
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
REVIEW article
Front. Microbiol. , 10 October 2023
Sec. Microbe and Virus Interactions with Plants
Volume 14 - 2023 | https://doi.org/10.3389/fmicb.2023.1271034
 Anamika Singh1
Anamika Singh1 Virendra Kumar Yadav2*
Virendra Kumar Yadav2* Hemant Gautam3
Hemant Gautam3 Lokendra Rathod4
Lokendra Rathod4 Rajendra Singh Chundawat1*
Rajendra Singh Chundawat1* Gulab Singh1
Gulab Singh1 Rakesh Kumar Verma1
Rakesh Kumar Verma1 Dipak Kumar Sahoo5*
Dipak Kumar Sahoo5* Ashish Patel2*
Ashish Patel2*A wide variety of bacteria are present in soil but in rhizospheric area, the majority of microbes helps plant in defending diseases and facilitate nutrient uptake. These microorganisms are supported by plants and they are known as plant growth-promoting rhizobacteria (PGPR). The PGPRs have the potential to replace chemical fertilizers in a way that is more advantageous for the environment. Fluoride (F) is one of the highly escalating, naturally present contaminants that can be hazardous for PGPRs because of its antibacterial capacity. The interactions of F with different bacterial species in groundwater systems are still not well understood. However, the interaction of PGPR with plants in the rhizosphere region reduces the detrimental effects of pollutants and increases plants’ ability to endure abiotic stress. Many studies reveal that PGPRs have developed F defense mechanisms, which include efflux pumps, Intracellular sequestration, enzyme modifications, enhanced DNA repair mechanism, detoxification enzymes, ion transporter/antiporters, F riboswitches, and genetic mutations. These resistance characteristics are frequently discovered by isolating PGPRs from high F-contaminated areas or by exposing cells to fluoride in laboratory conditions. Numerous studies have identified F-resistant microorganisms that possess additional F transporters and duplicates of the well-known targets of F. Plants are prone to F accumulation despite the soil’s low F content, which may negatively affect their growth and development. PGPRs can be used as efficient F bioremediators for the soil environment. Environmental biotechnology focuses on creating genetically modified rhizobacteria that can degrade F contaminants over time. The present review focuses on a thorough systemic analysis of contemporary biotechnological techniques, such as gene editing and manipulation methods, for improving plant-microbe interactions for F remediation and suggests the importance of PGPRs in improving soil health and reducing the detrimental effects of F toxicity. The most recent developments in the realm of microbial assistance in the treatment of F-contaminated environments are also highlighted.
Numerous types of bacteria live in the rhizosphere, most of which can protects plants from pathogenic invasions and make it easier for plants to absorb nutrients from the soil (Kour et al., 2019). Various studies have shown that the host plant considerably affects the bacterial community in its rhizosphere. Phylogenetically diverse microorganisms, such as viruses, nematodes, fungi, protists, bacteria, and archaea, can all be found in the rhizosphere (Ling et al., 2022). Rhizospheres of plants generally contain a wide range of soil microorganisms (Yang et al., 2022; Nie et al., 2023). Some of these are helpful to the plants since they encourage growth. These microorganisms are referred to as plant growth-promoting rhizobacteria (PGPR) and have the potential to replace chemical fertilizers (Bhattacharyya et al., 2020).
The majority of the PGPRs belong to genera like Pantoae, Alcaligenes, Arthrobacter, Pseudomonas, Rhodococcus, Serratia, Azoarcus, Azospirillum, Azotobacter, Herbaspirillum, Stenotrophomonas, Lactobacillus, Paenobacillus, Beijerinckia, Burkholderia, Derxia, and Zoogloea are some of the bacteria that have been identified (Vega-Celedón et al., 2021). Although tight interactions between soil, plants, and microbes are a complex phenomenon that occurs in natural ecosystems, it is challenging to measure how they affect plant growth, health, and production (Uruén et al., 2020; Nadarajah and Abdul Rahman, 2021).
Interactions of plants with PGPR in the rhizosphere area help to maintain soil fertility and plant health. A number of pollutants in the rhizosphere, have a detrimental effect on soil fertility and plant productivity (Vecchiato et al., 2021). In the rhizospheric region of soil, PGPR interacts with plants and protect them from the harmful effect of various pollutants by inducing their ability of abiotic stress tolerance (Khatoon et al., 2020).
Two studies shows that Bacillus sp. are suitable for abiotic environment, Bacillus subtilis ER-08 (BST) as a stress-resilient, multifunctional plant growth-promoting rhizobacterial isolate and this strain has also been found to increase the development of fenugreek (T. foenum-graecumL.) under salt and drought stress and solubilizing phosphate and producing ACC deaminase, siderophore, and IAA helps Bacillus sp. promote tomato growth in both non-stressed and salt-stressed environment. Increased amounts of osmoregulatory proline and soluble sugar, as well as ROS scavenging enzymes, were also connected to tomato seedlings’ ability to withstand salt stress (Patani et al., 2023; Patel et al., 2023a,b).
Several studies demonstrate the effect of the PGPR consortiums more efficient than single rhizobacteria for enhancing abiotic stress tolerance in plants (Singh et al., 2023; Wang et al., 2023; Patel et al., 2023a,b).
Among all the toxins, F is one of the toxic elements that cause detrimental effects to plants and other microorganisms. High F levels are a major threat due to their toxicological and geo-environmental concerns. A potent rhizobacterial species must be quickly developed in order to decrease the dangerous consequences of F accumulation in plants (Aggarwal and Bhushan, 2019; Chatterjee et al., 2020; Han et al., 2021).
Duffin et al. (2022) reported that F inhibits microbial development and organic matter decomposition, especially in quantities far higher than its natural values in soil.
F concentrations and the composition of the bacterial population in shallow groundwater were found to be correlated in the Qiji region of Northern China (Zhang et al., 2019). The amounts of Total Organic Carbon (TOC) and F in groundwater have a substantial impact on the bacterial communities. The result illuminates the biogeochemical processes of F and other elements in groundwater and suggests that F concentration should be considered while examining microbial response in an F-rich environment (Kashif et al., 2023).
Many organisms including PGPR have developed fluoride defense systems (Weerasooriyagedara et al., 2020). F riboswitch is one of the defense systems in PGPRs. In a study, Bacillus cereus F riboswitch was used in base-pair opening dynamics research with and without ligands to better understand the molecular mechanism of gene regulation in F riboswitch. The finding suggests that the F riboswitch controls gene expression via a two-step mechanism that includes conformational changes which is caused by Mg2+ (Lee H.-J. et al., 2021; Lee J. et al., 2021).
These resistance characteristics are typically discovered by separating organisms from F rich environment or in a laboratory setting, cells were exposed to F. Several studies have revealed F-resistant microorganisms with increased F transporters and copies of well-known targets for F (Faraj et al., 2019; Zhao et al., 2019; McIlwain et al., 2021).
Roshni, 2022 reported that F inhibited the activity of protease, alkaline phosphatase, and dehydrogenase enzymes in microorganisms.
PGPR from the genera Azotobacter, Flavobacterium, Micrococcus, Bacillus, Pseudomonas, Acinetobacter, Streptomyces, and Streptococcus have been found in the rhizosphere (Cochard et al., 2022). Streptococcus sp., Pseudomonas sp., Bacillus sp., and Acinetobacter sp. are a few prokaryotes that may survive in environments with high F compound concentrations. This is because of an old system made up of F-specific riboswitches and frequently linked proteins like CrcB (Li et al., 2018; Speed et al., 2018; Lee H.-J. et al., 2021; Lee J. et al., 2021).
The FEX membrane transport protein functions as the primary F defense mechanism in plants. Both prokaryotes (Fluoride channel; Fluc) and fungi (Fluoride Exporter; FEX) have F-specific ion transporters that effectively export fluoride to the extracellular environment. All around the plant kingdom, FEX homologs have been recognized. FEX is conserved in both yeast and plants (Ma et al., 2019; Tausta et al., 2021).
Zhu et al., 2019 reported an increase in germination rate in several lines when fluoride was used. Their finding indicates the optimal concentration or localization of FEX within the plant.
Although each plant has a different level of F tolerance by which they can escape its threatening progression.
According to Pelc et al. (2020) the Dalewar, Arkadia, and Tobak winter wheat cultivars were subjected to sodium fluoride (NaF) treatment, and the results revealed that NaF decreased germination, root growth, and the activity of antioxidant enzymes. These effects have been made worse at higher concentrations of NaF (Pelc et al., 2020; Li et al., 2021). For the last 6 years researchers have been working on the identification of F-tolerant PGPR and their diverse mechanism of tolerance, so that such PGPR can be used as a natural cure for F toxicity and protect crop health from the damaging effects of F (Liao et al., 2018; Mukherjee and Halder, 2018; Sahoo and Goli 2018; Ong et al., 2020; Pelc et al., 2020; Shanker et al., 2020; Chellaiah et al., 2021; Lee H.-J. et al., 2021; Lee J. et al., 2021; Mothe et al., 2021; Mushtaq et al., 2021).
In the current review, the authors have emphasized the current trends of in-depth systemic analysis of contemporary biotechnological approaches, for improving plant-microbe interactions for F degradation, such as gene manipulation and editing approaches, calling attention to the most recent advancements in the field of microbial-endorsed treatment of F contaminated ecosystems. Furthermore, here authors emphasized that with the help of additional investigation and knowledge of the molecular mechanisms behind F bioremediation, more capable strains will be identified for F-contaminated areas.
PGPR activity in the rhizosphere is predominantly caused by two different types of processes. Figure 1 depicts the direct and indirect categories. Both methods of PGPR activity are critical to long-term agricultural crop productivity (Patil et al., 2019; Aioub et al., 2022).
Constantia and Ferniah (2020) studied that, by releasing various beneficial compounds like phosphates, silicon, potassium, and zinc, as well as PGPR encourages plant growth by absorbing biologically fixed nitrogen, chelating iron, and other micronutrients, and increasing the amount of geospheric oxygen that is available. This stimulates the formation of aeriform biomass, growth of the roots, and lengthening of the stem. PGPR produces ethylene, auxins, cytokinin, indoleacetic acid, gibberellins, and auxins phytohormone (Del Orozco-Mosqueda et al., 2023).
Tiwari et al. (2018), reported that synthesis of the enzyme 1-aminocyclopropane 1-carboxylate deaminase (ACC), increases root density and length while lowering ethylene levels in crop roots. PGPR indirectly alters the rhizospheric environment, creates systemic resistance, and boosts the plant’s inherent resilience (Adrees et al., 2019).
PGPR released substances like siderophore, pigments, antibiotics, organic acids, water-soluble vitamins, and various volatile organic compounds like monoterpene alcohols and these substances activate the plant’s defense mechanism against various pathogenic microorganisms and promote the synthesis of physical and chemical barriers against abiotic stress (Chandran et al., 2021).
PGPR interferes with the quorum sensing signal and prevents harmful bacteria from developing biofilms around plant roots and in turn plants help PGPR to become more competitive in niche colonization (Hartmann, 2020). Additionally, contaminated soils can be cleaned up with PGPR as investigated (Vaishnav et al., 2022).
Phale (2018) describes the multifunctionality of PGPR and its high demand in agroforestry management. Furthermore, according to Lindemann (2019), PGPRs are significant ecosystem service providers because of their interaction with numerous microbial populations and multifunctional activities. Sylia et al. (2022) explore how the intricately interwoven PGPRs network influences the vegetational biome and soil microfauna by controlling the transmission and circulation of energy and resources across a whole ecosystem (Sylia et al., 2022).
Since all rhizobacteria have the capacity to fix atmospheric nitrogen, they are all referred to as rhizobacteria. Each of the direct and indirect mechanisms may be present or absent in all rhizobacteria (Singh et al., 2023) despite the fact that both the direct and indirect mechanisms (Figure 1) of PGPR benefit plant health (Mahmud et al., 2020).
PGPR is the finest nitrogen source for long-term crop production. Proteobacteria, Alphaproteobacteria, Rhodospirillales, Acetobacteraceae, Actinobacteria, Micrococcales, Microbacteraceae, and Roseomonas (Zhang et al., 2020), Burkholderiatropica, Achromobacterinsolitus, and Acetobacterdiazotrophicusare some nitrogen-fixing microorganisms (Singh et al., 2020).
F contamination of the environment, primarily caused by geological processes but occasionally also resulting from anthropogenic activity, F is absorbed by plants from the contaminated soil and water, which leads to abiotic stress and interference with vital physiological and biochemical processes (Makete et al., 2022).
F in soil-water systems is primarily caused by volcanic eruption, weathering, and rock leaching (Yadav et al., 2021). F has been identified as a mobile component in soil. Additionally, the relative movement showed that soil rather than rocks played a larger role in the release of fluoride into groundwater. Despite being primarily emitted into the atmosphere, water contamination is a significant issue. F contamination in water is due to its excess atmospheric emissions, dumping of contaminated wastewater in water bodies, and chemical weathering are some ways that brought F to the soil surface of plants (Singh et al., 2018; Kabir et al., 2019; Yadav et al., 2019).
A common phytotoxic plant pollutant is fluoride. Fluoride is taken by plants from soil and water through their roots and leaves (Gadi et al., 2020). There are numerous influences on fluoride accumulation in plants, including soil fluoride content, and soil and plant species characteristics (Makete et al., 2022). Root biochemistry, morphology, and physiological behavior are all affected by fluoride buildup in the soil. F toxicity in plants depends on the amount, frequency, and length of exposure as well as the genotype of the plant (Sharma and Kaur, 2018). On the other hand, F exposure, even in small doses, has a negative effect on crop species and other plants’ growth and productivity (Sahariya et al., 2022).
F accumulation in plants is a severe concern, impacting their growth and development (Sahariya et al., 2021). F accumulation in plants is affected by a number of factors, including soil F content, plant species, and soil qualities (Banerjee et al., 2020).
The ability of the tea plant to hyper-accumulate F in its leaves suggests that F addition can enhance pectin content and dimethyl esterification, resulting in greater absorption of metal cations and chelation of F in the cell wall via metal ion action (Luo et al., 2021).
Stomata or the root system of plants allow them to take up F from the air or soil. Through the secondary roots’ cortex and epidermis, F ions are transported directly into the xylem and phloem (Figure 2; Kumar et al., 2021). Following that, stomata on the plants allow F to diffuse out of the plants (Sharma and Kaur, 2018). Many agricultural plants are inhibited in their growth and metabolism by excess F, while some have an inbuilt capacity for F tolerance (Gadi et al., 2020).
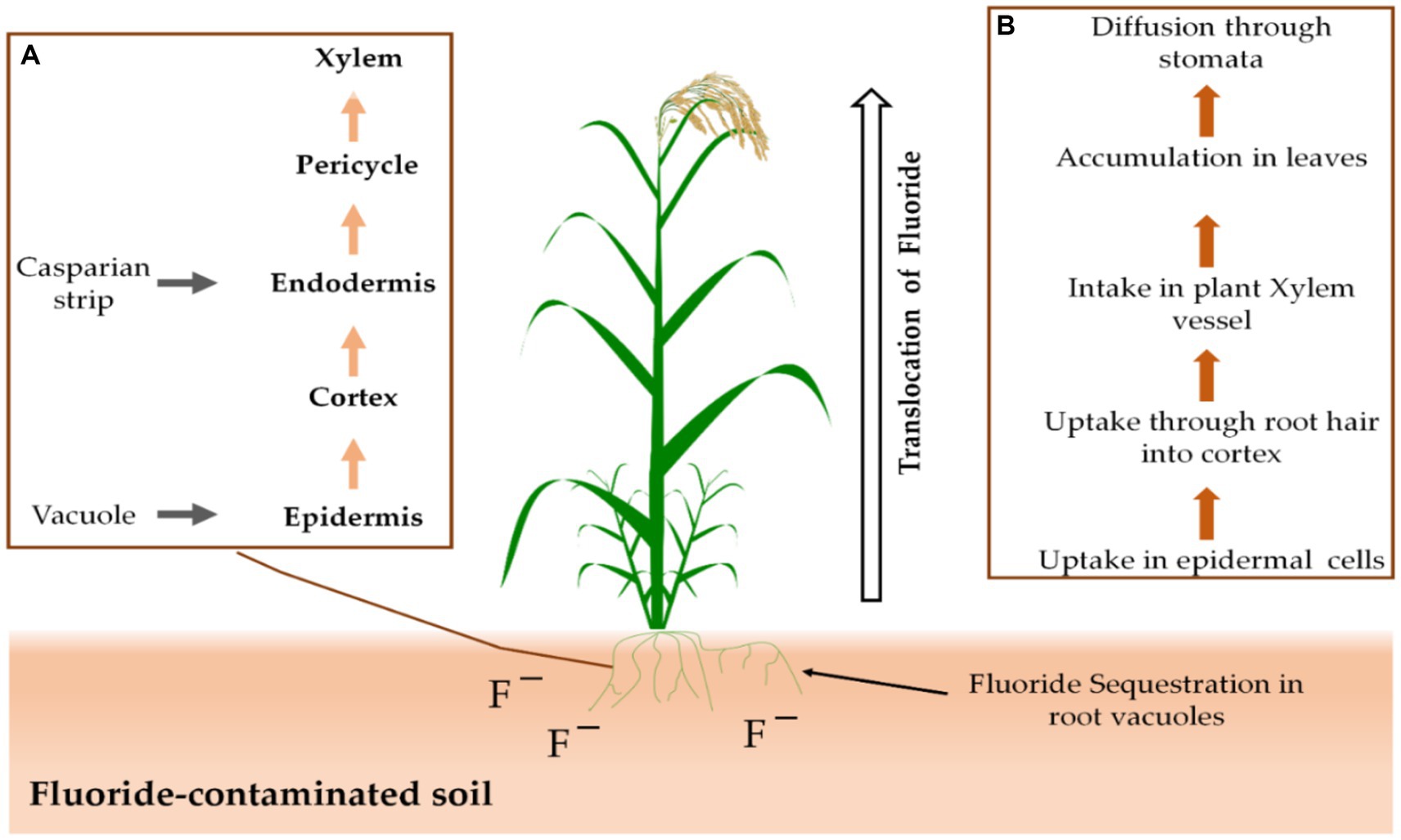
Figure 2. There are two distinct processes at play: The first (A) is the uptake mechanism of F that occurs in the roots, while the second (B) involves the overall movement of F from the roots up to the shoots.
Environmental F levels, particularly in surface and subsurface irrigation waters, can directly affect the germination of seeds and subsequent plant growth (Almeida Rodrigues et al., 2022). F toxicity leads to a decrease in root protrusion as well as impairment in the mobilization of carbohydrates that reduce healthy growth and embryonic axis development (Chatterjee et al., 2020). As a result, F may build up in vegetative tissues and edible cereal grains, posing a direct hazard to the food chain (Kadiresan and Khanal, 2018).
According to Singh and Roychoudhury (2020), seedlings experienced severe oxidative stress after being exposed to two different F concentrations, 25 and 50 mg L−1 NaF, which led to an increase in F accumulation and growth inhibition, as well as decreases in tissue biomass, the size of the roots and shoots, the amount of chlorophyll, the amount of H2O2 present, and the amount of lipid peroxidation (malondialdehyde content and lipoxygenase activity), and protein carbonylation.
Pelc et al. (2020) studied the effect of NaF on three winter wheat cultivars (Dalewar, Arkadia, and Tobak), and their results showed that NaF reduced germination, root development, and catalase (CAT) activity. The increase in NaF levels has exacerbated these consequences. Sodium fluoride inhibited catalase (CAT) activity significantly at all doses (Pelc et al., 2020).
Prolonged exposure to fluoride at a concentration of 40 mg/L, the photosynthetic pigments and antioxidant enzymes such as glutathione (GSH), ascorbic acid (AsA), and superoxide dismutase (SOD) were sharply decreased in Hydrilla verticillate (Gao et al., 2018). Furthermore, when the concentration of F increased, the observed effect became stronger.
According to Sharma and Kaur (2019) at high concentrations of 50 ppm for 24 and 72 h, maximum Glutathione reductase (GR) activity was found. However, for exposure times of 120 and 168 h, activity was decreased at 25 and 20 ppm concentrations of F. When under stress, H2O2 is scavenged via Ascorbate peroxidase (APOX) activity which is considerably increased at high concentrations of fluoride in Spirodela polyrhiza (Sharma and Kaur, 2019).
Although some plant species are naturally F-tolerant, excessive F impairs the growth and metabolism of many crop species (Figure 3; Gadi et al., 2020).
Plants can reduce the stress caused by metabolic disturbances to some extent by activating their defense mechanism. Plants defense system becomes hyperactive to reduce the harmful effects of Reactive Oxygen Species (ROS) (Caverzan et al., 2019; Huang et al., 2019; Hasanuzzaman et al., 2020).
The main F defense mechanism in plants is the action of the FEX (Fluoride Exporter) membrane transport protein. Plant FEX plays a conserved role in F tolerance. A CRISPR/Cas9-generated mutation in Arabidopsis thaliana, FEX makes the plant sensitive to low F concentrations (100 μM) at all stages of development. Pollen is particularly vulnerable and unable to develop even at extremely low F levels in the growing media (Tausta et al., 2021). Sequence alignment was used to find FEX homologous genes in nine plants (Banerjee and Roychoudhury, 2019).
Zhu et al. (2019) also studied the functions of the FEX protein in the Arabidopsis and tea plants and reported that the tea plant’s CsFEX fluoride export gene participates in F detoxification by heterologous expression. The homologous gene CsFEX, which is largely found in the plasma membrane, can be activated by exogenous fluorine treatment (Zhu et al., 2019). However, it is still unknown how fluoride outflow and buildup work at the molecular level, as well as whether regulatory systems are involved.
According to Do et al. (2021) ABCs (ATP-binding cassettes) transporters control cellular processes by binding ATP and hydrolyzing it to create energy for transport and other physiological and biochemical processes. Eight subfamilies make up the ABC transporter gene family, and the proteins they encode can move a wide range of substrates. ABCs bind ATP and hydrolyze it to provide energy that powers transport and controls additional cellular processes. There are eight subfamilies in the ABC transporter family of genes, and the proteins they code for can transport a wide variety of substrates. As a result, they control almost all physiological and biochemical processes in plants (Do et al., 2021).
A study showed that F increases the concentration of ABC transport proteins in tea trees and by RNA-sequence analysis, the CsABCB9 gene is explored whose expression is boosted by F treatment (Luo et al., 2022).
A study shows that exogenous salicylic acid (SA) has a critical role in mitigating the toxic effects of F in rice seedlings and demonstrates its significant role as a protective molecule against fluoride stress when supplied exogenously (Singh and Roychoudhury, 2021; Lu et al., 2022).
On the other hand, F circulation in the ecosystem of tea plantations is favorable, with more F being added than removed. The main sources of F absorption by tea plant root through active transmembrane transport and anion channels are magnesium chloride (MgCl2) extractable F and water extractable F in plantation soil. The majority of F is quickly transferred as F/F-Al complexes to the leaf cell walls and vacuole across the xylem. The results suggest that tea plants detoxify F and aluminum (Al) simultaneously through cell wall accumulation, vacuole compartmentalization, and F-Al complexes, which may be a mechanism of F tolerance that enables tea to withstand higher F concentrations than most plants (Peng et al., 2020; Luo et al., 2022).
According to Luo et al. (2022), tea plants (Camellia sinensis) collect a disproportionate quantity of F in their leaves when compared to other plants. However, it is uncertain how these plants tolerate F. A chloroplast F efflux gene (CsABCB9) was identified through transcriptome analysis, cloned from Camellia sinensis, and its function in F detoxication was proven in Escherichia coli and Arabidopsis thaliana. Tea leaves express the CsABCB9 efflux gene after F treatment.
Singh and Jajoo (2021) investigate the negative effects of F on photosynthesis, specifically in maize plants (Zea mays L.) Photosystem I (PSI) and Photosystem II (PSII) serve as the principal locations of energy conversion that convert light energy into chemical energy (Chadha et al., 2021). F has a negative effect on the activity of both photosystems, which serve as internal environmental monitors. In contrast to PSI, Y (I), the quantum yield of PSII, Y (I), was reduced at all NaF concentrations. The activation of cyclic electron flow (CEF) after F treatment was accompanied by the suppression of linear electron flow (LEF). PSI resistance to F poisoning appears to need LEF inhibition and CEF induction (Singh and Jajoo, 2021).
F removal is required before using fluoridated water because the content of F exceeds the allowable limits. F tolerance mechanisms in microorganisms vary depending on the specific organism and its adaptation to F-rich environments (Yan et al., 2022; Li et al., 2023). Many organisms have developed unique defense mechanisms for dealing with high F concentrations, for example, the synthesis of proteins capable of eliminating F from cells. However, these F transporters have not been found in all microorganisms and F transporters may vary in their tolerance capabilities between species, individuals, and even tissue types. This shows that the F tolerance capacity of PGPRs is also influenced by other factors.
The following sections describe various F tolerance mechanisms of PGPR including; Efflux pumps, Intracellular sequestration, enzyme modifications, enhanced DNA repair mechanism, detoxification enzymes, ion transporter/ antiporters, fluoride riboswitches, and genetic mutations.
According to the study, maintaining high bacterial biomass is critical in bacterial-based bioremediation for boosting bacterial species capacity and survival in the F environment, this is done by immobilizing bacterial cells (Mukherjee et al., 2018).
The most electronegative element is fluoride, which penetrates bacterial cells through diffusion as hydrogen fluoride, dissociating into H+ and F ions (Katiyar et al., 2020). These ions disrupt fluoride-ATPases and glycolysis enzymes. Microorganisms that have F tolerance capacity are thought to have altered enzymes (Waugh, 2019).
Numerous microorganisms have the potential to bioabsorb, biotransform, and bioaccumulate ligands like biosurfactants or siderophores, which impact the availability and solubility of pollutants in bacterial cells (Banat and Thavasi, 2019).
To avoid F toxicity, PGPR must be carried out in ionospheres, mineralization, metal intake, buildup, sorption, reduction, enzymatic oxidation extracellular precipitation, and xenobiotic outflow (Makete et al., 2022).
The bacterium Bacillus flexus (PN4) may be a promising strain for defluorinating drinking water, and it may offer a perfect opportunity to develop a novel bioremediation method, according to the findings of a study by Sakthi Thesai et al. (2018). The bacterium might be crucial in the process of removing F from aqueous medium. As a result, there might be a viable remediation method for F-containing water after fluoride accumulation and bacterial eradication.
According to Sakthi Thesai et al. (2018) F anion exporters are described by Fluc family F-specific ion channels. F hypersensitivity occurred in the bacteria due to a paucity of potential proteins. Their outcome was that the microbes developed F/H+ antiporters of CLC anion transporters, resulting in the emergence of the “Fluc” family of F-specific ion channels. Anionic F was able to enter the bacterial cell through the pore when stimulus molecules attached and stimulated the protein channel (fluc), causing the F ion to export into the bacterial cell (Figure 4). Rhizobacteria can develop fluoride resistance by changing the promoter of fluoride antiporters (Chiariello et al., 2020).
Li et al. (2022) studied F tolerance in Streptococcus mutans induced by high F concentrations via point mutations. They found three F-resistant S. mutans strains and termed SRR13846724, SRR13846723, and SRR13846722 for the FR300, FR600, and FR1000. Three types of variant studies were performed on the scaffolds of all strains using whole-genome sequencing: structural variations, insertion–deletion variant (InDel), and calling SNP. There was no evidence of chromosomal rearrangement in the resistant bacteria, despite their great similarity. These findings shed new light on the microbial F tolerance mechanism, F0F1-ATPase is necessary for antagonizing F inhibition and increasing F resistance in S. mutans (Li et al., 2022).
Pal et al. (2022) identified soil microorganisms and revealed that these microorganisms are very similar to Bacillus megaterium and resistant to solutions containing 35% (w/v) sodium chloride and 1,500 mg/L Fluoride. Bacillus megaterium (JF273850), an isolated microbe, may be useful for F protection.
A study suggests that soil bacterium Pseudomonas putida KT2440 reacts to F anion provided by Calero et al. (2022). The study uses a broad lens to illustrate how F affects the physiological response of the bacterium and the microorganism.
A study investigates the alteration in genes of Streptococcus mutans, which give rise to F-resistant strains, and also explains how genetic mutations cause pleiotropic consequences in the physiology of S. mutans. In the F-resistant genome, they discovered six mutations through sequencing analysis (Lee H.-J. et al., 2021; Lee J. et al., 2021). Fluoride stress causes a cumulative effect of genetic changes that may rewire intricate machinery to balance bacterial resistance and biological fitness in the best possible manner (Liao et al., 2018).
In order to determine which genetic mutations primarily cause the resistant phenotype and decreased fitness in S. mutans, complementation for each mutation should be carried out.
The pH of the aqueous solution and the functional groups on the microbe cell wall determine the degree of biosorption (El-Naggar et al., 2018). Increasing the pH concentration resulted in decreased fluoride adsorption. This might be because, at higher pH concentrations the amount of OH− is greatest and F, which is extremely electronegative, repels the negatively charged OH− (Nabbou et al., 2019). However, due to the low pH level, there may be more interaction between the negatively charged F and positively charged H+, which could result in the formation of hydrofluoric acid (HF) (Abraham and Acree, 2019).
A study suggests that F bioremediation capacity by Bacillus sp. subjected to varied pH. The effectiveness of bioremediation has risen from 40 to 55% at pH 7 to 60–78% at pH 6. When the pH is changed, the absorption efficiency increases from 200 ppm to 286 ppm. Thus, pH has an important function in minimizing environmental pollution and emphasizes the relevance of bacteria in an eco-friendly way (Sahoo et al., 2019).
Shanker et al. (2020) investigated the removal of F by Acinetobacter sp. and the findings gave a clear insight into the factors influencing bacterial growth and defluorination. The physical and chemical analysis of Acinetobacter sp. showed that after 10 h of incubation at 7.5 pH 57.3% of the F from the synthetic aqueous solutions (Shanker et al., 2020).
According to Mothe et al. (2021) from groundwater samples taken in Narketpally, a heavily fluoridated location in the Nalgonda district, three F-resistant bacteria (MB1, F, and G) with high F resistance (up to 500 mgL−1 NaF) were identified. After 8-day incubation, dextrose (10 g) was used as the carbon source per 100 ml of medium, and the concentration of F was 20 mgL−1 at 30°C and pH 7 The F and G strains showed the highest F degradation of 57, and 44%, respectively, while the MB1 strain showed maximum F elimination of 68%.
Several single nucleotide polymorphism (SNPs) were found in two different F-resistant Streptococcus mutans strains (Liao et al., 2018). In two different strains of the F-resistant S. mutans, they discovered overlapping chromosomal regions with SNPs by comparing genome sequences. In two intergenic areas (mutp and glpfp) and one pathway (glycolysis), validation discovered changes in gene expression and protein activities. New potential loci for inhibitor resistance are suggested by their findings. More investigation is required into the function and importance of these loci in the regulation of genes in the presence or absence of fluoride. These SNPs have the capacity to modify protein functions and gene expression.
In another study on the establishment of a collection containing mutated genes extracted from S. mutans, the identification of fluoride-related transcriptional regulator (FrtR) as a fundamental transcription factor that plays a significant role in the regulation of F. After analyzing the frtR mutant using RNA-sequencing, it was found that the fluoride related permease gene (frtP) was one of the downstream genes that are directly regulated by FrtR (Lu et al., 2020). The mechanism behind intrinsic F tolerance has yet to be fully understood.
In a recent study conducted by Su et al. (2020), to determine the effectiveness of the calcium-precipitating strain Acinetobacter sp. H12in eliminating F. The study showed that H12 was able to successfully decrease 85.24% of F at a consistent rate of 0.036 mg·L−1·h−1. The process was thoroughly analyzed using advanced imaging techniques and spectroscopy, which revealed that the bacterium acted as nucleation sites in forming biological crystal seeds. These seeds then proceeded to adsorb F. These findings demonstrate the significant potential of H12 in addressing the issue of F contamination (Su et al., 2020).
A study that delves into the properties and mechanisms of fluoride removal, highlights the use of biomineralizing bacteria, particularly Acinetobacter sp. H12, which has proven to be effective in eliminating F (Wu et al., 2021). Additionally, it was found that the carbon-to-nitrogen ratios affected the fluoride removal performance of Acinetobacter H12 in a quartz sand-filled biofilm reactor (Ali et al., 2021).
These findings have significant implications in advancing the treatment of F toxicity.
According to Chellaiah et al., 2021, eight F-resistant bacteria were isolated from the water samples of the South Indian region of Dindigul. This is the first study to identify fluoride-resistant bacteria, specifically in the Nathamtaluk of southern Tamil Nadu. F-resistant strains identified as Aeromonas caviae strains 31, 32, and 34, Enterobacter cloacae strain 3, E. hormaechei strain 14, Enterobacter sp. strain 21, E. hormaechei strain 22, and E. coli strain S2-9. The fluoride resistance of the selected isolates ranged between 200 and 300 mM NaF. F tolerance has also been associated with antibiotic resistance to Chloramphenicol, cefotaxime, doxycycline hydrochloride, fosfomycin, minocycline, nalidixic acid, nitrofurantoin, and trimethoprim. The ‘CrcB’ gene is revealed by them which is linked to bacterial F resistance. Furthermore, these identified microorganisms will be helpful for F bioremediation experiments (Chellaiah et al., 2021).
Twenty-two F-resistant bacteria in LB agar plates were resistant to 200 mM NaF and showed hemolytic activity on blood agar plates. Gene-specific PCR investigation verified the presence of virulence and biofilm-forming genes in Pseudomonas sp. Furthermore, a disc diffusion approach revealed that haemolytic Pseudomonas was resistant to various medications. By using gene-specific primers, it is concluded that crcB domain is responsible for F-resistant in Pseudomonas sp. (Edward Raja et al., 2022).
Defluoridation procedures could eliminate the difficulties associated with F contamination. Several technologies (reverse osmosis, nanofiltration, precipitation, adsorption, and so on) have been used to defluoridate water. Adsorption techniques are commonly used to remove excessive F from water Tea ash, Banana peel dust, coconut shell dust, Bsh scale dust (hot and semi-arid climate). However, due to significant capital and operating costs, these technologies could not be used in practice. Conventional F cleanup procedures are time-consuming, labor-intensive, and expensive, as a result, they are uneconomical for long-term cultivation. The solution is to reduce the use of modern, affordable, cost-effective, and economical methods to reduce this environmental pollution. In biological processes like bioremediation, the employment of fungi, bacteria, algae, and higher plants has the ability to drastically reduce F pollution, rehabilitate contaminated soil, and restore vegetation. The effectiveness of local natural agents in sustainable agriculture can be increased, improved, and chosen over harmful chemicals. Furthermore, these traditional procedure takes a lot of energy and chemical ingredients. Bioremediation approaches could help to solve these issues. The study of how PGPRs respond to toxicants is a crucial aspect of environmental protection and preservation. In this regard, the emergence of F resistance in PGPRs could potentially serve as a novel defense mechanism. Additionally, plasmid DNA may facilitate F tolerance in PGPRs. Furthermore, genetically engineered PGPRs could prove to be valuable tools for bioremediation efforts. However, it is important to evaluate the potential risks associated with these approaches and take appropriate measures to mitigate them.
The resilience of microorganisms in harsh environmental conditions is truly remarkable. However, the proliferation of natural F levels in soil can have devastating effects on plant health. Fortunately, recent research has indicated that PGPRs have the ability to endure high concentrations of fluoride, offering a promising solution for bioremediation. They pose protective mechanisms to overcome the high concentration of F along with increasing soil health and plant productivity, therefore they can be a promising solution for F bioremediation. Utilizing genetic engineering to enhance PGPR’s resistance against F and other harmful substances in soil represents a highly viable solution for tackling environmental issues. This approach has tremendous potential for success and warrants further exploration.
AS: Formal analysis, Investigation, Methodology, Writing – original draft, Writing – review & editing. VY: Conceptualization, Data curation, Investigation, Writing – original draft, Writing – review & editing. HG: Conceptualization, Data curation, Validation, Visualization, Writing – review & editing. LR: Conceptualization, Data curation, Project administration, Software, Writing – review & editing. RC: Conceptualization, Data curation, Formal analysis, Supervision, Writing – review & editing. GS: Conceptualization, Investigation, Validation, Visualization, Writing – review & editing. RV: Formal analysis, Project administration, Resources, Validation, Writing – review & editing. DS: Conceptualization, Funding acquisition, Methodology, Visualization, Writing – review & editing. AP: Conceptualization, Investigation, Visualization, Writing – original draft, Writing – review & editing.
The author(s) declare that no financial support was received for the research, authorship, and/or publication of this article.
The authors are thankful to the Department of Biosciences; Mody University of Science and Technology, Lakshmangarh Sikar, Rajasthan (SM/2019-20/003, SM/2020-21/008 and SM/2022-23/008) for providing financial support. The authors are also thankful to the Department of Life Sciences, Hemchandracharya North Gujarat University, Patan, Gujarat, for their technical support and providing the facilities.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Abraham, M. H., and Acree, W. E. (2019). Descriptors for the hydrogen halides, their solution properties and hydrogen- bonding acidity and basicity: comparison of the latter with gas phase data. J. Mol. Liq. 275, 667–673. doi: 10.1016/j.molliq.2018.11.110
Adrees, H., Haider, M., Anjum, T., and Akram, W. (2019). Inducing systemic resistance in cotton plants against charcoal root rot pathogen using indigenous rhizospheric bacterial strains and chemical elicitors. Crop Prot. 115, 75–83. doi: 10.1016/j.cropro.2018.09.011
Aggarwal, A., and Bhushan, P. (2019). Fluoride toxicity: a review article on adverse effects of fluoride. Santosh Univ. J. Health Sci. 5, 3–4. doi: 10.18231/j.sujhs.2019.002
Aioub, A. A., Elesawy, A. E., and Ammar, E. E. (2022). Plant growth promoting rhizobacteria (PGPR) and their role in plant-parasitic nematodes control: a fresh look at an old issue. J. Plant Dis Prot 129, 1305–1321. doi: 10.1007/s41348-022-00642-3
Ali, A., Wu, Z., Li, M., and Su, J. (2021). Carbon to nitrogen ratios influence the removal performance of calcium, fluoride, and nitrate by Acinetobacter H12 in a quartz sand-filled biofilm reactor. Bioresour. Technol. 333:125154. doi: 10.1016/j.biortech.2021.125154
Almeida Rodrigues, A., Almeida Rodrigues, D., de Fátima Sales, J., Carvalho Vasconcelos Filho, S., Carlos Costa, A., Lino Rodrigues, C., et al. (2022). Morphoanatomical, physiological, and biochemical indicators in Lactuca sativa L Germination and growth in response to fluoride. Plants 11:3406. doi: 10.3390/plants11233406
Banat, I. M., and Thavasi, R. (eds.) (2019). Microbial biosurfactants and their environmental and industrial applications. 1st Edn. CRC Press.
Banerjee, A., and Roychoudhury, A. (2019). Fluorine: a biohazardous agent for plants and phytoremediation strategies for its removal from the environment. Biol. Plant. 63, 104–112. doi: 10.32615/bp.2019.013
Banerjee, A., Samanta, S., and Roychoudhury, A. (2020). SPERMINE ameliorates prolonged fluoride toxicity in soil-grown rice seedlings by activating the antioxidant machinery and glyoxalase system. Ecotoxicol. Environ. Saf. 189:109737. doi: 10.1016/j.ecoenv.2019.109737
Bhattacharyya, C., Roy, R., Tribedi, P., Ghosh, A., and Ghosh, A. (2020). Biofertilizers as substitute to commercial agrochemicals. Agrochem. Detect. Treatment Remed., 263–290. doi: 10.1016/b978-0-08-103017-2.00011-8
Calero, P., Gurdo, N., and Nikel, P. I. (2022). Role of the crcb transporter of Pseudomonas putida in the multi-level stress response elicited by mineral fluoride. Environ. Microbiol. 24, 5082–5104. doi: 10.1111/1462-2920.16110
Caverzan, A., Piasecki, C., Chavarria, G., Stewart, C., and Vargas, L. (2019). Defenses against Ros in crops and weeds: the effects of interference and herbicides. Int. J. Mol. Sci. 20:1086. doi: 10.3390/ijms20051086
Chadha, S., Sharma, M., and Sayyed, A. (2021). Advances in sensing plant diseases by imaging and machine learning methods for precision crop protection. Microbial Manage Plant Stresses, 157–183. doi: 10.1016/b978-0-323-85193-0.00012-7
Chandran, H., Meena, M., and Swapnil, P. (2021). Plant growth-promoting rhizobacteria as a green alternative for sustainable agriculture. Sustainability 13:10986. doi: 10.3390/su131910986
Chatterjee, N., Sahu, G., Bag, A. G., Plal, B., and Hazra, G. C. (2020). Role of fluoride on soil, plant and human health: a review on its sources, toxicity and mitigation strategies. Int. J. Environ. Climate Change 10, 77–90. doi: 10.9734/ijecc/2020/v10i830220
Chellaiah, E. R., Ravi, P., and Uthandakalaipandian, R. (2021). Isolation and identification of high fluoride resistant bacteria from water samples of Dindigul district, Tamil Nadu, South India. Curr. Res. Microb. Sci. 2:100038. doi: 10.1016/j.crmicr.2021.100038
Chiariello, M. G., Bolnykh, V., Ippoliti, E., Meloni, S., Olsen, J. M., Beck, T., et al. (2020). Molecular basis of CLC antiporter inhibition by fluoride. J. Am. Chem. Soc. 142, 7254–7258. doi: 10.1021/jacs.9b13588
Cochard, B., Giroud, B., Crovadore, J., Chablais, R., Arminjon, L., and Lefort, F. (2022). Endophytic PGPR from tomato roots: isolation, in vitro characterization and in vivo evaluation of treated tomatoes (solanum lycopersicum L.). Microorganisms 10:765. doi: 10.3390/microorganisms10040765
Constantia, J., and Ferniah, R. (2020). Vegetative growth of rainbow chili (Capsicum annuum l.) in the treatment of PGPR (plant growth promoting rhizobacteria), PGPR-NPK fertilizer, and PGPR-compost combination. Agric 32, 95–104. doi: 10.24246/agric.2020.v32.i2.p95-104
Do, T. H., Martinoia, E., Lee, Y., and Hwang, J.-U. (2021). 2021 update on ATP-binding cassette (ABC) transporters: how they meet the needs of plants. Plant Physiol. 187, 1876–1892. doi: 10.1093/plphys/kiab193
Duffin, S., Duffin, M., and Grootveld, M. (2022). Revisiting fluoride in the twenty-first century: safety and efficacy considerations. Front. Oral Health 3:873157. doi: 10.3389/froh.2022.873157
Edward Raja, C., Pandeeswari, R., and Ramesh, U. (2022). Characterization of high fluoride resistant pseudomonas aeruginosa species isolated from water samples. Environ. Sci. Technol. 5, 325–339. doi: 10.35208/ert.1070624
El-Naggar, N. E.-A., Hamouda, R. A., Mousa, I. E., Abdel-Hamid, M. S., and Rabei, N. H. (2018). Biosorption optimization, characterization, immobilization and application of Gelidium amansii biomass for complete pb2+ removal from aqueous solutions. Sci. Rep. 8:13456. doi: 10.1038/s41598-018-31660-7
Faraj, S. E., Centeno, M., Rossi, R. C., and Montes, M. R. (2019). A kinetic comparison between E2P and the E2P-like state induced by a beryllium fluoride complex in the na, K-atpase. Interactions with RB+. Biochim. Biophys. Acta 1861, 355–365. doi: 10.1016/j.bbamem.2018.10.020
Gadi, B. R., Kumar, R., Goswami, B., Rankawat, R., and Rao, S. R. (2020). Recent developments in understanding fluoride accumulation, toxicity, and tolerance mechanisms in plants: An overview. J. Soil Sci. Plant Nutr. 21, 209–228. doi: 10.1007/s42729-020-00354-3
Gao, J., Liu, C., Zhang, J., Zhu, S., Shen, Y., and Zhang, R. (2018). Effect of fluoride on photosynthetic pigment content and antioxidant system of Hydrilla verticillata. Int. J. Phytoremediation 20, 1257–1263. doi: 10.1080/15226514.2017.1319328
Han, J., Kiss, L., Mei, H., Remete, A. M., Ponikvar-Svet, M., Sedgwick, D. M., et al. (2021). Chemical aspects of human and environmental overload with fluorine. Chem. Rev. 121, 4678–4742. doi: 10.1021/acs.chemrev.0c01263
Hartmann, A. (2020). Quorum sensing N-acyl-homoserine lactone signal molecules of plant beneficial gram-negative rhizobacteria support plant growth and resistance to pathogens. Rhizosphere 16:100258. doi: 10.1016/j.rhisph.2020.100258
Hasanuzzaman, M., Bhuyan, M. H. M., Zulfiqar, F., Raza, A., Mohsin, S., Mahmud, J., et al. (2020). Reactive oxygen species and antioxidant defense in plants under abiotic stress: revisiting the crucial role of a universal defense regulator. Antioxidants 9:681. doi: 10.3390/antiox9080681
Huang, H., Ullah, F., Zhou, D.-X., Yi, M., and Zhao, Y. (2019). Mechanisms of ROS regulation of plant development and stress responses. Front. Plant Sci. 10. doi: 10.3389/fpls.2019.00800
Kabir, H., Gupta, A. K., and Tripathy, S. (2019). Fluoride and human health: systematic appraisal of sources, exposures, metabolism, and toxicity. Crit. Rev. Environ. Sci. Technol. 50, 1116–1193. doi: 10.1080/10643389.2019.1647028
Kadiresan, K., and Khanal, P. R. (2018). “Rethinking irrigation for global food security,” in Irrigation and drainage. John Wiley and Sons Ltd, 8–11.
Kashif, M., Sang, Y., Mo, S., Rehman, S. U., Khan, S., Khan, M. R., et al. (2023). Deciphering the biodesulfurization pathway employing marine mangrove Bacillus aryabhattai strain NM1-A2 according to whole genome sequencing and transcriptome analyses. Genomics 115:110635. doi: 10.1016/j.ygeno.2023.110635
Katiyar, P., Pandey, N., and Sahu, K. K. (2020). Biological approaches of fluoride remediation: potential for environmental clean-up. Environ. Sci. Pollut. Res. 27, 13044–13055. doi: 10.1007/s11356-020-08224-2
Khatoon, Z., Huang, S., Rafique, M., Fakhar, A., Kamran, M. A., and Santoyo, G. (2020). Unlocking the potential of plant growth-promoting rhizobacteria on soil health and the sustainability of agricultural systems. J. Environ. Manag. 273:111118. doi: 10.1016/j.jenvman.2020.111118
Kour, D., Rana, K. L., Yadav, N., Yadav, A. N., Kumar, A., Meena, V. S., et al. (2019). “Rhizospheric microbiomes: biodiversity, mechanisms of plant growth promotion, and biotechnological applications for sustainable agriculture” in Plant growth promoting rhizobacteria for agricultural sustainability. eds. A. Kumar and V. Meena (Singapore: Springer), 19–65.
Kumar, R., Sinha, R., Sharma, P. K., Ivy, N., Kumar, P., Kant, N., et al. (2021). Bioaccumulation of fluoride in plants and its microbially assisted remediation: a review of biological processes and technological performance. PRO 9:2154. doi: 10.3390/pr9122154
Lee, H.-J., Song, J., and Kim, J. N. (2021). Genetic mutations that confer fluoride resistance modify gene expression and virulence traits of streptococcus mutans. Microorganisms 9:849. doi: 10.3390/microorganisms9040849
Lee, J., Sung, S.-E., Lee, J., Kang, J. Y., Lee, J.-H., and Choi, B.-S. (2021). Base-pair opening dynamics study of fluoride riboswitch in the bacillus cereus CrcB gene. Int. J. Mol. Sci. 22:3234. doi: 10.3390/ijms22063234
Liao, Y., Yang, J., Brandt, B. W., Li, J., Crielaard, W., van Loveren, C., et al. (2018). Genetic loci associated with fluoride resistance in streptococcus mutans. Front. Microbiol. 9. doi: 10.3389/fmicb.2018.03093
Li, C., Qi, C., Yang, S., Li, Z., Ren, B., Li, J., et al. (2022). F0F1-atpase contributes to the fluoride tolerance and cariogenicity of streptococcus mutans. Front. Microbiol. 12. doi: 10.3389/fmicb.2021.777504
Lindemann, S. (2019). Microbial ecology: functional ‘modules’ drive assembly of polysaccharide-degrading marine microbial communities. Curr. Biol. 29, R330–R332. doi: 10.1016/j.cub.2019.03.056
Ling, N., Wang, T., and Kuzyakov, Y. (2022). Rhizosphere bacteriome structure and functions. Nat. Commun. 13:836. doi: 10.1038/s41467-022-28448-9
Li, W., Shi, Y., Zhu, D., Wang, W., Liu, H., Li, J., et al. (2021). Fine root biomass and morphology in a temperate forest are influenced more by the nitrogen treatment approach than the rate. Ecol. Indic. 130:108031. doi: 10.1016/j.ecolind.2021.108031
Li, W., Wang, C., Liu, H., Wang, W., Sun, R., Li, M., et al. (2023). Fine root biomass and morphology in a temperate forest are influenced more by canopy water addition than by canopy nitrogen addition. Front. Ecol. Evol. 11. doi: 10.3389/fevo.2023.1132248
Li, X., Chen, F., Xiao, J., Chou, S.-H., Li, X., and He, J. (2018). Genome-wide analysis of the distribution of riboswitches and function analyses of the corresponding downstream genes in prokaryotes. Curr. Bioinform. 14, 53–61. doi: 10.2174/1574893613666180423145812
Lu, L., Zhai, X., Li, X., Wang, S., Zhang, L., Wang, L., et al. (2022). Met1-specific motifs conserved in OTUB subfamily of green plants enable rice OTUB1 to hydrolyse Met1 ubiquitin chains. Nat. Commun. 13:4672. doi: 10.1038/s41467-022-32364-3
Lu, M., Xiang, Z., Gong, T., Zhou, X., Zhang, Z., Tang, B., et al. (2020). Intrinsic fluoride tolerance regulated by a transcription factor. J. Dent. Res. 99, 1270–1278. doi: 10.1177/0022034520927385
Luo, B., Guang, M., Yun, W., Ding, S., Ren, S., and Gao, H. (2022). Camellia sinensis chloroplast fluoride efflux gene CSABCB9 is involved in the fluoride tolerance mechanism. Int. J. Mol. Sci. 23:7756. doi: 10.3390/ijms23147756
Luo, J., Ni, D., Li, C., Du, Y., and Chen, Y. (2021). The relationship between fluoride accumulation in tea plant and changes in leaf cell wall structure and composition under different fluoride conditions. Environ. Pollut. 270:116283. doi: 10.1016/j.envpol.2020.116283
Mahmud, K., Makaju, S., Ibrahim, R., and Missaoui, A. (2020). Current progress in nitrogen fixing plants and microbiome research. Plan. Theory 9:97. doi: 10.3390/plants9010097
Makete, N., Rizzu, M., Seddaiu, G., Gohole, L., and Otinga, A. (2022). Fluoride toxicity in cropping systems: mitigation, adaptation strategies and related mechanisms. A Review. Sci. Total Environ. 833:155129. doi: 10.1016/j.scitotenv.2022.155129
Ma, L., Wu, J., Liu, X., Tan, L., and Wang, X. (2019). The detoxification potential of ferric ions for bioleaching of the chalcopyrite associated with fluoride-bearing gangue mineral. Appl. Microbiol. Biotechnol. 103, 2403–2412. doi: 10.1007/s00253-018-09599-6
McIlwain, B. C., Ruprecht, M. T., and Stockbridge, R. B. (2021). Membrane exporters of fluoride ion. Annu. Rev. Biochem. 90, 559–579. doi: 10.1146/annurev-biochem-071520-112507
Mothe, T., Umashankar, P., and Sultanpuram, V. R. (2021). Isolation of fluoride resistant microorganisms from fluoride contaminated ground water samples of Nalgonda district and their role in bioremediation. Biosci. Biotechnol. Res. Asia 18, 107–112. doi: 10.13005/bbra/2900
Mukherjee, S., and Halder, G. (2018). A review on the sorptive elimination of fluoride from contaminated wastewater. J. Environ. Chem. Eng. 6, 1257–1270. doi: 10.1016/j.jece.2018.01.046
Mukherjee, S., Sahu, P., and Halder, G. (2018). Comparative assessment of the fluoride removal capability of immobilized and dead cells of Staphylococcus lentus (kx941098) isolated from contaminated groundwater. Environ. Prog. Sustain. 37, 1573–1586. doi: 10.1002/ep.12853
Mushtaq, I., Zulfiqar, A., Ali, M., and Saleem, A. (2021). Effect of sodium fluoride on the growth and productivity of Triticum aestivum L. in the presence of fluoride resistant bacteria. SSRN Electron. J. doi: 10.2139/ssrn.3922745
Nabbou, N., Belhachemi, M., Boumelik, M., Merzougui, T., Lahcene, D., Harek, Y., et al. (2019). Removal of fluoride from groundwater using natural clay (kaolinite): optimization of adsorption conditions. C. R. Chim. 22, 105–112. doi: 10.1016/j.crci.2018.09.010
Nadarajah, K., and Abdul Rahman, N. S. (2021). Plant–microbe interaction: above ground to belowground, from the good to the bad. Int. J. Mol. Sci. 22:10388. doi: 10.3390/ijms221910388
Nie, S., Mo, S., Gao, T., Yan, B., Shen, P., Kashif, M., et al. (2023). Coupling effects of nitrate reduction and sulfur oxidation in a subtropical marine mangrove ecosystem with Spartina alterniflora invasion. Sci. Total Environ. 862:160930. doi: 10.1016/j.scitotenv.2022.160930
Ong, S.-H., Kim, J., Baek, D.-H., and Yoo, S. (2020). Comparison of cariogenic characteristics between fluoride-sensitive and fluoride-resistant Streptococcus mutans. J. Korean Acad. Pediatr. Dent. 47, 397–405. doi: 10.5933/jkapd.2020.47.4.397
Orozco-Mosqueda, MaDel, Santoyo, G., and Glick, B. R. (2023). Recent advances in the bacterial phytohormone modulation of plant growth. Plan. Theory 12::606. doi: 10.3390/plants12030606
Pal, K. C., Mukhopadhyay, P., Chatterjee, S., and Mondal, N. K. (2022). A study on fluoride bioremediation via a novel bacterium Bacillus megaterium (JF273850) isolated from agricultural soil. J. Earth Syst. Sci. 131. doi: 10.1007/s12040-022-01931-z
Patani, A., Prajapati, D., Ali, D., Kalasariya, H., Yadav, V. K., Tank, J., et al. (2023). Evaluation of the growth-inducing efficacy of various bacillus species on the salt-stressed tomato (Lycopersicones culentum mill.). Front. Plant Sci. 14. doi: 10.3389/fpls.2023.1168155
Patel, M., Islam, S., Husain, F. M., Yadav, V. K., Park, H.-K., Yadav, K. K., et al. (2023a). Bacillus subtilis ER-08, a multifunctional plant growth-promoting rhizobacterium, promotes the growth of fenugreek (Trigonella foenum Graecum L.) plants under salt and drought stress. Front. Microbiol. 14. doi: 10.3389/fmicb.2023.1208743
Patel, M., Vurukonda, S. S., and Patel, A. (2023b). Multi-trait halotolerant plant growth-promoting bacteria mitigate induced salt stress and enhance growth of Amaranthus viridis. J. Soil Sci. Plant Nutr. 23, 1860–1883. doi: 10.1007/s42729-023-01143-4
Patil, A. S., Patil, S. R., and Sayyed, R. Z. (2019). “Interaction of rhizobacteria with soil microorganisms: an agro-beneficiary aspect” in Plant growth promoting rhizobacteria for sustainable stress management. ed. R. Sayyed , vol. 13 (Singapore: Springer)
Pelc, J., Śnioszek, M., Wróbel, J., and Telesiński, A. (2020). Effect of fluoride on germination, early growth and antioxidant enzymes activity of three winter wheat (Triticum aestivum L.) cultivars. Appl. Sci. 10:6971. doi: 10.3390/app10196971
Peng, C., Xu, X., Ren, Y., Niu, H., Yang, Y., Hou, R., et al. (2020). Fluoride absorption, transportation and tolerance mechanism in Camellia sinensis, and its bioavailability and health risk assessment: a systematic review. J. Sci. Food Agric. 101, 379–387. doi: 10.1002/jsfa.10640
Phale, S. (2018). Isolation and screening of multifunctional plant growth promoting rhizobacteria (PGPR) from onion rhizosphere (Allium cepa). J. Bioprocess. Biotech. 8. doi: 10.4172/2155-9821.1000335
Roshni, V. (2022). Effect of fluoride contamination on microbial enzymatic activity in soil. Int. J. Adv. Res. 10, 42–48. doi: 10.21474/ijar01/14855
Sahariya, A., Bharadwaj, C., Emmanuel, I., and Alam, A. (2021). Phytochemical profiling and GCMS analysis of two different varieties of barley (Hordeum vulgare L.) under fluoride stress. Sci. Temper 12. doi: 10.58414/scientifictemper.2021.12.1.01
Sahariya, A., Bharadwaj, C., Emmanuel, I., and Alam, A. (2022). Relative biochemical analysis of two cultivars of Hordeum vulgare L. under fluoride stress using Fourier transform-infrared spectroscopy (FT-IR). Plant Physiol. Rep. 27, 717–723. doi: 10.1007/s40502-022-00676-7
Sahoo, S., and Goli, D. (2018). De-fluoridation by Mangrove Microbe. Int. J. Sci. Res. 9, 1350–1355. doi: 10.21275/ART20204282
Sahoo, S. R., Goli, D., and Maringanti, B. (2019). Studies of two pigment producing halophilic bacteria from Karnataka mangrove soil. Indian J. Pharm. Sci. 81, 892–899. doi: 10.36468/pharmaceutical-sciences.584
Sakthi Thesai, A., Rajakumar, S., and Ayyasamy, P. M. (2018). Removal of fluoride in aqueous medium under the optimum conditions through intracellular accumulation in bacillus flexus (PN4). Environ. Technol. 41, 1185–1198. doi: 10.1080/09593330.2018.1523951
Shanker, A. S., Srinivasulu, D., and Pindi, P. K. (2020). A study on bioremediation of fluoride-contaminated water via a novel bacterium Acinetobacter sp. (gu566361) isolated from potable water. Results Chem. 2:100070. doi: 10.1016/j.rechem.2020.100070
Sharma, R., and Kaur, R. (2018). Insights into fluoride-induced oxidative stress and antioxidant defences in plants. Acta Physiol. Plant. 40. doi: 10.1007/s11738-018-2754-0
Sharma, R., and Kaur, R. (2019). Fluoride toxicity triggered oxidative stress and the activation of Antioxidative defence responses in Spirodela polyrhiza L. Schleiden. J. Plant Interact. 14, 440–452. doi: 10.1080/17429145.2019.1646826
Singh, A., and Roychoudhury, A. (2020). Silicon-regulated antioxidant and osmolyte defense and methylglyoxal detoxification functions co-ordinately in attenuating fluoride toxicity and conferring protection to Rice seedlings. Plant Physiol. Biochem. 154, 758–769. doi: 10.1016/j.plaphy.2020.06.023
Singh, A., and Roychoudhury, A. (2021). Salicylic acid–mediated alleviation of fluoride toxicity in rice by restricting fluoride bioaccumulation and strengthening the osmolyte, antioxidant and glyoxalase systems. Environ. Sci. Pollut. Res. 30, 25024–25036. doi: 10.1007/s11356-021-14624-9
Singh, A., Yadav, V. K., Chundawat, R. S., Soltane, R., Awwad, N. S., Ibrahium, H. A., et al. (2023). Enhancing plant growth promoting rhizobacterial activities through consortium exposure: a review. Front. Bioeng. Biotechnol. 11. doi: 10.3389/fbioe.2023.1099999
Singh, B., and Jajoo, A. (2021). Cyclic electron flow plays an important role in protecting PSI against fluoride stress in maize plant. J. Soil Sci. Plant Physiol.
Singh, G., Kumari, B., Sinam, G., Kriti,, Kumar, N., and Mallick, S. (2018). Fluoride distribution and contamination in the water, soil and plants continuum and its remedial technologies, an Indian perspective– a review. Environ. Pollut. 239, 95–108. doi: 10.1016/j.envpol.2018.04.002
Singh, R. K., Singh, P., Li, H.-B., Song, Q.-Q., Guo, D.-J., Solanki, M. K., et al. (2020). Diversity of nitrogen-fixing rhizobacteria associated with sugarcane: a comprehensive study of plant-microbe interactions for growth enhancement in Saccharum spp. BMC Plant Biol. 20:220. doi: 10.1186/s12870-020-02400-9
Speed, M. C., Burkhart, B. W., Picking, J. W., and Santangelo, T. J. (2018). An archaeal fluoride-responsive riboswitch provides an inducible expression system for hyperthermophiles. Appl. Environ. Microbio 84:e02306-17. doi: 10.1128/aem.02306-17
Su, J. F., Zhang, H., Huang, T. L., Hu, X. F., Chen, C. L., and Liu, J. R. (2020). The performance and mechanism of simultaneous removal of fluoride, calcium, and nitrate by calcium precipitating strain Acinetobacter sp. H12. Ecotoxicol. Environ. Saf. 187:109855. doi: 10.1016/j.ecoenv.2019.109855
Sylia, A., Corrêa, A., Cruz, C., Yadav, A., and Nabti, E. (2022). Plant growth promoting microbes as biofertilizers: Promising solutions for sustainable agriculture under climate change associated abiotic stresses. Plant Sci. Today 8, 60–76. doi: 10.14719/pst.1608
Tausta, S. L., Berbasova, T., Peverelli, M., and Strobel, S. A. (2021). The fluoride transporter fluoride exporter (FEX) is the major mechanism of tolerance to fluoride toxicity in plants1. Plant Physiol. 186, 1143–1158. doi: 10.1093/plphys/kiab131
Tiwari, G., Duraivadivel, P., Sharma, S., and Hariprasad, P. (2018). 1-aminocyclopropane-1-carboxylic acid deaminase producing beneficial rhizobacteria ameliorate the biomass characters of Panicum Maximum Jacq. By mitigating drought and salt stress. Sci. Rep. 8:17513. doi: 10.1038/s41598-018-35565-3
Uruén, C., Chopo-Escuin, G., Tommassen, J., Mainar-Jaime, R. C., and Arenas, J. (2020). Biofilms as promoters of bacterial antibiotic resistance and tolerance. Antibiotics 10:3. doi: 10.3390/antibiotics10010003
Vaishnav, A., Kumar, R., Singh, H., and Sarma, B. (2022). Extending the benefits of PGPR to bioremediation of nitrile pollution in crop lands for enhancing crop productivity. Sci. Total Environ. 826:154170. doi: 10.1016/j.scitotenv.2022.154170
Vecchiato, M., Bonato, T., Barbante, C., Gambaro, A., and Piazza, R. (2021). Organic pollutants in protected plain areas: the occurrence of pahs, musks, UV-filters, flame retardants and hydrocarbons in woodland soils. Sci. Total Environ. 796:149003. doi: 10.1016/j.scitotenv.2021.149003
Vega-Celedón, P., Bravo, G., Velásquez, A., Cid, F. P., Valenzuela, M., Ramírez, I., et al. (2021). Microbial diversity of psychrotolerant bacteria isolated from wild Flora of Andes Mountains and Patagonia of Chile towards the selection of plant growth-promoting bacterial consortia to alleviate cold stress in plants. Microorganisms 9:538. doi: 10.3390/microorganisms9030538
Wang, Y., Yang, L., Xu, J., Xin, F., and Jiang, L. (2023). Applications of synthetic microbial consortia in biological control of mycotoxins and fungi. Curr. Opin. Food Sci. 53:101074. doi: 10.1016/j.cofs.2023.101074
Waugh, D. T. (2019). Fluoride exposure induces inhibition of sodium-and potassium-activated adenosine triphosphatase (na+, k+−atpase) enzyme activity: molecular mechanisms and implications for public health. Int. J. Environ. Res. Public Health 16:1427. doi: 10.3390/ijerph16081427
Weerasooriyagedara, M., Ashiq, A., Rajapaksha, A. U., Wanigathunge, R. P., Agarwal, T., Magana-Arachchi, D., et al. (2020). Phytoremediation of fluoride from the environmental matrices: a review on its application strategies. Groundw. Sustain. Dev. 10:100349. doi: 10.1016/j.gsd.2020.100349
Wu, Z., Su, J., Ali, A., Hu, X., and Wang, Z. (2021). Study on the simultaneous removal of fluoride, heavy metals and nitrate by calcium precipitating strain Acinetobacter sp. H12. J. Hazard. Mater. 405:124255. doi: 10.1016/j.jhazmat.2020.124255
Yadav, K. K., Kumar, S., Pham, Q. B., Gupta, N., Rezania, S., Kamyab, H., et al. (2019). Fluoride contamination, health problems and remediation methods in Asian groundwater: a comprehensive review. Ecotoxicol. Environ. Saf. 182:109362. doi: 10.1016/j.ecoenv.2019.06.045
Yadav, M., Singh, G., and Jadeja, R. N. (2021). “Fluoride contamination in groundwater, impacts, and their potential remediation techniques,” in Groundwater geochemistry. eds. S. Madhav and P. Singh.
Yang, Y., Dou, Y., Wang, B., Xue, Z., Wang, Y., An, S., et al. (2022). Deciphering factors driving soil microbial life-history strategies in restored grasslands. iMeta. doi: 10.1002/imt2.66
Yan, Y., Jarvie, S., Liu, Q., and Zhang, Q. (2022). Effects of fragmentation on grassland plant diversity depend on the habitat specialization of species. Biol. Conserv. 275:109773. doi: 10.1016/j.biocon.2022.109773
Zhang, M., Yang, L., Hao, R., Bai, X., Wang, Y., and Yu, X. (2020). Drought-tolerant plant growth-promoting rhizobacteria isolated from jujube (ziziphus jujuba) and their potential to enhance drought tolerance. Plant Soil 452, 423–440. doi: 10.1007/s11104-020-04582-5
Zhang, X., Gao, X., Li, C., Luo, X., and Wang, Y. (2019). Fluoride contributes to the shaping of microbial community in high fluoride groundwater in Qiji County, Yuncheng City, China. Sci. Rep. 9:14488. doi: 10.1038/s41598-019-50914-6
Zhao, W., Wang, H., Liu, J., Zhang, Z., Zhu, S., and Zhou, B. (2019). Mitochondrial respiratory chain complex abnormal expressions and fusion disorder are involved in fluoride-induced mitochondrial dysfunction in ovarian granulosa cells. Chemosphere 215, 619–625. doi: 10.1016/j.chemosphere.2018.10.043
Keywords: rhizosphere, fluoride transporters, PGPR, defense systems, bioremediation, chemical fertilizers
Citation: Singh A, Yadav VK, Gautam H, Rathod L, Chundawat RS, Singh G, Verma RK, Sahoo DK and Patel A (2023) The role of plant growth promoting rhizobacteria in strengthening plant resistance to fluoride toxicity: a review. Front. Microbiol. 14:1271034. doi: 10.3389/fmicb.2023.1271034
Received: 01 August 2023; Accepted: 20 September 2023;
Published: 10 October 2023.
Edited by:
Rajarshi Kumar Gaur, Deen Dayal Upadhyaya Gorakhpur University, IndiaReviewed by:
Tara Chand Yadav, University of Rovira i Virgili, SpainCopyright © 2023 Singh, Yadav, Gautam, Rathod, Chundawat, Singh, Verma, Sahoo and Patel. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Rajendra Singh Chundawat, cnNjaHVuZGF3YXQuc2xhc0Btb2R5dW5pdmVyc2l0eS5hYy5pbg==; Dipak Kumar Sahoo, ZHNhaG9vQGlhc3RhdGUuZWR1; Virendra Kumar Yadav, eWFkYXZhOTRAZ21haWwuY29t; Ashish Patel, dW5pLmFzaGlzaEBnbWFpbC5jb20=
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.