
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Microbiol. , 26 October 2023
Sec. Microbe and Virus Interactions with Plants
Volume 14 - 2023 | https://doi.org/10.3389/fmicb.2023.1268333
 S. Vignesh1
S. Vignesh1 P. Renukadevi1
P. Renukadevi1 K. Nagendran2
K. Nagendran2 N. Senthil3
N. Senthil3 R. Vinoth Kumar4
R. Vinoth Kumar4 R. SwarnaPriya5
R. SwarnaPriya5 Tusar Kanti Behera2
Tusar Kanti Behera2 G. Karthikeyan1*†
G. Karthikeyan1*†Ash gourd (Benincasa hispida) is a cucurbitaceous crop cultivated as an edible vegetable rich in vitamins, minerals, dietary fibers and antioxidants. In a field survey conducted in the Udumalpet region of Tamil Nadu during 2019, the incidence of mosaic disease on ash gourd crop was observed to be 75%. The DNA-A and DNA-B components of begomovirus genome have been identified as associated with this disease. Both the cloned DNA-A and DNA-B genomic components shared highest pairwise sequence identities with the isolates of tomato leaf curl New Delhi virus (ToLCNDV), a bipartite begomovirus. Recombinant analysis showed that both the components are possibly evolved through intra-species recombination between ToLCNDV isolates. Tomato leaf curl Bangladesh betasatellite (ToLCBB) is not naturally associated with this sample. The results of infectivity studies on ash gourd and other cucurbitaceous crops demonstrates the Koch’s postulates, when co-inoculation of DNA-A and DNA-B of ToLCNDV was undertaken. However, the inoculation of non-cognate ToLCBB along with DNA-A and DNA-B enhances the symptom expression and reduces the time taken for symptom development. Thus, Koch’s postulates were proved for these virus complexes on cucurbitaceous crops. Furthermore, an enhanced accumulation of DNA-A component was detected in the cucurbits co-inoculated with ToLCNDV and ToLCBB. This report highlights the importance of investigating the spread of these disease complexes with other cucurbitaceous crops in India.
Cucurbits are an economically important crop grown in an area of around 7.89 million hectares with a global production of around 238.6 million tonnes. Among the different cucurbits, ash gourd [Benincasa hispida (Thunb.) Cogn.] is an important vegetable and unique melon mainly grown in India for its edible fruit. Fruits are not only consumed as a vegetable but also used in the confectionary industry for making candy, jam, ketchup, and cakes, among other dishes. The ash gourd crop is cultivated around the year both in tropical and subtropical regions of the Indian subcontinent. Viral infections of the plant pose a major threat to its productivity and the quality of the cucurbitaceous crops (Thresh, 2006; Damicone et al., 2007). Several viruses including begomoviruses (Riyaz et al., 2013; Roy et al., 2013; Kumari et al., 2022), potyviruses (Nagendran et al., 2017a), and tobamoviruses (Kumar et al., 2017) have been identified in ash gourd in India.
The Begomovirus (Family: Geminiviridae) comprises more than 450 species and causes diseases in a wide range of economically important crops as well as weeds in the “Old World” and “New World” regions (Brown et al., 2015; Kumar, 2019). Begomoviruses are transmitted by whiteflies (Bemisia tabaci) in a persistent and circulative manner. They are further subdivided into either bipartite consisting of two approximately similar sized DNA-A and DNA-B genomic components or monopartite with a homologous DNA-A-like genomic component. The DNA-A component encodes six proteins necessary for viral replication and encapsidation, whereas DNA-B has two proteins that mediate symptom expression and systemic virus movement (Kumar, 2019). These begomoviral genomic components were often associated with betasatellites to promote pathogenesis, virulence, and establishment of characteristic disease symptoms (Rouhibakhsh and Malathi, 2005; Sivalingam and Varma, 2012; Jyothsna et al., 2013; Ranjan et al., 2014; Kumar et al., 2015; Sharma et al., 2019; Singh et al., 2021). Begomoviruses undergo rapid evolution through mutation and recombination (Lefeuvre et al., 2007; Prasanna and Rai, 2007; Melgarejo et al., 2013; George et al., 2015; Kumar et al., 2015, 2017).
The whitefly-transmitted Tomato leaf curl New Delhi virus (ToLCNDV) is a limiting factor for cucurbit production in Asian and European countries (Srivastava et al., 1995; Hussain et al., 2004; Maruthi et al., 2005; Chang et al., 2010; Sangeetha et al., 2018). It has become a significant threat to numerous crops across plant families including the Apocyanaceae, Acanthaceae, Asteraceae, Cucurbitaceae, Caricaceae, Euphorbiaceae, Fabaceae, Malvaceae, Papaveraceae, Phyllanthaceae, and Solanaceae worldwide (Kumar et al., 2015; Moriones et al., 2017; Zaidi et al., 2017; Nagendran et al., 2017b; Ashwathappa et al., 2020; Gemert et al., 2020; Chakraborty and Kumar, 2021; Krishnan et al., 2023). The infection of ToLCNDV on cucurbits exhibits characteristic symptoms including yellow mosaic, reduction of internodal length, severe curling, vein swelling, rough skin, reduced fruit setting, etc. (Moriones et al., 2017; Zaidi et al., 2017). In this study, we have characterized the association of ToLCNDV infecting ash gourd from Tamil Nadu, India. The infectivity study highlighted the effect of either cognate DNA-B or non-cognate betasatellite for disease development on various cucurbitaceous crops.
Ash gourd leaf samples showing yellow mosaic, leaf puckering, chlorosis, and severe yellow mosaic symptoms were collected from the agricultural fields in the Udumalpet region (GPS co-ordinates: 10° 35′6.282″ N, 77°14′52.476″ E), a major ash gourd growing area of Tamil Nadu during November 2019. Totally eight samples from four different fields along with one apparently healthy sample were collected and stored at −20°C for further studies.
The total genomic DNA was isolated from all the ash gourd leaf samples using the CTAB method (Doyle and Doyle, 1990). The total DNAs were subjected to polymerase chain reaction (PCR) assay using the degenerate primers, PAR1v722/PAL1c1960 (Table 1) amplifying ~1.2 kb on DNA-A genome of begomovirus (Chatchawankanphanich and Maxwell, 2002).
Seventy to eighty ng of total DNA from all the PCR positive samples were subjected to rolling circle amplification (RCA) as per the manufacturer’s instructions (Thermo Fisher Scientific, United States; Supplementary Figure 2). The resultant RCA product was digested individually with five restriction endonucleases (BamHI, HindIII, KpnI, XbaI, and EcoRI; Supplementary Figure 3). The ~2.7 kb fragments were gel purified and cloned into a pUC18 vector (Thermo Fisher Scientific, United States) already linearized with the corresponding restriction endonucleases. The recombinant clones were screened by restriction analysis. The predicted full-length genome fragments were selected and sequenced using primer walking with M/S. Eurofins Genomics, Bangalore, India. A separate PCR assay was also done to examine RCA products for the association of alpha-satellites and beta-satellites with universal primer pairs, UN101/UN102 and Beta01/Beta02 (Table 1; Briddon et al., 2002; Bull et al., 2003).
The cloned viral genome sequences of the DNA-A (pUCND-A) and DNA-B (pUCND-B) with the previously reported isolates available in the NCBI database were aligned using the MUSCLE algorithm (Edgar, 2004). The Sequence Demarcation Tool (SDT) version 1.2 was used to assess the pairwise sequence identity among the viral sequences (Muhire et al., 2014). The maximum likelihood-based phylogenetic trees were generated using MEGA 11 software (Kumar et al., 2016). Using RDP4 software (Martin et al., 2015), the recombination analysis was performed as described earlier (George et al., 2015; Martin et al., 2015).
Partial tandem repeat DNA of the cloned viral genomic components were constructed to satisfy Koch’s postulates. A 750 bp fragment of pUCND-A [BamHI (125)-PstI (2110)] containing intergenic region (IR) was cloned into pCAMBIA2301 vector linearized with BamHI and PstI. The full-length monomer linearized with BamHI was then ligated to generate a tandem repeat construct of pUCND-A (referred to as A). For pUCND-B, a 2.1 kb [KpnI (1532)-BamHI (925)] fragment was cloned into pCAMBIA2301, followed by insertion of the full-length monomer (~2.7) linearized with KpnI was then ligated to generate tandem repeat construct of pUCND-B (referred as B; Supplementary Figures 4–7). Further, a dimeric construct of tomato leaf curl Bangladesh betasatellite (ToLCBB; OQ718502) available in our laboratory was also used (referred to as β).
The tandem repeat plasmid constructs of DNA-A and DNA-B were mobilized into Agrobacterium tumefaciens strain GV3101 (Supplementary Figure 8) by freeze–thaw technique (Chen et al., 1994). Agro-inoculation was performed on 10-days old ash gourd (cv. Suruchi), squash (cv. Green star), pumpkin (cv. Co2), bitter gourd (cv. Co1), ridge gourd (cv. CoH1), snake gourd (cv. Co2), cucumber (cv. Nazia F1), muskmelon (cv. Dulce F1), watermelon (cv. F1 555) and bottle gourd (cv. Co1) plants as described by Singh et al. (2021). Plant inoculation with agrobacterium carrying empty pCAMBIA2301 was considered a mock inoculation. After agro-inoculation, the plants were maintained at 28°C ± 2°C in an insect-proof growth chamber with 16/8 h of photoperiod.
The presence of DNA-A, DNA-B, and DNA-β components in the systemic leaves of agro-inoculated plants were analyzed with viral gene specific primers (Table 1). The viral genomic components in the agro-inoculated plants were quantified by quantitative PCR (qPCR). The qPCR was carried out in a CFX-96 real-time system (Bio-Rad) with a 20 μL reaction mixture consisting of 10 μL KAPA SYBR green fast qPCR Master Mix 2X (Sigma-Aldrich), 1.5 μL (10 pmol) of each forward and reverse primer and 2 μL template DNA (~36.5 ng). The optimized thermal cycling reactions were 94°C for 5 min, 40 cycles of 94°C for 30 s, 58°C for 30 s, and 72°C for 30s. All the reactions were performed triplicate in high-profile 96 well qPCR plates (Bio-Rad). The melt curve analysis was programmed from 65 to 95°C, with an increase of 0.5°C at every 5 s interval.
The total genomic DNA isolated from the systemic leaves of agro-inoculated plants were subjected to viral DNA detection using the partial coat protein specific primer. Real-time PCR was carried out using 10-fold serially diluted plasmid DNA as a template for the above. To construct the standard curves for plasmid DNA, a linear regression curve was plotted with the mean Ct values on the Y-axis and the log DNA dilution in ng on the X-axis.
To quantify the ToLCNDV DNA-A titer, systemically infected leaves were collected and total genomic DNA was isolated as described above. The absolute quantification of infectious viral copies were derived by the mean Ct values were fitted into the corresponding standard curve and the viral copy number was calculated by using the formula N = (X ng * 6.0221 × 1023 molecules/mole)/(n * 660 g/mole * 1 × 109 ng/g), where N indicates the number of viral copies; X represents the amount of amplicon in ng and n represents the number of bases of the recombinant plasmid (Roy et al., 2021).
Data analyses were performed in the R statistical software (R Core Team, 2023). To determine the statistical significance of ToLCNDV accumulation in agro-inoculated cucurbits samples at 28 dpi the mean ± standard error was calculated. The amount of the mock, A, A + B, and A + B + β components were compared using an ANOVA F-test (Hothorn et al., 2008).
The average disease incidence was 75% in the farmer’s field (data not shown). Based on the symptomatology and the presence of whiteflies in the surveyed fields, the samples were suspected of begomovirus infection.
The ash gourd plants bearing disease symptoms such as upward and downward cupping of leaves, yellow mosaic, chlorosis, leaf puckering, leaf curl, and malformation of fruits were observed in the cultivated fields at Udumalpet taluk of Tamil Nadu in India in 2019 (Figure 1). The preliminary diagnostic evaluation for begomovirus made through PCR assay using begomovirus universal primer pair (PAL1v722/PAL1c1960) has yielded an expected amplicon of ~1.2 kb from all the symptomatic samples (Supplementary Figure 1). However, these samples tested negative for the association of alphasatellite (UN101/UN102) and betasatellites (Beta01/Beta02) in the PCR assay.

Figure 1. Field symptoms of begomovirus infection on ash gourd. (A) Yellow mosaic; (B) Mosaic with puckering; (C) Yellow mosaic with downward cupping; (D) Chlorosis; (E) Leaf puckering; (F) Severe yellow mosaic pattern; (G) Healthy leaf.
Sequence analysis revealed the presence of both DNA-A (BamHI clone) and DNA-B (KpnI clone). The DNA-A sequence (MZ073374) identified here shared the highest pairwise sequence identity of 92.4% with a ridge gourd isolate (KT426905) of ToLCNDV from India (Figure 2A). Similarly, the DNA-B sequence (MZ073373) showed the highest nucleotide identity of 96.1% with the ToLCNDV isolate infecting zucchini (MH577612) from Spain (Figure 2B). According to begomovirus species demarcation guidelines of 91% (Brown et al., 2015), the cloned begomovirus can be considered as a distinct strain of ToLCNDV. In the phylogenetic dendrogram, this cloned DNA-A component from ash gourd was found to be clustered along with the isolates of ToLCNDV-5 species. In the case of DNA B, the ash gourd isolate had grouped with Spain (tomato and zucchini) isolates in the phylogenetic dendrogram (Figure 3).
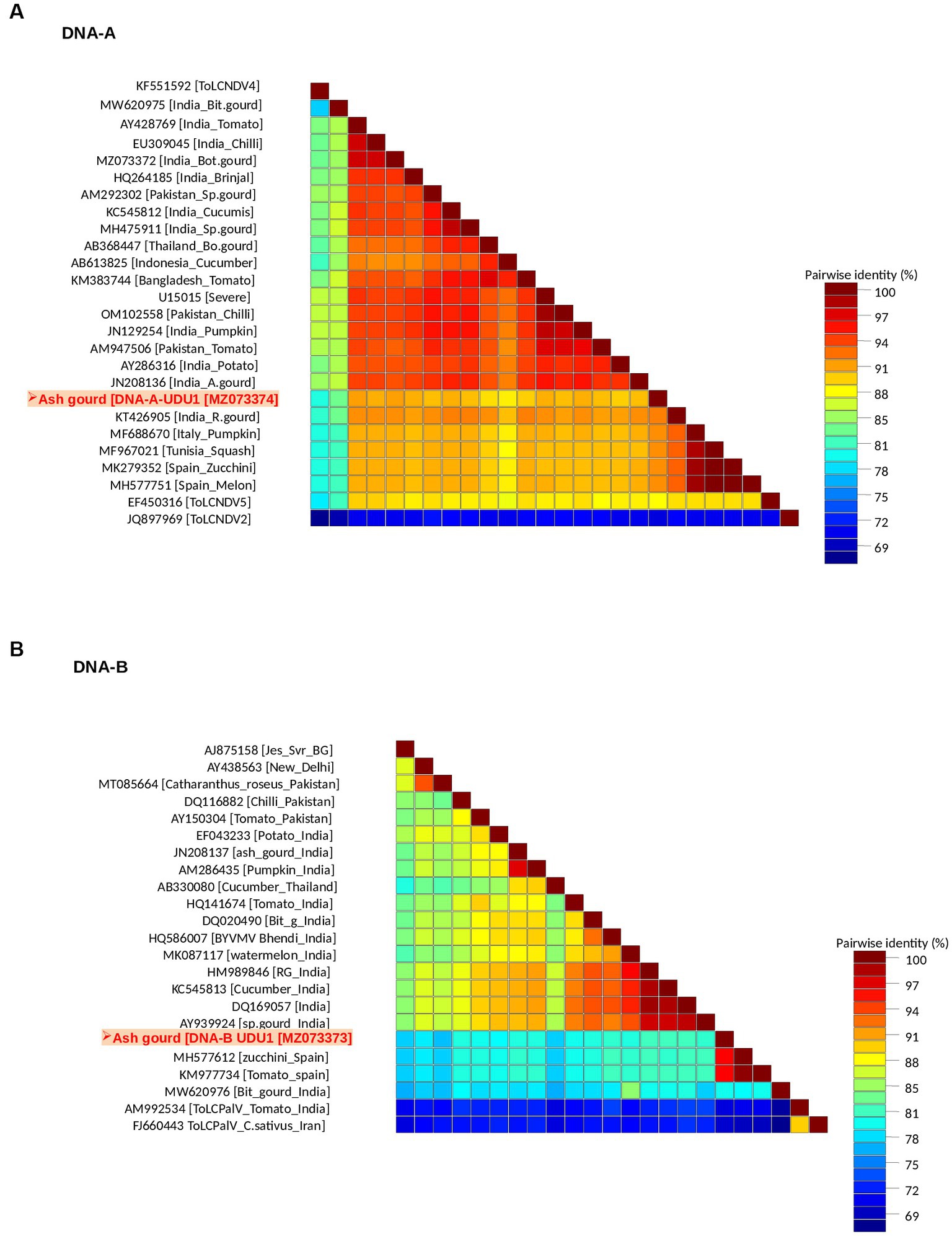
Figure 2. Association of a bipartite begomovirus, ToLCNDV, with the mosaic disease of ash gourd in India. Pairwise sequence identity matrices of begomovirus components, DNA-A (A) and DNA-B (B).
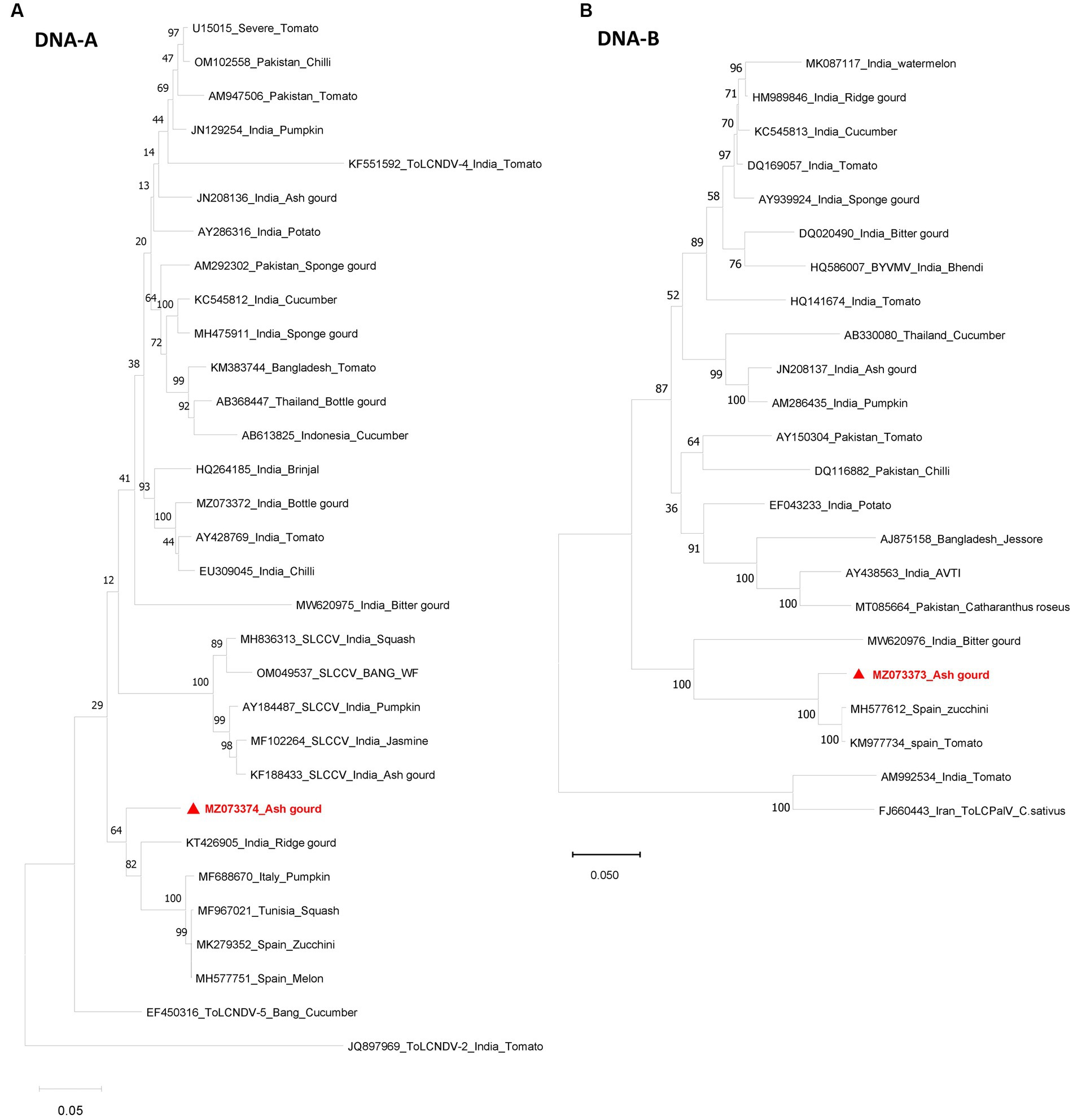
Figure 3. Phylogenetic dendrograms of DNA-A (A) and DNA-B (B) identified in this study. The numbers at the nodes indicate the percentage bootstrap values of 1,000 replicates. The scale bars represent the genetic distance. Viral genomes identified in this study are marked in red color.
Recombination analysis of the cloned ToLCNDV isolates revealed that the nucleotide sequence of the DNA-A component had evolved through intra-specific recombination between two ToLCNDV-tomato isolates as a major parent and a ToLCNDV-ridge gourd isolate as a minor parent with a breakpoint between 1,521–2,738 nucleotide positions. Similarly, DNA-B showed recombination between ToLCNDV isolate from a ridge gourd as a major parent and a sponge gourd as a minor parent (Table 2).
To ascertain the infectivity nature of the cloned viral infectious constructs, 10 different cucurbit plants (ash gourd, squash, pumpkin, bitter gourd, ridge gourd, snake gourd, cucumber, muskmelon, watermelon, and bottle gourd) were used for agro-inoculation study (Supplementary Figure 9). No symptoms were observed on any of the plants inoculated with DNA-A alone till 28 days post inoculation (dpi). However, when the bottle gourd, bitter gourd, cucumber, ridge gourd, and pumpkin plants were co-inoculated with DNA-A and DNA-B, mild mosaic symptoms were noticed (Figure 4). However, severe mosaic symptoms were observed on ash gourd, muskmelon, snake gourd, squash, and watermelon plants when co-inoculated with DNA-A and DNA-B. Interestingly, severe symptoms were observed on all the cucurbits species co-inoculated with DNA-A, DNA-B, and ToLCBB (Figure 4). Moreover, an additional symptom of leaf curling was developed on the bitter gourd, muskmelon, and squash plants co-inoculated with DNA-A, DNA-B, and ToLCBB. In addition to the elevated symptom severity, the reduced incubation period (of 2–4 dpi) and stunting were observed on the plants co-inoculated with DNA-A, DNA-B, and ToLCBB compared to DNA-A and DNA-B co-inoculated plants (Table 3). This phenomenon was noticed in all the cucurbits irrespective of the plant species tested (Supplementary Figure 10). The rate of infection on ash gourd and squash was found to be 100%, whereas the infectivity rate ranged between 45% and 85% on the other cucurbit crops (Table 3).

Figure 4. The cloned infectious viral constructs cause disease on various cucurbitaceous crops. The symptom appearance on the agro-inoculated cucurbits is at 28 dpi.
The presence of ToLCNDV genomic components and ToLCBB in the agro-inoculated plants were checked by conventional PCR using degenerate primers (Table 3). Further, qPCR using virus genome specific primers was employed to quantify the accumulation of ToLCNDV and ToLCBB in the systemic leaves of agro-inoculated plants. No viral DNA was detected in the mock or DNA-A alone inoculated plants whereas virus titer was detected in the co-inoculated plants at 28 dpi. A significant increase in the accumulation of DNA-A was observed in the co-inoculated plants of DNA-A + DNA-B and DNA-A + DNA-B + ToLCBB (Figure 5). Among these two treatments, the higher accumulation was observed in DNA-A + DNA-B + ToLCBB inoculated plants over DNA-A + DNA-B inoculated plants. Similar results were obtained in all inoculated cucurbits irrespective of the plant species tested.
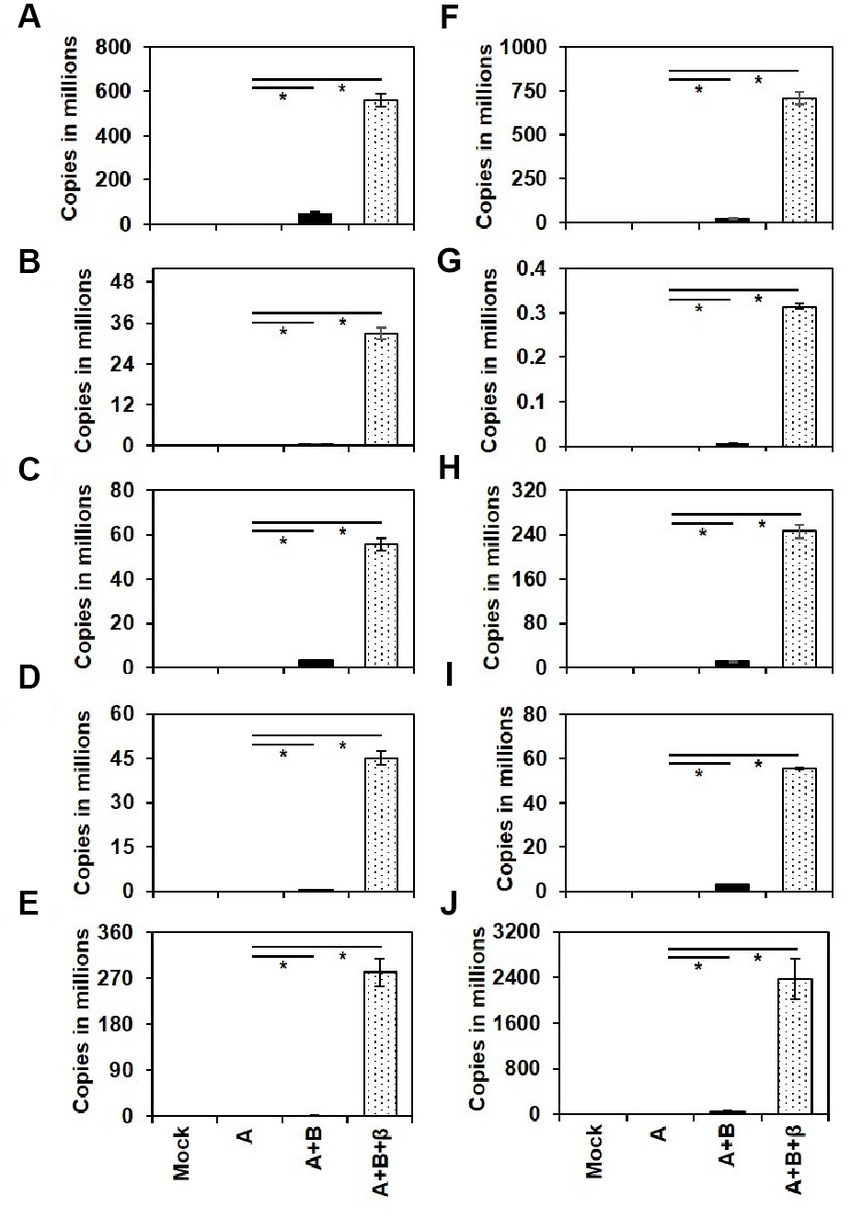
Figure 5. Viral DNA accumulation in the systemic leaves of the agro-inoculated cucurbits at 28 dpi. The cucurbit plants used are (A) ash gourd, (B) bottle gourd, (C) bitter gourd, (D) cucumber, (E) muskmelon, (F) pumpkin, (G) ridge gourd, (H) snake gourd, (I) squash and (J) watermelon. Each bar represents the mean of three individual replicates ± standard error and asterisks indicate statistical significance (p < 0.001) according to ANOVA F-test.
Among plant-infecting viruses, members of geminiviruses have emerged as a serious threat to cucurbit production globally. Initially, ToLCNDV was infecting tomato crops in India (Nagendran et al., 2019; Chakraborty and Kumar, 2021), but in recent decades it has widely expanded its host range to various vegetable crops such as chili (Kumar et al., 2015), okra (Venkataravanappa et al., 2012), potato (Jeevalatha et al., 2018), eggplant (Pratap et al., 2011), and several cucurbitaceous crops (Ito et al., 2008; Tiwari et al., 2010; Yamamoto et al., 2021; Kumari et al., 2022; Krishnan et al., 2023). Previously, the association of ToLCNDV with yellow stunt disease of ash gourd was reported from Northern India (Roy et al., 2013). Among various begomovirus species infecting cucurbits (Ito et al., 2008; Tiwari et al., 2010; Nagendran et al., 2017a; Sangeetha et al., 2018; Yamamoto et al., 2021; Kumari et al., 2022; Krishnan et al., 2023), Squash leaf curl China virus (SLCCNV) was reported with leaf curl and mosaic disease of ash gourd in India and Thailand, respectively (Sawangjit, 2009; Riyaz et al., 2013). Similarly, Samretwanich et al. (2000) have reported on the association of ToLCNDV with leaf yellowing disease of ash gourd in Thailand. In the present study, the association of ToLCNDV with this disease was characterized in ash gourd. The Koch’s postulates for this disease complex on various cucurbitaceous crops were satisfied. Furthermore, the influence of cognate DNA-B and non-cognate betasatellite on the disease development were also demonstrated.
The bipartite begomovirus (SLCCNV and ToLCNDV) complexes are associated with ash gourd diseases in India and Thailand (Samretwanich et al., 2000; Riyaz et al., 2013; Roy et al., 2013). Sequence analysis of the cloned genomes suggested the association of ToLCNDV with the mosaic disease of ash gourd in the Udumalpet region, India. Furthermore, several studies have documented the frequent association of ToLCNDV with mosaic disease of various cucurbits (excluding ash gourd) such as Coccinia grandis, Cucumis melo, Cucurbita pepo, Luffa cylindrica, Momordica charantia, Sechuimedule and Trichosanthes cucumerina, worldwide (Kumari et al., 2022). However, our study is the first report on the association of a begomovirus (ToLCNDV) with the mosaic disease of ash gourd. The cucurbit growing period likely coincides with the major tomato cultivation season, thus the potential spread of ToLCNDV from tomato to ash gourd cannot be ignored. This necessitates investigating the distribution of ToLCNDV across major ash gourd growing regions in the country. Collectively, the available evidence of such host range expansion by ToLCNDV to diverse plant families makes it one of the most devastating plant pathogens across the continents.
Recombination is a common phenomenon that occurs within and between virus species, and it plays a crucial role in the emergence and evolution of begomoviruses (Prasanna and Rai, 2007; Duffy and Holmes, 2008; George et al., 2015; Kumar and Chakraborty, 2018). In the present study, intraspecies recombination events were detected in the intergenic (IR), AC1, and BC1 regions. This result corroborates previous findings on these regions being recombination hotspots (Lefeuvre et al., 2007; Prasanna and Rai, 2007; George et al., 2015; Kumar et al., 2015, 2017). Moreover, the recombination event encompassing IR might have facilitated this ToLCNDV isolate in expanding its host range to ash gourd.
Koch’s postulates were confirmed for this disease complex on various cucurbitaceous crops including ash gourd. The DNA-A alone inoculated cucurbits were symptomless and no DNA-A was detected in the systemic leaves, whereas in the presence of cognate DNA-B component, mild symptoms were observed. This result highlights that this ToLCNDV isolate is a true bipartite begomovirus which requires DNA-B for its systemic movement in the tested cucurbitaceous crops. Our infectivity data is in agreement with previous findings of the indispensable nature of the DNA-B component in the pathogenicity of ToLCNDV (Padidam et al., 1995; Ranjan et al., 2014; Sangeeta Kumar et al., 2023). However, the cucurbits co-inoculated with DNA-A, DNA-B (cognate) and betasatellite (non-cognate) have developed severe symptoms with reduced latent period and multi-fold increased accumulation of DNA-A component (Figure 5; Table 3). This finding is consistent with the available literature, which demonstrated that the presence of betasatellite has resulted in enhanced symptom severity of ToLCNDV in tomato (Sivalingam and Varma, 2012; Jyothsna et al., 2013; Sangeeta Kumar et al., 2023). This data underscores the necessity of investigating the presence of betasatellite(s) with such disease complexes on cucurbitaceous crops. As this virus complex causes disease in various cucurbits, it can be utilized in identifying virus resistance genotypes among these cucurbitaceous crops.
The present study reports on the association of a bipartite begomovirus ToLCNDV with mosaic disease of ash gourd in India. The cloned genomic components were recombinant in nature. Furthermore, these cloned virus components cause disease in several cucurbitaceous crops including ash gourd, thereby fulfilling Koch’s postulates. Inoculation of DNA-A alone did not cause disease on cucurbits; however, the presence of either cognate DNA-B or non-cognate betasatellite resulted in enhanced symptom severity and helped begomovirus accumulation.
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/Supplementary material.
SV: Formal analysis, Methodology, Writing – original draft. PR: Methodology, Supervision, Writing – review & editing. KN: Methodology, Writing – review & editing, Formal analysis, Validation. NS: Supervision, Validation, Writing – review & editing. RK: Data curation, Formal analysis, Validation, Writing – review & editing. RS: Supervision, Validation, Writing – review & editing. TB: Supervision, Writing – review & editing. GK: Conceptualization, Data curation, Formal analysis, Methodology, Resources, Validation, Writing – review & editing.
The author(s) declare that no financial support was received for the research, authorship, and/or publication of this article.
The authors are thankful to the Science and Engineering Research Board (SERB), Department of Science and Technology, Government of India. We acknowledge V. G. Malathi for her guidance in conducting the research work.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2023.1268333/full#supplementary-material
Ashwathappa, K. V., Venkataravanappa, V., Reddy, C. N. L., and Reddy, M. K. (2020). Association of Tomato leaf curl New Delhi virus with mosaic and leaf curl disease of Chrysanthemum and its whitefly cryptic species. Ind. Phytopathol. 73, 533–542. doi: 10.1007/s42360-020-00214-1
Briddon, R. W., Bull, S. E., Mansoor, S., Amin, I., and Markham, P. G. (2002). Universal primers for the PCR-mediated amplification of DNA β: a molecule associated with some monopartite begomoviruses. Mol. Biotechnol. 20, 315–318. doi: 10.1385/MB:20:3:315
Brown, J. K., Zerbini, F. M., Navas-Castillo, J., Moriones, E., Ramos-Sobrinho, R., Silva, J. C. F., et al. (2015). Revision of Begomovirus taxonomy based on pairwise sequence comparisons. Arch. Virol. 160, 1593–1619. doi: 10.1007/s00705-015-2398-y
Bull, S. E., Briddon, R. W., and Markham, P. G. (2003). Universal primers for the PCR-mediated amplification of DNA 1: a satellite-like molecule associated with begomovirus-DNA β complexes. Mol. Biotechnol. 23, 83–86. doi: 10.1385/MB:23:1:83
Chakraborty, S., and Kumar, M. (2021). Tomato leaf curl New Delhi virus (Geminiviridae). Vol. 3. 4th edn. eds. D. H. Bamford and M. Zuckerman. San Diego, CA: Academic Press, 749–760.
Chang, H. H., Ku, H. M., Tsai, W. S., Chien, R. C., and Jan, F. J. (2010). Identification and characterization of a mechanical transmissible begomovirus causing leaf curl on oriental melon. Eur. J. Plant Pathol. 127, 219–228. doi: 10.1007/s10658-010-9586-0
Chatchawankanphanich, O., and Maxwell, D. P. (2002). Tomato leaf curl Karnataka virus from Bangalore, India, appears to be a recombinant begomovirus. Phytopathology 92, 637–645. doi: 10.1094/PHYTO.2002.92.6.637
Chen, H., Nelson, R. S., and Sherwood, J. L. (1994). Enhanced recovery of transformants of Agrobacterium tumefaciens after freeze-thaw transformation and drug selection. Biotechniques 16, 664–8, 670.
Damicone, J. P., Edelson, J. V., Sherwood, J. L., Myers, L. D., and Motes, J. E. (2007). Effects of border crops and intercrops on control of cucurbit virus diseases. Plant Dis. 91, 509–516. doi: 10.1094/PDIS-91-5-0509
Duffy, S., and Holmes, E. C. (2008). Phylogenetic evidence for rapid rates of molecular evolution in the single-stranded DNA begomovirus tomato yellow leaf curl virus. J. Virol. 82, 957–965. doi: 10.1128/JVI.01929-07
Edgar, R. C. (2004). MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 32, 1792–1797. doi: 10.1093/nar/gkh340
Gemert, J., Martijn, S., Candresse, T., Bottex, B., and Sybren, A. D. (2020). Pest survey card on tomato leaf curl New Delhi virus. EFSA Support. Publ. 1904, 7–17. doi: 10.2903/sp.efsa.2020.EN-1904
George, B., Alam, C. M., Kumar, R. V., Prabu, G., and Chakraborty, S. (2015). Potential linkage between compound microsatellites and recombination in geminiviruses: evidence from comparative analysis. Virology 482, 41–50. doi: 10.1016/j.virol.2015.03.003
Hothorn, T., Bretz, F., and Westfall, P. (2008). Simultaneous inference in general parametric models. Biom. J. 50, 346–363. doi: 10.1002/bimj.200810425
Hussain, M., Mansoor, S., Iram, S., Zafar, Y., and Briddon, R. W. (2004). First report of tomato leaf curl New Delhi virus affecting chilli pepper in Pakistan. Plant Pathol. 53:794. doi: 10.1111/j.1365-3059.2004.01073.x
Ito, T., Sharma, P., Kittipakorn, K., and Ikegami, M. (2008). Complete nucleotide sequence of a new isolate of tomato leaf curl New Delhi virus infecting cucumber, bottle gourd and muskmelon in Thailand. Arch. Virol. 153, 611–613. doi: 10.1007/s00705-007-0029-y
Jeevalatha, A., Kaundal, P., Kumar, R., Raigond, B., Kumar, R., Sharma, S., et al. (2018). Optimized loop-mediated isothermal amplification assay for tomato leaf curl New Delhi virus-[potato] detection in potato leaves and tubers. Eur. J. Plant Pathol. 150, 565–573. doi: 10.1007/s10658-017-1300-z
Jyothsna, P., Haq, Q. M. I., Singh, P., Sumiya, K. V., Praveen, S., Rawat, R., et al. (2013). Infection of tomato leaf curl New Delhi virus (ToLCNDV), a bipartite begomovirus with betasatellites, results in enhanced level of helper virus components and antagonistic interaction between DNA B and betasatellites. Appl. Microbiol. Biotechnol. 97, 5457–5471. doi: 10.1007/s00253-012-4685-9
Krishnan, N., Kumari, S., Chaubey, T., Kumar, R. V., Chinnappa, M., Dubey, V., et al. (2023). Study on mosaic disease of sponge gourd (Luffa cylindrica L.) caused by tomato leaf curl New Delhi virus. J. Plant Pathol. 105, 573–580. doi: 10.1007/s42161-023-01325-0
Kumar, R. V. (2019). Plant antiviral immunity against Geminiviruses and viral counter-defense for survival. Front. Microbiol. 10:1460. doi: 10.3389/fmicb.2019.01460
Kumar, R. V., and Chakraborty, S. (2018). “Evolution and emergence of geminiviruses: reasons and consequences” in Plant viruses: Diversity, interaction and management. eds. R. K. Gaur, S. M. P. Khurana, and Y. L. Dorokhov (Boca Raton: CRC Press), 97–116.
Kumar, R. V., Prasanna, H. C., Singh, A. K., Ragunathan, D., Garg, G. K., and Chakraborty, S. (2017). Molecular genetic analysis and evolution of begomoviruses and betasatellites causing yellow mosaic disease of bhendi. Virus Genes 53, 275–285. doi: 10.1007/s11262-016-1414-y
Kumar, R. V., Singh, A. K., Singh, A. K., Yadav, T., Basu, S., Kushwaha, N., et al. (2015). Complexity of begomovirus and betasatellite populations associated with chilli leaf curl disease in India. J. Gen. Virol. 96, 3143–3158. doi: 10.1099/jgv.0.000254
Kumar, S., Stecher, G., and Tamura, K. (2016). MEGA7: molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol. Biol. Evol. 33, 1870–1874. doi: 10.1093/molbev/msw054
Kumari, S., Nagendran, K., and Pandey, K. K. (2022). “Emergence of begomoviruses in cucurbits as a menace for its cultivation” in Geminivirus: Detection, diagnosis and management. eds. R. K. Gaur, P. Sharma, and H. Czosnek (London, UK: Academic Press), 107–124.
Lefeuvre, P., Lett, J. M., Reynaud, B., and Martin, D. P. (2007). Avoidance of protein fold disruption in natural virus recombinants. PLoS Pathog. 3:e181. doi: 10.1371/journal.ppat.0030181
Martin, D. P., Murrell, B., Golden, M., Khoosal, A., and Muhire, B. (2015). RDP4: detection and analysis of recombination patterns in virus genomes. Virus Evol. 1:vev003. doi: 10.1093/ve/vev003
Maruthi, M. N., Rekha, A. R., Cork, A., Colvin, J., Alam, S. N., and Kader, K. A. (2005). First report of tomato leaf curl New Delhi virus infecting tomato in Bangladesh. Plant Dis. 89:1011. doi: 10.1094/PD-89-1011C
Melgarejo, T. A., Kon, T., Rojas, M. R., Paz-Carrasco, L., Zerbini, F. M., and Gilbertson, R. L. (2013). Characterization of a new world monopartite begomovirus causing leaf curl disease of tomato in Ecuador and Peru reveals a new direction in geminivirus evolution. J. Virol. 87, 5397–5413. doi: 10.1128/JVI.00234-13
Moriones, E., Praveen, S., and Chakraborty, S. (2017). Tomato leaf curl New Delhi virus: an emerging virus complex threatening vegetable and fiber crops. Viruses 9:264. doi: 10.3390/v9100264
Muhire, B. M., Varsani, A., and Martin, D. P. (2014). SDT: a virus classification tool based on pairwise sequence alignment and identity calculation. PloS One 9:e108277. doi: 10.1371/journal.pone.0108277
Nagendran, K., Mohankumar, S., Aravintharaj, R., Balaji, C. G., Manoranjitham, S. K., Singh, A. K., et al. (2017a). The occurrence and distribution of major viruses infecting cucurbits in Tamil Nadu state, India. Crop Prot. 99, 10–16. doi: 10.1016/j.cropro.2017.05.006
Nagendran, K., Mohankumar, S., Mohammed Faisal, P., Bagewadi, B., and Karthikeyan, G. (2017b). Molecular evidence for the occurrence of tomato leaf curl New Delhi virus on chayote (Sechium edule) in southern India. Virus Dis. 28, 425–429. doi: 10.1007/s13337-017-0403-7
Nagendran, K., Venkataravanappa, V., Chauhan, N. S., Kodandaram, M. H., Rai, A. B., Singh, B., et al. (2019). Viral diseases: a threat for tomato cultivation in indo-Gangetic eastern plains of India. J. Plant Pathol. 101, 15–22. doi: 10.1007/s42161-018-0124-9
Padidam, M., Beachy, R. N., and Fauquet, C. M. (1995). Tomato leaf curl geminivirus from India has a bipartite genome and coat protein is not essential for infectivity. J. Gen. Virol. 76, 25–35. doi: 10.1099/0022-1317-76-1-25
Prasanna, H. C., and Rai, M. (2007). Detection and frequency of recombination in tomato-infecting begomoviruses of south and Southeast Asia. Virol. J. 4:111. doi: 10.1186/1743-422X-4-111
Pratap, D., Kashikar, A. R., and Mukherjee, S. K. (2011). Molecular characterization and infectivity of a tomato leaf curl New Delhi virus variant associated with newly emerging yellow mosaic disease of egg-plant in India. Virol. J. 8:305. doi: 10.1186/1743-422X-8-305
R Core Team. (2023). R development Core team R: A language and environment for statistical computing 2023. R Core Team: Vienna, Austria.
Ranjan, P., Singh, A. K., Kumar, R. V., Basu, S., and Chakraborty, S. (2014). Host-specific adaptation of diverse betasatellites associated with distinct Indian tomato-infecting begomoviruses. Virus Genes 48, 334–342. doi: 10.1007/s11262-013-1031-y
Riyaz, S. M., Deepan, S., Dharanivasan, G., Jesse, M. I., Raja, M., and Kathiravan, K. (2013). The first report on a variant of squash leaf curl China virus (SLCCNV) infecting Benincasa hispida in India. New Dis. Rep. 28:20. doi: 10.5197/j.2044-0588.2013.028.020
Rouhibakhsh, A., and Malathi, V. G. (2005). Severe leaf curl disease of cowpea–a new disease of cowpea in northern India caused by Mungbean yellow mosaic India virus and a satellite DNA β. Plant Pathol. 54:259. doi: 10.1111/j.1365-3059.2005.01139.x
Roy, B., Chakraborty, P., and Ghosh, A. (2021). How many begomovirus copies are acquired and inoculated by its vector, whitefly (Bemisia tabaci) during feeding? PloS One 16:e0258933. doi: 10.1371/journal.pone.0258933
Roy, A., Spoorthi, P., Panwar, G., Bag, M. K., Prasad, T. V., Kumar, G., et al. (2013). Molecular evidence for occurrence of tomato leaf curl New Delhi virus in ash gourd (Benincasa hispida) germplasm showing a severe yellow stunt disease in India. Ind. J. Virol. 24, 74–77. doi: 10.1007/s13337-012-0115-y
Samretwanich, K., Chiemsombat, P., Kittipakorn, K., and Ikegami, M. (2000). Yellow leaf disease of cantaloupe and wax gourd from Thailand caused by tomato leaf curl virus. Plant Dis. 84:200. doi: 10.1094/PDIS.2000.84.2.200C
Sangeeta Kumar, R. V., Yadav, B. K., Bhatt, B. S., Krishna, R., Krishnan, N., Karkute, S. G., et al. (2023). Diverse begomovirus-betasatellite complexes cause tomato leaf curl disease in the western India. Virus Res. 328:199079. doi: 10.1016/j.virusres.2023.199079
Sangeetha, B., Malathi, V. G., Alice, D., Suganthy, M., and Renukadevi, P. (2018). A distinct seed-transmissible strain of tomato leaf curl New Delhi virus infecting chayote in India. Virus Res. 258, 81–91. doi: 10.1016/j.virusres.2018.10.009
Sawangjit, S. (2009). The complete nucleotide sequence of squash leaf curl China virus-[wax gourd] and its phylogenetic relationship to other Geminiviruses. Sci. Asia 35, 131–136. doi: 10.2306/scienceasia1513-1874.2009.35.131
Sharma, D., Kulshreshtha, A., Roshan, P., and Hallan, V. (2019). Molecular characterization and infectivity analysis of a bipartite begomovirus associated with cotton leaf curl Multan betasatellite naturally infecting Rumex nepalensis in northern India. J. Plant Pathol. 101, 935–941. doi: 10.1007/s42161-019-00295-6
Singh, A. K., Yadav, B. K., Krishna, R., Kumar, R. V., Mishra, G. P., Karkute, S. G., et al. (2021). Bhendi yellow vein mosaic virus and bhendi yellow vein mosaic betasatellite cause enation leaf curl disease and alter host phytochemical contents in okra. Plant Dis. 105, 2595–2600. doi: 10.1094/PDIS-12-20-2655-RE
Sivalingam, P. N., and Varma, A. (2012). Role of betasatellite in the pathogenesis of a bipartite begomovirus affecting tomato in India. Arch. Virol. 157, 1081–1092. doi: 10.1007/s00705-012-1261-7
Srivastava, K. M., Hallan, V., Raizada, R. K., Chandra, G., Singh, B. P., and Sane, P. V. (1995). Molecular cloning of Indian tomato leaf curl vims genome following a simple method of concentrating the supercoiled replicative form of viral DNA. J. Virol. Methods 51, 297–304. doi: 10.1016/0166-0934(94)00122-W
Thresh, J. M. (2006). “Crop viruses and virus diseases: a global perspective” in Virus diseases and crop bio security. eds. J. I. Cooper, T. Kuehne, and V. P. Polischuk (Dordrecht, The Netherlands: Springer), 9–32.
Tiwari, A. K., Sharma, P. K., Khan, M. S., Snehi, S. K., Raj, S. K., and Rao, G. P. (2010). Molecular detection and identification of tomato leaf curl New Delhi virus isolate causing yellow mosaic disease in bitter gourd (Momordica charantia), a medicinally important plant in India. Med. Plants. 2, 117–123. doi: 10.5958/j.0975-4261.2.2.018
Venkataravanappa, V., Lakshminarayana, R. C. N., Salil, J., and Krishna, R. M. (2012). Molecular characterization of distinct bipartite begomovirus infecting bhendi (Abelmoschus esculentus L.) in India. Virus Genes 44, 522–535. doi: 10.1007/s11262-012-0732-y
Yamamoto, H., Wakita, Y., Kitaoka, T., Fujishiro, K., Kesumawati, E., and Koeda, S. (2021). Southeast Asian isolate of the tomato leaf curl New Delhi virus shows higher pathogenicity against tomato and cucurbit crops compared to that of the Mediterranean isolate. Hort. J. 90, 314–325. doi: 10.2503/hortj.UTD-269
Keywords: begomovirus, pathogenesis, evolution, agro-inoculation, cucurbits, qPCR
Citation: Vignesh S, Renukadevi P, Nagendran K, Senthil N, Kumar RV, SwarnaPriya R, Behera TK and Karthikeyan G (2023) A distinct strain of tomato leaf curl New Delhi virus that causes mosaic disease in ash gourd and other cucurbitaceous crops. Front. Microbiol. 14:1268333. doi: 10.3389/fmicb.2023.1268333
Received: 27 July 2023; Accepted: 05 October 2023;
Published: 26 October 2023.
Edited by:
Jesús Navas-Castillo, CSIC, SpainReviewed by:
Nagaraju Yalavarthi, National Bureau of Agriculturally Important Microorganisms (ICAR), IndiaCopyright © 2023 Vignesh, Renukadevi, Nagendran, Senthil, Kumar, SwarnaPriya, Behera and Karthikeyan. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: G. Karthikeyan, YWdyaWthcnRoaTIwMDNAZ21haWwuY29t
†ORCID: G. Karthikeyan, https://orcid.org/0000-0002-7587-6643
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.