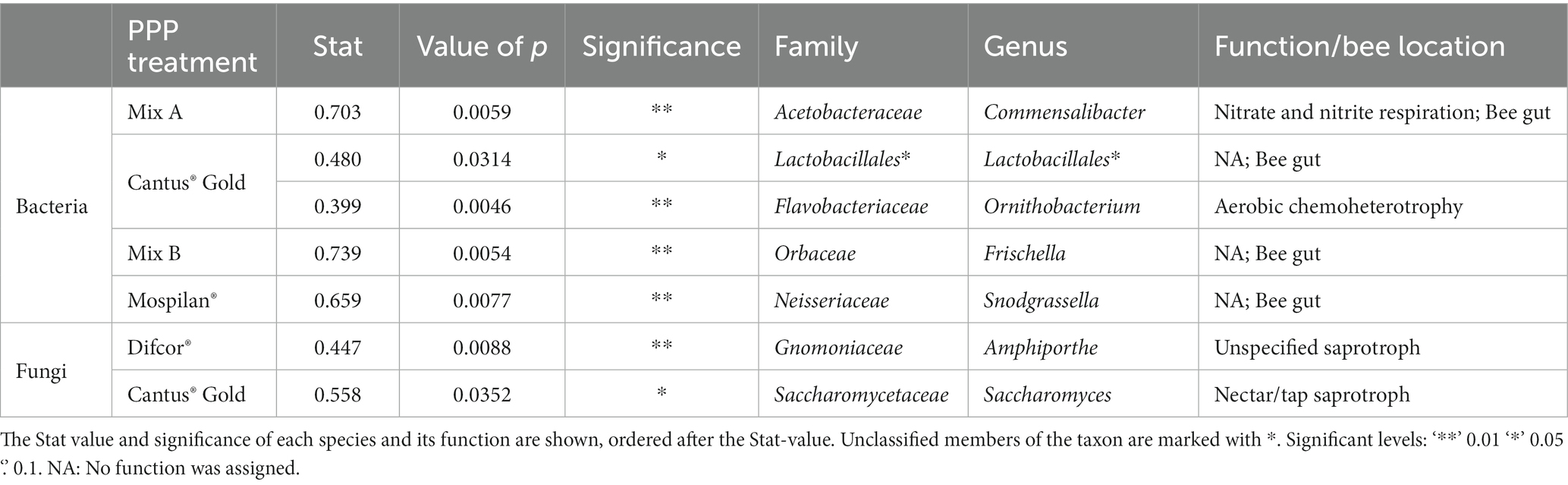
Table 3. Indicator species analysis of cuticular bacterial and fungal community members after PPP treatment.
In contrast to the fungal community composition, only the insecticide Mospilan® treatment led to significant differences in the bacterial functional composition compared to control B (p = 0.0165) (Supplementary Figure 4B). Indicator analysis of Difcor®, Steward®, and PPP mix A treatments showed that Commensalibacter was highly associated with the bacterial community of the PPP mix A treatment (Table 3). The fungicide Difcor® treatment, the insecticide Steward® treatment, and control A did not bear any bacterial indicators. The cuticular microbiome of Cantus® Gold-treated bees was significantly associated with unclassified members of the order Lactobacillales. Furthermore, a weaker association with Ornithobacterium could be observed (Table 3). The cuticular bacterial microbiome of Mospilan®-treated bees’ high associations with the genus Snodgrassela was observed. The cuticular bacterial microbiome of the combined treatment with PPP mix B was highly associated with the genus Frischella (Table 3). The fungicide Difcor®, insecticide Steward®, and PPP mix A treatments compared to control A (Supplementary Figure 4A), as well as the fungicide Cantus® Gold and PPP mix B treatments, did not affect the bacterial functional community composition compared to control B and the insecticide Mospilan® treatment (Supplementary Figure 4B).
4. Discussion
Tremendous effects on the fungal microbiome could be observed for all treatments. Moreover, single pesticide treatments such as the fungicide Difcor® significantly reduced the fungal gene copy numbers (Supplementary Figure 1B). The insecticide Steward® led to significant alterations in the fungal community composition and function (Figures 3B; Supplementary Figure 4A).
Fungicide Cantus® Gold, insecticide Mospilan®, and PPP mix B treatments increased bacterial and fungal gene copy numbers (Figures 2C,D; Supplementary Figure 1B), and all treatments significantly altered the bacterial community composition (Figure 3C; Supplementary Table 1). The insecticide Mospilan® led to significant changes in the bacterial functional composition (Supplementary Figure 4B). Fungicide Cantus® Gold had tremendous effects on the fungal cuticular community and functional composition (Figure 3D; Supplementary Figure 4B).
The fungicide Difcor® and insecticide Steward® treatments significantly impacted the cuticular bacterial community composition (Figure 3A; Supplementary Table 1A), while the bacterial gene copy numbers and bacterial community functions were unaffected (Figure 2A). The genus Commensalibacter was identified as an indicator taxa of the cuticular bacterial microbiome after PPP mix A treatment (Table 3). Members of the genus Commensalibacter were previously described as a core member of the honey bees’ gut microbiome (Martinson et al., 2011; Kwong and Moran, 2016) and as an essential part of the honey bees’ microbial ecosystem (Wu et al., 2022).
The fungicide Difcor®, insecticide Steward®, and PPP mix A treatments caused a reduction of the fungal gene copy numbers compared to the control A. Moreover, the fungicide Difcor®, insecticide Steward®, and PPP mix A treatments led to a higher diversity of the cuticular fungal community composition (Figure 3B). Similar results could be observed for the intestine microbiome of bees after treatment with pesticides (Syromyatnikov et al., 2020). Similarly, the fungicide Difcor® significantly reduced the fungal gene copy numbers (Supplementary Figure 1B), and members of the genus Amphiporthe were significantly associated with this treatment. However, a recent study found that members of the same family, Valsaceae, were described as sensitive toward difenoconazole (Silva-Campos et al., 2022), indicating an ambivalent biocide response in this family. Furthermore, Steward® significantly changed cuticular fungal community composition and function (Figures 3B, 4A; Supplementary Table 2). Saccharomyces was not found in the insecticide Steward®-treated groups, indicating that members of Saccharomyces were highly sensitive to the insecticide Steward®. However, Saccharomyces was the most abundant genus in control A (Figure 3). Unclassified members of the genera Dothioraceae and Capnodiales were the most abundant species in the Steward®-treated cuticular fungal microbiomes (Figure 3B). Members of the genus Dothioraceae were already found in the guts of nectar-collecting Apis cerana (Basukriadi et al., 2010). Capnodiales was previously described as increasing abundance of chlorothalonil-based fungicides in field-relevant level-treated hives (Steffan et al., 2017). Even though the active ingredient differs from the insecticide Steward®, this result indicates that Capnodiales benefits from the treatment with PPP either directly by the inactivation of competitors or their predators and/or indirectly by microbial metabolites or degradation products released from PPP-sensitive species. Nectar/tap saprotrophs were two-thirds the most abundant group in control A. Nectar/tap saprotrophic fungi were reduced in the fungicide Difcor®, insecticide Steward®, and PPP mix A treatments compared to control A (Figure 4A). Those were reduced by less than 1% in the insecticide Steward® treatment, indicating that this insecticide alone already caused this reduction of nectar/tap saprotrophic fungi. The active ingredient of Steward® is indoxacarb; the insecticidal activity occurs by blocking the sodium channels within the nervous system of insects (Wing et al., 2000). Even though mycorrhizal fungi are described as playing an important role in balancing salinity within the environment and the use of sodium for signaling, no voltage-gated sodium channels have been found for fungi (Scharnagl et al., 2017). Thereby, we are the first to describe this non-target effect of indoxacarb on nectar/tap saprotrophic fungi. To date, honey analyses have not shown any negative effects of the use of any of these fungicides. However, based on our study, existing data and set-ups should be revisited in detail if they do affect honey quality.
Interestingly, plant pathogens gained abundance in the cuticular fungal community of the fungicide Difcor® and insecticide Steward®-treated bees in comparison with control A (Figure 4A). Moreover, sooty mold fungi were increased in the cuticular fungal community of fungicide Difcor®, insecticide Steward®, and PPP mix A-treated bees. Those fungi are reported to show resistance against difenoconazole (Difcor®) (Yang et al., 2019). Moreover, those were described as showing cross-resistance even for fungicides with a different mode of action (Yang et al., 2019), which might be the reason for their high sequence read abundances in the insecticide Steward®-treated bees (Figure 4A). Similar to the cuticular fungal community composition, the diversity of functional composition was increased due to the fungicide Difcor®, insecticide Steward®, and PPP mix A treatments, which were already observed for the intestinal microbiome of bees (Silva-Campos et al., 2022).
The fungicide Cantus® Gold, insecticide Mospilan®, and PPP mix B treatments shifted the bacterial and fungal cuticular community composition. Fungicide Cantus® Gold significantly increased bacterial and fungal gene copy numbers (Figures 2C,D). Fungicide Cantus® Gold, insecticide Mospilan®, and PPP mix B treatments significantly changed cuticular bacterial community composition (Figure 3C; Supplementary Tables 1, 2). Although the composition of the bacterial community was altered by the treatments with the fungicide Cantus® Gold, the insecticide Mospilan®, and the PPP mixture B, only the insecticide Mospilan® showed a significant change in the functional composition of the cuticular bacteria (Supplementary Figure 4B). Alberoni et al. (2021) observed a significant decrease in the neonicotinoid-treated groups and a compirsed functionality of the gut microbiome of bees. This is in line with our results for the bacterial functional community composition after treatment with the insecticide Mospilan.
The indicator analysis for fungicide Cantus® Gold, insecticide Mospilan®, and PPP mix B-treated bees revealed indicator species for all treatments. For example, Snodgrassella was significantly associated with the insecticide Mospilan®-treated bees (Table 3), while Frischella was significantly associated with the PPP mix B. Both genera are dominant intestinal bacteria of bees (Babendreier et al., 2007). Ornithobacterium and unclassified members of the order Lactobacillales were significantly associated with the fungicide Cantus® Gold treatment. Lactobacillales are also known members of the bee’s gut microbiome (Babendreier et al., 2007; Martinson et al., 2011; Kwong and Moran, 2013). As gut microbiota are specialized to an ecological niche, it is likely to find those in another hive niche of its host species (Anderson et al., 2011). Furthermore, it was shown that grooming plays a role in implementing the bees’ gut microbiome (Powell et al., 2014). Thereby, the grooming processes of bees could also lead to the distribution of gut-associated bacteria on the cuticular.
The fungicide Cantus® Gold treatment significantly altered the cuticular fungal community composition of the bees compared to control B (Figure 3D; Supplementary Table 1). The insecticide Mospilan® and PPP mix B did not significantly differ from control B or the fungicide Cantus® Gold treatment. The fungal functional cuticular community composition of the fungicide Cantus® Gold treatment differed significantly from control B (Figure 4B). The relative sequence read abundance of sooty mold and nectar/tap saprotrophic fungi was significantly increased compared to the insecticide Mospilan® and PPP mix B treatments. Moreover, plant pathogenic fungi were significantly reduced by the fungicide Cantus® Gold from one-third to less than 1% (Figure 4B). It is known that fungicide treatment alters the hive fungal community composition by introducing residues from pollen or bees (Sammataro et al., 2012). Yoder et al. (2013) described that the mixture of boscalid and pyraclostrobin did alter the fungal community of bee bread. Pyraclostrobin is a strobilurin and belongs to the same chemical group as dimoxystrobin, which forms together with boscalid, the active ingredient of the fungicide Cantus® Gold. We already described that the in-hive microbiome is closely connected to the bees’ microbiome. Therefore, it is likely that this is also the case for fungi. Saccharomyces was identified as an indicator of the fungicide Cantus® Gold-treated cuticular fungal microbiomes (Table 3) and is described as nectar/tap saprotrophic. Hnátová et al. (2003) described that mutants of Saccharomyces show resistance against strobilurin fungicides. In our experiments, we did not analyze the effects of the fungicides, the insecticides, or their combinations on honey bee health or honey production. Based on studies by Degrandi-Hoffman et al. (2015), it can be assumed that the fungicide boscalid, which was also applied in our study, can increase pathogens such as deformed wing virus or black queen cell virus. Furthermore, Pettis et al. (2013) demonstrated the effect of boscalid ingestions on the probability of Nosema infections. Similar to fungicides, neonicotinoids can affect honey bee health. Harwood and Dolezal (2020) showed negative effects on hemocyte differentiation and function following neonicotinoid application. Brandt et al. (2016) demonstrated that neonicotinoids can reduce hemocyte density, encapsulation response, and antimicrobial activity in honey bee. In order to investigate the further effects of PPPs on bee health, experiments using a different experimental design compared to our study should be performed.
5. Conclusion
Our results have demonstrated for the first time that both insecticides and fungicides can have adverse effects on the microbiome on the cuticle of honey bees, which has to date been completely neglected when investigating the side effects of PPPs. The cuticle microbiome may serve important functions as a barrier against harmful microbes. A change in the composition of the microbiome may have severe effects on honey bee health, which might only become apparent long after the collection or consumption of the respective insecticides or fungicides. The insecticide Steward® with the active substance indoxacarb and the fungicide Cantus® Gold with the active substances boscalid and dimoxystrobin are the most frequent residues in beebread, altering the fungal community composition of honey bee cuticles significantly. The neonicotinoid Mospilan® with the active substance acetamiprid significantly affected bacterial functional community composition. Mixtures of fungicides and insecticides could enhance the side effects of single substances, which have rarely been observed because fungicides are generally believed to be harmless to bees and other pollinators. In particular, fungal cuticular community composition was affected, showing a phylogenetic diversification due to the PPP mix treatments and an increase in pathogenic fungi on the bees’ cuticle. Our results urge more studies on side effects on honey bees and other bees caused by the interaction of insecticides and fungicides and demonstrate that the microbiome of the cuticle is a promising site for investigation because it is susceptible to the actions of PPPs.
Data availability statement
The datasets presented in this study can be found in online repositories. The microbiome’s bacterial 16S rRNA and fungal ITS gene sequences were deposited in the NCBI nucleotide sequence databases (https://www.ncbi.nlm.nih.gov/) under accession PRJNA880009.
Ethics statement
Ethical approval was not required for the study involving animals in accordance with the local legislation and institutional requirements because our protocols comply with standard welfare practice in our field. The experiment involved honey bee from an apiary dedicated to research.
Author contributions
FR: Funding acquisition, Investigation, Supervision, Validation, Writing – original draft, Data curation, Formal analysis, Methodology, Visualization. AS: Data curation, Formal analysis, Investigation, Methodology, Writing – review & editing. LS: Data curation, Formal analysis, Investigation, Methodology, Writing – review & editing, Software, Validation, Visualization. MT: Writing – review & editing, Conceptualization. RS: Conceptualization, Writing – review & editing, Funding acquisition, Project administration, Resources, Supervision. MN: Conceptualization, Funding acquisition, Project administration, Resources, Supervision, Investigation, Validation, Writing – original draft.
Funding
The author(s) declare financial support was received for the research, authorship, and/or publication of this article. This research was financed by the Bavarian Environment Agency and supported by a grant from the Bavarian State Ministry of the Environment and Consumer Protection to MN and RS in the research network BayÖkotox [ID75253] (TP6 Bewertung Biozid-haltiger Baustoffe; TP 2 Honig- und Wildbienen unter Stress). Furthermore, the study was supported by a grant from the German Federal Foundation for the Environment (DBU) to AS (20021/748).
Acknowledgments
The authors would like to thank Julia Kenzel and Eva-Maria Wittmann for their excellent project organization and Felix Schneider for his help with the experiments.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
The author(s) declared that they were an editorial board member of Frontiers, at the time of submission. This had no impact on the peer review process and the final decision.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2023.1271498/full#supplementary-material
References
Aizenberg-Gershtein, Y., Izhaki, I., and Halpern, M. (2013). Do honeybees shape the bacterial community composition in floral nectar? PLoS One 8:e67556. doi: 10.1371/journal.pone.0067556
Alberoni, D., Favaro, R., Baffoni, L., Angeli, S., and Di Gioia, D. (2021). Neonicotinoids in the agroecosystem: in-field long-term assessment on honeybee colony strength and microbiome. Sci. Total Environ. 762:144116. doi: 10.1016/j.scitotenv.2020.144116
Almasri, H., Tavares, D. A., Pioz, M., Sené, D., Tchamitchian, S., Cousin, M., et al. (2020). Mixtures of an insecticide, a fungicide and a herbicide induce high toxicities and systemic physiological disturbances in winter Apis mellifera honey bees. Ecotoxicol. Environ. Saf. 203:111013. doi: 10.1016/j.ecoenv.2020.111013
Alvarez-Pérez, S., and Herrera, C. M. (2013). Composition, richness and nonrandom assembly of culturable bacterial-microfungal communities in floral nectar of Mediterranean plants. FEMS Microbiol. Ecol. 83, 685–699. doi: 10.1111/1574-6941.12027
Anderson, K. E., Sheehan, T. H., Eckholm, B. J., Mott, B. M., and DeGrandi-Hoffman, G. (2011). An emerging paradigm of colony health: microbial balance of the honey bee and hive (Apis mellifera). Insect. Soc. 58, 431–444. doi: 10.1007/s00040-011-0194-6
Anderson, K. E., Sheehan, T. H., Mott, B. M., Maes, P., Snyder, L., Schwan, M. R., et al. (2013). Microbial ecology of the hive and pollination landscape: bacterial associates from floral nectar, the alimentary tract and stored food of honey bees (Apis mellifera). PLoS One 8:e83125. doi: 10.1371/journal.pone.0083125
Babendreier, D., Joller, D., Romeis, J., Bigler, F., and Widmer, F. (2007). Bacterial community structures in honeybee intestines and their response to two insecticidal proteins. FEMS Microbiol. Ecol. 59, 600–610. doi: 10.1111/j.1574-6941.2006.00249.x
Babraham Bioinformatics FastQC a quality control tool for high throughput sequence data (2020). Available at: https://www.bioinformatics.babraham.ac.uk/projects/fastqc/ (Accessed April 14, 2022).
BASF, S. E. (2021). Gebrauchsanleitung Cantus. Available at: https://www.agrar.basf.de/Dokumente/Produkte/Cantus-Gold/ga-cantus-gold.pdf (Accessed May 23, 2021).
Basukriadi, A. D., Wellyzar, S., and Putra, B. B. (2010). Molecular identification and diversity of yeasts associated with Apis cerana foraging on flowers of Jatropha integerrima. Microbiol. Indones. 4, 44–48. doi: 10.5454/mi.4.1.9
Börner, H., Schlüter, K., and Aumann, J. (2009). Pflanzenkrankheiten und Pflanzenschutz. Berlin, Heidelberg: Springer
Brandt, A., Gorenflo, A., Siede, R., Meixner, M., and Büchler, R. (2016). The neonicotinoids thiacloprid, imidacloprid, and clothianidin affect the immunocompetence of honey bees (Apis mellifera L.). J. Insect Physiol. 86, 40–47. doi: 10.1016/j.jinsphys.2016.01.001
Corby-Harris, V., Maes, P., and Anderson, K. E. (2014). The bacterial communities associated with honey bee (Apis mellifera) foragers. PLoS One 9:e95056. doi: 10.1371/journal.pone.0095056
Cox, C., and Surgan, M. (2006). Unidentified inert ingredients in pesticides: implications for human and environmental health. Envrion. Health Perspect. 114, 1803–1806. doi: 10.1289/ehp.9374
de Cáceres, M., and Legendre, P. (2009). Associations between species and groups of sites: indices and statistical inference. Ecology 90, 3566–3574. doi: 10.1890/08-1823.1
Degrandi-Hoffman, G., Chen, Y., Watkins Dejong, E., Chambers, M. L., and Hidalgo, G. (2015). Effects of oral exposure to fungicides on honey bee nutrition and virus levels. J. Econ. Entomol. 108, 2518–2528. doi: 10.1093/jee/tov251
DOE Joint Genome Institute (2022). BBTools. Available at: https://jgi.doe.gov/data-and-tools/software-tools/bbtools/ (Accessed April 14, 2022).
Edgar, R. C. (2018). Updating the 97% identity threshold for 16S ribosomal RNA OTUs. Bioinformatics 34, 2371–2375. doi: 10.1093/bioinformatics/bty113
Edgar, R. C., Haas, B. J., Clemente, J. C., Quince, C., and Knight, R. (2011). UCHIME improves sensitivity and speed of chimera detection. Bioinformatics 27, 2194–2200. doi: 10.1093/bioinformatics/btr381
El Hassani, A. K., Dacher, M., Gary, V., Lambin, M., Gauthier, M., and Armengaud, C. (2008). Effects of sublethal doses of acetamiprid and thiamethoxam on the behavior of the honeybee (Apis mellifera). Arch. Environ. Contam. Toxicol. 54, 653–661. doi: 10.1007/s00244-007-9071-8
El-Nahhal, Y. (2020). Pesticide residues in honey and their potential reproductive toxicity. Sci. Total Environ. 741:139953. doi: 10.1016/j.scitotenv.2020.139953
Engel, P., Martinson, V. G., and Moran, N. A. (2012). Functional diversity within the simple gut microbiota of the honey bee. Proc. Natl. Acad. Sci. U. S. A. 109, 11002–11007. doi: 10.1073/pnas.1202970109
FMC Agricultural Solutions (2020). Produktinformation und Sicherheitsdatenblatt von Steward.. Available at: https://www.fmcagro.de/de/produkte/a-z/steward-im-obstbau.htm (Accessed July 5, 2020).
FMC Agricultural Solutions (2021). Mospilan SG. Available at: https://ag.fmc.com/de/de/produkte/insektizide/mospilan-sg (Accessed September 9, 2021).
Fridman, S., Izhaki, I., Gerchman, Y., and Halpern, M. (2012). Bacterial communities in floral nectar. Environ. Microbiol. Rep. 4, 97–104. doi: 10.1111/j.1758-2229.2011.00309.x
Friedle, C., Wallner, K., Rosenkranz, P., Martens, D., and Vetter, W. (2021). Pesticide residues in daily bee pollen samples (April–July) from an intensive agricultural region in Southern Germany. Environ. Sci. Pollut. Res. Int. 28, 22789–22803. doi: 10.1007/s11356-020-12318-2
Fungicide Resistance Action Commitee (2021). FRAC Code List 2022: Fungal control agents sorted by cross resistance pattern and mode of action (including coding for FRAC Groupson product labels). Available at: https://www.frac.info/docs/default-source/publications/frac-code-list/frac-code-list-2022--final.pdf?sfvrsn=b6024e9a_2 (Accessed May 23, 2021).
Galvan, T. L., Koch, R. L., and Hutchison, W. D. (2006). Toxicity of indoxacarb and spinosad to the multicolored Asian lady beetle, Harmonia axyridis (Coleoptera: Coccinellidae), via three routes of exposure. Pest Manag. Sci. 62, 797–804. doi: 10.1002/ps.1223
Gilliam, M., Prest, D. B., and Lorenz, B. J. (1989). Microbiology of pollen and bee bread: taxonomy and enzymology of molds. Apidologie 20, 53–68. doi: 10.1051/apido:19890106
Goulson, D., Nicholls, E., Botías, C., and Rotheray, E. L. (2015). Bee declines driven by combined stress from parasites, pesticides, and lack of flowers. Science 347:1255957. doi: 10.1126/science.1255957
Hammer, O., Harper, D., and Ryan, P. (2001). PAST: paleontological statistics software package for education and data analysis. Palaeontol. Electron. 4, 1–9.
Harwood, G. P., and Dolezal, A. G. (2020). Pesticide-virus interactions in honey bees: challenges and opportunities for understanding drivers of bee declines. Viruses 12:566. doi: 10.3390/v12050566
Hesselbach, H., and Scheiner, R. (2019). The novel pesticide flupyradifurone (Sivanto) affects honeybee motor abilities. Ecotoxicology 28, 354–366. doi: 10.1007/s10646-019-02028-y
Hnátová, M., Gbelská, Y., Obernauerová, M., Subíková, V., and Subík, J. (2003). Cross-resistance to strobilurin fungicides in mitochondrial and nuclear mutants of Saccharomyces cerevisiae. Folia Microbiol. 48, 496–500. doi: 10.1007/BF02931331
Holzschuh, A., Dormann, C. F., Tscharntke, T., and Steffan-Dewenter, I. (2013). Mass-flowering crops enhance wild bee abundance. Oecologia 172, 477–484. doi: 10.1007/s00442-012-2515-5
Ihrmark, K., Bödeker, I. T. M., Cruz-Martinez, K., Friberg, H., Kubartova, A., Schenck, J., et al. (2012). New primers to amplify the fungal ITS2 region-evaluation by 454-sequencing of artificial and natural communities. FEMS Microbial. Ecol. 82, 666–677. doi: 10.1111/j.1574-6941.2012.01437.x
Insecticide Resistance Action Committee (2021). IRAC - The IRAC Mode of Action Classification. Available at: https://irac-online.org/mode-of-action/ (Accessed May 23, 2021).
Keller, A., McFrederick, Q. S., Dharampal, P., Steffan, S., Danforth, B. N., and Leonhardt, S. D. (2021). (More than) Hitchhikers through the network: the shared microbiome of bees and flowers. Curr. Opin. Insect Sci. 44, 8–15. doi: 10.1016/j.cois.2020.09.007
Klindworth, A., Pruesse, E., Schweer, T., Peplies, J., Quast, C., Horn, M., et al. (2013). Evaluation of general 16S ribosomal RNA gene PCR primers for classical and next-generation sequencing-based diversity studies. Nucleic Acids Res. 41:e1. doi: 10.1093/nar/gks808
Kluser, S., and Peduzzi, P. (2007). Global pollinator decline: a literature review. Geneva: UNEP/GRID-Europe.
Kõljalg, U., Nilsson, H. R., Schigel, D., Tedersoo, L., Larsson, K.-H., May, T. W., et al. (2020). The taxon hypothesis paradigm-on the unambiguous detection and communication of taxa. Microorganisms 8:1910. doi: 10.3390/microorganisms8121910
Kwong, W. K., Mancenido, A. L., and Moran, N. A. (2017a). Immune system stimulation by the native gut microbiota of honey bees. Royal Soc. Open Sci. 4:170003. doi: 10.1098/rsos.170003
Kwong, W. K., Medina, L. A., Koch, H., Sing, K.-W., Soh, E. J. Y., Ascher, J. S., et al. (2017b). Dynamic microbiome evolution in social bees. Sci. Adv. 3:e1600513. doi: 10.1126/sciadv.1600513
Kwong, W. K., and Moran, N. A. (2013). Cultivation and characterization of the gut symbionts of honey bees and bumble bees: description of Snodgrassella alvi gen. nov., sp. nov., a member of the family Neisseriaceae of the Betaproteobacteria, and Gilliamella apicola gen. nov., sp. nov., a member of Orbaceae fam. nov., Orbales ord. nov., a sister taxon to the order ‘Enterobacteriales’ of the Gammaproteobacteria. Int. J. Syst. Evol. Microbiol. 63, 2008–2018. doi: 10.1099/ijs.0.044875-0
Kwong, W. K., and Moran, N. A. (2016). Gut microbial communities of social bees. Nat. Rev. Microbiol. 14, 374–384. doi: 10.1038/nrmicro.2016.43
Lasota, S., Stephan, I., Horn, M. A., Otto, W., and Noll, M. (2019). Copper in wood preservatives delayed wood decomposition and shifted soil fungal but not bacterial community composition. Appl. Environ. Microbiol. 85:e02391-18. doi: 10.1128/AEM.02391-18
Laurino, D., Porporato, M., Patteta, A., and Manino, A. (2011). Toxicity of neonicotinoid insecticides to honey bees: laboratory tests. Bull. Insectol. 64, 107–113.
Louca, S., Parfrey, L. W., and Doebeli, M. (2016). Decoupling function and taxonomy in the global ocean microbiome. Science 353, 1272–1277. doi: 10.1126/science.aaf4507
Lüken, D. J., and von der Ohe, W. (2018). A research about different residues in pollen and honey samples. Hazards Pestic. Bees 462, 198–202. doi: 10.5073/JKA.2018.462.064
Martinson, V. G., Danforth, B. N., Minckley, R. L., Rueppell, O., Tingek, S., and Moran, N. A. (2011). A simple and distinctive microbiota associated with honey bees and bumble bees. Mol. Ecol. 20, 619–628. doi: 10.1111/j.1365-294X.2010.04959.x
Mattila, H. R., Rios, D., Walker-Sperling, V. E., Roeselers, G., and Newton, I. L. G. (2012). Characterization of the active microbiotas associated with honey bees reveals healthier and broader communities when colonies are genetically diverse. PLoS One 7:e32962. doi: 10.1371/journal.pone.0032962
McArt, S. H., Urbanowicz, C., McCoshum, S., Irwin, R. E., and Adler, L. S. (2017). Landscape predictors of pathogen prevalence and range contractions in US bumblebees. Proc. Biol. Sci. 284:20172181. doi: 10.1098/rspb.2017.2181
Moran, N. A., Hansen, A. K., Powell, J. E., and Sabree, Z. L. (2012). Distinctive gut microbiota of honey bees assessed using deep sampling from individual worker bees. PLoS One 7:e36393. doi: 10.1371/journal.pone.0036393
Nilsson, R. H., Larsson, K.-H., Taylor, A. F. S., Bengtsson-Palme, J., Jeppesen, T. S., Schigel, D., et al. (2019). The UNITE database for molecular identification of fungi: handling dark taxa and parallel taxonomic classifications. Nucleic Acids Res. 47, D259–D264. doi: 10.1093/nar/gky1022
Noll, M., Matthies, D., Frenzel, P., Derakshani, M., and Liesack, W. (2005). Succession of bacterial community structure and diversity in a paddy soil oxygen gradient. Environ. Microbiol. 7, 382–395. doi: 10.1111/j.1462-2920.2005.00700.x
Nováková, E., Hypsa, V., and Moran, N. A. (2009). Arsenophonus, an emerging clade of intracellular symbionts with a broad host distribution. BMC Microbiol. 9:143. doi: 10.1186/1471-2180-9-143
Oksanen, J., Blanchet, F. G., Friedly, M., Kindt, R., Legendre, P., McGlinn, D., et al. (2022). Package ‘vegan’: Community ecology package. R package version 2. Available at: https://cran.r-project.org/package=vegan
Pettis, J. S., Lichtenberg, E. M., Andree, M., Stitzinger, J., Rose, R., and vanEngelsdorp, D. (2013). Crop pollination exposes honey bees to pesticides which alters their susceptibility to the gut pathogen Nosema ceranae. PLoS One 8:e70182. doi: 10.1371/journal.pone.0070182
Põlme, S., Abarenkov, K., Henrik Nilsson, R., Lindahl, B. D., Clemmensen, K. E., Kauserud, H., et al. (2021). Correction to: FungalTraits: a user friendly traits database of fungi and fungus-like stramenopiles. Fungal Divers. 107, 129–132. doi: 10.1007/s13225-021-00470-0
Potts, S. G., Biesmeijer, J. C., Kremen, C., Neumann, P., Schweiger, O., and Kunin, W. E. (2010). Global pollinator declines: trends, impacts and drivers. Trends Ecol. Evol. 25, 345–353. doi: 10.1016/j.tree.2010.01.007
Powell, J. E., Martinson, V. G., Urban-Mead, K., and Moran, N. A. (2014). Routes of acquisition of the gut microbiota of the honey bee Apis mellifera. Appl. Environ. Microbiol. 80, 7378–7387. doi: 10.1128/AEM.01861-14
R: The R Project for Statistical Computing (2022) Available at: https://www.r-project.org/
Raymann, K., and Moran, N. A. (2018). The role of the gut microbiome in health and disease of adult honey bee workers. Curr. Opin. Insect Sci. 26, 97–104. doi: 10.1016/j.cois.2018.02.012
Ribière, C., Hegarty, C., Stephenson, H., Whelan, P., and O'Toole, P. W. (2019). Gut and whole-body microbiota of the honey bee separate thriving and non-thriving hives. Microb. Ecol. 78, 195–205. doi: 10.1007/s00248-018-1287-9
Rortais, A., Arnold, G., Halm, M.-P., and Touffet-Briens, F. (2005). Modes of honeybees exposure to systemic insecticides: estimated amounts of contaminated pollen and nectar consumed by different categories of bees. Apidologie 36, 71–83. doi: 10.1051/apido:2004071
Rosenkranz, P., Ohe, W.von der, Schäfer, M., Genersch, E., Büchler, R., and Berg, S. (2019). Deutsches Bienenmonitoring: DeBiMo. Available at: https://bienenmonitoring.uni-hohenheim.de/fileadmin/einrichtungen/bienenmonitoring/Abschlussbericht_DeBiMo_2017-2019.pdf (Accessed September 21, 2022).
Sabree, Z. L., Hansen, A. K., and Moran, N. A. (2012). Independent studies using deep sequencing resolve the same set of core bacterial species dominating gut communities of honey bees. PLoS One 7:e41250. doi: 10.1371/journal.pone.0041250
Saccà, M. L., and Lodesani, M. (2020). Isolation of bacterial microbiota associated to honey bees and evaluation of potential biocontrol agents of Varroa destructor. Benef Microbes 11, 641–654. doi: 10.3920/BM2019.0164
Sammataro, D., Yoder, J. A., and Yoder, J., eds (2012). Honey bee colony health: Challenges and sustainable solutions. Boca Raton, FL: CRC Press.
Scharnagl, K., Scharnagl, A., and von Wettberg, E. (2017). Nature's potato chip: the role of salty fungi in a changing world. Am. J. Bot. 104, 641–644. doi: 10.3732/ajb.1700034
Scheiner, R., Abramson, C. I., Brodschneider, R., Crailsheim, K., Farina, W. M., Fuchs, S., et al. (2013). Standard methods for behavioural studies of Apis mellifera. J. Apic. Res. 52, 1–58. doi: 10.3896/IBRA.1.52.4.04
Schloss, P. D., Westcott, S. L., Ryabin, T., Hall, J. R., Hartmann, M., Hollister, E. B., et al. (2009). Introducing mothur: open-source, platform-independent, community-supported software for describing and comparing microbial communities. Appl. Environ. Microbiol. 75, 7537–7541. doi: 10.1128/AEM.01541-09
Schuhmann, A., Schmid, A. P., Manzer, S., Schulte, J., and Scheiner, R. (2022). Interaction of insecticides and fungicides in bees. Front. Insect Sci. 1:808335. doi: 10.3389/finsc.2021.808335
Shi, J., Liao, C., Wang, Z., Zeng, Z., and Wu, X. (2019). Effects of sublethal acetamiprid doses on the lifespan and memory-related characteristics of honey bee (Apis mellifera) workers. Apidologie 50, 553–563. doi: 10.1007/s13592-019-00669-w
Shi, J., Yang, H., Yu, L., Liao, C., Liu, Y., Jin, M., et al. (2020). Sublethal acetamiprid doses negatively affect the lifespans and foraging behaviors of honey bee (Apis mellifera L.) workers. Sci. Total Environ. 738:139924. doi: 10.1016/j.scitotenv.2020.139924
PLANTAN GmbH (2021). Produktinformation Difcor 250 EC. Available at: https://plantan.de/wp-content/uploads/2019/03/PI_DE_Difcor_250_EC_Maerz2021.pdf (Accessed May 23, 2021).
Silva-Campos, M., Islam, M. T., and Cahill, D. M. (2022). Fungicide control of Gnomoniopsis smithogilvyi, causal agent of chestnut rot in Australia. Australas. Plant Pathol. 51, 483–494. doi: 10.1007/s13313-022-00879-4
Sinpoo, C., Williams, G., and Chantawannakul, P. (2017). Dynamics of fungal communities in corbicular pollen and bee bread. Chiang Mai J. Sci. 44, 1244–1256.
Steffan, S. A., Dharampal, P. S., Diaz-Garcia, L., Currie, C. R., Zalapa, J., and Hittinger, C. T. (2017). Empirical, metagenomic, and computational techniques illuminate the mechanisms by which fungicides compromise bee health. J. Vis. Exp. 54631. doi: 10.3791/54631
Subotic, S., Boddicker, A. M., Nguyen, V. M., Rivers, J., Briles, C. E., and Mosier, A. C. (2019). Honey bee microbiome associated with different hive and sample types over a honey production season. PLoS One 14:e0223834. doi: 10.1371/journal.pone.0223834
Syromyatnikov, M. Y., Isuwa, M. M., Savinkova, O. V., Derevshchikova, M. I., and Popov, V. N. (2020). The effect of pesticides on the microbiome of animals. Agriculture 10:79. doi: 10.3390/agriculture10030079
Thompson, H. M., Fryday, S. L., Harkin, S., and Milner, S. (2014). Potential impacts of synergism in honeybees (Apis mellifera) of exposure to neonicotinoids and sprayed fungicides in crops. Apidologie 45, 545–553. doi: 10.1007/s13592-014-0273-6
Wernecke, A., and Castle, D. (2020). Auswirkungen von Pflanzenschutzmittel-Tankmischungen auf Honigbienen und mögliche physiologische Interaktionen. 154–161 Seiten / Journal für Kulturpflanzen, Bd. 72 Nr. 5 (2020): Themenheft Bienenschutz. doi: 10.5073/JfK.2020.05.05
Wernecke, A., Frommberger, M., Forster, R., and Pistorius, J. (2019). Lethal effects of various tank mixtures including insecticides, fungicides and fertilizers on honey bees under laboratory, semi-field and field conditions. J. Consum. Prot. Food Saf. 14, 239–249. doi: 10.1007/s00003-019-01233-5
Whelan, R. A., Doranalli, K., Rinttilä, T., Vienola, K., Jurgens, G., and Apajalahti, J. (2019). The impact of Bacillus subtilis DSM 32315 on the pathology, performance, and intestinal microbiome of broiler chickens in a necrotic enteritis challenge. Poult. Sci. 98, 3450–3463. doi: 10.3382/ps/pey500
White, T. J., Bruns, T., Lee, S., and Taylor, J. (1994). “Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics” in PCR protocols: a guide to methods and applications. ed. M. A. Innis (San Diego: California Academy Press), 315–322.
Wing, K. D., Sacher, M., Kagaya, Y., Tsurubuchi, Y., Mulderig, L., Connair, M., et al. (2000). Bioactivation and mode of action of the oxadiazine indoxacarb in insects. Crop Prot. 19, 537–545. doi: 10.1016/S0261-2194(00)00070-3
Wu, Y., Zheng, Y., Wang, S., Chen, Y., Tao, J., Chen, Y., et al. (2022). Genetic divergence and functional convergence of gut bacteria between the Eastern honey bee Apis cerana and the Western honey bee Apis mellifera. J. Adv. Res. 37, 19–31. doi: 10.1016/j.jare.2021.08.002
Yang, L.-N., He, M.-H., Ouyang, H.-B., Zhu, W., Pan, Z.-C., Sui, Q.-J., et al. (2019). Cross-resistance of the pathogenic fungus Alternaria alternata to fungicides with different modes of action. BMC Microbiol. 19:205. doi: 10.1186/s12866-019-1574-8
Yoder, J. A., Jajack, A. J., Rosselot, A. E., Smith, T. J., Yerke, M. C., and Sammataro, D. (2013). Fungicide contamination reduces beneficial fungi in bee bread based on an area-wide field study in honey bee, Apis mellifera, colonies. J. Toxicol. Environ. Health A 76, 587–600. doi: 10.1080/15287394.2013.798846
Zheng, H., Powell, J. E., Steele, M. I., Dietrich, C., and Moran, N. A. (2017). Honeybee gut microbiota promotes host weight gain via bacterial metabolism and hormonal signaling. Proc. Natl. Acad. Sci. U. S. A. 114, 4775–4780. doi: 10.1073/pnas.1701819114
Keywords: pesticides, bee, cuticular microbiome, fungi, bacteria, plant protection products, neonicotinoids
Citation: Reiß F, Schuhmann A, Sohl L, Thamm M, Scheiner R and Noll M (2023) Fungicides and insecticides can alter the microbial community on the cuticle of honey bees. Front. Microbiol. 14:1271498. doi: 10.3389/fmicb.2023.1271498
Edited by:
Ravindra Soni, Indira Gandhi Krishi Vishwavidyalaya, IndiaReviewed by:
Sanjay Kumar Rohaun, University of Illinois at Urbana-Champaign, United StatesHiren Patel, Anand Agricultural University, India
Shubha Banerjee, Indira Gandhi Krishi Vishwavidyalaya, India
Copyright © 2023 Reiß, Schuhmann, Sohl, Thamm, Scheiner and Noll. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Matthias Noll, bWF0dGhpYXMubm9sbEBocy1jb2J1cmcuZGU=
 Fabienne Reiß
Fabienne Reiß Antonia Schuhmann
Antonia Schuhmann Leon Sohl
Leon Sohl Markus Thamm
Markus Thamm Ricarda Scheiner
Ricarda Scheiner Matthias Noll
Matthias Noll