- 1State Key Laboratory of Bio-Control, Southern Marine Science and Engineering Guangdong Laboratory (Zhuhai), School of Life Sciences, Sun Yat-sen University, Guangzhou, China
- 2Laboratory for Marine Fisheries Science and Food Production Processes, Qingdao National Laboratory for Marine Science and Technology, Qingdao, China
- 3Department of Pediatrics, The Third Affiliated Hospital of Sun Yat-sen University, Guangzhou, China
- 4Guangdong Litai Pharmaceutical Co., Ltd., Jieyang, China
Introduction: Bacterial metabolic environment influences antibiotic killing efficacy. Thus, a full understanding for the metabolic resistance mechanisms is especially important to combat antibiotic-resistant bacteria.
Methods: Isobaric tags for relative and absolute quantification-based proteomics approach was employed to compare proteomes between ceftazidime-resistant and -sensitive Edwarsiella tarda LTB4 (LTB4-RCAZ and LTB4-S, respectively).
Results: This analysis suggested the possibility that the ceftazidime resistance mediated by depressed glucose is implemented through an inefficient metabolic flux from glycolysis, the pyruvate cycle, glutamate metabolism to purine metabolism. The inefficient flux was demonstrated by the reduced expression of genes and the decreased activity of enzymes in the four metabolic pathways. However, supplement upstream glucose and downstream guanosine separately restored ceftazidime killing, which not only supports the conclusion that the inefficient metabolic flux is responsible for the resistance, but also provides an effective approach to reverse the resistance. In addition, the present study showed that ceftazidime is bound to pts promoter in E. tarda.
Discussion: Our study highlights the way in fully understanding metabolic resistance mechanisms and establishing metabolites-based metabolic reprogramming to combat antibiotic resistance.
Introduction
Antibiotic resistance is a global health challenge involving human health, food safety, and the sustainable development of animal farming. A full understanding of antibiotic resistance mechanisms is especially important to explore ways for controlling these antibiotic-resistant pathogens. Four widely accepted antibiotic resistance mechanisms include decreased membrane permeability, increased abundance of efflux pumps, hydrolase/modifying enzymes (especially beta-lactamase), and modifications of the molecular targets (Peng et al., 2019; Tong et al., 2021). Besides them, metabolism-based resistance has been recently documented (Peng et al., 2015a,b; Lopatkin et al., 2021; Fan et al., 2023; Jiang et al., 2023), indicating that metabolic adaptation may represent a class of resistance mechanisms by which cells alter their metabolic response to mitigate downstream toxic aspects of antibiotic lethality (Lopatkin et al., 2021). A metabolic state-reprogramming system can be developed to reverse antibiotic resistance (Peng et al., 2015a,b; Cheng et al., 2019; Zhao et al., 2021; Zhou et al., 2022; Chen et al., 2023; Lei et al., 2023). Therefore, further exploration of the metabolic mechanism of antibiotic resistance is helpful in developing previously unknown approaches to eliminate antibiotic-resistant pathogens (Peng et al., 2023).
Ceftazidime is a third-generation cephalosporin administered intravenously or intramuscularly. This drug has a broad spectrum of activity against Gram-positive and Gram-negative aerobic bacteria, particularly Pseudomonas aeruginosa and Enterobacteriaceae (including beta-lactamase-positive strains) (Richards and Brogden, 1985). However, the long-term use and wide use of ceftazidime cause the development of drug resistance in clinical isolates (Sanz-García et al., 2018; Fröhlich et al., 2022). Not only in humans but also in untreated wastewater environments and aquaculture, ceftazidime-resistant bacteria are widespread (Adelowo et al., 2018; Salgueiro et al., 2021; Moya-Salazar et al., 2022), which poses a challenge to control infection caused by the resistant pathogens in hospitals and aquaculture. Recently, we have used gas chromatography-mass spectrometry (GC-MS) to profile the metabolic state of ceftazidime-resistant V. alginolyticus and identified an altered and inefficient pyruvate cycle (P cycle), increased biosynthesis of fatty acids, deregulation of Na+-translocating NADH:ubiquinone oxidoreductase (Na+-NQR), and low membrane proton motive force (PMF) as the greatest metabolic modulation in the resistance (Liu et al., 2019). Meanwhile, we have also shown that intracellular reactive oxygen species (ROS) production was lower in ceftazidime-resistant LTB4 than in its isogenic ceftazidime-sensitive LTB4, which attributed ROS to inactivation of the P cycle (Su et al., 2018; Ye et al., 2018a). The resistance can be reversed by exogenous Fe3+, which promotes the P cycle and increases ROS production (Ye et al., 2021). These findings identify an inactivated P cycle and reduced PMF and ROS as the consequences of ceftazidime resistance. However, why the P cycle was inefficient and whether its other downstream metabolic pathways contributed to the resistance are absent.
Isobaric tags for relative and absolute quantification (iTRAQ) are powerful tools to characterize global protein changes in antibiotic-resistant bacteria (Li et al., 2018; Ye et al., 2018b). Here, the iTRAQ-based approach was used to investigate proteomes in ceftazidime-resistant and ceftazidime-sensitive Edwarsiella tarda LTB4 (LTB4-RCAZ and LTB4-S, respectively). In combination with molecular biology and biochemistry technologies, the inefficiency of glycolysis, the P cycle, and purine metabolism was documented. This is attributed to depressed glucose. Finally, the mechanism by which glucose was depressed was explored.
Materials and methods
Bacterial strains and cultures
The bacteria used in the present study, E. tarda LTB4 and Escherichia coli K12 BW25113 (K12), were from the collection of our laboratory. LTB4-RCAZ originated from LTB4 and was sequentially propagated in an LB medium with 1/2 MIC of ceftazidime (Ye et al., 2021). Single colonies were cultured in fresh 50 ml TSB media placed into 250 ml flasks at 30°C, with 200 rpm shaking for 24 h. The overnight cultures were collected, washed three times with saline, and suspended in M9 medium to 0.2 of OD600 nm when metabolites or/and antibiotics were added if desired. These bacterial cells were cultured at 37°C (K12) or 30°C (LTB4 and LTB4-RCAZ) at 200 rpm for 6 h.
Sample preparation and iTRAQ labeling quantitative proteomics
Sample preparation and iTRAQ labeling analysis were described previously without any modifications (Ye et al., 2018a). In brief, 100 μg of proteins per sample were reduced by 200 mM Tris (2-carboxyethyl) phosphine and alkylated with 25 mM iodoacetamide. These proteins were digested by Promega trypsin and labeled with iTRAQ labeling kits. Liquid chromatography-tandem/mass spectrometry (LC-MS/MS) was adopted and performed on an AB SciexTripleTOF 5600 mass spectrometer (AB SCIEX; Concord, ON, Canada) with a NanoAcquity UPLC (ultraperformance liquid chromatography) (Waters, Milford, MA, USA) system. All LC–MS/MS data were processed using ProteinPilot version 4.5 software (Applied Biosystems, USA). Protein identification was performed by the MS/MS data searching against the E. tadar EIB202 database with at least two peptides matched. Differential abundances of proteins were selected using iTRAQ average reporter ion ratio threshold values of ≥1.5 (increased) or ≤ 0.667 (decreased), with both p-values between two biological replicates <0.05. Two biological repeat correlations were analyzed using IBM SPSS Statistics 19 software. The differential expression of proteins in the GO annotation and KEGG pathways was analyzed with OmicsBean (http://www.omicsbean.cn/). The functions of each protein were categorized according to the GO annotation based on the biological process hierarchy. The protein-protein interaction network was implemented using STRING and Cytoscape software. iPath analysis was carried out using a web-based tool (http://pathways.embl.de) for the visualization and analysis of cellular pathways.
Measurement of enzymatic activity
The activity of glucokinase (HK), 6-phosphofructokinase I (PFK), pyruvate kinase (PK), and glutamic-pyruvic transaminase (GPT) was measured according to kit instructions (Solarbio, Beijing, China). The activity of pyruvate dehydrogenase (PDH), α-ketoglutarate dehydrogenase (KGDH), succinate dehydrogenase (SDH), and malate dehydrogenase (MDH) was detected as described previously (Cheng et al., 2018). In brief, bacteria were cultured in an M9 medium with or without glucose at 30°C with shaking at 200 rpm for 6 h. Cells were collected, immediately snap-frozen in liquid nitrogen, and stored at−80°C. The cells were resuspended with 1 ml of PBS and disrupted by sonic oscillation for 10 min (200 W total power with 35% output, 2 s pulse, and 3 s pause over ice). The protein concentration of the supernatant was quantified with a bicinchoninic acid (BCA) kit (Beyotime, P0009, China), and the final protein concentration for each sample was diluted to the same concentration. The activity of glucokinase (HK), 6-phosphofructokinase I (PFK), and pyruvate kinase (PK) was determined at 340 nm using commercial assay kits (Solarbio, China; BC0745, BC0535, and BC0545, respectively). The activity of glutamic-pyruvic transaminase (GPT) was determined at 505 nm using a glutamic-pyruvic transaminase activity assay kit (Solarbio BC1555, China). For measurement of PDH and KGDH, a reaction mixture containing 0.15 mM 3-(4,5-dimethyl-2-thiazolyl)-2,5-diphenyl- 2H-tetrazolium bromide (MTT), 2.5 mM MgCl2, 6.5 mM phenazine methosulfate (PMS), 0.2 mM thiamine PPi (TPP), and 80 mM sodium pyruvate/alpha-ketoglutaric acid potassium salt was used. Another reaction mixture containing 0.15 mM MTT, 2.5 mM MgCl2, 13 mM PMS, and 80 mM sodium succinate/sodium malate was used for SDH and MDH activity measurement. Then, the reaction mixtures were incubated at 37°C for 30 min for MDH, PDH, and KGDH and 5 min for SDH and detected at 562 nm for colorimetric readings. Absorbance was measured using a microplate reader (Epoch2, BioTek Instruments, Inc., United States). All experiments were normalized by the protein concentration. Experiments were repeated in at least three independent biological replicates.
Quantitative real-time PCR
To investigate the effect of glucose on gene expression levels, quantitative real-time PCR (qRT-PCR) was performed as previously described (Cheng et al., 2017). In brief, after incubation in M9 with or without glucose at 30°C with 200 rpm for 6 h, the cells were collected and adjusted to 1.0 OD600 nm. Total RNA was isolated from 1 ml cell samples using TRIzol reagent (Invitrogen Life Technologies). cDNA was obtained from 1 μg of total RNA, and reverse transcription was performed according to a PrimeScript RT reagent kit with gDNA Eraser (Guangzhou IGE Biotechnology Ltd., China). The primers used for qRT-PCR are listed in Supplementary Table 1, where the 16S rRNA gene served as an internal control. The qRT-PCR was performed in 384-well plates with the SYBR Green Premix Pro Taq HS qPCR Kit (Guangzhou IGE Biotechnology Ltd., China) at a total volume of 10 ml. The reaction mixtures were run on a LightCycler 480 system (Roche, Germany). The cycling parameter values were set as follows: 95°C for 30 s to activate the polymerase, 40 cycles of 95°C for 5 s, and 58°C for 30 s. Fluorescence measurements were performed at 75°C for 1 s during each cycle. Cycling was terminated at 95°C with a calefactive velocity of 0.11°C s−1 to obtain a melting curve. Data were calculated as relative mRNA expression compared to the glucose group without the endogenous reference 16S rRNA gene.
Antibiotic-bactericidal assay
As previously described, an antibiotic bactericidal assay was performed (Jiang et al., 2020) in M9 with or without metabolites and/or antibiotics and incubated at 30°C with 200 rpm. After 6 h, 100 μl of samples was 10-fold continuously diluted, and 5 μl of each dilution was aliquoted on the 2% TSB agar plates and cultured at 30°C for ~24 h, and CFU per ml was determined. The percent survival was determined by dividing the CFU obtained from the treated sample by the CFU obtained from the control.
Measurement of glucose content
A series of experiments was performed according to the kit instructions (Solarbio, China; BC2505). In brief, bacteria were incubated in M9 with or without glucose at 30°C with 200 rpm for 6 h. Cells were collected and adjusted to 1.0 OD600 nm for subsequent assays. An aliquot of 10 ml cell samples was disrupted by sonic oscillation for 10 min (200 W total power with 35% output, a 2 s pulse, and a 3 s pause over ice). A portion of the supernatant was used to determine protein content using a bicinchoninic acid (BCA) kit (Beyotime, P0009, China). At the same time, the others were used for 505 nm absorbance measurement using a microplate reader (Epoch2, BioTek Instruments, Inc., United States). All experiments were normalized by the protein concentration.
Binding of ceftazidime to pts promoter
As previously described (Jiang et al., 2023), PCR products of pts promoter fragments were purified using a PCR purification kit (Sangon Biotech, Shanghai, China), and their concentration was quantified. Various concentrations of DNA, ranging from 100 to 3,200 ng, were mixed with 10 μl of ceftazidime with a concentration of 100 mg/ml, and finally, the total volume was made up to 100 μl with Tris–HCl buffer at 37°C for 3 h. A total of 20 tubes were pooled and precipitated by 1:1 isopropanol for 10 min and centrifuged at 14,000 g for 10 min. The precipitation was washed three times with 75% ethanol and dissolved in 10 μl of Tris–HCl buffer for subsequent assays. Overnight Escherichia coli K12 cultures were collected and washed with a saline solution. The cultures were adjusted to an OD600 of 0.6 with M9 medium and diluted to 1:100,000. Then, 50 μl of bacteria was incubated with or without pst promoter precipitated by isopropanol as described above at 30°C with shaking at 200 rpm for 6 h. CFU per milliliter was determined as described above.
Results
Differential abundances of proteins identified by itraq labeling technology
To understand E. tarda resistance to ceftazidime at the protein level, LTB4-RCAZ was generated from LTB4-S. There was a 16-fold difference between LTB4-RCAZ and LTB4-S (Supplementary Figure 1A). Protein samples of the two strains were analyzed using iTRAQ proteomics technology. In total, 2,013 proteins were identified in each sample. Linear correction was calculated with 0.81 between all proteins in LTB4-S and 0.911 between all proteins in LTB4-RCAZ (Supplementary Figure 1B), suggesting the high quality of the iTRAQ analysis. Among the 2,013 proteins, 150 showed differential abundances. Among them, 91 proteins were upregulated and 59 were downregulated (Supplementary Tables 2, 3).
Gene Ontology (GO) enrichment analysis was carried out on the upregulated and downregulated expressed proteins to classify molecular functions, biological processes, and KEGG pathways. For the analysis of molecular functions, proteins that were upregulated and downregulated were enriched in 16 and 11 genes, respectively. Almost all of them are involved in molecular activity and binding (Figure 1A). For the analysis of cellular components, more cellular components were documented in the upregulated proteins than in the downregulated proteins. Specifically, 11 were enriched (mostly belonging to organelles and protein complexes) in the upregulated proteins, while 2 (periplasmic space and extracellular region) were found in the downregulated proteins (Figure 1B). For the analysis of biological processes, equal numbers (20) were enriched in both the upregulated and downregulated proteins, but the involved biological processes were differential. The biological processes enriched by the upregulated proteins mostly belong to metabolic processes, ion homeostasis, and iron transport, whereas the biological processes enriched by the downregulated proteins mostly contribute to metabolic and catabolic processes. Although the metabolic processes of the general and detailed items overlap, they are not the same. Specifically, the enriched metabolic processes in the upregulated proteins mostly include macromolecule, protein, and peptide metabolic processes, whereas those in the downregulated proteins mostly contain nucleobase and amino acid metabolic processes, where, importantly, most contribute to purine metabolism (Figure 1C). These results indicate that a global change is determined in LTB4-RCAZ, where downregulated purine metabolism is the most characteristic feature of the strain.
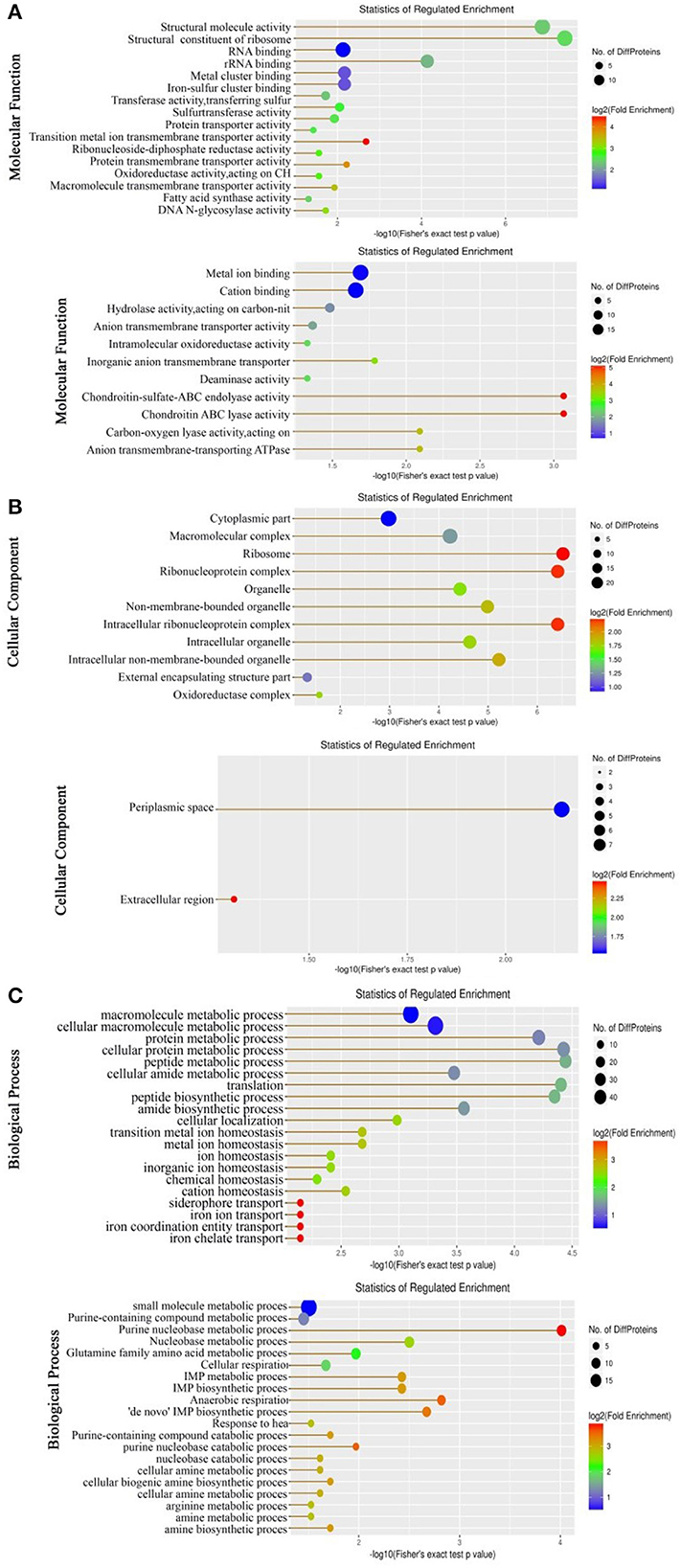
Figure 1. Molecular functional (A), cellular component (B), and biological process (C) of the differential abundance of proteins using GO (Gene Ontology) analysis. Up, Upregulation proteins. Down, Downregulation proteins. In biological processes, there are 10 pathways upregulated that are related to iron ions. Proteins in these 9 pathways were NDH, ETAC_08255 (HMUS), FECA, and ETAC_08250 (outer membrane receptor for ferrienterochelin and colicins). These four proteins cannot enrich any KEGG pathway.
Protein-protein interaction analysis and pathway enrichment
Protein-protein interaction (PPI) can help study the biological pathways and signal networks of cells (Ayub and Naveed, 2022). The 150 differential abundances of proteins were used to construct a PPI network using the web-based tool STRING. Among them, 53 were matched in the databases, resulting in the creation of a network based on seven functional categories, where 3 and 4 were upregulated and downregulated, respectively. Specifically, the ribosome, sulfur relay system, and protein export were upregulated, whereas purine metabolism, butanoate metabolism, the two-component system, and the TCA cycle were downregulated (Figure 2A). These were consistent with the data from the KEGG pathway analysis, except for the more reduced microbial metabolism in diverse environments and the biosynthesis of antibiotics in the KEGG pathway analysis (Figure 2B). Therefore, decreased glucose causes the negative modulation of central carbon metabolism and its downstream nucleotide metabolism, especially purine metabolism.
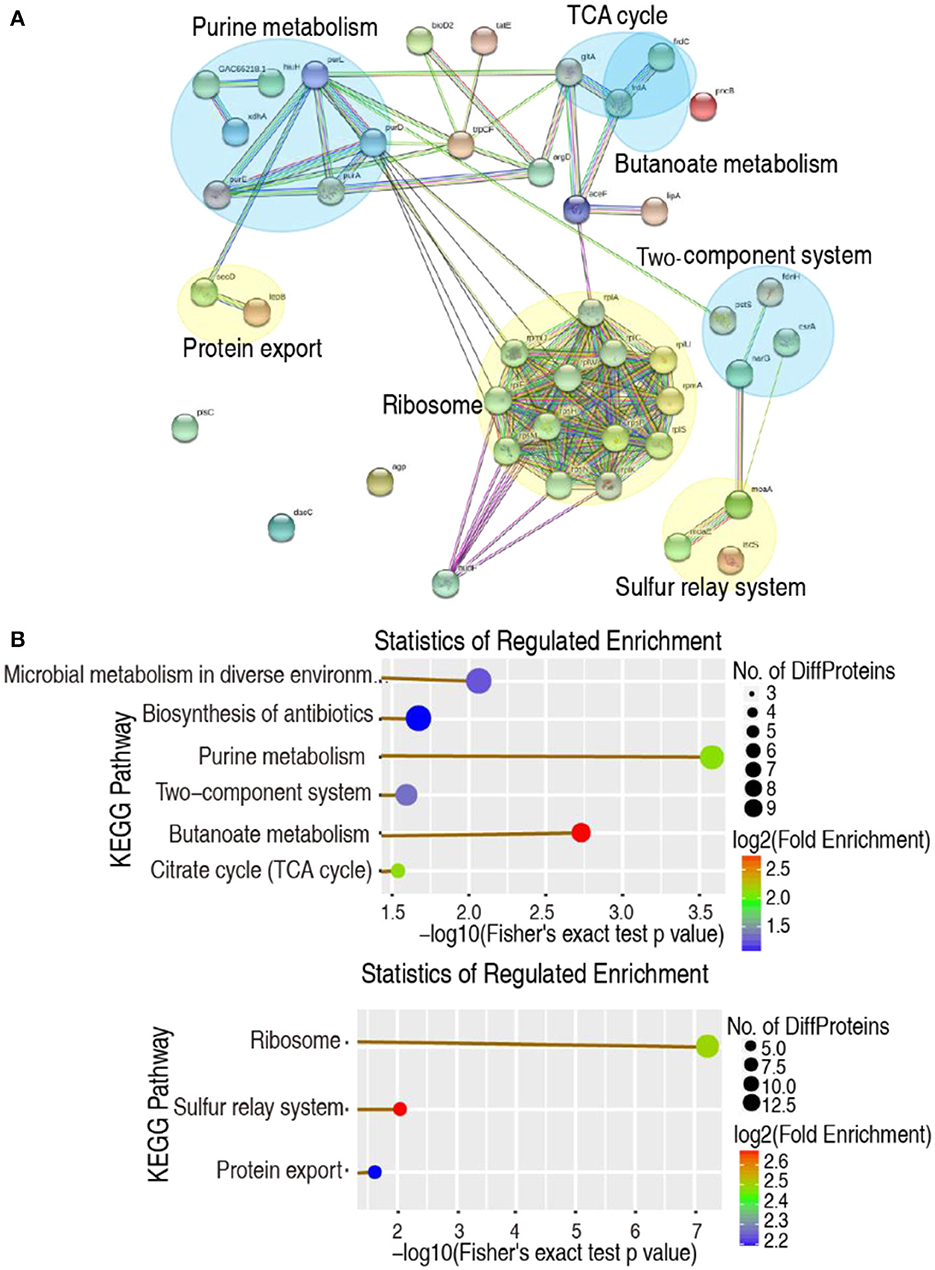
Figure 2. Functional proteins associated with metabolism. (A) The predicted physical and functional network of protein–protein interaction. Yellow, Upregulation pathway. Blue, Downregulation pathway. (B) KEGG function enrichment of metabolic pathway. Upper, Downregulation pathway. Lower, Upregulation pathway (company give-out).
Global overview and key metabolic pathway summary
Interactive Pathways Explorer (iPath) is a web-based tool for the visualization, analysis, and customization of various pathway maps (Darzi et al., 2018). Therefore, we used iPath to display a comparative global pathway analysis between LTB4-S and LTB4-RCAZ for a better insight into the effects of ceftazidime resistance on global metabolism. As shown in Figure 3A, red and green lines represent upregulated and downregulated pathways, respectively, in LTB4-RCAZ. Generally, carbohydrate metabolism, energy metabolism (especially the TCA cycle), and nucleotide metabolism were reduced. Furthermore, the mainly altered metabolism caused by the downregulated proteins is outlined in Figure 3B. These results suggest that inefficient glycolysis, the P cycle, and purine metabolism may be responsible for the resistance, which is related to the reduction of glucose.
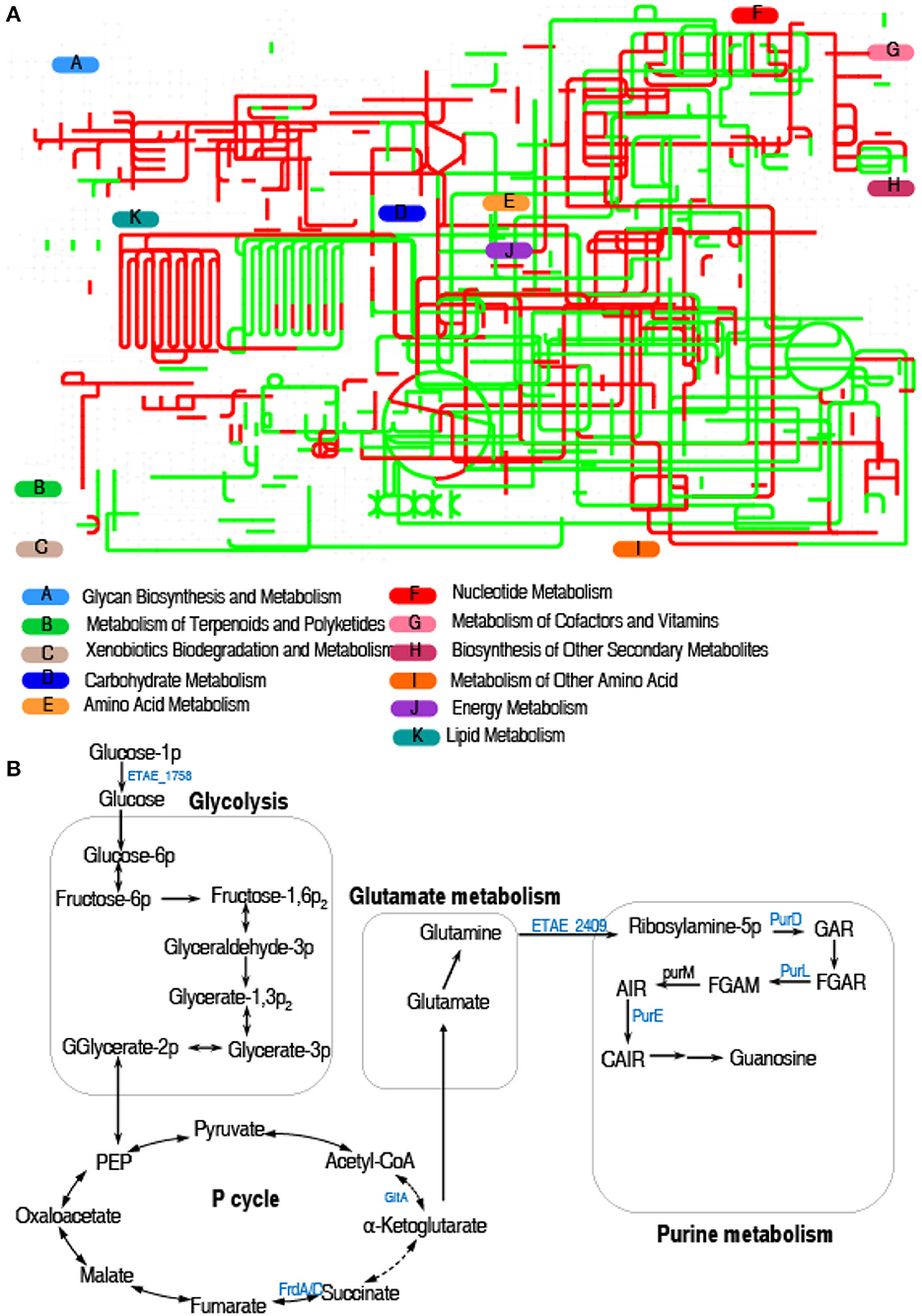
Figure 3. Metabolic pathway analysis. (A) Analysis for metabolic profiling showed a better insight into the effects of differential abundance proteins (p < 0.05). Based on the EggNOG database (http://eggnog5.embl.de), metabolic network pathways are further analyzed using iPath2.0 (http://pathways.embl.de). Red, increase; green, decrease. (B) Metabolic schematic diagram of differential proteins. The identified downregulated proteins (green) involved in metabolism are all shown in the flowchart.
Inactivation of glycolysis and the P cycle
To confirm the above findings on the inactivation of glycolysis and the P cycle that is attributed to the reduced glucose, glucose content, qRT-PCR, and enzyme activity measurements were performed in LTB4-S and LTB4-RCAZ. Lower glucose was measured in LTB4-RCAZ than in LTB4-S (Figure 4A). qRT-PCR was employed to detect nine genes encoding glycolysis. Among them, glk, pgi, pfkA, fbaA, gapA, and ETAE_2321 exhibited decreased expression, and the other three, ETAE_2957, pfkB, and gpmB, displayed increased and unchanged expression in LTB4-RCAZ compared with LTB4-S (Figure 4B). Further measurement of enzyme activity in glycolysis showed lower activity of glucokinase (HK, encoded by glk) and 6-phosphofructokinase I (PFK, encoded by pfkA and pfkB) in LTB4-RCAZ than LTB4-S (Figure 4C). Since the reduction of gene expression in the P cycle was reported in our previous report (Ye et al., 2021), only pykF was selected as a representative gene to confirm the reduction. Expression of pykF was lower in LTB4-RCAZ than in LTB4-S (Figure 4B). Consistently, pyruvate kinase (PK, encoded by pykF) was lower in LTB4-RCAZ compared with that in LTB4-S (Figure 4D). The activity of pyruvate dehydrogenase (PDH), α-ketoglutarate dehydrogenase (KGDH), succinate dehydrogenase (SDH), and malate dehydrogenase (MDH) was lower in LTB4-RCAZ than in LTB4-S (Figure 4E). Therefore, glycolysis and the P cycle were inefficient due to the depressed glucose (Figure 4F).
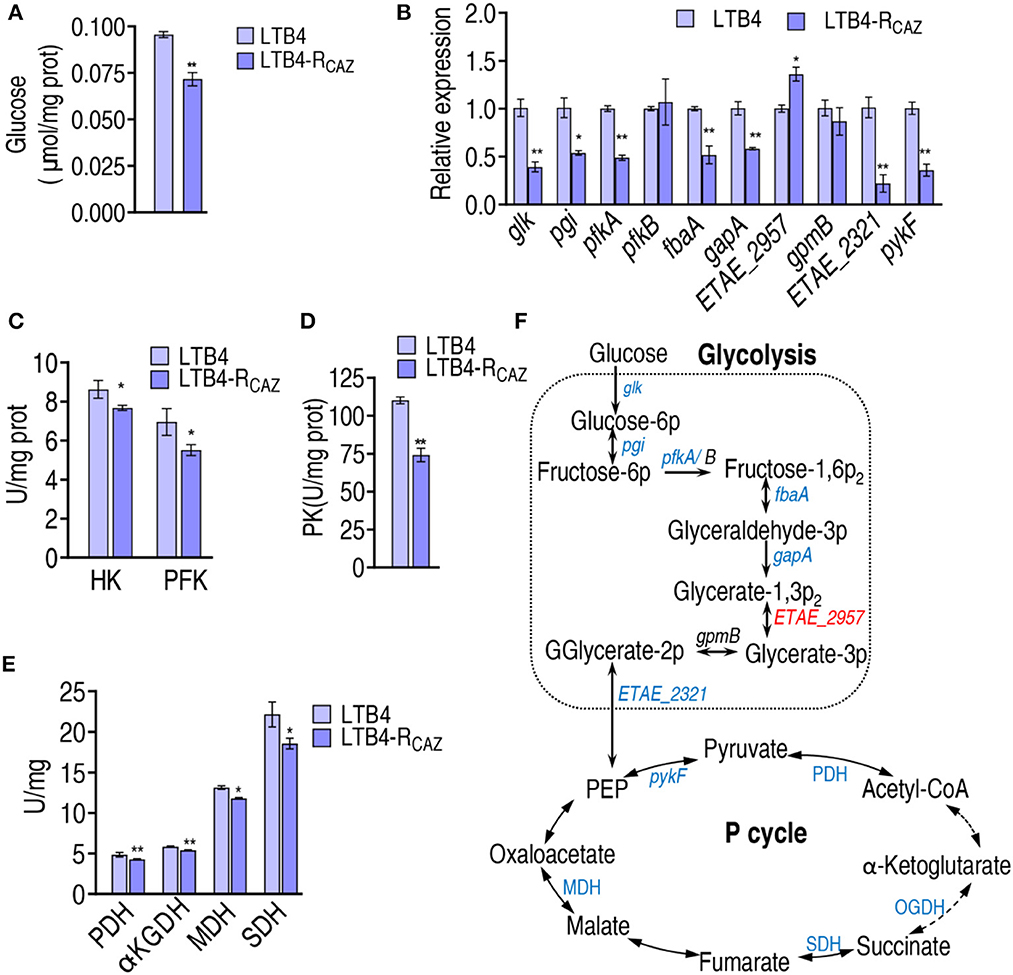
Figure 4. Alteration of glycolysis and the P cycle. (A) Quantification of glucose in LTB4-S and LTB4-RCAZ. (B) qRT-PCR for expression of genes encoding glycolysis and pykF in the P cycle. (C) Activity of HK and PFH in glycolysis. (D) Activity of PK in the P cycle. (E) Activity of enzymes in the P cycle. (F) A summarizing map showing the alteration of glycolysis and the P cycle. *, P < 0.05; **, P < 0.01.
Inactivation of glutamate metabolism and purine metabolism
The P cycle metabolism fluxes to purine metabolism via glutamate metabolism. The expression of gltD and gltB (encoding glutamate synthases) and glnA (catalyzing the ATP-dependent biosynthesis of glutamine from glutamate and ammonia) was reduced in LTB4-RCAZ (Figure 5A). Similarly, the activity of glutamic-pyruvic transaminase (GPT) converting 2-oxoglutarate to glutamate was decreased in LTB4-RCAZ (Figure 5B). Following the measurement of glutamate metabolism, the expression of genes encoding purine metabolism was quantified. A total of 14 genes were measured. Among them, 10 (ETAE_2409, purD, purN, purT, purl, purE, purH, guaB, guaA, and ETAE_1034) and 4 (purM, purK, purC, and purB2) displayed lower and unchanged expression in LTB4-RCAZ than LTB4-S, respectively (Figure 5C). These results confirm the inactivation of glutamate metabolism and purine metabolism (Figure 5D).
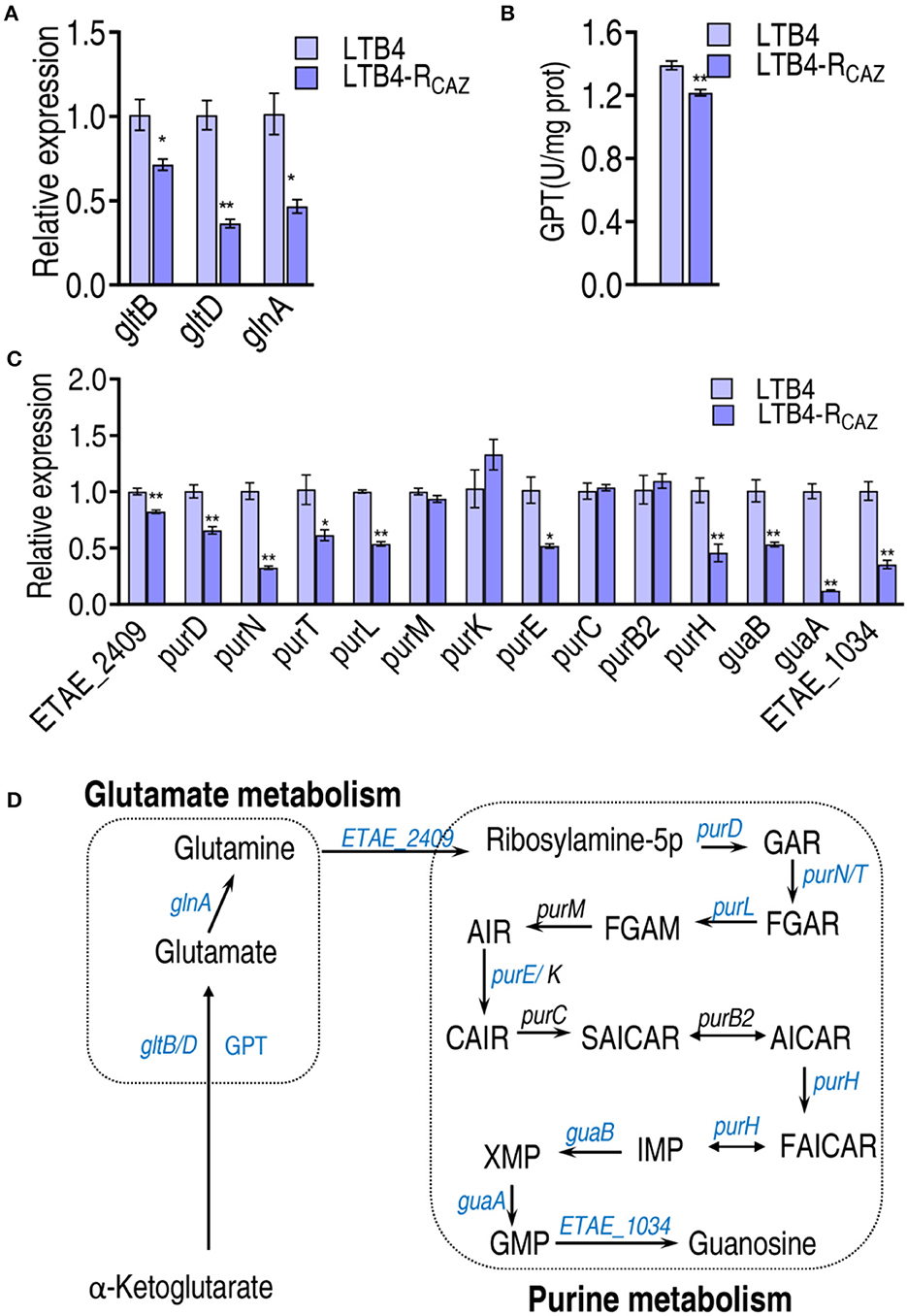
Figure 5. Alteration of glutamine metabolism and purine metabolism. (A) qRT-PCR for expression of genes encoding glutamate metabolism. (B) Activity of alanine transaminase (GPT). (C) qRT-PCR for expression of genes encoding purine metabolism. (D) A summarizing map showing the alteration of glutamate metabolism and purine metabolism. *, P < 0.05; **, P < 0.01.
Role of glucose in the flux from glycolysis to purine metabolism
The above results motivated us to suppose that the reduced glucose is responsible for the inefficient flux from glycolysis, the P cycle, and glutamate metabolism, to purine metabolism. To do this, exogenous glucose was complemented to test whether the inefficient flux was reversed. When glucose was added, the intracellular glucose of LTB4-RCAZ was elevated in a glucose dose-dependent manner (Figure 6A). Then, qRT-PCR was used to measure the expression of the 9 genes of glycolysis, the 1 gene (pykF) of the P cycle, the 3 genes of glutamate metabolism, and the 14 genes of purine metabolism in LTB4-RCAZ cultured in medium with and without glucose. Exogenous glucose promoted 6 genes of glycolysis, 1 gene of the P cycle, 2 genes of glutamate metabolism, and 9 genes of purine metabolism. The others kept stable except for weakly reduced pfkB (Figures 6B–D). Consistently, the activity of HK and PFK of glycolysis, PK, PDH, KGDH, and SDH of the P cycle, and GPT of glutamate metabolism were also elevated (Figures 6E–H). To test whether glucose reversed the ceftazidime resistance, the viability of LTB4-RCAZ was measured in the presence of different concentrations of glucose plus ceftazidime. The survival was reduced in a glucose dose-dependent manner between 1.25 mM and 5 mM glucose and kept stable between 5 mM and 40 mM (Figure 6I). Guanosine is an intermetabolite of purine metabolism that was repressed in LTB4-RCAZ (Liu et al., 2019). Thus, glucose was replaced with guanosine to explore whether the intermetabolite has the potential. A similar result was determined in the replacement (Figure 6J). Therefore, glucose reduction can cause guanosine-mediated antibiotic resistance through the metabolic flux from glycolysis, the P cycle, and glutamate metabolism to purine metabolism.
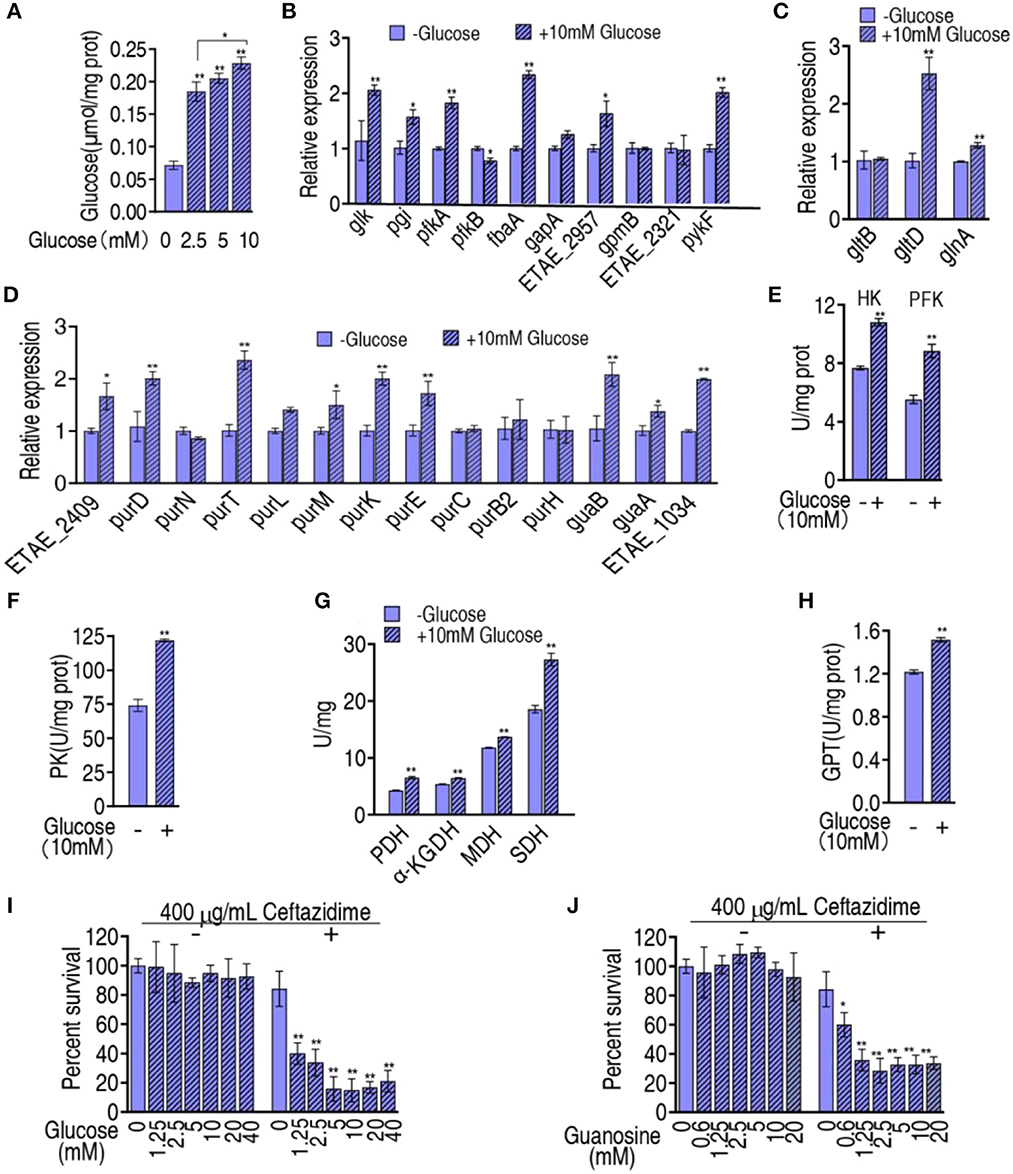
Figure 6. Effect of exogenous glucose on glycolysis, the P cycle, glutamate metabolism, and purine metabolism. (A) Intracellular glucose of LTB4-RCAZ in the presence of the indicated concentrations of exogenous glucose. (B–D) qRT-PCR for expression of genes encoding glycolysis, the P cycle (B), glutamate metabolism (C), and purine metabolism (D). (E–H), Activity of enzymes in glycolysis (E), the P cycle (F, G), glutamate metabolism. (I) Survival of LTB4-RCAZ in the presence of the indicated concentration of glucose plus ceftazidime. (J) Survival of LTB4-RCAZ in the presence of the indicated concentration of guanosine. *, P < 0.05; **, P < 0.01.
Mechanisms by which glucose was reduced
Our recent report has revealed that ampicillin binds with the pts promoter to negatively regulate the PTS system encoded by ptsL, ptsH, crr, and ptsG, which downregulates the glucose levels in antibiotic-resistant E. coli (Jiang et al., 2023). Ampicillin and ceftazidime belong to the same class of antibiotic β-lactam. Thus, we supposed that ceftazidime may have the same ability that binds to pts promoter. To explore this idea, qRT-PCR was used to detect the expression of ptsL, ptsH, crr, and ptsG. Lower expression of the four genes was quantified in LTB4-RCAZ than in LTB4-S (Figure 7A). To test whether the reduced expression of the four genes was also attributed to the binding of ceftazidime with the pts promoter, ceftazidime was incubated with different concentrations of pts promoter. The bound ceftazidime was used to kill E. coli K12. The viability of E. coli K12 was reduced in a pts promoter dose-dependent manner (Figure 7B). Meanwhile, the pts promoter was incubated with different concentrations of ceftazidime to capture ceftazidime and then used for an antibacterial test. Survival of E. coli K12 was decreased with increasing ceftazidime between 0.01 μg and 10 μg and kept stable between 10 μg and 1,000 μg (Figure 7C). These results are consistent with the finding that antibiotics control pts promoters to manipulate glucose-mediated antibiotic resistance (Jiang et al., 2023).
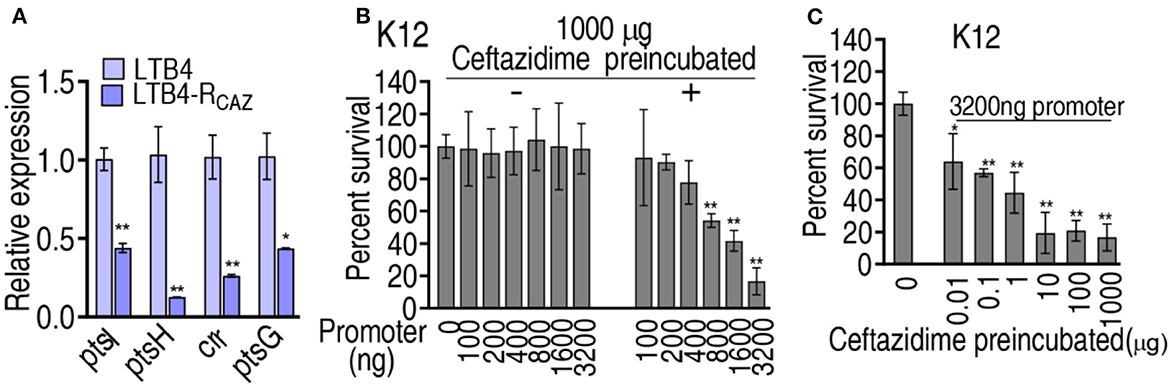
Figure 7. Expression of genes controlled by the pts promoter and the binding of pts promoter with ceftazidime. (A) qRT-PCR for expression of genes controlled by the pts promoter in LTB4-S and LTB4-RCAZ. (B) Survival of E. coli K12 exposed to ceftazidime isolated from the binding of 1,000 μg ceftazidime to the indicated concentration of pts promoter. (C) Survival of E. coli K12 exposed to ceftazidime isolated from the binding of 3,200 μg pts promoter with the indicated concentration of ceftazidime. *, P < 0.05; **, P < 0.01.
Discussion
Metabolic environments are changed to respond to antibiotic resistance, as has been documented (Stokes et al., 2019; Zhang et al., 2019; Li et al., 2020; Chen et al., 2022b), where intermetabolites of purine metabolism, such as inosine and guanosine, are reduced (Liu et al., 2019; Zhao et al., 2021). However, information regarding the reduction mechanisms is not clarified. The present study employs a proteomics approach to characterize the differential abundance of proteins between LTB4-RCAZ and LTB-S. This leads to the possibility that ceftazidime depresses glucose and the depressed glucose causes the reduction of purine metabolism through a glucose-mediated metabolic flux from glycolysis, the P cycle, and glutamate metabolism (Sauer and Eikmanns, 2005; Chen et al., 2022b). To demonstrate this possibility, the expression of genes and activity of enzymes in the four metabolic pathways are measured in LTB4-RCAZ and LTB4-S. Inactivation of the four metabolic pathways is documented in LTB4-RCAZ compared with LTB4-S. Moreover, glucose is complemented to elevate the glucose level that is reduced in LTB4-RCAZ. The complementation promotes the glucose-mediated metabolic flux from glycolysis, the P cycle, and glutamate metabolism to purine metabolism. Therefore, the inefficient purine metabolism can be attributed to the depressed glucose. To further demonstrate that the inactivation of purine metabolism mediated by glucose is responsible for antibiotic resistance, exogenous glucose and guanosine are used to potentiate ceftazidime killing. The elevated killing efficacy is similar as the glucose did. Finally, the combination of ceftazidime with pts promoter is explored. Similar to ampicillin (Jiang et al., 2023), ceftazidime is bound to the pts promoter. Notably, our previous report showed that higher, unchanged, and lower expression of the four genes (ptsI, ptsH, crr, and ptsG) controlled by the promoter is detected in antibiotic-tolerant cells, at the switch point, and in antibiotic-resistant cells during chronic intermittent exposure to ampicillin (Jiang et al., 2023). LTB4-RCAZ is a ceftazidime-resistant bacterium. Thus, a consistent result is a decrease in the expression of the four genes. Taken together, the present study identifies a glucose-mediated antibiotic resistance metabolic flux from glycolysis, the pyruvate cycle, and glutamate metabolism to purine metabolism, which is reversed by exogenous glucose and guanosine.
The core finding in the present study is that inefficient purine metabolism causes antibiotic resistance, which should be attributed to glucose depression. A recent metabolomics approach has shown that glucose and intermetabolites of purine metabolism are decreased simultaneously (Peng et al., 2015b; Zhang et al., 2019; Tang et al., 2022), but the relationship between them is absent. The present study provides robust proof that the intermetabolites of purine metabolism, such as guanosine, are a consequence of the depressed glucose. This finding not only clarifies the reason for reduced purine metabolism but also expands glucose-mediated antibiotic resistance mechanisms.
Equally importantly, following the first report that ampicillin binds with DNA, typically pts promoter, to manipulate the transition from tolerance to resistance in bacteria (Jiang et al., 2023), the present study further shows that ceftazidime has the same ability to bind to pts promoter. Since penicillin, ampicillin, and cephalosporin ceftazidime belong to β-lactams, this finding suggests that β-lactams have the ability to bind with pts promoters. In the evolution of antibiotic resistance, antibiotics are a trigger. Therefore, these reports on antibiotic-trigger resistance highlight the way to fully understand antibiotic resistance mechanisms.
Finally, the antibiotic resistance caused by the depressed glucose-mediated reduced purine metabolism can be reversed by not only triggering metabolite glucose but also lowering downstream metabolites such as guanosine. Metabolite-based metabolic reprogramming has been demonstrated to be effective in potentiating conventional antibiotics to kill antibiotic-resistant bacteria and promoting complement-mediated killing of serum-resistant bacteria (Cheng et al., 2019; Kou et al., 2022). More reprogramming metabolites are identified from upstream metabolites such as glucose and alanine (Peng et al., 2015b; Zhang et al., 2019; Chen et al., 2022a; Yin et al., 2022; Yang et al., 2023). The present study suggests that metabolic flux is an important clue to understanding the metabolic mechanisms underlying antibiotic resistance, which leads to downstream metabolites as reprogramming metabolites.
Conclusion
The present study identifies a glucose-mediated antibiotic resistance metabolic flux from glycolysis, the pyruvate cycle, and glutamate metabolism to purine metabolism, which can be reversed by glucose and guanosine. In addition, the binding of ceftazidime with the pts promoter is revealed. These results highlight the need to fully understand antibiotic resistance mechanisms from a metabolic perspective.
Data availability statement
The original contributions presented in the study are included in the article/Supplementary material, further inquiries can be directed to the corresponding authors.
Author contributions
JX: Investigation, Methodology, Writing—original draft. S-wW: Methodology, Writing—original draft. YT: Investigation, Methodology, Writing—original draft. J-zY: Data curation, Investigation, Methodology, Writing—original draft. YL: Investigation, Resources, Writing—original draft. X-xP: Conceptualization, Writing—review and editing. L-fY: Conceptualization, Writing—review and editing. HL: Conceptualization, Writing—review and editing.
Funding
The author(s) declare financial support was received for the research, authorship, and/or publication of this article. This work was financially supported by the grants from National Natural Science Foundation of China (42276125), Medical Scientific Research Foundation of Guangdong Province, China (A2021386), Natural Science Foundation of Guangdong Province (2022A1515012079), and Innovation Group Project of Southern Marine Science and Engineering Guangdong Laboratory (Zhuhai) (no. 311020006).
Conflict of interest
X-xP was employed by the Guangdong Litai Pharmaceutical Co., Ltd.
The remaining authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher's note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2023.1267729/full#supplementary-material
References
Adelowo, O. O., Caucci, S., Banjo, O. A., Nnanna, O. C., Awotipe, E. O., Peters, F. B., et al. (2018). Extended spectrum beta-lactamase (ESBL)-producing bacteria isolated from hospital wastewaters, rivers and aquaculture sources in Nigeria. Environ. Sci. Pollut. Res. Int. 25, 2744–2755. doi: 10.1007/s11356-017-0686-7
Ayub, U., and Naveed, H. (2022). BioAlign: an accurate global PPI network alignment algorithm. Evol. Bioinform. 18, 11769343221110658. doi: 10.1177/11769343221110658
Chen, X. W., Wu, J. H., Liu, Y. L., Munang'andu, H. M., and Peng, B. (2023). Fructose promotes ampicillin killing of antibiotic-resistant Streptococcus agalactiae. Virulence 14, 2180938. doi: 10.1080/21505594.2023.2180938
Chen, Y. T., Ma, Y. M., Peng, X. X., and Li, H. (2022a). Glutamine potentiates gentamicin to kill lab-evolved gentamicin-resistant and clinically isolated multidrug-resistant Escherichia coli. Front. Microbiol. 13, 1071278. doi: 10.3389/fmicb.2022.1071278
Chen, Y. T., Yang, K. X., Dai, Z. Y., Yi, H., Peng, X. X., Li, H., et al. (2022b). Repressed central carbon metabolism and its effect on related metabolic pathways in cefoperazone/sulbactam-resistant Pseudomonas aeruginosa. Front Microbiol. 13, 847634. doi: 10.3389/fmicb.2022.847634
Cheng, Z. X., Gong, Q. Y., Wang, Z., Chen, Z. G., Ye, J. Z., Li, J., et al. (2017). Edwardsiella tarda tunes tricarboxylic acid cycle to evade complement-mediated killing. Front. Immunol. 8, 1706. doi: 10.3389/fimmu.2017.01706
Cheng, Z. X., Guo, C., Chen, Z. G., Yang, T. C., Zhang, J. Y., Wang, J., et al. (2019). Glycine, serine and threonine metabolism confounds efficacy of complement-mediated killing. Nat. Commun. 10, 3325. doi: 10.1038/s41467-019-11129-5
Cheng, Z. X., Yang, M. J., Peng, B., Peng, X. X., Lin, X. M., Li, H., et al. (2018). The depressed central carbon and energy metabolisms is associated to the acquisition of levofloxacin resistance in Vibrio alginolyticus. J. Proteomics 181, 83–91. doi: 10.1016/j.jprot.2018.04.002
Darzi, Y., Letunic, I., Bork, P., and Yamada, T. (2018). iPath3.0: interactive pathways explorer v3. Nucleic Acids Res. 46, W510–W513. doi: 10.1093/nar/gky299
Fan, L., Pan, Z., Liao, X., Zhong, Y., Guo, J., Pang, R., et al. (2023). Uracil restores susceptibility of methicillin-resistant Staphylococcus aureus to aminoglycosides through metabolic reprogramming. Front. Pharmacol. 14, 1133685. doi: 10.3389/fphar.2023.1133685
Fröhlich, C., Sørum, V., Tokuriki, N., Johnsen, P. J., and Samuelsen, Ø. (2022). Evolution of β-lactamase-mediated cefiderocol resistance. J. Antimicrob. Chemother. 77, 2429–2436. doi: 10.1093/jac/dkac221
Jiang, M., Kuang, S. F., Lai, S. S., Zhang, S., Yang, J., Peng, B., et al. (2020). Na+-NQR Confers aminoglycoside resistance via the regulation of L-alanine metabolism. mBio. 11, e02086–e02020. doi: 10.1128/mBio.02086-20
Jiang, M., Su, Y. B., Ye, J. Z., Li, H., Kuang, S. F., Wu, J. H., et al. (2023). Ampicillin-controlled glucose metabolism manipulates the transition from tolerance to resistance in bacteria. Sci. Adv. 9, eade8582. doi: 10.1126/sciadv.ade8582
Kou, T. S., Wu, J. H., Chen, X. W., Chen, Z. G., Zheng, J., Peng, B., et al. (2022). Exogenous glycine promotes oxidation of glutathione and restores sensitivity of bacterial pathogens to serum-induced cell death. Redox Biol. 58, 102512. doi: 10.1016/j.redox.2022.102512
Lei, J., Huang, J., Xin, C., Liu, F., Zhang, J., Xie, Y., et al. (2023). Riboflavin targets the cellular metabolic and ribosomal pathways of Candida albicans in vitro and exhibits efficacy against oropharyngeal candidiasis. Microbiol. Spectr. 11, e0380122. doi: 10.1128/spectrum.03801-22
Li, L., Su, Y. B., Peng, B., and Peng, X. X. H. (2020). Metabolic mechanism of colistin resistance and its reverting in Vibrio alginolyticus. Environ. Microbiol. 22, 4295–4313. doi: 10.1111/1462-2920.15021
Li, W., Ali, F., Cai, Q., Yao, Z., Sun, L., Lin, W., et al. (2018). Quantitative proteomic analysis reveals that chemotaxis is involved in chlortetracycline resistance of Aeromonas hydrophila. J. Proteomics 172, 143–151. doi: 10.1016/j.jprot.2017.09.011
Liu, S. R., Peng, X. X., and Li, H. (2019). Metabolic mechanism of ceftazidime resistance in Vibrio alginolyticus. Infect. Drug Resist. 12, 417–429. doi: 10.2147/IDR.S179639
Lopatkin, A. J., Bening, S. C., Manson, A. L., Stokes, J. M., Kohanski, M. A., Badran, A. H., et al. (2021). Clinically relevant mutations in core metabolic genes confer antibiotic resistance. Science 371, eaba0862. doi: 10.1126/science.aba0862
Moya-Salazar, J., Díaz, C. R., Cañari, B., Badillo, R. X., Verano-Zelada, M., Chicoma-Flores, K., et al. (2022). Detection of pathogenic Aeromonas hydrophila from two rainbow trout (Oncorhynchus mykiss) farms in Peru. Braz. J. Vet. Med. 44, e000922. doi: 10.29374/2527-2179.bjvm000922
Peng, B., Li, H., and Peng, X. X. (2015a). Functional metabolomics: from biomarker discovery to metabolome reprogramming. Protein Cell 6, 628–37. doi: 10.1007/s13238-015-0185-x
Peng, B., Li, H., and Peng, X. X. (2019). Proteomics approach to understand bacterial antibiotic resistance strategies. Expert Rev. Proteomics 16, 829–839. doi: 10.1080/14789450.2019.1681978
Peng, B., Li, H., and Peng, X. X. (2023). Call for next-generation drugs that remove the uptake barrier to combat antibiotic resistance. J. Drug Dis. 2023, 103753. doi: 10.1016/j.drudis.2023.103753
Peng, B., Su, Y. B., Li, H., Han, Y., Guo, C., Tian, Y. M., et al. (2015b). Exogenous alanine and/or glucose plus kanamycin kills antibiotic-resistant bacteria. Cell Metab. 21, 249–262. doi: 10.1016/j.cmet.2015.01.008
Richards, D. M., and Brogden, R. N. (1985). Ceftazidime. A review of its antibacterial activity, pharmacokinetic properties and therapeutic use. Drugs 29, 105–161. doi: 10.2165/00003495-198529020-00002
Salgueiro, V., Reis, L., Ferreira, E., Botelho, M. J., Manageiro, V., Caniça, M., et al. (2021). Assessing the bacterial community composition of bivalve mollusks collected in aquaculture farms and respective susceptibility to antibiotics. Antibiotics 10, 1135. doi: 10.3390/antibiotics10091135
Sanz-García, F., Hernando-Amado, S., and Martínez, J. L. (2018). Mutation-driven evolution of Pseudomonas aeruginosa in the presence of either ceftazidime or ceftazidime-avibactam. Antimicrob. Agents Chemother. 62, e01379–18. doi: 10.1128/AAC.01379-18
Sauer, U., and Eikmanns, B. J. (2005). The PEP-pyruvate-oxaloacetate node as the switch point for carbon flux distribution in bacteria. BJ. FEMS. Microbiol. Rev. 29, 765–794. doi: 10.1016/j.femsre.2004.11.002
Stokes, J. M., Lopatkin, A. J., Lobritz, M. A., and Collins, J. J. (2019). Bacterial metabolism and antibiotic efficacy. Cell Metab. 30, 251–259. doi: 10.1016/j.cmet.2019.06.009
Su, Y. B., Peng, B., Li, H., Cheng, Z. X., Zhang, T. T., Zhu, J. X., et al. (2018). Pyruvate cycle increases aminoglycoside efficacy and provides respiratory energy in bacteria. Proc. Natl. Acad. Sci. U. S. A. 115, E1578–E1587. doi: 10.1073/pnas.1714645115
Tang, X. K., Su, Y. B., Ye, H. Q., Dai, Z. Y., Yi, H., Yang, K. X., et al. (2022). Glucose-potentiated amikacin killing of cefoperazone/sulbactam resistant Pseudomonas aeruginosa. Front. Microbiol. 12, 800442. doi: 10.3389/fmicb.2021.800442
Tong, C., Hu, H., Chen, G., Li, Z., Li, A., Zhang, J., et al. (2021). Chlorine disinfectants promote microbial resistance in Pseudomonas sp. Environ. Res. 199, 111296. doi: 10.1016/j.envres.2021.111296
Yang, H., Zhou, Y., Luo, Q., Zhu, C., and Fang, B. (2023). L-leucine increases the sensitivity of drug-resistant Salmonella to sarafloxacin by stimulating central carbon metabolism and increasing intracellular reactive oxygen species level. Front. Microbiol. 14, 1186841. doi: 10.3389/fmicb.2023.1186841
Ye, J., Su, Y., Peng, X., and Li, H. (2021). Reactive oxygen species-related ceftazidime resistance is caused by the pyruvate cycle perturbation and reverted by Fe3+ in Edwardsiella tarda. Front. Microbiol. 12, 654783. doi: 10.3389/fmicb.2021.654783
Ye, J. Z., Lin, X. M., Cheng, Z. X., Su, Y. B., Li, W. X., Ali, F. M., et al. (2018a). Identification and efficacy of glycine, serine and threonine metabolism in potentiating kanamycin-mediated killing of Edwardsiella piscicida. J. Proteomics 183, 34–44. doi: 10.1016/j.jprot.2018.05.006
Ye, J. Z., Su, Y. B., Lin, X. M., Lai, S. S., Li, W. X., Ali, F., et al. (2018b). Alanine enhances aminoglycosides-induced ROS production as revealed by proteomic analysis. Front. Microbiol. 9, 29. doi: 10.3389/fmicb.2018.00029
Yin, Y., Yin, Y., Yang, H., Chen, Z., Zheng, J., Peng, B., et al. (2022). Vibrio alginolyticus survives from ofloxacin stress by metabolic adjustment. Front. Microbiol. 13, 818923. doi: 10.3389/fmicb.2022.818923
Zhang, S., Wang, J., Jiang, M., Xu, D., Peng, B., Peng, X. X., et al. (2019). Reduced redox-dependent mechanism and glucose-mediated reversal in gentamicin-resistant Vibrio alginolyticus. Environ. Microbiol. 21, 4724–4739. doi: 10.1111/1462-2920.14811
Zhao, X. L., Chen, Z. G., Yang, T. C., Jiang, M., Wang, J., Cheng, Z. X., et al. (2021). Glutamine promotes antibiotic uptake to kill multidrug-resistant uropathogenic bacteria. Sci. Transl. Med. 13, eabj0716. doi: 10.1126/scitranslmed.abj0716
Keywords: antibiotic resistance, glucose, metabolic reprogramming, glycolysis, the pyruvate cycle, glutamate metabolism, purine metabolism, guanosine
Citation: Xiang J, Wang S-w, Tao Y, Ye J-z, Liang Y, Peng X-x, Yang L-f and Li H (2023) A glucose-mediated antibiotic resistance metabolic flux from glycolysis, the pyruvate cycle, and glutamate metabolism to purine metabolism. Front. Microbiol. 14:1267729. doi: 10.3389/fmicb.2023.1267729
Received: 27 July 2023; Accepted: 18 September 2023;
Published: 17 October 2023.
Edited by:
Rustam Aminov, University of Aberdeen, United KingdomReviewed by:
Xiangmin Lin, Fujian Agriculture and Forestry University, ChinaSrinivasan Ramanathan, Prince of Songkla University, Thailand
Copyright © 2023 Xiang, Wang, Tao, Ye, Liang, Peng, Yang and Li. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Hui Li, bGlodWkzMkBzeXN1LmVkdS5jbg==; Li-fen Yang, eWFuZ2xpZjhAbWFpbC5zeXN1LmVkdS5jbg==
 Jiao Xiang
Jiao Xiang Shi-wen Wang
Shi-wen Wang Yuan Tao1
Yuan Tao1 Jing-zhou Ye
Jing-zhou Ye Xuan-xian Peng
Xuan-xian Peng Hui Li
Hui Li