
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
REVIEW article
Front. Microbiol. , 02 October 2023
Sec. Microbiotechnology
Volume 14 - 2023 | https://doi.org/10.3389/fmicb.2023.1265139
Polyethylene terephthalate (PET) is a synthetic polymer in the polyester family. It is widely found in objects used daily, including packaging materials (such as bottles and containers), textiles (such as fibers), and even in the automotive and electronics industries. PET is known for its excellent mechanical properties, chemical resistance, and transparency. However, these features (e.g., high hydrophobicity and high molecular weight) also make PET highly resistant to degradation by wild-type microorganisms or physicochemical methods in nature, contributing to the accumulation of plastic waste in the environment. Therefore, accelerated PET recycling is becoming increasingly urgent to address the global environmental problem caused by plastic wastes and prevent plastic pollution. In addition to traditional physical cycling (e.g., pyrolysis, gasification) and chemical cycling (e.g., chemical depolymerization), biodegradation can be used, which involves breaking down organic materials into simpler compounds by microorganisms or PET-degrading enzymes. Lipases and cutinases are the two classes of enzymes that have been studied extensively for this purpose. Biodegradation of PET is an attractive approach for managing PET waste, as it can help reduce environmental pollution and promote a circular economy. During the past few years, great advances have been accomplished in PET biodegradation. In this review, current knowledge on cutinase-like PET hydrolases (such as TfCut2, Cut190, HiC, and LCC) was described in detail, including the structures, ligand–protein interactions, and rational protein engineering for improved PET-degrading performance. In particular, applications of the engineered catalysts were highlighted, such as improving the PET hydrolytic activity by constructing fusion proteins. The review is expected to provide novel insights for the biodegradation of complex polymers.
Poly(ethylene terephthalate) (PET) is used extensively worldwide in plastic products, and around 350 million tons have been produced in 2017 (Geyer et al., 2017). However, the accumulation of PET waste and its resistance to degradation in the environment have posed a serious threat and become a global concern (Choi et al., 2022; Li et al., 2022). PET has different crystallinities according to its usage, which leads to PET materials with different features and chemical properties, such as the fluctuation of polymer chains, surface area, functional groups, molecular weight, hydrophobicity, and melting temperature (Kawai et al., 2019). This has created another problem for scientists attempting to develop a strategy to efficiently degrade all kinds of PET wastes (Thomsen et al., 2022a).
To date, several strategies have been proposed for recycling PET waste, including primary recycling, mechanical recycling, chemical recycling, energy recycling, and biological recycling (Cao et al., 2022). Enzyme-based hydrolysis of postconsumer plastic (biological recycling) is an emerging strategy for closed-loop recycling of PET. This is a process catalyzed by PET-degrading enzymes (or microorganisms) that breaks the PET polymer into diverse products, such as terephthalic acid (TPA), ethylene glycol (EG), bis-(hydroxyethyl) terephthalate (BHET), and (mono-(2-hydroxyehyl)terephthalic acid (MHET)). This is the interactive result of active site accessibility to the polymer surface (Zumstein et al., 2017), and especially it was found that the endo-lytic activity (ability to hydrolyse internal bonds in the polymer) was a key parameter for overall enzyme performance (Schubert et al., 2023). Therefore, a circular carbon economy for PET is theoretically attainable through rapid enzymatic depolymerization followed by directed conversion into other high value-added products (Ru et al., 2020; Ellis et al., 2021; Kakadellis and Rosetto, 2021; Simon et al., 2021).
In a broad sense, the PET-degrading enzymes can be divided into the following categories according to their different functions: surface-modifying enzymes and PET hydrolases (Kawai et al., 2019). The former is usually applied for the enzymatic surface modification of polyester fibers to improve hydrophilicity (surface hydrophilization), including lipases, carboxylesterases, and cutinases (Biundo et al., 2018). These enzymes usually act on the ends or the loop (defined as a polymer chain, both ends of which are attached to the PET surface) on their surface of polymer chains, which are then hydrolysed to carboxylic acid and hydroxyl residues, increasing surface hydrophilicity (Kawai et al., 2017; Oda et al., 2018). The latter could significantly degrade the inner block of PET, as revealed by visible changes found through SEM. In addition, PET hydrolases are capable of performing surface hydrolysis, which would lead to the hydrolysis of the surface polymer chain instead of degrading the building blocks of PET.
The first enzymatic PET degradation study was reported by Rolf Joachim et al. (2005). It was shown that the obtained hydrolase hydrolysed two kinds of PET films at 55°C. Soon afterwards, the first purified poly(butylene terephthalate-co-adipate) (BTA) hydrolase 1 was purified and characterized from the culture supernatant of Thermobifida fusca (T. fusca) (Kleeberg et al., 2005), which further revealed that the sequence of Tfu0883 is consistent with that of the gene for BTA-1 (Lykidis et al., 2007). Since then, various effective enzymes have been observed to perform enzymatic PET hydrolysis. These enzymes with PET-degradation activity mainly belong to the subfamily of carboxylic ester hydrolases (EC 3.1.1.x), including carboxylesterase (EC 3.1.1.1) (von Haugwitz et al., 2022), lipases (Moyses et al., 2021), cutinases (EC 3.1.1.74) (Wei and Zimmermann, 2017), and arylesterase (EC 3.1.1.2) (Austin et al., 2018). In addition, several PETase-like enzymes have been identified, extending the scope of enzymatic PET recycling (Almeida et al., 2019; Karunatillaka et al., 2022; Li et al., 2022). Among them, great achievements have been accomplished in the fields of crystallographic and protein engineering of T. fusca cutinases with various released structures [e.g., PDB ID: 4CG1 (Roth et al., 2014), 6EQH (Austin et al., 2018), 6AID (Kitadokoro et al., 2019), 7VPB (Li et al., 2022)], rational protein engineering (Lu et al., 2022), and metabolic engineering or synthetic biology-driven studies (Brandenberg et al., 2022; Jia et al., 2022). Examples include the TfCut2 variant (Wei et al., 2016), Cut190 variant (Oda et al., 2018), HiC (Ronkvist et al., 2009), and leaf-branch compost cutinase (LCC) (Sulaiman et al., 2012; Xue et al., 2021). In particular, LCC was shown to be among the most effective enzymes and has a potential ability to recycle PET. The engineered LCC was shown to breakdown a minimum of 90% of PET into monomers over 10 h (Tournier et al., 2020). There are several excellent reviews or studies illustrating the molecular engineering, applications of cutinases and other biological strategies for PET hydrolysis (Zumstein et al., 2017; Ellis et al., 2021; Tamoor et al., 2021; Urbanek et al., 2021; Kumar et al., 2022; Brackmann et al., 2023).
In 2016, a novel bacterium, Ideonella sakaiensis 201-F6, was reported to proliferate on a low (1.9%)-crystallinity [The degree of PET crystallinity (XC) indicates the fraction of the total polymer chains being in the crystal structure state (Thomsen et al., 2022b)]. PET film at 30°C for 40 days and hydrolyse PET with catalysis by the two released enzymes [ISF6_4831 (IsPETase) and ISF6_0224 (IsMHETase)] (Yoshida et al., 2016). This led to the rapid development of IsPETase, an IsPETase-like enzyme, in the field of PET biodegradation. In addition, based on modern biotechnology, such as deep sequencing technologies, metagenomics, and AI-based bioscience, novel hydrolases with PET-degrading activity have been discovered and characterized, such as Ple629 (Meyer-Cifuentes et al., 2020), Mors1 and OaCut (Blázquez-Sánchez et al., 2022), MG8 (Eiamthong et al., 2022), RgPETase (Sagong et al., 2021) and BbPETase (Sagong et al., 2022).
To date, about 89 different enzymes [PAZy database (Buchholz et al., 2022)1] with PET-degrading ability have been discovered and characterized (Magalhães et al., 2021), and with the rapid development of bioinformatics and machine learning, an increasing number of PET hydrolases have been sourced from natural diversity (Erickson et al., 2022). This greatly expands the number and diversity of efficient scaffolds for enzymatic PET deconstruction. All effective PET hydrolases might belong to the family of cutinases or homologous enzymes (Kawai et al., 2019). Although the application of PET hydrolase is a green and sustainable strategy for PET degradation, it has been hampered by the lack of robustness to pH and temperature ranges, slow reaction rates and inability to directly use untreated postconsumer plastics. Therefore, research to improve the stability of PET-degrading enzymes and designing robust industrial catalysts is still facing great difficulties. It is worth noting that several biotech companies have long been committed to transforming plastic waste into recycled and recyclable plastic, such as the French company Carbios2 and the commercial hydrolase enzymes from Novozymes [e.g., CalB (Lipozyme® CALB L), RmL (Palatase® 20000L), CaL (Eversa® transform), TlL (Lipozyme® TL), HiC (Novozym® 51032)].
In this study, a brief overview of the recent advances in special cutinases and cutinase-like enzymes with PET-degrading activities is provided, including their structures, catalytic mechanisms, and applications. In particular, rational protein engineering based on macromolecule structures is highlighted. Finally, further perspectives on the industrial applications of enzyme-catalyzed PET degradation are also discussed.
Considering that polymer molecules tend to become flexible at temperatures above the glass transition temperature (Tg), the high Tg value of PET makes the thermostability of PET-degrading enzymes crucial for the efficient depolymerization of PET (Kawai, 2021). It is believed that the optimal temperature of a desired PET hydrolase needs to be >60 or preferably >70°C. Based on this recognition, a few thermophilic enzymes, including the TfH variant (TfCut2) (Mrigwani et al., 2023), LCC variant (LCCICCG) (Tournier et al., 2020), Cut190 variant (Kawai et al., 2022), HiC (Carniel et al., 2021), and BhrPETase (Xi et al., 2021), satisfy the requirements for thermostability above 60°C. In addition to thermophilic enzymes, the discovery of IsPETase from the mesophilic bacterium (Ideonella sakaiensis) caused mesophilic PET-hydrolysing enzymes to become a research hot spot. In this section, recent developments in special PET hydrolase with bright prospects are highlighted (Table 1).
The best enzymatic degradation reactions involving PET are carried out at (or near) PET’s glass transition temperature (~70°C), the temperature at which chains become mobile and accessible to the active site(s) of thermophilic hydrolases (Kawai, 2020). The following enzymes are notable due to their ability to function at temperatures between 60°C and 70°C (Sonnendecker et al., 2022): (i) metagenomically derived leaf branch compost cutinase (LCC) (Zheng et al., 2022); (ii) Humicola insolens cutinase (HiC) (Carniel et al., 2021); (iii) Thermobifida fusca cutinase (TfCut2) (Mrigwani et al., 2023); (iv) PES-H1 (or polyester hydrolase 7, PHL7), a close homolog of LCC that is twice as active as LCC (Richter et al., 2023); and (v) BhrPETase (from bacterium HR29), another close homolog of LCC that shows comparable activity.
Computational studies on the catalytic mechanism of PETase and LCC were performed using quantum mechanics/molecular mechanics (QM/MM) MD simulations with different algorithms (Boneta et al., 2021; Zheng et al., 2022). Based on the free energy surfaces, the established four-elementary-step mechanism was further confirmed, including mainly (1) the acylation of the serine of the catalytic triad, (2) breakage of the C-O bond, (3) nucleophilic attack by water molecules, and (4) deacylation. It was also revealed that the similar catalytic behavior of the active site might lead to a similar catalytic mechanism, and the better performance of LCCICCG than PETase might result from the higher optimal working temperature and the special structure–activity relationships with the PET polymer (Boneta et al., 2021).
Cutinases can usually hydrolyse both ester bonds found in aliphatic and aromatic polyesters, which display very large potential for biocatalytic plastic recycling (Carr et al., 2020; Ribitsch and Guebitz, 2021). In the hydrolysing process catalyzed by cutinases, the active-site serine residue is acylated by the substrate. Then, the substrates are transformed to carboxylic acids and alcohols through the formation of an acyl enzyme intermediate. This serine residue is located within a highly conserved GXSXG sequence motif and forms a catalytic triad with His and Asp.
Oda (2021) provided a great contribution to the development of Cut190. In 2014, a putative cutinase gene (cut190, from the thermophile Saccharomonospora viridis AHK190) was cloned and successfully expressed (Kawai et al., 2014). The mutant Cut190 (S226P/R228S) was shown to display optimal activity at 65–75°C, pH 6.5–8.0, and could remain stable at temperatures up to 65°C for 24 h; however, 60% of activity was lost after incubation for 1 h at 70°C. In particular, Ca2+ could clearly change the tertiary structure of Cut190, and the thermostability and activity could be greatly enhanced by high concentrations of calcium ions. To illustrate the structural basis for the Ca2+-enhanced thermostability, Takuya et al. reported the crystal structures of Cut190S226P in the Ca2+-bound and free states (Figure 1(1,2)) (Miyakawa et al., 2015). It mainly adopts a classical α/β hydrolase fold and contains 9 β-strands, 6 α-helices and a disulfide bond linking C287 and C302. The active-site Ser-His-Asp catalytic triad (S176, D222, and H254) was located on the surface without the lid structure. Ca2+ was located at the β1-β2 loop (Figure 1(3,4)) formed by residues from S76 to F81 (Figure 1(4)). Furthermore, the bound Ca2+ was coordinated by S76, A78, and F81 via their main-chain carbonyl groups. Once Ca2+ bound to Cut190S226P, it induced significant conformational changes, which mainly occurred in the following loops: the β1-β2 loop, the β3-α2 loop (F97-A121), and the β4-α3 loop (I126-E155). Upon Ca2+ binding, it will induce N133 to move toward the Ca2+ binding site (the β1-β2 loop, ~11.5Å), consequently enabling the movement of Q110 toward D131 and forming an additional hydrogen bond. The β3-α2 loop harbors the active-site sealing residue F106. Although Ca2+ binding induced the β3-α2 loop to the flexible open state, it might be helpful for substrate binding.
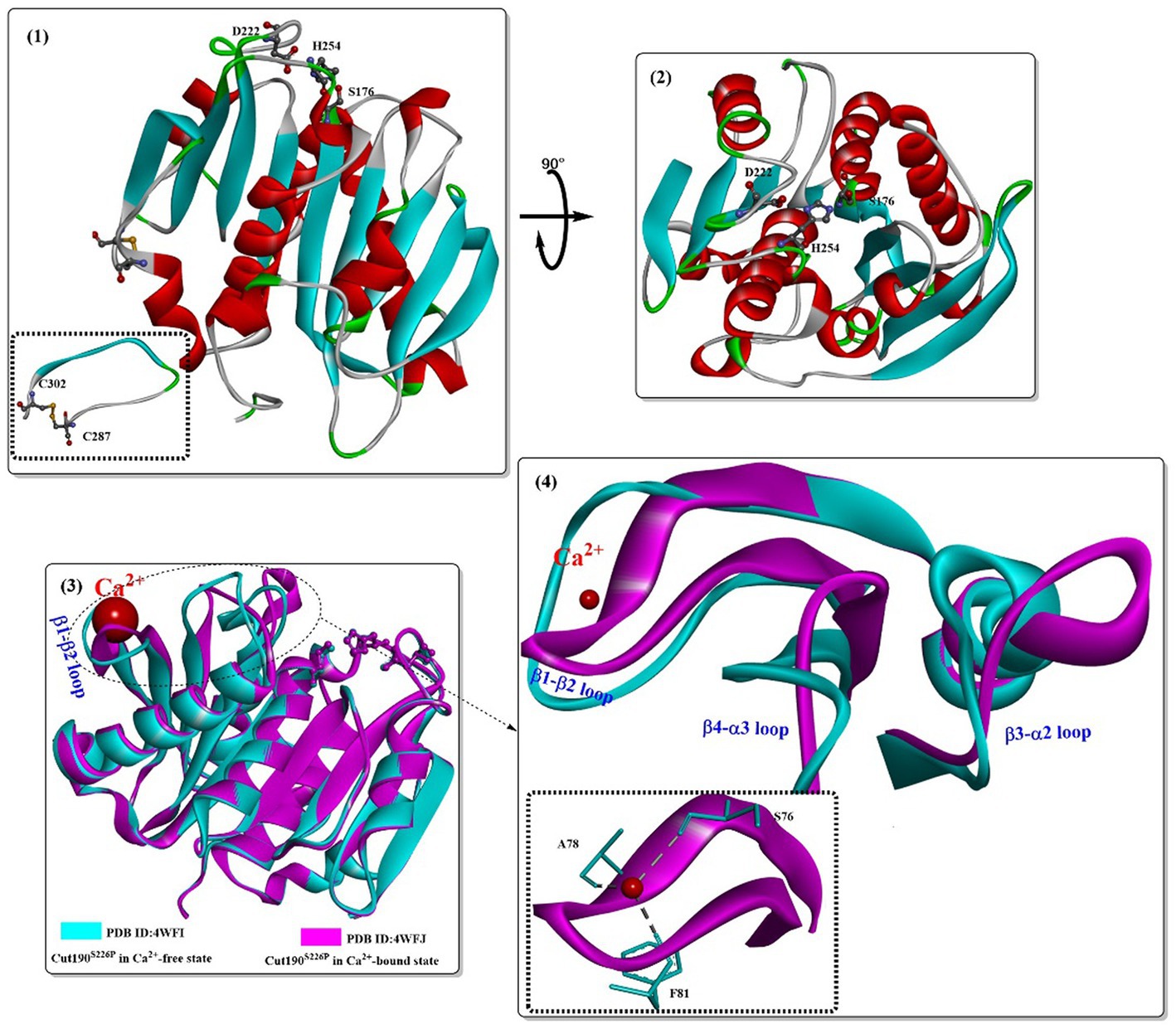
Figure 1. Structures of the Cut190 mutant (1,2; PDB ID: 4WFI) and the structure alignments of the Ca2+-free state and the Ca2+-bound state (3,4; PDB ID: 4WFJ). [The figures in this manuscript were generated using the program Discovery Studio 3.0; the special structural elements are shown in ball-and-stick model; and the dotted green (or light green) lines indicate the formed H-bonds].
Based on these findings, attention was focused on insights into the structure–activity relationship of substrate-Cut190 and rational protein engineering. Molecular modeling (ghecom: pocket finding; fkcombu: docking; the myPresto program package: energy minimization) was applied to investigate the reaction mechanism and improve the enzyme activity by Kawabata et al. (2017). The results showed that W201, F106, H175, M177 and F255 are important for substrate binding and enzymatic activities, especially for the aromatic polyester substrate. Numoto et al. (2018) further analyzed the substrate recognition mechanism of Cut190 with the crystal structure of the inactive Cut190 mutant (Cut190S176A). In this study, it was found that four Ca2+ ions were bound to Cut190S176A (Figure 2), which is different from that observed in a previous study (Miyakawa et al., 2015). From Figure 2, three of them are bound to the surface of the enzyme at site 1, site 2 and site 3. One more bound Ca2+ is located at the interface in site 4, which might be caused by the crystal packing that stabilizes the crystalline state of the protein. Structure alignment shows that site 1 is the same Ca2+ binding site, and Ca2+ binding displays a similar effect on the protein as that observed in the structure of Cut190S226P (Miyakawa et al., 2015) (Figure 2(1,2)). At site 2, G80 moved slightly away from the Ca2+-binding site upon Ca2+ binding, and Ca2+-binding showed little effect on site 3 (Figure 2(3,4)). Researchers reported that functions for the different Ca2+ are believed to be different, in which the Ca2+ at sites 1 and 3 might be responsible for the regulation of enzymatic activity, and Ca2+ at sites 2 and 3 can accommodate thermostability. It was further verified that Mn2+ and Mg2+ probably bind to site 2 with high affinity (responsible for stability improvement) and to sites 1 and 3 with low affinity (responsible for activity adjustment) (Senga et al., 2019). These results indicate that it might be possible to regulate the catalytic activity and thermostability by rational engineering of critical residues around site 1, site 2 and site 3.
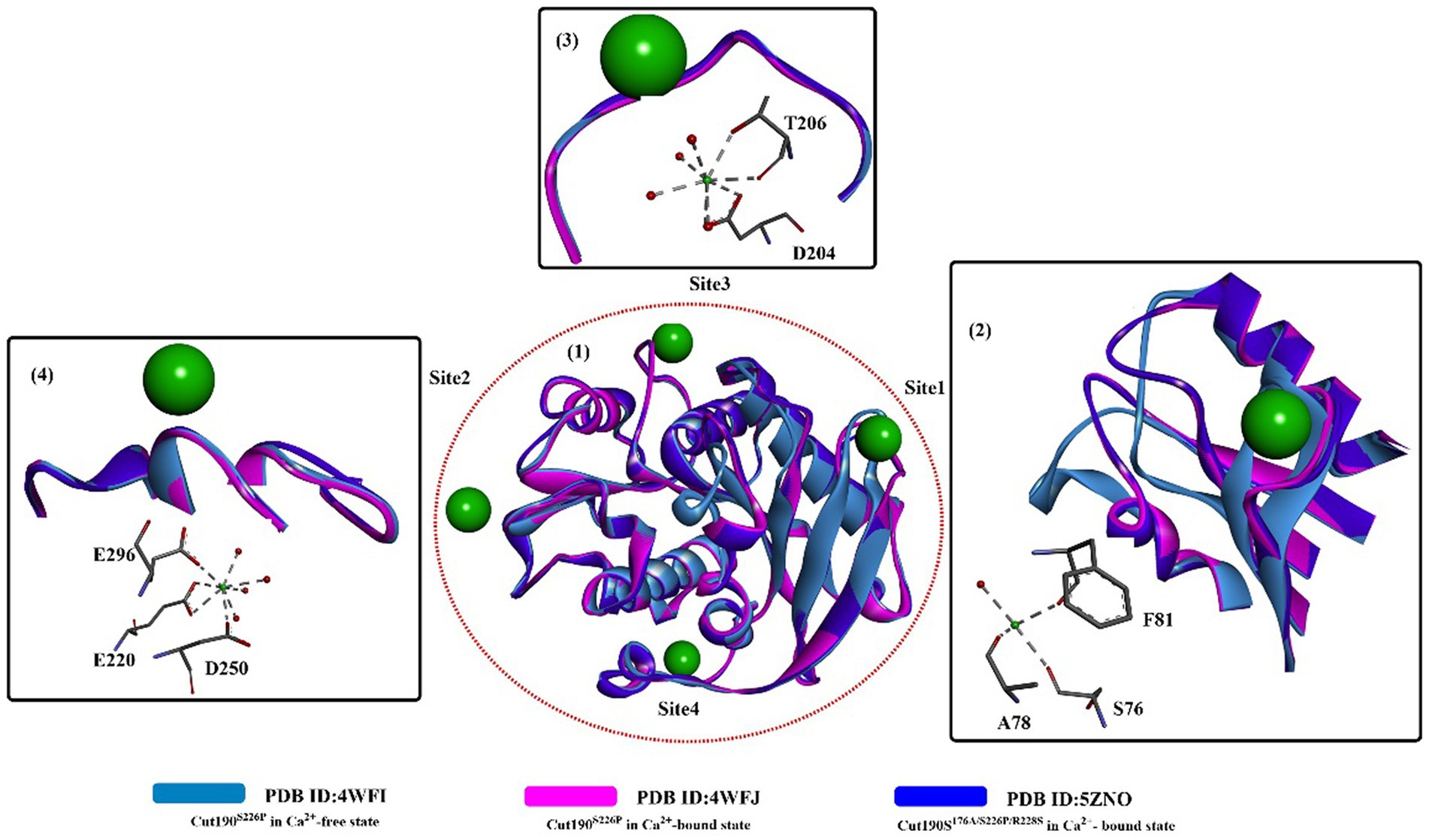
Figure 2. The structure alignment of the Ca2+-free state and the Ca2+-bound states (PDB ID: 4WFJ, PDB ID:5ZNO) (1) of Cut190S176A and the interactions of specific Ca2+ with local critical residues in the different sites (2,3,4). [The ball-and-stick models below in 2,3,4 mean the interactions formed and the dotted green (or light green) lines indicate the formed H-bonds].
Structure comparison of Cut190S176A bound with different substrates (Figure 3) clearly shows that two distinct states occur in the catalytic process, the prereaction state (engaged form, PDB ID: 5ZRR) and postreaction state (open form, PDB ID: 5ZRS). Interestingly, the structure of the postreaction state is almost identical to the structure bound with only one Ca2+. This might indicate that Ca2+ could greatly accelerate from the closed and the engaged form to the open form, which could also be illustrated by the weak Ca2+ binding interaction in site 1.
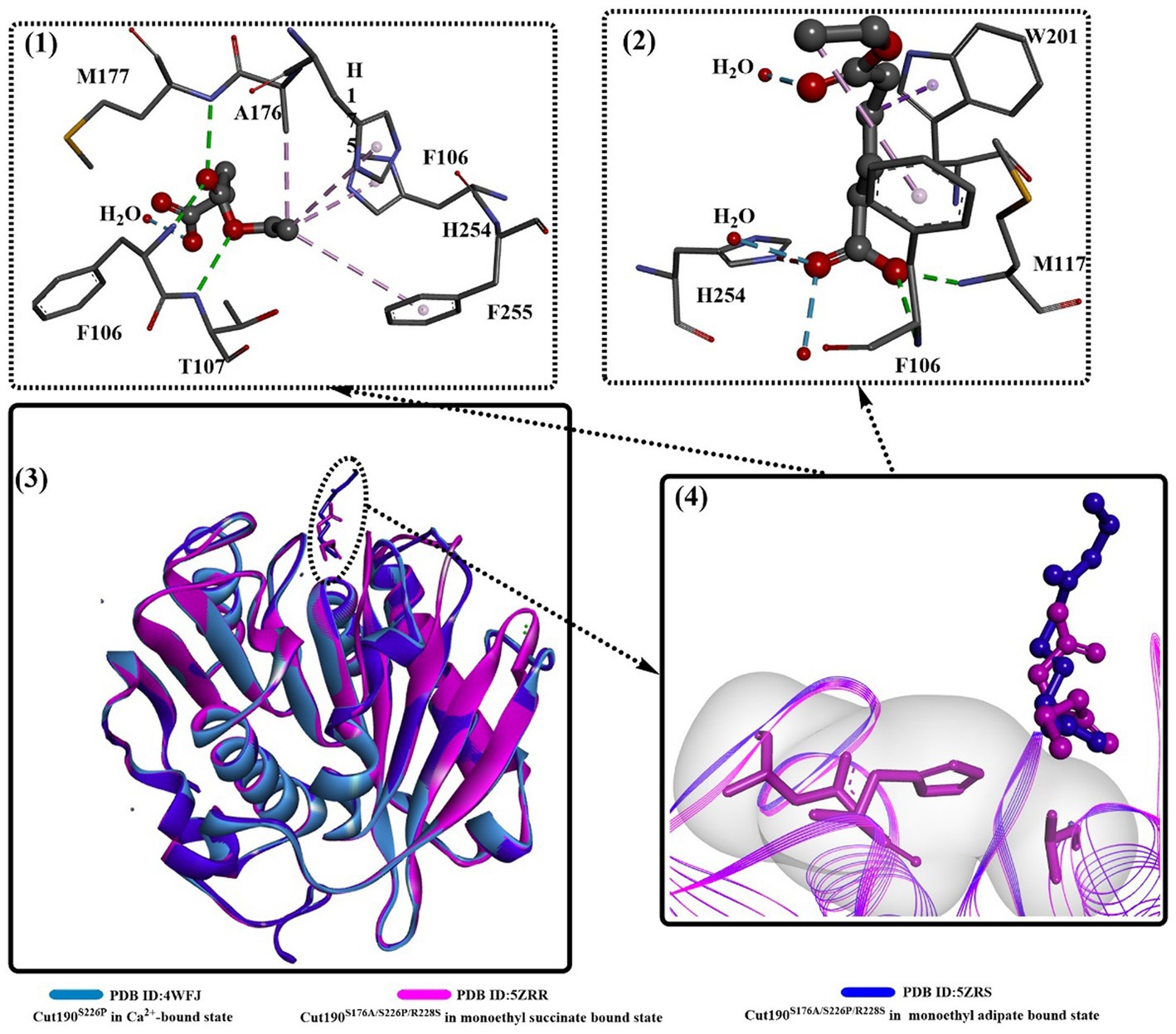
Figure 3. Structure alignment of different substrates (3), the local detailed structures (4) and the interaction modes of Cut190S176A with monoethyl adipate (1, open forms, PDB ID: 5ZRS) and monoethyl succinate (2, engaged forms, PDB ID: 5ZRR). [The dotted pink lines mean the formed hydrophobic interactions, the dotted green (or blue) lines indicate the formed H-bonds or weak H-bonds, and the line ribbons indicate the protein model].
Recently, it was reported that mutating one Ca2+-binding sites of Cut190D250C/E296C would increase its thermostability while causing little effect on its polyesterase activity (Emori et al., 2021). First, a disulfide bond would be introduced at the interface through the mutation of D250C/E296C (in site 2) without changing the overall structures (Figure 4(1)), which contributes greatly to the increase in thermostability. Furthermore, due to the weak binding of Ca2+ in site 1, the catalytic activity was unaffected when the conformational change was altered from the engaged state to the open state. The Ca2+ not bound to sites 2 and 3 is mainly caused by mutations in site 2 (D250C and E296C) and the residue nearby site 3 (N202H), which also reveals that Ca2+ binding has little effect on its overall structure (Figure 4(2)). Rapid release of the degrading products is a critical process for efficient PET-degrading, and this process has been already determined in the open form of Cut190 through molecular dynamics simulations (Numoto et al., 2018). One possibility is that the final products might be transported back to the engaged form and be tightly regulated. To deal with this issue, recently a novel state, the ejecting form at site 1 (Figure 4(3)) in the Cut190*-mediated catalytic process was determined with three C-terminal residues deleted in complex with metal ions (Senga et al., 2021). The closed-like and ligand-incompatible form displays a special conformation of F81 compared with the previous study (Numoto et al., 2018), in which either F77 and F81 all exhibit the inward conformations despite in the absence of bound Ca2+. This form is determined to allow the products to irreversibly dissociate, which should be properly controlled in the presence of an appropriate concentration of Ca2+. This phenomenon could also be illustrated by the finding that weak binding affinity of Ca2+ plays a critical role in the function of Cut190 (Senga et al., 2019). Once the products are irreversibly released, and the next cycle of reaction would be efficiently performed. In addition, deletion of three C-terminal residues contributes to the increased stability (Tm, 85°C), which is largely the consequence of the decreased fluctuation.
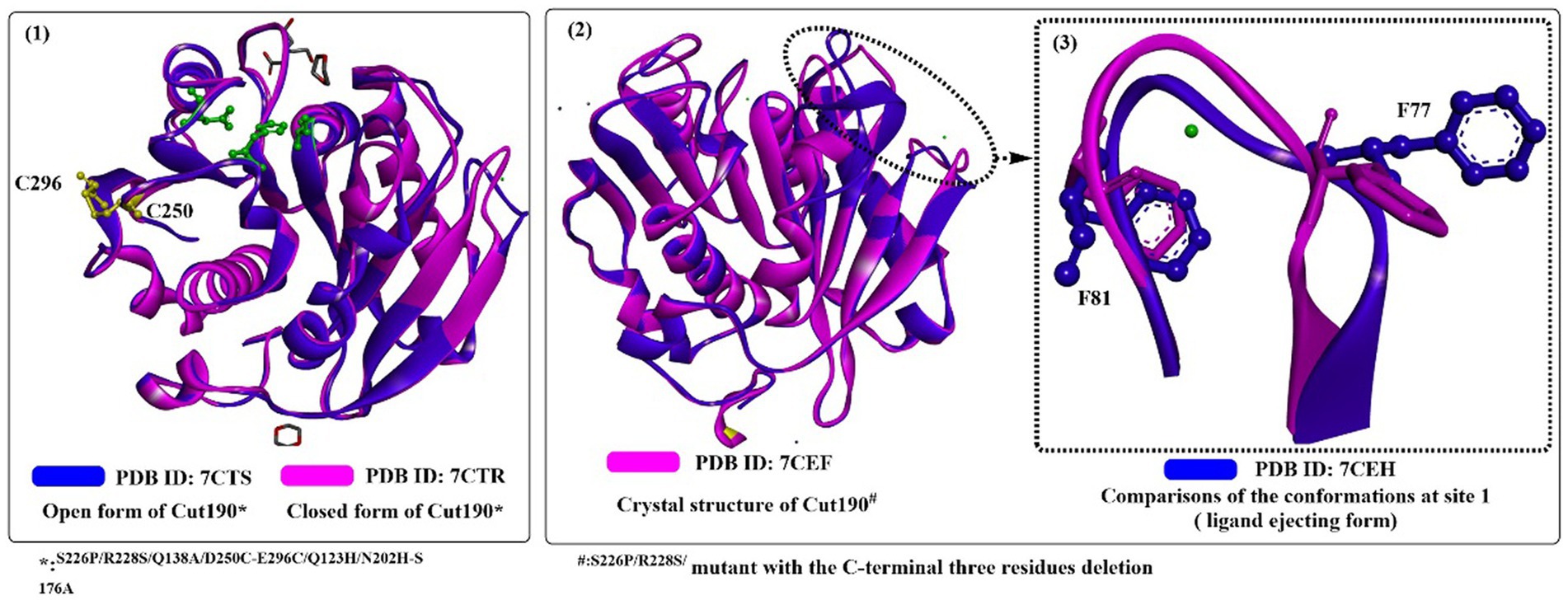
Figure 4. The structure alignment of Cut190*SS and Cut190*SS_S176A (1; PDB ID: 7CTR, PDB ID: 7CTS), Cut190** and Cut190**S176A (2; PDB ID: 7CEF, PDB ID: 7CEH) and the ejecting form of Cut190*(3). (The green residues mean the catalytic triads, and the yellow resides mean the introduced disulfide bond).
Recently, Cut190 (S226P/R228S) was further engineered (Kawai et al., 2022). Using the best performing Cut190 variant (L136F/Q138A/S226P/R228S/D250CE296C/Q123H/N202H/K305del/L306del/N307del) and amorphous PET powders, more than 90 mM degradation products were obtained in 3 days and approximately 80 mM in 1 day.
Sulaiman et al. (2012) reported the cloning and heterogeneous expression of a cutinase homolog, LC-cutinase, from a fosmid library of a leaf-branch compost metagenome. The enzyme shows 59.7% acid sequence identity to the Thermomonospora curvata (T. curvata) lipase and 57.4% identity to Thermobifida fusca (T. fusca) cutinase. The catalytic triad was formed by Ser165, Asp210, and His242, and an oxyanion hole (formed by Tyr95 and Met166) and a typical pentapeptide GxSxG sequence motif were also fully conserved in the LC-cutinase sequence (Figure 5(1,3)). A recent NMR study confirmed that Ca2+-binding site 2 (formed by D238-S283-E208, with higher affinity) and Ca2+-binding site 3 (formed by D193-T195, with weaker affinity) were also detected, which is similar to those of Cut190 (Charlier et al., 2022).
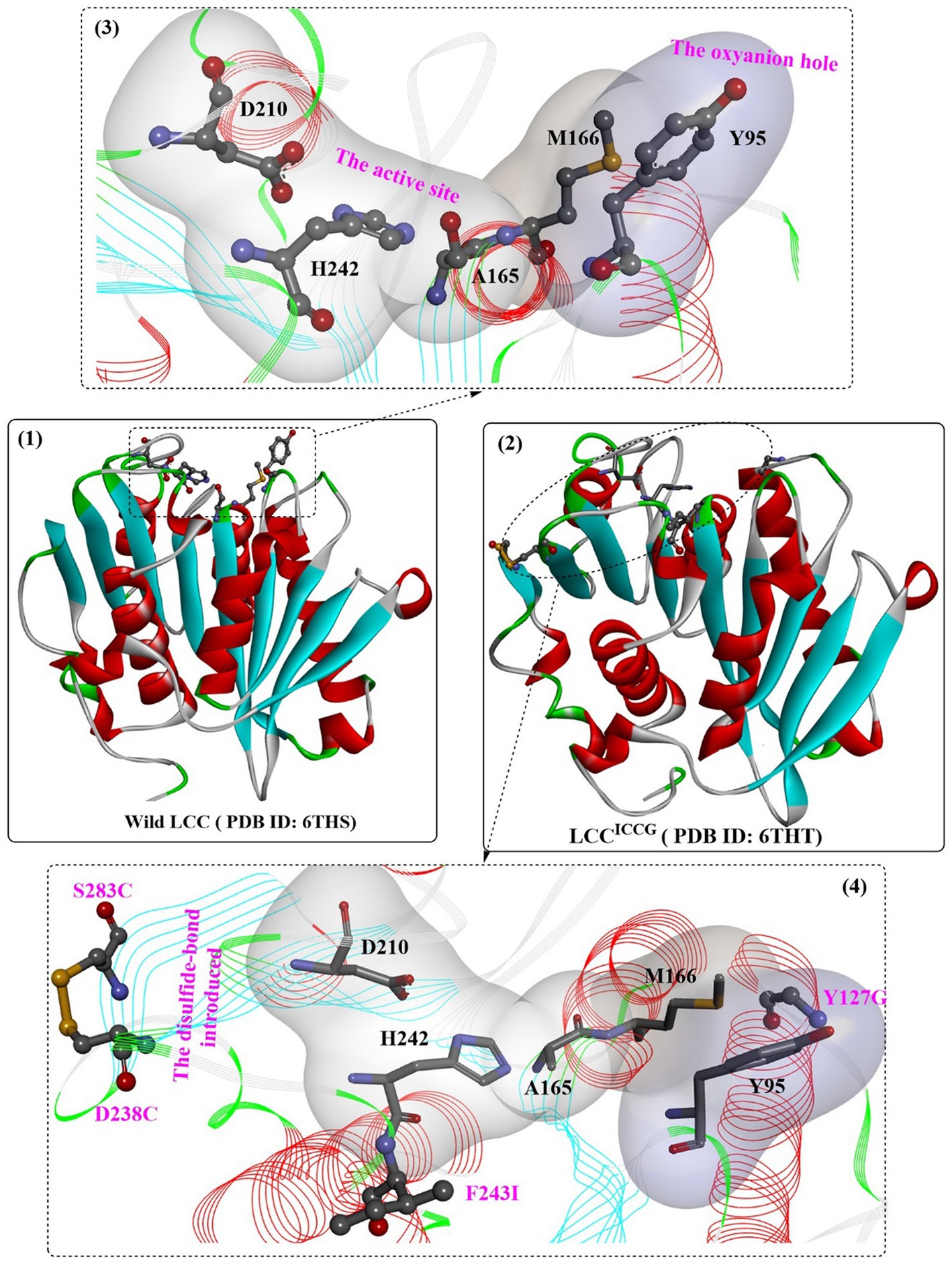
Figure 5. The structure of wild LCC (1), LCC ICCG (2) and the detailed structures of the substrate binding sites. (3: the local detailed structures of the catalytic triads and the formed oxyanion hole of wild LCC; 4: the detailed structures of the catalytic triads, the formed oxyanion hole and the introduced disulfide bond of wild LCC).
The enzyme could not only hydrolyse various fatty acid monoesters (with acyl chain lengths of 2–18) at pH 8.5 and 50°C but also degrade PET at pH 8.0 and 50°C with a specific degrading activity of 12 mg/h/mg of enzyme. At that time, the PET degradation rate of LC-cutinase was reported to be higher than the reported values for other cutinases by 230- to 970-fold. However, the thermostability is slightly less stable than that of T. fusca cutinase, with half-lives of 40 min at 70°C and 7 min at 80°C.
One of the most important variants of LCC [known as ICCG-LCC, Figure 5(2,4)] was rationally designed and characterized in 2020 by Tournier et al. and contains a four-site mutant, F243I, D238C, S283C, and Y127G (Tournier et al., 2020). In this study, molecular docking and enzyme contact-surface analysis were first performed to explore the structure–function relationship between LCC and the substrate 2-hydroxyethyl-[monohydroxyethyl terephthalate)3(2-HE(MHET)3]. Around the binding site, 15 critical amino acid residues were identified, among which 11 positions were further subjected to site-specific saturation mutagenesis, resulting in 209 variants. Although most displayed impaired depolymerization activities, F243I and F243W were identified to show improved activity, and T96M, Y127G, N246D and N246M were found to exhibit an increased melting temperature. Given that the disulfide bond enhanced thermostability, the D238C/S283C variant was designed with a disulfide bond. This led to an increase in melting temperature (94.5°C) by 9.8°C compared with that of wild-type LCC, but the catalytic activity decreased by approximately 28%. Then, F243I and F243W were added into this mutant, resulting in mutants F243I/D238C/S283C (ICC) and F243W/D238C/S283C (WCC) with higher melting temperatures (6.2°C and 10.1°C increased, respectively) but no activity losses. Finally, T96M, Y127G, N246D or N246M mutations were introduced into ICC and WCC, and the results showed that mutants ICCG, ICCM, WCCG and WCCM displayed improved melting temperatures ranging from 9.3°C to 13.4°C with little effect on specific activity compared with that of wild-type LCC. In the scale-up PET recycling process, ICCG and WCCG showed the best conversion level with 82 and 85% conversion in 20 h and 15 h, respectively. Moreover, under the same conditions, 90% depolymerization could be achieved for WCCG and ICCG after 10.5 h and 9.3 h, respectively, but a higher initial rate was observed for ICCG. The obtained ICCG mutant, as one of the most promising PET hydrolases (with a mean productivity of 16.7 g·L−1·h−1 TPA), displays great potential for future PET recycling and highlights the importance of computer-aided enzyme engineering in the field of PET degradation.
Recently, the substrate-binding mode of ICCG was investigated in detail (Zeng et al., 2022). MHET could bind to the surface cleft formed by the hydrophobic region of the active site and the oxyanion hole (Figure 6(1)). Two hydrogen bonds are formed between the carbonyl O of MHET and the main chain NH of Y95 and M166 to maintain the reaction status, and the benzene ring is further stabilized by the aromatic pi-pi interactions resulting from Y95, V212, M166 and W190 (Figure 6(2)). In addition, two water molecules are found to be associated with the formation of H-bonds with the hydroxyl at both ends of MHET. G127 and I243 (two critical mutations in ICCG) are located near the MHET-binding cleft (Figure 6(2)). G127 is located on the protein surface around the oxyanion hole region, and I243 is closer to the active site. The F243I substitution could further expand the substrate-binding site capable of increasing the PET-binding capacity, as mentioned by Tournier et al. (2020). Intriguingly, the substrate binding modes of ICCG are quite similar to those of IsPETase, with the different aromatic moiety conformations of the two bound ligands deviating by ∼30°.
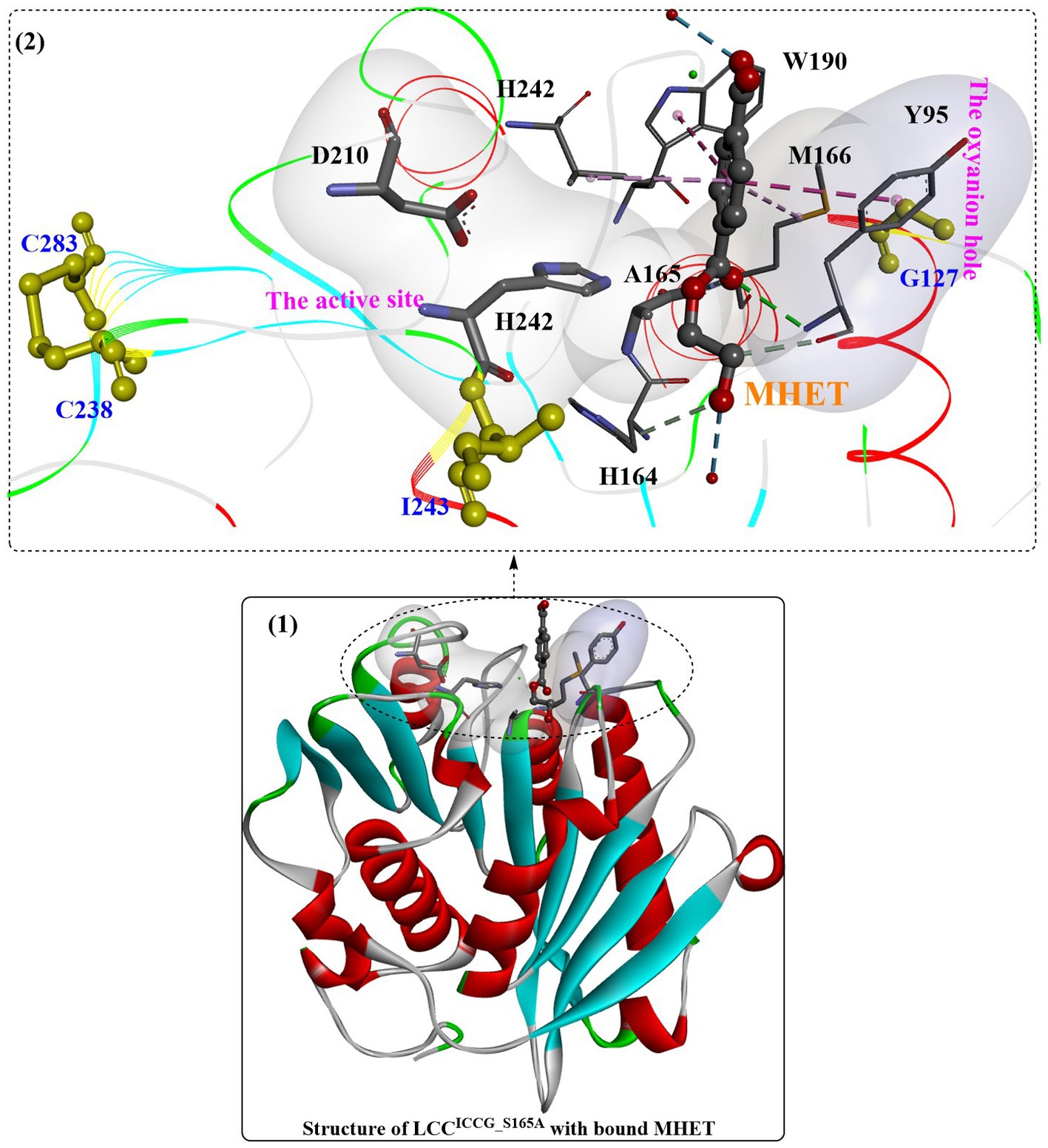
Figure 6. The complex structure of ICCG_S165A/MHET (1, PDB ID: 7VVE) and the interaction mode (2). [The line ribbons indicate the protein structure, the (or light) green and blue dotted lines mean the formed H-bonds, the pink dotes line mean the hydrophobic interactions and the yellow resides mean the formed disulfide bonds].
Based on these findings, structure-based rational design was conducted to improve the thermostability of ICCG by introducing proline or hydrophilic interactions on a protein surface or increasing internal hydrophobic interactions. Among the 27 obtained variants, mutants A59H, A59R, A59K, V63I, V75R, and N248P displayed improved activity at 90°C. Considering the functional roles of N248P, N248P combined with mutants A59R, A59K, V63I, and V75R was designed to generate seven triple mutants. The results showed that all obtained triple mutants displayed higher activity than that of ICCG at 90°C. When using reinforced PET as the substrate, mutants RIP (A59R/V63I/N248P), KIP (A59K/V63I/N248P) and KRP (A59K/V75R/N248P) exhibit more effective PET hydrolytic activity at optimal operating temperatures (80–85°C). In particular, mutant RIP showed the highest efficacy at 85°C.
Structure analysis showed that the crystal structures of KIP, RIP, and KRP are almost identical to the ICCG structure (Figure 7(1)), and the improved activities may be caused by the additional intramolecular interactions. N248P (located in the β8-α6 loop) could pack against the adjacent W104 to stabilize the β8-α6 loop and provide additional interactions between the loop and helix α1 (Figure 7(2)). For KRP, V75R forms two additional hydrogen bonds (shown in green line), and A59K could help form an additional hydrogen bond. Similarly, for RIP, A59R can pack against Y78 and form a hydrogen bond. The methyl group of the V63I mutant in RIP and KIP could stretch into a hydrophobic cluster sandwiched between the central β-sheet and helix α2 capable of strengthening the local hydrophobic interactions (Figure 7(4,5)). In summary, the hydrophilic interactions on the protein surface (N248P) and the intermolecular stabilization result from these mutations.
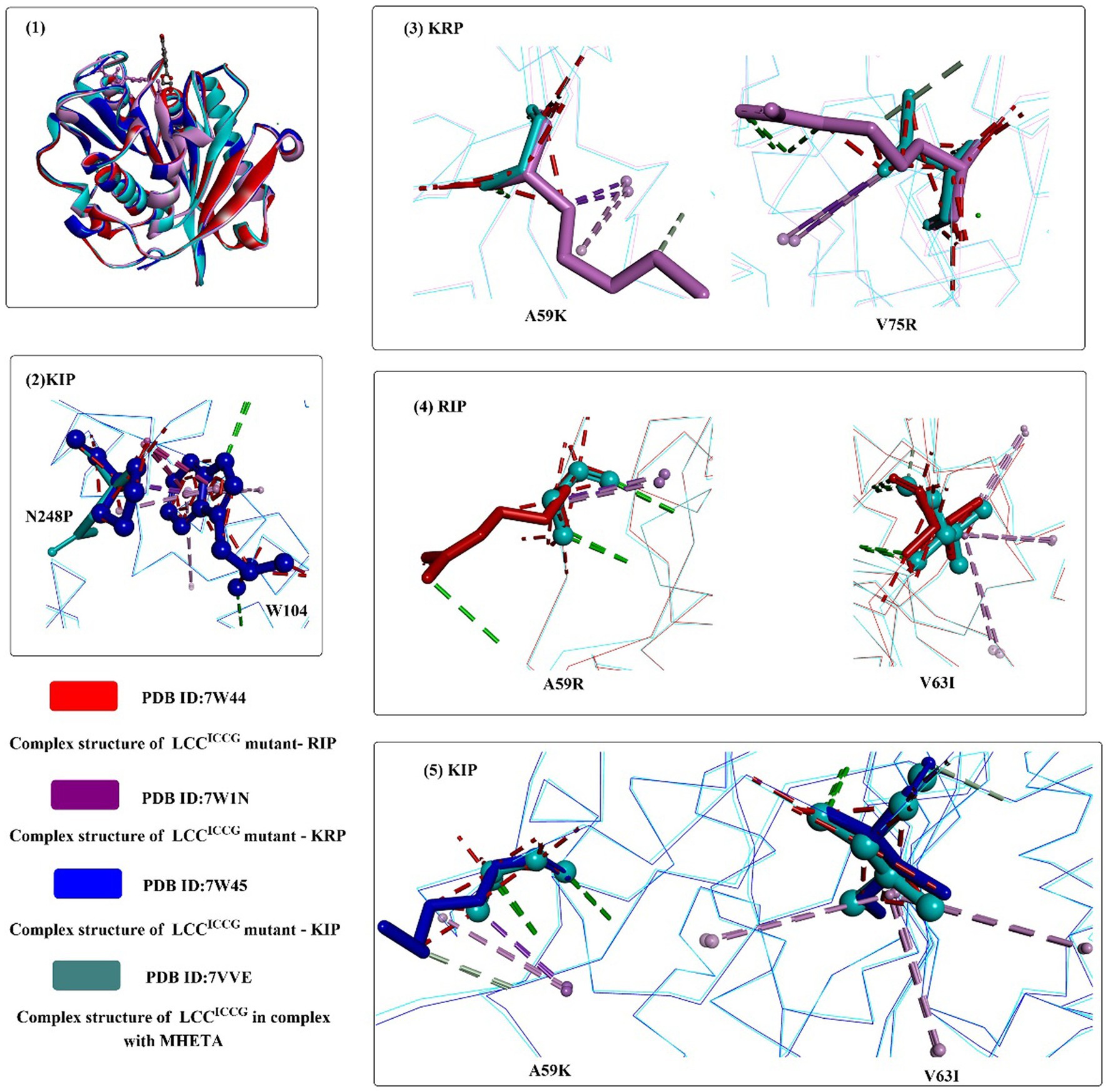
Figure 7. The structure alignment of LCCICCG-S165A with mutants RIP, KIP and KRP (1), and interaction modes of mutant amino acids with the surrounding residues of RIP, KIP and KRP compared with those of LCCICCG (2–5). (The mutant residues are shown in ball-and-stick models in different colors, the red dotted lines mean the steric bumps, and other dotted lines indicate the same as mentioned above).
In a recent study, a semirational protein engineering approach was used to improve the catalytic activities of LCC (Pirillo et al., 2023). LCC without the secretion signal (ΔLCC) was selected as the starting target. Molecular docking and molecular simulation were used to investigate the interactions between the substrate (3PET) and ΔLCC. Based on the individual ΔGbind values (≤ −1 kcal·mol−1) and the evolvability of each ΔLCC residue predicted, critical residues Y95, T96, Y127, M166, and V212 were selected as the binding hot spots capable of regulating the overall complex stability. The results obtained from site saturation mutagenesis (SSM) revealed that mutations at positions 212 and 243 lead to a significant fraction of improved variants, and F243T, F243I, V212T, V212M and T96H variants especially showed a significant increase in activity on PET nanoparticles at 50°C compared with ΔLCC (increased by ~1.75-fold, ~1.51-fold, ~1.34-fold, ~1.33-fold, and ~ 1.25-fold, respectively). For the biodegradation of PET film at 72°C, only the F243T variant showed an increased activity with a production of 21.9 mM of products with a reaction rate of 0.48 mmolproducts·mgenzyme−1·day−1, which is ~1.2-fold higher than that with wild-type ΔLCC. Then, mutations at A97 and S101 were introduced into ΔLCC_ F243T, which may play a role in the overall binding energy of the enzyme-PET complex. Four double variants (A97G/F243T, A97T/F243T, S101N/F243T and S101V/F243T) displaying significantly increased activity for PET degradation were obtained; furthermore, the S101N/F243T and A97T/F243T mutants showed the largest increase in activity for the degradation of PET nanoparticles (compared with ΔLCC_F243T and ΔLCC). S101N/F243T was more efficient than that of A97T/F243T mutants for the degradation of amorphous PET film at the tested temperatures and 2.0- and 3.5-fold faster than that of the wild ΔLCC at 55°C and 60°C, respectively. This study provides insights into the catalytic mechanism of LCC, especially the functional role played by F243, which should be further confirmed experimentally by crystal structures of the S101N/F243T complex.
In a very recent study (Fang et al., 2023), whole-cell screening assays combined with rational design and combinatorial mutagenesis were applied for the engineering of LCCICCG. It was revealed that the mutant RITK (LCCICCG-D53R/R143I/D193T/E208K) displayed better whole-cell biocatalytic performance with an 8.33-fold increase in biocatalytic activity compared to those expressing ICCG. Moreover, the thermostability was also enhanced with a 12.75-fold increase in depolymerization compared to ICCG at 85°C for 3 h. However, I believe further studies on the catalytic performance of RITK against different PET substrates with different crystallinities and the expression efficiency of different protein with the same expression vector. In a similar study, machine learning combined with evolutionary analysis was used for the rational design of LCCICCG (Ding et al., 2023). Results showed that mutants (S32L, D18T, S98R, T157P, E173Q, N213P) all displayed higher degrading-activities toward PET powder than wild LCCICCG at 75°C. Based on this finding, LCCICCG_I6M (carrying mutations S32L/D18T/S98R/T157P/E173Q/N213P) was constructed and was further determined to display the highest activity for PET powder (crystallinity 39.07%) at 75–80°C, which could release 4.028 ± 0.078 mM products with a 4.63-fold at 80°C compared with LCCICCG (0.871 ± 0.050 mM). In addition, the PET degrading performance against high crystallinity commercial PET materials (water bottle) was increased by 3.64-fold with 31.91 ± 0.99 mM soluble products detected compared with LCCICCG. Moreover, the stability was also improved with the optimal temperature increased from 65°C to 80°C. Molecular simulations suggested that N213P mutations might play a role in increasing the overall structural rigidity, and the E173Q mutation might be capable of enhancing the flexibility of the substrate-binding pocket of LCCICCG_I6M.
Two open reading frames encoding cutinases named Tfu_0882 (TfCut1) and Tfu_0883 (TfCut2) were identified in the thermophilic bacterium Thermobifida fusca (Chen et al., 2008). Although they share a high sequence identity of 93% in the amino acid sequence (261 aa), TfCut2 displayed much higher (~ two times higher) degradation activity than that of TfCut1 (Chen et al., 2010). TfCut2 displays 57.4% similarity in amino acid sequence with LCC. The homology model of Tfu_0883 displays a typical α/β-hydrolase fold with a canonical triad S170-H248-D216, a preformed oxyanion hole and a different nucleophilic serine exposed to the solvent. Although they show similar catalytic properties to fungal cutinases, TfCut1 and TfCut2 are a different subfamily of cutinases due to their sequence and structural differences. For example, they can hydrolyse insoluble triglycerides and soluble esters (e.g., pNPB) in addition to cutin. They also display superior thermostability compared with that of the F. solani pisi cutinase. A study showed that the higher linear degradation rates observed in the first 72 h of incubation are due to the high hydrolysis susceptibility of the mobile amorphous fraction (MAF, with a chain mobility similar to the purely amorphous polymer) of PET; however, a transition of MAF to the rigid amorphous fraction (RAF, with a lower chain mobility probably caused by its presence within the intraspherulitic or interlamellar regions in close vicinity to the crystalline domains) during further incubation resulted in drastically decreased degradation rates (Wei et al., 2019). The RAF could be cleaved only in an endo-type manner at a much lower rate.
To investigate the catalytic mechanisms of the bacteria-derived cutinases, several crystal structures of TfCut2 have been reported [e.g., PDB ID: 4CG2 (Roth et al., 2014), Figure 8, 5ZOA, 7QJR, 7XTT]. The structure of TfCut2 forms a classical α/β-hydrolase fold, contains conserved GXSXG motif, and contains a conserved catalytic triad of S-H-D residues is in a crevice on the surface. In the enzyme-inhibitor complex structure (Figure 8(2)), it was found that covalent modification of S130 would lead to the tighter interactions formed with the main chain of M131 and Y60 mainly through H-bonds interactions (Figure 8(3)). The phenyl ring is further stabilized mainly by hydrophobic interactions with I178 and Y60 (Figure 8(2)).
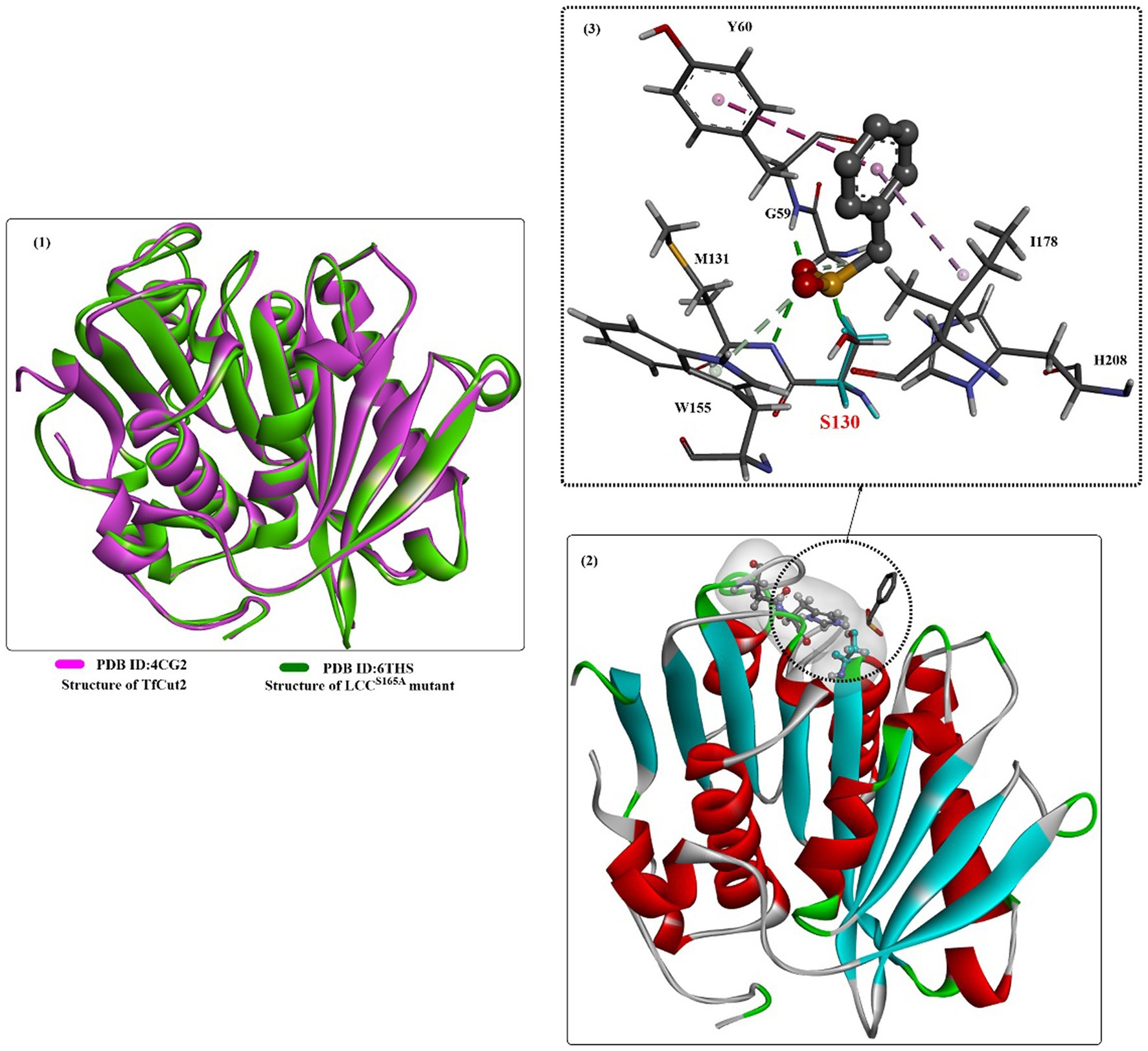
Figure 8. The structure alignment of TfCut2 with LCCS165A (1), the crystal structure of TfCut2 with the inhibitor-phenylmethanesulfonic acid and the hydrophobic surface around the active site (2), and the local detailed interaction mode of the complex.
Efforts have been made to further improve the catalytic performance of TfCut2 through directed evolution and rational mutagenesis. The critical residues associated with substrate recognition were engineered (Wei et al., 2016). First, critical residues around the active site close to (without direct interactions with) the docked 2PET were identified and genetically engineered by performing structure alignment with LCC and TfCut2. Four residues (G62, T63, I178, and I213) were selected for further protein engineering. Among the single mutants, G62A was determined to show markedly increased hydrolytic activity. The improved catalytic performance of G62A likely resulted from a reduction in product inhibition due to MHET rather than the altered surface hydrophobicity or the enlarged substrate binding pocket. Although the double variant G62A/I178V and the triple variant G62A/I178V/I213S showed much higher hydrolytic activity than the wild-type TfCut2, they all displayed slightly lower degrading activity than the single mutant G62A. The single mutant I213S showed the highest thermostability, followed by G62A and G62A/I213S. After 24 h at 65°C, G62A and G62A/I213S caused a 20.5% weight loss of PET films.
In a similar study, the functions of critical residues around the substrate binding site were investigated, which were thought to facilitate the accessibility of the enzyme’s active site (Dong et al., 2020). Eleven bulky residues were selected to increase the substrate binding efficiency by site-directed mutation to Ala, which are located around the catalytic triad, the oxygen hole and the substrate binding region. The results showed that Y60A, T207A, F209A, L90A and I213A exhibited increased pNPB hydrolysing activity, especially the cutin hydrolysing activities of L90A, I213A, and L90A/I213A, which were increased by 5-, 2.4-, and 3.2-fold, respectively. MD simulations revealed that the higher flexibility of the substrate binding site of mutants L90A and I213A might lead to increased hydrolysing activities toward cutin. However, a subsequent study showed that the L90A mutant showed ~11.2-fold reduced degradation activity on commercially sourced PET film, but the L90F mutation displayed considerably increased degradation (~1.5-fold increased) activity (Mrigwani et al., 2023). This might be caused by the more efficient binding of PET resulting from the replacement of the hydrophobic phenylalanine residue.
Based on the abovementioned findings, molecular rational engineering of TfCut2 was performed by docking and MD simulations to modify the space at the active site and streamline the hydrophobicity at the active site or other interacting regions (Mrigwani et al., 2023). The results showed that the selected residues H129W, W155F, H184A, and H184S for engineering the PET-binding cleft exhibited little positive effect on the catalytic activities. Residues (A173C) that were identified for the insertion of thiols/disulfides (A173C/A210C, and A173C/ A206C) might distort the PET-binding site, causing a complete loss of activity against solid PET. Interestingly, residues (L90) distal to the active site could adjust its catalytic activity, especially the L90F mutation, which could considerably increase TfCut2’s PET-degrading activity (~1.5-fold). F209I, F249A and F249R were constructed to modify the surface hydrophobicity proximal/distal to the active site. The results showed that the single mutant (F209I) caused an ~4.4-fold decrease in activity; however, F249R led to an ~1.27-fold increase in activity against solid PET. The double mutants displayed a remarkable increase in degradation activity (G62A/F209I with an ~6.4-fold improvement and G62A/F249R with an ~7.15-fold increase); however, their thermostability was reduced. These results also revealed that TfCut2 possesses high thermal stability, high kinetic-thermal stability, and high thermodynamic stability.
Based on the findings of the double mutation (DM) strategy (Chen et al., 2021), engineered TfCut was investigated for efficient biodegradation of poly(butylene adipate-co-terephthalate) (PBAT) (Yang et al., 2023). The results showed that the mutant (TfCut-DM) is a highly potent catalyst that can completely decompose PBAT films in 36 h and release 3-fold more TPA (the major product) than TfCut within 12 h. This strategy was also determined to be appropriate for BurPL and TcCut. The overall complex structure of the MHET-bound TfCut complex structure is almost identical to that of TfCut-DM (Figure 9(1)), and the DM residues (S224 and I228) are located below the catalytic triad. The ethylene glycol motif of MHET bound points outside the enzyme, and the aliphatic part stretches to fill a cleft across the protein surface (Figure 9(3)), which is lined by several critical residues, such as M171, W195, Y100, I218, and T223. Several water molecules are also shown to be associated with the binding of MHET through H-bond interactions. Interestingly, the side chain of W195 in TfCut-DM exhibits a B-type conformation [Figure 9(2) in purple]; however, wild-type TfCut harbors a TPA-binding Trp in the C-type conformation [Figure 9(2) in brown]. The DM mutation and the B-type W195 lead to a more open and flatter TPA-binding pocket (increased flexibility), which would probably cause less hindrance for the entrance of bulkier substrates. This may be the dominating factor behind the improved hydrolytic activity toward PBAT and PET.
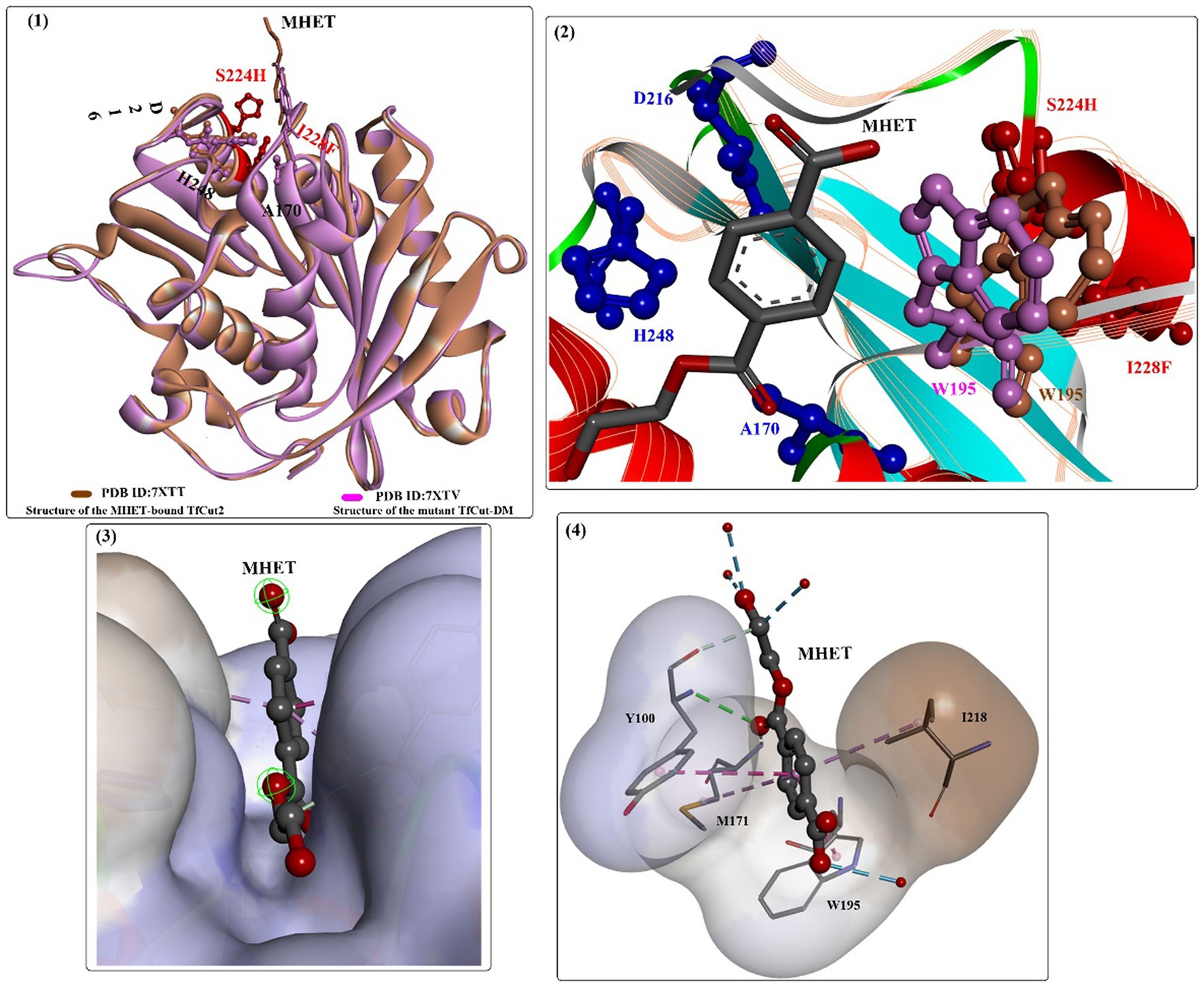
Figure 9. The structure alignment of MHET-bound TfCut (PDB ID: 7XTT) and TfCut-DM (PDB ID: 7XTV) (1), the different conformations of W195 associated with MHET binding (2, shown in purple and brown respectively), the binding mode of MHET in the binding cleft (3) and the detailed interaction modes (4).
Then et al. (2016) directly introduced a disulfide bridge (D204C-E253C) by substituting the calcium binding site of TfCut2 to eliminate dependence on calcium. The results showed that the thermal stability of the mutant was further improved, the melting point was increased to 94.7°C (TfCut2: 69.8°C), and the half-inactivation temperature was increased to 84.6°C (TfCut2: 67.3°C). Most curiously, the hydrolytic activity of the calcium-independent thermostable and efficient hydrolase was not affected when the calcium binding site was replaced. A similar study was carried out with the anchor peptide-cutinase mutant Tfuc2D204C/E253C (Liu et al., 2022). The PET degradation rate of the obtained fused protein was determined to be up to 57.9% at 70°C for 96 h, and the catalytic performance was improved by 22.7-fold compared with that of Tfuc2. Based on machine learning, Li et al. successfully designed a highly thermostable Tfuc2 mutant, S121P/D174S/D204P, by mining molecular dynamics simulation trajectories and rational introduction of hydrogen and ionic bonds (Li et al., 2022). The results showed that the PET degradation ratio was further increased by 46.42-fold at 70°C compared to that of wild-type TfCut2.
To further increase the thermostability of the polyester hydrolases from the thermophilic actinomycete Thermobifida fusca, rational engineering of the binding sites of specific metal ions (e.g., Mg2+ and Ca2+) was also investigated (Then et al., 2015). Based on the MD simulations, residues Asp174, Glu253, and Asp204 of TfCut2 might play roles in facilitating the binding site of Ca2+, which were then mutated to screen for stability-enhanced mutants. The results showed that mutants D204R and E253R increased by more than 14°C compared to that of wild-type TfCut2; however, the PET-degrading activities were determined to be slightly reduced. In addition, it was found that the addition of a cationic surfactant (C12-N(CH3)3+) greatly improved the degradation activity of TfCut2, which might be caused by the aggregation of enzymes near the PET film surface via electrostatic interactions (Arnling Bååth et al., 2022). The results showed that in the presence of cationic surfactant (30 ppm C12-N(CH3)3+), the activity of the TfCut2G62A/F209A mutant increased to 31 ± 0.1 nmol min−1 cm−2, which is approximately 12.7 times higher than that of wild-type TfCut2 in the absence of surfactant.
From the bacterium HR29, a close homolog of the LCC with 94% sequence identity was identified (BhrPETase, NCBI accession No. GBD22443) (Kato et al., 2018). Recently, high-level secretory expression of BhrPETase in Bacillus subtilis was successfully achieved with an expression titer of 0.66 g/L (Xi et al., 2021). More strikingly, a strong preference of BhrPETase for high temperature was determined for BhrPETase when the reaction temperature was increased from 30 to 90°C. Even at 90°C, the catalytic performance remained superior to that of wild LCC, which rapidly dropped to 55% of the maximum activity. The Tm value of BhrPETase was determined to be 101°C without the addition of Ca2+ ions, ~11°C higher than that of LCC. Against a crystallinity of 11.2% PET powder for 20 h at 70°C, approximately 6.3 mM degradation products were detected. Molecular modeling suggested that the increased thermostability might be caused by Ser175, Gln202, Pro199 and Pro248 in BhrPETase, which could either result in additional formed hydrogen bonds and/or increase the rigidity of the flexible loop.
In a recent study, molecular engineering of the hydrolase from the bacterium HR29 was reported by means of a protein language model and force-field-based algorithms (under review3). The obtained TurboPETase (BhrPETaseH218S/F222I/A209R/D238K/A251C/A281C/W104L/F243T, Tm of 84°C) exhibited a 4.4-fold increase in the degrading activities toward GF-PET films compared to that of wild-type BhrPETase. Moreover, the depolymerization rate of TurboPETase (21%) outperformed all the tested PET hydrolases, which was 4-, 4-, 8- and 33-folds higher than those of wild BhrPETase, LCCICCG, HotPETae and FastPETase at 65°C within 3 h, respectively. At 65°C after 24, approximately 100% depolymerization could be obtained. Especially, the full degradation of pretreated PET (up to 300 g L−1) could be accomplished within 10 h, with a maximum production rate of 77.3 gTPAeq L−1 h−1. It shows beyond doubt that this strategy and the obtained TurboPETase might serve as a promising option for the engineering of interfacial enzymes and efficient PET cycling.
Thermobifida alba strain AHK119 (AB298783) was shown to be capable of degrading aliphatic-aromatic copolyester film and polymer particles and releasing TPA (Hu et al., 2010). From AHK119, an esterase gene (est119) was cloned and identified, and the Est119 protein contains the highly conserved motif (G-X-S-X-G) of the serine hydrolase family, a catalytic triad (Ser129, His207, and Asp175) and the oxyanion hole formed by Tyr59 and Met130. The optimal reaction temperature and pH were found to be ~50°C and 6.0, respectively. Kitadokoro et al. determined the crystal structure of Est119 (Figure 10(1)), which shows a typical α/β-hydrolase fold that also consists of a central twisted β-sheet of 9 β-strands flanked by 9 α-helices on both sides (Kitadokoro et al., 2012).
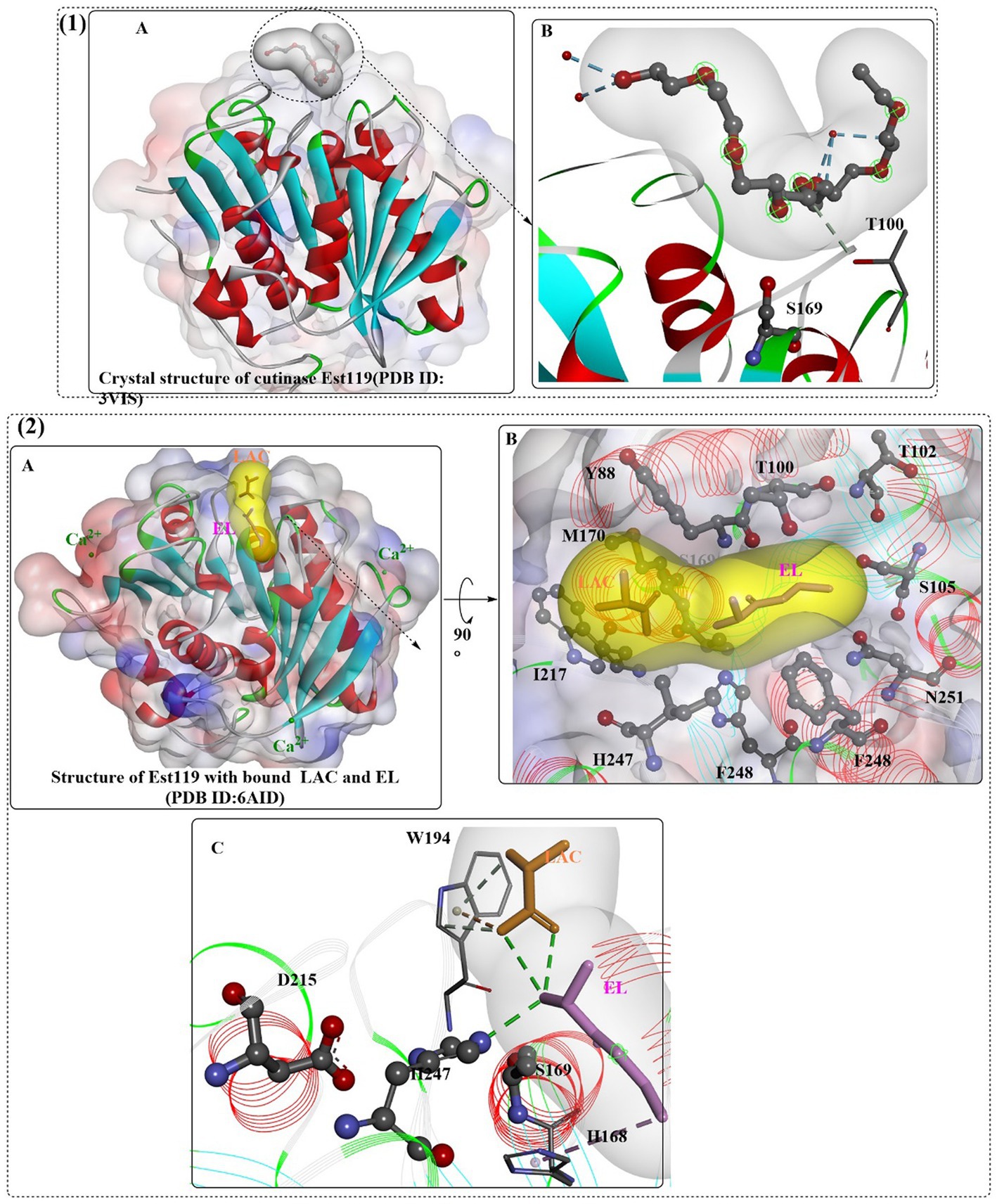
Figure 10. The crystal structures of Est119 with bound substrate PE4 (1A) and the local detailed interactions of the Est119- PE4 complex (1B); the crystal structures of Est119 with bound substrate LAC and EL (2A) and the local detailed interactions of the Est119- PE4 complex (2B,C). (The white and yellow surface mean the hydrophobic surfaces of the substrates in the active sites, and the colorful ones mean the hydrophobic surface of the whole protein).
In another study (Thumarat et al., 2012), it was further confirmed that Est119 is a typical cutinase-like enzyme, and Ca2+ was necessary to display full catalytic activity (300 mM) and maintain thermostability (200 mM, stable at 50°C for 16 h). Random mutagenesis analysis indicated that mutations with more hydrophobicity at the N-terminus would more likely result in enhanced activity, which led to mutants A68V and A68V/S219P showing the highest activity against pNPB. In particular, the degradation activity of A68V/S219P was further improved by ~50-fold and 1.7-fold compared with that of WT and A68V, respectively. In addition to est119, its homolog est1 was located upstream of est119 (Thumarat et al., 2015). Although showing 95% sequence identity and 98% sequence similarity with Est119, Est1 displays a higher (~2-fold higher) activity toward p-nitrophenyl butyrate. Moreover, Est1 displayed a much higher thermostability, with 300 mM Ca2+, and it could maintain nearly 100% of the residual activity at 50°C for 20 h. Est1 and Est119 could also recognize the aromatic polyester and produce EG and TPA. As mentioned above, Est1A68V/T253P was constructed, and the results showed that the Tm value was increased to 79°C and the specific activity (833 ± 22.5 U/mg) was also improved by ~57.4-fold against pNPB. The increased thermostability was estimated to be caused by the introduction of proline, which could decrease the flexibility and increase the structural rigidity, especially at sites 219 and 253. However, Est1A68V/T253P/M259K was shown to display a higher catalytic activity than Est1A68V/T253P, with a specific activity of 910 ± 42.0 U/mg, and the thermostability was lower than that of Est1A68V/T253P over 50°C.
In a recent study, the crystal structure of Est119 complexed with ethyl lactate (EL) and lactic acid (LAC) was obtained (Figure 10(2)) (Kitadokoro et al., 2019). As expected, three Ca2+ ions were found in Est119; moreover, LAC and EL are located in the active site cleft with different positions. Based on the crystal structure, it was found that the substrate-binding cleft was a long narrow groove lined with F248, I217, H247, S169, W194, M170, Y99, T100, T102, S105, and N251. In the binding site, EL was stabilized by the H-bonds formed with H247 and water and the hydrophobic interaction with H168. In addition, LAC was regulated by the hydrophobic interaction with W194 and the H-bonds formed with water. The catalytic S169 is located at the bottom of the cleft.
Recently, a novel directional-path modification (DPM) strategy was developed to improve the performance of PET-degrading enzymes, which mainly contains the introduction of positively charged amino acids and binding groove remodeling (Chen et al., 2022). First, the PET binding path was identified. Then, to decrease steric hindrance in the binding site at the exit of the PET binding path, neutral short side-chain amino acids were introduced. At the same time, positively charged amino acids with shorter flexible side chains (e.g., Lys) were selected to optimize the surface positively charged area. These modifications led to the identification of 13 potential residue sites and construction of 23 target mutants, and the results showed that the four-point mutant (4Mz, H184S/Q92G/F209I/I213K) displayed the highest degradation rate of ~90%. The degradation efficiency was increased by approximately 30-fold compared with that of the wild type. Molecular simulations revealed that the PET binding modes of 4Mz were extended to the left side, which was estimated to be more conducive to improving PET degradation. This finding was also confirmed by positive vacuum electrostatics on the left-side subbinding site. When applied to LCC, Est119, and BhrPETase, the constructed mutants LCCH218S/Y127G/F243I/S247K, Est119H248S/Q131G/F248I/I252K and BhrPETaseH184S/F93G/F209I/S213K all showed dramatically increased PET degradation efficiency, indicating the high efficiency and universality of the DPM strategy.
In a recent study, a novel thermophilic polyesterase from Deinococcus maricopensis (DmPETase) was characterized (Makryniotis et al., 2023). DmPETase, a phylogenetically distinct and thermophilic polyesterase, was found to display a similar overall structure with that of other bacterial cutinases, such as LCCICCG and IsPETase, and the catalytic triad is determined to be Ser185, His263 and Asp231. The optimal temperature for DmPETase was 50°C lower than that of LCCICCG, and display the highest catalytic efficiency toward pNPB. It is also capable of degrading various synthetic polymers, including PET, polyurethane, as well as four semi-crystalline aliphatic polyesters. On the whole, DmPETase shows a much lower catalytic activities compared with those of LCCICCG, for example against the PCL powder DmPETase displayed a degrading approximately 70% of the tested materials compared with 98.7% obtained by LCCICCG. However, at 50°C, it displays a comparable degrading activity compared with that of LCCICCG against the semi-crystalline sections of post-consumer PET bottles (crystallinity, 41%).
The cutinase from the fungus Humicola insolens (HIC, Figure 11) is a single-domain protein that shows high hydrolytic activity and stability at 70°C (Carniel et al., 2021), and sequence analysis showed that HiC exhibits high sequence identities to FsC (56%) (Kold et al., 2014).
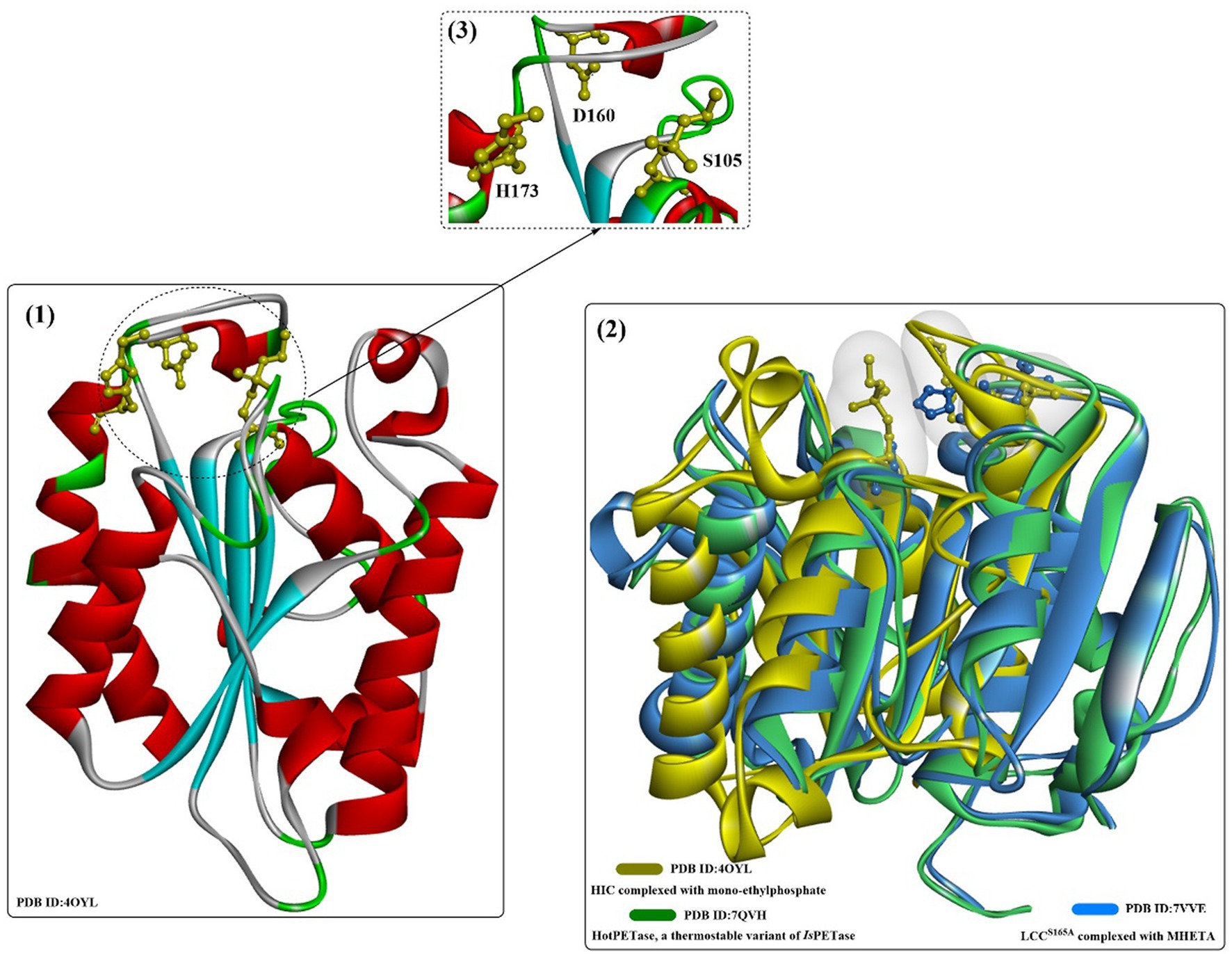
Figure 11. The structure and active site of HIC (1), the structure alignment of bacterial LCC (2, in blue) and IsPETase (2, in green), and the local detailed structure of the active site of HIC. [The hydrophobic regions of the substrate binding site are highlighted in white surface, and the catalytic triads are shown in ball-and-stick model in (3)].
When used in moist-solid reaction mixtures instead of the typical dilute aqueous solutions or slurries, cutinase from Humicola insolens can directly depolymerize amorphous and crystalline regions of PET equally without any pretreatmentl a 13-fold higher space–time yield and a 15-fold higher enzyme efficiency were achieved than those reported in prior studies with high crystallinity material. Furthermore, this process shows 26-fold selectivity for terephthalic acid over other hydrolysis products (Kaabel et al., 2021).
In an earlier study, it was found that cutinase from Humicola insolens (HIC) could hydrolyse PET, especially the low-crystallinity (7%) film (~10-fold higher than biaxially oriented PET, 35% crystallinity) (Ronkvist et al., 2009). Moreover, HiC shows good thermostability and functions best from 70 to 80°C, and the HiC-catalyzed degradation of the lcPET film resulted in a 97 ± 3% weight loss only releasing the water-soluble TPA after 96 h at 70°C. Based on this finding, a novel synergistic chemoenzymatic hydrolysis of PET was developed to produce terephthalic acid (TA) from textile waste (Quartinello et al., 2017). First, conversion of PET into TA (purity, 85%) and small oligomers was achieved by chemical treatment (T = 250°C, p = 40 bar). Afterwards, the obtained oligomers were further hydrolysed by HiC, yielding ~6.5 mM TA after 6 h of incubation at 50°C (purity, 97%). This is a practical strategy for the efficient recycling of PET in an environmentally friendly way under neutral conditions, especially for promising catalysts displaying lower catalytic activities toward high-crystallinity PET.
Carniel et al. (2021) investigated the HiC-assisted hydrolysis of postconsumed PET (PC-PET) through controlling the pH and performing fractionation of enzyme feeding in stirred reactors. The results showed that pH variation plays a critical role in the HiC-catalyzed reaction, and a 0.86 pH variation (optimal pH 8.95 ± 0.20) finally led to a decrease of 32% in the total of final products. Compared with the tris-buffered reaction, the soluble hydrolysis products from the unbuffered HiC-catalyzed reaction with pH control were increased by 81% at 50°C (26.3 mM). Although enzyme feeding fractionation contributed little activity for the improved PET hydrolysis, an increase of 2.41-fold in the final degrading products released (97 mM) per protein amount was obtained when only half of the enzyme was loaded. These findings are important for the industrial PET biorecycling process.
Additionally, in a recent study, HiC-catalyzed PC-PET degradation was optimized (PET particle size, pH, ionic strength, and enzyme concentrations) for improved performance with experimental and mathematical modeling approaches (Eugenio et al., 2021). The results revealed that the PET particle size might play the most critical role in HIC-catalytic PET hydrolysis, and with a greater surface area (semicrystalline PC-PET), an 8.5-fold increased hydrolysing rate (1.7 mmol·L−1·h−1) was achieved. After 4 days at 70°C, 10.9 mmol·L−1 TPA was detected against the semicrystalline PET (0.075–0.25 mm). The thermostability of wild HiC is much higher than that of the other reported cutinases, the half-life of which was determined to be 110 h at 70°C and pH 7.0. These results indicated that HiC displays great potential as a promising biocatalyst for efficient PET biodepolymerization.
SDS might be the most widely applied anionic surfactant and can regulate the bioactivities of cutinases or cutinase-like PET-degrading enzymes. The interactions of cutinase from Humicola insolens (HIC) with anionic surfactants (SDS) were investigated in several studies (Nielsen et al., 2005; Kold et al., 2014). The protein could specifically bind SDS at a very low molar ratio of SDS/HiC, and amino acids 160–163 and 168–176 (near the active site) were determined to be able to directly interact with SDS by MD simulations. Kold et al. (2014) investigated the detailed molecular interactions between SDS and HiC based on thermodynamic and structural studies. Crystal structures showed that the binding mode of SDS is quite similar to the previously reported substrate analogs to cutinases (Figure 12(2)), and the binding affinity (K) was determined to be ~105 M−1. In addition, mono-ethylphosphate (MEP) was found to covalently bind to S105 and could interact with F132, A108, A109, Q106, S28 and N69 by forming H-bonds (Figure 12(3)). Moreover, ethylated H173 was detected, probably resulting from the reaction with DNPP (diethyl p-nitrophenyl phosphate).
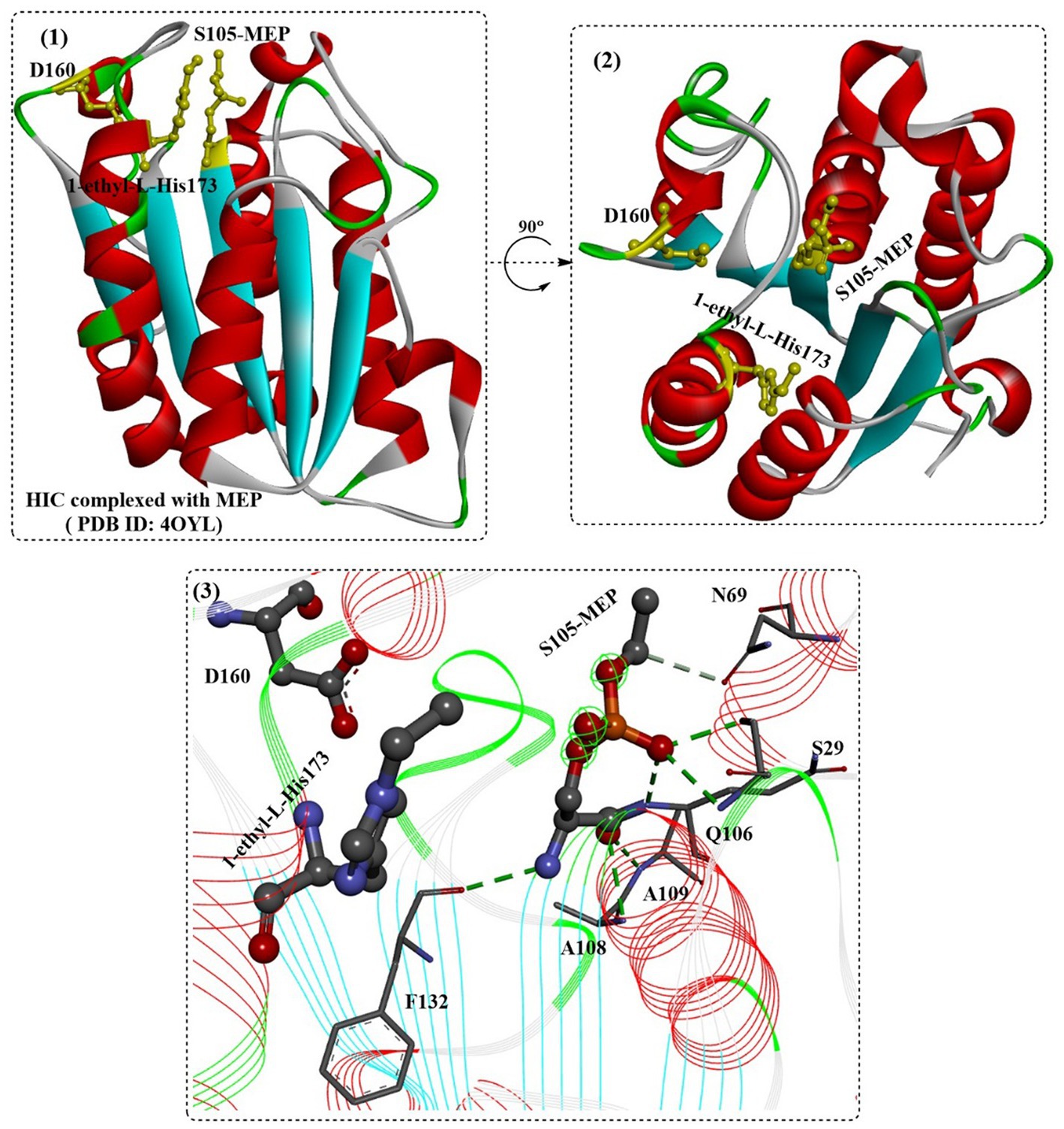
Figure 12. The structure of the binding mode of MEP in the active site of the HIC (1,2) and the local detailed interactions between MEP and HIC. [The catalytic triads are shown in ball-and-stick model, especially the ethylated H173, and the (or light) green dotted lines mean the formed H-bonds].
In a recent study, high-crystallinity PET in mixed PET/cotton textiles was reported to be directly and selectively depolymerized to TPA by using a commercial HIC (Novozym 51,032) and the Cellic CTec2 cellulases under moist-solid reaction conditions (Kaabel et al., 2023). Results revealed that TPA is the main degrading products with more than 40-fold selectivity over MHET after 7 days of static incubation at 55°C in the presence of HiC (0.65% w/w) and/or CTec2 (0.7% w/w), and 14.1% of TPA could be obtained when co-applied with CTec2. Interestingly, it was also found that the CTec2 enzymes were most efficient in the absence of HiC with up to 83.4% yield of glucose without any negative influence on the TPA yield.
FsC has long been explored for the hydrolysis of several industrially important esters, insoluble PET polymer films and poly(ε-caprolactone) (PCL) (Ronkvist et al., 2009). FsC is also a single-domain protein consisting of 197 amino acids (~20.8 kDa). The overall structure of FsC is similar to that of Hic, which is different from the bacterial-derived cutinase as a single-domain protein (Figure 13(1,2)). FsC also contains 5 slightly twisted parallel-stranded β-sheets that are covered by 4 α-helices on each side (Figure 13(3)). Among them, 2 flexible loop domains (residues 80–90 and 182–189) and 2 disulfide bridges (DS1:Cys31 and Cys109, DS2:Cys171 and Cys178) are highly conserved and may be associated with substrate binding and the stabilization of the overall structures (especially DS1) (Matak and Moghaddam, 2009).
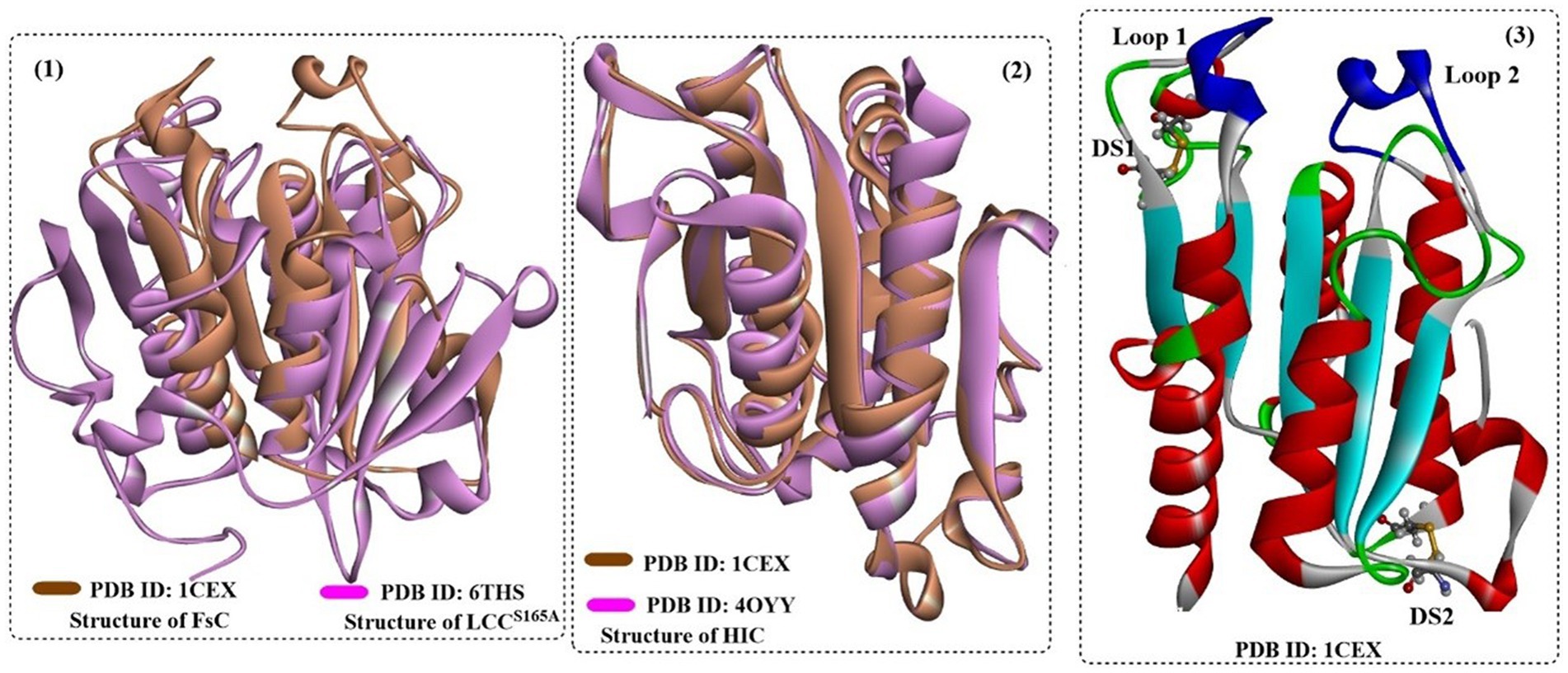
Figure 13. The structure alignment of FsC with LCC (1) and HIC (2) and the overall structure of FsC (3). [In (3), the formed two disulfide bonds are shown in ball-and-stick model and the typical loop1 and loop2 are highlighted in dark blue].
In a recent study, NMR spectroscopy was used to investigate FsC with PET-hydrolysing activity and the reaction, including the reaction conditions, interactions between FsC and BHET, and product release (Hellesnes et al., 2023). It was found that BHET binding is predominantly regulated by hydrophobic interactions (with adjacent polar residues and polar residues); in particular, a more extended binding cleft around G49 and G192 is needed. Given that FsCL81A (improved by 4-fold) and FsCL182A (improved by 5-fold) showed much higher hydrolytic activity on PET and polyamide 6,6 fibers, the enlarged binding site might more likely result in improved cutinase activity, as discovered by Araújo et al. (2007). The main product (80% of the products) from PET hydrolysis is MHET, and after ~400 min, the release of TPA was detected (Hellesnes et al., 2023). More importantly, it was found that the pH and pD conditions might regulate the relative amounts of degradation products, and the low degrading activity of FsC on PET is caused by poor substrate binding and the subsequent slow MHET hydrolysing activity. Previously, it was concluded that increasing accumulation of the final product EG around the active site reduced the overall flexibility of FsC (Groß et al., 2017), which certainly caused decreased thermostability and catalytic performance.
To further exploit the potential of FsC in the biodegradation of parabens, a novel biocatalyst (named SDFsC) was constructed by expressing FsC on the cell surface of Sacchromycese cerevisiae with Aga1p and Aga2p protein subunits and a glycosylphosphatidylinositol (GPI) attachment sequence (Zhu and Wei, 2019). Catalytic results indicated that SDFsC could be used for the efficient removal of parabens with Km values of 3.1 ± 0.5 mM (for pNPA) and 0.67 ± 0.15 mM (pNPB); thus, the catalytic performance of SDFsC was slightly affected. In addition, the side chain structures of parabens play a critical role in the degradation rates, and parabens with relatively long alkyl or aromatic side chains (propylparaben, butylparaben and benzylparaben) were more efficiently degraded, with efficiencies of 89, 97 and 93%, respectively, after 24 h at 30°C. Moreover, SDFsC showed excellent reusability with 93% residual enzyme activity after being used for paraben degradation 6 times. Altogether, it is believed that biocatalysis based on the enzyme cell surface display would be a green and efficient alternative for efficient PET biodegradation.
In 1997, two distinct serine esterases with cutinolytic activity (52 kDa and 26 kDa) were discovered (Trail and Köller, 1993), and a cutinase-encoding gene, CUTAB1, was then identified from an A. brassicicola cDNA library (Yao and Kller, 1994).
The cutinase CUTAB1 from Alternaria brassicicola (corresponding to the cutinase from the A. brassicicola culture) was successfully heterologously expressed in Pichia pastoris (Koschorreck et al., 2010). It displayed the highest enzymatic activity at 40°C and pH 7–9, and similarly, the chain lengths of different substrates showed significant effects on the hydrolysing activity; the highest catalytic activity was toward tributyrin and the lowest activity was toward p-nitrophenyl palmitate (p-NPP). In contrast to the findings for FsC (Hellesnes et al., 2023), the mutants AbCL80A, AbCL181A and AbCI183A showed a significant decrease in specific activity toward p-NPB and p-NPP. In contrast, the specific activity of AbCA84F against p-NPP was significantly improved by ~10-fold. This strange phenomenon was thought to be associated with increased interactions with longer chain and hydrophobic substrates caused by the bulkier and more hydrophobic phenylalanine.
Two cutinase genes from Fusarium solani (FsC) and Aspergillus fumigatus (AfC) were also successfully expressed in Pichia pastoris X33 (Ping et al., 2017). The results showed that FsC and AfC display significantly different optimal catalytic temperatures (40°C and 60°C, respectively) but similar pH values (7.5 and 8.0, respectively). This illustrated the superior thermostability of AfC. Both FsC and AfC can hydrolyse p-nitrophenyl substrates with different carbon chain lengths, and pNPD and pNPH were the preferred substrates for FsC and AfC, respectively. Moreover, AfC could completely hydrolyse the PCL film.
Aspergillus oryzae is a well-known filamentous fungus that has long been used in traditional microbial fermentation and the food industry. Recently, the fungus has attracted much attention for the biodegradation of poly(butylene succinate) (PBS) and poly(butylenes succinate-co-adipate) (PBSA) with the successful identification of a PBS-degrading enzyme (AoC) (Maeda et al., 2005). AoC (~21.6 kDa) displayed the highest catalytic activity at 55°C and could remain stable under a wide pH range (pH 6.0–11.0).
In a recent study, the biological properties of AoC were investigated, and the structure–activity relationship was analyzed based on the crystal structure (Liu et al., 2009). AoC showed better thermostability with a Tm value of 59°C. AoC showed a preference for pNPV with a Km value of 0.04 ± 0.01 μM and displayed the highest catalytic efficiency toward pNPB and pNPV with kcat/Km values of 3.49 ± 0.51 μM−1 min−1 and 3.32 ± 0.74 μM−1 min−1, respectively. At 40°C and pH 8.0, poly ε-(caprolactone) (PCL) was almost completely degraded (87% PCL) within 6 h; however, only 30% degradation was achieved catalyzed by FsC. The structure (Figure 14(1)), which is similar to that of FsC and HiC, contains a typical α/β fold characterized by a central β-sheet of 5 parallel strands; this sheet is surrounded by 10 α-helices with a highly conserved catalytic triad S126, D181, and H194. In addition, 3 disulfide bonds were identified, including DS1 between Cys37 and Cys115, DS2 between Cys177-Cys184, and DS3 between Cys63-Cys76. Among them, DS3 is unique to the cutinases from the Aspergillus family connecting helix 2 to strand 2 of the central β-sheet, which might lead to improved thermoactivity of AoC. Different from FsC and HiC, an extended groove was discovered around the substrate binding site that regulates the orientation of polymeric chains (Figure 14(3)), which resulted in a much higher hydrolysing activity of AoC toward the longer chain substrates. Furthermore, residues L87 and V190, similar to the “gate keepers,” may be associated with substrate recognition, especially the different alkyl chains.
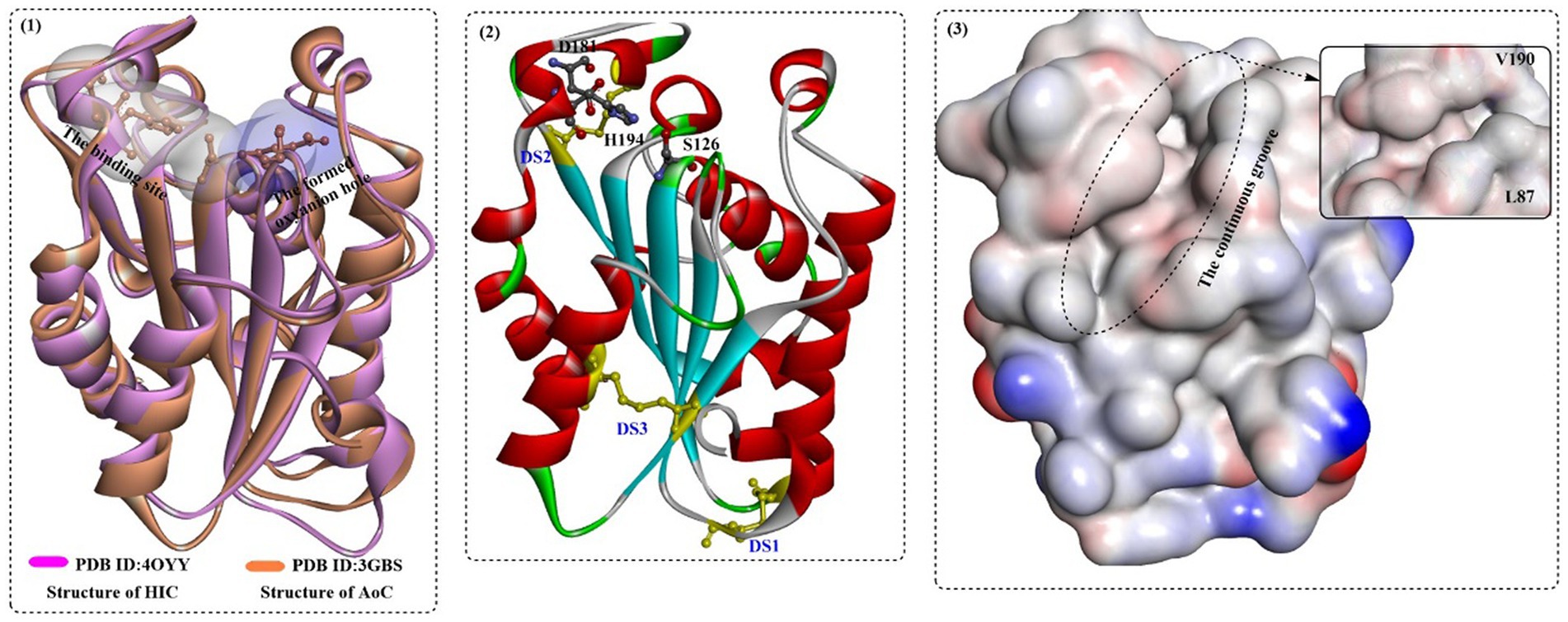
Figure 14. The structure alignment of AoC with HIC (1) and the special features of AoC (2,3). [In (1), the substrate binding site and the formed oxyanion hole are show in white and light blue surface; in (2), the formed three disulfide bonds and the catalytic triads are shown in in ball-and-stick model, and in (3) the hydrophobic surface of the HIC protein is displayed especially the continuous groove for substrate binding].
To further improve thermostability, Shirke et al. investigated the thermal inactivation mechanisms of AoC (Shirke et al., 2017). First, it was revealed that glycosylation plays a critical role in the improved thermostability of Thiellavia terrestris cutinase (TtC) expressed in Pichia pastoris by inhibiting its thermal aggregation. In view of this finding, kinetic models of cutinase thermal inactivation were created to account for thermal aggregation. However, the models also indicated that the glycosylation of TtC is helpful for the improved kinetic stability (5-15-fold) by inhibiting thermal aggregation at 61°C. Then, one glycosylation site (L185N) was designed and incorporated into AoC, which completely inhibited the thermal aggregation of AoC with a 4-fold improvement in thermostability. However, it should be noted that the undesirable catalytic efficiency of AoCL185N was lower (~70%) than that of wild-type AoC, and this may be avoided by rationally designed glycosylation sites.
Recently, Lewatit VP OC 1600 was used as the macroporous support for the immobilization of AoC, HIC and TfC for industrial applications of cutinases (Su et al., 2018). The results demonstrated that HiC displays the highest solvent tolerance with increased polarity, and TtC shows the best thermostability at 80°C. Furthermore, in nonane, the three obtained catalysts could maintain ~64% residual enzyme activity even at 90°C, which reveals that this strategy is efficient in improving the organic solvent tolerance.
In a study, the abovementioned cutinases were investigated in detail (Baker et al., 2012). HiC was shown to exhibit the highest stability with significantly improved poly(ε-caprolactone) hydrolysing activities at high temperatures under the tested conditions. Additionally, at low pH conditions, HiC could maintain the overall structure with little affected activity, but AbC exhibited the largest loss in structure. Similarly, FsC and AbC were found to be the least stable at high temperatures.
Recently, two highly similar thermophilic PET hydrolases (PES-H1 and PES-H2) were identified from a compost metagenome library, and PES-H1 and PES-H2 differ in only four residues (A/E1, L/F209, D/N232, and S/A254) (Sonnendecker et al., 2022). Although they contain quite similar amino acid sequences, PES-H1 displayed a much higher hydrolytic activity on amorphous PET films, even higher than that of the wild-type LCC under identical conditions. To investigate the catalytic mechanism, high-resolution crystal structures of PES-H1 complexed with various PET substrates were obtained, including citrate [PDB ID: 7E30, with 4-(2-hydroxyethylcarbamoyl) benzoic acid (MHETA), PDB ID:7W6C, 7W6O, and 7W6Q] and with bis(2-hydroxyethyl) terephthalate (BHET, PDB ID: 7W66) (Pfaff et al., 2022). Regarding the structures, PES-H1 adopts the canonical α/β-hydrolase fold consisting of 9 β-sheets and 10 α-helices and exhibits highly conserved features of the α/β-hydrolase superfamily similar to LCC. Overall, the binding modes of the different ligands in the active sites are quite different but similar to those observed for HEMT bound to IsPETase R103G/S131A28 and MHET bound to the LCC ICCG. Given that the multiple noncatalytic intermediate binding modes (e.g., MHETA) far from S130 are less likely to undergo nucleophilic attack, enzyme-mediated PET hydrolysis involves the dynamic reorientation of polymer chains in the substrate-binding site. The effect of W155 was also identified (Figure 15(2–7)), which is critical for PET binding and stabilization. Especially, from Figure 15(2–4), the different conformations of MHETA were obtained, which clearly revel the MHET degrading process. MHETA located away from the catalytic triad is stabilized by hydrophobic residues, including F62, I178, L209, M131 and F62. In addition, the TPA moiety is in the vicinity of F62 rather than close to the catalytic triads. Moreover, the special binding mode of MHETA located at the surface of PES-H1 (Figure 15(4)) indicates that for PET hydrolysis, it might first bind at the surface and be transferred into the deeper active site cavity.
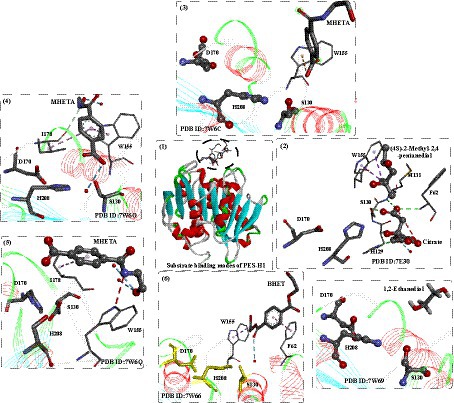
Figure 15. The binding modes of substrate analogs and degradation intermediates in the active site of PES-H1 (1), and the local detailed interactions modes of different substrates in the binding site (2–7). (2: detailed interactions of (4S)-2-Methyl-2,4-pentanediol in the active sits; 3: detailed interactions of MHETA in the active sits; 4: detailed interactions of MHETA in the active sits; 5: detailed interactions of BHET in the active sits; 6: detailed interactions of MHETA in the active sits; 7: detailed interactions of 1,2-Ethanediol in the active sits).
Given the crystal structures and the MD simulations, mutational hotspots were identified for rational engineering of PES-H1, such as the R204C/S250C variant, R6C/S49C variant, L92F/Q94Y, and L92F/Q94Y/R204C/S250C variant. The results showed that mutant PES-H1L92F/Q94Y displayed a 1.8°C higher Tm and could almost completely degrade the Gf-PET film after 24 h at 72°C, with the highest weight loss of 37.1 mg (62.4%). It outperforms the promising LCC ICCG mutant in the biohydrolysis of low-crystallinity (13%) PET powder with 2.2-fold more degradation products. It was believed that the enhanced activity was caused by the “aromatic tunnel” (substitutions of L90F) effect streaming the efficient binding of PET chains to the active site (Cui et al., 2021).
As a surface erosion process, low binding affinity with PET, as a critical factor, has been found to significantly restrict the further industrial application of various PET hydrolases. In addition to modulating protein evolution, the strategy involving constructing fusion proteins has also been proven to be effective in enhancing the binding affinity of enzymes and substrates with hydrophobic surface binding modules (Ribitsch et al., 2013; Várnai et al., 2014; Ribitsch et al., 2015).
Carbohydrate-binding modules (CBMs) are small noncatalytic protein domains that usually function as a part of modular enzymes (e.g., carbohydrate-active enzymes, CAZymes), which can enhance the binding affinity between enzymes and substrates (Armenta et al., 2017; Weber et al., 2019). Consequently, CBMs have been used as efficient affinity tags for protein immobilization owing to their high adsorption capacity for solid substrates (Oliveira et al., 2015; Zhou et al., 2020). Based on the topology of the ligand-binding site, CBMs can be classified into types A, B (endo-type), or C (exo-type) (Gilbert et al., 2013). Type A CBMs display a high ability to bind crystalline polysaccharides (e.g., cellulose and chitin) through hydrophobic interactions regulated by conserved aromatic triplets and are therefore potential candidates for PET-binding peptides (Zhang et al., 2013; Liu et al., 2022).
Weber et al. (2019) identified a novel PET-binding CBM and explored the CBM-PET interactions. First, a semiquantitative PET surface affinity assay was developed to detect CBMs bound to PET films. Then, eight CBMs from the carbohydrate active enzymes database4 (Drula et al., 2022) were screened for PET binding, and the results showed that BaCBM2 (GenBank accession numbers: ACQ50287/MK349005) possessed the strongest affinity toward PET. To investigate the CBM-PET interactions, molecular dynamics (MD) simulations identified an aromatic triad (Trp9/Trp44/Trp63) on the peptide surface that was stabilized by π-stacking interactions and hydrogen bonds. This was further verified by tryptophan quenching experiments and alanine point mutations; moreover, the strength of PET binding of CBMs was largely determined by the ratio of hydrophobic to polar contacts at the interface.
In a recent study, fusion of different CBMs to the C-terminus of LCCICCG was reported to improve the PET hydrolysing activities, including a ChBD from Chitinolyticbacter meiyuanensis SYBC-H1, the CBM from Hypocrea jecorina, the polyhydroxyalkanoate binding modules (PBM) from Alcaligenes faecalis and Trichoderma hydrophobins HFB4 (Xue et al., 2021). The results showed that LCCICCG-ChBD displayed a low Km value (131.9 μM) but a much higher kcat/Km (0.343 s−1/μM) compared to LCCICCG (171.8 μM, 0.178 s−1/μM), indicating better substrate affinity to amorphous PET film (GF-PET, crystalline 6.7%). The adsorption of LCCICCG-ChBD to GF-PET films revealed that a maximum adsorption efficiency of 30.2% was achieved after 70 min of incubation at 40°C. For pNPB hydrolysis, LCCICCG-ChBD showed a lower Km and slightly lower catalytic efficiency (kcat/Km, 0.404 s−1/μM) than LCCICCG (0.534 s−1/μM). However, for the ground GF-PET powder, depolymerization degrees of 87.5 and 98.5% were achieved with LCCICCG-ChBD and LCCICCG-CBM at 65°C, respectively, which are 19.6 and 30.6% higher than those obtained with LCCICCG.
In a similar study, to enhance the binding affinity of LCCICCG with PET, TrCBM and CfCBM were rationally selected from the CAZy database to construct fusion proteins with LCCICCG (Chen et al., 2023). Additional auxiliary domains TrCBM (PDB ID: 1AZ6) from Trichoderma reesei and CfCBM from Cellulomonas fimi were obtained from the CAZy database5 (Drula et al., 2022). CfCBM and TrCBM belong to the type A carbohydrate-binding modules (CBMs), which can bind hydrophobic substrates (e.g., chitin) through hydrophobic interactions. Then, blind molecular docking and molecular dynamics were applied to identify potential binding positions of 4-PET, and the results showed that five positions (2 for TrCBM and 3 for CfCBM) were favorable for the binding of PET-4 to CBMs, which occurred mainly through van der Waals interactions, conventional hydrogen bonding and pi-pi interactions. Then, the fusion proteins LCCICCG-TrCBM and CfCBM-LCCICCG were generated and characterized. The results showed that the optimal temperatures of LCCICCG-TrCBM and CfCBM-LCCICCG were further increased to 70 and 80°C, respectively. Compared with LCCICCG and CfCBM-LCCICCG, LCCICCG-TrCBM displayed the strongest thermal stability with 70.1% residual activity at 50°C for 24 h. CfCBM-LCCICCG and LCCICCG-TrCBM were found to more favorably bind to PET films, increasing by 1.4- and 1.3-fold in comparison to LCCICCG. The total products released from CfCBM-LCCICCG were shown to be 24.2% higher than those from LCCICCG. This study clearly describes the significant contributions achieved that enhance the binding ability of LCCICCG to PET films and significantly improve the degradation ability of the crystalline region of PET materials by constructing fusion proteins.
Rennison et al. (2023) investigated the binding of BaCBM2 for PET in detail and showed that the binding affinity is highly associated with temperature and crystallinity. With the increase in tested temperatures, the general trend of Kd increased first after declining. At ~40°C, a maximum Kd of 408 nM for BaCBM2 was detected, showing an obviously lower affinity for this PET substrate. At low temperatures, the increased binding affinity might be caused by a gain in entropy caused by dehydration of the binding surface; nevertheless, at higher temperatures, it might result from an increase in crystalline regions on the PET surface. At 20°C, a clear trend is observed in which the affinity increases at higher crystallinity, and a significant increase in affinity was seen as ~20% bulk crystallinity. This indicated that BaCBM2 might bind preferentially to crystalline areas on the PET surface. MD studies indicated that the binding is mainly driven by the functions of critical residues W9, W44 and W63 through a gain in entropy, which is caused by the dehydration of the binding surface. We believe that fusing a polymer-binding module to the PET hydrolase is a promising strategy to streamline the enzymatic hydrolysis of PET, however the binding capacity, affinity, and thermostability should be improved to satisfy the needs of industrial operation.
It has absolutely been accepted that biodegradation may create new opportunities for recycling PET materials. Research on PET biodegradation is still ongoing, and while progress has been achieved, the following challenges remain in developing efficient and scalable methods for biodegrading PET. (1) The relatively slow degradation rates: PET is a highly stable and resistant polymer, and it requires a long time for natural biodegradation processes to break it down. Enzymatic and microbial methods are also slow, and full biodegradation of PET can last weeks or even months. (2) Complexity of PET materials: PET is often used in complex products, such as multilayer packaging or textiles, which can make it more difficult to biodegrade. In some cases, only certain layers or components of the product may be biodegradable, while others are not. (3) Limited understanding of biodegradation mechanisms: While there has been significant progress in determining the mechanisms of PET biodegradation, much remains unknown. (4) Compatibility with recycling: Biodegradation of PET may pose challenges with traditional recycling methods, which often require separation of different types of plastics. (5) Cost: Developing and implementing effective and scalable methods for PET biodegradation can be expensive, which may limit their adoption by industry and governments. Overall, while challenges remain in developing effective and scalable methods for PET biodegradation, the potential benefits are significant and would certainly create new opportunities for sustainable materials management.
BS: Writing – original draft, Writing – review & editing. TW: Conceptualization, Supervision, Validation, Writing – original draft, Writing – review & editing. JF: Data curation, Writing – original draft. ZH: Data curation, Writing – original draft. TS: Data curation, Writing – original draft. ZL: Resources, Software, Writing – original draft. FL: Conceptualization, Funding acquisition, Writing – original draft. YZ: Supervision, Validation, Writing – original draft.
The author(s) declare financial support was received for the research, authorship, and/or publication of this article. The authors express their gratitude to the Research Fund for Academician Lin He New Medicine (No. JYHL2019MS12), the Supporting Fund for Teachers’ research of Jining Medical University (No. JYFC2019KJ037), the Shandong Provincial University Youth Innovation Team, China (No. 2022KJ102), and the National Natural Science Foundation of China (No. 32000194).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
2. ^ https://www.carbios.com/en/
3. ^ https://doi.org/10.21203/rs.3.rs-2465520/v1
4. ^http://www.cazy.org/, CAZy database.
Almeida, E. L., Carrillo Rincón, A. F., Jackson, S. A., and Dobson, A. D. W. (2019). In silico screening and heterologous expression of a polyethylene terephthalate hydrolase (PETase)-like enzyme (SM14est) with polycaprolactone (PCL)-degrading activity, from the marine sponge-derived strain Streptomyces sp. SM14. Front. Microbiol. 10:2187. doi: 10.3389/fmicb.2019.02187
Araújo, R., Silva, C., O'Neill, A., Micaelo, N., Guebitz, G., Soares, C. M., et al. (2007). Tailoring cutinase activity towards polyethylene terephthalate and polyamide 6,6 fibers. J. Biotechnol. 128, 849–857. doi: 10.1016/j.jbiotec.2006.12.028
Armenta, S., Moreno-Mendieta, S., Sánchez-Cuapio, Z., Sánchez, S., and Rodríguez-Sanoja, R. (2017). Advances in molecular engineering of carbohydrate-binding modules. Proteins 85, 1602–1617. doi: 10.1002/prot.25327
Arnling Bååth, J., Jensen, K., Borch, K., Westh, P., and Kari, J. (2022). Sabatier principle for rationalizing enzymatic hydrolysis of a synthetic polyester. JACS Au 2, 1223–1231. doi: 10.1021/jacsau.2c00204
Austin, H. P., Allen, M. D., Donohoe, B. S., Rorrer, N. A., Kearns, F. L., Silveira, R. L., et al. (2018). Characterization and engineering of a plastic-degrading aromatic polyesterase. Proc. Natl. Acad. Sci. U. S. A. 115, E4350–e4357. doi: 10.1073/pnas.1718804115
Baker, P. J., Poultney, C., Liu, Z., Gross, R., and Montclare, J. K. (2012). Identification and comparison of cutinases for synthetic polyester degradation. Appl. Microbiol. Biotechnol. 93, 229–240. doi: 10.1007/s00253-011-3402-4
Biundo, A., Ribitsch, D., and Guebitz, G. M. (2018). Surface engineering of polyester-degrading enzymes to improve efficiency and tune specificity. Appl. Microbiol. Biotechnol. 102, 3551–3559. doi: 10.1007/s00253-018-8850-7
Blázquez-Sánchez, P., Engelberger, F., Cifuentes-Anticevic, J., Sonnendecker, C., Griñén, A., Reyes, J., et al. (2022). Antarctic polyester hydrolases degrade aliphatic and aromatic polyesters at moderate temperatures. Appl. Environ. Microbiol. 88:e0184221. doi: 10.1128/AEM.01842-21
Boneta, S., Arafet, K., and Moliner, V. (2021). QM/MM study of the enzymatic biodegradation mechanism of polyethylene terephthalate. J. Chem. Inf. Model. 61, 3041–3051. doi: 10.1021/acs.jcim.1c00394
Brackmann, R., de Oliveira Veloso, C., de Castro, A. M., and Langone, M. A. P. (2023). Enzymatic post-consumer poly(ethylene terephthalate) (PET) depolymerization using commercial enzymes. 3 Biotech 13:135. doi: 10.1007/s13205-023-03555-6
Brandenberg, O. F., Schubert, O. T., and Kruglyak, L. (2022). Towards synthetic PETtrophy: engineering Pseudomonas putida for concurrent polyethylene terephthalate (PET) monomer metabolism and PET hydrolase expression. Microb. Cell Factories 21:119. doi: 10.1186/s12934-022-01849-7
Buchholz, P. C. F., Feuerriegel, G., Zhang, H., Perez-Garcia, P., Nover, L. L., Chow, J., et al. (2022). Plastics degradation by hydrolytic enzymes: the plastics-active enzymes database-PAZy. Proteins 90, 1443–1456. doi: 10.1002/prot.26325
Cao, F., Wang, L., Zheng, R., Guo, L., Chen, Y., and Qian, X. (2022). Research and progress of chemical depolymerization of waste PET and high-value application of its depolymerization products. RSC Adv. 12, 31564–31576. doi: 10.1039/D2RA06499E
Carniel, A., Gomes, A. D. C., Coelho, M. A. Z., and de Castro, A. M. (2021). Process strategies to improve biocatalytic depolymerization of post-consumer PET packages in bioreactors, and investigation on consumables cost reduction. Bioprocess Biosyst. Eng. 44, 507–516. doi: 10.1007/s00449-020-02461-y
Carr, C. M., Clarke, D. J., and Dobson, A. D. W. (2020). Microbial polyethylene terephthalate hydrolases: current and future perspectives. Front. Microbiol. 11:571265. doi: 10.3389/fmicb.2020.571265
Charlier, C., Gavalda, S., Borsenberger, V., Duquesne, S., Marty, A., Tournier, V., et al. (2022). An NMR look at an engineered PET depolymerase. Biophys. J. 121, 2882–2894. doi: 10.1016/j.bpj.2022.07.002
Chen, X. Q., Guo, Z. Y., Wang, L., Yan, Z. F., Jin, C. X., Huang, Q. S., et al. (2022). Directional-path modification strategy enhances PET hydrolase catalysis of plastic degradation. J. Hazard. Mater. 433:128816. doi: 10.1016/j.jhazmat.2022.128816
Chen, C. C., Han, X., Li, X., Jiang, P., and Guo, R. T. (2021). General features to enhance enzymatic activity of poly(ethylene terephthalate) hydrolysis. Nat. Catalysis 4, 425–430. doi: 10.1038/s41929-021-00616-y
Chen, S., Su, L., Billig, S., Zimmermann, W., Jian, C., and Jing, W. (2010). Biochemical characterization of the cutinases from Thermobifida fusca. J. Mol. Catal. B Enzymat. 63, 121–127. doi: 10.1016/j.molcatb.2010.01.001
Chen, S., Tong, X., Woodard, R. W., Du, G., Wu, J., and Chen, J. (2008). Identification and characterization of bacterial cutinase. J. Biol. Chem. 283, 25854–25862. doi: 10.1074/jbc.M800848200
Chen, Y., Zhang, S., Zhai, Z., Zhang, S., Ma, J., Liang, X., et al. (2023). Construction of fusion protein with carbohydrate-binding module and leaf-branch compost cutinase to enhance the degradation efficiency of polyethylene terephthalate. Int. J. Mol. Sci. 24:2780. doi: 10.3390/ijms24032780
Choi, J., Yang, I., Kim, S. S., Cho, S. Y., and Lee, S. (2022). Upcycling plastic waste into high value-added carbonaceous materials. Macromol. Rapid Commun. 43:e2100467. doi: 10.1002/marc.202100467
Cui, Y., Che, Y., Liu, X., Dong, S., Qiao, Y., Mitra, R., et al. (2021). Computational redesign of a PETase for plastic biodegradation under ambient condition by the GRAPE strategy. ACS Catal. 11, 1340–1350. doi: 10.1021/acscatal.0c05126
Ding, Z., Xu, G., Miao, R., Wu, N., Zhang, W., Yao, B., et al. (2023). Rational redesign of thermophilic PET hydrolase LCCICCG to enhance hydrolysis of high crystallinity polyethylene terephthalates. J. Hazard. Mater. 453:131386. doi: 10.1016/j.jhazmat.2023.131386
Dong, Q., Yuan, S., Wu, L., Su, L., and Zhou, J. (2020). Structure-guided engineering of a Thermobifida fusca cutinase for enhanced hydrolysis on natural polyester substrate. Bioresour. Bioprocess. 7:37. doi: 10.1186/s40643-020-00324-8
Drula, E., Garron, M. L., Dogan, S., Lombard, V., Henrissat, B., and Terrapon, N. (2022). The carbohydrate-active enzyme database: functions and literature. Nucleic Acids Res. 50, D571–d577. doi: 10.1093/nar/gkab1045
Eiamthong, B., Meesawat, P., Wongsatit, T., Jitdee, J., Sangsri, R., Patchsung, M., et al. (2022). Discovery and genetic code expansion of a polyethylene terephthalate (PET) hydrolase from the human saliva metagenome for the degradation and bio-functionalization of PET. Angew. Chem. Int. Ed. Engl. 61:e202203061. doi: 10.1002/anie.202203061
Ellis, L. D., Rorrer, N. A., Sullivan, K. P., Otto, M., Mcgeehan, J. E., Roman-Leshkov, Y., et al. (2021). Chemical and biological catalysis for plastics recycling and upcycling. Nat. Catal. 4, 539–556. doi: 10.1038/s41929-021-00648-4
Emori, M., Numoto, N., Senga, A., Bekker, G. J., Kamiya, N., Kobayashi, Y., et al. (2021). Structural basis of mutants of PET-degrading enzyme from Saccharomonospora viridis AHK190 with high activity and thermal stability. Proteins 89, 502–511. doi: 10.1002/prot.26034
Erickson, E., Gado, J. E., Avilán, L., Bratti, F., Brizendine, R. K., Cox, P. A., et al. (2022). Sourcing thermotolerant poly(ethylene terephthalate) hydrolase scaffolds from natural diversity. Nat. Commun. 13:7850. doi: 10.1038/s41467-022-35237-x
Eugenio, E. Q., Campisano, I. S. P., Castro, A. M., Coelho, M. A. Z., and Langone, M. A. P. (2021). Experimental and mathematical modeling approaches for biocatalytic post-consumer poly(ethylene terephthalate) hydrolysis. J. Biotechnol. 341, 76–85. doi: 10.1016/j.jbiotec.2021.09.007
Fang, Y., Chao, K., He, J., Wang, Z., and Chen, Z. (2023). High-efficiency depolymerization/degradation of polyethylene terephthalate plastic by a whole-cell biocatalyst. 3 Biotech 13:138. doi: 10.1007/s13205-023-03557-4
Geyer, R., Jambeck, J. R., and Law, K. L. (2017). Production, use, and fate of all plastics ever made. Sci. Adv. 3:e1700782. doi: 10.1126/sciadv.1700782
Gilbert, H. J., Knox, J. P., and Boraston, A. B. (2013). Advances in understanding the molecular basis of plant cell wall polysaccharide recognition by carbohydrate-binding modules. Curr. Opin. Struct. Biol. 23, 669–677. doi: 10.1016/j.sbi.2013.05.005
Groß, C., Hamacher, K., Schmitz, K., and Jager, S. (2017). Cleavage product accumulation decreases the activity of cutinase during PET hydrolysis. J. Chem. Inf. Model. 57, 243–255. doi: 10.1021/acs.jcim.6b00556
Hellesnes, K. N., Vijayaraj, S., Fojan, P., Petersen, E., and Courtade, G. (2023). Biochemical characterization and NMR study of a PET-Hydrolyzing Cutinase from Fusarium solani pisi. Biochemistry 62, 1369–1375. doi: 10.1021/acs.biochem.2c00619
Hu, X., Thumarat, U., Zhang, X., Tang, M., and Kawai, F. (2010). Diversity of polyester-degrading bacteria in compost and molecular analysis of a thermoactive esterase from Thermobifida alba AHK119. Appl. Microbiol. Biotechnol. 87, 771–779. doi: 10.1007/s00253-010-2555-x
Jia, Y., Samak, N. A., Hao, X., Chen, Z., Wen, Q., and Xing, J. (2022). Hydrophobic cell surface display system of PETase as a sustainable biocatalyst for PET degradation. Front. Microbiol. 13:1005480. doi: 10.3389/fmicb.2022.1005480
Kaabel, S., Arciszewski, J., Borchers, T. H., Therien, J. P. D., Friščić, T., and Auclair, K. (2023). Solid-state enzymatic hydrolysis of mixed PET/cotton textiles. ChemSusChem 16:e202201613. doi: 10.1002/cssc.202201613
Kaabel, S., Therien, J. P. D., Deschênes, C. E., Duncan, D., Friščić, T., and Auclair, K. (2021). Enzymatic depolymerization of highly crystalline polyethylene terephthalate enabled in moist-solid reaction mixtures. Proc. Natl. Acad. Sci. U. S. A. 118:e2026452118. doi: 10.1073/pnas.2026452118
Kakadellis, S., and Rosetto, G. (2021). Achieving a circular bioeconomy for plastics. Science 373, 49–50. doi: 10.1126/science.abj3476
Karunatillaka, I., Jaroszewski, L., and Godzik, A. (2022). Novel putative polyethylene terephthalate (PET) plastic degrading enzymes from the environmental metagenome. Proteins 90, 504–511. doi: 10.1002/prot.26245
Kato, S., Sakai, S., Hirai, M., Tasumi, E., Nishizawa, M., Suzuki, K., et al. (2018). Long-term cultivation and metagenomics reveal ecophysiology of previously uncultivated thermophiles involved in biogeochemical nitrogen cycle. Microbes Environ. 33, 107–110. doi: 10.1264/jsme2.ME17165
Kawabata, T., Oda, M., and Kawai, F. (2017). Mutational analysis of cutinase-like enzyme, Cut190, based on the 3D docking structure with model compounds of polyethylene terephthalate. J. Biosci. Bioeng. 124, 28–35. doi: 10.1016/j.jbiosc.2017.02.007
Kawai, T. (2020). Current state and perspectives related to the polyethylene terephthalate hydrolases available for biorecycling. ACS Sustain. Chem. Eng. 8, 8894–8908. doi: 10.1021/acssuschemeng.0c01638
Kawai, F. (2021). Emerging strategies in polyethylene terephthalate hydrolase research for biorecycling. ChemSusChem 14, 4115–4122. doi: 10.1002/cssc.202100740
Kawai, F., Furushima, Y., Mochizuki, N., Muraki, N., Yamashita, M., Iida, A., et al. (2022). Efficient depolymerization of polyethylene terephthalate (PET) and polyethylene furanoate by engineered PET hydrolase Cut190. AMB Express 12:134. doi: 10.1186/s13568-022-01474-y
Kawai, F., Kawabata, T., and Oda, M. (2019). Current knowledge on enzymatic PET degradation and its possible application to waste stream management and other fields. Appl. Microbiol. Biotechnol. 103, 4253–4268. doi: 10.1007/s00253-019-09717-y
Kawai, F., Kawase, T., Shiono, T., Urakawa, H., Sukigara, S., Tu, C., et al. (2017). Enzymatic hydrophilization of polyester fabrics using a recombinant cutinase cut 190 and their surface characterization. J. Fiber Sci. Technol. 73, 8–18. doi: 10.2115/fiberst.fiberst.2017-0002
Kawai, F., Oda, M., Tamashiro, T., Waku, T., Tanaka, N., Yamamoto, M., et al. (2014). A novel Ca2+-activated, thermostabilized polyesterase capable of hydrolyzing polyethylene terephthalate from Saccharomonospora viridis AHK190. Appl. Microbiol. Biotechnol. 98, 10053–10064. doi: 10.1007/s00253-014-5860-y
Kitadokoro, K., Kakara, M., Matsui, S., Osokoshi, R., Thumarat, U., Kawai, F., et al. (2019). Structural insights into the unique polylactate-degrading mechanism of Thermobifida alba cutinase. FEBS J. 286, 2087–2098. doi: 10.1111/febs.14781
Kitadokoro, K., Thumarat, U., Nakamura, R., Nishimura, K., Karatani, H., Suzuki, H., et al. (2012). Crystal structure of cutinase Est119 from Thermobifida alba AHK119 that can degrade modified polyethylene terephthalate at 1.76 resolution. Polymer Degradation Stability 97, 771–775. doi: 10.1016/j.polymdegradstab.2012.02.003
Kleeberg, I., Welzel, K., Vandenheuvel, J., Müller, R. J., and Deckwer, W. D. (2005). Characterization of a new extracellular hydrolase from Thermobifida fusca degrading aliphatic-aromatic copolyesters. Biomacromolecules 6, 262–270. doi: 10.1021/bm049582t
Kold, D., Dauter, Z., Laustsen, A. K., Brzozowski, A. M., Turkenburg, J. P., Nielsen, A. D., et al. (2014). Thermodynamic and structural investigation of the specific SDS binding of Humicola insolens cutinase. Protein Sci. 23, 1023–1035. doi: 10.1002/pro.2489
Koschorreck, K., Liu, D., Kazenwadel, C., Schmid, R. D., and Hauer, B. (2010). Heterologous expression, characterization and site-directed mutagenesis of cutinase CUTAB1 from Alternaria brassicicola. Appl. Microbiol. Biotechnol. 87, 991–997. doi: 10.1007/s00253-010-2533-3
Kumar, V., Sharma, N., Duhan, L., Pasrija, R., Thomas, J., Umesh, M., et al. (2022). Microbial engineering strategies for synthetic microplastics clean up: a review on recent approaches. Environ. Toxicol. Pharmacol. 98:104045. doi: 10.1016/j.etap.2022.104045
Li, N., Liu, H., Cheng, Z., Yan, B., Chen, G., and Wang, S. (2022). Conversion of plastic waste into fuels: a critical review. J. Hazard. Mater. 424:127460. doi: 10.1016/j.jhazmat.2021.127460
Li, Z., Zhao, Y., Wu, P., Wang, H., Li, Q., Gao, J., et al. (2022). Structural insight and engineering of a plastic degrading hydrolase Ple629. Biochem. Biophys. Res. Commun. 626, 100–106. doi: 10.1016/j.bbrc.2022.07.103
Li, Q., Zheng, Y., Su, T., Wang, Q., Liang, Q., Zhang, Z., et al. (2022). Computational design of a cutinase for plastic biodegradation by mining molecular dynamics simulations trajectories. Comput. Struct. Biotechnol. J. 20, 459–470. doi: 10.1016/j.csbj.2021.12.042
Liu, Z., Gosser, Y., Baker, P. J., Ravee, Y., Lu, Z., Alemu, G., et al. (2009). Structural and functional studies of Aspergillus oryzae cutinase: enhanced thermostability and hydrolytic activity of synthetic ester and polyester degradation. J. Am. Chem. Soc. 131, 15711–15716. doi: 10.1021/ja9046697
Liu, Y., Wang, P., Tian, J., Seidi, F., Guo, J., Zhu, W., et al. (2022). Carbohydrate-binding modules of potential resources: occurrence in nature, function, and application in fiber recognition and treatment. Polymers 14:1806. doi: 10.3390/polym14091806
Liu, Z., Zhang, Y., and Wu, J. (2022). Enhancement of PET biodegradation by anchor peptide-cutinase fusion protein. Enzym. Microb. Technol. 156:110004. doi: 10.1016/j.enzmictec.2022.110004
Lu, H., Diaz, D. J., Czarnecki, N. J., Zhu, C., Kim, W., Shroff, R., et al. (2022). Machine learning-aided engineering of hydrolases for PET depolymerization. Nature 604, 662–667. doi: 10.1038/s41586-022-04599-z
Lykidis, A., Mavromatis, K., Ivanova, N., Anderson, I., Land, M., DiBartolo, G., et al. (2007). Genome sequence and analysis of the soil cellulolytic actinomycete Thermobifida fusca YX. J. Bacteriol. 189, 2477–2486. doi: 10.1128/JB.01899-06
Maeda, H., Yamagata, Y., Abe, K., Hasegawa, F., Machida, M., Ishioka, R., et al. (2005). Purification and characterization of a biodegradable plastic-degrading enzyme from Aspergillus oryzae. Appl. Microbiol. Biotechnol. 67, 778–788. doi: 10.1007/s00253-004-1853-6
Magalhães, R. P., Cunha, J. M., and Sousa, S. F. (2021). Perspectives on the role of enzymatic biocatalysis for the degradation of plastic PET. Int. J. Mol. Sci. 22:20. doi: 10.3390/ijms222011257
Makryniotis, K., Nikolaivits, E., Gkountela, C., Vouyiouka, S., and Topakas, E. (2023). Discovery of a polyesterase from Deinococcus maricopensis and comparison to the benchmark LCCICCG suggests high potential for semi-crystalline post-consumer PET degradation. J. Hazard. Mater. 455:131574. doi: 10.1016/j.jhazmat.2023.131574
Matak, M. Y., and Moghaddam, M. E. (2009). The role of short-range Cys171-Cys178 disulfide bond in maintaining cutinase active site integrity: a molecular dynamics simulation. Biochem. Biophys. Res. Commun. 390, 201–204. doi: 10.1016/j.bbrc.2009.09.073
Meyer-Cifuentes, I. E., Werner, J., Jehmlich, N., Will, S. E., Neumann-Schaal, M., and Öztürk, B. (2020). Synergistic biodegradation of aromatic-aliphatic copolyester plastic by a marine microbial consortium. Nat. Commun. 11:5790. doi: 10.1038/s41467-020-19583-2
Miyakawa, T., Mizushima, H., Ohtsuka, J., Oda, M., Kawai, F., and Tanokura, M. (2015). Structural basis for the Ca(2+)-enhanced thermostability and activity of PET-degrading cutinase-like enzyme from Saccharomonospora viridis AHK190. Appl. Microbiol. Biotechnol. 99, 4297–4307. doi: 10.1007/s00253-014-6272-8
Moyses, D. N., Teixeira, D. A., Waldow, V. A., Freire, D. M. G., and Castro, A. M. (2021). Fungal and enzymatic bio-depolymerization of waste post-consumer poly(ethylene terephthalate) (PET) bottles using Penicillium species. 3 Biotech 11:435. doi: 10.1007/s13205-021-02988-1
Mrigwani, A., Pitaliya, M., Kaur, H., Kasilingam, B., Thakur, B., and Guptasarma, P. (2023). Rational mutagenesis of Thermobifida fusca cutinase to modulate the enzymatic degradation of polyethylene terephthalate. Biotechnol. Bioeng. 120, 674–686. doi: 10.1002/bit.28305
Nielsen, A. D., Arleth, L., and Westh, P. (2005). Analysis of protein–surfactant interactions—a titration calorimetric and fluorescence spectroscopic investigation of interactions between Humicola insolens cutinase and an anionic surfactant. Biochim. Biophys. Acta 1752, 124–132. doi: 10.1016/j.bbapap.2005.08.001
Numoto, N., Kamiya, N., Bekker, G. J., Yamagami, Y., Inaba, S., Ishii, K., et al. (2018). Structural dynamics of the PET-degrading cutinase-like enzyme from Saccharomonospora viridis AHK190 in substrate-bound states elucidates the ca(2+)-driven catalytic cycle. Biochemistry 57, 5289–5300. doi: 10.1021/acs.biochem.8b00624
Oda, M. (2021). Structural basis for Ca(2+)-dependent catalysis of a cutinase-like enzyme and its engineering: application to enzymatic PET depolymerization. Biophys. Physicobiol. 18, 168–176. doi: 10.2142/biophysico.bppb-v18.018
Oda, M., Yamagami, Y., Inaba, S., Oida, T., Yamamoto, M., Kitajima, S., et al. (2018). Enzymatic hydrolysis of PET: functional roles of three Ca(2+) ions bound to a cutinase-like enzyme, Cut190*, and its engineering for improved activity. Appl. Microbiol. Biotechnol. 102, 10067–10077. doi: 10.1007/s00253-018-9374-x
Oliveira, C., Carvalho, V., Domingues, L., and Gama, F. M. (2015). Recombinant CBM-fusion technology - applications overview. Biotechnol. Adv. 33, 358–369. doi: 10.1016/j.biotechadv.2015.02.006
Pfaff, L., Gao, J., Li, Z., Jäckering, A., Weber, G., Mican, J., et al. (2022). Multiple substrate binding mode-guided engineering of a thermophilic PET hydrolase. ACS Catal. 12, 9790–9800. doi: 10.1021/acscatal.2c02275
Ping, L. F., Chen, X. Y., Yuan, X. L., Zhang, M., Chai, Y. J., and Shan, S. D. (2017). Application and comparison in biosynthesis and biodegradation by Fusarium solani and Aspergillus fumigatus cutinases. Int. J. Biol. Macromol. 104, 1238–1245. doi: 10.1016/j.ijbiomac.2017.06.118
Pirillo, V., Orlando, M., Battaglia, C., Pollegioni, L., and Molla, G. (2023). Efficient polyethylene terephthalate degradation at moderate temperature: a protein engineering study of LC-cutinase highlights the key role of residue 243. FEBS J. 290, 3185–3202. doi: 10.1111/febs.16736
Quartinello, F., Vajnhandl, S., Volmajer Valh, J., Farmer, T. J., Vončina, B., Lobnik, A., et al. (2017). Synergistic chemo-enzymatic hydrolysis of poly(ethylene terephthalate) from textile waste. Microb. Biotechnol. 10, 1376–1383. doi: 10.1111/1751-7915.12734
Rennison, A. P., Westh, P., and Møller, M. S. (2023). Protein-plastic interactions: the driving forces behind the high affinity of a carbohydrate-binding module for polyethylene terephthalate. Sci. Total Environ. 870:161948. doi: 10.1016/j.scitotenv.2023.161948
Ribitsch, D., and Guebitz, G. M. (2021). Tuning of adsorption of enzymes to polymer. Methods Enzymol. 648, 293–315. doi: 10.1016/bs.mie.2020.12.010
Ribitsch, D., Herrero Acero, E., Przylucka, A., Zitzenbacher, S., Marold, A., Gamerith, C., et al. (2015). Enhanced cutinase-catalyzed hydrolysis of polyethylene terephthalate by covalent fusion to hydrophobins. Appl. Environ. Microbiol. 81, 3586–3592. doi: 10.1128/AEM.04111-14
Ribitsch, D., Yebra, A. O., Zitzenbacher, S., Wu, J., Nowitsch, S., Steinkellner, G., et al. (2013). Fusion of binding domains to Thermobifida cellulosilytica cutinase to tune sorption characteristics and enhancing PET hydrolysis. Biomacromolecules 14, 1769–1776. doi: 10.1021/bm400140u
Richter, P. K., Blázquez-Sánchez, P., Zhao, Z., Engelberger, F., Wiebeler, C., Künze, G., et al. (2023). Structure and function of the metagenomic plastic-degrading polyester hydrolase PHL7 bound to its product. Nat. Commun. 14:1905. doi: 10.1038/s41467-023-37415-x
Rolf Joachim, M., Hedwig, S., Jorn, P., Karolin, D., and Wolf, D. D. (2005). Enzymatic degradation of poly(ethylene terephthalate): rapid hydrolysis using a hydrolase from T. fusca. Macromol. Rapid Commun. 26, 1400–1405. doi: 10.1002/marc.200500410
Ronkvist, Å. M., Xie, W., Lu, W., and Gross, R. A. (2009). Cutinase-catalyzed hydrolysis of poly(ethylene terephthalate). Macromolecules 42, 5128–5138. doi: 10.1021/ma9005318
Roth, C., Wei, R., Oeser, T., Then, J., Föllner, C., Zimmermann, W., et al. (2014). Structural and functional studies on a thermostable polyethylene terephthalate degrading hydrolase from Thermobifida fusca. Appl. Microbiol. Biotechnol. 98, 7815–7823. doi: 10.1007/s00253-014-5672-0
Ru, J., Huo, Y., and Yang, Y. (2020). Microbial degradation and valorization of plastic wastes. Front. Microbiol. 11:442. doi: 10.3389/fmicb.2020.00442
Sagong, H. Y., Kim, S., Lee, D., Hong, H., Lee, S. H., Seo, H., et al. (2022). Structural and functional characterization of an auxiliary domain-containing PET hydrolase from Burkholderiales bacterium. J. Hazard. Mater. 429:128267. doi: 10.1016/j.jhazmat.2022.128267
Sagong, H. Y., Son, H. F., Seo, H., Hong, H., Lee, D., and Kim, K. J. (2021). Implications for the PET decomposition mechanism through similarity and dissimilarity between PETases from Rhizobacter gummiphilus and Ideonella sakaiensis. J. Hazard. Mater. 416:126075. doi: 10.1016/j.jhazmat.2021.126075
Schubert, S., Schaller, K., Bååth, J. A., Hunt, C., Borch, K., Jensen, K., et al. (2023). Reaction pathways for the enzymatic degradation of poly(ethylene terephthalate): what characterizes an efficient PET-hydrolase? Chembiochem 24:e202200516. doi: 10.1002/cbic.202200516
Senga, A., Hantani, Y., Bekker, G. J., Kamiya, N., Kimura, Y., Kawai, F., et al. (2019). Metal binding to cutinase-like enzyme from Saccharomonospora viridis AHK190 and its effects on enzyme activity and stability. J. Biochem. 166, 149–156. doi: 10.1093/jb/mvz020
Senga, A., Numoto, N., Yamashita, M., Iida, A., Ito, N., Kawai, F., et al. (2021). Multiple structural states of Ca2+−regulated PET hydrolase, Cut190, and its correlation with activity and stability. J. Biochem. 169, 207–213. doi: 10.1093/jb/mvaa102
Shirke, A. N., Su, A., Jones, J. A., Butterfoss, G. L., Koffas, M. A., Kim, J. R., et al. (2017). Comparative thermal inactivation analysis of Aspergillus oryzae and Thiellavia terrestris cutinase: role of glycosylation. Biotechnol. Bioeng. 114, 63–73. doi: 10.1002/bit.26052
Simon, N., Raubenheimer, K., Urho, N., Unger, S., Azoulay, D., Farrelly, T., et al. (2021). A binding global agreement to address the life cycle of plastics. Science 373, 43–47. doi: 10.1126/science.abi9010
Sonnendecker, C., Oeser, J., Richter, P. K., Hille, P., Zhao, Z., Fischer, C., et al. (2022). Low carbon footprint recycling of post-consumer PET plastic with a metagenomic polyester hydrolase. ChemSusChem 15:e202101062. doi: 10.1002/cssc.202101062
Su, A., Shirke, A., Baik, J., Zou, Y., and Gross, R. (2018). Immobilized cutinases: preparation, solvent tolerance and thermal stability. Enzym. Microb. Technol. 116, 33–40. doi: 10.1016/j.enzmictec.2018.05.010
Sulaiman, S., Yamato, S., Kanaya, E., Kim, J. J., Koga, Y., Takano, K., et al. (2012). Isolation of a novel cutinase homolog with polyethylene terephthalate-degrading activity from leaf-branch compost by using a metagenomic approach. Appl. Environ. Microbiol. 78, 1556–1562. doi: 10.1128/AEM.06725-11
Tamoor, M., Samak, N. A., Jia, Y., Mushtaq, M. U., Sher, H., Bibi, M., et al. (2021). Potential use of microbial enzymes for the conversion of plastic waste into value-added products: a viable solution. Front. Microbiol. 12:777727. doi: 10.3389/fmicb.2021.777727
Then, J., Wei, R., Oeser, T., Barth, M., Belisário-Ferrari, M. R., Schmidt, J., et al. (2015). Ca2+ and Mg2+ binding site engineering increases the degradation of polyethylene terephthalate films by polyester hydrolases from Thermobifida fusca. Biotechnol. J. 10, 592–598. doi: 10.1002/biot.201400620
Then, J., Wei, R., Oeser, T., Gerdts, A., Schmidt, J., Barth, M., et al. (2016). A disulfide bridge in the calcium binding site of a polyester hydrolase increases its thermal stability and activity against polyethylene terephthalate. FEBS Open Biol. 6, 425–432. doi: 10.1002/2211-5463.12053
Thomsen, T. B., Hunt, C. J., and Meyer, A. S. (2022a). Influence of substrate crystallinity and glass transition temperature on enzymatic degradation of polyethylene terephthalate (PET). New Biotechnol. 69, 28–35. doi: 10.1016/j.nbt.2022.02.006
Thomsen, T. B., Hunt, C. J., and Meyer, A. S. (2022b). Standardized method for controlled modification of poly (ethylene terephthalate) (PET) crystallinity for assaying PET degrading enzymes. MethodsX 9:101815. doi: 10.1016/j.mex.2022.101815
Thumarat, U., Kawabata, T., Nakajima, M., Nakajima, H., Sugiyama, A., Yazaki, K., et al. (2015). Comparison of genetic structures and biochemical properties of tandem cutinase-type polyesterases from Thermobifida alba AHK119. J. Biosci. Bioeng. 120, 491–497. doi: 10.1016/j.jbiosc.2015.03.006
Thumarat, U., Nakamura, R., Kawabata, T., Suzuki, H., and Kawai, F. (2012). Biochemical and genetic analysis of a cutinase-type polyesterase from a thermophilic Thermobifida alba AHK119. Appl. Microbiol. Biotechnol. 95, 419–430. doi: 10.1007/s00253-011-3781-6
Tournier, V., Topham, C. M., Gilles, A., David, B., Folgoas, C., Moya-Leclair, E., et al. (2020). An engineered PET depolymerase to break down and recycle plastic bottles. Nature 580, 216–219. doi: 10.1038/s41586-020-2149-4
Trail, F., and Köller, W. (1993). Diversity of cutinases from plant pathogenic fungi: purification and characterization of two cutinases from Alternaria brassicicola. Physiol. Mol. Plant Pathol. 42, 205–220. doi: 10.1006/pmpp.1993.1019
Urbanek, A. K., Kosiorowska, K. E., and Mirończuk, A. M. (2021). Current knowledge on polyethylene terephthalate degradation by genetically modified microorganisms. Front. Bioeng. Biotechnol. 9:771133. doi: 10.3389/fbioe.2021.771133
Várnai, A., Mäkelä, M. R., Djajadi, D. T., Rahikainen, J., Hatakka, A., and Viikari, L. (2014). Carbohydrate-binding modules of fungal cellulases: occurrence in nature, function, and relevance in industrial biomass conversion. Adv. Appl. Microbiol. 88, 103–165. doi: 10.1016/B978-0-12-800260-5.00004-8
von Haugwitz, G., Han, X., Pfaff, L., Li, Q., Wei, H., Gao, J., et al. (2022). Structural insights into (Tere)phthalate-Ester hydrolysis by a carboxylesterase and its role in promoting PET depolymerization. ACS Catal. 12, 15259–15270. doi: 10.1021/acscatal.2c03772
Weber, J., Petrović, D., Strodel, B., Smits, S. H. J., Kolkenbrock, S., Leggewie, C., et al. (2019). Interaction of carbohydrate-binding modules with poly(ethylene terephthalate). Appl. Microbiol. Biotechnol. 103, 4801–4812. doi: 10.1007/s00253-019-09760-9
Wei, R., Breite, D., Song, C., Gräsing, D., Ploss, T., Hille, P., et al. (2019). Biocatalytic degradation efficiency of postconsumer polyethylene terephthalate packaging determined by their polymer microstructures. Adv. Sci. 6:1900491. doi: 10.1002/advs.201900491
Wei, R., Oeser, T., Schmidt, J., Meier, R., Barth, M., Then, J., et al. (2016). Engineered bacterial polyester hydrolases efficiently degrade polyethylene terephthalate due to relieved product inhibition. Biotechnol. Bioeng. 113, 1658–1665. doi: 10.1002/bit.25941
Wei, R., and Zimmermann, W. (2017). Biocatalysis as a green route for recycling the recalcitrant plastic polyethylene terephthalate. Microb. Biotechnol. 10, 1302–1307. doi: 10.1111/1751-7915.12714
Wolfgang, Z., Ren, W., Patrick, H., Thorsten, O., and Juliane, S. (2019). New polypeptides having a polyester degrading activity and uses there of [P], EP3517608A1.
Xi, X., Ni, K., Hao, H., Shang, Y., Zhao, B., and Qian, Z. (2021). Secretory expression in Bacillus subtilis and biochemical characterization of a highly thermostable polyethylene terephthalate hydrolase from bacterium HR29. Enzym. Microb. Technol. 143:109715. doi: 10.1016/j.enzmictec.2020.109715
Xue, R., Chen, Y., Rong, H., Wei, R., Cui, Z., Zhou, J., et al. (2021). Fusion of chitin-binding domain from Chitinolyticbacter meiyuanensis SYBC-H1 to the leaf-branch compost cutinase for enhanced PET hydrolysis. Front. Bioeng. Biotechnol. 9:762854. doi: 10.3389/fbioe.2021.762854
Yang, Y., Min, J., Xue, T., Jiang, P., Liu, X., Peng, R., et al. (2023). Complete bio-degradation of poly(butylene adipate-co-terephthalate) via engineered cutinases. Nat. Commun. 14:1645. doi: 10.1038/s41467-023-37374-3
Yao, C., and Kller, W. (1994). Diversity of cutinases from plant pathogenic fungi: cloning and characterization of a cutinase gene from Alternaria brassicicola. Physiol. Mol. Plant Pathol. 44, 81–92. doi: 10.1016/S0885-5765(05)80103-1
Yoshida, S., Hiraga, K., Takehana, T., Taniguchi, I., Yamaji, H., Maeda, Y., et al. (2016). A bacterium that degrades and assimilates poly(ethylene terephthalate). Science 351, 1196–1199. doi: 10.1126/science.aad6359
Zeng, W., Li, X., Yang, Y., Min, J., Huang, J. W., Liu, W., et al. (2022). Substrate-binding mode of a thermophilic PET hydrolase and engineering the enzyme to enhance the hydrolytic efficacy. ACS Catal. 12, 3033–3040. doi: 10.1021/acscatal.1c05800
Zhang, Y., Wang, L., Chen, J., and Wu, J. (2013). Enhanced activity toward PET by site-directed mutagenesis of Thermobifida fusca cutinase-CBM fusion protein. Carbohydr. Polym. 97, 124–129. doi: 10.1016/j.carbpol.2013.04.042
Zheng, M., Li, Y., Dong, W., Feng, S., Zhang, Q., and Wang, W. (2022). Computational biotransformation of polyethylene terephthalate by depolymerase: a QM/MM approach. J. Hazard. Mater. 423:127017. doi: 10.1016/j.jhazmat.2021.127017
Zhou, J., Chen, J., Zhuang, N., Zhang, A., Chen, K., Xu, N., et al. (2020). Immobilization and purification of enzymes with the novel affinity tag ChBD-AB from Chitinolyticbacter meiyuanensis SYBC-H1. Front. Bioeng. Biotechnol. 8:579. doi: 10.3389/fbioe.2020.00579
Zhu, B., and Wei, N. (2019). Biocatalytic degradation of parabens mediated by cell surface displayed cutinase. Environ. Sci. Technol. 53, 354–364. doi: 10.1021/acs.est.8b05275
Zumstein, M. T., Rechsteiner, D., Roduner, N., Perz, V., Ribitsch, D., Guebitz, G. M., et al. (2017). Enzymatic hydrolysis of polyester thin films at the nanoscale: effects of polyester structure and enzyme active-site accessibility. Environ. Sci. Technol. 51, 7476–7485. doi: 10.1021/acs.est.7b01330
Keywords: biodegradation, cutinase, poly(ethylene terephthalate) (PET), PETase, protein engineering, rational design, metabolic engineering
Citation: Sui B, Wang T, Fang J, Hou Z, Shu T, Lu Z, Liu F and Zhu Y (2023) Recent advances in the biodegradation of polyethylene terephthalate with cutinase-like enzymes. Front. Microbiol. 14:1265139. doi: 10.3389/fmicb.2023.1265139
Received: 22 July 2023; Accepted: 15 September 2023;
Published: 02 October 2023.
Edited by:
Kenneth Jensen, Novozymes, DenmarkReviewed by:
Malene Billeskov Keller, Technical University of Denmark, DenmarkCopyright © 2023 Sui, Wang, Fang, Hou, Shu, Lu, Liu and Zhu. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Tao Wang, dGFvLXdhbmdAemp1LmVkdS5jbg==; Fei Liu, bGl1ZmVpMDkyNTMxQDE2My5jb20=; Youshuang Zhu, emh1eW91c2h1YW5nQDEyNi5jb20=
†These authors have contributed equally to this work
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.